Abstract
Endogenous retroviruses (ERV) are an abundant class of repetitive elements in mammalian genomes. To ensure genomic stability, ERVs are largely transcriptionally silent. However, these elements also feature physiological roles in distinct developmental contexts, under which silencing needs to be partially relieved. ERV silencing is mediated through a heterochromatic structure, which is established by histone modification and DNA methylation machineries. This heterochromatic structure is largely refractory to transcriptional stimulation, however, challenges to the heterochromatic state, such as DNA replication, require re-establishment of the heterochromatic state in competition with transcriptional activators. In this review, we discuss the major pathways leading to efficient establishment of robust and inaccessible heterochromatin across ERVs.
Keywords: ERV restriction, Setdb1, Trim28, Daxx, Atrx, H3K9me3, Retrotransposons, Transposable elements, TEs
Introduction
Transposable elements (TEs) are DNA sequences of ancient origin with the ability to jump into new locations within the genome. They are likely to have emerged as remnants of viral germ line infections that occurred during evolution and have accumulated to comprise a large fraction (40–60%) of mammalian genomes [17, 47, 93]. Transposable elements can be divided in two major classes: DNA transposons (class II) and retrotransposons (class I). DNA transposons move by the so-called “cut and paste” mechanism, driven by the encoded transposase enzyme. Hence, transposition does not result in copy number increase, and therefore, DNA transposons comprise only a very small part of mammalian genomes [27]. Retrotransposons, on the other hand, move by a “copy and paste” mechanism, which involves reverse transcription of an RNA intermediate and subsequent integration as an additional copy within the host genome [12]. This mechanism explains the high abundance of these elements that make up about 90% of all TEs present in humans and 95% in the mouse [3, 47, 93].
Mammalian retrotransposons can be further divided into two major groups defined by the presence or absence of flanking long terminal repeats (LTRs). Non-LTR retrotransposons include long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs). LINEs contain two open reading frames, orf1 and orf2, encoding proteins which mediate retrotransposition. A number of LINE elements, especially members of the LINE-1 (L1) subfamily, are still active and retrotransposition competent in mouse and human. Their movement was suggested to contribute to genome variation, but also to cause diseases [35]. SINEs on the other hand are non-autonomous and require LINE-encoded proteins for their retrotransposition [45]. In human, Alu elements are the most frequently transposing SINEs, with one new insertion in every 20th birth [15]. In mouse four SINE families (B1, B2, ID, B4) are expressed [45].
LTR containing retrotransposons closely resemble the proviral-integrated form of infectious retroviruses. Therefore, the term endogenous retrovirus (ERV) is often used synonymously [79]. Many ERV classes contain gag (group specific antigen), pro (protease) and pol (polymerase) genes derived from exogenous retroviruses. A small subset of retrotransposons furthermore encode envelope (env) proteins [12]. However, some ERV classes do not encode these genes or accumulated genomic mutations that left them non-autonomous and in the need to parasitize the small fraction of retrotransposons which is transposition competent [4, 82]. On the other hand, some elements in the mouse (e.g. MLV) are still able to generate functional retroviral particles [99]. In the human genome only HERV-K (HML2) elements seem to be retrotransposition competent [4].
ERVs are organized in three classes (I–III) based on sequence homology to the pol gene of exogenous retroviruses [30]. Class I ERVs are similar to gamma- and epsilonretroviruses, class II elements resemble alpha-, beta-, deltaretroviruses and lentiviruses, and class III ERV show similarity to spuma- and spumalike retroviruses [42].
Physiological roles of retrotransposons
Evolution
Since their discovery, transposable elements were considered harmful, parasitic or selfish [19, 62]. This view has changed recently with physiological roles of TEs for the host being increasingly appreciated. By now, numerous examples have been described in which retrotransposons were utilized by the host to fulfill new functions (exaptation). On the one hand, retroviral genes can give rise to new host genes. For instance, the placental syncytin gene was derived from a retroviral envelope protein [60], and also the evolution of the Xist gene was attributed to the integration of mobile elements [22]. On the other hand, TEs can contribute to gene regulation by serving as promoter or enhancer elements for host genes and by generating alternative splice sites or polyadenylation signals [31, 41, 69]. Thus, TEs are now considered an important source of genetic variation and an attractive force for genome evolution [14, 29, 59, 85].
Genome organization
A large fraction of transposable elements, in particular full length retrotransposons, reside in a heterochromatic chromatin state. Heterochromatin plays important roles for nuclear organization and maintenance of genomic stability [66]. For example, heterochromatic regions help to connect chromatin with the nuclear lamina [34] or buildup large nuclear domains in specific cell types [77]. As retrotransposons comprise a significant portion of the heterochromatic compartment, it is likely that these elements contribute to proper genome architecture. Future studies are necessary to specifically address functions for retrotransposons in this context. In particular, it would be important to clarify the higher order chromatin organization of retrotransposons. Due to their relatively short length of only several kilobases, in contrast to pericentric heterochromatin with megabase extension, their potency for forming large condensed chromatin areas is probably limited. However, it is possible that interactions between different retrotransposon copies based on their heterochromatic modification pattern may contribute to genome organization.
Transcriptional regulation
The most widely studied physiological function of retrotransposons is transcriptional regulation. Next-generation sequencing analyses revealed that 6–30% of mouse or human RNAs start within repetitive elements [26]. These data suggest that retrotransposon LTRs in the 5′ region of protein-coding genes, which act as alternative promoters, drive a significant proportion of host gene expression. Different retrotransposon classes can act in different developmental settings. One well described example is the two-cell stage-specific transcription of protein-coding genes in mouse embryos, regulated by the LTR of murine endogenous retrovirus-like (MERV-L) elements [54]. Members of the LTR class III retrotransposons, such as MaLR, are specifically active in mouse oocytes and can act as additional promoters for oocyte genes [65, 88]. In human preimplantation embryos ERVs are also systematically transcribed, and with the help of single cell RNA sequencing, expression of specific ERVs was shown to characterize distinct developmental stages [32]. In this study, human ERVs were found to display splice acceptor sites leading to fusion transcripts between ERV and non-ERV sequences. If these transcripts are functionally relevant in embryos remains to be tested.
Interestingly, stage-specific transcription from some ERVs appears to be crucial for differentiation. For example, altered transcriptional and post-transcriptional regulation of class II and III ERVs (IAP and MuERV-L) were observed to affect pluripotency and the differentiation potential of mouse ES cells [67]. More recently, specific retrotransposon-derived long non-coding RNAs were shown to influence pluripotency of mouse ES cells, as knock-down of these transcripts resulted in reduced pluripotency marker gene expression [28]. Also in human cells, ERV derived lncRNAs regulate the pluripotency transcriptome. HERV-H elements produce lncRNAs which potentially regulate genes in vicinity, and, HERV-H LTRs appear to function as enhancers for pluripotency-related genes [53]. A recent study could confirm HERV-H expression in human ES cells and found that transcription of these elements is enhanced in the primed compared to the naïve state [83]. Although physiological roles of ERVs were mainly investigated in embryonic stem cells or early embryonic development, ERVs are likely to affect also the transcriptional networks of differentiated cells. Evidence was provided in a recent study that showed modulation of RNA abundance and splicing by ERVs in different human cell types [44].
Unphysiological, adverse effects of uncontrolled retrotransposon expression
In all cell types investigated so far, the majority of retrotransposons reside in a repressive chromatin configuration, being largely transcriptionally inert. Repression of retrotransposons is crucial to prevent the “copy and paste” integration of potentially functional endogenous retroviruses. In the mouse, different retrotransposon classes are still functional, including IAP elements [18]. In humans LINE1 elements retain activity, whereas ERVs are generally considered inactive. Repression of retroviruses may not only be important to restrain activity of potentially functional copies, it may also prevent aberrant activation of host genes in the vicinity of ERVs [21, 43, 72].
Phenotypes which coincide with impaired retrotransposon repression have been investigated in mouse models lacking crucial factors for heterochromatin formation. All investigated mouse models displayed significant developmental phenotypes in connection with strong expression of different ERV classes. For example, Dnmt3l knock-out mice which display enhanced IAP expression in testis are infertile due to the loss of germ cells [8]. Dnmt1 knock-out embryos show strong IAP expression in somatic cells and die during early embryogenesis [75, 91]. Derepression of ERV classes due to loss of histone methylation has been observed in germ cells and somatic cells, always coinciding with impaired cell survival or proliferation [24, 51, 80]. These data strongly suggest that ERV derepression may cause such severe phenotypes. Still very little is known about mechanisms of ERV expression leading to impaired development. Derepression of ERVs often leads to over-expression of genes in their vicinity, which may adversely affect the transcriptional network of cells [21, 43, 72]. Strong expression of ERV transcripts is sensed by the innate immune system and can lead to activation of the interferon response pathway, resulting in elimination of cells by the immune system [11, 70]. We have recently found that in B cell development expression of retroviral proteins can induce activation of cellular stress pathways, such as the unfolded protein response, leading to apoptosis [63]. It is likely that the phenotypic outcomes of ERV derepression very much depend on the cell type and the ERV class which is derepressed. More analyses are necessary to better understand the mechanistic details of ERV derepression in the context of development.
In humans, complete abrogation of ERV silencing systems has not been observed. However, increased activity of HERVs was linked with different diseases, such as Amyotrophic lateral sclerosis (ALS) [50], Schizophrenia [76], autoimmune disorders [89] and cancer [6]. It is currently unclear if increased transcription of HERVs directly partakes in the development of these diseases. For example, in cancer cells enhanced transcriptional activity of HERVs may be due to the generally lower DNA methylation status. It was speculated that active transposable elements, able to integrate into new sites in the genome may contribute to cancer progression by mutating tumor suppressor genes, or by inducing genomic instability. However, analyses of cancer genomes have not revealed much new integration of such elements, rather suggesting that transposable elements have a minor role in cancer progression. It is interesting to note that enhanced levels of retrotransposon transcription in cancer cells can potentially be exploited in the context of immunstimulatory therapies. Retrotransposon transcripts can trigger an interferon response leading to removal of these cells by the immune system [11, 70].
Regulation of retrotransposon silencing by heterochromatin
The above-mentioned examples of physiological roles for “transcriptionally active” retrotransposons and adverse effects of overt retrotransposon activity highlight the fact that transcriptional activity of retrotransposons needs to be tightly controlled. The majority of retrotransposons display low transcriptional activity in most cell types, suggesting that silencing mechanisms for ERVs counteract transcriptional activation. In the following sections we will summarize the current knowledge of ERV silencing pathways which lead to establishment of a repressive, heterochromatic, chromatin architecture on these elements. Different classes of retrotransposons feature distinct chromatin configurations preventing transcriptional activity. In this review, we will focus on a wide-spread mechanism of ERV silencing by the combinatorial accumulation of H3K9me3, H4K20me3 and DNA methylation, the classical modification pattern of pericentric heterochromatin.
Targeting mechanisms for ERV silencing
To ensure retrotransposon silencing, these genomic elements need to be recognized and targeted for heterochromatin formation. In vitro silencing assays in which parts of ERVs were combined with a reporter gene revealed high silencing potential of specific ERV sequences. For example, two regions of IAP elements can confer reporter silencing: the 5′UTR region [71] and a small 160 bp region from the gag coding sequence [73]. These and other examples suggest that recognition of ERVs may happen on the level of the DNA sequence by specific binding proteins. The first example for this mechanism was Zfp809 which binds the primer binding site region of exogenous MuLV retrovirus [95]. The sequence of this primer binding site is also conserved in endogenous retroviruses and Zfp809 was shown to bind these elements [96]. Zfp809 belongs to the large family of KRAB zinc finger (KRAB-ZnF) proteins with hundreds of members in vertebrate genomes. These proteins feature varying numbers of zinc fingers mediating DNA binding specificity. They also contain a KRAB domain which binds the corepressor protein Trim28, facilitating recruitment of additional chromatin-modifying activities. These data raised the hypothesis that the family of KRAB-ZnF proteins may have evolved to recognize different retrotransposons in higher vertebrate genomes. Consistent with this hypothesis, additional KRAB-ZnF proteins could be identified to bind retrotransposon sequences in human and mouse cells. ZNF91/93 recognize human SVA and L1 elements, respectively [40]. Zfp819 is a KRAB-ZnF recognizing IAP elements [81]. Zfp932 and Gm15446 were recently identified to bind distinct sets of ERVK retroviruses in the mouse [21].
In addition to the sequence-specific recognition by DNA binding proteins, RNA-mediated targeting mechanisms appear to play crucial roles in targeting retrotransposon silencing. In this context, three different mechanisms are currently discussed: generation of siRNAs, production of antisense transcripts and piRNA-mediated silencing.
An siRNA-based silencing pathway was shown to affect silencing of human Line1 elements [78, 92, 98]. It is currently unclear how these siRNAs are being generated and if additional retrotransposon classes are affected by this pathway. It should also be noted that abrogation of siRNA production by knock-down of Dicer only resulted in subtle transcriptional activation of LINE1 elements [98], suggesting that the siRNA pathway is not the predominant targeting mechanism for silencing.
Another RNA-based mechanism which can result in heterochromatin-mediated silencing is the production of antisense RNAs. Initially shown for imprinted genes, antisense transcription appears to be a wide-spread mechanism to modulate gene expression [94]. Antisense transcripts are produced from different retrotransposon classes and for LINE1 elements it could already be demonstrated that their activity is affected by these antisense transcripts [49]. Also IAP retrotransposons appear to be regulated by antisense transcripts. In this case, asRNAs appear to mediate targeting of histone modifying activities to establish a heterochromatic structure across these elements [7].
Finally, the piRNA pathway plays major roles in targeting retrotransposons for silencing, specifically in germ cells of animals [2]. The piRNAs originate from retrotransposon transcripts and not only contribute to the degradation of retrotransposon mRNA, but also induce repressive chromatin marks like DNA methylation [1]. In fact, the piRNA pathway is necessary for establishment of DNA methylation on retrotransposons in mouse fetal testes [46].
These examples highlight that targeting mechanisms may differ in different cell types as initiation of ERV silencing takes place mainly in germ line cells and early embryos. Later in development, when heterochromatin is already established on ERVs, maintenance mechanisms which copy the heterochromatic state during replication may act in concert with initiation processes to ensure robust ERV repression.
Establishment of repressive chromatin modifications on ERVs
Transcriptional silencing of retrotransposons is mediated by the establishment of a repressive chromatin structure which prevents the access and/or function of transcriptional activators. The precise mechanism of transcriptional repression is not fully clear, but establishment of both H3K9me3 and DNA methylation across ERVs have been found to play crucial roles in silencing. Other modifications, such as H4K20me3, or chromatin inaccessibility are contributing factors to ERV repression. Current models assume that specific sites within ERVs mediate recruitment of chromatin-modifying activities through, for example, KRAB ZnF proteins. These proteins interact with the Trim28 corepressor, which then results in the recruitment of the histone methyltransferase Setdb1. From these nucleation sites Setdb1 establishes H3K9me3 across the ERV body. Spreading of H3K9me3 can even extend into neighboring regions [68]. A nucleation and looping mechanism, such as proposed for pericentric heterochromatin [61], could explain the spreading of H3K9me3 from the nucleation sites across a broader region.
Trim28 and Setdb1 are both required for ERV silencing, although to different extent. Trim28 knockout in ES cells results in derepression of several ERV classes, such as IAP and MERVL [71]. Interestingly, Setdb1 knock-out ES cells show derepression of additional ERV classes [43, 58]. These data suggest that Trim28-mediated targeting of Setdb1 is restricted to a subset of ERVs. The reason may be that in cells in which H3K9me3 heterochromatin is already established, the de novo KRAB ZnF–Trim28–Setdb1 pathway may no longer be necessary on some ERV classes. H3K9me3 reader proteins, such as HP1, could bind preexisting H3K9me3 and bridge to Setdb1 through direct interaction [74]. However, depletion of all three HP1 isoforms in ES cells did not result in strong derepression of selected ERVs [56]. The reason for this discrepancy could be that the tested ERVs allow Setdb1 targeting though the KRAB ZnF–Trim28–Setdb1 initiation pathway. More refined analyses are, therefore, necessary to understand which ERV classes in ES cells mainly depend on the maintenance pathway, in which HP1 proteins could be instrumental.
Other H3K9me3-specific methyltransferases with roles in ERV heterochromatin formation are Suv39h enzymes. Suv39h double knockout ES cells display reduced spreading of H3K9me3 on intact ERVs and overall reduced H3K9me3 on LINE1 elements [9]. Targeting of Suv39h enzymes to ERVs and spreading of H3K9me3 may be due to HP1 interaction, as in pericentric heterochromatin. If Suv39h is recruited to LINE1 elements through transcription factor-based targeting, as proposed for pericentric heterochromatin [10], remains to be tested. The second major repressive modification involved in ERV silencing is DNA methylation. During germ cell development DNA methylation is erased and then re-established. The latter process is mediated through complex interactions between the piRNA pathway, de novo DNA methyltransferases and auxiliary proteins [1, 5, 57, 64]. Probably all other cell types maintain DNA methylation on ERVs by the activity of Dnmt1. ES cells, deficient for all three DNA methyltransferases completely lack DNA methylation on ERVs, but only display subtle derepression of these elements [43]. Acute deletion of Dnmt1, however, resulted in transient ERV derepression, which was later compensated by enhanced Setdb1 activity [75]. Interestingly, upon differentiation or in somatic cells loss of DNA methylation severely impairs ERV silencing [37, 38]. In contrast, loss of Trim28 or Setdb1 in mouse embryonic fibroblast (MEF) cells did not lead to an upregulation of retrotransposon expression, indicating a minor role for retrotransposon silencing in differentiated cells [58, 71]. These data led to the idea that in ES cells, the Setdb1-H3K9me3 pathway is predominant for silencing, whereas differentiated cells largely depend on maintenance of DNA methylation for ERV repression. However, this view was challenged by recent studies that investigated deletion of Setdb1 or Trim28 in differentiated cell types. For example, deletion of Setdb1 or Trim28 in neural progenitor cells resulted in strong ERV derepression, mainly of the IAP and MMERVK10C class, with only subtle reduction in DNA methylation [25, 80]. Other examples include deletion of Setdb1 in B cell development, which resulted in strong derepression of endogenous murine leukemia virus copies, again with only subtle reduction in DNA methylation [13, 63]. Interestingly, IAP retrotransposons, the major Trim28/Setdb1 targets in mouse ES cells, were not derepressed in B cells.
These findings show that although H3K9me3 and DNA methylation mostly occur together on ERVs, their impact on silencing can vary in different cell types. How can this discrepancy be reconciled? As explained above, transcriptional activity of ERVs depends on both activation and silencing mechanism. First, different cell types feature distinct transcription factor activities. If in ES cells, transcription factors that can target IAP elements are expressed, these factors may be absent in B cells. In this scenario, B cells would be unable to express IAP elements with compromised heterochromatin. Second, transcription factors have differential sensitivity towards DNA methylation. Some TFs can tolerate 5mC in their binding motif, for others, DNA methylation compromises DNA binding. Third, transcription factors and the associated activation machineries compete with the establishment of repressive modifications and their binders. For example, high levels of DNA methylation and H3K9me3 lead to strong binding of methyl-DNA binding proteins and HP1 proteins which may compete with transcription factor binding. In addition, H3K9me3 and DNA methylation can prevent activity of chromatin modifiers which establish active modifications, such as H3K4me3. Based on these arguments, ERV activation would depend on the quality and the amount of transcription factors able to bind ERVs vs. the amount of repressive modifications and binding proteins. This can explain, why in some cell types, presence of DNA methylation or H3K9me3 may be enough to counteract TFs, in others, combined presence of these modifications (and binding proteins) is required to ensure silencing. This idea is supported by recent experiments in which combined deletion of Setbd1 and Dnmt1 in ES cells resulted in much stronger derepression of IAP retrotransposons than individual deletion of Dnmt1 or Setdb1 [75].
Additional players in heterochromatin formation
DNA and histone methyltransferases are essential components for heterochromatin formation. However, additional players are likely to aid in both establishment and spreading of chromatin modifications and in the formation of a chromatin state, non-permissive to transcriptional activation. Genome-wide genetic screens using ERV reporters have revealed additional components of the ERV silencing machinery [73, 97]. In this section, we propose models for how these factors can be integrated into the ERV silencing pathways.
Atrx/Daxx/H3.3
A function of Atrx in heterochromatin establishment on ERVs was initially identified through an shRNA screen for regulators of IAP silencing. The short heterochromatin inducing (SHIN) region within the gag coding sequence of IAP elements can induce heterochromatin formation by recruiting the Trim28/Setdb1 pathway, and knock-down of Atrx was found to impair SHIN silencing [73]. Atrx is a putative chromatin remodeler and interacts with the histone chaperone Daxx which deposits histone H3.3 into heterochromatic regions [20, 33, 48]. A series of subsequent studies revealed that Atrx and Daxx are crucial for proper heterochromatin formation on imprinted loci, telomeres and ERVs [36, 73, 87, 90]. Their role in heterochromatin formation seems linked with histone H3.3 deposition. Consistent with this hypothesis, knock-out of H3.3 resulted in reduced H3K9me3 on telomeric and ERV heterochromatin [23, 87]. However, ERV silencing is only marginally impaired in ES cells lacking Atrx, Daxx, or H3.3 [23, 73].
We think that functions of Atrx and Daxx mainly relate to the efficiency of Setdb1-mediated heterochromatin formation. In vitro assays using the SHIN reporter revealed that reporter silencing is not fully compromised, but works with delayed kinetics [73]. Switching the SHIN reporter to an active state and monitoring re-silencing revealed impaired spreading of H3K9me3 in absence of Atrx [73]. Importantly, re-silencing was completely impaired when in addition to Atrx knock-down, reporter activation was forced using an inducible transcription factor [73]. Together, these data suggest that expansion of heterochromatin from initiation sites, such as the SHIN sequence, require the Atrx/Daxx pathway. Still, endogenous IAP elements containing the SHIN sequence are not strongly derepressed in Atrx/Daxx knock-out cells. Redundancy by multiple initiation sites within IAP elements that ensure significant levels of H3K9me3 even in absence of Atrx/Daxx and the presence of DNA methylation which may act as backup mechanism for ERV silencing may ensure IAP repression. In support of this hypothesis, removing DNA methylation [36] or reducing Setdb1 activity [73] resulted in impaired IAP silencing. It is also interesting to note that Atrx activity is more critical in cell types other than ES cells. In this context, knock-down of Atrx in morula embryos resulted in enhanced IAP expression [36]. Furthermore, Atrx/Daxx may be more crucial for silencing of other ERV classes with less redundant silencing mechanisms. An example for this is derepression of MusD/ETn elements in Atrx knock-out ES cells [73], although very little is known how these elements are targeted for silencing.
Is histone H3.3 deposition critical for the ERV silencing function of Atrx/Daxx? Based on the currently available datasets this question cannot satisfactorily be answered. Atrx and Daxx are necessary to mediate H3.3 deposition on several heterochromatic regions, however, regarding IAP elements the data are a bit conflicting. In one dataset using epitope-tagged H3.3, in Atrx depleted cells H3.3 was not lost from IAP elements [33]. Two other studies, using antibodies against endogenous H3.3, could detect reduced H3.3 on these repeat elements [23, 90]. It is possible that the epitope tag alters the properties of histone H3.3 and ChIP studies using the tagged histone variant may not fully reflect the endogenous situation. This argument would be in support of a role of Atrx/Daxx in depositing H3.3 on ERVs. But is this deposition critical for silencing? H3.3 knock-out ES cells display only minor changes in IAP expression, just like Atrx ko ES cells [23], which may be due to the redundancy in silencing mechanisms, e.g. DNA methylation. Unfortunately, co-impairment of these redundancy mechanisms was not yet performed in H3.3 ko cells. However, initiation of SHIN silencing which is independent of DNA methylation was fully intact in H3.3 depleted cells [73]. Interestingly, derepression of MusD/ETn elements, observed in Atrx ko cells, was not detected in H3.3 ko [23]. Based on these evidences we think that Atrx/Daxx have additional functions in ERV regulation which are beyond H3.3 deposition and which may relate to more direct roles in organizing an inaccessible heterochromatic structure across ERVs [73].
Sumoylation pathway
Sumoylation of proteins is an important regulatory mechanism for protein–protein interactions [16]. Several studies have revealed the importance of the SUMO pathway for heterochromatin formation [55, 86]. Recently, a genome-wide screen for provirus silencing factors has confirmed an important role of this pathway for ERV repression [97]. Some of the major ERV silencing factors are either sumoylated or contain SUMO interaction motifs. For example, sumoylation of Trim28 is important for the interaction with Setdb1 [39]. Additional proteins, such as hnRNP K, may support Trim28 sumoylation and are required for the efficient recruitment of Setdb1 to Trim28 binding sites [84]. Sumoylation of Trim28 appears to be crucial not only for Setdb1 interaction, but also for stable binding to its target sites [97].
Atf7ip, an interaction partner of Setdb1 also features a SUMO interaction motif [86]. Sumoylation of the methyl-DNA binding protein Mbd1 mediates the interaction with Atf7ip and could provide a link of the DNA methylation pathway with histone modifying activities [86]. The SUMO pathway appears to play additional roles for the Atrx/Daxx pathway. Silencing of the SHIN reporter is impaired in Daxx ko ES cells, but can be rescued by expression of full length Daxx protein in these ko cells [73]. Interestingly, Daxx protein with mutated SUMO interaction motif fails in rescuing SHIN silencing [73].
Together these data demonstrate the broad implications of the SUMO pathway for heterochromatin formation and ERV silencing. However, depletion of Sumo2 in ES cells only resulted in minor transcriptional changes of ERVs [97] compared to a full Setdb1 knock-out in ES cells [43], rather suggesting that the SUMO pathway is a modulating factor for ERV silencing. More detailed studies are, therefore, necessary to identify the most critical roles of sumoylation in the context of ERV silencing.
Re-establishment of heterochromatin upon challenges
Heterochromatin across ERVs impairs transcription by generating an inaccessible chromatin structure, non-permissive to transcription. However, heterochromatin is not in a permanently stable configuration as it faces various challenges. The major challenge in proliferating cells is replication. Replication leads to incorporation of new histone molecules, lacking repressive modifications. Furthermore, DNA methylation is only present on the parental strand and needs to be re-established. Replication, therefore, makes chromatin more accessible to transcriptional activators, and re-establishment of heterochromatin needs to dominate transcriptional activation to ensure silencing. Other challenges may include transcription through heterochromatic domains. This happens, for example, when ERVs are located in introns of transcribed genes. Another challenge may be DNA damage, which requires opening of the chromatin structure to repair the damage. Finally, binding of transcription factors with pioneering activities may access binding sites within heterochromatic structures and needs to be counteracted by re-establishment of heterochromatin.
All factors for ERV silencing were so far investigated in the context of heterochromatin challenges, at least in the context of replicating cells. Therefore, most factors necessary for ERV silencing are likely to relate to the re-establishment of heterochromatin in such contexts (Fig. 1). We think that different scenarios need to be considered, which depend on the kind of challenge and on the individual ERV element. For example, an ERV integration with binding sites for a transcription factor may be kept silent by heterochromatin-mediated blocking of productive binding of this transcription factor. However, upon a challenge which alters this repressive chromatin environment, e.g. replication, these transcription factor binding sites may get more accessible. To ensure silencing synergistic activities of the major heterochromatin establishment pathways are necessary to counteract transcriptional activation (Fig. 1). The Trim28/Setdb1 pathway can recognize heterochromatin nucleation sites through KRAB-ZnF proteins or could be recruited through remaining H3K9me3/HP1. Additional factors, such as Atrx/Daxx help in facilitating H3K9me3 spreading and establishment of an inaccessible chromatin structure. Re-establishment of DNA methylation is driven by the recognition of hemimethylated DNA and/or H3K9me3 by Uhrf1, which then mediates the recruitment of Dnmt1 [52]. Depending on the intensity of the transcriptional stimulus (e.g. binding affinity of the TF, number of TF binding sites, strength of the TF transactivation domain), compromised heterochromatin re-establishment may lead to transcriptional activation of the underlying ERV.
Fig. 1.
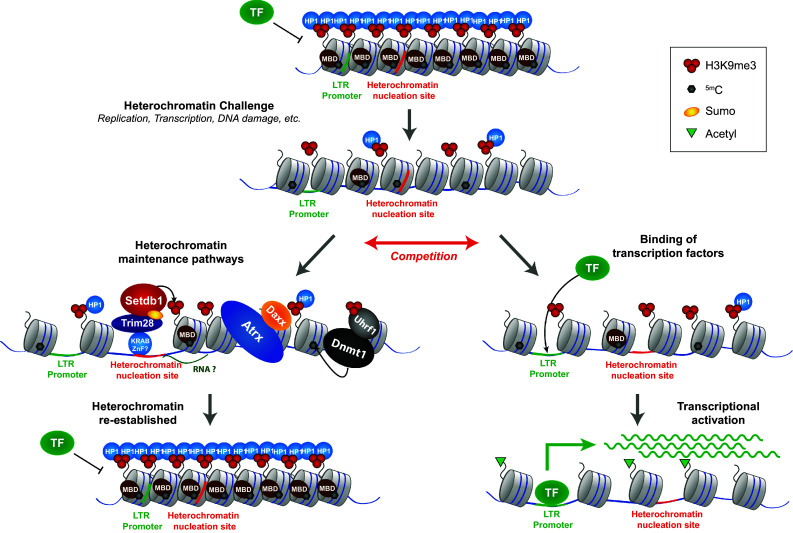
Heterochromatin establishment pathways act synergistically to re-establish heterochromatin upon challenges. The compact and less accessible state of heterochromatin can be compromised upon certain challenges, examples include replication, transcription through heterochromatin and DNA damage. The looser chromatin structure of challenged heterochromatin is more permissive to transcription factor binding. Competition between heterochromatin establishment pathways and transcription factor activity decides between re-establishment of heterochromatin or transcriptional activation of the ERV. If heterochromatin establishment pathways are compromised, e.g. upon Setdb1 or Dnmt1 knock-out, transcriptional activators dominate over heterochromatin re-establishment, leading to ERV derepression. MBD methyl DNA binding protein, TF transcription factor
A number of major questions remain. Heterochromatin nucleation sites are only known for very few ERV classes, and it is not clear which proteins (or RNA molecules) aid in recognition of these elements. The interplay between histone methylation and DNA methylation pathways is not fully understood. Transcriptional activation of ERVs also happens in a physiological context (e.g. B cell activation); what are the transcriptional activators and how can they counteract heterochromatin establishment factors? Is the transient derepression of ERVs really crucial for driving host gene expression during development? Can compromised heterochromatin and aberrant activation of ERVs lead to aberrant development or disease? Novel tools, such as genome-wide screens using CRISPR/Cas9, investigation of ERV silencing in developmental contexts and studies on the interplay between transcriptional activators and silencing factors will certainly reveal a more refined picture of the role of heterochromatin for ERV silencing in the near future.
Acknowledgements
Work in the lab of G.S. was funded by Deutsche Forschungsgemeinschaft (SFB 1064 and SPP1923).
References
- 1.Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Fejes Toth K, Bestor T, Hannon GJ. ) A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 3.Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci USA. 2004;101 Suppl 2:14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannert N, Kurth R. The evolutionary dynamics of human endogenous retroviral families. Annu Rev Genom Hum Genet. 2006;7:149–173. doi: 10.1146/annurev.genom.7.080505.115700. [DOI] [PubMed] [Google Scholar]
- 5.Barau J, Teissandier A, Zamudio N, Roy S, Nalesso V, Hérault Y, Guillou F, Bourc’his D. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science. 2016;354:909. doi: 10.1126/science.aah5143. [DOI] [PubMed] [Google Scholar]
- 6.Belancio VP, Roy-Engel AM, Deininger PL. All y’all need to know ‘bout retroelements in cancer. Semin Cancer Biol. 2010;20:200–210. doi: 10.1016/j.semcancer.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bierhoff H, Dammert MA, Brocks D, Dambacher S, Schotta G, Grummt I. Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol Cell. 2014;54:675–682. doi: 10.1016/j.molcel.2014.03.032. [DOI] [PubMed] [Google Scholar]
- 8.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 9.Bulut-Karslioglu A, De La Rosa-Velazquez IA, Ramirez F, Barenboim M, Onishi-Seebacher M, Arand J, Galan C, Winter GE, Engist B, Gerle B, et al. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol Cell. 2014;55:277–290. doi: 10.1016/j.molcel.2014.05.029. [DOI] [PubMed] [Google Scholar]
- 10.Bulut-Karslioglu A, Perrera V, Scaranaro M, de la Rosa-Velazquez IA, van de Nobelen S, Shukeir N, Popow J, Gerle B, Opravil S, Pagani M, et al. A transcription factor-based mechanism for mouse heterochromatin formation. Nat Struct Mol Biol. 2012;19:1023–1030. doi: 10.1038/nsmb.2382. [DOI] [PubMed] [Google Scholar]
- 11.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162:974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coffin JM, Hughes SH, Varmus HE. Retroviruses. NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 13.Collins PL, Kyle KE, Egawa T, Shinkai Y, Oltz EM. The histone methyltransferase SETDB1 represses endogenous and exogenous retroviruses in B lymphocytes. Proc Natl Acad Sci USA. 2015;112:8367–8372. doi: 10.1073/pnas.1422187112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordaux R, Hedges DJ, Herke SW, Batzer MA. Estimating the retrotransposition rate of human Alu elements. Gene. 2006;373:134–137. doi: 10.1016/j.gene.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 16.Cubenas-Potts C, Matunis MJ. SUMO: a multifaceted modifier of chromatin structure and function. Dev Cell. 2013;24:1–12. doi: 10.1016/j.devcel.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dewannieux M, Dupressoir A, Harper F, Pierron G, Heidmann T. Identification of autonomous IAP LTR retrotransposons mobile in mammalian cells. Nat Genet. 2004;36:534–539. doi: 10.1038/ng1353. [DOI] [PubMed] [Google Scholar]
- 19.Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 20.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ecco G, Cassano M, Kauzlaric A, Duc J, Coluccio A, Offner S, Imbeault M, Rowe HM, Turelli P, Trono D. Dev Cell. 2016;36:611–623. doi: 10.1016/j.devcel.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elisaphenko EA, Kolesnikov NN, Shevchenko AI, Rogozin IB, Nesterova TB, Brockdorff N, Zakian SM. A dual origin of the Xist gene from a protein-coding gene and a set of transposable elements. PloS One. 2008;3:e2521. doi: 10.1371/journal.pone.0002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elsasser SJ, Noh KM, Diaz N, Allis CD, Banaszynski LA. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015;522:240–244. doi: 10.1038/nature14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eymery A, Liu Z, Ozonov EA, Stadler MB, Peters AHFM. The methyltransferase Setdb1 is essential for meiosis and mitosis in mouse oocytes and early embryos. Development. 2016;143:2767. doi: 10.1242/dev.132746. [DOI] [PubMed] [Google Scholar]
- 25.Fasching L, Kapopoulou A, Sachdeva R, Petri R, Jonsson ME, Manne C, Turelli P, Jern P, Cammas F, Trono D, et al. TRIM28 represses transcription of endogenous retroviruses in neural progenitor cells. Cell Rep. 2015;10:20–28. doi: 10.1016/j.celrep.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 27.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fort A, Hashimoto K, Yamada D, Salimullah M, Keya CA, Saxena A, Bonetti A, Voineagu I, Bertin N, Kratz A, et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Genet. 2014;46:558–566. doi: 10.1038/ng.2965. [DOI] [PubMed] [Google Scholar]
- 29.Friedli M, Trono D. The developmental control of transposable elements and the evolution of higher species. Annu Rev Cell Dev Biol. 2015;31:429–451. doi: 10.1146/annurev-cellbio-100814-125514. [DOI] [PubMed] [Google Scholar]
- 30.Gifford R, Tristem M. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes. 2003;26:291–315. doi: 10.1023/A:1024455415443. [DOI] [PubMed] [Google Scholar]
- 31.Gifford WD, Pfaff SL, Macfarlan TS. Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol. 2013;23:218–226. doi: 10.1016/j.tcb.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Göke J, Lu X, Chan Y-S, Ng H-H, Ly L-H, Sachs F, Szczerbinska I. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell. 2015;16:135–141. doi: 10.1016/j.stem.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 33.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez-Sandoval A, Towbin Benjamin D, Kalck V, Cabianca Daphne S, Gaidatzis D, Hauer Michael H, Geng L, Wang L, Yang T, Wang X, et al. Perinuclear anchoring of H3K9-methylated chromatin stabilizes induced cell fate in C. elegans embryos. Cell. 2015;163:1333–1347. doi: 10.1016/j.cell.2015.10.066. [DOI] [PubMed] [Google Scholar]
- 35.Hancks DC, Kazazian HH. Active human retrotransposons: variation and disease. Curr Opin Genet Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He Q, Kim H, Huang R, Lu W, Tang M, Shi F, Yang D, Zhang X, Huang J, Liu D, et al. The Daxx/Atrx complex protects tandem repetitive elements during DNA hypomethylation by promoting H3K9 trimethylation. Cell Stem Cell. 2015;17:273–286. doi: 10.1016/j.stem.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hutnick LK, Golshani P, Namihira M, Xue Z, Matynia A, Yang XW, Silva AJ, Schweizer FE, Fan G. DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation. Hum Mol Genet. 2009;18:2875–2888. doi: 10.1093/hmg/ddp222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hutnick LK, Huang X, Loo TC, Ma Z, Fan G. Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism. J Biol Chem. 2010;285:21082–21091. doi: 10.1074/jbc.M110.125674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul GG, et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516:242–245. doi: 10.1038/nature13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev Genet. 2008;42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 42.Johnson WE. Endogenous retroviruses in the genomics era. Annu Rev Virol. 2015;2:135–159. doi: 10.1146/annurev-virology-100114-054945. [DOI] [PubMed] [Google Scholar]
- 43.Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelley DR, Hendrickson DG, Tenen D, Rinn JL. Transposable elements modulate human RNA abundance and splicing via specific RNA-protein interactions. Genome Biol. 2014;15:537. doi: 10.1186/s13059-014-0537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kramerov DA, Vassetzky NS. SINEs. Wiley Interdiscip Rev RNA. 2011;2:772–786. doi: 10.1002/wrna.91. [DOI] [PubMed] [Google Scholar]
- 46.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Consortium IHGS Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 48.Lewis PW, Elsaesser SJ, Noh K-M, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci USA. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Kannan M, Trivett AL, Liao H, Wu X, Akagi K, Symer DE. An antisense promoter in mouse L1 retrotransposon open reading frame-1 initiates expression of diverse fusion transcripts and limits retrotransposition. Nucleic Acids Res. 2014;42:4546–4562. doi: 10.1093/nar/gku091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li W, Lee M-H, Henderson L, Tyagi R, Bachani M, Steiner J, Campanac E, Hoffman DA, von Geldern G, Johnson K, et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci Transl Med. 2015;7:307ra153–307ra153. doi: 10.1126/scitranslmed.aac8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu S, Brind’Amour J, Karimi MM, Shirane K, Bogutz A, Lefebvre L, Sasaki H, Shinkai Y, Lorincz MC. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev. 2014;28:2041–2055. doi: 10.1101/gad.244848.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J, Koseki H, Wong J. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat Commun. 2013;4:1563. doi: 10.1038/ncomms2562. [DOI] [PubMed] [Google Scholar]
- 53.Lu X, Sachs F, Ramsay L, Jacques P-É, Göke J, Bourque G, Ng H-H. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol. 2014;21:423–425. doi: 10.1038/nsmb.2799. [DOI] [PubMed] [Google Scholar]
- 54.Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. ES cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maison C, Bailly D, Roche D, de Oca RM, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy J-P, et al. SUMOylation promotes de novo targeting of HP1[alpha] to pericentric heterochromatin. Nat Genet. 2011;43:220–227. doi: 10.1038/ng.765. [DOI] [PubMed] [Google Scholar]
- 56.Maksakova IA, Goyal P, Bullwinkel J, Brown JP, Bilenky M, Mager DL, Singh PB, Lorincz MC. H3K9me3-binding proteins are dispensable for SETDB1/H3K9me3-dependent retroviral silencing. Epigenet Chromatin. 2011;4:12–12. doi: 10.1186/1756-8935-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manakov SA, Pezic D, Marinov GK, Pastor WA, Sachidanandam R, Aravin AA. MIWI2 and MILI have differential effects on piRNA biogenesis and DNA methylation. Cell Rep. 2015;12:1234–1243. doi: 10.1016/j.celrep.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 59.Medstrand P, van de Lagemaat LN, Dunn CA, Landry JR, Svenback D, Mager DL. Impact of transposable elements on the evolution of mammalian gene regulation. Cytogenet Genome Res. 2005;110:342–352. doi: 10.1159/000084966. [DOI] [PubMed] [Google Scholar]
- 60.Mi S, Lee X, Li X-p, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang X-Y, Edouard P, Howes S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 61.Müller-Ott K, Erdel F, Matveeva A, Mallm JP, Rademacher A, Hahn M, Bauer C, Zhang Q, Kaltofen S, Schotta G, et al. Specificity, propagation, and memory of pericentric heterochromatin. Mol Syst Biol. 2014;10:746. doi: 10.15252/msb.20145377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Orgel LE, Crick FHC. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 63.Pasquarella A, Ebert A, Pereira de Almeida G, Hinterberger M, Kazerani M, Nuber A, Ellwart J, Klein L, Busslinger M, Schotta G. Retrotransposon derepression leads to activation of the unfolded protein response and apoptosis in pro-B cells. Development. 2016;143:1788–1799. doi: 10.1242/dev.130203. [DOI] [PubMed] [Google Scholar]
- 64.Pastor WA, Stroud H, Nee K, Liu W, Pezic D, Manakov S, Lee SA, Moissiard G, Zamudio N, Bourc’his D, et al. MORC1 represses transposable elements in the mouse male germline. Nat Commun. 2014;5:5795. doi: 10.1038/ncomms6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 66.Politz JC, Scalzo D, Groudine M. The redundancy of the mammalian heterochromatic compartment. Curr Opin Genet Dev. 2015;37:1–8. doi: 10.1016/j.gde.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramírez MA, Pericuesta E, Fernandez-Gonzalez R, Moreira P, Pintado B, Gutierrez-Adan A. Transcriptional and post-transcriptional regulation of retrotransposons IAP and MuERV-L affect pluripotency of mice ES cells. Reprod Biol Endocrinol. 2006;4:55–55. doi: 10.1186/1477-7827-4-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rebollo R, Karimi MM, Bilenky M, Gagnier L, Miceli-Royer K, Zhang Y, Goyal P, Keane TM, Jones S, Hirst M, et al. Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms. PLoS Genet. 2011;7:e1002301. doi: 10.1371/journal.pgen.1002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rebollo R, Romanish MT, Mager DL. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 2012;46:21–42. doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- 70.Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, Han H, Liang G, Jones PA, Pugh TJ, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell. 2015;162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 72.Rowe HM, Kapopoulou A, Corsinotti A, Fasching L, Macfarlan TS, Tarabay Y, Viville S, Jakobsson J, Pfaff SL, Trono D. TRIM28 repression of retrotransposon-based enhancers is necessary to preserve transcriptional dynamics in embryonic stem cells. Genome Res. 2013;23:452–461. doi: 10.1101/gr.147678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sadic D, Schmidt K, Groh S, Kondofersky I, Ellwart J, Fuchs C, Theis FJ, Schotta G. Atrx promotes heterochromatin formation at retrotransposons. EMBO Rep. 2015;16:836–850. doi: 10.15252/embr.201439937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sharif J, Endo TA, Nakayama M, Karimi MM, Shimada M, Katsuyama K, Goyal P, Brind’Amour J, Sun M-A, Sun Z, et al. Activation of endogenous retroviruses in dnmt1(/) ESCs involves disruption of SETDB1-mediated repression by NP95 binding to hemimethylated DNA. Cell Stem Cell. 2016;19:81–94. doi: 10.1016/j.stem.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 76.Slokar G, Hasler G. Human endogenous retroviruses as pathogenic factors in the development of schizophrenia. Front Psychiatry. 2015;6:183. doi: 10.3389/fpsyt.2015.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 78.Stein P, Rozhkov NV, Li F, Cárdenas FL, Davydenk O, Vandivier LE, Gregory BD, Hannon GJ, Schultz RM. Essential role for endogenous siRNAs during meiosis in mouse oocytes. PLoS Genet. 2015;11:e1005013. doi: 10.1371/journal.pgen.1005013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stocking C, Kozak CA. Murine endogenous retroviruses. Cell Mol Life Sci CMLS. 2008;65:3383–3398. doi: 10.1007/s00018-008-8497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tan SL, Nishi M, Ohtsuka T, Matsui T, Takemoto K, Kamio-Miura A, Aburatani H, Shinkai Y, Kageyama R. Essential roles of the histone methyltransferase ESET in the epigenetic control of neural progenitor cells during development. Development. 2012;139:3806–3816. doi: 10.1242/dev.082198. [DOI] [PubMed] [Google Scholar]
- 81.Tan X, Xu X, Elkenani M, Smorag L, Zechner U, Nolte J, Engel W, Pantakani DV. Zfp819, a novel KRAB-zinc finger protein, interacts with KAP1 and functions in genomic integrity maintenance of mouse embryonic stem cells. Stem Cell Res. 2013;11:1045–1059. doi: 10.1016/j.scr.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 82.Tanskanen JA, Sabot F, Vicient C, Schulman AH. Life without GAG: the BARE-2 retrotransposon as a parasite’s parasite. Gene. 2007;390:166–174. doi: 10.1016/j.gene.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 83.Theunissen TW, Friedli M, He Y, Planet E, O’Neil RC, Markoulaki S, Pontis J, Wang H, Iouranova A, Imbeault M, et al. molecular criteria for defining the naive human pluripotent state. Cell Stem Cell. 2016;19:502–515. doi: 10.1016/j.stem.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thompson PJ, Dulberg V, Moon K-M, Foster LJ, Chen C, Karimi MM, Lorincz MC. hnRNP K coordinates transcriptional silencing by SETDB1 in embryonic stem cells. PLoS Genet. 2015;11:e1004933. doi: 10.1371/journal.pgen.1004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thompson PJ, Macfarlan TS, Lorincz MC (2016) Long terminal repeats: from parasitic elements to building blocks of the transcriptional regulatory repertoire. Mol Cell 62:766–776 [DOI] [PMC free article] [PubMed]
- 86.Uchimura Y, Ichimura T, Uwada J, Tachibana T, Sugahara S, Nakao M, Saitoh H. Involvement of SUMO Modification in MBD1- and MCAF1-mediated Heterochromatin Formation. J Biol Chem. 2006;281:23180–23190. doi: 10.1074/jbc.M602280200. [DOI] [PubMed] [Google Scholar]
- 87.Udugama MM, Chang FT, Chan FL, Tang MC, Pickett HAR, McGhie JD, Mayne L, Collas P, Mann JR, Wong LH. Histone variant H3.3 provides the heterochromatic H3 lysine 9 tri-methylation mark at telomeres. Nucleic Acids Res. 2015;43:10227–10237. doi: 10.1093/nar/gkv847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Veselovska L, Smallwood SA, Saadeh H, Stewart KR, Krueger F, Maupetit-Méhouas S, Arnaud P, Tomizawa S-i, Andrews S, Kelsey G. Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape. Genome Biol. 2015;16:209. doi: 10.1186/s13059-015-0769-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Volkman HE, Stetson DB. The enemy within: endogenous retroelements and autoimmune disease. Nature Immunol. 2014;15:415–422. doi: 10.1038/ni.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Voon HP, Hughes JR, Rode C, De La R-V, Inti A, Jenuwein T, Feil R, Higgs DR, Gibbons RJ. ATRX plays a key role in maintaining silencing at interstitial heterochromatic loci and imprinted genes. Cell Rep. 2015;11:405–418. doi: 10.1016/j.celrep.2015.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 92.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 93.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 94.Werner A. Biological functions of natural antisense transcripts. BMC Biol. 2013;11:31. doi: 10.1186/1741-7007-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wolf G, Yang P, Fuchtbauer AC, Fuchtbauer EM, Silva AM, Park C, Wu W, Nielsen AL, Pedersen FS, Macfarlan TS. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 2015;29:538–554. doi: 10.1101/gad.252767.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang BX, El Farran CA, Guo HC, Yu T, Fang HT, Wang HF, Schlesinger S, Seah YF, Goh GY, Neo SP, et al. Systematic identification of factors for provirus silencing in embryonic stem cells. Cell. 2015;163:230–245. doi: 10.1016/j.cell.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang N, Kazazian HH. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 99.Young GR, Eksmond U, Salcedo R, Alexopoulou L, Stoye JP, Kassiotis G. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature. 2012;491:774–778. doi: 10.1038/nature11599. [DOI] [PMC free article] [PubMed] [Google Scholar]


