Abstract
Aptamers are small single-stranded DNA or RNA oligonucleotide fragments or small peptides, which can bind to targets by high affinity and specificity. Because aptamers are specific, non-immunogenic and non-toxic, they are ideal materials for clinical applications. Neurodegenerative disorders are ravaging the lives of patients. Even though the mechanism of these diseases is still elusive, they are mainly characterized by the accumulation of misfolded proteins in the central nervous system. So it is essential to develop potential measures to slow down or prevent the onset of these diseases. With the advancements of the technologies, aptamers have opened up new areas in this research field. Aptamers could bind with these related target proteins to interrupt their accumulation, subsequently blocking or preventing the process of neurodegenerative diseases. This review presents recent advances in the aptamer generation and its merits and limitations, with emphasis on its applications in neurodegenerative diseases including Alzheimer’s disease, Parkinson’s disease, transmissible spongiform encephalopathy, Huntington’s disease and multiple sclerosis.
Keywords: Aptamer, Neurodegenerative diseases, Alzheimer’s disease, Parkinson’s disease, Transmissible spongiform encephalopathy, Huntington’s disease, Multiple sclerosis
Introduction
Aptamers are single-stranded DNA or RNA nucleotides having a length of 40–100 mers. Recently, they have been extended to small peptides with 10–30 amino acid residues [1–4]. The name of aptamer originally comes from Latin “aptus” which means “fit” [5]. These single strands of nucleotides or peptides can specifically recognize and bind to a great deal of target molecules, such as metal ions, organic dyes and amino acids [6–8], antibodies [9], proteins [10, 11], whole cells [12–14], organs [15], viruses and bacteria [16, 17], by various intermolecular forces, such as van der Waals forces, hydrogen bonding, base stacking, etc. During the past 30 years, over 2000 aptamers have been developed to bind to about 141 target ligands [18]. Aptamers, considered as chemical antibodies, have more advantages over classical antibodies, since they have long half life, small size, low or no toxicity, and low immunogenicity, can be fastly and inexpensively produced [19]. Due to these merits, aptamers have attracted worldwide attentions for the diagnosis and therapy of various diseases.
Currently, aptamers have been applied in clinical diseases, such as cancer and inflammatory diseases, etc. [16, 20]. Pegaptanib, also called as Macugen, is the first aptamer approved by the US Food and Drug Administration to treat ocular neovascularization in December 2004. Meanwhile, it represents a milestone in drug development, because it is the first aptamer which is successfully used as a therapeutic agent in human beings [21–23]. Meanwhile, to be more interesting, aptamers have also been widely applied in food quality and safety monitoring [24].
Neurodegenerative disorders, such as Alzheimer’s disease (AD), Parkinson’s disease (PD), transmissible spongiform encephalopathy (TSE), Huntington’s disease (HD) and multiple sclerosis (MS), are plaguing people’s lives. Although the etiology of these neurodegenerative diseases is still elusive, there is a common pathological feature that is the accumulation of misfolded proteins. At present, which type of these misfolded protein, such as monomers, oligomers and fibrils, are toxic remains controversial. However, the predominant opinion is that oligomers are more toxic than others. Hence, preventing misfolded proteins accumulating as oligomers should be a good approach for treating the neurodegenerative diseases. Because aptamers just have the feature to bind with proteins to block their functions and accumulation, scientists are trying to use aptamers to treat disease or detect the pathologic processes. This paper mainly focuses on recent advances in the aptamer generation and its merits and limitations, with emphasis on its applications in neurodegenerative diseases.
Generation of aptamer
Aptamers were initially considered as DNA or RNA fragments. They were first developed almost simultaneously by two research groups of Tuerk and Ellington. In late 1990, Tuerk and Gold selected one eight-base RNA ligand which could interact with the bacteriophage T4 DNA polymerase [25]. Meanwhile, Ellington and Szostak isolated RNA molecules which could bind to a variety of organic dyes [5]. Recently, the term of aptamer has been extended to peptides which are selected by yeast-two-hybrid procedure or peptide array analysis [26–29]. In 1996, Pierre Colas et al. first generated peptide aptamers which could recognize and inhibit cuclin-dependent kinase 2 (Cdk2) [4].
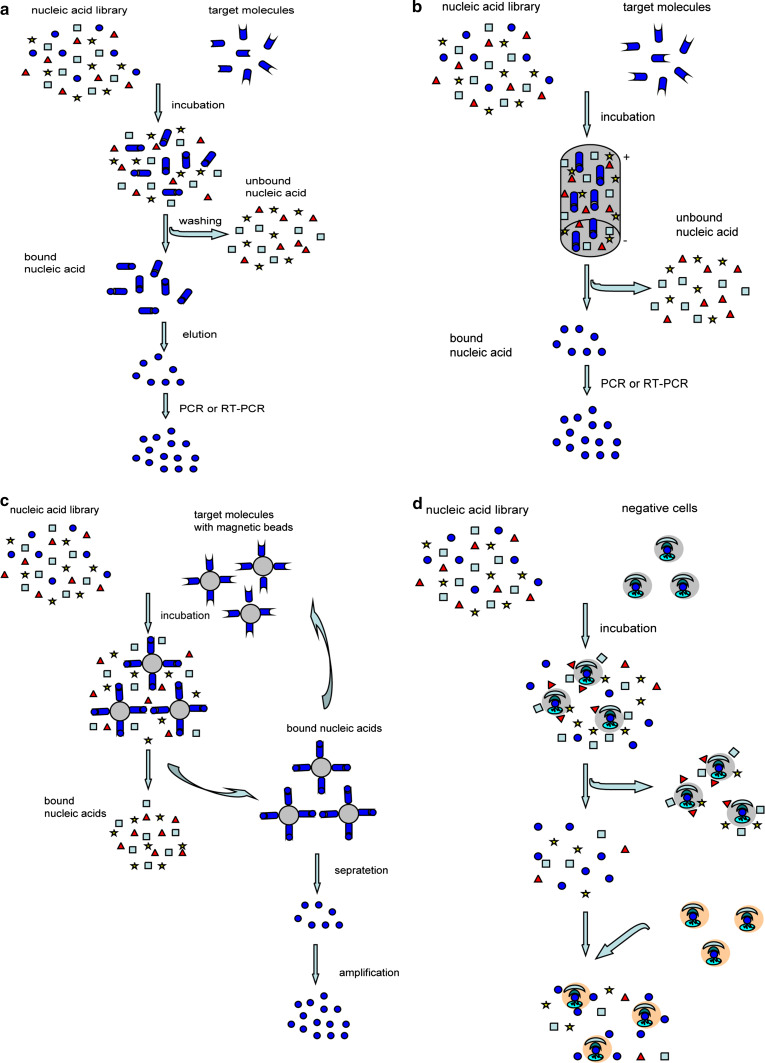
Aptamers are generally screened out by systematic evolution of ligands by exponential enrichment (SELEX), which is a combinatorial chemistry technique of screening specific ligands by repeated rounds of partition and amplification from a large nucleic acid library containing 1014–1016 different candidates [25, 30]. Although SELEX has emerged as the most commonly used name for the procedure, some researchers also referred to it as SAAB (selected and amplified binding site) or CAST (cyclic amplification and selection of targets) [31, 32]. The traditional SELEX largely includes two processes, namely selecting aptamers with affinity to targets, and amplifying the bound aptamers (Fig. 1a). Technically, the processes begin with the synthesis of a nucleic acid library, in which the oligonucleotide is composed of the random sequences in the middle, flanked by fixed sequences [33]. Then, the library is incubated with the target molecules for a period of time under appropriate buffer and temperature conditions. The candidates with affinity can bind to the targets, while others with no affinity are still in free state. After washing the unbound nucleic acids from the binding molecules, the bound molecules are amplified by PCR. In order to achieve single-stranded DNA, there is a plethora of methods, such as asymmetric PCR, lambda exonuclease digestion, biotin-streptavidin separation, size separation on denaturing-urea PAGE and so on. For RNA aptamers, a transcription step by RNA polymerase is needed [34, 35]. Then these single-stranded molecules are used in the next selecting cycle. In general, it needs about 8–20 cycles in the entire process, until the sequences with high affinity and specificity are enriched [36]. Many aptamers were selected by the traditional SELEX in fact, such as amyloid β-protein (Aβ) and alpha-synuclein (α-Syn) (which were the major markers of AD and PD, respectively) [37, 38].
Fig. 1.
The processes of traditional SELEX, CE-SELEX, magnetic bead-based SELEX, and cell-SELEX. a The processes of traditional SELEX. It begins with the synthesis of a nucleic acid library. Then, the library is incubated with the target molecules for a period of time. The candidates with affinity can bind to the targets. Washing the unbound nucleic acids, the remaining binding molecules are amplified by PCR or RT-PCR. Then the molecules achieved from PCR or RT-PCR are used in the next cycle. b The processes of CE-SELEX. The nucleic acid library is incubated with the target molecules to form the mixtures. Then the mixtures are injected into the capillary. The bound nucleic acids are separated from unbound sequences through free solution capillary electrophoresis based on a mobility shift. c The processes of magnetic bead-based SELEX. The target molecules were immobilized on magnetic beads. Nucleic acid library is incubated with the target molecules on magnetic beads. Then the bound sequences are separated from unbound by magnetic force. d The processes of cell-SELEX. The nucleic acid library is incubated with negative cells which are not related to the target cells. Then, the unbound nucleic acids are collected and incubated with positive cells
Since the classical SELEX method for aptamer selection was developed, various improvements and modifications have been introduced to greatly shorten selection time and improve binding affinity [39, 40]. Among these modified SELEX methods, CE-SELEX, magnetic bead-based SELEX, automated SELEX and cell-SELEX are most frequently used. CE-SELEX (Fig. 1b), was first developed by Mendosa and Bowser in 2004 [41, 42]. It can increase separation power and reduce the nonspecific binding because of performing in free solution. So far, many aptamers have been generated by CE-SELEX, including aptamers against IgE [42], protein kinase C and so on [10]. Magnetic bead-based SELEX (Fig. 1c), was first developed by Bruno in 1997 [43]. It can enhance the binding affinity owing to being performed on the surface of the magnetic beads. Besides, the sample volumes only need 50–100 μl of sample [24, 44]. Some aptamers were also developed by this method, such as aptamers against 4-chloroaniline, mammalian prion protein (the major protein of transmissible spongiform encephalopathy) and dopamine (a key molecule in Parkinson’s disease) [39, 45]. Automated SELEX was reported by the group of J. Colin Cox in 1998 based on the Beckman Biomek 2000 pipetting robot [46, 47]. It can reduce the number of selection round to eight, and can complete about 12 select rounds in only 2 days, which means the selection efficiency is greatly improved. Ellington et al. [48] first adapted automated workstations to select anti-protein aptamers. Spiegelmers, RNA aptamers against the mirror image configuration of substance P, were also selected by this method. During this process, the aptamers were selected among a normal RNA pool against d-substance P and then the final selected aptameric sequence was synthesized by l-ribonucleic acid units. Consequently, they could bind to the naturally occurring l-substance P with comparable affinity [46].
Above three SELEX methods are generally used to screen aptamers for definite moleculars, latterly the method to select aptamers against cells comes to be developed. Cell-SELEX (Fig. 1d), first reported in 2003, is designed to target whole living cells, such as cancer cells [24, 49]. The process of cell-SELEX is easy, fast, straightforward, and reproducible. Numerous aptamers against various cancer cells were selected by this method. Aptamers, selected through cell-SELEX by the group of Weihong Tan, could specifically recognize leukemia cells [14]. In addition, there are other aptamers which can recognize the target cells derived from mouse tumor endothelial cells, glioblastoma multi-forme, prostate cancer, colorectal cancer etc. [50–53]. However, these cell-SELEX is based on the two-dimensional (2D) cell culture which has a limitation that cells could not grow in all directions as in vivo. Recently the three-dimensional (3D) cell culture was developed, it could mimic the microenvironment in vivo and cells are permitted to grow in all dimensions, so a novel method, 3D cell-SELEX, was introduced to select aptamers against cellular and extracellular targets. Eight aptamers, against PC-3 prostate cancer cell line, were selected through 3D cell-SELEX by the group of Aline G. Souza in 2016 [54, 55].
Merits of aptamer
Generally, the aptamers are considered as “chemical antibodies” because of their similar functions between aptamers and classical antibodies. Despite their similarity in functions, aptamers have more advantages than classical antibodies (Table 1 for detail), which drives aptamers to be widely used in clinic [19, 56]. First of all, the molecular weight of aptamers is about 5–10 kDa, which is lower than that of classical antibodies (approximately 150 kDa), so aptamers can reach the intracellular target molecules more easily than antibodies [30, 57]. Second, they can be easily generated by chemical synthesis, while the classical antibodies may be obtained with tedious work. Third, they are neither immunogenic nor toxic, while the antibodies are immunogenic, which makes aptamers to be ideal strategies in the treatment of immune diseases. Fourth, aptamers can restore the original structure easily within minutes once denatured, while the antibodies cannot, which indicates aptamers are stable for long time and could be transported at ambient temperature [30]. Fifth, the dissociation constant of most aptamers is as low as the picomolar–femtomolar (pM) level, which suggests aptamers might have more affinity to targets [58]. Sixth, aptamers can be easily labeled and adjusted, which indicates that aptamers can be easily modified according to the specified requirement [59]. Moreover, aptamers can distinguish closely related, but not identical proteins based on their different functions and structures, while classical antibodies cannot [60].
Table 1.
Comparison of properties of aptamer with antibody
| Aptamer | Antibody |
|---|---|
| Smaller size allowing more efficiently entry into biological compartments | Large size limiting bioavailability or preventing access to many biological compartments |
| Produced chemically in a readily scalable process | Produced biologically in a difficult process |
| Non-immunogenic | Immunogenic |
| Able to select for specific targets | Limited ability to utilize negative selection pressure or to select against cell-surface |
| Production process is not prone to viral or bacterial contamination | Targets not available in functional recombinant form |
| Can usually be reversibly denatured | Susceptible to irreversible denaturation |
| Conjugation chemistries for the attachment of functional groups are orthogonal and can be readily introduced during synthesis | Chemistries required for the attachment of conjugation partners are stochastic and lead to product mixtures and reduced activity |
Due to these properties, aptamers might be potential tools broadly applied in clinic. To date, they have been used for the study of many diseases in which cancer is the most widely treated, as shown in Table 2.
Table 2.
Examples of aptamers applied in cancer
The limitations of aptamers and their optimization approaches
The limitations of aptamers
Despite the advantages mentioned above, aptamers still have obvious limitations which may hinder their use in clinical application [61–63]. The first serious limitation is the stability of unmodified aptamers, especially RNA aptamers, since these aptamers can be degraded by nucleases in blood [63]. The average half-life time ranges from several minutes to several tens of minutes. Such a short time range is not acceptable for most applications of aptamers in clinic. Then the fast clearance of aptamers also limits their therapeutic applications. The molecular weight of most aptamers ranges from 5 to 10 kDa, so they can rapidly penetrate into target tissue or tumor. However, they can also be rapidly cleared from the bloodstream before playing their roles in the clinical application [46, 64, 65].
The optimization approaches
To overcome the obstacles of aptamers in clinic, it is very necessary to develop approaches to enhance their stability as well as slow down their clearance. Recently, in order to ease the clinical application, various approaches have been developed, such as aptamer-chemical modification, aptamer–nanoparticle conjugation and so on.
Chemical modifications of aptamers
To improve the stability of aptamers, some chemical modifications have been used upon the aptamers such as 2′-fluoro-pyrimidine [66], 2′-O-methyl nucleotides [67, 68], locked nucleic acid (LNA) [67, 69], 2′-amino pyrimidines, 4′-thio pyrimidines [62, 65], or capping at the 3′- or 5′-termini with polyethylene glycol (PEG) [70], biotin-streptavidin [71], cholesterol [72] and so on. In 2003, 2′-fluoro-modified RNA aptamers, against prion protein which was an important protein in transmissible spongiform encephalopathy, were selected [66]. And in 2004, 2′-fluoro-pyrimidine was taken as a modification factor to develop Pegaptanib, which is the first aptamer approved by the US Food and Drug Administration to treat ocular age-related macular degeneration (AMD) [73, 74]. In 2006, two reports demonstrated aptamers with 2′-fluoro-pyrimidine against prostate-specific membrane antigen (PSMA) were used to target LNCaP cells expressing PSMA. The results indicated 2′-fluoro-pyrimidine can improve the aptamers stability [75, 76]. Moreover, LNA, a class of nucleic acid analogue with an extra bridge connecting the 2′-O and 4′-C in the ribose moiety, can improve the stability of aptameric sequences against nuclease digestion [69]. Kathrin S. Schmidt et al. demonstrated that LNA modification within aptamer TTA1 (recognizing human Tenascin-C) improved aptamers stability in 2004 [65].
Aptamer–nanoparticle conjugation
In addition to chemical modifications, nanoparticles conjugation to aptamers has also been tried to enhance the stability and affinity [77–79]. Nanoparticles are particles whose sizes are generally between 1 and 100 nm. They have unique virtues, such as the small size, the large ratio of surface area to mass, and the high reactivity [80, 81]. Due to these merits, nanoparticles have emerged as a novel tool to conjugate to aptamers to enhance aptamer stability and deliver aptamers to target tissue, such as liposomes, peptides, quantum dots, and virus-based vectors. Nanoparticle–aptamer (NP-Apt) bioconjugates can also improve the affinity of the aptamer. Farokhzad et al. identified that the conjugation of nanoparticle to A10 aptamer (which can bind PSMA transmembrane protein) could enhance the affinity between aptamer and the target. Moreover, they are the first to develop nanoparticle–aptamer bioconjugates to deliver targeted drugs in vitro [82, 83].
Besides, the aptamer–nanoparticle biosensor is another magical tool to recognize targets with high affinity. An aptamer–nanoparticle strip biosensor was developed by Liu et al. for the rapid, sensitive, and low-cost detection of cancer cells [84]. An AS1411 aptamer-functionalized nanoparticulate drug delivery system can prolong the circulation and accumulation of the drug in the target site of tumor, which can further inhibit tumor more efficiently and prolong mice survival [85]. A nanoelectronic biosensor for detecting dopamine (DA, a significant molecule in PD) modified by DNA–aptamer was developed by the group of Chen. This biosensor could improve and enhance the efficiency [86].
Other approaches
Except the two approaches mentioned above, there are also other means to overcome the limitations of aptamers. A great number of studies have proved that some vectors, such as polyethyleneimine (PEI), polycation, liposomes, chitosan, peptides, as well as quantum dots, could protect aptamer from degradation and improve its delivery efficiently [87–91]. Gong et al. first synthesized the complex of PEI/aptamer for cancer imaging in vivo. They were surprised to find PEI could successfully prevent TD05 aptamers from degradation at the N/P ratio from 3.8 to 15, without affecting the specific recognition of Ramos cells. Moreover, PEI/aptamer complex could more efficiently target passive tumor compared with free aptamer in mice bearing tumors. Hence, the PEI/aptamer complex may be a novel strategy to improve the stability of aptamers and to deliver aptamers to targets [87].
The applications of aptamer in neurodegenerative diseases
Neurodegenerative disorders have attracted worldwide attentions because of prevalent incidence in the upcoming aging society. These diseases, including AD, PD, TSE, HD and MS, are characterized by the accumulation of misfolded proteins in the central nervous system (CNS). Therefore, novel approaches to slow down or prevent these proteins accumulating would be potential approaches to treat these diseases. Aptamers could bind with these target proteins to prevent aggregating of the misfolded proteins or decrease the negative effects, which can further cure diseases or detect the pathologic processes.
Aptamers and AD
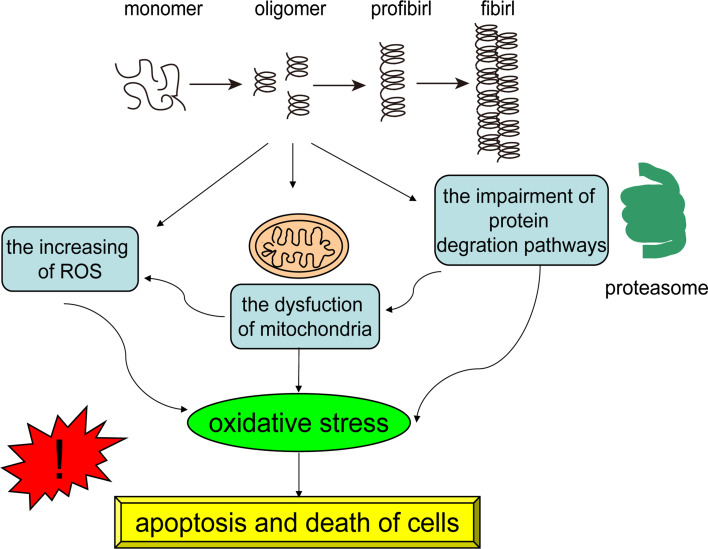
With the great increase of the human life span in the world, AD, one of the major diseases of the elderly progressive neurodegenerative diseases, is rapidly becoming a heavy burden and challenge in many countries [92]. The hallmark of AD is the misfolding of Aβ which is generated by sequential cleavage of amyloid precursor protein (APP). Aβ aggregation may disrupt the normal physiological functions of cells, inducing oxidative stress and nerve inflammation, which could further lead to the dysfunctions of cells, even apoptosis and death (Fig. 2). Therefore, Aβ has been a target for the diagnostic and therapeutic reagents of AD.
Fig. 2.
The damage of the aggregations of misfold proteins. Aβ and α-Syn are a class of proteins which are prone to aggregate and fold. The aggregations can disrupt the normal functions of cells and proteins, such as the impairment of the degradation systems, the dysfunction of mitochondria, the generation of oxidative stress, which could further lead to the disfunctions of cells, even apoptosis and death
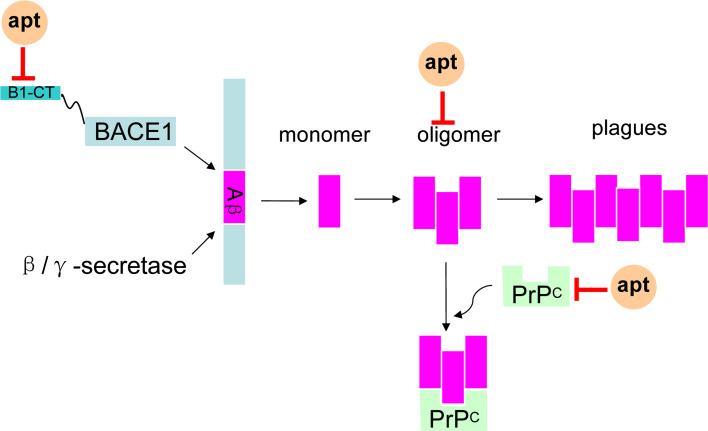
Since Aβ aggregation is toxic, firstly it is a good idea to reduce Aβ production. As is known to all, the generation of Aβ is triggered by β-site APP cleaving enzyme-1 (BACE1). Thus, BACE1 should be a worthy target of the interference of Aβ production and treatment of AD (Fig. 3) [93]. One DNA aptamer, called A1 selected by the group of Liang et al. may bind to BACE1 with high affinity and good specificity. Consequently, A1 had the potential to decrease the production of Aβ40 and Aβ42 [94]. Furthermore, RNA aptamers against a short cytoplasmic tail (B1-CT) of BACE1 also have been selected by Rentmeister et al. [95]. These RNA aptamers can specifically bind to B1-CT without affecting other important biological activities. So they could be potentially applied to prevent or slowdown the onset of AD.
Fig. 3.
The aptamers (apt) against the AD targets. The hallmark of AD is Aβ of which the oligomer will disrupt the normal functions of cells. BACE1 can trigger the generation of Aβ, and B1-CT is a tail of BACE1. PrPC is a receptor of Aβ oligomer. The aptamers against Aβ oligomers, B1-CT and PrPC might be used as tools to diagnosis the Aβ oligomers and even prevent the generation of Aβ, which can decrease the serious effects to the cells
As Aβ oligomer is toxic, the aptamer directly binding to oligomer may also prevent the toxicity (Fig. 3). RNA aptamers against Aβ40 oligomer were selected by Bitan et al. in 2009. However, the data showed that aptamers had low specificity to Aβ oligomer [96]. A DNA aptamer, which can specifically recognize and bind to Aβ oligomer, was generated in 2012. Since these aptamers are specific for oligomers, they could be more efficient and specific tools than antibodies in the application of recognizing the Aβ oligomer, preventing Aβ toxicity, as well as diagnosing AD [37, 38].
Since Aβ oligomer is toxic, there should be definite receptors to exert its toxic role. In this scenario, it may be a good choice to block the target receptor of Aβ oligomer (Fig. 3). It is found that the normal prion protein (PrPC) is one of the target receptors of Aβ oligomer, and Aβ-oligomer could bind to PrPC with affinity [97, 98]. So PrPC may be a mediator of Aβ-oligomer to induce synaptic dysfunction, and aptamers against PrPC can prevent Aβ-oligomer binding to PrPC and rescue synaptic plasticity, even treat AD [97].
Aptamers and PD
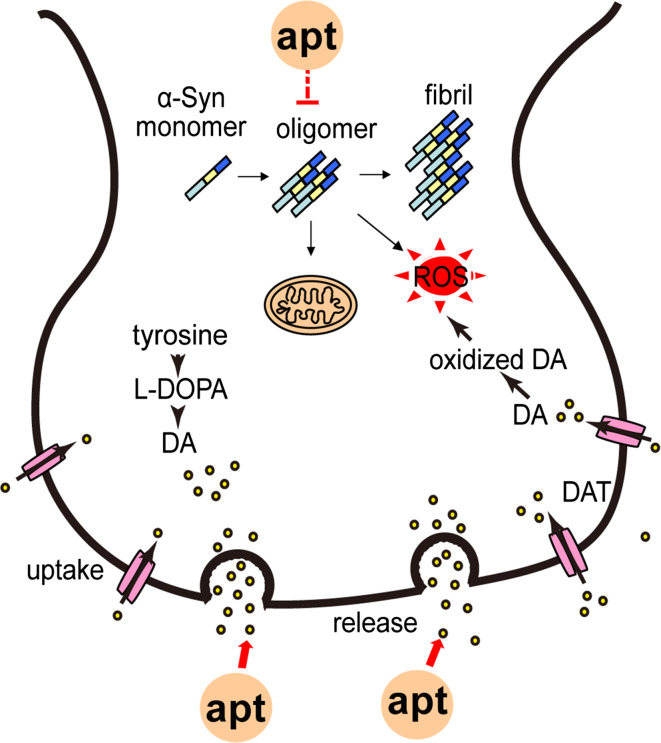
PD is the second most common neurodegenerative disease mainly affecting 1 % of individuals above the age of 60. It is primarily defined by symptoms of motor impairment, such as resting tremor, bradykinesia, postural instability, rigidity, and so on [99–102]. The pathology of PD is characterized by the presence of inclusions termed Lewy bodies (LBs) in neurons, and the selective loss of dopaminergic neurons in the substantia nigra pars compacta [103]. The major protein component of LBs is the presynaptic protein α-Syn which is misfolded and aggregated [104]. The aggregations of α-Syn, especially the oligomers, have negative roles in the neurons, such as the impairment of the degradation systems, the damage of mitochondria, the generation of oxidative stress and toxicity (Fig. 2) [105–108]. Hence, aptamers against α-Syn would be potential tools to diagnose or treat PD.
Predominantly, α-Syn oligomers are more toxic than monomers and fibrils, so oligomers are seen as the key targets of PD. The first aptamer against α-Syn oligomers, called “M5-15”, was selected by the group of Ikebukuro in 2010. But surprisingly, M5-15 could also slightly bind to α-Syn monomers, which indicates that M5-15 has low specificity to α-Syn oligomers [109]. Two years later, they developed another eight DNA aptamers against α-Syn oligomers. Interestingly, these aptamers definitely recognize the β-sheet structure [38], namely they can bind to not only α-Syn oligomer, but also Aβ oligomer, which indicated these aptamers could also be employed as magical drugs to treat even cure PD and AD (Fig. 4).
Fig. 4.
The aptamers (apt) against the targets of PD. The pathology of PD is characterized by the presence of inclusions termed LBs whose major protein component is α-Syn, and the selective loss of dopaminergic neurons in the SN. α-Syn oligomers can also disrupt the normal functions of cells. So aptamers targeting α-Syn oligomers could be developed as useful tools for diagnosing PD or preventing the onset of PD. DA is another target, and aptamers against DA can diagnose the concentration of DA
Interestingly, the peptide (GVLYVGSKTR), derived from β-Syn protein, could bind to α-Syn and inhibit its aggregating found by the group of Shaltiel-Karyo. This peptide could reduce α-Syn amyloid fibril and soluble oligomer in vitro. Strikingly, administering this peptide to a Drosophila PD model, expressing A53T α-Syn, could ameliorate the behavioral defects and reduce α-Syn accumulation in the brains [26]. Hence, this peptide should be another potential way to prevent the onset of PD.
Not only the presence of LBs, PD is also characterized by the selective loss of DA in the substantia nigra. DA is a small molecule with weight of 189 Da, and it is involved in neuronal signal transduction. The loss of DA would lead to the deregulation of basal ganglia which further causes motor symptoms and non-motor symptoms [110]. As we all know, the concentration of DA usually ranges from 10−8 to 10−6 M in the normal physiological environment [111]. The deficiency of DA neurotransmission may be implicated in PD [112, 113]. Accordingly, the aptamers against DA may be another tool to monitor DA concentration and study the pathological mechanism of PD. The first RNA aptamer against DA called dopa2, selected by Tocchini-Valentini et al. [45], could bind to DA with high affinity and specificity. Then a novel label-free electrochemical aptasensor for detecting DA was developed by Liu et al. in 2012. The aptasensor has successfully tested the DA of human serum [114]. Another RNA aptamer-based electrochemical biosensor for selective and label-free analysis of DA was reported by Farjami et al. [115]. To further enhance the detecting accuracy, Chen et al. developed a new nanoelectronic device as a biosensor for detecting DA by modifying the DNA–aptamers on a multiple-parallel-connected (MPC) silicon nanowire field-effect transistor. This biosensor can improve the detection limit, distinguish DA from analogues, as well as monitor the release of DA [86].
Aptamers and TSE
TSE is another class of neurodegenerative disorders, affecting human beings and other mammals [116]. The pathological characteristic of TSE is the conversion of normal cellular prion protein (PrPC), an α-helix-rich isoform, to abnormal PrPSC subtypes, a β-sheet-rich isoform, and accumulating in the brain [117]. Thus, diagnosing PrPC and PrPSC, as well as inhibiting the transformation from PrPC into PrPSC might be useful approaches to study the pathogenic of TSE and treat TSE. Recently, a great deal of aptamers have been selected against both PrPC and PrPSC and these aptamers are isoform- and species-specific.
SAF-93, 2′-fluoro-modified RNA oligomers, for PrPSc were selected in 2003 [66]. The affinity of these aptamers for PrPSc was more than tenfold higher than for PrPC because of the existence of two specific heparin-binding sites within the PrPSc molecule. Moreover, SAF-93 could inhibit the conversion of PrPC into PrPSc [66]. But because 2′-fluoro-modified SAF-93 are too long and complex, they are not ideal tools for the clinical diagnosis and therapy of TSE. Subsequently in 2004, James et al. identified the functional structures within SAF-93, and synthesized an aptamer SAF-93(1-34,35bioU,36-60) to detect the PrPSc [118]. These aptamers targeting PrPSc could be potentially used in diagnosing and treating TSE.
Because TSE is often occurring in various animals, the aptamers against PrP should also be species-specific (Table 3). Aptamers against the mouse prion protein (mPrP) were isolated by the team of Nishikawa [119, 120]. One DNA aptamer which could bind to both recombinant and mammalian PrPC but not PrPSc was selected by Takemura et al. [121]. In the same year, RM312, an RNA aptamer that could bind to sheep recombinant PrP, was selected by Mercey et al. [122]. Additionally, an RNA aptamer targeting bovine prion protein (bPrP) was selected by Mashima et al. [98]. So, species-specific aptamers might become novel tools to diagnose and treat TSE in various animals.
Table 3.
The species-specific aptamers against PrP
Aptamers and HD
HD is a dominantly inherited disease in neuronal tissue. It is suggested this disorder has affected about 5–10 individuals per 100,000 [123]. Affected individuals always suffer from progressive motor as well as cognitive declines, such as the loss of awareness, depression, dementia and anxiety. The pathogenesis of HD seems to be the misfolding and aggregation of mHTT which is a form of mutant huntingtin protein [123, 124]. Hence, we must make efforts to find agents which could destroy or prevent the pathway of mHtt aggregation.
In 2006, Skogen et al. reported that an aptamer, 20-m G-rich oligonucleotides, could effectively inhibit HTT aggregating. This aptamer could enhance the viability of PC12 cells which overexpressed the mHTT fragment gene, which suggested it would likely to be new agents to study the mechanism of HD and treat this disease [125].
Aptamers and MS
MS is an autoimmune neurodegenerative disease with a prevalence of around 0.1 % in the Western world. The clinical features include visual loss, gait and cognitive impairment, fatigue, and so on. And pathological characteristics are the destruction of myelin required for electrical insulation of neuronal axons in the CNS [126, 127]. So the myelin may be a key material for the research for MS. Several aptamers against myelin have been selected by the SELEX.
A 40-nucleotide DNA aptamer against murine myelin was selected by Nastasijevic et al. in 2012. This aptamer could promote remyelination of CNS lesions in the MS mice model [128]. Then LJM-3064, a guanosine-rich 40-m DNA aptamer, was found to mediate remyelination in the mouse model of MS in 2013. And this aptamer could undergo an ion-dependent conformational switch, which provided important thermodynamic insights that may be an optimizing tool for developing the therapeutic remyelinating agent [127]. Another two aptamers for myelin were obtained by Rozenblum et al. through SELEX method in 2014 [129]. Moreover, one selecting aptamer was more sensitive than the commercial antibody. Therefore, these aptamers might be useful tools for medical research and treatment of MS.
Conclusion and perspective
Aptamers have gained great attention since their introduction in the early 1990s. Aptamers are a class of substances having similar functions to classical antibodies, but they have unambiguous advantages over classical antibodies, including small size, low or no toxicity, and low immunogenicity. Despite the advantages over classical antibodies, there are several barriers which limit the use of aptamers in clinical application, such as rapid degradation, the susceptibility to nucleases, easy renal filtration and so on. Fortunately, various approaches have been proposed to overcome these limitations such as aptamer-chemical modification, aptamer–nanoparticle conjugation and others.
The obvious merits have driven aptamers usage in clinic, such as cancer, eye and inflammatory diseases. Pegaptanib is the first and sole aptamer that has been successfully applied in the clinic, which is a milestone of aptamers to be used in the diseases. Many other aptamers are being developed which indicates more and more aptamers will be applied in the neurodegenerative disorders.
Neurodegenerative disorders are characterized by the accumulation of misfolded proteins in the CNS, including AD, PD, TSE, HD and MS. Aptamers against these targets which are associated with these diseases have been developed, and they are being applied in clinical trials. Along with the advancements such as nanotechnology, microfluidics, microarray, and others in the field of clinical diseases, aptamers will gain the special area in clinic application. Moreover, the successful developments in aptamers over the last 30 years are notable, which reveals aptamers are likely to become valuable drugs to cure these neurodegenerative diseases.
Acknowledgments
This work was supported by Grants from the National Natural Science Foundation of China (31571202, 31271136, 81371398), the Young Talents Cultivation Plan in Beijing Local University (CIT&TCD201504087), the Beijing Natural Science Foundation Program and Scientific Research Key Program of Beijing Municipal Commission of Education (KZ201210025020, 7131001) and the Project of Construction of Innovative Teams and Teacher Career Development for Universities and Colleges Under Beijing Municipality (IDHT20140514). We also thank Ms. Gay Samuelson in the Banner Sun Health Research Institute for her help in proofreading the manuscript.
References
- 1.Bruno JG. Predicting the uncertain future of aptamer-based diagnostics and therapeutics. Molecules. 2015;20(4):6866–6887. doi: 10.3390/molecules20046866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santosh B, Yadava PK. Nucleic acid aptamers: research tools in disease diagnostics and therapeutics. Biomed Res Int. 2014;2014:540451. doi: 10.1155/2014/540451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pei X, Zhang J, Liu J. Clinical applications of nucleic acid aptamers in cancer. Mol Clin Oncol. 2014;2(3):341–348. doi: 10.3892/mco.2014.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colas P, Cohen B, Jessen T, Grishina I, McCoy J, Brent R. Genetic selection of peptide aptamers that recognize and inhibit cyclin-dependent kinase 2. Nature. 1996;380(6574):548–550. doi: 10.1038/380548a0. [DOI] [PubMed] [Google Scholar]
- 5.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 6.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002;62(14):4029–4033. [PubMed] [Google Scholar]
- 7.Mannironi C, Scerch C, Fruscoloni P, Tocchini-Valentini GP. Molecular recognition of amino acids by RNA aptamers: the evolution into an l-tyrosine binder of a dopamine-binding RNA motif. RNA. 2000;6(4):520–527. doi: 10.1017/S1355838200991763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geiger A, Burgstaller P, von der Eltz H, Roeder A, Famulok M. RNA aptamers that bind l-arginine with sub-micromolar dissociation constants and high enantioselectivity. Nucleic Acids Res. 1996;24(6):1029–1036. doi: 10.1093/nar/24.6.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams KP, Liu XH, Schumacher TN, Lin HY, Ausiello DA, Kim PS, Bartel DP. Bioactive and nuclease-resistant L-DNA ligand of vasopressin. Proc Natl Acad Sci USA. 1997;94(21):11285–11290. doi: 10.1073/pnas.94.21.11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mallikaratchy P, Stahelin RV, Cao Z, Cho W, Tan W. Selection of DNA ligands for protein kinase C-delta. Chem Commun. 2006;30:3229–3231. doi: 10.1039/b604778e. [DOI] [PubMed] [Google Scholar]
- 11.Nieuwlandt D, Wecker M, Gold L. In vitro selection of RNA ligands to substance P. Biochemistry. 1995;34(16):5651–5659. doi: 10.1021/bi00016a041. [DOI] [PubMed] [Google Scholar]
- 12.Chen HW, Medley CD, Sefah K, Shangguan D, Tang Z, Meng L, Smith JE, Tan W. Molecular recognition of small-cell lung cancer cells using aptamers. Chem Med Chem. 2008;3(6):991–1001. doi: 10.1002/cmdc.200800030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikratchy P, Chen HW, Li Y, Tan W. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal Chem. 2007;79(13):4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 14.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci USA. 2006;103(32):11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorger M, Engstler M, Homann M, Goringer HU. Targeting the variable surface of African trypanosomes with variant surface glycoprotein-specific, serum-stable RNA aptamers. Eukaryot Cell. 2003;2(1):84–94. doi: 10.1128/EC.2.1.84-94.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanwar JR, Mohan RR, Kanwar RK, Roy K, Bawa R. Applications of aptamers in nanodelivery systems in cancer, eye and inflammatory diseases. Nanomedicine. 2010;5(9):1435–1445. doi: 10.2217/nnm.10.115. [DOI] [PubMed] [Google Scholar]
- 17.Bruno JG, Kiel JL. In vitro selection of DNA aptamers to anthrax spores with electrochemiluminescence detection. Biosens Bioelectron. 1999;14(5):457–464. doi: 10.1016/S0956-5663(99)00028-7. [DOI] [PubMed] [Google Scholar]
- 18.Cowperthwaite MC, Ellington AD. Bioinformatic analysis of the contribution of primer sequences to aptamer structures. J Mol Evol. 2008;67(1):95–102. doi: 10.1007/s00239-008-9130-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toh SY, Citartan M, Gopinath SC, Tang TH. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens Bioelectron. 2015;64:392–403. doi: 10.1016/j.bios.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 20.McConnell EM, Holahan MR, DeRosa MC. Aptamers as promising molecular recognition elements for diagnostics and therapeutics in the central nervous system. Nucleic acid Ther. 2014;24(6):388–404. doi: 10.1089/nat.2014.0492. [DOI] [PubMed] [Google Scholar]
- 21.Feucht N, Matthias H, Lohmann CP, Maier M. Pegaptanib sodium treatment in neovascular age-related macular degeneration: clinical experience in Germany. Clin Ophthalmol. 2008;2(2):253–259. doi: 10.2147/OPTH.S2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Group VISiONCT. Chakravarthy U, Adamis AP, Cunningham ET, Jr, Goldbaum M, Guyer DR, Katz B, Patel M. Year 2 efficacy results of 2 randomized controlled clinical trials of pegaptanib for neovascular age-related macular degeneration. Ophthalmology. 2006;113(9):1508 e1501–e1525. doi: 10.1016/j.ophtha.2006.02.064. [DOI] [PubMed] [Google Scholar]
- 23.Gragoudas ES, Adamis AP, Cunningham ET, Jr, Feinsod M, Guyer DR, Group VISiONCT Pegaptanib for neovascular age-related macular degeneration. N Engl J Med. 2004;351(27):2805–2816. doi: 10.1056/NEJMoa042760. [DOI] [PubMed] [Google Scholar]
- 24.Wu J, Zhu Y, Xue F, Mei Z, Yao L, Wang X, Zheng L, Liu J, Liu G, Peng C, Chen W. Recent trends in SELEX technique and its application to food safety monitoring. Mikrochim Acta. 2014;181(5–6):479–491. doi: 10.1007/s00604-013-1156-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 26.Shaltiel-Karyo R, Frenkel-Pinter M, Egoz-Matia N, Frydman-Marom A, Shalev DE, Segal D, Gazit E. Inhibiting alpha-synuclein oligomerization by stable cell-penetrating beta-synuclein fragments recovers phenotype of Parkinson’s disease model flies. PLoS One. 2010;5(11):e13863. doi: 10.1371/journal.pone.0013863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenamyre JT, Hastings TG. Biomedicine. Parkinson’s—divergent causes, convergent mechanisms. Science. 2004;304(5674):1120–1122. doi: 10.1126/science.1098966. [DOI] [PubMed] [Google Scholar]
- 28.Crawford M, Woodman R, Ko Ferrigno P. Peptide aptamers: tools for biology and drug discovery. Brief Funct Genom Proteom. 2003;2(1):72–79. doi: 10.1093/bfgp/2.1.72. [DOI] [PubMed] [Google Scholar]
- 29.Buerger C, Groner B. Bifunctional recombinant proteins in cancer therapy: cell penetrating peptide aptamers as inhibitors of growth factor signaling. J Cancer Res Clin Oncol. 2003;129(12):669–675. doi: 10.1007/s00432-003-0489-8. [DOI] [PubMed] [Google Scholar]
- 30.Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem. 1999;45(9):1628–1650. [PubMed] [Google Scholar]
- 31.Wright WE, Binder M, Funk W. Cyclic amplification and selection of targets (CASTing) for the myogenin consensus binding site. Mol Cell Biol. 1991;11(8):4104–4110. doi: 10.1128/MCB.11.8.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blackwell TK, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250(4984):1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 33.Keefe AD, Szostak JW. Functional proteins from a random-sequence library. Nature. 2001;410(6829):715–718. doi: 10.1038/35070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marimuthu C, Tang TH, Tominaga J, Tan SC, Gopinath SC. Single-stranded DNA (ssDNA) production in DNA aptamer generation. Analyst. 2012;137(6):1307–1315. doi: 10.1039/c2an15905h. [DOI] [PubMed] [Google Scholar]
- 35.Svobodova M, Pinto A, Nadal P, O’Sullivan CK. Comparison of different methods for generation of single-stranded DNA for SELEX processes. Anal Bioanal Chem. 2012;404(3):835–842. doi: 10.1007/s00216-012-6183-4. [DOI] [PubMed] [Google Scholar]
- 36.Stoltenburg R, Reinemann C, Strehlitz B. SELEX—a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng. 2007;24(4):381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 37.Rahimi F, Bitan G. Selection of aptamers for amyloid beta-protein, the causative agent of Alzheimer’s disease. J Vis Exp. 2010 doi: 10.3791/1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsukakoshi K, Abe K, Sode K, Ikebukuro K. Selection of DNA aptamers that recognize alpha-synuclein oligomers using a competitive screening method. Anal Chem. 2012;84(13):5542–5547. doi: 10.1021/ac300330g. [DOI] [PubMed] [Google Scholar]
- 39.Darmostuk M, Rimpelova S, Gbelcova H, Ruml T. Current approaches in SELEX: an update to aptamer selection technology. Biotechnol Adv. 2015;33(6 Pt 2):1141–1161. doi: 10.1016/j.biotechadv.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 40.Xi Z, Huang R, Deng Y, He N. Progress in selection and biomedical applications of aptamers. J Biomed Nanotechnol. 2014;10(10):3043–3062. doi: 10.1166/jbn.2014.1979. [DOI] [PubMed] [Google Scholar]
- 41.Yang Y, Yang D, Schluesener HJ, Zhang Z. Advances in SELEX and application of aptamers in the central nervous system. Biomol Eng. 2007;24(6):583–592. doi: 10.1016/j.bioeng.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 42.Mendonsa SD, Bowser MT. In vitro selection of high-affinity DNA ligands for human IgE using capillary electrophoresis. Anal Chem. 2004;76(18):5387–5392. doi: 10.1021/ac049857v. [DOI] [PubMed] [Google Scholar]
- 43.Bruno JG. In vitro selection of DNA to chloroaromatics using magnetic microbead-based affinity separation and fluorescence detection. Biochem Biophys Res Commun. 1997;234(1):117–120. doi: 10.1006/bbrc.1997.6517. [DOI] [PubMed] [Google Scholar]
- 44.Bruno JG, Kiel JL (2002) Use of magnetic beads in selection and detection of biotoxin aptamers by electrochemiluminescence and enzymatic methods. BioTech 32 (1):178–180, 182–173 [DOI] [PubMed]
- 45.Mannironi C, Di Nardo A, Fruscoloni P, Tocchini-Valentini GP. In vitro selection of dopamine RNA ligands. Biochemistry. 1997;36(32):9726–9734. doi: 10.1021/bi9700633. [DOI] [PubMed] [Google Scholar]
- 46.Eulberg D, Buchner K, Maasch C, Klussmann S. Development of an automated in vitro selection protocol to obtain RNA-based aptamers: identification of a biostable substance P antagonist. Nucleic Acids Res. 2005;33(4):e45. doi: 10.1093/nar/gni044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cox JC, Rudolph P, Ellington AD. Automated RNA selection. Biotechnol Prog. 1998;14(6):845–850. doi: 10.1021/bp980097h. [DOI] [PubMed] [Google Scholar]
- 48.Cox JC, Ellington AD. Automated selection of anti-protein aptamers. Bioorg Med Chem. 2001;9(10):2525–2531. doi: 10.1016/S0968-0896(01)00028-1. [DOI] [PubMed] [Google Scholar]
- 49.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. A tenascin-C aptamer identified by tumor cell SELEX: systematic evolution of ligands by exponential enrichment. Proc Natl Acad Sci USA. 2003;100(26):15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Luo Y, Bing T, Chen Z, Lu M, Zhang N, Shangguan D, Gao X. DNA aptamer evolved by cell-SELEX for recognition of prostate cancer. PLoS One. 2014;9(6):e100243. doi: 10.1371/journal.pone.0100243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li WM, Bing T, Wei JY, Chen ZZ, Shangguan DH, Fang J. Cell-SELEX-based selection of aptamers that recognize distinct targets on metastatic colorectal cancer cells. Biomaterials. 2014;35(25):6998–7007. doi: 10.1016/j.biomaterials.2014.04.112. [DOI] [PubMed] [Google Scholar]
- 52.Aravind A, Yoshida Y, Maekawa T, Kumar DS. Aptamer-conjugated polymeric nanoparticles for targeted cancer therapy. Drug Del Transl Res. 2012;2(6):418–436. doi: 10.1007/s13346-012-0104-0. [DOI] [PubMed] [Google Scholar]
- 53.Bayrac AT, Sefah K, Parekh P, Bayrac C, Gulbakan B, Oktem HA, Tan W. In vitro selection of DNA aptamers to glioblastoma multiforme. ACS Chem Neurosci. 2011;2(3):175–181. doi: 10.1021/cn100114k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Souza AG, Marangoni K, Fujimura PT, Alves PT, Silva MJ, Bastos VA, Goulart LR, Goulart VA. 3D Cell-SELEX: development of RNA aptamers as molecular probes for PC-3 tumor cell line. Exp Cell Res. 2016;341(2):147–156. doi: 10.1016/j.yexcr.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 55.Li Z, Cui Z. Three-dimensional perfused cell culture. Biotechnol Adv. 2014;32(2):243–254. doi: 10.1016/j.biotechadv.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 56.Yu Y, Liang C, Lv Q, Li D, Xu X, Liu B, Lu A, Zhang G. Molecular selection, modification and development of therapeutic oligonucleotide aptamers. Int J Mol Sci. 2016 doi: 10.3390/ijms17030358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bunka DH, Platonova O, Stockley PG. Development of aptamer therapeutics. Curr Opin Pharmacol. 2010;10(5):557–562. doi: 10.1016/j.coph.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 58.Famulok M. Bringing picomolar protein detection into proximity. Nat Biotechnol. 2002;20(5):448–449. doi: 10.1038/nbt0502-448. [DOI] [PubMed] [Google Scholar]
- 59.Shamah SM, Healy JM, Cload ST. Complex target SELEX. Acc Chem Res. 2008;41(1):130–138. doi: 10.1021/ar700142z. [DOI] [PubMed] [Google Scholar]
- 60.Sassanfar M, Szostak JW. An RNA motif that binds ATP. Nature. 1993;364(6437):550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 61.Hu M, Zhang K. The application of aptamers in cancer research: an up-to-date review. Future Oncol. 2013;9(3):369–376. doi: 10.2217/fon.12.201. [DOI] [PubMed] [Google Scholar]
- 62.Yang Y, Ren X, Schluesener HJ, Zhang Z. Aptamers: selection, modification and application to nervous system diseases. Curr Med Chem. 2011;18(27):4159–4168. doi: 10.2174/092986711797189646. [DOI] [PubMed] [Google Scholar]
- 63.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9(7):537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lakhin AV, Tarantul VZ, Gening LV. Aptamers: problems, solutions and prospects. Acta Nat. 2013;5(4):34–43. [PMC free article] [PubMed] [Google Scholar]
- 65.Schmidt KS, Borkowski S, Kurreck J, Stephens AW, Bald R, Hecht M, Friebe M, Dinkelborg L, Erdmann VA. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 2004;32(19):5757–5765. doi: 10.1093/nar/gkh862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rhie A, Kirby L, Sayer N, Wellesley R, Disterer P, Sylvester I, Gill A, Hope J, James W, Tahiri-Alaoui A. Characterization of 2′-fluoro-RNA aptamers that bind preferentially to disease-associated conformations of prion protein and inhibit conversion. J Biol Chem. 2003;278(41):39697–39705. doi: 10.1074/jbc.M305297200. [DOI] [PubMed] [Google Scholar]
- 67.Meek KN, Rangel AE, Heemstra JM. Enhancing aptamer function and stability via in vitro selection using modified nucleic acids. Methods. 2016 doi: 10.1016/j.ymeth.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 68.Sun H, Zu Y. A highlight of recent advances in aptamer technology and its application. Molecules. 2015;20(7):11959–11980. doi: 10.3390/molecules200711959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Petersen M, Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 2003;21(2):74–81. doi: 10.1016/S0167-7799(02)00038-0. [DOI] [PubMed] [Google Scholar]
- 70.Watson SR, Chang YF, O’Connell D, Weigand L, Ringquist S, Parma DH. Anti-L-selectin aptamers: binding characteristics, pharmacokinetic parameters, and activity against an intravascular target in vivo. Antisense Nucleic Acid Drug Dev. 2000;10(2):63–75. doi: 10.1089/oli.1.2000.10.63. [DOI] [PubMed] [Google Scholar]
- 71.Dougan H, Lyster DM, Vo CV, Stafford A, Weitz JI, Hobbs JB. Extending the lifetime of anticoagulant oligodeoxynucleotide aptamers in blood. Nucl Med Biol. 2000;27(3):289–297. doi: 10.1016/S0969-8051(99)00103-1. [DOI] [PubMed] [Google Scholar]
- 72.de Smidt PC, Le Doan T, de Falco S, van Berkel TJ. Association of antisense oligonucleotides with lipoproteins prolongs the plasma half-life and modifies the tissue distribution. Nucleic Acids Res. 1991;19(17):4695–4700. doi: 10.1093/nar/19.17.4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ng EW, Shima DT, Calias P, Cunningham ET, Jr, Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov. 2006;5(2):123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 74.Siddiqui MA, Keating GM. Pegaptanib: in exudative age-related macular degeneration. Drugs. 2005;65(11):1571–1577. doi: 10.2165/00003495-200565110-00010. [DOI] [PubMed] [Google Scholar]
- 75.McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24(8):1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 76.Chu TC, Marks JW, 3rd, Lavery LA, Faulkner S, Rosenblum MG, Ellington AD, Levy M. Aptamer: toxin conjugates that specifically target prostate tumor cells. Cancer Res. 2006;66(12):5989–5992. doi: 10.1158/0008-5472.CAN-05-4583. [DOI] [PubMed] [Google Scholar]
- 77.Wang XW, Gao W, Fan H, Ding D, Lai XF, Zou YX, Chen L, Chen Z, Tan W. Simultaneous tracking of drug molecules and carriers using aptamer-functionalized fluorescent superstable gold nanorod-carbon nanocapsules during thermo-chemotherapy. Nanoscale. 2016;8(15):7942–7948. doi: 10.1039/C6NR00369A. [DOI] [PubMed] [Google Scholar]
- 78.Azhdarzadeh M, Atyabi F, Saei AA, Varnamkhasti BS, Omidi Y, Fateh M, Ghavami M, Shanehsazzadeh S, Dinarvand R. Theranostic MUC-1 aptamer targeted gold coated superparamagnetic iron oxide nanoparticles for magnetic resonance imaging and photothermal therapy of colon cancer. Colloids Surf B. 2016;143:224–232. doi: 10.1016/j.colsurfb.2016.02.058. [DOI] [PubMed] [Google Scholar]
- 79.Cansiz S, Zhang L, Wu C, Wu Y, Teng IT, Hou W, Wang Y, Wan S, Cai R, Jin C, Liu Q, Tan W. DNA aptamer based nanodrugs: molecular engineering for efficiency. Chem Asian J. 2015;10(10):2084–2094. doi: 10.1002/asia.201500434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liang H, Zhang XB, Lv Y, Gong L, Wang R, Zhu X, Yang R, Tan W. Functional DNA-containing nanomaterials: cellular applications in biosensing, imaging, and targeted therapy. Acc Chem Res. 2014;47(6):1891–1901. doi: 10.1021/ar500078f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang L, Gu FX, Chan JM, Wang AZ, Langer RS, Farokhzad OC. Nanoparticles in medicine: therapeutic applications and developments. Clin Pharmacol Ther. 2008;83(5):761–769. doi: 10.1038/sj.clpt.6100400. [DOI] [PubMed] [Google Scholar]
- 82.Tong R, Yala L, Fan TM, Cheng J. The formulation of aptamer-coated paclitaxel-polylactide nanoconjugates and their targeting to cancer cells. Biomaterials. 2010;31(11):3043–3053. doi: 10.1016/j.biomaterials.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Farokhzad OC, Jon S, Khademhosseini A, Tran TN, Lavan DA, Langer R. Nanoparticle-aptamer bioconjugates: a new approach for targeting prostate cancer cells. Cancer Res. 2004;64(21):7668–7672. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 84.Liu G, Mao X, Phillips JA, Xu H, Tan W, Zeng L. Aptamer-nanoparticle strip biosensor for sensitive detection of cancer cells. Anal Chem. 2009;81(24):10013–10018. doi: 10.1021/ac901889s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo J, Gao X, Su L, Xia H, Gu G, Pang Z, Jiang X, Yao L, Chen J, Chen H. Aptamer-functionalized PEG-PLGA nanoparticles for enhanced anti-glioma drug delivery. Biomaterials. 2011;32(31):8010–8020. doi: 10.1016/j.biomaterials.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 86.Li BR, Hsieh YJ, Chen YX, Chung YT, Pan CY, Chen YT. An ultrasensitive nanowire-transistor biosensor for detecting dopamine release from living PC12 cells under hypoxic stimulation. J Am Chem Soc. 2013;135(43):16034–16037. doi: 10.1021/ja408485m. [DOI] [PubMed] [Google Scholar]
- 87.Gong P, Shi B, Zheng M, Wang B, Zhang P, Hu D, Gao D, Sheng Z, Zheng C, Ma Y, Cai L. PEI protected aptamer molecular probes for contrast-enhanced in vivo cancer imaging. Biomaterials. 2012;33(31):7810–7817. doi: 10.1016/j.biomaterials.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 88.Li JM, Zhao MX, Su H, Wang YY, Tan CP, Ji LN, Mao ZW. Multifunctional quantum-dot-based siRNA delivery for HPV18 E6 gene silence and intracellular imaging. Biomaterials. 2011;32(31):7978–7987. doi: 10.1016/j.biomaterials.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 89.Piest M, Engbersen JF. Effects of charge density and hydrophobicity of poly(amido amine)s for non-viral gene delivery. J Control Rel. 2010;148(1):83–90. doi: 10.1016/j.jconrel.2010.07.109. [DOI] [PubMed] [Google Scholar]
- 90.Mok H, Park TG. Self-crosslinked and reducible fusogenic peptides for intracellular delivery of siRNA. Biopolymers. 2008;89(10):881–888. doi: 10.1002/bip.21032. [DOI] [PubMed] [Google Scholar]
- 91.Landen CN, Jr, Chavez-Reyes A, Bucana C, Schmandt R, Deavers MT, Lopez-Berestein G, Sood AK. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65(15):6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 92.Chakrabarti S, Khemka VK, Banerjee A, Chatterjee G, Ganguly A, Biswas A. Metabolic risk factors of sporadic Alzheimer’s disease: implications in the pathology, pathogenesis and treatment. Aging Dis. 2015;6(4):282–299. doi: 10.14336/AD.2014.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Das U, Wang L, Ganguly A, Saikia JM, Wagner SL, Koo EH, Roy S. Visualizing APP and BACE-1 approximation in neurons yields insight into the amyloidogenic pathway. Nat Neurosci. 2016;19(1):55–64. doi: 10.1038/nn.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liang H, Shi Y, Kou Z, Peng Y, Chen W, Li X, Li S, Wang Y, Wang F, Zhang X. Inhibition of BACE1 activity by a DNA aptamer in an Alzheimer’s disease cell model. PLoS One. 2015;10(10):e0140733. doi: 10.1371/journal.pone.0140733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rentmeister A, Bill A, Wahle T, Walter J, Famulok M. RNA aptamers selectively modulate protein recruitment to the cytoplasmic domain of beta-secretase BACE1 in vitro. RNA. 2006;12(9):1650–1660. doi: 10.1261/rna.126306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rahimi F, Murakami K, Summers JL, Chen CH, Bitan G. RNA aptamers generated against oligomeric Abeta40 recognize common amyloid aptatopes with low specificity but high sensitivity. PLoS One. 2009;4(11):e7694. doi: 10.1371/journal.pone.0007694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lauren J, Gimbel DA, Nygaard HB, Gilbert JW, Strittmatter SM. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-beta oligomers. Nature. 2009;457(7233):1128–1132. doi: 10.1038/nature07761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mashima T, Matsugami A, Nishikawa F, Nishikawa S, Katahira M. Unique quadruplex structure and interaction of an RNA aptamer against bovine prion protein. Nucleic Acids Res. 2009;37(18):6249–6258. doi: 10.1093/nar/gkp647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Du TT, Wang L, Duan CL, Lu LL, Zhang JL, Gao G, Qiu XB, Wang XM, Yang H. GBA deficiency promotes SNCA/alpha-synuclein accumulation through autophagic inhibition by inactivated PPP2A. Autophagy. 2015;11(10):1803–1820. doi: 10.1080/15548627.2015.1086055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Auluck PK, Caraveo G, Lindquist S. alpha-Synuclein: membrane interactions and toxicity in Parkinson’s disease. Annu Rev Cell Dev Biol. 2010;26:211–233. doi: 10.1146/annurev.cellbio.042308.113313. [DOI] [PubMed] [Google Scholar]
- 101.Zhao J, Yu S, Zheng Y, Yang H, Zhang J. Oxidative modification and its implications for the neurodegeneration of Parkinson’s disease. Mol Neurobiol. 2016 doi: 10.1007/s12035-016-9743-3. [DOI] [PubMed] [Google Scholar]
- 102.Bisaglia M, Filograna R, Beltramini M, Bubacco L. Are dopamine derivatives implicated in the pathogenesis of Parkinson’s disease? Ageing Res Rev. 2014;13:107–114. doi: 10.1016/j.arr.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 103.Kamel F. Epidemiology. Paths from pesticides to Parkinson’s. Science. 2013;341(6147):722–723. doi: 10.1126/science.1243619. [DOI] [PubMed] [Google Scholar]
- 104.Goedert M. NEURODEGENERATION. Alzheimer’s and Parkinson’s diseases: the prion concept in relation to assembled Abeta, tau, and alpha-synuclein. Science. 2015;349(6248):1255555. doi: 10.1126/science.1255555. [DOI] [PubMed] [Google Scholar]
- 105.Deleersnijder A, Gerard M, Debyser Z, Baekelandt V. The remarkable conformational plasticity of alpha-synuclein: blessing or curse? Trends Mol Med. 2013;19(6):368–377. doi: 10.1016/j.molmed.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 106.van Rooijen BD, Claessens MM, Subramaniam V. Membrane permeabilization by oligomeric alpha-synuclein: in search of the mechanism. PLoS One. 2010;5(12):e14292. doi: 10.1371/journal.pone.0014292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Karpinar DP, Balija MB, Kugler S, Opazo F, Rezaei-Ghaleh N, Wender N, Kim HY, Taschenberger G, Falkenburger BH, Heise H, Kumar A, Riedel D, Fichtner L, Voigt A, Braus GH, Giller K, Becker S, Herzig A, Baldus M, Jackle H, Eimer S, Schulz JB, Griesinger C, Zweckstetter M. Pre-fibrillar alpha-synuclein variants with impaired beta-structure increase neurotoxicity in Parkinson’s disease models. EMBO J. 2009;28(20):3256–3268. doi: 10.1038/emboj.2009.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kostka M, Hogen T, Danzer KM, Levin J, Habeck M, Wirth A, Wagner R, Glabe CG, Finger S, Heinzelmann U, Garidel P, Duan W, Ross CA, Kretzschmar H, Giese A. Single particle characterization of iron-induced pore-forming alpha-synuclein oligomers. J Biol Chem. 2008;283(16):10992–11003. doi: 10.1074/jbc.M709634200. [DOI] [PubMed] [Google Scholar]
- 109.Weng CH, Huang CJ, Lee GB. Screening of aptamers on microfluidic systems for clinical applications. Sensors. 2012;12(7):9514–9529. doi: 10.3390/s120709514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rodriguez-Oroz MC, Jahanshahi M, Krack P, Litvan I, Macias R, Bezard E, Obeso JA. Initial clinical manifestations of Parkinson’s disease: features and pathophysiological mechanisms. Lancet Neurol. 2009;8(12):1128–1139. doi: 10.1016/S1474-4422(09)70293-5. [DOI] [PubMed] [Google Scholar]
- 111.Zheng J, Zhou X. Sodium dodecyl sulfate-modified carbon paste electrodes for selective determination of dopamine in the presence of ascorbic acid. Bioelectrochemistry. 2007;70(2):408–415. doi: 10.1016/j.bioelechem.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 112.Swanson CJ, Perry KW, Koch-Krueger S, Katner J, Svensson KA, Bymaster FP. Effect of the attention deficit/hyperactivity disorder drug atomoxetine on extracellular concentrations of norepinephrine and dopamine in several brain regions of the rat. Neuropharmacology. 2006;50(6):755–760. doi: 10.1016/j.neuropharm.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 113.Kienast T, Heinz A. Dopamine and the diseased brain. CNS Neurol Disord Drug Targets. 2006;5(1):109–131. doi: 10.2174/187152706784111560. [DOI] [PubMed] [Google Scholar]
- 114.Liu S, Xing X, Yu J, Lian W, Li J, Cui M, Huang J. A novel label-free electrochemical aptasensor based on graphene-polyaniline composite film for dopamine determination. Biosens Bioelectron. 2012;36(1):186–191. doi: 10.1016/j.bios.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 115.Farjami E, Campos R, Nielsen JS, Gothelf KV, Kjems J, Ferapontova EE. RNA aptamer-based electrochemical biosensor for selective and label-free analysis of dopamine. Anal Chem. 2013;85(1):121–128. doi: 10.1021/ac302134s. [DOI] [PubMed] [Google Scholar]
- 116.Aguzzi A, Falsig J. Prion propagation, toxicity and degradation. Nat Neurosci. 2012;15(7):936–939. doi: 10.1038/nn.3120. [DOI] [PubMed] [Google Scholar]
- 117.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95(23):13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sayer NM, Cubin M, Rhie A, Bullock M, Tahiri-Alaoui A, James W. Structural determinants of conformationally selective, prion-binding aptamers. J Biol Chem. 2004;279(13):13102–13109. doi: 10.1074/jbc.M310928200. [DOI] [PubMed] [Google Scholar]
- 119.Ogasawara D, Hasegawa H, Kaneko K, Sode K, Ikebukuro K. Screening of DNA aptamer against mouse prion protein by competitive selection. Prion. 2014;1(4):248–254. doi: 10.4161/pri.1.4.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sekiya S, Noda K, Nishikawa F, Yokoyama T, Kumar PK, Nishikawa S. Characterization and application of a novel RNA aptamer against the mouse prion protein. J Biochem. 2006;139(3):383–390. doi: 10.1093/jb/mvj046. [DOI] [PubMed] [Google Scholar]
- 121.Takemura K, Wang P, Vorberg I, Surewicz W, Priola SA, Kanthasamy A, Pottathil R, Chen SG, Sreevatsan S. DNA aptamers that bind to PrP(C) and not PrP(Sc) show sequence and structure specificity. Exp Biol Med. 2006;231(2):204–214. doi: 10.1177/153537020623100211. [DOI] [PubMed] [Google Scholar]
- 122.Mercey R, Lantier I, Maurel MC, Grosclaude J, Lantier F, Marc D. Fast, reversible interaction of prion protein with RNA aptamers containing specific sequence patterns. Arch Virol. 2006;151(11):2197–2214. doi: 10.1007/s00705-006-0790-3. [DOI] [PubMed] [Google Scholar]
- 123.Spinney L. Uncovering the true prevalence of Huntington’s disease. Lancet Neurol. 2010;9(8):760–761. doi: 10.1016/S1474-4422(10)70160-5. [DOI] [PubMed] [Google Scholar]
- 124.Munoz-Sanjuan I, Bates GP. The importance of integrating basic and clinical research toward the development of new therapies for Huntington disease. J Clin Investig. 2011;121(2):476–483. doi: 10.1172/JCI45364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Skogen M, Roth J, Yerkes S, Parekh-Olmedo H, Kmiec E. Short G-rich oligonucleotides as a potential therapeutic for Huntington’s Disease. BMC Neurosci. 2006;7:65. doi: 10.1186/1471-2202-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li W, Lan X. Aptamer oligonucleotides: novel potential therapeutic agents in autoimmune disease. Nucleic acid Therap. 2015;25(4):173–179. doi: 10.1089/nat.2014.0529. [DOI] [PubMed] [Google Scholar]
- 127.Smestad J, Maher LJ., 3rd Ion-dependent conformational switching by a DNA aptamer that induces remyelination in a mouse model of multiple sclerosis. Nucleic Acids Res. 2013;41(2):1329–1342. doi: 10.1093/nar/gks1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nastasijevic B, Wright BR, Smestad J, Warrington AE, Rodriguez M, Maher LJ., 3rd Remyelination induced by a DNA aptamer in a mouse model of multiple sclerosis. PLoS One. 2012;7(6):e39595. doi: 10.1371/journal.pone.0039595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rozenblum GT, Kaufman T, Vitullo AD. Myelin basic protein and a multiple sclerosis-related MBP-peptide bind to oligonucleotides. Mol Ther Nucleic Acids. 2014;3:e192. doi: 10.1038/mtna.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chang M, Yang CS, Huang DM. Aptamer-conjugated DNA icosahedral nanoparticles as a carrier of doxorubicin for cancer therapy. ACS Nano. 2011;5(8):6156–6163. doi: 10.1021/nn200693a. [DOI] [PubMed] [Google Scholar]
- 131.Shi H, Tang Z, Kim Y, Nie H, Huang YF, He X, Deng K, Wang K, Tan W. In vivo fluorescence imaging of tumors using molecular aptamers generated by cell-SELEX. Chem Asian J. 2010;5(10):2209–2213. doi: 10.1002/asia.201000242. [DOI] [PubMed] [Google Scholar]
- 132.Nolte A, Klussmann S, Bald R, Erdmann VA, Furste JP. Mirror-design of l-oligonucleotide ligands binding to l-arginine. Nat Biotechnol. 1996;14(9):1116–1119. doi: 10.1038/nbt0996-1116. [DOI] [PubMed] [Google Scholar]
- 133.Hicke BJ, Marion C, Chang YF, Gould T, Lynott CK, Parma D, Schmidt PG, Warren S. Tenascin-C aptamers are generated using tumor cells and purified protein. J Biol Chem. 2001;276(52):48644–48654. doi: 10.1074/jbc.M104651200. [DOI] [PubMed] [Google Scholar]