Abstract
Colorectal cancer (CRC) is a leading cause of cancer-related deaths that is often associated with inflammation initiated by activation of pattern recognition receptors (PRRs). Nucleic acid sensing PRRs are one of the major subsets of PRRs that sense nucleic acid (DNA and RNA), mainly including some members of Toll-like receptors (TLR3, 7, 8, 9), AIM2-like receptors (AIM2, IFI16), STING, cGAS, RNA polymerase III, and DExD/H box nucleic acid helicases (such as RIG-I like receptors (RIG-I, MDA5, LPG2), DDX1, 3, 5, 7, 17, 21, 41, 60, and DHX9, 36). Activation of these receptors eventually leads to the release of cytokines and activation of immune cells, which are well known to play crucial roles in host defense against intracellular bacterial and virus infection. However, the functions of these nucleic acid sensing PRRs in the other diseases such as CRC and colitis remain largely unknown. Recent studies indicated that nucleic acid sensing PRRs contribute to CRC and/or colitis development, and therapeutic modulation of nucleic acid sensing PRRs may reduce the risk of CRC development. However, until now, a comprehensive review on the role of nucleic acid sensing PRRs in CRC and colitis is still lacking. This review provided an overview of the roles as well as the mechanisms of these nucleic acid sensing PRRs (AIM2, STING, cGAS, RIG-I and its downstream molecules, DDX3, 5, 6,17, and DHX9, 36) in CRC and colitis, which may aid the diagnosis, therapy, and prognostic prediction of CRC and colitis.
Keywords: Cell proliferation, Colitis, Colorectal cancer, Nucleic acid sensing pattern recognition receptors, Type I interferon
Introduction
Colorectal cancer (CRC) is one of the most common malignant digestive tract tumors, which is characterized by abdomen masses, bloody stool, and changes in stool character, etc. The incidence and mortality of CRC are about 1.4 million and 693,900 in worldwide in 2012, respectively [1]. Incidence and mortality rate trends of CRC commonly decreased in more developed countries, while increased in less developed countries [2]. According to data from the National Central Cancer Registry of China, it is estimated that there will be about 0.38 million new cases and about 0.19 million deaths caused by CRC in 2015 [3], indicating that CRC poses a serious threat to the health of Chinese people. Mechanisms of tumorigenesis and progression of CRC are very complicated and may include interactions among environmental exposures, diet, and heredity [4]. Inflammation is a putative risk factor for CRC, and emerging clinical studies show that patients with inflammatory bowel disease (IBD) have higher risk of developing CRC than controls [4].
Inflammation is normally initiated by activation of innate immune cells, which express a broad range of sensors termed pattern recognition receptors (PRRs) [5]. PRRs are localized to the plasma membrane, cytosol, or endosomes [5]. The main types of PRRs include Toll-like receptors (TLRs), Nod-like receptors (NLRs), RIG-I like receptors (RLRs), AIM2-like receptors (ALRs), and C-type Lectin receptors (CLRs). Upon activation of PRRs by pathogen-associated molecular patterns (PAMPs) and/or danger-associated molecular patterns (DAMPs), innate immune cells undergo a series of signaling cascades that lead to the production of diverse pro-inflammatory cytokines and activation of adaptive immune cells to protect the host from invading pathogens [6]. Additionally, PRRs (especially the TLRs and the NLRs) play crucial roles in maintaining intestinal homeostasis and regulating the development of IBD and CRC [7–9].
Many PRRs are involved in detecting pathogenic DNA and RNA or self-DNA and self-RNA in cells and are thus referred to as nucleic acid sensing PRRs. PRRs responsible for RNA recognition include TLRs (such as TLR3, 7, 8, and 13), RLRs [such as retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5)], and DExD/H box (the x can be any amino acid) nucleic acid helicases [such as Death Box 1, 3, 21, 41, 61 (DDX1, 3, 5, 6, 17, 21, 41, 60)] [10]. PRRs that recognize DNA include TLR9, stimulator of interferon genes (STING), ALRs [such as absent in melanoma 2 (AIM2), interferon-γ-inducible protein 16 (IFI16)], cGAMP synthase (cGAS), DDX41, RNA polymerase III (Pol III), Z-DNA binding protein 1 (DAI, ZBP1), DEAH-Box Helicase 9, 36(DHX9, 36), and Ku70 [11]. Activation of many of these receptors leads to the production of type I interferons (type I IFNs or IFN-α/β), chemokines, and inflammatory cytokines (Fig. 1) [10]. Some nucleic acid sensing PRRs, such as AIM2 and STING, are critical for host defense against intracellular bacteria and viruses [12, 13]. Although the underlying mechanisms are largely unknown, accumulating evidences suggest that many of these nucleic acid sensing PRRs are involved in the development of cancers such as CRC. In addition to mechanism, questions remain as to the importance of PRRs in the development of colitis and CRC. Here, we review the latest researches on the roles of nucleic acid sensing PRRs (Except TLRs that have been reviewed elsewhere [9, 14–17]) in the development of colitis and CRC, focusing on the intensively studied molecules– AIM2 and STING.
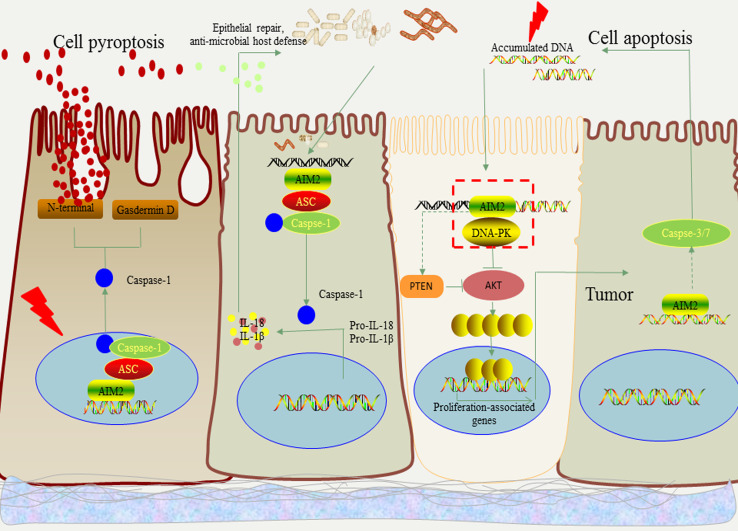
Fig. 1.
The model of AIM2 signaling pathway in the development of colitis and colorectal cancer. AIM2 in intestinal epithelial cells (IECs) can be activated by dsDNA in cytoplasm or nucleus. The activation of AIM2 can further assemble inflammasome by recruiting ASC and pro-caspase-1. AIM2 inflammasome maturates caspase-1 in turn. Caspase-1 maturated by cytoplasmic AIM2 inflammasome activated by microbiota and dying cell-derived dsDNA can further cleave pro-IL-1β and pro-IL-18 into IL-1β and IL-18, respectively. IL-18 promotes the repair of damaged IECs and induces an anti-microbial host defense, which potently alleviates the development of colitis. AIM2 can also assemble inflammasome in nucleus where AIM2 encounters damaged dsDNA induced by radiotherapy and chemotherapy. The activated caspase-1 cleaves the substrate gasdermin D and drives the pyroptosis of IECs by its N-terminal fragment. On the other hand, AIM2 binds to DNA-PK directly and inhibits the AKT-induced proliferation of IECs as well as intestinal stem cells to prevent the development of CRC. Moreover, AIM2 inhibits the activity of AKT by enhancing the level of its negative regulator PTEN. Additionally, AIM2 also accelerates the caspase3/7-dependent apoptosis of IECs
The DNA sensor AIM2 plays a critical role in the development of CRC and colitis
The ALRs family consists of AIM2 and IFI16 (murine p204) and resides in cytoplasm or nucleus. These proteins contain an N-terminal pyrin domain (PYD), which triggers intracellular signaling, and a one/two C-terminal HIN-200 domain, which binds to double-stranded DNA (dsDNA) [18]. After AIM2 binds to dsDNA in cytoplasm, the PYD interacts with apoptosis-associated speck-like protein containing CARD (ASC) to recruit pro-caspase-1, forming a multi-protein complex called the inflammasome [19]. IFI16 has been reported to induce the production of IFN-β via the adaptor STING upon sensing cytoplasmic DNA, while forming inflammasome in cell nucleus in response to Kaposi Sarcoma-associated herpesvirus (KSHV) infection[20, 21]. Activated caspase-1, on one hand, mediates the maturation of pro-IL1-β and pro-IL18 into IL-1β and IL-18, respectively [22]. On the other hand, activated caspase-1 can cleave the substrate gasdermin D, and promote cell pyroptosis, a new form of programmed cell death [12]. One recent study showed that AIM2 could also form inflammasome in nucleus in response to radiation-induced DNA damage and further promote the pyroptosis of intestinal epithelial cells (IECs)[23]. AIM2 inflammasome and its effector genes provide host immune surveillance to a range of bacteria, viruses, and fungi by detecting the pathogenic DNA components in host cell cytoplasm [12]. In addition to pathogen defense, AIM2 is also involved in tumorigenesis. However, the role of AIM2 in tumors may vary by the types of cancer. It was firstly found to be exclusively expressed in chromosome 1 of melanoma cells by cDNA subtractive hybridization technique [24]. Reduced expression of AIM2 was identified in prostate cancer and CRC patient tissues subsequently [25, 26]. However, increased expression of AIM2 was observed in nasopharyngeal carcinoma [27].
In recent years, accumulating studies have shown that AIM2 may suppress the development of CRC. The microsatellite instability phenotypes in tumors often arise from mutations in microsatellites-encoding genes that are attributed to the defect in mismatch repair [28]. Notably, more than half CRC tissues and cell lines with high level of microsatellite instability showed frameshift mutation in AIM2, and further genetic study showed that the mutations were located in the coding regions [29, 30]. Furthermore, the decreased level of AIM2 in CRC was associated with a poorer prognosis [26]. These studies indicated that AIM2 could be a CRC suppressor. However, until recently, the in vivo physiological role of AIM2 in CRC development was unknown, though inflammasome effectors induced by other inflammasome-associated PRRs, including NLR members NLRP3,NLRC4, and NLRP6, were shown to protect the host from colitis and CRC [31–35]. Two recent studies by the Kanneganti group from St. Jude Children’s Research Hospital, and the Ting group from the University of North Carolina, both located in the USA, revealed the critical role of AIM2 in the development of CRC in vivo [36, 37].
These two studies utilized a well-established colitis-associated CRC animal model to study host susceptibility of WT and Aim2 −/−mice to CRC. In this model, CRC is induced via the intraperitoneal injection of the carcinogenic chemical Azoxymethane (AOM) into mice and then the administration of a inflammation-inducing chemical, Dextran Sodium Sulfate (DSS), into the drinking water for three rounds [38]. Both studies found that Aim2 −/−mice harbor a dramatically increased colon tumor burden after AOM/DSS treatment compared to wild type (WT) mice despite similar changes in colon inflammatory infiltration, colon length, and the production of inflammatory mediators such as IL-6, tumor necrosis factor (TNF-α), and granulocyte colony stimulating factor (G-CSF) [36, 37]. Interestingly, the colonic expression levels of caspase-1 and the inflammasome effectors, IL-1β and IL-18, were comparable between Aim2 −/−and WT mice at various time points during CRC development [36, 37]. These results indicate that inflammation or an inflammasome defect in Aim2 −/−mice is probably not a major contributor to CRC susceptibility.
After excluding inflammation and inflammasome factors, these two studies found that, in comparison with WT mice, Aim2 −/−mice exhibited more proliferation of IECs at early time points after AOM/DSS treatment [37]. Microarray analysis revealed a number of upregulated proliferation-associated genes, such as S100a9, at day 14 post AOM injectionin Aim2 −/−mice [37]. To better understand the underlining mechanisms by which AIM2 inhibits enterocyte proliferation, the phosphorylation levels of signaling molecules in proliferation or apoptosis pathways, such as mitogen-activated protein kinase (MAPK) and phosphatidyl inositol 3-kinase-serine/threonine kinase (PI3K/AKT) signaling pathway, were measured in Aim2 −/−and WT mice. It was shown that the level of phosphorylated AKT (p-AKT) significantly increased whereas the PI3K-negative regulator tensin homology deleted on chromosome 10 (PTEN) decreased in Aim2 −/−mice [36, 37]. Loss- and gain-of-function experiments in vitro and in vivo confirmed the important role of AIM2 on AKT activation and the inhibition of colon tumorigenesis [36]. However, the negative effect of AIM2 on AKT is in cell type-dependent manner. The level of p-AKT in CRC cell line as well as intestinal stem cell can be suppressed by AIM2, while AKT activation was similar in Aim2 −/−and WTbone marrow-derived macrophages (BMDMs) upon stimulation [36]. To further understand which cell type contributes to AIM2-induced CRC restriction, bone marrow transplantation experiments were conducted. Consistently, Ting group identified that IECs were the main cell sources that were associated with AIM2-medicated CRC restriction [36]. However, Kanneganti group found that both AIM2 in bone marrow and IECs might be involved in this process [37]. AKT activation is regulated by a variety of kinases and phosphatases including DNA-dependent protein kinase (DNA-PK), and mTOR phosphorylates AKT at Ser473 [39, 40]. Co-immunoprecipitation experiments revealed that AIM2 interacts with DNA-PK but not with AKT or mTOR [36]. Although both AIM2 and DNA-PK bind DNA, AIM2 still interacts with DNA-PK after Ethidium Bromide (EtBr) treatment, which abolishes their interaction with DNA, indicating that there is a direct interaction between AIM2 and DNA-PK [36]. Further study suggested that AIM2 is a negative regulatory factor for DNA-PK, which is necessary for the increased AKT phosphorylation in Aim2 −/−CRC cells [36]. DNA-PK also promotes the stability of c-Myc, so loss of AIM2 and increased DNA-PK may contribute to the increased production of c-Myc in Aim2 −/− mice.
Persistent proliferation and differentiation of intestinal stem cells in crypts are essential to restore structure and function after damage to intestinal epithelial cells [41]. However, aberrant self-renewal of intestinal stem cells can be the source of intestinal tumors [42]. Indeed, the growth rates of intestinal stem cells derived from Aim2 −/−mice were significantly higher than those of WT mice [37]. The protective role of AIM2 in sporadic CRC was confirmed by higher tumor burden in the Aim2 −/−/Apc Min/+ mice than Apc Min/+ mice [36], which have aberrant activation of the Wnt/β-catenin signaling pathway in IECs, and have colonic neoplasia [43, 44]. However, whether this process is involved in intestinal stem cell proliferation and the exact association between AIM2 and adenomatosis polyposis coli gene (APC) is still unknown, though previous studies showed that aberrant activation of wnt/β-catenin signaling pathway exists in colon cancer stem cells [45]. To investigate the role of AIM2 in Wnt signaling, Aim2 −/−mice were crossed to a Prom1 CreERT2−LacZ (C−L)/+; Ctnnb1 lox(ex3)/+ ;RosaZsGreen mouse strain, and it was found that AIM2 is necessary for intestinal stem cell activation following aberrant β-catenin activation [37]. In this model, aberrant β-catenin activation is induced by tamoxifen, and Prom1 is used as a marker for intestinal stem cells. Although no macroscopic colon tumors were found, increased numbers of Ki67+ large intestinal cells were observed inAim2 −/−mice [37]. In addition, increased activation of AKT and c-Myc was found in the colon tissues of Prom1 C−L/+; Ctnnb1 lox (ex3)/+; RosaZsG; Aim2 −/−mice in comparison with controls [37]. On the other hand, the activation of the apoptosis indexes, such as caspase-3 and caspase-7, significantly decreased in Aim2 −/−mice colon tissues [36, 37]. Collectively, AIM2 may suppress colon tumorigenesis by inhibiting the proliferation and increasing their apoptosis of IECs or stem cells, which is consistent with the observation that upregulation of AIM2 expression in CRC cell lines results in cell cycle arrest [46].
The intestinal microbiota was also found to be an important etiological factor for CRC development. The Kanneganti’s study showed that AIM2 contributes to the maintenance of intestinal homeostasis by regulating gut microbiota. The gut microbial spectrum was different between Aim2 −/−and WT mice, and this difference could be diminished by co-housing [37]. When Aim2 −/−mice were co-housed with WT mice, the colonic tumor load decreased in Aim2 −/− mice and increased in WT mice [37]. These results suggest that AIM2 suppresses colon tumorigenesis partly due to its regulation of the gut microbiota though the mechanisms are still undefined.
Although the previous two studies showed that AIM2 protects host against CRC, the role of AIM2 or AIM2 inflammasome in intestinal inflammation is still unknown. Several recent studies investigated the role of AIM2 in the development of colitis [47–49]. Aim2 −/−mice developed more severe colitis than WT mice in a DSS-induced colitis model. The phenotype is associated with increased intestinal bacterial burden, especially Escherichia coli, because of the defective function of inflammasome effectors inAim2 −/−mice [47]. The further results suggested that the inflammasome effectors IL-1β and IL-18 are responsible for regulating the production of antimicrobial peptides that defend against Escherichia coli infection during DSS-induced colitis [47]. Consistently, another recent study found that AIM2 inflammasome in IECs prevent against colitis via IL-18/IL-22/STAT3/Reg3 axis. However, this study also found that AIM2 in myeloid cells is detrimental to the development of colitis, indicating that the effect of AIM2 in intestine is cell type dependent [49]. The discrepancy between this study and previous studies on the dependence of the inflammasome may be due to different sources of Aim2 −/−mice as discussed in another review [12]. Another study using a colitis model induced by Salmonella infection also showed a protective role for AIM2 in colitis [48]. In this model, there were less cell proliferation and more apoptosis in the intestinal tissues of Aim2 −/−mice. In addition, AIM2 restricts Salmonella diffusion by increasing the expression of tight junction proteins such as Claudin 3 and Occludin, rather than the inflammasome component caspase-1 [48].
In addition, one recent study showed that AIM2 inflammasome promotes the radiation-induced pyroptosis of IECs [23]. Compared to WT mice, Caspase-1 −/− mice suffered less radiation-induced gastrointestinal syndrome including intestinal damage, diarrhea, and malabsorption. Further results indicated that AIM2 inflammasome rather than Nlrp3 or Nlrc4 inflammasome was the activator of the death of IECs including intestinal stem cells, in a manner dependent on caspase-1 but not on cytokines, apoptosis, or AKT signaling [23]. The observation under immunofluorescence microscopy suggested that AIM2 detects blocked dsDNA in nucleus and form inflammasome [23]. These indicated that AIM2 could sense the damaged DNA in nucleus and form inflammasome to medicate radiation-induced intestinal damage and pyroptosis. These raised the question of that whether AIM2 inflammasome can also sense the damaged DNA in nucleus of tumor cells and further medicate pyroptosis, since tumor cells often contain genome instability [50].
Collectively, AIM2 is critical for the maintenance of intestinal homeostasis. On one hand, AIM2 in IECs directly binds to DNA-PK and negatively regulates the activation of AKT, thus restricting the proliferation of IECs as well as intestinal stem cells. On the other hand, AIM2 inflammasome promotes the apoptosis as well as pyroptosis of IECs. Furthermore, AIM2 in IECs sense the invasive DNA pathogens, and enhance the intestinal integrity via various mechanisms. These mechanisms may include AIM2 inflammasome-induced IL-18/IL-22BP/IL-22/STAT3/Reg3 axis activation and AIM2-induced AKT activation in the early time of colitis to enhance the integrity of intestinal epithelial barrier (Fig. 1; Table 1).
Table 1.
Main nucleic acid sensing pattern recognition receptors (PRRs) in colorectal cancer (CRC) or colitis
| PRRs | Location | Ligands | Adaptors | Main functions in CRC or colitis |
|---|---|---|---|---|
| AIM2 |
Cytoplasm Nucleus |
dsDNA | ASC |
In CRC: AIM2 suppresses DNA-PK/AKT signaling-induced proliferation of intestinal epithelial cells or stem cells, increases their apoptosis, and regulates the gut microbiota [36, 37] In colitis: AIM2 inflammasome and the related effectors protect mice from colitis by regulating the production of anti-microbial peptides [47]. AIM2 is required for the maintenance of the intestinal epithelial barrier by increasingAKT-induced production of tight junction proteins [48] |
| STING | Cytoplasm | CDNs | TBK1 | In CRC: STING restricts colon tumorigenesis by reducing inflammation in the colon, increasing inflammasome activity, and increasing type I IFNs-induced anti-tumor response of CD8+ T cells [60, 62, 67, 68] |
| cGAS | Cytoplasm | dsDNA | STING | In CRC: cGAS is undetectable in most CRC cell lines that partially contain CpG islands in cGAS promoter region. cGAS may be required for tumor-derived DNA recognition and the triggering of CD8+ T cell-induced anti-tumor response in CRC [67, 68, 71] |
| RIG-I | Cytoplasm | 5′PPP RNA, Short dsRNA | MAVS | In colitis: RIG-I plays a crucial role in colitis by regulating the activation of T cells as well as Gαi2 expression [88] |
| DDX3 |
Cytoplasm Nuclear |
dsRNA | MAVS | In CRC (Controversial): DDX3 serves as a CRC suppressor by regulating Snail and E-cadherin expression [110]. However, DDX3 may also serve as an oncogene for CRC through regulating Wnt/β-Catenin signaling[111–113] |
| DHX9 |
Cytoplasm Nucleus |
CpG-B DNA, dsRNA |
MAVS/ MyD88 |
In CRC: DHX9 is upregulated in human CRC tissues and cells [122, 123] |
| DDX5/17 | Cytoplasm | RNA | unidentified | In CRC: DDX5 and DDX17 serve as CRC promoter and enhance the proliferation of CRC cells via regulating oncogenes, such as c-Myc[126, 127] |
| DDX6 | Cytoplasm | RNA | unidentified | In CRC: DDX6 may promote the proliferation of CRC cell lines by downregulating Wnt/β-catenin signaling pathway[128, 129] |
| DHX36 | Cytoplasm | CPG-A DNA | MyD88 | In CRC: Overexpression of DHX36 can reduce the migration of CRC cells[132] |
However, works are still needed to understand the upstream mechanisms of decreased expression of AIM2 in human CRC. How and where AIM2 is activated in tumor setting is also unclear. In addition, more understanding of the function of AIM2 in various cell types in CRC microenvironment may potently aid to the regulation of AIM2 in CRC.
STING signaling pathway in colorectal cancer and colitis
STING is localized in the endoplasmic reticulum (ER) with 5 N-terminal transmembrane domains that anchor it to the ER membrane and a globular in C-terminal domain (CTD) that specifically binds to cyclic di-nucleotides (CDNs), such as cyclic-di-GMP, cyclic-di-AMP, and cyclic-GMP-AMP (cGAMP) [51]. CDNs are a class of second messengers from bacteria that are rarely expressed in eukaryotes [52]. A number of studies have shown that STING is required for the induction of type I IFN (IFNβ) in response to intracellular DNA following bacterial and viral infection [53]. However, further studies indicate that STING is also an adaptor for other DNA sensors based on the observation that STING binds to labeled CDNs directly, and unlabeled CDNs rather than DNA can compete with this binding [51]. Furthermore, cell lines with specific point mutations in regions that are involved in recognizing CDNs in STING showed intact response to cytoplasmic DNA [51]. Subsequently, in addition to DAI [54], DDX41 [55], and IFI16 [20], cGAS has been identified as another DNA sensor upstream of STING [56]. Upon cytoplasmic DNA recognition, cGAS catalyzes the synthesis of 2′3′-cGAMP or 2′5′-linked GAMP from ATP and GTP, which in turn act as second messengers to bind and activate STING in a manner different from the other CDNs[57, 58]. After activation, STING binds to TANK-binding kinase 1 (TBK1) in the ER and translocates to the perinuclear compartment where it interacts with Sec5 and induces the production of IFN-β via interferon regulatory factor 3 (IRF3) or nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling [53]. In addition to IFN-β, STING is also required for the signal transducer and activator of transcription 6 (STAT6)-induced production of Ccl2 and Ccl20, which are crucial for antiviral immune responses [59].
Previous studies on STING were mainly focused on its role in anti-viral immunity. Recent evidences suggest that STING also has a critical role in the development of CRC. Compared to WT mice, Sting −/−mice have a higher tumor load in colon tissues after AOM/DSS treatment [60]. Furthermore, histological analysis revealed an increased inflammatory cell infiltration and increased dysplasia in the colorectal tissues of Sting −/−mice [60]. Levels of pro-inflammatory factors such as IL-6 and KC were elevated in colon tissues and sera of Sting −/−mice in different stages of CRC development [60]. Moreover, the level of the phosphorylated signal transducer and activator of transcription 3 (STAT3), a downstream effector of IL-6R that contributes to colon tumorigenesis [61], was upregulated in colon tissues of Sting −/−mice [60]. These results suggest that STING restricts colon tumorigenesis by reducing inflammation in the colon. Notably, the main gut bacterial spectrum showed no significant difference between WT and Sting −/− mice, indicating that STING may suppress susceptibility to CRC independent of gut microbiota.
Interestingly, decreased maturation of pro-caspase1 and expression of IL-18 were observed in the colon tissues of Sting −/−mice [60], indicating that STING may upregulate inflammasome activation. Similarly, another study showed decreased production of IL-18 and IL-22 binding protein (IL-22BP) in AOM/DSS-induced CRC tumor tissues of Sting −/−mice [62]. However, this result seems contradictory to the previous observation that IL-18 is a negative regulator of IL-22BP in the presence of NLRP3 or NLRP6 inflammasomes [63]. To further understand the relationship between STING, IL-18 and IL-22BP, an in vitro experiment was conducted. The results confirmed that IL-18 upregulation is STING inducible that may further upregulate the expression of IL-22BP [62], a negative regulator of colon tumorigenesis promoter-IL-22 [63, 64]. It seems that moderate expression of IL-22 promotes the repair of injured intestinal cells, while aberrant and chronic activation of IL-22 leads to hyperproliferation of intestinal cells and tumor formation [63]. These data suggest that STING may also inhibit colon tumorigenesis through the activation of inflammasome activity.
In addition to suppressing inflammation and enhancing inflammasome activity, other studies suggest that STING may induce the production of type I IFNs and further activate anti-tumor CD8+ T cells. Increased CD8+T cells infiltration in CRC tissue is associated with a better prognosis and type I IFN is required for CD8+T cell priming [65, 66]. Two recent studies reported that the STING-IRF3-type I IFN signaling pathway, rather than other type I IFN-inducible signaling pathways, in dendritic cells (DCs) is required for tumor-specific CD8+T cellspriming [67, 68]. In addition, this process is mainly mediated in DCs by cGAS that specifically detects tumor-derived DNA [67–69]. Apart from DCs, recent study suggested that tumor cells themselves, such as CRC cells, could also induce the production of type I IFNs [70].
In summary, STING may suppress colon tumorigenesis through multiple mechanisms, including (but not limited to) inhibiting inflammation, activating the inflammasome, and/or inducing type I IFNs.
Due to the indispensible role of STING signaling in tumorigenesis, their expression profile and functional status in human colorectal cancer cell lines have been investigated. The expression of IFN-β and IL-1β was significantly reduced in most human CRC cell lines with a defect in STING compared to WT cell lines in the presence of dsDNA [71]. As previously discussed, upon activation, STING interacts with TBK1 and translocates from the ER to the perinuclear compartment to phosphorylate IRF3 or NF-κB. However, this process was defective to different extents in the human CRC cell lines in the presence of dsDNA [71]. To understand if mutations in STING were responsible for these observations, the genomes of the human CRC cell lines were sequenced. The results indicate that the STING signaling defects may be attributed to upstream DNA sensors [71]. Consistently, the expression of cGAS is undetectable in some CRC cell lines, including LS174T, SW480, SW1417, SW48, HT116, and colo205. However, gene sequence results showed there was no mutation in genome encoding cGAS and the suppression of cGAS may be due to epigenetic modification, such as hypermethylation that often leads to gene silencing [72]. Indeed, bisulfite sequencing analysis showed that there were CpG islands in cGAS promoter regions of these cell lines, and demethylation could rescue the defective STING signaling in these cells [71]. Additionally, the expression of STING and cGAS was detected in human CRC formalin-fixed paraffin-embedded tissues. Those samples exhibited low expression levels of STING and cGAS starting from stage II, although more studies are needed to understand the correlation between STING and CRC patients’ prognosis.
As a central adaptor for DNA sensing, STING mediates the production of IFN-β and plays an important role in host defense against viruses such as herpes simplex virus (HSV) [73]. However, HSV can be utilized to treat cancers [74] and thus STING signaling deficiency in tumor cells facilitates the killing of tumor cells by oncolytic viruses in turn. In vitro experiments showed that human CRC cell lines defective in STING exhibited less type I IFN production and underwent more cell death when receiving HSV treatment, partially due to massive HSV replication [71]. This indicates that HSV has greater efficacy against CRC in the absence of STING signaling. This notion was supported by in vivo nude mouse experiments that CRC cells with defective STING signaling were more sensitive to HSV therapy [71]. Consistently, it was shown that STING signaling was defective in melanoma cells, and these cells are highly susceptible to HSV treatment [75]. So the effective STING signaling in tumor cells seems a double-edged sword, and evaluation of STING signaling in CRC patients may be used to predict the potential antitumor efficacy of oncolytic viruses. On the other hand, many pre-clinical experiments tried to enhance STING signaling in CD8+ T cells to promote anti-tumor immunity. The results showed that STING agonists, along with traditional therapy managements, could effectively reduce tumor loads in mice, such as CRC, melanoma, and glioma [76–78]. Encouragingly, some agonists have been shown to activate human STING signaling, though more clinical trials are needed to confirm.
All of these depict the peculiar charm of STING signaling pathway in inhibiting the development of CRC. Tumor-derived DNA captured by cGAS in DCs as well as tumor cells themselves in tumor microenvironment (TEM) is the primary driving force to activate STING signaling pathway to induce the production of type I IFNs and the tumor-specific CD8+ T cell priming. Apart from reducing inflammation response in early stage of CRC formation, STING also interacts with inflammasome to activate the caspase1/IL-18/IL-22BP axis and control aberrant proliferation of IECs (Fig. 2; Table 1 ). Although STING silencing did not influence the gut microbiota, whether gut microbiota changes in colitis and CRC can signal via intestinal STING and influence the diseases progression in turn is still unknown. In addition, despite that STING signaling is defective in most CRC cell lines, more clinical studies are needed to understand the relationship between STING expression and CRC prognosis, and whether it can be more precisely to predict the CRC prognosis in combination of STING level and tumor stages. On the other hand, STING activation in TME may incite both antitumor responses and tolerogenic responses. Tolerogenic responses can reduce the risk of autoimmune disease, while it increases the risk of tumor immune escape somehow. Several studies have shown that STING activation in TME can induce the infiltration of immunosuppressive cells, such as regulatory T cells (Treg), myeloid derived suppressor cells (MDSC) via type I IFNs induced IDO [79, 80]. Therefore, more detailed studies of the role of STING in CRC TME are needed to optimize its application in clinic.
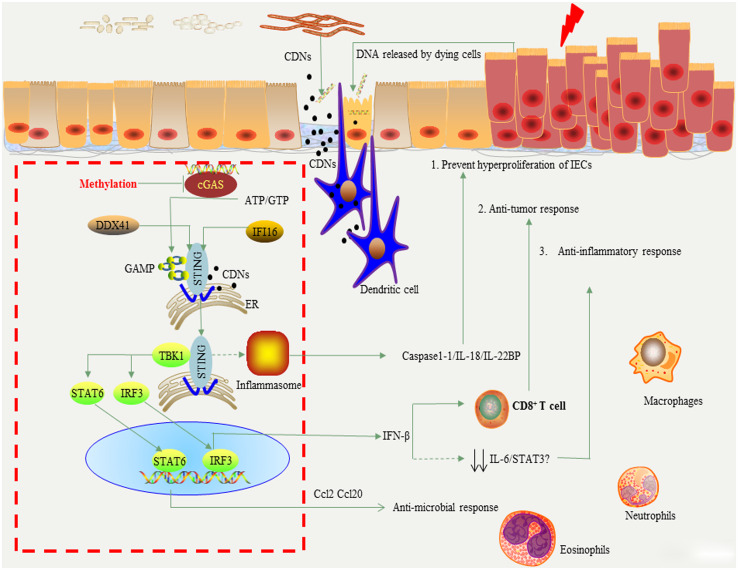
Fig. 2.
The model of cGAS-STING signaling pathway in the development of colorectal cancer.The ER-anchoring protein STING is an important adaptor for immune responses to DNA stimulation. STING in antigen presenting cells, especially in dendritic cells, directly binds to cyclic di-nucleotides (CDNs) that are specifically presented in bacteria or generated from ATP/GTP by cGAMP synthase (cGAS). The upstream candidate DNA receptors of STING include DDX41 and IFI16. STING is activated and shuttled to perinuclear compartment where it binds to and phosphorylates TBK1 when tumor-derived DNA is captured by cGAS. The phosphorylation of TBK1 leads to the activation of IRF3 and STAT6, which in turn leads to the production of type I IFN (IFN-β) and Ccl2/20, respectively. IFN-β is required for priming of tumor-specific CD8+T cell and can induce the tumor regression. On the other hand, STING may reduce the inflammatory response in intestine by inhibiting the expression of IL-6 and STAT3. Notably, STING may also induce the activation of inflammasome and suppress the proliferation of intestinal epithelial cells via activating inflammasome-derived IL-18
The role of RLRs signaling pathway in colitis or CRC
RNA helicases are a superfamily of proteins expressed by almost all living organisms and are characterized by harboring 7–8 conserved DExD/H domains [81]. They are involved in all aspects of genetic expression by unwinding folded RNAs and modifying the RNA–RNA interactions in an energy-dependent manner via hydrolysis of NTP in cytoplasm [81]. RLRs are a subfamily of DExD/H box RNA helicases composed of RIG-I, MDA5 and laboratory of genetics and physiology 2 (LGP2), which are widely expressed in most tissues and cells [82]. RIG-I and MDA5 share similar structures: a repressor domain (RD) embedded within the CTD, a central DEAD box involved in hydrolysis of NTP and unwinding of RNAs, and an N-terminal CARD [82]. LGP2 lacks the CARD modification and may serve as a positive regulator for RIG-I and a negative regulator for MDA5 [82]. Studies show that RIG-I can be activated by various RNA substrates, including double-stranded RNA (dsRNA) and single-stranded RNA (ssRNA) that contain an uncapped 5′-triphosphate (5′ppp) modification, as well as blunt-ended short dsRNA. The 5′ppp modification in RNA is often specifically present in virus genomes, which are quite different from human RNA structures [83]. Compared to RIG-I, MDA-5 is preferentially activated by long dsRNA with blunt end [82]. Upon activation, both RIG-I and MDA5 recruit the adaptor-mitochondrial antiviral signaling protein (MAVS) via their CARD domains and form a platform to activate TRAF. This in turn leads to the phosphorylation of IRF3 and NF-κB signaling and the production of pro-inflammatory genes and IFN-β, respectively [84]. Accumulating evidences showed that RLRs family plays an important role in anti-virus immune response, and mice lacking either RIG-I or MDA-5 were highly susceptible to virus infection [85]. In addition, different types of virus showed difference fashion on RIG-I and MDA-5 activation and some viruses, such as West Nile virus and rotavirus, can activate both of them [86, 87]. The mechanisms of RLRs in limiting the diffusion of virus may be partially attributed to the following factors: firstly, the activation of RLRs in different cells may lead to the production of IFN-β, which affects via the IFN receptors and induces the production of interferon-stimulated genes (ISG), vital molecules to initiate anti-virus immunity. Secondly, the RLRs themselves can directly hydrolyze the interaction among RNA–proteins, which are important for virus survival.
In addition to the roles of both RIG-I and MDA5 in the host defense against viruses, they are implicated in intestinal inflammation and cancer. Compared to WT mice, Rig-I −/−mice exhibit more intestinal inflammation in DSS-induced colitis [88]. Interestingly, it was found that both the number and size of Payer’s patches, the first line of immune defense in the gut, significantly decreased inRig-I −/−mice with increased number of apoptotic B220+ cells. The deregulation of T cells is associated with the development of colitis, and the Rig-I −/−mice also exhibited increased number of naïve T cells and decreased number of effector T cells in spleen [87, 88]. Further study found that associations may exist between RIG-I and Gαi2, a subunit of G protein which is involved in a range of biochemical activities since previous studies showed that Gαi2 −/− mice were also highly susceptible to colitis that was characterized by regression of Payer’s patches and deregulation of T cells subsets [89]. Indeed, further results confirmed that the expression of Gαi2 was significantly reduced in the colon tissues and lymphocytes of Rig-I −/−mice, and RIG-I promoted the activity of Gαi2 promoter [88], which may be incited by NF-κB [90]. One more recent study showed that RIG-I was mainly expressed in apical surface of the IECs, and RIG-I mRNA and protein levels were dramatically reduced in intestinal tissues of patients with Crohn’s disease (CD) [91]. All of these indicated that RIG-I can prevent the development of colitis, though the underlying mechanisms remain to be defined.
As downstream adaptors in RLR signaling pathways, MAVS and IRF3 also play roles in the development of colitis. Compared to WT mice, Mavs −/−andIrf3 −/− mice developed more severe colitis [92, 93]. Notably, Mavs −/−/MyD88 −/−mice died earlier than the Mavs −/− mice, indicating that RLR and TLR signaling may work cooperatively to protect mice from DSS-induced colitis [92]. The bone marrow transplantation experiments confirmed that MAVS in non-hematopoietic cells is crucial for the prevention of colitis [92]. In addition, apart from virus RNA, commensal bacteria-derived RNA can specifically activate RIG-I-MAVS signaling pathway in IECs and BMDMs and induce the production of IFN-β, inflammatory cytokines, and RegIIIγ. Both IFN-β and RegIIIγ are crucial molecules in anti-bacterial response [92]. As the downstream molecule of RLRs signaling, IRF3 can directly bind to and activate the promoter of thymic stromal lymphopoietin (TSLP), which is expressed both in hematopoietic cells and IECs and plays a preventive role against colitis [93]. All of these results suggest that the RIG-I-MAVS-IRF3 signaling pathway in IECs may protect mice from colitis. However, the underlying mechanisms by which RIG-I inhibits the development of colitis, and whether RIG-I is involved in CRC development are still unknown.
In summary, experiments in animals and IBD patients indicated that deregulation of RIG-I and MDA-5 signaling pathway may contribute to IBD development, despite that the specific contribution of RLRs in colitis is still poorly understood. It seems that RIG-I enhances the activation of Gαi2 and, thus, regulates peripheral T cells activation, while MDA5 senses invasive RNA microbiota in gut to maintain intestinal homeostasis by mediating the expression of type I IFNs and antimicrobial peptides. Apart from inducing the expression of type I IFNs, the downstream molecules of RIG-I/MDA5 may also interact with TSLP to prevent the progression of IBD (Fig. 3; Table 1). More animal and clinic experiments are needed to improve the limited understanding of the role of RLRs in colitis as well as CRC. Especially, issues about whether RLRs-induced DCs and NKs activation are involved in CRC development are worthy to be studied. Also, we should not ignore the relationship between RLRs and gut microbiota homeostasis. Moreover, whether the RIG-I and MDA5 induce similar phenotype remains to be unfolded in light of that the existence of RLRs member LGP2 leads to tumor cells including CRC cell lines insensitive to ionizing radiation-induced cytotoxic effects [94].
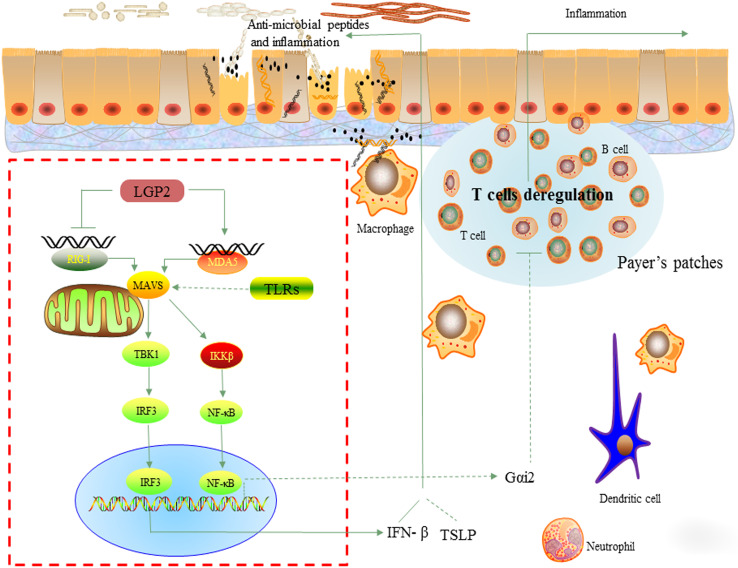
Fig. 3.
The model of RLR signaling pathway in the development of colitis. RIG-I-like receptors (RLRs) are required for cytoplasmic RNA detection. Upon activation, both RIG-I and MDA5 recruit the adaptor-mitochondrial antiviral signaling protein (MAVS) via their CARD domains and form a platform to activate TRAF. This in turn leads to the phosphorylation of IRF3 and NF-κB signaling to promote the production of IFN-β and pro-inflammatory genes, respectively. RIG-I enhances the promoter activity of Gαi2, partially through NF-κB, to reverses the deregulation of T cell subsets in Payer’s patches—an important instigator for colitis development. On the other hand, MDA5 senses RNA from invasive gut microbiota to maintain intestinal homeostasis by inducing the expression of IFN-β and antimicrobial peptides. Moreover, IRF3 can also directly bind to TSLP to protect host from colitis
Others nucleic acid sensing PRRs in colitis or CRC
The TLRs family is the first identified and the most studied PRRs. Nucleic acid sensing TLRs, including TLR3, TLR7, TLR8, and TLR9, are involved in the development of colitis and CRC, which have been reviewed elsewhere [9, 14–17]. Recent studies have showed that, in addition to TLRs, AIM2, STING, and RLRs, some other DExD/H box helicases, including DDX1, DDX3, DDX5, DDX6, DDX17, DDX21, DDX41, DHX9, DHX36, and DDX60, are also involved in pathogen recognition, immune responses, and cancers (including CRC) progression besides their functions on regulation of nucleic acid metabolism (Table 1) [95–98]. Among these receptors, DDX3, DDX5, DDX6, DHX9, DDX17, and DHX36 have been shown to be involved in the development of CRC. Furthermore, inherited and acquired mutation of DDX41, the DNA sensor upstream of STING that can activated by both cytoplasmic DNA and CDNs [55, 99], was frequently identified in acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS), indicating that DDX41 may serve as a tumor suppressor [100–104]. In this part, we will discuss the role of DDX3, DDX5, DDX6 and DHX9 in CRC development.
DDX3
DDX3, a member of DEAD-box proteins, is involved in diverse biological activities including binding RNA and acting as a transcriptional cofactor. DDX3 has two homologues, DDX3X and DDX3Y. The gene encoding DDX3X is located in X-chromosome in most tissues, while the gene encoding DDX3Y is located in the Y-chromosome in testis [105]. DDX3 is involved in regulation of several signaling pathways, including Wnt/β-catenin signaling pathway, epithelial-mesenchymal transition (EMT) signaling pathways, and IFN-β signaling pathway. The Wnt/β-catenin/T-cell factor (TCF) target genes, such as Snail1, matrix metalloproteinase 7 (MMP7), plasminogen activator inhibitor 1 (PAI1), and RAS, are often associated with tumor invasion [106]. DDX3 can directly interact with CK1ε to facilitate β-catenin trafficking to nuclear and trigger Wnt/β-catenin/TCF signaling [107]. EMT is a hallmark event that contributes to tumor invasion and is characterized by an increase in mesenchymal markers and a decrease in epithelial markers (such as E-cadherin). EMT may be induced by Wnt/β-catenin, notch, and some other signaling pathways [108]. DDX3 can promote EMT program by suppressing the expression of E-cadherin directly [109]. In addition, DDX3 is required for replication of RNA virus and is involved in inducing the production of IFN-β by interacting directly with the IFN-β promoter or interacting with IKKε, TBK1, or MAVS after binding to RNA virus [95].
The role of DDX3 in CRC is still controversial. One recent study indicated that the low level of DDX3 on CRC tissues is closely associated with a poorer prognosis and more severe metastasis [110]. In vivo and in vitro experiments confirmed that DDX3 silencing promotes the invasion of CRC that may be partially attributed to an increase in Snail expression and a decrease in E-cadherin expression [110]. These indicated that DDX3 might act as a CRC suppressor. However, another study demonstrated that the positive expression of DDX3 in CRC tissues is associated with increased level of β-catenin in nuclear [111]. In addition, DDX3 silencing in CRC cell lines resulted in decreased proliferation rates, cell cycle arrest, and reduced production of target genes of Wnt/β-catenin signaling [111]. These indicated that DDX3 serves as an oncogene for CRC that may be partially attributed to the regulation of Wnt/β-catenin signaling. Consistently, subsequent studies demonstrated that DDX3 could promote the CRC invasion by increasing activity of CK1ε/Dvl2/β-catenin/TCF signaling or by increasing activity of KRAS/ ERK/PTEN/AKT/β-catenin signaling [112, 113]. Another recent study showed that DDX3 inhibition is safe and effective for restriction of a range of virus infection, which provides potentials for targeting DDX3 in CRC [114].
DHX9
DHX9 can unwind both RNA and DNA and regulate the gene expression by interacting with transcription factors such as NF-κB, p53, and c-Myc [115]. In addition, DHX9 plays an important role in maintaining gene stability [116]. Recent study suggested that DHX9 can respond to both CpG-B DNA and dsRNA via MAVS and MyD88, respectively [95]. Since DHX9 is implicated in a range of cellular processes, deregulation of DHX9 may also have great effects on cell growth or viability, and even leads to various diseases. One previous study demonstrated that DHX9 level significantly increased in lung cancer tissues, and in comparison with non-small lung cancer, it was higher in small cell lung cancer, which was associated with a poorer prognosis [117, 118]. Subsequent studies showed that DHX9 is crucial for tumor survival, and DHX9 silencing restricts the tumor growth by regulating the transcript activities induced by p53 and c-Myc [119–121]. At the same time, DHX9 silencing in mice did not affect the vitality of normal cells, and DHX9 expression was upregulated in human CRC tissues and cells [98, 122, 123], indicating that DHX9 promotes the development and progression of CRC, and may serve as a safe and effective molecular target for CRC therapy. Better understanding on the expression pattern and function of DHX9 in human CRC and its association with the tumor growth, metastasis, and prognosis is urged.
DDX5/6/17 and DHX36
Compared to DDX3 and DHX9, the signaling pathways of other CRC-associated helicases are less studied. The role of DDX5/6/17 in tumors was discussed previously [124]. DDX5 (p68), a founding member of the DExD/H box family, forms complex with DDX17 and is involved in multiple cellular processes [125]. Studies showed that both the expression of DDX5 and DDX17 increased in human CRC cell lines and tissues, which were associated with more cellular differentiation [126, 127]. The detail mechanisms of increased DDX5 in tumor setting are still largely unknown, although there is a possible link with poly-ubiquitylation [127]. In addition, it was reported that DDX5 and DDX17 can promote the proliferation of CRC cells via regulating oncogenes, such as c-Myc [126]. DDX6 (p54) level is also increased in CRC tissues and cells [128]. Silencing of DDX6 may inhibit the proliferation of CRC cell lines, which is attributed to the downregulation of Wnt/β-catenin signaling pathway [129]. DHX36 was identified as a member of DExD/H box family in 2002. Later studies indicated that it forms complex with DDX1 and DDX21 to recognize dsRNA, and signals via TIR domain-containing adaptor-inducing IFNβ (TRIF), which eventually induces the production of type I IFN [130, 131]. However, the role of DHX36 in CRC is less studied, although one study reported that the overexpression of DHX36 can reduce the migration of CRC cells [132].
Taken together, the mechanisms of DExD/H box helicases on CRC are quite complicated. The DDX3 mainly interacts with Wnt-β-catenin signaling pathway and Snail/EMT signaling pathway to direct the development of CRC. DHX9 serves as an important role in tumor cell survival by regulating the transcript activities of oncogenes or tumor suppressor gene, such as p53 and c-Myc (Fig. 4; Table 1). Although studies indicated that the expression of DHX9 is increased in CRC tissues, additional studies are needed to characterize the underlying mechanisms. As for the DDX5/6/17 and DHX36, the mechanisms of their cancer promoting effects on CRC are more unshaped. Overall, some DExD/H box helicases exhibit a pro-tumor effect.
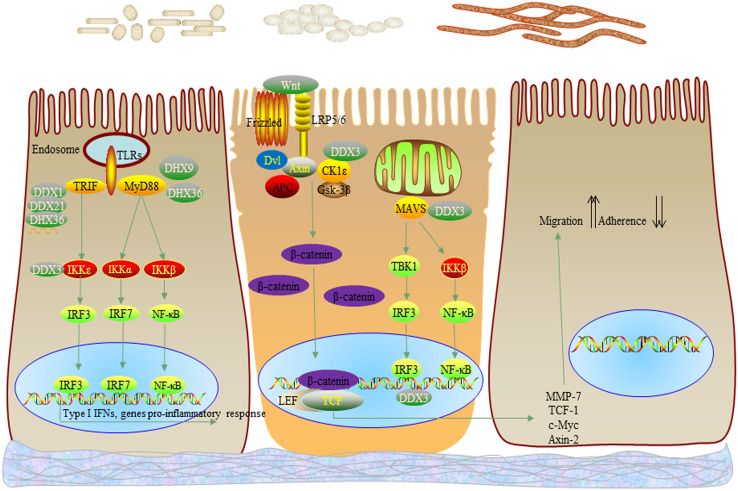
Fig. 4.
DExD/H box helicases signaling pathway in the development of colorectal cancer. DExD/H box helicases, including DDX1, DDX3, DDX5, DDX6, DDX17, DDX21, DDX41, DHX9, DHX36, DDX41, and DDX60, are involved in pathogen recognition, immune responses, and cancers (including CRC) progression, in addition to their functions in the regulation of nucleic acid metabolism. DHX36 responds to dsRNA and forms a complex with DDX1 and DDX21. The complex can signal via TRIF, or detect CpG-A DNA and trigger immune response via MyD88. DHX9 can respond to both CpG-B DNA and dsRNA, and signal via MAVS and MyD88, respectively. DDX60 interacts with RIG-I and sensitizes the binding of RIG-I and dsRNA. DDX3 can interact with dsRNA and signal via MAVS, in addition to acting as a mediator of downstream of TBK, IKKε and a transcriptional regulator of the IFN-β promoter. Furthermore, DDX3 can promote the CRC invasion by increasing activity of CK1ε/Dvl2/β-catenin/TCF signaling pathway. Except DDX3, the signaling pathways of DExD/H box helicases in CRC are largely unknown
Conclusions and future prospects
The immune cells and some other cells are armed with a range of nucleic acid sensing PRRs. Activation of these receptors by diverse nucleic acids of microbes leads to inflammatory responses and the activation of innate as well as adaptive immune cells, which plays critical roles in host defense against intracellular bacteria and virus infection. Aberrant activation of nucleic acid sensing PRRs signaling pathway by self-nucleic acids also contributes to inflammatory diseases. Additionally, recent accumulating studies from others and ours broaden the previous understanding on the function of nucleic acid sensing PRRs in tumors.
The nucleic acid sensing PRRs in IECs can be activated by invasive gut microbiota or the accumulated/damaged DNA in cytoplasmic or nucleus, and further influence the development of colitis and CRC by regulating the following aspects: inflammasome, gut microbiota, AKT, type I IFNs, Wnt-βcatenin, Snail/EMT, and oncogenes or tumor suppressor genes. Activation of AIM2 in IECs acids to maintain gut microbiota homeostasis in an inflammasome/IL-1β/IL-18/antimicrobial peptide-dependent manner to reduce pathogenic microflora such as Escherichia coli and thus protect against colitis. AIM2 binds to DNA-PK and inhibits the activation of AKT signaling pathway that is crucial for IECs proliferation. The STING signaling pathway mainly prevents the CRC development in a type I IFNs-CD8+T cell-dependent manner. Notably, the situation of DExD/H box helicases in CRC is much more complicated. Apart from RIG-I and MDA5 signaling pathway, the other members mentioned above exhibit an oncogenic activity though the underlying mechanisms are still largely unknown.
Despite accumulating evidences indicating that nucleic acid sensing PRRs are involved the development of CRC, more animal and clinical experiments are needed to illuminate their specific mechanisms in CRC and make their clinic application to be possible. Firstly, apart from AIM2 and DDX3, studies on the correction between the expression profile of the other nucleic acid sensing PRRs in CRC and tumor biological behaviors as well as prognosis are still lacking. In addition, it is of interest to known whether it could be more precise to predict the CRC prognosis when combined nucleic acid sensing PRRs with other clinical features such as tumor stages. Secondly, the signaling pathway of DExD/H box helicases is poorly defined thought they tend to interact with a range of transcription factors and modulate RNA metabolism throughout the whole life activities. Thirdly, most of the nucleic acid sensing PRRs shuttle between cytoplasm and nucleus, and the experiments are required to understand their function in different subcellular localization in the development of diseases. Fourthly, accumulated evidences showed that AIM2, STING, DHX9, LGP2, and DDX3 respond to chemoradiotherapy DNA damage and mediate different immunity responses. For instance, AIM2 responds to ionizing radiation-induced break DNA in nucleus and drives the pyroptosis of IECs, while STING senses dying tumor DNA after radiation potent leveraging radiotherapeutic management. These raise the questions that whether most of the nucleic acid sensing PRRs are involved in radiation-induced DNA responses. It is necessary to determine toxicity and synergy between chemoradiotherapy and nucleic acid sensing PRRs and, thus, optimize the treatment plans. Fifthly, in light of that most of the nucleic acid sensing PRRs play an important role in guarding pathogens invasion, whether most of them aid to maintain gut microbiota homeostasis like AIM2 and RLRs should be conceived. Lastly, it seems that some of the sensors such as AIM2 and STING can regulate each other and the cross-talk among these nucleic acid sensing PRRs is of interest. Overall, although some achievements have made in understanding the complicated role of nucleic acid sensing PRRs in colitis as well as CRC, there is still a long way to translate these discoveries into practice.
Acknowledgements
This work was supported by funds from Talents’ Start-up Fund of Gannan Medical University (QD201404), Natural Science Foundation of Jiangxi Province (20151BAB205061), Natural Science Foundation of China (31560260), and The Key Project from Department of Education of Jiangxi Province (150937) (All to Zhiping Liu).
Contributor Information
Zixiang Zhang, Phone: (86) 18079789122, Email: Zhangzixiang2001@163.com.
Zhiping Liu, Phone: (86) 13879793709, Email: Zhiping.Liu@gmu.cn.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2016 doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA-Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Brenner H, Kloor M, Pox CP. Colorectal cancer. The Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 6.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 7.Parlato M, Yeretssian G. NOD-like receptors in intestinal homeostasis and epithelial tissue repair. Int J Mol Sci. 2014;15:9594–9627. doi: 10.3390/ijms15069594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nunes T, de Souza HS (2013) Inflammasome in intestinal inflammation and cancer. Mediators Inflamm 2013:654963 [DOI] [PMC free article] [PubMed]
- 9.Li TT, Ogino S, Qian ZR. Toll-like receptor signaling in colorectal cancer: carcinogenesis to cancer therapy. World J Gastroenterol. 2014;20:17699–17708. doi: 10.3748/wjg.v20.i47.17699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurtler C, Bowie AG. Innate immune detection of microbial nucleic acids. Trends Microbiol. 2013;21:413–420. doi: 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Man SM, Karki R, Kanneganti TD. AIM2 inflammasome in infection, cancer, and autoimmunity: role in DNA sensing, inflammation, and innate immunity. Eur J Immunol. 2016;46:269–280. doi: 10.1002/eji.201545839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen GY. Role of Nlrp6 and Nlrp12 in the maintenance of intestinal homeostasis. Eur J Immunol. 2014;44:321–327. doi: 10.1002/eji.201344135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.So EY, Ouchi T. The application of Toll like receptors for cancer therapy. Int J Biol Sci. 2010;6:675–681. doi: 10.7150/ijbs.6.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slattery ML, Herrick JS, Bondurant KL, Wolff RK. Toll-like receptor genes and their association with colon and rectal cancer development and prognosis. Int J Cancer. 2012;130:2974–2980. doi: 10.1002/ijc.26314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furi I, Sipos F, Germann TM, Kalmar A, Tulassay Z, Molnar B, Muzes G. Epithelial toll-like receptor 9 signaling in colorectal inflammation and cancer: clinico-pathogenic aspects. World J Gastroenterol. 2013;19:4119–4126. doi: 10.3748/wjg.v19.i26.4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sipos F, Furi I, Constantinovits M, Tulassay Z, Muzes G. Contribution of TLR signaling to the pathogenesis of colitis-associated cancer in inflammatory bowel disease. World J Gastroenterol. 2014;20:12713–12721. doi: 10.3748/wjg.v20.i36.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ratsimandresy RA, Dorfleutner A, Stehlik C. An update on PYRIN domain-containing pattern recognition receptors: from immunity to pathology. Front Immunol. 2013;4:440. doi: 10.3389/fimmu.2013.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hornung V, Ablasser A, Charrel Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaki MH, Lamkanfi M, Kanneganti TD. The Nlrp3 inflammasome: contributions to intestinal homeostasis. Trends Immunol. 2011;32:171–179. doi: 10.1016/j.it.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu B, Jin C, Li HB, Tong J, Ouyang X, Cetinbas NM, Zhu S, Strowig T, Lam FC, Zhao C, Henao-Mejia J, Yilmaz O, Fitzgerald KA, Eisenbarth SC, Elinav E, Flavell RA. The DNA-sensing AIM2 inflammasome controls radiation-induced cell death and tissue injury. Science. 2016;354:765–768. doi: 10.1126/science.aaf7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeYoung KL, Ray ME, Su YA, Anzick SL, Johnstone RW, Trapani JA, Meltzer PS, Trent JM. Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene. 1997;15:453–455. doi: 10.1038/sj.onc.1201206. [DOI] [PubMed] [Google Scholar]
- 25.Ponomareva L, Liu H, Duan X, Dickerson E, Shen H, Panchanathan R, Choubey D. AIM2, an IFN-inducible cytosolic DNA sensor, in the development of benign prostate hyperplasia and prostate cancer. Mol Cancer Res. 2013;11:1193–1202. doi: 10.1158/1541-7786.MCR-13-0145. [DOI] [PubMed] [Google Scholar]
- 26.Dihlmann S, Tao S, Echterdiek F, Herpel E, Jansen L, Chang-Claude J, Brenner H, Hoffmeister M, Kloor M. Lack of Absent in Melanoma 2 (AIM2) expression in tumor cells is closely associated with poor survival in colorectal cancer patients. Int J Cancer. 2014;135:2387–2396. doi: 10.1002/ijc.28891. [DOI] [PubMed] [Google Scholar]
- 27.Chen LC, Wang LJ, Tsang NM, Ojcius DM, Chen CC, OuYang CN. Tumour inflammasome-derived IL-1β recruits neutrophils and improves local recurrence-free survival in EBV-induced nasopharyngeal carcinoma. EMBO Mol Med. 2012;12:1276–1293. doi: 10.1002/emmm.201201569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poulogiannis G, Frayling IM, Arends MJ. DNA mismatch repair deficiency in sporadic colorectal cancer and Lynch syndrome. Histopathology. 2010;56:167–179. doi: 10.1111/j.1365-2559.2009.03392.x. [DOI] [PubMed] [Google Scholar]
- 29.Woerner SM, Kloor M, Schwitalle Y, Youmans H, MvK Doeberitz, Gebert J, Dihlmann S. The putative tumor suppressor AIM2 is frequently affected by different genetic alterations in microsatellite unstable colon cancers. Gene Chromosome Canc. 2007;46:1080–1089. doi: 10.1002/gcc.20493. [DOI] [PubMed] [Google Scholar]
- 30.Kim TM, Laird PW, Park PJ. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell. 2013;155:858–868. doi: 10.1016/j.cell.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21:677–687. doi: 10.1038/nm.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen IC, TeKippe EM, Woodford RM, Uronis JM, Holl EK, Rogers AB, Herfarth HH, Jobin C, Ting JP-Y. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. Int J Clin Exp Med. 2010;207:1045–1056. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu B, Elinav E, Huber S, Booth CJ, Strowig T, Jin C, Eisenbarth SC, Flavell RA. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. Nat Immunol. 2010;107:21635–21640. doi: 10.1073/pnas.1016814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaki MH, Vogel P, Body-Malapel M, Lamkanfi M, Kanneganti T-D. IL-18 production downstream of the Nlrp3 inflammasome confers protection against colorectal tumor formation. J Immunol. 2010;185:4912–4920. doi: 10.4049/jimmunol.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu B, Elinav E, Flavell RA. Inflammasome-mediated suppression of inflammation-induced colorectal cancer progression is mediated by direct regulation of epithelial cell proliferation. Cell Cycle. 2011;10:1936–1939. doi: 10.4161/cc.10.12.16008. [DOI] [PubMed] [Google Scholar]
- 36.Wilson JE, Petrucelli AS, Chen L, Koblansky AA, Truax AD, Oyama Y, Rogers AB, Brickey WJ, Wang Y, Schneider M, Muhlbauer M, Chou WC, Barker BR, Jobin C, Allbritton NL, Ramsden DA, Davis BK, Ting JP. Inflammasome-independent role of AIM2 in suppressing colon tumorigenesis via DNA-PK and Akt. Nat Med. 2015;21:906–913. doi: 10.1038/nm.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Man SM, Zhu Q, Zhu L, Liu Z, Karki R, Malik A, Sharma D, Li L, Malireddi RK, Gurung P, Neale G, Olsen SR, Carter RA, McGoldrick DJ, Wu G, Finkelstein D, Vogel P, Gilbertson RJ, Kanneganti TD. Critical Role for the DNA Sensor AIM2 in Stem Cell Proliferation and Cancer. Cell. 2015;162:45–58. doi: 10.1016/j.cell.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snider AJ, Bialkowska AB, Ghaleb AM, Yang VW, Obeid LM, Hannun YA. Murine model for colitis-associated cancer of the colon. Methods Mol Biol. 2016;1438:245–254. doi: 10.1007/978-1-4939-3661-8_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng J, Park J, Cron P, Hess D, Hemmings BA. Identification of a PKB/Akt hydrophobic motif Ser-473 kinase as DNA-dependent protein kinase. J Biol Chem. 2004;279:41189–41196. doi: 10.1074/jbc.M406731200. [DOI] [PubMed] [Google Scholar]
- 40.Hresko RC, Mueckler M. mTOR· RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J Biol Chem. 2005;280:40406–40416. doi: 10.1074/jbc.M508361200. [DOI] [PubMed] [Google Scholar]
- 41.Van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 42.Barker N, Ridgway RA, Van Es JH, Van M, Begthel H, Van Den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 43.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 44.Vermeulen L, Melo FDSE, van der Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M, Merz C, Rodermond H, Sprick MR, Kemper K, Richel DJ, Stassi G, Medema JP. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 45.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 46.Patsos G, Germann A, Gebert J, Dihlmann S. Restoration of absent in melanoma 2 (AIM2) induces G2/M cell cycle arrest and promotes invasion of colorectal cancer cells. Int J Cancer. 2010;126:1838–1849. doi: 10.1002/ijc.24905. [DOI] [PubMed] [Google Scholar]
- 47.Hu S, Peng L, Kwak Y-T, Tekippe EM, Pasare C, Malter JS, Hooper LV, Zaki MH. The DNA sensor AIM2 maintains intestinal homeostasis via regulation of epithelial antimicrobial host defense. Cell Rep. 2015;13:1922–1936. doi: 10.1016/j.celrep.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu G, Song P, Li N, Chen W, Lei Q, Yu S, Zhang X, Du C, Deng X, Han W. AIM2 contributes to the maintenance of intestinal integrity via Akt and protects against Salmonella mucosal infection. Mucosal Immunol. 2016;9:1330–1339. doi: 10.1038/mi.2015.142. [DOI] [PubMed] [Google Scholar]
- 49.Ratsimandresy RA, Indramohan M, Dorfleutner A, Stehlik C. The AIM2 inflammasome is a central regulator of intestinal homeostasis through the IL-18/IL-22/STAT3 pathway. Cell Mol Immunol. 2016 doi: 10.1038/cmi.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pikor L, Thu K, Vucic E, Lam W. The detection and implication of genome instability in cancer. Cancer Metastasis Rev. 2013;32:341–352. doi: 10.1007/s10555-013-9429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schaap P. Cyclic di-nucleotide signaling enters the eukaryote domain. Lubmb Life. 2013;65:897–903. doi: 10.1002/iub.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Tanigui T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Röhl I, Hopfner K-P, Ludwig J, Hornung V. cGAS produces a 2 [prime]-5 [prime]-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen H, Sun H, You F, Sun W, Zhou X, Chen L, Yang J, Wang Y, Tang H, Guan Y. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell. 2011;147:436–446. doi: 10.1016/j.cell.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 60.Zhu Q, Man SM, Gurung P, Liu Z, Vogel P, Lamkanfi M, Kanneganti T-D. Cutting edge: STING mediates protection against colorectal tumorigenesis by governing the magnitude of intestinal inflammation. J Immunol. 2014;193:4779–4782. doi: 10.4049/jimmunol.1402051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corvinus FM. Orth C, Moriggl R, Tsareva SA, Wagner S, Pfitzner EB, Baus D, Kaufman R, Huber LA, Zatloukal K. Persistent STAT3 activation in colon cancer is associated with enhanced cell proliferation and tumor growth. Neoplasia. 2005;7:545–555. doi: 10.1593/neo.04571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahn J, Konno H, Barber G. Diverse roles of STING-dependent signaling on the development of cancer. Oncogene. 2015;34:5302–5308. doi: 10.1038/onc.2014.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huber S, Gagliani N, Zenewicz LA, Huber FJ, Bosurgi L, Hu B, Hedl M, Zhang W, O’Connor W, Murphy AJ. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature. 2012;491:259–263. doi: 10.1038/nature11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dumoutier L, Lejeune D, Colau D, Renauld J-C. Cloning and characterization of IL-22 binding protein, a natural antagonist of IL-10-related T cell-derived inducible factor/IL-22. J Immunol. 2001;166:7090–7095. doi: 10.4049/jimmunol.166.12.7090. [DOI] [PubMed] [Google Scholar]
- 65.Naito Y, Saito K, Shiiba K, Ohuchi A, Saigenji K, Nagura H, Ohtani H. CD8 + T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res. 1998;58:3491–3494. [PubMed] [Google Scholar]
- 66.Fuertes MB, Kacha AK, Kline J, Woo S-R, Kranz DM, Murphy KM, Gajewski TF. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8+ dendritic cells. J Exp Med. 2011;208:2005–2016. doi: 10.1084/jem.20101159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woo SR, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MY, Duggan R, Wang Y, Barber GN, Fitzgerald KA. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, Li X-D, Mauceri H, Beckett M, Darga T. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity. 2014;41:843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bode C, Fox M, Tewary P, Steinhagen A, Ellerkmann RK, Klinman D, Baumgarten G, Hornung V, Steinhagen F. Human plasmacytoid dentritic cells elicit a Type I Interferon response by sensing DNA via the cGAS-STING signaling pathway. Eur J Immunol. 2016;46:1615–1621. doi: 10.1002/eji.201546113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andzinski L, Spanier J, Kasnitz N, Kroger A, Jin L, Brinkmann MM, Kalinke U, Weiss S, Jablonska J, Lienenklaus S. Growing tumors induce a local STING dependent Type I IFN response in dendritic cells. Int J Cancer. 2016;139:1350–1357. doi: 10.1002/ijc.30159. [DOI] [PubMed] [Google Scholar]
- 71.Xia T, Konno H, Ahn J, Barber GN. Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis. Cell Rep. 2016;14:282–297. doi: 10.1016/j.celrep.2015.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peedicayil J. The role of DNA methylation in the pathogenesis and treatment of cancer. Curr Clin Pharmacol. 2012;7:333–340. doi: 10.2174/157488412803305858. [DOI] [PubMed] [Google Scholar]
- 73.Reinert LS, Lopusna K, Winther H, Sun C, Thomsen MK, Nandakumar R, Mogensen TH, Meyer M, Vaegter C, Nyengaard JR, Fitzgerald KA, Paludan SR. Sensing of HSV-1 by the cGAS-STING pathway in microglia orchestrates antiviral defence in the CNS. Nat Commun. 2016;7:13348. doi: 10.1038/ncomms13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang L, Tatsuya T, Nishiyama Y. Oncotarget Strategies For Herpes Simplex Virus-1. Curr Gene Ther. 2016;16:130–143. doi: 10.2174/1566523216666160331130153. [DOI] [PubMed] [Google Scholar]
- 75.Xia T, Konno H, Barber GN (2016) Recurrent loss of STING signaling in melanoma correlates with susceptibility to viral oncolysis. Cancer Res76: 6747-6759 [DOI] [PubMed]
- 76.Chandra D, Quispe-Tintaya W, Jahangir A, Asafu-Adjei D, Ramos I, Sintim HO, Zhou J, Hayakawa Y, Karaolis DK, Gravekamp C. STING ligand c-di-GMP improves cancer vaccination against metastatic breast cancer. Cancer Immunol Res. 2014;2:901–910. doi: 10.1158/2326-6066.CIR-13-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Corrales L, Glickman LH, McWhirter SM, Kanne DB, Sivick KE, Katibah GE, Woo S-R, Lemmens E, Banda T, Leong JJ. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 2015;11:1018–1030. doi: 10.1016/j.celrep.2015.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fu J, Kanne DB, Leong M, Glickman LH, McWhirter SM, Lemmens E, Mechette K, Leong JJ, Lauer P, Liu W. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci Transl Med. 2015;7:283ra52–283rara52. doi: 10.1126/scitranslmed.aaa4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lemos H, Mohamed E, Huang L, Ou R, Pacholczyk G, Arbab AS, Munn D, Mellor AL. STING Promotes the Growth of Tumors Characterized by Low Antigenicity via IDO Activation. Cancer Res. 2016;76:2076–2081. doi: 10.1158/0008-5472.CAN-15-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ding L, Huang XF, Dong GJ, Hu EL, Sheng C, Wang TT, Hu QG, Yan-HongN, Ni YH (2015) Activated STING enhances Tregs infiltration in the HPV-related carcinogenesis of tongue squamous cells via the c-jun/CCL22 signal. Biochim Biophys Acta 1852:2494–2503 [DOI] [PubMed]
- 81.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell. 2001;8:251–262. doi: 10.1016/S1097-2765(01)00329-X. [DOI] [PubMed] [Google Scholar]
- 82.Loo Y-M, Gale M. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet M-C, Besch R, Hopfner K-P. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. P NATL ACAD SCI USA. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hornung V. SnapShot: nucleic acid immune sensors, part 2. Immunity. 2014;41:1066–1067. doi: 10.1016/j.immuni.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 85.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 86.Broquet AH, Hirata Y, McAllister CS, Kagnoff MF. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J Immunol. 2011;186:1618–1626. doi: 10.4049/jimmunol.1002862. [DOI] [PubMed] [Google Scholar]
- 87.Errett JS, Suthar MS, McMillan A, Diamond MS, Gale M., Jr The essential, nonredundant roles of RIG-I and MDA5 in detecting and controlling West Nile virus infection. J Virol. 2013;87:11416–11425. doi: 10.1128/JVI.01488-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Y, Zhang HX, Sun YP, Liu ZX, Liu XS, Wang L, Lu SY, Kong H, Liu QL, Li XH. Rig-I–/– mice develop colitis associated with downregulation of Gαi2. Cell Res. 2007;17:858–868. doi: 10.1038/cr.2007.81. [DOI] [PubMed] [Google Scholar]
- 89.Wang Y, Zhang HX, Yue-Ping S, Liu X. Regression of Peyer’s patches in G alpha i2 deficient mice prior to colitis is associated with reduced expression of Bcl-2 and increased apoptosis. Cell Res. 2011;17:858–868. doi: 10.1038/cr.2007.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arinze IJ, Kawai Y. Transcriptional activation of the human Galphai2 gene promoter through nuclear factor-kappaB and antioxidant response elements. J Biol Chem. 2005;280:9786–9795. doi: 10.1074/jbc.M414006200. [DOI] [PubMed] [Google Scholar]
- 91.Funke B, Lasitschka F, Roth W, Penzel R, Meuer S, Saile M, Gretz N, Sido B, Schirmacher P, Autschbach F. Selective downregulation of retinoic acid-inducible gene I within the intestinal epithelial compartment in crohn’s disease. Inflamm Bowel Dis. 2011;17:1943–1954. doi: 10.1002/ibd.21572. [DOI] [PubMed] [Google Scholar]
- 92.Li XD, Chiu YH, Ismail AS, Behrendt CL, Wight-Carter M, Hooper LV, Chen ZJ. Mitochondrial antiviral signaling protein (MAVS) monitors commensal bacteria and induces an immune response that prevents experimental colitis. Proc Natl Acad Sci USA. 2011;108:17390–17395. doi: 10.1073/pnas.1107114108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Negishi H, Miki S, Sarashina H, Taguchi-Atarashi N, Nakajima A, Matsuki K, Endo N, Yanai H, Nishio J, Honda K. Essential contribution of IRF3 to intestinal homeostasis and microbiota-mediated Tslp gene induction. P Natl Acad Sci USA. 2012;109:21016–21021. doi: 10.1073/pnas.1219482110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ryan CW, Parekha AD, Rancka MC, Goldena DW, Kumara KA, Sooda RF, Pitrodaa SP, Liaoa Z, Huanga X, Dargaa TE, Xua D, Huangb L, Andradeb J. RIG-I–like receptor LGP2 protects tumor cells from ionizing radiation. Proc Natl Acad Sci USA. 2014;111:E484–E491. doi: 10.1073/pnas.1323253111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fullam A, Schröder M (2013) DExD/H-box RNA helicases as mediators of anti-viral innate immunity and essential host factors for viral replication. BBA-Gene Regul Mec 1829:854–865 [DOI] [PMC free article] [PubMed]
- 96.Moy RH, Cole BS, Yasunaga A, Gold B, Shankarling G, Varble A, Molleston JM, tenOever BR, Lynch KW, Cherry S. Stem-loop recognition by DDX17 facilitates miRNA processing and antiviral defense. Cell. 2014;158:764–777. doi: 10.1016/j.cell.2014.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen G, Liu CH, Zhou L, Krug RM. Cellular DDX21 RNA helicase inhibits influenza A virus replication but is counteracted by the viral NS1 protein. Cell Host Microbe. 2014;15:484–493. doi: 10.1016/j.chom.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yu SF, Lujan P, Jackson DL, Emerman M, Linial ML. The DEAD-box RNA helicase DDX6 is required for efficient encapsidation of a retroviral genome. PLoS Pathog. 2011;7:e1002303. doi: 10.1371/journal.ppat.1002303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, Liu ZJ, Modlin RL, Liu YJ, Cheng G. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol. 2012;13:1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kadono M, Kanai A, Nagamachi A, Shinriki S, Kawata J, Iwato K, Kyo T, Oshima K, Yokoyama A, Kawamura T, Nagase R, Inoue D, Kitamura T, Inaba T, Ichinohe T, Matsui H. Biological implications of somatic DDX41 p.R525H mutation in acute myeloid leukemia. Exp Hematol. 2016;745–54:e4. doi: 10.1016/j.exphem.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 101.Cardoso SR, Ryan G, Walne AJ, Ellison A, Lowe R, Tummala H, Rio-Machin A, Collopy L, Al Seraihi A, Wallis Y, Page P, Akiki S, Fitzgibbon J, Vulliamy T, Dokal I. Germline heterozygous DDX41 variants in a subset of familial myelodysplasia and acute myeloid leukemia. Leukemia. 2016;30:2083–2086. doi: 10.1038/leu.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li R, Sobreira N, Witmer PD, Pratz KW, Braunstein EM. Two novel germline DDX41 mutations in a family with inherited myelodysplasia/acute myeloid leukemia. Haematologica. 2016;101:e228–e231. doi: 10.3324/haematol.2015.139790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lewinsohn M, Brown AL, Weinel LM, Phung C, Rafidi G, Lee MK, Schreiber AW, Feng J, Babic M, Chong CE, Lee Y, Yong A, Suthers GK, Poplawski N, Altree M, Phillips K, Jaensch L, Fine M, D’Andrea RJ, Lewis ID, Medeiros BC, Pollyea DA, King MC, Walsh T, Keel S, Shimamura A, Godley LA, Hahn CN, Churpek JE, Scott HS. Novel germ line DDX41 mutations define families with a lower age of MDS/AML onset and lymphoid malignancies. Blood. 2016;127:1017–1023. doi: 10.1182/blood-2015-10-676098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Berger G, van den Berg E, Sikkema-Raddatz B, Abbott KM, Sinke RJ, Bungener LB, Mulder AB, Vellenga E. Re-emergence of acute myeloid leukemia in donor cells following allogeneic transplantation in a family with a germline DDX41 mutation. Leukemia. 2016;31:520–522. doi: 10.1038/leu.2016.310. [DOI] [PubMed] [Google Scholar]
- 105.Ditton HJ, Zimmer J, Kamp C, Rajpert-De Meyts E. The AZFa gene DBY (DDX3Y) is widely transcribed but the protein is limited to the male germ cells by translation control. Hum Mol Genet. 2004;13:2333–2341. doi: 10.1093/hmg/ddh240. [DOI] [PubMed] [Google Scholar]
- 106.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2016 doi: 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cruciat C-M, Dolde C, Groot REAd, Ohkawara B, Carmen Reinhard1 HCK, Christof Niehrs (2013) RNA helicase DDX3 is a regulatory subunit of casein kinase 1 in Wnt-β-catenin signaling. Science 339:1436–1441 [DOI] [PubMed]
- 108.Gurzu S, Silveanu C, Fetyko A, Butiurca V, Kovacs Z, Jung I. Systematic review of the old and new concepts in the epithelial-mesenchymal transition of colorectal cancer. World J Gastroenterol. 2016;22:6764–6775. doi: 10.3748/wjg.v22.i30.6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Botlagunta M, Vesuna F, Mironchik Y, Raman A, Lisok A, Winnard P, Jr, Mukadam S, Van Diest P, Chen JH, Farabaugh P, Patel AH, Raman V. Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene. 2008;27:3912–3922. doi: 10.1038/onc.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Su CY, Lin TC, Lin YF, Chen MH. DDX3 as a strongest prognosis marker and its downregulation promotes metastasis in colorectal cancer. Oncotarget. 2015;6:1860218612. doi: 10.18632/oncotarget.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Voss MRHv, Vesuna F, Trumpi K, Brilliant J. Identification of the DEAD box RNA helicase DDX3 as a therapeutic target in colorectal cancer. Oncotarget. 2015;6:28312–28326. doi: 10.18632/oncotarget.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wu DW, Lin PL, Cheng YW, Huang CC, Wang L, Lee H. DDX3 enhances oncogenic KRASinduced tumor invasion in colorectal cancer via the betacatenin/ZEB1 axis. Oncotarget. 2016;7:22687–22699. doi: 10.18632/oncotarget.8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.He TY, Wu DW, Lin PL, Wang L, Huang CC, Chou MC, Lee H. DDX3 promotes tumor invasion in colorectal cancer via the CK1epsilon/Dvl2 axis. Sci Rep. 2016 doi: 10.1038/srep21483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Brai A, Fazi R, Tintori C, Zamperini C, Sanguinetti M. Human DDX3 protein is a valuable target to develop broad spectrum antiviral agents. Proc Natl Acad Sci USA. 2016;113:53885393. doi: 10.1073/pnas.1522987113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang S, Grosse F. Nuclear DNA helicase II unwinds both DNA and RNA. Biochem. 1994;33:3906–3912. doi: 10.1021/bi00179a016. [DOI] [PubMed] [Google Scholar]
- 116.Jain A, Bacolla A, Del Mundo IM, Zhao J, Wang G, Vasquez KM. DHX9 helicase is involved in preventing genomic instability induced by alternatively structured DNA in human cells. Nucleic Acids Res. 2013;41:10345–10357. doi: 10.1093/nar/gkt804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wei X, Pacyna-Gengelbach M, Schluns K, An Q, Gao Y, Cheng S, Petersen I. Analysis of the RNA helicase A gene in human lung cancer. Oncol Rep. 2004;11:253–258. [PubMed] [Google Scholar]
- 118.Sun Z, Wang L, Eckloff BW, Deng B, Wang Y (2014) Conserved recurrent gene mutations correlate with pathway deregulation and clinical outcomes of lung adenocarcinoma in never-smokers. BMC Med Genomics 7:32. doi:10.1186/1755-8794-7-32 [DOI] [PMC free article] [PubMed]
- 119.Mills JR, Malina A, Lee T, Man SM. RNAi screening uncovers Dhx9 as a modifier of ABT-737 resistance in an Eμ-myc/Bcl-2 mouse model. Blood. 2013;121:3402–3412. doi: 10.1182/blood-2012-06-434365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee T, Di Paola D, Malina A, Mills JR, Kreps A, Grosse F, Tang H, Zannis-Hadjopoulos M, Larsson O, Pelletier J. Suppression of the DHX9 helicase induces premature senescence in human diploid fibroblasts in a p53-dependent manner. J Biol Chem. 2014;289:22798–22814. doi: 10.1074/jbc.M114.568535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fidaleo M, Svetoni F, Volpe E, Minana B, Caporossi D, Paronetto MP. Genotoxic stress inhibits Ewing sarcoma cell growth by modulating alternative pre-mRNA processing of the RNA helicase DHX9. Oncotarget. 2015;6:31740–31757. doi: 10.18632/oncotarget.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Albrethsen J, Knol JC, Piersma S, V T. Sub-nuclear proteomics in colorectal cancer: Identification of proteins enriched in the nuclear matrix fraction and regulation in adenoma to carcinoma progression. Mol Cell Proteomics. 2010;9:988–1005. doi: 10.1074/mcp.M900546-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mil J, Ray P, Liu J, Kuan C-T. In Vivo selection against human colorectal cancer xenografts identifies an aptamer that targets RNA helicase protein DHX9. MolTher-Nucl Acids. 2016;5:1–9. doi: 10.1038/mtna.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fuller-Pace FV. DEAD box RNA helicase functions in cancer. RNA Biol. 2013;10:121–132. doi: 10.4161/rna.23312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fuller-Pace FV, Ali S. The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochem Soc Trans. 2008;36:609–612. doi: 10.1042/BST0360609. [DOI] [PubMed] [Google Scholar]
- 126.Shin S, Rossow KL, Grande JP, Janknecht R. Involvement of RNA helicases p68 and p72 in colon cancer. Cancer Res. 2007;67:7572–7578. doi: 10.1158/0008-5472.CAN-06-4652. [DOI] [PubMed] [Google Scholar]
- 127.Causevic M, Hislop RG, Kernohan NM, Carey FA, Kay RA, Steele RJ, Fuller-Pace FV. Overexpression and poly-ubiquitylation of the DEAD-box RNA helicase p68 in colorectal tumours. Oncogene. 2001;20:7734–7743. doi: 10.1038/sj.onc.1204976. [DOI] [PubMed] [Google Scholar]
- 128.Nakagawa Y, Morikawa H, Hirata I, Shiozaki M, Matsumoto A, Maemura K, Nishikawa T, Niki M, Tanigawa N, Ikegami M, Katsu K, Akao Y. Overexpression of rck/p54, a DEAD box protein, in human colorectal tumours. Br J Cancer. 1999;80:914–917. doi: 10.1038/sj.bjc.6690441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lin F, Wang R, Shen JJ, Wang X, Gao P, Dong K, Zhang HZ. Knockdown of RCK/p54 expression by RNAi inhibits proliferation of human colorectal cancer cells in vitro and in vivo. Cancer Biol Ther. 2008;7:1669–1676. doi: 10.4161/cbt.7.10.6660. [DOI] [PubMed] [Google Scholar]
- 130.Zhang Z, Kim T, Bao M, Facchinetti V, Jung SY, Ghaffari AA, Qin J, Cheng G, Liu YJ. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34:866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fu JJ, Li LY, Lu GX. Molecular cloning and characterization of human DDX36 and mouse Ddx36 genes, new members of the DEAD/H box superfamily. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 2002;34:655–661. [PubMed] [Google Scholar]
- 132.Matsumura K, Kawasaki Y, Miyamoto M, Kamoshida Y, Nakamura J, Negishi L, Suda S, Akiyama T. The novel G-quadruplex-containing long non-coding RNA GSEC antagonizes DHX36 and modulates colon cancer cell migration. Oncogene. 2016 doi: 10.1038/onc.2016.282. [DOI] [PubMed] [Google Scholar]