Abstract
Rhamnolipids are surfactants produced by many Pseudomonad bacteria, including the species Pseudomonas aeruginosa. These rhamnolipids are known to aid and enable numerous phenotypic traits that improve survival of the bacteria that make them. These surfactants are also important for industrial products ranging from pharmaceuticals to cleaning supplies to cosmetics, to name a few. Rhamnolipids have structural diversity that leads to an array of congeners; however, little is known about the localization and/or distribution of these congeners in two-dimensional space. Differential distribution of congeners can reduce the uniformity of application in industrial settings and create heterogeneity within biological communities. We examined the distribution patterns of combinations of rhamnolipids in commercially available mixtures, cell-free spent media, and in colony biofilms using mass spectrometry. We found that even in the absence of cells, congeners exhibit different distribution patterns, leading to different rhamnolipid congener distributions on a surface. Congeners with shorter fatty acid chains were more centrally located while longer chains were more heterogeneous and distally located. We found that congeners with similar structures can distribute differently. Within developing colony biofilms we found rhamnolipid distribution patterns differed from cell-free environments, lacking simple trends noted in cell-free environments. Most strikingly, we found the distribution patterns of individual congeners in colony biofilms to be diverse. We note that congener distribution is far from homogeneous but composed of numerous local microenvironments of varied rhamnolipid congener composition.
Keywords: Pseudomonas aeruginosa, mass spectrometry, MALDI, biogeography, biofilm
INTRODUCTION
Rhamnolipids are a class of glycolipid surfactants made by the bacterium Pseudomonas aeruginosa and other Pseudomonads 1. For P. aeruginosa, rhamnolipids impact surface colonization, dispersal, carbon acquisition, and virulence of this opportunistic pathogen that transitions from environmental sources to cause infections in susceptible individuals. The production of rhamnolipids is a population-dependent phenomenon where transcription of the genes rhlA and rhlB, required for rhamnolipid synthesis, is regulated by intercellular quorum sensing signaling 2.
Rhamnolipids are well known as surfactants that decrease surface tension and promote motility and spreading of P. aeruginosa and the Pseudomonads 3. Rhamnolipids can alter biofilm architecture by facilitating the formation of open channels 4. They have also been noted for an ability to attack host immune cells and carry antibiotic properties 5–7. Because of their amphipathic traits, rhamnolipids are important for improving the bioavailability of hydrophobic molecules, such as alkyl quinolones 8, 9, this heightens alkyl quinolone induced virulence. Pseudomonas quinolone signal (PQS), one alkyl quinolone produced by P. aeruginosa, upregulates the production of several virulence factors and is associated with localized cell death within biofilms 10–12.
Rhamnolipids are also industrially significant. The global surfactants market is growing and expected to reach a valuation of $28.8 billion in 2023 13. Because traditional surfactants can persist in the environment and exhibit ecotoxic effects 14, rhamnolipids offer a greener alternative to these synthetic surfactants due to their reduced environmental toxicity and increased biodegradability 15. These rhamnolipids are potentially important tools for the remediation of petroleum products in the environment 5 and their ability to bind rare earth elements could prove useful in scavenging these elements for technology manufacturing 16.
The basic structure of rhamnolipid is formed by the linkage of one or two rhamnose moieties with one or two β-hydroxy fatty acids (Fig S1). The RhlA enzyme produces various hydroxyalkanoyl-hydroxyalkanoates (HAA) from the β-hydroxy fatty acids. Then, RhlB couples the first rhamnose sugar to HAA to form mono-rhamnolipids. Many Pseudomonads, including P. aeruginosa, also make the RhlC enzyme, which can couple a second rhamnose to mono-rhamnolipid and yield di-rhamnolipid 17–21. The fatty acid chains used to make HAA can be saturated or unsaturated and are generally 8 to 14 carbons in length. Varying the number of rhamnose or fatty acid units as well as the fatty acid chain lengths and location and number of double bonds within the hydrophobic chains leads to an array of possible rhamnolipid congeners 22.
The majority of prior research on rhamnolipids has focused on: 1) appreciating and characterizing the bacterial behavior resulting from the presence or absence of rhamnolipid mixtures, or 2) detailing bacterial culture conditions that yield abundant rhamnolipid production. Some of this work has distinguished the effects of mono- or di-rhamnolipid classes. For example, both mono- and di- rhamnolipid mixes have been reported to have greater antimicrobial activity in different environments against different microbes 23, 24 and with respect to environmental remediation, a mono-rhamnolipid dominant congener mix emulsified more crude oil than a di-rhamnolipid dominant mix 25. While this classification is structurally sound, there lacks evidence that mono-and di- rhamnolipids capture the diversity of physicochemical characteristics exhibited by Pseudomonas rhamnolipids. Indeed, research on specific rhamnolipid congeners has been limited by the ability to purify, synthesize, and identify individual congeners.
Rhamnolipid congeners function differently. For example, rhamnolipids with longer lipid chains (i.e. C14-C14) commonly produced by Burkholderia glumae or Burkholderia thailandensis exhibit different physicochemical properties in comparison to predominant rhamnolipids produced by P. aeruginosa (i.e. C10-C10) such as increased critical micelle concentration (CMC) and the capacity to reduce surface tension 26. Orientation of rhamnose moieties and fatty acid chains of surface adsorbed rhamnolipids differs by congener and is predicted to impact surface tension 27. In addition to micelles, some congeners can self-assemble into vesicles and lamellae 28.
A range of CMCs have been reported for different rhamnolipid mixtures 29–31. This is likely, in part, a result of the individual CMCs of congeners that compose each mixture. Pacheco et al. report that congeners with small structural differences can have quite different CMCs and impacts on surface tension while other, more structurally divergent congeners, can behave similarly 32. Currently, there are no parameters that have been characterized to enable accurate prediction of the CMC for a given rhamnolipid structure. It is known that rhamnolipid congeners vary in molecular surface area, ability to adsorb to the liquid air interface, and type of aggregates formed 32. However, such detail has only been reported for a handful of congeners. Modelling suggests that rhamnolipids can form multiple aggregate structures that vary from micelle to lipid bilayer to long tube-like structures in a manner that is both congener specific and concentration dependent 33.
While the specific properties of rhamnolipid congeners produced by Pseudomonads continues to be explored, the distribution of congeners that can result from surface-growing P. aeruginosa has not been explored. Understanding rhamnolipid congener heterogeneity in distribution is of interest in both industrial and biological settings. Therefore, we sought to map congener localization in cell-free and colony biofilm environments to understand how congeners naturally distribute.
In this work, we have probed the distribution of different rhamnolipid congeners added as a mixture to a single point in space. We examined the differential spreading of rhamnolipid from three commercial mixtures and from P. aeruginosa cell-free spent medium (CFSM) to explore the distribution of multiple rhamnolipid congeners in two-dimensional space. We accomplished this using matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI)34, 35. Use of high-resolution Fourier-transform ion cyclotron resonance mass spectrometry (FT-ICR MS) allowed us to differentiate congeners within mono- and di- rhamnolipid categories and estimate total carbons within the β-hydroxy fatty acid chains of the rhamnolipids based on high-resolution accurate masses (±7 ppm). A library covering 58 rhamnolipid congeners was used to identify peaks corresponding to rhamnolipids 22. Using this approach, we were able to distinguish distinct spatial distribution patterns of rhamnolipid congeners in both abiotic and biotic experiments.
RESULTS AND DISCUSSION
Rhamnolipid congeners from P. aeruginosa cultures display distribution patterns that primarily vary with fatty acid chain length.
We characterized the distribution and separation of rhamnolipid congeners produced by P. aeruginosa. For all experiments, we elucidated patterns using nutrient agar-based assays that are routinely used as a test substrate to examine P. aeruginosa behavior 17, 18, 36–39. However, to simplify the experiment and discriminate rhamnolipid properties from biological effects, we first examined distribution of rhamnolipids without P. aeruginosa cells using cell-free spent medium (CFSM) harvested from 48-hour planktonic P. aeruginosa cultures. While spreading of the 25 μL droplet appeared to occur within minutes, plate assays were left undisturbed for 18 h prior to subsequent preparation for matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI). Most rhamnolipids were observed as [M+Na]+, [M+K]+, and [M+H]+ adducts with high mass accuracy. We subsequently used the most intense [M+Na]+ adducts for comparisons across this study. The detected congeners in CFSM have both mono- and di-rhamnolipid and fatty acid chains with total carbons ranging from 10-24 (Table S1). Our results suggest most congeners had 2 fatty acid chains in combinations of C8, C10, or C12. While it is not possible to differentiate specific structures of equal mass by MSI (e.g. Rhamnose-Rhamnose-C8-C12 vs. Rhamnose-Rhamnose-C10-C10), prior characterization of rhamnolipids using LC/MS on this same P. aeruginosa strain has shown that fatty acid chain length of C20 are expected to be C10-C10 and not C8-C12 or C12-C8 40. We also identified 3 unsaturated congeners, with one degree of unsaturation, associated with total fatty acid chain length of C22 and C24 (e.g. C12-C10 and C12-C12).
To visualize differences in congener distribution patterns we measured intensity profiles along 3 cross-sections of each sample (Fig. 1). All rhamnolipids dispersed >10 mm radially from their initial central location of deposition. Previous studies have shown that rhamnolipids can be separated as mono- and di- rhamnolipids using thin-layer chromatography (e.g. 41). Given their physiochemical differences, we expected that the mono- and di- rhamnolipid congeners would exhibit divergent distribution patterns. However, there was no apparent spreading pattern associated with mono- versus di-rhamnose congeners. Most mono- and di-rhamnolipid congeners of the same lipid chain length distributed equivalently (e.g. Rha-C20 and dRha-C20). We noted two general rhamnolipid distribution profiles: (1) congeners that were more concentrated centrally, near the original deposition site, or (2) more distally.
Figure 1.
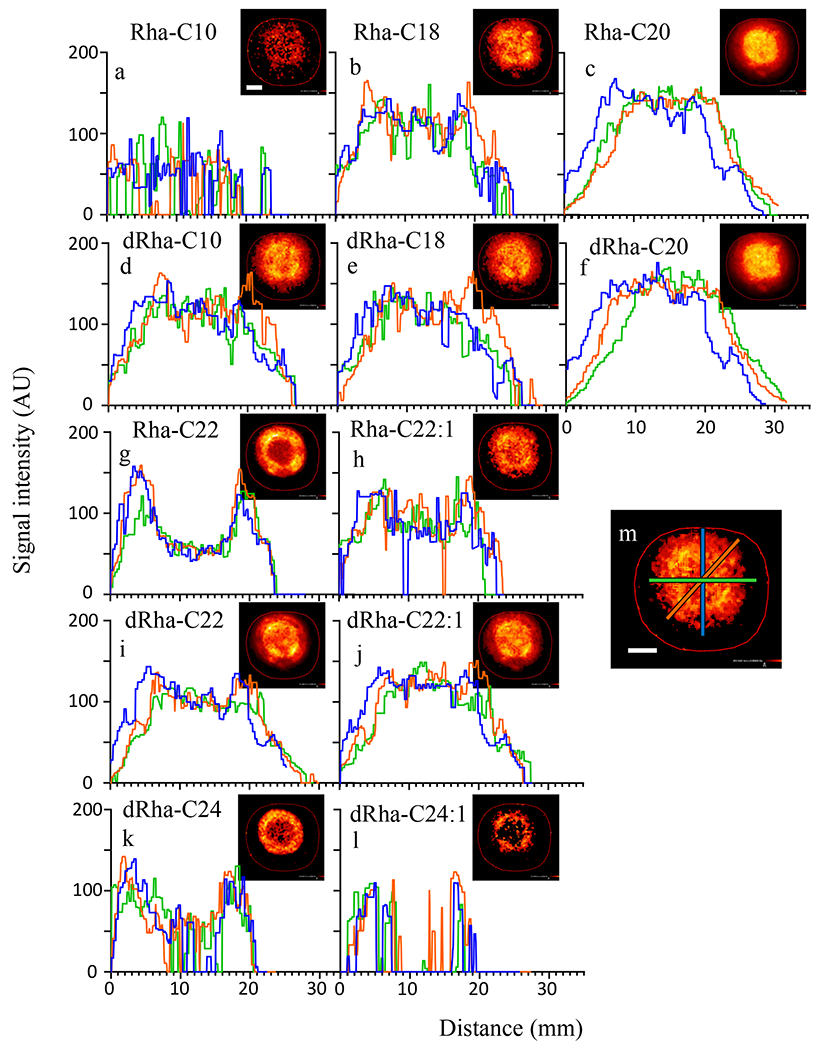
Radial distribution of rhamnolipid on agar varies with congener structure. The spatial MSI intensity profiles (a-l) from three cross sections of heat maps of congener distribution (insets) show diverse distribution patterns of rhamnolipid congeners of CFSM allowed to spread on 1.5% agar. Locations of cross-section signal intensity measurements correlate to blue, orange, and green lines, as depicted in image m, and indicate a diversity in both radial symmetry and small shifts in localization for the different congeners detected. Scale bar is 7 mm.
The majority of congeners were located centrally, this included all congeners with total fatty acid chains of 20 carbons or less. Of these centrally located rhamnolipids, dRha-C22 (di-rhamnolipid-C22) and dRha-C18 had highly similar distribution patterns. Rhamnolipids that were located more distally all possessed carbon chains containing 22 or more total carbons. We found that Rha-C22, dRha-C24 and dRha-C24:1 shared a similar distribution pattern with a pronounced presence outside of the central region. Select congener pairs with small differences in fatty acid chain length exhibited markedly distinct distribution patterns, such as Rha-C20 which was concentrated centrally while Rha-C22 was more concentrated distally. These congener distributions were consistent between replicates (Table S2).
To further identify and quantify similarity in distribution patterns we carried out a correlation analysis within each sample. This comparison normalizes the relative change between pairwise intensity patterns of congeners across the region of interest such that congeners with the same distribution patterns return the correlation coefficient (R2) as 1.0. The coefficient values decrease with the reduction in the similarity of spatial distribution patterns such that a completely unrelated distribution pattern has a coefficient value of 0.0. We assembled these resultant correlation coefficients for each pairwise combination of congeners in the form of a matrix plot and used these data to quantify trends from MSI results of Fig. 1 (Fig. 2 and Table S2). For example, similar distributions were exhibited for the mono- and di-rhamnolipid pair of Rha-C20 and dRha-C20 (R2 = 0.76-.97) as well as saturated- and unsaturated pair of dRha-C22 and dRha-C22:1 (R2 = 0.90-.94). (We note that these correlation coefficient comparisons highlight overall similarity in patterns of distribution and not the absolute amount of each congener. Because P. aeruginosa disproportionately produces select rhamnolipid congeners, parity in overall congener mass would never be expected.) Liquid chromatography- tandem mass spectrometry (LC-MS/MS) estimation of P. aeruginosa supernatant suggests that Rha-C10-C10 and dRha-C10-C10 are the most abundant congeners in mono-rhamnolipid and di-rhamnolipid classes, respectively 42, 43. We noted a general trend in correlation coefficients of saturated versus corresponding unsaturated form with other congeners (data not shown). In a pairwise comparison of three saturated congeners, Rha-C22, dRha-C22, and dRha-C24, with their respective unsaturated counterparts, we found correlation decreased consistently for Rha-C22 and dRha-C24 with unsaturation in CFSM samples (n=3). Though differences in the correlation values were observed across all samples, a similar pattern did not emerge in other environments. With Rha-C22 we see a shift from central to more distal localization occurs with desaturation, decreasing the correlation between localization patterns (R2 = 0.31-0.59), while desaturation of dRha-C22 maintains a similar, central localization to its unsaturated form (R2 = 0.90-0.94) (Table S2). We thus note that additional work is clearly needed to discern if any association between desaturation of fatty acids within these congeners can be used to predict rhamnolipid distribution.
Figure 2.
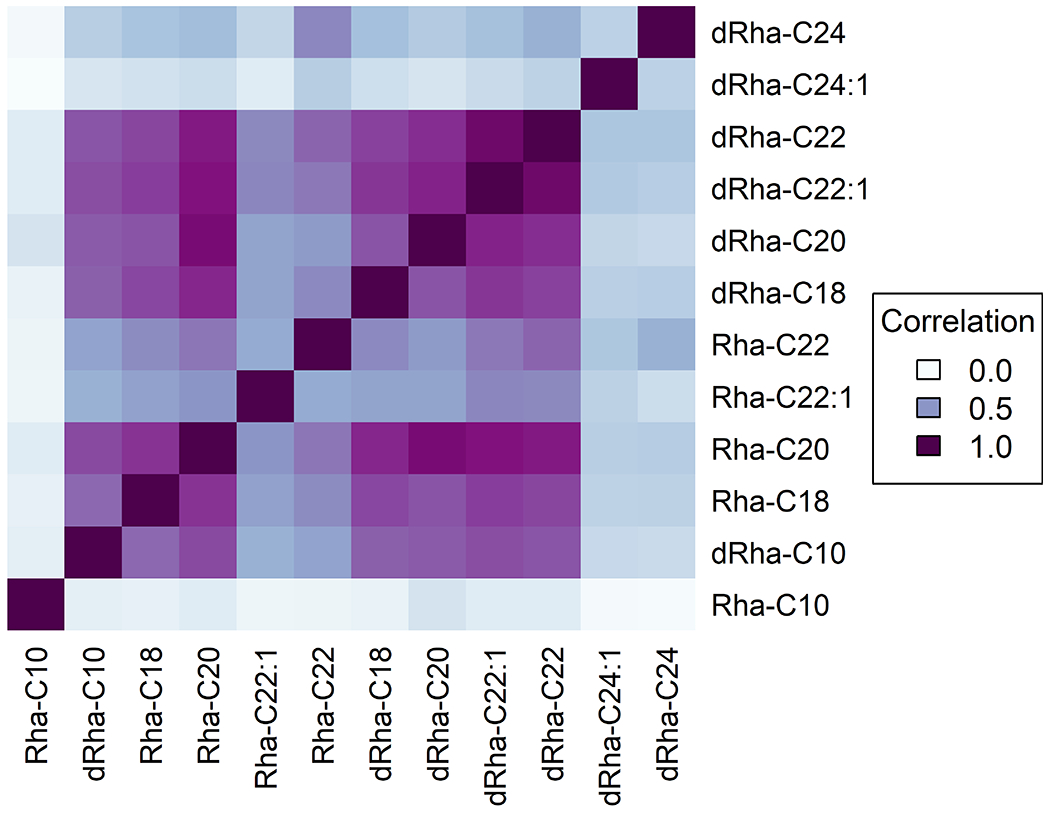
Correlation plot of CFSM rhamnolipid distribution on nutrient agar highlighting similarities in congener distribution. Correlation values derived from an average of three replicate samples.
The largest (dRha-C24 and dRha-C24:1) and the smallest (Rha-C10) of congeners (that would be expected to have the greatest differences in their physiochemical properties) exhibited the most distinct distribution patterns (dRha-C24 R2= 0.04-0.66 dRha-C24:1 R2= 0.02-0.46, Rha-C10 R2= 0.04-0.34). Congeners with mid-length fatty acid chains, particularly Rha-C20 and dRha-C20 had similar distributions (R2 = 0.897) irrespective of rhamnosylation. Previous work identifies a general trend of di-rhamnolipids to diffuse farther than HAAs, the rhamnolipid precursor, on a surface44. Looking at mono- and di-rhamnolipids we did not find a difference in these distances. Overall, we saw that congeners containing fatty acid chains of C22-24 total carbons were more likely to be distributed distally. Congeners with smaller fatty acid chains (≤C20) were centrally located. The majority of mono- and di- congeners with equivalent fatty acid chains had similar distributions, either central or distal.
Rhamnolipid distribution from commercial mixtures reflects trends observed in cell-free spent media.
Next, we studied whether the trends observed result from other constituents that could be present in CFSM by examining the distribution of commercial rhamnolipid mixtures on our nutrient agar plate assays. Because three different commercial rhamnolipid mixtures were readily available, this simultaneously enabled us to consider if select congeners influence overall spreading of multiple congeners. We reasoned that in the absence of other secreted factors from CFSM or absence of all di-rhamnolipids, differences in spreading of the rhamnolipids present in the mixture would be highlighted. The extracts consisted of a mono-rhamnolipid dominant (95%), di- rhamnolipid dominant (95%), and a general rhamnolipid mix (90%). Congener concentration can impact secondary structures formed by rhamnolipids33, however this study did not investigate the impact of concentration due to the lack of individual standards. In this study, we tested a concentration representative of the yields reported using glucose41, 45. Our internal analysis of these mono-rhamnolipid and di-rhamnolipid dominant mixes revealed that they share several congeners belonging to either category. However, we could not quantify each individual congener and estimate their percent content using the methods applied in this study.
We found congeners of the general rhamnolipid mix to have similar dispersal patterns with high correlation coefficient (R2 > 0.8), all localized centrally. However, both mono- and di- mixes exhibited more diversity (Fig. 3 and Tables S3–S5). The mono-rhamnolipid and di-rhamnolipid dominant mixes showed an overall trend of decreased correlation among congeners in comparison to the general mix. It could be that either a difference in relative congener concentration or presence of secondary components could be responsible for altering the rhamnolipid distribution in these mixtures.
Figure 3.
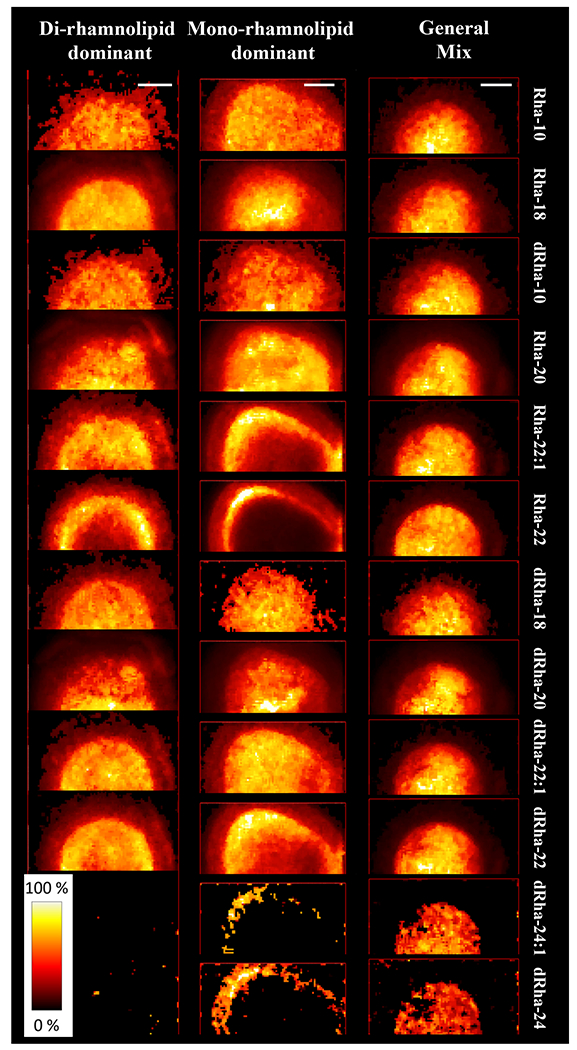
Congener distribution patterns observed in aqueous solutions of commercial rhamnolipid extracts applied to agar surfaces. Scale bars represent 5 mm.
As in CFSM, we found Rha-C22, dRha-C24, and dRha-C24:1 to distribute in an outer ring pattern. As before, these distally distributed congeners are likely to contain C12 or C14 chains, based on their total carbon numbers and previously reported abundance of rhamnolipid congeners 46. In the mono-dominant rhamnolipid mix dRha-C22 was distributed more distally yet, in contrast, the unsaturated dRha-C22:1 was more centrally located. As with CFSM, we found congeners with shorter carbon chains (C20 or less), to be centrally located (Fig. 3). We attribute the reduced intensity observed for dRha-C24 because of its low signal (and likely low abundance) in both the mono-rhamnolipid and the di-rhamnolipid dominant mixes. In a comparison of saturated and unsaturated congeners as carried out in CFSM, shifts in localization in commercial mixes were less consistent and unsaturated forms were not consistently divergent from general localization patterns. Compared to commercial mixes, CFSM is a complex mixture of passively and actively secreted compounds produced by the bacterial cell that include proteins, small organic acids, extracellular DNA, and polysaccharides. Certainly these proteins and other molecules present in CFSM could be reasonably predicted to influence rhamnolipid congener distribution and this is an important area of future research.
Congeners co-migrate in a complex distribution of colony biofilm rhamnolipids.
Because our overall goal is to understand the distribution of these molecules in the presence of P. aeruginosa communities, we probed rhamnolipid distributions in live cultures.
We observed that rhamnolipid distribution was more complex than that observed for CFSM or the commercial mixtures. P. aeruginosa colony biofilms exhibited multiple distinct regions of localization as seen from MALDI-MSI heatmaps (Fig. 4 and Table S6). Two identical circles are superimposed onto Figure 4 heat maps to highlight regions of intensity shifts in congener relative concentration. Unlike the abiotic experiments, we found that congeners with shorter fatty acid chains were not just centrally located. We found three unsaturated rhamnolipids, but unlike in experiments with CFSM, distributions were not consistently divergent from general localization patterns.
Figure 4.
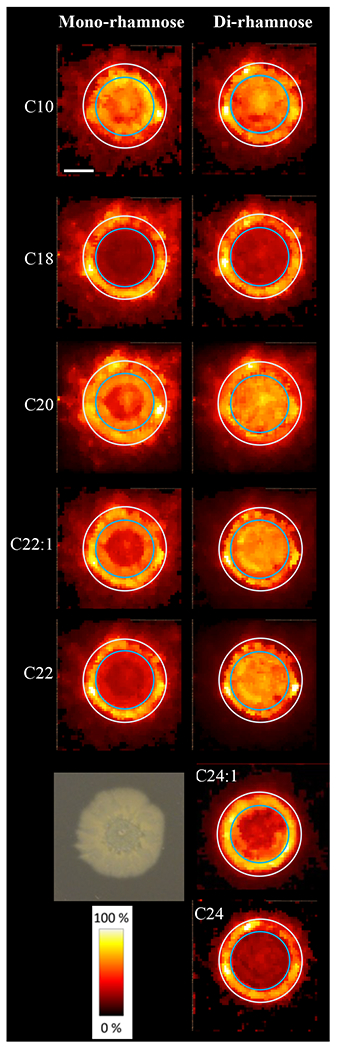
Rhamnolipid congener distribution patterns from 24-hour P. aeruginosa colony biofilms. A representative image of a 24-hour colony biofilm is shown lower left. Circles of 10 and 14 mm diameter are superimposed for perspective. Scale bar is 4 mm.
Looking at similarities with CSFM, biofilm saturated and unsaturated forms of dRha-C22 shared similar distribution patterns (R2 ≥ 0.86), exhibiting a high level of co-localization. Yet unlike CSFM and commercial mixes, Rha-C20 and dRha-C20 congeners exhibited distinct patterns with Rha-C20 less prominent in the center of the colony biofilm while outer regions the two congeners were highly co-localized. We saw similar distribution patterns between mono-rhamnose congeners with C20, C22, and C22:1 carbon chains though regions of highest relative concentration for both Rha-C22 congeners are slightly more centralized. We see this same trend with the corresponding di-rhamnose congeners.
Sempels et al. reported that in the absence of rhamnolipids, a drop of Pseudomonas culture dries in a ring formed at the edge of the droplet. This coffee ring is produced by capillary action that pulls cells and media components to the edge as the droplet dries. In the presence of rhamnolipids, however, the distribution of cells and media components becomes more homogeneous and centrally located because of the Marangoni effect, or recirculation of surfactants due to a surface tension gradient produced when surfactants are pulled to the edge by capillary action 47. However, it is unclear how these forces impact individual congeners within a growing colony biofilm.
The microbial community, originating from clonal cells, develops several radial zones of shifting rhamnolipid congener compositions. As with previous samples, we could see no evidence suggesting that mono- and di-rhamnolipid classes shared similar distribution patterns. However, taking total congener variation into account we see that this community exists within a heterogeneous landscape of rhamnolipid congener composition and concentration gradients that creates many unique microenvironments.
CONCLUSIONS
Rhamnolipid congeners present in CFSM harvested from active P. aeruginosa cultures exhibited a diversity of distribution patterns. Overall, however, two dominant distribution patterns emerged. Congeners with longer fatty acid chains (C22-24) were more likely to be distally located, while shorter congeners with shorter chains (C20 or less) were centrally located. Distribution patterns were less diverse in commercial rhamnolipid mixtures of 90-95% purity, indicating that components of CFSM increased the complexity of rhamnolipid distribution. Secreted factors, such as proteins, likely serve to diversify distribution patterns through interactions with rhamnolipids. The diversity in distribution of a general rhamnolipid mix was minor while both mono- and di- mixes exhibited distinct variation. In commercial mono- and di- rhamnolipid mixes, like CFSM, congeners containing fatty acid chains of C22-24 were more likely to be distally located, while those with shorter chains were centrally located. In 24-hour colony biofilms we found rhamnolipid congener distribution to occur in several distinct patterns. Unlike previous abiotic samples, congeners with shorter fatty acid chains were not necessarily centrally located. While most pairwise comparisons of various congeners did not show strong co-localization in these colony biofilms, some congeners, such as saturated and unsaturated forms of dRha-C22, were found to be colocalized in all samples examined in this study. We found that classifying rhamnolipids into mono- and di- classes does not encompass the diversity of localization patterns that accompany colony biofilm growth. Congener composition may be important for industrial purposes desiring homogeneous rhamnolipid distribution. These diverse distribution patterns also raise new questions concerning local roles of individual congeners in microbial communities such as colony expansion, defense, increasing bioavailability of secreted factors, and promoting virulence. More holistically, rhamnolipid congener localization is far from homogeneous, creating microenvironments of varied rhamnolipid profiles. This impacts the localization and movement of other, associated compounds. It is also not clear how dynamic rhamnolipid distribution is over time as colony biofilms expand and age. Future work is needed to further understand the role of rhamnolipid congener distribution in biofilm communities.
EXPERIMENTAL SECTION
Preparation of rhamnolipid and spent media samples.
Rhamnolipid mixtures of mono-dominant and di-dominant congeners, as well as a general mix (Sigma) were suspended in Nanopure water at 0.5 mg/mL and 25 μL spotted onto plates. Spent media was prepared from 48-h cultures of wild type P. aeruginosa (PAO1C) grown in modified Fastidious anaerobe broth (FAB) supplemented with 30 mM glucose at 37°C with shaking. Cultures were centrifuged at 7,500 rpms for 10 min, filtered using a 0.22 μm filter, and 25 μL of resulting filtrate was spotted onto media plates. All plates were prepared using modified FAB supplemented with 12 mM glucose and 1.5% Noble agar. Nutrient agar, containing samples, was removed from petri dishes, affixed to aluminum plates using copper tape, and dehydrated overnight under forced air 48. Colony biofilms were grown on FAB nutrient agar plates, spotted with 1 μL of overnight culture grown in modified FAB containing 30 mM glucose, and incubated for 24 h at 37 °C.
Mass spectrometry imaging analysis.
Dehydrated agar films were uniformly coated with a solution of 2,5-dihydroxy benzoic acid (DHB) (40 mg/mL in 50% Methanol) using overhead matrix sprayer (M5 sprayer, HTX). The spray parameters were maintained as follows, nozzle height - 40 mm, nozzle temperature - 70 °C, lateral nozzle speed - 1000 mm, lateral spacing - 3 mm, matrix flow rate - 0.05 mL/min and nitrogen pressure - 1 bar. Entire sample surface was coated with DHB in 4 passes with each layer sprayed at 90°C to the previous one and 10 s delay time between passes to allow drying. The plates were stored inside a nitrogen desiccator until analysis.
Mass spectrometry imaging was performed on Solarix FT-ICR mass spectrometer (Bruker, USA) equipped with matrix-assisted desorption/ionization (MALDI) source. To facilitate MSI, the samples were scanned using a flatbed document scanner at 1200 DPI. The MS parameters were set using ftmsControl (version 2.3.0, build 59, Bruker). The data acquisition was carried out in the positive ion mode with mass range 100-1600 m/z range, laser size set at “ultra-large”, and MS analyzer operated at 1M size (transient data points). Regions of interest for MSI were selected in FlexImaging (version 5.0, build 89, Bruker). The MSI data was processed in the SCILS lab (Version 2023a Pro). Extracted ion profiles were generated using ±16 ppm extraction window around each m/z. The profiles were normalized to the total ion counts (TIC) before the estimation of correlation values. Extracted ion intensities are represented using false fire color scaled to an intensity range of 0-100 % as dark to light color. Intensity profiles of CFSM were derived in ImageJ using the plot profiles function49. The correlation coefficient (R2) between spatial distribution patterns of congeners were estimated using the SCiLS software. All estimations are based on the same region of interest (ROI) as originally used to acquire the MS imaging data from the sample surface. The correlation values are generated from a scatter plot comparing the intensities of two analytes from each individual pixel of ROI.
Supplementary Material
Figure S1. Rhamnolipid general structure
Table S6. Correlation values for congener distributions in 24 hour colony biofilms
Table S2. Correlation values for congener distributions in CFSM
Table S3. Correlation values for congener distributions in general rhamnolipid mix
Table S4. Correlation values for congener distributions in di-rhamnolipid mix
Table S1. Rhamnolipid congeners and theoretical m/z values
Table S5. Correlation values for congener distributions in mono-rhamnolipid mix
ACKNOWLEDGMENT
Support for this work is provided by National Institutes of Health NIAID grant R01AI113219 (JDS and JVS)
ABBREVIATIONS
- CFSM
cell-free spent medium
- CMC
critical micelle concentration
- DHB
2,5-dihydroxy benzoic acid
- FT-ICR MS
Fourier-transform ion cyclotron resonance mass spectrometry
- HAA
hydroxyalkanoyl-hydroxyalkanoates
- MALDI-MSI
matrix-assisted laser desorption/ionization mass spectrometry imaging
- PQS
Pseudomonas quinolone signal
- TIC
total ion counts
Footnotes
REFERENCES
- (1).Soberon-Chavez G; Gonzalez-Valdez A; Soto-Aceves MP; Cocotl-Yanez M Rhamnolipids produced by Pseudomonas: from molecular genetics to the market. Microb Biotechnol 2021, 14 (1), 136–146. DOI: 10.1111/1751-7915.13700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Pearson JP; Pesci EC; Iglewski BH Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol 1997, 179 (18), 5756–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Mattingly AE; Weaver AA; Dimkovikj A; Shrout JD Assessing Travel Conditions: Environmental and Host Influences on Bacterial Surface Motility. J. Bacteriol 2018, 200 (11), 17 e00014-00018. DOI: 10.1128/jb.00014-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Davey ME; Caiazza NC; O’Toole GA Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. Journal of Bacteriology 2003, 185 (3), 1027–1036. DOI: 10.1128/JB.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Chrzanowski Ł; Ławniczak Ł; Czaczyk K Why do microorganisms produce rhamnolipids? World journal of microbiology & biotechnology 2012, 28 (2), 401–419. DOI: 10.1007/s11274-011-0854-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Jensen P; Bjarnsholt T; Phipps R; Rasmussen TB; Calum H; Christoffersen L; Moser C; Williams P; Pressler T; Givskov M; Høiby N Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology 2007, 153 (5), 1329–1338. DOI: 10.1099/mic.0.2006/003863-0. [DOI] [PubMed] [Google Scholar]
- (7).Lang S; Katsiwela E; Wagner F Antimicrobial Effects of Biosurfactants. Fat Science Technology 1989, 91 (9), 363–366. DOI: 10.1002/lipi.19890910908. [DOI] [Google Scholar]
- (8).Bru J.-l.; Rawson B; Trinh C; Whiteson K; Siryaporn A PQS Produced by the Pseudomonas aeruginosa Stress Response Repels Swarms Away from Bacteriophage and Antibiotics. Journal of Bacteriology 2019, 201 (23), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Heeb S; Fletcher MP; Chhabra SR; Diggle SP; Williams P; Cámara M Quinolones: from antibiotics to autoinducers. FEMS microbiology reviews 2011, 35 (2), 247–274. DOI: 10.1111/j.1574-6976.2010.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Argenio D. a. D.; Calfee MW; Rainey PB; Pesci EC. Autolysis and Autoaggregation in Pseudomonas aeruginosa Colony Morphology Mutants. Microbiology 2002, 184 (23), 6481–6489. DOI: 10.1128/JB.184.23.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Diggle SP; Winzer K; Chhabra SR; Worrall KE; Cámara M; Williams P The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Molecular Microbiology 2003, 50 (1), 29–43. DOI: 10.1046/j.1365-2958.2003.03672.x. [DOI] [PubMed] [Google Scholar]
- (12).Pesci EC; Milbank JB; Pearson JP; McKnight S; Kende a. S.; Greenberg EP; Iglewski BH. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences of the United States of America 1999, 96 (September), 11229–11234. DOI: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).TransparencyMarketResearch. Surfactants Market Rising at 4.20% CAGR from 2015-2023 due to Rising Demand for Detergents and Personal Care Products: Transparency Market Research. In GlobeNewsWire, 2016. [Google Scholar]
- (14).Kaczerewska O; Martins R; Figueiredo J; Loureiro S; Tedim J Environmental behaviour and ecotoxicity of cationic surfactants towards marine organisms. J Hazard Mater 2020, 392, 122299. DOI: 10.1016/j.jhazmat.2020.122299. [DOI] [PubMed] [Google Scholar]
- (15).Salek K; Euston SR; Janek T Phase Behaviour, Functionality, and Physicochemical Characteristics of Glycolipid Surfactants of Microbial Origin. Front Bioeng Biotechnol 2022, 10, 816613. DOI: 10.3389/fbioe.2022.816613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Hogan DE; Curry JE; Pemberton JE; Maier RM Rhamnolipid biosurfactant complexation of rare earth elements. J Hazard Mater 2017, 340, 171–178. DOI: 10.1016/j.jhazmat.2017.06.056. [DOI] [PubMed] [Google Scholar]
- (17).Caiazza NC; Shanks RM; O’Toole GA Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol 2005, 187 (21), 7351–7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Déziel E; Lépine F; Milot S; Villemur R rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 2003, 149 (8), 2005–2013. DOI: 10.1099/mic.0.26154-0. [DOI] [PubMed] [Google Scholar]
- (19).Soberón-Chávez G; Lépine F; Déziel E Production of rhamnolipids by Pseudomonas aeruginosa. Appl Microbiol Biotechnol 2005, 68 (6), 718–725. [DOI] [PubMed] [Google Scholar]
- (20).Ochsner UA; Fiechter A; Reiser J Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J. Biol. Chem 1994, 269 (31), 19787–19795. [PubMed] [Google Scholar]
- (21).Ochsner UA; Reiser J Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Nat. Acad. Sci. U.S.A 1995, 92 (14), 6424–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Abdel-Mawgoud AM; Lépine F; Déziel E Rhamnolipids: Diversity of structures, microbial origins and roles. Applied Microbiology and Biotechnology 2010, 86, 1323–1336. DOI: 10.1007/s00253-010-2498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zhao F; Wang B; Yuan M; Ren S Comparative study on antimicrobial activity of mono-rhamnolipid and di-rhamnolipid and exploration of cost-effective antimicrobial agents for agricultural applications. Microb Cell Fact 2022, 21 (1), 221. DOI: 10.1186/s12934-022-01950-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Elshikh M; Funston S; Chebbi A; Ahmed S; Marchant R; Banat IM Rhamnolipids from non-pathogenic Burkholderia thailandensis E264: Physicochemical characterization, antimicrobial and antibiofilm efficacy against oral hygiene related pathogens. N Biotechnol 2017, 36, 26–36. DOI: 10.1016/j.nbt.2016.12.009. [DOI] [PubMed] [Google Scholar]
- (25).Zhao F; Shi R; Ma F; Han S; Zhang Y Oxygen effects on rhamnolipids production by Pseudomonas aeruginosa. Microb Cell Fact 2018, 17 (1), 39. DOI: 10.1186/s12934-018-0888-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Dubeau D; Deziel E; Woods DE; Lepine F Burkholderia thailandensis harbors two identical rhl gene clusters responsible for the biosynthesis of rhamnolipids. BMC Microbiol 2009, 9, 263. DOI: 10.1186/1471-2180-9-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Euston SR; Banat IM; Salek K Congener-dependent conformations of isolated rhamnolipids at the vacuum-water interface: A molecular dynamics simulation. J Colloid Interface Sci 2021, 585, 148–157. DOI: 10.1016/j.jcis.2020.11.082. [DOI] [PubMed] [Google Scholar]
- (28).Baccile N; Poirier A; Perez J; Pernot P; Hermida-Merino D; Le Griel P; Blesken CC; Muller C; Blank LM; Tiso T Self-Assembly of Rhamnolipid Bioamphiphiles: Understanding the Structure-Property Relationship Using Small-Angle X-ray Scattering. Langmuir 2023, 39 (27), 9273–9289. DOI: 10.1021/acs.langmuir.3c00336. [DOI] [PubMed] [Google Scholar]
- (29).Abalos A; Pinazo A; Infante MR; Casals M; Garcia F; Manresa A Physicochemical and antimicrobial properties of new rhamnolipids produced by Pseudomonas aeruginosa AT10 from soybean oil refinery wastes. Langmuir 2001, 17 (5), 1367–1371. DOI: 10.1021/la0011735. [DOI] [Google Scholar]
- (30).Tiso T; Zauter R; Tulke H; Leuchtle B; Li WJ; Behrens B; Wittgens A; Rosenau F; Hayen H; Blank LM Designer rhamnolipids by reduction of congener diversity: production and characterization. Microb Cell Fact 2017, 16 (1), 225. DOI: 10.1186/s12934-017-0838-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Samadi N; Abadian N; Ahmadkhaniha R; Amini F; Dalili D; Rastkari N; Safaripour E; Mohseni FA Structural characterization and surface activities of biogenic rhamnolipid surfactants from Pseudomonas aeruginosa isolate MN1 and synergistic effects against methicillin-resistant Staphylococcus aureus. Folia Microbiol (Praha) 2012, 57 (6), 501–508. DOI: 10.1007/s12223-012-0164-z. [DOI] [PubMed] [Google Scholar]
- (32).Palos Pacheco R; Kegel LL; Pemberton JE Interfacial and Solution Aggregation Behavior of a Series of Bioinspired Rhamnolipid Congeners Rha-C14-Cx (x = 6, 8, 10, 12, 14). J Phys Chem B 2021, 125 (49), 13585–13596. DOI: 10.1021/acs.jpcb.1c09435. [DOI] [PubMed] [Google Scholar]
- (33).Lee MT Micellization of Rhamnolipid Biosurfactants and Their Applications in Oil Recovery: Insights from Mesoscale Simulations. J Phys Chem B 2021, 125 (34), 9895–9909. DOI: 10.1021/acs.jpcb.1c05802. [DOI] [PubMed] [Google Scholar]
- (34).Dunham SJB; Ellis JF; Li B; Sweedler JV Mass Spectrometry Imaging of Complex Microbial Communities. Accounts of Chemical Research 2017, 50 (1), 96–104. DOI: 10.1021/acs.accounts.6b00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Masyuko RN; Lanni EJ; Driscoll CM; Shrout JD; Sweedler JV; Bohn PW Spatial organization of Pseudomonas aeruginosa biofilms probed by combined matrix-assisted laser desorption ionization mass spectrometry and confocal Raman microscopy. The Analyst 2014, 139 (22), 5700–5708. DOI: 10.1039/c4an00435c. [DOI] [PubMed] [Google Scholar]
- (36).Madukoma CS; Liang P; Dimkovikj A; Chen J; Lee SW; Chen DZ; Shrout JD Single Cells Exhibit Differing Behavioral Phases during Early Stages of Pseudomonas aeruginosa Swarming. J. Bacteriol 2019, 201 (19). DOI: 10.1128/JB.00184-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Mattingly AE; Kamatkar NG; Morales-Soto N; Borlee BR; Shrout JD Multiple Environmental Factors Influence the Importance of the Phosphodiesterase DipA upon Pseudomonas aeruginosa Swarming. Appl Environ Microbiol 2018, 84 (7). DOI: 10.1128/AEM.02847-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Morales-Soto N; Cao T; Baig NF; Kramer KM; Bohn PW; Shrout JD Surface-Growing Communities of Pseudomonas aeruginosa Exhibit Distinct Alkyl Quinolone Signatures. Microbiol Insights 2018, 11, 1178636118817738. DOI: 10.1177/1178636118817738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Achermann Y; Goldstein EJ; Coenye T; Shirtliff ME Propionibacterium acnes: from commensal to opportunistic biofilm-associated implant pathogen. Clin Microbiol Rev 2014, 27 (3), 419–440. DOI: 10.1128/cmr.00092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Morris JD; Hewitt JL; Wolfe LG; Kamatkar NG; Chapman SM; Diener JM; Courtney AJ; Leevy WM; Shrout JD Imaging and analysis of Pseudomonas aeruginosa swarming and rhamnolipid production. Applied and environmental microbiology 2011, 77 (23), 8310–8317. DOI: 10.1128/AEM.06644-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Wittgens A; Tiso T; Arndt TT; Wenk P; Hemmerich J; Muller C; Wichmann R; Kupper B; Zwick M; Wilhelm S; Hausmann R; Syldatk C; Rosenau F; Blank LM Growth independent rhamnolipid production from glucose using the non-pathogenic Pseudomonas putida KT2440. Microb Cell Fact 2011, 10, 80. DOI: 10.1186/1475-2859-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Rudden M; Tsauosi K; Marchant R; Banat IM; Smyth TJ Development and validation of an ultra-performance liquid chromatography tandem mass spectrometry (UPLC-MS/MS) method for the quantitative determination of rhamnolipid congeners. Appl Microbiol Biotechnol 2015, 99 (21), 9177–9187. DOI: 10.1007/s00253-015-6837-1. [DOI] [PubMed] [Google Scholar]
- (43).Deziel E; Lepine F; Milot S; Villemur R Mass spectrometry monitoring of rhamnolipids from a growing culture of Pseudomonas aeruginosa strain 57RP. Biochimica et Biophysica Acta - Molecular and Cell Biology of Lipids 2000, 1485, 145–152. [DOI] [PubMed] [Google Scholar]
- (44).Tremblay J; Richardson AP; Lépine F; Déziel E Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environmental Microbiology 2007, 9, 2622–2630. DOI: 10.1111/j.1462-2920.2007.01396.x. [DOI] [PubMed] [Google Scholar]
- (45).Wang Q; Fang X; Bai B; Liang X; Shuler PJ; Goddard WA 3rd; Tang Y Engineering bacteria for production of rhamnolipid as an agent for enhanced oil recovery. Biotechnol Bioeng 2007, 98 (4), 842–853. DOI: 10.1002/bit.21462. [DOI] [PubMed] [Google Scholar]
- (46).Abdel-mawgoud AM; Hausmann R; Le F; De E; Markus MM Rhamnolipids: Detection, Analysis, Biosynthesis, Genetic Regulation and Bioengineering of Production; Springer Berlin Heidelberg, 2011. DOI: 10.1007/978-3-642-14490-5. [DOI] [Google Scholar]
- (47).Sempels W; De Dier R; Mizuno H; Hofkens J; Vermant J Auto-production of biosurfactants reverses the coffee ring effect in a bacterial system. Nat Commun 2013, 4, 1757. DOI: 10.1038/ncomms2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Dunham SJB; Ellis JF; Baig NF; Morales-soto N; Cao T; Shrout JD; Bohn PW; Sweedler JV Quantitative SIMS Imaging of Agar-Based Microbial Communities. Analytical chemistry 2018, (90), 5654–5663. DOI: 10.1021/acs.analchem.7b05180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Schneider C. a.; Rasband WS; Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 2012, 9 (7), 671–675. DOI: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Rhamnolipid general structure
Table S6. Correlation values for congener distributions in 24 hour colony biofilms
Table S2. Correlation values for congener distributions in CFSM
Table S3. Correlation values for congener distributions in general rhamnolipid mix
Table S4. Correlation values for congener distributions in di-rhamnolipid mix
Table S1. Rhamnolipid congeners and theoretical m/z values
Table S5. Correlation values for congener distributions in mono-rhamnolipid mix


