Abstract
The cystic fibrosis transmembrane conductance regulator (CFTR) protein is a member of the ATP-binding cassette (ABC) transporter superfamily that functions as an ATP-gated channel. Considerable progress has been made over the last years in the understanding of the molecular basis of the CFTR functions, as well as dysfunctions causing the common genetic disease cystic fibrosis (CF). This review provides a global overview of the theoretical studies that have been performed so far, especially molecular modelling and molecular dynamics (MD) simulations. A special emphasis is placed on the CFTR-specific evolution of an ABC transporter framework towards a channel function, as well as on the understanding of the effects of disease-causing mutations and their specific modulation. This in silico work should help structure-based drug discovery and design, with a view to develop CFTR-specific pharmacotherapeutic approaches for the treatment of CF in the context of precision medicine.
Keywords: ABCC7, ABC transporte, Ion channel, Cystic fibrosis, Modulators, Mutations
Introduction
The cystic fibrosis transmembrane conductance regulator (CFTR) protein belongs to the large ABC superfamily, which is characterized by a typical ATP-binding cassette (ABC) [1, 2]. This cassette, also designated as nucleotide binding domain (NBD), contains a number of specific sequences (the Walker A (P-loop) and Walker B motifs, the C-motif (also known as the signature sequence) and the A-, D-, Q- and H-loops). The vast majority of ABC proteins are transmembrane proteins, which function as ATP-dependent active transporters. Although the NBDs from ABC transporters share highly conserved structures, the membrane-spanning domains (MSDs) are much more diverse and thus form distinct families, distinguishing between importers and exporters.
CFTR, also called ABCC7, is a large protein comprising 1480 amino acids. It shares the typical and unique architecture of ABC exporters, referred to as the B-family ABC-exporter fold, which is made of two MSDs, each composed of six transmembrane helices and each followed by an NBD [3, 4]. The two halves (MSD1–NBD1 and MSD2–NBD2) are fused together to produce a heterodimeric organization. The CFTR protein is however unique in the ABC superfamily as it is the only known member that functions as a channel and is not believed to mediate active transport [5, 6]. The CFTR protein is indeed an ATP-gated chloride channel, the opening of which is directly linked to ATP-driven tight dimerization of the NBDs [7, 8]. The CFTR protein also possesses a unique additional domain (~195 amino acids), called the regulatory (R) region, linking the two halves of the protein, as well as N-terminal and C-terminal extensions (~65 and 33 amino acids, respectively). The R region contains multiple phosphorylation sites, which are the target of Protein Kinase A (PKA) and Protein Kinase C (PKC), and control the gating and trafficking of the chloride channel [9–12]. Most importantly, mutations in the CFTR protein cause cystic fibrosis (CF), one of the most common lethal autosomal recessive diseases in the Caucasian population [13–15].
Hence, considerable research has been undertaken to understand the normal function of CFTR and how CF mutations cause CFTR dysfunctions, with the ultimate goal of developing specific therapies targeting the root cause of disease [16]. In particular, there have been tremendous efforts to characterize the experimental 3D structure of the full-length CFTR protein, which is however difficult to express at high levels, purify and reconstitute in an active form [17]. In addition to the difficulties generally encountered for crystallizing membrane proteins, CFTR certainly suffers from the high flexibility of its R domain, which is commonly described as intrinsically disordered and acts as a hub for phosphorylation-dependent intra- and inter-molecular interactions [18]. So far, published information about the full-length protein was limited to low-resolution 3D structures [17, 19–24] and crystal structures at atomic resolution have only been obtained for individual NBDs, generally with the help of solubilizing mutations [25–29]. NMR studies have also been reported, focusing on the effects of perturbations (such as mutations and ligand binding), on the conformational fluctuations and interactions of isolated NBD1, as well as on the phosphorylation-dependent interaction landscape of the isolated NBD1 and R region [11, 18, 30–34]. Small angle X-ray scattering (SAXS) techniques have also been used to unveil structural changes occurring within the NBDs and the R region [35–37], as well as within detergent–CFTR complexes [38]. Detailed descriptions of most of these experimental studies have already been provided elsewhere (for reviews see [17, 39]).
In this context, where critical experimental information is still missing, several theoretical studies have been performed in order to gain insight into the structure and dynamics of the CFTR channel. Molecular modelling studies have been performed to understand the complex, multi-domain architecture of the CFTR protein, some of which have been coupled to molecular dynamics simulations for refining the proposed models, evaluating their stability and investigating conformational changes. Several theoretical approaches have also specifically addressed the dynamics of the individual NBD1 domain, which is affected by the most common CF mutation (deletion of F508). In an interesting review published 2 years ago [40], Odolczyk and Zielenkiewicz have summarized most of these molecular modelling studies already performed at that time with a view to gain insight into various features of the CFTR protein, especially as regards its 3D structure, its dynamics and its interactions with partners, including drug-like compounds. In particular, they emphasized the importance of sequence alignments with various 3D protein templates (on which the relevance of the final CFTR models depends quite largely) and they critically discussed the major differences between the models obtained so far. Since then, there were only a few theoretical studies that have provided additional information about the general MSD:NBD assembly [41–43] and about the dynamics of the NBD1 domain in presence of several perturbations [44].
Here, we have thus made the choice to concentrate on the whole set of molecular modelling studies, including molecular dynamics (MD) simulations, in light of specific features. Our first aim (see “From ABC exporters to the CFTR ion channel”) is to describe how theoretical studies, i.e. comparative modelling and MD simulations, allowed to gain insight into the CFTR protein 3D structure in terms of its specific evolution towards a channel function starting from an ABC-exporter framework. Next (“Disease-causing mutations: prediction of their impact and searching for modulators”), we also describe how these theoretical studies allowed estimation of the impact of disease-causing mutations, as well as prediction of potential 3D sites that can be targeted for correcting functional or folding defects. For the discussion of these different points, we took into account the CF literature (but obviously not in an exhaustive manner given its huge size) and combined our descriptions with numerous references to various experimental data that could provide support to the predictive studies or that were inspired by the modelling work. We believe that this last point is critical for assessing the quality of the theoretical studies, as the modelling of membrane proteins is generally a challenging task, due to the general low sequence identity observed with the templates used for modelling and the large conformational changes at play in the activity of such proteins.
From ABC exporters to the CFTR ion channel
Comparative modelling of the CFTR protein
The NBD1:NBD2 heterodimer
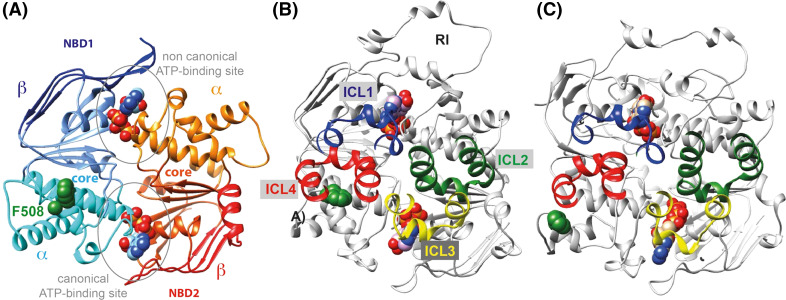
Comparative modelling could be applied for CFTR as experimental 3D structures, which may be used as reliable templates, had been solved. The first major milestone in this context was the publication in the early 2000s of several experimental 3D structures of ABC NBD dimers, in a head-to-tail conformation [45]. A first model of the CFTR NBD1:NBD2 architecture was then proposed [46, 47], based on the experimental 3D structure of the Methanoccus jannaschii ABC transporter MJ0796, solved in presence of two ATP-bound molecules [48]. This ATP-bound configuration was made possible via a mutation of the catalytic glutamate of the Walker B motif into glutamine (E-to-Q mutation), a change preventing activation of water for hydrolytic attack on ATP, whilst preserving high-affinity binding (see [17] for a review). Beyond the description of the global architecture of the heterodimer, the main lesson learned from this model was the description, at the atomic level, of the composite ATP-binding sites of CFTR at the interface of the dimer (with the Walker A and Walker B motifs and switch (H-loop) of one NBD and the ABC signature motif of the other NBD), one nucleotide binding site being canonical and the other one being non-canonical, degenerated (Fig. 1a). Experiments with purified and reconstituted NBD1 and NBD2 have provided direct evidence of a heterodimerization of the two domains, required for optimal catalytic activity [49]. Moreover, in vivo cross-linking has also demonstrated that NBD1 and NBD2 interact in a head-to-tail configuration [50]. Several other experimental studies have shown that the canonical ATP-binding site is catalytically competent, while ATP hydrolysis at the degenerated site is weaker, or even null [51–56]. This functional asymmetry of the ATPase site motifs is a common feature of several mammalian ABC transporters, such as TAP and MRP1 [57].
Fig. 1.
3D structure models (ribbon representations) of the CFTR NBD1:NBD2 heterodimer in a head-to-tail configuration. These were modelled from the experimental 3D structure a of MJ0796 (pdb 1L2T, 1.9 Å resolution) [46] and b and c of Sav1866 (pdb 2HYD, pdb 3 Å resolution) before (b) and after (c) short MD simulations [43]. NBDs are composed of three distinct subdomains: the catalytic core α/β subdomain, a α-helical subdomain and a β-sheet subdomain. The positions of the coupling helices of ICL1 to ICL4 from MSD1 and MSD2 are shown on the Sav1866-based models, as well as the RI. A particular plasticity of the NBD1 α-subdomain was observed during these MD simulations (c), resulting in new contacts between F508 and ICL4. Figures were prepared using UCSF Chimera [200]
In the model of the CFTR NBD1:NBD2 heterodimer, the large regulatory insertion (RI) of about 35 amino acids, which is located between the two first β-strands of NBD1 and contains a phosphorylatable serine residue (S422), covers, like a cap, the degenerated ATP-binding site, suggesting that it may reinforce its non-canonical feature and help to limit dissociation of the heterodimer. This hypothesis is consistent with the results of two independent studies that have suggested that the NBD dimer interface remains formed around the non-canonical binding site, throughout several gating cycles [58, 59]. Existence of the RI at this location in the primary structure was correctly identified through the sequence analysis underlying the model construction and was validated by the experimental 3D structure of individual NBD1, in which it is highly disordered [26, 27]. As positioned in these experimental 3D structures, the CFTR RI as well as its regulatory extension (RE) (located at the extremity of NBD1 and likely corresponding to the first segment of the R region) might prevent the association of NBD1 with NBD2, as they occupy the interface in the modelled NBD dimer. Interestingly, it has been shown that removal of the RI strongly stabilizes the NBD1 in vitro [60, 61] and improves biogenesis of F508del-CFTR in cells [62]. Even though no data at atomic resolution is yet available for the CFTR NBD1:NBD2 assembly (only the homodimeric structure of NBD1 has been reported [25]), SAXS data have shown that the shape of the heterodimer (in solution and in presence of ATP) does well fit the conformation predicted from modelling studies, whilst in absence of ATP, the heterodimer has a bi-lobular arrangement [35, 36, 39], similar to that proposed in a model of the closed form of CFTR (vide infra). Such a configuration has also been supported by experimental 3D structures of full ABC transporters (vide infra). According to this scheme, ATP binding to both sites was thought to promote dimerization and conformational changes leading to channel opening, whereas hydrolysis of the nucleotide at the conventional site precedes channel closing [52].
Similar models of the NBD1:NBD2 heterodimer were subsequently built using (i) an hybrid strategy (models of the individual NBDs build on the mouse NBD1 template [26] and then fitted onto the MJ0796 dimeric conformation) [63] or (ii) a protein–protein docking approach, without any a priori knowledge of the dimeric architecture [64]. These two models were subsequently used for docking of activators (vide infra).
The MSD:NBD assembly
A second turning point in the ABC-exporter knowledge was reached in 2006, when the Locher’s group published the experimental 3D structure of the Sav1866 ABC exporter from Staphylococcus aureus, obtained at 3.0 Å resolution in an outward-facing conformation [65, 66]. This solved the uncertainties regarding the previously published 3D structures of the bacterial MsbA proteins, which were proven to be misinterpreted and were subsequently corrected [67]. These first 3D structures of ABC exporters have thus provided templates for the first homology modelling studies of the CFTR MSD:NBD assembly [68–70]. The first 3D models built by two independent groups using the Sav1866 3D structure as a template [68, 70] were in adequacy with the general topology of ABC exporters, in which domain swapping is observed in the MSDs (Fig. 2a). Indeed, for crossing the membrane, the MSDs form two bundles of six transmembrane helices (TM), in which the two first TMs of one MSD are in close association with the four last TMs of the other MSD. As in ABC exporters, four of the six TMs largely protrude into the cytosol (by approximately 25 Å) to form intracellular loops (ICLs), which end by short coupling helices running parallel to the plane of the membrane and contacting the NBDs. ICL1 (located between TM2 and TM3) and ICL3 (between TM8 and TM9) contact the NBDs at the level of the ATP-binding sites (the non-canonical and canonical ones, respectively), whereas ICL2 (between TM4 and TM5) and ICL4 (between TM10 and TM11) bind in a groove located at the surface of NBD2 and NBD1, respectively (Figs. 1b, 2a). The inner helices of ICLs associate together into a compact four-helix bundle. Even though the modelling procedures followed by Mornon et al. [68] and Serohijos et al. [70] were both based on the Sav1866 architecture, there were however substantial discrepancies between the two models, because of (i) alignment differences in the MSDs (that are particularly difficult to align due to the low levels (~10–15 %) of sequence identity with the template) and (ii) the use of either the sole Sav1866 template [68] or multiple templates (Sav1866 and experimental CFTR NBD1 [70]), the result being a different topology of the ICLs:NBDs interfaces. All the discrepancies observed between the different models (these first ones and the following other ones) and their sources were described in details elsewhere [40]. Among all the CFTR models published so far, only the two first ones tentatively proposed distinct models of the R region, which were constructed ab initio [70] or based on hydrophobic cluster analysis (HCA) [68]. These R models were however very divergent and did not match all the available experimental data. Low-energy conformations of the isolated R domain were explored later using computational methods, generating an ensemble of accessible R region conformations [71].
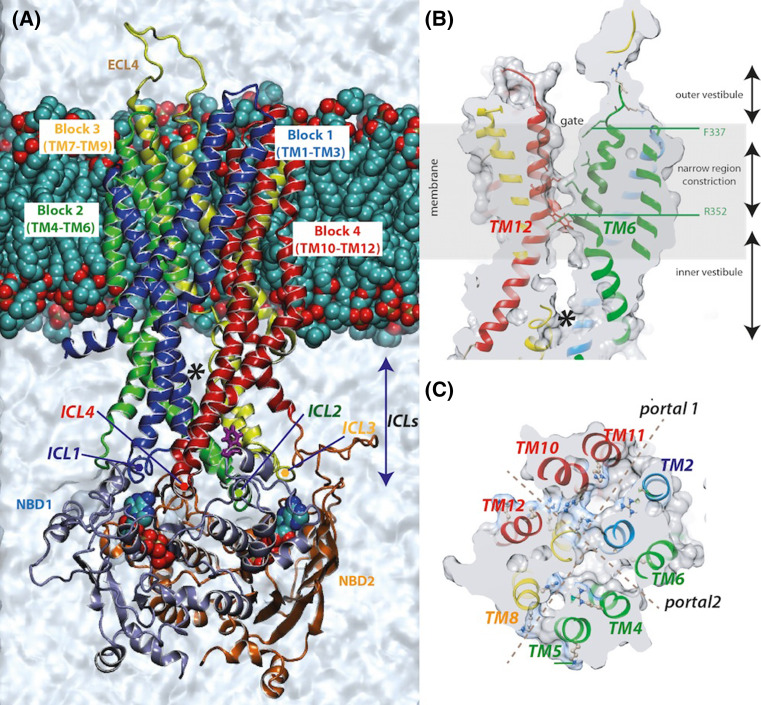
Fig. 2.
3D structure model (ribbon representation) of the CFTR MSD:NBD assembly. a 3D structure model, in a lipid bilayer, of a possible open form of the CFTR MSD:NBD assembly, built using the Sav1866 experimental 3D structure as template and refined by short MD simulations [43]. One of the lateral cytoplasmic openings is clearly visible (asterisk). b Longitudinal cross-section of the model, illustrating the global shape of the pore, with the inner and outer vestibules, separated by a narrow constriction formed as TM6 (green) and TM12 (red) are close to each other. The channel is open over its whole length. c Transversal view at the level of the inner vestibule, highlighting two perpendicular tunnels at the level of the ICLs, allowing access from the cytoplasm through lateral portals
The predicted NBDs/ICLs interfaces revealed in the two first models of the MSD:NBD assembly were verified by cysteine cross-linking experiments, especially highlighting the importance of clusters of aromatic amino acids, within a highly dynamic network, which implicate multiple regions in the NBDs [70, 72]. These first 3D models of the CFTR MSD-NBD assembly, based on the outward-facing Sav1866 3D structure, were thus proposed to correspond to the open form of the channel, as NBDs are closely associated in a head-to-tail configuration, similar to that observed in the ATP-bound conformation of MJ0796 (vide supra), although ADP was observed in the ATP-binding sites of Sav1866 [65].
The inward-facing conformations observed in the corrected MsbA 3D structures [67] were used shortly afterwards for modelling a first, potential closed form of the channel [69]. The experimental 3D structure chosen as template was that of the closed-apo VcMsbA, in which the NBDs are not fully separated. Transition from the open configuration was obtained by a twisting of the transmembrane helices and a sliding motion of the NBDs, the contact points between the ICLs coupling helices and the NBDs remaining however unchanged. In the NBDs, the RI was proposed to play the role of a “safety catch” which hinders the complete disassembly of the NBDs. A similar role in preventing the dissociation of the NBDs might be hypothesized for the regulatory R region. The overall configuration of the NBDs proposed at that time was supported by the bi-lobular arrangement observed in SAXS experiments performed for the NBD1:NBD2 heterodimer [35, 36, 39].
From an ABC exporter to an ion channel
The prevailing model for active transport in ABC exporters is the “alternating access” model, in which the pathway defined by the membrane-spanning domains is alternatively exposed to the internal and external sides of the membrane, with large conformational changes being involved in the translocation mechanism [73]. There is however some controversy about this mechanism, as there is substantial evidence suggesting that the separation of ABC NBDs is limited, and that the large physical separation observed in crystal structures of ABC transporters might be an artefact [74]. As discussed above, the first models of the CFTR channel have been built based on the assumption that the ATP-bound, outward-facing conformation is likely to be similar to that adopted by CFTR when the channel is open, whereas the inward-facing conformation should correspond to the closed form of the channel. However, the models built in this manner do not match all the expected features of a channel-like structure, comprising outer and inner vestibules linked by a narrow region containing the selectivity filter [75]. Moreover, as also mentioned above, experimental evidence exists for a “constant contact-like” model in which the CFTR NBD1:NBD2 dimer interface remains formed around the non-canonical binding site, throughout several gating cycles [58, 59]. In fact, the CFTR protein shows rapid transitions between open and closed states, with subconductance states, on a timescale much faster than the ATP hydrolysis rate, suggesting that small conformational shifts may occur, with conservation of an ATP-bound state [76, 77]. Thus, an important question here is: if the alternating access model does not really apply, what would then be a correct model and what are the key structural features that have enabled CFTR to evolve towards an ion channel? Molecular dynamics performed on homology models as well as the recently solved 3D structures of ABC exporters that may be used as templates for new modelling studies provided several insightful clues to address this important issue.
Molecular dynamics simulations
Asymmetric hourglass conformation of the channel
Thus, molecular dynamics (MD) has been used to optimize the channel pore architecture in the open conformation [43, 78–80]. Indeed, if molecular modelling (which critically depends on the accuracy of the alignment with the templates’ sequences) allows to get a starting conformer of the 3D structure, MD simulations allow in relatively short simulation times, not only to explore the short time scale conformational stability of the model and refine it, but also to sample potential conformational states that lie nearby the starting point in the energy landscape. In particular, despite differences in the sequence alignments and modelling strategies that were already detailed elsewhere [40], all these studies converged towards an asymmetric hourglass conformation of the channel in the open conformation, with a shallow outward-facing outer vestibule and a wide inner vestibule, the two vestibules being linked by a narrow bottleneck (Fig. 2b). Such a shape fits well with that of two-dimensional CFTR crystals observed by electron crystallography [21] and is also in good agreement with experimental data, especially numerous substituted cysteine accessibility mutagenesis (SCAM) studies (reviewed in [75]), as well as cross-linking studies [81, 82]. These functional studies have precisely defined amino acids lining the pore and the extracellular vestibule in the different conformational states and have indicated that TM6 and TM12, but also TM1 and TM11 play a central role in the CFTR continuous central chloride permeation pathway, with evidence of an asymmetric channel pore [78, 79, 83–98]. In contrast, other SCAM investigations failed to identify key residues in either TM5 (analogous to TM11 in MSD1) or TM7 (analogous to TM1 in MSD2) [83]. The model of the “full-open” conformation of the CFTR MSD:NBD assembly, observed after short MD [43] may account for such an observed asymmetry. Indeed, establishment of a salt-bridge between the residues R352 (TM6) and D993 (TM9), which is a key marker of the “full-open” conformation [99], resulted in a central narrow pore, lined by TM1, TM3, TM6, TM12, TM11, TM12, which excludes TM5, TM8, TM7, TM9 confined to a pentagonal duct (formed together with TM6). The formation of this salt-bridge has also been reported in other simulations [78, 79]. Worth noting here is the frequent occurrence of salt bridges in the CFTR MSDs, which was supported by different modelling studies [43, 78, 80, 100], some of which having proved to play a key role in channel gating [99, 101, 102]. A critical salt-bridge between E267 and K1060 was also highlighted in the four-helix bundle formed by the ICLs internal helices [103, 104].
Location of the external gate
The boundary between the constriction of the pore and the outer vestibule was suggested to determine the anion selectivity filter of CFTR, which exhibits characteristics of a lyotropic series [75, 105]. These features imply that anions with the lowest free energy of hydratation tend to show a higher permeability, i.e. are more likely to pass through the channel. The model of a possible closed form of the CFTR MSD:NBD assembly, obtained when starting from the “closed-apo” conformation of MsbA [69], as well as more recent models (i) built starting from the TM287/288 3D structure [42] (vide infra) and (ii) obtained after further MD simulations of a refined Sav1866 3D model [43], support the fact that this boundary may be a plausible location of the gate, which controls the anion flow through the channel [106]. The last model [43] is believed to correspond to a C2 state, where the NBDs are fully dimerized but the channel is nevertheless still closed for conduction (as opposed to the so-called closed C1 or C0 state, where one or both nucleotide-binding sites are empty, respectively). Interestingly, this suggests that it is possible that relatively small, localized conformational changes within the MSDs might be sufficient to open and close the CFTR channel. In all these three models of closed forms, an aromatic amino acid, F337, notably contributes to the closure of the gate, together with a set of mainly hydrophobic amino acids from the different transmembrane helices (Fig. 3a, b—bottom). Mutation of this conserved F337 had gain-of-function (GOF) effects, which were observed in the absence of any exogenous activator (i.e. ATP or PKA), suggesting that the aromatic cycle of this amino acid may help to stabilize the closed form of the channel [107]. Interestingly, results of 30 ns MD simulations made with both the MsbA-based and Sav1866-based models of the closed and open forms [68, 69] suggested that, in the closed conformation, the conserved residue P355 in TM6 exists in a stable cis conformer within a type IV β-turn and stabilized by the adjacent F354, whereas in the open conformation, P355 exists in a trans conformer [108]. P355 could thus constitute a stable element also stabilizing the closed state of the channel, as assessed again by GOF mutations at this particular location [108]. A recent analysis of energetic coupling between residues across the dimer interface in single CFTR channels with GOF mutations (P355A and K978C) associated with spontaneous pore opening/closure showed that the NDB dimer is formed when the pore spontaneously opens, demonstrating a strict coupling between NBD and MSD movements [109].
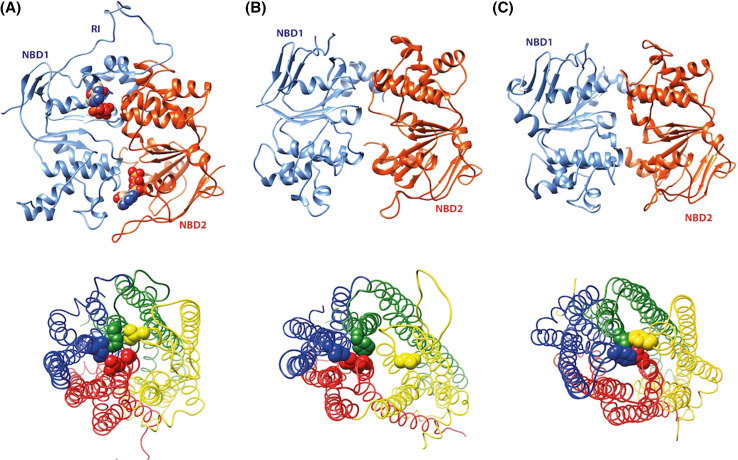
Fig. 3.
3D structure models (ribbon representations) of two possible closed forms of the CFTR MSD:NBD assembly. a Sav1866-based model, after short MD simulations, leading to a possible model of the C2 form [43], b TM286/287-based model [42]. These models were compared to the experimental 3D structure of ATP-free PCAT1 (pdb 4RY2, [117]) (c). Near the extracellular surface, hydrophobic amino acids form similar closed gates, which involve (i) in CFTR: I106 (TM1, blue), F337 (TM6, green), L883 (TM7, yellow), G1130 and L1133 (TM12, red) and (ii) in PCAT1 : F194 (TM1 and TM1′, blue and yellow) and L426 (TM6 and TM6′, green and red)
Lateral cytoplasmic portals towards the channel
The electrodiffusional transmembrane movement of ions in a channel implies that the protein in the open state has a continuous pathway between the intracellular and extracellular solutions. However, an important unanswered issue of all the models published before 2015 was the features of the cytoplasmic entrance to the central pore, the chloride permeation pathway seeming indeed sealed on the cytoplasmic side of the membrane at the level of the ICLs. According to Gadsby et al. [110], the cytoplasmic-side gate of CFTR should have become atrophied or at least uncoupled from the outer gate, making CFTR a “broken” ABC exporter. The recent MD simulation performed on a refined model of the MSD:NBD assembly, based on the Sav1866 3D structure, has suggested that chloride ions enter the central pore via at least one and potentially several lateral portals that are displayed at the level of the ICLs (between TM2 and TM11, TM5 and TM8 (principal lateral tunnel), TM4 and TM6, TM2 and TM11 (secondary lateral tunnel)) and communicate with the inner vestibule [43] (Figs. 2c, 4a). Such horizontal lateral cytoplasmic portals have already been observed in several other ion channels [111, 112] and seem also to be present in CFTR, as suggested by electron crystallography [21]. Multiple positively charged residues line these putative lateral portals and may play a role in attracting negatively charged ions towards the pore. The existence of such portals, at least that formed between TM4 and TM6 (Figs. 2c, 4a) (and perhaps TM10 and TM12), was supported by recent experimental data using cysteine substitutions and in situ modifications by negatively and positively charged methanethiosulfonate (MTS) reagents [113, 114]. K190, R248, R303 and K370 (TM4/TM6 entrance), as well as K1041 and R1048 (TM10/TM12 entrance) may contribute to the electrostatic attraction of cytoplasmic Cl− ions to the pore. Worth noting is that a similar cytoplasmic portal between TM4 and TM6 has been also highlighted in a recent model of the CFTR open channel based on the McjD 3D structure [42] (vide infra). Lateral portals also tend to be formed in order to allow to reach an internal cavity (inner vestibule), when the role of ATP binding as a driving force for NBD dimerization was analysed by 100 ns MD simulations, starting from a model constructed on the basis of an inward-facing configuration (multi-template-based model, combining the global architecture of mouse P-glycoprotein (PgP) and crystal structures of NBDs [115]). In this study, it was shown that ATP binding was a driving force not only for NBD dimerization, which occurred within only 10 ns and was similar that observed in the closed-apo model of MsbA [67], but also for NBD–MSD concerted motions (although the pore did not reach an open form at the end of this MD simulation). Rahman et al. [100] have also explored the conformational transition between a model of the CFTR open channel, built on the Sav1866 template, and a model of the CFTR closed channel, built on the mouse PgP, using targeted molecular dynamics. Here, a progressive conformational wave was observed during opening transition, but no clear entrance from the cytoplasmic milieu could be detected in the model of the open form.
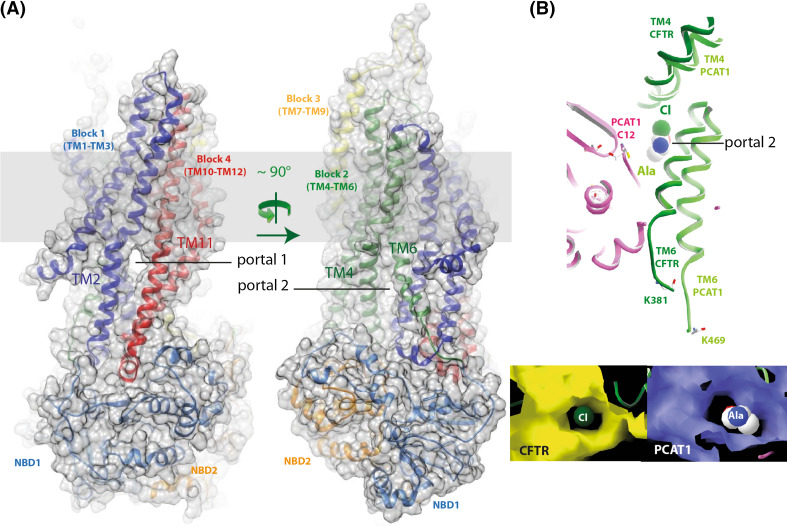
Fig. 4.
Lateral portals. a 3D structure model (ribbon and surface representation) of the possible open form of the CFTR MSD:NBD assembly, also shown in Fig. 2 [43]. The two portals are clearly visible on the two views, rotated by ~90°. b Comparison of the modelled CFTR TM4/TM6 lateral cytoplasmic portal with that observed in the ATP-free PCAT1 experimental 3D structure (pdb 4RY2 [117]), onto which the peptidase domain docks and which allows the substrate to access the central cavity. Cl- and an amino acid (alanine) are shown at the level of the openings. These two lateral portals occupy nearly the same position in the two structures
New experimental 3D structures that might be used as templates
A second set of data is based on very recent experimental 3D structures of type 1 ABC exporters, which revealed highly original features related to the MSD and NBD assemblies and may thus be considered for further modelling of the CFTR-specific structure.
The first important observation deduced from three recent crystal structures is the occurrence of a new conformation of the MSDs, called nucleotide-bound outward occluded, which has similarities with both the outward-facing and inward-facing conformations. This conformation was first observed in the Escherichia coli exporter McjD (bound to the ATP analogue AMP-PNP [116]), and then in the Clostridium thermocellum PCAT1 (ATPγS-bound [117]). A similar, but ADP-bound, outward-occluded form has also been observed in the Campylobacter jejuni flippase PlgK [118]. These transition intermediates may thus represent realistic frameworks for the conformation of the CFTR channel, in agreement with what was observed after molecular dynamics simulations (see above). In line with these arguments, Corradi et al. [42] suggested, after structural analysis of models based on different templates, that a McjD-based model could indeed account at best for the open state of the CFTR channel.
Of note, an interesting feature of the intermediate conformations observed in the PCAT1 3D structure in absence of ATP is that lateral openings of the TM pathway are observed at the level of ICLs, onto which the N-terminal peptidase domain of this ABC exporter docks and which perfectly superimpose with the lateral portals which were previously described before in the CFTR open channel model obtained after short MD [43] (Fig. 4b). This observation thus provides additional support to the plausible existence of such structures in CFTR, a likelihood which was already suggested by electron crystallography data [21]. Such a lateral opening was also recently observed in the subnanometric 3D structure of TrmA observed by electronic microscopy [118].
Interestingly, the closure of these experimental 3D structures in the vicinity of the extracellular surface does also provide additional support to the models of the closed forms that were published so far (MD refined Sav1866-based [43], closed-apo MsbA-based [69], TM287/288-based [42] (vide infra)). Indeed, rings of hydrophobic residues, displayed by both MSDs, are formed through van der Waals contacts, forming closed gates. These rings, as well as the positions of the transmembrane helices are strikingly similar to those observed in the CFTR models, in which F337 plays a critical role (see top and bottom of Fig. 3c).
The second observation deduced from recent crystal structures is that the NBDs are not necessarily disengaged or twisted in inward-facing conformations, in contrast to what was first observed in experimental 3D structures of MsbA (from Vibrio cholerae and Escherichia coli) [67], and later in the experimental 3D structures of the mouse, Caenorhabditis elegans and Cyanidioschyzon merolae PgP [119–123], of the Saccharomyces cerevisiae and Novosphingobium aromaticivorans Atm1 [124, 125], of the human mitochondrial ABCB10 [126] and of the Campylobacter jejuni PlgK [127]. In fact, it is at present generally admitted that inward-facing conformations without NBD contact are not likely to be physiological [4].
This was first evidenced in the heterodimeric TM287/288 ABC exporter from Thermotoga maritima [128, 129] which, like CFTR, has a degenerate ATP-binding site and in which a separation of the coupling helices ICL2 and ICL4 by 15 Å is sufficient to open the substrate-binding cavity, without complete NBD dimer disengagement. These 3D structures, solved in an apo (nucleotide-free) form and with AMP–PNP bound at the degenerate catalytic site, respectively, have evidenced the critical importance of D-loops, running antiparallel between the ATP-binding sites [130], for association of the NBDs, as well as for allosteric communication between the two asymmetrical ATP-binding sites. These structures may thus provide reliable 3D templates for modelling the CFTR NBD heterodimer in the closed form, in agreement with experimental data showing little separation between the NBDs [58, 59]. A 3D model of a fully closed form of the CFTR channel, based on this TM287/288 3D structure, has recently been proposed by Corradi et al. [42] (Fig. 3b, top). Worth noting here is that such a limited dissociation of the NBD dimer was also highlighted in the nucleotide-free, but homodimeric 3D structure of Clostridium thermocellum PCAT1, with inter-domain contacts also mediated by the conserved D-loop [117] (Fig. 3c, top).
Disease-causing mutations: prediction of their impact and searching for modulators
Prediction of the impact of CFTR mutations
More than 2000 CF mutations have been described, conferring a wide range of phenotypes, with variable disease severity [15]. Six major classes are currently defined, according to the observed primary biological defects [13]. A refinement of this widely used classification is however at present proposed in order to take into account the complex molecular and cellular phenotypes of the various CFTR alleles [15]. Moreover, mutations can also be described in light of their responsiveness to drugs, thereby defining “theratypes” [16] and paving the way to personalized medicine. In this context, information gained by structural biology, either at the experimental level or from in silico modelling studies, should be useful to better understand the molecular basis of the mutation-caused defects and also to identify or design specific therapeutic approaches. Over the last decades, extensive efforts have been made with a view to identify small-molecule CFTR modulators, which include activators, potentiators and correctors, developed for treating CF, as well as inhibitors (blockers), which may be used for treating enterotoxin-induced diarrhoea and autosomal dominant polycystic kidney disease (ADPKD) [15, 131, 132]. Whereas CFTR activators stimulate the channel functions by increasing intracellular cAMP/cGMP levels, CFTR potentiators and correctors act by increasing the channel gating activity and the amount of protein at the cell surface, respectively. Current approaches are based on high-throughput screening (HTS), but they provide little information on the molecular mechanisms in which the identified molecules are involved. Consideration of structural data may thus provide useful information not only for understanding these mechanisms and eventually improving the activity of the identified molecules, but, most importantly, also for rational structure-based design of novel CFTR modulators, in order to target specific mutations.
Several theoretical studies have addressed the important question of the consequences of the most common CF mutation, the deletion of F508, a phenylalanine residue located in the NBD1 alpha-subdomain. Most of the F508del mutant protein is misfolded and targeted to the endoplasmic reticulum-associated degradation pathway, whereas the few F508del molecules escaping degradation suffer from a lower activity and a lower membrane stability ([133] for a review). Moreover, the silent codon change for isoleucine 507 (ATC → ATT), caused by the three nucleotide deletion responsible for the F508del variant, alters the F508del CFTR mRNA structure, with a reduction of translation efficacy [134]. Comparison of a series of static crystal structures of isolated NBD1 [25–29, 135] showed very localized differences between the wild-type (wt) and F508del constructs, primarily localized to the F508 region, whereas NMR studies of murine NBD1 highlighted non-local differences in the dynamic profiles of the structures in solution [30]. On the other hand, the first models of the CFTR MSD:NBD assembly have indicated that the side and main chains of F508 interact with amino acids from ICL4 [68, 70]. Accordingly, the F508del mutation compromises both the NBD1 thermodynamic stability and domain assembly [29, 60, 81, 135–141]. While two independent studies [136, 137] have indicated that an optimal correction might require both the stabilization of F508del NBD1 and the restoration of its interactions with ICL4, a more recent study has suggested that the restoration of the thermal stability might be sufficient to promote CFTR maturation [142]. However, the current correctors, including VX-809, appear unable to restore thermodynamic stability, while allowing however global conformational maturation [143].
The dynamics profiles of isolated wt and F508del NBD1 have been thoroughly analysed using MD simulations. A first study used classical, short (2 ns) MD simulations on models (based on the X-ray structures of mouse NBD1) of the human wt and mutated NBD1 [144]. It permitted to observe substantial differences in the dynamics properties, differences which were smaller when F508del NBD1 is in complex with the small ligand CPX (8-cyclopentyl-1,3-dipropylxanthine), bound in the ATP-binding site. Next, a second study using a slightly different approach, considering longer MD simulation times (20 ns), confirmed the existence of an increased conformational freedom of the mutated NBD1, with a disruption of the conformation of the linker between the ABCα and ABCβ subdomains [145]. In contrast, Bisignano and Moran observed no significant difference after shorter MD simulations (5 ns) at room temperature, with no change in enthalpy or entropy [146]. A slight variation in the energy of interaction with the solvent was observed, as well as a decreased affinity for ATP. The effect of the deletion of the RI has also been investigated and a reduction of the fluctuations of the F508del NBD1 was observed, a finding consistent with the rescue effect observed experimentally [62]. Besides these studies focusing on the fluctuations observed between wt and mutated domains, another work has indicated that F508del alters the folding dynamics and kinetics of NBD1 [147]. By showing that meta-stable intermediate states appearing along the wt and mutant folding pathways were differently populated, these authors identified sensitive regions in NBD1 in which rescue mutations were designed to increase folding propensity. Graph theoretical methods applied to the study of dynamic couplings also identified S492 as a key residue for understanding the allosteric interactions between RI and the F508del and I507del mutations [148]. Finally, Replica Exchange MD (REMD) simulations were also applied to a series of NBD1 constructs from which the RI was removed and including F508del as well as some mutations (tested or not in the F508del background) which are known to partially rescue the folding and function of F508del CFTR [44]. The REMD-generated root mean square fluctuations (RMSF) profiles consistently indicated the higher fluctuations of the CFTR F508del NBD1, with both local and non-local differences in the dynamic profile, within a dynamically coupled network; these local and non-local fluctuations, which highlight the high allostericity of the NBD1 domain, were proposed to affect the thermostability, the maturation and the function of F508del CFTR by increasing the aggregation propensity (exposure of hydrophobic amino acids such as V510 and W496), by reducing the ATP binding capacity (enhanced flexibility in the vicinity of the ATP-binding site) and by impairing interactions with other domains (enhanced flexibility of regions in contact with ICL1, ICL2 and ICL4). These REMD simulations have also permitted to gain insight into the rescuing mechanism, as suppressor mutations (V510D [149]) and Teem mutations (G550E, R553Q, R555K [135]) reduce the fluctuations in multiple areas of NBD1, both in the vicinity of the mutation and in remote regions. This effect has also been observed when introducing proline residues at key positions in NBD1 [150]. Overall, all these MD studies performed on isolated NDB1 have thus provided insightful information that cannot be derived from its static 3D structures, especially by defining fluctuating regions, that can be targeted for designing specific therapeutic approaches (vide infra). Most of the MD simulations applied to models of the CFTR MSD:NBD assembly were focused on the study of the channel pore and the conformational transitions between the open and closed forms; they have thus not addressed the dynamics properties of the NBD1 (either in the wt form or with the F508del mutation) in this particular context of the MSD:NBD assembly [43, 78–80, 100, 115].
Recent work of the Oscar Moran group [41] has however specifically focused on the properties of the NBDs in wt constructs of the NBD:MSD assembly, as well as in the context of F508del and G551D mutations. Of note, the G551D mutation is the third most common mutation in CF; it impairs the gating mechanisms of CFTR and can be rescued by the potentiator VX-770 [151]. The authors have performed MD simulations on a previously published model of the MSD:NBD assembly [68, 69], from which the RI was removed. Analysis of the MD trajectories (over simulation times of 15–20 ns) indicated that, although no significant conformational transitions were observed for the NBDs or the MSDs, the NBD1–NBD2 interactions were however severely affected in both mutants. Higher internal energy was observed in the F508del mutant, with increased contacts between NBD1 and NBD2; as regards the G551D construct, no modification of the internal energy was observed relative to the wt construct, but there was a significant reduction of close NBD1–NBD2 contacts. A modification of the interface between NBDs and ICL2–ICL4 was also observed, which could affect channel gating. Interestingly, one may also want to put these results in the context of NMR studies of an F508del NBD1 construct, which suggested that NBD dimerization may be inhibited, highlighting the importance of the NBD dimer interface [33].
Finally, our own recent MD work has also suggested that the part of the NBD1 α-subdomain containing the residue F508 exhibits a particular structural plasticity, which appears to be driven by rearrangements at the MSD level and to be associated with the full-open channel configuration [43]. The segment bearing F508 remains in contact with ICL4, but within a modified environment, involving contacts with T1076 and L1077, as well as a salt-bridge between K1080 and E504, which contributes to stabilize this conformation (Fig. 1c).
It should also be stressed here that this analysis of this MD-refined 3D model of the MSD:NBD assembly [43] indicated that the well-characterized missense mutations reported in the Clinical and Functional Translation of CFTR (CFTR2) database (http://www.cftr2.org/) tend to cluster around host spots. Mutations encountered in the MSDs mainly affect residues lining the pore (G85E, E92K, D110H, P205S, R334W, I336K, T338I, S341P, R347H/R347P, R352Q). Some mutations affect residues involved in the assembly of the ICL bundle (G178E/G178R, G970R), whereas a large hotspot is located at the NBD1:ICL4 interface (S492F, I507del, F508del, V520F, A559T, R560K/R560T, A561E, H1054D, G2061R, L1065P, R1066H/R1066C, F1074L, L1077P). Finally, a last hotspot is found within the canonical ATP-binding site (S549N, S549R, G551D/G551S, G1244E, S1251N, S1255P). G551D, the third most common CF mutation, disrupts the canonical ATP-binding site normally involved in ATP-dependent gating, converting it into an inhibitory site [152].
In silico studies and CFTR modulators
In silico studies were performed for identifying potential binding sites of CFTR modulators, the assumption here being that modulators directly bind to the CFTR protein. In general, there is however only indirect evidence and very few experimental studies have demonstrated a direct binding of modulators [153, 154]. Even though limitations of in silico approaches can be mentioned [such as (i) limited resolution and accuracy of the 3D models, (ii) limited consideration of the conformational flexibility and plasticity of the interfaces, and (iii) consideration of the folded, but not intermediate states], these may nevertheless allow rational structure-based discovery of small modulators, with increased hit rates when compared to the high-throughput screening (HTS) campaigns.
The first structure-based study was performed by Moran and colleagues [63], who have used a model of the NBD1:NBD2 heterodimer for predicting the binding sites of CFTR potentiators by molecular docking. This model was built on the basis of experimental 3D structures of NBD1, fitted into the MJ0796 dimer. A blind rigid-docking strategy was used, which revealed three hotspots with low interaction energy, two (sites 2 and 3) being compatible with the experimental results. Binding of genistein to site 2, at the NBD1:NBD2 interface, was particularly relevant, when comparing the theoretical binding free energy in the model to that estimated from the apparent dissociation constants KD. The genistein binding site was also predicted in an independent study, starting from a model of the NBD1:NBD2 dimer built using a protein–protein docking approach [64]. This study identified five putative binding sites, located at the ATP-binding sites, on NBD1 and NBD2 and at the NBD1:NBD2 interface.
To the best of our knowledge, there is yet no published data about structure-based predictions of the binding site of the well-characterized hydrophobic potentiator VX-770 (Ivacaftor) [155, 156], which is already used for the treatment of CF patients with the G551D mutation [157] and other class III mutations [158]. Some indirect evidence (such as hydrophobic nature of the compound, kinetics data, chemical modification of engineered cysteine in TM6; reviewed in [155, 156, 159]) have led to the hypothesis that it may be located at the interface between the TMDs and the lipid core of the membrane bilayer. The VX-770 binding site is likely to be distinct from that of some permeant anions (nitrate), which also modulate gating and should bind the pore [159], in agreement with the gating properties also observed with some channel blockers [160–162].
Structure-based studies (based on experimental 3D structures of isolated NBD1 and models of MSD:NBD architecture) were also performed with a view to identify potential binding sites for correctors of the F508del defect. It can indeed be hypothesized that small molecules that bind to and stabilize NBD1 and/or inter-domain regions of CFTR may promote increased maturation of the F508del protein.
The first report was published by Kalid and colleagues [163], who have considered several F508del models [NBD1:NBD2, full cytoplasmic domains (ICLs + NBDs)] to perform structure-based virtual screening. This allowed to identify three cavities, located at (i) the interface between the two NBDs, (ii) at the interface between NBD1 and ICL4 and (iii) at the multi-domain interface between NBD1, NBD2, ICL1, ICL2 and ICL4. Fifteen correctors were identified out of the 496 candidate compounds selected for in vitro testing, as well as compounds with potentiator activity and compounds with a dual potentiator–corrector activity. However, none of these hits were potent enough to show corrector activity in human bronchial epithelial cells.
Another structure-based virtual screening was reported [164], the starting point for molecular docking being here the MD frame of the isolated F508del NBD1 which was the most distinct from the wt NBD1 [145]. Two cavities were identified, which were screened against the National Cancer Institute diversity set I (NCIDS). The underlying assumption here was that the small molecules identified could prevent the interaction between the misfolded F508del and housekeeping proteins, an interaction limiting F508del CFTR delivery to the plasma membrane. The selected compounds, which disrupted the interaction of F508del CFTR with keratin8, induced functional expression in transfected HeLa cells, in human bronchial CF cells in primary cultures and in the nasal epithelium of homozygous F508del mice in vivo. The direct effect of the compounds on NBD1 was supported by hydrogen–deuterium exchange reaction coupled with mass spectrometry (HDex-MS). One of the two pockets identified (pocket 2), in which the binding of the most promising molecule (407882) was observed, is occupied by ICL4 in the model of the MSD:NBD assembly [68], whereas the other one (pocket 1) is localized in the deep interaction interface between NBD1 and NBD2.
Several docking experiments were also performed on F508del MSD:NBD assemblies, in order to identify putative binding sites for VX-809 (lumacaftor), which is now used in combination with VX-770 (ivacaftor) in CF patients homozygous for the F508del mutation [165, 166]. A flexible-receptor, flexible-ligand docking of VX-809 focused on the NBD1:ICL4 interface of the F508del MSD:NBD assembly derived from the Serohijos model [70]; it showed that this site was a possible binding pocket for VX-809 [143]. A second study, in which the model was build based on the Sav1866 architecture, combined with NBD1 X-ray data, also highlighted the NBD1:ICL4 interface as a plausible binding site for VX-809, whereas VRT-325 appeared to be best accommodated within the ATP-binding site and corr-4A would fit into none of the two [167]. The likelihood of the VX-809 binding site at the NBD1:ICL4 interface is further supported by the fact that VX-809 also significantly restores folding of disease-related mutations in CFTR ICL4 [143]. Finally, a third docking experiment of VX-809 onto several interfaces (NBD1:ICL1, NBD1:ICL4, NBD1:NBD2) was performed using two distinct models [70, 80]; it also suggested that the NBD1:ICL1:ICL4 interface is a primary target for VX-809 [168]. However, dissecting further the VX-809 binding site using wt CFTR domain combinations suggested that VX-809 targeting of MSD1 is a prerequisite for stabilizing the domain interface composed of ICL1, ICL4 and NBD1 [168]. This suggestion was supported by other studies that have shown that VX-809 stabilizes an N-terminal domain of CFTR containing only the MSD1 [169, 170] and therefore regulates in an allosteric manner the NBD1:ICL4 interface through interdomain communication. It should be noted here that the relatively moderate effect of Orkambi, a combinational treatment containing the corrector VX-809 and the potentiator VX-770, may be explained by the VX-770-mediated destabilization of VX-809-rescued F508del CFTR, as suggested by the observation that chronic administration of these compounds destabilized the low temperature (27 °C) and VX-809 corrected F508del [171, 172]. This does obviously strongly invite to identify new potentiators that do not interfere with correctors [173], or new correctors that do not interfere with potentiators and also which may act in a synergistic way [174], as well as dual activity compounds, such as aminoarylthiazole derivatives [175]. In this context, models of the F508del MSD:NBD assembly can be used for the in silico design of novel compounds which may fit the pocket at the interface between NBD1 and ICL4, but also other pockets found within the 3D structure (our unpublished results). Worth noting is that class II correctors, such as the bithiazole Corr-4 [176], which has an additive effect to VX-809 [177], appeared to bind NBD1–NBD2 and/or NBD2–MSD1/2 interfaces, thus distinct sites from that targeted by VX-809 (which has been classified as a class-I corrector), as supported by both experimental and docking studies [168]. However, there is still controversy about this Corr-4a binding site, as other authors have reported that it should rather bind to the MSDs [178–180]. Finally, complementary strategies are also be developed to stabilize F508del CFTR, for instance by designing peptides inhibiting the binding of the rescued F508del CFTR to components of the cell surface protein quality control machinery [181].
Worth noting is that none of the small-molecule correctors of F508del CFTR identified so far in cell-based screens appear to act on NBD1, a fact which is likely to explain the lack of effect on thermal stability of the full-length protein [142, 143]. Only chemical chaperones, surrogates of class III correctors, are able to stabilize F508del-NBD1 [168]. This contrasts with the effect of “revertant” mutations in NBD1 (such as deletion of the RI (amino acids 405–436) or 3S mutants (F429S, F494N, Q637R) [62, 135]), which are able to rescue both thermal instability of NBD1 and decrease of trafficking toward the cell surface. Interestingly, thermal stability of F508del NBD1 can be increased by small compounds BIA (5-bromoindole-3-acetic acid) and BEIA (5-bromo-3-ethoxyindole-acetic acid), which bind to a specific site on NBD1 in co-crystallization experiments (SGX Pharmaceuticals) [142] and have an additive effect with VX-809. BIA is buried within the NBD1 structure, between the α/β and α-subdomains, and bound by H-bonds and cation–π interactions. REMD simulations performed with an F508del NBD1 construct in complex with BIA showed an overall attenuation of the fluctuations observed in the F508del NBD1, similar to that observed with rescuing mutations [44]. The BIA site is distinct from that of the dual corrector/potentiator compound CFFT-001, for which flexible docking or induced fit procedures (conducted onto higher energy conformations of NBD1 (F508del ∆RI∆RE) accessed in replica exchange simulations) indicated multiple possible binding modes to the surface of β-strands S3, S9 and S10, with a few specific interactions [31]. Two classes of compounds, causing chemical shift perturbations at the level of the BIA site and CFFT-001 subdomains, were identified after NMR-based binding screen performed against a fragment library of approximately 2600 compounds using the 3S form (mutations at F429S, F494N and Q637R) of F508 and F508del NBD1. However, these NBD1 binders did not show sufficient affinity, a fact obviously limiting the development of such molecules [182]. Worth noting in this context is the recent discovery of the dual corrector/potentiator activity of rattlesnake PLA2CB, which binds with nanomolar affinity to both NBD1 and F508del-NBD1 [183]. Docking experiments revealed a putative PLA2CB binding site on NBD1, which was supported by HDX-MS experiments. Examination of the full-length CFTR model (our unpublished data) indicated that binding of PLA2CB is possible at the surface of the whole complex. Thus, the beneficial protein–protein interaction observed in the PLA2CB:NBD1 complex may open new perspectives for the development of modulators with dual activity and directly acting on NBD1.
Docking experiments have also been performed for predicting binding sites of small-molecule inhibitors, which have potential interest for therapy of (i) enterotoxin-mediated secretory diarrheas such as cholera and (ii) for autosomal dominant polycystic kidney disease (ADPKD) [184–186]. Docking of (R)-BPO-27 on the MD-refined model of the CFTR MSD:NBD assembly in its closed conformation [43] suggested several potential binding sites, the best one (which showed a perfect consensus of scoring functions) being located at the interface between the two NBDs [187]. Such a location, which may lead to interference with ATP-mediated gating in the NBDs, was further supported by ATP competition studies with inside-out membrane patches [187]. This probable binding site is different from the chloride ion-binding sites, targeted by a diverse group of organic ions, which occlude the pore and inhibit thereby chloride transport.
Computational studies have suggested a binding site for the glycine hydrazide GlyH-101 within the MSDs, near F337, a result in agreement with experimental data [78]. GlyH-101 might thus directly occlude the CFTR pore near its extracellular entrance. Docking of seven open channel blockers (including the sulfonylurea glibenclamide) on a model (built using a combination of restrained homology modelling and ROSETTA refinement and introducing a chloride column into the modelling scheme), revealed highly overlapping blocking binding sites, involving ionic interactions with K95 [80], in agreement with experimental data [188]. All of the docked open channel blockers also share an aromatic/hydrophobic interaction with W1145 and a cation–π/hydrophobic interaction with R352. A second blocker binding site in the inner pore vestibule identified R303 as being involved in the binding of suramin, a larger negatively charged organic molecule [189]. The binding site of the thiazolidinone derivative CFTRinh-172 [190], which does apparently involve the residue R347 [191], remains relatively poorly characterized at the computational level. In addition to the basic residues that are conserved among CFTR proteins from various species, it has been shown that the local environment might also play a critical role in the specificity of the channel to specific inhibitors [192].
Concluding remarks
Over the last several years, substantial progress has been made in the understanding of the molecular mechanisms underlying the CFTR functions and dysfunctions by combining experimental and theoretical studies. As shown here, these last ones, including MD simulations, have been instrumental in deciphering many functional features of the CFTR protein, from the definition of the active sites within the asymmetric NBD1:NBD2 heterodimer to the understanding of the shape and specific properties of the channel. Moreover, even though they are by essence approximate, molecular modelling and MD simulations may also provide useful insights into the possible binding sites of CFTR modulators. Such rational approaches, compared to “brute-force” methods, may also be beneficial for developing new CFTR-specific modulators [193].
However, it must be acknowledged that the intrinsic dynamics of the large transmembrane CFTR protein, that MD simulation can help to decipher, introduces an additional degree of complexity for developing specific therapeutic approaches, as these approaches have to preserve the CFTR functional features associated with this intrinsic dynamics while correcting defects introduced by CF mutations. Another challenging issue is related to the understanding of the CFTR folding pathway, in particular for the development of pharmacological chaperones that might guide NBD1 on its way to a stable 3D structure. Of note, only 20–40 % of the wt CFTR nascent chains attain their native conformation, highlighting the complex, cooperative folding behaviour of the protein [133, 194, 195]. This cooperative folding mechanism however offers multiple interfaces that could be targeted by small molecules, but it also points out the probable necessity of using a combination of several drugs for efficient corrector therapy. Along these lines, it should be stressed here that the targeting of transient conformations encountered during the folding process represents an important challenge for treating folding diseases in general.
Regarding the modelling of the CFTR native 3D structure, there are still unresolved questions that need to be addressed in the near future. In particular, a relevant model of the full-length CFTR protein, including N- and C-terminal extremities, as well as the R region, is still lacking; it should allow to better understand the key role that these regions play in the regulation of CFTR functions. This cannot be made without taking into account the post-translational modifications (phosphorylations, palmitoylations, etc.), which involve specific sites of the CFTR protein [196]. Even though this modelling is particularly tricky to achieve in absence of relevant 3D templates and in the general context of disordered regions, it should be recalled here that the isolated R region retains a degree of secondary structure organization, as assessed by SAXS and NMR data [11, 18, 30, 34, 37]. By exploiting such experimental data and using improved predictive tools, better new models of these “extra” regions can now be built, providing new hypotheses about their specific roles in CFTR functioning. Finally, one can also hope that the recent significant developments in high-resolution 3D structures of large macromolecular complexes using cryo-electron microscopy may also open new perspectives for the understanding of the structure of the CFTR full-length protein, even though CFTR lies at the lower limit of the macromolecular structures size recently obtained using such techniques [197–199]. One can also expect that MD simulations performed on longer time periods, as well as metadynamics approaches, should allow to give further clues about the different states of the CFTR protein during the gating cycle.
In conclusion, despite the fact that there are still many unanswered questions, important information has already be gained on the CFTR structure and dynamics, helping to define working hypotheses that could be tested experimentally and to effectively design CFTR-specific strategies for the development of drug therapies for the treatment of CF.
Acknowledgments
We thank the Association “Vaincre La Mucoviscidose” (Paris, France) for constant support. This work was granted access to the HPC resources of IDRIS/CINES under the allocations 2014-077206, 2015-077206 and 2016-077206 made by GENCI.
References
- 1.Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11:1156–1566. doi: 10.1101/gr.GR-1649R. [DOI] [PubMed] [Google Scholar]
- 2.ter Beek J, Guskov A, Slotboom DJ. Structural diversity of ABC transporters. J Gen Physiol. 2014;143:419–435. doi: 10.1085/jgp.201411164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 4.Locher KP. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat Struct Mol Biol. 2016;23:487–493. doi: 10.1038/nsmb.3216. [DOI] [PubMed] [Google Scholar]
- 5.Linsdell P. Mechanism of chloride permeation in the cystic fibrosis transmembrane conductance regulator chloride channel. Exp Physiol. 2006;91:123–129. doi: 10.1113/expphysiol.2005.031757. [DOI] [PubMed] [Google Scholar]
- 6.McCarty NA. Permeation through the CFTR chloride channel. J Exp Biol. 2000;203:1947–1962. doi: 10.1242/jeb.203.13.1947. [DOI] [PubMed] [Google Scholar]
- 7.Muallem D, Vergani P. ATP hydrolysis-driven gating in cystic fibrosis transmembrane conductance regulator. Philos Trans R Soc Lond B Biol Sci. 2009;364:247–255. doi: 10.1098/rstb.2008.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang TC, Sheppard DN. Gating of the CFTR Cl-channel by ATP-driven nucleotide-binding domain dimerisation. J Physiol. 2009;587:2151–2161. doi: 10.1113/jphysiol.2009.171595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winter MC, Welsh MJ. Stimulation of CFTR activity by its phosphorylated R domain. Nature. 1997;389:294–296. doi: 10.1038/38514. [DOI] [PubMed] [Google Scholar]
- 10.Liang X, Da Paula AC, Bozóky Z, Zhang H, Bertrand CA, Peters KW, et al. Phosphorylation-dependent 14-3-3 protein interactions regulate CFTR biogenesis. Mol Biol Cell. 2012;23:996–1009. doi: 10.1091/mbc.E11-08-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bozoky Z, Krzeminski M, Chong P, Forman-Kay JD. Structural changes of CFTR R region upon phosphorylation: a plastic platform for intramolecular and intermolecular interactions. FEBS J. 2013;280:4407–4416. doi: 10.1111/febs.12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chappe V, Irvine T, Liao J, Evagelidis A, Hanrahan JW. Phosphorylation of CFTR by PKA promotes binding of the regulatory domain. EMBO J. 2005;24:2730–2740. doi: 10.1038/sj.emboj.7600747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rowe SM, Miller S, Sorscher EJ. Cystic fibrosis. N Engl J Med. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Wrennall JA, Cai Z, Li H, Sheppard DN. Understanding how cystic fibrosis mutations disrupt CFTR function: from single molecules to animal models. Int J Biochem Cell Biol. 2014;52:47–57. doi: 10.1016/j.biocel.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Veit G, Avramescu RG, Chiang AN, Houck SA, Cai Z, Peters KW, et al. From CFTR biology toward combinatorial pharmacotherapy: expanded classification of cystic fibrosis mutations. Mol Biol Cell. 2016;27:424–433. doi: 10.1091/mbc.E14-04-0935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cutting GR. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet. 2015;16:45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunt JF, Wang C, Ford RC. Cystic fibrosis transmembrane conductance regulator (ABCC7) structure. Cold Spring Harb Perspect Med. 2013;3:a009514. doi: 10.1101/cshperspect.a009514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bozoky Z, Krzeminski M, Muhandiram R, Birtley JR, Al-Zahrani A, Thomas PJ, et al. Regulatory R region of the CFTR chloride channel is a dynamic integrator of phospho-dependent intra- and intermolecular interactions. Proc Natl Acad Sci USA. 2013;110:E4427–E4436. doi: 10.1073/pnas.1315104110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Aleksandrov LA, Riordan JR, Ford RC. Domain location within the cystic fibrosis transmembrane conductance regulator protein investigated by electron microscopy and gold labelling. Biochim Biophys Acta. 2011;1808:399–404. doi: 10.1016/j.bbamem.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Aleksandrov LA, Z Z, Birtley JR, Riordan JR, Ford RC (2009) Architecture of the cystic fibrosis transmembrane conductance regulator protein and structural changes associated with phosphorylation and nucleotide binding. J Struct Biol 167:242–251 [DOI] [PubMed]
- 21.Rosenberg MF, O’Ryan LP, Hughes G, Zhao Z, Aleksandrov LA, Riordan JR, et al. The cystic fibrosis transmembrane conductance regulator (CFTR): three-dimensional structure and localization of a channel gate. J Biol Chem. 2011;286:42647–42654. doi: 10.1074/jbc.M111.292268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg MF, Kamis AB, Aleksandrov LA, Ford RC, Riordan JR. Purification and crystallization of the cystic fibrosis transmembrane conductance regulator (CFTR) J Biol Chem. 2004;279:39051–39057. doi: 10.1074/jbc.M407434200. [DOI] [PubMed] [Google Scholar]
- 23.Mio K, Ogura T, Mio M, Shimizu H, Hwang TC, Sato C, et al. Three-dimensional reconstruction of human cystic fibrosis transmembrane conductance regulator chloride channel revealed an ellipsoidal structure with orifices beneath the putative transmembrane domain. J Biol Chem. 2008;283:30300–30310. doi: 10.1074/jbc.M803185200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Awayn NH, Rosenberg MF, Kamis AB, Aleksandrov LA, Riordan JR, Ford RC. Crystallographic and single-particle analyses of native- and nucleotide-bound forms of the cystic fibrosis transmembrane conductance regulator (CFTR) protein. Biochem Soc Trans. 2005;33:996–999. doi: 10.1042/BST0330996. [DOI] [PubMed] [Google Scholar]
- 25.Atwell S, Brouillette CG, Conners K, Emtage S, Gheyi T, Guggino WB, et al. Structures of a minimal human CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant. Protein Eng Des Sel. 2010;23:375–384. doi: 10.1093/protein/gzq004. [DOI] [PubMed] [Google Scholar]
- 26.Lewis HA, Buchanan SG, Burley SK, Conners K, Dickey M, Dorwart M, et al. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 2004;23:282–293. doi: 10.1038/sj.emboj.7600040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis HA, Wang C, Zhao X, Hamuro Y, Conners K, Kearins MC, et al. Structure and dynamics of NBD1 from CFTR characterized using crystallography and hydrogen/deuterium exchange mass spectrometry. J Mol Biol. 2010;396:406–430. doi: 10.1016/j.jmb.2009.11.051. [DOI] [PubMed] [Google Scholar]
- 28.Lewis HA, Zhao X, Wang C, Sauder JM, Rooney I, Noland BW, et al. Impact of the deltaF508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure. J Biol Chem. 2005;280:1346–1353. doi: 10.1074/jbc.M410968200. [DOI] [PubMed] [Google Scholar]
- 29.Thibodeau PH, Brautigam CA, Machius M, Thomas PJ. Side chain and backbone contributions of Phe508 to CFTR folding. Nat Struct Mol Biol. 2005;12:10–16. doi: 10.1038/nsmb881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanelis V, Hudson RP, Thibodeau PH, Thomas PJ, Forman-Kay JD. NMR evidence for differential phosphorylation-dependent interactions in WT and DeltaF508 CFTR. EMBO J. 2010;29:263–277. doi: 10.1038/emboj.2009.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hudson RP, Chong PA, Protasevich II, Vernon R, Noy E, Bihler H, et al. Conformational changes relevant to channel activity and folding within the first nucleotide binding domain of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 2012;287:28480–28494. doi: 10.1074/jbc.M112.371138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dawson JE, Farber PJ, Forman-Kay JD. Allosteric coupling between the intracellular coupling helix 4 and regulatory sites of the first nucleotide-binding domain of CFTR. PLoS One. 2013;8:e74347. doi: 10.1371/journal.pone.0074347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chong PA, Farber PJ, Vernon RM, Hudson RP, Mittermaier AK, Forman-Kay JD. Deletion of phenylalanine 508 in the first nucleotide-binding domain of the cystic fibrosis transmembrane conductance regulator increases conformational exchange and inhibits dimerization. J Biol Chem. 2015;290:22862–22878. doi: 10.1074/jbc.M115.641134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker JM, Hudson RP, Kanelis V, Choy WY, Thibodeau PH, Thomas PJ, et al. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat Struct Mol Biol. 2007;14:738–745. doi: 10.1038/nsmb1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galeno L, Galfrè E, Moran O. Small-angle X-ray scattering study of the ATP modulation of the structural features of the nucleotide binding domains of the CFTR in solution. Eur Biophys J. 2011;40:811–824. doi: 10.1007/s00249-011-0692-5. [DOI] [PubMed] [Google Scholar]
- 36.Galfrè E, Galeno L, Moran O. A potentiator induces conformational changes on the recombinant CFTR nucleotide binding domains in solution. Cell Mol Life Sci. 2012;69:3701–3713. doi: 10.1007/s00018-012-1049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marasini C, Galeno L, Moran O. A SAXS-based ensemble model of the native and phosphorylated regulatory domain of the CFTR. Cell Mol Life Sci. 2013;70:923–933. doi: 10.1007/s00018-012-1172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollock NL, Satriano L, Zegarra-Moran O, Ford RC, Moran O. Structure of wild type and mutant F508del CFTR: a small-angle X-ray scattering study of the protein-detergent complexes. J Struct Biol. 2016;194:102–111. doi: 10.1016/j.jsb.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Moran O. On the structural organization of the intracellular domains of CFTR. Int J Biochem Cell Biol. 2014;52:7–14. doi: 10.1016/j.biocel.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 40.Odolczyk N, Zielenkiewicz P. Molecular modelling approaches for cystic fibrosis transmembrane conductance regulator studies. Int J Biochem Cell Biol. 2014;52:39–46. doi: 10.1016/j.biocel.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 41.Belmonte L, Moran O. On the interactions between nucleotide binding domains and membrane spanning domains in cystic fibrosis transmembrane regulator: a molecular dynamic study. Biochimie. 2015;111:19–29. doi: 10.1016/j.biochi.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 42.Corradi V, Vergani P, Tieleman DP. Cystic fibrosis transmembrane conductance regulator (CFTR): closed and open state channel models. J Biol Chem. 2015;290:22891–22906. doi: 10.1074/jbc.M115.665125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mornon J-P, Hoffmann B, Jonic S, Lehn P, Callebaut I. Full-open and closed CFTR channels, with lateral tunnels from the cytoplasm and an alternative position of the F508 region, as revealed by molecular dynamics. Cell Mol Life Sci. 2015;72:1377–1403. doi: 10.1007/s00018-014-1749-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhenin M, Noy E, Senderowitz H. REMD simulations reveal the dynamic profile and mechanism of action of deleterious, rescuing, and stabilizing perturbations to NBD1 from CFTR. J Chem Inf Model. 2015;55:2349–2364. doi: 10.1021/acs.jcim.5b00312. [DOI] [PubMed] [Google Scholar]
- 45.Schmitt L, Tampé R. Structure and mechanism of ABC transporters. Curr Opin Struct Biol. 2002;12:754–760. doi: 10.1016/S0959-440X(02)00399-8. [DOI] [PubMed] [Google Scholar]
- 46.Callebaut I, Eudes R, Mornon J-P, Lehn P. Nucleotide-binding domains of human cystic fibrosis transmembrane conductance regulator: detailed sequence analysis and three-dimensional modeling of the heterodimer. Cell Mol Life Sci. 2004;61:230–242. doi: 10.1007/s00018-003-3386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eudes R, Lehn P, Férec C, Mornon J-P, Callebaut I. Nucleotide binding domains of human CFTR: a structural classification of critical residues and disease-causing mutations. Cell Mol Life Sci. 2005;62:2112–2123. doi: 10.1007/s00018-005-5224-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith PC, Karpowich N, Millen L, Moody JE, Rosen J, Thomas PJ, et al. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol Cell. 2002;10:139–149. doi: 10.1016/S1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kidd JF, Ramjeesingh M, Stratford F, Huan LJ, Bear CE. A heteromeric complex of the two nucleotide binding domains of cystic fibrosis transmembrane conductance regulator (CFTR) mediates ATPase activity. J Biol Chem. 2004;279:41664–41669. doi: 10.1074/jbc.M407666200. [DOI] [PubMed] [Google Scholar]
- 50.Mense M, Vergani P, White DM, Altberg G, Nairn AC, Gadsby DC. In vivo phosphorylation of CFTR promotes formation of a nucleotide-binding domain heterodimer. EMBO J. 2006;25:4728–4739. doi: 10.1038/sj.emboj.7601373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stratford FL, Ramjeesingh M, Cheung JC, Huan LJ, Bear CE. The Walker B motif of the second nucleotide-binding domain (NBD2) of CFTR plays a key role in ATPase activity by the NBD1–NBD2 heterodimer. Biochem J. 2007;401:581–586. doi: 10.1042/BJ20060968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vergani P, Lockless SW, Nairn AC, Gadsby DC. CFTR channel opening by ATP-driven tight dimerization of its nucleotide-binding domains. Nature. 2005;433:876–880. doi: 10.1038/nature03313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramjeesingh M, Li C, Garami E, Huan LJ, Galley K, Wang Y, et al. Walker mutations reveal loose relationship between catalytic and channel-gating activities of purified CFTR (cystic fibrosis transmembrane conductance regulator) Biochemistry. 1999;38:1463–1468. doi: 10.1021/bi982243y. [DOI] [PubMed] [Google Scholar]
- 54.Aleksandrov L, Aleksandrov AA, Chang XB, Riordan JR. The first nucleotide binding domain of cystic fibrosis transmembrane conductance regulator is a site of stable nucleotide interaction, whereas the second is a site of rapid turnover. J Biol Chem. 2002;277:15419–15425. doi: 10.1074/jbc.M111713200. [DOI] [PubMed] [Google Scholar]
- 55.Basso C, Vergani P, Nairn AC, Gadsby DC. Prolonged nonhydrolytic interaction of nucleotide with CFTR’s NH2-terminal nucleotide binding domain and its role in channel gating. J Gen Physiol. 2003;122:333–348. doi: 10.1085/jgp.200308798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou Z, Wang X, Liu HY, Zou X, Li M, Hwang TC. The two ATP binding sites of cystic fibrosis transmembrane conductance regulator (CFTR) play distinct roles in gating kinetics and energetics. J Gen Physiol. 2006;128:413–422. doi: 10.1085/jgp.200609622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Procko E, Ferrin-O’Connell I, Ng SL, Gaudet R. Distinct structural and functional properties of the ATPase sites in an asymmetric ABC transporter. Mol Cell. 2006;24:51–62. doi: 10.1016/j.molcel.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 58.Szollosi A, Muallem DR, Csanády L, Vergani P. Mutant cycles at CFTR’s non-canonical ATP-binding site support little interface separation during gating. J Gen Physiol. 2011;137:549–562. doi: 10.1085/jgp.201110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsai MF, Li M, Hwang TC. Stable ATP binding mediated by a partial NBD dimer of the CFTR chloride channel. J Gen Physiol. 2010;135:399–414. doi: 10.1085/jgp.201010399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Protasevich I, Yang Z, Wang C, Atwell S, Zhao X, Emtage S, et al. Thermal unfolding studies show the disease causing F508del mutation in CFTR thermodynamically destabilizes nucleotide-binding domain 1. Protein Sci. 2010;19:1917–1931. doi: 10.1002/pro.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang C, Protasevich I, Yang Z, Seehausen D, Skalak T, Zhao X, et al. Integrated biophysical studies implicate partial unfolding of NBD1 of CFTR in the molecular pathogenesis of F508del cystic fibrosis. Protein Sci. 2010;19:1932–1947. doi: 10.1002/pro.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aleksandrov AA, Kota P, Aleksandrov LA, He L, Jensen T, Cui L, et al. Regulatory insertion removal restores maturation, stability and function of DeltaF508 CFTR. J Mol Biol. 2010;401:194–210. doi: 10.1016/j.jmb.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moran O, Galietta LJ, Zegarra-Moran O. Binding site of activators of the cystic fibrosis transmembrane conductance regulator in the nucleotide binding domains. Cell Mol Life Sci. 2005;62:446–460. doi: 10.1007/s00018-004-4422-3. [DOI] [PubMed] [Google Scholar]
- 64.Huang S-Y, Bolser D, Liu H-Y, Hwang TC. Molecular modeling of the heterodimer of human CFTR’s nucleotide-binding domains using a protein–protein docking approach. J Mol Graph Model. 2009;27:822–828. doi: 10.1016/j.jmgm.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 66.Dawson RJ, Locher KP. Structure of the multidrug ABC transporter Sav 1866 from Staphylococcus aureus in complex with AMP–PNP. FEBS Lett. 2007;581:935–938. doi: 10.1016/j.febslet.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 67.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc Natl Acad Sci USA. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mornon J-P, Lehn P, Callebaut I. Atomic model of human cystic fibrosis transmembrane conductance regulator: membrane-spanning domains and coupling interfaces. Cell Mol Life Sci. 2008;65:2594–2612. doi: 10.1007/s00018-008-8249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mornon J-P, Lehn P, Callebaut I. Molecular models of the open and closed states of the whole human CFTR protein. Cell Mol Life Sci. 2009;66:3469–3486. doi: 10.1007/s00018-009-0133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Serohijos AW, Hegedus T, Aleksandrov AA, He L, Cui L, Dokholyan NV, et al. Phenylalanine-508 mediates a cytoplasmic-membrane domain contact in the CFTR 3D structure crucial to assembly and channel function. Proc Natl Acad Sci USA. 2008;105:3256–3261. doi: 10.1073/pnas.0800254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hegedűs T, Serohijos AW, Dokholyan NV, He L, Riordan JR. Computational studies reveal phosphorylation-dependent changes in the unstructured R domain of CFTR. J Mol Biol. 2008;378:1052–1063. doi: 10.1016/j.jmb.2008.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He L, Aleksandrov AA, Serohijos AW, Hegedus T, Aleksandrov LA, Cui L, et al. Multiple membrane-cytoplasmic domain contacts in the cystic fibrosis transmembrane conductance regulator (CFTR) mediate regulation of channel gating. J Biol Chem. 2008;283:26383–26390. doi: 10.1074/jbc.M803894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Higgins CF, Linton KJ. The ATP switch model for ABC transporters. Nat Struct Mol Biol. 2004;11:918–926. doi: 10.1038/nsmb836. [DOI] [PubMed] [Google Scholar]
- 74.Jones PM, George AM. A reciprocating twin-channel model for ABC transporters. Q Rev Biophys. 2014;47:189–220. doi: 10.1017/S0033583514000031. [DOI] [PubMed] [Google Scholar]
- 75.Linsdell P. Functional architecture of the CFTR chloride channel. Mol Membr Biol. 2014;31:1–16. doi: 10.3109/09687688.2013.868055. [DOI] [PubMed] [Google Scholar]
- 76.Cai Z, Scott-Ward TS, Sheppard DN. Voltage-dependent gating of the cystic fibrosis transmembrane conductance regulator Cl-channel. J Gen Physiol. 2003;122:605–620. doi: 10.1085/jgp.200308921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Da Paula AC, Sousa M, Xu Z, Dawson ES, Boyd AC, Sheppard DN, et al. Folding and rescue of a cystic fibrosis transmembrane conductance regulator trafficking mutant identified using human-murine chimeric proteins. J Biol Chem. 2010;285:27033–27044. doi: 10.1074/jbc.M110.120352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Norimatsu Y, Ivetac A, Alexander C, O’Donnell N, Frye L, Sansom MS, et al. Locating a plausible binding site for an open-channel blocker, GlyH-101, in the pore of the cystic fibrosis transmembrane conductance regulator. Mol Pharmacol. 2012;82:1042–1055. doi: 10.1124/mol.112.080267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alexander C, Ivetac A, Liu X, Norimatsu Y, Serrano JR, Landstrom A, et al. Cystic fibrosis transmembrane conductance regulator: using differential reactivity toward channel-permeant and channel-impermeant thiol-reactive probes to test a molecular model for the pore. Biochemistry. 2009;48:10078–10088. doi: 10.1021/bi901314c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dalton J, Kalid O, Schushan M, Ben-Tal N, Villà-Freixa J. New model of cystic fibrosis transmembrane conductance regulator proposes active channel-like conformation. J Chem Inf Model. 2012;52:1842–1853. doi: 10.1021/ci2005884. [DOI] [PubMed] [Google Scholar]
- 81.Chen EY, Bartlett MC, Loo TW, Clarke DM. The DeltaF508 mutation disrupts packing of the transmembrane segments of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 2004;279:39620–39627. doi: 10.1074/jbc.M407887200. [DOI] [PubMed] [Google Scholar]
- 82.Gao X, Hwang TC. Spatial positioning of CFTR’s pore-lining residues affirms an asymmetrical contribution of transmembrane segments to the anion permeation pathway. J Gen Physiol. 2016;147:407–422. doi: 10.1085/jgp.201511557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang W, El Hiani Y, Rubaiy HN, Linsdell P. Relative contribution of different transmembrane segments to the CFTR chloride channel pore. Pflugers Arch. 2014;466:477–490. doi: 10.1007/s00424-013-1317-x. [DOI] [PubMed] [Google Scholar]
- 84.Wang W, El Hiani Y, Linsdell P. Alignment of transmembrane regions in the cystic fibrosis transmembrane conductance regulator chloride channel pore. J Gen Physiol. 2011;138:165–178. doi: 10.1085/jgp.201110605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Quian F, El Hiani Y, Linsdell P. Functional arrangement of the 12th transmembrane region in the CFTR chloride channel pore based on functional investigation of a cysteine-less CFTR variant. Eur J Physiol. 2011;462:559–571. doi: 10.1007/s00424-011-0998-2. [DOI] [PubMed] [Google Scholar]
- 86.McCarty NA, Zhang ZR. Identification of a region of strong discrimination in the pore of CFTR. Am J Physiol Lung Cell Mol Physiol. 2001;281:L852–L867. doi: 10.1152/ajplung.2001.281.4.L852. [DOI] [PubMed] [Google Scholar]
- 87.Gupta J, Evagelidis A, Hanrahan JW, Linsdell P. Asymmetric structure of the cystic fibrosis transmembrane conductance regulator chloride channel pore suggested by mutagenesis of the twelfth transmembrane region. Biochemistry. 2001;40:6620–6627. doi: 10.1021/bi002819v. [DOI] [PubMed] [Google Scholar]
- 88.Ge N, Muise CN, Gong X, Linsdell P. Direct comparison of the functional roles played by different transmembrane regions in the cystic fibrosis transmembrane conductance regulator chloride channel pore. J Biol Chem. 2004;279:55283–55289. doi: 10.1074/jbc.M411935200. [DOI] [PubMed] [Google Scholar]
- 89.Gao X, Bai Y, Hwang TC. Cysteine scanning of CFTR’s first transmembrane segment reveals its plausible roles in gating and permeation. Biophys J. 2013;104:786–797. doi: 10.1016/j.bpj.2012.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fatehi M, Linsdell P. Novel residues lining the CFTR chloride channel pore identified by functional modification of introduced cysteines. J Membr Biol. 2009;228:151–164. doi: 10.1007/s00232-009-9167-3. [DOI] [PubMed] [Google Scholar]
- 91.Cui G, Song B, Turki HW, McCarty NA. Differential contribution of TM6 and TM12 to the pore of CFTR identified by three sulfonylurea-based blockers. Pflugers Arch. 2012;463:405–418. doi: 10.1007/s00424-011-1035-1. [DOI] [PubMed] [Google Scholar]
- 92.Cheung M, Akabas MH. Identification of cystic fibrosis transmembrane conductance regulator channel-lining residues in and flanking the M6 membrane-spanning segment. Biophys J. 1996;70:2688–2699. doi: 10.1016/S0006-3495(96)79838-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bai Y, Li M, Hwang TC. Structural basis for the channel function of a degraded ABC transporter, CFTR (ABCC7) J Gen Physiol. 2011;138:495–507. doi: 10.1085/jgp.201110705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bai Y, Li M, Hwang TC. Dual roles of the sixth transmembrane segment of the CFTR chloride channel in gating and permeation. J Gen Physiol. 2010;136:293–309. doi: 10.1085/jgp.201010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Akabas MH, Kaufmann C, Cook TA, Archdeacon P. Amino acid residues lining the chloride channel of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1994;269:14865–14868. [PubMed] [Google Scholar]
- 96.Beck EJ, Yang Y, Yaemsiri S, Raghuram V. Conformational changes in a pore-lining helix coupled to cystic fibrosis transmembrane conductance regulator channel gating. J Biol Chem. 2008;283:4957–4966. doi: 10.1074/jbc.M702235200. [DOI] [PubMed] [Google Scholar]
- 97.El Hiani Y, Linsdell P. Changes in accessibility of cytoplasmic substances to the pore associated with activation of the cystic fibrosis transmembrane conductance regulator chloride channel. J Biol Chem. 2010;285:32126–32140. doi: 10.1074/jbc.M110.113332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qian F, Liu L, Liu Z, Lu C. The pore architecture of the cystic fibrosis transmembrane conductance regulator channel revealed by co-mutation in pore-forming transmembrane regions. Physiol Res. 2016;65:505–515. doi: 10.33549/physiolres.933143. [DOI] [PubMed] [Google Scholar]
- 99.Cui G, Freeman CS, Knotts T, Prince CZ, Kuang C, McCarty NA. Two salts bridges differentially contribute to the maintenance of cystic fibrosis transmembrane conductance regulator (CFTR) channel function. J Biol Chem. 2013;288:20758–20767. doi: 10.1074/jbc.M113.476226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rahman KS, Cui G, Harvey SC, McCarty NA. Modeling the conformational changes underlying channel opening in CFTR. PLoS One. 2013;8:e74574. doi: 10.1371/journal.pone.0074574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cui G, Khazanov N, Stauffer B, Infield DT, Imhoff BR, Senderowitz H, et al. Potentiators exert distinct effects on human, murine, and Xenopus CFTR. Am J Physiol Lung Cell Mol Physiol. 2016;311:L192–L207. doi: 10.1152/ajplung.00056.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Infield DT, Cui G, Kuang C, McCarty NA. Positioning of extracellular loop 1 affects pore gating of the cystic fibrosis transmembrane conductance regulator. Am J Physiol Lung Cell Mol Physiol. 2016;310:L403–L414. doi: 10.1152/ajplung.00259.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Billet A, Mornon J-P, Jollivet M, Lehn P, Callebaut I, Becq F. CFTR: effect of ICL2 and ICL4 amino acids in close spatial proximity on the current properties of the channel. J Cyst Fibros. 2013;12:737–745. doi: 10.1016/j.jcf.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 104.Wang W, Roessler BC, Kirk KL. An electrostatic interaction at the tetrahelix bundle promotes phosphorylation-dependent cystic fibrosis transmembrane conductance regulator (CFTR) channel opening. J Biol Chem. 2014;289:30364–30378. doi: 10.1074/jbc.M114.595710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Linsdell P. Anion conductance selectivity mechanism of the CFTR chloride channel. Biochim Biophys Acta. 2016;1858:740–747. doi: 10.1016/j.bbamem.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 106.Gao X, Hwang TC. Localizing a gate in CFTR. Proc Natl Acad Sci USA. 2015;112:2461–2466. doi: 10.1073/pnas.1420676112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wei S, Roessler BC, Icyuz M, Chauvet S, Tao B, Hartman JLT, et al. Long-range coupling between the extracellular gates and the intracellular ATP binding domains of multidrug resistance protein pumps and cystic fibrosis transmembrane conductance regulator channels. FASEB J. 2016;30:1247–1262. doi: 10.1096/fj.15-278382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wei S, Roessler BC, Chauvet S, Guo J, Hartman JL, Kirk KL. Conserved allosteric hot spots in the transmembrane domains of cystic fibrosis transmembrane conductance regulator (CFTR) channels and multidrug resistance protein (MRP) pumps. J Biol Chem. 2014;289:19942–19957. doi: 10.1074/jbc.M114.562116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mihályi C, Töröcsik B, Csanády L. Obligate coupling of CFTR pore opening to tight nucleotide-binding domain dimerization. Elife. 2016;5:e18164. doi: 10.7554/eLife.18164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gadsby DC. Ion channels versus ion pumps: the principal difference, in principle. Nat Rev Mol Cell Biol. 2009;10:344–352. doi: 10.1038/nrm2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Samways DS, Khakh BS, Dutertre S, Egan TM. Preferential use of unobstructed lateral portals as the access route to the pore of human ATP-gated ion channels (P2X receptors) Proc Natl Acad Sci USA. 2011;108:13800–13805. doi: 10.1073/pnas.1017550108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.El Hiani Y, Linsdell P. Functional architecture of the cytoplasmic entrance to the cystic fibrosis transmembrane conductance regulator chloride channel pore. J Biol Chem. 2015;290:15855–15865. doi: 10.1074/jbc.M115.656181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.El Hiani Y, Negoda A, Linsdell P. Cytoplasmic pathway followed by chloride ions to enter the CFTR channel pore. Cell Mol Life Sci. 2016;73:1917–1925. doi: 10.1007/s00018-015-2113-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Furukawa-Hagiya T, Furuta T, Chiba S, Sohma Y, Sakurai M. The power stroke driven by ATP binding in CFTR as studied by molecular dynamics simulations. J Phys Chem B. 2013;117:83–93. doi: 10.1021/jp308315w. [DOI] [PubMed] [Google Scholar]
- 116.Choudhury HG, Tong Z, Mathavan I, Li Y, Iwata S, Zirah S, et al. Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc Natl Acad Sci USA. 2014;111:9145–9150. doi: 10.1073/pnas.1320506111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lin DY, Huang S, Chen J. Crystal structures of a polypeptide processing and secretion transporter. Nature. 2015;523:425–430. doi: 10.1038/nature14623. [DOI] [PubMed] [Google Scholar]
- 118.Kim J, Wu S, Tomasiak TM, Mergel C, Winter MB, Stiller SB, et al. Subnanometre-resolution electron cryomicroscopy structure of a heterodimeric ABC exporter. Nature. 2015;517:396–400. doi: 10.1038/nature13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, et al. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ward AB, Szewczyk P, Grimard V, Lee CW, Martinez L, Doshi R, et al. Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain. Proc Natl Acad Sci USA. 2013;110:13386–13391. doi: 10.1073/pnas.1309275110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li J, Jaimes KF, Aller SG. Refined structures of mouse P-glycoprotein. Protein Sci. 2014;23:34–46. doi: 10.1002/pro.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jin MS, Oldham ML, Zhang Q, Chen J. Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans . Nature. 2012;490:566–569. doi: 10.1038/nature11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kodan A, Yamaguchi T, Nakatsu T, Sakiyama K, Hipolito CJ, Fujioka A, Hirokane R, Ikeguchi K, Watanabe B, Hiratake J, Kimura Y, Suga H, Ueda K, Kato H. Structural basis for gating mechanisms of a eukaryotic P-glycoprotein homolog. Proc Natl Acad Sci. 2014;111(11):4049–4054. doi: 10.1073/pnas.1321562111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Srinivasan V, Pierik AJ, Lill R. Crystal structures of nucleotide-free and glutathione-bound mitochondrial ABC transporter Atm1. Science. 2014;343:1137–1140. doi: 10.1126/science.1246729. [DOI] [PubMed] [Google Scholar]
- 125.Lee JY, Yang JG, Zhitnitsky D, Lewinson O, Rees DC. Structural basis for heavy metal detoxification by an Atm1-type ABC exporter. Science. 2014;343:1133–1136. doi: 10.1126/science.1246489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shintre CA, Pike AC, Li Q, Kim JI, Barr A, Goubin S, et al. Structures of ABCB10, a human ATP-binding cassette transporter in apo- and nucleotide-bound states. Proc Natl Acad Sci USA. 2013;110:9710–9715. doi: 10.1073/pnas.1217042110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Perez C, Gerber S, Boilevin J, Bucher M, Darbre T, Aebi M, et al. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature. 2015;524:433–438. doi: 10.1038/nature14953. [DOI] [PubMed] [Google Scholar]
- 128.Hohl M, Briand C, Grütter MG, Seeger MA. Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation. Nat Struct Mol Biol. 2012;19:395–402. doi: 10.1038/nsmb.2267. [DOI] [PubMed] [Google Scholar]
- 129.Hohl M, Hürlimann LM, Böhm S, Schöppe J, Grütter MG, Bordignon E, et al. Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter. Proc Natl Acad Sci USA. 2014;111:11025–11030. doi: 10.1073/pnas.1400485111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jones PM, George AM. Role of the D-loops in allosteric control of ATP hydrolysis in an ABC transporter. J Phys Chem A. 2012;116:3004–3013. doi: 10.1021/jp211139s. [DOI] [PubMed] [Google Scholar]
- 131.Liu J, Cami-Kobeci G, Wang Y, Kuituan P, Cai Z, Li H et al (2014) The therapeutic potential of small-molecule modulators of the cystic fibrosis transmembrane conductance regulator (CFTR) Cl-channel. In: Cox B, Gosling M (eds) Ion channel drug discovery, Royal Society of Chemistry, pp 156–185
- 132.Farinha CM, Matos P. Repairing the basic defect in cystic fibrosis—one approach is not enough. FEBS J. 2016;283:246–264. doi: 10.1111/febs.13531. [DOI] [PubMed] [Google Scholar]
- 133.Lukacs GL, Verkman AS. CFTR: folding, misfolding and correcting the DeltaF508 conformational defect. Trends Mol Med. 2012;18:81–91. doi: 10.1016/j.molmed.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lazrak A, Fu L, Bal IV, Bartoszewski R, Rab A, Havasi V et al (2013) The silent codon change I507-ATC → ATT contributes to the severity of the ΔF508 CFTR channel dysfunction. FASEB J 27:4630–4645 [DOI] [PMC free article] [PubMed]
- 135.Thibodeau PH, Richardson JMr, Wang W, Millen L, Watson J, Mendoza JL et al (2010) The cystic fibrosis-causing mutation deltaF508 affects multiple steps in cystic fibrosis transmembrane conductance regulator biogenesis. J Biol Chem 285:35825–35835 [DOI] [PMC free article] [PubMed]
- 136.Rabeh WM, Bossard F, Xu H, Okiyoneda T, Bagdany M, Mulvihill CM, et al. Correction of both NBD1 energetics and domain interface is required to restore ΔF508 CFTR folding and function. Cell. 2012;148:150–163. doi: 10.1016/j.cell.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mendoza JL, Schmidt A, Li Q, Nuvaga E, Barrett T, Bridges RJ, et al. Requirements for efficient correction of DF508 CFTR revealed by analyses of evolved sequences. Cell. 2012;148:164–174. doi: 10.1016/j.cell.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rosser MF, Grove DE, Chen L, Cyr DM. Assembly and misassembly of cystic fibrosis transmembrane conductance regulator: folding defects caused by deletion of F508 occur before and after the calnexin-dependent association of membrane spanning domain (MSD) 1 and MSD2. Mol Biol Cell. 2008;19:4570–4579. doi: 10.1091/mbc.E08-04-0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cui L, Aleksandrov L, Chang XB, Hou YX, He L, Hegedus T, et al. Domain interdependence in the biosynthetic assembly of CFTR. J Mol Biol. 2007;365:981–994. doi: 10.1016/j.jmb.2006.10.086. [DOI] [PubMed] [Google Scholar]
- 140.He L, Aleksandrov LA, Cui L, Jensen TJ, Nesbitt KL, Riordan JR. Restoration of domain folding and interdomain assembly by second-site suppressors of the DeltaF508 mutation in CFTR. FASEB J. 2010;24:3103–3112. doi: 10.1096/fj.09-141788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Du K, Sharma M, Lukacs GL. The DeltaF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nat Struct Mol Biol. 2005;12:17–25. doi: 10.1038/nsmb882. [DOI] [PubMed] [Google Scholar]
- 142.He L, Aleksandrov AA, An J, Cui L, Yang Z, Brouillette CG, et al. Restoration of NBD1 thermal stability is necessary and sufficient to correct DF508 CFTR folding and assembly. J Mol Biol. 2015;427:106–120. doi: 10.1016/j.jmb.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.He L, Kota P, Aleksandrov AA, Cui L, Jensen T, Dokholyan NV, et al. Correctors of ΔF508 CFTR restore global conformational maturation without thermally stabilizing the mutant protein. FASEB J. 2013;27:536–545. doi: 10.1096/fj.12-216119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Warner DJ, Vadolia MM, Laughton CA, Kerr ID, Doughty SW. Modelling the restoration of wild-type dynamic behaviour in DeltaF508-CFTR NBD1 by 8-cyclopentyl-1,3-dipropylxanthine. J Mol Graph Model. 2007;26:691–699. doi: 10.1016/j.jmgm.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 145.Wieczorek G, Zielenkiewicz P. DeltaF508 mutation increases conformational flexibility of CFTR protein. J Cyst Fibros. 2008;7:295–300. doi: 10.1016/j.jcf.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 146.Bisignano P, Moran O. Molecular dynamics analysis of the wild type and dF508 mutant structures of the human CFTR-nucleotide binding domain 1. Biochimie. 2010;92:51–57. doi: 10.1016/j.biochi.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 147.Serohijos AW, Hegedus T, Riordan JR, Dokholyan NV. Diminished self-chaperoning activity of the DeltaF508 mutant of CFTR results in protein misfolding. PLoS Comput Biol. 2008;4:e1000008. doi: 10.1371/journal.pcbi.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Proctor EA, Kota P, Aleksandrov AA, He L, Riordan JR, Dokholyan NV. Rational coupled dynamics network manipulation rescues disease-relevant mutant cystic fibrosis transmembrane conductance regulator. Chem Sci. 2015;6:1237–1246. doi: 10.1039/C4SC01320D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Loo TW, Barlett MC, Clarke DM. The V510D suppressor mutation stabilizes DeltaF508-CFTR at the cell surface. Biochemistry. 2010;49:6352–6357. doi: 10.1021/bi100807h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Aleksandrov AA, Kota P, Cui L, Jensen T, Alekseev AE, Reyes S, et al. Allosteric modulation balances thermodynamic stability and restores function of DF508 CFTR. J Mol Biol. 2012;419:41–60. doi: 10.1016/j.jmb.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ramsey BW, Davies J, McElvaney NG, Tullis E, Bell SC, Dřevínek P, et al. A CFTR potentiator in patients with cystic fibrosis and the G551D mutation. N Engl J Med. 2011;365:1663–1672. doi: 10.1056/NEJMoa1105185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lin WY, Jih KY, Hwang TC. A single amino acid substitution in CFTR converts ATP to an inhibitory ligand. J Gen Physiol. 2014;144(4):311–320. doi: 10.1085/jgp.201411247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Eckford PD, Ramjeesingh M, Molinski S, Pasyk S, Dekkers JF, Li C, et al. VX-809 and related corrector compounds exhibit secondary activity stabilizing active F508del-CFTR after its partial rescue to the cell surface. Chem Biol. 2014;21:666–678. doi: 10.1016/j.chembiol.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 154.Sampson HM, Robert R, Liao J, Matthes E, Carlile GW, Hanrahan JW, et al. Identification of a NBD1-binding pharmacological chaperone that corrects the trafficking defect of F508del-CFTR. Chem Biol. 2011;18:231–242. doi: 10.1016/j.chembiol.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 155.Eckford PD, Li C, Ramjeesingh M, Bear CE. Cystic fibrosis transmembrane conductance regulator (CFTR) potentiator VX-770 (ivacaftor) opens the defective channel gate of mutant CFTR in a phosphorylation-dependent but ATP-independent manner. J Biol Chem. 2012;287:36639–36649. doi: 10.1074/jbc.M112.393637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jih KY, Hwang TC. Vx-770 potentiates CFTR function by promoting decoupling between the gating cycle and ATP hydrolysis cycle. Proc Natl Acad Sci USA. 2013;110:4404–4409. doi: 10.1073/pnas.1215982110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Van Goor F, Hadida S, Grootenhuis PD, Burton B, Cao D, Neuberger T, et al. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc Natl Acad Sci USA. 2009;106:18825–18830. doi: 10.1073/pnas.0904709106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yu H, Burton B, Huang CJ, Worley J, Cao D, Johnson JPJ, et al. Ivacaftor potentiation of multiple CFTR channels with gating mutations. J Cyst Fibros. 2012;11:237–245. doi: 10.1016/j.jcf.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 159.Yeh HI, Yeh JT, Hwang TC. Modulation of CFTR gating by permeant ions. J Gen Physiol. 2015;145:47–60. doi: 10.1085/jgp.201411272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Linsdell P. State-dependent blocker interactions with the CFTR chloride channel: implications for gating the pore. Pflugers Arch. 2014;466:2243–2255. doi: 10.1007/s00424-014-1501-7. [DOI] [PubMed] [Google Scholar]
- 161.Csanády L, Töröcsik B. Catalyst-like modulation of transition states for CFTR channel opening and closing: new stimulation strategy exploits nonequilibrium gating. J Gen Physiol. 2014;143:269–287. doi: 10.1085/jgp.201311089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Csanády L, Töröcsik B. Structure-activity analysis of a CFTR channel potentiator: distinct molecular parts underlie dual gating effects. J Gen Physiol. 2014;144:321–336. doi: 10.1085/jgp.201411246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kalid O, Mense M, Fischman S, Shitrit A, Bihler H, Ben-Zeev E, et al. Small molecule correctors of F508del-CFTR discovered by structure-based virtual screening. J Comput Aided Mol Des. 2010;24:971–991. doi: 10.1007/s10822-010-9390-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Odolczyk N, Fritsch J, Norez C, Servel N, da Cunha MF, Bitam S, et al. Discovery of novel potent ΔF508-CFTR correctors that target the nucleotide binding domain. EMBO Mol Med. 2013;5:1484–1501. doi: 10.1002/emmm.201302699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wainwright CE, Elborn JS, Ramsey BW, Marigowda G, Huang X, Cipolli M, et al. Lumacaftor–ivacaftor in patients with cystic fibrosis homozygous for Phe508del CFTR. N Engl J Med. 2015;373:220–231. doi: 10.1056/NEJMoa1409547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Elborn JS, Ramsey BW, Boyle MP, Konstan MW, Huang X, Marigowda G, et al. Efficacy and safety of lumacaftor/ivacaftor combination therapy in patients with cystic fibrosis homozygous for Phe508del CFTR by pulmonary function subgroup: a pooled analysis. Lancet Respir Med. 2016;4:617–626. doi: 10.1016/S2213-2600(16)30121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Farinha CM, King-Underwood J, Sousa M, Correia AR, Henriques BJ, Roxo-Rosa M, et al. Revertants, low temperature, and correctors reveal the mechanism of F508del-CFTR rescue by VX-809 and suggest multiple agents for full correction. Chem Biol. 2013;20:943–955. doi: 10.1016/j.chembiol.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 168.Okiyoneda T, Veit G, Dekkers JF, Bagdany M, Soya N, Xu H, et al. Mechanism-based corrector combination restores DF508-CFTR folding and function. Nat Chem Biol. 2013;9:444–454. doi: 10.1038/nchembio.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ren HY, Grove DE, Houck SA, Sopha P, van Goor F, Hoffman BJ, et al. VX-809 corrects folding defects in cystic fibrosis transmembrane conductance regulator protein through action on membrane-spanning domain 1. Mol Biol Cell. 2013;24:3016–3024. doi: 10.1091/mbc.E13-05-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Loo TW, Bartlett MC, Clarke DM. Corrector VX-809 stabilizes the first transmembrane domain of CFTR. Biochem Pharmacol. 2013;86:612–619. doi: 10.1016/j.bcp.2013.06.028. [DOI] [PubMed] [Google Scholar]
- 171.Veit G, Avramescu RG, Perdomo D, Phuan PW, Bagdany M, Apaja PM, Borot F, Szollosi D, Wu YS, Finkbeiner WE, Hegedus T, Verkman AS, Lukacs GL. Some gating potentiators, including VX-770, diminish ΔF508-CFTR functional expression. Sci Transl Med. 2014;6(246):246ra97. doi: 10.1126/scitranslmed.3008889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cholon DM, Quinney NL, Fulcher ML, Esther CR, Jr, Das J, Dokholyan NV, Randell SH, Boucher RC, Gentzsch M. Some gating potentiators, including VX-770, diminish ΔF508-CFTR functional expression. Sci Transl Med. 2014;6(246):246ra97. doi: 10.1126/scitranslmed.3008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Phuan PW, Veit G, Tan JA, Finkbeiner WE, Lukacs GL, Verkman AS. Potentiators of defective ΔF508-CFTR gating that do not interfere with corrector action. Mol Pharmacol. 2015;88:791–799. doi: 10.1124/mol.115.099689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Phuan PW, Veit G, Tan J, Roldan A, Finkbeiner WE, Lukacs GL, et al. Synergy-based small-molecule screen using a human lung epithelial cell line yields ΔF508-CFTR correctors that augment VX-809 maximal efficacy. Mol Pharmacol. 2014;86:42–51. doi: 10.1124/mol.114.092478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Pesce E, Bellotti M, Liessi N, Guariento S, Damonte G, Cichero E, et al. Synthesis and structure-activity relationship of aminoarylthiazole derivatives as correctors of the chloride transport defect in cystic fibrosis. Eur J Med Chem. 2015;99:14–35. doi: 10.1016/j.ejmech.2015.05.030. [DOI] [PubMed] [Google Scholar]
- 176.Pedemonte N, Lukacs GL, Du K, Caci E, Zegarra-Moran O, Galietta LJ, et al. Small-molecule correctors of defective DeltaF508-CFTR cellular processing identified by high-throughput screening. J Clin Invest. 2005;115:2564–2571. doi: 10.1172/JCI24898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Van Goor F, Hadida S, Grootenhuis PD, Burton B, Stack JH, Straley KS, et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc Natl Acad Sci USA. 2011;108:18843–18848. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Loo TW, Bartlett MC, Clarke DM. Bithiazole correctors rescue CFTR mutants by two different mechanisms. Biochemistry. 2013;52:5161–5163. doi: 10.1021/bi4008758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wang Y, Loo TW, Bartlett MC, Clarke DM. Correctors promote maturation of cystic fibrosis transmembrane conductance regulator (CFTR)-processing mutants by binding to the protein. J Biol Chem. 2007;282:33247–33251. doi: 10.1074/jbc.C700175200. [DOI] [PubMed] [Google Scholar]
- 180.Grove DE, Rosser MF, Ren HY, Naren AP, Cyr DM. Mechanisms for rescue of correctable folding defects in CFTRDelta F508. Mol Biol Cell. 2009;20:4059–4069. doi: 10.1091/mbc.E08-09-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Roberts KE, Cushing PR, Boisguerin P, Madden DR, Donald BR. Computational design of a PDZ domain peptide inhibitor that rescues CFTR activity. PLoS Comput Biol. 2012;8:e1002477. doi: 10.1371/journal.pcbi.1002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hall JD, Wang H, Byrnes LJ, Shanker S, Wang K, Efremov IV, et al. Binding screen for cystic fibrosis transmembrane conductance regulator correctors finds new chemical matter and yields insights into cystic fibrosis therapeutic strategy. Protein Sci. 2016;25:360–373. doi: 10.1002/pro.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Faure G, Bakouh N, Lourdel S, Odolczyk N, Premchandar A, Servel N, et al. Rattlesnake phospholipase A2 increases CFTR-chloride channel current and corrects ∆F508CFTR dysfunction: impact in cystic fibrosis. J Mol Biol. 2016;428:2898–2915. doi: 10.1016/j.jmb.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 184.Thiagarajah JR, Ko EA, Tradtrantip L, Donowitz M, Verkman AS. Discovery and development of antisecretory drugs for treating diarrheal diseases. Clin Gastroenterol Hepatol. 2014;12:204–209. doi: 10.1016/j.cgh.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Li H, Sheppard D. Therapeutic potential of cystic fibrosis transmembrane conductance regulator (CFTR) inhibitors in polycystic kidney disease. BioDrugs. 2009;23:203–216. doi: 10.2165/11313570-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 186.Linsdell P. Cystic fibrosis transmembrane conductance regulator chloride channel blockers: pharmacological, biophysical and physiological relevance. World J Biol Chem. 2014;5:26–39. doi: 10.4331/wjbc.v5.i1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kim Y, Anderson MO, Park J, Lee MG, Namkung W, Verkman AS. Benzopyrimido-pyrrolo-oxazine-dione (R)-BPO-27 inhibits CFTR chloride channel gating by competition with ATP. Mol Pharmacol. 2015;88:689–696. doi: 10.1124/mol.115.098368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Linsdell P. Location of a common inhibitor binding site in the cytoplasmic vestibule of the cystic fibrosis transmembrane conductance regulator chloride channel pore. J Biol Chem. 2005;280:8945–8950. doi: 10.1074/jbc.M414354200. [DOI] [PubMed] [Google Scholar]
- 189.St Aubin CN, Zhou JJ, Linsdell P. Identification of a second blocker binding site at the cytoplasmic mouth of the cystic fibrosis transmembrane conductance regulator chloride channel pore. Mol Pharmacol. 2007;71:1360–1368. doi: 10.1124/mol.106.031732. [DOI] [PubMed] [Google Scholar]
- 190.Kopeikin Z, Sohma Y, Li M, Hwang TC. On the mechanism of CFTR inhibition by a thiazolidinone derivative. J Gen Physiol. 2010;136:659–671. doi: 10.1085/jgp.201010518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Caci E, Caputo A, Hinzpeter A, Arous N, Fanen P, Sonawane N, et al. Evidence for direct CFTR inhibition by CFTRinh-172 based on Arg347 mutagenesis. Biochem J. 2008;413:135–142. doi: 10.1042/BJ20080029. [DOI] [PubMed] [Google Scholar]
- 192.Stahl M, Stah lK, Brubacher MB, Forrest JNJ (2012) Divergent CFTR orthologs respond differently to the channel inhibitors CFTRinh-172, glibenclamide, and GlyH-101. Am J Physiol Cell Physiol 302:C67–C76 [DOI] [PMC free article] [PubMed]
- 193.Noy E, Senderowitz H. Combating cystic fibrosis: in search for CF transmembrane conductance regulator (CFTR) modulators. ChemMedChem. 2011;6:243–251. doi: 10.1002/cmdc.201000488. [DOI] [PubMed] [Google Scholar]
- 194.Kleizen B, van Vlijmen T, de Jonge HR, Braakman I. Folding of CFTR is predominantly cotranslational. Mol Cell. 2005;20:277–287. doi: 10.1016/j.molcel.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 195.Hoelen H, Kleizen B, Schmidt A, Richardson J, Charitou P, Thomas PJ, et al. The primary folding defect and rescue of ΔF508 CFTR emerge during translation of the mutant domain. PLoS One. 2010;5:e15458. doi: 10.1371/journal.pone.0015458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.McClure M, DeLucas LJ, Wilson L, Ray M, Rowe SM, Wu X, et al. Purification of CFTR for mass spectrometry analysis: identification of palmitoylation and other post-translational modifications. Protein Eng Des Sel. 2012;25:7–14. doi: 10.1093/protein/gzr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bai XC, McMullan G, Scheres SH. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci. 2015;40:49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 198.Nogales E, Scheres SH. Cryo-EM: a unique tool for the visualization of macromolecular complexity. Mol Cell. 2015;58:677–689. doi: 10.1016/j.molcel.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cant N, Pollock N, Ford RC (2014) CFTR structure and cystic fibrosis. Int J Biochem Cell Biol 52:15–25 [DOI] [PubMed]
- 200.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]