Abstract
Purpose
microRNA (miRNA) regulates diverse biological mechanisms and metabolisms in plants and animals. Thus, the discoveries of miRNA has revolutionized the life sciences and medical research.The miRNA represses and cleaves the targeted mRNA by binding perfect or near perfect or imperfect complementary base pairs by RNA-induced silencing complex (RISC) formation during biogenesis process. One miRNA interacts with one or more mRNA genes and vice versa, hence takes part in causing various diseases. In this paper, the different microRNA target databases and their functional annotations developed by various researchers have been reviewed. The concurrent research review aims at comprehending the significance of miRNA and presenting the existing status of annotated miRNA target resources built by researchers henceforth discovering the knowledge for diagnosis and prognosis.
Methods and results
This review discusses the applications and developmental methodologies for constructing target database as well as the utility of user interface design. An integrated architecture is drawn and a graphically comparative study of present status of miRNA targets in diverse diseases and various biological processes is performed. These databases comprise of information such as miRNA target-associated disease, transcription factor binding sites (TFBSs) in miRNA genomic locations, polymorphism in miRNA target, A-to-I edited target, Gene Ontology (GO), genome annotations, KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways, target expression analysis, TF–miRNA and miRNA–mRNA interaction networks, drugs–targets interactions, etc.
Conclusion
miRNA target databases contain diverse experimentally and computationally predicted target through various algorithms. The comparison of various miRNA target database has been performed on various parameters. The computationally predicted target databases suffer from false positive information as there is no common theory for prediction of miRNA targets. The review conclusion emphasizes the need of more intelligent computational improvement for the miRNA target identification, their functional annotations and datasbase development.
Keywords: Bioinformatics, Data mining, Genome, Biomarker, Pathology, High-throughput
Introduction
The microRNAs (miRNAs) are small non-coding RNAs (ncRNAs) which play dominant role in post-transcriptional process as a gene repressor or a destroyer of targeted mRNA by binding with complementary base pair [1]. The miRNA is 20–24 nt in length binding sequence with specific order such as partial or near perfect complement on 3′UTR, 5′UTR or seed regions and coding regions of targeted mRNA to cleave and repress translation in plants, animals and viruses [1, 2]. The large number of protein coding genes is under control of miRNA which is regulated tightly [2]. Before 1993, scientific community had been concentrating only on coding sequence for controlling expression behaviour of genes and had kept non-coding RNAs aside. The first miRNA, lin-4 was discovered in 1993 by Lee et al. in the study of larval development of nematode (C. elegance) [3]. Then after seven years (2000) another 21 nt length miRNA Let-7 was discovered by Reinhart et al. in L4-to-adult evolution of larval growth [4]. Since 2000, the thousands of miRNAs have been discovered that take critical part in biological process of plants, animals and viruses. Afterwards revolution occurred in the research of new class named as small ncRNAs. The ncRNAs have three classes, miRNAs, small interfering RNA (siRNAs) and piwi-interacting RNA (piRNAs).
The miRNA controls diverse biological mechanisms such as cell differentiation, proliferation, growth, mobility, apoptosis and fat metabolism in animals and protein degradation, response to environmental stress, pathogen invasive in plants [5–9]. The miRNA deregulation is tissue specific such as one miRNA deregulates in breast cancer while regulates in lung cancer. This is because of genetic diversity of tumours and cancer cell lines. The deregulation or cleavage of genes by miRNA causes various viral or infectious diseases, immune diseases, neurons diseases, cancer, cardio vascular and myogenesis in animals and stress resistance degradation in plants [10–23]. Target identification is important step in the biomarker and drug discovery process for diagnosis and cure of the diseases [24, 25]. The miRNA can be used as biomarker because they are also found in plasma and serum [26, 27].
microRNA biogenesis
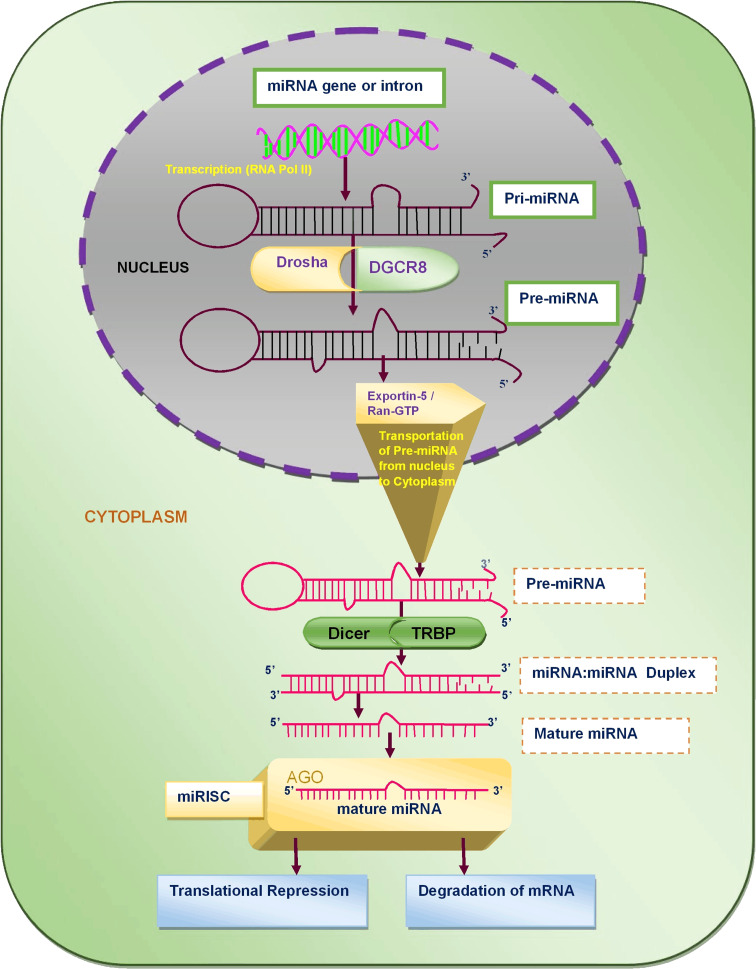
miRNA biogenesis (Fig. 1) includes numerous chronological processes from nucleus to cytoplasm for the breeding of mature miRNA. Initially, the primary miRNA (pri-miRNA) is generated in the nucleus by transcription of miRNA gene through RNA pol-II enzyme. The pri-miRNA is converted into precursor miRNA (hair-pin premature miRNA) through Drosha and DGCR8 (DiGeorge syndrome critical region gene 8). Afterwards, the pre-miRNA is transported into cytoplasm through Exportine-5 and Ran-GTP. The pre-miRNA arrived in cytoplasm is processed by DICER and TRBP (transactivating response RNA-binding protein) to form miRNA duplex. Further, one strand decomposed to lead mature miRNA. It is loaded into argonaute proteins to form miRISC (mi-RNA-induced silencing complex). In this complex, miRNA bind with mRNA using imperfect or perfect base pairing cause translation repression or mRNA degradation [28–33].
Fig. 1.
The diagrammatic representation of miRNA biogenesis
Identification and prediction techniques of miRNA target
This section has discussed experimental and computational techniques for the target identification which are used in miRNA target database development. The miRNA targets are identified by various low- and high-throughput expression technologies such as qRT-PCR, luciferase reporter assays, western blot, microarray, proteomics, CLIP-seq, degradome and others [34, 35]. The predicted target is validated with gene-specific studies by PCR, reporter assays and western blot techniques. Using the above technologies, researchers can experimentally identify miRNA targets by various profiling of components such as gene-specific target validations, conditions of miRNA over-expression, gene expression analysis, immunoprecipitation of the RISC components, Argonaute (AGO)or TNRC6, high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation (HITS-CLIP), photoactivatableribonucleoside-enhanced crosslinking and immunoprecipitation(PAR-CLIP), Biotin tagged miRNA study, direct cleavage targets, reverse transcription of targets, stable isotope labelling with amino acids in cell culture (SILAC), translation profiling and CLASH (Cross-linking Ligation and Sequencing of Hybrids). These methods have won merit and demerit mentioned in references [34–37].
The miRNA binds near perfect or perfect complementary in miRNA:mRNA duplex in plants and near perfect or imperfect complementary in miRNA:mRNA duplex in animals [36, 37]. Animals have more complex predictions of target identification than plants due to imperfect complementarity among binding of targets. The several computational tools have been reported for target predictions are based on evolutionary conserved regions and target binding sites such as 3′UTR and 5′UTR sites and coding regions in plants as well as animals [36–38]. The miRNA target identified by various methods such as Ab-initio, machine learning (support vector machine (SVM), genetic algorithm (GA), genetic programming (GP), artificial neural network (ANN), ensemble learning, Bayesian data learning Boosting, etc.) and hybrid approaches. These methods used to predict targets based on diverse features such as conservation, free energy of duplex and miRNA target accessibility, seed match, expression behaviour, target inhibitions, etc. are reported by many scientists [37–39]. The Ab-initio approach indirectly identifies miRNA target from experimental data through computational models. The machine learning approach directly predicts target from experimental data through supervised learning or semi-supervised learning of known target data. The hybrid approach is combination of both the approaches. The various computational tools such as psRNATarget [40], psRobot [41], TAPIR [42], patscan [43], miRU [44], TargetFinder [45, 46], TargetAling [47], Target Prediction [48], pTAREE [49], TargetScan [50–53], RNAhybrid [54], miRanda [55], miRchek [56], Find miRNA [57] are used to predict miRNA targets in plants. Each and every tool uses different scoring and prediction techniques. The animal targets are predicted by various tools such as PicTar [58–60],RNA22 [61], RNAhybrid [54], miRanda [55], TargetScan [50–53], PITA [62], EIMMO [63], DIANA-microT [64], MicroInspector [65], EMBL [66], miRwip [67], miRTar [68] along with machine learning based techniques such as TargetBoost (GP with boosting algorithm) [69], GenMIR++ (Bayesian Learning) [70], miTarget (SVM) [71], Targetspy (MultiBoost and rank feature by ReliefF Algorithms) [72], TragetMiner (SVM) [73], miRtif (SVM) [74],Ensemble (Ensemble learning) [75], miRtarget2(SVM) [76], Mtar(ANN) [77], miRSVR(SVM) [78], MiRror (Ensemble learning) [79, 80], miREE(GA and SVM) [81]. These tools use different scoring mechanisms for target predictions.
Computational and experimental miRNA target database with annotations
This review has studied the experimentally as well as computationally predicted target databases furnished with the annotation information (Tables 1, 2, 3). This study is focused on methodology of database development tools and techniques along with applications of databases and user interface design of different target databases.
Table 1.
List of miRNA target databases with experimentally validated targets
| Database name | Number of entry | Version/year | Implementation back/front end | Reference |
|---|---|---|---|---|
| TarBase |
miRNA–gene interactions:65 814 Species :24 Paper curate: 10,000 |
TarBase (2006) Tarbase 5 (2008) Tarbase 6.0 (2012) TarBase (2014) |
Sql Server PHP, Apache |
http://diana.imis.athenainnovation.gr/DianaTools/index.php?r=tarbase/index |
| miRecords |
miRNA-gene interaction: 2705 miRNA: 664 gene:1907 from 9 animal Curated from low through put experiment:2028 |
miRecords (2008) last updated April 2013 |
MySQL, Fedora Core 6 Linux PHP project on Apache 2.0 |
http://mirecords.biolead.org/ |
| miRGator |
Mature miRNA: 1856 Validated target:4745 Predicted target :6 218 792 |
miRGator (2008) miRGator (2011) miRGator (2013) |
MySQL Java script J-Query |
|
| miRWalk | 858,750,070 interactions between 11,748 miRNAs and 308,700 genes |
miRwalk 1.0 (2010) miRWalks2.0 (2014) |
MySQL, JavaScript Perl and BioPerl Scripts, bw GRID (High Performance Cluster) |
http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/ |
| miRTarBase |
Species: 18 Target genes: 22,563 miRNAs: 3786 miRNA–target interactions: 366,181 Articles: 4966 |
miRTarBase 1.0 (2010) miRTarBase 4.0 (2014) miRTarBase 5.0(2015) miRTarBase 6.0(2016) |
MySQL PHP |
http://mirtarbase.mbc.nctu.edu.tw/index.php |
| miRSel |
Human (2112 pairs) Mouse (895 pairs) Rat (231pairs) Other organism: (452 pairs) |
miRSel (2010) |
JAVA (JDK 1.6, ZKoss framework) Tomcat v 6.0 |
http://services.bio.ifi.lmu.de/mirsel/ |
| starBase |
miRNA-circRNA: 9000 miRNA-pseudogene: 16 000 protein–RNA: 285 000 |
StarBase (2011) Starbase (2014) |
MySQL PHP on Apache |
http://starbase.sysu.edu.cn |
| PMTED |
miRNA:1897 Target: 5449 Species:12 Experiment:311 Assay:3492 |
PMTED (2013) |
MySql PHP HTML |
http://pmted.agrinome.org/ |
| ComiRNet |
5 million MTIs miRNA: 934 mRNA:30,835 biclusterhierarchies:15 |
ComiRNet(2015) |
PostgreSql Play2.2 framework runs on server Netty (netty.io) |
http://www.comirnet.di.uniba.it |
| MtiBase |
Gene: 15,546 miRNA: 4420 4 400 000 CDS - 470 000 5′UTR miRNA target sites SNP influence 290,000 CDS and 28,000 5′UTR miRNA target sites |
MtiBase(2015) |
MySql PHP |
http://mtibase.sysu.edu.cn |
Table 2.
List of Computationally predicted microRNA target database
| Database name | Number of entry | Version/year | Implementation back/ffront end | Machine learning tools / Techniques | Reference |
|---|---|---|---|---|---|
| TargetScan |
Unique gene Human = 18,393 (30,858 transcripts) Mouse = 18,615 (23,795 transcripts) Worm = 16,257 Fly = 14,053 |
Release ver 1.0. 2.0, 2.1, 3.0, 3.1, 4.0–4.2, 5.0–5.2, 6.0 7.1 (2003–2016) |
MySQL, CGI apache server |
- | http://www.targetscan.org/ |
| miRDB |
miRNA with target, Gene target Unique Target gene of species Human:2588, 947,941, 17,925 Mouse:1912, 634,009, 18,639 Rat: 764,179539, 15,489 Dog:453, 128,703, 13,150 Chicken 992, 214,816, 12,911 |
miRDB ver 1,2,3,4,5 (2006,2007,2009,2012 2016) |
MySQL Linux apache Server PERL PHP5 |
MirTarget | http://mirdb.org/miRDB/ |
| microRNA.org |
miRNA,H. sapiens: 1100 M.musculus: 717 R.norvegicus: 387 D. melanogaster: 186 C.elegans: 233 |
microRNA.org.1(2008) microRNA.2 Ver-2 (2010) |
MySQL Java Struts, JSPs and Javascript |
mirSVR algorithm | http://www.microrna.org/microrna/getMirnaForm.do |
| HOCTAR |
miRNA:290 miRNA target:265,136 Total gene: 9963 Host:230 |
HOCTAR 1.0(2009) HOCTAR 2.0 (2011) |
MySQL ver. 5.0.44 Perl v5.8.8 PHP 4.4.7 Linux server |
– | http://hoctar.tigem.it/ |
| RepTar | 4000 records | RepTar (2010) |
MySql Java sript |
http://reptar.ekmd.huji.ac.il/ | |
| microPIR |
miRNA:3310 gene = 47,090 Human unique target sense & antisense: 40079646,40334822 Mouse target: 21,256,927, 21,431,773 |
microPIR(2012) microPIR(2014) |
MySQL version 5.5.1 Python scripts and toolkits |
– | http://www4a.biotec.or.th/micropir/About |
| miRNA targets |
Target- miRanda-RNAhybrid Human:18,340,081, 25,772,789 Mouse:9,889,849, 6,573,689 Cheken:2,099,138, 984,979 Zebra fish:1,627,051, 709,606 Cow:3,117,593, 733,956 C. elegans :1,375,889, 765,604 D. Melanogaster:1,359,496, 1,327,560 |
miRNA_Targets (2012) |
MySQL PHP |
– | http://mamsap.it.deakin.edu.au/~amitkuma/mirna_targetsnew/find_gene.html |
| Pharmaco-miR |
(VerSe): miRNAs : 105 miRNA-gene: 190 Gene -drugs:210, dugs:72 Pharmaco genomic annotation 269 from 149 article Total miRNA-gene: 1,026,667 |
Pharmaco-miR(2014) | MySql, PHP | – | http://www.pharmaco-mir.org/about |
| multiMiR |
50 million records compile from 14 database |
mutiMiR (July 2014) |
MySql R package PHP |
– |
Table 3.
List of miRNA target databases having TF-miRNAs, SNP, A to I editing target and Virus target
| Database Name | Number of Entry | Version/Year | Implementation Back/Front End |
Reference |
|---|---|---|---|---|
| TransmiR |
Entries:735, TFs ~201 miRNA: ~209, species: 16 publications:268 |
TransmiR 1.1 (2009) TransmiR1.2 (2013) |
SQLite,Django a Python web framework |
http://www.cuilab.cn/transmir |
| PuTmiR |
TF binding to 10 kb USR of miRNA: 601 TF-10 kb DSR-miRNA: 613 Relationship both: 11,485 and 12,251 |
PuTmiR 1.0 (2010) PuTmiR 1.1(2012) PuTmiR 2.0(2016) |
MySql PHP |
http://www.isical.ac.in/~bioinfo_miu/TF-miRNA/TF-miRNA.html [32] |
| miRdSNP |
3′UTR Genes:19,834 3′UTRSNPs:175,351 dSNPs:630 Disease-SNP:786 Diseases:204 Paper:754 |
miRdSNP 11.03 (2012) |
MySql PHP |
http://mirdsnp.ccr.buffalo.edu/index.php |
| PolymiRTS |
SNPs and INDEL in miRNA target sites: 22 979, 1047 miRNA (Human):2578 |
PolymiRTS 2.0(2012) PolymiRTS 3.0(2014) |
MySql Apache 2.2.15 (CentOS) PHP |
http://compbio.uthsc.edu/miRSNP/ |
| miR-EdiTar |
Edited 3′UTRs: 10571 Edited predicted miRNA binding sites: 9532, miRNAs target affected by editing:1664 Novel sites created by editing:1400 |
miE- ediTar(2012) | MySQL (v5.1) ,Apache server (v2.2.15).Ruby on Rails (v2.3.5), Ajax, MVC desing | http://microrna.osumc.edu/mireditar/ |
| ViTa |
Total virus: 28 670 Species: 2156 |
ViTa(2006) |
MySQL PHP on apache |
http://vita.mbc.nctu.edu.tw/index.php |
| Vir-Mir db |
Hairpin candidates: 33 691 Genome: 1491 |
Vir-Mir db (2006) Vir-Mir db(2007) |
MySQL Java script CGI |
http://alk.ibms.sinica.edu.tw/cgi-bin/miRNA/miRNA.cgi |
Experimentally verified microRNA target databases
TarBase
TarBase [82–84] is experimentally verified (gene specific and high-throughput) miRNA target database which includes information about miRNAs-gene, experimental proof along with other functional related data information integrated with UniProt, Ensemble and RefSeq databases. The miRNA target information curated by automated text mining based three segment pipelines such as (1) name entity recognition, (2 miRNA target association recognition, (3) score, text markup and enhancement from index literature in PubMed library which is experimentally verified by Reporter genes, qPCR, blotting, microarray, proteomics (pSILAC), sequencing (RNA sequence, HITS-CLIP, PAR-CLIP), Degradome-Seq and other [enzyme linked immuno sorbent assay (ELISA), rapid amplification of cDNA ends (RACE),immune-histo-chemistry(IHC)] techniques reported by various researchers. The name entity recognition segment includes gene recognition with AIIAGMT, miRNA regular expression and miRNA–gene association recognized by regular expression (based on experimental outcome (+/−), regulation (up/down), tissues specific methodology information).TarBase facilitates interactive user interface for browsing miRNA target information and outcome of hundreds of experimental methods. Each entry has ample information such as description of gene and miRNA, experimental evidence references and gene–miRNA information by KEGG pathways. The user can filter the miRNA- and gene-associated information by advance search and it is also integrated with various experimental target databases (miRTarBase, miR2Diseaseand miRecords). The database is updated periodically along with experimentally validated miRNA targets as well as its experimental evidences. User interface is facing with the problem of multiple clicking in exploring the miRNA functional annotation information [85].
miRecords
miRecords [86] accommodates information of experimentally validated miRNA–target and computationally predicted miRNA–target interaction database of animals accompanied with strong documentation of experimental evidence of miRNA–target interactions. The predicted targets are computed by eleven existing tools (DIANA-microT, MicroInspector, miRanda, mi-Target, Mir-Target2, NBmir-Tar, PicTar, PITA, RNA22, RNAhybrid, TargetScan/TargetScanS). The experimentally validated targets and their documentation are manually retrieved from published literature (PubMed). The miRecords has categorical description of experimentally validated miRNA–target interactions such as endogenous vs exogenous miRNA, corresponding to miRNA (target genes, target regions, and target sites), expression behavior (over or/and under), experimental level (Reporter assays, mRNA or proteins level measurements and others) and target sites mutations. User can download and search validated targets and predicted targets with various search options such as species, miRNA, targets gene from user interface.
miRGator
miRGator [87–89] is a portal of miRNA targets, gene expression, functional and genome annotation of microRNA including enrichment analysis in terms of gene ontology(GO), pathway and disease annotations as well as integration of other expression databases for analytics. This database contains predicted miRNA target by miRanda, PicTar, TargetScanS, PITA, microcosm by genome wide sequencing of mouse and human. miRNA and genome sequences have been drawn out from miRBase and UCSC genome, respectively. Few predicted targets are also extracted from miRDB and microRNA.org. The functional annotation of predicted targets have been extracted from GO, KEGG, GenMAPP/BioCarta and ArrayXpath in terms of gene ontology, pathways and disease ingenuity pathway analysis, respectively. The expression correlation of miRNA, miRNA target, proteins is computed from downloaded expression datasets (GEO expression data set) which results in visualization of key issues pertaining to differentially expressed components in tissues and cell lines of diseases. miRGator user interface has two modules one is target-functional expression and other is miRNA-expression profiles. There are two more versions of miRGator namely miRGator 2.0 and miRGator3.0 [88, 89]. The miRGator 2.0 comprises information on miRNA expression obtained under various investigational conditions, miRNA–miRNA pair expression, miRNA-perturbation (miRNA knock-out or knock-in) and various associations (miRNA–miRNA, miRNA–target, miRNA disease based on expression profiles). This version has mainly three modules (1) data set browser, (2) miRNA sets and gene sets analysis and (3) disease association analysis including associated network visualization. The miRGator 3.0 includes information on diseases caused by miRNA. It has also incorporated miRNA–target deep sequencing data which have been extracted from gene expression omnibus (GEO), sequence read archive (SRA), and the cancer genome atlas (TCGA) database. Gene set analysis of miRNA target has been performed by gene expression comparison. This database has three modules for data analysis such as miR-seq browser, miR target and expression and gene set analysis including search options.
miRWalk
miRWalk [90] is a predicted and validated miRNA target database along with mRNA, mitochondrial genes and 10 kb upstream flanking region of human, mouse and rat genes. The targets predicted by new algorithm “miRWalk” (run on bwGRID server) work by walking on three positions of genome (3′UTR, 5′UTR and Coding regions). miRWalk also includes predicted target information obtained via eight putative miRNA programs such as DIANA-microT, miRanda, miRDB, PicTar, PITA, RNA22, TargetScan, and RNA hybrid. The validated targets are extracted from published literature along with miRNA–miRNA interaction of 15 species (human, orangutan, chimpanzee, monkey, mouse, rat, pig, chicken, dog, cow, frog, zebra fish, opossum, fruit fly and worm). The sequences of miRNA, mRNA, information on 10 kb upstream flanking regions of genes along with promoter region and mitochondrial genes can be accessed from miRBase20, Gene Bank, RefSeq and Ensembl databases, respectively. The nomenclature- and keyword-related information such as gene, miRNAs, (disease, organs), Mendelian Inheritance in Man (OMIM) disorder, proteins participates in miRNA processing and gene pathways are obtained from (Human Genome Organization (HUGO) gene nomenclature database, mouse genome database, Rates genome), miRBase, medical subject heading, OMIM, PubMed and (KEGG, Biocarta) databases, respectively. The validation of curated genes and miRNA is performed with the help of automated text mining tools in PubMed. The miRWalk user interface has two modules one is predicted target and another is validated target module. The validated target modules have following links such as Gene–miRNA targets, miRNAs–Gene Targets, Gene–miRNAs Pathways, Gene–miRNA–GO, Gene class targets, cell lines based miRNAs, Organs-based miRNAs, miRNA base search in organs, Gene–miRNA–OMIM targets, Disease Ontology, human phenotypes ontology, Disease base miRNAs, literature on miRNAs and miRNAs processing Proteins. The predicted modules contain links regarding miRNAs–Gene targets, miRNA–miRNA targets, Gene–miRNA Pathways, Gene–miRNA–GO, Gene class targets, chromosomes targets, Gene–miRNAs–OMIM targets, disease search, Human Phenotypes search, mitochondrial targets, Genomic locations and customize data sets and holistic views. miRWalk is unstable and unreliable database as it produces inaccurate results with maximum searches [85].
miRTarBase
miRTarBase [91, 92] has information on experimentally validated miRNA-target interactions (MTIs) including microRNA target identification, miRNA target molecular network along with system biology, expression data of miRNA with its target and miRNA target-associated diseases. The experimentally validated miRNA-target interactions and its evidence of experimental techniques reporter assays, Western blot, northern blot, qRT-PCR, micro array, pSILAC, NGS (CLIP-seq, Degradome-Seq, and CLASH-Seq) is manually curated from index literature in PubMed library. Molecular network of MTIs can be visualized with the help of Cytoscape web incorporated tools. Functional annotation can be explored through gene ontology and KEGG pathways (DAVID gene tools for annotation). The associated diseases of miRNA target are integrated with HMMD2.0 and miR2Disease along with some miRNA-disease-associated data from literature. The MTIs expression profile data on various phenotype specifics are downloaded from NCBI GEO datasets and analyzed for miRNA regulatory pathways (Phenotype specific) and miRNA–mRNA correlation. The miRTarBase user interface has searching, browsing and downloading options for the data exploration, as well as links for other database. The searching of MTIs information is performed by selecting various options such as miRNA, target gene, disease, experiment technique, pathways and literature and by advance search from user interface. The user can easily download spreadsheets of miRNA target-associated function from download link.
miRSel
miRSel [93] is a database of experimentally verified association of genes and miRNA along with other miRNA–gene association resources such as TarBase, miRecord and miR2Disease. miRNA–gene association is extracted with the help of automated text mining tools (by 70 terminology) according to miRBase naming convention of miRNA equipped with several relationship features such as miRNA target, physical target, co-expression, repression, induction and cleavage from PubMed abstracted literature. The other annotated information (name, symbol, official names, synonyms, abbreviations) of gene, miRNA and proteins is curated from various databases such as gene databases (HUGO Gene nomenclature committee, Mouse Genome Database and Entrez Gene), miRNA database (miRGen, miRBase) and Protein Database (Swiss-Prot), respectively. The selection of 70 terminologies is made after preprocessing of the above dictionaries for miRNA–gene relationships computations (text mining). The miRSel also provides user interface with various searching options (search by gene ID, miRNA ID, PubMed ID and Gene Ontology) for gene–miRNA association on selection of databases and species. The Gene search option requires gene name, symbol, etc. as an input. The miRNA option requires miRNA name, gene family and PubMed ID for searching key words in particular paper as inputs. The Gene Ontology (GO) option requires miRNA–gene association with GO terminology and GO ID.
StarBase
StarBase [94, 95] is miRNA–circRNA, miRNA–pseudogene, miRNA–mRNA, miRNA–lncRNA and miRNA–ceRNA interaction database along with miRFunction and ceRNAFunction tools for function annotation of miRNA and ncRNA, respectively. These interactions are analyzed by identifying AGO (Argonaute) and RNA binding proteins from HITS-CLIP, PAR-CLIP, iCLIP and CLASH dataset (GEO dataset) by identifying binding clusters/peaks. The CLIP-based miRNA targets (high and low conserve) are extracted from TargetScan and miRNA from miRBase (20). The Genomic coordinate of extracted target are computed with the help of various programs such as TargetScan, miRanda/miSVR, PITA, PicTar and RNA22. The collected coordinates are converted into genome assembly (hg19, mm9/mm10, ce6/ce7) by LiftOver tool and then CLIP-supported target is predicted. The miRNA target or gene annotations of various species are extracted from different sources such as human from GENCODE v17, mouse from Ensembl and C. elegans from Liftover (mm9/mm10, ce6/ce10) while circRNA annotations of mouse, human, C. elegans from circBase. The ceRNA pairs are predicted with hyper geometric test. The functional annotations of miRNA and gene with different words such as gene ontology, pathways, protein analysis by evolutionary relationships are drawn from NCBI RefSeq, KEGG pathway database, ((PANTHER) pathways, Reactome Pathway database, Molecular Signatures Database (MSigDB)), respectively. Enrichment analysis of pathways analyzed with hyper geometric test by Bonferroni Correction and FDR (False Discovery Rate). The user interface of StarBase has links for miRNA–mRNA interaction prediction which is obtained by entering gene ID. It also includes other links such as miRNA–lncRNA, Pan-Cancer, Protein–RNA, ceRNA networks, downloads and functional annotation information of miRNA mediated regulatory networks by selecting one or multiple miRNA as per requirement.
PMTED
PMTED [96] is experimentally validated miRNA target expression database of plants including miRNA and their target sequence, gene ontology and differential expression profiles. The miRNA sequences are downloaded from plant microRNAs database (PMRD) for predicting the targets on CDS regions by in-house written PERL script which produces better results in comparison to psRNATarget program. The extracted microarray expression datasets (from GEO) are classified into five groups (subgroup) such as stimulus, development, mutation, small RNA and epigenetic besides others according to the experimental interest (biotic stress, abiotic stress, tissues and developmental stages). This also contains future extracted 55 annotation terminologies on the basis of sub grouping of their experimental conditions among which 25 annotations are from abiotic stress (subgroup: drought, salt, acid), 20 annotations from biotic stress (subgroup: rice stripe virus, powdery mildew, fungus) and 10 annotations from development (subgroup: leaf, root, stem). All extracted terminologies share common experimental platform and process pipeline to help in building of meta-networks. The preprocessing of raw data is carried out with the help of Robin and R (BioConductor)-based program and normalized by multi array average of MAS 5.0. The differential expression of genes under various experimental conditions is determined by linear modeling (one log-fold and p = 0.5 in Robin) and linking common biological terms employed in experiment for analysis meta-data linking [96]. In the user interface, user can search basic miRNA, expression behavior and targets information by selecting option miRNA, experiment, Target ID and target prediction. User can also see the meta-networks of bioprocesses, species, miRNAs family by direct linking with web-cytoscape program.
ComiRNet
ComiRNet (Co-clustered miRNA Regulatory Networks) [97–102] is an online database containing efficient tools for miRNA–gene target interaction discovery and for detection of miRNA function as well as its mechanism. The database is integrated with miRBase, GeneCards, Entrez gene and miRTarBase for the functional annotations of miRNA and gene. ComiRNet adopted two computing approaches such as semi supervised ensemble-based classifier for miRNA–gene target Interaction (MTIs) prediction and bi-clustering algorithm HOCCLUS2 for prediction of miRNA–gene target regulatory networks (MGRNs). Here, Semi-supervised learning is acquired with experimentally validated miRNA-target interactions of miRTarBase (Positive/Known interaction) and predicted target of miRDIP which is not present in miRTarBase (negative/unknown interaction). Semi-supervised learning has been carried out in three steps: (1) representation of the vector score of predicted interaction by each algorithms along with associated label (known/unknown interaction) that shows whether the label is an experimentally validated interaction or not. (2) LIBSVM is non-traditional probabilistic classifier to learn and predict the likelihood that signifies whether the interaction is known or unknown. In case of disproportionate ratio of known or unknown, interaction is 2/25,000 then LIBSVM always predicts the unknown interaction. This problem is resolved in step (3) Learning of a weight-aware classifiers: another probabilistic classifier used to predict likelihood as in step (2), to learn and predict class along with an associated score, which decides whether the interaction is true or false. HOCCLUS2 predicts highlycohesive, possibly overlapping and hierarchically organized bi-cluster by utilising the above predicted set of MTIs for discovering MGRNs. HOCCLUS2 uses three steps to discover bi-clusters: In first step, it generates bi-clusters as bi-cliques based on bidirectional interaction (miRNA to mRNA and mRNA to miRNA). It produces total number of bi-cliques on the basis of selected direction of interaction and β defined threshold value and then merge in a single bi-cliques. In Second step, it predicts the overlapping bi-cluster by SVM model with the help of α defined threshold value and merges it. In third step, it ranks the bi-cluster on the basis of ρ value of statistical test, which evaluates the functional similarity of mRNA in intra bi-cluster over inter bi-cluster. The biological significance of extracted miRNA–gene networks is examined through various ways such as score averaging-three best (SA-3B), Weighted score averaging-three best (WSA-3B) and Score averaging (SA). ComiRNet contains user friendly web interface for query, browsing of MTIs, browsing and exploring of MGRNs. User interface takes list of gene symbol or miRNA ID as an input for searching miRNA interaction and shows results in table; moreover, various file format options are also available as per interest of user. Output display in the table of searched MTIs contains miRNA ID, gene symbol, Entrez gene ID, Interaction score, and validation of MTIs associated information. This table also encompasses hyper links for miRBase, GeneCards, Entrez gene ID, and experimentally validated target link with miRTarBase. ComiRNet stores 15 different types of predicted MGRNs on different combination of α and β score. MGRNs are visualised by the bi-cluster in the form of dynamic graph based network as per predicted bi-cluster by HOCCLUS2.
MtiBase
MtiBase [103] is a web based database that contains information of miRNA target located on coding sequences (CDS) and 5′UTR genes of human and mouse as well as incorporates regulatory effect of miRNA on mRNA stability and translation. It also contains Single Nucleotide polymorphisms (SNPs) that affect CDS and 5′UTR located miRNA target sites. The 61 Ago CLIP–Seq dataset has been downloaded from GEO Expression (NCBI) or starBase. Ago-binding region for 45 datasets out of 61 has been identified from star-Base and for remaining datasets Ago-binding region is predicted as per the process given in star-Base. Later, the miRNA targets are predicted by miRanda, RNA22, miRWalk, microT-CDS and STarMir program. The results of above target prediction algorithms are obtained from respective websites except miRanda. miRanda predicts the target by mature miRNA sequences downloaded from miRBase and the reference sequences of CDS and 5′UTR are downloaded from BioMart of Ensembl. The obtained sequences of the target sites are then mapped to the reference genome to obtain its genomic coordinates using Bowtie. The corresponding CDS and 5′UTR located on miRNA targets that overlap with Ago regions are designated as putative miRNA targets. Species conservation analysis has been carried out using PhastCons conservation scores which have been downloaded from UCSC Bioinformatics website. The Expression profile and CLIP-Seq are extracted from different resources mention in [103]. The experimentally verified CDS and 5′UTR located target sites are curated from published literature. The miRNA affects stability of mRNA which has been analyzed by 222 gene expression dataset in response to miRNAs over-expression or under-expression. There are several steps used in the study of stability of mRNA such as normalised microarray data extracted from GEO and normalised probe intensities extracted through GEO-query package of R software and log twofold changes have been calculated. The effect of miRNA on translational efficiency and proteins synthesis are evaluated by log twofold change of RPF data (from GEO) and pSILAC dataset (pSILAC database). The SNPs are downloaded from dbSNP and their functional annotation from NHGRI-GWAS Catalog. Then lift-over tools are used to convert the genome coordinate of SNPs for comparing the coordinate sites of CDS and 5′UTR target with those SNPs to identify target site intersecting in SNP and to extract overlapping target site from reference genome to generate Ref-Sequence(RNA sequence). Alt-Sequence generated by converting corresponding alleles to Alternative alleles. SNPs associated with miRNA target predicted by RNA-hybrid using Ref-sequence and Alt-Sequence. GO Enrichment of SNPs are analyzed by PANTHER. MtiBase contains user friendly interface having many links for CDS and 5′UTR target analysis including miRNA target, miRNA effect, SNPs, validated miRNA target, downloads and submit link. User search CDS and 5′UTR target sites from miRNA target links and result shows in optimal search such as prediction program details, conservation score and CLIP-Seg details. User can search miRNA effect on the stability of mRNA and translation analysis from miRNA effect module by choosing gene expression and PSILAC dataset option. The SNP associated target search from SNP modules and validated CDS and 5′UTR target data from validated target modules. User can also download and upload the Ago-CLIP-seq, gene expression, RPF and pSILAC dataset from download and submit links.
Computationally predicted microRNA target databases
TargetScan
TargetScan [50–53] is a database of predicted miRNA target of human, mouse, fly, worm, and fish. The TargetScan algorithm predicts the target with the help of comparison of seed region in presence of conserved 8mer and 7mer, and ORF region conserve and non-conserve target site predicted with including features such as AU-rich regions near sites, co-express miRNA, 13–16 residue pairing, 15 nt within 3′UTR from stop codon and position away from long UTR centre. The identification of target sites is predicted by comparison of mismatch in seed region and 3′UTR combination. Here, predicted targets are ranked on the basis of four features such as site-type contribution, 3′UTR pairing contribution, local AU contribution and position contribution (base is context + Score). The annotation of 3′UTR and its orthologue matches are considered as defined by UCSC genome alignment. TargetScan user interface has miRNA target search option according to the selected link of species. The searching of miRNA target by selecting input options such as gene, conserve family and miRNA name.
miRDB
miRDB [76, 104] is a database of miRNA targets and their functional annotations based on mature RNA (included multiple precursors) with the help of miRNA-mediated gene regulation and wiki interface. The prediction of targets by a machine learning based algorithms i.e., MirTarget (used SVM) including down regulation feature of miRNA selected from miR-124 dataset (miRBase) and selected genes from TarBase. The annotation of miRNA sequences, functions, precursor genome location and nomenclature is the same as mentioned in miRBase, along with validated target links with TarBase and pathways from PANTHER pathway. This database also has tissue expression profiles as for (Liang et al. 2007) RT-PCR and seed sequence as for mentioned in (Levis et al. 2005 and Linsley et al. 2007) as well as secondary structure predicted by RNA fold. miRDB (2016) version includes miRNA target predicted by CLIP-ligation study. New version user interface of miRDB contains Target-Search module for searching miRNA target from database, Target-Mining module of searching miRNA Target by choosing multiple option, Custom-Prediction module for prediction of miRNA target of user provided gene or miRNA, FuncMir-collection module for browsing of functional annotation of pre-miRNA and download module for downloading the text file for storing miRNA in database. The old version has wiki interface which provides a facility to user to edit functional annotation of miRNA. User can update with internet access. This process can be performed in two ways such as transcluding template files of miRNAs (produced by automated pipeline and update by admin) or miRNAs page description including history associated tabs (user can rollbacks the annotation done by previous persons).
MicroRNA.org
MicroRNA.org [105, 106] is a resource of miRNA targets along with information on normal and abnormal tissues or cell lines of miRNA expression profile database. The miRNA target has been predicted by miRanda algorithm with complementary sequence comparison between mature miRNA sequence and mRNA sequence by weighted dynamic programming (Smith–Waterman Algorithm) with help of extracted datasets of miRNA and 3′UTR sequence of gene from miRBase 10.0, UCSC genome table, respectively. The miRNA expressions of normal vs disease tissues or cell lines have been extracted from published literature. The microRNA.org (version 2010) contains ranking of miRNA target sites by mirSVR algorithm based on down regulation score through identifying non-canonical and non-conserved sites. This resource provides interface for searching of miRNA target with fuzzy option, input as miRNA and gene, based on user interests and results shows annotated information of target genes and detailed binding sites. User can search tissue specific expression profile of miRNA by selected species from target expression module. We can also download miRNA target score and expression files of miRNA.
HOCTAR
HOCTAR (Host Gene Opposite Correlated Targets) [107, 108] is an annotated database containing ranking information between intragenic miRNA and host gene, based on inverse correlation of gene expression patterns corresponding to the human disorder as well as gene ontology relationship. It is also linked with HUGO gene database. The host gene and intragenic miRNA relationship is manually verified from UCSE genome browser. The intragenic miRNA corresponding to host genes are downloaded from miRBase [by including many features such as sequence overlapping of pre-miRNA on exons and introns, 3′UTR region of gene transcribed from the same strand as the miRNA and gene represented in HG-U133A (Affymetrix platform)] and predicted miRNA Target on the basis of corresponding miRNA by PicTar (conserved in mammalian genome), TargetScan and miRanda algorithm. The Computing of inverse correlation between gene expression (independently normalized and pre-process) data of host gene and corresponding putative miRNA target expression [downloaded 217 datasets from GEO repository on the same experimental platforms (HG-U133A GeneChip)], examining the anti-correlated probe host gene from each experimental data set based on the inverse correlation and ranking the target probe with respect to the number of occurrences vs mean ranks in sign list are done. The gene ontology analysis (30th, 50th, and 100th percentiles) of 290 miRNA in HOCTAR rank list is performed using DIANA web-tools with default arguments. The validation of HOCTAR miRNA targets by evaluating 135 host genes from curated 28 miRNAs gene from PubMed library and TarBase, and ranking of 135 gene on the basis of 50th, 30th and 10th percentile, is resultant to 125 (91%), 112(83%) and 65(48%), respectively. HOCTARdb provides user interface having search options with intragenic miRNA or host gene name and views the result in an interactive output page (gene ontology, rank list of inverse correlation of miRNA target and host gene relationships).
RepTar
RepTar [109] is a predicted (RepTar algorithms) cellular miRNA target database, based on various repetitive elements (binding sites) such as seed binding sites, 3′ compensatory sites, wobbles seed match, center sites and full match binding sites. It also contains genome wide predicted miRNA and cellular viral targets of human and mouse. The miRNA sequence and 3′UTR sequence (gene) of human, mouse and virus are downloaded from miRBase and UCSE genome browser (hg 18, mm9). The RepTar algorithm has many steps for predicting miRNA targets with single and repetitive sites (conservational and non-conservational). At first, search of the repetitive element (motifs) in 3′UTR sequence and resultant objects is evaluated in hidden markov model (HMM) and further, next reverse compliment HMM to predict matching (full or partial) miRNA sequence. The Viena package RNAcofold program is used to analyze the binding patterns (seed, 3 compensatory, full match) and thermodynamic (<=−10 kcal/mole) properties of miRNA–mRNA interaction and HMM combined for center sites targets prediction. These multiple and single binding sites targets are stored in database. RepTar provides interface for searching (simple search, advance search and load file search), browsing and downloading files for miRNA and its target information by various search option (miRNAs, species, repeats elements, etc.).
MicroPIR
micro-RNA–promoter interaction resource (microPIR) [110] database contains miRNA target located in promoter sequence and its genomic annotation in human along with other annotations (repetitive element, transposable elements, transcription factors binding sites, CpG islands and SNPs) by genome sequences obtained from UCSC genome table and from various SNP databases. This also contains associated information such as gene ontology, kyoto encyclopedia of genes and genomes (KEGG) pathways and Online Mendelian Inheritance in Man (OMIM association) of target gene, coupled with supporting information AGO binding sites and EST, sequence conservation scores and PCR design units used for investigational validations. The promoter sequence located on miRNA target is predicted by RNA hybrid program corresponding to miRNA (miRBase) along with miRNA target identified with 5000 bp upstream sequences from RefSeq genes (UCSC genome Table). The AGO proteins–miRNA target interaction, expression sequence tag (EST), sequence conservation information are extracted from CLIPZ database and UCSC Genome Browser, respectively. This database provides user interface for searching (basic or advanced options), downloading and browsing genome browser to analyze miRNA target. The basic search option takes input as miRNA or gene name or ID with default parameter setting and gives interactive output list of targets. Advance search option takes user defined parameters such as length of upstream regions, strands, mean of conservation binding sites, miRNAs target binding parameters along with user choice of minimum free energy, internal loops, bulges, G-U, internal loops, etc. The genome browser link is used for visualization of the target binding sites within promoter genes and, relationships between binding sites and other genome sequences.
miRNA_targets
miRNA_targets [111] is a predicted miRNA target database of human, mouse, cow, chicken, Zebra fish, fruit fly and C. elegans on 3′UTR, 5′UTR and coding region of mRNA by miRanda, RNAHybrid program. It also includes goprofiler (R package) for gene ontology (molecular function, cellular location, biological process of gene (second level of GO classification)) and visualization of miRNA targets. The developmental process of this database is taken to mRNA sequence from Ensembl database by BioMart tools and mature sequence of miRNAs from miRBase and then miRNA target is computed by miRanda and RNAhybrid. The user interface of this database has three search options: First is miRNA-targeted gene search by entering miRNA ID with selected species and energy cut-offs, second is gene-targeted miRNA search by entering GeneID along with selected species and energy cut-offs and third is mRNA sequence and miRNA sequence of 15 species by entering mRNA sequence with selected miRNA along with particular species and cut-offs.
Pharmaco-miR
Pharmaco-miR [112] is miRNA target-associated drug interaction database including proteins–drugs annotation and experimentally verified interaction (miRNA–gene–drugs) drawn from available literature, created as VerSet. The basic concept behind the development of this database is to interact one miRNA with multiple or one gene and vice versa, and gene interaction with drugs. The miRNAs repress the protein encoding gene which increases the drug’s effectiveness. Pharmaco-miR contains experimentally verified target from miRBase, miRecords in addition to predicted target from TargetScan, miRanda and PITA. It has gene-drugs annotated information from PharmGKB. The user can search interaction between miRNA targeted gene and drug from VerSet and PharmGKB. The user can search miRNA–gene–drug relationship from interactive user interface of Pharmaco-miR, by entering optional inputs such as miRNA, gene, drug name or upload the files containing miRNA gene and drug. Users can select various options for searching miRNA-gene-drug relationships all associations or overlapping associations, experimentally miRNA gene association (VerSet) or predicted target (TargetScan, miRTarbase, miRecords, PITA, miRanda) gene association or selected all, drugs-associations from VerSet or PharmGKB or both. It also contains annotated details or reduced association table for result visualization in limited search.
multiMiR
multiMiR [113] is R package and miRNA target database containing computationally and experimentally predicted target interactions, equipped with user defined cutoff selection of binding strength, case study of mouse alcohol consumption, chronic obstructive pulmonary disease in human and bladder cancer metastasis in human cell line and produce testable hypothesis of experiment. In this database, the query performed by user is processed through R package with validated target or predicted target and associated disease-drug associations. There are two types of miRNA targets, one is validated target from miRrecords, miRTarBase, TarBase databases and another predicted target from DIANA-microT, EIMMo, MicroCosm, miRanda, miRDB, PicTar, PITA and TargetScan databases. The miRNAs target, disease and drug association data are accessed from three databases such as miR2Disease, PharmacomiR and PhenomiR. The extracted data such as mature miRNA (from miRBase) and gene symbols (Entrez and EnsEmble GeneID) are preprocessed by base.db package and biomaRt R package respectively. These packages are employed to clean and characterize (typographical error, redundancy, consistency) the extracted information. The multiMiR has a number of functions with respective packages such as mutimir-db Schema to view schema of database, list.multimir to visualize gene, disease and drugs entry. The get.multimir is main interface among these databases which interacts with client for input and output. The get.multimir function takes input as several optional parameters such as miRNA name and Accession number, gene symbols, disease or drug terms with a defined cutoff score (target strength) to generate the miRNAs target–disease–drug association. There are three case studies performed by qRT-PCR, Luciferage assay and microarray.
microRNA target databases having TF-miRNAs, SNPs, edited A to I target and Viruses target
TransmiR
TransmiR [114] is a database of transcription factor that regulates miRNA including TFs–miRNAs regulatory networks, experimental evidence of TFs–miRNAs along with annotations of TFs–miRNAs, and disease relationships. The TFs–miRNA influences the gene regulation and causes the disease and phenotype variations. The annotations of TFs–miRNAs such as functions, cellular elements and biological processes are manually curated from index literature in PubMed repository and its relationships with disease from human miRNAs disease database (HMDD).The function of each miRNA is taken from UCbase. The analysis of TFs–miRNAs networks (one TF connects with multiple miRNAs vice versa) on the basis of node containing high degree > = 3 and low degree <3 and selected the groups of TF–miRNA suited to the degree in a graph represents highly conserved during evolution. The user interface has download link for text or excel files, submit option for submitting new entry of TFs–miRNAs and search option for TFs and miRNA in integrated database.
PuTmiR
PuTmiR [115] is putative TFs-miRNAs database that is based on TFs binding within genomic location of miRNA [10 kb upstream regions (USR) and downstream regions (DSR)] corresponding to the starting or ending bases on chromosome [109]. The miRNA details and TFs details are extracted from miRBase and UCSC genome browser, respectively. The extracted data is preprocessed and the TFs binding (10 kb upstream and downstream regions) is identified for a particular miRNA through retrieval tools from UCSC genome table (by setting parameter clade: as Mammal, Genome: as mouse, assembly date: mar 2006, track: TFBS, table: tfbsConsSites and tfbsConsFactors). The TFs binding sites are predicted by matching score corresponding to binding matrix (TRANSFACT 7.0 public available matrix) for each genomic location of human, mouse and rat from UCSC genome. The predicted TFs–miRNA relationship is stored in 10 kb USR and DSR table in relational database. PuTmiR interface has options for downloading the downstream and upstream data, and Search the genomic location of TFs-miRNA binding sites by entering miRNAs name.
miRdSNP
miRdSNP [116] is single nucleotides polymorphism (SNP) in miRNA binding on 3′UTR gene database along with nucleotide distance into miRNA target and dSNP, SNP sequence, refSeq gene and disease–SNP association. The dSNPs on 3′UTR region of human genes are manually curated from published literature (PubMed article) corresponding to 3′UTR refSeq of human (UCSC genome browser) gene and SNP dataset (NCBI:dbSNP build 130). The miRdSNP contains information such as min one and max three dSNP among breast cancers (52 dSNPs), type 2 diabetes (42 dSNPs), schizophrenia (38 dSNP), rheumatoid arthritis (24 dSNP), obesity (21 dSNP) and colorectal cancer (20 dSNP). The miRNAs target sites are predicted by Target-Scan 5.1 and PicTar. The genomic coordinates are indexed or mapped with RefSeq gene after converting hg17 into hg18 assembly by Lift-Over tools (UCSC Genome) and nucleotides’ distance between dSNP and miRNA target are predicted by computing the exon index of miRNA target sites. The user interface of miRdSNP has links for searching, browsing by chromosome (SNP, miRNA targets and Disease), browsing by gene (gene details), searching of disease SNP and download the files (CSV, BED format). The user can search experimentally validated target from TarBase, miRTarBase, miRecords and miR2disease by entering gene name along with selected options such as miRNAs, prediction methods, disease, etc. as a searching criteria.
PolymiRTS
Polymorphism in microRNAs and their TargetSites (PolymiRTS) [117–119] is polymorphism database of SNPs and INDEL (small insertion and deletion) on seed region of miRNA including polymorphism in miRNA sites associated genes cause human disease as well as phenotypic traits of genome wide association study is also an inbuilt feature. This database also contains polymorphisms in target sites on seed regions along with experimental evidence, cis-acting expression quantitative trait loci (e-QTLs) of mRNA, miRNA–mRNA interaction predicted by CLASH experiments and resultant interaction compared with predicted target by TargetScan program (context + score deference) and its biological pathways. The SNP in miRNA target sites (experimentally validated or predicted) are predicted by mapping of dbSNP dataset on genome 3′UTR sequence (mm9, hg19) and scan miRNA target sites according to mature miRNA sequence of miRBase by TargetScan. The target sites are classified into four classes such as O: ancestral allele not predicted, D: identify allele disrupts more vertebrate genome, N: derived allele disrupts non conserve sites and C: predicted allele of new miRNA target sites. The experimentally validated SNPs in target sites are extracted from various target databases such as TarBase, miRecords and miRTarBase. Binding sites are classified as high throughput (HT), low throughput (LT), specific binding into HTL, and LTL on the basis of experimental evidence of miRNA–mRNA interaction. The SNPs in miRNA seed regions are predicted through 3′UTR gene sequence (extracted from Ensembl genome) of human and mouse [117–119]. The tissues specific cis-eQTLs of mouse and human are integrated into this database from Gene Networks and GTEx-eQTL browser. The gene polymorphisms which affect phenotype traits and diseases related information are extracted from NHGRI-GWAS Catalog. PolymiRTS 3.0 is added with new feature like miRNA–mRNA interaction by CLASH experiment data as well as INDELS in miRNA along with its targets predicted through dbSNP 137 mapping genome and selected feature from Target scan (Context + Score) for miRNA-mRNA interaction. The biological Pathways of polymorphism associated gene are accessed from KGGE. The PolymiRTS interface has browsing, searching (simple and batch search), SNP in gene chromosomal locations and downloading options for user to analyze the SNP influence target gene.
miR-EdiTar
miR-EdiTar [120] contains miRNAs target sites affected by A-to-I edit that become novel miRNAs target sites after the A-to-I edit and also integration with miRo (web environments for annotation). When inosine replaces adenosine that affects the miRNA target binding sites as well as influences the miRNA facilitated gene regulation results to lead generate the novel binding sites. The A-to-I editing process done by adenosine deaminase acting on RNA (ADAR) enzyme in eukaryotes that result in causing infections, neurological diseases and cancers. The prediction of miRNA target which participates in editing the sites by miRiam tools, is based on structural accessibility of the target site and the energy of the miRNA–mRNA duplex, from A-to-I edit of 3′UTR sequence (DARNED database). The miRNA sequences from miRBase-18 have been classified into two categories based on edited position at seed regions (6mer, 7mer-A-1, 7mer-m8, and 8 mer) and non-seed regions. User interface of this database provides search option based on miRNA and based on target gene for miRNA edited binding sites details. User can access information such as edited sites on the 3′UTRs, edited sites on a predicted miRNA binding site, targets 3′UTR sequences affected by editing, target 3′UTR sequences with at least one edited base in a predicted miRNA binding site, target 3′UTR sequences with all their edited bases in a predicted miRNA binding site, target 3′UTR sequences with at least one novel predicted binding site created by editing, miRNA with predicted sites affected by editing and miRNA with predicted novel sites created by editing from miR-EdiTar.
ViTa
ViTa [121] is viral miRNA target database of host (human, mice, rat, and chicken) along with virus annotations such as virus infected organ tissues, tissues specificity of host miRNA, conserved region of virus (BLAST) and disease relationship. This database has been developed after investigation of host regulatory associations among host miRNA from miRBase and associated virus from literature and Virus data collected from four locations such as ICTVdB, VBRC, VirGen and through websites surfing. The various virus targeted miRNA genes of host were predicted by miRanda and TargetScan algorithms based on minimum free energy. The expressions profiles of miRNA are obtained from database (Lu et al.) and then regulatory relationships among virus and host are determined for annotation classification of viruses (dsDNA virus, ssDNA virus, dsRNA virus, (+ssRNA virus, (−) ssRNA virus, retro-transcribing virus). The ViTa user interface has an arbitrary search option (target virus or host) with given inputs as (virus name, hostname, miRNAs, minimum free energy, etc.) and got outputs (miRNA details annotation, order, family, etc.). User can also browse detailed annotation of target genes and download target files according to species and algorithm.
Vir-Mir db
Vir-Mir db [122, 123] is a viral miRNA hairpin database developed after genome wide study of viral-host interaction relationship along with hairpins classification, host-target prediction by RNAhybrid and 3′UTR sequence of human, rat, mouse, Arabidopsis, rice and zebra fish. The viral miRNAs hairpin secondary structures are predicted from 2266 virus genome (NCBI: genomic.fna files) by Srnaloop (90 bases long and score minimum 17) and filter the sequence and structure based on the defined different referenced range value of GC content, minimum free energy of core and whole hairpin structure with its ratio. The prediction of viral miRNA hairpins are optimized by hairpin ORFs (open reading frame) overlapping analysis (fromNCBI: viral.protein.gbff). The above pipeline steps have shown 98, 84, and 78% sensitivity on miRNAs after Srnaloop filter, miRNA after structure and sequence filter and miRNA after ORF filter, respectively. The user interface provides links for specific viral hairpin structure as well as links for RNA hybrid algorithms for targets prediction.
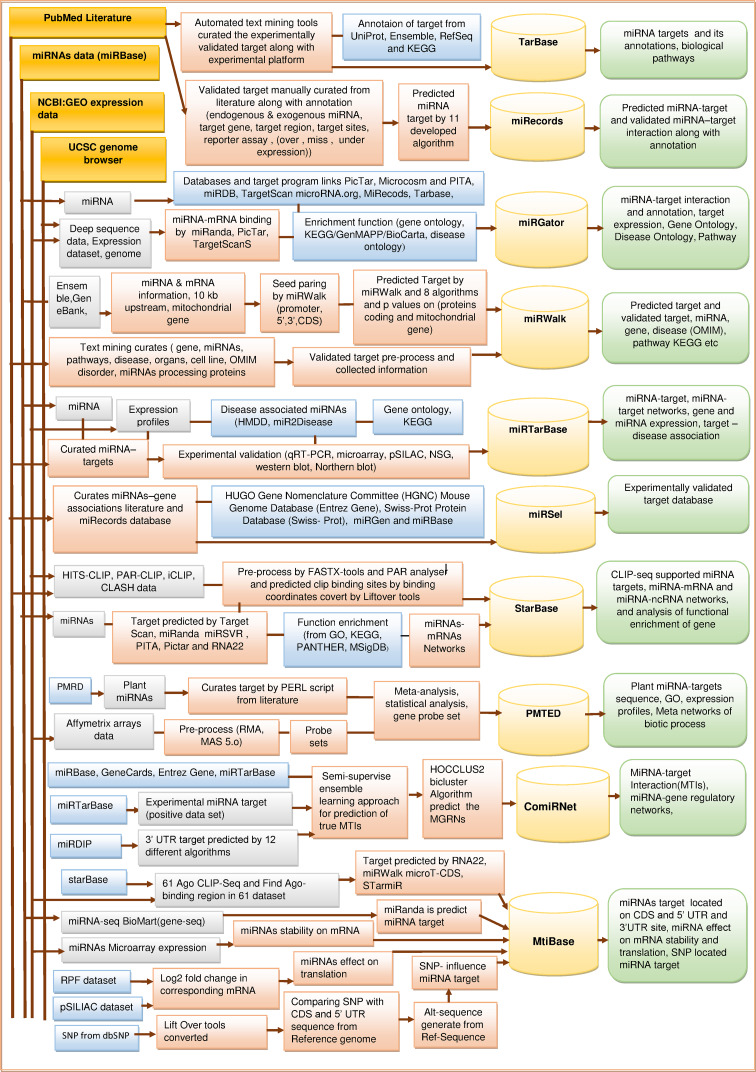
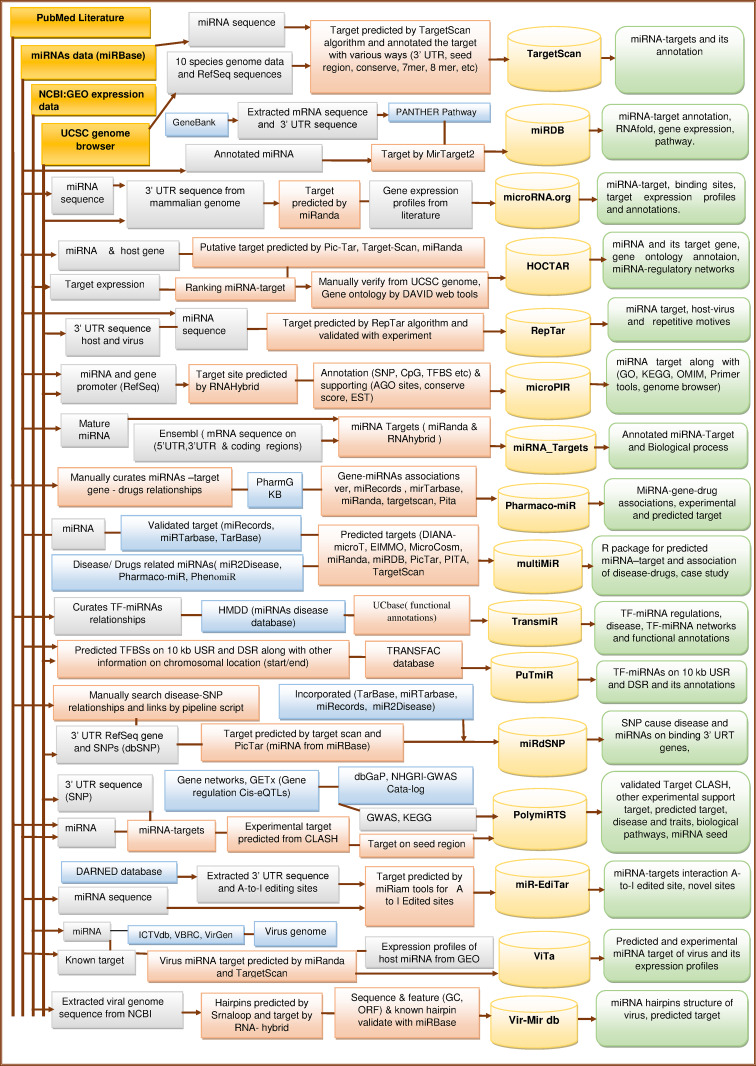
Integrated architecture of miRNAs target databases
The Figs. 2, 3 exhibit the schematic overview of developmental processes of several miRNA target databases. The architecture is divided into three parts. (1) Extraction or computation of miRNA informatics from different gene and genome resources (UCSC genome, PubMed literature, NCBI, miRNAs database, etc.) along with bioinformatics tools. The annotation of gene, disease and target, etc. is retrieved from HUGO Gene Nomenclature Committee, GWAS, GO database, OMIM, Chemdb, KEGG and many more. (2) A relational database for implementation of web resources along with user interface. (3) This part describes the types of information user can access from the database (right most rounded rectangular boxes).
Fig. 2.
Integrated architecture of experimentally validated miRNA target databases (in this figure, four dark golden colored rectangles represent the common databases which is used for extraction of various datasets or information. Lavender colored rectangles represent the type of dataset or information extracted from other databases or resources for the purpose of processing. Light pink colored rectangles represent the processing techniques. Light blue colored rectangles represent the databases used for miRNA target annotation or target prediction. Lighter golden colored magnetic disks represent the implementation of database storage after final complete process of miRNA target database development and lighter green colored rounded rectangles represent the types of information access from each, miRNA target database)
Fig. 3.
Integrated architecture of predicted miRNA target and TF–miRNAs, SNPs in target, edited A to I target and viruses miRNA target databases ((In this figure, four dark golden colored rectangles represent the common databases which is used for extraction of various datasets or information. Lavender colored rectangles represent the type of dataset or information extracted from other databases or resources for the purpose of processing. Light pink colored rectangles represent the processing techniques. Light blue colored rectangles represent the databases used for miRNA target annotation or target prediction. Lighter golden colored magnetic disks represent the implementation of database storage after final complete process of miRNA target database development and lighter green colored rounded rectangles represent the types of information access from each, miRNA target database)
Graphical comparative study
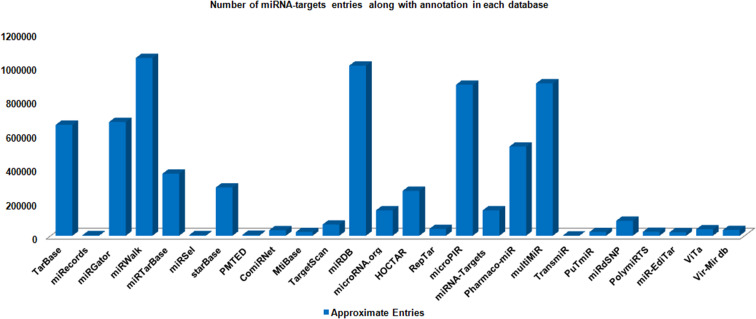
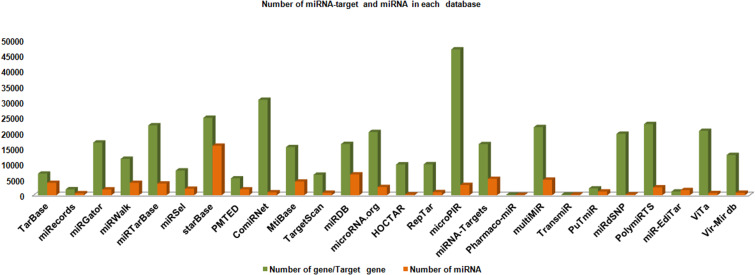
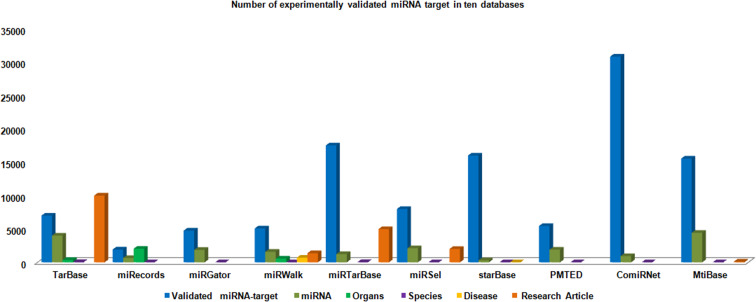
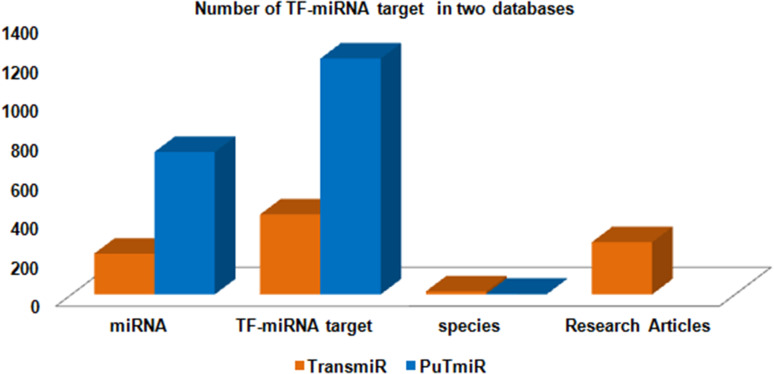
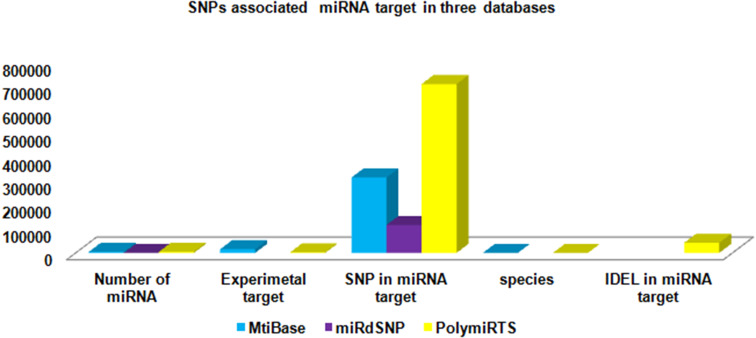
The various miRNAs target entries have been represented into different databases with the help of column chart (Figs. 4, 5, 6, 7, 8). The Fig. 4 demonstrates the average maximum number of miRNA target along with annotation entries that contain six databases, namely TarBase, miRGator, miRWalk, miRDB, microPIR and multimiR. The miRecords miRSel, PMTED and TransmiR have an average minimum number of entries. Figure 5 demonstrates the TransmiR containing minimum number of entries of miRNA target among reviewed databases and starBase having maximum number of both miRNA target as well as miRNA entries and microPIR maximum number of target gene. The Fig. 6 demonstrates the TarBase and miRTarBase which contain maximum number of experimentally validated miRNA targets curated from the literature. TheTarbase and miRTarbase contain maximum number of species and miRWalk has maximum number of organs and disease contained in experimentally validated miRNA target database. The Fig. 7 illustrates TransmiR which contains maximum number of curated TF-miRNA, of different species from the literature. The Fig. 8 demonstrates that PolymiRTS contains maximum number of SNPs and INDELs in miRNA targets.
Fig. 4.
The graph makes a comparative study of different target databases including miRNA target and its annotations
Fig. 5.
The graph shows the number of miRNA target and miRNA entries in different databases
Fig. 6.
The graph makes a comparative study of experimentally validated targets in different databases
Fig. 7.
The graph shows TF-miRNA relationships in different databases
Fig. 8.
The graph shows SNP in miRNA target associations into three databases
Discussion and conclusion
This research review discusses the experimental and computational prediction of miRNA targets database along with several functional annotations of miRNA target. Among validated databases, TarBase, miRwalk and miRSel are curated miRNA target from published literature through text mining tools while miRecords, miRTarBase and MtiBase are curated manually. miRSel is an inconsistent database because it is not an updated database and new feature of miRNA target prediction are six years old. miRWalk has several annotations of predicted and validated miRNA target database but this database is unstable and unreliable. TarBase, miRecords and miRTarbase have highest coverage of experimental techniques (reporter assays, Western blot, northern blot, qRT-PCR, micro array, pSILAC, NGS (CLIP-seq, Degradome-Seq, and CLASH-Seq)) of miRNA target prediction from literature. MtiBase contains validated target by only three experimental techniques (reporter assays, PCR, western blot). miRecords have strong experimental evidence of miRNA target and it also contains predicted target with twelve existing program but is not updated from last three years. The KEGG pathway is a common database for biological pathway annotation in TarBase and miRTarBase and both databases are updated timely. The current user interface of TarBase contains search and downloadable file option for licensed user. miTarBase user interface is also incorporated with Cytoscape tools for MTIs networks visualisation. The PMTED is a special database of miRNA target expression of plant that incorporates cytoscape tools for visualising networks of several biotic processes. ComiRNet is a recently developed special database through the training of miRTarBase and miRDIP data with the machine learning algorithm. User interface of ComiRNet searches MTIs and visualises by bi-cluster networks of MGRNs without any prior Knowledge. miRGator shares common experimentally validated miRNA target from Tarbase, miRecords, miRTarBase rather than separate prediction and database is more focused on computing various annotations (miRNA disease expression, gene set and miRNA–mRNA networks, miRNA–mRNA Pathways) of miRNA target. miRGator is not updated from last three years. starBase is special portal not only having experimentally validated (By CLIP-seq) miRNA target but also have various types of RNA–Proteins, miRNA–lncRNA, and Pan-Cancer relational information. StarBase is having strong evidence for predicted miRNA target, validated with CILP-Seq and good user interface with bioinformatics tools such as Cytoscape, miRfunction, ceRNAfunction for the analysis of miRNA- arget, proteins and cancer relation.
TargetScan is widely shared database among predicted target databases and algorithms of TargetScan are widely used in miRecords, miRGator, miRwalk, HOCTAR, Pharmaco-miR, multiMiR, miRdSNP, PolymiRTS and ViTA. TargetScan user interface searches and provides fast result of 3′UTR conservations which are visualised on genome browser. The database is updated yearly. Highest numbers of predicted tools for miRNA target are used in miRecords database. miRDB also have predicted conserved and non conserved miRNA target by analyzing down regulation properties of microarray data expression through SVM based model. miRDB user interface shows strong evidence of functional annotation of miRNA target and it is updated recently but suffer from multiple click for exploring of annotation. MicroRNA.org is a database of functional non-conserved and non-canonical predicted miRNA target by miRSVR and target expression data. Currently, Search option of MicroRNA.org user interface is not working properly and database has not been updated from last six years. HOCTAR database is focused on functional analysis of miRNA target by incorporating ranking of predicted targets based on gene ontology (HUGO genes). This user interface does not have download option and database has also not been updated from last three years. RepTar database has 3′UTR predicted target of human and mouse along with virus by special HMM based model but the user interface is not working properly and database has not been updated from last six years. microPIR is a special types of miRNA predicted target database contains information on miRNA-promoter interaction of human and various annotations. The database contains strong evidence of target and annotation. microPIR is updated in every two years and user interface have special genome browser for the analysis, interaction and annotation. The Search results from user interface shows miRNA–promoter interaction details along with overlapping feature (AGO, EST, TFBSs, and SNPs). MtiBase and miRNA_Targets databases having predicted target (match on 3′UTR, 5′UTR and CDS) but MiRNA_Targets database has not been updated from last four years and user interface does not have download option. Pharmaco-miR is special database of miRNA–gene–drugs interaction and shares experimentally validated target of the miRecords and miRTarBase and predicted target by TargetScan, miRanda and PITA. User interface of Pharmaco-miR does not have download option. multiMiR is integrated resource of validated and predicted target databases along with R package.
TransmiR and PuTmiR are TFs-miRNA interaction databases. TransmiR has validated TFs–miRNAs from literature while PuTmiR has predicted TFs–miRNA on specific 10 kb USR and DSR of genome. Search results on user interface of TransmiR visualize the TFs–miRNA regulatory networks as well as the disease annotations but the database has not been updated from last three years. User interface of PuTmiR has search results showing in tabular / file format. miRdSNP, PolymiRTS and MtiBase are databases of SNP influenced miRNA target. miRdSNP database contains target predicted on 3′UTR and SNP–miRNA target along with disease association. miRdSNP has evidence of SNP–disease associations and search results show in density plot and the details of polymorphism in the browser but database has not been updated from last three years. MtiBase and PolymiRTS both contain experimentally validated SNPs in miRNA target. MtiBase has 3′UTR, 5′UTR and CDS located on SNP in target while PolymiRTS has only 3′UTR located on SNP in target. PolymiRTS also contains INDEL in miRNA target which is not available in other database. PolymiRTS and MtiBase use PANTHER database and NHGRI-GWAS catalogue for SNP biological pathway and Gene set analysis, respectively. MtiBase user interface does not have browse option. PolymiRTS user interface browse the SNP in miRNA by various categories of prediction and annotation but database has not been updated from last three years. miR-EditTar is a special database that contains experimentally validated A to I edited miRNA target sites but database has not been updated from four years. ViTa is a database of predicted miRNA target of virus and Vir-Mir db is a database of classified virus miRNA hairpins along with predicted targets but both the databases have not been updated from last ten years. Vir-Mir database user interface does not contain search and download option. It only contains hyper links for browsing details of virus target and annotation according to the classification of virus. User interface of ViTa is currently slow and search target results can be visualised in browser, along with functional annotation of various viruses.
The experimental techniques for target identification analyze the expression of miRNA followed by downstream gene expression or proteomics examination. Here, various limitations of experimental target identification are discussed. The experimental techniques suffer from low coverage, high cost of re-agent and time requirement. The gene specific target validations can be performed by expression of miRNAs at proteins label and mRNAs label by western blot and qRT-PCR, respectively. This method cannot distinguish the direct and secodary miRNAs target. The miRNA over-expression in different physiological conditions techniques for the target identification cannot diffrentiate the miRNA belonging to the same family with similar sequences.The gene expression analysis techniques of the target identifcation by microarray do not differentiate direct targets without seed match. HITS-CLIP and PAR-CLIP techniques do not identify specifc binding position, they only identify target region and suffer from ultra voilet intensity of light. The direct cleavage target methods for target identification do not predict accurate target due to limited cleavage in mammals. Reverse transcription of target methods only applied on C. elegans. The proteomics assays, translation profiling and CLASH are used little due to their technological limitations for target identifcation.
The computational target prediction tools reported by various researchers have some common features such as seed matching, sequence conservation, the calculation of free energy and site accessibility. But these features do not gurrantee the accuracy of true positive target prediction because some non-conserved binding sites are also functional and there may be error in free energy calculation of data source. There are a small number of features of target sites such as AU, GU, etc. that occurs on 3′ compensatory sites, center sites match, 3′UTR, 5′UTR, and CDS contents have been used by some researchers for target prediction. The recent research findings have found out that the target gene is also co-regulated by common transcription factor. In future, this feature can also be incorporated into target prediction. The miRNA regulation is tissue specific. Therefore, the tissue specific features such as highly expressed miRNAs or miRNA isoforms, tissue-specific mRNA transcript variants and lists of highly upregulated or downregulated genes miRNAs regulation can be included in the target prediction. The Combined features of more than one tool can be included into the target prediction tools and improve the computational speed and accuracy using soft-computing approach. The TargetScan, DIANA-microT-CDS, RNAhybrid and miRanda are widely used target prediction tools. These tools have various advantages and limitations. It is concluded that more intelligent computational and experimental techniques are required for miRNAs target identification, prediction, database designing, implementation and interactive user interface.
Acknowledgements
The author would like to avail the opportunity to thank the MANIT BHOPAL and Ministry of HRD, Government of India who furnished the support infrastructure to make this article a reality. The author expresses sincere, warm and heartfelt thanks towards Mr. Dinesh Sharma, English Language coach, Professional Trainer, and mentor of English and Business Communication, who put sincere and concentrated efforts to edit the language of this manuscript with utmost dedication and patience.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;16:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Almeida MI, Reis RM, Calin GA. MicroRNA history: discovery, recent applications and next frontiers. Mutat Res. 2011;717:1–8. doi: 10.1016/j.mrfmmm.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 4.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditiselegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 5.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/S0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal micrornas. Nature. 2004;431(350):355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.Du T, Zamore PD. Beginning to understand microrna function. Cell Res. 2007;17:661–663. doi: 10.1038/cr.2007.67. [DOI] [PubMed] [Google Scholar]
- 8.Bushati N, Cohen SM. Microrna functions. Annu Rev Cell Dev Bi. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 9.Khraiwesh B, Zhu JK, Zhu J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochimica Biophysica Acta. 2012;1819:137–148. doi: 10.1016/j.bbagrm.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kloosterman WP, Plasterk RH. The diverse functions of MicroRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Jacobsen A, Silber J, Harinath G, Huse JT, Schultz N, Sander C. Analysis of microRNA-target interactions across diverse cancer types. Nat Struct Mol Biol. 2013;20:1325–1332. doi: 10.1038/nsmb.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckardt NA. A microRNA cascade in plant defense. Plant Cell. 2012;24:840–840. doi: 10.1105/tpc.112.240311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh Y, Kaul V, Mehra A, Chatterjee S, Tousif S, Dwivedi VP, et al. Mycobacterium tuberculosis controls microRNA-99b (miR-99b) expression in infected murine dendritic cells to modulate host immunity. J Biol Chem. 2013;288(7):5056–5061. doi: 10.1074/jbc.C112.439778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar P, Dezso Z, MacKenzie C, Oestreicher J, Agoulnik S, et al. Circulating miRNA Biomarkers for Alzheimer’s disease. PLoS ONE. 2013;8:e69807. doi: 10.1371/journal.pone.0069807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bej S, Basak J. MicroRNAs: the potential biomarkers in plant stress response. Am J Plant Sci. 2014;5:748–759. doi: 10.4236/ajps.2014.55089. [DOI] [Google Scholar]
- 17.Nelson PT, Wang WX, Bernard WR. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu Z, Li W, Fu B. MicroRNAs in Autoimmune Diseases. BioMed Res Int. 2014;2014:8. doi: 10.1155/2014/527895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shantikumar S, Caporali A, Emanueli C. Role of miRNA in diabetes and its cardiovascular complications. Cardiovasc Res. 2011;93:583–593. doi: 10.1093/cvr/cvr300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao Y, Mohan R, Zhang S, Tang X. MicroRNAs as pharmacological targets in diabetes. Pharmaco Res. 2013;75:37–47. doi: 10.1016/j.phrs.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pauley KM, Cha S, Chan EK. MicroRNA in autoimmunity and autoimmune diseases. J Autoimmun. 2009;32:189–194. doi: 10.1016/j.jaut.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farazi TA, Hoell JI, Morozov, et al. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duskova K, Nagilla P, Le H, Iyer P, Thalamuthu A, et al. MicroRNA regulation and its effects on cellular transcriptome in human immunodeficiency virus-1 (HIV-1) infected individuals with distinct viral load and CD4 cell counts. BMC Infect Dis. 2013;13:250. doi: 10.1186/1471-2334-13-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindsay MA. Target discovery. Nat Rev Drug Discov. 2003;2:831–838. doi: 10.1038/nrd1202. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, Adelstein SJ, Kassis AI. Target discovery from data mining approaches. Drug Discov Today. 2009;14:147–154. doi: 10.1016/j.drudis.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR-208 as biomarker of myocardial injury. Clin Chem. 2009;55:1944–1949. doi: 10.1373/clinchem.2009.125310. [DOI] [PubMed] [Google Scholar]
- 27.Hulanicka M, Garncarz M, Parzeniecka-Jaworska M, Jank M. Plasma miRNAs as potential biomarkers of chronic degenerative valvular disease in Dachshunds. BMC Vet Res. 2014;10:205–208. doi: 10.1186/s12917-014-0205-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spadaro PA, Bredy TW. Emerging role of non-coding RNA in neural plasticity cognitive function and neuropsychiatric disorders. Front Genet. 2012;3:132. doi: 10.3389/fgene.2012.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol. 2013;48:51–68. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ekimler S, Sahin K. Computational methods for MicroRNA target prediction. Genes. 2014;5:671–68. doi: 10.3390/genes5030671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dimitrov R. microRNA gene finding and target prediction—basic principles and challenges. MOJ Proteomics Bioinform. 2014;1:00024. doi: 10.15406/mojpb.2014.01.00024. [DOI] [Google Scholar]
- 32.Dónal C, Schaefer A. General principals of miRNA biogenesis and regulation in the brain. Neuropsychopharmacol. 2012;38:39–54. doi: 10.1038/npp.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voinnet O. Origin, biogenesis and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 34.Martinez-Sanchez A, Murphy CL. MicroRNA target identification—experimental approaches. Biology. 2013;2:189–205. doi: 10.3390/biology2010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2014;39:6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmadi H, Ahmadi A, Azimzadeh-Jamalkandi S, Shoorehdeli MA, Salehzadeh-Yazdi A, et al. Homotarget: a new algorithm for prediction of microRNA targets in Homo sapiens. Genomics. 2013;101:94–100. doi: 10.1016/j.ygeno.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Reyes-Herrera PH, Ficarra E. One Decade of development and evolution of MicroRNA target prediction algorithms. Genom Proteom Bioinform. 2012;10:254–263. doi: 10.1016/j.gpb.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srivastava PK, Moturu TR, Pandey P, Baldwin IT, Pandey SP (2014) A comparison of performance of plant miRNA target prediction tools and the characterization of features for genome-wide target prediction. BMC Genom 15 :348.doi:10.1186/1471-2164-15-348 [DOI] [PMC free article] [PubMed]
- 39.Peterson SM, Thompson JA, Ufkin ML, Sathyanarayana P, et al. Common features of microRNA target prediction tools. Front Genet. 2014;5:13. doi: 10.3389/fgene.2014.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39:W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu HJ, Ma YK, Chen T, Wang M, Wang XJ. PsRobot: a web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 2012;40:W22–W28. doi: 10.1093/nar/gks554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonnet E, He Y, Billiau K, Van de Peer Y(2010) TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics 26:1566–1568. doi:10.1093/bioinformatics/btq233 [DOI] [PubMed]
- 43.Bonnet E, Wuyts J, Rouzé P, Van de Peer Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativaidentifies important target genes. PNAS. 2004;101:11511–11516. doi: 10.1073/pnas.0404025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 2005;33:W701–W704. doi: 10.1093/nar/gki383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 47.Xie F, Zhang B. Target-align: a tool for plant microRNA target identification. Bioinformatics. 2010;26(23):3002–3003. doi: 10.1093/bioinformatics/btq568. [DOI] [PubMed] [Google Scholar]
- 48.Sun YH, Lu S, Shi R, Chiang VL. Computational Prediction of Plant miRNATargets. Methods Mol Biol. 2011;744:175–186. doi: 10.1007/978-1-61779-123-9_12. [DOI] [PubMed] [Google Scholar]
- 49.Jha A, Shankar R. Employing machine learning for reliable miRNA target identification in plants. BMC Genomics. 2011;12:636. doi: 10.1186/1471-2164-12-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are MicroRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 51.Friedman RC, Farh KK, Burge CB, Bartel David P. Most mammalian mRNAs are conserved targets of MicroRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.García DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other miRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krüger. Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnes-Rahoades MW, Bartel DP. Computational identification of plant microRNAsand their targets, inducing a stress-induced miRNA. Mol Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 57.Adai A, Cameron J, Sizolwenkosi M, Sarah A, Varun M, Vicki V, Venkatesan S. Computational prediction of miRNAs in Arabidopsis thaliana . Genome Res. 2005;15:78–91. doi: 10.1101/gr.2908205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grün D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. microRNA target predictions across seven drosophila species and comparison to mammalian targets. PLoS Comput Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders G, Mackowiak SD, Jens M, Maaskola J, Kuntzagk A, Rajewsky N, et al. doRiNA: a database of RNA interactions in post-transcriptional regulation. Nucleic Acids Res. 2012;40:D180–D186. doi: 10.1093/nar/gkr1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krek A, Grun D, Poy M, Wolf R, Rosenberg L, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 61.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 62.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 63.Gaidatzis D, Van Nimwegen E, Hausser J, Zavolan M. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics. 2007;8:69. doi: 10.1186/1471-2105-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maragkakis M, Reczko M, Simossis VA, Alexiou P P, Papadopoulos GL, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37:W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.RusIN. ov V, Baev V, Minkov IN, Tabler M. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 2005;33:W696–W700. doi: 10.1093/nar/gki364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 67.Hammell M, Dang L, Zhang L, Lee A, Carmack CS, et al. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein–enriched transcripts. Nat methods. 2008;5:813–819. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hsu JB, Chiu CM, Hsu SD, Huang WY, Chien CH, Lee TY, Huang HD. miRTar: an integrated system for identifying miRNA-target interactions in human. BMC Bioinform. 2011;12:300. doi: 10.1186/1471-2105-12-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saetrom O, Snove O, Jr, Saetrom P. Weighted sequence motifs as an improved seeding step in microRNA target prediction algorithms. RNA. 2005;11:995–1003. doi: 10.1261/rna.7290705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang JC, Babak T, Corson TW, Chua G, Khan S, et al. Using expression profiling data to identify human microRNA, targets. Nat Methods. 2007;4:1045–1049. doi: 10.1038/nmeth1130. [DOI] [PubMed] [Google Scholar]
- 71.Kim SK, Nam JW, Rhee JK, Lee WJ, Zhang BT. miTarget: microRNA target gene prediction using a support vector machine. BMC Bioinform. 2006;7:411. doi: 10.1186/1471-2105-7-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sturm M, Hackenberg M, Langenberger D, Frishman D. TargetSpy: a supervised machine learning approach for microRNA target prediction. BMC Bioinform. 2010;11:292. doi: 10.1186/1471-2105-11-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bandyopadhyay S, Mitra R. TargetMiner: microRNA target prediction with systematic identification of tissue-specific negative examples. Bioinformatics. 2009;25:2625–2631. doi: 10.1093/bioinformatics/btp503. [DOI] [PubMed] [Google Scholar]
- 74.Yang Y, Wang YP, Li KB. MiRTif: a support vector machine-based microRNA target interaction filter. BMC Bioinform. 2008;9:S4. doi: 10.1186/1471-2105-9-S12-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yan X, Chao T, Tu K, Zhang Y, Xie Lu, Gong Y, Yuan J, Qiang B, Peng X. Improving the prediction of human microRNA target genes by using ensemble algorithm. FEBS Lett. 2007;581:1587–1593. doi: 10.1016/j.febslet.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 76.Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24:325–332. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- 77.Chandra V, devi GR, Nair AS, Pillai SS, Pillai RM. MTar: a computational microRNA target prediction architecture for human transcriptome. BMC Bioinform. 2010;11:S2. doi: 10.1186/1471-2105-11-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Friedman Y, Karsenty S, Linial M (2014) miRror-Suite: decoding coordinated regulation by microRNAs. Database(Oxford): bau043 doi:10.1093/database/bau043 [DOI] [PMC free article] [PubMed]
- 80.Friedman I, Naamati G, Linial M. MiRror: a combinatorial analysis web tool for ensembles of microRNAs and their targets. Bioinformatics. 2010;26(15):1920–1921. doi: 10.1093/bioinformatics/btq298. [DOI] [PubMed] [Google Scholar]
- 81.Reyes-Herrera PH, Ficarra E, Acquaviva A, Macii E. miREE: miRNA recognition elements ensemble. BMC Bioinform. 2011;12:454. doi: 10.1186/1471-2105-12-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vergoulis T, Vlachos IS, Alexiou P, Georgakilas G, et al. Tarbase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 2012;40:D222–D229. doi: 10.1093/nar/gkr1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37:D155–D158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.LeeYJ, Kim V, Muth DC, Witwer KW. Validated MicroRNA target databases: an evaluation. Drug Dev Res. 2015;76:389–396. doi: 10.1002/ddr.21278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA–target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nam S, Kim B, Shin S, Lee S. miRGator: an integrated system for functional annotation of microRNAs. Nucleic Acids Res. 2008;36(Database issue):D159–D164. doi: 10.1093/nar/gkm829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cho S, Jun Y, Lee S, Choi HS, Jung S, Jang Y, Park C, Kim S, Lee S, Kim W. miRGator v2.0: an integrated system for functional investigation of microRNAs. Nucleic Acids Res. 2011;39(Database issue):D158–D162. doi: 10.1093/nar/gkq1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cho S, Jang I, Jun Y, Yoon S, et al. miRGator v3.0: a microRNA portal for deep sequencing, expression profiling, and mRNA targeting. Nucleic Acids Res. 2013;41(Database issue):D252–D257. doi: 10.1093/nar/gks1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk—database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 91.Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, et al. miRTarBase: a database curates experimentally validated microRNA—target interactions. Nucleic Acids Res. 2011;39:D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH, Chu CF, et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42:D78–D85. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Naeem H, Kuffner R, Csaba G, Zimmer R. miRSel: automated extraction of associations between microRNAs and genes from the biomedical literature. BMC Bioinformatics. 2010;11:135. doi: 10.1186/1471-2105-11-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang JH, Li, JH Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA–mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202–D209. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(D1):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sun X, Dong B, Yin L, Zhang R, Du W, Liu D, et al. PMTED: a plant microRNA target expression database. BMC Bioinform. 2013;14:174. doi: 10.1186/1471-2105-14-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pio G, Ceci M, Malerba D, D Elia D. ComiRNet: a web-based system for the analysis of miRNA-gene regulatory networks. BMC Bioinform. 2015;16:S7. doi: 10.1186/1471-2105-16-S9-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pio G, Ceci M, D’Elia D, Malerba D. Integrating microRNA target predictions for the discovery of gene regulatory networks: a semi-supervised ensemble learning approach. BMC Bioinform. 2014;15:S4. doi: 10.1186/1471-2105-15-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pio G, Ceci M, D’Elia D, Loglisci C, Malerba D. A novel biclustering algorithm for the discovery of meaningful biological correlations between miRNAs and mRNAs. BMC Bioinform. 2013;14:S8. doi: 10.1186/1471-2105-14-S7-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pio G, Ceci M, D’Elia D, Loglisci C, Malerba D (2013) HOCCLUS2: a biclustering algorithm for the discovery of miRNA:mRNA regulatory modules. Italian Symposium on Advanced Database Systems (SEBD)
- 101.Pio G, Ceci M, D’Elia D, Loglisci C, Malerba D 2012 Hierarchical and Overlapping Co-Clustering of mRNA:miRNA. Interactions European Conference on Artificial Intelligence (ECAI)
- 102.Pio G, Ceci M, D’Elia D, Malerba D (2014) Network reconstruction for the identification of miRNA:mRNA interaction networks. Machine Learning and Knowledge Discovery in Databases, ECML-PKDD-LNCS 8726:pp 508–511.
- 103.Guo ZW, Xie C, Yang JR, Li JH, Yang JH, Zheng L. MtiBase: a database for decoding microRNA target sites located within CDS and 5′UTR regions from CLIP-Seq and expression profile datasets. Database: (Journal of Biological Databases Curation) 2015:bav102. 2015 doi: 10.1093/database/bav102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Betel D, Koppal A, Agius P, Sander C, Leslie C. mirSVR predicted target site scoring method: comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gennarino VA, Sardiello M, Mutarelli M, Dharmalingam G, Maselli V, et al. HOCTAR database: a unique resource for microRNA target prediction. Gene. 2011;480:51–58. doi: 10.1016/j.gene.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gennarino VA, Sardiello M, Avellino R, Meola N, Maselli V, Anand S, Cutillo L, Ballabio A, Banfi S. MicroRNA target prediction by expression analysis of host genes. Genome Res. 2008;1916:481–4902008. doi: 10.1101/gr.084129.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Elefant N, Berger A, Shein H, Hofree M, Margalit H, Altuvia Y. RepTar: a database of predicted cellular targets of host and viral miRNAs. Nucleic Acids Res. 2011;39:D188–D194. doi: 10.1093/nar/gkq1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Piriyapongsa J, Bootchai C, Ngamphiw C, Tongsima S. microPIR: an integrated database of MicroRNA target sites within human promoter sequences. PLoS One. 2012;7:e33888. doi: 10.1371/journal.pone.0033888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kumar A, Wong AK, Tizard ML, Moore RJ, Lefèvre C. miRNA_targets: a database for miRNA target predictions in coding and non-coding regions of mRNAs. Genomics. 2012;100:352–356. doi: 10.1016/j.ygeno.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 112.Rukov JL, Wilentzik R, Jaffe I, Vinther J, Shomron N. Pharmaco-miR: linking microRNAs and drug effects. Brief Bioinform. 2013;15:648–659. doi: 10.1093/bib/bbs082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ru Y, Kechris KJ, Tabakoff B, Hoffman P, Radcliffe RA, Bowler B, et al. The multiMiR R package and database: integration of microRNA–target interactions along with their disease and drug associations. Nucleic Acids Res. 2014;42:e133. doi: 10.1093/nar/gku631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang J, Lu M, Qiu C, Cui Q. TransmiR: a transcription factor–microRNA regulation database. Nucleic Acids Res. 2010;38:D119–D122. doi: 10.1093/nar/gkp803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bandyopadhyay S, Bhattacharyya M. PuTmiR: a database for extracting neighboring transcription factors of human microRNAs. BMC Bioinform. 2010;11:190. doi: 10.1186/1471-2105-11-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bruno AE, Li L, Kalabus JL, Pan Y, Yu A, Hu Z. miRdSNP: a database of disease-associated SNPs and microRNA target sites on 3′UTRs of human genes. BMC Genomics. 2012;13:44. doi: 10.1186/1471-2164-13-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bao L, Zhou M, Wu L, Lu L, Goldowitz D, Williams RW, Cui Y. PolymiRTS Database: linking polymorphisms in microRNA target sites with complex traits. Nucleic Acids Res. 2007;35:D51–D54. doi: 10.1093/nar/gkl797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ziebarth JD, Bhattacharya A, Chen A, Cui Y. PolymiRTS Database 2.0: linking polymorphisms in microRNA target sites with human diseases and complex traits. Nucleic Acids Res. 2012;40:D216–D221. doi: 10.1093/nar/gkr1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bhattacharya A, Ziebarth JD, Cui Y. PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014;42:D86–D91. doi: 10.1093/nar/gkt1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Laganà A, Paone A, Veneziano D, Cascione L, Gasparini P, et al. miR-EdiTar: A database of predicted A-to-I edited miRNA target sites. Bioinformatics. 2012;28:3166–3168. doi: 10.1093/bioinformatics/bts589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hsu PW, Lin LZ, Hsu SD, Hsu JB, Huang HD. ViTa: prediction of host microRNAs targets on viruses. Nucleic Acids Res. 2007;35:D381–D385. doi: 10.1093/nar/gkl1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li SC, Shiau CK, Lin WC. Vir-Mir db: prediction of viral microRNA candidate hairpins. Nucleic Acids Res. 2008;36:D184–D189. doi: 10.1093/nar/gkm610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li SC, Pan CY, Lin WC. Bioinformatic discovery of microRNA precursors from human ESTs and introns. BMC Genomics. 2006;7(1):164. doi: 10.1186/1471-2164-7-164. [DOI] [PMC free article] [PubMed] [Google Scholar]