Abstract
After the characterization of the central pacemaker in the suprachiasmatic nucleus, the expression of clock genes was identified in several peripheral tissues including the immune system. The hierarchical control from the central clock to peripheral clocks extends to other functions including endocrine, metabolic, immune, and mitochondrial responses. Increasing evidence links the disruption of the clock genes expression with multiple diseases and aging. Chronodisruption is associated with alterations of the immune system, immunosenescence, impairment of energy metabolism, and reduction of pineal and extrapineal melatonin production. Regarding sepsis, a condition coursing with an exaggerated response of innate immunity, experimental and clinical data showed an alteration of circadian rhythms that reflects the loss of the normal oscillation of the clock. Moreover, recent data point to that some mediators of the immune system affects the normal function of the clock. Under specific conditions, this control disappears reactivating the immune response. So, it seems that clock gene disruption favors the innate immune response, which in turn induces the expression of proinflammatory mediators, causing a further alteration of the clock. Here, the clock control of the mitochondrial function turns off, leading to a bioenergetic decay and formation of reactive oxygen species that, in turn, activate the inflammasome. This arm of the innate immunity is responsible for the huge increase of interleukin-1β and entrance into a vicious cycle that could lead to the death of the patient. The broken clock is recovered by melatonin administration, that is accompanied by the normalization of the innate immunity and mitochondrial homeostasis. Thus, this review emphasizes the connection between clock genes, innate immunity and mitochondria in health and sepsis, and the role of melatonin to maintain clock homeostasis.
Keywords: Clock genes, Innate immunity, Oxidative stress, Mitochondria, Melatonin
Introduction
Sepsis is a type of inflammation produced by an exaggerated response of the immune system that may lead to septic shock and multiorgan failure (MOF) and, eventually, to death [1]. A third international consensus definitions have been proposed for sepsis and septic shock [2]. Sepsis is now defined as life-threatening organ dysfunction caused by a dysregulated host response to infection. Organ dysfunction should be identified as an acute change in total Sequential Organ Failure Assessment (SOFA) score ≥2 points consequent to the infection. Hospital mortality exceeds 40% in patients with septic shock with persisting hypotension requiring vasopressors to maintain mean artrial pressure ≥65 mmHg and having a serum lactate level >2 mM despite adequate volume resuscitation. As a result, even a modest degree of organ dysfunction is associated with an in-hospital mortality in excess of 10%. Although rapid intervention is required for a better diagnoses and management of sepsis, this disease is currently the first cause of death in the ICUs of any hospital worldwide [3, 4]. Better understanding the pathophysiology of sepsis and search for more effective therapies against this disease is, therefore, mandatory.
The initial response in sepsis: the NF-κB pathway
Sepsis involves the activation of the host innate immunity through a wide variety of physiological and pathological processes. Activators of the immune response are a series of antigenic structures named pathogen-associated molecular patterns (PAMPs); they are conserved patterns present in different microorganisms that activate specific receptors of the innate system [5]. Toll-like receptors (TLRs) play an essential role in the initiation of adaptive immune responses by recognizing these PAMPs, initiating the activation of NF-kappaB and other signaling pathways [6, 7]. TLRs are proteins belonging to the family of interleukin-1 (IL-1) receptors that play a key role in the recognition of microbial proteins, lipids, and nucleic acids existing in bacteria, parasites, and fungi, and other components derived from the host cell damage. Specifically, TLR4 is constitutively active and represent the main pathway to activate the nuclear factor kappa B (NF-κB) that, in turn, induces the transcription of a series of proinflammatory cytokines, adhesion molecules, and other molecules required for the activation of T cells [8].
TLR4 is expressed in the cell membrane of macrophages and neutrophils and, with CD14 and MD-2, form a receptor complex for bacterial lipopolysaccharides (LPS), which are major stimuli for triggering inflammation [9]. Canonical signaling of TLR4 depends on MyD88, an adaptor protein that connects the TLR4 complex domain Toll/IL-1 Receptor (TIR) with the active forms of the IL-1 receptor-associated kinases (IRAKs), inducing the activity of IRAK when TLR4 is activated. The phosphorylating activity of IRAK reduces its own affinity to MyD88, increasing that of TNF receptor-activated factor 6 (TRAF6) [9]. The IRAK/TRAF6 complex dissociates from the receptor, IRAK is eliminated through proteasomal degradation, and TRAF6 activates NF-κB in the cytosol [10]. An MyD88-independent pathway of TLR4 signaling may release cytosolic responses independent of NF-κB through other transcriptional factors and they may induce the expression of proinflammatory proteins [11–13].
NF-κB is constitutively present in the cytosol in an inactive form due to its binding to the inhibitor of κB (IκB). So, only after specific signals able to release IκB, NF-κB can be translocated to the nucleus and exert its transcriptional activity. There are five members of NF-κB/Rel family of transcription factors, coded by different genes, with a common N-terminal domain that includes a nuclear localization sequence, and different C-terminal domains [14, 15]. These features yield different homo- and heterodimers, with the p50/p65 heterodimer the most prominent pair of the NF-κB/Rel family. Whereas p50 facilitates the binding to DNA, p65 activates the gene transcription [16]. NF-κB activation and translocation to the nucleus require the dissociation of the NF-κB/IκB complex and inactivation of IκB.
Two pathways, canonical and non-canonical, have been described to explain the activation of NF-κB. Canonical pathway involves a series of steps after TLR4 activation that include the recruitment of serine/threonine specific IL-1 receptor-associated kinases-1 and 4 (IRAK-1 and IRAK-4, respectively). The interaction between MyD88 and these kinases induces the phosphorylation of the latter that, in subsequential steps, leads to the IκB kinase (IKK) complex activation [17]. IKK complex, constituted by IKKα and IKKβ, phosphorylates IκB that is further ubiquitinated and degraded by the proteasome, releasing NF-κB to the nucleus [14, 16, 18]. Once in the nucleus, NF-κB undergoes a series of phosphorylations and acetylations that modify the regulatory gene activity of the former due to its binding to coactivators such as CREB-binding protein and p300 (CBP/p300). Of note, deacetylation of NF-κB by deacetylases including histone deacetylase type 3 (HDAC3) and silent mating type information regulation 2 homolog (Sirt1) enhances the binding of NF-κB with IκB, forming again an NF-κB/IκB complex that is now translocated to the cytosol [19].
NF-κB transcriptional activity involves the regulation of more than 200 genes [20], yielding the expression of proinflammatory cytokines, adhesion molecules, and antioxidant enzymes. NF-κB also controls the expression of the NLRP3, a protein related to the activation of the inflammasome in sepsis and other inflammatory diseases. A variety of signals may trigger the inflammatory response to sepsis and, so, products from Gram− and Gram+ bacteria are able to interact with TLR4 (and also other TLRs such as TLR2) leading to a rapid NF-κB translocation to the nucleus and activation of gene transcription [21]. Greater levels of NF-κB in the nucleus are associated with higher rates of mortality in sepsis and a worse clinical outcome. The products of gene regulation by NF-κB involved in sepsis include cytokines (IL-1α, IL-1β, TNFα, IG-CSF, GM-CSF, etc.); chemokines (MIP-1α, MIP-2, IL-8, etc.); adhesion molecules (ICAM-I, VCAM-1, etc.); coagulation factors (tissue factor, etc.); proinflammatory and pro-oxidant enzymes (iNOS, COX-2, LOX, etc.); and antioxidant enzymes (SOD, GPx, etc.), among others [21, 22]. Some of these products positively feedback and activate the NF-κB pathway, enhancing the inflammatory response in sepsis. Among them, coagulation factors, which induce the formation of inflammatory mediators, IL-1β, and TNFα, participate in the amplification of the inflammatory response [23].
Consistent with the inflammatory response, there is an increase in the production of reactive oxygen (ROS) and nitrogen (RNS) species leading to a crosstalk of these molecules and NF-κB signaling [24]. On the one hand, activated leukocytes during the inflammatory response enhance the production of ROS, mainly superoxide anion (O·−2), which is dismutated to hydrogen peroxide (H2O2) by superoxide dismutases (SODs) [25], whereas TNFα further enhances the production of ROS [26]. Mitochondria are the greatest source of ROS, mainly O·−2 and H2O2 produced by electron leak during electron transfer system activity. The ROS produced suppress NF-κB activation, reducing NF-κB-dependent survival signaling and favoring the ROS-dependent cell death [26]. On the other hand, it was shown that mitochondrial ROS can promote, rather than inhibit, TNF-mediated NF-κB activation [24, 27, 28]. Although these ROS are controlled by the antioxidant system, mainly SOD and the glutathione cycle [29], H2O2 has important functions as an intracellular messenger [30]. An impaired mitochondrial function and an exaggerate response of the immune cells lead to an accumulation of ROS able to activate NF-κB in a dose-dependent manner [31, 32].
The production of RNS, mainly nitric oxide (NO·) and peroxynitrites (ONOO−), is formed by the activation of both cytosolic (iNOS) and mitochondrial (i-mtNOS) inducible nitric oxide synthases under the control of NF-κB. Whereas NO· is the primary product of these enzymes, it causes deleterious effects because it produces a systemic vasodilatation and hypotension, and increased cell damage [33, 34]. But NO· rapidly reacts with O·−2 yielding the highly toxic ONOO− [35]. This reaction is specially significant in the mitochondria, where ONOO− irreversibly inhibits the mitochondrial ETS complexes and the ATP synthase, reducing ATP formation [36–38] and favoring cell death.
Although the excess of ROS produced during sepsis triggers both apoptotic and necrotic cell death, depending on the severity of the oxidative stress [39], NF-κB targeted genes may promote cell survival. In fact, whereas ROS modulate NF-κB that also induces the expression of antioxidant genes, a crosstalk between NF-κB, TNFα, and JNK modulates the expression of ROS, promoting cell survival [40]. Collectively, the data support a main role of NF-κB in the inflammatory response to sepsis. However, the inhibition of NF-κB only may probably not be beneficial, because of its intervention at the same time in pro-survival pathways and in promoting cell death, depending on the mitochondrial involvement in the response. So, mitochondria may play a central role in the cell survival in sepsis.
The complementary response in sepsis: the NLRP3 inflammasome pathway
During the activation of the NF-κB pathway during sepsis, the subsequent damage to the cell releases a series of molecules or danger signals, named damage-associated molecular patterns (DAMPs) that, different from PAMPs, are recognized by a family of NLRs cytosolic receptors (nucleotide-binding domain and leucine-rich repeat containing receptors) [41]. Twenty-three members of this family of receptors have been identified in humans, and they share a structure composed by a leucine-rich repeat (LRR) C-terminal domain, which are repetitive enriched leucine sequences involved in the recognition of the ligand; a central NACHT domain, which is common to the NLRs with an ATPase activity required for protein oligomerization and formation of the active complexes named inflammasomes, and an effector N-terminal domain, related to the activation of caspases [42, 43]. NLRs have been classified into three main subfamilies: NALPs/NLRPs subfamily, which contains 14 proteins (NLRP1–14), all with a pyrin (PYR) domain N-terminal and involved in the formation of inflammasomes and in caspase-1 activation; IPAF subfamily, including a member with a caspase activation and recruitment domain (CARD) N-terminal domain, and the NOD subfamily including NOD1–4 proteins with an N-terminal CARD domain also [43]. NOD is a cytosolic receptor able to sense peptidoglycans after bacterial phagocytosis. NOD1 and NOD2 trigger a response involving in last instance the phosphorylation of IκBα by IKKs and its ubiquitination and proteasomal degradation, releasing NF-κB to the nucleus [44, 45].
One of the inflammasomes directly related with the inflammatory response during sepsis depends on the NLRP3 protein. The NLRP3 inflammasome complex is activated upon signs of cellular ‘danger’ that triggers innate immune defenses through the maturation of proinflammatory cytokines such as IL-1β. Some mutations of the nlrp3 gene yield huge amounts of this cytokine typical of the so-called autoinflammatory diseases or cryopyrinopathies [45, 46]. The NLRP3 inflammasome is a multiproteic complex constituted by the member of the NLR family, NLRP3 protein, the apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC), and a pro-caspase-1, which results in caspase-1 activation and cleavage of proinflammatory cytokines pro-IL-1β, pro-IL-18, and pro-IL33 to their mature forms [47]. NLRP3 protein is constitutively expressed in the cytosol under an inactive form due to the interaction between its NACHT-NAD and LRR domains, but a variety of danger signals can activate it to the inflammasome.
NLRP3 inflammasome activation occurs through molecular modifications that allow the oligomerization and further interaction of the NLRP3 with ASC through a PYR domain. Moreover, ASC has a CARD domain that interacts with the CARD domain of a pro-caspase-1, yielding an active caspase-1 [47]. The signals that trigger the inflammasome are, among other PAMPs and DAMPs, uric-acid crystals, ROS and mtDNA released from damaged mitochondria, and ATP from extracellular sources, [47, 48]. Extracellular ATP is associated with cell damage and necrosis, and it acts through the ATP-operated ion channels (P2X7) [43]. Although ROS produced by NADPH oxidases were initially related to the inflammasome activation [49], the mitochondria, as the main source of cellular ROS, were soon after identified as the main inducers of the inflammasome [50]. In parallel, it was shown that autophagy, which induces elimination of damaged mitochondria, prevents from ROS and mtDNA release and, thus, prevents inflammasome activation [51].
Physiology of mitochondrial function
From the data reported here, the connection between the NLRP3 inflammasome activation and mitochondrial dysfunction emerges as the main mechanism involved in the activation of this pathway of the innate immunity.
Mitochondria are the major providers of energy to the cell, and they make it in form of ATP through the basic process of respiration that is controlled by the availability of ATP itself, calcium, oxygen, nitric oxide, and proton leak, among others [52, 53]. The process of energy generation requires two main steps: one, the rapid oxidation of the specific substrates NADH and FADH produced during metabolic pathways including glycolysis, Krebs cycle and β-oxidation of fatty acids, and subsequent transport of electrons from these substrates to oxygen through the electron transfer system (ETS); two, the oxidative phosphorylation (OXPHOS) that involves the phosphorylation of one molecule of ADP and formation of one molecule of water [54]. The ETS is a multiprotein system involved in oxide-reduction reactions through the complexes I, II, III, and IV, and two electron carriers, coenzyme Q (CoQ) and cytochrome c (cyt c). Following the chemiosmosis hypothesis [54], during electron transfer, CI, CIII, and CIV pump protons against an electrochemical gradient from the mitochondrial matrix to the intermembrane space, generating a proton gradient along the inner mitochondrial membrane (IMM). These protons re-enter to the matrix through the CV or ATP synthase, and the energy of the gradient is dissipated and used to phosphorylate ADP. During these steps, the electrons reaching CIV reduce oxygen to water. OXPHOS produces more than the 90% of the ATP used by the cell. The energy released through the CV depends on the electrochemical gradient, i.e., the mitochondrial inner membrane potential (∆+μH); this means that the greater ∆+μH the more energy available for ATP production. But high ∆+μH favors electron leak that easily reduced the oxygen present in the inner mitochondrial space to O·−2 by increasing the formation of ROS.
The proton gradient can be used for other purposes besides the ATP production as it can be dissipated to heat to maintain the body temperature. This occurs when the protons pass through specialized uncoupling proteins (UCPs) to the mitochondrial matrix through the MIM instead of CV [55]. UCPs are proton transports across the IMM, driven only by the membrane potential with well-conserved regulation that includes free fatty acids and CoQ, among others [52, 56]. There are five members of the UCP family (UCP1–5); UCP1 is involved in the heat production in collaboration with thyroid hormones [57]; UCP2 has been related to neuroprotective properties [58, 59], and the roles for the other UCPs are not yet well established.
Besides their well-recognized roles, UCPs are involved in the control of ROS formation by the mitochondria. An excess of ROS inside these organelles may blunt their capacity to energy conservation, impairment of IMM, depletion of glutathione (GSH) and opening the mitochondrial permeability transition pore (MPTP); the latter releases death signals to the cytosol to induce apoptosis or necrosis [55, 60]. Mechanistically, UCPs act by opening a channel across the IMM that allows the transports of protons from the intermembrane space to the matrix; this process drops the electrochemical proton gradient across the IMM and, thus, the ∆+μH, reducing the formation of ROS. Besides ROS, other free radicals also produced in the mitochondria are RNS. Mitochondria contain NO·, which can come from the cytosol [61].
A series of studies demonstrated the presence of at least two NOS isoforms in the mitochondria, i.e., nNOS and iNOS. The former is a Ca2+-calmodulin-dependent enzyme that is constitutively expressed, whereas iNOS is an inducible enzyme. Both mitochondrial isoenzymes come from the cytosol, and they are coded by the same genes as the cytosolic forms [62–64]. Mitochondrial iNOS (i-mtNOS) that, unlike in the cytosol, is also constitutively expressed in the mitochondria contributes under physiological conditions to the intramitochondrial pool of NO· and, thus, participates with nNOS in the mitochondrial homeostasis, competing with oxygen for the binding site at CIV [36, 53, 65–67]. But under pathophysiological conditions such as inflammation, cytosolic and mitochondrial iNOS forms are significantly induced, yielding huge amounts of NO· and ONOO− that not only reduce the efficiency of the OXPHOS thereby reducing ATP formation, but also produce nitration of tyrosine residues that irreversibly inhibit the ETS complexes [37, 63–65, 67–70].
Mitochondria have their own antioxidant system to protect them against oxidative stress. A first line of defense may be the so-called mild uncoupling that prevents a strong rise in ∆+μH and, hence O·−2 production. Mitochondrial SOD dismutases O·−2 to H2O2, which can be transformed to the hydroxyl radical (·HO), the most toxic ROS. Moreover, the enzymes of the GSH cycle, glutathione peroxidase (GPx) and reductase (GRd) maintain the intramitochondrial pool of GSH preventing a rise in the GSSG/GSH ratio, which is critical to maintain the reduced state of the MPTP. Here, NADP(H) is of particular importance because it is required for recovering GSH from its oxidized form, GSSG. The magnitude of ∆+μH becomes critical again because if it is too high it favors the formation of ROS, whereas if it is too low, it reduces ATP production and oxidizes NADP(H) [58].
An additional component of the mitochondria to be consider here is the mtDNA. Mitochondria contain their own genome although with a modified genetic code. The mtDNA and multiple copies are located in the mitochondrial matrix. The mtDNA molecule is circular and double stranded of about 16.6 kpb. The heavy (H) strand contains most of the genes for two rRNAs, 14 tRNAs, and 12 proteins. The light (L) strand codes eight tRNAs and a polypeptide [71]. The genes do not contain introns and intergenic sequences are almost absent. The polypeptides coded by the mtDNA correspond to subunits of the CI, CIII, CIV, and CV. Replication of mtDNA is unidirectional and an mtDNA polymerase γ is required for the synthesis of mtDNA.
Mitochondria do not contain histones and the transcription factor mtTFA binds at regularly spaced intervals [71]. Due to its location in the mitochondrial matrix and the absence of histones, mtDNA is exposed to ROS that cause oxidative damage requiring mechanisms of repair. In this regard, mitochondria contain an efficient base excision repair mechanism that removes oxidized bases from the mtDNA. Adducts formed by exposure to UV light are removed by a nucleotide excision repair mechanism. Nevertheless, the constant exposure to ROS causes mtDNA base modifications including 8-hydroxydeoxyguanosine (8-HOdG), a G to C transversion, which constitutes a mutagenic lesion that is not recognized by mtDNA polymerase γ. This situation favors the continuous damage to mtDNA leading to more inefficient mitochondria along the time [72].
A last question to be considered here is that ROS in general, and particularly mitochondrial ROS, are not only deleterious molecules that should be removed from the body, but they play multiple physiological functions [73]. As it is known, the most oxygen consumption occurs during OXPHOS and most of the O·−2 formed is generated by the mitochondria [74]. Experimental data indicate that some of these ROS contribute to the maintenance of cellular homeostasis, although if they are produced in excess they can negatively affect survival [73]. ROS are produced in huge amounts by macrophages to eliminate bacteria [75]; they trigger programmed cell pathways and kill damaged and mutagenic cells; ROS also regulate cell proliferation [76]; they function as second messengers in the gut microbiota [77], and they control insulin release in pancreatic β-cells [78].
Mitochondria, the link between NF-κB and NLRP3 inflammasome in sepsis
Mitochondria are much more than simple powerhouses of the cell. In recent years, more functions were added to their ability to synthesize ATP and/or produce heat. Among them, mitochondria play a key role in Ca2+ homeostasis [79], and regulate the fate of the cell triggering apoptosis/necrosis pathways [80]. Mitochondria also are in constant dynamic change that affects the cell physiology and pathology [81], and produce RNS and ROS which are used also as signaling messengers. Recently, the finding that the NLRP3 inflammasome can be activated by mitochondrial ROS and mtDNA [50, 51] yields an unexpected new function for mitochondria in the control of the innate immunity. This directly connects mitochondria to severe inflammatory diseases including neurodegenerative diseases, metabolic diseases, and sepsis [82, 83].
A relationship between ROS and TLRs signaling is known. Mitochondrial ROS contribute to macrophage bactericidal activity, and it was shown that TLR4 activation enhances mitochondrial recruitment to macrophage phagosomes, increasing their production of ROS [75]. These data further demonstrate the importance of ROS in the antibacterial response of the innate immune system. ROS also regulate the NF-κB response, whereas the latter enhances the production of ROS [24]. Moreover, caspase-1 can be modulated by SOD in macrophages, linking ROS with sepsis and this piece of the NLRP3 inflammasome [84]. Other component of the NLRP3 inflammasome, ASC, regulates the interaction between caspase-1 and receptor interacting protein-2 (RIP2), leading to NF-κB activation [85]. A role for the NLRP3-dependent caspase-1 activation in the steps leading to proinflammatory cytokine maturation was elsewhere reported [86]. These and other data clarified the role of the NLRP3 inflammasome as a sensor of danger signals for the immune response [87], and the role of ROS/RNS in the inflammasome control [88]. In turn, the protective effects of reducing ROS production [84] or eliminating NLRP3 against liver ischemia–reperfusion or acetaminophen-induced injury in mice were reported [89, 90]. Thus, the relationship between NLRP3 inflammasome activation as the final point of ROS production was suggested [48]. Soon thereafter, thioredoxin was found to link ROS with NLRP3 inflammasome activation [91]; the former suppresses sepsis through the inhibition of the apoptotic signals trigger by mitochondria [92]. In addition to ROS, ATP was related also to the NLRP3 inflammasome activation [93]. Considering the data, it was not surprising that a link between mitochondria and NLRP3 inflammasome stimulation was proposed [94]. Soon after, the connection between mitochondria and NLRP3 inflammasome was documented [50]. At the same time, mitophagy, the process involved in the elimination of damaged mitochondria, was also related to NLRP3 inflammasome activation [51]. These data were consistent with the view that accumulation of damaged mitochondria favors the opening of the MPTP, releasing cyt c, ROS and mtDNA to the cytosol; all are confirmed as triggers of the inflammasome [95].
With these data, the relationship between danger signals that activates TLR4 inducing the NF-κB response of the innate immunity, mitochondrial impairment and ROS production, and the subsequent activation of the NLRP3 inflammasome, became evident (Fig. 1). Working together, the two pathways of the innate immunity, i.e., NF-κB and NLRP3 inflammasome, explain the exaggerated response of the innate immunity in some circumstances such as sepsis, due to the positive feedback between some cytokines matured by caspase-1 including IL-1β and the NF-κB pathway. This complex pathophysiological condition also explains the lack of efficacy of anti-inflammatory drugs in many of the diseases in which the NF-κB/NLRP3 inflammasome pathways are simultaneously activated [82, 96]. Consequently, the development of a new class of drugs that target NLRP3 will be very welcome [97].
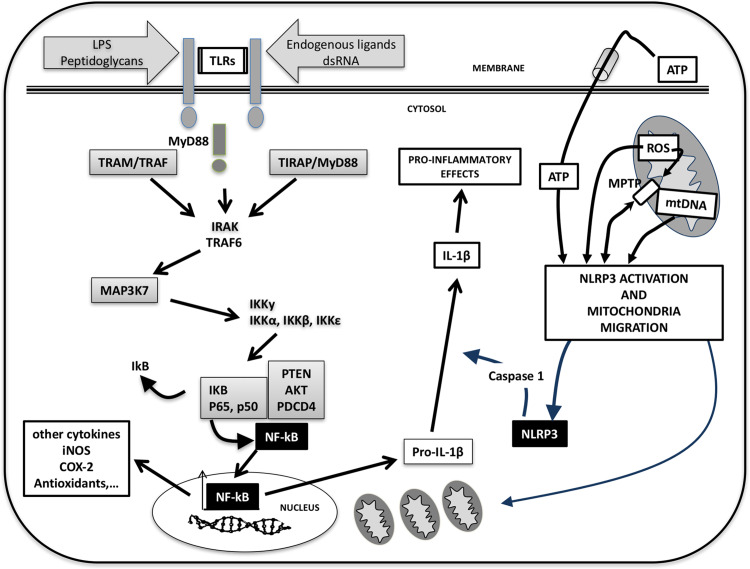
Fig. 1.
The connection between NF-κB and NLRP3 inflammasome during sepsis. Signals released by invaders microorganisms activate specific receptors (TLRs) in the cell membrane. This leads to changes in a series of kinases and adaptor proteins that ultimately release free NF-κB to the cytosol. Once in the nucleus, NF-κB activates the expression of multiple inflammatory proteins and proinflammatory cytokines including pro-IL-1β. But NF-κB also causes mitochondrial oxidative stress causing opening of the mitochondrial permeability transition (MPT), releasing free radicals and mtDNA to the cytosol. These molecules trigger the NLRP3 inflammasome that in turn activates a pro-caspase-1 inducing the maturation of IL-1β. Among others, IL-1β further activates the NF-κB response, closing the vicious cycle of the innate immunity during sepsis
From clock genes to mitochondrial dysfunction in sepsis
From a phylogenetic point of view, all living organisms adapted their vital functions to the environment. This constituted an important issue for species survival because it allows them to anticipate the climatic changes along the seasons. In this way, animals are able to regulate their mating behavior to favor offspring birth in spring or summer, when the availability of water and food, and the environment temperature are optimal for their survival. But seasonal reproductive behavior is only one of the biological rhythms conditioned by the environment. Most of them run under a period of 24 h and they are named “circadian” rhythms, which adapt physiological events to the daily photoperiod or light:dark cycle. Virtually every function in a living organism changes rhythmically along the 24 h period. The machinery underlying this type of adaptation constitutes the so-called “biological clock” and the photoperiod is the Zeitgeber that synchronizes the clock with the length of the day. Initially, the control of our circadian rhythms was attributed to the oscillations of activity in a group of about 20,000 neurons in the suprachiasmatic nucleus (SCN) of the hypothalamus. The SCN clock, entrained by the photoperiod, dictates the body’s 24-h rhythms [98]. This point of view has changed and we know that most peripheral cells also contains clocks working in a similar manner as the central clock, but they are influenced by other environmental cues or Zeitgebers, such as the timing of food consumption and exercise. It is believed that the central clock acts hierarchically to synchronize all peripheral clocks through the daily production of melatonin, which impart both clock and calendar information [99].
Circadian system organization
The light:dark cycle is the main cue for circadian entrainment in most animals. The retina possesses specialized photosensitive ganglionic cells using melanopsin as a visual pigment, which projects through the retinohypothalamic tract to the SCN for photic entrainment of the circadian clock [100, 101]. Retinohypothalamic tract releases glutamate in the SCN increasing clock gene expression. A secondary input to the SCN comes from the geniculohypothalamic tract that mainly releases GABA, thus modulating the excitatory signals from the retinohypothalamic tract [102]. Other pathways including serotoninergic afferents from the raphe nucleus and noradrenergic inputs from the locus coeruleus reach the SCN and may modulate the main photoperiodic entrainment signal [103]. In turn, the SCN sends two main efferent signals, chronobiotic and homeostatic ones [104]. The latter project mainly to the hypothalamus targeting the autonomic and neuroendocrine systems [105], and the former constitute the main signal to control the pineal synthesis of melatonin [106], which in turn feedback on clock genes [107]. By this means, the pineal production of melatonin follows a circadian rhythm peaking at night, when it is rapidly released to the blood and cerebrospinal fluid reaching all cells of the body [99].
The discovery of period (per) mutants in Drosophila melanogaster that yielded long- and short-period phenotypes was the starting point of clock genetics [108]. From a molecular point of view, four main genes, clock (circadian locomotor output cycles kaput), bmal1 (brain and muscle arnt like protein 1), per1 and per2 (periods), and cry1 and cry2 (cryptochromes) constitute the core of the biological clock. Moreover, rors (retinoid -related orphan receptors) and rev-erbα (reversed-viral erythroblastosis α) act as positive and negative modulators. Furthermore, chrono and its product, CHONO, have been identified as a core component of the circadian clock [109]. These genes and their products work in a transcriptional/translational feedback loops [110]. Here, BMAL1 interacts with CLOCK or NPAS2 (neuronal PAS domain-containing protein 2) and RORs favoring transcription genes, whereas CRY, PERs and REV-ERBα negatively regulate transcription. The dimers BMAL1:CLOCK or BMAL1:NPSS2 bind to over 6000 sites in the chromatin, which correspond to approximately 3000 genes [111]. Among them, BMAL1:CLOCK or BMAL1:NPSS2 (it has been proposed that NPSS2 is a redundant gene) upregulate the expression of cry and per genes. Cry and Per proteins form homo- and heterodimers in the cytosol and then enter the nucleus where they bind to and inactivate BMAL1:CLOCK dimer, downregulating the expression of the formers. In this situation, a further reduction in the relative levels of CRY and PER does not inhibit BMAL1:CLOCK, starting a new cycle of the clock [112]. In this loop, RORα and REV-ERBα act to repress and enhance BMAL1 transcription, respectively [113, 114].
Once the clock genes were identified in the SCN, their expression was reported in most of the tissues with similar pattern of functioning [115]. This means that almost all cells of the body may express a clock to control their own biological cycle. However, the existence of billions of clocks working freely without coordination between them is not understandable from a homeostatic point of view. Thus, the circadian organization of the body suggested that the central biological clock located in the SCN works in a hierarchical fashion as the master circadian pacemaker controlling the peripheral clocks [103, 116]. Thus, we can consider the circadian system as a multioscillatory timing system controlling multiple central and peripheral rhythms; the SCN clock being the central pacemaker that uses homeostatic and endocrine signals to synchronize the peripheral clocks. In turn, peripheral clocks may yield rhythmic signals that could reinforce the circadian rhythm generating system [117].
Clock genes and the innate immunity
The immune system activity oscillates with a 24 h period, suggesting that a connection with the circadian system should exist. Daily variations in blood T and B lymphocytes, monocytes, macrophages, and other cells of the immune system have been described. This is also applicable to the daily oscillation in blood levels of proinflammatory cytokines [118]. These rhythms are controlled by the central pacemaker, and cortisol seems to be the mediator, because changes in cortisol levels disrupt the T lymphocyte rhythm [119]. Circadian changes in the immune system occur also after its activation by inflammatory signals such as LPS [120].
These data point to the control of the innate immunity by the clock proteins CLOCK, BMAL1, RORα, and REV-ERBα (Fig. 2). We can consider the immune system in two states: an state of anticipation (a typical feature that the biological clock allows) and increased activity, and a subsequent state of repair and regeneration [121]. In mice, during the transition to their active phase (at night), the immune system anticipates an elevated risk of infection. In these conditions, however, there exists an elevated risk of sepsis due to an exaggerated response of the immune system and greater induction of proinflammatory cytokines after LPS. During the remaining phase of mice, i.e., in the day, the ability of LPS to induce an inflammatory response is decreased, leading to a smaller capacity of the innate immune response [121]. This situation depends in part on BMAL1, which is the central mediator of the innate immunity. Thus, BMAL1 inhibits the expression of the chemokine (C-C motif) ligand 2 (Ccl2), which in turn reduces the proinflammatory monocytes [122]. Another two players are involved in this game. In fact, BMAL1 induces the expression of rorα, increasing the content of RORα that binds to an RORE sequence in the promotor of bmal1 enhancing its expression. Moreover, RORα also induces the expression of IκB, the inhibitor of NF-κB, blocking its activation and reducing its translocation to the nucleus. The importance of RORα in the control of the immune response was confirmed since mice deficient in RORα have an enhanced activity of their immunity and they are more susceptible to sepsis [123]. But BMAL1 also induces rev-erbα expression and content of REV-ERBα, which binds to the same RORE in the promoter of bmal1 suppressing its expression, thus triggering the NF-κB-dependent inflammatory pathway [124]. Although it was suggested that REV-ERBα may also repress CCL2, acting as an anti-inflammatory molecule [120], there is consensus regarding its proinflammatory roles. The consequence of BMAL1 activity is the reduction of the NF-κB-dependent immune response, decreasing the expression of proinflammatory molecules including iNOS, COX-2, cytokines, and other mediators of the inflammation. Thus, BMAL1 functions as an anti-inflammatory molecule controlling the rhythmic activity of the innate immunity under basal conditions, but also preventing an exaggerated immune response against infection.
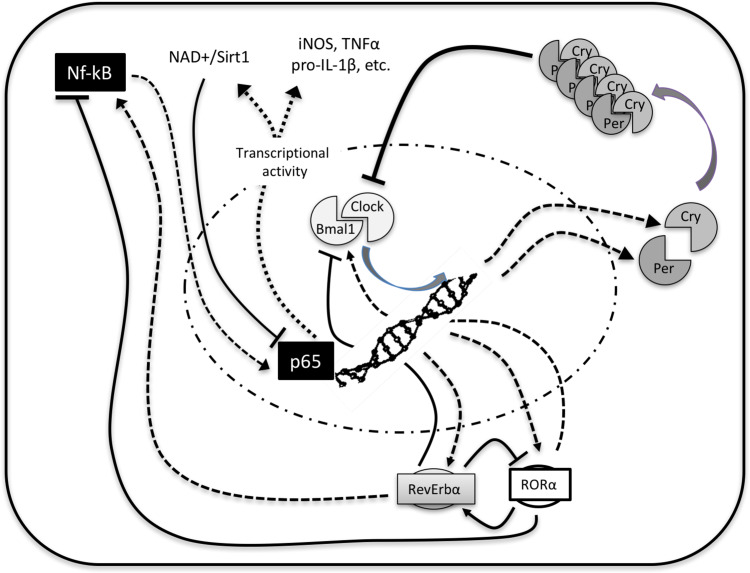
Fig. 2.
A simplified scheme of the clock-NF-κB connection. BMAL1:CLOCK constitutes a loop controlled by PER:CRY heterodimers. Over them, RORα enhances BMAL1 and REV-ERBα represses it. Among other mechanisms, BMAL1 regulates nampt expression and the NAMPT protein that synthesizes NAD+, the cofactor for SIRT1. The deacetylase activity of SIRT1 inactivates NF-κB, controlling the immune response. When REV-ERBα dominates, blocks BMAL1 releasing the inflammatory activity of NF-κB
CLOCK coexists in the protein complex with the p65 subunit of NF-κB, enhancing its phosphorylation and acetylation, and increasing its transcriptional activity [125]. Thus, CLOCK and BMAL1 exert opposite effects on immunity and BMAL1, recruiting CLOCK, prevents the proinflammatory effects of the latter. Another two proteins of the clock, PER and CRY, also modulate the immune system. Although there are three PER proteins, PER2 seems to be the most significant in controlling the immune system. PER2 controls the production of INFγ and IL-1β, and the absence of PER2 significantly reduces these cytokines in response to sepsis. But PER2 binds to block the BMAL1:CLOCK activity, enhancing the inflammatory response [126]. PER2 may also inhibit the activity of REV-ERBα, thereby having a more complex role in the immunity [113]. Thus, whereas PER2 controls the circadian production of some cytokines, it promotes inflammation by reducing the activity of BMAL1. The cryptochromes CRY1 and CRY2 are other supervisors of the BMAL1:CLOCK function. In this case, the lack of CRY1 and CRY2 triggers the production of iNOS, IL-6, and TNFα, reflecting a proinflammatory condition. The effects of these cryptochromes depend on their activation of cAMP-dependent NF-κB phosphorylation and subsequently activation [127].
The fact that CLOCK is a histone acetylase (HAT) that modulates NF-κB transcriptional activity through the acetylation of its p65 unit suggests that other acetylases/deacetylases may also influence NF-κB and/or CLOCK itself. Class III mammalian histone deacetylases (HDACs) include sirtuins 1–7, and they sense cellular energy metabolism [128]. The most studied sirtuins is SIRT1, a nuclear protein involved in metabolism control. It is a deacetylase that uses NADH/NAD+ to remove an acetyl group from its substrates and it is involved in metabolic control and aging [128]. Because metabolism and SIRT1 deacetylase activity oscillate with a daily period, a relationship between the pacemaker and SIRT1 was proposed [129]. BMAL1, CLOCK, and SIRT1 associate together and locate at the promoters of clock-controlled genes favor the circadian gene expression. It was proposed that SIRT1 interacts directly with CLOCK and interacts with the BMAL1:CLOCK complex, controlling the circadian histone acetylation by CLOCK. The circadian function of BMAL1 is regulated by its acetylation by CLOCK, and it has been suggested that deacetylation of BMAL1 by SIRT1 contributes to its activity [129]. But SIRT1 activity depends on the circadian production of nicotinamide phosphoribosyl transferase (NAMPT), the limiting enzyme required for the synthesis of NAD+ that is under the control of BMAL1. SIRT1 controls its own activity inducing the expression of NAMPT due to its interaction with BMAL1:CLOCK, which means that NAMPT, NAD+, and SIRT1 activity are under circadian control, constituting a loop with BMAL1:CLOCK complex [130].
Inflammation disrupts the molecular clock through multiple pathways. On the one hand, inflammation represses both expression and oscillation of the clock genes, effects related to the inhibitory roles of TNFα and IL-1β, and probably NF-κB itself, on the transcriptional activity of the BMAL1:CLOCK complex [131]. The feedback connection between clock genes and inflammation further explains the exaggerated response of the innate immunity in some conditions such as sepsis.
Clock genes and mitochondrial function
Although mitochondria are related commonly to the main source of ATP in the cell, they have many other functions including the energy-to-heat production balance, calcium homeostasis, and apoptosis/necrosis control of the cell death [60]. These functions are directly linked to the cell metabolism and, so, it is not surprising that mitochondrial activity could also fluctuate in a clock-dependent fashion [132, 133]. Rhythms in some genes related to glucose and lipid metabolism, among others, are rhythmically expressed in mouse liver [134]. The circadian expression of hepatic metabolic enzymes is of high importance because clock gene dysfunction leads to metabolic disorders. As an example, bmal1 deletion produces hypoglycemia and clock mutations induce obesity and metabolic syndrome [135]. Because mitochondria are involved in some of these metabolic pathways, a relationship between clock genes, metabolism, and mitochondria is emerging. The expression of pgc1α (peroxisome proliferator-activated receptor gamma coactivator 1-alpha) oscillates with per2 in peripheral tissues including the liver and skeletal muscle, regulating energy metabolism. PGC1α through coactivation of RORα induces the expression of bmal1 and rev-erbα [136]. In this loop of clock genes modulation SIRT1 participates; it deacetylates PGC-1α promoting its activation [137]. So, PGC-1α constitutes a link between clock genes and metabolism, with RORα as an intermediary signal. But SIRT1 and its substrate PGC-1α are present in the mitochondrial where they regulate energy metabolism [138]. They activate the mitochondrial transcription factor A (TFAM) that regulates mtDNA copy number and its transcriptional activity [139].
Taking into account the data above mentioned, the connection between clock genes and mitochondrial function can be established at several levels. On the one hand, in its active form, CRY is able to inhibit the expression of some enzymes of the gluconeogenesis pathway, whereas AMP-activated kinase (AMPK) phosphorylates and inactivates CRY. This situation leads to a modification of the clock function because CRY is a repressor of the BMAL1/CLOCK complex. Here, BMAL1 enhances rorα expression, favoring the expression of bmal1 and rev-erbα. In a following step, BMAL1:CLOCK dimer enhances the expression of cry, again initiating the cycle. In this loop, SIRT1/PGC1α participates in regulating mitochondrial functional capacity. So, it can be summarized that CRY influences mitochondrial activity reducing gluconeogenesis and favoring the substrates for OXPHOS through glycolysis, enhancing ATP production. AMPK senses the energy increase, reducing CRY activity and, thus, ATP production by the mitochondria. Now, CRY inactivation releases BMAL:CLOCK that initiates a new cycle enhancing cry, nampt, and rorα expression, whereas the latter, with SIRT1/PGC1α, further induces the expression of bmal1 and mitochondrial biogenesis. REV-ERBα acts as a repressor of BMAL1, modulating clock genes function. Thus, mitochondrial dynamics, biogenesis, and function are connected in a circadian manner with clock oscillations [140].
When speaking about mitochondria, we must have in mind that they produce ATP because they consume oxygen. Approximately, 95% of the oxygen entering the body is used by the mitochondria during OXPHOS. But some of the O2 is partially reduced by one, two or three electrons yielding O·−,2 H2O2, and ·HO [141, 142], a group of ROS that may impair mitochondrial function. Mitochondria also produce RNS, mainly NO· and ONOO−, a product formed through the reaction of NO· with O·−2 [143]. ONOO− is an extremely reactive compound that irreversibly damages the ETC complexes leading to mitochondrial failure [144]. Moreover, normal mitochondria are the main source of O·−2 in the cytosol, where it can also react with NO· forming ONOO− in circumstances where there is an excess of O·−2 production [145]. However, NO· plays a central role in mitochondrial bioenergetics through the modulation of the oxygen consumption. In fact, NO· competes with O2 for the same site in the cytochrome c oxidase [65]. The excess of these ROS/RNS is normally eradicated by the antioxidative defense system. In some circumstances, however, when the production of ROS is elevated or the antioxidant defense is insufficient, the levels or ROS rise generating oxidative damage. This is particularly significant during inflammatory processes, when ROS and RNS increase.
Because mitochondrial production of ROS is a normal consequence of mitochondrial metabolism, one wonders whether ROS formation also oscillates along the 24 h period. There is evidence showing that ROS levels change along the day in several tissues of mice, and bmal1-deficient mice produce high levels of ROS that have been related to the short life-span of these animals [146]. In human red blood cells, rhythms in the peroxiredoxin oxidation have been reported [147]. Moreover, changes in ROS formation are also related to a similar pattern of changes in LPO and protein oxidation [148]. The antioxidant defense is also under the control of the clock that regulates the expression of the antioxidant enzymes, probably acting through NF-κB and Nrf2 expression. ROS produce damage at multiple levels, and besides its elimination, the molecules, specially DNA, should be repaired. In this regard, the nucleotide excision repair systems oscillate also with a 24 h period [149]. If the daily changes in ROS production directly depend on the pacemaker or they are a consequence of the metabolic rhythms, remains to be clarified. However, ROS can damage sufficiently the cell to trigger cell death events. Among others, autophagy is related to remove damaged organelles from the cell when they are malfunctioning, normally due to oxidative damage. It is known that genes associated with autophagy and autophagic activity itself are under the control of the biological clock [110, 150]. Thus, the disruption of clock genes impedes a normal program of autophagy, leading to damaged mitochondrial accumulation, contributing to disease [82, 83, 96].
Central vs. peripheral clocks
It has been mentioned that the oscillations in SCN clock genes expression lead to a parallel protein rhythm and function. The transcription translation oscillating (TTO) model served to explain the oscillatory regulation of the clock-controlled genes [133, 151]. However, post-translational modifications of the clock proteins, mainly phosphorylation/dephosphorylation of PER proteins, yield an additional regulatory control of the clock [152]. Rhythmic clock gene expression is not limited to the central pacemaker, and it is also detected in most of the cells in the organism where they control metabolic and other rhythms [151]. Studies on cardiomyocytes showed the presence of a circadian clock in the heart [153], leading to identify its response to lipid metabolism. In fact, food intake serves as synchronizer for peripheral clocks but lesions of the SCN impede this effect [154].
The presence and functions of the circadian clock in the immune system are now an important matter of study. It oscillates in several immune cells including macrophages, B cells, and dendritic cells [155]. Importantly, the expression of some TLR receptors that recognize PAMPs such as TLR9 shows a circadian rhythm that is dependent on PER2 [155]. The response of the immune cells also changes along the day, and mice show increased T cell response when they are immunized during the day than at night [156]. These peripheral clocks hierarchically depend on the central pacemaker. Lesions in the SCN lead to a loss in the rhythm of peripheral clocks, suggesting that the latter are not autonomously rhythmic but require its regulation by the SCN clock [157]. Also, the SCN lesion leads to the impairment of the melatonin rhythm, blunting its role as endogenous synchronizer and loss of peripheral rhythms. Even more, deletion of a clock gene may affect the function of several organs [158]. Thus, the central pacemaker, through multiple signals including neurotransmitters signal inside the brain, endocrine (melatonin and corticoids), and nervous (through the autonomic nervous system) signals to peripheral organs, orchestrated the peripheral clocks maintaining them in synchrony [151, 157].
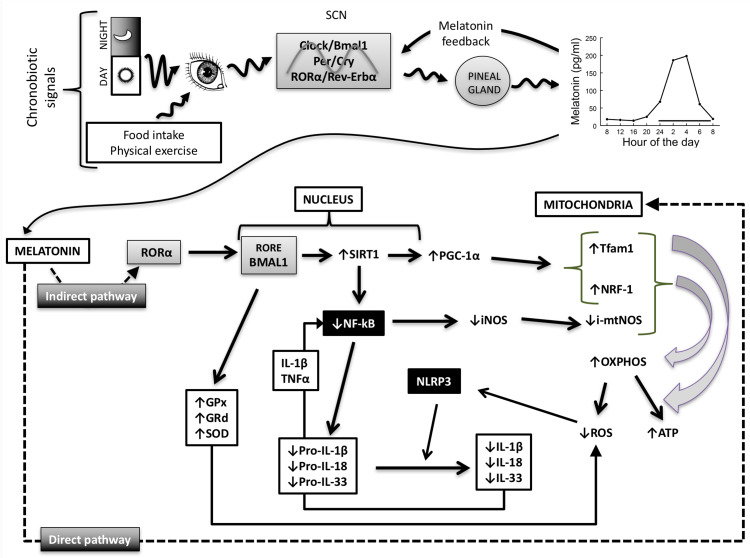
Melatonin-mitochondria interplay in sepsis
Melatonin and clock genes
Melatonin, N-acetyl-5-methoxytryptamine, is a product of tryptophan and derived from 5-hydroxytryptamine (serotonin). Initially reported as a product of the pineal gland [159, 160], it is now evident that melatonin is synthesized in most organs and tissues via the same enzymatic machinery as in the pineal gland [161]. Unlike pineal melatonin, extrapineal melatonin does not follow a circadian rhythm, and its concentrations are one or two orders of magnitude higher than pineal melatonin. Moreover, it is unknown to date if its synthesis is controlled by clock genes [162]. Pineal melatonin production is under the control of the central pacemaker that transforms the photoperiodic signal in an endocrine one. Melatonin mechanisms of action include its effect through specific MT1 and MT2 membrane receptors that mediated chronobiotic properties of the indoleamine [163]; genomic effects, through nuclear receptors belonging to the ROR/RZR family of nuclear transcription factors [164], and regulatory effects through its binding to calmodulin and calreticulin, two proteins related to calcium homeostasis. The binding of melatonin to these proteins shows a K d in the low nanomolar range, and it fulfills pharmacological features of a receptor [165, 166]. Melatonin also regulates mitochondrial homeostasis by several mechanisms including nuclear and mitochondrial DNA transcriptional activity [60, 167]. Mitochondria have not been proven definitively to possess melatonin receptors; however, they may be a site of melatonin synthesis [168].
Perhaps the most intriguing feature of melatonin is the differences in the way it is handled by the pineal and extrapineal sources. Pineal melatonin is not stored in the gland, where it is released into the blood and cerebrospinal fluid soon after it is produced; this occurs with the typical nocturnal peak. This rhythm conveys a chronobiotic signal that synchronizes multiple functions of the body to a 24 h day/night cycle [99]. The maximal concentration of melatonin in blood at night rarely exceeds 1 nM, although it is in much higher levels in the CSF [161, 169].
Pineal melatonin synthesis is controlled by the central biological clock in the SCN, which generates circadian rhythms through the transcriptional/translational feedback loop described above. This clock machinery is entrained by the light:dark cycle that induces the expression of bmal1 and clock at night, with the corresponding translation into BMAL1 and CLOCK proteins some hours later. Then, the BMAL1/CLOCK complex triggers the expression of per and cry yielding the proteins PER and CRY that accumulates during the day and, once they reached a critical levels, forms heterodimers that enter the nucleus to negatively feedback the BMAL1/CLOCK complex [170]. During the first phase of the clock genes loop, i.e., at night, is when melatonin synthesis in the pineal gland starts. The signals coming from the SCN to the pineal gland constitute a polysynaptic pathway culminating in the release of norepinephrine on the pinealocyte that induces the expression of arylalkylamine N-acetyltransferase (AANAT) [171, 172]. Although it was suggested for years that AANAT is the limiting enzyme in melatonin synthesis, it is currently suggested that acetylserotonin-O-methyltransferase (ASMT) is actually the enzyme that limits the synthesis of the indoleamine [173].
Melatonin also can influence the SCN pacemaker through a feedback pathway that closes the loop of the central regulation of the circadian rhythms. One hypothesis suggests that melatonin stabilizes clock proteins in the SCN transcribed during the dark phase of the light:dark cycle via the ubiquitin–proteasome system [107]. This feedback pathway implies melatonin to maintain rhythm stability [174]. Some possibilities arise for this effect of melatonin. On the one hand, melatonin exerts an inhibitory role on glutamatergic activity in several areas of the brain independently of MT1 and MT2 receptors [175–177] and, thus, the indoleamine may modulate the glutamatergic activation of the SCN conveyed by the retinohypothalamic tract. On the other hand, it was shown that melatonin phase-shifts bmal1 and rev-erbα expression [178], which could be produced through its own receptors described in the SCN [179]. These double mechanisms of action of melatonin in the SCN may account for the two distinct actions of the indoleamine elsewhere reported [180]: One of them, an acute inhibitory effect on neuronal firing mediated by glutamatergic and or glutamatergic-related events, and the second that involves phase-shifting of the clock. In any event, the activity of melatonin on the central pacemaker could serve to fine tunning the circadian clock, informing it of the correct time and amplitude of its rhythm [181].
Melatonin and the NF-κB pathway during sepsis
The beneficial effects of melatonin in sepsis was shown in our pioneer study demonstrating that melatonin administration inhibited in a dose-dependent manner the inflammatory response triggered by LPS administration to rats [182]. Three-month-old animals received 10 mg/kg bw of LPS serotype 0127:B8, a dose that produces acute sepsis and MOF, and the rats die 6 h later. Groups of animals received melatonin at different doses from 0 to 60 mg/kg bw. The mRNA expression, protein content, and enzymatic activity of iNOS, and NO· production were totally counteracted by melatonin in the liver and lungs of the rats. Even more, melatonin absolutely prevented liver, kidney, and cardiovascular dysfunction in these animals, restoring the normal carbohydrate, lipid, and protein metabolism. Of note, septic rats were only treated with melatonin, which was able to recover rat from septic shock-mediated MOF.
Because both the incidence and mortality of sepsis further increase with age, the next step was to evaluate the antiseptic properties of melatonin in aged rats injected with LPS. For this purpose, 18 months old rats were treated following the same protocol as before, and iNOS expression and activity, NO· production, and LPO levels were analyzed in liver and lungs. The inflammatory reaction was higher in aged rats than previously recorded in young animals. Melatonin administration to aged rats at the same doses as injected to young animals totally prevented the inflammatory reaction and the MOF [70]. Compared with the effects in young rats, the efficacy of melatonin to recover aged animals from sepsis was higher. These data imply that melatonin is more efficient in recovering to a healthy condition when the injury is greater.
The experimental model of acute sepsis induced by LPS is not completely comparable with clinical sepsis, and the effects of melatonin were analyzed in the model of sepsis induced by CLP in mice. With this model, the development of sepsis is slower than with LPS, and the maximal survival of animals reaches 35 h. We also used inos +/+ and inos −/− mice strains to analyze the effects of NF-κB activation on the septic process and melatonin effects. Melatonin was given at dose of 120 mg/kg bw, since in preliminary experiments this dose blunted absolutely iNOS activity and expression. Moreover, 120 mg/kg bw in mice correspond to 60 mg/kg bw of melatonin in rats, in accordance with the dose translation calculations [183]. We analyzed here the expression of inos, and the levels of nitrotyrosine residues and carbonyl groups in the liver of septic mice. The results showed that the activation of NOS isoforms, i.e., iNOS and nNOS, was related to liver damage during sepsis. The absence of iNOS in inos −/− mice blunted the septic response, with no contribution of nNOS to liver damage after CLP. In contrast, nnos −/− mice did not affect the response to CLP. We demonstrated that the effects of sepsis were unrelated to nNOS [38]. Moreover, melatonin treatment blunted the septic response in inos +/+ mice, reducing to normal levels the expression of iNOS and the levels to nitrotyrosine residues and protein oxidation. Moreover, skeletal and cardiac muscles failure during clinical sepsis is critical for the survival of septic patients. So, we analyzed with the CLP model the effects of sepsis on these tissues and the antiseptic properties of melatonin. Diaphragm, skeletal muscle, and heart of mice were analyzed in inos +/+ and inos −/− mice strains. In all cases, it was shown that inos gene was required for triggering the septic response, which produced a severe damage to these tissues, in a similar extend as that reported previously in liver and lungs. As usual, melatonin treatment counteracted the effects of sepsis [62, 63, 67].
These data reflected a role of melatonin to reduce the effects of activation of the NF-κB pathway during the inflammatory response during sepsis, but they do not demonstrate a direct effect of melatonin on NF-κB activation. Some reports showed also that in different circumstances, melatonin antagonized NF-κB [184, 185], whereas others suggested that, in turn, NF-κB could modulate the synthesis of melatonin [186, 187].
To get more insight into the mechanisms involved in the inhibitory effects of melatonin on NF-κB pathway, we performed a series of experiments to evaluate the complete pathway of its activation from TLR4 signaling to the NF-κB binding to DNA and subsequent gene expression [82]. For this study, we used C57/Bl6 and rorα sg/sg mice, the latter deficient in rorα gene, because of the participation of RORα nuclear receptor in the genomic effects of melatonin. Mice became septic after CLP and the results showed for the first time that melatonin administration blunted the NF-κB activation through SIRT1-dependent NF-κB deacetylation in septic mice. Melatonin also counteracted the NF-κB-dependent gene expression, reducing to normal values the levels of iNOS and of the proinflammatory cytokines including TNFα and IL-1β. Melatonin also restored the redox balance in septic mice. Because NF-κB also induces the expression of antioxidant enzymes, the effect of melatonin could be mediated through its binding to RORα. Interestingly, the inhibition of NF-κB by melatonin was blunted in rorα sg/sg mice, demonstrating also by the first time that functional RORα nuclear receptor modulates the NF-κB-dependent innate immune activation in sepsis, and it is the target for the anti-inflammatory actions of melatonin [82].
Using NF-κB-RE-luc [BALB/c-Tg(NFkB-RE-luc)-Xen] mice, a transgenic strain that expresses a luciferase reporter under transcriptional control of NF-κB, we followed the inflammatory response during the first 24 h of sepsis in vivo, when most of the animals die if they are not treated. We showed that whole body and, specifically the heart, was subjected to the inflammatory response, which disappeared when the animals were treated with melatonin. Importantly, the bacterial load was analyzed in vivo in BALB/c mice after injection of Bacteriosense 645 (5 nmol/100 μl PBS/mouse i.v.), a probe that binds to negatively charged lipids on the bacterial cell membrane. We found the same bacterial load in the presence and absence of melatonin. With the results showing that melatonin did not affect the cytosolic signaling events triggered by TLR4 stimulation, our data demonstrated that melatonin blocks the NF-κB response at nuclear level without removal of the bacterial stimulus, thus preventing the bacterial LPS triggering the innate immunity [82]. An inhibitory role of melatonin on NF-κB binding to DNA was also shown in cancer metastasis [188]. In a model of aging, we further demonstrated that the activation of NF-κB with age follows the same signaling pathways that sepsis in young mice, and melatonin administration affected the same molecular targets [96]. Thus, the connection of melatonin with NF-κB in sepsis and aging was solved.
Mitochondrial function during sepsis
A recent review addressed some key questions related to the role of mitochondria in sepsis [189]. A major question is whether mitochondrial (dys)function during sepsis is a primary event or a secondary player in the disease. A second question is that sometimes mitochondria are not excessively damaged during sepsis, opening the discussion on to what extent they contribute to the organ system dysfunction during the inflammatory response. Related to these findings, the third question is a methodological one, and it is linked to the measurement of mitochondrial bioenergetics in the tissues of septic patients and in experimental models of sepsis.
During sepsis, there is an increased production of ROS and RNS, which can originate in the cytosol and contribute to mitochondrial impairment. But ROS and RNS are also produced by mitochondria and so the cellular pool of these radicals is composed by the sum of both sources, cytosolic and mitochondrial. The extent to which each source of ROS/RNS contributes to the total pool of free radicals is a matter of debate. For mitochondria, increased ROS/RNS production over the capacity of their antioxidant defense leads to an impairment of their function; so, mitochondrial damage and subsequent ROS/RNS formation should be secondary to sepsis.
To address these questions, we performed a series of experiments conducted in healthy conditions and in experimental sepsis. We found that during the first steps of sepsis mitochondria function may not be excessively dysfunctional, and the degree of mitochondrial failure depends on the tissue [37]. Here, young (3 mo.) and aged (18 mo.) rats were injected with LPS (serotype 0127:B78, 10 mg/kg b.w.) and inos/nnos/enos expression in the mitochondria, iNOS activity, and complex I and IV activities of the mitochondrial ETS were measured in liver and lungs. Rats were killed 6 h after LPS administration. The results showed for the first time that iNOS was constitutively present in the mitochondria, together with nNOS, whereas no eNOS was detected. Mitochondrial iNOS (i-mtNOS), but not nNOS (n-mtNOS), expression and activity were significantly increased during sepsis, parallel to the changes in cytosolic iNOS. However, complex I and IV activities were more significantly reduced in the lung mitochondria than in liver. These results reflect that mitochondrial damage is secondary to sepsis; it depends, at least in part, on the iNOS induction, and it is tissue dependent [37].
These results prompt a new query regarding mitochondria in sepsis: what was the source of mitochondrial NOS isoforms and what are the role(s) of the NO• produced by each isoform in mitochondria. Since n-mtNOS was identified as a cytosolic nNOS that enters mitochondria after post-translational modifications, and it is coded by the same gene as nNOS [190, 191], we conducted other experiments in inos −/− and nnos −/− mice to address this question. These experiments were carried out in the model of sepsis induced by CLP, and mice were evaluated at 24 h after CLP. Due to the tissue-specific response of mitochondria to sepsis, different organs and tissues were analyzed, including diaphragm, skeletal muscle, heart, and liver [62, 63, 67, 82, 192, 193]. The results showed that CLP induced the expression of iNOS and i-mtNOS at 24 h after CLP, with a parallel increased in mitochondrial NO· and LPO levels, and a rise in the GSSG/GSH ratio. The antioxidant enzyme GRd decreased drastically during sepsis, whereas GPx activity increased. The results, supporting an oxidative/nitrosative stress in mitochondria, coincided with a reduction in the activity of the four ETS complexes. On the contrary, inos −/− mice were protected against mitochondrial damage during sepsis. They did not display any activation of i-mtNOS and had no oxidative damage. These data confirmed that i-mtNOS comes from cytosolic iNOS and it is coded by the same gene.
We were able to measure the proportion of mitochondrial NO· yielded by i-mtNOS and its role on mitochondrial bioenergetics [67]. Under physiological conditions, the intramitochondrial pool of NO· comes from both i-mtNOS and c-mtNOS, which account for the 32 and 68% of the total NO· pool, respectively. Here, the constitutive isoforms of NOS in the mitochondria produce NO· giving an [O2]/[NO] ratio of 500/1000, which competitively and reversibly inhibits cytochrome c oxidase by 16–26% [53]. Mice lacking iNOS and, thus, deficient in i-mtNOS, showed the equivalent reduction in NO· of 0.9 nmol/mg prot, which was enough to increase two-fold the activity of the complex IV. So, increasing levels of NO· during sepsis clearly inhibit complex IV activity in a dose-dependent manner leading to OXPHOS impairment, electron leak, and formation of ROS [68]. The parallel increase in O·−2 and NO· favors the formation of ONOO−, causing nitration and inactivation of the four respiratory complexes [144]. However, in the presence of an excess of NO·, in addition to complex IV inhibition, NO· and ONOO− also interact with ubiquinol, inducing its oxidation to ubiquinone preventing nitration of tyrosine residues by ONOO− [194]. These apparent paradoxical effects of NO· in mitochondria help to explain the different degrees of energetic failure during sepsis, and it may depend on the experimental conditions of sepsis, the animal and type of strain, but also on the type and evolution of the septic patient.
We then decided to explore whether mitochondrial preparation method affected the bioenergetics measurement. Mice were made septic by CLP and the evolution of the mitochondrial bioenergetics was assessed at 8 and 24 h after CLP. Heart permeabilized fibers were prepared for high-resolution respirometry (HRR). This preparation maintained the full mitochondrial pool and maintained also mitochondrial ultrastructure and interactions, resembling the in vivo conditions. Moreover, ETS, OXPHOS, cytochromes redox states, and OXPHOS supercomplexes were also measured. Our results showed a significant time-dependent impairment in ETS and OXPHOS during sepsis, which were apparent at 8 h after CLP. Sepsis induced a significant time-dependent increase in leak and in coupling control ratio, and altered the CIII supercomplexes molecular structure [193]. These data were compared with our previous experiments measuring mitochondrial function in isolated mitochondria from hearts of septic mice under the same experimental protocol of sepsis [63]. We observed that ETS complexes were less damaged when analyzed isolated mitochondrial preparation than when used permeabilized fibers.
These data collectively indicate that the extensive mitochondrial damage that occurs during sepsis is a time-dependent phenomenon. Together with energetic malfunction, there is a significantly oxidative and nitrosative stress that participates in the mitochondrial malfunction [195]. Moreover, the data point to a major methodological issue: when mitochondria are isolated from any tissue, most of the damaged mitochondria are lost or broken during the procedure, and the resulting pool contains relatively well-coupled mitochondria. However, using permeabilized fibers we work with the complete pool of mitochondria. Using HRR and mitochondria isolated from liver of septic mice, we further corroborated this hypothesis, because these mitochondria were apparently less damaged than those of permeabilized fibers [192]. These differences account for a major mitochondrial impairment detected in permeabilized fibers than in isolated mitochondria, raising concerns regarding the methods used to detect mitochondrial dysfunctions during sepsis.
Melatonin and mitochondria in sepsis
From in vitro studies in isolated mitochondria to cell culture as well as in vivo reports, the relationship between melatonin and mitochondria has been thoroughly analyzed; the data document that mitochondria are a major target of melatonin in the cell. This connection started with our paper in which we showed the effects of melatonin on isolated mitochondria from rat liver and brain. Mitochondria were incubated with or without 100 μM t-butyl hydroperoxide (t-BHP) and in the presence or absence of 10–100 nM melatonin [167]. t-BHP increased mitochondrial hydroperoxides, leading to an oxidation of the 90% of the GSH pool, and blocking the activities of GPx and GRd. This situation produced mitochondrial swelling and their destruction. Melatonin, at doses of 100 nM, totally prevented the damage induced by t-BHP, recovering the GSH pool and the GPx and GRd activities, and normalizing the mitochondrial redox status. These changes were accompanied with a recovery by melatonin of the ETS complexes inhibited by the toxin. To further analyze the specificity of the effects of melatonin, N-acetyl-cysteine (NAC), vitamin C and vitamin E were also tested for their antioxidant activity. These antioxidants, even at doses of 1 mM, i.e., 10,000 times higher than the dose of melatonin, had not ability to counteract the damage induced by the hydroperoxides to mitochondria. Melatonin also increased the activity of the ETS complexes in mitochondria not exposed to t-BHP. The results support melatonin as the best antioxidant and mitochondrial protector against oxidative damage, and raised serious concerns regarding the value of the other antioxidants in clinical practice [167]. In parallel, we injected ruthenium red to normal rats to induce similar mitochondrial damage and oxidative stress in vivo. Melatonin administration at doses of 10 mg/kg bw increased the activity of the complexes I and IV in brain and liver mitochondria of untreated rat; it counteracted the inhibitory effect of ruthenium red in these respiratory complexes, and restored the GSH pool [196]. This was the first demonstration that melatonin in vivo exerts direct beneficial effects on mitochondria.
With these results in mind, the next experiments were designed to clarify the effects of melatonin on mitochondria. Rat liver and brain mitochondria were cultured in vitro and the activity of the complex I and IV, the synthesis of ATP, was analyzed. Melatonin enhanced the activity of complex I and IV from 1 nM to 10 nM, and the actions of the indoleamine reflected an allosteric regulation of these enzymes [197]. The specificity of melatonin was further analyzed. Blue native-PAGE and histochemical analysis of the complex I demonstrated an increase in its activity. Moreover, titration studies showed that melatonin counteracted the inhibition of the complex IV by cyanide. Finally, melatonin also increased the production of ATP. The next experiments showed that melatonin binds specifically to a matrix mitochondrial protein, and increased the transcriptional activity of mtDNA of the subunits 1, 2, and 3 of the complex IV in vitro and in vivo [198].
Collectively, these data speak in favor of high sensitivity of mitochondria to the regulatory effects of melatonin that can be considered the intracellular targets of the indoleamine. With the use of HRR, fluorimeter, and spectrophotometry, mitochondrial bioenergetics was evaluated in vitro in the presence of increasing amounts of melatonin added to the respiratory chamber. Mitochondria were obtained from mouse liver and melatonin was added from 1 nM to 1 mM [199]. The results showed that melatonin decreased oxygen consumption in a dose-dependent manner, without changes in oxygen flux when ADP was added. Melatonin also had a mild-uncoupling effect, reducing the ∆+μH and leading to a reduction in the formation of O·−2 and H2O2. Melatonin also enhanced the activity of the four ETS complexes, maintaining OXPHOS and ATP synthesis. Pharmacokinetic studies revealed that mitochondria take up melatonin in a dose-dependent manner, thus supporting that the effects of the indoleamine here recorded depend on a direct effect inside the organelle. Together, the data above summarized further confirm melatonin as the major endogenous hormone involved in the control of mitochondrial homeostasis.
With the demonstration of the ability of melatonin to boost both normal and oxidatively damaged mitochondrial function, it was expected that the indoleamine should maintain mitochondrial efficiency in situations such as sepsis, where these organelles are severely damaged. The connection between melatonin and mitochondrial function in sepsis came from our paper published in 2003 reporting for the first time the existence of an i-mtNOS, a mitochondrial form of iNOS [37]. Here, we found that sepsis induced an age-dependent increase of mitochondrial NO· in rats injected with LPS that was dependent on the induction of i-mtNOS. The high levels of NO· reduced the activity of the ETS complexes I and IV in the mitochondria of lungs and liver, which was counteracted by melatonin administration at doses of 60 mg/kg bw. The deleterious effects of sepsis on mitochondria were significantly higher in aged than in young rats, which also showed higher levels of i-mtNOS. The effects of melatonin were also more pronounced in aged rats, which was related to the lower levels of melatonin in the latter. The efficacy of melatonin to restore the normal mitochondrial function preventing their failure during sepsis was then identified.
The failure of skeletal and cardiac muscles is the main cause of mortality during sepsis and their mitochondria show an elevated oxidative and nitrosative stress with a parallel drop in the ETS complexes activity [62, 67]. These changes follow the increased activity of i-mtNOS during endotoxemia. Melatonin administration at doses of 120 mg/kg bw to septic mice prevented the mitochondrial dysfunction and the development of septic shock and MOF in these mice, reflecting the high efficacy of the antiseptic properties of melatonin. Of note, the dose of 120 mg/kg bw of melatonin in mice corresponds to that of 60 mg/kg bw in rats, according to the equivalence of dose elsewhere published [183]. Moreover, melatonin therapy doubled the survival time of the animals [63]. Importantly, mitochondrial dysfunction does not occur in mice lacking iNOS/i-mtNOS isoforms, with the effects of melatonin being less marked, supporting the idea that these NOS isoforms are responsible for the bioenergetic impairment during sepsis. Studies with human endothelial cells in culture showed that melatonin counteracted LPS-dependent induction of NF-κB, reduced IL-6 and IL-8 levels, and improved mitochondrial function [200].
The specific effects of melatonin to improve the ETS activity should be accompanied with a significant increase in their ability to produce ATP; this hypothesis was further analyzed in the same experimental model of septic mice. Here, we measured the content of the adenine nucleotides and the ATP production. The results showed that the mitochondrial ATPase activity did not change during sepsis or melatonin treatment, but it increased the production of ATP that may explain the reduction of the mortality [63]. The absence of changes in the complex V activity indicates that the drop in the ATP formation depends on the ETS damage during sepsis [201]. Moreover, the lack of effects of melatonin on the activity of the complex V was further demonstrated in isolated mitochondria analyzed by HRR [199]. The ability of melatonin to enhance the bioenergetic capacity of the mitochondria reducing their oxygen consumption protects this organelle from oxidative damage favoring their recovery during sepsis [202].
Melatonin and the NLRP3 inflammasome pathway during sepsis
The sequence of events during sepsis as revised herein is summarized as follows: (1) stimulation of TLR4 (and others) receptors by the bacterial LPS; (2) release of free NF-κB to the cytosol and its translocation to the nucleus; (3) activation of NF-κB transcriptional activity; (4) production of proinflammatory molecules including iNOS and both mature and immature proinflammatory cytokines such as TNF and pro-IL-1β; (5) positive feedback of these cytokines on NF-κB; (6) increased oxidative damage to mitochondria that lose their capacity to produce ATP, and (7) eventually cell death. These events correspond to the classical NF-κB pathway of the innate immunity activated during sepsis. As a secondary but necessary component of the innate immunity that is triggered during sepsis is the NLRP3 inflammasome. The existence of a mitochondrial failure during sepsis and the fact that the NLRP3 inflammasome is activated by ROS and mtDNA from impaired mitochondria, close the loop and explain the exaggerated inflammatory response in this disease.
The fact that mitochondria are the main intracellular targets of melatonin suggests that melatonin may protect against NLRP3 inflammasome activation preventing the release of ROS and mtDNA from these organelles. Moreover, since NF-κB transcriptional activity enhances the cytosolic levels of NLRP3, the activity of melatonin to blunt NF-κB yields an additional pathway of inhibition of the NLRP3 inflammasome activation by the indoleamine.
To address the mechanism of action of melatonin in the regulation of the NLRP3 inflammasome, we performed a series of experiments that confirmed for the first time that melatonin inhibits the NLRP3 inflammasome activation during sepsis by a mechanism independent of its inhibition of NF-κB [82]. We showed that the NF-κB induction by sepsis depends on the inhibition of the expression of RORα and activation of REV-ERBα, thus reducing the activation of SIRT1. In these conditions, p65 was not deacetylated maintaining its binding to DNA and subsequent gene transcriptional activity. These data explain why in ror sg/sg mice we found a major NF-κB activation and immune response. Melatonin was able to block the NF-κB activation in response to sepsis in rorα +/+ mice, but it lacks of effect in ror sg/sg mice. However, melatonin blunted the activation of the NLRP3 inflammasome in both rorα +/+ and ror sg/sg mice. Further experiments confirmed that the effect of melatonin to counteract the oxidative damage to mitochondria in septic mice, reducing the cytosolic levels of ROS and preventing mtDNA release from mitochondria, was the mechanism involved in the reduction of the NLRP3 inflammasome activity [82].
In other conditions including age and mucositis, where the activation of the NF-κB/NLRP3 inflammasome pathways was demonstrated, melatonin administration counteracts both of them through the same mechanisms as here reported in sepsis [96, 203]. Additional experiments were done to assess the melatonin-NLRP3 inflammasome connection. Mice lacking nlrp3 gene are expected to respond to sepsis with lower innate immune reactivity than wild type counterparts. We compared the septic response to CLP in these mice. The results showed that nlrp3 −/− mice had an inhibition of the septic response comparable to that found after melatonin treatment to septic wild-type mice. Mice lacking nlrp3 −/− gene, however, have a disruption of the expression of clock genes, which was restored when they were treated with melatonin [204]. The lack of nlrp3 gene converts sepsis in a moderate inflammatory disease, and it is comparable with the therapeutic effect of melatonin administration. These data identify NLRP3 inflammasome as the main therapeutic objective in sepsis, which may be considered a target for the antiseptic properties of melatonin. The results also established dual roles of melatonin to control the innate immune response: an RORα/REVERBα-dependent mechanism to curb NF-κB activation during sepsis, and a mitochondrial-dependent mechanism to inhibit the activation of the inflammasome. These data also identified melatonin as the first drug able to blunt these two pathways of the innate immunity.
Melatonin, clock genes and the NF-κB/NLRP3/mitochondrial connection in sepsis
It is known that the circadian system is directly involved in the control of the immune system, which shows a day/night changes compatible with a circadian rhythm. Clock genes play important roles in the control of the immune responses, and ablation of the SCN enhances the immune response [205]. In turn, cytokines mediate the immune-circadian loop. Proinflammatory cytokines and the expression of their receptors display circadian rhythms. INFγ affects the clock gene expression [206], whereas TNFα, which is under rhythmic release in normal conditions, inhibits BMAL1/CLOCK-dependent transactivation of clock genes [131]. TNFα also phase-shifts per2 expression affecting the central clock [207]. Under inflammatory conditions, these and others cytokines may promote chronodisruption leading to a major immune response. This and other information have been revised here. Interesting, melatonin affects the SCN clock by different pathways including the transcription of per1 and per2 at night [208], which is related to the feedback effect of the indoleamine to induce advance phases of the clock. Also, melatonin antagonizes NF-κB, alters the effect of NF-κB to repress BMAL1/CLOCK [209], and the role of CLOCK to enhance NF-κB activation, and supports a more complex connection between the anti-inflammatory properties of melatonin to reduce proinflammatory cytokines and the roles of both melatonin and these cytokines on the central clock (Fig. 3).
Fig. 3.
A model to explain the anti-inflammatory activity of melatonin. Melatonin is released in a rhythmic manner controlled by the central clock, which in turn exerts a feedback control of the clock. The effects of melatonin also include its binding to RORα receptors enhancing BMAL1-dependent SIRT1 activity and reducing the NF-κB activity. Moreover, melatonin directly acts on the mitochondria, boosting the production of ATP and reducing the formation of free radicals. These effects maintain the integrity of the mitochondria preventing opening the MPT. Thus, melatonin impedes the activation of the NLRP3 inflammasome. These three effects of melatonin blunt the inflammatory response during sepsis and recover the clock/NF-κB/NLRP3/mitochondria normality
Whether sepsis drives clock gene expression towards a proinflammatory rearrangement or clock genes themselves change their expression favoring the immune response in sepsis is yet unclear. To look for further insights that could help to decipher this connection, we performed a series of experiments in septic and aged mice. Septic mice constitute an acute model of inflammation that also acutely influences the clock, whereas aging is a chronic, subclinical model of inflammation that results in low level expression of proinflammatory molecules. In the model of sepsis induced by CLP in mice, the induction of the innate immune response in terms of NF-κB/NRLP3 inflammasome activation and proinflammatory molecules including TNFα, iNOS, INFγ, and IL1-β was synchronous with the disruption of the BMAL1/CLOCK/RORα/REV-ERBα loop. In parallel with the immune response, the RORα/REV-ERBα ratio was quickly reversed, favoring REV-ERBα. This change conditioned the reduction in BMAL1 and CLOCK expression, which in turn diminishes the SIRT1-dependent deacetylation of NF-κB, enhancing its transcriptional activity [82]. These events close the loop of the innate immunity as it has been shown in this review.
Aged animals display the same activation of the innate immunity that young animals with sepsis, although with a low grade of NF-κB/NRLP3 inflammasome induction [96]. In the aged animals, there is a small, but significant reduction in the RORα/REV-ERBα ratio, leading to the activation of the full pathway of the NF-κB/NRLP3 inflammasome although at much lower extent than in sepsis. The induction of sepsis in aged mice worsens this situation, promoting the high-level of innate immune response, with a major alteration in the BMAL1/CLOCK/RORα/REV-ERBα loop. In both cases, i.e., sepsis and aging, therapeutic administration of melatonin restored the clock and blunted the inflammation. Of note, acute administration of melatonin to aged and septic mice blunted the innate immune response to the level of non-septic aged mice; a similar response was recorded in relation to the BMAL1/CLOCK/RORα/REV-ERBα [96]. These data support the fact that chronodisruption is the condition that, during age, leads to the activation of the innate immunity.
In conclusion, it seems that age-dependent chronodisruption may release the innate response, entering in a feedback cycle that favors the aging process itself. These data led us to propose the chronoinflammatory theory of aging [96]. In the presence of an inflammatory signal such as bacterial LPS, however, the release of some proinflammatory molecules may be the first signal to induce loss of clock homeostasis, driving it towards an increased inflammatory condition that continues in a vicious cycle and ends in an uncontrolled response of the innate immunity, leading eventually to death. Both conditions, sepsis and aging, courses with diminished production of melatonin, allowing major chronodisruption and immune reactivity. In sepsis and aging, melatonin restored the pacemaker homeostasis, reducing the innate immunity and enhancing survival. Thus, we can consider melatonin as the link between the central pacemaker and the innate immunity. The outstanding properties of melatonin to revert the BMAL1/CLOCK/RORα/REV-ERBα/NF-κB/NLRP3 inflammasome activation make it an ideal candidate for its therapeutic application in sepsis and aging [210]. The lack of adverse effects of melatonin elsewhere reported [211, 212], and its broad antiseptic properties [213], supports the use of melatonin in humans. These inti-inflammatory properties of melatonin are being currently assayed in a Phase II clinical trial in septic patients (Eudract # 2008-006782-83). The patients have been treated with a new patented injectable formulation of melatonin (PCT/ES2015070236). From the preliminary results, similar beneficial effects of melatonin have been obtained in septic patients as in experimental models of the disease.
Acknowledgements
This study was partially supported by Grants from the Ministerio de Economía, Industria y Competitividad, y por el Fondo de Desarrollo Regional FEDER, Spain # RD12/0043/0005; PI13-00981; PI16-00519; PI16-00CB16-10-00238), and from the Consejería de Innovación, Ciencia y Empresa, Junta de Andalucía (P07-CTS-03135, P10-CTS-5784, and CTS-101), Spain. CA-F is a Resident Medical Intern, Hospital Universitario de Canarias, San Cristóbal de La Laguna, Santa Cruz de Tenerife, Spain; MF-O is supported by a FPU fellowship from the Ministerio de Educación, Cultura y Deporte, Spain; MED-C is supported by a postdoctoral fellowship from the Consejería de Innovación, Ciencia y Empresa, Junta de Andalucía, Spain, and LCL is supported by the “Ramón y Cajal” National Program (Ministerio de Economía y Competitividad, Spain).
References
- 1.Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, Ferrer R, Kumar A, Sevransky JE, Sprung CL, Nunnally ME, Rochwerg B, Rubenfeld GD, Angus DC, Annane D, Beale RJ, Bellinghan GJ, Bernard GR, Chiche JD, Coopersmith C, De Backer DP, French CJ, Fujishima S, Gerlach H, Hidalgo JL, Hollenberg SM, Jones AE, Karnad DR, Kleinpell RM, Koh Y, Lisboa TC, Machado FR, Marini JJ, Marshall JC, Mazuski JE, McIntyre LA, McLean AS, Mehta S, Moreno RP, Myburgh J, Navalesi P, Nishida O, Osborn TM, Perner A, Plunkett CM, Ranieri M, Schorr CA, Seckel MA, Seymour CW, Shieh L, Shukri KA, Simpson SQ, Singer M, Thompson BT, Townsend SR, Van der Poll T, Vincent JL, Wiersinga WJ, Zimmerman JL, Dellinger RP. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Crit Care Med. 2017;45:486–552. doi: 10.1097/CCM.0000000000002255. [DOI] [PubMed] [Google Scholar]
- 2.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, van der Poll T, Vincent JL, Angus DC. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fleischmann C, Scherag A, Adhikari NK, Hartog CS, Tsaganos T, Schlattmann P, Angus DC, Reinhart K, International Forum of Acute Care Trialists Assessment of global incidence and mortality of hospital-treated sepsis. current estimates and limitations. Am J Respir Crit Care Med. 2016;193:259–272. doi: 10.1164/rccm.201504-0781OC. [DOI] [PubMed] [Google Scholar]
- 4.Hattori Y, Hattori K, Suzuki T, Matsuda N. Recent advances in the pathophysiology and molecular basis of sepsis-associated organ dysfunction: novel therapeutic implications and challenges. Pharmacol Ther. 2017 doi: 10.1016/j.pharmthera.2017.02.040. [DOI] [PubMed] [Google Scholar]
- 5.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 6.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 7.Schnare M, Barton GM, Holt AC, Takeda K, Akira S, Medzhitov R. Toll-like receptors control activation of adaptive immune responses. Nat Immunol. 2001;2:947–950. doi: 10.1038/ni712. [DOI] [PubMed] [Google Scholar]
- 8.Poltorak A, Smirnova I, He X, Liu MY, Van Huffel C, McNally O, Birdwell D, Alejos E, Silva M, Du X, Thompson P, Chan EK, Ledesma J, Roe B, Clifton S, Vogel SN, Beutler B. Genetic and physical mapping of the Lps locus: identification of the toll-4 receptor as a candidate gene in the critical region. Blood Cells Mol Dis. 1998;24:340–355. doi: 10.1006/bcmd.1998.0201. [DOI] [PubMed] [Google Scholar]
- 9.Fujihara M, Muroi M, Tanamoto K, Suzuki T, Azuma H, Ikeda H. Molecular mechanisms of macrophage activation and deactivation by lipopolysaccharide: roles of the receptor complex. Pharmacol Ther. 2003;100:171–194. doi: 10.1016/j.pharmthera.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Zingarelli B, Hake PW, O’Connor M, Denenberg A, Kong S, Aronow BJ. Absence of poly(ADP-ribose)polymerase-1 alters nuclear factor-kappa B activation and gene expression of apoptosis regulators after reperfusion injury. Mol Med. 2003;9:143–153. doi: 10.2119/2003-00011.Zingarelli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaisho T, Takeuchi O, Kawai T, Hoshino K, Akira S. Endotoxin-induced maturation of MyD88-deficient dendritic cells. J Immunol. 2001;166:5688–5694. doi: 10.4049/jimmunol.166.9.5688. [DOI] [PubMed] [Google Scholar]
- 12.Seki E, Tsutsui H, Nakano H, Tsuji N, Hoshino K, Adachi O, Adachi K, Futatsugi S, Kuida K, Takeuchi O, Okamura H, Fujimoto J, Akira S, Nakanishi K. Lipopolysaccharide-induced IL-18 secretion from murine Kupffer cells independently of myeloid differentiation factor 88 that is critically involved in induction of production of IL-12 and IL-1beta. J Immunol. 2001;166:2651–2657. doi: 10.4049/jimmunol.166.4.2651. [DOI] [PubMed] [Google Scholar]
- 13.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nat Immunol. 2011;12:695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 15.Perkins ND, Gilmore TD. Good cop, bad cop: the different faces of NF-kappaB. Cell Death Differ. 2006;13:759–772. doi: 10.1038/sj.cdd.4401838. [DOI] [PubMed] [Google Scholar]
- 16.Gilmore TD. Introduction to NF-kappaB: players, pathways, perspectives. Oncogene. 2006;25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 17.Chen ZJ. Ubiquitination in signaling to and activation of IKK. Immunol Rev. 2012;246:95–106. doi: 10.1111/j.1600-065X.2012.01108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acharyya A, Villalta A, Bakkar N, Bupha-Intr T, Janssen PML, Carathers M, Li ZW, Beg AA, Ghosh S, Sahenk Z, Weinstein M, Gardner KL, Rafael-Fortney JA, Karin M, Tidball JG, Baldwin AS, Guttridge DC. Interplay of IKK/NFkB signaling in macrophages and myofibers promotes muscle degeneration in Duchenne muscular dystrophy. J Clin Investig. 2007;117:889–901. doi: 10.1172/JCI30556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunsch C, Ruben SM, Rosen CA. Selection of optimal kappa B/Rel DNA-binding motifs: interaction of both subunits of NF-kappa B with DNA is required for transcriptional activation. Mol Cell Biol. 1992;12:4412–4421. doi: 10.1128/MCB.12.10.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abraham E. Nuclear factor-kappaB and its role in sepsis-associated organ failure. J Infect Dis. 2003;187:S364–S369. doi: 10.1086/374750. [DOI] [PubMed] [Google Scholar]
- 22.Tak PP, Firestein GS. NF-kappaB: a key role in inflammatory diseases. J Clin Investig. 2001;107:7–11. doi: 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu SF, Malik AB. NF-kappa B activation as a pathological mechanism of septic shock and inflammation. Am J Physiol Lung Cell Mol Physiol. 2006;290:L622–L645. doi: 10.1152/ajplung.00477.2005. [DOI] [PubMed] [Google Scholar]
- 24.Morgan MJ, Liu ZG. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res. 2011;21:103–115. doi: 10.1038/cr.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webster NR, Nunn JF. Molecular structure of free radicals and their importance in biological reactions. Br J Anaesth. 1988;60:98–108. doi: 10.1093/bja/60.1.98. [DOI] [PubMed] [Google Scholar]
- 26.Shen HM, Pervaiz S. TNF receptor superfamily-induced cell death: redox-dependent execution. FASEB J. 2006;20:1589–1598. doi: 10.1096/fj.05-5603rev. [DOI] [PubMed] [Google Scholar]
- 27.Hughes G, Murphy MP, Ledgerwood EC. Mitochondrial reactive oxygen species regulate the temporal activation of nuclear factor kappaB to modulate tumour necrosis factor-induced apoptosis: evidence from mitochondria-targeted antioxidants. Biochem J. 2005;389:83–89. doi: 10.1042/BJ20050078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blaser H, Dostert C, Mak TW, Brenner D. TNF and ROS crosstalk in inflammation. Trends Cell Biol. 2016;26:249–261. doi: 10.1016/j.tcb.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Rhee SG, Yang KS, Kang SW, Woo HA, Chang TS. Controlled elimination of intracellular H(2)O(2): regulation of peroxiredoxin, catalase, and glutathione peroxidase via post-translational modification. Antioxid Redox Signal. 2005;7:619–626. doi: 10.1089/ars.2005.7.619. [DOI] [PubMed] [Google Scholar]
- 30.Rhee SG, Kang SW, Jeong W, Chang TS, Yang KS, Woo HA. Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr Opin Cell Biol. 2005;17:183–189. doi: 10.1016/j.ceb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 31.Zhou LZ, Johnson AP, Rando TA. NF kappa B and AP-1 mediate transcriptional responses to oxidative stress in skeletal muscle cells. Free Radic Biol Med. 2001;31:1405–1416. doi: 10.1016/S0891-5849(01)00719-5. [DOI] [PubMed] [Google Scholar]
- 32.Horton JW. Free radicals and lipid peroxidation mediated injury in burn trauma: the role of antioxidant therapy. Toxicology. 2003;189:75–88. doi: 10.1016/S0300-483X(03)00154-9. [DOI] [PubMed] [Google Scholar]
- 33.Lanone S, Mebazaa A, Heymes C, Henin D, Poderoso JJ, Panis Y, Zedda C, Billiar T, Payen D, Aubier M, Boczkowski J. Muscular contractile failure in septic patients: role of the inducible nitric oxide synthase pathway. Am J Respir Crit Care Med. 2000;162:2308–2315. doi: 10.1164/ajrccm.162.6.2001097. [DOI] [PubMed] [Google Scholar]
- 34.Lundberg JO, Weitzberg E. NO generation from nitrite and its role in vascular control. Arterioscler Thromb Vasc Biol. 2005;25:915–922. doi: 10.1161/01.ATV.0000161048.72004.c2. [DOI] [PubMed] [Google Scholar]
- 35.Lipton SA, Choi YB, Pan ZH, Lei SZ, Chen HS, Sucher NJ, Loscalzo J, Singel DJ, Stamler JS. A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature. 1993;364:626–632. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- 36.Cadenas E, Poderoso JJ, Antunes F, Boveris A. Analysis of the pathways of nitric oxide utilization in mitochondria. Free Radic Res. 2000;33:747–756. doi: 10.1080/10715760000301271. [DOI] [PubMed] [Google Scholar]
- 37.Escames G, Leon J, Macias M, Khaldy H, Acuña-Castroviejo D. Melatonin counteracts lipopolysaccharide-induced expression and activity of mitochondrial nitric oxide synthase in rats. FASEB J. 2003;17:932–934. doi: 10.1096/fj.02-0692fje. [DOI] [PubMed] [Google Scholar]
- 38.Garcia JA, Ortiz F, Miana J, Doerrier C, Fernandez-Ortiz M, Rusanova I, Escames G, Garcia JJ, Acuna-Castroviejo D. Contribution of inducible and neuronal nitric oxide synthases to mitochondrial damage and melatonin rescue in LPS-treated mice. J Physiol Biochem. 2017;73:235–244. doi: 10.1007/s13105-017-0548-2. [DOI] [PubMed] [Google Scholar]
- 39.Saito Y, Nishio K, Ogawa Y, Kimata J, Kinumi T, Yoshida Y, Noguchi N, Niki E. Turning point in apoptosis/necrosis induced by hydrogen peroxide. Free Radic Res. 2006;40:619–630. doi: 10.1080/10715760600632552. [DOI] [PubMed] [Google Scholar]
- 40.Nakajima S, Kitamura M. Bidirectional regulation of NF-kappaB by reactive oxygen species: a role of unfolded protein response. Free Radic Biol Med. 2013;65:162–174. doi: 10.1016/j.freeradbiomed.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 41.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/S1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 42.Inohara Chamaillard, McDonald C, Nunez G. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- 43.Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 44.Tattoli I, Travassos LH, Carneiro LA, Magalhaes JG, Girardin SE. The Nodosome: Nod1 and Nod2 control bacterial infections and inflammation. Semin Immunopathol. 2007;29:289–301. doi: 10.1007/s00281-007-0083-2. [DOI] [PubMed] [Google Scholar]
- 45.Escames G, Lopez LC, Garcia JA, Garcia-Corzo L, Ortiz F, Acuña-Castroviejo D. Mitochondrial DNA and inflammatory diseases. Hum Genet. 2012;131:161–173. doi: 10.1007/s00439-011-1057-y. [DOI] [PubMed] [Google Scholar]
- 46.de Torre-Minguela C, Mesa Del Castillo P, Pelegrin P. The NLRP3 and pyrin inflammasomes: implications in the pathophysiology of autoinflammatory diseases. Front Immunol. 2017;8:43. doi: 10.3389/fimmu.2017.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 48.Tschopp J, Schroder K. NLRP3 inflammasome activation: the convergence of multiple signalling pathways on ROS production? Nat Rev Immunol. 2010;10:210–215. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- 49.Dostert C, Petrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 51.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, Choi AM. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown GC. Control of respiration and ATP synthesis in mammalian mitochondria and cells. Biochem J. 1992;284:1–13. doi: 10.1042/bj2840001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boveris A, Costa LE, Poderoso JJ, Carreras MC, Cadenas E. Regulation of mitochondrial respiration by oxygen and nitric oxide. Ann N Y Acad Sci. 2000;899:121–135. doi: 10.1111/j.1749-6632.2000.tb06181.x. [DOI] [PubMed] [Google Scholar]
- 54.Mitchell P. Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biol Rev Camb Philos Soc. 1966;41:445–502. doi: 10.1111/j.1469-185X.1966.tb01501.x. [DOI] [PubMed] [Google Scholar]
- 55.Diehl AM, Hoek JB. Mitochondrial uncoupling: role of uncoupling protein anion carriers and relationship to thermogenesis and weight control “the benefits of losing control”. J Bioenerg Biomembr. 1999;31:493–506. doi: 10.1023/A:1005452624640. [DOI] [PubMed] [Google Scholar]
- 56.Woyda-Ploszczyca AM, Jarmuszkiewicz W. The conserved regulation of mitochondrial uncoupling proteins: from unicellular eukaryotes to mammals. Biochim Biophys Acta. 2017;1858:21–33. doi: 10.1016/j.bbabio.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 57.Solmonson A, Mills EM. Uncoupling proteins and the molecular mechanisms of thyroid thermogenesis. Endocrinology. 2016;157:455–462. doi: 10.1210/en.2015-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicholls DG, Budd SL. Mitochondria and neuronal survival. Physiol Rev. 2000;80:315–360. doi: 10.1152/physrev.2000.80.1.315. [DOI] [PubMed] [Google Scholar]
- 59.Cardoso S, Correia S, Carvalho C, Candeias E, Placido AI, Duarte AI, Seica RM, Moreira PI. Perspectives on mitochondrial uncoupling proteins-mediated neuroprotection. J Bioenerg Biomembr. 2015;47:119–131. doi: 10.1007/s10863-014-9580-x. [DOI] [PubMed] [Google Scholar]
- 60.Acuña Castroviejo D, Lopez LC, Escames G, Lopez A, Garcia JA, Reiter RJ. Melatonin-mitochondria interplay in health and disease. Curr Top Med Chem. 2011;11:221–240. doi: 10.2174/156802611794863517. [DOI] [PubMed] [Google Scholar]
- 61.Bombicino SS, Iglesias DE, Zaobornyj T, Boveris A, Valdez LB. Mitochondrial nitric oxide production supported by reverse electron transfer. Arch Biochem Biophys. 2016;607:8–19. doi: 10.1016/j.abb.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 62.Escames G, Lopez LC, Tapias V, Utrilla P, Reiter RJ, Hitos AB, Leon J, Rodriguez MI, Acuña-Castroviejo D. Melatonin counteracts inducible mitochondrial nitric oxide synthase-dependent mitochondrial dysfunction in skeletal muscle of septic mice. J Pineal Res. 2006;40:71–78. doi: 10.1111/j.1600-079X.2005.00281.x. [DOI] [PubMed] [Google Scholar]
- 63.Escames G, Lopez LC, Ortiz F, Lopez A, Garcia JA, Ros E, Acuña-Castroviejo D. Attenuation of cardiac mitochondrial dysfunction by melatonin in septic mice. FEBS J. 2007;274:2135–2147. doi: 10.1111/j.1742-4658.2007.05755.x. [DOI] [PubMed] [Google Scholar]
- 64.Ortiz F, Garcia JA, Acuña-Castroviejo D, Doerrier C, Lopez A, Venegas C, Volt H, Luna-Sanchez M, Lopez LC, Escames G. The beneficial effects of melatonin against heart mitochondrial impairment during sepsis: inhibition of iNOS and preservation of nNOS. J Pineal Res. 2014;56:71–81. doi: 10.1111/jpi.12099. [DOI] [PubMed] [Google Scholar]
- 65.Brown GC. Regulation of mitochondrial respiration by nitric oxide inhibition of cytochrome c oxidase. Biochim Biophys Acta. 2001;1504:46–57. doi: 10.1016/S0005-2728(00)00238-3. [DOI] [PubMed] [Google Scholar]
- 66.Acuña-Castroviejo D, Escames G, Lopez LC, Hitos AB, Leon J. Melatonin and nitric oxide: two required antagonists for mitochondrial homeostasis. Endocrine. 2005;27:159–168. doi: 10.1385/ENDO:27:2:159. [DOI] [PubMed] [Google Scholar]
- 67.Lopez LC, Escames G, Tapias V, Utrilla P, Leon J, Acuña-Castroviejo D. Identification of an inducible nitric oxide synthase in diaphragm mitochondria from septic mice: its relation with mitochondrial dysfunction and prevention by melatonin. Int J Biochem Cell Biol. 2006;38:267–278. doi: 10.1016/j.biocel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 68.Poderoso JJ, Carreras MC, Lisdero C, Riobo N, Schopfer F, Boveris A. Nitric oxide inhibits electron transfer and increases superoxide radical production in rat heart mitochondria and submitochondrial particles. Arch Biochem Biophys. 1996;328:85–92. doi: 10.1006/abbi.1996.0146. [DOI] [PubMed] [Google Scholar]
- 69.Brown GC, Borutaite V. Inhibition of mitochondrial respiratory complex I by nitric oxide, peroxynitrite and S-nitrosothiols. Biochim Biophys Acta. 2004;1658:44–49. doi: 10.1016/j.bbabio.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 70.Escames G, Lopez LC, Ortiz F, Ros E, Acuña-Castroviejo D. Age-dependent lipopolysaccharide-induced iNOS expression and multiorgan failure in rats: effects of melatonin treatment. Exp Gerontol. 2006;41:1165–1173. doi: 10.1016/j.exger.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 71.Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta. 1999;1410:103–123. doi: 10.1016/S0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 72.Bohr VA, Anson RM. Mitochondrial DNA repair pathways. J Bioenerg Biomembr. 1999;31:391–398. doi: 10.1023/A:1005484004167. [DOI] [PubMed] [Google Scholar]
- 73.Sanz A. Mitochondrial reactive oxygen species: do they extend or shorten animal lifespan? Biochim Biophys Acta. 2016;1857:1116–1126. doi: 10.1016/j.bbabio.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 74.Boveris A, Cadenas E. Mitochondrial production of hydrogen peroxide regulation by nitric oxide and the role of ubisemiquinone. IUBMB Life. 2000;50:245–250. doi: 10.1080/15216540051080912. [DOI] [PubMed] [Google Scholar]
- 75.West AP, Brodsky IE, Rahner C, Woo DK, Erdjument-Bromage H, Tempst P, Walsh MC, Choi Y, Shadel GS, Ghosh S. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Diebold L, Chandel NS. Mitochondrial ROS regulation of proliferating cells. Free Radic Biol Med. 2016;100:86–93. doi: 10.1016/j.freeradbiomed.2016.04.198. [DOI] [PubMed] [Google Scholar]
- 77.Jones RM, Neish AS. Redox signaling mediated by the gut microbiota. Free Radical Biol Med. 2016;105:41–47. doi: 10.1016/j.freeradbiomed.2016.10.495. [DOI] [PubMed] [Google Scholar]
- 78.Sakai K, Matsumoto K, Nishikawa T, Suefuji M, Nakamaru K, Hirashima Y, Kawashima J, Shirotani T, Ichinose K, Brownlee M, Araki E. Mitochondrial reactive oxygen species reduce insulin secretion by pancreatic beta-cells. Biochem Biophys Res Commun. 2003;300:216–222. doi: 10.1016/S0006-291X(02)02832-2. [DOI] [PubMed] [Google Scholar]
- 79.Nicholls DG. Mitochondria and calcium signaling. Cell Calcium. 2005;38:311–317. doi: 10.1016/j.ceca.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 80.Dawson TM, Dawson VL. Mitochondrial mechanisms of neuronal cell death: potential therapeutics. Annu Rev Pharmacol Toxicol. 2017;57:437–454. doi: 10.1146/annurev-pharmtox-010716-105001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Horbay R, Bilyy R. Mitochondrial dynamics during cell cycling. Apoptosis. 2016;21:1327–1335. doi: 10.1007/s10495-016-1295-5. [DOI] [PubMed] [Google Scholar]
- 82.Garcia JA, Volt H, Venegas C, Doerrier C, Escames G, Lopez LC, Acuña-Castroviejo D. Disruption of the NF-kappaB/NLRP3 connection by melatonin requires retinoid-related orphan receptor-alpha and blocks the septic response in mice. FASEB J. 2015;29:3863–3875. doi: 10.1096/fj.15-273656. [DOI] [PubMed] [Google Scholar]
- 83.Diaz-Casado ME, Lima E, Garcia JA, Doerrier C, Aranda P, Sayed RK, Guerra-Librero A, Escames G, Lopez LC, Acuña-Castroviejo D. Melatonin rescues zebrafish embryos from the parkinsonian phenotype restoring the parkin/PINK1/DJ-1/MUL1 network. J Pineal Res. 2016;61:96–107. doi: 10.1111/jpi.12332. [DOI] [PubMed] [Google Scholar]
- 84.Meissner F, Molawi K, Zychlinsky A. Superoxide dismutase 1 regulates caspase-1 and endotoxic shock. Nat Immunol. 2008;9:866–872. doi: 10.1038/ni.1633. [DOI] [PubMed] [Google Scholar]
- 85.Sarkar A, Duncan M, Hart J, Hertlein E, Guttridge DC, Wewers MD. ASC directs NF-kappaB activation by regulating receptor interacting protein-2 (RIP2) caspase-1 interactions. J Immunol. 2006;176:4979–4986. doi: 10.4049/jimmunol.176.8.4979. [DOI] [PubMed] [Google Scholar]
- 86.Watanabe A, Sohail MA, Gomes DA, Hashmi A, Nagata J, Sutterwala FS, Mahmood S, Jhandier MN, Shi Y, Flavell RA, Mehal WZ. Inflammasome-mediated regulation of hepatic stellate cells. Am J Physiol Gastrointest Liver Physiol. 2009;296:G1248–G1257. doi: 10.1152/ajpgi.90223.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cassel SL, Joly S, Sutterwala FS. The NLRP3 inflammasome: a sensor of immune danger signals. Semin Immunol. 2009;21:194–198. doi: 10.1016/j.smim.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Latz E. NOX-free inflammasome activation. Blood. 2010;116:1393–1394. doi: 10.1182/blood-2010-06-287342. [DOI] [PubMed] [Google Scholar]
- 89.Imaeda AB, Watanabe A, Sohail MA, Mahmood S, Mohamadnejad M, Sutterwala FS, Flavell RA, Mehal WZ. Acetaminophen-induced hepatotoxicity in mice is dependent on Tlr9 and the Nalp3 inflammasome. J Clin Investig. 2009;119:305–314. doi: 10.1172/JCI35958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhu P, Duan L, Chen J, Xiong A, Xu Q, Zhang H, Zheng F, Tan Z, Gong F, Fang M. Gene silencing of NALP3 protects against liver ischemia-reperfusion injury in mice. Hum Gene Ther. 2011;22:853–864. doi: 10.1089/hum.2010.145. [DOI] [PubMed] [Google Scholar]
- 91.Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol. 2010;11:136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- 92.Chen G, Li X, Huang M, Zhou X, Li Y, Mao X, Bai J (2016) The Role of thioredoxin-1 in suppression sepsis through inhibiting mitochonrial-induced apoptosis in spleen. Shock (in press) [DOI] [PubMed]
- 93.Hoegen T, Tremel N, Klein M, Angele B, Wagner H, Kirschning C, Pfister HW, Fontana A, Hammerschmidt S, Koedel U. The NLRP3 inflammasome contributes to brain injury in pneumococcal meningitis and is activated through ATP-dependent lysosomal cathepsin B release. J Immunol. 2011;187:5440–5451. doi: 10.4049/jimmunol.1100790. [DOI] [PubMed] [Google Scholar]
- 94.Tschopp J. Mitochondria: sovereign of inflammation? Eur J Immunol. 2011;41:1196–1202. doi: 10.1002/eji.201141436. [DOI] [PubMed] [Google Scholar]
- 95.Martinon F. Dangerous liaisons: mitochondrial DNA meets the NLRP3 inflammasome. Immunity. 2012;36:313–315. doi: 10.1016/j.immuni.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 96.Volt H, Garcia JA, Doerrier C, Diaz-Casado ME, Guerra-Librero A, Lopez LC, Escames G, Tresguerres JA, Acuña-Castroviejo D. Same molecule but different expression: aging and sepsis trigger NLRP3 inflammasome activation, a target of melatonin. J Pineal Res. 2016;60:193–205. doi: 10.1111/jpi.12303. [DOI] [PubMed] [Google Scholar]
- 97.Haneklaus M, O’Neill LA, Coll RC. Modulatory mechanisms controlling the NLRP3 inflammasome in inflammation: recent developments. Curr Opin Immunol. 2013;25:40–45. doi: 10.1016/j.coi.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 98.Buhr ED, Takahashi JS. Molecular components of the mammalian circadian clock. Handb Exp Pharmacol. 2013;217:3–27. doi: 10.1007/978-3-642-25950-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reiter RJ. The melatonin rhythm: both a clock and a calendar. Experientia. 1993;49:654–664. doi: 10.1007/BF01923947. [DOI] [PubMed] [Google Scholar]
- 100.Moore RY, Speh JC, Card JP. The retinohypothalamic tract originates from a distinct subset of retinal ganglion cells. J Comp Neurol. 1995;352:351–366. doi: 10.1002/cne.903520304. [DOI] [PubMed] [Google Scholar]
- 101.Hattar S, Liao HW, Takao M, Berson DM, Yau KW. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science. 2002;295:1065–1070. doi: 10.1126/science.1069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Harrington ME. The ventral lateral geniculate nucleus and the intergeniculate leaflet: interrelated structures in the visual and circadian systems. Neurosci Biobehav Rev. 1997;21:705–727. doi: 10.1016/S0149-7634(96)00019-X. [DOI] [PubMed] [Google Scholar]
- 103.Rosenwasser AM, Turek FW. Neurobiology of circadian rhythm regulation. Sleep Med Clin. 2015;10:403–412. doi: 10.1016/j.jsmc.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 104.Perreau-Lenz S, Pevet P, Buijs RM, Kalsbeek A. The biological clock: the bodyguard of temporal homeostasis. Chronobiol Int. 2004;21:1–25. doi: 10.1081/CBI-120027984. [DOI] [PubMed] [Google Scholar]
- 105.Kalsbeek A, Palm IF, La Fleur SE, Scheer FA, Perreau-Lenz S, Ruiter M, Kreier F, Cailotto C, Buijs RM. SCN outputs and the hypothalamic balance of life. J Biol Rhythms. 2006;21:458–469. doi: 10.1177/0748730406293854. [DOI] [PubMed] [Google Scholar]
- 106.Perreau-Lenz S, Kalsbeek A, Garidou ML, Wortel J, van der Vliet J, van Heijningen C, Simonneaux V, Pevet P, Buijs RM. Suprachiasmatic control of melatonin synthesis in rats: inhibitory and stimulatory mechanisms. Eur J Neurosci. 2003;17:221–228. doi: 10.1046/j.1460-9568.2003.02442.x. [DOI] [PubMed] [Google Scholar]
- 107.Vriend J, Reiter RJ. Melatonin feedback on clock genes: a theory involving the proteasome. J Pineal Res. 2015;58:1–11. doi: 10.1111/jpi.12189. [DOI] [PubMed] [Google Scholar]
- 108.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster . Proc Natl Acad Sci USA. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Goriki A, Hatanaka F, Myung J, Kim JK, Yoritaka T, Tanoue S, Abe T, Kiyonari H, Fujimoto K, Kato Y, Todo T, Matsubara A, Forger D, Takumi T. A novel protein, CHRONO, functions as a core component of the mammalian circadian clock. PLoS Biol. 2014;12:e1001839. doi: 10.1371/journal.pbio.1001839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kondratova AA, Kondratov RV. The circadian clock and pathology of the ageing brain. Nat Rev Neurosci. 2012;13:325–335. doi: 10.1038/nrn3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Takahashi JS. Molecular components of the circadian clock in mammals. Diabetes Obes Metab. 2015;17(Suppl 1):6–11. doi: 10.1111/dom.12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 113.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/S0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 114.Hirayama J, Sassone-Corsi P. Structural and functional features of transcription factors controlling the circadian clock. Curr Opin Genet Dev. 2005;15:548–556. doi: 10.1016/j.gde.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 115.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genom Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hughes ME, Hong HK, Chong JL, Indacochea AA, Lee SS, Han M, Takahashi JS, Hogenesch JB. Brain-specific rescue of Clock reveals system-driven transcriptional rhythms in peripheral tissue. PLoS Genet. 2012;8:e1002835. doi: 10.1371/journal.pgen.1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Buijs FN, Guzman-Ruiz M, Leon-Mercado L, Basualdo MC, Escobar C, Kalsbeek A, Buijs RM. Suprachiasmatic nucleus interaction with the arcuate nucleus; essential for organizing physiological rhythms. eNeuro. 2017;4:e0028. doi: 10.1523/ENEURO.0028-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cermakian N, Westfall S, Kiessling S. Circadian clocks and inflammation: reciprocal regulation and shared mediators. Arch Immunol Ther Exp (Warsz) 2014;62:303–318. doi: 10.1007/s00005-014-0286-x. [DOI] [PubMed] [Google Scholar]
- 119.Dimitrov S, Benedict C, Heutling D, Westermann J, Born J, Lange T. Cortisol and epinephrine control opposing circadian rhythms in T cell subsets. Blood. 2009;113:5134–5143. doi: 10.1182/blood-2008-11-190769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sato S, Sakurai T, Ogasawara J, Takahashi M, Izawa T, Imaizumi K, Taniguchi N, Ohno H, Kizaki T. A circadian clock gene, Rev-erbalpha, modulates the inflammatory function of macrophages through the negative regulation of Ccl2 expression. J Immunol. 2014;192:407–417. doi: 10.4049/jimmunol.1301982. [DOI] [PubMed] [Google Scholar]
- 121.Curtis AM, Bellet MM, Sassone-Corsi P, O’Neill LA. Circadian clock proteins and immunity. Immunity. 2014;40:178–186. doi: 10.1016/j.immuni.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 122.Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, Chawla A. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science. 2013;341:1483–1488. doi: 10.1126/science.1240636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stapleton CM, Jaradat M, Dixon D, Kang HS, Kim SC, Liao G, Carey MA, Cristiano J, Moorman MP, Jetten AM. Enhanced susceptibility of staggerer (RORalphasg/sg) mice to lipopolysaccharide-induced lung inflammation. Am J Physiol Lung Cell Mol Physiol. 2005;289:L144–L152. doi: 10.1152/ajplung.00348.2004. [DOI] [PubMed] [Google Scholar]
- 124.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24:90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Spengler ML, Kuropatwinski KK, Comas M, Gasparian AV, Fedtsova N, Gleiberman AS, Gitlin II, Artemicheva NM, Deluca KA, Gudkov AV, Antoch MP. Core circadian protein CLOCK is a positive regulator of NF-kappaB-mediated transcription. Proc Natl Acad Sci USA. 2012;109:E2457–E2465. doi: 10.1073/pnas.1206274109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu J, Malkani G, Shi X, Meyer M, Cunningham-Runddles S, Ma X, Sun ZS. The circadian clock Period 2 gene regulates gamma interferon production of NK cells in host response to lipopolysaccharide-induced endotoxic shock. Infect Immun. 2006;74:4750–4756. doi: 10.1128/IAI.00287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Narasimamurthy R, Hatori M, Nayak SK, Liu F, Panda S, Verma IM. Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proc Natl Acad Sci USA. 2012;109:12662–12667. doi: 10.1073/pnas.1209965109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 129.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Grimaldi B, Nakahata Y, Kaluzova M, Masubuchi S, Sassone-Corsi P. Chromatin remodeling, metabolism and circadian clocks: the interplay of CLOCK and SIRT1. Int J Biochem Cell Biol. 2009;41:81–86. doi: 10.1016/j.biocel.2008.08.035. [DOI] [PubMed] [Google Scholar]
- 131.Cavadini G, Petrzilka S, Kohler P, Jud C, Tobler I, Birchler T, Fontana A. TNF-alpha suppresses the expression of clock genes by interfering with E-box-mediated transcription. Proc Natl Acad Sci USA. 2007;104:12843–12848. doi: 10.1073/pnas.0701466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]
- 134.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 135.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 137.Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Aquilano K, Vigilanza P, Baldelli S, Pagliei B, Rotilio G, Ciriolo MR. Peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC-1alpha) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis. J Biol Chem. 2010;285:21590–21599. doi: 10.1074/jbc.M109.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Campbell CT, Kolesar JE, Kaufman BA. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim Biophys Acta. 2012;1819:921–929. doi: 10.1016/j.bbagrm.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 140.Tang BL. Sirt1 and the mitochondria. Mol Cells. 2016;39:87–95. doi: 10.14348/molcells.2016.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lee I, Bender E, Arnold S, Kadenbach B. New control of mitochondrial membrane potential and ROS formation—a hypothesis. Biol Chem. 2001;382:1629–1636. doi: 10.1515/BC.2001.198. [DOI] [PubMed] [Google Scholar]
- 142.Broussard SR, McCusker RH, Novakofski JE, Strle K, Shen WH, Johnson RW, Dantzer R, Kelley KW. IL-1beta impairs insulin-like growth factor I-induced differentiation and downstream activation signals of the insulin-like growth factor I receptor in myoblasts. J Immunol. 2004;172:7713–7720. doi: 10.4049/jimmunol.172.12.7713. [DOI] [PubMed] [Google Scholar]
- 143.Radi R, Cassina A, Hodara R, Quijano C, Castro L. Peroxynitrite reactions and formation in mitochondria. Free Radic Biol Med. 2002;33:1451–1464. doi: 10.1016/S0891-5849(02)01111-5. [DOI] [PubMed] [Google Scholar]
- 144.Radi R, Cassina A, Hodara R. Nitric oxide and peroxynitrite interactions with mitochondria. Biol Chem. 2002;383:401–409. doi: 10.1515/BC.2002.044. [DOI] [PubMed] [Google Scholar]
- 145.Li Y, Zhu H, Kuppusamy P, Zweier JL, Trush MA. Mitochondrial electron transport chain-derived superoxide exits macrophages: implications for mononuclear cell-mediated pathophysiological processes. React Oxyg Species. 2016;1:81–98. doi: 10.20455/ros.2016.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hardeland R, Coto-Montes A, Poeggeler B. Circadian rhythms, oxidative stress, and antioxidative defense mechanisms. Chronobiol Int. 2003;20:921–962. doi: 10.1081/CBI-120025245. [DOI] [PubMed] [Google Scholar]
- 149.Kang TH, Sancar A. Circadian regulation of DNA excision repair: implications for chrono-chemotherapy. Cell Cycle. 2009;8:1665–1667. doi: 10.4161/cc.8.11.8707. [DOI] [PubMed] [Google Scholar]
- 150.Sachdeva UM, Thompson CB. Diurnal rhythms of autophagy: implications for cell biology and human disease. Autophagy. 2008;4:581–589. doi: 10.4161/auto.6141. [DOI] [PubMed] [Google Scholar]
- 151.Richards J, Gumz ML. Advances in understanding the peripheral circadian clocks. FASEB J. 2012;26:3602–3613. doi: 10.1096/fj.12-203554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lee HM, Chen R, Kim H, Etchegaray JP, Weaver DR, Lee C. The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc Natl Acad Sci USA. 2011;108:16451–16456. doi: 10.1073/pnas.1107178108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Durgan DJ, Young ME. The cardiomyocyte circadian clock: emerging roles in health and disease. Circ Res. 2010;106:647–658. doi: 10.1161/CIRCRESAHA.109.209957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sabath E, Salgado-Delgado R, Guerrero-Vargas NN, Guzman-Ruiz MA, del Carmen Basualdo M, Escobar C, Buijs RM. Food entrains clock genes but not metabolic genes in the liver of suprachiasmatic nucleus lesioned rats. FEBS Lett. 2014;588:3104–3110. doi: 10.1016/j.febslet.2014.06.045. [DOI] [PubMed] [Google Scholar]
- 155.Silver AC, Arjona A, Hughes ME, Nitabach MN, Fikrig E. Circadian expression of clock genes in mouse macrophages, dendritic cells, and B cells. Brain Behav Immun. 2012;26:407–413. doi: 10.1016/j.bbi.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Fortier EE, Rooney J, Dardente H, Hardy MP, Labrecque N, Cermakian N. Circadian variation of the response of T cells to antigen. J Immunol. 2011;187:6291–6300. doi: 10.4049/jimmunol.1004030. [DOI] [PubMed] [Google Scholar]
- 157.Buijs RM, Scheer FA, Kreier F, Yi C, Bos N, Goncharuk VD, Kalsbeek A. Organization of circadian functions: interaction with the body. Prog Brain Res. 2006;153:341–360. doi: 10.1016/S0079-6123(06)53020-1. [DOI] [PubMed] [Google Scholar]
- 158.Paschos GK, Ibrahim S, Song WL, Kunieda T, Grant G, Reyes TM, Bradfield CA, Vaughan CH, Eiden M, Masoodi M, Griffin JL, Wang F, Lawson JA, Fitzgerald GA. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med. 2012;18:1768–1777. doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lerner AB, Case JD, Takahasi Y, Lee TH, Mori W. Isolation of melatonin, the pineal gland factor that lightens melanocytes. J Am Chem Soc. 1958;80:2587. doi: 10.1021/ja01543a060. [DOI] [Google Scholar]
- 160.Lerner AB, Case JD, Takahashy Y, Lee TH. Structure of melatonin and 5-methoxy-indole-3-acetic acid from bovine pineal gland. J Biol Chem. 1960;235:1992–1997. [PubMed] [Google Scholar]
- 161.Acuña-Castroviejo D, Escames G, Venegas C, Diaz-Casado ME, Lima-Cabello E, Lopez LC, Rosales-Corral S, Tan DX, Reiter RJ. Extrapineal melatonin: sources, regulation, and potential functions. Cell Mol Life Sci. 2014;71:2997–3025. doi: 10.1007/s00018-014-1579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Venegas C, Garcia JA, Escames G, Ortiz F, Lopez A, Doerrier C, Garcia-Corzo L, Lopez LC, Reiter RJ, Acuña-Castroviejo D. Extrapineal melatonin: analysis of its subcellular distribution and daily fluctuations. J Pineal Res. 2012;52:217–227. doi: 10.1111/j.1600-079X.2011.00931.x. [DOI] [PubMed] [Google Scholar]
- 163.Jockers R, Delagrange P, Dubocovich ML, Markus RP, Renault N, Tosini G, Cecon E, Zlotos DP. Update on melatonin receptors: IUPHAR review 20. Br J Pharmacol. 2016;173:2702–2725. doi: 10.1111/bph.13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Becker-Andre M, Wiesenberg I, Schaeren-Wiemers N, Andre E, Missbach M, Saurat JH, Carlberg C. Pineal gland hormone melatonin binds and activates an orphan of the nuclear receptor superfamily. J Biol Chem. 1994;269:28531–28534. [PubMed] [Google Scholar]
- 165.Romero MP, Garcia-Perganeda A, Guerrero JM, Osuna C. Membrane-bound calmodulin in Xenopus laevis oocytes as a novel binding site for melatonin. FASEB J. 1998;12:1401–1408. doi: 10.1096/fasebj.12.13.1401. [DOI] [PubMed] [Google Scholar]
- 166.Macias M, Escames G, Leon J, Coto A, Sbihi Y, Osuna A, Acuña-Castroviejo D. Calreticulin–melatonin. An unexpected relationship. Eur J Biochem. 2003;270:832–840. doi: 10.1046/j.1432-1033.2003.03430.x. [DOI] [PubMed] [Google Scholar]
- 167.Martin M, Macias M, Escames G, Leon J, Acuña-Castroviejo D. Melatonin but not vitamins C and E maintains glutathione homeostasis in t-butyl hydroperoxide-induced mitochondrial oxidative stress. FASEB J. 2000;14:1677–1679. doi: 10.1096/fj.99-0865fje. [DOI] [PubMed] [Google Scholar]
- 168.Tan DX, Manchester LC, Liu X, Rosales-Corral SA, Acuña-Castroviejo D, Reiter RJ. Mitochondria and chloroplasts as the original sites of melatonin synthesis: a hypothesis related to melatonin’s primary function and evolution in eukaryotes. J Pineal Res. 2013;54:127–138. doi: 10.1111/jpi.12026. [DOI] [PubMed] [Google Scholar]
- 169.Tan DX, Manchester LC, Sanchez-Barcelo E, Mediavilla MD, Reiter RJ. Significance of high levels of endogenous melatonin in Mammalian cerebrospinal fluid and in the central nervous system. Curr Neuropharmacol. 2010;8:162–167. doi: 10.2174/157015910792246182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 171.Teclemariam-Mesbah R, Ter Horst GJ, Postema F, Wortel J, Buijs RM. Anatomical demonstration of the suprachiasmatic nucleus-pineal pathway. J Comp Neurol. 1999;406:171–182. doi: 10.1002/(SICI)1096-9861(19990405)406:2<171::AID-CNE3>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 172.Klein DC. Arylalkylamine N-acetyltransferase: “the timezyme”. J Biol Chem. 2007;282:4233–4237. doi: 10.1074/jbc.R600036200. [DOI] [PubMed] [Google Scholar]
- 173.Ribelayga C, Pevet P, Simonneaux V. HIOMT drives the photoperiodic changes in the amplitude of the melatonin peak of the Siberian hamster. Am J Physiol Regul Integr Comp Physiol. 2000;278:R1339–R1345. doi: 10.1152/ajpregu.2000.278.5.R1339. [DOI] [PubMed] [Google Scholar]
- 174.Cassone VM. Effects of melatonin on vertebrate circadian systems. Trends Neurosci. 1990;13:457–464. doi: 10.1016/0166-2236(90)90099-V. [DOI] [PubMed] [Google Scholar]
- 175.Castillo-Romero JL, Vives-Montero F, Reiter RJ, Acuña-Castroviejo D. Pineal modulation of the rat caudate-putamen spontaneous neuronal activity: roles of melatonin and vasotocin. J Pineal Res. 1993;15:147–152. doi: 10.1111/j.1600-079X.1993.tb00522.x. [DOI] [PubMed] [Google Scholar]
- 176.Escames G, Macias M, Leon J, Garcia J, Khaldy H, Martin M, Vives F, Acuña-Castroviejo D. Calcium-dependent effects of melatonin inhibition of glutamatergic response in rat striatum. J Neuroendocrinol. 2001;13:459–466. doi: 10.1046/j.1365-2826.2001.00656.x. [DOI] [PubMed] [Google Scholar]
- 177.Escames G, Leon J, Lopez LC, Acuña-Castroviejo D. Mechanisms of N-methyl-d-aspartate receptor inhibition by melatonin in the rat striatum. J Neuroendocrinol. 2004;16:929–935. doi: 10.1111/j.1365-2826.2004.01250.x. [DOI] [PubMed] [Google Scholar]
- 178.Agez L, Laurent V, Pevet P, Masson-Pevet M, Gauer F. Melatonin affects nuclear orphan receptors mRNA in the rat suprachiasmatic nuclei. Neuroscience. 2007;144:522–530. doi: 10.1016/j.neuroscience.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 179.Reppert SM, Weaver DR, Rivkees SA, Stopa EG. Putative melatonin receptors in a human biological clock. Science. 1988;242:78–81. doi: 10.1126/science.2845576. [DOI] [PubMed] [Google Scholar]
- 180.Liu C, Weaver DR, Jin X, Shearman LP, Pieschl RL, Gribkoff VK, Reppert SM. Molecular dissection of two distinct actions of melatonin on the suprachiasmatic circadian clock. Neuron. 1997;19:91–102. doi: 10.1016/S0896-6273(00)80350-5. [DOI] [PubMed] [Google Scholar]
- 181.Reiter RJ, Tan DX, Kim SJ, Cruz MH. Delivery of pineal melatonin to the brain and SCN: role of canaliculi, cerebrospinal fluid, tanycytes and Virchow-Robin perivascular spaces. Brain Struct Funct. 2014;219:1873–1887. doi: 10.1007/s00429-014-0719-7. [DOI] [PubMed] [Google Scholar]
- 182.Crespo E, Macias M, Pozo D, Escames G, Martin M, Vives F, Guerrero JM, Acuña-Castroviejo D. Melatonin inhibits expression of the inducible NO synthase II in liver and lung and prevents endotoxemia in lipopolysaccharide-induced multiple organ dysfunction syndrome in rats. FASEB J. 1999;13:1537–1546. [PubMed] [Google Scholar]
- 183.Reagan-Shaw S, Nihal M, Ahmad N. Dose translation from animal to human studies revisited. FASEB J. 2008;22:659–661. doi: 10.1096/fj.07-9574LSF. [DOI] [PubMed] [Google Scholar]
- 184.Shi D, Xiao X, Wang J, Liu L, Chen W, Fu L, Xie F, Huang W, Deng W. Melatonin suppresses proinflammatory mediators in lipopolysaccharide-stimulated CRL1999 cells via targeting MAPK, NF-kappaB, c/EBPbeta, and p300 signaling. J Pineal Res. 2012;53:154–165. doi: 10.1111/j.1600-079X.2012.00982.x. [DOI] [PubMed] [Google Scholar]
- 185.Nopparat C, Sinjanakhom P, Govitrapong P. Melatonin reverses H2O2-induced senescence in SH-SY5Y cells by enhancing autophagy via sirtuin 1 deacetylation of the RelA/p65 subunit of NF-kappaB. J Pineal Res. 2017 doi: 10.1111/jpi.12407. [DOI] [PubMed] [Google Scholar]
- 186.Muxel SM, Pires-Lapa MA, Monteiro AW, Cecon E, Tamura EK, Floeter-Winter LM, Markus RP. NF-kappaB drives the synthesis of melatonin in RAW 264.7 macrophages by inducing the transcription of the arylalkylamine-N-acetyltransferase (AA-NAT) gene. PLoS One. 2012;7:e52010. doi: 10.1371/journal.pone.0052010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Muxel SM, Laranjeira-Silva MF, Carvalho-Sousa CE, Floeter-Winter LM, Markus RP. The RelA/cRel nuclear factor-kappaB (NF-kappaB) dimer, crucial for inflammation resolution, mediates the transcription of the key enzyme in melatonin synthesis in RAW 264.7 macrophages. J Pineal Res. 2016;60:394–404. doi: 10.1111/jpi.12321. [DOI] [PubMed] [Google Scholar]
- 188.Lin YW, Lee LM, Lee WJ, Chu CY, Tan P, Yang YC, Chen WY, Yang SF, Hsiao M, Chien MH. Melatonin inhibits MMP-9 transactivation and renal cell carcinoma metastasis by suppressing Akt-MAPKs pathway and NF-kappaB DNA-binding activity. J Pineal Res. 2016;60:277–290. doi: 10.1111/jpi.12308. [DOI] [PubMed] [Google Scholar]
- 189.Arulkumaran N, Deutschman CS, Pinsky MR, Zuckerbraun B, Schumacker PT, Gomez H, Gomez A, Murray P, Kellum JA, Workgroup AX. Mitochondrial function in sepsis. Shock. 2016;45:271–281. doi: 10.1097/SHK.0000000000000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ghafourifar P, Richter C. Nitric oxide synthase activity in mitochondria. FEBS Lett. 1997;418:291–296. doi: 10.1016/S0014-5793(97)01397-5. [DOI] [PubMed] [Google Scholar]
- 191.Kanai AJ, Pearce LL, Clemens PR, Birder LA, VanBibber MM, Choi SY, de Groat WC, Peterson J. Identification of a neuronal nitric oxide synthase in isolated cardiac mitochondria using electrochemical detection. Proc Natl Acad Sci USA. 2001;98:14126–14131. doi: 10.1073/pnas.241380298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Doerrier C, Garcia JA, Volt H, Diaz-Casado ME, Lima-Cabello E, Ortiz F, Luna-Sanchez M, Escames G, Lopez LC, Acuña-Castroviejo D. Identification of mitochondrial deficits and melatonin targets in liver of septic mice by high-resolution respirometry. Life Sci. 2015;121:158–165. doi: 10.1016/j.lfs.2014.11.031. [DOI] [PubMed] [Google Scholar]
- 193.Doerrier C, Garcia JA, Volt H, Diaz-Casado ME, Luna-Sanchez M, Fernandez-Gil B, Escames G, Lopez LC, Acuña-Castroviejo D. Permeabilized myocardial fibers as model to detect mitochondrial dysfunction during sepsis and melatonin effects without disruption of mitochondrial network. Mitochondrion. 2016;27:56–63. doi: 10.1016/j.mito.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 194.Schopfer F, Riobo N, Carreras MC, Alvarez B, Radi R, Boveris A, Cadenas E, Poderoso JJ. Oxidation of ubiquinol by peroxynitrite: implications for protection of mitochondria against nitrosative damage. Biochem J. 2000;349:35–42. doi: 10.1042/bj3490035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Neri M, Riezzo I, Pomara C, Schiavone S, Turillazzi E. Oxidative-nitrosative stress and myocardial dysfunctions in sepsis: evidence from the literature and postmortem observations. Mediat Inflamm. 2016;2016:3423450. doi: 10.1155/2016/3423450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Martin M, Macias M, Escames G, Reiter RJ, Agapito MT, Ortiz GG, Acuña-Castroviejo D. Melatonin-induced increased activity of the respiratory chain complexes I and IV can prevent mitochondrial damage induced by ruthenium red in vivo. J Pineal Res. 2000;28:242–248. doi: 10.1034/j.1600-079X.2000.280407.x. [DOI] [PubMed] [Google Scholar]
- 197.Martin M, Macias M, Leon J, Escames G, Khaldy H, Acuña-Castroviejo D. Melatonin increases the activity of the oxidative phosphorylation enzymes and the production of ATP in rat brain and liver mitochondria. Int J Biochem Cell Biol. 2002;34:348–357. doi: 10.1016/S1357-2725(01)00138-8. [DOI] [PubMed] [Google Scholar]
- 198.Acuña-Castroviejo D, Escames G, Leon J, Carazo A, Khaldy H. Mitochondrial regulation by melatonin and its metabolites. Adv Exp Med Biol. 2003;527:549–557. doi: 10.1007/978-1-4615-0135-0_63. [DOI] [PubMed] [Google Scholar]
- 199.Lopez A, Garcia JA, Escames G, Venegas C, Ortiz F, Lopez LC, Acuña-Castroviejo D. Melatonin protects the mitochondria from oxidative damage reducing oxygen consumption, membrane potential, and superoxide anion production. J Pineal Res. 2009;46:188–198. doi: 10.1111/j.1600-079X.2008.00647.x. [DOI] [PubMed] [Google Scholar]
- 200.Lowes DA, Almawash AM, Webster NR, Reid VL, Galley HF. Melatonin and structurally similar compounds have differing effects on inflammation and mitochondrial function in endothelial cells under conditions mimicking sepsis. Br J Anaesth. 2011;107:193–201. doi: 10.1093/bja/aer149. [DOI] [PubMed] [Google Scholar]
- 201.Lopez LC, Escames G, Ortiz F, Ros E, Acuña-Castroviejo D. Melatonin restores the mitochondrial production of ATP in septic mice. Neuro Endocrinol Lett. 2006;27:623–630. [PubMed] [Google Scholar]
- 202.Reyes-Toso CF, Rebagliati IR, Ricci CR, Linares LM, Albornoz LE, Cardinali DP, Zaninovich A. Effect of melatonin treatment on oxygen consumption by rat liver mitochondria. Amino Acids. 2006;31:299–302. doi: 10.1007/s00726-005-0280-z. [DOI] [PubMed] [Google Scholar]
- 203.Ortiz F, Acuña-Castroviejo D, Doerrier C, Dayoub JC, Lopez LC, Venegas C, Garcia JA, Lopez A, Volt H, Luna-Sanchez M, Escames G. Melatonin blunts the mitochondrial/NLRP3 connection and protects against radiation-induced oral mucositis. J Pineal Res. 2015;58:34–49. doi: 10.1111/jpi.12191. [DOI] [PubMed] [Google Scholar]
- 204.Rahim I, Djerdjouri B, Sayed RK, Fernández-Ortiz M, Fernández-Gil B, Hidalgo-Gutiérrez A, López LC, Escames G, Reiter RJ, Acuña-Castroviejo D (2017) Melatonin administration to wild-type mice and non-treated NLRP3 mutant mice share similar inhibition of the inflammatory response during sepsis J Pineal Res (in press) [DOI] [PubMed]
- 205.Guerrero-Vargas NN, Salgado-Delgado R, Basualdo Mdel C, Garcia J, Guzman-Ruiz M, Carrero JC, Escobar C, Buijs RM. Reciprocal interaction between the suprachiasmatic nucleus and the immune system tunes down the inflammatory response to lipopolysaccharide. J Neuroimmunol. 2014;273:22–30. doi: 10.1016/j.jneuroim.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 206.Ohdo S, Koyanagi S, Suyama H, Higuchi S, Aramaki H. Changing the dosing schedule minimizes the disruptive effects of interferon on clock function. Nat Med. 2001;7:356–360. doi: 10.1038/85507. [DOI] [PubMed] [Google Scholar]
- 207.Duhart JM, Leone MJ, Paladino N, Evans JA, Castanon-Cervantes O, Davidson AJ, Golombek DA. Suprachiasmatic astrocytes modulate the circadian clock in response to TNF-alpha. J Immunol. 2013;191:4656–4664. doi: 10.4049/jimmunol.1300450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kandalepas PC, Mitchell JW, Gillette MU. Melatonin signal transduction pathways require e-box-mediated transcription of Per1 and Per2 to reset the SCN Clock at dusk. PLoS One. 2016;11:e0157824. doi: 10.1371/journal.pone.0157824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bellet MM, Zocchi L, Sassone-Corsi P. The RelB subunit of NFkappaB acts as a negative regulator of circadian gene expression. Cell Cycle. 2012;11:3304–3311. doi: 10.4161/cc.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Escames G, Acuña-Castroviejo D, Lopez LC, Tan DX, Maldonado MD, Sanchez-Hidalgo M, Leon J, Reiter RJ. Pharmacological utility of melatonin in the treatment of septic shock: experimental and clinical evidence. J Pharm Pharmacol. 2006;58:1153–1165. doi: 10.1211/jpp.58.9.0001. [DOI] [PubMed] [Google Scholar]
- 211.Galley HF, Lowes DA, Allen L, Cameron G, Aucott LS, Webster NR. Melatonin as a potential therapy for sepsis: a phase I dose escalation study and an ex vivo whole blood model under conditions of sepsis. J Pineal Res. 2014;56:427–438. doi: 10.1111/jpi.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Andersen LP, Gogenur I, Rosenberg J, Reiter RJ. The safety of melatonin in humans. Clin Drug Investig. 2016;36:169–175. doi: 10.1007/s40261-015-0368-5. [DOI] [PubMed] [Google Scholar]
- 213.Esteban-Zubero E, Alatorre-Jimenez MA, Lopez-Pingarron L, Reyes-Gonzales MC, Almeida-Souza P, Cantin-Golet A, Ruiz-Ruiz FJ, Tan DX, Garcia JJ, Reiter RJ. Melatonin’s role in preventing toxin-related and sepsis-mediated hepatic damage: a review. Pharmacol Res. 2016;105:108–120. doi: 10.1016/j.phrs.2016.01.018. [DOI] [PubMed] [Google Scholar]