Abstract
DNA methylation is the major epigenetic modification and it is involved in the negative regulation of gene expression. Its alteration can lead to neoplastic transformation. Several biomolecular approaches are nowadays used to study this modification on DNA, but also on RNA molecules, which are known to play a role in different biological processes. RNA methylation is one of the most common RNA modifications and 5-methylcytosine presence has recently been suggested in mRNA. However, an analysis of nucleic acid methylation at electron microscope is still lacking. Therefore, we visualized DNA methylation status and RNA methylation sites in the interphase nucleus of HeLa cells and rat hepatocytes by ultrastructural immunocytochemistry and cytochemical staining. This approach represents an efficient alternative to study nucleic acid methylation. In particular, this ultrastructural method makes the visualization of this epigenetic modification on a single RNA molecule possible, thus overcoming the technical limitations for a (pre-)mRNA methylation analysis.
Keywords: 5mC, mRNA methylation, Methylation gradient, Epigenetics, EM immunolabelling, Electron microscopy
Introduction
In eukaryotes DNA methylation is the major epigenetic modification. It is mediated by DNA-methyltransferases (DNMTs) at position 5 of cytosine followed by guanosine (CpG island) and induces transcriptional gene silencing. This modification is involved in regulation of gene expression, X-chromosome inactivation, genomic imprinting and chromosome stability [1, 2]. It also plays an essential role in mammalian embryonic development [3]. Alterations of DNA methylation are known to occur in several diseases [2], especially in cancer [4].
Different methods have been developed for the discrimination of CpG methylation status [5]. Current approaches can be essentially divided in separation techniques [6] and restriction endonuclease-based [7] or bisulfite conversion-based methods [8]. Moreover, DNA microarray technology [9–11] and sequencing-based approaches [12, 13] allow to obtain a complete epigenetic profile. Global DNA methylation levels can also be detected at fluorescence microscopy [14, 15]. Recently, methylation-specific fluorescence in situ hybridization (MeFISH) was developed for the visualization of DNA methylation at specific sequences [16]. Although an electron microscope (EM) determination of DNA methylation was carried out on plant programmed cell death (PCD) process [17], an EM analysis of DNA methylation dynamics and distribution is still lacking.
Several methods for DNA methylation analysis have been adapted to detect modified nucleotides on RNAs [18] and, to our knowledge, no data are available at EM about RNA methylation. The role of RNAs as important factors in different biological processes has become even clearer [19]. About 100 post-transcriptional modifications were found [20] among which RNA methylation is one of the most commonly, occurring in different types of RNAs [21]. In particular, 5-methylcytosine (5mC) is widely studied: placed in the variable region and in the anticodon loop, it stabilizes tRNA secondary structure affecting Mg2+ binding and is involved in codon recognition [22]; in rRNA it seems to be implicated in tRNA identification and peptidyl-transferase activity [23]. 5mC has only been recently suggested to be localized in the untranslated regions of mRNA [22]. Its function is still unknown: due to the proximity of mRNA methylation sites to the binding ones of Argonaute protein (the central component of miRNA/RISC complex), mRNA 5mC could be involved in miRNA degradation pathway [24] but there is no evidence about methylation involvement in mRNA stability [22].
Here, we propose an analysis of nucleic acid methylation at transmission electron microscope (TEM) to visualize DNA methylation status and RNA methylation sites in cell interphase nuclei at the ultrastructural level. This study has been carried out on different tissue and cell models. Particularly, in liver samples the peripheral condensed chromatin regions are generally evident allowing to better analyse DNA 5mC distribution. For the analysis of RNA methylation, proliferating HeLa cells were also chosen for their high transcription level.
Materials and methods
Cells, tissues and treatments
Cells in vitro
HeLa cells were grown in Dulbecco’s Minimal Essential Medium (DMEM) supplemented with 10% foetal bovine serum, 1% glutamine, 100 U/mL penicillin and streptomycin at 37 °C in a 5% CO2 humidified atmosphere.
For electron microscopy, the cells were grown in 25 cm2 plastic flasks, detached by a mild trypsinization and fixed with 4% paraformaldehyde in the culture medium for 2 h at 4 °C to allow for a good preservation of antigen integrity. The cells were then centrifuged at 2000 rpm for 10 min and rinsed thoroughly with phosphate buffered saline (PBS). They were incubated in 0.5 M NH4Cl in PBS for 30 min at room temperature (RT) to block free aldehyde groups and rinsed again with several changes of PBS. The cell pellets were pre-embedded in 2% Agar in H2O, dehydrated in graded ethanol and embedded in LRWhite resin. Thin sections of 70–80 nm were obtained with a Reichert OM3 ultramicrotome and collected on formvar-carbon-coated nickel grids (200 mesh).
Tissues
Samples from rat liver were fixed with 4% paraformaldehyde in Sörensen phosphate buffer pH 7.2 for 2 h at 4 °C and then processed for ultrastructural histochemistry, as described above.
Treatments
To label transcribed RNA, some cell samples were incubated with 5 mM Fluoro-Uridine (FU; Sigma-Aldrich) for 15 min at 37 °C [25, 26]. These samples were then processed for ultrastructural cytochemistry, as previously described.
EM ultrastructural analysis
EM immunocytochemistry
The grids were floated on normal goat serum (NGS) diluted 1:50 in PBS for 5 min at RT and incubated with mouse monoclonal or rabbit polyclonal anti-5mC antibody (GeneTex, GT4111; GeneTex, GTX128455) overnight at 4 °C. The primary antibody was diluted 1:500 in PBS containing 0.1% Bovine Serum Albumin (BSA) and 0.05% Tween 20. The samples were rinsed with PBS-Tween two times for 5 min and equally with PBS. After incubation in NGS, the grids were treated with the specific secondary antibody (Jackson ImmunoResearch) coupled with colloidal gold of 12 nm diluted 1:20 in PBS for 30 min at RT. The sections were rinsed with PBS for 5 min twice and then with H2O.
Some sections were double labelled. In addition to anti-5mC labelling, a rat anti-FU antibody (Techno Genetics) and a chicken anti-hnRNPs antibody (courtesy of Dr. T. Martin) binding to hnRNP core proteins [27] were used. The primary antibodies were diluted 1:10 and 1:500, respectively, in PBS/BSA/TWEEN20. Moreover, a mouse anti-7 methylguanosine (7mG; courtesy of Dr. R. Lührmann) [28] was used to recognize the 5′ cap of mRNA: it was diluted 1:200 in PBS/BSA/TWEEN20. For this double labelling a rabbit anti-5mC antibody diluted 1:500 in PBS/BSA/TWEEN20 was used. The anti-5mC was recognized by the specific secondary antibody coupled with colloidal gold of 6 nm (Jackson ImmunoResearch) while other antigens were revealed with 12 nm specific secondary antibodies (Jackson ImmunoResearch).
As a control of the specificity, some grids were incubated in parallel in PBS/BSA/Tween20 mixture from which the primary antibody was excluded and then processed as above.
As a further control, some grids were incubated with both DNase (500 U/mL) and RNase (1 mg/mL) or RNase alone (1 mg/mL) for 2 h at 37 °C and Proteinase K (PK; 1 mg/mL) for 15 min at 37 °C.
EM in situ hybridization
Electron microscope in situ hybridization (EMISH) was performed to recognize the poly(A) tail of mRNA. The sections were incubated with a pre-hybridization solution containing 20% baker RNA, 20% dextran and 4× saline sodium citrate (SSC) for 15 min at RT to allow the following hybridization for 3 h at 37 °C. The hybridization mixture was prepared by adding biotin-labelled poly-d(T) probe (Sigma-Aldrich) to the pre-hybridization solution to give a final concentration of 1 μM of the oligonucleotide. The grids were floated onto 4× SSC for 5 min two times at 37 °C; stringency washings were done in 4× SSC, 2× SSC and 1× SSC at RT. The incubation with NGS (1:100 in PBS) for 3 min was followed by the anti-biotin antibody coupled with 10-nm colloidal gold (Aurion) for 30 min at RT (1:10 in PBS). The samples were rinsed with PBS and H2O several times.
Immuno-labelling using anti-5mC antibody was performed on these samples, as described above, identifying the 5mC by a 6 nm gold secondary antibody.
Staining procedures
Sections were stained for ribonucleoproteins (RNPs) or nucleic acids with one of the following procedures:
regressive EDTA technique for RNPs [29]: the grids were incubated in uranyl acetate for 2 min, in EDTA for 30 s to remove uranyl from DNA and finally in lead citrate for other 2 min;
terbium citrate for RNA [30, 31]: the specimens were floated on terbium citrate drops for 30 min and quickly washed in H2O for 10 and then 5 s—this staining method gives a very low contrast despite its accuracy;
osmium ammine for DNA [32, 33]: the sections were hydrolyzed with 5 N HCl for 30 min, washed with H2O several times, incubated in osmium ammine (Polysciences, Inc.) for 1 h and rinsed thoroughly with H2O.
After the enzymatic digestion the specimens were stained with uranyl acetate (2 min) and lead citrate (2 min) only.
All the samples were observed on a Zeiss EM900 electron microscope operating at 80 kV.
Statistical analysis
The statistical analysis was performed to confirm the results of enzymatic digestions. Ten nuclei were selected showing similarities in size and condensed chromatin areas. The operator counted the gold grains on the condensed chromatin regions in hepatocyte nuclei without any treatment and after PK digestion or DNase and RNase digestion.
The data were organized and analyzed in Excel; a t test was carried out between the untreated sample and each sample after enzymatic digestion.
Results
DNA methylation analysis
EM immuno-gold labelling of thin sections from rat liver showed the localization of 5mC on the condensed chromatin. The areas of condensed chromatin near the nuclear envelope are bleached by the EDTA staining technique. The labelling is mainly present near the surface of chromatin, facing the inner part of the nucleus (Fig. 1a). An abundant 5mC labelling is also detectable on the nucleolus associated chromatin (Fig. 1b). Few gold grains are visible on the nucleolus itself. After specific DNA staining with osmium ammine and 5mC labelling, the signal was more clearly visible on chromatin areas both at the periphery of the nucleus (Fig. 1c) and surrounding the nucleolus (Fig. 1d). The scanty labelling on the nucleolus can be referred to methylated DNA fibres (arrow) or possibly to methylated rRNA, in the cases where no DNA is visible nearby.
Fig. 1.
a, b The samples are stained with EDTA regressive technique for RNPs. 5mC labelling is localized on condensed chromatin region, delimited by the hatching, near the nuclear envelope (a) and around the nucleolus (b). A constant 5mC labelling is also present in the perichromatin region where transcription normally occurs. Rat liver; 150 and 250 nm, respectively. c, d After the specific DNA staining with osmium ammine, 5mC labelling is observed more clearly on condensed DNA nearby the nuclear envelope (c) and around the nucleolus (d). In d, some gold grains inside the nucleolus and lying on thin DNA fibres (arrow) have been detected. Rat liver; 200 and 150 nm, respectively
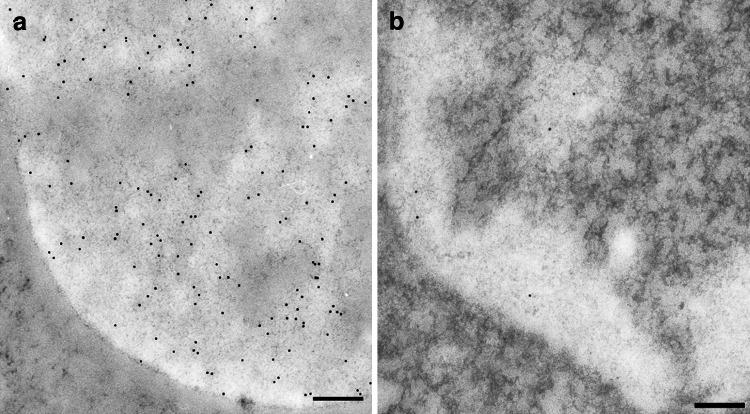
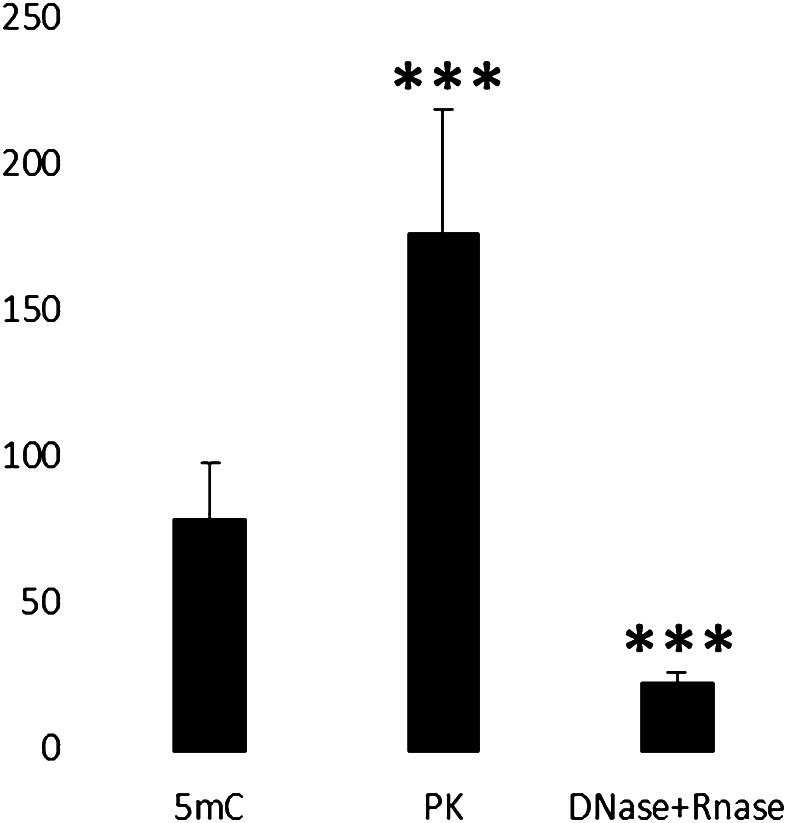
To further confirm the localization of the 5mC immunolabelling on DNA, the results of enzymatic digestions were considered. The signal continued to be present after PK digestion (Fig. 2a), thus demonstrating that proteins did not contribute to the labelling but their removal seemed to increase the yield. This could be due to the unmasking of epitopes before covered by proteins. The labelling pattern, however, remained the same, i.e., more present at the chromatin surface. The immunopositivity, on the contrary, drastically decreased from chromatin areas after digestion of both DNA and RNA (Fig. 2b). The statistical analysis confirmed the significant immuno-gold labelling increase after PK digestion and reduction after DNase and RNase treatment (Fig. 3).
Fig. 2.
a Digestion by proteinase K was carried out to demonstrate that proteins did not contribute to the labelling: in fact, the 5mC signal continues to be present after the removal of proteins, reproducing the distribution also in the perichromatin regions. Heterochromatin appears unstained. Rat liver; 200 nm. b After removal of both DNA and RNA, the labelling is significantly decreased on chromatin and in the perichromatin region, confirming that the signal depends on DNA but also on nascent RNA molecules confined in the sites of active transcription. Mouse liver; 200 nm
Fig. 3.
The statistical analysis of the number of gold grains of the 5mC immuno-gold labelling on condensed chromatin areas reveals a significant difference in the hepatocyte nuclei before and after the specific enzymatic digestion. The PK digestion significantly increases the immuno-gold labelling thus demonstrating not only that proteins do not contribute to the signal but also that the yield of the immuno-reaction could be increased removing chromatin associated proteins (p < 0.01). After the removal of both DNA and RNA the gold grains are significantly reduced on the condensed chromatin regions confirming that the signal depends on DNA (p < 0.01)
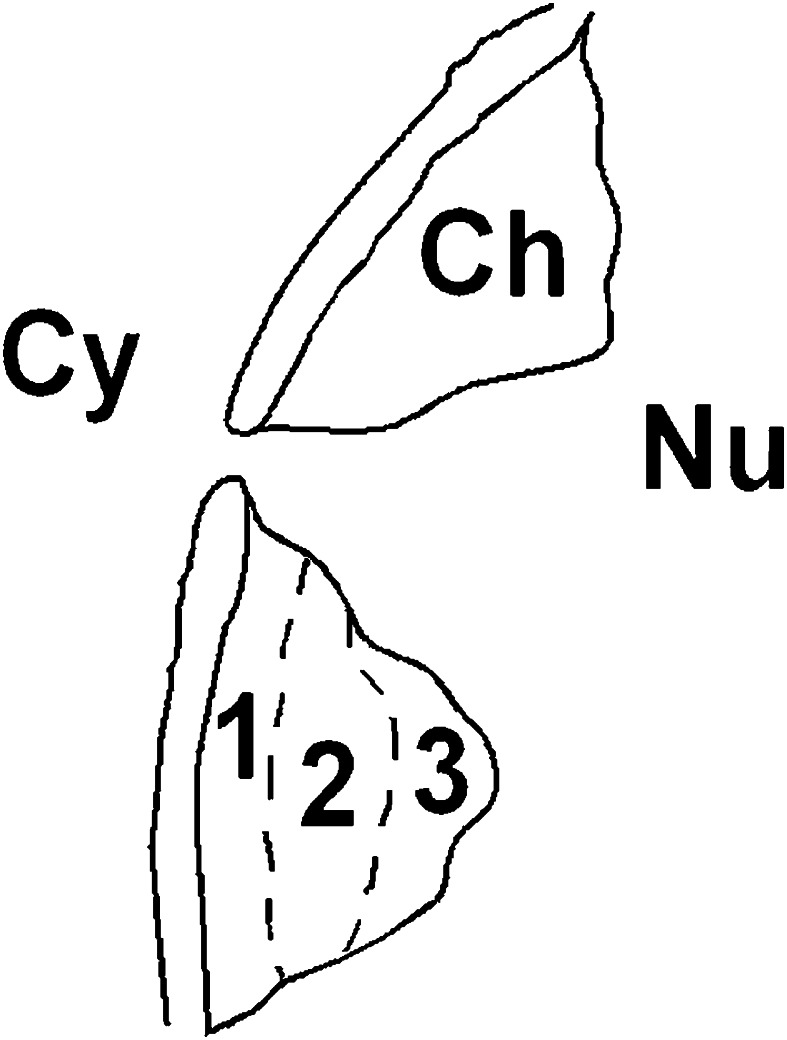
When considering the EM feature of a cell nucleus, according to the data in literature [34], one can arbitrarily subdivide the condensed chromatin areas in three regions (Fig. 4): zone 1_peripheral region near the nuclear envelope; zone 2_a central region; zone 3_inner peripheral region toward the interchromatin space. The analysis of hepatocyte nuclei seemed to reveal a particular distribution of 5mC labelling, changing from the surface of condensed chromatin to the nuclear envelope. The 5mC labelling was generally abundant on the zone 3 while its density gradually decreased towards the nuclear envelope (zone 1), where the signal was often absent (Fig. 5).
Fig. 4.
The condensed chromatin areas are arbitrarily subdivided in three regions according to EM feature of a cell nucleus. Zone 1: it is the peripheral region nearby the nuclear envelope. Zone 2: this is the central region of condensed chromatin area, changing the zone 1 in the last region. Zone 3: the inner region toward the interchromatin space represents the superficial part of the condensed chromatin area; it is surrounded by the perichromatin region. The artwork was realized by Paint Shop Pro 7
Fig. 5.
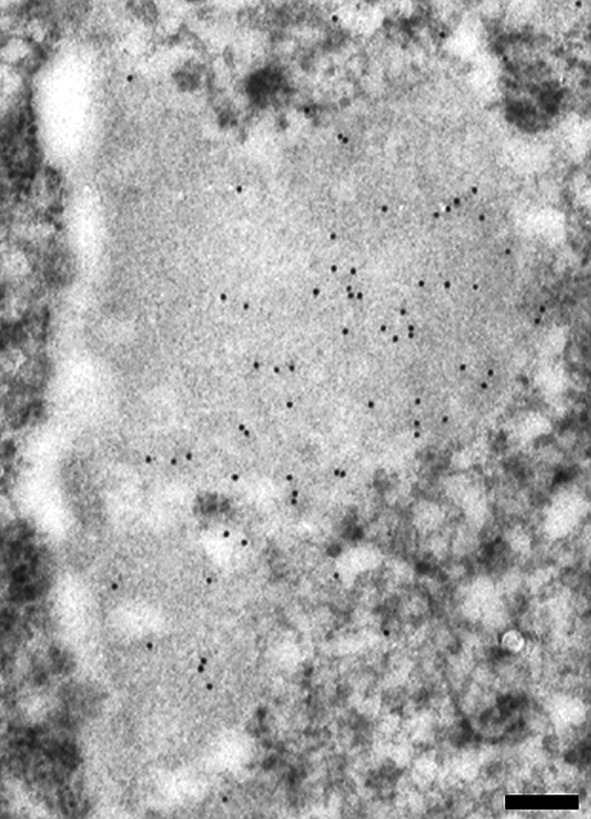
5mC labelling on the condensed chromatin changes moving towards the nuclear envelope. The chromatin appears less stained after EDTA technique. At the surface of the heterochromatin (zone 3), the gold grains (12 nm) are abundant gradually decreasing in the zone 2; nearby the nuclear envelope (zone 1) they are almost absent. Rat liver; 250 nm
5mC detection on RNA fibrils and RNA-containing granules
Interestingly, we found a constant and not negligible 5mC signal at the border of heterochromatin areas (Fig. 1a), in the so called perichromatin region where transcription by RNA polymerase II (Pol II) normally occurs [35]. 5mC labelling in the perichromatin region was difficult to detect after a RNase treatment alone (not shown) and in combination with DNase (Fig. 2b).
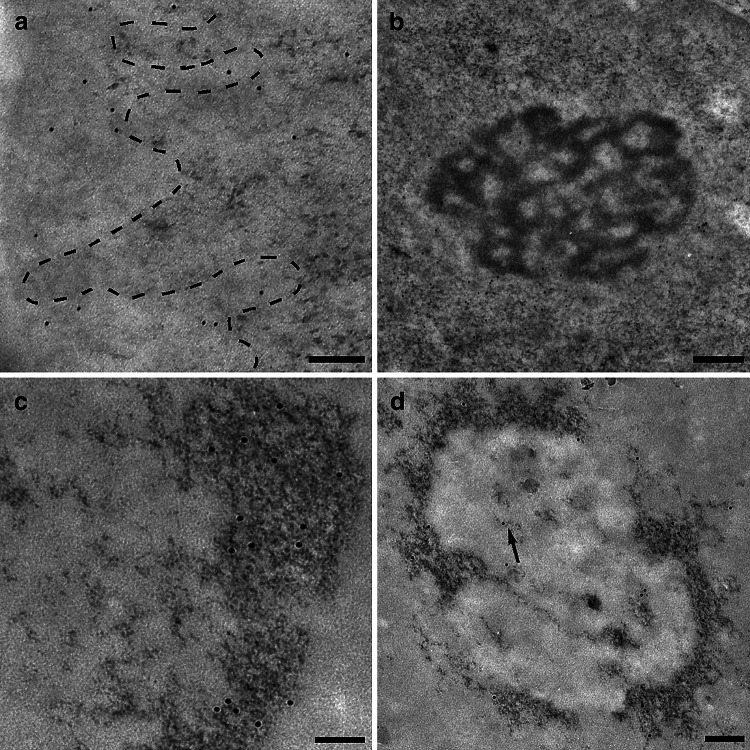
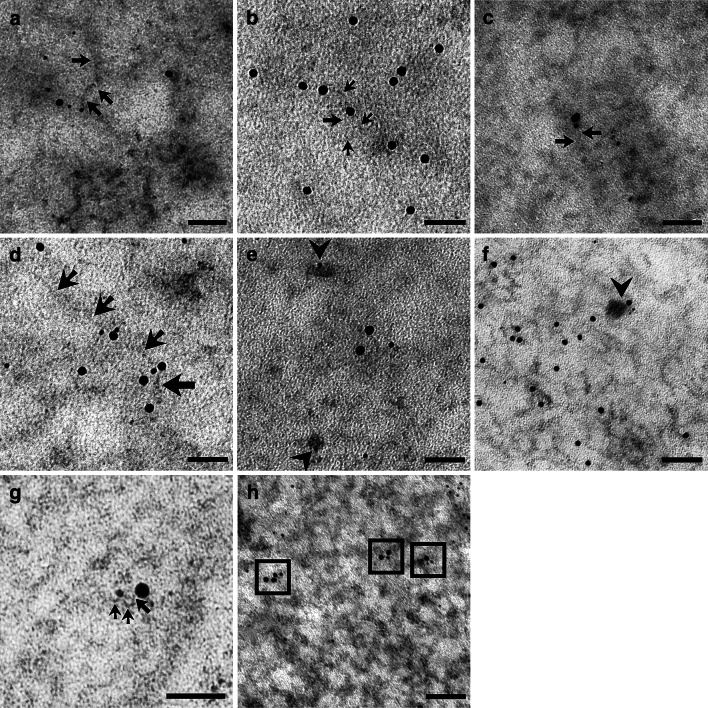
After EDTA staining, RNPs were preferentially contrasted. In Fig. 6a, the gold labelling for 5mC was present over RNA fibrils. When terbium staining was used, only RNA is contrasted in the specimen. 5mC can be labelled on the stained RNA fibril (Fig. 6b). To further confirm the presence of 5mC on nascent RNA fibrils, we performed an immunolabelling after an RNA precursor incorporation. In Fig. 6c, a double labelling for 5mC and FU is shown. Several double labelled PF were found, thus showing that RNA methylation can occur precociously. Moreover, double labelling for 5mC and hnRNP core proteins was found to be present on PF (Fig. 6d): hnRNPs are considered markers of PF as in situ forms of nascent transcripts [36]. Despite the presence of 5mC on nascent RNA fibrils, the modified nucleotide was also detected on perichromatin granules (PG; Fig. 6e). To corroborate our finding, double labelled PG for 5mC and hnRNPs were also found several times (Fig. 6f). PG are considered to be a form of stored mRNA leaving the nucleus later [37]. We found labelled PF both near the nuclear pore and in the cytoplasm close to ribosomes (not shown). These data could suggest that this modification is not only an early event, but it remains during the RNA fibril lifespan.
Fig. 6.
a 6 nm gold labels 5mC on RNA fibril stained with EDTA technique (12 nm gold grains label FU): PF are indicated by arrows. HeLa cell; 50 nm. b After terbium staining, 5mC is revealed by a 12 nm secondary antibody on PF (arrows). HeLa cell; 50 nm. c After EDTA staining, FU (12 nm) and 5mC (6 nm) colocalize on a nascent RNA fibril, indicated by arrows, suggesting methylation as an early modification on a nascent transcript. HeLa cell; 50 nm. d Terbium stained PF (arrows) are labelled both by anti-hnRNPs antibody (12 nm) and anti-5mC antibody (6 nm). HeLa cell; 50 nm. e The arrowheads show the 6 nm 5mC signal on PG, stained specifically with terbium citrate. As forms of (pre-)mRNA, labelled PG confirm the methylation of this subset of RNA. HeLa cell; 50 nm. f PG double labelled for both 5mC and hnRNPs were found. Rat liver; 50 nm. g A perichromatin fibril (arrows) is detected in the perichromatin region near the nuclear pore by EDTA staining: it is labelled both by anti-5mC (6 nm) and anti-7mG (12 nm) demonstrating that (pre-)mRNA could be methylated. Rat liver; 50 nm. h 5mC immunopositivity is detected on poly-adenylated RNA fibrils: in the squares 5mC is labelled by 6 nm gold while poly(A) tail by 12 nm grain. This colocalization seems to confirm mRNA methylation. HeLa cell; 50 nm
RNA perichromatin fibrils were double labelled for 5mC and 7-methylguanosine (Fig. 6g). Even if this capping is present on other RNA products of Pol II, it is considered specific for mRNA. Finally, the poly(A) tail was recognized by in situ hybridization while 5mC was labelled immunocytochemically: the poly(A) tail and 5mC frequently colocalized (Fig. 6h). As well as 7mG, poly(A) tail is one of mRNA marker despite its presence on other Pol II products. Taken together, 7mG and poly(A) labelling strongly suggest (pre-)mRNA as the target.
Discussion
This paper describes a different approach for the analysis of nucleic acid methylation. So far, to our knowledge, no data are available on the detection at electron microscope of DNA methylation and especially of RNA modification.
The EM analysis of DNA methylation represents an effective method for characterizing 5mC distribution on DNA. This analysis also allows its localization in the different regions of condensed chromatin. Since the latter are characterized by a different functionality in terms of transcription, 5mC detection could be of interest in correlating DNA modifications, chromatin structure and chromatin functionality. Although until now this modification was considered to directly silence genes involving DNA-binding proteins [38, 39] and to lead to a more compact chromatin structure [40], recent works have revealed that 5mC increases DNA stiffness through the restriction of the conformational fluctuations caused by the methyl group. This fact explains the suppression of DNA looping by 5mC and the consequent loosening of DNA ends around a nucleosome [41]. Moreover, other works have excluded that this modification is the predominant pathway for gene silencing [42]. Here, we showed that the more peripheral condensed regions of chromatin, transcriptionally inactive, are almost unlabelled for 5mC. The more superficial region, on the contrary, is the most labelled. This EM gradient distribution could be the starting point for elucidating the role of 5mC in chromatin condensation/functionality in the new light of what previously cited in the literature.
Finally, EM immunolabelling could be used to detect different level of DNA methylation through a semi-quantitative analysis. In this view, a DNA methylation analysis at TEM could be conducted in tumour cell lines to verify the possible effect of a specific treatment.
As for RNA, we have found 5mC labelled and terbium stained fibrils in the perichromatin region, in close proximity to the condensed chromatin area where transcription occurs [35]. This prompted us to hypothesize that 5mC modification could be an early event. To confirm this, we used FU incorporation to label nascent RNA and hnRNP core proteins labelling as a marker of PF. The identification of double labelled and RNA-specifically stained PF supports that RNA methylation is not only an early event but probably occurs cotranscriptionally.
Since in this thin perichromatin region the RNAs are synthesized by Pol II [35], we decided to verify if it could be possible to detect 5mC presence on (pre-)mRNA by TEM because this modified nucleotide was recently detected on mRNA [24]. Nascent PF were co-labelled for 5mC and 7mG and poly-adenylated RNA fibrils were also methylated. The spatial localization of PF, the double labelling and the relative abundance of each Pol II product strongly suggest that the majority of the 5mC-labelled PF pertain to (pre-)mRNA.
Moreover, we found 5mC labelling on PG which are known as forms of (pre-)mRNA [37] and to contain poly-adenylated RNAs [43]. They are present in all cells and most probably represent a storage form of (pre-)mRNA which will leave the nucleus at a later time [37]. In this view, RNA methylation is also a long-lasting RNA modification. This data is supported by the finding of labelled PF in close proximity of the nuclear pore or ribosomes in the cytoplasm which might indicate that methylated RNAs could be exported from the nucleus whereby this modification remains present during the total RNA lifespan.
In conclusion, this TEM study allows the detailed visualization of a single RNA molecule verifying its possible epigenetic modification via ultrastructural immunocytochemistry. This approach represents an in situ biomolecular characterization of RNA methylation at high resolution level which could have several applications in epigenetic research.
Acknowledgements
The authors are grateful for the technical assistance of Ms. Francine Flach and Ms. Paola Veneroni and for the critical review of the manuscript of Prof. Carlo Pellicciari and Prof. Antonella Forlino.
Compliance with ethical standards
Funding
This work was supported by Fondi di Ateneo per la Ricerca (F.A.R. 2013–2014) from the University of Pavia.
Integrity of research
The experiments described in this manuscript comply with the current Italian laws.
Conflict of interest
The authors declare that no competing interests exist.
References
- 1.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/MCB.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 3.Li E, Bestor TH, Jaenisch R. Targeted mutation of DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- 4.Bird AP. The relationship of DNA methylation to cancer. Cancer Surv. 1996;28:87–101. [PubMed] [Google Scholar]
- 5.Harrison A, Parle-McDermott A. DNA methylation: a timeline of methods and applications. Front Genet. 2011;2:74. doi: 10.3389/fgene.2011.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gehrke CW, McCune RA, Gama-Sosa MA, Ehrlich M, Kuo KC. Quantitative reversed-phase high-performance liquid chromatography of major and modified nucleosides in DNA. J Chromatogr. 1984;301:199–219. doi: 10.1016/S0021-9673(01)89189-5. [DOI] [PubMed] [Google Scholar]
- 7.Bestor TH, Hellewell SB, Ingram VM. Differentiation of two mouse cell lines is associated with hypomethylation of their genomes. Mol Cell Biol. 1984;4:1800–1806. doi: 10.1128/MCB.4.9.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang TH, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Hum Mol Genet. 1999;8:459–470. doi: 10.1093/hmg/8.3.459. [DOI] [PubMed] [Google Scholar]
- 10.Gitan RS, Shi H, Chen CM, Yan PS, Huang TH. Methylation-specific oligonucleotide microarray: a new potential for high-throughput methylation analysis. Genome Res. 2002;12:158–164. doi: 10.1101/gr.202801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber M, Davies JJ, Wittig D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 12.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulfite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJ, Haussler D, Marra MA, Hirst M, Wang T, Costello JF. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santos F, Hendrich B, Reik W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 15.Kobayakawa S, Miike K, Nakao M, Abe K. Dynamic changes in the epigenomic state and nuclear organization of differentiating mouse embryonic stem cells. Genes Cells. 2007;12:447–460. doi: 10.1111/j.1365-2443.2007.01063.x. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Miyanari Y, Shirane K, Nitta H, Kubota T, Ohashi H, Okamoto A, Sasaki H. Sequence-specific microscopic visualization of DNA methylation status at satellite repeats in individual cell nuclei and chromosomes. Nucleic Acids Res. 2013;41:e186. doi: 10.1093/nar/gkt766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solís MT, Chakrabarti N, Corredor E. Epigenetic changes accompany developmental programmed cell death in tapetum cells. Plant Cell Physiol. 2014;55:16–29. doi: 10.1093/pcp/pct152. [DOI] [PubMed] [Google Scholar]
- 18.Hussain S, Aleksic J, Blanco S, Dietmann S, Frye M. Characterizing 5-methylcytosine in the mammalian epitranscriptome. Genome Biol. 2013;14:215. doi: 10.1186/gb4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Liu N, Pan T. RNA epigenetics. Transl Res. 2015;165:28–35. doi: 10.1016/j.trsl.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kellner S, Burhenne J, Helm M. Detection of RNA modifications. RNA Biol. 2010;7:237–247. doi: 10.4161/rna.7.2.11468. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Jia G. Methylation modifications in eukaryotic messenger RNA. J Genet Genomics. 2014;41:21–33. doi: 10.1016/j.jgg.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Motorin Y, Lyko F, Helm M. 5-Methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010;38:1415–1430. doi: 10.1093/nar/gkp1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Squires JE, Patel HR, Nousch M. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dundr M, Raska I. Nonisotopic ultrastructural mapping of transcription sites within the nucleolus. Exp Cell Res. 1993;208:275–281. doi: 10.1006/excr.1993.1247. [DOI] [PubMed] [Google Scholar]
- 26.Trentani A, Testillano PS, Risueño MC, Biggiogera M. Visualization of transcription sites at electron microscope. Eur J Histochem. 2003;47:195–200. doi: 10.4081/827. [DOI] [PubMed] [Google Scholar]
- 27.Jones RE, Okamura CS, Martin TE. Immunofluorescent localization of the proteins of nuclear ribonucleoprotein complexes. J Cell Biol. 1980;86:235–243. doi: 10.1083/jcb.86.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bochnig P, Reuter R, Bringmann P, Lührmann R. A monoclonal antibody against 2,2,7-trimethylguanosine that reacts with intact, class U, small nuclear ribonucleoproteins as well as with 7-methylguanosine-capped RNAs. Eur J Biochem. 1987;168:461–467. doi: 10.1111/j.1432-1033.1987.tb13439.x. [DOI] [PubMed] [Google Scholar]
- 29.Bernhard W. A new staining procedure for electron microscopical cytology. J Ultrastruct Res. 1969;27:250–265. doi: 10.1016/S0022-5320(69)80016-X. [DOI] [PubMed] [Google Scholar]
- 30.Biggiogera M, Fakan S. Fine structural specific visualization of RNA on ultrathin sections. J Histochem Cytochem. 1998;46:389–395. doi: 10.1177/002215549804600313. [DOI] [PubMed] [Google Scholar]
- 31.Biggiogera M, Masiello I. Visualizing RNA at electron microscopy by terbium citrate. In: Pellicciari C, Biggiogera M, editors. Histochemistry of single molecules. 1. Pavia: Springer; 2017. pp. 277–283. [DOI] [PubMed] [Google Scholar]
- 32.Vazquez-Nin GH, Biggiogera M, Echeverrìa OM. Activation of osmium ammine by SO2-generating chemicals for EM Feulgen-type staining of DNA. Eur J Histochem. 1995;39:101–106. [PubMed] [Google Scholar]
- 33.Masiello I, Biggiogera M. Osmium ammine for staining DNA in electron microsocopy. In: Pellicciari C, Biggiogera M, editors. Histochemistry of single molecules. 1. Pavia: Springer; 2017. pp. 261–267. [DOI] [PubMed] [Google Scholar]
- 34.Cmarko D, Verschure PJ, Otte AP, van Driel R, Fakan S. Polycomb group gene silencing proteins are concentrated in the perichromatin compartment of the mammalian nucleus. J Cell Sci. 2003;116:335–343. doi: 10.1242/jcs.00225. [DOI] [PubMed] [Google Scholar]
- 35.Cmarko D, Verschure PJ, Martin TE, Dahmus ME, Krause S, Fu XD, Van Driel R, Fakan S. Ultrastructural analysis of transcription and splicing in the cell nucleus after BrUTP-microinjection. Mol Biol Cell. 1999;10:211–223. doi: 10.1091/mbc.10.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fakan S. Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol. 1994;4:86–90. doi: 10.1016/0962-8924(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 37.Fakan S. The functional architecture of the nucleus as analysed by ultrastructural cytochemistry. Histochem Cell Biol. 2004;122:83–93. doi: 10.1007/s00418-004-0681-1. [DOI] [PubMed] [Google Scholar]
- 38.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. doi: 10.1126/science.1111098. [DOI] [PubMed] [Google Scholar]
- 39.Riggs AD. X chromosome inactivation, differentiation, and DNA methylation revisited, with a tribute to Susumu Ohno. Cytogenet Genome Res. 2002;99:17–24. doi: 10.1159/000071569. [DOI] [PubMed] [Google Scholar]
- 40.Geiman TM, Robertson KD. Chromatin remodeling, histone modifications, and DNA methylation-how does it all fit together? J Cell Biochem. 2002;87:117–125. doi: 10.1002/jcb.10286. [DOI] [PubMed] [Google Scholar]
- 41.Ngo TTM, Yoo J, Dai Q, Zhang Q, He C, Aksimentiev A. Effects of cytosine modifications on DNA flexibility and nucleosome mechanical stability. Nat Commun. 2016;7:10813. doi: 10.1038/ncomms10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Visa N, Puvion-Dutilleul F, Harper F, Bachellerie JP, Puvion E. Intranuclear distribution of poly(A) RNA determined by electron microscope in situ hybridization. Exp Cell Res. 1993;208:19–34. doi: 10.1006/excr.1993.1218. [DOI] [PubMed] [Google Scholar]