Abstract
The RAS genes encode for members of a large superfamily of guanosine-5′-triphosphate (GTP)-binding proteins that control diverse intracellular signaling pathways to promote cell proliferation. Somatic mutations in the RAS oncogenes are the most common activating lesions found in human cancers. These mutations invariably result in the gain-of-function of RAS by impairing GTP hydrolysis and are frequently associated with poor responses to standard cancer therapies. In this review, we summarize key findings of past and present landmark studies that have deepened our understanding of the RAS biology in the context of oncogenesis. We also discuss how emerging areas of research could further bolster a renewed global effort to target the largely undruggable oncogenic RAS and/or its activated downstream effector signaling cascades to achieve better treatment outcomes for RAS-mutant cancer patients.
Keywords: KRAS, NRAS, HRAS, Small GTPases, Kinases, Autophagy, Signaling, Cancer therapeutics
Introduction
A brief history of RAS
The field of RAS biology debuted more than four decades ago when eminent scientist Harvey first observed that the passaging of murine leukemia virus (MLV) induced sarcomas in newborn BALB/c mice [1]. In less than 3 years, serial passaging of the Kirsten murine sarcoma virus in Wister–Furth rats was similarly shown to drive sarcoma genesis [2]. The transforming property of these single-stranded RNA viruses stems from their ability to convert normal mammalian genes into potent oncogenes by incorporating these newly acquired genes into their own viral genomes [3]. As a result of their unique ability to cause rat sarcomas, these viral oncogenes were thereafter known as the RAS genes. Conveniently, the names of their discoverers became a part of the scientific nomenclature to distinguish them. For instance, the Harvey and Kirsten viral RAS oncogenes were named HRAS and KRAS, respectively. It took almost a decade before another major breakthrough in this field was made. The laboratories of Robert Weinberg, Michael Wigler and Mariano Barbacid successively identified key mammalian genes that are critical to cell transformation and oncogenesis [4–6]. Remarkably, these three research groups raced each other to the finishing line to show that the mutant oncogene in human T24 and EJ bladder carcinoma cell lines is in fact the cellular homologue of the well-characterized viral HRAS gene [7–12]. These landmark studies set the stage for an intense, ongoing global quest to target mutant RAS and its aberrant signaling cascades in human cancers of diverse tissue origin.
RAS family members
There are three major isoforms of RAS presently known to be frequently mutated in human cancers: HRAS, KRAS and NRAS. KRAS has two splicing isoforms, a minor isoform 4A (KRAS-4A) and a major isoform 4B (KRAS-4B). NRAS, not identified in any of the early pioneering retroviruses work, was discovered in human neuroblastoma-derived DNA [13, 14]. These RAS isoforms are highly identical in their N-terminal amino acid residues that make up the G domain for GTP binding and hydrolysis. The remaining C-terminal amino acid residues, however, are highly varied among these isoforms and have been shown to be critical for their localization to the cell membrane.
While HRAS, KRAS-4B and NRAS are ubiquitously expressed, KRAS-4A is predominantly expressed in renal, hepatic and gastrointestinal tissues [15]. These highly conserved RAS isoforms carry out similar but non-redundant cellular functions that govern apoptosis, proliferation and tumorigenesis [16]. Studies using whole body gene knockout mouse models suggest that KRAS is more important for mouse embryonic development than the other RAS isoforms, since Kras-ablated mice die during embryogenesis [17, 18]. On the contrary, Hras- or Nras-ablated mice, as well as Hras and Nras double knockout mice, are viable and exhibit no overt abnormalities [19]. Although the replacement of Kras with Hras at its gene locus (Hras Knock-in; HrasKI) could rescue the embryonic developmental defects, adult HrasKI mice exhibit dilated cardiomyopathy associated with arterial hypertension [20]. Notably, the specificity for Kras mutations in lung and Hras mutations in skin tumors are also controlled by local regulatory elements in the target Ras genes [21]. Hence, cis-acting gene transcriptional control elements, the timing of gene expression and, to some extent, functional differences in the encoded RAS proteins, could induce differential effects of RAS isoforms on the cancer progression of diverse tissue origins.
Mutant RAS and cancers
Frequency of RAS activating mutations in human cancers
A recent analysis of the Catalogue of Somatic Mutations in Cancer (COSMIC) database (http://www.sanger.ac.uk/genetics/CGP/cosmic/) revealed that KRAS is the most frequently mutated RAS family member in human cancers, followed by NRAS and HRAS. Activating mutations of KRAS-4B, the predominant isoform of KRAS, occurs in approximately 21 % of all human cancers, and accounts for approximately 90 % of pancreatic cancers, 45 % of colon cancers, and 30 % of lung cancers. NRAS-activating mutations occur in approximately 8 % of all human cancers, with as much as 15–30 % and 5–10 % of these mutations found in cutaneous melanomas and acute myeloid leukemia, respectively. HRAS, the first oncogenic RAS gene to be characterized, has the lowest frequency of activating mutations in human cancers (3 %), and is most notably found in 5–10 % of bladder urothelial carcinomas, as well as 5 % of squamous cell carcinomas of the head and neck. The frequency and distribution of RAS mutations in human cancers have recently been updated and reviewed [22, 23].
One of the most puzzling conundrums in the field of RAS biology is the mismatch between the relative abundance of these oncoproteins and their mutation frequencies in human cancers. Although KRAS is frequently mutated, the mutant KRAS oncoprotein that it encodes is only weakly expressed in cancers and thus results in a smaller tumor size. On the contrary, HRAS is the least mutated RAS gene but its mutant HRAS oncoprotein is highly expressed in cancers thereby promoting larger tumors [24]. Owing to the recent work of Counter and colleagues, we now appreciate that the variation in K/N/HRAS protein expression (despite their high sequence similarity) is mainly due to differential codon bias in the translation of these oncoproteins [24]. KRAS has been shown to be highly enriched in genomically underrepresented or rare codons and thus more poorly translated, as compared to HRAS. This allow human cancers harboring KRAS mutations to escape tumor suppressor surveillance mechanisms such as oncogene-induced senescence, thus driving cancer cells en route to more efficient oncogenesis. Conversely, converting rare codons back to common codons increases KRAS expression and reverted tumorigenicity to levels comparable to those observed for HRAS. More recently, the same research group performed a series of elegant experiments via transgenic mouse models and showed that the effects of codon bias on mutant RAS-driven tumorigenesis could indeed be recapitulated in vivo [25]. However, it remains an open question whether rare codon bias at specific hotspots (e.g., G12, G13 and Q61 amino acid residues) could increase the probability of KRAS, NRAS and HRAS activating mutations found in human cancers. Alternatively, since mutant KRAS induces smaller tumors because it is weakly expressed in the tumor cells, does KRAS undergo codon optimization/modification by yet-to-be-elucidated factors in human tumor cells such that it is robustly expressed to drive larger tumors in the presence of other cooperating somatic mutations?
Consequences of RAS mutation
Upon different growth factors stimulation, the proto-oncogenic RAS proteins oscillates between an inactive GDP-bound state and an active GTP-bound state. While the activation of RAS is induced by guanidine exchange factors (GEFs), its inactivation is catalyzed by GTPase-activating proteins (GAPs) that enhance the intrinsic GTPase activity of RAS proteins to hydrolyze GTP. GEFs bind inactive RAS-GDP, leading to a decreased affinity of RAS for GDP. By releasing GDP, GTP is able to re-enter the G domain of RAS proteins preferentially, and signaling cycles are renewed. Intrinsically, RAS proteins have slow GTPase activity, and their associations with GAPs could greatly accelerate the hydrolysis of GTP (Table 1 for KRAS as an example) [26–28]. The fold change in GAP-stimulated GTP hydrolysis rates appeared to vary dramatically among the RAS isoforms. It remains unclear whether these apparent variations in GAP-stimulated GTP hydrolysis rates are attributed to the different RAS isoforms or experimental approaches used in these studies. While NRAS and KRAS GTP hydrolysis rates were determined by various in vitro assays [26, 28], the GTP hydrolysis rate of HRAS was calculated by kinetic parameter-based equations [27]. The presence of a conserved ‘arginine finger’ domain on GAPs helps orientate the position of GTP on RAS-GTP for a more efficient nucleophilic attack by water, thus making GTP a better substrate to be hydrolyzed more readily.
Table 1.
GTP hydrolysis rates of wild-type (WT) and mutant KRAS (intrinsic versus GAP-stimulated). The intrinsic/GAP-stimulated GTP hydrolysis rate of WT/mutant KRAS was derived from [26] and expressed as fold change relative to the intrinsic GTP hydrolysis rate of WT KRAS
| Fold change in GTP hydrolysis rate of wild-type and mutant KRAS | ||
|---|---|---|
| Intrinsic | GAP-stimulated | |
| WT | 1.00 | 63.24 |
| P-loop mutants | ||
| G12A | 0.02 | 0.47 |
| G12C | 0.72 | 0.29 |
| G12D | 0.28 | 1.03 |
| G12R | 0.03 | 0.29 |
| G12V | 0.06 | 0.35 |
| G13D | 0.14 | 0.29 |
| Switch 2 mutants | ||
| Q61L | 0.01 | 0.49 |
| Q61H | 0.02 | 3.85 |
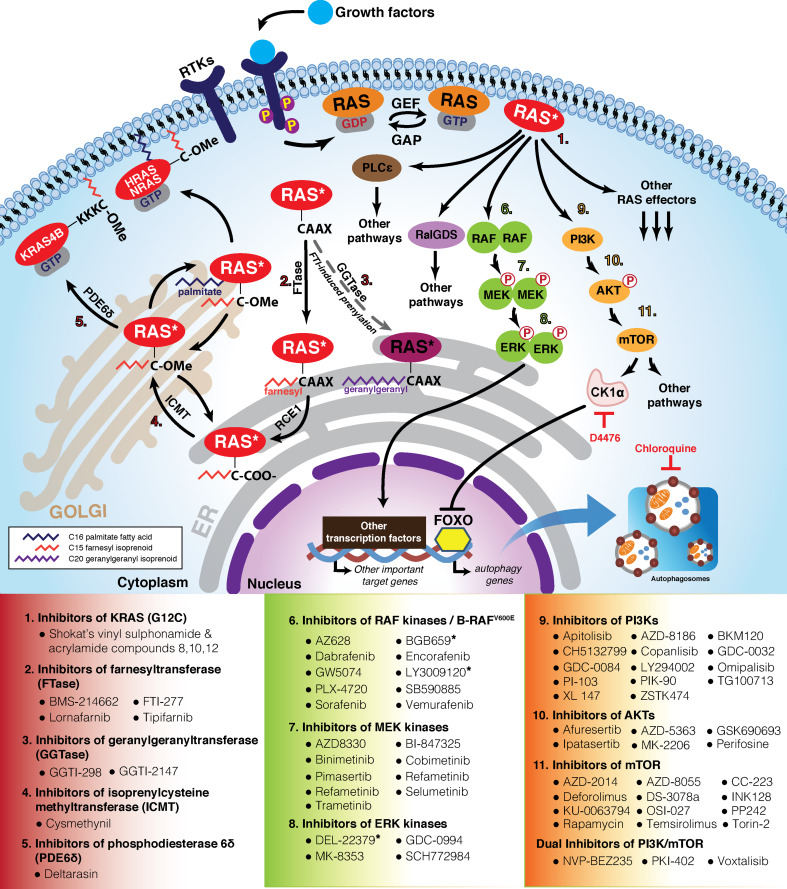
In the GTP-bound state, the activated RAS proteins are able to bind several downstream effector proteins, such as rapidly accelerated fibrosarcoma (RAF) kinase, phosphoinositide-3 kinase (PI3K), and Ral guanine nucleotide dissociation stimulator (RalGDS), leading to further initiation of a wide cellular signaling network downstream. In normal cells, these RAS signaling cascades are only transiently activated as the cytoplasmic GAPs serve as powerful rheostats to inactivate the proto-oncogenic RAS proteins. Nevertheless, a strong selective pressure to overcome the tight control of GAPs exists. As such, human cancer cells with activating mutations of RAS invariably result in mutant RAS oncoproteins that are locked in the active GTP-bound form, thereby constitutively activating downstream effectors in the absence of extracellular growth factors. Although the mechanism underlying how RAS-GTP activates its downstream effectors remains largely elusive, it has been previously reported that the specific binding of RAS-GTP to RAF helps recruit RAF protein from the cytosol to the plasma membrane for its subsequent activation [29, 30]. Hence, we speculate that RAS-GTP could engage in a similar approach to activate other normally cytoplasmic effector proteins like PI3K and RalGDS via their relocalization to the plasma membrane.
The aberrant activation of oncogenic RAS downstream effectors in RAS-mutant cancers has been intensely characterized and extensively reviewed [16, 23]. These downstream signaling cascades could operate linearly or cross talk with each other to regulate a broad spectrum of biological processes. For instance, the two major RAS effector branches, the RAF/mitogen or extracellular signal-regulated kinases (MEK/ERK) and PI3K/AKT/mammalian target of rapamycin (mTOR) pathways, are found to interact at various nodes in the complex RAS signaling network to promote cancer cell survival [31]. The active rewiring of these downstream signaling cascades is best demonstrated in the paradoxical activation of epidermal growth factor (EGF)-induced AKT activity by the MEK inhibitors [32, 33]. In addition, ERK is found to promote mTOR Complex 1 (mTORC1) activity via phosphorylation of the regulatory-associated protein of mTOR (RAPTOR) and tuberous sclerosis complex 2 (TSC2) [34–36]. AKT have also been shown to block MEK activity via phosphorylation of RAF at specific inhibitory sites [37, 38]. In neurofibromatosis type 1 (NF1)-mutant cells, cross talk between the mitogen-activated protein kinase (MAPK) and AKT/mTOR pathways are dependent on differential expression of musculoaponeurotic fibrosarcoma (MAF) proteins and DEP domain-containing mTOR-interacting (DEPTOR) proteins [39]. Importantly, inhibition of mTORC1 after Everolimus treatment induces MAPK activation through a PI3K-dependent feedback loop in multiple human cancers, such as melanoma, breast and colon cancers [40].
While acute activation of RAS signaling has been shown to trigger cell death [41], chronic activation of RAS signaling could lead to persistently high metabolic demands that require a constant supply of new biosynthetic intermediates to drive robust cancer cell growth. In both contexts, autophagy appears to be a key determinant of cell fate. Autophagy is a tightly controlled intracellular self-degradation process that salvages key metabolites to sustain metabolic or nutrient homeostasis, thereby promoting survival of cells that are growing under metabolic stress or nutrient depletion [42]. Recent reports demonstrate that RAS-mutant cancers, particularly those of colonic, lung and pancreatic origin, critically depend on elevated level of basal autophagy for survival [43–46]. For instance, genetic depletion of essential autophagy proteins, ATG5 or ATG7, sensitizes RAS-mutant cancer cells to growth arrest or apoptosis [43]. In addition, we recently reported that oncogenic RAS upregulates the ubiquitously expressed serine/threonine kinase, casein kinase 1α (CK1α), to attenuate autophagy at the level of autophagy gene transcription [47]. This transcriptional repression of autophagy genes is governed via CK1α-dependent phosphoregulation of a transcription factor known as Forkhead box O3A (FOXO3A). Our findings provide further insights into the complex interplay between autophagy and mutant RAS, and identifies CK1α as a potential biomarker to further stratify patients with RAS-mutant cancers for therapeutic approaches that target the autophagy pathway.
Notably, the autophagy protein LC3B was found to bind directly with the nuclear lamina protein, lamin B1, in the nucleus and interacts with lamin-associated domains (LADs) on chromatin in the presence of mutant RAS [48]. This LC3B-lamin B1 interaction leads to lamin B1 degradation and activation of mutant RAS-induced cellular senescence in primary BJ fibroblasts that possess wild-type TP53 (commonly known as p53), thus preventing these cells from undergoing oncogenesis. Collectively, these studies indicate that activation of autophagy could result in different cell fate in pre-malignant and malignant cells. The juxtaposition of findings from our lab and Dou et al. might also provide some conceptual link between CK1α, autophagy and cellular senescence in RAS-mutant cancers. We speculate that oncogenic RAS promotes senescence of pre-malignant cells by increasing CK1α protein abundance, thereby reducing the rate of LC3B synthesis. The slow but continual buildup of LC3B in these cells over time might degrade nuclear lamin B1 and eventually induces cellular senescence. This could be one of the few plausible reasons why it takes days for cultured primary fibroblasts with wild-type p53 to undergo cellular senescence upon induction of mutant RAS expression [49].
Cell-based and animal models of mutant RAS-induced tumorigenesis
Cancer development is a multi-step process in which cells acquire a series of genetic modifications that accumulatively program a malignant phenotype. While past and current approaches of studying cancer biology have been informative and useful, our understanding of the complex molecular basis of cancer initiation and development could not be further enhanced without technological innovations that create new experimental models of cancer.
Over a decade ago, Hahn, Counter and colleagues demonstrated that they could transform normal human epithelial and fibroblastic cells by introducing defined genetic elements, such as telomerase catalytic subunit hTERT, the simian virus 40 (SV40) large-T antigen and oncogenic HRAS [50]. Since then, other groups have also co-expressed these genetic factors in astrocytes [51], airway epithelial cells [52], ovarian epithelial cells [53], human mammary epithelial cells [54], mesothelial cells [55] and endothelial cells [56], and recapitulated the observation that multiple mutations cooperate to promote tumor formation. These discoveries led to the creation of a plethora of cell-based models of mutant RAS-driven tumorigenesis that facilitates many scientific investigations in a cell- and tissue-specific manner. A major drawback of cell-based models, however, is the lack of supporting stroma and extracellular matrix influencing tumor vasculature and progression, and hence less physiological.
With the advent of gene targeting techniques, animal tumor models that closely encapsulate cancer development in its native microenvironment could be created at will. Genetically modified mouse tumor models have been particularly useful in helping us understand the detailed molecular mechanisms that promote tumor development and drive cancer metastasis. In addition, they also serve as invaluable tools for pre-clinical evaluation of novel anti-cancer therapeutics. The first attempt to create a mutant RAS-driven lung cancer model in mice was undertaken by Jacks, Tuveson and colleagues. This targeted latent “hit and run” lung cancer mouse model (Kras LA2 strain), however, was difficult to generate because the integration of the ectopic mutant Kras oncogene (“hit”) into the mouse genome is highly dependent on a stochastic recombination event (“run”) and almost impossible to control experimentally [57]. These investigators then developed a conditional/inducible lung cancer mouse model (Kras +/LSLG12D strain) that permits controlled timing of lung tumor initiation and multiplicity upon intranasal infection with Cre recombinase-containing adenoviruses. These mice carry Cre-mediated recombination of a lox-STOP-lox (LSL) cassette inserted in the first intron of the Kras locus, thus allowing control and prevention of untimely expression of the Kras +/LSLG12D allele [58]. Since then, the Kras +/LSLG12D mouse strain has become the “gold standard” for most oncogenic Kras-driven animal tumor models. Importantly, the development of such controllable systems in which expression of mutant Ras could be turned on and off at will has greatly accelerated the pace of RAS research.
By crossing these Kras +/LSLG12D mice with mice that carry Cre recombinase that are controlled by the Pdx 1- or Ptf1a/P48-specific pancreatic promoters, an inducible-Ras mouse model of pancreatic cancer was engineered a few years later [59]. These mice faithfully reproduce the natural progression of human pancreatic ductal adenocarcinomas (PDACs), where precursor lesions like pancreatic intraepithelial neoplasias (PanINs) first occurred with complete penetrance, followed by full-blown development of PDACs. However, a caveat of this cancer model is that the progression to PDAC usually occurs with low frequencies. To overcome this problem, others engineered a more robust pancreatic cancer mouse model by further mutating key tumor suppression genes that are usually found to be altered or lost in advanced human PDACs. These commonly include the inactivation of p16Ink4a/p19Arf, p53, Lkb1, p21 Cip1, or Smad4 [60–64]. All mice with these genetic alterations quickly developed advanced pancreatic cancers, where accelerated progression of PanINs into invasive PDACs and induction of metastatic spreading was documented. In recent years, many other inducible mouse models for PDACs were developed, such as one that allows reversible controlled expression of the Kras G12D by addition or withdrawal of doxycycline treatment [65–67].
Apart from these transgenic mice, we noted that other animal models of oncogenic Ras-induced tumorigenesis have also been created. In search of an ideal animal model that is cost-effective and exhibits short latency in tumor initiation, some researchers opted to modify zebrafishes. A transgenic Ras-mutant zebrafish liver cancer model with mifepristone-inducible oncogenic Kras [68] as well as a transgenic Ras-mutant zebrafish rhabdomyosarcoma cancer model with conditional expression of oncogenic Kras [69] has been reported. Most recently, a transgenic ‘oncopig’ that possesses Cre recombinase-inducible transgenes like Kras G12D and TP53 R167H was engineered [70]. A porcine Ras-mutant model was chosen because of its large physical size and its close resemblance to humans in terms of anatomy, physiology, metabolism and genetics. Besides serving as a genetically malleable model, this “oncopig” experiment provides the roadmap to create other large animal models of cancer in the future.
Targeting oncogenic RAS and/or its aberrant signaling networks in cancers
The undruggable mutant RAS
Given that mutant RAS is a key driver of at least one-third of all cancers, much attention has been focused on the identification of chemical compounds that could bind directly to mutant RAS to alter its activity since the 1980s. However, efforts to find the elusive RAS-binding compound have largely been futile for many years due to the lack of good drug-binding pockets in RAS. Unlike the identification and use of small ATP-competitive compounds to effectively inhibit the function of oncogenic kinases, the GDP/GTP-binding pocket of RAS was ruled out as a suitable drug target site because of its picomolar affinity for guanine nucleotides and the presence of relatively high intracellular concentration of GTP. Consequently, small molecules would not be able to outcompete endogenous GTP for binding to RAS. Activating mutations in amino acid residues 12, 13 or 61 of the G domain also limits the access of RAS to GTPase-activating proteins (GAPs), thus keeping mutant RAS invariably in the active GTP-bound state to trigger downstream signaling pathways. The reader is referred to other excellent reviews on why mutant RAS is considered by many to be undruggable [22, 23, 71–77].
Targeting lipid modification of RAS proteins—early setbacks
It took almost two decades after the discovery of RAS before researchers elucidated the lipid modification pathways that are critical for the intracellular trafficking of RAS. The search for compounds that block lipidation of RAS was intensely pursued over a number of years. Post-translational lipidation of RAS at its carboxy-terminal CAAX (C: cysteine, A: aliphatic amino acids and X: any amino acids; in RAS, X is usually S or M) motif is essential for its association with the plasma membrane and with other intracellular membranes to activate signaling cascades [78, 79]. In the first and obligate step, the cysteine residue of the CAAX motif of RAS is modified with a 15-carbon (C15) farnesyl polyisoprene lipid moiety through a stable thioether linkage by farnesyltransferase (FTase). This C15 modification promotes the association of RAS with endoplasmic reticulum (ER), where the modified CAAX motif is cleaved by RAS-converting CAAX endopeptidase 1 (RCE) to remove the AAX residues and further carboxymethylated by isoprenylcysteine carboxylmethyltransferase (ICMT) [80, 81]. Pharmacologic targeting of FTase thus appears to be a sensible move because inhibitors of FTase (FTIs) could prevent the maturation of RAS and thus perturb RAS signaling. The intensive screen for FTIs by many research labs in the academia and industry identified a number of potent FTIs, such as Lonafarnib [82] and Tipifarnib [83]. These FTIs were shown to be efficacious against mutant HRAS-driven cancer models in the pre-clinical setting. However, they ultimately failed to show efficacy against mutant KRAS-driven cancers in the advanced human clinical trials. Even though these compounds might still be useful for combating an extremely small subset of mutant HRAS-driven cancers, many viewed it as a major setback in the quest to cure RAS-mutant cancers. It was subsequently demonstrated that bypass prenylation of KRAS and NRAS at the cysteine residue of CAAX motif by geranylgeranyltransferase 1 (GGTase 1) occurs in the cancer cell to promote resistance to FTI [84]. The dual inhibition of FTase and GGTase 1 might potentially overcome the problem of alternative prenylation but it failed to progress further due to toxicity issues. This is likely due to the blockade of farnesylation and geranylgeranylation of many proteins that are required for the survival of normal cells.
Apart from targeting CAAX prenylation of RAS, others focused their attention on the identification of compounds that would block post-prenylation of CAAX-processing enzymes like RCE1 and ICMT [85–87]. Since these enzymes may also modify other CAAX motif-containing proteins, their inhibitors are likely to elicit undesirable cytotoxicity issues.
Drugging RAS effector pathways—unexpected complexity
A compelling body of evidence demonstrates that mutant RAS promotes tumor initiation and progression, in part, via the activation of its three major downstream effector pathways. These include the RAF, RalGEF and PI3K signaling pathways. Mutant RAS recruits RAF, a cytosolic serine/threonine kinase, to the plasma membrane to activate the latter for subsequent initiation of its downstream MEK/ERK signaling cascade. This RAF/MEK/ERK effector branch of RAS induces anchorage-independent growth, angiogenesis and epithelial–mesenchymal transition (EMT). Mutant RAS has also been shown to activate the RalGEFs family of five guanine exchange factors (GEFs) in a similar manner. RalGEFs, in turn, activate RalA and RalB to drive tumorigenesis. Lastly, mutant RAS recruits and activates the PI3K via its p85 regulatory subunit, resulting in the phosphorylation of phosphoinositides and induction of signaling pathways that enable angiogenesis, transformed morphologies and cancer cell survival.
Kinases appear to be critical second messengers in at least two of these effector pathways of mutant RAS signaling. Owing to the absolute reliance on adenosine triphosphate (ATP) for kinase activity, many drug-screening efforts have been focused on the identification and development of small molecules that have higher affinity for the nucleotide-binding pocket of these kinases than intracellular ATP. Alternative screens that are based on allosteric regulation of kinase activity have also been pursued to find non-ATP-competitive inhibitors of kinases implicated in mutant RAS signaling. To date, many potent and selective small molecule inhibitors of RAF, MEK1/2, ERK1/2, PI3K, AKT and mTOR have been identified and developed (Fig. 1; reviewed and summarized in [22]). A good number of these compounds have been evaluated in the pre-clinical and clinical settings, with a few of them even approved for use by the United States (US) Food and Drug Administration (FDA). However, the blockade of any one effector pathway turned out to be necessary but insufficient to curb the growth of RAS-mutant cancer cells due to undesirable induction of compensatory mechanisms. For instance, the first-generation BRAF-mutant inhibitors (e.g., Vemurafenib) paradoxically activate ERK signaling by inducing wild-type RAF dimerization to promote sustained RAS-mutant cancer cell growth [88–90]. Similarly, MEK inhibitors are efficacious for the treatment of BRAF-mutant melanoma but they have limited efficacy against RAS-mutant human tumor cell lines [91] as well as mouse models of Ras-mutant cancer [92]. This is attributed in part to reprogramming events that activate expression of receptor tyrosine kinases (RTKs), such as PDGFRβ, or promote NRAS-activating mutations [93]. The utility of ERK inhibitors is also limited by its unexpected activation of MEK activity, since ERK feedback phosphorylation is required to inactivate RAF [94]. Lastly, sole targeting of the PI3K/AKT/mTOR effector branch also fell short because small molecule inhibitors of this signaling cascade failed to block the growth of Kras-mutant mouse lung adenocarcinomas in vivo [92].
Fig. 1.
Activated RAS signaling in cancers and its targeted therapeutic strategies. A subset of small molecule inhibitors that targets mutant RAS, its post-translational modifications and/or its activated downstream effector pathways is listed. Drug* denotes new inhibitors that block effector dimerization or effector dimer function. Wild-type RAS (orange); mutant RAS (red); RTKs receptor tyrosine kinases
Intriguingly, the full potential of inhibitors of various RAS effector branches appears to be unleashed when used in the right combination. The dual pan PI3K and mTOR inhibitor, NVP-BEZ235, acts synergistically with the MEK inhibitor, ARRY-142886, to shrink Kras-mutant mouse lung tumors [92]. This observation led to the initiation of a slew of clinical trials to evaluate the combined blockade of the RAF/MEK/ERK and PI3K/AKT/mTOR effector signaling in the treatment of RAS-mutant cancers [95]. According to ClinicalTrials.gov, many of the Phase 1 clinical trials of dual inhibition of PI3K/mTOR and MEK in patients with advanced cancers (NCT01476137, NCT01392521, NCT01390818 and NCT01347866) were recently completed but their results have not been publicly posted. In a Phase 2 clinical trial of dual AKT and MEK inhibition (NCT01333475), a significant reduction in phosphorylated ERK (pERK) and phosphorylated AKT (pAKT) levels in tumor biopsies from patients with advanced colorectal cancer was observed.
Manipulating autophagy in mutant RAS-driven cancers: a viable alternative approach?
Hijacking the intracellular autophagic recycling process provides RAS-mutant cancer cells an avenue to deal with the proteotoxic stress and dynamic changes in energy metabolism driven by oncogenic RAS. Persistant oncogenic RAS signaling not only results in high rates of protein misfolding but also high energy demand for its anabolic programs, which rapidly deplete biosynthetic intermediates from the tricarboxylic acid (TCA) cycle. This leads to aggregation of proteins, loss of mitochondrial respiration and accumulation of dysfunctional mitochondria. To maintain proper protein homeostasis and a healthy pool of mitochondria, RAS-mutant cancer cells rely on the catabolic nature of the autophagy-lysosomal pathway to clear bulky protein aggregates and damaged mitochondria. Biosynthetic intermediates from these cargoes could then be recycled to support the robust growth of RAS-mutant cancer cells [96, 97]. This process appears to be highly calibrated to prevent self-cannibalism, as the activation of PI3K/AKT/mTOR effector branch of RAS signaling has been shown to suppress autophagy initiation [98–102]. Notably, growth of BRAF-mutant cancer cells has also been shown to be critically dependent on elevated basal and induced autophagy [103–105]. Importantly, inhibition of effective autophagy by chloroquine (CQ) acts synergistically with the RAF inhibitor Vemurafenib or standard chemotherapeutics to regress BRAFV600E-driven brain tumors [103, 106].
The driving forces behind a heightened state of basal autophagy in RAS-mutant cancer cells remain poorly understood. In the case of BRAF-mutant melanoma cells, high basal autophagy appears to be driven by BRAFV600E-induced chronic ER stress [105]. Although oncogenic RAS has been reported to promote an ER stress-associated unfolded protein response (UPR) and that ER stress has been shown to be a potent inducer of autophagy [107], there is lack of compelling evidence to show that basal autophagy elevation is directly caused by mutant RAS-induced chronic ER stress. We and others have recently discovered that Casein Kinase 1 alpha (CK1α) is a critical factor for the survival of RAS-mutant cancer cells [47, 108]. We demonstrate that dual inhibition of CK1α and lysosomal/autophagy function synergistically triggers death of mutant RAS-driven human colon and bladder cancer cells [47]. The result of this paired autophagy inducer and inhibitor therapy is consistent with an earlier finding that PI3K or AKT inhibitors synergize with lysosomotropic agents like CQ to trigger death of phosphatase and tensin homolog (PTEN)-null cancer cells [109]. The recently concluded Phase 1 clinical trials, which evaluated the combination of hydroxychloroquine (HCQ) with mTOR inhibitor or other standard-of-care chemotherapeutics, also suggest that these paired therapies are safe and efficacious for the treatment of solid tumors and melanomas [110–115].
Mutant RAS synthetic lethal screens—a glass half empty or full
A number of mutant RAS synthetic lethal genetic screens have been undertaken over the past few years with the hope of uncovering new mutant RAS-cooperating protein partners that are tractable drug targets (as reviewed and summarized in [72]). However, these studies yield data with low degree of overlap and they have not led to the development of novel clinical approaches to combat RAS-mutant cancers thus far. The lack of reproducibility of experimental findings from these screens could be attributed to wide-ranging differences from diverse methodologies/reagents to the gene mutational landscapes in tumor cells of specific tissue lineages [116–118]. Notably, the proteasome was identified as a rare common hit in multiple screens, indicating that the maintenance of protein homeostasis may be critical for RAS-mutant cancer cells [119–121]. However, the enthusiasm to pursue further application of this finding was quenched by subsequent observations that cultured RAS-mutant cells are relatively insensitive to single-agent proteasomal inhibitors [122, 123]. In addition, these RAS synthetic lethal screens were almost completely driven by the RNA interference (RNAi) technology that results in various hypomorphic effects due to partial loss of gene expression. To minimize RNAi-associated artifacts, we propose that robust gene editing systems like zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) or clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 should be applied to RAS synthetic lethal screens. These screens could also incorporate challenges to tumor cells in addition to ongoing two-dimensional growth, so as to identify genes important for cell survival following matrix deprivation (anoikis), DNA damage, or other forms of stress. In addition, assays might be performed in high-throughput three-dimensional cell culture systems, such as spheroid or organoid cultures, that better mimic the physiologic state of tumors growing in vivo.
RAS mutation profiling of human cancers
Existing standard-of-care therapies targeting epithelial growth factor receptor (EGFR), such as the use of chimeric monoclonal antibodies (panitumumab or cetuximab) or tyrosine kinase inhibitors (Erlotinib), have been prescribed to patients suffering from EGFR-overexpressed, advanced colorectal as well as non-small-cell lung cancers. However, anti-EGFR therapeutics tend to fail in cancers that are no longer EGFR dependent, such as those with activating mutations of KRAS [124]. Approximately 45 % of colon cancers and 30 % of lung cancers harbor somatic missense mutations in the KRAS gene at codons 12 and 13, and they are often associated with poor response to anti-EGFR therapies. As such, diagnosis of RAS mutational status in these cancers is recommended prior to anti-EGFR therapies.
Although RAS mutational testing has been widely adopted in recent years, it has been constantly revised to take into account of new clinical evidences. For instance, the previous standard-of-care mutation profiling for colon cancers was originally performed to identify exon 2 (codons 12 and 13) mutations in the KRAS gene. Recent data, however, suggests that colon cancer patients carrying NRAS gene mutations also displayed lower response rate to anti-EGFR therapeutics [125, 126]. As such, the recommended clinical guidelines for the use of anti-EGFR therapies on metastatic colorectal cancers now require an expanded mutational testing of KRAS and NRAS exon 2 (codons 12 and 13), exon 3 (codons 59 and 61), and exon 4 (codons 117 and 146). In addition, recent breakthroughs have also been made to improve sensitivity of molecular diagnostic tools developed for RAS mutation testing. Using high-depth targeted next-generation sequencing (NGS) and robust bioinformatics algorithms, Tan and colleagues reported somatic variants across more than 750 cancer-related genes in liver-limited metastatic colon cancers. These include KRAS, NRAS, BRAF and PIK3CA, as well as other genes that are present at low allele frequency [127]. While the contribution of these somatic variants to reduced therapeutic efficacy needs to be definitively determined, we predict that such a highly sensitive approach would become the new benchmark to better stratify cancer patients for personalized anti-cancer therapies.
The RAS renaissance: recent breakthroughs in direct inhibition of mutant RAS and dimerization of downstream effectors
A major breakthrough in the direct targeting of mutant RAS was first achieved by the Shokat lab 2 years ago when they developed of small molecules that bind irreversibly to KRASG12C [128]. These compounds rely on the mutated cysteine residue for selective binding to this species of oncogenic RAS. They allosterically alter the nucleotide preference of KRASG12C to favor GDP over GTP as well as impair the recruitment of RAF to KRASG12C. These direct inhibitors of KRASG12C may be particularly effective against a subset of lung adenocarcinoma, colorectal and pancreatic carcinomas, since the KRASG12C mutation was frequently found in these solid tumors, respectively [22]. Although vast majority of RAS mutations in human cancers do not involve glycine-to-cysteine substitution [16], the development of KRASG12C inhibitors provides a useful conceptual framework to develop compounds that could selectively target other commonly found RAS mutations.
Another emerging area of research that holds great promise is the recent development of ERK and RAF dimer inhibitors [129–131]. The dimerization of ERK is triggered by activation of the RAS–RAF–MEK signaling axis and it is required for the extranuclear function of ERK. In a study to find small molecules capable of blocking ERK dimerization, Herrero et al. identified a water-soluble 3-arylidene-2-oxindole derivative, DEL-22379 [129]. Using in silico docking analyses and microscale thermophoresis, they showed that DEL-22379 effectively blocks ERK2 homodimerization without affecting its phosphorylation by binding to ERK2 at a groove within the dimerization interface. Importantly, DEL-22379 inhibited growth and induced apoptosis in cultured RAS- or BRAF-mutant human cancer cell lines and tumor xenografts in mice. DEL-22379 also blocked metastasis of BRAF-mutant patient-derived xenografts, as shown in the same study. On the other hand, dimerization of RAF has recently been shown to be required for normal RAS-dependent RAF activation and for the cellular function of disease-associated RAF mutants with weakened or impaired kinase activity [132]. Perhaps “unity is strength” in these contexts is sufficient to drive RAF signaling forward, since dimerization is not required in RAF mutants with high catalytic activity, such as BRAFV600E. To explore this vulnerability in RAF signaling, a proof-of-concept RAF dimer interface peptide was developed and shown to be efficacious in the inhibition of aberrant RAF signaling [132]. Although mutant BRAFV600E monomers are effectively inhibited by Vemurafenib, this FDA-approved drug has limited efficacy against tumors that possess constitutive, RAS-independent, BRAF-activating mutant dimers [131]. Remarkably, Yao and coworkers identified a type II, ATP-competitive RAF inhibitor (BGB659) that could bind RAF dimers and effectively block all RAF-mutant tumor growth in mice [131]. In a back-to-back report published in the same issue of Cancer Cell, Peng and coworkers identified and characterized a novel pan-RAF inhibitor of three RAF isoforms, LY3009120. Although LY3009120 induces RAF dimerization, it inhibits the kinase activity of the induced dimers as well as pre-formed dimers. LY3009120 was also shown to elicit little paradoxical pathway activation and is active for RAS- or BRAF-mutant cancer cells [130].
Notably, mutant RAS has also been shown to form dimers for the activation of downstream effector pathways both in vivo [133] and in vitro [134]. However, it took researchers more than a decade to get hold of further concrete experimental evidence of RAS dimerization-driven RAF/MAPK signaling in cancer cells [135, 136], as well as understanding the structural basis of how GTP-dependent α-helical dimerization of KRAS favors RAF dimerization [137]. Hence, we envisage that the development of mutant RAS dimerization inhibitors is likely to be pursued by academic labs and the industry with much enthusiasm in the immediate future.
Concluding remark
Although over four decades of intense research is yet to yield a drug that could safely and effectively curb the aberrant activity of oncogenic RAS in human cancers, our deepened understanding of the RAS biology has led to the development of novel innovative approaches that target mutant RAS directly or its over-stimulated downstream effector pathways (Fig. 1). The timely announcement of the RAS initiative by the National Cancer Institute (NCI) in early 2015 could lend a crucial support to fuel a newly gained momentum in making this notorious oncoprotein a truly tractable target in cancer therapy.
Acknowledgments
Space limitations preclude our manuscript from being a comprehensive review, and this unfortunately limits appropriate recognition of many of our colleagues worldwide, who have contributed immeasurably to the development of the RAS field. We thank David Virshup (Duke-NUS) for his critical review of our manuscript. This work was supported by an NMRC-CBRG New Investigator Grant (NMRC/BNIG/1078/2012) and a Duke-NUS-St. Baldrick’s Foundation Pediatric Cancer Research Fund (Duke-NUS-SBF/2015/0004) to JKC.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Harvey JJ. An unidentified virus which causes the rapid production of tumours in mice. Nature. 1964;204:1104–1105. doi: 10.1038/2041104b0. [DOI] [PubMed] [Google Scholar]
- 2.Kirsten WH, Mayer LA. Morphologic responses to a murine erythroblastosis virus. J Natl Cancer Inst. 1967;39(2):311–335. [PubMed] [Google Scholar]
- 3.Scolnick EM, Rands E, Williams D, Parks WP. Studies on the nucleic acid sequences of Kirsten sarcoma virus: a model for formation of a mammalian RNA-containing sarcoma virus. J Virol. 1973;12(3):458–463. doi: 10.1128/jvi.12.3.458-463.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shih C, Shilo BZ, Goldfarb MP, Dannenberg A, Weinberg RA. Passage of phenotypes of chemically transformed cells via transfection of DNA and chromatin. Proc Natl Acad Sci USA. 1979;76(11):5714–5718. doi: 10.1073/pnas.76.11.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shih C, Padhy LC, Murray M, Weinberg RA. Transforming genes of carcinomas and neuroblastomas introduced into mouse fibroblasts. Nature. 1981;290(5803):261–264. doi: 10.1038/290261a0. [DOI] [PubMed] [Google Scholar]
- 6.Perucho M, Goldfarb M, Shimizu K, Lama C, Fogh J, Wigler M. Human-tumor-derived cell lines contain common and different transforming genes. Cell. 1981;27(3 Pt 2):467–476. doi: 10.1016/0092-8674(81)90388-3. [DOI] [PubMed] [Google Scholar]
- 7.Shih C, Weinberg RA. Isolation of a transforming sequence from a human bladder carcinoma cell line. Cell. 1982;29(1):161–169. doi: 10.1016/0092-8674(82)90100-3. [DOI] [PubMed] [Google Scholar]
- 8.Goldfarb M, Shimizu K, Perucho M, Wigler M. Isolation and preliminary characterization of a human transforming gene from T24 bladder carcinoma cells. Nature. 1982;296(5856):404–409. doi: 10.1038/296404a0. [DOI] [PubMed] [Google Scholar]
- 9.Pulciani S, Santos E, Lauver AV, Long LK, Robbins KC, Barbacid M. Oncogenes in human tumor cell lines: molecular cloning of a transforming gene from human bladder carcinoma cells. Proc Natl Acad Sci USA. 1982;79(9):2845–2849. doi: 10.1073/pnas.79.9.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Der CJ, Krontiris TG, Cooper GM. Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proc Natl Acad Sci USA. 1982;79(11):3637–3640. doi: 10.1073/pnas.79.11.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parada LF, Tabin CJ, Shih C, Weinberg RA. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature. 1982;297(5866):474–478. doi: 10.1038/297474a0. [DOI] [PubMed] [Google Scholar]
- 12.Santos E, Tronick SR, Aaronson SA, Pulciani S, Barbacid M. T24 human bladder carcinoma oncogene is an activated form of the normal human homologue of BALB- and Harvey-MSV transforming genes. Nature. 1982;298(5872):343–347. doi: 10.1038/298343a0. [DOI] [PubMed] [Google Scholar]
- 13.Shimizu K, Goldfarb M, Perucho M, Wigler M. Isolation and preliminary characterization of the transforming gene of a human neuroblastoma cell line. Proc Natl Acad Sci USA. 1983;80(2):383–387. doi: 10.1073/pnas.80.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall A, Marshall CJ, Spurr NK, Weiss RA. Identification of transforming gene in two human sarcoma cell lines as a new member of the ras gene family located on chromosome 1. Nature. 1983;303(5916):396–400. doi: 10.1038/303396a0. [DOI] [PubMed] [Google Scholar]
- 15.Pells S, Divjak M, Romanowski P, Impey H, Hawkins NJ, Clarke AR, Hooper ML, Williamson DJ. Developmentally-regulated expression of murine K-ras isoforms. Oncogene. 1997;15(15):1781–1786. doi: 10.1038/sj.onc.1201354. [DOI] [PubMed] [Google Scholar]
- 16.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11(11):761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E, Bronson RT, Umanoff H, Edelmann W, Kucherlapati R, Jacks T. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11(19):2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koera K, Nakamura K, Nakao K, Miyoshi J, Toyoshima K, Hatta T, Otani H, Aiba A, Katsuki M. K-ras is essential for the development of the mouse embryo. Oncogene. 1997;15(10):1151–1159. doi: 10.1038/sj.onc.1201284. [DOI] [PubMed] [Google Scholar]
- 19.Esteban LM, Vicario-Abejon C, Fernandez-Salguero P, Fernandez-Medarde A, Swaminathan N, Yienger K, Lopez E, Malumbres M, McKay R, Ward JM, Pellicer A, Santos E. Targeted genomic disruption of H-ras and N-ras, individually or in combination, reveals the dispensability of both loci for mouse growth and development. Mol Cell Biol. 2001;21(5):1444–1452. doi: 10.1128/MCB.21.5.1444-1452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Potenza N, Vecchione C, Notte A, De Rienzo A, Rosica A, Bauer L, Affuso A, De Felice M, Russo T, Poulet R, Cifelli G, De Vita G, Lembo G, Di Lauro R. Replacement of K-Ras with H-Ras supports normal embryonic development despite inducing cardiovascular pathology in adult mice. EMBO Rep. 2005;6(5):432–437. doi: 10.1038/sj.embor.7400397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.To MD, Wong CE, Karnezis AN, Del Rosario R, Di Lauro R, Balmain A. Kras regulatory elements and exon 4A determine mutation specificity in lung cancer. Nat Genet. 2008;40(10):1240–1244. doi: 10.1038/ng.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: mission possible? Nat Rev Drug Discov. 2014;13(11):828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25(3):272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 24.Lampson BL, Pershing NL, Prinz JA, Lacsina JR, Marzluff WF, Nicchitta CV, MacAlpine DM, Counter CM. Rare codons regulate KRas oncogenesis. Curr Biol. 2013;23(1):70–75. doi: 10.1016/j.cub.2012.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pershing NL, Lampson BL, Belsky JA, Kaltenbrun E, MacAlpine DM, Counter CM. Rare codons capacitate Kras-driven de novo tumorigenesis. J Clin Invest. 2015;125(1):222–233. doi: 10.1172/JCI77627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter JC, Manandhar A, Carrasco MA, Gurbani D, Gondi S, Westover KD. biochemical and structural analysis of common cancer-associated KRAS mutations. Mol Cancer Res. 2015;13(9):1325–1335. doi: 10.1158/1541-7786.MCR-15-0203. [DOI] [PubMed] [Google Scholar]
- 27.Wey M, Lee J, Jeong SS, Kim J, Heo J. Kinetic mechanisms of mutation-dependent Harvey Ras activation and their relevance for the development of Costello syndrome. Biochemistry. 2013;52(47):8465–8479. doi: 10.1021/bi400679q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trahey M, McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987;238(4826):542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- 29.Leevers SJ, Paterson HF, Marshall CJ. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature. 1994;369(6479):411–414. doi: 10.1038/369411a0. [DOI] [PubMed] [Google Scholar]
- 30.Marais R, Light Y, Paterson HF, Marshall CJ. Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. EMBO J. 1995;14(13):3136–3145. doi: 10.1002/j.1460-2075.1995.tb07316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36(6):320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu CF, Liu ZX, Cantley LG. ERK negatively regulates the epidermal growth factor-mediated interaction of Gab1 and the phosphatidylinositol 3-kinase. J Biol Chem. 2002;277(22):19382–19388. doi: 10.1074/jbc.M200732200. [DOI] [PubMed] [Google Scholar]
- 33.Hoeflich KP, O’Brien C, Boyd Z, Cavet G, Guerrero S, Jung K, Januario T, Savage H, Punnoose E, Truong T, Zhou W, Berry L, Murray L, Amler L, Belvin M, Friedman LS, Lackner MR. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin Cancer Res. 2009;15(14):4649–4664. doi: 10.1158/1078-0432.CCR-09-0317. [DOI] [PubMed] [Google Scholar]
- 34.Carriere A, Romeo Y, Acosta-Jaquez HA, Moreau J, Bonneil E, Thibault P, Fingar DC, Roux PP. ERK1/2 phosphorylate Raptor to promote Ras-dependent activation of mTOR complex 1 (mTORC1) J Biol Chem. 2011;286(1):567–577. doi: 10.1074/jbc.M110.159046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foster KG, Acosta-Jaquez HA, Romeo Y, Ekim B, Soliman GA, Carriere A, Roux PP, Ballif BA, Fingar DC. Regulation of mTOR complex 1 (mTORC1) by raptor Ser863 and multisite phosphorylation. J Biol Chem. 2010;285(1):80–94. doi: 10.1074/jbc.M109.029637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121(2):179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 37.Zimmermann S, Moelling K. Phosphorylation and regulation of Raf by Akt (protein kinase B) Science. 1999;286(5445):1741–1744. doi: 10.1126/science.286.5445.1741. [DOI] [PubMed] [Google Scholar]
- 38.Guan KL, Figueroa C, Brtva TR, Zhu T, Taylor J, Barber TD, Vojtek AB. Negative regulation of the serine/threonine kinase B-Raf by Akt. J Biol Chem. 2000;275(35):27354–27359. doi: 10.1074/jbc.M004371200. [DOI] [PubMed] [Google Scholar]
- 39.Brundage ME, Tandon P, Eaves DW, Williams JP, Miller SJ, Hennigan RH, Jegga A, Cripe TP, Ratner N. MAF mediates crosstalk between Ras-MAPK and mTOR signaling in NF1. Oncogene. 2014;33(49):5626–5636. doi: 10.1038/onc.2013.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, Egia A, Sasaki AT, Thomas G, Kozma SC, Papa A, Nardella C, Cantley LC, Baselga J, Pandolfi PP. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118(9):3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elgendy M, Sheridan C, Brumatti G, Martin SJ. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol Cell. 2011;42(1):23–35. doi: 10.1016/j.molcel.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 42.Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol. 2015;16(8):461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- 43.Guo JYCH-Y, Mathew R, Fan J, Strohecker AM, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim MJWS, Yoon CH, Lee JS, An S, et al. Involvement of autophagy in oncogenic K-Ras-induced malignant cell transformation. J Biol Chem. 2011;286:12924–12932. doi: 10.1074/jbc.M110.138958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lock RRS, Kenific CM, Su JS, Salas E, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22:165–178. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang SWX, Contino G, Liesa M, Sahin E, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheong JK, Zhang F, Chua PJ, Bay BH, Thorburn A, Virshup DM. Casein kinase 1alpha-dependent feedback loop controls autophagy in RAS-driven cancers. J Clin Invest. 2015;125(4):1401–1418. doi: 10.1172/JCI78018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dou Z, Xu C, Donahue G, Shimi T, Pan JA, Zhu J, Ivanov A, Capell BC, Drake AM, Shah PP, Catanzaro JM, Daniel Ricketts M, Lamark T, Adam SA, Marmorstein R, Zong WX, Johansen T, Goldman RD, Adams PD, Berger SL. Autophagy mediates degradation of nuclear lamina. Nature. 2015;527(7576):105–109. doi: 10.1038/nature15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. doi: 10.1016/S0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 50.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400(6743):464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 51.Rich JN, Guo C, McLendon RE, Bigner DD, Wang XF, Counter CM. A genetically tractable model of human glioma formation. Cancer Res. 2001;61(9):3556–3560. [PubMed] [Google Scholar]
- 52.Lundberg AS, Randell SH, Stewart SA, Elenbaas B, Hartwell KA, Brooks MW, Fleming MD, Olsen JC, Miller SW, Weinberg RA, Hahn WC. Immortalization and transformation of primary human airway epithelial cells by gene transfer. Oncogene. 2002;21(29):4577–4586. doi: 10.1038/sj.onc.1205550. [DOI] [PubMed] [Google Scholar]
- 53.Liu J, Yang G, Thompson-Lanza JA, Glassman A, Hayes K, Patterson A, Marquez RT, Auersperg N, Yu Y, Hahn WC, Mills GB, Bast RC., Jr A genetically defined model for human ovarian cancer. Cancer Res. 2004;64(5):1655–1663. doi: 10.1158/0008-5472.CAN-03-3380. [DOI] [PubMed] [Google Scholar]
- 54.Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, Popescu NC, Hahn WC, Weinberg RA. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15(1):50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu J, Boyapati A, Rundell K. Critical role for SV40 small-t antigen in human cell transformation. Virology. 2001;290(2):192–198. doi: 10.1006/viro.2001.1204. [DOI] [PubMed] [Google Scholar]
- 56.MacKenzie KL, Franco S, Naiyer AJ, May C, Sadelain M, Rafii S, Moore MA. Multiple stages of malignant transformation of human endothelial cells modelled by co-expression of telomerase reverse transcriptase, SV40 T antigen and oncogenic N-ras. Oncogene. 2002;21(27):4200–4211. doi: 10.1038/sj.onc.1205425. [DOI] [PubMed] [Google Scholar]
- 57.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410(6832):1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 58.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15(24):3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4(6):437–450. doi: 10.1016/S1535-6108(03)00309-X. [DOI] [PubMed] [Google Scholar]
- 60.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17(24):3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF, Feng B, Brennan C, Weissleder R, Mahmood U, Hanahan D, Redston MS, Chin L, Depinho RA. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci USA. 2006;103(15):5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7(5):469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 63.Ijichi H, Chytil A, Gorska AE, Aakre ME, Fujitani Y, Fujitani S, Wright CV, Moses HL. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev. 2006;20(22):3147–3160. doi: 10.1101/gad.1475506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morton JP, Jamieson NB, Karim SA, Athineos D, Ridgway RA, Nixon C, McKay CJ, Carter R, Brunton VG, Frame MC, Ashworth A, Oien KA, Evans TR, Sansom OJ (2010) LKB1 haploinsufficiency cooperates with Kras to promote pancreatic cancer through suppression of p21-dependent growth arrest. Gastroenterology 139(2):586–597, 597e1–6 [DOI] [PMC free article] [PubMed]
- 65.Collins MA, Bednar F, Zhang Y, Brisset JC, Galban S, Galban CJ, Rakshit S, Flannagan KS, Adsay NV, Pasca di Magliano M. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest. 2012;122(2):639–653. doi: 10.1172/JCI59227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Collins MA, Brisset JC, Zhang Y, Bednar F, Pierre J, Heist KA, Galban CJ, Galban S, Pasca di Magliano M. Metastatic pancreatic cancer is dependent on oncogenic Kras in mice. PLoS One. 2012;7(12):e49707. doi: 10.1371/journal.pone.0049707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, Yan H, Wang W, Chen S, Viale A, Zheng H, Paik JH, Lim C, Guimaraes AR, Martin ES, Chang J, Hezel AF, Perry SR, Hu J, Gan B, Xiao Y, Asara JM, Weissleder R, Wang YA, Chin L, Cantley LC, DePinho RA. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149(3):656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen AT, Emelyanov A, Koh CH, Spitsbergen JM, Parinov S, Gong Z. An inducible kras(V12) transgenic zebrafish model for liver tumorigenesis and chemical drug screening. Dis Model Mech. 2012;5(1):63–72. doi: 10.1242/dmm.008367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langenau DM, Keefe MD, Storer NY, Guyon JR, Kutok JL, Le X, Goessling W, Neuberg DS, Kunkel LM, Zon LI. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007;21(11):1382–1395. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schook LB, Collares TV, Hu W, Liang Y, Rodrigues FM, Rund LA, Schachtschneider KM, Seixas FK, Singh K, Wells KD, Walters EM, Prather RS, Counter CM. A Genetic Porcine Model of Cancer. PLoS One. 2015;10(7):e0128864. doi: 10.1371/journal.pone.0128864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cox AD, Der CJ, Philips MR. Targeting RAS membrane association: back to the future for anti-RAS drug discovery? Clin Cancer Res. 2015;21(8):1819–1827. doi: 10.1158/1078-0432.CCR-14-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Downward J. RAS synthetic lethal screens revisited: still seeking the elusive prize? Clin Cancer Res. 2015;21(8):1802–1809. doi: 10.1158/1078-0432.CCR-14-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kimmelman AC. Metabolic dependencies in RAS-driven cancers. Clin Cancer Res. 2015;21(8):1828–1834. doi: 10.1158/1078-0432.CCR-14-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marcus K, Mattos C. Direct attack on RAS: intramolecular communication and mutation-specific effects. Clin Cancer Res. 2015;21(8):1810–1818. doi: 10.1158/1078-0432.CCR-14-2148. [DOI] [PubMed] [Google Scholar]
- 75.McCormick F. KRAS as a therapeutic target. Clin Cancer Res. 2015;21(8):1797–1801. doi: 10.1158/1078-0432.CCR-14-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Singh H, Longo DL, Chabner BA. Improving prospects for targeting RAS. J Clin Oncol. 2015;33:3650–3659. doi: 10.1200/JCO.2015.62.1052. [DOI] [PubMed] [Google Scholar]
- 77.Spiegel J, Cromm PM, Zimmermann G, Grossmann TN, Waldmann H. Small-molecule modulation of Ras signaling. Nat Chem Biol. 2014;10(8):613–622. doi: 10.1038/nchembio.1560. [DOI] [PubMed] [Google Scholar]
- 78.Bivona TG, Perez De Castro I, Ahearn IM, Grana TM, Chiu VK, Lockyer PJ, Cullen PJ, Pellicer A, Cox AD, Philips MR. Phospholipase Cgamma activates Ras on the Golgi apparatus by means of RasGRP1. Nature. 2003;424(6949):694–698. doi: 10.1038/nature01806. [DOI] [PubMed] [Google Scholar]
- 79.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL, 2nd, Cox AD, Philips MR. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4(5):343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 80.Bergo MO, Leung GK, Ambroziak P, Otto JC, Casey PJ, Young SG. Targeted inactivation of the isoprenylcysteine carboxyl methyltransferase gene causes mislocalization of K-Ras in mammalian cells. J Biol Chem. 2000;275(23):17605–17610. doi: 10.1074/jbc.C000079200. [DOI] [PubMed] [Google Scholar]
- 81.Kim E, Ambroziak P, Otto JC, Taylor B, Ashby M, Shannon K, Casey PJ, Young SG. Disruption of the mouse Rce1 gene results in defective Ras processing and mislocalization of Ras within cells. J Biol Chem. 1999;274(13):8383–8390. doi: 10.1074/jbc.274.13.8383. [DOI] [PubMed] [Google Scholar]
- 82.Liu M, Bryant MS, Chen J, Lee S, Yaremko B, Lipari P, Malkowski M, Ferrari E, Nielsen L, Prioli N, Dell J, Sinha D, Syed J, Korfmacher WA, Nomeir AA, Lin CC, Wang L, Taveras AG, Doll RJ, Njoroge FG, Mallams AK, Remiszewski S, Catino JJ, Girijavallabhan VM, Bishop WR, et al. Antitumor activity of SCH 66336, an orally bioavailable tricyclic inhibitor of farnesyl protein transferase, in human tumor xenograft models and wap-ras transgenic mice. Cancer Res. 1998;58(21):4947–4956. [PubMed] [Google Scholar]
- 83.End DW, Smets G, Todd AV, Applegate TL, Fuery CJ, Angibaud P, Venet M, Sanz G, Poignet H, Skrzat S, Devine A, Wouters W, Bowden C. Characterization of the antitumor effects of the selective farnesyl protein transferase inhibitor R115777 in vivo and in vitro. Cancer Res. 2001;61(1):131–137. [PubMed] [Google Scholar]
- 84.James GL, Goldstein JL, Brown MS. Polylysine and CVIM sequences of K-RasB dictate specificity of prenylation and confer resistance to benzodiazepine peptidomimetic in vitro. J Biol Chem. 1995;270(11):6221–6226. doi: 10.1074/jbc.270.11.6221. [DOI] [PubMed] [Google Scholar]
- 85.Manandhar SP, Hildebrandt ER, Schmidt WK. Small-molecule inhibitors of the Rce1p CaaX protease. J Biomol Screen. 2007;12(7):983–993. doi: 10.1177/1087057107307226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Porter SB, Hildebrandt ER, Breevoort SR, Mokry DZ, Dore TM, Schmidt WK. Inhibition of the CaaX proteases Rce1p and Ste24p by peptidyl (acyloxy)methyl ketones. Biochim Biophys Acta. 2007;1773(6):853–862. doi: 10.1016/j.bbamcr.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Winter-Vann AM, Baron RA, Wong W, dela Cruz J, York JD, Gooden DM, Bergo MO, Young SG, Toone EJ, Casey PJ. A small-molecule inhibitor of isoprenylcysteine carboxyl methyltransferase with antitumor activity in cancer cells. Proc Natl Acad Sci USA. 2005;102(12):4336–4341. doi: 10.1073/pnas.0408107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, Morales T, Aliagas I, Liu B, Sideris S, Hoeflich KP, Jaiswal BS, Seshagiri S, Koeppen H, Belvin M, Friedman LS, Malek S. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464(7287):431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 89.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, Marais R. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140(2):209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464(7287):427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gilmartin AG, Bleam MR, Groy A, Moss KG, Minthorn EA, Kulkarni SG, Rominger CM, Erskine S, Fisher KE, Yang J, Zappacosta F, Annan R, Sutton D, Laquerre SG. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin Cancer Res. 2011;17(5):989–1000. doi: 10.1158/1078-0432.CCR-10-2200. [DOI] [PubMed] [Google Scholar]
- 92.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, Chirieac LR, Kaur R, Lightbown A, Simendinger J, Li T, Padera RF, Garcia-Echeverria C, Weissleder R, Mahmood U, Cantley LC, Wong KK. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14(12):1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, Chodon T, Nelson SF, McArthur G, Sosman JA, Ribas A, Lo RS. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468(7326):973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dougherty MK, Muller J, Ritt DA, Zhou M, Zhou XZ, Copeland TD, Conrads TP, Veenstra TD, Lu KP, Morrison DK. Regulation of Raf-1 by direct feedback phosphorylation. Mol Cell. 2005;17(2):215–224. doi: 10.1016/j.molcel.2004.11.055. [DOI] [PubMed] [Google Scholar]
- 95.Britten CD. PI3K and MEK inhibitor combinations: examining the evidence in selected tumor types. Cancer Chemother Pharmacol. 2013;71(6):1395–1409. doi: 10.1007/s00280-013-2121-1. [DOI] [PubMed] [Google Scholar]
- 96.Liu EY, Ryan KM. Autophagy and cancer—issues we need to digest. J Cell Sci. 2012;125(Pt 10):2349–2358. doi: 10.1242/jcs.093708. [DOI] [PubMed] [Google Scholar]
- 97.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12(6):401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ganley IG, du Lam H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284(18):12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20(7):1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273(7):3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 102.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7(2):167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 103.Levy JM, Thompson JC, Griesinger AM, Amani V, Donson AM, Birks DK, Morgan MJ, Mirsky DM, Handler MH, Foreman NK, Thorburn A. Autophagy inhibition improves chemosensitivity in BRAF(V600E) brain tumors. Cancer Discov. 2014;4(7):773–780. doi: 10.1158/2159-8290.CD-14-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, McMahon M, White E. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 2013;3(11):1272–1285. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Corazzari M, Rapino F, Ciccosanti F, Giglio P, Antonioli M, Conti B, Fimia GM, Lovat PE, Piacentini M. Oncogenic BRAF induces chronic ER stress condition resulting in increased basal autophagy and apoptotic resistance of cutaneous melanoma. Cell Death Differ. 2015;22(6):946–958. doi: 10.1038/cdd.2014.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ma XH, Piao SF, Dey S, McAfee Q, Karakousis G, Villanueva J, Hart LS, Levi S, Hu J, Zhang G, Lazova R, Klump V, Pawelek JM, Xu X, Xu W, Schuchter LM, Davies MA, Herlyn M, Winkler J, Koumenis C, Amaravadi RK. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest. 2014;124(3):1406–1417. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Milani M, Rzymski T, Mellor HR, Pike L, Bottini A, Generali D, Harris AL. The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with bortezomib. Cancer Res. 2009;69(10):4415–4423. doi: 10.1158/0008-5472.CAN-08-2839. [DOI] [PubMed] [Google Scholar]
- 108.Bowman BM, Sebolt KA, Hoff BA, Boes JL, Daniels DL, Heist KA, Galban CJ, Patel RM, Zhang J, Beer DG, Ross BD, Rehemtulla A, Galban S. Phosphorylation of FADD by the kinase CK1alpha promotes KRASG12D-induced lung cancer. Sci Signal. 2015;8(361):ra9. doi: 10.1126/scisignal.2005607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Degtyarev M, De Maziere A, Orr C, Lin J, Lee BB, Tien JY, Prior WW, van Dijk S, Wu H, Gray DC, Davis DP, Stern HM, Murray LJ, Hoeflich KP, Klumperman J, Friedman LS, Lin K. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol. 2008;183(1):101–116. doi: 10.1083/jcb.200801099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Barnard RA, Wittenburg LA, Amaravadi RK, Gustafson DL, Thorburn A, Thamm DH. Phase I clinical trial and pharmacodynamic evaluation of combination hydroxychloroquine and doxorubicin treatment in pet dogs treated for spontaneously occurring lymphoma. Autophagy. 2014;10(8):1415–1425. doi: 10.4161/auto.29165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mahalingam D, Mita M, Sarantopoulos J, Wood L, Amaravadi RK, Davis LE, Mita AC, Curiel TJ, Espitia CM, Nawrocki ST, Giles FJ, Carew JS. Combined autophagy and HDAC inhibition: a phase I safety, tolerability, pharmacokinetic, and pharmacodynamic analysis of hydroxychloroquine in combination with the HDAC inhibitor vorinostat in patients with advanced solid tumors. Autophagy. 2014;10(8):1403–1414. doi: 10.4161/auto.29231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rangwala R, Chang YC, Hu J, Algazy KM, Evans TL, Fecher LA, Schuchter LM, Torigian DA, Panosian JT, Troxel AB, Tan KS, Heitjan DF, DeMichele AM, Vaughn DJ, Redlinger M, Alavi A, Kaiser J, Pontiggia L, Davis LE, O’Dwyer PJ, Amaravadi RK. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy. 2014;10(8):1391–1402. doi: 10.4161/auto.29119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rangwala R, Leone R, Chang YC, Fecher LA, Schuchter LM, Kramer A, Tan KS, Heitjan DF, Rodgers G, Gallagher M, Piao S, Troxel AB, Evans TL, DeMichele AM, Nathanson KL, O’Dwyer PJ, Kaiser J, Pontiggia L, Davis LE, Amaravadi RK. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy. 2014;10(8):1369–1379. doi: 10.4161/auto.29118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rosenfeld MR, Ye X, Supko JG, Desideri S, Grossman SA, Brem S, Mikkelson T, Wang D, Chang YC, Hu J, McAfee Q, Fisher J, Troxel AB, Piao S, Heitjan DF, Tan KS, Pontiggia L, O’Dwyer PJ, Davis LE, Amaravadi RK. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy. 2014;10(8):1359–1368. doi: 10.4161/auto.28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vogl DT, Stadtmauer EA, Tan KS, Heitjan DF, Davis LE, Pontiggia L, Rangwala R, Piao S, Chang YC, Scott EC, Paul TM, Nichols CW, Porter DL, Kaplan J, Mallon G, Bradner JE, Amaravadi RK. Combined autophagy and proteasome inhibition: a phase 1 trial of hydroxychloroquine and bortezomib in patients with relapsed/refractory myeloma. Autophagy. 2014;10(8):1380–1390. doi: 10.4161/auto.29264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Carretero J, Shimamura T, Rikova K, Jackson AL, Wilkerson MD, Borgman CL, Buttarazzi MS, Sanofsky BA, McNamara KL, Brandstetter KA, Walton ZE, Gu TL, Silva JC, Crosby K, Shapiro GI, Maira SM, Ji H, Castrillon DH, Kim CF, Garcia-Echeverria C, Bardeesy N, Sharpless NE, Hayes ND, Kim WY, Engelman JA, Wong KK. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer Cell. 2010;17(6):547–559. doi: 10.1016/j.ccr.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kim HS, Mendiratta S, Kim J, Pecot CV, Larsen JE, Zubovych I, Seo BY, Kim J, Eskiocak B, Chung H, McMillan E, Wu S, De Brabander J, Komurov K, Toombs JE, Wei S, Peyton M, Williams N, Gazdar AF, Posner BA, Brekken RA, Sood AK, Deberardinis RJ, Roth MG, Minna JD, White MA. Systematic identification of molecular subtype-selective vulnerabilities in non-small-cell lung cancer. Cell. 2013;155(3):552–566. doi: 10.1016/j.cell.2013.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ou YH, Torres M, Ram R, Formstecher E, Roland C, Cheng T, Brekken R, Wurz R, Tasker A, Polverino T, Tan SL, White MA. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Mol Cell. 2011;41(4):458–470. doi: 10.1016/j.molcel.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, Frohling S, Chan EM, Sos ML, Michel K, Mermel C, Silver SJ, Weir BA, Reiling JH, Sheng Q, Gupta PB, Wadlow RC, Le H, Hoersch S, Wittner BS, Ramaswamy S, Livingston DM, Sabatini DM, Meyerson M, Thomas RK, Lander ES, Mesirov JP, Root DE, Gilliland DG, Jacks T, Hahn WC. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kumar MS, Hancock DC, Molina-Arcas M, Steckel M, East P, Diefenbacher M, Armenteros-Monterroso E, Lassailly F, Matthews N, Nye E, Stamp G, Behrens A, Downward J. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell. 2012;149(3):642–655. doi: 10.1016/j.cell.2012.02.059. [DOI] [PubMed] [Google Scholar]
- 121.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, Elledge SJ. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137(5):835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P, Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, Liu Q, Iorio F, Surdez D, Chen L, Milano RJ, Bignell GR, Tam AT, Davies H, Stevenson JA, Barthorpe S, Lutz SR, Kogera F, Lawrence K, McLaren-Douglas A, Mitropoulos X, Mironenko T, Thi H, Richardson L, Zhou W, Jewitt F, Zhang T, O’Brien P, Boisvert JL, Price S, Hur W, Yang W, Deng X, Butler A, Choi HG, Chang JW, Baselga J, Stamenkovic I, Engelman JA, Sharma SV, Delattre O, Saez-Rodriguez J, Gray NS, Settleman J, Futreal PA, Haber DA, Stratton MR, Ramaswamy S, McDermott U, Benes CH. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, Papadimitriou CA, Murray S. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9(10):962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 125.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, De Dosso S, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Di Fiore F, Gangloff AO, Ciardiello F, Pfeiffer P, Qvortrup C, Hansen TP, Van Cutsem E, Piessevaux H, Lambrechts D, Delorenzi M, Tejpar S. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 126.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocakova I, Ruff P, Blasinska-Morawiec M, Smakal M, Canon JL, Rother M, Williams R, Rong A, Wiezorek J, Sidhu R, Patterson SD. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369(11):1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 127.Tan IB, Malik S, Ramnarayanan K, McPherson JR, Ho DL, Suzuki Y, Ng SB, Yan S, Lim KH, Koh D, Hoe CM, Chan CY, Ten R, Goh BK, Chung AY, Tan J, Chan CX, Tay ST, Alexander L, Nagarajan N, Hillmer AM, Tang CL, Chua C, Teh BT, Rozen S, Tan P. High-depth sequencing of over 750 genes supports linear progression of primary tumors and metastases in most patients with liver-limited metastatic colorectal cancer. Genome Biol. 2015;16:32. doi: 10.1186/s13059-015-0589-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503(7477):548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Herrero A, Pinto A, Colon-Bolea P, Casar B, Jones M, Agudo-Ibanez L, Vidal R, Tenbaum SP, Nuciforo P, Valdizan EM, Horvath Z, Orfi L, Pineda-Lucena A, Bony E, Keri G, Rivas G, Pazos A, Gozalbes R, Palmer HG, Hurlstone A, Crespo P. Small molecule inhibition of ERK dimerization prevents tumorigenesis by RAS-ERK pathway oncogenes. Cancer Cell. 2015;28(2):170–182. doi: 10.1016/j.ccell.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 130.Peng SB, Henry JR, Kaufman MD, Lu WP, Smith BD, Vogeti S, Rutkoski TJ, Wise S, Chun L, Zhang Y, Van Horn RD, Yin T, Zhang X, Yadav V, Chen SH, Gong X, Ma X, Webster Y, Buchanan S, Mochalkin I, Huber L, Kays L, Donoho GP, Walgren J, McCann D, Patel P, Conti I, Plowman GD, Starling JJ, Flynn DL. Inhibition of RAF isoforms and active dimers by LY3009120 leads to anti-tumor activities in RAS or BRAF mutant cancers. Cancer Cell. 2015;28(3):384–398. doi: 10.1016/j.ccell.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 131.Yao Z, Torres NM, Tao A, Gao Y, Luo L, Li Q, de Stanchina E, Abdel-Wahab O, Solit DB, Poulikakos PI, Rosen N. BRAF mutants evade ERK-dependent feedback by different mechanisms that determine their sensitivity to pharmacologic inhibition. Cancer Cell. 2015;28(3):370–383. doi: 10.1016/j.ccell.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Freeman AK, Ritt DA, Morrison DK. Effects of Raf dimerization and its inhibition on normal and disease-associated Raf signaling. Mol Cell. 2013;49(4):751–758. doi: 10.1016/j.molcel.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Santos E, Nebreda AR, Bryan T, Kempner ES. Oligomeric structure of p21 ras proteins as determined by radiation inactivation. J Biol Chem. 1988;263(20):9853–9858. [PubMed] [Google Scholar]
- 134.Inouye K, Mizutani S, Koide H, Kaziro Y. Formation of the Ras dimer is essential for Raf-1 activation. J Biol Chem. 2000;275(6):3737–3740. doi: 10.1074/jbc.275.6.3737. [DOI] [PubMed] [Google Scholar]
- 135.Lin WC, Iversen L, Tu HL, Rhodes C, Christensen SM, Iwig JS, Hansen SD, Huang WY, Groves JT. H-Ras forms dimers on membrane surfaces via a protein-protein interface. Proc Natl Acad Sci USA. 2014;111(8):2996–3001. doi: 10.1073/pnas.1321155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nan X, Tamguney TM, Collisson EA, Lin LJ, Pitt C, Galeas J, Lewis S, Gray JW, McCormick F, Chu S. Ras-GTP dimers activate the mitogen-activated protein kinase (MAPK) pathway. Proc Natl Acad Sci USA. 2015;112(26):7996–8001. doi: 10.1073/pnas.1509123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Muratcioglu S, Chavan TS, Freed BC, Jang H, Khavrutskii L, Freed RN, Dyba MA, Stefanisko K, Tarasov SG, Gursoy A, Keskin O, Tarasova NI, Gaponenko V, Nussinov R. GTP-dependent K-Ras dimerization. Structure. 2015;23(7):1325–1335. doi: 10.1016/j.str.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]