Highlights
-
•
HMGB2 could be an independent prognostic indicator leading to a shorter survival time and a higher risk of local progression in glioma patients.
-
•
HMGB2 potentially related to the BER pathway and could enhanced the radio-resistance of glioma cells.
-
•
HMGB2 expression level had an inverse association with ADCmean but positively with the thickness of enhancing margin.
Keywords: HMGB2, Glioma, Prognosis, Base excision repair, Radio-resistance
Abstract
Background
High mobility group box 2 (HMGB2) is considered as a biomarker of poor prognosis in various cancers.This study aims to investigate the effect and mechanism of HMGB2 in gliomas.
Methods
With the glioma related on-line and our local hospital databases, the expression differences of HMGB2,Kaplan-Meier survival analysis and COX regression analysis were performed.The correlation analysis between the clinicopathological features and imaging parameters with the HMGB2 expression had been done. Then GSEA and PPI networks were carried out to find out the most significant pathway. The pathway inhibitor was applied to verify HMGB2’s participation. CCK8,EDU assays,γ-H2AX immunofluorescence staining and colony formation assay were conducted to observe effects on glioma cells.
Results
Available datasets showed that HMGB2 was highly expressed in glioma and patients with high expression of HMGB2 had poorer prognosis and molecular characteristics. Protein level evidence of western blot and immunohistochemistry from our center supported the conclusions above. Analysis on imaging features suggested that HMGB2 expression level had an inverse association with ADCmean but positively with the thickness of enhancing margin. Results from GSEA and PPI network analysis exhibited that HMGB2 was involved in base excision repair (BER) signaling pathway. Experimental evidence demonstrated that the overexpression of HMGB2 promoted the proliferation of glioma cells and enhanced the radio-resistance.
Conclusions
HMGB2 could promote glioma development and enhance the radioresistance of glioma cells, potentially related to the BER pathway, suggesting it may serve as an underlying biomarker for patients with glioma.
Introduction
Gliomas, originating from glial cells, account for the majority of primary malignant brain tumors and it occurs in approximately 6 people per 100,000 each year [[1], [2], [3]]. Despite the continual improvement of the treatment strategy, the efficacy has not improved significantly. Especially those with high-grades, the 5-year overall survival rate does not exceed 5 % [4,5].Although the addition of tumor treating fields (TTFields) to maintenance temozolomide (TMZ) chemotherapy can increase the overall survival by nearly 5 months in glioma [6], it is not widely accepted because of high treatment costs, differences in long-term survival patterns, a burden of patients having to wear the device with high compliance, and other reasons [7,8].The alternative treatment options such as targeted therapy and immunotherapy have not yet demonstrated a clear benefit of survival.Thus,the glioma patients are still exposed to high risks of relapse and mortality.
In most cases, tumors are difficult to remove completely by surgical excision due to the infiltrating growth. The ionizing radiation and chemotherapy drugs induce DNA single or double strand breaks in glioma cells, which consequently leads to cell apoptosis and death. Meanwhile glioma cells initiate a series of DNA repair machineries to prevent cells death after DNA damage, resulting in high intrinsic radio-resistance and chemo-resistance [[9], [10], [11], [12]], which may lead to tumor progression and poor clinical outcome.Therefore, exploring effective strategies to reduce the resistance of glioma to chemo-radiotherapy is imperative.
Among the nonhistone chromosomal proteins, the high mobility group (HMG) proteins are the most abundant and ubiquitous participating in several biological processes. The high mobility group box 1(HMGB1),an prototypical member of HMGs, plays a significant role in DNA recombination, transcription, replication and repair [13,14]. As a signal factor, HMGB1 promotes the progression in multiple cancers and is closely related to tumor drug resistance [15,16].HMGB2 which has gradually attracted the attention of researchers in recent years has 80 % homology with HMGB1 and promotes the development of various malignant tumors like pancreatic cancer, cervical cancer, gastric cancer and liver cancer [[17], [18], [19], [20], [21]].Only a previous study using cell experiments found that HMGB2 might play a role in cells invasion and temozolomide induced chemotherapy resistance in glioblastoma patients [22], but whether it is involved in radiotherapy resistance and the specific mechanism has not yet been reported.The correlations of HMGB2 expression with conventional MRI characteristics and more comprehensive clinical biological indicators of glioma patients are not investigated. Therefore, an in-depth study of the role of HMGB2 in the progression of gliomas is of great significance for the evaluation of therapeutic efficacy and prognosis monitoring of gliomas.
In this study, the HMGB2 expression and its association with clinical outcomes and clinicopathological features especially the imaging features were investigated. The purpose is to guide clinical treatment and imaging differentiation diagnosis based the expression of glioma biomarker.Moreover, it was suggested that HMGB2 might be implicated in DNA damage repair pathways through potential interactions with other proteins. Furthermore, in vitro assays, we assessed the effects of HMGB2 overexpression and knockdown on glioma cell's proliferation and apoptosis, as well as radio-resistance.In conclusion,the role of HMGB2 in gliomas was thoroughly dissected, potentially suggesting a new therapeutic target in glioma therapy.
Materials and methods
Data download and differential expression analysis
We analyzed the level of HMGB2 expression in multiple cancers and their matched normal tissues using GEPIA website based on the Cancer Genome Atlas (TCGA,https://portal.gdc.cancer.gov/) database. The GSE 4290,GSE 7696,GSE 50,161 datasets were downloaded from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database. Similarly, the different expression of HMGB2 between 257 glioma samples and 20 controls was analyzed based on the data of the Chinese Glioma Genome Atlas (CGGA,http://www.cgga.org.cn/). We used GraphPad Prism 8 software to compare the differential expression level of HMGB2 between glioma samples and normal brain tissues.
Patient samples in our center
This study included 64 patients with glioma who underwent surgical resection and other comprehensive treatment in the Second Hospital of Hebei Medical University from January 2016 to March 2021. Western Blotting was performed in 12 pairs of glioma patients with their adjacent normal tissues to verify the differential HMGB2 expression between glioma and non-tumor tissues. The other 52 patients were used to observe the expression of HMGB2 in tumor tissues by IHC, and the pre-treatment magnetic resonance images (MRI) data were analyzed. All those were approved by the Ethics Committee of the Second Hospital of Hebei Medical University.
HMGB2 expression and its correlations with clinicopathological features
According to the expression level of HMGB2, 301 cases of mRNA microarray data from CGGA database were divided into high expression group and low expression group. The clinical prognosis of different groups was analyzed by R software. The mRNA sequencing data of 325 cases in CGGA database were used as internal verification, and the data of 52 cases in our hospital were used as external verification.The survival curves of different HMGB2 expression levels were drawn with "Survival" and "Survivminer" software packages in R version 4.0.5. We used the “Survival ROC” package with Kaplan-Meier method to calculate the 1-, 3-, and 5-year receiver operator characteristic (ROC) curves for HMGB2. Univariate and multivariate COX regression analysis were used to predict the prognosis at the significant level of P < 0.001. Then, based on the CGGA data, we used the "beeswarm" package in R version 3.6.3 to analyze the correlation between the expression of HMGB2 and clinicopathological features.
Relationship between HMGB2 expression and pre-treatment MRI imaging parameters
In our center,there were a total of 52 patients with full MRI images which were acquired within 1 week before operation.All these MRI data included four images:T1-weighted, T1-weighted postgadolinium, T2-weighted and T2 fluid attenuated inversion recovery (FLAIR).Of these,11 had diffusion weighted imaging (DWI) and the apparent diffusion coefficient (ADC) values (ADCmean, ADCmin, ADCmax) were extracted from ADC maps.Two neuroradiologists blinded to clinicopathological features analyzed the pre-treatment MRI imaging, if the opinions were different a consensus decision was made after their discussion. MRI image assessment mainly consisting of 11 qualitative features based on VASARI lexicon: (1)tumor epicenter (left,right); (2)largest diameter (< = 4.5 cm,>4.5 cm); (3)enhancing margin (smooth,irregular); (4)non contrast-enhancing tumor (nCET) margin (smooth,irregular); (5)Proportion Enhancing%(0–33,34–66,67–100); (6)Proportion necrosis%(0–33,34–66,67–100); (7)Proportion nCET%(0–33,34–66,67–100); (8)Thickness of enhancing margin (< = 3 mm,>3 mm); (9)Pial invasion (yes,no); (10)Deep WM invasion (yes,no) and (11)T1/FLAIR (T1<F,T1<<F,T1 = F). The exact description of these MRI imaging features can be found at the National Cancer Institute's Cancer Imaging Archive <https://wiki.cancerimagingarchive.net/display/Public/VASARI+Research+Project>. The details are listed in the supplementary file of VASARI MR Feature KEY. Based on these pre-treatment MRI imaging data, the relationship between HMGB2 expression with MRI imaging features was analyzed with GraphPad Prism 8 software.
Transcriptome analysis for pathway identification
The data were divided into high and low groups based on the median HMGB2 expression by the mRNAseq_325 and mRNA-array_301 glioma data downloaded from CGGA database.In order to explore the signal pathway involved in HMGB2, Gene set enrichment analysis (GSEA) website was used to analyze the data of high expression group and low expression group of HMGB2. The top twenty significant enrichment pathways of Kyoto Encyclopedia of Genes and Genomes (KEGG) were exhibited on the basis of |NES| (net enrichment score)>1,NOM p < 0.05, and FDR q < 0.25.On the basis of KEGG enrichment pathways, the interaction between HMGB2 and pathway proteins was established by STRING website (https://string-db.org/) website, and the correlation among these proteins was analyzed by Cytoscape (https://cytoscape.org/).
Western blot
Cells and tissues were lysed in RIPA buffer containing protein phosphatase inhibitor. After denaturation, protein extracts were separated by 10 % SDS-polyacrylamide gel and then transferred to PVDF membrane (Millipore). Then the membranes were incubated with primary antibodies (HMGB2: Proteintech, 1:1000; HMGB1: ZENBIO, 1:1000; GAPDH: Proteintech, 1:8000) overnight at 4 °C. In the second day, they were incubated with secondary antibodies bound to HRP and imaged with an enhanced chemiluminescence (ECL) system (BIO-RAD, USA).ImageJ software was used to analyze the images.
Immunohistochemical staining and analysis
The paraffin-embedded blocks were cut into slices of 4 mm thickness. Antigen repair is accomplished by boiling water in Tris-EDTA buffer (pH 9.0) for 60 minutes. Then the tissue slides were incubated with an anti-human rabbit HMGB2 (14,597–1-AP Proteintech,1:200) antibody. All immunohistochemical sections were evaluated and scored by two pathologists who did not know the clinical parameters with a semi-quantitative scoring system [23] containing the staining intensity (-:no staining; +:weak staining; ++:moderate staining; +++:strong staining) and the percentage of stained cells (0: < 5 %; 1:5 %-25 %; 2:26 %-50 %; 3:51 %-75 %; 4:>75 %). The staining intensity scores and the percentage of positive cells scores were then multiplied for each case [24]. All cases were categorized into high expression group and low expression group based on the immune response score.
Cell culture and Irradiation
In this study, the human glioma cell lines U87MG and U251MG were used for functional study which were obtained from the national cell line resource infrastructure of China. All cells were cultured in Dulbecco modified Iger medium (DMEM) supplemented with 10 % fetal bovine serum and 1 % penicillin/streptomycin with a 37 °C moist environment containing 5 % CO2. 5MW X-ray linear accelerator (Elekta Synergy, England Elekta Limited) was used to irradiate the cells with 2 Gy,4 Gy,6 Gy and 10 Gy.
Gene silencing and overexpression
The small interference RNA (siRNA), the overexpression vector (OE) for HMGB2 and the negative controls were provided by GenePharma (Shanghai, China). Using Lipofectamine 3000 kit (Thermo Fisher Scientific) according to the manufacturer's instructions, the siRNAs of HMGB2 were respectively transfected into U251MG and the overexpression vectors were respectively transfected into U87 MG cells. After incubated in DMEM for 48 hours, the protein expression of HMGB2 was verified, and then the cell experiments were carried out. The sequences of HMGB2-siRNA were CUGAACAUCGCCCAAAGAUTT (sence) and AUCUUUGGGCGAUGUUCAGTT (anti-sence).
RNA extraction and quantitative PCR
Total RNA was extracted from glioma cells using TRIzol (Invitrogen) reagent, then the complementary DNA (cDNA) was synthesized from RNA samples with reverse transcription kit (Thermo Fisher Scientific,Waltham,MA,United States). Quantitative PCR analysis was performed with YESEN (Shanghai, China) PCR kit using a qPCR system (Bio-Rad, United States). The HMGB2 primer sequences were as follows CCGGACTCTTCCGTCAATTTC (forward primer sequence) and GTCATAGCGAGCTTTGTCACT (reverse primer sequence). An internal control was conducted using GAPDH.
EDU Assay
The cells were maintained in 96-well plate at the density of 8000–10,000/well and incubated for 24 hours at 37 °C Celsius with 5 % CO2. Then the EdU reagent (Ribobio,Guangzhou,China) was added to the cells. Two hours later, the cells were fixed with 4 % paraformaldehyde for 30 minutes, and then punched with 0.2 % Triton X-100 for 20 minutes. Finally, the cells were stained by 1 × Apollo® 488. We used fluorescence microscope (Leica, Wetzlar, Germany) to observe the experimental results and the proportions of cell proliferation were the ratio of EdU-positive cells to DAPI-positive cells.
CCK-8 assay
According to the instructions we used the cell counting kit 8 to detect the proliferation of U251 and U87 cells after 0 Gy or 6 Gy irradiation. A seeding density of 2000–3000 cells per well was applied to 96-well plates and there were three duplicate holes in each group. The cells were incubated in the medium containing CCK-8 reagent (0.5 mg/mL) at 37 °C for 1 h, then the sample's absorption was measured at 450 nm by using a microplate reader (BioTek) at the same time each day for 7 days.
TUNEL assay
The TUNEL apoptosis assay kit (KeyGEN BioTECH, China) was used to detect the apoptotic cells according to the manufacturer's instructions. We fixed all cells with 4 % paraformaldehyde for 30 min, followed by permeabilization with 0.2 % Triton X-100 for 20 min. Then these cells were stained with terminal deoxynucleotidyl transferase (TDT) reaction mixture for 60 min. Nuclei of the cells were counterstained with DAPI for 10 min. Finally, the proportion of apoptotic cells was determined as the ratio of TUNEL positive cells to DAPI positive cells.
Clonogenic survival assay
Appropriate numbers of cells were seeded in 6-well plates and left to adhere overnight. Then exposed the cells to 0, 2, 4, 6, 8 and 10 Gy of X-rays respectively and cultured them for another two weeks. After that the cells were fixed with anhydrous ethanol containing 1 % methyl violet for 20 minutes and the number of surviving colonies (defined as those with > 50 cells) could be counted. SF was calculated using prism 9.0 software (Graphpad software,Inc.USA) based on the multitarget/single-hit model (SF = 1-[1-e−D/D0]N). The extrapolation number (N), the average lethal dose (D0), and the quasi‑threshold dose (DQ) were also calculated.
γ-H2AX Immunofluorescence Staining
After receiving the X-ray irradiation (5 Gy) the cells were seeded on a 24-well plate and incubated at 37 °C for 24 hours. At 0.5 h, 2 h, 6 h and 24 h after the irradiation, 4 % paraformaldehyde was applied to all cells for 30 minutes, followed by 0.2 % Triton X-100 for 20 minutes.Then these cells were incubated with anti-γ-H2AX antibody (Cell Signaling Technology,1:200) for 2 hours and stained with secondary antibodies (Abcam, 1:200) for 1 hour at room temperature. The nuclei were stained with DAPI (Solarbio) for 30 minutes. Finally, the fluorescence microscope was used to visualize the images.The γ-H2AX fluorescent spots in the nucleus were counted by Image J software.
Statistical analysis
The statistical analyses were carried out using R software (version: 4.0.1, http://www.r-project.org/), and Prism 8 (GraphPad Software,Inc). Significant differences between two groups were compared by Student's t test or χ2 test. To calculate the statistical significance of differences between experimental groups, one-way ANOVAs with multiple comparison tests were used.The correlation between HMGB2 expression levels and ADC values was calculated with Spearman's correlation coefficient. P < 0.05 was considered statistically significant.
Results
HMGB2 is highly expressed in gliomas
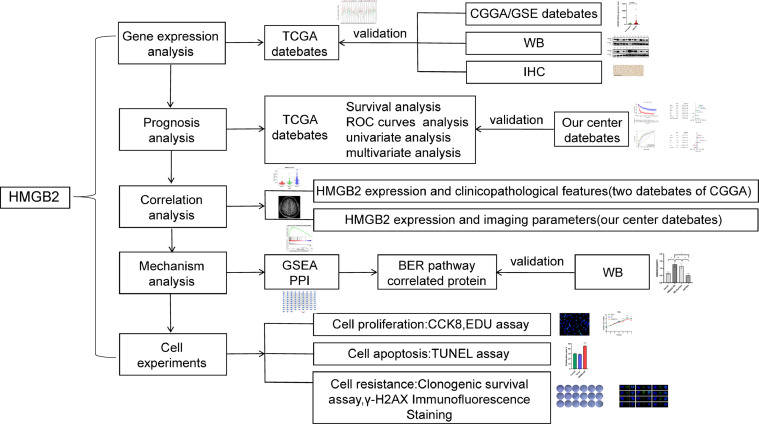
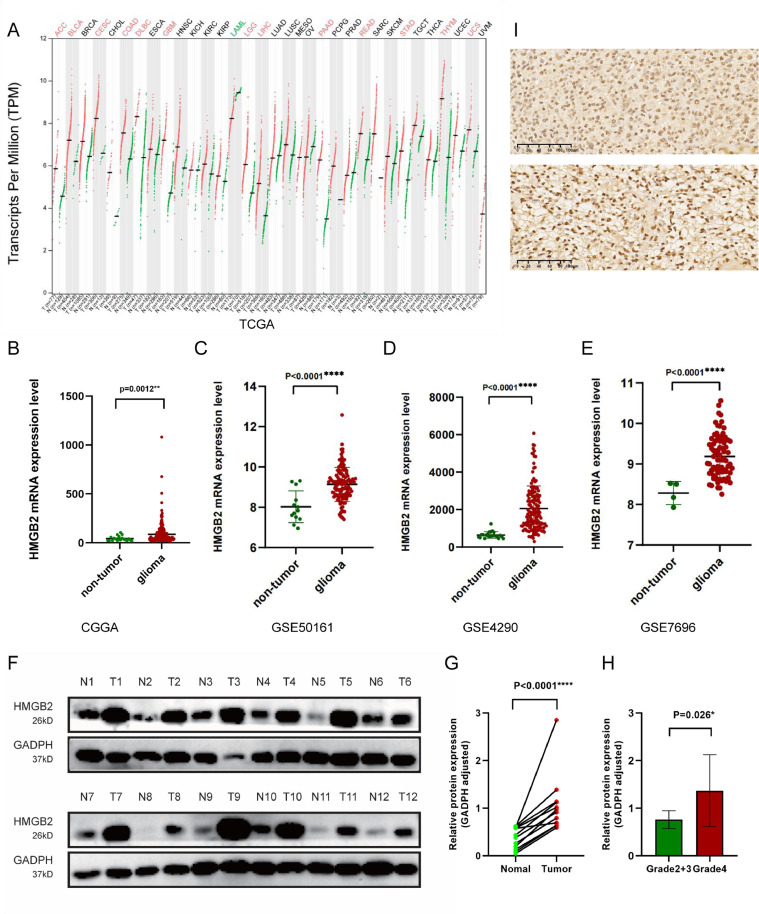
Fig. 1 shows the design flow chart of the study. According to the pan-cancer analysis of HMGB2 gene with TCGA datasets by Gene Expression Profiling Interactive Analysis (GEPIA), the HMGB2 expression levels were significantly higher in lower-grade gliomas (LGG) and glioblastoma multiforme (GBM) than non-tumor tissues (Fig. 2A). The similar results were verified by CGGA and GEO database (Fig. 2B-E). We selected 12 pairs of gliomas in our center and their para-cancerous tissues for WB analysis and the paired-sample T-test result showed that the expression of HMGB2 in gliomas was remarkably higher than that in corresponding adjacent tissues (p < 0.0001)(Fig. 2F-G), and the HMGB2 expression was greatly higher in Grade 4 gliomas than in LGG (Fig. 2H). Two GBM patients of IHC images revealed that HMGB2 was expressed in the nucleus (Fig. 2I).
Fig. 1.
Study flow diagram.
Fig. 2.
HMGB2 gene expression in glioma and non-tumor tissue.(A)The gene expression profile of HMGB2 across different cancer types in the TCGA database.In both LGG and GBM, HMGB2 shows statistically significant differences in expression between gliomas and non-tumor tissue.(B)CGGA mRNAseq_325 dataset. (C)GSE50161 dataset. (D)GSE4290 dataset. (E) GSE7696 dataset.(F)Western-blot in 12 pairs of gliomas and their adjacent tissues.(G)Paired-sample t-test of 12 glioma and their corresponding paracancerous tissues (4 grade II astrocytoma with IDH mutant, 2 grade III astrocytoma with IHD mutant, 1 grade IV IDH mutant astrocytoma and 5 glioblastoma). (H)Paired-sample t-test of Grade 4 gliomas and LGG tumor samples. (I)Immunohistochemical staining of two patients with Grade 4 gliomas.
High HMGB2 expression predicted poor prognosis in gliomas
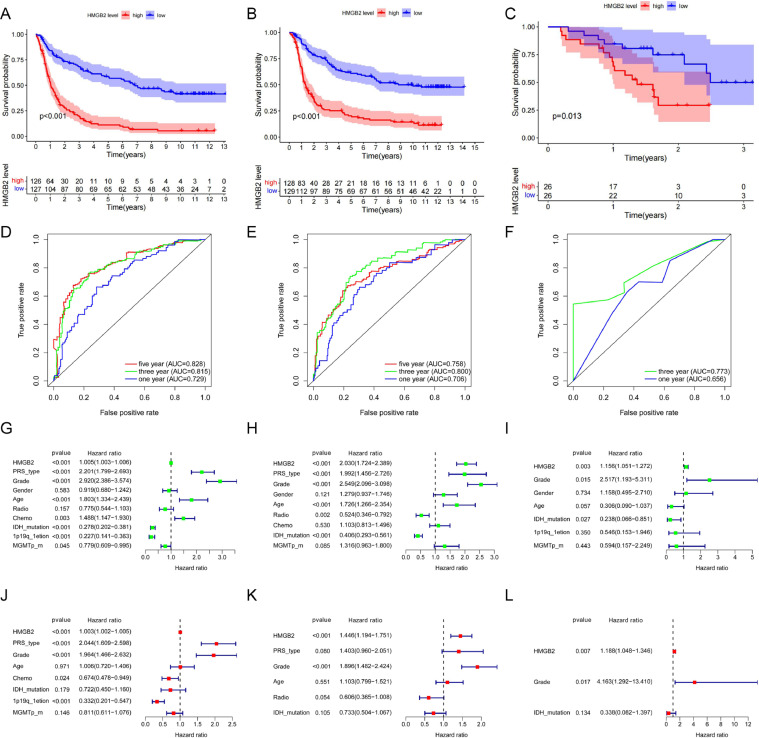
Kaplan-Meier survival analysis of CGGA-mRNA-array_301 and CGGA-mRNAseq_325 datasets showed that high expression of HMGB2 was associated with poor overall survival of gliomas (p < 0.001, Fig. 3 A, B).The ROC curves revealed that HMGB2 was a predictive marker for 1-, 3-and 5-year survival with the AUC values greater than 0.7 (Fig. 3D, E).Similarly, with the clinical information of 52 glioma patients in our center, the same results were verified. The survival time of these patients with high HMGB2 expression was shorter than the low expression (p = 0.013, Fig. 3C).The areas under the curves of OS are 0.656 for 1 year and 0.773 for 3 year (Fig. 3F). All the training, internal validation, and external validation datasets indicated that HMGB2 expression was an independent prognostic factor of OS in gliomas with univariate (P < 0.01) and multivariate (P < 0.01) cox regression analysis (Fig. 3G-L). According to the WHO 5th brain tumor classification,similar results were obtained for the LGG subgroup and Grade 4 glioma subgroup (see Supplement Fig. B.1).The clinicopathological data of patients were listed in Table 1.
Fig. 3.
The effect of HMGB2 expression on survival and prognosis of patients with glioma.(A-C) Overall survival analysis of HMGB2. (D-F) Survival ROC curve of HMGB2 at 1,3,and 5 years. (G-I)Univariate analysis of HMGB2. (G-L) Multivariate analysis of HMGB2.
Table 1.
Clinicopathological features of gliomas.
| Characteristics | CGGA-mRNAseq_325 | CGGA-mRNA-array_301 | Our center | |
|---|---|---|---|---|
| Gender | Male | 158 | 146 | 33 |
| Female | 95 | 111 | 19 | |
| Age | <42 | 120 | 123 | 11 |
| ≥42 | 133 | 134 | 41 | |
| Radiotherapy | Yes | 195 | 222 | 46 |
| No | 51 | 35 | 6 | |
| NA | 7 | 0 | 0 | |
| Chemotherapy | Yes | 146 | 124 | 46 |
| No | 96 | 133 | 6 | |
| NA | 11 | 0 | 0 | |
| WHO grade | II | 85 | 93 | 6 |
| III | 31 | 51 | 17 | |
| IV | 137 | 113 | 29 | |
| IDH mutation | Yes | 155 | 115 | 11 |
| No | 98 | 142 | 21 | |
| NA | 0 | 0 | 20 | |
| 1p19q codeletion | Yes | 51 | 14 | 7 |
| No | 199 | 67 | 20 | |
| NA | 3 | 176 | 25 | |
| MGMT methylation | Yes | 128 | 90 | 9 |
| No | 111 | 167 | 8 | |
| NA | 14 | 0 | 35 | |
The association between HMGB2 expression and clinicopathological features or imaging characteristics
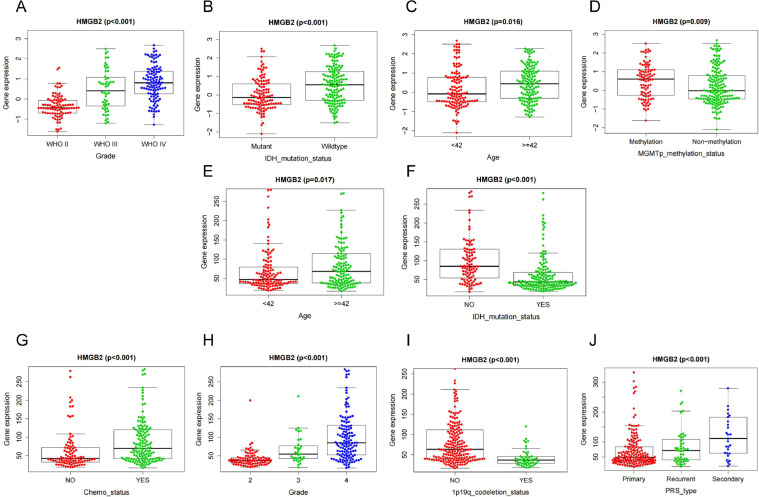
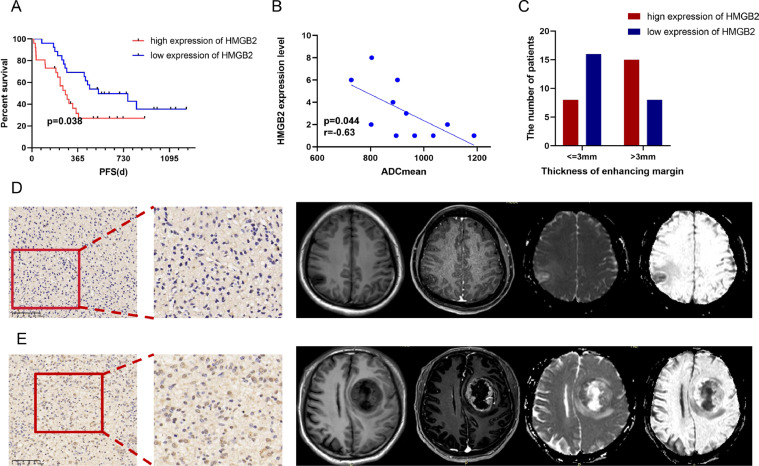
Based on the CGGA-mRNA-array_301 and CGGA-mRNAseq_325 online datasets, we found that the expression of HMGB2 was related to some clinicopathological features (Fig. 4A-D, E-J).Simultaneously the HMGB2 expression of IDH mutant status was lower than that of IDH wildtype (P < 0.001)(Fig. 4B, F) and higher HMGB2 expression indicated higher tumor grade (P < 0.001)(Fig. 4A, H) and older age (P < 0.05)(Fig. 4C,E). According to 52 glioma patients in our center it was found that the higher HMGB2 expression was associated with poor progression-free survival (PFS) (Fig. 5A) which means higher HMGB2 expression had a higher risk of relapse.We analyzed the clinical and imaging characteristics of these patients and it was identified that the expression of HMGB2 was related to apparent diffusion coefficient mean (ADCmean)(Fig. 5B) and thickness of enhancing margin (Fig. 5C). The expression of HMGB2 was higher in patients with the lower ADCmean (P = 0.044,r = -0.63) and the larger thickness of enhancing margin (P = 0.042). Comparison of the imaging characteristics of HMGB2 between high expression group and low group were shown in Table 2. Fig. 5D-E shows the features of IHC and MRI from two representative cases of glioma.
Fig. 4.
The association between HMGB2 gene and clinicopathological features based on CGGA-mRNA-array_301 database (A-D) and CGGA-mRNAseq_325 database (E-J). (A,H)Expression of HMGB2 gene in different grade.(B,F)Expression of HMGB2 gene in different IDH status.(C,E)Expression of HMGB2 gene in age.(D)Expression of HMGB2 gene in different MGMT status.(I)Expression of HMGB2 gene in different 1p19q status.(G)Expression of HMGB2 gene in different chemotherapy status.(J)Expression of HMGB2 gene in glioma types.
Fig. 5.
The association between HMGB2 and imaging characteristics.(A)progression-free survival analyse of HMGB2 based on 52 patients in our center.(B)The correlations between HMGB2 expression and ADCmean.(C)The correlations between HMGB2 expression and thickness of enhancing margin.(D)An IHC map with low HMGB2 expression and the corresponding MRI images of a 44-year-old male glioma patient of Grade 3 with ADCmean of 933.9 and thin thickness of enhancing margin.The patient's PFS was 556 days.(E)An IHC map with high HMGB2 expression of a 51-year-old male glioma patient of grade 4 with ADCmean of 727.4 and solid thickness of enhancing margin. The patient's PFS was 325 days.
Table 2.
The imaging parameters based on HMGB2 expression in our center.
| Characteristics | High group (n) | Low group (n) | P value (χ2) | |
|---|---|---|---|---|
| Tumor Epicenter | left | 11 | 12 | >0.999 |
| right | 15 | 14 | ||
| Largest diameter | < = 4.5cm | 10 | 14 | 0.404 |
| >4.5cm | 16 | 12 | ||
| Enhancing margin | Smooth | 16 | 19 | 0.538 |
| Irregular | 9 | 6 | ||
| noncon-trast-enhancing tumor (nCET) margin | Smooth | 18 | 11 | 0.093 |
| Irregular | 8 | 15 | ||
| Proportion Enhancing % | 0–33 | 23 | 22 | >0.999 |
| 34–100 | 3 | 4 | ||
| Proportion necrosis% | 0–33 | 20 | 20 | >0.999 |
| 34–100 | 6 | 6 | ||
| Proportion nCET % | 0–33 | 11 | 14 | 0.307 |
| 34–66 | 10 | 5 | ||
| 67–100 | 5 | 7 | ||
| Thickness of enhancing margin | < = 3mm | 8 | 16 | 0.042 |
| >3mm | 15 | 8 | ||
| Pial invasion | Yes | 4 | 5 | >0.999 |
| No | 22 | 21 | ||
| Deep WM invasion | Yes | 3 | 3 | >0.999 |
| No | 23 | 23 | ||
| T1/FLAIR | T1<F | 14 | 12 | 0.784 |
| T1<<F | 5 | 7 | ||
| T1 = F | 7 | 7 | ||
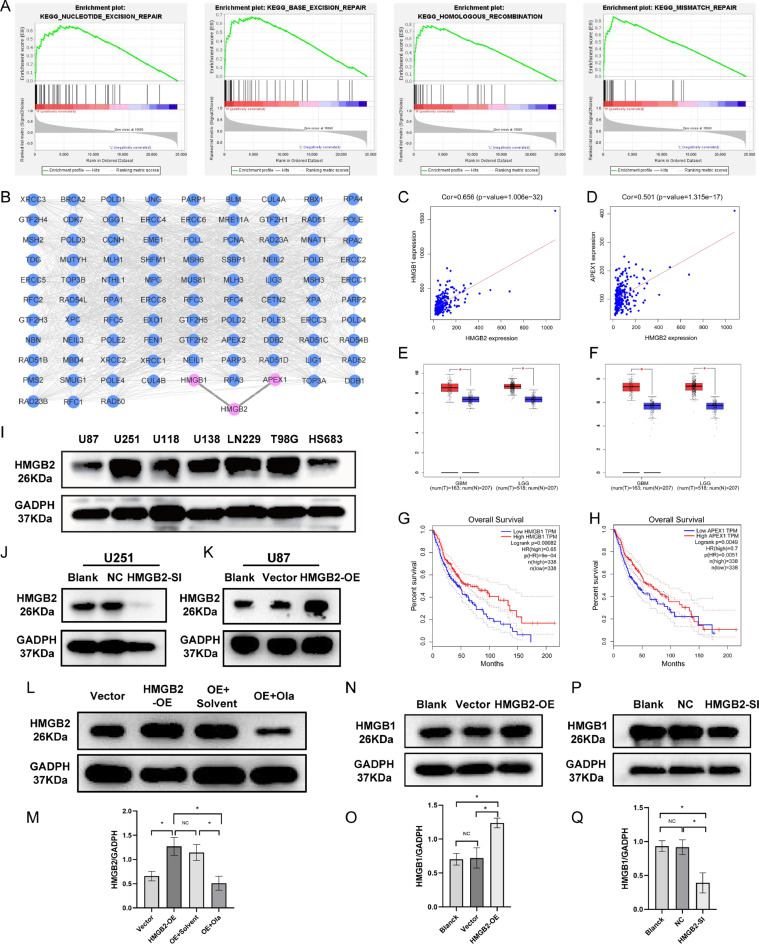
Gene set enrichment analysis,protein–protein interaction network and the application of pathway inhibitor
GSEA analysis showed that HMGB2 was both enriched in the base excision repair (BER),nucleotide excision repair (NER),mismatch repair (MMR),homologous recombination (HR) pathways according to the datasets of CGGA-mRNA-array_301(Supplement Fig.A.1A) and CGGA-mRNAseq_325 (Fig. 6A). In order to further clarify the pathway HMGB2 involved in, the interactions between protein HMGB2 and BER,NER,MMR,HR pathways related proteins were analyzed. The proteins with P> 0.7 were screened and visualized by Cytoscape. The results showed that the proteins with the strongest correlation with HMGB2 were apurinic/apyrimidinic endodeoxyribonuclease 1(APEX1) and HMGB1 (Fig. 6B).Through gene co-expression analysis, it was found that HMGB2 was closely related to APEX1 and HMGB1(Fig. 6C-D). Moreover in gliomas, the expression of APEX1 and HMGB1 in tumor tissues was significantly higher than that in adjacent tissues (Fig. 6E-F), and the high expression of APEX1 and HMGB1 has shorter survival time (Fig. 6G-H). There are seven different glioma cell lines were selected and their HMGB2 expression levels were detected by western blotting analysis.The highest expression level of HMGB2 was in U251 cells and the lowest was in U87 cells (Fig. 6I).So U251 cells were transfected with siRNAs for HMGB2 knockdown study and HMGB2 overexpression plasmids were constructed and introduced into U87 cells for gene overexpression study. Which were both verified by WB (Fig. 6J-K) (Supplement Fig.A.1B) and qPCR (Supplement Fig.A.1C). According to the genes involved in KEGG DNA damage repair (DDR) pathways proposed by GSEA website, both APEX1 and HMGB1 are only involved in BER pathway, so a BER pathway inhibitor (olaparib 5μM [25]) was applied to the HMGB2 overexpression glioma cells. The results showed that the expression of HMGB2 decreased greatly after the application of olaparib, (Fig. 6L-M).In the glioma cells experiments, it was found that the content of HMGB1 protein increased in HMGB2 overexpression cells (Fig. 6N-O) and decreased in HMGB2 knockdown cells (Fig. 6P-Q), but this phenomenon was not verified in APEX1, indicating a close relationship between HMGB2 and HMGB1.
Fig. 6.
Enrichment pathways of HMGB2 and co-expression protein analysis.(A) Enriched DDR pathways by GSEA based on CGGA-mRNAseq_325 dataset.(B)PPI between HMGB2 and the proteins of enriched DDR pathways constructed by the STRING and cytoscape visualization software. (C-D)Co-expression analysis of HMGB2 with APEX1 and HMGB1 with CGGA-mRNAseq_325 dataset.(E-F)The expression levels of APEX1 and HMGB1 in glioma and non-glioma tissues in GBM and LGG with TCGA datebase. (G-H)In the TCGA database, the survival differences between the high expression and low expression groups of HMGB1 and APEX1.(I)Expression of HMGB2 proteins in glioma cell lines (U87,U251,U118,U138,LN229,T98G,Hs683)by western blotting.(J-K)WB verification of HMGB2 knock-down and overexpression.(L-M)WB verification was performed after HMGB2 overexpression cells were added with solvent and OLA.(N,P)Verification of HMGB1 expression in HMGB2 overexpression and knockdown cells.(O,Q)HMGB1 expression histogram with prism8.
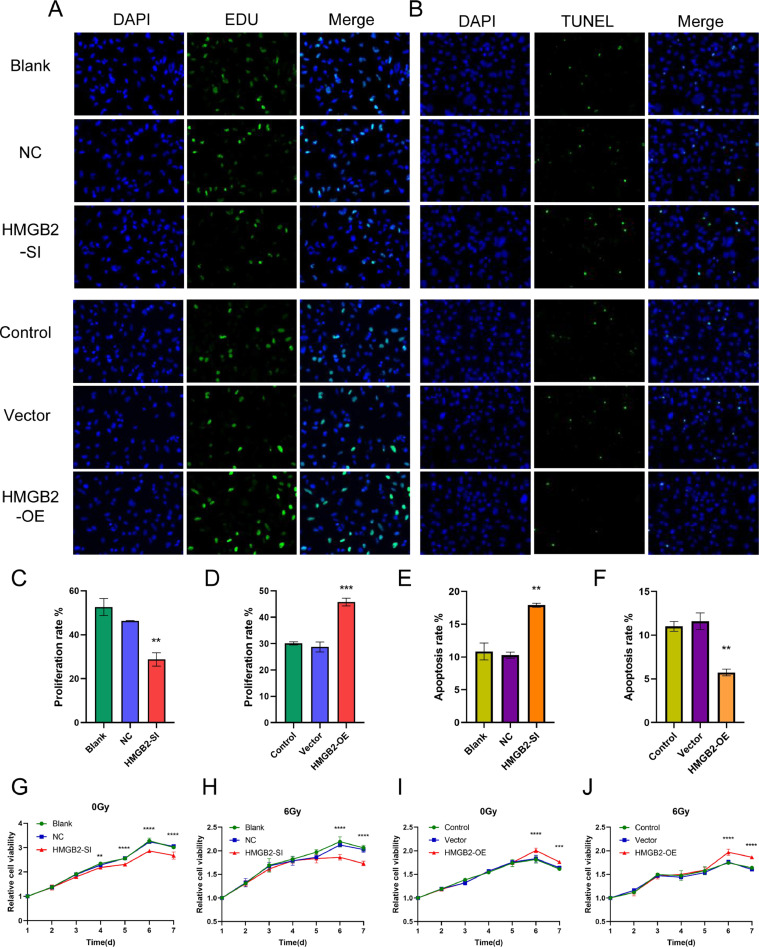
HMGB2 promotes cell proliferation and inhibits apoptosis in glioma cells
The results of EDU showed that the proliferation rate of HMGB2 overexpression cells was significantly higher than that of control and blank groups, while HMGB2 knockdown cells was lower than that of control and blank ones (Fig. 7A,C,D).The same cellular proliferation results were confirmed by CCK-8 assay.Glioma cells with higher expression of HMGB2 were found to have stronger proliferative ability both under 0 Gy and 6 Gy doses of irradiation (Fig. 7G-J).The TUNEL results illustrated that the apoptosis rate of HMGB2 overexpression cells was greatly lower than that of control group and blank group, while the HMGB2 knockdown cells were higher (Fig. 7B,E,F). All above results were reported as statistically significant at p < 0.05.
Fig. 7.
HMGB2 factor promotes cell proliferation and inhibits apoptosis.(A)Effect of HMGB2 siRNA on cell proliferation.Cells were detected by staining with DAPI (blue), and the proliferating cells were recognized by staining with EdU (green). (B)Effect of HMGB2 overexpression on cell apoptosis.Cells were detected by staining with DAPI (blue), and the apoptosis cells were recognized by staining with TUNEL (green).(C-D)Percentages of EdU-positive cells were graphed with Histogram.(E-F)Percentages of TUNEL-positive cells were graphed with Histogram.CCK-8 assays were carried out in HMGB2 knockdown cells (G-H) and overexpression cells (I-J) after treated with doses of 0 and 6 Gy. *p < 0.05, **p < 0.01, ***p < 0.001,****p < 0.0001.
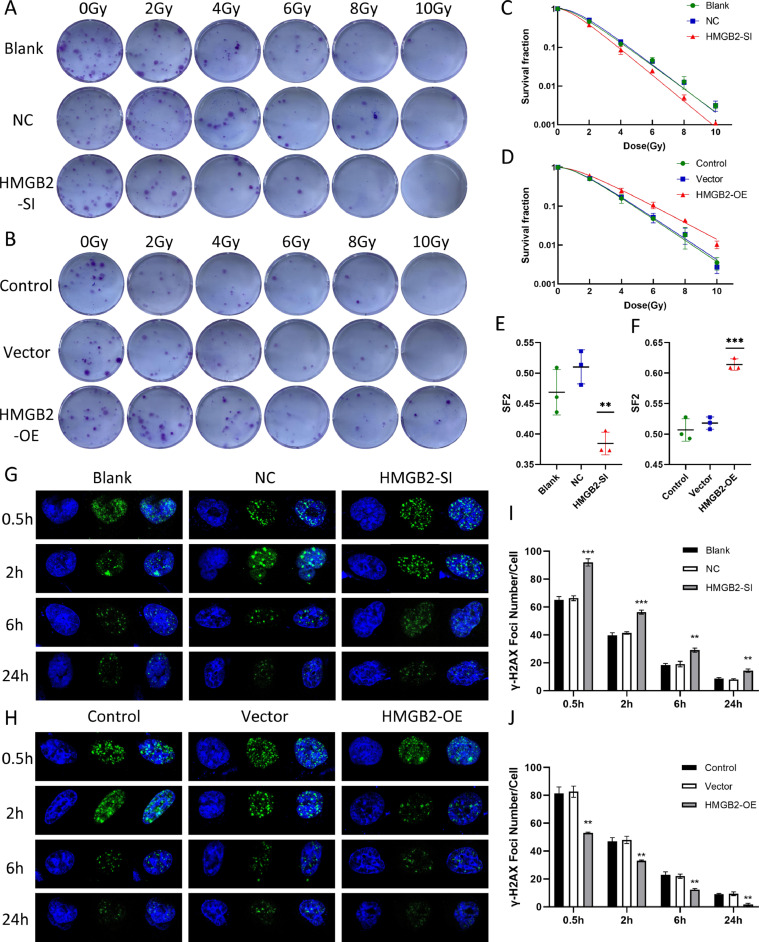
HMGB2 promoted colony formation and enhanced radiation resistance of glioma cells
Compared with the control groups,the number of colonies and the γ-H2AX foci at 0.5 h, 2 h, 6 h and 24 h after 5 Gy radiation formed in gliomas in response to HMGB2 overexpression was significantly increased (Fig. 8B,H).On the contrary, Knockdown of HMGB2 reduced colony formation in soft agar and radiation-induced γ-H2AX foci in U251 cells versus the control group. (Fig. 8A,G). The values of SF2, D0, Dq and N decreased while the SER increased, indicating the increased radiosensitivity of glioma cells with HMGB2-SI (P < 0.05)(Fig. 8C,E). In HMGB2-OE group, the radio-resistance increased with the decreased SER and the increased SF2,N,D0 as well as Dq (P < 0.05)(Fig. 8D,F). The detailed values were displayed in Table 3.The numbers of γ-H2AX foci in the HMGB2-SI group were much more than that in the control groups at 0.5 h, 2 h, 6 h and 24 h after irradiation (Fig. 8I) and in the HMGB2-OE group were significantly less than that in the control groups (Fig. 8J). It was indicated that the DNA damage repair of HMGB2 overexpression cells was obviously activated, and the sensitivity to radiation of these cells decreased and the radio-resistance increased.
Fig. 8.
The radioresistant effect of HMGB2 in gliomas. (A)HMGB2-SI and (B)HMGB2-OE cells separately displayed weaker and stronger colony formation capability than control groups under the same radiation dose, the representative colony images of U251 and U87 cells exposed to 0, 2, 4,6,8,10 Gy were given. clonogenic survivals of (C)U251 and (D)U87 cells with HMGB2 knockdown and overexpression.SF2 of (E)U251 and (F)U87 cells. Images of γ-H2AX foci formed in (H)U87 cells and (G)U251 cells at 0.5 h, 2 h, 6 h and 24 h after 5 Gy radiation. Histogram of γ-H2AX foci number in (J)U87 and (I)U251 cells. D0,mean lethal dose; Dq,quasi-threshold dose; SF2,surviving fraction at 2 Gy; N,extrapolation number; SER,sensitization enhancement ratio. *p < 0.05, **p < 0.01, ***p < 0.001,****p < 0.0001.
Table 3.
The results of colony formation.
| U251Blank | U251NC | U251SI | U87Blank | U87Vector | U87OE | |
|---|---|---|---|---|---|---|
| K | 0.6934 | 0.7132 | 0.7773 | 0.6355 | 0.6239 | 0.4986 |
| N | 2.188 | 2.59 | 2.041 | 2.143 | 2.153 | 2.055 |
| D0 | 1.442169 | 1.402131 | 1.286505 | 1.573564 | 1.602821 | 2.005616 |
| Dq | 1.129201 | 1.334349 | 0.917844 | 1.199381 | 1.229143 | 1.444597 |
Discussion
HMGB2 is an important nuclear protein, which is up-regulated in many tumors.Previous studies have found that HMGB2 has been identified as a biomarker for the diagnosis of gastric cancer.The knocking-down of HMGB2 could inhibit the proliferation and glycolysis of cells [20]. In the breast and cervical cancer, the expression of HMGB2 in cancer cells was higher than in adjacent normal tissues, which promoted the tumor progression.HMGB2 promoted proliferation by activating AKT signaling pathway and inhibited the expression of p21 and p27 in cervical cancer. And by regulating the Warburg effect in breast cancer [19,26].HMGB2 was overexpressed in head and neck squamous cell carcinoma (HNSCC), and the silencing of HMGB2 increased the sensitivity of HNSCC cell lines to cisplatin and 5-FU [27].HMGB2 is necessary to protect cells from DNA damage and effective DNA repair, which is related to poor response to preoperative radiotherapy and chemotherapy in patients of colorectal cancer [28].In gliomas, only a few studies described the increased expression of HMGB2 in GBM. A simple verification was conducted in vitro cellular experiments that high expression of HMGB2 could make glioblastoma cells resistant to TMZ [22].However, the correlation between HMGB2 and radiosensitivity and related mechanisms in glioma patients, as well as the relationship between HMGB2 and clinical biology and imaging manifestations, has not been reported.Whether HMGB2 is a factor with targeted therapeutic value closely related to the key biological behaviors and treatment responses in glioma needs to be further explored.In our study the difference of HMGB2 expression and clinical prognosis between glioma tissues and normal tissues were analyzed. According to the mechanism analysis and in vitro experiments, it was first found that HMGB2 promoted the radiation resistance of glioma. The relationship between HMGB2 and MRI imaging features was compared for the first time including precontrast T1WI, contrast-enhanced T1WI, T2WI,T2-FLAIR and DWI.
By analyzing the databases of TCGA, GEO and CGGA, it was found that the HMGB2 expression levels in glioma tissues are significantly higher than those in adjacent normal tissues and the expression of HMGB2 was closely related to tumor grade (P < 0.001) and IDH status (P < 0.001). Therefore, with the data resources in our center a comprehensive analysis of clinical biology was performed. Firstly, by comparing the HMGB2 expression of 12 pairs isolated tissues of glioma patients with WB, it was discovered that the expression level of HMGB2 was significantly increased in both low-grade glioma and glioblastoma, and the higher the tumor grade, the higher the HMGB2 expression level. Then a total of 52 glioma patients were selected for HMGB2 immunohistochemistry staining.Prognostic analysis was carried out according to different expression levels. It was observed that the OS and PFS were poor in patients with high expression of HMGB2. Univariate and multivariate analysis showed that HMGB2 was a risk factor for overall survival of glioma patients. At the same time, the correlation between the HMGB2 expression level by immunohistochemistry and the corresponding preoperative MRI images was analyzed for the first time. The results showed that the expression level of HMGB2 was significantly correlated with ADCmean (P = 0.044) and enhanced tumor edge thickness (P = 0.042). The expression of HMGB2 was higher in gliomas with lower ADC value and thicker marginal enhancement. Finally, the cell experiments showed that HMGB2 promoted the proliferation and inhibited the apoptosis of glioma cells, and the radiation resistance of glioma cells with high HMGB2 expression was significantly increased. To sum up, our study strongly showed that HMGB2 is a prognostic factor of gliomas, the possible mechanism is worthy of further discussion.Previous studies have found that wild-type p53 can inhibit the transcription of the HMGB2 gene [28,29]. The P53 gene status of the U251 and U87 cell lines is mutant and wild-type [30,31]. In this study, HMGB2 is downregulated in U87 cells (P53-wild type) and upregulated in U251 cells (mutant), which reflects the role of P53 in inhibiting HMGB2. This further confirms the pivotal role of HMGB2 in gliomas.
Several factors can lead to DNA damage,cells have evolved a variety of mechanisms to detect and repair DNA damage [32,33].These mechanisms called DNA damage repair (DDR) which can be divided into a series of different but functionally intertwined pathways includes BER, NER, MMR, HR, and non-homologous end joining (NHEJ) and so on [[34], [35], [36], [37], [38]]. Tumor cells can also repair their own DNA through the above pathway to avoid apoptosis and increase invasiveness. Previous studies have found that glioblastoma contains stem cell-like subsets, showing high expression of DNA damage response factors, resulting in therapeutic resistance and disease recurrence [39].At present, the clinical trials of DDR inhibitors in gliomas are very popular, and the therapeutic targets include:PARP/ DNA-PK/Chk1/ATM [40]. However, there is no targeted drugs with clear benefits have been found so far, we speculate that it may be because the specific DDR pathway inhibitors were not selected for treatment based on the corresponding high expression biomarkers. According to the GSEA analysis of two CGGA glioma databases, it was found that HMGB2 is enriched in multiple DDR pathways, including BER, NER, MMR and HR. It is suggested that HMGB2 promotes tumor progression through abnormal DNA damage repair in gliomas. Then, through PPI network analysis, it was discovered that HMGB2 is closely related to HMGB1 and APEX1 in all DDR proteins. According to the DDR pathways gene data of KEGG in GSEA, HMGB1 and APEX1 were both confirmed to be involved in BER pathway (Supplement Table A.1). After that, the experiments of mutual expression relationship were carried out in vitro and the results indicated that the expressions of HMGB1 were decreased in HMGB2 knockdown group and increased in HMGB2 overexpressed cells, but this result was not found in APEX1. This suggests that HMGB2 may potentially play a role, possibly through HMGB1, in the BER pathway.PARP1 is a single strand break receptor protein, which is recruited to the site of DNA strand breaks at very early time.PARP1 interacts with various nuclear components of the BER complexes, and plays a key role in DNA break repair. PARP-1 regulates the metabolism of DNA through physical binding or polymerization (ADP- ribosyl) with partner proteins, including histones, HMG proteins, BER factors and multiple transcription factors [41,42].Olaparib is an effective PARP inhibitor which can inhibit the activity of BER pathway [[43], [44], [45]].So finally, olaparib was added to the HMGB2 overexpressed glioma cells. It was found that after BER pathway was inhibited the HMGB2 expression level significantly decreased, indicating that HMGB2 might be implicated in BER pathway to some extent which has not been reported in previous studies.It has been reported in previous literature that HMGB2 overexpression enhances the chemotherapy resistance of GBM [22]. However, it is not reported whether HMGB2 enhances the glioma cells’ resistance to radiation. Through the experiment with glioma cells, it was found that HMGB2 could promote the proliferation of glioma cells and reduce apoptosis. After radiation, the number of cell colonies and γ-H2AX foci of HMGB2 overexpression glioma cells was significantly higher than that of the control group, indicating that the DNA damage repair was significantly activated, and the sensitivity of these cells to radiation decreased. With the clinical analysis of 52 gliomas patients in our hospital, it was demonstrated that the patients with high HMGB2 expression had shorter local recurrence time and higher recurrence rates, which proved that the effect of radiotherapy with high HMGB2 expression was relatively poor and the radiotherapy resistance was obvious. This suggests that in order to reduce the risk of progression in gliomas, we could appropriately expand the radiotherapy target area, increase the radiation dose and temozolomide administration time for patients with high HMGB2 expression. Therefore, HMGB2 is a very important prognostic factor for glioma which can guide the treatments of glioma according to its expression level.
Magnetic resonance imaging is widely used in the diagnosis and treatment of gliomas, and the functional imaging and imaging radiomics have developed rapidly in recent years. MRI imaging analysis has been reported in preoperative diagnosis of glioma, prediction of common gene expression and evaluation of curative effect. In this study, the differences and correlation between MRI imaging features before surgery and HMGB2 expression in 52 patients with glioma were analyzed.It was found that the patients with lower ADCmean score and larger thickness of enhancing margin had higher expression of HMGB2, suggesting that these patients had a higher degree of malignancy and a poorer prognosis. Similarly, previous researches showed that lower ADC values of glioma tumor indicated the worse prognosis [46,47]. Our study showed that according to the correlation between some usual characteristics of nuclear magnetic resonance imaging and biomarkers, the biological behavior of tumor could be evaluated, the recurrence and progress status could be predicted. On the other hand, when the MR imaging metrics of tumor progression are not clear, the expression level of HMGB2 can be used as a reference to distinguish between disease recurrence and pseudo-progression. This is a new research direction, and the clinical value is worthy of in-depth exploration. Therefore, it is necessary to increase the sample size for further summary and analysis.
In conclusion, it was found that HMGB2 was related to the progression of glioma and its overexpression led to a worse OS and PFS in glioma patients. The overexpression of HMGB2 could promote cells proliferation and radio-resistance. For the first time, HMGB2 was suggested to be somewhat related to the BER pathway, possibly in association with the HMGB1 protein, and it may improve radiation resistance by enhancing the repair ability of DNA damage.
Funding
This work was supported by Hebei Natural Science Foundation (Grant Number H2022206430; H2022206572) and the S&T Program of Hebei (Grant Number 236Z7718G).
Ethics approval
The study was approved by the Ethics Committee of the Second Hospital of Hebei Medical University, the publication of patient information was approved by all of the patients after giving informed written consent.
CRediT authorship contribution statement
Wei Han: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data curation, Visualization, Writing – original draft. Huandi Zhou: Conceptualization, Methodology, Writing – review & editing. Xinyuan Zhang: Investigation, Validation. Haonan Li: Software, Investigation. Xuetao Han: Formal analysis. Linlin Su: Validation. Lei Tian: Software. Xiaoying Xue: Conceptualization, Resources, Project administration, Funding acquisition, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank all the online databases and the patients participating in this study.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2024.101977.
Appendix. Supplementary materials
References
- 1.Ricard D, Idbaih A, Ducray F, Lahutte M, Hoang-Xuan K, Delattre JY. Primary brain tumours in adults. Lancet. 2012;379(9830):1984–1996. doi: 10.1016/S0140-6736(11)61346-9. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Cioffi G, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2014-2018. Neuro. Oncol. 2021;23(12 Suppl 2):iii1–iii105. doi: 10.1093/neuonc/noab200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang T, Tang GF, Lin Y, Peng XX, Zhang X, Zhai XW, et al. Prevalence estimates for primary brain tumors in China: a multi-center cross-sectional study. Chin. Med. J. (Engl.) 2011;124(17):2578–2583. [PubMed] [Google Scholar]
- 4.Weller M, Wick W, Aldape K, Brada M, Berger M, Pfister SM, et al. Glioma. Nat. Rev. Dis. Primers. 2015;1:15017. doi: 10.1038/nrdp.2015.17. [DOI] [PubMed] [Google Scholar]
- 5.Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, et al. The epidemiology of glioma in adults: a "state of the science" review. Neuro Oncol. 2014;16(7):896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stupp R, Taillibert S, Kanner A, Read W, Steinberg D, Lhermitte B, et al. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: A randomized clinical trial. JAMA. 2017;318(23):2306–2316. doi: 10.1001/jama.2017.18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rominiyi O, Vanderlinden A, Clenton SJ, Bridgewater C, Al-Tamimi Y, Collis SJ. Tumour treating fields therapy for glioblastoma: current advances and future directions. Br. J. Cancer. 2021;124(4):697–709. doi: 10.1038/s41416-020-01136-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stupp R, Wong ET, Kanner AA, Steinberg D, Engelhard H, Heidecke V, et al. NovoTTF-100A versus physician's choice chemotherapy in recurrent glioblastoma: a randomised phase III trial of a novel treatment modality. Eur. J. Cancer. 2012;48(14):2192–2202. doi: 10.1016/j.ejca.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 9.Fang C, Wang K, Stephen ZR, Mu Q, Kievit FM, Chiu DT, et al. Temozolomide nanoparticles for targeted glioblastoma therapy. ACS Appl. Mater. Interfaces. 2015;7(12):6674–6682. doi: 10.1021/am5092165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee SY. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016;3(3):198–210. doi: 10.1016/j.gendis.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Stevens MF, Bradshaw TD. Temozolomide: mechanisms of action, repair and resistance. Curr Mol Pharmacol. 2012;5(1):102–114. doi: 10.2174/1874467211205010102. [DOI] [PubMed] [Google Scholar]
- 12.Han X, Xue X, Zhou H, Zhang G. A molecular view of the radioresistance of gliomas. Oncotarget. 2017;8(59):100931–100941. doi: 10.18632/oncotarget.21753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pallier C, Scaffidi P, Chopineau-Proust S, Agresti A, Nordmann P, Bianchi ME, et al. Association of chromatin proteins high mobility group box (HMGB) 1 and HMGB2 with mitotic chromosomes. Mol. Biol. Cell. 2003;14(8):3414–3426. doi: 10.1091/mbc.E02-09-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czura CJ, Wang H, Tracey KJ. Dual roles for HMGB1: DNA binding and cytokine. J. Endotoxin Res. 2001;7(4):315–321. doi: 10.1177/09680519010070041401. [DOI] [PubMed] [Google Scholar]
- 15.Liu W, Zhang Z, Zhang Y, Chen X, Guo S, Lei Y, et al. HMGB1-mediated autophagy modulates sensitivity of colorectal cancer cells to oxaliplatin via MEK/ERK signaling pathway. Cancer Biol. Ther. 2015;16(4):511–517. doi: 10.1080/15384047.2015.1017691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen SY, Hsu YH, Wang SY, Chen YY, Hong CJ, Yen GC. Lucidone inhibits autophagy and MDR1 via HMGB1/RAGE/PI3K/Akt signaling pathway in pancreatic cancer cells. Phytother. Res. 2022;36(4):1664–1677. doi: 10.1002/ptr.7385. [DOI] [PubMed] [Google Scholar]
- 17.Chen R, Kang R, Tang D. The mechanism of HMGB1 secretion and release. Exp. Mol. Med. 2022;54(2):91–102. doi: 10.1038/s12276-022-00736-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai X, Ding H, Liu Y, Pan G, Li Q, Yang Z, et al. Expression of HMGB2 indicates worse survival of patients and is required for the maintenance of Warburg effect in pancreatic cancer. Acta Biochim Biophys Sin (Shanghai) 2017;49(2):119–127. doi: 10.1093/abbs/gmw124. [DOI] [PubMed] [Google Scholar]
- 19.Han X, Zhong S, Zhang P, Liu Y, Shi S, Wu C, et al. Identification of differentially expressed proteins and clinicopathological significance of HMGB2 in cervical cancer. Clin Proteomics. 2021;18(1):2. doi: 10.1186/s12014-020-09308-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui G, Cai F, Ding Z, Gao L. HMGB2 promotes the malignancy of human gastric cancer and indicates poor survival outcome. Hum. Pathol. 2019;84:133–141. doi: 10.1016/j.humpath.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 21.Kwon JH, Kim J, Park JY, Hong SM, Park CW, Hong SJ, et al. Overexpression of high-mobility group box 2 is associated with tumor aggressiveness and prognosis of hepatocellular carcinoma. Clin. Cancer Res. 2010;16(22):5511–5521. doi: 10.1158/1078-0432.CCR-10-0825. [DOI] [PubMed] [Google Scholar]
- 22.Wu ZB, Cai L, Lin SJ, Xiong ZK, Lu JL, Mao Y, et al. High-mobility group box 2 is associated with prognosis of glioblastoma by promoting cell viability, invasion, and chemotherapeutic resistance. Neuro. Oncol. 2013;15(9):1264–1275. doi: 10.1093/neuonc/not078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q, Wu J, Wei P, Xu Y, Zhuo C, Wang Y, et al. Overexpression of forkhead Box C2 promotes tumor metastasis and indicates poor prognosis in colon cancer via regulating epithelial-mesenchymal transition. Am. J. Cancer Res. 2015;5(6):2022–2034. [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Jiang Z, Zhang Y, Wang X, Liu L, Fan Z. RNA sequencing analysis reveals protective role of kruppel-like factor 3 in colorectal cancer. Oncotarget. 2017;8(13):21984–21993. doi: 10.18632/oncotarget.15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghorai A, Mahaddalkar T, Thorat R, Dutt S. Sustained inhibition of PARP-1 activity delays glioblastoma recurrence by enhancing radiation-induced senescence. Cancer Lett. 2020;490:44–53. doi: 10.1016/j.canlet.2020.06.023. [DOI] [PubMed] [Google Scholar]
- 26.Fu D, Li J, Wei J, Zhang Z, Luo Y, Tan H, et al. HMGB2 is associated with malignancy and regulates Warburg effect by targeting LDHB and FBP1 in breast cancer. Cell Commun. Signal. 2018;16(1):8. doi: 10.1186/s12964-018-0219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Syed N, Chavan S, Sahasrabuddhe NA, Renuse S, Sathe G, Nanjappa V, et al. Silencing of high-mobility group box 2 (HMGB2) modulates cisplatin and 5-fluorouracil sensitivity in head and neck squamous cell carcinoma. Proteomics. 2015;15(2-3):383–393. doi: 10.1002/pmic.201400338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin YJ, Kim MS, Kim MS, Lee J, Kang M, Jeong JH. High-mobility group box 2 (HMGB2) modulates radioresponse and is downregulated by p53 in colorectal cancer cell. Cancer Biol. Ther. 2013;14(3):213–221. doi: 10.4161/cbt.23292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stros M, Ozaki T, Bacikova A, Kageyama H, Nakagawara A. HMGB1 and HMGB2 cell-specifically down-regulate the p53- and p73-dependent sequence-specific transactivation from the human Bax gene promoter. J. Biol. Chem. 2002;277(9):7157–7164. doi: 10.1074/jbc.M110233200. [DOI] [PubMed] [Google Scholar]
- 30.Afshar G, Jelluma N, Yang X, Basila D, Arvold ND, Karlsson A, et al. Radiation-induced caspase-8 mediates p53-independent apoptosis in glioma cells. Cancer Res. 2006;66(8):4223–4232. doi: 10.1158/0008-5472.CAN-05-1283. [DOI] [PubMed] [Google Scholar]
- 31.Hara S, Nakashima S, Kiyono T, Sawada M, Yoshimura S, Iwama T, et al. p53-Independent ceramide formation in human glioma cells during gamma-radiation-induced apoptosis. Cell Death Differ. 2004;11(8):853–861. doi: 10.1038/sj.cdd.4401428. [DOI] [PubMed] [Google Scholar]
- 32.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481(7381):287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 33.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447(7147):941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat. Rev. Genet. 2009;10(11):756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 36.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 2010;11(3):196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006;7(5):335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 39.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 40.Bonm A, Kesari S. DNA damage response in glioblastoma: mechanism for treatment resistance and emerging therapeutic strategies. Cancer J. 2021;27(5):379–385. doi: 10.1097/PPO.0000000000000540. [DOI] [PubMed] [Google Scholar]
- 41.Jagtap P, Szabó C. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat. Rev. Drug Discov. 2005;4(5):421–440. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- 42.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7(7):517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 43.Dianov GL, Hübscher U. Mammalian base excision repair: the forgotten archangel. Nucleic. Acids. Res. 2013;41(6):3483–3490. doi: 10.1093/nar/gkt076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ström CE, Johansson F, Uhlén M, Szigyarto CA, Erixon K, Helleday T. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic. Acids. Res. 2011;39(8):3166–3175. doi: 10.1093/nar/gkq1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith G, Alholm Z, Coleman RL, Monk BJ. DNA damage repair inhibitors-combination therapies. Cancer J. 2021;27(6):501–505. doi: 10.1097/PPO.0000000000000561. [DOI] [PubMed] [Google Scholar]
- 46.Villanueva-Meyer JE, Wood MD, Choi BS, Mabray MC, Butowski NA, Tihan T, et al. MRI Features and IDH mutational status of grade II diffuse gliomas: impact on diagnosis and prognosis. AJR Am. J. Roentgenol. 2018;210(3):621–628. doi: 10.2214/AJR.17.18457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feraco P, Bacci A, Ferrazza P, van den Hauwe L, Pertile R, Girlando S, et al. Magnetic resonance imaging derived biomarkers of idh mutation status and overall survival in grade iii astrocytomas. Diagnostics (Basel) 2020;10(4) doi: 10.3390/diagnostics10040247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.