Abstract
MicroRNAs (miRNAs) coordinate vascular repair by regulating injury-induced gene expression in vascular smooth muscle cells (SMCs) and promote the transition of SMCs from a contractile to a proliferating phenotype. However, the effect of miRNA expression in SMCs on neointima formation is unclear. Therefore, we studied the role of miRNA biogenesis by Dicer in SMCs in vascular repair. Following wire-induced injury to carotid arteries of Apolipoprotein E knockout (Apoe −/−) mice, miRNA microarray analysis revealed that the most significantly regulated miRNAs, such as miR-222 and miR-21-3p, were upregulated. Conditional deletion of Dicer in SMCs increased neointima formation by reducing SMC proliferation in Apoe −/− mice, and decreased mainly the expression of miRNAs, such as miR-147 and miR-100, which were not upregulated following vascular injury. SMC-specific deletion of Dicer promoted growth factor and inflammatory signaling and regulated a miRNA–target interaction network in injured arteries that was enriched in anti-proliferative miRNAs. The most connected miRNA in this network was miR-27a-3p [e.g., with Rho guanine nucleotide exchange factor 26 (ARHGEF26)], which was expressed in medial and neointimal SMCs in a Dicer-dependent manner. In vitro, miR-27a-3p suppresses ARHGEF26 expression and inhibits SMC proliferation by interacting with a conserved binding site in the 3′ untranslated region of ARHGEF26 mRNA. We propose that Dicer expression in SMCs plays an essential role in vascular repair by generating anti-proliferative miRNAs, such as miR-27a-3p, to prevent vessel stenosis due to exaggerated neointima formation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-016-2349-0) contains supplementary material, which is available to authorized users.
Keywords: MicroRNA, Dicer, Smooth muscle cells, Cell proliferation, ARHGEF26
Introduction
Smooth muscle cells (SMCs) form the medial layer of arteries and regulate the vascular tone via their contractile apparatus. Transcriptional master regulators, such as the serum response factor and myocardin, promote a contractile SMC phenotype associated with a low proliferation rate [1]. Because arterial injury may disrupt the integrity of the vascular wall, risking fatal hemorrhage, during arterial wound healing, SMCs form neointimal tissue by switching to a synthetic, proliferative phenotype [1]. Stent implantation into arteries with atherosclerotic stenosis is accompanied by arterial injury that frequently results in excessive neointima formation due to SMC proliferation, ultimately causing re-narrowing of the arterial lumen (also known as restenosis) [2].
Growth factors, such as the platelet-derived growth factor (PDGF) and the epidermal growth factor (EGF), mediate SMC proliferation following vascular injury by activating mitogenic signaling pathways, e.g., AKT/mTOR and the Ras/RAF/ERK1/2 pathways [3, 4]. Moreover, cytokines, such as interleukin-1β (IL-1β) and tumor necrosis factor α (TNFα), activate pro-inflammatory NF-κB signaling in neointimal SMCs, which engages in cross-talk with mitogenic pathways and augments the SMC proliferative response during neointima formation [5, 6].
Transcriptional changes during cell fate transitions are intrinsically noisy, and microRNAs (miRNAs) confer phenotypic robustness by decreasing variability in protein expression [7, 8]. Nearly, all mature miRNAs are generated by the RNase III endonuclease Dicer that cleaves hairpin-structured precursor miRNAs. This cleavage occurs near the terminal loop of the hairpin to yield miRNA duplexes 21–25 nucleotides in length [9]. One strand of the miRNA duplex is retained in the miRNA-induced-silencing complexes (miRISCs), which contains the Argonaute and trinucleotide repeat containing 6A (TNRC6A, also known as GW182) proteins. In the miRISC, nucleotides 2–7 at the 5′ end of the miRNA bind to complementary sequences in the 3′-untranslated regions (UTRs) of target mRNAs, leading to their degradation or translational inhibition. Because one miRNA can bind hundreds of target mRNAs, and conversely, one mRNA can be targeted by multiple miRNAs, large and diverse miRNA–mRNA interaction networks regulate gene expression in a cell-type-specific manner [10, 11]. In SMCs, Dicer is essential for the development and maintenance of the contractile phenotype [12, 13]. Although miRNAs can inhibit (e.g., miR-143-3p and -145-5p) or promote (e.g., miR-21-5p) neointima formation [14–17], the impact of miRNA expression in SMCs on arterial repair remains unclear.
In mice, the deletion of Dicer in SMCs decreased neointima formation by suppressing SMC proliferation. This effect of Dicer could be attributed to the downregulation of several anti-proliferative miRNAs, such as miR-27a-3p, which inhibited SMC proliferation by targeting growth-factor-signaling activator Rho guanine nucleotide exchange factor 26 (ARHGEF26) mRNA.
Materials and methods
Animals
Male myosin, heavy chain 11, smooth muscle (MYH11)-Cre+ mice were crossed with female Dicer +/+ /Apoe −/− and Dicer flox/flox/Apoe −/− mice (The Jackson Laboratory, Bar Harbor, ME, USA) to obtain MYH11-Cre+/Dicer +/+ /Apoe −/− (SM-Dicer +/+) mice (as control group) and MYH11-Cre+/Dicer flox/flox /Apoe −/− (SM-Dicer −/−) mice [18, 19]. Littermates (6–8 weeks) were used for experiments. Cre recombinase activity was induced in SM-Dicer +/+ and SM-Dicer −/− mice by intraperitoneal injection with tamoxifen (2 mg per 20 g body weight; Sigma-Aldrich GmbH, Munich, Germany) dissolved in neutral oil (Miglyol, Sasol, Hamburg, Germany) for five consecutive days. Subsequently, mice were fed a high-fat diet (HFD) comprising 21 % crude fat, 0.15 % cholesterol, and 19.5 % casein (Altromin GmbH, Lage, Germany). After 1 week of HFD feeding, mice were anesthetized with ketamin (Pfizer, Berlin, Germany) and xylazine (Serumwerk, Bernburg, Germany), and vascular injury was induced to the left common carotid artery by advancing a 0.36-mm flexible angioplasty guide wire retrograde through the external carotid artery [20]. After immersion fixation (PAXgene, Qiagen GmbH, Hilden, Germany), carotid arteries were embedded in paraffin for histological experiments. In addition, carotid arteries were harvested after perfusion with RNA later (Thermo Scientific, Braunschweig, Germany) for RNA isolation using the mirVana Isolation Kit (Thermo Scientific). All animal experiments were reviewed and approved by the local authorities (State Agency for Nature, Environment and Consumer Protection of North Rhein-Westphalia) in accordance with German animal protection laws.
MiRNA expression profile
Carotid arteries from Apoe −/− mice were harvested before (0 day) and 1, 7, 14, and 28 days after wire-induced injury following in situ perfusion with RNA later (Thermo Scientific). The RNA was isolated using the mirVana miRNA isolation kit (Thermo Scientific). The RNA integrity number of each sample determined by Bioanalyzer Agilent 2100 (Agilent Technologies, Santa Clara, CA, USA) was higher than 7.5. A one-color-based hybridization protocol was applied (DNAvision, Gosselies, Belgium) using mouse miRNA microarrays 2.0 (miRBase v12; Agilent Technologies). The microarray data were analyzed using the Genespring GX13 software (Agilent Technologies). The microarray data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database under accession number GSE34054.
MiRNA real-time PCR array
Total RNA was isolated from carotid arteries using the mirVana miRNA isolation kit (Thermo Scientific), and the RNA quality was determined using an Agilent 2100 Bioanalyzer (RIN > 7.5) (Agilent Technologies). Reverse transcription and pre-amplification were performed using the Megaplex RT and Preamp Rodent Pool Set (Thermo Scientific) according to the manufacturer’s instructions. Samples were loaded onto preconfigured 384-well microfluidic cards (TaqMan Array miRNA Cards) for the real-time analysis of 518 mouse miRNAs (Sanger miRBase v10) using a 7900HT RT-PCR System (Thermo Scientific). Data were analyzed using the StatMiner software (Integromics, Granada, Spain) according to the ΔΔCt method using multiple internal control genes. The most stable combination of internal controls was determined using the Genorm algorithm. The fold change compared with the control group was calculated and logarithmically transformed (log10).
Laser-capture microdissection system (LCM)
PAXgen (Qiagen)-fixed serial sections (4-μm thick) of the uninjured carotid arteries (100–120 sections per mouse) were collected on UV-sterilized and RNase-free POL-membrane 0.9 µm FrameSlides (Leica Microsystem, Wetzlar, Germany) and dried at 40 °C (Thermostat plus, Eppendorf, Hamburg, Germany). Endothelial cells (ECs) and SMCs were collected using laser microdissection (LMD7000, Leica Microsystem) equipped with an inverted camera (Leica DFC365 FX) to enable a sample visualization on a computer. Total RNA was isolated using the PAXgene RNA MinElute kit (Qiagen).
Global gene expression analysis
Carotid arteries were harvested at day 14 after vascular injury following in situ perfusion with RNA later (Thermo Scientific). A one-color-based hybridization protocol was applied (IMGM Laboratories GmbH, Munich, Germany) to 8 × 60K SurePrint G3 Mouse GE Microarrays (Agilent Technologies). The analysis of the raw data was performed using the GeneSpring GX13 software (Agilent Technologies). The microarray data were analyzed by the Ingenuity Pathway Analysis software (Qiagen) to predict the upstream regulators of the differentially expressed genes.
Integrative target prediction analysis
The integrative target prediction analysis of 66 miRNAs downregulated (P < 0.05; fold change ≥2) and 217 annotated genes upregulated (P < 0.05, fold change ≥2) in SM-Dicer −/− mice compared with SM-Dicer +/+ mice using the web tool Magia2 (http://gencomp.bio.unipd.it/magia2/start/) with the miRanda prediction algorithm (http://www.microrna.org/microrna/home.do), including the top 50 % predictions (prediction score cutoff = −0.3) [21]. First, a meta-analysis approach based on a P value calculation according to LIMMA was applied separately for genes and miRNAs in the two groups and combined with the inverse Chi-square distribution to identify oppositely regulated miRNA–gene pairs. In addition, the false positive discovery rates for each mRNA–miRNA interaction were calculated following the Benjamini and Hochberg estimation method, and the interactions with an adjusted P value <0.05 were selected. The network of the top 70 interactions was graphically depicted using the Cytoscape software (http://www.cytoscape.org/). The conservation of miRNA binding sites in the mRNA targets between human and mouse was analyzed with TargetScan (http://www.targetscan.org) and microRNA.org.
Histology and immunostaining
Serial sections (5-μm thick) of the common carotid arteries (3–5 sections per mouse) were stained with elastic van Gieson stain. The lesion area was quantified by the image analysis software (ImageJ).
Quantitative immunostaining was performed using primary antibodies for α-smooth muscle actin (SMA; 1:200, clone 1A4, Dako, Hamburg, Germany), macrophage specific Mac2 (Mac2; 1:200, clone M3/38, Cedarlane, Burlington, Canada), Ki67 (1:1500, rabbit polyclonal, ab15580, Abcam), ARHGEF26 (1:20, rabbit polyclonal, ab129265, Abcam), activated Caspase 3 (1:400, Rabbit polyclonal antibody, Cell Signaling, Danvers, MA, USA), CD31 (1:75, goat polyclonal Antibody, Santa Cruz Biotechnology, Santa Cruz, CA, USA), and CD68 (1:100, Rabbit polyclonal, ab125212, Abcam). Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Vectashield, Vector Laboratories, Peterborough, UK). The primary antibody was detected with fluorescently labeled secondary antibodies (Jackson ImmunoResearch, Hamburg, Germany). Digital images were acquired using a Leica-DM6000 B light microscope (Leica Microsystem) connected to a CCD camera (Leica DFC365FX) and LAS AF software (version 3.2.0.9652, Leica). The number of positive cells and the percentage of the positively stained area per total plaque area (2–3 sections/mouse; 50–100 μm distance between sections) were determined using the image analysis software (ImageJ).
Cell culture
Human aortic smooth muscle cells (HASMCs) (passage 2–5, Promocell, Heidelberg, Germany) were seeded on six-well plates (Sigma-Aldrich) at a density of 1.5 × 105 cells per well and grown in SMC growth medium 2 (Promocell). HASMCs transfected for 48 h with locked nucleic acid (LNA)-modified miR-27a-3p inhibitors (CGGAACTTAGCCACTGTGA), non-targeting LNA-modified oligonucleotides (GTGTAACACGTCTATACGCCCA) (50 nM each, Exiqon, Vedbaek, Denmark), miR-27a-3p mimics (UUCACAGUGGCUAAGUUCCGC) (50 nM each, Thermo Scientific), or non-targeting mimic oligonucleotides (50 nM each, Thermo Scientific) using Lipofectamine 2000 (Thermo Scientific). ARHGEF26 was silenced in HASMCs using GapmeR oligonucleotides (GTAATGCAAGGATAGA) (50 nM, Exiqon) and the results compared with cells treated with control oligonucleotides (AACACGTCTATACGC) (50 nM, Exiqon). In addition, HASMCs were co-transfected with LNA-modified miR-27a-3p inhibitors (50 nM, Exiqon) and GapmeR oligonucleotides (50 nM, Exiqon) or the respective control oligonucleotides. HASMCs were also transfected with target site blockers (TSBs, TTCACAGGATTCAAATAG) that specifically block the interaction between miR-27a-3p and ARHGEF26, or control-TSBs (GCTCCCTTCAATCCAA) (50 nM each, miRCURY LNATM miRNA Target Site Blockers; Exiqon) (Online Resource Supplemental Fig. 6B). HASMCs were stimulated with IL-1β (5 ng/ml, Thermo Scientific). Total RNA was isolated using the RNeasy Mini Kit (Qiagen) or mirVana RNA Isolation Kit.
In vitro immunostaining
HASMCs (passage 2–5) were plated on glass coverslips (Neuvitro, Vancouver, WA, USA) in 24-well tissue culture plates (Sigma-Aldrich) for 24 h at a density of 3 × 104 cells per well. HASMCs were fixed in ice-cold methanol (50 % v/v in acetone) for 5 min.
Quantitative immunostaining was performed using Ki67 (1:1500, rabbit polyclonal, ab15580, Abcam). Cell nuclei were counterstained with DAPI. The primary antibody was detected with fluorescently labeled secondary antibodies (Jackson ImmunoResearch, Hamburg, Germany). Images were acquired using a Leica-DM6000 B light microscope, and the numbers of positive cells were counted using the image analysis software (ImageJ).
Combined in situ PCR and immunostaining
PAXgene-fixed carotid artery sections (5 μm thick) were treated with DNase (Roche, Basel, Switzerland) overnight at 37 °C. One-step reverse transcriptase in situ PCR (Mastercycler nexus, Eppendorf) was performed using gene-specific Taq in situ primers (Sigma-Aldrich) (Online Resource Supplemental Table 1), SuperScript One-Step RT-PCR with PlatinumTaq (Thermo Scientific), and digoxigenin-11-dUTPs (Roche) [22]. After stringent washing with SSC buffer and blockade of non-specific binding sites using nitroblue tetrazolium chloride (PerkinElmer, Waltham, MA, USA) and biotin/avidin binding sites using a blocking kit (Vector Laboratories), sections were incubated with peroxidase-conjugated anti-digoxigenin sheep Fab fragments (Fab fragments from sheep, 1:100 dilution; Roche) for 1 h at 37 °C. A tyramide-based amplification system (TSA Plus Biotin, PerkinElmer) and Dylight 549-conjugated streptavidin (1:200, KPL, Gaithersburg, MD, USA) were used to visualize the probe. Sections were subsequently incubated with SMA antibody (1:200, clone 1A4, Dako, Hamburg, Germany) followed by a FITC-conjugated secondary antibody (Jackson ImmunoResearch).
Quantitative real-time PCR
Expression levels of miRNAs were quantified using the TaqMan miRNA (Thermo Scientific) or miScript (Qiagen) primer assays. Specific primer sets were designed (Sigma-Aldrich) (Online Resource Supplemental Table 1), and TaqMan assays were used to determine mRNA and miRNA expression levels. The PCRs were run on a 7900HT thermocycler (Thermo Scientific). Relative expression levels were normalized to either single or multiple reference genes (snoRNA-135 or RNU44 for miRNAs and B2M or GAPDH for mRNA), scaled to the sample with the lowest expression using the QbasePLUS software (Biogazelle NV, Zwijnaarde, Belgium), and logarithmically transformed (log10).
MiRNA target identification and quantification
HASMCs were co-transfected with miR-27a-3p mimics (50 nM, Thermo Scientific) and the pMirTrap vector using the XfectTM miRNA transfection reagent in combination with Xfect Polymer (all from Clontech, aint-Germain-en-Laye, France). The pMirTrap vector expresses a DYKDDDDK-tagged GW182 protein, which enables locking of the miRNA/mRNA complex in the miRISC [23]. HASMCs were harvested after 24 h, washed in ice-cold phosphate-buffered saline, and incubated in lysis buffer (MirTrap System) supplemented with protease inhibitors (Complete Protease Inhibitor Cocktail Tablets; Roche). The input RNA was harvested from the cell lysates. Anti-DYKDDDDK-conjugated magnetic beads were washed twice with 1× lysis/wash buffer containing 1 mM DTT, 0.1 unit/μl RNase inhibitor and protease inhibitors, and blocked for 3 h at 4 °C with tRNA solution and bovine serum albumin. Immunoprecipitation was performed by incubating anti-DYKDDDDK beads with the cell lysate for 2 h at 4 °C. RNA from the input and immunoprecipitated samples was reverse transcribed using the high-capacity cDNA reverse transcription kit (Thermo Scientific), and the target genes were amplified with gene-specific primers (Online Resource Supplemental Table 1) and SYBR Green PCR Master Mix (Thermo Scientific) using a 7900HT fast real-time PCR system (Applied Biosystems, Darmstadt, Germany). Transfection efficiency was determined by the transfection of miR-132 mimics, the empty pMirTrap vector, or the pMirTrap positive control vector, which expresses an AcGFP1 fluorescein protein containing the miR-132 target sequence. The fold enrichment of the target genes in the GW182 immunoprecipitates was normalized to that of GAPDH according to the manufacturer’s protocol.
Luciferase reporter assay
HEK293 cells cultured in complete DMEM (PAA Laboratories GmbH, Cölbe, Germany) were co-transfected with the Gaussia luciferase (GLuc) expressing pEZX-MT05 vector with or without the full-length 3′-UTR of the human ARHGEF26 (500 ng, GeneCopoeia, Vienna, Austria), and miR-27a-3p mimic or control mimic oligonucleotides using Lipofectamine 2000 for 48 h. The miR-27a-3p binding site was mutated using the QuickChange site-directed mutagenesis kit (Agilent Technologies), specific primers (Sigma-Aldrich) (Online Resource Supplemental Table 1), PfuTurbo DNA polymerase (Thermo Scientific), and a PCR cycler (Mastercycler nexus, Eppendorf). The product was treated with DpnI endonuclease to digest the parental DNA template and to select for mutation-containing-synthesized DNA [24]. The vector DNA containing the desired mutation was transformed into XL10-Gold Ultra component cells (Agilent Technologies), and the plasmid was isolated using the EndoFree Plasmid Maxi Kit (Qiagen). The GLuc and secreted alkaline phosphatase (SEAP) activities were measured 48 h after the transfection using the Secrete Pair Dual Luminescent Assay (GeneCopoeia) and a microplate reader (Tecan Group Ltd., Männedorf, Switzerland). The GLuc luminescence signal was normalized to that of SEAP.
Western blot analysis
HASMCs were lysed in RIPA buffer (Sigma-Aldrich), including protease inhibitors. Cell lysates were resolved on SDS-PAGE gels and then transferred to nitrocellulose membranes. Proteins were detected using primary antibodies against ARHGEF26 (1:150, rabbit polyclonal, ab129265, Abcam) and β-actin (1:1000, rabbit polyclonal, ab8227, Abcam), and a secondary HRP-conjugated antibody (1:1000, anti-rabbit IgG HRP, Cell signaling). Protein bands were visualized using an enhanced chemiluminescence detection system (ECL Advance, GE Healthcare Life Sciences) and an LAS 3000 Imager (Fuji Photo Film Co., Ltd., Tokyo, Japan), and quantified using the Multigauge software (Fuji Photo Film). Intensities of the ARHGEF26 band were normalized to those of the β-actin bands.
Human carotid lesion samples
Human atherosclerotic lesion samples were obtained during carotid endarterectomy and fixed with 4 % paraformaldehyde (Carl Roth, Karlsruhe, Germany). The Ethics Committee of the Medical Faculty at RWTH Aachen University approved the study protocol for the collection of human atherosclerotic plaque specimens, and all participants gave their written informed consent.
Statistical analysis
Quantitative PCR miRNA array data are presented as means, and groups were compared using an unpaired, moderated two-tailed t test (Statminer 4.2, Integromics). All other data are presented as mean ± SEM. Two-group comparisons were performed using the two-tailed t test, and multi-group comparisons were performed using the one-way ANOVA (GeneSpring GX, Agilent) or two-way ANOVA followed by Tukey’s multiple comparison test (Prism 6.0, GraphPad). P < 0.05 was considered to indicate a statistically significant difference.
Results
miRNAs expression patterns during neointima formation
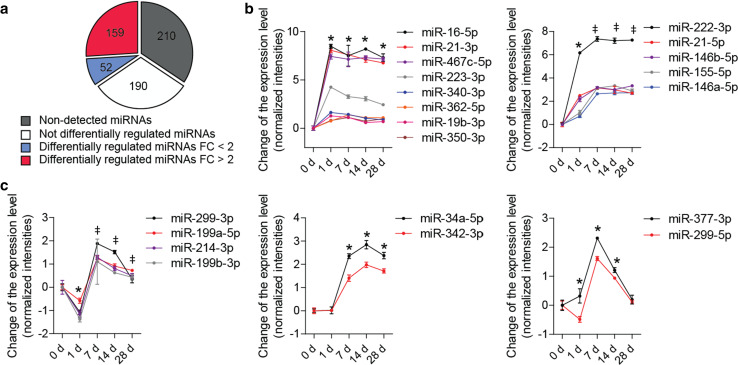
We performed the miRNA microarray analysis to assess the expression of 611 miRNAs in carotid arteries from apolipoprotein E−/− (Apoe −/−) mice before (0 day) and 1, 7, 14, and 28 days after wire-induced injury. A total of 401 miRNAs were detected, of which 211 miRNAs were differentially expressed during neointima formation (P ≤ 0.05, n = 3–4 mice per group) (Fig. 1a; Online Resource Supplemental Table 2). The expression levels of 159 differentially expressed miRNAs changed more than twofold (Fig. 1a; Online Resource Supplemental Table 2). Notably, 21 of the 25 miRNAs regulated with the greatest statistical significance were upregulated at day 7 and 14 after injury (Fig. 1b, c), although their regulation patterns differed at day 1. The expression of six miRNAs (e.g., miR-16-5p) peaked at day 1, whereas the expression of five miRNAs (e.g., miR-222-3p) increased stepwise between days 1 and 7 (Fig. 1b). The expression of two miRNAs (e.g., miR-34a-5p) remained unchanged, and four miRNAs (e.g., miR-299-3p) were transiently downregulated at day 1 (Fig. 1c). By contrast, 4 of the 25 miRNAs were not upregulated on days 7 and 14. The expression of miR-654-3p peaked at day 1, but returned to baseline levels thereafter (Online Resource Supplemental Fig. 1A). Three miRNAs, including miR-122-5p, were downregulated on days 7 and 14 after injury (Online Resource Supplemental Fig. 1B).
Fig. 1.
Differential expression of miRNAs during neointima formation. a–c miRNA microarray analysis was performed in carotid arteries from Apoe −/− mice before (0 day) and 1, 7, 14, and 28 days after wire-induced injury (n = 3–4 mice per group). a Number of miRNAs differentially expressed (P ≤ 0.05) or not differentially expressed, among all detectable miRNAs. FC fold change. b, c Expression patterns of 21 out of the 25 most significantly regulated miRNAs (FC ≥ 2). Error bars represent ±SEM. *P < 0.05 compared with 0 day; ‡ P < 0.05 compared with 1 day
Effect of Dicer expression in SMCs on neointima formation
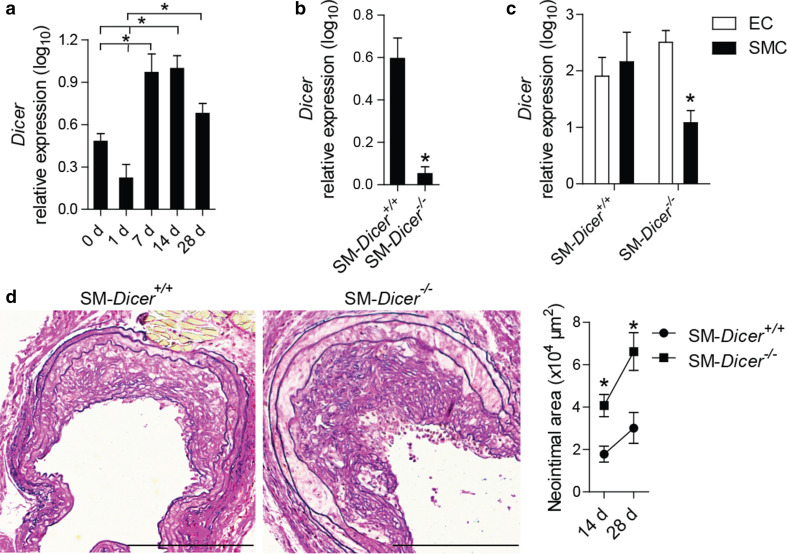
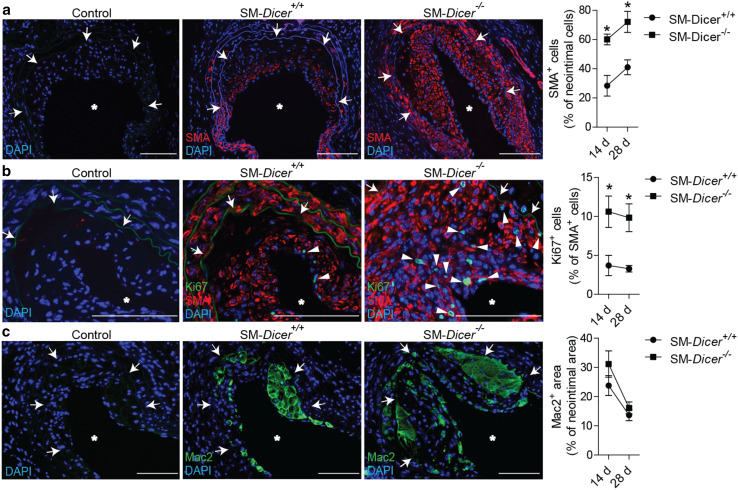
In parallel with the prevailing expression time course of miRNAs, the expression of Dicer was elevated in the carotid arteries of Apoe −/− mice on days 7 and 14 after injury (Fig. 2a). To investigate the role of Dicer in SMCs in neointima formation, we deleted the Dicer gene in SMCs by the tamoxifen treatment of myosin, heavy chain 11, smooth muscle (MYH11)-Cre+ Dicer flox/flox Apoe −/− (SM-Dicer −/−) mice. In comparison with MYH11-Cre+ Dicer +/+ Apoe −/− (SM-Dicer +/+) mice, Dicer mRNA expression was reduced in carotid arteries of SM-Dicer −/− mice 14 days after injury (Fig. 2b). In contrast to SM-Dicer +/+ mice, Dicer mRNA expression was lower in SMCs than in ECs isolated by laser microdissection from uninjured carotid artery of SM-Dicer −/− mice (Fig. 2c). Neointimal area (Fig. 2d), neointimal SMA-positive cell content (Fig. 3a), and proliferation of neointimal SMCs (Fig. 3b) were elevated at days 14 and 28 after injury in SM-Dicer −/− mice, whereas neointimal Mac2- and CD68-positive macrophage content (Fig. 3c; Online Resource Supplemental Fig. 2), rate of apoptosis in neointimal SMCs (Online Resource Supplemental Fig. 3), and the endothelial recovery (Online Resource Supplemental Fig. 4) did not differ significantly between the groups.
Fig. 2.
Dicer deletion in SMCs increased neointima formation. a Dicer mRNA expression in injured carotid arteries from Apoe −/− mice, determined by qPCR (n = 3–4 mice per group). b, c Quantitation of Dicer mRNA expression in injured carotid arteries 14 days after injury (b), in SMCs and ECs of uninjured carotid arteries (c) from MYH11-Cre+ Dicer flox/flox Apoe −/− (SM-Dicer −/−) mice and MYH11-Cre+ Dicer +/+ Apoe −/− (SM-Dicer +/+) mice (n = 2–4 mice per group). d Lesion areas 28 days after injury in carotid artery sections stained with elastic van Gieson’s stain (n = 5–7 mice per group). Scale bars 200 μm. Error bars represent ±SEM. *P < 0.05
Fig. 3.
SMC specific Dicer deletion increased neointimal SMC proliferation. a SMC content in the neointima, determined by SMA immunostaining. b Neointimal SMC proliferation, determined by Ki67/SMA double immunostaining. Nuclei were counterstained with DAPI. The arrowheads indicate Ki67+ SMA+ cells. c Neointimal macrophage content, determined by Mac2 immunostaining. Control represents the non-specific antibody for the primary antibody. The arrows indicate the neointimal area. The asterisks indicate the lumen. Scale bars 100 μm. Error bars represent ±SEM (n = 5–7 mice per group). *P < 0.05
Transcriptional changes mediated by SMC-specific Dicer deletion
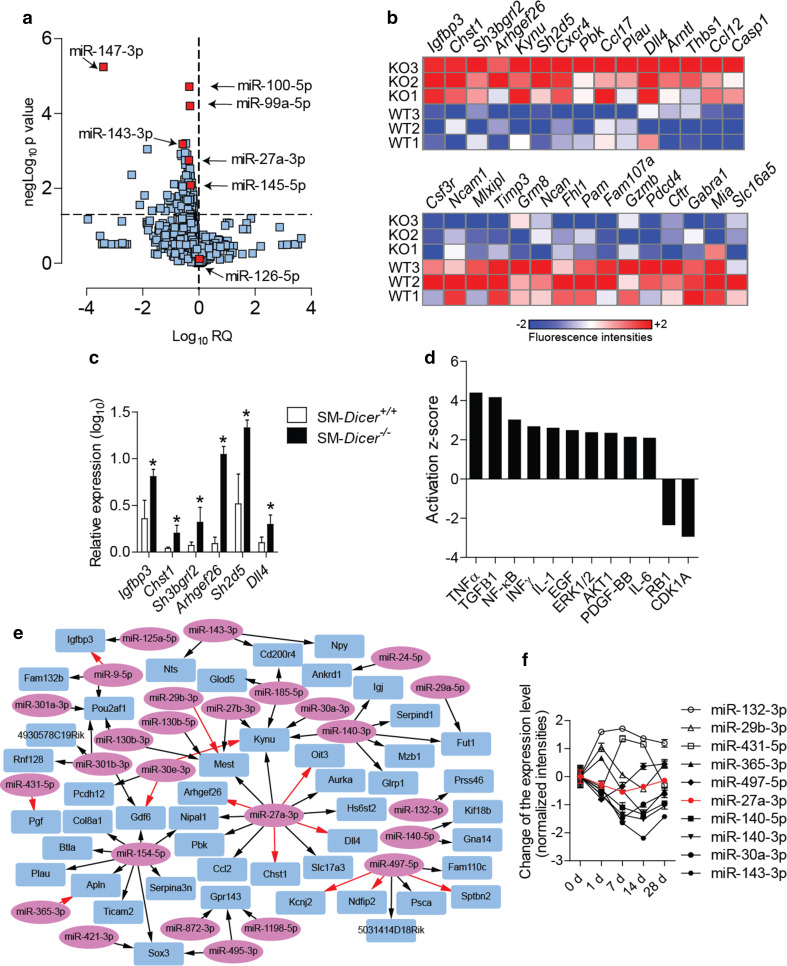
Next, to identify miRNAs regulated by Dicer in SMCs during neointima formation, we compared the miRNA expression profile in carotid arteries 14 days after injury between SM-Dicer −/− mice and SM-Dicer +/+ mice. Among the 92 miRNAs downregulated in SM-Dicer −/− mice, the levels of miR-147-3p, miR-143-3p, miR-100-5p, miR-99a-5p, and miR-27a-3p were most significantly reduced (Fig. 4a; Online Resource Supplemental Table 3). The expression of the endothelial miRNA miR-126-5p did not differ between the groups (Fig. 4a). In addition, 484 annotated genes were upregulated, and 294 genes were downregulated, in the injured carotid arteries of SM-Dicer −/− mice after 14 days determined by the mRNA microarray analysis (fold change ≥1.5, P ≤ 0.05, n = 3 mice per group; Fig. 4b; Online Resource Supplemental Table 4). The upregulation of insulin-like growth-factor-binding protein 3 (Igfbp3), SH2 domain containing 5 (Sh2d5), Arhgef26, carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 (Chst1), SH3 domain-binding glutamate-rich protein-like 2 (Sh3bgrl2), and delta-like 4 (Dll4) in SM-Dicer −/− mice was confirmed by qPCR (Fig. 4c). The pathway analysis of differentially expressed genes indicated elevated growth factor (e.g., via EGF, PDGF, ERK1/2, and AKT) and inflammatory signaling (e.g., via NF-κB and IL-1β) in SM-Dicer −/− mice (Fig. 4d).
Fig. 4.
Effect of SMC-specific Dicer deletion on gene expression in injured arteries. a Expression profile of miRNAs in carotid lesions from SM-Dicer −/− mice compared to SM-Dicer +/+ mice 14 days after vascular injury (n = 4 mice per group). b Heat map of genes differentially expressed between SM-Dicer −/− mice (KO) and SM-Dicer +/+ mice (WT) in carotid arteries 14 days after vascular injury (P < 0.05). c Quantitation of gene expression in carotid arteries 14 days after injury by qPCR (n = 4–6 mice per group). Error bars represent ±SEM. *P < 0.05. d Main upstream regulators of differential gene expression between SM-Dicer −/− mice and SM-Dicer +/+ mice as predicted by the Ingenuity Pathway Analysis software. e Integrative target prediction analysis of the downregulated miRNAs (ellipse) and upregulated mRNAs (rectangular) in SM-Dicer −/− mice compared with SM-Dicer +/+ mice, performed using the web tool Magia2 software. The 70 predicted interactions with the greatest statistical significance are shown. Red arrows indicate that a predicted interaction is conserved between mouse and humans. f Expression time course of miRNAs predicted to target mRNAs during neointima formation in Apoe −/− mice (n = 3–4 mice per group). Black and white symbols, P < 0.05; red symbols, P > 0.05; error bars represent ±SEM
The integrative target prediction analysis of the downregulated miRNAs and upregulated mRNAs in SM-Dicer −/− mice revealed 521 significant interactions between 51 miRNAs and 126 mRNAs (Fig. 4e; Online Resource Supplemental Table 5). Among the 70 interactions with the greatest statistical significance (between 26 miRNAs and 47 mRNAs), the largest number of targets was identified for miR-27a-3p (12 mRNAs), miR-154-5p (nine mRNAs), miR-140-3p (six mRNAs), and miR-497-5p (six mRNAs). Moreover, 4 of the 12 putative miR-27a-3p targets, including Arhgef26, Chst1, Dll4, and oncoprotein-induced transcript 3 (Oit3), contained highly conserved binding sites for miR-27a-3p (Fig. 4e). Among the 26 miRNAs involved in the 70 most significant interactions, ten miRNAs, including miR-27a-3p, were not differentially regulated during neointima formation, whereas 11 miRNAs, including miR-143-3p and miR-140-3p/-5p, were downregulated (Fig. 4f; Online Resource Supplemental Table 3). In addition, five miRNAs, such as miR-132-3p, were upregulated following vascular injury (Fig. 4f; Online Resource Supplemental Table 3).
MiR-27a-3p regulates ARHGEF26 expression in SMCs
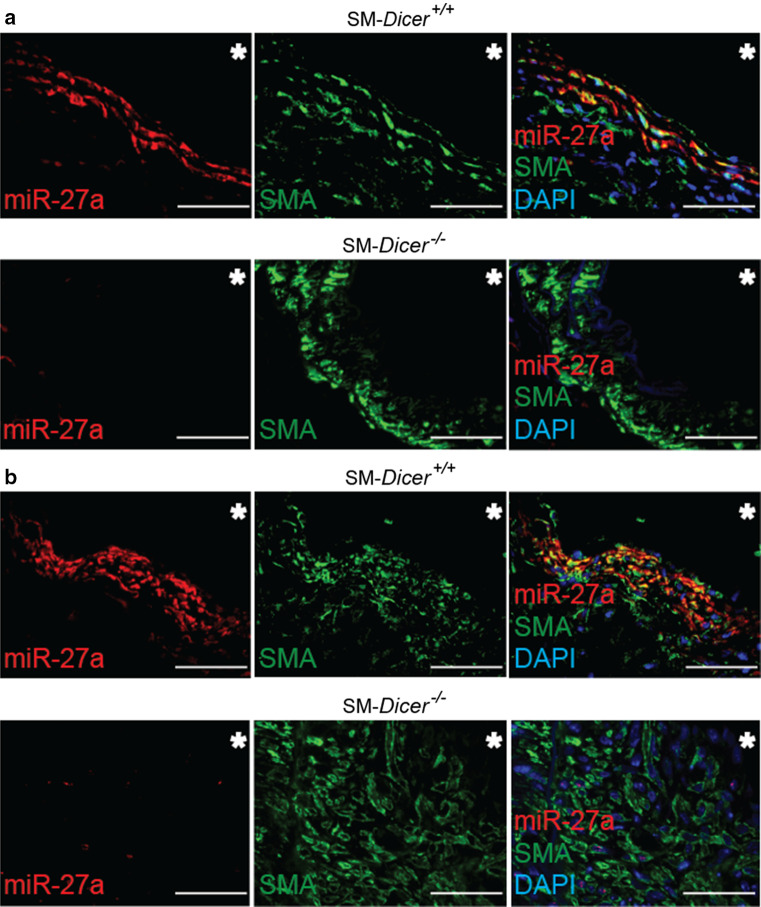
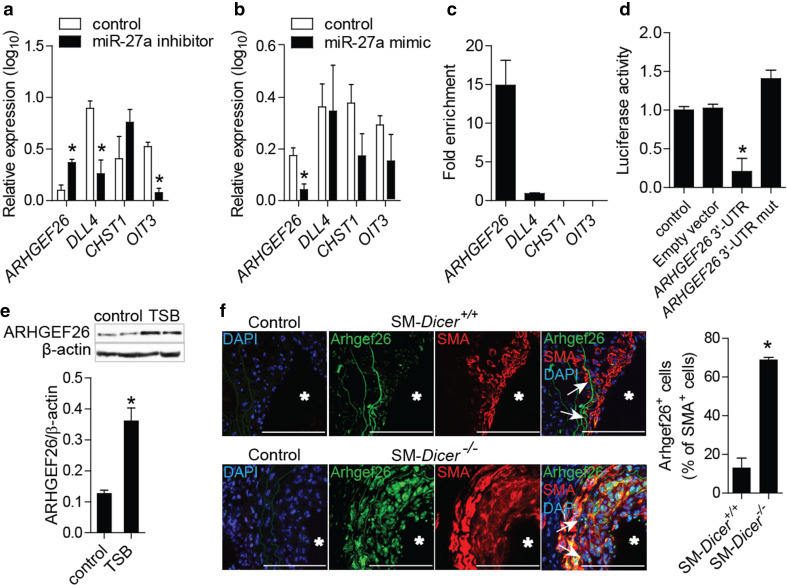
In contrast to SM-Dicer −/− mice, medial and neointimal SMCs from SM-Dicer +/+ mice expressed miR-27a-3p, as determined by in situ PCR of miR-27a-3p combined with SMA immunostaining (Fig. 5). In cultured HASMCs, gain- and loss-of-function studies revealed that miR-27a-3p suppressed ARHGEF26, but not of CHST1 (Fig. 6a, b; Online Resource Supplemental Fig. 5A, B). The treatment of HASMCs with miR-27a-3p inhibitors decreased the expression of DLL4 and OIT3, whereas the overexpression of miR-27a-3p did not alter DLL4 and OIT3 expression levels (Fig. 6a, b). To identify targets of miR-27a-3p, we treated HASMCs overexpressing MYC-tagged TNRC6A with miR-27a-3p mimics. In contrast to DLL4, CHST1, and OIT3, ARHGEF26 was enriched 15-fold in the miRISC of HASMCs after treatment with miR-27a-3p mimics, as determined by TNRC6A immunoprecipitation (Fig. 6c). To study the binding site of miR-27a-3p in the ARHGEF26 mRNA, luciferase reporter assays were performed in HEK293 cells co-transfected with miR-27a-3p mimics and a vector containing the 3′-UTR of ARHGEF26 mRNA (Fig. 6d). The mutation of the predicted miR-27a-3p binding site abrogated the inhibition of luciferase activity by miR-27a-3p mimics (Fig. 6d; Online Resource Supplemental Fig. 6A), demonstrating that miR-27a-3p represses ARHGEF26 by binding to this site. The treatment of HASMCs with LNA-modified oligonucleotides (target site blockers, TSBs) that specifically block the interaction between the miR-27a-3p and the ARHGEF26 3′-UTR (Online Resource Supplemental Fig. 6B), upregulated the expression of ARHGEF26 at the mRNA (Online Resource Supplemental Fig. 6C) and protein levels (Fig. 6e). Moreover, the number of Arhgef26-expressing neointimal SMCs was higher in SM-Dicer −/− mice than in SM-Dicer +/+ mice (Fig. 6f). Notably, Arhgef26 expression was detected in arterial SMCs of uninjured carotid arteries from SM-Dicer −/− mice, but not in SM-Dicer +/+ mice (Online Resource Supplemental Fig. 7). In human atherosclerotic lesions, miR-27a-3p and ARHGEF26 were co-expressed in SMCs (Online Resource Supplemental Fig. 8), as determined by combined SMA immunostaining and in situ PCR of miR-27a-3p.
Fig. 5.
Expression of miR-27a-3p in vascular SMCs. a In situ PCR of miR-27a-3p combined with SMA immunostaining in uninjured carotid arteries from SM-Dicer −/− mice and SM-Dicer +/+ mice. b In situ PCR of miR-27a-3p combined with SMA immunostaining in carotid arteries 14 days after injury from SM-Dicer −/− mice and SM-Dicer +/+ mice. Nuclei were counterstained with DAPI. The asterisks indicate the lumen. Scale bars 50 μm
Fig. 6.
Effect of miR-27a-3p on the expression of ARHGEF26 in human and mouse aortic SMCs. a, b Gene expression quantitated in HASMCs by qPCR after treatment with miR-27a-3p inhibitors (a) or mimics (b). c Effect of miR-27a-3p mimics on enrichment of predicted targets in the miRISC of HASMCs overexpressing a MYC-tagged GW182 protein, determined by GW182 immunoprecipitation and qPCR (n = 2). Results are expressed as enrichment in miR-27a-3p mimic-treated vs control mimic-treated HASMCs. d Luciferase reporter assays was performed using HEK293 cells co-transfected with miR-27a-3p mimics and the pEZX-MT05 vector containing the wild-type ARHGEF26 3′-UTR or a ARHGEF26 3′-UTR mutated in the predicted miR-27a-3p binding site. e ARHGEF26 protein levels determined by the western blot analysis in HASMCs treated with target site blockers (TSBs) that block the interaction between miR-27a-3p and ARHGEF26, or with a control oligonucleotide. f Arhgef26 expression in carotid arteries of SM-Dicer +/+ mice and SM-Dicer −/− mice 14 days after injury, determined by double immunostaining of Arhgef26 and SMA (n = 5 mice per group). Control represents the non-specific antibody for the primary antibody. Nuclei were counterstained with DAPI. The arrows indicate the neointimal area. The asterisks indicate the lumen. Scale bars 100 μm. Error bars represent ±SEM (n = 4–5). *P < 0.05
MiR-27a-3p inhibits SMC proliferation by targeting ARHGEF26
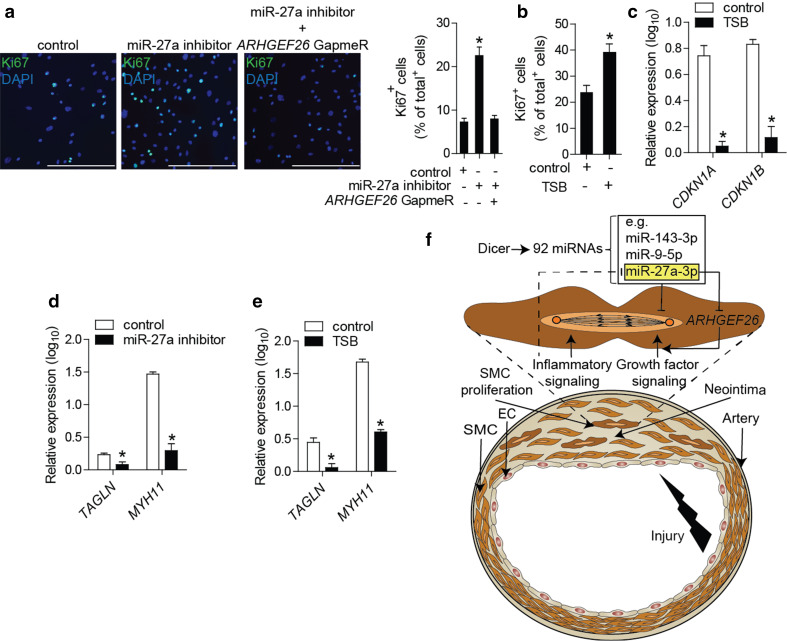
To study the effect of miR-27a-3p on SMC proliferation, we treated HASMCs with miR-27a-3p mimics or inhibitors. The inhibition or overexpression of miR-27a-3p increased or reduced HASMC proliferation, respectively, as determined by Ki67 immunostaining (Fig. 7a; Online Resource Supplemental Fig. 9). Silencing of ARHGEF26 prevented the induction of HASMC proliferation induced by miR-27a-3p inhibition (Fig. 7a; Online Resource Supplemental Fig. 10). Treatment with TSBs increased HASMC proliferation (Fig. 7b) and suppressed the expression of cell-cycle inhibitors, such as CDKN1A and CDKN1B (Fig. 7c). Moreover, the inhibition of miR-27a-3p or treatment with TSBs decreased the expression of the contractile genes transgelin (TAGLN) and MYH11 (Fig. 7d, e). Inflammatory activation by IL-1β increased the expression of ARHGEF26 and reduced the expression of miR-27a-3p in HASMCs (Online Resource Supplemental Fig. 11).
Fig. 7.
MiR-27a-3p inhibits SMC proliferation by targeting ARHGEF26. a Proliferation of HASMCs after treatment with miR-27a-3p inhibitors and silencing of ARHGEF26, determined by Ki67 immunostaining. Nuclei were counterstained with DAPI. Scale bars 250 μm. b Proliferation of HASMCs after treatment with TSBs or control oligonucleotides, determined by Ki67 immunostaining. c Expression levels of cell-cycle inhibitors in HASMCs after treatment with TSBs or control oligonucleotides, determined by qPCR. d, e Expression levels of contractile genes in HASMCs after treatment with miR-27a-3p inhibitors (d) or TSBs (e). Non-targeting oligonucleotides were used in the control groups (d, e). f Schematic illustration of the findings of this study showing the role of Dicer in limiting vascular repair by producing anti-proliferative miRNAs, such as miR-27a-3p, which reduces inflammation-induced SMC proliferation by suppressing ARHGEF26. Error bars represent ±SEM (n = 4–5). *P < 0.05
Discussion
We found that the SMC-specific loss of Dicer enhanced neointima formation and neointimal SMC proliferation, and induced growth factor and inflammatory signaling in injured arteries. By integrating miRNA and mRNA expression profiling data, we predicted a miRNA–mRNA interaction network that may contribute to the effects of Dicer on SMCs during neointima formation. The most connected miRNA in this network was miR-27a-3p, which inhibited SMC proliferation by suppressing one of its predicted targets, the guanine nucleotide exchange factor ARHGEF26. Thus, miR-27a-3p may contribute to neointimal growth control by Dicer in SMCs (Fig. 7f).
Dicer impairs the differentiation of ECs [25, 26], but promotes a contractile SMC phenotype in uninjured arteries [13]. Our findings indicate that the biogenesis of miRNAs by Dicer in SMCs after arterial injury limits SMC proliferation by inhibiting growth factor and inflammatory signaling. By contrast, during development, Dicer increases SMC proliferation [12], indicating that Dicer plays different roles in SMCs during development and arterial repair. This difference may be due to variations in the mechanisms of SMC proliferation under these two conditions. Notably, the combinatorial activation of inflammatory and growth factor signaling that causes neointimal SMC proliferation is specific to arterial repair [5, 6, 27, 28]. In line with the previous findings in rats, we found that the expression of certain miRNAs, such as miR-21 and miR-146a/b, was elevated during neointima formation [15]; however, the deletion of Dicer in SMCs largely reduced expression of miRNAs that were not upregulated after vascular injury. This result suggests that during neointima formation, many miRNAs are upregulated in other cell types, such as leukocytes, rather than in SMCs. In fact, Dicer appears to be essential for maintaining the expression levels of miRNAs and limiting the downregulation of miRNAs in SMCs following vascular injury.
Among the miRNAs downregulated following Dicer deletion, only miR-26a-5p was previously reported to increase SMC proliferation [29]. However, 31 miRNAs, including miR-143-3p, miR-9-5p, miR-27a-3p, and miR-27b-3p, inhibit SMC proliferation in vitro [30, 31] (Online Resource Supplemental Table 2). Notably, 11 of these 31 anti-proliferative miRNAs, including miR-132-3p, which reduces neointima formation [32], were involved in the 70 most significant interactions predicted between downregulated miRNAs and upregulated mRNAs in SM-Dicer −/− mice. This result indicates that a network of miRNA–mRNA interactions negatively regulates SMC proliferation after vascular injury. MiR-27a-3p had the largest number of predicted targets, suggesting that this miRNA plays an important role in neointima formation. MiR-27a is expressed in a serum response factor-regulated polycistronic transcript that also contains miR-23a and miR-24-2 [33, 34]. Although miR-27a-3p inhibits SMC proliferation in vitro [30], the underlying mechanism remains unclear.
Among its targets predicted during neointima formation, miR-27a-3p suppressed ARHGEF26 in SMCs. ARHGEF26 encodes a guanine nucleotide exchange factor that activates the components of growth-factor-signaling pathways, such as the Rho GTPase RhoG, Akt, and ERK1/2, and also promotes cancer cell proliferation [35–37]. Accordingly, miR-27a-3p inhibited the proliferation and promoted differentiation of SMCs by targeting ARHGEF26. Moreover, our data suggest that the downregulation of miR-27a-3p in SMCs by inflammatory stimuli plays a crucial role in inflammation-induced SMC proliferation during neointima formation by mediating the NF-κB-induced upregulation of ARHGEF26 [38].
In summary, we showed that Dicer activity controls neointimal hyperplasia by reducing SMC proliferation after vascular injury. In addition to other anti-proliferative miRNAs, miR-27a-3p-mediated targeting of Arhgef26 may contribute to the effect of Dicer in SMCs on neointima formation by reducing inflammation-induced growth factor signaling. Thus, an increasing Dicer activity in SMCs represents a potential approach to prevent restenosis due to neointimal hyperplasia.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work has been funded by the German Research Foundation (DFG) as part of the Collaborative Research Center 1123 (B04) and by the German Center for Cardiovascular Research (MHA VD1.2). The authors declare no competing financial interests.
Abbreviations
- Apoe
Apolipoprotein E
- ARHGEF26
Rho guanine nucleotide exchange factor 26
- CHST1
Carbohydrate (keratan sulfate Gal-6) sulfotransferase 1
- DAPI
4’,6-Diamidino-2-phenylindole
- DLL4
Delta-like 4
- DMEM
Dulbecco′s modified eagle medium
- EC
Endothelial cell
- EGF
Epidermal growth factor
- HASMC
Human aortic smooth muscle cell
- HEK293
Human embryonic kidney 293 cell
- HFD
High-fat diet
- IGFBP3
Insulin-like growth-factor-binding protein 3
- IL-1β
Interleukin-1β
- KO
Knockout
- LNA
Locked nucleic acid
- microRNA
miRNA, miR
- MYH11
Myosin, heavy chain 11, smooth muscle
- NF-κB
Nuclear factor of kappa light polypeptide gene enhancer in B-cells
- OIT3
Oncoprotein-induced transcript 3
- PDGF
Platelet-derived growth factor
- miRISC
miRNA-induced-silencing complex
- SH3BGRL2
SH3-domain-binding glutamate-rich protein-like 2
- SMA
Smooth muscle actin
- SMC
Smooth muscle cell
- TAGLN
Transgelin
- TNFα
Tumor necrosis factor α
- TNRC6A
Argonaute and trinucleotide repeat containing 6A
- TSB
Target site blocker
- UTR
Untranslated region
- WT
Wild type
Footnotes
F. Zahedi and M. Nazari-Jahantigh are co-first authors.
References
- 1.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 2.Alfonso F, Byrne RA, Rivero F, Kastrati A. Current treatment of in-stent restenosis. J Am Coll Cardiol. 2014;63:2659–2673. doi: 10.1016/j.jacc.2014.02.545. [DOI] [PubMed] [Google Scholar]
- 3.Liu B, Fisher M, Groves P. Down-regulation of the ERK1 and ERK2 mitogen-activated protein kinases using antisense oligonucleotides inhibits intimal hyperplasia in a porcine model of coronary balloon angioplasty. Cardiovasc Res. 2002;54:640–648. doi: 10.1016/S0008-6363(02)00335-8. [DOI] [PubMed] [Google Scholar]
- 4.Stabile E, Zhou YF, Saji M, Castagna M, Shou M, Kinnaird TD, Baffour R, Ringel MD, Epstein SE, Fuchs S. Akt controls vascular smooth muscle cell proliferation in vitro and in vivo by delaying G1/S exit. Circ Res. 2003;93:1059–1065. doi: 10.1161/01.RES.0000105086.31909.1B. [DOI] [PubMed] [Google Scholar]
- 5.Peppel K, Zhang L, Orman ES, Hagen PO, Amalfitano A, Brian L, Freedman NJ. Activation of vascular smooth muscle cells by TNF and PDGF: overlapping and complementary signal transduction mechanisms. Cardiovasc Res. 2005;65:674–682. doi: 10.1016/j.cardiores.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida T, Yamashita M, Horimai C, Hayashi M. Smooth muscle-selective inhibition of nuclear factor-kappaB attenuates smooth muscle phenotypic switching and neointima formation following vascular injury. J Am Heart Assoc. 2013;2:e000230. doi: 10.1161/JAHA.113.000230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siciliano V, Garzilli I, Fracassi C, Criscuolo S, Ventre S, di Bernardo D. MiRNAs confer phenotypic robustness to gene networks by suppressing biological noise. Nat Commun. 2013;4:2364. doi: 10.1038/ncomms3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebert MS, Sharp PA. Roles for MicroRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 10.Clark PM, Loher P, Quann K, Brody J, Londin ER, Rigoutsos I. Argonaute CLIP-Seq reveals miRNA targetome diversity across tissue types. Sci Rep. 2014;4:5947. doi: 10.1038/srep05947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albinsson S, Suarez Y, Skoura A, Offermanns S, Miano JM, Sessa WC. MicroRNAs are necessary for vascular smooth muscle growth, differentiation, and function. Arterioscler Thromb Vasc Biol. 2010;30:1118–1126. doi: 10.1161/ATVBAHA.109.200873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albinsson S, Skoura A, Yu J, DiLorenzo A, Fernandez-Hernando C, Offermanns S, Miano JM, Sessa WC. Smooth muscle miRNAs are critical for post-natal regulation of blood pressure and vascular function. PLoS One. 2011;6:e18869. doi: 10.1371/journal.pone.0018869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu Q, Deitch EA, Huo Y, Delphin ES, Zhang C. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res. 2009;105:158–166. doi: 10.1161/CIRCRESAHA.109.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 16.Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, Richardson JA, Bassel-Duby R, Olson EN. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonald RA, Hata A, MacLean MR, Morrell NW, Baker AH. MicroRNA and vascular remodelling in acute vascular injury and pulmonary vascular remodelling. Cardiovasc Res. 2012;93:594–604. doi: 10.1093/cvr/cvr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wirth A, Benyo Z, Lukasova M, Leutgeb B, Wettschureck N, Gorbey S, Orsy P, Horvath B, Maser-Gluth C, Greiner E, Lemmer B, Schutz G, Gutkind JS, Offermanns S. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat Med. 2008;14:64–68. doi: 10.1038/nm1666. [DOI] [PubMed] [Google Scholar]
- 20.Schober A, Knarren S, Lietz M, Lin EA, Weber C. Crucial role of stromal cell-derived factor-1alpha in neointima formation after vascular injury in apolipoprotein E-deficient mice. Circulation. 2003;108:2491–2497. doi: 10.1161/01.CIR.0000099508.76665.9A. [DOI] [PubMed] [Google Scholar]
- 21.Bisognin A, Sales G, Coppe A, Bortoluzzi S, Romualdi C. MAGIA2: from miRNA and genes expression data integrative analysis to microRNA-transcription factor mixed regulatory circuits (2012 update) Nucleic Acids Res. 2012 doi: 10.1093/nar/gks460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nuovo GJ, Elton TS, Nana-Sinkam P, Volinia S, Croce CM, Schmittgen TD. A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nat Protoc. 2009;4:107–115. doi: 10.1038/nprot.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cambronne XA, Shen R, Auer PL, Goodman RH. Capturing microRNA targets using an RNA-induced silencing complex (RISC)-trap approach. Proc Natl Acad Sci USA. 2012;109:20473–20478. doi: 10.1073/pnas.1218887109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson M, Mcclelland M. Use of DNA methyltransferase endonuclease enzyme combinations for megabase mapping of chromosomes. Method Enzymol. 1992;216:279–303. doi: 10.1016/0076-6879(92)16027-H. [DOI] [PubMed] [Google Scholar]
- 25.Hartmann P, Zhou Z, Natarelli L, Wei Y, Nazari-Jahantigh M, Zhu M, Grommes J, Steffens S, Weber C, Schober A. Endothelial Dicer promotes atherosclerosis and vascular inflammation by miRNA-103-mediated suppression of KLF4. Nat Commun. 2016;7:10521. doi: 10.1038/ncomms10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schober A, Nazari-Jahantigh M, Weber C. MicroRNA-mediated mechanisms of the cellular stress response in atherosclerosis. Nat Rev Cardiol. 2015;12:361–374. doi: 10.1038/nrcardio.2015.38. [DOI] [PubMed] [Google Scholar]
- 27.Cook CL, Weiser MC, Schwartz PE, Jones CL, Majack RA. Developmentally timed expression of an embryonic growth phenotype in vascular smooth muscle cells. Circ Res. 1994;74:189–196. doi: 10.1161/01.RES.74.2.189. [DOI] [PubMed] [Google Scholar]
- 28.Chen CN, Li YS, Yeh YT, Lee PL, Usami S, Chien S, Chiu JJ. Synergistic roles of platelet-derived growth factor-BB and interleukin-1beta in phenotypic modulation of human aortic smooth muscle cells. Proc Natl Acad Sci USA. 2006;103:2665–2670. doi: 10.1073/pnas.0510973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leeper NJ, Raiesdana A, Kojima Y, Chun HJ, Azuma J, Maegdefessel L, Kundu RK, Quertermous T, Tsao PS, Spin JM. MicroRNA-26a is a novel regulator of vascular smooth muscle cell function. J Cell Physiol. 2011;226:1035–1043. doi: 10.1002/jcp.22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fiedler J, Stohr A, Gupta SK, Hartmann D, Holzmann A, Just A, Hansen A, Hilfiker-Kleiner D, Eschenhagen T, Thum T. Functional microRNA library screening identifies the hypoxamir miR-24 as a potent regulator of smooth muscle cell proliferation and vascularization. Antioxid Redox Signal. 2014;21:1167–1176. doi: 10.1089/ars.2013.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choe N, Kwon JS, Kim JR, Eom GH, Kim Y, Nam KI, Ahn Y, Kee HJ, Kook H. The microRNA miR-132 targets Lrrfip1 to block vascular smooth muscle cell proliferation and neointimal hyperplasia. Atherosclerosis. 2013;229:348–355. doi: 10.1016/j.atherosclerosis.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernandez-Torres F, Aranega AE, Franco D. Identification of regulatory elements directing miR-23a-miR-27a-miR-24-2 transcriptional regulation in response to muscle hypertrophic stimuli. Biochim Biophys Acta. 2014;1839:885–897. doi: 10.1016/j.bbagrm.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 35.Ellerbroek SM, Wennerberg K, Arthur WT, Dunty JM, Bowman DR, DeMali KA, Der C, Burridge K. SGEF, a RhoG guanine nucleotide exchange factor that stimulates macropinocytosis. Mol Biol Cell. 2004;15:3309–3319. doi: 10.1091/mbc.E04-02-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Wu R, Yu L, Wu F, Li S, Zhao Y, Li H, Luo G, Wang J, Zhou J. SGEF is overexpressed in prostate cancer and contributes to prostate cancer progression. Oncol Rep. 2012;28:1468–1474. doi: 10.3892/or.2012.1917. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Li S, Li H, Wang P, Huang F, Zhao Y, Yu L, Luo G, Zhang X, Wang J, Zhou J. Grb2 interacts with SGEF and antagonizes the ability of SGEF to enhance EGF-induced ERK1/2 activation. Mol Cell Biochem. 2014;389:239–247. doi: 10.1007/s11010-013-1945-7. [DOI] [PubMed] [Google Scholar]
- 38.Tang RH, Zheng XL, Callis TE, Stansfield WE, He J, Baldwin AS, Wang DZ, Selzman CH. Myocardin inhibits cellular proliferation by inhibiting NF-kappaB(p65)-dependent cell cycle progression. Proc Natl Acad Sci USA. 2008;105:3362–3367. doi: 10.1073/pnas.0705842105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.