Abstract
The ability to maintain cells in a differentiated state and to prevent them from reprogramming into a multipotent state has recently emerged as a central theme in neural development as well as in oncogenesis. In the developing central nervous system (CNS) of the fruit fly Drosophila, several transcription factors were recently identified to be required in postmitotic cells to maintain differentiation, and in their absence, mature neurons undergo dedifferentiation, giving rise to proliferative neural stem cells and ultimately to tumor growth. In this review, we will highlight the current understanding of dedifferentiation and cell plasticity in the Drosophila CNS.
Keywords: Neural stem cells, Neuroblast, Dedifferentiation, Neurons, Post-mitotic cells
Introduction
The cellular diversity of the central nervous system (CNS) is the result of the activity of neural stem cells which are multipotent progenitors capable of self-renewing and generating the various cell types in the brain largely during development. The process of differentiation, i.e., the acquisition of a specialized cell fate, often associated with cell cycle exit, was once thought to be strictly unidirectional from stem cell to differentiated cell. However, this dogma has been challenged. From the first reprogramming of somatic cells by nuclear transfer in frogs [1] to the successful generation of induced pluripotent stem cells (iPSC) from terminally differentiated fibroblasts [2], we now know that differentiation is a highly plastic process. Indeed, as opposed to differentiation, dedifferentiation occurs when specialized cell types, such as neurons, lose their committed cell fate gene expression signature, in favor of the expression of more primitive markers and regain stem cell-like characteristics.
Studying how differentiation is maintained at the molecular level is of pivotal importance for understanding not only the therapeutic potential of iPSCs but also the pathogenesis of diseases such as cancer, where specialized, postmitotic cells re-acquire a stem cell identity and unlimited self-renewal potential [3].
The fruit fly Drosophila has been extensively utilized to investigate stem cell biology [4, 5]. Using this model organism, we are now beginning to gain valuable insight into the transcriptional programs that are in place to maintain differentiation during development. In this review, we will discuss recent advances gained in the field using the Drosophila CNS as a model system.
Maintenance of neuronal differentiation in the Drosophila CNS
The CNS of the fruit fly Drosophila has recently served as a model for studies of differentiation maintenance in the context of neurogenesis as well as tumorigenesis. Many aspects of mammalian stem cell biology are fully recapitulated in the fly brain and the system is very well characterized and easily genetically amenable [4]. Drosophila neural stem cells, the neuroblasts (NBs), populate the developing larval brain and are responsible for the generation of the adult CNS. NBs divide asymmetrically to self-renew, and to give rise to specialized cell types of the brain, namely neurons and glial cells. The NBs of the larval brain can be classified based on the mode of cell division: type I NBs, and the NBs of the optic lobe (OL, the presumptive visual center of the fly) produce a NB and a daughter cell called the ganglion mother cell (GMC) that will divide only once to generate two neurons or glial cells (Fig. 1a). Type II NBs, on the other hand, produce a transit-amplifying daughter cell, called an INP (intermediate neural progenitor), that will undergo several rounds of asymmetric cell division to generate another INP and a GMC that will then divide to give rise to two differentiated progeny [5].
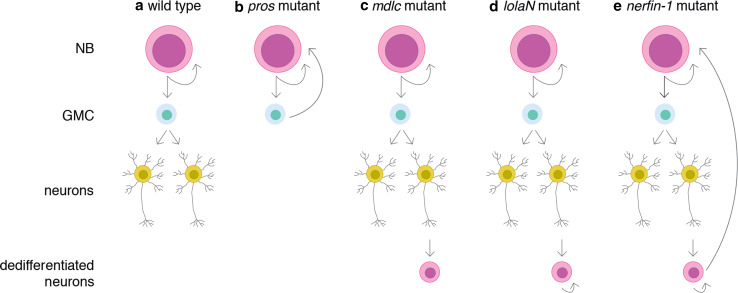
Fig. 1.
Dedifferentiation in the Drosophila brain. a Wild-type neuroblasts (NBs) divide asymmetrically to self-renew and to generate a smaller daughter cell, the ganglion mother cell (GMC) that divide once to generate differentiated neurons. b pros mutant GMCs are unable to differentiate and re-acquire a NB fate [6]. c mdlc mutant neurons express NB markers but are unable to proliferate [20]. d lolaN mutant post-mitotic neurons in the optic lobes dedifferentiate and re-enter the cell cycle [34]. e Nerfin-1-deficient post-mitotic neurons dedifferentiate, re-enter the cell cycle and grow back to give rise to a fully functional NB [32]
Asymmetric cell division is a key process in stem cell function. It ensures that specific cell fate determinants are exclusively inherited by the daughter cell committed to differentiation, allowing the other cell to retain its multipotent state. One of such cell fate determinants is the atypical homeodomain transcription factor Prospero (Pros). In the NB, Pros is sequestered in the cytoplasm and during asymmetric cell division it is specifically localized to the cortex of the budding GMC. In the newly formed GMC, Pros translocates to the nucleus where it acts as a ‘binary switch’ shutting down self-renewal and proliferation-promoting genes and activating the transcription of differentiation-promoting factors [6].
Not surprisingly most of the genes involved in asymmetric cell division and cell fate determination behave as tumor suppressors in the fly brain. Their loss of function or impairment causes abnormal expansion of the stem cell population and thus tumor growth [7–18]. GMCs that are mutant for Pros, for instance, are unable to exit the cell cycle and undergo differentiation, and instead revert to a NB-like fate (Fig. 1b) producing large, fast proliferating tumors [6, 8–10, 19]. Many examples also exist of mutations that cause the INPs produced by type II lineages to revert to a NB-like and thus more multipotent, less mature state [8–18]. However, only recently have genes been identified that function more downstream in the differentiation process. These genes act to maintain the differentiated status of the post-mitotic neurons, and, when lost, cause neuron-to-NB reversion.
The first hint of the existence of such a category of genes came in 2013 when Carney and colleagues identified a factor, named Midlife crisis (Mdlc), that represses NB-specific genes in differentiated neurons of both type I and type II lineages (Fig. 1c). Mdlc is a conserved zinc finger-containing protein involved in RNA splicing of the transcription factor Pros [20]. mdlc mutant neurons show ectopic expression of NB genes, likely due to aberrant splicing of Pros and its consequent loss. However, these ectopic NB-like cells are ‘locked’ into a neuron/NB intermediate state, incapable of proliferating, suggesting that other factors may function to inhibit proliferation in these cells [20].
In 2014, Southall and colleagues found that a specific isoform of the BTB-Zn finger transcription factor Longitudinal lacking (Lola), LolaN, is required to maintain the neurons of the optic lobes in a differentiated post-mitotic state [21]. LolaN is expressed in the neurons shortly after GMC division. There, it binds to both stem cell-specific and neuronal-specific genes, a significant subset of which are also targets of Pros. Loss of LolaN in optic lobe neurons causes derepression of NB factors and triggers their reentry in the cell cycle (Fig. 1d). This results in large tumors that persist into adulthood. Loss of LolaN is, however, not required to maintain differentiation of neurons in other regions of the brain (type I and II lineages), where Pros is expressed. This observation led the authors to hypothesize that Lola acts redundantly with Pros to maintain the differentiated status of post-mitotic neurons. While the role of Pros in the GMC to turn off stem cell genes and turn on differentiation genes is well defined, its exact role in maintaining differentiation during later stages of neuronal life is, however, still unclear.
Finally, the zinc finger transcription factor nervous fingers 1 (Nerfin-1) was recently identified as a key factor in the maintenance of neuronal differentiation in the fly brain [19]. nerfin-1 mutant neurons undergo step-wise reversion to NBs, in a Myc- and Tor signaling-dependent manner. nerfin-1 mutant cells first switch on NB markers, while maintaining neuronal identity. As they further increase cellular growth, nerfin-1 mutant cells gradually switch off neuronal genes altogether, to become fully functional NBs capable of asymmetric cell division (Fig. 1e). Eventually they give rise to large, highly proliferative tumors which persist in adult life and are invasive when transplanted into naïve hosts [19]. Nerfin-1 loss-dependent dedifferentiation was found to affect both type I and type II NB lineages [19], suggesting that Nerfin-1 likely represents a general factor required for the maintenance of differentiation in the fly CNS. Chromatin immunoprecipitation studies revealed that, similarly to Pros, Nerfin-1 is able to repress stem cell genes and activate neuronal genes. Nevertheless, nerfin-1 mutant neurons are found to revert to a stem cell-like state even in the presence of Pros, suggesting that Pros cannot repress NB genes in the absence of Nerfin-1. Interestingly, nerfin-1 is a Pros transcriptional target [16], further confirming the high level of cross-regulation between the transcription factors involved in maintaining neuronal differentiation in this system.
Pros function appears to be conserved in vertebrates [22, 23], and its mammalian orthologue, PROX1, plays a key role in the development of various organs such as the CNS, eye, liver, heart, and lymphatic system (for a review see [24]). In the CNS, PROX1 has been shown to be involved in regulating neurogenesis both during development and in adulthood by promoting cell cycle exit and differentiation of progenitor cells [25–29]. In mouse and chick embryos, for example, PROX1 represses Notch target genes allowing neural progenitor cells to exit the cell cycle and differentiate [27]. Similarly in the mouse adult hippocampus PROX1 functions downstream of Wnt signaling in promoting the generation of neurons [28]. Not surprisingly PROX1 has also been implicated in many types of cancers, such as colorectal and hepatocellular cancer, playing both a tumor suppressive and an oncogenic role depending on the context (for a review see [24]).
Nerfin-1 function is also conserved throughout evolution: its orthologues have been shown to be involved in neuronal differentiation in C. elegans [30], medaka [31], zebrafish [32] and mouse [33]. Remarkably, mice lacking the Nerfin-1 orthologue Insulinoma-Associated 1 (INSM1) show an increase in the number of apical progenitors at the expense of the more cell fate-restricted basal progenitors and neurons in the mouse neocortex [33]. INSM1 also plays a role in the maintenance of differentiation in neuroendocrine cells: adult mouse pancreatic β-cells that are mutant for INSM1 revert to a more primitive state as they resemble the immature β-cells of newborn mice both in gene expression and function [34]. In contrast to its transient expression in the developing neuroendocrine tissues, INSM1 is abundantly expressed in a variety of neuroendocrine tumors, including insulinoma, small cell lung carcinoma, pituitary tumor, pheochromocytoma, medullary thyroid carcinoma, medulloblastoma, neuroblastoma and retinoblastoma [35]. However, its role in tumor growth has so far not been explored in depth in mammalian models.
Together, these data highlight a role for a network of transcription factors in actively maintaining neurons in a differentiated state. Loss of members of this network can lead to re-acquisition of stem cell characteristics, re-entry into the cell cycle and ultimately tumor formation.
Conclusions
Over 50 years ago, Conrad Waddington proposed the model of “the epigenetic landscape”, which likens the process of cellular differentiation to a marble rolling down a hill, its fate becoming restricted as differentiation occurs, implying that the stem cell-to-differentiated cell transition occurs in an unidirectional fashion. However, a still growing body of evidence favors a model of inter-convertibility between stem cells and their differentiated progeny. Studies in the Drosophila CNS have begun to uncover the role of a series of conserved transcription factors, which function in the repression of pluripotency in postmitotic cells. When the function of these transcription factors is lost, neurons dedifferentiate and re-enter the cell cycle. Dedifferentiation has indeed emerged as a key driver in the development of cancers [3]. In glioblastoma multiforme (GBM), an aggressive human primary brain tumor, loss of differentiation markers, acquisition of stem cell properties, and increased plasticity, are associated with a worse prognosis [36]. Within GBMs, the neoplastic stem cells, commonly referred to as glioblastoma stem cells can perpetuate indefinitely and are responsible for the generation of the heterogeneity found within tumors. These cancer stem cells have been shown to originate from different cell types including terminally differentiated neurons and astrocytes [37, 38]; however, the mechanisms that drive this process are still largely unknown. Understanding the molecular bases of dedifferentiation is therefore of pivotal importance if we hope to gain a deeper insight into the pathogenesis of these diseases.
Acknowledgments
We thank Dr. Joep Vissers for critical reading of the manuscript.
References
- 1.Gurdon JB. Adult frogs derived from the nuclei of single somatic cells. Dev Biol. 1962;4:256–273. doi: 10.1016/0012-1606(62)90043-X. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Friedmann-Morvinski D, Verma IM. Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO Rep. 2014;15:244–253. doi: 10.1002/embr.201338254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brand AH, Livesey FJ. Neural stem cell biology in vertebrates and invertebrates: more alike than different? Neuron. 2011;70:719–729. doi: 10.1016/j.neuron.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 5.Homem CCF, Knoblich JA. Drosophila neuroblasts: a model for stem cell biology. Development. 2012;139:4297–4310. doi: 10.1242/dev.080515. [DOI] [PubMed] [Google Scholar]
- 6.Choksi SP, et al. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 7.Caussinus E, Gonzalez C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster . Nat Genet. 2005;37:1125–1129. doi: 10.1038/ng1632. [DOI] [PubMed] [Google Scholar]
- 8.Bello B, Reichert H, Hirth F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila . Development. 2006;133:2639–2648. doi: 10.1242/dev.02429. [DOI] [PubMed] [Google Scholar]
- 9.Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 10.Lee C-Y, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev Cell. 2006;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, et al. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 2006;20:3453–3463. doi: 10.1101/gad.1487506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowman SK, et al. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila . Dev Cell. 2008;14:535–546. doi: 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin S, et al. Lineage-specific effects of Notch/Numb signaling in post-embryonic development of the Drosophila brain. Development. 2009;137:43–51. doi: 10.1242/dev.041699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weng M, Golden KL, Lee C-Y. dFezf/Earmuff maintains the restricted developmental potential of intermediate neural progenitors in Drosophila . Dev Cell. 2010;18:126–135. doi: 10.1016/j.devcel.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.San-Juán BP, Baonza A. The bHLH factor deadpan is a direct target of Notch signaling and regulates neuroblast self-renewal in Drosophila . Dev Biol. 2011;352:70–82. doi: 10.1016/j.ydbio.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 16.Song Y, Lu B. Regulation of cell growth by Notch signaling and its differential requirement in normal vs. tumor-forming stem cells in Drosophila . Genes Dev. 2011;25:2644–2658. doi: 10.1101/gad.171959.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao Q, Komori H, Lee C-Y. klumpfuss distinguishes stem cells from progenitor cells during asymmetric neuroblast division. Development. 2012;139:2670–2680. doi: 10.1242/dev.081687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zacharioudaki E, Magadi SS, Delidakis C. bHLH-O proteins are crucial for Drosophila neuroblast self-renewal and mediate Notch-induced overproliferation. Development. 2012;139:1258–1269. doi: 10.1242/dev.071779. [DOI] [PubMed] [Google Scholar]
- 19.Froldi F, et al. The transcription factor Nerfin-1 prevents reversion of neurons into neural stem cells. Genes Dev. 2015;29:129–143. doi: 10.1101/gad.250282.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carney TD, Struck AJ, Doe CQ. midlife crisis encodes a conserved zinc-finger protein required to maintain neuronal differentiation in Drosophila . Development. 2013;140:4155–4164. doi: 10.1242/dev.093781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Southall TD, Davidson CM, Miller C, Carr A, Brand AH. Dedifferentiation of neurons precedes tumor formation in Lola mutants. Dev Cell. 2014;28:685–696. doi: 10.1016/j.devcel.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliver G, et al. Prox 1, a prospero-related homeobox gene expressed during mouse development. Mech Dev. 1993;44:3–16. doi: 10.1016/0925-4773(93)90012-M. [DOI] [PubMed] [Google Scholar]
- 23.Zinovieva RD, et al. Structure and chromosomal localization of the human homeobox gene Prox 1. Genomics. 1996;35:517–522. doi: 10.1006/geno.1996.0392. [DOI] [PubMed] [Google Scholar]
- 24.Elsir T, Smits A, Lindström MS, Nistér M. Transcription factor PROX1: its role in development and cancer. Cancer Metastasis Rev. 2012;31:793–805. doi: 10.1007/s10555-012-9390-8. [DOI] [PubMed] [Google Scholar]
- 25.Misra K, Mishra K, Gui H, Matise MP. Prox1 regulates a transitory state for interneuron neurogenesis in the spinal cord. Dev Dyn. 2008;237:393–402. doi: 10.1002/dvdy.21422. [DOI] [PubMed] [Google Scholar]
- 26.Elkouris M, et al. Sox1 maintains the undifferentiated state of cortical neural progenitor cells via the suppression of Prox1-mediated cell cycle exit and neurogenesis. Stem Cells. 2011;29:89–98. doi: 10.1002/stem.554. [DOI] [PubMed] [Google Scholar]
- 27.Kaltezioti V, et al. Prox1 regulates the notch1-mediated inhibition of neurogenesis. PLoS Biol. 2010;8:e1000565. doi: 10.1371/journal.pbio.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karalay O, Jessberger S. Translating niche-derived signals into neurogenesis: the function of Prox1 in the adult hippocampus. Cell Cycle. 2011;10:2239–2240. doi: 10.4161/cc.10.14.15850. [DOI] [PubMed] [Google Scholar]
- 29.Stergiopoulos A, Elkouris M, Politis PK. Prospero-related homeobox 1 (Prox1) at the crossroads of diverse pathways during adult neural fate specification. Front Cell Neurosci. 2014;8:454. doi: 10.3389/fncel.2014.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu J. Inhibition of touch cell fate by egl-44 and egl-46 in C. elegans . Genes Dev. 2001;15:789–802. doi: 10.1101/gad.857401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Candal E, et al. Ol-insm1b, a SNAG family transcription factor involved in cell cycle arrest during medaka development. Dev Biol. 2007;309:1–17. doi: 10.1016/j.ydbio.2007.04.038. [DOI] [PubMed] [Google Scholar]
- 32.Forbes-Osborne MA, Wilson SG, Morris AC. Developmental biology. Dev Biol. 2013;380:157–171. doi: 10.1016/j.ydbio.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farkas LM, et al. Insulinoma-associated 1 has a panneurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex. Neuron. 2008;60:40–55. doi: 10.1016/j.neuron.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 34.Jia S, et al. Insm1 cooperates with Neurod1 and Foxa2 to maintain mature pancreatic β-cell function. EMBO J. 2015;34:1417–1433. doi: 10.15252/embj.201490819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breslin MB, Zhu M, Lan MS. NeuroD1/E47 regulates the E-box element of a novel zinc finger transcription factor, IA-1, in developing nervous system. J Biol Chem. 2003;278:38991–38997. doi: 10.1074/jbc.M306795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuller GN, Scheithauer BW. The 2007 revised World Health Organization (WHO) classification of tumours of the central nervous system: newly codified entities. Brain Pathol. 2007;17:304–307. doi: 10.1111/j.1750-3639.2007.00084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedmann-Morvinski D, et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bachoo RM, et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–277. doi: 10.1016/S1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]