Abstract
PLK1 regulates almost every aspect of mitotic events, including mitotic entry, spindle assembly, chromosome alignment, sister chromatid segregation, metaphase-anaphase transition, cytokinesis, etc. In regulating the chromosome alignment and sister chromatid segregation, PLK1 has to be localized to and removed from kinetochores at the right times, and the underlying mechanism that regulates PLK1 both spatially and temporally only became clearer recently. It has been found that deubiquitination and ubiquitination of PLK1 are responsible for its localization to and dissociation from the kinetochores, respectively. The equilibrium of this ubiquitination and deubiquitination plays an important role in regulating proper chromosome alignment and timely sister chromatid segregation. Here, we summarize and discuss the recent findings in investigating the spatial and temporal regulation of PLK1 during chromosome alignment and sister chromatid segregation.
Keywords: Polo-like kinase 1, Mitosis, Usp16, Kinetochore, Cullin 3, E3 ligase
Introduction
Polo-like kinases are a group of evolutionarily conserved serine/threonine protein kinases. It consists of five members, and polo-like kinase 1 (PLK1) is the best-studied kinase that plays a pivotal role in mitosis [1, 2]. PLK1 regulates almost every aspect of mitotic events, including mitotic entry, spindle assembly, chromosome alignment, sister chromatid segregation, metaphase-anaphase transition, cytokinesis, etc. Downregulation of PLK1 or inhibition of its kinase activity often leads to mitotic defects that ultimately result in activation of the spindle assembly checkpoint (SAC) and apoptosis [3, 4].
One of the important functions of PLK1 is to regulate the alignment of chromosomes and the segregation of sister chromatids. It is well established that outer kinetochore phosphorylation by Aurora B decreases upon bi-orientation [5], suggesting that correction is occurring during prometaphase, but it was unclear how the initial stable kinetochore–microtubule (KT–MT) attachments are formed in prometaphase. It was shown recently that in addition to recruiting PP2A to prometaphase kinetochores leading to the dephosphorylation of Aurora B substrates [6], PLK1 specifically promotes the initial establishment of KT–MT attachments during prometaphase by suppressing KT–MT dynamics to balance the destabilizing activity of Aurora B [7]. Kinetochores represent a major point of contact between mitotic spindle microtubules and chromosomes [8, 9]. Numerous proteins accumulate at kinetochores to generate a “wait anaphase” signal to maintain SAC activity until all chromosomes have completed bipolar attachment.
Liu et al. showed that maintaining high level of PLK1 at kinetochores after all chromosomes were aligned at metaphase plate resulted in a failure to establish interkinetochore tension, i.e., spindle microtubules failed to exert normal pulling forces on sister kinetochores, and decreased intrakinetochore stretch [7]. Dynamic microtubules are required for the establishment of interkinetochore tension and increase of intrakinetochore stretch [10–12]. This result suggests that PLK1 activity at kinetochores regulates microtubule dynamics, and it must be removed in metaphase to maintain dynamic microtubules and allow the successful separation of sister chromatids [7]. It should be noted that constitutively active PLK1T201D was used in this study [7]. It is, therefore, possible that if wild-type PLK1 was used, the removal of PLK1 from the kinetochores might have been unnecessary as dephosphorylation on PLK1 substrates might provide adequate regulation. In addition, the use of wild-type polo box domain (PBD)-containing Hec1-PLK1T201D might have recruited other PBD interactors to the outer kinetochore and potentially created a non-physiologic function. Furthermore, the PLK1T201D was localized by Hec1 tag to outer kinetochore, which may not necessarily be the right physiologic location for PLK1 as it is difficult to reach many of its substrates from this location.
Microtubules must be dynamic to allow correction of any errors in KT–MT attachment [13, 14]. Increased PLK1 level at metaphase also results in increased attachment errors [7, 15]. In metaphase, PLK1 substrates are dephosphorylated at kinetochores [16, 17], and the dephosphorylation of the PLK1 substrates is likely a result of both the recruitment of protein phosphatase 1 [18] and reduction of PLK1 levels [7]. Thus, the localization of PLK1 on the kinetochores must be regulated in a timely manner to ensure a smooth transition of mitotic events. The molecular mechanism underlying the regulation of PLK1 both spatially and temporally remained unknown until recently.
PLK1 is regulated spatially and temporally during prometaphase/metaphase transition
The multifaceted role of PLK1 in mitosis is preceded by dynamic changes of its sub-cellular localization. Recent results suggest that a fine balance of PLK1 protein levels and its kinase activity is required for chromosome alignment and faithful chromosome segregation [7, 17, 19]. Initially, PLK1 localizes to centromeres in G1 [20] and G2 phases [21]. Later, a portion of PLK1 accumulates at the kinetochores during the prometaphase stage to promote the initial establishment of KT–MT attachments. Once the KT–MT attachments have been established, most PLK1 needs to be removed from the kinetochores in metaphase to allow for the stabilization of KT–MT interactions, SAC silencing, and anaphase onset. The spatial and temporal regulation of PLK1 from prometaphase to metaphase relies on a delicate molecular mechanism.
The initial association of PLK1 with kinetochores was thought to depend on its binding to PBD-interacting protein 1 (PBIP1), a kinetochore scaffold protein [21]. This binding appears to be self-regulated as PLK1 phosphorylation at Thr-78 of PBIP1 generates a binding motif for PBD. PBIP1 forms a stable complex with another kinetochore component, CENP-Q [22]. It is shown that the PBIP1-CENP-Q complex becomes hyperphosphorylated and rapidly delocalized from kinetochores as cells enter mitosis. PLK1 phosphorylates the CENP-Q subunit of the PBIP1-CENP-Q complex at nine sites to promote the dissociation of CENP-Q from chromatin and prevent the CENP-Q from localizing to interphase constitutive centromere-associated network (CCAN). Interestingly, both the 9 A and 9D/E mutants of CENP-Q induce a defect in proper chromosome segregation, suggesting that both timely localization of the PBIP1-CENP-Q complex to CCAN and delocalization from kinetochores are critical for normal mitosis progression. Although PLK1 did not alter the level of PBIP1 and CENP-Q ubiquitination, PLK1-dependent phosphorylation and dissociation of these proteins from kinetochores appeared to indirectly regulate their degradation in the cytosol.
After PBIP1 is degraded in early mitosis, some PLK1 is retained on the kinetochores and centromere through its binding to certain kinetochore-localized proteins, possibly including BubR1, and inner centromere protein (INCENP) [23], respectively. At kinetochores, PLK1 phosphorylates BubR1 [17, 24], and this phosphorylation allows the recruitment of a phosphatase, PP2A-B56, which counteracts the function of microtubule-destabilizing kinase Aurora B [24] and, therefore, stabilizes the initial KT–MT attachments. Testis expressed 14 (Tex14) was also identified as a kinetochore-localized protein that binds to PLK1 in a CDK1-dependent manner [25]. PLK1 phosphorylates Tex14 and recruits it to the kinetochores, and this recruitment appears to be essential for the formation of stable KT–MT attachments [25]. During metaphase, the PLK1-dependent phosphorylation of Tex14 promotes anaphase-promoting complex/cyclosome (APC/C)-mediated Tex14 degradation and metaphase-anaphase transition. Inhibition of this phosphorylation event causes retention of Tex14 at kinetochores and defects in chromosome segregation and delayed metaphase-anaphase transition. However, Tex14 is unlikely generally required for the formation of stable KT–MT attachments, because Tex14 is not ubiquitously expressed [26], and the defects in Tex14 knockout mice are restricted to germ cells [27]. Recently, chromatin remodeler RSF1 has been identified as an essential protein for the recruitment of PLK1 to kinetochores [28]. CDK1 phosphorylates kinetochore-localized RSF1 at S1375 and generates a PBD-binding motif. PLK1, in turn, further phosphorylates RSF1 and stabilizes the localization of PLK1 on kinetochores, and hence, promotes the initiation of KT–MT attachment.
Dynactin is a protein that forms a complex with dynein. During mitosis, they are required for spindle pole focusing, helping chromosomes engage with and move on spindle microtubules, and removing SAC proteins from kinetochores to facilitate the silence of SAC. Dynactin also helps recruiting PLK1 to kinetochores. CDK1 phosphorylates dynactin at Thr186 of its p27 subunit and generates a binding motif for PBD at kinetochores. Removal of p27 from dynactin results in reduced levels of PLK1 and its phosphorylated substrates at kinetochores in prometaphase, leading to aberrant KT–MT interactions, improper chromosome alignment, and abbreviated mitosis.
It has become very clear that PBD plays an important role in the localization of PLK1 to kinetochores. Recent studies have shown that deubiquitination and ubiquitination of residue(s) in PBD play a major role in regulating the recruitment of PLK1 to the kinetochores in prometaphase and the dissociation of PLK1 from kinetochores in metaphase, respectively.
PBD and its function
There are five PLK members (from PLK1 to PLK5) in humans, all share a closely related catalytic domain at the amino terminus and a characteristic sequence motif, the PBD, in the carboxy-terminal region [29]. The PBD of PLK1-3 is composed of two structurally similar polo box (PB) motifs, PB1 and PB2. The two PBs form a module binding to phospho-Ser/Thr motifs [30]. PLK4 only has one PB that exhibits a lower level of homology with PB1 or PB2. The PB of PLK4 homodimerizes to form a stable dimer [31, 32], and the dimerized PB binds to a target in a way that does not require a phosphorylated motif [33]. It is unclear whether the PBD of PLK5 also binds to a phospho-Ser/Thr motif at this stage.
PLK1 is the most evolutionarily conserved and the best-studied member of the PLKs [34]. The PBD of PLK1 serves as an essential molecular mediator that brings the kinase domain of PLK1 into proximity with its substrates, and targets PLK1 to specific sub-cellular locations. The binding motif on the substrates usually needs to be pre-phosphorylated [35], and the motif contains [Pro/Phe]-[Φ/Pro]-[Φ]-[Thr/Gln/His/Met]-Ser-[pThr/pSer]-[Pro/X], where Φ represents hydrophobic residues and X means any residues [30, 36]. The phosphorylated motif can be generated by PLK1 itself (self-priming), but in most of the cases, by a kinase other than PLK1, such as CDK1 and CaMKII [37, 38] (non-self-priming). As an example of self-priming, PLK1 phosphorylates the T78 residue of PBIP1 and binds to the resulting phosphorylated motif to recruit PLK1 itself to kinetochores [21], whereas in the case of non-self-priming, CaMK II phosphorylates Emi2 to generate a binding motif for PLK1 during the release of cytostatic factor-induced meiotic cell cycle arrest [37]. Regardless of the mode of priming, by binding to these phosphorylated motifs, PLK1 is targeted to various sub-cellular locations [33].
Usp16 is a novel substrate of PLK1
In an effort to identify novel PLK1 substrates, ubiquitin-specific peptidase 16 (Usp16) was identified as a PLK1 interacting protein in a co-immunoprecipitation (co-IP) assay in a PBD-dependent manner [15]. Usp16 is a deubiquitinase in the USP family, and is able to deubiquitinate mono-ubiquitinated histone H2A at K119 at the execution phase of apoptosis [39]. Its inactivation blocks progression in cell cycle [40]. Primary structure analysis shows that Usp16 contains a BUZ domain at its N-terminus and a catalytic domain at its C-terminus.
PLK1 is able to phosphorylate Usp16 at S330, S386, and S486 in vitro, and this phosphorylation has been verified by mass spectrometry analysis of peptides derived from endogenous Usp16 isolated from HeLa cells. Meanwhile, Usp16 is also a substrate of CDK1 that phosphorylates Usp16 at S552, which is within the PBD-interacting region. The phosphorylation of Usp16 by CDK1 creates a binding motif for PBD, and hence, promotes further phosphorylation of Usp16 by PLK1. Importantly, the phosphorylation by PLK1 activates Usp16, as the sequential phosphorylation of Usp16 by CDK1 and PLK1 significantly increases the deubiquitination activity of Usp16 in vitro, and decreases the ubiquitination level of histone H2A in mitotic cells [15].
Deubiquitination and cell cycle
Like phosphorylation, ubiquitination is a reversible process of protein modification, and the reverse process is catalyzed by a group of enzymes call deubiquitinase (DUB). There are five subfamilies of DUBs: (1) the ubiquitin C-terminal hydrolases (UCHs), (2) the ubiquitin-specific proteases/ubiquitin-specific processing proteases (USPs/UBPs), (3) the ovarian tumor proteases (OTUs), (4) the Josephin or Machado–Joseph disease protein domain proteases (MJDs), and (5) the Jab1/MPN domain-associated metalloisopeptidase (JAMM) domain proteins. So far, more than 100 DUBs have been identified in human genome [41]. Ubiquitin ligases and DUBs participate in cell cycle control at almost every level [42]. A main function of the DUBs is to maintain the intracellular ubiquitin level by cleaving ubiquitin off substrates before the tagged substrates being translocated into the proteasome, so the ubiquitin can be recycled for the next ubiquitination. Failure to do so will lead to depletion of intracellular ubiquitin and could cause delay in cell cycle progression [43, 44]. In addition, failure to cleave ubiquitin off the substrates that are already engaged with the proteasome and targeted for degradation would impair proteasome function and, consequently, cell cycle progression [45].
Interestingly, DUBs are often found in the same complex with ubiquitin ligase, such as Usp7 and Mdm2 [46–48]; and Brca1/Bard1 and two DUBs: UCH Bap1 and the JAMM-domain DUB Brcc36 [49, 50]. By regulating the stability or activity of the E3 ligases, DUBs control cell cycle. In the case of Usp7 and Mdm2, depletion of Usp7 results in premature degradation of Mdm2 mediated by Mdm2 itself, and causes the accumulation of p53, a target of Mdm2, leading to cell cycle arrest in G1 or G2 phase.
DUBs also control cell cycle by regulating the stability or activity of some key cell cycle regulators, including transcription factors and cyclins. For example, Usp28 protects c-Myc from SCFFbw7α-dependent ubiquitination and degradation and, therefore, promotes c-Myc-induced cell proliferation [51]. Cell cycle progression also depends on the accessibility of chromatin by transcription factors, and the accessibility is directly related to modification of histones. One of these modifications is the transient ubiquitination of histones. The progression through mitosis requires the deubiquitination of histone H2A, and suppression of Usp16 causes elevated level of histone H2A ubiquitination in mitosis and accumulation of mitotic cells [52]. DUBs also regulate cell cycle checkpoints. In mitosis, the SAC suppresses the activity of the APC/C to allow the attachment of microtubules to kinetochores. Once chromosomes are aligned properly, the APC/C is activated, which leads to the ubiquitination of Cdc20 and the dissociation of its inhibitor Mad2 [53]. Usp44, however, counteracts the activity of the APC/C by deubiquitinating Cdc20, thereby preventing premature Mad2-dissociation and SAC silencing [54]. Thus, Usp44 enhances the SAC by directly counteracting APC/C-dependent ubiquitination.
Usp16 deubiquitinates PLK1 and promotes the recruitment of PLK1 to the kinetochores
In the presence of mitotic stress, PLK1 is targeted by Chfr E3 ligase for degradation during G2-M transition, which results in a delay in CDK1 activation, representing a novel checkpoint pathway [55]. PLK1 is also ubiquitinated by cullin 3 (CUL3)-based E3 ubiquitin ligase in mitosis, but this ubiquitination does not lead to PLK1 proteolysis [15, 56]. Surprisingly, it was discovered that while Usp16 is a substrate of PLK1, it can, in turn, deubiquitinate PLK1. Importantly, the deubiquitination of PLK1 by Usp16 promotes the localization of PLK1 to the kinetochores and the proper alignment of chromosome on metaphase plate. It is thought that the localization of PLK1 on the kinetochores depends on its binding to kinetochore-localized proteins, and the deubiquitination of PLK1 most likely facilitates its binding to these proteins. BubR1 is a kinetochore-localized protein, though its interaction with PLK1 seems to not relate to the recruitment of PLK1 to kinetochores, it was found that deubiquitination of PLK1 enhanced it binding towards BubR1, suggesting an increased binding towards other kinetochore-localized PLK1 substrates. Knockdown of Usp16 results in a decrease of PLK1 level on the kinetochores and ~40% chromosomal misalignment [15]. Though the direct connection between the loss of PLK1 on the kinetochores and misalignment of chromosomes has not been firmly established, and the possibility that Usp16 deubiqutinates other substrate(s) on the kintetochores to regulate chromosome alignment in mitosis is not excluded, this result is consistent with the previous report that PLK1 is required for the initial establishment of KT–MT attachment [7].
CUL3-based E3 ligase ubiquitinates PLK1 and promotes the removal of PLK1 from the kinetochores
The E3 ubiquitin ligases can be divided into three major families on the basis of their assembly and mechanism of action: the HECT (homologous with E6-associated protein C-terminus) domain E3s, the RING finger E3s, and RBR (RING-between RING–RING) E3s. HECT E3s accept ubiquitin from E2 ~ ubiquitin to form a covalent thioester intermediate via a conserved cysteine residue of the E3 itself before transferring the ubiquitin on to the substrate. In contrast, RING E3s directly transfer ubiquitin to the substrate by bringing both E2 ~ ubiquitin and the substrate in close proximity to each other. The RBR ligases represent an additional family of E3s that combine characteristics of both HECT and RING families, as they recruit E2 ~ ubiquitin conjugates by an N-terminal RING domain and then transfer ubiquitin on to a HECT-type C-terminal catalytic cysteine residue of the E3 before final transfer on to the substrate [57].
Cullin-based E3 ligases are a group of RING finger E3s. Cullins are proteins that play a role in post-translational modification of proteins including ubiquitination. The cullin family is highly conserved among species [58]; seven different cullins have been identified in mammals (Cul1, 2, 3, 4A, 4B, 5 and 7) [59, 60]. Each cullin forms a distinct class of cullin-based E3 ligase complex consisting of different adapters and/or substrate recognition subunits [59, 61]. This diversity of functions is given by each of the adapters present in the complex.
Compared to other cullin-based complexes, the cullin complex 3 (CUL3) does not require different adapters to recognize its target protein, but only requires a protein with a bric-a-brac/tramtrack/broad-complex (BTB) domain to recognize it. BTB domains were originally found in transcription factors of Drosophila melanogaster, but it is now known that all eukaryotic species express a variety of BTB domain proteins [62]. In recent years, the complex based on CUL3 has been implicated in processes, such as the cell cycle regulation.
CUL3 overexpression promotes the ubiquitination of Aurora-A both in vivo and in vitro. Thus, CUL3 is able to regulate the entrance to mitosis in an Aurora-A-dependent manner by interacting with KLHL18 protein, therefore, mediating the activation of Aurora-A in centrosome. PLK1 has been proven to be a target for CUL3-based E3 ligase complex, and it is also recognized by a BTB protein called KLHL22 that functions as an adapter for the ligase. In the absence of KLHL22, PLK1 is accumulated at kinetochores, promoting the activation of the SAC to ensure the KT–MT attachment and proper chromosome alignment [56, 63].
CUL3/KLHL22 was reported to directly bind PLK1 and ubiquitinates it at Lys 492 and Lys 19. While the function of Lys 19 ubiquitination is unclear, the ubiquitination of Lys 492 located within the PBD leads to the dissociation of PLK1 from kinetochore-localized PBD-interacting proteins [56]. However, a recent report shows that the ubiquitination of Lys 492 may not be so crucial for cell cycle progression [64]. Though the difference in result could be attributed to different cell lines used in the experiments, the exact function of Lys 492 ubiquitination remains to be further investigated. Thus, it is possible that ubiquitination of PLK1 at other site(s) is also important for the dissociation of PLK1 from the kinetochores. KLHL22 associates with the mitotic spindle and its interaction with PLK1 increases as cells achieve proper chromosome alignment. Together, this data suggest that CUL3/KLHL22-mediated ubiquitination signals that are degradation-independent remove PLK1 from kinetochores to satisfy SAC, a process required for faithful progression through mitosis.
The ubiquitination status of PLK1 regulates its kinetochore localization, and hence, the proper chromosome alignment and timely sister chromatid segregation
CUL3-based E3 ligase and Usp16 seem to have antagonized functions on the kinetochore localization of PLK1, as Usp16 knockdown results in the reduction of kinetochore-localized PLK1 level, and KLHL22 knockdown leads to increased level of kinetochore-localized PLK1. Since Lys 492 is located within PBD, it is, therefore, possible that the equilibrium of Usp16-mediated deubiquitination and Cul3-based E3 ligase-mediated ubiquitination at this site regulates the interaction between PBD and its kinetochore-localized binding partners, and, hence, the localization of PLK1 on kinetochores. As the localization of PLK1 on the kinetochores in early mitosis is important for the establishment of the initial KT–MT attachments and proper chromosome alignment, and the removal of PLK1 from the kinetochores at metaphase is required for sister chromatid segregation, the deubiquitination/ubiquitination equilibrium, therefore, appears to be a key mechanism regulating proper chromosome alignment and timely sister chromatid segregation.
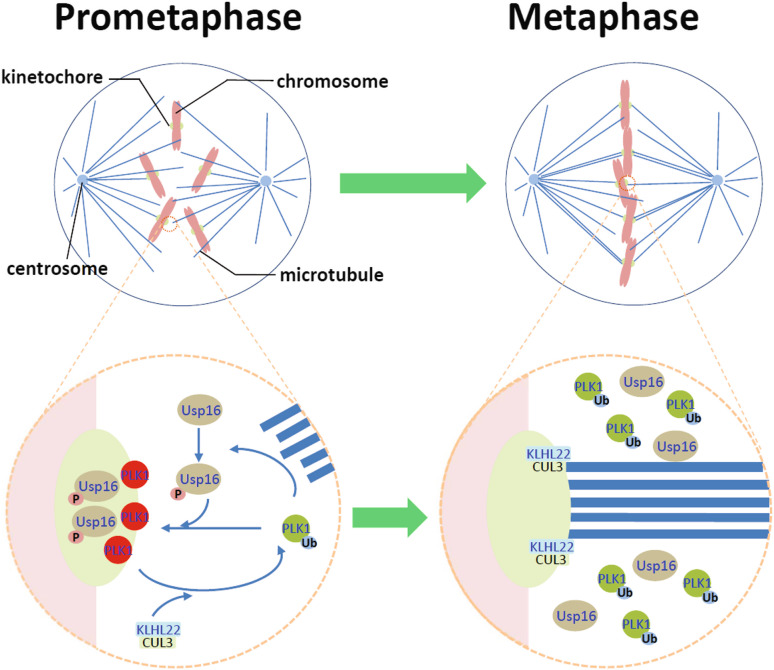
Studies from Zhou et al. and Beck et al. have deciphered the molecular mechanism underlying the regulation of this deubiquitination/ubiquitination equilibrium. As illustrated in the model (Fig. 1), in early mitosis when PLK1 activity increases, Usp 16, mainly in cytosol, is phosphorylated and activated, which, in turn, deubiquitinates a portion of PLK1 and promotes the binding of PLK1 to kinetochore-localized proteins. Though the identity of these kinetochore-localized PLK1 binding partners has not be revealed, Usp16 and BubR1 are two probable candidates. The deubiquitination of PLK1 by Usp16 not only promotes the recruitment of PLK1 to kinetochores, but also retains PLK1 on the kinetochores to ensure the establishment of KT–MT attachment. Once chromosomes are properly aligned and SAC is satisfied, this portion of PLK1 is then removed from the kinetochores to allow the timely segregation of sister chromatids. This is achieved by CUL3-based E3 ligase that ubiquitinates PLK1 at Lys 492 and possibly other sites as well, which is likely to disrupt the interaction between PBD and its binding partners, resulting in the dissociation of PLK1 from the kinetochores. How exactly the CUL3-based E3 ligase is recruited to the kinetochores at metaphase is unclear at moment, but it was shown that the association of KLHL22 with the mitotic spindle and kinetochore-localized PLK1 increases as cells achieve chromosome bi-orientation [56], suggesting that KLHL22 may be involved in the recruitment. It was reported recently that a portion of mitotic PLK1 localizes close to the inner kinetochore, internal to the histone variant CENPA [65], and also binds to inner centromere-localized proteins INCENP and MCAK [23, 66]. As a portion of PLK1 remains at the kinetochores during anaphase [20], it is possible that this portion of inner kinetochore and/or inner centromere-localized PLK1 may not subject to ubiquitination mediated by CUL3-based E3 liagase.
Fig. 1.
Equilibrium of ubiquitination and deubiquitination at PLK1 regulates its localization on the kinetochores and KT–MT attachment. In prometaphase, activated PLK1 phosphorylates and activates Usp16, which, in turn, deubiquitinates PLK1, leading to the localization of PLK1 to the kinetochores. The kinetochore-localized PLK1 promotes the establishment of initial KT–MT attachment and proper chromosome alignment. Once the SAC is satisfied at metaphase, CUL3-based E3 ligase is enriched on the kinetochores and ubiquitinates PLK1, resulting in the dissociation of PLK1 from the kinetochores. The dissociation of PLK1 stabilizes the KT–MT attachment and allows the segregation of sister chromatids. As detailed in the text, it should be noted that a portion of PLK1 localized at inner kinethochore and centromere may not subject to this regulation
Concluding remarks
Regulating proper chromosome alignment and timely sister chromatid segregation are two important functions of PLK1. It requires PLK1 being recruited to and removed from kinetochores at particular stages of mitosis. It is now evident that the recruitment and retention of PLK1 on kinetochores is regulated by Usp16-mediated deubiquitination, and the removal of PLK1 from kinetochores is promoted by CUL3-based E3 ligase that ubiquitinates PLK1 at Lys 492 located in PBD. Usp16 is phosphorylated and activated by PLK1 in early mitosis, but this phosphorylation does not regulate the localization of Usp16 on kinetochores. When and how Usp16 is recruited to and removed from kinetochores remain to be investigated. On the other hand, CUL3-based E3 ligase seems to be present throughout the cell cycle, but only becomes enriched at kinetochores when the chromosomes are properly aligned. What is the mechanism that senses the timing and recruits the E3 ligase complex to kinetochores? These are some questions need to be addressed to completely understand the spatial and temporal regulation of PLK1, and hence, the mechanism underlying the regulation of chromosome alignment and sister chromatid segregation.
Acknowledgements
We thank other members of our laboratories for helpful comments and critical discussion on this work. Work in Liu laboratory is supported by a NIH Grant (5SC2GM089622-03), and in Zhang laboratory by the National Natural Science Foundation of China (NSFC) (31520103906, 31371365, and 31430051) and the Ministry of Science and Technology of China (2016YFA0100501 and 2016YFA0500201).
Abbreviations
- PLK1
Polo-like kinase 1
- Usp16
Ubiquitin-specific peptidase 16
- CUL3
Cullin 3
- SAC
Spindle assembly checkpoint
- KT–MT
Kinetochore–microtubule
- PB
Polo box
- PBD
Polo box domain
- PBIP1
Polo box domain-interacting protein 1
- INCENP
Inner centromere protein
- APC/C
Anaphase-promoting complex/cyclosome
- CCAN
Constitutive centromere-associated network
Contributor Information
Junjun Liu, Email: junjunliu@cpp.edu.
Chuanmao Zhang, Email: zhangcm@pku.edu.cm.
References
- 1.Glover DM, Hagan IM, Tavares AA. Polo-like kinases: a team that plays throughout mitosis. Genes Dev. 1998;12:3777–3787. doi: 10.1101/gad.12.24.3777. [DOI] [PubMed] [Google Scholar]
- 2.Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 3.Sumara I, Gimenez-Abian JF, Gerlich D, Hirota T, Kraft C, de la Torre C, Ellenberg J, Peters JM. Roles of polo-like kinase 1 in the assembly of functional mitotic spindles. Curr Biol. 2004;14:1712–1722. doi: 10.1016/j.cub.2004.09.049. [DOI] [PubMed] [Google Scholar]
- 4.Lenart P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters JM. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 5.DeLuca KF, Lens SM, DeLuca JG. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J Cell Sci. 2011;124:622–634. doi: 10.1242/jcs.072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foley EA, Maldonado M, Kapoor TM. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat Cell Biol. 2011;13:1265–1271. doi: 10.1038/ncb2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu D, Davydenko O, Lampson MA. Polo-like kinase-1 regulates kinetochore-microtubule dynamics and spindle checkpoint silencing. J Cell Biol. 2012;198:491–499. doi: 10.1083/jcb.201205090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maiato H, DeLuca J, Salmon ED, Earnshaw WC. The dynamic kinetochore-microtubule interface. J Cell Sci. 2004;117:5461–5477. doi: 10.1242/jcs.01536. [DOI] [PubMed] [Google Scholar]
- 9.Chan GK, Liu ST, Yen TJ. Kinetochore structure and function. Trends Cell Biol. 2005;15:589–598. doi: 10.1016/j.tcb.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 10.Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uchida KS, Takagaki K, Kumada K, Hirayama Y, Noda T, Hirota T. Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol. 2009;184:383–390. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khodjakov A, Pines J. Centromere tension: a divisive issue. Nat Cell Biol. 2010;12:919–923. doi: 10.1038/ncb1010-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol CB. 2009;19:1937–1942. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhuo X, Guo X, Zhang X, Jing G, Wang Y, Chen Q, Jiang Q, Liu J, Zhang C. Usp16 regulates kinetochore localization of Plk1 to promote proper chromosome alignment in mitosis. J Cell Biol. 2015;210:727–735. doi: 10.1083/jcb.201502044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 17.Elowe S, Hummer S, Uldschmid A, Li X, Nigg EA. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 2007;21:2205–2219. doi: 10.1101/gad.436007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu D, Vleugel M, Backer CB, Hori T, Fukagawa T, Cheeseman IM, Lampson MA. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol. 2010;188:809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maia AR, Garcia Z, Kabeche L, Barisic M, Maffini S, Macedo-Ribeiro S, Cheeseman IM, Compton DA, Kaverina I, Maiato H. Cdk1 and Plk1 mediate a CLASP2 phospho-switch that stabilizes kinetochore-microtubule attachments. J Cell Biol. 2012;199:285–301. doi: 10.1083/jcb.201203091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKinley KL, Cheeseman IM. Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell. 2014;158:397–411. doi: 10.1016/j.cell.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang YH, Park JE, Yu LR, Soung NK, Yun SM, Bang JK, Seong YS, Yu H, Garfield S, Veenstra TD, et al. Self-regulated Plk1 recruitment to kinetochores by the Plk1-PBIP1 interaction is critical for proper chromosome segregation. Mol Cell. 2006;24:409–422. doi: 10.1016/j.molcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 22.Park CH, Park JE, Kim TS, Kang YH, Soung NK, Zhou M, Kim NH, Bang JK, Lee KS. Mammalian Polo-like kinase 1 (Plk1) promotes proper chromosome segregation by phosphorylating and delocalizing the PBIP1.CENP-Q complex from kinetochores. J Biol Chem. 2015;290:8569–8581. doi: 10.1074/jbc.M114.623546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goto H, Kiyono T, Tomono Y, Kawajiri A, Urano T, Furukawa K, Nigg EA, Inagaki M. Complex formation of Plk1 and INCENP required for metaphase-anaphase transition. Nat Cell Biol. 2006;8:180–187. doi: 10.1038/ncb1350. [DOI] [PubMed] [Google Scholar]
- 24.Suijkerbuijk SJ, Vleugel M, Teixeira A, Kops GJ. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev Cell. 2012;23:745–755. doi: 10.1016/j.devcel.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Mondal G, Ohashi A, Yang L, Rowley M, Couch FJ. Tex14, a Plk1-regulated protein, is required for kinetochore-microtubule attachment and regulation of the spindle assembly checkpoint. Mol Cell. 2012;45:680–695. doi: 10.1016/j.molcel.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu MH, Rajkovic A, Burns KH, Yan W, Lin YN, Matzuk MM (2003)Sequence and expression of testis-expressed gene 14 (Tex14): a gene encoding a protein kinase preferentially expressed during spermatogenesis. GEP 3:231–236 [DOI] [PubMed]
- 27.Greenbaum MP, Iwamori N, Agno JE, Matzuk MM. Mouse TEX14 is required for embryonic germ cell intercellular bridges but not female fertility. Biol Reprod. 2009;80:449–457. doi: 10.1095/biolreprod.108.070649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee HS, Park YY, Cho MY, Chae S, Yoo YS, Kwon MH, Lee CW, Cho H. The chromatin remodeller RSF1 is essential for PLK1 deposition and function at mitotic kinetochores. Nat Commun. 2015;6:7904. doi: 10.1038/ncomms8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nigg EA. Polo-like kinases: positive regulators of cell division from start to finish. Curr Opin Cell Biol. 1998;10:776–783. doi: 10.1016/S0955-0674(98)80121-X. [DOI] [PubMed] [Google Scholar]
- 30.Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115:83–95. doi: 10.1016/S0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 31.Leung GC, Hudson JW, Kozarova A, Davidson A, Dennis JW, Sicheri F. The Sak polo-box comprises a structural domain sufficient for mitotic subcellular localization. Nat Struct Biol. 2002;9:719–724. doi: 10.1038/nsb848. [DOI] [PubMed] [Google Scholar]
- 32.Slevin LK, Nye J, Pinkerton DC, Buster DW, Rogers GC, Slep KC. The structure of the plk4 cryptic polo box reveals two tandem polo boxes required for centriole duplication. Structure. 2012;20:1905–1917. doi: 10.1016/j.str.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee KS, Park JE, Kang YH, Kim TS, Bang JK. Mechanisms underlying Plk1 polo-box domain-mediated biological processes and their physiological significance. Mol Cells. 2014;37:286–294. doi: 10.14348/molcells.2014.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Carcer G, Manning G, Malumbres M. From Plk1 to Plk5: functional evolution of polo-like kinases. Cell Cycle. 2011;10:2255–2262. doi: 10.4161/cc.10.14.16494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park JE, Soung NK, Johmura Y, Kang YH, Liao C, Lee KH, Park CH, Nicklaus MC, Lee KS. Polo-box domain: a versatile mediator of polo-like kinase function. CMLS. 2010;67:1957–1970. doi: 10.1007/s00018-010-0279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 37.Liu J, Maller JL. Calcium elevation at fertilization coordinates phosphorylation of XErp1/Emi2 by Plx1 and CaMK II to release metaphase arrest by cytostatic factor. Curr Biol. 2005;15:1458–1468. doi: 10.1016/j.cub.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, Chen Q, Feng J, Hou J, Yang F, Liu J, Jiang Q, Zhang C. Sequential phosphorylation of Nedd1 by Cdk1 and Plk1 is required for targeting of the gammaTuRC to the centrosome. J Cell Sci. 2009;122:2240–2251. doi: 10.1242/jcs.042747. [DOI] [PubMed] [Google Scholar]
- 39.Mimnaugh EG, Kayastha G, McGovern NB, Hwang SG, Marcu MG, Trepel J, Cai SY, Marchesi VT, Neckers L. Caspase-dependent deubiquitination of monoubiquitinated nucleosomal histone H2A induced by diverse apoptogenic stimuli. Cell Death Differ. 2001;8:1182–1196. doi: 10.1038/sj.cdd.4400924. [DOI] [PubMed] [Google Scholar]
- 40.Cai SY, Babbitt RW, Marchesi VT. A mutant deubiquitinating enzyme (Ubp-M) associates with mitotic chromosomes and blocks cell division. Proc Natl Acad Sci USA. 1999;96:2828–2833. doi: 10.1073/pnas.96.6.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Song L, Rape M. Reverse the curse–the role of deubiquitination in cell cycle control. Curr Opin Cell Biol. 2008;20:156–163. doi: 10.1016/j.ceb.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanna J, Leggett DS, Finley D. Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol Cell Biol. 2003;23:9251–9261. doi: 10.1128/MCB.23.24.9251-9261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson SM, Bhattacharyya B, Rachel RA, Coppola V, Tessarollo L, Householder DB, Fletcher CF, Miller RJ, Copeland NG, Jenkins NA. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat Genet. 2002;32:420–425. doi: 10.1038/ng1006. [DOI] [PubMed] [Google Scholar]
- 45.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 46.Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 48.Liu X, Zhou T, Kuriyama R, Erikson RL. Molecular interactions of Polo-like-kinase 1 with the mitotic kinesin-like protein CHO1/MKLP-1. J Cell Sci. 2004;117:3233–3246. doi: 10.1242/jcs.01173. [DOI] [PubMed] [Google Scholar]
- 49.Jensen DE, Rauscher FJ., 3rd BAP1, a candidate tumor suppressor protein that interacts with BRCA1. Ann N Y Acad Sci. 1999;886:191–194. doi: 10.1111/j.1749-6632.1999.tb09414.x. [DOI] [PubMed] [Google Scholar]
- 50.Dong Y, Hakimi MA, Chen X, Kumaraswamy E, Cooch NS, Godwin AK, Shiekhattar R. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol Cell. 2003;12:1087–1099. doi: 10.1016/S1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- 51.Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, Moll R, Elledge SJ, Eilers M. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9:765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 52.Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, Chang C, Wang H. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- 53.Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- 54.Stegmeier F, Rape M, Draviam VM, Nalepa G, Sowa ME, Ang XL, McDonald ER, 3rd, Li MZ, Hannon GJ, Sorger PK et al (2007) Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature 446:876–881 [DOI] [PubMed]
- 55.Kang D, Chen J, Wong J, Fang G. The checkpoint protein Chfr is a ligase that ubiquitinates Plk1 and inhibits Cdc2 at the G2 to M transition. J Cell Biol. 2002;156:249–259. doi: 10.1083/jcb.200108016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beck J, Maerki S, Posch M, Metzger T, Persaud A, Scheel H, Hofmann K, Rotin D, Pedrioli P, Swedlow JR, et al. Ubiquitylation-dependent localization of PLK1 in mitosis. Nat Cell Biol. 2013;15:430–439. doi: 10.1038/ncb2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bulatov E, Ciulli A. Targeting Cullin-RING E3 ubiquitin ligases for drug discovery: structure, assembly and small-molecule modulation. Biochem J. 2015;467:365–386. doi: 10.1042/BJ20141450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12:220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 60.Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marin I. Diversification of the cullin family. BMC Evol Biol. 2009;9:267. doi: 10.1186/1471-2148-9-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maerki S, Olma MH, Staubli T, Steigemann P, Gerlich DW, Quadroni M, Sumara I, Peter M. The Cul3-KLHL21 E3 ubiquitin ligase targets aurora B to midzone microtubules in anaphase and is required for cytokinesis. J Cell Biol. 2009;187:791–800. doi: 10.1083/jcb.200906117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lasek AL, McPherson BM, Trueman NG, Burkard ME. The functional significance of posttranslational modifications on polo-like kinase 1 revealed by chemical genetic complementation. PLoS One. 2016;11:e0150225. doi: 10.1371/journal.pone.0150225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lera RF, Potts GK, Suzuki A, Johnson JM, Salmon ED, Coon JJ, Burkard ME. Decoding Polo-like kinase 1 signaling along the kinetochore-centromere axis. Nat Chem Biol. 2016;12:411–418. doi: 10.1038/nchembio.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang L, Shao H, Huang Y, Yan F, Chu Y, Hou H, Zhu M, Fu C, Aikhionbare F, Fang G, et al. PLK1 phosphorylates mitotic centromere-associated kinesin and promotes its depolymerase activity. J Biol Chem. 2011;286:3033–3046. doi: 10.1074/jbc.M110.165340. [DOI] [PMC free article] [PubMed] [Google Scholar]