Abstract
Transthyretin (TTR) is an extracellular protein able to deposit into well-defined protein aggregates called amyloid, in pathological conditions known as senile systemic amyloidosis, familial amyloid polyneuropathy, familial amyloid cardiomyopathy and leptomeningeal amyloidosis. At least three distinct partially folded states have been described for TTR, including the widely studied amyloidogenic state at mildly acidic pH. Here, we have used fluorescence resonance energy transfer (FRET) experiments in a monomeric variant of TTR (M-TTR) and in its W41F and W79F mutants, taking advantage of the presence of a unique, solvent-exposed, cysteine residue at position 10, that we have labelled with a coumarin derivative (DACM, acceptor), and of the two natural tryptophan residues at positions 41 and 79 (donors). Trp41 is located in an ideal position as it is one of the residues of β-strand C, whose degree of unfolding is debated. We found that the amyloidogenic state at low pH has the same FRET efficiency as the folded state at neutral pH in both M-TTR and W79F-M-TTR, indicating an unmodified Cys10–Trp41 distance. The partially folded state populated at low denaturant concentrations also has a similar FRET efficiency, but other spectroscopic probes indicate that it is distinct from the amyloidogenic state at acidic pH. By contrast, the off-pathway state accumulating transiently during refolding has a higher FRET efficiency, indicating non-native interactions that reduce the Cys10–Trp41 spatial distance, revealing a third distinct conformational state. Overall, our results clarify a negligible degree of unfolding of β-strand C in the formation of the amyloidogenic state and establish the concept that TTR is a highly plastic protein able to populate at least three distinct conformational states.
Keywords: Protein aggregation, Protein misfolding, Protein folding, Folding intermediate, SSA, FAP, FAC
Introduction
Transthyretin (TTR) is a homotetrameric protein with a total molecular mass of 55 kDa that is mainly synthesized in the liver, choroid plexus of the brain, and retina [1, 2], albeit further studies have demonstrated significant synthesis in the pancreas, kidneys, Schwann cells and neurons [3–7]. In the plasma TTR transports thyroxine (T4) and the retinol binding protein (RBP), whereas in the cerebrospinal fluid (CSF) TTR is the primary transporter of T4 [8, 9].
TTR is also one of the few proteins associated with systemic amyloidoses, a group of disorders resulting from the extracellular deposition of well-defined protein aggregates characterized by a fibrillar morphology, cross-β structure and peculiar tinctorial properties in the presence of specific dyes such as Congo red or thioflavin S [10, 11]. Amyloid deposition of wild-type TTR occurs in the heart of 10–25% of humans older than 80 years, resulting in senile systemic amyloidosis (SSA), often leading to congestive heart failure [12, 13]. Amyloid deposition by TTR is accelerated by the presence of any of the approximately 100 different amyloidogenic mutations of TTR responsible for early-onset TTR amyloidosis with autosomal dominant inheritance, such as familial amyloid polyneuropathy (FAP), familial amyloid cardiomyopathy (FAC) and leptomeningeal amyloidosis [14, 15]. A list of all TTR mutations associated with such disorders is reported in a database named “Mutations of Hereditary Amyloidosis” (http://www.amyloidosismutations.com/mut-attr.php).
While the main component of amyloid fibrils in patients suffering from sporadic SSA is wild-type TTR and its fragments, the main component of amyloid fibrils in TTR familial amyloidoses is mutant TTR, with the wild-type counterpart only scarcely represented [10, 16–18]. The single-site mutations that cause such diseases do not significantly change the tertiary or quaternary structure of tetrameric TTR; instead they appear to be responsible for the reduced thermodynamic and/or kinetic stability of native tetrameric TTR in favor of the monomeric, partially unfolded amyloidogenic intermediate, which self-assembles into amyloid fibrils under mildly denaturing conditions, at pH 3.9–5.0 [19–21].
Considerable effort has been expended to determine the structure of the monomeric amyloidogenic state of TTR populated at weakly acidic pH values. Biophysical and limited proteolysis experiments have led to the conclusion that upon acidification, tetrameric TTR dissociates into monomers with the subsequent unfolding of the β-strand C, the β-strand D and the connecting CD loop [22, 23]. In another report it was proposed, on the basis of hydrogen/deuterium exchange experiments coupled to NMR, that the β-sheet CBEF is the most destabilized upon acidification of TTR at pH 4.5 [24]. Significant advances have been reached with a double mutant of TTR carrying two single mutations (F87M/L110M) designed to destabilise the molecular interface between the subunits that compose the tetramer [25]. This double mutant, generally referred to as M-TTR, was shown to be stable as a monomer at neutral pH and has proved a unique tool for characterizing, at the molecular level, the transition from the folded non-amyloidogenic monomer populated at neutral pH to the amyloidogenic state populated at low pH [25]. Solution NMR studies have shown that M-TTR is folded at neutral pH, with many of the resonances of the β-sheet DAGH exhibiting line broadening, particularly the β-strand H, indicative of structural fluctuations [26]. By contrast, the β-sheet CBEF is not just folded, but also substantially packed. Upon acidification to pH values promoting amyloid fibril formation, line broadening extends to the β-strand D, the D–E loop, the E–F helix, and the residues of the A–B loop forming interactions with the helix, whereas most residues of the β-sheet CBEF maintain sharp, non-broadened resonances [26]. These observations have been confirmed later on tetrameric TTR at mildly low pH [27]. Hence, it is not yet clear if the partial unfolding of the monomeric folded unit of TTR to produce the amyloidogenic state involves the β-strands C and D and the interconnecting loop [22, 23], the CBEF β-sheet [24] or only the peripheral portion of the DAGH β-sheet [26].
In an independent study, the folding process of M-TTR was characterized kinetically and at equilibrium using a number of biophysical probes, leading to the identification of a molten globule state populated at equilibrium at low urea concentrations and to an off-pathway partially folded state populated transiently during the folding process of M-TTR [28]. The correspondence between any of such conformational states and the amyloidogenic state of M-TTR populated at weakly acidic pH has not yet been established.
To clarify these unresolved issues, we have carried out fluorescence resonance energy transfer (FRET) experiments, taking advantage of the presence of a unique, solvent-exposed, cysteine residue at position 10 and of the two naturally occurring tryptophan residues at positions 41 and 79 in M-TTR. We have labelled the Cys10 residue of M-TTR with a coumarin derivative that acts as a FRET acceptor of the intrinsic fluorescence emitted by the Trp41 and Trp79 residues that thus act as FRET donors. Trp41 is located in an ideal position as it is one of the residues of β-strand C which has been reported to be either unfolded or fully folded in the various structural characterizations described above. Cys10 is solvent-exposed and its labelling is expected to have only minor influence on the structure of M-TTR. Hence, labelled M-TTR is expected to contain a new solvent-exposed environmental probe at position 10, thus permitting the measurement of the spatial distance between the probe linked to Cys10 and Trp41/Trp79. We have also produced and labeled with coumarin two single mutants of M-TTR, namely W41F and W79F, to monitor the spatial distance between Cys10 and the two tryptophan residues separately. The utility of FRET results from the strong dependence of the efficiency of energy transfer on the sixth power of the spatial distance between donor and acceptor, which has led to the often quoted reference to FRET as a ‘spectroscopic ruler’ [29].
We will report the FRET efficiency values of the various labelled mutants under various experimental conditions to shed light on the structure of the various conformational states of M-TTR so far characterized, including the fully folded and fully unfolded states at neutral pH, the amyloidogenic state at low pH, the equilibrium partially folded state at low urea concentrations and the kinetically trapped state transiently populated during folding.
Materials and methods
Materials
Dimethyl sulfoxide (DMSO), glutathione (GSH), Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), trifluoroacetic acid (TFA) and urea were purchased from Sigma-Aldrich (St Louis, MO, USA). N-(7-Dimethylamino-4-methylcoumarin-3-yl)maleimide (DACM) was purchased from Thermo Fisher Scientific (Waltham, MA, USA).
Protein expression, purification and mutagenesis
Production of M-TTR and its mutants was carried out as previously described [9], except for the bacterial growth temperature of the W41F-M-TTR which was an overnight incubation at 25 °C. Purified proteins were stored at −20 °C in 20 mM phosphate buffer, pH 7.4. Mutations in the gene coding for M-TTR were generated using the QuickChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA). The DNA sequences of the wild-type and mutated genes were checked with DNA sequencing. The molecular masses of the purified variants were checked with MALDI mass spectrometry. Protein purity was found by SDS-PAGE to be >95% in all cases.
Labeling with DACM
Each protein variant was diluted to 0.2 mM in 20 mM phosphate buffer at pH 7.4, 25 °C. Aliquots of DACM dissolved in pure DMSO were added to a tenfold molar excess of dye. The sample was wrapped with aluminium foil and incubated under shaking for 1 h at 37 °C. The reaction was quenched with 5 μl of TFA. The unbound dye was removed by extensive dialysis, using membranes with a 3.0 kDa molecular weight cut off, and the sample was then centrifuged to remove any precipitate. DACM concentration of the resulting labelled protein sample was determined using ε 381 = 27,000 M−1 cm−1. Protein concentration was measured at 280 nm using ε 280 = 18,450 M−1 cm−1 for labelled M-TTR, ε 280 = 12,950 M−1 cm−1 for labelled W79F and W41F-M-TTR, and after subtraction of the contribution of an equimolar concentration of DACM-GSH. An ε 280 = 77,600 M−1 cm−1 for labelled WT-TTR was used as previously reported [19], To this purpose, the optical absorption spectra of DACM-GSH and DACM-M-TTR (or its mutants) were normalised to match identical maximum coumarin absorbance and have fully comparable spectra.
Dynamic light scattering (DLS) measurements
Samples were prepared at a final protein concentration of 15 μM in 20 mM phosphate buffer, pH 7.4, 25 °C or 20 mM acetate buffer, pH 4.4, 25 °C. Before the measurements, the protein samples were filtered with Anotop filters having a cutoff of 20 nm (Whatman, Little Chalfont, UK). DLS measurements were performed using a Zetasizer Nano S device from Malvern Instruments (Malvern, Worcestershire, UK) thermostated with a Peltier system. Low-volume 10 × 4 mm disposable cells were used. The values of refractive index and viscosity set on the instrument were determined using the software provided with the instrument, based on the information of buffer and temperature provided by the user. The presented size distributions were the average of three consecutive measurements.
X-ray crystallography
DACM-WT-TTR and DACM-M-TTR were concentrated and buffer-exchanged at 4 °C with Amicon Ultra-4 Centrifugal Filter Unit with Ultracel-3 membrane, Ultracel®-3K (Merck Millipore, Billerica, MA, USA) down to a final concentration of 12 mg ml−1, in 100 mM KCl, 20 mM Sodium Phosphate, pH 7.4. Both were crystallized by sitting-drops technique at 20 °C: DACM-WT-TTR crystallized in 0.2 M of CaCl2, 0.1 M of HEPES sodium, pH 7.5, 28% v/v polyethylene glycol 400; DACM-M-TTR crystallized in 0.2 M of CaCl2, 0.1 M of sodium acetate, pH 4.6, 20% v/v 2-propanol. Both were cryoprotected adding glycerol to the crystallization solution (final concentration 30% v/v) and flash-frozen in liquid nitrogen. Diffraction data for DACM-WT-TTR and DACM-M-TTR were collected at the European Synchrotron Radiation Facility (ESRF, Grenoble, France) at the ID23-1 and at the ID23-2 beam line to the resolution of 1.42 and 1.7 Å, respectively.
To assess if DACM was damaged during the X-ray data-collection, crystal absorption spectra were collected before and after the diffraction experiment at ID29S (ESRF): both spectra display the maximum absorption at 384 nm typical of DACM and their intensities are comparable ruling out major radiation damage (data not shown).
The diffraction data were integrated and processed using MOSFLM [30], POINTLESS and SCALA [31–33]. The crystal structure of DACM-WT-TTR was determined using BALBES [34] while that of DACM-M-TTR was determined by MOLREP [35] using the structure of M-TTR (pdb code: 1GKO [25]) as a search model. Refinement was performed using Refmac5 and Phenix Refine [36, 37]. Manual model building, visual inspection, addition of water molecules were carried out with Coot [38]. Images of all TTR structures were generated by CCP4mg [33].
Molecular dynamics (MD) simulations
M-TTR was modelled with the Amber99sb force field [39], while DACM was modelled with the GAFF force field [40]. Starting from the X-ray crystallographic structure of DACM-M-TTR, the system was solvated with 5171 TIP3P water molecules in a volume of 1750 Å3. A MD simulation was carried out with Gromacs 4.5.5 for 20 ns at the temperature of 300 K, maintained by a Nosè–Hoover thermostat. Atomic coordinates and structure factors for the TTR variants DACM-WT-TTR and DACM-M-TTR have been deposited with the Protein Data Bank, with accession codes 5LLL and 5LLV, respectively.
Fluorescence spectroscopy
Fluorescence emission spectra (excitation 290 nm) were recorded using a PerkinElmer LS 55 spectrofluorimeter (Waltham, MA, USA) equipped with a thermostated cell holder attached to a Haake F8 water bath (Karlsruhe, Germany). For typical FRET measurements protein samples were diluted to a concentration of 3 µM in 0.5 ml, in 20 mM phosphate buffer, 0.0–7.8 M urea, pH 7.4 25 °C, unless stated otherwise. Pipetting of the protein mother solution for the various protein samples was carried out using a Handy Step from Brand (Wertheim, Germany) to secure accuracy. Each sample was analysed in triplicate (three samples) and three spectra were recorded in each case, implying that each spectrum is the average of nine spectra. For FRET measurements during aggregation protein samples were diluted to a concentration of 1, 3, 7, 14, 28 µM in 0.7 ml under the conditions reported in the “Turbidimetry” section. A 2 × 10 mm and a 4 × 10 mm quartz cuvette were used for the two sets of experiments, respectively.
Equilibrium urea unfolding
28–34 samples were prepared containing 3 µM protein in 20 mM phosphate buffer, pH 7.4, with urea concentrations ranging from 0 to 7.8 M and were incubated for 1 h at 25 °C. Pipetting of the protein mother solution for the various protein samples was carried out using a Handy Step from Brand (Wertheim, Germany) to secure accuracy. Each sample was analysed in triplicate (three samples) and three spectra were recorded in each case, implying that each spectrum is the average of nine spectra. Fluorescence spectra were acquired at 25 °C from 300 to 550 nm (excitation 290 nm) using the spectrofluorimeter, water bath and cuvette described above. Plots of fluorescence at a given wavelength (either 348, 362 or 462 nm) versus urea concentration were analyzed with the two-state unfolding model provided by Santoro and Bolen [25, 41] to obtain quantitative measurements of the free energy change upon denaturation in the absence of denaturant (), the dependence of the free energy change upon denaturation on urea concentration (m value) and the concentration of middle denaturation (C m).
Stopped-flow measurements
Unfolding and refolding reactions were followed using a Bio-Logic (Claix, France) SFM-3 stopped-flow device equipped with an FC-08 cuvette, coupled to a fluorescence detection system and thermostated with a Haake F8 water bath (Karlsruhe, Germany). Excitation wavelength was 290 nm. Band-pass filters cutting emission below 385 and 320 nm were used to monitor DACM and tryptophan fluorescence, respectively. All the experiments were performed in 20 mM phosphate buffer at pH 7.4 and 25 °C, at final protein concentrations of 1.5–2.9 μM. For the unfolding experiments, native proteins in 0.5 M urea were diluted into solutions containing urea at final concentrations ranging from 3.0 to 6.5 M. Refolding reactions were initiated by 10- to 20-fold dilutions of the protein denatured with 5 M urea into solutions containing low urea at final concentrations ranging from 0.25 to 3.5 M. The dead time was generally 10.4 ms. Folding/unfolding kinetic traces were analysed as previously described [28]. In another set of experiments, the equilibrium signal measured at the end of unfolding kinetics was plotted versus urea concentration to linearly extrapolate the fluorescence of the labelled or unlabelled unfolded protein under native conditions (0.5 and 2.1 M urea).
Turbidimetry
15 µM M-TTR and 15 µM DACM-M-TTR were incubated in 20 mM acetate buffer, pH 4.4, 37 °C, in the presence of 0, 30, 60, 90 or 137 mM NaCl. 15 µM W79F-M-TTR and 15 µM DACM-W79F-M-TTR were incubated in 20 mM acetate buffer, pH 5.6, 37 °C, in the presence of 0, 30, 60, 90 or 137 mM NaCl. The time dependent development of turbidity at 450 nm was followed at 37 °C using a Jasco V-630 spectrophotometer (Tokyo, Japan), thermostated within ±0.1 °C by a Haake F8 water bath (Karlsruhe, Germany) and using a 10 mm path length cell. All turbidity values were blank subtracted.
Far-UV CD spectroscopy
Far-UV CD spectra of samples containing 16 µM non-mutated, W41F and W79F-M-TTR in 20 mM sodium phosphate buffer, pH 7.4, 25 °C were collected from 185 to 250 nm using a J-810 Spectropolarimeter from Jasco (Tokyo, Japan) equipped with a thermostated cell holder attached to a Thermo Haake C25P water bath (Karlsruhe, Germany). A 0.1 mm path-length cell (Hellma, Müllheim, Germany) was used. All spectra were blank subtracted and converted to mean residue ellipticity per residue [Θ].
Results
Purification and labeling of M-TTR
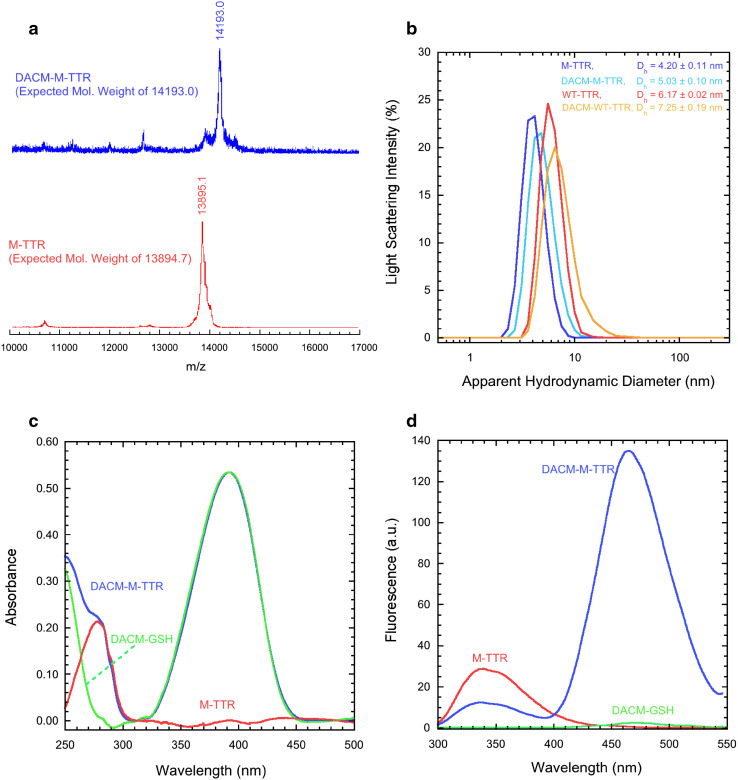
We first purified M-TTR and labelled it with N-(7-dimethylamino-4-methylcoumarin-3-yl)maleimide (DACM), as described in the “Materials and methods” section. The samples containing the purified unlabelled and labelled proteins were analysed with MALDI mass spectrometry (Fig. 1a). A single peak at 13,895 ± 10 and 14,193 ± 10 Da were detected for unlabelled and labelled M-TTR, respectively, in agreement with the expected molecular weights of the two protein samples, i.e. 13,894.7 and 14,193.0 Da, respectively. In the mass spectrum of DACM-M-TTR we could not detect peaks corresponding to the molecular weights of M-TTR with double or triple DACM labelling, excluding the presence of M-TTR labelled with two or more DACM moieties. We could not detect any peak corresponding to unlabelled M-TTR, indicating that no detectable unlabelled protein remained at the end of the labeling reaction.
Fig. 1.
Labelling of M-TTR. a MALDI mass spectrometry analysis of M-TTR (red) and DACM-M-TTR (blue). The expected molecular weights for human M-TTR and DACM-M-TTR are 13,894.7 and 14,193.0, respectively. b Size distributions of M-TTR (blue), DACM-M-TTR (pale blue), WT-TTR (red) and DACM-WT-TTR (orange) samples obtained with DLS at pH 7.4, 25 °C. c Optical absorption spectra of DACM-M-TTR (blue) and DACM-GSH (green) at pH 7.4, 25 °C and at the same probe concentration. The difference spectrum obtained by subtracting the latter from the former is also shown (red). d Fluorescence spectra of M-TTR (red), DACM-M-TTR (blue) and DACM-GSH (green) at pH 7.4, 25 °C (excitation 290 nm)
As an evidence that unlabelled and labelled M-TTR are monomeric, we assessed the size distributions of M-TTR, DACM-M-TTR and wild-type tetrameric TTR (WT-TTR) by means of dynamic light scattering (DLS) at pH 7.4, 25 °C (Fig. 1b). An apparent hydrodynamic diameter of 4.20 ± 0.11 nm was observed for M-TTR, which is consistent with the hydrodynamic diameter of monomeric folded M-TTR, as reported previously [25, 28, 42]. An apparent hydrodynamic diameter of 5.03 ± 0.10 nm was observed for DACM-M-TTR. This diameter is higher than that of unlabelled M-TTR, possibly due to the presence of the covalently attached and solvent-exposed DACM moiety. A diameter of 5.03 ± 0.10 nm is, however, significantly lower than that measured for wild-type tetrameric TTR (6.17 ± 0.02 nm) and of that expected for labelled dimeric M-TTR (>5.2 nm) and labelled tetrameric M-TTR (>6.5 nm) based on purely geometric considerations and of the diffusion of dimeric and tetrameric species in solution in the three dimensions. Overall, the DLS results indicate that both M-TTR and DACM-M-TTR are monomeric.
Next, to gauge the labelling degree of DACM-M-TTR, we acquired optical absorption spectra of DACM-M-TTR and of DACM covalently attached to glutathione (DACM-GSH) at pH 7.4, 25 °C and at the same probe concentration (Fig. 1c). The concentration of DACM bound to M-TTR was determined by measuring the optical absorption at 381 nm (A 381), using a molar extinction coefficient (ε 381) of 27,000 cm−1 M−1 (Fig. 1c, blue spectrum). Protein optical absorption in the DACM-M-TTR sample was then reconstructed by subtracting the spectrum of DACM-GSH (Fig. 1c, green spectrum) from that of DACM-M-TTR sample (Fig. 1c, blue spectrum). In the resulting difference spectrum (Fig. 1c, red spectrum), protein concentration was measured at 280 nm (A 280), using a molar extinction coefficient (ε 280) of 18,450 cm−1 M−1 (an ε 280 value of 12,950 cm−1 M−1 was used for the W79F mutant described below). Labelling degree d was then calculated using:
| 1 |
The spectroscopic investigation described below was carried out with samples having d ≥ 95%.
The fluorescence spectrum of M-TTR at pH 7.4, 25 °C (excitation at 290 nm) is dominated by a major large band at 330–350 nm (Fig. 1d, red spectrum), resulting largely from Trp41 as the fluorescence of Trp79 is largely quenched [23]. The fluorescence spectrum of an equimolar quantity of DACM-GSH, acquired under the same conditions (excitation at 290 nm), is very low in intensity and negligible (Fig. 1d, green spectrum). By contrast, the fluorescence spectrum of an equimolar quantity of DACM-M-TTR under the same conditions (excitation at 290 nm) is characterized by a significant decrease of tryptophan fluorescence intensity, relative to the spectrum of M-TTR, and a substantial enhancement of the DACM emission intensity, relative to the spectrum of DACM-GSH, indicating that FRET is occurring. FRET is largely due to the energy transfer from Trp41 to DACM, both because the fluorescence of Trp79 is quenched and because Trp79 is more distant from DACM compared to Trp41. Importantly, the fluorescence intensity of the tryptophan band is not completely cancelled, leading to a FRET efficiency value (E) intermediate between 0 (no FRET) and 1 (full FRET), making it possible to monitor changes of the distance between donor (Trp41) and acceptor (DACM) by FRET measurements.
Overall, these data indicate that DACM-M-TTR is monomeric, singly labelled with an efficiency close to 100% and with donor/acceptor distance ideal for FRET measurements.
X-ray crystal structures of DACM-M-TTR and DACM-WT-TTR
To determine if DACM labeling on Cys10 affects M-TTR structure, the X-ray crystal structures of DACM-WT-TTR and DACM-M-TTR were determined at high resolution and compared with that of WT-TTR and M-TTR previously deposited in the Protein Data Bank [25, 43]. Data collection and refinement statistics are reported in Table 1. In both structures, from Cys10 to Asn124, all polypeptide chains are well traceable into the electron density map and, in the case of DACM-M-TTR, the F87M/L110M mutations are clearly detectable. As observed in most of the previous TTR structures, the first nine residues are flexible and not present in the electron density. In both DACM-WT-TTR and DACM-M-TTR, the presence of some extra density around Cys10 suggests the presence of DACM. The quality of such density is too poor to allow the building of DACM in the model, suggesting a high degree of conformational flexibility of the DACM moiety.
Table 1.
Data collection and refinement statistics for DACM-WT-TTR and DACM-M-TTR
| Crystala | DACM-WT-TTR | DACM-M-TTR |
|---|---|---|
| Space group | P 21221 | P 212121 |
| Unit cell constants (Å) |
a = 43.68, b = 64.49, c = 85.27 α, β, γ = 90 °C |
a = 64.44, b = 83.76, c = 86.53 α, β, γ = 90 °C |
| Resolution (Å) | 22.35–1.42 (1.50–1.42) | 22.04–1.70 (1.79–1.70) |
| R merge (%)b | 9.7 (73.0) | 11.1 (77.2) |
| I/σI | 8.1 (2.1) | 8.1 (1.9) |
| Completeness (%) | 99.1 (99.2) | 99.5 (99.8) |
| Multiplicity | 5.0 (5.0) | 4.1 (4.3) |
| Molecules per asymmetric unit | 2 | 4 |
| Refinement | ||
| R work (%)c | 16.0 | 15.0 |
| R free (%) | 21.7 | 22.3 |
| Ramachandran plot, n (residues) | ||
| Most favored region | 200 | 421 |
| Allowed region | 3 | 8 |
| Outliers | 0 | 2 |
| RMSDd | 0.31 Å 114/114 Cα | 0.60 Å 115/115 Cα |
aValues given in parenthesis refer to the high-resolution shell
b R merge = Σhkl ΣjIhkl,j − /ΣhklΣjIhkl,j where I is the observed intensity and is the average intensity
c R work = ΣhklFo − Fc/ΣhklFo for all data except 5–10%, which were used for the R free calculation
dRMSD values calculated from the structural superposition of the DACM-WT-TTR and DACM-M-TTR structures with the non-labeled WT-TTR (pdb code: 1BMZ) and M-TTR (pdb code: 1GKO)
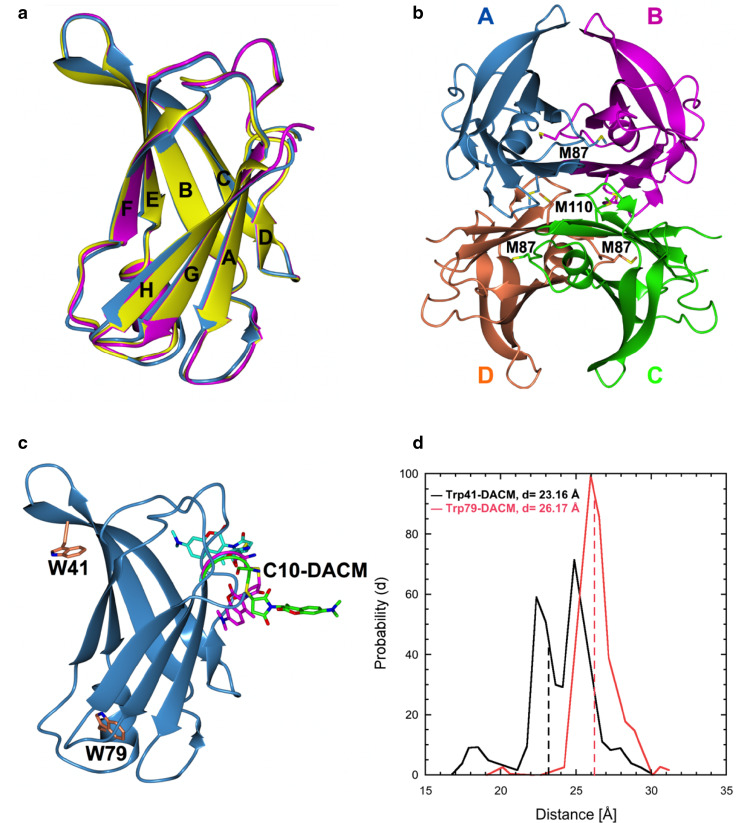
DACM-WT-TTR and DACM-M-TTR crystallised in P21221 and P212121 space group, respectively: DACM-WT-TTR has two molecules in the asymmetric unit (AU), whereas the AU of the DACM-M-TTR structure contains four molecules, as already observed in the M-TTR structure [25]. Superimposition of the DACM-labelled variants with the corresponding non-labeled variants shows that the structures of the DACM-WT-TTR and DACM-M-TTR are perfectly superimposable onto those of WT-TTR and M-TTR, respectively, indicating that DACM labeling does not interfere with the TTR native fold in the crystal (Fig. 2a, b; Table 1).
Fig. 2.
X-ray crystal structures of DACM-M-TTR and DACM-WT-TTR. a Superposition of the structures of DACM-M-TTR (blue), DACM-WT-TTR (magenta) and WT-TTR (yellow, pdb code: 1BMZ [43]). b Tetrameric structure of DACM-M-TTR. The residues Met87 and Met110, that destabilize the tetrameric structure in solution, are shown as sticks. c MD simulations of DACM-M-TTR: Trp41, Trp79 and the most populated conformation of Cys10-DACM are shown as sticks. d Distributions of the spatial distances between the centers of mass of Trp41 and DACM (black) and those of Trp79 and DACM (red), calculated using MD simulations starting from the crystal structure of DACM-M-TTR. The dashed lines indicate the mean spatial distances assuming a single average conformer for the DACM moiety
As mentioned above, DACM was not traceable in the electron density, due to its high mobility. Thus, MD simulations of the solvated DACM-M-TTR were performed, as described in the Materials and Methods, to provide the correct distribution of the DACM conformers, weighted according to Boltzmann statistics. The DACM moiety was not found to be randomly distributed in space during the simulation, with certain conformations more populated than others (Fig. 2c). The mean Trp41–DACM and Trp79–DACM spatial distances, calculated over the centers of mass, were found to be 23.2 ± 2.0 and 26.2 ± 2.0 Å, respectively (Fig. 2d).
FRET efficiency of native M-TTR and Förster distance of the Trp/DACM pair
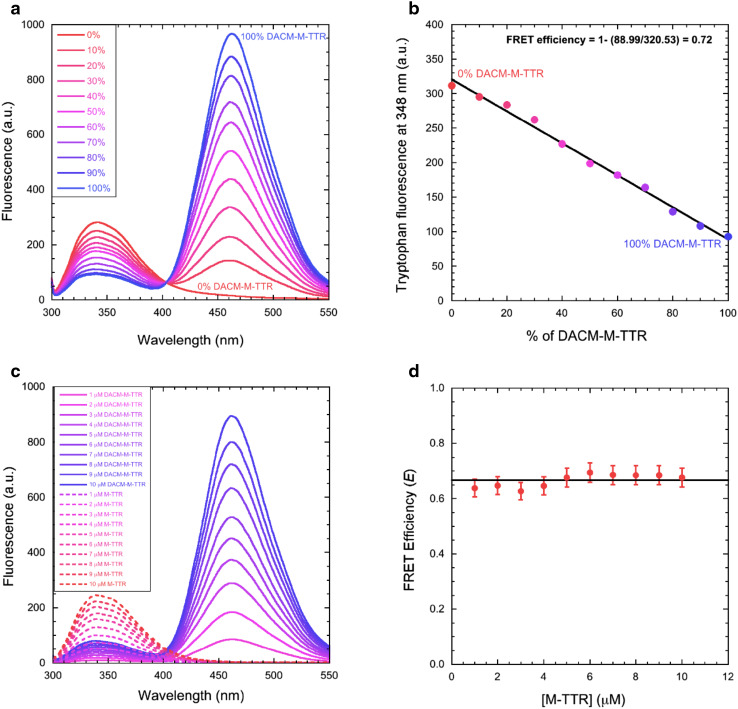
We first determined the FRET efficiency of DACM-M-TTR under conditions in which the protein is folded, at pH 7.4, 25 °C. To this aim, we mixed different volumes of samples containing M-TTR and DACM-M-TTR, each at the same concentration of 3 µM, to achieve different percentages of labelled M-TTR, from 0 to 100%. Fluorescence emission spectra were obtained for all the resulting samples using an excitation wavelength of 290 nm (Fig. 3a). The emission spectrum of unlabeled M-TTR (0% DACM-M-TTR) showed an intense peak at 330–350 nm, resulting from tryptophan fluorescence emission, and no emission at 465 nm due to the absence of the DACM moiety (Fig. 3a, red spectrum). By contrast, the emission spectrum of labeled M-TTR (100% DACM-M-TTR) shows a low intensity peak at 330–350 nm and an intense peak at 465 nm, due to the energy transfer from the excited tryptophan residues to the DACM group, which fluoresces at 465 nm (Fig. 3a, blue spectrum). The samples presenting intermediate degrees of labelling have an intermediate behaviour, with the donor and acceptor peaks decreasing and increasing in intensity with labelling degree, respectively (Fig. 3a, red to blue spectra).
Fig. 3.
FRET of native M-TTR. a Fluorescence spectra of mixtures of M-TTR and DACM-M-TTR at the indicated percentages of the latter, at 3 µM total protein concentration, pH 7.4, 25 °C. b Tryptophan fluorescence emission at 348 nm versus the percentage of DACM-M-TTR. The straight line represents the best fit of the data points to a linear function. The equation indicates how E was determined. c Fluorescence spectra of M-TTR (dashed lines) and DACM-M-TTR (continuous lines) at various protein concentrations ranging from 1 to 10 µM, pH 7.4, 25 °C (excitation 290 nm). d Plot of E versus M-TTR concentration. The straight line represents the average value
The FRET efficiency can be calculated using [44]:
| 2 |
where E is the FRET efficiency, F DA and F D are the fluorescence emission values of the donor in the presence and absence of acceptor, respectively, i.e. the tryptophan emission values at 348 nm measured for the samples containing 100 and 0% DACM-M-TTR, respectively. This led to E = 0.70 ± 0.02. To achieve more accurate measurements of F DA, F D and E we plotted the tryptophan fluorescence emission at 348 nm versus the percentage of DACM-M-TTR and analyzed the resulting plot with a procedure of best fitting using a linear equation (Fig. 3b). This provided more accurate values of F DA and F D and a value of E = 0.72 ± 0.02.
We then measured the fluorescence spectra of M-TTR and DACM-M-TTR at various protein concentrations ranging from 1 to 10 µM (Fig. 3c) and measured the E values at each of these concentrations (Fig. 3d). E was found to be independent of protein concentration, indicating that M-TTR and DACM-M-TTR remain largely monomeric under this range of protein concentration. In addition, the value of E remains lower than that measured for tetrameric DACM-WT-TTR (data not shown), corroborating the absence of protein tetramerisation under the conditions studied here.
To determine the distance between Trp41 (donor) and DACM (acceptor) in the folded monomer using the E value, we first determined experimentally the Förster distance using:
| 3 |
where R 0 is the Förster distance (in Å), κ 2 is the orientation factor describing the relative orientation of the transition dipole moments of the donor and the acceptor, n is the refractive index of the aqueous solution, Q D is the quantum yield of an isolated donor, and J(λ) is the integral expressing the degree of spectral overlap between donor emission and acceptor absorption (units of nm4 M−1 cm−1) [44, 45]. A k 2 value of 0.87 was used as a good approximation of a donor–acceptor orientation factor of a folded protein [46], whereas n, Q D and J(λ) were determined experimentally (Table 2). This allowed the R 0 parameter to be determined for folded M-TTR as 24.9 ± 1.4 Å (Table 2). We then used these R 0 and E values determined experimentally for folded DACM-M-TTR to determine the donor–acceptor distance (R) using the equation [44]:
| 4 |
where R is the distance between the centers of mass of Trp41 and DACM (in Å). This led to a value of R of 21.3 ± 1.6 Å, in good agreement with that determined from X-ray crystallography (R = 23.2 ± 2.0 Å).
Table 2.
FRET parameters of M-TTR adopting different conformational states
| Conformational state | R (Å) | E | R 0 (Å) | Q D | n | J(λ) (nm4 M−1 cm−1) | κ 2 |
|---|---|---|---|---|---|---|---|
| Folded (0 M urea) | 21.3 (±1.6) | 0.72 (±0.02) | 24.9 (±1.4) | 0.050 (±0.008) | 1.332 (±0.001) | 1.96 × 1014 (±0.20 × 1014) | 0.87 (±0.2) |
| Unfolded (6 M urea) | 30.8 (±1.7) | 0.29 (±0.01) | 26.5 (±1.4) | 0.070 (±0.004) | 1.384 (±0.002) | 3.09 × 1014 (±0.15 × 1014) | 0.67 (±0.2) |
| Equilibrium partially folded (1 M urea) | 24.0 (±1.6) | 0.63 (±0.02) | 26.2 (±1.3) | 0.054 (±0.003) | 1.358 (±0.001) | 2.66 × 1014 (±0.13 × 1014) | 0.87 (±0.2) |
| Amyloidogenic partially folded (0 M urea, pH 4.5) | 21.1 (±1.2) | 0.73 (±0.02) | 24.9 (±1.4) | 0.049 (±0.003) | 1.332 (±0.001) | 2.03 × 1014 (±0.10 × 1014) | 0.87 (±0.2) |
E values were determined experimentally from the fluorescence spectra of DACM-M-TTR and M-TTR, respectively. Q D was determined experimentally using free tryptophan as a reference. n was determined experimentally using a 2WAJ ABBE bench refractometer from Optika Microscopes (Bergamo, Italy). J(λ) was determined experimentally as the integral expressing the degree of spectral overlap between donor emission and acceptor absorption [44, 45]. κ 2 was assumed to be 0.87 ± 0.2 for folded proteins [46] and 2/3 (0.67 ± 0.2) for unfolded proteins [44]. R 0 values were determined using Eq. 3. R values were determined from E and R 0 values using Eq. 4
FRET efficiency of urea-induced unfolded and molten globule states of M-TTR
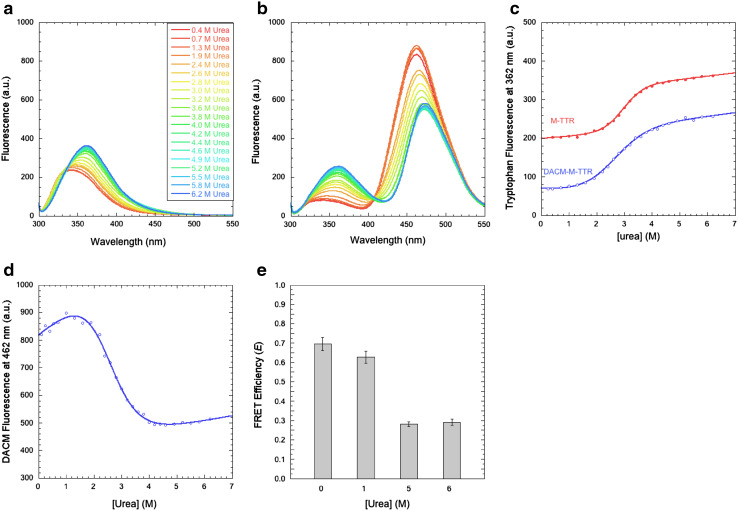
Next, we acquired fluorescence spectra of M-TTR and DACM-M-TTR (excitation 290 nm) at urea concentrations ranging from 0 to 7 M at pH 7.4, 25 °C (Fig. 4a, b). The corresponding urea denaturation curves (spectroscopic signal versus urea concentration at equilibrium) were obtained using tryptophan fluorescence at 362 nm as a spectroscopic probe for both the labelled and unlabeled proteins (Fig. 4c). The analysis yielded values of conformational stability in the absence of denaturant () of 20.4 ± 1.5 and 11.1 ± 1.5 kJ mol−1, dependencies of on urea concentration (m value) of 6.9 ± 0.5 and 4.3 ± 0.5 kJ mol−1 M−1 and midpoint of denaturation (C m) of 3.0 ± 0.1 and 2.6 ± 0.1 M for M-TTR and DACM-M-TTR, respectively. The values obtained for M-TTR are in reasonable agreement with those obtained previously under slightly different conditions [28]. The urea denaturation curve of DACM-M-TTR was also obtained using DACM fluorescence at 462 nm as a spectroscopic probe (Fig. 4d). This led to values of 12.4 ± 1.3 kJ mol−1, 4.9 ± 0.4 kJ mol−1 M−1 and 2.5 ± 0.1 M for , m and C m, respectively, in agreement with those obtained using tryptophan fluorescence as a spectroscopic probe. This analysis indicates that the conformational stability of DACM-M-TTR is significantly lower than that of M-TTR, indicating that the labeling has an effect on protein conformational stability, as it is often observed for mutations.
Fig. 4.
FRET of urea-unfolded and molten globule states of M-TTR. a, b Fluorescence spectra of M-TTR (a) and DACM-M-TTR (b) at urea concentrations ranging from 0 to 6.2 M at pH 7.4, 25 °C (excitation 290 nm). c Urea denaturation curves (spectroscopic signal versus urea concentration) using tryptophan fluorescence at 362 nm as a spectroscopic probe for both M-TTR and DACM-M-TTR. d Urea denaturation curve of DACM-M-TTR, using DACM fluorescence at 462 nm as a spectroscopic probe. e FRET E values at the indicated urea concentrations, pH 7.4, 25 °C
In principle, one can obtain a urea denaturation curve by plotting E versus urea concentration. However, such a curve suffers from different conformational stabilities of labelled and unlabelled M-TTR which makes it difficult to compare fluorescence spectra for the two proteins at the same urea concentration and determine E at each urea concentration. In spite of these difficulties, it is possible to determine the E values at low (<1.2 M) and high (>4.8 M) urea concentrations when the major conformational transition has not yet and has completely occurred, respectively, for both labelled and unlabeled proteins. The FRET E values at 5.0 and 6.0 M urea, when DACM-M-TTR is unfolded, are remarkably lower than those determined at 0.0 and 1.0 M urea, indicating a significantly higher distance between the DACM moiety attached to Cys10 and the two tryptophan residues (Fig. 4e). The R 0 and R values, determined as described above using Eqs. 3 and 4, confirm a larger mean distance between donor and acceptor in the unfolded state (Table 2). The large decrease of the FRET E value upon unfolding indicates that such distance can be monitored with high sensitivity. The DACM fluorescence at 462 nm also decreases from 0.0–1.0 M to 5.0–6.0 M urea (Fig. 4d), confirming that the spatial distance between DACM and the two tryptophan residues increases upon unfolding and gives us the opportunity to observe the movement of DACM from Trp41 in the transition region in which the FRET E value could not be calculated.
Importantly, the FRET E value at 1.0 M urea is similar, within experimental error, to that determined at 0.0 M urea (Fig. 4e). This also results into a similar R value, determined as described above using Eqs. 3 and 4 (Table 2). The DACM fluorescence also does not change significantly (Fig. 4d). This indicates that the equilibrium molten globule state previously detected at low urea concentrations and having a transition at 0.0–2.4 M urea [28] has an unmodified distance between the DACM moiety and Trp41.
FRET efficiency of the transiently populated partially folded state of M-TTR
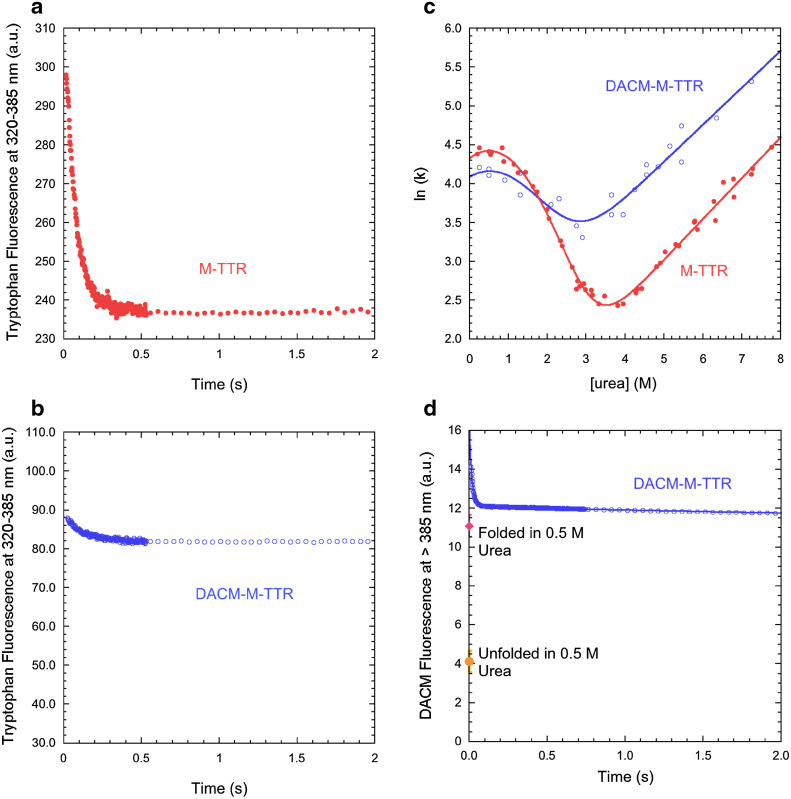
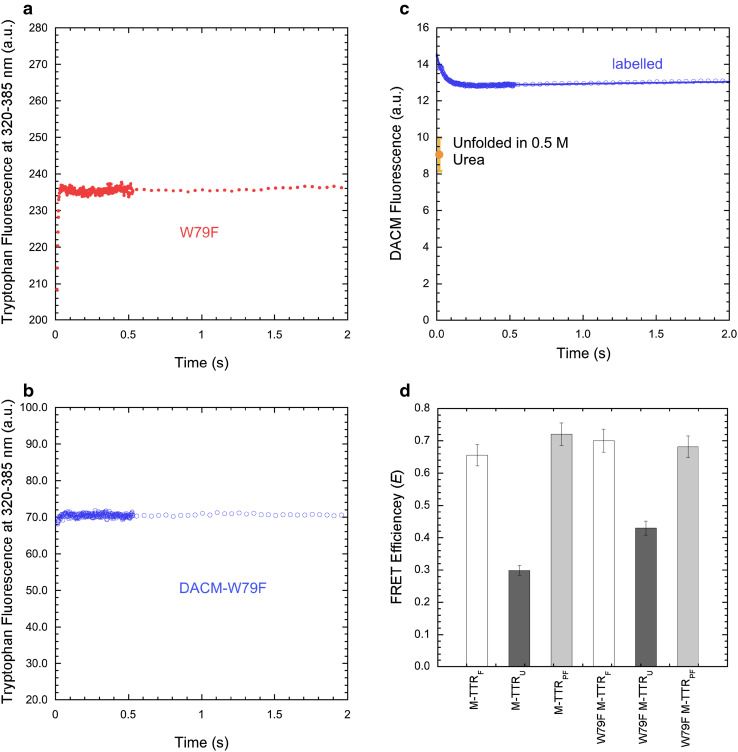
We then monitored the folding and unfolding processes of M-TTR and DACM-M-TTR in real time by diluting the urea-unfolded proteins into solutions containing low urea concentrations (folding) and by diluting the folded protein into solutions containing high urea concentrations (unfolding) using a stopped-flow device coupled to a fluorescence detection system. Representative folding traces recorded in 0.5 M urea, pH 7.4, 25 °C and monitored with intrinsic tryptophan fluorescence at 320–385 nm are shown for M-TTR and DACM-M-TTR (Fig. 5a, b). In addition to the major folding phase, two more low-amplitude exponential phases were observed for M-TTR, which were complete on a longer time scale, as observed previously [28]. Moreover, in addition to the major unfolding phase, one additional low-amplitude unfolding phase was apparent on a long time scale, again as observed previously [28]. This behaviour was also observed for the labelled protein.
Fig. 5.
FRET during M-TTR refolding. a, b Refolding time course of M-TTR (red filled circles) and DACM-M-TTR (blue open circles) monitored by tryptophan fluorescence (excitation 290 nm, emission 320–385 nm) in 0.5 M urea at pH 7.4, 25 °C. c Natural logarithm of the observed folding/unfolding microscopic rate constants for DACM-M-TTR (blue open circles) compared to data previously published for M-TTR (red filled circles) [28], plotted as a function of urea concentration (chevron plot), at pH 7.4, 25 °C. d Refolding time course of DACM-M-TTR (blue open circles) monitored by DACM fluorescence (excitation 290 nm, emission >385 nm) in 0.5 M urea at pH 7.4, 25 °C. The signals of folded and unfolded DACM-M-TTR are also indicated
The analysis of the resulting kinetic traces of folding and unfolding with a procedure of best fit using exponential functions allowed the rate constants for the major folding/unfolding transition (k) to be determined at the various urea concentrations (see “Materials and methods” for further details). These values were used to build the plot of ln(k) versus urea concentration, generally referred to as Chevron plot, for both the labelled and unlabelled proteins (Fig. 5c). The C m values obtained from the analysis of these plots, calculated as the urea concentration at which extrapolated refolding and unfolding rate constants are equal (3.2 ± 0.2 and 2.2 ± 0.2 M for M-TTR and DACM-M-TTR, respectively), were in good agreement with those determined from equilibrium experiments (3.0 ± 0.1 and 2.5 ± 0.1 M, respectively). The downward curvature in the folding branch of the M-TTR plot results from a rapidly formed, off-pathway, partially folded species that forms rapidly and accumulates during the folding process [28]. Such a curvature was also observed for DACM-M-TTR (Fig. 5c). The refolding rate constant did not vary as the concentration of DACM-M-TTR ranged from 0.005 to 0.080 mg ml−1, ruling out that the downward curvature arises from DACM-M-TTR aggregation during folding (data not shown). The extrapolated unfolding rate constants in the absence of denaturant were 1.82 ± 0.18 s−1 [28] and 6.73 ± 0.70 s−1 for M-TTR and DACM-M-TTR, respectively. Given the conformational stabilities obtained by equilibrium experiments, the folding rate constants under the same conditions would be ~7100 s−1 [28] and ~890 s−1, respectively, on the assumption they were two-state folders. By contrast, the refolding rate constants extrapolated by experimental data were orders of magnitude lower than the calculated values, i.e. 81 ± 8 s−1 [28] and 60 ± 5 s−1 for M-TTR and DACM-M-TTR, respectively. This corroborates the hypothesis that a partially folded species accumulates for both M-TTR and DACM-M-TTR.
The spectroscopic signals at 320–385 nm recorded for M-TTR and DACM-M-TTR at the beginning and at the end of the kinetic traces in 0.5 M urea, pH 7.4, 25 °C (Fig. 5a, b) were thus used to calculate transient and equilibrium FRET E values of the folded and partially folded states under these conditions. These were found to be 0.66 ± 0.03 and 0.72 ± 0.03, indicating a slightly higher FRET E value for the partially folded state relative to the folded state. We also monitored the DACM fluorescence emission of such a partially folded state by inspecting the kinetic trace for folding in 0.5 M urea using a band-pass filter that cuts the signal below 385 nm, to monitor the fluorescence of the DACM moiety and exclude that of the tryptophan indole groups (Fig. 5d). Such a kinetic trace involved a decrease in DACM fluorescence, complete in ~50 ms in 0.5 M urea, as DACM-M-TTR underwent refolding (Fig. 5d). The signal of the folded protein and that of the unfolded protein extrapolated down to 0.5 M urea by linear extrapolation from measurements at high urea concentrations, where the protein is 100% unfolded, were remarkably lower (Fig. 5d). This analysis suggests that the transient partially folded state of M-TTR has a DACM fluorescence higher not just than the unfolded state, but also than the fully folded state. The observation of an increase of FRET efficiency by monitoring both tryptophan and DACM fluorescence and using the same stopped-flow apparatus and experimental conditions indicates that the increase is significant. As a control, the kinetic trace for folding in the presence of 2.1 M urea, where the partially folded state cannot be detected, did not show such a burst-phase increase of DACM fluorescence (data not shown). Phenomena different from FRET may concur to the hyperfluorescence exhibited by DACM in the partially folded state, including a different local environment experienced by the coumarin moiety.
DACM fluorescence emission was also followed during unfolding. A decrease in DACM emission was observed, in agreement with an increase in distance between donor and acceptor during the process. As previously observed using intrinsic fluorescence as an optical probe [28], such decrease involved two phases, lasting ca. 0.1 and 30 s, respectively, in the trace recorded in 4.5 M urea taken here as an example (data not shown).
FRET efficiency of the amyloidogenic state of M-TTR
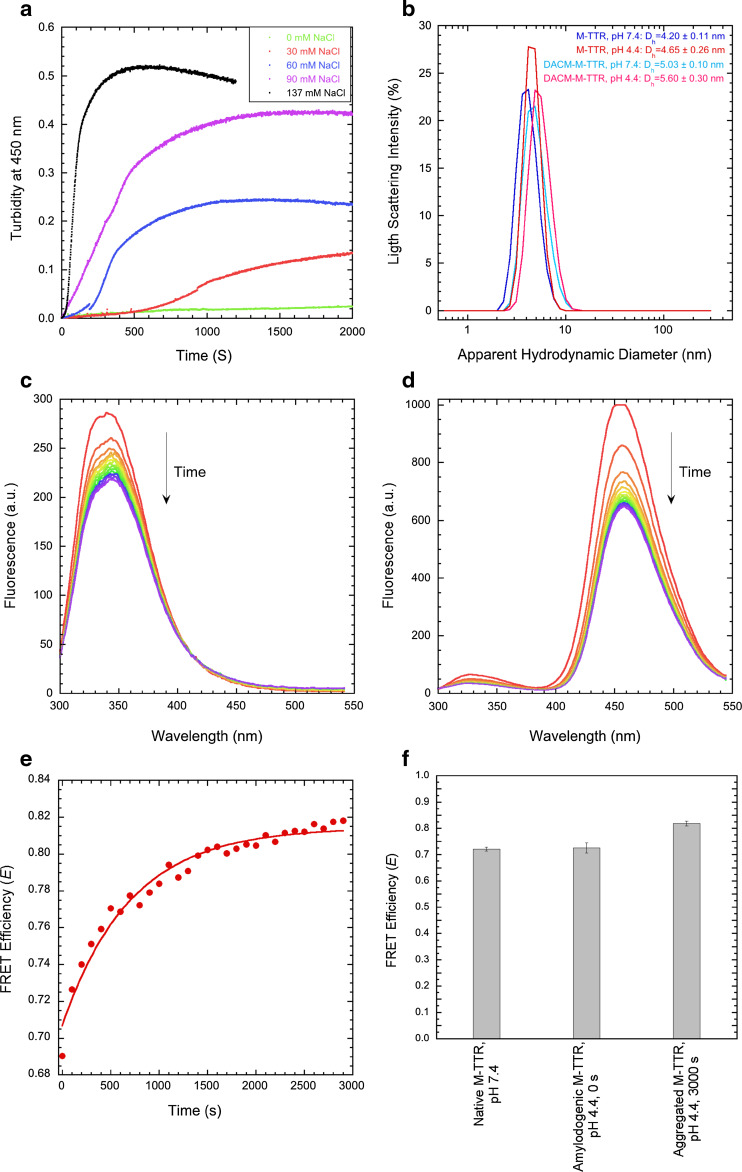
The FRET analysis was also used to investigate the structure of M-TTR under conditions in which the protein is initially monomeric but slowly converts into amyloid fibrils, i.e. at pH 4.4, 37 °C [23, 25]. To check that these conditions of pH and temperature also promoted aggregation of DACM-M-TTR and identify conditions suitable for slow aggregation (so that the initial monomeric DACM-M-TTR can be monitored), DACM-M-TTR aggregation was studied at a protein concentration of 15 µM, in 20 mM acetate buffer, pH 4.4, 37 °C, under the effect of different ionic strengths, that is in the presence of 0, 30, 60, 90 and 137 mM NaCl. The time courses of turbidity at 450 nm show a gradual increase of the rate of turbidity development with the increase of ionic strength (Fig. 6a). The sigmoidal pattern of the intermediate time courses indicates that formation of amyloid fibrils involves the typical three phases of the amyloid fibril formation process: an initial lag phase, a subsequent growth phase and a final saturation phase (plateau). At 30 mM NaCl the time length of the first phase was relatively large but aggregation could still be observed, so that we selected this condition of ionic strength to study the aggregation of the protein.
Fig. 6.
FRET of the amyloidogenic and aggregated states of M-TTR. a Effect of different NaCl concentrations on the kinetics of 15 µM DACM-M-TTR aggregation in 20 mM acetate buffer, pH 4.4, 37 °C, monitored with turbidimetry at 450 nm. b DLS size distributions of M-TTR and DACM-M-TTR at both pH 7.4 and 4.4. c, d Fluorescence spectra at different time points for M-TTR (c) and DACM-M-TTR (d) in 20 mM acetate buffer, 30 mM NaCl, pH 4.4, 37 °C. e Change of E during aggregation. Conditions as in c, d. f FRET E values for native M-TTR at pH 7.4, 25 °C (first bar), amyloidogenic monomeric M-TTR at pH 4.4, 37 °C, 0 s (second bar) and aggregated M-TTR at pH 4.4, 37 °C, 3000 s (third bar)
DLS measurements confirmed that DACM-M-TTR was monomeric immediately after incubation at pH 4.4 (Fig. 6b). Indeed, the size distributions of M-TTR and DACM-M-TTR at pH 7.4, used here as references of monomeric conformations, had major peaks of hydrodynamic diameters at 4.20 ± 0.11 and 5.03 ± 0.10 nm, respectively, with the slightly higher increase of the latter due to the presence of the DACM moiety (see above). The size distributions acquired immediately after incubation at pH 4.4 were found to have major peaks at 4.65 ± 0.26 and 5.60 ± 0.30 nm, respectively (Fig. 6b). These were both slightly higher than those of the corresponding proteins at pH 7.4, most likely as a consequence of the partial unfolding of the proteins at pH 4.4 [23], but were still lower than those of partially unfolded dimers or higher order oligomers.
Hence, both M-TTR and DACM-M-TTR were incubated at 1, 3, 7, 14, 28 µM in 20 mM acetate buffer, 30 mM NaCl, pH 4.4, 37 °C, and fluorescence spectra were collected at different time points, to measure the FRET efficiency (E) values as aggregation proceeded (Fig. 6c–e). Since M-TTR and DACM-M-TTR aggregate with different rates under the selected conditions, the FRET E values are meaningful only immediately after incubation (time 0 s) and at the plateau of the aggregation process (time 3000 s) when both proteins are completely monomeric and aggregated, respectively. The FRET E value of the amyloidogenic state at 0 s from the five different kinetic traces and the resulting R value determined with Eqs. 3 and 4 were found to be 0.73 ± 0.02 and 21.1 ± 1.2 Å, respectively, which are very similar to those measured for the folded proteins at pH 7.4, 25 °C, indicating an unchanged distance between Trp41 and Cys10 upon partial unfolding at pH 4.4, 37 °C (Fig. 6f). The FRET E value at 3000 s was found to be 0.82 ± 0.01. This value is, however, difficult to interpret because both tryptophan and coumarin fluorescences are quenched in the aggregated state, the Förster distance can change following aggregation and is difficult to determine and a population of different aggregates may exist in the sample.
Purification and analysis of W41F and W79F mutants of M-TTR
Although Trp79 is quenched and more distant than Trp41 to Cys10 in the folded state, doubts remain in partially or fully unfolded states, where Trp79 may not be quenched and may be placed closer to Cys10 than Trp41. We therefore, inserted single point mutations in the gene encoding M-TTR and we purified and labelled two single mutants of M-TTR, replacing either Trp41 or Trp79 by a phenylalanine residue (W79F and W41F) and repeated the FRET experiments to assess the involvement of individual inter-residue distances in the overall measured FRET.
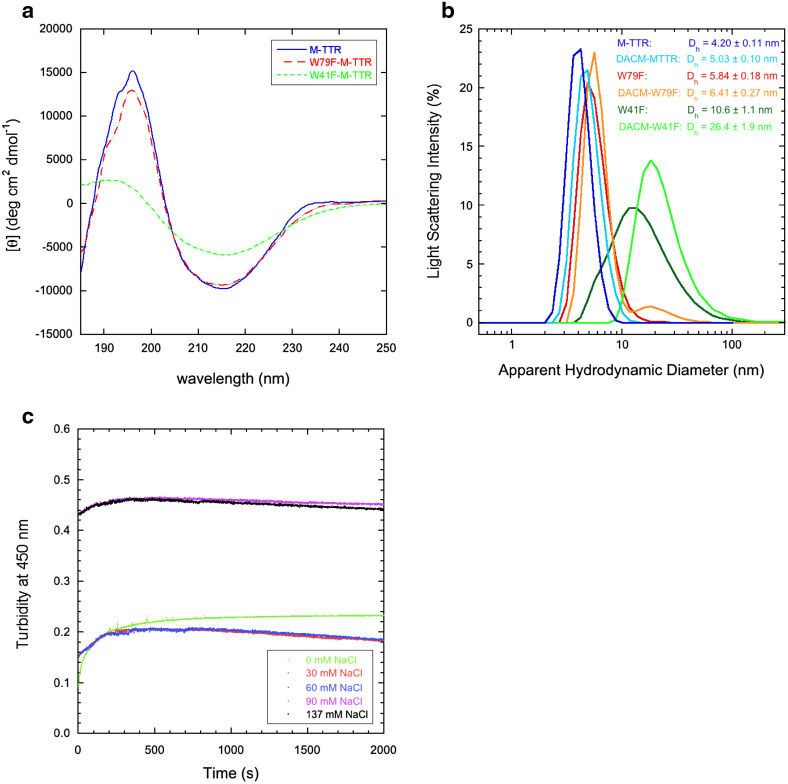
The W79F mutant has a far-UV circular dichroism (CD) spectrum very similar to that of non-mutated M-TTR in 20 mM phosphate buffer, pH 7.4, 25 °C, with a negative peak around 215 nm and a positive peak at 196 nm, typical of β-sheet structure (Fig. 7a). By contrast, the W41F mutant is remarkably different, with mean residue ellipticity peaks characterized by a low intensity, that is reminiscent of protein aggregation (Fig. 7a). The DLS analysis at pH 7.4, 25 °C confirmed that both W79F-M-TTR and DACM-W79F-M-TTR are monomeric, whereas both W41F-M-TTR and DACM-W41F-M-TTR are aggregated (Fig. 7b). As a further evidence that W41F-M-TTR aggregates rapidly under native conditions, the time courses of turbidity at 15 µM protein concentration, 20 mM phosphate buffer, 0–137 mM NaCl, pH 7.4, 37 °C indicate that W41F-M-TTR has a high turbidity at time 0 s in all kinetic traces under native conditions, including at 0 mM NaCl (Fig. 7c).
Fig. 7.
Structural and aggregation properties of W41F and W79F-M-TTR mutants. a Far-UV CD spectra of the non-mutated and mutant forms of M-TTR obtained in 20 mM phosphate buffer, pH 7.4, 25 °C. b Size distributions by DLS of unlabelled and labelled non-mutated, W79F- and W41F-M-TTR at pH 7.4, 25 °C. c Aggregation time courses of W41F-M-TTR at pH 7.4, 37 °C in the presence of 0–137 mM NaCl
Comparison of the FRET efficiency values of mutant and non-mutated M-TTR
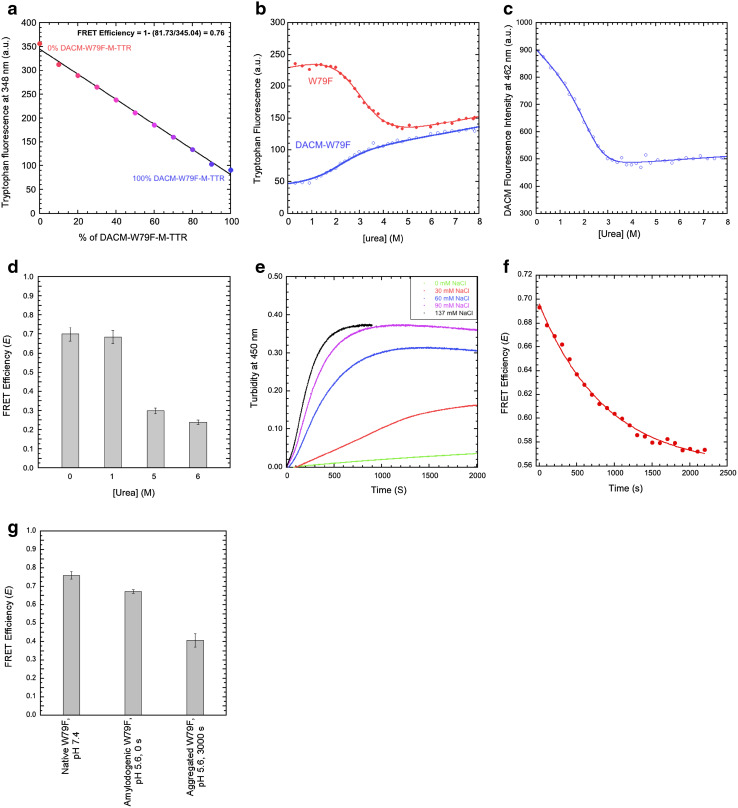
Since W79F-M-TTR, unlike the W41F variant, appears to be soluble and monomeric at pH 7.4, 25 °C, we repeated the same experiments carried out with non-mutated M-TTR, as the mutant provides the opportunity to measure the distance between DACM and Trp41 in the absence of possible artefacts (Fig. 8). The FRET E value of the folded state of W79F-M-TTR at pH 7.4, 25 °C was found to be 0.76 ± 0.02 (Fig. 8a), which is close to the value of non-mutated M-TTR (E = 0.72 ± 0.02). This indicates that the mutation does not alter significantly the inter-residue distance between Cys10 and Trp41 in native M-TTR and reinforces the view that the FRET E value determined for wild-type M-TTR mainly reports on Trp41 and its distance from DACM.
Fig. 8.
FRET of W79F-M-TTR under different conditions. a Tryptophan fluorescence emission at 348 nm versus the percentage of DACM-W79F-M-TTR, at pH 7.4, 25 °C. The straight line represents the best fit of the data points to a linear function. b Urea denaturation curves (spectroscopic signal versus urea concentration at equilibrium) using tryptophan fluorescence at 330 and 365 nm as a spectroscopic probe for W79F-M-TTR and DACM-W79F-M-TTR, respectively, pH 7.4, 25 °C. c Urea denaturation curve of DACM-W79F-M-TTR, using DACM fluorescence at 462 nm as a spectroscopic probe, pH 7.4, 25 °C. d FRET E values at the indicated urea concentrations. e Effect of different NaCl concentrations on the kinetics of 15 µM DACM-W79F-M-TTR aggregation in 20 mM acetate buffer, pH 5.6, 37 °C, monitored with turbidimetry at 450 nm. f FRET E during aggregation. Conditions were 20 mM acetate buffer, 30 mM NaCl, pH 5.6, 37 °C. g FRET E values for native W79F-M-TTR at pH 7.4 (first bar), amyloidogenic monomeric W79F-M-TTR at pH 5.6, 0 s (second bar) and aggregated W79F-M-TTR at pH 5.6, 3000 s (third bar)
The analysis of the urea denaturation curves of the unlabelled and labelled W79F mutant, monitored with tryptophan fluorescence, yielded values of of 13.0 ± 1.4 and 8.6 ± 0.9 kJ mol−1, respectively, m values of 4.3 ± 0.4 and 4.2 ± 0.4 kJ mol−1 M−1, respectively, C m values of 3.0 ± 0.1 and 2.0 ± 0.2 M, respectively (Fig. 8b). The values of W79F-M-TTR and DACM-W79F-M-TTR are significantly lower than those of the corresponding wild-type proteins, indicating that the mutation lowers the conformational stability of the folded state. Moreover, the value of DACM-W79F-M-TTR is significantly lower than that of W79F-M-TTR, indicating that the DACM-labelling lowers the conformational stability of the folded state, as observed for non-mutated M-TTR. The FRET E values measured after denaturation in 5 and 6 M urea (E = 0.30 ± 0.05 and E = 0.24 ± 0.02, respectively) are lower than those in 0 and 1 M urea (E = 0.70 ± 0.04 and E = 0.68 ± 0.03, respectively), indicating a higher distance between the DACM group and the Trp41 residue (Fig. 8d). The FRET E value in 1.0 M urea is similar or just slightly lower than that determined in 0.0 M urea, confirming that the equilibrium molten globule state detectable at low urea concentrations has an unmodified distance between the DACM moiety and Trp41. The plot of DACM fluorescence versus urea concentration confirms that the distance between the DACM moiety and the Trp41 residue increases upon unfolding (Fig. 8c).
Folding of W79F-M-TTR and DACM-W79F-M-TTR was monitored in real time using the stopped-flow apparatus, similarly to the non-mutated protein (Fig. 9a–c). The spectroscopic signals analysed by imposing an off-pathway model [28] were thus used to calculate transient and equilibrium FRET E values of the folded (F), unfolded (U), and partially folded (PF) states (Fig. 9d). Interestingly, the folded and partially folded states of W79F-M-TTR were found to have similar FRET E values, unlike non-mutated M-TTR (Fig. 9d). In addition, the transiently populated partially folded state for W79F-M-TTR has a DACM fluorescence significantly higher than those of both the folded and unfolded states (Fig. 9c), but this effect is less dramatic than that observed for the non-mutated protein (Fig. 5d). Kinetic data analysis and calculation of the FRET E values highlighted several interesting features (Fig. 9a–d). Apparent refolding rates in 0.5 M urea were found equal to 16.3 ± 0.2 and 172 ± 20 s−1 for M-TTR and W79F-M-TTR, respectively, suggesting a strongly destabilized partially folded state following the W79F mutation. However, when the DACM label is attached to M-TTR, refolding rates in 0.5 M urea were found to be 20.2 ± 0.2 for both DACM-M-TTR and DACM-W79F-M-TTR, suggesting a higher energy barrier for refolding when the label is attached to the W79F mutant. The FRET E value (Fig. 9d) of the W79F-M-TTR partially folded state in 0.5 M urea is slightly lower than that of M-TTR (0.68 ± 0.03, as opposed to 0.72 ± 0.03), an observation that, together with that obtained with the refolding rates, is reminiscent of a slightly less compact partially folded state in the mutant.
Fig. 9.
RET of W79F-M-TTR during refolding. a, b Refolding time course of W79F-M-TTR (red filled circles) and DACM-W79F-M-TTR (blue open circles) monitored by tryptophan fluorescence (excitation 290 nm, emission 320–385 nm) in 0.5 M urea at pH 7.4, 25 °C. c Refolding time course of DACM-W79F-M-TTR (blue open circles) monitored by DACM fluorescence (excitation 290 nm, emission >385 nm) in 0.5 M urea at pH 7.4, 25 °C. d Comparison between the FRET E values calculated, from the spectroscopic data, for the folded (F), unfolded (U) and partially folded (PF) states of M-TTR and W79F-M-TTR in 0.5 M urea at pH 7.4, 25 °C
We then attempted to study the aggregation process of the labelled and unlabeled W79F mutant at pH 4.4, 37 °C, as we did for the non-mutated M-TTR. However, the aggregation process was too fast and DLS measurements showed that the protein was aggregated immediately after incubation under these conditions. We therefore, performed a preliminary study with DLS to identify a pH value and ionic strength ideal for slow aggregation of the labelled and unlabeled W79F mutant at a concentration of 15 µM and we found that at pH 5.6, both W79F-M-TTR and DACM-W79F-M-TTR aggregated sufficiently slowly to allow the detection and analysis of both the monomeric amylodogenic state and the aggregated state (data not shown). We therefore, acquired time courses of aggregation for 15 µM W79F-M-TTR and DACM-W79F-M-TTR in 20 mM acetate buffer, pH 5.6, 37 °C in the presence of 0, 30, 60, 90 and 137 mM NaCl, using turbidimetry at 450 nm as a probe for aggregation (Fig. 8e). We selected a NaCl concentration of 30 mM for FRET measurements and performed the experiments as described above for the wild-type protein (Fig. 8f) determining a FRET E value of 0.67 ± 0.01 for the amyloidogenic state (Fig. 8g). A FRET E value of 0.41 ± 0.04 was obtained for the aggregated state (Fig. 8g), although this value is difficult to interpret for the reasons explained above. Overall, the FRET E values measured for the native states at pH 7.4 and for the amyloidogenic states at mildly acidic pH for M-TTR and W79F-M-TTR are all similar, within experimental error, indicating that the distance between the DACM moiety attached to Cys10 and Trp41 does not change significantly in the transition from the native to the amyloidogenic state.
Discussion
The production of a mutant of human wild-type TTR that is stable as a monomer, generally referred to as M-TTR [25], has greatly facilitated the study of the folding process of this protein, of the structural perturbations occurring in the conversion of the fully folded monomer to the amyloidogenic state at weakly acidic pH values, of the aggregation process from such a state and of the mechanism of action of TTR as a toxin detoxifier [25, 26, 28, 47–49]. Following the observation that M-TTR contains only a solvent-exposed cysteine residue at position 10 that can be potentially labelled with a coumarin derivative able to act as an acceptor of the fluorescence of the naturally present tryptophan residues at positions 41 and 79, we generated a coumarin-labelled M-TTR molecule (DACM-M-TTR) to investigate the spatial distance between the coumarin moiety at position 10 and Trp41/Trp79, particularly Trp41, which is non-quenched and spatially closer to Cys10.
Five conformational states of M-TTR have been investigated, namely (1) the fully folded state at pH 7.4, (2) the urea-unfolded state at pH 7.4, (3) the molten globule state populated at equilibrium at low urea concentrations, pH 7.4, (4) the partially folded state accumulating transiently during folding, pH 7.4 and (5) the amyloidogenic state at pH 4.4.
The first study performed at pH 7.4, 25 °C, no added denaturants, indicated that DACM-M-TTR is folded, monomeric in solution, singly and uniformly labelled, and maintains a structure superimposable to that of unlabeled M-TTR. The labelled protein is less stable than the unlabeled one, as indicated by the lower free energy change of unfolding (), but this is attributable to the high sensitivity of the value to covalent modifications, as indicated by the many observations accumulated so far that most mutants of TTR are indeed less stable than the wild-type protein, regardless of the position and type of the mutation [20]. A FRET E value of 0.72 ± 0.02 was obtained for DACM-M-TTR at pH 7.4. Such a value is largely due to the energy transfer from Trp41 to DACM, both because the fluorescence of Trp79 is quenched [23] and because Trp79 is more distant than Trp41 from Cys10 [25, 43]. The observation that the W79F mutant of DACM-M-TTR features a similar FRET E value under the same conditions, supports this hypothesis. A FRET E value of 0.72 ± 0.02 results into a spatial distance of 21.3 ± 1.6 nm, between Trp41 and DACM.
The conversion of the non-mutated and W79F mutant forms of M-TTR from their folded state to the amyloidogenic state at pH 4.4 (or 5.6 in the case of the W79F-M-TTR) does not change significantly the FRET E value and the related distance between DACM and Trp41, indicating that β-strand C, where Trp41 is located, does not increase its distance from Cys10 and does not unfold upon such conversion. This observation is particularly important as previous structural investigations on WT-TTR, carried out with spectroscopic methods, limited proteolysis and hydrogen/deuterium exchange monitored with NMR, attributed such a structural conversion mainly to the unfolding of β-strands C/D and the interconnecting loop [23] or to the entire β-sheet CBEF [24]. If this was the case, a decrease of the FRET E value should have been observed upon acidification, given the high sensitivity of FRET to even minor structural perturbations or increased dynamics. Our observation is indeed in agreement with more recent observations obtained with solution NMR that the whole CBEF β-sheet, including β-strand C, does not unfold, nor does it increase its structural dynamics, upon conversion of native M-TTR and WT-TTR into the amyloidogenic state at mildly acidic pH [26, 27, 50]. A unifying picture of the amyloidogenic state of TTR at mildly low pH (pH 3.9–5.5) that is consistent with all recent data obtained so far with different techniques is that of a largely native-like state with the entire DAGH β-sheet, the D–E loop, the E–F α-helix and its associated residues from the A–B loop, exhibiting structural fluctuations, in the presence of a substantially folded and packed CBEF β-sheet.
The equilibrium molten globule state of M-TTR, or W79F-M-TTR, populated at low urea concentrations, pH 7.4, has a FRET E value and a resulting R value similar to those of the folded and amyloidogenic states of the same protein isoforms, suggesting that the Cys10–Trp41 distance is not significantly altered in such a conformational state. It is interesting to compare the structural information collected so far on such molten globule state of M-TTR [28] with that obtained for the amyloidogenic state of M-TTR and WT-TTR at pH 3.9-5.0 [23, 25–27, 50], to assess whether the two conformational states are essentially similar albeit populated under different conditions. The two conformational states share similar FRET E and R values and similar near-UV CD spectra, but different intrinsic fluorescence and far-UV CD spectra. The molten globule state of M-TTR in 2.0 M urea, pH 7.4, has a far-UV CD spectrum different from that of the folded/amyloidogenic states [27], which appear to be similar between them [23, 28]. Indeed, the molten globule state indicates a substantial unfolding of some portions of the protein [28]. The molten globule state in 2.0 M urea, pH 7.4, unlike the amyloidogenic state at pH 4.4, has an intrinsic fluorescence spectrum similar to that of the folded state at pH 7.4 [23, 28]. Hence, it appears that the two conformational states are structurally distinct. It will be important to establish the amyloidogenic nature of the molten globule state, which may be as important as the amyloidogenic state populated at low pH.
As far as the off-pathway, partially folded state formed transiently during folding at pH 7.4 is concerned, it has a FRET E value significantly higher than that of the folded and amyloidogenic states, suggesting that Trp41 is closer to Cys10 than it is in the folded/amyloidogenic states. This result reinforces the view that such a conformational state is an off-pathway species with some non-native contacts that need to be unfolded before folding can proceed [28]. The kinetic and amyloidogenic conformational states share similar values of mean residue ellipticity, at least at 219 nm, and both induce an increase and blue-shift of the ANS dye [23, 25, 28]. However, in addition to showing different FRET E values, they also differ for the intrinsic fluorescence spectra, exhibiting a fluorescence intensity higher and lower than the folded state at pH 7.4, respectively [23, 28]. Again, it appears that the two conformational states are structurally distinct.
Another interesting observation emerging from these results is the high aggregation propensity exhibited by the W41F mutant, which was indeed found to be insoluble even under native conditions, limiting our FRET analysis. Among the many mutants of TTR associated with TTR diseases, one has been reported to involve Trp41 (W41L) and to cause vitreous opacity with amyloid-material positive for TTR in the vitreous fluid of the index patient [51, 52]. W41F and W41L TTRs share similarly high aggregation propensities, observed in vivo and in vitro, respectively, suggesting that Trp41 is a very important amino acid residue for maintaining the stability and solubility of native TTR.
Conclusions
In conclusion, the FRET study described here provides insight into the structural changes occurring during the folding, unfolding and aggregation of TTR, a protein whose aggregation has been linked to systemic amyloidoses. The most important achievements of our results is the clarification of the degree of unfolding and dynamics of β-strand C in the formation of the amyloidogenic state and the establishment of the concept that both the molten globule state and kinetically trapped species accumulating during folding are structurally distinct from such a state, indicating that TTR is a highly plastic protein able to populate a number of structurally discrete conformational states.
Acknowledgements
We thank the Iranian Ministry of Science Research and Technology for providing the studentship for S.A.G. and the University of Florence for Fondi di Ateneo. F.B.’s research is funded by the Italian MIUR (Programma per Giovani Ricercatori Rita Levi Montalcini 2010). We also thank Martino Bolognesi for insightful discussion. We thank Joel Buxbaum and Xinyi Li for providing the gene coding M-TTR.
Abbreviations
- FRET
Fluorescence resonance energy transfer
- TTR
Transthyretin
- M-TTR
Monomeric variant of TTR
- RBP
Retinol binding protein
- CSF
Cerebrospinal fluid
- SSA
Senile systemic amyloidosis
- FAP
Familial amyloid polyneuropathy
- FAC
Familial amyloid cardiomyopathy
- DMSO
Dimethyl sulfoxide
- GSH
Glutathione
- TCEP
Tris(2-carboxyethyl)phosphine hydrochloride
- TFA
Trifluoroacetic acid
- DACM
N-(7-Dimethylamino-4-methylcoumarin-3-yl)maleimide
- DLS
Dynamic light scattering
- MD
Molecular dynamics
- AU
Asymmetric unit
- CD
Circular dichroism
References
- 1.Soprano DR, Herbert J, Soprano KJ, Schon EA, Goodman DS. Demonstration of transthyretin mRNA in the brain and other extrahepatic tissues in the rat. J Biol Chem. 1985;260(21):11793–11798. [PubMed] [Google Scholar]
- 2.Stauder AJ, Dickson PW, Aldred AR, Schreiber G, Mendelsohn FA, Hudson P. Synthesis of transthyretin (pre-albumin) mRNA in choroid plexus epithelial cells, localized by in situ hybridization in rat brain. J Histochem Cytochem. 1986;34(7):949–952. doi: 10.1177/34.7.3458812. [DOI] [PubMed] [Google Scholar]
- 3.Herbert J, Wilcox JN, Pham KT, Fremeau RT, Jr, Zeviani M, Dwork A, Soprano DR, Makover A, Goodman DS, Zimmerman EA, et al. Transthyretin: a choroid plexus-specific transport protein in human brain. The 1986 S. Weir Mitchell award. Neurology. 1986;36(7):900–911. doi: 10.1212/WNL.36.7.900. [DOI] [PubMed] [Google Scholar]
- 4.Jacobsson B. Localization of transthyretin-mRNA and of immunoreactive transthyretin in the human fetus. Virchows Arch A Pathol Anat Histopathol. 1989;415(3):259–263. doi: 10.1007/BF00724913. [DOI] [PubMed] [Google Scholar]
- 5.Jacobsson B, Collins VP, Grimelius L, Pettersson T, Sandstedt B, Carlstrom A. Transthyretin immunoreactivity in human and porcine liver, choroid plexus, and pancreatic islets. J Histochem Cytochem. 1989;37(1):31–37. doi: 10.1177/37.1.2642294. [DOI] [PubMed] [Google Scholar]
- 6.Murakami T, Ohsawa Y, Zhenghua L, Yamamura K, Sunada Y. The transthyretin gene is expressed in Schwann cells of peripheral nerves. Brain Res. 2010;1348:222–225. doi: 10.1016/j.brainres.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 7.Wakasugi S, Maeda S, Shimada K. Structure and expression of the mouse prealbumin gene. J Biochem. 1986;100(1):49–58. doi: 10.1093/oxfordjournals.jbchem.a121705. [DOI] [PubMed] [Google Scholar]
- 8.Buxbaum JN, Reixach N. Transthyretin: the servant of many masters. Cell Mol Life Sci. 2009;66(19):3095–3101. doi: 10.1007/s00018-009-0109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reixach N, Foss TR, Santelli E, Pascual J, Kelly JW, Buxbaum JN. Human-murine transthyretin heterotetramers are kinetically stable and non-amyloidogenic. A lesson in the generation of transgenic models of diseases involving oligomeric proteins. J Biol Chem. 2008;283(4):2098–2107. doi: 10.1074/jbc.M708028200. [DOI] [PubMed] [Google Scholar]
- 10.Sekijima Y. Transthyretin (ATTR) amyloidosis: clinical spectrum, molecular pathogenesis and disease-modifying treatments. J Neurol Neurosurg Psychiatry. 2015;86(9):1036–1043. doi: 10.1136/jnnp-2014-308724. [DOI] [PubMed] [Google Scholar]
- 11.Sipe JD, Benson MD, Buxbaum JN, Ikeda SI, Merlini G, Saraiva MJ, Westermark P. Amyloid fibril proteins and amyloidosis: chemical identification and clinical classification International Society of Amyloidosis 2016 Nomenclature Guidelines. Amyloid. 2016;23(4):209–213. doi: 10.1080/13506129.2016.1257986. [DOI] [PubMed] [Google Scholar]
- 12.Lie JT, Hammond PI. Pathology of the senescent heart: anatomic observations on 237 autopsy studies of patients 90 to 105 years old. Mayo Clin Proc. 1988;63(6):552–564. doi: 10.1016/S0025-6196(12)64885-X. [DOI] [PubMed] [Google Scholar]
- 13.Tanskanen M, Peuralinna T, Polvikoski T, Notkola IL, Sulkava R, Hardy J, Singleton A, Kiuru-Enari S, Paetau A, Tienari PJ, Myllykangas L. Senile systemic amyloidosis affects 25% of the very aged and associates with genetic variation in alpha2-macroglobulin and tau: a population-based autopsy study. Ann Med. 2008;40(3):232–239. doi: 10.1080/07853890701842988. [DOI] [PubMed] [Google Scholar]
- 14.Connors LH, Lim A, Prokaeva T, Roskens VA, Costello CE. Tabulation of human transthyretin (TTR) variants, 2003. Amyloid. 2003;10(3):160–184. doi: 10.3109/13506120308998998. [DOI] [PubMed] [Google Scholar]
- 15.Garzuly F, Vidal R, Wisniewski T, Brittig F, Budka H. Familial meningocerebrovascular amyloidosis, Hungarian type, with mutant transthyretin (TTR Asp18Gly) Neurology. 1996;47(6):1562–1567. doi: 10.1212/WNL.47.6.1562. [DOI] [PubMed] [Google Scholar]
- 16.Cornwell GG, 3rd, Sletten K, Johansson B, Westermark P. Evidence that the amyloid fibril protein in senile systemic amyloidosis is derived from normal prealbumin. Biochem Biophys Res Commun. 1988;154(2):648–653. doi: 10.1016/0006-291X(88)90188-X. [DOI] [PubMed] [Google Scholar]
- 17.Saraiva MJ, Costa PP, Goodman DS. Transthyretin (prealbumin) in familial amyloidotic polyneuropathy: genetic and functional aspects. Adv Neurol. 1988;48:189–200. [PubMed] [Google Scholar]
- 18.Westermark P, Sletten K, Johansson B, Cornwell GG., 3rd Fibril in senile systemic amyloidosis is derived from normal transthyretin. Proc Natl Acad Sci USA. 1990;87(7):2843–2845. doi: 10.1073/pnas.87.7.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mccutchen SL, Colon W, Kelly JW. Transthyretin mutation Leu-55-Pro significantly alters tetramer stability and increases amyloidogenicity. Biochemistry. 1993;32(45):12119–12127. doi: 10.1021/bi00096a024. [DOI] [PubMed] [Google Scholar]
- 20.Sekijima Y, Wiseman RL, Matteson J, Hammarstrom P, Miller SR, Sawkar AR, Balch WE, Kelly JW. The biological and chemical basis for tissue-selective amyloid disease. Cell. 2005;121(1):73–85. doi: 10.1016/j.cell.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 21.Steinrauf LK, Hamilton JA, Braden BC, Murrell JR, Benson MD. X-ray crystal structure of the Ala-109 → Thr variant of human transthyretin which produces euthyroid hyperthyroxinemia. J Biol Chem. 1993;268(4):2425–2430. [PubMed] [Google Scholar]
- 22.Kelly JW, Colon W, Lai ZH, Lashuel HA, McCulloch J, McCutchen SL, Miroy GJ, Peterson SA. Transthyretin quaternary and tertiary structural changes facilitate misassembly into amyloid. Adv Protein Chem. 1997;50:161–181. doi: 10.1016/S0065-3233(08)60321-6. [DOI] [PubMed] [Google Scholar]
- 23.Lai Z, Colon W, Kelly JW. The acid-mediated denaturation pathway of transthyretin yields a conformational intermediate that can self-assemble into amyloid. Biochemistry. 1996;35(20):6470–6482. doi: 10.1021/bi952501g. [DOI] [PubMed] [Google Scholar]
- 24.Liu K, Cho HS, Hoyt DW, Nguyen TN, Olds P, Kelly JW, Wemmer DE. Deuterium–proton exchange on the native wild-type transthyretin tetramer identifies the stable core of the individual subunits and indicates mobility at the subunit interface. J Mol Biol. 2000;303(4):555–565. doi: 10.1006/jmbi.2000.4164. [DOI] [PubMed] [Google Scholar]
- 25.Jiang X, Smith CS, Petrassi HM, Hammarstrom P, White JT, Sacchettini JC, Kelly JW. An engineered transthyretin monomer that is nonamyloidogenic, unless it is partially denatured. Biochemistry. 2001;40(38):11442–11452. doi: 10.1021/bi011194d. [DOI] [PubMed] [Google Scholar]
- 26.Lim KH, Dyson HJ, Kelly JW, Wright PE. Localized structural fluctuations promote amyloidogenic conformations in transthyretin. J Mol Biol. 2013;425(6):977–988. doi: 10.1016/j.jmb.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim KH, Dasari AK, Hung I, Gan Z, Kelly JW, Wemmer DE. Structural changes associated with transthyretin misfolding and amyloid formation revealed by solution and solid-state NMR. Biochemistry. 2016;55(13):1941–1944. doi: 10.1021/acs.biochem.6b00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conti S, Li X, Gianni S, Ghadami SA, Buxbaum J, Cecchi C, Chiti F, Bemporad F. A complex equilibrium among partially unfolded conformations in monomeric transthyretin. Biochemistry. 2014;53(27):4381–4392. doi: 10.1021/bi500430w. [DOI] [PubMed] [Google Scholar]
- 29.Clegg RM. Fluorescence resonance energy transfer. Curr Opin Biotechnol. 1995;6(1):103–110. doi: 10.1016/0958-1669(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 30.Leslie AGW. Joint CCP4 and ESF-EACMB newsletter on protein crystallography. Warrington: SERC Daresbury Laboratory; 1992. [Google Scholar]
- 31.Collaborative Computational Project N The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 32.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 1):72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 33.McNicholas S, Potterton E, Wilson KS, Noble ME. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Long F, Vagin AA, Young P, Murshudov GN. BALBES: a molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 1):125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. doi: 10.1107/S0021889897006766. [DOI] [Google Scholar]
- 36.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65(3):712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 41.Santoro MM, Bolen DW. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry. 1988;27(21):8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 42.Pires RH, Karsai A, Saraiva MJ, Damas AM, Kellermayer MS. Distinct annular oligomers captured along the assembly and disassembly pathways of transthyretin amyloid protofibrils. PLoS One. 2012;7(9):e44992. doi: 10.1371/journal.pone.0044992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peterson SA, Klabunde T, Lashuel HA, Purkey H, Sacchettini JC, Kelly JW. Inhibiting transthyretin conformational changes that lead to amyloid fibril formation. Proc Natl Acad Sci USA. 1998;95(22):12956–12960. doi: 10.1073/pnas.95.22.12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lakowicz JR. Principles of fluorescence spectroscopy. 3. New York: Springer; 2006. [Google Scholar]
- 45.Nazarov PV, Koehorst RB, Vos WL, Apanasovich VV, Hemminga MA. FRET study of membrane proteins: simulation-based fitting for analysis of membrane protein embedment and association. Biophys J. 2006;91(2):454–466. doi: 10.1529/biophysj.106.082867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Visser NV, Westphal AH, van Hoek A, van Mierlo CP, Visser AJ, van Amerongen H. Tryptophan-tryptophan energy migration as a tool to follow apoflavodoxin folding. Biophys J. 2008;95(5):2462–2469. doi: 10.1529/biophysj.108.132001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hurshman AR, White JT, Powers ET, Kelly JW. Transthyretin aggregation under partially denaturing conditions is a downhill polymerization. Biochemistry. 2004;43(23):7365–7381. doi: 10.1021/bi049621l. [DOI] [PubMed] [Google Scholar]
- 48.Cascella R, Conti S, Mannini B, Li X, Buxbaum JN, Tiribilli B, Chiti F, Cecchi C. Transthyretin suppresses the toxicity of oligomers formed by misfolded proteins in vitro. Biochim Biophys Acta 1832. 2013;12:2302–2314. doi: 10.1016/j.bbadis.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 49.Cappelli S, Penco A, Mannini B, Cascella R, Wilson MR, Ecroyd H, Li X, Buxbaum JN, Dobson CM, Cecchi C, Relini A, Chiti F. Effect of molecular chaperones on aberrant protein oligomers in vitro: super-versus sub-stoichiometric chaperone concentrations. Biol Chem. 2016;397(5):401–415. doi: 10.1515/hsz-2015-0250. [DOI] [PubMed] [Google Scholar]
- 50.Das JK, Mall SS, Bej A, Mukherjee S. Conformational flexibility tunes the propensity of transthyretin to form fibrils through non-native intermediate states. Angew Chem Int Ed Engl. 2014;53(47):12781–12784. doi: 10.1002/anie.201407323. [DOI] [PubMed] [Google Scholar]
- 51.Yazaki M, Connors LH, Eagle RC, Jr, Leff SR, Skinner M, Benson MD. Transthyretin amyloidosis associated with a novel variant (Trp41Leu) presenting with vitreous opacities. Amyloid. 2002;9(4):263–267. doi: 10.3109/13506120209114104. [DOI] [PubMed] [Google Scholar]
- 52.Yazaki M, Varga J, Dyck PJ, Benson MD. A new transthyretin variant Leu55Gln in a patient with systemic amyloidosis. Amyloid. 2002;9(4):268–271. doi: 10.3109/13506120209114105. [DOI] [PubMed] [Google Scholar]