Abstract
Immune checkpoint blockade (ICB) can induce durable cancer remission. However, only a small subset of patients gain benefits. While tumor mutation burden (TMB) differentiates responders from nonresponders in some cases, it is a weak predictor in tumor types with low mutation rates. Thus, there is an unmet need to discover a new class of genetic aberrations that predict ICB responses in these tumor types. Here, we report analyses of pan-cancer whole genomes which revealed that intragenic rearrangement (IGR) burden is significantly associated with immune infiltration in breast, ovarian, esophageal, and endometrial cancers, particularly with increased M1 macrophage and CD8+ T-cell signatures. Multivariate regression against spatially counted tumor-infiltrating lymphocytes in breast, endometrial, and ovarian cancers suggested that IGR burden is a more influential covariate than other genetic aberrations in these cancers. In the MEDI4736 trial evaluating durvalumab in esophageal adenocarcinoma, IGR burden correlated with patient benefits. In the IMVigor210 trial evaluating atezolizumab in urothelial carcinoma, IGR burden increased with platinum exposure and predicted patient benefit among TMB-low, platinum-exposed tumors. Altogether, we have demonstrated that IGR burden correlates with T-cell inflammation and predicts ICB benefit in TMB-low, IGR-dominant tumors, and in platinum-exposed tumors.
Keywords: Intragenic rearrangements, neoantigen burden, immune infiltration, immunotherapy response, cancer
Introduction
One of the most important breakthroughs in cancer treatment was the development of immune checkpoint blockade (ICB) (1,2). However, only up to 12.5% of cancer patients receive benefit from such treatments (3). In addition, 10–25% of patients receiving ICB experience severe or even lethal immune-related adverse events (irAEs) (4,5) and 4–29% of patients experience ICB-induced dramatic acceleration of disease, which is known as ‘hyperprogressive disease’ (HPD) (6). Thus, predictive biomarkers of ICB responses are in high demand to ensure patient benefits and to avoid adverse clinical outcomes.
Besides the commonly used PD-L1 tests, several genetic biomarkers are associated with T-cell inflammation and ICB effectiveness, including tumor mutation burden (TMB) (7), somatic copy number alternation (SCNA) (8), gene fusions (9), insertion–deletion mutations (Indels) (10), and microsatellite instability (MSI) (11). However, none of these genetic markers are sufficient to distinguish ICB responders from nonresponders across a broad spectrum of cancer types. For example, in triple-negative breast cancer (TNBC), defined as breast tumors that lack expression of estrogen receptor, progesterone receptor, and HER2, only a small fraction of tumors have high TMB (12), but a much larger fraction of tumors are inflamed (13,14). Moreover, TNBC tumors with high T-cell inflammation show lower TMB and fewer SCNA (15,16), but respond well to ICB treatment (17). This suggests the existence of an unidentified source of neoantigens in TNBC. Similarly, most ovarian tumors have low TMB and the IMagyn050 trial showed that TMB was not predictive of ICB response in ovarian cancer (18). In esophageal adenocarcinoma (ESAD), TMBhigh and MSIhigh tumors account for only 2.4% and <2% of tumors, respectively (19), and TMB is neither associated with T-cell infiltration nor does it predict response to ICB (20). Additionally, biomarker analysis of the CheckMate649 trial suggested that nivolumab (anti-PD1) plus chemotherapy resulted in a higher overall survival compared to chemotherapy alone in ESAD, regardless of TMB levels (21). Thus, the identification of a new class of genetic aberrations that can predict ICB benefits in these cancers is an unmet need in oncology.
Here, we propose a new class of biomarker, which estimates the number of cryptic intragenic rearrangements (IGRs) from the tumor genome. IGRs result in exon duplications or deletions through rearrangements of a genomic distance of tens of kilobases (Supplementary Fig. S1). While there are sporadic reports of specific IGRs as oncogenic events (22–25), there is a paucity of landscape studies of IGRs in cancer. We conducted analyses of pan-cancer whole genomes (26), genomic datasets for breast (27), ovarian (28), and esophageal cancers (29), as well as densities of tumor-infiltrating lymphocytes (TILs) in histological images from 13 tumor types within TCGA. We demonstrate a strong association between IGR burden and T-cell inflammation in breast, ovarian, endometrial, and esophageal cancers. Furthermore, we analyzed datasets from two clinical trials evaluating ICB: MEDI4736 for esophageal cancer (30) and IMvigor210 for urothelial carcinoma (31). IGR burden was associated with ICB responses in patients with esophageal cancer and in TMB-low urothelial carcinoma patients who received platinum-based therapy.
Materials and Methods
Data and preprocessing
The data processing workflow and datasets used in this study are described in Supplementary Fig. S1A and Supplementary Table S1 respectively. The IGR burden was estimated based on the total number of exon deletions or duplications as detailed below (Supplementary Fig. S1B). Fusions were estimated based on the total number of exon junctions between two different genes. TMB was estimated based on the total number of missense mutations. As shown in Supplementary Fig. S1C, the total number of missense mutations was almost linearly correlated with the total number of single nucleotide variants (Pearson’s R=0.997, p<2.2e-16, Supplementary Fig. S1C). SCNA was assessed based on a previous method (8). Square root transformation was performed for all the genetic biomarkers discussed in this study to maintain the outlier distribution within a smaller range.
Estimation of IGR burden
IGRs were estimated from the somatic structural variations in the variant calling format files from WGS data and chimeric junction files from RNAseq data. For the ICGC WGS data (26), the structural variant (SV) calling files from dRanger-snowman and svfix were used in our analyses. For RNAseq data, chimeric junction files are generated using STAR aligner v1.8.1 (32). SV junctions were mapped to the exon annotation files of genome build GRCh37 for WGS data, or GRCh38 for RNAseq data to identify the aberrant exon junctions generated by an IGR. The IGR burden was calculated as the square root of the total number of IGRs. Among the datasets analyzed, we found that the size factor, the total number of reads per library, showed strong correlation to IGR burden in the datasets with read length less than 75bp. This may be due to the fact that to detect IGR, reads that span the IGR junction are required to identify the precise IGR junctions. When the read length is short, the coverage will have a more substantial effect on detecting the junction reads. Thus, for the datasets with short read length (less than 75bp), we scaled the IGR burden through dividing by the total number of uniquely mapped reads. For stratification of IGR or TMB high/low levels, we used the median as the binary cutoff if skewness was below 3 (a skewness greater than 3 indicates absolute non-normal distribution). In the ICGC pan-cancer dataset, we used median plus median absolute deviation (MAD) as the binary cutoff as the distributions of IGR and TMB values become highly right-skewed (the skewness are 9.69 and 4.99 respectively) when different cancers were pooled together. This method has been previously proven effective in highly skewed data (33).
Expression analysis
CIBERSORT was applied on transcript per million (TPM) expression data to estimate populations of infiltrating immune cell types (34). For the estimation of T-inflamed signature, we first retrieved the expressions of inflammatory genes (Supplementary Table S2). We then used R package singscore/1.14.0 to analyze the single sample signature score by rank-based statistics. The cell cycle signature was calculated as the mean of the cell cycle gene set (Supplementary Table S2), as previously described (8). Differentially expressed genes were calculated using R package limma/v3.50.0 with default parameters. The significant genes (p<0.05) were used for pathway analysis. The gene set enrichment analysis (GSEA) using hallmark database was conducted using R packages msigdbr/v7.4.1 and fgsea/v1.21.0. Hallmark pathways with significantly adjusted p-value are shown in the figures.
Statistical analysis
To perform survival analysis on metastatic urothelial carcinoma data (31), we applied the Kaplan-Meier plot in R package survival/v3.2.7. In the survival analysis for both TMB and IGR, we used multivariable Cox-proportional hazard models. In the survival analysis for TMB alone, log-rank test was employed to compare the survival distributions between two groups. For comparing the means between two groups, one-sided Wilcox rank-sum test was employed to calculate the p values. Receiver operating characteristic (ROC) curve was used to evaluate the performances of biomarkers in predicting binary response. The composite marker in the IMVigor210 data analysis represents a linear combination of normalized IGR burden and TMB. Pearson’s correlation was used for analyzing biomarker associations. Benjamini-Hochberg method was used for false discovery rate adjustment in pathway enrichment analysis.
Data availability
The genomic data and clinical data used in this study can be retrieved through the links provided in Supplementary Table S1. The IGR burden quantification tool and the scripts used in this study are available through Github (https://github.com/wangxlab/intragenic-rearrangement-burden). The dataset, gene list and IGR burden used in this study are provided in Supplementary Tables S1, S2, and S3, respectively. All other data generated in this study are available within the article and its Supplementary Data Files or upon request from the corresponding author.
Results
High IGR burden defines a group of TMB-low cancers entities.
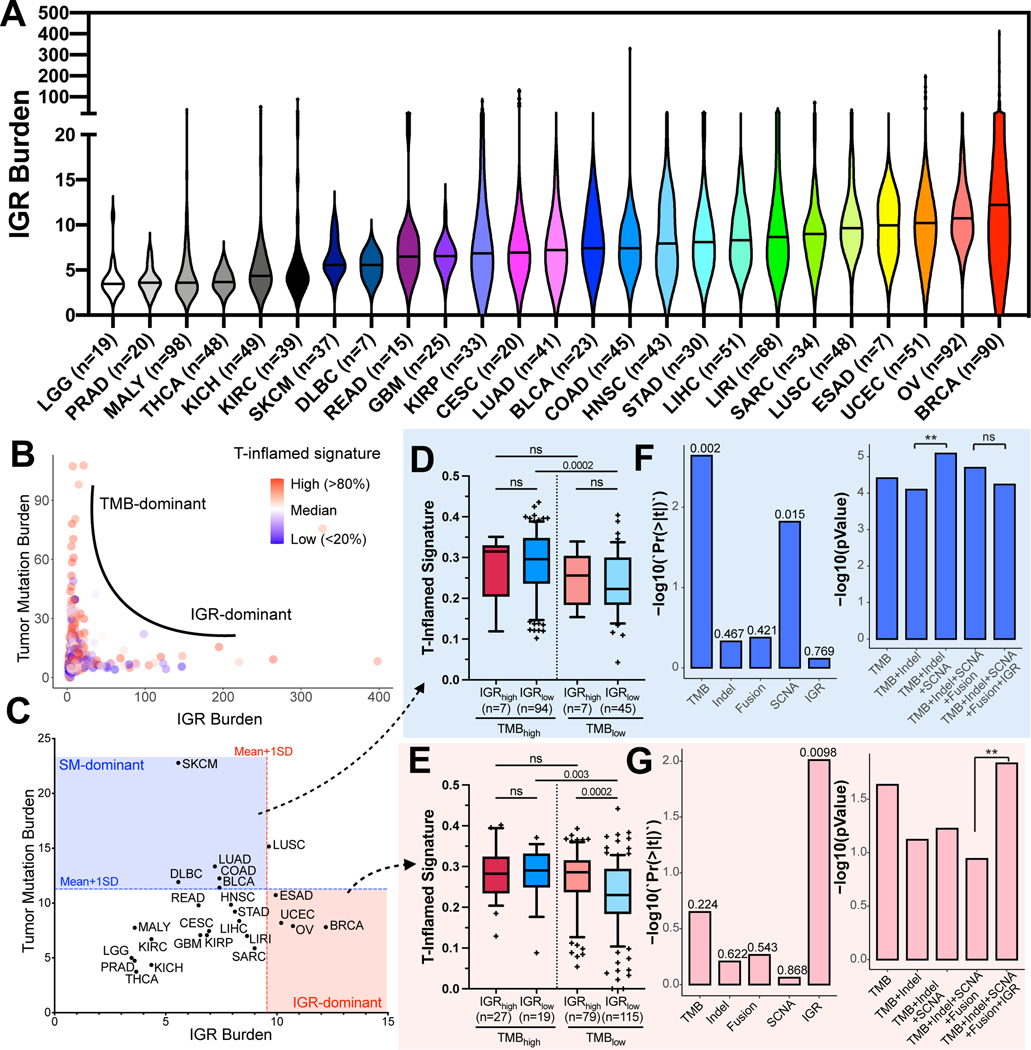
To quantitate IGR burden, we calculated the total number of IGRs from the structural variant calls of the pan-cancer WGS dataset provided by ICGC (n = 1,033). An assessment of the distributions of IGR burden across distinct cancer types revealed that Breast Cancer (BRCA) and Ovarian Cancer (OV) showed the highest median IGR burden, while Low Grade Glioma (LGG) exhibited the lowest median IGR burden (Fig. 1A). Next, we examined the associations between TMB and IGR burdens in the pan-cancer dataset (Fig. 1B). We found that tumors showed either a high TMB or a high IGR burden (R=0.09), suggesting the existence of two distinct classes of cancers driven by either point mutations or IGRs. We then assessed the correlations of TMB and IGR with other classes of genetic aberrations (Supplementary Figs S2 and S3). We observed a moderate correlation between TMB and Indels (R=0.59, Supplementary Fig. S2A), but independence between high IGR and Indels (R=0.05, Supplementary Fig. S2B). In addition, IGR-high tumors tended to have a modest level of SCNA, whereas SCNA-high tumors showed low IGR burden (R=0.22, Supplementary Fig. S3B). Furthermore, gene fusions resulting from IGRs showed a strong correlation with SCNA (R=0.85), but a weak correlation with IGR burden (R=0.30) (Supplementary Figs S3C and S3D).
Figure 1. The distribution of IGR burden and its association with T-inflamed signature in pan-cancer analysis.
(A) The landscape of IGR burden across all ICGC cancer types are shown in violin plots based on ascending order of the median of IGR burden in each cancer type. The median for each type is indicated as the solid line in the violin plot. (B) Scatter plot of IGR burden (x-axis) and tumor mutation burden (y-axis) for all ICGC samples (n=1,033). Tumors were colored according to their T-inflamed signatures. (C) Median of IGR burden and TMB in each cancer type. Cancer types with their median IGR burden or TMB levels above the cutoffs shown as dotted lines (mean + one standard deviation of all cancer types) on the X or Y axis are considered IGR-dominant (red area) or simple mutation-dominant (SM-dominant, blue area). (D, E) T-inflamed signatures of four subgroups stratified based on IGR and TMB levels in SM-dominant cancers (D) and IGR-dominant cancers (E) The median and whisker interval of 10%−90% of each group are shown in the boxplot. P-values were calculated using one-sided Wilcox rank-sum test. (F-G) Contributions of different classes of genetic markers to T-inflamed signature in SM-dominant cancers (F) and IGR-dominant cancers (G). Left panels of F-G, the p-value for each genetic marker in the multivariate model containing all genetic markers, when the confounding effects from other genetic variables are removed. Right panels of F-G, comparing the composite models containing different genetic markers: Y-axis refers to the transformed p-value of the F-test of each composite model and different multivariable models are compared using ANOVA. **p<0.01; ‘ns’ denotes p>=0.05.
We then examined the distribution of TMB and IGR burden in different tumor types based on their median levels (Fig. 1C). Using the cutoff of one standard deviation above the median, selected cancer entities can be grouped into IGR-dominant cancers, such as Breast Cancer (BRCA), Ovarian Cancer (OV), Uterine Corpus Endometrial Carcinoma (UCEC), and Esophageal Adenocarcinoma (ESAD) or simple mutation (SM)-dominant cancers, such as Skin Cutaneous Melanoma (SKCM), Lung Adenocarcinoma (LUAD), Colon Adenocarcinoma (COAD), and Bladder Cancer (BLCA). The distribution of Indels versus TMB or IGR in different cancer types is shown in Supplementary Figs S2C and S2D.
Genetic aberrations in cancer cells can result in altered protein sequences that can be recognized as foreign by T cells, leading to T-cell inflammation (35). To determine the associations of TMB and IGR burden with T-inflamed signature, we compared the T-inflamed signatures of SM-dominant and IGR-dominant cancers stratified into four groups according to their IGR and TMB levels (Fig. 1D, E). In SM-dominant cancer types, the vast majority of tumors had low IGR burden, among which the T-inflamed signatures of tumors with high TMB were significantly higher than those with low TMB (p<0.001, Fig. 1D). Conversely, in IGR-dominant cancer types, the vast majority of tumors had low TMB, among which tumors with high IGR burden showed significantly elevated T-inflamed signature compared to those with low IGR burden (p<0.001, Fig. 1E). Next, we calculated the associations of different types of genetic alterations with T-cell inflammation using a multivariate linear regression that includes TMB, Indels, SCNA, fusions, and IGR. This analysis revealed that TMB was the most significant covariate to T-cell inflammation in SM-dominant cancers (p<0.01), while IGR burden was the most influential covariate on T-cell inflammation in IGR-dominant cancers (p<0.01) (Fig. 1F–G, left panels). Analysis of variance (ANOVA) demonstrated that when adding IGR on top of other biomarkers, the explanation of the variance of T-cell inflammation significantly improved in IGR-dominant cancers (p<0.01) but not in SM-dominant cancers (Fig. 1F–G, right panels). To test removing the effect of tumor types, we regressed out tumor types and observed that TMB or IGR remained the most significant covariates of T-cell inflammation in SM-dominant or IGR-dominant cancers, respectively (Supplementary Fig. S4). These data suggest IGR burden as the most influential covariate of T-cell inflammation in IGR-dominant cancers.
IGR burden correlates with TILs in TNBC.
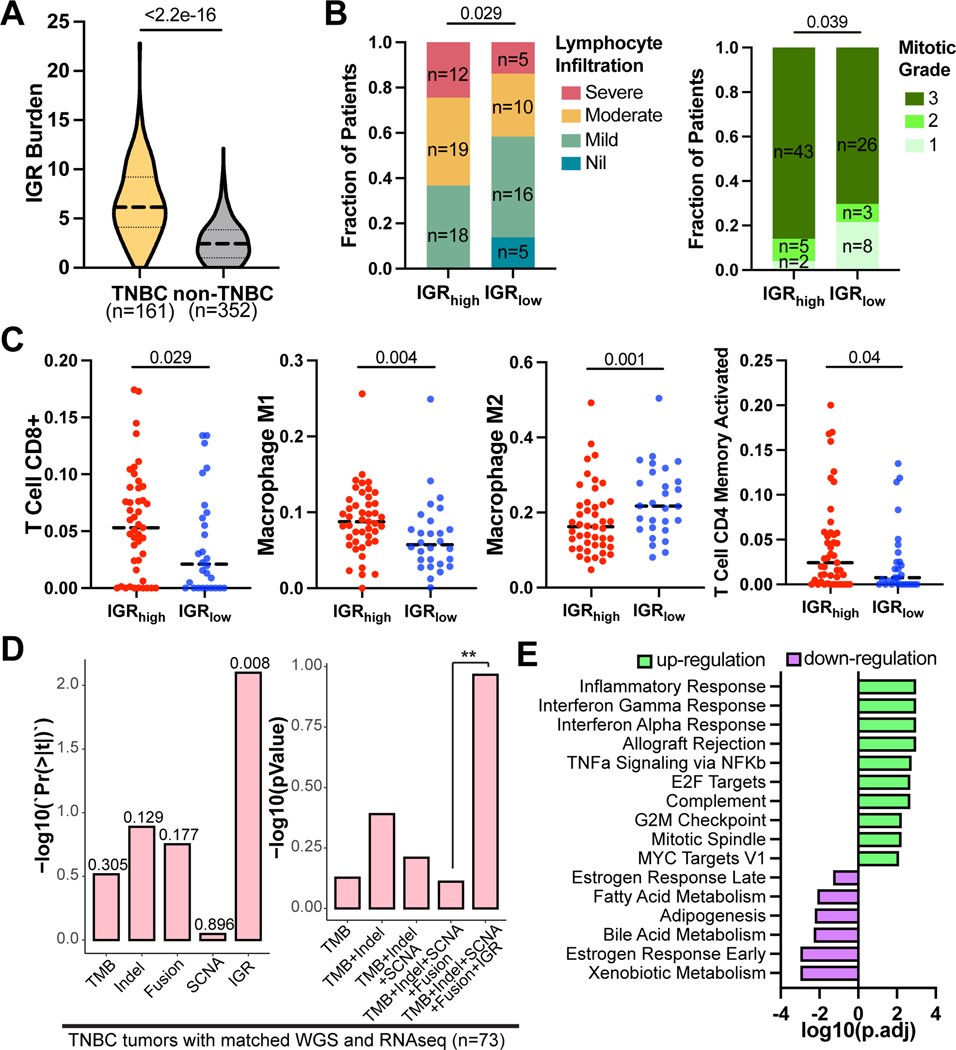
To verify our findings that IGR was associated with T-cell inflammation in IGR-dominate tumors we analyzed data from an independent breast cancer dataset (n=513, Nik-Zainal et al. (27)). We found that TNBC tumors showed a much higher overall IGR burden than non-TNBC breast cancers (Fig. 2A). Among TNBC subtypes, basal-like 1 (BL1), immunomodulatory (IM), and mesenchymal (M) subtypes showed higher IGR burden than basal-like 2 (BL2) and luminal androgen receptor (LAR) subtypes (Supplementary Fig. S5A). TNBC tumors with high IGR burden showed significantly higher lymphocyte infiltration levels (p<0.05) and mitotic grades (p<0.05), based on histopathological evaluations from the original study (27) (Fig. 2B). We then examined immune cell compositions in the tumor microenvironment deconvoluted from RNAseq data. Compared to tumors with low IGR burden, tumors with high IGR burden showed higher levels of CD8+ T cells, M1 macrophages, and effector-memory CD4+ T cells, but a lower level of M2 macrophages, indicating a type-1 antitumor immune response (Fig. 2C). In addition, IGRhigh tumors also showed a higher T-inflamed signature, histopathological mitotic counts, and homologous recombination deficiency (HRD) scores compared to IGRlow tumors (Supplementary Fig. S5B). These data suggest that increased mitosis and HRD may contribute to the increased IGR burden in TNBC tumors. Although IGR burden positively correlated with HRD score (R=0.58), HRD score showed a modest correlation with T-inflamed signature (R=0.11, Supplementary Fig. S5C). On the other hand, SCNA showed a positive correlation with HRD (R=0.263), but not with T-inflamed signature (R=0.037, Supplementary Fig. S5D). Multivariable linear regression including TMB, Indels, SCNA, Fusion, and IGR against T-inflamed signature in TNBC showed that IGR burden was the only significant association (p = 0.008), when the confounding effects from other genetic variables were removed (Fig. 2D, left panel). On the other hand, TMB and SCNA showed minimal association with the T-inflamed signature, which is consistent with previous reports (15,16). Next, we tested if adding HRD into the multivariate model would diminish the predictive effect of IGR burden. Our result showed that IGR burden was still the only significant predictive variable in the model (p<0.05, Supplementary Fig. S5E). This suggests that while HRD may associate with both increased SCNA and IGR burden, IGR burden appears to be the most important covariate of T-inflammation in TNBC. Furthermore, the inclusion of the IGR burden in the composite model containing TMB, Indels, SCNA, and fusion significantly improved fitting the model to the T-inflamed signature (Fig. 2D, right panel).
Figure 2. IGR burden correlates with T-inflamed signature in TNBC.
(A) Violin plot showing the distribution of IGR burden in TNBC and non-TNBC breast tumors. The dashed line refers to the median and dotted line indicates first and third quantile. (B) Barplots illustrating fractions of TNBC patients with different TIL levels (left) and mitotic grades (right). The p-values are calculated using Chi-squared test. (C) Jitter plots, with the medians shown in dotted horizontal lines, demonstrate the distributions of immune cell fractions deconvoluted using CIBERSORT in IGRhigh (n=45) and IGRlow (n=28) TNBC tumors. P-values were calculated using one-sided Wilcox rank-sum test. (D) Correlations of different types of genetic markers with T-inflamed signature in the TNBC tumors that have matched WGS and RNAseq data (n=73). Left panel, the p-values for each genetic marker in the multivariate model containing all genetic markers. Right panel, comparing the composite models containing different genetic markers. **p<0.01. (E) Pathway enrichment results from GSEA. The pathways are sorted by the direction and logarithm of adjusted p-values from GSEA.
Next, we selected important genes for tumor immune responses and examined their differential expression in IGR high and low TNBC tumors (Supplementary Fig. S5F). The resulting heatmap was stratified by IGR levels and TIL groups. Overall, IGRhigh TNBC tumors showed higher expression of these immunologically relevant genes compared to IGRlow TNBC tumors. Finally, GSEA comparing IGR high and low tumors showed upregulation of pro-inflammatory pathways, such as inflammatory response, IFNγ and IFNα responses, and TNFα signaling via NFκB pathways (Fig. 2E, Supplementary Fig. S5G).
IGR burden correlates with immune infiltration and durvalumab response in esophageal adenocarcinoma.
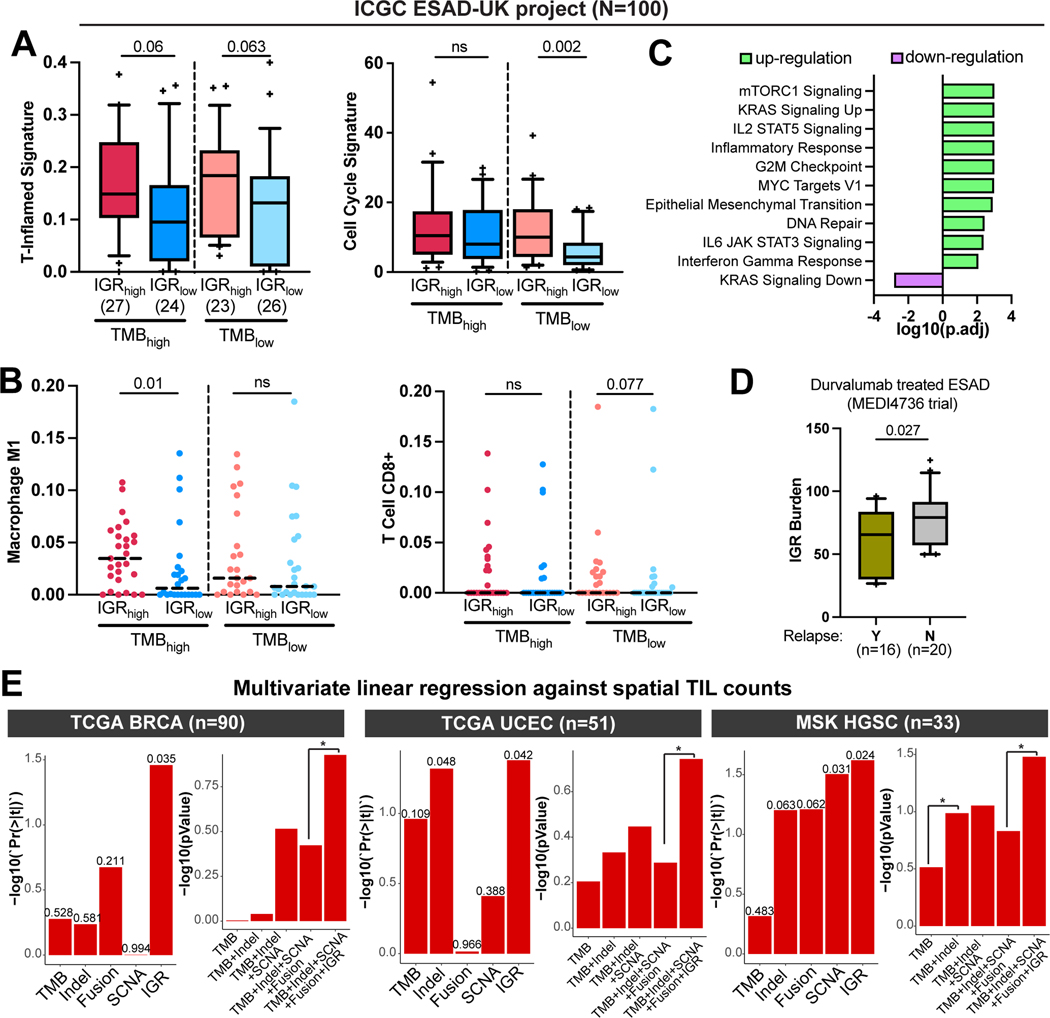
To confirm these results in another IGR-dominant tumor type, we examined the association of IGR burden with the immune microenvironment in an ESAD dataset from the ICGC ESAD-UK project (n=100) (29). As in the ICGC analysis, we categorized patients’ tumors into four groups based on their TMB and IGR burden values. A trend of increased T-inflamed signature was observed in IGRhigh tumors compared to IGRlow tumors irrespective of TMB levels (Fig. 3A). In addition, IGRhigh tumors had a higher cell cycle signature and higher M1 Macrophage and CD8+ T-Cell signatures in selected TMB groups (Fig. 3A–B). Furthermore, the markers of TILs and key immune checkpoints were significantly upregulated in tumors with high IGR burdens in both TMB levels, and this trend was more evident in the TMBlow group (Supplementary Fig. S6). Moreover, GSEA revealed upregulation of proliferation pathways such as mTORC1 and KRAS signaling, MYC targets, G2M checkpoint, and immune response pathways, such as IL2 signaling, inflammatory response, and IFNγ response in IGRhigh tumors (Fig. 3C). Together, these results suggest that increased IGR burden is associated with type 1 immune response in ESAD.
Figure 3. IGR burden correlates with T-inflamed signature in esophageal adenocarcinoma and with spatial TIL abundance in breast, endometrial, and ovarian cancers.
(A) Boxplots showing the distribution of T-inflamed signature and cell cycle signature in IGRhigh and IGRlow groups. The median and whisker interval of 10%−90% of each group are shown. (B) Dot-plots with the medians shown in dotted horizontal lines, demonstrating the distributions of CD8+ T Cell and Macrophage M1 deconvoluted using CIBERSORT in IGRhigh and IGRlow tumors. P-values were calculated using one-sided Wilcox rank-sum test. (C) Pathway enrichment results from GSEA comparing IGRhigh vs IGRlow tumors. The pathways are sorted by the direction and logarithm of adjusted p-values from GSEA. (D) Boxplot with 10%−90% interval comparing IGR burdens in the ESAD patients with or without cancer relapse in the MEDI4736 trial testing durvalumab. P-value of one-sided Wilcox sum-rank test is shown. (E) The correlations of genetic markers with spatial TIL counts in TCGA breast cancer (BRCA) (left), TCGA uterine corpus endometrial carcinoma (UCEC) (middle), or high-grade serous carcinomas (HGSC) of the MSK dataset (right). Tumors with matched WGS and spatial TIL count data are shown in the figure. Left panels, the p-value for each genetic marker in the multivariate model containing all genetic markers. Right panels, comparisons of the composite models containing different genetic markers. *p<0.05.
To investigate if IGR burden correlated with ICB response in ESAD, we examined data from the phase II MEDI4736 trial in which durvalumab, a PD-L1 inhibitor, was evaluated in ESAD patients who received prior chemoradiation therapy (30). Our results indicate that IGR burden was significantly lower in patients who relapsed, which suggests that IGR burden could predict ICB benefits in ESAD (Fig. 3D).
IGR burden is an important correlate of the abundance of TILs in IGR-dominant cancer types.
To examine the correlation of IGR burden with abundance of TILs, we gathered spatially determined TIL count data for 13 TCGA tumor types, which were quantitated using convolutional neural network based on histological images (28). Within the TCGA tumors for which both WGS data and spatial TIL counts were available, there were 90 breast cancers and 51 endometrial carcinomas, but few or no esophageal and ovarian tumors. Multivariate linear regression using all genetic markers against spatial TIL counts revealed that IGR burden was one of the most influential predictors of TIL abundance among all genetic markers in breast and endometrial cancers. The inclusion of IGR burden in the composite model containing TMB, Indels, SCNA, and fusions significantly increased the predictive value of the model (Fig. 3E, left and middle panels).
Next, we sought to examine the association of IGR burden with spatial TIL counts in ovarian cancer based on a WGS dataset for high-grade serous carcinoma (HGSC) matched with spatially determined TIL counts from the MSK-IMPACT cohort (n = 33) (28). Similar to the above results, IGR burden was the most significant predictor of TIL abundance among all genetic markers and the inclusion of IGR burden to the composite model containing TMB, Indels, SCNA, and fusions significantly increased the predictive value of the model (Fig. 3E, right panel).
Association of IGR burden with ICB benefit in platinum-exposed metastatic urothelial carcinoma.
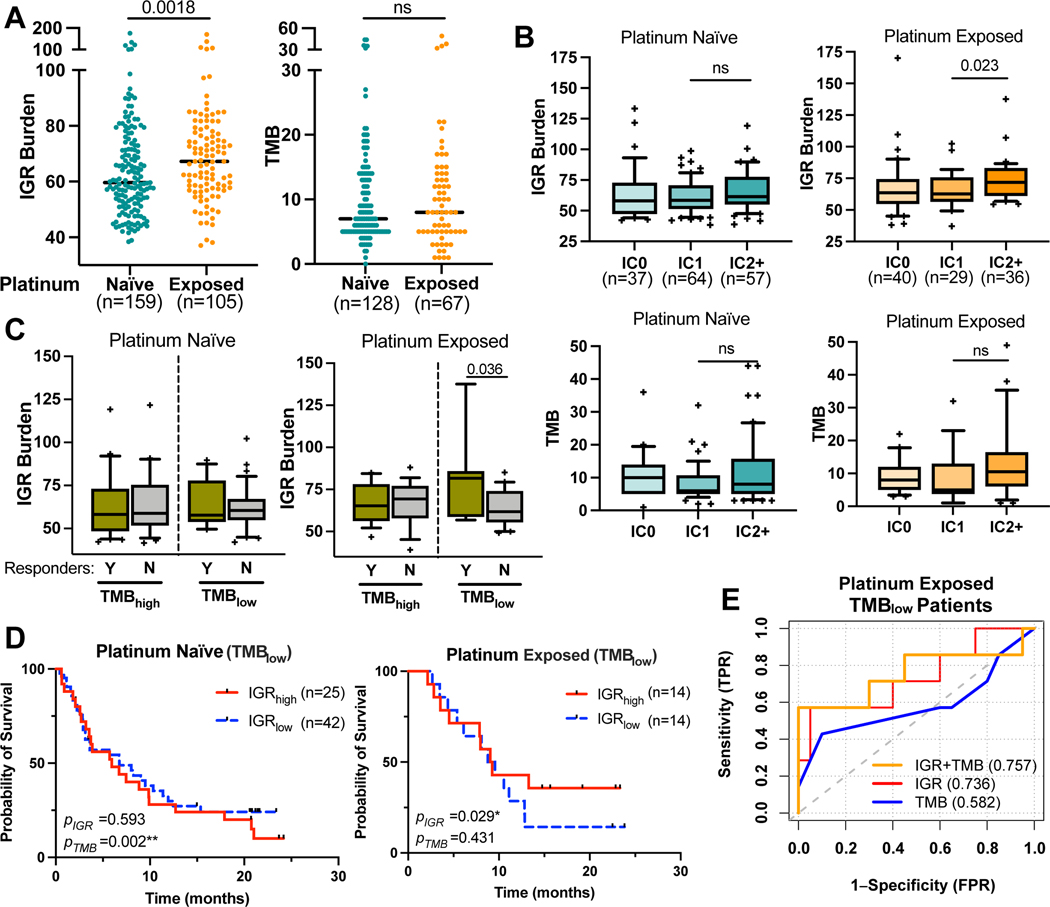
To further examine whether IGR burden is associated with patient response to ICB treatment, we accessed a large clinical trial dataset in which patients with metastatic urothelial carcinoma received atezolizumab, mostly following platinum induction (31). While urothelial carcinoma is classified as a TMBhigh cancer (Fig. 1C), previous research has demonstrated that platinum induction leads to extensive DNA damage and activation of the DNA damage repair, which may induce IGRs. We thus compared the distributions of IGR burden and TMB in tumor samples collected before or after platinum treatment (platinum-naïve or exposed). IGR burden after platinum exposure was indeed higher than before treatment, whereas no difference in TMB was observed before and after treatment (Fig. 4A). We then correlated IGR burden with PD-L1 expression as measured by immunohistochemistry (IHC) in the original study (31). IGR burden was significantly increased in the PD-L1 IC2+ group in platinum-exposed tumors but not in platinum-naïve tumors (Fig. 4B).
Figure 4. IGR burden predicts ICB responses in metastatic urothelial carcinoma patients with low TMB, who received prior platinum therapy.
(A) Dot-plots, with the medians shown in dotted horizontal lines, showing the distribution of IGR burden and TMB in tumor samples collected before platinum treatment (platinum-naïve) or after platinum treatment (platinum-exposed). (B) Boxplots with 10%−90% interval showing IGR burden and TMB in platinum naïve and exposed tumors with different PD-L1 immunohistochemistry levels of immune cells (IC levels). The IC classes were provided by the IMVigor210 publication(31).: IC0 (<1%), IC1 (≥1% and <5%) and IC2+ (≥5%). (C) Pairwise boxplots comparing the IGR burden in responders (n=16 and 7 for TMBhigh and TMBlow, respectively) and non-responders (n=19 and 20 for TMBhigh and TMBlow, respectively), stratified by TMB levels. (D) Kaplan-Meier curves of TMBlow patients who received prior platinum stratified by median IGR levels of all patients in the IMVigor210 dataset. Tumor samples were either platinum-naïve (left) or platinum-exposed (right). P-values of each biomarker based on multivariate Cox-proportional hazard regression are shown in the bottom left for each panel. (E) ROC curves of TMB, IGR and composite biomarker scores in platinum-exposed tumors for determining patient response to immune checkpoint inhibition. **p<0.01; *p<0.05; ‘ns’ denotes p>=0.05.
We next categorized the patients with progressive disease under ICB treatment as non-responders, and the rest as responders. For platinum-exposed tumors, responders showed a significantly higher IGR burden compared to non-responders in TMBlow tumors (Fig. 4C). Survival analysis suggested that TMB was predictive of overall survival for platinum-naïve tumors but not for platinum-exposed tumors (Supplementary Fig. S7), whereas IGR was predictive in platinum-exposed TMBlow tumors (Fig. 4D). No correlation was observed between IGR burden and TMB in either the platinum-naïve or exposed tumors (R=0.057 and −0.109, respectively). Receiver operating characteristic (ROC) curves revealed that IGR burden has a higher predictive value (AUROC = 0.736) than TMB (AUROC = 0.582) in the platinum-exposed, TMBlow patients. Linear combination of IGR burden with TMB resulted in a small increase of predictive value compared to IGR burden alone (AUROC = 0.757) (Fig. 4E). Furthermore, we also analyzed a clinical dataset for patients with advanced melanoma treated with nivolumab (n = 49) (36) and our result showed that IGR burden was not predictive of ICB benefit in melanoma, which that had the highest TMB level among all cancer entities (Supplementary Fig. S8, Fig. 1C). These data suggest that IGR burden is a potential new biomarker for ICB response in certain cancer types when other genetic biomarkers such as TMB are not applicable.
Discussion
In this study, we have identified a biomarker of ICB response called IGR burden, designed to estimate a category of poorly studied cryptic rearrangements that alter the exon structure within a gene. To our knowledge, the association of IGRs with immunotherapy response has not been reported in cancer. In fact, large WGS studies cataloging genomic rearrangements in cancer have not examined the importance of IGRs rigorously (27,37–39). The key difference between IGR and SCNA is that IGR detects cryptic intragenic rearrangements that affect exons, which are both balanced and unbalanced. In contrast, SCNA only detects unbalanced rearrangements. In addition, the limited coverage of the genomic array is deemed insufficient to comprehensively detect IGRs. Thus, SCNA burden based on genomic arrays mostly reflects larger-sized unbalanced rearrangements.
Based on our analyses of pan-cancer whole genomes, we identified two groups of cancer types that are dominated by either simple mutations or IGRs. We demonstrated that IGR burden was a key predictor of T-inflamed signature in IGR-dominant cancers, whereas TMB was a pivotal correlate of the T-inflamed signature in SM-dominant cancers. Further analyses of breast cancer (Nik-Zainal et al (27)) and esophageal adenocarcinoma (ESAD-UK (29)) datasets revealed that a high IGR burden correlated with increased T-inflamed signature and a type-I immune cellular contexture, including increased CD8+ T cells and M1 macrophage. Moreover, through analyses of spatial TIL count data based on histopathological images, we showed that IGR burden was a pivotal correlate of TIL abundance in breast, endometrial, and ovarian cancers.
Finally, we tested the association of IGR burden with ICB response in two clinical trial datasets. We found that IGR burden correlated with patient benefits in the phase II MEDI4736 trial for durvalumab in esophageal adenocarcinoma. IGR also predicted ICB response and overall survival in TMBlow, platinum-exposed bladder cancer patients, which might be attributed to the increased IGR burden from platinum-induced de novo rearrangements. Consistent with this hypothesis, data from some clinical trials suggest that ICB following platinum improved patient survival (40). Future experimental studies will be required to elucidate the DNA repair defects leading to increased IGR burden.
Taken together, our data showed that IGR burden is a pivotal correlate of T-cell inflammation in IGR-dominant cancers. The correlation of IGR burden with ICB response provided substantial evidence that IGRs might be immunogenic. It is feasible that de novo exon junctions generated by IGRs give rise to neoantigen epitopes, which could be recognized by the host immune system. It is notable that current HLA affinity–based neoantigen prediction models based on simple mutations have a low accuracy. A recent consortium study suggested that only 6% of predicted peptides were found to be recognized by patient-matched T cells (41). In addition, another study suggested that the inclusion of HLA neoepitope prediction in the TMB model minimally improved the estimates of neoantigen burden (42). Thus, we did not include HLA subtyping and HLA neoepitope prediction in our IGR burden model. Future experimental studies will be required to identify the TILs that react to the neoantigen peptides generated by IGRs in order to establish the immunogenicity of IGRs.
Supplementary Material
SYNOPSIS.
IGRs are an ill-studied class of cryptic genomic rearrangements. The authors suggest that IGR burden is a pivotal contributor to immune infiltration and a new genetic biomarker for immunotherapy response in TMB-low, IGR-dominant tumors, and in platinum-exposed tumors.
Funding Information:
This study was supported by NIH grant 1R01CA181368 (X-S.W.), and 1R01CA183976 (X-S.W.), 1R21CA237964 (X-S.W.), CDMRP BCRP W81XWH-22-1-1036 (X-S.W.) and W81XWH-22-1-1037 (B-F.L.), PA breast cancer coalition (X-S.W.), the Shear Family Foundation, and the Hillman Foundation. This research was supported in part by the University of Pittsburgh Center for Research Computing, RRID:SCR_022735, through the resources provided. Specifically, this work used the HTC cluster, which is supported by NIH award number S10OD028483. This work also used the Extreme Science and Engineering Discovery Environment (XSEDE)(43), which is supported by National Science Foundation grant number ACI-1548562. Specifically, it used the Bridges-2 system, which is supported by NSF award number ACI-1928147, at the Pittsburgh Supercomputing Center (PSC).
Footnotes
Competing Interests Statement: The authors declare no competing interests.
References
- 1.Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 2019;19(3):133–50 doi 10.1038/s41568-019-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishino M, Ramaiya NH, Hatabu H, Hodi FS. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol 2017;14(11):655–68 doi 10.1038/nrclinonc.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haslam A, Prasad V. Estimation of the Percentage of US Patients With Cancer Who Are Eligible for and Respond to Checkpoint Inhibitor Immunotherapy Drugs. JAMA Netw Open 2019;2(5):e192535 doi 10.1001/jamanetworkopen.2019.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martins F, Sofiya L, Sykiotis GP, Lamine F, Maillard M, Fraga M, et al. Adverse effects of immune-checkpoint inhibitors: epidemiology, management and surveillance. Nat Rev Clin Oncol 2019;16(9):563–80 doi 10.1038/s41571-019-0218-0. [DOI] [PubMed] [Google Scholar]
- 5.Reid PD, Cifu AS, Bass AR. Management of Immunotherapy-Related Toxicities in Patients Treated With Immune Checkpoint Inhibitor Therapy. JAMA 2021;325(5):482–3 doi 10.1001/jama.2020.17308. [DOI] [PubMed] [Google Scholar]
- 6.Adashek JJ, Subbiah IM, Matos I, Garralda E, Menta AK, Ganeshan DM, et al. Hyperprogression and Immunotherapy: Fact, Fiction, or Alternative Fact? Trends Cancer 2020;6(3):181–91 doi 10.1016/j.trecan.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348(6230):124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017;355(6322) doi 10.1126/science.aaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang W, Lee KW, Srivastava RM, Kuo F, Krishna C, Chowell D, et al. Immunogenic neoantigens derived from gene fusions stimulate T cell responses. Nat Med 2019;25(5):767–75 doi 10.1038/s41591-019-0434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turajlic S, Litchfield K, Xu H, Rosenthal R, McGranahan N, Reading JL, et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. The Lancet Oncology 2017;18(8):1009–21 doi 10.1016/s1470-2045(17)30516-8. [DOI] [PubMed] [Google Scholar]
- 11.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372(26):2509–20 doi 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Collet L, Rediti M, Debien V, De Caluwe A, Venet D, et al. Predictive Biomarkers for Response to Immunotherapy in Triple Negative Breast Cancer: Promises and Challenges. J Clin Med 2023;12(3) doi 10.3390/jcm12030953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bareche Y, Buisseret L, Gruosso T, Girard E, Venet D, Dupont F, et al. Unraveling Triple-Negative Breast Cancer Tumor Microenvironment Heterogeneity: Towards an Optimized Treatment Approach. J Natl Cancer Inst 2020;112(7):708–19 doi 10.1093/jnci/djz208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammerl D, Martens JWM, Timmermans M, Smid M, Trapman-Jansen AM, Foekens R, et al. Spatial immunophenotypes predict response to anti-PD1 treatment and capture distinct paths of T cell evasion in triple negative breast cancer. Nat Commun 2021;12(1):5668 doi 10.1038/s41467-021-25962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karn T, Jiang T, Hatzis C, Sanger N, El-Balat A, Rody A, et al. Association Between Genomic Metrics and Immune Infiltration in Triple-Negative Breast Cancer. JAMA Oncol 2017;3(12):1707–11 doi 10.1001/jamaoncol.2017.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zerdes I, Simonetti M, Matikas A, Harbers L, Acs B, Boyaci C, et al. Interplay between copy number alterations and immune profiles in the early breast cancer Scandinavian Breast Group 2004-1 randomized phase II trial: results from a feasibility study. NPJ Breast Cancer 2021;7(1):144 doi 10.1038/s41523-021-00352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas R, Al-Khadairi G, Decock J. Immune Checkpoint Inhibitors in Triple Negative Breast Cancer Treatment: Promising Future Prospects. Front Oncol 2020;10:600573 doi 10.3389/fonc.2020.600573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landen CN, Molinero L, Hamidi H, Sehouli J, Miller A, Moore KN, et al. Influence of Genomic Landscape on Cancer Immunotherapy for Newly Diagnosed Ovarian Cancer: Biomarker Analyses from the IMagyn050 Randomized Clinical Trial. Clin Cancer Res 2023. doi 10.1158/1078-0432.CCR-22-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vivaldi C, Catanese S, Massa V, Pecora I, Salani F, Santi S, et al. Immune Checkpoint Inhibitors in Esophageal Cancers: are we Finally Finding the Right Path in the Mist? Int J Mol Sci 2020;21(5) doi 10.3390/ijms21051658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGrail DJ, Pilie PG, Rashid NU, Voorwerk L, Slagter M, Kok M, et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol 2021;32(5):661–72 doi 10.1016/j.annonc.2021.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lei M, Janjigian YY, Ajani JA, Moehler M, Wang X, Shen L, et al. Nivolumab (NIVO) plus chemotherapy (chemo) vs chemo as first-line (1L) treatment for advanced gastric cancer/gastroesophageal junction cancer/esophageal adenocarcinoma (GC/GEJC/EAC): CheckMate 649 biomarker analyses. Cancer Research 2022;82:CT023 doi 10.1158/1538-7445.AM2022-CT023. [DOI] [Google Scholar]
- 22.Wong AJ, Ruppert JM, Bigner SH, Grzeschik CH, Humphrey PA, Bigner DS, et al. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci U S A 1992;89(7):2965–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenstermaker RA, Ciesielski MJ. Deletion and tandem duplication of exons 2 - 7 in the epidermal growth factor receptor gene of a human malignant glioma. Oncogene 2000;19(39):4542–8 doi 10.1038/sj.onc.1203802. [DOI] [PubMed] [Google Scholar]
- 24.Gallant JN, Sheehan JH, Shaver TM, Bailey M, Lipson D, Chandramohan R, et al. EGFR Kinase Domain Duplication (EGFR-KDD) Is a Novel Oncogenic Driver in Lung Cancer That Is Clinically Responsive to Afatinib. Cancer Discov 2015;5(11):1155–63 doi 10.1158/2159-8290.CD-15-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castiglioni F, Tagliabue E, Campiglio M, Pupa SM, Balsari A, Menard S. Role of exon-16-deleted HER2 in breast carcinomas. Endocr Relat Cancer 2006;13(1):221–32 doi 10.1677/erc.1.01047. [DOI] [PubMed] [Google Scholar]
- 26.Goldman MJ, Zhang J, Fonseca NA, Cortes-Ciriano I, Xiang Q, Craft B, et al. A user guide for the online exploration and visualization of PCAWG data. Nat Commun 2020;11(1):3400 doi 10.1038/s41467-020-16785-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016;534(7605):47–54 doi 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, et al. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep 2018;23(1):181–93 e7 doi 10.1016/j.celrep.2018.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mourikis TP, Benedetti L, Foxall E, Temelkovski D, Nulsen J, Perner J, et al. Patient-specific cancer genes contribute to recurrently perturbed pathways and establish therapeutic vulnerabilities in esophageal adenocarcinoma. Nat Commun 2019;10(1):3101 doi 10.1038/s41467-019-10898-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mamdani H, Schneider B, Perkins SM, Burney HN, Kasi PM, Abushahin LI, et al. A Phase II Trial of Adjuvant Durvalumab Following Trimodality Therapy for Locally Advanced Esophageal and Gastroesophageal Junction Adenocarcinoma: A Big Ten Cancer Research Consortium Study. Front Oncol 2021;11:736620 doi 10.3389/fonc.2021.736620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554(7693):544–8 doi 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21 doi 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung N, Zhang XD, Kreamer A, Locco L, Kuan P-F, Bartz S, et al. Median absolute deviation to improve hit selection for genome-scale RNAi screens. Journal of biomolecular screening 2008;13(2):149–58. [DOI] [PubMed] [Google Scholar]
- 34.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12(5):453–7 doi 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Efremova M, Finotello F, Rieder D, Trajanoski Z. Neoantigens Generated by Individual Mutations and Their Role in Cancer Immunity and Immunotherapy. Front Immunol 2017;8:1679 doi 10.3389/fimmu.2017.01679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 2017;171(4):934–49 e16 doi 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 2009;462(7276):1005–10 doi 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staaf J, Glodzik D, Bosch A, Vallon-Christersson J, Reutersward C, Hakkinen J, et al. Whole-genome sequencing of triple-negative breast cancers in a population-based clinical study. Nat Med 2019;25(10):1526–33 doi 10.1038/s41591-019-0582-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Angus L, Smid M, Wilting SM, van Riet J, Van Hoeck A, Nguyen L, et al. The genomic landscape of metastatic breast cancer highlights changes in mutation and signature frequencies. Nat Genet 2019;51(10):1450–8 doi 10.1038/s41588-019-0507-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tripathi A, MacDougall K, Sonpavde GP. Therapeutic Landscape Beyond Immunotherapy in Advanced Urothelial Carcinoma: Moving Past the Checkpoint. Drugs 2022;82(17):1649–62 doi 10.1007/s40265-022-01802-3. [DOI] [PubMed] [Google Scholar]
- 41.Wells DK, van Buuren MM, Dang KK, Hubbard-Lucey VM, Sheehan KCF, Campbell KM, et al. Key Parameters of Tumor Epitope Immunogenicity Revealed Through a Consortium Approach Improve Neoantigen Prediction. Cell 2020;183(3):818–34 e13 doi 10.1016/j.cell.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wood MA, Weeder BR, David JK, Nellore A, Thompson RF. Burden of tumor mutations, neoepitopes, and other variants are weak predictors of cancer immunotherapy response and overall survival. Genome Med 2020;12(1):33 doi 10.1186/s13073-020-00729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genomic data and clinical data used in this study can be retrieved through the links provided in Supplementary Table S1. The IGR burden quantification tool and the scripts used in this study are available through Github (https://github.com/wangxlab/intragenic-rearrangement-burden). The dataset, gene list and IGR burden used in this study are provided in Supplementary Tables S1, S2, and S3, respectively. All other data generated in this study are available within the article and its Supplementary Data Files or upon request from the corresponding author.