Highlights
-
•
The low expression of RBP4 was observed in liver tissues of patients with hepatocellular carcinoma (HCC).
-
•
Serum RBP4 demonstrates superior diagnostic value compared to AFP in multi-layer and large-scale validation cohorts.
-
•
Combining AFP with RBP4 may enhance the detection rate for surveillance of high-risk populations at risk for liver cancer.
-
•
RBP4 serves as a diagnostic and prognostic biomarker for HCC.
Keywords: RBP4, Biomarkers, HCC, Proteomic, Diagnosis model
Abstract
Background
The prognosis of hepatocellular carcinoma (HCC) is universally poor. Early diagnosis plays a pivotal role in determining the outcome of HCC.
Methods
We employed a comparative proteomics approach to identify potential biomarkers and validated the application of retinol-binding protein 4 (RBP4) as a biomarker for HCC. RBP4 protein expression was examined in liver tissues from 80 HCC patients through immunohistochemical analysis. Serum RBP4 concentrations were measured by ELISA in a cohort comprising 290 HCC patients, matched 202 chronic hepatitis B patients and 269 healthy controls. Survival data were collected from HCC patients. The diagnostic and prognostic values of RBP4 were evaluated using receiver operating curve (ROC) analysis.
Results
The validation results demonstrated a significant reduction in RBP4 levels in both liver tissues and serum samples from HCC patients. ROC analysis of the diagnostic value of RBP4 revealed an AUC of 0.879 (95 % CI: 0.854∼0.903) for HCC. When combined with AFP, the AUC increased to 0.919, with a sensitivity of 87.9 % and specificity of 80 %. Survival analysis revealed significantly reduced overall survival time in individuals with low-expression of RBP4 compared to those with high-expression. The joint prognostic model exhibited an AUC of 0.926 (95 % CI: 0.888∼0.964), which was significantly higher than that of AFP alone (AUC=0.809; P <0.0001).
Conclusions
RBP4 shows a great potential as a biomarker with appreciable diagnostic value, complementing the AFP in HCC diagnosis. Additionally, it holds promise as a prognostic biomarker that, when integrated into a combined prognostic model, could greatly improve HCC prognosis efficiency.
Introduction
Liver cancer ranks as the second (among males) or sixth (among females) leading cause of cancer-related mortality worldwide [1]. The regions with the highest incidence of primary liver cancer are Asia and Africa [2]. Hepatocellular carcinoma (HCC), accounting for 80∼90 % of all primary liver cancers, exhibits a rapid increase in its occurrence and remains a significant global public health burden [3]. In China, the 5-year survival rate for HCC patients diagnosed between 2010∼2014 was merely 14.1 % [4]. In America, the average 5-year survival rate for HCC was reported as 29.6 %, but it plummeted to a mere 2.5 % when diagnosed at an advanced stage [5]. Hence, early and accurate diagnosis plays a pivotal role in improving the prognosis of HCC.
The current diagnosis of HCC relies on a combination of serological tumor biomarkers, medical imaging, and invasive pathological [6] tissue biopsy, among which the biomarkers play a crucial role in diagnostic accuracy. As the most direct, rapid, and effective diagnostic method, the use of serum biomarkers is the main means of early detection, early treatment, and therapeutic effect monitoring. At present, the major serum biomarker used for clinical diagnosis of HCC is alpha-fetoprotein (AFP). Under normal conditions, AFP is produced by embryo liver cells and drops to trace amounts after birth. Under pathological conditions, AFP occurs at high levels in the liver and blood of adults with hepatoma and is also found elevated in chronic hepatitis and cirrhosis [7,8]. The diagnostic accuracy of AFP is, however, unsatisfactory with a sensitivity of only 41∼65 % (at the cut-off of 20 ng/mL) for HCC at any stage [9,10]. Particularly in the early stage liver cancer with the tumor size less than 2 cm in diameter, the proportion of AFP-positive HCC was less than 33.3 % [11]. Therefore, the specificity and reliability of AFP biomarkers are of low value for the early stages of HCC [12,13]. Even in individuals with advanced HCC, their AFP serum levels remain normal in 15.5 % of patients [14]. Therefore, the search and discovery of supplemental serum biomarkers for AFP-negative HCC (serum AFP < 25 ng/ml) has become an important research focus.
In general, the clinical application of cancer biomarkers demands highly sensitive and specific, non-invasive, and cost-effective molecular measures. The most desirable biomarker needs to be tumor-specific and easily detectable in biofluids, such as serum or plasma. The proteomics approach has been shown to promise the discovery of serum biomarkers for early HCC detection [[15], [16], [17]]. Currently, some candidate biomarkers have been proposed to discriminate HCC based on serum proteomics studies. For example, serum paraoxonase 1 (PON1), a calcium-dependent hydrolase synthesized mainly in the liver, is a novel diagnostic biomarker for the microvascular invasion in HCC [18]. Two large-scale and multicenter studies reported that Dickkopf-1 (DKK1), a secretory antagonist of the Wnt signaling pathway, could be used as the potential biomarkers to detect HCC [19]; Plasma heat shock protein 90 alpha (HSP90a), an important molecular chaperone, showed remarkable sensitivities and specificities in detecting liver cancer patients, even at the early stage from non-liver cancers [20,21]. Nevertheless, many of these candidate biomarkers display limited sensitivity and specificity when used alone, especially for the early stages of HCC. Furthermore, most studies are in the early phase of biomarker discovery or were performed with small sample size. Several newly identified markers are yet to be evaluated further in phase II, followed by phase III studies, which will retrospectively determine whether they can be used to detect preclinical diseases [22]. More prospective studies are thus necessary to validate the broad utility of biomarker candidates for HCC surveillance among patients of different etiologies. In the meanwhile, their use as a tumor biomarker for AFP-negative HCC remains to be evaluated by clinical trials before being applied in clinical practice. Finding a new reliable serum biomarker and assessing its diagnostic value for AFP-negative HCC is of pressing need.
It should be noted that chronic inflammation of the liver predisposes individuals to HCC. Most cases of HCC are associated with cirrhosis developed from chronic hepatitis B virus (HBV) or hepatitis C virus (HCV) infections and HBV was the most common risk factor of liver cancer in China [23]. Therefore, in this study, we first conducted a comparative proteome analysis to identify differentially expressed proteins (DEPs) among patients with hepatocellular carcinoma (HCC), chronic hepatitis B and healthy controls. Subsequently, we investigated the potential application of retinol binding protein 4 (RBP4) as a biomarker for HCC in a large validation cohort.
Patients and methods
Study subjects
The numbers and source of study subjects are summarized in Table 1. HCC patients were diagnosed by B-ultrasound, Computed Tomography (CT) or magnetic resonance imaging (MRI), AFP and liver enzyme serology, and histopathological confirmation by two board-certified pathologists after surgical resection. Secondary liver cancer or primary cancer at other sites were excluded. Chronic hepatitis B virus infection (CHB) patients were confirmed by positive HBsAg serology and screened for liver fibrosis, cirrhosis, and tumor absence via ultrasound or CT. Liver cirrhosis (LC) or Liver fibrosis (LF) patients were conformed to the clinical diagnostic criteria of cirrhosis or were pathologically diagnosed as Liver fibrosis, excluding malignant tumors including liver cancer. Healthy controls (HC) were recruited from a routine physical examination. Patients with liver fibrosis or cirrhosis and tumors were excluded from CHB group. The above four groups of subjects were matched in age and sex, and the relevant data were complete. Follow-up visits were conducted for HCC patients (enrolled cohort) from June 31, 2014 to December 31, 2020 through phone calls, SMS messages, wechat communication, and other means. The longest follow-up period was five years. These studies were approved by the Ethics Committee of the Guangxi Medical University. All patients provided written consent to participate in these studies.
Table 1.
Sample sources.
| assay | sample | case | source | time |
|---|---|---|---|---|
| iTRAQ analysis | HCC cases | 10 | a | 2013 |
| chronic hepatitis B cases | 10 | |||
| liver fibrosis cases | 10 | |||
| health controls | 10 | |||
| MRM verification | HCC cases | 24 | b | 2014 |
| chronic hepatitis B cases | 10 | a | ||
| liver cirrhosis cases | 10 | a | ||
| health controls | 13 | a | ||
| Immunohistochemistry assay | HCC and adjacent tissues | 80 | b | 2014 |
| ELISA verification | HCC cases | 290 | b | 2013–2018 |
| chronic hepatitis B cases | 202 | c | ||
| health controls | 269 | a |
iTRAQ, isobaric tags for the relative and absolute quantization; MRM, multiple reaction monitoring; ELISA, enzyme-linked immunosorbent assay; a, the First Affiliated Hospital of Guangxi Medical University; b, the Affiliated Tumor Hospital of Guangxi Medical University; c, the 8th Affiliated Hospital of Guangxi Medical University.
Serum preparation
Whole blood was collected into a serum separator tube. After clot formation, the samples were centrifuged at 2000 × g for 10 min, and the serums were collected. Aliquots of undiluted serum were stored at –80 °C before proteomics and ELISA analyses.
Serum proteomic profiling by iTRAQ-MALDI-TOF-MS/MS
A total of 40 study subjects of age- and sex-matched HCC patients, CHB patients, LF patients, and healthy controls (N = 10 per group) were enrolled in the First Affiliated Hospital of Guangxi Medical University (Nanning, China) in 2013 (Table 1). Ten serum samples from each group were respectively pooled, diluted three times, and transferred to a spin filter to remove particles. The 14 most abundant proteins were depleted using Agilent Human 14 MARS columns (manufacturer), after which the samples were desalted and resuspended in digestion buffer. Protein samples (100 μg/group) were digested and labeled with iTRAQ (isobaric tags for the relative and absolute quantization) reagents (AB Sciex, Framingham, MA, USA). The healthy control, CHB, LF and HCC samples were labeled with iTRAQ and assigned as 113,114,115 and 116, respectively. The labeled peptides were mixed, desalted, and fractionated with a strong cation exchange chromatography on a polysulfoethyl column (PolyLC, Columbia, MD, USA) and nano-HPLC separation. Twenty fractions were collected, and each was spotted automatically onto the matrix-assisted laser desorption/ionization (MALDI) plates. The MS/MS analyses of offline spotted peptide samples were performed using the 5800 MALDI-TOF/TOF Analyzer (Applied Biosystems, MA, USA). The iTRAQ experiment was performed with three technical replicates to gather reliable quantitative information.
Protein identification and quantification were performed with ProteinPilotTM 4.0 software (version 4.0; Applied Biosystems, MA, USA) using the Paragon databaseTM [24]. A minimum of 3 peptides per protein identified with >95 % confidence and unused ProtScore >1.3 was used as the cut-off criteria for qualification. Differentially expressed proteins (DEPs) relative to health control or CHB group were considered up- or down-regulated when their iTRAQ ratios were >1.2 or <0.8, respectively.
UALCAN gene expression, oncolnc cox regression and KM plotter survival analysis
Systematic analyses of the mRNA expression pattern and prognostic value of target genes in liver carcinoma were performed using UALCAN and OncoLnc. The web-portal UALCAN [25) (http://ualcan.path.uab.edu/) was used to perform TCGA gene expression data analyses. The Cox regression for survival analysis of the mRNA levels of the RBP4 gene in patients with LIHC was assessed using the online database OncoLnc (http://www.oncolnc.org/). The expression of RBP4 gene was collected for each case and was divided into the high- and low-expression groups, and the survival difference between the two subgroups was determined using the Kaplan-Meier Plotter (https://kmplot.com/analysis/).
Targeted quantitation of serum RBP4 by multiple reaction monitoring (MRM)
The MRM-initiated detection and sequencing process was used to optimize the target protein MRM quantitative peptide, MRM ion pair, and the collision energy of each ion pair. Serum samples from 48 age- and sex-matched study subjects of HCC patients (N = 24), CHB patients (N = 10), LC patients (N = 10), and healthy controls (N = 13) were collected in the Affiliated Tumor Hospital and the First Affiliated Hospital of Guangxi Medical University (Nanning, China) in 2014 (Table 1). Serum levels of RBP4 protein were quantified using the MRM approach [26] on a 4000 QTRAP mass spectrometer (AB SCIEX, Foster City, CA) equipped with NanoLC 2D High-Performance Liquid Chromatography System (AB SCIEX, Foster City, CA). The MRM workflow includes: i) the Skyline Software was used to select peptides specific for RBP4, ii) MRM analysis monitored the sequence-specific transitions derived from intense productions in the MS/MS spectrum, and iii) the integrated peak areas of the signal corresponding to transitions were used to calculate the target protein concentration. Sample pretreatment, liquid-phase separation gradient, and mass matrix effect reduction were optimized to establish a quantitative method for the target protein. The parameters of AB Sciex 4000 QTRAP mass spectrometer were set as follows: curtain gas, 25; ion source gas, 20; ion spray voltage, 2300 V; interface heater temperature, 150 °C; Collision gas, High; Declustering potential, 100 and entrance potential, 10. The data for each peptide segment were quantitatively analyzed using Skyline2.5.0 software (University of Washington) to obtain the peak area value. The integral peak area of the healthy control group was taken as a parameter (the value was set as 1), and the ratio of the integrated peak area of the other groups was the relative expression concentration of the protein.
Immunohistochemistry (IHC)
For the IHC study, a total of 80 pairs of HCC cancerous and adjacent normal tissues were obtained from the Affiliated Tumor Hospital of Guangxi Medical University (Nanning, China) in 2014. IHC analysis was performed on paired formalin-fixed and paraffin-embedded cancerous and adjacent normal tissues from HCC patients according to standard procedures. Briefly, paraffin sections were hydrated, followed by microwave antigen retrieval and blocking of endogenous peroxidase activity with 3 % H2O2. Tissue sections were blocked for non-specific binding sites with 10 % goat serum and then incubated with RBP4 primary antibody (1:150, Anti-RBP4 antibody, Abcam, Cambridge, MA, USA) at 4 °C overnight. Thereafter, the sections were incubated with biotinylated secondary antibody (Envision™ Detection kit; Gene Tech, Shanghai, China) for 2 h and stained with diaminobenzidine. Positive immunoreactivity (brown staining) were analyzed semi-quantitatively into 4 ranks (from low to high: -, +, ++, and +++) as described previously [27].
Enzyme-linked immunosorbent assay (ELISA)
For the ELISA verification, a total of 761 serum samples from age- and sex-matched HCC patients (N = 290), CHB patients (N = 202), and healthy controls (N = 269) were collected in the Affiliated Tumor Hospital, the First Affiliated Hospital of Guangxi Medical University (Nanning, China) and the 8th Affiliated Hospital of Guangxi Medical University (Guigang, China) between 2013 and 2018. ELISA measurement of serum RBP4 was measured using a commercial RBP4 in vitro Simple-Step ELISA kit (Abcam, Cambridge, MA, USA) according to the manufacturer's protocol.
Receiver operating characteristic (ROC) curves
ROC curves were generated by plotting sensitivity against the false positive rate for RBP4 and AFP using the IBM SPSS software v26.0 (IBM Corp., Armonk, USA). Diagnostic performance was evaluated via ROC curve analysis and quantified using the area under the curve (AUC) with 95 % confidence interval (CI). Optimal cut-off values were selected at concentrations exhibiting the highest sum of sensitivity and specificity (Youden Index (J)). Statistical analysis datasets were compared using the IBM SPSS software v26.0 (IBM Corp., Armonk, USA).
Five-years follow-up and overall survival analysis
A total of 221 liver cancer patients were included in the follow-up cohort at the Affiliated Hospital of Guangxi Medical University. The case information system was utilized for research and data collection, encompassing variables such as initial admission details, gender, age, ethnicity, AFP levels, CEA levels, CA125 levels, CA153 levels, CA199 levels, etc. Survival time was measured in months from the first day of hospitalization until death occurred. Data for patients still alive at the end of follow-up were censored. The correlation between overall survival time and serum RBP4 expression level in HCC patients was analysed. Univariate logistic regression and multivariate Cox proportional hazards regression models were employed to analyse the survival time and identify prognostic factors influencing HCC prognosis among enrolled patients. Furthermore, a combined prognostic diagnostic model for HCC was established using Cox regression analysis with an evaluation of its diagnostic value through ROC curve analysis.
Statistical analysis
Statistical analyses were performed with IBM SPSS Statistics 26.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism v8.0. For continuous data, the comparison between groups was performed by the two-tailed unpaired Welch's t-test or by one-way ANOVA followed by Duncan's new multiple range test. Data are shown as the means ± standard error of the means. For discrete data, nonparametric, two-sided Mann–Whitney U tests were used for single comparisons and nonparametric Kruskal–Wallis tests for multiple comparisons. P < 0.05 was considered statistically significant. R 4.2.1 Software was used to build the nomogram.
Results
Screening for candidate serum biomarkers of HCC
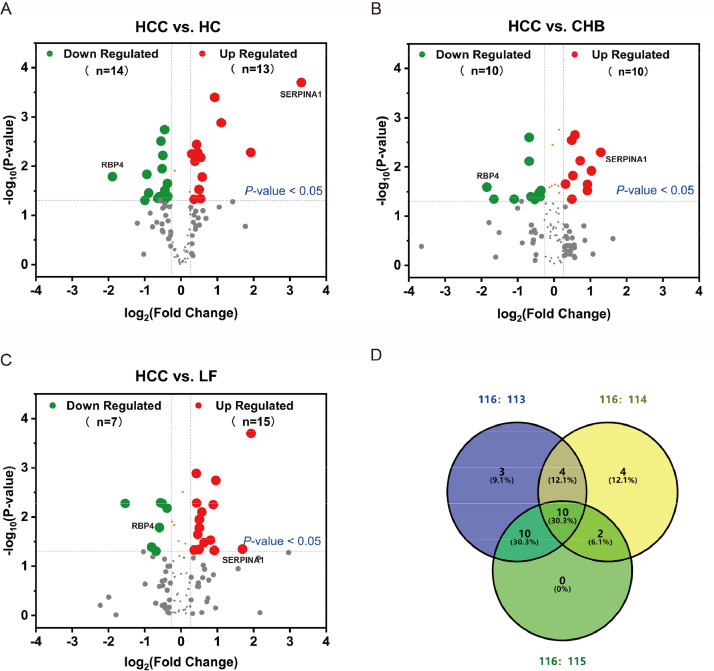
We performed the initial screening for candidate HCC biomarkers using pooled serum samples derived from HCC patients (designated as 116), LF patients (designated as 115), CHB patients (designated as 114), and healthy controls (designated as 113) by iTRAQ-MALDI-TOF-MS/MS proteomics analysis (N = 10 per group). A total of 103 serum proteins met the MS identification criteria, among which 27, 20 and 22 DEPs (abundance ratio > 1.2 or < 0.8) were identified in HCC patients when compared to healthy controls (Fig. 1A), CHB patients (Fig. 1B), and LF patients (Fig. 1C) respectively; additionally, 10 serum proteins (SERPINA1, A2M, APOA1, C4B, TF, LRG1, CFB, APOA4, C7 and RBP4) were differentially expressed in HCC patients relative to all other non-tumor controls (Fig. 1D, Supplementary Table 1), wherein serpin family A member 1 (SERPINA1) and RBP4 were the top up-regulated (10-fold of healthy controls) and down-regulated (0.27-fold of healthy controls) DEPs, respectively. And then, utilizing public databases UALCAN, we analyzed the mRNA expression levels of SERPINA1 and RBP4 in a large number of liver tumors and normal liver tissues. This result agreed with the observed trend for RBP4, but not for SERPINA1, in our serum proteomics screening study. Cox regression analysis using OncoLnc revealed a significant inverse association between RBP4 expression and cumulative hazard ratio (HR) in LIHC (Cox Coefficient = −0.323, P < 0.01). Additionally, survival analyses conducted on liver cancer patients using the Kaplan-Meier Plotter demonstrated that the overall survival rate (HR=0.32; P < 0.0001) was significantly lower in the RBP4 low-expression group (n = 92) compared to the high-expression group (n = 272).
Fig. 1.
Screening of HCC DEPs via iTRAQ-MALDI-TOF-MS/MS analysis and functional enrichment analyses of DEGs (A-C) The scatter plot for global serum proteins. The red and green dots indicate up- (fold change > 1.2) and down-regulated (fold change < 0.8) DEPs, respectively, when comparing 116 (HCC) versus 113 (HC), 114(CHB) or versus 115(LF). P < 0.05. HC, healthy controls; CHB, chronic hepatitis B virus infection; LF, liver fibrosis; HCC, hepatocellular carcinoma. (D) Venn diagram illustrating 27 DEPs were identified from HC (113) and HCC (116) comparison, 20 DEPs from CHB (114) and HCC (116) comparison, 22 DEPs from LF (115) and HCC (116) comparison and 10 DEPs were shared. HC, healthy controls; CHB, chronic hepatitis B virus infection; LC, liver cirrhosis; HCC, hepatocellular carcinoma; RBP4, retinol-binding protein 4; SERPINA1, serpin family A member 1.
Validation studies on RBP4 as a candidate biomarker for HCC
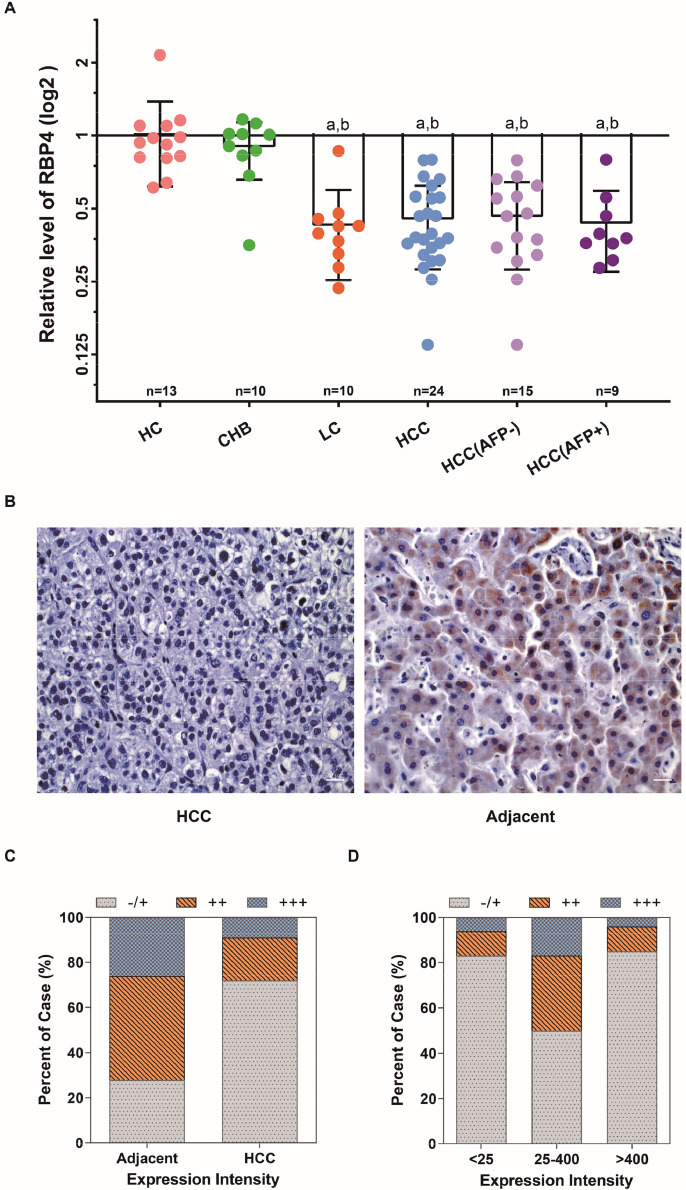
We employed two approaches to re-evaluate the pattern of RBP4 expression associated with HCC. First, serum levels of RBP4 were quantified by targeted MRM in the second cohort comprising 24 HCC patients (including 9 AFP+ and 15 AFP- cases), 10 CHB patients and 13 healthy controls (Table 1). The MRM result confirmed that serum RBP4 levels decreased exclusively in HCC patients when compared to CHB patients and healthy controls (P < 0.0001); in this cohort, there was no difference in serum RBP4 levels between AFP+ and AFP- HCC (Fig. 2A). Second, RBP4 expressions in HCC cancerous and adjacent noncancerous tissues derived from the third cohort of 80 HCC patients (Table 1) were examined by immunohistochemical (IHC) staining (Fig. 2B). Semiquantitative IHC of RBP4 showed >70 % of HCC cancerous tissues were marginally RBP4-positive (-/+), whereas >70 % of adjacent noncancerous tissues were moderately (++) or highly (+++) RBP4-positive (Fig. 2C) indicating significant down-regulation of RBP4 protein in HCC tumors. Besides, correlation analyses were performed to investigate the association between RBP4 expression and demographic features (age and gender) and clinical characterizations of HCC patients (including serum AFP levels, tumor size, and portal vein metastasis) (Table 2). Among these features, only serum levels of AFP were found correlated with the expression levels of RBP4 (Table 2); specifically, cancer tissues from > 80 % of APF-negative (serum AFP < 25 ng/mL) or AFP-high (serum AFP > 400 ng/mL) HCC patients showed low RBP4 immunostaining (-/+) (Fig. 2D). Collectively, these verification studies revealed that RBP4 was significantly low in both HCC tumor tissues and patients’ serum.
Fig. 2.
RBP4 expression in HCC serum and tissue samples. (A) Relative expression of RBP4 protein in serum as verified by MRM. a, compared with HC, P < 0.0001; b, compared with CHB, P < 0.001. (B) Representative images (400 ×) of positive RBP4 staining in HCC tissues (right panel) and negative staining in adjacent non-tumorous liver tissues (left panel) by IHC staining. Differential expression of RBP4 between (C) cancerous and adjacent non-tumorous tissues (P < 0.0001) of HCC patients, and between (D) cancerous tissues of AFP-negative (serum AFP < 25 ng/mL) and AFP-positive (serum AFP = 25–400 ng/mL and AFP >400 ng/mL) HCC patients (P < 0.01) based on semi-quantification of RBP4 IHC. HC, healthy controls; CHB, chronic hepatitis B virus infection; LC, liver cirrhosis; HCC, hepatocellular carcinoma.
Table 2.
Clinical data of 80 cases of RBP4 for HCC.
| Characteristics | No. of cases | RBP4 |
Z/χ2 | P-Val | |||
|---|---|---|---|---|---|---|---|
| – | + | ++ | +++ | ||||
| Age(year) | |||||||
| < 40 | 18 | 1 | 10 | 7 | 0 | 5.111 | 0.078 |
| 40–60 | 49 | 9 | 28 | 7 | 5 | ||
| > 60 | 13 | 4 | 8 | 1 | 0 | ||
| AFP (ng/ml) | |||||||
| < 25 | 23 | 6 | 13 | 3 | 1 | 9.434 | 0.009 |
| 25–400 | 26 | 2 | 12 | 8 | 4 | ||
| > 400 | 31 | 7 | 20 | 3 | 1 | ||
| Gender | |||||||
| Male | 69 | 10 | 43 | 12 | 4 | −0.328 | 0.743 |
| Female | 11 | 4 | 3 | 3 | 1 | ||
| Tumor size (T) (cm) | |||||||
| < 5 | 19 | 4 | 9 | 5 | 1 | 0.302 | 0.860 |
| 5–10 | 43 | 7 | 25 | 8 | 3 | ||
| > 10 | 18 | 3 | 12 | 2 | 1 | ||
| Metastasis | |||||||
| + | 21 | 1 | 13 | 6 | 1 | −1.598 | 0.110 |
| − | 49 | 13 | 33 | 9 | 4 | ||
| capsule integrity | |||||||
| + | 33 | 4 | 18 | 9 | 2 | −1.494 | 0.135 |
| − | 47 | 10 | 28 | 6 | 3 | ||
AFP, alpha-fetoprotein;RBP4, retinol-binding protein 4; #, by Kruskai-Wallis H test; *, by Mann-Whitney U test.
Evaluation of diagnostic application of serum RBP4 for HCC
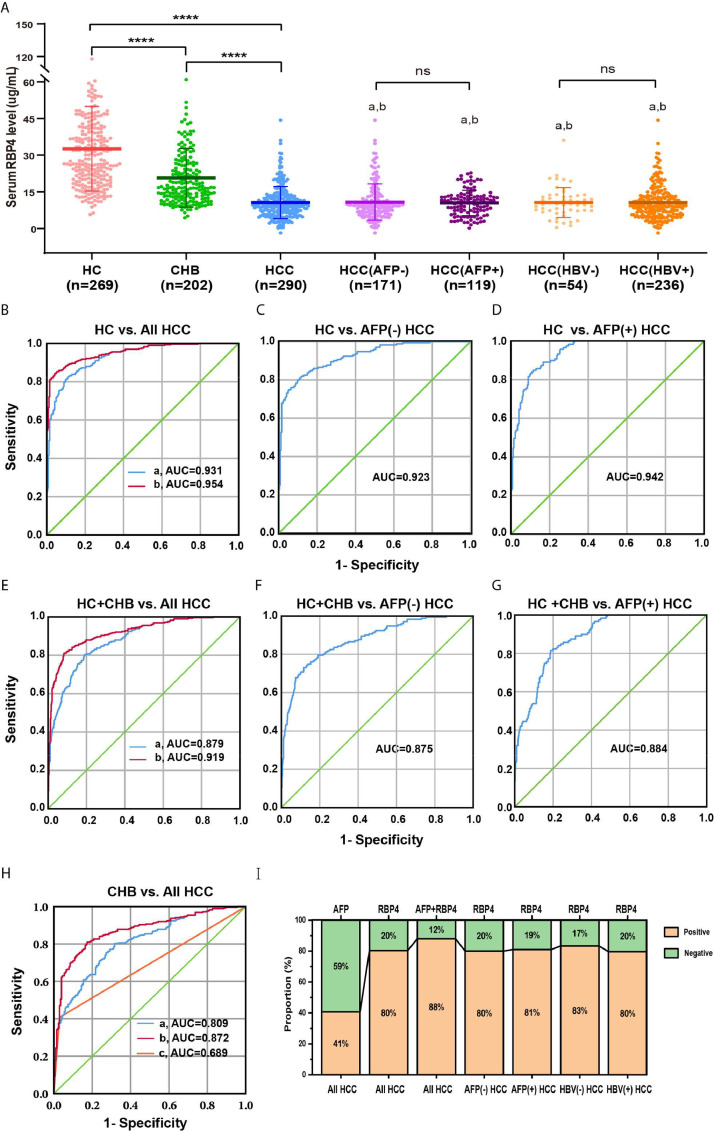
RBP4 levels were measured in 761 serum samples collected from HCC (N = 290) and CHB patients (N = 202), and HC (N = 269) by ELISA. The demographic and clinical characteristics of 290 HCC patients are summarized in Table 3. HCC patients included 249 males and 41 females, with a mean age of 50.02 years (range, 19∼76 years) at the time of the collection. The 290 HCC patients were subdivided into AFP-negative (serum AFP < 25 ng/mL) and AFP-positive (serum AFP > 25 ng/mL) groups, or into HBV-negative and HBV-positive groups. AFP-negative and HBsAg-positive HCC account for 58.97 % and 81.38 %, respectively. In this large cohort, serum levels of RBP4 were significantly lower in HCC patients (10.64 ± 6.56 μg/mL) than in CHB patients (20.71 ± 11.95 μg/mL, P < 0.0001) or in healthy controls (32.61 ± 17.32 μg/mL, P < 0.0001) (Fig. 3A). However, serum RBP4 levels were not different between different subgroups of HCC (Fig. 3A). Additionally, the ROC curves were plotted to analyze and compare the diagnostic values when RBP4 and AFP were used for HCC diagnosis (Fig. 3B-G, Table 4). A cut-off value of 14.06 μg/mL for serum RBP4 was selected at concentrations exhibiting the highest Youden index (J). The sensitivity and specificity of RBP4 for HCC diagnosis in all HCC subjects was 80.3 % and 80.9 %, respectively. The sensitivity and specificity of RBP4 in combination with AFP for HCC diagnosis was 87.9 % and 80.0 %, respectively. For AFP-negative HCC diagnosis, the sensitivity and specificity of RBP4 was 79.5 % and 80.7 %, respectively. The ROC curves showed that, when HC is used as the control, the AUC of RBP4 was 0.931 (95 % CI: 0.911∼0.950) for all HCC (Fig. 3B), 0.923 (95 % CI: 0.897∼0.948) for AFP-negative HCC (Fig. 3C) and 0.942 (95 % CI: 0.921∼0.963) for AFP-positive HCC (Fig. 3D), respectively. When HC and CHB are used as the control, AUC of RBP4 was 0.879 (95 % CI: 0.854∼0.903) for all HCC (Fig. 3E), 0.875 (95 % CI: 0.843∼0.906) for AFP-negative HCC (Fig. 3F) and 0.884 (95 % CI: 0.855∼0.913) for AFP-positive HCC (Fig. 3G), respectively. When CHB is used as the control, AUC of RBP4 was 0.809 (95 % CI: 0.771∼0.846) for all HCC (Fig. 3H). AUC for RBP4 was higher than AFP (0.689, 95% CI: 0.643∼0.735, Z = −4.901, P < 0.0001). Importantly, the combination of AFP and RBP4 further increased AUC to 0.954 (95 % CI 0.939∼0.970, P < 0.001) when HC is used as the control or 0.919 (95 % CI 0.898∼0.940, P < 0.001) when HC and CHB are used as the control) (Fig. 3B & E). A total of 233 (80.3 %) out of 290 HCC patients had low serum RBP4 (<14.06 μg/mL), among which more than half (58.4 %) patients were AFP-negative (Fig. 3I). Therefore, the true positive rate of RBP4 is higher than that of AFP, and the missed diagnosis rate of AFP is higher than that of RBP4. Collectively, these results showed that RBP4 had a higher diagnostic accuracy than AFP for all HCC including AFP-negative HCC.
Table 3.
HCC patient demographic and clinical characteristics.
| Features | Reference value | N (%) | RBP4 (µg/mL) |
|---|---|---|---|
| Age(year), mean ± SD | |||
| < 40 | 50(17.2 %) | 12.04±6.31 | |
| 40–60 | 187(64.5 %) | 10.58±6.72 | |
| > 60 | 53(18.3 %) | 9.55±6.11 | |
| Gender | |||
| Male | 249(85.86 %) | 10.68±6.44 | |
| Female | 41(14.14 %) | 10.43±7.35 | |
| TP(g/L) | ≥60 g/L | a | |
| < 60 | 25(8.62 %) | 7.98±4.94 | |
| ≥ 60 | 265(91.38 %) | 10.89±6.65 | |
| ALT(U/L) | ≤40 U/L | ||
| > 40 | 98(33.79 %) | 10.58±5.53 | |
| ≤ 40 | 192(66.21 %) | 10.67±7.05 | |
| AST(U/L) | ≤40 U/L | ||
| > 40 | 132(45.52 %) | 10.13±5.57 | |
| ≤ 40 | 158(54.48 %) | 11.07±7.28 | |
| ALB(g/L) | ≥35 g/L | b | |
| < 35 | 69(23.79 %) | 8.97±5.16 | |
| ≥35 | 221(76.21 %) | 11.17±6.88 | |
| GLO(g/L) | ≤30 g/L | ||
| > 30 | 132(45.52 %) | 10.59±7.19 | |
| ≤ 30 | 158(54.48 %) | 10.68±6.01 | |
| AFP (ng/ml) | <25 ng/ml | ||
| < 25 | 171(58.97 %) | 10.8 ± 7.39 | |
| ≥ 25 | 119(41.03 %) | 10.47±5.15 | |
| HBsAg(ng/ml) | ≤0.5 ng/ml | ||
| ≥ 0.5(+) | 236(81.38 %) | 10.65±6.13 | |
| < 0.5(-) | 54(18.62 %) | 10.64±6.67 | |
| Anti-HCV | |||
| Positive | 8(2.76 %) | 10.51±3.74 | |
| Negative | 282(97.24 %) | 10.65±6.63 | |
| Liver cirrhosis | |||
| Present | 38(13.10 %) | 9.73±6.28 | |
| Absence | 252(86.90 %) | 10.78±6.6 | |
| Metastasis | c | ||
| Present | 28(9.66 %) | 8.47±4.15 | |
| Absence | 262(90.34 %) | 10.88±6.72 |
TP, total protein; ALT, alanine transaminase; AST, aspertate transaminase; anti-HCV, Hepatitis C virus antibody; HBsAg, hepatitis Bs antigen; ALB, albumin; GLO, globulin; AFP, alpha-fetoprotein. Data are N (%) or means ± SD. a, serum RBP4 level of TG < 60 g/L group vs. TG ≥60 g/L group, t = 2.730, P = 0.0101; b, serum RBP4 level of ALB < 35 g/L group vs. ALB ≥ 35 g/L group, t = 2.445, P = 0.015; c, serum RBP4 level of present metastasis group vs. absence metastasis group, t = 2.715, P < 0.01. by Unpaired t-test.
Fig. 3.
Serum concentrations of RBP4 by ELISA and receiver operating characteristic (ROC) curve analysis of diagnostic value (A) Serum levels of RBP4 were measured by ELISA in patients with HCC, CHB and healthy controls. a, compared with HC, P < 0.001; b, compared with CHB, P < 0.001. ROC curves for (B) RBP4 alone and RBP4 combined with AFP for all HCC patients versus HC (a, RBP4, AUC = 0.931, 95 % CI: 0.911∼0.950; b, RBP4+AFP, AUC = 0.954, 95 % CI: 0.939∼0.970,R, reference line), (C) AFP negative HCC versus healthy controls (AUC = 0.923, 95 % CI: 0.897∼0.948), (D) AFP positive HCC versus healthy controls (AUC = 0.942, 95 %CI: 0.921∼0.963), (E) RBP4 alone and RBP4 combined with AFP for all HCC patients versus all controls (a, RBP4, AUC = 0.879, 95 % CI: 0.854∼0.903; b,RBP4+AFP, AUC = 0.919, 95 % CI: 0.898∼0.940), (F)AFP negative HCC versus all controls (AUC = 0.875, 95 % CI: 0.843∼0.906) and (G) AFP positive HCC versus all controls (AUC = 0.884, 95 % CI: 0.855∼0.913). (H) CHB versus all HCC (a, RBP4, AUC = 0.809; 95 % CI: 0.771–0.846; b, RBP4+AFP, AUC = 0.872, 95 % CI: 0.840∼0.903; c, AFP, AUC = 0.689, 95 % CI: 0.643∼0.735) (I) Proportion of positive results for AFP, RBP4 or both in patients with HCC. The cutoff value of RBP4 was determined by area under receiver operating characteristic curve (cutoff = 14.06 μg/mL). AFP positive, AFP > 25 ng/mL; AFP negative, AFP < 25 ng/mL; RBP4 positive, RBP4 < 14.06 μg/mL; RBP4 negative, RBP4 > 14.06 μg/mL.
Table 4.
Results for measurement of serum RBP4, AFP, or both * in the diagnosis of HCC.
| AUC# (%) | 95% CI | Sensitivity (%) | Specificity (%) | Youden's index (%) | |
|---|---|---|---|---|---|
| HCC vs HC+CHB | |||||
| RBP4 | 87.9 | 85.4∼90.3 | 80.3 | 80.9 | 61.2 |
| AFP | 69.7 | 65.6∼73.8 | 40.7 | 98.7 | 39.4 |
| RBP4+AFP | 91.9 | 89.8∼94.0 | 87.9 | 80.0 | 67.9 |
| HCC (AFP-) vs HC+CHB | |||||
| RBP4 | 87.5 | 84.3∼90.6 | 79.5 | 80.7 | 60.4 |
| HCC (AFP+) vs HC+CHB | |||||
| RBP4 | 88.4 | 85.5∼91.3 | 81.4 | 80.7 | 62.2 |
| HCC vs CHB | |||||
| RBP4 | 80.9 | 77.2∼84.7 | 80.3 | 68.3 | 48.6 |
| AFP | 68.9 | 64.3∼73.5 | 40.8 | 97.0 | 37.9 |
| RBP4+AFP | 87.2 | 84.0∼90.3 | 81.0 | 82.7 | 63.6 |
RBP4, retinol-binding protein 4; AFP, alpha-fetoprotein; HCC, Hepatocellular carcinoma; HC, healthy controls; CHB, chronic hepatitis B virus infection; AUC, area under the ROC curve. *, The diagnostic cutoff values for serum RBP4 and AFP were 14.06 μg/mL and 25 ng/mL respectively. # P < 0.05 by ROC curve analysis compared to the reference AUC.
Evaluation of prognosis application of RBP4 for HCC
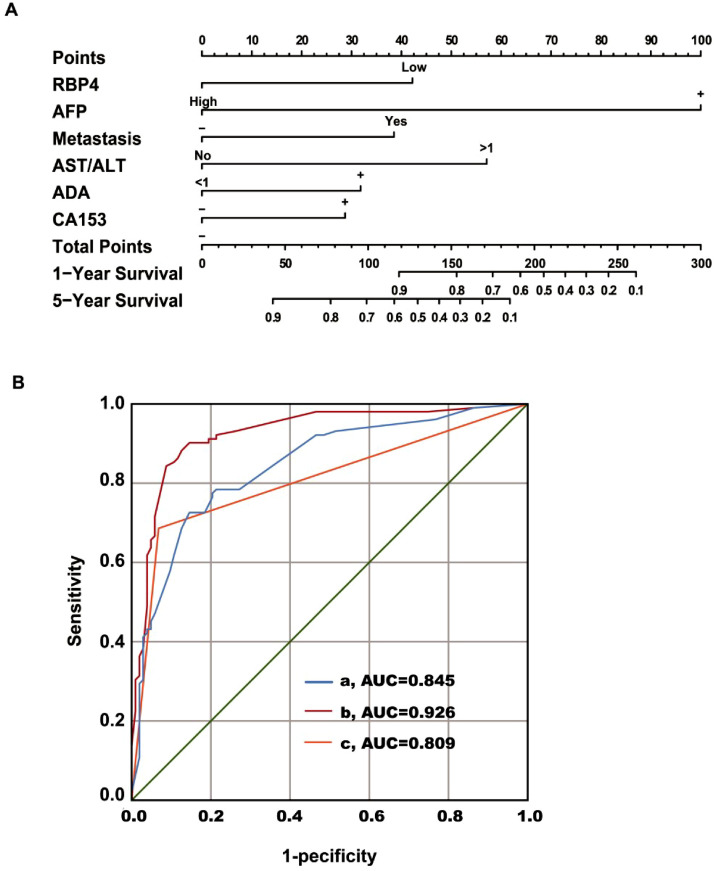
This study followed a total of 221 patients diagnosed with liver cancer. At the end of the follow-up period, there were 200 patients (177 males and 23 females), while 21 cases were lost to follow-up, resulting in a loss rate below 10 %. In the successfully followed cohort, 98 patients succumbed to mortality and 102 survived. A binary logistic regression analysis was conducted to investigate the relationship between factors such as age, gender, RBP4, AFP, metastasis, treatment, TP, AST/ALT, ALB/GLO (A/G), ADA, CA724, CA199, CA153, CA125 and the prognosis of liver cancer with overall survival time as the dependent variable (Table 5). The results revealed that elevated AFP levels and AST/ALT ratio along with the presence of metastasis were identified as independent risk factors. Conversely, normal A/G ratio levels and high RBP4 levels were found to be protective factors for patients' prognosis in liver cancer Additionally, treatment was also observed to have a positive impact on patient outcomes. Multivariate Cox proportional risk model regression analysis (Forward LR) demonstrated that RBP4 level along with AFP levels, metastasis status, AST/ALT ratio values, ADA levels, CA153 levels and intervention treatment were independent risk factors for HCC patients' prognosis (Table 5). Notably, the serum RBP4 level exhibited a negative correlation with overall survival time (HR = 0.447; 95 % CI: 0.256∼0.708; P = 0.005). Furthermore, a nomogram based on the multi-factor combined prognostic diagnosis model incorporating RBP4 was developed for predicting overall survival in HCC patients (Fig. 4A). The area under curve (AUC) of this combined prognostic model using RBP4 was calculated at 0.926 (95 % CI: 0.888∼0.964; Fig. 4B), yielding sensitivity and specificity rates of 89.2 % and 87 %, respectively. Compared to using AFP alone (AUC = 0.809), the combined prognostic model showed significantly higher predictive accuracy (Z = 5.288, P < 0.0001).
Table 5.
Univariable Logistic and Multivariable Cox regression analysis for prognosis of HCC.
| Variable | Threshold | Univariable |
Multivariable |
||
|---|---|---|---|---|---|
| P value | HR (95 %CI) | P value | HR (95 %CI) | ||
| RBP4 | 14.06 μg/mL | 0.001 | 0.090 (0.023∼0.359) | 0.005 | 0.447 (0.256∼0.708) |
| AFP | 25 ng/mL | <0.001 | 44.812 (11.878∼169.057) | <0.001 | 7.193 (4.464∼11.592 |
| Metastasis | 0.008 | 8.758 (1.742∼44.044) | 0.002 | 2.396 (1.376∼4.173) | |
| AST/ALT | 1 | 0.002 | 9.204 (2.310∼36.683) | <0.001 | 3.176 (1.732∼5.823) |
| Treatment | 0.143 | ||||
| Surgery | 0.041 | 5.947 (1.074∼32.919) | 0.766 | 0.908 (0.481∼1.714) | |
| Intervention | 0.070 | 7.112 (0.849∼59.555) | 0.017 | 2.085 (1.139∼3.819) | |
| Others | 0.187 | 2.768 (0.610∼12.548) | 0.185 | 0.688 (0.395∼1.196) | |
| Age | 0.875 | ||||
| Gender | 0.569 | ||||
| HBsAg | 0.5 ng/ml | 0.112 | |||
| A/G | 1.3 | 0.002 | 0.123 (0.032∼0.477) | 0.107 | |
| ADA | 22 U/L | 0.494 | 0.003 | 1.948 (1.249∼3.037) | |
| TP | 60 g/L | 0.916 | |||
| CA724 | 6 U/mL | 0.800 | |||
| CA199 | 37 U/mL | 0.111 | |||
| CA153 | 28 U/mL | 0.590 | 0.021 | 1.814 (1.094∼3.008 | |
| CA125 | 35 U/mL | 0.474 | |||
RBP4, retinol-binding protein 4; AFP, alpha-fetoprotein; AST, alanine transaminase; ALT, aspertate transaminase; HBsAg, hepatitis Bs antigen; A/G, albumin/globulin; ADA, adenosine deaminase; TP, total protein; CA724, carbohydrate antigen 724; CA199, carbohydrate antigen 199; CA153, carbohydrate antigen 153; CA125, carbohydrate antigen 125.
Fig. 4.
Diagnostic value of RBP4 combined prognostic diagnosis model (A) Nomogram to predict overall survival time of HCC. To use this nomogram, the specific point for each variable of the patient lies on each variable axis. Draw a vertical line upward to determine the point at which each variable accepts; the sum of these points is located on the Total Points axis, and draw a vertical line down to the survival axis to determine the probability of 1- and 5- year overall survival time. The length of the line segment reflected the contribution of the factor to the final event. The sum of all individual scores corresponded to the corresponding 1-year and 5-year survival time. (B) ROC curve for HCC prognostic model (a, RBP4+other, AUC = 0.845, 95 % CI: 0.791∼0.899; b, RBP4+AFP+other, AUC = 0.926, 95 % CI: 0.888∼0.964; c, AFP, AUC = 0.809, 95 % CI: 0.747∼0.871).
Discussion
Although omics studies, including proteomics and transcriptomics, have provided us with a broad knowledge of HCC, the molecular events accompanying HCC development are still largely undefined. One big challenge of liver cancer management is the lack of highly sensitive biomarkers for early-stage or AFP-negative HCC. The focus of the current study was to identify and validate novel HCC biomarkers by employing multi-layer approaches including: [1] iTRAQ-MALDI- TOF-MS/MS proteomic screening for DEPs in serums from HCC patients; [2] mRNA expression and survival analysis of top target DEPs using the UALCAN database to predict the correlation between DEPs and HCC; [3] validation studies using MRM, IHC and ELISA; and [4] evaluation of the diagnostic value of candidate biomarker. Through these studies, we identified 10 HCC-associated DEPs in the serum, among which RBP4 is the top down-regulated protein; liver cancer samples from the UALCAN database show low RBP4 mRNA expression and tumor RBP4 mRNA levels are positively correlated with cancer survival; low expression of RBP4 protein in HCC patients is confirmed in cancerous tissues by IHC and in serums by MRM and ELISA. Based on these results, we conclude that low levels of RBP4 may represent a signature molecular feature associated with HCC.
Validation of putative protein biomarkers is a major challenge in clinical proteomic samples. MRM, a well-established targeted proteomics technology, can quantify proteins in biological specimens with high specificity and selectivity. MRM is emerging to bridge the gap between biomarker discovery and clinical validation [28,29]. In the current study, we used the MRM assay to measure RBP4 protein levels in the serum of HCC patients and controls, which greatly improved the credibility of proteomics verification.
The current study represents an advance in evaluating RBP4 as a promising biomarker for HCC by validation study in a large-scale cohort, particularly for AFP-negative liver tumors. In this cohort of a total of 761 study subjects, the AUC and sensitivity of RBP4 were higher than that of AFP in differentiating HCC cases from controls. AFP is a biomarker most commonly used in the screening of early HCC. To date, it is the only serum biomarker that has been evaluated in a randomized and controlled trial [30]. However, the diagnostic power of AFP has been continuously debated [[31], [32], [33]]. A large-scale study showed that elevated serum AFP was only observed in 57.8 %–67.0 % of overall patients with HCC and 18.8 %∼61 % of patients with chronic hepatitis or cirrhosis [19]. Indeed, ∼59 % (171/290) of HCC patients recruited for our ELISA study were AFP-negative (< 25 ng/mL). In contrast, we observed that ∼80 % of HCC patients and ∼80 % of the AFP-negative HCC patients had low serum RBP4 (< 14.06 μg/mL). More importantly, the sensitivity and specificity of using RBP4 in combination with AFP for HCC diagnosis is a significant improvement over AFP alone. Therefore, serum RBP4 has a great potential to be used as a complementary biomarker for AFP-negative HCC diagnosis. Nevertheless, further analyses are warranted in the future to translate these observations to clinical applications.
RBP4 protein is a monomeric-binding protein of 21 kDa and participates in the transport of retinol vitamin A in blood [34]. RBP4 is mainly secreted by the liver and adipose tissue, wherein the liver has the highest expression of RBP4. GO annotations linked to RBP4 include negative regulation of cardiac muscle cell proliferation, digestive system process, complement, and coagulation cascades, retinol transporter activity, female genitalia morphogenesis, lipid transport, and localization, alcohol metabolic process, and positive regulation of insulin and immunoglobulin secretion. In 2005, one study found that adipocyte-derived RBP4 levels were elevated in insulin-resistant patients with obesity; it suggested that RBP4 was a new fat factor involved in insulin resistance, which may affect glucose homeostasis and lead to inflammation in mice [35]. Latter studies have identified RBP4 as being associated with other metabolic diseases including type 2 diabetes, hepatic steatosis, and steatohepatitis [[36], [37], [38]], all of which are also linked to primary liver cancer. However, some clinical studies show a lack of association between RBP4 levels and obesity or insulin-resistant states, which was proposed to be a consequence of increased expression, altered secretion or clearance of RBP4 from the circulation [39,40]. Also, the RBP4 level is differentially expressed in certain tumors, such as ovarian cancer, colorectal cancer, breast cancer, and acute lymphocytic leukemia [[41], [42], [43], [44], [45], [46]]. There are few reports on RBP4 in the context of liver cancer. Kinoshita et al. performed differential gene display analysis (DGDA) to identify DEGs in HCC relative to chronic viral hepatitis and found the under-expression of the RBP4 gene in cancer tissues of 12 HCC patients [47], which is consistent with our results. At the molecular level, one study indicates that increased unbound RBP4 can activate JAK2/STAT5, a receptor-mediated signaling pathway, leading to changes in cell proliferation, apoptosis, and malignancy [48]. Additionally, HCC progression is associated with the cross-talk between tumor cells and the surrounding stroma including activated hepatic stellate cells (HSC); in this context, the expression of hepatotropic cytokines, including RBP4, DKK1, and CCL5, was found to be negatively regulated by endosialin-expressing HSCs, thereby impacting the progression of HCC [49]. Also, RBP4 can induce IL-6 and TNF-α through the TLR4/NF-κB pathway through adipose tissue exosome-mediated activation of macrophage-induced insulin resistance [50]. Recently, the bioinformatics analysis of RBP4 conducted by Li M et al. utilizing online analysis tools revealed a significant down-regulation of RBP4 expression in hepatocellular carcinoma (HCC) and cholangiocarcinoma, the down-regulation is indicative of a poor prognosis for HCC and is closely associated with immune cell infiltration within the tumor microenvironment [51]. Taken together, we propose that RBP4 may be involved in precancerous hepatic lesions through exosome-mediated macrophage activation. More experimental evidence should be collected in future studies to test this hypothesis.
HBV infection has been one of the most common causes of liver cancer in China. Epidemiological studies have shown that about 60 %∼80 % of liver cancer cases in China are associated with HBV carrier status [52]. Given the high incidence of HBV infection in Chinese HCC patients, we have included CHB patients in every study cohort as controls that are recommended for HCC surveillance. In the ELISA validation cohort, ∼80 % of the liver cancer patients are HBsAg positive. We found that serum concentrations of RBP4 were lower in patients with chronic hepatitis B relative to healthy controls, whereas there was no difference in serum RBP4 levels between HBV (−) HCC and HBV (+) HCC patients, suggesting low serum RBP4 noted in HCC patients is not likely due to HBV infection. In addition to HBV infection, other important etiological factors of liver cancer include hepatitis C infection, liver cirrhosis, alcoholic liver disease, consumptions of aflatoxin-contaminated food, fatty liver disease, and diabetes [53]. One study revealed that partial knockdown of RBP4 had a positive impact on HCV replication, implying a link between HCV infection and RBP4 [54]. However, in our study, there was no difference in serum RBP4 levels between HCV (−) HCC and HCV (+) HCC patients. Similarly, an inverse association between serum RBP4 levels and liver fibrosis stage has been reported [55]. In our study, there were no significant differences observed in serum RBP4 levels between HCC patients with cirrhosis and those without cirrhosis. This lack of difference may attribute to the relatively low prevalence of HCV infection (2.76 %) and cirrhosis (13.1 %) within our HCC cohort. Consequently, further investigation is warranted to elucidate the intricate relationship among HCV infection, liver cirrhosis, RBP4, and HCC development. Future studies focusing on exploring the association between RBP4 levels and liver cancers originating from diverse etiologies would be highly valuable.
Notably, stratified analysis of serum RBP4 levels in HCC patients with different clinicopathological characteristics revealed a potential association between serum TP level and serum RBP4 level, while patients with metastasis exhibited lower levels of serum RBP4. Cox regression survival analysis demonstrated that low expression of RBP4 was correlated with reduced 5-year survival rate, suggesting its potential as a prognostic marker for liver cancer. The serum levels of RBP4 in HCC patients exhibited significant correlations with liver cirrhosis, tumor size, venous invasion, disease stage, and an unfavorable prognosis [56]. further multicenter studies are warranted to validate the prognostic significance of RBP4 in HCC patients.
Collectively, through extensive cohort validation, we demonstrate that RBP4 represents a superior diagnostic marker for HCC when compared to AFP. Additionally, combining RBP4 with AFP can greatly improve the detection rate of HCC in high-risk populations under surveillance. Furthermore, RBP4 holds promise as a prognostic biomarker, and integrating it into a combined prognostic model has great potential for enhancing the efficiency of predicating HCC prognosis. Therefore, RPB4 may serve as a reliable serum marker for both diagnosis and prediction of the prognosis of HCC, complementing AFP and offering a potential target for future liver cancer treatment.
CRediT authorship contribution statement
Fengjie Wan: Writing – original draft, Visualization, Validation, Methodology, Investigation, Data curation. Yujia Zhu: Investigation, Data curation. Feixiang Wu: Validation, Investigation, Data curation. Xuejing Huang: Validation, Investigation. Ying Chen: Writing – review & editing. Yi Zhou: Methodology. Hongtao Li: Methodology. Lifang Liang: Data curation. Lirong Qin: Data curation. Qi Wang: Project administration. Min He: Writing – review & editing, Supervision, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81760612, 81260445); Program of Key Laboratory of High-Incidence-Tumor Prevention and Treatment, Guangxi Medical University, Ministry of Education, China (GKE-ZZ202002, GKE-ZZ202108, GKE-ZZ202212); Science and Technology Department of Guangxi (AB16380184); International Communication of Guangxi Medical University Graduate Education.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2024.101979.
Contributor Information
Qi Wang, Email: wangqi@stu.gxmu.edu.cn.
Min He, Email: hemin@gxmu.edu.cn.
Appendix. Supplementary materials
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.McGlynn K.A., Petrick J.L. El-Serag HB. epidemiology of hepatocellular carcinoma. Hepatology. 2021;73 Suppl 1(Suppl 1):4–13. doi: 10.1002/hep.31288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akinyemiju T., Abera S., Ahmed M., Alam N., Alemayohu M.A., Allen C., et al. The Burden of Primary Liver Cancer and Underlying Etiologies From 1990 to 2015 at the Global, Regional, and National Level: results From the Global Burden of Disease Study 2015. JAMa Oncol. 2017;3(12):1683–1691. doi: 10.1001/jamaoncol.2017.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allemani C., Matsuda T., Di Carlo V., Harewood R., Matz M., Niksic M., et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391(10125):1023–1075. doi: 10.1016/S0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chidambaranathan-Reghupaty S., Fisher P.B., Sarkar D. Hepatocellular carcinoma (HCC): epidemiology, etiology and molecular classification. Adv. Cancer Res. 2021;149:1–61. doi: 10.1016/bs.acr.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li M.X., Wang Z.H., Zhu L.X., Shui Y.F., Zhang S.J., Guo W.Z. Down-regulation of RBP4indicates a poor prognosis and correlates with immune cell infiltration in hepatocellular carcinoma. Biosci. Rep. 2021;41(4):12. doi: 10.1042/bsr20210328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arrieta O., Cacho B., Morales-Espinosa D., Ruelas-Villavicencio A., Flores-Estrada D., Hernandez-Pedro N. The progressive elevation of alpha fetoprotein for the diagnosis of hepatocellular carcinoma in patients with liver cirrhosis. BMC Cancer. 2007;7:28. doi: 10.1186/1471-2407-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tong M.J., Huynh T.T., Siripongsakun S., Chang P.W., Tong L.T., Ha Y.P., et al. Predicting clinical outcomes in patients with HBsAg-positive chronic hepatitis. Hepatol. Int. 2015;9(4):567–577. doi: 10.1007/s12072-015-9651-z. [DOI] [PubMed] [Google Scholar]
- 9.Tsuchiya N. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J. Gastroenterol. 2015;21(37):10573. doi: 10.3748/wjg.v21.i37.10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta S., Bent S., Kohlwes J. Test characteristics of alpha-fetoprotein for detecting hepatocellular carcinoma in patients with hepatitis C. A systematic review and critical analysis. Ann. Intern. Med. 2003;139(1):46–50. doi: 10.7326/0003-4819-139-1-200307010-00012. [DOI] [PubMed] [Google Scholar]
- 11.Wang C.S., Lin C.L., Lee H.C., Chen K.Y., Chiang M.F., Chen H.S., et al. Usefulness of serum des-gamma-carboxy prothrombin in detection of hepatocellular carcinoma. World J. Gastroenterol. 2005;11(39):6115–6119. doi: 10.3748/wjg.v11.i39.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tateishi R., Yoshida H., Matsuyama Y., Mine N., Kondo Y., Omata M. Diagnostic accuracy of tumor markers for hepatocellular carcinoma: a systematic review. Hepatol. Int. 2008;2(1):17–30. doi: 10.1007/s12072-007-9038-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen D.S., Sung J.L., Sheu J.C., Lai M.Y., How S.W., Hsu H.C., et al. Serum alpha-fetoprotein in the early stage of human hepatocellular carcinoma. Gastroenterology. 1984;86(6):1404–1409. [PubMed] [Google Scholar]
- 14.Bai D.S., Zhang C., Chen P., Jin S.J., Jiang G.Q. The prognostic correlation of AFP level at diagnosis with pathological grade, progression, and survival of patients with hepatocellular carcinoma. Sci. Rep. 2017;7(1):12870. doi: 10.1038/s41598-017-12834-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zinkin N.T., Grall F., Bhaskar K., Otu H.H., Spentzos D., Kalmowitz B., et al. Serum proteomics and biomarkers in hepatocellular carcinoma and chronic liver disease. Clin. Cancer Res. 2008;14(2):470–477. doi: 10.1158/1078-0432.CCR-07-0586. [DOI] [PubMed] [Google Scholar]
- 16.Chignard N., Beretta L. Proteomics for hepatocellular carcinoma marker discovery. Gastroenterology. 2004;127(5 Suppl 1):S120–S125. doi: 10.1053/j.gastro.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 17.Tsai T.H., Song E., Zhu R., Di Poto C., Wang M., Luo Y., et al. LC-MS/MS-based serum proteomics for identification of candidate biomarkers for hepatocellular carcinoma. Proteomics. 2015;15(13):2369–2381. doi: 10.1002/pmic.201400364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang C., Wang Y., Liu S., Ding G., Liu W., Zhou J., et al. Quantitative proteomic analysis identified paraoxonase 1 as a novel serum biomarker for microvascular invasion in hepatocellular carcinoma. J. Proteome Res. 2013;12(4):1838–1846. doi: 10.1021/pr3011815. [DOI] [PubMed] [Google Scholar]
- 19.Shen Q., Fan J., Yang X.R., Tan Y., Zhao W., Xu Y., et al. Serum DKK1 as a protein biomarker for the diagnosis of hepatocellular carcinoma: a large-scale, multicentre study. Lancet Oncol. 2012;13(8):817–826. doi: 10.1016/S1470-2045(12)70233-4. [DOI] [PubMed] [Google Scholar]
- 20.Fu Y., Xu X., Huang D., Cui D., Liu L., Liu J., et al. Plasma heat shock protein 90alpha as a biomarker for the diagnosis of liver cancer: an official, large-scale, and multicenter clinical trial. EBioMedicine. 2017;24:56–63. doi: 10.1016/j.ebiom.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sourbier C. Plasma HSP90alpha and liver cancer: a potential biomarker? EBioMedicine. 2017;25:7–8. doi: 10.1016/j.ebiom.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pepe M.S., Etzioni R., Feng Z., Potter J.D., Thompson M.L., Thornquist M., et al. Phases of biomarker development for early detection of cancer. J. Natl. Cancer Inst. 2001;93(14):1054–1061. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- 23.Park J.W., Chen M., Colombo M., Roberts L.R., Schwartz M., Chen P.J., et al. Global patterns of hepatocellular carcinoma management from diagnosis to death: the BRIDGE study. Liver. Int. 2015;35(9):2155–2166. doi: 10.1111/liv.12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He X., Wang Y., Zhang W., Li H., Luo R., Zhou Y., et al. Screening differential expression of serum proteins in AFP-negative HBV-related hepatocellular carcinoma using iTRAQ -MALDI-MS/MS. Neoplasma. 2014;61(1):17–26. [PubMed] [Google Scholar]
- 25.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B., et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eshghi A., Borchers C.H. Multiple reaction monitoring using double isotopologue peptide standards for protein quantification. Methods Mol. Biol. 2018;1788:193–214. doi: 10.1007/7651_2017_112. [DOI] [PubMed] [Google Scholar]
- 27.Liu X.K., Zhang X.R., Zhong Q., Li M.Z., Liu Z.M., Lin Z.R., et al. Low expression of PTK6/Brk predicts poor prognosis in patients with laryngeal squamous cell carcinoma. J. Transl. Med. 2013;11:59. doi: 10.1186/1479-5876-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chambers A.G., Percy A.J., Simon R., Borchers C.H. MRM for the verification of cancer biomarker proteins: recent applications to human plasma and serum. Expert. Rev. Proteomics. 2014;11(2):137–148. doi: 10.1586/14789450.2014.877346. [DOI] [PubMed] [Google Scholar]
- 29.Meng Z., Veenstra T.D. Targeted mass spectrometry approaches for protein biomarker verification. J. Proteomics. 2011;74(12):2650–2659. doi: 10.1016/j.jprot.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 30.Zhang B.H., Yang B.H., Tang Z.Y. Randomized controlled trial of screening for hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2004;130(7):417–422. doi: 10.1007/s00432-004-0552-0. [DOI] [PubMed] [Google Scholar]
- 31.Mehinovic L., Islamagic E., Husic-Selimovic A., Kurtovic-Kozaric A., Vukobrat-Bijedic Z., Suljevic D. Evaluation of diagnostic efficiency of alpha-fetoprotein in patients with liver cirrhosis and hepatocellular carcinoma: single-center experience. open access maced. J. Med. Sci. 2018;6(9):1668–1673. doi: 10.3889/oamjms.2018.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen M.H., Garcia R.T., Simpson P.W., Wright T.L., Keeffe E.B. Racial differences in effectiveness of alpha-fetoprotein for diagnosis of hepatocellular carcinoma in hepatitis C virus cirrhosis. Hepatology. 2002;36(2):410–417. doi: 10.1053/jhep.2002.34744. [DOI] [PubMed] [Google Scholar]
- 33.Farinati F., Marino D., De Giorgio M., Baldan A., Cantarini M., Cursaro C., et al. Diagnostic and prognostic role of alpha-fetoprotein in hepatocellular carcinoma: both or neither? Am. J. Gastroenterol. 2006;101(3):524–532. doi: 10.1111/j.1572-0241.2006.00443.x. [DOI] [PubMed] [Google Scholar]
- 34.Blaner W.S. Retinol-binding protein: the serum transport protein for vitamin A. Endocr. Rev. 1989;10(3):308–316. doi: 10.1210/edrv-10-3-308. [DOI] [PubMed] [Google Scholar]
- 35.Yang Q., Graham T.E., Mody N., Preitner F., Peroni O.D., Zabolotny J.M., et al. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 2005;436(7049):356–362. doi: 10.1038/nature03711. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y., Sun L., Lin X., Yuan J.M., Koh W.P., Pan A. Retinol binding protein 4 and risk of type 2 diabetes in Singapore Chinese men and women: a nested case-control study. Nutr. Metab. 2019;16:3. doi: 10.1186/s12986-018-0329-0. (Lond) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaker O., El-Shehaby A., Zakaria A., Mostafa N., Talaat S., Katsiki N., et al. Plasma visfatin and retinol binding protein-4 levels in patients with type 2 diabetes mellitus and their relationship to adiposity and fatty liver. Clin. Biochem. 2011;44(17–18):1457–1463. doi: 10.1016/j.clinbiochem.2011.08.1148. [DOI] [PubMed] [Google Scholar]
- 38.Nderitu P., Bosco C., Garmo H., Holmberg L., Malmstrom H., Hammar N., et al. The association between individual metabolic syndrome components, primary liver cancer and cirrhosis: a study in the Swedish AMORIS cohort. Int. J. Cancer. 2017;141(6):1148–1160. doi: 10.1002/ijc.30818. [DOI] [PubMed] [Google Scholar]
- 39.Kotnik P., Fischer-Posovszky P., Wabitsch M. RBP4: a controversial adipokine. Eur. J. Endocrinol. 2011;165(5):703–711. doi: 10.1530/EJE-11-0431. [DOI] [PubMed] [Google Scholar]
- 40.Ma X., Zhou Z., Chen Y., Wu Y., Liu Y. RBP4 functions as a hepatokine in the regulation of glucose metabolism by the circadian clock in mice. Diabetologia. 2016;59(2):354–362. doi: 10.1007/s00125-015-3807-1. [DOI] [PubMed] [Google Scholar]
- 41.Kwak J.Y., Ma T.Z., Yoo M.J., Choi B.H., Kim H.G., Kim S.R., et al. The comparative analysis of serum proteomes for the discovery of biomarkers for acute myeloid leukemia. Exp. Hematol. 2004;32(9):836–842. doi: 10.1016/j.exphem.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Noy N., Li L., Abola M.V., Berger N.A. Is retinol binding protein 4 a link between adiposity and cancer? Horm. Mol. Biol. Clin. Investig. 2015;23(2):39–46. doi: 10.1515/hmbci-2015-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y., Wang Y., Zhang Z. Adipokine RBP4 drives ovarian cancer cell migration. J. Ovarian. Res. 2018;11(1):29. doi: 10.1186/s13048-018-0397-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fei W., Chen L., Chen J., Shi Q., Zhang L., Liu S., et al. RBP4 and THBS2 are serum biomarkers for diagnosis of colorectal cancer. Oncotarget. 2017;8(54):92254–92264. doi: 10.18632/oncotarget.21173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karunanithi S., Levi L., DeVecchio J., Karagkounis G., Reizes O., Lathia J.D., et al. RBP4-STRA6 pathway drives cancer stem cell maintenance and mediates high-fat diet-induced colon carcinogenesis. Stem Cell Rep. 2017;9(2):438–450. doi: 10.1016/j.stemcr.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiao C., Cui L., Ma A., Li N., Si H. Elevated serum levels of retinol-binding protein 4 are associated with breast cancer risk: a case-control study. PLoS One. 2016;11(12) doi: 10.1371/journal.pone.0167498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kinoshita M., Miyata M. Underexpression of mRNA in human hepatocellular carcinoma focusing on eight loci. Hepatology. 2002;36(2):433–438. doi: 10.1053/jhep.2002.34851. (Baltimore, Md) [DOI] [PubMed] [Google Scholar]
- 48.Chen C.H., Hsieh T.J., Lin K.D., Lin H.Y., Lee M.Y., Hung W.W., et al. Increased unbound retinol-binding protein 4 concentration induces apoptosis through receptor-mediated signaling. J. Biol. Chem. 2012;287(13):9694–9707. doi: 10.1074/jbc.M111.301721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mogler C., Konig C., Wieland M., Runge A., Besemfelder E., Komljenovic D., et al. Hepatic stellate cells limit hepatocellular carcinoma progression through the orphan receptor endosialin. EMBo Mol. Med. 2017;9(6):741–749. doi: 10.15252/emmm.201607222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng Z.B., Poliakov A., Hardy R.W., Clements R., Liu C., Liu Y., et al. Adipose tissue exosome-like vesicles mediate activation of macrophage-induced insulin resistance. Diabetes. 2009;58(11):2498–2505. doi: 10.2337/db09-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li M., Wang Z., Zhu L., Shui Y., Zhang S., Guo W. Down-regulation of RBP4 indicates a poor prognosis and correlates with immune cell infiltration in hepatocellular carcinoma. Biosci. Rep. 2021;41(4) doi: 10.1042/bsr20210328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka M., Katayama F., Kato H., Tanaka H., Wang J., Qiao Y.L., et al. Hepatitis B and C virus infection and hepatocellular carcinoma in China: a review of epidemiology and control measures. J. Epidemiol. 2011;21(6):401–416. doi: 10.2188/jea.JE20100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J.D., Hainaut P., Gores G.J., Amadou A., Plymoth A., Roberts L.R. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019;16(10):589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gouthamchandra K., Kumar A., Shwetha S., Mukherjee A., Chandra M., Ravishankar B., et al. Serum proteomics of hepatitis C virus infection reveals retinol-binding protein 4 as a novel regulator. J. Gen. Virol. 2014;95(Pt 8):1654–1667. doi: 10.1099/vir.0.062430-0. [DOI] [PubMed] [Google Scholar]
- 55.Kataria Y., Deaton R.J., Enk E., Jin M., Petrauskaite M., Dong L., et al. Retinoid and carotenoid status in serum and liver among patients at high-risk for liver cancer. BMC. Gastroenterol. 2016;16:30. doi: 10.1186/s12876-016-0432-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang D.D, Zhao Y.M., Wang L., Ren G., Wang F., Xia Z.G., et al. Preoperative serum retinol-binding protein 4 is associated with the prognosis of patients with hepatocellular carcinoma after curative resection. J. Cancer Res. Clin. Oncol. 2011;137(4):651–658. doi: 10.1007/s00432-010-0927-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.