Abstract
Numerous studies have demonstrated a link between genetic markers on chromosome 13 and schizophrenia, bipolar affective disorder, and other psychiatric phenotypes. The G72/G30 genes (transcribed in opposite directions) are located on chromosome 13q33, a region demonstrating strong evidence for linkage with various neuropsychiatric disorders. G72/G30 was identified in 2002 as a schizophrenia susceptibility locus; however, subsequent association studies did not reach consensus on single SNPs within the locus. Simultaneously, a new vision for the genetic architecture of psychiatric disorders suggested that schizophrenia was a quantitative trait, therefore ascribable to potentially hundreds of genes and subjected to the vagaries of the environment. The main protein product of G72 gene is named pLG72 or d-amino acid oxidase activator DAOA (153 amino acids) and its function is still debated. Functional analyses, also showing controversial results, indicate that pLG72 contributes to N-methyl-d-aspartate receptor modulation by affecting activity of the flavoprotein d-amino acid oxidase, the enzyme responsible for degrading the neuromodulator d-serine. In this review we, for the first time, summarize findings from molecular genetic linkage and association studies concerning G72 gene, cellular and molecular studies on pLG72, and investigations performed on G72/G30 transgenic mice. This will help elucidate the role of psychosis susceptibility genes, which will have a major impact on our understanding of disease pathophysiology and thus change classification and treatment.
Keywords: pLG72, DAOA, Schizophrenia, d-Serine, Molecular basis, NMDA receptors
Introduction
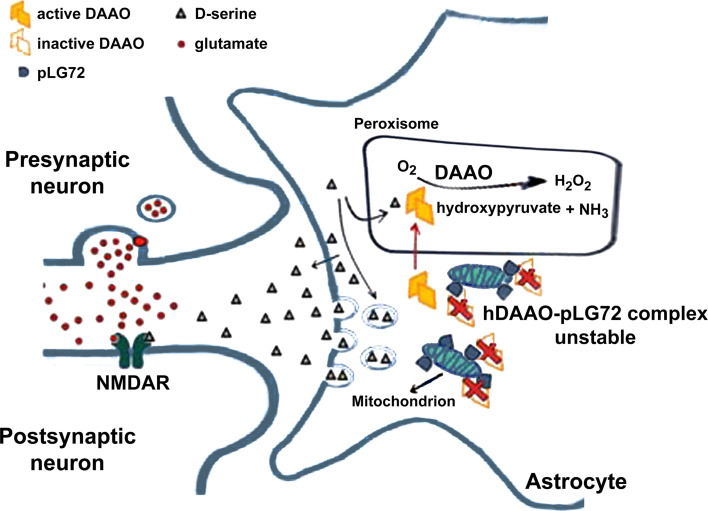
In 2002, a case–control study of French-Canadian families by single nucleotide polymorphisms (SNP) linkages, found a link between schizophrenia and a 5-Mb region on the long arm of chromosome 13, where the new genes G72 and G30 were identified [1]. G72 encodes pLG72 (153 amino acids), a rare case of primate-specific protein. The best-known SNP in G72 rs2391191 (M15) induces Arg30Lys substitution: this mutation has been correlated with the decreased thickness of the brain cortex in schizophrenic patients [2] and with poorer episodic memory function [3]. An association of polymorphisms in G72 with schizophrenia could not be confirmed in a recent study, however [4]. pLG72 was identified as an interacting partner of the flavoenzyme d-amino acid oxidase (DAAO), which degrades d-serine [5, 6], a key endogenous coagonist of the N-methyl-d-aspartate type glutamate receptor (NMDAR) [7–9] which is critically involved in synaptic plasticity, learning, memory, and excitotoxicity [10, 11]. Abnormal variations in pLG72 levels might affect DAAO activity and thus, by altering d-serine levels, NMDAR function at synapsis (Fig. 1). Indeed, pLG72 is present in mitochondria and thus was also reported to be responsible for mitochondrial dysfunction and fragmentation [12].
Fig. 1.
Proposed role of hDAAO–pLG72 interaction in the modulation of d-serine availability at glutamatergic synapses [16]. d-Serine is released by astrocytes, either from a cytosolic pool by a transporter, or from a vesicular pool by a calcium-dependent mechanism. Once in the synaptic cleft, d-serine binds to NMDAR concomitantly with glutamate, activating the receptors at the membrane of postsynaptic neurons [7–9]. In glial cells cytosolic hDAAO can interact with pLG72 on the mitochondrial outer membrane. pLG72 binding negatively affects the amount of active hDAAO, thus preventing an excessive degradation of d-serine. Under pathological conditions, alterations in pLG72 expression levels would result in an overactivation of hDAAO and a consequent decrease in d-serine cellular concentration yielding an NMDAR hypofunctionality [16]
In this paper, we aim for the first time to summarize the results of 15 years of investigations on the biological function of pLG72 as obtained by a number of different approaches.
G72 transcript and protein distribution
G72 represents a rare case of primate-specific gene which encodes a protein characterized by a rapidly changing structure during evolution [1]. No orthologs were identified in rodents or other species and the deduced open-reading frames (ORFs) in ape genomes differ from their human counterparts.
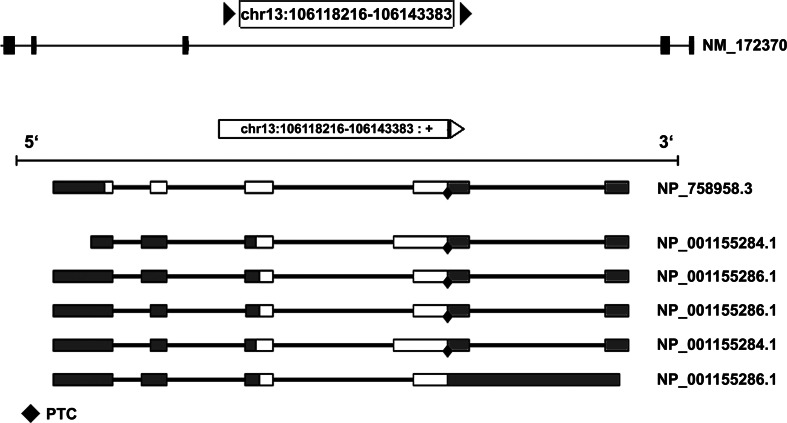
In humans, the longest G72 ORF was detected at low levels in the amygdala, caudate nucleus, spinal cord, brain, and testis [1, 13]. However, the full-length pLG72 has never been detected in these tissues. This could reflect a complex splicing history: in 2003 Hattori and colleagues [13] proposed that up to nine exons could be recognized in the G72 gene, resulting in different proteins due to differential splicing. Actually, current annotation of the gene suggests a maximum of seven exons and three predicted proteins (Fig. 2). The picture is still fuzzy indeed, also because the only isoform identified endogenously corresponds to the 153 amino acids protein (RefSeq NM_172370) we discuss in this review.
Fig. 2.
Gene structure and differential splicing of G72. The exon–intron structure of the G72 gene (top) and the mRNA products of differential splicing (bottom). Ten different splicing products are reported by ASPicDB [53], coding for six different protein isoforms. We chose to present only the mRNA coding for the three protein isoforms present in GenBank (accession nos. NP_758958.3, NP_001155284.1 and NP_001155286.1). Out of these, isoform 1 (NP_758958.3) is the one corresponding to the full-length 153 amino acids protein. Coding exons are in white, non-coding exons in grey. PTC: predicted termination codon
Notably, upregulated levels of G72 gene transcription and protein expression were detected in schizophrenic patients compared to healthy controls [14] as well as higher pLG72 plasma content in affected individuals [15]. Further western blot analyses and immunohistochemical studies revealed that pLG72 is present in human prefrontal cortex, in primary astrocytes [16], and in the membrane fraction (enriched in mitochondria) from human amygdala and caudate nucleus [12]. However, the presence of both G72 transcript and pLG72 protein has repeatedly been questioned [17].
In the near future accumulating expression data from tissue/primary cells [18] and individuals [19], as well as resources allowing analysis across tissues and individuals simultaneously, e.g., the Genome-Tissue Expression project, GTEx [20, 21] will assist researchers on this subject. Unfortunately, very few data on G72 gene expression is currently available on the GTEx portal (at http://www.gtexportal.org), due to the low number of samples processed.
pLG72 protein and its interaction with hDAAO
Recombinant expression
The untagged 18-kDa full-length pLG72 containing the R30K substitution was initially expressed in a soluble form in E. coli cells (and COS-7 cells) [1]. Unexpectedly, two products at 24 and 13.5 kDa were detected by western blot analysis (consistently observed by using different expression systems): the authors supposed they arose from a particular secondary structure and from proteolytic cleavage of the full-length protein, respectively.
A dramatic increase in pLG72 production (>95 mg/L of fermentation broth) was obtained by designing a synthetic gene to optimize codon usage for E. coli expression [22]. Here, the recombinant R30K pLG72 was recovered from the inclusion bodies by a refolding process using 0.5 % N-lauroylsarcosine (NLS, an anionic detergent) at alkaline pH and in the presence of a reducing agent, producing a soluble pLG72 preparation with a >90 % purity and the expected molecular mass. SDS-PAGE analysis of the refolded recombinant pLG72 shows two bands with a unique and identical N-terminal sequence (MLEKLMGADSL…), corresponding to the intact protein. Mass spectroscopy analysis showed that both bands correspond to the full-length sequence (18,079.4 Da), the heavier one to a complex with one molecule of NLS. Indeed, two tagged pLG72 variants carrying a 6xHis sequence at either the N- or C-terminal were also expressed [22].
In our opinion, the quality of the recombinant pLG72 produced by employing different procedures can explain the discrepancy observed in functional experiments (see below).
Biochemical properties
Presently no three-dimensional structure of pLG72 is available. Near- and far-UV CD spectra of the refolded pLG72 clearly demonstrate that it contains a high content of α-helices and acquired the tertiary structure [22]. Notably, the absorbance spectrum of pLG72 is a useful probe for protein conformation: an absorbance maximum at ≤267 nm indicates alterations in protein fold, negatively affecting its functional properties. The oligomerization state of pLG72 strongly depends on the added detergent concentration: in the absence of detergent, soluble pLG72 forms oligomers [1] while at an NLS concentration >0.025 % refolded pLG72 is monodispersed [22].
Soluble pLG72 binds β-d-galactopyranoside residues [1] and large, hydrophobic molecules such as FAD, FMN, and riboflavin (K d values of 25–65 μM), as well as chlorpromazine (CPZ, K d ~5 μM), an aliphatic phenothiazine widely used as an antipsychotic drug to treat schizophrenia [16, 22].
The hDAAO–pLG72 interaction
A nearly full-length clone of d-amino acid oxidase (DAAO) was isolated from a human brain cDNA library using a two-hybrid yeast system and G72 as bait [1]. The ability of pLG72 to specifically interact with human DAAO (hDAAO) was subsequently demonstrated using purified recombinant proteins [16] by: (1) far western blot analysis using both denaturing and native conditions; (2) quantitative pull-down experiments; (3) spectroscopic methods; and (4) size-exclusion chromatography. By the latter, a complex of ~200 kDa containing two pLG72 monomers and two hDAAO homodimers (2 × 18 kDa + 2 × 80 kDa) was produced. Surface plasmon resonance using the BIAcore technology yielded an apparent K d of 8.3 µM for the hDAAO–pLG72 complex. Notably, pLG72 interacts with both the holoenzyme, active hDAAO, and the apoprotein, inactive form of the enzyme: recognition is strictly specific for the mammalian DAAO [16].
Although there is common agreement that pLG72 and hDAAO interact, the effect of the complex formation on flavoenzyme functionality has long been debated, and two opposite effects were reported.
pLG72 acts as a DAAO activator. This assumption is based on the data reported by Chumakov et al. [1] who measured d-serine oxidation by porcine DAAO, which significantly differs from the human counterpart [23], in the presence of increasing concentrations of pLG72, showing activation at a 5:1 pLG72:DAAO molar ratio. However, these results were not reproduced employing similar experimental conditions and our pLG72 preparation [16].
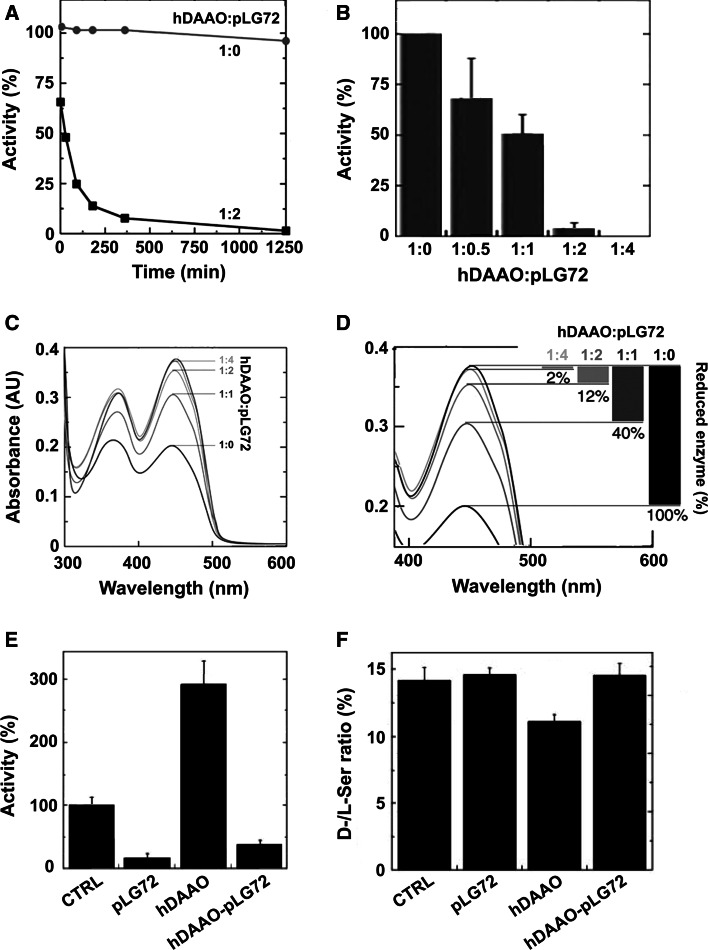
pLG72 negatively affects hDAAO activity. In detail, by mixing a fixed amount of hDAAO with increasing concentrations of pLG72 (at 0.06 % NLS) a time-dependent inactivation was apparent (Fig. 3a, b), an effect not due to proteolysis or protein precipitation [16]. Neither the apparent kinetic parameters of hDAAO on d-serine nor coenzyme binding were altered by adding pLG72 [16, 24]. Instead, pLG72 binding decreased the amount of active hDAAO holoenzyme, i.e., the form that can be reduced by the substrate (Fig. 3c, d). The regulatory role of pLG72 was confirmed at the cellular level in a human glioblastoma cell line (U87 cells): decreased d-serine levels were observed in U87 cells overexpressing hDAAO, while cotransfection with pLG72 decreased the hDAAO activity and restored d-serine levels (Fig. 3e, f) [16].
Fig. 3.
Effect of pLG72 binding on hDAAO activity and d-serine cellular concentration [16]. a Effect of pLG72 on the time course of hDAAO enzymatic activity. A slow inactivation is evident in the presence of a twofold molar excess of pLG72 (filled squares) in buffer containing 0.06 % NLS. b Effect of pLG72 on hDAAO activity following 30 min of incubation of a fixed amount of hDAAO (0.1 nmol/mL) with increasing amounts of pLG72. c, d Effect of pLG72 on the substrate-induced FAD reduction in hDAAO. Flavin reduction was monitored spectroscopically in anaerobic conditions by detecting the change in absorbance at 455 nm after the addition of 1 mM d-serine to hDAAO solutions (15 μM) containing free FAD and different amounts of pLG72 (hDAAO:pLG72 molar ratio 1:0, 1:1, 1:2, 1:4). d Spectral changes at 455 nm at higher magnification. The percentage of hDAAO flavin reduction (i.e., the amount of active hDAAO) at different concentrations of pLG72 is reported on the right. e DAAO activity was assayed in control U87 cells (CTRL, fixed as 100 %) and in the same transfected cells expressing hDAAO, pLG72 or both proteins. A significant increase in activity was evident in hDAAO transfected cells with respect to the control (p = 0.012) while a decrease was observed when pLG72 was overexpressed. f d-Serine concentration in control and transfected U87 cell extracts. d- and l-serine cellular content was determined by HPLC analysis. A significant decrease in the d-/l-serine ratio is apparent in hDAAO transfected cells with respect to controls (p = 0.004) while no difference is observed in pLG72 and hDAAO–pLG72 co-transfected cells
In contrast, the observation that pLG72 failed to modify basal enzymatic activity when human U251 glioma cells, endogenously expressing DAAO, were transfected with G72 and incubated in a buffer containing d-proline was used to suggest that pLG72 likely does not regulate DAAO activity [12]. However, this conclusion is biased by overlooking the fact that d-proline is also a substrate of human d-aspartate oxidase [25].
Additional studies using physiological levels of pLG72 (and hDAAO) protein will be required to conclusively determine whether pLG72 is a hDAAO-positive or -negative modulator.
Subcellular localization and turnover studies
Several lines of experimental evidence suggest that pLG72 is a mitochondrial protein. Its cellular distribution was investigated in several cell types transfected with G72: immunocytochemical studies in COS-7 cells transfected with a myc-tagged pLG72 expression construct showed distinct punctuate and tubular staining localized to mitochondria, a result confirmed by western blotting [12]. No alteration in the observed cellular localization was apparent using the R30K pLG72 variant. Mitochondrial targeting sequences are usually located at the N-terminal end of a protein: accordingly, pLG72 variants lacking the first 25 or 35 amino acids showed a diffuse cytoplasmic distribution of the corresponding signal, although partial localization to mitochondria was still observed (in ~40 % of the cells).
pLG72 mitochondrial localization was confirmed by confocal microscopy in U87 cells co-transfected with pLG72–ECFP or pLG72–FLAG constructs and EYFP–hDAAO or untagged hDAAO vectors: hDAAO was mainly targeted to peroxisomes, thus indicating that EYFP–hDAAO and pLG72–ECFP fusion proteins localized to distinct cellular compartments [26]. However, fluorescence signals of both proteins colocalized in the perinuclear region at 24 h from transfection, after which hDAAO distribution becomes targeted to peroxisomes. FRET analysis confirmed that the overexpressed pLG72 binds the newly synthesized hDAAO [26]. Notably, the time course of d-serine cellular concentration following hDAAO transient expression was different in U87 control vs. U87 pLG72–ECFP cells since the binding of pLG72 inactivates the newly synthesized hDAAO, thus preventing d-serine degradation [26].
According to this evidence and that from [12, 17], we suggest that pLG72 and hDAAO could (transiently) interact on the mitochondrial surface.
Cellular studies in U87-transfected cells indicated that pLG72 is a short-lived protein with an estimated half-life of ~30 min, while hDAAO is a long-lived protein (half-life of ~60 h). Moreover, pLG72 is largely degraded via the proteasome pathway, although the protein is not ubiquitinated [27], while hDAAO degradation is mainly mediated by the lysosome/endosome system, likely via autophagy processes. However, ubiquitin can label newly synthesized cytosolic hDAAO, which is then degraded through the ubiquitin–proteasome system. pLG72 speeds up hDAAO degradation by significantly decreasing its half-life (from ~60 to 6 h) [26, 27]. In this regard, we suggested that pLG72 might trigger degradation of the cytosolic form of hDAAO via the ubiquitin–proteasome system, thus contributing to the regulation of d-serine cellular levels [27].
Additional hypothesis about the physiological role of pLG72
In addition to its role in regulating DAAO activity, pLG72 might represent a primate-specific protein involved in modulating the morphology of the mitochondrial network, indirectly affecting key mitochondrial functions. Following prolonged expression of wild-type or K30R pLG72 in COS-7 cells, U251 human glioblastoma cells, or human astrocytes, mitochondria were vesicular and more fragmented at the expense of tubular and mixed mitochondrial shapes, while maintaining their functionality [12]. In primary rat hippocampal neurons transfected with pLG72, mitochondrial fragmentation was accompanied by an increase in dendritic complexity: the number of mitochondria/mm of the dendritic tree increased along the dendritic shafts, and the vesicular mitochondria were more mobile, possibly delivered more rapidly to sites of intense growth. Importantly, pLG72 overexpression seemed to promote dendritic branching only within the period of active dendritogenesis in synaptically immature neurons [12]. Furthermore, the mitochondrial methionine-R-sulfoxide reductase B2 (MRSB2, responsible for the elimination of cellular ROS) was identified as a binding partner of pLG72 [28]. pLG72 was thus suggested to impair MRSB2 function in oxidative stress defense—a link with ROS production was also recently suggested by Wang et al. [29]—although functional analyses were not provided.
Indeed, subcellular localization of pLG72 is of utmost importance since the presence of NMDAR was reported on the inner membrane of mitochondria [30]. These receptors were insensitive to glycine as coagonist (pointing to a role of d-serine as the physiological modulator) and might represent a further conduit for calcium entry in mitochondria. In a paper on rare genetic variants affecting schizophrenia [31], an effect on the membrane-associated guanylate kinase family of proteins was detected. These proteins are responsible among others for the correct assembly of NMDAR and other calcium channels and it is suggested that the following possible alteration of the calcium-dependent signaling network is connected to schizophrenia.
G72 transgenic mice
To address pLG72 in vivo function, the consequences of its expression were evaluated for behavior in “humanized” transgenic mice carrying the bacterial artificial chromosome (BAC) RP11-111A8 of the entire human G72/G30 locus [32] and, recently, a different BAC clone RP11-166E2 [33]. Two different mouse strains, CD1 and B6CBAF1/J, were used, thus establishing two transgenic lines, G72TgA and G72TgB, respectively. Mice of both lines appeared healthy and showed stable transgene transmission between generations and significant levels of transcripts corresponding to the reported human pLG72 splice variants. In G72TgA three further novel splice variants encoding for N-terminally truncated forms of pLG72 were detected. The highest G72/G30 transcripts levels were apparent in the brain, in particular in cerebellum and hippocampus (a significant amount was also detected in cortex) [32]. In the cerebral cortex of G72TgB mice, 19 splice variants were detected [33], suggesting that in the transgenic mice G72 has a more complex splicing profile than that reported in humans [13]. Moreover, differently from G72TgA animals, the highest levels of the G72 mRNA in G72TgB mice were observed in testis followed by brain, where G72 transcript was enriched in cerebral cortex, striatum, and hippocampus, while barely detectable in the cerebellum. The stability of the cortical G72 transcript content at different postnatal stages in G72TgB mice, not showing significant sex-related differences, suggested that G72 transcription is not developmentally regulated [33].
Consistently with G72 transcript distribution, low expression of pLG72 in the cerebellum, hippocampus, and cerebral cortex of G72TgA mice was apparent; pLG72 could also be detected in the dentate gyrus and olfactory bulb [32]. In contrast, no evidence was found for the presence of the protein in the mitochondrial fraction or total tissue lysates of G72TgB mice, probably because of the intrinsic instability, see above and [27].
In light of these findings, it is not clear whether pLG72 expression levels in the generated transgenic mice might reflect those in humans.
On the one hand, multiple BAC integration was observed in the G72TgA mouse model; therefore, pLG72 content is likely elevated, thus reproducing the increased levels reported in the brain of schizophrenia-affected individuals [14]. The mice expressing the human G72/G30 locus displayed several behavioral alterations relevant to psychiatric disorders, e.g., G72TgA mice were characterized by an abnormal motor coordination phenotype depending on cerebellum deficit [32]. Moreover, sensorimotor gating was affected since a significant deficit in pre-pulse inhibition (PPI) was evident in G72TgA mice that was normalized by administering the antipsychotic haloperidol, a D2 receptor antagonist. G72TgA mice were also more sensitive to treatments with drugs affecting glutamatergic neurotransmission (i.e., phencyclidine, PCP) and displayed increased compulsive behaviors [32]. On the other hand, the G72TgB mice exhibit different phenotypes: an altered locomotor activity and a higher base startle reactivity, and no genotype-dependent effect on PPI was reported [33].
A proteomics analysis of G72TgA mice showed that expression levels of several proteins involved in myelination-related processes (i.e., Mog, Plp, and Cld11), mitochondrial function, oxidative stress, cytoskeletal organization, and calcium transport were altered in the transgenic mice cerebellum [34]. The downregulation of such altered proteins components of the CNS myelin has been reported in schizophrenic patients [35–37] that also showed elevated cortical levels of sphingolipids and plasmalogens in the cortex: these results suggest that myelin is dysfunctional in schizophrenia [38].
Schizophrenia is highly comorbid with tobacco smoking, and nicotine is assumed to improve cognitive symptoms in individuals affected by the disease [39]. Notably, chronic administration of high doses of free-base nicotine (24 mg/kg a day for 24 days) completely restored working memory deficits in G72Tg mice, a process probably mediated via upregulation of α7nACh receptor density in the dentate gyrus [40].
According to the role of pLG72 in regulating mitochondrial functions [12], G72Tg mice showed reduced activity of aconitase and complex I of the respiratory chain, increased oxidative stress with elevated cerebellar peroxidation and expression of detoxifying glutathione transferases, and reduced levels of glutathione [41]. The increased oxidative stress likely contributes to some of the behavioral abnormalities reported in this mouse model, as they were rescued by the increase in glutathione levels induced by treatment with N-acetyl cysteine.
Genetic studies
Genome-wide association studies (or GWAS) and the search for variation in candidate genes, or association mapping, have been studied investigating association genotype–phenotype in humans and in plant and animal species for several decades already; however, none of the mapping techniques have come to terms with psychiatric disorders yet. This is partially due to the inherent complexity of psychiatric disorders.
Starting from the linkage between schizophrenia and the G72 locus found by Chumakov et al. [1], the association between this gene and the disorder was investigated by genotyping 16 SNPs on more than 500 persons from two pedigrees characterized by bipolar disorder [13]. Transmission/disequilibrium testing gave significant results in both pedigrees, finding a haplotype associated to affected individuals and encompassing the G72 region. However, no single SNP was significantly associated with the disorder in this analysis.
In 2006, the search was extended to normal controls by analyzing a large number of families to detect a relationship between cognitive measures (attention, working, and episodic memory) and a set of G72 SNPs [42]. Mixed-model ANOVA failed to identify a significant association between genotype and any cognitive variable. However, a significant genotype × diagnosis interaction regarding those cognitive variables assessing working memory was found for G72 SNP10 and a strong genotype effect at this locus was recorded for activation of the hippocampus by fMRI, functional magnetic resonance imaging, suggesting that mutations in G72 may be particularly penetrant “at the level of brain information processing implicated in cognitive impairments” [42].
A meta-analysis published in 2006 [43] concluded that the evidence of an association between several markers used and schizophrenia was highly significant, with a less significant effect for bipolar disorder. However, only the SNP rs1421292 (M24) was consistently found to be associated with the phenotype in all studies. At that time, the number of studies on the association between schizophrenia, bipolar disorder, and other psychiatric disorders and genetic factors was skyrocketing: the availability of SzGene database [44] spurred even more research. In an association mapping study on progression from prodromal syndromes to schizophrenia, nine common SNPs in the G72 gene were genotyped on 82 persons showing prodromal states [45]. The genotype at two SNPs, rs1341402 and rs778294 (M19), was significantly associated with progression using χ 2 test. Interestingly, the previous SNP did not show linkage disequilibrium with the character studied.
The use of GWAS by high-multiplexed SNPs arrays or deep sequencing produced another burst in genetic studies of psychiatric disorders so that in 2011 a two-stage meta-analysis based on total of about 9400 cases and 12,500 controls (8400 and 21,400 in the second stage) over 1,252,901 autosomal SNPs [46] identified seven significant SNP loci for schizophrenia, plus an additional three for the composite phenotype schizophrenia–bipolar disorder. None of the significant SNPs is, however, localized close to G72, actually not even on chromosome 13. Sullivan [4] listed 14 SNPs for schizophrenia (among them the seven previously mentioned), but still no connection to G72 or chromosome 13 (where G72 localizes) was present.
Now it seems that schizophrenia is influenced by genetic variation at hundreds of different loci [47].
In the effort to obtain more statistical power, the Psychiatric Genomics Consortium launched a second multi-stage meta-GWAS based on case–control and parent–offspring data, reinforced by results from deCODE genetics, with a grand total of about 37,000 cases and 113,000 controls. In this largest of the studies 108 genomic regions were found associated with the character, 75 % of which contain at least one protein-coding gene [48]. Eighty-three of these regions were newly found connected to schizophrenia, thus shaping a picture in which all chromosomes harbor many loci relevant for the character, with the notable exception again of chromosome 13. In this study, however, many genes involved in glutamatergic neurotransmission are present in the significant genomic regions (such as GRM3, GRIN2A, SRR, GRIA1). The main issue here is that if schizophrenia is influenced by genetic variation at hundreds of different loci, even the most powerful GWAS will unveil only those genes showing variability in the population studied among all the genes contributing to the character, because each population will display a different genetic structure for the loci involved and will cope with different environmental conditions. Besides, if the frequency of the causative allele at an important locus is low in a population, the locus will not be identified. Thus, even though GWAS are very important tools to shed light on the causative pathways of a multi-factorial pathology, they will not always be conclusive. Several studies have identified G72 as involved in schizophrenia, therefore the interest in the availability of pLG72 for functional analyses.
Genetic/MRI studies
Functional magnetic resonance imaging has been used by various groups to investigate the effect of G72 SNPs on brain functionality for many years. Initially, study of 61 subjects at high risk of schizophrenia showed that genetic variation in the G72 gene (SNP M23—rs3918342, and M24—rs1421292) modulates both hippocampal and frontal lobe functions [49]. Concerning the M23 genotype, the blood oxygen level-dependent (BOLD) signal in the imaging analysis exhibited a significant cluster of activation in the left hippocampus by contrasting sentence completion versus rest and increased activation in the right inferior frontal gyrus/sub-gyral region with increasing difficulty in completing sentences. Healthy subjects were also genotyped for the same SNPs and recorded speech was analyzed [50]. The fMRI group analysis of the contrast “semantic verbal fluency” > “reading aloud” revealed activations in the left inferior frontal gyrus, left middle temporal gyrus (MTG), and the cingulate gyrus and showed a linear effect of minor alleles in M24 in the right MTG and the right precuneus, i.e., regions which are prominent areas for speech and psychosis. Regression analysis showed a relationship between increase in BOLD response in verbal fluency and reading aloud and the number of minor alleles in M24: a linear increase in activation was found in the MTG, showing hyperactivations in patients with schizophrenia. These findings imply that brain activation is altered in the right MTG in all tested individuals with a G72 genotype that has been found to be associated with schizophrenia.
Most recently, the R30K pLG72 variant was reported to significantly impact on cortical thickness in schizophrenia but not in healthy controls: carriers with schizophrenia had a thinner cortex in an area covering the temporoparietal junction and lateral occipital regions in both hemispheres [2]. The temporoparietal junction has been indicated to be an anatomical node of the theory of mind (ToM) network [51, 52]: impairments in the ToM network can be regarded as a trait marker for schizophrenia; therefore, individuals showing G72 risk allele carriers might be predominantly vulnerable to developing ToM deficits.
Later on, four G72 polymorphisms previously associated with schizophrenia (rs3696165, rs3696167, rs2391191, and rs778294) were genotyped in 110 subjects with schizophrenia or schizoaffective disorder from whom two brain MRI scans on average 3 years apart were recorded [53]. The four polymorphisms captured three haplotypes, one of which was strongly associated with an increased rate of frontal lobe volume decrement, and with more severe psychotic symptoms at the time of the second scan. Schizophrenic individuals homozygous for haplotype 1 experienced an accelerated rate of frontal lobe tissue loss (4.1 %) compared to subjects with other G72 haplotypes and, at the time of the second scan, showed more severe psychotic symptoms. Previous meta-analyses by Detera-Wadleigh and McMahon [43] and Li and He [54] supported the findings of a detrimental effect of haplotype 1 on brain structure volumes in schizophrenia. The main limitations of these studies were that only individuals with schizophrenia were examined; thus, whether the identified associations are specific to schizophrenia or are more generalized was not investigated [53].
Very recently, the Kircher’s group for the first time reported an association between a G72 haplotype and fiber tract anomalies [55]. Forty-seven healthy subjects underwent diffusion tensor imaging, a technique useful to evaluate changes in white matter architecture: the fractional anisotropy (FA) index, which quantifies the directional level of the local tract structure, was evaluated. Higher FA values in right peninsular region and in the right inferior parietal lobe were found in subjects homozygous for the M23 risk T allele or for the M24 risk A allele; notably, both regions have been implicated in schizophrenia and bipolar disease. Increased FA values might indicate alterations in dendritic morphology, in good agreement with the observations from [12].
Caution is required in considering imaging genetics studies [56]. Actually, even the robust association reported between the V158M substitution in catechol-O-methyltransferase (affecting the enzymatic activity and altering the hippocampal gray matter volume) and many psychiatric traits is biased because numerous studies have low statistical power or misconsidered the gender contribution [57]. Indeed, brain volume may be altered by environmental factors, e.g., medications [56]. As stated by Medland et al. [58] “genetic analyses of brain images still require thousands of individuals to find credible genetic associations that replicate across cohorts”: the combination of a large sample size and appropriate statistical techniques might overcome the problem in identifying a true signal of small effect.
Conclusions
In the last few years, a new paradigm has emerged: as in other psychiatric disorders, schizophrenia is a complex trait [59]. Actually, when analyzing a quantitative trait we study the average value for the trait in the population; thus, we can expect that truly relevant alleles will be rare because the phenotype would be distant from the population mean. If the frequency of the causative allele at an important locus is low in a population, the locus will not be identified. Therefore, linking a single, albeit important gene, to a complex disease such as schizophrenia can be a bold assumption indeed. In particular, for schizophrenia no Mendelian forms have ever been found [47], with few exceptions (neurexin-1 and vasoactive intestinal peptide receptor-2) [59, 60, 61].
However, owing to the correlation with schizophrenia and other psychiatric disorders, the novel mammalian G72 gene and the encoded pLG72 protein have been the object of multidisciplinary investigations for the past 15 years. Genetic analyses did not reach a conclusion concerning association of G72 with schizophrenia susceptibility. However, fMRI findings on minor alleles in SNP M24 reported alteration in brain activation in the right MTG in all tested individuals with a G72 genotype associated with schizophrenia [50]. Furthermore, the SNP M15, resulting in Arg30Lys substitution, significantly impacted on cortical thickness in schizophrenia but not in healthy controls [2], pointing to M15 as a novel marker to determine memory performance in patients with schizophrenia.
The longest G72 transcript was detected at low levels in the amygdala, caudate nucleus, spinal cord, brain, and testis although the full-length pLG72 protein has never been detected in these tissues. Notably, upregulated levels of G72 gene transcription and protein expression were reported in schizophrenic patients compared to healthy controls [14, 15]. pLG72 is a small-size mitochondrial protein that we suggest could (transiently) interact with hDAAO on the mitochondrial surface: the binding negatively affects hDAAO activity and in turn its ability to degrade d-serine [5, 6], an NMDAR ligand required for the receptor activation [7–9]. pLG72 expression might also modulate the morphology of the mitochondrial network (its overexpression seemed to promote dendritic branching within the period of active dendritogenesis in synaptically immature neurons) [12], thus indirectly affecting key mitochondrial functions.
In order to elucidate the role of G72, two distinct transgenic mice carrying the entire human G72/G30 locus were generated: the two strains showed the highest transcripts levels in the testis and in different regions of the brain [32, 33]. In G72TgA transgenic mice, the pLG72 content is likely elevated, thus reproducing the increased levels reported in the brain of schizophrenia-affected individuals: actually, the G72TgA mice displayed several behavioral alterations relevant to psychiatric disorders, such as an abnormal motor coordination phenotype depending on cerebellum deficit [32].
We now know more details about this system, although a number of open issues hamper our efforts to define its physiological role. Among the still unanswered questions are:
Does NMDAR modulation represent the connection between pLG72 and d-serine metabolism?
Since NMDAR is critical for the development and modifiability of neuronal contacts, are G72 and DAAO expressed during both prenatal and postnatal development?
What targeting process brings pLG72 to mitochondria?
Notably, the studies so far performed on G72 did not identify a definite correlation with schizophrenia as well as with physio-pathological processes related to such a disease. Facing the need to address individual risk, which is not a task for genetics alone [62], availability of the protein might represent a feasible starting point. On this side, resolving the pLG72 three-dimensional structure will give more detailed knowledge of its structure–function relationships: by identifying the residues involved in the interaction between pLG72 and hDAAO we can begin to design molecules affecting cellular d-serine levels by acting on pLG72 [63, 64].
Acknowledgments
The experimental work about pLG72 was supported by a grant from Fondo di Ateneo per la Ricerca to SS and LP.
Abbreviations
- BAC
Bacterial artificial chromosome
- BOLD
Blood oxygen level-dependent
- CNS
Central nervous system
- CPZ
Chlorpromazine
- DAAO
d-Amino acid oxidase
- FA
Fractional anisotropy
- fMRI
Functional magnetic resonance imaging
- GWAS
Genome-wide association studies
- hDAAO
Human d-amino acid oxidase
- MRSB2
Mitochondrial methionine-R-sulfoxide reductase B2
- MTG
Middle temporal gyrus
- NLS
N-Lauroylsarcosine
- NMDAR
N-Methyl-d-aspartate type glutamate receptor
- ORF
Open-reading frames
- PCP
Phencyclidine
- PPI
Pre-pulse inhibition
- SNP
Single nucleotide polymorphism
- ToM
Theory of mind
References
- 1.Chumakov I, Blumenfeld M, Guerassimenko O, Cavarec L, Palicio M, Abderrahim H, et al. Genetic and physiological data implicating the new human gene G72 and the gene for d-amino acid oxidase in schizophrenia. Proc Natl Acad Sci USA. 2002;99:13675–13680. doi: 10.1073/pnas.182412499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schultz CC, Nenadic I, Koch K, Wagner G, Roebel M, Schachtzabel C, et al. Reduced cortical thickness is associated with the glutamatergic regulatory gene risk variant DAOA Arg30Lys in schizophrenia. Neuropsychopharmacology. 2011;36:1747–1753. doi: 10.1038/npp.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donohoe G, Morris DW, Robertson IH, McGhee KA, Murphy K, Kenny N, et al. DAOA ARG30LYS and verbal memory function in schizophrenia. Mol Psychiatry. 2007;12:795–796. doi: 10.1038/sj.mp.4002026. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan PF. Puzzling over schizophrenia: schizophrenia as a pathway disease. Nat Med. 2012;18:210–212. doi: 10.1038/nm.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sacchi S, Caldinelli L, Cappelletti P, Pollegioni L, Molla G. Structure–function relationships in human d-amino acid oxidase. Amino Acids. 2012;43:1833–1850. doi: 10.1007/s00726-012-1345-4. [DOI] [PubMed] [Google Scholar]
- 6.Pollegioni L, Sacchi S. Metabolism of the neuromodulator d-serine. Cell Mol Life Sci. 2010;67:2387–2404. doi: 10.1007/s00018-010-0307-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mothet JP, Parent AT, Wolosker H, Brady RO, Jr, Linden DJ, Ferris CD, et al. d-serine is an endogenous ligand for the glycine site of the N-methyl-d-aspartate receptor. Proc Natl Acad Sci USA. 2000;97:4926–4931. doi: 10.1073/pnas.97.9.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panatier A, Theodosis DT, Mothet JP, Touquet B, Pollegioni L, Poulain DA, et al. Glia-derived d-serine controls NMDA receptor activity and synaptic memory. Cell. 2006;125:775–784. doi: 10.1016/j.cell.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 9.Papouin T, Ladépêche L, Ruel J, Sacchi S, Labasque M, Hanini M, et al. Synaptic and extrasynaptic NMDA receptors are gated by different endogenous coagonists. Cell. 2012;150:633–646. doi: 10.1016/j.cell.2012.06.029. [DOI] [PubMed] [Google Scholar]
- 10.Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 11.Bear MF, Malenka RC. Synaptic plasticity: LTP and LTD. Curr Opin Neurobiol. 1994;4:389–399. doi: 10.1016/0959-4388(94)90101-5. [DOI] [PubMed] [Google Scholar]
- 12.Kvajo M, Dhilla A, Swor DE, Karayiorgou M, Gogos JA. Evidence implicating the candidate schizophrenia/bipolar disorder susceptibility gene G72 in mitochondrial function. Mol Psychiatry. 2008;13:685–696. doi: 10.1038/sj.mp.4002052. [DOI] [PubMed] [Google Scholar]
- 13.Hattori E, Liu C, Badner JA, Bonner TI, Christian SL, Maheshwari M, et al. Polymorphisms at the G72/G30 gene locus, on 13q33, are associated with bipolar disorder in two independent pedigree series. Am J Hum Genet. 2003;72:1131–1140. doi: 10.1086/374822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korostishevsky M, Kaganovich M, Cholostoy A, Ashkenazi M, Ratner Y, Dahary D, et al. Is the G72/G30 locus associated with schizophrenia? single nucleotide polymorphisms, haplotypes, and gene expression analysis. Biol Psychiatry. 2004;56:169–176. doi: 10.1016/j.biopsych.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Lin CH, Chang HT, Chen YJ, Lin CH, Huang CH, Tun R, et al. Distinctively higher plasma G72 protein levels in patients with schizophrenia than in healthy individuals. Mol Psychiatry. 2014;19:636–637. doi: 10.1038/mp.2013.80. [DOI] [PubMed] [Google Scholar]
- 16.Sacchi S, Bernasconi M, Martineau M, Mothet JP, Ruzzene M, Pilone MS, et al. pLG72 modulates intracellular d-serine levels through its interaction with d-amino acid oxidase: effect on schizophrenia susceptibility. J Biol Chem. 2008;283:22244–22256. doi: 10.1074/jbc.M709153200. [DOI] [PubMed] [Google Scholar]
- 17.Benzel I, Kew JN, Viknaraja R, Kelly F, de Belleroche J, Hirsch S, et al. Investigation of G72 (DAOA) expression in the human brain. BMC Psychiatry. 2008;8:94–107. doi: 10.1186/1471-244X-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.FANTOM Consortium and the RIKEN PMI and CLST (DGT) Forrest AR, Kawaji H, Rehli M, Baillie JK, de Hoon MJ, et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lappalainen T, Sammeth M, Friedländer MR, ’t Hoen PA, Monlong J, Rivas MA, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ardlie KG, Deluca DS, Segrè AV, Sullivan TJ, Young TR, Gelfand ET, et al. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molla G, Bernasconi M, Sacchi S, Pilone MS, Pollegioni L. Expression in Escherichia coli and in vitro refolding of the human protein pLG72. Protein Expr Purif. 2006;46:150–155. doi: 10.1016/j.pep.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Pollegioni L, Piubelli L, Sacchi S, Pilone MS, Molla G. Physiological functions of d-amino acid oxidases: from yeast to humans. Cell Mol Life Sci. 2007;64:1373–1394. doi: 10.1007/s00018-007-6558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caldinelli L, Molla G, Bracci L, Lelli B, Pileri S, Cappelletti P, et al. Effect of ligand binding on human d-amino acid oxidase: implications for the development of new drugs for schizophrenia treatment. Protein Sci. 2010;19:1500–1512. doi: 10.1002/pro.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katane M, Kawata T, Nakayama K, Saitoh Y, Kaneko Y, Matsuda S, et al. Characterization of the enzymatic and structural properties of human d-aspartate oxidase and comparison with those of the rat and mouse enzymes. Biol Pharm Bull. 2015;38:298–305. doi: 10.1248/bpb.b14-00690. [DOI] [PubMed] [Google Scholar]
- 26.Sacchi S, Cappelletti P, Giovannardi S, Pollegioni L. Evidence for the interaction of d-amino acid oxidase with pLG72 in a glial cell line. Mol Cell Neurosci. 2011;48:20–28. doi: 10.1016/j.mcn.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Cappelletti P, Campomenosi P, Pollegioni L, Sacchi S. The degradation (by distinct pathways) of human d-amino acid oxidase and its interacting partner pLG72—two key proteins in d-serine catabolism in the brain. FEBS J. 2014;281:708–723. doi: 10.1111/febs.12616. [DOI] [PubMed] [Google Scholar]
- 28.Otte DM, Raskó T, Wang M, Dreiseidler M, Drews E, Schrage H, Wojtalla A, Höhfeld J, Wanker E, Zimmer A. Identification of the mitochondrial MSRB2 as a binding partner of LG72. Cell Mol Neurobiol. 2014;34:1123–1130. doi: 10.1007/s10571-014-0087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang M, Chen H-J, Zhang J, Li W, Xie X, Chang H-T (2015) Identification of pLG72-induced oxidative stress using systemic approaches. BioMed Res Int (Article ID 429253) [DOI] [PMC free article] [PubMed]
- 30.Korde AS, Maragos WF. Identification of an N-methyl-d-aspartate receptor in isolated nervous system mitochondria. J Biol Chem. 2012;287:35192–35200. doi: 10.1074/jbc.M111.322032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otte DM, Bilkei-Gorzó A, Filiou MD, Turck CW, Yilmaz O, Holst MI, et al. Behavioral changes in G72/G30 transgenic mice. Eur Neuropsychopharmacol. 2009;19:339–348. doi: 10.1016/j.euroneuro.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 33.Cheng L, Hattori E, Nakajima A, Woehrle NS, Opal MD, Zhang C, et al. Expression of the G72/G30 gene in transgenic mice induces behavioral changes. Mol Psychiatry. 2014;19:175–183. doi: 10.1038/mp.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filiou MD, Teplytska L, Otte DM, Zimmer A, Turck CW. Myelination and oxidative stress alterations in the cerebellum of the G72/G30 transgenic schizophrenia mouse model. J Psychiatr Res. 2012;46:1359–1365. doi: 10.1016/j.jpsychires.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Tkachev D, Mimmack ML, Ryan MM, Wayland M, Freeman T, Jones PB, et al. Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet. 2003;362:798–805. doi: 10.1016/S0140-6736(03)14289-4. [DOI] [PubMed] [Google Scholar]
- 36.Martins-de-Souza D, Gattaz WF, Schmitt A, Maccarrone G, Hunyadi-Gulyás E, Eberlin MN, et al. Proteomic analysis of dorsolateral prefrontal cortex indicates the involvement of cytoskeleton, oligodendrocyte, energy metabolism and new potential markers in schizophrenia. J Psychiatr Res. 2009;43:978–986. doi: 10.1016/j.jpsychires.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 37.Martins-de-Souza D, Maccarrone G, Wobrock T, Zerr I, Gormanns P, Reckow S, et al. Proteome analysis of the thalamus and cerebrospinal fluid reveals glycolysis dysfunction and potential biomarkers candidates for schizophrenia. J Psychiatr Res. 2010;44:1176–1189. doi: 10.1016/j.jpsychires.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 38.Wood PL, Filiou MD, Otte DM, Zimmer A, Turck CW. Lipidomics reveals dysfunctional glycosynapses in schizophrenia and the G72/G30 transgenic mouse. Schizophr Res. 2014;159:365–369. doi: 10.1016/j.schres.2014.08.029. [DOI] [PubMed] [Google Scholar]
- 39.Harris JG, Kongs S, Allensworth D, Martin L, Tregellas J, Sullivan B, et al. Effects of nicotine on cognitive deficits in schizophrenia. Neuropsychopharmacology. 2004;29:1378–1385. doi: 10.1038/sj.npp.1300450. [DOI] [PubMed] [Google Scholar]
- 40.Hambsch B, Keyworth H, Lind J, Otte DM, Racz I, Kitchen I, et al. Chronic nicotine improves short-term memory selectively in a G72 mouse model of schizophrenia. Br J Pharmacol. 2014;171:1758–1771. doi: 10.1111/bph.12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otte DM, Sommersberg B, Kudin A, Guerrero C, Albayram O, Filiou MD, et al. N-acetyl cysteine treatment rescues cognitive deficits induced by mitochondrial dysfunction in G72/G30 transgenic mice. Neuropsychopharmacology. 2011;36:2233–2243. doi: 10.1038/npp.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goldberg TE, Straub RE, Callicott JH, Hariri A, Mattay VS, Bigelow L, et al. The G72/G30 gene complex and cognitive abnormalities in schizophrenia. Neuropsychopharmacology. 2006;31:2022–2032. doi: 10.1038/sj.npp.1301049. [DOI] [PubMed] [Google Scholar]
- 43.Detera-Wadleigh SD, McMahon FJ. G72/G30 in schizophrenia and bipolar disorder: review and meta-analysis. Biol Psychiatry. 2006;60:106–114. doi: 10.1016/j.biopsych.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 44.Allen NC, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 45.Mössner R, Schuhmacher A, Wagner M, Quednow BB, Frommann I, Kühn KU, et al. DAOA/G72 predicts the progression of prodromal syndromes to first episode psychosis. Eur Arch Psychiatry Clin Neurosci. 2010;260:209–225. doi: 10.1007/s00406-009-0044-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schizophrenia Psychiatric Genome-Wide Association Study Consortium Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schizophrenia working group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hall J, Whalley HC, Moorhead TW, Baig BJ, McIntosh AM, Job DE, et al. Genetic variation in the DAOA (G72) gene modulates hippocampal function in subjects at high risk of schizophrenia. Biol Psychiatry. 2008;64:428–433. doi: 10.1016/j.biopsych.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 50.Krug A, Markov V, Krach S, Jansen A, Zerres K, Eggermann T, et al. Genetic variation in G72 correlates with brain activation in the right middle temporal gyrus in a verbal fluency task in healthy individuals. Hum Brain Mapp. 2011;32:118–126. doi: 10.1002/hbm.21005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andreasen NC, Calarge CA, O’Leary DS. Theory of mind and schizophrenia: a positron emission tomography study of medication-free patients. Schizophr Bull. 2008;34:708–719. doi: 10.1093/schbul/sbn034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brüne M, Ozgürdal S, Ansorge N, von Reventlow HG, Peters S, Nicolas V, et al. An fMRI study of “theory of mind” in at-risk states of psychosis: comparison with manifest schizophrenia and healthy controls. Neuroimage. 2011;55:329–337. doi: 10.1016/j.neuroimage.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 53.Hartz SM, Ho BC, Andreasen NC, Librant A, Rudd D, Epping EA, et al. G72 influences longitudinal change in frontal lobe volume in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:640–647. doi: 10.1002/ajmg.b.31033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li D, He L. G72/G30 genes and schizophrenia: a systematic meta-analysis of association studies. Genetics. 2007;175:917–922. doi: 10.1534/genetics.106.061796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nickl-Jockschat T, Stöcker T, Krug A, Markov V, Maximov II, Huang R, et al. Genetic variation in the G72 gene is associated with increased frontotemporal fiber tract integrity. Eur Arch Psychiatry Clin Neurosci. 2015;265:291–301. doi: 10.1007/s00406-014-0516-6. [DOI] [PubMed] [Google Scholar]
- 56.Hashimoto R, Ohi K, Yamamori H, Yasuda Y, Fujimoto M, Umeda-Yano S, et al. Imaging genetics and psychiatric disorders. Curr Mol Med. 2015;15:168–175. doi: 10.2174/1566524015666150303104159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sumner JA, Powers A, Jovanovic T, Koenen K. Genetic influences on the neural and physiological bases of acute threat: a research domain criteria (RDoC) perspective. Am J Med Genet B Neuropsychiatr Part B. 2015;1718:44–64. doi: 10.1002/ajmg.b.32384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Medland SE, Jahanshad N, Neale BM, Thompson PM. Whole-genome analyses of whole-brain data: working within an expanded search space. Nat Neurosci. 2014;17:791–800. doi: 10.1038/nn.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rujescu D, Ingason A, Cichon S, Pietiläinen OP, Barnes MR, Toulopoulou T, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vacic V, McCarthy S, Malhotra D, Murray F, Chou HH, Peoples A, et al. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature. 2011;471:499–503. doi: 10.1038/nature09884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weiss KM. Tilting at Quixotic Trait Loci (QTL): an evolutionary perspective on genetic causation. Genetics. 2008;179:1741–1756. doi: 10.1534/genetics.108.094128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sacchi S, Rosini E, Pollegioni L, Molla G. d-amino acid oxidase inhibitors as a novel class of drugs for schizophrenia therapy. Curr Pharm Des. 2013;19:2499–2511. doi: 10.2174/1381612811319140002. [DOI] [PubMed] [Google Scholar]
- 64.Terry-Lorenzo RT, Masuda K, Sugao K, Fang QK, Orsini MA, Sacchi S, et al. High-throughput screening strategy identifies allosteric, covalent human d-amino acid oxidase inhibitor. J Biomol Screen. 2015;20:1218–1231. doi: 10.1177/1087057115600413. [DOI] [PubMed] [Google Scholar]