Abstract
Pluripotent stem cells differentiate into almost any specialized adult cell type of an organism. PSCs can be derived either from the inner cell mass of a blastocyst—giving rise to embryonic stem cells—or after reprogramming of somatic terminally differentiated cells to obtain ES-like cells, named induced pluripotent stem cells. The potential use of these cells in the clinic, for investigating in vitro early embryonic development or for screening the effects of new drugs or xenobiotics, depends on capability to maintain their genome integrity during prolonged culture and differentiation. Both human and mouse PSCs are prone to genomic and (epi)genetic instability during in vitro culture, a feature that seriously limits their real potential use. Culture-induced variations of specific chromosomes or genes, are almost all unpredictable and, as a whole, differ among independent cell lines. They may arise at different culture passages, suggesting the absence of a safe passage number maintaining genome integrity and rendering the control of genomic stability mandatory since the very early culture passages. The present review highlights the urgency for further studies on the mechanisms involved in determining (epi)genetic and chromosome instability, exploiting the knowledge acquired earlier on other cell types.
Keywords: Embryonic stem cells, Induced pluripotent stem cells, Aneuploidy, Regenerative medicine, Tissue engineering, Cell therapy
Introduction
Pluripotent stem cells (PSCs) have the potential to differentiate into almost any specialized adult cell type of an organism. PSCs can be derived from the inner cell mass (ICM) of a preimplantation blastocyst [1–3], giving rise to embryonic stem cell (ESC) lines. ESCs are undifferentiated and self-renewing cells that, once successfully established in vitro, preserve features and developmental potential (pluripotency) of the founder cells of the embryo. Almost a decade ago, Yamanaka’s group, enforcing the ectopic expression of four key transcription factors Oct-4, Sox2, Klf4, and c-Myc, reprogrammed for the first time somatic terminally differentiated fibroblast cells to pluripotency [4, 5]. These cells, called induced pluripotent stem cells (iPSCs), show similar self-renewal and differentiation potential of ESCs [4, 6, 7]. Since then, many other reprogramming methods have been proposed which entail either the genome integration of reprogramming genes (single cassette reprogramming vectors with Cre-Lox mediated transgene excision) or not (Adenovirus, Sendai Virus, PiggyBac, minicircle vectors, episomal plasmids), the overexpression of reprogramming proteins (proteins and mRNA delivery), the modulation of the expression of pluripotency or reprogramming genes (miRNA) and epigenetic chromatin remodeling (e.g., hydroxamic acid, trichostatin A and valproic acid) [8].
The potential use of these cells in regenerative medicine and tissue replacement, for investigating in vitro early embryonic development, screening the effects of new drugs or xenobiotics, relies on their capability to maintain genetic integrity during prolonged culture and differentiation [9]. Although PSCs have specialized mechanisms to preserve their genome integrity, such as facilitated apoptosis, a ~100 times lower mutation frequency compared to differentiated cells, high efficient DNA repair mechanisms and antioxidant defense [10, 11], they accumulate genetic and epigenetic abnormalities when maintained in long-term culture in vitro [for reviews, see 12, 13]. The diversity of culture protocols among independent laboratories perplexes the identification of the causes of such variations, although the presence or absence of a feeder layer, the animal or artificial source of serum and the different methods used for cell passaging seem to be major factors affecting the maintenance of a correct genome integrity [14].
In this review, we present a detailed overview of the alterations observed during long-term culture of human or mouse ESCs and iPSCs. We grouped these numerous alterations into five main types, such as chromosome number and structural variations, microsatellite instability, epigenetic instability and aberrations of mitochondrial DNA; also, for each of them we gave a synthesis of the known causal mechanisms.
Chromosome number variations
The loss of euploidy in PSCs during long-term culture has been widely reported by several independent laboratories. Using classical and molecular cytogenetic techniques, gain or loss of entire chromosomes has been described in both human and mouse ESC lines, with a strong species specificity. Gain of specific chromosomes appears to confer growth advantage and higher culture adaptation to cells carrying numerical variations (see paragraph “Chromosome structural variations”).
IPSC lines maintain a normal karyotype during the very early culture passages [5, 15], but, as for ESCs, they display chromosome number variability during prolonged culture.
Gain of entire chromosomes 12, 17, 20 or, to a lesser extent, 16 and X (particularly in female cell lines) frequently occurs in human ESCs (hESCs) [16–22], representing about 50 % of all aberrations observed [18]. A progressive tendency to chromosome gain occurs during culture and thus it is advisable to avoid their use beyond passage 70 [23].
Since the first significant study reporting the presence of alterations in the chromosome make-up of mouse ESC (mESC) lines [24], it has become evident that chromosome number variations are a general feature of mESCs. More recently, cytogenetic analyses performed on 540 [25] and 97 [26] independent mESC lines show, by 20 passages, that about half of the cell lines has modal chromosomal numbers of 41, 42 or 39 and about 25 % of all aberrations are trisomies of chromosomes 8 and 11. These same recurrent abnormalities are also described in other cell lines together with other numerical random aberrations [25–32].
As for ESCs, genetic variations in iPSCs may originate either early from individual starting cells or during prolonged culture, but they may be caused also by the reprogramming process itself. Large scale screenings of more than 320 lines show that about 10–20 % of either mouse or human iPSC lines contain numerous chromosomal aberrations [20, 21, 33–35] and of these, about 40-45 % are recurrent and involve full trisomy of chromosomes 8, 12 or X or aneuploidy of chromosomes 6 (in 1.4 % of the cell lines analyzed), 11 (16.9 %), 12 (5.63 %), 19 (1.41 %) and Y (7.04 %) [21, 30, 33, 36–40]. The recurrence of these specific aberrations suggests a process of culture adaptation during reprogramming [33], perhaps conferring a selective and/or proliferative advantage.
Mechanisms involved in the generation of chromosome number variations
In eukaryotic cells, four main mechanisms have been described to control spindle assembly, microtubule/kinetochore coupling, centrosome duplication and sister chromatid cohesion during cell replication (“Box 1”). The failure of one or more of them is the main cause of chromosome number variations described in cancer and differentiated cells [41, 42]. Up to date, an abnormal number of centrosomes, with the formation of multipolar spindles, has been the sole mechanism described as possible cause of chromosome number variability in hESCs [43–45]. To this regard, an abnormal number of centrosomes, associated with an altered expression level of mitotic spindle-related molecules such as Aurora A, CDK2 [44], BUB1, CENPE and MAD2 [45] results in the formation of multipolar spindles and centrosome number variability in 12 hESC lines derived from normal or parthenogenetic embryos, respectively.
Chromosome structural variations
The presence of structural variations (SVs; duplications, deletions, insertions, inversions, translocations, and complex rearrangements) in PSCs has been extensively reported making use of array comparative genomic hybridization. The most recurrent SVs detected are chromosome deletions, which cause loss of heterozygosity, and duplications, also defined as copy number variations (CNVs) [46]. Importantly, the presence of an additional copy of some specific genes (see below) confers to cells resistance to apoptosis, growth and/or survival advantage, leading to culture adaptation.
Following prolonged culture, hESC lines display several SVs, among which frequent additional segments of chromosomes 1, 12, 17 or 20 and deletions of chromosomes 8, 10, 16, 18 and 22 have been described (Table 1). Interestingly, additional SVs of chromosomes 1, 12, 17 or 20 have also been described in cancer cells in association with growth and/or survival advantage. Specifically, the duplication of the 12p13 band, containing the cell-cycle regulator CCND2, the pluripotency marker genes DPPA3 (Stella-Related), GDF3 and NANOG, has been associated with proliferative advantage and culture adaptation [17]. Proliferative advantage and culture adaptation in cancer cells is also associated with gain of chromosome 17q, containing BIRC5, encoding for the anti-apoptotic gene Survivin [47], and the miRNA has-mir-21, which regulates the anti-apoptotic gene Bcl2 [48, 49]. The duplicated 20q11.21 region contains the stemness gene DNMT3B, TPX2 and KIF3B, involved in the cell cycle, ID1 in cell growth, BCL2L1 and PDRG1 in cell survival and the immature form of the micro-RNA miR-1825 [50–53].
Table 1.
Summary of SVs described in hESC lines during prolonged in vitro culture
| Structural variations | Aberration | Putative candidates | References | |
|---|---|---|---|---|
| Genes | miRNA | |||
|
1q11–1p36 1q31.3 |
Gain | MCL1, IL6R, PSMD4, PSMB4, UBE2Q, UBAP2L, RBM8A, RPS27, PIAS3, POLR3C, HIST2H2AA, LASS2, MRPL9, JTB, HAX1, SHC1, APH-1A, BCL9, ZNF364, SSR2, MAPBPIP, CCT3, MEF2D, C1orf85 | hsa-mir-9-1 | [22, 66, 182] |
| Partial chromosome 12 or isochromosome 12p | Gain | NANOG, DPPA3 (Stella-Related), GDF3, CCND2 | n.d. | [17, 22, 183] |
|
17q11–q12, 17q21–17q23.2, 17q25–qter |
Gain | BIRC5 | miRNA has-mir-21 | [17, 22] |
| 20q11.21 | Gain | DNMT3B, TPX2, KIF3B, BCL2L1 PDRG1 | n.d. | [22, 66–68] |
| 8q24.3 | Loss | n.d. | n.d. | [18, 21, 46, 53, 184] |
| 10p13pter | Loss | n.d. | n.d. | |
| 16q | Loss | n.d. | n.d. | |
| 18q21qter | Loss | n.d. | n.d. | |
| 22q13qter | Loss | n.d. | n.d. | |
n.d. not detected
Several random SVs were detected in 49 (38.0 %) out of 129 mESC samples, revealing a high frequency of chromosomal aberrations. In 5 out of 129 mESC lines, the most recurrent aberration, acquired within 12 passages of culture, is the gain of 11qE2, a region that is fully syntenic with human 17q25, the latter a common aberration detected in hPSCs. Also, deletions of chromosomes 10qB and 14qC-14qE have been observed in 13.5 and 4.8 % of the mESC lines analyzed, respectively [30]. In addition, duplications of specific regions on chromosome 1, 2 and 12 were described in other mESC lines [54].
Comprehensive genome-wide analyses revealed that both hiPSCs and miPSCs accumulate a number of SVs higher than ESCs (during more than 50 passages) [18, 40, 55–59]. Whilst certain SVs may arise in both ESCs and iPSCs, some are specific of iPSCs and are recurrent (as found in ESCs) and shared by more that 25 % of all hiPSC lines analyzed; instead, others result from the amplification of 1q31.1, 8q24.3 and 17q21.1 [60] unique to hiPSCs. Loss of 8q24.3 region, observed in 12 % of cell lines analyzed, is found specifically in hiPSC lines [21].
In miPSCs, both gain and loss of SVs are variable among different cell lines and distributed along the majority of chromosomes [59].
Mechanisms involved in the generation of structural variations
The main processes, whose failure leads to chromosome deletions or duplications, are the homologous recombination (HR)—a repair mechanism that requires the presence of DNA sequence identity between chromosomes—and the non-homologous recombination (NHR), which, by contrast, either employs micro-homology or does not require homology. These processes are highly conserved in higher eukaryotes and among different species and are active in PSCs [61, 62]. Repair of DNA double strand breaks (DSBs) by HR could be the source of deletions arising in PSCs during prolonged culture, whereas several NHR sub-pathways (“Box 2”) [non-allelic homologous recombination (NAHR), non-homologous end-joining (NHEJ), microhomology-mediated break-induced replication (MMBIR) and fork stalling and template switching (FoSTeS)] seem to be specifically involved in the generation of duplications [63, 64].
One or more of these mechanisms are directly involved in the onset of SVs in PSCs. The recurrent gain of the 20q11-13 region, in several hESC lines (i.e., SA01, H1, VUB05, H7 and HSF1), might be caused by error-prone HR events, as reported for the synthenic mouse 2H region, ‘hypermutable’ for the presence of microsatellite D2MIT140 [65–68]. In addition, NAHR and NHEJ in hESM01, hESM01f and HS27 hESC lines are responsible for the mis-rejoining of DSBs, resulting in SVs [46].
In E14, AB2.2 and JM8 mESC lines, NAHR is responsible of the appearance of frequent CNVs during routine culture [69]. More recently, in TC1 and Xrcc4 −/− mESC lines, FoSTeS and MMBIR were proposed as candidate mechanisms for CNVs, as well as other more complex rearrangements [70].
Both hiPSC and miPSC lines harbor higher levels of SVs than ESCs [21, 33, 40, 55–57, 59], likely originated by NHEJ, FoSTeS or MMBIR, which appear the prevalent mechanisms during reprogramming and culture adaptation [57, 58].
Microsatellites instability
Microsatellites are highly polymorphic stretches of repetitive DNA regions, disseminated throughout the genome and very abundant in eukaryotes. Microsatellite instability (MSI) is a common hallmark observed in tumor cells, which display additions or deletions of repeated units [71]. To this regard, only a very recent paper tackles the occurrence of MSI in PSCs when maintained in culture for a long period. Specifically, ten hESC lines that were analyzed for 122 microsatellites dispersed over the whole genome, screened at early, medium and late passages from passage 7 to 334, showed that 2 out of 10 hESC lines accumulate altered (both deletions and duplications) microsatellites patterns, involving about 1–20 % of the total number of alleles, indicating very low and fluctuating frequency of MSI. The novel alleles observed could take over during prolonged culture as passenger mutations, without conferring a selective advantage [53].
Mechanisms involved in microsatellite instability
In cancer cells, the main cause of MSI is the inactivation or deficiency in the post-replicative mismatch repair (MMR) process [72] (“Box 3”), due to the failure of some key MMR genes. This failure was not observed in hESC lines, whose expression of MMR genes and proteins resulted unaltered [53].
Epigenetic instability
DNA methylation and histone tail modification are the major epigenetic processes that contribute to regulate gene expression in early developmental phases [for reviews, see 73, 74]. During prolonged in vitro culture, an increasing number of studies reported the accumulation of epigenetic alterations in PSCs [18, 66, 75–77]. These alterations dramatically affect their pluripotency status [78, 79] and, persisting during differentiation, are associated with aberrant phenotypes [80]. Following, we will summarize some alterations of the epigenetic status of PSCs, comprising modifications in imprinted genes, DNA and histone methylation and X-chromosome inactivation (XCI) [81, 82].
Imprinted genes
In several hESC and hiPSC lines, an epigenetic drift in the methylation pattern of some paternally (e.g., H19, PEG1, SNRPN and IGF2) or maternally (e.g., MEG3, IGF2R and SLC22A18) imprinted genes has been observed during prolonged culture in many independent laboratories [83–87]. These data were recently reinforced by a very comprehensive study that, analyzing more than 200 hPSC lines, demonstrated an overall variable allelic expression of several imprinted loci (DIRAS3, NAP1L5, MEST, H19, ZIM2/PEG3, PLAGL1, GRB10 and GNAS) compared to somatic cells [88].
In several mESC lines, the epigenetic status of H19, Peg1, Snrpn and Igf2r imprinted genes significantly changes during about 30 in vitro passages [89]. Epigenetic changes have also been reported in individual mESC subclones derived from the same cell line and independently expanded [90].
Altered epigenetic profile of imprinted genes, e.g. H19, MEST, IGF2R, SNRPN, which during mammalian development are involved in the regulation of cell division [91, 92] may confer to ESCs a selective growth advantage [81].
DNA and histone methylation
Cytosines methylation changes in CpG dinucleotides may arise both during the earliest stages of derivation and during in vitro expansion [76]. The RASSF1, DcR1 and PTPN6 genes, selected for their altered methylation in cancer, showed methylation modifications in their promoter during long-term hESCs culture (from 22 to 175 passages) [66]. Variations of DNA methylation levels of the same and of other genes (CBLN4, GPC3, RASSF1, RalGDS/AF-6 and glypican 3) were reported in other hESC lines by other groups, suggesting a high susceptibility of these loci to culture-induced changes [18, 75]. More recently, high-resolution techniques brought to light that 20 independent hESC lines have an almost identical DNA methylation pattern at both single CpG sites and CpG islands, but displayed high variability when the analysis was extended to global DNA methylation and transcription. Various genes exhibited transcriptional variations, while having demethylated promoters, whereas others showed high variability in DNA methylation of their promoters but expression stability [79].
Aberrant DNA methylation was also reported for few mESC lines. Abnormal hypermethylation pattern of various CpG islands was shown to cause dysregulated expression of genes involved in various processes of differentiation. Also, a strong correlation between abnormal methylation and density of H3K27me3 (associated with transcription repression; [93]) at specific loci was evidenced, H3K27me3 that mediates the recruitment of Dnmts to these specific sites [94]. Fractions of both h- and m-ESC populations have high levels of both H3K4me3 (associated with transcription activation; [93]) and H3K27me3 (bivalent chromatin) at lineage-specific loci [95], strongly influencing the differentiation potential of the cell line. This heterogeneity complicates the analysis of the epigenetic regulation in PSCs, in particular during prolonged culture.
The analysis of DNA methylation in iPSCs is complicated by the difficulty to discriminate between the impact of the reprogramming process itself, which requires a global remodeling of the epigenetic status, and the culture conditions. When compared to ESC line profiles, many loci or CpG islands resulted hypomethylated, whereas others, being incompletely reprogrammed, were variable among different iPSC lines [82]. Interestingly, these differences are reduced following long-term culture (>40 passages), suggesting continuous reprogramming of the iPSC population [96].
X-chromosome inactivation
X-chromosome inactivation (XCI) is the functional heterochromatinization and silencing of one of the two X chromosomes in female cells, an event that occurs very early during mammalian female embryonic development. The X-inactive specific transcript (XIST) drives X chromosome silencing, regulating the initiation of random XCI in the inner cell mass of an embryo [97].
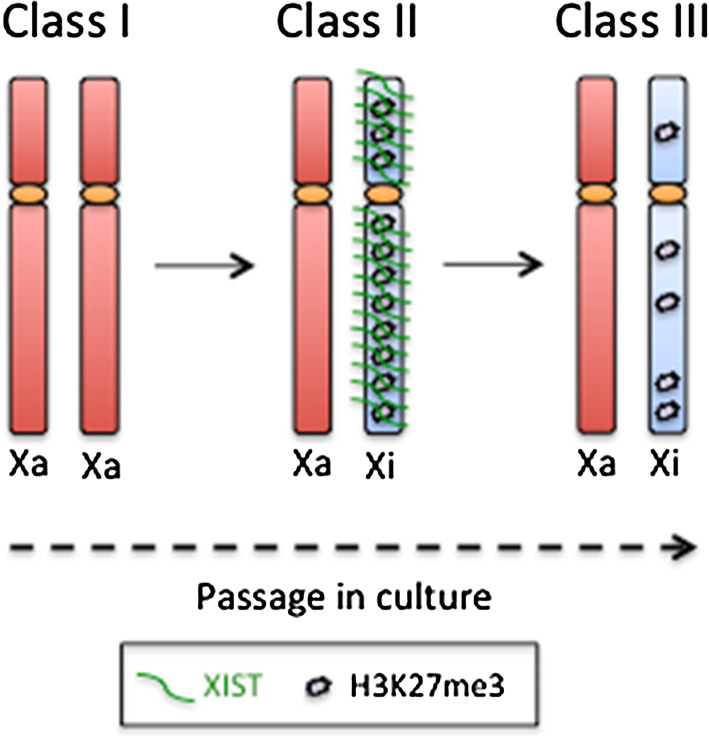
Several papers report variable status of XCI in female hESCs. Numerous hESC lines exhibit different states of XCI, together with a heterogeneity among cells of the same cell line [77, 99]. In brief, in all hESC lines analyzed, three different classes of the X inactivation status were identified (Fig. 1). Cells with both active X chromosomes (Xa) (Class I) express very low or undetectable XIST RNA. During differentiation they upregulate XIST and undergo XCI, as described also for mESCs. These cells easily switch to class II, in which the expression of the XIST RNA leads to the random inactivation of one of the two X chromosomes (Xi). Class II cells may switch to class III where the expression of XIST is lost, leading to partial reactivation of some Xi-linked genes [77]. However, XIST is not the sole player involved in the XCI process. XCI and XIST up-regulation occurs in combination with trimethylation of H3K27 on the inactive X [100]. In hESCs, Class I cells show neither H3K27me3 nor XIST foci, class II cells have H3K27me3 domains in all XIST+ cells, whereas class III cells do not display H3K27me3 at all. Thus, XIST clouds correlate perfectly with hypermethylation of H3K27, and loss of XIST in class III cells always induces the depletion of H3K27me3 on the inactive X [77]. The X chromosome status was found to be hyper-variable both in different hESC lines and among subculture or different culture passages in the same hESC line [77, 101]. It has been clearly demonstrated that the transit along the three classes of the X chromosome status typically occurs with continuous passage and during long-term culture. Under optimal culture conditions, active X chromosome status can be stably maintained in female hESCs for more than 100 passages [98]; however, suboptimal culture conditions (i.e., 20 % O2 concentration, harsh freeze–thaw cycles, high level of cell death during passages) induce XCI instability [102, 103].
Fig. 1.
Variations in X chromosome inactivation status in hESC lines during long-term culture
During reprogramming of somatic cells to iPSCs, the acquisition of pluripotency should be coupled with X chromosome reactivation. As for hESCs, hiPSC showed variable XCI pattern [82] with non-random X inactivation in low passage female hiPSCs, suggesting the maintenance of the X-inactivation pattern of the somatic founder cells [104, 105] or the presence of two active X chromosomes [106, 107]. Erosion of the XCI status during culture, due to loss of both XIST expression and foci with H3K27me3, has also been reported [105].
The variable pattern of XCI observed in hPSCs is probably an adaptive response of cells to the suboptimal in vitro culture conditions. However, the skewed pattern leads to an altered expression of X-linked genes, event that dramatically affects the undifferentiated status and the differentiation potential of the cell lines [102].
To our knowledge, no specific alterations in XCI have been described in mESC or miPSC lines. Female mESCs normally have two active X chromosomes (Xa) [108, 109] likely maintained by the Oct4, Sox2 and Nanog pluripotency regulating factors which bind to intron 1 of Xist and repress its activity [110, 111].
Mechanisms involved in epigenetic instability
The several enzymes in DNA/histone methylation (“Box 4”) and their cofactors [for a review 112] can be causal to the epigenetic instability. Nevertheless, up to date, the mechanisms producing aberrant epigenetic patterns observed in PSCs during long-term culture are unknown. DNA methylation, although critical during differentiation, does not appear to be essential for the maintenance of the undifferentiated status, since the triple knockout Dnmt1−/−/Dnmt3a−/−/Dnmt3b−/− mESC line proliferates, maintains pluripotency and retains their ability to differentiate [113]. It has been suggested that ESCs maintain stable heterochromatin and chromosomes by an epigenetic mechanism independent of CpG methylation. To this regard, previous studies have shown that ESCs have specific epigenetic regulatory mechanisms, namely hyperdynamic architectural chromatin proteins that bind loosely to chromatin. This state may contribute to maintain chromatin in a global open and plastic state [114, 115].
Aberrations of mitochondrial DNA
Human and mouse PSCs possess few and immature mitochondria, similar to those found in preimplantation embryos [116]. The integrity of mitochondrial DNA (mtDNA) has been scarcely investigated and its alterations have been described in few hPSC lines. Specifically, heteroplasmic missense or nonsense point mutations in NADH dehydrogenases 1, 2 and 4 and in ATPase genes were found in two out of nine (22 %) late-passage hESC lines, mutations that impair the proteins function [66]. More recently, large mtDNA deletions (ranged between 4973 and 8876 bp) were identified in 16 hESC lines cultivated from passage 3–334. The deletions observed may be present since the early passages after derivation and dispersed over the whole mitochondrial genome. Also, several mutated mtDNA molecules can be present in a single hESC line [117].
Homoplasmic and heteroplasmic single base mutations (substitutions, insertions and deletions) were also identified in four hiPSC lines reprogrammed from fibroblast of young and healthy donors [118]. Up to date, no information is currently available on the mtDNA integrity in mouse ESCs and iPSCs.
Mechanisms involved in the generation of mtDNA alterations
mtDNA deletions can arise during the replication phase, through slipped-strand mispairing between direct homologous base-pairs [119]. Although mitochondria possess active repair mechanisms, mtDNA has a high mutation rate, 100- to 1000-fold higher than nuclear DNA [120], probably due to the stressor environment of the mitochondria. The occurrence of point mutations in mtDNA has been extensively described, due to a failure of Base Excision Repair pathway that does not repair the oxidative DNA lesions [121].
Conclusions
Following the derivation of the first hESC line in 1998 [3] and, in 2006, of iPSCs [4], PSCs have represented a great promise for future therapeutic and clinical applications. However, PSCs are prone to genomic and (epi)genetic instability during in vitro culture [31, 122, 123], a feature that seriously limits their real potential use. Culture-induced aberrations of specific chromosomes or genes are almost all unpredictable. Although aberrations of chromosomes 12, 17 and X for hPSCs or 8 and 11 for mPSCs are recurrent in several different cell lines, as a whole, aberrations differ among independent cell lines and may arise at different culture passages. Regarding the effects that aberrations might have on PSCs, several studies indicate that their stemness is not affected [29, 31, 124–126]. However, upon differentiation, in the presence of specific abnormalities such as trisomy of chromosome 12, transplanted hPSCs increase their tumorigenicity [127]. Similarly, in the presence of trisomy of chromosome 21, the electrophysiological features of hESCs-derived cardiomyocytes are impaired [128] or the degeneration of mESCs-derived neurons is increased [129].
For successful therapeutical applications, the goal is the generation of Good Manufacturing Practice-grade PSCs. Two are the levels of intervention that are urgently needed: (1) to establish protocols that eliminate or minimize (epi)genetic and chromosome instability during culture, by acting, for iPSCs, on the reprogramming process itself (e.g., episomal plasmids, mRNA and microRNA technologies) and, for all PSCs, on the culture environmental conditions (e.g., animal feeder- and serum-free culture, reduction of ROS levels by inducing HIF pathway, increased levels of CHK1 or nucleoside or vitamin C supplementation, to reduce replication stress) [130, 131]; promising methods rely on engineered in vitro culture techniques that result in a downstream improvement of the karyotype stability [132–135] and quality of cell differentiation [136, 137]. (2) The absence of a safe passage number makes mandatory the monitoring of the genome integrity in PSCs, with the establishment of efficient and systematic methods, beyond the conventional banding techniques, spectral karyotyping, multicolor fluorescent in situ hybridization and array-based comparative genomic hybridization [32]. To this regard, an important contribution might derive from high-throughput microscopy, which allows the rapid and automated acquisition of numerous images, and the exome sequencing technique, the latter a cost-effective alternative to whole-genome sequencing [130]. Furthermore, the present review highlights the urgency for further studies on the mechanisms involved in determining (epi)genetic and chromosome instability, exploiting the knowledge acquired earlier on other cell types.
Box 1
The spindle assembly checkpoint (SAC) [for a review see 138] controls erroneous attachments of chromosomes to the mitotic spindle, to ensure proper chromosome segregation [139]. Defects or dysfunction in proteins involved in SAC lead to chromosome instability (onset of aneuploidies or deletions) in daughter cells [140, 141].
Similarly, microtubule/kinetochore coupling (attachment of spindle microtubules to the kinetochore) is crucial for the correct segregation of chromosomes during anaphase. Erroneous coupling (monotelic, syntelic or merotelic attachment) is the most common cause of genomic instability [142, 143], resulting in chromosome missegregation, mitotic delay and in aneuploid cell progeny [144–146]. Merotelic kinetochore attachment is the most frequent cause of aneuploidy in mitotic cells [147] and it is the primary described mechanism of chromosome instability in tumor cells [148]. The chromosomal passenger complex is responsible for the correction of these anchoring errors and its abolishment induces accumulation of errors in chromosome anchoring, with the consequent onset of abnormalities [149–151].
Aberrant centrosome duplication may adversely affect the maintenance of cell polarity and an abnormal number of centrosomes lead to multipolar spindles, resulting in chromosomal segregation abnormalities and aneuploidy of daughter cells [152, 153]. Several cell cycle kinases operate during the centriole duplication [154–156]. Among these, the Polo-like kinase 4 (Plk4) plays a key role in centrosome cycle [157], a process whose disruption causes aneuploidy [158, 159].
Sister chromatid cohesion is the process by which sister chromatids are paired and held together for alignment on the metaphase spindle. Cohesin is a chromosomal protein complex which regulates the correct distribution of the chromatid into the two daughter cells [160]. The lack or altered expression of cohesins induces premature sister separation or failed chromosome disjunction at the beginning of anaphase [161, 162], leading to cell death or aneuploidy [42].
Box 2
NAHR is one of the best-characterized recombination-based mechanisms in primates and it is responsible for recurrent CNVs. In this process, two regions of DNA with high sequence similarity are used for the repair of a DNA double strand break [163]. NAHR is mostly mediated by 10–300 kb long interspersed low-copy repeats [highly homologous sequences (95–97 %)] [164, 165] or short microsatellite with recombination hotspots that facilitate the occurrence of a misalignment during the strand invasion step [63].
NHEJ, MMBIR, FoSTeS are involved in the generation of CNVs, particularly in the formation of non-recurrent variation in copy number of portions of DNA [166]. NHEJ, together with HR, is one of the major mechanisms used by eukaryotic cells to repair DSBs [167], but some authors proposed this mechanism to explain chromosome duplication [168, 169]. MMBIR is involved in repairing single double-strand ends when portions of single-stranded DNA are present and share microhomology with the 3′ single-strand end from the collapsed fork [63]. Similarly, the replication-based mechanism FoSTeS acts when a replication fork stalls for the presence of a DNA lesion, allowing the lagging strand to detach and move to a region of micro-homology on a neighboring replication fork [170].
Box 3
Repetitive DNA regions are more prone than others to accumulate DNA replication errors and the failure of some key MMR genes (MLH1, MSH2, MSH6 and PMS2) or the minor accuracy of the process cause the accumulation of unrepaired mitotic errors. A replication slippage of the DNA polymerase occurs through five main steps: (1) DNA polymerase tackles a direct repeat during the DNA replication process; (2) DNA polymerase arrests and is, for a short time, released from the template strand; (3) the newly synthesized strand detaches from the template strand and pairs with another direct repeat upstream, forming a loop; (4) DNA polymerase, returning on the template strand, restarts with the normal replication process, but during the course of reassembling, the polymerase slips and repeats the insertion of dNTPs that were previously added. This faulty event causes the insertion of new nucleotides. Usually, MMR proteins recognize the temporary insertion–deletion loop, but when they do not work properly, this loop results in frame-shift mutations [171–173].
Box 4
The principal molecules involved in cytosines methylation are the DNA methyltransferases (Dnmt). Dnmt1, Dnmt3a, Dnmt3b and Dnmt3L are a family of proteins implicated in the maintenance of the correct DNA methylation status. Dnmt1, localizing to DNA replication foci and showing a strong affinity for hemi-methylated DNA, plays a major role in the maintenance of methylation in dividing cells [174, 175]. Dnmt3a and Dnmt3b, working on unmethylated as well as on hemi-methylated substrates, possess a de novo methylation activity [176, 177]. Dnmt3L (Dnmt3-like) methyltransferase is a regulator of DNA methylation, it lacks the catalytic functional domains and stimulates de novo methylation by Dnmt3a and Dnmt3b [178, 179]. In mESCs, Dnmt3a and Dnmt3b proteins are physically associated and act synergistically to methylate the promoters of pluripotency genes (i.e., Oct4, Nanog) during differentiation processes [180].
Covalent histone tails modifications govern the conformation of the chromatin and influence the gene transcription by promoting or restricting the recruitment of gene regulatory proteins [181]. Histones can be methylated on lysine (K) and arginine (R) residues and H3 and H4 histone tails are the main targets of methylation [for a review, 112]. Several histone-modifying enzymes including lysine or arginine methyltransferases, serine-threonine kinases, and acetyltransferases are involved in this process [93].
Contributor Information
Paola Rebuzzini, Phone: +39 0382 986323, Email: paola.rebuzzini@unipv.it.
Maurizio Zuccotti, Phone: +39 0382 986323, Email: maurizio.zuccotti@unipr.it.
Silvia Garagna, Phone: +39 0382 986323, Email: silvia.garagna@unipv.it.
References
- 1.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 7.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 8.González F, Boué S, Izpisúa Belmonte JC. Methods for making induced pluripotent stem cells: reprogramming à la carte. Nat Rev Genet. 2011;12:231–242. doi: 10.1038/nrg2937. [DOI] [PubMed] [Google Scholar]
- 9.Zapata AG, Alfaro D, García-Ceca J. Biology of stem cells: the role of microenvironments. Adv Exp Med Biol. 2012;741:135–151. doi: 10.1007/978-1-4614-2098-9_10. [DOI] [PubMed] [Google Scholar]
- 10.Tichy ED, Stambrook PJ. DNA repair in murine embryonic stem cells and differentiated cells. Exp Cell Res. 2008;314:1929–1936. doi: 10.1016/j.yexcr.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giachino C, Orlando L, Turinetto V. Maintenance of genomic stability in mouse embryonic stem cells: relevance in aging and disease. Int J Mol Sci. 2013;14:2617–2636. doi: 10.3390/ijms14022617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson SE, Loring JF. Genomic instability in pluripotent stem cells: implications for clinical applications. J Biol Chem. 2014;289:4578–4584. doi: 10.1074/jbc.R113.516419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliveira PH, da Silva CL, Cabral JM. Concise review: genomic instability in human stem cells: current status and future challenges. Stem Cells. 2014;32:2824–2832. doi: 10.1002/stem.1796. [DOI] [PubMed] [Google Scholar]
- 14.Rebuzzini P, Redi CA, Zuccotti M, Garagna S (2011) Genome stability in embryonic stem cells. In: Embryonic stem cells– recent advantages in pluripotent stem cell-based regenerative medicine Edited by Craig S, Atwood, p 399. doi: 10.5772/15091
- 15.Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Draper JS, Smith K, Gokhale P, Moore HD, Maltby E, Johnson J, Meisner L, Zwaka TP, Thomson JA, Andrews PW. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol. 2004;22:53–54. doi: 10.1038/nbt922. [DOI] [PubMed] [Google Scholar]
- 17.Baker DE, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, Heath PR, Holden H, Andrews PW. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- 18.Amps K, Andrews PW, Anyfantis G, et al. Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Int Stem Cell Initiat Nat Biotechnol. 2011;29:1132–1144. doi: 10.1038/nbt.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben-David U, Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer. 2011;11:268–277. doi: 10.1038/nrc3034. [DOI] [PubMed] [Google Scholar]
- 20.Taapken SM, Nisler BS, Newton MA, Sampsell-Barron TL, Leonhard KA, McIntire EM, Montgomery KD. Karotypic abnormalities in human induced pluripotent stem cells and embryonic stem cells. Nat Biotechnol. 2011;29:313–314. doi: 10.1038/nbt.1835. [DOI] [PubMed] [Google Scholar]
- 21.Martins-Taylor K, Nisler BS, Taapken SM, Compton T, Crandall L, Montgomery KD, Lalande M, Xu RH. Recurrent copy number variations in human induced pluripotent stem cells. Nat Biotechnol. 2011;29:488–491. doi: 10.1038/nbt.1890. [DOI] [PubMed] [Google Scholar]
- 22.Garitaonandia I, Amir H, Boscolo FS, Wambua GK, Schultheisz HL, Sabatini K, Morey R, Waltz S, Wang YC, Tran H, Leonardo TR, Nazor K, Slavin I, Lynch C, Li Y, Coleman R, Gallego Romero I, Altun G, Reynolds D, Dalton S, Parast M, Loring JF, Laurent LC. Increased risk of genetic and epigenetic instability in human embryonic stem cells associated with specific culture conditions. PLoS One. 2015;10:e0118307. doi: 10.1371/journal.pone.0118307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olariu V, Harrison NJ, Coca D, Gokhale PJ, Baker D, Billings S, Kadirkamanathan V, Andrews PW. Modeling the evolution of culture-adapted human embryonic stem cells. Stem Cell Res. 2010;4:50–56. doi: 10.1016/j.scr.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Longo L, Bygrave A, Grosveld FG, Pandolfi PP. The chromosome make-up of mouse embryonic stem cells is predictive of somatic and germ cell chimaerism. Transgenic Res. 1997;6:321–328. doi: 10.1023/A:1018418914106. [DOI] [PubMed] [Google Scholar]
- 25.Sugawara A, Goto K, Sotomaru Y, Sofuni T, Ito T. Current status of chromosomal abnormalities in mouse embryonic stem cell lines used in Japan. Comp Med. 2006;56:31–34. [PubMed] [Google Scholar]
- 26.Kim YM, Lee JY, Xia L, Mulvihill JJ, Li S. Trisomy 8: a common finding in mouse embryonic stem (ES) cell lines. Mol Cytogenet. 2013;6:3. doi: 10.1186/1755-8166-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rebuzzini P, Neri T, Mazzini G, Zuccotti M, Redi CA, Garagna S. Karyotype analysis of the euploid cell population of a mouse embryonic stem cell line revealed a high incidence of chromosome abnormalities that varied during culture. Cytogenet Genome Res. 2008;121:18–24. doi: 10.1159/000124377. [DOI] [PubMed] [Google Scholar]
- 28.Rebuzzini P, Neri T, Zuccotti M, Redi CA, Garagna S. Chromosome number variation in three mouse embryonic stem cell lines during culture. Cytotechnology. 2008;58:17–23. doi: 10.1007/s10616-008-9164-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rebuzzini P, Pignalosa D, Mazzini G, Di Liberto R, Coppola A, Terranova N, Magni P, Redi CA, Zuccotti M, Garagna S. Mouse embryonic stem cells that survive γ-rays exposure maintain pluripotent differentiation potential and genome stability. J Cell Physiol. 2012;227:1242–1249. doi: 10.1002/jcp.22908. [DOI] [PubMed] [Google Scholar]
- 30.Ben-David U, Benvenisty N. High prevalence of evolutionarily conserved and species-specific genomic aberrations in mouse pluripotent stem cells. Stem Cells. 2012;30:612–622. doi: 10.1002/stem.1057. [DOI] [PubMed] [Google Scholar]
- 31.Gaztelumendi N, Nogués C. Chromosome instability in mouse embryonic stem cells. Sci Rep. 2014;4:5324. doi: 10.1038/srep05324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rebuzzini P, Zuccotti M, Redi CA, Garagna S. Chromosomal abnormalities in embryonic and somatic stem cells. Cytogenet Genome Res. 2015;147:1–9. doi: 10.1159/000441645. [DOI] [PubMed] [Google Scholar]
- 33.Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 34.Chen Q, Shi X, Rudolph C, Yu Y, Zhang D, Zhao X, Mai S, Wang G, Schlegelberger B, Shi Q. Recurrent trisomy and Robertsonian translocation of chromosome 14 in murine iPS cell lines. Chromosome Res. 2011;19:857–868. doi: 10.1007/s10577-011-9239-y. [DOI] [PubMed] [Google Scholar]
- 35.Pasi CE, Dereli-Öz A, Negrini S, Friedli M, Fragola G, Lombardo A, Van Houwe G, Naldini L, Casola S, Testa G, Trono D, Pelicci PG, Halazonetis TD. Genomic instability in induced stem cells. Cell Death Differ. 2011;18:745–753. doi: 10.1038/cdd.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilić J, Pekarik V, Tiscornia G, Edel M, Boué S, Izpisúa Belmonte JC. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 37.Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, Khvorostov I, Ott V, Grunstein M, Lavon N, Benvenisty N, Croce CM, Clark AT, Baxter T, Pyle AD, Teitell MA, Pelegrini M, Plath K, Lowry WE. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ, Williams DJ, Kahler DJ, Yamaki M, Davidow L, Rodolfa CT, Dimos JT, Mikkilineni S, MacDermott AB, Woolf CJ, Henderson CE, Wichterle H, Eggan K. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, Lee JH, Loh YH, Manos PD, Montserrat N, Panopoulos AD, Ruiz S, Wilbert ML, Yu J, Kirkness EF, Izpisua Belmonte JC, Rossi DJ, Thomson JA, Eggan K, Daley GQ, Goldstein LS, Zhang K. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu P, Kaplan A, Yuan B, Hanna JH, Lupski JR, Reiner O. Passage number is a major contributor to genomic structural variations in mouse iPSCs. Stem Cells. 2014;32:2657–2667. doi: 10.1002/stem.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Curr Biol. 2010;20:R285–R295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holland AJ, Cleveland DW. Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep. 2012;13:501–514. doi: 10.1038/embor.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Lan W, Ems-McClung SC, Stukenberg PT, Walczak CE. Aurora B phosphorylates multiple sites on mitotic centromere-associated kinesin to spatially and temporally regulate its function. Mol Biol Cell. 2007;18:3264–3276. doi: 10.1091/mbc.E07-01-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holubcová Z, Matula P, Sedláčková M, Vinarský V, Doležalová D, Bárta T, Dvořák P, Hampl A. Human embryonic stem cells suffer from centrosomal amplification. Stem Cells. 2011;29:46–56. doi: 10.1002/stem.549. [DOI] [PubMed] [Google Scholar]
- 45.Brevini TA, Pennarossa G, Maffei S, Tettamanti G, Vanelli A, Isaac S, Eden A, Ledda S, de Eguileor M, Gandolfi F. Centrosome amplification and chromosomal instability in human and animal parthenogenetic cell lines. Stem Cell Rev. 2012;8:1076–1087. doi: 10.1007/s12015-012-9379-2. [DOI] [PubMed] [Google Scholar]
- 46.Närvä E, Autio R, Rahkonen N, Kong L, Harrison N, Kitsberg D, Borghese L, Itskovitz-Eldor J, Rasool O, Dvorak P, Hovatta O, Otonkoski T, Tuuri T, Cui W, Brüstle O, Baker D, Maltby E, Moore HD, Benvenisty N, Andrews PW, Yli-Harja O, Lahesmaa R. High-resolution DNA analysis of human embryonic stem cell lines reveals culture-induced copy number changes and loss of heterozygosity. Nat Biotechnol. 2010;28:371–377. doi: 10.1038/nbt.1615. [DOI] [PubMed] [Google Scholar]
- 47.Azuhata T, Scott D, Takamizawa S, Wen J, Davidoff A, Fukuzawa M, Sandler A. The inhibitor of apoptosis protein survivin is associated with high-risk behavior of neuroblastoma. J Pediatr Surg. 2001;36:1785–1791. doi: 10.1053/jpsu.2001.28839. [DOI] [PubMed] [Google Scholar]
- 48.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caldas C, Brenton JD. Sizing up miRNAs as cancer genes. Nat Med. 2005;11:712–714. doi: 10.1038/nm0705-712. [DOI] [PubMed] [Google Scholar]
- 50.Tanner MM, Tirkkonen M, Kallioniemi A, Isola J, Kuukasjärvi T, Collins C, Kowbel D, Guan XY, Trent J, Gray JW, Meltzer P, Kallioniemi OP. Independent amplification and frequent co-amplification of three nonsyntenic regions on the long arm of chromosome 20 in human breast cancer. Cancer Res. 1996;56:3441–3445. [PubMed] [Google Scholar]
- 51.Midorikawa Y, Yamamoto S, Ishikawa S, Kamimura N, Igarashi H, Sugimura H, Makuuchi M, Aburatani H. Molecular karyotyping of human hepatocellular carcinoma using single-nucleotide polymorphism arrays. Oncogene. 2006;25:5581–5590. doi: 10.1038/sj.onc.1209537. [DOI] [PubMed] [Google Scholar]
- 52.Koynova DK, Jordanova ES, Milev AD, Dijkman R, Kirov KS, Toncheva DI, Gruis NA. Gene-specific fluorescence in situ hybridization analysis on tissue microarray to refine the region of chromosome 20q amplification in melanoma. Melanoma Res. 2007;17:37–41. doi: 10.1097/CMR.0b013e3280141617. [DOI] [PubMed] [Google Scholar]
- 53.Nguyen HT, Geens M, Mertzanidou A, Jacobs K, Heirman C, Breckpot K, Spits C. Gain of 20q11.21 in human embryonic stem cells improves cell survival by increased expression of Bcl-xL. Mol Hum Reprod. 2014;20:168–177. doi: 10.1093/molehr/gat077. [DOI] [PubMed] [Google Scholar]
- 54.Liang G, Zhang Y. Embryonic stem cell and induced pluripotent stem cell: an epigenetic perspective. Cell Res. 2013;23:49–69. doi: 10.1038/cr.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, Barrero MJ, Ku S, Martynova M, Semechkin R, Galat V, Gottesfeld J, Izpisua Belmonte JC, Murry C, Keirstead HS, Park HS, Schmidt U, Laslett AL, Muller FJ, Nievergelt CM, Shamir R, Loring JF. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Närvä E, Ng S, Sourour M, Hämäläinen R, Olsson C, Lundin K, Mikkola M, Trokovic R, Peitz M, Brüstle O, Bazett-Jones DP, Alitalo K, Lahesmaa R, Nagy A, Otonkoski T. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 57.Quinlan AR, Boland MJ, Leibowitz ML, Shumilina S, Pehrson SM, Baldwin KK, Hall IM. Genome sequencing of mouse induced pluripotent stem cells reveals retroelement stability and infrequent DNA rearrangement during reprogramming. Cell Stem Cell. 2011;9:366–373. doi: 10.1016/j.stem.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abyzov A, Mariani J, Palejev D, Zhang Y, Haney MS, Tomasini L, Ferrandino AF, Rosenberg Belmaker LA, Szekely A, Wilson M, Kocabas A, Calixto NE, Grigorenko EL, Huttner A, Chawarska K, Weissman S, Urban AE, Gerstein M, Vaccarino FM. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012;492:438–442. doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Y, Guo L, Chen J, Zhao X, Zhou W, Zhang C, Wang J, Jin L, Pei D, Zhang F. Genome-wide CNV analysis in mouse induced pluripotent stem cells reveals dosage effect of pluripotent factors on genome integrity. BMC Genom. 2014;15:79. doi: 10.1186/1471-2164-15-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martins-Taylor K, Xu RH. Concise review: genomic stability of human induced pluripotent stem cells. Stem Cells. 2012;30:22–27. doi: 10.1002/stem.705. [DOI] [PubMed] [Google Scholar]
- 61.Adams BR, Golding SE, Rao RR, Valerie K (2010a) Dynamic dependence on ATR and ATM for double-strand break repair in human embryonic stem cells and neural descendants. PLoS One 5:e10001. doi: 10.1371/journal.pone.0010001 [DOI] [PMC free article] [PubMed]
- 62.Adams BR, Hawkins AJ, Povirk LF, Valerie K. ATM-independent, high-fidelity nonhomologous end joining predominates in human embryonic stem cells. Aging (Albany NY) 2010;2:582–596. doi: 10.18632/aging.100197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bogomazova AN, Lagarkova MA, Tskhovrebova LV, Shutova MV, Kiselev SL. Error-prone nonhomologous end joining repair operates in human pluripotent stem cells during late G2. Aging (Albany NY) 2011;3:584–596. doi: 10.18632/aging.100336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rithidech K, Bond VP, Cronkite EP, Thompson MH, Bullis JE. Hypermutability of mouse chromosome 2 during the development of x-ray-induced murine myeloid leukemia. Proc Natl Acad Sci USA. 1995;92:1152–1156. doi: 10.1073/pnas.92.4.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maitra A, Arking DE, Shivapurkar N, Ikeda M, Stastny V, Kassauei K, Sui G, Cutler DJ, Liu Y, Brimble SN, Noaksson K, Hyllner J, Schulz TC, Zeng X, Freed WJ, Crook J, Abraham S, Colman A, Sartipy P, Matsui S, Carpenter M, Gazdar AF, Rao M, Chakravarti A. Genomic alterations in cultured human embryonic stem cells. Nat Genet. 2005;37:1099–1103. doi: 10.1038/ng1631. [DOI] [PubMed] [Google Scholar]
- 67.Lefort N, Feyeux M, Bas C, Féraud O, Bennaceur-Griscelli A, Tachdjian G, Peschanski M, Perrier AL. Human embryonic stem cells reveal recurrent genomic instability at 20q11.21. Nat Biotechnol. 2008;26:1364–1366. doi: 10.1038/nbt.1509. [DOI] [PubMed] [Google Scholar]
- 68.Spits C, Mateizel I, Geens M, Mertzanidou A, Staessen C, Vandeskelde Y, Van der Elst J, Liebaers I, Sermon K. Recurrent chromosomal abnormalities in human embryonic stem cells. Nat Biotechnol. 2008;26:1361–1363. doi: 10.1038/nbt.1510. [DOI] [PubMed] [Google Scholar]
- 69.Liang Q, Conte N, Skarnes WC, Bradley A. Extensive genomic copy number variation in embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:17453–17456. doi: 10.1073/pnas.0805638105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arlt MF, Rajendran S, Birkeland SR, Wilson TE, Glover TW. De novo CNV formation in mouse embryonic stem cells occurs in the absence of Xrcc4-dependent nonhomologous end joining. PLoS Genet. 2012;8:e1002981. doi: 10.1371/journal.pgen.1002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Richards RI, Sutherland GR. Dynamic mutations: a new class of mutations causing human disease. Cell. 1992;70:709–712. doi: 10.1016/0092-8674(92)90302-S. [DOI] [PubMed] [Google Scholar]
- 72.Umar A, Kunkel TA. DNA-replication fidelity, mismatch repair and genome instability in cancer cells. Eur J Biochem. 1996;238:297–307. doi: 10.1111/j.1432-1033.1996.0297z.x. [DOI] [PubMed] [Google Scholar]
- 73.Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol. 2010;28:1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- 74.Ferguson-Smith AC. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet. 2011;12:565–575. doi: 10.1038/nrg3032. [DOI] [PubMed] [Google Scholar]
- 75.Bibikova M, Chudin E, Wu B, Zhou L, Garcia EW, Liu Y, Shin S, Plaia TW, Auerbach JM, Arking DE, Gonzalez R, Crook J, Davidson B, Schulz TC, Robins A, Khanna A, Sartipy P, Hyllner J, Vanguri P, Savant-Bhonsale S, Smith AK, Chakravarti A, Maitra A, Rao M, Barker DL, Loring JF, Fan JB. Human embryonic stem cells have a unique epigenetic signature. Genome Res. 2006;16:1075–1083. doi: 10.1101/gr.5319906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Allegrucci C, Young LE. Differences between human embryonic stem cell lines. Hum Reprod Update. 2007;13:103–120. doi: 10.1093/humupd/dml041. [DOI] [PubMed] [Google Scholar]
- 77.Silva SS, Rowntree RK, Mekhoubad S, Lee JT. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:4820–4825. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nagy A, Gócza E, Diaz EM, Prideaux VR, Iványi E, Markkula M, Rossant J. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- 79.Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, Gnirke A, Eggan K, Meissner A. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dean W, Bowden L, Aitchison A, Klose J, Moore T, Meneses JJ, Reik W, Feil R. Altered imprinted gene methylation and expression in completely ES cell-derived mouse fetuses: association with aberrant phenotypes. Development. 1998;125:2273–2282. doi: 10.1242/dev.125.12.2273. [DOI] [PubMed] [Google Scholar]
- 81.Lund RJ, Närvä E, Lahesmaa R. Genetic and epigenetic stability of human pluripotent stem cells. Nat Rev Genet. 2012;13:732–744. doi: 10.1038/nrg3271. [DOI] [PubMed] [Google Scholar]
- 82.Nguyen HT, Geens M, Spits C. Genetic and epigenetic instability in human pluripotent stem cells. Hum Reprod Update. 2013;19:187–205. doi: 10.1093/humupd/dms048. [DOI] [PubMed] [Google Scholar]
- 83.Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Human embryonic stem cells as a model for studying epigenetic regulation during early development. Cell Cycle. 2005;4:1323–1326. doi: 10.4161/cc.4.10.2076. [DOI] [PubMed] [Google Scholar]
- 84.Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Status of genomic imprinting in human embryonic stem cells as revealed by a large cohort of independently derived and maintained lines. Hum Mol Genet. 2007;16:R243–R251. doi: 10.1093/hmg/ddm245. [DOI] [PubMed] [Google Scholar]
- 85.Pick M, Stelzer Y, Bar-Nur O, Mayshar Y, Eden A, Benvenisty N. Clone- and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells. 2009;27:2686–2690. doi: 10.1002/stem.205. [DOI] [PubMed] [Google Scholar]
- 86.Li SS, Yu SL, Singh S. Epigenetic states and expression of imprinted genes in human embryonic stem cells. World J Stem Cells. 2010;2:97–102. doi: 10.4252/wjsc.v2.i4.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nishino K, Toyoda M, Yamazaki-Inoue M, Fukawatase Y, Chikazawa E, Sakaguchi H, Akutsu H, Umezawa A. DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet. 2011;7:e1002085. doi: 10.1371/journal.pgen.1002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nazor KL, Altun G, Lynch C, Tran H, Harness JV, Slavin I, Garitaonandia I, Müller FJ, Wang YC, Boscolo FS, Fakunle E, Dumevska B, Lee S, Park HS, Olee T, D’Lima DD, Semechkin R, Parast MM, Galat V, Laslett AL, Schmidt U, Keirstead HS, Loring JF, Laurent LC. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Horii T, Hatada I. Epigenetic instability in embryonic stem cells in “pluripotent stem cells”, edited by Deepa Bhartiya and Nibedita Lenka. INTECH Open Access Publisher. 2013 [Google Scholar]
- 90.Humpherys D, Eggan K, Akutsu H, Hochedlinger K, Rideout WM, 3rd, Biniszkiewicz D, Yanagimachi R, Jaenisch R. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- 91.Piedrahita JA. The role of imprinted genes in fetal growth abnormalities. Birth Defects Res A Clin Mol Teratol. 2011;91:682–692. doi: 10.1002/bdra.20795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ishida M, Moore GE. The role of imprinted genes in humans. Mol Aspects Med. 2013;34:826–840. doi: 10.1016/j.mam.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 93.Kouzarides T. SnapShot: histone-modifying enzymes. Cell. 2007;131:822. doi: 10.1016/j.cell.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 94.Ludwig G, Nejman D, Hecht M, Orlanski S, Abu-Remaileh M, Yanuka O, Sandler O, Marx A, Roberts D, Benvenisty N, Bergman Y, Mendelsohn M, Cedar H. Aberrant DNA methylation in ES cells. PLoS ONE. 2014;9:e96090. doi: 10.1371/journal.pone.0096090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 96.Planello AC, Ji J, Sharma V, Singhania R, Mbabaali F, Müller F, Alfaro JA, Bock C, De Carvalho DD, Batada NN. Aberrant DNA methylation reprogramming during induced pluripotent stem cell generation is dependent on the choice of reprogramming factors. Cell Regen (Lond) 2014;3:4. doi: 10.1186/2045-9769-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pollex T, Heard E. Recent advances in X-chromosome inactivation research. Curr Opin Cell Biol. 2012;24:825–832. doi: 10.1016/j.ceb.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 98.Shen Y, Matsuno Y, Fouse SD, Rao N, Root S, Xu R, Pellegrini M, Riggs AD, Fan G. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc Natl Acad Sci USA. 2008;105:4709–4714. doi: 10.1073/pnas.0712018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dvash T, Lavon N, Fan G. Variations of X chromosome inactivation occur in early passages of female human embryonic stem cells. PLoS ONE. 2010;5:e11330. doi: 10.1371/journal.pone.0011330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- 101.Liu W, Yin Y, Jiang Y, Kou C, Luo Y, Huang S, Zheng Y, Li S, Li Q, Guo L, Gao S, Sun X. Genetic and epigenetic X-chromosome variations in a parthenogenetic human embryonic stem cell line. J Assist Reprod Genet. 2011;28:303–313. doi: 10.1007/s10815-010-9517-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, Santagata S, Powers D, Barrett CB, Young RA, Lee JT, Jaenisch R, Mitalipova M. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 103.Diaz Perez SV, Kim R, Li Z, Marquez VE, Patel S, Plath K, Clark AT. Derivation of new human embryonic stem cell lines reveals rapid epigenetic progression in vitro that can be prevented by chemical modification of chromatin. Hum Mol Genet. 2012;21:751–764. doi: 10.1093/hmg/ddr506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tchieu J, Kuoy E, Chin MH, Trinh H, Patterson M, Sherman SP, Aimiuwu O, Lindgren A, Hakimian S, Zack JA, Clark AT, Pyle AD, Lowry WE, Plath K. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell. 2010;7:329–342. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mekhoubad S, Bock C, de Boer AS, Kiskinis E, Meissner A, Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bermejo-Alvarez P, Ramos-Ibeas P, Gutierrez-Adan A. Solving the “X” in embryos and stem cells. Stem Cells Dev. 2012;21:1215–1224. doi: 10.1089/scd.2011.0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Minkovsky A, Patel S, Plath K. Concise review: pluripotency and the transcriptional inactivation of the female Mammalian X chromosome. Stem Cells. 2012;30:48–54. doi: 10.1002/stem.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Navarro P, Chambers I, Karwacki-Neisius V, Chureau C, Morey C, Rougeulle C, Avner P. Molecular coupling of Xist regulation and pluripotency. Science. 2008;321:1693–1695. doi: 10.1126/science.1160952. [DOI] [PubMed] [Google Scholar]
- 111.van Bemmel JG, Mira-Bontenbal H, Gribnau J. Cis- and trans-regulation in X inactivation. Chromosoma. 2015 doi: 10.1007/s00412-015-0525-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Putiri EL, Robertson KD. Epigenetic mechanisms and genome stability. Clin Epigenetics. 2011;2:299–314. doi: 10.1007/s13148-010-0017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, Nakayama J, Okano M. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- 114.Kimura H, Tada M, Nakatsuji N, Tada T. Histone code modifications on pluripotential nuclei of reprogrammed somatic cells. Mol Cell Biol. 2004;24:5710–5720. doi: 10.1128/MCB.24.13.5710-5720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 116.Lonergan T, Bavister B, Brenner C. Mitochondria in stem cells. Mitochondrion. 2007;7:289–296. doi: 10.1016/j.mito.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Van Haute L, Spits C, Geens M, Seneca S, Sermon K. Human embryonic stem cells commonly display large mitochondrial DNA deletions. Nat Biotechnol. 2013;31:20–23. doi: 10.1038/nbt.2473. [DOI] [PubMed] [Google Scholar]
- 118.Prigione A, Lichtner B, Kuhl H, Struys EA, Wamelink M, Lehrach H, Ralser M, Timmermann B, Adjaye J. Human induced pluripotent stem cells harbor homoplasmic and heteroplasmic mitochondrial DNA mutations while maintaining human embryonic stem cell-like metabolic reprogramming. Stem Cells. 2011;29:1338–1348. doi: 10.1002/stem.683. [DOI] [PubMed] [Google Scholar]
- 119.Pitceathly RD, Rahman S, Hanna MG. Single deletions in mitochondrial DNA-molecular mechanisms and disease phenotypes in clinical practice. Neuromuscul Disord. 2012;22:577–586. doi: 10.1016/j.nmd.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 120.Wallace DC, Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Med. 2013;3:a021220. doi: 10.1101/cshperspect.a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Alexeyev MF. Is there more to aging than mitochondrial DNA and reactive oxygen species? FEBS J. 2009;276:5768–5787. doi: 10.1111/j.1742-4658.2009.07269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Catalina P, Montes R, Ligero G, Sanchez L, de la Cueva T, Bueno C, Leone PE, Menendez P. Human ESCs predisposition to karyotypic instability: is a matter of culture adaptation or differential vulnerability among hESC lines due to inherent properties? Mol Cancer. 2008;7:76. doi: 10.1186/1476-4598-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Peterson SE, Westra JW, Rehen SK, Young H, Bushman DM, Paczkowski CM, Yung YC, Lynch CL, Tran HT, Nickey KS, Wang YC, Laurent LC, Loring JF, Carpenter MK, Chun J. Normal human pluripotent stem cell lines exhibit pervasive mosaic aneuploidy. PLoS ONE. 2011;6:e23018. doi: 10.1371/journal.pone.0023018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Biancotti JC, Narwani K, Buehler N, Mandefro B, Golan-Lev T, Yanuka O, Clark A, Hill D, Benvenisty N, Lavon N. Human embryonic stem cells as models for aneuploid chromosomal syndromes. Stem Cells. 2010;28:1530–1540. doi: 10.1002/stem.483. [DOI] [PubMed] [Google Scholar]
- 125.Rebuzzini P, Fassina L, Mulas F, Bellazzi R, Redi CA, Di Liberto R, Magenes G, Adjaye J, Zuccotti M, Garagna S. Mouse embryonic stem cells irradiated with γ-rays differentiate into cardiomyocytes but with altered contractile properties. Mutat Res. 2013;756:37–45. doi: 10.1016/j.mrgentox.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 126.Luft S, Pignalosa D, Nasonova E, Arrizabalaga O, Helm A, Durante M, Ritter S. Fate of D3 mouse embryonic stem cells exposed to X-rays or carbon ions. Mutat Res, Genet Toxicol Environ Mutagen. 2014;760:56–63. doi: 10.1016/j.mrgentox.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 127.Ben-David U, Arad G, Weissbein U, Mandefro B, Maimon A, Golan-Lev T, Narwani K, Clark AT, Andrews PW, Benvenisty N, Carlos Biancotti J. Aneuploidy induces profound changes in gene expression, proliferation and tumorigenicity of human pluripotent stem cells. Nat Commun. 2014;5:4825. doi: 10.1038/ncomms5825. [DOI] [PubMed] [Google Scholar]
- 128.Bosman A, Letourneau A, Sartiani L, Del Lungo M, Ronzoni F, Kuziakiv R, Tohonen V, Zucchelli M, Santoni F, Guipponi M, Dumevska B, Hovatta O, Antonarakis SE, Jaconi ME. Perturbations of heart development and function in cardiomyocytes from human embryonic stem cells with trisomy 21. Stem Cells. 2015;33:1434–1446. doi: 10.1002/stem.1961. [DOI] [PubMed] [Google Scholar]
- 129.Wang CC, Kazuki Y, Oshimura M, Ikeo K, Gojobori T. Gene dosage imbalance of human chromosome 21 in mouse embryonic stem cells differentiating to neurons. Gene. 2011;481:93–101. doi: 10.1016/j.gene.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 130.Ruiz S, Lopez-Contreras AJ, Gabut M, Marion RM, Gutierrez-Martinez P, Bua S, Ramirez O, Olalde I, Rodrigo-Perez S, Li H, Marques-Bonet T, Serrano M, Blasco MA, Batada NN, Fernandez-Capetillo O. Limiting replication stress during somatic cell reprogramming reduces genomic instability in induced pluripotent stem cells. Nat Commun. 2015;6:8036. doi: 10.1038/ncomms9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gao Y, Han Z, Li Q, Wu Y, Shi X, Ai Z, Du J, Li W, Guo Z, Zhang Y. Vitamin C induces a pluripotent state in mouse embryonic stem cells by modulating microRNA expression. FEBS J. 2015;282:685–699. doi: 10.1111/febs.13173. [DOI] [PubMed] [Google Scholar]
- 132.Otsuji TG, Bin J, Yoshimura A, Tomura M, Tateyama D, Minami I, Yoshikawa Y, Aiba K, Heuser JE, Nishino T, Hasegawa K, Nakatsuji N. A 3D sphere culture system containing functional polymers for large-scale human pluripotent stem cell production. Stem Cell Reports. 2014;2:734–745. doi: 10.1016/j.stemcr.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lou YR, Kanninen L, Kuisma T, Niklander J, Noon LA, Burks D, Urtti A, Yliperttula M. The use of nanofibrillar cellulose hydrogel as a flexible three-dimensional model to culture human pluripotent stem cells. Stem Cells Dev. 2014;23:380–392. doi: 10.1089/scd.2013.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lei Y, Jeong D, Xiao J, Schaffer DV. Developing defined and scalable 3D culture systems for culturing human pluripotent stem cells at high densities. Cell Mol Bioeng. 2014;7:172–183. doi: 10.1007/s12195-014-0333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xu K, Narayanan K, Lee F, Bae KH, Gao S, Kurisawa M. Enzyme-mediated hyaluronic acid-tyramine hydrogels for the propagation of human embryonic stem cells in 3D. Acta Biomater. 2015;24:159–171. doi: 10.1016/j.actbio.2015.06.026. [DOI] [PubMed] [Google Scholar]
- 136.Takayama K, Kawabata K, Nagamoto Y, Kishimoto K, Tashiro K, Sakurai F, Tachibana M, Kanda K, Hayakawa T, Furue MK, Mizuguchi H. 3D spheroid culture of hESC/hiPSC-derived hepatocyte-like cells for drug toxicity testing. Biomaterials. 2013;34:1781–1789. doi: 10.1016/j.biomaterials.2012.11.029. [DOI] [PubMed] [Google Scholar]
- 137.Berger DR, Ware BR, Davidson MD, Allsup SR, Khetani SR. Enhancing the functional maturity of induced pluripotent stem cell-derived human hepatocytes by controlled presentation of cell-cell interactions in vitro. Hepatology. 2015;61:1370–1381. doi: 10.1002/hep.27621. [DOI] [PubMed] [Google Scholar]
- 138.Lew DJ, Burke DJ. The spindle assembly and spindle position checkpoints. Annu Rev Genet. 2003;37:251–282. doi: 10.1146/annurev.genet.37.042203.120656. [DOI] [PubMed] [Google Scholar]
- 139.Musacchio A. Spindle assembly checkpoint: the third decade. Philos Trans R Soc Lond B Biol Sci. 2011;366:3595–3604. doi: 10.1098/rstb.2011.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 141.Rao CV, Yamada HY, Yao Y, Dai W. Enhanced genomic instabilities caused by deregulated microtubule dynamics and chromosome segregation: a perspective from genetic studies in mice. Carcinogenesis. 2009;30:1469–1474. doi: 10.1093/carcin/bgp081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tanaka K, Hirota T. Chromosome segregation machinery and cancer. Cancer Sci. 2009;100:1158–1165. doi: 10.1111/j.1349-7006.2009.01178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Silkworth WT, Nardi IK, Paul R, Mogilner A, Cimini D. Timing of centrosome separation is important for accurate chromosome segregation. Mol Biol Cell. 2012;23:401–411. doi: 10.1091/mbc.E11-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gregan J, Polakova S, Zhang L, Tolić-Nørrelykke IM, Cimini D. Merotelic kinetochore attachment: causes and effects. Trends Cell Biol. 2011;21:374–381. doi: 10.1016/j.tcb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Adams RR, Carmena M, Earnshaw WC. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 2001;11:49–54. doi: 10.1016/S0962-8924(00)01880-8. [DOI] [PubMed] [Google Scholar]
- 150.Kallio MJ, Beardmore VA, Weinstein J, Gorbsky GJ. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J Cell Biol. 2002;158:841–847. doi: 10.1083/jcb.200201135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Guerrero AA, Martínez AC, van Wely KH. Merotelic attachments and non-homologous end joining are the basis of chromosomal instability. Cell Div. 2010;5:13. doi: 10.1186/1747-1028-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Salisbury JL, Whitehead CM, Lingle WL, Barrett SL. Centrosomes and cancer. Biol Cell. 1999;91:451–460. doi: 10.1111/j.1768-322X.1999.tb01100.x. [DOI] [PubMed] [Google Scholar]
- 153.Krämer A, Neben K, Ho AD. Centrosome replication, genomic instability and cancer. Leukemia. 2002;16:767–775. doi: 10.1038/sj.leu.2402454. [DOI] [PubMed] [Google Scholar]
- 154.Strnad P, Gönczy P. Mechanisms of procentriole formation. Trends Cell Biol. 2008;18:389–396. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 155.Loncarek J, Khodjakov A. Ab ovo or de novo? Mechanisms of centriole duplication. Mol Cells. 2009;27:135–142. doi: 10.1007/s10059-009-0017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Nigg EA, Stearns T. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J Cell Biol. 2010;188:191–198. doi: 10.1083/jcb.200911102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nigg EA. Polo-like kinases: positive regulators of cell division from start to finish. Curr Opin Cell Biol. 1998;10:776–783. doi: 10.1016/S0955-0674(98)80121-X. [DOI] [PubMed] [Google Scholar]
- 159.McCoy RC, Demko Z, Ryan A, Banjevic M, Hill M, Sigurjonsson S, Rabinowitz M, Fraser HB, Petrov DA. Common variants spanning PLK4 are associated with mitotic-origin aneuploidy in human embryos. Science. 2015;348:235–238. doi: 10.1126/science.aaa3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/S0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 161.Iwaizumi M, Shinmura K, Mori H, Yamada H, Suzuki M, Kitayama Y, Igarashi H, Nakamura T, Suzuki H, Watanabe Y, Hishida A, Ikuma M, Sugimura H. Human Sgo1 downregulation leads to chromosomal instability in colorectal cancer. Gut. 2009;58:249–260. doi: 10.1136/gut.2008.149468. [DOI] [PubMed] [Google Scholar]
- 162.Zhang N, Ge G, Meyer R, Sethi S, Basu D, Pradhan S, Zhao YJ, Li XN, Cai WW, El-Naggar AK, Baladandayuthapani V, Kittrell FS, Rao PH, Medina D, Pati D. Overexpression of Separase induces aneuploidy and mammary tumorigenesis. Proc Natl Acad Sci USA. 2008;105:13033–13038. doi: 10.1073/pnas.0801610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. Pathogenetics. 2008;1:4. doi: 10.1186/1755-8417-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/S0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 165.Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- 166.Arlt MF, Wilson TE, Glover TW. Replication stress and mechanisms of CNV formation. Curr Opin Genet Dev. 2012;22:204–210. doi: 10.1016/j.gde.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lieber MR, Gu J, Lu H, Shimazaki N, Tsai AG. Nonhomologous DNA end joining (NHEJ) and chromosomal translocations in humans. Subcell Biochem. 2010;50:279–296. doi: 10.1007/978-90-481-3471-7_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Woodward KJ, Cundall M, Sperle K, Sistermans EA, Ross M, Howell G, Gribble SM, Burford DC, Carter NP, Hobson DL, Garbern JY, Kamholz J, Heng H, Hodes ME, Malcolm S, Hobson GM. Heterogeneous duplications in patients with Pelizaeus-Merzbacher disease suggest a mechanism of coupled homologous and nonhomologous recombination. Am J Hum Genet. 2005;77:966–987. doi: 10.1086/498048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lee JH, Kim KH, Morio T, Kim H. Ataxia-telangiectasia-mutated-dependent activation of Ku in human fibroblasts exposed to hydrogen peroxide. Ann N Y Acad Sci. 2006;1091:76–82. doi: 10.1196/annals.1378.056. [DOI] [PubMed] [Google Scholar]
- 170.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 171.Wells RD. Molecular basis of genetic instability of triplet repeats. J Biol Chem. 1996;271:2875–2878. doi: 10.1074/jbc.271.6.2875. [DOI] [PubMed] [Google Scholar]
- 172.Viguera E, Canceill D, Ehrlich SD. Replication slippage involves DNA polymerase pausing and dissociation. EMBO J. 2001;20:2587–2595. doi: 10.1093/emboj/20.10.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138(2073–2087):e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bestor TH. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 1992;11:2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-P. [DOI] [PubMed] [Google Scholar]
- 176.Liang G, Chan MF, Tomigahara Y, Tsai YC, Gonzales FA, Li E, Laird PW, Jones PA. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol. 2002;22:480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yokochi T, Robertson KD. Preferential methylation of unmethylated DNA by Mammalian de novo DNA methyltransferase Dnmt3a. J Biol Chem. 2002;277:11735–11745. doi: 10.1074/jbc.M106590200. [DOI] [PubMed] [Google Scholar]
- 178.Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci USA. 2002;99:16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 180.Li JY, Pu MT, Hirasawa R, Li BZ, Huang YN, Zeng R, Jing NH, Chen T, Li E, Sasaki H, Xu GL. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol Cell Biol. 2007;27:8748–8759. doi: 10.1128/MCB.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 182.Yang RF, Li CM, Qiu HR, Lu H, Wu HX, Xu JR, Li JY, Chen LJ. Investigation of chromosome 1 aberrations in patients with multiple myeloma using cIg-FISH method and its significance. Zhonghua Xue Ye Xue Za Zhi. 2010;31:804–808. [PubMed] [Google Scholar]
- 183.Imreh MP, Gertow K, Cedervall J, Unger C, Holmberg K, Szöke K, Csöregh L, Fried G, Dilber S, Blennow E, Ahrlund-Richter L. In vitro culture conditions favoring selection of chromosomal abnormalities in human ES cells. J Cell Biochem. 2006;99:508–516. doi: 10.1002/jcb.20897. [DOI] [PubMed] [Google Scholar]
- 184.Avery S, Hirst AJ, Baker D, Lim CY, Alagaratnam S, Skotheim RI, Lothe RA, Pera MF, Colman A, Robson P, Andrews PW, Knowles BB. BCL-XL mediates the strong selective advantage of a 20q11.21 amplification commonly found in human embryonic stem cell cultures. Stem Cell Reports. 2013;1:379–386. doi: 10.1016/j.stemcr.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]