Abstract
Cytochrome P450 2U1 (CYP2U1) exhibits several distinctive characteristics among the 57 human CYPs, such as its presence in almost all living organisms with a highly conserved sequence, its particular gene organization with only five exons, its major location in thymus and brain, and its protein sequence involving an unusually long N-terminal region containing 8 proline residues and an insert of about 20 amino acids containing 5 arginine residues after the transmembrane helix. Few substrates, including fatty acids, N-arachidonoylserotonin (AS), and some drugs, have been reported so far. However, its biological roles remain largely unknown, even though CYP2U1 mutations have been involved in some pathological situations, such as complicated forms of hereditary spastic paraplegia. These data together with its ability to hydroxylate some fatty acids and AS suggest its possible role in lipid metabolism.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-016-2443-3) contains supplementary material, which is available to authorized users.
Keywords: Orphan cytochrome P450, Brain, Thymus, Fatty acid hydroxylation, N-Arachidonoylserotonin, Hereditary spastic paraplegia
Introduction
The cytochromes P450 (CYPs) constitute a very large superfamily of hemeproteins that are widely distributed in living organisms [1]. Most often, they are involved as catalysts for the insertion of one atom of the dioxygen molecule into a great number of substrates (monooxygenase reaction), but some of them catalyze other various reactions [1–3]. In the human genome, 57 genes encoding CYPs have been identified (http://drnelson.utmem.edu/CytochromeP450.html). Most of them are known to be involved in the oxidative metabolism of xenobiotics, such as drugs, pesticides, and other environmental chemicals, or in the metabolism of endogenous compounds, such as steroid hormones, fatty acids, eicosanoids, and vitamins [1, 4]. About 1/4 of the human CYPs have been called “orphans” as their biological functions are still largely unknown [5–7]. Among those “orphans”, CYP2U1 is particularly intriguing because of its singularities in terms of gene organization, protein sequence, and tissue expression [8, 9].
It has been proposed that CYP2U1 has a pre-vertebrate origin and that the CYP2U subfamily is very old and highly conserved across species [10–13]. Thus, the sea urchin Strongylocentrotus purpuratus genome contains 73 CYP2-like genes, but it was possible to only assign a single CYP2 gene, CYP2U, to a subfamily represented in vertebrates [14]. Moreover, zebrafish has 47 CYP2 genes, but only two of them, CYP2U1 and CYP2R1, were recognized as orthologs of human CYP2 genes [11]. Human CYP2U1 shares 99, 89, 88, 87, and 83% amino-acid sequence identity with chimpanzee (Pan troglodytes), rat (Rattus norvegicus), dog (Canis lupus familiaris), horse (Equus caballus), and mouse (Mus musculus) CYP2U1, respectively (Table 1). It also shares 67, 64, and 53% amino-acid sequence identity with bird (Taeniopygia guttata), xenope (Xenopus tropicalis), and fish (Takifugu rubripes) CYP2U1 (Table 1). Finally, CYP2U1 is presently the only reported member of the CYP2U subfamily; in other terms, there is no CYP2U2 reported so far.
Table 1.
Protein sequence identity between human CYP2U1 and CYP2U1 from various species
| Species | Identity with human CYP2U1 | References |
|---|---|---|
| Chimpanzee (Pan troglodytes) | 99.8 | [5] |
| Rat (Rattus norvegicus) | 88.9 | [9] |
| Dog (Canis lupus familiaris) | 88 | [18] |
| Horse (Equus caballus) | 87 | [18] |
| Mouse (Mus musculus) | 78.9 | [8] |
| Bird (Taeniopygia guttata) | 67 | [18] |
| Xenope (Xenopus tropicalis) | 64 | [18] |
| Fish (Gasterosteus aculeatus) | 58 | [18] |
| Zebra fish (Danio rerio) | 56 | [18] |
| Fish (Takifugu rubripes) | 52.6a | [9] |
a58% in [8]
Distinctive characteristics of the human CYP2U1 gene within the human CYP2 family
The CYP2U1 gene is located on chromosome 4q25 and spans 18 kb [15]. It contains five exons, whereas all the members of subfamily 2, except CYP2R1, have nine exons [15, 16]. Interestingly, CYP2R1 is the closest human relative of CYP2U1 (39% amino-acid sequence identity) [8, 9, 16]. Figure 1 shows that CYP2U1 and 2R1 have three introns (1, 3, and 4) equivalent to those of the other members of family 2 (2, 6, and 8), that should be present on an ancestral gene common to this family. Starting from this ancestral gene, CYP2U1 and 2R1 should have acquired a new intron (intron 2 of CYP2U1 in Fig. 1). In the case of CYP2U1, the appearance of this intron would be linked to a GC splicing site specific to this gene [15]. Phylogenetic trees of CYP2U1 and other human CYPs showed CYP2U1 clusters together with the other members of family 2 [7–9, 15, 16] with the closest similarity to CYP2R1 [7–9, 16].
Fig. 1.
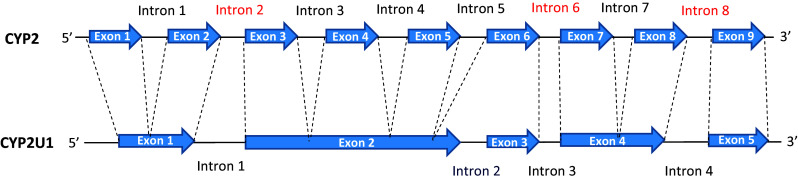
Schematic comparison between the intron/exon organization of the CYP2U1 gene with that generally found for the genes of the human CYP2 family (except CYP2R1), from [8, 15, 18]
Singularities of the human CYP2U1 protein sequence
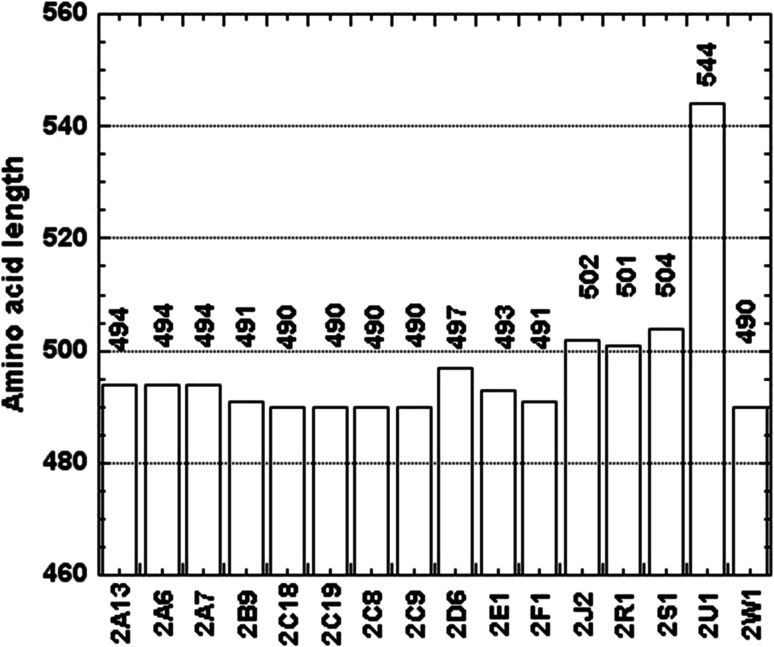
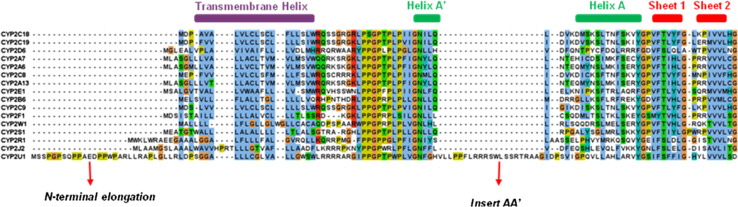
Alignment of the CYP2U1 protein sequence to the closest human relatives CYP2R1 and CYP2D6 and to rabbit CYP2C5 showed that CYP2U1 contains about 50 extra amino acids compared to these other CYP2s. Those extra amino acids appeared to be located at the N-terminus of the protein and after the N-terminal membrane spanning domain [7, 8]. A comparison of the primary sequence length of the 16 members of the human CYP2 family showed that there is a striking gap between CYP2U1 (544 residues) and the other CYP2s (490–504 residues) (Fig. 2) [17]. A sequence alignment of the 16 human CYP2 isoforms clearly showed that the CYP2U1 extra amino acids are located in the N-terminal region of the protein (Fig. 3) [17, 18]. As shown in Fig. 3, the N-terminal region of CYP2U1 located before the predicted transmembrane helix (TMH) involves 20 amino acids more than that of the longer member of family 2 (in that region). Moreover, the CYP2U1 region located after TMH, between the predicted helices A and A′, contains an extra insert of about 20 amino acids (called insert AA′ hereafter). Interestingly, insert AA′ contains five arginine residues. The C-terminal block (104–544) of CYP2U1 aligns pretty well with those of all the other members of human family 2 [17]. A BLASTP search [19] showed that these two regions (the N-terminal region before TMH and insert AA′) are present in all CYP2U1 orthologs, but their length and composition can vary. A BLAST search of human CYP2U1 insert AA′ failed to detect any other similar sequences neither in PDB, nor in UniProtKB databases [18].
Fig. 2.
Amino-acid length of the members of human CYP2 family, from [18]
Fig. 3.
Alignment of the N-terminal regions of the 16 members of human CYP2 family, from [17]
A sequence alignment of the 57 human CYPs (Figure S1 of Supplementary Information) showed that CYP2U1 is the longest one. It has at least 40 amino acids more than the CYPs of families 2 and 3, and 13–33 amino acids more than the CYPs of family 4. As previously mentioned from the alignment of the 16 human CYPs of family 2, it mainly differs from the other human CYPs in its N-terminal part. The region before TMH in CYP2U1 is the longest one, with 14–25 amino acids more than those of CYPs of families 2 and 3 (Figure S1 of Supplementary Information). The CYPs having the longest region before TMH, after CYP2U1, are some mitochondrial CYPs, some CYPs belonging to families 1, such as CYP1B1, and 4, such as CYP4F22, and CYP2R1 and CYP2J2, the two CYPs having the largest sequence identity with CYP2U1 [8, 9, 16, 20]. A first intriguing singularity of the CYP2U1 sequence is the presence in the region before TMH of eight proline residues, whereas in all the other human P450s, one finds not more than four prolines in the same region (Figure S1 of Supplementary Information). Another singularity of the CYP2U1 sequence is the above-mentioned presence of an about 20 amino acids insert, called AA′, that cannot be found in any of the other human CYPs. Interestingly, as mentioned above, this insert contains five arginine residues.
In summary, when compared to the other 56 human CYPs, CYP2U1 exhibits the following singularities: a longer N-terminal region with eight proline residues before TMH and the unique presence of an about 20 amino acids insert containing five arginine residues located after TMH.
Expression of CYP2U1 in the various organs and tissues
First data published in 2004 showed that the level of human CYP2U1 mRNA was particularly high in thymus and cerebellum [8, 9]. In a more general manner, such RNA studies in humans, rats, and mice showed highest expression in thymus followed by brain and heart [8, 9, 21]. A further detailed study in 22 human tissues revealed the presence of human CYP2U1 mRNA in almost all these tissues with the highest expression in thymus followed by bladder, prostate, uterus, testis, and kidney [22]. In the human brain, the major CYP transcripts corresponded to CYP1B1, CYP2D6, CYP2E1, CYP2J2, CYP46A1, and CYP2U1; they were found in all brain structures with an unequal distribution in the different cerebral regions. CYP2U1 represented about 10% of the CYP transcripts expressed in total human brain [23]. Moreover, the main CYP gene expressed in the hCMEC/D3 immortalized human cerebral microvascular endothelial cell line, a promising in vitro model of the human blood brain barrier, is the CYP2U1 gene [24]. A review article of 2006 [25] indicated that the CYP2U1 gene was expressed in the skin of human individuals, in epidermis or cultured keratinocytes, and it was reported that CYP2U1 was expressed at the highest level in undifferentiated keratinocytes [25].
An analysis of CYPs by western blotting in microsomal and mitochondrial purified fractions from human liver and brain samples, including frontal lobe, hippocampus, substantia nigra, and cerebellum, showed the presence of the CYP2U1 protein in all the analyzed brain regions [23]. The subcellular location of CYP2U1 both in the microsomal and mitochondrial fractions was found in all these regions. Interestingly, whereas CYP2U1 transcripts were detected in brain and liver samples, the CYP2U1 protein was only found in the brain samples [23]. This protein was present in the frontal lobe at higher levels than in the cerebellum; a similar result was reported for CYP2U1 in rat [8]. In a more general manner, in rat, the CYP2U1 protein was detected in brain (limbic structures, cortex, olfactory bulbs, pons, and medulla) and thymus, but not in spleen, lung, intestine, liver, and kidney [7, 8]. The topographical and cellular distribution of CYP1B1, CYP2D6, CYP2J2, CYP2U1, and CYP46A1 in three human brain regions, the frontal lobe, hippocampus, and cerebellum, was determined by immunohistochemical staining [23]. Moreover, this study showed an intense staining at the level of the astrocytes and a particularly strong detection in the astrocytes that contact microvessels. CYP2U1 and CYP1B1 were the main CYPs detected in freshly isolated human brain microvessels by RT-PCR and LC–MS/MS [26, 27]. CYP2U1 was also detected at the mRNA and protein levels in human platelets [28] and in human megakaryocytic Dami cells [29]. It is also expressed in white adipose tissue, and it is noteworthy that expression of CYPs is considerably lower in white adipose tissue than in liver, except for CYP2U1 and CYP1B1 that are the most highly expressed adipose tissue CYPs in all the studied individuals [30].
Regulation of CYP2U1
Few data are presently available about the regulation of CYP2U1. In mouse liver, mRNA levels decrease after activation of Nrf2 [31]. Recently, a study of the effects of TCPOBOP, a ligand of the constitutive androstane receptor, on mouse liver showed a slight decrease of the CYP2U1 mRNA levels [32]. In a study of the expression of several CYP2 proteins in prefrontal cortex and amygdala from alcoholic smokers and nonsmokers, and nonalcoholic smokers and nonsmokers, it was found that CYP2U1 and CYP2E1 were expressed in all analyzed samples and elevated in alcoholics [33, 34]. Expression of CYP2U1 was also slightly increased in smokers. These effects of alcohol and tobacco on CYP2U1 expression may have consequences on the metabolism of drugs and endogenous compounds by this cytochrome [33, 34].
Few articles on the effects of pathological situations on CYP2U1 expression in humans have been published. Thus, CYP2U1 was found to be expressed at a significantly higher level in primary colorectal [35], primary ovarian [36], and breast [37] cancer, compared with normal tissues. Moreover, in a study of the effects of major blunt trauma on gene expression of oxidative enzymes in circulating leukocytes, it was found that the expression of CYP2U1 is significantly reduced [38].
Pathological situations associated with CYP2U1 mutations
A study of the genetic polymorphism of CYP2U1 in 70 individuals showed that no mutation could be observed at the exons level and that the most frequent mutations occurring in the 5′-flanking region and in intron 2 did not seem to alter the expression of CYP2U1 in lung [39]. These data indicate that CYP2U1 exhibits few genetic variations and support a probable endogenous role of this protein [39, 40]. However, some rare CYP2U1 mutations have recently been associated with several pathological situations. Such mutations in the CYP2U1 gene were found to be related to the appearance of hereditary spastic paraplegia (HSP), a neurological disorder [41–45]. Quite recently, three cases of pigmentary degenerative maculopathy associated with progressive spastic paraplegia were found to be linked to a CYP2U1 homozygous mutation [44]. In a completely different area, the search for genetic variants associated with lung function, it was shown that nine SNPs in strong linkage disequilibrium in the CYP2U1 gene were associated with “forced expiratory volume in one second” which seems to be a good predictor of mortality [46].
Enzymatic activities of human CYP2U1
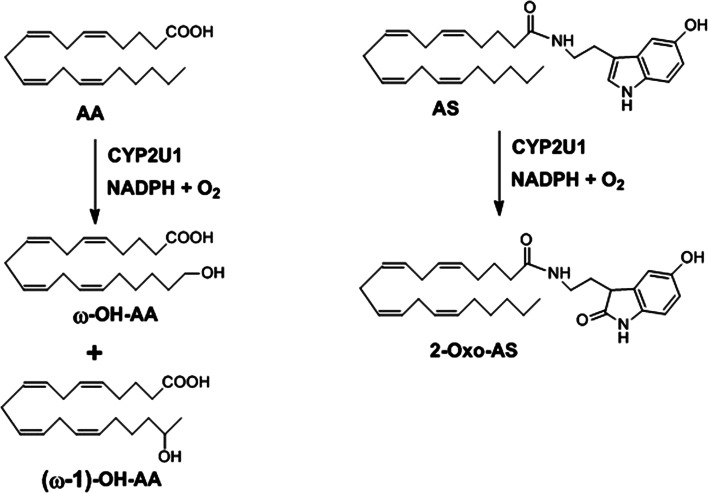
Recombinant human CYP2U1 has been expressed in human embryonic kidney cells (HEK 293) [8], baculovirus-infected Sf9 insect cells [9], E. coli [20, 47], and yeast [17]. CYP2U1 expressed in insect cells was found to catalyze the hydroxylation of fatty acids [9]. Arachidonic acid (AA) was oxidized, with an apparent K m value of 2.7 µM, with formation of ω- and ω-1-hydroxy-AA that were completely identified (Fig. 4). Other unsaturated fatty acids (linolenic, palmitoleic, vaccenic, eicosatrienoic, eicosapentaenoic, EPA, and docosahexaenoic, DHA, acids) as well as long saturated fatty acids (palmitic and stearic acids) were also found to be oxidized by CYP2U1. LC–MS studies showed the insertion of an oxygen atom with the possible formation of hydroxy- or epoxy-fatty acid metabolites, but the precise structures of the metabolites were not established. By contrast, lauric acid and linoleic acid were not substrates of CYP2U1 [9].
Fig. 4.
Regioselectivity of the CYP2U1-catalyzed oxidation of AA and AS, from [9, 20]
Recombinant human CYP2U1 expressed in E. coli was also found to hydroxylate AA at the ω- and ω-1 positions with a rate of 19 ± 5 pmol products min−1 nmol P450−1 [20]. Another CYP2U1 substrate was identified in a metabolomic search for substrates and products of this cytochrome using bovine brain extracts. This substrate, N-arachidonoyl serotonin (AS), was oxidized by E. coli expressed human CYP2U1 at position 2 of its indole ring [20] (Fig. 4). The k cat and K m values found for this CYP2U1 reaction were 32 ± 9 pmol min−1 nmol CYP2U1−1 and 82 ± 2 µM, respectively.
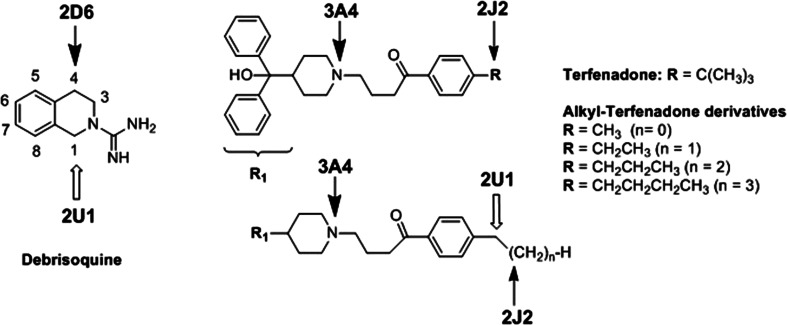
Another search for CYP2U1 substrates was performed using microsomes from the W(hR) yeast expressing human CYP2U1 [17]. About 40 compounds most of which were known to be transformed by other members of the CYP2 family were tested. This included known substrates of CYP2J2, 2D6, and 2R1 that are the closest CYPs phylogenetically related to CYP2U1. Among these compounds that were of exogenous or endogenous origin, a drug, debrisoquine (Deb), and a series of xenobiotics related to the drug terfenadine were found to be hydroxylated by CYP2U1 (Fig. 5).
Fig. 5.
Formula of debrisoquine, terfenadone, and terfenadone derivatives bearing an alkyl group in place of the tertiobutyl one. Arrows show the main sites of oxidations of Deb catalyzed by CYP2D6 and 2U1 [17, 51], and of Terf and alkyl-Terf by CYP3A4, 2J2, and 2U1 [17, 55, 56]
Deb is an anti-hypertensive drug [48] that is a reference substrate of CYP2D6 used for phenotyping human genetic polymorphism of CYP2D6 [49]. Its main metabolite formed upon oxidation with CYP2D6 and found in human urine is 4-hydroxy-Deb [50]. CYP2U1 expressed in yeast was found to hydroxylate Deb with a regioselectivity clearly different from that observed for CYP2D6 [17]. Actually, CYP2U1-catalyzed oxidation of Deb mainly leads to 1-hydroxy-Deb, to smaller amounts of 4-hydroxy- and 6-hydroxy-Deb, and to very small amounts of 8-hydroxy-Deb [17] (Fig. 5), whereas CYP2D6- catalyzed oxidation of Deb mainly leads to 4-hydroxy-Deb and 6-hydroxy-Deb [51]. As shown in Table 2, the k cat values calculated for the CYP2U1-catalyzed 1-, 4-, and 6- hydroxylations of Deb were between 0.2 and 0.5 min−1, and are comparable to that reported for the CYP2D6-catalyzed 4-hydroxylation of Deb (0.8 min−1) [52]. However, the K m values found for these CYP2U1 reactions were clearly higher (about 400 µM) than those reported for CYP2D6- catalyzed 4-hydroxylation of Deb (12 µM) [51]. A study of the structural elements that are important for recognition of Deb as a CPY2U1 substrate showed that the presence of a charged guanidine function was necessary, as suppression of the C(NH2)=NH moiety or replacement of the guanidine function by urea or N-hydroxyguanidine functions led to compounds that were not substrates of CYP2U1 [17].
Table 2.
Enzymatic activities reported for CYP2U1
| Substrates | Product(s) | k cat (min−1) | K m (μM) |
|---|---|---|---|
| AA [9, 20] | ω- and ω-1 hydroxy-AA | 0.019 ± 0.005 | 2.7 |
| AS [20] | 2-Oxo-AS | 0.032 ± 0.009 | 82 ± 2 |
| Deb [17] | 1-OH-Deb | 0.5 ± 0.05 | 480 ± 20 |
| Terf analogs [17] | Chain hydroxylationa | ||
| R=CH3 | Chain hydroxylationa | 0.9 ± 0.3 | 1600 ± 500 |
| R=CH2CH3 | Chain hydroxylationa | 0.4 ± 0.07 | 1200 ± 300 |
| R=(CH2)2CH3 | Chain hydroxylationa | 0.6 ± 0.3 | 800 ± 300 |
| R=(CH2)3CH3 | Chain hydroxylationa | 0.4 ± 0.12 | 1100 ± 400 |
aSee Fig. 5 for structures
Terfenadone (Terf) is a derivative of terfenadine, an antihistaminic drug [53]. Terf and alkyl-Terf, Terf derivatives, in which the tertiobutyl substituent was replaced by various linear alkyl chains (Fig. 5), are oxidized by CYP2J2 with the major formation of products resulting from an hydroxylation in the homobenzylic position [54, 55]. By contrast, the oxidation of Terf and alkyl-Terf by CYP3A4 mainly leads to products derived from an N-dealkylation reaction [55, 56]. All the tested alkyl-Terf are substrates of CYP2J2 or CYP3A4, whereas only alkyl-Terf involving a terminal linear alkyl chain R were found as substrates of CYP2U1 [17]. Moreover, CYP2J2-catalyzed hydroxylation of alkyl-Terf mainly occurs on the homobenzylic position, whereas their hydroxylation catalyzed by CYP2U1 mainly occurs on the benzylic position (Fig. 5). The kcat values found for these CYP2U1-catalyzed hydroxylations (between 0.4 and 0.9 min−1) were similar to that found for Deb hydroxylation and higher than those found for AA and AS hydroxylation (Table 2). However, their K m values (around 1 mM) were also much higher than those found for AA and AS hydroxylation (Table 2). The very different regioselectivities of Deb hydroxylation by CYP2U1 and CYP2D6 and those of Terf derivatives by CYP2U1 and CYP2J2 will be explained in the following.
Construction of a 3D model of CYP2U1 and docking of substrates in its active site
A 3D model of CYP2U1 deprived of its membrane spanning domain (residues 57–544) was constructed by homology modeling on the basis of template X-ray structures of CYP2 family members closely related to CYP2U1 in terms of sequence identity [17]. As expected, the secondary structure elements of the obtained CYP2U1 3D model were identical to those of the other member of the CYP2 family. Its SAMS (solvent accessible molecular surface) active site volume, 931 A3, is intermediate between those of CYP2E1 or CYP2A6 that are known for their small active site, and those of CYPS that bind large substrates, such as CYP2C8 [57]. It is comparable to those of CYP2B6, 2C5, or 2J2 [17].
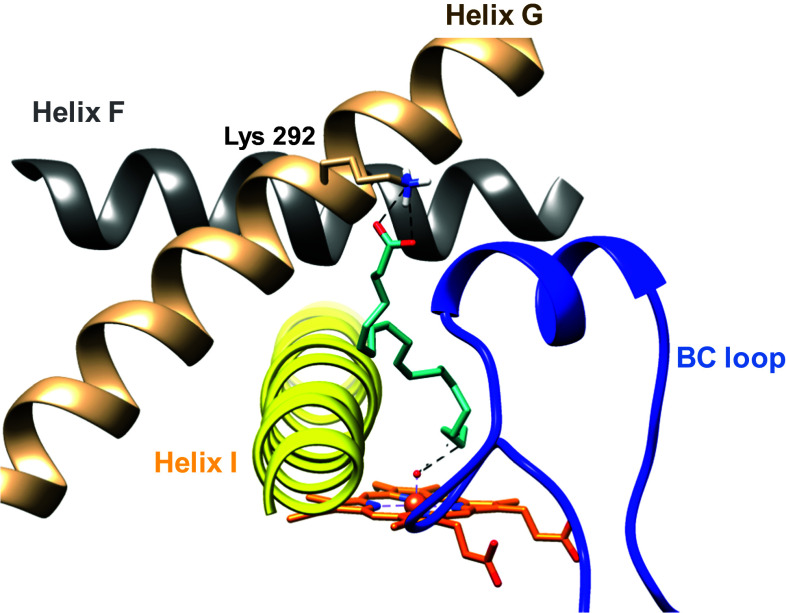
Docking of the reported CYP2U1 substrates into the 3D model of this cytochrome allowed one to explain the observed regioselectivities of the CYP2U1-catalyzed hydroxylation of these substrates at the molecular level [17, 47]. Thus, docking of AA in the aforementioned CYP2U1 3D model led to most energetically favored poses with AA in an extended conformation stretching through channel 2ac (according to the P450 channel nomenclature of Wade et al. [58, 59]) up to the protein surface [17] (Fig. 6). Its carboxyl group interacts with Lys 292 at the entrance of channel 2ac and its alkyl chain establishes hydrophobic interactions with several hydrophobic residues of channel 2ac. The distances between the heme iron-oxo oxygen atom and the ω and ω-1 carbons of AA (between 2.5 and 3.5 Å) are in good agreement with the previously reported regioselectivity of CYP2U1-catalyzed hydroxylation of AA [9].
Fig. 6.
Docking of AA in the CYP2U1 3D model, from [17]. The carboxylate group of AA interacts with εN of Lys292 (distance 3.0 Å). The BC loop is colored blue, the helix F grey, helix G brown, and helix I yellow. The heme (orange), AA (magenta), and side-chain of Lys292 are shown in stick representation, with N atoms in blue and O atoms in red
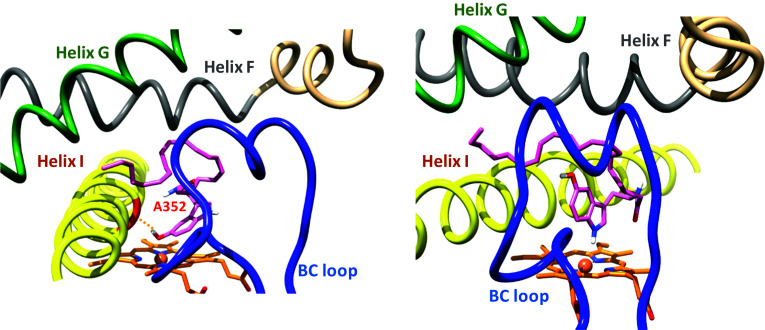
In the most frequently found poses corresponding to the lowest energy observed in the docking of AS in this 3D model [47], it is the indole ring of AS that is closest to the heme (Fig. 7). Either its phenol-OH substituent or its indole nitrogen atom is closest to iron as a function of the considered pose, with Fe–O and Fe–N distances of 2.2 and 4.1 Å, respectively. This different positioning of AA and AS in the CYP2U1 active site allowed one to understand the different regioselectivity of their hydroxylation. In the case of AA, the terminal part position of the AA alkyl chain, close to iron, explains its ω and ω-1 hydroxylations, whereas, in the case of AS, the indole ring is closest to iron explaining the formation of 2-oxo-AS. Formation of the latter product could result either from an epoxidation of the indole C2–C3 double bond followed by an isomerization of the intermediate epoxide, or from an oxidation of the AS phenol function leading to an intermediate quinone-imine followed by an addition of H2O on this intermediate [47]. The results of Siller et al. [20] that used a 1/1 mixture of 16/18O2 would tend to favor the former mechanism.
Fig. 7.
Docking of AS in the CYP2U1 3D model, from [47]. Poses illustrating the two most energetically favored situations. In the pose on the left, the Ala352 backbone C=O group interacts with the phenol-OH group (C=O···HO distance 2.2 Å). In the pose on the right, the indole N-atom is at 4.1 Å from the heme-FeIII atom. The BC loop is colored blue, the helix F grey, helix G green, and helix I yellow. The heme (orange) and AS (pink) are shown in stick representation, with N atoms blue and O atoms red
Docking of Deb in the CYP2U1 model led to energetically favored clusters of poses in which the C-1 and C-6 carbons of Deb are closest to iron in good agreement with the regioselectivity observed for CYP2U1-catalyzed hydroxylation of Deb (Table 2) [17].
Docking of Ethyl-Terf in CYP2U1 led to two clusters of poses in which this substrate is positioned either in channel S or in channel 2ac [17]. In both situations, the ethyl carbons of Ethyl-Terf are close to iron. Docking of the other alkyl-Terf substrates shown in Fig. 5 indicated binding modes and interactions with channel S or 2ac residues similar to those found in the case of Ethyl-Terf. For all these substrates, there is a constraint between their phenylalkyl moiety and the protein residues close to the heme. The distances between the iron and each of the substrate alkyl chain carbons were in the range of distances expected for hydroxylation by the CYP iron-oxo species. These docking data also explained the very different regioselectivities observed for alkyl-Terf substrates oxidation by CYP2U1, 2J2, and 3A4 (Fig. 5). Because of the very wide CYP3A4 active site cavity, the regioselectivity of its reaction should be mainly dictated by the intrinsic chemical reactivity of the different functions of Terf analogs, explaining that the main observed oxidation is an N-dealkylation following N-oxidation. By contrast, the highly regioselective hydroxylation of alkyl-Terf derivatives on their poorly reactive homobenzylic C–H bonds by CYP2J2 was attributed to the very strict positioning of their terminal alkyl group in the CYP2J2 active site in which a crown of protein residues just above the heme leaves a little room for substrate access to CYP2J2 iron [55, 60]. In CYP2U1, this crown of protein residues above the heme leaves more room for substrate access [17]. This would explain the regioselectivity in favor of the hydroxylation of the more reactive benzylic position of the studied alkyl-Terf by CYP2U1.
Spectroscopic studies on CYP2U1
Few data were available on the spectroscopic characteristics of CYP2U1 and its interactions with substrates or ligands. This included the UV–Vis difference spectra of its FeII–CO complex (λ max = 450 nm) [8, 9, 17, 20, 47], the UV–Vis spectrum of native CYP2U1 expressed in E. coli (λ max = 418 nm) indicating that native CYP2U1 mainly exists in a low-spin hexacoordinate Fe(III) state, and the UV–Vis difference spectrum of its complex with AS (“reverse type I”: λ max = 390 nm and λ min = 420 nm) [20]. Recently, a detailed spectroscopic study of human CYP2U1 expressed in E. coli and of its complexes with various substrates and ligands [47] confirmed that native CYP2U1 mainly exists as a low-spin hexacoordinate iron complex characterized by a Soret peak at 418 nm and α and β bands at 534 and 566 nm, and EPR g values at 2.42, 2.25, and 1.92 (at 35 K), presumably involving a water molecule in trans position to the axial cysteinate ligand. At 11 K, its EPR spectrum showed a weak band at g = 8. This indicates that it also exists as a high-spin pentacoordinate complex, the equilibrium between the hexa and pentacoordinate complexes being greatly displaced towards the former. Upon addition of imidazole, these values shifted to 424, 538, and 570 nm, and g values at 2.50, 2.26 and 1.83, as expected for the replacement of the axial H2O ligand with imidazole [61].
A study by UV–Vis difference spectroscopy of the interaction of several imidazole and pyridine derivatives, such as miconazole, with CYP2U1 showed that these derivatives bind to CYP2U1 iron with K s values between 1.5 and 520 µM [47]. The previously described substrates, AS and Deb, led to reverse Type 1 difference spectra, as previously reported in the case of AS [20]. These spectral data could be explained at the molecular level by docking of those substrates and ligands on the aforementioned CYP2U1 3D model [47].
Conclusion and perspectives
Human CYP2U1 is a very peculiar member of the CYP2 family, because of:
its presence in almost all living species with a highly conserved protein sequence (there is so far only one member of subfamily 2U, CYP2U1, reported in the literature),
the particular organization of its gene with 5 exons instead of 9 for the other human CYP2 genes, if one excepts CYP2R1,
its major location in thymus and brain,
its protein sequence involving a long N-terminal region containing 8 proline residues and an insert of about 20 amino acids containing 5 arginine residues after the TMH that are not found in the other human CYPs.
Its two activities with K m values at the µM level, known so far, are the ω- and ω-1-hydroxylation of AA and the oxidation of AS on position 2. However, these activities correspond to low k cat around 0.02 min−1. CYP2U1 is also able to hydroxylate xenobiotics, such as Deb and Terf derivatives, with higher K m (at the mM level) and higher k cat (around 1 min−1).
A 3D homology model for CYP2U1 deprived of its transmembrane spanning domain (residues from 57 to 544) was constructed and used for docking several substrates and ligands. This allowed one to interpret the regioselectivities of the hydroxylations of AA, AS, Deb, and alkylTerf derivatives. Such 3D models should be useful to discover new substrates and selective inhibitors of CYP2U1.
Concerning the biological functions of CYP2U1, it has been proposed that, through the formation of 20- and 19-hydroxy-AA from AA, CYP2U1 could play a role in controlling ion transport and blood pressure in some organs or tissues [9]. CYP2U1 was also proposed to play a role in modulating signal transduction and/or gene expression within immune cells because of its ability to hydroxylate AA, DHA, and EPA in thymus [9]. Moreover, somatic mutations in arachidonic acid metabolism pathway genes, including Cyp2U1, were recently related to enhance oral cancer post-treatment disease-free survival [62].
Several biological activities have been reported for endogenous AS such as its inhibitory properties towards fatty acid amide hydrolase that hydrolyzes anandamide agonists of cannabinoid receptors [63]. Thus, it was proposed that another function of CYP2U1 could be to down-regulate the biological activities of AS [20]. Because of its ability to hydroxylate xenobiotics, such as Deb or alkylTerf derivatives and its location in brain, CYP2U1 could also be involved in the metabolism of drugs and other xenobiotics in brain [17].
As above mentioned, CYP2U1 mutations are involved in complicated forms of hereditary spastic paraplegia that are characterized by a variable combination of neurologic and extra-neurologic symptoms [41–45, 64]. Such HSPs related to CYP2U1 mutations have been associated with thin corpus callosum, white matter abnormalities and/or calcification of the basal ganglia, axonal peripheral neuropathy [41], hydromyelia [43], or pigmentary degenerative maculopathy [44]. Alteration of mitochondrial architecture and bioenergetics with increased oxidative stress were associated with the HSP pathophysiology; this would suggest a role of CYP2U1 in lipid metabolism [41].
Anyway, we are presently far to understand all the biological functions of CYP2U1 in humans or in other species. Moreover, the functional consequences of the CYP2U1 protein sequence singularities mentioned above (long N-terminal region with 8 proline residues and presence of a 20 amino-acid insert after the transmembrane helix) remain to be determined. An X-ray structure and/or a 3D model of membrane-bound full-length CYP2U1 would be quite helpful to determine the roles of the insert that is only found in CYP2U1.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank the French “Ministère de l’Education et de la Recherche” for a fellowship to L.D.s and University Paris Descartes for a collaborative grant.
Abbreviations
- AA
Arachidonic acid
- AS
N-Arachidonoylserotonin
- CYP
Cytochrome P450
- Deb
Debrisoquine
- DHA
Docosahexaenoic acid
- EPA
Eicosapentaenoic acid
- HSP
Hereditary spastic paraplegia
- SAMS
Solvent accessible molecular surface
- Terf
Terfenadone
- TMH
Transmembrane helix
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Ortiz de Montellano PR. Cytochrome P450: structure, mechanism and biochemistry. 4. New York: Kluwer Academic/Plenum Publishers; 2015. [Google Scholar]
- 2.Ortiz de Montellano PR (2015) Substrate oxidation by cytochrome P450 enzymes. In: Ortiz de Montellano PR (ed) Cytochrome P450: structure, mechanism and biochemistry, 4th edn. Kluwer Academic/Plenum Publishers, New York, pp 111–176
- 3.Guengerich FP, Munro AW. Unusual cytochrome P450 enzymes and reactions. J Biol Chem. 2013;288:17065–17073. doi: 10.1074/jbc.R113.462275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guengerich FP (2015) Human cytochrome P450 enzymes. In: Ortiz de Montellano PR (ed), Cytochrome P450: structure, mechanism and biochemistry, 4th edn. Kluwer Academic/Plenum Publishers, New York, pp 523–785
- 5.Stark K, Guengerich FP. Characterization of orphan human cytochromes P450. Drug Metab Rev. 2007;39:627–637. doi: 10.1080/03602530701467708. [DOI] [PubMed] [Google Scholar]
- 6.Guengerich FP, Cheng Q. Orphans in the human cytochrome P450 superfamily: approaches to discovering functions and relevance in pharmacology. Pharmacol Rev. 2011;63:684–699. doi: 10.1124/pr.110.003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karlgren M, Miura SI, Ingelman-Sundberg M. Novel extrahepatic cytochrome P450s. Toxicol Appl Pharmacol. 2005;207:S57–S61. doi: 10.1016/j.taap.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 8.Karlgren M, Backlund M, Johansson I, Oscarson M, Ingelman-Sundberg M. Characterization and tissue distribution of a novel human cytochrome P450-CYP2U1. Biochem Biophys Res Commun. 2004;315:679–685. doi: 10.1016/j.bbrc.2004.01.110. [DOI] [PubMed] [Google Scholar]
- 9.Chuang SS, Helvig C, Taimi M, Ramshaw HA, Collop AH, Amad M, White JA, Petkovich M, Jones G, Korczak B. CYP2U1, a novel human thymus- and brain-specific cytochrome P450, catalyzes omega- and (omega-1)-hydroxylation of fatty acids. J Biol Chem. 2004;279:6305–6314. doi: 10.1074/jbc.M311830200. [DOI] [PubMed] [Google Scholar]
- 10.Nelson DR. Comparison of P450s from human and fugu: 420 million years of vertebrate P450 evolution. Arch Biochem Biophys. 2003;409:18–24. doi: 10.1016/S0003-9861(02)00553-2. [DOI] [PubMed] [Google Scholar]
- 11.Goldstone JV, McArthur AG, Kubota A, Zanette J, Parente T, Jönsson ME, Nelson DR, Stegeman JJ. Identification and developmental expression of the full complement of cytochrome P450 genes in Zebrafish. BMC Genom. 2010;11:643–663. doi: 10.1186/1471-2164-11-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirischian N, McArthur AG, Jesuthasan C, Krattenmacher B, Wilson JY. Phylogenetic and functional analysis of the vertebrate cytochrome p450 2 family. J Mol Evol. 2011;72:56–71. doi: 10.1007/s00239-010-9402-7. [DOI] [PubMed] [Google Scholar]
- 13.Kubota A, Stegeman JJ, Goldstone JV, Nelson DR, Kim EY, Tanabe S, Iwata H. Cytochrome P450 CYP2 genes in the common cormorant: evolutionary relationships with 130 diapsid CYP2 clan sequences and chemical effects on their expression. Comp Biochem Physiol C Toxicol Pharmacol. 2011;153:280–289. doi: 10.1016/j.cbpc.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldstone JV, Hamdoun A, Cole BJ, Howard-Ashby M, Nebert DW, Scally M, Dean M, Epel D, Hahn ME, Stegeman JJ. The chemical defensome: environmental sensing and response genes in the Strongylocentrotus purpuratus genome. Dev Biol. 2006;300:366–384. doi: 10.1016/j.ydbio.2006.08.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2004;14:1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- 16.Cheng JB, Motola DL, Mangelsdorf DJ, Russell DW. De-orphanization of cytochrome P450 2R1: a microsomal vitamin D 25-hydroxylase. J Biol Chem. 2003;278:38084–38093. doi: 10.1074/jbc.M307028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ducassou L, Jonasson G, Dhers L, Pietrancosta N, Ramassamy B, Xu-Li Y, Loriot MA, Beaune P, Bertho G, Lombard M, Mansuy D, André F, Boucher JL. Expression in yeast, new substrates, and construction of a first 3D model of human orphan cytochrome P450 2U1: interpretation of substrate hydroxylation regioselectivity from docking studies. Biochim Biophys Acta. 2015;1850:1426–1437. doi: 10.1016/j.bbagen.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 18.Ducassou L. Study of a human orphan cytochrome P450, CYP2U1. PhD-Thesis. France: University Paris Descartes; 2012. [Google Scholar]
- 19.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siller M, Goyal S, Yoshimoto FK, Xiao Y, Wei S, Guengerich FP. Oxidation of endogenous N-arachidonoylserotonin by human cytochrome P450 2U1. J Biol Chem. 2014;289:10476–10487. doi: 10.1074/jbc.M114.550004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choudhary D, Jansson I, Stoilov I, Sarfarazi M, Schenkman JB. Expression patterns of mouse and human CYP orthologs (families 1-4) during development and in different adult tissues. Arch Biochem Biophys. 2005;436:50–61. doi: 10.1016/j.abb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Bièche I, Narjoz C, Asselah T, Vacher S, Marcellin P, Lidereau R, Beaune P, de Waziers I. Reverse transcriptase-PCR quantification of mRNA levels from cytochrome (CYP)1, CYP2 and CYP3 families in 22 different human tissues. Pharmacogenet Genomics. 2007;17:731–742. doi: 10.1097/FPC.0b013e32810f2e58. [DOI] [PubMed] [Google Scholar]
- 23.Dutheil F, Dauchy S, Diry M, Sazdovitch V, Cloarec O, Mellottée L, Bièche I, Ingelman-Sundberg M, Flinois JP, de Waziers I, Beaune P, Declèves X, Duyckaerts C, Loriot MA. Xenobiotic-metabolizing enzymes and transporters in the normal human brain: regional and cellular mapping as a basis for putative roles in cerebral function. Drug Metab Dispos. 2009;37:1528–1538. doi: 10.1124/dmd.109.027011. [DOI] [PubMed] [Google Scholar]
- 24.Dauchy S, Miller F, Couraud PO, Weaver RJ, Weksler B, Romero IA, Scherrmann JM, De Waziers I, Declèves X. Expression and transcriptional regulation of ABC transporters and cytochromes P450 in hCMEC/D3 human cerebral microvascular endothelial cells. Biochem Pharmacol. 2009;77:897–909. doi: 10.1016/j.bcp.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Du L, Neis MM, Ladd PA, Lanza DL, Yost GS, Keeney DS. Effects of the differentiated keratinocyte phenotype on expression levels of CYP1-4 family genes in human skin cells. Toxicol Appl Pharmacol. 2006;213:135–144. doi: 10.1016/j.taap.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Shawahna R, Uchida Y, Declèves X, Ohtsuki S, Yousif S, Dauchy S, Jacob A, Chassoux F, Daumas-Duport C, Couraud PO, Terasaki T, Scherrmann JM. Transcriptomic and quantitative proteomic analysis of transporters and drug metabolizing enzymes in freshly isolated human brain microvessels. Mol Pharm. 2011;8:1332–1341. doi: 10.1021/mp200129p. [DOI] [PubMed] [Google Scholar]
- 27.Decleves X, Jacob A, Yousif S, Shawahna R, Potin S, Scherrmann JM. Interplay of drug metabolizing CYP450 enzymes and ABC transporters in the blood-brain barrier. Curr Drug Metab. 2011;12:732–741. doi: 10.2174/138920011798357024. [DOI] [PubMed] [Google Scholar]
- 28.Jarrar YB, Cho SA, Oh KS, Kim DH, Shin JG, Lee SJ. Identification of cytochrome P450s involved in the metabolism of arachidonic acid in human platelets. Prostaglandins Leukot Essent Fatty Acids. 2013;89:227–234. doi: 10.1016/j.plefa.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Jarrar YB, Shin JG, Lee SJ. Expression of arachidonic acid-metabolizing cytochrome P450s in human megakaryocytic Dami cells. In Vitro Cell Dev Biol Anim. 2013;49:492–500. doi: 10.1007/s11626-013-9633-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellero S, Chakhtoura G, Barreau C, Langouët S, Benelli C, Penicaud L, Beaune P, de Waziers I. Xenobiotic-metabolizing cytochromes p450 in human white adipose tissue: expression and induction. Drug Metab Dispos. 2010;38:679–686. doi: 10.1124/dmd.109.029249. [DOI] [PubMed] [Google Scholar]
- 31.Wu KC, Cui JY, Klaassen CD. Effect of graded Nrf2 activation on phase-I and -II drug metabolizing enzymes and transporters in mouse liver. PLoS One. 2012;7:e39006. doi: 10.1371/journal.pone.0039006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moriya N, Kataoka H, Nishikawa JI, Kugawa F. Identification of candidate target Cyp genes for microRNAs whose expression is altered by PCN and TCPOBOP, representative ligands of PXR and CAR. Biol Pharm Bull. 2016;39:1381–1386. doi: 10.1248/bpb.b16-00279. [DOI] [PubMed] [Google Scholar]
- 33.Toselli F, de Waziers I, Dutheil M, Vincent M, Wilce PA, Dodd PR, Beaune P, Loriot MA, Gillam E. Gene expression profiling of cytochromes P450, ABC transporters and their principal transcription factors in the amygdala and prefrontal cortex of alcoholics, smokers and drug-free controls by qRT-PCR. Xenobiotica. 2015;45:1129–1137. doi: 10.3109/00498254.2015.1040102. [DOI] [PubMed] [Google Scholar]
- 34.Toselli F, Booth Depaz IM, Worrall S, Etheridge N, Dodd PR, Wilce PA, Gillam EM. Expression of CYP2E1 and CYP2U1 proteins in amygdala and prefrontal cortex: influence of alcoholism and smoking. Alcohol Clin Exp Res. 2015;39:790–797. doi: 10.1111/acer.12697. [DOI] [PubMed] [Google Scholar]
- 35.Kumarakulasingham M, Rooney PH, Dundas SR, Telfer C, Melvin WT, Curran S, Murray GI. Cytochrome p450 profile of colorectal cancer: identification of markers of prognosis. Clin Cancer Res. 2005;11:3758–3765. doi: 10.1158/1078-0432.CCR-04-1848. [DOI] [PubMed] [Google Scholar]
- 36.Downie D, McFadyen MC, Rooney PH, Cruickshank ME, Parkin DE, Miller ID, Telfer C, Melvin WT, Murray GI. Profiling cytochrome P450 expression in ovarian cancer: identification of prognostic markers. Clin Cancer Res. 2005;11:7369–7375. doi: 10.1158/1078-0432.CCR-05-0466. [DOI] [PubMed] [Google Scholar]
- 37.Murray GI, Patimalla S, Stewart KN, Miller ID, Heys SD. Profiling the expression of cytochrome P450 in breast cancer. Histopathology. 2010;57:202–211. doi: 10.1111/j.1365-2559.2010.03606.x. [DOI] [PubMed] [Google Scholar]
- 38.Brandfellner HM, Ruparel SB, Gelfond JA, Hargreaves KM. Major blunt trauma evokes selective upregulation of oxidative enzymes in circulating leukocytes. Shock. 2013;40:182–187. doi: 10.1097/SHK.0b013e31829de02f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Devos A, Lino Cardenas CL, Glowacki F, Engels A, Lo-Guidice JM, Chevalier D, Allorge D, Broly F, Cauffiez C. Genetic polymorphism of CYP2U1, a cytochrome P450 involved in fatty acids hydroxylation. Prostaglandins Leukot Essent Fatty Acids. 2010;83:105–110. doi: 10.1016/j.plefa.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 40.Leclerc J, Tournel G, Courcot-Ngoubo Ngangue E, Pottier N, Lafitte JJ, Jaillard S, Mensier E, Lhermitte M, Broly F, Lo-Guidice JM. Profiling gene expression of whole cytochrome P450 superfamily in human bronchial and peripheral lung tissues: differential expression in non-small cell lung cancers. Biochimie. 2010;92:292–306. doi: 10.1016/j.biochi.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 41.Tesson C, Nawara M, Salih MA, Rossignol R, Zaki MS, Al Balwi M, Schule R, Mignot C, Obre E, Bouhouche A, Santorelli FM, Durand CM, Oteyza AC, El-Hachimi KH, Al Drees A, Bouslam N, Lamari F, Elmalik SA, Kabiraj MM, Seidahmed MZ, Esteves T, Gaussen M, Monin ML, Gyapay G, Lechner D, Gonzalez M, Depienne C, Mochel F, Lavie J, Schols L, Lacombe D, Yahyaoui M, Al Abdulkareem I, Zuchner S, Yamashita A, Benomar A, Goizet C, Durr A, Gleeson JG, Darios F, Brice A, Stevanin G. Alteration of fatty-acid-metabolizing enzymes affects mitochondrial form and function in hereditary spastic paraplegia. Am J Hum Genet. 2012;91:1051–1064. doi: 10.1016/j.ajhg.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Citterio A, Arnoldi A, Panzeri E, D’Angelo MG, Filosto M, Dilena R, Arrigoni F, Castelli M, Maghini C, Germiniasi C, Menni F, Martinuzzi A, Bresolin N, Bassi MT. Mutations in CYP2U1, DDHD2 and GBA2 genes are rare causes of complicated forms of hereditary spastic paraparesis. J Neurol. 2014;261:373–381. doi: 10.1007/s00415-013-7206-6. [DOI] [PubMed] [Google Scholar]
- 43.Masciullo M, Tessa A, Perazza S, Santorelli FM, Perna A, Silvestri G. Hereditary spastic paraplegia: novel mutations and expansion of the phenotype variability in SPG56. Eur J Paediatr Neurol. 2016;20:444–448. doi: 10.1016/j.ejpn.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 44.Leonardi L, Ziccardi L, Marcotulli C, Rubegni A, Longobardi A, Serrao M, Storti E, Pierelli F, Tessa A, Parisi V, Santorelli FM, Carlo C. Pigmentary degenerative maculopathy as prominent phenotype in an Italian SPG56/CYP2U1 family. J Neurol. 2016;263:781–783. doi: 10.1007/s00415-016-8066-7. [DOI] [PubMed] [Google Scholar]
- 45.Kariminejad A, Schöls L, Schüle R, Tonekaboni SH, Abolhassani A, Fadaee M, Rosti RO, Gleeson JG. CYP2U1 mutations in two Iranian patients with activity induced dystonia, motor regression and spastic paraplegia. Eur J Paediatr Neurol. 2016;20:782–787. doi: 10.1016/j.ejpn.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thyagarajan B, Wojczynski M, Minster RL, Sanders J, Barral S, Christiansen L, Barr RG, Newman A, CHARGE consortium, SpiroMeta consortium Genetic variants associated with lung function: the long life family study. Respir Res. 2014;15:134–142. doi: 10.1186/s12931-014-0134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dhers L, Pietrancosta N, Ducassou L, Ramassamy B, Dairou J, Jaouen M, André F, Mansuy D, Boucher JL. Spectral and 3D model studies of the interaction of orphan human cytochrome P450 2U1 with substrates and ligands. Biochim Biophys Acta. 2016;1861:3144–3153. doi: 10.1016/j.bbagen.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 48.Abrams WB, Pocelinko R, Klausner M, Hanauer L, Whitman EN. Clinical pharmacological studies with debrisoquin sulfate, a new antihypertensive agent. J New Drugs. 1964;4:268–283. doi: 10.1002/j.1552-4604.1964.tb00372.x. [DOI] [PubMed] [Google Scholar]
- 49.Fuhr U, Jetter A, Kirchheiner J. Appropriate phenotyping procedures for drug metabolizing enzymes and transporters in humans and their simultaneous use in the “cocktail” approach. Clin Pharmacol Ther. 2007;81:270–283. doi: 10.1038/sj.clpt.6100050. [DOI] [PubMed] [Google Scholar]
- 50.Lanz M, Theurillat R, Thormann W. Characterization of stereoselectivity and genetic polymorphism of the debrisoquine hydroxylation in man via analysis of urinary debrisoquine and 4-hydroxydebrisoquine by capillary electrophoresis. Electrophoresis. 1997;18:1875–1881. doi: 10.1002/elps.1150181025. [DOI] [PubMed] [Google Scholar]
- 51.Lightfoot T, Ellis SW, Mahling J, Ackland MJ, Blaney FE, Bijloo GJ, De Groot MJ, Vermeulen NP, Blackburn GM, Lennard MS, Tucker GT. Regioselective hydroxylation of debrisoquine by cytochrome P4502D6: implications for active site modelling. Xenobiotica. 2000;30:219–233. doi: 10.1080/004982500237622. [DOI] [PubMed] [Google Scholar]
- 52.Ellis SW, Hayhurst GP, Lightfoot T, Smith G, Harlow J, Rowland-Yeo K, Larsson C, Mahling J, Lim CK, Wolf CR, Blackburn MG, Lennard MS, Tucker GT. Evidence that serine 304 is not a key ligand-binding residue in the active site of cytochrome P450 2D6. Biochem J. 2000;345:565–571. doi: 10.1042/bj3450565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kinsolving CR, Munro NL. The objective and timing of drug disposition studies, appendix V. A comparison of the bioavailability of three dosage forms of terfenadine. Drug Metab Rev. 1975;4:285–290. doi: 10.3109/03602537508993762. [DOI] [PubMed] [Google Scholar]
- 54.Lafite P, Dijols S, Zeldin DC, Dansette PM, Mansuy D. Selective, competitive and mechanism-based inhibitors of human cytochrome P450 2J2. Arch Biochem Biophys. 2007;464:155–168. doi: 10.1016/j.abb.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lafite P, André F, Zeldin DC, Dansette PM, Mansuy D. Unusual regioselectivity and active site topology of human cytochrome P450 2J2. Biochemistry. 2007;46:10237–10247. doi: 10.1021/bi700876a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yun CH, Okerholm RA, Guengerich FP. Oxidation of the antihistaminic drug terfenadine in human liver microsomes. Role of cytochrome P-450 3A(4) in N-dealkylation and C-hydroxylation. Drug Metab Dispos. 1993;21:403–409. [PubMed] [Google Scholar]
- 57.Schoch GA, Yano JK, Sansen S, Dansette PM, Stout CD, Johnson EF. Determinants of cytochrome P450 2C8 substrate binding: structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid. J Biol Chem. 2008;283:17227–17237. doi: 10.1074/jbc.M802180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cojocaru V, Winn PJ, Wade RC. The ins and outs of cytochrome P450s. Biochim Biophys Acta. 2007;1770:390–401. doi: 10.1016/j.bbagen.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 59.Yu X, Cojocaru V, Wade RC. Conformational diversity and ligand tunnels of mammalian cytochrome P450s. Biotechnol Appl Biochem. 2013;60:134–145. doi: 10.1002/bab.1074. [DOI] [PubMed] [Google Scholar]
- 60.Li W, Tang Y, Liu H, Cheng J, Zhu W, Jiang H. Probing ligand binding modes of human cytochrome P450 2J2 by homology modeling, molecular dynamics simulation, and flexible molecular docking. Prot Struct Func Genet. 2008;71:938–949. doi: 10.1002/prot.21778. [DOI] [PubMed] [Google Scholar]
- 61.Dawson JH, Andersson LA, Sono M. Spectroscopic investigations of ferric cytochrome P-450-CAM ligand complexes. J Biol Chem. 1982;257:3606–3617. [PubMed] [Google Scholar]
- 62.Biswas NK, Das S, Maitra A, Sarin R, Majumder PP. Somatic mutations in arachidonic acid metabolism pathway genes enhance oral cancer post-treatment disease-free survival. Nat Commun. 2014;5:5835. doi: 10.1038/ncomms6835. [DOI] [PubMed] [Google Scholar]
- 63.Bisogno T, Melck D, De Petrocellis L, Bobrov M, Gretskaya NM, Bezuglov VV, Sitachitta N, Gerwick WH, Di Marzo V. Arachidonoylserotonin and other novel inhibitors of fatty acid amide hydrolase. Biochem Biophys Res Commun. 1998;248:515–522. doi: 10.1006/bbrc.1998.8874. [DOI] [PubMed] [Google Scholar]
- 64.Fink JK. Hereditary spastic paraplegia: clinico-pathologic features and emerging molecular mechanisms. Acta Neuropathol. 2013;126:307–328. doi: 10.1007/s00401-013-1115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.