Abstract
Research of the past two decades has proved the relevance of single cell biology in basic research and translational medicine. Successful detection and isolation of specific subsets is the key to understand their functional heterogeneity. Antibodies are conventionally used for this purpose, but their relevance in certain contexts is limited. In this review, we discuss some of these contexts, posing bottle neck for different fields of biology including biomedical research. With the advancement of chemistry, several methods have been introduced to overcome these problems. Even though microfluidics and microraft array are newer techniques exploited for single cell biology, fluorescence-activated cell sorting (FACS) remains the gold standard technique for isolation of cells for many biomedical applications, like stem cell therapy. Here, we present a comprehensive and comparative account of some of the probes that are useful in FACS. Further, we illustrate how these techniques could be applied in biomedical research. It is postulated that intracellular molecular markers like nucleostemin (GNL3), alkaline phosphatase (ALPL) and HIRA can be used for improving the outcome of cardiac as well as bone regeneration. Another field that could utilize intracellular markers is diagnostics, and we propose the use of specific peptide nucleic acid probes (PNPs) against certain miRNAs for cancer surgical margin prediction. The newer techniques for single cell biology, based on intracellular molecules, will immensely enhance the repertoire of possible markers for the isolation of cell types useful in biomedical research.
Keywords: Aptamer, Molecular beacon (MB), Targeting cell-penetrating peptide (targeting CPP), Cardiac regeneration, miR-21, miR-143/145
Introduction
Contrary to our earlier view that all the cells grown in a Petri dish are alike, latest evidences prove the existence of cellular heterogeneity in cell populations in organs and in vitro culture systems. For example, in Drosophila ovary, a few of the Notch ligand expressing cap cells are the determinants of the location and number of germinal stem cells [1]. Functional heterogeneity in morphologically similar cells is observed in stem cell pool, where the committed cells, which are morphologically similar, acquire different fate. Likewise, in the case of tumorigenesis or immunological responses to pathogens, a few cells may drive the overall processes, which are indistinguishable from bulk population. Thus, many of our output measurements based on cell population are misleading averages, highlighting the need of single cell biology. Identification of subpopulations that differ in size, protein/RNA content, and exhibit functional heterogeneity are the key to answer unsolved questions in stem cell biology, developmental biology and cancer research.
To decipher the differential response of heterogeneous population, we have to enhance the resolution of the underlying biology through single cell analysis. The different high throughput-analyses, like Single cell-omics, allow us to gain insight into the unique processes occurring on multiple functional levels of the single cell. Several attempts to understand the microenvironment and cell–cell interactions have unraveled the basis of functional heterogeneity in several contexts [2]. The basic requirement for single cell biology is the isolation of single cells of interest using different techniques like microfluidics, microraft array (MRA) or fluorescence-activated cell sorting (FACS) (Table 1).
Table 1.
Methods employed in single cell biology
| Technique | Instrumentation for detection | Method of detection | Applications | Biomedical applications |
|---|---|---|---|---|
|
Microfluidics (a) Droplet-based Microfluidics (b) Digital microfluidics (c) Quantum dots |
Special microfluidic device coupled with optical device (a) Dielectrophoresis (DEP) (b) Fluorescence-activated droplet sorting (FADS) |
Based on the property of encapsulated cells Using probes (antibodies) for secreted molecules |
High throughput screening Proteomic/transcriptomic/genomic analyses of single cells |
Biomarker detection Drug screening Disease diagnosis |
| Microrafts | Microwell plates with magnetic nanoparticles | Conventional microscopy/flowcytometry |
Functional studies of single cells Proteomic/transcriptomic/genomic analyses of single cells |
|
| Fluorescence-activated cell sorting (FACS) | Fluorescence activated-cell sorter (FACS machine) |
Using probes (a) Antibodies (b) Aptamers (c) Molecular beacons (d) Peptide nucleic acids (e) Targeting CPPs |
Detection of heterogenic populations Characterization of single cells Functional studies of single cells |
Diagnosis Screening Stem cell therapy |
The different techniques, their features, and applications are summarized in the table
Microfluidics is a multidisciplinary field with applications to design systems in which low volumes of fluids are processed to achieve high throughput screening. Droplet-based microfluidics utilizes droplets of very small volume, which function as bioreactors for cells, and these cells can be analyzed or sorted based on their properties, like secreted molecules [3]. Recently, digital microfluidics, where the manipulation of droplets is by electric forces, has received attention for single cell analysis and screening [4]. Another advancing field is Quantum dots, which makes use of various biofunctionalized nanoparticles, is used for cancer biomarker detection [5]. Microrafts are arrays of microwells loaded with raft molds containing magnetic nanoparticles, where cells are dispersed in 1 cell/well dilution. This device provides a method to sort, isolate, analyze, and/or generate clonal populations. This technique is a powerful tool to isolate clones of cells based on their biological function [6, 7]. FACS is the conventional method for detection and purification of cells based on fluorescence. In many instances, the functional heterogeneity can be marked by differential expression of certain proteins, or markers. Detection probes for those markers tagged to fluorescent molecules can be used to isolate cells using FACS. Currently, the use of microfluidics is mainly for single cell analysis, while microrafts can be used for both analysis and enrichment of a population of interest. For many biomedical applications including stem cell therapy and regenerative medicine, number of cells required is more than what is grown in a microraft, and hence the cells are sorted using FACS.
The conventional probe used for the identification of single cell in FACS is antibody. Thus, single cell enrichment of live cells is limited to populations that have distinct surface markers, since antibody requires cell permeabilization to detect internal molecules. Studies using engineered reporter cells have shown that many intracellular molecules can function as potential markers for pleuripotent cells, committed cells for a lineage, cancer stem cells, metastatic malignant cells, etc. [8–10]. A proper methodology to isolate and trace these cells will revolutionize regenerative and translational medicine. Now chemistry has advanced, and integration of chemistry and bioinformatics to molecular biology has resulted in many probes that can identify and purify single cells based on intracellular molecules from heterogeneous populations, which can be used for FACS. Here, we review the relevance of intracellular markers in biomedical research, and the methodologies available for the isolation and purification of single cells based on them.
Relevance of intracellular markers in single cell biology
Single cell biology relevant to biomedical research focuses on the cell types involved in stem cell therapy, regenerative medicine, and diagnostics of various diseases including cancer. Currently these cell types are identified and isolated mainly based on surface marker profiles. Even though there are other more useful markers, they are not exploited due to technical limitation. Some of the intracellular markers that can be relevant in various fields of biomedical research are discussed in the present review (Table 2).
Table 2.
Examples of intracellular molecules relevant in biomedical research
| Markers | For identification of |
|---|---|
| HSC 70 and cytoplasmic Cyclin D1) | G0 phase of cell cycle |
| CDK4/6 | G1 phase of cell cycle |
| ALDH | Self-renewing cells |
| SOX2, OCT4, NANOG | Self-renewing cells |
| Unchanged HIRA and ASf1a | Myoblast committed to myogenic differentiation |
| Decreased HIRA and ASf1a | Myoblast committed to osteogenic differentiation |
| ID1 | Metastatic marker |
| Increased miR-21 | Cancer cells/cancer-associated stroma |
| Decreased miR-143/145 | Cancer cells |
| Rab7/LAMP1 | Formation of phagolysosome |
Stem cells and regenerative medicine
In regenerative medicine, the success of the protocol depends on the purity of cells used for therapy. Many protocols tend to isolate pleuripotent cells, and differentiate them to specific cell types using defined cytokine cocktail. Another fact which should be noted is when a pleuripotent cell differentiates; it can go to different cell fates. A committed cell to a particular cell fate will not respond to cues for differentiation to another cell type. So, for the success of therapy, the source of cells used should be either pleuripotent or a committed cell to the desired cell fate. One of the major challenges for this approach is identifying pleuripotent cells with long-term self-renewal capacity. Majority of the stem cell marker cocktails currently used fail to distinguish pleuripotent cells from committed cells. Recently, it has been shown that cell fate decisions are closely related to cell-cycle stages. So identification of cells in the correct cell-cycle stage for the required cell type will be beneficial to get maximum output in cytokine mediated differentiation. Taken together, identification of quiescent pleuripotent cells and cells committed for the desired cell type is the key to obtain maximum clinical output in stem cell therapy.
Even though several markers are reported for stem cells of different origin, quiescence (arrest in G0 phase or exit from cell cycle) is the common feature for all long-term self-renewing pleuripotent cells [11]. It has been shown that very high expression of heat shock cognate protein 70 (HSC70), and its cytoplasmic localization with Cyclin D1 marks quiescent stem cells [12]. One of the most commonly used markers of self-renewing cells is the cytosolic enzyme aldehyde dehydrogenase (ALDH), and it is identified to be a potential candidate for isolating long-term repopulating cells in clinical application [13]. SOX2, OCT-4, and NANOG are transcription factors that regulate self-renewal property in many cell types, and can be used as markers for self-renewing cells [14].
When pleuripotent quiescent cells differentiate, they enter cell cycle. Cells in early G1 phase preferably go to the fate of endoderm or mesoderm, and late G1 cells differentiate to neuroectoderm, while cells in the G2/S/M state do not respond to differentiation signals [9]. The cell-cycle marker CDK4 or CDK6 could be used to pick up cells in the G1 phase. Cell differentiation is a process by which a less-specialized cell undergoes reprogramming to acquire specialized functions, which involves epigenetic reprogramming, and requires factors like histones and histone chaperones. A pleuripotent mesenchymal stem cell has the potential to differentiate to osteoblasts, chondrocytes, adipocyte or myoblasts. Depending on the growth factor profile, C2C12 myoblast cell line differentiates either to myotubes or to osteoblasts. It has been reported that histones and histone chaperones (like CAF1, Asf1, and HIRA) are important factors regulating skeletal muscle formation [15]. The expression of HIRA and Asf1a is unchanged during myogenic differentiation, while their expression considerably decreases during osteogenic conversion [16]. This information could be used to identify committed cell types to muscle cell or bone cell during differentiation, provided we have a means to isolate cells based on the expression of these nuclear chaperones.
Cancer biology
Cancer consists of heterogeneous population varying in their tumor initiation ability, invasive property or metastatic ability. A number of molecules are identified to be regulating these properties and considered to be markers for cancer stem cells, chemoresistant cells or metastatic cells. In such cases, validation of the markers for the respective function depends on fractionation of cells based on these markers. Majority of these are done in cell lines modified with reporters. An actual validation of these markers in primary cells from clinical samples is not possible for many intracellular markers. Though research in the past decade has proven the importance of cancer stem cells in the progression and treatment response of different forms of cancer, identification of cancer stem cell is still a hurdle due to lack of reliable surface markers. But consistently in all cancer types, these self-renewing cancer cells express high levels of stemness-associated molecules, like SOX2, OCT4, NANOG, ALDH, etc., which show intracellular expression [14, 17–19]. ID1 expression is one of the parameters that dictate metastasis of breast cancer cells to lung [20]. Validation of metastatic potential of a subpopulation of tumor cells over-expressing ID1 in a primary tumor sample is not possible due to lack of a method to isolate live cells based on ID1 expression.
MicroRNAs (miRNAs) as biomarkers
miRNAs are short, single-stranded, non-coding sequences that regulate gene expression. These molecules are identified to regulate many cellular functions, and deregulation of many of the miRNAs has been shown to associate with several diseases. Many of these miRNAs are circulated in the body, and they are identified to be markers of several diseases including diabetes, heart diseases, and cancer [21–26]. Apart from the circulating miRNAs, their tissue level expressions are shown to mark several forms of cancer. The loss of expression of Let-7 is shown to be a marker of less differentiated, advanced cancer in ovarian carcinoma tissues [27]. The decreased expressions of miR-143 and miR-145 mark the initiation of tumorigenesis, while their loss marks malignant cancer in colorectal carcinoma [28]. One of the miRNAs reported to be over-expressing in different forms of cancer is miR-21 [29]. Interestingly, its presence in tumor stroma of oral cancer and triple negative breast cancer marks high-risk group for relapse [30, 31], or biochemical failure in breast cancer and prostate cancer, respectively [32]. This observation suggests miR-21 as an attractive molecule to mark surgical margins, which is a decisive factor in disease-free survival. But the usual method of miRNA detection by RT-PCR or in situ hybridization is not applicable in live tissues. A rapid and sensitive method to detect miRNA that work in live tissue will be suitable for predicting accurate surgical margins during surgery.
Host–pathogen interaction in infection
Phagosome–lysosome fusion in the macrophage is considered to be an important step in the clearance of many pathogens. Some viruses like influenza and HIV, and a spectrum of other microbes including mycobacterium alter this phagosome maturation pathway to evade host immune machinery [33, 34]. Consistent with that, phagosome maturation pathway offers a suitable target for host-directed therapeutics for diseases like tuberculosis [35]. For many of the drugs used for the treatment of tuberculosis, exact method of action is still unknown. To confirm whether the drug is targeting the phagosome maturation pathway, and whether a patient is responding to the drug, the fusion of phagosome and lysosome in the macrophages should be confirmed which can be marked by co-localization of Rab 7 and LAMP1.
Taken together, currently there are many areas where identification and characterization of cells at a single cell level is not possible due to limitations of methods for their purification. So far, we have discussed some intracellular molecules that can mark important subpopulations relevant in regenerative medicine, diagnostics, and therapy. Currently, their clinical use is hampered by lack of techniques for live detection of these intracellular molecules. In this scenario, we describe some of the methods introduced to biology for single cell isolation, which are summarized in Table 3.
Table 3.
Comparative account of probes other than antibodies for single cell identification
| Method | Target | Type of probe | Applications | Advantages | Limitations |
|---|---|---|---|---|---|
| Molecular beacons (MBs) | DNA/RNA | DNA/RNA | Pathogen detection | Sensitive, high signal to background ratio | It needs a reagent to introduce MBs to living cell |
| Stem cell isolation | Endonucleases cleave MBs, limiting live cell application | ||||
| Cancer detection | Affected by temperature and pH shift | ||||
| Aptamers | Inorganic molecules | DNA/RNA | Diagnostics | Wide range of targets | SELEX protocol is time consuming and laborious |
|
Toxins Metabolites |
Therapy | High affinity binding to targets | Probes selected in vitro condition do not work well under physiological condition | ||
| Nucleic acids | Modifications possible | Relatively unstable in biological fluids | |||
| Carbohydrates | Synthesis is cheaper and reproducible | SELEX protocol varies with target | |||
| Amino acids | No immunogenicity | ||||
| Peptides | Aptamers may not require reagents to introduce to living cells | ||||
| Proteins | |||||
| Complex biological structures | |||||
| Peptide nucleic acid probes (PNPs) | Nucleic acids miRNA | PNA | Diagnostics | High specificity | Cannot cross cell membranes, limiting live cell application |
| Therapy | Not detected by nucleases or proteases | ||||
| Stable over a wide pH range | |||||
| Targeting cell-penetrating peptides (targeting CPPs) | Proteins | Peptides | Translational medicine | Easy to synthesis | Degradation by proteases |
| Therapy | Easy to modify | Limited to targets, whose interacting partners and interacting regions are known | |||
| High affinity binding to targets | |||||
| Less immunogenicity |
Methods of single cell identification
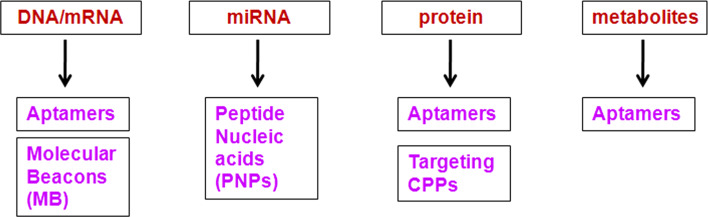
Depending on the context, several intracellular molecules, like DNA, RNA, miRNA, proteins and metabolites, and other complex biological structures act as targets for identification. The different probes, like, Aptamers, Molecular beacons (MB), Peptide Nucleic acid Probes (PNPs), and targeting CPPs, that can be applied for the identification of these molecules are summarized in Fig. 1. Figure 2 depicts the methods for single cell identification using FACS utilizing these probes. Each of these approaches has advantages and limitations as briefed in the table (Table 3).
Fig. 1.
Flow chart for detection methods for various intracellular molecules using probes other than antibodies
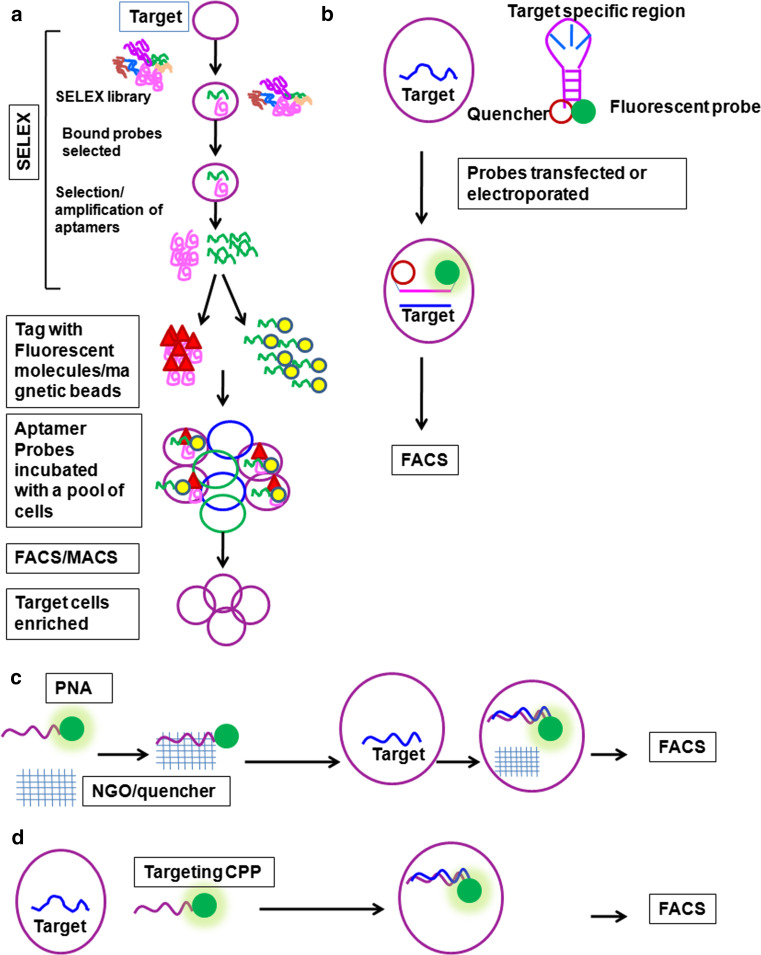
Fig. 2.
Methods of intracellular target detection. Strategies employed in detection and purification of live cells. a Aptamers can be used for subpopulation enrichment as depicted. First, a SELEX library is made which is introduced to the cells. Only a few of the molecules bind to the target population. Those Aptamers are selected and tagged with specific fluorophores, which then can be used for FACS. b Molecular beacons (MBs) are nucleotide sequences with tagged fluorophore and quencher separated by single-stranded sequence, which is complementary to the target molecule. MBs have to be introduced to the cell by transfection. When unbound, the MB is not fluorescent due to the activity of quencher. On the other hand, when it is bound to the target, the quencher is specially removed from fluorophore, and it starts to fluoresce. c Peptide nucleic acid probes (PNPs) are tagged with fluorophores, the activity of which is inhibited by a quencher. When these PNPs are introduced to the cell, the target displaces the quencher and they start to fluoresce. d Targeting cell-penetrating peptides (CPPs) are tagged with fluorophores, which are internalized by the living cells. When they bind to target they remain in the cell and those cells can be detected and purified by FACS
Aptamers
Aptamers are oligonucleotide molecules raised in vitro from large combinatorial libraries of nucleic acids that bind to targets with high affinity and specificity like antibody. Thus, they are considered as oligonucleotide analogs to antibodies, and can be used to identify and isolate cells based on a variety of molecules as reviewed elsewhere [36]. Combinatorial chemistry, which is an important technology that helps in the discovery of new drugs and probes, is used to design Aptamers. It is achieved by the synthesis and simultaneous screening of large libraries of related, but structurally distinct compounds to identify and isolate functional molecules. Systematic Evolution of Ligands by Exponential enrichment (SELEX) is one of those technologies, which allows the synthesis of single-stranded DNA or RNAs for target identification. The initial step of SELEX is chemical synthesis of random nucleotides (of the order 1014–1015 molecules that can form a three-dimensional confirmation) and in vitro target binding. Then the bound molecules are amplified and further selected to get specific Aptamer(s) for the target, the sequence of which is confirmed by sequencing [36]. Several Aptamers have been reported for a variety of purposes including cancer detection [37–40].
Aptamers can be tagged with suitable fluorescent molecules for detection or isolation of cells expressing target molecules like conventional antibody, using FACS [41, 42]. Even though conventional microfluidics does not involve specific probes, modifications have been incorporated for certain applications. Such a device utilizing cancer specific Aptamers were used to purify cancer cells with 96 % purity [43]. An added advantage is that the samples do not require any treatment; it is a simple run through the column, which resulted in 135-fold enrichment of rare tumor cells [43], offering a convenient method for detection of tumor cell, specifically the circulating tumor cells. Another modification reported for Aptamers is magnetic bead tagging, which then can be used for magnetic separation (MACS) of the target cells [44].
Aptamers are considered superior to antibodies in clinical applications since they are generally non-immunogenic and non-toxic in vivo. Further, they show high cell-penetrating capability, improving the bioavailability in clinical applications. Added to that, in commercial synthesis, they are thermostable and offer high reproducibility [40]. Even though Aptamers possess several attractive features as mentioned, they have certain limitations as well. One of the major disadvantages is the selection process, which is time consuming and laborious, and also needs standardization for each target molecule [36]. Added to that, it has short stability in biological fluids [36]. Since the whole selection process is done in vitro, the specificity may not translate well in the complex biological systems [36].
Molecular beacons
Molecular beacons (MBs) are oligonucleotide hybridization probes that generate fluorescent signals only when they are hybridized to a complementary target sequence. They are hairpin-shaped molecules with an internally quenched fluorophore whose fluorescence is restored when they bind to a target nucleic acid sequence [45]. Majority of these beacons are introduced into the cell by electroporation, microinjection or streptolysin O, and they are used to detect mRNA in living cells [45]. Using this tool, real-time imaging of viral replication was experimentally proved in mammalian cells [46]. Modified versions of MBs were used to purify pleuripotent embryonic stem cells and osteogenic mesenchymal stem cells by FACS. For the isolation of embryonic stem cells, a dual FRET molecular beacon was designed for OCT4. The embryonic stem cells transfected with the construct were used for sorting pleuripotent population from differentiated cells by FACS [47]. Since alkaline phosphatase (ALPL) expression is more in stem cell fraction committed for osteogenic differentiation, an oligodeoxynucleotide molecular beacon probe specific for ALPL mRNA was designed to purify osteogenic-committed adipose-derived stem cells. Stromal vascular fraction cells that have undergone osteogenic priming were electroporated with the probe, and the cells were sorted using FACS [48].
Most of the applications of MBs in single cell identification are limited to in vitro systems, since it requires a probe delivery system to introduce MBs into the cell. This problem can be overcome by tagging it with cell-penetrating peptides to introduce the MBs [46]. Another problem arising in the application of MB is the false positivity arising either due to degradation of the stem of MB by an endogenous nuclease in the cell or interaction of MB with other intracellular proteins [49]. The false positivity in the application of MBs could be counteracted by duel FRET MBs [49].
Peptide nucleic acid (PNA) probes (PNPs)
Peptide nucleic acid is an artificial DNA-mimic in which the sugar phosphate backbone is replaced with a pseudopeptide backbone composed of N-(2-aminoethyl) glycine units. PNA binds to target miRNA with high specificity and sensitivity. PNA can be easily tagged with a cell-penetrating peptide to improve the intracellular delivery. Recently, a method for detection and quantification of miRNA in living cells using a nanosized graphene oxide (NGO) miRNA sensor was reported [50]. A specific PNA was synthesized against an miRNA (miR-21), tagged it with a fluorescent dye, and then it was bound to the surface of NGO, which quenches the fluorescence. This PNA-NGO complex is internalized and the target miRNA displaces the NGO and binds to the PNA-tagged to fluorophore, recovering the fluorescence. The fluorescence intensity obtained in flowcytometric analysis corresponds to the quantity of miRNA present in living cells. This probe showed sensitivity up to 1 pM level [50]. It is a useful tool for detection, quantification, and isolation of populations based on miR-21.
Targeting cell-penetrating peptides (targeting CPPs)
Cell-penetrating peptides (CPPs) are peptide sequences that possess the capability to cross cell membrane. These peptides like, TAT and penetratin, are not targeted toward any specific intracellular molecule, and when they are tagged to other molecules or cargos, they are useful in cargo delivery to cytoplasm [51]. There are certain properties like cationicity, helicity, and amphipathicity, required for a CPP, which can be exploited to design CPPs. Another class of peptides is cell-targeting peptides (CTPs) that can bind to specific markers in a cell, like RGD peptide that detects integrins on cancer cells [52]. This targeting ability of peptides has been widely exploited for cancer detection and therapeutics. One of the classic examples for peptide probe for imaging is RGD peptide, used for cancer detection [53, 54]. Followed by RGD peptides, several other peptides were synthesized and tested for optical imaging [53, 54]. Many of the CTPs currently used are targeting cell surface molecules. The targeting peptides for intracellular molecules can be designed in such a way that they behave as CPPs, which then can be used for isolation and detection of live cells. Recently, we have reported a targeting peptide for a nuclear chaperone, HIRA, which can be used for live cell detection and purification by FACS [55].
The success of the method of detection by targeting CPP depends on the specificity of the probe. Like Aptamers, targeting peptides can be developed by combinatorial chemistry. There are bioinformatic programs like PeptideMine (http://caps.ncbs.res.in/peptidemine), which predict interacting peptide sequences. An alternative strategy is to select the interacting proteins of the protein of interest, to make targeting peptides. For this, proteins with minimum interactions to other proteins should be selected further, using Human Protein Reference Database (http://www.hprd.org/). From the short listed molecules, a protein has to be selected, for which the residues of interaction with the proteins of interest are proved experimentally. Based on the region of interaction, Peptides can be designed by PeptGen peptide generator (http://www.hiv.lanl.gov/content/sequence/PEPTGEN/peptgen.html). The peptides that meet the requirements of CPP can be selected. The different methods of design and synthesis of CPPs, and essential features of it are reviewed elsewhere [51, 56]. The specificity of the peptide can be evaluated using western blot with a modified protocol, as we have reported [55].
Peptides are physiological compounds and are intrinsically non-toxic, which are easy to synthesize and modify chemically by radiolabeling or conjugating dyes. They exhibit high affinity receptor binding and are usually rapidly excreted from the body, making it suitable for in vivo applications. Major disadvantage of this technique is the degradation of peptides by peptidases in vivo, which can be overcome by linking suitable chelator molecules to the synthetic peptides. This can be used in, in vitro cultured cells or in vivo system or primary cells without any modification. CPPs can be designed and synthesized easily compared to other systems like Aptamers. However, the success of CPP depends on its design to get specific interaction. Even though bioinformatic tools help to design a targeting peptide, its performance has to be ascertained by wet-lab experiments. A more reliable approach can be used, as discussed, for molecules where the interacting residues are reported. Thus, developing targeting CPPs may not be possible for all intracellular proteins.
Future directions
Since FACS is the conventionally used cell isolation modality for biomedical research, there are certain limitations for the markers that could be used for the isolation of the specific cell types for those applications. We have also summarized some of the molecules that can be used as potential markers to isolate cell populations of interest. Taking this further, we propose some of the strategies that can improve regenerative medicine and cancer diagnostics. We portray the protocols that are currently used in cardiac regeneration as well as bone regeneration. Further, the limitations of the current clinical practice are described, which demand application of new methodologies relying on intracellular molecules, unexplored previously, due to technical limitations. We put forward strategies using Aptamers, Molecular beacons or targeting CPPs to overcome those technical problems, to improve clinical outcome in regenerative medicine. A new strategy for surgical margin prediction using PNPs against miRNAs, which are not currently used in clinical practice, is also recommended.
Cardiac regeneration
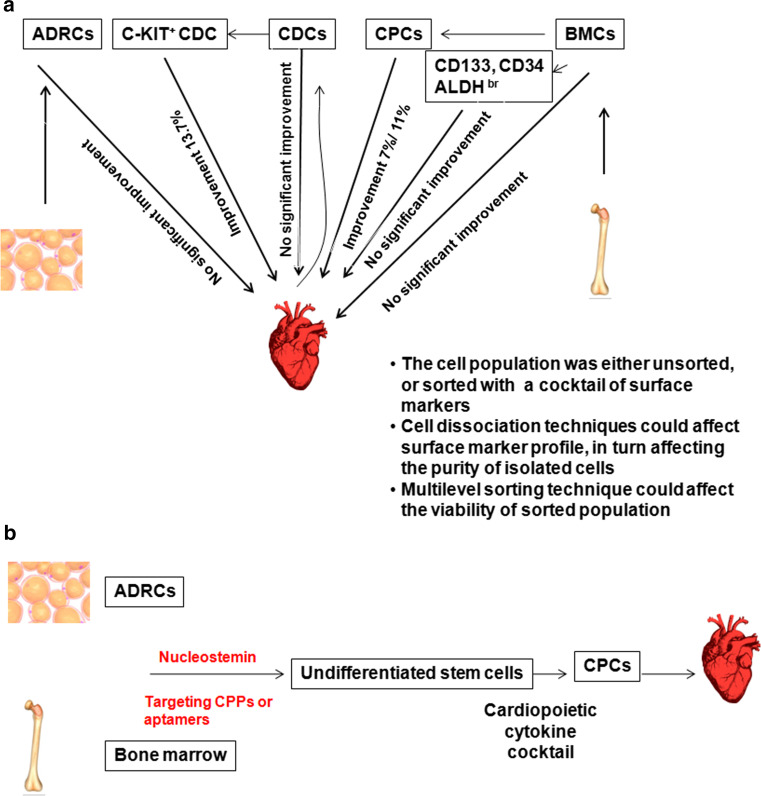
Cardio vascular disease is still a major health concern despite the improved medical care. The main issue of current therapies is their disability to compensate the irreversible loss of functional cardiomyocytes. Hence, the future challenge of cardiovascular therapies will be the functional regeneration of cardiomyocytes. Although use of ES cells, induced pleuripotent cells (iPSCs), and direct reprogramming of fibroblast cells for cardiac regeneration are preclinically evaluated, their clinical use is not yet started due to concerns of safety aspects [57, 58]. Some of the techniques tested in clinical evaluation are summarized in Fig. 3a [57].
Fig. 3.
Application of probes for intracellular molecules in cardiac regeneration. a The strategies used currently in clinics are summarized. Adipose tissue-derived regenerative cells (ADRCs) or bone marrow cells (BMCs) are isolated from adipose tissue or bone marrow, respectively, from the same patient. These cells are used either directly or after enrichment using certain markers for therapy. Similarly, cardiosphere-derived cells (CDCs) are isolated from cardiospheres generated from cardiomyocytes obtained from the same patient heart. b A new method that could be adopted for cardiac regeneration. We propose nucleostemin as a marker for undifferentiated stem cells, which could be detected using cell-penetrating Aptamers or targeting CPPs
Adipose tissue-derived regenerative cells (ADRCs), like mesenchymal stem cells (MSCs), possess the capability to differentiate to different lineages. These cells can be isolated from the patient by liposuction and processing of the fat tissue to obtain stromal vascular fraction, which contains the stem cell fraction that can be enriched as adherent population. Depending on the conditions provided, this fraction can have up to 60 % colony forming units corresponding to stem cells [59]. Another source of stem cells for cardiac regeneration is bone marrow cells (BMCs) consisting of BM-HSCs and BM-MSCs along with other cell types. Conventional method of isolation of BMCs for therapy is either collection of adherent population, or gelatine-polysuccinate density gradient sedimentation that removes only platelets and erythrocytes. Clinical trials using these non-enriched populations have not produced significant improvement in cardiac regeneration [57]. Isolation of CD34+, CD133+ or ALDH bright cells from bone marrow also did not significantly improved the clinical outcome compared to control groups, although ALDH bright cells showed a trend toward significant improvement in reversibility [60–63]. Another approach that showed a significant improvement in clinical outcome is the direct reprogramming of BMCs to cardiopoietic stem cells (CPCs) using specified cytokines and using them for transplantation without further selection. Several reports suggest that direct reprogramming could significantly enhance treatment outcome [57, 58]. Another source of stem cells for cardiac transplantation is cardiosphere-derived cells (CDCs), derived from the same patient heart and maintained as sphere culture. Even though the clinical studies show that unsorted cells do not give any promising results, sorting the population for C-KIT expressing CDCs from a mixture of stromal, mesenchymal, and progenitor cells enhances the clinical outcome [57].
When we analyze the factors reducing clinical outcome, the important limitation of ADRCs and BMCs is that it is only a small fraction within them are capable of regenerating to cardiomyocytes. The multilevel sorting enrichment for the small fraction within them capable of regenerating to cardiomyocytes decreases the viability of the enriched fraction, again reducing the success rate. The outcome of transplantation using cardiac progenitor cell is high, but it involves a major surgery to collect the cells. In view of these facts, one of the strategies that could be adopted is depicted in Fig. 3b.
Nucleostemin (GNL3) is a nucleolar GTP-binding protein that is involved in stem cell proliferation, embryonic development, and ribosome biogenesis in mammals, which is an established stem cell marker expressed only in the undifferentiated stem cells [64]. Using knock-outs, it has been shown that stemness of CPCs is dependent on nucleostemin [65]. Since this protein is localized in the nucleolar region, it was not explored for its possibility as a marker of stem cells for isolation and purification. We propose that targeting CPPs or Aptamers could be employed to purify the undifferentiated cell fraction from ADRCs or BMCs. The sorted nuleostemin positive, undifferentiated cells can be treated with cardiopoietic cytokines to convert to CPCs in vitro, and then can be used without further purification.
Bone regeneration
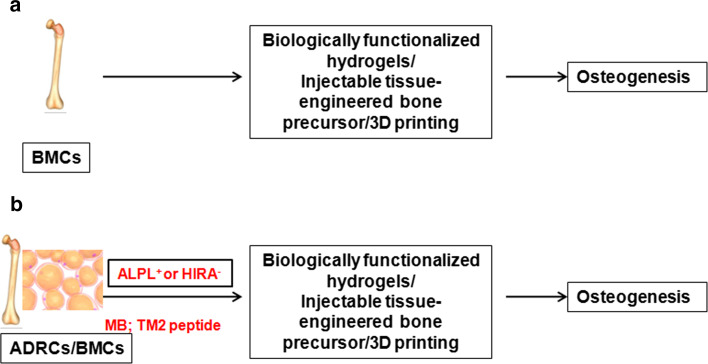
Even though bone is capable of regeneration, in pathological fractures and massive bone defects, where natural regeneration fails, tissue engineering helps in bone regeneration. Bone grafts are implanted materials that promote bone healing through regeneration. Tissue engineering involves the use of scaffolds, growth factors that promote healing and stem cells. Figure 4a represents the current strategies in bone tissue engineering. Several methods are adopted to improve the scaffold of the graft to enhance the regenerative capacity [66–68]. Stem cells, BMCs, and ADRCs are used in bone regeneration. BMP-2-functionalized hydrogels incorporating BMCs or ADRCs were shown to have a significantly higher bone regenerative capacity [69]. A clinical investigation of injectable tissue engineered bone comprising mesenchymal stem cells isolated from bone marrow showed significant improvement in functional regeneration of alveolar defects [70]. 3D-printing (3DP) is another new technique that improves the clinical outcome of bone regeneration. Using recently established functional 3D-printable particle-laden biomaterial inks contains hydroxyapatite microspheres or graphene nanoflakes, it has been shown that mesenchymal stem cells can be used for organ reconstruction [71]. Since isolation of BMCs is more invasive than isolation of ADRCs, the later has been tested for its efficacy for bone regeneration [72]. It has been shown that a molecular beacon can be used to isolate ALPL+ cells with osteogeneic potential from stromal vascular fraction of fat tissues. Here we propose that this method can be adopted to enrich ADRCs with osteogeneic potential for tissue engineering (Fig. 4b). Another marker that could be used to demarcate stem cells with osteogeneic potential is HIRA. The loss of HIRA expression is shown to mark the myoblasts committed to osteogenic differentiation [16]. Recently we have reported a targeting CPP, TM2 that can be used to detect and isolate HIRA expressing cells [55]. A combination of these two probes to isolate osteogenic-committed ADRC might be a better therapeutic option for bone regeneration.
Fig. 4.
Application of probes for intracellular molecules in Bone regeneration. a The strategies currently used for bone tissue engineering are summarized. Bone marrow cells (BMCs) isolated from the same patient bone marrow is generally used for osteogenesis. b A new strategy proposed for bone tissue engineering after sorting with molecular beacons for alkaline phosphatase (ALPL) or targeting CPP for HIRA
Surgical margin prediction in cancer
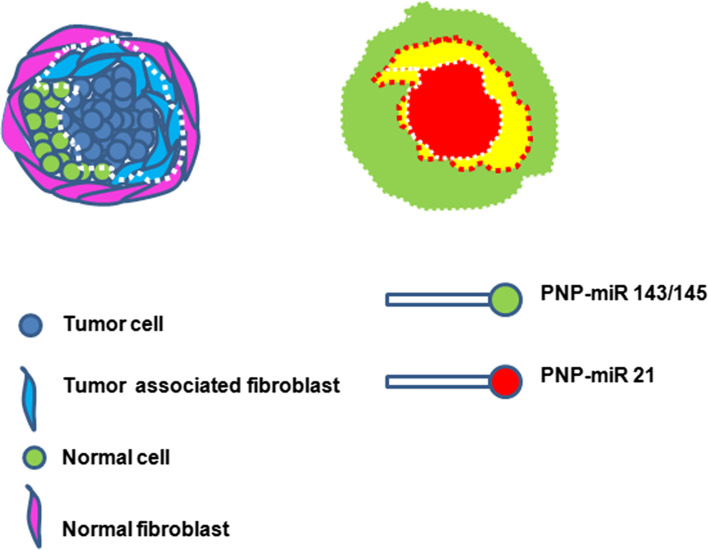
Prediction of surgical margin is a critical factor deciding the disease-free survival after surgery in cancer treatment. Many a time, some tumor cells are left out due to inadequate margin prediction, which results in relapse of the disease. Yet in some other cases, the altered tumor stroma, which favors the growth of tumor, might lead to relapse. So, recent reports suggest that removal of tumor cells as well as tumor-associated stroma (comprised of fibroblasts) is important in disease-free survival. Over-expression of miR-21 is reported to mark tumor cells as well as tumor-associated fibroblasts. Moreover, it has been shown that stromal expression of miR-21 is associated with poor prognosis [31]. Loss of expression of miR-143 and -145 is reported to associate with cancer progression. So up-regulated miR-21 and down-regulated miR-143/145 mark the tumor cells as well as tumor stroma, which needs to be surgically removed for disease-free survival. As discussed earlier, PNPs can be developed for this purpose and used for surgical margin prediction as summarized in Fig. 5.
Fig. 5.
PNPs for surgical margin prediction. A diagrammatic representation of tumor surrounded by normal cells, normal fibroblasts, and tumor fibroblasts. The two PNPs tagged with different fluorescent tags detecting distinct miRNAs can be used for imaging. The merge of the two probes are shown by yellow color. Removal of red and yellow region will ensure disease-free survival
Conclusions
Although antibodies remain the most popular and widely used probe to detect and isolate single cells in biology, there are certain situations, where they cannot be used for single cell isolation. A comprehensive account of such contexts, and some of the probes that can be employed in such situations, were described. A comparative account of the different modalities used (Fig. 2), and their advantages and limitations are summarized (Table 3). From the literature it is evident that Aptamers and molecular beacons can be used in vitro and in vivo for single cell identification based on the expression of mRNA or protein. PNAs are extremely useful for target miRNA identification. Though the currently used CTPs are against surface molecules, they can be designed with characteristics of CPPs. Targeting CPPs will be useful in identification of intracellular protein molecules. Each of these techniques has limitations, and research in the field might improve their application in basic research and translational medicine. In this review, we have proposed the application of Aptamers, Molecular beacons or targeting CPPs that could bind to intracellular molecules, like nucleostemin, ALPL or HIRA, which might be useful markers to identify subpopulations relevant for applications in regenerative medicine. Another field where the diagnostic probes can be applied is disease diagnosis, as we have illustrated for cancer diagnostics using PNPs. Taken together, it appears that there is considerable advancement in the field of chemistry to offer probes and techniques to detect and isolate single cells based on intracellular molecules. In other words, molecular markers used in biomedical research should no longer be limited to surface molecules. If this concept is accepted in scientific community, it will prompt identifying better markers for various fields of biomedical research, including stem cell therapy as well as disease diagnosis and therapeutic applications.
Acknowledgments
We acknowledge the funding from Department of Biotechnology, India (BT/PR14379/Med/30/536/2010). We also sincerely thank the help of Praveen Chandran and Avinash Kumar in manuscript preparation.
Compliance with ethical standards
Conflict of interest
Author declares no conflict of interest.
References
- 1.Ward EJ, Shcherbata HR, Reynolds SH, Fischer KA, Hatfield SD, Ruohola-Baker H. Stem cells signal to the niche through the Notch pathway in the Drosophila ovary. Curr Biol. 2006;16(23):2352–2358. doi: 10.1016/j.cub.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 2.Tsioris K, Torres AJ, Douce TB, Love JC. A new toolbox for assessing single cells. Annu Rev Chem Biomol Eng. 2014;5:455–477. doi: 10.1146/annurev-chembioeng-060713-035958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazutis L, Gilbert J, Ung WL, Weitz DA, Griffiths AD, Heyman JA. Single-cell analysis and sorting using droplet-based microfluidics. Nat Protoc. 2013;8(5):870–891. doi: 10.1038/nprot.2013.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He JL, Chen AT, Lee JH, Fan SK. Digital microfluidics for manipulation and analysis of a single cell. Int J Mol Sci. 2015;16(9):22319–22332. doi: 10.3390/ijms160922319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Liu L, Fu X, Zhu Z. Microfluidic beads-based immunosensor for sensitive detection of cancer biomarker proteins using multienzyme-nanoparticle amplification and quantum dots labels. Biosens Bioelectron. 2013;42:23–30. doi: 10.1016/j.bios.2012.10.076. [DOI] [PubMed] [Google Scholar]
- 6.Welch JD, Williams LA, DiSalvo M, Brandt AT, Marayati R, Sims CE, Allbritton NL, Prins JF, Yeh JJ, Jones CD. Selective single cell isolation for genomics using microraft arrays. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gach PC, Wang Y, Phillips C, Sims CE, Allbritton NL. Isolation and manipulation of living adherent cells by micromolded magnetic rafts. Biomicrofluidics. 2011;5(3):32002–3200212. doi: 10.1063/1.3608133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreu Z, Khan MA, Gonzalez-Gomez P, Negueruela S, Hortiguela R, San Emeterio J, Ferron SR, Martinez G, Vidal A, Farinas I, Lie DC, Mira H. The cyclin-dependent kinase inhibitor p27 kip1 regulates radial stem cell quiescence and neurogenesis in the adult hippocampus. Stem Cells. 2015;33(1):219–229. doi: 10.1002/stem.1832. [DOI] [PubMed] [Google Scholar]
- 9.Pauklin S, Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155(1):135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh AM. Cell cycle-driven heterogeneity: on the road to demystifying the transitions between “poised” and “restricted” pluripotent cell states. Stem Cells Int. 2015;2015:219514. doi: 10.1155/2015/219514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhawan J, Laxman S. Decoding the stem cell quiescence cycle—lessons from yeast for regenerative biology. J Cell Sci. 2015;128(24):4467–4474. doi: 10.1242/jcs.177758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou P, Yoshihara H, Hosokawa K, Tai I, Shinmyozu K, Tsukahara F, Maru Y, Nakayama K, Nakayama KI, Suda T. p57(Kip2) and p27(Kip1) cooperate to maintain hematopoietic stem cell quiescence through interactions with Hsc70. Cell Stem Cell. 2011;9(3):247–261. doi: 10.1016/j.stem.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Paczkowska E, Kawa M, Klos P, Staniszewska M, Sienko J, Dabkowska E. Aldehyde dehydrogenase (ALDH)—a promising new candidate for use in preclinical and clinical selection of pluripotent very small embryonic-like stem cells (VSEL SCs) of high long-term repopulating hematopoietic potential. Ann Transplant. 2011;16(3):59–71. doi: 10.12659/AOT.881996. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Liu Y, Zhou K, Zhang G, Wang F, Ren J. Isolation and characterization of CD105+/CD90+ subpopulation in breast cancer MDA-MB-231 cell line. Int J Clin Exp Pathol. 2015;8(5):5105–5112. [PMC free article] [PubMed] [Google Scholar]
- 15.Yang JH, Song Y, Seol JH, Park JY, Yang YJ, Han JW, Youn HD, Cho EJ. Myogenic transcriptional activation of MyoD mediated by replication-independent histone deposition. Proc Natl Acad Sci USA. 2011;108(1):85–90. doi: 10.1073/pnas.1009830108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song TY, Yang JH, Park JY, Song Y, Han JW, Youn HD, Cho EJ. The role of histone chaperones in osteoblastic differentiation of C2C12 myoblasts. Biochem Biophys Res Commun. 2012;423(4):726–732. doi: 10.1016/j.bbrc.2012.06.026. [DOI] [PubMed] [Google Scholar]
- 17.Zimmerer RM, Korn P, Demougin P, Kampmann A, Kokemuller H, Eckardt AM, Gellrich NC, Tavassol F. Functional features of cancer stem cells in melanoma cell lines. Cancer Cell Int. 2013;13(1):78. doi: 10.1186/1475-2867-13-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kemmerling R, Alinger B, Dietze O, Bosmuller HC, Ocker M, Wolkersdorfer GW, Berr F, Neureiter D, Kiesslich T. Association of stem cell marker expression pattern and survival in human biliary tract cancer. Int J Oncol. 2012;41(2):511–522. doi: 10.3892/ijo.2012.1477. [DOI] [PubMed] [Google Scholar]
- 19.He QZ, Luo XZ, Wang K, Zhou Q, Ao H, Yang Y, Li SX, Li Y, Zhu HT, Duan T. Isolation and characterization of cancer stem cells from high-grade serous ovarian carcinomas. Cell Physiol Biochem. 2014;33(1):173–184. doi: 10.1159/000356660. [DOI] [PubMed] [Google Scholar]
- 20.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin VY, Ng EK, Chan VW, Kwong A, Chu KM. A three-miRNA signature as promising non-invasive diagnostic marker for gastric cancer. Mol Cancer. 2015;14(1):202. doi: 10.1186/s12943-015-0473-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skrha P, Hajer J, Andel M, Horinek A, Korabecna M. miRNA as a new marker of diabetes mellitus and pancreatic carcinoma progression. Cas Lek Cesk. 2015;154(3):122–126. [PubMed] [Google Scholar]
- 23.Hsu CM, Lin PM, Wang YM, Chen ZJ, Lin SF, Yang MY. Circulating miRNA is a novel marker for head and neck squamous cell carcinoma. Tumour Biol. 2012;33(6):1933–1942. doi: 10.1007/s13277-012-0454-8. [DOI] [PubMed] [Google Scholar]
- 24.Luo X, Burwinkel B, Tao S, Brenner H. MicroRNA signatures: novel biomarker for colorectal cancer? Cancer Epidemiol Biomarkers Prev. 2011;20(7):1272–1286. doi: 10.1158/1055-9965.EPI-11-0035. [DOI] [PubMed] [Google Scholar]
- 25.Li X, Yang W, Lou L, Chen Y, Wu S, Ding G. microRNA: a promising diagnostic biomarker and therapeutic target for hepatocellular carcinoma. Dig Dis Sci. 2014;59(6):1099–1107. doi: 10.1007/s10620-013-3006-1. [DOI] [PubMed] [Google Scholar]
- 26.Kishore A, Borucka J, Petrkova J, Petrek M. Novel insights into miRNA in lung and heart inflammatory diseases. Mediators Inflamm. 2014;2014:259131. doi: 10.1155/2014/259131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, Feig C, Lengyel E, Peter ME. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA. 2007;104(27):11400–11405. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitade Y, Akao Y. MicroRNAs and their therapeutic potential for human diseases: microRNAs, miR-143 and -145, function as anti-oncomirs and the application of chemically modified miR-143 as an anti-cancer drug. J Pharmacol Sci. 2010;114(3):276–280. doi: 10.1254/jphs.10R12FM. [DOI] [PubMed] [Google Scholar]
- 29.Pan X, Wang ZX, Wang R. MicroRNA-21: a novel therapeutic target in human cancer. Cancer Biol Ther. 2010;10(12):1224–1232. doi: 10.4161/cbt.10.12.14252. [DOI] [PubMed] [Google Scholar]
- 30.MacKenzie TA, Schwartz GN, Calderone HM, Graveel CR, Winn ME, Hostetter G, Wells WA, Sempere LF. Stromal expression of miR-21 identifies high-risk group in triple-negative breast cancer. Am J Pathol. 2014;184(12):3217–3225. doi: 10.1016/j.ajpath.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hedback N, Jensen DH, Specht L, Fiehn AM, Therkildsen MH, Friis-Hansen L, Dabelsteen E, von Buchwald C. MiR-21 expression in the tumor stroma of oral squamous cell carcinoma: an independent biomarker of disease free survival. PLoS One. 2014;9(4):e95193. doi: 10.1371/journal.pone.0095193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melbo-Jorgensen C, Ness N, Andersen S, Valkov A, Donnem T, Al-Saad S, Kiselev Y, Berg T, Nordby Y, Bremnes RM, Busund LT, Richardsen E. Stromal expression of MiR-21 predicts biochemical failure in prostate cancer patients with Gleason score 6. PLoS One. 2014;9(11):e113039. doi: 10.1371/journal.pone.0113039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dumas A, Le-Bury G, Marie-Anais F, Herit F, Mazzolini J, Guilbert T, Bourdoncle P, Russell DG, Benichou S, Zahraoui A, Niedergang F. The HIV-1 protein Vpr impairs phagosome maturation by controlling microtubule-dependent trafficking. J Cell Biol. 2015;211(2):359–372. doi: 10.1083/jcb.201503124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith LM, May RC. Mechanisms of microbial escape from phagocyte killing. Biochem Soc Trans. 2013;41(2):475–490. doi: 10.1042/BST20130014. [DOI] [PubMed] [Google Scholar]
- 35.Hawn TR, Matheson AI, Maley SN, Vandal O. Host-directed therapeutics for tuberculosis: can we harness the host? Microbiol Mol Biol Rev. 2013;77(4):608–627. doi: 10.1128/MMBR.00032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoltenburg R, Reinemann C, Strehlitz B. SELEX—a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng. 2007;24(4):381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 37.Champanhac C, Teng IT, Cansiz S, Zhang L, Wu X, Zhoa Z, Fu T, Tan W. Development of a panel of DNA Aptamers with high affinity for pancreatic ductal adenocarcinoma. Sci Rep. 2015;5:16788. doi: 10.1038/srep16788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hung LY, Wang CH, Che YJ, Fu CY, Chang HY, Wang K, Lee GB. Screening of aptamers specific to colorectal cancer cells and stem cells by utilizing On-chip Cell-SELEX. Sci Rep. 2015;5:10326. doi: 10.1038/srep10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Z, Lu Y, Pu Y, Liu J, Liu B, Yu B, Chen K, Fu T, Yang CJ, Liu H, Tan W. Using aptamers to elucidate esophageal cancer clinical samples. Sci Rep. 2015;5:18516. doi: 10.1038/srep18516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun H, Zu Y. A highlight of recent advances in aptamer technology and its application. Molecules. 2015;20(7):11959–11980. doi: 10.3390/molecules200711959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayer G, Ahmed MS, Dolf A, Endl E, Knolle PA, Famulok M. Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat Protoc. 2010;5(12):1993–2004. doi: 10.1038/nprot.2010.163. [DOI] [PubMed] [Google Scholar]
- 42.Meyer S, Maufort JP, Nie J, Stewart R, McIntosh BE, Conti LR, Ahmad KM, Soh HT, Thomson JA. Development of an efficient targeted cell-SELEX procedure for DNA aptamer reagents. PLoS One. 2013;8(8):e71798. doi: 10.1371/journal.pone.0071798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y, Phillips JA, Yan J, Li Q, Fan ZH, Tan W. Aptamer-based microfluidic device for enrichment, sorting, and detection of multiple cancer cells. Anal Chem. 2009;81(17):7436–7442. doi: 10.1021/ac9012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo KT, SchAfer R, Paul A, Gerber A, Ziemer G, Wendel HP. A new technique for the isolation and surface immobilization of mesenchymal stem cells from whole bone marrow using high-specific DNA aptamers. Stem Cells. 2006;24(10):2220–2231. doi: 10.1634/stemcells.2006-0015. [DOI] [PubMed] [Google Scholar]
- 45.Bratu DP. Molecular beacons: fluorescent probes for detection of endogenous mRNAs in living cells. Methods Mol Biol. 2006;319:1–14. doi: 10.1007/978-1-59259-993-6_1. [DOI] [PubMed] [Google Scholar]
- 46.Yeh HY, Yates MV, Mulchandani A, Chen W. Visualizing the dynamics of viral replication in living cells via Tat peptide delivery of nuclease-resistant molecular beacons. Proc Natl Acad Sci USA. 2008;105(45):17522–17525. doi: 10.1073/pnas.0807066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.King FW, Liszewski W, Ritner C, Bernstein HS. High-throughput tracking of pluripotent human embryonic stem cells with dual fluorescence resonance energy transfer molecular beacons. Stem Cells Dev. 2011;20(3):475–484. doi: 10.1089/scd.2010.0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marble HD, Sutermaster BA, Kanthilal M, Fonseca VC, Darling EM. Gene expression-based enrichment of live cells from adipose tissue produces subpopulations with improved osteogenic potential. Stem Cell Res Ther. 2014;5(5):145. doi: 10.1186/scrt502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han SX, Jia X, Ma JL, Zhu Q. Molecular beacons: a novel optical diagnostic tool. Arch Immunol Ther Exp (Warsz) 2013;61(2):139–148. doi: 10.1007/s00005-012-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryoo SR, Lee J, Yeo J, Na HK, Kim YK, Jang H, Lee JH, Han SW, Lee Y, Kim VN, Min DH. Quantitative and multiplexed microRNA sensing in living cells based on peptide nucleic acid and nano graphene oxide (PANGO) ACS Nano. 2013;7(7):5882–5891. doi: 10.1021/nn401183s. [DOI] [PubMed] [Google Scholar]
- 51.Copolovici DM, Langel K, Eriste E, Langel U. Cell-penetrating peptides: design, synthesis, and applications. ACS Nano. 2014;8(3):1972–1994. doi: 10.1021/nn4057269. [DOI] [PubMed] [Google Scholar]
- 52.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 53.Raucher D, Ryu JS. Cell-penetrating peptides: strategies for anticancer treatment. Trends Mol Med. 2015;21(9):560–570. doi: 10.1016/j.molmed.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 54.Kersemans V, Kersemans K, Cornelissen B. Cell penetrating peptides for in vivo molecular imaging applications. Curr Pharm Des. 2008;14(24):2415–2447. doi: 10.2174/138161208785777432. [DOI] [PubMed] [Google Scholar]
- 55.Kochurani KJ, Suganya AA, Nair MG, Louis JM, Majumder A, Kumar KS, Abraham P, Dutta D, Maliekal TT. Live detection and purification of cells based on the expression of a histone chaperone, HIRA, using a binding peptide. Sci Rep. 2015;5:17218. doi: 10.1038/srep17218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hansen M, Kilk K, Langel U. Predicting cell-penetrating peptides. Adv Drug Deliv Rev. 2008;60(4–5):572–579. doi: 10.1016/j.addr.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 57.Doppler SA, Deutsch MA, Lange R, Krane M. Cardiac regeneration: current therapies-future concepts. J Thorac Dis. 2013;5(5):683–697. doi: 10.3978/j.issn.2072-1439.2013.08.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doppler SA, Deutsch MA, Lange R, Krane M. Direct reprogramming—the future of cardiac regeneration? Int J Mol Sci. 2015;16(8):17368–17393. doi: 10.3390/ijms160817368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Russo V, Yu C, Belliveau P, Hamilton A, Flynn LE. Comparison of human adipose-derived stem cells isolated from subcutaneous, omental, and intrathoracic adipose tissue depots for regenerative applications. Stem Cells Transl Med. 2014;3(2):206–217. doi: 10.5966/sctm.2013-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manginas A, Goussetis E, Koutelou M, Karatasakis G, Peristeri I, Theodorakos A, Leontiadis E, Plessas N, Theodosaki M, Graphakos S, Cokkinos DV. Pilot study to evaluate the safety and feasibility of intracoronary CD133(+) and CD133(−) CD34(+) cell therapy in patients with nonviable anterior myocardial infarction. Catheter Cardiovasc Interv. 2007;69(6):773–781. doi: 10.1002/ccd.21023. [DOI] [PubMed] [Google Scholar]
- 61.Patel AN, Geffner L, Vina RF, Saslavsky J, Urschel HC, Jr, Kormos R, Benetti F. Surgical treatment for congestive heart failure with autologous adult stem cell transplantation: a prospective randomized study. J Thorac Cardiovasc Surg. 2005;130(6):1631–1638. doi: 10.1016/j.jtcvs.2005.07.056. [DOI] [PubMed] [Google Scholar]
- 62.Stamm C, Kleine HD, Choi YH, Dunkelmann S, Lauffs JA, Lorenzen B, David A, Liebold A, Nienaber C, Zurakowski D, Freund M, Steinhoff G. Intramyocardial delivery of CD133+ bone marrow cells and coronary artery bypass grafting for chronic ischemic heart disease: safety and efficacy studies. J Thorac Cardiovasc Surg. 2007;133(3):717–725. doi: 10.1016/j.jtcvs.2006.08.077. [DOI] [PubMed] [Google Scholar]
- 63.Perin EC, Silva GV, Zheng Y, Gahremanpour A, Canales J, Patel D, Fernandes MR, Keller LH, Quan X, Coulter SA, Moore WH, Herlihy JP, Willerson JT. Randomized, double-blind pilot study of transendocardial injection of autologous aldehyde dehydrogenase-bright stem cells in patients with ischemic heart failure. Am Heart J. 2012;163(3):415–421. doi: 10.1016/j.ahj.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 64.Tsai RY, McKay RD. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16(23):2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hariharan N, Quijada P, Mohsin S, Joyo A, Samse K, Monsanto M, De La Torre A, Avitabile D, Ormachea L, McGregor MJ, Tsai EJ, Sussman MA. Nucleostemin rejuvenates cardiac progenitor cells and antagonizes myocardial aging. J Am Coll Cardiol. 2015;65(2):133–147. doi: 10.1016/j.jacc.2014.09.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng MQ, Wahafu T, Jiang GF, Liu W, Qiao YQ, Peng XC, Cheng T, Zhang XL, He G, Liu XY. A novel open-porous magnesium scaffold with controllable microstructures and properties for bone regeneration. Sci Rep. 2016;6:24134. doi: 10.1038/srep24134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee JH, Shin YC, Lee SM, Jin OS, Kang SH, Hong SW, Jeong CM, Huh JB, Han DW. Enhanced osteogenesis by reduced graphene oxide/hydroxyapatite nanocomposites. Sci Rep. 2015;5:18833. doi: 10.1038/srep18833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang X, Zeng D, Li N, Wen J, Jiang X, Liu C, Li Y. Functionalized mesoporous bioactive glass scaffolds for enhanced bone tissue regeneration. Sci Rep. 2016;6:19361. doi: 10.1038/srep19361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dosier CR, Uhrig BA, Willett NJ, Krishnan L, Li MT, Stevens HY, Schwartz Z, Boyan BD, Guldberg RE. Effect of cell origin and timing of delivery for stem cell-based bone tissue engineering using biologically functionalized hydrogels. Tissue Eng Part A. 2015;21(1–2):156–165. doi: 10.1089/ten.tea.2014.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamada Y, Nakamura S, Ito K, Umemura E, Hara K, Nagasaka T, Abe A, Baba S, Furuichi Y, Izumi Y, Klein OD, Wakabayashi T. Injectable bone tissue engineering using expanded mesenchymal stem cells. Stem Cells. 2013;31(3):572–580. doi: 10.1002/stem.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jakus AE, Shah RN. Multi- and mixed 3D-printing of graphene-hydroxyapatite hybrid materials for complex tissue engineering. J Biomed Mater Res A. 2016 doi: 10.1002/jbm.a.35684. [DOI] [PubMed] [Google Scholar]
- 72.Correia CR, Pirraco RP, Cerqueira MT, Marques AP, Reis RL, Mano JF. Semipermeable capsules wrapping a multifunctional and self-regulated co-culture microenvironment for osteogenic differentiation. Sci Rep. 2016;6:21883. doi: 10.1038/srep21883. [DOI] [PMC free article] [PubMed] [Google Scholar]