Abstract
Enterovirus infections are implicated in the development of type 1 diabetes (T1D). MicroRNAs as regulators of gene expression are involved in many physiological and pathological processes. Given that viral infections dysregulate cellular microRNAs, we investigated the impact of persistent coxsackievirus B4 infection on microRNA expression of human pancreatic cells. Next-generation sequencing was used to determine microRNA expression in PANC-1 cells persistently infected (for several weeks) with coxsackievirus B4 and uninfected control cells. Target prediction restricted to T1D risk genes was performed with miRWalk2.0. Functional annotation analysis was performed with DAVID6.7. Expression of selected microRNAs and T1D risk genes was measured by quantitative reverse-transcription polymerase chain reaction. Eighty-one microRNAs were dysregulated in persistently infected PANC-1 cells. Forty-nine of the known fifty-five T1D risk genes were predicted as putative targets of at least one of the dysregulated microRNAs. Most functional annotation terms that were enriched in these 49 putative target genes were related to the immune response or autoimmunity. mRNA levels of AFF3, BACH2, and IL7R differed significantly between persistently infected cells and uninfected cells. This is the first characterization of the microRNA expression profile changes induced by persistent coxsackievirus B4 infection in pancreatic cells. The predicted targeting of genes involved in the immune response and autoimmunity by the dysregulated microRNAs as well as the dysregulated expression of diabetes risk genes shows that persistent coxsackievirus B4 infection profoundly impacts the host cell. These data support the hypothesis of a possible link between persistent coxsackievirus B4 infection and the development of T1D.
Keywords: miRNA, Persistence, Latency, Gene expression, Latent infection
Introduction
Type 1 diabetes (T1D) is thought to be caused by an interplay between genetic factors, the immune system, and environmental factors [1]. Among the later, enterovirus infections, especially with type B coxsackieviruses (CVB), have been linked to the development of T1D [2, 3]. Signs of enteroviral infection have frequently been detected in pancreatic cells of T1D patients [4]. Therefore, a persistent enteroviral infection of the pancreas could be one of the causes responsible for the development of T1D [1, 3, 5, 6].
CVB are small non-enveloped RNA viruses that belong to the Enterovirus genus of the Picornaviridae family. CVB can establish persistent infections in vitro as well as in vivo [7]. Persistence of CVB4 in human pancreatic ductal cells resulted in an impaired formation and viability of islet-like cell aggregates [8]. CVB might initiate or accelerate the development of T1D by different mechanisms, including direct destruction of beta-cells, inflammatory cytokine induction, molecular mimicry, or dysregulation of host cell microRNA (miRNA) expression [1, 9].
miRNAs are small 19–23 nucleotide RNA molecules that regulate gene expression by inducing translational arrest and/or degradation of messenger RNAs [10]. miRNAs are involved in many key regulatory processes in cells and have also been shown to play an important role in beta-cell function [11]. Dysregulation of miRNA expression has been found in many diseases, including viral infections and T1D [9, 12].
The impact of enterovirus infection on host cell miRNA expression has been described in several recent studies [13–16]. However, these studies focused on acute enterovirus infection. To our knowledge, the impact of persistent enterovirus infection on pancreatic cell miRNA expression has not been investigated to date. Given that persistent enterovirus infection of the pancreas may be one underlying mechanism of T1D development, the aim of the current study was to investigate the impact of persistent CVB4 infection on the miRNA expression profile of pancreatic cells. To this end, we took advantage of pancreatic ductal cells persistently infected with CVB4, in which a stable persistent infection was obtained beyond 5 weeks post-inoculation of the virus [8, 17].
We further investigated whether the dysregulated miRNAs potentially target T1D risk genes.
Methods
Cells and virus
The human pancreatic ductal cell line PANC-1 (ATCC, LGC Standards, Molsheim, France) was cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% of heat inactivated fetal calf serum (FCS), 1% of l-glutamine, penicillin, and streptomycin (Thermofisher Scientific, Courtaboeuf, France). The diabetogenic CVB4E2 strain (kindly provided by Ji-Won Yoon, Julia McFarlane Diabetes Research Center, Calgary, Alberta, Canada) was propagated in Hep-2 cells (BioWhittaker, Vervier, Belgium). The CVB4E2 strain was isolated by Yoon et al. from the pancreas of a child with recently diagnosed type 1 diabetes and is diabetogenic in mice [18]. Briefly, after three freeze–thaw cycles, the suspension was collected and clarified at 2000 g for 10 min at 4 °C. Aliquots of virus preparations were stored at −80 °C.
Persistent CVB4 infection in PANC-1 cells
A persistent CVB4 infection was established in PANC-1 cells [17]. Briefly, a 25 cm2 Nunc® cell culture flask (Thermofisher Scientific, Villebon, France) containing an average of 106 cells was inoculated with CVB4E2 at a multiplicity of infection (MOI) of 0.01. During the acute lytic infection, cells were regularly washed to remove excess virus. A stable equilibrium developed between viral replication and cell proliferation. The CVB4 infected and uninfected PANC-1 cells were scraped and subcultured once a week.
Cells were collected from four independent biological replicates of PANC-1 cells persistently infected with CVB4 (after 13 passages, i.e., 13 weeks (n = 3), and after 20 passages, i.e., 20 weeks (n = 1), of persistent CVB4 infection) and four replicates of uninfected PANC-1 cells. Cells were washed with PBS. RNA was extracted with the miRNeasy Mini kit (Qiagen, Courtaboeuf, France) with on-column DNA digestion using DNase I (Qiagen, Courtaboeuf, France). Extracted RNA was quantified with a Nanodrop® spectrophotometer (Thermofisher Scientific, Courtaboeuf, France) and RNA quality was assessed using the 2100 Bioanalyzer (Agilent technologies, Courtaboeuf, France).
Sequencing of microRNAs
Small RNA libraries were prepared from 1 µg of total RNA using the Ion Total RNA-Seq Kit v2.0 (Life Technologies, Carlsbad, CA, USA). Each sample was ligated to two barcodes to detect and bypass PCR biases. Barcoded libraries were quantified and assessed for quality using the Agilent 2100 BioAnalyzer (Agilent technologies France, Courtaboeuf, France). Libraries were pooled in equimolar amounts and sequenced on an Ion PROTON™ Platform using an Ion P1™ Chip Kit v2 and the Ion P1™ Sequencing 200 kit v3 (Life Technologies, Thermofisher Scientific, Courtaboeuf, France).
Primary analysis transforming signal to DNA sequences was done with the default parameters on a Torrent Server 4.0.2 (Life Technologies, Thermofisher Scientific, Courtaboeuf, France). Demultiplexing was done with 0 errors allowed in barcodes. Raw reads were analyzed with ncPRO-Seq 1.5.1 [19]. For each sample, reproducibility of counts within the two barcodes was investigated by Spearman correlation and visual inspection of counts showing no biases between the barcodes. Each pair of barcodes was then pooled and a new ncPRO-Seq analysis was performed on all samples (n = 8). The differential expression of the raw counts obtained from ncPRO-Seq was performed with DESeq2 [20].
Quantification of miRNAs by RT-PCR
The expression of hsa-miR-663b, hsa-miR-1913, hsa-miR-10a-5p, hsa-miR-23a-3p, hsa-miR-23b-3p, hsa-miR-125b-5p, hsa-miR-146a-5p, and hsa-miR-138-5p was quantified using the TaqMan® MicroRNA kits (Life Technologies, Thermofisher Scientific, Courtaboeuf, France). The Taqman MicroRNA Reverse Transcription Kit was used for cDNA synthesis, and real-time PCRs were performed with the Taqman Small RNA assays (primers and probe) and the Taqman Universal PCR Master Mix II according to the manufacturer’s recommendations, on a Mx3000p® thermocycler (Agilent technologies France, Courtaboeuf, France).The expression of hsa-miR-6087 and hsa-miR-4516 was quantified using the miScript PCR Starter Kit (Qiagen, Courtaboeuf, France) according to the manufacturer’s recommendations, on a Mx3000p® thermocycler (Agilent technologies France, Courtaboeuf, France). RNU6B was used for normalization and the relative expression was determined using the 2−ΔΔCt formula [21].
MiRNA target prediction
miRWalk 2.0 was used to predict potential target genes of the 81 dysregulated miRNAs [22]. The following settings were applied: “species: human”; “restricted to 3′UTR”; “minimum seed length = 7”; “p value = 0.05”; and using the databases: miRWalk, RNA22, miRanda, and Targetscan. The list of genes predicted as targets by at least three of the four databases was compared with the list of type 1 diabetes risk genes obtained from T1Base [23] to identify T1D risk genes that are potentially targeted by the dysregulated miRNAs.
Functional annotation analysis
The 49 T1D risk genes that were potential targets of the 81 dysregulated miRNAs were functionally annotated using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) 6.7 [24]. This analysis yields functional annotations that are enriched in these genes and indicates an enrichment p value.
Gene expression analysis
2 µg of total RNA from PANC-1 cells persistently infected with CVB4 and uninfected control cells were DNAse treated (RQ1 RNase-Free DNase, Promega France, Charbonnières-les-Bains, France). cDNA synthesis was performed using the Superscript III first strand synthesis system for RT-PCR kit (Thermofisher Scientific, Courtaboeuf, France) and random hexamer primers. cDNA was diluted 1:10 and quantitative PCR was performed with the Power SYBR green master mix (Applied Biosystems, Thermofisher Scientific, Courtaboeuf, France). The following primers were used (Eurofins Genomics, Ebersberg, Germany): CLEC16A: CATCAAGACGAGTGGGGAGAGT and TCCTCGTCCGTGGTGTTCTG; GLIS3: CAACCAGATCAGTCCTAGCTTACA and GCGAAATAAGGGACCTGGTATC; IKZF1: CACAGTGAAATGGCAGAAGACC and GGCCCTTGTCCCCAAGAAAT; SH2B3: AACCACCAGGTTCCTGCAAC and GGACAGCCAGAAGAACTAAGGTG [16]; BACH2: GACTTTGATCGTGGAGAGGAA and GCAAGCTGACCACCAAATC; beta-Actin: ACCGAGCGCGGCTACAG and CTTAATGTCACGCACGATTTCC; AFF3: ACTCAACAGGATGATGGCACG and TGCCTAAAGTGTTCTGGATCCG; IKZF4: ACGAAATACGTGACCTGGAGATG and CTTGCGTTTGGTGAGGCTATTG; IL7R: CTGGTTTGTAGGCAGCAGAAGA and GGAGCCTCCTATTTGAGACTTGAC.
Cycling was performed on an ABI7500 (Applied Biosystems, Thermofisher Scientific, Courtaboeuf, France) with the following thermal profile: 10 min at 95 °C followed by 40 cycles of 15 s at 95 °C, 30 s at 60 °C, and 30 s at 72 °C. Cycle thresholds (Cts) were normalized to beta-Actin levels. Relative mRNA expression was expressed as fold change in PANC-1 cells persistently infected with CVB4 compared to uninfected control cells. Relative mRNA expression in persistently infected versus non-infected cells was compared using the Mann–Whitney test. A p value <0.05 was considered statistically significant. Statistical analyses and graphs were performed with the Graph Pad Prism 6.03 software (GraphPad Software, La Jolla, CA).
Results
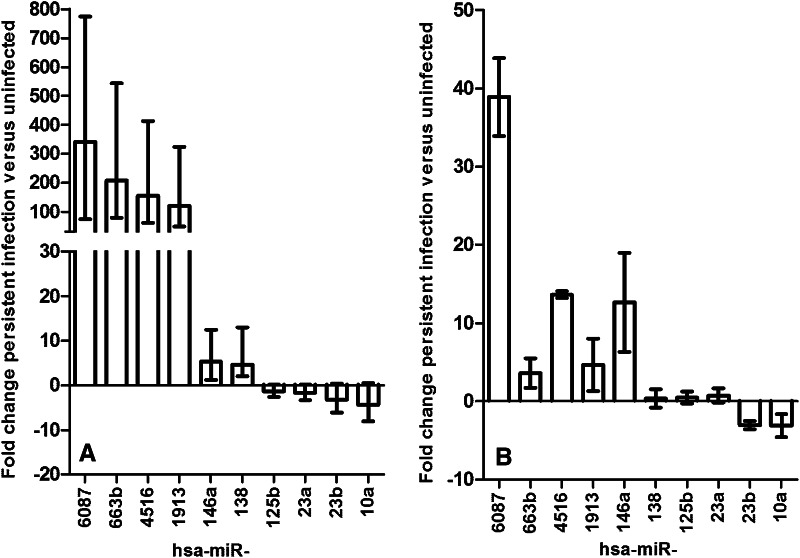
The miRNA expression profile differed between PANC-1 cells with persistent CVB4 infection and uninfected PANC-1 cells. A total of 81 miRNAs showed a significant differential expression (defined as a fold change ≥3, p < 0.05), and among them, 65 miRNAs were up-regulated and 16 were down-regulated in persistently infected cells (Table 1). The highest induction was seen for hsa-miR-6087, hsa-miR-663b, hsa-miR-4516, hsa-miR-4532, hsa-miR-1913, and hsa-miR-3621 that showed a more than hundredfold higher expression in persistently infected compared to uninfected cells. In contrast, the down-regulated miRNAs displayed less than tenfold down-regulation in persistently infected cells compared to uninfected cells. The expression of selected miRNAs that showed a high fold change in the sequencing data or that are known to be implicated in enteroviral infection (hsa-miR-6087, hsa-miR-663b, hsa-miR-4516, hsa-miR-1913, hsa-miR-10a-5p, hsa-miR-23a-3p, hsa-miR-23b-3p, hsa-miR-125b-5p, and hsa-miR-146a-5p, hsa-miR-138-5p) [13–15, 25] was additionally analyzed by RT qPCR. Five of the six miRNAs analyzed that showed an up-regulation in the sequencing data (Fig. 1a) and also showed an up-regulation by RT qPCR, whereas one showed no change (Fig. 1b). Two of the four miRNAs analyzed that showed a down-regulation in the sequencing data (Fig. 1a) and also showed a down-regulation by RT qPCR (Fig. 1b). The extent of the dysregulation was different between the sequencing and the RT qPCR data, for example hsa-miR-6087 showed a 341-fold up-regulation by sequencing and a 39-fold up-regulation by RT qPCR (Fig. 1).
Table 1.
MiRNAs differentially expressed in PANC-1 cells persistently infected with CVB4 compared to uninfected cells
| miRNA (Mirbase-v20) | Dysregulationa | Fold changeb | Adjusted p valuec |
|---|---|---|---|
| hsa-miR-6087 | Up | 340.51 | 1.37E−125 |
| hsa-miR-663b | Up | 208.55 | 1.53E−27 |
| hsa-miR-4516 | Up | 156.42 | 2.16E−22 |
| hsa-miR-4532 | Up | 133.15 | 9.88E−22 |
| hsa-miR-1913 | Up | 120.21 | 6.09E−18 |
| hsa-miR-3621 | Up | 115.92 | 9.22E−16 |
| hsa-miR-6730-3p | Up | 95.54 | 3.41E−15 |
| hsa-miR-4492 | Up | 85.40 | 1.08E−14 |
| hsa-miR-6800-3p | Up | 78.41 | 4.11E−12 |
| hsa-miR-6087 | Up | 73.40 | 1.19E−11 |
| hsa-miR-1292-3p | Up | 65.85 | 9.49E−21 |
| hsa-miR-3687 | Up | 58.50 | 3.22E−16 |
| hsa-miR-6858-3p | Up | 46.37 | 1.17E−08 |
| hsa-miR-4449 | Up | 41.62 | 1.92E−14 |
| hsa-miR-663a | Up | 37.51 | 1.79E−07 |
| hsa-miR-885-3p | Up | 32.61 | 6.28E−36 |
| hsa-miR-7641 | Up | 32.51 | 4.84E−13 |
| hsa-miR-4488 | Up | 31.76 | 1.51E−06 |
| hsa-miR-3656 | Up | 30.96 | 8.46E−10 |
| hsa-miR-1470 | Up | 25.14 | 4.34E−18 |
| hsa-miR-4497 | Up | 23.67 | 2.36E−06 |
| hsa-miR-6858-5p | Up | 23.14 | 1.64E−14 |
| hsa-miR-4508 | Up | 21.92 | 9.26E−25 |
| hsa-miR-3182 | Up | 17.61 | 9.72E−05 |
| hsa-miR-3654 | Up | 16.42 | 3.86E−04 |
| hsa-miR-4477a | Up | 16.14 | 1.83E−06 |
| hsa-miR-664b-5p | Up | 13.57 | 1.17E−08 |
| hsa-miR-6821-5p | Up | 11.83 | 2.70E−03 |
| hsa-miR-6808-3p | Up | 11.18 | 2.65E−03 |
| hsa-miR-4758-5p | Up | 11.05 | 2.74E−04 |
| hsa-miR-664a-5p | Up | 10.88 | 1.77E−04 |
| hsa-miR-922 | Up | 10.60 | 4.21E−03 |
| hsa-miR-6834-3p | Up | 10.51 | 4.65E−03 |
| hsa-miR-4710 | Up | 9.17 | 8.41E−03 |
| hsa-miR-6892-5p | Up | 8.79 | 9.80E−03 |
| hsa-miR-204-5p | Up | 8.54 | 2.24E−04 |
| hsa-miR-582-5p | Up | 7.94 | 1.17E−02 |
| hsa-miR-1268b | Up | 7.59 | 4.39E−03 |
| hsa-miR-551a | Up | 7.17 | 1.05E−02 |
| hsa-miR-4417 | Up | 6.76 | 2.73E−02 |
| hsa-miR-668-5p | Up | 6.72 | 2.77E−02 |
| hsa-miR-483-3p | Up | 6.65 | 4.19E−03 |
| hsa-miR-8072 | Up | 6.59 | 1.92E−02 |
| hsa-miR-137 | Up | 6.07 | 3.80E−02 |
| hsa-miR-30d-3p | Up | 5.61 | 4.11E−02 |
| hsa-miR-15a-3p | Up | 5.59 | 1.38E−03 |
| hsa-miR-483-5p | Up | 5.58 | 2.96E−02 |
| hsa-miR-5701 | Up | 5.57 | 3.43E−02 |
| hsa-miR-146a-5p | Up | 5.41 | 2.06E−09 |
| hsa-miR-1908-3p | Up | 5.31 | 2.02E−02 |
| hsa-miR-412-5p | Up | 5.30 | 4.14E−02 |
| hsa-miR-1227-3p | Up | 5.23 | 2.20E−02 |
| hsa-miR-380-5p | Up | 5.03 | 1.27E−02 |
| hsa-miR-147b | Up | 4.91 | 6.80E−08 |
| hsa-miR-138-5p | Up | 4.61 | 4.34E−02 |
| hsa-miR-7704 | Up | 4.13 | 7.64E−08 |
| hsa-miR-668-3p | Up | 3.73 | 4.21E−03 |
| hsa-miR-376b-3p | Up | 3.66 | 2.06E−15 |
| hsa-miR-222-3p | Up | 3.60 | 3.85E−18 |
| hsa-miR-3117-3p | Up | 3.55 | 7.65E−03 |
| hsa-miR-665 | Up | 3.50 | 3.88E−08 |
| hsa-miR-1248 | Up | 3.42 | 5.00E−04 |
| hsa-miR-376a-3p | Up | 3.30 | 4.66E−08 |
| hsa-miR-675-3p | Up | 3.23 | 2.19E−02 |
| hsa-miR-1307-5p | Up | 3.04 | 1.61E−10 |
| hsa-let-7f-1-3p | Down | −8.20 | 7.85E−03 |
| hsa-miR-23b-5p | Down | −7.90 | 9.65E−03 |
| hsa-let-7b-3p | Down | −6.78 | 4.86E−07 |
| hsa-miR-335-3p | Down | −6.17 | 2.96E−02 |
| hsa-miR-194-5p | Down | −5.53 | 4.41E−02 |
| hsa-miR-3178 | Down | −5.39 | 2.22E−02 |
| hsa-miR-34c-5p | Down | −5.31 | 2.45E−05 |
| hsa-miR-187-3p | Down | −5.25 | 7.60E−06 |
| hsa-let-7d-3p | Down | −5.21 | 9.63E−04 |
| hsa-miR-132-5p | Down | −4.64 | 4.17E−02 |
| hsa-miR-10a-5p | Down | −4.28 | 1.66E−33 |
| hsa-miR-10b-5p | Down | −3.55 | 1.35E−20 |
| hsa-miR-98-3p | Down | −3.44 | 2.91E−02 |
| hsa-miR-23b-3p | Down | −3.22 | 4.62E−22 |
| hsa-miR-335-5p | Down | −3.11 | 1.53E−03 |
| hsa-miR-1260b | Down | −3.07 | 2.17E−02 |
amiRNAs up-regulated or down-regulated in PANC-1 cells persistently infected with CVB4 compared to uninfected cells are listed
bFold change of miRNA expression in PANC-1 cells persistently infected with CVB4 compared to uninfected cells
cAdjusted p value calculated between PANC-1 cells persistently infected with CVB4 and uninfected cells obtained with DESeq2 (based on negative binomial generalized linear models)
Fig. 1.
Changes of miRNA expression in PANC-1 cells persistently infected with CVB4. a Expression of selected miRNAs expressed as fold changes of persistently infected compared to uninfected control cells. Data of four independent biological replicates are shown as mean ± standard error obtained from sequencing data analyzed by DESeq2. b Expression of selected miRNAs determined by RT qPCR expressed as fold changes of persistently infected compared to uninfected control cells. Data of six independent biological replicates are shown except for hsa-miR-6087 and hsa-miR-4516, where data of two independent biological replicates are shown as mean ± standard error of the mean
We next investigated the potential impact of the miRNA expression changes induced by persistent CVB4 infection on genes that are known to be implicated in T1D. To this end, miRNA target prediction was performed using miRWalk and target genes were restricted to the 55 T1D risk genes contained in the T1D database [23]. Forty-nine of the fifty-five T1D risk genes were predicted as putative targets of at least one of the dysregulated miRNAs (Table 2). Most of the genes were potentially targeted by several of the dysregulated miRNAs, and the genes targeted by the highest number of dysregulated miRNAs (n = 24) were AFF3 and BACH2. Most of the dysregulated miRNAs were also predicted to target several of the T1D risk genes (Table 2).
Table 2.
Type 1 diabetes risk genes predicted as targets of the 81 dysregulated miRNAs
| Type 1 diabetes risk gene | Up-regulation | Down-regulation | Number of miRNAsa |
|---|---|---|---|
| AFF3 | hsa-miR-4492, hsa-miR-6087, hsa-miR-6800-3p, hsa-miR-4417, hsa-miR-4710, hsa-miR-582-5p, hsa-miR-1227-3p, hsa-miR-138-5p, hsa-miR-146a-5p, hsa-miR-222-3p, hsa-miR-30d-3p, hsa-miR-380-5p, hsa-miR-1227-3p, hsa-miR-222-3p, hsa-miR-30d-3p, hsa-miR-1248, hsa-miR-665, hsa-miR-675-3p | hsa-let-7b-3p, hsa-let-7f-1-3p, hsa-miR-194-5p, hsa-miR-23b-3p, hsa-miR-335-3p, hsa-miR-98-3p | 24 |
| BACH2 | hsa-miR-4492, hsa-miR-4516, hsa-miR-663b, hsa-miR-885-3p, hsa-miR-204-5p, hsa-miR-582-5p, hsa-miR-922, hsa-miR-137, hsa-miR-147b, hsa-miR-3117-3p, hsa-miR-380-5p, hsa-miR-483-3p, hsa-miR-5701, hsa-miR-668-3p, hsa-miR-137, hsa-miR-147b, hsa-miR-380-5p, hsa-miR-483-3p, hsa-miR-1248 | hsa-miR-23b-3p, hsa-miR-23b-5p, hsa-miR-335-3p, hsa-miR-335-5p, hsa-miR-34c-5p | 24 |
| BAD | hsa-miR-4449, hsa-miR-663a | hsa-miR-132-5p, hsa-miR-34c-5p | 4 |
| C1QTNF6 | hsa-miR-4492, hsa-miR-4516, hsa-miR-1913, hsa-miR-3656, hsa-miR-4532, hsa-miR-6087, hsa-miR-663a, hsa-miR-663b, hsa-miR-6858-5p | hsa-miR-132-5p, hsa-miR-23b-5p | 11 |
| CCR5 | hsa-miR-15a-3p, hsa-miR-204-5p, hsa-miR-3182, hsa-miR-6800-3p, hsa-miR-6834-3p | hsa-miR-335-3p, hsa-miR-335-3p | 7 |
| CD226 | hsa-miR-204-5p | hsa-miR-335-3p | 2 |
| CD69 | hsa-miR-23b-3p | 1 | |
| CLEC16A | hsa-miR-4492, hsa-miR-1248, hsa-miR-138-5p, hsa-miR-1913, hsa-miR-1914-3p, hsa-miR-222-3p, hsa-miR-3182, hsa-miR-380-5p, hsa-miR-4488, hsa-miR-4497, hsa-miR-4508, hsa-miR-4710, hsa-miR-4758-5p, hsa-miR-483-3p, hsa-miR-663b, hsa-miR-664a-5p, hsa-miR-6800-3p, hsa-miR-922 | hsa-miR-3178, hsa-miR-335-3p | 20 |
| COBL | hsa-miR-3117-3p, hsa-miR-664a-5p, hsa-miR-668-3p, hsa-miR-6808-3p | hsa-miR-23b-3p | 5 |
| CTLA4 | hsa-miR-1248, hsa-miR-664a-5p | hsa-miR-23b-5p, hsa-miR-335-3p | 4 |
| CTSH | hsa-miR-4710 | 1 | |
| CYP27B1 | hsa-miR-146a-5p, hsa-miR-15a-3p | hsa-miR-335-3p | 3 |
| DEXI | hsa-miR-137, hsa-miR-15a-3p, hsa-miR-3182, hsa-miR-4497, hsa-miR-6800-3p | hsa-miR-1260b, hsa-miR-23b-5p | 7 |
| EFR3B | hsa-miR-1268b, hsa-miR-15a-3p, hsa-miR-1913, hsa-miR-4492, sa-miR-4516, hsa-miR-6087, hsa-miR-663a, hsa-miR-664a-5p, hsa-miR-664b-5p, hsa-miR-665, hsa-miR-6730-3p, hsa-miR-6858-5p, hsa-miR-922 | hsa-miR-10a-5p, hsa-miR-10b-5p, hsa-miR-1260b, hsa-miR-34c-5p | 17 |
| ERBB3 | hsa-miR-137, hsa-miR-204-5p, hsa-miR-3182, hsa-miR-4492, hsa-miR-4516, hsa-miR-664a-5p, hsa-miR-665 | hsa-miR-335-5p | 8 |
| FUT2 | hsa-miR-1268b, hsa-miR-664a-5p, hsa-miR-675-3p | hsa-miR-1260b, hsa-miR-23b-5p | 5 |
| GLIS3 | hsa-miR-137, hsa-miR-138-5p, hsa-miR-146a-5p, hsa-miR-204-5p, hsa-miR-3117-3p, hsa-miR-3182, hsa-miR-380-5p, hsa-miR-4710, hsa-miR-582-5p, hsa-miR-665 | hsa-let-7b-3p, hsa-let-7f-1-3p, hsa-miR-335-3p, hsa-miR-98-3p | 14 |
| GPR183 | hsa-miR-582-5p | 1 | |
| GSDMB | hsa-miR-34c-5p | 1 | |
| HLA-A | hsa-miR-6800-3p | hsa-miR-23b-3p | 2 |
| HLA-B | hsa-miR-30d-3p, hsa-miR-4417, hsa-miR-6800-3p | hsa-miR-23b-3p | 4 |
| HLA-DQB1 | hsa-miR-137, hsa-miR-1913, hsa-miR-30d-3p, hsa-miR-6858-3p, hsa-miR-7641 | hsa-miR-335-3p | 6 |
| HLA-DRB1 | hsa-miR-15a-3p | 1 | |
| IKZF1 | hsa-miR-137, hsa-miR-138-5p, hsa-miR-146a-5p, hsa-miR-1470, hsa-miR-15a-3p, hsa-miR-3117-3p, hsa-miR-3182, hsa-miR-3654, hsa-miR-376a-3p, hsa-miR-376b-3p, hsa-miR-380-5p, hsa-miR-4488, hsa-miR-4516, hsa-miR-4710, hsa-miR-483-3p, hsa-miR-582-5p, hsa-miR-663a, hsa-miR-6858-3p, hsa-miR-665, hsa-miR-664a-5p | hsa-miR-3178, hsa-miR-335-3p, hsa-miR-34c-5p | 23 |
| IKZF4 | hsa-miR-137, hsa-miR-146a-5p, hsa-miR-1913, hsa-miR-1914-3p, hsa-miR-30d-3p, hsa-miR-3117-3p, hsa-miR-3182, hsa-miR-4516, hsa-miR-4710, hsa-miR-483-3p, hsa-miR-582-5p, hsa-miR-663b, hsa-miR-665, hsa-miR-6858-3p, hsa-miR-6858-5p, hsa-miR-885-3p, hsa-miR-922 | hsa-miR-10a-5p, hsa-miR-10b-5p, hsa-miR-34c-5p, hsa-miR-98-3p | 21 |
| IL10 | hsa-miR-3182, hsa-miR-4492 | hsa-miR-194-5p, hsa-miR-23b-5p | 4 |
| IL21 | hsa-miR-194-5p | 1 | |
| IL27 | hsa-miR-30d-3p, hsa-miR-4492 | 2 | |
| IL2RA | hsa-miR-1248, hsa-miR-138-5p, hsa-miR-1913, hsa-miR-1914-3p, hsa-miR-204-5p, hsa-miR-3182, hsa-miR-6808-3p, hsa-miR-922 | 8 | |
| IL7R | hsa-miR-1227-3p, hsa-miR-1248, hsa-miR-137, hsa-miR-138-5p, hsa-miR-146a-5p, hsa-miR-1913, hsa-miR-1914-3p, hsa-miR-204-5p, hsa-miR-3182, hsa-miR-483-3p, hsa-miR-5701, hsa-miR-665 | hsa-miR-194-5p, hsa-miR-23b-3p, hsa-miR-335-3p | 15 |
| INS | hsa-miR-3656, hsa-miR-8072 | 2 | |
| ITGB7 | hsa-miR-668-3p | 1 | |
| NAA25 | hsa-miR-15a-3p, hsa-miR-3656, hsa-miR-6800-3p, hsa-miR-922 | hsa-let-7b-3p, hsa-let-7f-1-3p, hsa-miR-194-5p, hsa-miR-23b-5p, hsa-miR-335-5p | 9 |
| NRP1 | hsa-miR-1248, hsa-miR-137, sa-miR-1914-3p, hsa-miR-204-5p, hsa-miR-3654, hsa-miR-376a-3p, hsa-miR-376b-3p, hsa-miR-4492, hsa-miR-483-3p, hsa-miR-5701, hsa-miR-582-5p, hsa-miR-668-3p | hsa-miR-194-5p, hsa-miR-335-3p, hsa-miR-98-3p | 15 |
| ORMDL3 | hsa-miR-15a-3p, hsa-miR-1913, hsa-miR-204-5p, hsa-miR-3182, hsa-miR-4516, hsa-miR-4710, hsa-miR-663b, hsa-miR-665, hsa-miR-6800-3p | hsa-miR-23b-3p, hsa-miR-34c-5p | 11 |
| PHTF1 | hsa-miR-7641 | 1 | |
| PRKCQ | hsa-miR-137, hsa-miR-30d-3p, hsa-miR-4488, hsa-miR-4492, hsa-miR-4516, hsa-miR-665, hsa-miR-668-5p, hsa-miR-7641 | hsa-miR-34c-5p | 9 |
| PTPN2 | hsa-miR-137, hsa-miR-138-5p, hsa-miR-1913, hsa-miR-4417, hsa-miR-582-5p, hsa-miR-675-3p, hsa-miR-7641 | hsa-miR-194-5p, hsa-miR-23b-5p, hsa-miR-335-3p | 10 |
| PTPN22 | hsa-miR-30d-3p | hsa-miR-335-5p | 2 |
| RAC2 | hsa-miR-146a-5p, hsa-miR-1914-3p, hsa-miR-4492, hsa-miR-922 | 4 | |
| RASGRP1 | hsa-miR-146a-5p, hsa-miR-222-3p, hsa-miR-30d-3p, hsa-miR-376a-3p, hsa-miR-582-5p | hsa-let-7f-1-3p | 6 |
| RBM17 | hsa-miR-1248, hsa-miR-15a-3p | hsa-miR-23b-3p | 3 |
| RNLS | hsa-miR-137, hsa-miR-204-5p, hsa-miR-222-3p, hsa-miR-4516 | hsa-miR-194-5p, hsa-miR-335-3p | 6 |
| SH2B3 | hsa-miR-1248, hsa-miR-138-5p, sa-miR-147b, hsa-miR-1913, hsa-miR-204-5p, hsa-miR-30d-3p, hsa-miR-3656, hsa-miR-380-5p, hsa-miR-4417, hsa-miR-668-3p, hsa-miR-6858-3p, hsa-miR-6892-5p | hsa-miR-10a-5p, hsa-miR-10b-5p, hsa-miR-3178 | 15 |
| STAT4 | hsa-miR-1227-3p, hsa-miR-30d-3p, hsa-miR-3182, hsa-miR-6858-5p | hsa-miR-335-3p | 5 |
| TAGAP | hsa-miR-6858-5p | hsa-miR-335-3p | 2 |
| TNFAIP3 | hsa-miR-1248, hsa-miR-15a-3p, hsa-miR-1914-3p, hsa-miR-204-5p, hsa-miR-30d-3p, hsa-miR-380-5p, hsa-miR-4477a, hsa-miR-4516, hsa-miR-664a-5p, hsa-miR-6808-3p, hsa-miR-6858-3p, hsa-miR-885-3p, hsa-miR-922 | hsa-miR-23b-3p, hsa-miR-335-3p | 15 |
| TYK2 | hsa-miR-4710, hsa-miR-922 | 1 | |
| UBASH3A | hsa-miR-138-5p, hsa-miR-4492 | hsa-miR-132-5p, hsa-miR-23b-3p | 4 |
Type 1 diabetes risk genes are shown with the miRNAs that are predicted to target them and that have been found up-regulated or down-regulated by persistent CVB4 infection in the current study
aThe total number of miRNAs found dysregulated in the current study that are predicted to target the type 1 diabetes risk genes
mRNA levels of the house-keeping gene beta-Actin and selected T1D risk genes were measured by RT qPCR in PANC-1 cells with persistent CVB4 infection and uninfected PANC-1 cells. Beta-Actin mRNA levels were similar in persistently infected and uninfected cells (mean difference of 0.5 Cts, data not shown). mRNA levels of AFF3 and IL7R were significantly higher in persistently infected cells, whereas BACH2 levels were significantly lower (Fig. 2). The mRNA levels of the other analyzed genes were similar in infected and uninfected cells.
Fig. 2.
mRNA expression of T1D risk genes in PANC-1 cells persistently infected with CVB4. mRNA expression of selected T1D risk genes was measured by RT qPCR and expressed as fold changes in cells persistently infected with CVB4 compared to uninfected control cells. Data of five independent biological replicates are shown as mean ± standard error of the mean. **p < 0.01
Functional annotation analysis of the 49 putative target genes was studied with DAVID [26]. “Autoimmune thyroid disease” showed that the highest enrichment score and genes associated with viral myocarditis and T1D were also enriched. Most functional annotation terms that were enriched in these 49 putative target genes were related to the immune response or autoimmunity (Table 3).
Table 3.
Functional annotation analysis of 49 T1D risk genes that are potential targets of 81 dysregulated miRNAs
| Functional annotation term | p value |
|---|---|
| Autoimmune thyroid disease | 4.66E−06 |
| Allograft rejection | 2.83E−05 |
| Cell adhesion molecules | 4.10E−05 |
| Type I diabetes mellitus | 5.27E−05 |
| Viral myocarditis | 4.14E−04 |
| Graft-versus-host disease | 9.60E−04 |
| Jak-STAT signaling pathway | 9.66E−04 |
| MHC class II receptor activity | 1.24E−03 |
| Intestinal immune network for IgA production | 1.87E−03 |
| Growth factor binding | 3.02E−03 |
| Antigen processing and presentation | 8.32E−03 |
| Asthma | 9.38E−03 |
| Endocytosis | 1.33E−02 |
| T cell receptor signaling pathway | 1.70E−02 |
| Cytokine binding | 3.66E−02 |
| Cytokine-cytokine receptor interaction | 4.21E−02 |
| MHC class I receptor activity | 4.61E−02 |
The 49 type 1 diabetes risk genes that were potential targets of the 81 dysregulated miRNAs were functionally annotated using DAVID 6.7. Functional annotations that are enriched with an enrichment p value <0.05 are shown
Discussion
miRNAs are major regulators of gene expression and are involved in many physiological and pathological processes. Inhibition of miRNA production by disruption of the gene Dicer leads to development of diabetes in mice [27]. Disruption of Dicer leads to altered islet morphology, marked decreased beta-cell mass, and reduced insulin production [27]. Several miRNAs have been described to be involved in pancreas development, glucose sensing, insulin production, and diabetes [9, 28, 29]. miRNA expression changes induced by persistent enterovirus infection could thus impact on the function or regeneration of pancreatic beta-cells.
We, therefore, investigated the miRNA expression profile changes associated with persistent CVB4 infection in a human pancreatic cell line (PANC-1). To our knowledge, our study is the first to describe the impact of persistent enterovirus infection on miRNA expression in pancreatic cells. As far as acute enterovirus infection is concerned, two studies have analyzed the impact of CVB infection on pancreatic miRNA expression [16, 30]. Kim et al. reported a dysregulation of 33 miRNAs out of a total 754 analyzed in human pancreatic islets during acute CVB5 infection [16], and Lam et al. investigated the impact of acute CVB4 infection on miRNA expression in rat beta-cells [30]. When we compared the miRNAs that were reported to be dysregulated in these two studies with the list of 81 dysregulated miRNAs identified in the study presented here, there was no overlap with the study by Lam et al. and only one miRNA was dysregulated in our study and the study by Kim et al.: hsa-miR-663b. The function and targets of hsa-miR-663b are unknown, and to date, no reports on its involvement in viral infection or T1D have been published. The small overlap of dysregulated miRNAs between our study and the two previous studies might partly be due to methodological differences of the miRNA expression analysis [31] and/or the use of different cells. Furthermore, the effect on cellular miRNA expression may depend on the virus strain. The small overlap between the miRNA expression changes identified in our study and the two previous studies also demonstrate that acute CVB infection has a different impact on the cellular miRNA expression profile and thus on the resulting gene expression changes than persistent CVB infection. Concerning persistent CVB infection, a recent study compared miRNA expression profiles in endomyocardial biopsies of patients with CVB3 cardiomyopathy who spontaneously eliminated CVB3 with those who had virus persistence on follow-up. 16 miRNAs were differentially expressed and were proposed as markers for the risk of CVB3 persistence in CVB3 cardiomyopathy [32]. Interestingly, none of these 16 miRNAs was found dysregulated in our study, underlining again that the changes in miRNA expression profiles are cell-specific and virus-specific.
MiRNAs play an important role in the regulation of insulin expression and glucose homeostasis. Hsa-miR-204 is highly expressed in beta-cells and overexpressed in diabetes. Its overexpression reduces insulin expression in human islets by targeting a key insulin transcription factor, MafA [33]. We found that hsa-miR-204 is induced by persistent CVB4 infection (Table 1). Induction of this miRNA by persistent CVB4 infection may, therefore, reduce insulin expression and contribute to the insulin deficiency observed in T1D. The let-7 family is also involved in the control of glucose homeostasis and insulin sensitivity [34]. Several members of the let-7 family were found down-regulated in our study (Table 1), suggesting that persistent CVB4 infection may have an impact on glucose homeostasis. Hsa-miR-10a-5p was also found down-regulated during persistent CVB4 infection in our study (Table 1; Fig. 1a). The mouse homolog of this miRNA is expressed to a lower level in regulatory T cells of non-obese diabetic mice (NOD) as compared to autoimmunity-resistant mice [35] suggesting a potential link of this miRNA to autoimmunity. The link of the miRNAs dysregulated during persistent CVB4 infection to autoimmunity is further supported by the results of the functional annotation analysis, because most of the enriched functional annotation terms were related to the immune response or autoimmunity (Table 3). Hsa-miR-10a-5p was also shown to be up-regulated during differentiation of human induced pluripotent stem cells to insulin-producing cells [36]. The down-regulation of hsa-miR-10a-5p that we observed during persistent CVB4 infection (Table 1) might thus be implicated in the impaired formation of islet-like cell aggregates of PANC-1 cells reported previously [8]. A recent study found that EV-71 infection induced expression of miR-146a in human cells and in a mouse model. MiR-146a was further shown to target TRAF6 and IRAK1 [37]. Inhibition of mir-146a restored IRAK1 and TRAF6 expression, increased IFN-α production, inhibited viral propagation, and improved mouse survival. The authors concluded that miR-146a facilitates enteroviral pathogenesis by suppressing IFN production [37]. Interestingly, proinflammatory cytokines induce miR-146a in MIN6 cells and human islets via the NfκB pathway [38, 39]. Moreover, induction of miR-146 promotes beta-cell apoptosis, while miR-146 inhibition reduces beta-cell death [38, 40]. Expression of this miRNA is increased in islets of prediabetic NOD mice [38]. Taken together, these findings suggest that overexpression of miR-146a during persistent CVB4 infection as shown in the current study (Table 1; Fig. 1) might link enteroviral infection to the development of T1D. In agreement with this hypothesis, hsa-miR-146a has been reported to be dysregulated in the plasma of prediabetic patients and in PBMCs of patients with T1D [41].
Persistent CVB4 infection induced dysregulation of 81 miRNAs in pancreatic cells (Table 1). A single miRNA can potentially regulate hundreds of genes [42]; therefore, the changes in miRNA expression are expected to have a strong impact on the transcriptome and biology of these cells. Importantly, the miRNAs dysregulated by persistent CVB4 infection were found to potentially target several T1D risk genes (Table 2). Indeed, gene expression of selected genes investigated by RT qPCR analysis confirmed dysregulation of AFF3, BACH2, and IL7R in PANC-1 cells persistently infected with CVB4 (Fig. 2), whereas the other investigated genes showed similar mRNA expression. However, the expression of these genes might be impacted at the protein level.
Basic leucine zipper transcription factor 2 (BACH2) is a key regulation factor in B and T cell differentiation and function [43]. BACH2 is also involved in the regulation of apoptosis. Inhibition of BACH2 sensitized beta-cells to cytokine-induced apoptosis [44]. BACH2 is a predicted target of 24 of the dysregulated miRNAs in our study and dysregulation of its expression has been found by RT qPCR (Table 2; Fig. 2). Cells persistently infected with CVB4 might, therefore, be more susceptible to apoptosis.
AF4/FMR2 family member 3 (AFF3) is a nuclear transcriptional activator that is preferentially expressed in lymphoid tissue [45]. AFF3 polymorphisms have been found associated with autoimmune diseases [46]. AFF3 is a predicted target of 24 of the dysregulated miRNAs and dysregulation of its expression has been found by RT qPCR (Table 2; Fig. 2).
Interleukin-7 receptor (IL7R) is indispensable for normal lymphocyte development and IL7R polymorphisms have been found associated with autoimmune diseases [47, 48]. Binding of IL7 to the IL7R activates multiple pathways that regulate lymphocyte survival, glucose uptake, proliferation, and differentiation [49]. Blocking of IL7R in non-obese diabetic (NOD) mice prevented autoimmune diabetes and reversed disease in new-onset diabetic mice by modulation of effector/memory T cell function [50]. The soluble form of IL7R antagonizes IL7 signaling and is increased in patients at the onset of T1D [51]. IL7R is a predicted target of 15 of the dysregulated miRNAs and dysregulation of its expression has been found by RT qPCR in our study (Table 2; Fig. 2). Given that dysregulation of IL7/IL7R signaling is involved in the development of T1D, our results suggest that persistent CVB4 infection has an impact on IL7/IL7R signaling in pancreatic cells that might be linked to the development of T1D.
Our previous studies showed that during persistent CVB4 infection, approximately 50% of cells harbored CVB4 RNA and 1–5% cells were VP1 positive [8]. Therefore, it is noteworthy, that although only approximately 50% of the cells are infected during persistent infection, expression of some miRNAs and mRNAs changed strongly. In fact, miRNAs can be secreted and taken up by surrounding cells and these miRNAs can be functional [52, 53]. Therefore, it is possible that the persistently infected cells influence the miRNA expression profile of surrounding non-infected cells and consequently also their gene expression profile.
In conclusion, our study presents the first characterization of the miRNA expression profile changes induced by persistent CVB4 infection in pancreatic cells. The predicted targeting of genes involved in the immune response and autoimmunity by the dysregulated miRNAs as well as the dysregulated expression of the diabetes risk genes AFF3, IL7R, and BACH2 show that persistent CVB4 infection profoundly impacts the host cell and link persistent CVB4 infection to the development of T1D. To fully understand the impact of miRNA dysregulation induced by persistent CVB4 infection on the cell physiology, it will be interesting to study the global gene and protein expression profiles of these cells using transcriptomic and proteomic approaches.
Acknowledgements
This work was supported by Ministère de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche, Université Lille 2 (Equipe d’accueil 3610) and Centre Hospitalier Régional et Universitaire de Lille, and by EU FP7 (GA-261441-PEVNET): Persistent virus infection as a cause of pathogenic inflammation in type 1 diabetes—an innovative research program of biobanks and expertise).
Abbreviations
- CVB
Type B coxsackieviruses
- CVB4
Coxsackievirus B4
- Ct
Cycle threshold
- FCS
Fetal calf serum
- miRNA
MicroRNA
- T1D
Type 1 diabetes
Compliance with ethical standards
Conflict of interest
The authors declare that no conflict of interest exists.
References
- 1.Hober D, Sauter P. Pathogenesis of type 1 diabetes mellitus: interplay between enterovirus and host. Nat Rev Endocrinol. 2010;6(5):279–289. doi: 10.1038/nrendo.2010.27. [DOI] [PubMed] [Google Scholar]
- 2.Yeung WC, Rawlinson WD, Craig ME. Enterovirus infection and type 1 diabetes mellitus: systematic review and meta-analysis of observational molecular studies. BMJ. 2011;342:d35. doi: 10.1136/bmj.d35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alidjinou EK, Sane F, Engelmann I, Geenen V, Hober D. Enterovirus persistence as a mechanism in the pathogenesis of type 1 diabetes. Discov Med. 2014;18(100):273–282. [PubMed] [Google Scholar]
- 4.Dotta F, Censini S, van Halteren AG, Marselli L, Masini M, Dionisi S, Mosca F, Boggi U, Muda AO, Prato SD, Elliott JF, Covacci A, Rappuoli R, Roep BO, Marchetti P. Coxsackie B4 virus infection of beta cells and natural killer cell insulitis in recent-onset type 1 diabetic patients. Proc Natl Acad Sci USA. 2007;104(12):5115–5120. doi: 10.1073/pnas.0700442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hober D, Sane F. Enteroviral pathogenesis of type 1 diabetes. Discov Med. 2010;10(51):151–160. [PubMed] [Google Scholar]
- 6.Jaidane H, Sauter P, Sane F, Goffard A, Gharbi J, Hober D. Enteroviruses and type 1 diabetes: towards a better understanding of the relationship. Rev Med Virol. 2010;20(5):265–280. doi: 10.1002/rmv.647. [DOI] [PubMed] [Google Scholar]
- 7.Pinkert S, Klingel K, Lindig V, Dorner A, Zeichhardt H, Spiller OB, Fechner H. Virus-host coevolution in a persistently coxsackievirus B3-infected cardiomyocyte cell line. J Virol. 2011;85(24):13409–13419. doi: 10.1128/JVI.00621-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sane F, Caloone D, Gmyr V, Engelmann I, Belaich S, Kerr-Conte J, Pattou F, Desailloud R, Hober D. Coxsackievirus B4 can infect human pancreas ductal cells and persist in ductal-like cell cultures which results in inhibition of Pdx1 expression and disturbed formation of islet-like cell aggregates. Cell Mol Life Sci. 2013;70(21):4169–4180. doi: 10.1007/s00018-013-1383-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandez-Valverde SL, Taft RJ, Mattick JS. MicroRNAs in beta-cell biology, insulin resistance, diabetes and its complications. Diabetes. 2011;60(7):1825–1831. doi: 10.2337/db11-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filios SR, Shalev A. Beta-cell microRNAs: small but powerful. Diabetes. 2015;64(11):3631–3644. doi: 10.2337/db15-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91(3):827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 13.Ho BC, Yu SL, Chen JJ, Chang SY, Yan BS, Hong QS, Singh S, Kao CL, Chen HY, Su KY, Li KC, Cheng CL, Cheng HW, Lee JY, Lee CN, Yang PC. Enterovirus-induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe. 2011;9(1):58–69. doi: 10.1016/j.chom.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Tong L, Lin L, Wu S, Guo Z, Wang T, Qin Y, Wang R, Zhong X, Wu X, Wang Y, Luan T, Wang Q, Li Y, Chen X, Zhang F, Zhao W, Zhong Z. MiR-10a* up-regulates coxsackievirus B3 biosynthesis by targeting the 3D-coding sequence. Nucleic Acids Res. 2013;41(6):3760–3771. doi: 10.1093/nar/gkt058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wen BP, Dai HJ, Yang YH, Zhuang Y, Sheng R. MicroRNA-23b inhibits enterovirus 71 replication through downregulation of EV71 VPl protein. Intervirology. 2013;56(3):195–200. doi: 10.1159/000348504. [DOI] [PubMed] [Google Scholar]
- 16.Kim KW, Ho A, Alshabee-Akil A, Hardikar AA, Kay TW, Rawlinson WD, Craig ME. Coxsackievirus B5 infection induces dysregulation of microRNAs predicted to target known type 1 diabetes risk genes in human pancreatic islets. Diabetes. 2016;65(4):996–1003. doi: 10.2337/db15-0956. [DOI] [PubMed] [Google Scholar]
- 17.Alidjinou EK, Engelmann I, Bossu J, Villenet C, Figeac M, Romond MB, Sane F, Hober D. Persistence of coxsackievirus B4 in pancreatic ductal-like cells results in cellular and viral changes. Virulence. 2017 doi: 10.1080/21505594.2017.1284735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon JW, Austin M, Onodera T, Notkins AL. Isolation of a virus from the pancreas of a child with diabetic ketoacidosis. N Engl J Med. 1979;300(21):1173–1179. doi: 10.1056/NEJM197905243002102. [DOI] [PubMed] [Google Scholar]
- 19.Chen CJ, Servant N, Toedling J, Sarazin A, Marchais A, Duvernois-Berthet E, Cognat V, Colot V, Voinnet O, Heard E, Ciaudo C, Barillot E. ncPRO-seq: a tool for annotation and profiling of ncRNAs in sRNA-seq data. Bioinformatics. 2012;28(23):3147–3149. doi: 10.1093/bioinformatics/bts587. [DOI] [PubMed] [Google Scholar]
- 20.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dweep H, Gretz N, Sticht C. miRWalk database for miRNA-target interactions. Methods Mol Biol. 2014;1182:289–305. doi: 10.1007/978-1-4939-1062-5_25. [DOI] [PubMed] [Google Scholar]
- 23.Burren OS, Adlem EC, Achuthan P, Christensen M, Coulson RM, Todd JA. T1DBase: update 2011, organization and presentation of large-scale data sets for type 1 diabetes research. Nucleic Acids Res. 2011;39(Database issue)):D997–D1001. doi: 10.1093/nar/gkq912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8(9):R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Q, Xiao Z, He F, Zou J, Wu S, Liu Z. MicroRNAs regulate the pathogenesis of CVB3-induced viral myocarditis. Intervirology. 2013;56(2):104–113. doi: 10.1159/000343750. [DOI] [PubMed] [Google Scholar]
- 26.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 27.Kalis M, Bolmeson C, Esguerra JL, Gupta S, Edlund A, Tormo-Badia N, Speidel D, Holmberg D, Mayans S, Khoo NK, Wendt A, Eliasson L, Cilio CM. Beta-cell specific deletion of Dicer1 leads to defective insulin secretion and diabetes mellitus. PLoS One. 2011;6(12):e29166. doi: 10.1371/journal.pone.0029166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, German MS. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56(12):2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 29.Isaacs SR, Wang J, Kim KW, Yin C, Zhou L, Mi QS, Craig ME. MicroRNAs in type 1 diabetes: complex interregulation of the immune system, beta cell function and viral infections. Curr Diabetes Rep. 2016;16(12):133. doi: 10.1007/s11892-016-0819-2. [DOI] [PubMed] [Google Scholar]
- 30.Lam WY, Cheung AC, Tung CK, Yeung AC, Ngai KL, Lui VW, Chan PK, Tsui SK. miR-466 is putative negative regulator of coxsackie virus and adenovirus receptor. FEBS Lett. 2015;589(2):246–254. doi: 10.1016/j.febslet.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 31.Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, Cheo D, D’Andrade P, DeMayo M, Dennis L, Derveaux S, Feng Y, Fulmer-Smentek S, Gerstmayer B, Gouffon J, Grimley C, Lader E, Lee KY, Luo S, Mouritzen P, Narayanan A, Patel S, Peiffer S, Ruberg S, Schroth G, Schuster D, Shaffer JM, Shelton EJ, Silveria S, Ulmanella U, Veeramachaneni V, Staedtler F, Peters T, Guettouche T, Wong L, Vandesompele J. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014;11(8):809–815. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- 32.Kuehl U, Lassner D, Gast M, Stroux A, Rohde M, Siegismund C, Wang X, Escher F, Gross M, Skurk C, Tschoepe C, Loebel M, Scheibenbogen C, Schultheiss HP, Poller W. Differential cardiac microRNA expression predicts the clinical course in human enterovirus cardiomyopathy. Circ Heart Fail. 2015;8(3):605–618. doi: 10.1161/CIRCHEARTFAILURE.114.001475. [DOI] [PubMed] [Google Scholar]
- 33.Xu G, Chen J, Jing G, Shalev A. Thioredoxin-interacting protein regulates insulin transcription through microRNA-204. Nat Med. 2013;19(9):1141–1146. doi: 10.1038/nm.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci USA. 2011;108(52):21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeker LT, Zhou X, Gershberg K, de Kouchkovsky D, Morar MM, Stadthagen G, Lund AH, Bluestone JA. MicroRNA 10a marks regulatory T cells. PLoS One. 2012;7(5):e36684. doi: 10.1371/journal.pone.0036684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sebastiani G, Valentini M, Grieco GE, Ventriglia G, Nigi L, Mancarella F, Pellegrini S, Martino G, Sordi V, Piemonti L, Dotta F. MicroRNA expression profiles of human iPSCs differentiation into insulin-producing cells. Acta Diabetol. 2016 doi: 10.1007/s00592-016-0955-9. [DOI] [PubMed] [Google Scholar]
- 37.Ho BC, Yu IS, Lu LF, Rudensky A, Chen HY, Tsai CW, Chang YL, Wu CT, Chang LY, Shih SR, Lin SW, Lee CN, Yang PC, Yu SL. Inhibition of miR-146a prevents enterovirus-induced death by restoring the production of type I interferon. Nat Commun. 2014;5:3344. doi: 10.1038/ncomms4344. [DOI] [PubMed] [Google Scholar]
- 38.Roggli E, Britan A, Gattesco S, Lin-Marq N, Abderrahmani A, Meda P, Regazzi R. Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic beta-cells. Diabetes. 2010;59(4):978–986. doi: 10.2337/db09-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lovis P, Roggli E, Laybutt DR, Gattesco S, Yang JY, Widmann C, Abderrahmani A, Regazzi R. Alterations in microRNA expression contribute to fatty acid-induced pancreatic beta-cell dysfunction. Diabetes. 2008;57(10):2728–2736. doi: 10.2337/db07-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seyhan AA, Nunez Lopez YO, Xie H, Yi F, Mathews C, Pasarica M, Pratley RE. Pancreas-enriched miRNAs are altered in the circulation of subjects with diabetes: a pilot cross-sectional study. Sci Rep. 2016;6:31479. doi: 10.1038/srep31479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 43.Richer MJ, Lang ML, Butler NS. T cell fates zipped up: how the Bach2 basic leucine zipper transcriptional repressor directs T cell differentiation and function. J Immunol. 2016;197(4):1009–1015. doi: 10.4049/jimmunol.1600847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marroqui L, Santin I, Dos Santos RS, Marselli L, Marchetti P, Eizirik DL. BACH2, a candidate risk gene for type 1 diabetes, regulates apoptosis in pancreatic beta-cells via JNK1 modulation and crosstalk with the candidate gene PTPN2. Diabetes. 2014;63(7):2516–2527. doi: 10.2337/db13-1443. [DOI] [PubMed] [Google Scholar]
- 45.Ma C, Staudt LM. LAF-4 encodes a lymphoid nuclear protein with transactivation potential that is homologous to AF-4, the gene fused to MLL in t(4;11) leukemias. Blood. 1996;87(2):734–745. [PubMed] [Google Scholar]
- 46.Wallace C, Rotival M, Cooper JD, Rice CM, Yang JH, McNeill M, Smyth DJ, Niblett D, Cambien F, Tiret L, Todd JA, Clayton DG, Blankenberg S. Statistical colocalization of monocyte gene expression and genetic risk variants for type 1 diabetes. Hum Mol Genet. 2012;21(12):2815–2824. doi: 10.1093/hmg/dds098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weber F, Fontaine B, Cournu-Rebeix I, Kroner A, Knop M, Lutz S, Muller-Sarnowski F, Uhr M, Bettecken T, Kohli M, Ripke S, Ising M, Rieckmann P, Brassat D, Semana G, Babron MC, Mrejen S, Gout C, Lyon-Caen O, Yaouanq J, Edan G, Clanet M, Holsboer F, Clerget-Darpoux F, Muller-Myhsok B. IL2RA and IL7RA genes confer susceptibility for multiple sclerosis in two independent European populations. Genes Immun. 2008;9(3):259–263. doi: 10.1038/gene.2008.14. [DOI] [PubMed] [Google Scholar]
- 48.Mazzucchelli RI, Riva A, Durum SK. The human IL-7 receptor gene: deletions, polymorphisms and mutations. Semin Immunol. 2012;24(3):225–230. doi: 10.1016/j.smim.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmer MJ, Mahajan VS, Trajman LC, Irvine DJ, Lauffenburger DA, Chen J. Interleukin-7 receptor signaling network: an integrated systems perspective. Cell Mol Immunol. 2008;5(2):79–89. doi: 10.1038/cmi.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Penaranda C, Kuswanto W, Hofmann J, Kenefeck R, Narendran P, Walker LS, Bluestone JA, Abbas AK, Dooms H. IL-7 receptor blockade reverses autoimmune diabetes by promoting inhibition of effector/memory T cells. Proc Natl Acad Sci USA. 2012;109(31):12668–12673. doi: 10.1073/pnas.1203692109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monti P, Brigatti C, Krasmann M, Ziegler AG, Bonifacio E. Concentration and activity of the soluble form of the interleukin-7 receptor alpha in type 1 diabetes identifies an interplay between hyperglycemia and immune function. Diabetes. 2013;62(7):2500–2508. doi: 10.2337/db12-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Horizontal transfer of microRNAs: molecular mechanisms and clinical applications. Protein Cell. 2012;3(1):28–37. doi: 10.1007/s13238-012-2003-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu X, Odenthal M, Fries JW. Exosomes as miRNA carriers: formation-function-future. Int J Mol Sci. 2016 doi: 10.3390/ijms17122028. [DOI] [PMC free article] [PubMed] [Google Scholar]