Abstract
DNA double-strand breaks (DSBs) are a nasty form of damage that needs to be repaired to ensure genome stability. The DSB ends can undergo a strand-biased nucleolytic processing (resection) to generate 3′-ended single-stranded DNA (ssDNA) that channels DSB repair into homologous recombination. Generation of ssDNA also triggers the activation of the DNA damage checkpoint, which couples cell cycle progression with DSB repair. The checkpoint response is intimately linked to DSB resection, as some checkpoint proteins regulate the resection process. The present review will highlight recent works on the mechanism and regulation of DSB resection and its interplays with checkpoint activation/inactivation in budding yeast.
Keywords: Double-strand break, Resection, MRX, Mec1, Tel1, Nucleases
Introduction
DNA double-strand breaks (DSBs) are dangerous forms of DNA damage that need to be accurately repaired to preserve genome stability. Failure to repair them can result in genome instability that is a hallmark of cancer cells. DSBs can form accidentally as a result of failure in DNA replication, as well as of exposure to ionizing radiations or chemicals. Moreover, they can arise when telomeres undergo extensive erosion, which leads to the activation of a DNA damage response and to the onset of apoptosis and/or senescence. DSBs can also form in a programmed manner during physiological cellular processes, such as the prophase of the first meiotic division or the rearrangement of the immunoglobulin genes in lymphocytes.
DSB occurrence triggers the activation of a highly conserved pathway, called DNA damage checkpoint, which couples DSB repair with cell cycle progression [1]. Key checkpoint players include the protein kinases Mec1 and Tel1, whose mammalian orthologs are ATR and ATM, respectively [1]. Once Mec1/ATR and/or Tel1/ATM are activated, their checkpoint signals are propagated through the protein kinases Rad53 and Chk1 (Chk2 and Chk1 in mammals, respectively), whose activation requires the conserved protein Rad9 (53BP1 in mammals) [2–4].
DSBs are repaired by two major conserved pathways: non-homologous end-joining (NHEJ) and homologous recombination (HR). NHEJ directly rejoins together the two broken ends [5], whereas HR uses intact homologous duplex DNA sequences (sister chromatids or homologous chromosomes) as a template for accurate repair [6, 7]. The first step in HR is the degradation of the 5′ DNA strands on either side of the DSB to generate 3′ single-stranded DNA (ssDNA) tails through a process termed DNA end resection. The ssDNA tails are first coated by the ssDNA binding complex replication protein A (RPA), which is then replaced by the evolutionarily conserved recombinase Rad51. This replacement leads to the formation of a right-handed helical filament that searches for homologous duplex DNA molecules and catalyzes their invasion. The invading 3′ end serves to prime DNA synthesis using the intact homologous DNA sequence as a template, followed by the resolution of the resulting DNA structures and DNA ligation [6, 7].
Much of our knowledge about the DNA end resection mechanism and its regulation has come from genetic and biochemical studies in the budding yeast Saccharomyces cerevisiae. Here, we will review the mechanism and regulation of end resection in this organism, as well as its interplays with the checkpoint response.
End resection by MRX, Exo1 and Dna2 nucleases
Genetic studies in the budding yeast S. cerevisiae identified the highly conserved MRX (Mre11-Rad50-Xrs2) complex, Exo1 and Dna2 as key nucleases for DSB resection. The MRX complex had long been known to be required for the processing of meiotic DSBs generated by the topoisomerase-like protein Spo11 [8–12]. MRX has a DNA binding activity with a preference toward DNA ends [13] and localizes very close to the DSB ends [14, 15]. The Rad50 subunit has an ATPase activity that induces conformational changes that regulate MRX functions in DNA damage signaling, resection and maintenance of the DSB ends tethered to each other [16–20]. Mre11 has a 3′–5′ dsDNA exonuclease activity, whose polarity is opposite to that needed to generate the 3′-ended ssDNA at the DSB ends [21, 22]. However, Mre11 has also a weak endonuclease activity on 5′-terminated DNA strands and on other DNA structures [23, 24]. This endonuclease activity is dependent on the ATPase activity of Rad50 and on the physical interaction between MRX and the Sae2 protein (CtIP in mammals), which strongly promotes the endonuclease activity of Mre11 within the context of the MRX complex [25]. The ability of MRX to cleave 5′-terminated DNA ends suggested that this complex initiates DNA resection via its endonuclease, rather than its exonuclease activity, by creating a nick that provides the access for nucleases capable of degrading DNA in a 5′–3′ direction [26].
In both yeast and mammals, the 5′–3′ exonuclease activity is provided by two partially overlapping pathways dependent on Dna2 and Exo1 nucleases [27, 28]. In yeast, inactivation of each single pathway results in only minor resection defects, whereas severe resection defects are observed when the two pathways are inactivated simultaneously [27, 28]. The MRX complex not only provides the access for Dna2 and Exo1 through its endonuclease activity, but it has also a structural role in promoting their stable association to DSBs [29]. This nuclease-independent role of MRX can explain why the lack of Mre11 causes a resection defect more severe than that caused by the lack of either Sae2 or Mre11 nuclease activity.
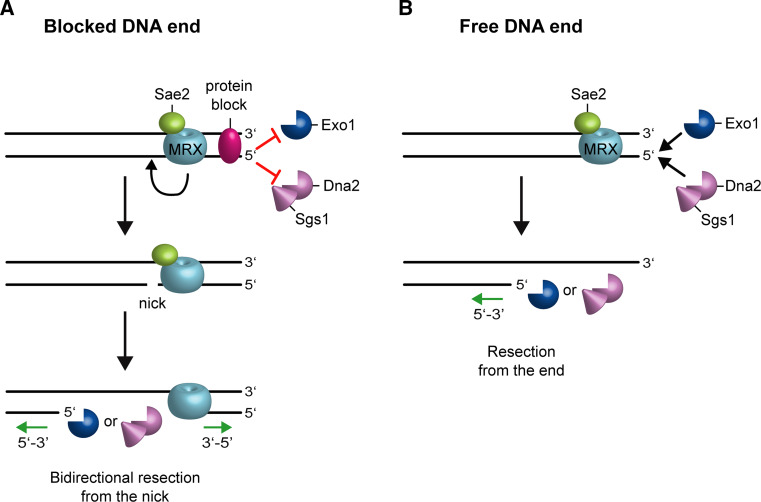
Although Dna2 is recruited on ssDNA ends, it has an endonuclease activity that can cleave both 5′ and 3′ ssDNA overhangs adjoining a duplex DNA, giving rise to degradation products of ~5 to 10 oligonucleotides in length [30, 31]. The separation of DNA strands in the Dna2-mediated nucleolytic processing is carried out by the RecQ helicase Sgs1 (BLM in humans), which unwinds double-stranded DNA (dsDNA) in a 3′–5′ polarity [27, 28, 32]. Unlike Dna2, Exo1 degrades the 5′-terminated strand within duplex DNA and therefore it does not require a helicase activity to unwind DNA [33–35]. Altogether, these data support a model in which MRX-Sae2 provides an initial endonucleolytic cleavage of the 5′ strand at both DSB ends. Then, the nick enables resection in a bidirectional manner, using Exo1 and Dna2-Sgs1 in the 5′–3′ direction away from the DSB end, and Mre11 in the 3′–5′ direction towards the DSB end (Fig. 1a).
Fig. 1.
Model for resection of blocked and free DNA ends. The MRX complex and Sae2 are recruited to DNA ends. a The 5′ strand of a DSB blocked by a covalent adduct or a tightly bound protein is not accessible to Exo1 and Sgs1-Dna2. MRX-Sae2-dependent incision of the 5′ strand allows bidirectional processing by Exo1/Sgs1-Dna2 in the 5′–3′ direction from the nick and by MRX in the 3′–5′ direction toward the DSB ends. b The processing of DNA ends with no covalent modifications can occur directly by Dna2-Sgs1 or Exo1. MRX is required to promote the association of Exo1 and Sgs1-Dna2 at both blocked and free DNA ends
Biochemical reconstitution studies have revealed that the RPA complex regulates DSB resection by preventing nonspecific binding of Exo1 to ssDNA, promoting the unwinding activity of Sgs1, stimulating the 5′–3′ resection polarity of Dna2 and attenuating the degradation of the 3′-terminated DNA strand [36, 37]. These in vitro findings have been supported in vivo, as RPA depletion in S. cerevisiae eliminates both the Sgs1-Dna2 and Exo1-dependent resection pathways [38].
The requirement for MRX-Sae2 nuclease activity in end resection is dependent on how well Exo1 and Sgs1-Dna2 can access the DSB ends. MRX and Sae2 are important to resect DSBs that possess either chemical modifications or proteins covalently bound at their 5′ ends that restrict the access of Exo1 and Sgs1-Dna2 (Fig. 1a). For example, sae2∆ and mre11 nuclease defective mutants are defective in resecting meiotic DSBs, where Spo11 cleaves DNA by a topoisomerase-like transesterase mechanism, forming a covalent bond between a tyrosine residue of the enzyme and the 5′ ends of the DSB [8–12, 39–42]. Furthermore, the same mutants exhibit a marked sensitivity to ionizing radiations, which can generate double- and single-strand breaks and DNA–protein crosslinks [43, 44], as well as to camptothecin, which extends the half-life of DNA-topoisomerase cleavage complexes [45]. Consistent with this model, the endonuclease activity of MRX-Sae2 is strongly stimulated by the presence of dsDNA substrates in which one DNA end is blocked by a protein adduct [25].
Both Sae2 and Dna2 have been shown to be targets of cyclin-dependent kinase (Cdk1 in yeast)-Clb complexes [46, 47], which allows DSB resection to take place only during the S and G2 phases of the cell cycle, when sister chromatids or homologous chromosomes are present as repair templates [48, 49]. Substitution of Sae2 serine 267 with a nonphosphorylatable residue impairs DSB processing, whereas the same process takes place quite efficiently when Sae2 serine 267 is replaced by a glutamic residue mimicking constitutive phosphorylation [46]. Furthermore, substitution of three Cdk1 consensus sites of Dna2 with nonphosphorylatable residues reduces extensive resection [47].
Inhibition of end resection by Ku and Rad9
The initial endonucleolytic cleavage catalyzed by MRX and Sae2 is critical to resect DSBs whose DNA ends are not accessible to Exo1 and Sgs1-Dna2 due to the presence of chemical modifications or covalently bound-proteins [8–12, 39–42, 45] (Fig. 1a). By contrast, DNA ends (such as those generated by endonucleases) with free 3′ hydroxyl and 5′ phosphate groups are resected by Exo1 and Sgs1-Dna2 even in the absence of MRX-Sae2-mediated cleavage of the 5′ strand (Fig. 1b) [12]. However, initiation of resection of endonuclease-induced DSBs in sae2∆ cells occurs less efficiently than in wild type cells [50], suggesting that MRX-Sae2 can be important even at these DSBs to relieve possible inhibition of Dna2 and/or Sgs1-Dna2 activity.
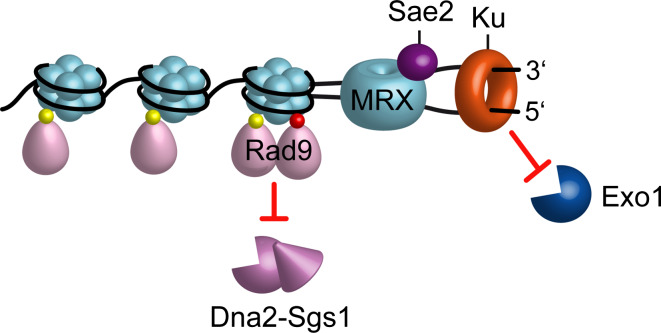
The nucleolytic processing catalyzed by Exo1 is inhibited by the presence of the Ku complex bound at the DSB ends (Fig. 2) [29, 51–54]. The absence of Ku suppresses the resection defect of mre11∆ and sae2∆ cells in an Exo1-dependent manner [29, 53, 54], indicating that Ku restricts Exo1-mediated resection. As Ku is bound very close to the DSB ends, the MRX-Sae2 clipping could allow Exo1 to initiate resection from a nick and this, in turn, would overcome the inhibition exerted by Ku on Exo1 activity.
Fig. 2.
Inhibition of DSB resection by Ku and Rad9. Ku is bound very close to the DSB end. Rad9 is bound to chromatin even in the absence of DSBs via interaction with methylated histone H3 (yellow dots). Rad9 association at DSBs is enhanced by γH2A generation (red dots). Ku and Rad9 inhibit DSB resection by limiting Exo1 and Sgs1-Dna2 access to DNA ends, respectively
By contrast, Sgs1-Dna2 is unable to initiate end resection without MRX even in the absence of Ku, suggesting the existence of another inhibitory pathway. The resection activity of Sgs1-Dna2 is inhibited mainly by the Rad9 protein (Fig. 2) [55, 56], which was originally identified as an adaptor in the DNA damage checkpoint pathway, linking the checkpoint kinases Mec1 and Tel1 to the activation of the effector kinases Rad53 and Chk1 [57]. Rad9 is already bound to chromatin even in the absence of DNA damage by an interaction with methylated lysine 79 of histone H3 (H3-K79) [58–61]. Rad9 binding to the sites of damage is further strengthened by an interaction between its BRCT domain and histone H2A that has been phosphorylated on serine 129 (γH2A) by the checkpoint kinases Mec1 and Tel1 (Fig. 2) [62–64]. Inhibition of DSB resection by Rad9 requires its chromatin association, as the lack of the H3-K79 methyltransferase Dot1 or the presence of a H2A variant where serine 129 is substituted by a non-phosphorylatable alanine residue increases the resection efficiency [60, 65, 66]. Elimination of the ATP-dependent chromatin remodeler Fun30 increases Rad9 accumulation at DSBs, suggesting that Fun30 can overcome the barrier to resection imposed by Rad9-bound chromatin [65, 67, 68].
Several lines of evidence indicate that Rad9 acts as a barrier toward end processing enzymes by restricting the access of Sgs1-Dna2 to the DSB ends. The lack of Rad9 suppresses the resection defect of Sae2-deficient cells, which show an increased amount of Rad9 bound very close to the DSB ends [55, 56]. The lack of Rad9 increases the resection efficiency also in a wild type context [60, 69] and this rapid resection is mainly dependent on Sgs1, whose recruitment at DSBs is inhibited by Rad9 [55, 56]. Further support for a Rad9-mediated inhibition of Sgs1 comes from the recent identification of a hypermorphic allele of SGS1 (SGS1-SS) that behaves like a rad9∆ phenocopy with respect to resection [55]. The Sgs1-ss variant, which suppresses both the hypersensitivity to DNA damaging agents and the resection defect of sae2∆ cells, is robustly associated to the DSB ends both in the presence and in the absence of Rad9 and resects the DSB more efficiently than wild type Sgs1 [55]. Altogether, these findings indicate that Rad9 inhibits the activity of Sgs1-Dna2 by limiting Sgs1 binding/persistence at DSB ends and that the Sgs1-ss mutant variant escapes this inhibition possibly because it is more tightly bound to DNA and exerts its helicase activity through Rad9-containing chromatin more efficiently than wild type Sgs1.
End resection and checkpoint activation
Generation of DSBs elicits the activation of the DNA damage checkpoint, whose key players include the S. cerevisiae protein kinases Mec1 and Tel1, as well as their mammalian orthologs ATR and ATM, respectively [1]. In both yeast and mammals, Mec1 physically interacts with Ddc2 (ATRIP in mammals), which helps the recruitment of Mec1 to the DSB ends [70–73]. By contrast, Tel1 activation depends on the MRX complex, which is required for Tel1 recruitment to the site of damage through direct interaction between Tel1 with Xrs2, as well as for Tel1 kinase activity [74–80]. Whereas Tel1 is recruited on blunt DSB ends or DNA ends with short ssDNA tails, Mec1 recognizes RPA-coated ssDNA that results from resection of the DSB ends [81–84].
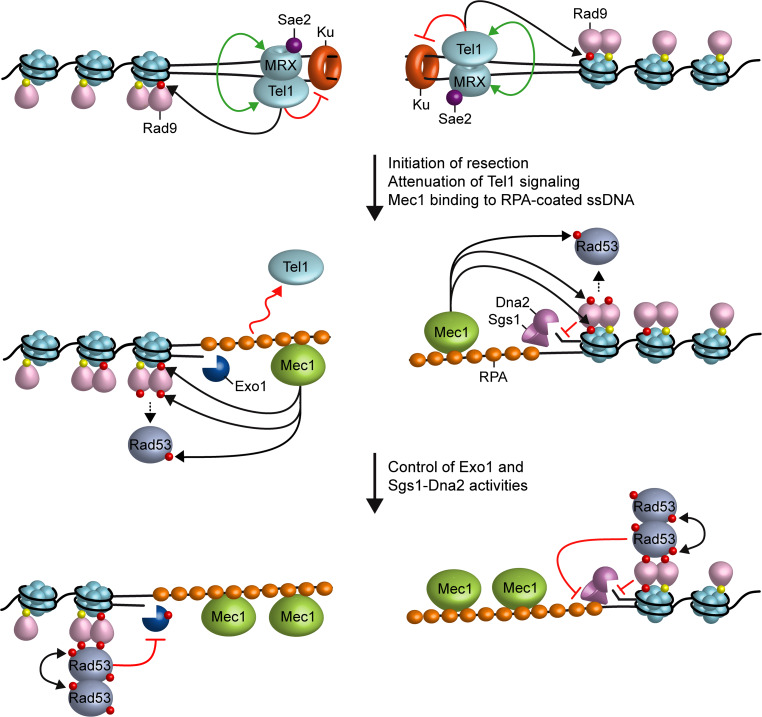
Mec1 and Tel1 themselves regulate the generation of 3′-ended ssDNA at the DSB ends. Cells lacking Mec1 accelerate the generation of ssDNA at the DSBs, whereas the same process is impaired in mec1-ad cells that carry an hypermorphic mec1 allele [66]. Mec1 inhibits DSB resection at least in two ways: (1) by inducing Rad53-dependent phosphorylation of Exo1 that leads to the inhibition of Exo1 activity [85, 86] and (2) by promoting the binding of the resection inhibitor Rad9 close to DNA lesions through phosphorylation of H2A on serine 129 (Fig. 3) [66]. In fact, while the lack of Mec1 reduces Rad9 recruitment at the DSB ends by impairing γH2A generation, the Mec1-ad variant enhances the association of Rad9 at DNA ends by increasing the efficiency of γH2A generation [66]. Furthermore, eliminating Rad9 or decreasing its binding to the DSB by preventing γH2A formation suppresses the resection defect of mec1-ad cells [66]. Consistent with a role of Mec1 in inhibiting DSB resection, the ring-shaped Ddc1-Mec3-Rad17 checkpoint complex, which is required for full Mec1 activation [87], inhibits resection by promoting the recruitment of Rad9 near DSBs [88]. In any case, the rapid resection in mec1∆ cells is not as efficient as in rad9∆ cells [66], suggesting that Mec1 also positively controls DSB resection. Consistent with this hypothesis, Mec1 is known to phosphorylate Sae2 and this phosphorylation is important for Sae2 function in DSB resection [89, 90].
Fig. 3.
Interplays between end resection and checkpoint. Rad9 is bound to methylated histone H3 (yellow dots) even in the absence of DSBs. When a DSB occurs, the MRX complex and Sae2 localize to the DSB ends. MRX is required for the recruitment at DSBs of Tel1, which in turn stabilizes MRX retention at DSBs in a positive feedback loop (double green arrows). Tel1 promotes the removal of Ku from the DSB and the initiation of resection. Furthermore, it contributes to the recruitment of Rad9 to the DSB ends through γH2A generation (red dots). When DSB resection takes place, the resulting 3′-ended ssDNA attenuates Tel1 signaling activity and, once coated by RPA, allows activation of Mec1. Activated Mec1 contributes to γH2A generation that leads to a further enrichment of Rad9 at DSBs, which provides a barrier to the resection activity of Sgs1-Dna2. Mec1 also phosphorylates Rad9 and these phosphorylation events create binding sites for Rad53 molecules, which then undergo in-trans autophosphorylation and activation (double black arrows). Mec1-dependent phosphorylation of Rad53 allows further autoactivation. Once activated by Mec1, Rad53 counteracts DSB resection by phosphorylating and inhibiting Exo1 and by restricting the access to the DSB of Sgs1-Dna2 possibly by reducing Sgs1 binding to RPA-coated DNA. Phosphorylation events are indicated by black arrows and red dots
In contrast to Mec1-deficient cells, cells lacking Tel1 slightly reduce the efficiency of resection [91] and concomitantly increases both precise and imprecise NHEJ events [92]. Interestingly, tel1∆ cells show an abnormally high persistence of Ku at the DSBs [93]. Since an increase of Ku level delays DSB processing [52], the high Ku association at DSBs in tel1∆ cells might explain the resection defect observed in the same cells.
Tel1, once loaded at DSBs by MRX, supports MRX function in a positive feedback loop (Fig. 3). In fact, the lack of Tel1 was shown to impair MRX association at DNA ends flanked by telomeric DNA repeats [94]. Furthermore, a synthetic phenotype screen has isolated a rad50-V1269M allele that sensitizes tel1∆ cells to genotoxic agents [95]. The rad50-V1269M mutation impairs MRX association at DSBs and the lack of Tel1 reduces further the amount of MRV1269MX bound at DSBs. As a consequence, tel1∆ rad50-V1269M cells are severely defective in keeping the DSB ends tethered to each other and in repairing a DSB by either HR or NHEJ [95]. Interestingly, MRX association to DNA has been shown to induce parallel orientation of the Rad50 coiled coils that favors intercomplex association needed for DNA tethering [96]. These data suggest that Tel1, once loaded at DSBs by MRX, promotes a proper MRX-DNA association needed for the tethering of broken DNA ends and DSB repair. Tel1 exerts this function independent of its kinase activity [94, 95], suggesting that it plays a structural role in promoting/stabilizing MRX retention to DSBs.
Altogether these data support a model wherein the binding of MRX to DNA ends drives the recruitment of Tel1, which facilitates the removal of Ku from the DSB ends to prevent Ku-mediated end-joining and to facilitate resection of the DSB ends (Fig. 3). Tel1 also promotes proper MRX association at DSBs needed for end tethering. When DSB resection takes place, the resulting ssDNA-coated by RPA is recognized by Mec1 that phosphorylates H2A on serine 129. γH2A generation promotes the enrichment of Rad9 to the DSB ends, which inhibits DSB resection by counteracting Sgs1-Dna2 activity. Mec1 also phosphorylates Rad9 and these phosphorylation events create a binding site for Rad53, which then undergoes in-trans autophosphorylation events required for Rad53 activation as a kinase. Once activated, Rad53 in turn inhibits DSB resection by phosphorylating and inhibiting Exo1 and by promoting Rad9-mediated inhibition of Sgs1-Dna2 activity (see below). Because Mec1 is activated by RPA-loaded ssDNA, this Mec1-mediated inhibition of DSB resection contributes to keep under control Mec1 itself in a negative feedback loop, thus coupling resection with checkpoint activation.
End resection and checkpoint inactivation
In both yeast and mammals, Mec1 activation is coupled with ssDNA-dependent loss of Tel1 activation, suggesting that the increase in length of the single-stranded 3′ overhangs drives a switch from a Tel1- to a Mec1-dependent checkpoint [91, 97]. Defects in DSB resection caused by either dysfunctions of the resection machinery or Rad9 excess at DSBs causes unscheduled Tel1-mediated cell cycle arrest because it leads to a persistent MRX occupancy at DSBs [65, 66, 98, 99]. As the mammalian counterpart of MRX has been shown to bind ss/dsDNA junctions [100], one possibility is that the slowing down of DSB resection can generate stable ss/dsDNA junctions that are recognized by MRX and this, in turn, can lead to a persistent Tel1 signaling activity. Alternatively, the ssDNA at the dsDNA junction can be prone to breakage to generate a second DSB that activates MRX-Tel1, as suggested in [101]. Modulation of MRX-Tel1 activity by DSB resection can be important also at stalled replication forks, where Tel1 is involved in preventing rearrangements and accumulation of cruciform DNA structures [102, 103].
Persistent checkpoint activation caused by enhanced MRX signaling activity at DSBs contributes to the DNA damage hypersensitivity of Sae2-deficient cells. In fact, mre11 mutant alleles that reduce MRX binding to DSBs restore DNA damage resistance in sae2∆ cells [104, 105]. Furthermore, impairment of Rad53 activity either by affecting its interaction with Rad9 or by abolishing its kinase activity suppresses the sensitivity to DNA damaging agents of sae2∆ cells [106]. Similarly, reduction in Tel1 binding to DNA ends or abrogation of its kinase activity restores DNA damage resistance in sae2∆ cells [106].
Defects in Rad53 or Tel1 signaling also suppress the resection defect of sae2∆ cells [106]. The bypass of Sae2 function in DSB resection by Rad53 and Tel1 impairment is due to decreased amount of Rad9 bound at the DSBs [106]. As Rad9 inhibits Sgs1-Dna2 [55, 56], reduced Rad9 association at DSBs likely bypasses Sae2 function in DNA damage resistance and DSB resection by relieving the inhibition of the Sgs1-Dna2 resection machinery. Altogether, these findings suggest that the primary cause of the resection defect caused by the lack of Sae2 is an enhanced Rad9 binding to DSBs that is promoted by the persistent MRX-dependent Tel1 and Rad53 signaling activities. While Tel1 can control Rad9 association through γH2A generation [106], how Rad53 can facilitate Rad9-mediated inhibition of Sgs1-Dna2 activity remains to be determined. Because Rad53 and RPA compete for binding to Sgs1 [107], it is tempting to propose that activation of Rad53 signaling activity may shift Sgs1 binding preference from RPA to Rad53, leading to decreased Sgs1 association to RPA-coated ssDNA that in turn can potentiate the barrier to resection imposed by Rad9-bound chromatin (Fig. 3).
In any case, the bypass of Sae2 function in DSB resection by Rad53 or Tel1 dysfunction needs a sufficient amount of MRX to be present at DSBs to promote stable association of Sgs1-Dna2 to DNA ends. In fact, attenuation of Rad53 signaling by mre11 mutant alleles that reduce MRX binding to DSBs is not capable to restore wild type levels of resection in sae2∆ cells [104, 105]. Furthermore, TEL1 deletion, which reduces the association of MRX to DSBs [94, 95], is not capable to suppress the resection defect of sae2∆ cells, whereas the same process is restored by the lack of Tel1 kinase activity via generation of a kinase-dead allele [104–106].
Conclusion
It is clear that the nucleolytic processing of the DSB ends generates stretches of 3′ ended ssDNA that are necessary to initiate HR and to elicit a checkpoint response. On the other hand, this process has to be strictly regulated. In fact, long ssDNA formed at DSBs, at dysfunctional replication forks or after break-induced DNA replication can be the source of clustered mutations (kataegis) frequently occurring during carcinogenesis [108–112]. Furthermore, extensive DNA end resection may induce error-prone repair events that can cause DNA deletions and translocations. Given the importance of DSB repair mechanisms in tumor biology, further understanding of their positive and negative regulators as well as of their connections with the checkpoint mechanisms is strongly relevant to human disease.
Acknowledgments
We thank Giovanna Lucchini and Michela Clerici for critical reading of the manuscript. This work was supported by grants from Associazione Italiana per la Ricerca sul Cancro (AIRC) (Grant IG15210) to MPL. CC was supported by a fellowship from Fondazione Italiana Ricerca sul Cancro (FIRC).
Abbreviations
- DSB
Double-strand break
- ssDNA
Single-stranded DNA
- dsDNA
Double-stranded DNA
- MRX
Mre11-Rad50-Xrs2
- RPA
Replication protein A
- HR
Homologous recombination
- NHEJ
Non-homologous end joining
- Ku
Ku70–Ku80
References
- 1.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert CS, Green CM, Lowndes NF. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol Cell. 2001;8:129–136. doi: 10.1016/S1097-2765(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 3.Sweeney FD, Yang F, Chi A, et al. Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr Biol. 2005;15:1364–1375. doi: 10.1016/j.cub.2005.06.063. [DOI] [PubMed] [Google Scholar]
- 4.Usui T, Foster SS, Petrini JHJ. Maintenance of the DNA-damage checkpoint requires DNA-damage-induced mediator protein oligomerization. Mol Cell. 2009;33:147–159. doi: 10.1016/j.molcel.2008.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiruvella KK, Liang Z, Wilson TE. Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol. 2013;5:a012757. doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mehta A, Haber JE. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2014;6:a016428. doi: 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 8.Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-I. [DOI] [PubMed] [Google Scholar]
- 9.Nairz K, Klein F. mre11S-a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 1997;11:2272–2290. doi: 10.1101/gad.11.17.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsubouchi H, Ogawa H. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol Cell Biol. 1998;18:260–268. doi: 10.1128/MCB.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furuse M, Nagase Y, Tsubouchi H, et al. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/MCB.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trujillo KM, Roh DH, Chen L, et al. Yeast Xrs2 binds DNA and helps target Rad50 and Mre11 to DNA ends. J Biol Chem. 2003;278:48957–48964. doi: 10.1074/jbc.M309877200. [DOI] [PubMed] [Google Scholar]
- 14.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 15.Shroff R, Arbel-Eden A, Pilch D, et al. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lammens K, Bemeleit DJ, Möckel C, et al. The Mre11:rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell. 2011;145:54–66. doi: 10.1016/j.cell.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim HS, Kim JS, Park YB, et al. Crystal structure of the Mre11-Rad50-ATPgS complex: understanding the interplay between Mre11 and Rad50. Genes Dev. 2011;25:1091–1104. doi: 10.1101/gad.2037811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Möckel C, Lammens K, Schele A, et al. ATP driven structural changes of the bacterial Mre11:rad50 catalytic head complex. Nucleic Acids Res. 2012;40:914–927. doi: 10.1093/nar/gkr749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams RS, Moncalian G, Williams JS, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deshpande RA, Williams GJ, Limbo O, et al. ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. EMBO J. 2014;33:482–500. doi: 10.1002/embj.201386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/S1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 22.Trujillo KM, Yuan S-SF, Lee EY-HP, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 23.Trujillo KM, Sung P. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50·Mre11 complex. J Biol Chem. 2001;276:35458–35464. doi: 10.1074/jbc.M105482200. [DOI] [PubMed] [Google Scholar]
- 24.Hopkins BB, Paull TT. The P. furiosus Mre11/Rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell. 2008;135:250–260. doi: 10.1016/j.cell.2008.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cannavo E, Cejka P. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature. 2014;514:122–125. doi: 10.1038/nature13771. [DOI] [PubMed] [Google Scholar]
- 26.Garcia V, Phelps SEL, Gray S, Neale MJ. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011;479:241–244. doi: 10.1038/nature10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Z, Chung W-H, Shim EY, et al. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shim EY, Chung W-H, Nicolette ML, et al. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 2010;29:3370–3380. doi: 10.1038/emboj.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Budd ME, Choe W-C, Campbell JL. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J Biol Chem. 1995;270:26766–26769. doi: 10.1074/jbc.270.45.26766. [DOI] [PubMed] [Google Scholar]
- 31.Kao H-I, Campbell JL, Bambara RA. Dna2p helicase/nuclease is a tracking protein, like FEN1, for flap cleavage during Okazaki fragment maturation. J Biol Chem. 2004;279:50840–50849. doi: 10.1074/jbc.M409231200. [DOI] [PubMed] [Google Scholar]
- 32.Niu H, Chung W-H, Zhu Z, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae . Nature. 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee B-I, Wilson DM. The RAD2 domain of human exonuclease 1 exhibits 5′ to 3′ exonuclease and flap structure-specific endonuclease activities. J Biol Chem. 1999;274:37763–37769. doi: 10.1074/jbc.274.53.37763. [DOI] [PubMed] [Google Scholar]
- 34.Tran PT, Erdeniz N, Dudley S, Liskay RM. Characterization of nuclease-dependent functions of Exo1p in Saccharomyces cerevisiae . DNA Repair. 2002;1:895–912. doi: 10.1016/S1568-7864(02)00114-3. [DOI] [PubMed] [Google Scholar]
- 35.Cannavo E, Cejka P, Kowalczykowski SC. Relationship of DNA degradation by Saccharomyces cerevisiae Exonuclease 1 and its stimulation by RPA and Mre11-Rad50-Xrs2 to DNA end resection. Proc Natl Acad Sci. 2013;110:E1661–E1668. doi: 10.1073/pnas.1305166110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cejka P, Cannavo E, Polaczek P, et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nimonkar AV, Genschel J, Kinoshita E, et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H, Lisby M, Symington LS. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol Cell. 2013;50:589–600. doi: 10.1016/j.molcel.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/S0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 40.McKee AH, Kleckner N. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2 . Genetics. 1997;146:797–816. doi: 10.1093/genetics/146.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prinz S, Amon A, Klein F. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae . Genetics. 1997;146:781–795. doi: 10.1093/genetics/146.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henner WD, Grunberg SM, Haseltine WA. Enzyme action at 3′ termini of ionizing radiation-induced DNA strand breaks. J Biol Chem. 1983;258:15198–15205. [PubMed] [Google Scholar]
- 44.Barker S, Weinfeld M, Zheng J, et al. Identification of mammalian proteins cross-linked to DNA by ionizing radiation. J Biol Chem. 2005;280:33826–33838. doi: 10.1074/jbc.M502477200. [DOI] [PubMed] [Google Scholar]
- 45.Hartsuiker E, Neale MJ, Carr AM. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell. 2009;33:117–123. doi: 10.1016/j.molcel.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huertas P, Cortés-Ledesma F, Sartori AA, et al. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X, Niu H, Chung W-H, et al. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat Struct Mol Biol. 2011;18:1015–1019. doi: 10.1038/nsmb.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ira G, Pellicioli A, Balijja A, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem. 2005;280:38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- 51.Tomita K, Matsuura A, Caspari T, et al. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol. 2003;23:5186–5197. doi: 10.1128/MCB.23.15.5186-5197.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clerici M, Mantiero D, Guerini I, et al. The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep. 2008;9:810–818. doi: 10.1038/embor.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foster SS, Balestrini A, Petrini JHJ. Functional interplay of the Mre11 nuclease and Ku in the response to replication-associated DNA damage. Mol Cell Biol. 2011;31:4379–4389. doi: 10.1128/MCB.05854-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29:3358–3369. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonetti D, Villa M, Gobbini E, et al. Escape of Sgs1 from Rad9 inhibition reduces the requirement for Sae2 and functional MRX in DNA end resection. EMBO Rep. 2015;16:351–361. doi: 10.15252/embr.201439764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferrari M, Dibitetto D, De Gregorio G, et al. Functional interplay between the 53BP1-ortholog Rad9 and the Mre11 complex regulates resection, end-tethering and repair of a double-strand break. PLoS Genet. 2015;11:e1004928. doi: 10.1371/journal.pgen.1004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pellicioli A, Foiani M. Signal transduction: how Rad53 kinase is activated. Curr Biol. 2005;15:R769–R771. doi: 10.1016/j.cub.2005.08.057. [DOI] [PubMed] [Google Scholar]
- 58.Grenon M, Costelloe T, Jimeno S, et al. Docking onto chromatin via the Saccharomyces cerevisiae Rad9 Tudor domain. Yeast. 2007;24:105–119. doi: 10.1002/yea.1441. [DOI] [PubMed] [Google Scholar]
- 59.Wysocki R, Javaheri A, Allard S, et al. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lazzaro F, Sapountzi V, Granata M, et al. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J. 2008;27:1502–1512. doi: 10.1038/emboj.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Granata M, Lazzaro F, Novarina D, et al. Dynamics of Rad9 chromatin binding and checkpoint function are mediated by its dimerization and are cell cycle-regulated by CDK1 activity. PLoS Genet. 2010;6:e1001047. doi: 10.1371/journal.pgen.1001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hammet A, Magill C, Heierhorst J, et al. Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO Rep. 2007;8:851–857. doi: 10.1038/sj.embor.7401036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Downs JA, Lowndes NF, Jackson SP. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- 64.Redon C, Pilch DR, Rogakou EP, et al. Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep. 2003;4:678–684. doi: 10.1038/sj.embor.embor871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eapen VV, Sugawara N, Tsabar M, et al. The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation. Mol Cell Biol. 2012;32:4727–4740. doi: 10.1128/MCB.00566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clerici M, Trovesi C, Galbiati A, et al. Mec1/ATR regulates the generation of single-stranded DNA that attenuates Tel1/ATM signaling at DNA ends. EMBO J. 2014;33:198–216. doi: 10.1002/embj.201386041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen X, Cui D, Papusha A, et al. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature. 2012;489:576–580. doi: 10.1038/nature11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Costelloe T, Louge R, Tomimatsu N, et al. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature. 2012;489:581–584. doi: 10.1038/nature11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lydall D, Weinert T. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- 70.Paciotti V, Clerici M, Lucchini G, et al. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- 71.Cortez D, Guntuku S, Qin J, et al. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 72.Nakada D, Hirano Y, Tanaka Y, et al. Role of the C terminus of Mec1 checkpoint kinase in its localization to sites of DNA damage. Mol Biol Cell. 2005;16:5227–5235. doi: 10.1091/mbc.E05-05-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen X, Zhao R, Glick GG, et al. Function of the ATR N-terminal domain revealed by an ATM/ATR chimera. Exp Cell Res. 2007;313:1667–1674. doi: 10.1016/j.yexcr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakada D, Matsumoto K, Sugimoto K. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 2003;17:1957–1962. doi: 10.1101/gad.1099003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 76.Lee J-H, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 77.You Z, Chahwan C, Bailis J, et al. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berkovich E, Monnat RJ, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 79.Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 80.Dupré A, Boyer-Chatenet L, Gautier J. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat Struct Mol Biol. 2006;13:451–457. doi: 10.1038/nsmb1090. [DOI] [PubMed] [Google Scholar]
- 81.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 82.Adams KE, Medhurst AL, Dart DA, Lakin ND. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene. 2006;25:3894–3904. doi: 10.1038/sj.onc.1209426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jazayeri A, Falck J, Lukas C, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 84.Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem. 2006;281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jia X, Weinert T, Lydall D. Mec1 and Rad53 inhibit formation of single-stranded DNA at telomeres of Saccharomyces cerevisiae cdc13-1 mutants. Genetics. 2004;166:753–764. doi: 10.1534/genetics.166.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morin I, Ngo H-P, Greenall A, et al. Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J. 2008;27:2400–2410. doi: 10.1038/emboj.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Paciotti V, Lucchini G, Plevani P, Longhese MP. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 1998;17:4199–4209. doi: 10.1093/emboj/17.14.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ngo GHP, Lydall D. The 9-1-1 checkpoint clamp coordinates resection at DNA double strand breaks. Nucleic Acids Res. 2015;43:5017–5032. doi: 10.1093/nar/gkv409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baroni E, Viscardi V, Cartagena-Lirola H, et al. The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Mol Cell Biol. 2004;24:4151–4165. doi: 10.1128/MCB.24.10.4151-4165.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cartagena-Lirola H, Guerini I, Viscardi V, et al. Budding yeast Sae2 is an in vivo target of the Mec1 and Tel1 checkpoint kinases during meiosis. Cell Cycle. 2006;5:1549–1559. doi: 10.4161/cc.5.14.2916. [DOI] [PubMed] [Google Scholar]
- 91.Mantiero D, Clerici M, Lucchini G, Longhese MP. Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep. 2007;8:380–387. doi: 10.1038/sj.embor.7400911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee K, Zhang Y, Lee SE. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature. 2008;454:543–546. doi: 10.1038/nature07054. [DOI] [PubMed] [Google Scholar]
- 93.Iwasaki D, Hayashihara K, Shima H, et al. The MRX complex ensures NHEJ fidelity through multiple pathways including Xrs2-FHA-dependent Tel1 activation. PLoS Genet. 2016;12:e1005942. doi: 10.1371/journal.pgen.1005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hirano Y, Fukunaga K, Sugimoto K. Rif1 and rif2 inhibit localization of tel1 to DNA ends. Mol Cell. 2009;33:312–322. doi: 10.1016/j.molcel.2008.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cassani C, Gobbini E, Wang W, et al. Tel1 and Rif2 regulate MRX functions in end-tethering and repair of DNA double-strand breaks. PLoS Biol. 2016;14:e1002387. doi: 10.1371/journal.pbio.1002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moreno-Herrero F, de Jager M, Dekker NH, et al. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 97.Shiotani B, Zou L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Usui T, Ogawa H, Petrini JH. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol Cell. 2001;7:1255–1266. doi: 10.1016/S1097-2765(01)00270-2. [DOI] [PubMed] [Google Scholar]
- 99.Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep. 2006;7:212–218. doi: 10.1038/sj.embor.7400593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Duursma AM, Driscoll R, Elias JE, Cimprich KA. A role for the MRN complex in ATR activation via TOPBP1 recruitment. Mol Cell. 2013;50:116–122. doi: 10.1016/j.molcel.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Beyer T, Weinert T. Mec1 and Tel1: an arresting dance of resection. EMBO J. 2014;33:176–178. doi: 10.1002/embj.201387440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Doksani Y, Bermejo R, Fiorani S, et al. Replicon dynamics, dormant origin firing, and terminal fork integrity after double-strand break formation. Cell. 2009;137:247–258. doi: 10.1016/j.cell.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kaochar S, Shanks L, Weinert T. Checkpoint genes and Exo1 regulate nearby inverted repeat fusions that form dicentric chromosomes in Saccharomyces cerevisiae . Proc Natl Acad Sci. 2010;107:21605–21610. doi: 10.1073/pnas.1001938107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen H, Donnianni RA, Handa N, et al. Sae2 promotes DNA damage resistance by removing the Mre11–Rad50–Xrs2 complex from DNA and attenuating Rad53 signaling. Proc Natl Acad Sci. 2015;112:E1880–E1887. doi: 10.1073/pnas.1503331112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Puddu F, Oelschlaegel T, Guerini I, et al. Synthetic viability genomic screening defines Sae2 function in DNA repair. EMBO J. 2015;34:1509–1522. doi: 10.15252/embj.201590973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gobbini E, Villa M, Gnugnoli M, et al. Sae2 function at DNA double-strand breaks is bypassed by dampening Tel1 or Rad53 activity. PLoS Genet. 2015;11:e1005685. doi: 10.1371/journal.pgen.1005685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hegnauer AM, Hustedt N, Shimada K, et al. An N-terminal acidic region of Sgs1 interacts with Rpa70 and recruits Rad53 kinase to stalled forks: sgs1 binds RPA and Rad53 during replicative stress. EMBO J. 2012;31:3768–3783. doi: 10.1038/emboj.2012.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roberts SA, Sterling J, Thompson C, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nik-Zainal S, Alexandrov LB, Wedge DC, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sakofsky CJ, Roberts SA, Malc E, et al. Break-induced replication is a source of mutation clusters underlying kataegis. Cell Rep. 2014;7:1640–1648. doi: 10.1016/j.celrep.2014.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Taylor BJ, Nik-Zainal S, Wu YL, et al. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]