Abstract
Targeted genome modifications using techniques that alter the genomic information of interest have contributed to multiple studies in both basic and applied biology. Traditionally, in gene targeting, the target-site integration of a targeting vector by homologous recombination is used. However, this strategy has several technical problems. The first problem is the extremely low frequency of gene targeting, which makes obtaining recombinant clones an extremely labor intensive task. The second issue is the limited number of biomaterials to which gene targeting can be applied. Traditional gene targeting hardly occurs in most of the human adherent cell lines. However, a new approach using designer nucleases that can introduce site-specific double-strand breaks in genomic DNAs has increased the efficiency of gene targeting. This new method has also expanded the number of biomaterials to which gene targeting could be applied. Here, we summarize various strategies for target gene modification, including a comparison of traditional gene targeting with designer nucleases.
Keywords: Homologous recombination, Designer nucleases, Double-stranded DNA breaks
Introduction
Genetic modification by gene targeting has contributed to the advancement of research in both basic and applied biology [1–4]. Gene targeting involves the replacement of an endogenous gene by homologous recombination (HR) [5, 6]. Standard gene targeting vectors contain two regions that are homologous to the target genome locus, called “targeting arms”, flanking a selective marker gene [2, 7]. For successful gene replacement, nucleotide sequences between targeting arms and the target genomic locus should be identical, because a single mismatch during recombination could lead to the elimination of the targeting vector from the chromosomal DNA by initiating the mismatch repair mechanism [8]. Standard gene targeting involves target-site integration using a selective marker gene [9] (Fig. 1a). To remove the selective marker gene, the Flp-FRT system is often used. Flp is a site-specific recombinase that can recombine the two FRT sites, resulting in the removal of the drug resistance gene cassette. To remove the specific exon located between two loxP sites, as well as the selective marker gene, the Cre-loxP system is often used [9] (Fig. 1a, right). Cre is a site-specific recombinase that can recombine the two LoxP sites, resulting in the removal of the drug resistance gene cassette. Gene targeting is also used for gene modifications at target loci, called “knockin” (Fig. 1b). The knockin approach is used to investigate the function of a target gene along with its endogenous gene expression status [2]. One or several nucleotides are substituted with other nucleotides, and the biological functions of these substituted residues are investigated. This strategy is often used for complementation tests against specific mutations. The typical knockin approach involves fusing the carboxy-terminal end of the cDNA in-frame at a specific exon on the targeting vector (Fig. 1b, left). This results in the expression of C-terminal fused proteins. In the case where the cDNA contains small nucleotide substitutions, the function of the modified residues can be investigated. Another application is to introduce visualization marker genes, such as lacZ, fluorescent protein genes (GFPs and others), and luciferase genes [2]. Visualization of the target gene is achieved through two strategies. One is to introduce the marker gene with an internal ribosomal entry site (IRES) sequence [2]. This knockin results in bicistronic expression and allows investigation of the promoter activity of the target gene. In the second approach, the knockin results in the expression of a fusion protein with a visualization marker [10]. This allows for the study of the dynamics of the target protein, as well as the promoter activity. Another gene knockin strategy is to introduce a single-nucleotide substitution. For this approach, two methods have been reported: (1) using a selective marker gene (Fig. 1b, center) and (2) without using a selective marker (Fig. 1b, right). This approach is often used to study the functions of mutated genes. Gene targeting can also be used to induce a large deletion in the gene of interest (Fig. 1c). In the case where adjacent repeats are present, gene targeting results in two products, with or without the large deletion (Fig. 1c, upper). However, this approach is not very efficient, except in Saccharomyces cerevisiae. Therefore, the preferred strategy to remove a target locus is the Cre-LoxP-based system. In this approach, two loxP sites should be introduced by two independent gene targeting events. After the two loxP sites have been introduced on the same chromosome, the deletion is carried out using the Cre-recombinase (Fig. 1c, lower).
Fig. 1.
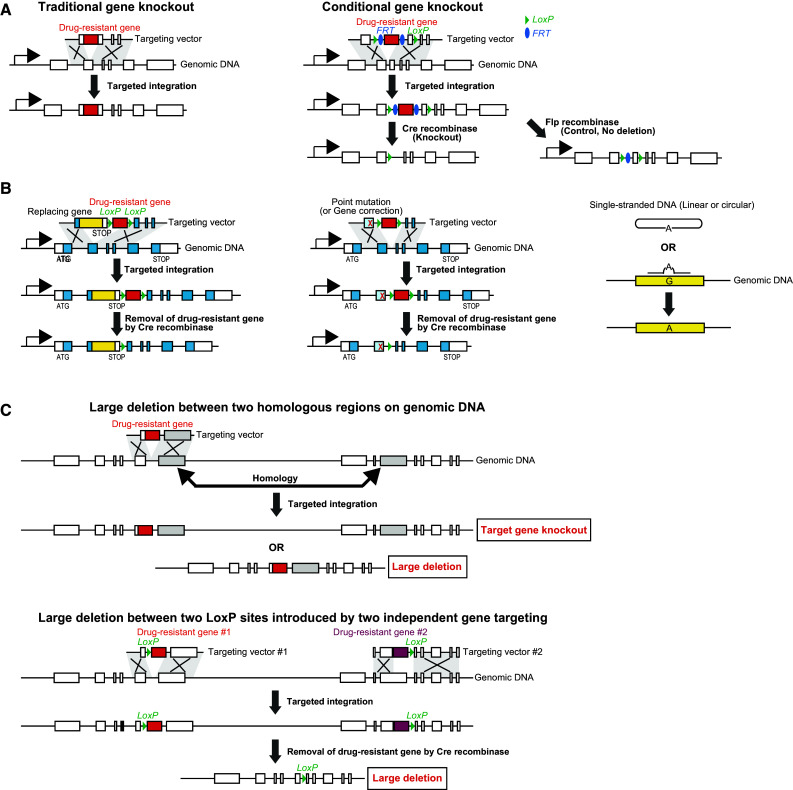
Various applications of gene targeting. a Traditional gene targeting strategy. Standard gene targeting vectors contain two regions homologous to the target genome locus, called “targeting arms”, flanking a selective marker gene, which is often a drug-resistance gene cassette. This vector is integrated into the target genome by homologous recombination. To remove the selective marker gene, the Flp-FRT system is often used. Flp is a site-specific recombinase that can recombine the two FRT sites, resulting in the removal of the drug resistance gene cassette. To remove the specific exon located between two loxP sites, the Cre-loxP system is often used. Cre is a site-specific recombinase that can recombine the two LoxP sites, resulting in the removal of the drug resistance gene cassette. b Strategy for nucleotide-level modification by gene targeting. One or several nucleotides are replaced with others, and the biological effect of the point mutation is investigated. The carboxy-terminus of the cDNA carrying the nucleotide substitution is introduced into the target genome locus. This results in the expression of C-terminally fused proteins (left). Gene targeting also allows single-nucleotide substitutions. For this, two methods are proposed. One involves the use of a selective marker gene (center) and the other does not use a selective marker (right). c Large deletions can be also induced by gene targeting. In the case where adjacent repeats are present on the genomic DNA, gene targeting results in two products (upper). The conventional strategy is to remove the target locus between two LoxP sites introduced by two independent gene targeting events. The large deletion is then introduced by the expression of Cre-recombinase (lower)
Recently, several methods for the site-specific cleavage of chromosomal DNA using artificial nucleases have been developed [4, 11]. The specific DNA sequence-recognition modules of these nucleases are designed; therefore, these nucleases are often called “designer nucleases” [4, 12]. The development of designer nucleases has greatly increased the frequency of successful gene targeting events, and expanded the number of biomaterials that can be genetically modified. Traditional gene targeting strategies were successful only in limited types of cell lines, such as mouse embryonic stem (ES) cells [2], chicken B lymphocyte cell line DT40 [7], and some human cancer cell lines [13, 14]. By contrast, gene modification using designer nucleases can be applied to a wide variety of cell types, such as primary cells isolated from plants and humans [4]. Although gene targeting has been used in numerous studies, its mechanism is poorly understood. In this review, we summarize the various strategies for gene targeting, with an emphasis on their mechanisms in higher eukaryotes, such as mammals, birds, and plants.
Mechanisms of traditional gene targeting by HR
HR is defined as a genome rearrangement that occurs between two homologous regions on the DNA. The biological roles of HR are the repair of double-stranded DNA breaks (DSBs), re-start of stalled DNA replication forks, and meiotic recombination [15]. Only the DSB repair function of HR contributes to gene targeting [5, 6, 16]; therefore, we only describe this mechanism in this article. The first step in this process is the DNA-end processing, also called “resection” [17]. Broken DNA-ends are processed by certain exonucleases and/or endonucleases, such as the MRE11-RAD50-NBS1 (MRN) complex, EXO1, and CtIP, and this action results in the 3′-single-stranded tails, often called “3′-overhangs” (Fig. 2). After 3′-overhangs have been generated on the two broken DNA-ends, the HR pathway encounters its first branch: it can proceed by either the single-strand annealing (SSA) pathway or the RAD51-dependent pathway (Fig. 2). In the case where the two 3′-overhangs generated from one DSB possess complimentary sequences to each other, such DNA ends would anneal and their homologous regions would re-ligated through the SSA pathway (Fig. 1a). In this pathway, small deletions would occur, but DSBs are efficiently repaired. In addition, SSA requires homologous regions on both ends. Therefore, SSA occurs only when a DSB occurs between direct repeats. Its locus-dependent nature means that SSA cannot be the major pathway for HR. To date, RAD52 and XPF-ERCC1 have been identified as required factors for SSA [18]. However, the complete mechanism of SSA in mammalian cells has not been revealed.
Fig. 2.
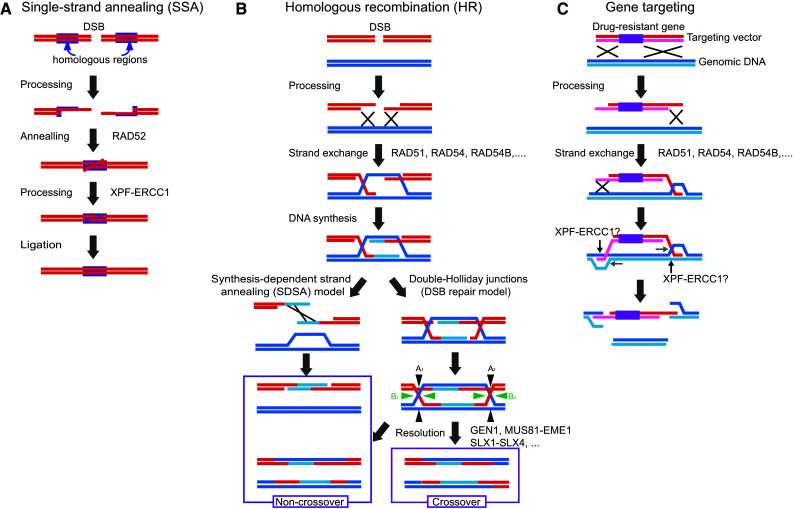
Proposed models of double-stranded DNA break (DSB) repair by homologous recombination (HR) and gene targeting. a Model of the single-strand annealing (SSA) model. The DSB on the genomic DNA is processed by exonucleases and/or endonucleases, resulting in 3′-single-stranded tails. In the case of two 3′-overhangs generated from one DSB that possess complimentary sequences to each other, such DNA-ends would anneal and their homologous regions would be re-ligated through the SSA pathway. b The synthesis-dependent single-strand annealing (SDSA) model and DSB repair model. These are the major pathways of HR. RAD51 proteins bind to the 3′-overhangs and a protein-DNA complex. Then, RAD51 proteins catalyze homology search and strand exchange reactions. After the resynthesis of the broken part of the DNA using complimentary sequences, invaded strands are released, and the resynthesized strands anneal. This is the mechanism for the SDSA model. Contrastingly, in the case where the invaded strands are converted to Holliday junctions (four-way junction structures), Holliday junction resolvases complete the HR process, which is termed the DSB repair model. This process results in two distinct recombination products: crossover and non-crossover products. c Model of gene targeting. The two strands of a targeting vector are processed, resulting in 3′-overhangs. Strand invasion by both 3′-overhangs of the targeting vector into the homologous sequences of the genomic DNA to create two D-loops. Such D-loops are resolved by XPF-ERCC1 structure-specific endonucleases
The major pathway for HR is the RAD51-dependent pathway [15, 19]. Multiple RAD51 proteins bind to the 3′-overhangs and form RAD51-filament structures on the single-stranded DNAs [20]. For RAD51-loading, BRCA2 and RAD51 paralogs, such as XRCC2, XRCC3, RAD51B, RAD51C, and RAD51D, play essential roles [21]. Without BRCA2 and RAD51 paralogs, RAD51 cannot be localized to the damaged sites [21]. Nonessential factors include RAD54 and RAD54B, which support RAD51’s function [16]. The RAD51-DNA complex promotes the homology search, using the 3′-overhang sequence. If homologous regions are found nearby, RAD51 catalyzes strand exchange with the homologous double-stranded DNA target and promotes the annealing of the 3′-overhang with its complimentary sequence (Fig. 2b) [15, 20]. The triple-stranded structures created by the strand exchange reaction are called D-loops, which are important intermediate structures in HR. After this step, two distinct pathways have been suggested for HR. One is the synthesis-dependent single-strand annealing (SDSA) model [22]. The invaded 3′-overhang can initiate DNA synthesis using the complimentary sequence in the D-loop, and the genetic information lost because of the DSB is restored. After DNA synthesis, the invaded strands are released from the D-loop by reverse branch migration. The released single-stranded DNA strands anneal to their homologous regions located in the newly synthesized DNA (Fig. 2b). In this model, the flanking genetic loci will not be crossed-over in the HR event (Fig. 2b). HR in somatic cells hardly involves crossovers; therefore, it is thought to be carried out by SDSA [22]. The other pathway is the Holliday junction-mediated pathway, also known as the DSB repair model [22]. To initiate this pathway, D-loops should be converted into Holliday junctions, defined as four-way junctions comprising four nucleotide strands. During DSB repair by HR, double-Holliday junctions are frequently formed, and the direction in which they are resolved determines the crossover and non-crossover products. A schematic model of double-Holliday junctions is shown in Fig. 2b. If the resolution of the double-Holliday junction occurs in either the A1–A2 or B1–B2 directions, a non-crossover product would be formed; if the resolution occurs in either the A1–B2 or B1–A2 directions, a crossover product would be formed (Fig. 2b). As mentioned above, most somatic cells show HR with no crossovers; however, cells isolated from individuals with certain genetic disorders, such as Bloom’s syndrome, show higher frequencies of crossover events, indicating that the DSB repair pathway of HR is activated [23]. Thus, gene targeting in Blm-deficient ES cells, which show this phenotype, were used to study the frequency of gene targeting. The results showed not only a higher frequency of gene targeting but also inter-allelic gene conversion between homologous chromosomes [23]. Therefore, Blm-deficient ES cells were used for genetic engineering, such as generating an insertion mutant library [23, 24]. Holliday junctions are resolved by the structure-specific endonuclease, Holliday junction resolvase [25]. MUS81-EME1 was identified as the first eukaryotic Holliday junction resolvase [26]. In addition, purified MUS81-EME1 showed asymmetric cleavage of Holliday junctions [26]. Recently, another structure-specific endonuclease, SLX1–SLX4, was identified [27, 28]. SLX1 contains a nuclease motif, and SLX4 is a scaffold protein that interacts with various structure-specific endonucleases, such as SLX1, MUS81–EME1, and XPF–ERCC1 [27, 28]. Unlike MUS81–EME1, SLX1–SLX4 catalyzes the symmetrical cleavage of Holliday junctions, and co-incubation with MUS81–EME1 enhanced the cleavage of Holliday junctions in vitro, indicating that SLX4 must be a director of structure-specific nucleases acting on recombination intermediates [27, 28]. Another important discovery was GEN1, a Holliday junction resolvase in mammalian cells [29]. GEN1 forms a homodimer and cleaves Holliday junctions symmetrically in vitro [29]. Cellular studies suggested that GEN1 acts independently from MUS81–EME1 and SLX1–SLX4 [25]. These Holliday junction resolvases complete the HR process, which is particularly important for meiotic recombination.
Gene targeting largely overlaps with HR; however, the resolution steps seem to be different for the two events [30–32]. Previous studies suggested that at least NBS1, RAD51 paralogs, RAD52, RAD54, and RAD54B are involved in gene targeting, although not all HR factors have been studied [5, 33, 34]. The ends of the transfected targeting vector are processed by NBS1, probably through the MRN complex [34], and D-loop formation occurs, similar to that in the conventional HR pathways [5]. However, neither Mus81- nor Eme1-deficient ES cells showed a decreased frequency of gene targeting, while Ercc1-deficient ES cells were completely defective [31, 32]. This result suggests that the resolution of recombination intermediates in gene targeting is probably carried out by XPF–ERCC1 [30]. Based on these observations, Niedernhofer and colleagues proposed a mechanism of gene targeting (Fig. 2c) [30]. In their model, both processed 3′-ends of the targeting vector are utilized for the homology search, with the aid of the general HR machinery, such as RAD51, RAD51 paralogs, RAD52, and RAD54, and this action results in two D-loops (Fig. 2c). The D-loops are then resolved by the structure-specific endonuclease XPF–ERCC1. The substrate of XPF–ERCC1 is the single-stranded branched arms, and its preferential cleavage sites are located in double-stranded DNA just 5′- to a 3′-single-stranded domain. In gene targeting, the cleavage of the targeting vector by XPF–ERCC1 results in the favored outcome of target gene integration (Fig. 2c). A similar model was proposed from studies in yeasts [6, 35].
Problems with traditional gene targeting strategies
Saccharomyces cerevisiae and chicken DT40 cells show an extremely high frequency of gene targeting. Therefore, these cell types are used widely in reverse genetics studies [6, 7, 35]. The gene targeting efficiency in mouse ES cells is generally very low, although a few gene targeting vectors have shown improved efficiency. Thus, gene targeting in mouse ES cells is labor intensive. However, the greatest advantage of using mouse ES cells is that they permit the generation of genetically modified mice [2]. One disadvantage of using chicken DT40 cells and mouse ES cells is that not many primary antibodies are commercially available. Most commercially available antibodies are usually produced using human antigens, and do not always cross-react with mouse or chicken antigens. Therefore, researchers need to determine whether these commercially available antibodies cross-react with the mouse and/or chicken antigens of interest. With regard to gene targeting in human cells, although some human cell types can be used for genome modifications by gene targeting, a number of problems still remain. One is that the efficiency of the gene targeting in human cells is extremely low, particularly in adherent cells. Another problem is that many human cell lines used in reverse genetics were established from cancerous tumors [13]. Cancer cells possess many mutations; therefore, these cell lines are not always suitable for basic science studies. To overcome these problems, new methods have been developed. Site-directed DSBs induced by designer nucleases allows targeted gene mutations as well as increasing the frequency of targeting vector integration [36]. Using this system, the efficiency of genome modifications in mammalian cells improved considerably, resulting in increased applications of targeted gene modifications to a variety of mammalian cell lines. The designer nuclease-induced genome modifications will be summarized in the next section. Moreover, the main advantage of genome editing methodologies based on designer nucleases is that the approach could be used to alter the genome of any whole organism, not only humans. Genome modification by designer nucleases may allow the genetic engineering of model animals such as Drosophila [37], zebrafish [38], Xenopus [39, 40], rat [41], rabbit [42], and cow [43], which is not possible through the classical ES cell approach.
Targeted induction of genomic DSBs by designer nucleases has improved the frequency of gene targeting
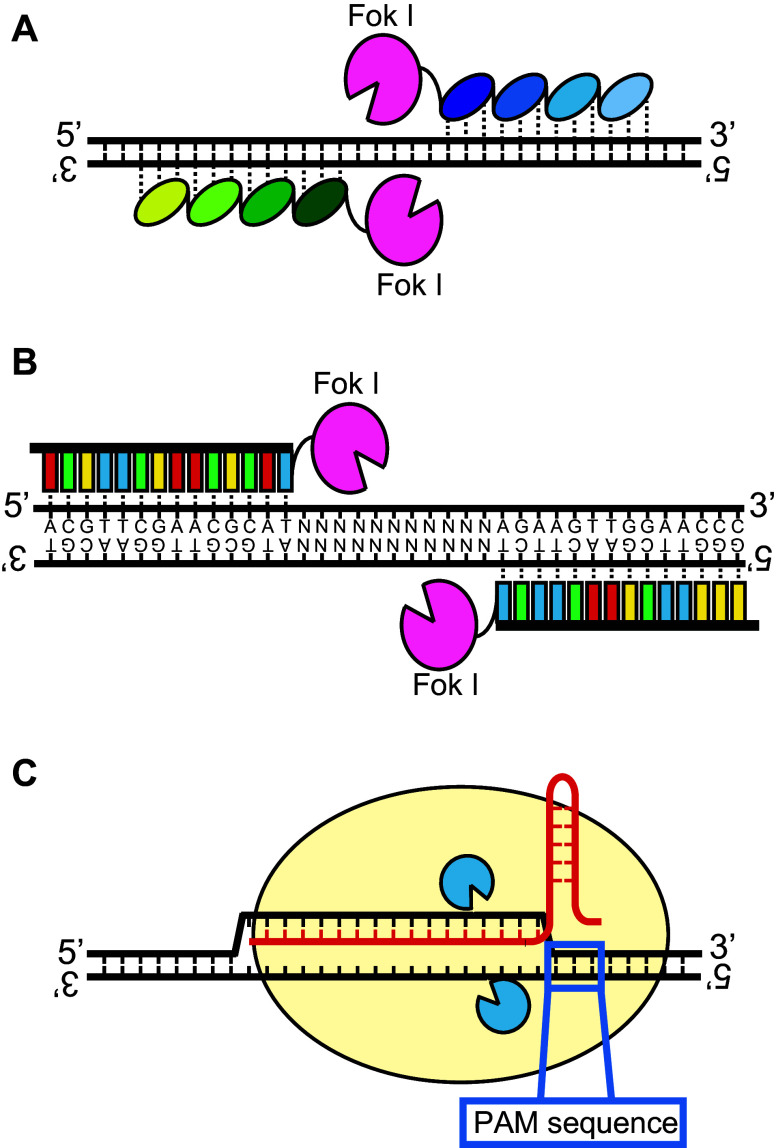
Various strategies to create a site-specific DSB have been investigated to improve the efficiency of gene targeting. To date, several artificial nuclease-based systems have been developed [4, 36]. The first designer nuclease is the zinc finger nuclease (ZFN). ZFNs contain zinc finger (ZF) domains for the recognition of the target sequence and a nuclease domain, which is typically the catalytic domain of the restriction enzyme FokI, for DNA cleavage [44]. Notably, this innovation enables the engineering of enzymes that cleave desired sequences on genomic DNA. The DNA recognition site is composed of C2H2 type ZF domain of a transcription factor, which recognizes a specific triplet sequence on the DNA [44, 45]. The structure-based mechanism of nucleotide recognition by ZFs is beyond the topic of this review; therefore, we recommend a comprehensive review on the biochemical aspects of target sequence recognition [46]. A large number of ZFs that recognize various nucleotide triplet sequences have been identified, and it has become possible to engineer ZFNs that recognize specific sequences [44, 47]. For example, to recognize the particular 12-bp target sequence, a sequential combination of four ZFs corresponding to each triplet nucleotide sequences of the target would be required (Fig. 3a). Currently, ZFNs are typically designed to contain three to six zinc finger domains, and are therefore capable of recognizing specific sequences of between nine and 18 base pairs (bps) (Fig. 3a). One ZFN cleaves only one strand of the target DNA; therefore, the introduction of DSBs requires the design of another ZFN that cleaves the complementary sequence nearby (Fig. 3a). This technique is currently used in genetic studies, as well as in anti-retroviral therapy against human immunodeficiency viruses, to knockout the gene encoding the CCR5 chemokine receptor, the main means of viral access to the host cells. However, ZFNs have several problems. One is the difficulty of designing ZFNs, which requires knowledge of various ZF domains, making it relatively complex to design customized ZFNs. The second problem is that the genome engineering by ZFNs is barely applicable to regions lacking GNN trinucleotides, the preferential target of C2H2 ZFs. The third problem is non-specific cleavage of the genome, termed off-target cleavage. Some ZFNs cleave not only the target sequence of interest but also other sites with similar sequences [48].
Fig. 3.
Principle of site-specific DSB formations by designer nucleases. a DNA sequence recognition by zinc finger nuclease (ZFN). Each ellipse represents a zinc finger (ZF) domain that recognizes the specific triplet nucleotides. To recognize the particular 12-bp target sequence for example, the sequential combination of four ZFs corresponding to each triplet nucleotide sequences of the target would be required. Dotted lines represent the target sequence recognitions by ZFN. b DNA sequence recognition by transcription activator-like effector nuclease (TALEN). Red boxes represent the NI at RVD that targets adenine on the DNA. Green boxes represent HD at RVD that targets cytosine. Blue boxes represent NG at RVD that targets thymine. Yellow boxes represent NN, NK, or NH, which target guanine. c DNA sequence recognition by the CLISPR/cas9 system. The guide RNA determines the target sites, and the protein subunit cleaves the DNA strands
The second designer nuclease is the transcription activator-like effector nuclease (TALEN) [12]. TALEN also comprises a DNA recognition domain and a DNA cleavage domain. Similar to ZFNs, the nuclease domain often used is a FokI catalytic domain (Fig. 3b), whereas the DNA recognition domain is composed of multiple transcription activator-like (TAL) domains, which were originally isolated from the Xanthomonas genus of plant bacterial pathogens [49]. The central TAL targeting domain comprises typically 1.5–35 repeats of a 34 amino-acid core unit. Typically, the 12th and 13th residues of the core units are variable, and are also known as the repeat-variable di-residue (RVD) [12]. The RVD determines the single nucleotide that a TAL domain will recognize. If the amino-acid sequence of the RVD is asparagine and isoleucine (NI), it targets adenine, represented by red-boxes in Fig. 3b; if the sequence is histidine and glutamate (HD), it targets cytosine (green-boxes in Fig. 3b); if the sequence is asparagine and glycine (NG), it targets thymine (blue-boxes in Fig. 3b); and if the sequence is asparagine and asparagine (NN), it targets guanine, (yellow boxes in Fig. 3b). However, because NN can also bind adenine with lower specificity, this may increase the risk of off-target cleavages. However, recent studies showed that owing to the reduced specificity of NN-di-residues for guanines, NN di-residues, NK or NH di-residues are preferred because they show a stronger affinity for their targets [50, 51]. Similar to ZFN, one unit of TALEN cleaves only one strand of the target DNA; thus, the introduction of DSBs requires the design of another enzyme that cleaves the complementary sequence nearby (Fig. 3b).
The third designer nuclease is the CRISPR (clustered regularly interspaced short palindromic repeats) and cas (CRISPR-associated) 9 gene system [52]. This system is essential for immunity in selected bacteria, acting as a defense mechanism against invading genetic elements, such as bacteriophages and plasmids [52]. Invading genetic elements from bacteriophages or plasmids are cleaved to small fragments, typically about 20 bps, and integrated into the CRISPR locus [52]. These sequences are then used by bacteria as a memory of previous infections. When additional genetic invasions occur, the CRISPR locus is transcribed and the RNA molecules are then processed to small RNAs, called CRISPR RNAs. CRISPR RNAs, often called guide RNAs, are used to guide effectors for Cas endonucleases that cleave complementarity sequences of CRISPR RNAs, resulting in the degradation of invading genetic materials. Thus, bacteria protect their genome from invasion by genetic elements. Recently, this system was used in a genome modification technique for targeted site-specific DSB formation in genomic DNA. The cleavage of target DNA sites is typically carried out by Cas 9, an endonuclease encoded by cas genes. The target gene of interest can be selected by choosing guide RNAs that are complementary to the DNA sequence of the genomic target. An important point is that unique target sequences need to be selected with the protospacer adjacent motif (PAM) next to the 3′-end (Fig. 3c). The first PAM sequence, the NGG sequence, was derived from Streptomyces pyogenes and the CRISPR/cas9 systems can utilize different PAM sequences derived from other bacteria, thus increasing the targeting repertoire.
Co-transfection of CRISPR/cas9 expression vectors with guide RNAs or guide RNA-expression vectors induces site-specific DSBs in mammalian cells. The advantage of the CRISPR/cas9 system is that the construction and processing of target sites is very easy. However, a disadvantage is that off-target cleavage is higher than with TALENs. To increase the site-specificity, the CRISPR/nickase system is often used [52]. In this case, two guide RNAs are required for efficient DSB formation, resulting in site-specific induction of DSBs. The CRISPR/cas9 or CRISPR/nickase systems are very simple to design and are now used widely.
Alternatively, it was reported that meganucleases could enhance gene targeting at the cleaved locus by up to 1000-fold. Megaendonucleases, often referred to as homing endonucleases, are enzymes that recognize and cleave specific target sequences in the genome. In nature, they are encoded by mobile introns or inteins, and function as endonucleases to promote lateral transfer of these intervening sequences. I-CreI has become the most extensively engineered homing endonuclease. Site-directed mutagenesis downstream of the recognition sequence of the meganuclease allows induction of locus-specific DSBs in the genome [53–55]. Using I-CreI-derived proteins, genome modification by meganuclease-mediated gene targeting was conducted in the human genome, for example at the RAG1 and XPC loci [11, 56, 57]. Meganuclease-mediated gene targeting is highly efficient if the target site of the meganuclease is present. However, a disadvantage of using meganucleases is that designing the target sequence is not as flexible as that of the other designer nucleases described above. Therefore, this approach is not widely used currently.
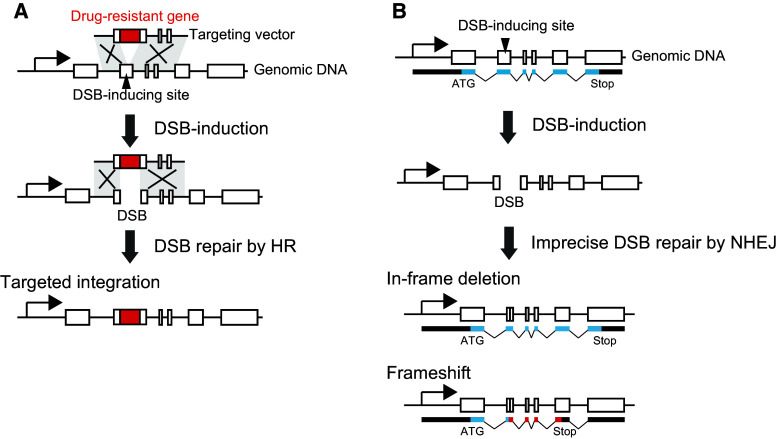
Introduction of site-specific DSBs increases the efficiency of gene targeting. This probably reflects DSB repair by HR using the targeting vector as the homologous template (Fig. 4a). The locus-specific DSB is introduced in the genomic DNA by the action of designer nucleases. Broken DNA ends are processed by certain exonucleases and/or endonucleases, producing 3′-overhangs. At the same time, the ends of the targeting vector are also processed to produce 3′-overhangs. The 3′-overhangs produced on the genomic DNA and the targeting vector are complimentary to each other; therefore, they will anneal and be ligated. However, the detailed mechanism of this process has not been determined. Nevertheless, even without gene targeting vectors, targeted gene mutations have been introduced [4]. In mammalian cells, the dominant pathway for DSB repair is the non-homologous end-joining (NHEJ) pathway, which involves direct re-ligation of two DNA ends [19]. Imprecise DSB repairs by NHEJ result in small genome rearrangements at the site of the break, leading to deletions and insertions (Fig. 4b). Such genome rearrangements result in an in-frame deletion, which may result in the expression of proteins with small truncations, or a frameshift mutation, which may result in abnormal proteins (Fig. 4b). Even such small mutations can often abolish the gene function completely, thereby enabling investigation of gene function. Because NHEJ is active throughout the cell cycle, imprecise DSB repairs by NHEJ may occur at a higher frequency than targeting vector integration by HR [58]. HR is active only in S-phase and mostly acts on DNA replication sites [58]. Cell lines with higher frequencies of traditional gene targeting show greater percentages of cells in S-phase, (approximately 50 %). However, most cell lines do not show such a high percentage of cells in the S-phase, lowering the chances of successful gene targeting through the traditional approach. Thus, the development of gene modification techniques using designer nucleases will contribute to the expanded application of reverse genetics in basic and applied biology.
Fig. 4.
Models of targeted gene mutations induced by designer nucleases. a Designer nuclease-induced gene targeting. Co-transfection of designer nucleases with the gene targeting vector results in higher efficiency of vector integration. b Designer nuclease-induced gene mutations. If a designer nuclease acts on the target genomic DNA, a site-specific DSB will occur. Imprecise DSB repair by non-homologous end-joining (NHEJ) then induces small DNA rearrangements, such as deletions. This results in either an in-frame deletion or a frameshift mutation
Perspective
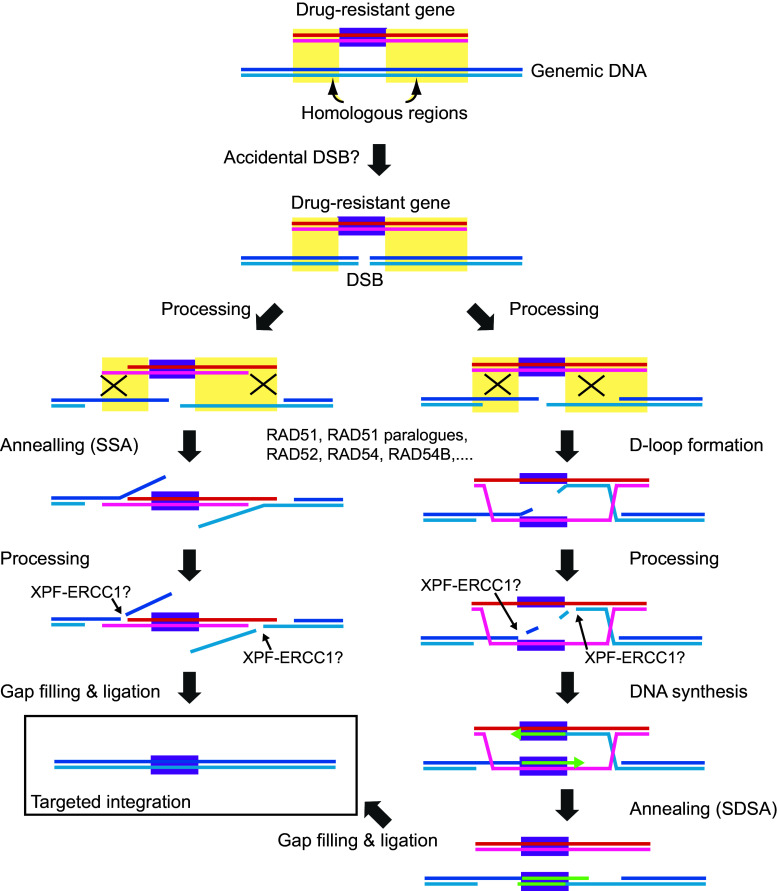
Designer nucleases have contributed to the increased use of reverse genetics for gene modifications. However, several problems still remain. As mentioned above, these nucleases introduce off-target mutations. The risk of off-target mutations varies depending on the sequence of the targets. Detailed knowledge of the target sequences is essential to reduce the risk of off-target cleavages. First, the most important improvement required is to increase the DNA-binding specificity of the DNA-recognition modules. One possibility is to optimize the target sequence to increase the sequence-specific DNA binding. Similar to RNA interference, such as with siRNAs and shRNAs, the construction of a designer nuclease requires consideration of the prevalence of a target sequences in the whole genome, using in silico identification. The activity of the designer nuclease is another important point. If too much designer nuclease is expressed, the risk of off-target cleavage could be increased, while too little nuclease reduces the frequency of target gene modification. At present, lipofection is often used to deliver transgenes. This introduces a large number of DNA molecules, thereby increasing the expression of the nucleases. By contrast, electroporation introduces fewer transgenes into the cells, and the efficiency of transfection is lower than that of other methods, such as lipofection and lentiviral infection. Further studies are required to overcome these problems. In addition, even if a designer nuclease is used, the frequency of target gene modification is not always high. In contrast, even in traditional gene targeting strategy, some targeting vectors show high efficiency of targeting vector integration, typically more than 30 %. The reason for this increased frequency using certain vectors is not well understood; however, the high frequency is not simply correlated with the length of the homologous regions on the targeting arms. It is likely that some genomic loci are hotspots for gene targeting. How do these hotspots of the gene targeting appear? One explanation could be the chromatin states. Gene targeting of loci within euchromatin, a loosely packed state of chromatin, should be easier than that of loci within heterochromatin, a tightly packed state of chromatin. Another possibility is that the hotspots for gene targeting might also be hotspots for spontaneous DSBs. For example, mammalian chromosomes have fragile sites [59]. The origins and termination sites for DNA replication are also potential loci that show higher incidence of DSBs [59]. The interaction between transcription and DNA replication machineries may also induced a higher incidence of DSBs [60]. When transcription and replication machineries traveling towards each other on the same DNA substrate collide, the DNA replication forks are usually stalled, thereby inducing HR [60]. In these cases, the broken ends of the chromosomal DNAs initiate the homology search reaction and are ligated through either SSA or SDSA using the targeting vector as a homologous template (Fig. 5). Furthermore, experimental detection of DSB and HR intermediates at the target locus are difficult in mammals and chicken DT40 cells; however, if these possibilities were experimentally proven, it would contribute substantially to our understanding of genomic modification techniques. Overall, recent innovations in genomic modification techniques have contributed greatly to increasing the efficiency of gene targeting. However, further improvements are still needed. To expand the applications of this innovative technology, a deeper understanding of the molecular mechanisms of gene targeting and genetic recombination is required.
Fig. 5.
Perspectives for alternative mechanisms of gene targeting. Gene targeting might be initiated from a DSB on the genomic DNA. The broken ends of the chromosomal DNAs initiate the homology search reaction and ligate through either SSA or SDSA, using the targeting vector as the homologous template
Acknowledgments
KH is funded by a Grant-in-Aid for Young Scientists (A) (25710010), Japan Society for the Promotion of Science (JSPS), The Ministry of Education, Culture, Sports, Science and Technology (MEXT). The authors declare that no conflict of interest exists.
References
- 1.Madyagol M, Al-Alami H, Levarski Z, Drahovska H, Turna J, Stuchlik S. Gene replacement techniques for Escherichia coli genome modification. Folia Microbiol Praha. 2011;56(3):253–263. doi: 10.1007/s12223-011-0035-z. [DOI] [PubMed] [Google Scholar]
- 2.Bouabe H, Okkenhaug K. Gene targeting in mice: a review. Methods Mol Biol. 2013;1064:315–336. doi: 10.1007/978-1-62703-601-6_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puchta H, Fauser F. Gene targeting in plants: 25 years later. Int J Dev Biol. 2013;57(6–8):629–637. doi: 10.1387/ijdb.130194hp. [DOI] [PubMed] [Google Scholar]
- 4.Maggio I, Goncalves MA. Genome editing at the crossroads of delivery, specificity, and fidelity. Trends Biotechnol. 2015;33(5):280–291. doi: 10.1016/j.tibtech.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 5.Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol. 2001;21(8):2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langston LD, Symington LS. Gene targeting in yeast is initiated by two independent strand invasions. Proc Natl Acad Sci USA. 2004;101(43):15392–15397. doi: 10.1073/pnas.0403748101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamazoe M, Sonoda E, Hochegger H, Takeda S. Reverse genetic studies of the DNA damage response in the chicken B lymphocyte line DT40. DNA Repair Amst. 2004;3(8–9):1175–1185. doi: 10.1016/j.dnarep.2004.03.039. [DOI] [PubMed] [Google Scholar]
- 8.Abuin A, Zhang H, Bradley A. Genetic analysis of mouse embryonic stem cells bearing Msh3 and Msh2 single and compound mutations. Mol Cell Biol. 2000;20(1):149–157. doi: 10.1128/MCB.20.1.149-157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhn R, Schwenk F. Advances in gene targeting methods. Curr Opin Immunol. 1997;9(2):183–188. doi: 10.1016/S0952-7915(97)80133-1. [DOI] [PubMed] [Google Scholar]
- 10.Agarwal S, van Cappellen WA, Guenole A, Eppink B, Linsen SE, Meijering E, Houtsmuller A, Kanaar R, Essers J. ATP-dependent and independent functions of Rad54 in genome maintenance. J Cell Biol. 2011;192(5):735–750. doi: 10.1083/jcb.201011025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munoz IG, Prieto J, Subramanian S, Coloma J, Redondo P, Villate M, Merino N, Marenchino M, D’Abramo M, Gervasio FL, Grizot S, Daboussi F, Smith J, Chion-Sotinel I, Paques F, Duchateau P, Alibes A, Stricher F, Serrano L, Blanco FJ, Montoya G. Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus. Nucleic Acids Res. 2011;39(2):729–743. doi: 10.1093/nar/gkq801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39(12):e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanez RJ, Porter AC. Therapeutic gene targeting. Gene Ther. 1998;5(2):149–159. doi: 10.1038/sj.gt.3300601. [DOI] [PubMed] [Google Scholar]
- 14.Adachi N, Nishijima H, Shibahara K. Gene targeting using the human Nalm-6 pre-B cell line. Biosci Trends. 2008;2(5):169–180. [PubMed] [Google Scholar]
- 15.Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol. 2013;5(11):a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wesoly J, Agarwal S, Sigurdsson S, Bussen W, Van Komen S, Qin J, van Steeg H, van Benthem J, Wassenaar E, Baarends WM, Ghazvini M, Tafel AA, Heath H, Galjart N, Essers J, Grootegoed JA, Arnheim N, Bezzubova O, Buerstedde JM, Sung P, Kanaar R. Differential contributions of mammalian Rad54 paralogs to recombination, DNA damage repair, and meiosis. Mol Cell Biol. 2006;26(3):976–989. doi: 10.1128/MCB.26.3.976-989.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferretti LP, Lafranchi L, Sartori AA. Controlling DNA-end resection: a new task for CDKs. Front Genet. 2013;4:99. doi: 10.3389/fgene.2013.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Motycka TA, Bessho T, Post SM, Sung P, Tomkinson AE. Physical and functional interaction between the XPF/ERCC1 endonuclease and hRad52. J Biol Chem. 2004;279(14):13634–13639. doi: 10.1074/jbc.M313779200. [DOI] [PubMed] [Google Scholar]
- 19.Huhn D, Bolck HA, Sartori AA. Targeting DNA double-strand break signalling and repair: recent advances in cancer therapy. Swiss Med Wkly. 2013;143:w13837. doi: 10.4414/smw.2013.13837. [DOI] [PubMed] [Google Scholar]
- 20.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87(4):757–766. doi: 10.1016/S0092-8674(00)81394-X. [DOI] [PubMed] [Google Scholar]
- 21.van Veelen LR, Essers J, van de Rakt MW, Odijk H, Pastink A, Zdzienicka MZ, Paulusma CC, Kanaar R. Ionizing radiation-induced foci formation of mammalian Rad51 and Rad54 depends on the Rad51 paralogs, but not on Rad52. Mutat Res. 2005;574(1–2):34–49. doi: 10.1016/j.mrfmmm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 22.Andersen SL, Sekelsky J. Meiotic versus mitotic recombination: two different routes for double-strand break repair: the different functions of meiotic versus mitotic DSB repair are reflected in different pathway usage and different outcomes. BioEssays. 2010;32(12):1058–1066. doi: 10.1002/bies.201000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, Youssoufian H, Vogel H, Schultz RA, Bradley A. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat Genet. 2000;26(4):424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- 24.Yamanishi A, Yusa K, Horie K, Tokunaga M, Kusano K, Kokubu C, Takeda J. Enhancement of microhomology-mediated genomic rearrangements by transient loss of mouse Bloom syndrome helicase. Genome Res. 2013;23(9):1462–1473. doi: 10.1101/gr.152744.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarbajna S, West SC. Holliday junction processing enzymes as guardians of genome stability. Trends Biochem Sci. 2014;39(9):409–419. doi: 10.1016/j.tibs.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Chen XB, Melchionna R, Denis CM, Gaillard PH, Blasina A, Van de Weyer I, Boddy MN, Russell P, Vialard J, McGowan CH. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol Cell. 2001;8(5):1117–1127. doi: 10.1016/S1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- 27.Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, 3rd, Russell P, Fuchs RP, McGowan CH, Gaillard PH. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138(1):78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138(1):63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456(7220):357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- 30.Niedernhofer LJ, Essers J, Weeda G, Beverloo B, de Wit J, Muijtjens M, Odijk H, Hoeijmakers JH, Kanaar R. The structure-specific endonuclease Ercc1–Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J. 2001;20(22):6540–6549. doi: 10.1093/emboj/20.22.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abraham J, Lemmers B, Hande MP, Moynahan ME, Chahwan C, Ciccia A, Essers J, Hanada K, Chahwan R, Khaw AK, McPherson P, Shehabeldin A, Laister R, Arrowsmith C, Kanaar R, West SC, Jasin M, Hakem R. Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J. 2003;22(22):6137–6147. doi: 10.1093/emboj/cdg580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McPherson JP, Lemmers B, Chahwan R, Pamidi A, Migon E, Matysiak-Zablocki E, Moynahan ME, Essers J, Hanada K, Poonepalli A, Sanchez-Sweatman O, Khokha R, Kanaar R, Jasin M, Hande MP, Hakem R. Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science. 2004;304(5678):1822–1826. doi: 10.1126/science.1094557. [DOI] [PubMed] [Google Scholar]
- 33.Rijkers T, Van Den Ouweland J, Morolli B, Rolink AG, Baarends WM, Van Sloun PP, Lohman PH, Pastink A. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol Cell Biol. 1998;18(11):6423–6429. doi: 10.1128/MCB.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakahara M, Sonoda E, Nojima K, Sale JE, Takenaka K, Kikuchi K, Taniguchi Y, Nakamura K, Sumitomo Y, Bree RT, Lowndes NF, Takeda S. Genetic evidence for single-strand lesions initiating Nbs1-dependent homologous recombination in diversification of Ig v in chicken B lymphocytes. PLoS Genet. 2009;5(1):e1000356. doi: 10.1371/journal.pgen.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langston LD, Symington LS. Opposing roles for DNA structure-specific proteins Rad1, Msh2, Msh3, and Sgs1 in yeast gene targeting. EMBO J. 2005;24(12):2214–2223. doi: 10.1038/sj.emboj.7600698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menke DB. Engineering subtle targeted mutations into the mouse genome. Genesis. 2013;51(9):605–618. doi: 10.1002/dvg.22422. [DOI] [PubMed] [Google Scholar]
- 37.Lin SC, Chang YY, Chan CC. Strategies for gene disruption in Drosophila . Cell Biosci. 2014;4(1):63. doi: 10.1186/2045-3701-4-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzales AP, Yeh JR. Cas9-based genome editing in zebrafish. Methods Enzymol. 2014;546:377–413. doi: 10.1016/B978-0-12-801185-0.00018-0. [DOI] [PubMed] [Google Scholar]
- 39.Van Nieuwenhuysen T, Naert T, Tran HT, Van Imschoot G, Geurs S, Sanders E, Creytens D, Van Roy F, Vleminckx K. TALEN-mediated apc mutation in Xenopus tropicalis phenocopies familial adenomatous polyposis. Oncoscience. 2015;2(5):555–566. doi: 10.18632/oncoscience.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakayama T, Blitz IL, Fish MB, Odeleye AO, Manohar S, Cho KW, Grainger RM. Cas9-based genome editing in Xenopus tropicalis . Methods Enzymol. 2014;546:355–375. doi: 10.1016/B978-0-12-801185-0.00017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mashimo T. Gene targeting technologies in rats: zinc finger nucleases, transcription activator-like effector nucleases, and clustered regularly interspaced short palindromic repeats. Dev Growth Differ. 2014;56(1):46–52. doi: 10.1111/dgd.12110. [DOI] [PubMed] [Google Scholar]
- 42.Honda A, Hirose M, Sankai T, Yasmin L, Yuzawa K, Honsho K, Izu H, Iguchi A, Ikawa M, Ogura A. Single-step generation of rabbits carrying a targeted allele of the tyrosinase gene using CRISPR/cas9. Exp Anim. 2015;64(1):31–37. doi: 10.1538/expanim.14-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu H, Wang Y, Zhang Y, Yang M, Lv J, Liu J, Zhang Y. TALE nickase-mediated SP110 knockin endows cattle with increased resistance to tuberculosis. Proc Natl Acad Sci USA. 2015;112(13):E1530–E1539. doi: 10.1073/pnas.1421587112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 45.Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33(18):5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu F, Voytas DF. Zinc Finger Database (ZiFDB) v2.0: a comprehensive database of C(2)H(2) zinc fingers and engineered zinc finger arrays. Nucleic Acids Res. 2013;41(Database issue):D452–D455. doi: 10.1093/nar/gks1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta A, Meng X, Zhu LJ, Lawson ND, Wolfe SA. Zinc finger protein-dependent and -independent contributions to the in vivo off-target activity of zinc finger nucleases. Nucleic Acids Res. 2011;39(1):381–392. doi: 10.1093/nar/gkq787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- 50.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Streubel J, Blucher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nat Biotechnol. 2012;30(7):593–595. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- 52.Li K, Wang G, Andersen T, Zhou P, Pu WT. Optimization of genome engineering approaches with the CRISPR/cas9 system. PLoS One. 2014;9(8):e105779. doi: 10.1371/journal.pone.0105779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seligman LM, Chisholm KM, Chevalier BS, Chadsey MS, Edwards ST, Savage JH, Veillet AL. Mutations altering the cleavage specificity of a homing endonuclease. Nucleic Acids Res. 2002;30(17):3870–3879. doi: 10.1093/nar/gkf495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Epinat JC, Arnould S, Chames P, Rochaix P, Desfontaines D, Puzin C, Patin A, Zanghellini A, Paques F, Lacroix E. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 2003;31(11):2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosen LE, Morrison HA, Masri S, Brown MJ, Springstubb B, Sussman D, Stoddard BL, Seligman LM. Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucleic Acids Res. 2006;34(17):4791–4800. doi: 10.1093/nar/gkl645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arnould S, Perez C, Cabaniols JP, Smith J, Gouble A, Grizot S, Epinat JC, Duclert A, Duchateau P, Paques F. Engineered I-CreI derivatives cleaving sequences from the human XPC gene can induce highly efficient gene correction in mammalian cells. J Mol Biol. 2007;371(1):49–65. doi: 10.1016/j.jmb.2007.04.079. [DOI] [PubMed] [Google Scholar]
- 57.Grizot S, Smith J, Daboussi F, Prieto J, Redondo P, Merino N, Villate M, Thomas S, Lemaire L, Montoya G, Blanco FJ, Paques F, Duchateau P. Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Res. 2009;37(16):5405–5419. doi: 10.1093/nar/gkp548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saleh-Gohari N, Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32(12):3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ozeri-Galai E, Tur-Sinai M, Bester AC, Kerem B. Interplay between genetic and epigenetic factors governs common fragile site instability in cancer. Cell Mol Life Sci. 2014;71(23):4495–4506. doi: 10.1007/s00018-014-1719-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gottipati P, Helleday T. Transcription-associated recombination in eukaryotes: link between transcription, replication and recombination. Mutagenesis. 2009;24(3):203–210. doi: 10.1093/mutage/gen072. [DOI] [PubMed] [Google Scholar]