Abstract
The XPF/ERCC1 heterodimeric complex is essentially involved in nucleotide excision repair (NER), interstrand crosslink (ICL), and double-strand break repair. Defects in XPF lead to severe diseases like xeroderma pigmentosum (XP). Up until now, XP-F patient cells have been utilized for functional analyses. Due to the multiple roles of the XPF/ERCC1 complex, these patient cells retain at least one full-length allele and residual repair capabilities. Despite the essential function of the XPF/ERCC1 complex for the human organism, we successfully generated a viable immortalised human XPF knockout cell line with complete loss of XPF using the CRISPR/Cas9 technique in fetal lung fibroblasts (MRC5Vi cells). These cells showed a markedly increased sensitivity to UVC, cisplatin, and psoralen activated by UVA as well as reduced repair capabilities for NER and ICL repair as assessed by reporter gene assays. Using the newly generated knockout cells, we could show that human XPF is markedly involved in homologous recombination repair (HRR) but dispensable for non-homologous end-joining (NHEJ). Notably, ERCC1 was not detectable in the nucleus of the XPF knockout cells indicating the necessity of a functional XPF/ERCC1 heterodimer to allow ERCC1 to enter the nucleus. Overexpression of wild-type XPF could reverse this effect as well as the repair deficiencies.
Keywords: CRISPR/Cas9, Xeroderma pigmentosum group F, Nucleotide excision repair, Interstrand crosslink repair, Homologous recombination repair
Introduction
The nucleotide excision repair (NER) pathway eliminates UV-induced DNA photoproducts and other bulky DNA lesions [1]. It is a major contributor to genomic integrity and cancer prevention. Xeroderma pigmentosum (XP) patients with defective NER already develop first skin cancers at a median age of 8 years [2]. Seven XP complementation groups (XP-A to XP-G) according to the respective disease-causing mutated genes (XPA-XPG) and a variant form with a defect in translesion synthesis (Pol H gene) have been identified [3]. The human XPF (ERCC4) gene (OMIM: 278760) encodes for a 916 amino-acid (aa) protein that is part of the heterodimer XPF/ERCC1 which cleaves 5′ of UV-induced lesions. It is located on chromosome 16p13.2-p13.1 [4]. The ERCC1 gene (OMIM: 126380) encodes for a 297 aa protein and is located on chromosome 19q13.32 [5]. Patients with defective ERCC1 have been described with COFS (cerebro-oculo-facio-skeletal) syndrome [6, 7]. In addition to its core function in NER, the endonuclease complex XPF/ERCC1 plays a key role in interstrand crosslink (ICL) repair [8]. In this context, it is also involved in homologous recombination at DNA replication forks [9]. For decades, extensive studies of the complex functions of XPF/ERCC1—besides NER and ICL repair—have been done to assess its role in subpathways of homology-directed double-strand repair via single-strand annealing (SSA), gene conversion, and homologous gene targeting [10–12].
So far, a key limitation to study the functions of the XPF/ERCC1 complex has been the lack of an appropriate human XPF- or ERCC1-defective cell line. Up until now, these XP patient cell lines were utilized, however, as due to the essential role of these genes, all mutations characterized in patients so far retained at least one full-length allele with residual functional capabilities [13]. Most of these XP-F patients show relatively mild symptoms of photosensitivity with occasional neurological abnormalities. To investigate the multiple roles of the XPF/ERCC1 complex in different repair pathways, a suitable host cell line with a complete knockout is of great interest.
Quite recently, considerable attention has been paid to genome editing tools as zinc-finger nucleases (ZFNs) or bacterial transcription activator-like type III effector nucleases (TALENs) have been used for targeted gene knockout (KO) [14]. These technologies use a strategy of tethering endonuclease catalytic domains to modular DNA-binding proteins for inducing targeted DNA double-stranded breaks (DSBs) at specific genomic loci. However, these techniques are often inefficient, time consuming, laborious, and expensive [15]. By contrast, the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/ CRISPR associated (Cas) nuclease 9 system, which is a part of the microbial immune system, is guided by small RNAs to target specific DNA sequences [16, 17]. A DSB is induced by Cas9 cleavage and the target locus typically undergoes one of two major DNA repair pathways: the error-prone non-homologous end-joining (NHEJ) or the error-free homologous recombination repair (HRR). In the absence of a repair template, DSBs are re-ligated through the NHEJ pathway leading to insertion/deletion mutations resulting in frameshift mutations and premature stop codons [18]. In comparison to ZFNs and TALENs, the CRISPR/Cas 9 system is markedly easier to design, highly specific, efficient, and well suited for high-throughput and multiplexed gene editing for a variety of cell types and organisms.
The main purpose of our study was to create and characterise a human XPF KO cell line originating from wildtype (WT) immortalised fetal lung fibroblasts (MRC5Vi) using the CRISPR/Cas9 technique. We successfully generated an XPF CRISPR/Cas9 KO in MRC5Vi cells despite the important role of XPF in the human organism. These cells are defective in NER and ICL, as expected. We found that HRR but not NHEJ is impaired, emphasizing the importance of human XPF in this subpathway of double-strand break (DSB) repair. Without its heterodimer partner XPF, ERCC1 is retained in the cytoplasm. Co-transfection of a plasmid containing the cDNA of the full-length WT XPF could rescue these effects.
Materials and methods
Cell culture
The WT immortalised MRC5Vi cell line was kindly supplied by Sarah Sertic (University of Milan, Department of Life Sciences, Milan, Italy). Cells were cultivated in Dulbecco’s modified Eagle’s medium (DMEM) high glucose culture media (Lonza) supplemented with 10% fetal bovine serum (Biochrom) and 1% penicillin and streptomycin (Lonza) in a humified atmosphere at 37 °C and 5% CO2. Cells were passaged 1:10 when they reached confluency. Therefore, cells were rinsed with 10 ml phosphate buffered saline (PBS, pH 7.4) and dissociated from the culture flask using 4 ml trypsin/EDTA (Lonza) and 5 min incubation at 37 °C. The reaction was stopped by addition of 10 ml complete culture medium. Then, cells were centrifuged at 1000 rpm for 5 min and the supernatant was discarded. For antibiotic selection, cells were treated with 0.25 µg/ml puromycin (InvivoGen).
CRISPR construct generation
The pSpCas9n(BB)-2A-Puro (pX462) vector containing the cDNA encoding Streptococcus pyogenes Cas9 (hSpCas9) and a puromycin resistance cassette was purchased from Addgene (plasmid #48141) [19]. The guide sequence oligonucleotides (5′-CACCatttcatttgttacacggcgaggg-3′, 5′-AAACccctcgccgtgtaacaaatgaaat-3′), including the BbsI restriction site overhangs, and the PAM sequence targeting exon two within XPF were annealed (95 °C, 5 min, cool down overnight) and cloned into the pX462 vector. The plasmid was digested with BbsI (NEB) according to the manufacturer’s instruction. The oligonucleotides were phosphorylated by T4 Kinase (Thermo Scientific) and ligated into the vector using the T4 Ligase (Thermo Scientific) according to the manufacturer’s instructions. Constructs were transformed into Escherichia coli strain DH5α cells and selected using ampicillin (Sigma–Aldrich, 100 µg/ml). To validate the correct sequence, we used the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems); the product was loaded onto a 3100-Avant Genetic Analyzer (Applied Biosystems) and analysed with the Chromas Lite version 2.01 software (Technelysium Pty Ltd) as described in Schäfer et al. [20] using the U6 fwd primer 5′-actatcatatgcttaccgtaac-3′. The guide sequence was preselected from six different XPF guideRNA pairs using in silico on- and off-target predictions (http://crispr.mit.edu/, http://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design, http://crispr.cos.uni-heidelberg.de/index.html) and T7EI analysis of polyclonal populations in respect of XPF gene targeting in MRC5Vi cells (data not shown).
CRISPR/Cas9 transfection and single clone expansion
MRC5Vi cells were seeded in 100 mm tissue culture dishes (Greiner Bio-One) at a density of 750 000 cells and transfected using Attractene (Qiagen) and the fast-forward transfection protocol according to the manufacturer’s instructions (4 µg DNA (pX462 containing the guide sequence, and a puromycin resistance gene, or no DNA for the control), filled up to a total volume of 300 µl DMEM without supplements and 15 µl Attractene). On the next day, puromycin was added to the dishes and cells were cultured in puromycin containing medium (see above) until the control cells had died. For single clone selection, cells were separated using the serial dilution method in a 96-well plate (Greiner Bio-One) after coating with superfibronectin (Sigma–Aldrich) and cells were further cultured in FibroLife fibroblast medium (LifeLine) without puromycin. After 2 days, the plate was evaluated under the microscope (Axiovert A1, Zeiss); single cells were marked and expanded for 2 weeks to form colonies. Then, the colonies were transferred into 6-well plates (Greiner Bio-One) and further expanded for genomic DNA isolation.
Sequencing and clone evaluation
Cells were harvested as described above and genomic DNA was isolated using the QIAamp DNA Blood Mini Kit (Qiagen) according to the manufacturer’s instructions. The genomic region of XPF exon two was amplified using the PCR primers 5′-tgtagactggttggctgaagttac-3′ and 5′-tgatgtagggagctgagtccttc-3’. PCR products were purified using an ExoSAP (Affymetrix) digestion and sequenced with the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) as described in Schäfer et al. [20] with the PCR primer 5′-tgtagactggttggctgaagttac-3′. In addition to exon two, the entire XPF gene of KO clones was completely sequenced to exclude further mutations using the following primer pairs for PCR and the forward primer also for sequencing: exon one (5′-cacgatcatctcagtctcagctc-3′) (5′-cctagcgaccccttacatacgtc-3′), exon three (5′-ctctgttctgtgcgtggctatatg-3′) (5′-gaaaagcaaccatcaaattgctctc-3′), exon four (5′-gcttttcgtgttgtttgtagca-3′) (5′-gtgatgcttatatgccaatccacat-3′), exon five (5′-tagccacttccttgaataatgcttg-3′) (5′-acgttaagtaggcggaaacattagc-3′), exon six (5′-aagacttgccatgctgtatacttcg-3′) (5′-gccagttacgtatgtaggtcatgtg-3′), exon seven (5′-ttcaggaaagcagattccatctaac-3′) (5′-cactaggatctcagtgttcatttgc-3′), exon eight (5′-gatttaagtaatcttgccagagagg-3′) (5′-agcagcatcgtaacggatattaaag-3′), exon nine (5′-cctgtgtggtaacgagacctttaac-3′) (5′-ggacaattcagaccacaggtttatc-3′), exon ten (5′-cttcctttacccatcatttgtcttg-3′) (5′-ctggaacataacccattctaagctg-3′), and exon eleven (5′-ctttcctattagctcggtttccttc-3′) (5′-ctgaaaagtacaggcatgggataa-3′). For allele-specific sequencing of the XPF KO clone, exon two was amplified and cloned into the pJET1.2/blunt plasmid according to the manufacturer’s instruction of the CloneJET PCR Cloning Kit (Thermo Fisher Scientific), transformed in DH5α cells, and single colonies were sequenced as described above.
T7 Endonuclease I assay, surveyor nuclease mutation detection assay
The T7 endonuclease I (T7EI) and surveyor nuclease assay were performed according to the suggested manufacturer’s protocol for genome targeting and surveyor mutation detection (NEB, IDT). These enzyme mismatch cleavage (EMC) methods make use of nucleases that cleave heteroduplex DNA at mismatches and extrahelical loops (single or multiple nucleotides). The bacteriophage T7EI cleaves cruciform DNA structures, Holliday structures or junctions, heteroduplex DNA, and more slowly, nicked double-stranded DNA. T7EI is proficient at recognising insertions and deletions of ≥2 bases that are generated by NHEJ activity. The surveyor nuclease is a member of the celery endonuclease (CEL) family, which recognises and cleaves mismatches due to the presence of single-nucleotide polymorphisms (SNPs) or small insertions or deletions.
mRNA expression quantification
We performed mRNA expression quantification as described in Schäfer et al. [21]. XPF cDNA levels were normalised to GAPDH. For both genes, Qiagen Quantitect Primer Assays (Qiagen) were used (QT00063091, QT000792479).
Western blot analysis
WT or XPF KO MRC5Vi cells were harvested as described above. For overexpression of XPF cDNA, XPF KO MRC5Vi cells were seeded in 100 mm tissue culture dishes (Greiner Bio-One) at a density of 750 000 cells and transfected on the next day using Attractene (Qiagen) and the protocol according to the manufacturer’s instructions (4 µg XPF cDNA, filled up to a total volume of 300 µl DMEM without supplements and 15 µl Attractene). Cells were harvested after 48 h. Cell pellets were washed twice with 10 ml ice-cold PBS. Cytosolic and nuclear protein extracts were prepared using buffer A (10 mM Hepes, 10 mM KCl, 0.1 mM EGTA, 0.1 mM EDTA, 1 mM DTT, 0.5mM PMSF) and B (20mM Hepes, 400mM NaCl, 1 mM EGTA, 1 mM EDTA, 1 mM DTT, and 1 mM PMSF). Cell pellets were resuspended in an appropriate volume of buffer A and incubated on ice for 15 min. A tenth of the volume of 1% Nonidet NP40 in buffer A was added to the suspension and vortexed for 20 s. Afterwards, cells were centrifuged for 10 min at 14,000 U/min. The supernatant was transferred into a new reaction tube (cytosolic fraction) and kept on ice. The nuclear pellet was washed three times with 500 µl buffer A. Then, 20–50 µl buffer B was added and the reaction tubes were put in an ultrasonic bath with ice for 30 min. Thereupon, the mixture, was pelleted at 14,000 U/min for 30 min. The supernatant was transferred into a new reaction tube (nuclear fraction). Protein concentration was examined with a ready to use Bradford solution (Roti-Quant, Bio-Rad) and photometrically quantified. A BSA calibration curve was established using a standard serial dilution (0–1500 μg/ml). Each sample was diluted 1:20 and 1:40 in the respective buffer and 150 µl Bradford solution (Roti-Quant 1:5 in Aqua Bidest) and incubated shaking for 5 min at room temperature. The OD595 was measured with a GloMax® Discover Multimode Detection System (Promega).
For SDS–PAGE, the Mini-PROTEAN Tetra cell and precast 4–12% polyacrylamide gradient gels (Bio-Rad) were used applying the semidry-blot method in a Trans-Blot® Turbo™ Transfer System (Bio-Rad). Equal amounts of WT and XPF KO MRC5Vi protein extracts were loaded onto the gel together with a prestained protein marker (Marker VI, AppliChem). The WesternBreeze Chemiluminescent Immunodetection System for rabbit or mouse (Invitrogen) was used according to the manufacturer’s instructions. The anti-XPF mouse monoclonal antibody clone 3F2/3 (Santa Cruz), anti-ERCC1 rabbit polyclonal antibody FL-297 (Santa Cruz), and anti-β-actin mouse monoclonal clone AC-74 (Sigma–Aldrich) were diluted 1:250, 1:250, and 1:5000 in blocking solution, respectively. Incubation with the specific antibodies was performed overnight at 4 °C. Finally, incubation with the secondary antibodies (anti-mouse or anti-rabbit) was performed for 30 min. Chemiluminescence was developed and quantified using the Chemo Cam Imager 3.2 and the LabImage 1D software (Intas).
Post-toxin cell survival
For analysis of post-UV cell survival, the CellTiter96® Non-Radioactive Cell Proliferation Assay (Promega) was used as described in Schäfer et al. [21]. 2000 cells were seeded in 96-well plates (Greiner Bio-One) and after 24 h irradiated with UVC doses of 0–160 J/m2 (UVC 500 Crosslinker at 254 nm, Amersham), treated with cis-diammineplatinum(II) dichloride (CP) (Sigma–Aldrich), 0–8 µg/ml, 4,5′,8-trimethylpsoralen (TMP) (Sigma–Aldrich), 0–364.5 ng/ml (30 min preincubation), followed by irradiation with 1 J/cm2 UVA (Biolink BLX UVA crosslinker at 365 nm, Vilber), camptothecin (CPT) 0–512 nM, etoposide 0–16 µM, or bleomycin 0–16 µg/ml. After treatment, cells were cultured for 48 h until the substrate was added. As the compounds were solved in DMSO, the DMSO volume of the maximal toxin amount applied was used as a control and survival was set to 100%. The assay was repeated at least four times in quadruplicates.
Reporter gene assays
Analyses of NER repair capacity and complementation of the XPF KO cells with WT XPF, XPG, or ERCC1 cDNA via host cell reactivation (HCR) were performed according to the descriptions in Schäfer et al. [20]. Liposomal transient transfections were carried out in a 24-well format (Greiner Bio-One) using Attractene transfection reagent (Qiagen) according to the manufacturer’s instructions. Unirradiated Renilla luciferase was used for normalisation. Luminescence was measured 48 h after transfection with the GloMax® Discover Multimode Detection System (Promega). The relative repair capability [%] was calculated as the quotient of irradiated (1000 J/m2 UVC) to unirradiated firefly/Renilla ratios. The assay was repeated at least four times in triplicates.
For the analysis of the ICL repair capability, the same reporter gene assay was utilized and the firefly plasmid was treated with CP (Sigma–Aldrich) in a molecular ratio of 1:40 (vector:CP) or with TMP (Sigma–Aldrich) 1:50 (vector:TMP) followed by irradiation with 1 J/cm² UVA, respectively.
For the measurement of HRR and NHEJ capabilities, the DRGFP and pEGFP-Pem1-Ad2 reporter gene assay were utilized [22]. Cells were seeded on glass coverslips in 24-well plates and transfected with 100 ng DRGFP or pEGFP-Pem1-Ad2 plasmid, 100 ng pcDNA3.1(+) (used as empty control) or pCBAI-SceI plasmid and 50 ng pcDNA3.1(+) mCherry for normalisation using Attractene transfection reagent (Qiagen) according to the manufacturer’s instructions. After 48 h, cells were washed with PBS three times and fixed with 4% paraformaldehyde (PFA) in PBS for 15 min at room temperature. After fixation, the slides were washed again and then stained using Hoechst33342 (LifeTechnologies), diluted 1:1000 in PBS, for 20 min at room temperature covered in aluminum foil. Thereafter, the slides were washed and mounted with Fluoromount fluorescent mounting medium (Dako). Results were documented using an Axiovert A1 microscope (Zeiss) with a 200x/400x magnification. Subsequently, approximately 100 mCherry positive cells per condition were then assessed (blinded) for additional GFP positivity. Hence, the repair capability was calculated as the percentage of GFP positive cells compared to mCherry positive cells. The assay was repeated at least five times. The plasmids were a kind gift from Prof. Matthias Dobbelstein (Department of Molecular Oncology, Georg-August-University, Goettingen).
Results
Establishment of a complete XPF CRISPR/Cas9 knockout in MRC5Vi cells
We started by investigating the feasibility of a complete XPF KO in human fibroblasts. The major disadvantage has been that fibroblasts from XP-F patients still retain relatively high repair activities [7, 23, 24].
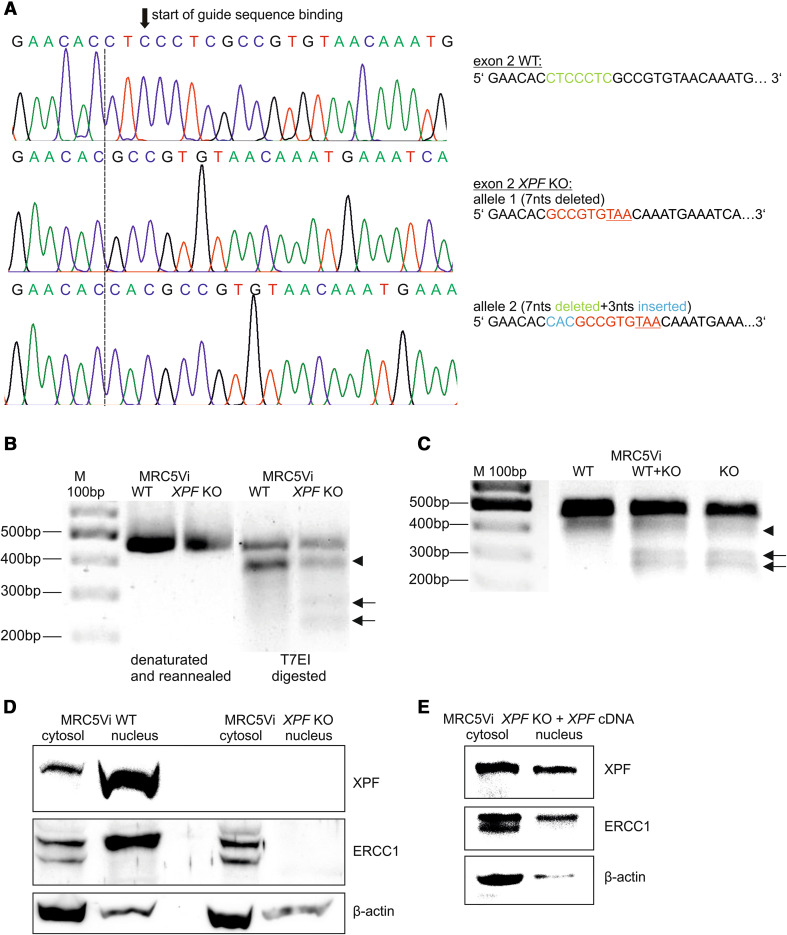
Drawing on these previous findings, we aimed to create an XPF KO cell line from immortalised fetal lung fibroblasts (MRC5Vi) that have already been used for DNA repair studies [25, 26], by employing the CRISPR/Cas9 nuclease system targeting one of the first exons (one-three) of XPF. As the long-term presence of artificial nucleases in cells is undesirable, we applied a transient transfection approach with a puromycin resistance cassette containing plasmid for selection of positive clones. Following intensive on and off-target analyses of different guide RNAs, we identified a suitable target sequence in exon two of XPF. After single clone expansion, we analysed the success of the KO by amplifying and sequencing exon two. At this point, cells were sensitive to puromycin again. As shown in Fig. 1a (part of the XPF exon two sequencing results), we were able to successfully generate a compound heterozygous complete and still viable XPF KO cell line. The section of the XPF exon two sequence indicates the binding of the guide RNA (black arrow), and as expected, the Cas9 nuclease induces a DSB ~3 bp upstream of the PAM sequence, where in our case a part of the guide sequence was deleted during the NHEJ process [19]. Seven nucleotides (nts) of the WT sequence were deleted resulting in one allele with a premature stop codon nine nucleotides downstream and a second allele with an additional insertion of three nucleotides and a premature stop after twelve nucleotides, respectively. This suggests a second cutting event by the Cas9 nuclease at the same position. Sequencing of the entire XPF gene of the KO clone revealed no additional mutations (data not shown).
Fig. 1.
Structural analyses of XPF CRISPR/Cas9 KO and WT MRC5Vi cells. a XPF exon two Sanger sequencing of WT (upper panel) and KO (middle and lower panel) cells. The black arrow illustrates the start of the guide RNA binding sequence, while the dashed line indicates the start of the sequence alterations in the XPF KO cells. The green part of the sequence to the right is deleted in the KO cells, while the blue nucleotides are inserted. The red nucleotides indicate the continuation of the WT sequence after the CRISPR event up until the stop codon, resulting from the frameshift (underlined). The middle and lower panels show the two different alleles of the KO cells originated from the CRISPR/Cas9 genome editing. b XPF exon two was PCR amplified from genomic DNA of WT and KO cells, denaturated, reannealed, and T7EI-digested or c subject to a surveyor nuclease assay. Black arrows indicate the positions of digested DNA; the arrow head marks an additional digestion product to the undigested PCR product, also seen in the WT cells (polymorphism). d Protein expression was assessed by horizontal SDS–PAGE followed by Western blot analyses. Equal amounts of cytosolic and nuclear extracts of WT and KO cells were loaded onto SDS-gels and stained with an anti-XPF or anti-ERCC1 antibody. The gel was stripped and reprobed for anti-β-actin to serve as loading control. e Same procedure was performed after complementation of the XPF KO cells with a plasmid containing XPF cDNA
For further analysis of the mutated KO clone, we utilized the T7EI and surveyor nuclease assays. In a first step, PCR products were produced from genomic DNA of WT and XPF KO MRC5Vi cells, using the primers for XPF exon two as described above. The PCR products were denatured, followed by re-annealing, leading to a population of double-strand fragments, some of them containing mismatches due to the afore mentioned compound heterozygous nature of the KO. These mismatches were detected by T7EI. In the event of successful CRISPR/Cas9 targeting in the cells, cleavage products can be visualised on an agarose gel. Indeed, the WT cells did not yield amplicons susceptible to site-specific cleavage by T7EI (Fig. 1b). The KO cells, on the other hand, showed efficient cleavage of the endogenous chromosomal target sequence, evidenced by T7EI digestion products (arrows), confirming the presence of indels at the locus. Interestingly, we observed an additional cleavage product, also in the WT cells (arrow head). This can be explained by a physiologically appearing already described three nucleotide deletion in exon two (polymorphismus rs771203473 (NCBI), deletion AGA/-, chromosome 16:13922057–13922059).
For the surveyor nuclease assay, we prepared PCR amplicons from mutant (KO) and WT DNA, mixed equal amounts of KO and WT DNA, and hybridized them by heating and cooling the mixture to form hetero- and homoduplexes. The annealed heteroduplex/homoduplex mixture was treated with surveyor nuclease. The reference DNA, alone treated similarly, serves as a negative control. The formation of new cleavage products, due to the presence of one or more mismatches, is indicated by the presence of additional bands. In accordance with the T7EI assay, the WT cells showed no cleavage products. As anticipated, we detected cleavage products not only in the WT + KO mixture, but also in the KO alone, which is in concurrence with the compound heterozygous state of the KO (Fig. 1c, arrows). Again, we could detect an additional cleavage product, due to the previously mentioned polymorphism (arrow head).
Finally, we aimed to confirm the generation of a complete XPF KO on protein level. Therefore, we isolated cytosolic and nuclear protein fractions of WT and KO cells. The full-length XPF molecular weight is predicted to be 112 kDa. With the KO inducing a premature stop codon after 86/87 aa, it would result in a theoretic molecular weight of approximately 10 kDa. In the WT cells, we detected an accumulation of the XPF protein in the nucleus. In contrast, XPF protein could not be detected in either the cytosol or the nucleus of KO cells (Fig. 1d), also at lower sections of the gel, where a 10 kDa protein would be visible (section of the gel not shown). XPF protein expression could be restored in the XPF KO cells by overexpression of a plasmid containing XPF cDNA (Fig. 1e). Furthermore, there was an about 50% reduction in XPF mRNA expression between WT and KO cells most likely due to nonsense-mediated messenger decay (WT XPF normalised to GAPDH 100 ± 4.71%, KO 54.76 ± 13.32%, data are presented as mean ± SEM).
In addition, we also assessed the protein expression of ERCC1, with which XPF forms a functional heterodimer for its catalytic activity in DNA repair. ERCC1 was detected as a double band at 38 kDa in the cytosol which is probably caused by post-translational modifications [27, 28]. Notably, the WT cells displayed an accumulation of the modified form of ERCC1 in the nucleus (Fig. 1d). Interestingly, ERCC1 could not be detected in the nucleus of the XPF KO cells. This indicates the inability of ERCC1 to enter the nucleus without its binding partner XPF. Nuclear localisation could be rescued by transfecting the XPF KO cells with a plasmid containing XPF cDNA (Fig. 1e).
The XPF KO cells show an increased sensitivity to several DNA damaging toxins
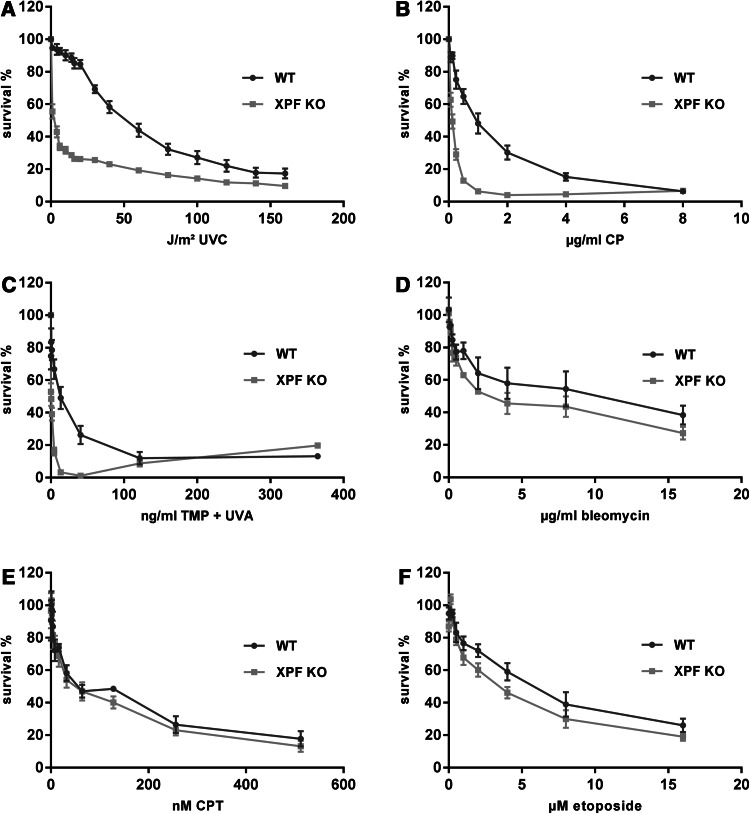
To further characterise the newly generated XPF KO cells, we investigated the cell survival after treatment with UVC irradiation, the main trigger of NER, and treatment with ICL inducing toxins (CP and TMP plus UVA). Cisplatin, which is a frequently used chemotherapeutic, predominantly produces intrastrand crosslinks between guanine residues or adenine and guanine (90%), which are also predominantly repaired by NER pathway. Interstrand crosslinks are formed between guanines to a minor extent of 2–5% [29]. On the other hand, psoralen and its derivatives, like TMP, mainly form interstrand crosslinks with DNA when activated by irradiation with long-wavelength UV light (UVA) [30].
We analysed the post-toxin cell survival using the MTT method [31] to determine the lethal dose 50 (LD50), which indicates the dose of a toxin that kills 50% of the cells. Therefore, this value gives an impression of the cellular sensitivity towards a special toxin. The XPF KO cells showed a significantly enhanced sensitivity against UVC irradiation compared to WT cells (LD50WT = 50 J/m², LD50KO < 1 J/m²) (***P < 0.001, n = 4, one-tailed, unpaired student’s t test) (Fig. 2a). Likewise, after treatment with intra- and interstrand crosslink inducing agents like CP or TMP activated by UVA irradiation, the XPF KO cells exhibited a significantly reduced survival (CP: LD50WT = 1.5 µg/ml, LD50KO = 0.125 µg/ml; TMP: LD50WT = 13.5 ng/ml, LD50KO = 0.5 ng/ml) (***P < 0.001, n = 4) (Fig. 2b, c). Treatment with TMP or UVA alone did not markedly affect the cell survival (data not shown).
Fig. 2.
Post-toxin survival analyses of XPF CRISPR/Cas9 KO and WT MRC5Vi cells. A total of 2000 cells were seeded in 96-well plates and a irradiated with increasing doses of UVC (0–160 J/m²), b treated with increasing doses of cisplatin (0–8 µg/ml), c trimethylpsoralen (0–364.5 ng/ml) activated by 1 J/cm² UVA, d bleomycin (0–16 µg/ml), e camptothecin (0–512nM), or f etoposide (0–16 µM). Cell survival was determined after 48 h by using an MTT assay, and survival of the DMSO control cells was set to 100 %. Data are presented as the mean ± SEM. At least four independent experiments in quadruplicates were performed
As the XPF/ERCC1 complex is also involved in the different subpathways of mammalian DSB repair, we investigated the effects of different DSB creating toxins [32, 33]. Although the XPF KO cells appeared to be slightly more sensitive to the bleomycin treatment, there was no significant reduction in survival in comparison to WT cells (LD50WT = 3 µg/ml, LD50KO = 2.5 µg/ml) (n.s. P > 0.05, n = 4) (Fig. 2d). In addition, we looked at the effects of CPT, a topoisomerase I (TOP1) inhibitor, and etoposide, a topoisomerase II (TOP2) inhibitor. As with the previously tested toxin, only a slight, but non-significant, decrease in cell survival was measurable after drug treatment (CPT: LD50WT = 50 nM, LD50KO = 50 nM; etoposide: LD50WT = 6 µM, LD50KO = 3.5 µM) (n.s. P > 0.05, n = 4) (Fig. 2e, f).
Overall, the results imply that XPF is not as essential for coping with DSB inducing drugs, as for protecting cells against UVC irradiation or ICL inducing agents.
Loss of XPF reduces the cellular repair capability for NER, ICL, and HRR
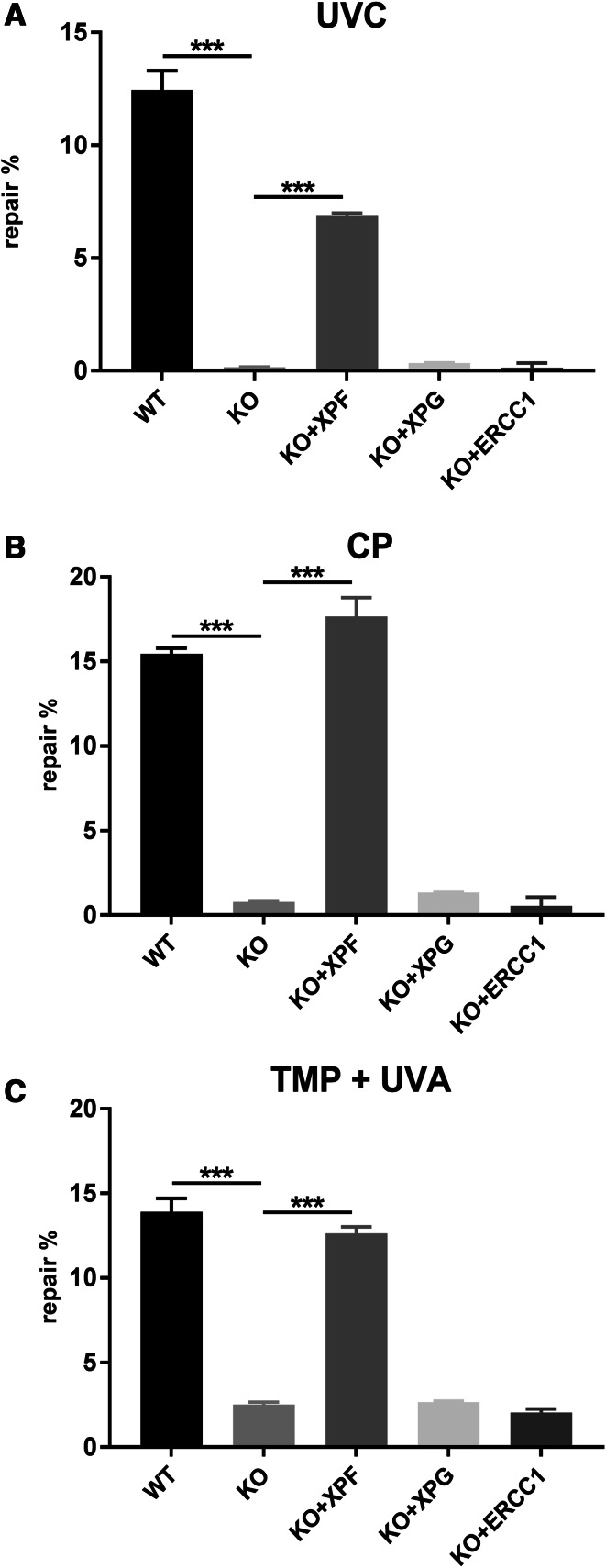
The XPF/ERCC1 complex is involved in multiple repair pathways and cellular processes. Therefore, we tested the effect of the XPF KO on NER, ICL, and DSB repair in more detail by utilizing reporter gene assays. We analysed the correct repair of UVC induced DNA lesions using the HCR assay. XPF KO MRC5Vi cells showed a significant reduction in NER capability (0.15 ± 0.02%) in comparison to WT cells (12.45 ± 0.86%) (***P < 0.001, n = 4). This effect could be rescued by complementation with a plasmid containing the cDNA of full-length XPF (6.87 ± 0.13%) (***P < 0.001, n = 4), but neither XPG (0.35 ± 0.01%) nor ERCC1 (0.13 ± 0.21%) used as controls (n.s. P > 0.05, n = 4) (Fig. 3a).
Fig. 3.
Reactivation of a reporter gene after treatment with UVC, cisplatin, or trimethylpsoralen activated by UVA in XPF CRISPR/Cas9 KO and WT MRC5Vi cells. Firefly plasmids containing specific lesions due to treatment with a UVC irradiation, b cisplatin (intrastrand crosslinks), or c trimethylpsoralen activated by 1 J/cm2 UVA irradiation (interstrand crosslinks) were transfected into MRC5Vi WT and XPF KO cells and complemented with plasmids containing XPF, XPG, or ERCC1 cDNA. The relative repair capability is calculated as the percentage (repair%) of the reporter gene activity (firefly luciferase) compared to the untreated plasmid, after normalisation to an internal co-transfected control (Renilla luciferase). Data are presented as the mean ± SEM. The one-tailed, unpaired student’s t test was applied, ∗∗∗P < 0.001. At least four independent experiments in triplicates were performed
ICLs are challenging to repair, because they obstruct DNA replication and transcription [34, 35]. XPF-deficient cells are exceptionally sensitive to agents that can form ICLs, such as CP or TMP plus UVA [36]. We adapted the HCR assay to plasmids treated with CP or TMP activated by UVA irradiation to investigate the repair of inter- and intrastrand crosslinks. The repair of intrastrand crosslinks caused by CP, as well as interstrand crosslinks produced by TMP treatment of the firefly luciferase plasmid followed by irradiation with UVA, was impaired in the KO cells. XPF KO MRC5Vi cells also showed a significant reduction in ICL repair capability (CP 0.78 ± 0.08%, TMP 2.50 ± 0.15%) compared to WT cells (CP 15.43 ± 0.035 %, TMP 13.90 ± 0.81 %) (***P < 0.001, n = 4). This effect could be rescued by co-transfecion of a full-length XPF containing plasmid (CP 17.65 ± 1.13 %, TMP 12.65 ± 0.37 %) (***P < 0.001, n = 4), but neither by XPG (CP 1.34 ± 0.02 %, TMP 2.65 ± 0.07 %) nor ERCC1 (CP 0.56 ± 0.51 % TMP 2.04 ± 0.22 %) (n.s. P > 0.05, n = 4) (Fig. 3b, c).
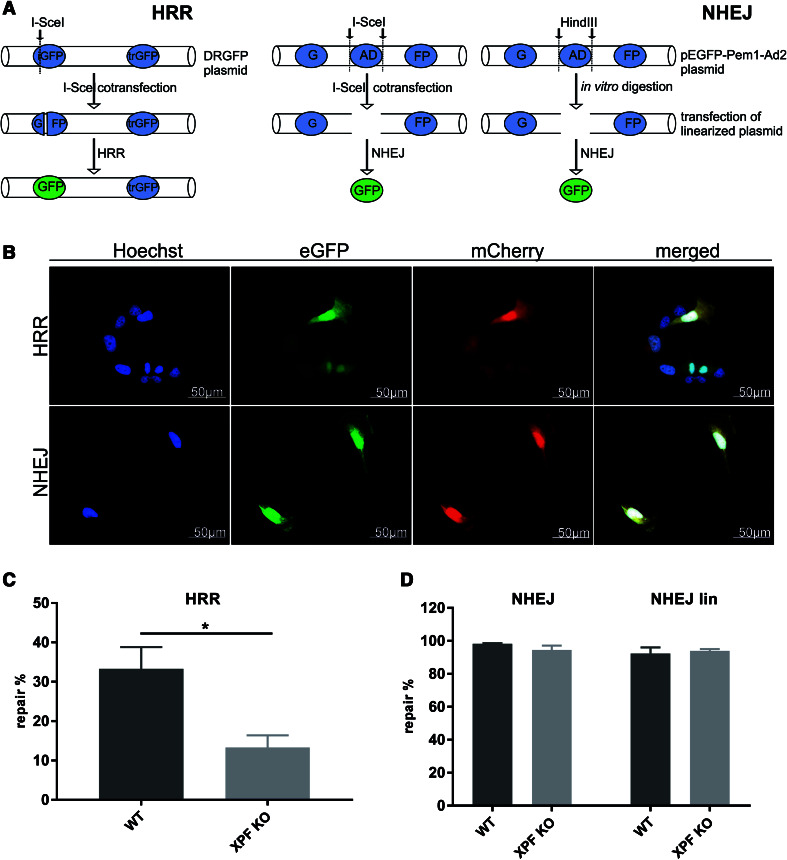
We used the DRGFP reporter assay to investigate the HRR capability of our cells. In addition, we implemented a reporter gene assay to assess NHEJ using the pEGFP-Pem1-Ad2 plasmid (Fig. 4a, b) [22]. The XPF KO MRC5Vi cells showed a significant reduction in HRR capability (13.31 ± 3.07 %) in comparison to WT cells (33.27 ± 5.52 %) (*P < 0.05, n = 5). In contrast, we could not detect a reduction in NHEJ activity, either for the I-SceI (WT 98.29 ± 0.36 %, KO 94.49 ± 2.70 %) or the in vitro HindIII digestion, which we used as an additional control (WT 92.25 ± 3.81 %, KO 93.94 ± 1.12 %) (n.s. P > 0.05, n = 4) (Fig. 4c, d). For quality assurance, the overall level of HRR capability was lower (up to 33.27 %) than the NHEJ activity (up to 98.92 %) in the WT cells reflecting the exclusivity of HRR during M or G2 cell cycle phases where a repair template is present.
Fig. 4.
Analyses of different DSB repair pathways in XPF CRISPR/Cas9 KO and WT MRC5Vi cells. a Schematic illustration of the assay principle. The left panel depicts the HRR assay. The GFP plasmid contains two GFP cassettes. One cassette (iGFP) with a deletion in the first exon of GFP combined with the insertion of I-SceI restriction sites is followed by a promoter-less/ATG-less first exon and intron of GFP (trGFP). The DRGFP plasmid was co-transfected with pCBASceI and pcDNA3.1(+) mCherry for internal normalisation. Upon expression of I-SceI (from pCBAScel), a DSB is produced in the DRGFP plasmid. If the cells repair the DSB using the trGFP as template, a functional GFP cassette is formed and HRR proficient cells can express GFP, while deficient cells do not. The middle panel shows that the way NHEJ was assessed using the pEGFP-Pem1-Ad2 plasmid. It contains a GFP gene with an engineered 3 kb intron from the Pem1 gene with an adenoviral (AD) exon flanked by recognition sequences for HindIII and I-SceI to induce DSBs. The intact NHEJ cassette is GFP negative as the adenoviral exon disrupts the GFP ORF. We co-transfected the plasmid with pCBASceI and pcDNA3.1(+) mCherry for normalisation. In NHEJ proficient cells, the I-Scel-induced DSBs eliminating the intron are repaired by NHEJ, so that the cell can transcribe and express functional GFP. Furthermore, as an additional analyses of NHEJ, we performed an in vitro digestion of the plasmid using HindIII and then transfected the linearized plasmid (NHEJlin) together with pcDNA3.1(+) (to maintain the same DNA rations) and pcDNA3.1(+) mCherry for normalisation (right panel) b Representative fluorescent images acquired for the HRR (upper panel) and NHEJ (lower panel) assay in WT cells. For both assays, approximately 100 mCherry positive cells per condition were counted (blinded) for GFP positive cells after c HRR or d NHEJ assay. Hence, the repair capability was calculated as percentage (repair%) of the GFP compared to mCherry positive cells. Data are presented as the mean ± SEM. The one-tailed, unpaired student’s t test was applied, ∗P < 0.05. At least five independent experiments were performed
Discussion
Manipulating DNA has been of great interest since the discovery of the DNA double helix. Various site-specific techniques for the modification of genomes have emerged over the past years using the site-directed ZFNs or TALENs. Recently, the simplicity of programming the CRISPR/Cas9 to modify specific genomic loci presents a new and customisable way to disrupt gene function and generate specific knockout cells, being easier to design, highly specific and efficient. The endonuclease XPF has been implicated in various DNA repair pathways among them NER, ICL, and DSB repair. The multiple roles of the protein are also reflected by the variety of phenotypes and diseases arising from different mutations in the XPF gene. XP-F is characterized by mild clinical features and patients show minor sun sensitivity, but lack of neurological abnormalities [37]. Missense mutations are present in at least one allele, none of which affect the nuclease activity of the enzyme and, therefore, do not completely abolish XPF function resulting in residual repair capability of UV damage (up to 20 %) [38, 39]. Mutations leading to premature stop codons or missense mutations in essential residues result in the more severe phenotypes of Cockayne syndrome (CS), XP/CS complex phenotype, XFE progeroid syndrome, or fanconi anemia [7, 40, 41]. Nevertheless, there are no suitable patient cells with a complete loss of the XPF protein available to be used in molecular and mechanistic studies. Tian et al. described an Xpf-deficient mouse model with a mutated exon 8, leading to a premature stop codon and no Xpf mRNA due to nonsense-mediated decay. The animals showed defective postnatal growth and a shortened life span (of about three weeks). Embryonic fibroblasts isolated from these mice were hypersensitive to UV and mitomycin C treatment [42].
To the best of our knowledge, this is a unique study to generate a human XPF KO cell line using the CRISPR/Cas9 technique targeting exon two of XPF to obtain a massively truncated protein with no residual activity. By applying this technique, a viable compound heterozygous XPF KO cell line with a premature stop codon in exon two was established (Fig. 1a–d). Our results describe that it is possible to generate viable human fibroblasts lacking the essential endonuclease XPF, while germline mutations to this extent are not compatible with life in neither humans nor mice. Knockout strategies are limited in studying genes critical for embryogenesis, but conditional gene inactivation can circumvent this limitation. This has recently been achieved using conditional alleles generated by also using CRISPR/Cas9 in mice [43]. Further studies also showed that the CRISPR/Cas9 system is a useful tool to identify sets of essential and non-essential genes [44, 45].
The immortalised XPF KO fibroblasts generated in this study showed no further mutations in the XPF gene, and XPF protein expression could not be detected, while mRNA levels were only reduced by 50 %. This is consistent with the observations reported in a subset of XP-F patients, indicating that mutant XPF protein might be unstable [24]. Furthermore, the non-detectability of the XPF protein can be explained by the lack of the antibody epitope mapping aa 629–905. Notably, we observed that ERCC1 was not detectable in the nuclear fraction of the KO cells. The previous studies reported that ERCC1 and XPF are unstable in the absence of their respective binding partner [5, 46, 47]. Furthermore, XP-F patients frequently show low levels of ERCC1 protein in total protein extracts [48] and extracts from ERCC1- and XPF-defective cells do not complement one another in vitro [46, 47]. This highlights the importance of the formation of the XPF/ERCC1 complex to maintain protein stability, while alone either protein forms aggregates that are subject to proteolytic degradation, or protects it from being subject to nuclear export [49]. In contradiction to this, we showed that ERCC1 is stable in the cytosol in our KO cells; however, it was not able to enter or remain in the nucleus without its heterodimer partner XPF as seen in the WT cells. The latter can be explained by the lack of a nuclear localization signal (NLS) in ERCC1 [50], rendering it unable to enter the nucleus by itself [38]. Another explanation might be a rapid nuclear export of ERCC1 without protection in the prematurely formed XPF/ERCC1 heterodimer, due to the absence of the XPF protein in our XPF KO cells. After overexpression of XPF in the XPF KO cells, ERCC1 could be found in the nucleus again, because of the premature complex formation prior to nuclear transport. As the previous results stem from experiments with whole cell protein extracts, it remains unclear whether ERCC1 can be stabilized in the cytosol by other interaction partners than XPF and merits further investigations. In a study by Jordheim et al. [51], a compound was identified that inhibits XPF-ERCC1 protein–protein interaction. Applying this compound to two different cancer cell lines sensitized those cells to CP, mitomycin C, and UV irradiation, and led to reduced DSB removal. This confirms the findings in our XPF KO cells in which we disrupted the heterodimer as well.
Functional analyses revealed that NER capability was almost completely ablated in the XPF KO cells as indicated by their high sensitivity against UVC irradiation and reduced host cell reactivation of an UVC-treated reporter gene plasmid (Figs. 2a, 3a). XPF/ERCC1 is an essential structure-specific endonuclease for the repair of helix-distorting DNA lesions, such as UV-photoproducts, in both replicating and non-replicating cells. It makes incisions on the damaged DNA strand on the 5′ side of the open “denaturation-bubble” intermediate formed during NER [52, 53]. Only complementation with full-length XPF, but neither XPG nor ERCC1 could rescue the repair capability of the cells up to WT levels.
Moreover, our cells are very well suited for the analysis of CP induced lesions. Enoiu et al. [54] previously reported a 20 % residual repair of CP crosslinks using GM08437 (XP-F) cells for their studies, that still harbour one full-length allele with a missense mutation. In our XPF KO cells, the repair activity decreased to 0.78 %, rendering them an exquisite model system for mechanistic analysis of ICL repair (Fig. 3b).
In agreement with the results for CP removal, the XPF KO cells exhibited a high sensitivity against TMP plus UVA irradiation induced lesions (Figs. 2b, c, 3b, c). Psoralen compounds are also used for the treatment of psoriasis, vitiligo, as well as cutaneous T-cell lymphoma and are repaired by the ICL repair pathway [55–57]. XPF functions in ICL repair in a NER-independent manner and is essential for the incision or “unhooking” step of ICL repair [35, 36, 58–60]. It remains to be investigated, whether other endonucleases like Mus81/Eme1, SLX4/SLX1, or FAN1 can also perform incisions that are normally conducted by XPF, however, in a much less efficient manner [35, 61–66].
Ahmad et al., Mogi and Oh, Wood et al., and Murray et al. [32, 67–69] reported about the moderate sensitivity of XPF/ERCC1-deficient mammalian cells to DSB inducing agents and ionizing radiation, indicating the important role of XPF/ERCC1 in one or more subpathways of DSB repair. In mammalian cells, XPF/ERCC1, or the yeast homolog Rad1/Rad10, respectively, has been accounted for HRR as well as NHEJ due to its ability to remove non-homologous 3′ single-stranded flaps [11, 70, 71]. In HRR, the heterodimeric complex functions through the error-prone single-strand annealing (SSA) subpathway, gene conversion, and homologous gene targeting in yeast and mammals [10–12, 72, 73], while for NHEJ, the complex is only involved in the Rad52- and Ku70/Ku86-independent microhomology-mediated end-joining (MMEJ) subpathway [32, 33, 74–76]. All these studies primarily focus on mice, yeast, and hamster cells, while only implicating the importance of the human XPF/ERCC1 complex in HRR. Nevertheless, the studies never used a complete human knockout cell line. Our results of a 60 % reduced HHR capability and unchanged NHEJ capability (Fig. 4c, d) indicate that human XPF is important for error-prone single-strand annealing, gene conversion, and homologous gene targeting, rather than microhomology-mediated end-joining. This consolidates the function of human XPF in the HRR subpathway of DSB repair, as already implicated by the yeast-based studies. As XPF/ERCC1 is only involved in subpathways of HRR as well as NHEJ, the loss of XPF may not have such a marked effect on DSB repair as in NER or ICL repair, where the endonuclease is essential for repair. This was also reflected by the mild sensitivity against DSB inducing agents (Fig. 2d, f). Bleomycin is a DSB inducing agent with an unsolved mode of action. Hypotheses suggest that it chelates metal ions resulting in a pseudoenzyme that reacts with oxygen to produce superoxide and hydroxide free radicals that cleave DNA [77]. CPT irreversibly traps TOP1 to the DNA, resulting in replication or transcription-induced DSBs [78]. Etoposide forms a ternary complex with DNA and TOP2, preventing re-ligation of the DNA strands and thereby resulting in DSBs [79]. Since XPF is involved in three different HR-related repair mechanisms, these effects might add up to render the KO cells less capable of repair in terms of HRR substrates that cannot be repaired by NHEJ. Furthermore, as discussed before, XPF plays a key role in ICL repair not only via its endonuclease function, but also in the late stage of HR during ICL repair [9]. On the other hand, XPF seems to be dispensable for NHEJ, because it is involved in one minor subpathway only, which apparently can be compensated for (Fig. 4d).
In conclusion, we applied a novel and innovative technique to successfully generate a viable human XPF KO cell line that is useful for further analyses regarding XPF’s various functions in different cellular processes. We characterized the cells and used them as a tool to analyse the involvement of XPF in different repair pathways. Thereby, we could show that XPF is dispensable for ERCC1 protein stability but essential for ERCC1 nuclear localization. Furthermore, in addition to its essential role in NER and ICL repair, XPF’s endonuclease activity appears to be partially essential in HR processes, a subpathway of DSB repair.
On the basis of these newly generated cells it may be possible to shed light onto more general questions. For example, there is still an ongoing discussion about the essential nuclease for incising the DNA around an interstrand crosslink. Whether or not it might exclusively be XPF could now be investigated using this cell line. In addition, detailed mechanistic questions of repair pathways like NER can be studied in a setting without residual XPF activity. Moreover, the proposed method has great potential for applications on other genes of the DNA repair machinery and can be successfully used for a number of in vitro and in vivo studies.
Acknowledgements
The authors wish to acknowledge the assistance and excellent technical support of Antje Apel (Department of Dermatology, Venereology and Allergology, University Medical Center Goettingen) and Sarah Sertic (University of Milan) for supply of the MRC5Vi cells. We also thank Andreas Ohlenbusch (Department of Child and Adolescent Health, University Medical Center Goettingen) for his assistance with Sanger sequencing of the constructs and clones.
Compliance with Ethical Standards
Funding
This work was supported by the German Cancer Aid (Deutsche Krebshilfe e.V.) [111377] and the Heinz and Heide Duerr Foundation [2014−2.2.1/02 to S.E.].
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Scharer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5(10):a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kraemer KH, DiGiovanna JJ. Forty years of research on xeroderma pigmentosum at the US National Institutes of Health. Photochem Photobiol. 2015;91(2):452–459. doi: 10.1111/php.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehmann J, Schubert S, Emmert S. Xeroderma pigmentosum: diagnostic procedures, interdisciplinary patient care, and novel therapeutic approaches. J Dtsch Dermatol Ges. 2014;12(10):867–872. doi: 10.1111/ddg.12419. [DOI] [PubMed] [Google Scholar]
- 4.Brookman KW, Lamerdin JE, Thelen MP, Hwang M, Reardon JT, Sancar A, Zhou ZQ, Walter CA, Parris CN, Thompson LH. ERCC4 (XPF) encodes a human nucleotide excision repair protein with eukaryotic recombination homologs. Mol Cell Biol. 1996;16(11):6553–6562. doi: 10.1128/MCB.16.11.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sijbers AM, van der Spek PJ, Odijk H, van den Berg J, van Duin M, Westerveld A, Jaspers NG, Bootsma D, Hoeijmakers JH. Mutational analysis of the human nucleotide excision repair gene ERCC1. Nucleic Acids Res. 1996;24(17):3370–3380. doi: 10.1093/nar/24.17.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaspers NG, Raams A, Silengo MC, Wijgers N, Niedernhofer LJ, Robinson AR, Giglia-Mari G, Hoogstraten D, Kleijer WJ, Hoeijmakers JH, Vermeulen W. First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. Am J Hum Genet. 2007;80(3):457–466. doi: 10.1086/512486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kashiyama K, Nakazawa Y, Pilz DT, Guo C, Shimada M, Sasaki K, Fawcett H, Wing JF, Lewin SO, Carr L, Li TS, Yoshiura K, Utani A, Hirano A, Yamashita S, Greenblatt D, Nardo T, Stefanini M, McGibbon D, Sarkany R, Fassihi H, Takahashi Y, Nagayama Y, Mitsutake N, Lehmann AR, Ogi T. Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, xeroderma pigmentosum, and Fanconi anemia. Am J Hum Genet. 2013;92(5):807–819. doi: 10.1016/j.ajhg.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defects in interstrand cross-link uncoupling do not account for the extreme sensitivity of ERCC1 and XPF cells to cisplatin. Nucleic Acids Res. 2002;30(17):3848–3856. doi: 10.1093/nar/gkf479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Minawi AZ, Lee YF, Hakansson D, Johansson F, Lundin C, Saleh-Gohari N, Schultz N, Jenssen D, Bryant HE, Meuth M, Hinz JM, Helleday T. The ERCC1/XPF endonuclease is required for completion of homologous recombination at DNA replication forks stalled by inter-strand cross-links. Nucleic Acids Res. 2009;37(19):6400–6413. doi: 10.1093/nar/gkp705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adair GM, Rolig RL, Moore-Faver D, Zabelshansky M, Wilson JH, Nairn RS. Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. EMBO J. 2000;19(20):5552–5561. doi: 10.1093/emboj/19.20.5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niedernhofer LJ, Essers J, Weeda G, Beverloo B, de Wit J, Muijtjens M, Odijk H, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J. 2001;20(22):6540–6549. doi: 10.1093/emboj/20.22.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sargent RG, Meservy JL, Perkins BD, Kilburn AE, Intody Z, Adair GM, Nairn RS, Wilson JH. Role of the nucleotide excision repair gene ERCC1 in formation of recombination-dependent rearrangements in mammalian cells. Nucleic Acids Res. 2000;28(19):3771–3778. doi: 10.1093/nar/28.19.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schubert S, Lehmann J, Kalfon L, Slor H, Falik-Zaccai TC, Emmert S (2014) Clinical utility gene card for: Xeroderma pigmentosum. Eur J Hum Genet 22 (7). doi:10.1038/ejhg.2013.233 [DOI] [PMC free article] [PubMed]
- 14.Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Meyer BJ. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333(6040):307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei C, Liu J, Yu Z, Zhang B, Gao G, Jiao R. TALEN or Cas9 - rapid, efficient and specific choices for genome modifications. Journal of genetics genomics = Yi chuan xue bao. 2013;40(6):281–289. doi: 10.1016/j.jgg.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 16.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Zhang Y, Chen KL, Shan QW, Wang YP, Liang Z, Gao CX. [CRISPR/Cas: a novel way of RNA-guided genome editing] Yi Chuan. 2013;35(11):1265–1273. doi: 10.3724/SP.J.1005.2013.01265. [DOI] [PubMed] [Google Scholar]
- 18.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schafer A, Schubert S, Gratchev A, Seebode C, Apel A, Laspe P, Hofmann L, Ohlenbusch A, Mori T, Kobayashi N, Schurer A, Schon MP, Emmert S. Characterization of three XPG-defective patients identifies three missense mutations that impair repair and transcription. J Invest Dermatol. 2013;133(7):1841–1849. doi: 10.1038/jid.2013.54. [DOI] [PubMed] [Google Scholar]
- 21.Schafer A, Hofmann L, Gratchev A, Laspe P, Schubert S, Schurer A, Ohlenbusch A, Tzvetkov M, Hallermann C, Reichrath J, Schon MP, Emmert S. Molecular genetic analysis of 16 XP-C patients from Germany: environmental factors predominately contribute to phenotype variations. Exp Dermatol. 2013;22(1):24–29. doi: 10.1111/exd.12052. [DOI] [PubMed] [Google Scholar]
- 22.Seluanov A, Mao Z, Gorbunova V. Analysis of DNA double-strand break (DSB) repair in mammalian cells. J Vis Exp. 2010 doi: 10.3791/2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cleaver JE, Thompson LH, Richardson AS, States JC. A summary of mutations in the UV-sensitive disorders: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. Hum Mutat. 1999;14(1):9–22. doi: 10.1002/(SICI)1098-1004(1999)14:1<9::AID-HUMU2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 24.Matsumura Y, Nishigori C, Yagi T, Imamura S, Takebe H. Characterization of molecular defects in xeroderma pigmentosum group F in relation to its clinically mild symptoms. Hum Mol Genet. 1998;7(6):969–974. doi: 10.1093/hmg/7.6.969. [DOI] [PubMed] [Google Scholar]
- 25.Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, Mullenders LH, Yamashita S, Fousteri MI, Lehmann AR. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell. 2010;37(5):714–727. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Sertic S, Pizzi S, Cloney R, Lehmann AR, Marini F, Plevani P, Muzi-Falconi M. Human exonuclease 1 connects nucleotide excision repair (NER) processing with checkpoint activation in response to UV irradiation. Proc Natl Acad Sci U S A. 2011;108(33):13647–13652. doi: 10.1073/pnas.1108547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nowotny M, Gaur V. Structure and mechanism of nucleases regulated by SLX4. Curr Opin Struct Biol. 2016;36:97–105. doi: 10.1016/j.sbi.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Perez-Oliva AB, Lachaud C, Szyniarowski P, Munoz I, Macartney T, Hickson I, Rouse J, Alessi DR. USP45 deubiquitylase controls ERCC1-XPF endonuclease-mediated DNA damage responses. EMBO J. 2015;34(3):326–343. doi: 10.15252/embj.201489184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones JC, Zhen WP, Reed E, Parker RJ, Sancar A, Bohr VA. Gene-specific formation and repair of cisplatin intrastrand adducts and interstrand cross-links in Chinese hamster ovary cells. J Biol Chem. 1991;266(11):7101–7107. [PubMed] [Google Scholar]
- 30.McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2(8):483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- 31.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65(1–2):55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 32.Ahmad A, Robinson AR, Duensing A, van Drunen E, Beverloo HB, Weisberg DB, Hasty P, Hoeijmakers JH, Niedernhofer LJ. ERCC1-XPF endonuclease facilitates DNA double-strand break repair. Mol Cell Biol. 2008;28(16):5082–5092. doi: 10.1128/MCB.00293-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 2008;24(11):529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hodskinson MR, Silhan J, Crossan GP, Garaycoechea JI, Mukherjee S, Johnson CM, Scharer OD, Patel KJ. Mouse SLX4 is a tumor suppressor that stimulates the activity of the nuclease XPF-ERCC1 in DNA crosslink repair. Mol Cell. 2014;54(3):472–484. doi: 10.1016/j.molcel.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein Douwel D, Boonen RA, Long DT, Szypowska AA, Raschle M, Walter JC, Knipscheer P. XPF-ERCC1 acts in Unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell. 2014;54(3):460–471. doi: 10.1016/j.molcel.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wood RD. Mammalian nucleotide excision repair proteins and interstrand crosslink repair. Environ Mol Mutagen. 2010;51(6):520–526. doi: 10.1002/em.20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayakawa H, Ishizaki K, Inoue M, Yagi T, Sekiguchi M, Takebe H. Repair of ultraviolet radiation damage in xeroderma pigmentosum cells belonging to complementation group F. Mutat Res. 1981;80(2):381–388. doi: 10.1016/0027-5107(81)90110-X. [DOI] [PubMed] [Google Scholar]
- 38.Ahmad A, Enzlin JH, Bhagwat NR, Wijgers N, Raams A, Appledoorn E, Theil AF, JH JH, Vermeulen W, NG JJ, Scharer OD, Niedernhofer LJ. Mislocalization of XPF-ERCC1 nuclease contributes to reduced DNA repair in XP-F patients. PLoS Genet. 2010;6(3):e1000871. doi: 10.1371/journal.pgen.1000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gregg SQ, Robinson AR, Niedernhofer LJ. Physiological consequences of defects in ERCC1-XPF DNA repair endonuclease. DNA Repair (Amst) 2011;10(7):781–791. doi: 10.1016/j.dnarep.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bogliolo M, Schuster B, Stoepker C, Derkunt B, Su Y, Raams A, Trujillo JP, Minguillon J, Ramirez MJ, Pujol R, Casado JA, Banos R, Rio P, Knies K, Zuniga S, Benitez J, Bueren JA, Jaspers NG, Scharer OD, de Winter JP, Schindler D, Surralles J. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am J Hum Genet. 2013;92(5):800–806. doi: 10.1016/j.ajhg.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, Theil AF, Vermeulen W, van der Horst GT, Meinecke P, Kleijer WJ, Vijg J, Jaspers NG, Hoeijmakers JH. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444(7122):1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 42.Tian M, Shinkura R, Shinkura N, Alt FW. Growth retardation, early death, and DNA repair defects in mice deficient for the nucleotide excision repair enzyme XPF. Mol Cell Biol. 2004;24(3):1200–1205. doi: 10.1128/MCB.24.3.1200-1205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evers B, Jastrzebski K, Heijmans JP, Grernrum W, Beijersbergen RL, Bernards R. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat Biotechnol. 2016;34(6):631–633. doi: 10.1038/nbt.3536. [DOI] [PubMed] [Google Scholar]
- 45.Hart T, Brown KR, Sircoulomb F, Rottapel R, Moffat J. Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Mol Syst Biol. 2014;10:733. doi: 10.15252/msb.20145216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biggerstaff M, Szymkowski DE, Wood RD. Co-correction of the ERCC1, ERCC4 and xeroderma pigmentosum group F DNA repair defects in vitro. EMBO J. 1993;12(9):3685–3692. doi: 10.1002/j.1460-2075.1993.tb06043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Vuuren AJ, Appeldoorn E, Odijk H, Yasui A, Jaspers NG, Bootsma D, Hoeijmakers JH. Evidence for a repair enzyme complex involving ERCC1 and complementing activities of ERCC4, ERCC11 and xeroderma pigmentosum group F. EMBO J. 1993;12(9):3693–3701. doi: 10.1002/j.1460-2075.1993.tb06044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yagi T, Wood RD, Takebe H. A low content of ERCC1 and a 120kDa protein is a frequent feature of group F xeroderma pigmentosum fibroblast cells. Mutagenesis. 1997;12(1):41–44. doi: 10.1093/mutage/12.1.41. [DOI] [PubMed] [Google Scholar]
- 49.Gaillard PH, Wood RD. Activity of individual ERCC1 and XPF subunits in DNA nucleotide excision repair. Nucleic Acids Res. 2001;29(4):872–879. doi: 10.1093/nar/29.4.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boulikas T (1997) Nuclear import of DNA repair proteins. Anticancer Res 17 (2 A):843–863 [PubMed]
- 51.Jordheim LP, Barakat KH, Heinrich-Balard L, Matera EL, Cros-Perrial E, Bouledrak K, El Sabeh R, Perez-Pineiro R, Wishart DS, Cohen R, Tuszynski J, Dumontet C. Small molecule inhibitors of ERCC1-XPF protein–protein interaction synergize alkylating agents in cancer cells. Mol Pharmacol. 2013;84(1):12–24. doi: 10.1124/mol.112.082347. [DOI] [PubMed] [Google Scholar]
- 52.Mu D, Hsu DS, Sancar A. Reaction mechanism of human DNA repair excision nuclease. J Biol Chem. 1996;271(14):8285–8294. doi: 10.1074/jbc.271.14.8285. [DOI] [PubMed] [Google Scholar]
- 53.Sijbers AM, de Laat WL, Ariza RR, Biggerstaff M, Wei YF, Moggs JG, Carter KC, Shell BK, Evans E, de Jong MC, Rademakers S, de Rooij J, Jaspers NG, Hoeijmakers JH, Wood RD. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86(5):811–822. doi: 10.1016/S0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 54.Enoiu M, Jiricny J, Scharer OD. Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucleic Acids Res. 2012;40(18):8953–8964. doi: 10.1093/nar/gks670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cimino GD, Gamper HB, Isaacs ST, Hearst JE. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu Rev Biochem. 1985;54:1151–1193. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]
- 56.Gupta AK, Anderson TF. Psoralen photochemotherapy. J Am Acad Dermatol. 1987;17(5 Pt 1):703–734. doi: 10.1016/S0190-9622(87)70255-2. [DOI] [PubMed] [Google Scholar]
- 57.Hearst JE. Psoralen photochemistry. Annu Rev Biophys Bioeng. 1981;10:69–86. doi: 10.1146/annurev.bb.10.060181.000441. [DOI] [PubMed] [Google Scholar]
- 58.Fisher LA, Bessho M, Bessho T. Processing of a psoralen DNA interstrand cross-link by XPF-ERCC1 complex in vitro. J Biol Chem. 2008;283(3):1275–1281. doi: 10.1074/jbc.M708072200. [DOI] [PubMed] [Google Scholar]
- 59.Kuraoka I, Kobertz WR, Ariza RR, Biggerstaff M, Essigmann JM, Wood RD. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J Biol Chem. 2000;275(34):26632–26636. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- 60.Rahn JJ, Adair GM, Nairn RS. Multiple roles of ERCC1-XPF in mammalian interstrand crosslink repair. Environ Mol Mutagen. 2010;51(6):567–581. doi: 10.1002/em.20583. [DOI] [PubMed] [Google Scholar]
- 61.Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006;25(20):4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pizzolato J, Mukherjee S, Scharer OD, Jiricny J. FANCD2-associated nuclease 1, but not exonuclease 1 or flap endonuclease 1, is able to unhook DNA interstrand cross-links in vitro. J Biol Chem. 2015;290(37):22602–22611. doi: 10.1074/jbc.M115.663666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takahashi D, Sato K, Hirayama E, Takata M, Kurumizaka H. Human FAN1 promotes strand incision in 5′-flapped DNA complexed with RPA. J Biochem. 2015;158(3):263–270. doi: 10.1093/jb/mvv043. [DOI] [PubMed] [Google Scholar]
- 64.Wang R, Persky NS, Yoo B, Ouerfelli O, Smogorzewska A, Elledge SJ, Pavletich NP. DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1. Science. 2014;346(6213):1127–1130. doi: 10.1126/science.1258973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamamoto KN, Kobayashi S, Tsuda M, Kurumizaka H, Takata M, Kono K, Jiricny J, Takeda S, Hirota K. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc Natl Acad Sci U S A. 2011;108(16):6492–6496. doi: 10.1073/pnas.1018487108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J, Walter JC. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair (Amst) 2014;19:135–142. doi: 10.1016/j.dnarep.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mogi S, Oh DH. gamma-H2AX formation in response to interstrand crosslinks requires XPF in human cells. DNA Repair (Amst) 2006;5(6):731–740. doi: 10.1016/j.dnarep.2006.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murray D, Macann A, Hanson J, Rosenberg E. ERCC1/ERCC4 5′-endonuclease activity as a determinant of hypoxic cell radiosensitivity. Int J Radiat Biol. 1996;69(3):319–327. doi: 10.1080/095530096145878. [DOI] [PubMed] [Google Scholar]
- 69.Wood RD, Burki HJ, Hughes M, Poley A. Radiation-induced lethality and mutation in a repair-deficient CHO cell line. Int J Radiat Biol Relat Stud Phys Chem Med. 1983;43(2):207–213. doi: 10.1080/09553008314550241. [DOI] [PubMed] [Google Scholar]
- 70.Al-Minawi AZ, Saleh-Gohari N, Helleday T. The ERCC1/XPF endonuclease is required for efficient single-strand annealing and gene conversion in mammalian cells. Nucleic Acids Res. 2008;36(1):1–9. doi: 10.1093/nar/gkm888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sargent RG, Rolig RL, Kilburn AE, Adair GM, Wilson JH, Nairn RS. Recombination-dependent deletion formation in mammalian cells deficient in the nucleotide excision repair gene ERCC1. Proc Natl Acad Sci U S A. 1997;94(24):13122–13127. doi: 10.1073/pnas.94.24.13122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fishman-Lobell J, Haber JE. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258(5081):480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 73.Ivanov EL, Haber JE. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15(4):2245–2251. doi: 10.1128/MCB.15.4.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4(6):e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol Cell Biol. 2003;23(23):8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, Rajewsky K, Alt FW. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449(7161):478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 77.Hecht SM. Bleomycin: new perspectives on the mechanism of action. J Nat Prod. 2000;63(1):158–168. doi: 10.1021/np990549f. [DOI] [PubMed] [Google Scholar]
- 78.Tomicic MT, Kaina B. Topoisomerase degradation, DSB repair, p53 and IAPs in cancer cell resistance to camptothecin-like topoisomerase I inhibitors. Biochim Biophys Acta. 2013;1835(1):11–27. doi: 10.1016/j.bbcan.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 79.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17(5):421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]