Abstract
Periostin is a matricellular protein that is composed of a multi-domain structure with an amino-terminal EMI domain, a tandem repeat of four FAS 1 domains, and a carboxyl-terminal domain. These distinct domains have been demonstrated to bind to many proteins including extracellular matrix proteins (Collagen type I and V, fibronectin, tenascin, and laminin), matricellular proteins (CCN3 and βig-h3), and enzymes that catalyze covalent crosslinking between extracellular matrix proteins (lysyl oxidase and BMP-1). Adjacent binding sites on periostin have been suggested to put the interacting proteins in close proximity, promoting intermolecular interactions between each protein, and leading to their assembly into extracellular architectures. These extracellular architectures determine the mechanochemical properties of connective tissues, in which periostin plays an important role in physiological homeostasis and disease progression. In this review, we introduce the proteins that interact with periostin, and discuss how the multi-domain structure of periostin functions as a scaffold for the assembly of interacting proteins, and how it underlies construction of highly sophisticated extracellular architectures.
Keywords: EMI, FAS 1, Fibronectin, Tenascin, Collagen, BMP-1
Introduction
Periostin is a secretory protein of around 90 kDa with a multi-domain structure that is composed of an amino-terminal EMI domain, a tandem repeat of four FAS 1 domains, and a carboxyl-terminal domain (CTD) [1–3]. The CTD amino acid sequence varies as a result of alternative splicing [1, 4]. Periostin has been suggested to function as a scaffold for the assembly of several extracellular matrix proteins as well as its accessory proteins, which underlie highly sophisticated extracellular meshwork architectures [5–8]. This scaffold function is likely due to periostin’s multi-domain structure, which puts the interacting proteins in close proximity and assembles them into a large complex. In accordance with the requirements of the mechanochemical properties of living tissues, the adjacent domains of periostin interact with different kinds of proteins, which serves to maintain pathophysiological states.
Kudo and colleagues developed rabbit polyclonal antibodies against the first FAS 1 domain and the carboxyl-terminal end of the CTD [1, 9], and have revealed the spatiotemporal distribution of periostin in mouse and human tissue. Commercially available antibodies against periostin were also used to investigate periostin localization. Immunohistochemical analyses using these antibodies demonstrated that periostin physiologically localizes at collagen-dense areas in connective tissue, including in the periodontal ligament [1, 10–12], periosteum [1, 11], cardiac valve [13–15], and alveolar wall in the lung [16–18]. Periostin has also been found to pathologically localize in infarcted myocardium [9, 19], fibrosis [18, 20–23], the wound healing process [24–27], and cancer-associated stroma [28–40]. Thus, expression of periostin is also closely associated with tissue regeneration post-injury [41].
Immuno-electron microscopic analyses verified that periostin is localized on the cytoplasmic processes of fibroblasts in the mouse periodontal ligament, and is largely confined to their cell membranes tightly associated with the bundles of surrounding collagen fibers [10, 12]. The cell-surface localization of periostin was also observed in pericryptal fibroblasts in mouse colonic mucosa [30]. Immuno-reactivity for periostin was also detected on collagen bundles in the mouse periodontal ligament [10, 12]. Additionally, immuno-gold transmission electron microscopic localization of periostin was observed on collagen bundles of the mouse atria-ventricular valve [25]. Taken together, these reports suggest that periostin binds to the proteins localized in the extracellular matrix and on the cell surface.
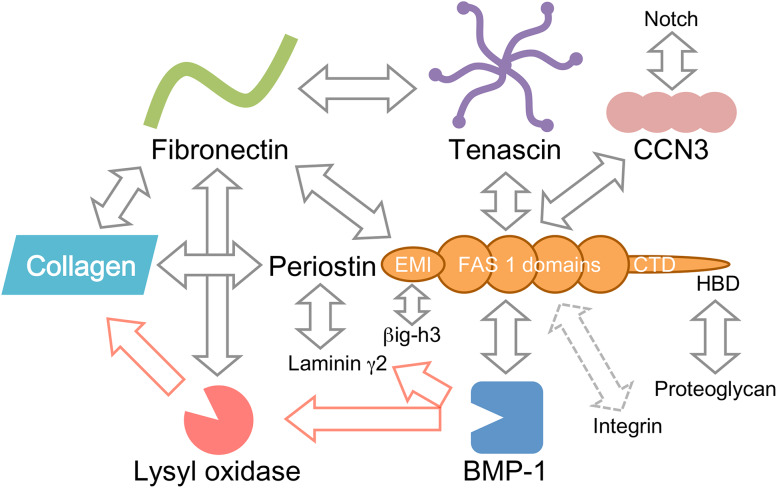
In this review, we introduce the proteins that interact with each of the adjacent domains in periostin (Fig. 1), and discuss that the multi-domain structure of periostin functions as a scaffold for the assembly of these interacting proteins. This scaffold function of periostin likely underlies the construction of highly sophisticated extracellular architectures.
Fig. 1.
Periostin and its interacting proteins. The known interacting proteins of periostin are depicted based on the multi-domain structure of periostin. CTD carboxyl-terminal domain, HBD heparin-binding domain
The amino-terminal EMI domain of periostin interacts with extracellular matrix proteins
The EMI domain, named after its presence in the EMILIN family [42], is composed of approximately 80 amino acid residues, and includes six highly conserved cysteine residues. These cysteine residues are likely to be responsible for multimer formation via a disulfide bond. In a similar manner, periostin forms disulfide-bonded multimers (dimer to hexamer) [6]. The EMI domain is always a single copy located at the amino-terminus of secretory proteins [43]. EMILIN-1 is an extracellular matrix protein involved in elastin deposition [44]. The EMI domain of EMILIN-1 was reported to interact with the C1q domain of EMILIN-2 [45, 46], indicating heterogeneous multimerization between EMILIN-1 and EMILIN-2. It has been demonstrated that the EMI domain of EMILIN-1 and EMILIN-3 binds to pro-TGF-β1 [47, 48]. These reports suggest that the EMI domain is a protein–protein interaction module.
The EMI domain of periostin has been demonstrated to interact with fibronectin. Purified periostin bound to fibronectin-coated microtiter plates [6, 25], indicating a direct interaction. The EMI domain that fused to Fc was co-immunoprecipitated with fibronectin in 293T cells transfected with the expression vectors of EMI-Fc and fibronectin [6]. In addition, a close proximity between periostin and fibronectin was detected inside fibroblastic C3H10T12 cells [49], possibly indicating that the interaction between periostin and fibronectin occurs before its secretion. Furthermore, this interaction in the secretory pathway enhanced secretion of fibronectin into the extracellular milieu [49], suggesting an undetermined role for periostin in protein secretion. Periostin was localized preferentially in the Golgi apparatus and the endoplasmic reticulum, indicating its function in the secretory pathway [6, 49, 50]. The EMI domain possesses a conserved tryptophan residue between the second and third cysteine residue [6, 42]. In periostin, this conserved tryptophan residue locates to position 65. The substitution of this tryptophan to alanine caused loss of the interaction with fibronectin [6], clearly demonstrating that the EMI domain is the interaction domain for fibronectin and that this conserved tryptophan residue is essential for the interaction.
Periostin has also been shown to interact with collagens, which is consistent with the localization of periostin on collagen bundles [10, 12, 25]. Periostin was co-immunoprecipitated with collagen type I [25], and purified periostin protein bound to collagen type V-coated microtiter plates [22]. However, the binding site for collagen has not yet been identified and further studies of this interaction are required.
Fibronectin has several binding regions for collagen, possibly indicating that periostin directly and indirectly interacts with collagens via the EMI domain. As consequence of the interaction of periostin with collagen and fibronectin that is an essential factor for collagen fibrillogenesis [51], periostin plays a role in promoting collagen fibrillogenesis [25]. Collagen fibrillogenesis is a complicated multi-step process that is still poorly understood [52]. Molecular studies on the interaction of periostin with collagen and fibronectin, both inside and outside cells, would contribute to clarifying the mechanism of collagen fibrillogenesis.
The EMI domain of periostin interacts with the EMI domain of βig-h3 (also known as keratoepithelin and RGD-CAP), which is a protein with structural and sequence homology to periostin [53]. βig-h3 is coded in the TGFBI (transforming growth factor-β-induced) gene, mutations of which are associated with corneal dystrophies, progressive eye disorders [54]. The interaction between periostin and βig-h3 likely results from hetero-multimerization via their EMI domains [50]. This interaction was found to be essential for the proper secretion of a periostin/βig-h3 hetero-multimer [50]. Further, it has been demonstrated that βig-h3 directly binds to collagens type I, II, and IV [55], and localizes to the Golgi apparatus as periostin [50]. These similarities between periostin and βig-h3 suggest redundancy in the molecular functions of these matricellular proteins.
The tandem repeat of four FAS 1 domains binds to tenascin, BMP-1, and CCN3
The FAS 1 domain was originally identified in an insect neural cell adhesion molecule, Drosophila fasciclin I, consisting of the tandem repeat of four FAS 1 domains [56, 57]. The crystal structure of the third and fourth FAS 1 domains of Drosophila fasciclin I revealed a unique domain fold, consisting of a seven-stranded β wedge and a number of α helices [57]. Nuclear magnetic resonance spectroscopy was also used to solve the structure of the FAS 1 domain of Mycobacterium bovis secretory protein MPB70, which has structural homology to the FAS 1 domain of fasciclin I [58].
The tandem repeat of four FAS 1 domains of periostin has been demonstrated to bind to tenascin [6]. Tenascin is an extracellular matrix protein, the amino-terminus of which forms a disulfide-bonded hexamer, resulting in a six-armed oligomer, termed a hexabrachion [59]. Co-immunoprecipitation analysis confirmed the interaction of the FAS 1 domains of periostin and tenascin [6]. The purified FAS 1 domains of periostin bound to tenascin-coated microtiter plates [6], clearly demonstrating direct binding. Interestingly, the interaction between the FAS 1 domains and tenascin required cleavage of the CTD of periostin [6]. The CTD of periostin is likely to be cleaved in the secretory pathway, especially in the Golgi apparatus [6]. Although the inhibitory mechanism of the CTD on the FAS 1 domain-tenascin interaction is not currently well understood, inter- and intra-molecular interactions in periostin have been reported [22, 60]. It was shown that recombinant periostin CTD bound to the recombinant tandem repeat of four FAS 1 domains in a solid-phase binding assay [60], suggesting that this intra- or inter-molecular interaction in periostin inhibits interaction of the FAS 1 domains and tenascin. Structural analysis of periostin would clarify this auto-inhibitory mechanism.
The four FAS 1 domains of periostin have been reported to interact with bone morphogenetic protein-1 (BMP-1), which is the pro-collagen C-proteinase that cleaves the carboxyl-terminal propeptides of procollagens I, II, and III [61]. Co-immunoprecipitation analysis revealed that the tandem repeat of the four FAS 1 domains of periostin interacted with the metalloproteinase domain of BMP-1 [7]. In addition, a solid-phase binding assay using purified proteins confirmed direct binding between periostin and BMP-1 [7, 62]. Furthermore, three-dimensional docking simulation was performed between the FAS 1 domains of periostin and BMP-1, implicating their binding sites [62].
This direct interaction between the FAS 1 domains and BMP-1 promoted proteolytic activation of lysyl oxidase (LOX), which was indirectly associated with the EMI domain of periostin through fibronectin [7, 63]. LOX is an enzyme that catalyzes the formation of highly reactive aldehydes from peptidyl lysine residues in collagen molecules [64]. These aldehydes spontaneously react with other aldehydes or with intact lysine residues intermolecularly, resulting in the crosslinking of collagen molecules, which is essential for the stabilization of collagen fibrils. Consequently, targeted deletion of the periostin gene in mice caused the reduction of cross-links in collagens [6, 9, 25]. Thus, the periostin-BMP-1-LOX axis underlies the mechanochemical property of the collagen matrix.
The tandem repeat of four FAS 1 domains in periostin has also been reported to interact with CCN3 [65]. CCN3 is a matricellular protein that belongs to the CCN family. CCN3 consists of four domains: the insulin-like growth factor-binding protein-like domain (IGFBP), the von Willebrand type C-like domain (VWC), the thrombospondin type 1-like domain (TSP1), and the carboxyl-terminal domain (CT). A co-immunoprecipitation experiment revealed that the four FAS 1 domains of periostin interacted with TSP1 and CT of CCN3 [65]. Periostin acted as an anchor of CCN3 for its localization in the extracellular matrix in the mouse periodontal ligament [65]. Similarly to tenascin, BMP-1, and CCN3, periostin is likely to act as an anchor for the FAS 1 domain-interacting proteins to the extracellular matrix through the EMI domain.
The CTD possesses an Arg-rich region responsible for binding to proteoglycans
The CTD of periostin has been demonstrated to interact with heparin [3]. Heparin is a highly acidic polysaccharide with a sulfate group, which is conjugated on transmembrane proteins and secretory proteins called heparan sulfate proteoglycans. The CTD contains basic amino acid residues in its terminal end (Arg–Arg–Arg–Leu–Arg in human) [3], which is a motif common in heparin-binding proteins as B1–B2–X–B3, where B represents a basic residue. This basic amino acid sequence is highly conserved in periostin among species [4], indicating its importance. Thus, periostin is likely to interact with cell-surface heparan sulfate proteoglycans and to regulate cellular processes such as cell migration and growth factor signaling [66]. These interactions have not been extensively investigated, but are likely involved in connective tissue development and disease progression.
The CTD has a variation in amino acid sequence as a result of alternative splicing [1, 4]. The alternatively spliced region in the CTD has not been a target of biological analyses, and thus its molecular function is not well understood. The periostin paralogue βig-h3 lacks an extended region equivalent to the CTD, suggesting that the CTD is of functional relevance and could modulate periostin function. The CTD was shown to inhibit the interaction of the FAS 1 domains with tenascin-C, and cleavage of the CTD was required for the interaction [6]. This cleavage mechanism would underlie the spatiotemporal regulation of the interaction between periostin and tenascin in the secretory pathway. Identification of the enzymes that catalyze this CTD cleavage would clarify the periostin-specific regulation mechanism for the assembly of extracellular matrix proteins.
The predicted protein interactome of periostin
A system-wide functional association for the specific and functionally productive interaction of proteins has been investigated with knowledge-based approaches based on known and predicted protein–protein association data [67]. STRING is a database of protein–protein interactions, including physical and functional associations from computational prediction, knowledge transfer, and interactions of other databases [68]. The interactome map of periostin that was generated by STRING is shown in Fig. 2. This map shows the known interacting proteins of periostin such as fibronectin (fn1), collagen (Col1a1), tenascin (Tnc), and BMP-1 (Bmp1). The map also indicates strong associations between periostin and integrin (Itga2b, Itga3, Itgam, Itgax, Itgb1, Itgb3, and Itgb5). Interestingly, these interactions seem to congregate at Akt1, which is a down-stream cell-survival signal mediator of integrin, consistent with the hypothesis that periostin activates Akt signaling [9, 39, 69–74]. Thus, this knowledge-based approach would be useful for molecular studies of periostin.
Fig. 2.
The interactome map for periostin in STRING. Protein interactome map of periostin, as obtained from the STRING database. STRING analysis of known and predicted protein–protein associations is shown. STRING Version 10.5 was used
Other databases are also useful. The BioPlex is a network of the building blocks based on the immunopurification of tagged proteins and the detection of associated proteins by mass spectrometry [75, 76]. The BioPlex database gives us a physical interaction dataset for proteins of interest. Unfortunately, periostin was not found in the BioPlex. However, the interacting proteins of periostin (fibronectin, BMP-1 and so on) were found, which are useful for understanding the functions of periostin. It would be valuable to identify all of the interacting proteins of periostin by quantitative mass spectrometry-based proteomics in combination with affinity purification protocols, as this would accelerate molecular studies of periostin.
Concluding remarks
In addition to the proteins described in this review, periostin has been reported to interact with the integrin superfamily. Although periostin is thought to interact with integrins, including αVβ3, αVβ5, and α6β4, which promote cell proliferation, cell migration, epithelial to mesenchymal transformation, and modulation of the biomechanical properties of connective tissues [39, 69, 77–82], evidence of the direct interaction between periostin and integrins has not been demonstrated. Periostin has also been found to interact with notch and laminin γ2 [24, 83], and to function as a regulator of these proteins; however, the interacting domain of periostin has not been identified. To understand the relationship between the interacting proteins and periostin, it is necessary to map the interaction sites on the multi-domain structure of periostin.
The pathological roles of periostin have been the focus of a number of published papers. In most cases, inhibition of periostin function has shown a beneficial effect on the prevention and treatment of disease, such as cardiac remodeling, fibrosis, and cancer progression [16, 19, 20, 23, 74, 82, 84–96]. Development of a small organic molecule drug targeting periostin could be useful for disease prevention and treatment. However, structural analyses have not identified a pocket for drug binding in periostin. In addition, drug discovery targeting the protein–protein interaction surface is a promising approach; however, this is still difficult to perform. The interacting proteins of periostin described in this review may be potential drug discovery targets. For example, BMP-1 is an enzyme that has been targeted by small molecule inhibitors [97]. Inhibitors of LOX have been tested as a drug for cancer metastasis [98–100]. Enzymes such as BMP-1 and LOX are involved in the large protein complex based on periostin, and are attractive targets of drug discovery for periostin-related diseases.
Acknowledgements
This work was supported by the Project for Cancer Research and Therapeutic Evolution (P-CREATE) (IK) from the Japan Agency for Medical Research and Development (AMED).
References
- 1.Horiuchi K, Amizuka N, Takeshita S, Takamatsu H, Katsuura M, Ozawa H, Toyama Y, Bonewald LF, Kudo A. Identification and characterization of a novel protein, periostin, with restricted expression to periosteum and periodontal ligament and increased expression by transforming growth factor beta. J Bone Miner Res. 1999;14(7):1239–1249. doi: 10.1359/jbmr.1999.14.7.1239. [DOI] [PubMed] [Google Scholar]
- 2.Kudo A. Periostin in fibrillogenesis for tissue regeneration: periostin actions inside and outside the cell. Cell Mol Life Sci. 2011;68(19):3201–3207. doi: 10.1007/s00018-011-0784-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sugiura T, Takamatsu H, Kudo A, Amann E. Expression and characterization of murine osteoblast-specific factor 2 (OSF-2) in a baculovirus expression system. Protein Expr Purif. 1995;6(3):305–311. doi: 10.1006/prep.1995.1040. [DOI] [PubMed] [Google Scholar]
- 4.Hoersch S, Andrade-Navarro MA. Periostin shows increased evolutionary plasticity in its alternatively spliced region. BMC Evol Biol. 2010;10:30. doi: 10.1186/1471-2148-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliott CG, Hamilton DW. Deconstructing fibrosis research: do pro-fibrotic signals point the way for chronic dermal wound regeneration? J Cell Commun Signal. 2011;5(4):301–315. doi: 10.1007/s12079-011-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kii I, Nishiyama T, Li M, Matsumoto K, Saito M, Amizuka N, Kudo A. Incorporation of tenascin-C into the extracellular matrix by periostin underlies an extracellular meshwork architecture. J Biol Chem. 2010;285(3):2028–2039. doi: 10.1074/jbc.M109.051961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maruhashi T, Kii I, Saito M, Kudo A. Interaction between periostin and BMP-1 promotes proteolytic activation of lysyl oxidase. J Biol Chem. 2010;285(17):13294–13303. doi: 10.1074/jbc.M109.088864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snider P, Hinton RB, Moreno-Rodriguez RA, Wang J, Rogers R, Lindsley A, Li F, Ingram DA, Menick D, Field L, Firulli AB, Molkentin JD, Markwald R, Conway SJ. Periostin is required for maturation and extracellular matrix stabilization of noncardiomyocyte lineages of the heart. Circ Res. 2008;102(7):752–760. doi: 10.1161/CIRCRESAHA.107.159517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimazaki M, Nakamura K, Kii I, Kashima T, Amizuka N, Li M, Saito M, Fukuda K, Nishiyama T, Kitajima S, Saga Y, Fukayama M, Sata M, Kudo A. Periostin is essential for cardiac healing after acute myocardial infarction. J Exp Med. 2008;205(2):295–303. doi: 10.1084/jem.20071297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kii I, Amizuka N, Minqi L, Kitajima S, Saga Y, Kudo A. Periostin is an extracellular matrix protein required for eruption of incisors in mice. Biochem Biophys Res Commun. 2006;342(3):766–772. doi: 10.1016/j.bbrc.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 11.Rios H, Koushik SV, Wang H, Wang J, Zhou HM, Lindsley A, Rogers R, Chen Z, Maeda M, Kruzynska-Frejtag A, Feng JQ, Conway SJ. Periostin null mice exhibit dwarfism, incisor enamel defects, and an early-onset periodontal disease-like phenotype. Mol Cell Biol. 2005;25(24):11131–11144. doi: 10.1128/MCB.25.24.11131-11144.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki H, Amizuka N, Kii I, Kawano Y, Nozawa-Inoue K, Suzuki A, Yoshie H, Kudo A, Maeda T. Immunohistochemical localization of periostin in tooth and its surrounding tissues in mouse mandibles during development. Anat Rec A Discov Mol Cell Evol Biol. 2004;281(2):1264–1275. doi: 10.1002/ar.a.20080. [DOI] [PubMed] [Google Scholar]
- 13.Hakuno D, Kimura N, Yoshioka M, Mukai M, Kimura T, Okada Y, Yozu R, Shukunami C, Hiraki Y, Kudo A, Ogawa S, Fukuda K. Periostin advances atherosclerotic and rheumatic cardiac valve degeneration by inducing angiogenesis and MMP production in humans and rodents. J Clin Investig. 2010;120(7):2292–2306. doi: 10.1172/JCI40973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Norris RA, Moreno-Rodriguez R, Hoffman S, Markwald RR. The many facets of the matricellular protein periostin during cardiac development, remodeling, and pathophysiology. J Cell Commun Signal. 2009;3(3–4):275–286. doi: 10.1007/s12079-009-0063-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norris RA, Potts JD, Yost MJ, Junor L, Brooks T, Tan H, Hoffman S, Hart MM, Kern MJ, Damon B, Markwald RR, Goodwin RL. Periostin promotes a fibroblastic lineage pathway in atrioventricular valve progenitor cells. Dev Dyn. 2009;238(5):1052–1063. doi: 10.1002/dvdy.21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bozyk PD, Bentley JK, Popova AP, Anyanwu AC, Linn MD, Goldsmith AM, Pryhuber GS, Moore BB, Hershenson MB. Neonatal periostin knockout mice are protected from hyperoxia-induced alveolar simplication. PLoS One. 2012;7(2):e31336. doi: 10.1371/journal.pone.0031336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondoh H, Nishiyama T, Kikuchi Y, Fukayama M, Saito M, Kii I, Kudo A. Periostin deficiency causes severe and lethal lung injury in mice with bleomycin administration. J Histochem Cytochem. 2016;64(7):441–453. doi: 10.1369/0022155416652611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okamoto M, Hoshino T, Kitasato Y, Sakazaki Y, Kawayama T, Fujimoto K, Ohshima K, Shiraishi H, Uchida M, Ono J, Ohta S, Kato S, Izuhara K, Aizawa H. Periostin, a matrix protein, is a novel biomarker for idiopathic interstitial pneumonias. Eur Respir J. 2011;37(5):1119–1127. doi: 10.1183/09031936.00059810. [DOI] [PubMed] [Google Scholar]
- 19.Oka T, Xu J, Kaiser RA, Melendez J, Hambleton M, Sargent MA, Lorts A, Brunskill EW, Dorn GW, 2nd, Conway SJ, Aronow BJ, Robbins J, Molkentin JD. Genetic manipulation of periostin expression reveals a role in cardiac hypertrophy and ventricular remodeling. Circ Res. 2007;101(3):313–321. doi: 10.1161/CIRCRESAHA.107.149047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishikawa K, Yoshida S, Nakao S, Nakama T, Kita T, Asato R, Sassa Y, Arita R, Miyazaki M, Enaida H, Oshima Y, Murakami N, Niiro H, Ono J, Matsuda A, Goto Y, Akashi K, Izuhara K, Kudo A, Kono T, Hafezi-Moghadam A, Ishibashi T. Periostin promotes the generation of fibrous membranes in proliferative vitreoretinopathy. FASEB J. 2014;28(1):131–142. doi: 10.1096/fj.13-229740. [DOI] [PubMed] [Google Scholar]
- 21.Naik PK, Bozyk PD, Bentley JK, Popova AP, Birch CM, Wilke CA, Fry CD, White ES, Sisson TH, Tayob N, Carnemolla B, Orecchia P, Flaherty KR, Hershenson MB, Murray S, Martinez FJ, Moore BB, Investigators Consortium Periostin promotes fibrosis and predicts progression in patients with idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2012;303(12):L1046–L1056. doi: 10.1152/ajplung.00139.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takayama G, Arima K, Kanaji T, Toda S, Tanaka H, Shoji S, McKenzie AN, Nagai H, Hotokebuchi T, Izuhara K. Periostin: a novel component of subepithelial fibrosis of bronchial asthma downstream of IL-4 and IL-13 signals. J Allergy Clin Immunol. 2006;118(1):98–104. doi: 10.1016/j.jaci.2006.02.046. [DOI] [PubMed] [Google Scholar]
- 23.Uchida M, Shiraishi H, Ohta S, Arima K, Taniguchi K, Suzuki S, Okamoto M, Ahlfeld SK, Ohshima K, Kato S, Toda S, Sagara H, Aizawa H, Hoshino T, Conway SJ, Hayashi S, Izuhara K. Periostin, a matricellular protein, plays a role in the induction of chemokines in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2012;46(5):677–686. doi: 10.1165/rcmb.2011-0115OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishiyama T, Kii I, Kashima TG, Kikuchi Y, Ohazama A, Shimazaki M, Fukayama M, Kudo A. Delayed re-epithelialization in periostin-deficient mice during cutaneous wound healing. PLoS One. 2011;6(4):e18410. doi: 10.1371/journal.pone.0018410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norris RA, Damon B, Mironov V, Kasyanov V, Ramamurthi A, Moreno-Rodriguez R, Trusk T, Potts JD, Goodwin RL, Davis J, Hoffman S, Wen X, Sugi Y, Kern CB, Mjaatvedt CH, Turner DK, Oka T, Conway SJ, Molkentin JD, Forgacs G, Markwald RR. Periostin regulates collagen fibrillogenesis and the biomechanical properties of connective tissues. J Cell Biochem. 2007;101(3):695–711. doi: 10.1002/jcb.21224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ontsuka K, Kotobuki Y, Shiraishi H, Serada S, Ohta S, Tanemura A, Yang L, Fujimoto M, Arima K, Suzuki S, Murota H, Toda S, Kudo A, Conway SJ, Narisawa Y, Katayama I, Izuhara K, Naka T. Periostin, a matricellular protein, accelerates cutaneous wound repair by activating dermal fibroblasts. Exp Dermatol. 2012;21(5):331–336. doi: 10.1111/j.1600-0625.2012.01454.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhou HM, Wang J, Elliott C, Wen W, Hamilton DW, Conway SJ. Spatiotemporal expression of periostin during skin development and incisional wound healing: lessons for human fibrotic scar formation. J Cell Commun Signal. 2010;4(2):99–107. doi: 10.1007/s12079-010-0090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukushima N, Kikuchi Y, Nishiyama T, Kudo A, Fukayama M. Periostin deposition in the stroma of invasive and intraductal neoplasms of the pancreas. Mod Pathol. 2008;21(8):1044–1053. doi: 10.1038/modpathol.2008.77. [DOI] [PubMed] [Google Scholar]
- 29.Kashima TG, Nishiyama T, Shimazu K, Shimazaki M, Kii I, Grigoriadis AE, Fukayama M, Kudo A. Periostin, a novel marker of intramembranous ossification, is expressed in fibrous dysplasia and in c-Fos-overexpressing bone lesions. Hum Pathol. 2009;40(2):226–237. doi: 10.1016/j.humpath.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 30.Kikuchi Y, Kashima TG, Nishiyama T, Shimazu K, Morishita Y, Shimazaki M, Kii I, Horie H, Nagai H, Kudo A, Fukayama M. Periostin is expressed in pericryptal fibroblasts and cancer-associated fibroblasts in the colon. J Histochem Cytochem. 2008;56(8):753–764. doi: 10.1369/jhc.2008.951061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kikuchi Y, Kunita A, Iwata C, Komura D, Nishiyama T, Shimazu K, Takeshita K, Shibahara J, Kii I, Morishita Y, Yashiro M, Hirakawa K, Miyazono K, Kudo A, Fukayama M, Kashima TG. The niche component periostin is produced by cancer-associated fibroblasts, supporting growth of gastric cancer through ERK activation. Am J Pathol. 2014;184(3):859–870. doi: 10.1016/j.ajpath.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 32.Liu AY, Zheng H, Ouyang G. Periostin, a multifunctional matricellular protein in inflammatory and tumor microenvironments. Matrix Biol. 2014;37:150–156. doi: 10.1016/j.matbio.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Nitsche U, Stangel D, Pan Z, Schlitter AM, Esposito I, Regel I, Raulefs S, Friess H, Kleeff J, Erkan M. Periostin and tumor–stroma interactions in non-small cell lung cancer. Oncol Lett. 2016;12(5):3804–3810. doi: 10.3892/ol.2016.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oskarsson T, Massague J. Extracellular matrix players in metastatic niches. EMBO J. 2012;31(2):254–256. doi: 10.1038/emboj.2011.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin X, Yan M, Zhang J, Wang X, Shen Z, Lv Z, Li Z, Wei W, Chen W. TGFbeta3-mediated induction of Periostin facilitates head and neck cancer growth and is associated with metastasis. Sci Rep. 2016;6:20587. doi: 10.1038/srep20587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruan K, Bao S, Ouyang G. The multifaceted role of periostin in tumorigenesis. Cell Mol Life Sci. 2009;66(14):2219–2230. doi: 10.1007/s00018-009-0013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sung PL, Jan YH, Lin SC, Huang CC, Lin H, Wen KC, Chao KC, Lai CR, Wang PH, Chuang CM, Wu HH, Twu NF, Yen MS, Hsiao M, Huang CY. Periostin in tumor microenvironment is associated with poor prognosis and platinum resistance in epithelial ovarian carcinoma. Oncotarget. 2016;7(4):4036–4047. doi: 10.18632/oncotarget.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian Y, Choi CH, Li QK, Rahmatpanah FB, Chen X, Kim SR, Veltri R, Chia D, Zhang Z, Mercola D, Zhang H. Overexpression of periostin in stroma positively associated with aggressive prostate cancer. PLoS One. 2015;10(3):e0121502. doi: 10.1371/journal.pone.0121502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Underwood TJ, Hayden AL, Derouet M, Garcia E, Noble F, White MJ, Thirdborough S, Mead A, Clemons N, Mellone M, Uzoho C, Primrose JN, Blaydes JP, Thomas GJ. Cancer-associated fibroblasts predict poor outcome and promote periostin-dependent invasion in oesophageal adenocarcinoma. J Pathol. 2015;235(3):466–477. doi: 10.1002/path.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z, Ouyang G. Periostin: a bridge between cancer stem cells and their metastatic niche. Cell Stem Cell. 2012;10(2):111–112. doi: 10.1016/j.stem.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 41.Conway SJ, Izuhara K, Kudo Y, Litvin J, Markwald R, Ouyang G, Arron JR, Holweg CT, Kudo A. The role of periostin in tissue remodeling across health and disease. Cell Mol Life Sci. 2014;71(7):1279–1288. doi: 10.1007/s00018-013-1494-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doliana R, Bot S, Bonaldo P, Colombatti A. EMI, a novel cysteine-rich domain of EMILINs and other extracellular proteins, interacts with the gC1q domains and participates in multimerization. FEBS Lett. 2000;484(2):164–168. doi: 10.1016/S0014-5793(00)02140-2. [DOI] [PubMed] [Google Scholar]
- 43.Callebaut I, Mignotte V, Souchet M, Mornon JP. EMI domains are widespread and reveal the probable orthologs of the Caenorhabditis elegans CED-1 protein. Biochem Biophys Res Commun. 2003;300(3):619–623. doi: 10.1016/S0006-291X(02)02904-2. [DOI] [PubMed] [Google Scholar]
- 44.Zanetti M, Braghetta P, Sabatelli P, Mura I, Doliana R, Colombatti A, Volpin D, Bonaldo P, Bressan GM. EMILIN-1 deficiency induces elastogenesis and vascular cell defects. Mol Cell Biol. 2004;24(2):638–650. doi: 10.1128/MCB.24.2.638-650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bot S, Andreuzzi E, Capuano A, Schiavinato A, Colombatti A, Doliana R. Multiple-interactions among EMILIN1 and EMILIN2N- and C-terminal domains. Matrix Biol. 2015;41:44–55. doi: 10.1016/j.matbio.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 46.Doliana R, Canton A, Bucciotti F, Mongiat M, Bonaldo P, Colombatti A. Structure, chromosomal localization, and promoter analysis of the human elastin microfibril interfase located proteIN (EMILIN) gene. J Biol Chem. 2000;275(2):785–792. doi: 10.1074/jbc.275.2.785. [DOI] [PubMed] [Google Scholar]
- 47.Schiavinato A, Becker AK, Zanetti M, Corallo D, Milanetto M, Bizzotto D, Bressan G, Guljelmovic M, Paulsson M, Wagener R, Braghetta P, Bonaldo P. EMILIN-3, peculiar member of elastin microfibril interface-located protein (EMILIN) family, has distinct expression pattern, forms oligomeric assemblies, and serves as transforming growth factor beta (TGF-beta) antagonist. J Biol Chem. 2012;287(14):11498–11515. doi: 10.1074/jbc.M111.303578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zacchigna L, Vecchione C, Notte A, Cordenonsi M, Dupont S, Maretto S, Cifelli G, Ferrari A, Maffei A, Fabbro C, Braghetta P, Marino G, Selvetella G, Aretini A, Colonnese C, Bettarini U, Russo G, Soligo S, Adorno M, Bonaldo P, Volpin D, Piccolo S, Lembo G, Bressan GM. Emilin1 links TGF-beta maturation to blood pressure homeostasis. Cell. 2006;124(5):929–942. doi: 10.1016/j.cell.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 49.Kii I, Nishiyama T, Kudo A. Periostin promotes secretion of fibronectin from the endoplasmic reticulum. Biochem Biophys Res Commun. 2016;470(4):888–893. doi: 10.1016/j.bbrc.2016.01.139. [DOI] [PubMed] [Google Scholar]
- 50.Kim BY, Olzmann JA, Choi SI, Ahn SY, Kim TI, Cho HS, Suh H, Kim EK. Corneal dystrophy-associated R124H mutation disrupts TGFBI interaction with Periostin and causes mislocalization to the lysosome. J Biol Chem. 2009;284(29):19580–19591. doi: 10.1074/jbc.M109.013607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kadler KE, Hill A, Canty-Laird EG. Collagen fibrillogenesis: fibronectin, integrins, and minor collagens as organizers and nucleators. Curr Opin Cell Biol. 2008;20(5):495–501. doi: 10.1016/j.ceb.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Canty EG, Kadler KE. Procollagen trafficking, processing and fibrillogenesis. J Cell Sci. 2005;118(Pt 7):1341–1353. doi: 10.1242/jcs.01731. [DOI] [PubMed] [Google Scholar]
- 53.Mosher DF, Johansson MW, Gillis ME, Annis DS. Periostin and TGF-beta-induced protein: two peas in a pod? Crit Rev Biochem Mol Biol. 2015;50(5):427–439. doi: 10.3109/10409238.2015.1069791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kannabiran C, Klintworth GK. TGFBI gene mutations in corneal dystrophies. Hum Mutat. 2006;27(7):615–625. doi: 10.1002/humu.20334. [DOI] [PubMed] [Google Scholar]
- 55.Hashimoto K, Noshiro M, Ohno S, Kawamoto T, Satakeda H, Akagawa Y, Nakashima K, Okimura A, Ishida H, Okamoto T, Pan H, Shen M, Yan W, Kato Y. Characterization of a cartilage-derived 66-kDa protein (RGD-CAP/beta ig-h3) that binds to collagen. Biochim Biophys Acta. 1997;1355(3):303–314. doi: 10.1016/S0167-4889(96)00147-4. [DOI] [PubMed] [Google Scholar]
- 56.Clout NJ, Hohenester E. A model of FAS1 domain 4 of the corneal protein beta(ig)-h3 gives a clearer view on corneal dystrophies. Mol Vis. 2003;9:440–448. [PubMed] [Google Scholar]
- 57.Clout NJ, Tisi D, Hohenester E. Novel fold revealed by the structure of a FAS1 domain pair from the insect cell adhesion molecule fasciclin I. Structure. 2003;11(2):197–203. doi: 10.1016/S0969-2126(03)00002-9. [DOI] [PubMed] [Google Scholar]
- 58.Carr MD, Bloemink MJ, Dentten E, Whelan AO, Gordon SV, Kelly G, Frenkiel TA, Hewinson RG, Williamson RA. Solution structure of the Mycobacterium tuberculosis complex protein MPB70: from tuberculosis pathogenesis to inherited human corneal desease. J Biol Chem. 2003;278(44):43736–43743. doi: 10.1074/jbc.M307235200. [DOI] [PubMed] [Google Scholar]
- 59.Midwood KS, Chiquet M, Tucker RP, Orend G. Tenascin-C at a glance. J Cell Sci. 2016;129(23):4321–4327. doi: 10.1242/jcs.190546. [DOI] [PubMed] [Google Scholar]
- 60.Takayama I, Kii I, Kudo A. Expression, purification and characterization of soluble recombinant periostin protein produced by Escherichia coli . J Biochem. 2009;146(5):713–723. doi: 10.1093/jb/mvp117. [DOI] [PubMed] [Google Scholar]
- 61.Vadon-Le Goff S, Hulmes DJ, Moali C. BMP-1/tolloid-like proteinases synchronize matrix assembly with growth factor activation to promote morphogenesis and tissue remodeling. Matrix Biol. 2015;44–46:14–23. doi: 10.1016/j.matbio.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 62.Hwang EY, Jeong MS, Park EK, Kim JH, Jang SB. Structural characterization and interaction of periostin and bone morphogenetic protein for regulation of collagen cross-linking. Biochem Biophys Res Commun. 2014;449(4):425–431. doi: 10.1016/j.bbrc.2014.05.055. [DOI] [PubMed] [Google Scholar]
- 63.Garnero P. The contribution of collagen crosslinks to bone strength. Bonekey Rep. 2012;1:182. doi: 10.1038/bonekey.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trackman PC. Enzymatic and non-enzymatic functions of the lysyl oxidase family in bone. Matrix Biol. 2016;52–54:7–18. doi: 10.1016/j.matbio.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takayama I, Tanabe H, Nishiyama T, Ito H, Amizuka N, Li M, Katsube KI, Kii I, Kudo A. Periostin is required for matricellular localization of CCN3 in periodontal ligament of mice. J Cell Commun Signal. 2017;11(1):5–13. doi: 10.1007/s12079-016-0371-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ratajczak-Wielgomas K, Dziegiel P. The role of periostin in neoplastic processes. Folia Histochem Cytobiol. 2015;53(2):120–132. doi: 10.5603/FHC.a2015.0014. [DOI] [PubMed] [Google Scholar]
- 67.Snider J, Kotlyar M, Saraon P, Yao Z, Jurisica I, Stagljar I. Fundamentals of protein interaction network mapping. Mol Syst Biol. 2015;11(12):848. doi: 10.15252/msb.20156351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ghatak S, Misra S, Norris RA, Moreno-Rodriguez RA, Hoffman S, Levine RA, Hascall VC, Markwald RR. Periostin induces intracellular cross-talk between kinases and hyaluronan in atrioventricular valvulogenesis. J Biol Chem. 2014;289(12):8545–8561. doi: 10.1074/jbc.M113.539882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Masuoka M, Shiraishi H, Ohta S, Suzuki S, Arima K, Aoki S, Toda S, Inagaki N, Kurihara Y, Hayashida S, Takeuchi S, Koike K, Ono J, Noshiro H, Furue M, Conway SJ, Narisawa Y, Izuhara K. Periostin promotes chronic allergic inflammation in response to Th2 cytokines. J Clin Investig. 2012;122(7):2590–2600. doi: 10.1172/JCI58978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsuzawa M, Arai C, Nomura Y, Murata T, Yamakoshi Y, Oida S, Hanada N, Nakamura Y. Periostin of human periodontal ligament fibroblasts promotes migration of human mesenchymal stem cell through the alphavbeta3 integrin/FAK/PI3K/Akt pathway. J Periodontal Res. 2015;50(6):855–863. doi: 10.1111/jre.12277. [DOI] [PubMed] [Google Scholar]
- 72.Utispan K, Sonongbua J, Thuwajit P, Chau-In S, Pairojkul C, Wongkham S, Thuwajit C. Periostin activates integrin alpha5beta1 through a PI3K/AKT dependent pathway in invasion of cholangiocarcinoma. Int J Oncol. 2012;41(3):1110–1118. doi: 10.3892/ijo.2012.1530. [DOI] [PubMed] [Google Scholar]
- 73.Xiao ZM, Wang XY, Wang AM. Periostin induces chemoresistance in colon cancer cells through activation of the PI3K/Akt/survivin pathway. Biotechnol Appl Biochem. 2015;62(3):401–406. doi: 10.1002/bab.1193. [DOI] [PubMed] [Google Scholar]
- 74.Yang L, Serada S, Fujimoto M, Terao M, Kotobuki Y, Kitaba S, Matsui S, Kudo A, Naka T, Murota H, Katayama I. Periostin facilitates skin sclerosis via PI3K/Akt dependent mechanism in a mouse model of scleroderma. PLoS One. 2012;7(7):e41994. doi: 10.1371/journal.pone.0041994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H, Szpyt J, Tam S, Zarraga G, Pontano-Vaites L, Swarup S, White AE, Schweppe DK, Rad R, Erickson BK, Obar RA, Guruharsha KG, Li K, Artavanis-Tsakonas S, Gygi SP, Harper JW. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017;545(7655):505–509. doi: 10.1038/nature22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, Dong R, Guarani V, Vaites LP, Ordureau A, Rad R, Erickson BK, Wuhr M, Chick J, Zhai B, Kolippakkam D, Mintseris J, Obar RA, Harris T, Artavanis-Tsakonas S, Sowa ME, De Camilli P, Paulo JA, Harper JW, Gygi SP. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015;162(2):425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bao S, Ouyang G, Bai X, Huang Z, Ma C, Liu M, Shao R, Anderson RM, Rich JN, Wang XF. Periostin potently promotes metastatic growth of colon cancer by augmenting cell survival via the Akt/PKB pathway. Cancer Cell. 2004;5(4):329–339. doi: 10.1016/S1535-6108(04)00081-9. [DOI] [PubMed] [Google Scholar]
- 78.Gillan L, Matei D, Fishman DA, Gerbin CS, Karlan BY, Chang DD. Periostin secreted by epithelial ovarian carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5) integrins and promotes cell motility. Cancer Res. 2002;62(18):5358–5364. [PubMed] [Google Scholar]
- 79.Khurana S, Schouteden S, Manesia JK, Santamaria-Martinez A, Huelsken J, Lacy-Hulbert A, Verfaillie CM. Outside-in integrin signalling regulates haematopoietic stem cell function via Periostin–Itgav axis. Nat Commun. 2016;7:13500. doi: 10.1038/ncomms13500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Orecchia P, Conte R, Balza E, Castellani P, Borsi L, Zardi L, Mingari MC, Carnemolla B. Identification of a novel cell binding site of periostin involved in tumour growth. Eur J Cancer. 2011;47(14):2221–2229. doi: 10.1016/j.ejca.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 81.Shao R, Bao S, Bai X, Blanchette C, Anderson RM, Dang T, Gishizky ML, Marks JR, Wang XF. Acquired expression of periostin by human breast cancers promotes tumor angiogenesis through up-regulation of vascular endothelial growth factor receptor 2 expression. Mol Cell Biol. 2004;24(9):3992–4003. doi: 10.1128/MCB.24.9.3992-4003.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sugiyama A, Kanno K, Nishimichi N, Ohta S, Ono J, Conway SJ, Izuhara K, Yokosaki Y, Tazuma S. Periostin promotes hepatic fibrosis in mice by modulating hepatic stellate cell activation via alphav integrin interaction. J Gastroenterol. 2016;51(12):1161–1174. doi: 10.1007/s00535-016-1206-0. [DOI] [PubMed] [Google Scholar]
- 83.Tanabe H, Takayama I, Nishiyama T, Shimazaki M, Kii I, Li M, Amizuka N, Katsube K, Kudo A. Periostin associates with Notch1 precursor to maintain Notch1 expression under a stress condition in mouse cells. PLoS One. 2010;5(8):e12234. doi: 10.1371/journal.pone.0012234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ashley SL, Wilke CA, Kim KK, Moore BB. Periostin regulates fibrocyte function to promote myofibroblast differentiation and lung fibrosis. Mucosal Immunol. 2017;10(2):341–351. doi: 10.1038/mi.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hong L, Shejiao D, Fenrong C, Gang Z, Lei D. Periostin down-regulation attenuates the pro-fibrogenic response of hepatic stellate cells induced by TGF-beta1. J Cell Mol Med. 2015;19(10):2462–2468. doi: 10.1111/jcmm.12636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huang Y, Liu W, Xiao H, Maitikabili A, Lin Q, Wu T, Huang Z, Liu F, Luo Q, Ouyang G. Matricellular protein periostin contributes to hepatic inflammation and fibrosis. Am J Pathol. 2015;185(3):786–797. doi: 10.1016/j.ajpath.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 87.Kaur H, Takefuji M, Ngai CY, Carvalho J, Bayer J, Wietelmann A, Poetsch A, Hoelper S, Conway SJ, Mollmann H, Looso M, Troidl C, Offermanns S, Wettschureck N. Targeted ablation of periostin-expressing activated fibroblasts prevents adverse cardiac remodeling in mice. Circ Res. 2016;118(12):1906–1917. doi: 10.1161/CIRCRESAHA.116.308643. [DOI] [PubMed] [Google Scholar]
- 88.Lorts A, Schwanekamp JA, Baudino TA, McNally EM, Molkentin JD. Deletion of periostin reduces muscular dystrophy and fibrosis in mice by modulating the transforming growth factor-beta pathway. Proc Natl Acad Sci USA. 2012;109(27):10978–10983. doi: 10.1073/pnas.1204708109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lorts A, Schwanekamp JA, Elrod JW, Sargent MA, Molkentin JD. Genetic manipulation of periostin expression in the heart does not affect myocyte content, cell cycle activity, or cardiac repair. Circ Res. 2009;104(1):e1–e7. doi: 10.1161/CIRCRESAHA.108.188649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nakama T, Yoshida S, Ishikawa K, Kobayashi Y, Zhou Y, Nakao S, Sassa Y, Oshima Y, Takao K, Shimahara A, Yoshikawa K, Hamasaki T, Ohgi T, Hayashi H, Matsuda A, Kudo A, Nozaki M, Ogura Y, Kuroda M, Ishibashi T. Inhibition of choroidal fibrovascular membrane formation by new class of RNA interference therapeutic agent targeting periostin. Gene Ther. 2015;22(2):127–137. doi: 10.1038/gt.2014.112. [DOI] [PubMed] [Google Scholar]
- 91.Nakama T, Yoshida S, Ishikawa K, Kubo Y, Kobayashi Y, Zhou Y, Nakao S, Hisatomi T, Ikeda Y, Takao K, Yoshikawa K, Matsuda A, Ono J, Ohta S, Izuhara K, Kudo A, Sonoda KH, Ishibashi T. Therapeutic effect of novel single-stranded RNAi agent targeting periostin in eyes with retinal neovascularization. Mol Ther Nucleic Acids. 2017;6:279–289. doi: 10.1016/j.omtn.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nam BY, Park JT, Kwon YE, Lee JP, Jung JH, Kim Y, Kim S, Park J, Um JE, Wu M, Han SH, Yoo TH, Kang SW. Periostin-binding DNA aptamer treatment ameliorates peritoneal dialysis-induced peritoneal fibrosis. Mol Ther Nucleic Acids. 2017;7:396–407. doi: 10.1016/j.omtn.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schwanekamp JA, Lorts A, Vagnozzi RJ, Vanhoutte D, Molkentin JD. Deletion of periostin protects against atherosclerosis in mice by altering inflammation and extracellular matrix remodeling. Arterioscler Thromb Vasc Biol. 2016;36(1):60–68. doi: 10.1161/ATVBAHA.115.306397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Taniyama Y, Katsuragi N, Sanada F, Azuma J, Iekushi K, Koibuchi N, Okayama K, Ikeda-Iwabu Y, Muratsu J, Otsu R, Rakugi H, Morishita R. Selective blockade of periostin exon 17 preserves cardiac performance in acute myocardial infarction. Hypertension. 2016;67(2):356–361. doi: 10.1161/HYPERTENSIONAHA.115.06265. [DOI] [PubMed] [Google Scholar]
- 95.Yokota K, Kobayakawa K, Saito T, Hara M, Kijima K, Ohkawa Y, Harada A, Okazaki K, Ishihara K, Yoshida S, Kudo A, Iwamoto Y, Okada S. Periostin promotes scar formation through the interaction between pericytes and infiltrating monocytes/macrophages after spinal cord injury. Am J Pathol. 2017;187(3):639–653. doi: 10.1016/j.ajpath.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 96.Zhu M, Saxton RE, Ramos L, Chang DD, Karlan BY, Gasson JC, Slamon DJ. Neutralizing monoclonal antibody to periostin inhibits ovarian tumor growth and metastasis. Mol Cancer Ther. 2011;10(8):1500–1508. doi: 10.1158/1535-7163.MCT-11-0046. [DOI] [PubMed] [Google Scholar]
- 97.Turtle E, Chow N, Yang C, Sosa S, Bauer U, Brenner M, Solow-Cordero D, Ho WB. Design and synthesis of procollagen C-proteinase inhibitors. Bioorg Med Chem Lett. 2012;22(24):7397–7401. doi: 10.1016/j.bmcl.2012.10.067. [DOI] [PubMed] [Google Scholar]
- 98.Chang J, Lucas MC, Leonte LE, Garcia-Montolio M, Singh LB, Findlay AD, Deodhar M, Foot JS, Jarolimek W, Timpson P, Erler JT, Cox TR. Pre-clinical evaluation of small molecule LOXL2 inhibitors in breast cancer. Oncotarget. 2017;8(16):26066–26078. doi: 10.18632/oncotarget.15257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hutchinson JH, Rowbottom MW, Lonergan D, Darlington J, Prodanovich P, King CD, Evans JF, Bain G. Small molecule lysyl oxidase-like 2 (LOXL2) inhibitors: the identification of an inhibitor selective for LOXL2 over LOX. ACS Med Chem Lett. 2017;8(4):423–427. doi: 10.1021/acsmedchemlett.7b00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rowbottom MW, Bain G, Calderon I, Lasof T, Lonergan D, Lai A, Huang F, Darlington J, Prodanovich P, Santini AM, King CD, Goulet L, Shannon KE, Ma GL, Nguyen K, MacKenna DA, Evans JF, Hutchinson JH. Identification of 4-(aminomethyl)-6-(trifluoromethyl)-2-(phenoxy)pyridine derivatives as potent, selective, and orally efficacious inhibitors of the copper-dependent amine oxidase, lysyl oxidase-like 2 (LOXL2) J Med Chem. 2017;60(10):4403–4423. doi: 10.1021/acs.jmedchem.7b00345. [DOI] [PubMed] [Google Scholar]