Abstract
Coxsackievirus B3 (CVB3) is the primary pathogen of viral myocarditis. Upon infection, CVB3 exploits the host cellular machineries, such as chaperone proteins, to benefit its own infection cycles. Inducible heat shock 70-kDa proteins (Hsp70s) are chaperone proteins induced by various cellular stress conditions. The internal ribosomal entry site (IRES) within Hsp70 mRNA allows Hsp70 to be translated cap-independently during CVB3 infection when global cap-dependent translation is compromised. The Hsp70 protein family contains two major members, Hsp70-1 and Hsp70-2. This study showed that Hsp70-1, but not Hsp70-2, was upregulated during CVB3 infection both in vitro and in vivo. Then a novel mechanism of Hsp70-1 induction was revealed in which CaMKIIγ is activated by CVB3 replication and leads to phosphorylation of heat shock factor 1 (HSF1) specifically at Serine 230, which enhances Hsp70-1 transcription. Meanwhile, phosphorylation of Ser230 induces translocation of HSF1 from the cytoplasm to nucleus, thus blocking the ERK1/2-mediated phosphorylation of HSF1 at Ser307, a negative regulatory process of Hsp70 transcription, further contributing to Hsp70-1 upregulation. Finally, we demonstrated that Hsp70-1 upregulation, in turn, stabilizes CVB3 genome via the AU-rich element (ARE) harbored in the 3′ untranslated region of CVB3 genomic RNA.
Keywords: RNA decay, CaMKII, ARE, AUF1
Introduction
Coxsackievirus B3 (CVB3), a member of Enteroviral genus in the family of Picornaviridae, is one of the primary causative pathogens of viral myocarditis, which often leads to dilated cardiomyopathy (DCM) at the end stage [1–3]. CVB3 genome is a positive, single-strand RNA molecule encoding a single long open reading frame flanked by the 5′ and 3′ untranslated regions (UTRs), which can be translated directly using eukaryotic translational machinery. The 5′ end of the genome does not contain a 7-methylguanylate cap structure but instead is linked to a viral protein VPg. The 5′UTR harbors an internal ribosome entry site (IRES) for viral cap-independent, IRES-driven translational initiation [4, 5]. During infection, CVB3 inhibits the cap-dependent translation of the host cell to release translational machinery for its own translation [6, 7]. The 3′UTR harbors an AU-rich element (ARE) attached with a poly-A tail [8]. This structure is believed to play an important role in facilitating viral transcription and translation.
Inducible heat shock 70-kDa protein (Hsp70) is a key component of cellular chaperone system involved in response to various cellular stresses [9]. The Hsp70 protein family contains two major members, Hsp70-1 and Hsp70-2, which are encoded by gene HSPA1A and HSPA1B, respectively. The proteins of Hsp70-1 and Hsp70-2 differ on two amino acids and both of them are induced in heat shock, but only Hsp70-2 is stimulated in response to hypertonicity since only HSPA1B contains a tonicity-responsive enhancer (TonE) [10]. Hsp70 has been reported to be induced in viral infection [11] and acts as a regulatory factor, either negative or positive, in viral life cycle [12–15]. Similar to CVB3 genomic RNA, Hsp70 mRNA also contains an IRES element, which makes it possible for Hsp70 to be translated during CVB3 infection when the cap-dependent translation is compromised. Heat shock factor 1 (HSF1) is the major transcriptional factor of Hsp70-1 reported in various stress conditions [16, 17]. The transcriptional activity of HSF1 is modulated at multiple phosphorylation sites. Two serine (Ser) sites, Ser230 and Ser307, are phosphorylated and involved in the modulation of the activity of HSF1 primarily [18]. The phosphorylation at Ser230 is catalyzed by active calcium/calmodulin-dependent protein kinase II (CaMKII) [18], while activation of CaMKII is initialized by elevated intracellular calcium/calmodulin (Ca2+/CaM) and then sustained by threonine (Thr) phosphorylation [19–21]. During CVB3 infection, free cytosolic calcium is elevated [22, 23], which may result in persistent CaMKII activation. Unlike Ser230, phosphorylation at Ser307 represses transcriptional activity of HSF1 [24]. Ser307 phosphorylation is catalyzed by extracellular signal-regulated kinase (ERK) [25] and ERK has been reported to be activated during CVB3 infection [26, 27]. However, to our knowledge, the mechanism by which the Hsp70-1 upregulation during CVB3 infection and its effect on CVB3 replication have not been studied yet. As for Hsp70-2, its transcription is stimulated by nuclear factor of activated T-cells (NFAT5) via the interaction between NFAT5 and TonE [10].
Hsp70 has been reported to stabilize messenger RNAs (mRNAs) containing adenosine-uridine-rich elements (AREs) by preventing mRNA degradation. One explanation for Hsp70-mediated stabilization of ARE-containing mRNAs focuses on the interactions between Hsp70 and ARE/poly(U)-binding/degradation factor 1 (AUF1). It was reported that degradation of ARE-containing mRNAs is induced by the binding of AUF1 on the ARE site [28]. AUF1 has four isoforms (p37, p40, p42 and p45) due to alternative splicing, among which p37AUF1 and p40AUF1 are most strongly associated with ARE-mRNA decay [29]. Hsp70 can sequester AUF1 in nucleus to avoid the AUF1 and mRNA interaction in the cytoplasm [30]. Another explanation is that Hsp70-1, but not Hsp70-2, binds to ARE directly and stabilize the mRNA, which is supported by the highly selective binding of Hsp70-1 and ARE [31]. The genomic RNA of CVB3 contains an ARE site within the 3′UTR and it has been reported that AUF1 can bind to CVB3 genomic RNA (24); however, it is still unclear whether Hsp70 can modulate the stability of CVB3 genomic RNA during infection.
In this study, we demonstrated that Hsp70-1, but not Hsp70-2, was upregulated during CVB3 infection. We further revealed a novel mechanism by which CVB3 infection activates CaMKIIγ and selectively phosphorylates HSF1 at Ser230, leading to enhanced Hsp70-1 transcription. Meanwhile, phosphorylation of Ser230 induces translocation of HSF1 from the cytosol to nucleus, thus blocking the ERK1/2-mediated phosphorylation of HSF1 at Ser307, a negative regulatory process on Hsp70-1 transcription, further contributing to upregulation of Hsp70-1. Finally, we demonstrated that Hsp70-1 upregulation, in turn, facilitates CVB3 replication by stabilizing viral genomic RNA via AUF1 and the ARE of CVB3 genome. These data indicate that Hsp70-related cellular chaperone system may be hijacked by CVB3 to favor the viral replication.
Methods
Animals, cell culture and viral infection
HeLa cells (ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10 % fetal bovine serum (FBS) (Sigma). The HL-1 cell line, a mouse cardiac muscle cell line established from a cardiomyocyte tumor lineage, was a gift from Dr. William C. Claycomb (Louisiana State University Health Science Center). HL-1 cells were maintained in Claycomb medium (Sigma) supplemented with 10 % fetal bovine serum (FBS) (JRH Biosciences), 0.1 mM norepinephrine (Sigma) and 2 mM l-glutamine (Invitrogen). SV40 immortalized human cardiomyocytes were purchased from Applied Biological Materials (Richmond, BC, Canada) and cultured in Prigrow I medium with 10 % FBS. Primary rat neonatal cardiomyocytes were isolated from rat pups using commercial Neonatal Cardiomyocyte Isolation Kit (Cellutron) according to the protocol provided. Briefly, hearts were dissected from 1-day-old Sprague–Dawley rats and transferred into a sterile beaker. Each heart was digested and stirred in the beaker at 37 °C for 12 min. The supernatant was then transferred to a new sterile tube and spun at 1200×g for 1 min. The cell pellets were then resuspended in D3 buffer and the cells were seeded onto an uncoated plate, which was incubated at 37 °C for 1 h in a CO2 incubator to allow the attaching of cardiac fibroblasts. The unattached cardiomyocytes were transferred onto precoated plates with NS medium supplemented with 10 % FBS. After overnight culturing, the NS medium was replaced with a serum-free NW (without serum) medium. The cardiomyocyte cultures were ready for experiments 48 h after the initial plating. All these cells were sustained in a humidified incubator supplemented with 5 % CO2 at 37 °C. For heat shock, HeLa cells were incubated at 42 °C for different durations.
CVB3 (CG) strain was obtained from Dr. Charles Gauntt (University of Texas Health Science Center) and propagated in HeLa cells. Viral stock was prepared from the cells by three freeze–thaw cycles followed by centrifugation to remove cell debris and stored at −80 °C. The titer of virus stock was determined by plaque assay as described below. Cell cultures were infected with CVB3 for 1 h (HeLa) or 1.5 h (HL-1, SV40 and primary rat neonatal cardiomyocytes) in serum-free medium, washed with phosphate-buffered saline (PBS), and then replenished with fresh medium containing FBS. The total proteins of CVB3-infected cells were extracted by lysing the cells with RIPA buffer (Santa Cruz) at different time points post-infection (pi). Male A/J mice (4 weeks old) were purchased from Jackson Laboratory. Mice were infected by intraperitoneal inoculation with 5 × 103 plaque-forming unit (pfu) of CVB3 or sham-infected with PBS. Heart tissues were collected at day 7 pi for immunostaining.
Viral plaque assay
Samples were freeze-thawed and then centrifuged (4000×g) to isolate viruses. HeLa cells were seeded onto 6-well plates (8 × 105 cells/well) and incubated at 37 °C for 20 h to a confluence of approximately 90 % and then washed with PBS and overlaid with 800 µl of virus-containing samples serially diluted in cell culture medium. After a viral adsorption period of 60 min at 37 °C, the supernatant was removed and the cells overlaid with 2 ml of sterilized soft Bacto-agar-minimal essential medium, cultured at 37 °C for 72 h, fixed with Carnoy’s fixative for 30 min, and stained with 1 % crystal violet. The plaques were counted and viral pfu per ml calculated.
UV irradiation of CVB3
One mL of CVB3 stock in a 2-mL tube was kept on ice. UV irradiation was conducted in a UV Stratalinker 1800 (Stratagene) for 30 min with the virus tube kept 5-cm from the UV bulb. The viruses were tested for successful irradiation by infection of HeLa cells and then Western blot detection of the absence of CVB3 VP1 protein.
RNA extraction and quantitative real-time PCR
Total cellular RNAs were extracted using RNeasy mini kit (Qiagen) according to the manufacturer’s instructions. cDNAs were then synthesized by reverse transcription using SuperScript III First-Strand Synthesis System (Invitrogen) and detected by quantitative real-time PCR (qPCR) using QuantiTect SYBR Green PCR kit (Qiagen). The mRNA level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was detected as the endogenous control. All qPCR experiments were performed in triplicates with the no-template as a negative control. The primers for the q-RT-PCR are shown in Table 1.
Table 1.
Primers used in the study
| Experiment | Primer | Sequence |
|---|---|---|
| Q-RT-PCR | Human Hsp70-1 forward | TGCATTTCCTAGTATTTCTGTTTG |
| Human Hsp70-1 reverse | AGAAATAGTCGTAAGATGGCAG | |
| Human Hsp70-2 forward | TGTTTGTCTTTGAGGTGGAC | |
| Human Hsp70-2 reverse | AAGAATTCTAATGAACATATCGGTTG | |
| Mouse Hsp70 forward | GCCTGATCGGCCGCAAGTT | |
| Mouse Hsp70 reverse | GGAAGGGCCAGTGCTTCAT | |
| Renilla luciferase forward | GCAGAAGTTGGTCGTGAGG | |
| Renilla luciferase reverse | TCATCCGTTTCCTTTGTTCTG | |
| Human GAPDH forward | AATCCCATCACCATCTTCCA | |
| Human GAPDH reverse | TGGACTCCACGACGTACTCA | |
| Mouse GAPDH forward | GGCAAATTCAACGGCACAGT | |
| Mouse GAPDH reverse | AGATGGTGATGGGCTTCCC | |
| CVB3 2A forward | GCTTTGCAGACATCCGTGATC | |
| CVB3 2A reverse | CAAGCTGTGTTCCACATAGTCCTTCA | |
| Molecular cloning | CVB3 3′UTR forward/EcoRI | GCCTTAAGAAGTGGTTGGACTCCTTTTAG |
| CVB3 3′UTR reverse/EcoRI | GCCTTAAGTTTTTTTTTTCCGCACCGAATGCGGAG | |
| Mutant CVB3 3′UTR forward/EcoRI | GCCTTAAGAAGTGGTTGGACTCCTTTTAGATTAGAGACAATTTGAAATACGGGAGATTGGCTTAACCCTAC |
Western blot analysis
Cells were washed with cold PBS before the addition of an appropriate volume of RIPA lysis buffer (Santa Cruz). After incubation for 20 min on ice, the cell lysates were centrifuged at 13,000×g for 15 min at 4 °C, and protein-containing supernatant was collected. The isolated proteins were separated by 10 % SDS-PAGE and transferred onto nitrocellulose membranes. Membranes were blocked with 5 % skim milk in TBST and incubated with one of the following primary antibodies overnight: monoclonal mouse anti-VP1 (Novocastra); monoclonal mouse anti-Hsp70 (Enzo Life Sciences); monoclonal mouse anti-β-Actin, monoclonal mouse anti-Hsp70/Hsc70, monoclonal mouse anti-HSF1, polyclonal rabbit anti-Histone H1, polyclonal rabbit anti-phosphorylated-HSF1 (Ser230), polyclonal rabbit anti-phosphorylated-HSF1 (Ser307), polyclonal rabbit anti-ERK1/2 and polyclonal rabbit anti-NFAT5 (Santa Cruz); monoclonal rabbit anti-phosphorylated-ERK1/2 (Thr202/Tyr204), monoclonal rabbit anti-CaMKII (pan) and polyclonal rabbit anti-pCaMKII-Thr286 (Cell Signaling). After several washes with TBST, each blot was further incubated with an appropriate secondary antibody (goat anti-mouse or donkey anti-rabbit) conjugated to horseradish peroxidase (Amersham). Detection was carried out by enhanced chemiluminescence (Amersham) as per the manufacturer’s instructions. β-actin was detected as a loading control. Signal intensities were quantified by using the ImageJ (NIH) program and normalized to the control samples (set as 1.00).
Immunohistochemistry (IHC)
The collected mouse hearts were fixed in 10 % buffered formalin and embedded in paraffin. The IHC analysis was performed using the MACH 4™ Universal AP Polymer Kit (Biocare Medical) as per the manufacturer’s instructions. Brief, a set of 4-μm-thick serial sections of the ventricular wall from each sample were dewaxed in two changes of CitriSolution (Biocare Medical) and hydrated by sequential incubation with 100, 90, 70 % isopropanol and distilled water. The sections were then immersed in Tris–EDTA (pH 6.0) and autoclaved at 120 °C for 20 min to retrieve the antigen. Non-specific antibody binding was blocked with 1 % bovine serum albumin (BSA) (Sigma) in Tris-buffered saline (TBS, pH 7.6) for 15 min at room temperature. After blocking, the sections were incubated overnight at 4 °C with the Hsp70 antibody (Cell Signaling, 1:500 dilution) mentioned above. After the first incubation, the sections were washed with TBS, incubated with MACH 4™ Probe for 10 min and then with MACH 4™ MR AP-Polymer for another 10 min at room temperature. Finally, the sections were again rinsed in TBS. The slides were developed with Vulcan Fast Red Chromogen 2 (Biocare Medical). After development, the slides were rinsed, counterstained with Mayer Haematoxylin for 5 s and dipped in saturated lithium carbonate for 5 s. The images were captured using a Nikon Eclipse E600 microscope.
Transfection of DNA plasmids and siRNAs
All the siRNAs were purchased from Santa Cruz Biotechnology and transfected into cells using Oligofectamine™ (Life Technologies) according to the manufacturer’s instructions. Briefly, 2 × 105 HeLa cells were grown at 37 °C overnight to 30–40 % confluence in 6-well plates, washed with PBS and overlaid for 6 h with transfection complex containing siRNAs and Oligofectamine. The transfection medium was then replaced with DMEM containing 10 % FBS and the incubation was continued for 48 h. The plasmids pEGFP-Hsp70-1 and pEGFP-Hsp70-1 (K71E) were a gift from Dr. Lois Greene (Addgene plasmid # 15216) [32]. The plasimds pFLAG-AUF1s was a gift from Dr. Robert J. Schneider [33]. These plasmids were transfected using the same procedures as those described for siRNAs except Lipofectamine™ 2000 (Life Technologies) was used as transfection reagent and the initial cell confluence was 80–90 %. The following analyses were performed at 24 h or 36 h post-transfection (pt).
Immunofluorescence and confocal microscopy
Cells cultured on glass cover slips (Thermo Fisher) were washed with PBS and fixed and permeabilized with methanol/acetone (1:1) for 20 min at −20 °C. Cells were then washed with TBS twice and blocked with 2.5 % BSA in TBS for 1 h at room temperature followed by incubation with monoclonal mouse anti-HSF1 antibody (Santa Cruz) diluted in blocking buffer overnight at 4 °C. Cells were then washed with TBS five times at room temperature. Slides were stained with goat anti-rabbit IgG (H + L) labeled with ALEXA Fluor 488 and then incubated for 1 h at room temperature. After final wash with TBS, the slides were stained with DAPI (DAKO) and mounted onto microscope glass slides (Thermo Fisher) with nail oil. Images were captured using a Leica AOBS SP2 confocal microscope (Leica, Allendale, NJ) and analyzed by using the Volocity software as described previously [34].
Reporter construction, dual-luciferase assay and mRNA turnover assay
The corresponding DNA fragments of wild-type (WT) or mutant CVB3 3′UTR were amplified by PCR using specific primers (Table 1) targeting the cDNA template of the CVB3-genome. The synthesized DNA fragments were inserted into the EcoRI restriction site of the Dual-Luciferase Expression Vector C49, a kind gift from Dr. Joanna Floros’s laboratory. C49 plasmid contains two tandem open reading frames encoding firefly luciferase and Renilla luciferase, respectively, with an EcoR I restriction site in between. We constructed our reporter plasmids by inserting the WT or mutant CVB3 3′UTR to the downstream region of Renilla luciferase coding sequence and the SV40 promoter to the upstream region of Renilla luciferase coding sequence, generating C49-WT-CVB3-3′UTR and C49-WT-CVB3-3′UTR, respectively. The reporter plasmids were co-transfected with Hsp70-1 siRNAss into HeLa cells. At 48 h pt, the cell lysates were used for luciferase assay to determine the relative luciferase activity (Renilla/Firefly) by using the Dual-Luciferase® Reporter Assay System (Promega) as per the manufacturer’s instructions. Meanwhile, another batch of cells were subjected to the same transfection and then treated by actinomycin D at a dose of 2.5 μg/mL (Santa Cruz) for different durations or by 0.1 % DMSO as a control, in order to do mRNA turnover assay. Then the total cellular RNAs were extracted, reversely transcribed and subjected to qPCR to detect the mRNA levels of Renilla luciferase as described above. Each treatment was verified by three biological repeats.
Statistical analysis
The Student’s t test was employed to analyze the data. The results are expressed as means ± standard deviations (SD) of three independent experiments. A p value less than 0.05 was considered statistically significant.
Results
Protein level of Hsp70 is increased during CVB3 infection in vitro and in vivo
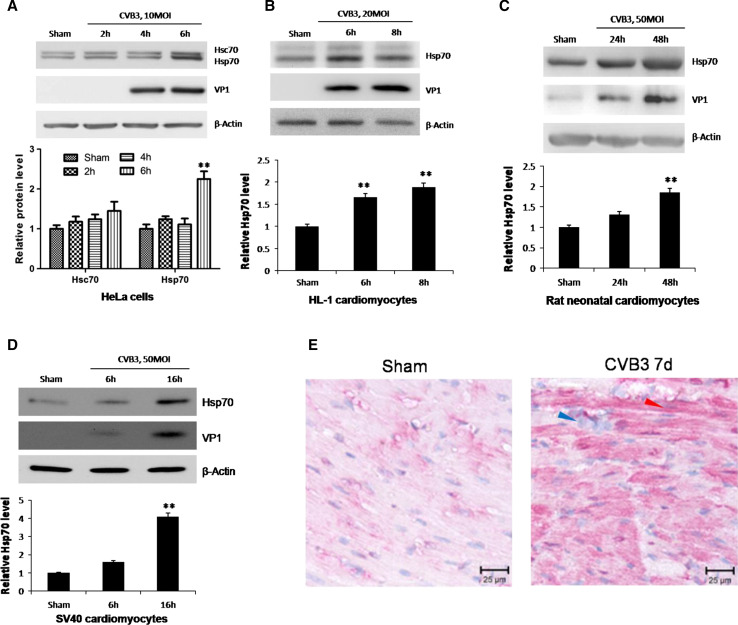
To examine the association between viral infection phases and Hsp70 expression, we detected mRNA and protein levels of Hsp70 in HeLa cells at different time points pi of CVB3 at a multiplicity of infection (MOI) of 10. Hsp70 protein was evaluated by Western blot analysis using an Hsp70/Hsc70-specific antibody (Fig. 1a, the upper panel). The quantification of Hsp70 was conducted by densitometric analysis and normalized to β-actin. We observed a 100 % increase of Hsp70 protein at 6 h pi. However, Hsc70, a constitutively expressed cognate of Hsp70, showed less than 50 % changes during CVB3 infection (Fig. 1a, the lower panel). To further verify the change of Hsp70 expression in cardiomyocytes, we detected the Hsp70 protein levels in HL-1 mouse cardiomyocytes, SV40 immortalized human cardiomyocytes and primary neonatal rat cardiomyocytes after infection with CVB3 at 20, 50 and 50 MOI, respectively. Similarly, we found an approximately 100 % increase of Hsp70 in HL-1 cells at 8 h pi and neonatal rat cardiomyocytes at 48 h pi (Fig. 1b, c), and a more than 3-time increase in human cardiomyocytes (Fig. 1d). The time points of significant upregulation of Hsp70 varied due to different sensitivities of cell types to viral infection, which can be indicated by the expression levels of viral capsid protein VP1 (Fig. 1a–d). To test whether CVB3 infection can induce upregulation of Hsp70 in vivo, we performed CVB3 infection in A/J mice (a well-established viral myocarditis model) and IHC analysis using the ventricular wall tissue to detect Hsp70 protein expression (Fig. 1e). In the IHC images, the blue dots indicate cellular nuclei and the red signal represents Hsp70 protein. At 7 days pi, there appeared immune infiltration featured by accumulation of immunocyte nuclei in the heart issue (Fig. 1e, blue arrow), indicating that the heart was infected successfully. At the same point, we observed more red signals in cardiomyocytes compared with sham-infected tissue (Fig. 1e, red arrow), indicating a higher expression level of Hsp70 in CVB3-infected heart.
Fig. 1.
CVB3 infection induces upregulation of Hsp70 both in vitro and in vivo. HeLa cells (a), HL-1 cardiomyocytes (b), neonatal rat cardiomyocytes (c) and SV40 human cardiomyocytes (d) were infected by CVB3 at 10 MOI, 20 MOI, 50 MOI and 50 MOI, respectively. Cell lysates were collected for Western blot to detect the protein levels of Hsp70. Hsc70, a constitutively expressed cognate of Hsp70, was detected by a monoclonal mouse anti-Hsp70/Hsc70 antibody simultaneously (a). β-actin was detected as a loading control and cells treated with PBS (Sham) were used as a negative control. Band intensities were quantified using the ImageJ program, normalized against β-actin and shown as mean ± SD (n = 3) (see lower panels of a, b, c and d; **p < 0.01). e 4-week old A/J mice were infected with 105 pfu of CVB3 or sham-infected with saline. At 4 days pi, mice were killed and the ventricular wall tissue was fixed and subjected to immunohistochemical staining using an anti-Hsp70 antibody. The blue arrow indicates immune cell nuclei in the myocardium. The red arrow indicates a typical cardiomyocyte with Hsp70 protein upregulation (red)
CVB3 upregulates the expression of Hsp70-1 but not Hsp70-2
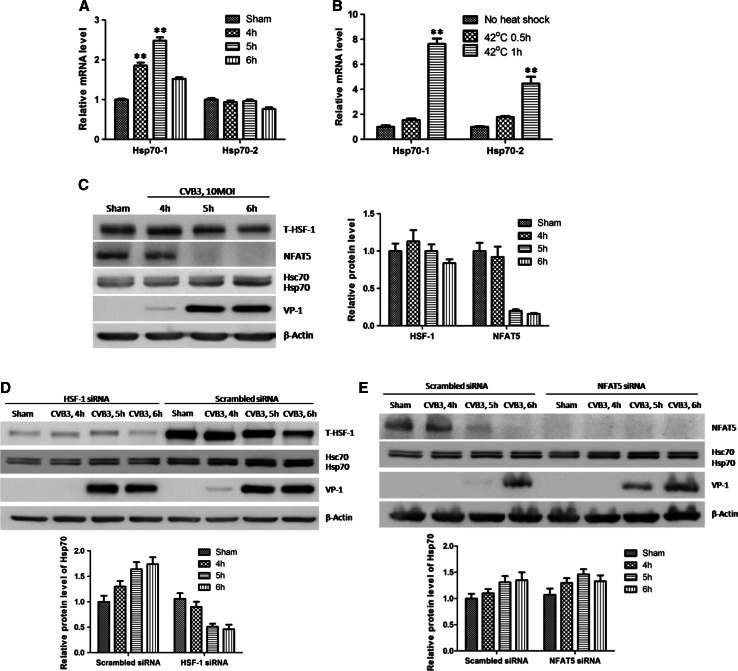
Our results showed that the protein level of Hsp70 was increased during CVB3 infection. However, Hsp70 family contains two major members, Hsp70-1 and Hsp70-2. We attempted to further confirm which isoform contributes to the increase. Hsp70-1 and Hsp70-2 differ from each other only on two amino acids; hence it is hard to distinguish them with antibodies. To detect these two isoforms specifically, we designed two sets of qPCR primers targeting them, respectively, based on the sequence difference in their 3′UTRs. Then we detected the mRNA levels of Hsp70-1 and Hsp70-2 in HeLa cells infected by CVB3. The qPCR results showed an approximately 100 % increase of Hsp70-1 mRNA while there is no significant change of Hsp70-2 mRNA (Fig. 2a), indicating a transcriptional enhancement of Hsp70-1 but not Hsp70-2 during CVB3 infection. To confirm that the primers we used were specific for the Hsp70 mRNAs, we extracted the RNAs from cells heat-shocked at 42 °C for 0.5 h and 1 h and then used the same primers to detect the mRNA of Hsp70-1 and Hsp70-2. The qPCR results showed that Hsp70-1 had a eightfold induction after 1 h of heat shock while Hsp70-2 was upregulated threefold (Fig. 2b), indicating that the primers we used are specific for heat-shock-induced proteins.
Fig. 2.
CVB3 infection stimulates the transcription of Hsp70-1 but not Hsp70-2. a HeLa cells were infected by CVB3 at 10 MOI for different time points and cellular total RNA was extracted. mRNA levels of Hsp70-1 and Hsp70-2 were detected by q-RT-PCR using primers targeting Hsp70-1 and Hsp70-2 specifically, and were normalized against those of GAPDH and shown as mean ± SD (n = 6). b HeLa cells were heat-shocked at 42 °C for 0.5 and 1 h and cellular total RNA was extracted. mRNA levels of Hsp70-1 and Hsp70-2 were detected by qPCR. c HeLa cells were infected with CVB3 as described above. Cell lysates were extracted for Western blot analysis using indicated antibodies and the results were quantified as described (lower panel). d, e HeLa cells were transfected with specific siRNAs to knock down HSF1 (d) or NFAT5 (e) and subjected to CVB3 infection at 10 MOI. Then the cell lysates were extracted for Western blot analysis of indicated proteins. The quantification of Hsp70 is shown in the right panel (c) or the lower panels (d, e)
Since the transcription of Hsp70-1 and Hsp70-2 are stimulated by HSF1 and NFAT5, respectively [10, 35], we attempted to evaluate the transcriptional alteration of Hsp70-1 and Hsp70-2 by detecting the protein levels of HSF1 and NFAT5 in sham- and CVB3-infected HeLa cells. Unexpectedly, HSF1 showed modest increase at 4 h pi and slightly decreased thereafter. As for NFAT5, it underwent a robust decrease at those time points compared with sham-infected cells (Fig. 2c), corresponding to no induction of Hsp70-2 transcription. Besides, the HSF1 bands showed a slow mobility on the gel, probably due to the post-translational modification of the protein, which may still affect the activity of HSF1 regardless of no change in total protein level. To confirm it, we detected the roles of HSF1 and NFAT5 in Hsp70 transcription by siRNA-mediated knocking-down of the genes. We observed a decrease of Hsp70 expression during CVB3 infection when HSF1 was knocked-down (Fig. 2d). Conversely, in the case of NFAT5 knocking-down, the expression level of Hsp70 seemed not changed (or slightly increased) compared to cells transfected with scrambled siRNAs, further confirming that the Hsp70-2 is not inducible in CVB3 infection. The slight increase of Hsp70 protein is probably attributed to the enhancement of viral replication, which is evidenced by the increased VP1 production at 5 h pi (Fig. 2e). These results indicate that CVB3 infection enhances the transcription of Hsp70-1 but not Hsp70-2.
Phosphorylation of HSF1 at Ser230 is responsible for Hsp70-1 upregulation
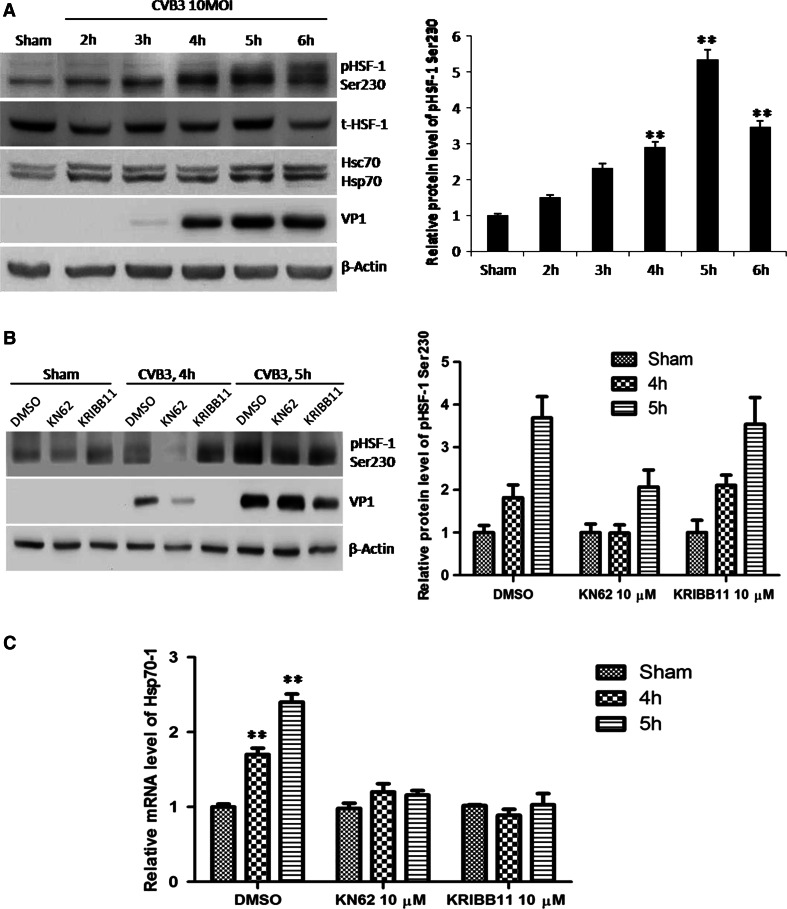
Noting that the expression level of HSF1 was barely changed and there was a little shift of HSF1 band at 5 and 6 h pi (Fig. 2a), we speculated that HSF1 was phosphorylated and led to the enhancement of the transcriptional activity of HSF1. Ser230 is a potential phosphorylation site in this case since such phosphorylation enhances the transcriptional activity of HSF1 [18]. To verify this hypothesis, we determined Ser230 phosphorylation during CVB3 infection by Western blot using an antibody against phosphorylated HSF1 at Ser230 (p-HSF1 Ser230). Coinciding with Hsp70 upregulation, HSF1 showed a significant increase of phosphorylation at 4 and 5 h pi (Fig. 3a).
Fig. 3.
Phosphorylation of HSF1 at Ser230 upregulates Hsp70-1 during CVB3 infection. a HeLa cells were infected with CVB3 and expression levels of phosphorylated HSF1 at Ser230 and Hsp70 were detected at different time points pi by Western blot analysis. β-actin was used as a loading control. Quantification of band intensities was conducted as described in Fig. 1. b, c HeLa cells were treated with KN62, a CaMKII inhibitor and KRIBB11, an HSF1 inhibitor, 1 h before infection. Then the cells were infected with CVB3. At different time points pi, the cellular proteins and total RNA were extracted for Western blot analysis of HSF1 phosphorylation at Ser 230 (b) and q-RT-PCR detection of Hsp70-1 mRNA (c), respectively
To further substantiate the role of such phosphorylation, we treated HeLa cells with 10 µM of KN62 (1-[N,O-bis-(5-Isoquinolinesulfonyl)-N-methyl-l-tyrosyl]-4-phenylpiperazine, Santa Cruz), a chemical that inhibits the phosphorylation of HSF1 at Ser230 [36], and then detect the phosphorylation of HSF1 at Ser230 as well as the mRNA level of Hsp70-1. Upon KN62 treatment, we observed that both phosphorylation of HSF1 at Ser230 and increase of Hsp70-1 mRNA were diminished during CVB3 infection (Fig. 3b, c). In order to exclude the possibility that phosphorylated HSF-1 enhances transcription of Hsp70-1 via other mediators, we treated HeLa cells with 2 µM of KRIBB11 (N(2)-(1H-indazole-5-yl)-N(6)-methyl-3-nitropyridine-2,6-diamine), a specific inhibitor for the transcriptional activity of HSF1 but not the phosphorylation of HSF1 at Ser230 [37]. According to the results, though the treatment of HeLa cells with KRIBB11 did not inhibit the phosphorylation of HSF1 at Ser230 (Fig. 3b), it diminished the increase of Hsp70-1 mRNA induced by CVB3 (Fig. 3c). Together, our results demonstrate that HSF1 is phosphorylated at Ser230 and such phosphorylation is responsible for transcriptional enhancement of Hsp70-1 during CVB3 infection.
CVB3-induced phosphorylation of HSF1 at Ser230 is via phosphorylation of CaMKIIγ
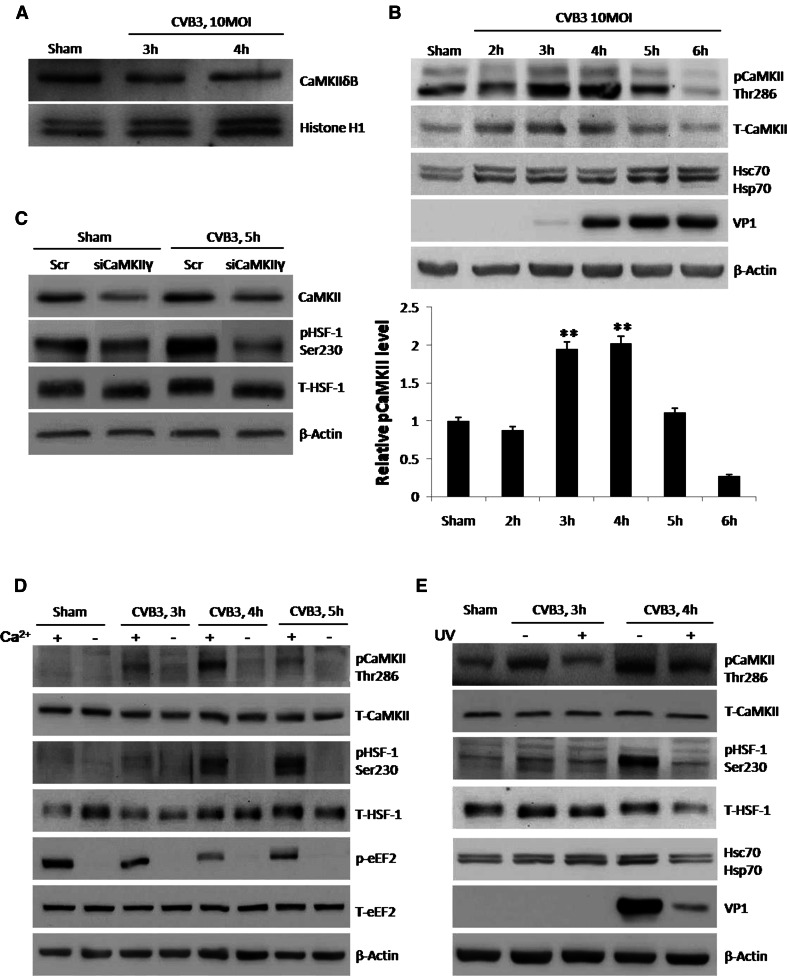
Previous studies showed that phosphorylation of HSF1 at Ser230 is catalyzed by CaMKII [18]. The family of CaMKIIs contains four isoforms [38], among which isoforms γ and δ are the predominant isoforms in the heart [39], thus we focused our studies on these two isoforms. CaMKIIδB is a sub-isoform of CaMKIIδ accumulated in the nucleus in response to stresses and contributing to phosphorylation of HSF1 [40]. However, to our surprise, Western blot analysis showed a decrease of CaMKIIδB in HeLa cells during CVB3 infection (Fig. 4a). Then we changed our focus onto CaMKIIγ. Activation of CaMKIIγ is initialized by binding to Ca2+/CaM and retained by autophosphorylation at Thr286 due to persistent high level of Ca2+/CaM [19]. Therefore, we hypothesized that CVB3 infection activated CaMKIIγ via Thr286 phosphorylation. We detected the phosphorylated CaMKII by Western blot using a specific antibody and found that CaMKII was indeed phosphorylated at 3 h pi, a time point ahead of phosphorylation of HSF1 at Ser230 (Fig. 4b). Furthermore, we knocked-down CaMKIIγ in CVB3-infected HeLa cells using specific siRNAs and found that phosphorylation of HSF1 at Ser230 was diminished (Fig. 4c). These results indicate that CaMKIIγ activation is required for HSF1 phosphorylation at Ser230 during CVB3 infection.
Fig. 4.
CaMKII is activated by phosphorylation during CVB3 infection to phosphorylate HSF1 at Ser230. a HeLa cells were infected with CVB3 and expression levels of CaMKIIδB were detected by Western blot using nuclear extracts. Histone H1 was detected as the loading control. b HeLa cells were infected with CVB3 and phosphorylation of CaMKII at Thr286 was detected at different time points pi. c HeLa cells were transfected with siRNAs targeting CaMKIIγ and then infected with CVB3 at 10 MOI. Cell lysates were used for Western blot analysis of HSF1 phosphorylation at Ser230 and other indicated proteins. d HeLa cells were cultured in medium with or without calcium, a key factor of CaMKII phosphorylation (activation). Then the cells were infected with CVB3 or sham-infected with PBS for 5 h. Phosphorylation of CaMKII and HSF1 was detected by Western blot analysis using the cell lysates. Phosphorylation of eEF2 was detected as a control indicating calcium-free environment. e HeLa cells were infected with alive or UV-irradiated CVB3 at 10 MOI and phosphorylation of CaMKII and HSF1 was detected in the cell lysate
Upon determining the role of CaMKIIγ, we next studied the upstream signal molecules. Since cytosolic calcium is a prerequisite for CaMKII phosphorylation, we cultured HeLa cells in calcium-free medium and detected the change of downstream signals during CVB3 infection. As expected, less phosphorylation of CaMKII at Thr286 and HSF1 at Ser230 was observed compared with cells cultured in normal calcium-containing medium (Fig. 4d). Here we used phosphorylation of eukaryotic elongation factor 2 (p-eEF2) as a control for Ca2+-free condition since eEF2 phosphorylation requires the presence of calcium. It has been reported that active CVB3 replication induces increase of cytosolic calcium [41], thus we speculated that viral replication is required for activation of CaMKII and its downstream signals. Hence, we used UV-irradiated CVB3, which is capable of receptor binding and internalization but not able to replicate, to infect HeLa cells. Indeed, modest phosphorylation of CaMKII at Thr286 and HSF1 at Ser230 was observed upon infection of UV-irradiated CVB3 (Fig. 4e). In all, these results reveal a signaling cascade in which CVB3 replication raises cytosolic calcium, activates CaMKII, and phosphorylates HSF1 at Ser230 and finally upregulates Hsp70-1.
Phosphorylation of HSF1 at Ser307 is blocked during CVB3 infection
Unlike Ser230, phosphorylation of HSF1 at Ser307 inhibits the transcriptional activity of HSF1 [24]. Phosphorylation of HSF1 at Ser307 is catalyzed by active ERK1/2. However, ERK1/2 is substantially activated by phosphorylation at 5 h post-CVB3 infection [27], the exact same time point when Hsp70-1 is upregulated, thus we cannot neglect the potential adverse effect of Ser307 phosphorylation of HSF1 on Hsp70-1 induction during CVB3 infection. To determine the possible contributions of Ser307 phosphorylation to the HSF-1 activity during CVB3 infection, we detected it as well as its upstream signal, phosphorylated ERK1/2, by Western blot using specific antibodies. Surprisingly, though dramatic phosphorylation of ERK1/2 was observed at 5 h pi, coinciding with previous study, phosphorylated HSF1 at Ser307 could be barely detected (Fig. 5a), indicating that ERK1/2-mediated phosphorylation of HSF1 at Ser307 was blocked. However, when we detected the phosphorylation of HSF1 at Ser307 at very early time points of CVB3 infection (0.5–2 h pi, the time for CVB3 binding and internalization) during which ERK1/2 was also transiently phosphorylated, we observed phosphorylation of HSF1 at Ser307 (Fig. 5a). We suspected that blocking of Ser307 phosphorylation at 4 h pi was resulted from translocation of HSF1 into the nucleus and isolation from ERK1/2 after phosphorylation at Ser230 [40]. This speculation was verified by Western blot analysis using protein extracts isolated from nucleus and cytoplasm separately. We found that HSF1 increased dramatically at 4 h pi in the nucleus, coinciding with the time of HSF1 phosphorylation at Ser230, whereas ERK1/2 was mainly located in the cytoplasm (Fig. 5b). This finding was solidified by immunostaining and confocal imaging, showing the accumulation of HSF1 in the nucleus at 4 h pi (Fig. 5c), the time point when Ser230 was phosphorylated. These data indicate that phosphorylation of Ser230 isolates HSF1 from ERK1/2 and avoids the negative regulation of Hsp70.
Fig. 5.
Phosphorylation of HSF1 at Ser307 was blocked by nuclear translocation of HSF1. a HeLa cells were infected with CVB3 at 10 MOI. Phosphorylation of both HSF1 at Ser307 and its upstream signal ERK1/2 was detected by Western blot using indicated antibodies. b HeLa cells were infected with CVB3 or sham-infected with PBS. Nuclear and cytosolic proteins were isolated for detection of HSF1 and ERK1/2, respectively. Histone H1 protein was used as a purity control for nuclear fraction. β-actin was the loading control. c HeLa cells were infected with CVB3 at 10 MOI. At different time points pi, the cells were fixed for the immunostaining of HSF1. Nuclei were counterstained with DAPI. The cellular localization of HSF1 was visualized by confocal microscopy
Hsp70 favors CVB3 replication
To investigate the effect of Hsp70-1 on CVB3 infection, we first silenced Hsp70-1 expression by transfection of HeLa cells for 48 h with siRNAs targeting Hsp70-1 or scrambled control siRNAs and then infected with CVB3 for 2–6 h. Total cellular RNAs were extracted at different time points pi and then the levels of CVB3 genome were detected by q-RT-PCR using the primers targeting the coding region of one of 2A, one of the viral genes. We observed a dramatic decrease of 2A RNA in Hsp70-1 siRNAs-transfected cells compared to the control (Fig. 6a). This negative effect of Hsp70-1 silencing on CVB3 replication was further solidified by Western blot analysis of CVB3 VP1 protein using cell. As shown in Fig. 6b, transfection of siRNA targeting Hsp70-1 resulted in a decrease in VP1 production.
Fig. 6.
Hsp70-1 upregulation benefits CVB3 replication. HeLa cells were transfected with specific siRNAs to knock down endogenous Hsp70-1 and then infected with CVB3 at 10 MOI. Viral replication was evaluated by q-RT-PCR detection of 2A RNA (a) and Western blot analysis of VP1 (b). Cells transfected with scrambled siRNAs served as the negative control. Meanwhile, HeLa cells were transfected with plasmid pEGFP-Hsp70-1 to overexpress Hsp70-1 and then infected with CVB3 at 10 MOI. Viral replication was measured by q-RT-PCR detection of 2A RNA (c) and Western blot analysis of VP1 (d). Cells transfected with the empty vector pEGFP and pEGFP-Hsp70-1 (K71E) mutant plasmid served as negative controls (e). In both q-RT-PCR results, the RNA levels of CVB3 2A were normalized against the mRNA levels of GAPDH and shown as mean ± SD (n = 6, p < 0.01)
To further confirm the positive effects of Hsp70-1 on CVB3 replication, we next overexpressed Hsp70-1 in HeLa cells using the plasmid pEGFP-Hsp70-1. Meanwhile, we transfected the cells with the empty vector pEGFP and pEGFP-Hsp70-1(K71E), a loss-of-function mutant of Hsp70-1, as negative controls. After transfection for 20 h and then infected with CVB3, viral RNA and VP1 levels were detected as described above. As shown in Fig. 6c, d particularly at 4 h pi, compared with controls, overexpression of WT Hsp70-1 enhanced viral replication at levels of transcription and translation, while overexpression of pEGFP-Hsp70-1(K71E) barely changed the levels of viral RNA and protein (Fig. 6c, e).
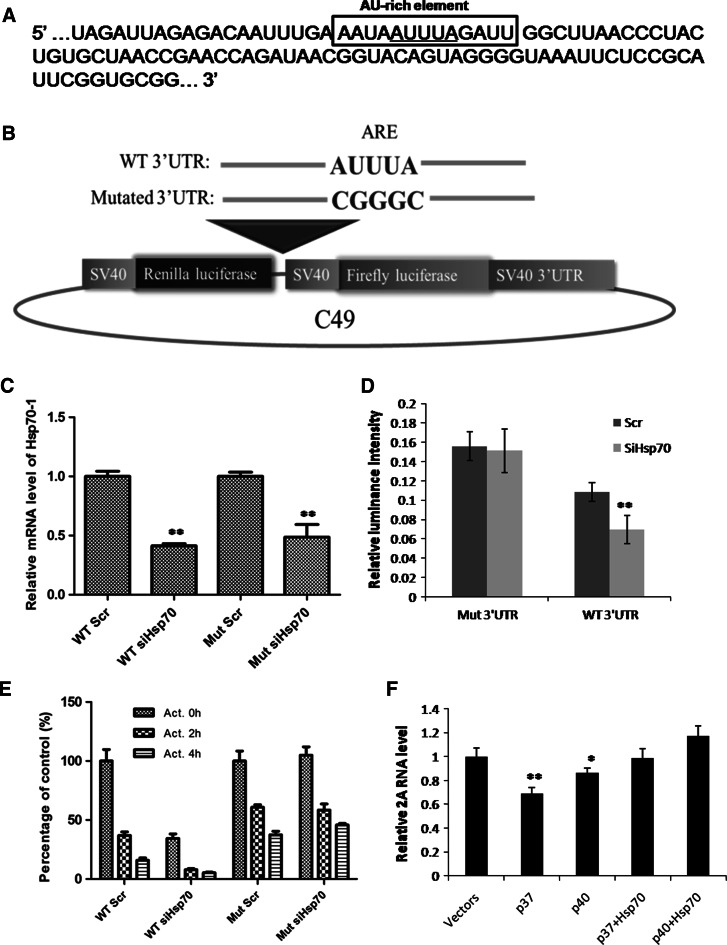
Hsp70-1 stabilizes CVB3 genomic RNA via the ARE site of CVB3 3′UTR
Hsp70-1 is known to play a role in stabilizing mRNAs containing an ARE site in its 3′UTR [30]. As shown in Fig. 7a, CVB3 genomic RNA also contains an ARE sequence within its 3′UTR. Thus, we focused our attention to investigate whether Hsp70-1 enhances CVB3 replication via stabilizing viral genome. To this end, we constructed the dual-luciferase reporter plasmids, C49-WT-CVB3-3′UTR and C49-Mut-CVB3-3′UTR, by inserting WT and mutant 3′UTR of CVB3 RNA at the downstream of Renilla luciferase coding region, so that these two reporters contain a WT and a mutant ARE site, respectively (Fig. 7b). We co-transfected the reporter plasmids and Hsp70-1 siRNAs or scrambled siRNAs into HeLa cells. The knockdown efficiency of Hsp70-1 was confirmed by qPCR detecting Hsp70-1 mRNA, which showed that the mRNA levels of Hsp70-1 decreased more than 50 % in Hsp70-siRNA-transfected cells compared to scrambled-siRNA-transfected controls (Fig. 7c). Then we measured the activity of Renilla luciferase by luciferase assay, and the data were normalized to the activity of firefly luciferase. The results showed that knocking-down of Hsp70-1 decreased the expression of the luciferase with the wild-type ARE site, whereas the luciferase gene with the mutant ARE site was not sensitive to Hsp70-1 knocking-down (Fig. 7d). To further confirm that Hsp70-1 is critical for the stability of mRNAs modulated by CVB3 3′UTR, we treated the reporter cells with 2.5 μg/mL of actinomycin D for 2 and 4 h to inhibit new transcription, and detected the mRNA levels of Renilla luciferase by qPCR using specific primers (Table 1). All the qPCR results were normalized by setting the mRNA level of groups treated with scrambled siRNAs but no actinomycin D as 100 %. As shown in Fig. 7e, the mRNA of Renilla luciferase modulated by WT CVB3 3′UTR has a decrease of 60 % at 2 h and 85 % at 4 h after actinomycin D treatment, compared with 40 % decrease at 2 h and 60 % decrease at 4 h of the reporter modulated by mutant CVB3 3′UTR, indicating a more rapid decay of the mRNA with the WT ARE site when the new transcription is blocked. Moreover, when Hsp70-1 was knocked-down, a lower level of Renilla luciferase mRNA was detected in cells transfected with C49-WT-CVB3-3′UTR, but no significant changes were observed in cells transfected with C49-Mut-CVB3-3′UTR, supporting the previous finding that the mRNA with the WT ARE site is more sensitive to Hsp70-1 change and decay faster when Hsp70-1 level is low. These results indicate that Hsp70-1 stabilizes CVB3 genomic RNA and this function requires an ARE sequence.
Fig. 7.
Hsp70-1 stabilizes CVB3 genomic RNA via the ARE site in the 3′UTR of CVB3 genome. a An AU-rich element is located in the 3′UTR of CVB3 genome. b Schematic structures of C49 luciferase reporter plasmid. WT and mutated CVB3 3′UTRs were amplified by PCR using specific primers and inserted into the downstream of Renilla luciferase coding region on the reporter plasmid, generating C49-WT-CVB3-3′UTR and C49-Mut-CVB3-3′UTR. c HeLa cells were co-transfected with different combinations of plasmids and siRNA: C49-WT-CVB3-3′UTR and scrambled siRNAs (WT Scr), C49-WT-CVB3-3′UTR and Hsp70-1 siRNAs (WT siHsp70), C49-Mut-CVB3-3′UTR and scrambled siRNAs (Mut Scr) or C49-Mut-CVB3-3′UTR and Hsp70-1 siRNAs (Mut siHsp70). The knockdown efficiency of Hsp70-1 by siRNA was determined by qPCR detection of Hsp70-1 mRNA. The results were normalized by GAPDH mRNA and the relative mRNA levels of Hsp70-1 in Scr siRNA-treated cells were set as 1.0. d Luciferase assay. HeLa cells were co-transfected with the C49 plasmids containing WT or mutated CVB3 3′UTR and Hsp70-1 siRNAs (siHsp70) or scrambled siRNAs (Scr). At 48 h pt, cell lysates were collected and subjected to luciferase assay. Relative luciferase activities (Renilla/firefly) are shown as mean ± SD (n = 9). e HeLa cells were transfected with the same plasmids and siRNAs as those in (c) and (d). The transfected cells were treated with 2.5 μg/mL of actinomycin D for 2 h (Act. 2 h) or 4 h (Act. 4 h). The same cells treated with 0.1 % DMSO (Act. 0 h) served as a control. The rate of mRNA decay was determined by qPCR detection of the Hsp70-1 mRNA. The results were normalized by GAPDH mRNA and the relative mRNA levels of Hsp70-1 in Scr siRNA-treated cells without actinomycin D treatment were set as 100 %. f HeLa cells were co-transfected with different plasmid combinations: vectors (pFLAG + pEGFP), p37 (pFLAG-p37AUF1 + pEGFP), p40 (pFLAG-p40AUF1 + pEGFP), p37 + Hsp70 (pFLAG-p37AUF1 + pEGFP-Hsp70-1), and p40 + Hsp70 (pFLAG-p40AUF1 + pEGFP-Hsp70-1). At 36 h pt, the cells were infected with CVB3 at 10 MOI for 6 h and then viral genomic RNA was measured by q-RT-PCR detection of CVB3 2A RNA. The RNA levels of 2A were normalized against the mRNA levels of GAPDH and shown as mean ± SD (n = 6, p < 0.01)
It has been reported that Hsp70-1 stabilizes ARE-mRNA via sequestering AUF1 in the nucleus [30], and AUF1 isoforms p37AUF1 and p40AUF1 are involved in this process [42]. Thus, we further tested whether these two AUF1 isoforms play a role in stabilizing CVB3 genomic RNA during infection. To this end, we expressed either p37AUF1 or p40AUF1 by plasmid transfection in CVB3-infected HeLa cells and then measured viral genomic RNA during infection. The q-RT-PCR results showed that CVB3 genomic RNA was significantly decreased upon expression of these AUF1 isoforms although p40AUF1 caused less decrease than p37AUF1 (Fig. 7f). However, when Hsp70-1 was co-transfected with AUF1, the decrease of viral genome was diminished (Fig. 7f). These results demonstrate that p37AUF1 or p40AUF1 reduces the abundance of CVB3 genomic RNA while Hsp70-1 reverses such effect, indicating that Hsp70-1 stabilizes CVB3 genomic RNA via p37AUF1 and p40AUF1.
Discussion
Hsp70 is one of the best-characterized chaperon proteins in the Hsp70 family. Its expression is regulated by various cellular stress conditions including viral infection. Accumulating evidence indicates that the mutual regulation of viruses and Hsp70 is critical for viral replication and the pathogenesis of virus-induced diseases. On one hand, viral infections, such as foot-and-mouth disease virus (FMDV) [43] and rotavirus [44], induce upregulation of Hsp70. On the other hand, Hsp70 expression plays different roles in viral replication, for example, the positive regulation in rabies virus [44], Japanese encephalitis virus [45] and porcine circovirus [46] and the negative regulation in vesicular stomatitis virus [47], human immunodeficiency virus-1 [48] and rotavirus [44]. However, such study has not been conducted in coxsackievirus infection and particularly has never distinguished the roles of Hsp70-1 and Hsp70-2 in viral replication.
In this study, we aim to understand the mutual regulation between Hsp70 and CVB3. Our results showed that Hsp70-1, not Hsp70-2, is upregulated during CVB3 infection in cells of different origins, though the induction level is much lower than that in heat shock, which is probably due to the expression inhibition of some transcriptional and translational factors during CVB3 infection. In searching for the underlying mechanism, we focused our attention on HSF1, the transcriptional factor of Hsp70-1. By siRNA-mediated knocking-down of HSF1, we found that HSF1 is responsible for CVB3-induced upregulation of Hsp70-1. However, no significant upregulation except a molecular-weight shift of HSF1 band was observed in the virus-infected samples, indicating that the transcriptional activity of HSF1 may be enhanced by certain post-translational modifications. Thus, we tried to determine HSF1 phosphorylation and found that HSF1 was indeed phosphorylated at Ser230. This result is consistent with previous report that phosphorylation of Ser230 activates transcriptional activity of HSF1 [18]. CaMKII is the upstream kinase of HSF1 [18], which is activated via phosphorylation at Thr286 in an elevated Ca2+/CaM condition and responsible for phosphorylation of HSF1. Thus we drew our attention on CaMKII. We found that CaMKII was phosphorylated at Thr286 during CVB3 infection. Correspondingly, our results showed that KN62, a specific inhibitor of CaMKII-calcium/calmodulin binding [36], inhibited phosphorylation of HSF1 at Ser230 and thus Hsp70-1 production during CVB3 infection. The family of CaMKIIs contains four isoforms, α, β, γ and δ [38]. Since α and β isoforms are almost exclusively expressed in the brain and the γ and δ are the predominant isoforms in the heart [39], we focused our identification on γ and δ isoforms. By siRNA-targeted gene silencing, we finally identified CaMKIIγ as the responsible isoform for HSF1 phosphorylation at Ser230 during CVB3 infection. These data suggest that CVB3-induced Hsp70-1 upregulation is via selective phosphorylation of HSF1 at Ser230, which is catalyzed by CaMKIIγ.
There are several other phosphorylation sites on HSF-1, among which we focused on the ERK1/2-regulated phosphorylation of HSF1 at Ser307 as this phosphorylation can inhibit the transcriptional activity of HSF1 [24]. However, this event was blocked during later time points of CVB3 infection, contributing to the enhancement of HSF1 activity and Hsp70-1 upregulation. Our previous study showed that ERK1/2 is activated at two separate phases although the later phase is dominant during CVB3 infection [27]. Here, we showed that ERK1/2 activation at the first phase was capable of inducing phosphorylation of HSF1 at Ser307 but the second phase of ERK1/2 activation was not. This is likely due to nuclear translocation of HSF1, which isolates HSF1 from ERK1/2, at late time points of CVB3 infection.
Having determined the upregulation of Hsp70-1 during CVB3 infection, we next invested the effects of Hsp70-1 upregulation on CVB3 infection. By both the siRNA-mediated gene silencing and plasmid-mediated overexpression of Hsp70-1, we confirmed that Hsp70-1 expression benefits CVB3 replication at the levels of transcription and translation. These data suggest that Hsp70-1 upregulation generates a positive feedback loop on CVB3 replication. This signaling pathway is summarized in Fig. 8. Then the question was how Hsp70-1 favors CVB3 replication. One of our hypotheses was that Hsp70-1 protects viral genomic RNA from exonuclease-driven degradation, considering Hsp70-1 is closely related to mRNA turnover [49]. Many cellular mRNAs contain an ARE sequence in their 3′UTR. Association of ARE-binding protein (AUPBs) with these mRNAs promotes rapid mRNA degradation. AUF1, one of the best-characterized AUBPs, binds to many ARE-mRNAs and assembles other factors necessary to recruit the mRNA degradation machineries. These factors include eIF4G, PABP, Hsp70-1 and many unknown proteins [50]. During normal physiological conditions, AUF1 interacts simultaneously with eIF4G and ARE, while PABP binds both eIF4G and poly(A) tail. However, during the active normal translation of ARE-containing mRNAs, the ribosome might relocate AUF1, which could bind PABP, thus causing exposure of the poly-A tail to deadenylases to initiate decay. During cellular stress or infection, the association of Hsp70-1 with AUF1 may disrupt or block the AUF-PABP interaction, leaving PABP free to remain bound to poly-A tail, thus masking it from ribonuclease [51]. This mechanism for stabilizing the viral mRNA has been reported in DNA virus infection [52]. By sequence screening, we found that the genomic RNA of CVB3 harbors an ARE site within its 3′UTR. To verify whether this ARE site plays a role in Hsp70-1-mediated stabilization of CVB3 genomic RNA, we performed luciferase assay and mRNA turnover assay using reporter plasmids containing CVB3 3′UTR with a WT or mutant ARE site. In the luciferase activity assay, we found that only the reporter regulated by the WT CVB3 ARE site is sensitive to Hsp70-1 level, supported by the result that the reporter signal was significantly reduced when Hsp70-1 was knocked-down but no significant change was observed in the mutant control. In the mRNA turnover assay, the decay of the luciferase mRNA was detected directly and the results showed that the mRNA containing CVB3 3′UTR with WT ARE had a more rapid decay rate and its level was positively correlated with Hsp70-1 level, supporting that Hsp70-1 stabilizes the RNA with CVB3 3′UTR via the ARE site. We further determined whether AUF1 is involved in the process via ectopic expression of different AUF1 isoforms. We found that overexpression of p37AUF1 and p40AUF1 decreased the abundance of CVB3 genomic RNA; however, such decrease was suppressed by the overexpression of Hsp70-1, indicating that the stability of CVB3 genomic RNA is negatively regulated, at least in part, by AUF1-mediated RNA decay, whereas Hsp70-1 plays a role in counteracting this process.
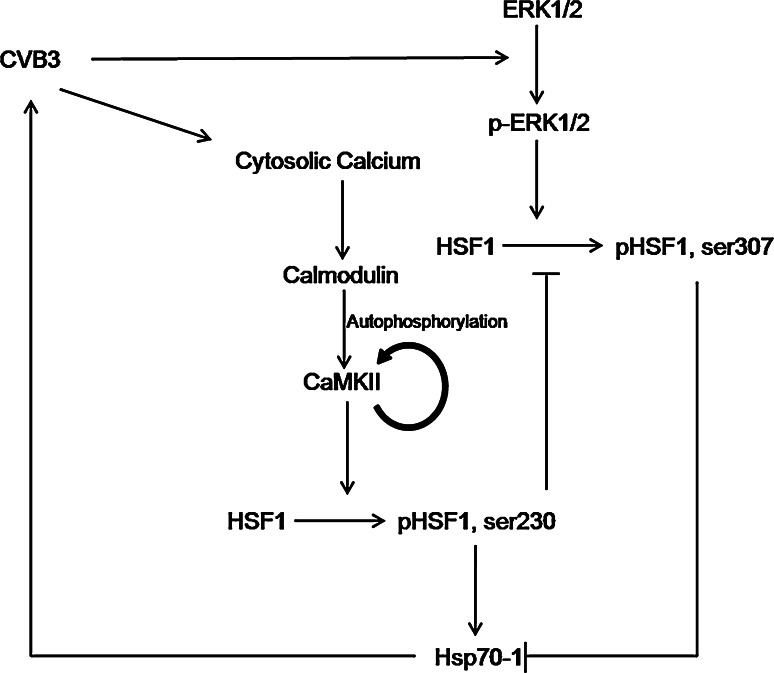
Fig. 8.
A putative model of the mutually beneficial regulation of CVB3 and Hsp70. CVB3 infection elevates the level of cytosolic calcium and activates CaMKII via inducing autophosphorylation of CaMKII at Thr286. The active CaMKII further phosphorylates HSF1 at Ser230 to enhance the transcription of Hsp70-1. On the other hand, HSF1 activation causes the nuclear translocation of HSF1 and thus blocks the ERK1/2-catalyzed phosphorylation of HSF1 at Ser307, a negative regulatory process of Hsp70-1 transcription, thus further contributing to Hsp70-1 upregulation. Finally, the upregulated Hsp70 in turn positively feedbacks on CVB3 replication
CVB3 infection is the primary cause of viral myocarditis, an inflammatory heart disease. Lesions of the heart caused by CVB3 infection are not only due to viral replication in cardiomyocytes, but also resulted from virus-induced exaggerated immune responses. Hsp70-1 has been reported to be secreted upon induction and act as an immune stimulator leading to immune infiltration [47, 53, 54]. Thereby, we believe that CVB3-induced upregulation of Hsp70-1 not only favors viral replication, but also enhances immune infiltration during the development of myocarditis, both of which lead to exacerbation of the disease. Thus, Hsp70-1 chaperone is a rationale pharmaceutical target for viral myocarditis.
Acknowledgments
This work was supported by a grant from the Canadian Institutes of Health Research (Grant # MOP-125995). Xin Ye is a recipient of the UGF Award from the University of British Columbia. Jeff Zong and Brian Cho were supported by the Pathology Summer Student Program of the University. We would like to thank Dr. Lois Greene at NIH for providing us the Hsp70-1 plasmids and Dr. Robert J. Schneider at New York University for providing us the AUF1 plasmids.
References
- 1.Beck MA, Chapman NM, McManus BM, Mullican JC, Tracy S. Secondary enterovirus infection in the murine model of myocarditis. Pathologic and immunologic aspects. Am J Pathol. 1990;136(3):669–681. [PMC free article] [PubMed] [Google Scholar]
- 2.McManus BM, Chow LH, Radio SJ, Tracy SM, Beck MA, Chapman NM, Klingel K, Kandolf R. Progress and challenges in the pathological diagnosis of myocarditis. Eur Heart J. 1991;12(Suppl D):18–21. doi: 10.1093/eurheartj/12.suppl_D.18. [DOI] [PubMed] [Google Scholar]
- 3.McManus BM, Yanagawa B, Rezai N, Luo H, Taylor L, Zhang M, Yuan J, Buckley J, Triche T, Schreiner G, Yang D. Genetic determinants of coxsackievirus B3 pathogenesis. Ann N Y Acad Sci. 2002;975:169–179. doi: 10.1111/j.1749-6632.2002.tb05950.x. [DOI] [PubMed] [Google Scholar]
- 4.Joachims M, Van Breugel PC, Lloyd RE. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J Virol. 1999;73(1):718–727. doi: 10.1128/jvi.73.1.718-727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chau DH, Yuan J, Zhang H, Cheung P, Lim T, Liu Z, Sall A, Yang D. Coxsackievirus B3 proteases 2A and 3C induce apoptotic cell death through mitochondrial injury and cleavage of eIF4GI but not DAP5/p97/NAT1. Apoptosis. 2007;12(3):513–524. doi: 10.1007/s10495-006-0013-0. [DOI] [PubMed] [Google Scholar]
- 6.Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JW. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem. 1982;257(24):14806–14810. [PubMed] [Google Scholar]
- 7.Lamphear BJ, Yan R, Yang F, Waters D, Liebig HD, Klump H, Kuechler E, Skern T, Rhoads RE. Mapping the cleavage site in protein synthesis initiation factor eIF-4 gamma of the 2A proteases from human Coxsackievirus and rhinovirus. J Biol Chem. 1993;268(26):19200–19203. [PubMed] [Google Scholar]
- 8.van Ooij MJ, Polacek C, Glaudemans DH, Kuijpers J, van Kuppeveld FJ, Andino R, Agol VI, Melchers WJ. Polyadenylation of genomic RNA and initiation of antigenomic RNA in a positive-strand RNA virus are controlled by the same cis-element. Nucleic Acids Res. 2006;34(10):2953–2965. doi: 10.1093/nar/gkl349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snoeckx LH, Cornelussen RN, Van Nieuwenhoven FA, Reneman RS, Van Der Vusse GJ. Heat shock proteins and cardiovascular pathophysiology. Physiol Rev. 2001;81(4):1461–1497. doi: 10.1152/physrev.2001.81.4.1461. [DOI] [PubMed] [Google Scholar]
- 10.Woo SK, Lee SD, Na KY, Park WK, Kwon HM. TonEBP/NFAT5 stimulates transcription of HSP70 in response to hypertonicity. Mol Cell Biol. 2002;22(16):5753–5760. doi: 10.1128/MCB.22.16.5753-5760.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iordanskiy S, Zhao Y, Dubrovsky L, Iordanskaya T, Chen M, Liang D, Bukrinsky M. Heat shock protein 70 protects cells from cell cycle arrest and apoptosis induced by human immunodeficiency virus type 1 viral protein R. J Virol. 2004;78(18):9697–9704. doi: 10.1128/JVI.78.18.9697-9704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agostini I, Popov S, Li J, Dubrovsky L, Hao T, Bukrinsky M. Heat-shock protein 70 can replace viral protein R of HIV-1 during nuclear import of the viral preintegration complex. Exp Cell Res. 2000;259(2):398–403. doi: 10.1006/excr.2000.4992. [DOI] [PubMed] [Google Scholar]
- 13.Choukhi A, Ung S, Wychowski C, Dubuisson J. Involvement of endoplasmic reticulum chaperones in the folding of hepatitis C virus glycoproteins. J Virol. 1998;72(5):3851–3858. doi: 10.1128/jvi.72.5.3851-3858.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh WK, Song J. Hsp70 functions as a negative regulator of West Nile virus capsid protein through direct interaction. Biochem Biophys Res Commun. 2006;347(4):994–1000. doi: 10.1016/j.bbrc.2006.06.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oglesbee MJ, Kenney H, Kenney T, Krakowka S. Enhanced production of morbillivirus gene-specific RNAs following induction of the cellular stress response in stable persistent infection. Virology. 1993;192(2):556–567. doi: 10.1006/viro.1993.1072. [DOI] [PubMed] [Google Scholar]
- 16.Padmini E, Lavanya S. Over expression of HSP70 and HSF1 in endothelial cells during pre-eclamptic placental stress. Aust N Z J Obstet Gynaecol. 2011;51(1):47–52. doi: 10.1111/j.1479-828X.2010.01246.x. [DOI] [PubMed] [Google Scholar]
- 17.Dai Q, Zhang C, Wu Y, McDonough H, Whaley RA, Godfrey V, Li HH, Madamanchi N, Xu W, Neckers L, Cyr D, Patterson C. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003;22(20):5446–5458. doi: 10.1093/emboj/cdg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmberg CI, Hietakangas V, Mikhailov A, Rantanen JO, Kallio M, Meinander A, Hellman J, Morrice N, MacKintosh C, Morimoto RI, Eriksson JE, Sistonen L. Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. EMBO J. 2001;20(14):3800–3810. doi: 10.1093/emboj/20.14.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erickson JR, Joiner ML, Guan X, Kutschke W, Yang J, Oddis CV, Bartlett RK, Lowe JS, O’Donnell SE, Aykin-Burns N, Zimmerman MC, Zimmerman K, Ham AJ, Weiss RM, Spitz DR, Shea MA, Colbran RJ, Mohler PJ, Anderson ME. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 2008;133(3):462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hudmon A, Schulman H. Structure-function of the multifunctional Ca2+/calmodulin-dependent protein kinase II. Biochem J. 2002;364(Pt 3):593–611. doi: 10.1042/bj20020228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenberg OS, Deindl S, Sung RJ, Nairn AC, Kuriyan J. Structure of the autoinhibited kinase domain of CaMKII and SAXS analysis of the holoenzyme. Cell. 2005;123(5):849–860. doi: 10.1016/j.cell.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 22.Liu G, Yang Y, Gu Q. The effect of coxsackie virus B3 on intracellular calcium homeostasis in rat cardiomyocytes. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2000;14(2):113–116. [PubMed] [Google Scholar]
- 23.Bozym RA, Morosky SA, Kim KS, Cherry S, Coyne CB . Release of intracellular calcium stores facilitates coxsackievirus entry into polarized endothelial cells. PLoS Pathog. 2010;6(10):e1001135. doi: 10.1371/journal.ppat.1001135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melling CW, Krause MP, Noble EG. PKA-mediated ERK1/2 inactivation and hsp70 gene expression following exercise. J Mol Cell Cardiol. 2006;41(5):816–822. doi: 10.1016/j.yjmcc.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Chu B, Soncin F, Price BD, Stevenson MA, Calderwood SK. Sequential phosphorylation by mitogen-activated protein kinase and glycogen synthase kinase 3 represses transcriptional activation by heat shock factor-1. J Biol Chem. 1996;271(48):30847–30857. doi: 10.1074/jbc.271.48.30847. [DOI] [PubMed] [Google Scholar]
- 26.Esfandiarei M, Luo H, Yanagawa B, Suarez A, Dabiri D, Zhang J, McManus BM. Protein kinase B/Akt regulates coxsackievirus B3 replication through a mechanism which is not caspase dependent. J Virol. 2004;78(8):4289–4298. doi: 10.1128/JVI.78.8.4289-4298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo H, Yanagawa B, Zhang J, Luo Z, Zhang M, Esfandiarei M, Carthy C, Wilson JE, Yang D, McManus BM. Coxsackievirus B3 replication is reduced by inhibition of the extracellular signal-regulated kinase (ERK) signaling pathway. J Virol. 2002;76(7):3365–3373. doi: 10.1128/JVI.76.7.3365-3373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henics T, Nagy E, Oh HJ, Csermely P, von Gabain A, Subjeck JR. Mammalian Hsp70 and Hsp110 proteins bind to RNA motifs involved in mRNA stability. J Biol Chem. 1999;274(24):17318–17324. doi: 10.1074/jbc.274.24.17318. [DOI] [PubMed] [Google Scholar]
- 29.He C, Schneider R. 14-3-3sigma is a p37 AUF1-binding protein that facilitates AUF1 transport and AU-rich mRNA decay. EMBO J. 2006;25(16):3823–3831. doi: 10.1038/sj.emboj.7601264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284(5413):499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 31.Kishor A, Tandukar B, Ly YV, Toth EA, Suarez Y, Brewer G, Wilson GM. Hsp70 is a novel posttranscriptional regulator of gene expression that binds and stabilizes selected mRNAs containing AU-rich elements. Mol Cell Biol. 2013;33(1):71–84. doi: 10.1128/MCB.01275-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng XC, Bhasin S, Wu X, Lee JG, Maffi S, Nichols CJ, Lee KJ, Taylor JP, Greene LE, Eisenberg E. Hsp70 dynamics in vivo: effect of heat shock and protein aggregation. J Cell Sci. 2004;117(Pt 21):4991–5000. doi: 10.1242/jcs.01373. [DOI] [PubMed] [Google Scholar]
- 33.Lund N, Milev MP, Wong R, Sanmuganantham T, Woolaway K, Chabot B, Abou Elela S, Mouland AJ, Cochrane A. Differential effects of hnRNP D/AUF1 isoforms on HIV-1 gene expression. Nucleic Acids Res. 2012;40(8):3663–3675. doi: 10.1093/nar/gkr1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye X, Zhang HM, Qiu Y, Hanson PJ, Hemida MG, Wei W, Hoodless PA, Chu F, Yang D. Coxsackievirus-induced miR-21 disrupts cardiomyocyte interactions via the downregulation of intercalated disk components. PLoS Pathog. 2014;10(4):e1004070. doi: 10.1371/journal.ppat.1004070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D, Kim SH, Li GC. Proteasome inhibitors MG132 and lactacystin hyperphosphorylate HSF1 and induce hsp70 and hsp27 expression. Biochem Biophys Res Commun. 1999;254(1):264–268. doi: 10.1006/bbrc.1998.9840. [DOI] [PubMed] [Google Scholar]
- 36.Tokumitsu H, Chijiwa T, Hagiwara M, Mizutani A, Terasawa M, Hidaka H. KN-62, 1-[N, O-bis(5-isoquinolinesulfonyl)-N-methyl-l-tyrosyl]-4-phenylpiperazi ne, a specific inhibitor of Ca2+/calmodulin-dependent protein kinase II. J Biol Chem. 1990;265(8):4315–4320. [PubMed] [Google Scholar]
- 37.Yoon YJ, Kim JA, Shin KD, Shin DS, Han YM, Lee YJ, Lee JS, Kwon BM, Han DC. KRIBB11 inhibits HSP70 synthesis through inhibition of heat shock factor 1 function by impairing the recruitment of positive transcription elongation factor b to the hsp70 promoter. J Biol Chem. 2011;286(3):1737–1747. doi: 10.1074/jbc.M110.179440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tombes RM, Faison MO, Turbeville JM. Organization and evolution of multifunctional Ca(2+)/CaM-dependent protein kinase genes. Gene. 2003;322:17–31. doi: 10.1016/j.gene.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 39.Backs J, Backs T, Neef S, Kreusser MM, Lehmann LH, Patrick DM, Grueter CE, Qi X, Richardson JA, Hill JA, Katus HA, Bassel-Duby R, Maier LS, Olson EN. The delta isoform of CaM kinase II is required for pathological cardiac hypertrophy and remodeling after pressure overload. Proc Natl Acad Sci USA. 2009;106(7):2342–2347. doi: 10.1073/pnas.0813013106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng W, Zhang Y, Zheng M, Cheng H, Zhu W, Cao CM, Xiao RP. Cardioprotection by CaMKII-deltaB is mediated by phosphorylation of heat shock factor 1 and subsequent expression of inducible heat shock protein 70. Circ Res. 2010;106(1):102–110. doi: 10.1161/CIRCRESAHA.109.210914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Kuppeveld FJ, Hoenderop JG, Smeets RL, Willems PH, Dijkman HB, Galama JM, Melchers WJ. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 1997;16(12):3519–3532. doi: 10.1093/emboj/16.12.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics. 1998;48(2):195–202. doi: 10.1006/geno.1997.5142. [DOI] [PubMed] [Google Scholar]
- 43.Gulbahar MY, Kabak YB, Karayigit MO, Yarim M, Guvenc T, Parlak U. The expressions of HSP70 and alphaB-crystallin in myocarditis associated with foot-and-mouth disease virus in lambs. J Vet Sci. 2011;12(1):65–73. doi: 10.4142/jvs.2011.12.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Broquet AH, Lenoir C, Gardet A, Sapin C, Chwetzoff S, Jouniaux AM, Lopez S, Trugnan G, Bachelet M, Thomas G. Hsp70 negatively controls rotavirus protein bioavailability in caco-2 cells infected by the rotavirus RF strain. J Virol. 2007;81(3):1297–1304. doi: 10.1128/JVI.01336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye J, Chen Z, Zhang B, Miao H, Zohaib A, Xu Q, Chen H, Cao S. Heat shock protein 70 is associated with replicase complex of Japanese encephalitis virus and positively regulates viral genome replication. PLoS One. 2013;8(9):e75188. doi: 10.1371/journal.pone.0075188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, Bai J, Zhang L, Jiang Z, Wang X, Li Y, Jiang P. Hsp70 positively regulates porcine circovirus type 2 replication in vitro. Virology. 2013;447(1–2):52–62. doi: 10.1016/j.virol.2013.08.025. [DOI] [PubMed] [Google Scholar]
- 47.Kim MY, Ma Y, Zhang Y, Li J, Shu Y, Oglesbee M. hsp70-dependent antiviral immunity against cytopathic neuronal infection by vesicular stomatitis virus. J Virol. 2013;87(19):10668–10678. doi: 10.1128/JVI.00872-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar M, Rawat P, Khan SZ, Dhamija N, Chaudhary P, Ravi DS, Mitra D. Reciprocal regulation of human immunodeficiency virus-1 gene expression and replication by heat shock proteins 40 and 70. J Mol Biol. 2011;410(5):944–958. doi: 10.1016/j.jmb.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 49.Gratacos FM, Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip Rev RNA. 2010;1(3):457–473. doi: 10.1002/wrna.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson GM, Sutphen K, Bolikal S, Chuang KY, Brewer G. Thermodynamics and kinetics of Hsp70 association with A + U-rich mRNA-destabilizing sequences. J Biol Chem. 2001;276(48):44450–44456. doi: 10.1074/jbc.M108521200. [DOI] [PubMed] [Google Scholar]
- 51.Lu JY, Bergman N, Sadri N, Schneider RJ. Assembly of AUF1 with eIF4G-poly(A) binding protein complex suggests a translation function in AU-rich mRNA decay. RNA. 2006;12(5):883–893. doi: 10.1261/rna.2308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan W, Schwartz S. The Rev protein of human immunodeficiency virus type 1 counteracts the effect of an AU-rich negative element in the human papillomavirus type 1 late 3′ untranslated region. J Virol. 1995;69(5):2932–2945. doi: 10.1128/jvi.69.5.2932-2945.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Juhasz K, Lipp AM, Nimmervoll B, Sonnleitner A, Hesse J, Haselgruebler T, Balogi Z. The complex function of hsp70 in metastatic cancer. Cancers. 2013;6(1):42–66. doi: 10.3390/cancers6010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Juhasz K, Thuenauer R, Spachinger A, Duda E, Horvath I, Vigh L, Sonnleitner A, Balogi Z. Lysosomal rerouting of Hsp70 trafficking as a potential immune activating tool for targeting melanoma. Curr Pharm Des. 2013;19(3):430–440. doi: 10.2174/138161213804143644. [DOI] [PMC free article] [PubMed] [Google Scholar]