Abstract
Chemotaxis is the directed motility by means of which microbes sense chemical cues and relocate towards more favorable environments. Methyl-accepting chemotaxis proteins (MCPs) are the most common receptors in bacteria and archaea. They are arranged as trimers of dimers that, in turn, form hexagonal arrays in the cytoplasmic membrane or in the cytoplasm. Several different classes of MCPs have been identified according to their ligand binding region and membrane topology. MCPs have been further classified based on the length and sequence conservation of their cytoplasmic domains. Clusters of membrane-embedded MCPs often localize to the poles of the cell, whereas cytoplasmic MCPs can be targeted to the poles or distributed throughout the cell body. MCPs play an important role in cell survival, pathogenesis, and biodegradation. Bacterial adaptation to diverse environmental conditions promotes diversity among the MCPs. This review summarizes structure, classification, and structure–activity relationship of the known MCP receptors, with a brief overview of the signal transduction mechanisms in bacteria and archaea.
Keywords: Chemoreceptor, ligand-binding domain; Signaling domain; Sensory domain; Structure–activity relationship; Protein structure
Introduction
Chemotaxis is the directed motility that allows microbes to sense environmental cues and move towards factors that favor survival and away from toxic chemicals. Microbes have a diverse group of chemoreceptors that sense intracellular and environmental signals and relay them to the downstream signaling pathways in the cytoplasm [1, 2]. The methyl-accepting chemotaxis proteins (MCPs) are the predominant chemoreceptors in bacteria and archaea. They are involved in regulation of diverse aspects of cellular activities including biofilm formation [3], flagellum biosynthesis [1], degradation of xenobiotic compounds [4], encystment, fruiting body formation [1, 5–8], exopolysaccharide production [9], and production of toxins [10]. For example, an MCP is required for encystment of the photosynthetic bacterium, Rhodospirillum centenum, because this receptor senses harsh environmental conditions, where the cyst formation would allow survival [1]. Pseudomonas aeruginosa has membrane-bound MCP receptors which modulate biofilm formation by regulating the cellular cyclic dimeric guanosine monophosphate level [11]. MCPs play an important role in the pathogenicity of many bacteria including P. aeruginosa [12], Campylobacter jejuni [13], Cronobacter sakazakii [14], and Vibrio cholerae [15]. Some MCPs function as photoreceptors and direct bacterial movement in response to light (exemplified by the phototaxis of halophilic marine archaea Halobacterium salinarum [16, 17]).
Genes encoding MCPs were mainly found in genomes of motile microbes [18]. Currently, the InterPro database contains more than 102,346 MCP signaling domains (accession number IPR004089) [19]. Among them, 100,550 are from bacteria (76,295 in Proteobacteria, 20,335 in Terrabacteria, 2,459 in Spirochaetes, 250 in Thermotogae, 227 in PVC group, 203 in Nitrospirae, 195 in FCB group, 108 in Synergistetes, 72 in Deferribacteres, 52 in Aquificae, 43 in Thermodesulfobacteria, 28 in Chrysiogenetes, 24 in Acidobacteria and 259 in others, respectively), 1,596 from archaea, and 200 from eukaryotes (88 in Opisthokonta, 57 in Alveolata, 23 in Viridiplantae, 9 in Euglenozoa, 8 in Amoebozoa, 7 in Haptophyceae, 7 in Stramenopiles and 1 in Apusozoa, respectively). Bacteria living in a stable, nutrient-rich environment harbor a lower number of MCPs and response regulatory proteins than soil and aquatic bacteria [20, 21]. For example, many pathogenic bacteria (such as Helicobacter pylori [22, 23] and C. jejuni [24]) have fewer MCPs than bacteria that form a mutualistic/symbiotic relationship with other organisms [21]. Bacterial adaptation to diverse environmental conditions promotes diversity among their signaling proteins including MCPs [20, 21]. For example, Myxococcus xanthus is a soil bacterium that has a complex lifestyle which includes the development of biofilm and multicellular fruiting bodies. Its genome encodes 21 MCPs [6, 7]. Magnetospirillum magnetotacticum exhibits a unique feature—magnetotaxis—and is able to migrate along the geomagnetic field lines [25, 26]. It uses ferric iron as the terminal electron acceptor in the electron transport chain and harbors 65 putative MCP genes [27]. Symbionts commonly harbor 9–90 MCP genes in their chromosomes [21, 28, 29]. For example, Sinorhizobium meliloti that forms a symbiotic relationship with the alfalfa plant harbors 9 MCP genes in its chromosome [21, 27]. On the other end, the spectrum is Azospirillum sp. B510, an N2-fixing soil bacterium that promotes the growth and disease resistance of its symbiotic partner Oryza sativa [30]. Its genome harbors 89 MCP genes [29]. MCPs are less common in archaea than in bacteria. Archaeal MCPs are believed to have originated via the horizontal gene transfer from bacteria [18].
Characteristic structural features of MCPs
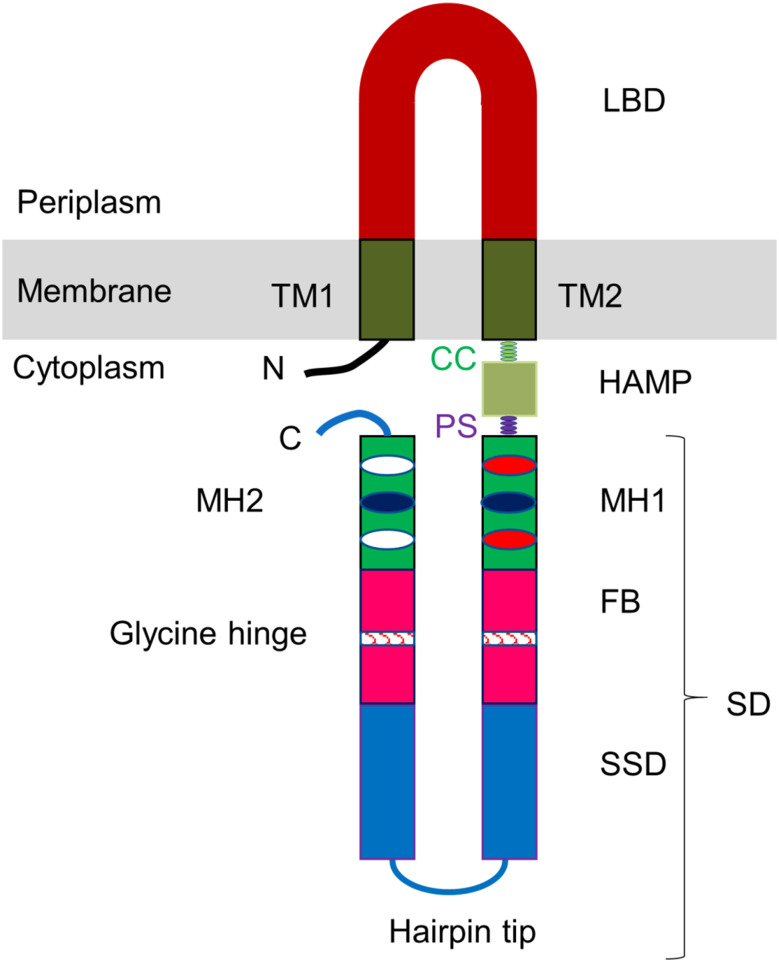
A typical MCP receptor consists of a ligand-binding domain (LBD), transmembrane (TM) helices, and a cytoplasmic signaling domain (SD) that interact with the downstream regulatory proteins including histidine kinase CheA, receptor coupling protein CheW, methyltransferase CheR, and methylesterase CheB [31, 32] (Fig. 1). The cytoplasmic domain harbors a histidine kinase, adenyl cyclase, methyl-accepting chemotaxis protein and phosphatase (HAMP) region, methylation helices (MH), flexible bundles (FB), and a signaling subdomain (SSD) [27, 33]. Some MCPs recognize their ligands directly [34, 35], while sensing by others may be mediated by the interaction with ligand-binding proteins (periplasmic binding proteins) [36, 37]. Upon interaction with signal molecules, MCPs transduce the signal to their cytoplasmic SDs that regulate the activity of CheA, the central kinase that phosphorylates its response regulator protein CheY [38]. The levels of phosphorylated CheY control the rotation of flagellar motors [31]. MCPs are arranged as trimers of dimers that form hexagonal arrays in the cytoplasmic membrane or the cytoplasm [38, 39]; molecules of CheA and CheW bound to the MCP’s SSD stabilize this lattice. The assembly of the MCPs, CheA, and CheW into ordered arrays increases their local concentration and allows positive cooperativity, thus enhancing the sensitivity of the receptors and the speed of the response. Methylation/demethylation of glutamate residues within the MH subdomain by CheR and CheB, respectively, serves as the mechanism of sensory adaptation in many bacteria [31, 40].
Fig. 1.
Overall topology of typical methyl-accepting chemotaxis protein. LBD ligand binding domain, TM transmembrane helix, CC control cable, HAMP histidine kinase, adenyl cyclase, methyl-accepting chemotaxis protein and phosphatase region, PS phase stutter, SD signaling domain, MH methylation helix, FB flexible bundle, SSD signaling subdomain
Classification of MCPs based on LBD and membrane topology
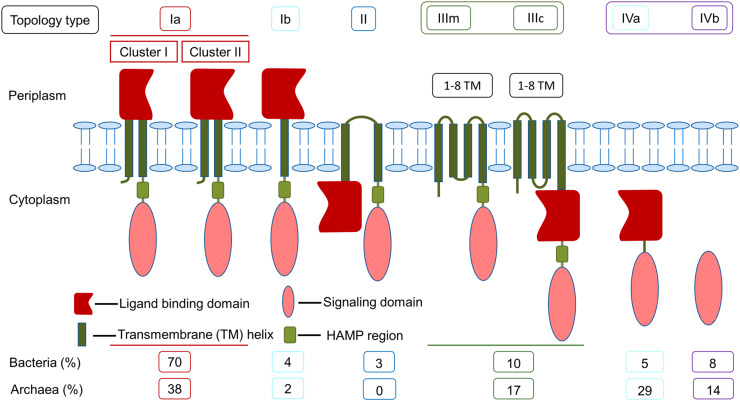
The ligand binding domains (LBDs) are involved in recognition of a broad range of extracellular or intracellular chemical and physical cues, and accordingly, LBDs can be located in the periplasm or the cytoplasm. Furthermore, MCPs can be membrane-embedded or cytoplasmic (soluble) [21, 41]. Bacterial receptors that sense extracellular signals commonly have a TM domain and a periplasmic LBD, whereas entirely cytoplasmic chemoreceptors often sense the energy state of the cell; the latter group of MCPs are more common in archaea than bacteria. Zhulin has classified MCPs into four major classes (I–IV) by analyzing their membrane topology [18]. Later, Wuichet and colleagues have subdivided class III into two (IIIm and IIIc) based on the presence of an LBD domain [42]. Subsequently, Lacal et al. [21] analyzed more than 3500 sequences of MCPs and elaborated the classification into seven different topologies (Ia, Ib, II, IIIm, IIIc, IVa, and IVb), where classes I and IV were subdivided based on the number of TM helices and the presence or absence of the LBD domain (Fig. 2). Based on the length of the primary sequence of the LBD, subclass Ia MCPs have been further grouped into two clusters (clusters I and II) [21].
Fig. 2.
Classification of methyl-accepting chemotaxis proteins based on membrane topology and LBD. The larger size of the LBD represents the longer amino acid sequence
Class I MCP receptors
A typical class I MCP harbors a periplasmic LBD, a TM domain, and a cytosolic SD [21]. This is a predominant class of MCPs both in bacteria and in archaea [21]. Based on the number of TM helices, they are categorized into two subclasses: Ia (two TM helices) and Ib (one TM helix). The subclass Ia MCPs are more common than subclass Ib. As shown in Fig. 2, the subclass Ia proteins consist of an N-terminal TM helix followed by a periplasmic LBD, a second TM helix, and a C-terminal cytoplasmic SD (Fig. 2). The subclass Ib MCPs differ from subclass Ia in that they lack an N-terminal TM helix (Fig. 2).
The class I MCPs have been further subdivided into two clusters according to the size of their LBDs: (1) cluster I (the predominant one) with an LBD of approximately 120–215 amino acids (aa) and (2) cluster II with an LBD of approximately 215–299 aa [21]. About 80% of the cluster I LBDs are α-helical, with the remainder adopting an α/β fold. Cluster I LBDs are very diverse in their three-dimensional structures and can adopt a variety of folds including a four-helix bundle (4HB), Per-Arnt-Sim (PAS), cGMP-specific phosphodiesterases, adenylyl cyclases and FhlA (GAF), calcium channels and chemotaxis receptors (Cache), and cyclases/histidine kinases associated sensory extracellular (CHASE) fold. For example, the well-characterized Escherichia coli taxis to aspartate and repellents (Tar) and taxis to serine and repellents (Tsr) receptors are cluster I proteins with an LBD that possess a 4HB fold [43–45]. The acid-sensing chemoreceptor TlpB from H. pylori is a cluster I protein, the LBD of which adopts a PAS-like Cache fold [46]. Geobacter sulfurreducens has two cluster I MCPs that contain a periplasmic LBD with a PAS fold harboring a c-type heme-binding motif [47]. Desulfovibrio vulgaris DcrA is also a c-type heme-binding protein with a 158-aa LBD that adopts a PAS fold [48]. DcrA is the cluster I MCP that serves as a redox and/or oxygen sensor [48].
The cluster II LBDs are found mainly in archaea. 90% of the archaeal cluster II LBDs are α-helical and 10% are α/β protein [21]. In contrast, most of the bacterial cluster II LBDs have an α/β fold (78%), with only 22% adopting an α-helical structure [21]. The structurally characterized LBDs of the cluster II receptors include the helical bimodular (HBM) and double Cache [previously named tandem-PAS or double PDC (PhoQ, DucS, CitA)] domains [34, 36, 49–52]. For example, P. putida McpS is a cluster II receptor with a 258-aa LBD that adopts an HBM fold. This receptor mediates chemotaxis towards tricarboxylic acid cycle intermediates and butyrate [51]. C. jejuni Tlp3 is a different cluster II receptor that contains a double Cache LBD (250 aa) and is responsible for directly sensing the branched-chain amino acid isoleucine [34].
Structural studies of cluster-I and cluster-II LBDs suggested that the former have one ligand-binding pocket [43, 46, 53, 54], whereas the latter are composed of two subdomains, each containing a putative ligand-binding site [34, 51]. There appears to be no correlation between the ligand specificity and whether the LBD belongs to cluster I or cluster II. For example, the cluster I LBD of CtpL and cluster II LBD of CtpH from P. aeruginosa are both involved in sensing inorganic phosphate, despite sharing no significant sequence homology [55].
Class II MCP receptors
Class II receptors harbor an N-terminal cytoplasmic LBD followed by two TM helices, a cytoplasmic HAMP region and a cytoplasmic SD (Fig. 2). The class II receptors are present solely in bacteria and are much less common (3%) in comparison with other classes of receptors [21]. They are known to be involved in aerotaxis or sensing of the cellular redox status. Examples of class II MCPs are E. coli aerotaxis receptor (Aer) [56] and Azotobacter vinelandii redox sensor NifL [57], both harboring an N-terminal cytoplasmic sensing PAS domain [57, 58]. In archaea, aerotaxis is mediated by receptors with a different topology [59].
Class III MCP receptors
Class III MCPs have a variable number of TM helices (1–8), with the sensor element located either within the membrane part (subclass IIIm), or in the cytoplasm, after the last TM helix (subclass IIIc), and followed by the cytoplasmic HAMP region and SD (Fig. 2) [21, 42]. They are more common in archaea than in bacteria [21]. For example, HtrVIII (halobacterial transducer of rhodopsin) from the archaeon H. salinarum is a subclass IIIm MCP that has six TM helices harboring a heme-binding site that serves as an oxygen sensor [59]. The H. salinarum sensory rhodopsin-I is a phototaxis receptor that has seven TM helices at the N-terminus, with a covalently bound light-sensitive retinal chromophore and a cytoplasmic SD [60]. Examples of bacterial class III MCPs include Bacillus subtilis KinB, a subclass IIIm receptor with six TM helices which is involved in spore formation, and B. subtilis KinD, a subclass IIIc receptor that plays a major role in bacterial survival in harsh environmental conditions [61, 62].
Class IV MCP receptors
Class IV MCPs are cytoplasmic (soluble) proteins with or without an identifiable LBD (subclasses IVa or IVb, respectively) [21]. They are more common in archaea than in bacteria [21]. Well-studied examples of archaeal and bacterial subclass IVa MCPs are aerotaxis receptor HemAT (haem-based aerotaxis transducer) from H. salinarum [17] and B. subtilis [63], respectively, that contain a globin motif in their LBD. The mechanism of ligand recognition by subclass IVb MCPs remains to be established. An example of an archaeal receptor from subclass IVb is Tm14 from Thermotoga maritima (278 aa) [64, 65]. The class IV chemoreceptors are also found in bacteria including Rhodobacter sphaeroides [66] and V. cholerae [41].
Subcellular location of MCPs
Subcellular localization of MCPs is determined by the type of signals they sense. Membrane-embedded MCPs with periplasmic LBD are involved in sensing environmental cues, while cytosolic MCPs and membrane-embedded MCPs with cytoplasmic LBD sense intracellular signals. Both bacterial and archaeal membrane-embedded MCPs exist as trimers of dimers that are arranged in highly ordered one-layer hexagonal arrays [39, 67, 68]. These arrays are stabilized by the interactions between the cytoplasmic tips of the receptors and the CheW and CheA proteins [41]. Cytoplasmic MPCs are arranged in hexagonal arrays in a fashion similar to that of transmembrane MCPs. However, in contrast to transmembrane MCPs, the cytoplasmic MCP arrays in bacteria [41] and archaea [68] form a two-layer structure, sandwiched between two CheA-CheW plates. LBDs in the cytoplasmic MCP arrays are thought to be positioned in the middle of the sandwich [41]. This hexagonal packaging of MCPs facilitates cooperative interactions between the receptors and thereby plays an important role in signal transduction [69].
The location of the hexagonal arrays of transmembrane and soluble MCPs within the cell varies between species, and also between different MCPs within one organism. In bacteria, most transmembrane MCPs are localized to the pole(s) of the cell [67], although membrane-embedded MCP with lateral localization has also been found [70]. Polar localization of transmembrane receptor arrays (reported as polar organelle) has also been found in the archaeon H. salinarum [71]. Bacterial soluble MCPs can be targeted to the poles or distributed throughout the cell body [41], while the archaeal cytosolic MCPs are often located near the mid-section of the cell [68]. For example, all four MCPs from E. coli are membrane proteins that are clustered at the cell poles [72]. Both membrane-embedded and cytosolic MCPs of V. cholerae [73] localize to polar regions. Membrane-embedded MCPs of R. sphaeroides are mainly localized at cell pole, while its soluble MCPs form clusters in the cytoplasm, grouped around the mid-section or ¼ and ¾ positions, depending on the cell-cycle stage [74]. The WspA protein in P. aeruginosa is a membrane-embedded MCP that forms clusters both at the poles and lateral locations [70]. The methane-producing archaeon Methanoregula formicica has six cytosolic MCPs which are arranged as bilayer hexagonal arrays near the mid-cell [68].
In many well-characterized species, there appears to be no direct link between the position of the chemoreceptor arrays and the cellular localization of flagella. For example, the polar location of E. coli MCPs does not correlate with the random distribution of its flagella around the cell. Similarly, in R. sphaeroides, the predominantly polar location of its membrane receptor clusters is distinct from the location of its single flagellum that is randomly positioned on the side of the cell body. It is believed that the subcellular location and distribution of the chemoreceptor arrays, which often changes in different stages of the bacterial cell cycle, ensures receptor inheritance on division, so that both progeny cells possess the complete chemosensory apparatus [75].
HAMP region
The HAMP region is composed of one or more ~50 aa long signal relay modules that play a crucial role in the propagation of the signal from the sensing domain to the cytoplasmic SD [76, 77]. In most MCPs, one end of the HAMP region is connected to a TM helix by a five-residue control cable, while the other end is linked to the methylation helix (sensory-adaptation subdomain) of SD via a four-residue phase stutter [78]. The single HAMP module has a parallel four-helix bundle fold and is a dimer of two pairs of parallel amphipathic α-helices (AS1 and AS2) joined by a 14-amino-acid long flexible connector [79, 80]. The amino acid sequence of aliphatic α-helices AS1 and AS2 contains a typical seven-residue heptad repeat (a–b–c–d–e–f–g), where one heptad repeat corresponds exactly to two α-helical turns. The hydrophobic side chains of the residues at positions a and d form “knobs” that are directed inwards and are buried in the core of the molecule [80, 81]. In most characterized HAMP modules, the flexible connector between AS1 and AS2 harbors a conserved motif with a consensus sequence G-x-HR1-x-x-x-HR2, where HR1 and HR2 are conserved hydrophobic residues [76, 80]. Removal of the HAMP domain results in inactivation of the receptor [33, 76]. Although the sequence identity among HAMP domains of different proteins is very low [82], they can be swapped between different MCPs without altering their cellular functions which suggest a common mechanism of the signal relay by this domain [76]. In addition, the number of HAMP modules varies among different receptor proteins [83]. For example, P. aeruginosa cytosolic aerotaxis receptor harbors five HAMP domains [81], while the E. coli Aer receptor contains only one domain [84]. Hulko and colleagues [80] reported the first 3D structure of a HAMP domain from Archaeoglobus fulgidus Af1503 which was solved by nuclear magnetic resonance. Later, the structure of a polyHAMP domain of P. aeruginosa Aer2 receptor has been reported [81, 84].
The role of an HAMP domain is to transmit signal received at the LBD to the SD. A canonical HAMP domain serves as a universal signal converter which tunes a broad range of input signals into conformational changes in the downstream SD via concerted axial rotation of all helices [80, 83]. Zhou and colleagues [85] proposed a three-state dynamic bundle model for the signal transmission by the E. coli HAMP domain. In the presence of an attractant, HAMP helices form a stable bundle, which results in a loose packing of the downstream four-helix methylation bundle and a kinase-off state of the receptor. In contrast, interaction with a repellent result in destabilization of the HAMP bundle and a more stable packing of the methylation bundle, which induces the kinase-on conformation of the SD. The detailed signal transduction mechanism by the HAMP modules has been described elsewhere [77, 81, 85].
Cytoplasmic signaling domain
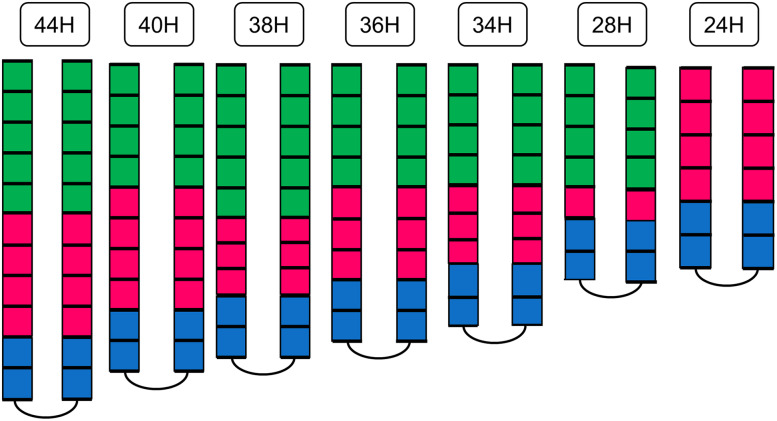
The cytoplasmic signaling domain (SD) is the most conserved element of MCP receptors, consisting of adaptation (methylation) subdomain, connected to the signaling subdomain (SSD) via a flexible bundle (Fig. 1). Alexander and Zhulin [27] have categorized MCPs into seven major (44, 40, 38, 36, 34, 28, and 24 H) and five minor (48, 42, 52, 58, and 64 H) classes based on the analysis of sequence conservation and on the number of heptads in the SD (Fig. 3). The seven major classes contained ~90% of the ~2000 analyzed MCPs. The amino-acid sequence of the SSD is highly conserved; the adaptation subdomain shows a moderate degree of conservation, while the flexible bundle is the least conserved part of the SD (Figs. 1, 3) [27].
Fig. 3.
Seven major classes of cytoplasmic signaling domains of methyl-accepting chemotaxis proteins. The methylation bundle, flexible bundle, and signaling subdomain are colored green, purple, and blue, respectively. Each box represents two heptads. The names of MCP classes were assigned according to the number of heptads (24–44 H) [27]
Methylation subdomain
The methylation subdomain [often referred to as methylation helices (MH)] harbors several crucial residues that, in most characterized receptors, undergo signal-dependent reversible methylation; changes to the extent of the methylation lead to sensory adaptation. In many bacteria, the MH subdomains of different MCPs contain the consensus sequence-[A/S/T/G]-[A/S/T/G]-X-X-[E/Q]-[E/Q]-X-X-[A/S/T/G]-[A/S/T/G]-, where one of the two consecutive Glx (E/Q) residues is the methylation site [27, 86]. The methylation is catalyzed by CheR, while CheB and CheD are responsible for demethylation and deamidation of the site, respectively [87–89]. The small conserved residues flanking the EQ pairs are believed to be important for proper docking of the helical turn in the active site of the methylation modification enzymes [90]. An example of a bacterium that deviates from the above-mentioned MH consensus sequence is T. maritima, the six transmembrane chemoreceptors of which have the methylation sites -[A/S]-[A/S/T/G]-X-[E/Q]-[E/Q]-X-[A/S/T/G]-[A/S]- [91]. The number and location of the methylation sites vary among different classes of MCP receptors.
Flexible bundle subdomain
The flexible bundle (FB) subdomain connects the methylation/adaptation subdomain MH with the signaling subdomain (SSD) [27]. The only conserved amino acid sequence features are the glycine hinge at midpoint that plays an important role in the formation of a trimer of dimers in signaling complexes [78] and the hydrophobic knob residues a and d of the heptads (a–b–c–d–e–f–g) that form interactions that define the conformation of the four-helical bundle. The precise mechanism of how the FB subdomain transmits signal is yet to be established.
Signaling subdomain
The signaling subdomain (SSD) located at the tip of the cytoplasmic SD directly interacts with CheA and CheW for the modulation of bacterial flagellum rotation [78]. The kinase-activating hairpin tip of the SSD, showing the highest amino acid sequence conservation among MCPs (Alexander and Zhulin [27]), fluctuates between two stable conformations in a signal-dependent manner [92]. The interactions between the hairpin tip, CheA and CheW stabilize the clusters of trimers of dimers, and thereby, modulate the formation and activity of the receptor signaling complexes [78, 93]. The tip contains two conserved residues (F396 and E391 in E. coli Tsr) that play a crucial role in switching between the two output states of the MCP receptor [78, 92, 93]. Substitutions of the surface-exposed E391 residue with an apolar residue resulted in fast switching between the kinase-on and off states which suggested that this tip dynamics is likely to be involved in regulation of the kinase activity [93]. The two buried side chain of F396 on the two halves of the receptor stack against each other which stabilizes the dimer interface; flipping of the stacking arrangement due to rotameric freedom of the two rings results in an alternative tip conformation and hence, alternative signaling state [92]. Crosslinking studies confirmed the proximity between E391 of MCP and CheW [94] and identified the MCP tip residues that interact with CheA [95].
Ligand sensing and signal transduction mechanisms
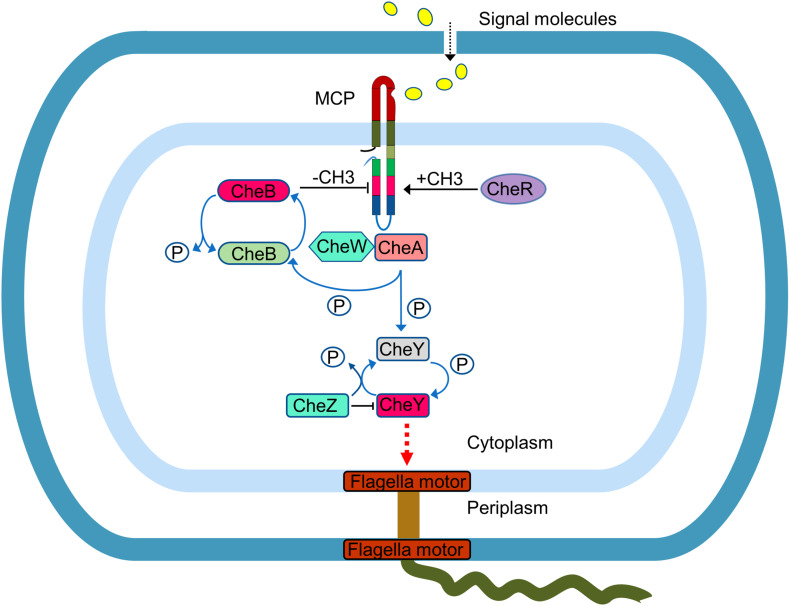
MCP receptors allow the cell to navigate in gradients of various chemical (e.g. pH, osmolarity, concentration) and physical (e.g., light, temperature, magnetic field) cues. Ligand sensing mechanism has been extensively studied in E. coli MCPs Tar and Tsr, where ligand-binding to the LBD was shown to induce a small (~2 Å) piston-like sliding of one of the TM helices (TM2) in the MCP dimer, in the direction normal to the plane of the membrane [96]. This inward movement of the TM2 helix is believed to modulate the control cable helicity (by creating a kink or break in a α-helix) that enhances the HAMP bundle packing stability [97]. The phase stutter that connects the HAMP region with the MH bundle propagates the signal in a counter-phase manner: tight packing of the HAMP bundle causes loose packing of the MH subdomain and vice versa [98, 99]. The modulation of the packing geometry or axial motions of the MHs controls the output state of the SSD. Bending at the glycine hinge of the flexible bundle subdomain that links the MH subdomain with the SSD is thought to play an important role during the assembly of trimers of dimers or during the switching of SSD between ‘kinase ON’ and ‘kinase OFF’ states [67, 100]. The SSD’s interaction with kinase CheA is mediated by the receptor coupling protein CheW; the three proteins function as the MCP/CheA/CheW signaling complex [31]. The ‘kinase ON’ and ‘kinase OFF’ states of SSD control the activity of CheA, thus influencing the level of phosphorylation of its cognate response regulator CheY (Fig. 4). The CheY-P shuttles between the receptor-signaling complexes and the flagellar motors [31, 78]; interaction of CheY-P with the motors controls the rotation of the flagella. In E. coli, the direction of flagella rotation (counter-clockwise or clockwise) depends on the ratio of CheY:CheY-P. Removal of a phosphate group from CheY-P by a phosphatase CheZ terminates the signal.
Fig. 4.
Signal recognition and transduction mechanism by a typical MCP receptor in bacteria
Response to a temporal variation of ligand (attractants or repellents) concentration is controlled by a system consisting of methylesterase CheB and methyltransferase CheR. The CheR is a constitutively active; it methylates the conserved glutamate or deamidated glutamine residues in the MH subdomain of the chemoreceptors using S-adenosylmethionine as a cofactor. Elevated methylation results in inhibition of MCP signaling [31, 78], which is the core adaptation mechanism. The activity of methylesterase CheB is controlled by the levels of activated CheA: the CheA phosphorylates and thus activates CheB. The activated CheB removes methyl groups from MCPs and thereby restores their signaling capacity (Fig. 4). CheB and CheR act in coordination to allow the cell to modulate its stimulation level. This adaptation system enables a cell to navigate in gradients of signals (attractants or repellents) over a broad range of concentrations, from nanomolar to millimolar [40]. Interestingly, a different signaling paradigm has been found in B. subtilis. While increasing concentration of attractants decreases the kinase activity in E. coli, the opposite effect is found in B. subtilis [101]. Examples of archaea have been reported, where the chemotaxis systems use an additional protein, CheF, to relay the signal from CheY-P to the motors [16].
Conclusions and future directions
Chemotaxis plays an important role in the ecology of bacterial populations [32, 102]. Signal recognition by MCPs and a cell response in the form of modulation of its motile behavior underpin the ability of bacteria and archaea to colonize nutrient-rich microenvironmental niches—the process central to symbiosis and pathogenesis. Chemotaxis is essential for the host colonization and virulence of many pathogenic bacteria that cause diseases in humans, animals, and plants [103, 104]. For example, chemotaxis towards chemicals released by corals and their symbionts plays an important role in the infection of corals by pathogenic bacteria associated with coral disease [104]. Sustainable and renewable production of nitrogen for agriculture via a symbiotic association between Rhizobium bacteria and legumes is dependent on the ability of bacteria to sense and move to legume roots [105]. Furthermore, bacterial chemotaxis plays a key role in large-scale biogeochemical fluxes, including carbon, nitrogen, and sulfur cycling [106]. Owing to its ubiquitous nature, detailed understanding of how bacteria and archaea sense attractants and repellents is important.
The cognate signal molecules are known for few chemoreceptors, and ligand identification represents a major research need in this area. Many other challenges lie ahead including answering the question of how the same chemoreceptor can sense, and differentiate between, attractants and repellents [43, 107]. The current understanding of ligand sensing mechanism mainly comes from the studies on the E. coli chemoreceptors. However, the signal transduction mechanisms in microorganisms that lead a more complex lifestyle utilize additional proteins and chemoreceptors with a distinctly different signaling domain. It remains to be established how chemoreceptors with the sensing domain that differs from the four-helix bundle (such as HBM, Cache, and so on) transmit signals across the membrane. The mechanisms of signal amplification in many bacteria and archaea that deviate from the E. coli CheA/CheW/CheY paradigm are not yet fully defined.
Currently, only low-resolution cryo-electron microscopy structures of a full-length chemoreceptor are available. No crystal structure of a receptor/repellent complex has been elucidated so far. In addition, the complete three-dimensional structure of the signaling complex is yet to determined. It remains to be established if the patterns of receptor arrangement are conserved across different bacteria and archaea. Furthermore, how the clusters of chemoreceptors are targeted to their subcellular localization is not yet understood.
Knowledge derived through the extensive structural and biochemical analysis of chemoreceptors would have many biotechnological applications, including the design of biofertilizers that are based on the chemoattractants for the symbiotic Rhizobia, and bioengineering of chemoreceptors to enhance the bacterial ability for bioremediation of xenobiotic compounds [108]. As chemotaxis was shown to be required for the long-term persistence of many pathogenic bacteria in the host, inhibiting chemoreceptors with small molecules offers an alternative strategy for treatment of bacterial infections. MCPs represent a convenient target for drug design as they are absent in humans. Inhibition of MCPs would have a detrimental effect on the survival and virulence of bacteria, helping the host to clear the infection, while not creating conditions that are likely to give rise to resistance. Thus, detailed structural studies of the mechanisms of sensing and signaling by chemoreceptors may pave the way to the structure-guided design of novel therapeutics.
Compliance with ethical standards
Conflict of interest
The authors have no conflict interest.
References
- 1.Berleman JE, Bauer CE. Involvement of a Che-like signal transduction cascade in regulating cyst cell development in Rhodospirillum centenum . Mol Microbiol. 2005;56:1457–1466. doi: 10.1111/j.1365-2958.2005.04646.x. [DOI] [PubMed] [Google Scholar]
- 2.He K, Bauer CE. Chemosensory signaling systems that control bacterial survival. Trends Microbiol. 2014;22:389–398. doi: 10.1016/j.tim.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Nat Acad Sci USA. 2005;102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luu RA, Kootstra JD, Nesteryuk V, Brunton CN, Parales JV, Ditty JL, Parales RE. Integration of chemotaxis, transport and catabolism in Pseudomonas putida and identification of the aromatic acid chemoreceptor PcaY. Mol Microbiol. 2015;96:134–147. doi: 10.1111/mmi.12929. [DOI] [PubMed] [Google Scholar]
- 5.Kirby JR. Chemotaxis-like regulatory systems: unique roles in diverse bacteria. Annu Rev Microbiol. 2009;63:45–59. doi: 10.1146/annurev.micro.091208.073221. [DOI] [PubMed] [Google Scholar]
- 6.Kirby JR, Zusman DR. Chemosensory regulation of developmental gene expression in Myxococcus xanthus . Proc Nat Acad Sci USA. 2003;100:2008–2013. doi: 10.1073/pnas.0330944100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moine A, Agrebi R, Espinosa L, Kirby JR, Zusman DR, Mignot T, Mauriello EM. Functional organization of a multimodular bacterial chemosensory apparatus. PLoS Genet. 2014;10:e1004164. doi: 10.1371/journal.pgen.1004164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z, Geng Y, Shi W. A DnaK homolog in Myxococcus xanthus is involved in social motility and fruiting body formation. J Bacteriol. 1998;180:218–224. doi: 10.1128/jb.180.2.218-224.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Black WP, Yang Z. Myxococcus xanthus chemotaxis homologs DifD and DifG negatively regulate fibril polysaccharide production. J Bacteriol. 2004;186:1001–1008. doi: 10.1128/JB.186.4.1001-1008.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harkey CW, Everiss KD, Peterson KM. The Vibrio cholerae toxin-coregulated-pilus gene tcpI encodes a homolog of methyl-accepting chemotaxis proteins. Infect Immun. 1994;62:2669–2678. doi: 10.1128/iai.62.7.2669-2678.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotter PA, Stibitz S. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Opin Microbiol. 2007;10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Sampedro I, Parales RE, Krell T, Hill JE. Pseudomonas chemotaxis. FEMS Microbiol Rev. 2014;153:119–128. doi: 10.1111/1574-6976.12081. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Lou H, Ojcius DM, Sun A, Sun D, Zhao J, Lin Xa, Yan J. Methyl-accepting chemotaxis proteins 3 and 4 are responsible for Campylobacter jejuni chemotaxis and jejuna colonization in mice in response to sodium deoxycholate. J Med Microbiol. 2014;63:343–354. doi: 10.1099/jmm.0.068023-0. [DOI] [PubMed] [Google Scholar]
- 14.Choi Y, Kim S, Hwang H, Kim KP, Kang DH, Ryu S. Plasmid-encoded MCP is involved in virulence, motility, and biofilm formation of Cronobacter sakazakii ATCC 29544. Infect Immun. 2015;83:197–204. doi: 10.1128/IAI.02633-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishiyama S-i, Takahashi Y, Yamamoto K, Suzuki D, Itoh Y, Sumita K, Uchida Y, Homma M, Imada K, Kawagishi I. Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants. Sci Rep. 2016;6:20866. doi: 10.1038/srep20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlesner M, Miller A, Streif S, Staudinger WF, Müller J, Scheffer B, Siedler F, Oesterhelt D. Identification of Archaea-specific chemotaxis proteins which interact with the flagellar apparatus. BMC Microbiol. 2009;9:1. doi: 10.1186/1471-2180-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou S, Freitas T, Larsen RW, Piatibratov M, Sivozhelezov V, Yamamoto A, Meleshkevitch EA, Zimmer M, Ordal GW, Alam M. Globin-coupled sensors: a class of heme-containing sensors in archaea and bacteria. Proc Nat Acad Sci USA. 2001;98:9353–9358. doi: 10.1073/pnas.161185598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhulin IB. The superfamily of chemotaxis transducers: from physiology to genomics and back. Adv Microb Physiol. 2001;45:157–198. doi: 10.1016/S0065-2911(01)45004-1. [DOI] [PubMed] [Google Scholar]
- 19.InterPro Database. http://www.ebi.ac.uk/interpro. Accessed on 1 Feb 2017
- 20.Ashby MK. Survey of the number of two-component response regulator genes in the complete and annotated genome sequences of prokaryotes. FEMS Microbiol Lett. 2004;231:277–281. doi: 10.1016/S0378-1097(04)00004-7. [DOI] [PubMed] [Google Scholar]
- 21.Lacal J, García-Fontana C, Muñoz-Martínez F, Ramos JL, Krell T. Sensing of environmental signals: classification of chemoreceptors according to the size of their ligand binding regions. Environ Microbiol. 2010;12:2873–2884. doi: 10.1111/j.1462-2920.2010.02325.x. [DOI] [PubMed] [Google Scholar]
- 22.Croxen MA, Sisson G, Melano R, Hoffman PS. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J Bacteriol. 2006;188:2656–2665. doi: 10.1128/JB.188.7.2656-2665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andermann TM, Chen YT, Ottemann KM. Two predicted chemoreceptors of Helicobacter pylori promote stomach infection. Infect Immun. 2002;70:5877–5881. doi: 10.1128/IAI.70.10.5877-5881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchant J, Wren B, Ketley J. Exploiting genome sequence: predictions for mechanisms of Campylobacter chemotaxis. Trends Microbiol. 2002;10:155–159. doi: 10.1016/S0966-842X(02)02323-5. [DOI] [PubMed] [Google Scholar]
- 25.Blakemore R. Magnetotactic Bacteria. Science. 1975;190:377–379. doi: 10.1126/science.170679. [DOI] [PubMed] [Google Scholar]
- 26.Zhu X, Ge X, Li N, Wu L-F, Luo C, Ouyang Q, Tu Y, Chen G. Angle sensing in magnetotaxis of Magnetospirillum magneticum AMB-1. Integr Biol. 2014;6:706–713. doi: 10.1039/C3IB40259B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Nat Acad Sci USA. 2007;104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krell T, Lacal J, Muñoz-Martínez F, Reyes-Darias JA, Cadirci BH, García-Fontana C, Ramos JL. Diversity at its best: bacterial taxis. Environ Microbiol. 2011;13:1115–1124. doi: 10.1111/j.1462-2920.2010.02383.x. [DOI] [PubMed] [Google Scholar]
- 29.Alexandre G. Handbook for Azospirillum. Switzerland: Springer; 2015. Chemotaxis in Azospirillum; pp. 101–114. [Google Scholar]
- 30.Yasuda M, Isawa T, Shinozaki S, Minamisawa K, Nakashita H. Effects of colonization of a bacterial endophyte, Azospirillum sp. B510, on disease resistance in rice. Biosci Biotechnol Biochem. 2009;73:2595–2599. doi: 10.1271/bbb.90402. [DOI] [PubMed] [Google Scholar]
- 31.Bi S, Lai L. Bacterial chemoreceptors and chemoeffectors. Cell Mol Life Sci. 2015;72:691–708. doi: 10.1007/s00018-014-1770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 33.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu YC, Machuca MA, Beckham SA, Gunzburg MJ, Roujeinikova A. Structural basis for amino-acid recognition and transmembrane signalling by tandem Per-Arnt-Sim (tandem PAS) chemoreceptor sensory domains. Acta Crystallogr D Biol Crystallogr. 2015;71:2127–2136. doi: 10.1107/S139900471501384X. [DOI] [PubMed] [Google Scholar]
- 35.Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE, Kim S-H. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991;254:5036. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- 36.Machuca MA, Liu YC, Beckham SA, Gunzburg MJ, Roujeinikova A. The crystal structure of the tandem-PAS sensing domain of Campylobacter jejuni chemoreceptor Tlp1 suggests indirect mechanism of ligand recognition. J Struct Biol. 2016;194:205–213. doi: 10.1016/j.jsb.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 37.Kossmann M, Wolff C, Manson M. Maltose chemoreceptor of Escherichia coli: interaction of maltose-binding protein and the tar signal transducer. J Bacteriol. 1988;170:4516–4521. doi: 10.1128/jb.170.10.4516-4521.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Briegel A, Ortega DR, Tocheva EI, Wuichet K, Lia Z, Chen S, Muller A, Iancu CV, Murphy GE, Dobro MJ, Zhulin IB, Jensen GJ. Universal architecture of bacterial chemoreceptor arrays. Proc Nat Acad Sci USA. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Hu B, Morado DR, Jani S, Manson MD, Margolin W. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc Nat Acad Sci USA. 2012;109:E1481–E1488. doi: 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Endres RG, Wingreen NS. Precise adaptation in bacterial chemotaxis through “assistance neighborhoods”. Proc Nat Acad Sci USA. 2006;103:13040–13044. doi: 10.1073/pnas.0603101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Briegel A, Ladinsky MS, Oikonomou C, Jones CW, Harris MJ, Fowler DJ, Chang Y-W, Thompson LK, Armitage JP, Jensen GJ. Structure of bacterial cytoplasmic chemoreceptor arrays and implications for chemotactic signaling. Elife. 2014;3:e02151. doi: 10.7554/eLife.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wuichet K, Alexander RP, Zhulin IB. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol. 2007;422:3–31. doi: 10.1016/S0076-6879(06)22001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mise T. Structural analysis of the ligand-binding domain of the aspartate receptor Tar from Escherichia coli . BioChemistry. 2016;55:3708–3713. doi: 10.1021/acs.biochem.6b00160. [DOI] [PubMed] [Google Scholar]
- 44.Kim KK, Yokota H, Kim S-H. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 45.Krikos A, Mutoh N, Boyd A, Simon MI. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell. 1983;33:615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- 46.Sweeney EG, Henderson JN, Goers J, Wreden C, Hicks KG, Foster JK, Parthasarathy R, Remington SJ, Guillemin K. Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB. Structure. 2012;20:1177–1188. doi: 10.1016/j.str.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pokkuluri P, Pessanha M, Londer Y, Wood S, Duke N, Wilton R, Catarino T, Salgueiro C, Schiffer M. Structures and solution properties of two novel periplasmic sensor domains with c-type heme from chemotaxis proteins of Geobacter sulfurreducens: implications for signal transduction. J Mol Biol. 2008;377:1498–1517. doi: 10.1016/j.jmb.2008.01.087. [DOI] [PubMed] [Google Scholar]
- 48.Yoshioka S, Kobayashi K, Yoshimura H, Uchida T, Kitagawa T, Aono S. Biophysical properties of a c-type heme in chemotaxis signal transducer protein DcrA. BioChemistry. 2005;44:15406–15413. doi: 10.1021/bi0513352. [DOI] [PubMed] [Google Scholar]
- 49.Upadhyay AA, Fleetwood AD, Adebali O, Finn RD, Zhulin IB. Cache domains that are homologous to, but different from pas domains comprise the largest superfamily of extracellular sensors in prokaryotes. PLoS Comput Biol. 2016;12:e1004862. doi: 10.1371/journal.pcbi.1004862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Z, Hendrickson WA. Structural characterization of the predominant family of histidine kinase sensor domains. J Mol Biol. 2010;400:335–353. doi: 10.1016/j.jmb.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pineda-Molina E, Reyes-Darias J-A, Lacal JA, Ramos JL, García-Ruiz JM, Gavira JA, Krell T. Evidence for chemoreceptors with bimodular ligand-binding regions harboring two signal-binding sites. Proc Nat Acad Sci USA. 2012;109:18926–18931. doi: 10.1073/pnas.1201400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ortega Á Krell T. The HBM domain: introducing bimodularity to bacterial sensing. Protein Sci. 2014;23:332–336. doi: 10.1002/pro.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reinelt S, Hofmann E, Gerharz T, Bott M, Madden DR. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J Biol Chem. 2003;278:39189–39196. doi: 10.1074/jbc.M305864200. [DOI] [PubMed] [Google Scholar]
- 54.Brewster JL, McKellar JL, Finn TJ, Newman J, Peat TS, Gerth ML. Structural basis for ligand recognition by a Cache chemosensory domain that mediates carboxylate sensing in Pseudomonas syringae . Sci Rep. 2016;6:35198. doi: 10.1038/srep35198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu H, Kato J, Kuroda A, Ikeda T, Takiguchi N, Ohtake H. Identification and characterization of two chemotactic transducers for inorganic phosphate in Pseudomonas aeruginosa . J Bacteriol. 2000;182:3400–3404. doi: 10.1128/JB.182.12.3400-3404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watts KJ, Ma Q, Johnson MS, Taylor BL. Interactions between the PAS and HAMP domains of the Escherichia coli aerotaxis receptor Aer. J Bacteriol. 2004;186:7440–7449. doi: 10.1128/JB.186.21.7440-7449.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Key J, Hefti M, Purcell EB, Moffat K. Structure of the redox sensor domain of Azotobacter vinelandii NifL at atomic resolution: signaling, dimerization, and mechanism. BioChemistry. 2007;46:3614–3623. doi: 10.1021/bi0620407. [DOI] [PubMed] [Google Scholar]
- 58.Amin DN, Taylor BL, Johnson MS. Topology and boundaries of the aerotaxis receptor Aer in the membrane of Escherichia coli . J Bacteriol. 2006;188:894–901. doi: 10.1128/JB.188.3.894-901.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brooun A, Bell J, Freitas T, Larsen RW, Alam M. An archaeal aerotaxis transducer combines subunit I core structures of eukaryotic cytochrome c oxidase and eubacterial methyl-accepting chemotaxis proteins. J Bacteriol. 1998;180:1642–1646. doi: 10.1128/jb.180.7.1642-1646.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoff WD, Jung KH, Spudich JL. Molecular mechanism of photosignaling by archaeal sensory rhodopsins. Annu Rev Biophys Biomol Struct. 1997;26:223–258. doi: 10.1146/annurev.biophys.26.1.223. [DOI] [PubMed] [Google Scholar]
- 61.Wu R, Gu M, Wilton R, Babnigg G, Kim Y, Pokkuluri P, Szurmant H, Joachimiak A, Schiffer M. Insight into the sporulation phosphorelay: crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD. Protein Sci. 2013;22:564–576. doi: 10.1002/pro.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fabret C, Feher VA, Hoch JA. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J Bacteriol. 1999;181:1975–1983. doi: 10.1128/jb.181.7.1975-1983.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang W, Phillips GN. Structure of the oxygen sensor in Bacillus subtilis: signal transduction of chemotaxis by control of symmetry. Structure. 2003;11:1097–1110. doi: 10.1016/S0969-2126(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 64.Pollard AM, Bilwes AM, Crane BR. The structure of a soluble chemoreceptor suggests a mechanism for propagating conformational signals. BioChemistry. 2009;48:1936–1944. doi: 10.1021/bi801727m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li X, Fleetwood AD, Bayas C, Bilwes AM, Ortega DR, Falke JJ, Zhulin IB, Crane BR. The 3.2Å resolution structure of a receptor: CheA:CheW signaling complex defines overlapping binding sites and key residue interactions within bacterial chemosensory arrays. BioChemistry. 2013;52:3852–3865. doi: 10.1021/bi400383e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wadhams G, Martin AC, Porter S, Maddock J, Mantotta J, King H, Armitage J. TlpC, a novel chemotaxis protein in Rhodobacter sphaeroides, localizes to a discrete region in the cytoplasm. Mol Microbiol. 2002;46:1211–1221. doi: 10.1046/j.1365-2958.2002.03252.x. [DOI] [PubMed] [Google Scholar]
- 67.Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Nat Acad Sci USA. 2012;109:3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Briegel A, Ortega DR, Huang AN, Oikonomou CM, Gunsalus RP, Jensen GJ. Structural conservation of chemotaxis machinery across Archaea and Bacteria. Environ Microbiol Rep. 2015;7:414–419. doi: 10.1111/1758-2229.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sourjik V. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 2004;12:569–576. doi: 10.1016/j.tim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 70.O’Connor JR, Kuwada NJ, Huangyutitham V, Wiggins PA, Harwood CS. Surface sensing and lateral subcellular localization of WspA, the receptor in a chemosensory-like system leading to c-di-GMP production. Mol Microbiol. 2012;86:720–729. doi: 10.1111/mmi.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Metlina A. Bacterial and archaeal flagella as prokaryotic motility organelles. BioChemistry. 2004;69:1203–1212. doi: 10.1007/s10541-005-0065-8. [DOI] [PubMed] [Google Scholar]
- 72.Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1717. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 73.Ringgaard S, Schirner K, Davis BM, Waldor MK. A family of ParA-like ATPases promotes cell pole maturation by facilitating polar localization of chemotaxis proteins. Genes Dev. 2011;25:1544–1555. doi: 10.1101/gad.2061811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chiu SW, Roberts MA, Leake MC, Armitage JP. Positioning of chemosensory proteins and FtsZ through the Rhodobacter sphaeroides cell cycle. Mol Microbiol. 2013;90:322–337. doi: 10.1111/mmi.12366. [DOI] [PubMed] [Google Scholar]
- 75.Jones CW, Armitage JP. Positioning of bacterial chemoreceptors. Trends Microbiol. 2015;23:247–256. doi: 10.1016/j.tim.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 76.Ames P, Zhou Q, Parkinson JS. HAMP domain structural determinants for signalling and sensory adaptation in Tsr, the Escherichia coli serine chemoreceptor. Mol Microbiol. 2014;91:875–886. doi: 10.1111/mmi.12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stewart V. The HAMP signal-conversion domain: static two-state or dynamic three-state? Mol Microbiol. 2014;91:853–857. doi: 10.1111/mmi.12516. [DOI] [PubMed] [Google Scholar]
- 78.Parkinson JS, Hazelbauer GL, Falke JJ. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 2015;23:257–266. doi: 10.1016/j.tim.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Butler SL, Falke JJ. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. BioChemistry. 1998;37:10746–10756. doi: 10.1021/bi980607g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 81.Airola MV, Sukomon N, Samanta D, Borbat PP, Freed JH, Watts KJ, Crane BR. HAMP domain conformers that propagate opposite signals in bacterial chemoreceptors. PLoS Biol. 2013;11:e1001479. doi: 10.1371/journal.pbio.1001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Appleman JA, Stewart V. Mutational analysis of a conserved signal-transducing element: the HAMP linker of the Escherichia coli nitrate sensor NarX. J Bacteriol. 2003;185:89–97. doi: 10.1128/JB.185.1.89-97.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dunin-Horkawicz S, Lupas AN. Comprehensive analysis of HAMP domains: implications for transmembrane signal transduction. J Mol Biol. 2010;397:1156–1174. doi: 10.1016/j.jmb.2010.02.031. [DOI] [PubMed] [Google Scholar]
- 84.Ma Q, Johnson MS, Taylor BL. Genetic analysis of the HAMP domain of the Aer aerotaxis sensor localizes flavin adenine dinucleotide-binding determinants to the AS-2 helix. J Bacteriol. 2005;187:193–201. doi: 10.1128/JB.187.1.193-201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou Q, Ames P, Parkinson JS. Biphasic control logic of HAMP domain signalling in the Escherichia coli serine chemoreceptor. Mol Microbiol. 2011;80:596–611. doi: 10.1111/j.1365-2958.2011.07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Terwilliger T, Wang JY, Koshland D. Kinetics of receptor modification. The multiply methylated aspartate receptors involved in bacterial chemotaxis. J Biol Chem. 1986;261:10814–10820. [PubMed] [Google Scholar]
- 87.Sherris D, Parkinson JS. Posttranslational processing of methyl-accepting chemotaxis proteins in Escherichia coli . Proc Nat Acad Sci USA. 1981;78:6051–6055. doi: 10.1073/pnas.78.10.6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kristich CJ, Ordal GW. Bacillus subtilis CheD is a chemoreceptor modification enzyme required for chemotaxis. J Biol Chem. 2002;277:25356–25362. doi: 10.1074/jbc.M201334200. [DOI] [PubMed] [Google Scholar]
- 89.Callahan A, Parkinson JS. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: cheD mutations affect the structure and function of the Tsr transducer. J Bacteriol. 1985;161:96–104. doi: 10.1128/jb.161.1.96-104.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pérez-Rueda E, Collado-Vides J, Segovia L. Phylogenetic distribution of DNA-binding transcription factors in bacteria and archaea. Comput Biol Chem. 2004;28:341–350. doi: 10.1016/j.compbiolchem.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 91.Perez E, Zheng H, Stock AM. Identification of methylation sites in Thermotoga maritima chemotaxis receptors. J Bacteriol. 2006;188:4093–4100. doi: 10.1128/JB.00181-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ortega DR, Yang C, Ames P, Baudry J, Parkinson JS, Zhulin IB (2013) A phenylalanine rotameric switch for signal-state control in bacterial chemoreceptors. Nat Commun 4:2881. doi:10.1038/ncomms3881 [DOI] [PMC free article] [PubMed]
- 93.Mowery P, Ostler JB, Parkinson JS. Different signaling roles of two conserved residues in the cytoplasmic hairpin tip of Tsr, the Escherichia coli serine chemoreceptor. J Bacteriol. 2008;190:8065–8074. doi: 10.1128/JB.01121-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pedetta A, Parkinson JS, Studdert CA. Signalling-dependent interactions between the kinase-coupling protein CheW and chemoreceptors in living cells. Mol Microbiol. 2014;93:1144–1155. doi: 10.1111/mmi.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Piasta KN, Ulliman CJ, Slivka PF, Crane BR, Falke JJ. Defining a key receptor–CheA kinase contact and elucidating its function in the membrane-bound bacterial chemosensory array: a disulfide mapping and TAM-IDS Study. BioChemistry. 2013;52:3866–3880. doi: 10.1021/bi400385c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26:257–265. doi: 10.1016/S0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ames P, Hunter S, Parkinson JS. Evidence for a helix-clutch mechanism of transmembrane signaling in a bacterial chemoreceptor. J Mol Biol. 2016;428:3776–3788. doi: 10.1016/j.jmb.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qea Zhou. Mutational analyses of HAMP helices suggest a dynamic bundle model of input–output signalling in chemoreceptors. Mol Microbiol. 2009;73:801–814. doi: 10.1111/j.1365-2958.2009.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Park H, Im W, Seok C. Transmembrane signaling of chemotaxis receptor Tar: insights from molecular dynamics simulation studies. Biophys J. 2011;100:2955–2963. doi: 10.1016/j.bpj.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Coleman MD, Bass R, Mehan RS, Falke JJ. Conserved glycine residues in the cytoplasmic domain of the aspartate receptor play essential roles in kinase coupling and on–off switching. BioChemistry. 2005;44:7687–7695. doi: 10.1021/bi0501479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Szurmant H, Ordal GW. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Falke JJ, Bass RB, Butler SL, Chervitz SA, Danielson MA. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ottemann KM, Lowenthal AC. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect Immun. 2002;70:1984–1990. doi: 10.1128/IAI.70.4.1984-1990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol. 2007;5:355–362. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- 105.Fox JE, Gulledge J, Engelhaupt E, Burow ME, McLachlan JA. Pesticides reduce symbiotic efficiency of nitrogen-fixing rhizobia and host plants. Proc Nat Acad Sci USA. 2007;104:10282–10287. doi: 10.1073/pnas.0611710104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stocker R, Seymour JR. Ecology and physics of bacterial chemotaxis in the ocean. Microbiol Mol Biol Rev. 2012;76:792–812. doi: 10.1128/MMBR.00029-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brennan CA, DeLoney-Marino CR, Mandel MJ. Chemoreceptor VfcA mediates amino acid chemotaxis in Vibrio fischeri . Appl Environ Microbiol. 2013;79:1889–1896. doi: 10.1128/AEM.03794-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shao Z, Wang W. Enzymes and genes involved in aerobic alkane degradation. Front Microbiol. 2013;4:116. doi: 10.3389/fmicb.2013.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]