Abstract
The idea of internal initiation is frequently exploited to explain the peculiar translation properties or unusual features of some eukaryotic mRNAs. In this review, we summarize the methods and arguments most commonly used to address cases of translation governed by internal ribosome entry sites (IRESs). Frequent mistakes are revealed. We explain why “cap-independent” does not readily mean “IRES-dependent” and why the presence of a long and highly structured 5′ untranslated region (5′UTR) or translation under stress conditions cannot be regarded as an argument for appealing to internal initiation. We carefully describe the known pitfalls and limitations of the bicistronic assay and artefacts of some commercially available in vitro translation systems. We explain why plasmid DNA transfection should not be used in IRES studies and which control experiments are unavoidable if someone decides to use it anyway. Finally, we propose a workflow for the validation of IRES activity, including fast and simple experiments based on a single genetic construct with a sequence of interest.
Keywords: CITE, Cellular IRES, RNA transfection, Cap-independent translation, RRL, Bicistronic vector
Introduction
Although IRESs were first discovered more than a quarter of a century ago, a common standardized methodology to detect and validate novel IRES elements has yet to be adopted by the research community. Consequently, the required control experiments are rarely performed, and the number of questionable reports continues to increase. This has detrimental effects on research in the field of eukaryotic protein synthesis as well as on bordering areas of the life sciences, e.g., crucial aspects of translation in cancer are currently (mis)linked to cellular IRESs.
Much has been already said about the inherent limitations of the workhorse of IRES research: the bicistronic assay [1–5]. Therefore, we mainly focus on what has remained behind the veil, the critical controls that are required when using this technique appropriately, and address several rarely discussed ideas about how internal initiation should (not) be studied. Specifically, not each and every cap-independent translation equals the internal initiation, especially in in vitro assays. Thus, careful interpretation (in peer-reviewing process also) is required to avoid false-positive IRES identification. Finally, we provide a simple workflow utilizing a standardized set of methods that can robustly prove or disprove internal initiation.
Modes of translation initiation in eukaryotes: more than just two
The translation efficiency and stability of a particular mRNA species are regulated by elements that are usually located in untranslated regions (UTRs). Some of these regulatory elements can ensure the efficient translation of the given mRNA under specific conditions when the bulk mRNA translation is repressed. One of the proposed regulatory mechanisms effectuating such differential translation is internal ribosome entry governed by internal ribosome entry sites (IRESs) of mRNA.
First, we should define what IRES is (and what it is not) since a misunderstanding of the basic definitions leads to confusion. IRES is a segment of mRNA that permits the recruitment of preinitiation complex to the initiation codon without the involvement of the 5′ end of the mRNA. This requirement is extremely important because it segregates internal initiation from a larger clade of all possible cap-independent initiation pathways (Table 1). Consequently, only IRESs are able to function if mRNA is circularized.
Table 1.
Manifestations of the three mechanisms of translation initiation
| Cap-dependent | IRES | CITE | |
|---|---|---|---|
| eIF4G cleavage | Sensitive | Resistanta | Resistanta |
| mTORC1 inhibition | Sensitive | Resistant | Resistanta |
| 4EBP or m7GTP | Sensitive | Resistanta | Resistant |
| Bicistronic mRNA activity | No | Yes | No |
| Cap-dependence | Strong or intermediate | Weakb | Weak |
| A-capped mRNA activity | Weak or intermediate | Weak or intermediate or strong | Strong or intermediate |
A comparison of the translation initiation mechanisms’ manifestations in commonly used assays. Note that IRES and CITE mechanisms only differ in their activity in bicistronic mRNAs
aHAV IRES requires intact eIF4G, and many plant virus CITEs bind eIF4E; thus, their translation is sensitive to a disruption of the m7G-eIF4E-eIF4G link
bThe translation of relatively weak HCV IRES in a monocistronic context is stimulated by a 5′ cap, presumably due to activating the cap-dependent translation by which a ribosome scans through the IRES
Unlike many viral mRNAs, all cytoplasmic mRNAs are capped and are thus destined to attract ribosomes to their 5′ ends. “Presumption of innocence” should be applied to any naturally capped mRNA that it is scanned by ribosomes unless compelling proof of the opposite is provided.
In this section, we briefly outline the most important features of the translation initiation pathways in mammals. For a detailed description, we refer readers to excellent reviews of the cap-dependent mechanism [6–8] or internal initiation [4, 9–11], as we only stress some major points and facets of the mechanisms that are relevant to our topic.
5′ end-dependent translation initiation: cap- and CITE-assisted translation
The most studied and perhaps most widely used variant of 5′ end-dependent translation is cap-dependent translation. In this pathway the 7-methylguanosine moiety at the mRNA’s 5′ end is recognized by eIF4F, which consists of eIF4E, the cap-binding subunit; eIF4A, an RNA helicase; and eIF4G, a scaffold for the former two subunits and other proteins, such as PABP, eIF3 and Mnk1/2 kinases. Once bound to the cap, eIF4F recruits the 43S complex, which consists of 40S ribosome, eIF1, eIF1A, eIF2*GTP*Met-tRNAMeti ternary complex, eIF3, and eIF5 (eIF5 is not necessary for 43S recruitment and may be absent from certain scanning complexes). Then, the ribosome starts unidirectional 5′–3′ movement in search of an initiator codon. This process is thus termed “scanning” [6, 8]. After the scanning ribosome locates the initiator codon in a suitable nucleotide context, GTP hydrolysis is accomplished, the initiation factors leave the 40S subunit, and the 60S subunit joins in the eIF5B-dependent reaction. The availability of eIF4E is mainly controlled by mTOR kinase, which is believed to be a major signalling hub for cell survival and proliferation [12–14]. However, other kinases that phosphorylate 4E-BPs (at least in vitro) are also known [15–18]. When hypophosphorylated, eIF4E-binding proteins (4E-BPs) displace eIF4G from eIF4E and expectedly inhibit cap-dependent translation. In fact, eIF4E interacts with other proteins which may also disrupt eIF4E–eIF4G link [19, 20].
Concurrently with the discovery of m7G cap, it was shown that although capping strongly stimulates ribosome binding, it is not always a prerequisite for the latter [21, 22]. This notion has been complemented by dozens of reports, the most remarkable of which employed uncapped transcripts made by RNA polymerase III to demonstrate the 5′ end dependence of scanning-based translation initiation in living cells [23, 24]. Accordingly, the translation of A-capped mRNAs (i.e., capped with ApppN, which does not bind eIF4E) is inhibited by the insertion of the 5′ end proximal stem-loops, by annealing antisense oligos to the 5′ end or by the insertion of uAUGs [25–29]. This means that even uncapped mRNAs drive translation in a 5′ end-dependent manner. In addition, certain viral mRNAs that lack IRESs are effectively translated under conditions of severely impaired cap-dependent translation [30, 31]. Another well-known example of cap-independent translation is the translation of certain mRNAs from plant viruses mediated by cap-independent translation enhancers, mostly found in 3′UTRs and thus called 3′CITEs (for recent reviews, see [32–34]). The question remains as to whether there are CITEs in mRNAs of organisms other than plants or their viruses; some candidates with such properties have been reported [35, 36]. In an attempt to demonstrate the feasibility of CITEs in mammals, we showed that a high-affinity eIF4G-binding site placed in either the 5′UTR or 3′UTR of an uncapped mRNA significantly stimulated its 5′ end-dependent translation to the extent that rendered it almost cap-independent [27, 37]. Importantly, such artificial CITE does not function as an IRES. The binding of eIF3 to certain mRNAs also affects their 5′ end-dependent translation [38, 39].
If m7G cap were the only attractor of the ribosome, the cap-dependence (i.e., the degree of stimulation of translation by the m7G cap) of all mRNAs would be similar. We know, however, that cap-dependence varies significantly among different mRNA species [27, 40], and the current view suggests that the enhanced translation of certain mRNAs in the uncapped state depends on the mRNA’s affinity to some mRNA-interacting initiation factors, presumably eIF4G [4, 27, 35, 41]. However, the premise that a higher level of translation of a non-capped mRNA is always relevant to a weaker stimulation of its translation by a 5′ cap does not seem to be correct (also see below).
Thus, the widely accepted dichotomy “IRES-dependent translation vs. cap-dependent translation” is a dangerous oversimplification. The logically correct dichotomy should be “5′ end-dependent translation vs. 5′ end-independent translation”.
5′ end-independent (internal) initiation
The lifecycle of certain viruses takes place exclusively in the cytoplasm. These viruses, therefore, cannot rely on capping by nuclear transcription machinery; some of them encode their own capping enzymes, while some others snatch m7G cap from cellular mRNAs. Some plant viruses have adopted 3′CITEs (see above), relieving the need for the cap altogether. However, picornaviruses, certain flaviviruses and representatives of the Dicistroviridae family utilize a completely different approach. Their mRNAs possess highly specific binding sites for eIF4G (in the case of certain picornaviruses), eIF3 and 40S (in the case of flaviviruses and some picornaviruses), or the 40S and 60S ribosomal subunits (in the case of Dicistroviridae IRESs). These binding sites place the ribosome near to or immediately to the initiator codon; such a “ribosome entry window” (or “ribosome landing pad”) near the authentic initiator codon is usually narrow [42–44]. In certain cases (FMDV, HRV, PV), the initial binding of the preinitiation 43S complex is followed by a limited scanning to locate the initiator codon.
These features enable IRESs to promote translation in a 5′ end- and, thus, m7G-independent fashion and, expectedly, to operate efficiently under conditions when the cap-dependent translation is compromised. A feature that is shared between IRESs and CITEs is that they bind components of the translational apparatus and promote translation in a cap-independent manner. The discriminatory attribute of IRESs is their independence on the 5′ terminus in stark contrast to the strict dependence of the CITE-mediated translation on the free 5′ end.
There is a striking difference between the results of the mutagenesis of viral or putative cellular IRESs. Bona fide IRESs of viral origin may be easily inactivated by point mutations, not to mention larger deletions [45–49]. In contrast, deletion analysis of many cellular IRESs apparently showed that their non-overlapping segments could separately provide internal initiation, though usually less efficiently than the complete IRESs. This sort of findings led to an idea that cellular IRESs may have a modular structure, in which separate modules bind initiation factors and work more or less independently. Current view is that such “IRES” modular structure discovered by plasmid transfections most probably reflects the modular structure of the cryptic promoters [4] (also see below).
The mutagenesis of viral IRESs suggests that the binding of initiation factors itself is not sufficient to promote internal initiation and that the spatial organization of the binding sites and “landing pads” is equally important. Our knowledge of viral IRESs suggests that IRESs not only bind translational machinery but also position an mRNA initiation region into the mRNA-binding cleft of the ribosome. The latter is true hallmark of internal initiation. It is not clear how modules designed to land a ribosome on specific point within the mRNA do not interfere with each other and can work separately in an unnatural nucleotide context.
A notable contradiction to these considerations is the case of RhPV or HalV IRESs, which are indeed modular and cannot be inactivated by partial deletions [50–53]. This seems to be the other extreme of the IRES story. These long and A/U-rich single-stranded IRESs can apparently occupy the mRNA-binding cleft in an eIF3-dependent fashion [50, 53] without the involvement of specific high-order structural elements [54]. Consequently, they bind ribosomes from evolutionary distant organisms. Thus, modular IRESs do exist, but known cases have no or little secondary structures.
“Downstream” IRESs and leaderless mRNAs
Several reports have suggested the existence of IRESs located within coding regions. The most studied case is gag-pol mRNA of HIV-1 [55, 56] or HIV-2 [55, 57–59]. The presence of tentative IRESs in these mRNAs was initially deduced from the translation of the corresponding unnatural model mRNAs that lacked any 5′UTR. The rationale was that such a leaderless mRNA is not capable of efficient translation unless it contains an IRES. However, this is clearly not the case [60]. We have shown that a leaderless mRNA efficiently binds 43S complexes and also can bind pre-assembled 80S ribosomes without the involvement of any initiation factor [61]. Another route of translation initiation of leaderless mRNAs involving eIF2D instead of eIF2 has been described [62]. Since neither of these two eIF2-independent pathways requires eIF4F, the resistance of leaderless gag-pol mRNA translation to inhibition by eIF4A dominant negative mutant [56] or to eIF4G cleavage [57] is not a valid reason to propose the existence of an IRES-dependent translation. Experiments with “downstream IRES” in bicistronic mRNAs (see below) have not been performed.
However, not each and every leaderless mRNA is translated efficiently [59], indicating that there is indeed something special about the leaderless mRNAs that are translated. The clue may come from prokaryotes, where the lack of a secondary structure near the 5′ terminus of leaderless mRNAs makes 70S binding possible [63], although sequence-specific effects may not be neglected.
Leaky scanning, 43S sliding and reinitiation events
Many viral mRNAs code for more than one protein but do not use IRESs. For example, HIV-1 Env expression from polycistronic mRNA depends on leaky scanning through the Vpu initiator codon [64], and Hepatitis B P-protein is expressed from a bicistronic mRNA also via leaky scanning [65].
It seems that the eukaryotic ribosome’s ability to reach 5′ distally located start codons is underestimated [66]. Although leaky scanning usually requires a suboptimal nucleotide context for the 5′-proximal AUG or its location within a short distance from the 5′ terminus, another recently discovered phenomenon, 43S sliding, is not limited to such AUGs [67]. Thus, 43S sliding may be used to explain alternative start codon usage in such complex cases.
More frequently, the synthesis of two proteins from a single mRNA is ensured by translation reinitiation, when a ribosome is not released from the mRNA after completing the first ORF translation but rather resumes scanning and thus initiates the translation on the next AUG codon. In cases of subgenomic mRNAs of mammalian caliciviruses or Influenza B M2 mRNA, a highly efficient reinitiation relies on TURBS motifs found upstream of the reinitiation site (reviewed in [68]) and occurs between overlapping (A UG A) or closely spaced stop and start codons. In contrast, a regular reinitiation, represented by the case of bicistronic LINE-1 retrotransposon mRNA, is rather inefficient although apparently sufficient to provide physiologically relevant levels of second cistron expression [69, 70]. Addressing the reinitiation possibility is especially important in the case of natural bicistronic mRNAs.
The dark side of IRES research
Several features of an mRNA are frequently considered to draw a foregone conclusion that internal initiation takes place. These are long 5′UTRs, the presence of multiple uORFs, translation under stressful conditions and the direct interaction of a ribosome or other component(s) of the translational machinery with certain mRNAs. In this chapter, we successively examine all of these arguments and explain why they may not be considered as a proof of IRES presence in a particular 5′UTR.
Why may one decide that a message under study has an IRES?
Minimal free energy of RNA folding, 5′UTR length and multiple uAUGs
Quite often, a long and highly structured 5′UTR is thought to be an insurmountable obstacle for a scanning ribosome. As a matter of fact, in living cells, ribosomes rather easily penetrate long 5′UTRs (see, e.g., [70–72] and references therein), and the HCV-like IRESs inactivated by point mutations in 40S ribosome binding is scanned through, regardless of the presence of extended stem-loops and a pseudoknot [27, 73]. The processivity of a scanning ribosome is, however, significantly reduced in cell-free translation systems prepared from the same cultured cells [74] and dramatically reduced in the rabbit reticulocyte lysate [71].
Secondary structures inhibit translation drastically when positioned close to the 5′ terminus [75–77], thus interfering with the initial attachment of 40S rather than with scanning itself. Moreover, the common practice of calculating the cumulative 5′UTR energy implies the necessity to melt all of these structures at once during scanning and ignores the possibility that scanning ribosome melts them separately, i.e., stem by stem [5].
Multiple uAUGs do not indicate an internal initiation either (see above). The frequency of uAUGs in 5′UTRs of cytoplasmic mRNA with or without reported IRESs is roughly the same [78]. Ribosomal profiling data clearly show that many uORFs in cytoplasmic mRNAs are translated [79], indicating scanning rather than internal initiation. The 5′UTR of human SLC35A4 mRNA is a notable example: it is ~750 nt long and has 11 uAUGs although it drives translation in a cap-dependent fashion (S.E. Dmitriev, unpublished).
Translation under unfavourable conditions
eIF4F is crucial for efficient ribosome accommodation to 5′ termini of the conventional cap-dependent mRNAs. Numerous viruses use this feature to inhibit host translation by eIF4G proteolysis and/or 4E-BPs activation. Obviously, if a cellular mRNA withstands the inhibition of cap-dependent translation, it has to use some special route of ribosome binding. However, several points here are usually overlooked. The most malicious mistake is seeing translation regulation as a simple combination of on/off switches.
First, knocking down eIF4E does not drastically reduce translation. One of the possible reasons for this is a concomitant reduction of 4E-BP level [80]. In addition, 4E-BPs never turn translation off completely [81, 82], although different mRNAs do react unequally to the activation of 4E-BPs in cells or the supplementation of in vitro translation reactions with purified 4E-BPs [41, 83, 84]. In fact, recent genome-wide studies show that the translation of the vast majority of cytoplasmic mRNAs is not so sensitive to mTOR inhibition and subsequent 4E-BPs activation as might be expected [83, 84]. It seems that eIF4E is present in excess and is only critical for a limited subset of mRNAs [41]. This does not mean, however, that the remaining mRNAs use IRESs to operate under these conditions.
This relaxed dependence on eIF4E is usually missed, and the resistance of translation to the action of eIF4F inhibitors is considered an omen of internal initiation. For example, translation of monocistronic mRNA with the 5′UTR from Drosophila insulin-like receptor is not inhibited in vitro by the addition of m7GTP or 4E-BP, nor it is stimulated by mRNA capping [85]; amyloid precursor protein (APP) synthesis is resistant to mTOR inhibition or eIF4E siRNA-mediated knockdown [86]; and c-jun expression is sustained in glioblastoma after LY294002 (PI3K inhibitor) treatment [87]. However, the authors of these papers have not considered alternatives to internal initiation. Similarly, a low degree of translation stimulation of Aurora A kinase mRNA by m7G cap (low cap-dependence in our terms) in transfected cells [88] was read into an IRES-dependent translation. Not that rare, the translation of an A-capped monocistronic mRNA is somewhat crudely referred to as IRES-dependent [89]. Notably, certain mRNA species are translated extremely well in the uncapped state and nevertheless have a high cap-dependence [90]. Therefore, the efficient translation of A-capped mRNAs should not misguide researchers (or reviewers).
Next, the estimation of eIF4G-dependence of translation is often unconvincing. For example, the effect of poliovirus 2A protease-induced eIF4G cleavage seems to be specific for the cell line and/or method of 2Apro gene delivery. The transfection-based approach is usually rather slow in terms of eIF4G cleavage [91, 92] and only has a modest short-term effect on cellular translation [92]. At its best, transient transfection-based delivery of CBV 2Apro cDNA to HeLa cells leads to rapid death, but surprisingly in this case, eIF4G cleavage is far from complete [93], indicating that events other than eIF4G cleavage may contribute to translation inhibition and cell death after 2Apro expression. On the other hand, inducible 2Apro expression leads to fast eIF4G cleavage and cell death [91]. Moreover, siRNA-driven eIF4GI knockdown in mammalian cells for yet unknown reasons has little impact on bulk protein synthesis [94, 95].
During apoptosis, 4E-BPs are activated and eIF4G is cleaved, but at the point of the complete eIF4G1 cleavage, cytoplasmic translation is only inhibited by ~2/3 [96]. Contrary to viral proteases, caspase-3 does not detach the eIF4E-binding site from the eIF4G core and the implications for cap-dependent translation are only remotely known. In fact, the translation of reporters in apoptotic 293T cells still exhibits reasonable cap-dependence despite the cleavage of eIF4G and dephosphorylation of 4E-BPs [97].
Cell cycle progression is also linked to translation regulation via IRESs. Frequently, the translational status of mitotic cells is considered “negative”. A recent study, however, suggests that the observed 60–75% translation inhibition (which can hardly be called drastic) during mitosis [98–101] is not a natural result of cell cycle progression, but rather a consequence of treatment with nocodazole or colcemid used for cell culture synchronization, both of which disassemble microtubules and cause stress [102]. Other data are less coherent. Some researchers found no inhibition of translation in HeLa cells during G2/M-transition after thymidine block [102], while others reported ~2-fold inhibition [103], and there is no agreement on the phosphorylation status of 4E-BPs during M-phase [102–104]. Besides, the inhibition of translation during mitosis was also observed at the level of elongation [105, 106]. However, translation most probably remains cap-dependent even after nocodazole treatment [25], and there is no published evidence for a global switch from cap-dependent translation to cap-independent translation.
In general, when characterizing particular conditions, it is important to explicitly demonstrate the cellular translational status with respect to cap-dependence and the efficiency of translation of certain reference mRNAs.
Binding of initiation factors or the 40S ribosome to an mRNA
There is a commonly used abduction that the direct binding of translation initiation machinery component(s) to a certain mRNA is a clear indication of internal initiation. Indeed, each and every validated IRES functions by the binding of eIF3, eIF4G or the 40S ribosomal subunit itself [10, 107, 108].
40S or eIF3 can bind RNA in a sequence-specific manner [109–111]. RNAs lacking developed secondary structure also can bind 40S, at least in vitro [112]. 40S and eIF3 are capable of forming stable ternary complexes with U-rich RNAs [111]. The formation of such ternary complexes was reported in vitro on the HCV mRNA 3′UTR, which has a U-rich tract [113], HIV-2 gag-pol ORF [57], the 5′UTR of RhPV mRNA [50], the 5′UTR of HalV mRNA [53], the 5′ IRES of CDV [114], and Sindbis virus subgenomic 26S RNA [115]. Ribosomal subunits themselves were reported to bind HoxA9 [116], HIV-2 [57], c-src [117] or KSHV vFLIP [84] mRNAs. One should remember that 40S binding per se does not mean that the mRNA is successfully loaded into the mRNA-binding channel. Likewise, whether such 40S*mRNA or 40S*eIF3*mRNA complexes are true 48S complex formation reaction intermediates has never been shown.
The major question is whether the binding of eIF3 or eIF4G or 40S to an mRNA is sufficient to promote internal initiation. Current knowledge indicates that the answer is definitely “no”. The first line of evidence emerges from the mutagenesis of viral IRESs. Point mutations in EMCV IRES that do not affect eIF4G binding are able to strongly inhibit the IRES activity [45, 46]. HCV IRES can be mutated in a way that precludes the binding of either eIF3 or 40S with a fatal effect on translation initiation despite the fact that the binding of another component is not affected [118, 119]. We showed recently that, reciprocally, inserting the eIF4G-binding JK domain from EMCV IRES into the 5′UTR of an mRNA reporter does not change its initiation mode from 5′ end-dependent to 5′ end-independent, although it dramatically alters dependence on m7G cap [27]. Strictly speaking, tethering eIF4G1 or eIF4G2 to an intercistronic position does positively affect second cistron expression [27, 120, 121], although it remains pretty low. The lack of secondary structure in mRNA may also alter requirements for certain initiation factors [122–125], but it cannot switch the translation initiation mode. The recent discovery that eIF3 can bind methylated adenosines in mRNA to promote 5′ end-dependent (but not internal) translation [39] is another argument for the notion that although the binding of the translation machinery to mRNA is necessary for IRES functioning, this is simply not enough to promote efficient internal initiation.
The other concern over ribosome binding to an mRNA is its physiological relevance. The intracellular concentration of free magnesium ions is estimated to be 0.5–0.9 mM, depending on cell type [126, 127]. A higher Mg2+ concentration inhibits the translation of both cap- and IRES-dependent mRNAs [128] but does not necessarily preclude ribosomal binding to IRESs [129, 130]. Nevertheless, 40S ribosome binding to mRNA is performed at 5 mM [117] or even 10 mM Mg2+ [116]. The binding at high magnesium concentrations may not reflect the physiological pattern and may lead to artefacts, including non-specific interactions.
Several reports have used the toe-printing assay to map the binding of ribosomes and/or initiation factors to mRNAs of viral and cellular origin [53, 131–134]. However, samples of initiation factors or ribosomes are not guaranteed to be free of RNase activity. It may not be a priori excluded that certain bands that appear on a gel arise from mRNA cleavage rather than represent authentic SHAPE signals or toe-print/foot-print bands. In our experience, the control reverse transcription of a deproteinized RNA sample after incubation is obligatory to exclude unanticipated degradation events, which represent in fact a minor fraction of the bands [135], but it is implemented only slightly more often than never. Once, this control denied our “discovery” of the eIF4G-binding site within the RhPV IRES (I.M. Terenin, unpublished).
The search for complementarity between 18S rRNA and mRNA, a magnetic reminiscence of the bacterial mode of ribosome recruitment, is also worth mentioning. While such interaction does seem possible [136, 137], there is a huge difference between bacterial translation initiation and cases of mRNA–rRNA interaction from eukaryotes. An initiator codon of mRNA bound to the 30S via Shine–Dalgarno sequence is placed close to the P-site, and upon initiator factors binding ribosome accommodates the mRNA into its mRNA-binding cleft, exactly following the prescription for internal initiation. No other example of mRNA–rRNA complementarity has been unambiguously shown to guide mRNA to occupy the ribosome in a translation-competent manner. Therefore, in most cases, ribosome binding to an mRNA should probably activate the CITE mode of translation (ribosome-binding CITEs from plant viruses have been documented [138, 139]), but should not lead to internal initiation. In line with this idea, the proposed mRNA–rRNA interactions were found to enhance the translation of corresponding monocistronic mRNAs [140–142]. Unfortunately, no adequate mRNA identity tests have been performed for the bicistronic reporters [143, 144] to draw a conclusion about internal initiation.
Mapping authentic 5′ termini
The first troublesome step in studying the translational properties of a particular 5′UTR is the determination of its 5′ boundary, i.e., transcription start site. A shorter-than-authentic 5′UTR may represent a truncated (thus non-functional) IRES or CITE, while a longer sequence will include the promoter parts, absent from the authentic transcripts. The latter can spoil both DNA transfection (as it may lead to undesired incorrect transcription) and RNA transfection (as these sequences may contain AUG codons). Arguably, a huge fraction of mammalian mRNAs is still poorly annotated with respect to their authentic 5′ termini. Here, an analysis of expressed sequence tags (EST) and next-generation mRNA sequencing combined with CAGE data may be useful. Several interactive databases have been developed that incorporate EST, CAGE, RNA-seq, DHS-seq, and ChIP-Seq data to delineate transcription start sites, e.g., ZENBU [145] or EPD [146].
There are numerous cases when such analysis could have helped. For example, translation inhibitory elements (TIE) as well as IRESs have been recently reported for mouse Hox mRNAs [116]. A search in the ZENBU or EPD databases suggests that most (if not virtually all) of the naturally occurring mouse Hox mRNAs’ 5′UTRs are much shorter than annotated in the RefSeq database or determined by 5′RACE [116]. Another example is the XIAP 5′UTR: contemporary data do not support the existence of the longer XIAP 5′UTR that was originally described in the paper in which the XIAP IRES was identified [147]. However, one may not exclude that in certain tissues these isoforms are indeed expressed.
A very reasonable idea is to analyse the 5′UTRs of mRNAs from polysomal fractions [148–150] rather than address the total RNA, as the latter can be the source of unspliced or other aberrant non-translated transcripts. The same approach can be used to test the 5′UTR integrity of ribosome-bound mRNAs in in vitro assays [50].
Bicistronic assay in cultured cells
The only discriminative property of internal initiation is its independence from the 5′ end of an mRNA, as follows from the definition. Therefore, to address internal initiation, one needs to block 5′ end-dependent translation. Generally, there are three ways to do this. The most striking way is to circularize an mRNA so that it simply lacks any termini. The second way is to use a bicistronic mRNA, in which the second cistron translation is driven by a sequence under study and, in the absence of a translational coupling between the two cistrons (e.g., by means of reinitiation), should be translated strictly in a 5′ end-independent manner. The third approach is to block the 5′ end by a stable stem-loop structure that precludes ribosome recruitment. None of the three is free of shortcomings, the most deleterious of which will be addressed in this section.
Bicistronic assay
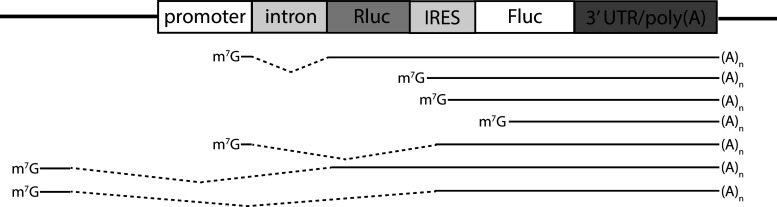
The bicistronic assay’s advantage is that it is less technically challenging than RNA circularization and that bicistronic mRNA already contains an internal control, i.e., the first cistron, translation of which should not be affected by alternating downstream sequences. However, the bicistronic assay requires plenty of controls. It is not always appreciated that any bicistronic mRNA is capable of the expression of a second cistron (at least at a level close to the background). The question is, therefore, how to assess it unambiguously. Obviously, readout values must be compared to those derived from control mRNAs, and here we face a problem of which mRNAs should be used as the controls? Any intermediate value of the second cistron expression provokes questions of whether it is closer to a positive or a negative control and how much over the negative control is meaningful? (Fig. 1). In fact, such hesitation may also arise from a circular mRNA analysis. Certain sequences do promote the internal initiation at a level that exceeds a negative control over an order of magnitude and may be named IRESs, following the definition. However, if an mRNA under study is naturally monocistronic (which is the case for the vast majority of reported IRESs), we must consider the contribution of the 5′ end-dependent translation.
Fig. 1.
a When different bicistronic mRNAs are compared to each other, a choice of whether or not to call the sequence an IRES strongly relies on what the latter is compared to. Inevitably, the choice becomes very subjective, especially when different negative controls are used. Note that the y-axis is split to show the relative efficiencies of EMCV and HCV IRESs. b A comparison of m7G- and A-capped mRNAs discriminates cap-dependent from cap-independent translation, while a comparison of monocistronic and bicistronic mRNAs reveals the contribution of internal (5′ end-independent) initiation to the overall level of translation. Such segregation should be made for a particular condition under which IRES is thought to be active
There are two principle ways of bicistronic mRNA delivery into a cell, i.e., plasmid transfection when an mRNA is synthesized within the nucleus and the transfection of in vitro transcribed mRNA [151]. The two approaches require different sets of controls and are thus discussed separately. In general, all controls aim to address two major issues, i.e., the authenticity and stability of bicistronic mRNAs and the independence of the second cistron translation from that of the first one. Although these issues were realized long ago [10, 144], the jury is still out on what is a credible experimental set-up.
DNA transfection: promoterless and RNAi controls
It is well-documented that the transfection of bicistronic plasmids results in aberrant transcripts due to cryptic promoter activity and/or unanticipated splicing events (Fig. 2; reviewed in [4, 144]). Among tests for mRNA integrity are RNase protection assay, RT-PCR, and Northern blot. The two former approaches suffer from the shortcoming that possible undesirable mRNA species may be easily missed if their boundaries are not correctly anticipated. A recent rigorous analysis of cryptic transcripts generated from pGL3 or pGL4 vectors, which are the most common backbones for bicistronic reporters, suggests that these plasmids are intrinsically prone to the production of aberrant transcripts [152], not to mention transcription from within Firefly luciferase itself [153]. Northern blots (without overexposition) and RNase protection assays may not provide enough sensitivity. Even trace amounts of undesired capped monocistronic mRNAs may account for the total yield of the second cistron expression. For example, if a cistron in a capped monocistronic mRNA is translated ~50 times more than in the corresponding bicistronic one (which is a deliberate underestimation), then the presence of ~2% aberrant transcripts is sufficient to double second cistron expression. Sometimes the integrity of a bicistronic reporter is addressed via a real-time PCR comparison of both cistrons levels [154]. However, this approach is only suitable for demonstrating a substantial difference in the cistrons’ levels.
Fig. 2.
Plasmid transfection leads to spurious transcription and splicing. When assayed by RT-PCR, most of these aberrant mRNA species will pass unnoticed
An important and absolutely essential control is the expression of a bicistronic reporter from a promoterless plasmid (i.e., plasmid without a promoter that would guide the transcription of the bicistronic template). This can reveal cryptic promoter activity, including transcription from a plasmid backbone. However, cases of aberrant splicing are obviously missed with this approach.
Very elegant assay for bicistronic RNA authenticity was suggested by Richard Lloyd and colleagues. It is based on the siRNA-targeting of the first cistron of a bicistronic mRNA [155]. Obviously, the expression of both reporter genes should be inhibited to a similar extent unless they are expressed from different mRNA species.
The problem with RNAi control implementation is that a rare bicistronic vector unambiguously passes this stringent test [40, 70, 155–158], which is particularly true for natural mammalian 5′UTRs, especially long and GC-rich ones. This can be expected because mammalian promoter-enhancer motifs are usually not limited to regions upstream of transcription start sites and often expand into the 5′UTRs.
Among well-characterized IRESs, only HRV IRES lacks a promoter/splicing activity [25, 70], while the IRESs from CrPV, HCV and EMCV demonstrate appreciable levels of undesired monocistronic transcripts in accordance with the discovery of cryptic promoters in these sequences [159–161]. This is not surprising since these viruses never exist as an DNA, and hence, sequences that may act as weak promoters or weak splice sites are not under negative selection. The EMCV case deserves a separate passage. Due to its credibility and efficiency, the EMCV IRES’s essence is often taken for granted, and the necessity for the controls is ignored. In a recent epitome of the plasmid-based approach, certain deletion variants of the EMCV IRES were found to be as active as the wild-type IRES [154]. It is likely that in any RNA-based experiment, these mutants would most probably be as dead as a doornail. This representative example provides probably the most convincing arguments against using plasmid transfection for IRES detection. Overall, the DNA-based bicistronic assay should be used with great care and only to complement mRNA transfection. Another problem with DNA transfection is that different promoters may distinctly guide splicing patterns [162], making it difficult to compare results obtained using different vectors.
The idea that mRNA should experience some nuclear events to become active as an IRES has emerged to explain why certain putative cellular IRESs were completely inactive when corresponding bicistronic mRNAs were transfected into cells. Presumably, there should be an array of RNA-binding proteins that bind to mRNAs in the nucleus and then orchestrate their behaviour in the cytoplasm. The existence of nuclear events for IRES activation has never been shown; thus, it always seems to be similar to Russell’s teapot (reviewed in [4]). However, this possibility may never been ruled out. In fact, recent findings that promoter sequence may affect the subcellular localization of heat-shock proteins mRNAs and their selective translation under stress conditions in yeast [163], as well as mRNA degradation kinetics [164], resurrect this concept. It is known that human Hsp70 and Hsp90 mRNAs avoid localization to stress granules upon stress [165, 166]. The N6-methylation of adenosines within mRNAs of stress-inducible heat-shock proteins may promote the binding of eIF3 [39], which in turn drives cap-independent (but not internal!) translation [38, 39]. Thus, it may appear that in mammals, mRNAs for heat-shock proteins (and probably for some other proteins as well) are similarly “imprinted” by the corresponding promoters and methylation. In such cases, the transfection of in vitro transcribed unmethylated mRNAs would indeed be irrelevant, as would be the transfection of plasmids lacking authentic promoters. Anyway, the 5′ end-dependent translation of Hsp70 mRNA [38–40, 90] contrasts the idea of cellular IRESs.
Internal initiation has been reported not only for mammalian mRNAs but also for certain mRNAs from other organisms, e.g., in Drosophila or yeasts. The most commonly used method for IRES research in yeast is DNA transfection, and many of the abovementioned considerations about it are applicable. Unluckily, S. cerevisiae had lost its RNA-interference pathway [167], making it impossible to perform RNAi tests on this particular species. The inability to transfect intact yeast cells with RNA prevents addressing the cap-dependence of yeast mRNAs in vivo, but in electroporated spheroplasts, cap-dependence values exceed one hundred [168, 169]. How yeast translational machinery responds to spheroplasts preparation and electroporation has not been studied. Thus, it is unclear if cytoplasmic extracts correctly reproduce cap-dependence in this case. Strains with temperature-sensitive eIF4E mutants may be useful for determining the real contribution of the 5′ cap to translation in yeasts.
HCV IRES has been used as a positive control for internal initiation in Drosophila (e.g., in [170]) or in Danio rerio [171], but is it relevant? The inability of HCV IRES to bind plant 40S ribosomes has been documented [118], and to the best of our knowledge, it has not been reported whether 40S ribosomes or eIF3 from insects, yeasts or fishes are able to bind HCV IRES. In fact, in S. cerevisiae, HCV IRES worked ~2 times worse than a negative control and only ~2 times better than itself in the antisense orientation [172, 173]. Similarly, HCV IRES drove internal translation worse than a negative control mRNA in extracts from Drosophila embryo cells [170]. Poliovirus IRES does not function in yeast [174]; EMCV IRES is inactive in S. cerevisiae [175] and in plants [176], most probably because they do not bind the corresponding eIF4G orthologues [177]. The suitable “universal” mRNA is one of the CrPV-like IRESs, which have been documented to function properly (albeit not very efficiently) in S. cerevisiae [178]. Most probably, RhPV 5′IRES and HalV 5′IRES, which both were shown to function in mammals, insects and plants [52, 53], can also operate in yeasts, but this has not been shown yet.
In vitro transcribed RNA-based bicistronic assay
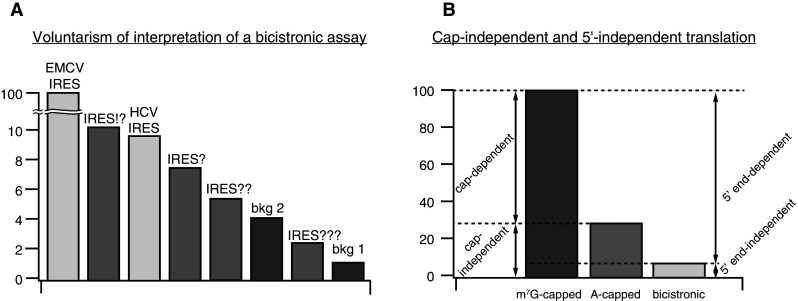
It is possible to avoid almost all of the abovementioned shortcomings by transfecting cells with in vitro transcribed mRNAs. Similar to plasmid transfection, the positive control here is usually a bicistronic mRNA that has a well-characterized viral IRES in the intercistronic position. However, certain IRESs are “more equal” with regard to using them as positive controls. When bicistronic mRNAs are transfected into HEK293T cells, HCV IRES works ~10 times worse than EMCV IRES [40]. By coincidence the weaker HCV IRES is usually used as a positive control. This means that if, e.g., HTLV-1 IRES (which was found to function only 2 times weaker than HCV IRES [157]) were compared to EMCV IRES, it would work 20 times less efficiently than the positive control, which in this case would not be convincing (Fig. 3a). It is important, therefore, to vindicate such low levels of expression.
Fig. 3.
a Single bicistronic plasmid is sufficient to provide PCR-made templates for the transcription of monocistronic and the bicistronic mRNAs. b Four sets of mRNAs are sufficient to dissect translation initiation mechanisms operating on a particular 5′UTR
Data from the bicistronic assay, when presented as FLuc/RLuc ratios, may be inherently misleading for several reasons. EMCV and FMDV IRESs enhance the translation of the upstream cistron in a bicistronic mRNA 2–4-fold, functioning as a 3′CITE for it [27, 179]. This obviously leads to a decreased FLuc/RLuc ratio, which does not reflect the full potential of these IRESs. Hence, they actually function 2–4 times better than the observed FLuc/RLuc ratios tell, and the apparent activity of IRES under study is inflated when compared to EMCV IRES, unless the former also acts as a 3′CITE.
Activation of an IRES-mediated translation under certain conditions is commonly inferred from increase of FLuc/RLuc ratio, but it increases in G2/M-arrested cells compared to asynchronous cells, during apoptosis, after 2Apro treatment [180] for almost any bicistronic mRNA, irrespectively of whether it bears an IRES or not. The cell culture density strongly affects the FLuc/RLuc ratio in the case of HCV IRES bicistronic plasmids [181]. Our data suggest this is frequently true for the RNA-based bicistronic assay. However, does this reflect IRES activation? This consideration is especially relevant to “weak IRESs”, which provide a rather inefficient translation of the downstream cistron, and our experience is that low reporter activity tends to be much more resistant to almost any treatment.
The choice of negative control may also affect interpretations. The FLuc/RLuc ratio of “empty” bicistronic mRNA may easily vary within an order of magnitude, depending on the particular sequence of the intergenic spacer [182] and its length [183]. Functional IRES expression, therefore, can be buried in high background values, while a too low background translation may result in a false-positive identification of an IRES (Fig. 3a).
An analysis of dozens of published papers shows that this problem is not fictional. To overcome it, we and others have suggested that the only relevant and reliable control is the nucleotide sequence under study [40, 158, 161, 184]. In fact, this sort of control was proposed by Marilyn Kozak back in 1989 when she discussed poliovirus IRES [185]. The rationale for the monocistronic control is that putative cellular IRESs are located in the 5′UTRs of naturally m7G-capped monocistronic mRNAs and are doomed to recruit the ribosome to their 5′ ends. The crux is, therefore, how to discriminate contributions of IRES-dependent, cap-dependent and cap-independent (but 5′ end-dependent) mechanisms. We believe that only the comparison of the monocistronic mRNA, which is presumably translated via a both 5′ end-dependent and IRES-dependent mechanisms, with the respective bicistronic or circular mRNA, which are only able to use the latter mode of translation initiation, can reveal the true contribution of the putative IRES to the overall translation level (Fig. 3b; see also the “Workflow” section). Here lies one of the reasons why plasmid transfection may be inapplicable to monocistronic mRNA translation studies: ambiguity of viral promoter transcription start sites. For instance, the early SV40 promoter has four major TSSs, all residing within the promoter sequence. How these sequences affect the translation of a particular mRNA species is simply unknown and needs to be investigated.
Circular RNA can be used instead of a bicistronic reporter. It has been shown that the artificially circularized mRNA of tobacco mosaic virus or UGA25–110 polymers do not bind ribosomes [186, 187]. Consequently, Chang-you Chen and Peter Sarnow used this approach to show that EMCV IRES indeed efficiently functioned in RRL as a part of a circular mRNA molecule [188, 189]. While many reviews pinpoint the circular mRNA test as the most stringent assay, they quite rarely mention that not a single IRES apart from the EMCV case has ever been reported to be addressed by these means. In practice, however, this test should only suffice for observing qualitative effects. Low (but above the background) expression levels will pose the problem of physiological relevance, as they do for the conventional bicistronic assay. Moreover, in some cases, circular mRNAs can produce enough protein to be detected by Western blotting in the absence of any IRES activity [190]. Thus, this test is not a palatable substitute for the composite strategy described below.
Weak downstream cistron expression may originate from either reinitiation, which is rarely efficient, or weak internal initiation. It is important, therefore, to discriminate between these two possibilities. In the case of reinitiation, the expression of the second cistron should correlate with that of the first one. The translation of the upstream cistron may be artificially blunted by substituting A cap for m7G cap, the introduction of a stable hairpin at the 5′ end, or the inclusion of uAUGs. It may also be useful to increase first cistron translation by exchanging 5′UTR and/or initiator codon context improvement to see if the second cistron responds. True IRES-driven translation should be insensitive to these manipulations.
All of these considerations are equally applicable to natural bicistronic mRNAs. Kaposi's sarcoma-associated Herpes Virus (KSHV) and related cytomegaloviruses encode several polycistronic mRNAs [191, 192], the most studied of which are those encoding v-Cyclin and v-FLIP (ORF72 and ORF71, respectively) [193] or UL136 and UL138 in the case of HHV5 [192]. These two ORFs are separated by ~80 nucleotides, a distance that matches the reported optimum for reinitiation in mammalian cells [194]. Although numerous reports have attempted to demonstrate internal initiation [89, 195, 196], none of them have eliminated the possibility of reinitiation, as the translation of bicistronic mRNAs in RRL does not count (see below). Moreover, in all in vitro experiments, the translation levels of the two cistrons in v-FLIP bicistronic mRNA correlated with one another [89], which agrees with reinitiation rather than with internal initiation.
While experiments aiming to demonstrate an IRES driving v-FLIP expression suffer from many of the described shortcomings, there is a conceptual issue that is worth discussing. In the case of naturally bicistronic mRNA, we lack any relevant monocistronic control and thus must rely on the (absence of) correlation between the expression of two cistrons. To avoid the use of tricistronic mRNAs, it is possible to affect first cistron expression in a manner discussed above. Surprisingly, we could not find reports of a direct measurement of the v-Cyclin to v-FLIP ratio in infected cells. The recent ribosomal profiling of KSHV-infected iSLK-219 cells [191] or HHV5-infected human fibroblasts [197] shows that the downstream genes are expressed at an evidently lower level, which is compatible with the possibility of reinitiation as well.
In contrast, PITSLRE case, regarded sometimes as the strongest candidate for a true cellular IRES [144], has an extensive physiological background [198]. A shorter form, p58PITSLRE, accumulates during the G2/M phase, and its translation relies on the AUG codon residing within the p110PITSLRE ORF. None of the seven in-frame AUGs lying between AUG1 (p110) and AUG18 (p58) is apparently seen by the initiating ribosome, which might support the idea of internal initiation. Probably, a single experiment with transfection of A- and m7G-capped monocistronic reporters with or without AUG1 in G2/M-synchronized cells would be sufficient to discriminate between the 5′ end-dependent mechanism (which may include ribosome shunting [199] or ribosome tethering [200]) and true internal initiation.
What to expect from an uncapped monocistronic mRNA?
It is not uncommon to study IRES-dependent translation by means of non-capped (or A-capped) monocistronic mRNAs. For example, A-capped APP mRNA is translated ~3 times better than A-capped β-globin mRNA, while the opposite is observed for m7G-capped mRNAs [86]; this observation was used as a proof of internal initiation. Such interpretation is based on the premise that in the absence of m7G cap, an mRNA can only be translated if it possesses an IRES. As we have already mentioned, this is not correct. Uncapped mRNAs can be translated with a reasonable efficiency in a 5′ end- and scanning-dependent manner. Moreover, certain A-capped 5′UTRs drive scanning-dependent translation more efficiently than, e.g., HCV IRES [40].
Under conditions of impaired cap-dependent translation (if eIF2 remains active), the translation efficiency of many uncapped mRNA increases. This may result from a trivial relief of competition with more translationally active capped mRNAs. It is not, therefore, surprising that eIF4G deletion variants that lack the eIF4E-binding site are able to stimulate the translation of uncapped mRNAs both in vitro [26, 28, 201] and in vivo [202], while the translation of m7G-capped mRNAs is hardly affected. Similarly, the release of eIF4G from eIF4F complexes after mTOR inhibition by torin also stimulates uncapped mRNAs translation [25]. This apparently mirrors the non-specific activation of uncapped mRNAs translation in vitro upon the addition of 4E-BPs or m7GTP rather than the activation of a putative IRES [27, 203]. In summary, the inhibition of cap-dependent translation seems to equivocally increase the translation of uncapped (A-capped) monocistronic mRNAs irrespectively of the mechanism that they use.
The next step in eliminating the 5′ end-dependent translation in the context of monocistronic mRNA is the inclusion of a stable stem-loop to the 5′ terminus of an A-capped mRNA [26, 141, 169, 201, 204]. The residual translation observed in these cases is rather weak and is thought to be solely IRES-dependent. Unfortunately, the bicistronic test is frequently omitted and this probably valid assumption remains unjustified. However, if an A-capped mRNA with a stem-loop provides translation at a level comparable to that of the bicistronic mRNA, it indeed operates in the 5′ end-independent mode. This situation highlights the importance of a physiological reference for any reporter mRNA. Researchers should determine whether they deal with a promising IRES or just play with the background activity. It is important to have an idea about the relevance of the internal initiation contribution. If it is much lower than that of the 5′ end-dependent translation of the respective 5′UTR in the monocistronic context under the same conditions and in the same cells, then, in our opinion, it makes little sense to perform further experiments.
Antisense 5′UTR: an unreliable control
Frequently, the reverse complement of a 5′UTR under study is used as a control for both mono- and bicistronic reporters. For example, the A-capped 5′UTR of FLO8 mRNA from S. cerevisiae bearing a 5′-terminal stem-loop supports the translation of monocistronic mRNA ~20 times better than its antisense variant [169]. Notably, the latter contains five uAUGs compared to none in the sense orientation, which expectedly inhibits translation. The reversed c-myc 5′UTR is unable to drive the significant expression of the reporter in the case of the non-capped monocistronic variant compared to itself in the sense orientation [202]. This observation can be explained similarly: the antisense variant contains three uAUGs that inhibit translation. The p27/Kip1 5′UTR acquires two additional AUG codons after flipping [156]. Therefore, these antisense 5′UTRs have deliberately diminished potential for translation. Reversed sequence may only be used if neither sense nor antisense sequences contain uAUGs or near-cognate start codons. In all other cases, different uORFs will unequally affect reporter expression, making the results incomparable.
This sort of control encroaches on other fields of mRNA research. For instance, the binding of eIF3 to BTG1 mRNA was postulated to inhibit translation on the basis of the comparison to the reversed sequence [38]. The authors ignored a simpler explanation that the authentic sequence contained one uAUG, while the reversal lacked any.
Dangers of in vitro translation
Despite the fact that rabbit reticulocyte lysate is an extremely bad choice to study translation in vitro for several well-documented reasons, the practice of using RRL for internal initiation research is still inexorcizable.
The pattern of gene expression in reticulocytes is narrow, and translated endogenous mRNAs do not possess long and/or highly structured 5′UTRs; therefore, it is not surprising that the translation of exogenous mRNAs with long 5′UTRs in RRL is compromised [40, 71]. Importantly, when RRL is supplemented with m7G cap analogues or 4E-BP, weakly translated mRNAs do not respond properly, and their translation is sustained [88]. Accordingly, their translation is not adequately stimulated by 5′ cap or poly(A)-tail (see [71, 205] and references therein), and in these cases, the degree of inhibition by cap analogue is reciprocal with translation efficiency. This may not always be evident for readers because the concentration curves of inhibition by cap analogue are often provided with all of the mRNAs normalized to 100%, and no absolute values are given [86, 117]. The apparent resistance is routinely interpreted as a reflection of IRES-driven translation. In other words, many sequences seem to work as weak IRESs in RRL (also in our hands), but this is due to some (yet mechanistically unclear) bug of this particular cell-free system. Generally, these effects cannot be reproduced in other cell-free systems and, most importantly, in cultured cells [40, 71]. RRL produces especially miraculous results if treated with picornavirus proteases. It permits anomalously efficient translation under conditions of eIF4G cleavage, particularly at higher mRNA concentrations [206]. “HIV-1 IRES” is a showcase: eIF4G cleavage does not inhibit HIV-1 5′UTR-driven translation in RRL [207–209] but does so in HeLa S10 or Krebs-2 extracts [25, 210, 211]. Moreover, commercially available RRL is rapidly (15–20 min) inactivated by means of eIF2 phosphorylation [212, 213]. Therefore, translation in RRL occurs under conditions of limiting eIF2, making the interpretation of the results even more complicated.
All of these flaws of RRL are strongly dissuasive; thus, we believe that in vitro assays should be performed in cytoplasmic extracts prepared from cultured cells only, as they much more closely reproduce what is observed in cells. Unfortunately, such extracts are still unavailable commercially. However, they can be easily prepared from regular mammalian cell cultures (see below).
A workflow
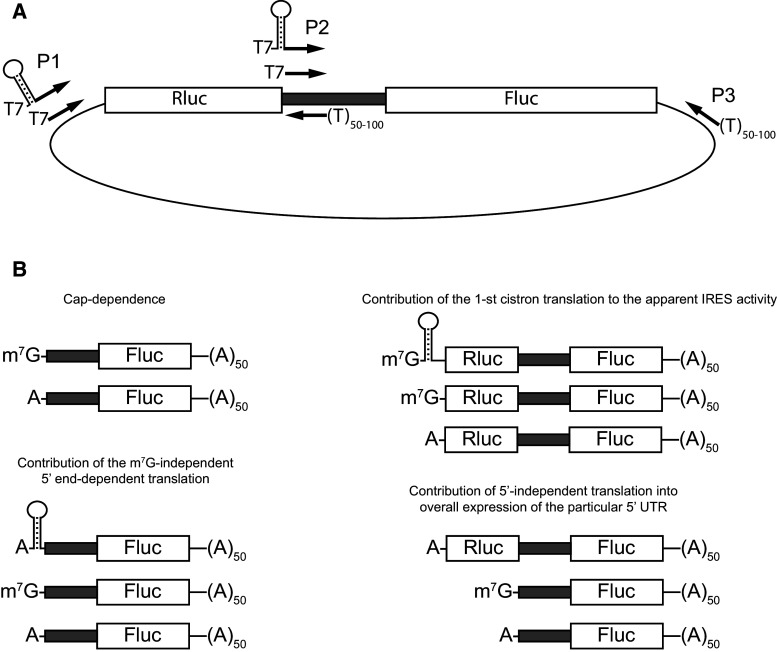
We suggest a simple experimental design to validate the IRES activity in a nucleotide sequence of interest. The creation of only one plasmid is sufficient for all downstream applications (Fig. 3a). Basically, one should clone the sequence of interest into the intercistronic position of a suitable bicistronic plasmid (for example, pGL3R [214]). Optionally, the 3′UTR of the gene of interest may be cloned downstream of the second cistron to more relevantly emulate the mRNA under study. Additionally, it is up to the researcher to decide whether to clone an AUG-proximal part of the CDS in frame with FLuc because the downstream context of AUG may strongly influence internal initiation, as shown for HCV-like IRESs [215, 216]. In this case, the deletion of the in-frame FLuc initiator codon is a very reasonable idea.
Next, PCR is carried out to prepare templates for T7 transcription. The universal reverse primer P3 containing a poly(T) tail allows the production of polyadenylated mRNA. According to our experience, a poly(A)-tail as long as 90 nt can be easily introduced in this way. Usually, the primer annealing site is located approximately 200 nt downstream of the stop codon to create a relevant 3′UTR (mRNAs lacking any 3′UTR are unstable and deficient in adequate translation termination). Primer P2, which contains the T7 promoter, is used for monocistronic mRNA production. It can be either a universal primer annealing to the intercistronic sequence from the vector backbone or a gene-specific primer designed to create the natural 5′ end of mRNA. Primer P1 is used to create a bicistronic template. Additionally, a template from the control monocistronic RLuc plasmid should be obtained. Generally, the presence of a T7 promoter in the plasmid yields a higher mRNA specific activity. It is also necessary to use an optimized PCR regime and/or a gel-purified PCR product. In our hands, the transcripts obtained with either approach give similar results. It is reasonable to provide all T7-containing primers with an additional 5–6 nucleotides at their 5′ ends to ensure efficient T7 polymerase binding.
Additional primers that may be useful are P1-sl and P2-sl, which introduce 5′ terminal stem-loop structures to inhibit 5′ end-dependent initiation. We routinely use the sequence GGGAGTGGACTTCGGTCCACTCCC, where the first GGG constitute the transcription start site for T7 RNA polymerase. This stem-loop inhibits the translation of cap-dependent mRNAs 5–10-fold and can be easily extended to produce a more pronounced effect on translation, if desired. Purified PCR products are then converted to m7G- and A-capped mRNAs during co-translational capping with ARCA cap or ApppG non-functional cap. mRNAs are then purified by 2 M LiCl precipitation and analyzed for integrity. Typically, we obtain 10–20 µg of mRNA from 10 µl reaction, which is sufficient for 50–100 transfections. The same mRNAs may be used in a cell-free translation system.
When only the m7G-capped transcript is needed, post-transcriptional capping with commercially available vaccinia virus capping system may be used. However, one should avoid simultaneous using of different mRNA preparations. Ideally, all transcripts should be obtained in parallel.
It is possible to follow the generalized workflow presented below (Fig. 4). If starting with plasmid transfection, it is necessary to test a promoterless plasmid to address the cryptic promoter activity and perform the RNAi test. However, as it is almost impossible to come to an unambiguous conclusion on the existence of internal initiation by means of plasmid transfection, the next step is inevitably RNA-based, even if the controls are successfully passed. Using in vitro transcribed mRNAs, a comparison of only three experimental points may be enough for a preliminary (and sometimes ultimate) conclusion (Figs. 1b, 3b). These are (a) bicistronic mRNA, (b) m7G-capped monocistronic mRNA (along with m7G-RLuc normalization control), and (c) A-capped monocistronic mRNA (with the same RLuc mRNA). For a standard cap-dependent mRNA, the ratio between m7G- and A-capped mRNA translation (cap-dependence) usually reaches 40–80 depending on the particular cell line and growth conditions, while the ratio between A-capped and bicistronic mRNA usually exceeds an order of magnitude. Consequently, an m7G-capped monocistronic mRNA (which represents the natural context for eukaryotic mRNAs) is usually translated 2–3 orders of magnitude better than the same sequence in the bicistronic context. These data reveal the contribution of either mechanism (cap-dependent, internal or cap-independent 5′ end-dependent) and in most cases rule out the presence of any meaningful impact of internal initiation on the overall translation under particular experimental conditions. The same approach can be easily applied to conditions where an IRES is thought to be activated [25, 97, 217]. Data obtained from transfections can be complemented with in vitro assays. These may include tests with 4E-BPs or proteases that cleave eIF4G.
Fig. 4.
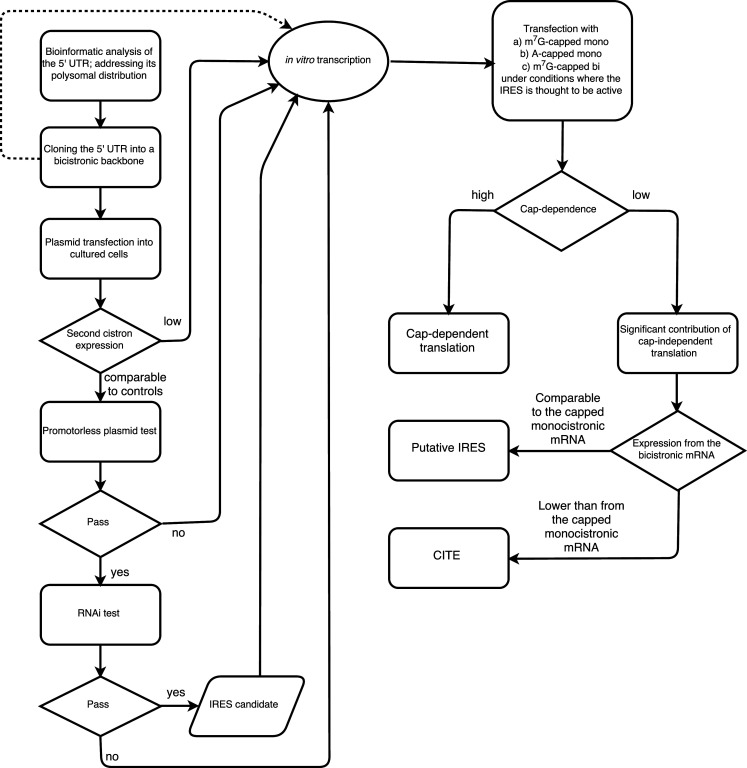
A generalized workflow of how to address internal initiation. Notably, all DNA-based experiments are inevitably followed by RNA-based experiments; thus, it is advisable to start with the latter
A complex issue of mRNA capping
Apart from the guanine methylation, mRNAs of higher eukaryotes generally have one or two first nucleotides that are also methylated at 2′-hydroxyls [218, 219], i.e., cap-1 and cap-2. In addition, when the first nucleotide of mRNA is adenine, it can be methylated at the N6 position [220]. The 2′-O methylation of cap-0 significantly stimulates the translation of non-polyadenylated mRNA in Xenopus oocytes [221] but hardly affects the ribosome recruitment in RRL [222]. The knockdown of the CMTR1, enzyme responsible for 2′-O cap methylation does not exhibit a substantial repression of general translation in HeLa cells [223]. This is not an issue for yeasts, which have only monomethylated caps [224].
However, cap-0 containing mRNAs (used in the vast majority of the reports) are subject to the innate immune response driven by IFITs [225, 226]. In commonly used 293T cells, cap-0 and cap-1 work similarly (I.M. Terenin, unpublished), which may be explained by a cytoplasmic cap-0 to cap-1 transformation [223] or by the absence of expressed IFITs, but this situation should not be taken for granted for other cell lines, which may call for the authentically methylated cap. Luckily, the cotranscriptionally added cap can be additionally methylated by commercially available 2′-O-methylases. Primary cells may even require extensive mRNA modification by 5-methylcytidines or pseudouridines to reduce the mRNA immunogenicity and increase the expression level [227, 228]. The bad news is that modified RNA most probably folds unnaturally, which could be critical for translation research. A recent study, however, claimed that unmodified mRNAs having cap-1 on 100% of molecules are well tolerated by primary cells [229].
A different approach that rightly has not acquired popularity is the in-cell cytoplasmic capping of T7 polymerase-synthesized mRNA by the vaccinia virus capping enzyme. This method results in a huge overload of the cell with T7 promoter-driven transcripts, which have quite a low degree of accomplished capping [230].
Sometimes, a cap analogue is ambiguously called unmethylated (e.g., in [231]), leaving a reader to guess if it was unmethylated GpppG or inactive ApppG? At least in some in vitro systems, methylase activity may be easily revealed if GpppG-capped mRNA is translated in the presence of the methylase inhibitor sinefungin (I.M. Terenin, unpublished). To avoid a misunderstanding, GpppG cap analogue should be referred to as unmethylated and not be used, while ApppG cap analogue (the A-cap) should be called nonfunctional.
Finally, the use of unstable uncapped mRNAs should be avoided. Moreover, 5′-triphosphate moiety activates RIG-1 [232] or even PKR [233], but transfected uncapped mRNAs are most probably degraded before these proteins exert their actions.
Transfection and stability issue
When either bicistronic RLuc/putative IRES/FLuc or an equimolar mixture of FLuc and control RLuc mRNAs are transfected, usually 0.05–0.2 µg of each mRNAs per well is sufficient in the 24-well plate format. Then, 2–4 h post transfection, cells are lysed and luciferase activities are addressed. For some “easy-to-transfect” cell lines (such as 293T, Huh7, RKO, BHK21) and m7G-capped mRNAs, the expected values easily reach millions of light units, which is more than enough for reliable and reproducible measurements. For “hard-to-transfect” cell lines, an increase in the mRNA amount, the use of brighter luciferases (e.g., Gaussia or NanoLuc) and the use of unorthodox transfection reagents may solve the problem [25, 229].
There are several ways to deliver an in vitro transcribed mRNA to the cytoplasm. Electroporation is known to rapidly induce eIF2 phosphorylation via the GCN2 and PERK pathways [234]. Numerous polyethyleneimide-based transfection reagents are poorly suitable for mRNA transfection in our hands, but others have exploited it successfully [229]. A magnet-assisted mRNA transfection is also possible [35]. Lipofection works well and does not induce eIF2 phosphorylation [234]. However, a failure to release most of the RNA from vesicles into the cytoplasm makes the subsequent RT-PCR or Northern blot analyses of mRNA integrity pointless [235].
Faute de mieux, a kinetic approach has been proposed that addresses the accumulation of the reporter over time [35, 40, 70, 158]. Obviously, if an mRNA is stable in the cytoplasm, the accumulation of the reporter (i.e., luciferase) should be linear or demonstrate acceleration. In contrast, the translation of unstable mRNA should slow down. The latter is the case for uncapped mRNAs, while m7G- or A-capped species show no deceleration until 4–6 h post transfection in various cell lines [35, 40, 70, 236]. This underscores importance of short-time transfections, as sometimes expression of transfected RNA reporters is measured 16 or even 24 h after transfection, when the significant portion of the transfected RNA may be degraded (see, e.g., [237] for stability analysis).
When the kinetic approach was used to study translation in MEFs, the accumulation of the reporter luciferase expressed from an mRNA with the 5′UTR from the murine Hsp70 mRNA plateaued as early as 1.5 h post transfection, while the translation of a reporter with short unspecific 5′UTR lasted for at least 3 h [90]. It is not clear whether this reflects some translational properties of the Hsp70 mRNA or indeed its instability, but what it definitely shows is that transfection with a single time-point readout is meaningless when you compare two mRNAs of a different nature.
Real-time analyses of transfection can be performed with luciferin added to the cell culture medium [90], but there most likely is a problem of substrate uptake and enzyme turnover. Since it is rarely needed to obtain a highly precise curve, it is possible to stop translation at the desired time points by the addition of 100 µg/ml cycloheximide and to measure all of the experimental points later.
An important but rarely discussed issue is whether mRNA transfection combined with a drug treatment correctly reflects the behaviour of endogenous mRNAs. At least in vitro, during the course of translation, mRNA becomes heavily loaded with ribosomes [238, 239], and the mode of translation presumably shifts from initiation de novo to recycling. Recycling or reinitiation mode was shown to be less sensitive to inhibition by cap analogue [240, 241]. This could be explained if heavier polysomes had a slower translation elongation rate [239, 242], although translation in yeast extracts directly points to the insensitivity of translation initiation on circularized polysomes to the cap analogue action [241]. After transfection, apart from entering into polysomes, mRNA acquires a set of RNA-binding proteins that may also affect translation and/or stability.
Hence, one may predict that transfection into stressed cells and the application of stress to the cells transfected beforehand may have different outcomes. The latter approach, although rarely implemented, should more closely reflect the behaviour of endogenous mRNAs [217]. Moreover, drug treatment may affect transfection efficiency (S.E. Dmitriev, unpublished). In all such cases, cells should be transfected first and only then treated. In this approach, addressing the kinetics of reporter accumulation is essential.
In vitro assays
Because RRL is a very artefact-prone system, we strongly recommend using easily cooked cell-free systems from cultured cells. There is a number of published protocols ([27, 71, 243–245] and references therein) that yield efficient translation systems. Our experience is that the protocol described in [27, 246] is applicable to various adherent or suspension cell cultures. In certain cases, supplementation of a lysate with creatine phosphokinase is strictly required. RRL supplemented with the commercially available HeLa cytoplasmic fraction (20% v/v) also works well (see [247] and references therein). These in vitro systems more adequately respond to mRNA capping, treatment with picornaviral proteases or inhibitors of cap-dependent translation, i.e., m7GTP, 4E-BPs or eIF4A R362Q dominant negative mutant [27, 40, 71, 180, 206, 244, 245, 247].
Batches of lysates may easily differ from each other; therefore, it is advisable to make larger batches and optimize ionic conditions. Importantly, one should pay more attention to the adequate cap-dependence of the reference mRNAs and their reasonable response to 4E-BP or m7GTP than to the level of translation efficiency itself. It is always possible to inadvertently create artificial conditions under which a highly cap-dependent mRNA is translated efficiently but only marginally stimulated by the 5′ cap or even where the capped mRNA translation is stimulated by the addition of a cap analogue to the lysate simply by varying ionic conditions. Thus, it is critically important to use an appropriate control mRNA, i.e., an mRNA that only utilizes the canonical cap-dependent mechanism (e.g., β-globin or β-actin), under the same conditions.
As with translation in cultured cells, the real-time measurement of the reporter accumulation may have its merits. An aggregate difference between the translation levels certain mRNAs may accumulate over time; thus, saying that one mRNA is translated quantitatively better than another may not always make perfect sense.
A few words for peer reviewers
No single experiment or criterion is sufficient to unambiguously demonstrate (or reject) internal initiation. Individually, they say little. For example, HCV IRES works somewhat better in monocistronic mRNA than in bicistronic mRNA, and its translation is stimulated by capping [40], although not dramatically. One might conclude that HCV IRES is not a true IRES, but we are confident that it is, for we know how it functions. Arguably, only the established mechanism of how a putative IRES works can provide incontrovertible proof of its existence. Several major points from above are worth summarizing because addressing these key issues is obligatory. These points represent the core of the experimental set-up.
The translation of an uncapped mRNA is not necessarily IRES-dependent. Cap-independent translation may be represented by two distinct mechanisms: internal initiation and 5′ end-dependent initiation (Table 1; Fig. 1b). Therefore, two indispensable tests must be performed: the determination of the cap-dependence of a monocistronic mRNA (m7G cap vs. A cap) and the explicit comparison of monocistronic and bicistronic reporters (Figs. 1b, 3b), while a comparison of different bicistronic mRNAs is misleading (Fig. 1a). Moreover, one should expect that the translation of any uncapped monocistronic mRNA is stimulated upon the inhibition of cap-dependent translation. After these experiments are performed, it should become clear whether the contribution of either specific or sporadic internal initiation is of any significance. It is important to provide not only fold changes or ratios but also absolute values, at least normalized to a reference because, e.g., a tenfold excess over a very low background of second cistron expression in bicistronic mRNA is still a very low value that may only represent a small fraction of the monocistronic mRNA translation.
Does the cloned sequence correctly correspond to the major form of a naturally occurring mRNA in particular cells under particular conditions? Is the 5′ terminus mapped unambiguously? If alternative transcripts are present in a cell, what is the contribution of other isoforms to the overall level of the gene expression?
Although DNA transfection should generally be avoided, it can be considered if RNA identity tests are passed unambiguously. In particular, it is obligatory to perform the RNAi test, and expression from a promoterless plasmid must be addressed. Attempts should be made to detect shorter transcripts rather than to demonstrate the presence of the full-length transcript, i.e., several pairs of primers for RT-PCR should be used. Importantly, qRT-PCR may not be used to show that only the full-length transcript is present in cells. Ideally, 5′RACE should be performed to detect the boundaries of the reporter mRNAs synthesized in cell. The cell line specificity, if observed, should correspond at the DNA-transfection level to that at the RNA-transfection level. Anyway, the sole use of plasmids for transfection is unacceptable: DNA transfection may only complement mRNA transfection tests rather than be the key experiment.
The description of the translational status of cells undergoing apoptosis, mitosis, viral infection, etc., in terms of 4E-BPs or eIF2 phosphorylation is insufficient. These switches are not binary; they do not provide all-or-nothing regulation. A wide spectrum of cases lays between fully resistant and highly sensitive mRNAs and there is no evidence that cap-dependent translation can by switched off completely. Therefore, translation under conditions of, e.g., mitosis or apoptosis is hardly an evidence of IRES-dependent initiation.
In all cases of bicistronic mRNAs and especially for those mRNAs that are naturally bicistronic, the possibility of reinitiation must be addressed.
Regarding mRNA transfection, single time-point readouts may be misleading as they neglect potentially distinct kinetics of mRNA entry into polysomes and/or stability issues. The kinetics of reporter accumulation should become de rigueur to compare the translational efficiencies of different mRNAs both in vitro and in cultured cells. It is extremely important to perform a short-time transfection, typically 2–4 h, to avoid RNA degradation.
Among in vitro translation systems, nuclease-treated RRL is clearly the worst choice, and its use should generally be avoided. Cases in which only RRL is used in in vitro tests are alarming, especially when RRL is used to demonstrate insensitivity to 4E-BPs or m7GTP or to evaluate dependence on the m7G cap. Cytoplasmic extracts from cultured cells or RRL supplemented with a commercially available HeLa cytoplasmic extract are a much better option.
Reverse complements of 5′UTRs may contain (additional) uAUGs compared to the sense orientation, thus use of reverse complements as controls is not reasonable.
Binding of the translational apparatus component(s) to an mRNA under study is not an evidence of internal initiation but may indeed indicate unusual translational properties.
Once a putative IRES has successfully passed all of the abovementioned tests, establishing its mechanism of functioning by biochemical experiments [248].
An afterword
Many things in cancer aetiology, such as apoptosis, regulation of oncogenes or tumour suppressors expression, are currently (mis)linked to cellular IRESs [249–251]. As pharmaceutical companies have to rely on basic science, investigations have been undertaken to screen for potential inhibitors of cancer-relevant cellular IRESs. For instance, pharmaceutical companies [252, 253] have recently performed screenings for drugs against c-myc IRES expressed from a bicistronic plasmid. The result was predictably uninspiring since the use of DNA transfection in the case of putative c-myc IRES has been disapproved, and c-myc IRES existence itself has been challenged. This shows that the problem runs really deep.
In recent years, translation control has become attractive for medical researchers. They enter the field to learn the conventional wisdom that says that cellular IRESs are firmly established. This hampers the development of new ideas in translational control, i.e., alternative mechanisms of translation initiation and its regulation.
Acknowledgements
We thank Vasili Hauryliuk for critical reading of the manuscript. The work was supported by grants from Russian Science Foundation to Ivan N. Shatsky (16-14-10065) and Russian Foundation for Basic Research to Ilya M. Terenin (13-04-00903a and 16-04-0162816).
Abbreviations
- 4E-BP
eIF4E-binding protein
- CAGE
Cap analysis of gene expression
- CDS
Coding DNA sequence
- CITE
Cap-independent translation enhancer
- CrPV
Cricket paralysis virus
- eIF
Eukaryotic translation initiation factor
- EMCV
Encephalomyocarditis virus
- FMDV
Foot and mouth decease virus
- HalV
Halastavi árva virus
- HCV
Hepatitis C virus
- HIV
Human immunodeficiency virus
- HRV
Human rhinovirus
- IFIT
Interferon-induced proteins with tetratricopeptide repeats
- IRES
Internal ribosome entry site
- KSHV
Kaposi's sarcoma-associated Herpes Virus
- PV
Poliovirus
- RhPV
Rhopalosiphum padi virus
- RRL
Rabbit reticulocyte lysate
- SHAPE
Selective 2′-hydroxyl acylation analyzed by primer extension
- TSS
Transcription start site
- uORF
Upstream open reading frame
- UTR
Untranslated region
Contributor Information
Ilya M. Terenin, Phone: +74959394857, Email: terenin@genebee.msu.ru
Ivan N. Shatsky, Email: shatsky@genebee.msu.ru
References
- 1.Thompson SR. So you want to know if your message has an IRES? Wiley Interdiscip Rev RNA. 2012;3:697–705. doi: 10.1002/wrna.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert WV. Alternative ways to think about cellular internal ribosome entry. J Biol Chem. 2010;285:29033–29038. doi: 10.1074/jbc.R110.150532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozak M. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 2005;33:6593–6602. doi: 10.1093/nar/gki958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson RJ. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb Perspect Biol. 2013;5:a011569. doi: 10.1101/cshperspect.a011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shatsky IN, Dmitriev SE, Terenin IM, Andreev DE. Cap- and IRES-independent scanning mechanism of translation initiation as an alternative to the concept of cellular IRESs. Mol Cells. 2010;30:285–293. doi: 10.1007/s10059-010-0149-1. [DOI] [PubMed] [Google Scholar]
- 6.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 7.Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol. 2012;4:a011544. doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filbin ME, Kieft JS. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol. 2009;19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 11.Lozano G, Martínez-Salas E. Structural insights into viral IRES-dependent translation mechanisms. Curr Opin Virol. 2015;12:113–120. doi: 10.1016/j.coviro.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Morita M, Gravel S-P, Hulea L, et al. mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle. 2015;14:473–480. doi: 10.4161/15384101.2014.991572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puertollano R. mTOR and lysosome regulation. F1000 Prime Rep. 2014;6:52. doi: 10.12703/P6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albert V, Hall MN. mTOR signaling in cellular and organismal energetics. Curr Opin Cell Biol. 2015;33:55–66. doi: 10.1016/j.ceb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Heesom KJ, Gampel A, Mellor H, Denton RM. Cell cycle-dependent phosphorylation of the translational repressor eIF-4E binding protein-1 (4E-BP1) Curr Biol. 2001;11:1374–1379. doi: 10.1016/S0960-9822(01)00422-5. [DOI] [PubMed] [Google Scholar]
- 16.Shang Z-F, Yu L, Li B, et al. 4E-BP1 participates in maintaining spindle integrity and genomic stability via interacting with PLK1. Cell Cycle. 2012;11:3463–3471. doi: 10.4161/cc.21770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin S, Wolgamott L, Tcherkezian J, et al. Glycogen synthase kinase-3β positively regulates protein synthesis and cell proliferation through the regulation of translation initiation factor 4E-binding protein 1. Oncogene. 2014;33:1690–1699. doi: 10.1038/onc.2013.113. [DOI] [PubMed] [Google Scholar]
- 18.Herbert TP, Tee AR, Proud CG. The extracellular signal-regulated kinase pathway regulates the phosphorylation of 4E-BP1 at multiple sites. J Biol Chem. 2002;277:11591–11596. doi: 10.1074/jbc.M110367200. [DOI] [PubMed] [Google Scholar]
- 19.Topisirovic I, Borden KLB. Homeodomain proteins and eukaryotic translation initiation factor 4E (eIF4E): an unexpected relationship. Histol Histopathol. 2005;20:1275–1284. doi: 10.14670/HH-20.1275. [DOI] [PubMed] [Google Scholar]
- 20.Kamenska A, Simpson C, Standart N. eIF4E-binding proteins: new factors, new locations, new roles. Biochem Soc Trans. 2014;42:1238–1245. doi: 10.1042/BST20140063. [DOI] [PubMed] [Google Scholar]
- 21.Rose JK, Lodish HF. Translation in vitro of vesicular stomatitis virus mRNA lacking 5′-terminal 7-methylguanosine. Nature. 1976;262:32–37. doi: 10.1038/262032a0. [DOI] [PubMed] [Google Scholar]
- 22.Kozak M, Shatkin AJ. Identification of features in 5′ terminal fragments from reovirus mRNA which are important for ribosome binding. Cell. 1978;13:201–212. doi: 10.1016/0092-8674(78)90150-2. [DOI] [PubMed] [Google Scholar]
- 23.Gunnery S, Mäivali U, Mathews MB. Translation of an uncapped mRNA involves scanning. J Biol Chem. 1997;272:21642–21646. doi: 10.1074/jbc.272.34.21642. [DOI] [PubMed] [Google Scholar]
- 24.Gunnery S, Mathews MB. Functional mRNA can be generated by RNA polymerase III. Mol Cell Biol. 1995;15:3597–3607. doi: 10.1128/MCB.15.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smirnova VV, Terenin IM, Khutornenko AA, et al. Does HIV-1 mRNA 5′-untranslated region bear an internal ribosome entry site? Biochimie. 2015;121:228–237. doi: 10.1016/j.biochi.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Hundsdoerfer P, Thoma C, Hentze MW. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc Natl Acad Sci USA. 2005;102:13421–13426. doi: 10.1073/pnas.0506536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terenin IM, Andreev DE, Dmitriev SE, Shatsky IN. A novel mechanism of eukaryotic translation initiation that is neither m7G-cap-, nor IRES-dependent. Nucleic Acids Res. 2013;41:1807–1816. doi: 10.1093/nar/gks1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Gregorio E, Preiss T, Hentze MW. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end-dependent. RNA. 1998;4:828–836. doi: 10.1017/S1355838298980372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thoma C, Hasselblatt P, Köck J, et al. Generation of stable mRNA fragments and translation of N-truncated proteins induced by antisense oligodeoxynucleotides. Mol Cell. 2001;8:865–872. doi: 10.1016/S1097-2765(01)00364-1. [DOI] [PubMed] [Google Scholar]
- 30.Dolph PJ, Racaniello V, Villamarin A, et al. The adenovirus tripartite leader may eliminate the requirement for cap-binding protein complex during translation initiation. J Virol. 1988;62:2059–2066. doi: 10.1128/jvi.62.6.2059-2066.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frolov I, Schlesinger S. Comparison of the effects of Sindbis virus and Sindbis virus replicons on host cell protein synthesis and cytopathogenicity in BHK cells. J Virol. 1994;68:1721–1727. doi: 10.1128/jvi.68.3.1721-1727.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon AE, Miller WA. 3′ cap-independent translation enhancers of plant viruses. Annu Rev Microbiol. 2013;67:21–42. doi: 10.1146/annurev-micro-092412-155609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicholson BL, White KA. 3′ Cap-independent translation enhancers of positive-strand RNA plant viruses. Curr Opin Virol. 2011;1:373–380. doi: 10.1016/j.coviro.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Miller WA, Wang Z, Treder K. The amazing diversity of cap-independent translation elements in the 3′-untranslated regions of plant viral RNAs. Biochem Soc Trans. 2007;35:1629–1633. doi: 10.1042/BST0351629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andreev DE, Dmitriev SE, Terenin IM, Shatsky IN. Cap-independent translation initiation of apaf-1 mRNA based on a scanning mechanism is determined by some features of the secondary structure of its 5′ untranslated region. Biochem Mosc. 2013;78:157–165. doi: 10.1134/S0006297913020041. [DOI] [PubMed] [Google Scholar]
- 36.Edgil D, Polacek C, Harris E. Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited. J Virol. 2006;80:2976–2986. doi: 10.1128/JVI.80.6.2976-2986.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paek KY, Hong KY, Ryu I, et al. Translation initiation mediated by RNA looping. Proc Natl Acad Sci USA. 2015;112:1041–1046. doi: 10.1073/pnas.1416883112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee ASY, Kranzusch PJ, Cate JHD. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature. 2015;522:111–114. doi: 10.1038/nature14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyer KD, Patil DP, Zhou J, et al. 5′ UTR m(6)A promotes cap-independent translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andreev DE, Dmitriev SE, Terenin IM, et al. Differential contribution of the m7G-cap to the 5′ end-dependent translation initiation of mammalian mRNAs. Nucleic Acids Res. 2009;37:6135–6147. doi: 10.1093/nar/gkp665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shatsky IN, Dmitriev SE, Andreev DE, Terenin IM. Transcriptome-wide studies uncover the diversity of modes of mRNA recruitment to eukaryotic ribosomes. Crit Rev Biochem Mol Biol. 2014;49:164–177. doi: 10.3109/10409238.2014.887051. [DOI] [PubMed] [Google Scholar]
- 42.Kaminski A, Belsham GJ, Jackson RJ. Translation of encephalomyocarditis virus RNA: parameters influencing the selection of the internal initiation site. EMBO J. 1994;13:1673–1681. doi: 10.1002/j.1460-2075.1994.tb06431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rijnbrand RC, Abbink TE, Haasnoot PC, et al. The influence of AUG codons in the hepatitis C virus 5′ nontranslated region on translation and mapping of the translation initiation window. Virology. 1996;226:47–56. doi: 10.1006/viro.1996.0626. [DOI] [PubMed] [Google Scholar]
- 44.Pilipenko EV, Gmyl AP, Maslova SV, et al. Starting window, a distinct element in the cap-independent internal initiation of translation on picornaviral RNA. J Mol Biol. 1994;241:398–414. doi: 10.1006/jmbi.1994.1516. [DOI] [PubMed] [Google Scholar]
- 45.López de Quinto S, Martinez-Salas E. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J Virol. 1997;71:4171–4175. doi: 10.1128/jvi.71.5.4171-4175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robertson ME, Seamons RA, Belsham GJ. A selection system for functional internal ribosome entry site (IRES) elements: analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA. 1999;5:1167–1179. doi: 10.1017/S1355838299990301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rijnbrand R, Bredenbeek P, van der Straaten T, et al. Almost the entire 5′ non-translated region of hepatitis C virus is required for cap-independent translation. FEBS Lett. 1995;365:115–119. doi: 10.1016/0014-5793(95)00458-L. [DOI] [PubMed] [Google Scholar]
- 48.Wilson JE, Powell MJ, Hoover SE, Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol Cell Biol. 2000;20:4990–4999. doi: 10.1128/MCB.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoffman MA, Palmenberg AC. Mutational analysis of the J-K stem-loop region of the encephalomyocarditis virus IRES. J Virol. 1995;69:4399–4406. doi: 10.1128/jvi.69.7.4399-4406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terenin IM, Dmitriev SE, Andreev DE, et al. A cross-kingdom internal ribosome entry site reveals a simplified mode of internal ribosome entry. Mol Cell Biol. 2005;25:7879–7888. doi: 10.1128/MCB.25.17.7879-7888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Groppelli E, Belsham GJ, Roberts LO. Identification of minimal sequences of the Rhopalosiphum padi virus 5′ untranslated region required for internal initiation of protein synthesis in mammalian, plant and insect translation systems. J Gen Virol. 2007;88:1583–1588. doi: 10.1099/vir.0.82682-0. [DOI] [PubMed] [Google Scholar]
- 52.Woolaway KE, Lazaridis K, Belsham GJ, et al. The 5′ untranslated region of Rhopalosiphum padi virus contains an internal ribosome entry site which functions efficiently in mammalian, plant, and insect translation systems. J Virol. 2001;75:10244–10249. doi: 10.1128/JVI.75.21.10244-10249.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abaeva IS, Pestova TV, Hellen CUT. Attachment of ribosomal complexes and retrograde scanning during initiation on the Halastavi árva virus IRES. Nucleic Acids Res. 2016;44:2362–2377. doi: 10.1093/nar/gkw016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sogorin EA, Agalarov SC, Spirin AS. Internal initiation of polyuridylic acid translation in bacterial cell-free system. Biochem Mosc. 2013;78:1354–1357. doi: 10.1134/S0006297913120055. [DOI] [PubMed] [Google Scholar]
- 55.Weill L, James L, Ulryck N, et al. A new type of IRES within gag coding region recruits three initiation complexes on HIV-2 genomic RNA. Nucleic Acids Res. 2010;38:1367–1381. doi: 10.1093/nar/gkp1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Breyne S, Chamond N, Decimo D, et al. In vitro studies reveal that different modes of initiation on HIV-1 mRNA have different levels of requirement for eukaryotic initiation factor 4F. FEBS J. 2012;279:3098–3111. doi: 10.1111/j.1742-4658.2012.08689.x. [DOI] [PubMed] [Google Scholar]
- 57.Locker N, Chamond N, Sargueil B. A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40S subunit and eIF3. Nucleic Acids Res. 2011;39:2367–2377. doi: 10.1093/nar/gkq1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ricci EP, Herbreteau CH, Decimo D, et al. In vitro expression of the HIV-2 genomic RNA is controlled by three distinct internal ribosome entry segments that are regulated by the HIV protease and the Gag polyprotein. RNA. 2008;14:1443–1455. doi: 10.1261/rna.813608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herbreteau CH, Weill L, Decimo D, et al. HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nat Struct Mol Biol. 2005;12:1001–1007. doi: 10.1038/nsmb1011. [DOI] [PubMed] [Google Scholar]
- 60.Hughes MJ, Andrews DW. A single nucleotide is a sufficient 5′ untranslated region for translation in an eukaryotic in vitro system. FEBS Lett. 1997;414:19–22. doi: 10.1016/S0014-5793(97)00965-4. [DOI] [PubMed] [Google Scholar]
- 61.Andreev DE, Terenin IM, Dunaevsky YE, et al. A leaderless mRNA can bind to mammalian 80S ribosomes and direct polypeptide synthesis in the absence of translation initiation factors. Mol Cell Biol. 2006;26:3164–3169. doi: 10.1128/MCB.26.8.3164-3169.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dmitriev SE, Terenin IM, Andreev DE, et al. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem. 2010;285:26779–26787. doi: 10.1074/jbc.M110.119693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Balakin AG, Skripkin EA, Shatsky IN, Bogdanov AA. Unusual ribosome binding properties of mRNA encoding bacteriophage lambda repressor. Nucleic Acids Res. 1992;20:563–571. doi: 10.1093/nar/20.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwartz S, Felber BK, Pavlakis GN. Mechanism of translation of monocistronic and multicistronic human immunodeficiency virus type 1 mRNAs. Mol Cell Biol. 1992;12:207–219. doi: 10.1128/MCB.12.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fouillot N, Tlouzeau S, Rossignol JM, Jean-Jean O. Translation of the hepatitis B virus P gene by ribosomal scanning as an alternative to internal initiation. J Virol. 1993;67:4886–4895. doi: 10.1128/jvi.67.8.4886-4895.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X-Q, Rothnagel JA. 5′-untranslated regions with multiple upstream AUG codons can support low-level translation via leaky scanning and reinitiation. Nucleic Acids Res. 2004;32:1382–1391. doi: 10.1093/nar/gkh305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Terenin IM, Akulich KA, Andreev DE, et al. Sliding of a 43S ribosomal complex from the recognized AUG codon triggered by a delay in eIF2-bound GTP hydrolysis. Nucleic Acids Res. 2016;44:1882–1893. doi: 10.1093/nar/gkv1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jackson RJ, Hellen CUT, Pestova TV. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- 69.Alisch RS, Garcia-Perez JL, Muotri AR, et al. Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev. 2006;20:210–224. doi: 10.1101/gad.1380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dmitriev SE, Andreev DE, Terenin IM, et al. Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5′ untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap dependent rather than internal ribosome entry site mediated. Mol Cell Biol. 2007;27:4685–4697. doi: 10.1128/MCB.02138-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dmitriev SE, Andreev DE, Adyanova ZV, et al. Efficient cap-dependent in vitro and in vivo translation of mammalian mRNAs with long and highly structured 5′-untranslated regions. Mol Biol (Mosk) 2009;43:108–113. doi: 10.1134/S0026893309010154. [DOI] [PubMed] [Google Scholar]
- 72.Berkhout B, Arts K, Abbink TEM. Ribosomal scanning on the 5′-untranslated region of the human immunodeficiency virus RNA genome. Nucleic Acids Res. 2011;39:5232–5244. doi: 10.1093/nar/gkr113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Belsham GJ, Nielsen I, Normann P, et al. Monocistronic mRNAs containing defective hepatitis C virus-like picornavirus internal ribosome entry site elements in their 5′ untranslated regions are efficiently translated in cells by a cap-dependent mechanism. RNA. 2008;14:1671–1680. doi: 10.1261/rna.1039708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vassilenko KS, Alekhina OM, Dmitriev SE, et al. Unidirectional constant rate motion of the ribosomal scanning particle during eukaryotic translation initiation. Nucleic Acids Res. 2011;39:5555–5567. doi: 10.1093/nar/gkr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol. 1989;9:5134–5142. doi: 10.1128/MCB.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Babendure JR, Babendure JL, Ding J-H, Tsien RY. Control of mammalian translation by mRNA structure near caps. RNA. 2006;12:851–861. doi: 10.1261/rna.2309906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muckenthaler M, Gray NK, Hentze MW. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol Cell. 1998;2:383–388. doi: 10.1016/S1097-2765(00)80282-8. [DOI] [PubMed] [Google Scholar]
- 78.Baird SD, Turcotte M, Korneluk RG, Holcík M. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ji Z, Song R, Regev A, Struhl K. Many lncRNAs, 5′UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife. 2015;4:e08890. doi: 10.7554/eLife.08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yanagiya A, Suyama E, Adachi H, et al. Translational homeostasis via the mRNA cap-binding protein, eIF4E. Mol Cell. 2012;46:847–858. doi: 10.1016/j.molcel.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Svitkin YV, Herdy B, Costa-Mattioli M, et al. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol Cell Biol. 2005;25:10556–10565. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mothe-Satney I, Yang D, Fadden P, et al. Multiple mechanisms control phosphorylation of PHAS-I in five (S/T)P sites that govern translational repression. Mol Cell Biol. 2000;20:3558–3567. doi: 10.1128/MCB.20.10.3558-3567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thoreen CC, Chantranupong L, Keys HR, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hsieh AC, Liu Y, Edlind MP, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marr MT, D’Alessio JA, Puig O, Tjian R. IRES-mediated functional coupling of transcription and translation amplifies insulin receptor feedback. Genes Dev. 2007;21:175–183. doi: 10.1101/gad.1506407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Beaudoin ME, Poirel V-J, Krushel LA. Regulating amyloid precursor protein synthesis through an internal ribosomal entry site. Nucleic Acids Res. 2008;36:6835–6847. doi: 10.1093/nar/gkn792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blau L, Knirsh R, Ben-Dror I, et al. Aberrant expression of c-Jun in glioblastoma by internal ribosome entry site (IRES)-mediated translational activation. Proc Natl Acad Sci USA. 2012;109:E2875–E2884. doi: 10.1073/pnas.1203659109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dobson T, Chen J, Krushel LA. Dysregulating IRES-dependent translation contributes to overexpression of oncogenic Aurora A Kinase. Mol Cancer Res. 2013;11:887–900. doi: 10.1158/1541-7786.MCR-12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Othman Z, Sulaiman MK, Willcocks MM, et al. Functional analysis of Kaposi’s sarcoma-associated herpesvirus vFLIP expression reveals a new mode of IRES-mediated translation. RNA. 2014;20:1803–1814. doi: 10.1261/rna.045328.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun J, Conn CS, Han Y, et al. PI3K-mTORC1 attenuates stress response by inhibiting cap-independent Hsp70 translation. J Biol Chem. 2011;286:6791–6800. doi: 10.1074/jbc.M110.172882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barco A, Feduchi E, Carrasco L. A stable HeLa cell line that inducibly expresses poliovirus 2A(pro): effects on cellular and viral gene expression. J Virol. 2000;74:2383–2392. doi: 10.1128/JVI.74.5.2383-2392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Amorim R, Costa SM, Cavaleiro NP, et al. HIV-1 transcripts use IRES-initiation under conditions where Cap-dependent translation is restricted by poliovirus 2A protease. PLoS One. 2014;9:e88619. doi: 10.1371/journal.pone.0088619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chau DHW, Yuan J, Zhang H, et al. Coxsackievirus B3 proteases 2A and 3C induce apoptotic cell death through mitochondrial injury and cleavage of eIF4GI but not DAP5/p97/NAT1. Apoptosis. 2007;12:513–524. doi: 10.1007/s10495-006-0013-0. [DOI] [PubMed] [Google Scholar]
- 94.Badura M, Braunstein S, Zavadil J, Schneider RJ. DNA damage and eIF4G1 in breast cancer cells reprogram translation for survival and DNA repair mRNAs. Proc Natl Acad Sci USA. 2012;109:18767–18772. doi: 10.1073/pnas.1203853109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ramírez-Valle F, Braunstein S, Zavadil J, et al. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J Cell Biol. 2008;181:293–307. doi: 10.1083/jcb.200710215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bushell M, Poncet D, Marissen WE, et al. Cleavage of polypeptide chain initiation factor eIF4GI during apoptosis in lymphoma cells: characterisation of an internal fragment generated by caspase-3-mediated cleavage. Cell Death Differ. 2000;7:628–636. doi: 10.1038/sj.cdd.4400699. [DOI] [PubMed] [Google Scholar]
- 97.Andreev DE, Dmitriev SE, Zinovkin R, et al. The 5′ untranslated region of Apaf-1 mRNA directs translation under apoptosis conditions via a 5′ end-dependent scanning mechanism. FEBS Lett. 2012;586:4139–4143. doi: 10.1016/j.febslet.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 98.Venkatesan A, Sharma R, Dasgupta A. Cell cycle regulation of hepatitis C and encephalomyocarditis virus internal ribosome entry site-mediated translation in human embryonic kidney 293 cells. Virus Res. 2003;94:85–95. doi: 10.1016/S0168-1702(03)00136-9. [DOI] [PubMed] [Google Scholar]
- 99.Qin X, Sarnow P. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J Biol Chem. 2004;279:13721–13728. doi: 10.1074/jbc.M312854200. [DOI] [PubMed] [Google Scholar]
- 100.Fan H, Penman S. Regulation of protein synthesis in mammalian cells. II. Inhibition of protein synthesis at the level of initiation during mitosis. J Mol Biol. 1970;50:655–670. doi: 10.1016/0022-2836(70)90091-4. [DOI] [PubMed] [Google Scholar]
- 101.Stumpf CR, Moreno MV, Olshen AB, et al. The translational landscape of the mammalian cell cycle. Mol Cell. 2013;52:574–582. doi: 10.1016/j.molcel.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Coldwell MJ, Cowan JL, Vlasak M, et al. Phosphorylation of eIF4GII and 4E-BP1 in response to nocodazole treatment: a reappraisal of translation initiation during mitosis. Cell Cycle. 2013;12:3615–3628. doi: 10.4161/cc.26588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pyronnet S, Dostie J, Sonenberg N. Suppression of cap-dependent translation in mitosis. Genes Dev. 2001;15:2083–2093. doi: 10.1101/gad.889201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shuda M, Velásquez C, Cheng E, et al. CDK1 substitutes for mTOR kinase to activate mitotic cap-dependent protein translation. Proc Natl Acad Sci USA. 2015;112:5875–5882. doi: 10.1073/pnas.1505787112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sivan G, Kedersha N, Elroy-Stein O. Ribosomal slowdown mediates translational arrest during cellular division. Mol Cell Biol. 2007;27:6639–6646. doi: 10.1128/MCB.00798-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sivan G, Aviner R, Elroy-Stein O. Mitotic modulation of translation elongation factor 1 leads to hindered tRNA delivery to ribosomes. J Biol Chem. 2011;286:27927–27935. doi: 10.1074/jbc.M111.255810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kieft JS. Viral IRES RNA structures and ribosome interactions. Trends Biochem Sci. 2008;33:274–283. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Malys N, McCarthy JEG. Translation initiation: variations in the mechanism can be anticipated. Cell Mol Life Sci. 2011;68:991–1003. doi: 10.1007/s00018-010-0588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Williamson AR. The attachment of polyuridylic acid to reticulocyte ribosomes. Biochem J. 1969;111:515–520. doi: 10.1042/bj1110515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Iwasaki K. Translation of poly(A) in eukaryotic cell-free systems. J Biochem. 1982;91:1617–1627. doi: 10.1093/oxfordjournals.jbchem.a133853. [DOI] [PubMed] [Google Scholar]
- 111.Kolupaeva VG, Unbehaun A, Lomakin IB, et al. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA. 2005;11:470–486. doi: 10.1261/rna.7215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Seal SN, Schmidt A, Marcus A. Ribosome binding to inosine-substituted mRNAs in the absence of ATP and mRNA factors. J Biol Chem. 1989;264:7363–7368. [PubMed] [Google Scholar]
- 113.Bai Y, Zhou K, Doudna JA. Hepatitis C virus 3′UTR regulates viral translation through direct interactions with the host translation machinery. Nucleic Acids Res. 2013;41:7861–7874. doi: 10.1093/nar/gkt543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Asnani M, Pestova TV, Hellen CUT. Initiation on the divergent Type I cadicivirus IRES: factor requirements and interactions with the translation apparatus. Nucleic Acids Res. 2016;44:3390–3407. doi: 10.1093/nar/gkw074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Skabkin MA, Skabkina OV, Dhote V, et al. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xue S, Tian S, Fujii K, et al. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature. 2015;517:33–38. doi: 10.1038/nature14010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Allam H, Ali N. Initiation factor eIF2-independent mode of c-Src mRNA translation occurs via an internal ribosome entry site. J Biol Chem. 2010;285:5713–5725. doi: 10.1074/jbc.M109.029462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pestova TV, Shatsky IN, Fletcher SP, et al. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sizova DV, Kolupaeva VG, Pestova TV, et al. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nousch M, Reed V, Bryson-Richardson RJ, et al. The eIF4G-homolog p97 can activate translation independent of caspase cleavage. RNA. 2007;13:374–384. doi: 10.1261/rna.372307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.De Gregorio E, Preiss T, Hentze MW. Translation driven by an eIF4G core domain in vivo. EMBO J. 1999;18:4865–4874. doi: 10.1093/emboj/18.17.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kozak M. Influence of mRNA secondary structure on binding and migration of 40S ribosomal subunits. Cell. 1980;19:79–90. doi: 10.1016/0092-8674(80)90390-6. [DOI] [PubMed] [Google Scholar]
- 123.Morgan MA, Shatkin AJ. Initiation of reovirus transcription by inosine 5′-triphosphate and properties of 7-methylinosine-capped, inosine-substituted messenger ribonucleic acids. Biochemistry. 1980;19:5960–5966. doi: 10.1021/bi00567a003. [DOI] [PubMed] [Google Scholar]
- 124.Seal SN, Schmidt A, Sonenberg N, Marcus A. Initiation factors eIF4A and C1 from wheat germ and the formation of mRNA X ribosome complexes. Arch Biochem Biophys. 1985;238:146–153. doi: 10.1016/0003-9861(85)90150-X. [DOI] [PubMed] [Google Scholar]
- 125.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang J, Zhao F, Zhao Y, et al. Hypoxia induces an increase in intracellular magnesium via transient receptor potential melastatin 7 (TRPM7) channels in rat hippocampal neurons in vitro. J Biol Chem. 2011;286:20194–20207. doi: 10.1074/jbc.M110.148494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tashiro M, Inoue H, Konishi M. Physiological pathway of magnesium influx in rat ventricular myocytes. Biophys J. 2014;107:2049–2058. doi: 10.1016/j.bpj.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shenvi CL, Dong KC, Friedman EM, et al. Accessibility of 18S rRNA in human 40S subunits and 80S ribosomes at physiological magnesium ion concentrations—implications for the study of ribosome dynamics. RNA. 2005;11:1898–1908. doi: 10.1261/rna.2192805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pestova TV, Borukhov SI, Hellen CU. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 130.Otto GA, Lukavsky PJ, Lancaster AM, et al. Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. RNA. 2002;8:913–923. doi: 10.1017/S1355838202022057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Thakor N, Holcík M. IRES-mediated translation of cellular messenger RNA operates in eIF2α-independent manner during stress. Nucleic Acids Res. 2012;40:541–552. doi: 10.1093/nar/gkr701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chamond N, Deforges J, Ulryck N, Sargueil B. 40S recruitment in the absence of eIF4G/4A by EMCV IRES refines the model for translation initiation on the archetype of Type II IRESs. Nucleic Acids Res. 2014;42:10373–10384. doi: 10.1093/nar/gku720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kolupaeva VG, Lomakin IB, Pestova TV, Hellen CUT. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol Cell Biol. 2003;23:687–698. doi: 10.1128/MCB.23.2.687-698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Soto Rifo R, Rubilar PS, Limousin T, et al. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 2012;31:3745–3756. doi: 10.1038/emboj.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pisarev AV, Chard LS, Kaku Y, et al. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J Virol. 2004;78:4487–4497. doi: 10.1128/JVI.78.9.4487-4497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pánek J, Kolár M, Vohradský J, Shivaya Valásek L. An evolutionary conserved pattern of 18S rRNA sequence complementarity to mRNA 5′ UTRs and its implications for eukaryotic gene translation regulation. Nucleic Acids Res. 2013;41:7625–7634. doi: 10.1093/nar/gkt548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Deforges J, Locker N, Sargueil B. mRNAs that specifically interact with eukaryotic ribosomal subunits. Biochimie. 2015;114:48–57. doi: 10.1016/j.biochi.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 138.Stupina VA, Yuan X, Meskauskas A, et al. Ribosome binding to a 5′ translational enhancer is altered in the presence of the 3′ untranslated region in cap-independent translation of turnip crinkle virus. J Virol. 2011;85:4638–4653. doi: 10.1128/JVI.00005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stupina VA, Meskauskas A, McCormack JC, et al. The 3′ proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits. RNA. 2008;14:2379–2393. doi: 10.1261/rna.1227808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chappell SA, Edelman GM, Mauro VP. Biochemical and functional analysis of a 9-nt RNA sequence that affects translation efficiency in eukaryotic cells. Proc Natl Acad Sci USA. 2004;101:9590–9594. doi: 10.1073/pnas.0308759101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dresios J, Chappell SA, Zhou W, Mauro VP. An mRNA–rRNA base-pairing mechanism for translation initiation in eukaryotes. Nat Struct Mol Biol. 2006;13:30–34. doi: 10.1038/nsmb1031. [DOI] [PubMed] [Google Scholar]
- 142.Panopoulos P, Mauro VP. Antisense masking reveals contributions of mRNA–rRNA base pairing to translation of Gtx and FGF2 mRNAs. J Biol Chem. 2008;283:33087–33093. doi: 10.1074/jbc.M804904200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chappell SA, Edelman GM, Mauro VP. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc Natl Acad Sci USA. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kozak M. New ways of initiating translation in eukaryotes? Mol Cell Biol. 2001;21:1899–1907. doi: 10.1128/MCB.21.6.1899-1907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Severin J, Lizio M, Harshbarger J, et al. Interactive visualization and analysis of large-scale sequencing datasets using ZENBU. Nat Biotechnol. 2014;32:217–219. doi: 10.1038/nbt.2840. [DOI] [PubMed] [Google Scholar]
- 146.Dreos R, Ambrosini G, Périer RC, Bucher P. The Eukaryotic Promoter Database: expansion of EPDnew and new promoter analysis tools. Nucleic Acids Res. 2015;43:D92–D96. doi: 10.1093/nar/gku1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Riley A, Jordan LE, Holcík M. Distinct 5′ UTRs regulate XIAP expression under normal growth conditions and during cellular stress. Nucleic Acids Res. 2010;38:4665–4674. doi: 10.1093/nar/gkq241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Arribere JA, Gilbert WV. Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome Res. 2013;23:977–987. doi: 10.1101/gr.150342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang X, Hou J, Quedenau C, Chen W. Pervasive isoform-specific translational regulation via alternative transcription start sites in mammals. Molecular Systems Biology. 2016;12:875. doi: 10.15252/msb.20166941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Floor SN, Doudna JA. Tunable protein synthesis by transcript isoforms in human cells. Elife. 2016;5:e10921. doi: 10.7554/eLife.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Andreev DE, Terenin IM, Dmitriev SE, Shatsky IN. Pros and cons of pDNA and mRNA transfection to study mRNA translation in mammalian cells. Gene. 2016;578:1–6. doi: 10.1016/j.gene.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 152.Lemp NA, Hiraoka K, Kasahara N, Logg CR. Cryptic transcripts from a ubiquitous plasmid origin of replication confound tests for cis-regulatory function. Nucleic Acids Res. 2012;40:7280–7290. doi: 10.1093/nar/gks451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Vopalensky V, Masek T, Horvath O, et al. Firefly luciferase gene contains a cryptic promoter. RNA. 2008;14:1720–1729. doi: 10.1261/rna.831808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Weingarten-Gabbay S, Elias-Kirma S, Nir R et al (2016) Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351:aad4939–aad4939. doi:10.1126/science.aad4939 [DOI] [PubMed]
- 155.van Eden ME, Byrd MP, Sherrill KW, Lloyd RE. Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA. 2004;10:720–730. doi: 10.1261/rna.5225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jiang H, Coleman J, Miskimins R, et al. Cap-independent translation through the p27 5′-UTR. Nucleic Acids Res. 2007;35:4767–4778. doi: 10.1093/nar/gkm512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Olivares E, Landry DM, Cáceres CJ, et al. The 5′ untranslated region of the human T-cell lymphotropic virus type 1 mRNA enables cap-independent translation initiation. J Virol. 2014;88:5936–5955. doi: 10.1128/JVI.00279-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bert AG, Grépin R, Vadas MA, Goodall GJ. Assessing IRES activity in the HIF-1alpha and other cellular 5′ UTRs. RNA. 2006;12:1074–1083. doi: 10.1261/rna.2320506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.de Almeida RA, Heuser T, Blaschke R, et al. Control of MYEOV protein synthesis by upstream open reading frames. J Biol Chem. 2006;281:695–704. doi: 10.1074/jbc.M511467200. [DOI] [PubMed] [Google Scholar]
- 160.Dumas E, Staedel C, Colombat M, et al. A promoter activity is present in the DNA sequence corresponding to the hepatitis C virus 5′ UTR. Nucleic Acids Res. 2003;31:1275–1281. doi: 10.1093/nar/gkg199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Elango N, Li Y, Shivshankar P, Katz MS. Expression of RUNX2 isoforms: involvement of cap-dependent and cap-independent mechanisms of translation. J Cell Biochem. 2006;99:1108–1121. doi: 10.1002/jcb.20909. [DOI] [PubMed] [Google Scholar]
- 162.Gendra E, Colgan DF, Meany B, Konarska MM. A sequence motif in the simian virus 40 (SV40) early core promoter affects alternative splicing of transcribed mRNA. J Biol Chem. 2007;282:11648–11657. doi: 10.1074/jbc.M611126200. [DOI] [PubMed] [Google Scholar]
- 163.Zid BM, O’Shea EK. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature. 2014;514:117–121. doi: 10.1038/nature13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bregman A, Avraham-Kelbert M, Barkai O, et al. Promoter elements regulate cytoplasmic mRNA decay. Cell. 2011;147:1473–1483. doi: 10.1016/j.cell.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 165.Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- 166.Stöhr N, Lederer M, Reinke C, et al. ZBP1 regulates mRNA stability during cellular stress. J Cell Biol. 2006;175:527–534. doi: 10.1083/jcb.200608071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Drinnenberg IA, Weinberg DE, Xie KT, et al. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Benard L. Inhibition of 5′ to 3′ mRNA degradation under stress conditions in Saccharomyces cerevisiae: from GCN4 to MET16. RNA. 2004;10:458–468. doi: 10.1261/rna.5183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gilbert WV, Zhou K, Butler TK, Doudna JA. Cap-independent translation is required for starvation-induced differentiation in yeast. Science. 2007;317:1224–1227. doi: 10.1126/science.1144467. [DOI] [PubMed] [Google Scholar]
- 170.Olson CM, Donovan MR, Spellberg MJ, Marr MT. The insulin receptor cellular IRES confers resistance to eIF4A inhibition. Elife. 2013;2:e00542. doi: 10.7554/eLife.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhao Y, Qin W, Zhang J-P, et al. HCV IRES-mediated core expression in zebrafish. PLoS One. 2013;8:e56985. doi: 10.1371/journal.pone.0056985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Masek T, Vopalensky V, Horvath O, et al. Hepatitis C virus internal ribosome entry site initiates protein synthesis at the authentic initiation codon in yeast. J Gen Virol. 2007;88:1992–2002. doi: 10.1099/vir.0.82782-0. [DOI] [PubMed] [Google Scholar]
- 173.Rosenfeld AB, Racaniello VR. Components of the multifactor complex needed for internal initiation by the IRES of hepatitis C virus in Saccharomyces cerevisiae. RNA Biol. 2010;7:596–605. doi: 10.4161/rna.7.5.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Coward P, Dasgupta A. Yeast cells are incapable of translating RNAs containing the poliovirus 5′ untranslated region: evidence for a translational inhibitor. J Virol. 1992;66:286–295. doi: 10.1128/jvi.66.1.286-295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Evstafieva AG, Beletsky AV, Borovjagin AV, Bogdanov AA. Internal ribosome entry site of encephalomyocarditis virus RNA is unable to direct translation in Saccharomyces cerevisiae. FEBS Lett. 1993;335:273–276. doi: 10.1016/0014-5793(93)80745-G. [DOI] [PubMed] [Google Scholar]
- 176.Dorokhov YL, Skulachev MV, Ivanov PA, et al. Polypurine (A)-rich sequences promote cross-kingdom conservation of internal ribosome entry. Proc Natl Acad Sci USA. 2002;99:5301–5306. doi: 10.1073/pnas.082107599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Pestova TV, Shatsky IN, Hellen CU. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/MCB.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Thompson SR, Gulyas KD, Sarnow P. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc Natl Acad Sci USA. 2001;98:12972–12977. doi: 10.1073/pnas.241286698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jünemann C, Song Y, Bassili G, et al. Picornavirus internal ribosome entry site elements can stimulate translation of upstream genes. J Biol Chem. 2007;282:132–141. doi: 10.1074/jbc.M608750200. [DOI] [PubMed] [Google Scholar]
- 180.Byrd MP, Zamora M, Lloyd RE. Translation of eukaryotic translation initiation factor 4GI (eIF4GI) proceeds from multiple mRNAs containing a novel cap-dependent internal ribosome entry site (IRES) that is active during poliovirus infection. J Biol Chem. 2005;280:18610–18622. doi: 10.1074/jbc.M414014200. [DOI] [PubMed] [Google Scholar]
- 181.Honda M, Kaneko S, Matsushita E, et al. Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology. 2000;118:152–162. doi: 10.1016/S0016-5085(00)70424-0. [DOI] [PubMed] [Google Scholar]
- 182.van Eden ME, Byrd MP, Sherrill KW, Lloyd RE. Translation of cellular inhibitor of apoptosis protein 1 (c-IAP1) mRNA is IRES mediated and regulated during cell stress. RNA. 2004;10:469–481. doi: 10.1261/rna.5156804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gallie DR, Ling J, Niepel M, et al. The role of 5′-leader length, secondary structure and PABP concentration on cap and poly(A) tail function during translation in Xenopus oocytes. Nucleic Acids Res. 2000;28:2943–2953. doi: 10.1093/nar/28.15.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mardanova ES, Zamchuk LA, Skulachev MV, Ravin NV. The 5′ untranslated region of the maize alcohol dehydrogenase gene contains an internal ribosome entry site. Gene. 2008;420:11–16. doi: 10.1016/j.gene.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 185.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Konarska M, Filipowicz W, Domdey H, Gross HJ. Binding of ribosomes to linear and circular forms of the 5′-terminal leader fragment of tobacco-mosaic-virus RNA. Eur J Biochem. 1981;114:221–227. doi: 10.1111/j.1432-1033.1981.tb05139.x. [DOI] [PubMed] [Google Scholar]
- 187.Kozak M. Inability of circular mRNA to attach to eukaryotic ribosomes. Nature. 1979;280:82–85. doi: 10.1038/280082a0. [DOI] [PubMed] [Google Scholar]
- 188.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 189.Chen CY, Sarnow P. Internal ribosome entry sites tests with circular mRNAs. Methods Mol Biol. 1998;77:355–363. doi: 10.1385/0-89603-397-X:355. [DOI] [PubMed] [Google Scholar]
- 190.Abe N, Matsumoto K, Nishihara M, et al. Rolling circle translation of circular RNA in living human cells. Sci Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Arias C, Weisburd B, Stern-Ginossar N, et al. KSHV 2.0: a comprehensive annotation of the Kaposi’s sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog. 2014;10:e1003847. doi: 10.1371/journal.ppat.1003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Grainger L, Cicchini L, Rak M, et al. Stress-inducible alternative translation initiation of human cytomegalovirus latency protein pUL138. J Virol. 2010;84:9472–9486. doi: 10.1128/JVI.00855-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Talbot SJ, Weiss RA, Kellam P, Boshoff C. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology. 1999;257:84–94. doi: 10.1006/viro.1999.9672. [DOI] [PubMed] [Google Scholar]
- 194.Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987;7:3438–3445. doi: 10.1128/MCB.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bieleski L, Talbot SJ. Kaposi’s sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J Virol. 2001;75:1864–1869. doi: 10.1128/JVI.75.4.1864-1869.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Low W, Harries M, Ye H, et al. Internal ribosome entry site regulates translation of Kaposi’s sarcoma-associated herpesvirus FLICE inhibitory protein. J Virol. 2001;75:2938–2945. doi: 10.1128/JVI.75.6.2938-2945.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Stern-Ginossar N, Weisburd B, Michalski A, et al. Decoding human cytomegalovirus. Science. 2012;338:1088–1093. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Cornelis S, Bruynooghe Y, Denecker G, et al. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol Cell. 2000;5:597–605. doi: 10.1016/S1097-2765(00)80239-7. [DOI] [PubMed] [Google Scholar]
- 199.Haimov O, Sinvani H, Dikstein R. Cap-dependent, scanning-free translation initiation mechanisms. Biochim Biophys Acta. 2015;1849:1313–1318. doi: 10.1016/j.bbagrm.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 200.Chappell SA, Edelman GM, Mauro VP. Ribosomal tethering and clustering as mechanisms for translation initiation. Proc Natl Acad Sci USA. 2006;103:18077–18082. doi: 10.1073/pnas.0608212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Thoma C, Fraterman S, Gentzel M, et al. Translation initiation by the c-myc mRNA internal ribosome entry sequence and the poly(A) tail. RNA. 2008;14:1579–1589. doi: 10.1261/rna.1043908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Kaiser C, Dobrikova EY, Bradrick SS, et al. Activation of cap-independent translation by variant eukaryotic initiation factor 4G in vivo. RNA. 2008;14:2170–2182. doi: 10.1261/rna.1171808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Carrington JC, Freed DD. Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J Virol. 1990;64:1590–1597. doi: 10.1128/jvi.64.4.1590-1597.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wellensiek BP, Larsen AC, Stephens B, et al. Genome-wide profiling of human cap-independent translation-enhancing elements. Nat Methods. 2013;10:747–750. doi: 10.1038/nmeth.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Soto Rifo R, Ricci EP, Décimo D, et al. Back to basics: the untreated rabbit reticulocyte lysate as a competitive system to recapitulate cap/poly(A) synergy and the selective advantage of IRES-driven translation. Nucleic Acids Res. 2007;35:e121. doi: 10.1093/nar/gkm682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Novoa I, Martínez-Abarca F, Fortes P, et al. Cleavage of p220 by purified poliovirus 2A(pro) in cell-free systems: effects on translation of capped and uncapped mRNAs. Biochemistry. 1997;36:7802–7809. doi: 10.1021/bi9631172. [DOI] [PubMed] [Google Scholar]
- 207.Monette A, Valiente-Echeverría F, Rivero M, et al. Dual Mechanisms of Translation Initiation of the Full-Length HIV-1 mRNA Contribute to Gag Synthesis. PLoS One. 2013;8:e68108. doi: 10.1371/journal.pone.0068108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Brasey A, Lopez-Lastra M, Ohlmann T, et al. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J Virol. 2003;77:3939–3949. doi: 10.1128/JVI.77.7.3939-3949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ventoso I, Blanco R, Perales C, Carrasco L. HIV-1 protease cleaves eukaryotic initiation factor 4G and inhibits cap-dependent translation. Proc Natl Acad Sci USA. 2001;98:12966–12971. doi: 10.1073/pnas.231343498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Perales C, Carrasco L, Ventoso I. Cleavage of eIF4G by HIV-1 protease: effects on translation. FEBS Lett. 2003;533:89–94. doi: 10.1016/S0014-5793(02)03764-X. [DOI] [PubMed] [Google Scholar]
- 211.Castelló A, Franco D, Moral-López P, et al. HIV- 1 protease inhibits Cap- and poly(A)-dependent translation upon eIF4GI and PABP cleavage. PLoS One. 2009;4:e7997. doi: 10.1371/journal.pone.0007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat Struct Mol Biol. 2008;15:836–841. doi: 10.1038/nsmb.1445. [DOI] [PubMed] [Google Scholar]
- 213.Anastasina M, Terenin I, Butcher SJ, Kainov DE. A technique to increase protein yield in a rabbit reticulocyte lysate translation system. Biotechniques. 2014;56:36–39. doi: 10.2144/000114125. [DOI] [PubMed] [Google Scholar]
- 214.Stoneley M, Paulin FE, Le Quesne JP, et al. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 215.Reynolds JE, Kaminski A, Kettinen HJ, et al. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Fletcher SP, Ali IK, Kaminski A, et al. The influence of viral coding sequences on pestivirus IRES activity reveals further parallels with translation initiation in prokaryotes. RNA. 2002;8:1558–1571. [PMC free article] [PubMed] [Google Scholar]
- 217.Andreev DE, O’Connor PBF, Fahey C, et al. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. Elife. 2015;4:e03971. doi: 10.7554/eLife.03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell. 1975;4:379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 219.Furuichi Y, Morgan M, Shatkin AJ, et al. Methylated, blocked 5 termini in HeLa cell mRNA. Proc Natl Acad Sci USA. 1975;72:1904–1908. doi: 10.1073/pnas.72.5.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Keith JM, Ensinger MJ, Mose B. HeLa cell RNA (2′-O-methyladenosine-N6-)-methyltransferase specific for the capped 5′-end of messenger RNA. J Biol Chem. 1978;253:5033–5039. [PubMed] [Google Scholar]
- 221.Kuge H, Brownlee GG, Gershon PD, Richter JD. Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis. Nucleic Acids Res. 1998;26:3208–3214. doi: 10.1093/nar/26.13.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Muthukrishnan S, Moss B, Cooper JA, Maxwell ES. Influence of 5′-terminal cap structure on the initiation of translation of vaccinia virus mRNA. J Biol Chem. 1978;253:1710–1715. [PubMed] [Google Scholar]
- 223.Bélanger F, Stepinski J, Darzynkiewicz E, Pelletier J. Characterization of hMTr1, a human Cap1 2′-O-ribose methyltransferase. J Biol Chem. 2010;285:33037–33044. doi: 10.1074/jbc.M110.155283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sripati CE, Groner Y, Warner JR. Methylated, blocked 5′ termini of yeast mRNA. J Biol Chem. 1976;251:2898–2904. [PubMed] [Google Scholar]
- 225.Daffis S, Szretter KJ, Schriewer J, et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kumar P, Sweeney TR, Skabkin MA, et al. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5′ ppp-mRNAs. Nucleic Acids Res. 2014;42:3228–3245. doi: 10.1093/nar/gkt1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Preskey D, Allison TF, Jones M, et al. Synthetically modified mRNA for efficient and fast human iPS cell generation and direct transdifferentiation to myoblasts. Biochem Biophys Res Commun. 2016;437:743–751. doi: 10.1016/j.bbrc.2015.09.102. [DOI] [PubMed] [Google Scholar]
- 228.Warren L, Wang J (2013) Feeder-free reprogramming of human fibroblasts with messenger RNA. Curr Protoc Stem Cell Biol 27:4A.6.1–4A.6.27. doi:10.1002/9780470151808.sc04a06s27 [DOI] [PubMed]
- 229.Rohani L, Fabian C, Holland H, et al. Generation of human induced pluripotent stem cells using non-synthetic mRNA. Stem Cell Res. 2016;16:662–672. doi: 10.1016/j.scr.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 230.Fuerst TR, Moss B. Structure and stability of mRNA synthesized by vaccinia virus-encoded bacteriophage T7 RNA polymerase in mammalian cells. Importance of the 5′ untranslated leader. J Mol Biol. 1989;206:333–348. doi: 10.1016/0022-2836(89)90483-X. [DOI] [PubMed] [Google Scholar]
- 231.Bahar Halpern K, Veprik A, Rubins N, et al. GPR41 gene expression is mediated by internal ribosome entry site (IRES)-dependent translation of bicistronic mRNA encoding GPR40 and GPR41 proteins. J Biol Chem. 2012;287:20154–20163. doi: 10.1074/jbc.M112.358887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Hornung V, Ellegast J, Kim S, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 233.Nallagatla SR, Hwang J, Toroney R, et al. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 234.Anderson BR, Karikó K, Weissman D. Nucleofection induces transient eIF2α phosphorylation by GCN2 and PERK. Gene Ther. 2013;20:136–142. doi: 10.1038/gt.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Barreau C, Dutertre S, Paillard L, Osborne HB. Liposome-mediated RNA transfection should be used with caution. RNA. 2006;12:1790–1793. doi: 10.1261/rna.191706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Chiu W-W, Kinney RM, Dreher TW. Control of translation by the 5′- and 3′-terminal regions of the dengue virus genome. J Virol. 2005;79:8303–8315. doi: 10.1128/JVI.79.13.8303-8315.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Bradrick SS, Walters RW, Gromeier M. The hepatitis C virus 3′-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res. 2006;34:1293–1303. doi: 10.1093/nar/gkl019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Kopeina GS, Afonina ZA, Gromova KV, et al. Step-wise formation of eukaryotic double-row polyribosomes and circular translation of polysomal mRNA. Nucleic Acids Res. 2008;36:2476–2488. doi: 10.1093/nar/gkm1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Afonina ZA, Myasnikov AG, Khabibullina NF, et al. Topology of mRNA chain in isolated eukaryotic double-row polyribosomes. Biochem Mosc. 2013;78:445–454. doi: 10.1134/S0006297913050027. [DOI] [PubMed] [Google Scholar]
- 240.Asselbergs FA, Peters W, Venrooij WJ, Bloemendal H. Diminished sensitivity of re-initiation of translation to inhibition by cap analogues in reticulocyte lysates. Eur J Biochem. 1978;88:483–488. doi: 10.1111/j.1432-1033.1978.tb12473.x. [DOI] [PubMed] [Google Scholar]
- 241.Amrani N, Ghosh S, Mangus DA, Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Nelson EM, Winkler MM. Regulation of mRNA entry into polysomes. Parameters affecting polysome size and the fraction of mRNA in polysomes. J Biol Chem. 1987;262:11501–11506. [PubMed] [Google Scholar]
- 243.Thoma C, Ostareck-Lederer A, Hentze MW. A poly(A) tail-responsive in vitro system for cap- or IRES-driven translation from HeLa cells. Methods Mol Biol. 2004;257:171–180. doi: 10.1385/1-59259-750-5:171. [DOI] [PubMed] [Google Scholar]
- 244.Bergamini G, Preiss T, Hentze MW. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA. 2000;6:1781–1790. doi: 10.1017/S1355838200001679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Svitkin YV, Sonenberg N. An efficient system for cap- and poly(A)-dependent translation in vitro. Methods Mol Biol. 2004;257:155–170. doi: 10.1385/1-59259-750-5:155. [DOI] [PubMed] [Google Scholar]
- 246.Lyons SM, Achorn C, Kedersha NL, et al. YB-1 regulates tiRNA-induced Stress Granule formation but not translational repression. Nucleic Acids Res. 2016;44:6949–6960. doi: 10.1093/nar/gkw418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Dmitriev SE, Bykova NV, Andreev DE, Terenin IM. Adequate system for studying translation initiation on the human retrotransposon L1 mRNA in vitro. Mol Biol. 2006;40:20–24. doi: 10.1134/S0026893306010043. [DOI] [PubMed] [Google Scholar]
- 248.Pestova TV, Kolupaeva VG, Lomakin IB, et al. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Stumpf CR, Ruggero D. The cancerous translation apparatus. Curr Opin Genet Dev. 2011;21:474–483. doi: 10.1016/j.gde.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Ruggero D. Translational control in cancer etiology. Cold Spring Harb Perspect Biol. 2013;5:a012336. doi: 10.1101/cshperspect.a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Horvilleur E, Wilson LA, Bastide A, et al. Cap-independent translation in hematological malignancies. Front Oncol. 2015;5:293. doi: 10.3389/fonc.2015.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Didiot M-C, Hewett J, Varin T, et al. Identification of cardiac glycoside molecules as inhibitors of c-Myc IRES-mediated translation. J Biomol Screen. 2013;18:407–419. doi: 10.1177/1087057112466698. [DOI] [PubMed] [Google Scholar]
- 253.Shi Y, Yang Y, Hoang B, et al. Therapeutic potential of targeting IRES-dependent c-myc translation in multiple myeloma cells during ER stress. Oncogene. 2016;35:1015–1024. doi: 10.1038/onc.2015.156. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]