Abstract
Primary cilia are solitary, microtubule-based protrusions of the cell surface that play fundamental roles as photosensors, mechanosensors and biochemical sensors. Primary cilia dysfunction results in a long list of developmental and degenerative disorders that combine to give rise to a large spectrum of human diseases affecting almost any major body organ. Depending on the cell type, primary ciliogenesis is initiated intracellularly, as in fibroblasts, or at the cell surface, as in renal polarized epithelial cells. In this review, we have focused on the routes of primary ciliogenesis placing particular emphasis on the recently described pathway in renal polarized epithelial cells by which the midbody remnant resulting from a previous cell division event enables the centrosome for initiation of primary cilium assembly. The protein machinery implicated in primary cilium formation in epithelial cells, including the machinery best known for its involvement in establishing cell polarity and polarized membrane trafficking, is also discussed.
Keywords: Primary ciliogenesis, Protein machinery, Fibroblasts, Polarized epithelial cells, Midbody remnant, Intracellular pathway, Alternative pathway
Types and general functions of cilia
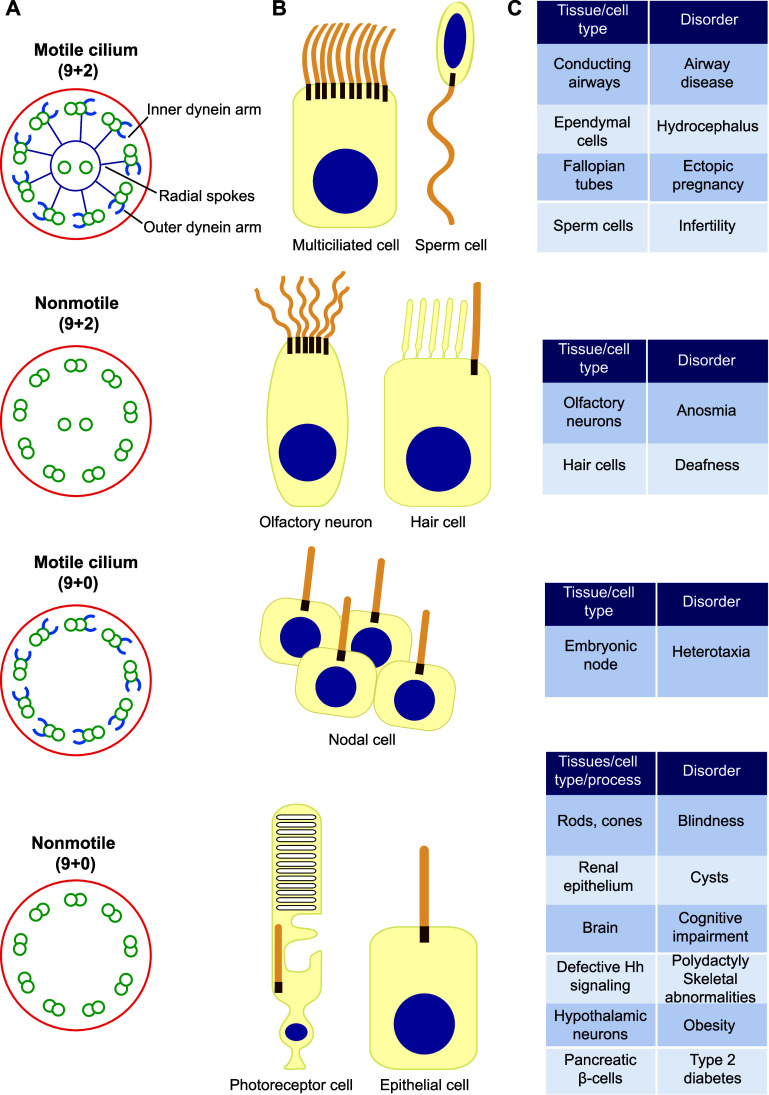
Cilia are highly conserved microtubule-based membrane extensions that protrude from the cell surface [1–3]. They are organized around a central microtubular scaffold, termed the axoneme, which derives from the centrosome and is surrounded by the ciliary membrane. Cilia are classified as 9 + 2 and 9 + 0, according to the number of microtubules associated with the axoneme. The numbers indicate the number of peripheral doublets (nine) and the presence (two) or absence (zero) of central microtubule singlets. In the case of 9 + 2 cilia, protein complexes known as radial spokes connect the central pair and the outer doublets. Mammalian cilia have also customarily been divided into two categories: motile and nonmotile cilia. Motile cilia contain outer and inner arms formed by the motor protein dynein in each microtubule doublet. 9 + 2 cilia are usually called flagella when they are motile and long (>10 μm). Nonmotile cilia lack dynein arms and can adopt the 9 + 2 or 9 + 0 configuration (Fig. 1a).
Fig. 1.
Types of cilium. a Schematic of distinct types of cilium as seen in cross-section. b Examples of cell types harboring each type of cilium. c Examples of disorders associated with the dysfunction of the distinct types of cilium
Cilia whose function is to move mucus or other fluids (Fig. 1b), such as multiciliated cells of conducting airways, ependymal cells and the fallopian tubes [4], and those involved in cell motility, such as the single flagellum of spermatozoa and trypanosomes [5, 6] or the two flagella of the green algae Chlamydomonas, contain motile 9 + 2 cilia with dynein arms [7]. Nonmotile 9 + 2 cilia without dynein arms are found in some sensory cells, such as mammalian olfactory neurons, which have 10–30 cilia, and the hair cells of the inner ear whose cilia, known as kinocilia, are involved in mechanotransduction (Fig. 1a, b) [8]. 9 + 2 cilia vary in length ranging from 3 to 10 μm in multiciliated cells, from 50 to 150 μm for sperm flagella and to 200 μm in the case of olfactory cilia.
Cells in the ventral node, which is an embryonic cavity at the midline filled with extra-embryonic fluid, contain a single motile cilium, referred to as the nodal cilium, which has a 9 + 0 pattern and contains dynein arms (Fig. 1a, b). Nodal cilia rotate to generate unidirectional leftward fluid flow, which is essential for breaking the left–right symmetry of internal organs in vertebrates during embryogenesis [9, 10].
Cells of almost all mammalian tissues have a single copy of a nonmotile cilium, referred to as the primary cilium, which has a 9 + 0 structure and no dynein arms (Fig. 1a, b). The primary cilium typically attains a length of 3–10 μm and is found in quiescent and differentiated cells [11]. This review will focus on primary cilia, with particular emphasis on the biogenesis of the primary cilium of polarized epithelial cells, and on the machinery underlying the process of primary ciliogenesis specific to these cells.
Primary cilium function
The role of the primary cilium is well known in photoreceptor cells, in which the cilium adopts a specialized structure that concentrates visual pigments for photon absorption [12]. Although the primary cilium was first described more than a century ago [13], its function in all other cells has been an enigma for a long time. Nowadays, in addition to photosensors, a fundamental role has been established for primary cilia as mechanosensors and biochemical sensors [3, 14, 15].
Mechanosensation refers to the physical sensation of flow, pressure, touch or vibration. Much of our understanding of the mechanosensory functions associated with cilia derives from studies of renal epithelial cells, in which the force of luminal fluid flow is sensed by primary cilia [16]. Polycystin-2 is a transient receptor potential family Ca2+ channel that associates with polycystin-1 at the ciliary membrane. Mutations of polycystin-1 and polycystin-2 both cause autosomal dominant polycystic kidney disease [17]. The primary cilia of epithelial Madin–Darby canine kidney (MDCK) cells become deflected through a combination of bending and pivoting [18]. Ciliary bending and pivoting can both trigger membrane channels, and are able to induce Ca2+ influx through the action of polycystin-2 [18]. It has been reported that, upon shear stress, cilia import extracellular Ca2+, raising their ciliary concentration [19–22]. Increased Ca2+ ciliary levels have been proposed to act as a second messenger to regulate multiple downstream processes in primary cilia [15, 22–25]. However, although the role of Ca2+ in mechanotransduction had been generally accepted, it has recently been challenged on the grounds that cilia-specific Ca2+ influxes have not been observed in physiological or even highly supraphysiological levels of fluid flow in primary cilia of cultured kidney epithelial cells, the thick limb of the ascending kidney tubule, crown cells of the embryonic node, hair cells, and several cell lines [26]. Therefore, the induction of Ca2+ flow as the mechanism of primary cilia in transducing mechanosensation is being reexamined [27].
Primary cilia act as biochemical sensors when they respond to hormones or other soluble factors capable of triggering a number of signaling cascades. Primary cilia transduce environmental stimuli through surface receptors specifically localized on the ciliary membrane, and regulate signaling pathways important for development, cell proliferation, differentiation, survival and migration [3, 14, 15]. Hedgehog (Hh) proteins regulate the development of a wide range of metazoan embryonic and adult structures, and disruption of Hh signaling pathways results in human disease [28, 29]. In response to Hh, its receptor, Patched, which normally resides in the primary cilium, leaves the cilium, and Smoothened (Smo), which is normally excluded, enters the cilium, and the downstream effectors of the Hh pathway assemble for signaling [30, 31]. Platelet-derived growth factor (PDGF)-α signaling, which controls cell migration, proliferation and survival, also occurs in the primary cilia since its receptor localizes to primary cilia in fibroblasts, where PDFG-α activates it and triggers the activation of downstream signaling machinery [32]. Canonical and non-canonical/planar cell polarity Wnt pathways regulate developmental and homeostatic processes [33]. The Hippo pathway controls organ size and proliferation [34]. The primary cilium seems to play a role in dictating the outcome (canonical or non-canonical) of Wnt signaling [35, 36] and also controls the Hippo pathway [37]. In addition, G protein-coupled receptors (GPCRs) of hormones, peptides, lipids and neurotransmitters, including for instance those of dopamine, serotonin, neuropeptide Y and somatostatin, reside in primary cilia and use cilia for signaling [25, 38].
Given the importance of cilia, it is not surprising that ciliary dysfunction by mutation in cilia-related genes causes a great variety of disorders in humans (Fig. 1c). If the mutated gene affects motile 9 + 2 cilia the disorders caused are related to mucociliary clearance (bronchitis, rhinosinusitis), hydrocephalus and infertility; defective functioning of nodal cilia causes heterotaxia (the abnormal arrangement of internal organs in the chest and/or abdomen). Dysfunction of nonmotile cilia produces defects in signaling that result in a large variety of symptoms (renal and liver cysts, blindness, cognitive impairment, deafness, anosmia, polydactyly, skeletal abnormalities, obesity, etc.). Depending on the gene(s) affected, these alterations combine to produce numerous heterogeneous human developmental and degenerative genetic diseases, collectively known as ciliopathies, that affect nearly every major body organ (for extensive reviews, see Refs. [39–41]). Ciliopathies are characterized by overlapping phenotypes that may include multiple symptoms caused by the dysfunction of motile and/or primary cilia. For instance, primary cilia dyskinesia and Kartagener syndrome are caused by defective motile cilia [42], whereas in other ciliopathies, such as nephronophthisis (NPHP), Meckel (MKS) and Joubert syndromes (JBTS), only primary cilia function tends to be affected [43]. Largely due to their medical relevance, there has been an immense increase in research in recent times aimed at better understanding the structure, mechanisms of assembly, maintenance and function of primary cilia.
Structure of primary cilia
The general structure of cilia is evolutionarily conserved, despite the obvious difference between the distinct types of axoneme [44]. In contrast to specialized cilia such as those in photoreceptor cells, the primary cilium of epithelial cells, fibroblasts, muscle cells, neurons, etc., adopts a similar morphology and size (Fig. 2) [2, 45].
Fig. 2.
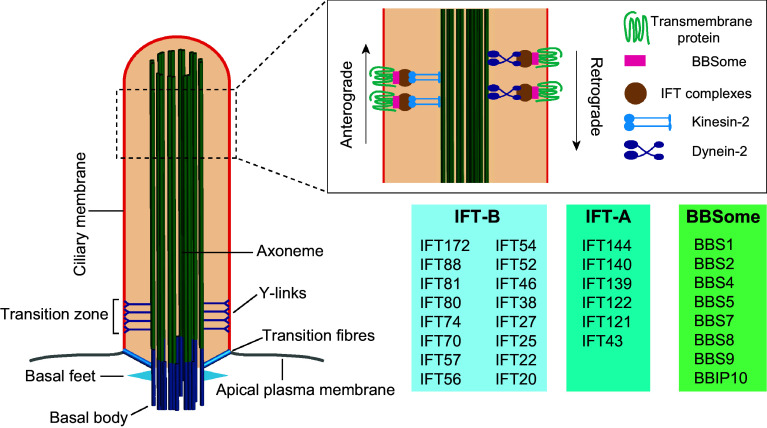
General structure of the primary cilium. The basal body is attached to the ciliary membrane by the transition fibers. The axoneme constitutes the backbone of the cilium and is surrounded by the ciliary membrane, which is continuous with the plasma membrane. IFT is carried out by the IFT-A and IFT-B complexes powered by dynein-2 and kinesin-2 motors, respectively, and with the participation of the BBSome complex
Primary cilia have a basal body, which consists of the older of the two centrioles, also known as the mother centriole, in the centrosome and the associated accessory structures. These accessory structures include transition fibers, basal feet and ciliary rootlets [46–48]. Transition fibers and basal feet are ultrastructurally similar to the distal and subdistal appendages, respectively, of the mother centriole. Transition fibers emerge from the central microtubule of each triplet of the basal body just before the end of the most external microtubule, and are involved in docking the basal body to the plasma membrane [49]. The distal appendages possess proteins, such as Cep164, Cep89 and Cep83/Ccdc41, that are important for ciliogenesis. Basal bodies have nine transition fibers, but only one or two basal feet. The basal feet further differ from the subdistal appendages in that they are larger and more electron dense [50]. The outer dense fiber protein 2 (Odf2)/Cenexin, a component of the distal and subdistal appendages, is essential for the formation of transition fibers and basal feet [51]. The rootlet is a thick (80–100 nm) striated bundle of filaments made of the protein rootletin [52]. Basal feet, which anchor microtubules, and striated rootlets, which project from the proximal end of the basal body and extend close to the nucleus, provide structural support to the cilium.
Beyond the basal body is the transition zone, which is an intermediate region between the basal body and the axoneme [53, 54]. The transition zone is distinguished by the shift from triplet microtubules in the basal body to axonemal doublets, and by the presence of characteristic scallop-like structures at the ciliary surface and of inner structures, known as Y-links, that appear Y-shaped under electron microscopy and connect the outer doublet microtubules to the overlying ciliary membrane [55–57]. The transition zone houses a network of two biochemically distinct protein complexes involved in ciliopathies. One of the modules spans the membrane and contains many of the proteins (Tctn1–3, MKS1, B9d1, B9d2, Cep290, Ahi and the transmembrane proteins Tmem67, Tmem216, Tmem17, Tmem231, Tmem107, etc.) involved in MKS and JBTS. The second module, called the NPHP module, includes three proteins (Nphp1, Nphp4 and Rpgrip1l) encoded by genes mutated in NPHP, and is proximal to the axoneme. The collaboration of the two modules in which Rpgrip1l and Cep290 play an important role explains the overlapping phenotypes seen in MKS, JBTS and NPHP, in which proteins belonging to these modules are involved [58, 59].
Following the transition zone, the axoneme of primary cilia is constructed from the elongation of the nine parallel doublet microtubules formed at the transition zone [60, 61]. As the axoneme becomes longer, it loses microtubules and the doublets transform into singlets. Singlets are also lost gradually in such a way that the tip of the cilium often contains only a few of them. The axoneme is subject to numerous post-translational modifications, including acetylation, detyrosination, glutamylation and glycylation, which are related to microtubule structure, flexibility and function [62–64].
Cilia have no machinery for protein synthesis, so all ciliary proteins must be synthesized elsewhere in the cell and imported selectively into the cilium. Although the ciliary compartment lacks a limiting membrane that separates it from the cytosol, the base of the primary cilium selectively regulates the entry of proteins. Protein segregation is made possible by a functional gate at the ciliary base that is responsible for the selective entry of proteins into the primary cilium. This gate encompasses the transition fibers, the ciliary base and the transition zone and also involves importins, the GTPase Ran and nucleoporins, similar to nuclear import, and septin polymers (for extensive reviews see Refs. [65–68]).
Ciliary growth is regulated at the tip by the receipt of tubulin and other axonemal precursors that elongates the axoneme and by their removal during axoneme disassembly. Defined structures at the tip were reported for the 9 + 2 flagella of Chlamydomonas and of other organisms [69–72]. These structures cap the central singlet microtubules at their tip and, it has been proposed, are involved in cargo loading and unloading, and signal transduction [73]. However, such well-defined structures have not been detected in primary cilia of mammalian cells. Not only is the tip of primary cilia the place where ciliary growth is regulated, as in 9 + 2 cilia, but also it is involved in cell signaling. Kif7, a kinesin-4 family protein that is a conserved regulator of the Hh signaling pathway and a human ciliopathy protein, binds to the distal end of axonemal microtubules and organizes a specialized compartment where the activity of the Gli family of Hh transcription factors is regulated [30, 74, 75]. Recent evidence from Chlamydomonas flagella and Caenorhabditis elegans and mammalian primary cilia showed that ciliary signaling is also regulated at the ciliary tip by shedding receptors and other material in the form of extracellular vesicles [76–81]. Cilia typically begin to form during the Go phase of the cell cycle and begin to disassemble as cells re-enter the cell cycle to free the centrosome [82, 83]. While cilia disassembly has for some time been considered to occur solely through resorption [84], release of extracellular vesicles also helps to regulate ciliary disassembly and size [77, 78, 80]. In conclusion, although the nature of the tip of primary cilia is not well understood, it appears to act as a hub that coordinates many important ciliary functions.
Protein machinery for ciliary growth, targeting and transport
Cilia require general machinery for the processes of licensing the centrosome to initiate cilium formation, and protein transport along the ciliary membrane. These general tasks are performed by the regulators of the centriolar protein Cp110 and by intraflagellar transport (IFT) machinery, respectively. The BBsome complex is also responsible for the traffic of certain receptors at the primary cilium. Rab-family proteins control membrane trafficking during primary cilium initiation and the targeting of cargo to the ciliary base once the cilium has formed. In addition to the general machinery, the Par complex, which is involved in acquisition of cell polarity, and the exocyst, which is a protein complex implicated in polarized transport in epithelial cells, are also important for ciliogenesis in polarized epithelial MDCK cells and kidney tubules. Since the functions performed by the IFT and the BBSome complex have been extensively reviewed recently [85, 86], they will be mentioned only briefly below, whereas particular emphasis will be placed on the machinery specific to polarized epithelial cells.
The protein CP110 and its regulators
The well-known negative regulators of ciliogenesis, such as centriolar protein Cp110 and its network of interacting partners, have been studied in the human bone osteosarcoma U2OS cell line, as well as in NIH-3T3 fibroblasts and retinal pigment epithelial (RPE)-1 cells [87–89]. Cp110 localizes at the mother and the daughter centrioles, blocking primary cilium formation. At the beginning of ciliogenesis, Cp110 is removed from the basal body to allow axoneme extension, but remains at the daughter centriole. The transition zone protein Cep290 interacts with Cp110 and this interaction is essential for suppressing primary cilium formation [88]. Another complex containing Cp110 and Cep97 also serves to suppress primary cilium assembly [87]. Cp110 removal requires the activity of positive ciliary regulators, such as Tau tubulin kinase-2 (Ttbk2), whose knockout inhibits Cp110 removal from the basal bodies in mouse embryonic fibroblasts (MEFs) [90]. Cep164, which is present in transition fibers, is essential for ciliogenesis and for recruiting Ttbk2 to the basal body [91]. Microtubule affinity regulating kinase 4 (Mark4) is also required for Cp110 removal and accumulates at the basal body as Cp110 is displaced [92]. These findings indicate that disappearance of Cp110 from the mother centriole is crucial for initiating primary cilium biogenesis. Cp110 removal seems universally required for initiating the ciliation process, since cells that normally do not form a primary cilium, such as T lymphocytes, assemble one when Cp110 expression is knocked down [93]. The Aurora A kinase is necessary for the organization and alignment of the chromosomes during prometaphase and for their separation, and contributes to the completion of cytokinesis [94]. Ciliogenesis is inhibited by Aurora A, which is bound and activated by trichoplein at the centriole [95]. Cul3-RING E3 ligases, aided by the protein KCTD17, ubiquitinate trichoplein and target it for degradation by the proteasome, allowing initiation of axoneme extension [96]. Therefore, given its role in regulating the cell cycle, the ubiquitin–proteasome system has emerged as an important coordinator of cell cycle progression and primary ciliogenesis initiation (for extensive reviews, see Refs. [97–99]).
IFT machinery
Soluble and membrane proteins are transported along the primary cilium by the IFT machinery [7, 85, 100]. IFT is highly conserved and is required for the assembly of cilia. It is formed of two multisubunit complexes: IFT-A and IFT-B, comprising 6 and 16 subunits, respectively (Fig. 2). Mutations in IFT proteins can cause several ciliopathies [101]. With the exception of the two small GTPases IFT22 and IFT27, none of the other IFT proteins is predicted to have enzymatic activity. IFT-complex polypeptides are largely composed of α-solenoids and β-propeller domains that predominate in COPI, COPII and clathrin cage components and form planar coats resembling flat COPI, COPII and clathrin coats at the ultrastructural level [102]. IFT-B mediates anterograde movement (from the cell body to the cilium) of ciliary proteins, whereas IFT-A directs retrograde transport (from the cilium to the cell body) and anterograde transport of certain proteins such as Arl13b and Smo. The IFT-B complex transports cargo to the ciliary tip with the participation of kinesin-2 motors—heterotrimeric kinesin-2, composed of Kif3a, Kif3b, and Kap, or homodimeric Kif17—whereas turnover products or signaling components destined for internalization are returned to the cell body via the IFT-A complex propelled by cytoplasmic dynein-2 [85]. The switch of the machinery for anterograde and retrograde IFT and their respective motors takes place at the ciliary tip. In contrast to trimeric kinesin-2, Kif17 is not required for ciliogenesis and its ciliary entry requires it to interact with both importin-β2 and Rab23 [103]. In Chlamydomonas, it has been demonstrated that each microtubule doublet is used as a bidirectional double-track railway in such a way that anterograde and retrograde IFT trains move along the exterior and the interior microtubules, respectively, of each doublet [104]. Thus, the microtubule doublet geometry provides direction-specific rails to coordinate the bidirectional transport of ciliary components.
Small Rab GTPases
In a screen of 39 human GTPase-activating proteins, GAPs for Rab8a, Rab17 and Rab23 were identified as necessary for primary cilium formation in RPE-1 cells [105]. Rab8a was the only one of the three GTPases in that study found to localize to the cilium. Rab8a is recruited to the centrosome by a direct interaction with Odf2/Cenexin and is required for primary ciliogenesis [105, 106]. Although they were not identified in the original screen, Rab11 and Rab10 have also been implicated in this process [106–108]. Two centrosome appendage proteins, centriolin and Odf2/Cenexin, regulate the association of Rab11 vesicles with the distal part of the mother centriole [109]. The GTP-bound form of Rab11 interacts directly with its downstream effector Rabin8 to target it to the centriole and stimulates its guanine nucleotide exchange factor (GEF) activity toward Rab8 in RPE-1 cells [107]. Rabin8, in turn, interacts with the membrane-tethering transport protein particle II complex, which is a GEF for Rab11 [110]. It has been proposed that, similar to the Rab5–Rab7 switch [111], Rab11 vesicles are converted into a Rab8 preciliary vesicle with the participation of Rabin8 and transport protein particle II complex [106]. Knockdown of Rab11, Rabin8 or Rab8 inhibits ciliogenesis, highlighting the importance of this signaling cascade [106, 107]. Despite evidence supporting a crucial role for Rab8 in primary cilium formation in cultured cells, the absence of its two isoforms, Rab8a and Rab8b, in Rab8a and Rab8b double-knockout mice does not disturb ciliogenesis of olfactory epithelium, photoreceptors and MEFs [112]. However, the additional knockdown of Rab10, but not of Rab13, in MEFs from these mice greatly reduces the percentage of ciliated cells [112]. This finding suggests that the Rab8a, Rab8b and Rab10 proteins are simultaneously, rather than individually, involved in ciliogenesis.
Rab23 has been shown to localize to the cilium of MDCK cells, thereby mediating the ciliary turnover of Smo and possibly of other membrane receptors [113]. Coimmunoprecipitation and affinity-binding studies revealed that Rab23 exists in a complex with Kif17 and importin β2, implying that Kif17 needs to bind to regulatory proteins like Rab23 for its ciliary transport. Although a ciliary-cytoplasmic gradient of nuclear Ran is necessary to regulate the ciliary transport of Kif17, Rab23 and Ran appear to have differing roles in regulating the ciliary entry of Kif17 [114]. On the other hand, consistent with a role of Rab23 in ciliary transport, Rab23 and Kif17 have been observed to collaborate with IFT-B to deliver specific receptors to primary cilia [115].
The BBSome
Bardet–Biedl syndrome (BBS) is a compound phenotype disorder exhibiting cystic kidneys, obesity, mental retardation, hypogonadism, heterotaxia, polydactyly and retinal degeneration [86, 116]. The BBSome is a multimeric protein complex composed of seven highly conserved BBS proteins (BBS1, BBS2, BBS4, BBS5, BBS7, BBS8 and BBS9) and BBIP10, each of which is present in stoichiometric amounts [117]. In humans, defects in the BBSome result in BBS [116]. The BBSome contains coat-like structural elements common to COPI, COPII, and clathrin coats, and is the main effector of the Arf-like GTPase Arl6/BBS3 [118]. Rabin8 interacts directly with the BBS1 subunit and recruits the BBSome. Since Rabin8 activates Rab8, it has been proposed that the BBSome acts upstream of Rab8 [119]. Silencing BBS2 or BBS4 greatly reduces the number of ciliated RPE-1 cells. Live analysis of cargo transport in olfactory sensory neurons revealed that the BBSome complex moves in association with IFT trains and cargo through cilia, suggesting that the BBsome acts as a cargo adaptor between membrane cargoes and the IFT machinery [119].
The BBSome was initially implicated in GPCR delivery to cilia [118, 120, 121]. However, it is now known to be important in retrograde trafficking. The BBsome regulates the removal of GPCRs [122–124], polycystin-2 [125], and membrane-associated proteins from cilia [126, 127]. The conflict resulting from the role of the BBsome in anterograde transport is explained by the observation that when membrane receptors fail to undergo BBSome-mediated retrieval from the cilium back into the cell, they are removed by ectocytosis giving the impression that their sorting to the cilium was defective [77].
The exocyst complex and Arl13b
Tethering complexes are large protein complexes that establish long-range interactions between donor and acceptor membranes to capture transport vesicles and enable their fusion with acceptor organelles before contacts between v- and t-SNARES occur [128]. In addition to capturing vesicles, tethering complexes appear to regulate the spatial and temporal assembly of the SNARE complex. The exocyst is an eight-subunit (Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, and Exo84) complex [129, 130] that delineates its function as a tethering complex in the trafficking of vesicles from a post-Golgi compartment, the recycling endosome, to the basolateral plasma membrane in polarized epithelial cells [131]. Exocyst levels are higher in patients with autosomal dominant polycystic kidney disease [132].
The exocyst has been shown to be a downstream effector of exocytic Rab GTPases [131]. The exocyst subunit Sec15 directly interacts with Rab11 and Rabin8 and allows activation of Rab8 [133, 134]. The exocyst subunit Sec10 localizes to the primary cilium [135, 136]. Consistent with the role of the exocyst in primary ciliogenesis, knockdown of the exocyst component Sec10 leads to shorter cilia, whereas its overexpression leads to elongated cilia in MDCK cells. In addition, confirming the importance of Sec10, Sec10-knockout mice have defects in primary cilium assembly. These mice also show abnormal epithelial cell extrusion in renal tubes indicating a role for the exocyst and Sec10 in tubulogenesis [136]. As a further evidence of the role of the exocyst, the Sec6/8 subunits localize in apical ring-like structures at the base of nascent cilia of MDCK cells and colocalize and interact with Rab10, which is a Rab GTPase that collaborates with Rab8, which in turn, is involved in primary cilium assembly [108]. Sec10 colocalizes with polycystin-2 at the axoneme, and associates with polycystin-2, as well as with the IFT proteins IFT88 and IFT20. This interaction enables polycystin-2 to reach the primary cilium since, in the absence of Sec10, the ciliary localization of polycystin-2 is impaired [137]. This was corroborated by the demonstration that the effects of Sec10 knockdown in vitro and in vivo partially resemble those of polycystin-2 knockdown [137].
Arl13b is a member of the ADP-ribosylation factor family and the Ras superfamily of GTPases. Arl13b concentrates in primary cilia and attaches to the ciliary membrane through a palmitoyl residue [138]. Arl13b mutation results in JBTS, a ciliopathy characterized by mental retardation, congenital cerebellar ataxia and hypotonia [139]. Arl13b has been found to play multiple independent roles, such as modulation of the ciliary length and post-translational modifications of ciliary tubulin [140], and the correct recruitment of Hh signaling components, such as Smo, to the cilium [140, 141]. It is of note that Arl13b directly interacts with the exocyst subunits Sec8 and Sec5, which, together with Arl13b, have been detected along the axoneme of NIH-3T3 fibroblasts, epithelial RPE-1 cells and polarized epithelial MDCK cells [142]. Additionally, a synergistic genetic interaction between Arl13b and Sec10 has been demonstrated in zebrafish morphants showing cilia-related phenotypes [142]. Consistent with this role, the conditional deletion of Arl13b and Sec10 in mouse kidney causes cyst development and reduces ciliogenesis [142]. The observation that the exocyst complex preferentially binds to Arl13b coupled to GTP suggests that the exocyst is an effector of Arl13b.
The Par complex and Crumbs3
In polarized epithelial cells, tight junctions allow the generation of well-defined apical and basal membrane domains. Cilium elongation is the final event of the polarization process in these cells. Thus, many components of the polarity machinery, such as those involved in apical membrane biogenesis, establishment of cell junctions, and lumen formation, are directly linked to cilium formation. The Par complex consists of Par3, Par6, which are PDZ domain proteins, atypical protein kinase C (aPKC), and the small Rho-family GTPase Cdc42. In epithelial cells, the Par complex has been shown to play a role in regulating tight junction formation [143]. Par3, Par6 and aPKC localize to the cilia of MDCK and inner medullary collecting duct 3 cells, and Par3 and aPKC activity have been shown to be essential for efficient primary cilium formation in MDCK cells [144, 145]. In mammalian epithelia, 14-3-3 binds Par3, and disruption of the interaction leads to loss of cell polarity [146]. 14-3-3η, a 14-3-3 isoform, localizes to the primary cilium of MDCK cells and its depletion inhibits ciliogenesis [145], indicating that Par3 and 14-3-3η are essential for Par complex function in primary ciliogenesis. In addition, Par3, Par6, aPKC and 14-3-3η associate with the axoneme through the microtubule motor Kif3a [145]. Consistent with this finding, the coiled-coil domain of Par3, which is required for interaction with Kif3a, was demonstrated to be essential for ciliogenesis [144].
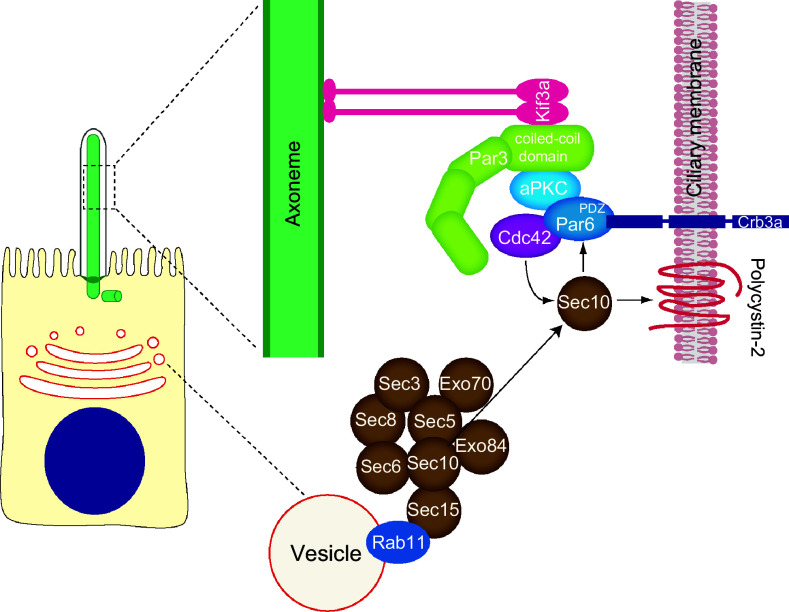
Cdc42, a component of the Par complex, interacts with exocyst subunit Sec10 and colocalizes with Sec10 at the primary cilium [147]. Cdc42 or Tuba, a GEF of Cdc42, knockdown inhibits ciliogenesis and ciliary targeting of polycystin-2. Moreover, depletion of Cdc42 affects the ciliary localization of Sec8, indicating that Cdc42 function is necessary for targeting the exocyst to the primary cilium. In addition, Sec10 directly interacts with the Par complex protein Par6, which itself associates with Cdc42. This observation led to the proposal that the exocyst complex is targeted to the primary cilium by Cdc42 and is then stabilized by binding to the Par complex via the Sec10–Par6 interaction (Fig. 3). Once it has become stabilized at the primary cilium, the exocyst targets and docks vesicles carrying ciliary proteins, such as polycystin-2, by interacting with Rab8 [147].
Fig. 3.
Multi-protein complexes involved in polarized trafficking and cell polarity implicated in primary cilium formation by polarized epithelial cells. In addition to their role in polarization process, the exocyst and the Par complexes participate in primary cilium assembly. Par complex consists of Par3, Par6, aPKC and Cdc42. The motor Kif3a targets the Par complex to the ciliary axoneme through interaction with the coiled-coil domain of Par3. The Par complex has been proposed to be an adaptor for targeting the transmembrane protein Crumbs3a to the ciliary membrane through association with the Par6 PDZ domain. Cdc42 is responsible for recruitment of exocyst complex, which targets important cargo, such as polycystin-2, to the cilium
Crumbs3, which is a transmembrane protein that plays an important role in the biogenesis of apical membrane and that is a key component in cell polarization [146], localizes to the primary cilium of MDCK and IMCD3 cells and is critical for primary cilium assembly [145]. Crumbs3 directly interacts with the PDZ domain of Par6 [144, 148]. This interaction is necessary for targeting Crumbs3 to the primary cilium, indicating that Par complex might act as an adaptor for targeting membrane proteins to the ciliary membrane via Kif3a [144]. Therefore, the Par complex is required for targeting both Crumbs3 and the exocyst complex to the primary cilium via Par6 and Cdc42, respectively (Fig. 3). The sphingolipid ceramide, which is a component of condensed membranes, localizes at the ciliary base in a compartment called the apical ceramide-enriched compartment, where it regulates a lipid–protein molecular network that sustains the primary cilium [74, 149]. Ceramide binds and activates aPKC, which, in turn, colocalizes with Rab11 in the apical ceramide-enriched compartment. Disruption of the ceramide-aPKC interaction results in impaired primary cilium formation and loss of the association of Rab11 vesicles with Cdc42, the exocyst subunit Sec8, and Rab8 [74]. Supporting the important role of ceramide, inhibition of ceramide biosynthesis by fumonisin B1 severely impairs primary ciliogenesis in MDCK cells [149].
An alternate splice form of Crumbs3, known as Crumbs3–CLPI, localizes at the primary cilium of MDCK cells and interacts with importin β-1 in a Ran-regulated fashion. Depletion of Crumbs3–CLPI disrupts primary cilium formation without affecting the junctional complex. However, unlike Crumbs3, Crumbs–CLPI does not interact with Par6 [150]. Therefore, the exact role of Crumbs–CLPI and how it is targeted to the ciliary membrane remain unknown. Other proteins involved in apical transport, including annexin-13, caveolin-1, galectin-3, syntaxin-3, syntaxin-2 and the MAL protein, are also shown to be involved in ciliogenesis [151–153]. This finding is further evidence of the participation of the machinery for apical membrane morphogenesis in primary cilium biogenesis and strengthens the relationship between the two processes.
Pathways of primary ciliogenesis
Despite the evolutionary conservation of the ciliary structure and ciliogenic machinery, cilia in different cell types and tissues are not created equal. The pioneering work of Sorokin [154] established that primary ciliogenesis proceeds by two distinct pathways, depending whether the position of the centrosome in the cell is near the nucleus or close to its apex.
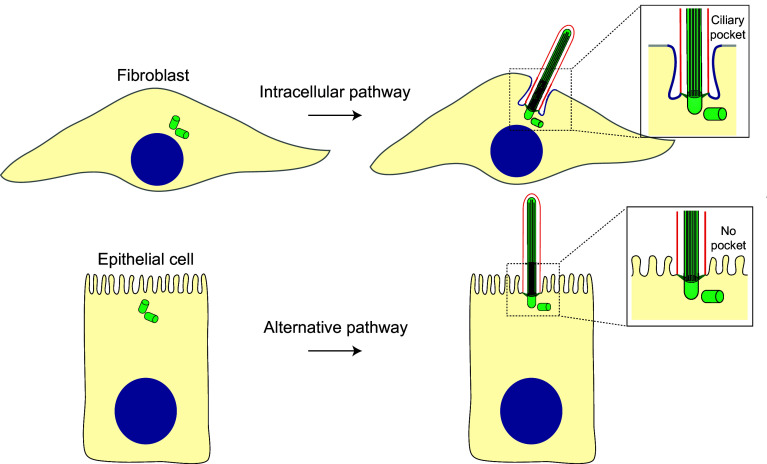
In cells of connective tissues, such as fibroblasts and chondrocytes, the primary cilium is present within an invagination of the plasma membrane, known as the ciliary pocket, whereas in other cell types it directly protrudes from the plasma membrane [155–158]. The ciliary pocket is characterized by containing budding clathrin-coated pits, and is thought to mediate ciliary endocytic activity and vesicular trafficking [156, 159]. It has also been proposed as a compartment of signal transduction including transforming growth factor-β signaling, which plays critical roles in cell cycle control, migration and differentiation [160]. In some cell types with ciliary pocket, the pocket is deep and the cilium is almost completely submerged, whereas in other cell types the pocket is shallow and the cilium is mostly exposed to the extracellular environment [2, 161–163]. RPE-1 cells, which generally form cilia with deep pockets, can be forced to form shallow pockets under specific confinement conditions [164]. The subdistal appendages, which presumably keep the mother centriole anchored to the Golgi apparatus, determine whether the pockets are shallow or deep [162]. Cells without a ciliary pocket or with a shallow one are free to sense motion, a process crucial for mechanosensation [165].
The presence or absence of the ciliary pocket appears to be a consequence of the route of primary cilium assembly used and, therefore, of the position of centrosome [157, 158]. When the centrosome is near the nucleus ciliogenesis starts intracellularly and finishes at the plasma membrane, generating a pocket, whereas when the centrosome is close to the plasma membrane the process takes place entirely at the plasma membrane and no pocket appears. The first route is referred to as the intracellular or “classic” pathway; the second route is known as the alternative pathway (Fig. 4).
Fig. 4.
Routes of primary ciliogenesis. The position of the centrosome, near the nucleus or close to the plasma membrane, and the presence or absence of a ciliary pocket predicts the type of pathway used for primary ciliogenesis. Fibroblasts and polarized epithelial cells are shown as examples of cells that use the intracellular and alternative routes, respectively
The intracellular pathway
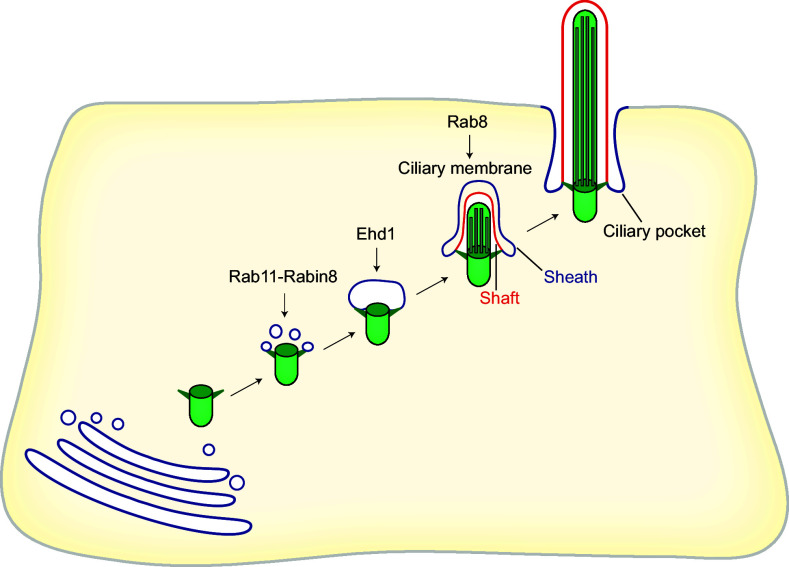
The process of primary ciliogenesis by the intracellular route was investigated in great detail in the seminal electron microscopy work of Sorokin [161], who analyzed the process in fibroblasts and smooth muscle cells. The ciliogenic process has been recapitulated entirely by electron microscopic analysis of cultured cells such as RPE-1 cells and NIH-3T3 fibroblasts [91, 92, 166–168]. Primary cilium formation in these cells starts intracellularly with the docking of small cytoplasmic vesicles to the mother centriole (Fig. 5). The distal appendage protein Cep164 and the distal centriolar protein Talpid3 are indispensable for the docking of these vesicles [169]. The origin of those vesicles is unclear, although they are presumably generated in the Golgi and recycling endosomes [170], and in embryonic neocortical stem cells they appear to derive from a previous ciliary membrane [171]. The vesicles associated with the mother centriole then fuse, generating a large ciliary vesicle that encapsulates the nascent axoneme. EHD1 and EHD3, two membrane-shaping proteins, have been identified as being crucial for the fusion of the small cytoplasmic vesicles in RPE-1 cells and zebrafish [166, 172]. SNAP-29, a SNARE membrane fusion regulator and EHD1-binding protein, also intervenes in the fusion process. EHD1 is required for Cp110 loss from the mother centriole, and in its absence the mother centriole fails to recruit the transition zone protein Cep290 and IFT20, suggesting an important role for EHD1 in the early steps of primary cilium biogenesis. It is of note that Rabin8 colocalizes with EHD1 on preciliary vesicles but does not require EHD1 for accumulation at the mother centriole. Rabin8 activates Rab8 for ciliary extension only after ciliary vesicle assembly [166]. Hook2, a member of the Hook family of adaptor proteins, is also necessary for the formation of the large ciliary vesicle at the mother centriole [167]. During its conversion to a basal body, at the time that the centriole migrates towards the cell surface for docking, the two internal microtubules of each of the nine triplets at the distal tip of the mother centriole elongate, and the centriolar appendages mature into transition fibers. Ttbk2 or Mark4 knockdown blocks axoneme extension at this stage by impeding removal of Cp110 [91, 92]. The axoneme then elongates and deforms the ciliary vesicle in such a way that an outer membrane (sheath) and an inner membrane (shaft) surround the incipient axoneme and the distal part of the mother centriole. After transitional fiber-mediated docking of the mother centriole to the plasma membrane, the ciliary vesicle is exocytosed and fuses with the plasma membrane, exposing the nascent cilium to the extracellular milieu. Upon fusion, the sheath gives rise to the ciliary pocket, while the shaft forms the ciliary membrane. Finally, the axoneme continues elongating from its tip to reach its final size and the part proximal to the basal body remains structurally distinct from the rest of the cilium, forming the transition zone [158] (Fig. 5).
Fig. 5.
The intracellular pathway. Ciliogenesis initiates intracellularly with the formation of a large ciliary vesicle at the distal end of the appendages of the mother centriole by fusion of smaller vesicles. The axoneme starts forming intracellularly and, as it grows, deforms the ciliary vesicle and establishes an inner membrane (shaft) and an outer membrane (sheath). The incipient cilium is finally exocytosed and the cilium becomes exposed in the plasma membrane. The sheath gives rise to the ciliary pocket, and the shaft forms the ciliary membrane
The alternative pathway
When renal epithelial cells polarize, the centrosome localizes at the center of their apical membrane. Therefore, according to Sorokin’s proposal [154], the assembly of the primary cilium in these cells takes place entirely at the plasma membrane. The fact that the primary cilium of renal tubule epithelial cells lacks a ciliary pocket [173] is consistent with their function of sensing liquid flow [174] and with the use of the alternative pathway to assemble a primary cilium.
Most of the work on primary cilium biogenesis has focused on cell models that rely on the intracellular pathway, even though the primary cilium is also of pivotal importance in cells that use the alternative route. The importance of cilia in renal epithelial cells is exemplified by the ciliary defects that cause kidney cystic diseases, which are the most common of the many abnormalities associated with ciliopathies [175]. Renal epithelial MDCK cells have been used over the last 40 years as a paradigmatic cell model to study polarized membrane trafficking since they are considered to represent bona fide distal tubule epithelial cells [176]. Similar to renal tubular epithelial cells, MDCK cells polarize the centrosome to the center of the apical membrane and have no pocket at the base of the primary cilium [135, 153, 177]. Consistent with sensing liquid flow, bending of the primary cilium of epithelial MDCK cells results in an increase of intracellular Ca2+ [20], whereas removal of the cilium inhibits it [178]. Unlike RPE-1 cells, which use the intracellular route, a large ciliary vesicle is not assembled at the distal part of the mother centriole in MDCK cells, although Rab11, Rab8, exocyst subunit Sec8, and BBS1 accumulate in the vicinity of the centrosome at the apical plasma membrane [74]. The absence of such a vesicle is consistent with MDCK cells following a route of ciliogenesis different from that of fibroblasts. Given their relevance, MDCK cells have also been adopted to study primary cilia and to identify machinery important for its assembly [74, 108, 135, 144, 147, 149, 151, 153, 179–182] and constitute a suitable cell model to study the alternative pathway of primary cilium biogenesis [177].
Several proteins known to have a role in ciliogenesis, such as Sec10, Cep97 and IFT88, are also known to participate in the proper orientation of the mitotic spindle and/or cytokinesis, indicating that primary cilium biogenesis, which takes place in quiescent cells, and mitosis share protein machinery [87, 183, 184]. In animal cells, cytokinesis begins with the formation of an actomyosin ring at the equator of the two spindle poles [185, 186]. Contraction of the actomyosin ring leads to the ingression of a cleavage furrow that splits the cytoplasm in half. The two halves remain connected by an intercellular bridge containing antiparallel microtubule bundles, which, at least in part, arises from compressed mitotic spindle microtubules covered by plasma membrane [186, 187]. The amorphous electron-dense structure situated in the middle of the bridge is referred to as the midbody or Flemming body, which is 1.0–1.5-μm in size. The physical cleavage of the membrane bridge by the endosomal sorting complexes required for transport (ESCRT) machinery with the help of transport vesicle fusion separates the two daughter cells in a process known as abscission [186, 188]. Physical separation of the daughter cells requires severing the intercellular bridge at a single site on either side of the midbody. In this case, one of the daughter cells receives the midbody remnant [189]. The inherited remnant can then be conserved on the cell surface as a microtubule-rich membrane protrusion, degraded by autophagy, or released later on if the remnant is cleaved on the other side. The intercellular bridge is often severed on both sides and the remnant is released. Whether the midbody remnant is released, conserved or degraded depends on cell type and status [190–193]. Increasingly, studies are revealing non-cytokinetic implications for the post-mitotic midbody [189, 194]. It is of note that the daughter cell with the older mother centriole tends to inherit the midbody more frequently than its sister cell, implying the existence of communication between the centrosome and the severing machinery [190, 195]. Stem cell-like and cancer cells often accumulate more than one remnant. Accumulation of remnants is associated with increased cell reprogramming efficiency and in vitro tumorigenicity, respectively, in these cells [190, 195]. The midbody remnant also provides polarity cues for the place of the initiation of lumen formation in epithelial MDCK cells and for the formation of the first neurite in D. melanogaster neurons and the dorsoventral axis during C. elegans development [196–198].
Proteomic analyses have shown that intercellular bridge midbodies [199] and primary cilia [200] have a wide spectrum of shared components [201]. It is of note that Rab11, Rab8, IFT20, IFT88, exocyst complex subunits, acetylated microtubules, BBS6, ESCRT components and septins have been identified in both structures [202]. Some of the shared proteins (e.g., Rab11, Rab8, the exocyst and septins) are known to function in both cytokinesis and primary cilium formation [109, 183, 203], whereas the function of others had been traditionally assigned only to one of the two processes. For instance, IFT20 and IFT88 have been found in the intercellular bridge and midbody remnants [177, 204], although the IFT machinery has long been thought to be exclusive to cilia. Conversely, the ESCRT machinery, which has a role in severing the intercellular bridge, is also present in primary cilia. These observations raise the interesting possibility that a considerable part of the machinery is used for both primary cilium formation and cytokinesis.
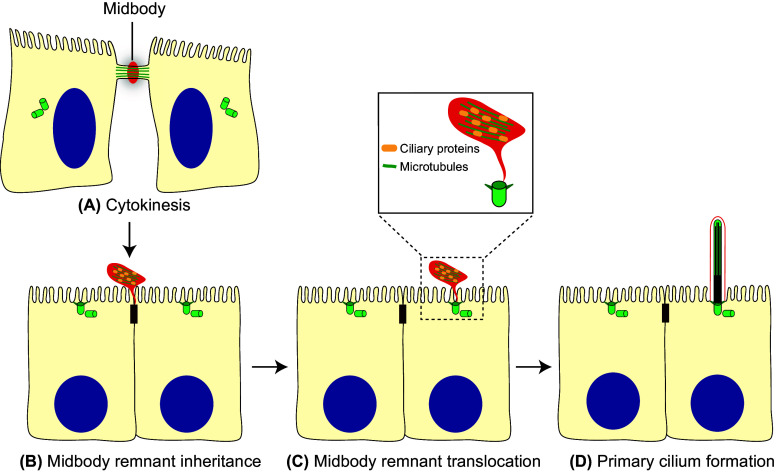
The cleavage furrow of MDCK cells and of other polarized epithelial cells initiates coincidently at the apical and basal surfaces but, since the rate of furrow ingression is more rapid from the basal surface, the intercellular bridge becomes located apically [205, 206] (Fig. 6a). When abscission occurs only on one side, this location of the bridge causes the midbody remnant to become positioned at the periphery of the apical surface of the cell, close to the tight junction (Fig. 6b). The remnant remains physically tethered to the surface of the cell that inherits it by a thin plasma membrane stalk that originates from the unresolved side of the bridge.
Fig. 6.
The alternative route. a In polarized epithelial cells, the intercellular bridge containing ciliary proteins forms at the apical cell surface during cytokinesis. b When abscission occurs, one of the two daughter cells inherits the midbody remnant, which localizes apically at the cell periphery, near the tight junctions. c The remnant subsequently moves over the apical surface towards the centrosome, which is docked at the center of the apical membrane. d When the midbody meets the centrosome, the initiation of primary cilium assembly is facilitated. The entire process of primary cilium formation takes place in the plasma membrane
After abscission, the midbody remnant can remain tethered to the cell for a long period, moving across the cell surface [177, 183, 207]. In polarized MDCK cells, the remnant, which carries Rab8, IFT20, IFT88, exocyst subunits and, probably, other ciliary machinery, traffics to the central part of the apical surface to meet the centrosome (Fig. 6c). Although it is not clear how the remnant reaches the center of the apical surface, it is known that its journey is dependent on Rab8 expression. The encounter between the midbody remnant and the centrosome is essential for primary cilium formation, since ciliogenesis is severely impaired in cells whose remnant has been removed [177]. Ultrastructural analysis of serial sections shows that the membrane of the midbody remnant is still connected to the adjacent plasma membrane by a membranous stalk. The establishment of a short microtubular connection between the midbody remnant and the centrosome has been detected before primary cilium starts forming, but the function of such a connection is currently unknown. The physical continuity of the remnant membrane and the plasma membrane raises the possibility that the remnant could transfer to the centrosome materials required for ciliogenesis [177]. Another possibility is that the remnant signals to the basal body to start primary cilium assembly. Further studies are required to understand the mechanism by which the midbody remnant licenses the centrosome for primary ciliogenesis (Fig. 6d).
Primary ciliogenesis is regulated by cell confinement in non-polarized cells, as shown by RPE-1 cells cultured on adhesive micropatterns, in which high spatial confinement results in a greater percentage of ciliated cells [164]. This is also true in epithelial MDCK cells, since cell–cell contacts are crucial to ciliogenesis [177]. It is of note that the area of MDCK cells governs the conservation of the midbody remnant, its movement to the center of the apical membrane and the beginning of primary cilium assembly. When cells proliferate, the availability of space becomes limited and cells are progressively constrained by their neighbors. Under these conditions, MDCK grow in height and reduce their area of attachment to the substrate, and the midbody remnant is conserved. Successive cycles of cell division increase the number of cells with a midbody remnant and, subsequently, the percentage of ciliated cells [177]. As this process progresses, compressive stress replaces tensile stress [208, 209]. These gradual changes in stress forces probably trigger the conservation of the remnant, its transition to the center of the apical surface to meet the centrosome and the beginning of ciliogenesis in polarized epithelial cells.
A negative correlation is known to exist between mitosis and primary cilium formation [210]. The dual function of the centrosome as a basal body in ciliogenesis and as MTOC in mitosis provides a point of connection between the two processes. Whereas a centriole is essential for primary cilium formation, mitosis can take place in the absence of centrioles [211, 212]. These observations suggest that centrioles may be more important for the first process than for the latter. The finding that a structure arising in the last stage of the cell division cycle, such as the midbody remnant, licenses the centrosome for primary ciliogenesis reveals a new mechanism by which these two processes are coupled [177]. This mechanism of coupling might be particularly important in epithelial cells that form cilia when they become quiescent as a part of the epithelial polarization/differentiation pathway of cells, such as kidney tubule cells and cholangiocytes. In these cells, primary cilia need to be exposed to the lumen to sense changes in fluid flow [163, 213, 214] and, therefore, they need to use a pathway of ciliogenesis that leads to primary cilia deprived of their ciliary pocket.
Conclusions and future directions
Despite Sorokin’s proposal nearly 50 years ago of the existence of two distinct pathways for ciliogenesis, most of the work on primary cilium biogenesis has focused on cell models that rely on the intracellular pathway, even though the primary cilium is also of pivotal importance in cells that were postulated to use an alternative pathway, as is the case of renal epithelial cells. The great efforts made to investigate the intracellular route have driven advances in the field, revealing many molecular details of the cellular events and the machinery involved. However, there is still a long way to go before the process is fully understood. In polarized epithelial cells, primary cilium biogenesis seems to be a sequential process by which the establishment of tight cell junctions and subsequent cell polarization modulates the conservation of the midbody remnant, its movement to meet the centrosome, and the beginning of primary cilium assembly at the middle of the apical surface. This route of primary ciliogenesis establishes a new biological mechanism that links the three major microtubule-based organelles—the centrosome, the cilium and the midbody—in the same process. Since this mechanism is entirely new, it raises many interesting questions that, it is to be hoped, will stimulate research on this pathway and help us understand the function of the midbody remnant, whose relevance to cellular processes other than cytokinesis has only recently begun to be considered.
One of the most important questions that arises is whether there is a transfer of materials from the midbody remnant to the centrosome to feed primary ciliogenesis. If this is the case, this material will need to be characterized in order to appreciate how it potentiates the centrosome for cilium formation. A second point is to understand how the midbody remnant moves towards the center of the apical surface until it meets the centrosome. How is the remnant propelled? How does the remnant “know” where to go and how is it able to arrive at the cell center despite the conspicuous presence of microvilli at the apical surface? What causes it to stop? Another important matter concerns the possible involvement of the midbody in the intracellular pathway. Is the participation of the midbody remnant exclusive to polarized epithelial cells or does it also intervene in cells using the intracellular route? In this regard, the midbody might also participate, for instance, by providing materials for the formation of the ciliary vesicles that surround the intracellular, nascent cilium. The mother centriole approaches the intercellular bridge of dividing mouse L929 fibroblasts and HeLa cells [215, 216], so it is possible that this contact could serve the centriole to obtain materials used subsequently for primary cilium formation. Activation of autophagy and initiation of ciliogenesis are simultaneous events in which some proteins participate direct or indirectly in both processes [217, 218]. For instance, Ofd1, which is a repressor of ciliogenesis, is removed from the centriolar satellites by autophagy, enabling primary cilium biogenesis [218]. It would also be fascinating to examine whether there is a transfer of ciliary material to the centrosome during autophagy of the midbody remnant in cells relying on the intracellular pathway and in which the remnant is internalized. Finally, it will be interesting to identify the protein machinery specific to each of the two pathways of ciliogenesis. Therefore, further research is needed to elucidate the cellular and molecular basis that controls the process of primary cilium biogenesis in different cell types.
Acknowledgements
We thank members of M. A. Alonso and I. Correas laboratories for helpful discussions. We also thank Dr. Phil Mason for revising the English language of the manuscript. Research in the laboratory of Miguel A. Alonso is supported by a Grant (BFU2015-67266-R) from the Ministerio de Economía y Competitividad MINECO, Spain, and the Fondo Europeo de Desarrollo Regional (FEDER), European Union. MB-R is the holder of a predoctoral contract from the MINECO.
Abbreviations
- aPKC
Atypical protein kinase C
- BBS
Bardet–Biedl syndrome
- GEF
Guanine nucleotide exchange factor
- GPCR
G protein-coupled receptor
- Hh
Hedgehog
- IFT
Intraflagellar transport
- JBTS
Joubert syndrome
- MDCK
Madin–Darby canine kidney
- MKS
Meckel syndrome
- MEF
Mouse embryonic fibroblast
- NPHP
Nephronophthisis
- PDGF
Platelet-derived growth factor
- RPE
Retinal pigment epithelial
- Smo
Smoothened
References
- 1.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu Rev Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- 3.Ishikawa H, Marshall WF. Ciliogenesis: building the cell’s antenna. Nat Rev Mol Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- 4.Brooks ER, Wallingford JB. Multiciliated cells: a review. Curr Biol. 2014;24:R973–R982. doi: 10.1016/j.cub.2014.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindemann CB, Lesich KA. Functional anatomy of the mammalian sperm flagellum. Cytoskeleton. 2016;73:652–669. doi: 10.1002/cm.21338. [DOI] [PubMed] [Google Scholar]
- 6.Oberholzer M, Bregy P, Marti G, Minca M, Peier M, Seebeck T. Trypanosomes and mammalian sperm: one of a kind? Trends Parasitol. 2007;23:71–77. doi: 10.1016/j.pt.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 8.Falk N, Lösl M, Schöeder N, Giebl A. Specialized cilia in mammalian sensory systems. Cells. 2015;4:500–519. doi: 10.3390/cells4030500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shinohara K, Chen D, Nishida T, Misaki K, Yonemura S, Hamada H. Absence of radial spokes in mouse node cilia is required for rotational movement but confers ultrastructural instability as a trade-off. Dev Cell. 2015;35:236–246. doi: 10.1016/j.devcel.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Yoshiba S, Hamada H. Roles of cilia, fluid flow, and Ca2+ signaling in breaking of left-right symmetry. Trends Genet. 2014;30:10–17. doi: 10.1016/j.tig.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Wheatley DN, Wang AM, Strugnell GE. Expression of primary cilia in mammalian cells. Cell Biol Int. 1996;20:73–81. doi: 10.1006/cbir.1996.0011. [DOI] [PubMed] [Google Scholar]
- 12.Pearring JN, Salinas RY, Baker SA, Arshavsky VY. Protein sorting, targeting and trafficking in photoreceptor cells. Prog Retina Eye Res. 2013;36:24–51. doi: 10.1016/j.preteyeres.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zimmerman KW. Beiträge zur Kenntniss einiger drüsen und epithelien. Arch Mikr Anat. 1898;52:552–706. doi: 10.1007/BF02975837. [DOI] [Google Scholar]
- 14.Malicki JJ, Johnson CA. The cilium: cellular antenna and central processing unit. Trends Cell Biol. 2017;27:126–140. doi: 10.1016/j.tcb.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmerman K, Yoder BK. Snapshot: sensing and signaling by cilia. Cell. 2015;161(692–692):e691. doi: 10.1016/j.cell.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishikawa H, Marshall WF. Mechanobiology of ciliogenesis. Bioscience. 2014;64:1084–1091. doi: 10.1093/biosci/biu173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J. Polycystins and primary cilia: primers for cell cycle progression. Annu Rev Physiol. 2009;71:83–113. doi: 10.1146/annurev.physiol.70.113006.100621. [DOI] [PubMed] [Google Scholar]
- 18.Battle C, Ott CM, Burnette DT, Lippincott-Schwartz J, Schmidt CF. Intracellular and extracellular forces drive primary cilia movement. Proc Natl Acad Sci USA. 2015;112:1410–1415. doi: 10.1073/pnas.1421845112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Praetorius HA, Frokiaer J, Nielsen S, Spring KR. Bending the primary cilium opens Ca2+-sensitive intermediate-conductance K+ channels in MDCK cells. J Membr Biol. 2003;191:193–200. doi: 10.1007/s00232-002-1055-z. [DOI] [PubMed] [Google Scholar]
- 20.Praetorius HA, Spring KR. Bending the MDCK cell primary cilium increases intracellular calcium. J Membr Biol. 2001;184:71–79. doi: 10.1007/s00232-001-0075-4. [DOI] [PubMed] [Google Scholar]
- 21.DeCaen PG, Delling M, Vien TN, Clapham DE. Direct recording and molecular identification of the calcium channel of primary cilia. Nature. 2013;504:315–318. doi: 10.1038/nature12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delling M, DeCaen PG, Doerner JF, Febvay S, Clapham DE. Primary cilia are specialized calcium signalling organelles. Nature. 2013;504:311–314. doi: 10.1038/nature12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takao D, Nemoto T, Abe T, Kiyonari H, Kajiura-Kobayashi H, Shiratori H, Nonaka S. Asymmetric distribution of dynamic calcium signals in the node of mouse embryo during left–right axis formation. Dev Biol. 2013;376:23–30. doi: 10.1016/j.ydbio.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 24.Praetorius HA. The primary cilium as sensor of fluid flow: new building blocks to the model. Am J Physiol Cell Physiol. 2015;308:C198. doi: 10.1152/ajpcell.00336.2014. [DOI] [PubMed] [Google Scholar]
- 25.Doerner JF, Delling M, Clapham DE. Ion channels and calcium signaling in motile cilia. eLife. 2015;4:e11066. doi: 10.7554/eLife.11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delling M, Indzhykulian AA, Liu X, Liu Y, Xie T, Corey DP, Clapham DE. Primary cilia are not calcium-responsive mechanosensors. Nature. 2016;531:656–660. doi: 10.1038/nature17426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofherr A, Kottgen M. Polycystic kidney disease: cilia and mechanosensation revisited. Nat Rev Nephrol. 2016;12:318–319. doi: 10.1038/nrneph.2016.61. [DOI] [PubMed] [Google Scholar]
- 28.Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 29.Robbins DJ, Fei DL, Riobo NA. The Hedgehog signal transduction network. Sci Signal. 2012;5:re6. doi: 10.1126/scisignal.2002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorojankina T. Hedgehog signaling pathway: a novel model and molecular mechanisms of signal transduction. Cell Mol Life Sci. 2016;73:1317–1332. doi: 10.1007/s00018-015-2127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 32.Schneider L, Clement CA, Teilmann SC, Pazour GJ, Hoffmann EK, Satir P, Christensen ST. PDGFRα signaling is regulated through the primary cilium in fibroblasts. Curr Biol. 2005;15:1861–1866. doi: 10.1016/j.cub.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 33.Wilcockson SG, Sutcliffe C, Ashe HL. Control of signaling molecule range during developmental patterning. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2433-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao B, Li L, Lei Q, Guan K-L. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.May-Simera HL, Kelley MW. Cilia, Wnt signaling, and the cytoskeleton. Cilia. 2012;1:7. doi: 10.1186/2046-2530-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wallingford JB, Mitchell B. Strange as it may seem: the many links between Wnt signaling, planar cell polarity, and cilia. Genes Dev. 2011;25:201–213. doi: 10.1101/gad.2008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Habbig S, Bartram MP, Müller RU, Schwarz R, Andriopoulos N, Chen S, Sägmüller JG, Hoehne M, Burst V, Liebau MC, Reinhardt HC, Benzing T, Schermer B. NPHP4, a cilia-associated protein, negatively regulates the Hippo pathway. J Cell Biol. 2011;193:633–642. doi: 10.1083/jcb.201009069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hilgendorf KI, Johnson CT, Jackson PK. The primary cilium as a cellular receiver: organizing ciliary GPCR signaling. Curr Opin Cell Biol. 2016;39:84–92. doi: 10.1016/j.ceb.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 41.Novarino G, Akizu N, Gleeson JG. Modeling human disease in humans: the ciliopathies. Cell. 2011;147:70–79. doi: 10.1016/j.cell.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zariwala MA, Knowles MR, Omran H. Genetic defects in ciliary structure and function. Annu Rev Physiol. 2007;69:423–450. doi: 10.1146/annurev.physiol.69.040705.141301. [DOI] [PubMed] [Google Scholar]
- 43.Tobin JL, Beales PL. The nonmotile ciliopathies. Genet Med. 2009;11:386–402. doi: 10.1097/GIM.0b013e3181a02882. [DOI] [PubMed] [Google Scholar]
- 44.Mitchell DR. Evolution of cilia. Cold Spring Harb Perspect Biol. 2017 doi: 10.1101/cshperspect.a028290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizuno N, Taschner M, Engel BD, Lorentzen E. Structural studies of ciliary components. J Mol Biol. 2012;422:163–180. doi: 10.1016/j.jmb.2012.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia G, Reiter JF. A primer on the mouse basal body. Cilia. 2016;5:17. doi: 10.1186/s13630-016-0038-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vertii A, Hung H-F, Hehnly H, Doxsey S. Human basal body basics. Cilia. 2016;5:13. doi: 10.1186/s13630-016-0030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vertii A, Hehnly H, Doxsey S. The centrosome, a multitalented renaissance organelle. Cold Spring Harb Perspect Biol. 2016 doi: 10.1101/cshperspect.a025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei Q, Ling K, Hu J. The essential roles of transition fibers in the context of cilia. Curr Opin Cell Biol. 2015;35:98–105. doi: 10.1016/j.ceb.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14:25–34. doi: 10.1016/S0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- 51.Tateishi K, Yamazaki Y, Nishida T, Watanabe S, Kunimoto K, Ishikawa H, Tsukita S. Two appendages homologous between basal bodies and centrioles are formed using distinct Odf2 domains. J Cell Biol. 2013;203:417–425. doi: 10.1083/jcb.201303071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang J, Liu X, Yue G, Adamian M, Bulgakov O, Li T. Rootletin, a novel coiled-coil protein, is a structural component of the ciliary rootlet. J Cell Biol. 2002;159:431–440. doi: 10.1083/jcb.200207153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benzing T, Schermer B. Transition zone proteins and cilia dynamics. Nat Genet. 2011;43:723–724. doi: 10.1038/ng.896. [DOI] [PubMed] [Google Scholar]
- 54.Szymanska K, Johnson CA. The transition zone: an essential functional compartment of cilia. Cilia. 2012;1:10. doi: 10.1186/2046-2530-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garcia-Gonzalo FR, Reiter JF. Scoring a backstage pass: mechanisms of ciliogenesis and ciliary access. J Cell Biol. 2012;197:697–709. doi: 10.1083/jcb.201111146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garcia-Gonzalo FR, Reiter JF. Open sesame: how transition fibers and the transition zone control ciliary composition. Cold Spring Harb Perspect Biol. 2017 doi: 10.1101/cshperspect.a028134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reiter JF, Blacque OE, Leroux MR. The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 2012;13:608–618. doi: 10.1038/embor.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yee LE, Garcia-Gonzalo FR, Bowie RV, Li C, Kennedy JK, Ashrafi K, Blacque OE, Leroux MR, Reiter JF. Conserved genetic interactions between ciliopathy complexes cooperatively support ciliogenesis and ciliary signaling. PLoS Genet. 2015;11:e1005627. doi: 10.1371/journal.pgen.1005627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li C, Jensen VL, Park K, Kennedy J, Garcia-Gonzalo FR, Romani M, De Mori R, Bruel A-L, Gaillard D, Brn Doray, Lopez E, Rivière J-B, Faivre L, Thauvin-Robinet C, Reiter JF, Blacque OE, Valente EM, Leroux MR. MKS5 and CEP290 dependent assembly pathway of the ciliary transition zone. PLoS Biol. 2016;14:e1002416. doi: 10.1371/journal.pbio.1002416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li S, Fernandez J-J, Marshall WF, Agard DA. Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. EMBO J. 2012;31:552–562. doi: 10.1038/emboj.2011.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jana SC, Marteil G, Bettencourt-Dias M. Mapping molecules to structure: unveiling secrets of centriole and cilia assembly with near-atomic resolution. Curr Opin Cell Biol. 2014;26:96–106. doi: 10.1016/j.ceb.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 62.Portran D, Schaedel L, Xu Z, Thery M, Nachury MV. Tubulin acetylation protects long-lived microtubules against mechanical ageing. Nat Cell Biol. 2017;19:391–398. doi: 10.1038/ncb3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wloga D, Joachimiak E, Louka P, Gaertig J. Posttranslational modifications of tubulin and cilia. Cold Spring Harb Perspect Biol. 2016 doi: 10.1101/cshperspect.a028159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu Z, Schaedel L, Portran D, Aguilar A, Gaillard J, Marinkovich MP, Théry M, Nachury MV. Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science. 2017;356:328. doi: 10.1126/science.aai8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takao D, Verhey KJ. Gated entry into the ciliary compartment. Cell Mol Life Sci. 2016;73:119–127. doi: 10.1007/s00018-015-2058-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Verhey KJ, Yang W. Permeability barriers for generating a unique ciliary protein and lipid composition. Curr Opin Cell Biol. 2016;41:109–116. doi: 10.1016/j.ceb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nachury MV, Seeley ES, Jin H. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu Rev Cell Dev Biol. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu Q, Nelson WJ. The ciliary diffusion barrier: the gatekeeper for the primary cilium compartment. Cytoskeleton (Hoboken, NJ) 2011;68:313–324. doi: 10.1002/cm.20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dentler WL, Rosenbaum JL. Flagellar elongation and shortening in Chlamydomonas. III. Structures attached to the tips of flagellar microtubules and their relationship to the directionality of flagellar microtubule assembly. J Cell Biol. 1977;74:747–759. doi: 10.1083/jcb.74.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Satir P. Studies on cilia: III. Further studies on the cilium tip and a “sliding filament” model of ciliary motility. J Cell Biol. 1968;39:77–94. doi: 10.1083/jcb.39.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dentler WL. Structures linking the tips of ciliary and flagellar microtubules to the membrane. J Cell Sci. 1980;42:207–220. doi: 10.1242/jcs.42.1.207. [DOI] [PubMed] [Google Scholar]
- 72.Portman RW, LeCluyse EL, Dentler WL. Development of microtubule capping structures in ciliated epithelial cells. J Cell Sci. 1987;87:85–94. doi: 10.1242/jcs.87.1.85. [DOI] [PubMed] [Google Scholar]
- 73.Sloboda RD. Intraflagellar transport and the flagellar tip complex. J Cell Biochem. 2005;94:266–272. doi: 10.1002/jcb.20323. [DOI] [PubMed] [Google Scholar]
- 74.He M, Subramanian R, Bangs F, Omelchenko T, Liem KF, Kapoor TM, Anderson KV. The kinesin-4 protein KIF7 regulates mammalian hedgehog signaling by organizing the cilia tip compartment. Nat Cell Biol. 2014;16:663–672. doi: 10.1038/ncb2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pusapati GV, Rohatgi R. Location, location, and location: compartmentalization of Hedgehog signaling at primary cilia. EMBO J. 2014;33:1852–1854. doi: 10.15252/embj.201489294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cao M, Ning J, Hernandez-Lara CI, Belzile O, Wang Q, Dutcher SK, Liu Y, Snell WJ. Uni-directional ciliary membrane protein trafficking by a cytoplasmic retrograde IFT motor and ciliary ectosome shedding. eLife. 2015;4:e05242. doi: 10.7554/eLife.05242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nager AR, Goldstein JS, Herranz-Pérez V, Portran D, Ye F, Garcia-Verdugo JM, Nachury MV. An actin network dispatches ciliary GPCRs into extracellular vesicles to modulate signaling. Cell. 2017;168:1–12. doi: 10.1016/j.cell.2016.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Phua SC, Chiba S, Suzuki M, Su E, Roberson EC, Pusapati GV, Setou M, Rohatgi R, Reiter JF, Ikegami K, Inoue T. Dynamic remodeling of membrane composition drives cell cycle through primary cilia excision. Cell. 2017;168(264–279):e215. doi: 10.1016/j.cell.2016.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wood CR, Huang K, Diener DR, Rosenbaum JL. The cilium secretes bioactive ectosomes. Curr Biol. 2013;23:906–911. doi: 10.1016/j.cub.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wood CR, Rosenbaum JL. Ciliary ectosomes: transmissions from the cell’s antenna. Trends Cell Biol. 2015;25:276–285. doi: 10.1016/j.tcb.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang J, Silva M, Haas L, Morsci N, Nguyen KCQ, Hall DH, Barr MM. C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr Biol. 2014;24:519–525. doi: 10.1016/j.cub.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kobayashi T, Dynlacht BD. Regulating the transition from centriole to basal body. J Cell Biol. 2011;193:435–444. doi: 10.1083/jcb.201101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim S, Tsiokas L. Cilia and cell cycle re-entry: more than a coincidence. Cell Cycle. 2011;10:2683–2690. doi: 10.4161/cc.10.16.17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liang Y, Meng D, Zhu B, Pan J. Mechanism of ciliary disassembly. Cell Mol Life Sci. 2016;73:1787–1802. doi: 10.1007/s00018-016-2148-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Taschner M, Lorentzen E. The intraflagellar transport machinery. Cold Spring Harb Perspect Biol. 2016 doi: 10.1101/cshperspect.a028092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hernandez-Hernandez V, Henkins D. Advances in the understanding of the BBSome complex structure and function. Res Rep Biol. 2015;6:191–201. [Google Scholar]
- 87.Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130:678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 88.Tsang WY, Bossard C, Khanna H, Peranen J, Swaroop A, Malhotra V, Dynlacht BD. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev Cell. 2008;15:187–197. doi: 10.1016/j.devcel.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tsang WY, Dynlacht BD. CP110 and its network of partners coordinately regulate cilia assembly. Cilia. 2013;2:9. doi: 10.1186/2046-2530-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goetz SC, Liem KF, Anderson KV. The spinocerebellar ataxia-associated gene Tau tubulin kinase 2 (TTBK2) controls the initiation of ciliogenesis. Cell. 2012;151:847–858. doi: 10.1016/j.cell.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cajanek L, Nigg EA. Cep164 triggers ciliogenesis by recruiting Tau tubulin kinase 2 to the mother centriole. Proc Natl Acad Sci USA. 2014;111:E2841–E2850. doi: 10.1073/pnas.1401777111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kuhns S, Schmidt KN, Jr Reymann, Gilbert DF, Neuner A, Hub B, Carvalho R, Wiedemann P, Zentgraf H, Erfle H, Klingmüller U, Boutros M, Pereira G. The microtubule affinity regulating kinase MARK4 promotes axoneme extension during early ciliogenesis. J Cell Biol. 2013;200:505–522. doi: 10.1083/jcb.201206013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Prosser SL, Morrison CG. Centrin2 regulates CP110 removal in primary cilium formation. J Cell Biol. 2015;208:693–701. doi: 10.1083/jcb.201411070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nikonova AS, Astsaturov I, Serebriiskii IG, Dunbrack RL, Golemis EA. Aurora-A kinase (AURKA) in normal and pathological cell growth. Cell Mol Life Sci. 2016;70:661–687. doi: 10.1007/s00018-012-1073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Inoko A, Matsuyama M, Goto H, Ohmuro-Matsuyama Y, Hayashi Y, Enomoto M, Ibi M, Urano T, Yonemura S, Kiyono T, Izawa I, Inagaki M. Trichoplein and Aurora A block aberrant primary cilia assembly in proliferating cells. J Cell Biol. 2012;197:391–405. doi: 10.1083/jcb.201106101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kasahara K, Kawakami Y, Kiyono T, Yonemura S, Kawamura Y, Era S, Matsuzaki F, Goshima N, Inagaki M. Ubiquitin-proteasome system controls ciliogenesis at the initial step of axoneme extension. Nat Commun. 2014;5:5081. doi: 10.1038/ncomms6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Izawa I, Goto H, Kasahara K, Inagaki M. Current topics of functional links between primary cilia and cell cycle. Cilia. 2015;4:12. doi: 10.1186/s13630-015-0021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shearer RF, Saunders DN. Regulation of primary cilia formation by the ubiquitin-proteasome system. Biochem Soc Trans. 2016;44:1265–1271. doi: 10.1042/BST20160174. [DOI] [PubMed] [Google Scholar]
- 99.Goto H, Inaba H, Inagaki M. Mechanisms of ciliogenesis suppression in dividing cells. Cell Mol Life Sci. 2016;74:881–890. doi: 10.1007/s00018-016-2369-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bhogaraju S, Engel BD, Lorentzen E. Intraflagellar transport complex structure and cargo interactions. Cilia. 2013;2:10. doi: 10.1186/2046-2530-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Braun DA, Hildebrandt F. Ciliopathies. Cold Spring Harb Perspect Biol. 2017 doi: 10.1101/cshperspect.a028191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van Dam TJP, Townsend MJ, Turk M, Schlessinger A, Sali A, Field MC, Huynen MA. Evolution of modular intraflagellar transport from a coatomer-like progenitor. Proc Natl Acad Sci USA. 2013;110:6943–6948. doi: 10.1073/pnas.1221011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dishinger JF, Kee HL, Jenkins PM, Fan S, Hurd TW, Hammond JW, Truong YN-T, Margolis B, Martens JR, Verhey KJ. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-β2 and Ran-GTP. Nat Cell Biol. 2010;12:703–710. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stepanek L, Pigino G. Microtubule doublets are double-track railways for intraflagellar transport trains. Science. 2016;352:721–724. doi: 10.1126/science.aaf4594. [DOI] [PubMed] [Google Scholar]
- 105.S-i Yoshimura, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Westlake CJ, Baye LM, Nachury MV, Wright KJ, Ervin KE, Phu L, Chalouni C, Beck JS, Kirkpatrick DS, Slusarski DC, Sheffield VC, Scheller RH, Jackson PK. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc Nat Acad Sci USA. 2011;108:2759–2764. doi: 10.1073/pnas.1018823108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Knodler A, Feng S, Zhang J, Zhang X, Das A, Peränen J, Guo W. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc Natl Acad Sci USA. 2010;107:6346–6351. doi: 10.1073/pnas.1002401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Babbey CM, Bacallao RL, Dunn KW. Rab10 associates with primary cilia and the exocyst complex in renal epithelial cells. Am J Physiol -Renal Physiol. 2010;299:F495–F506. doi: 10.1152/ajprenal.00198.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hehnly H, Chen C-T, Powers CM, Liu H-L, Doxsey S. The centrosome regulates the Rab11- dependent recycling endosome pathway at appendages of the mother centriole. Curr Biol. 2012;22:1944–1950. doi: 10.1016/j.cub.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Thomas LL, Fromme JC. GTPase cross talk regulates TRAPPII activation of Rab11 homologues during vesicle biogenesis. J Cell Biol. 2016;215:499–513. doi: 10.1083/jcb.201608123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 112.Sato T, Iwano T, Kunii M, Matsuda S, Mizuguchi R, Jung Y, Hagiwara H, Yoshihara Y, Yuzaki M, Harada R, Harada A. Rab8a and Rab8b are essential for several apical transport pathways but insufficient for ciliogenesis. J Cell Sci. 2014;127:422–431. doi: 10.1242/jcs.136903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Boehlke C, Bashkurov M, Buescher A, Krick T, John A-K, Nitschke R, Walz G, Kuehn EW. Differential role of Rab proteins in ciliary trafficking: Rab23 regulates Smoothened levels. J Cell Sci. 2010;123:1460. doi: 10.1242/jcs.058883. [DOI] [PubMed] [Google Scholar]
- 114.Lim YS, Tang BL. A role for Rab23 in the trafficking of Kif17 to the primary cilium. J Cell Sci. 2015;128:2996–3008. doi: 10.1242/jcs.163964. [DOI] [PubMed] [Google Scholar]
- 115.Leaf A, Von Zastrow M. Dopamine receptors reveal an essential role of IFT-B, KIF17, and Rab23 in delivering specific receptors to primary cilia. eLife. 2015;4:e06996. doi: 10.7554/eLife.06996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sheffield VC. The blind leading the obese: the molecular pathophysiology of a human obesity syndrome. Trans Am Clin Climatol Assoc. 2010;121:172–182. [PMC free article] [PubMed] [Google Scholar]
- 117.Jin H, Nachury MV. The BBSome. Curr Biol. 2009;19:R472–R473. doi: 10.1016/j.cub.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 118.Jin H, White SR, Shida T, Schulz S, Aguiar M, Gygi SP, Bazan JF, Nachury MV. The conserved Bardet–Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peränen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, Jackson PK. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 120.Berbari NF, Lewis JS, Bishop GA, Askwith CC, Mykytyn K. Bardet–Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Prooc Natl Acad Sci USA. 2008;105:4242–4246. doi: 10.1073/pnas.0711027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Loktev AV, Jackson PK. Neuropeptide Y family receptors traffic via the Bardet–Biedl syndrome pathway to signal in neuronal primary cilia. Cell Rep. 2013;5:1316–1329. doi: 10.1016/j.celrep.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 122.Domire JS, Green JA, Lee KG, Johnson AD, Askwith CC, Mykytyn K. Dopamine receptor 1 localizes to neuronal cilia in a dynamic process that requires the Bardet–Biedl syndrome proteins. Cell Mol Life Sci. 2011;68:2951–2960. doi: 10.1007/s00018-010-0603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Eguether T, San Agustin JT, Keady BT, Jonassen JA, Liang Y, Francis R, Tobita K, Johnson CA, Abdelhamed ZA, Lo CW, Pazour GJ. IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev Cell. 2014;31:279–290. doi: 10.1016/j.devcel.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liew GM, Ye F, Nager AR, Murphy JP, Lee JS, Aguiar M, Breslow DK, Gygi SP, Nachury MV. The intraflagellar transport protein IFT27 promotes BBSome exit from cilia through the GTPase ARL6/BBS3. Dev Cell. 2014;31:265–278. doi: 10.1016/j.devcel.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Xu Q, Zhang Y, Wei Q, Huang Y, Li Y, Ling K, Hu J. BBS4 and BBS5 show functional redundancy in the BBSome to regulate the degradative sorting of ciliary sensory receptors. Sci Rep. 2015;5:11855. doi: 10.1038/srep11855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lechtreck K-F, Johnson EC, Sakai T, Cochran D, Ballif BA, Rush J, Pazour GJ, Ikebe M, Witman GB. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J Cell Biol. 2009;187:1117–1132. doi: 10.1083/jcb.200909183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lechtreck KF, Brown JM, Sampaio JL, Craft JM, Shevchenko A, Evans JE, Witman GB. Cycling of the signaling protein phospholipase D through cilia requires the BBSome only for the export phase. J Cell Biol. 2013;201:249–261. doi: 10.1083/jcb.201207139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yu IM, Hughson FM. Tethering factors as organizers of intracellular vesicular traffic. Annu Rev Cell Dev Biol. 2010;26:137–156. doi: 10.1146/annurev.cellbio.042308.113327. [DOI] [PubMed] [Google Scholar]
- 129.Hertzog M, Chavrier P. Cell polarity during motile processes: keeping on track with the exocyst complex. Biochem J. 2011;433:403–409. doi: 10.1042/BJ20101214. [DOI] [PubMed] [Google Scholar]
- 130.Munson M, Novick P. The exocyst defrocked, a framework of rods revealed. Nat Struct Mol Biol. 2006;13:577–581. doi: 10.1038/nsmb1097. [DOI] [PubMed] [Google Scholar]
- 131.Heider MR, Munson M. Exorcising the exocyst complex. Traffic. 2012;13:898–907. doi: 10.1111/j.1600-0854.2012.01353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rogers KK, Wilson PD, Snyder RW, Zhang X, Guo W, Burrow CR, Lipschutz JH. The exocyst localizes to the primary cilium in MDCK cells. Biochem Biophys Res Commun. 2004;319:138–143. doi: 10.1016/j.bbrc.2004.04.165. [DOI] [PubMed] [Google Scholar]
- 133.Zhang X-M, Ellis S, Sriratana A, Mitchell CA, Rowe T. Sec15 is an effector for the Rab11 GTPase in mammalian cells. J Biol Chem. 2004;279:43027–43034. doi: 10.1074/jbc.M402264200. [DOI] [PubMed] [Google Scholar]
- 134.Wu S, Mehta SQ, Pichaud F, Bellen HJ, Quiocho FA. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nat Struct Mol Biol. 2005;12:879–885. doi: 10.1038/nsmb987. [DOI] [PubMed] [Google Scholar]
- 135.Zuo X, Guo W, Lipschutz JH. The exocyst protein sec10 is necessary for primary ciliogenesis and cystogenesis in vitro. Mol Biol Cell. 2009;20:2522–2529. doi: 10.1091/mbc.E08-07-0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Polgar N, Lee AJ, Lui VH, Napoli JA, Fogelgren B. The exocyst gene Sec10 regulates renal epithelial monolayer homeostasis and apoptotic sensitivity. Am J Physiol Cell Physiol. 2015;309:C190–C201. doi: 10.1152/ajpcell.00011.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fogelgren B, Lin S-Y, Zuo X, Jaffe KM, Park KM, Reichert RJ, Bell PD, Burdine RD, Lipschutz JH. The exocyst protein Sec10 interacts with polycystin-2 and knockdown causes PKD-phenotypes. PLoS Gen. 2011;7:e1001361. doi: 10.1371/journal.pgen.1001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li Y, Ling K, Hu J. The emerging role of Arf/Arl small GTPases in cilia and ciliopathies. J Cell Biochem. 2012;113:2201–2207. doi: 10.1002/jcb.24116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cantagrel V, Silhavy JL, Bielas SL, Swistun D, Marsh SE, Bertrand JY, Audollent S, Attié-Bitach T, Holden KR, Dobyns WB, Traver D, Al-Gazali L, Ali BR, Lindner TH, Caspary T, Otto EA, Hildebrandt F, Glass IA, Logan CV, Johnson CA, Bennett C, Brancati F, The International Joubert Syndrome Related Disorders Study Group. Valente EM, Woods CG, Gleeson JG. Mutations in the cilia Gene ARL13B lead to the classical form of Joubert syndrome. Am J Hum Genet. 2008;83:170–179. doi: 10.1016/j.ajhg.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Larkins CE, Aviles GDG, East MP, Kahn RA, Caspary T. Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol Biol Cell. 2011;22:4694–4703. doi: 10.1091/mbc.E10-12-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Caspary T, Larkins CE, Anderson KV. The graded response to sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 142.Cl Seixas, Choi SY, Polgar N, Umberger NL, East MP, Zuo X, Moreiras H, Ghossoub R, Benmerah A, Kahn RA, Fogelgren B, Caspary T, Lipschutz JH, Barral DC. Arl13b and the exocyst interact synergistically in ciliogenesis. Mol Biol Cell. 2016;27:308–320. doi: 10.1091/mbc.E15-02-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Joberty G, Petersen C, Gao L, Macara IG. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- 144.Sfakianos J, Togawa A, Maday S, Hull M, Pypaert M, Cantley L, Toomre D, Mellman I. Par3 functions in the biogenesis of the primary cilium in polarized epithelial cells. J Cell Biol. 2007;179:1133–1140. doi: 10.1083/jcb.200709111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fan S, Hurd TW, Liu C-J, Straight SW, Weimbs T, Hurd EA, Domino SE, Margolis B. Polarity proteins control ciliogenesis via kinesin motor interactions. Curr Biol. 2004;14:1451–1461. doi: 10.1016/j.cub.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 146.Hurd TW, Fan S, Liu C-J, Kweon HK, Hakansson K, Margolis B. Phosphorylation-dependent binding of 14-3-3 to the polarity protein Par3 regulates cell polarity in mammalian epithelia. Curr Biol. 2003;13:2082–2090. doi: 10.1016/j.cub.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 147.Zuo X, Fogelgren B, Lipschutz JH. The small GTPase Cdc42 is necessary for primary ciliogenesis in renal tubular epithelial cells. J Biol Chem. 2011;286:22469–22477. doi: 10.1074/jbc.M111.238469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lemmers C, Michel D, Lane-Guermonprez L, Delgrossi M-H, Médina E, Arsanto J-P, Le Bivic A. CRB3 binds directly to Par6 and regulates the morphogenesis of the tight junctions in mammalian epithelial cells. Mol Biol Cell. 2004;15:1324–1333. doi: 10.1091/mbc.E03-04-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang G, Krishnamurthy K, Bieberich E. Regulation of primary cilia formation by ceramide. J Lipid Res. 2009;50:2103–2110. doi: 10.1194/jlr.M900097-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fan S, Fogg V, Wang Q, Chen X-W, Liu C-J, Margolis B. A novel Crumbs3 isoform regulates cell division and ciliogenesis via importin β interactions. J Cell Biol. 2007;178:387–398. doi: 10.1083/jcb.200609096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Torkko JM, Manninen A, Schuck S, Simons K. Depletion of apical transport proteins perturbs epithelial cyst formation and ciliogenesis. J Cell Sci. 2008;121:1193–1203. doi: 10.1242/jcs.015495. [DOI] [PubMed] [Google Scholar]
- 152.Takiar V, Mistry K, Carmosino M, Schaeren-Wiemers N, Caplan MJ. VIP17/MAL expression modulates epithelial cyst formation and ciliogenesis. Am J Physiol Cell Physiol. 2012;303:C862–C871. doi: 10.1152/ajpcell.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Reales E, Bernabé-Rubio M, Casares-Arias J, Rentero C, Fernández-Barrera J, Rangel L, Correas I, Enrich C, Andrés G, Alonso MA. The MAL protein is crucial for proper membrane condensation at the ciliary base, which is required for primary cilium elongation. J Cell Sci. 2015;128:2261–2270. doi: 10.1242/jcs.164970. [DOI] [PubMed] [Google Scholar]
- 154.Sorokin SP. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci. 1968;3:207–230. doi: 10.1242/jcs.3.2.207. [DOI] [PubMed] [Google Scholar]
- 155.Ghossoub R, Molla-Herman A, Bastin P, Benmerah A. The ciliary pocket: a once-forgotten membrane domain at the base of cilia. Biol Cell. 2011;103:131–144. doi: 10.1042/BC20100128. [DOI] [PubMed] [Google Scholar]
- 156.Molla-Herman A, Ghossoub R, Blisnick T, Meunier A, Serres C, Silbermann F, Emmerson C, Romeo K, Bourdoncle P, Schmitt A, Saunier S, Spassky N, Bastin P, Benmerah A. The ciliary pocket: an endocytic membrane domain at the base of primary and motile cilia. J Cell Sci. 2010;123:1785–1795. doi: 10.1242/jcs.059519. [DOI] [PubMed] [Google Scholar]
- 157.Benmerah A. The ciliary pocket. Curr Opin Cell Biol. 2013;25:78–84. doi: 10.1016/j.ceb.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 158.Rohatgi R, Snell WJ. The ciliary membrane. Curr Opin Cell Biol. 2010;22:541–546. doi: 10.1016/j.ceb.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Rattner JB, Sciore P, Ou Y, van der Hoorn FA, Lo IK. Primary cilia in fibroblast-like type B synoviocytes lie within a cilium pit: a site of endocytosis. Histol Histopathol. 2010;25:865–875. doi: 10.14670/HH-25.865. [DOI] [PubMed] [Google Scholar]
- 160.Clement CA, Ajbro KD, Koefoed K, Vestergaard ML, Veland IR, Henriques de Jesus MP, Pedersen LB, Benmerah A, Andersen CY, Larsen LA, Christensen ST. TGF-β signaling is associated with endocytosis at the pocket region of the primary cilium. Cell Rep. 2013;3:1806–1814. doi: 10.1016/j.celrep.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 161.Sorokin S. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J Cell Biol. 1962;15:363–377. doi: 10.1083/jcb.15.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mazo G, Soplop N, Wang W-J, Uryu K, Tsou M-FB. Spatial control of primary ciliogenesis by subdistal appendages alters sensation-associated properties of cilia. Dev Cell. 2016;39:424–437. doi: 10.1016/j.devcel.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.LaRusso NF, Masyuk TV. The role of cilia in the regulation of bile flow. Dig Dis. 2011;29:6–12. doi: 10.1159/000324121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pitaval A, Tseng Q, Bornens M, Théry M. Cell shape and contractility regulate ciliogenesis in cell cycle-arrested cells. J Cell Biol. 2010;191:303–312. doi: 10.1083/jcb.201004003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Galati DF, Mitchell BJ, Pearson CG. Subdistal appendages stabilize the ups and downs of ciliary life. Dev Cell. 2016;39:387–389. doi: 10.1016/j.devcel.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 166.Lu Q, Insinna C, Ott C, Stauffer J, Pintado PA, Rahajeng J, Baxa U, Walia V, Cuenca A, Hwang Y-S, Daar IO, Lopes S, Lippincott-Schwartz J, Jackson PK, Caplan S, Westlake CJ. Early steps in primary cilium assembly require EHD1- and EHD3-dependent ciliary vesicle formation. Nat Cell Biol. 2015;17:228–240. doi: 10.1038/ncb3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Baron Gaillard CL, Pallesi-Pocachard E, Massey-Harroche D, Richard F, Arsanto J-P, Chauvin J-P, Lecine P, Krämer H, Borg J-P, Le Bivic A. Hook2 is involved in the morphogenesis of the primary cilium. Mol Biol Cell. 2011;22:4549–4562. doi: 10.1091/mbc.E11-05-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tucker RW, Pardee AB, Fujiwara K. Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell. 1979;17:527–535. doi: 10.1016/0092-8674(79)90261-7. [DOI] [PubMed] [Google Scholar]
- 169.Schmidt KN, Kuhns S, Neuner A, Hub B, Zentgraf H, Pereira G. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J Cell Biol. 2012;199:1083–1101. doi: 10.1083/jcb.201202126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Pedersen LB, Veland IR, Schrøder JM, Christensen ST. Assembly of primary cilia. Dev Dyn. 2008;237:1993–2006. doi: 10.1002/dvdy.21521. [DOI] [PubMed] [Google Scholar]
- 171.Paridaen JTML, Wilsch-Bräuninger M, Huttner WB. Asymmetric inheritance of centrosome-associated primary cilium membrane directs ciliogenesis after cell division. Cell. 2013;155:333–344. doi: 10.1016/j.cell.2013.08.060. [DOI] [PubMed] [Google Scholar]
- 172.Yee LE, Reiter JF. Ciliary vesicle formation: a prelude to ciliogenesis. Dev Cell. 2015;32:665–666. doi: 10.1016/j.devcel.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Latta H, Maunsbach AB, Madden SC. Cilia in different segments of the rat nephron. J Biophys Biochem Cytol. 1961;11:248–252. doi: 10.1083/jcb.11.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Praetorius HA, Spring KR. A physiological view of the primary cilium. Annu Rev Physiol. 2005;67:515–529. doi: 10.1146/annurev.physiol.67.040403.101353. [DOI] [PubMed] [Google Scholar]
- 175.Zhang Q, Taulman PD, Yoder BK. Cystic kidney diseases: all roads lead to the cilium. Physiology. 2004;19:225. doi: 10.1152/physiol.00003.2004. [DOI] [PubMed] [Google Scholar]
- 176.Rodriguez-Boulan E, Kreitzer G, Musch A. Organization of vesicular trafficking in epithelia. Nat Rev Mol Cell Biol. 2005;6:233–247. doi: 10.1038/nrm1593. [DOI] [PubMed] [Google Scholar]
- 177.Bernabé-Rubio M, Andrés G, Casares-Arias J, Fernández-Barrera J, Rangel L, Reglero-Real N, Gershlick DC, Fernández JJ, Millán J, Correas I, Miguez DG, Alonso MA. Novel role for the midbody in primary ciliogenesis by polarized epithelial cells. J Cell Biol. 2016;214:259–273. doi: 10.1083/jcb.201601020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Praetorius HA, Spring KR. Removal of the MDCK cell primary cilium abolishes flow sensing. J Membr Biol. 2003;191:69–76. doi: 10.1007/s00232-002-1042-4. [DOI] [PubMed] [Google Scholar]
- 179.Francis SS, Sfakianos J, Lo B, Mellman I. A hierarchy of signals regulates entry of membrane proteins into the ciliary membrane domain in epithelial cells. J Cell Biol. 2011;193:219–233. doi: 10.1083/jcb.201009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DYR, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 181.Praetorius HA, Praetorius J, Nielsen S, Frokiaer J, Spring KR. β-Integrins in the primary cilium of MDCK cells potentiate fibronectin-induced Ca2+ signaling. Am J Physiol Renal Physiol. 2004;287:F969–F978. doi: 10.1152/ajprenal.00096.2004. [DOI] [PubMed] [Google Scholar]
- 182.Vieira OV, Gaus K, Verkade P, Fullekrug J, Vaz WLC, Simons K. FAPP2, cilium formation, and compartmentalization of the apical membrane in polarized Madin–Darby canine kidney (MDCK) cells. Proc Nat Acad Sci USA. 2006;103:18556–18561. doi: 10.1073/pnas.0608291103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gromley A, Yeaman C, Rosa J, Redick S, Chen C-T, Mirabelle S, Guha M, Sillibourne J, Doxsey SJ. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123:75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 184.Delaval B, Bright A, Lawson ND, Doxsey S. The cilia protein IFT88 is required for spindle orientation in mitosis. Nat Cell Biol. 2011;13:461–468. doi: 10.1038/ncb2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fededa JP, Gerlich DW. Molecular control of animal cell cytokinesis. Nat Cell Biol. 2012;14:440–447. doi: 10.1038/ncb2482. [DOI] [PubMed] [Google Scholar]
- 186.Green RA, Paluch E, Oegema K. Cytokinesis in animal cells. Annu Rev Cell Dev Biol. 2012;28:29–58. doi: 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- 187.Mullins J, Biesele JJ. Terminal phase of cytokinesis in D-98S cells. J Cell Biol. 1977;73:672–684. doi: 10.1083/jcb.73.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Mierzwa B, Gerlich DW. Cytokinetic abscission: molecular mechanisms and temporal control. Dev Cell. 2014;31:525–538. doi: 10.1016/j.devcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 189.Dionne LK, Wang X-J, Prekeris R. Midbody: from cellular junk to regulator of cell polarity and cell fate. Curr Opin Cell Biol. 2015;35:51–58. doi: 10.1016/j.ceb.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kuo T-C, Chen C-T, Baron D, Onder TT, Loewer S, Almeida S, Weismann C, Xu P, Houghton J-M, Gao F-B, Daley GQ, Doxsey S. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat Cell Biol. 2011;13:1214–1223. doi: 10.1038/ncb2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Marzesco A-M, Janich P, Wilsch-Bräuninger M, Dubreuil V, Langenfeld K, Corbeil D, Huttner WB. Release of extracellular membrane particles carrying the stem cell marker prominin-1 (CD133) from neural progenitors and other epithelial cells. J Cell Sci. 2005;118:2849–2858. doi: 10.1242/jcs.02439. [DOI] [PubMed] [Google Scholar]
- 192.Pohl C, Jentsch S. Midbody ring disposal by autophagy is a post-abscission event of cytokinesis. Nat Cell Biol. 2009;11:65–70. doi: 10.1038/ncb1813. [DOI] [PubMed] [Google Scholar]
- 193.Salzmann V, Chen C, Chiang CYA, Tiyaboonchai A, Mayer M, Yamashita YM. Centrosome-dependent asymmetric inheritance of the midbody ring in Drosophila germline stem cell division. Mol Biol Cell. 2014;25:267–275. doi: 10.1091/mbc.E13-09-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chen C-T, Ettinger AW, Huttner WB, Doxsey SJ. Resurrecting remnants: the lives of post-mitotic midbodies. Trends Cell Biol. 2012;23:118–128. doi: 10.1016/j.tcb.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ettinger AW, Wilsch-Brauninger M, Marzesco A-M, Bickle M, Lohmann A, Maliga Z, Karbanova J, Corbeil D, Hyman AA, Huttner WB. Proliferating versus differentiating stem and cancer cells exhibit distinct midbody-release behaviour. Nat Commun. 2011;2:503. doi: 10.1038/ncomms1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Li D, Mangan A, Cicchini L, Margolis B, Prekeris R. FIP5 phosphorylation during mitosis regulates apical trafficking and lumenogenesis. EMBO Rep. 2014;15:428–437. doi: 10.1002/embr.201338128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Singh D, Pohl C. Coupling of rotational cortical flow, asymmetric midbody positioning, and spindle rotation mediates dorsoventral axis formation in C. elegans . Dev Cell. 2014;28:253–267. doi: 10.1016/j.devcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 198.Pollarolo G, Schulz JG, Munck S, Dotti CG. Cytokinesis remnants define first neuronal asymmetry in vivo. Nat Neurosci. 2011;14:1525–1533. doi: 10.1038/nn.2976. [DOI] [PubMed] [Google Scholar]
- 199.Skop AR, Liu H, Yates J, Meyer BJ, Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 2004;305:61–66. doi: 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ishikawa H, Thompson J, Yates JR, Marshall WF. Proteomic analysis of mammalian primary cilia. Curr Biol. 2012;22:414–419. doi: 10.1016/j.cub.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Smith KR, Kieserman EK, Wang PI, Basten SG, Giles RH, Marcotte EM, Wallingford JB. A role for central spindle proteins in cilia structure and function. Cytoskeleton (Hoboken, NJ) 2011;68:112–124. doi: 10.1002/cm.20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ott CM. Midbody remnant licenses primary cilia formation in epithelial cells. J Cell Biol. 2016;214:237–239. doi: 10.1083/jcb.201607046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Fielding AB, Schonteich E, Matheson J, Wilson G, Yu X, Hickson GRX, Srivastava S, Baldwin SA, Prekeris R, Gould GW. Rab11-FIP3 and FIP4 interact with Arf6 and the exocyst to control membrane traffic in cytokinesis. EMBO J. 2005;24:3389–3399. doi: 10.1038/sj.emboj.7600803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wood CR, Wang Z, Diener D, Zones JM, Rosenbaum J, Umen JG. IFT proteins accumulate during cell division and localize to the cleavage furrow in Chlamydomonas . PLoS One. 2012;7:e30729. doi: 10.1371/journal.pone.0030729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Morais-de-Sá E, Sunkel C. Adherens junctions determine the apical position of the midbody during follicular epithelial cell division. EMBO Rep. 2013;14:696–703. doi: 10.1038/embor.2013.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Reinsch S, Karsenti E. Orientation of spindle axis and distribution of plasma membrane proteins during cell division in polarized MDCKII cells. J Cell Biol. 1994;126:1509–1526. doi: 10.1083/jcb.126.6.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Crowell EF, Gaffuri A-L, Gayraud-Morel B, Tajbakhsh S, Echard A. Engulfment of the midbody remnant after cytokinesis in mammalian cells. J Cell Sci. 2014;127:3840–3851. doi: 10.1242/jcs.154732. [DOI] [PubMed] [Google Scholar]
- 208.Bazellières E, Conte V, Elosegui-Artola A, Serra-Picamal X, Bintanel-Morcillo M, Roca-Cusachs P, Muñoz JJ, Sales-Pardo M, Guimerá R, Trepat X. Control of cell-cell forces and collective cell dynamics by the intercellular adhesome. Nat Cell Biol. 2015;17:409–420. doi: 10.1038/ncb3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Trepat X, Wasserman MR, Angelini TE, Millet E, Weitz DA, Butler JP, Fredberg JJ. Physical forces during collective cell migration. Nat Phys. 2009;5:426–430. doi: 10.1038/nphys1269. [DOI] [Google Scholar]
- 210.Dingemans KP. The relation between cilia and mitoses in the mouse adenohypophysis. J Cell Biol. 1969;43:361–367. doi: 10.1083/jcb.43.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 212.Marshall WF. Basal bodies: platforms for building cilia. Curr Topics Dev Biol. 2008;85:1–22. doi: 10.1016/S0070-2153(08)00801-6. [DOI] [PubMed] [Google Scholar]
- 213.Masyuk AI, Masyuk TV, LaRusso NF. Cholangiocyte primary cilium in liver health and disease. Dev Dyn. 2008;237:2007–2012. doi: 10.1002/dvdy.21530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Davenport JR, Yoder BK. An incredible decade for the primary cilium: a look at a once-forgotten organelle. Am J Physiol Renal Physiol. 2005;289:F1159. doi: 10.1152/ajprenal.00118.2005. [DOI] [PubMed] [Google Scholar]
- 215.Piel M, Nordberg J, Euteneuer U, Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–1553. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- 216.Jonsdottir AB, Dirks RW, Vrolijk J, Ögmundsdottir HM, Tanke HJ, Eyfjörd JE, Szuhai K. Centriole movements in mammalian epithelial cells during cytokinesis. BMC Cell Biol. 2010;11:1–9. doi: 10.1186/1471-2121-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Pampliega O, Orhon I, Patel B, Sridhar S, Diaz-Carretero A, Beau I, Codogno P, Satir BH, Satir P, Cuervo AM. Functional interaction between autophagy and ciliogenesis. Nature. 2013;502:194–200. doi: 10.1038/nature12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Tang Z, Lin MG, Stowe TR, Chen S, Zhu M, Stearns T, Franco B, Zhong Q. Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature. 2013;502:254–257. doi: 10.1038/nature12606. [DOI] [PMC free article] [PubMed] [Google Scholar]