Abstract
Conformational flexibility conferred though regions of intrinsic structural disorder allows proteins to behave as dynamic molecules. While it is well-known that intrinsically disordered regions can undergo disorder-to-order transitions in real-time as part of their function, we also are beginning to learn more about the dynamics of disorder-to-order transitions along evolutionary time-scales. Intrinsically disordered regions endow proteins with functional promiscuity, which is further enhanced by the ability of some of these regions to undergo real-time disorder-to-order transitions. Disorder content affects gene retention after whole genome duplication, but it is not necessarily conserved. Altered patterns of disorder resulting from evolutionary disorder-to-order transitions indicate that disorder evolves to modify function through refining stability, regulation, and interactions. Here, we review the evolution of intrinsically disordered regions in eukaryotic proteins. We discuss the interplay between secondary structure and disorder on evolutionary time-scales, the importance of disorder for eukaryotic proteome expansion and functional divergence, and the evolutionary dynamics of disorder.
Keywords: Intrinsic disorder, Protein evolution, Neutrality, Dosage, Gene duplication, Functional divergence, Disorder-to-order transition, Evolutionary dynamics
Introduction
Proteins tend to evolve through an intricate interplay between sequence divergence, protein structure stability, and functional constraint. In general, protein structure is assumed to be mostly maintained as sequences diverge in order for proteins to fold properly [1]. If a protein does not fold properly, its functional properties are often negatively affected. Based on the PDB collection, protein secondary structure elements and their topology (or fold) are often highly conserved, implying that the topology of protein secondary structure elements can remain very similar even after sequences have diverged beyond recognition.
Nonetheless, most proteins in PDB are shown as static snapshots that belie their structural flexibility. Proteins with highly flexible regions are not amenable to traditional experimental structure determination and, in many cases, such methods are not even attempted [2]. Multidomain proteins are frequently truncated to bypass high flexibility or size restrictions. Examples of shape-shifting or metamorphic proteins [3] that can refold upon changes in domain contacts or by changes in environmental conditions are still rare in PDB, but one interesting example is RfaH, where the C-terminal domain can refold from an all alpha-helical fold to a fold containing only beta strands in response to altered interdomain contacts [4]. This extreme case of fold transition and conformational flexibility illustrates that protein structure is not always conserved among homologous proteins and emphasizes the importance of domain context for our understanding of protein fold space. However, conformational flexibility is not always as dramatic as for metamorphic proteins and smaller changes are more common.
Many proteins exist as conformational ensembles rather than a single conformation. This enables the rapid sampling of multiple conformations in a flattened, rugged energy landscape where certain conformations may predominate, even for proteins with global intrinsic disorder and for proteins with intrinsically disordered regions (IDRs) [5]. Some IDRs can act as dynamic switches in response to various signals such as pH, temperature, ligands, allosteric effectors, and post-translational modifications [6], allowing the conformational ensemble to re-equilibrate, ultimately causing a population shift [7]. In accordance with the extended conformational selection processes, binding events ranging from lock-and-key to induced fit are plausible, including IDRs that participate in important interactions mediated by fold-upon-binding events [8], or bind without folding [9] Furthermore, the conformational ensemble undergoes population shifts in response to point mutations and sequence variability [10]. Consequently, amino acid replacement will ultimately impact processes of conformational selection in response to different stimuli in a lineage-specific manner, as a mutation-driven conformational selection process [11]. For ambiguous sites that are disordered in some PDB structures but ordered in others, the amount of ambiguity depends on exposure to different environments, implying that regions with conflicting disorder assignments should not be regarded as lacking intrinsic disorder entirely [12]. By definition, a region of ambiguous disorder can be either disordered or ordered, depending on the environmental conditions, similar to previously described dual-personality regions [13].
Structurally disordered proteins are often found to interact with many different cellular targets and to perform promiscuous or moonlighting functions [14–16]. As the conformational ensemble transitions from one favored conformation to another, it may pass through unintended opportunistic functional conformations. Thus, mutation-driven conformational selection provides a mechanism for functional divergence among related proteins and conformational flexibility in proteins may play an important role in the evolutionary innovation and fluctuation of protein functions mediated through IDRs (e.g., [17–20]). Importantly, mutation-driven conformational selection may mostly be driven by genetic drift in a near-neutral, perhaps deleterious, manner that at times offers a rapid way to adapt to altered environmental conditions or signals.
Contemporary work on intrinsically disordered proteins has illuminated the profound functional importance of disorder, particularly in regard to high-level eukaryotic cellular complexity and the expansion of the eukaryotic proteome. Recent work by Chakrabortee et al. entitled Intrinsically Disordered Proteins Drive Emergence and Inheritance of Biological Traits, describes disordered yeast proteins with the capacity to induce heritable molecular memories with specific biological traits, stable over generations and transmissible from individual to individual [20]. The inheritance of these protein-driven traits is prion-like but, importantly, amyloid formation is not detected, and the inheritance-inducing proteins are conserved from human to yeast [20]. Additionally, the relaxed selective pressure experienced by many IDRs may allow for the emergence of parallel, nucleotide-level functionality within the coding regions of disordered or partially disordered proteins [21]. Here, we review fundamental evolutionary underpinnings that have influenced intrinsic disorder content in eukaryotic genomes, with an emphasis on the importance of disorder for eukaryotic proteome expansion and functional divergence, the interplay between secondary structure and disorder on evolutionary time-scales, and the evolutionary dynamics of intrinsic disorder.
Distribution of intrinsic disorder
According to proteome-wide disorder predictions, eukaryotes have a significantly larger fraction of intrinsic disorder in their proteomes than prokaryotes [22, 23]. On average, the disorder content is 7.4, 8.5, and 20.5% in Archaea, Bacteria, and Eukaryotes, respectively [24]. Despite the sharp increase in disorder content from prokaryotes to eukaryotes, the notion that disorder is correlated with organismal complexity (as measured by number of cell types) has not been strongly supported [22, 23]. However, many characteristic features of eukaryotic genomes appear to be linked to intrinsic disorder, particularly those with perplexing evolutionary origins. It is widely noted that concepts of organismal complexity are tightly linked with small effective population sizes, suggesting some type of drift barrier driving complexity in an expanded genome and/or simple relaxed selection on structure, as described below.
Most prokaryotic genomes are densely packed (“wall-to-wall”) with transcribed DNA, containing relatively few intergenic regions or non-coding spacers within their protein-coding genes, whereas eukaryote genome sizes are largely decoupled from their biological information content, and in many taxa, only a small fraction of the total genomic DNA is evidently transcribed [25, 26]. A compelling explanation for this disparity in genome architecture relates to the fundamental theorem of natural selection originally derived by Fisher, namely, that the efficiency of natural selection is directly related to the diversity (and by extension, the effective size) of a population [27]. Recent work suggests that complex genomic features in eukaryotes, including the emergence of large protein families and the presence of intronic regions within protein-coding genes, are the result of “non-adaptive” evolution: persistently low selective pressure maintained by small effective population size [28–30]. Interestingly, many hallmark features of eukaryotic proteins such as intronic DNA insertions, large functional domain architectures and complex molecular interaction networks are often associated with or even dependent upon intrinsic disorder (further discussed below).
Structural evidence has confirmed that the non-coding DNA fragments within eukaryotic protein-coding genes (introns) are in fact derived from ancient bacterial Group II selfish elements that were introduced to the nuclear genome by endosymbionts during eukaryogenesis [31–33]. This unique evolutionary event facilitated the emergence of the eukaryotic splicesome, allowing for introns to be removed, as well as for exons (the remaining coding regions) to be rearranged, prior to translation. Nilsen and Graveley contend that alternative splicing has enabled a crucial expansion of the effective eukaryotic proteome [34] and, notably, protein regions associated with alternative splicing are often intrinsically disordered [35, 36].
Laboratory simulations have demonstrated that under strong, efficient selective pressure, genomes become minimally short, and even mildly deleterious genes are eliminated [37, 38]. Consequently, there is mounting support for the notion that rapid eukaryotic genome expansion, and the resulting low-information-density architecture, is a “syndrome” brought about by pervasive genetic drift and low purifying selective pressure [39, 40] Importantly, this expansion has occurred alongside several other genomic features (some of which were discussed above), and Koonin [41] asserts that the common ancestor of all eukaryotes was of comparable complexity to many modern protists, indicating that expansive, complex genomes are an enduring trait within Eukaryota. Given the close connection that intrinsic disorder has to several defining features of eukaryotes, it is likely that the sharp rise in disordered proteins observed in this lineage is yet another “symptom” of their genomic “syndrome.”
Eukaryote proteome expansion (and what disorder has to do with it)
Gene and genome duplications
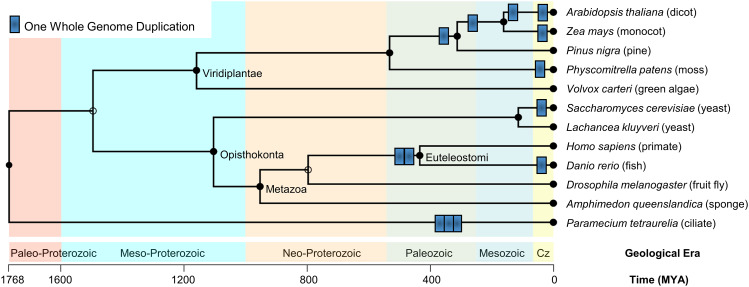
During the course of eukaryotic evolution, multiple whole genome duplication (WGD) events are known to have occurred in major eukaryotic lineages. Based on sequence comparison, only more recent WGD events can be detected, but earlier WGD events are probable [42]. A selection of known WGD events (Fig. 1) show that Paramecium tetraurelia has undergone three rounds of WGD [43], WGD is common in plants, with both more ancient [44] and recent WGD especially in flowering plants [45], but also in moss [46]. In fungi, one WGD occurred in the Saccharomyces cerevisiae lineage [47], in animals, two rounds of WGD occurred at the origin of vertebrates [48], followed by numerous WGD in teleosts, e.g., Danio rerio has undergone one round of WGD [49], while Salmo salar has undergone a fourth round [50] (not shown). In addition to WGD, small-scale gene duplications (SSDs) whereby one gene or chromosome segment is duplicated, also constitute a major mechanism driving functional divergence in protein family evolution. The evolutionary dynamics of genes that emerged after WGD versus SSD are different and this has been analyzed in detail [42, 51].
Fig. 1.
A selection of known whole genome duplication (WGD) events in eukaryotes. One round of WGD is illustrated by a blue rectangle. The background is colored by geological era. Time axis and geological eras are from TimeTree [113–115]
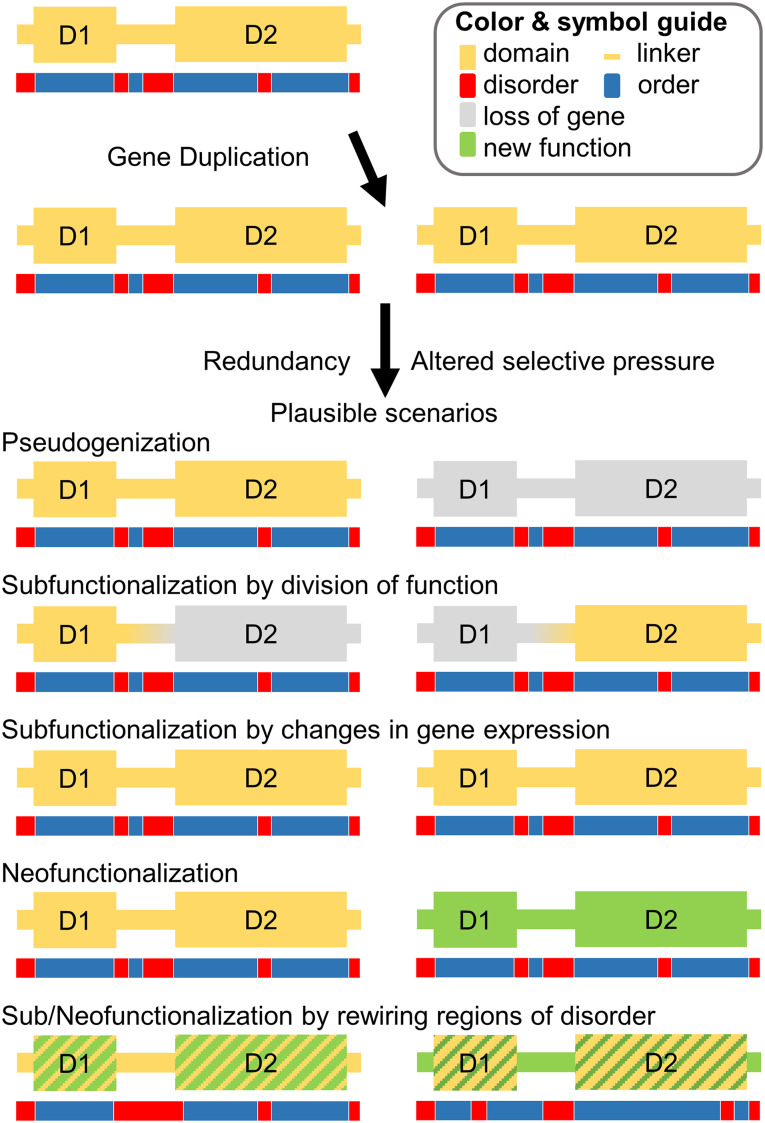
Gene duplications generate redundancy, enabling the exploration of novel functions [52]. Through accumulation of mutations, different evolutionary fates are plausible for the two different copies [28, 53]. The most common scenario after gene duplication is that one copy loses its function and becomes pseudogenized [28]. Retention rates are higher for duplicates that stem from WGD than from SSD, especially for gene copies that are sensitive to altered gene stoichiometry (dosage effects) [42]. For genes that are retained in duplicate, functional divergence between the two copies often results [54]. Proposed models for retention are neofunctionalization [52] and subfunctionalization [55] (Fig. 2). In the neofunctionalization model, one domain copy is able to retain its original function while the duplicated domain can explore new functions. In the subfunctionalization model, the ancestral function is divided amongst the resulting duplicates. Subfunctionalization has been computationally shown to be a neutral process that can result in neofunctionalization [56]. In addition, subfunctionalization in gene expression (dosage) between two duplicated copies contributes to their pattern of retention [57]. Recent work has described the expected interplay of gene dosage with neofunctionalization and subfunctionalization [58].
Fig. 2.
Gene duplication generates two copies of the same gene and consequently functional redundancy. Different scenarios after gene duplication include pseudogenization (one copy is lost), subfunctionalization (the two copies subdivide function or gene expression), and neofunctionalization (at least one copy gains a new function)
In vertebrates, the retention rate for ohnologs (proteins related by WGD) from the WGD events at the origin of vertebrates is significantly higher than for SSD for genes involved in protein binding, signal transduction, development, DNA binding, receptor activity, ion transport, and protein modifications [59]. In plants, genes with functions in signal transductions and transcriptional regulations follow a similar pattern [60]. Copies retained after WGD are often dosage-sensitive (sensitive to unbalanced stoichiometry of gene copies) [42]. Many of these protein functions are known to depend on intrinsic disorder [61]. Indeed, intrinsically disordered proteins have been found to be dosage-sensitive (sensitive to unbalanced gene expression) and it was postulated that the promiscuous interactions that disordered proteins frequently partake in could explain the need to maintain stoichiometry [62]. On evolutionary time-scales, multiple interaction partners provide multiple opportunities to subfunctionalize and each partner can neofunctionalize, increasing the selective pressure for both copies from both partners to be retained [42]. Furthermore, after WGD in yeast, proteins enriched in post-translational modification sites are retained at a greater rate [63]. The post-translational modification sites are often found within IDRs [64, 65] and indeed, yeast ohnologs are more intrinsically disordered than singletons (for which the other copy was lost after WGD) [66]. Further comparison of the yeast ohnologs with pre-duplication orthologs shows that 29% of the duplicates and 25% of the singletons have gained disorder, while 37% of the duplicates and 25% of the singletons have lost disorder [66]. The ohnologs that gained disorder were also found to have a higher number of interactions, suggesting that disorder facilitates divergence and innovation [66]. Comparing interactomes of human, fly, and yeast, structurally disordered networks are rewired significantly faster than ordered networks, leading to a speculation that disordered proteins have a higher capacity to rapidly rewire their interactions [67].
Domain rearrangements
Eukaryotic proteins are significantly longer and have more domains than prokaryotic proteins [68]. Domains are the main unit of protein evolution [69]. In addition to sequence divergence, proteins also diverge by rearranging domain architectures and through loss and gain of domains [70, 71]. Eukaryotic multidomain proteins are frequently the result of stepwise insertions of a single domain, but occasionally, several domains are added in tandem [71]. Mostly, established domains that already exist in the proteome are added to proteins and many domains are found in numerous different domain architectures. Gain of a novel (emerging) domain may occur by, e.g., acquisition of novel genetic material, converting non-protein coding genetic material into protein-coding genes and this novel genetic material is often intrinsically disordered [72]. Disordered, emerging domains were found to be rapidly spread across Drosophila lineages [73] and in plants [74]. Domains can also be lost from multidomain proteins [73]. Altered domain architecture may impact the amount of disorder that a protein can withstand, as is the case in the p53 DNA-binding domain [75]. In the p53 family, a choanoflagellate has three of four domains found in the vertebrate p53 family and the four domain protein is present in gastropods. All but one domain are missing from the p53 protein in Neoptera. For the p53 DNA-binding domain that is shared from choanoflagellates to vertebrates, the disorder content is positively correlated with the number of domains in its domain architecture. The neopteran proteins have not only lost the other domains but also disorder content, while the early choanoflagellate and four domain gastropod proteins have disorder content similar to the 3–4 domain proteins in vertebrates [75]. In addition, for the three vertebrate paralogs in the p53 family (p53, p63, and p73), p53 has lost one domain and for the p53 DNA-binding domain, some of the secondary elements have lower conservation of disorder for the p53 clade than in the p63 and p73 clades, while others, e.g., one of the main beta strands in the central beta sheet are conserved in disorder for the p53 clade, but are not disordered in the p63 and p73 clades [75].
Expansions of eukaryotic proteins are often due to insertion of disordered sequence [76]. A common event in protein evolution is the occurrence of insertions and deletions (indels) [77]. Indels have been demonstrated to have high disorder content, with longer indels being particularly disordered [78]. However, indels do not induce disorder but rather appear to accumulate in regions that are already disordered [76]. Repeat sequences, which are often disordered [79–81], have been associated with increased indels [82]. At the gene level, indels often occur in multiples of three, an indication that there may be selective pressure to maintain the reading frame, as a frameshift mutation may be deleterious [83]. Predictions on the effect of known frameshift mutations showed that the majority were gene-damaging [84]. Deleterious mutations caused by a frameshift indel may be compensated for by another indel that restores the reading frame [83, 84].
Sequence divergence rate in disordered sites
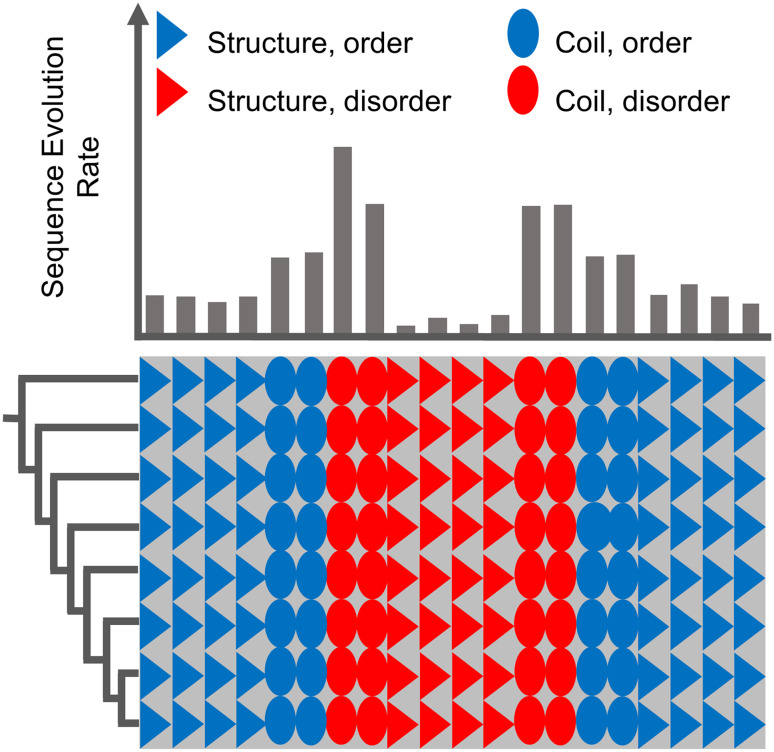
Early research has suggested that intrinsically disordered regions diverge rapidly in sequence [85, 86]. However, in a later study, disorder-promoting residues were found to have higher conservation in disordered regions than in ordered regions, and more than 25% of the disordered sites evolved more slowly than the ordered sites [87]. A possible reason for such conflicting results is that, in general, the relationship between sequence divergence and intrinsic disorder has been conceptualized in a “one-way” statistical framework, without direct consideration of the possible interaction among the multiple structural factors that drive sequence divergence. To address this, a large-scale study of metazoan protein families investigating the interaction of disorder, secondary structure, and functional domains on site-specific sequence divergence rates was recently performed [88]. Focusing only on gap-free sites, with 100% conserved structural predictions across all sequences in each alignment, statistically significant shifts in the rate distributions of opposing structural properties were found: ordered sites tended to be more conserved than disordered sites, sites in secondary structures tended to be more conserved than sites in random coils, and sites within functional domains tended to be more conserved than sites in linkers [88]. However, a considerable overlap between each of these rate distribution pairs was found, and factorial analysis indicated a strong confounding interaction between disorder propensity and secondary structure involvement: sites that were predicted to be disordered, but also involved in secondary structure, were the most evolutionarily constrained at the residue level, even more so than sites within ordered secondary structures [88] (Fig. 3).
Fig. 3.
Hypothetical multiple sequence alignment illustrating the relationship between structural properties and the rate of sequence evolution: sites with propensity for both intrinsic disorder and secondary structure tend to evolve slowly
In silico simulations have also found that disorder is more difficult to maintain than secondary structure elements on evolutionary time-scales [89]. The dataset from [88] described above had a total of ~5.9 million gap free alignment sites, about ~29% of which show a mixture of disorder and order among sequences. This result corroborates the notion that disorder is not necessarily a conserved trait among members of a protein family. Other researchers have argued that there are actually distinct types of intrinsic disorder, some of which are retained across lineages and have highly conserved amino acid sequences [90, 91].
Together, these findings are compatible with the realization that different IDRs play diverse and often important functional roles in vivo [92]. For example, whereas some IDRs simply function as entropic chains or flexible linker regions around domains, others act as recognition sites that mediate protein–protein interactions by undergoing disorder-to-order transitions upon binding to their one or many different interaction partners [61].
Disorder-to-order transitions
Real-time disorder-to-order transitions
Regions in proteins that are involved in disorder-to-order transitions are commonly referred to as molecular recognition features (MoRFs) that upon interaction with another protein or nucleic acid can fold into an alpha helical structure, a beta strand, a fixed coil, or a complex mixture of all [93]. Eukaryotic proteins contain about 2.5 disordered regions. Of these disordered regions, about one-fifth contains at least 1 MoRF [94]. Also embedded in disordered regions are small linear motifs (SLiMs) and low complexity regions. Altogether, these contribute to function and functional promiscuity mediated through disordered regions with both beneficial and some less beneficial effects [95]. Notably, MoRFs are known to form transient secondary structural elements in their bound state [93, 96], and it is possible that the highly conserved protein regions described by [88], which are predicted to be both intrinsically disordered and involved in secondary structures, are actually MoRFs. Furthermore, proteins may also contain ordered regions that are activated by unfolding in response to a certain trigger [97]. The triggers range from biomolecular interactions to global environmental factors such as temperature, pH, or light causing these proteins to undergo functionally important order-to-disorder transitions in real time [97].
Evolutionary time-scale disorder-to-order transitions
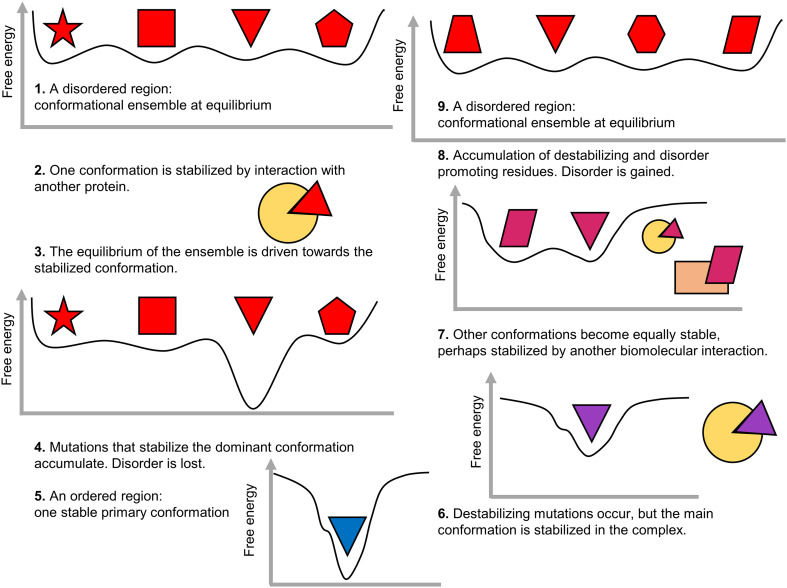
Disorder evolves in patterns that suggest it contributes to fine-tuning regulation, stability, and interactions, especially after gene duplication. Some of these functions are induced through post-translational modification, such as phosphorylation. As noted above, post-translationally modified genes are retained at a higher rate after WGD [63]. Importantly, sites enabling post-translational regulation have been found to systematically contribute to functional divergence after gene duplication [98]. SLiMs that promote transient interactions with other proteins are abundant in disordered regions. While some SLiMs are conserved, others are rapidly gained and lost in different lineages, as well as after gene duplication [99]. Beneficial motifs that have an adaptive phenotype are thought to (1) become fixed more frequently and (2) optimize the motif binding pocket, sometimes at the expense of the motif itself [99]. A similar scenario can be envisaged for disorder. Disordered regions are present as a conformational ensemble at an equilibrium, but when a non-functional disordered region gains a conformation with a possibly beneficial function (e.g., displaying a SLiM, sometimes by chance), mutations may stabilize that conformation further, driving the initial conformational equilibrium towards that conformation and eventually, the disordered region will become ordered (Fig. 4). By becoming ordered, the protein can undergo a neostructuralization event, where it obtains ordered structured regions not present in ancestral homologs [19]. By gaining an ordered region, homeostasis can be at risk since loss of disorder increases the protein’s half-life and disorder content can potentially fine-tune protein turnover rate on evolutionary time-scales [100]. One can speculate that the previously disordered, now ordered, segment has increased its fitness, allowing another region to become less structurally constrained. Thus, an ordered region can transition towards disorder, perhaps through transient functional conformations and motifs. Eventually, a transition from order-to-disorder has occurred on evolutionary time-scales. It should be noted that even if the same region transitions from disorder to order and back to disorder, the conformational ensemble will likely have a different composition (Fig. 4).
Fig. 4.
Disorder-to-order transition and order-to-disorder transition on evolutionary time-scales. Disorder-to-order: a region from a hypothetical disordered protein becomes, e.g., preferentially stabilized, driving the equilibrium of the conformational ensemble towards solely the preferred conformation. The preferred conformation is further stabilized by mutations and becomes incrementally predominant. Over time, the region becomes ordered displaying only the predominant conformation. Order-to-disorder: a region from a hypothetical order protein starts to become more flexible, but is stabilized under certain conditions. Flexibility is beneficial and mutations to promote disorder accumulate, perhaps additional functions arise, and additional preferred conformations may become more predominant. Over time, the region becomes disordered, existing as a conformational ensemble
Evolutionary transitions from disorder-to-order and from order-to-disorder were observed in a large-scale study of 17 kinase paralogous clades. Looking at patterns of disorder conservation within and between clades, disorder-prone regions are apparent [101]. The disorder-prone regions have conserved regions of disorder in multiple clades, but not necessarily in closely related clades. This suggests that even if disorder is found for the same region in two different clades, the disorder may be a homoplasic trait (due to convergence) with important differences in the conformational ensemble and consequently, function may not be the same. Notably, no disorder-prone region is conserved across all 17 clades [101]. Within orthologs, certain sites are undergoing disorder-to-order transitions on evolutionary time-scales in a lineage-specific manner, characterized by a moderate disorder-to-order transition rate. Lineage-specific changes in conserved disorder are also present in the p53 family: the p63 and p73 clades have strong signals of regions that have become ordered in the ray-finned fish lineage implying functional divergence [75]. Similar results are observed in Arabidopsis NAC transcriptions factors, where intrinsic disorder is not conserved across the entire family though subgroup-specific patterns can be found [102]. Additional examples of protein families where disorder prediction implies that evolutionary disorder-to-order transitions have occurred are the mediator complex [103], the vertebrate Prion protein family [104], the clusterin family [19], the synuclein family [19] and in emerin, various phylogenetic groups showed differential tendencies towards being disordered [105].
The evolutionary disorder-to-order transitions are potentially biased from disorder to order since disorder is difficult to maintain on evolutionary time-scales [89], but transitions in both directions must occur. When different models of evolution were constructed for disordered versus ordered proteins, the resulting disordered and ordered matrices showed that substitutions from order-promoting residues to disorder-promoting residues were unlikely for both matrices, though they were slightly more likely for disordered proteins [106]. Considering that different studies have found that the degree of sequence conservation in disordered regions depends on structural and functional properties of the disordered sites [85, 87, 88, 107], e.g., sites with both disordered and structured properties are more conserved than other disordered sites [88, 107], it is necessary to carefully construct such models considering additional properties of the disordered sites. In addition, even if disorder may be found in disorder-prone regions, these are not necessarily conserved, and care must be taken to ensure disorder conservation across compared sites. Disorder patterns that seem conserved between two paralogous clades can arise from convergent evolution [101], but further research is needed in this area. Nevertheless, patterns of disorder can be informative in finding remote homologs that are difficult to detect with sequence-based methods alone, and have been found to identify remote Myc homologs [108] and remotely related E3 ubiquitin-protein ligases [109], but clustering of sequences based on such patterns may be more informative for functional inference than for phylogenetic signal.
Conservation of functional disorder
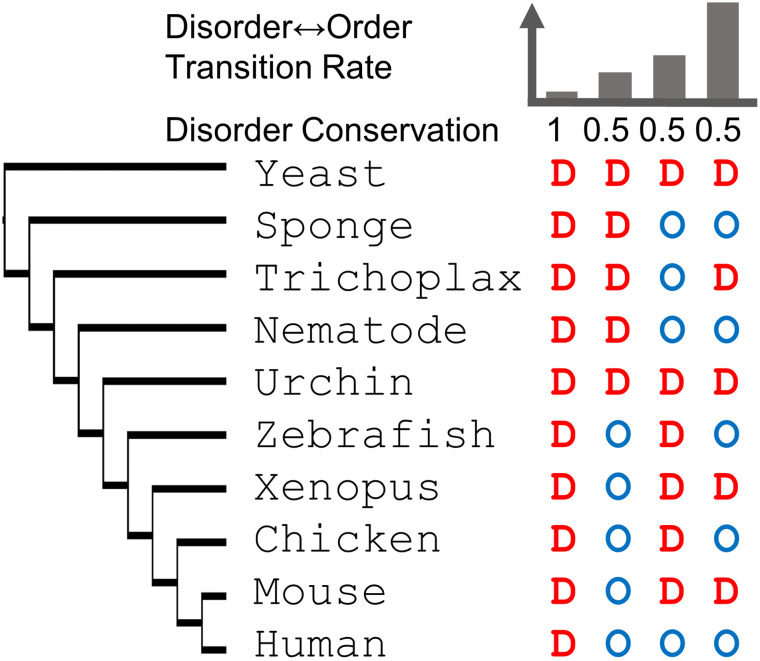
Bellay et al. classified disordered sites among yeast orthologs into functionally constrained disorder, considering disorder to be conserved if at least 50% of sequences at an alignment site were predicted to be disordered and constrained if sequence was conserved at 50% [90]. Furthermore, sites were classified to have functionally flexible disorder if at least 50% of sequences at a site were predicted to be disordered but with sequence conservation below 50%. Last, sites with few disorder predictions were classified as non-functional disorder. Using slightly more generous cutoffs across metazoa, constrained disorder was allowed to be less conserved (>30%) in disorder but highly conserved (>90%) in sequence while flexible disorder was less conserved in sequence (<90%) showing that approximately 30% of sites were disordered (constrained or flexible) and that more constrained disorder is found for human proteins that lack yeast orthologs (8%) than for human proteins with yeast orthologs (5%) [91]. While this may indicate that the older orthologs have lost disorder or that more disordered domains have emerged or spread after the divergence of yeast and metazoa, the arbitrary cutoffs in these studies are concerning since an arbitrary cutoff of 50% disorder conservation at a site could mean that the state changed one time or that it is changing between every other species with high explicit impact on the evolutionary dynamics (or rate) by which disorder is lost or gained (Fig. 5).
Fig. 5.
Superimposing disorder prediction onto a multiple sequence alignment enables rate inference of disorder and order for homologous sites over a phylogenetic tree based on the corresponding multiple sequence alignment. Conservation of intrinsic disorder versus rate of disorder-order transition for four hypothetical sites: while the first site is conserved in disorder, sites 2–4 have a conservation of 0.5 but the rate varies from slow to fast depending on the pattern of disorder and order in the evolutionary context
Protein evolvability and disorder
Examining the fold distribution according to the CATH database, about 1300 folds describe the experimentally determined protein structure space [110]. More than half of the non-redundant domains in CATH can be described by the 100 most frequently found CATH superfamily domains [110]. Many of these domains have folds that display regular secondary structure architectures with supersecondary structures forming a stable core [110]. These are folds with high evolvability. Like disordered regions, these are characterized by high sequence divergence and a plethora of functional contexts. One important distinction must be made; while the common folds can promote various functions, proteins that assume these folds typically only have one function, whereas disorder enables functional versatility within the same protein. The amount of disorder is positively correlated with robustness to withstand mutations while still maintaining structure and both are negatively correlated with fold complexity [111]. In this context, fold complexity is defined as average contact order based on the linear distance in the sequence between two contacting residues. Alpha-helices have low contact order due to their local contacts [112] and consequently, several of the most common CATH folds, with regular secondary structure architectures and rich in supersecondary structures, may also have low contact order and thus low fold complexity. The disordered sites that also have propensity to form secondary structure are more conserved [88]. Thus, this category of disorder appears to have lower robustness that may be due to an increased constraint to fold under certain conditions.
Evolution of disorder drives biological diversity
Using Bellay’s criteria [90], only a small fraction of protein sequence space contains functional disorder. Indeed, most disordered regions appear to experience relaxed selective pressure, and thus, high amino acid substitution rates [85, 88]. However, it is now also clear that intrinsically disordered sequences should be considered in a larger structural and functional context to evaluate the evolutionary pressures that act upon them. Moreover, the interplay between intrinsic disorder and other structural/functional properties is likely to have unforeseen, confounding effects that can only be detected using appropriately complex analyses [88].
What Bellay et al. [90] classify as non-functional disorder may in fact contribute significantly to natural variation within a species and to biological diversity between species. While some disordered regions may need to perform predictable, reliable functions, others may be important for generating subtle changes in response to a signal. By accumulating tiny changes in function affecting protein dynamics, binding affinities, and promiscuous and moonlighting functions, subtle variation and diversity can emerge within a population or protein family. Ultimately, such small changes in disorder content can greatly impact a population’s response to changes in the environment.
If disorder can be used to prime or seed molecular memories that promote a heritable and beneficial trait [20], can that trait be selected for, in the sense that disorder-prone residues will start to become replaced with order-prone residues that can fold into the beneficial conformation without the original primer or seed if the environmental trigger remains? Additionally, if IDRs tend to occur in evolutionarily labile sequence regions, can they serve as hotbeds for the novel acquisition of parallel, nucleotide-level biological function [21]? Hopefully, future work will shed more light on the increasingly broad functional capacity of intrinsic disorder in eukaryotes. Still, what has been discovered so far provides compelling evidence for the notion that protein disorder is an indispensable component of the seemingly non-adaptive evolutionary processes responsible for the striking complexities and functional novelties observed throughout the eukaryotic lineage.
Acknowledgements
JNC was supported by NIH/NIGMS R25 GM061347. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health.
References
- 1.Kolodny R, Pereyaslavets L, Samson AO, Levitt M. On the universe of protein folds. Annu Rev Biophys. 2013;42:559–582. doi: 10.1146/annurev-biophys-083012-130432. [DOI] [PubMed] [Google Scholar]
- 2.Slabinski L, Jaroszewski L, Rodrigues APC, et al. The challenge of protein structure determination—lessons from structural genomics. Protein Sci. 2007;16:2472–2482. doi: 10.1110/ps.073037907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murzin AG. Biochemistry: metamorphic proteins. Science. 2008;320:1725–1726. doi: 10.1126/science.1158868. [DOI] [PubMed] [Google Scholar]
- 4.Burmann BM, Knauer SH, Sevostyanova A, et al. An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell. 2012;150:291–303. doi: 10.1016/j.cell.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai CJ, Ma B, Sham YY, et al. Structured disorder and conformational selection. Proteins. 2001;44:418–427. doi: 10.1002/prot.1107. [DOI] [PubMed] [Google Scholar]
- 6.Smock RG, Gierasch LM. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma B, Kumar S, Tsai CJ, Nussinov R. Folding funnels and binding mechanisms. Protein Eng. 1999;12:713–720. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- 8.Csermely P, Palotai R, Nussinov R. Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem Sci. 2010;35:539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borg M, Mittag T, Tony P, et al. Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc Natl Acad Sci. 2007;104:9650–9655. doi: 10.1073/pnas.0702580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinha N, Nussinov R. Point mutations and sequence variability in proteins: redistributions of preexisting populations. Proc Natl Acad Sci USA. 2001;98:3139–3144. doi: 10.1073/pnas.051399098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siltberg-Liberles J, Grahnen JA, Liberles DA. The evolution of protein structures and structural ensembles under functional constraint. Genes (Basel) 2011;2:748–762. doi: 10.3390/genes2040748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeForte S, Uversky VN. Resolving the ambiguity: making sense of intrinsic disorder when PDB structures disagree. Protein Sci. 2016;25:676–688. doi: 10.1002/pro.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Stec B, Godzik A. Between order and disorder in protein structures: analysis of “dual personality” fragments in proteins. Structure. 2007;15:1141–1147. doi: 10.1016/j.str.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oldfield CJ, Meng J, Yang JY, et al. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genom. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tompa P, Szász C, Buday L. Structural disorder throws new light on moonlighting. Trends Biochem Sci. 2005;30:484–489. doi: 10.1016/j.tibs.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 16.Hsu W-L, Oldfield CJ, Xue B, et al. Exploring the binding diversity of intrinsically disordered proteins involved in one-to-many binding. Protein Sci. 2013;22:258–273. doi: 10.1002/pro.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.James LC, Tawfik DS. Conformational diversity and protein evolution—a 60-year-old hypothesis revisited. Trends Biochem Sci. 2003;28:361–368. doi: 10.1016/S0968-0004(03)00135-X. [DOI] [PubMed] [Google Scholar]
- 18.Sikosek T, Chan HS, Bornberg-Bauer E. Escape from adaptive conflict follows from weak functional trade-offs and mutational robustness. Proc Natl Acad Sci USA. 2012;109:14888–14893. doi: 10.1073/pnas.1115620109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siltberg-Liberles J. Evolution of structurally disordered proteins promotes neostructuralization. Mol Biol Evol. 2011;28:59–62. doi: 10.1093/molbev/msq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakrabortee S, Byers JS, Jones S, et al. Intrinsically disordered proteins drive emergence and inheritance of biological traits. Cell. 2016;167(369–381):e12. doi: 10.1016/j.cell.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pancsa R, Tompa P. Coding regions of intrinsic disorder accommodate parallel functions. Trends Biochem Sci. 2016;41:898–906. doi: 10.1016/j.tibs.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 22.Xue B, Dunker AK, Uversky VN. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn. 2012;30:137–149. doi: 10.1080/07391102.2012.675145. [DOI] [PubMed] [Google Scholar]
- 23.Schad E, Tompa P, Hegyi H. The relationship between proteome size, structural disorder and organism complexity. Genome Biol. 2011;12:R120. doi: 10.1186/gb-2011-12-12-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng Z, Yan J, Fan X, et al. Exceptionally abundant exceptions: comprehensive characterization of intrinsic disorder in all domains of life. Cell Mol Life Sci. 2014;72:137–151. doi: 10.1007/s00018-014-1661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch M. The origins of genome architecture. Sunderland: Sinauer Associates Inc; 2007. [Google Scholar]
- 26.Koonin EV. The logic of chance: the nature and origin of biological evolution. Upper Saddle River: FT Press Science; 2011. [Google Scholar]
- 27.Fisher RA. The genetical theory of natural selection. 1930 [Google Scholar]
- 28.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 29.Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Natl Acad Sci USA. 2007;4:8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lynch M. The evolution of genetic networks by non-adaptive processes. Nat Rev Genet. 2007;8:803–813. doi: 10.1038/nrg2192. [DOI] [PubMed] [Google Scholar]
- 31.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keating KS, Toor N, Perlman PS, Pyle AM. A structural analysis of the group II intron active site and implications for the spliceosome. RNA. 2010;16:1–9. doi: 10.1261/rna.1791310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharp PA. On the origin of RNA splicing and introns. Cell. 1985;42:397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- 34.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buljan M, Chalancon G, Dunker AK, et al. Alternative splicing of intrinsically disordered regions and rewiring of protein interactions. Curr Opin Struct Biol. 2013;23:443–450. doi: 10.1016/j.sbi.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Romero PR, Zaidi S, Fang YY, et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc Natl Acad Sci. 2006;103:8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mills D, Peterson R, Spiegelman S. An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule. Proc Natl Acad Sci USA. 1967;58:217–224. doi: 10.1073/pnas.58.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spiegelman S, Haruna I, Holland I, et al. The synthesis of a self-propagating and infectious nucleic acid with a purified enzyme. Proc Natl Acad Sci USA. 1965;54:919–927. doi: 10.1073/pnas.54.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lynch M, Bobay L-M, Catania F, et al. The repatterning of eukaryotic genomes by radom genetic drift. Annu Rev Genomics Hum Genet. 2011;12:347–366. doi: 10.1146/annurev-genom-082410-101412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lynch M. The origins of eukaryotic gene structure. Mol Biol Evol. 2006;23:450–468. doi: 10.1093/molbev/msj050. [DOI] [PubMed] [Google Scholar]
- 41.Koonin EV. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 2010;11:209. doi: 10.1186/gb-2010-11-5-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hughes T, Liberles DA. Whole-genome duplications in the ancestral vertebrate are detectable in the distribution of gene family sizes of tetrapod species. J Mol Evol. 2008;67:343–357. doi: 10.1007/s00239-008-9145-x. [DOI] [PubMed] [Google Scholar]
- 43.Aury J-M, Jaillon O, Duret L, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia . Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 44.Jiao Y, Wickett NJ, Ayyampalayam S, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Wang X, Paterson AH. Genome and gene duplications and gene expression divergence: a view from plants. Ann N Y Acad Sci. 2012;1256:1–14. doi: 10.1111/j.1749-6632.2011.06384.x. [DOI] [PubMed] [Google Scholar]
- 46.Rensing SA, Ick J, Fawcett JA, et al. An ancient genome duplication contributed to the abundance of metabolic genes in the moss Physcomitrella patens . BMC Evol Biol. 2007;7:130. doi: 10.1186/1471-2148-7-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 48.Smith JJ, Kuraku S, Holt C, et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 2013;45:415–421. doi: 10.1038/ng.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Postlethwait JH. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–1902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- 50.Lien S, Koop BF, Sandve SR, et al. The Atlantic salmon genome provides insights into rediploidization. Nature. 2016;533:200–205. doi: 10.1038/nature17164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hughes T, Liberles DA. The power-law distribution of gene family size is driven by the pseudogenisation rate’s heterogeneity between gene families. Gene. 2008;414:85–94. doi: 10.1016/j.gene.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 52.Ohno S. Evolution by gene duplication. New York: Springer; 1970. [Google Scholar]
- 53.Lupas AN, Ponting CP, Russell RB. On the evolution of protein folds: are similar motifs in different protein folds the result of convergence, insertion, or relics of an ancient peptide world? J Struct Biol. 2001;134:191–203. doi: 10.1006/jsbi.2001.4393. [DOI] [PubMed] [Google Scholar]
- 54.Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 2010;11:4. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- 55.Force A, Lynch M, Pickett FB, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rastogi S, Liberles DA. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol Biol. 2005 doi: 10.1186/1471-2148-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gout JF, Lynch M. Maintenance and loss of duplicated genes by dosage subfunctionalization. Mol Biol Evol. 2015;32:2141–2148. doi: 10.1093/molbev/msv095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teufel AI, Liu L, Liberles DA. Models for gene duplication when dosage balance works as a transition state to subsequent neo- or sub-functionalization. BMC Evol Biol. 2016;16:45. doi: 10.1186/s12862-016-0616-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blomme T, Vandepoele K, De Bodt S, et al. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7:R43. doi: 10.1186/gb-2006-7-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maere S, De Bodt S, Raes J, et al. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van der Lee R, Buljan M, Lang B, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114:6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138:198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 63.Amoutzias GD, He Y, Gordon J, et al. Posttranslational regulation impacts the fate of duplicated genes. Proc Natl Acad Sci USA. 2010;107:2967–2971. doi: 10.1073/pnas.0911603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iakoucheva LM, Radivojac P, Brown CJ, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pejaver V, Hsu W-L, Xin F, et al. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 2014;23:1077–1093. doi: 10.1002/pro.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Montanari F, Shields DC, Khaldi N. Differences in the number of intrinsically disordered regions between yeast duplicated proteins, and their relationship with functional divergence. PLoS One. 2011;6:e24989. doi: 10.1371/journal.pone.0024989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mosca R, Pache RA, Aloy P. The role of structural disorder in the rewiring of protein interactions through evolution. Mol Cell Proteomics. 2012;11(M111):014969. doi: 10.1074/mcp.M111.014969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brocchieri L, Karlin S. Protein length in eukaryotic and prokaryotic proteomes. Nucleic Acids Res. 2005;33:3390–3400. doi: 10.1093/nar/gki615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 70.Weiner J, Bornberg-Bauer E. Evolution of circular permutations in multidomain proteins. Mol Biol Evol. 2006;23:734–743. doi: 10.1093/molbev/msj091. [DOI] [PubMed] [Google Scholar]
- 71.Björklund ÅK, Ekman D, Light S, et al. Domain rearrangements in protein evolution. J Mol Biol. 2005;353:911–923. doi: 10.1016/j.jmb.2005.08.067. [DOI] [PubMed] [Google Scholar]
- 72.Buljan M, Frankish A, Bateman A. Quantifying the mechanisms of domain gain in animal proteins. Genome Biol. 2010;11:R74. doi: 10.1186/gb-2010-11-7-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moore AD, Bornberg-Bauer E. The dynamics and evolutionary potential of domain loss and emergence. Mol Biol Evol. 2012;29:787–796. doi: 10.1093/molbev/msr250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kersting AR, Bornberg-Bauer E, Moore AD, Grath S. Dynamics and adaptive benefits of protein domain emergence and arrangements during plant genome evolution. Genome Biol Evol. 2012;4:316–329. doi: 10.1093/gbe/evs004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dos Santos HG, Nunez-Castilla J, Siltberg-Liberles J. Functional diversification after gene duplication: paralog specific regions of structural disorder and phosphorylation in p53, p63, and p73. PLoS One. 2016;11:e0151961. doi: 10.1371/journal.pone.0151961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Light S, Sagit R, Sachenkova O, et al. Protein expansion is primarily due to indels in intrinsically disordered regions. Mol Biol Evol. 2013;30:2645–2653. doi: 10.1093/molbev/mst157. [DOI] [PubMed] [Google Scholar]
- 77.Grishin NV. Fold change in evolution of protein structures. J Struct Biol. 2001;134:167–185. doi: 10.1006/jsbi.2001.4335. [DOI] [PubMed] [Google Scholar]
- 78.Light S, Sagit R, Ekman D, Elofssson A. Long indels are disordered: a study of disorder and indels in homologous eukaryotic proteins. Biochim Biophys Acta Proteins Proteomics. 2013;1834:890–897. doi: 10.1016/j.bbapap.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 79.Tompa P. Intrinsically unstructured proteins evolve by repeat expansion. BioEssays. 2003;25:847–855. doi: 10.1002/bies.10324. [DOI] [PubMed] [Google Scholar]
- 80.Jorda J, Xue B, Uversky VN, Kajava AV. Protein tandem repeats—the more perfect, the less structured. FEBS J. 2010;277:2673–2682. doi: 10.1111/j.1742-4658.2010.07684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Simon M, Hancock JM. Tandem and cryptic amino acid repeats accumulate in disordered regions of proteins. Genome Biol. 2009;10:R59. doi: 10.1186/gb-2009-10-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McDonald MJ, Wang W-C, Huang H-D, Leu J-Y. Clusters of nucleotide substitutions and insertion/deletion mutations are associated with repeat sequences. PLoS Biol. 2011;9:e1000622. doi: 10.1371/journal.pbio.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Williams LE, Wernegreen JJ. Sequence context of indel mutations and their effect on protein evolution in a bacterial endosymbiont. Genome Biol Evol. 2013;5:599–605. doi: 10.1093/gbe/evt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu J, Ng PC. Predicting the effects of frameshifting indels. Genome Biol. 2012;13:R9. doi: 10.1186/gb-2012-13-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brown CJ, Takayama S, Campen AM, et al. Evolutionary rate heterogeneity in proteins with long disordered regions. J Mol Evol. 2002;55:104–110. doi: 10.1007/s00239-001-2309-6. [DOI] [PubMed] [Google Scholar]
- 86.Brown CJ, Johnson AK, Dunker AK, Daughdrill GW. Evolution and disorder. Curr Opin Struct Biol. 2011;21:441–446. doi: 10.1016/j.sbi.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Szalkowski AM, Anisimova M. Markov models of amino acid substitution to study proteins with intrinsically disordered regions. PLoS One. 2011;6:e20488. doi: 10.1371/journal.pone.0020488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ahrens J, Dos Santos HG, Siltberg-Liberles J. The nuanced interplay of intrinsic disorder and other structural properties driving protein evolution. Mol Biol Evol. 2016 doi: 10.1093/molbev/msw092. [DOI] [PubMed] [Google Scholar]
- 89.Schaefer C, Schlessinger A, Rost B. Protein secondary structure appears to be robust under in silico evolution while protein disorder appears not to be. Bioinformatics. 2010;26:625–631. doi: 10.1093/bioinformatics/btq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bellay J, Han S, Michaut M, et al. Bringing order to protein disorder through comparative genomics and genetic interactions. Genome Biol. 2011;12:R14. doi: 10.1186/gb-2011-12-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Colak R, Kim T, Michaut M, et al. Distinct types of disorder in the human proteome: functional implications for alternative splicing. PLoS Comput Biol. 2013;9:e1003030. doi: 10.1371/journal.pcbi.1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dunker AK, Brown CJ, Lawson JD, et al. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 93.Mohan A, Oldfield CJ, Radivojac P, et al. Analysis of molecular recognition features (MoRFs) J Mol Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 94.Yan J, Dunker AK, Uversky VN, Kurgan L. Molecular recognition features (MoRFs) in three domains of life. Mol BioSyst. 2016;12:697–710. doi: 10.1039/C5MB00640F. [DOI] [PubMed] [Google Scholar]
- 95.Cumberworth A, Lamour G, Babu MM, Gsponer J. Promiscuity as a functional trait: intrinsically disordered regions as central players of interactomes. Biochem J. 2013;454:361–369. doi: 10.1042/BJ20130545. [DOI] [PubMed] [Google Scholar]
- 96.Vacic V, Oldfield CJ, Mohan A, et al. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res. 2007;6:2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jakob U, Kriwacki R, Uversky VN. Conditionally and transiently disordered proteins: awakening cryptic disorder to regulate protein function. Chem Rev. 2014;114:6779. doi: 10.1021/cr400459c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nguyen Ba AN, Strome B, Hua JJ, et al. Detecting functional divergence after gene duplication through evolutionary changes in posttranslational regulatory sequences. PLoS Comput Biol. 2014;10:e1003977. doi: 10.1371/journal.pcbi.1003977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Davey NE, Cyert MS, Moses AM. Short linear motifs—ex nihilo evolution of protein regulation. Cell Commun Signal. 2015;13:43. doi: 10.1186/s12964-015-0120-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van der Lee R, Lang B, Kruse K, et al. Intrinsically disordered segments affect protein half-life in the cell and during evolution. Cell Rep. 2014;8:1832–1844. doi: 10.1016/j.celrep.2014.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dos Santos HG, Siltberg-Liberles J. Paralog-specific patterns of structural disorder and phosphorylation in the vertebrate SH3–SH2–tyrosine kinase protein family. Genome Biol Evol. 2016;8:2806–2825. doi: 10.1093/gbe/evw194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Stender EG, O’Shea C, Skriver K. Subgroup-specific intrinsic disorder profiles of arabidopsis NAC transcription factors: identification of functional hotspots. Plant Signal Behav. 2015;10:e1010967. doi: 10.1080/15592324.2015.1010967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nagulapalli M, Maji S, Dwivedi N, et al. Evolution of disorder in mediator complex and its functional relevance. Nucleic Acids Res. 2016;44:1591–1612. doi: 10.1093/nar/gkv1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Richmond K, Masterson P, Ortiz JF, Siltberg-Liberles J. Did the prion protein become vulnerable to misfolding after an evolutionary divide and conquer event? J Biomol Struct Dyn. 2014;32:1074–1084. doi: 10.1080/07391102.2013.809022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yuan J, Xue B. Role of structural flexibility in the evolution of emerin. J Theor Biol. 2015;385:102–111. doi: 10.1016/j.jtbi.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 106.Brown CJ, Johnson AK, Daughdrill GW. Comparing models of evolution for ordered and disordered proteins. Mol Biol Evol. 2010;27:609–621. doi: 10.1093/molbev/msp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Narasumani M, Harrison PM. Bioinformatical parsing of folding-on-binding proteins reveals their compositional and evolutionary sequence design. Sci Rep. 2015;5:18586. doi: 10.1038/srep18586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mahani A, Henriksson J, Wright APH. Origins of Myc proteins—using intrinsic protein disorder to trace distant relatives. PLoS One. 2013;8:e75057. doi: 10.1371/journal.pone.0075057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Boomsma W, Nielsen SV, Lindorff-Larsen K, et al. Bioinformatics analysis identifies several intrinsically disordered human E3 ubiquitin-protein ligases. PeerJ. 2016;4:e1725. doi: 10.7717/peerj.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sillitoe I, Dawson N, Thornton J, Orengo C. The history of the CATH structural classification of protein domains. Biochimie. 2015;119:209–217. doi: 10.1016/j.biochi.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ferrada E, Wagner A. Protein robustness promotes evolutionary innovations on large evolutionary time-scales. Proc R Soc B Biol Sci. 2008;275:1595–1602. doi: 10.1098/rspb.2007.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J Mol Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 113.Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- 114.Kumar S, Hedges SB. TimeTree2: species divergence times on the iPhone. Bioinformatics. 2011;27:2023–2024. doi: 10.1093/bioinformatics/btr315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hedges SB, Marin J, Suleski M, et al. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 2015;32:835–845. doi: 10.1093/molbev/msv037. [DOI] [PMC free article] [PubMed] [Google Scholar]