Abstract
A systematic understanding of different factors influencing cell type specific microRNA profiles is essential for state-of-the art biomarker research. We carried out a comprehensive analysis of the biological variability and changes in cell type pattern over time for different cell types and different isolation approaches in technical replicates. All combinations of the parameters mentioned above have been measured, resulting in 108 miRNA profiles that were evaluated by next-generation-sequencing. The largest miRNA variability was due to inter-individual differences (34 %), followed by the cell types (23.4 %) and the isolation technique (17.2 %). The change over time in cell miRNA composition was moderate (<3 %) being close to the technical variations (<1 %). Largest variability (including technical and biological variance) was observed for CD8 cells while CD3 and CD4 cells showed significantly lower variations. ANOVA highlighted that 51.5 % of all miRNAs were significantly influenced by the purification technique. While CD4 cells were least affected, especially miRNA profiles of CD8 cells were fluctuating depending on the cell purification approach. To provide researchers access to the profiles and to allow further analyses of the tested conditions we implemented a dynamic web resource.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-016-2154-9) contains supplementary material, which is available to authorized users.
Keywords: microRNA, Next-generation sequencing, Blood cells, FACS
Introduction
Understanding cell subtype specific molecular pattern contributes to our understanding of normal physiological processes and pathological alterations. The first step for profiling of molecules in cell subtypes is the purification of the respective cell types. Already this first step is known to influence the generated molecular profiles substantially. Especially for gene expression, different extraction techniques including positive immune-magnetic selection, negative immune-magnetic selection or fluorescent activated cell sorting (FACS) have been explored and compared to each other. In a study led by Woelk, authors hypothesized that positive selection and FACS show similar expression profiles because of their comparable higher purity [1]. Contrary to this hypothesis, the authors found that positive selection deviated more from FACS as compared to negative selection. As final conclusion, the usage of FACS for gene expression profiling was recommended. In an earlier study on gene expression differences depending on cell purification, Lyons and co-workers came to the conclusion that a rapid positive selection should be applied for microarray gene expression studies [2]. In their work, however, FACS was not included as purification technology. Already these—at the first glance contradicting—results highlight that a systematic exploration of cell purification is essential to analyze the relevance of molecular variability for pathogenic processes independent of technical factors.
Beyond protein coding genes, miRNAs have been claimed as minimally invasive biomarkers that can be measured from body fluids. For multiple oncological, cardio-vascular and neurological disorders, discriminatory miRNA patterns have been suggested [3–10]. As biological source, blood cells, serum and plasma are most commonly used. Especially if peripheral blood mononuclear cells (PBMC) are profiled, signatures of complex mixtures of different blood components are generated. Comparable to protein coding gene expression, the cell types contributing most to the pathological patterns are now searched. In a recent study we explored differential expression of miRNAs in cell types of lung cancer patients and unaffected controls [11]. We used positive immuno-magnetic selection to extract CD3, CD19, CD15, CD14, and CD56 cells and generated miRNA patterns for patients and controls using microarray technology. Thereby we discovered discriminatory patterns in each cell type. Especially cells of the innate immune system allowed almost perfect classification between cases and controls. In another study, we aimed to improve the understanding of the technical reproducibility and the variability of miRNA cell subtypes over time [12]. Again, we applied positive immuno-magnetic selection and performed microarray measurements, indicating an overall high stability of blood cell components’ miRNA profiles.
However a systematic investigation of miRNA blood compounds is still missing. In the present study, we analyzed the most likely sources for variations in miRNA abundance of blood cell subtypes in a systematic manner. Since the so-far applied microarray technology covers just a part of all miRNAs we exchanged the measurement system and performed our study using next-generation-sequencing (NGS) to decipher the blood cell types’ miRNomes. As putative factors contributing to the variations in miRNA expression we considered (a) the cell purification process, (b) technical variations of the whole process (i.e. template/cell preparation, library preparation and NGS), (c) changes of the miRNA composition over time, (d) differences between the cell types and (e) differences between individuals (inter-individual). To systematically explore the source of variations, all combinations of these factors were investigated, resulting in an exponentially growing number of samples to be profiled. We measured three blood donors for three different cell types extracted by three different approaches and repeated all measurements a second time after 2–3 weeks. Finally, all calculations were done as technical replicates such that 3 × 3 × 3 × 2 × 2 = 108 samples had to be processed. These were then evaluated by bioinformatics approaches and the most important sources for variations were explored.
Results
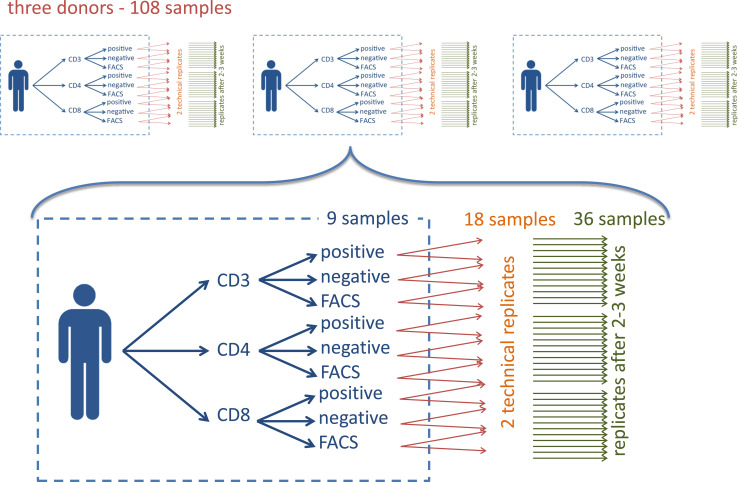
In the present study, we present a comprehensive analysis of the most likely sources for variations including different purification technologies to obtain blood cell type specific miRNA patterns in a systematic manner. We deep-sequenced a total of 108 whole miRNomes from 108 independent library preparations from three individuals without known disease affection (healthy blood donors). The large number of samples is due to the combinatorial complexity of different purification types from different cell types as well as technical and biological replicates that were required to assess the variation introduced by different technologies as well as the biological variability. Specifically, we included n = 3 individuals, for each of them j = 3 different cell populations were considered (CD3, CD4, CD8), which were purified using k = 3 different approaches (positive selection, negative selection and FACS). Each experiment was done in l = 2 technical replicates and was repeated after 2–3 weeks with samples from the same individuals, resulting in m = 2 measurements at different time points of the same donors. Thus, we collected from the three individuals n × j × k × l × m = 3 × 3 × 3 × 2 × 2 = 108 samples. This approach allows us to quantify five important sources of variation including (a) the cell purification process, (b) technical variations of the whole process (i.e. template/cell preparation, library preparation and NGS), (c) changes of the miRNA composition over time, (d) differences between the cell populations and (e) differences between individuals. An overview of the sample collection procedure is presented in Fig. 1.
Fig. 1.
Schema of the study set-up. The blue box presents the nine different combinations of three cell types purified with three approaches. Orange arrows indicate the technical replicates and green arrows the biological replicates leading to 36 samples per patient
For the 108 samples a total of 2.4 billion reads were generated by Illumina HiSeq NGS. On average 22.4 million reads per sample were sequenced. To provide a high quality data set and limit the influence of miRNAs close to background noise, we excluded low abundant miRNAs as well as experimental outliers. In the analysis we included those miRNAs with a total of at least 100 counts from which at least 5 samples had to show at least 10 counts of the respective miRNAs. Altogether, 726 miRNAs fulfilled these criteria. Additionally, we excluded 11 samples where less than 500,000 reads mapped to the miRBase [13]. The number of reads per sample including the ones below threshold are presented in Supplemental Figure 1.
Purity of different extraction approaches
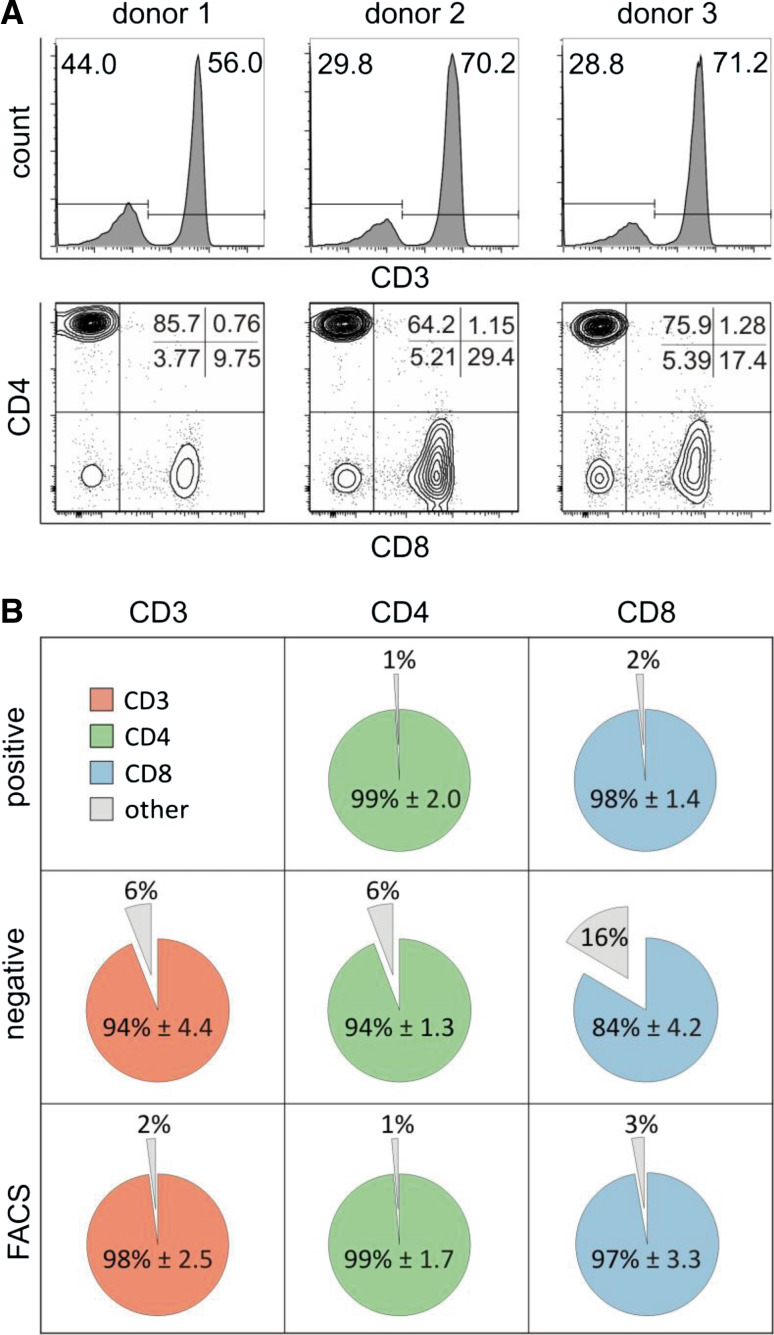
First, we analyzed the CD3+ subpopulation using PBMC as starting material. Figure 2a shows a representative staining of each donor indicating variable percentages of CD3 subpopulation in the PBMC preparation between the different donors. The same is true for CD3+/CD4+ and CD3+/CD8+ subpopulations, uncovering for instance a high CD3+/CD8+ content in donor 2 compared to the other donors (29.4 %, Fig. 2a, lower panel). Next, we assessed the purity of the different cell isolation technologies (Fig. 2b). The highest purity was reached for CD4 cells using positive selection with mean/standard deviation of 99 and 2 %. A similar high purity was found for CD4 cells following FACS selection (98.6, 1.7 %), for CD8 positive selected cells (98.3, 1.4 %), for CD3 cells following FACS selection (98, 2.5 %) and for CD8 FACS purified cells (97.2, 3.3 %). The remaining purifications showed slightly decreased purity around 94 %. The only outlier was CD8+ cells following negative selection with a purity of only 83.5 % and standard deviation of 4.2 % (Grubbs test p value of 0.004). The purity of CD3 cells after positive selection could not be analyzed by cytometry due to the anti-CD3 antibody that cannot be removed after magnetic isolation. For this sample, purity was therefore determined by fluorescence microscopy through a staining with anti-CD4 and anti-CD8 antibodies assuming that these cells are CD3 positive cells. By this presumption, which probably underestimated the real number of CD3 positive cells, purity was 88.1 % with standard deviation of 5.1 %. The numbers of each individual sample are summarized in Supplemental Table 1.
Fig. 2.
Cell purity. a Analysis of PBMC samples before isolation of subtypes. The CD3 cell subsets within PBMC of each donor are shown as histograms. CD3+ cells were selected to differentiate into CD4+ and CD8+ subtypes. Average percentages are displayed for each donor. b Purities of isolated CD3+, CD4+ or CD8+ subtypes are shown for positive selection, negative selection and FACS. Pie charts display the average percentages of CD3+, CD4+ or CD8+ subtypes across donors for each isolation method except for CD3 positively isolated (mean ± standard deviation)
NGS expression profiling and technical variability
As outlined above we performed all analyses with 726 miRNAs that were expressed in the measured profiles and removed 11 samples with low read counts matching to the miRBase. We first analyzed whether these 11 samples (highlighted in Supplemental Figure 1) specifically belong to a certain cell type, individual or purification technology. We did not observe a significant enrichment for any of the tested characteristics. The smallest p value was reached for the purification technology, of the 11 samples two were from positive selection, two from FACS and seven from negative selection, indicating a slight tendency for drop out of samples purified by negative selection (p = 0.105). The remaining 97 samples were normalized such that the read counts of all samples showed the same distribution (quantile normalization) before further analysis.
Next, we investigated the technical reproducibility of different cell types and different extraction approaches. To this end, we calculated for all technical replicates the Pearson correlation coefficient, which reached a high value of 0.982 on average. We ranked all correlation coefficients from highest to lowest and then performed an enrichment analysis to find conditions that have specifically high or low technical reproducibility. The generated running sum statistics for the different conditions indicate that technical reproducibility is similar for the three different donors (donor 1, 0.978; donor 2, 0.979; donor 3, 0.989). Similarly, no difference between positive sorting, negative sorting and FACS was found in terms of technical reproducibility (correlation of 0.98, 0.988 and 0.978, respectively). For the different cell types, we however found enrichment for CD3 and CD4 cells with respect to high correlation and thus high technical reproducibility. In line with this observation, the average correlation for CD3 and CD4 cells was 0.989 and 0.998, while for CD8 cells just average correlation of 0.959 was reached. Thus, CD8 cells showed a significantly (p = 0.015) lower technical reproducibility as compared to CD3 and CD4 cells.
Variability of cell type patterns over time
Our study set-up also allows quantification of the stability of patterns over time since the same individuals were re-profiled 2–3 weeks after the first analysis. On average, the correlation almost reached the values calculated for the technical reproducibility, reaching a value of 0.971. Since we observed large technical variations for CD8 cells, which may partially depend on the purity of CD8 cells (compare Fig. 2 and Supplemental Table 1), we also expected a higher variation with respect to the different time points compared to CD3 or CD4 cells. Indeed, CD3 cells showed a correlation of 0.988, CD4 cells of 0.997 and CD8 cells of only 0.932, again corresponding to a significantly increased variability (p = 0.0004). Importantly, this variability is the consequence of technical as well as biological variations. In contrast to the technical reproducibility analysis presented above we here also observed larger deviations with respect to the purification technology: while positive and negative selection showed correlation values of 0.976 and 0.982, FACS sorting values were slightly decreased (0.959). Finally, also divergent stability over time for the three different individuals was discovered. The three individuals showed correlation values of 0.958, 0.974 and 0.985. The overall lowest deviation after 2–3 weeks was found for CD4 cells of donor 3, with a correlation of 0.999, while the largest divergence was observed for CD8 cells of donor 1 with correlation of 0.918.
Cluster and principal component analysis
The results presented in the previous sections demonstrate that miRNA cell type pattern are generally stable over time and can be measured reproducible by NGS. Technical replicates and the time course of the respective individuals matched very well to each other further validating the stability of miRNA cell type pattern. The technical reproducibility is also a necessary basis to analyze different sources for variation in a quantitative manner.
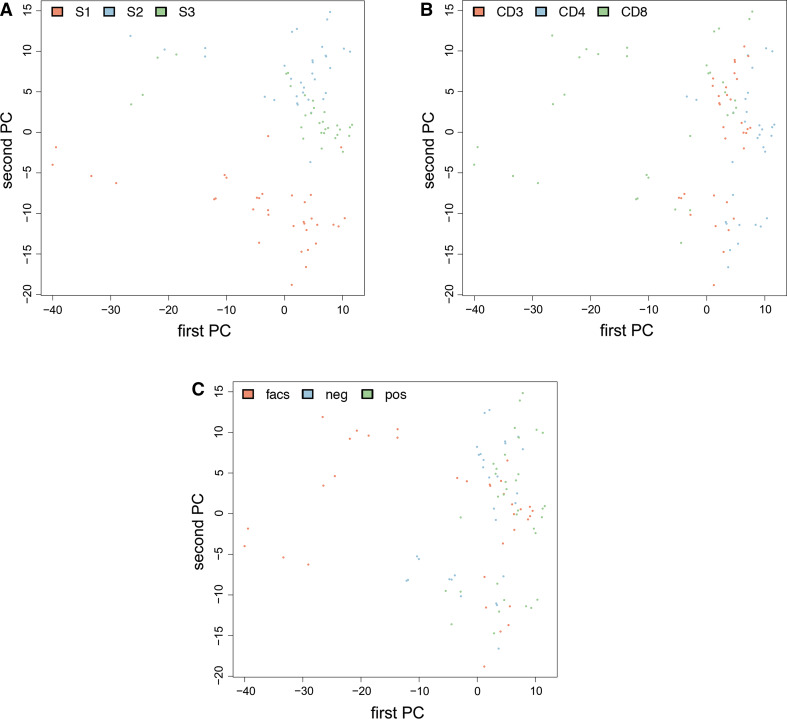
So far, we considered different sources for variation only separately from each other. To understand the overall influence of the combination of different factors, we applied cluster and principal component analysis. Figure 3 presents the second vs. first principal component (PC) as scatter plot. The identical plot is presented three times only the color scheme has been changed in each variant. Figure 3a is colored with respect to the three donors, Fig. 3b with respect to the different cell types and Fig. 3c with respect to the isolation technique. We observe that the three donors can be well distinguished, especially focusing on the second PC. Donor 1 has the smallest values (approx. −5 to −20), followed by donor 3 (which clusters around 2) and donor 2 (with most values between +5 and +15). In Fig. 3b, c it is evident that all dots in the left part of the images belong to the CD8 sample purified by FACS, which are in consequence most different compared to the other purification techniques/cell populations.
Fig. 3.
Scatter plot for the first vs. second principal component colored in red, blue and green with respect to the three donors (a), the different cell types (b) and the different purification techniques (c)
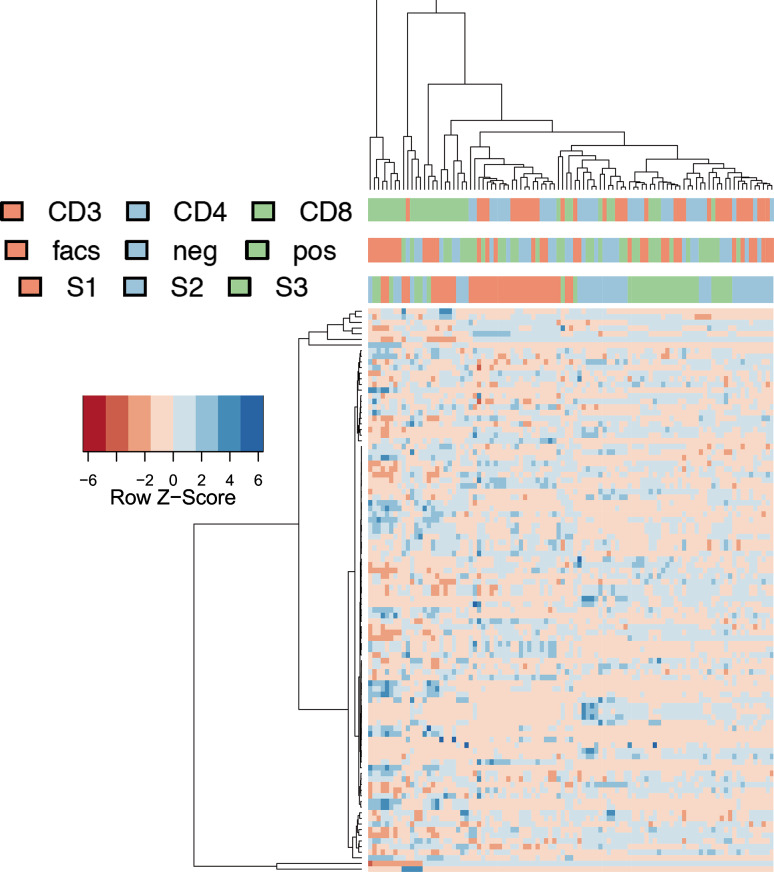
To further explore the variability we performed hierarchical clustering and colored the clusters with respect to individuals, cell types and purification technologies. The resulting heat map is presented in Fig. 4. Here, we can see that individuals’ pattern generally cluster well together, but again FACS-purified CD8 cells (leftmost cluster of 8 samples) and CD8 cells in general showed substantial divergence from other samples. As indicated by the previous analyses, technical replicates and time courses revealed a very good clustering, underlining the high reproducibility of the profiling in general.
Fig. 4.
Heat map as result of a cluster analysis of 50 miRNAs with highest variability. Color scheme is used analogously to Fig. 3
Given the five factors: different individuals, different cell types, different purification technologies, different time points and technical replicates, we asked on the percentage of variability that the factors—either alone or in combination—contribute to the overall variance. An approach that is well suited for such analyses is principal variance component analysis (PVCA), yielding the percentile of variability for each factor and combinations of factors. The highest value of 34 % was reached for the donor, followed by the different cell types (23.4 %) and different isolation techniques (17.2 %). Already these three factors alone explain 74.7 % of the variance. The fourth most relevant factor was the composition of cell type and isolation technique (10.6 %). While already the change over time in cell miRNA composition was moderate (<3 %), the technical variations were even smaller (<1 %). A bar chart detailing the composition of the variance for the different factors is presented in Fig. 5.
Fig. 5.
Contribution of different factors to the overall variability sorted from highest to lowest
Analysis of variance of cell types and purification technique
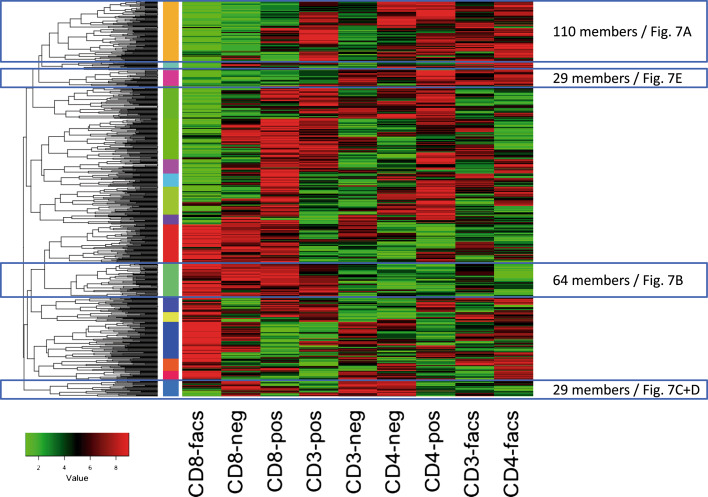
One central question of our study was to explore differences in the cell types and depending on the purification techniques. Thus, we performed an ANOVA with the respective nine groups, i.e. each of the three cell types and each of the three purification techniques. Following adjustment for multiple testing, 476 miRNAs reached a significant deviation in at least one of the nine groups. The lowest p values were as small as 10−46 (miR-4772-5p). Inspecting the patterns of the miRNAs and by clustering the median expression in the nine respective groups we observed almost all possible patterns. The cluster heat map for a complete linkage hierarchical clustering on the ranks of the median expression for the nine groups per miRNA using the Euclidean distance is presented in Fig. 6. Using 26 different common measures for estimating the number of clusters we observed between the minimal and maximal values of 9 (7 of 26 approaches) and 50 clusters (3 of 26 approaches) on average 23.75 clusters in the data (median of 18.5 clusters). Cutting the dendrogram at 1/1.2 of the maximal height we finally extracted 16 clusters that are presented as vertical color bar between the dendrogram and heat map in Fig. 6.
Fig. 6.
Clustering of the ranks of median expression in the nine cell type and purification technique combinations. Vertical color bar between the heat map and the dendrogram indicates the 16 most dominant clusters
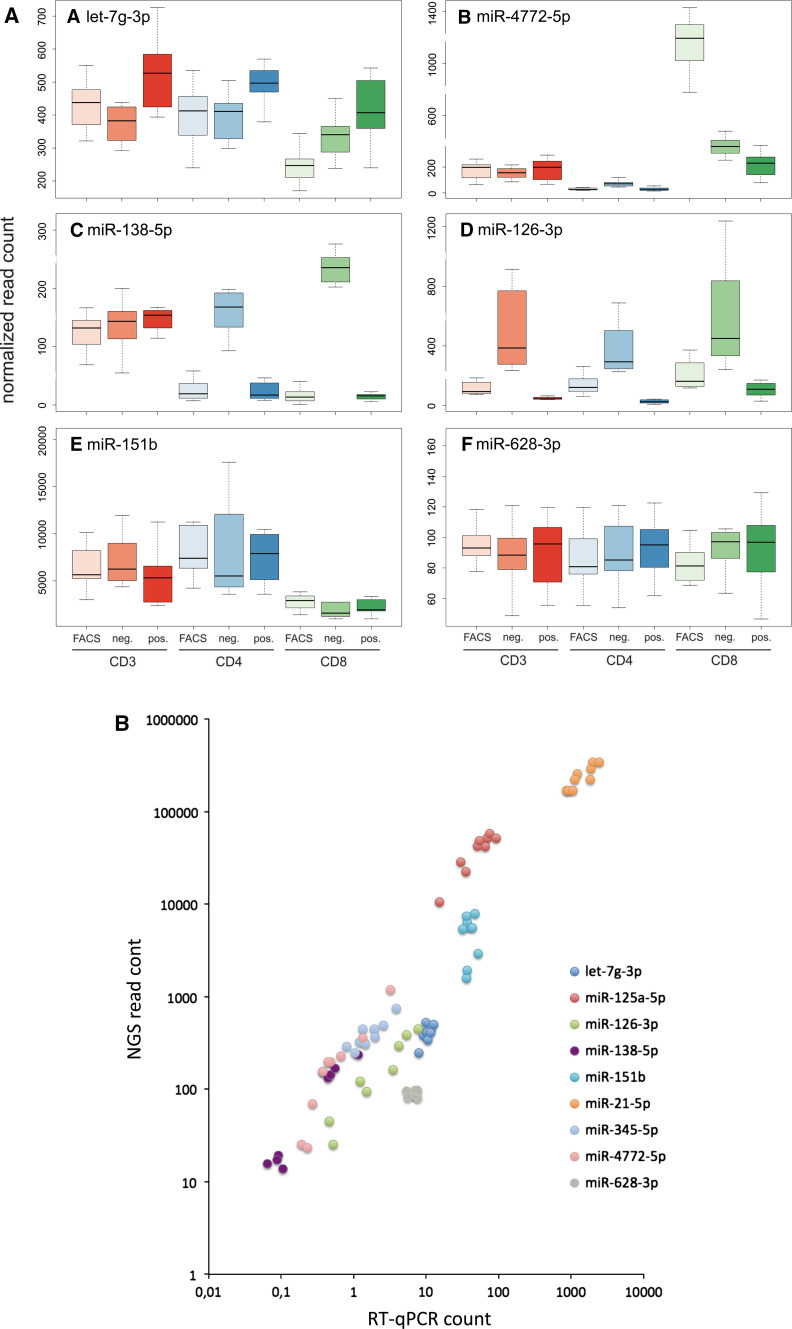
The largest cluster contained 110 miRNAs (cluster 1, highlighted in Fig. 6). In this cluster we observe miRNAs with overall lowest expression in CD8 FACS patterns, followed by increased expression in CD8 negatively selected cells while most other cell types and purification approaches show higher miRNA counts for markers in this cluster. One example of this cluster is let-7g-3p (Fig. 7a) representing the pattern most frequently observed. In case of let-7g-3p (p < 10−10), positive immuno-magnetic selection of cells showed in general a higher expression as compared to negative selection and FACS sorting.
Fig. 7.
a Box-plots for the nine combinations of three cell types and three purification techniques. CD3 cells are colored in shades of orange, CD4 cells in shades of blue and CD8 cells in shades of green. y-axis describes the normalized NGS read count per miRNA. a–f Present each an own miRNA. b Scatter plot that directly compares the RT-qPCR measurements to the NGS data. Each miRNA is represented by an own color. For each miRNA, nine data points representing the different combinations of cell types and purification techniques are included
In contrast to the expression of miRNAs in this largest cluster, we observed 64 miRNAs with overall highest expression in CD8 cells obtained by FACS while all other approaches show lower abundance. One representative example is miR-4772-5p (Fig. 7b). With the 64 cluster members, this pattern was among the most frequently observed ones.
miR-138-5p (p < 10−37) shows a comparable high expression in all CD3 cell purification techniques (Fig. 7c) but is predominantly high in negative selected cell populations. This miRNAs represents 29 other miRNAs. However, while miR-138-5p is comparably low expressed in CD4 and CD8 isolated by FACS or positive selection, its expression in negatively purified CD4 and CD8 cell populations exceeded the other purification techniques significantly and was even higher compared to all isolation methods of CD3 cells. Another more extreme example for high expression just in negative selected cell populations is miR-126-3p (p < 10−13, Fig. 7d).
Nevertheless, we also found miRNAs that only minimally varied between the different purification approaches but strongly varied between different cell types. E.g. one representative of the group of 29 miRNAs lower expressed in the CD8 cell population as compared to the CD3 and CD4 population independently of the purification technique is miR-151b (p < 10−5, see Fig. 7e).
Beyond the significantly affected miRNAs with representative examples in panels a–e, we identified miRNAs that were not affected at all, either by cell type or by purification approach. Exemplarily, miR-628-3p (p = 0.96) is presented in panel f. The patterns for all miRNAs can be accessed from the web resource described at the end of the results section.
Finally, we analyzed the direct influence of purification technique on the different cell types by considering the cell types separately from each other and doing the ANOVA for each of the cell types. After adjustment for multiple testing, 77 miRNAs were significantly affected by the purification approach for CD3 cells, 17 for CD4 cells, and 354 for CD8 cells. Especially for CD8 cells, the large number may be explained by the deviation resulting from FACS purification.
RT-qPCR validation
Validation of high-throughput results is essential to discover potentially false positive findings. We thus carried out RT-qPCR of the miRNAs described in the previous section. Since miRNAs with larger delta Cq values are lower expressed (in contrast to NGS where large values correspond to high expression) and delta Cq values are on log2 scale we calculated pseudo expression counts (see “Methods”). These pseudo counts facilitate the direct comparison between NGS and RT-qPCR results. In the cluster analysis presented above let-7g-3p was in the largest cluster containing miRNAs being lowest expressed in CD8 FACS cells. This finding was replicated in RT-qPCR, where CD8 FACS cells were lowest (7.8 counts), followed by CD3 negative selected cells (9.1 counts). The results for all cell types are presented as bar chart in Supplemental Figure 2A.
As example of 64 miRNAs with overall highest expression in CD8 cells obtained by FACS followed by the other CD8 cells while all other approaches show lower abundance we described miR-4772-5p. Again, this result was reproduced. CD8 FACS cells had 3.2 counts, followed by negative (2.6) and positive selection (1.3) while all other results were below 1 (Supplemental Figure 2B). Moreover, melting curve analysis of this miRNA identified different T m (temperature of melting) peaks that potentially correspond to iso-forms of this miRNA. Inspecting the NGS reads we also observed respective iso-forms with length between 24 and 27 bases.
miR-138-5p was highest expressed in all CD3 cell types, and negative selected CD4 and CD8 cells. As for the first two examples this pattern was reproduced by RT-qPCR (Supplemental Figure 2C). An example for a miRNA with exceptional high expression in negative selected cells was miR-126-3p. Also in RT-qPCR, the negative selected cell types had highest expression. Also concordant to the NGS results, positive selected cell types had lowest expression (Supplemental Figure 2D).
miR-151b was originally reported to be lower expressed in all CD8 cells independent of the purification approach. This result was just partially confirmed, indeed CD8 cells selected by FACS showed very high expression (Supplemental Figure 2E), altogether the variation observed for this miRNA with respect to the different experimental set-ups was substantially smaller as compared to the variations observed by NGS. Since this replication did not matched we investigated two further examples following this pattern, miR-125-5p and miR-21-5p. Expression pattern in NGS and RT-qPCR for the first miRNA are presented in Supplemental Figure 3A and B and for miR-21-5p in Supplemental Figure 3C and D. In both case NGS and qRT-PCR approaches matched well and the original results were confirmed. As counter example to this expression pattern we picked miR-345-5p, which was highest expressed in CD8 cells compared to the other cell types. Indeed, this miRNA had average counts of 2.8 in CD8 cells and 1.3 counts in the other cell types, independently of the purification technique (Supplemental Figure 3E and F).
As marker that was not significantly affected we reported miR-628-3p. This miRNA also showed the overall lowest variation of expression values in the RT-qPCR experiments (maximal expression 7.6 and minimal expression 5.4 across all cell types and purification techniques). The expression values for this miRNA are presented in Supplemental Figure 2F.
To quantify the overall degree of reproducibility we calculated Pearson correlation coefficient for the RT-qPCR measurements and NGS expression for each miRNA separately and then for all data points together. The worst correlation was calculated for the previously described outlier miR-151b (correlation coefficient of 0.04). For all other miRNAs significant correlation was observed. Notably, five of the tested miRNAs had correlation values exceeding 0.9, reaching up to 0.99. In detail, correlation values sorted in increasing order were 0.66 for let-7g-3p, 0.88 for miR-21-5p, 0.91 for miR-125a-5p, 0.93 for miR-345-5p, 0.93 for miR-138-5p, 0.97 for miR-126-3p and 0.99 for miR-4772-5p. Taking all single data points together, correlation was 0.98 and highly significant (p < 10−10). To allow for direct comparison of all measurements for the different miRNAs we show the values as scatter plot in Fig. 7b.
Novel miRNAs that are differentially regulated between cell types
NGS data also allow for discovering novel miRNA candidates [14]. Altogether, miRDeep2 using default parameters proposed 648 novel miRNA precursors. Of those, we discarded 263 candidates since they matched other RNA resources in a BLAST analysis. For the remaining miRNAs we focused on those that are differentially abundant in different cell types. Following Bonferroni adjustment for multiple testing, four miRNAs remained significant. These are detailed in Table 1. In two cases, CD8 cells showed highest abundance with average reads of 4.4 and 2.2 reads per sample. CD4 cells had values of 0.3 and 0 reads and CD3 cells of 2 and 1.1 reads for both miRNAs. The two other candidates had highest values in CD3 cells, namely 1.4 and 8.3 reads. In contrast, CD4 cells had values of 0.8 and 1.9 and CD8 cells of 0.1 and 2.3 counts. Generally, highest abundance was discovered for the positively selected cells.
Table 1.
Novel miRNA candidates
| miRNA | Sequence | av cd3 | av cd4 | av cd8 | ttest cd3 vs. cd4 | ttest cd3 vs. cd8 | ttest cd4 vs. cd8 |
|---|---|---|---|---|---|---|---|
| cell-mir-29 | UCUGAUGGGGAAUGGCCUGCCU | 2.0 | 0.3 | 4.4 | 2.2−05 | 5.2−03 | 6.6−07 |
| cell-mir-1099* | ACUUCGCACUGACUGUUUAGCA | 1.1 | 0.0 | 2.2 | 9.8−04 | 4.7−02 | 1.1−05 |
| cell-mir-359* | UCAGGCGCGGAGGGGAACAGC | 1.4 | 0.8 | 0.1 | 6.7−02 | 2.7−05 | 4.7−03 |
| cell-mir-633 | GCGCGAUGGACCGAGGGC | 8.3 | 1.9 | 2.3 | 4.9−05 | 1.7−04 | 6.1−01 |
Interactive web service to browse and compare profiles
To provide easy access to the measured cell type profiles and to enable further analyses we set up a web repository, which can be accessed at http://www.ccb.uni-saarland.de/cf. Here, the analyses that are described in the manuscript can be inspected in detail. Moreover, we implemented capabilities for interactive comparison of cell types and purification methods. Users can select two arbitrary groups describing cell subtype and isolation technique. For example, “CD4 FACS” vs. “CD4-pos” or “CD4 FACS” vs. “CD8 FACS” can be selected. Then, the respective groups are compared to each other and graphical output as well as tables are generated and presented to the user. The interface and results are shown in Supplemental Figure 4 for the comparison of CD3 positively selected vs. CD4 positively selected cells. The middle panels present a selection of the graphical output that is generated, including a volcano plot, a scatter plot and a heat map. Already the volcano plot indicates a higher expression of 13 miRNAs in CD3 positively selected cells (red dots in left panel) while just six miRNAs were down-regulated (green dots in left panel). The same can be seen in the scatter plot that details median expression in CD3 positively vs. CD4 positively selected cells on a logarithmic scale. Here, blue points are higher expressed in CD3 positively selected cells while orange points are higher expressed in CD4 positively selected cells. Additionally, the 20 most significant miRNAs are presented as heat map, the color scheme on top indicates that CD3 positively selected and CD4 positively selected cells can be perfectly separated from each other. Below the graphical output, an interactive table is generated. This contains besides the miRNA name, the median and standard deviation of expression in both groups, fold change, logarithm of fold change, raw and adjusted Wilcoxon–Mann–Whitney and t test p values as well as the area under the receiver characteristics curve (AUC). In the presented example we have discovered 24 miRNAs with significant dys-regulation (adjusted t test p value of <0.05). Of these, 9 were higher expressed in CD4 positively selected cells while 15 were higher abundant in CD3 positively selected cells. The most significant miRNA was miR-138-5p, with median count of 154 in CD3 positively selected cells compared to only 16 in CD4 positively selected cells. The adjusted t test p value was <10−8. The miRNA, which was most significantly higher expressed in CD4 positively selected cells is miR-222-5p, having median read counts of 9 (CD3 positively selected) and 45 (CD4 positively selected) and a respective adjusted t test p value of 0.003 (see Supplemental Figure 4, table for selected miRNAs).
To further demonstrate the usability of our tool, we searched the literature for other studies addressing the same topic and compared the results. You and co-workers previously published that for CD4 cells no influence of positive vs. negative selected cells has been observed by using OpenArray technology [15]. In their study, 110 miRNAs were expressed but for none of them differential expression was discovered. The main difference between both studies includes the bead size (nanometer scale in You et al. vs. micrometer scale in our study) and measurement of miRNAs (open array in You et al. vs. NGS in our study). We used the data generated in this study and the web resource to search for differences in CD4 positive vs. CD4 negative selected cells. Thereby, we discovered two miRNAs that were significantly affected by the selection process, including the afore-mentioned miR-222-5p as well as miR-138-5p. The fold change for the latter miRNA was as high as tenfold. The same miRNAs and multiple other ones have been detected in the comparison of CD3 positive vs. CD3 negative selected samples.
Conclusions
While cell type specific gene expression patterns have already been systematically explored and also different purification techniques were compared to each other, for miRNAs just few aspects have been investigated.
We thus set out to explore the miRNA repertoire of common blood cell types and common purification techniques using next-generation-sequencing in a systematic manner. Additionally, we performed technical replicates as well as a time course of blood donors. While our analysis suggested a stable miRNA repertoire over time that can be very reproducibly measured, inter-individual variations were substantial. Moreover, we also detected cell type specific miRNA repertoires and found strong variations dependent on the purification technique. Since the presented data set opens many possibilities for addressing further questions beyond those covered in this work we set up an interactive web resource. Using this resource, researches can explore the details of the analyses presented in this work but can also perform own comparisons and analyses. While it is not possible to generally determine the best purification strategy, our results can aid researchers in selecting the best purification technique for the requirements and prerequisites in their studies.
Methods
Study set up
From three blood donors without known disease affection 108 samples were collected (36 samples per donor). These 36 samples are due to combinatorial complexity of our systematic analysis. For each of the n = 3 donors, j = 3 different cell types were considered (CD3, CD4, CD8), which were purified using k = 3 different approaches (positive selection, negative selection and FACS sorting). Each experiment was done in l = 2 technical replicates and at m = 2 different time points (2–3 weeks apart). Thus, we had to collect j × k × l × m = 3 × 3 × 2 × 2 = 36 samples for each of the donors. The study set up is sketched in Fig. 1. The local ethics committee approved the study and all participants gave informed consent.
Cell isolation and cell separation
Peripheral blood mononuclear cells (PBMC) were isolated as described previously [16]. Shortly, the leucocyte reduction chamber systems (LRS chamber, cones) from platelet apheresis of healthy donors were provided by the local blood bank. Cones were flushed with 8–10 ml HBSS (PAA) and loaded on a standard density gradient [leucocyte separation medium (LSM 1077, PAA)]. PBMC were isolated by a density gradient centrifugation (450g, 30 min, at room temperature with lowest acceleration and no break used). The PBMC layer was washed in HBSS and remaining red blood cells were removed by a 1–2 min incubation in lysis buffer (155 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA, pH = 7.3). Cells were washed again in HBSS, counted in a Z2 cell counter (Beckman&Coulter) and until further use kept on ice in PBS/0.5 % BSA.
CD3+ T (=CD3) cells, CD4+ T (=CD4) cells and CD8+ T (=CD8) cells were isolated using three different methods: (1) Dynabeads Untouched Human Cell isolation kit (CD3+, #11344D; CD4+, #11346D or CD8+ T cells #11348D, Life Technologies), (2) Dynabeads® positive Human Cell isolation kit (CD3+, #11151D; CD4+, #11331D; CD8+ T cells, #11333D, Life Technologies), (3) FACS (for details see section “FACS of CD3+, CD4+ and CD8+ T cells”). Bead isolations were done as described by the manufacturer, but PBS/0.5 % BSA was used instead of PBS/0.1 % BSA. To guarantee comparable time periods for isolation as good as possible, different isolations were done in parallel by three different operators. Isolated cells were counted either by Z2 cell counter (Beckman&Coulter) or MoxiZ (VWR).
FACS of CD3+, CD4+ and CD8+ T cells
A density of 1.6 × 107 PBMC/ml was used to isolate CD3+ T cells (2.0 × 107 cells), CD4+ T cells (4.0 × 107 cells) and CD8+ T cells (1.0–1.4 × 108 cells) using anti-CD3-PerCP (SK7; Biolegend), anti-CD4-PE (MT310, DAKO) and anti-CD8-FITC (DK25; DAKO) antibodies. Cells were stained 40 min at 4 °C with the corresponding antibody, washed twice and re-suspended in PBS/0.5 % BSA. T-lymphocyte subsets were sorted in a FACSAriaIII (BD Biosciences) fitted with a 70 µm nozzle. Sorting gates were set as follows: doublets were excluded and living cells were selected using FSC vs. SSC. The remaining cells were separated based on the CD3, CD4 or CD8 expression. Sorted cells were immediately centrifuged and pellets were re-suspended in Trizol (Life Technologies) and stored at −80 °C until RNA isolation.
Cytometry analysis of isolated cell populations
Purity of positively (except for CD3 positively selected population, see below), negatively or sorted subpopulations was determined by using the same antibodies as for the sorting. If available, 1.0 × 106 cells were stained with anti-CD3-PerCP, anti-CD4-PE and anti-CD8-FITC antibodies for 20 min at RT. Cytometry analysis was done at the FACSAriaIII (BD Biosciences) with the same gating strategy used for FACS. The analyses of all probes were done as soon as possible after sorting was finished.
Antibody staining of CD3 positively immunomagnetic selected populations
Cells were stained as described above with anti-CD4 and anti CD8 antibodies. Samples were adhered to poly-l-ornithine-coated (0.1 mg/ml) glass coverslips and analyzed on the stage of an Olympus IX 70 microscope equipped with a 20× objective (UApo/340, N.A. 0.75) and appropriate filter sets from AHF (HQ-Cy3, #F41-007; HQ-EGFP, #F41-017). For acquisition and analyses the software TILL Vision was used.
RNA isolation
Total RNA from 3.0 × 106 cells were isolated using TRIzol® Reagent (Life Technologies, #15596018) including 1 µl Glycogen (5 µg/µl, Life Technologies, #10814-010) following the manufacturer’s protocol.
Library prep and next-generation sequencing
For the library preparation, 100 ng of total RNA was used per sample, as determined with a DNF-489 Standard Sensitivity RNA Analysis Kit on the Fragment Analyzer (Advanced Analytical Technologies). Preparation was performed following the protocol of the TruSeq Small RNA Sample Prep Kit (Illumina). Concentration of the ready prepped libraries was measured with the Qiagen Gene Read Library Quant Kit on a ViiA 7 Real Time PCR system (Life Technologies). Libraries were then pooled in batches of six samples in equal amounts and clustered with a concentration of 14 pmol in one lane each of a single read flowcell using the cBot (Illumina). Sequencing of 50 cycles was performed on a HiSeq 2000 (Illumina). Demultiplexing of the raw sequencing data and generation of the fastq files was done using CASAVA v.1.8.2.
In silico analysis
The de-multiplexed samples have been analyzed using the miRDeep2 pipeline using standard parameters [17]. As output an expression matrix has been calculated containing miRNAs in rows and samples in columns. All downstream calculations were done using R version 3.0.2. First, low abundant miRNAs and experimental outliers were filtered. Only miRNAs with a total of at least 100 counts and at least five samples showing at least ten counts of these miRNAs were considered. Additionally, samples where less than 500,000 reads mapped to the miRBase have been excluded. The resulting reduced matrix was quantile normalized and normalized expression intensity values were used for further calculations. To detect differences between the tested conditions, principal component analysis (PCA) and hierarchical clustering has been carried out. To calculate p values, analysis of variance (ANOVA) was performed. We also used principal variance component analysis (PVCA) to discover the conditions adding largest part of variance to the expression profiles. To assess differences between groups, student t test as well as Wilcoxon–Mann–Whitney tests were carried out and significance values were adjusted for multiple testing using the Benjamini–Hochberg approach [18]. Additionally, the area under the receiver characteristics curve (AUC) was calculated. For clustering, the hclust and the heatmap.2 function from R have been applied. Complete Linkage hierarchical clustering on the Euclidean distance has been performed. To assess the number of clusters, 30 different common approaches have been carried out using the NbClust package and a consensus vote has been carried out. In detail we asked for between 9 and up to 50 clusters which cluster number is most supported by the respective approaches given the miRNA expression data as input. For the detection and coloring of matching patterns in the cluster analysis the dendrogram has been cut at 1/1.2 of the maximal height.
RT-qPCR validation
Since the variance introduced by technical replicates and the second measurement was small compared to the inter-individual and cell type specific differences, we analyzed one replicate of the samples from the first blood draw, i.e. 27 samples, using RT-qPCR. Quantitative RT-PCR was performed using the miScript PCR system of Qiagen after manufacturers instructions. In detail, 50 ng RNA was reverse transcribed using miScript RT II kit (Qiagen, Hilden, Germany) and diluted 1:10. We analyzed expression of 9 miRNAs (let-7g-3p, miR-4772-5p, miR-138-5p, miR-126-3p, miR-151b, miR-628-3p, miR-125a-5p, miR-21-5p, miR-345-5p) and RNU6B as endogenous control. Primers were purchased from Qiagen (miScript Primer Assays). PCR reactions were set up in duplicates in 20 µl total volume using miScript SYBR Green PCR kit with 2 µl of diluted cDNA as input and run on a StepOnePlus cycler (AppliedBiosystems). Threshold cycle (Cq) values were calculated by StepOne Software v2.3 using a fixed threshold of 0.2 and automatic baseline setting. All melting curves were reviewed to ensure specific amplification. For miR-4772-5p, different Tm peaks were observed that potentially correspond to iso-forms of this miRNA. To make qRT-PCR results better comparable to NGS results we transformed the log2 RT-qPCR delta Cq values as follows:
Research carried out for this study with human material (leukocyte reduction system chambers from human blood donors) is authorized by the local ethic committee [declaration from 16.4.2015 (84/15; Prof. Dr. Rettig-Stürmer)].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Figure 1: log10 of mapped reads per sample, samples below the threshold of 0.5 million reads are highlighted
Supplemental Figure 2: RT-qPCR results for the same miRNAs as presented in Figure 7. The bar height corresponds to the pseudo counts as defined in the Methods
Supplemental Figure 3: Additional RT-qPCR validation experiments for three miRNAs. Left hand side presents the NGS results (panels A, C, E) while right hand side presents RT-qPCR based pseudo counts (panels B, D, F)
Supplemental Figure 4: Upper part shows the interface of the web resource with the example input CD3 positive selected vs. CD4 positive selected cells. Middle part presents a volcano plot of the analysis CD3 positive selected vs. CD4 positively selected, detailing the negative logarithm of the p-value vs. the logarithm of fold change. For the same comparison a scatter plot shows the logarithm of median expression for CD3 positive selected cells on the x-axis and for the CD4 positive selected cells on the y-axis. The hierarchical clustering of the 20 most significant miRNAs graphically demonstrates that the respective miRNAs are differentially regulated between both groups. The lower part presents the tabular output
Acknowledgments
The work of AK, BM is supported by the European Union FP7 (BestAgeing). BM is grateful for support from the German Center for Cardiovascular Research (DZHK). We thank Elmar Krause for support within the FACS facility. We acknowledge funding by the Deutsche Forschungsgemeinschaft (DFG), the collaborative research centers SFB 1027 (Project A2 to MH) and SFB 894 (Project A1 to MH) and the research training group GK 1326 (to ECS and MH). We thank Stephanie Deutscher for excellent technical assistance.
Footnotes
E. C. Schwarz and C. Backes contributed equally as first authors.
References
- 1.Beliakova-Bethell N, et al. The effect of cell subset isolation method on gene expression in leukocytes. Cytometry A. 2014;85(1):94–104. doi: 10.1002/cyto.a.22352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyons PA, et al. Microarray analysis of human leucocyte subsets: the advantages of positive selection and rapid purification. BMC Genom. 2007;8:64. doi: 10.1186/1471-2164-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leidinger P, et al. The blood-borne miRNA signature of lung cancer patients is independent of histology but influenced by metastases. Mol Cancer. 2014;13(1):202. doi: 10.1186/1476-4598-13-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meder B, et al. MicroRNA signatures in total peripheral blood as novel biomarkers for acute myocardial infarction. Basic Res Cardiol. 2011;106(1):13–23. doi: 10.1007/s00395-010-0123-2. [DOI] [PubMed] [Google Scholar]
- 5.Leidinger P, et al. A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 2013;14(7):R78. doi: 10.1186/gb-2013-14-7-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keller A, et al. Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS One. 2009;4(10):e7440. doi: 10.1371/journal.pone.0007440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bloomston M, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. J Am Med Assoc. 2007;297(17):1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 8.Roth P, et al. Differentially regulated miRNAs as prognostic biomarkers in the blood of primary CNS lymphoma patients. Eur J Cancer. 2015;51(3):382–390. doi: 10.1016/j.ejca.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 9.Keller A, et al. Comprehensive analysis of microRNA profiles in multiple sclerosis including next-generation sequencing. Mult Scler. 2014;20(3):295–303. doi: 10.1177/1352458513496343. [DOI] [PubMed] [Google Scholar]
- 10.Joerger M, et al. Circulating microRNA profiling in patients with advanced non-squamous NSCLC receiving bevacizumab/erlotinib followed by platinum-based chemotherapy at progression (SAKK 19/05) Lung Cancer. 2014;85(2):306–313. doi: 10.1016/j.lungcan.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 11.Leidinger P, et al. What makes a blood cell based miRNA expression pattern disease specific?—a miRNome analysis of blood cell subsets in lung cancer patients and healthy controls. Oncotarget. 2014;5(19):9484–9497. doi: 10.18632/oncotarget.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leidinger P, et al. The human miRNA repertoire of different blood compounds. BMC Genom. 2014;15:474. doi: 10.1186/1471-2164-15-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths-Jones S, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Backes C et al (2015) Prioritizing and selecting likely novel miRNAs from NGS data. Nucleic Acids Res. doi:10.1093/nar/gkv1335 [DOI] [PMC free article] [PubMed]
- 15.Yu IW, et al. OpenArray profiling reveals no differential modulation of miRNA by positive and negative CD4+ T cell immunoselection. Exp Hematol. 2014;42(1):11–13. doi: 10.1016/j.exphem.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kummerow C, et al. A simple, economic, time-resolved killing assay. Eur J Immunol. 2014;44(6):1870–1872. doi: 10.1002/eji.201444518. [DOI] [PubMed] [Google Scholar]
- 17.Friedländer MR, et al. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57(1):289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.