ABSTRACT
Piscine lactococcosis is a significant threat to cultured and wild fish populations worldwide. The disease typically presents as a per-acute to acute hemorrhagic septicemia causing high morbidity and mortality, recalcitrant to antimicrobial treatment or management interventions. Historically, the disease was attributed to the gram-positive pathogen Lactococcus garvieae. However, recent work has revealed three distinct lactococcosis-causing bacteria (LCB)—L. garvieae, L. petauri, and L. formosensis—which are phenotypically and genetically similar, leading to widespread misidentification. An update on our understanding of lactococcosis and improved methods for identification are urgently needed. To this end, we used representative isolates from each of the three LCB species to compare currently available and recently developed molecular and phenotypic typing assays, including whole-genome sequencing (WGS), end-point and quantitative PCR (qPCR) assays, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), API 20 Strep and Biolog systems, fatty acid methyl ester analysis (FAME), and Sensititre antimicrobial profiling. Apart from WGS, sequencing of the gyrB gene was the only method capable of consistent and accurate identification to the species and strain level. A qPCR assay based on a putative glycosyltransferase gene was also able to distinguish L. petauri from L. garvieae/formosensis. Biochemical tests and MALDI-TOF MS showed some species-specific patterns in sugar and fatty acid metabolism or protein profiles but should be complemented by additional analyses. The LCB demonstrated overlap in host and geographic range, but there were relevant differences in host specificity, regional prevalence, and antimicrobial susceptibility impacting disease treatment and prevention.
IMPORTANCE
Lactococcosis affects a broad range of host species, including fish from cold, temperate, and warm freshwater or marine environments, as well as several terrestrial animals, including humans. As such, lactococcosis is a disease of concern for animal and ecosystem health. The disease is endemic in European and Asian aquaculture but is rapidly encroaching on ecologically and economically important fish populations across the Americas. Piscine lactococcosis is difficult to manage, with issues of vaccine escape, ineffective antimicrobial treatment, and the development of carrier fish or biofilms leading to recurrent outbreaks. Our understanding of the disease is also widely outdated. The accepted etiologic agent of lactococcosis is Lactococcus garvieae. However, historical misidentification has masked contributions from two additional species, L. petauri and L. formosensis, which are indistinguishable from L. garvieae by common diagnostic methods. This work is the first comprehensive characterization of all three agents and provides direct recommendations for species-specific diagnosis and management.
KEYWORDS: aquaculture, fish, Lactococcus, septicemia
INTRODUCTION
The global aquaculture industry plays an increasingly critical role in food security, economic growth, and ecosystem health. Fisheries and aquaculture generate billions in annual revenue and provide 4.3 billion people worldwide with at least 15% of their daily protein intake, in addition to supporting over 600 million livelihoods (1). As the human population continues to grow, so too does our reliance on fish, but addressing this need in an environmentally, socially, and economically sustainable way remains a challenge. Contributions from wild fish stocks have effectively plateaued, and aquaculture is now the major source of food fish globally (1, 2). Further development of both inland and marine aquaculture is anticipated, but infectious diseases continue to limit the expansion of the industry (3, 4). Intensified production practices provide an environment conducive to disease outbreaks, and rising surface temperatures associated with global climate change may amplify the impact of “warmwater” diseases like piscine lactococcosis (2, 3).
Lactococcosis is a bacterial disease that typically presents as a hyper-acute or acute hemorrhagic septicemia in fish, causing morbidity and mortality rates between 20% and 50% (5–7). The earliest published case of piscine lactococcosis was in Japanese amberjack (Seriola quinqueradiata) in the 1970s (8), followed by the first European outbreak in rainbow trout (Oncorhynchus mykiss) in 1998 (9). Since then, lactococcosis has become a major disease in freshwater and marine aquaculture across Asia, Europe, Australia, and the Middle East (10–13). Prior to 2016, lactococcosis was reported only sporadically in the Americas (14). Recently, however, it has become a disease of significant concern, with outbreaks and epizootics in Brazil, México, Canada, and multiple regions across the United States (US). The disease has been particularly devastating to cultured rainbow trout (O. mykiss), the second largest sector in the US food fish industry (15). It also poses a notable risk to the leading sector, catfish (Ictalurus spp.) farming, with recent infections occurring in the Southeastern US. Clinical or sub-clinical infections have been reported in a wide range of hosts, including cultured and wild fish from fresh and marine environments, marine mammals, terrestrial ungulates, dogs, rabbits, reptiles, birds, and humans (6, 16–19). Human lactococcosis is considered an emerging disease globally (20–22). It is most commonly associated with infective endocarditis and has a fatality rate of 16% (20). Other clinical presentations, including septicemia, meningitis, and peritonitis, have been reported, often attributed to zoonotic transmission from fish (20, 22, 23). Lactococcosis is therefore a disease of interest for aquaculture, conservation, terrestrial agriculture, and human health.
Preventing and controlling piscine lactococcosis has proven difficult. Recurrent outbreaks are common and thought to be associated with bacterial persistence in carrier fish, avian piscivores, terrestrial animals, and environmental biofilms (6, 24–26). Control of active outbreaks with medicated feed often fails, due to the acute nature of the disease, population inappetence, or antimicrobial resistance (6, 20, 26–28). Autogenous or commercial vaccines are available in some regions but have shown mixed success in limiting disease (26, 29, 30). A large contributor to the variability in treatment success is that, until very recently, piscine lactococcosis was thought to be caused by only a single species: Lactococcus garvieae. However, advances in molecular methodologies have precipitated the discovery of widespread misidentification, masking contributions from two other species—L. petauri and L. formosensis (7, 13, 31–33).
Lactococcus garvieae, the historical agent of lactococcosis, was described from a bovine mastitis case published in the 1980s (34) and went on to become a notorious pathogen of wild and cultured fish. It has undergone several taxonomic adjustments, having previously been included in the Enterococcus and then Streptococcus genera, before becoming a defining member of the genus Lactococcus (9, 35). The genetic heterogeneity of “L. garvieae” isolates from different environmental niches has long been appreciated (36). Genotyping studies over the last two decades have noted multiple genetic groups, and there has been cumulative progress in recognizing that at least two of these groups should be partitioned into different species. Zhang et al. (37) proposed that L. garvieae subsp. bovis strains belong to L. formosensis, a species described from fermented broccoli in 2014 (33). More recently, Mahmoud et al. (13) added to this conclusion, using genomic evidence to show that L. garvieae serotype II, an emerging problem in Asian mariculture, is synonymous with L. formosensis. In a parallel trajectory, L. petauri was isolated from a sugar glider abscess in 2017 (31), in a seminal paper that redefined L. garvieae subgroup A as L. petauri. In their partial phylogenomic analysis, Goodman et al. (31) also noted the fish isolate PAQ2015-99 (14) was more closely related to the L. petauri type strain than L. garvieae and suggested re-assignment. Retrospective analysis by our collaborative research group in North America and several groups in Europe and Brazil have added further evidence that many outbreaks attributed to L. garvieae were actually caused by L. petauri or L. formosensis (7, 32, 38–42). The full implications of these new insights on the field of fish health are not yet appreciable, as until now, there has not been a united discussion or comparison of all three agents. However, these early studies already demonstrate species-specific differences in serotype, regional prevalence, host virulence, and antimicrobial susceptibility (13, 24, 25, 40). Widespread application of updated terminology and typing will be necessary to evaluate the relevance of each species more completely.
The widespread misidentification of L. petauri and L. formosensis almost a decade after their description is due to their high similarity to L. garvieae in disease presentation, phenotype, and genetics. The few recent papers that use correctly identified L. petauri or L. formosensis demonstrate that clinical signs in fish are consistent with traditional L. garvieae: exophthalmia, melanosis, irregular swimming, anorexia, and hemorrhage of the eyes, skin, fin bases, opercula, and other tissues (13, 24, 39). Unlike the pigmented type strain of L. petauri (31), all fish isolates of this species look essentially identical to isolates of L. garvieae and L. formosensis, with white, circular, α- or non-hemolytic colonies on 5% sheep blood agar. They form short gram-positive chains or pairs, and are facultatively anaerobic, non-motile, and catalase and oxidase negative. In addition, initial analyses indicate that common diagnostic methods such as partial 16S rDNA sequencing, rapid biochemical tests (API, Vitek, or Biolog), MALDI-TOF, and currently available “species-specific” primers cannot differentiate the lactococcosis-causing bacteria (LCB) (7, 20, 31, 32, 39, 40, 43–45). No study, however, has comprehensively and systematically assessed methods of identification for all three species of LCB, nor compared clinically relevant phenotypes. This paper addresses a pressing need to resolve our fractured understanding of the LCB, to provide treatment recommendations, and to identify diagnostic options that are consistent, accurate, practical, and disseminable, to curtail the spreading effects of this devastating disease.
RESULTS
Molecular identification
Genome characteristics
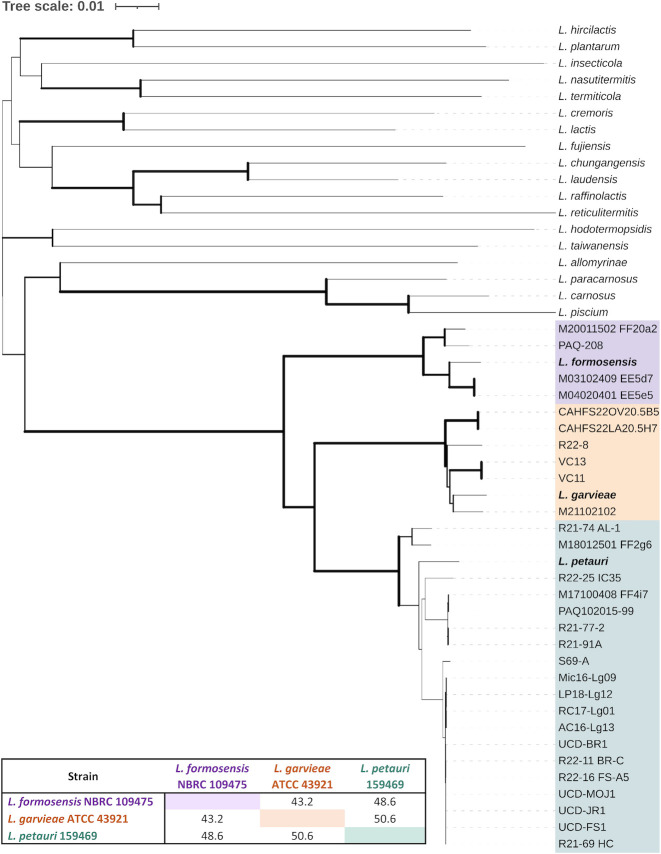
In all, 29 representative North American isolates were selected for full genetic and phenotypic characterization (Table S1), collected from fish (n = 27) and non-fish (n = 2) hosts in different regions of the US, México, or Canada. Genome sequencing of these isolates confirmed 19 as L. petauri, 6 as L. garvieae, and 4 as L. formosensis. Genome characteristics were similar across the three species (Table 1). Chromosomes were between 2,016,716–2,358,485 bp in size with 38.0%−38.9% GC content. All isolates had one tmRNA gene and 16 rRNA genes, apart from PAQ2015-99, which was previously sequenced by Nelson et al. (14). The number of tRNA genes (47 – 69) and predicted protein-coding genes (1,881 – 2,369) differed by isolate. Digital DNA-DNA hybridization (dDDH) values showed high similarity at the genomic level, while still meeting cutoffs for speciation (Fig. 1; Table S3). Interspecies dDDH values were between 42.6% and 54.3%, compared to the intraspecies ranges of 82%–89.1% (L. formosensis), 85.1%–87.7% (L. garvieae), and 80.8%–87.1% (L. petauri). Speciation was confirmed with OrthoANI (46), with 97.77%–98.67% genome similarity between isolates and the corresponding type strain, and 90.73%– 93.66% between the different species (Table S4).
TABLE 1.
Genome characteristics for representative isolates from the Americasc
| Species | Isolate | Length (bp) | %GC | CDS | rRNA | tRNA | tmRNA | ncRNA | Plasmids | ARG |
|---|---|---|---|---|---|---|---|---|---|---|
| L. formosensis | M20011502 FF20a2 | 2,124,569 | 38.1 | 2049 | 16 | 59 | 1 | 10 | 0 | lsa(D), vanTG, vanYB, tet(S) |
| PAQ-208 | 2,111,913 | 38.1 | 1981 | 16 | 61 | 1 | 10 | 0 | lsa(D), vanTG, vanYB, tet(S) | |
| M03102409 EE5d7 | 2,024,587 | 38.3 | 1883 | 16 | 56 | 1 | 9 | 1 | lsa(D), vanTG, vanYB, tet(S),a tet(L)a | |
| M04020401 EE5e5 | 2,024,581 | 38.3 | 1881 | 16 | 56 | 1 | 9 | 1 | lsa(D), vanTG, vanYB, tet(S),a tet(L)a | |
| L. garvieae | CAHFS22OV20.5B5 | 2,016,716 | 38.8 | 1894 | 16 | 58 | 1 | 5 | 0 | lsa(D), vanTG, vanYB |
| CAHFS22LA20.5H7 | 2,016,721 | 38.8 | 1894 | 16 | 58 | 1 | 5 | 0 | lsa(D), vanTG, vanYB | |
| R22-8 | 2,358,485 | 38.3 | 2369 | 16 | 71 | 1 | 6 | 1 | lsa(D), vanTG, vanYB | |
| VC11 | 2,190,521 | 38.8 | 2145 | 16 | 68 | 1 | 7 | 0 | lsa(D), vanTG, vanYB | |
| VC13 | 2,190,543 | 38.8 | 2145 | 16 | 68 | 1 | 7 | 0 | lsa(D), vanTG, vanYB | |
| M21102102 | 2,050,570 | 38.9 | 1961 | 16 | 62 | 1 | 5 | 1 | lsa(D), vanTG, vanYB | |
| L. petauri | R21-74 AL-1 | 2,059,061 | 38.1 | 2068 | 16 | 61 | 1 | 9 | 3 | lsa(D), vanTG, vanYB |
| M18012501 FF2g6 | 2,089,801 | 38.2 | 2115 | 16 | 66 | 1 | 9 | 1 | lsa(D), vanTG, vanYB, tet(S) | |
| R22-25 IC35 | 2,082,218 | 38.2 | 1956 | 16 | 58 | 1 | 9 | 0 | lsa(D), vanTG, vanYB | |
| M17100408 FF4i7 | 2,090,887 | 38.1 | 1953 | 16 | 59 | 1 | 9 | 0 | lsa(D), vanTG, vanYB | |
| PAQ102015-99b | 2,068,357b | 38b | 1947b | 5b | 47b | 1 | 6 | 0 | lsa(D), vanTG, vanYB | |
| R21-91A | 2,146,082 | 38 | 2007 | 16 | 59 | 1 | 10 | 0 | lsa(D), vanTG, vanYB, qacJ | |
| R21-77-2 | 2,146,072 | 38 | 2008 | 16 | 60 | 1 | 10 | 0 | lsa(D), vanTG, vanYB, qacJ | |
| S69-A | 2,145,353 | 38.1 | 2077 | 16 | 60 | 1 | 10 | 0 | lsa(D), vanTG, vanYB | |
| Mic16-Lg09 | 2,050,723 | 38.2 | 1883 | 16 | 56 | 1 | 9 | 0 | lsa(D), vanTG, vanYB | |
| LP18-Lg12 | 2,050,735 | 38.2 | 1935 | 16 | 60 | 1 | 9 | 0 | lsa(D), vanTG, vanYB | |
| RC17-Lg01 | 2,050,764 | 38.2 | 1936 | 16 | 60 | 1 | 9 | 0 | lsa(D), vanTG, vanYB | |
| AC16-Lg13 | 2,050,746 | 38.2 | 1939 | 16 | 60 | 1 | 9 | 0 | lsa(D), vanTG, vanYB | |
| UCD-BR1 | 2,112,235 | 38.1 | 2019 | 16 | 62 | 1 | 10 | 0 | lsa(D), vanTG, vanYB, tet(S), tet(L) | |
| R22-11 BR-C | 2,112,193 | 38.1 | 2021 | 16 | 62 | 1 | 10 | 0 | lsa(D), vanTG, vanYB, tet(S), tet(L) | |
| R22-16 FS-A5 | 2,112,204 | 38.1 | 2022 | 16 | 62 | 1 | 10 | 0 | lsa(D), vanTG, vanYB, tet(S), tet(L) | |
| UCD-MOJ1 | 2,112,084 | 38.1 | 2020 | 16 | 61 | 1 | 10 | 0 | lsa(D), vanTG, vanYB, tet(S), tet(L) | |
| UCD-JR1 | 2,112,161 | 38.1 | 2020 | 16 | 62 | 1 | 10 | 0 | lsa(D), vanTG, vanYB, tet(S), tet(L) | |
| UCD-FS1 | 2,112,152 | 38.1 | 2021 | 16 | 62 | 1 | 10 | 0 | lsa(D), vanTG, vanYB, tet(S), tet(L) | |
| R21-69 HC | 2,112,152 | 38.1 | 2024 | 16 | 62 | 1 | 10 | 0 | lsa(D), vanTG, vanYB, tet(S), tet(L) |
Fig 1.
Whole-genome phylogeny of 29 representative isolates and type strains of currently accepted Lactococcus species. The tree was inferred with FastME 2.1.6.1 [47] from GBDP distances calculated from genome sequences. The branch lengths are scaled in terms of the GBDP distance formula d5. Increasing branch width reflects higher pseudo-bootstrap support. The tree was rooted at the midpoint [48]. Colors represent the three piscine-pathogenic species: Purple—L. formosensis, Orange—L. garvieae, and Teal—L. petauri. The table shows type strain in silico DNA-DNA hybridization (dDDH) values based on the Genome-to-Genome Distance Calculator (GGDC 3.0). An interactive, annotated version of this tree can be accessed at https://itol.embl.de/tree/9942121104151391702399750.
Single plasmids were present for L. formosensis isolates M03102409 EE5d7 and M04020401 EE5e5 (~67 kb), L. garvieae isolates R22-8 and M21102102 (~68–94 kb), and L. petauri isolate M18012501 FF2g6 (~23 kb). Lactococcus petauri isolate R21-74 AL-1 had three plasmids between ~10 and 42 kb. The Comprehensive Antibiotic Resistance Database (CARD; (51)) identified putative antimicrobial resistance genes (ARG) lsa(D), vanTG, and vanYB in all genomes. The tet(S) gene was present on the chromosome or plasmid of all L. formosensis isolates, and almost half of the L. petauri, including all isolates from California. Californian L. petauri and two of the L. formosensis isolates also encoded tet(L). Canadian L. petauri isolates had a unique gene, qacJ, for a putative small multidrug efflux pump.
Phylogenetics
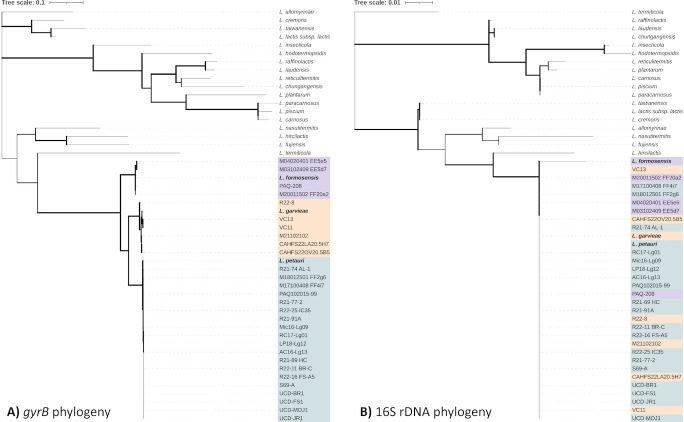
The representative isolates and type strains of L. petauri, L. garvieae, and L. formosensis formed discrete clades by whole-genome phylogenetic analysis and were more closely related to each other than any other species of Lactococcus (Fig. 1) — forming an LCB genetic group. Phylogenetic analysis based on a previously described multilocus sequence analysis (MLSA) scheme consisting of the 16S rRNA, gyrB, pheT, and rpoB genes (52) was also able to discriminate between clades (Fig. S1), but the gyrB gene alone was sufficient for accurate species separation (Fig. 2A). The partial 16S rRNA gene sequences were nearly identical between isolates [99.9% pairwise identity (PI)], and subsequently typed all LCB into a single homogenous clade (Fig. 2B).
Fig 2.
Comparative gyrB (A) and 16S rRNA (B) phylogenies of representative isolates and type strains of currently accepted Lactococcus species. Phylogenies were annotated in iTOL [53], and color denotes species determined by whole-genome analysis (Fig. 1). Branch width is proportional to bootstrap support with increasing thickness. The gyrB gene tree (A) was inferred using the Maximum Likelihood method and Tamura 3-parameter model [54] in MEGA-X [55] with a discrete Gamma distribution and a rate variation model allowing some sites to be evolutionarily invariable. The 16S rRNA gene tree (B) was similarly inferred but with the Kimura 2-parameter model [56]. Interactive, annotated versions can be accessed at (A) https://itol.embl.de/tree/168150115248209731686089834 and (B) https://itol.embl.de/tree/168150115248206121686089807.
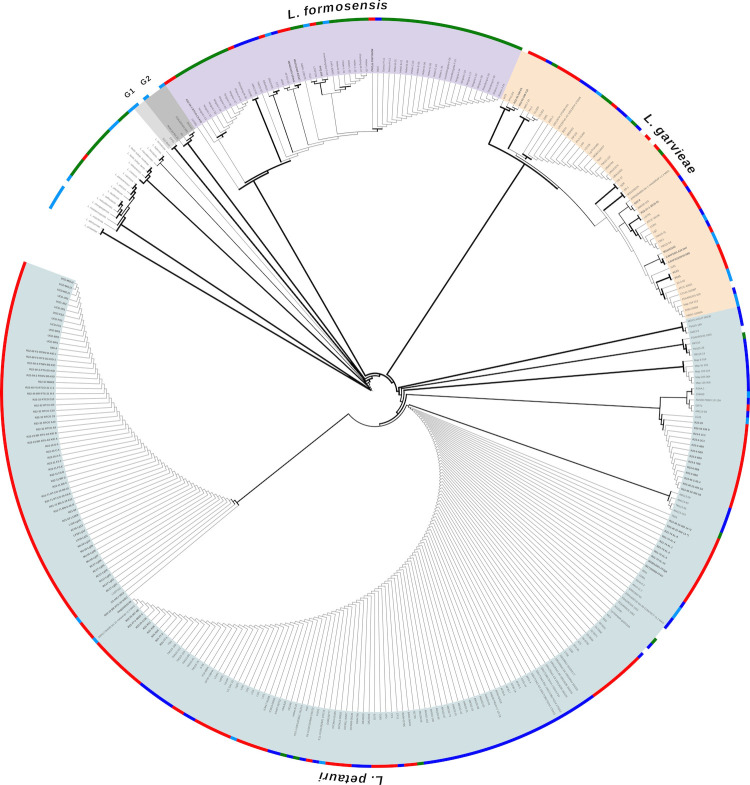
An expanded gyrB analysis was performed to include an additional 78 clinical isolates (Table S1) and 223 LCB genomes currently available on the National Center for Biotechnology Information (NCBI) database (Table S2). The expanded phylogeny revealed all LCB species contained isolates from fish, humans, non-human animals, and food products, and originated from Asia, Europe, and the Americas, but with unequal representation (Fig. 3). When comparing all LCB, 66% of strains were L. petauri, 17% L. garvieae, and 17% L. formosensis. When analyzing only fish-associated strains, the representation of L. petauri increased to 82%, while L. garvieae and L. formosensis were reduced to 13% and 5%, respectively. Lactococcus petauri contained several internal genetic groups, including a subclade with 95% of clinical fish isolates from California and all isolates from México. Six of the “L. garvieae” reference genomes fell into two additional clades (G1 and G2) distinct from any of the currently described LCB.
Fig 3.
Extended gyrB phylogeny of LCB isolates and type strains of other currently accepted Lactococcus species. Phylogenies were annotated in iTOL [53], and clade color denotes species determined by whole-genome analysis (Fig. 1). Branch width is proportional to bootstrap support with increasing thickness. Colored strips denote strain origin: Red—Fish, Dark blue—Human, Light blue—Other animal, Green—Food. The tree was inferred using the Maximum Likelihood method and Tamura 3-parameter model [54] in MEGA-X [55] with a discrete Gamma distribution and a rate variation model allowing some sites to be evolutionarily invariable. An interactive, annotated version of this tree can be accessed athttps://itol.embl.de/tree/9942121104197701693846730.
Quantitative PCR
Representative genomes were used to identify species-specific regions for qPCR primer development. A hypothetical protein “P6” was determined to be unique to piscine L. petauri, with the highest sequence homology to other hypothetical proteins from Lactococcus, followed by glycotransferase proteins from Weissella and Streptococcus species. The putative glycosyltransferase gene was present in 80/87 L. petauri genomes investigated, with high homology in the target sequence (>98.5% pairwise identity). Genomes without a match were all mammalian strains. The targeted gene was not found in L. garvieae, L. formosensis, or any other Lactococcus species (Table 2). The designed primer-probe set “Lp-p6” targets a 126 bp region within the gene and binding was again specific to L. petauri genomes.
TABLE 2.
Quantitative PCR summary table with primer and probe sequences for the novel Lp-p6 (ΔRn= 0.35) and previously published LG-ITS assay (ΔRn= 0.60) (44) and results for in silico and in vitro specificity for L. petauri
| LG-ITSa | Lp-p6 | |
|---|---|---|
| Forward primer | 5′-CAA GAT AGA GAA GAT TGC GTT GAG-3′ | 5′-CGT CTT CCA TAT TCT GAA ATT TGT ATC AGG-3′ |
| Reverse primer | 5′-CCG TAT CTT ATG GAG CCT AGC-3′ | 5′-GCC AAT TCT ACC AAT GCT AAC AC-3′ |
| Probe | 5′-FAM- TGC TTT GCA CGC AGG AGG TCA -BHQ-3′ | 5′-NED- AGC GTA AGG TAG AGC TTT CTT GTG CC -NFQ-3′ |
| In silico binding | ||
| L. formosensis | 5/5 | 0/5 |
| L. garvieae | 7/7 | 0/7 |
| L. petauri | 87/87 | 80/87 |
| Other Lactococcus | 0/13 | 0/13 |
| DNA amplification | ||
| L. formosensis | 4/4 | 0/4 |
| L. garvieae | 8/8 | 0/8 |
| L. petauri | 26/26 | 26/26 |
| Other Lactococcus | 0/21 | 0/21 |
Shahin et al. (44).
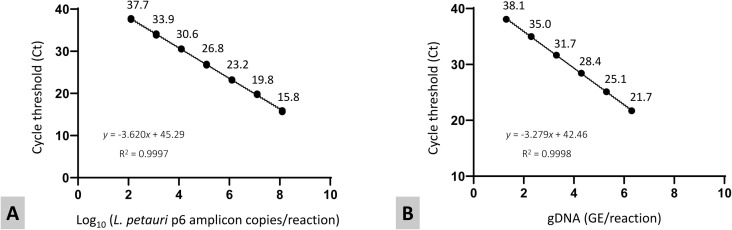
Specificity was confirmed in vitro, with the Lp-p6 assay showing high sensitivity for fish-associated L. petauri under all tested conditions: purified gDNA, p6 PCR products, spiked water or tissue, and experimentally infected fish samples. Positive amplification by Lp-p6 was detected only for L. petauri isolates, while LG-ITS amplified all strains of LCB indiscriminately (Table S5). Using Lp-p6 PCR products, a linear correlation between qPCR cycle threshold (Ct) values and the log of amplicon copies was obtained (R2 = 0.9997) with a high assay efficiency of 88.87% (Fig. 4A). A strong linear correlation was maintained in L. petauri spiked tissue samples (R2 = 0.9993–0.9998) and in triplicate runs of spiked water samples (R2 = 0.9993–0.9998) (Fig. 4B) with efficiencies of 98.4%–101.8% and 99.3%–103.3%, respectively. No inhibition was detected using IPC controls in any of the tested samples. Amplification was also compared between tissue collected from L. petauri infected fish (mortalities) and sham-infected negative controls using both Lp-p6 and a previously developed LG-ITS assay that amplifies all three LCB (Table 2; (25, 44)). There was 100% agreement between assays, with positive amplification (Ct ≤35) from 100% of spleen and brain samples, 60% of heart samples, 25% of gill, and 20% of liver and kidney samples from experimentally induced mortalities (Table S6). Average Ct values for the Lp-p6 assay were consistently lower than comparable averages for the LG-ITS assay. There was no amplification from any of the sham-infected negative control fish.
Fig 4.
Quantitative PCR standard curves for (A) diluted P6 primer amplification products from L. petauri UCD-JR1 and (B) spiked water samples with gDNA from the same isolate. Spiked water samples were run in quadruplicate in three separate experiments. The figure is a composite of the three trials. PCR product dilutions were run once in triplicate.
Biochemical identification
API 20 Strep System
The API database identified most LCB as L. lactis subsp. lactis with low confidence (Fig. 5; Table S7). Lactococcus formosensis isolates M20011502 FF20a2 and M03102409 EE5d7 were called as Enterococcus durans, while L. petauri isolate R21-74 AL-1 was called as E. avium and R22-16 FS-A5 as Aerococcus viridans. All species tested were positive for acetoin production (VP), β-glucosidase hydrolysis (ESC), and pyrolidonyl arylamidase (PYRA) reactions, and negative for α- and β- galactosidase (αGAL & βGAL), β-glucuronidase (βGUR), alkaline phosphatase (PAL), sorbitol (SOR), lactose (LAC), and inulin (INU) reactions (Table S7). None of the reactions included on the API 20 STREP strip were discriminatory between LCB species.
Fig 5.
Microbial phenotypic identification by API 20 STREP, BIOLOG GEN III Microplate, MALDI-TOF MS, and sucrose metabolism tests for 29 Lactococcus isolates based on individual phenotypic biochemical profiles. Correct identification by whole-genome analysis (Fig. 1) is indicated by the dendrogram and colored shading.
BIOLOG GEN III microplate assay
BIOLOG software identified all LCB isolates as L. garvieae, except for L. petauri R21-74 AL-1 which was identified as E. casseliflavus (Fig. 5). The phenotypic profiles of individual isolates of L. petauri, L. formosensis, and L. garvieae were combined to generate composite species profiles (Table S8). A comparison of the L. petauri, L. formosensis, and L. garvieae summary characteristics demonstrated a homogeneous phenotypic profile with the exception that L. petauri was the only species to utilize sucrose (Fig. 5; Table S8).
MALDI-TOF MS
MALDI-TOF MS was performed using custom profiles generated from six representative L. petauri strains and five L. formosensis strains, with a success rate of 93.1% (Fig. 5). In duplicate runs of 29 isolates, there were four mis-calls, and all were between L. petauri vs L. garvieae (Table S9). Lactococcus petauri isolate R21-74 AL-1 and L. garvieae isolate R22-8 were misidentified as the opposing species on one of two replicate runs, with similar confidence levels between calls. Lactococcus garvieae isolate M21102102 was incorrectly identified as L. petauri in both replicates. No L. formosensis isolates were misidentified.
Phenotypic characterization
Bacterial growth and morphology
All isolates formed round, smooth, non-pigmented, and non-hemolytic colonies after 24 h of incubation at 28°C on trypticase soy agar supplemented with 5% sheep’s blood (SBA) (Fig. S2). Growth was also observed on solid media at 37°C and 40°C. Growth kinetics in brain heart infusion (BHI) liquid media were similar between isolates and species at 28°C, reaching the mid-exponential stage around 12 h (Fig. S2). Increasing temperatures to 37°C increased variability between isolates, and one L. petauri isolate (UCD-MOJ1) had severely limited growth. All L. garvieae isolates were able to grow at pH 6, while growth varied by isolate for L. petauri (11/19) and L. formosensis (3/4). There was no growth at pH 5. Growth in 1%–8% NaCl was also variable, though all L. garvieae grew at 1% NaCl, and none of the L. formosensis grew at 8% NaCl (Table S8).
FAME analysis
The fatty acid profiles of L. petauri, L. formosensis, and L. garvieae exhibited similar percentages of the fatty acids 14:0, 16:0 iso, 16:0, 17:0 anteiso, 17:0, 18:1 ω9c, 18:0, 19:0 cyclo ω8c, 20:4 ω6,9,12,15c, Summed Feature 3, Summed Feature 5, and Summed Feature 8 (Table 3). However, L. petauri was the only species to produce the fatty acids 15:1 ω5c and 17:1 iso ω5c and in contrast to L. formosensis and L. garvieae did not produce the fatty acids 18:0 iso and 20:1 ω9c.
TABLE 3.
Fatty acid composition summary comparison between Lactococcus petauri (n = 19), L. formosensis (n = 4), and L. garvieae (n = 6)d
| Fatty acid | Lactococcus petauri | Lactococcus formosensis | Lactococcus garvieae |
|---|---|---|---|
| 14:00 | 19.92 ± 2.83 | 19.54 ± 1.27 | 20.58 ± 1.64 |
| 15:1 ω5c | 0.14 ± 0.35 | ND | ND |
| 16:0 iso | 0.02 ± 0.08 | 0.42 ± 0.34 | 0.17 ± 0.27 |
| 16:00 | 25.20 ± 1.58 | 26.80 ± 1.80 | 26.52 ± 0.47 |
| 17:1 iso ω5c | 0.17 ± 0.55 | ND | ND |
| 17:0 anteiso | 0.03 ± 0.08 | 0.11 ± 0.22 | 0.03 ± 0.09 |
| 17:00 | 0.16 ± 0.22 | 0.27 ± 0.32 | 0.32 ± 0.25 |
| 18:0 iso | ND | 0.09 ± 0.17 | 0.94 ± 2.30 |
| 18:1 ω9c | 5.04 ± 1.03 | 5.64 ± 1.01 | 4.70 ± 2.46 |
| 18:00 | 4.06 ± 0.52 | 3.59 ± 0.30 | 3.33 ± 0.24 |
| 19:0 cyclo ω8c | 9.76 ± 1.48 | 8.04 ± 3.03 | 8.08 ± 1.87 |
| 20:4 ω6,9,12,15c | 0.84 ± 0.36 | 1.04 ± 0.17 | 1.05 ± 0.06 |
| 20:1 ω9c | ND | 0.14 ± 0.29 | 0.03 ± 0.07 |
| Summed Feature 3a | 10.54 ± 0.70 | 10.47 ± 0.84 | 10.22 ± 0.60 |
| Summed Feature 5b | 4.27 ± 1.58 | 3.85 ± 1.44 | 3.95 ± 1.21 |
| Summed Feature 8c | 19.87 ± 2.43 | 19.98 ± 1.81 | 20.08 ± 1.05 |
The fatty acids (16:1 ω7c/16:1 ω6c, 16:1 ω6c/16:1 ω7c) could not be separated from each other and together were considered Summed Feature 3.
The fatty acids (18:0 ante/18:2 ω6,9c, 18:2 ω6,9c/18:0 ante) could not be separated from each other and together were considered Summed Feature 5.
The fatty acids (18:1 ω7c, 18:1 ω6c) could not be separated from each other and together were considered Summed Feature 8.
No detection is denoted by “ND.” fatty acids specific to L. petauri are in bold.
Antimicrobial profiling
The minimum inhibitory concentration (MIC) profiles for common antimicrobials used in veterinary medicine differed between isolates and showed some species-specific trends (Table 4). All LCB showed MICs at or above the highest concentrations tested for clindamycin (>4 µg/mL), tylosin tartrate (≥20 µg/mL), sulfadimethoxine (>256 µg/mL), and sulphathiazole (≥ 256 µg/mL). Growth was also similar for all LCB in florfenicol, penicillin, amoxicillin, spectinomycin, and novobiocin. Lactococcus formosensis had comparatively high MICs for erythromycin, but low MICs for trimethoprim/sulfamethoxazole (< 0.5/9.5 µg/mL) compared to the other LCB species. Lactococcus garvieae had comparatively low MICs for enrofloxacin, tetracycline, and oxytetracycline. Lactococcus petauri had more variation and a larger sample size, but at least 50% of isolates displayed the highest observed MICs to trimethoprim/sulfamethoxazole (2/38 µg/mL) and streptomycin (64–128 μg/mL). All Californian L. petauri isolates also had MICs outside of the testing range (≥8 µg/mL) for both oxytetracycline and tetracycline. Reference strain profiles for Escherichia coli ATCC 25922 and Streptococcus pneumoniae ATCC 49619 were within ranges established by CLSI (57, 58).
TABLE 4.
Minimum inhibitory concentration profiles for common antimicrobials used in veterinary medicinea
| Enrofloxacin | Gentamicin | Ceftiofur | Neomycin | Erythromycin | Oxytetracycline | Tetracycline | Amoxicillin | Spectinomycin | Sulfadimethoxine | Trimethoprim/Sulfamethoxazole | Florfenicol | Sulfathiazole | Penicillin | Streptomycin | Novobiocin | Tylosin tartrate | Clindamycin | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L. formosensis | M20011502 FF20a2 | 1 | 4 | 1 | ≤2 | ≤0.12 | >8 | >8 | 0.5 | 64 | >256 | ≤0.5/9.5 | 2 | >256 | 1 | 16 | 4 | >20 | >4 |
| PAQ-208 | 1 | 2 | 1 | 4 | >4 | >8 | >8 | 2 | 64 | >256 | ≤0.5/9.5 | 2 | >256 | 1 | 32 | 2 | >20 | >4 | |
| M03102409 EE5d7 | 1 | 1 | 2 | ≤2 | 0.25 | >8 | >8 | 1 | 32 | >256 | ≤0.5/9.5 | ≤1 | >256 | 1 | 16 | 4 | >20 | >4 | |
| M04020401 EE5e5 | 2 | 1 | 2 | ≤2 | 0.5 | >8 | >8 | 0.5 | 64 | >256 | ≤0.5/9.5 | 2 | >256 | 1 | 16 | 2 | >20 | >4 | |
| L. garvieae | CAHFS22OV20.5B5 | 0.25 | 2 | 2 | ≤2 | ≤0.12 | ≤0.25 | ≤0.25 | 2 | 32 | >256 | 1/19 | 2 | >256 | 1 | 16 | >4 | 20 | >4 |
| CAHFS22LA20.5H7 | 0.5 | ≤0.5 | 2 | ≤2 | ≤0.12 | 0.5 | ≤0.25 | 1 | 16 | >256 | 1/19 | 2 | >256 | 1 | ≤8 | >4 | >20 | >4 | |
| R22-8 | 0.5 | 2 | 2 | 8 | ≤0.12 | ≤0.25 | ≤0.25 | 0.5 | 64 | >256 | 1/19 | 2 | >256 | 1 | 16 | 4 | >20 | >4 | |
| VC11 | 0.25 | 2 | 2 | 8 | ≤0.12 | ≤0.25 | ≤0.25 | 0.5 | 32 | >256 | 1/19 | 2 | >256 | 2 | 32 | 4 | >20 | >4 | |
| VC13 | 0.25 | 2 | 2 | 8 | ≤0.12 | ≤0.25 | ≤0.25 | 0.5 | 32 | >256 | 1/19 | 2 | >256 | 1 | 32 | 4 | >20 | >4 | |
| M21102102 | ≤0.12 | ≤0.5 | 2 | ≤2 | ≤0.12 | 0.5 | 0.5 | 1 | 32 | >256 | 1/19 | 2 | 256 | 2 | ≤8 | 4 | 20 | >4 | |
| L. petauri | R21-74 AL-1 | 0.5 | 4 | 1 | 4 | ≤0.12 | 0.5 | 0.5 | 0.5 | 64 | >256 | 2/38 | 2 | >256 | 1 | 64 | 2 | >20 | >4 |
| M18012501 FF2g6 | 1 | 4 | 2 | 16 | ≤0.12 | >8 | >8 | 0.5 | >64 | >256 | 1/19 | 2 | >256 | 1 | 32 | >4 | >20 | >4 | |
| R22-25 IC35 | 1 | 4 | 1 | 8 | ≤0.12 | 0.5 | 0.5 | 0.5 | 64 | >256 | 1/19 | 2 | >256 | 1 | 64 | 2 | >20 | >4 | |
| M17100408 FF4i7 | 1 | 4 | 1 | 8 | ≤0.12 | 1 | 0.5 | 1 | 64 | >256 | 2/38 | 2 | >256 | 0.5 | 32 | 4 | >20 | >4 | |
| PAQ102015-99 | 1 | 8 | 1 | 16 | ≤0.12 | 0.5 | 0.5 | 1 | 64 | >256 | 2/38 | 2 | >256 | 1 | 64 | 4 | >20 | >4 | |
| R21-91A | 1 | 2 | 1 | 4 | ≤0.12 | 0.5 | 0.5 | 0.5 | 64 | >256 | 1/19 | 2 | >256 | 1 | 32 | 4 | >20 | >4 | |
| R21-77-2 | 1 | 2 | 1 | 16 | ≤0.12 | 0.5 | 0.5 | 0.5 | 64 | >256 | 1/19 | 2 | >256 | 1 | 64 | 2 | >20 | >4 | |
| S69-A | 1 | 2 | 2 | 16 | ≤0.12 | ≤0.25 | ≤0.25 | 1 | 64 | >256 | 1/19 | 2 | >256 | 1 | 64 | 4 | >20 | >4 | |
| Mic16-Lg09 | 1 | 8 | 1 | 16 | ≤0.12 | 0.5 | 0.5 | 1 | >64 | >256 | 2/38 | 2 | >256 | 1 | 32 | 4 | >20 | >4 | |
| LP18-Lg12 | 1 | 8 | 1 | 16 | ≤0.12 | 0.5 | 0.5 | 1 | 64 | >256 | 2/38 | 2 | >256 | 1 | 64 | 4 | >20 | >4 | |
| RC17-Lg01 | 1 | 1 | 2 | 8 | ≤0.12 | 0.5 | 0.5 | 1 | 64 | >256 | 2/38 | 2 | >256 | 1 | 64 | 4 | >20 | >4 | |
| AC16-Lg13 | 1 | 2 | 2 | 8 | ≤0.12 | 1 | 1 | 1 | 32 | >256 | 2/38 | 2 | >256 | 1 | 32 | >4 | >20 | >4 | |
| UCD-BR1 | 1 | 2 | 2 | 8 | ≤0.12 | >8 | >8 | 1 | 64 | >256 | 1/19 | 2 | >256 | 1 | 16 | 4 | >20 | >4 | |
| R22-11 BR-C | 1 | 2 | 2 | 16 | ≤0.12 | >8 | >8 | 0.5 | 64 | >256 | 1/19 | ≤1 | >256 | 0.25 | ≤8 | 2 | >20 | >4 | |
| R22-16 FS-A5 | 1 | 1 | 2 | 16 | ≤0.12 | >8 | >8 | 0.5 | >64 | >256 | 2/38 | 2 | >256 | 1 | 64 | 4 | >20 | >4 | |
| UCD-MOJ1 | 1 | 2 | ≤0.25 | 4 | ≤0.12 | >8 | >8 | 0.5 | 64 | >256 | 1/19 | 2 | >256 | 0.5 | 16 | 2 | 20 | >4 | |
| UCD-JR1 | 1 | 1 | 2 | 8 | ≤0.12 | >8 | >8 | 0.5 | 64 | >256 | 2/38 | 2 | >256 | 1 | 128 | 2 | >20 | >4 | |
| UCD-FS1 | 1 | 2 | 1 | 16 | ≤0.12 | >8 | >8 | 0.5 | 64 | >256 | 2/38 | 2 | >256 | 1 | 64 | 2 | >20 | >4 | |
| R21-69HC | 1 | 1 | 1 | 8 | ≤0.12 | >8 | >8 | 1 | 64 | >256 | 1/19 | 2 | >256 | 1 | 64 | 2 | >20 | >4 | |
|
Escherichia coli ATCC 25922 |
≤0.12 | 1 | 1 | 4 | >4 | 0.5 | 0.5 | 8 | 16 | >256 | ≤0.5/9.5 | 4 | 64 | >8 | 16 | >4 | >20 | >4 | |
|
Streptococcus pneumoniae ATCC 49619 |
0.25 | 8 | ≤0.25 | 32 | ≤0.12 | ≤0.25 | ≤0.25 | ≤0.25 | 16 | ≤32 | ≤0.5/9.5 | ≤1 | ≤32 | 0.25 | 32 | ≤0.5 | ≤2.5 | ≤4 | |
| Lactococcus spp. S | ≤0.5 | ≤2 | ≤2/38 | ≤1 | ≤0.5 | ||||||||||||||
| Lactococcus spp. R | ≥8 | ≥8 | ≥4/76 | ≥4 | ≥4 | ||||||||||||||
Concentrations are in μg/mL. When available, Lactococcus spp. susceptible (S) and resistant (R) ranges (57) are included, and values above the established R-range are in bold.
DISCUSSION
The genus Lactococcus represents a heterogeneous collection of lactic acid bacteria with important, but sometimes contradictory roles in food production, biotechnology, and human and animal health (59). When separated out from Streptococcus and Lactobacillus in 1985, the genus consisted only of four species, including archetypical Lactococcus lactis, L. plantarum, L. raffinolactis, and L. garvieae (35). With the advent of the molecular era, whole-genome sequencing has become more accessible and routine in microbial typing, leading to a taxonomic surge of newly described species. The application and implications of taxonomic rearrangements and systemic reorganizations, however, face a period of delay. Significant work is necessary to understand the biological relevance of speciation and to widely disseminate and integrate updated terminology in contemporary research. The possibility that “L. garvieae” represented more than one species has been a decade-long conversation, advanced piecemeal by researchers around the globe (7, 13, 31–33, 38–40). In this paper, we bring together the building evidence to show there are three distinct agents of piscine lactococcosis—L. garvieae, L. petauri, and L. formosensis.
Widespread historical misidentification, both before and after descriptions of L. formosensis in 2014 (33) and L. petauri in 2017 (31), is primarily due to the phenotypic and genetic similarities between the three LCB. Previous studies have used a range of different methods to compare subsets of L. petauri to L. garvieae, or L. garvieae to L. formosensis, but a comprehensive comparison was necessary to unite the field. We used whole-genome sequencing as the gold standard for typing and established a set of 29 representative L. petauri (n = 19), L. formosensis (n = 4), and L. garvieae (n = 6) for further analysis (Fig. 1). Partial sequencing of the 16S rRNA gene is the most commonly employed method for microbial identification worldwide (60), but often lacks the phylogenetic power to resolve closely related species (60) and was unable to distinguish the LCB (Fig. 2). The amplified 16S rRNA fragment was nearly identical between strains from the three species, as has previously been reported in studies on L. petauri and L. garvieae (40). In our description of the first lactococcosis epizootic in the United States (6), we included 16S rRNA sequencing in an MLSA scheme adapted from the piscine pathogen Streptococcus iniae (52). The improved resolution demonstrated a unique US clade of “L. garvieae” that was later determined to belong to L. petauri (41). In this study, we found that MLSA was still accurate to the species level, but the gyrB gene alone offered similar discriminatory power and is more practical for routine identification (Fig. 2).
Gyrase B gene sequencing was therefore used to type an expanded set of isolates from the Americas (n = 106) and all assembled LCB genomes available on NCBI GenBank at the time of writing (n = 226). The three LCB species formed clearly distinct clades with high bootstrap support (99%; Fig. 3). The majority of NCBI genomes were correctly typed, but 15% of “L. garvieae” still need reassignment at the time of writing (Table S2). In addition to misidentified L. petauri and L. formosensis, there were two other distinct groups (G1 and G2) from human or animal gut microbiomes that may represent new species. They did not contain any clinical fish isolates, so further genomic analysis was not pursued. In analyzing trends in host and geographic origins, sampling bias must be considered, as this study is limited to isolates submitted to our research team or NCBI. Still, suggestive evidence is apparent in the current data set. Lactococcus formosensis contained 5% of known fish and human strains, either from clinical or microbiome samples. It had the largest proportion of ungulate and food-based isolates. The majority of these were from the same study by Lin et al. (61), sequencing “L. garvieae” from bovine mastitis milk samples, but this association is also supported by recent bovine mastitis cases in Canada caused by L. formosensis (Slavic et al., unpublished). All but two strains were from Asia or the US. Bonafide L. garvieae was still identified in 14% of fish cases worldwide and 16% of human cases. It maintained its reputation for diverse isolation sources, including food products, rodents, insects, reptiles, and environmental samples (Table S2). Lactococcus garvieae was the only fish-associated LCB collected in Italy. Lactococcus petauri also displayed a wide host range and encompassed a sizeable 66% of all LCB strains. This is due, in part, to the high representation of isolates from California, where it is a pervasive problem in state and private trout facilities (62, 63). However, it also represented 81% of global fish strains, including the majority of isolates from the United States, México, Spain, Greece, and Turkey. This overrepresentation jumps to 90% when considering rainbow trout samples and 100% for catfish isolates, clearly indicating its importance for these industries and making it the LCB of most concern for aquaculture in the United States.
This observed importance of L. petauri presented a sub-aim for the development of diagnostic tools. Whole-genome or gyrB sequencing were the only accurate methods for discriminating all three species, but a specific and less labor-intensive method for confirmatory identification of L. petauri is also highly desirable. Quantitative PCR facilitates more rapid detection of pathogens from environmental or tissue samples (25, 44). Several “species-specific” qPCR assays have been developed for L. garvieae (44), but in light of new genomic evidence, none are, indeed, specific. To improve on existing options, we found a putative glycosyltransferase gene (p6) unique to L. petauri and generated a partially validated TaqMan probe-based qPCR assay. When compared to our previously developed LG-ITS assay (44), the Lp-p6 primer-probe set showed specific amplification of L. petauri and no amplification of L. garvieae, L. formosensis or other Lactococcus species by both in silico and in vitro testing (Table 2). Strong amplification was consistent between purified PCR products and spiked water or tissue samples, without evidence of inhibition from host or environmental DNA (Fig. 4). Tissues collected during experimental challenges, previously performed by Littman et al. (24), were also used as models for a natural outbreak scenario. There were no false positives, and brain and spleen results were 100% congruent between qPCR assays and the original bacterial culture reports [Table S6; (24)]. These tissues are target organs for lactococcal and streptococcal infection and are regarded as the best sampling sites for diagnostics (17, 25, 45). Putative glycosyltransferase and ABC transporter permease genes have also recently been used in a multiplex PCR assay for L. garvieae and L. petauri (40), which could offer an additional non-quantitative option for identification. However, specificity should still be verified against L. formosensis by this method. Furthermore, in silico analysis has also indicated that the P6 gene was absent in a handful of L. petauri mammalian strains and may not be essential for bacterial survival in these hosts. For use in clinical aquaculture, however, it is a promising starting point and useful substitute for non-specific assays.
In facilities where phenotypic and biochemical profiling are more feasible, FAME analysis or MALDI-TOF MS combined with metabolic testing could be implemented. The fatty acid profile of L. petauri was distinct from L. garvieae and L. formosensis, but similar between the latter species (Table 3). Contrastingly, MALDI-TOF MS was able to differentiate L. formosensis from L. garvieae and L. petauri, but occasionally made mis-calls between the latter (Fig. 5; Table S9). It also required the generation of a custom library, otherwise all LCB were called L. garvieae. The inclusion of L. petauri and L. formosensis in Bruker MALDI Biotyper, API, and BIOLOG reference databases may improve species identification, but high interspecific phenotypic similarity and intraspecific variability will likely continue to make reliability an issue. Further testing with additional strains will be necessary to strengthen the observed trends, but pairing MALDI-TOF MS with a simple sucrose test could be an efficient and effective method for identification. All L. petauri isolates investigated to date, in our study (Fig. 5; Tables S8 and 9) or in existing literature (31, 40, 42, 45), are positive for sucrose metabolism, while verified L. garvieae and L. formosensis strains are negative. In the same vein, non-discriminatory molecular tests like 16S rRNA gene sequencing or the LG-ITS qPCR assay could be paired with a sucrose test when L. petauri is suspected.
Accurate, reliable, and practical diagnostic methods are crucial for disease mitigation. The etiologic agents of lactococcosis overlap in range, and multiple species may be present even at the same facility [Tables S1 and S2; (13, 32, 39, 42)]. To a certain degree, the general risk of lactococcosis can be reduced by universally beneficial practices in aquaculture: maintaining good water quality, appropriate rearing conditions, and strict biosecurity measures. However, the realities of high-intensity production systems, complex bacterial transmission routes, and external environmental influences make complete prevention by these efforts impossible, necessitating specific and appropriate strategies for limiting disease spread and severity. The three LCB may present different risks dependent on the susceptibility of fish populations and conducive environmental conditions. Lactococcus petauri was overrepresented in trout samples, and this host specificity is supported by recent studies on LCB virulence in different fish species. Abraham et al. (25) compared LCB infections in rainbow trout (O. mykiss) and largemouth bass (Micropterus salmoides) and found L. petauri caused significantly higher mortality (66%) in rainbow trout than L. formosensis (7.5%) and L. garvieae (0%). Bass did not exhibit mortality from any LCB tested. In addition, Littman et al. (24) investigated the effect of temperature and host species in L. petauri infections. Only rainbow trout and chinook salmon (O. tshawytscha) were susceptible to L. petauri, and only at the higher temperature of 18°C. White sturgeon (Acipenser transmontanus), ornamental koi (Cyprinus rubrofuscus), and Nile tilapia (Oreochromis niloticus) showed no mortality under the tested conditions. However, natural outbreaks of verified L. petauri and L. garvieae in tilapia have occurred in Brazil and Singapore [Table S2; (39)] Lack of experimental mortality may reflect optimal host temperature, as 18°C is warm for salmonids but relatively cool for tilapia culture.
Both Littman and Abraham et al. noted high bacterial persistence in surviving fish across host types (24, 25) and Littman et al. further demonstrated that apparently healthy tilapia survivors could transmit L. petauri to naïve rainbow trout in cohabitation challenges. Such subclinical infections have been widely reported for “L. garvieae” (6, 20, 27, 45, 64–67). Algöet et al. (67), for example, investigated the virulence of “L. garvieae” in salmonids—rainbow trout (O. mykiss), Atlantic salmon (Salmo salar), grayling (Thymallus thymallus), and brown trout (Salmo trutta)—and in cyprinids—carp (Cyprinus carpio), tench (Tinca tinca), rudd (Scardinius erythrophthalmus), barbel (Barbus barbus), chub (Leuciscus cephalus), dace (Leuciscus leuciscus), and roach (Rutilus rutilus). They found higher mortality rates in salmonid hosts, particularly rainbow trout and grayling, but viable bacteria persisted in all fish species. This record of persistence by LCB species in diverse host types strongly suggests the involvement of carrier fish in the direct or indirect transmission of lactococcosis. Infectious bacteria may also persist in biofilms, adding another layer of complexity. In our preliminary analysis in Shahin et al. (6), we found that isolates now identified as L. petauri and L. formosensis formed strong biofilms with increased resistance to hydrogen peroxide, and biofilm-associated “L. garvieae” have been previously shown to be infective of mullet (68). Environmental reservoirs make it difficult to eradicate virulent bacteria from afflicted systems, making proactive vaccination and rapid response to outbreaks critical.
Currently, treatment by antimicrobial feed is the main method used to control lactococcosis, but often fails under field conditions and antimicrobial resistance is already an established concern (28, 64). Species-specific trends in antimicrobial susceptibility are evident in our genomic data (Table 1), in vitro profiles (Table 4), and isolate case histories (6). Particularly relevant were MIC distributions for drugs approved in aquaculture in the United States: florfenicol, oxytetracycline, and a representative potentiated sulfa, trimethoprim/sulfamethoxazole (69). Florfenicol MICs were similar across LCB species, ranging from <1 to 2 µg/mL. Lactococcus formosensis was unable to grow in any of the tested concentrations of the potentiated sulfa, while MICs for L. garvieae and L. petauri were between 1/19 and 2/38 µg/mL, but still within the “susceptible range” by CLSI guidelines for Lactococcus species (57). Oxytetracycline values had a notable dichotomy. All L. formosensis isolates, and approximately half of L. petauri isolates, had MICs above the tested range (>8 µg/ml), while all L. garvieae and the remainder of the L. petauri were <0.25–1 µg/mL. There are no CLSI guidelines for oxytetracycline in Lactococcus, but the MICs for the comparable drug tetracycline were also above the tested range (>8 µg/mL) and the threshold to be considered “resistant.”
High MICs for oxytetracycline and tetracycline correlated with the presence of chromosomally or plasmid-encoded ARGs tet(S) and tet(L) (Table 1). The tet(L) gene encodes a tetracycline-specific efflux pump, while tet(S) encodes a ribosomal protection protein (70). These genes were typically chromosomally encoded but were found on plasmids instead for two L. formosensis isolates. Putative transposon genes were proximal to tet(S) and tet(L), similar to descriptions of the tet(S) and tet(M) genetic regions in oxytetracycline-resistant LCB from Japan (71). Currently, the Californian isolates are the only American L. petauri that contain tet(S) and tet(L), but the potentially mobile nature of these ARG indicates that continued spread is possible. Lactococcus species may also be a reservoir of ARG for other bacteria, and resistant bacteria selected on food-producing animals may contaminate milk or meat and persist in fermented foods such as cheeses and sausages. For example, Amkal et al. (72) demonstrated that plasmid-encoded erm(B) from “L. garvieae serotype II” could be transferred to E. faecalis. The erythromycin resistance gene erm(B) was not present in any American strains, which exhibited low MICs to the antimicrobial, but the potential for horizontal acquisition from other gram-positive bacteria should be acknowledged. Another regionally restricted gene was qacJ, solely in Canadian L. petauri isolates. This gene codes for a small efflux pump mediating resistance to quaternary ammonium compounds (QACs) and other cationic biocides (73). It is a gene of interest for methicillin-resistant Staphylococcus aureus (MRSA) and impacts disinfection efficacy in veterinary and human medicine. No relevant QACs or surface disinfectants were included in this study, so any effects on L. petauri resistance are not yet ascertained.
In addition to species- or strain-specific ARG, all LCB genomes contained the lsa(D), vanTG, and vanYB genes. The lsa(D) gene encodes an ABC-F subfamily protein, described first in “L. garvieae serotype II”. Functional Lsa-D confers resistance to streptogramin A, pleuromutilin, and lincosamide antimicrobial classes, and may help explain intrinsic clindamycin resistance characteristic to “L. garvieae” (64, 74, 75). Vancomycin resistance gene vanTG is a vanT variant found in the vanG gene cluster, and vanYB is a vanY variant in the vanB gene cluster (51). Both impart glycopeptide antibiotic resistance, which was not assessed by our antimicrobial panel. Vancomycin and clindamycin are rarely used in fish medicine but may be implemented in human cases, alongside penicillin, amoxicillin, or gentamicin in monotherapy or combination (20, 22). The MICs for penicillin and amoxicillin were consistent between LCB and within the CLSI “susceptible” range for Lactococcus spp. (57) but resistance or intermediate resistance to penicillins has also been reported (21, 22). Clinical human strains were found in all three LCB, and were not genetically distinct from fish, food, or other terrestrial animal isolates, making zoonoses a concern. Zoonotic transmission from consumption of seafood has been verified in at least one case and was suspected in a majority of cases reviewed by Gibello et al. (20). Given the increasing importance of lactococcosis to human and veterinary medicine, broadening antimicrobial analysis to include additional LCB will improve our understanding of regional and species-specific characteristics, and encourage prudent and appropriate use of antibiotics in both public health and agriculture.
Proactive intervention through informed management and vaccination can reduce the current reliance on antimicrobials to mitigate lactococcosis. Developing cross-protective mucosal vaccines will be a priority moving forward, as little work has been done in this area. De Rutyer et al. (2023) investigated a whole-cell killed L. petauri vaccine by immersion and injection and found injection, or a combination approach of immersion with an injection booster, was necessary for adequate protection (29). To our knowledge, no studies have yet tested cross-protection or serological cross-reactions between properly identified LCB species. The existence of multiple “L. garvieae” serotypes is well known, but only serotype II has been clearly linked to a specific LCB—L. formosensis (13). The molecular basis of serotype and its effect on virulence and immune response is also poorly understood. It has been proposed that surface carbohydrates differ between serotypes, while surface proteins were inferred to be similar (76, 77). Immunoproteomics and pangenome comparisons between typed strains will be useful in determining future shared antigen candidates for vaccine design.
In conclusion, piscine lactococcosis caused by L. petauri, L. formosensis, and L. garvieae is an emerging disease in the Americas. At present, only whole-genome or gyrB sequencing can consistently and accurately distinguish the three agents to the species level. However, our recently developed qPCR assay or a two-step approach combining non-discriminatory methods with a sucrose test is sufficient for specific identification of L. petauri from L. garvieae or L. formosensis. Sampling of the brain or spleen is recommended for bacterial culture or qPCR analysis. While all three LCB can be found worldwide in multiple aquatic and terrestrial animals, there are differences in host specificity and geographic prevalence relevant to disease management. Lactococcus petauri represents the greatest threat to wild and cultured trout in the Americas and may carry tetracycline resistance genes impacting antimicrobial efficacy. Understanding the local circulating diversity of LCB will enable better resource allocation, biosecurity measures, vaccination, and treatment interventions to limit disease spread and animal and economic losses.
MATERIALS AND METHODS
Bacteria
A total of 29 isolates were included in all aspects of this study, recovered from 27 fish and 2 mammalian cases in different regions of North America (Table S1). Up to 78 additional isolates were included in the expanded gyrB genotyping analysis or in qPCR assay validation. Most isolates (88/107) were recovered from diagnostic cases submitted to the Aquatic Animal Health Laboratory (AAHL) in the School of Veterinary Medicine at the University of California, Davis. Bacteria were stored in 1 mL aliquots in porcine brain heart infusion broth (BHI; MP Biomedicals) with 20% glycerol at −80°C. Before each assay, isolates revived from frozen stocks were cultured at 28°C for 24 h on trypticase soy agar supplemented with 5% sheep’s blood (SBA; University of California, Biological Media Services). Isolate growth in liquid media was compared at different temperatures to determine optimal conditions for further analysis. Briefly, revived bacteria were resuspended in sterile phosphate-buffered saline (PBS) to an OD600 of 0.15, equivalent to ~1.5×108 colony forming units (CFU) per mL, read on a UV/Vis photometer (BioPhotometer Plus, Eppendorf AG). Suspensions were diluted 1:1,000 in BHI and 150 µL aliquoted into a 96-well plate. Plates were incubated at 28 or 37°C with shaking and OD600 was recorded every hour for 40 h using a Cytation 5 Imaging Reader (BioTek, USA). Endpoint growth at 40°C was investigated on solid media.
DNA extraction
Bacterial strains used in whole-genome sequencing, conventional PCR, or qPCR analyses were revived from freezer stocks on SBA at 28°C for 24 h. A single isolated colony was then transferred into 5 mL BHI broth media for expansion at 28°C for 24 h with shaking. One milliliter of the expanded bacterial suspension was centrifuged for 10 min at 5,000 × g (7,500 rpm) and genomic DNA (gDNA) extracted from the concentrated pellet or directly from plate colonies using the DNeasy Blood and Tissue or PowerSoil Pro Kits (Qiagen, USA), following the manufacturer’s recommendations for gram-positive bacteria. The quality and quantity of recovered DNA were assessed using a NanoDrop One Microvolume UV-Vis Spectrophotometer (Thermo Fisher Scientific, USA), and samples with 260/280 ratios of 1.8–2.0 were cryogenically stored (−20°C) until further analysis.
Whole-genome analysis
Sequencing and assembly
Purified gDNA was submitted to GENEWIZ (Azenta Life Sciences, USA) for Illumina MiSeq sequencing (2 × 150 PE). Quality checking was performed with FastQC (v0.11.9, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) followed by quality trimming and adapter removal with Trimmomatic v0.39 (78). Oxford Nanopore sequencing was done in-house at the USDA ARS Warmwater Aquaculture Research Unit using the Native Barcoding Kit 24 (Oxford Nanopore, UK) and a GridION. Basecalling was performed with Guppy using the super-accurate model and sequence statistics were calculated in NanoStat (v1.6.0 (79)). Sequences were filtered with NanoFilt (min q = 10, min length = 1,000; v2.8.0 (79)), and assembled using Canu (v2.2 (80)) with consensus sequence correction by Medaka (v1.7.2, https://github.com/nanoporetech/medaka). Long-read assemblies were polished with Illumina reads using two rounds of Pilon (v1.24, (81)). Chromosomes were annotated using Proksee (49) integrating the CARD Resistance Gene Identifier (51) to identify known antimicrobial resistance genes, the Bakta light gram-positive database (50) to identify putative proteins, and mobileOG-db (82) for mobile genetic elements, under default settings. Plasmids were annotated with pLannotate (83) (<50 kb) or Bakta (≥50 kb), CARD, and mobileOG-db in Proksee when present.
Phylogenetic analysis
Genome assemblies and 21 reference genomes (Table S2) from Lactococcus species validly published under the International Code of Nomenclature of Prokaryotes (ICNP) (84) were uploaded to the Type Strain Genome Server (TYGS) for whole-genome-based taxonomic analysis ((85); https://tygs.dsmz.de). For the phylogenomic inference, all pairwise comparisons among the set of genomes were conducted using GBDP and accurate intergenomic distances were inferred under the algorithm “trimming” and distance formula d5 (86). Digital DDH values and confidence intervals were calculated using the recommended settings of the GGDC 4.0 (86, 87), OrthoANI values were calculated using OAT 0.93.1 (Orthologous Average Nucleotide Identity Tool) (46). The resulting intergenomic distances were used to infer a balanced minimum evolution tree with branch support via FASTME 2.1.6.1 including SPR postprocessing (47). Branch support was inferred from 100 pseudo-bootstrap replicates each. The trees were rooted at the midpoint (48) and visualized with PhyD3 (88), then exported as Newick files for annotation in iTOL (53).
Conventional PCR
Sequencing and assembly
Amplification of 16S rRNA, gyrB, pheT, and rpoB genes was performed using previously published primers (6) and Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific, USA) following the manufacturer’s instructions. Reaction mixtures (20 µL) consisted of 4 µL 5× Phusion HF Buffer, 0.4 µL dNTPs (10 mM), 1 µL each of the respective forward and reverse primers (10 µM), 0.2 µL Phusion polymerase (2 U/µL), 1–2 µL of template DNA and up to 20 µL diethyl pyrocarbonate (DEPC) water. Thirty seconds of initial denaturation at 98°C was followed by 35 cycles of 98°C for 10 s, 57°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 5 min. For difficult templates, 3% DMSO was included in Phusion reaction mixtures, and annealing temperatures were adjusted to 6°C below the suggested melting temperature for each primer set.
Amplification reactions were passed by electrophoresis through 1% agarose gels supplemented with SYBR Safe DNA gel stain (Invitrogen, USA; 1 µL/mL) alongside concurrently run molecular weight standards (Quick-Load Purple 100 bp DNA Ladder, New England BioLabs, USA) and visualized under ultraviolet light to confirm the presence of appropriately sized bands. PCR products were purified using the QIAquick PCR Purification Kit (Qiagen, USA), and their concentration and purity were assessed by Nanodrop. Purified products and corresponding forward and reverse primers were diluted and submitted for Sanger sequencing through GENEWIZ (Azenta, USA). Forward and reverse sequences were imported into Geneious Prime (v 2023.2.1), trimmed to a 0.01 error probability limit, and de novo assembled. Isolate sequences for each gene were aligned by MUSCLE using default settings and trimmed to the region of quality bases shared by all isolate sequences. A representative trimmed sequence of the 16S rRNA, arcC, or pheT genes was used in BLAST searches of a localized database populated by 21 Lactococcus type strain genomes (Table S2). A database including any additional LCB genomes currently available on NCBI (n = 223; Table S2) was used for the expanded gyrB sample set. For MLSA, the sequences were concatenated in alphabetical order before phylogenetic analysis.
Phylogenetic analysis
Trimmed 16S rRNA, gryB, or concatenate sequences wre exported to MEGA-X (55) and aligned by MUSCLE using the default settings. The best-fit substitution model for each data set was selected based on Bayesian and Akaike information criteria; the Kimura 2-parameter model (56) was used for the 16S rRNA tree and the Tamura 3-parameter (54) for the gyrB and MLSA trees. Maximum likelihood trees were generated independently for each data set. The percentage bootstrap confidence levels were calculated from 1,000 re-samplings of the original data. Phylogenetic trees were exported from MEGA as Newick files and formatted and annotated in iTOL (53).
Quantitative PCR
Primer design
Whole-genome sequences using Illumina MiSeq were obtained for six isolates of L. petuari and two of L. formosensis that were recovered from clinical cases submitted to the UC Davis AAHL for testing. The genome assembly was done at the Animal Health Laboratory, University of Guelph, Ontario, using BioNumerics software, version 7.6 (BioMerieux) and upon visual inspection, a few DNA regions unique to L. petuari were detected, with putative protein 6 showing to be unique to L. petauri based on initial GenBank BLASTN analysis. A standard amino acid translation for this gene was generated from the full coding sequence (CDS) of a fish strain reference genome (LG_SAV_20) and submitted for a protein-protein BLAST against the NCBI non-redundant protein sequence database ((24, 89), https://blast.ncbi.nlm.nih.gov/Blast.cgi). The primers and the probe were designed using the PrimerQuest tool (Integrated DNA Technologies) and target a 126 bp region within the putative protein CDS (Table 2).
Primer specificity was investigated in silico against whole-genome sequences of the 29 representative isolates (Table S1) and 21 Lactococcus type strain genomes, in addition to 67 currently available L. petauri assemblies on Genbank (Table S2). Specificity was compared between the newly generated Lp-p6 assay and a previously published LCB TaqMan qPCR assay targeting the 16S–23S rRNA ITS region (Table 2 (44)). Binding was tested for each primer-probe set against each genome using Geneious Prime (v 2023.2.1) allowing for up to five mismatches in the binding region.
In vitro testing
Primer specificity was confirmed using gDNA from 59 typed clinical isolates of LCB (n = 36) and other related Lactococcus spp. (n = 21) extracted as described previously. Procedures for the LG-ITS assay were adapted from Shahin et al. (44). The reaction mixture for the Lp-P6 assay consisted of 1 × TaqMan Environmental PCR Master Mix (Applied Biosystems, USA), 0.4 µM of each primer, 0.08 µM of TaqMan probe, 5 µL of template DNA, and nuclease-free water to a final reaction volume of 12 µL. A concentration of 1 ng/µL was used for pure bacterial DNA and 50 ng/µL for tissue samples. The same reaction mixtures but with PCR-grade water were used as a negative non-template control. The amplification conditions for both assays were as follows: 60°C for 30 s, 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s and annealing at 60°C for 1 min. All samples were run in triplicate in MicroAmp 96-well reaction plates (Applied Biosystems) using a QuantStudio 3 Real-time PCR thermal cycler (Thermo Fisher Scientific, USA). A cycle threshold value (Ct) of <35 in all replicate wells was considered a positive detection.
PCR products from the Lp-p6 primer set were used to evaluate the analytical sensitivity of the qPCR assay using 5 µL of a dilution range of 105 to 1 copies/µL. The qPCR was repeated three times using individual master mixes. Each run included triplicate reactions and a negative control (PCR grade water only). Amplification efficacy was obtained from the standard curve using the equation E = 10(−1/S) − 1, where “S” is the standard curve slope (90). To evaluate the analytical specificity of the qPCR reaction, the assay was tested using 5 µL of gDNA of a representative of each LCB species—L. petauri (UCD-JR1), L. formosensis (PAQ-208), and L. garvieae (VC11)—serially diluted in DPEC water and using the same conditions mentioned above.
The diagnostic performance of the newly developed Lp-p6 assay was compared to the LG-ITS assay using both spiked rainbow trout (O. mykiss) DNA and experimentally infected fish samples. Isolate UCD-JR1 gDNA was added to trout posterior kidney, spleen, liver, gill, and heart DNA at a concentration ratio of 1:50 ng/µL and serially diluted. Samples previously generated by Littman et al. (24) were used to investigate the detection of infected fish tissues. Briefly, rainbow trout (O. mykiss) were experimentally infected with L. petauri UCD-JR1 or sham-infected with sterile PBS by intracoelomic injection at 18°C. Fish were monitored for 21 days, and tissues were collected from mortalities and survivors. Reaction conditions followed those described above. The TaqMan Exogenous Internal Positive Control (IPC) Reagents (Catalogue no. 4308323, Thermo Fisher Scientific) were included for each sample to evaluate potential PCR inhibition. Each sample was run in triplicate including a positive control (L. petauri gDNA) and a non-template negative control (PCR-grade water only) per plate.
Phenotypical characterization
Rapid biochemical test systems
Phenotypic fingerprints for the 29 representative isolates of L. petuari (n = 19), L. formosensis (n = 4), and L. garvieae (n = 6) were generated using BIOLOG GEN III MicroPlate and API 20 STREP systems. BIOLOG testing followed the manufacturer’s protocol C1. Isolates were streaked onto BIOLOG BUG+B agar plates (BIOLOG, USA) and incubated at 35°C for 24 h. The inoculum for the microplates was prepared by adjusting the cell density to a transmittance of 95% in inoculating fluid C and then used for inoculation. Microplates were incubated at 35°C for 24 h, and then analyzed using a Biolog MicroStation and Biolog’s Microbial Identification Systems software. Test results identified as borderline by the software were considered negative.
API testing was similarly performed and interpreted according to the manufacturer’s protocol but with temperature adjusted to 28°C. Bacteria were revived on SBA incubated at 28°C for 24 h, then resuspended in 2 mL API Suspension Medium to a visual turbidity greater than a 4 McFarland standard (≥ 1.2 x 109 CFU/mL). The suspension was accordingly distributed into test strip cupules, and mineral oil was added when relevant. The test strip in its humidified incubation box was incubated for 4 hours at 28°C, and then respective reagents were added for the VP, HIP, PYRA, αGAL, ßGUR, ßGAL, PAL, and LAP tests. The strip was read after 10 minutes, returned to incubation, then read again at 24 h. The numerical profile for each isolate was determined following manufacturer guidelines and entered into the APIWEB database for identification.
MALDI-TOF mass spectrometry (MS)
Custom MALDI-TOF profiles for L. petauri and L. formosensis were built following the instrument manufacturer’s protocol (91). Of the 29 characterized strains (Table S1), 6 L petauri (UCD-JR1, Mic16-Lg09, S69-A, M17100408 FF4i7, R22-11 BR-C, and R21-74 AL-1) and 4 L. formosensis (PAQ-208, M20011502 FF20a2, M04020401 EE5e5, and M03102409 EE5d7) were selected to build each custom profile. Isolates were grown at 30°C for 24 h on 5% Columbia SBA (Hardy Diagnostics, USA); colonies were then prepared using the Bruker methods for the extended direct preparation and the full extraction procedure. For the extended direct method, one colony was spotted onto each of 4 target plate spots, allowed to dry, overlaid with 1 µL of 70% formic acid (LC-MS grade, Sigma-Aldrich, USA), allowed to dry, and then overlaid with 1 µL of matrix [2-Cyano-3-(4-hydroxyphenyl) acrylic acid (HCCA), Bruker Daltonics, USA]. For the extraction procedure, five individual isolated colonies on the Colombia SBA culture plate for each isolate were extracted by suspending in 300 µL of UHPLC Grade water (Sigma-Aldrich, USA) and vortexing to create a homogeneous suspension, then adding 900 µL of 100% Ethanol (Sigma-Aldrich, USA). The suspension was vortexed thoroughly, then centrifuged at 12,100 × g for 2 min at room temperature. The supernatant was decanted completely, and a second centrifugation was performed to facilitate the removal of residual alcohol with a Pasteur pipette. The pellet was air-dried for 2–3 min, then 25 µL of 70% Formic acid (LC-MS grade Sigma-Aldrich, USA) was added to the pellet and the suspension was mixed by carefully pipetting 7–10 times, after which 25 µL of 100% Acetonitrile (UHPLC Grade > 99.9%, Sigma-Aldrich, USA) was added. The resulting suspension was centrifuged for 2 min at 12,100 × g, a volume of 1 µL of the supernatant was applied to four spots on the target plate, air-dried, and then overlaid with 1 µL of the HCCA matrix. The target plate was analyzed in a MALDI Biotyper sirius one (Bruker Scientific LLC, USA) by interrogating each of the eight spots (four extracted, four extended) for each isolate with the laser three times to collect 24 profiles. Each profile underwent a quality check to search for outliers, flat lines, or dramatic shifts in peaks. Using the MBT Compass Explorer software, a minimum of 20 profiles from extended direct and extracted spots were combined to create the custom Main Spectra Profiles (MSPs) for each isolate which were then uploaded as separate strains into the 2022 MBT Compass Library (Research Use Only) database previously installed on the Center for Animal Health and Food Safety (CAHFS) internal network. To probe the 29 isolates against the custom library, each isolate was grown at 30°C for 24 h on 5% Columbia SBA and prepared using the extraction method then spotted in duplicate on the target plate. The spots were analyzed with the laser in a MALDI Biotyper sirius one and resultant spectra were matched to profiles in the custom library 2022 MBT Compass Library modified with MSPs for L. petauri and L formosensis.
Fatty acid methyl ester analysis
The 29 LCB isolates were streaked onto SBA plates (TSA + 5% sheep blood, Remel, USA) and incubated for 24 h at 28°C. An average of 70 mg of bacterial wet weight was harvested in duplicate from each isolate and utilized to attain FAME profiles. Saponification of bacteria, methylation, FAME extraction, and gas chromatography of FAMEs were performed as previously described (92, 93). Following gas chromatography, samples were analyzed using the Sherlock Microbial Identification System (MIS) RCLIN6 6.2 library (version 6.2. MIDI, Inc). The duplicate FAME results for each isolate were averaged and then used for calculating the average for each species.
Antimicrobial susceptibility profiling
Antimicrobial susceptibility was compared between the 29 selected strains using the Sensititre Avian AVIAN1F AST Plate system (Thermo Fisher Scientific, USA) following the manufacturer’s protocol and CLSI recommendations (57), as previously described in Shahin et al. (6). Briefly, a 0.5 McFarland suspension of each strain was diluted 1:1,000 (~1.5 × 105 CFU/mL) into cation-adjusted Mueller-Hinton broth with 5% lysed horse blood (CAMHB) then 50 µL of suspension distributed to Sensititre plate wells. Plates were incubated at 28°C and the MIC for each antimicrobial was determined by the absence of bacterial growth in the relevant wells after 24-h incubation. At least one strain from each LCB species was repeated to confirm reproducibility. Escherichia coli ATCC 25922 and Streptococcus pneumoniae ATCC 49619 were used as reference quality control (QC) strains, incubated at 28°C.
ACKNOWLEDGMENTS
We would like to acknowledge past and present members of the Aquatic Animal Health Laboratory and the California Department of Wildlife for their roles in responding to initial lactococcosis outbreaks and sample collection. We would also like to acknowledge Cyndi Ware at Mississippi State University for sample processing and Marissa Simpson-Roberts from the UC Davis VMTH Microbiology Laboratory for her assistance in using the APIWEB database. Additional thanks to Joe Marcino from the Arizona Game and Fish Department, and Coja Yamashita from the Pennsylvania Fish and Boat Commission for providing LCB isolates.
The California Department of Wildlife supported this project through Statewide Fish Health Research Contract 2022–2025. This research used resources provided by the SCINet project and/or the AI Center of Excellence of the USDA Agricultural Research Service, ARS project numbers 0201-88888-003-000D and 0201-88888-002-000D.
Contributor Information
Esteban Soto, Email: sotomartinez@ucdavis.edu.
Knut Rudi, Norwegian University of Life Sciences, Ås, Norway.
DATA AVAILABILITY
Whole-genome sequence data are accessible on NCBI under BioProject PRJNA1055960. All phylogenetic trees generated in this study can be accessed at https://itol.embl.de/shared/tiheckman under the project “Redefining Piscine Lactococcosis.”
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.02349-23.
Tables S1 to S9; Figures S1 and S2.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. FAO . 2020. The state of world fisheries and aquaculture 2020: sustainability in action. FAO, Rome, Italy. doi: 10.4060/ca9229en. [DOI] [Google Scholar]
- 2. Lafferty KD, Harvell CD, Conrad JM, Friedman CS, Kent ML, Kuris AM, Powell EN, Rondeau D, Saksida SM. 2015. Infectious diseases affect marine fisheries and aquaculture economics. Ann Rev Mar Sci 7:471–496. doi: 10.1146/annurev-marine-010814-015646 [DOI] [PubMed] [Google Scholar]
- 3. Rodger HD. 2016. Fish disease causing economic impact in global aquaculture, p 1–34. In Adams A (ed), Fish vaccines. Springer, Basel. doi: 10.1007/978-3-0348-0980-1_1. [DOI] [Google Scholar]
- 4. Tavares-Dias M, Martins ML. 2017. An overall estimation of losses caused by diseases in the Brazilian fish farms. J Parasit Dis 41:913–918. doi: 10.1007/s12639-017-0938-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Avci H, Birincioglu SS, Tanrikul TT, Epikmen ET, Metin N, Avsever ML. 2014. Experimental Lactococcus garvieae infection in rainbow trout, Oncorhynchus mykiss, Walbaum 1792: a comparative histopathological and immunohistochemical study. J Fish Dis 37:481–495. doi: 10.1111/jfd.12132 [DOI] [PubMed] [Google Scholar]
- 6. Shahin K, Veek T, Heckman TI, Littman E, Mukkatira K, Adkison M, Welch TJ, Imai DM, Pastenkos G, Camus A, Soto E. 2021. Isolation and characterization of Lactococcus garvieae from rainbow trout, Oncorhynchus mykiss, from California, USA. Transbound Emerg Dis 69:2326–2343. doi: 10.1111/tbed.14250 [DOI] [PubMed] [Google Scholar]
- 7. Kotzamanidis C, Malousi A, Bitchava K, Vafeas G, Chatzidimitriou D, Skoura L, Papadimitriou E, Chatzopoulou F, Zdragas A. 2020. First report of isolation and genome sequence of L. petauri strain from a rainbow trout lactococcosis outbreak. Curr Microbiol 77:1089–1096. doi: 10.1007/s00284-020-01905-8 [DOI] [PubMed] [Google Scholar]
- 8. Kusuda R, Salati F. 1993. Major bacterial diseases affecting mariculture in Japan. Annu Rev Fish dis 3:69–85. doi: 10.1016/0959-8030(93)90029-B [DOI] [Google Scholar]
- 9. Eldar A, Ghittino C, Asanta L, Bozzetta E, Goria M, Prearo M, Bercovier H. 1996. Enterococcus seriolicida is a junior synonym of Lactococcus garvieae, a causative agent of septicemia and meningoencephalitis in fish. Curr Microbiol 32:85–88. doi: 10.1007/s002849900015 [DOI] [PubMed] [Google Scholar]
- 10. Eldar A, Ghittino C. 1999. Lactococcus garvieae and Streptococcus iniae infections in rainbow trout Oncorhynchus mykiss: similar, but different diseases. Dis Aquat Organ 36:227–231. doi: 10.3354/dao036227 [DOI] [PubMed] [Google Scholar]
- 11. Evans JJ, Klesius PH, Shoemaker CA. 2009. First isolation and characterization of Lactococcus garvieae from Brazilian Nile tilapia, Oreochromis niloticus (L.), and pintado, Pseudoplathystoma corruscans (Spix & Agassiz). J Fish Dis 32:943–951. doi: 10.1111/j.1365-2761.2009.01075.x [DOI] [PubMed] [Google Scholar]
- 12. Meyburgh CM, Bragg RR, Boucher CE. 2017. Lactococcus garvieae: an emerging bacterial pathogen of fish. Dis Aquat Organ 123:67–79. doi: 10.3354/dao03083 [DOI] [PubMed] [Google Scholar]
- 13. Mahmoud MM, Abdelsalam M, Kawato S, Harakawa S, Kawakami H, Hirono I, Kondo H. 2023. Comparative genome analyses of three serotypes of Lactococcus bacteria isolated from diseased cultured striped jack (Pseudocaranx dentex). J Fish Dis 46:829–839. doi: 10.1111/jfd.13792 [DOI] [PubMed] [Google Scholar]
- 14. Nelson MC, Varney JS, Welch TJ, Graf J. 2016. Draft genome sequence of Lactococcus garvieae strain PAQ102015-99, an outbreak strain isolated from a commercial trout farm in the Northwestern United States. Genome Announc 4:00781–16. doi: 10.1128/genomeA.00781-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. United States Department of Agriculture . 2019. 2018 census of aquaculture.
- 16. Evans JJ, Pasnik DJ, Klesius PH, Al-Ablani S. 2006. First report of Streptococcus agalactiae and Lactococcus garvieae from a wild bottlenose dolphin (Tursiops truncatus). J Wildl Dis 42:561–569. doi: 10.7589/0090-3558-42.3.561 [DOI] [PubMed] [Google Scholar]
- 17. Vendrell D, Balcázar JL, Ruiz-Zarzuela I, de Blas I, Gironés O, Múzquiz JL. 2006. Lactococcus garvieae in fish: a review. Comp Immunol Microbiol Infect Dis 29:177–198. doi: 10.1016/j.cimid.2006.06.003 [DOI] [PubMed] [Google Scholar]
- 18. Neupane S, Rao S, Yan W-X, Wang P-C, Chen S-C. 2023. First identification, molecular characterization, and pathogenicity assessment of Lactococcus garvieae isolated from cultured pompano in Taiwan. J Fish Dis 46:1295–1309. doi: 10.1111/jfd.13848 [DOI] [PubMed] [Google Scholar]
- 19. Rao S, Chen M, Sudpraseart C, Lin P, Yoshida T, Wang P, Chen S. 2022. Genotyping and phenotyping of Lactococcus garvieae isolates from fish by pulse-field gel electrophoresis (PFGE) and electron microscopy indicate geographical and capsular variations. J Fish Dis 45:771–781. doi: 10.1111/jfd.13601 [DOI] [PubMed] [Google Scholar]
- 20. Gibello A, Galán-Sánchez F, Blanco MM, Rodríguez-Iglesias M, Domínguez L, Fernández-Garayzábal JF. 2016. The zoonotic potential of Lactococcus garvieae: an overview on microbiology, epidemiology, virulence factors and relationship with its presence in foods. Res Vet Sci 109:59–70. doi: 10.1016/j.rvsc.2016.09.010 [DOI] [PubMed] [Google Scholar]
- 21. Colagrossi L, Costabile V, Scutari R, Agosta M, Onori M, Mancinelli L, Lucignano B, Onetti Muda A, Del Baldo G, Mastronuzzi A, Locatelli F, Trua G, Montanari M, Alteri C, Bernaschi P, Perno CF. 2022. Evidence of pediatric sepsis caused by a drug resistant Lactococcus garvieae contaminated platelet concentrate.. Emerg Microbes Infect 11:1325–1334. doi: 10.1080/22221751.2022.2071174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Malek A, De la Hoz A, Gomez-Villegas SI, Nowbakht C, Arias CA. 2019. Lactococcus garvieae, an unusual pathogen in infective endocarditis: case report and review of the literature. BMC Infect Dis 19:301. doi: 10.1186/s12879-019-3912-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Desiderato CK, Hasenauer KM, Reich SJ, Goldbeck O, Holivololona L, Ovchinnikov KV, Reiter A, Oldiges M, Diep DB, Eikmanns BJ, Riedel CU. 2022. Garvicin Q: characterization of biosynthesis and mode of action. Microb Cell Fact 21:236. doi: 10.1186/s12934-022-01952-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Littman EM, Heckman TI, Yazdi Z, Veek T, Mukkatira K, Adkison M, Powell A, Camus A, Soto E. 2023. Temperature-associated virulence, species susceptibility and interspecies transmission of a Lactococcus petauri strain from rainbow trout. Dis Aquat Organ 155:147–158. doi: 10.3354/dao03747 [DOI] [PubMed] [Google Scholar]
- 25. Abraham T, Yazdi Z, Littman E, Shahin K, Heckman TI, Quijano Cardé EM, Nguyen DT, Hu R, Adkison M, Veek T, Mukkatira K, Richey C, Kwak K, Mohammed HH, Ortega C, Avendaño-Herrera R, Keleher W, LePage V, Gardner I, Welch TJ, Soto E. 2023. Detection and virulence of Lactococcus garvieae and L. petauri from four lakes in southern California. J Aquat Anim Health 35:187–198. doi: 10.1002/aah.10188 [DOI] [PubMed] [Google Scholar]
- 26. Soltani M, Baldisserotto B, Hosseini Shekarabi SP, Shafiei S, Bashiri M. 2021. Lactococcosis a re-emerging disease in aquaculture: disease significant and phytotherapy. Vet Sci 8:181. doi: 10.3390/vetsci8090181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Savvidis G, Anatoliotis C, Kanaki Z, Vafeas G. 2007. Epizootic outbreaks of Lactococcosis disease in rainbow trout, Oncorhynchus mykiss (Walbaum), culture in Greece. Bull Eur Assoc Fish Pathol 27:223–228. [Google Scholar]
- 28. Torres-Corral Y, Santos Y. 2022. Predicting antimicrobial resistance of Lactococcus garvieae: PCR detection of resistance genes versus MALDI-TOF protein profiling. Aquaculture 553:738098. doi: 10.1016/j.aquaculture.2022.738098 [DOI] [Google Scholar]
- 29. de Ruyter T, Littman E, Yazdi Z, Adkison M, Camus A, Yun S, Welch TJ, Keleher WR, Soto E. 2023. Comparative evaluation of booster vaccine efficacy by Intracoelomic injection and immersion with a whole-cell killed vaccine against Lactococcus petauri infection in rainbow trout (Oncorhynchus mykiss). Pathogens 12:632. doi: 10.3390/pathogens12050632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nishiki I, Oinaka D, Iwasaki Y, Yasuike M, Nakamura Y, Yoshida T, Fujiwara A, Nagai S, Katoh M, Kobayashi T. 2016. Complete genome sequence of nonagglutinating Lactococcus garvieae strain 122061 isolated from yellowtail in Japan. Genome Announc 4:e00592-16. doi: 10.1128/genomeA.00592-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goodman LB, Lawton MR, Franklin-Guild RJ, Anderson RR, Schaan L, Thachil AJ, Wiedmann M, Miller CB, Alcaine SD, Kovac J. 2017. Lactococcus petauri sp. nov., isolated from an abscess of a sugar glider. Int J Syst Evol Microbiol 67:4397–4404. doi: 10.1099/ijsem.0.002303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Altinok I, Ozturk RC, Ture M. 2022. NGS analysis revealed that Lactococcus garvieae Lg-Per was Lactococcus petauri in Türkiye. J Fish Dis 45:1839–1843. doi: 10.1111/jfd.13708 [DOI] [PubMed] [Google Scholar]
- 33. Chen Y-S, Otoguro M, Lin Y-H, Pan S-F, Ji S-H, Yu C-R, Liou M-S, Chang Y-C, Wu H-C, Yanagida F. 2014. Lactococcus formosensis sp. nov., a lactic acid bacterium isolated from yan-tsai-shin (fermented broccoli stems). Int J Syst Evol Microbiol 64:146–151. doi: 10.1099/ijs.0.052811-0 [DOI] [PubMed] [Google Scholar]
- 34. Collins MD, Farrow JA, Phillips BA, Kandler O. 1983. Streptococcus garvieae sp. nov. and Streptococcus plantarum sp. nov. J Gen Microbiol 129:3427–3431. doi: 10.1099/00221287-129-11-3427 [DOI] [PubMed] [Google Scholar]
- 35. Schleifer KH, Kraus J, Dvorak C, Kilpper-Bälz R, Collins MD, Fischer W. 1985. Transfer of Streptococcus lactis and related streptococci to the genus Lactococcus gen. nov. Syst Appl Microbiol 6:183–195. doi: 10.1016/S0723-2020(85)80052-7 [DOI] [Google Scholar]
- 36. Ferrario C, Ricci G, Milani C, Lugli GA, Ventura M, Eraclio G, Borgo F, Fortina MG. 2013. Lactococcus garvieae: where is it from? A first approach to explore the evolutionary history of this emerging pathogen. PLOS One 8:e84796. doi: 10.1371/journal.pone.0084796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang HX, Tian WL, Gu CT. 2021. Proposal of Lactococcus garvieae subsp. bovis Varsha and Nampoothiri 2016 as a later heterotypic synonym of Lactococcus formosensis Chen et al. 2014 and Lactococcus formosensis subsp. bovis comb. nov. Int J Syst Evol Microbiol 71:004980. doi: 10.1099/ijsem.0.004980 [DOI] [PubMed] [Google Scholar]
- 38. Stoppani N, Colussi S, Pastorino P, Prearo M, Sciuto S, Altinok I, Öztürk RÇ, Ture M, Vela AI, Blanco MDM, Kotzamanidis C, Bitchava K, Malousi A, Fariano L, Volpatti D, Acutis PL, Fernández-Garayzábal JF. 2023. 16S-23S rRNA internal transcribed spacer region (ITS) sequencing: a potential molecular diagnostic tool for differentiating Lactococcus garvieae and Lactococcus petauri. Microorganisms 11:1320. doi: 10.3390/microorganisms11051320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Egger RC, Rosa JCC, de Pádua SB, de Oliveira Barbosa F, Zerbini MT, Tavares GC, Figueiredo HCP. 2022. Emerging fish pathogens Lactococcus petauri and L. garvieae in Nile tilapia (Oreochromis niloticus) farmed in Brazil. bioRxiv. doi: 10.1101/2022.08.19.504548 [DOI]
- 40. Saticioglu IB, Onuk EE, Ay H, Ajmi N, Demirbas E, Altun S. 2023. Phenotypic and molecular differentiation of Lactococcus garvieae and Lactococcus petauri isolated from trout. Aquaculture 577:739933. doi: 10.1016/j.aquaculture.2023.739933 [DOI] [Google Scholar]
- 41. Heckman TI, Yazdi Z, Griffin MJ, Waldbieser GC, Welch TJ, Abraham T, Littman EM, Chow AM, Shahin K, Mukkatira K, Richey C, Kwak K, Keleher B, Avendaño-Herrera R, Ortega C, Mitchell H, Mohammed HM, LePage V, Risatti G, Hyatt MW, Frasca S, Adkison M, Soto E. 2022. “For LAC of a better name: Redefining Piscine Lactococcosis” Proceedings of the 9th International Aymposium on Aquatic Animal Health; Santiago, Chile [Google Scholar]
- 42. Vela AI, del Mar Blanco M, Colussi S, Kotzamanidis C, Prearo M, Altinok I, Acutis PL, Volpatti D, Alba P, Feltrin F, Ianzano A, Domínguez L, Fernández-Garayzábal JF. 2024. The association of Lactococcus petauri with lactococcosis is older than expected. Aquaculture 578:740057. doi: 10.1016/j.aquaculture.2023.740057 [DOI] [Google Scholar]
- 43. Dang HT, Park HK, Myung SC, Kim W. 2012. Development of a novel PCR assay based on the 16S–23S rRNA internal transcribed spacer region for the detection of Lactococcus garvieae. J Fish Dis 35:481–487. doi: 10.1111/j.1365-2761.2012.01382.x [DOI] [PubMed] [Google Scholar]
- 44. Shahin K, Mukkatira K, Yazdi Z, Richey C, Kwak K, Heckman TI, Mohammed HH, Ortega C, Avendaño-Herrera R, Keleher B, Hyatt MW, Drennan JD, Adkison M, Griffin MJ, Soto E. 2022. Development of a quantitative polymerase chain reaction assay for detection of the aetiological agents of piscine lactococcosis. J Fish Dis 45:847–859. doi: 10.1111/jfd.13610 [DOI] [PubMed] [Google Scholar]
- 45. Ortega C, Irgang R, Valladares-Carranza B, Collarte C, Avendaño-Herrera R. 2020. First identification and characterization of Lactococcus garvieae isolated from rainbow trout (Oncorhynchus mykiss) cultured in Mexico. Animals (Basel) 10:1609. doi: 10.3390/ani10091609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee I, Ouk Kim Y, Park S-C, Chun J. 2016. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103. doi: 10.1099/ijsem.0.000760 [DOI] [PubMed] [Google Scholar]
- 47. Lefort V, Desper R, Gascuel O. 2015. FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol Biol Evol 32:2798–2800. doi: 10.1093/molbev/msv150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Farris JS. 1972. Estimating phylogenetic trees from distance matrices. Am Nat 106:645–668. doi: 10.1086/282802 [DOI] [Google Scholar]
- 49. Grant JR, Enns E, Marinier E, Mandal A, Herman EK, Chen C-Y, Graham M, Van Domselaar G, Stothard P. 2023. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res 51:W484–W492. doi: 10.1093/nar/gkad326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schwengers O, Jelonek L, Dieckmann MA, Beyvers S, Blom J, Goesmann A. 2021. Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb Genom 7:000685. doi: 10.1099/mgen.0.000685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alcock BP, Huynh W, Chalil R, Smith KW, Raphenya AR, Wlodarski MA, Edalatmand A, Petkau A, Syed SA, Tsang KK, et al. 2023. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res 51:D690–D699. doi: 10.1093/nar/gkac920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Heckman T, Griffin M, Camus A, LaFrentz B, Morick D, Smirnov R, Ofek T, Soto E. 2020. Multilocus sequence analysis of diverse Streptococcus iniae isolates indicates an underlying genetic basis for phenotypic heterogeneity. Dis Aquat Org. 141:53–69. doi: 10.3354/dao03521 [DOI] [PubMed] [Google Scholar]
- 53. Letunic I, Bork P. 2019. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47:W256–W259. doi: 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tamura K. 1992. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol 9:678–687. doi: 10.1093/oxfordjournals.molbev.a040752 [DOI] [PubMed] [Google Scholar]
- 55. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- 57. Clinical and Laboratory Standards Institute (CLSI) . 2015. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria. 3rd ed. Clinical and Laboratory Standards Institute, West Valley Road, Suite 2500, Wayne, Pennsylvania 19087 USA. [Google Scholar]
- 58. Clinical and Laboratory Standards Institute (CLSI) . 2020. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. 5th ed. Vol. 40. Clinical and Laboratory Standards Institute. [Google Scholar]
- 59. Kim W. 2014. The genus Lactococcus, p 429–443. In Lactic acid bacteria. John Wiley & Sons, Ltd. doi: 10.1002/9781118655252.ch26. [DOI] [Google Scholar]
- 60. Janda JM, Abbott SL. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 45:2761–2764. doi: 10.1128/JCM.01228-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lin Y, Han J, Barkema HW, Wang Y, Gao J, Kastelic JP, Han B, Qin S, Deng Z. 2023. Comparative genomic analyses of Lactococcus garvieae isolated from bovine mastitis in China. Microbiol Spectr 11:e02995-22. doi: 10.1128/spectrum.02995-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. California Department of Fish and Wildlife . 2021. FAQ for Lactococcus Garvieae outbreak in Southern California fish Hatcheries. https://nrm.dfg.ca.gov/FileHandler.ashx?DocumentID=180707&inline.
- 63. California Department of Fish and Wildlife . 2022. “New bacterial outbreak confirmed at two Eastern Sierra fish hatcheries” CDFW news. https://wildlife.ca.gov/News/new-bacterial-outbreak-confirmed-at-two-eastern-sierra-fish-hatcheries.
- 64. Feito J, Araújo C, Gómez-Sala B, Contente D, Campanero C, Arbulu S, Saralegui C, Peña N, Muñoz-Atienza E, Borrero J, del Campo R, Hernández PE, Cintas LM. 2022. Antimicrobial activity, molecular typing and in vitro safety assessment of Lactococcus garvieae isolates from healthy cultured rainbow trout (Oncorhynchus mykiss, Walbaum) and rearing environment. LWT 162:113496. doi: 10.1016/j.lwt.2022.113496 [DOI] [Google Scholar]
- 65. Salogni C, Bertasio C, Accini A, Gibelli LR, Pigoli C, Susini F, Podavini E, Scali F, Varisco G, Alborali GL. 2024. The characterisation of Lactococcus garvieae isolated in an outbreak of septicaemic disease in farmed sea bass (Dicentrarchus labrax, Linnaues 1758) in Italy. Pathogens 13:49. doi: 10.3390/pathogens13010049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pastorino P, Vela Alonso AI, Colussi S, Cavazza G, Menconi V, Mugetti D, Righetti M, Barbero R, Zuccaro G, Fernández-Garayzábal JF, Dondo A, Acutis PL, Prearo M. 2019. A summer mortality outbreak of Lactococcosis by Lactococcus garvieae in a raceway system affecting farmed rainbow trout (Oncorhynchus mykiss) and brook trout (Salvelinus fontinalis). Animals (Basel) 9:1043. doi: 10.3390/ani9121043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Algöet M, Bayley AE, Roberts EG, Feist SW, Wheeler RW, Verner-Jeffreys DW. 2009. Susceptibility of selected freshwater fish species to a UK Lactococcus garvieae isolate. J Fish Dis 32:825–834. doi: 10.1111/j.1365-2761.2009.01058.x [DOI] [PubMed] [Google Scholar]
- 68. Su F-J, Periyasamy T, Chen M-M. 2022. Comparative Transcriptomic immune responses of mullet (Mugil cephalus) infected by planktonic and biofilm Lactococcus garvieae Front Cell Infect Microbiol 12:887921. doi: 10.3389/fcimb.2022.887921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Food and Drug Administration . Approved Aquaculture Drugs. Available from: https://www.fda.gov/animal-veterinary/aquaculture/approved-aquaculture-drugs. Retrieved 1011 NovNovember 2023. Accessed , 1011 NovNovember 2023
- 70. Grossman TH. 2016. Tetracycline antibiotics and resistance. Cold Spring Harb Perspect Med 6:a025387. doi: 10.1101/cshperspect.a025387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kim S-R, Nonaka L, Suzuki S. 2004. Occurrence of tetracycline resistance genes tet(M) and tet(S) in bacteria from marine aquaculture sites. FEMS Microbiol Lett 237:147–156. doi: 10.1016/j.femsle.2004.06.026 [DOI] [PubMed] [Google Scholar]
- 72. Akmal M, Akatsuka M, Nishiki I, Yoshida T. 2023. Resistance and genomic characterization of a plasmid Pkh2101 harbouring Erm(B) isolated from emerging fish pathogen Lactococcus garvieae serotype II in Japan. J Fish Dis 46:841–848. doi: 10.1111/jfd.13793 [DOI] [PubMed] [Google Scholar]
- 73. Bjorland J, Steinum T, Sunde M, Waage S, Heir E. 2003. Novel Plasmid-borne gene qacJ mediates resistance to quaternary ammonium compounds in equine Staphylococcus aureus, Staphylococcus simulans, and Staphylococcus intermedius. Antimicrob Agents Chemother 47:3046–3052. doi: 10.1128/AAC.47.10.3046-3052.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Elliott JA, Facklam RR. 1996. Antimicrobial susceptibilities of Lactococcus lactis and Lactococcus garvieae and a proposed method to discriminate between them. J Clin Microbiol 34:1296–1298. doi: 10.1128/jcm.34.5.1296-1298.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shi Y-Z, Yoshida T, Fujiwara A, Nishiki I. 2021. Characterization of lsa(D), a novel gene responsible for resistance to lincosamides, streptogramins A, and pleuromutilins in fish pathogenic Lactococcus garvieae serotype II. Microb Drug Resist 27:301–310. doi: 10.1089/mdr.2020.0218 [DOI] [PubMed] [Google Scholar]
- 76. Barnes AC, Ellis AE. 2004. Role of capsule in serotypic differences and complement fixation by Lactococcus garvieae. Fish Shellfish Immunol 16:207–214. doi: 10.1016/S1050-4648(03)00079-2 [DOI] [PubMed] [Google Scholar]
- 77. Barnes AC, Guyot C, Hanse BG, Mackenzie K, Horn MT, Ellis AE. 2002. Resistance to serum killing may contribute to differences in the abilities of capsulate and non-capsulated isolates of Lactococcus garvieae to cause disease in rainbow trout (Oncorhynchus mykiss L.). Fish Shellfish Immunol 12:155–168. doi: 10.1006/fsim.2001.0361 [DOI] [PubMed] [Google Scholar]
- 78. Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. De Coster W, D’Hert S, Schultz DT, Cruts M, Van Broeckhoven C. 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34:2666–2669. doi: 10.1093/bioinformatics/bty149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. doi: 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLOS One 9:e112963. doi: 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Brown CL, Mullet J, Hindi F, Stoll JE, Gupta S, Choi M, Keenum I, Vikesland P, Pruden A, Zhang L. 2022. mobileOG-db: a manually curated database of protein families mediating the life cycle of bacterial mobile genetic elements. Appl Environ Microbiol 88:e0099122. doi: 10.1128/aem.00991-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. McGuffie MJ, Barrick JE. 2021. pLannotate: engineered plasmid annotation. Nucleic Acids Res. 49:W516–W522. doi: 10.1093/nar/gkab374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Parte AC, Sardà Carbasse J, Meier-Kolthoff JP, Reimer LC, Göker M. 2020. List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol 70:5607–5612. doi: 10.1099/ijsem.0.004332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Meier-Kolthoff JP, Göker M. 2019. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:2182. doi: 10.1038/s41467-019-10210-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Göker M. 2022. TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res 50:D801–D807. doi: 10.1093/nar/gkab902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kreft L, Botzki A, Coppens F, Vandepoele K, Van Bel M. 2017. PhyD3: a phylogenetic tree viewer with extended phyloXML support for functional genomics data visualization. Bioinformatics 33:2946–2947. doi: 10.1093/bioinformatics/btx324 [DOI] [PubMed] [Google Scholar]
- 89. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 90. Azinheiro S, Carvalho J, Prado M, Garrido-Maestu A. 2020. Multiplex detection of Salmonella spp., E.coli O157 and L. monocytogenes by qPCR melt curve analysis in spiked infant formula. Microorganisms 8:1359. doi: 10.3390/microorganisms8091359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Microbiology & Diagnostics . Creation of library entries tutorial 2020. Available from: https://www.bruker.com/en/services/training/microbiology-and-diagnostics.html. Retrieved 11 Dec 2023.
- 92. Reichley SR, Ware C, Steadman J, Gaunt PS, García JC, LaFrentz BR, Thachil A, Waldbieser GC, Stine CB, Buján N, Arias CR, Loch T, Welch TJ, Cipriano RC, Greenway TE, Khoo LH, Wise DJ, Lawrence ML, Griffin MJ. 2017. Comparative phenotypic and genotypic analysis of Edwardsiella isolates from different hosts and geographic origins, with emphasis on isolates formerly classified as E. tarda, and evaluation of diagnostic methods. J Clin Microbiol 55:3466–3491. doi: 10.1128/JCM.00970-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Soto E, Griffin MJ, Morales JA, Calvo EB, de Alexandre Sebastião F, Porras AL, Víquez-Rodríguez X, Reichley SR, Rosser TG, Ware C, Byrne BA, García JC, LaFrentz BR, Camus AC. 2018. Francisella Marina sp. nov., etiologic agent of systemic disease in cultured spotted rose snapper (Lutjanus guttatus) in Central America. Appl Environ Microbiol 84:e00144-18. doi: 10.1128/AEM.00144-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S9; Figures S1 and S2.
Data Availability Statement
Whole-genome sequence data are accessible on NCBI under BioProject PRJNA1055960. All phylogenetic trees generated in this study can be accessed at https://itol.embl.de/shared/tiheckman under the project “Redefining Piscine Lactococcosis.”