Abstract
The Streptococcus-derived CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas9 (CRISPR-associated protein 9) system has emerged as a very powerful tool for targeted gene modifications in many living organisms including plants. Since the first application of this system for plant gene modification in 2013, this RNA-guided DNA endonuclease system has been extensively engineered to meet the requirements of functional genomics and crop trait improvement in a number of plant species. Given its short history, the emphasis of many studies has been the optimization of the technology to improve its reliability and efficiency to generate heritable gene modifications in plants. Here we review and analyze the features of customized CRISPR/Cas9 systems developed for plant genetic studies and crop breeding. We focus on two essential aspects: the heritability of gene modifications induced by CRISPR/Cas9 and the factors affecting its efficiency, and we provide strategies for future design of systems with improved activity and heritability in plants.
Keywords: CRISPR-Cas, sgRNA, Gene editing, Mutagenesis, Inheritance
Introduction
Since 1996, when the first genetically modified seeds were planted in the United States for commercial use, genetically modified crops have been grown on an accumulated area of more than 1.8 billion hectares worldwide. So far, about 80 % of the area used to grow soybeans and 70 % of cotton growing area used genetically modified cultivars [1]. Some of the benefits brought by genetic modification in agriculture to the world’s growing population are increased crop yields, reduced costs for food production, reduced need for pesticides, improved food quality, resistance to biotic and abiotic stress and increased health benefits [2]. Over the years, plant genetic modification has been based on the incorporation of foreign DNA into the plant genome by plant transformation but even though transformation methods have become quite efficient, the insertion site of the transferred DNA fragments as well as the number of copies cannot be controlled [3]. With the widespread adoption of transgenics, genetically modified crops have triggered an intense debate about their potential or perceived health and environmental risks and appeals for more precise gene modification technologies are increasing [4]. In contrast to the established genetic modification methods, targeted gene modification is able to manipulate plant genes in a site-specific manner making them very appealing for functional genomics research and crop improvement [5].
The concept of targeted gene modification encompasses precise gene editing and targeted gene mutagenesis. Precise gene modification, also known as gene targeting, was first achieved in mammalian cells via homology-directed repair (HDR). The HDR process is naturally activated during the pairing of inherited chromosomes in fertilized cells, allowing the exchange of parental DNA fragments and thus generating new genetic variation in the population [6]. To harness this process for precise gene modification, the most popular strategy is to introduce DNA templates flanked with sequences homologous to the target site into reproductive cells [7]. The first successful report of targeted gene modification in plants was reported in 1988 [8]. Using tobacco protoplasts, a DNA fragment containing part of the neomycin phosphotransferase (NPTII) gene conferring kanamycin resistance was integrated into the targeted genomic locus via HDR, though at a very low frequency (0.5–4.2 × 10−4). This frequency is at least ten times lower than the one reported for mammalian cells, suggesting that it would be almost impossible to perform gene modification in plant cells without the use of a selectable marker. Agrobacterium-mediated gene transformation of plant explants was later used to create heritable gene modifications in Arabidopsis whole plants using a partially duplicated beta-d-glucuronidase (GUS) gene as reporter [9]. Successful recombination events were observed in somatic and meristematic cells of the developing shoot apex. This outstanding work also suggested that in the absence of selection, plant somatic recombination frequencies ranged between 10−6 and 10−7 events/genome.
An important step towards the practical development of HDR in plant cells was reported in 1993, when the meganuclease I-SceI was introduced into Nicotiana plumbaginifolia protoplasts for site-specific generation of double strand breaks (DSBs) in the partially duplicated GUS reporter gene [10]. The frequency of extrachromosomal HDR was increased 4.5–10 times when I-SceI was co-expressed with the two non-functional GUS genes, strongly suggesting that DSBs promote the incidence of HDR in plants. To pave the way for the use of DSBs as a method for genome manipulation in plants, the frequency of DSB-induced HDR was analyzed in transgenic tobacco calli and Arabidopsis seedlings [11, 12]. In both cases, approximately 1 % of the transformants were successively targeted, increasing the frequency of gene modification in excess of 100 times and highlighting the potential of harnessing the DSB repair pathways for targeted gene modification in plants.
The application of site-specific endonucleases for DSB generation also contributed to the development of targeted mutagenesis in plants. In addition to the homology directed repair pathway, DSBs in eukaryotes can also be repaired via the error-prone non-homologous end joining (NHEJ) pathway [13]. It was shown that in the absence of repair templates, the DSBs induced by I-SceI within a negative selectable marker gene could be repaired by nucleotide insertions and deletions. Such incorrect DNA repairs resulted in loss-of-function mutations of the toxic marker gene that facilitated the identification of targeted mutations by screening for survival [14]. However, meganucleases can only recognize a specific DNA sequence, seriously limiting their application for targeted mutagenesis.
Two alternatives to meganucleases for targeted mutagenesis are zinc finger nucleases (ZFNs) and ‘transcription activator-like effector nucleases’ (TALEN). Both are chimeric proteins consisting of an engineered DNA-binding domain and the DNA cleavage domain from the FokI restriction endonuclease [15]. Compared to meganucleases, the DNA-binding domains used for target recognition in ZFNs and TALENs are more versatile. Each DNA-binding domain is composed of individual modules that can be engineered to recognize either a specific tri-nucleotide (ZFN) or mono-nucleotide (TALEN). In each case, a number of individual DNA-binding modules are usually assembled to form an array recognizing specific DNA sequences ranging from 12 to 20 nucleotides. [16–18]. The non-specific FokI DNA cleavage domain requires dimerization to cut double stranded DNA; therefore, two ZFNs or TALEN proteins are usually designed that each contain a DNA-binding domain targeting DNA sequences on opposite genomic strands [19, 20]. Both ZFNs and TALENs have been shown to generate targeted gene modifications in a number of plant species, but the targeted gene mutagenesis efficiency of ZFN was reported to be lower (1.7–19.6 %) than that of TALEN (30–48 %) [21–25]. Moreover, the specificity of ZFNs was not satisfactory [26]. Although TALENs seem to be superior to ZFNs in some aspects, the vector assembly process for both systems is quite laborious.
Emergence of the CRISPR/Cas9 system for targeted gene modification
Compared to ZFN and TALEN, the recently developed CRISPR/Cas9 system has established itself as the most efficient and versatile tool available for targeted gene modifications (Table 1). This endonuclease system was derived from the adaptive immune system of bacteria and archaea to fight against the attacks of invasive viruses and plasmids [27, 28]. A typical Type II CRISPR/Cas9 system is composed of a Cas9 endonuclease, capable of cleaving double stranded DNA (dsDNA) and two short RNAs, the CRISPR RNA (crRNA) and the trans-activating crRNA (tracrRNA) for target recognition [29]. In bacterial cells, the tracrRNA hybridizes to the repeat regions of the crRNA precursor directing crRNA processing by RNaseIII [30]. The mature crRNA forms a duplex with the tracrRNA to guide the associated Cas9 protein to its target via base-pairing. The three-component CRISPR/Cas9 system can be simplified by artificially fusing the tracrRNA to the 3′ end of crRNA to create a chimeric single guide RNA (sgRNA), streamlining the construction of CRISPR/Cas9 vectors and removing the need for the formation of the RNA duplex in vivo [29].
Table 1.
Comparison of ZFN, TALEN and CRISPR/Cas9 gene editing technologies
| Property/Tools | ZFN | TALEN | CRISPR/Cas9 |
|---|---|---|---|
| Type of recognition | Protein–DNA | Protein–DNA | RNA–DNA |
| Cleavage activity | Paired nickase | Paired nickase | Nuclease |
| Methylation sensitivity | Sensitive | Sensitive | Insensitive |
| Module assembly | Complicated | Somewhat | Simple |
| Off-target effects | More | Less | Variable |
| Multiplexing | Rarely used | Rarely used | Capable |
The target specificity of the Cas9 endonuclease is determined by the first 20 nt of the chimeric sgRNA and the presence of a Protospacer Adjacent Motif (PAM), a 3-nt sequence (NGG) located immediately downstream of the sgRNA target site and required for Cas9-mediated DNA cleavage. In the bacterial immune system, the PAM motif serves as a tag allowing Cas9 to discriminate non-infectious “self” from infectious “non-self” [31]. Although every base within the 20 nt guide sequence contributes to the overall specificity, mismatches close to the PAM-distal end of the guide RNA are usually tolerated to a greater degree than those at the PAM-proximal end [29, 32]. The remaining 65 nt of the full length sgRNA (85 nt) form a secondary structure that allows its incorporation into the Cas9 protein. Analysis of the crystal structure of the Streptococcus pyogenes Cas9 protein in complex with the sgRNA and target DNA, has revealed the presence of two main lobes: the recognition lobe is responsible for binding the sgRNA:DNA duplex and the nuclease lobe for target cleavage [33]. The nuclease lobe contains two distinct nuclease domains, HNH and RuvC, for cleavage of the complementary and non-complementary strands of the target DNA, respectively. In addition, a carboxyl-terminal domain in the nuclease lobe interacts with the PAM. To produce a functional system in eukaryotic cells, additional sequences such as nuclear localization signals must be artificially added to the original Cas9 gene [34, 35]. Conventionally, sgRNA transcription is controlled by RNA polymerase III dependent promoters, while Cas9 expression is driven by RNA polymerase II dependent promoters (Fig. 1). In some instances, RNA polymerase II dependent promoters, such as the cauliflower mosaic virus (CaMV) 35S promoter, has also been used to drive the expression of sgRNAs in combination with the Nos terminator [36, 37], which suggested the flexibility of sgRNA engineering.
Fig. 1.
A schematic showing the conventional structure of the CRISPR/Cas9 cassette and the mechanism of action of the engineered CRISPR/Cas9 system. To achieve efficient gene expression in plant cells, a Pol III-dependent promoter and a poly T terminator are used to drive transcription of the sgRNA, while Cas9 transcription is controlled by a Pol II-dependent promoter and a corresponding terminator. Once transcribed, the 85 nt sgRNA is folded and incorporated into the Cas9 protein, guiding the complex to the targeted gene locus by RNA-DNA base pairing. The DNA cleavage activity of the Cas9 endonuclease complex also requires the presence of a PAM motif located immediately downstream of the 20 nt guide sequence. T poly T terminator, NLS nuclear localization signal
Successful application of the engineered CRISPR/Cas9 system for targeted gene modification was first reported in mammalian cells [34, 35]. Shortly after the initial report, the CRISPR/Cas9 system was successfully used for gene modification in Arabidopsis, tobacco, rice and wheat [38]. Subsequently, a number of reports highlighted the potential of this system for multiple applications in numerous plant species (Table 2). Most of the developments have been focused on two aspects: the activity of CRISPR/Cas9 system in plant cells and the heritability of the CRISPR/Cas9 induced gene modifications and to some extent, these two factors are correlated.
Table 2.
Summary of the adaption of CRISPR/Cas9 system for targeted gene modification in various plant species
| Species | CRISPR/Cas9 system | Transformation methods | References |
|---|---|---|---|
| Arabidopsis | DNA-free | Protoplast transfection | [62] |
| Constitutive | Protoplast transfection | [43, 70, 72] | |
| Constitutive | Agroinfiltration | [43, 140] | |
| Constitutive | Agrobacterium mediated in planta | [55, 70–72, 95, 105, 106, 141, 142] | |
| Meristem specific | Agrobacterium mediated in planta | [55, 119, 120] | |
| Germline specific | Agrobacterium mediated in planta | [112, 116] | |
| Tobacco | DNA-free | Protoplast transfection | [62] |
| Constitutive | Protoplast transfection | [43] | |
| Constitutive | Agroinfiltration | [36, 42, 140, 142] | |
| Constitutive | Agroinfiltration followed by tissue regeneration | [42] | |
| Viral-based | Agroinfiltration | [61, 84, 132] | |
| Viral-based | Agroinfiltration followed by tissue regeneration | [132] | |
| Rice | DNA-free | Protoplast transfection | [62] |
| Constitutive | Protoplast transfection | [50, 101, 109, 140, 142, 143] | |
| Constitutive | Agrobacterium-mediated | [54, 70, 72, 79, 104, 106, 109, 128, 142, 143] | |
| Constitutive | Biolistic-mediated | [50] | |
| Wheat | Constitutive | Protoplast transfection | [50] |
| Constitutive | Agrobacterium-mediated | [36] | |
| Constitutive | Biolistic-mediated | [81] | |
| MAIZE | Constitutive | Protoplast transfection | [102, 105, 144, 145], |
| Constitutive | Agrobacterium-mediated | [102, 105, 145, 146], | |
| Constitutive | Biolistic-mediated | [146] | |
| Barley | Constitutive | Agrobacterium-mediated | [103] |
| Soybean | Constitutive | Protoplast transfection | [44] |
| Constitutive | Hairy root transformation | [44, 45, 53, 75, 77] | |
| Constitutive | Agrobacterium-mediated | [53] | |
| Constitutive | Biolistic-mediated | [75, 76] | |
| Tomato | Constitutive | Hairy root transformation | [147] |
| Constitutive | Agrobacterium-mediated | [46, 148] | |
| Viral-based | Protoplast transfection | [133] | |
| Viral-based | Agrobacterium-mediated | [133] | |
| Potato | Constitutive | Agrobacterium-mediated | [47, 149] |
| Viral-based | Agrobacterium-mediated | [47] | |
| Petunia | DNA-free | Protoplast transfection | [150] |
| Constitutive | Agrobacterium-mediated | [48] | |
| Brassica oleracea | Constitutive | Agrobacterium-mediated | [103] |
| Lettuce | DNA-free | Protoplast transfection followed by tissue regeneration | [62] |
| Sorghum | Constitutive | Agrobacterium-mediated | [140] |
| Populus | Constitutive | Agrobacterium-mediated | [49] |
| Sweet organe | Constitutive | Agroinfiltration | [37] |
| Medicago truncatula | Constitutive | Hairy root transformation | [45] |
| Liverwort | Constitutive | Agrobacterium-mediated | [151] |
CRISPR/Cas9 systems engineered to generate heritable gene modifications in plants
The activity of the CRISPR/Cas9 system is affected by many factors, including the transcript levels of the transgenes, the translational efficiency of Cas9 and the accessibility to the gene targets. The expression of the sgRNA has been traditionally driven by RNA polymerase III dependent promoters, such as the U3 or U6 promoters [39]. In eukaryotic cells, RNA polymerase III transcribes a number of non-coding RNAs, such as the ribosomal 5S rRNA, tRNAs and other small RNAs [40]. These Pol III-transcribed housekeeping genes are expressed in all cell types and under most environmental conditions, therefore reasonably high transcription levels of the sgRNA in transgenic plants can be expected when using their promoters. In addition, the regulatory elements and transcriptional characteristics of these genes are well defined. For example, the transcription of U6 genes is initiated 26 bp downstream of the TATA box and terminated by a small polyT stretch [41]. Using this knowledge, the exact sequence and length of the sgRNA transcripts can be controlled. Importantly, the U6 promoters from dicotyledonous and monocotyledonous plants are exchangeable within classes, but not between classes. For example, sgRNAs driven by an Arabidopsis U6 promoter are efficiently transcribed in tobacco [42, 43], soybean [44, 45], tomato [46], potato [47], petunia [48] and poplar [49], but not in monocotyledonous plants, such as rice and wheat [50]. Nevertheless, endogenous promoters seem to drive higher levels of sgRNAs transcription than exogenous ones [44]. Strong constitutive RNA Polymerase II dependent promoters have been the most popular choice to achieve high levels of Cas9 expression in plant cells [39]. The CaMV 35S promoter is the most widely used promoter to drive transgene expression in plants, specially dicot species although it is relatively less effective in monocots for which ubiquitin promoters such as the maize ZmUBI have been favored [51, 52]. Multiple studies have shown that the level of promoter activity plays a critical role in the efficiency of the customized CRISPR/Cas9 system for plant gene modification [53–55].
Plant codon optimized Cas9 genes have been used to further increase the expression levels of the CRISPR/Cas9 system with over ten different versions engineered so far in various plant species [56]. Unlike ZFN and TALEN, which were derived from nuclear localized transcription factors, the endonuclease activity of the native Cas9 protein occurs in the cytoplasm to target invading DNA. Therefore, usually one or two nuclear localization signal peptides are fused to the ends of the codon optimized Cas9 proteins to direct their transport into the nucleus [38].
Conventionally, CRISPR/Cas9 expression cassettes are introduced into the plant genome using either Agrobacterium-mediated or biolistic transformation methods. Although both methods can cause integration of multiple copies of the transgenes, particle bombardment generally results in much higher transgene copy numbers than Agrobacterium-mediated transformation [57]. Multiple integration events, often coupled with inverted repeat T-DNA integration patterns, increase the frequency of transgene silencing [58] and will complicate the removal of the transgenes from the progeny by backcrossing or simple Mendelian segregation. Removal of the transgenes after induction of the intended gene modification is extremely important as it will minimize the chances of non-target effects. In addition, as long as there are no transgenes present, in some cases the progeny of CRISPR/Cas9 edited plants might be classified as non-GM, exempting them from regulatory approval, a fact that has immense commercial implications [59]. As an alternative to the physical integration of the CRISPR/Cas9 sequences into the genome, DSBs could be achieved by transient expression of Cas9 and the sgRNA using non-integrative vectors thus avoiding the GM intermediate step [60, 61]. DNA-free systems have also been used to induce DSBs by introducing preassembled sgRNA and Cas9 ribonucleoproteins directly into the cells [62]. Therefore, heritable gene modifications can be obtained in plants by either stable or non-stable transformation methods as summarized in Table 3.
Table 3.
Summary of the applicable strategies used to generate heritable gene modifications in plants
| CRISPR/Cas9 systems | Genetic materials | Expressional regulation | Plant materials | Transformation methods | sgRNA multiplexing | Heritable modifications |
|---|---|---|---|---|---|---|
| Integrative | T-DNA based | Constitutive, spatial-temporal or inducible | Seedlings (flowering or root tissues) | Agrobacterium-mediated | Tandem repeats | Via fertilization |
| T-DNA based | Constitutive or inducible | Cultured embryonic cells | Agrobacterium-mediated | Tandem repeats or mixture | Via tissue regeneration | |
| Plasmid-based or linearized DNA | Constitutive or inducible | Cultured embryonic cells | Biolistic or microinjection | Mixture | Via tissue regeneration | |
| Non-integrative | Plasmid-based or linearized DNA | Constitutive | Protoplasts | PEG or electroporation | Mixture | Via tissue regeneration |
| Virus-based | Rep binding motifs or viral promoter | Seedlings (leaf tissues) | Leaf infiltration | Mixture | Via fertilization or tissue regeneration | |
| DNA-free (mRNA or ribonucleoprotein) | Naked or chemical modified | Protoplasts | PEG or electroporation | Mixture | Via tissue regeneration |
Heritable gene modifications obtained via sexual propagation using integrative CRISPR/Cas9 systems
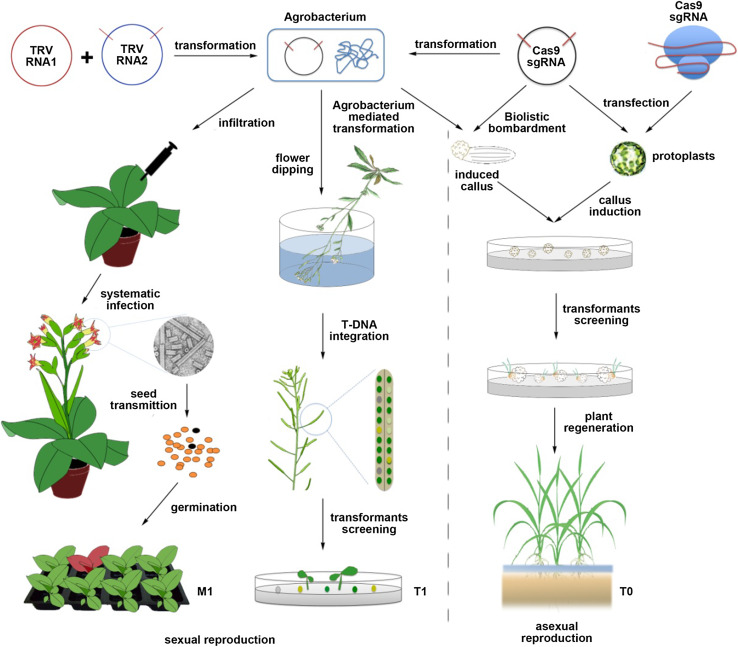
Two different strategies are usually applied to generate heritable gene modifications in plants, depending on the method used for plant reproduction (Fig. 2). Generally speaking, the production of new offspring can be accomplished via sexual or asexual reproduction methods [63]. Sexual reproduction produces offspring by the fusion of gametes involving two fundamental processes: meiosis, which rearranges fragments of genomic DNA and halves the number of chromosomes, and fertilization, which restores the chromosome to the pre-meiosis number. The offspring of sexually reproduced plants are genetically different from their parents due to the recombination of homologous chromosomes [63]. Theoretically, to generate heritable gene modifications via sexual reproduction, targeted gene modifications should occur either in gametes during or after meiosis, or in zygotes during fertilization. In plants, however, somatic cells within the meristems can differentiate into reproductive cells opening the door to alternative strategies aiming to achieve targeted gene modification in meristematic cells during vegetative growth [64, 65].
Fig. 2.
Methods used to generate heritable gene modifications in plants by CRISPR/Cas9. To obtain heritable gene modifications by CRISPR/Cas9, at least three different transformation methods can be used. In planta transformation by flower dipping is the method of choice for Arabidopsis. Heritable gene modifications can be created in the T1 generation by early expression of CRISPR/Cas9 in egg cells. Targeted gene modification is also possible using viruses as vehicles for CRISPR/Cas9 delivery, although only those that can infect meristematic tissues or be transmitted through seeds are suitable for this application. Tissue regeneration from genetically modified calli is suitable for most crop species with delivery being provided by plasmids or preassembled ribonucleoproteins. Both Agrobacterium-mediated and biolistic methods can be used to deliver the CRISPR/Cas9 DNA constructs into calli, whereas the preassembled CRISPR/Cas9 complex can only be delivered into protoplasts by cell transfection before calli induction. In all cases, homozygous mutants can be obtained in the first transformed generation; however, the frequency is far higher in plants regenerated from protoplasts
Agrobacterium-mediated transformation is the prevalent method for genetic modification in plants, being efficient for a wide spectrum of plant species [66]. Although Agrobacterium-mediated transformation usually involves a laborious tissue culture step, the development of in planta transformation methods in Arabidopsis thaliana incredibly simplified the process making this species the preferred model for genetic studies over the last 20 years [67, 68]. The use of in planta transformation for CRISPR/Cas9 delivery has been widely adopted for the routine and efficient generation of heritable gene modifications via sexual reproduction [69]. Using constitutive promoters for Cas9 expression, such as the CaMV 35S, targeted gene modifications have been achieved in T1 Arabidopsis plants at a frequency of 30–84 % although the frequency was reduced to 20–54 % in the T2 progeny [70, 71]. In most cases, only 15–25 % of the mutations present in the T2 generation were also found in the T1 generation, suggesting a high degree of chimerism in T1 plants and the possibility that many of the somatic mutations were not inherited. In contrast, use of the AtUBQ1 promoter for Cas9 expression resulted in a mutagenesis frequency of 74–92 % in T1 plants without a noticeable reduction in the subsequent generation (58–96 %) with a relatively large proportion of the T2 mutations (37–66 %) present in the T1 generation [71, 72]. Although the mutagenesis efficiency of these two CRISPR/Cas9 systems is not comparable due to the difference in target sites, the AtUBQ1 promoter appears to be more efficient than the CaMV 35S promoter for generating heritable gene mutations. These differences might be attributable to the expression characteristics of each promoter as there is evidence suggesting that even though both are considered to be constitutive, neither the CaMV 35S promoter nor the AtUBQ1 promoter are uniformly expressed in plants. Studies using the GUS reporter gene have shown that the expression patterns of these two promoters are different in tobacco transgenic lines with the AtUBQ1 promoter generating strong staining in pollens and ovules of immature flowers, whereas adjacent vascular tissues showed lower staining levels. In contrast, weak staining was observed in pollens of CaMV 35S transgenic lines, whereas the vascular tissues exhibited the strongest signal [73]. In addition, the CaMV 35S promoter was unable to drive gene expression in young zygotic embryos [74]. The differences observed in reproductive cells suggest that the AtUBQ1 promoter might be more suitable than the CaMV 35S promoter to generate heritable gene modifications in plants like Arabidopsis. Besides, it was also found that neither promoter generated homozygous, bi-allelic or heterozygous mutants in the T1 generation, with all of the analyzed T1 mutants being chimeras (i.e. individuals containing a mix of mutated and non-mutated cells). In such plants, the mutations induced by the incorporated CRISPR/Cas9 cassette occurred after the division of zygotes, explaining why the genotypes of the targeted sites are not uniform within the same individual plant. However, mutants with all zygotic types were identified in the T2 generation at variable frequencies. In the absence of the Cas9 gene, progeny from T2 mutants segregated according to the Mendel’s law, whereas in the presence of Cas9, new mutation types were obtained from heterozygous, chimeric and wild type seedlings, but not from homozygous or bi-allelic ones, suggesting that the CRISPR/Cas9-induced gene mutations in Arabidopsis were faithfully inherited [71]. In summary, the ideal scenario to obtain stably heritable gene modifications using integrative CRISPR/Cas9 systems, is to remove the CRISPR/Cas9 cassette as soon as possible from the transgenic lines. But even if the transgenes are not removed, the specificity of CRISPR/Cas9 system for target recognition will prevent it from introducing new mutations in the modified gene loci.
Heritable gene modifications obtained via asexual propagation using integrative CRISPR/Cas9 systems
For most crop species, the in planta transformation method is not feasible and it is necessary to regenerate transgenic plants from explants via asexual reproduction [66]. To perform targeted gene modification in those species, both Agrobacterium-mediated transformation and the biolistic particle system can be used to deliver the CRISPR/Cas9 cassette into the plant cells (Fig. 2). In these cases, the heritability of the targeted gene modifications almost equals the mutagenesis frequency of transformed cells. Assuming that no cellular toxicity is conferred by the induced gene modification, the gene-edited seedlings will be regenerated from transformed cells without frequency change. To maximize expression levels, and the mutagenesis efficiency of the CRISPR/Cas9 system, the endogenous U6 and the CaMV 35S promoters have been routinely used to drive sgRNA and Cas9 transcription, respectively, while high genetic transformation efficiency has been achieved by the use of suitable transformation vectors, Agrobacteria strains and explant materials [56].
Six different studies have used the CRISPR/Cas9 system to perform targeted gene modifications in soybean, five of which used Agrobacterium rhizogenes-mediated gene transformation for quick detection of CRISPR/Cas9-induced gene mutations in hairy roots [44, 45, 53, 75–77]. This bacterium can infect a number of dicotyledonous plants upon wounding and generate “composite plants”, i.e., plants with wild-type shoots and transgenic roots, by transferring its T-DNA to the plant genome [78]. Considering that each transgenic hairy-root represents an independent transformation event, high numbers of transformants can be obtained and analyzed with this method. One study developed a soybean codon optimized CRISPR/Cas9 system to perform targeted gene editing in hairy root of both soybean and Medicago truncatula [45]. Another reported targeted mutagenesis with efficiencies as high as 95 % using a combination of the M. truncatula U6.6 polymerase III promoter to drive sgRNA transcription and the double-enhanced CaMV 35S promoter for Cas9 expression in hairy roots [75]. Two other studies suggested that the soybean endogenous U6 promoter is more effective than the Arabidopsis U6 promoter to generate targeted gene modifications in soybean [44, 53]. The rest showed that two endogenous soybean genes can be targeted simultaneously using only one AtU6 promoter-regulated sgRNA module [77]. Although hairy root transformation allows for the quick identification of CRISPR/Cas9-induced gene modifications and facilitates optimization of the system, regeneration of whole plants from transgenic hairy roots is difficult, making stable transformation the method of choice to obtain heritable mutations in soybean. Therefore, biolistic-mediated transformation was applied to deliver the CRISPR/Cas9 system into soybean somatic embryo cultures [75, 76]. These embryonic cell cultures were usually obtained from non-zygotic immature plant tissues and can be cultured to form undifferentiated cell masses before tissue regeneration. Jacobs et al. showed that only 5 of the 31 recovered transgenic lines contained a complete Cas9 gene, but continuous Cas9 expression leads to additional mutations during the development of somatic embryos [75]. Li et al. reported that a high mutagenesis frequency at the targeted gene loci (up to 76 %) was achieved by using a CRISPR/Cas9 system driven by soybean elongation factor gene promoter (EF1A2). Targeted gene integration was also achieved at a frequency of 3.8–4.6 % and the integrated genes were successfully transmitted to the T1 generation and segregated according to Mendelian laws, demonstrating the feasibility of the CRISPR/Cas9 system in soybean [76]. Besides, Agrobacterium tumefaciens-mediated transformation was also used to target the phytoene desaturase (PDS) gene in soybean. The expected dwarf and albino phenotype was observed in adventitious buds regenerated from the inoculated cotyledon node [53]. Together, these studies demonstrated the feasibility of obtaining heritable gene modification using integrative CRSIPR/Cas9 systems.
A more detailed study of the heritability of CRISPR/Cas9-induced gene mutations was performed in rice using a constitutive promoter to drive Cas9 expression and Agrobacterium-mediated transformation of somatic embryos [79]. Targeted gene mutagenesis was detected at an average frequency of 44.4 % in the T0 generation and the presence of homozygous, bi-allelic, heterozygous and chimeric individuals confirmed. Notably, 3.8 % of the analyzed T0 plants were homozygous, suggesting that targeted gene modifications in these seedlings occurred before the first round of embryonic cell division and the mutation types of the two gene alleles happened to be the same. In contrast, Arabidopsis studies using Cas9 driven by constitutive promoters and Agrobacterium-mediated in planta transformation produced only chimeras in the first generation, perhaps reflecting the fact that this transformation method targets the ovule and therefore the only chance of obtaining homozygous and bi-allelic mutants with this transformation method is by inducing mutations before the first division of the zygote.
Polyploid crop species pose an additional challenge as their genomes are larger and highly repetitive compared to diploid species. A recent report described the application of TALEN and CRISPR tools in bread wheat, an allohexaploid in which most of the genes have three similar but not identical copies [80, 81]. While TALEN was used to simultaneously edit all three homoeoalleles that encode the MILDEW-RESISTANCE LOCUS (TaMLO) protein, CRISPR constructs targeted just one allele. Genetic transformation of a winter wheat variety was performed using immature embryos via particle bombardment. Twenty seven mutations were identified in 450 independent T0 transgenic TALEN lines (6.0 %), with only one line containing heterozygous mutations for all the three homoeoalleles. The mutagenesis frequency induced by CRISPR/Cas9 was very similar to that observed in TALEN lines with four mutations generated in the TaMLO-A1 allele out of 72 T0 transgenic lines (5.6 %). Two homozygous mlo triple mutants lacking the TALEN transgene cassette were obtained by self-pollination and segregation in two generations. As expected, the triple mutants exhibited broad-spectrum resistance to powdery mildew [81].
Heritable gene modifications obtained via asexual propagation using non-integrative CRISPR/Cas9 systems
Integration of the CRISPR/Cas9 coding sequences into the plant genome is not an absolute requirement to perform in vivo gene modification. In animals, in vitro transcribed RNAs encoding Cas9 and sgRNA can be directly injected into zygotes to generate homozygous and biallelic mutants with high frequency [82]. However, the plant cell wall provides a strong physical barrier that handicaps the application of mechanical approaches such as microinjection, electroporation and PEG-mediated DNA transfection. To overcome this obstacle, plant protoplasts can be used as the target for transfection with the added advantage that the delivered cargos can be DNA, RNA or even proteins. In the case of CRISPR/Cas9, the delivery of preassembled ribonucleoproteins rather than DNA plasmids can appease concerns about non-intended incorporation of plasmid fragments into the host genome [62, 83]. Using this delivery method, targeted mutagenesis was achieved at a frequency ranging from 8.4 to 44 % in transfected Arabidopsis thaliana, tobacco, lettuce and rice protoplasts. The resulting gene modifications were maintained after the regeneration of microcalli from transfected protoplasts and the overall mutation frequency in lettuce calli was up to 46 %. It was also shown that the targeted gene modifications present in regenerated plants can be transmitted to the germline and inherited by their offspring. Although promising, the main drawback of this approach is that regeneration of plants from protoplasts is feasible in only a few plant species.
Heritable gene modifications obtained via sexual propagation using non-integrative CRISPR/Cas9 systems
As an alternative, viral vectors have been used to attempt gene modification using non-integrative expression of CRISPR/Cas9 in host cells. Systemic infection of N. benthamiana leaves with tobacco rattle virus (TRV) has been used to deliver sgRNA by incorporating it into a RNA2 replicon-based vector [84]. TRV is a bipartite, positive-sense RNA virus and its RNA2 genome can be modified to carry foreign gene fragments (usually less than 2–3 Kb) into plant cells by agroinoculation. The incorporated sgRNAs was under the control of the pea early browning virus promoter to facilitate its transcription from the virus replicon. Systemic expression of the engineered sgRNA modules was achieved by co-inoculating the TRV1- and TRV2-carrying Agrobacterium strains into 3-week-old plant leaves [85]. However, this viral system does not have the capacity to include the Cas9 gene due to the large size of Cas9 gene, and thus the TRV inoculation must be performed on stable transgenic plants expressing the Cas9 protein. Targeted gene mutagenesis was detected 10 days post-inoculation in inoculated and systemic leaves at a frequency of 56 and 30 %, respectively. Notably, two seedlings carrying targeted gene mutations were recovered from the progeny suggesting that the virus-based system is able to generate heritable gene modifications in plants. In a similar approach, another study attempted to use the bipartite begomovirus Cabbage Leaf Curl virus (CaLCuV) as a vector for sgRNA delivery [61]. CaLCuV is a DNA virus in the Geminiviridae family that replicates in the nucleus and encodes seven proteins in two separate genomes. T-DNA vectors based on these two genomes have been developed to deliver foreign genes into plant cells by agroinoculation [86, 87]. In order to adapt the viral replicons for targeted gene modification in host plants, the AtU6-sgRNA expression module was constructed in the genome A-based vector to inoculate Cas9-expressing transgenic plants along with the genome B-based vector. Although mutagenesis frequency of the endogenous PDS gene in systemically infected leaves was up to 85 %, no heritable mutations were produced in the offspring of inoculated plants [61]. A possible explanation for this apparent disparity is that the CaLCuV virus does not move to all plant tissues and often exists in mature cells which do not divide, whereas TRV has a low chance to be transmitted to the offspring through the germline [88]. Therefore, to induce heritable gene modifications using the CaLCuV-based system it is necessary to regenerate plants from systemically infected tissues.
Target specificity of the CRISPR/Cas9 system in plants
Target specificity is arguably the most important feature of the CRISPR/Cas9 technology and multiple studies have been performed to systematically examine the specificity of the CRISPR/Cas9 system both in vivo and in vitro [32, 89–91]. In general, the nucleotides in the PAM and the PAM-proximal sgRNA sequence are crucial for target recognition while PAM-distal sequences can tolerate some, but not many, mismatches. However, the number and position of tolerable mismatches between the DNA target and the guide sequence seems to be target-dependent, making it difficult to define simple rules for sgRNA design based on the available data from these studies [92]. In addition to the positional effects, a trade-off between specificity and activity has been found in mammalian cells, where shorter guide RNAs can be more specific than longer ones and high concentrations of CRISPR/Cas9 complex can cleave off-target sites that are not usually cleaved when the complex concentration is low [32, 90, 93]. To minimize the incidence of off-target effects, a nickase version of Cas9 (Cas9n) was engineered by mutating its N terminal RuvC-like nuclease domain. In order to produce DSBs with Cas9n it is necessary to express two different sgRNAs complementary to opposite strands of the target site, effectively extending the target recognition site from 20 to 40 nt [91, 94]. Application of this strategy in mammalian cells was reported to reduce the off-target activity by 50–1500 folds without sacrificing on-target cleavage efficiency [94]. However, use of the nickase in Arabidopsis resulted in a 740-fold reduction of NHEJ-mediated mutagenesis compared to the regular CRISPR/Cas9 system but similar HDR frequencies [95].
In plants systems, the potential off-target effects of the CRISPR/Cas9 system have been analyzed using two strategies. The first strategy uses the BLASTN algorithm to predict candidate off-target sites by sequence similarity and later determines the existence of mutations on the predicted off-target candidates. Although the parameters used for candidate selection are somewhat hard to determine due to the lack of strong rules to predict mismatch tolerance of the CRISPR/Cas9 system, a number of webtools have been developed to assist the prediction of sgRNAs off-targets based on the number and position of potential mismatches in plant genomes [96–98]. The second approach is to perform whole genome re-sequencing followed by evaluation of the genome editing outcomes using analytic tools [98, 99]. Employing these strategies, no off-target effects were detected in Arabidopsis T1 and T2 plants obtained using the Agrobacterium-mediated flower dipping method [71, 100]. A limited number of off-target mutations were identified in rice, soybean, Brassica olerecea, barley and maize transgenic plants produced via either biolisitic or Agrobacterium-mediated transformation of cultured cells [44, 50, 79, 101–103]. The observed differences in off-target effects among species could be the result of differences in the expression levels of the CRISPR/Cas9 components, which is also correlated with the dosage of CRISPR/Cas9 reagents delivered into plant cells. In summary, the CRISPR/Cas9 system seems to have very low incidence of off-target effects in plants and even in the cases when they occurred, they could be easily removed by backcrossing.
Developing multiplex CRISPR/Cas9 systems in plants
Compared to Zinc-finger and TAL-effector proteins, the innate multiplex crRNA array makes the recognition of multiple DNA targets much simpler for CRISPR/Cas9 systems. The use of CRISPR/Cas9 to simultaneously edit two endogenous gene loci was first reported in mammalian cells by co-expressing either two crRNAs or two sgRNAs [34, 35]. However, the dual-crRNA constructs seemed to be less efficient than the sgRNA constructs for generating targeted mutagenesis in plants probably due to the inefficient processing of the mature crRNA by the tracerRNA in vivo [104]. A more efficient way to achieve multiplex gene modifications in plants via agrobacterium-mediated methods is to assemble multiple sgRNA modules into one binary vector for co-expression in transgenic plants [105–108]. Using this approach, homozygous individuals with multiple targeted mutations were successfully identified in the progenies of Arabidopsis, rice and maize transformants, suggesting that these multiple mutations are heritable. Alternatively, multiple targeting can be achieved in plants by adopting an interstitial tRNA architecture for sgRNA expression, with up to eight individual sgRNAs precisely excised from a single transcript by the plant native tRNA processing machinery [109]. Using this approach, very high efficiencies (up to 100 %) have been reported to simultaneously mutagenize four different loci in stable transgenic rice plants, although the possible quadruple mutants were not presented. A major concern when designing multiplex gene editing systems is to reduce the risk of triggering the plant gene silencing pathways, which are usually activated by highly repetitive DNA sequences. To reduce the repetitiveness of sgRNA modules, different RNA Polimerase III-dependent promoters have been used to drive the expression of the multiple sgRNAs [105–107].
In addition, multiplexing can also be achieved at the initial stage by providing multiple sgRNAs cloned into separate vectors or transcribed as sgRNAs in vitro for simultaneous delivery by biolisitic bombardment or cell transfection [43, 50]. Individual plants with heritable multiple mutations can be generated by tissue regeneration afterwards. In summary, selection of the ideal strategy for multiple gene modification largely depends on the plant species and the transformation methods available for the generation of transgenic lines.
Improving the heritability of CRISPR/Cas9-induced gene modifications
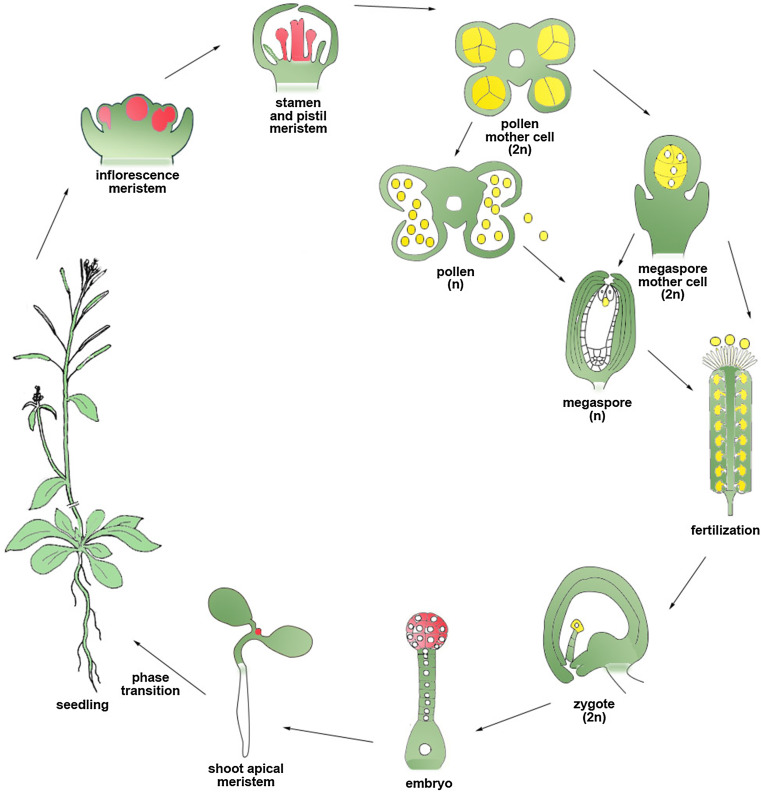
In CRISPR/Cas9 systems developed for stable transformation via sexual reproduction, the heritability of CRISPR/Cas9-induced gene modifications is strongly affected by the sgRNA and Cas9 promoter activities in reproductive cells. Two types of promoters can be used to improve heritability by optimizing expression of Cas9 in reproductive cells (Fig. 3). Germline-specific promoters are specifically expressed during gamete formation from gametocytes. Gametes are mature haploid cells generated from the successive division of diploid gametocytes. In flowering plants, the male gamete is produced inside the pollen grain and the female gamete is produced inside the embryo sac of the ovule. The Arabidopsis SPOROCYTELESS/NOZZLE (SPL) gene is a central regulator of cell division and differentiation in both anther and ovule [110, 111]. Early expression of this gene was observed in both anther and ovule primordia, but its transcription is much stronger in pollen mother cells (PMCs) than in megaspore mother cells (MMCs). Expression of Cas9 in the genomic context of the SPL gene was able to constrain the activity of the CRISPR/Cas9 system to germline cells [112]. Compared to a constitutively expressed CRISPR/Cas9 system, both the abundance and the diversity of heritable gene modifications were increased by this germline specific CRISPR/Cas9 system. However, due to the preferential expression of the SPL promoter in PMCs, only heterozygous mutants were obtained in the T2 population. Considering the large quantity of pollen cells in a single individual plant, application of this germline specific system will facilitate the screening of specific mutation types in offspring derived from a limited number of transgenic plants.
Fig. 3.
Optimal expression patterns to generate heritable gene modifications in Arabidopsis with the CRISPR/Cas9 system. During the plant life cycle, heritable gene modifications can be generated in dividing cells at the vegetative and reproductive growth stages. These dividing cells can be classified into two classes, meristematic cells (red) and germline cells (yellow). The meristematic cells including young embryo cells, shoot apical meristem, inflorescence meristems, floral meristems, stamen and pistil meristems can differentiate into germline cells. Generally speaking, the earlier the mutations are induced, the more uniform the mutation types are. Germline cells include ancestor cells that will give rise to pollen and megaspores as well as zygotes (egg cells after fertilization). Mutations induced in either of the two gametes can only generate heterozygotes in the progeny, whereas those induced in zygotes will produce homozygous or bi-allelic mutants
In addition to the SPL promoter, the same study reported the use of Arabidopsis DD45 and tomato LAT52 promoters to attempt germline specific gene modifications in Arabidopsis. The DD45 gene encodes an egg-cell-specific protein expressed in zygotes and early embryos [113, 114], while the tomato LAT52 gene is transcribed during the late stage of pollen development [115]. While the mutagenesis frequency of these two systems in the T1 population was comparable, the DD45 system produced more mutations in the T2 population, suggesting that it was more efficient than the LAT52 system in generating heritable gene modifications. Moreover, the presence of bi-allelic and homozygous gene modifications in the T1 population of the DD45 system indicated that targeted gene modifications were successfully induced in both alleles of the targeted genes before the first cell division of zygotes, though at a low frequency (1 of 36 for one of the targeted loci and 2 of 36 for the other). Interestingly, this study used the NOS terminator in the Cas9 genetic construct in combination with each promoter and a previous report had suggested that the choice of terminator had a strong influence on the efficiency in producing non-mosaic T1 mutants when using egg cell-specific promoters [116]. According to Wang et al., the mutagenesis efficiency of the CRISPR/Cas9 system when using the DD45/EC1.2 promoter to drive Cas9 could be sharply improved by the rbcS-E9t terminator compared to the NOS terminator [116]. The EC1.2 system was reported to produce homozygous etc2 try cpc triple mutants at a frequency of 8.3 % in T1 plants and the resulting mutations could be reliably passed to T2 plants; shortening the time needed to produce homozygous or bi-allelic mutants to one generation. In addition, the frequency of homozygous T1 triple mutants was further increased to 17 % by adding an additional enhancer to the native EC1.2 promoter. These studies clearly illustrate the importance of the regulatory elements used in the Cas9 expression cassette and the possibility to improve performance by manipulating them. Further improvements using similar strategies are still achievable as extensive expression studies of angiosperm male and female gametogenesis have provided a stock of additional regulatory elements yet to be tested [117, 118].
Besides germline-specific promoters, meristem-specific promoters are logical candidates to improve the heritability of targeted gene modifications induced by stably transformed CRISPR/Cas9 systems. All aerial organs in higher plants are generated from meristematic tissues in shoot apices, therefore any mutation induced in shoot meristematic cells can produce mutated somatic sectors in the plant throughout shoot development. Some of those sectors can develop into floral primordia and lead to the transmission of gene modifications through the gametes. A number of meristem-active promoters, such as INCURVATA2 (ICU2), APETALA (AP1), YAOZHE (YAO), and Histon H4 (AtH4) have been used to drive the expression of Cas9 in Arabidopsis [55, 100, 119, 120]. All of these systems were highly efficient in generating targeted gene modifications in dividing cells of T1 plants and heritable gene mutations were obtained in the T2 populations, but the severity and time of onset of mutations in T1 plants were different for each system. The ICU2 and YAO promoters are highly expressed in proliferating cells, including those in meristems, primordia, vegetative shoot, inflorescences and flowers [121–123]. Early expression of these two genes was identified in egg cells and young embryos after fertilization, therefore the resulting T1 mutants were usually chimeras produced during vegetative growth. In contrast, AP1 encodes a homeobox protein involved in the establishment of floral meristem [124], thus T1 mutations will be created after the transition from vegetative growth to reproductive growth. In summary, the earlier and broader the induction of mutagenesis occurs in the meristematic tissues of T1 plants, the more progeny with heritable gene mutations will be obtained.
Although a number of tissue-specific promoters have been tested to improve the heritability of CRISPR/Cas9-induced targeted gene modifications in plants, most efforts were focused on the Cas9 gene rather than the sgRNA. An innovative approach has used ribozyme-flanked sgRNAs (RGRs) as part of the CRISPR/Cas9 system [125]. Primary RGR transcripts undergo self-catalyzed cleavage to generate the desired gRNA containing 2′,3′-cyclic phosphate and 5′-hydroxyl modified termini. It has been suggested that such an RNA structure may be more stable in vivo because some nucleases require the 5′-terminal phosphate group for specific cleavage [126]. Application of this approach in yeast and mammalian cells resulted in high cleavage activity both in vitro and in vivo but it has not been tested in plants thus far [125, 127].
In the case of stable transformation via asexual reproduction, the heritability of CRISPR/Cas9-induced gene modifications can also be affected by the length of the culture period in addition to the expression level and transformation efficiency. Considering that that the promoters routinely used to drive the expression of the CRISPR/Cas9 system in transformed cells are already very strong, any improvements will need to address the remaining variable, culture length. In fact, extension of the culture period increased the proportion of mutated cells in transformed rice and soybean calli [53, 128]. However, prolonged tissue culture also increases the incidence of somaclonal mutations and decreases regeneration ability, which may hamper its application for crop improvement.
The heritability of targeted gene modifications in virus-based systems largely depends on the virus species used for delivery. In the case of TRV, this virus can infect roots and move to aerial parts of several plant species in Solanaceae, but can also infect meristematic tissues and achieve seed transmission, whereas many other virus species cannot [88, 129, 130]. TRV can infect pollen and fertilized flowers and can then enter the seed through the megaspore mother cell and egg, or through the pollen mother cells and pre-meiotic pollen allowing the delivery of sgRNA and the generation of heritable gene modification in gametes [131]. However, most viruses cannot infect the meristems and therefore alternative strategies need to be devised. The DNA replicons of Geminiviral species can be modified to carry the CRISPR/Cas9 system in plants although the reformed replicon cannot achieve systemic infection [132]. In compensation, Agrobacterium or particle bombardment can be used to deliver the viral vector to cells. This geminivirus-based system takes advantage of the virus replication initiator protein (Rep) to amplify the CRISPR/Cas9 coding sequences or DNA repair templates to very high copy numbers in plant cells increasing the probability of obtaining heritable gene modifications via homology-directed repair. The geminivirus-based system has been applied to generate precise gene modifications in tomato and potato [47, 133]. In this application, a strong promoter was inserted upstream of the anthocyanin synthesis gene along with an antibiotic selection marker to allow the screening of recombinant cells. The frequency of targeted gene replacement was as high as ~10 % and whole plants recovered from these recombinant cells segregated according to Mendel’s laws. However, compared to the conventional T-DNA vector approach, the geminivirus-based system seemed to be less efficient for generating targeted mutagenesis in potato [47]. The reduction of targeted mutations supports the use of the geminiviral vector system for promoting HR purposes rather than NHEJ-induced mutagenesis.
Future prospects
The development of CRISPR/Cas9 systems used for genetic screening
Over the past thirty years, forward genetics approaches have been the primary choice for gene functional studies. Random mutagenesis approaches using chemical or physical agents were extensively used to produce large mutant populations that were subsequently screened for phenotypes of interests. However, the work load of this approach is enormous and is only applicable to a few self-pollinated plant species. With the emergence of the CRISPR/Cas9 system, targeted gene mutagenesis in plants is becoming routine and the development of methods to generate genome-scale knockout libraries is now conceivable and has already been reported in bacteria and mammalian cells [50, 134]. The nature of these libraries can be controlled as it is determined by the combination of sgRNA inputs, a huge advantage over random mutagenesis. If used for genetic screening using negative or positive selection strategies, they can strongly facilitate the validation of candidate genes obtained from “Omics” approaches. In Arabidopsis, constitutively expressed CRISPR/Cas9 systems usually generate chimeras in the first generation of transgenic plants with homozyogous or biallelic plants not available until the T2 generation, thus increasing the complexity of genetic screenings [71, 112]. In contrast, germline specific, especially egg-cell specific systems, produce multiple homozygous and biallelic mutations in the T1 generation, saving labor and time. However, the available egg-cell specific systems still generate a high proportion of chimeras in the T1 generation, which may hamper its application for large scale genetic screenings, underpinning the need for novel promoters exhibiting early and continuous expression in egg-cells.
The development of CRISPR/Cas9 systems for crop breeding
To generate heritable gene modifications in crop species, both the integrative and the non-integrative CRISPR/Cas9 approaches can be applied. In integrated systems, the transcription level of CRISPR/Cas9 in somatic embryos during tissue regeneration is critical and new promoter combinations could increase the efficiency of the system to obtain heritable modifications. In addition, it is important to remove the CRISPR/Cas9 sequences from the genome by conventional crossing to produce varieties with a “clean” background, a time consuming procedure especially for asexually propagated crops. These limitations could be overcome by using non-integrative CRISPR/Cas9 systems. Integration of the Agrobacterium T-DNA into the plant chromosome requires the activity of the VirD2 protein and strains lacking the C-terminal ω domain of VirD2 are unable to integrate the T-DNA into the plant genome [135]. Tobacco cells infected with the ω mutant strain, show transient expression of a GUS gene contained in the T-DNA for at least 2 days after infection, although such a short time might not be enough for the CRISPR/Cas9 system to induce mutations. An interesting alternative is the introduction of preassembled CRISPR/Cas9 complexes into protoplasts [62]. Transient expression of these ribonucleoproteins in plant cells can induce heritable gene mutations with an efficiency directly related to the transfection rate. The relative proportions and dosage of the synthesized sgRNA and Cas9 proteins could be finely adjusted before transfection in order to achieve maximal efficiency. The microcalli regenerated from mutated cells are either heterozygous or biallelic, therefore the probability of obtaining heritable gene modifications equals the overall mutagenesis frequency in transfected protoplasts. The main drawback of this approach is the difficulty to regenerate whole plants from protoplasts. Future modifications in the nature of the components of the CRISPR/Cas9 system could further increase efficiency with over 600 Cas9 orthologues identified so far, some of which show improved properties for targeted gene modifications in mammalian cells [136].
Targeted gene modification in plants is rapidly becoming routine owing to the rapid development of multiple CRISPR systems. These powerful tools can help to create a large variety of mutations in otherwise inaccessible genes, and can simultaneously target multiple loci or produce large fragment rearrangements, thus accelerating plant breeding programs. The generated genetic modifications are stably inherited and any introduced transgenes can be easily removed by segregation or backcrossing. In many cases, crops engineered using CRISPR/Cas9 are virtually indistinguishable from plants in which the mutation might have happened naturally and may not be regulated as GMO plants. In fact, the US Department of Agriculture has recently decided that a CRISPR/Cas9 engineered mushroom in which a polyphenol oxidase gene had been mutagenized does not require special regulation [59]. This mushroom is the first CRSIPR/Cas9-edited crop exempted from regulatory approval. Several previous examples of crops bypassing regulatory approval were obtained using other gene-editing techniques such as ZFN and TALEN systems [137]. This is definitely encouraging news to scientists trying to apply the CRISPR/Cas9 system to other crops. Moreover, it was shown that mutations in the centromere-specific histone H3 variant CENH3 could be harnessed for the induction of haploids in Arabidopsis and several crop species [138, 139]. On self-pollination, these induced haploids can produce normal diploid seeds. The possibility of combining the CRISPR/Cas9 technology with this centromere-mediated genome elimination method for plant genetic engineering is certainly exciting. Considering that the production of new varieties with stacked traits can be accomplished in one generation by simultaneously editing multiple gene loci by CRISPR/Cas9, the speed of genetic research and crop breeding could undergo a quantum leap. We anticipate that widespread adoption of the CRISPR/Cas9 technology will revolutionize basic and applied research in plant biology.
Acknowledgments
We thank members of the Zhu lab for helpful discussion and insights with this work. Our work was supported by the Chinese Academy of Sciences. JRB acknowledges the award of a Visiting Professorship for Senior International Scientists by the Chinese Academy of Sciences.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest with respect to this work.
Contributor Information
Yanfei Mao, Email: yfmao@sibs.ac.cn.
Jian-Kang Zhu, Email: jkzhu@purdue.edu.
References
- 1.Compass G (2016) Cultivation of GM plants: Increase worldwide, no great change in Europe. http://www.gmo-compassorg/eng/agri_biotechnology/gmo_planting/
- 2.Phillips T. Genetically modified organisms (GMOs): transgenic crops and recombinant DNA technology. Nat Educ. 2008;1(1):213. [Google Scholar]
- 3.Grunewald W, Bury J, Inze D. Biotechnology: thirty years of transgenic plants. Nature. 2013;497(7447):40. doi: 10.1038/497040a. [DOI] [PubMed] [Google Scholar]
- 4.Compass G (2016) Genetically modified plants and the environment. http://www.gmo-compassorg/eng/safety/environmental_safety/
- 5.Voytas DF, Gao C. Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol. 2014;12(6):e1001877. doi: 10.1371/journal.pbio.1001877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radding CM. Homologous pairing and strand exchange in genetic recombination. Annu Rev Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- 7.Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6(6):507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- 8.Paszkowski J, Baur M, Bogucki A, Potrykus I. Gene targeting in plants. EMBO J. 1988;7(13):4021–4026. doi: 10.1002/j.1460-2075.1988.tb03295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swoboda P, Gal S, Hohn B, Puchta H. Intrachromosomal homologous recombination in whole plants. EMBO J. 1994;13(2):484–489. doi: 10.1002/j.1460-2075.1994.tb06283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puchta H, Dujon B, Hohn B. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 1993;21(22):5034–5040. doi: 10.1093/nar/21.22.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puchta H, Dujon B, Hohn B. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc Natl Acad Sci USA. 1996;93(10):5055–5060. doi: 10.1073/pnas.93.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiurazzi M, Ray A, Viret JF, Perera R, Wang XH, Lloyd AM, Signer ER. Enhancement of somatic intrachromosomal homologous recombination in Arabidopsis by the HO endonuclease. Plant Cell. 1996;8(11):2057–2066. doi: 10.1105/tpc.8.11.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79(2010):181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salomon S, Puchta H. Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J. 1998;17(20):6086–6095. doi: 10.1093/emboj/17.20.6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93(3):1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang B, Sugio A, White FF. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc Natl Acad Sci USA. 2006;103(27):10503–10508. doi: 10.1073/pnas.0604088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326(5959):1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 18.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 19.Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S, Carroll D. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28(17):3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459(7245):442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lloyd A, Plaisier CL, Carroll D, Drews GN. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc Natl Acad Sci USA. 2005;102(6):2232–2237. doi: 10.1073/pnas.0409339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, Choi VM, Rock JM, Wu YY, Katibah GE, Zhifang G, McCaskill D, Simpson MA, Blakeslee B, Greenwalt SA, Butler HJ, Hinkley SJ, Zhang L, Rebar EJ, Gregory PD, Urnov FD. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459(7245):437–441. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Zhang F, Li X, Baller JA, Qi Y, Starker CG, Bogdanove AJ, Voytas DF. Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol. 2012;161(1):20–27. doi: 10.1104/pp.112.205179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li T, Liu B, Spalding MH, Weeks DP, Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol. 2012;30(5):390–392. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- 26.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8(9):765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 28.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 29.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155(Pt 3):733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 32.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Upadhyay SK, Kumar J, Alok A, Tuli R. RNA-guided genome editing for target gene mutations in wheat. G3 (Bethesda) 2013;3(12):2233–2238. doi: 10.1534/g3.113.008847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia H, Wang N. Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS One. 2014;9(4):e93806. doi: 10.1371/journal.pone.0093806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V. Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods. 2013;9(1):39. doi: 10.1186/1746-4811-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar V, Jain M. The CRISPR-Cas system for plant genome editing: advances and opportunities. J Exp Bot. 2014;66(1):47–57. doi: 10.1093/jxb/eru429. [DOI] [PubMed] [Google Scholar]
- 40.Willis IM. RNA polymerase III. Genes, factors and transcriptional specificity. Eur J Biochem. 1993;212(1):1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- 41.Paule MR, White RJ. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28(6):1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(8):691–693. doi: 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- 43.Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31(8):688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun X, Hu Z, Chen R, Jiang Q, Song G, Zhang H, Xi Y. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci Rep. 2015;5:10342. doi: 10.1038/srep10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michno JM, Wang X, Liu J, Curtin SJ, Kono TJ, Stupar RM. CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web-tool and a modified Cas9 enzyme. GM Crops Food. 2015;6(4):243–252. doi: 10.1080/21645698.2015.1106063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brooks C, Nekrasov V, Lippman ZB, Van Eck J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014;166(3):1292–1297. doi: 10.1104/pp.114.247577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Butler NM, Atkins PA, Voytas DF, Douches DS. Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PLoS One. 2015;10(12):e0144591. doi: 10.1371/journal.pone.0144591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang B, Yang X, Yang C, Li M, Guo Y. Exploiting the CRISPR/Cas9 system for targeted genome mutagenesis in petunia. Sci Rep. 2016;6:20315. doi: 10.1038/srep20315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan D, Liu T, Li C, Jiao B, Li S, Hou Y, Luo K. Efficient CRISPR/Cas9-mediated targeted mutagenesis in populus in the first generation. Sci Rep. 2015;5:12217. doi: 10.1038/srep12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL, Gao C. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31(8):686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- 51.Patro S, Kumar D, Ranjan R, Maiti IB, Dey N. The development of efficient plant promoters for transgene expression employing plant virus promoters. Mol Plant. 2012;5(4):941–944. doi: 10.1093/mp/sss028. [DOI] [PubMed] [Google Scholar]
- 52.Christensen AH, Sharrock RA, Quail PH. Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol. 1992;18(4):675–689. doi: 10.1007/BF00020010. [DOI] [PubMed] [Google Scholar]
- 53.Du H, Zeng X, Zhao M, Cui X, Wang Q, Yang H, Cheng H, Yu D. Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J Biotechnol. 2015;217:90–97. doi: 10.1016/j.jbiotec.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 54.Mikami M, Toki S, Endo M. Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol Biol. 2015;88(6):561–572. doi: 10.1007/s11103-015-0342-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yan L, Wei S, Wu Y, Hu R, Li H, Yang W, Xie Q. High-efficiency Genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol Plant. 2015;8(12):1820–1823. doi: 10.1016/j.molp.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 56.Bortesi L, Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv. 2014;33(1):41–52. doi: 10.1016/j.biotechadv.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 57.Shou HX, Frame BR, Whitham SA, Wang K. Assessment of transgenic maize events produced by particle bombardment or Agrobacterium-mediated transformation. Mol Breeding. 2004;13(2):201–208. doi: 10.1023/B:MOLB.0000018767.64586.53. [DOI] [Google Scholar]
- 58.Jorgensen RA, Cluster PD, English J, Que Q, Napoli CA. Chalcone synthase cosuppression phenotypes in petunia flowers: comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA sequences. Plant Mol Biol. 1996;31(5):957–973. doi: 10.1007/BF00040715. [DOI] [PubMed] [Google Scholar]
- 59.Waltz E. Gene-edited CRISPR mushroom escapes US regulation. Nature. 2016;532(7599):293. doi: 10.1038/nature.2016.19754. [DOI] [PubMed] [Google Scholar]
- 60.Ali Z, Abul-faraj A, Li L, Ghosh N, Piatek M, Mahjoub A, Aouida M, Piatek A, Baltes NJ, Voytas DF, Dinesh-Kumar S, Mahfouz MM. Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol Plant. 2015;8(8):1288–1291. doi: 10.1016/j.molp.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 61.Yin K, Han T, Liu G, Chen T, Wang Y, Yu AY, Liu Y. A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci Rep. 2015;5:14926. doi: 10.1038/srep14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woo JW, Kim J, Kwon SI, Corvalan C, Cho SW, Kim H, Kim SG, Kim ST, Choe S, Kim JS. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol. 2015;33(11):1162–1164. doi: 10.1038/nbt.3389. [DOI] [PubMed] [Google Scholar]
- 63.Schmidt A, Schmid MW, Grossniklaus U. Plant germline formation: common concepts and developmental flexibility in sexual and asexual reproduction. Development. 2015;142(2):229–241. doi: 10.1242/dev.102103. [DOI] [PubMed] [Google Scholar]
- 64.Forner J, Pfeiffer A, Langenecker T, Manavella PA, Lohmann JU. Germline-transmitted genome editing in Arabidopsis thaliana Using TAL-effector-nucleases. PLoS One. 2015;10(3):e0121056. doi: 10.1371/journal.pone.0121056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walbot V. Sources and consequences of phenotypic and genotypic plasticity in flowering plants. Trends Plant Sci. 1996;1(1):27–32. doi: 10.1016/S1360-1385(96)80020-3. [DOI] [Google Scholar]
- 66.Wang K. Agrobacterium protocols. Methods Mol Biol. 2015;1224:vii–viii. doi: 10.1007/978-1-4939-1658-0. [DOI] [PubMed] [Google Scholar]
- 67.Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci. 1993;316:1194–1199. [Google Scholar]
- 68.Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc. 2006;1(2):641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- 69.Liu W, Zhu X, Lei M, Xia Q, Botella J, Zhu J-K, Mao Y. A detailed procedure for CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana . Sci Bull. 2015;60(15):1332–1347. doi: 10.1007/s11434-015-0848-2. [DOI] [Google Scholar]
- 70.Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y, Zhu JK. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23(10):1229–1232. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L, Zeng L, Liu X, Zhu JK. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA. 2014;111(12):4632–4637. doi: 10.1073/pnas.1400822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu JK. Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant. 2013;6(6):2008–2011. doi: 10.1093/mp/sst121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Callis J, Raasch JA, Vierstra RD. Ubiquitin extension proteins of Arabidopsis thaliana. Structure, localization, and expression of their promoters in transgenic tobacco. J Biol Chem. 1990;265(21):12486–12493. [PubMed] [Google Scholar]
- 74.Custers JBMSS, Jansen HJ, Zhang L, van Lookeren Campagne MM. The 35S-CaMV promoter is silent during early embryogenesis but activated during non-embryo-genic sporophytic development in microspore culture. Protoplasma. 1999;208:257–264. doi: 10.1007/BF01279097. [DOI] [Google Scholar]
- 75.Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA. Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 2015;15(1):16. doi: 10.1186/s12896-015-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Z, Liu ZB, Xing A, Moon BP, Koellhoffer JP, Huang L, Ward RT, Clifton E, Falco SC, Cigan AM. Cas9-guide RNA directed genome editing in soybean. Plant Physiol. 2015;169(2):960–970. doi: 10.1104/pp.15.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cai Y, Chen L, Liu X, Sun S, Wu C, Jiang B, Han T, Hou W. CRISPR/Cas9-mediated genome editing in soybean hairy roots. PLoS One. 2015;10(8):e0136064. doi: 10.1371/journal.pone.0136064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Collier R, Fuchs B, Walter N, Kevin Lutke W, Taylor CG. Ex vitro composite plants: an inexpensive, rapid method for root biology. Plant J. 2005;43(3):449–457. doi: 10.1111/j.1365-313X.2005.02454.x. [DOI] [PubMed] [Google Scholar]
- 79.Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Xu N, Zhu JK. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J. 2014;12(6):797–807. doi: 10.1111/pbi.12200. [DOI] [PubMed] [Google Scholar]
- 80.Slade AJ, Fuerstenberg SI, Loeffler D, Steine MN, Facciotti D. A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat Biotechnol. 2005;23(1):75–81. doi: 10.1038/nbt1043. [DOI] [PubMed] [Google Scholar]
- 81.Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32(9):947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- 82.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24(6):1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ali Z, Abul-Faraj A, Li L, Ghosh N, Piatek M, Mahjoub A, Aouida M, Piatek A, Baltes NJ, Voytas DF, Dinesh-Kumar S, Mahfouz MM. Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol Plant. 2015 doi: 10.1016/j.molp.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 85.Ratcliff F, Martin-Hernandez AM, Baulcombe DC. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 2001;25(2):237–245. doi: 10.1046/j.0960-7412.2000.00942.x. [DOI] [PubMed] [Google Scholar]
- 86.Turnage MA, Muangsan N, Peele CG, Robertson D. Geminivirus-based vectors for gene silencing in Arabidopsis. Plant Journal. 2002;30(1):107–114. doi: 10.1046/j.1365-313X.2002.01261.x. [DOI] [PubMed] [Google Scholar]
- 87.Tang Y, Wang F, Zhao J, Xie K, Hong Y, Liu Y. Virus-based microRNA expression for gene functional analysis in plants. Plant Physiol. 2010;153(2):632–641. doi: 10.1104/pp.110.155796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Senthil-Kumar M, Mysore KS. Virus-induced gene silencing can persist for more than 2 years and also be transmitted to progeny seedlings in Nicotiana benthamiana and tomato. Plant Biotechnol J. 2011;9(7):797–806. doi: 10.1111/j.1467-7652.2011.00589.x. [DOI] [PubMed] [Google Scholar]
- 89.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31(9):839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31(9):833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Carroll D. Staying on target with CRISPR-Cas. Nat Biotechnol. 2013;31(9):807–809. doi: 10.1038/nbt.2684. [DOI] [PubMed] [Google Scholar]
- 93.Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, Joung JK. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2014;33(2):187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fauser F, Schiml S, Puchta H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana . Plant J. 2014;79(2):348–359. doi: 10.1111/tpj.12554. [DOI] [PubMed] [Google Scholar]
- 96.Xie K, Zhang J, Yang Y. Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol Plant. 2014;7(5):923–926. doi: 10.1093/mp/ssu009. [DOI] [PubMed] [Google Scholar]
- 97.Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant. 2014;7(9):1494–1496. doi: 10.1093/mp/ssu044. [DOI] [PubMed] [Google Scholar]
- 98.Chuai GH, Wang QL, Liu Q. In Silico meets in vivo: towards computational CRISPR-based sgRNA design. Trends Biotechnol. 2016 doi: 10.1016/j.tibtech.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 99.Guell M, Yang L, Church GM. Genome editing assessment using CRISPR genome analyzer (CRISPR-GA) Bioinformatics. 2014;30(20):2968–2970. doi: 10.1093/bioinformatics/btu427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Osakabe Y, Watanabe T, Sugano SS, Ueta R, Ishihara R, Shinozaki K, Osakabe K. Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci Rep. 2016;6:26685. doi: 10.1038/srep26685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xie K, Yang Y. RNA-guided genome editing in plants using a CRISPR-Cas system. Mol Plant. 2013;6(6):1975–1983. doi: 10.1093/mp/sst119. [DOI] [PubMed] [Google Scholar]
- 102.Feng C, Yuan J, Wang R, Liu Y, Birchler JA, Han F. Efficient targeted genome modification in maize using CRISPR/Cas9 system. J Genet Genom. 2016;43(1):37–43. doi: 10.1016/j.jgg.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 103.Lawrenson T, Shorinola O, Stacey N, Li C, Ostergaard L, Patron N, Uauy C, Harwood W. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 2015;16:258. doi: 10.1186/s13059-015-0826-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu LJ. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013;23(10):1233–1236. doi: 10.1038/cr.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014;14:327. doi: 10.1186/s12870-014-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Guo J, Chen L, Zhao X, Dong Z, Liu YG. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8(8):1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 107.Zhang Z, Mao Y, Ha S, Liu W, Botella JR, Zhu JK. A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep. 2015;35(7):1519–1533. doi: 10.1007/s00299-015-1900-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang C, Shen L, Fu Y, Yan C, Wang K. A simple CRISPR/Cas9 system for multiplex genome editing in rice. J Genet Genom. 2016;42(12):703–706. doi: 10.1016/j.jgg.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 109.Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA. 2015;112(11):3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang WC, Ye D, Xu J, Sundaresan V. The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 1999;13(16):2108–2117. doi: 10.1101/gad.13.16.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K. Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana . Proc Natl Acad Sci USA. 1999;96(20):11664–11669. doi: 10.1073/pnas.96.20.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mao Y, Zhang Z, Feng Z, Wei P, Zhang H, Botella JR, Zhu JK. Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol J. 2015;14(2):519–532. doi: 10.1111/pbi.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Steffen JG, Kang IH, Macfarlane J, Drews GN. Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 2007;51(2):281–292. doi: 10.1111/j.1365-313X.2007.03137.x. [DOI] [PubMed] [Google Scholar]
- 114.Sprunck S, Rademacher S, Vogler F, Gheyselinck J, Grossniklaus U, Dresselhaus T. Egg cell-secreted EC1 triggers sperm cell activation during double fertilization. Science. 2012;338(6110):1093–1097. doi: 10.1126/science.1223944. [DOI] [PubMed] [Google Scholar]
- 115.Eady C, Lindsey K, Twell D. Differential activation and conserved vegetative cell-specific activity of a late pollen promoter in species with bicellular and tricellular pollen. Plant J. 1994;5:543–550. doi: 10.1046/j.1365-313X.1994.5040543.x. [DOI] [Google Scholar]
- 116.Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015;16:144. doi: 10.1186/s13059-015-0715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rutley N, Twell D. A decade of pollen transcriptomics. Plant Reprod. 2015;28(2):73–89. doi: 10.1007/s00497-015-0261-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Drews GN, Wang D, Steffen JG, Schumaker KS, Yadegari R. Identification of genes expressed in the angiosperm female gametophyte. J Exp Bot. 2010;62(5):1593–1599. doi: 10.1093/jxb/erq385. [DOI] [PubMed] [Google Scholar]
- 119.Hyun Y, Kim J, Cho SW, Choi Y, Kim JS, Coupland G. Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta. 2014;241(1):271–284. doi: 10.1007/s00425-014-2180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci USA. 2015;112(7):2275–2280. doi: 10.1073/pnas.1500365112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hyun Y, Yun H, Park K, Ohr H, Lee O, Kim DH, Sung S, Choi Y. The catalytic subunit of Arabidopsis DNA polymerase alpha ensures stable maintenance of histone modification. Development. 2012;140(1):156–166. doi: 10.1242/dev.084624. [DOI] [PubMed] [Google Scholar]
- 122.Li H-J, Liu N-Y, Shi D-Q, Liu J, Yang W-C. YAO is a nucleolar WD40-repeat protein critical for embryogenesis and gametogenesis in Arabidopsis. BMC Plant Biol. 2010;10(1):1–12. doi: 10.1186/1471-2229-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Atanassova R, Chaubet N, Gigot C. A 126 bp fragment of a plant histone gene promoter confers preferential expression in meristems of transgenic Arabidopsis. Plant J. 1992;2(3):291–300. doi: 10.1111/j.1365-313x.1992.00291.x. [DOI] [PubMed] [Google Scholar]
- 124.Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360(6401):273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- 125.Gao Y, Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol. 2014;56(4):343–349. doi: 10.1111/jipb.12152. [DOI] [PubMed] [Google Scholar]
- 126.Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475(7355):201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell. 2014;54(4):698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mikami M, Toki S, Endo M. Parameters affecting frequency of CRISPR/Cas9 mediated targeted mutagenesis in rice. Plant Cell Rep. 2015;34(10):1807–1815. doi: 10.1007/s00299-015-1826-5. [DOI] [PubMed] [Google Scholar]
- 129.Senthil-Kumar M, Mysore KS. New dimensions for VIGS in plant functional genomics. Trends Plant Sci. 2011;16(12):656–665. doi: 10.1016/j.tplants.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 130.Liu H, Reavy B, Swanson M, MacFarlane SA. Functional replacement of the tobacco rattle virus cysteine-rich protein by pathogenicity proteins from unrelated plant viruses. Virology. 2002;298(2):232–239. doi: 10.1006/viro.2002.1421. [DOI] [PubMed] [Google Scholar]
- 131.Maule AJ, Wang D. Seed transmission of plant viruses: a lesson in biological complexity. Trends Microbiol. 1996;4(4):153–158. doi: 10.1016/0966-842X(96)10016-0. [DOI] [PubMed] [Google Scholar]
- 132.Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF. DNA replicons for plant genome engineering. Plant Cell. 2014;26(1):151–163. doi: 10.1105/tpc.113.119792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cermak T, Baltes NJ, Cegan R, Zhang Y, Voytas DF. High-frequency, precise modification of the tomato genome. Genome Biol. 2015;16:232. doi: 10.1186/s13059-015-0796-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509(7501):487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 135.Mysore KS, Bassuner B, Deng XB, Darbinian NS, Motchoulski A, Ream W, Gelvin SB. Role of the Agrobacterium tumefaciens VirD2 protein in T-DNA transfer and integration. Mol Plant Microbe Interact. 1998;11(7):668–683. doi: 10.1094/MPMI.1998.11.7.668. [DOI] [PubMed] [Google Scholar]
- 136.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.USDA (2016) Regulated article letters of inquiry. https://www.aphis.usda.gov/aphis/ourfocus/biotechnology/am-i-regulated/Regulated+Article+Letters+of+Inquiry
- 138.Ravi M, Chan SW. Haploid plants produced by centromere-mediated genome elimination. Nature. 2010;464(7288):615–618. doi: 10.1038/nature08842. [DOI] [PubMed] [Google Scholar]
- 139.Britt AB, Kuppu S. Cenh3: an emerging player in haploid induction technology. Front Plant Sci. 2016;7:357. doi: 10.3389/fpls.2016.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41(20):e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jiang W, Yang B, Weeks DP. Efficient CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS One. 2014;9(6):e99225. doi: 10.1371/journal.pone.0099225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lowder LG, Zhang D, Baltes NJ, Paul JW, 3rd, Tang X, Zheng X, Voytas DF, Hsieh TF, Zhang Y, Qi Y. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015;169(2):971–985. doi: 10.1104/pp.15.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhou H, Liu B, Weeks DP, Spalding MH, Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014;42(17):10903–10914. doi: 10.1093/nar/gku806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liang Z, Zhang K, Chen K, Gao C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genom. 2014;41(2):63–68. doi: 10.1016/j.jgg.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 145.Zhu J, Song N, Sun S, Yang W, Zhao H, Song W, Lai J. Efficiency and inheritance of targeted mutagenesis in maize using CRISPR-Cas9. J Genet Genom. 2016;43(1):25–36. doi: 10.1016/j.jgg.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 146.Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015;169(2):931–945. doi: 10.1104/pp.15.00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ron M, Kajala K, Pauluzzi G, Wang D, Reynoso MA, Zumstein K, Garcha J, Winte S, Masson H, Inagaki S, Federici F, Sinha N, Deal RB, Bailey-Serres J, Brady SM. Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol. 2014;166(2):455–469. doi: 10.1104/pp.114.239392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ito Y, Nishizawa-Yokoi A, Endo M, Mikami M, Toki S. CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem Biophys Res Commun. 2015;467(1):76–82. doi: 10.1016/j.bbrc.2015.09.117. [DOI] [PubMed] [Google Scholar]
- 149.Wang S, Zhang S, Wang W, Xiong X, Meng F, Cui X. Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep. 2015;34(9):1473–1476. doi: 10.1007/s00299-015-1816-7. [DOI] [PubMed] [Google Scholar]
- 150.Subburaj S, Chung SJ, Lee C, Ryu SM, Kim DH, Kim JS, Bae S, Lee GJ. Site-directed mutagenesis in Petunia × hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 2016 doi: 10.1007/s00299-016-1937-7. [DOI] [PubMed] [Google Scholar]
- 151.Sugano SS, Shirakawa M, Takagi J, Matsuda Y, Shimada T, Hara-Nishimura I, Kohchi T. CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol. 2014;55(3):475–481. doi: 10.1093/pcp/pcu014. [DOI] [PubMed] [Google Scholar]