Abstract
Sirtuins are an evolutionary conserved family of NAD+-dependent protein lysine deacylases. Mammals have seven Sirtuin isoforms, Sirt1–7. They contribute to regulation of metabolism, stress responses, and aging processes, and are considered therapeutic targets for metabolic and aging-related diseases. While initial studies were focused on Sirt1 and 2, recent progress on the mitochondrial Sirtuins Sirt3, 4, and 5 has stimulated research and drug development for these isoforms. Here we review the roles of Sirtuins in regulating mitochondrial functions, with a focus on the mitochondrially located isoforms, and on their contributions to disease pathologies. We further summarize the compounds available for modulating the activity of these Sirtuins, again with a focus on mitochondrial isoforms, and we describe recent results important for the further improvement of compounds. This overview illustrates the potential of mitochondrial Sirtuins as drug targets and summarizes the status, progress, and challenges in developing small molecule compounds modulating their activity.
Keywords: Deacylase, Metabolic regulation, Inhibitor, Activator, Drug development, Sirt3, Sirt4, Sirt5
The sirtuin family of NAD+-dependent protein deacylases
Reversible (de)acetylation of protein Lys side-chains, initially observed in histones, is now known to affect all types of cellular proteins [1]. With more than 6800 protein acetylation sites in mammals, acetylation rivals phosphorylation in prevalence and appears to be the more ancient posttranslational modification, dominant in some processes such as regulation of mitochondrial metabolism [2–5]. Attachment and removal of acetyl groups are catalyzed by enzymes of the protein lysine acetyl transferases (KAT) and protein lysine deacetylases (KDAC) family, respectively. Of the four KDAC classes [6], Sirtuins form class III, which is unique in using NAD+ as a co-substrate (see below) [7]. This dependence on oxidized NAD+ (NADH acts as a weak inhibitor) renders Sirtuins metabolic sensors and leads to formation of the co-products (besides deacetylated protein) 2′-O-acetyl-ADP-ribose and nicotinamide [7, 8], which in turn acts as a product inhibitor. Furthermore, several of the seven mammalian Sirtuin isoforms, Sirt1–7, appear to preferentially remove protein lysine acylations different from acetylation. More and more acyl groups are found as protein lysine modifications, for example succinylation and crotonylation [9], and while Sirt1–3 prefer acetylated substrates, Sirt5’s dominant activity is desuccinylation. The Sirtuins thus are a family of protein deacylases. Sirtuin family members contribute to the regulation of metabolism and stress responses [10, 11] and to the beneficial health effects of calorie restriction (CR), including lifespan extension, in lower organisms and mammals [8, 12–14]. Sirtuins are thus emerging as potential targets for pharmacological treatment of metabolic disorders and aging-related diseases such as cancer and Parkinson’s disease [15–17].
We will now shortly review mitochondria related functions and disease relevance for the nuclear Sirtuin isoforms Sirt1, Sirt6, and Sirt7, and we will then comprehensively describe the current knowledge on function and disease contributions of the mitochondrial Sirtuins Sirt3, 4, and 5. We will finally describe the architecture and catalysis of Sirtuins and how the activity of mitochondrial Sirtuin isoforms can be modulated with small molecules.
Sirtuins in mitochondrial metabolism: Sirt1, Sirt6, and Sirt7
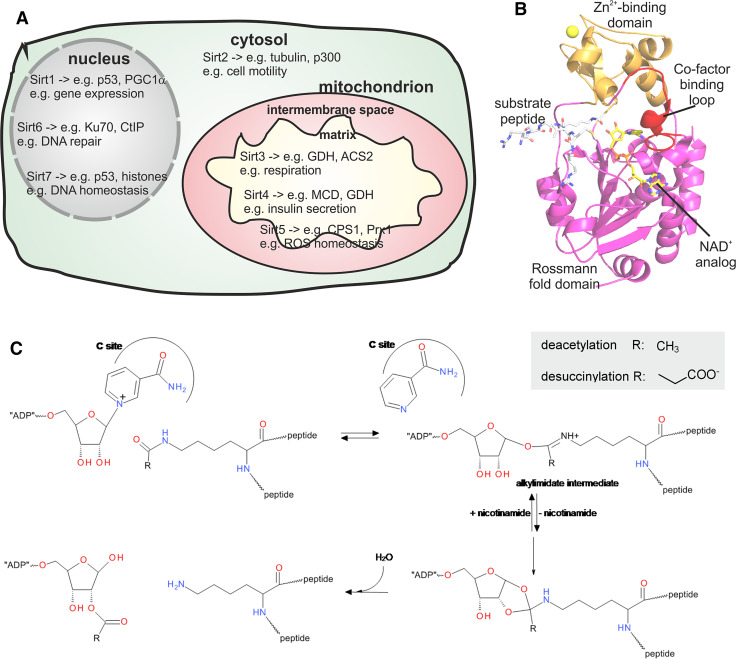
The mammalian Sirtuin isoforms Sirt1, 6, and 7 are located in the nucleus (Fig. 1a) and modify substrates involved in, e.g., chromatin stability and transcriptional regulation, although cytosolic functions have also been described, under specific circumstances, for Sirt1 and Sirt6 [16, 18–21]. Sirt2 is mainly cytosolic and was reported to deacetylate tubulin and p300 [22, 23], but it can accumulate in the nucleus during the cell cycle G2/M transition or in response, e.g., to ionizing radiation [24, 25]. The Sirtuins Sirt3, 4, and 5 are mainly located in mitochondria [18, 26] and directly target and modulate various mitochondrial proteins and processes (Fig. 1a; see below). However, functions in regulating mitochondrial metabolism have also been found for Sirt1, Sir6, and Sirt7 [27, 28].
Fig. 1.
Localization and functions of mammalian Sirtuin isoforms and architecture and mechanism of their catalytic domains. a Schematic view on the main subcellular localization of mammalian Sirtuin isoforms. Examples for major functions and substrates are indicated. b Cartoon presentation of the structure of human Sirt3 with bound AceCS2 substrate peptide and NAD+ analog (ligands as sticks; PDB ID 4FVT [232]). The large Rossmann fold domain, the small Zn2+ binding domain, and the cofactor binding loop are indicated; the Zn2+ ion is shown as a sphere. c Reaction mechanism of Sirtuin-catalyzed, NAD+-dependent deacylations
Sirt1 is involved in multiple pathways of DNA damage recognition and response [29], but it also has a central role in metabolic adaption through deacetylation of transcriptional regulators [27]. During fasting, Sirt1 expression and activity are increased. In the liver, Sirt1 deacetylates the transcription factor FOXO1 and the transcriptional coactivator PGC-1α [30], leading to upregulation of gluconeogenesis and repression of glycolysis [29]. In muscle, Sirt1-dependent PGC-1α deacetylation further activates peroxisome proliferator-activated receptor a (PPARα), which upregulates genes for fatty-acid catabolism [31]. Concomitantly, PGC-1α upregulates the genes for the mitochondrial respiratory chain and thus supports a shift from glucose to fatty-acid oxidation for generating ATP [29]. Sirt1 also induces, via PPARγ repression in white adipocytes, release of fatty acids from lipid storage [32]. Sirt1 thus coordinates, via regulation of gene expression, an adaption of several metabolic processes, including mitochondrial ones such as cellular respiration and citric acid cycle, to fatty acid oxidation instead of storage. Consistent with this metabolic shift, Sirt1 protects against negative effects of high fat diet [29].
Sirt1 also regulates mitochondrial metabolism globally by influencing biogenesis of the organelle through a complex and only partly characterized regulatory network involving the kinase AMPK, the acetyl transferase GCN5, and the transcription factors Foxo3, PGC-1α, NRF-1 and CREB [33]. For example, Sirt1 increases expression of respiratory chain components (see above) and, via NRF-1, expression of the mitochondrial transcription factor A (mtTFA) [33], which appear to be key triggers for increases in mitochondrial mass.
For Sirt6, like for Sirt1, functions in DNA homeostasis but also in metabolic adaption have been described [29]. Sirt6 protects against genomic instability by activating several DNA repair complexes and pathways [29], and Sirt6 loss leads to a progeroid syndrome-like phenotype [34]. Consistently, Sirt6 was shown to contribute to lifespan regulation in male mice [14]. The most dominant phenotype of Sirt6 knockout mice is, however, a severe hypoglycaemia [34] through increased glucose uptake and glycolysis without increased mitochondrial respiration. This metabolic change resembles the Warburg effect, which enables anoxic growth of cancer cells. It appears to be based on a Sirt6 function as co-repressor of the transcription factor Hif-1α, which also mediates Sirt3 effects (see below), and the Sirt6-dependent deacetylation of histone H3-Lys9 in promoters of glycolytic genes [35]. Under low nutrient conditions, Sirt6 seems to be inactivated, resulting in increased transcription of glycolytic genes.
For Sirt7, a regulatory role in mitochondrial homeostasis was reported via deacetylation of GABPβ1, which leads to activation of the GABPα/GABPβ complex as a key transcriptional regulator of nuclear-encoded mitochondrial genes [36].
Sirt1 and 6 are considered potential targets for various aging-related diseases [16, 27, 29]. Accumulating evidence supports in particular the idea that activation of nuclear Sirtuins can be a therapeutic approach for metabolic and neurodegenerative diseases [15, 27]. Sirt1 and its homologs contribute to beneficial health effects of caloric restriction [8, 13], and although it has long been controversially discussed, Sirt1 activation by small molecules is now confirmed and has yielded promising physiological effects (see [12, 37–40] and below). The most prominent Sirt1 activator resveratrol can, e.g., induce mitochondrial mass accumulation through SIRT1-dependent mechanisms [41], and more potent compounds have been described [12, 40]. For Sirt6, the identification of fatty acids as activators for its low basal deacetylation activity indicates that this isoform should also be amenable to small molecule activation (see [42] and below). The biology of the nuclear isoforms Sirt1 and 6 and their implications in disease pathologies are complex, however, and for a detailed discussion of these isoforms we have to refer the reader to some of the excellent reviews that cover these isoforms in more detail [16, 27, 29].
Sirtuins in mitochondrial metabolism: mitochondrial isoforms Sirt3, 4, and 5
Several hundred mitochondrial proteins are known to be acylated [1, 4, 43–45]. The absolute modification levels for most acetylation sites appear to be low [46], but many of these modifications change dynamically [43, 47] and a functional relevance has been shown for a fast increasing number of them (see below). Also, mitochondria appear to harbor many acylated proteins but fewer enzymes modulating this PTM than other cellular compartments [26]. No mitochondrial PATs are known, only the protein GCN5L1 was reported to support overall mitochondrial acetylation as a putative transferase subunit [48], and non-enzymatic mechanisms based on activated acyl metabolites appear to contribute to mitochondrial acylation or might even be the only mechanism [49–51]. In the latter case, acylation would be regulated globally through the level of acyl donors, such as acetyl-CoA, and possibly more specifically through the mitochondrial deacylases. The Sirtuin isoforms Sirt3, 4 and 5 are mitochondrial, while no members of the other HDAC/KDAC classes have been identified in this compartment. These Sirtuin isoforms appear responsible for regulation of a large variety of metabolic enzymes and stress response mechanisms [11, 26], and they might be suitable targets for a variety of metabolic and aging-associated diseases (Table 1).
Table 1.
Disease types with contributions from mitochondrial Sirtuins
| Isoform | Disease implications | Exemplary references |
|---|---|---|
| Sirt3 | Insulin signalling, type II diabetes | [73, 76] |
| Obesity, metabolic syndrome | [74] | |
| Aging-related hearing loss | [60, 82] | |
| Infertility | [84, 85] | |
| Liver damage induced, e.g., by alcohol abuse or drugs | [55, 101] | |
| Cardiovascular and heart diseases | [103, 104] | |
| Various cancer types | [117, 131] | |
| Sirt4 | Insulin signalling, type II diabetes | [140, 141] |
| Obesity, metabolic syndrome | [139, 143] | |
| Various cancer types | [147, 148] | |
| Sirt5 | Metabolic dysfunctions | [169] |
| Neurodegeneration | [162, 163] | |
| Infertility | [157] | |
| Lung cancer | [174] |
Sirt3
Most of the recent proteomic studies on protein acetylation have focused on Sirt3 [52–55], likely because of the prominent occurrence of protein Lys acetylation in this compartment and the fact that Sirt3 is the only mitochondrial Sirtuin with significant basal deacetylase activity. Sirt3 appears to deacetylate a wide variety of proteins, with diverse functions in mitochondrial physiology from energy metabolism to ROS detoxification and apoptosis regulation [3, 4, 54]. In an early proteomics study, Sirt3 was shown to be the major mitochondrial deacetylase with many targets in this compartment, but a Sirt3 k.o. revealed remarkably little consequences under basal conditions [56]. Only part of these substrates, and the effects of reversible acetylation on their function, have now been studied in detail, but it is evident that Sirt3 mediates concerted changes in metabolic pathways for adaption to nutrient levels and types, and that it can activate stress response mechanisms. It thus appears to be an attractive target for metabolic and degenerative diseases.
Sirt3 was first shown to deacetylate, and thereby activate mitochondrial acetyl-CoA synthetase 2 [57, 58]. Interestingly, Sirt1 can deacetylate the cytoplasmic isoform [57], consistent with a general Sirtuin function in mobilizing alternative nutrient sources during starvation. Along that line, Sirt3 was shown to deacetylate glutamate dehydrogenase (GDH) in vitro and in vivo [56, 59], and thereby to activate GDH [59], which allows anaplerosis of the tricarboxylic acid cycle (TCA) from protein degradation. Sirt3 was further shown to activate mitochondrial isocitrate dehydrogenase 2 (ICDH2) [59], a major TCA regulation point. Interestingly, ICDH2 and GDH are among those dehydrogenases that generate, instead of NADH, NADPH, which is required for reducing antioxidants, and the contribution of this mechanism to ROS detoxification, and its relevance in aging-associated hearing loss, has been shown [60].
Sirt3 promotes switching from glycolysis to oxidative phosphorylation [61]. Sirt3 deacetylates CypD, which decreases its activity and induces detachment of hexokinase II from mitochondria, which is necessary for oxidative phosphorylation [61]. Furthermore, Sirt3 deacetylates and activates the pyruvate dehydrogenase (PDH) component E1a and thereby regulates a key step in oxidative phosphorylation [62]. The Sirt3-regulated switch to oxidation is the opposite of the adaptation under anoxic conditions known in fast growing tumors as the Warburg effect, and Sirt3 downregulation indeed seems to mediate the Warburg effect in cancer cells (see below). Consistent with activation of oxidative metabolism, Sirt3 increases ATP production through upregulation of the electron transfer chain (ETC) [63], at least through activation of Complex I, II and V via deacetylation of specific subunits [e.g., complex V subunits α, β, and OSCP (oligomycin sensitivity conferral protein)] [63–67]. Sirt3 is upregulated in liver and brown adipose tissue during fasting, and it deacetylates and activates long-chain acyl coenzyme A dehydrogenase (LCAD) to upregulate fatty acid catabolism [68]. It also deacetylates HMG-CoA synthase 2 in mitochondria, which functions in formation of ketone bodies for the transport of acetyl groups for energy production to brain and muscle. Interestingly, Sirt1 appears to deacetylate the cytosolic isoforms of this Sirt3 substrate [69], similar to the ACS isoforms (see above). In addition, Sirt3 was found to deacetylate and activate ornithine transcarbamoylase, thereby stimulating the urea cycle allowing amino acid catabolism [70]. Sirt3 further shows a downregulating effect on mitochondrial protein synthesis via deacetylation of a subunit of the mitochondrial ribosome [71], and it affects protein folding through deacetylation of the chaperone Hsp10 [72]. Thus, Sirt3-regulated deacetylation affects proteins from pathways such as β-oxidation, amino acid catabolism, tricarboxy acid cycle, ketogenesis, respiration, protein synthesis and redox stress responses, and Sirt3 thereby can coordinate metabolic reprogramming by regulating various anabolic and catabolic pathways [4, 54].
Consistent with a function as key coordinator of mitochondrial energy metabolism, Sirt3 expression responds to nutrient intake [73, 74], is reduced in diabetes models [73], and is regulated by exercise in skeletal muscle [75]. Knocking down Sirt3 in myoblasts and animals decreased mitochondrial oxidation in skeletal muscle and impaired insulin signaling [73]. Consistently, Sirt3 was found to be important for proper mouse pancreatic beta cell function, and Sirt3 expression was significantly decreased in islets from human type 2 diabetes patients [76]. Sirt3(-/-) mice are also more susceptible to high fat diet (HFD)-induced obesity, hyperlipidemia, and steatohepatitis [74], consistent with disturbed metabolic adaptation when Sirt3 is lacking. Upregulation of Sirt3 through treatment with the natural compound berberine, in contrast, protected hepatic cells and muscle mitochondria against HFD-induced metabolic dysfunctions [77, 78]. Sirt3 expression is also upregulated in response to metformin and appears to contribute to the drug’s effect on glucose uptake [79]. These findings support a key function of Sirt3 in maintaining homeostasis of mitochondrial metabolism.
A major function of Sirt3 is the activation of stress response mechanisms. Mitochondrial energy metabolism produces the majority of cellular reactive oxygen species (ROS), and Sirt3 can activate several enzymes involved in containing cellular levels of the potentially cytotoxic ROS. Sirt3 can deacetylate and activate superoxide dismutase (SOD) 2 (also known as MnSOD) [80], and CR-dependent Sirt3 upregulation thereby reduces oxidative stress [81]. A decrease of SOD2 activation due to decreasing Sirt3 expression during aging was also suggested to contribute to ROS accumulation in the auditory cortex and thereby to aging-related auditory dysfunctions [82]. Sirt3 activates SOD2 through deacetylation at Lys122, which acts as a nutrient- and ionizing radiation-dependent switch, and the SOD2 activity inhibits superoxide accumulation [83]. Furthermore, Sirt3-dependent upregulation of NADPH, and thereby antioxidant levels—at least partially via ICDH2 and GDH activation [59]—was confirmed in vivo, and loss of this mechanism appears to contribute to ROS-mediated, aging-dependent retardations leading to hearing loss [60]. A decrease of Sirt3 expression and an increase of GDH acetylation was also observed in granulosa and cumulus cells from women with advanced maternal age [84], and an associated decrease in progesterone secretion suggests a Sirt3 role in folliculogenesis and luteinization and a mechanism contributing to aging-dependent decline in fertility [85]. CR also reduces oxidative damage and prevents aging-related hearing loss, and these effects are lost in Sirt3 k.o. mice [60]. Consistently, upregulation of Sirt3 activity using the NAD+ precursor nicotinamide riboside protected against noise-induced hearing loss [86], and Sirt3 also contributes to the protective effect of adjudin on cochlear hair cells treated with the ototoxic drug gentamicin [87]. Sirt3 further protects against hepatic lipotoxicity, apparently by preventing a lipid-induced raise of ROS levels, possibly through its regulatory effect on the ETC [88] and through activation of antioxidant systems. HFD reduces hepatic Sirt3 expression, downregulating this protective mechanism [88]. Sirt3-dependent upregulation of redox stress responses also appears to mediate protective effects in cortical neurons and hematopoietic stem cells, and reversal of the enzyme’s aging-associated downregulation improved regenerative capacity of hematopoietic stem cells [89, 90]. Sirt3 prevents oxidative stress-induced keratinocyte differentiation [91], and UVB-induced changes in Sirt3 expression in keratinocytes was suggested to cause the observed changes in ATP and peroxide levels [92]. Sirt3 thus appears to be a key regulator of redox homeostasis.
Sirt3 seems to mediate nutrient sensing and coordination of metabolism with redox processes, through direct regulation of the enzymes involved as well as through more indirect mechanism. Sirt3 expression is upregulated by fasting and by physical exercise [75, 93], which was suggested to further upregulate phosphorylation of AMPK and CREB and the expression of PGC-1α [75]. The other way round, PGC-1α was reported to stimulate Sirt3 expression in mouse muscle cells and hepatocytes, and Sirt3 was essential for PGC-1α-dependent induction of ROS-detoxifying and ETC enzymes, indicating Sirt3 and its targets as mediators in a ROS decreasing, PGC-1α-regulated pathway [94]. Sirt3 also interacts with the transcriptional activator FOXO3a and increases FOXO3a-dependent gene expression [95]. Glucose restriction, which increases Sirt3 expression (see above), also triggers FOXO3a accumulation in mitochondria in an AMPK-dependent manner and formation of a FOXO3a/Sirt3/mitochondrial RNA polymerase complex, which causes increased mitochondrial transcription and respiration [96]. Sirt3-dependent FOXO3 deacetylation and activation is also induced by peroxide and activates genes protecting against oxidative damage [97]. Furthermore, expression of genes affecting mitochondrial quantity and quality are thereby upregulated [97]. Sirt3 was further shown to modulate mitochondrial dynamics through deacetylation and activation of the optic atrophy 1 (OPA1), a mitochondrial fusion protein [98], and Sirt3-dependnet improvement of mitochondrial dynamics mediates protective effects during acute kidney injury [99].
In a proteomics study on liver mitochondria in a mouse model of chronic ethanol consumption, this regime induced hyperacetylation of various known Sirt3 substrates, and Sirt3 ablation led to the same result, indicating that impaired Sirt3-dependent deacetylation of targets in metabolic and antioxidant pathways contributes to alcoholic liver disease [55]. Along that line, ethanol was found to inactivate Sirt3 in cell culture, leading to aldehyde dehydrogenase (ADH) 2 hyperacetylation, which in turn led to activation of endothelial NO synthase (eNOS) [100]. Sirt3-dependent deacetylation of aldehyde dehydrogenase 2 also promoted acetaminophen-induced ADH2 inactivation and thereby liver injury induced by this drug [101]. In contrast, Sirt3 seems to protect against acute radiation-induced liver injury, apparently through lowering superoxide levels [102].
The functions of Sirt3 in energy metabolism and cell growth regulation (see below) makes it a candidate for contributions to heart diseases, and it indeed appears to have protective functions against heart failure. Sirt3 expression is regulated by pressure overload in the heart, and various mitochondrial Sirt3 targets have been implicated in heart function and pathology [103]. A proteomics study identifying hyperacetylated proteins in hypertensive heart failure models revealed several mitochondrial metabolic enzymes [104] including, e.g., the confirmed Sirt3 substrates LCAD and malate dehydrogenase (MDH) [3, 68]. Also, Sirt3 expression in murine cardiomyocytes is increased by cellular stressors and protects against cell death [105]. The effect was proposed to be mediated by Sirt3-dependent deacetylation of the DNA repair protein Ku70 [105]. However, the long Sirt3 isoform identified in nuclei and cytosol, which still carries the N-terminal mitochondrial localization sequence (MLS), was described as catalytically inactive [106]. In another report, full-length enzyme was reported to locate to the nucleus and to deacetylate H4-K16 in HEK cells, and to relocate to mitochondria upon cellular stress [107]. Also, nuclear full-length Sirt3 protein was reported to associate with chromatin and to be degraded under cellular stress, leading to de-repression of target genes [108]. More recently, Sirt3-dependent decrotonylation of histone protein was also shown to affect gene transcription [88]. However, other studies found an influence of Sirt5 expression on Sirt3’s localization [109], or no nuclear Sirt3 localization at all [110, 111]. It thus remains to be further clarified in which systems and under which conditions the reported Sirt3 effects outside mitochondria contribute to its physiological function and to its involvement in heart disease. As a mitochondrial regulator protein, Sirt3 appears to contribute to protection against pulmonary arterial hypertension [112] and to hypoxic and ischemic conditions [113, 114].
As for other Sirtuins, in particular Sirt1, the role of Sirt3 in cancer biology is complex [115] and its function as a tumor suppressor or as a tumor promoter might depend on the exact type of tumor. For Sirtuin targeted cancer therapy, further studies appear necessary and a very precise targeting might be required to avoid unwanted side-effects.
HDACs/KDACs in general are long recognized as potential anti-cancer targets, and several inhibitors for HDACs/KDACs from other classes than Sirtuins are approved for therapy or in clinical trials [116]. Expression analysis for 18 HDACs/KDACs, including Sirtuins, in normal B cells and 200 chronic lymphocytic leukemia (CLL) patients showed significant deregulation for several HDACs/KDACs, mainly upregulation [117]. Among the Sirtuins, Sirt2, 3, and 6 were correlated with overall survival, and Sirt3 downregulation was associated with poor prognosis and suitable as a predictor [117]. In general, Sirt3 appears to oppose the metabolic change typically associated with cancer cell growth, the high level of glycolysis allowing oxygen-independent biomass generation and known as Warburg effect [118]. Sirt3 activates PDH and destabilizes hypoxia-inducible factor-1α (HIF-1α), which controls expression of genes for glycolytic enzymes [118, 119]. Consistently, the loss of Sirt3 observed in breast cancer cells increased ROS production and thereby HIF-1α stability. Sirt3 overexpression in these cells indeed repressed glycolysis and proliferation [118]. Consistent with this report, Sirt3 downregulation in MEFs and several cancer cell lines increased proliferation, and a Sirt3 knockdown increased tumorigenesis in mouse xenografts, which could be abolished by feeding an anti-oxidant [120]. Sirt3 overexpression in turn inhibited HIF-1α stabilization and decreased tumorigenesis [120], suggesting that Sirt3 acts as a tumor suppressor at least partly by suppressing ROS formation and HIF-1a stabilization [120]. Sirt3(-/-) MEFs indeed showed abnormal mitochondrial physiology and an increase in stress-induced ROS formation, accompanied by increased genomic instability [121]. Sirt3(-/-) mice developed ER/PR-positive mammary tumors, and tissues from human breast and other cancer types showed reduced Sirt3 levels [121]. Clinical analyses indeed suggest that lowered Sirt3 expression can serve as a biomarker for aggressive tumors and increased relapse probability after treatment for breast and gastric cancers [122–124]. Sirt3 also appears to function as a tumor suppressor in lung adenocarcinoma [125]. Sirt3 is downregulated in lung adenocarcinoma tissue compared to healthy tissue, and Sirt3 overexpression inhibited growth of the A549 lung adenocarcinoma cell line through upregulation of the mitochondrial apoptosis pathway [125], consistent with Sirt3’s reported pro-apoptotic function [126] but contradicting another study reporting an anti-apoptotic Sirt3 function [127]. The exact role of Sirt3 to apoptosis regulation might in fact depend on other regulators involved and remains to be established in more detail. Sirt3 overexpression in the A549 lung adenocarcinoma cells upregulated the stress response proteins p53 and p21 and decreased intracellular ROS levels, which likely contributed to its growth inhibiting effect. Sirt3 was also found to be downregulated in ~67 % of 248 tissue samples from hepatocellular carcinoma (HCC) cases, which correlated with several clinical parameters [128]. Low Sirt3 expression indeed correlated well with unfavorable overall and recurrence-free survival, respectively, and thus appears to be a marker for unfavorable prognosis of HCC disease progression [128]. Sirt3 overexpression inhibited hepatocellular carcinoma cell growth in vitro, partly due to increased apoptosis induction and upregulation of p53 through Mdm2 (homolog of murine double minute 2) downregulation [129]. Hepatic Sirt3-dependent SOD2 activation also inhibited superoxide accumulation and oncogene-dependent immortalization in cell culture [83]. In pancreatic cancers Sirt3 has also tumor suppressor function, apparently via regulation of iron homeostasis [130].
Despite this evidence for a tumor suppressor function for Sirt3, the enzyme appears to act as a tumor promoter in other tumor types [115]. Increased Sirt3 expression in colon cancers appears to correlate with lower survival rate, and Sirt3 downregulation inhibited proliferation, invasion, and migration of cultured colon cell lines [131]. Sirt3 is overexpressed in oral squamous cell carcinoma (OSCC) cells compared to normal human oral keratinocytes [132], and a clinical analysis suggests that Sirt3 expression can serve as a prognostic biomarker for oesophageal squamous cell carcinoma [133]. Down-regulation of Sirt3 inhibited OSCC cell growth and proliferation, increased the sensitivity of these cells to treatment with radiation and cis-platin in vitro, and reduced tumor burden in vivo [132]. The authors concluded that Sirt3 acts as a promoter of cell proliferation and survival in oral cancer carcinogenesis and thus constitutes a potential target for treatment of oral cancer. Another study also observed Sirt3 overexpression in OSCC cell lines, but at the same time its activity appeared to be reduced through a point mutation close to the active site [134]. It thus remains to be seen whether increased activity is required for Sirt3’s contribution to growth of this tumor type. Sirt3 also contributes to resistance of breast cancer cells to tamoxifen [135], a widely used therapeutic anti-estrogen. Sirt3 was quickly upregulated in MCF-7 cells when continuously treated with tamoxifen, and vector-mediated Sirt3 overexpression likewise decreased tamoxifen sensitivity and blocked tamoxifen-induced apoptosis [135]. Consistently, silencing Sirt3 expression in MTR-3 cells increased mitochondrial estrogen receptor β, ROS levels, and apoptosis, and Sirt3 silencing in tamoxifen resistant cells sensitized them again for tamoxifen-induced apoptosis [135].
In summary, Sirt3 appears to be a key coordinator in mitochondrial metabolism and redox regulation. It orchestrates the activities of central metabolic pathways, through posttranslational modification of their key enzymes, for adaptation to metabolic changes in response to factors such as exercise, nutrients, and cellular stress. Sirt3 mainly mediates, e.g., circadian clock regulation of acetylation patterns on mitochondrial enzymes [47] and remodeling of these patterns during CR in mice [54]. Besides oxidative catabolism, which generates ROS, Sirt3 also regulates enzymes responsible for ROS quenching, and the enzyme thereby appears to protect against oxidative stress-dependent pathologies. Increasing Sirt3 activity thus appears promising for ameliorating metabolic syndrome and other redox stress-related diseases [136]. Sirt3 activation by exogenous NAD+ or the NAD+ precursor nicotinamide riboside indeed led to Sirt3-dependent anti-hypertrophic effects [137] and Sirt1 and 3-mediated protection against HFD induced metabolic changes [138]. Since no potent and Sirt3-specific activators are yet available (see below), stimulating Sirt3 expression, e.g., through Sirt1 activation [33], is currently the only available approach. In some cancer types, however, Sirt3 inhibition rather than activation appears desirable, and first potent Sirt3 inhibitors are now available. Due to its prominent effect on energy metabolism, Sirt3 can also affect the activity of nuclear isoforms through modulation of the NAD+/NADH ratio (see below), and these Sirtuins thus appear to form a complex regulatory network, which will have to be studied further for a complete understanding and for proper intervention with drugs.
Sirt4
Despite a recent report on a first deacetylation substrate for Sirt4 [139], the low basal in vitro deacetylation activity of the enzyme suggests that major aspects of its function—such as other acyl substrates or activating ligands or modifications—are not yet understood. However, several physiological functions regulated by Sirt4 have already been identified.
In contrast to Sirt3-dependent GDH activation, Sirt4 inhibits this key metabolic enzyme in the pancreas and thereby downregulates insulin secretion in response to amino acids [140]. Sirt4 also interacts with insulin-degrading enzyme and alleviates insulin secretion in response to glucose [141], indicating that it acts as a general nutrient sensor controlling insulin release. Furthermore, Sirt4 can induce mitochondrial permeability transition pore (PTP) opening [142], a key step in mitochondria-mediated apoptosis, and it interacts with Adenine nucleotide translocator (ANT) proteins [141], known contributors to PTP opening. Inhibition of Sirt4, along with Sirt3, was shown to mediate the protective effect of nicotinamide on high glucose/palmitate-induced cell death in INS-1 beta cells [143], indicating that it regulates mitochondria-mediated apoptosis in the pancreas and suggesting that Sirt4 inhibition might be helpful for the treatment of metabolic disorders. This conclusion is supported by the finding that Sirt4 deacetylates and inhibits malonyl-CoA decarboxylase (MCD), which shifts fatty acid metabolism toward accumulation [139].
Sirt4 appears to oppose the effects of CR in the pancreas [140], suggesting that Sirt4 inhibitors might be attractive compounds to elicit CR-like effects, different to the other isoforms that would need to be activated. These initial reports suggested Sirt4 to regulate its targets through NAD+-dependent ADP-ribosylation, an activity observed for most Sirtuins but likely representing a side reaction of the main deacylase activity without physiological relevance [144]. More recently, deacetylase activity has been described for Sirt4 [3, 139], albeit with low catalytic efficiency. In light of the recently discovered Sirt5 (see below) and Sirt6 (see above) deacylase activities, however, it is tempting to speculate that Sirt4’s major physiological deacylase activity also targets Lys acylations other than acetylation. Sirt4 was in fact reported to inhibit pyruvate dehydrogenase (PDH) through deliponylation, but despite an increase in k cat/K M compared to deacetylation this modification is still an inefficient substrate and better ones might exist [145]. Other possibilities are that basal Sirt4 activity is increased in vivo similar to the potential activation of Sirt6 by free fatty acids, or that proper Sirt4 substrate sequences have not yet been identified. In a microarray approach testing in parallel the deacetylation of ~6500 peptides representing physiological acetylation sites, new candidates for Sirt4 deacetylation substrates were identified, such as NAD(P) transhydrogenase (Nnt), acetyl-CoA acetyltransferase, and the heat shock protein Stress-70 [3]. It remains to be seen whether these proteins are in vivo substrates efficiently deacetylated by Sirt4.
In liver and muscle tissue, Sirt4 inhibition increases mitochondrial gene expression and in particular fatty acid oxidation, suggesting a therapeutic application of Sirt4 inhibitors for metabolic diseases such as type 2 diabetes [146]. Malonyl CoA decarboxylase (MCD) was recently identified as a Sirt4 target mediating this function of Sirt4 as a key regulator of lipid homeostasis. Sirt4 deacetylates and inhibits MCD, which converts a major building block of lipogenesis and inhibitor of lipid oxidation [139], in skeletal muscle and white adipose tissue. Thereby, Sirt4 activity promotes lipid anabolism, and suppressing Sirt4 activity increases exercise tolerance and protects against diet-induced obesity, reinforcing the idea to use Sirt4 inhibitors for the treatment of metabolic disorders.
On the other hand, Sirt4 appears to have tumor suppressor activity. Sirt4 expression is induced by various genotoxic agents and Sirt4-dependent inhibition of mitochondrial glutamine metabolism is required for proper execution of cellular DNA damage response programs [147]. Consistently, loss of Sirt4 leads to increased proliferation and sensitivity to genotoxic stresses, and Sirt4 knockout mice spontaneously develop lung tumors [147]. Sirt4 expression is reported to be reduced in human cancers such as colon and gastric cancer, an effect that can be induced through activation of the mammalian target of rapamycin complex 1 (mTORC1), and Sirt4 expression in xenografts reduced cell proliferation and tumor development in nude mice [148]. These results suggest a potential for Sirt4 activating compounds in cancer therapy and indicate that application of Sirt4 inhibitors for metabolic diseases (see above) has to be analyzed carefully to avoid harmful side-effects.
Sirt4 is also highly expressed in brain early during development, specifically in the astrocytes of glia, and Sirt4-dependent inhibition of GDH, a key regulator of gliogenesis development harboring an activating mutation in congenital hyperinsulinism/hyperammonemia, has been implicated in regulating the development of glia cells [149].
In contrast to Sirt3, our understanding of functions and targets of Sirt4 is still very limited. Sirt4 appears to contribute to regulation of the metabolic functions of mitochondria and might serve as a therapeutic target for diseases such as diabetes, but more studies on this isoform are currently required.
Sirt5
The seven mammalian Sirtuin isoforms can be assigned to slightly different Sirtuin classes based on sequence similarities [150]. The mitochondrial Sirt5 belongs to class III, and class III Sirtuins are the ones most widely distributed in prokaryotes, suggesting Sirt5 might be an ancestor Sirtuin and/or differ in some features from other mammalian isoforms. Sirt5 indeed has small differences from Sirt1, 2, and 3 in its active site, rendering it primarily a desuccinylase rather than a deacetylase [151], a difference that can also cause a unique response to small molecule modulators (see below) [152].
Sirt5 was initially reported to localize to the mitochondrial matrix [18]. Subsequent studies indicated that Sirt5 also resides in the mitochondrial intermembrane space (IMS) [59] and possibly also outside mitochondria in the cytoplasm [44, 153]. In the mitochondrial matrix of liver cells, Sirt5 deacetylates and thereby activates carbamoyl phosphate synthetase 1 (CPS1), the enzyme catalyzing the committed step of ammonia detoxification [154, 155]. Sirt5-dependent CPS1 activation is increased when amino acid metabolism is upregulated during fasting and under CR [154], and Sirt5 levels are elevated through CR [156], indicating that Sirt5 might contribute to CR effects. A decline in Sirt5 expression in granulosa and cumulus cells at advanced maternal age was also suggested to cause the associated increase in follicular-fluid ammonia via CPS1 regulation, and thereby to influence oocyte quality [157]. Sirt5 regulates ammonia metabolism in additional non-liver cells, via desuccinylation of glutaminase, and it thereby controls ammonia-induced autophagy and mitophagy [158]. In mice, Sirt5 also appears to deacetylate urate oxidase, an enzyme involved in ammonia detoxification in this organism but without a functional analog in humans [159]. Sirt5 can further deacetylate cytochrome c [59] and Lys197 of peroxiredoxin 1 (Prx1) [3, 152], which regulates the activity of this redox scavenger protein that appears to reside in cytosol and IMS [160], but the relevance of these proteins and the functions they mediate, apoptosis, respiration, and redox regulation, as physiological Sirt5 targets remain to be confirmed. Nevertheless, Sirt5 activation appears to be a viable approach for treatment of certain metabolic dysfunctions and for eliciting beneficial effects known to be induced by CR. Sirt5 was reported to be able to provide neuroprotection [161], and decreased Sirt5 expression correlates with molecular brain aging and was suggested to contribute to aging-related mitochondrial diseases, such as Parkinson’s [162]. On the other hand, Sirt5 levels increase during Morbus Alzheimer progression, apparently due to its appearance in microglia cells, but its disease contribution remains to be analyzed [163]. Interestingly, Sirt5 expression can also be reduced through chronic alcohol consumption [164], suggesting its repression as a potential molecular mechanism for alcohol effects.
More recently, Sirt5 was found to display much higher deacylation activity against malonyl, succinyl, and glutaryl modifications of protein Lys residues [44, 151, 165, 166]. These Lys modifications occur in mammalian cells and bacteria [44, 45, 167], as well as further modifications such as propionylation and crotonylation [9]. It is tempting to speculate that further acylations might exist, especially with groups that are available as activated metabolites such as CoA thioesters [51, 168]. Succinylation, malonylation, and glutarylation in CPS1 appear to overlap with acetylation sites but are removed much more efficiently [45, 151, 152], indicating these deacylation activities as the major physiological function for Sirt5 and probably also as the switch regulating CPS1 activity. Characterization of a Sirt5(-/-) mouse model confirmed global protein hypersuccinylation and elevated ammonia levels in serum during fasting [169]. Regular diet yielded no metabolic abnormalities, indicating that Sirt5 is dispensable for metabolic homeostasis under basal conditions, but Sirt5 expression and mitochondrial succinylation appear to be under the control of the metabolic sensors and regulators AMPK and PGC-1a [170]. Recent proteomics studies on protein succinylation yielded novel leads for Sirt5 targets and functions. One study identified 2565 succinylation sites on 779 proteins with functions in various mitochondrial metabolic pathways, such as fatty acid metabolism and respiration, but also in cytosolic and nuclear proteins, indicating potential Sirt5 targets outside mitochondria [44]. And while this succinylome showed only moderate overlap between acetylation and succinylation [44], two subsequent succinylome studies identified more than 1100 and 2100 succinylation sites, respectively, many of them also serving as acetylation sited [45, 171]. Many of the succinylations identified by these proteomics studies are substrate sites for Sirt5 [44, 45]. Such MS based studies suffer from the general technical problem that trypsin is by far the best protease for sample preparation, but it does not cleave at acylated Lys residues, and using other proteases indeed enables identification of additional acylation sites [172], suggesting that even more Sirt5-regulated succinylation sites should be identified in the future. In another approach, testing ~6500 peptides representing already known physiological acylation sites identified additional Sirt5 substrate candidates, such as ATP synthase, a voltage-dependent anion channel (VDAC), and Stress-70, suggesting Sirt5 functions in energy metabolism but also in other functions such as stress response and apoptosis [3]. These breakthroughs in Sirt5 research, and the progress in analytical methods for studying deacylation activities [173], have largely improved our understanding of Sirt5 function and promise exciting new insights in targets, function, and regulation of this long neglected isoform.
One of the identified succinylome sites is SOD1-Lys123, which is close to the active site and conserved in vertebrate SOD1 (also known as Cu/ZnSOD), and it appears to be a main target of SOD1 succinylation [174]. SOD1 is a SOD isoform localized in cytoplasm and mitochondrial intermembrane space (IMS), and Sirt5 is the only deacylase known to partially localize to the IMS where it was speculated to regulate its in vitro substrate cytochrome c [59]. Sirt5 can bind and desuccinylate SOD1 at Lys123, thereby increasing SOD1 activity and thus reducing cellular ROS levels. Succinylation of SOD1 appeared to support growth of a lung cancer cell line [174], which might indicate that Sirt5-dependent desuccinylation of this target promotes cancer growth.
Taken together, the function of Sirt3 as the major mitochondrial deacetylase and as a coordinator of mitochondrial functions in response to various signals, such as nutrient availability and exercise, is well established. Also studied in less detail, Sirt4 and Sirt5 seem to contribute to similar functions, indicated, e.g., through common target pathways such as the urea cycle for Sirt5 (CPS1) and Sirt3 (ornithine transcarbamoylase). In light of the Sirt5 preference for succinylated substrates, it is tempting to speculate that the three mitochondrial Sirtuins regulate these pathways through three different deacylation activities, since basal Sirt4 deacetylation activity is also low. Since the respective Lys acylation levels depend on the corresponding acyl-CoA levels, Sirt3, 4, and 5 would thereby contribute differently to the regulation of these common target pathways. For studying these and other aspects of the functions of mitochondrial Sirtuins in physiology and diseases (Table 1), small molecules for specifically modulating their activity are urgently required. We will now shortly review Sirtuin architecture and catalysis and how it is regulated by physiological ligands, and we will then describe the current status of Sirtuin-targeted drug development with a focus on the mitochondrial isoforms.
Sirtuin structure and catalysis
Sirtuins consist of an evolutionary conserved catalytic core of ~275 amino acids. In the different Sirtuin enzymes, it is flanked by N- and C-terminal extensions that vary in length and sequence and that contribute to cellular localization and activity regulation [106, 175, 176]. In contrast to the large N- and C-terminal extensions of Sirt1, which appear to contribute to Sirt1 stability and activity regulation [177, 178], the mitochondrial Sirtuins Sirt3, 4, 5 have only short extensions. They feature N-terminal MLS of varying length, 28 aa in Sirt4, 36 residues in Sirt5, and ~100 aa in Sirt3. In Sirt3, proteolytic processing of the N-terminus upon mitochondrial import was reported to activate the enzyme [22], but this autoinhibitory function of the Sirt3 N-terminus has been challenged [107]. For Sirt5, N-terminal processing has also been observed and occurs after residue 36 [154]. A short sequence at the Sirt5 C-terminus was reported also to contribute to mitochondrial localization by acting as a mitochondrial membrane insertion signal [153], but its physiological function remains to be worked out. Furthermore, localization of a Sirt5 fraction to the mitochondrial IMS has been reported [59], and also localization in the nucleus [44]. For Sirt4, removal of the 28 N-terminal residues has been reported [140].
The Sirtuin active site is located in a cleft between two subdomains of the catalytic core (Fig. 1b), a Rossmann-fold subdomain typical for NAD+ binding proteins and a smaller Zn2+-binding domain. The zinc ion has a structural function and is not involved in catalysis, in contrast to the other KDAC classes, which catalyze hydrolysis of the Lys-ε-N-acylamide through Zn2+-dependent activation of a water molecule. The unique Sirtuin mechanism is instead based on conversion of the NAD+ co-substrate (Fig. 1c). In a first step, nicotinamide is released from the 1′-ribose carbon atom, which establishes a covalent bond to the acyl oxygen resulting in a 1′-O-alkylamidate intermediate. The intermediate then rearranges into a bicyclic 1′-2′-acetal intermediate, which is finally hydrolyzed into deacetylated polypeptide and 2′-O-acetyl-ADP-ribose [10]. This mechanism only involves the substrate carbamide moiety and thus is assumed to equally apply to all deacylations catalyzed by Sirtuins, independent of the modification’s acyl chain (Fig. 1c). Crystal structures of several Sirtuins, including human Sirt1, 2, 3, 5, and 6, and of Sirtuin complexes with different ligands including acetylated and succinylated substrates, co-substrate and intermediate mimics, and a native alkylamide intermediate, helped to establish and characterize this unusual catalytic mechanism [10, 179–183]. The relative orientation of small and Rossmann-fold domain changes upon binding of the acyl-Lys containing substrate to a groove between these domains [182, 184], and a flexible so-called “cofactor binding loop” (Fig. 1b) adopts different conformations during the catalytic steps [181–183]. It gets ordered upon substrate binding and transforms in a closed conformation during acyl transfer (see, e.g., [185, 186]), possibly contributing to ejection of the co-product of the first reaction step, nicotinamide. These rearrangements demonstrate the complexity of Sirtuin catalysis and the conformational dynamics of these enzymes, which makes characterization of inhibition mechanisms more challenging (see [182, 187] and below). Sirtuins recognize a wide variety of substrate sequences but show moderate, isoform-specific sequence preferences due to differences in details of their substrate binding grooves [3]. These differences, together with different conformational dynamics, also cause the isoforms-specific acyl selectivities (see above) and inhibitor sensitivities (see below) [151, 152, 165, 188].
Physiological regulation of Sirtuin activity
Protein acylation and Sirtuins regulate mitochondrial homeostasis and function through mechanisms outside as well as inside this organelle, together with other emerging mitochondrial signalling systems such as redox signalling [189]. Thus, physiological and pharmacological regulators targeting Sirt1 or Sirt6 as well as those targeting the mitochondrial isoforms Sirt3, 4, and 5, can modulate mitochondria-mediated functions. We will shortly review physiological Sirtuin regulators, since their analysis can provide insights in isoform-specific features helpful for drug development, and we will then summarize pharmacological approaches for Sirtuin modulation, with a focus on mitochondrial isoforms.
So far, regulatory proteins have only been described for Sirt1 and Sirt2. AROS (Active Regulator of Sirt1) was reported to enhance Sirt1-dependent p53 deacetylation [190], but AROS alone is unable to activate Sirt1 in vitro [177] and AROS expression was found not to correlate with Sirt1 activity [191], indicating that it belongs to a more complex regulatory network. Dbc1 (Deleted in Breast Cancer 1), in contrast, binds and inhibits Sirt1 [192].
A natural small molecule activator of Sirt1 [39, 193], the plant stilbene resveratrol (Fig. 2), is assumed to exploit a binding site and mechanism relevant for a yet to be identified physiological Sirt1 modulator. For Sirt6, free fatty acids have recently been reported as physiological metabolites with a regulatory effect. Sirt6 has a large, hydrophobic acyl binding pocket and indeed shows much stronger deacylation activity against Lys modified by long-chain acyl groups. Free fatty acids appear also able to occupy this pocket, since they can inhibit Sirt6 competitively with myristoyl substrate. Interestingly, fatty acid binding in presence of acetylated substrate increased the low basal in vitro deacetylation activity of the enzyme [42], possibly explaining the observation that in vivo, Sirt6 displays substantial deacetylation activity [194]. It remains to be seen whether specific acyl substrates and/or physiological modulators exist for other isoforms. Insights in acyl specificity have been important for progress in understanding Sirt6 and Sirt5 function, and although a first deacetylation substrate has been reported for Sirt4, it will be essential to find out whether the enzyme is an activatable deacetylase, similar to Sirt6, or has a unique acyl specificity like Sirt5.
Fig. 2.
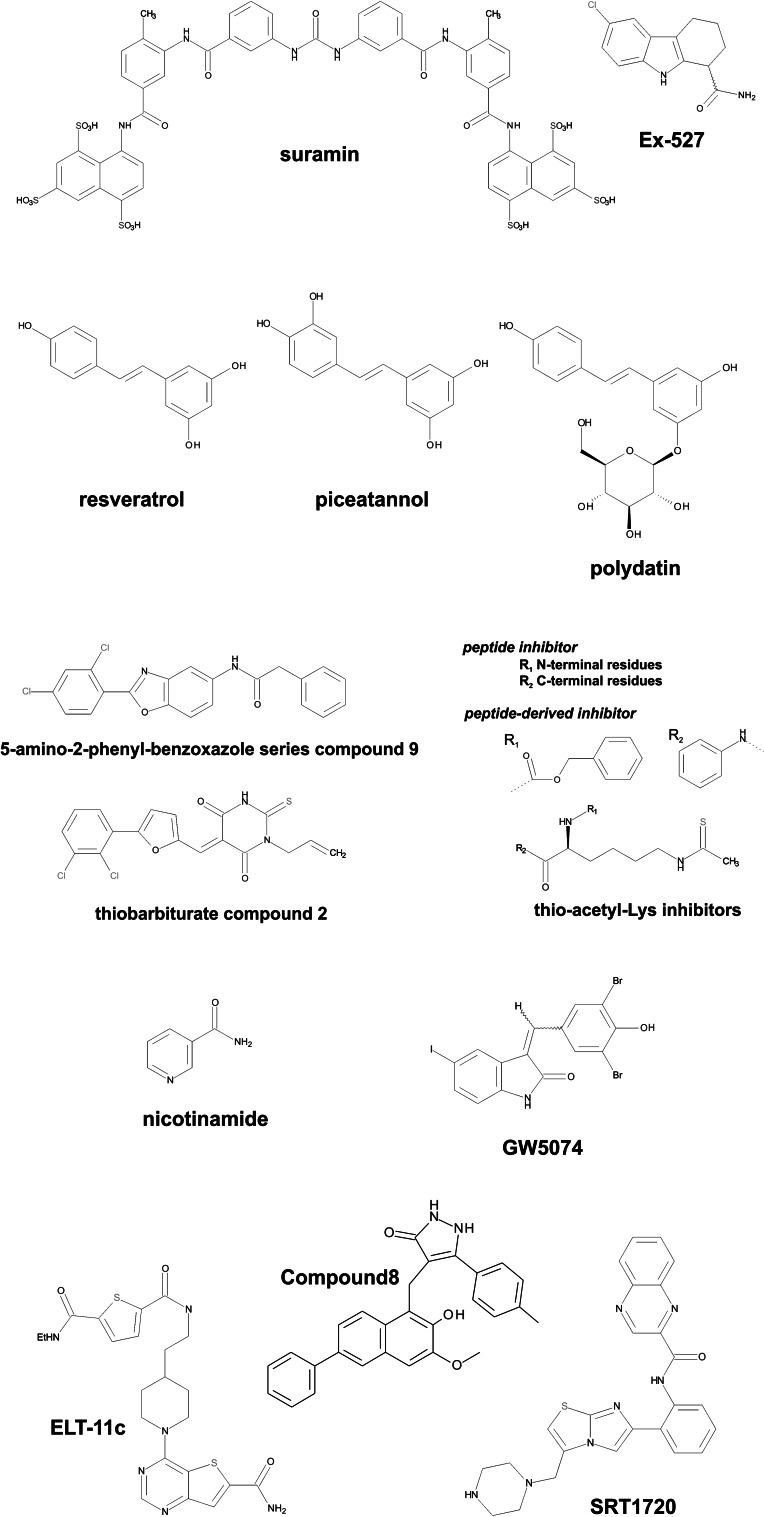
Chemical structures of compounds with significant effects on the activities of mitochondrial Sirtuins
Sirtuin activity requires NAD+ as an essential co-substrate. NAD+ is quickly reduced to NADH, which acts as a weak Sirtuin inhibitor [195] under normal physiological conditions. The NAD+/NADH ratio can increase in a tissue-specific manner, e.g., during fasting or exercise [196], leading to increased Sirtuin activity. NAD+ levels, and thus Sirtuin activity, can also be influenced via NAD+ de novo biosynthesis and by salvage pathways reusing NAD+ degradation products [28]. Another metabolite with an established function in regulating Sirtuins is nicotinamide, released from NAD+ as the first product of the Sirtuin catalyzed reaction during formation of the alkylimidate intermediate. Rebinding of nicotinamide to the Sirtuin/intermediate complex leads to reversal of the initial reaction step, reforming the substrate NAD+ and thus inhibiting protein deacetylation [197]. This un-competitive mechanism is assumed to inhibit all Sirtuins, with K i values of about 0.05–0.2 mM [197]. Sirtuins are thus sensitive to variations in physiologic nicotinamide concentrations, which are assumed to reach up to 0.1 mM. A nicotinamide role as endogenous Sirtuin regulator is supported by in vivo studies in various organisms [197, 198]. Interestingly, for Sirt5 a differential inhibition of its deacetylase and desuccinylase activities by nicotinamide was found, apparently caused by the Sirt5-specific residues Arg105/Tyr102, which also mediate the enzyme’s preference for distal carboxylate groups (see [152] and below). It remains to be seen whether this substrate acyl specific effect has a physiological relevance for the regulation of Sirt5 functions.
Pharmacological Sirtuin modulation
Due to their physiological functions and pathophysiological roles, Sirtuins are considered attractive drug targets. Isoform specific small molecule modulators would also be excellent tools for in vivo investigations on Sirtuin function and regulation. Therefore, huge efforts have aimed at developing potent and specific small molecule modulators for mammalian Sirtuins [199, 200]. Although a large number of inhibitors, and to a smaller extend also activators, are now available, the majority suffers from weak potency and/or low specificity. Missing or limited mechanistic information—a Sirt5/suramin complex (Figs. 2, 3a; see below) was the only Sirtuin/drug complex structure for many years, and kinetic characterizations are available only for few compounds—have hampered their improvement. More recently, substrate and assay improvements have increased the available data on compound action and specificity [173], and additional Sirtuin/drug complex structures revealed the intricate interaction of compounds with the complex Sirtuin catalytic cycle (see [179, 187, 201–203] and below). We will summarize available compounds, shortly for Sirt1 and Sirt6 and comprehensively for mitochondrial Sirt3, 4, and 5, and we will summarize recent progress in understanding Sirtuin modulator mechanisms and how it supports further development of Sirtuin-targeting drugs.
Fig. 3.
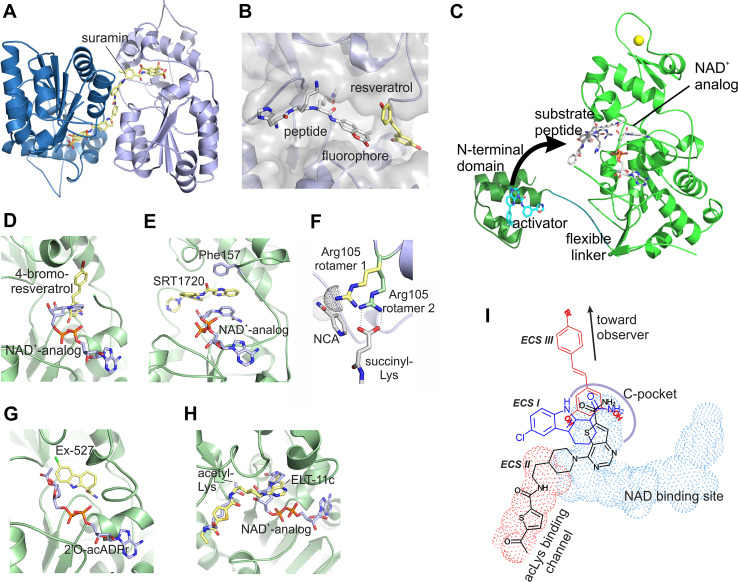
Structural analysis of Sirt3 and Sirt5 modulation by small molecule drugs. a Crystal structure of Sirt5 in complex with suramin (PDB ID 2NYR; [222]), which inhibits Sirtuins non-specifically by blocking the active site and artificially crosslinks two monomers. b Crystal structure of Sirt5 in complex with resveratrol and fluorophore-modified peptide (PDB ID 4HDA; [201]) showing the direct contact between substrate and activator. c Crystal structure of Sirt1 with substrate peptide and activator (grey sticks) bound to the active site of the catalytic core (green), and with an activator (cyan sticks) bound to the Sirt1-specific N-terminal domain (dark green). The N-terminal domain is speculated to swing, via the flexible linker, toward the active site (arrow) and to cover it lid-like with the exposed surface of the activator, resulting in a direct activator/substrate contact. d Crystal structure of Sirt3 in complex with 4-bromo-resveratrol (PDB ID 4C7B [203]). The NAD+-analog carba-NAD+ was modeled through an overlay with a Sirt3/ACS2-peptide/carba-NAD+ structure (PDB ID 4FVT; [239]) to illustrate that the inhibitor blocks the binding pocket for the NAD+ nicotinamide group. e Crystal structure of Sirt3 in complex with SRT1720 (PDB ID 4BN5; [187]) and the NAD+-analog carba-NAD+ showing the extensive interaction between these two uncompetitive ligands. f Model for the molecular basis of Sirt5’s differential nicotinamide sensitivity. The structure of Sirt5 in complex with succinylated peptide and NAD+ (PDB ID 3RIY; [151]) shows an interaction between Arg105 (green, rotamer 2) and the substrate succinylate. In absence of this interaction, for example when an acetylated substrate is bound, a preferred conformation of Arg105 (yellow, rotamer 1) would clash with nicotinamide (NCA; modelled into its known binding pocket). g Crystal structure of Sirt3 in complex with Ex-527 and the co-product 2′-O-acetyl-ADP-ribose (PDB ID 4BVH; [179]), which increases inhibitor affinity apparently by stabilizing a closed active site conformation. h Crystal structure of Sirt3 in complex with ELT-11c (PDB ID 4JSR; [202]). Acetyl-Lys and the NAD+-analog carba-NAD+ were modeled in through an overlay with the complex structure Sirt3/ACS2-peptide/carba-NAD+ (PDB ID 4FVT; [239]) to illustrate that the inhibitor occupies the acetyl-Lys channel und the binding region for the NAD+ nicotinamide riboside. i Scheme for the binding modes of Sirtuin inhibitors exploiting the C site
Initial modulator development efforts were mostly focused on Sirt1 and Sirt2, and most of the first compounds showed limited potency and isoform discrimination. Cambinol, e.g., inhibits Sirt1 and 2 equally and with limited potency (IC50 values 50–500 μM). Despite many development efforts, only few Sirt1 compounds are now available that feature improved properties. The most promising and best studied Sirtuin inhibitor is Ex-527 (Fig. 2), a potent Sirt1 inhibitor used in many physiological studies. Ex-527 inhibits Sirt1 with an IC50 of 0.1 µM, about two orders of magnitude more potently than Sirt2 (IC50 = 19.6 µM) and Sirt3 (IC50 = 22.4 µM), and it does not affect Sirt5 [179, 204]. Moreover, it is Sirtuin-specific [204] as it exploits their unique NAD+-dependent deacetylation mechanism (see [179] and below). Even though Ex527 is most potent against Sirt1, its binding site and inhibition mechanism have been studied with several Sirtuin isoforms, including Sirt3, and provide mechanistic information helpful for development efforts for all Sirtuins. We therefore describe details for Ex527 action below, where we review effects on mitochondrial Sirtuins. Likewise, the so-called ELT (extended library technology) inhibitors, which are the most potent Sirtuin inhibitors reported but equally affect Sirt1, 2, and mitochondrial Sirt3 [202], will be described below. For Sirt6, no potent and specific compounds are available, only weak effects of non-specific modulators such as quercetin have been described [205]. This situation might change now, however. Sirt4, 6, and 7 have been neglected so far due to the lack of suitable deacylation substrates, but using the identified Sirt6 substrates and activators should allow identification and development of better Sirt6 inhibitors. Also, the inhibition of Sirt6-dependent long-chain deacylation and activation of deacetylation by physiological fatty acids [42] shows the potential of the Sirt6 acyl binding site for the development of Sirt6-selective inhibitors and activators.
Sirtuins are special targets because they can be stimulated with small molecules. Sirtuin activating compounds—first compounds were resveratrol-related polyphenols (Fig. 2), but other compound classes were described subsequently [12, 193]—can extend lifespan in various species and have beneficial effects in mammals such as promotion of cell survival and protection against insulin resistance [8]. Since the Sirt1 activators resveratrol (and related compounds) and SRT1720 also affect the activities of mitochondrial Sirtuins, they will be reviewed below. New and more potent pharmacological Sirt1 activators have been developed but appear to use similar mechanisms and are thus not treated in more detail here [12, 40].
For the majority of Sirtuin inhibitors, no comprehensive specificity, potency and mechanistic data comparable to those for Ex-527 are available. Initially, compounds were mostly tested on Sirt1 and Sirt2 [199, 200], but Sirt3 and Sirt5 are now more and more often included in selectivity tests due to improvements of substrates and assays [173]. While Sirt3 shows a tendency to respond to compounds comparable to Sirt1 and Sirt2, Sirt5 revealed a surprising insensitivity to most common Sirtuin inhibitors (see [206] and below). We will now summarize Sirtuin regulators with effects on mitochondrial Sirtuins and describe mechanistic insights on their action from kinetics, affinity, and structural data. No compounds are yet available that target mitochondrial Sirtuins with both, high potency and high specificity, but the insights from the available modulators will now support the more efficient development of such drugs for subsequent use for in vivo studies and therapy.
Resveratrol-related Sirtuin activators and inhibitors, other Sirtuin activators
The stilbene resveratrol (Fig. 2), a plant metabolite naturally occurring, e.g., in grapes, was the first Sirtuin activator to be described [193, 207]. The compound was reported to activate yeast Sir2 and human Sirt1 and thereby to induce lifespan extension and beneficial health effects similar to caloric restriction [12, 208, 209]. Resveratrol also affects Sirt3 and Sirt5 activity in vitro with potencies comparable to Sirt1 [201], and these isoforms thus might contribute to physiological resveratrol effects. Initially, direct Sirtuin activation was discussed controversially, since resveratrol affects a broad variety of cellular targets, which likely contributes to its in vivo effects, and it can indeed also influence Sirtuins through indirect mechanisms such as regulation of their expression levels [182, 210, 211]. Moreover, resveratrol-dependent Sirt1 activation in vitro appeared to require the fluorophore of the artificial Fluor-de-Lys (FdL) peptide substrate used in initial studies [212, 213]. Consistently, we found that resveratrol and its metabolite piceatannol (Fig. 2) directly contact the FdL fluorophore in complex structures with Sirt3 and Sirt5 (Fig. 3b), respectively, thereby influencing the binding mode of the substrate peptide [201]. Nevertheless, regular polypeptide substrates would also be compatible with this compound binding mode depending on their amino acid sequence. Indeed, modulation of Sirt1, 3, and 5 activity against fluorophore-free peptide and protein substrates could be shown [39, 40, 201, 214]. Most strikingly, peptide experiments on ~6500 physiological acetylation motifs revealed an influence of the substrate sequence on the compound effect as expected from the observed direct contact between substrate and drug [39]. Resveratrol significantly activated Sirt1-dependent deacetylation of about 150–200 peptides, but it also inhibited deacetylation of a similar number of peptides, while deacetylation of the majority of peptides was not affected. Interestingly, resveratrol-dependent modulation of Sirt3 and 5 requires only the enzyme’s catalytic domain, independent of the substrate used, whereas activation of Sirt1 can be achieved with the catalytic core in the FdL assay [177] but requires the Sirt1-specific N-terminal domain with regular substrates [40, 215]. Crystal structures of Sirt1/activator complexes (Fig. 3c) indeed showed that the N-terminal domain provides a binding patch for a large set of “Sirt1 activating compounds” (STACs) [215] that are not chemically related to resveratrol but appear to trigger the same activation mechanism [40]. The binding site appears incomplete and exposed, however, since half of the compound surface remains uncovered and engages in non-physiological crystal contacts. On the other hand, crystal structure analysis of Sirt1 in complex with FdL substrate and resveratrol showed the direct substrate/activator interaction previously seen with Sirt3 and Sirt5 [201, 216] and simultaneously an interaction with the N-terminal domain. It is tempting to speculate that Sirt1 activation normally proceeds via such a sandwich-like interaction with compound between N-terminal domain and the active site-bound substrate (Fig. 3c). In case of the FdL substrate, the interaction with the substrate fluorophore appears sufficient, however, explaining the effects on Sirtuin catalytic cores in the FdL assay. Also for some regular substrates this interaction, possibly supported by some weaker interactions with active site loops, might be sufficient and thus explain the effects observed with Sirt5, which lacks a Sirt1-like N-terminal domain. However, further studies will be required to test, refine or modify such a unifying Sirtuin activation mechanism.
For Sirt3, exclusively resveratrol-dependent inhibition has been reported so far, whereas Sirt1 and Sirt5 can either be activated or inhibited depending on the peptide or protein substrate used [201, 39]. Moreover, resveratrol effects can differ depending on the Sirt5 deacylation activity tested, similar to the observation that inhibitor potency against Sirt5 can strongly be influenced by the substrate acyl modification (see [152, 206] and below). For the same peptide substrate sequence, Sirt5-dependent deacylation was either activated (deacetylation) or inhibited (desuccinylation) by resveratrol [201]. Therefore, studying resveratrol-dependent effects on Sirtuins in vivo appears challenging—substrate-dependent effects on several isoforms have to be considered. On the other hand, these findings show that Sirtuin isoforms and even certain Sirtuin/substrate pairs can potentially be targeted selectively, indicating an opportunity for the development of small molecule drugs with very specific effects [182].
Resveratrol itself appears not suitable for developing selective Sirtuin modulators. Its bioavailability is limited by its moderate solubility, its sensitivity to oxidation, and its metabolic conversion. The low potency of resveratrol (EC50 Sirt1 0.05–0.1 mM [12, 193]; EC50 Sirt5 0.07–0.21 mM [201]), in combination with its low specificity, further questions its suitability for eliciting defined in vivo effects. Therefore, resveratrol derivatives have been investigated as Sirtuin modulators. Piceatannol, a resveratrol metabolite with an additional hydroxyl group, and the glucose-modified analog polydatin (Fig. 2), both show improved solubility and are still capable of affecting Sirt1, 3, and 5 [193, 201, 203]. Furthermore, we found that 4′-bromo-resveratrol (Fig. 2), a derivative with a bromine replacing a resveratrol hydroxyl group, inhibits Sirt1 and 3 with strongly increased potency (K i = 8 µM for Sirt3), whereas Sirt5 is only weakly affected [203]. Through structural studies on Sirt3, supported by activity and binding assays, we identified the nicotinamide-accommodating C-site and a neighboring site as the inhibitor binding region (Fig. 3d). The additional binding site region accommodates the bromophenyl moiety and shows significant variation between Sirtuin isoforms [203], making it a promising target side for the development of isoforms-selective compounds. Interestingly, a second 4′-bromo-resveratrol molecule could weakly bind to the outer surface of Sirt3, and we speculated that it might indicate an allosteric binding site contributing to Sirtuin activation [40, 203]. SDX-437, a compound more distantly related to resveratrol, was recently described as a potent Sirt3-over-Sirt1-specific inhibitor (IC50 0.7 μM) that resulted from a high-throughput screen conducted with a new substrate peptide and mass spectrometry-based assay [217]. It will be interesting to see how this compound inhibits Sirt3 and whether it affects other Sirtuin isoforms. Likewise, several approved drugs were found to inhibit Sirt5 through a screen using a microchip electrophoresis assay system, with the most potent effect for the polyphenol anthralin, but selectivity and mechanistic information remain to be reported [218]. Mitochondria-targeted resveratrol derivatives have also been developed, through coupling to the membrane-permeable cation triphenylphosphonium [219]. These mitochondriotropic analogs benefit from increased solubility and stability and appear to display cytotoxic effects specifically on fast growing cells [219], but it remains to be seen whether these compounds act via modulation of the mitochondrial Sirtuin isoforms.
More recently, a chemically diverse set of compounds has been described to activate Sirt1 via binding to its unique N-terminal domain (see above) [40, 215]. Also, 1,4-dihydropyridine (DHP)-derived compounds were described as Sirt1 activators, and some of them also increased Sirt2 and Sirt3 activity to slightly lower extent in the FdL assay [220]. DHP-dependent activation might proceed via a resveratrol-like mechanism (see above), but no mechanistic information is available yet, and DHP-dependent activation of Sirt2 and Sirt3 remain to be confirmed with substrates other than FdL. More recently, the lignan honokiol was reported to activate Sirt3-dependent deacetylation of a protein substrate [221], but a more detailed characterization of this effect remains to be awaited.
Substrate mimics and other peptide-competitive inhibitors
Many Sirtuin inhibitors are based on the approach to target either the polypeptide substrate binding cleft or the NAD+ binding pocket for competitive inhibition [199, 200]. The huge, symmetric diarylurea suramin in fact blocks both binding sites (Figs. 2, 3a) in the crystal structure of its complex with Sirt5 [222]. Suramin inhibits, however, Sirt1, 2, and 5—and possibly other, not yet tested isoforms—with comparable, low micromolar potencies [206, 222, 223], and it also affects other cellular targets [224]. This lack of selectivity, and its poor pharmacological properties, such as strong charge and high molecular weight, make its improvement challenging. Whereas NAD+ blocking compounds are generally expected to suffer from low isoform selectivity due to the significant sequence conservation in the Sirtuin NAD+ binding sites, targeting polypeptide substrate site and acyl-Lys binding channel appear more promising for selective Sirtuin inhibition. Virtual screening of pseudopeptidic inhibitors on Sirt3 indeed revealed two new inhibitor scaffolds and structure activity relationship analysis resulted in compounds with a certain selectivity for Sirt2 and Sirt3 over Sirt1 but with modest potency (~70 % inhibition of Sirt2 and 3 at 200 µM of the best compound) [225]. Another docking study targeted at the acyl-Lys peptide sites of Sirt2, 3, 5, and 6 resulted in a number of potent, Sirt2 selective inhibitors (best hit CSC8; Sirt2 IC50 4.8 µM) but no selective compounds for Sirt3 and 5 [226]. Inhibition by blocking the acyl-Lys peptide site can also be achieved with modified peptides. Introduction of a thio modification in the substrate acetyl group (Fig. 2) results in formation of a stable thioalkyl-imidate during catalysis, thereby trapping the Sirtuin in the reaction intermediate state [179, 227, 228]. This approach can yield low micromolar inhibitors with certain selectivity for Sirtuin isoforms, including Sirt3 [229]. Addressing the acyl-Lys peptide binding site with drugs appears especially promising for Sirt5 due to its unique succinyl substrate preference. Thio-succinylated peptides have successfully been used for Sirt5 inhibition with an IC50 of 5 µM, and 100 µM of these peptides showed no inhibition of Sirt1-3 [230]. Also, variation of the acyl moiety allowed development of Sirt5-inhibiting peptides, such as Z-glutaryl-CPS1 and 3-methyl-3-phenyl-succinyl-CPS1; the latter one inhibited Sirt5 most potently (K i = 4.3 μM) and showed high isoform selectivity [165]. However, peptides usually are rapidly degraded in a cellular environment and are often incapable of crossing biological membranes, limiting their use for in vivo applications. In successful attempts to convert peptidic Sirtuin inhibitors into cell active compounds, the N- and C-terminal amino acid extensions to the thio-acetyl-Lys have been exchanged, e.g., by anilino and benzyloxycarbonyl groups (Fig. 2), respectively [231, 232]. For inhibition of Sirt5, the best compromise between potency and bioavailability so far appears to be provided by a truncated thio-succinyl penta-peptide and a protected Z-glutaryl-Lys, respectively, which both appear to inhibit with an IC50 of 25 µM [158, 230]. They might be attractive starting points for further development into potent Sirt5 inhibitors for in vivo applications.
In another approach to find Sirt5-selective small molecules, different thioureas were screened and yielded a thiobabiturate inhibiting with an IC50 of 2.3 and possibly exploiting the Sirt5-specific acyl-Lys pocket (Fig. 2) [233]. Computational docking suggests that the thiobarbiturate ring mimics the substrate succinyl by forming H-bonds to the Sirt5-specific residues Tyr102 and Arg105. N-alkylation of these compounds led to selectivity for Sirt5 over Sirt3 and, partially, Sirt1. Sirt2, however, was inhibited with an IC50 similar to Sirt5, but the IC50 comparison might not well reflect relative potencies due to the different substrate concentrations used for Sirt5 and Sirt1-3. It will be interesting to see whether these compounds can be further improved into selective compounds suitable for in vivo applications.
A moderately potent and Sirt3-specific small molecule inhibitor (Compound8, Fig. 2; IC50 6 μM, fivefold selective over Sirt2) was recently developed based on the non-specific, peptide-competitive Sirtuin inhibitor cambinol [234]. A more potent compound with pronounced isoform specificity for Sirt3 is SRT1720 (Fig. 2). Initially described as a Sirt1 activator [12], SRT1720 turned out to potently inhibit Sirt3 (K i of ~0.6 µM) but not Sirt5 [187]. Activity and binding assays revealed un-competitive Sirt3 inhibition with respect to the co-substrate NAD+ in combination with competition with the acetyl-Lys substrate [187, 235]. A Sirt3/SRT1720 complex structure showed a unique binding mode to the Sirtuin active site (Fig. 3e) [187]. SRT1720 occupies part of the acetyl-Lys binding channel with its piperizine and imidazothiazole groups, rationalizing the competition with the peptide substrate. Unlike other peptide competitive Sirtuin inhibitors, such as the chemically related ELT (“extended library technology”; see below) compounds, SRT1720 does not compete with the NAD+ cosubstrate but instead interacts tightly with this co-ligand. The SRT1720 quinoxaline forms a tight, sandwich-like π-electron stacking interaction with the NAD+ nicotinamide moiety on one side and Phe157 from the cofactor binding loop on its other side. This interaction leads to a major, and so far unique, rearrangement of the cofactor binding loop, and SRT1720 thus appears to induce, together with NAD+, formation of a protein conformation favorable for its binding and stabilizing an inactive enzyme-cosubstrate-inhibitor complex. The insensitivity of Sirt5 to SRT1720 inhibition might be caused by Sirt5-specific features such as the succinyl-recognizing Arg105, which also appears to cause Sirt5 insensitivity to nicotinamide and other C-pocket ligands (see below). Alternatively, a long, Sirt5-specific loop of Sirt5 was speculated to interfere indirectly with the cofactor binding loop rearrangements necessary for SRT1720 binding [187].
Nicotinamide and extended C-site (ECS) inhibitors
NAD+ binding to the Sirtuin active side in a productive conformation places its nicotinamide moiety inside the so-called C-pocket [179, 183]. In this conformation, the glycosidic bond to nicotinamide is distorted, facilitating nicotinamide release and formation of the alkylimidate intermediate. At this point, free nicotinamide (Fig. 2) can rebind to the C-pocket and induce the reverse reaction [197], thereby inhibiting deacetylation (so-called base-exchange-reaction). Nicotinamide is generally considered a pan Sirtuin inhibitor, but the Sirt5 deacetylase activity shows unexpected nicotinamide insensitivity. Whereas Sirt3 is inhibited with an IC50 of 43 µM, typical for Sirtuin enzymes, Sirt5 deacetylation activity does not respond to the nicotinamide concentrations of ~0.1 mM assumed to be physiological (IC50 0.7–1.6 mM) [152]. Only at high nicotinamide concentrations, Sirt5 deacetylation activity is inhibited through competition with NAD+. Structure comparisons and mutagenesis indicated that the Sirt5-specific Arg105, responsible for the enzyme’s unique desuccinylase specificity, mediates the nicotinamide insensitivity through sterical hindrance in the case of acetyl substrates, where the Arg cannot interact with the acyl group (Fig. 3f, rotamer 1) [152]. Consistently, nicotinamide affected Sirt5’s deacylation activity against succinylated substrate potently (Fig. 3f, rotamer 2), with an IC50 of 21 µM. Sequence analyzes and testing other Sirtuins indicate an Arg105-centered sequence pattern in a class III Sirtuin subfamily with Sirt5-like properties [152]. Thus, there are molecular differences close to the Sirtuin C-pocket that cause sub-class specific features and should allow the development of selective inhibitors.
An inhibitor indeed exploiting the C-pocket and its environment is Ex-527 (Fig. 2), a potent Sirt1 inhibitor (IC50 = 0.1 µM) that inhibits more weakly Sirt2 (IC50 = 19.6 µM) and Sirt3 (IC50 = 22.4 µM) but has no effect on Sirt5 [179, 204]. Structural analyses, in combination with activity and affinity measurements, on the bacterial Sirt1 homolog Sir2Tm (Sir2 from Thermotoga maritima) and on human Sirt1 and 3 show that Ex-527 exploits the unique NAD+-dependent deacetylation mechanism of Sirtuins [179]. The compound binds to the Sirtuin C-pocket and a neighboring, hydropbobic cleft only after formation of the reaction intermediate and inhibits by stabilizing a Sirtuin complex with the co-product 2′-O-acetyl-ADP-ribose (Fig. 3g). Consistent with the Sirt5-specific molecular features close to the C-site, Sirt5 is insensitive to Ex527. However, the Ex527 binding pockets are identical in several Sirtuin isoforms such as Sirt1 and 3, and the compound’s selectivity between these isoforms appears to be caused by the different kinetics of co-product formation and release [179]. Improvement of the selectivity of Ex527-based compounds thus will require extension into additional, more isoform-specific regions.
Other compounds also appear to exploit the C-pocket and/or its environment. Testing known Sirtuin inhibitors against the deacetylase and desuccinylase activities of Sirt5 showed that exclusively GW5074 (Fig. 2) inhibited Sirt5-dependent desuccinylation efficiently (IC50 19.5 µM) [206]. GW5074 is a potent kinase and Sirt2 inhibitor [236] and thus not suitable for Sirt5 in vivo studies. However, GW5074 inhibitory potency depends on the substrate acyl modification, similar to nicotinamide inhibition, and apparently also on the peptide sequence, and a docking study with a GW5074-related compound suggested the C-pocket as potential binding site [237]. GW5074 thus might be helpful as a starting point for the development of compounds exploiting Sirt5-specific features, which cause insensitivity to nicotinamide and Ex527, for the specific inhibition of Sirt5.
The C-pocket has also been used for potent Sirt3 inhibition. Synthesis and screening of a huge compound library using an “encoded library technology” (ELT) yielded thieno[3,2-d]pyrimidine-6-carboxamides that potently inhibited Sirt3, with an IC50 of 4 nM for the best hit ELT-11c (Fig. 2) [202]. All identified compounds also inhibited Sirt1 and 2 with potencies comparable to those against Sirt3, however, so that they are not suitable for in vivo studies and reveal approaches for potent inhibition, but not for isoforms selection. Sirt3 structures in complex with ELT compounds show that the inhibitors occupy parts of acetyl-Lys binding channel and C-pocket (Fig. 3h) [202]. The compound carboxamide forms H-bonds to C-pocket residues comparable to nicotinamide and Ex-527. Additionally, π-stacking interactions of the thieno[3,2-d]pyrimidine with a conserved Phe within the cofactor binding loop (Phe157 in Sirt3) appear important for compound binding. This interaction resembles this residue’s contribution to SRT1720 binding, but in the ELT-11c complex the Phe157-containing loop stays in a regular conformation also seen in other Sirtuin structures and appears not to exploit isoform-specific features. Although ELT compounds are pan Sirtuin inhibitors, they appear to inhibit this enzyme family rather selectively since they show no pronounced effects on other cellular targets such as kinases, ion channels or nuclear receptors. ELT compounds thus appear promising starting points for the development of Sirtuin inhibitors, and the required improvement of their isoform selectivity might be supported by the structural insights in the binding sites and mechanisms of these and other compounds.
Outlook for drug development
Analyzing the now available structural data for Sirtuin inhibition indicates promising design approaches for their improvement. For example, comparing the Sirt3 complexes with the C-pocket exploiting inhibitors nicotinamide, Ex-527, ELT compounds, and 4-bromo-resveratrol reveals several different binding pockets in close proximity (Fig. 3i). All C-site ligands except nicotinamide exploit additional, neighboring active site pockets and we thus call them “extended C-site” (ECS) inhibitors [179]. Since different pockets around the C-site are used by different compounds, they can be sub-classified into ECS-I compounds using the pocket accommodating the Ex527 A-ring [179]; ECS-II compounds such as ELT-11c exploiting the nicotinamide-carrying ribose and acyl lysine pockets [202]; and ECS-III compounds using the pocket accommodating the bromophenyl moiety of 4′-bromo-resveratrol [203] (Fig. 3i). Due to the significant conservation in C-site and ADP-ribose binding-pockets, combining moieties for different pockets, in particular the ECS-III pocket with its isoform-specific features, with each other and/or with C-site ligands should be helpful to create new Sirtuin inhibitors with improved isoform selectivity. Combining a bromo-phenyl group with the Ex527 scaffold, e.g., should increase specificity for Sirt1. Including moieties recognizing the Sirt5-specific acyl-Lys binding pocket (Fig. 3e) should also enable to obtain potent, Sirt5-selective inhibitors. Including features from the more complex Ex-527 and SRT1720 inhibition mechanisms might also be helpful, since these compounds exploit unique properties of Sirtuins and should enable the development of compounds with little effects on other cellular targets. The accumulating structural information on Sirtuin inhibition should also improve the success rates from virtual screening, since dynamic parts of the active site, isoform differences, and the various conformations accessible to Sirtuins can now be incorporated. Although no compounds are yet available that target mitochondrial Sirtuins with high potency and, at the same time, with high selectivity, the recent progress in characterizing binding sites and mechanisms of available compounds has opened outstanding opportunities for the efficient development of improved drugs for in vitro studies and eventually for therapy.
Conclusions
Mitochondrial functions are controlled by regulatory networks inside and outside the organelle itself, and Sirtuins contribute to both networks [33, 189]. Nuclear and mitochondrial Sirtuin isoforms thereby contribute to functions such as metabolic adaptation and stress responses. Recent progress in techniques for studying reversible acylation [173] and identifying activities and targets for the mitochondrial isoforms Sirt3, 4, and 5 revealed various contributions from these long neglected isoforms and have stimulated extensive new research efforts. The mitochondrial Sirtuins regulate and coordinate various metabolic pathways, such as glucose versus fat consumption, and they also adapt cellular redox containment systems to the ROS levels generated by this metabolic activity or by external stressors. Thereby, mitochondrial Sirtuins also contribute to various disease pathologies, such as obesity or hypoxic cell growth. Although many open questions require further studies, such as the difference in physiological effect between tissue-specific and germline Sirt3 k.o. [238] or the exact function in tumor biology [115], mitochondrial Sirtuins appear to be attractive therapeutic targets for diseases such as metabolic syndrome and cancer. Potent and isoforms-specific small molecule modulators for Sirt3, 4, and 5 would thus be highly desirable, as leads for the development of therapeutic drugs but also as chemical tools for studying these Sirtuins in vivo. Recent insights in the enzymatic activities of Sirtuins, such as their varying acyl and sequence selectivities [3, 151], have improved our understanding of their functions and also facilitate the development of small molecule modulators. They allow the reliable and comprehensive characterization of drug effects on Sirtuins [173], which is so far lacking for most compounds, and the now recognized isoforms-specific active site features can also be exploited for the improvement of compound selectivity. Our improved understanding of the biology of mitochondrial Sirtuins and of small molecule effects on Sirtuin activity thus promise that specific modulators will soon be obtained and will be highly useful for the treatment of mitochondria-mediated diseases.
Acknowledgments
We are grateful to members of the Steegborn lab and to many colleagues in the field for helpful discussions, and we apologize to those whose publications could not be covered in this review due to length limitations. Work on Sirtuins in the authors’ lab was supported by Deutsche Forschungsgemeinschaft grants STE1701/5 and STE1701/15, Alzheimer Forschung Initiative, and Oberfrankenstiftung (to CS).
Abbreviations
- ADH
Aldehyde dehydrogenase
- ANT
Adenine nucleotide transporter
- ECS
Extended C-site
- ELT
Encoded library technology
- ETC
Electron transport chain
- FdL
Fluor-de-Lys
- GDH
Glutamate dehydrogenase
- ICDH2
Isocitrate dehydrogenase 2
- IMS
Intermembrane space
- MCD
Malonyl CoA decarboxylase
- MDH
Malate dehydrogenase
- MLS
Mitochondrial localization sequences
- SOD
Superoxide dismutase
- Prx
Peroxiredoxin
- PTP
Permeability transition pore
- ROS
Reactive oxygen species
- TCA
Tricarboxylic acid cycle
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 2.Norvell A, McMahon SB. Cell biology. Rise of the rival. Science. 2010;327:964–965. doi: 10.1126/science.1187159. [DOI] [PubMed] [Google Scholar]
- 3.Rauh D, Fischer F, Gertz M, Lakshminarasimhan M, Bergbrede T, Aladini F, Kambach C, Becker CFW, Zerweck J, Schutkowski M, Steegborn C. An acetylome peptide microarray reveals specificities and deacetylation substrates for all human sirtuin isoforms. Nat Commun. 2013;4:2327. doi: 10.1038/ncomms3327. [DOI] [PubMed] [Google Scholar]
- 4.Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, Schilling B, Mooney SD, Kahn CR, Verdin E, Gibson BW. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc Natl Acad Sci USA. 2013;110:6601–6606. doi: 10.1073/pnas.1302961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Still AJ, Floyd BJ, Hebert AS, Bingman CA, Carson JJ, Gunderson DR, Dolan BK, Grimsrud PA, Dittenhafer-Reed KE, Stapleton DS, Keller MP, Westphall MS, Denu JM, Attie AD, Coon JJ, Pagliarini DJ. Quantification of mitochondrial acetylation dynamics highlights prominent sites of metabolic regulation. J Biol Chem. 2013;288:26209–26219. doi: 10.1074/jbc.M113.483396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 7.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 8.Guarente L, Picard F. Calorie restriction-the SIR2 connection. Cell. 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 9.Lin H, Su X, He B. Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS Chem Biol. 2012;7:947–960. doi: 10.1021/cb3001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cen Y, Youn DY, Sauve AA. Advances in characterization of human sirtuin isoforms: chemistries, targets and therapeutic applications. Curr Med Chem. 2011;18:1919–1935. doi: 10.2174/092986711795590084. [DOI] [PubMed] [Google Scholar]
- 11.Bell EL, Guarente L. The SirT3 divining rod points to oxidative stress. Mol Cell. 2011;42:561–568. doi: 10.1016/j.molcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baur JA, Chen D, Chini EN, Chua K, Cohen HY, de Cabo R, Deng C, Dimmeler S, Gius D, Guarente LP, Helfand SL, Imai S, Itoh H, Kadowaki T, Koya D, Leeuwenburgh C, McBurney M, Nabeshima Y, Neri C, Oberdoerffer P, Pestell RG, Rogina B, Sadoshima J, Sartorelli V, Serrano M, Sinclair DA, Steegborn C, Tatar M, Tissenbaum HA, Tong Q, Tsubota K, Vaquero A, Verdin E. Dietary restriction: standing up for sirtuins. Science. 2010;329:1012–1013. doi: 10.1126/science.329.5995.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 15.Donmez G, Outeiro TF. SIRT1 and SIRT2: emerging targets in neurodegeneration. EMBO Mol Med. 2013;5:344–352. doi: 10.1002/emmm.201302451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavu S, Boss O, Elliott PJ, Lambert PD. Sirtuins—novel therapeutic targets to treat age-associated diseases. Nat Rev Drug Discov. 2008;7:841–853. doi: 10.1038/nrd2665. [DOI] [PubMed] [Google Scholar]
- 18.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jedrusik-Bode M, Studencka M, Smolka C, Baumann T, Schmidt H, Kampf J, Paap F, Martin S, Tazi J, Muller KM, Kruger M, Braun T, Bober E. The sirtuin SIRT6 regulates stress granule formation in C. elegans and mammals. J Cell Sci. 2013;126:5166–5177. doi: 10.1242/jcs.130708. [DOI] [PubMed] [Google Scholar]
- 21.Byles V, Chmilewski LK, Wang J, Zhu L, Forman LW, Faller DV, Dai Y. Aberrant cytoplasm localization and protein stability of SIRT1 is regulated by PI3 K/IGF-1R signaling in human cancer cells. Int J Biol Sci. 2010;6:599–612. doi: 10.7150/ijbs.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/S1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 23.Black JC, Mosley A, Kitada T, Washburn M, Carey M. The SIRT2 deacetylase regulates autoacetylation of p300. Mol Cell. 2008;32:449–455. doi: 10.1016/j.molcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoue T, Hiratsuka M, Osaki M, Yamada H, Kishimoto I, Yamaguchi S, Nakano S, Katoh M, Ito H, Oshimura M. SIRT2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene. 2007;26:945–957. doi: 10.1038/sj.onc.1209857. [DOI] [PubMed] [Google Scholar]
- 25.Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gertz M, Steegborn C. Function and regulation of the mitochondrial Sirtuin isoform Sirt5 in Mammalia. Biochim Biophys Acta. 2010;1804:1658–1665. doi: 10.1016/j.bbapap.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Chalkiadaki A, Guarente L. Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat Rev Endocrinol. 2012;8:287–296. doi: 10.1038/nrendo.2011.225. [DOI] [PubMed] [Google Scholar]
- 28.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrn3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Pastor B, Mostoslavsky R. Sirtuins, metabolism, and cancer. Front Pharmacol. 2012;3:22. doi: 10.3389/fphar.2012.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 31.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, De Oliveira RM, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brenmoehl J, Hoeflich A. Dual control of mitochondrial biogenesis by sirtuin 1 and sirtuin 3. Mitochondrion. 2013;13:755–761. doi: 10.1016/j.mito.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 34.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 35.Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, Clish CB, Vaitheesvaran B, Iliopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryu D, Jo YS, Lo Sasso G, Stein S, Zhang H, Perino A, Lee JU, Zeviani M, Romand R, Hottiger MO, Schoonjans K, Auwerx J. A SIRT7-dependent acetylation switch of GABPbeta1 controls mitochondrial function. Cell Metab. 2014;20:856–869. doi: 10.1016/j.cmet.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Baur JA, Ungvari Z, Minor RK, Le Couteur DG, de Cabo R. Are sirtuins viable targets for improving healthspan and lifespan? Nat Rev Drug Discov. 2012;11:443–461. doi: 10.1038/nrd3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Libri V, Brown AP, Gambarota G, Haddad J, Shields GS, Dawes H, Pinato DJ, Hoffman E, Elliot PJ, Vlasuk GP, Jacobson E, Wilkins MR, Matthews PM. A pilot randomized, placebo controlled, double blind phase I trial of the novel SIRT1 activator SRT2104 in elderly volunteers. PLoS One. 2012;7:e51395. doi: 10.1371/journal.pone.0051395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lakshminarasimhan M, Rauth D, Schutkowski M, Steegborn C. Sirt1 activation by resveratrol is substrate sequence-selective. Aging Albany NY. 2013;5:151–154. doi: 10.18632/aging.100542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hubbard BP, Gomes AP, Dai H, Li J, Case AW, Considine T, Riera TV, Lee JE, Yen S, Lamming DW, Pentelute BL, Schuman ER, Stevens LA, Ling AJ, Armour SM, Michan S, Zhao H, Jiang Y, Sweitzer SM, Blum CA, Disch JS, Ng PY, Howitz KT, Rolo AP, Hamuro Y, Moss J, Perni RB, Ellis JL, Vlasuk GP, Sinclair DA. Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science. 2013;339:1216–1219. doi: 10.1126/science.1231097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price NL, Gomes AP, Ling AJ, Duarte FV, Martin-Montalvo A, North BJ, Agarwal B, Ye L, Ramadori G, Teodoro JS, Hubbard BP, Varela AT, Davis JG, Varamini B, Hafner A, Moaddel R, Rolo AP, Coppari R, Palmeira CM, de Cabo R, Baur JA, Sinclair DA. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012;15:675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 44.Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans BM, Skinner ME, Lombard DB, Zhao Y. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50:919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, Guo A, Gut P, Sahu AK, Li B, Uppala R, Fitch M, Riiff T, Zhu L, Zhou J, Mulhern D, Stevens RD, Ilkayeva OR, Newgard CB, Jacobson MP, Hellerstein M, Goetzman ES, Gibson BW, Verdin E. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013;18:920–933. doi: 10.1016/j.cmet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weinert BT, Moustafa T, Iesmantavicius V, Zechner R, Choudhary C. Analysis of acetylation stoichiometry suggests that SIRT3 repairs nonenzymatic acetylation lesions. EMBO J. 2015;34:2620–2632. doi: 10.15252/embj.201591271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peek CB, Affinati AH, Ramsey KM, Kuo HY, Yu W, Sena LA, Ilkayeva O, Marcheva B, Kobayashi Y, Omura C, Levine DC, Bacsik DJ, Gius D, Newgard CB, Goetzman E, Chandel NS, Denu JM, Mrksich M, Bass J. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science. 2013;342:1243417. doi: 10.1126/science.1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott I, Webster BR, Li JH, Sack MN. Identification of a molecular component of the mitochondrial acetyltransferase programme: a novel role for GCN5L1. Biochem J. 2012;443:655–661. doi: 10.1042/BJ20120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner GR, Payne RM. Widespread and enzyme-independent Nepsilon-acetylation and Nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J Biol Chem. 2013;288:29036–29045. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinert BT, Iesmantavicius V, Wagner SA, Scholz C, Gummesson B, Beli P, Nystrom T, Choudhary C. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli . Mol Cell. 2013;51:265–272. doi: 10.1016/j.molcel.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 51.Simic Z, Weiwad M, Schierhorn A, Steegborn C, Schutkowski M. The epsilon-amino group of protein lysine residues is highly susceptible to nonenzymatic acylation by several physiological Acyl-CoA thioesters. ChemBioChem. 2015;16:2337–2347. doi: 10.1002/cbic.201500364. [DOI] [PubMed] [Google Scholar]
- 52.Rardin MJ, Held JM, Gibson BW. Targeted quantitation of acetylated lysine peptides by selected reaction monitoring mass spectrometry. Methods Mol Biol. 2013;1077:121–131. doi: 10.1007/978-1-62703-637-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sol EM, Wagner SA, Weinert BT, Kumar A, Kim HS, Deng CX, Choudhary C. Proteomic investigations of lysine acetylation identify diverse substrates of mitochondrial deacetylase sirt3. PLoS One. 2012;7:e50545. doi: 10.1371/journal.pone.0050545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, Westphall MS, Pagliarini DJ, Prolla TA, Assadi-Porter F, Roy S, Denu JM, Coon JJ. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell. 2013;49:186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fritz KS, Galligan JJ, Hirschey MD, Verdin E, Petersen DR. Mitochondrial acetylome analysis in a mouse model of alcohol-induced liver injury utilizing SIRT3 knockout mice. J Proteome Res. 2012;11:1633–1643. doi: 10.1021/pr2008384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci USA. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci USA. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 60.Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shulga N, Wilson-Smith R, Pastorino JG. Sirtuin-3 deacetylation of cyclophilin D induces dissociation of hexokinase II from the mitochondria. J Cell Sci. 2010;123:894–902. doi: 10.1242/jcs.061846. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Ozden O, Park SH, Wagner BA, Yong Song H, Zhu Y, Vassilopoulos A, Jung B, Buettner GR, Gius D. SIRT3 deacetylates and increases pyruvate dehydrogenase activity in cancer cells. Free Radic Biol Med. 2014;76:163–172. doi: 10.1016/j.freeradbiomed.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci USA. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Finley LW, Haas W, Desquiret-Dumas V, Wallace DC, Procaccio V, Gygi SP, Haigis MC. Succinate dehydrogenase is a direct target of sirtuin 3 deacetylase activity. PLoS One. 2011;6:e23295. doi: 10.1371/journal.pone.0023295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rahman M, Nirala NK, Singh A, Zhu LJ, Taguchi K, Bamba T, Fukusaki E, Shaw LM, Lambright DG, Acharya JK, Acharya UR. Drosophila Sirt2/mammalian SIRT3 deacetylates ATP synthase beta and regulates complex V activity. J Cell Biol. 2014;206:289–305. doi: 10.1083/jcb.201404118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vassilopoulos A, Pennington JD, Andresson T, Rees DM, Bosley AD, Fearnley IM, Ham A, Flynn CR, Hill S, Rose KL, Kim HS, Deng CX, Walker JE, Gius D. SIRT3 deacetylates ATP synthase F1 complex proteins in response to nutrient- and exercise-induced stress. Antioxid Redox Signal. 2014;21:551–564. doi: 10.1089/ars.2013.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu YT, Lee HC, Liao CC, Wei YH. Regulation of mitochondrial F(o)F(1)ATPase activity by Sirt3-catalyzed deacetylation and its deficiency in human cells harboring 4977 bp deletion of mitochondrial DNA. Biochim Biophys Acta. 2013;1832:216–227. doi: 10.1016/j.bbadis.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 68.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr, Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hirschey MD, Shimazu T, Capra JA, Pollard KS, Verdin E. SIRT1 and SIRT3 deacetylate homologous substrates: AceCS1,2 and HMGCS1,2. Aging Albany NY. 2011;3:635–642. doi: 10.18632/aging.100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hallows WC, Yu W, Smith BC, Devries MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, Zhao S, Guan KL, Denu JM. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang Y, Cimen H, Han MJ, Shi T, Deng JH, Koc H, Palacios OM, Montier L, Bai Y, Tong Q, Koc EC. NAD+-dependent deacetylase SIRT3 regulates mitochondrial protein synthesis by deacetylation of the ribosomal protein MRPL10. J Biol Chem. 2010;285:7417–7429. doi: 10.1074/jbc.M109.053421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu Z, Chen Y, Aponte AM, Battaglia V, Gucek M, Sack MN. Prolonged fasting identifies heat shock protein 10 as a Sirtuin 3 substrate: elucidating a new mechanism linking mitochondrial protein acetylation to fatty acid oxidation enzyme folding and function. J Biol Chem. 2015;290:2466–2476. doi: 10.1074/jbc.M114.606228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jing E, Emanuelli B, Hirschey MD, Boucher J, Lee KY, Lombard D, Verdin EM, Kahn CR. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proc Natl Acad Sci USA. 2011;108:14608–14613. doi: 10.1073/pnas.1111308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stancakova A, Goetzman E, Lam MM, Schwer B, Stevens RD, Muehlbauer MJ, Kakar S, Bass NM, Kuusisto J, Laakso M, Alt FW, Newgard CB, Farese RV, Jr, Kahn CR, Verdin E. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell. 2011;44:177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Palacios OM, Carmona JJ, Michan S, Chen KY, Manabe Y, Ward JL, 3rd, Goodyear LJ, Tong Q. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1alpha in skeletal muscle. Aging Albany NY. 2009;1:771–783. doi: 10.18632/aging.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Caton PW, Richardson SJ, Kieswich J, Bugliani M, Holland ML, Marchetti P, Morgan NG, Yaqoob MM, Holness MJ, Sugden MC. Sirtuin 3 regulates mouse pancreatic beta cell function and is suppressed in pancreatic islets isolated from human type 2 diabetic patients. Diabetologia. 2013;56:1068–1077. doi: 10.1007/s00125-013-2851-y. [DOI] [PubMed] [Google Scholar]
- 77.Teodoro JS, Duarte FV, Gomes AP, Varela AT, Peixoto FM, Rolo AP, Palmeira CM. Berberine reverts hepatic mitochondrial dysfunction in high-fat fed rats: a possible role for SirT3 activation. Mitochondrion. 2013;13:637–646. doi: 10.1016/j.mito.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 78.Gomes AP, Duarte FV, Nunes P, Hubbard BP, Teodoro JS, Varela AT, Jones JG, Sinclair DA, Palmeira CM, Rolo AP. Berberine protects against high fat diet-induced dysfunction in muscle mitochondria by inducing SIRT1-dependent mitochondrial biogenesis. Biochim Biophys Acta. 2012;1822:185–195. doi: 10.1016/j.bbadis.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Song Y, Shi J, Wu Y, Han C, Zou J, Shi Y, Liu Z. Metformin ameliorates insulin resistance in L6 rat skeletal muscle cells through upregulation of SIRT3. Chin Med J (Engl) 2014;127:1523–1529. [PubMed] [Google Scholar]
- 80.Chen Y, Zhang J, Lin Y, Lei Q, Guan KL, Zhao S, Xiong Y. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Rep. 2011;12:534–541. doi: 10.1038/embor.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 82.Zeng L, Yang Y, Hu Y, Sun Y, Du Z, Xie Z, Zhou T, Kong W. Age-related decrease in the mitochondrial sirtuin deacetylase Sirt3 expression associated with ROS accumulation in the auditory cortex of the mimetic aging rat model. PLoS One. 2014;9:e88019. doi: 10.1371/journal.pone.0088019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, Olivier AK, Spitz DR, Gius D. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pacella-Ince L, Zander-Fox DL, Lan M. Mitochondrial SIRT3 and its target glutamate dehydrogenase are altered in follicular cells of women with reduced ovarian reserve or advanced maternal age. Hum Reprod. 2014;29:1490–1499. doi: 10.1093/humrep/deu071. [DOI] [PubMed] [Google Scholar]
- 85.Fu H, Wada-Hiraike O, Hirano M, Kawamura Y, Sakurabashi A, Shirane A, Morita Y, Isono W, Oishi H, Koga K, Oda K, Kawana K, Yano T, Kurihara H, Osuga Y, Fujii T. SIRT3 positively regulates the expression of folliculogenesis- and luteinization-related genes and progesterone secretion by manipulating oxidative stress in human luteinized granulosa cells. Endocrinology. 2014;155:3079–3087. doi: 10.1210/en.2014-1025. [DOI] [PubMed] [Google Scholar]
- 86.Brown KD, Maqsood S, Huang JY, Pan Y, Harkcom W, Li W, Sauve A, Verdin E, Jaffrey SR. Activation of SIRT3 by the NAD(+) precursor nicotinamide riboside protects from noise-induced hearing loss. Cell Metab. 2014;20:1059–1068. doi: 10.1016/j.cmet.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Quan Y, Xia L, Shao J, Yin S, Cheng CY, Xia W, Gao WQ. Adjudin protects rodent cochlear hair cells against gentamicin ototoxicity via the SIRT3-ROS pathway. Sci Rep. 2015;5:8181. doi: 10.1038/srep08181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bao J, Scott I, Lu Z, Pang L, Dimond CC, Gius D, Sack MN. SIRT3 is regulated by nutrient excess and modulates hepatic susceptibility to lipotoxicity. Free Radic Biol Med. 2010;49:1230–1237. doi: 10.1016/j.freeradbiomed.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dai SH, Chen T, Wang YH, Zhu J, Luo P, Rao W, Yang YF, Fei Z, Jiang XF. Sirt3 protects cortical neurons against oxidative stress via regulating mitochondrial Ca2+ and mitochondrial biogenesis. Int J Mol Sci. 2014;15:14591–14609. doi: 10.3390/ijms150814591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brown K, Xie S, Qiu X, Mohrin M, Shin J, Liu Y, Zhang D, Scadden DT, Chen D. SIRT3 reverses aging-associated degeneration. Cell Rep. 2013;3:319–327. doi: 10.1016/j.celrep.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bause AS, Matsui MS, Haigis MC. The protein deacetylase SIRT3 prevents oxidative stress-induced keratinocyte differentiation. J Biol Chem. 2013;288:36484–36491. doi: 10.1074/jbc.M113.472324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dong K, Pelle E, Yarosh DB, Pernodet N. Sirtuin 4 identification in normal human epidermal keratinocytes and its relation to sirtuin 3 and energy metabolism under normal conditions and UVB-induced stress. Exp Dermatol. 2012;21:231–233. doi: 10.1111/j.1600-0625.2011.01439.x. [DOI] [PubMed] [Google Scholar]
- 93.Hokari F, Kawasaki E, Sakai A, Koshinaka K, Sakuma K, Kawanaka K. Muscle contractile activity regulates Sirt3 protein expression in rat skeletal muscles. J Appl Physiol. 2010;109:332–340. doi: 10.1152/japplphysiol.00335.2009. [DOI] [PubMed] [Google Scholar]
- 94.Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, Fang F, Chang Y. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One. 2010;5:e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jacobs KM, Pennington JD, Bisht KS, Aykin-Burns N, Kim HS, Mishra M, Sun L, Nguyen P, Ahn BH, Leclerc J, Deng CX, Spitz DR, Gius D. SIRT3 interacts with the daf-16 homolog FOXO3a in the mitochondria, as well as increases FOXO3a dependent gene expression. Int J Biol Sci. 2008;4:291–299. doi: 10.7150/ijbs.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peserico A, Chiacchiera F, Grossi V, Matrone A, Latorre D, Simonatto M, Fusella A, Ryall JG, Finley LW, Haigis MC, Villani G, Puri PL, Sartorelli V, Simone C. A novel AMPK-dependent FoxO3A-SIRT3 intramitochondrial complex sensing glucose levels. Cell Mol Life Sci. 2013;70:2015–2029. doi: 10.1007/s00018-012-1244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tseng AH, Shieh SS, Wang DL. SIRT3 deacetylates FOXO3 to protect mitochondria against oxidative damage. Free Radic Biol Med. 2013;63:222–234. doi: 10.1016/j.freeradbiomed.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 98.Samant SA, Zhang HJ, Hong Z, Pillai VB, Sundaresan NR, Wolfgeher D, Archer SL, Chan DC, Gupta MP. SIRT3 deacetylates and activates OPA1 to regulate mitochondrial dynamics during stress. Mol Cell Biol. 2014;34:807–819. doi: 10.1128/MCB.01483-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Morigi M, Perico L, Rota C, Longaretti L, Conti S, Rottoli D, Novelli R, Remuzzi G, Benigni A. Sirtuin 3-dependent mitochondrial dynamic improvements protect against acute kidney injury. J Clin Invest. 2015;125:715–726. doi: 10.1172/JCI77632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xue L, Xu F, Meng L, Wei S, Wang J, Hao P, Bian Y, Zhang Y, Chen Y. Acetylation-dependent regulation of mitochondrial ALDH2 activation by SIRT3 mediates acute ethanol-induced eNOS activation. FEBS Lett. 2012;586:137–142. doi: 10.1016/j.febslet.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 101.Lu Z, Bourdi M, Li JH, Aponte AM, Chen Y, Lombard DB, Gucek M, Pohl LR, Sack MN. SIRT3-dependent deacetylation exacerbates acetaminophen hepatotoxicity. EMBO Rep. 2011;12:840–846. doi: 10.1038/embor.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Coleman MC, Olivier AK, Jacobus JA, Mapuskar KA, Mao G, Martin SM, Riley DP, Gius D, Spitz DR (2014) Superoxide mediates acute liver injury in irradiated mice lacking sirtuin 3. Antioxid Redox Signal 20:1423-1435 [DOI] [PMC free article] [PubMed]
- 103.Sack MN. Emerging characterization of the role of SIRT3-mediated mitochondrial protein deacetylation in the heart. Am J Physiol Heart Circ Physiol. 2011;301:H2191–H2197. doi: 10.1152/ajpheart.00199.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Grillon JM, Johnson KR, Kotlo K, Danziger RS. Non-histone lysine acetylated proteins in heart failure. Biochim Biophys Acta. 2012;1822:607–614. doi: 10.1016/j.bbadis.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol Cell Biol. 2008;28:6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920–928. doi: 10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Iwahara T, Bonasio R, Narendra V, Reinberg D. SIRT3 functions in the nucleus in the control of stress-related gene expression. Mol Cell Biol. 2012;32:5022–5034. doi: 10.1128/MCB.00822-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nakamura Y, Ogura M, Tanaka D, Inagaki N. Localization of mouse mitochondrial SIRT proteins: shift of SIRT3 to nucleus by co-expression with SIRT5. Biochem Biophys Res Commun. 2008;366:174–179. doi: 10.1016/j.bbrc.2007.11.122. [DOI] [PubMed] [Google Scholar]
- 110.Cooper HM, Spelbrink JN. The human SIRT3 protein deacetylase is exclusively mitochondrial. Biochem J. 2008;411:279–285. doi: 10.1042/BJ20071624. [DOI] [PubMed] [Google Scholar]
- 111.Gurd BJ, Holloway GP, Yoshida Y, Bonen A. In mammalian muscle, SIRT3 is present in mitochondria and not in the nucleus; and SIRT3 is upregulated by chronic muscle contraction in an adenosine monophosphate-activated protein kinase-independent manner. Metabolism. 2012;61:733–741. doi: 10.1016/j.metabol.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 112.Paulin R, Dromparis P, Sutendra G, Gurtu V, Zervopoulos S, Bowers L, Haromy A, Webster L, Provencher S, Bonnet S, Michelakis ED. Sirtuin 3 deficiency is associated with inhibited mitochondrial function and pulmonary arterial hypertension in rodents and humans. Cell Metab. 2014;20:827–839. doi: 10.1016/j.cmet.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 113.Wang Q, Li L, Li CY, Pei Z, Zhou M, Li N. SIRT3 protects cells from hypoxia via PGC-1alpha- and MnSOD-dependent pathways. Neuroscience. 2015;286:109–121. doi: 10.1016/j.neuroscience.2014.11.045. [DOI] [PubMed] [Google Scholar]
- 114.Tseng AH, Wu LH, Shieh SS, Wang DL. SIRT3 interactions with FOXO3 acetylation, phosphorylation and ubiquitinylation mediate endothelial cell responses to hypoxia. Biochem J. 2014;464:157–168. doi: 10.1042/BJ20140213. [DOI] [PubMed] [Google Scholar]
- 115.Alhazzazi TY, Kamarajan P, Verdin E, Kapila YL. SIRT3 and cancer: tumor promoter or suppressor? Biochim Biophys Acta. 2011;1816:80–88. doi: 10.1016/j.bbcan.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.New M, Olzscha H, La Thangue NB. HDAC inhibitor-based therapies: can we interpret the code? Mol Oncol. 2012;6:637–656. doi: 10.1016/j.molonc.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Van Damme M, Crompot E, Meuleman N, Mineur P, Bron D, Lagneaux L, Stamatopoulos B. HDAC isoenzyme expression is deregulated in chronic lymphocytic leukemia B-cells and has a complex prognostic significance. Epigenetics. 2012;7:1403–1412. doi: 10.4161/epi.22674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, Pandolfi PP, Haigis MC. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fan J, Shan C, Kang HB, Elf S, Xie J, Tucker M, Gu TL, Aguiar M, Lonning S, Chen H, Mohammadi M, Britton LM, Garcia BA, Aleckovic M, Kang Y, Kaluz S, Devi N, Van Meir EG, Hitosugi T, Seo JH, Lonial S, Gaddh M, Arellano M, Khoury HJ, Khuri FR, Boggon TJ, Kang S, Chen J. Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Mol Cell. 2014;53:534–548. doi: 10.1016/j.molcel.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Desouki MM, Doubinskaia I, Gius D, Abdulkadir SA. Decreased mitochondrial SIRT3 expression is a potential molecular biomarker associated with poor outcome in breast cancer. Hum Pathol. 2014;45:1071–1077. doi: 10.1016/j.humpath.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Huang KH, Hsu CC, Fang WL, Chi CW, Sung MT, Kao HL, Li AF, Yin PH, Yang MH, Lee HC. SIRT3 expression as a biomarker for better prognosis in gastric cancer. World J Surg. 2014;38:910–917. doi: 10.1007/s00268-013-2359-0. [DOI] [PubMed] [Google Scholar]
- 124.Yang B, Fu X, Shao L, Ding Y, Zeng D. Aberrant expression of SIRT3 is conversely correlated with the progression and prognosis of human gastric cancer. Biochem Biophys Res Commun. 2014;443:156–160. doi: 10.1016/j.bbrc.2013.11.068. [DOI] [PubMed] [Google Scholar]
- 125.Xiao K, Jiang J, Wang W, Cao S, Zhu L, Zeng H, Ouyang R, Zhou R, Chen P. Sirt3 is a tumor suppressor in lung adenocarcinoma cells. Oncol Rep. 2013;30:1323–1328. doi: 10.3892/or.2013.2604. [DOI] [PubMed] [Google Scholar]
- 126.Allison SJ, Milner J. SIRT3 is pro-apoptotic and participates in distinct basal apoptotic pathways. Cell Cycle. 2007;6:2669–2677. doi: 10.4161/cc.6.21.4866. [DOI] [PubMed] [Google Scholar]
- 127.Pellegrini L, Pucci B, Villanova L, Marino ML, Marfe G, Sansone L, Vernucci E, Bellizzi D, Reali V, Fini M, Russo MA, Tafani M. SIRT3 protects from hypoxia and staurosporine-mediated cell death by maintaining mitochondrial membrane potential and intracellular pH. Cell Death Differ. 2012;19:1815–1825. doi: 10.1038/cdd.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang CZ, Liu L, Cai M, Pan Y, Fu J, Cao Y, Yun J. Low SIRT3 expression correlates with poor differentiation and unfavorable prognosis in primary hepatocellular carcinoma. PLoS One. 2012;7:e51703. doi: 10.1371/journal.pone.0051703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang YY, Zhou LM. Sirt3 inhibits hepatocellular carcinoma cell growth through reducing Mdm2-mediated p53 degradation. Biochem Biophys Res Commun. 2012;423:26–31. doi: 10.1016/j.bbrc.2012.05.053. [DOI] [PubMed] [Google Scholar]
- 130.Jeong SM, Lee J, Finley LW, Schmidt PJ, Fleming MD, Haigis MC. SIRT3 regulates cellular iron metabolism and cancer growth by repressing iron regulatory protein 1. Oncogene. 2015;34:2115–2124. doi: 10.1038/onc.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu C, Huang Z, Jiang H, Shi F. The sirtuin 3 expression profile is associated with pathological and clinical outcomes in colon cancer patients. Biomed Res Int. 2014;2014:871263. doi: 10.1155/2014/871263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Alhazzazi TY, Kamarajan P, Joo N, Huang JY, Verdin E, D’Silva NJ, Kapila YL. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer. 2011;117:1670–1678. doi: 10.1002/cncr.25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yan SM, Han X, Han PJ, Chen HM, Huang LY, Li Y. SIRT3 is a novel prognostic biomarker for esophageal squamous cell carcinoma. Med Oncol. 2014;31:103. doi: 10.1007/s12032-014-0103-8. [DOI] [PubMed] [Google Scholar]
- 134.Chen IC, Chiang WF, Liu SY, Chen PF, Chiang HC. Role of SIRT3 in the regulation of redox balance during oral carcinogenesis. Mol Cancer. 2013;12:68. doi: 10.1186/1476-4598-12-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang L, Ren X, Cheng Y, Huber-Keener K, Liu X, Zhang Y, Yuan YS, Yang JW, Liu CG, Yang JM. Identification of Sirtuin 3, a mitochondrial protein deacetylase, as a new contributor to tamoxifen resistance in breast cancer cells. Biochem Pharmacol. 2013;86:726–733. doi: 10.1016/j.bcp.2013.06.032. [DOI] [PubMed] [Google Scholar]
- 136.Giralt A, Villarroya F. SIRT3, a pivotal actor in mitochondrial functions: metabolism, cell death and aging. Biochem J. 2012;444:1–10. doi: 10.1042/BJ20120030. [DOI] [PubMed] [Google Scholar]
- 137.Pillai VB, Sundaresan NR, Kim G, Gupta M, Rajamohan SB, Pillai JB, Samant S, Ravindra PV, Isbatan A, Gupta MP. Exogenous NAD blocks cardiac hypertrophic response via activation of the SIRT3-LKB1-AMP-activated kinase pathway. J Biol Chem. 2010;285:3133–3144. doi: 10.1074/jbc.M109.077271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, Gademann K, Rinsch C, Schoonjans K, Sauve AA, Auwerx J. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Laurent G, German NJ, Saha AK, de Boer VC, Davies M, Koves TR, Dephoure N, Fischer F, Boanca G, Vaitheesvaran B, Lovitch SB, Sharpe AH, Kurland IJ, Steegborn C, Gygi SP, Muoio DM, Ruderman NB, Haigis MC. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol Cell. 2013;50:686–698. doi: 10.1016/j.molcel.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 141.Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, Maechler P, Verdin E. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. 2007;282:33583–33592. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- 142.Verma M, Shulga N, Pastorino JG. Sirtuin-4 modulates sensitivity to induction of the mitochondrial permeability transition pore. Biochim Biophys Acta. 2013;1827:38–49. doi: 10.1016/j.bbabio.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 143.Lee SJ, Choi SE, Jung IR, Lee KW, Kang Y. Protective effect of nicotinamide on high glucose/palmitate-induced glucolipotoxicity to INS-1 beta cells is attributed to its inhibitory activity to sirtuins. Arch Biochem Biophys. 2013;535:187–196. doi: 10.1016/j.abb.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 144.Du J, Jiang H, Lin H. Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogues and 32P-NAD. Biochemistry. 2009;48:2878–2890. doi: 10.1021/bi802093g. [DOI] [PubMed] [Google Scholar]
- 145.Mathias RA, Greco TM, Oberstein A, Budayeva HG, Chakrabarti R, Rowland EA, Kang Y, Shenk T, Cristea IM. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell. 2014;159:1615–1625. doi: 10.1016/j.cell.2014.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nasrin N, Wu X, Fortier E, Feng Y, Bare OC, Chen S, Ren X, Wu Z, Streeper RS, Bordone L. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J Biol Chem. 2010;285:31995–32002. doi: 10.1074/jbc.M110.124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, Pierce K, Li YH, Wang X, Laurent G, German NJ, Xu X, Li C, Wang RH, Lee J, Csibi A, Cerione R, Blenis J, Clish CB, Kimmelman A, Deng CX, Haigis MC. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. 2013;23:450–463. doi: 10.1016/j.ccr.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T, Henske EP, Haigis MC, Cantley LC, Stephanopoulos G, Yu J, Blenis J. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013;153:840–854. doi: 10.1016/j.cell.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Komlos D, Mann KD, Zhuo Y, Ricupero CL, Hart RP, Liu AY, Firestein BL. Glutamate dehydrogenase 1 and SIRT4 regulate glial development. Glia. 2013;61:394–408. doi: 10.1002/glia.22442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 151.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fischer F, Gertz M, Suenkel B, Lakshminarasimhan M, Schutkowski M, Steegborn C. Sirt5 deacylation activities show differential sensitivities to nicotinamide inhibition. PLoS One. 2012;7:e45098. doi: 10.1371/journal.pone.0045098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Matsushita N, Yonashiro R, Ogata Y, Sugiura A, Nagashima S, Fukuda T, Inatome R, Yanagi S. Distinct regulation of mitochondrial localization and stability of two human Sirt5 isoforms. Genes Cells. 2011;16:190–202. doi: 10.1111/j.1365-2443.2010.01475.x. [DOI] [PubMed] [Google Scholar]
- 154.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ogura M, Nakamura Y, Tanaka D, Zhuang X, Fujita Y, Obara A, Hamasaki A, Hosokawa M, Inagaki N. Overexpression of SIRT5 confirms its involvement in deacetylation and activation of carbamoyl phosphate synthetase 1. Biochem Biophys Res Commun. 2010;393:73–78. doi: 10.1016/j.bbrc.2010.01.081. [DOI] [PubMed] [Google Scholar]
- 156.Geng YQ, Li TT, Liu XY, Li ZH, Fu YC. SIRT1 and SIRT5 activity expression and behavioral responses to calorie restriction. J Cell Biochem. 2011;112:3755–3761. doi: 10.1002/jcb.23315. [DOI] [PubMed] [Google Scholar]
- 157.Pacella-Ince L, Zander-Fox DL, Lane M. Mitochondrial SIRT5 is present in follicular cells and is altered by reduced ovarian reserve and advanced maternal age. Reprod Fertil Dev. 2014;26:1072–1083. doi: 10.1071/RD13178. [DOI] [PubMed] [Google Scholar]
- 158.Polletta L, Vernucci E, Carnevale I, Arcangeli T, Rotili D, Palmerio S, Steegborn C, Nowak T, Schutkowski M, Pellegrini L, Sansone L, Villanova L, Runci A, Pucci B, Morgante E, Fini M, Mai A, Russo MA, Tafani M. SIRT5 regulation of ammonia-induced autophagy and mitophagy. Autophagy. 2015;11:253–270. doi: 10.1080/15548627.2015.1009778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nakamura Y, Ogura M, Ogura K, Tanaka D, Inagaki N. SIRT5 deacetylates and activates urate oxidase in liver mitochondria of mice. FEBS Lett. 2012;586:4076–4081. doi: 10.1016/j.febslet.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 160.Gertz M, Fischer F, Leipelt M, Wolters D, Steegborn C. Identification of Peroxiredoxin 1 as a novel interaction partner for the lifespan regulator protein p66Shc. Aging Albany NY. 2009;1:254–265. doi: 10.18632/aging.100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pfister JA, Ma C, Morrison BE, D’Mello SR. Opposing effects of sirtuins on neuronal survival: SIRT1-mediated neuroprotection is independent of its deacetylase activity. PLoS One. 2008;3:e4090. doi: 10.1371/journal.pone.0004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Glorioso C, Oh S, Douillard GG, Sibille E. Brain molecular aging, promotion of neurological disease and modulation by sirtuin 5 longevity gene polymorphism. Neurobiol Dis. 2011;41:279–290. doi: 10.1016/j.nbd.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lutz MI, Milenkovic I, Regelsberger G, Kovacs GG. Distinct patterns of sirtuin expression during progression of Alzheimer’s disease. Neuromolecular Med. 2014;16:405–414. doi: 10.1007/s12017-014-8288-8. [DOI] [PubMed] [Google Scholar]
- 164.Lieber CS, Leo MA, Wang X, Decarli LM. Alcohol alters hepatic FoxO1, p53, and mitochondrial SIRT5 deacetylation function. Biochem Biophys Res Commun. 2008;373:246–252. doi: 10.1016/j.bbrc.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 165.Roessler C, Nowak T, Pannek M, Gertz M, Nguyen GT, Scharfe M, Born I, Sippl W, Steegborn C, Schutkowski M. Chemical probing of the human sirtuin 5 active site reveals its substrate acyl specificity and peptide-based inhibitors. Angew Chem Int Ed Engl. 2014;53:10728–10732. doi: 10.1002/anie.201402679. [DOI] [PubMed] [Google Scholar]
- 166.Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, Ro J, Wagner GR, Green MF, Madsen AS, Schmiesing J, Peterson BS, Xu G, Ilkayeva OR, Muehlbauer MJ, Braulke T, Muhlhausen C, Backos DS, Olsen CA, McGuire PJ, Pletcher SD, Lombard DB, Hirschey MD, Zhao Y. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, Zwaans BM, Tishkoff D, Ho L, Lombard D, He TC, Dai J, Verdin E, Ye Y, Zhao Y. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011;10(M111):012658. doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pougovkina O, Te Brinke H, Wanders RJ, Houten SM, de Boer VC. Aberrant protein acylation is a common observation in inborn errors of acyl-CoA metabolism. J Inherit Metab Dis. 2014;37:709–714. doi: 10.1007/s10545-014-9684-9. [DOI] [PubMed] [Google Scholar]
- 169.Yu J, Sadhukhan S, Noriega LG, Moullan N, He B, Weiss RS, Lin H, Schoonjans K, Auwerx J. Metabolic Characterization of a Sirt5 deficient mouse model. Sci Rep. 2013;3:2806. doi: 10.1038/srep02806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Buler M, Aatsinki SM, Izzi V, Uusimaa J, Hakkola J. SIRT5 is under the control of PGC-1alpha and AMPK and is involved in regulation of mitochondrial energy metabolism. FASEB J. 2014;28:3225–3237. doi: 10.1096/fj.13-245241. [DOI] [PubMed] [Google Scholar]
- 171.Weinert BT, Scholz C, Wagner SA, Iesmantavicius V, Su D, Daniel JA, Choudhary C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013;4:842–851. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 172.Fränzel B, Fischer F, Steegborn C, Wolters DA (2013) Proteinase K has deep impact on quantitative acetylation studies. J Proteome Res (in press)
- 173.Schutkowski M, Fischer F, Roessler C, Steegborn C (2014) New assays and approaches for discovery and design of Sirtuin modulators. Exp Opin Drug Disc (in press) [DOI] [PubMed]
- 174.Lin ZF, Xu HB, Wang JY, Lin Q, Ruan Z, Liu FB, Jin W, Huang HH, Chen X. SIRT5 desuccinylates and activates SOD1 to eliminate ROS. Biochem Biophys Res Commun. 2013;441:191–195. doi: 10.1016/j.bbrc.2013.10.033. [DOI] [PubMed] [Google Scholar]
- 175.Zhao K, Chai X, Marmorstein R. Structure of the yeast Hst2 protein deacetylase in ternary complex with 2′-O-acetyl ADP ribose and histone peptide. Structure. 2003;11:1403–1411. doi: 10.1016/j.str.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 176.Tennen RI, Berber E, Chua KF. Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization. Mech Ageing Dev. 2010;131:185–192. doi: 10.1016/j.mad.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lakshminarasimhan M, Curth U, Moniot S, Mosalaganti S, Raunser S, Steegborn C. Molecular architecture of the human protein deacetylase Sirt1 and its regulation by AROS and resveratrol. Biosci Rep. 2013;33:e00037. doi: 10.1042/BSR20120121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pan M, Yuan H, Brent M, Ding EC, Marmorstein R. SIRT1 contains N- and C-terminal regions that potentiate deacetylase activity. J Biol Chem. 2012;287:2468–2476. doi: 10.1074/jbc.M111.285031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gertz M, Fischer F, Nguyen GT, Lakshminarasimhan M, Schutkowski M, Weyand M, Steegborn C. Ex-527 inhibits sirtuins by exploiting their unique NAD+-dependent deacetylation mechanism. Proc Natl Acad Sci USA. 2013;110:E2772–E2781. doi: 10.1073/pnas.1303628110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhou Y, Zhang H, He B, Du J, Lin H, Cerione RA, Hao Q. The bicyclic intermediate structure provides insights into the desuccinylation mechanism of human sirtuin 5 (SIRT5) J Biol Chem. 2012;287:28307–28314. doi: 10.1074/jbc.M112.384511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Moniot S, Schutkowski M, Steegborn C. Crystal structure analysis of human Sirt2 and its ADP-ribose complex. J Struct Biol. 2013;182:136–143. doi: 10.1016/j.jsb.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 182.Moniot S, Weyand M, Steegborn C. Structures, substrates, and regulators of Mammalian sirtuins—opportunities and challenges for drug development. Front Pharmacol. 2012;3:16. doi: 10.3389/fphar.2012.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sanders BD, Jackson B, Marmorstein R. Structural basis for sirtuin function: what we know and what we don’t. Biochim Biophys Acta. 2010;1804:1604–1616. doi: 10.1016/j.bbapap.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Jin L, Wei W, Jiang Y, Peng H, Cai J, Mao C, Dai H, Choy W, Bemis JE, Jirousek MR, Milne JC, Westphal CH, Perni RB. Crystal structures of human SIRT3 displaying substrate-induced conformational changes. J Biol Chem. 2009;284:24394–24405. doi: 10.1074/jbc.M109.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Chang JH, Kim HC, Hwang KY, Lee JW, Jackson SP, Bell SD, Cho Y. Structural basis for the NAD-dependent deacetylase mechanism of Sir2. J Biol Chem. 2002;277:34489–34498. doi: 10.1074/jbc.M205460200. [DOI] [PubMed] [Google Scholar]
- 186.Hoff KG, Avalos JL, Sens K, Wolberger C. Insights into the sirtuin mechanism from ternary complexes containing NAD+ and acetylated peptide. Structure. 2006;14:1231–1240. doi: 10.1016/j.str.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 187.Nguyen GT, Schaefer S, Gertz M, Weyand M, Steegborn C. Structures of human sirtuin 3 complexes with ADP-ribose and with carba-NAD(+) and SRT1720: binding details and inhibition mechanism. Acta Crystallogr D Biol Crystallogr. 2013;69:1423–1432. doi: 10.1107/S0907444913015448. [DOI] [PubMed] [Google Scholar]
- 188.Feldman JL, Dittenhafer-Reed KE, Kudo N, Thelen JN, Ito A, Yoshida M, Denu JM. Kinetic and structural basis for acyl-group selectivity and NAD(+) dependence in sirtuin-catalyzed deacylation. Biochemistry. 2015;54:3037–3050. doi: 10.1021/acs.biochem.5b00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lakshminarasimhan M, Steegborn C. Emerging mitochondrial signaling mechanisms in physiology, aging processes, and as drug targets. Exp Gerontol. 2011;46:174–177. doi: 10.1016/j.exger.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 190.Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 191.Knight JR, Allison SJ, Milner J. Active regulator of SIRT1 is required for cancer cell survival but not for SIRT1 activity. Open Biol. 2013;3:130130. doi: 10.1098/rsob.130130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 193.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 194.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sauve AA. Sirtuin chemical mechanisms. Biochim Biophys Acta. 1804;2010:1591–1603. doi: 10.1016/j.bbapap.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae . Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Chen L. Medicinal chemistry of sirtuin inhibitors. Curr Med Chem. 2011;18:1936–1946. doi: 10.2174/092986711795590057. [DOI] [PubMed] [Google Scholar]
- 200.Cen Y. Sirtuins inhibitors: the approach to affinity and selectivity. Biochim Biophys Acta. 2010;1804:1635–1644. doi: 10.1016/j.bbapap.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 201.Gertz M, Nguyen GT, Fischer F, Suenkel B, Schlicker C, Franzel B, Tomaschewski J, Aladini F, Becker C, Wolters D, Steegborn C. A molecular mechanism for direct sirtuin activation by resveratrol. PLoS One. 2012;7:e49761. doi: 10.1371/journal.pone.0049761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Disch JS, Evindar G, Chiu CH, Blum CA, Dai H, Jin L, Schuman E, Lind KE, Belyanskaya SL, Deng J, Coppo F, Aquilani L, Graybill TL, Cuozzo JW, Lavu S, Mao C, Vlasuk GP, Perni RB. Discovery of thieno[3,2-d]pyrimidine-6-carboxamides as potent inhibitors of SIRT1, SIRT2, and SIRT3. J Med Chem. 2013;56:3666–3679. doi: 10.1021/jm400204k. [DOI] [PubMed] [Google Scholar]
- 203.Nguyen GT, Gertz M, Steegborn C. Crystal structures of sirt3 complexes with 4′-bromo-resveratrol reveal binding sites and inhibition mechanism. Chem Biol. 2013;20:1375–1385. doi: 10.1016/j.chembiol.2013.09.019. [DOI] [PubMed] [Google Scholar]
- 204.Napper AD, Hixon J, McDonagh T, Keavey K, Pons JF, Barker J, Yau WT, Amouzegh P, Flegg A, Hamelin E, Thomas RJ, Kates M, Jones S, Navia MA, Saunders JO, DiStefano PS, Curtis R. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J Med Chem. 2005;48:8045–8054. doi: 10.1021/jm050522v. [DOI] [PubMed] [Google Scholar]
- 205.Yasuda M, Wilson DR, Fugmann SD, Moaddel R. Synthesis and characterization of SIRT6 protein coated magnetic beads: identification of a novel inhibitor of SIRT6 deacetylase from medicinal plant extracts. Anal Chem. 2011;83:7400–7407. doi: 10.1021/ac201403y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Suenkel B, Fischer F, Steegborn C. Inhibition of the human deacylase Sirtuin 5 by the indole GW5074. Bioorg Med Chem Lett. 2013;23:143–146. doi: 10.1016/j.bmcl.2012.10.136. [DOI] [PubMed] [Google Scholar]
- 207.Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- 208.Barger JL, Kayo T, Vann JM, Arias EB, Wang J, Hacker TA, Wang Y, Raederstorff D, Morrow JD, Leeuwenburgh C, Allison DB, Saupe KW, Cartee GD, Weindruch R, Prolla TA. A low dose of dietary resveratrol partially mimics caloric restriction and retards aging parameters in mice. PLoS One. 2008;3:e2264. doi: 10.1371/journal.pone.0002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Pirola L, Frojdo S. Resveratrol: one molecule, many targets. IUBMB Life. 2008;60:323–332. doi: 10.1002/iub.47. [DOI] [PubMed] [Google Scholar]
- 211.Baur JA. Biochemical effects of SIRT1 activators. Biochim Biophys Acta. 2010;1804:1626–1634. doi: 10.1016/j.bbapap.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, Bedalov A, Kennedy BK. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 213.Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 214.Yang H, Baur JA, Chen A, Miller C, Adams JK, Kisielewski A, Howitz KT, Zipkin RE, Sinclair DA. Design and synthesis of compounds that extend yeast replicative lifespan. Aging Cell. 2007;6:35–43. doi: 10.1111/j.1474-9726.2006.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Dai H, Case AW, Riera TV, Considine T, Lee JE, Hamuro Y, Zhao H, Jiang Y, Sweitzer SM, Pietrak B, Schwartz B, Blum CA, Disch JS, Caldwell R, Szczepankiewicz B, Oalmann C, Yee Ng P, White BH, Casaubon R, Narayan R, Koppetsch K, Bourbonais F, Wu B, Wang J, Qian D, Jiang F, Mao C, Wang M, Hu E, Wu JC, Perni RB, Vlasuk GP, Ellis JL. Crystallographic structure of a small molecule SIRT1 activator-enzyme complex. Nat Commun. 2015;6:7645. doi: 10.1038/ncomms8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Cao D, Wang M, Qiu X, Liu D, Jiang H, Yang N, Xu RM. Structural basis for allosteric, substrate-dependent stimulation of SIRT1 activity by resveratrol. Genes Dev. 2015;29:1316–1325. doi: 10.1101/gad.265462.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Patel K, Sherrill J, Mrksich M, Scholle MD. Discovery of SIRT3 Inhibitors Using SAMDI Mass Spectrometry. J Biomol Screen. 2015;20:842–848. doi: 10.1177/1087057115588512. [DOI] [PubMed] [Google Scholar]
- 218.Guetschow ED, Kumar S, Lombard DB, Kennedy RT. Identification of sirtuin 5 inhibitors by ultrafast microchip electrophoresis using nanoliter volume samples. Anal Bioanal Chem. 2016;408:721–731. doi: 10.1007/s00216-015-9206-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Biasutto L, Mattarei A, Marotta E, Bradaschia A, Sassi N, Garbisa S, Zoratti M, Paradisi C. Development of mitochondria-targeted derivatives of resveratrol. Bioorg Med Chem Lett. 2008;18:5594–5597. doi: 10.1016/j.bmcl.2008.08.100. [DOI] [PubMed] [Google Scholar]
- 220.Mai A, Valente S, Meade S, Carafa V, Tardugno M, Nebbioso A, Galmozzi A, Mitro N, De Fabiani E, Altucci L, Kazantsev A. Study of 1,4-dihydropyridine structural scaffold: discovery of novel sirtuin activators and inhibitors. J Med Chem. 2009;52:5496–5504. doi: 10.1021/jm9008289. [DOI] [PubMed] [Google Scholar]
- 221.Pillai VB, Samant S, Sundaresan NR, Raghuraman H, Kim G, Bonner MY, Arbiser JL, Walker DI, Jones DP, Gius D, Gupta MP. Honokiol blocks and reverses cardiac hypertrophy in mice by activating mitochondrial Sirt3. Nat Commun. 2015;6:6656. doi: 10.1038/ncomms7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Schuetz A, Min J, Antoshenko T, Wang CL, Allali-Hassani A, Dong A, Loppnau P, Vedadi M, Bochkarev A, Sternglanz R, Plotnikov AN. Structural basis of inhibition of the human NAD+-dependent deacetylase SIRT5 by suramin. Structure. 2007;15:377–389. doi: 10.1016/j.str.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 223.Trapp J, Meier R, Hongwiset D, Kassack MU, Sippl W, Jung M. Structure-activity studies on suramin analogues as inhibitors of NAD+-dependent histone deacetylases (sirtuins) ChemMedChem. 2007;2:1419–1431. doi: 10.1002/cmdc.200700003. [DOI] [PubMed] [Google Scholar]
- 224.McGeary RP, Bennett AJ, Tran QB, Cosgrove KL, Ross BP. Suramin: clinical uses and structure-activity relationships. Mini Rev Med Chem. 2008;8:1384–1394. doi: 10.2174/138955708786369573. [DOI] [PubMed] [Google Scholar]
- 225.Salo HS, Laitinen T, Poso A, Jarho E, Lahtela-Kakkonen M. Identification of novel SIRT3 inhibitor scaffolds by virtual screening. Bioorg Med Chem Lett. 2013;23:2990–2995. doi: 10.1016/j.bmcl.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 226.Schlicker C, Boanca G, Lakshminarasimhan M, Steegborn C. Structure-based development of novel sirtuin inhibitors. Aging Albany NY. 2011;3:852–857. doi: 10.18632/aging.100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Smith BC, Denu JM. Mechanism-based inhibition of Sir2 deacetylases by thioacetyl-lysine peptide. Biochemistry. 2007;46:14478–14486. doi: 10.1021/bi7013294. [DOI] [PubMed] [Google Scholar]
- 228.Hawse WF, Hoff KG, Fatkins DG, Daines A, Zubkova OV, Schramm VL, Zheng W, Wolberger C. Structural insights into intermediate steps in the Sir2 deacetylation reaction. Structure. 2008;16:1368–1377. doi: 10.1016/j.str.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Fatkins DG, Zheng W. Substituting N(epsilon)-thioacetyl-lysine for N(epsilon)-acetyl-lysine in peptide substrates as a general approach to inhibiting human NAD(+)-dependent protein deacetylases. Int J Mol Sci. 2008;9:1–11. doi: 10.3390/ijms9010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.He B, Du J, Lin H. Thiosuccinyl peptides as Sirt5-specific inhibitors. J Am Chem Soc. 2012;134:1922–1925. doi: 10.1021/ja2090417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Asaba T, Suzuki T, Ueda R, Tsumoto H, Nakagawa H, Miyata N. Inhibition of human sirtuins by in situ generation of an acetylated lysine-ADP-ribose conjugate. J Am Chem Soc. 2009;131:6989–6996. doi: 10.1021/ja807083y. [DOI] [PubMed] [Google Scholar]
- 232.Suzuki T, Asaba T, Imai E, Tsumoto H, Nakagawa H, Miyata N. Identification of a cell-active non-peptide sirtuin inhibitor containing N-thioacetyl lysine. Bioorg Med Chem Lett. 2009;19:5670–5672. doi: 10.1016/j.bmcl.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 233.Maurer B, Rumpf T, Scharfe M, Stolfa DA, Schmitt ML, He W, Verdin E, Sippl W, Jung M. Inhibitors of the NAD dependent protein desuccinylase and demalonylase Sirt5. ACS Med. Chem. Lett. 2012;3:1050–1053. doi: 10.1021/ml3002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Mahajan SS, Scian M, Sripathy S, Posakony J, Lao U, Loe TK, Leko V, Thalhofer A, Schuler AD, Bedalov A, Simon JA. Development of pyrazolone and isoxazol-5-one cambinol analogues as sirtuin inhibitors. J Med Chem. 2014;57:3283–3294. doi: 10.1021/jm4018064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Jin L, Galonek H, Israelian K, Choy W, Morrison M, Xia Y, Wang X, Xu Y, Yang Y, Smith JJ, Hoffmann E, Carney DP, Perni RB, Jirousek MR, Bemis JE, Milne JC, Sinclair DA, Westphal CH. Biochemical characterization, localization, and tissue distribution of the longer form of mouse SIRT3. Protein Sci. 2009;18:514–525. doi: 10.1002/pro.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Trapp J, Jochum A, Meier R, Saunders L, Marshall B, Kunick C, Verdin E, Goekjian P, Sippl W, Jung M. Adenosine mimetics as inhibitors of NAD+-dependent histone deacetylases, from kinase to sirtuin inhibition. J Med Chem. 2006;49:7307–7316. doi: 10.1021/jm060118b. [DOI] [PubMed] [Google Scholar]
- 237.Huber K, Schemies J, Uciechowska U, Wagner JM, Rumpf T, Lewrick F, Suss R, Sippl W, Jung M, Bracher F. Novel 3-arylideneindolin-2-ones as inhibitors of NAD+ -dependent histone deacetylases (sirtuins) J Med Chem. 2010;53:1383–1386. doi: 10.1021/jm901055u. [DOI] [PubMed] [Google Scholar]
- 238.Fernandez-Marcos PJ, Jeninga EH, Canto C, Harach T, de Boer VC, Andreux P, Moullan N, Pirinen E, Yamamoto H, Houten SM, Schoonjans K, Auwerx J. Muscle or liver-specific Sirt3 deficiency induces hyperacetylation of mitochondrial proteins without affecting global metabolic homeostasis. Sci Rep. 2012;2:425. doi: 10.1038/srep00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Szczepankiewicz BG, Dai H, Koppetsch KJ, Qian D, Jiang F, Mao C, Perni RB. Synthesis of carba-NAD and the structures of its ternary complexes with SIRT3 and SIRT5. J Org Chem. 2012;77:7319–7329. doi: 10.1021/jo301067e. [DOI] [PubMed] [Google Scholar]