Abstract
Mouse embryonic stem cells (mESCs), characterized by their pluripotency and capacity for self-renewal, are driven by a complex gene expression program composed of several regulatory mechanisms. These mechanisms collaborate to maintain the delicate balance of pluripotency gene expression and their disruption leads to loss of pluripotency. In this review, we provide an extensive overview of the key pillars of mESC pluripotency by elaborating on the various essential transcription factor networks and signaling pathways that directly or indirectly support this state. Furthermore, we consider the latest developments in the role of epigenetic regulation, such as noncoding RNA signaling or histone modifications.
Keywords: Pluripotency, Mouse embryonic stem cells, Transcriptional regulation, Epigenetic regulation
Introduction
Mouse embryonic stem cells (mESCs) are derived from the inner cell mass (ICM) of the blastocyst during development [1]. The ability to capture this otherwise transitional state in vitro comes as a result of culture techniques which maintain mESCs in a state of unlimited proliferation, i.e., self-renewal, whilst retaining their pluripotency [1]. Potency refers to the differentiation potential of a cell, ranging from totipotency to unipotency. Totipotent cells are able to give rise to any cell type, whereas unipotent cells are restricted to one linage [2, 3]. Pluripotent cells are capable of differentiating into any of the three germ layers (mesoderm, endoderm, and ectoderm), but are unable to differentiate into extra-embryonic (placental) tissue [4]. Accordingly, chimera formation using mESCs is the ultimate demonstration of their pluripotency, reflecting the developmental end-point of the ICM during embryogenesis [4]. Loss of pluripotency can be regarded as occurring not only upon directed or spontaneous differentiation, but also upon loss of range of potency or ability to differentiate. As such, the regulatory mechanisms that maintain mESCs in their pluripotent state must balance stability, to maintain pluripotency, with plasticity to allow entry into specific programs of differentiation. In this review, we integrate the different mechanisms supporting the maintenance of pluripotency in mESCs, from regulatory transcription factors and signaling pathways to small RNA signaling and epigenetic regulation, to thoroughly understand how this balance is maintained.
Transcription factors for pluripotency maintenance
There are many reported transcription factors acting as a regulatory network that directly or indirectly drives the mESC identity. In particular, the transcription factors Oct4, Sox2, and Nanog form a core regulatory circuit that is controlled by an auto-regulatory feedback loop [5, 6]. The members of this core circuit are not necessarily restricted to complexing with each other. Several auxiliary transcription factors have also recently been found to be essential. Oct4 itself can also form its own network centered around secondary transcription factors that also play a role in pluripotency maintenance and differentiation.
Core regulatory circuit: Oct4
Oct4 (officially denoted as Pou5f1) belongs to the POU family, as defined by their bipartite DNA-binding POU domain, and can regulate gene expression either positively or negatively to maintain mESC pluripotency. For example, Oct4 synergizes with Sox2 to maintain mESC pluripotency [7–9] or acts as a repressor of ESC differentiation by interacting with lineage-specific transcription factors, such as FoxD3 [10]. The balance of Oct4 expression level itself is also critical for maintenance of pluripotency, and its disruption cripples the ability to derive mESCs from the ICM [11]. Moderate expression of Oct4 enables derivation and maintenance of mESCs, whereas high expression promotes differentiation into mesoderm or endoderm lineages and low expression leads to trophectoderm formation [12, 13]. This balance is fine-tuned by the interaction of Oct-4 with secondary transcription factors through three cis-elements, a distal enhancer, a proximal enhancer, and a proximal promoter [14]. As such, chromatin structure becomes important for Oct4 transcription; methylation in both enhancer regions has been shown to inhibit Oct4 expression [15]. Indeed, such chromatin modifications play an important role in pluripotency maintenance and will be discussed later in this review.
Core regulatory circuit: Sox2
Sox2, which belongs to the Sox family as characterized by their conserved high-mobility-group (HMG) DNA-binding domain, notably synergizes with Oct4 to support the maintenance of ESC pluripotency [16, 17]. This cooperation is established by structural interaction between their DNA-binding domains [19]. Depletion of Sox2 results in loss of pluripotency, although this phenomenon can be rescued by forced expression of Oct4, suggesting that Sox2 plays a secondary role to Oct4 in pluripotency [9, 18]. During embryogenesis, Sox2 expression persists during the development of the central nervous system, whilst the expressions of other pluripotency factors are lost [18]. Therefore, stringent spatiotemporal regulation of Sox2 expression is essential for pluripotency maintenance. As such, for ESC pluripotency, Sox2 not only plays a synergistic role with Oct4 but also maintains a certain expression level to avoid inducing differentiation.
Core regulatory circuit: Nanog
Nanog was first defined by the early embryo-specific NK (ENK) gene, by virtue of its homeodomain bearing similarities to the NK family [19]. However, due to low conservation of DNA sequence with the other members of the NK family, Nanog is regarded as a unique homeodomain transcription factor [20]. Nanog plays a role in maintaining mESC pluripotency and during mouse embryo development. Its expression starts in the morulae and gradually concentrates in the ICM before ultimately halting in the trophectoderm [21]. Some of the downstream targets of Nanog include inhibition of Trp53, a negative regulator of pluripotency [22]. However, the role of Trp53 in maintaining mESC pluripotency is not absolute, since Trp53-null ES cells fail to differentiate in vitro but retain pluripotency of contributing to chimeric embryos [23]. The Oct4-Sox2 complex, in addition to secondary transcription factors, such as FoxD3, binds to the proximal promoter of Nanog to modulate its high expression [24]. As the expression of FoxD3 is also regulated by Oct4, this is a robust means by which Oct4 can regulate Nanog [25]. On the other hand, Tcf3 is reported to negatively regulate Nanog expression; its depletion would support high expression of Nanog for pluripotency maintenance [26]. Interestingly, it has also been observed that Nanog can regulate its own expression through auto-repression independently from Oct4–Sox2 [27]. Reflecting this, it has been shown that Nanog is dispensable for mESC pluripotency; Nanog deficient mESCs and iPSCs retain several hallmarks of pluripotency, such as self-renewal and potency, including the ability to form chimeric mice [21, 28].
Core Klf circuitry
Several Krüppel-like factors are also important for pluripotency maintenance [29]. Klf4 in combination with Oct4/Sox2/cMyc can transform terminally differentiated somatic cells to a pluripotent state thus giving rise to induced pluripotent stem cells (iPSCs). The core Klf circuitry connects to the core regulatory circuit of Oct4/Sox2/Nanog to prevent ESC differentiation and support pluripotency maintenance [30]. As members of the core KLF circuitry, Klf2, Klf4, and Klf5 together increase Oct4/Sox2/Nanog expression by binding to their distal enhancer. Conversely, Klf2 expression is then activated by Oct4, whilst Klf4 and Klf5 are activated by Nanog to form a feedback loop [31]. Klfs share a functional relationship with Nanog and both regulate similar targets [29].
Other transcription factors
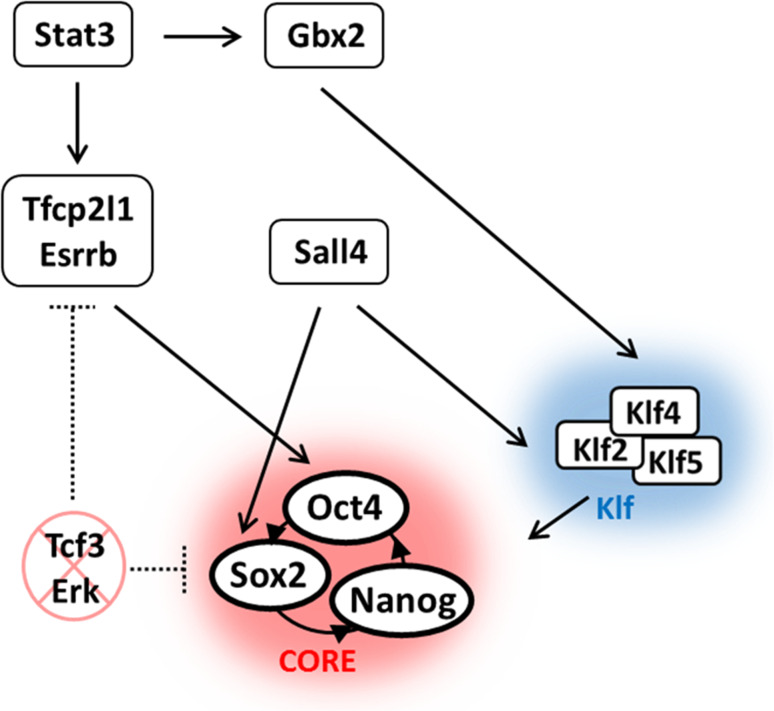
To establish a clear picture of the molecular mechanisms regulating pluripotency, computational approaches have been utilized to elucidate the essential components and interactions sufficient maintaining ESC pluripotency [32]. Currently, 12 components and 16 interactions have been established and constitute the known regulatory network [32]. For example, Stat3 signaling increases the activity of the Klf circuitry and supports the expression of Tfcp2l1 with either Esrrb or Sall4 to facilitate core or Klf regulatory circuits [32]. Inhibition of two transcription factors, Tcf3 and Erk, is necessary to prevent differentiation and maintain the core regulatory circuit Oct4/Sox2/Nanog [32]. Together, these additional components form the known regulatory network which interacts with the core circuitry to maintain mESC pluripotency (Fig. 1).
Fig. 1.
Regulation of key transcription factors for pluripotency maintenance of mouse embryonic stem cells
Signaling pathways for pluripotency maintenance
Several signaling pathways are involved in the integration of external cues and induction of the mESC identity through modulation of the key transcription factors driving pluripotency and self-renewal. These pathways may furthermore crosstalk to maintain pluripotency.
LIF signaling
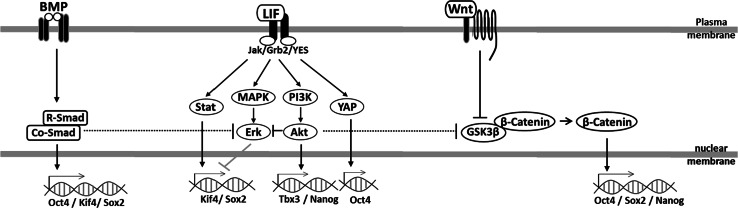
Leukemia Inhibitory factor (LIF), which belongs to the IL6 family, is secreted from murine embryonic fibroblasts and plays a crucial role in the maintenance of the mESC state [33]. LIF binds to the heterodimeric gp130 and LIF receptor beta, resulting in the activation of a broad range of downstream intracellular signaling pathways which regulate different aspects of the core transcriptional circuitry. Some of these are pro-pluripotency, such as Jak-Stat3, PI3K-Akt, and YES-YAP pathways, whereas others are actually pro-differentiation, such as MAPK-Erk [34, 35].
Jak-Stat3 signaling begins with the phosphorylation of Jak upon binding with the SH2 domain of the gp130 receptor. pJak then phosphorylates and activates Stat3, resulting in its homodimerization and translocation into the nucleus [36]. Both pStat3 itself and its downstream intracellular targets, such as Bcl3, are reported to play important roles in regulating gene expression for the maintenance of pluripotency [37, 38]. pStat3 regulates the core pluripotency regulatory circuit Oct4/Sox2/Nanog through activation of Klf4 [39], whereas we have shown that Bcl3 interacts with Oct4 protein to regulate Oct4 and Nanog promoter activity. Forced expression of Bcl3 partially maintains alkaline phosphatase activity, an indicator of stemness, after induction of differentiation [38].
PI3K-Akt contributes to the maintenance of pluripotency through two contrasting mechanisms. First, PI3K-Akt blocks MAPK-Erk signaling, which is a driver of endoderm differentiation [40, 41]. Second, Akt significantly increases Tbx3 activity and Nanog expression for ESC pluripotency and proliferation [39].
MAPK/Erk signaling negatively affects Nanog activity through antagonizing the nuclear localization of Tbx3 [39], whilst MEK activates the downstream Erk signaling to repress Nanog expression for primitive endoderm differentiation [42].
The YES-YAP pathway begins with activation of YES through binding with gp130 receptor [43]. Upon phosphorylation of gb130, YAP then translocates into the nucleus and binds to TEAD2, enabling it to bind the Oct4 promoter and induce its expression [43].
BMP signaling
BMP, a member of the TGFβ family, acts through binding to and activation of heteromeric type I and II BMP receptors [44]. The BMP signaling pathway mainly acts through the Smad complex, which consists of three categories: receptor-regulated Smads (R-Smads), cooperating Smad (Co-Smad), and inhibitory Smads (I-Smads). BMP activation leads to phosphorylation of the R-Smads, two of which will then complex with one Co-Smad. This complex then translocates to nucleus to directly regulate pluripotent gene expression. I-Smad competes with Co-Smad for R-Smad binding to drive ubiquitin degradation of R-Smads, thereby blocking signaling [45]. BMP4 signaling also cooperates with LIF to support pluripotency maintenance of ESCs during in vitro culture, since LIF alone promotes neural differentiation of ESCs under serum-free condition—this differentiation can be halted by induction of BMP4 signaling [46].
Wnt signaling
Similar to the inhibition of MAPK/Erk signaling by LIF, Wnt signaling contributes to pluripotency maintenance by acting as a repressor of ESC differentiation [47]. More specifically, it has been shown that mutations in Wnt3a lead to ectopic neural tube formation in the gastrulating embryo, suggesting that Wnt signaling mainly inhibits neural differentiation [48]. Wnt binds and activates the heterodimeric receptor Frizzled and LRP, resulting in the phosphorylation of GSK3β. This leads to the release of β-catenin, thereby preventing its degradation. Upon translocation into the nucleus, β-catenin binds directly to activate Oct4 or repress Tcf3 to regulate their transcriptional activity [49, 50]. Cytosolic β-catenin can also associate with the membrane and complexes with Oct4 and E-cadherin. This complex is destroyed upon differentiation [51].
Crosstalk between signaling pathways
Crosstalk between these diverse pathways is also essential for the regulation of pluripotency maintenance in mESCs. Within the LIF pathway, Akt (PI3K-Akt signaling) can inhibit both Erk (MAPK/Erk signaling) and GSK3β (thereby activating Wnt signaling) to prevent ESC differentiation [52–54]. The Co-Smad/R-Smad complex from BMP signaling also inhibits Erk (LIF signaling) to keep ESCs in undifferentiated state [55]. Therefore, each signaling pathway not only possesses its own function but also connects with others in an integrated system for the maintenance of pluripotency (Fig. 2).
Fig. 2.
Regulation of major signaling pathways for pluripotency maintenance of mouse embryonic stem cells
Reflecting the importance of this integration to stabilize the pluripotent state is the landmark development of 2i culture conditions, which target two separate pathways: PD03 (PD0325901) to inhibit MEK (upstream of Erk signaling) and CHIRON (CHIR99021) to inhibit GSK3 [56]. Crucially, the transition from using serum to 2i culture conditions enabled the derivation of mESCs capable of forming chimeric mice regardless of genetic background [57]. Therefore, the development of 2i culture conditions demonstrates that pluripotency could be maintained solely by inhibition of intrinsic signalings.
Epigenetic regulation of pluripotency maintenance
It has emerged that noncoding RNAs and the regulation of chromatin packing dynamics by histone modifications and DNA methylation play an important role in pluripotency maintenance. These factors provide epigenetic regulation of gene expression and cellular functions both positively and negatively.
miRNAs in transcriptional and post-transcriptional regulation
MicroRNAs (miRNAs) are small (22–24 nt) noncoding RNAs which modulate gene expression through negative post-transcriptional regulation. miRNAs first emerge as primary miRNAs (pri-miRNAs) generated by RNA polymerase II. These are processed into precursor miRNAs (pre-miRNAs), with their stem-loop hairpin structures, in the nucleus upon cleavage by the Drosha-DGCR8 endonuclease [58] and then exported to the cytoplasm to be further cleaved by another endonuclease, Dicer, to generate double-strand RNAs (dsRNAs) [59]. One of the RNA strands becomes the mature miRNA and binds to AGO proteins, and becomes integrated into the RNA-induced silencing complex (RISC) which targets the 3′UTR of mRNAs as directed by sequence complementarity with the guide miRNA [60]. This targeting silences gene expression through mRNA degradation and deadenylation or inhibition of mRNA translation [61, 62].
Studies of Dicer and Dgcr8 knockout mESCs have shown that miRNAs play an important role in both maintaining pluripotency and the ability to transition into differentiation. Whilst Dicer knockout mESCs remain viable, they constitutively express Oct4 and fail to properly differentiate in both in vitro and in vivo differentiation assays [63]. Similarly, DGCR8-deficient mESCs are also unable to fully downregulate pluripotency markers upon attempted differentiation and retain the ability to produce ESC colonies. This is again due to impaired silencing of mESC self-renewal that normally occurs with the induction of differentiation [64]. Interestingly, the phenotype of Dicer and Dgcr8 knockout mESCs differs and it has been suggested that the processing machinery itself may play a miRNA-independent role in ESC function [64].
To identify the miRNAs involved in mESC self-renewal, Wang et al. transfected miRNA mimics in an attempt to rescue proliferation defects in Dgcr8 knockout mESCs. A group of miRNAs involved in regulating G1-S transition that also shared similar seed sequences were identified: miR-20a, miR-20b, miR-93, miR-106a, miR-291a-3p, miR-291b-3p, miR-294, miR-295, miR-302b, miR-302c, and miR-302d [65].
Of these, the miR-290 family (miR-291a-3p, miR-291b-3p, miR-294, and miR-295) was singled out for investigation due to being an embryonic-specific group of microRNAs found to be enriched in undifferentiated mESCs and rapidly decrease upon differentiation [66–68]. Their transcription is regulated by key pluripotent transcription factors [69]. Rbl2 and Lats2 are considered the potential targets by which this miRNA family promotes G1-S transition given that they are the inhibitors of the cyclinE-Cdk2 pathway [65] (Table 1).
Table 1.
Noncoding RNAs regulate pluripotency maintenance in mESCs
| Name | Function | References | |
|---|---|---|---|
| miRNA | miR-290 family |
To maintain self-renewal property, especially G1-S transition To regulate DNA methylation in the pluripotency state |
[65, 70] |
| lncRNA | Xist RepA | Negatively regulate naïve pluripotency status by recruitment of PRC2 complex and X chromosome inactivation | [82, 83] |
| lncRNA | AK028326 | Oct4 co-transcriptional factor and activate Oct4 expression | [87] |
Rbl2 is also a means by which the miR-290 family regulates pluripotency through control of DNA methylation. Transcriptome analysis of Dicer null mESCs indicates that this may be due to the downregulation of DNA de novo methyltransferase (Dmnt) genes Dnmt3a, Dnmt3b, and Dnmt3l, likely by Rbl2, which leads to decreased methylation of the Oct4 promoter during differentiation. The silencing of Oct4 in differentiating Dicer null mESCs relies on repressive histone marks [70]. Accordingly, this can be rescued by ectopic expression of DNA de novo methyltransferases or miR-290 family microRNAs [70]. As can be seen, DNA methylation, therefore, plays an important role in the regulation of pluripotency in mESCs and will be discussed later in this review.
Interestingly, the miR-290 family can also enhance the generation of mouse iPSCs through somatic reprogramming using by Oct4, Sox2, and Klf4 [71]. As such, these embryonic-specific miRNAs function in maintaining the pluripotency of mESCs, somatic reprogramming, and sustain expression of DNA de novo methyltransferases for Oct4 promoter methylation during differentiation.
lncRNAs in transcriptional and post-transcriptional regulation
Long noncoding RNAs (lncRNAs) are arbitrarily defined as RNAs longer than 200 nt. Similar to mRNAs, they are transcribed by RNA polymerase II. Nascent transcripts are processed with 5′-capping, splicing and 3′ polyadenylation. Several functions of lncRNAs have been reported, such as modulation of chromatin structure, regulation of transcription, and post-transcriptional regulation. This is through acting as signals to recruit transcription factors, as molecular decoys titrating proteins away from chromatin, as scaffolds to stabilize protein complex, as RNA guides to recruit chromatin modifiers or targeting miRNAs for degradation [72–75].
Xist is a well-known lncRNA which mediates X chromosome inactivation in female mammals during embryogenesis to balance the dosage of X-linked gene expression [76–79]. In female embryo, the paternal X chromosome is inactivated during cleavage and then transiently reactivated in the inner cell mass of pre-implanting embryo. After implantation, one of the X chromosomes becomes randomly inactivated again [80]. Reflecting this cycle of inactivation, X chromosome inactivation status of mESCs is one of the characteristics defining naïve and primed states of pluripotency. mESCs, which are defined as displaying naïve ground pluripotency, are characteristics of the ICM and possess two activated X chromosomes. In contrast, in vitro epiblast stem cells (EpiSCs), which are derived from primed epiblasts and defined as displaying primed pluripotency, carry one inactivated X chromosome [81]. RepA is an lncRNA that is also transcribed from the Xist locus and recruits polycomb repressive complex 2 (PRC2) to form heterochromatin [82]. In mESCs, Oct4, Sox2, and Nanog repress Xist expression [83] (Table 1). Notably, during somatic reprogramming, it has been reported that reactivation of X chromosome is necessary and occurs through several mechanisms, such as DNA demethylation and induction of endogenous pluripotency transcription factors [84].
Many other lncRNAs have now also been identified in mESCs using chromatin IP sequencing. Through identification of potential transcriptionally active domains, conserved large noncoding genes, and pluripotency transcription factor binding potential, more than 100 lncRNAs have been discovered in mESCs [85]. Guttmman et al. identified 30 lncRNAs related to repression of lineage-specific differentiation through a loss of function screening study knocking down mESC enriched lncRNAs. Using a Nanog promoter driven luciferase as a pluripotency reporter, they found 26 lncRNAs involved in regulating mESC pluripotency. Subsequently, through mapping of transcription factors in the genome, they also found that most of these lncRNAs are, in turn, regulated by pluripotency associated transcription factors and bind with several chromatin protein complexes, such as polycomb repression complexes, histone modifiers, and DNA-binding proteins [86]. The function of several other pluripotency related lncRNAs has also been studied. LncRNA AK028326 transcription is activated by and collaborates with Oct4. LncRNA AK141205 overexpression positively regulates Oct4 expression [87]. Inhibition of AK028326 or AK141205 results in downregulation of Oct4 expression, with abrogation of AK141205 reducing cell proliferation, and promotes differentiation [87] (Table 1).
One kind of lncRNA named pRNA, mediating heterochromatin formation especially in ribosomal DNA (rDNA) region. To exit the pluripotent state, pRNA maturation is required. In undifferentiated mESCs, chromatin structure, including rDNA, mostly remains open. The proceeding of pRNA maturation from IGS-rRNA (intergenic spacer rRNA) is restricted. Upon differentiation, mature pRNA interacts with a transcription terminator factor, TTF1, and a TTF1 interacting protein TIP5, on rDNA region, and this recruitment of TIP50 initiates rDNA heterochromatin formation. rDNA heterochromatin triggers genome-wild heterochromatinization. Ectopic expression of mature pRNA to induce heterochromatin formation leads to loss of pluripotency [88], suggesting that chromatin remodeling by lncRNA is able to drive the exit of pluripotency state.
Chromatin remodeling and histone modification complexes in pluripotent stem cells
Chromatin modifiers function through mediation of the post-translational modification (PTM) of histone proteins or via ATP-dependent chromatin modifier release of DNA from histone binding for transcription, DNA repair, and replication [89].
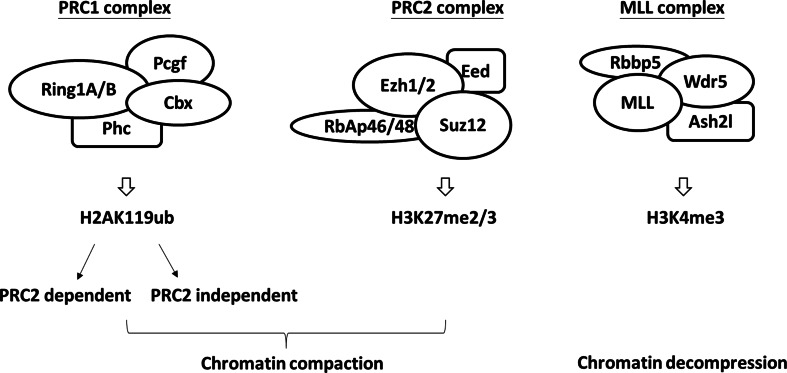
Two important complexes mediating PTM modifications of histone proteins are polycomb repressive complex 1/2 (PRC1/2) and the MLL complex, which are responsible for epigenetic repression and activation of gene transcription, respectively (Fig. 3). Study of these PRC complexes originates from work carried out on Drosophila polycomb (Pc) mutants, which displays abnormal body segmentation due to aberrant Homeotic (Hox) gene silencing, [90, 91] and several other mutants displaying the same phenotype. Collectively, the proteins involved in this patterning are referred to as the polycomb group (PcG) proteins.
Fig. 3.
Complexes-mediating histone modifications for pluripotency maintenance in mouse embryonic stem cells. Key subunits of PRC1 complexes are Cbx (Cbx2/4/6/7/8), Ring1A/B, Phc (Phc1/2/3), and Pcgf1/6. Ring1A/B are ubiquitin ligases responsible for ubiquitylization of lysine 119 of Histone 2A. Key subunits of PRC2 complexes are Ezh1/2, Suz12, Eed, and RbAp46/48. Ezh1/2 are methyltransferases responsible for di- or tri-methylation of lysine 27 of Histone 3. MLL is composed of Wdr5, Ash2l, and Rbbp5. MLL is a histone methyltransferase responsible for tri-methylation of lysine 4 of Histone 3
PRC1 and PRC2 were defined based on the nature of their post-transcriptional modification. PRC1, whose core components include Cbx proteins, Ring1A/B, Phc proteins and Pcgf proteins (Fig. 3), and monoubiquitylates lysine 119 of Histone 2A (H2AK119ub) via its Ring1A and Ring1B ubiquitin ligase subunits [91]. A double knockout of Ring1A/B impairs mESC proliferation and self-renewal [92]. Notably, PRC1 can also mediate gene silencing with or without its enzymatic activity [93]. Meanwhile, PRC2, the key components of which are Ezh1/2, Suz12, Eed, and RbAp46/48 (Fig. 3), mediates lysine 27 di- or tri-methylation of histone 3 (H3K27me2/3) via its Ezh1 and Ezh2 methyltransferase subunits [94]. Other components, such as Aebp2, Pcl, and Jarid2, are important in the positive regulation or negative regulation of this enzymatic activity [95–99]. Genome-wide analysis of protein binding shows that PRC1 and PRC2 complex proteins co-occupy on the promoters of several transcription factors related to development, such as the Gata family [100]. Although the PRC2 function is dispensable in pluripotency maintenance [101], Eed-deficient cells de-repress developmental gene expression [100]. This suggests a role of PRC2 in gene silencing in mESCs [100]. Inactivation of PRC2 components delays the reduction of Oct4 and Nanog expression during differentiation [97, 102, 103]. PRC2 complexes are, therefore, crucial for the repression of developmental regulators during maintenance of pluripotency and then silencing pluripotency upon mESC differentiation through its histone modification activity.
In mESCs, around half of H3K27 chains are dimethylated, 20% of H3K27 are monomethylated, and 10–20% of H3K27 are trimethylated [104]. Trimethylated H3K27 is enriched in repressive chromatin regions or bivalent domains in mESCs [105–107]. PRC1 can be considered downstream of PRC2 given that the PRC1 component, Cbx proteins, targets to H3K27me3, a catalytic product of PRC2 [108]. As such, both PRC2 and PRC1 co-occupy H3K27me3-modified gene domains encoding developmental regulatory factors. However, PRC1 can act independently from PRC2 and, furthermore, can take over PRC2′s targets in its absence [94, 109]. The binding of PRC1 to target genes is mediated by Oct4 in this case [92]. L3mbtl2, a PcG protein, is an atypical member of the PRC1 complex and plays a crucial role in regulating mESC self-renewal during the G1-S transition through a noncanonical PRC1-mediated repression mechanism—its knockout results in embryonic lethality [110]. L3mbtl2 targets gene loci characterized by lysine 9 dimethylation of histone 3 (H3K9me2), low histone acetylation, and lysine 199 monoubiquitination of histone 2A. These target genes are not bound by canonical PRC1 and PRC2 complexes, so their modifications are dependent upon the recruitment of PRC1-related components, such as G9a methyltransferase, Hdac1 histone deacetylase, and Ring1B ubiquitin ligase.
In contrast to PcG proteins, trithorax group (TrxG) proteins play a role in epigenetic gene activation through histone 3 lysine 4 tri-methylation. During development, TrxG and PcG proteins have opposite functions but often target similar chromatin regions in mESCs [107, 111, 112]. The mammalian TrxG proteins form the mixed-lineage leukemia (MLL) complex, whose key components include Mll1/2/3, Wdr5, Ash2l, and Rbbp5 [113] (Fig. 3). Wdr5 is a downstream target of Oct4 and Nanog, and its reduction is correlated with a decrease in H3K4me3 status during differentiation. Knockdown of Wdr5 causes mESCs to lose stemness properties, including cell morphology, alkaline phosphatase activity, and self-renewal ability [114]. Using genome-wide mapping of Wdr5, Rbbp5, H3K4me3, and Oct4 binding, Ang et al. found that pluripotency factors cooperate with the MLL complex to activate the transcription of regulators of self-renewal [114], suggesting a crucial role in mESC pluripotency.
General ATP-dependent chromatin-remodeling protein complexes include SWI/SNF, CHD, and INO80. esBAF is a unique ESC-specific SWI/SNF complex which regulates mESC self-renewal and pluripotency. It has been found to colocalize with Oct4, Sox2, Nanog, Stat3, and Smad1, indicating that esBAF has a wide ranging involvement in the core transcription circuitry and the LIF and BMP signaling pathways [115, 116].
Chd1 is an ATP-dependent DNA helicase maintaining euchromatin for gene activation. Although Chd1 knockdown cells remain in an undifferentiated state, these cells show the defect to differentiate into primitive ectoderm and show a bias in differentiation towards the ectoderm lineage [117]. This suggests that chromatin compaction can lead to a reduction in potency.
Ino80 has been shown to target pluripotency gene promoters through interaction with Oct4 and Wdr5, and its knockdown reduces pluripotency gene expression and promotes cell differentiation [118]. Tip60-400, a complex belonging to the INO80 family, was implicated in the maintenance of mESC pluripotency in an RNAi screening study [119]. More specifically, the Tip60-400 complex binds to H3K4me3 marked chromatin and enables access to Nanog targeted promoters [119]. Accordingly, knockdown of its subunits alters cell morphology and cell cycle profile, suggesting a loss of pluripotency. As such, the INO80 family as a whole can be considered to facilitate DNA binding of the core circuitry.
Bivalent histone modifications
Chromatin packing into either ‘active’ euchromatin or ‘inactive’ heterochromatin is dependent on the type of post-translational modification present on histone tails, which affects their charge. Therefore, the pattern of modification is considered to ‘code’ the epigenetic regulation of gene expression. For example, H3K4me3, H3K9ac, and H3K14ac are associated with euchromatin, whereas H3K27me3 and H3K9me3 are associated with heterochromatin [120, 121]. Interestingly, H3K27me3 (an ‘inactive’ code) is often accompanied with H3K4me3 (an ‘active’ histone code) in the promoters of developmental genes in mESCs. This pairing is referred to as a bivalent histone modification and it is proposed that these promoters are “poised” for further activation [107]. Furthermore, bivalent modifications are established and maintained by a combination of PRC2 and MLL complexes as mentioned above in combination with DNA methylation, transcription factors, and noncoding RNAs [107, 122].
DNA methylation
The impact of DNA methylation on gene expression is widely known [123–125]. This form of modification typically occurs at cytosines in CpG dinucleotide, resulting in the formation of 5-methylcytosine (5mC) which then recruits methyl-DNA-binding (MBD) proteins and methyl-CpG binding protein 2 (MeCP2). These proteins further recruit histone modifiers and chromatin regulators for higher order chromatin organization to repress gene expression [126].
DNA methyltransferase 3A and 3B (Dnmt3A and 3B) are responsible for de novo methylation, whereas Dnmt1 maintains methylation during DNA replication; knockout of Dnmt in mESCs causes DNA hypo-methylation, which can have an impact on differentiation and lineage determination. Dnmt1-deficient mESCs die through apoptosis upon attempted induction of differentiation [127]. When cells are cultured in trophoblast stem cell medium, they are able to generate 25% of trophoblast giant cells [127]. DNA hypo-methylation de-represses Elf5, a trophoblast-specific transcription factor, which contributes to trophoblast cell lineage [128]. Those Dnmt3a- or 3b-deficient ES cells with severe global DNA hypo-methylation, which possess just 0.6% CpG methylation, are not able to initiate differentiation but remain viable whilst retaining stemness characteristics [129]. Mesoderm cells derived from Dnmt3a- and Dnmt3b-deficient ES cells retain their ability to convert into endoderm lineage by Gata4 induction [130].
Tet proteins, which are responsible for DNA demethylation, convert 5mC to 5-hydroxymethylcytosine (5hmC), which then becomes 5-formylcytosine (5fC) and 5-carboxylcytosine (5-caC) [131, 132]. Tet1 and Tet2 are enriched in undifferentiated cells and become downregulated after differentiation. Although it has been shown that depletion of Tet1 and Tet2 reduces global 5hmC, no effect on mESC self-renewal ability was observed. On the other hand, differentiation capability was restricted [133]. In vivo, Tet1 and Tet2 double knockout mice showed partial perinatal lethality and abnormal DNA imprinting [134].
Others
Several other factors also contribute indirectly to the maintenance of the mESC state. For example, the histone protein variant H2A.Z interacts with and maintains Nanog protein levels through inhibition of protein degradation [135]. RNA polymerase-mediated transcription machinery assists core transcription factors in pluripotency maintenance and somatic cell reprogramming [136]. Cell cycle protein Geminin antagonizes chromatin-remodeling proteins during the S phase to maintain the expression of Oct4, Sox2, and Nanog [137]. Geminin also restrains mesodermal lineage commitment and is associated with antagonism of Wnt signaling and enhanced repressive polycomb-mediated repression [138]. The THO protein complex functions in coupling mRNA transcription and export to cytoplasm. Knockdown of Thoc2 and Thoc5, two subunits of THO complexes, inhibits export of pluripotency gene transcripts. This highlights the importance of mRNA export system in the regulation of mESC pluripotency [139].
Concluding remarks
In the past few decades, scientists have successfully managed to maintain pluripotency in vitro using LIF treatment and signaling inhibitors [4, 31, 54, 140–142] to enable the study into this remarkable state. What has emerged is a picture, whereby pluripotency in mESCs is an embodiment of several levels of balance, from the balance of pluripotency gene expression to that of different regulatory mechanisms that can both agonize and antagonize each other. Offset of just one element can result in a shift in the equilibrium towards differentiation, or, indeed, the inability to make this shift. Intriguing questions about these regulatory mechanisms still exist and the complexity of the involved regulation networks continues to be constructed. Indeed, epigenetics has emerged as a crucial player, with elements such as chromatin modeling capable of overriding signaling induced gene expression programs. It is hoped that this review, which presents a thorough, up-to-date aggregation of the reported regulatory mechanisms in maintenance of mESC pluripotency, will represent a comprehensive aid to the study of pluripotency.
Compliance with ethical standards
Funding
This review was supported by the Ministry of Science and Technology (MOST 104-2811-B-001-036 and 105-2325-B-001-009), and the Academia Sinica Translational Medicine Program.
Footnotes
C.-Y. Chen and Y.-Y. Cheng contributed equally to this work.
References
- 1.Solter D. From teratocarcinomas to embryonic stem cells and beyond: a history of embryonic stem cell research. Nat Rev Genet. 2006;7:319–327. doi: 10.1038/nrg1827. [DOI] [PubMed] [Google Scholar]
- 2.Mintz B, Illmensee K. Normal genetically mosaic mice produced from malignant teratocarcinoma cells. Proc Natl Acad Sci USA. 1975;72:3585–3589. doi: 10.1073/pnas.72.9.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Illmensee K, Mintz B. Totipotency and normal differentiation of single teratocarcinoma cells cloned by injection into blastocysts. Proc Natl Acad Sci USA. 1976;73:549–553. doi: 10.1073/pnas.73.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 5.Loh KM, Lim B. A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell. 2011;8:363–369. doi: 10.1016/j.stem.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Thomson M, Liu SJ, Zou LN, Smith Z, Meissner A, Ramanathan S. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomioka M, Nishimoto M, Miyagi S, Katayanagi T, Fukui N, Niwa H, Muramatsu M, Okuda A. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res. 2002;30:3202–3213. doi: 10.1093/nar/gkf435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- 9.Ambrosetti DC, Scholer HR, Dailey L, Basilico C. Modulation of the activity of multiple transcriptional activation domains by the DNA binding domains mediates the synergistic action of Sox2 and Oct-3 on the fibroblast growth factor-4 enhancer. J Biol Chem. 2000;275:23387–23397. doi: 10.1074/jbc.M000932200. [DOI] [PubMed] [Google Scholar]
- 10.Guo Y, Costa R, Ramsey H, Starnes T, Vance G, Robertson K, Kelley M, Reinbold R, Scholer H, Hromas R. The embryonic stem cell transcription factors Oct-4 and FoxD3 interact to regulate endodermal-specific promoter expression. Proc Natl Acad Sci USA. 2002;99:3663–3667. doi: 10.1073/pnas.062041099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/S0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 12.Niwa H. Molecular mechanism to maintain stem cell renewal of ES cells. Cell Struct Funct. 2001;26:137–148. doi: 10.1247/csf.26.137. [DOI] [PubMed] [Google Scholar]
- 13.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 14.Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, Hubner K, Scholer HR. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- 15.Ben-Shushan E, Pikarsky E, Klar A, Bergman Y. Extinction of Oct-3/4 gene expression in embryonal carcinoma × fibroblast somatic cell hybrids is accompanied by changes in the methylation status, chromatin structure, and transcriptional activity of the Oct-3/4 upstream region. Mol Cell Biol. 1993;13:891–901. doi: 10.1128/MCB.13.2.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- 17.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dailey L, Basilico C. Coevolution of HMG domains and homeodomains and the generation of transcriptional regulation by Sox/POU complexes. J Cell Physiol. 2001;186:315–328. doi: 10.1002/1097-4652(2001)9999:9999<000::AID-JCP1046>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 19.Wang SH, Tsai MS, Chiang MF, Li H. A novel NK-type homeobox gene, ENK (early embryo specific NK), preferentially expressed in embryonic stem cells. Gene Expr Patterns. 2003;3:99–103. doi: 10.1016/S1567-133X(03)00005-X. [DOI] [PubMed] [Google Scholar]
- 20.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/S0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 21.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/S0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 22.Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 23.Shigeta M, Ohtsuka S, Nishikawa-Torikai S, Yamane M, Fujii S, Murakami K, Niwa H. Maintenance of pluripotency in mouse ES cells without Trp53 . Sci Reports. 2013;3:2944. doi: 10.1038/srep02944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 25.Hanna LA, Foreman RK, Tarasenko IA, Kessler DS, Labosky PA. Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Genes Dev. 2002;16:2650–2661. doi: 10.1101/gad.1020502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan G, Li J, Zhou Y, Zheng H, Pei D. A negative feedback loop of transcription factors that controls stem cell pluripotency and self-renewal. FASEB J. 2006;20:1730–1732. doi: 10.1096/fj.05-5543fje. [DOI] [PubMed] [Google Scholar]
- 27.Navarro P, Festuccia N, Colby D, Gagliardi A, Mullin NP, Zhang W, Karwacki-Neisius V, Osorno R, Kelly D, Robertson M, Chambers I. OCT4/SOX2-independent Nanog autorepression modulates heterogeneous Nanog gene expression in mouse ES cells. EMBO J. 2012;31(24):4547–4562. doi: 10.1038/emboj.2012.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwarz BA, Bar-Nur O, Silva JC, Hochedlinger K. Nanog is dispensable for the generation of induced pluripotent stem cells. Curr Biol. 2014;24(3):347–350. doi: 10.1016/j.cub.2013.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 30.Bourillot PY, Savatier P. Kruppel-like transcription factors and control of pluripotency. BMC Biol. 2010;8:125. doi: 10.1186/1741-7007-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall J, Guo G, Wray J, Eyres I, Nichols J, Grotewold L, Morfopoulou S, Humphreys P, Mansfield W, Walker R, Tomlinson S, Smith A. Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5:597–609. doi: 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 32.Dunn SJ, Martello G, Yordanov B, Emmott S, Smith AG. Defining an essential transcription factor program for naive pluripotency. Science. 2014;344:1156–1160. doi: 10.1126/science.1248882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 34.Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boeuf H, Hauss C, Graeve FD, Baran N, Kedinger C. Leukemia inhibitory factor-dependent transcriptional activation in embryonic stem cells. J Cell Biol. 1997;138:1207–1217. doi: 10.1083/jcb.138.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stahl N, Farruggella TJ, Boulton TG, Zhong Z, Darnell JE, Jr, Yancopoulos GD. Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science. 1995;267:1349–1353. doi: 10.1126/science.7871433. [DOI] [PubMed] [Google Scholar]
- 37.Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T, Yokota T. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen CY, Lee DS, Yan YT, Shen CN, Hwang SM, Lee ST, Hsieh PC. Bcl3 Bridges LIF-STAT3 to Oct4 Signaling in the Maintenance of Naive Pluripotency. Stem Cells. 2015;33:3468–3480. doi: 10.1002/stem.2201. [DOI] [PubMed] [Google Scholar]
- 39.Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- 40.Jirmanova L, Afanassieff M, Gobert-Gosse S, Markossian S, Savatier P. Differential contributions of ERK and PI3-kinase to the regulation of cyclin D1 expression and to the control of the G1/S transition in mouse embryonic stem cells. Oncogene. 2002;21:5515–5528. doi: 10.1038/sj.onc.1205728. [DOI] [PubMed] [Google Scholar]
- 41.Burdon T, Stracey C, Chambers I, Nichols J, Smith A. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev Biol. 1999;210:30–41. doi: 10.1006/dbio.1999.9265. [DOI] [PubMed] [Google Scholar]
- 42.Hamazaki T, Kehoe SM, Nakano T, Terada N. The Grb2/Mek pathway represses Nanog in murine embryonic stem cells. Mol Cell Biol. 2006;26:7539–7549. doi: 10.1128/MCB.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamm C, Bower N, Anneren C. Regulation of mouse embryonic stem cell self-renewal by a Yes-YAP-TEAD2 signaling pathway downstream of LIF. J Cell Sci. 2011;124:1136–1144. doi: 10.1242/jcs.075796. [DOI] [PubMed] [Google Scholar]
- 44.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/S0092-8674(03)00432-X. [DOI] [PubMed] [Google Scholar]
- 45.Murakami G, Watabe T, Takaoka K, Miyazono K, Imamura T. Cooperative inhibition of bone morphogenetic protein signaling by Smurf1 and inhibitory Smads. Mol Biol Cell. 2003;14:2809–2817. doi: 10.1091/mbc.E02-07-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. doi: 10.1016/S0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 47.Aubert J, Dunstan H, Chambers I, Smith A. Functional gene screening in embryonic stem cells implicates Wnt antagonism in neural differentiation. Nat Biotechnol. 2002;20:1240–1245. doi: 10.1038/nbt763. [DOI] [PubMed] [Google Scholar]
- 48.Yoshikawa Y, Fujimori T, McMahon AP, Takada S. Evidence that absence of Wnt-3a signaling promotes neuralization instead of paraxial mesoderm development in the mouse. Dev Biol. 1997;183:234–242. doi: 10.1006/dbio.1997.8502. [DOI] [PubMed] [Google Scholar]
- 49.Nguyen H, Rendl M, Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127:171–183. doi: 10.1016/j.cell.2006.07.036. [DOI] [PubMed] [Google Scholar]
- 50.Kelly KF, Ng DY, Jayakumaran G, Wood GA, Koide H, Doble BW. beta-catenin enhances Oct-4 activity and reinforces pluripotency through a TCF-independent mechanism. Cell Stem Cell. 2011;8:214–227. doi: 10.1016/j.stem.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Faunes F, Hayward P, Descalzo SM, Chatterjee SS, Balayo T, Trott J, Christoforou A, Ferrer-Vaquer A, Hadjantonakis AK, Dasgupta R, Arias AM. A membrane-associated beta-catenin/Oct4 complex correlates with ground-state pluripotency in mouse embryonic stem cells. Development. 2013;140:1171–1183. doi: 10.1242/dev.085654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guan KL, Figueroa C, Brtva TR, Zhu T, Taylor J, Barber TD, Vojtek AB. Negative regulation of the serine/threonine kinase B-Raf by Akt. J Biol Chem. 2000;275:27354–27359. doi: 10.1074/jbc.M004371200. [DOI] [PubMed] [Google Scholar]
- 53.Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11:11–23. doi: 10.1016/S1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 54.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rajan P, Panchision DM, Newell LF, McKay RD. BMPs signal alternately through a SMAD or FRAP-STAT pathway to regulate fate choice in CNS stem cells. J Cell Biol. 2003;161:911–921. doi: 10.1083/jcb.200211021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanda A, Sotomaru Y, Shiozawa S, Hiyama E. Establishment of ES cells from inbred strain mice by dual inhibition (2i) J Reprod Dev. 2012;58(1):77–83. doi: 10.1262/jrd.10-178A. [DOI] [PubMed] [Google Scholar]
- 58.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 59.Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin SL, Chang D, Ying SY. Asymmetry of intronic pre-miRNA structures in functional RISC assembly. Gene. 2005;356:32–38. doi: 10.1016/j.gene.2005.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 62.Fazi F, Nervi C. MicroRNA: basic mechanisms and transcriptional regulatory networks for cell fate determination. Cardiovasc Res. 2008;79:553–561. doi: 10.1093/cvr/cvn151. [DOI] [PubMed] [Google Scholar]
- 63.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific microRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/S1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 67.Houbaviy HB, Dennis L, Jaenisch R, Sharp PA. Characterization of a highly variable eutherian microRNA gene. RNA. 2005;11:1245–1257. doi: 10.1261/rna.2890305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen C, Ridzon D, Lee CT, Blake J, Sun Y, Strauss WM. Defining embryonic stem cell identity using differentiation-related microRNAs and their potential targets. Mamm Genome. 2007;18:316–327. doi: 10.1007/s00335-007-9032-6. [DOI] [PubMed] [Google Scholar]
- 69.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 71.Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 73.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2015;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 74.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-M. [DOI] [PubMed] [Google Scholar]
- 77.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-I. [DOI] [PubMed] [Google Scholar]
- 78.Borsani G, Tonlorenzi R, Simmler MC, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, Willard HF, Avner P, Ballabio A. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 79.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 80.Heard E. Recent advances in X-chromosome inactivation. Curr Opin Cell Biol. 2004;16:247–255. doi: 10.1016/j.ceb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 81.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 82.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Navarro P, Chambers I, Karwacki-Neisius V, Chureau C, Morey C, Rougeulle C, Avner P. Molecular coupling of Xist regulation and pluripotency. Science. 2008;321:1693–1695. doi: 10.1126/science.1160952. [DOI] [PubMed] [Google Scholar]
- 84.Pasque V, Tchieu J, Karnik R, Uyeda M, Sadhu Dimashkie A, Case D, Papp B, Bonora G, Patel S, Ho R, Schmidt R, McKee R, Sado T, Tada T, Meissner A, Plath K. X chromosome reactivation dynamics reveal stages of reprogramming to pluripotency. Cell. 2014;159:1681–1697. doi: 10.1016/j.cell.2014.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang XP, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:U260–U295. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2010;16:324–337. doi: 10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Savic N, Bar D, Leone S, Frommel SC, Weber FA, Vollenweider E, Ferrari E, Ziegler U, Kaech A, Shakhova O, Cinelli P, Santoro R. lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell. 2014;15:720–734. doi: 10.1016/j.stem.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 89.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 91.Struhl G. A gene product required for correct initiation of segmental determination in Drosophila. Nature. 1981;293:36–41. doi: 10.1038/293036a0. [DOI] [PubMed] [Google Scholar]
- 92.Endoh M, Endo TA, Endoh T, Fujimura Y, Ohara O, Toyoda T, Otte AP, Okano M, Brockdorff N, Vidal M, Koseki H. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135(8):1513–1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- 93.Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, Bickmore WA. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrn2731. [DOI] [PubMed] [Google Scholar]
- 95.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 96.Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Kocher T, Cohen A, Stunnenberg HG, Wilm M, Muller J. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 2007;26:4078–4088. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Walker E, Chang WY, Hunkapiller J, Cagney G, Garcha K, Torchia J, Krogan NJ, Reiter JF, Stanford WL. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2010;6:153–166. doi: 10.1016/j.stem.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol. 2008;28:2718–2731. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Savla U, Benes J, Zhang J, Jones RS. Recruitment of Drosophila Polycomb-group proteins by Polycomblike, a component of a novel protein complex in larvae. Development. 2008;135:813–817. doi: 10.1242/dev.016006. [DOI] [PubMed] [Google Scholar]
- 100.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 101.Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26(6):1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, Brookes E, Pombo A, Fisher C, Skarnes WC, Snoek T, Bezstarosti K, Demmers J, Klose RJ, Casanova M, Tavares L, Brockdorff N, Merkenschlager M, Fisher AG. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol. 2010;12:618–624. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/S1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 105.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 106.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 107.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 108.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, Adli M, Kasif S, Ptaszek LM, Cowan CA, Lander ES, Koseki H, Bernstein BE. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, Wutz A, Vidal M, Elderkin S, Brockdorff N. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qin J, Whyte WA, Anderssen E, Apostolou E, Chen HH, Akbarian S, Bronson RT, Hochedlinger K, Ramaswamy S, Young RA, Hock H. The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell. 2012;11:319–332. doi: 10.1016/j.stem.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, Bourque G, Oh S, Ruan Y, Ng HH, Wei CL. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 112.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 114.Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, Wang J, Rendl M, Bernstein E, Schaniel C, Lemischka IR. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci U S A. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, Ramalho-Santos M. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–868. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang L, Du Y, Ward JM, Shimbo T, Lackford B, Zheng X, Miao YL, Zhou B, Han L, Fargo DC, Jothi R, Williams CJ, Wade PA, Hu G. INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell. 2014;14:575–591. doi: 10.1016/j.stem.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. 2014;15:703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 122.Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Christman JK, Price P, Pedrinan L, Acs G. Correlation between hypomethylation of DNA and expression of globin genes in Friend erythroleukemia cells. Eur J Biochem. 1977;81:53–61. doi: 10.1111/j.1432-1033.1977.tb11926.x. [DOI] [PubMed] [Google Scholar]
- 124.McGhee JD, Ginder GD. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature. 1979;280:419–420. doi: 10.1038/280419a0. [DOI] [PubMed] [Google Scholar]
- 125.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 126.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 127.Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 128.Ng RK, Dean W, Dawson C, Lucifero D, Madeja Z, Reik W, Hemberger M. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol. 2008;10(11):1280–1290. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, Nakayama J, Okano M. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- 130.Oda M, Kumaki Y, Shigeta M, Jakt LM, Matsuoka C, Yamagiwa A, Niwa H, Okano M. DNA methylation restricts lineage-specific functions of transcription factor Gata4 during embryonic stem cell differentiation. PLoS Genet. 2013;9(6):e1003574. doi: 10.1371/journal.pgen.1003574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Koh KP, Rao A. DNA methylation and methylcytosine oxidation in cell fate decisions. Curr Opin Cell Biol. 2013;25:152–161. doi: 10.1016/j.ceb.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, Lahesmaa R, Orkin SH, Rodig SJ, Daley GQ, Rao A. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, Gao Q, Powell BE, Li Z, Xu M, Faull KF, Lyko F, Jaenisch R. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang J, Qiao M, He Q, Shi R, Loh SJ, Stanton LW, Wu M. Pluripotency activity of nanog requires biochemical stabilization by variant histone protein H2A.Z. Stem Cells. 2015;33:2126–2134. doi: 10.1002/stem.2011. [DOI] [PubMed] [Google Scholar]
- 136.Pijnappel WW, Esch D, Baltissen MP, Wu G, Mischerikow N, Bergsma AJ, van der Wal E, Han DW, Bruch H, Moritz S, Lijnzaad P, Altelaar AF, Sameith K, Zaehres H, Heck AJ, Holstege FC, Scholer HR, Timmers HT. A central role for TFIID in the pluripotent transcription circuitry. Nature. 2013;495:516–519. doi: 10.1038/nature11970. [DOI] [PubMed] [Google Scholar]
- 137.Yang VS, Carter SA, Hyland SJ, Tachibana-Konwalski K, Laskey RA, Gonzalez MA. Geminin escapes degradation in G1 of mouse pluripotent cells and mediates the expression of Oct4, Sox2, and Nanog. Curr Biol. 2011;21:692–699. doi: 10.1016/j.cub.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Caronna EA, Patterson ES, Hummert PM, Kroll KL. Geminin restrains mesendodermal fate acquisition of embryonic stem cells and is associated with antagonism of Wnt signaling and enhanced polycomb-mediated repression. Stem Cells. 2013;31:1477–1487. doi: 10.1002/stem.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang L, Miao YL, Zheng X, Lackford B, Zhou B, Han L, Yao C, Ward JM, Burkholder A, Lipchina I, Fargo DC, Hochedlinger K, Shi Y, Williams CJ, Hu G. The THO complex regulates pluripotency gene mRNA export and controls embryonic stem cell self-renewal and somatic cell reprogramming. Cell Stem Cell. 2013;13:676–690. doi: 10.1016/j.stem.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Smith AG, Hooper ML. Buffalo rat liver cells produce a diffusible activity which inhibits the differentiation of murine embryonal carcinoma and embryonic stem cells. Dev Biol. 1987;121:1–9. doi: 10.1016/0012-1606(87)90132-1. [DOI] [PubMed] [Google Scholar]
- 141.Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 142.Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]