Abstract
Stress is among the primary causes of mental health disorders, which are the most common reason for disability worldwide. The ubiquity of these disorders, and the costs associated with them, lends a sense of urgency to the efforts to improve prediction and prevention. Down-stream metabolic changes are highly feasible and accessible indicators of pathophysiological processes underlying mental health disorders. Here, we show that remote and cumulative ancestral stress programs central metabolic pathways linked to mental health disorders. The studies used a rat model consisting of a multigenerational stress lineage (the great-great-grandmother and each subsequent generation experienced stress during pregnancy) and a transgenerational stress lineage (only the great-great-grandmother was stressed during pregnancy). Urine samples were collected from adult male F4 offspring and analyzed using 1H NMR spectroscopy. The results of variable importance analysis based on random variable combination were used for unsupervised multivariate principal component analysis and hierarchical clustering analysis, as well as metabolite set enrichment analysis (MSEA) and pathway analysis. We identified distinct metabolic profiles associated with the multigenerational and transgenerational stress phenotype, with consistent upregulation of hippurate and downregulation of tyrosine, threonine, and histamine. MSEA and pathway analysis showed that these metabolites are involved in catecholamine biosynthesis, immune responses, and microbial host interactions. The identification of metabolic signatures linked to ancestral programming assists in the discovery of gene targets for future studies of epigenetic regulation in pathogenic processes. Ultimately, this research can lead to biomarker discovery for better prediction and prevention of mental health disorders.
Keywords: Multigenerational stress, Transgenerational stress, Disease etiology, Metabolic profile, 1H nuclear magnetic resonance spectroscopy, Pathway analysis, Metabolite set enrichment analysis
Introduction
Mental health disorders are the leading cause of disability worldwide. A recent survey of representative communities from 28 countries conducted by the World Health Organization (WHO) World Mental Health initiative found the prevalence of mental health disorders to be up to 36 % in the population [1]. Among these, anxiety was the most common condition, followed by mood disorders, externalizing disorders, such as attention deficit hyperactive disorder and oppositional defiant disorder, and substance abuse disorders [1]. Although these disorders differ in their symptoms, all of them are commonly influenced by the experience of stress or exposure to an adverse environment [2, 3]. Treatments for these disorders are often ineffective, and the lack of understanding concerning their etiology is hindering the ability to develop more effective cures.
Lifetime stress experienced by the individual has long been considered as a possible cause for mental health disorders. In addition, recent discoveries have emphasized that stress experienced by the mother during pregnancy can impact the developing fetal brain and increase the probability of the offspring developing mental illness [4–6]. Support for this argument comes from human cohort studies of individuals whose mothers were pregnant during a traumatic event or natural disaster [7, 8]. Results from these studies showed that maternal exposure to adversity during pregnancy was associated with accelerated cognitive decline in later life [9], higher lifetime incidences of schizophrenia [10], poor cognitive health [11, 12], altered immune function [13], and unique DNA-methylation signatures [14]. Other studies on prenatal stress during human development support a link between the mother’s mood during pregnancy and increased risk of attention deficit hyperactive disorder, anxiety, and cognitive impairment [15]. Experimental studies have isolated the influence of prenatal stress (PS) on higher lifetime incidence of altered stress response, anxiety, reduced attention, learning deficits [15], and that these changes may propagate to subsequent generations [16–18].
Recent studies have documented that an early adverse environment affects more than one generation of offspring [17–19]. For example, human studies involving the Dutch Famine Birth Cohort have suggested that prenatal undernutrition can significantly affect the health of children (F1) and grandchildren (F2) [20]. Experimental data have shown that prenatal exposure to toxins [19, 21, 22] and stress [17] alter behavior and stress response across several generations (F1–F3). Long-term outcomes include altered affective state and physiology [6, 16–18, 23] and the development of new behavioral traits based on multigenerational stress programming in the F4 generation [24–26]. Potential mechanisms of transgenerational programming involve variations in maternal care [27, 28], epigenetic regulation by DNA methylation [5, 14], and microRNAs [17, 18].
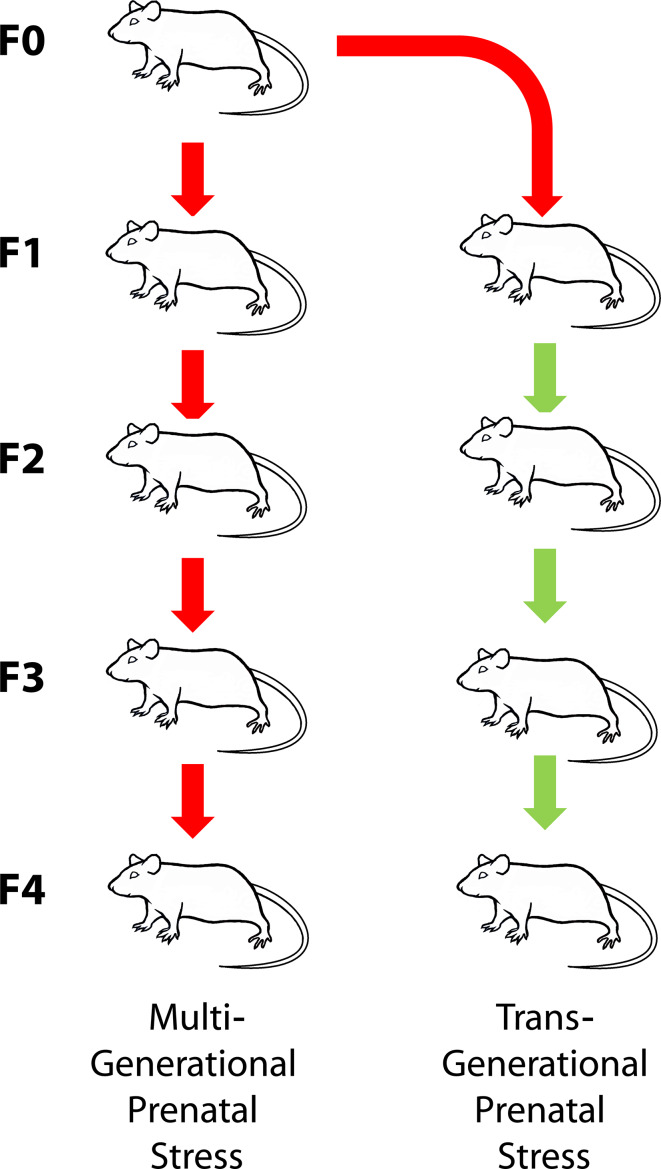
Metabolic profiles reflect cellular functioning; therefore, upstream epigenetically regulated gene and protein expression will be detectable in metabolic profiles [29–33]. Clearly identifiable metabolic signatures linked to mental health disorders have been identified using 1H nuclear magnetic resonance (NMR) spectroscopy [34, 35]. Connecting altered epigenetic states, due to PS, to the functional pathways reveals that its neurodevelopmental consequences are linked to altered brain [36–38] and amniotic fluid [39] metabolic signatures. Here, we used 1H NMR spectroscopy to determine if epigenetic programming caused by a single exposure to experimental PS four generations removed permanently alters metabolic activity. In addition, we identified clearly distinguishable metabolic fingerprints in urine to discriminate the consequences of multigenerational versus transgenerational prenatal stress in the filial F4 generation (Fig. 1). These metabolic profiles were used to determine metabolic pathways that are implicated in mental health disorders. These findings are an important demonstration that ancestral origins of altered mental health can be indicated by metabolic signatures that are of clinical predictive and diagnostic value.
Fig. 1.
Illustration of the experimental design that tested the F4 offspring of a lineage in which stress occurred in each generation (multigenerational stress, SSSS) or in which stress was limited to the first parental generation (transgenerational stress, SNNN). A lineage of non-stressed rats (NNNN) served as control
Materials and methods
Animals
This study involved Long–Evans rats raised at the Canadian Centre for Behavioural Neuroscience, University of Lethbridge vivarium. The animals were housed in pairs under a 12:12-h light/dark cycle with light starting at 07:30 h and the room temperature set at 22 °C. Rat chow food and water were available at libitum. All procedures were approved by the University of Lethbridge Animal Care Committee in compliance with the guidelines of the Canadian Council on Animal Care.
Experimental design
To investigate unambiguous transgenerational programming of an adult metabolic phenotype [40], this study focuses on 6-month-old males from the F4 generation. The use of males minimized the potential impact of female hormonal fluctuations [41]. Four successive generations of timed-pregnant female rats were bred under consistent laboratory conditions. Parental female rats (F0) were exposed to stress during pregnancy. For the multigenerational stress lineage (F4-SSSS; n = 6), the pregnant daughters (F1), granddaughters (F2), and great granddaughters (F3) were also stressed during pregnancy. The transgenerational stress lineage (F4-SNNN; n = 6) was bred by stressing only the F0 mothers and not the F1–F3 mothers (see Fig. 1; S = stress, N = non-stress condition). Yolked controls were bred in parallel for each generation (control, n = 7).
Stress procedure
Pregnant dams were subjected to stress daily from gestational day (GD) 12 to GD 18. This timeframe was used as it corresponds to the third trimester in human pregnancy when substantial neural development occurs [42]. Stressors included restraint in a Plexiglas cylinder for 20 min and forced swimming in warm water at 21 °C for 5 min. Stressors were administered each day in a semi-random alternating order, in the morning between 8:00 and 9:00 h or in the afternoon between 16:00 and 17:00 h.
Behavioral testing
Open-field locomotor activity was used to measure the exploratory behavior, which serves as a standard parameter in the assessment of stress-induced emotional state [25, 43]. Animals were placed individually into Accuscan activity monitoring Plexiglas boxes (length 42 cm, width 42 cm, and height 30 cm) and recorded for 10 min. The boxes attached to the computer recorded the activity based on sensor beam breaks. The horizontal beam breaks were recorded on the computer with the VersaMax™ program, and converted to spread sheets using VersaDat™ software (AccuScan Instruments Inc., OH, USA). Distance traveled in the center of the open field arena was averaged to obtain the total distance traveled (in cm) per minute.
Sample collection and preparation
Subjects for analysis were euthanized with an injection of Euthanosol (Merck, QC, Canada) and perfused transcardially with phosphate-buffered saline (approximately 200 ml) followed by a transcardial injection of approximately 200 ml of 4 % paraformaldahyde (Sigma-Aldrich, MO, USA). Urine samples were obtained via bladder puncture with a 1.5-ml sterile syringe at time of euthanization and stored at −80 °C. To prepare the samples for NMR spectroscopy, urine samples were thawed at room temperature and 450 μl aliquots were transferred into 1.5-ml centrifuge tubes with 250 μl of phosphate buffer. The phosphate buffer was prepared as a 4:1 ratio of KH2PO4:K2HPO4 in a 4:1 H2O:D2O solution to a final concentration of 0.5 M. The D2O came with 0.05 % by weight trimethylsilyl propanoic acid (TSP) as a chemical shift reference. To protect the metabolite profile integrity, 0.02 % w/v of sodium azide was added to the buffer solution as an antimicrobial agent. Once the sample was mixed with the buffer, it was centrifuged at 12,000 rpm for 10 min to precipitate any particulate matter. After centrifugation, 550 μl of the supernatant was transferred to a 5-mm NMR tube for NMR analysis.
NMR data acquisition and processing
NMR spectra were collected on a 700 MHz Bruker Avance III HD spectrometer (Bruker, ON, Canada). The Bruker 1-D NOESY gradient water suppression pulse sequence ‘noesygppr1d’ was used. Each sample was run for 128 scans to a total acquisition size of 128 k. The spectra were zero filled to 256 k, automatically phased, baseline corrected, and line-broadened by 0.3 Hz. The processed spectra were then exported to MATLAB (The MathWorks, MA, USA) for statistical analysis. Spectra were manually binned to reduce the size of the dataset. Each spectrum had the areas corresponding to water and urea removed before being normalized to remove effects of imperfect water signal suppression. The data set was then autoscaled.
Statistical analysis
Data visualization to determine sample structure and the presence of distinct groups within the dataset was conducted using the principal component analysis (PCA). Hierarchical clustering analysis was also performed and is presented as a dendrogram coupled to a heat map. Both PCA and the clustering analysis were performed using the online chemometrics software Metaboanalyst [44–46]. Percent differences for each metabolite in each comparison group were also calculated using MATLAB (The MathWorks, MA, USA).
Variable importance analysis based on random variable combination (VIAVC) [47] is a new feature selection method and was used for this analysis. Briefly, VIAVC provides a method for systematically resampling variables to determine if potentially synergetic effects exist between seemingly unimportant variables. The algorithm combines random permutation of variable inclusion or exclusion with a tenfold cross validation (CV) of models to determine the optimal subset of variables that provide the most information about the differences between groups. It should be noted that even though the VIAVC script only employs CV, the way the algorithm is structured produces results that are no different from the method of double cross validation (DCV) that has been recommended elsewhere for the validation of metabolomics data sets [48]. The p values reported are calculated from a t test of the distribution of scores based on whether the particular metabolite was included or excluded from the model. All of the VIAVC tests were carried out using MATLAB. A more detailed description of this complex method is available by Yun et al. [47]. Furthermore, for the behavioral analysis, the average center distance traveled in an open field was analyzed using an unpaired t test, and its relationship to metabolic outcomes was determined using Pearson R correlations in MATLAB.
The biological significance of the important metabolites was investigated using two tools offered by Metaboanalyst. Metabolite set enrichment analysis (MSEA) uses a set of predefined metabolic pathways to identify significant and meaningful changes in functionally related metabolites within a biologically relevant context [49]. Data were entered into the MSEA as a list of metabolites and the over-representation analysis (ORA) algorithm was used. Pathway analysis was also conducted, which combines the results of pathway enrichment analysis with pathway topology to aid in the identification of the most relevant pathways involved in the conditions of the study [50]. The data were input as a list of metabolites, the Rat pathway library was chosen, and the ORA algorithm was selected using hypergeometric test. These two analysis tools were used in the identification of pathways to discuss in connection with significant metabolites.
Metabolite identification
An in-house spectral database of pure metabolite substances was used to identify the majority of the metabolites in the spectra. Any metabolites not in our database were referenced using the online Human Metabolome Database [51–53].
Results and discussion
Metabolite identification and selection
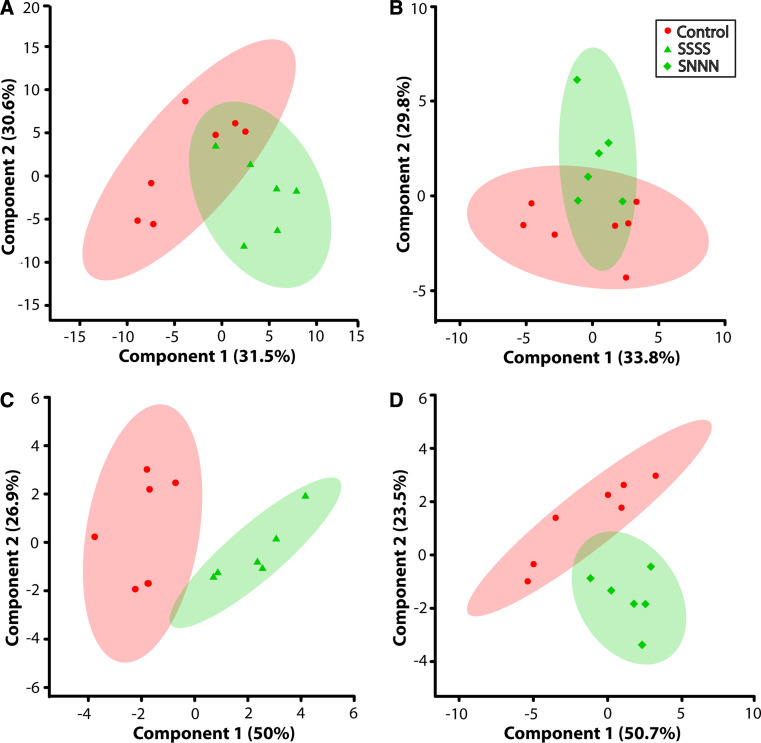
The purpose of this research was to identify urine metabolic signatures generated by cumulative or transgenerational ancestral stress that associate with or predict mental health outcomes. Urine represents the combined collection and concentration of metabolic wastes from an organism, and thus, it reflects individual phenotype, which is frequently affected by adverse experiences, such as stress [54]. The initial results of principal component analysis (PCA), when all metabolites were included, are shown in Fig. 2a, b and revealed no separation of groups. The present approach used variable importance analysis based on random variable combination (VIAVC) [47] as the feature selection method, because it incorporates tenfold cross validation (CV) and random permutation methods to address the tendency of partial least squares discriminant analysis (PLS-DA) to overfit data [48, 55]. Furthermore, VIAVC method also allows the extraction of informative metabolites, exploits synergetic effects between metabolites, and excludes metabolites that vary randomly between samples. The p value of each metabolite identified as significant by VIAVC, along with the percent differences for each, are displayed in Table 1. Once the important metabolites were identified, they were used to rerun PCA (Fig. 2c, d) and hierarchical clustering (Fig. 3a, b) analysis. Both of these unsupervised grouping methods correctly separated the experimental groups from the controls when considering only the variables identified by VIAVC. Not all of the metabolites identified by the VIAVC method for the two experimental groups were identical; however, there were a number of metabolites that were similar and these will be discussed later.
Fig. 2.
Scores plots showing components 1 (X-axis) and 2 (Y-axis) of the PCA analysis for a, c F4-SSSS multigenerationally stressed lineage versus controls and b, d F4-SNNN transgenerationally stressed lineage versus controls. a, b The result of the analysis when all metabolites were considered, while c, d the result of the analysis when only the metabolites identified by VIAVC were considered. The percentages shown along each axis indicate the amount of the variance in the data set given by each component, and the shaded ellipses designate the 95 % confidence interval for each group
Table 1.
Display of the percent differences and p values of metabolites that were determined to be significant for at least one of the stress lineages based on the VIAVC analysis (control, n = 7; F4-SSSS, n = 6; F4-SNNN, n = 6)
| SSSS versus control | SNNN versus control | |||
|---|---|---|---|---|
| p value | Percent difference | p value | Percent difference | |
| Formate | 3.22 × 10−18 | −25.06 | 3.08 × 10−12 | −31.19 |
| IMP | −6.98 | 1.61 × 10−9 | −41.78 | |
| 1-MN | 6.64 × 10−13 | −11.66 | 6.64 × 10−13 | −29.02 |
| Histamine | 3.30 × 10−24 | −16.82 | 5.03 × 10−16 | −39.14 |
| Hippurate | 1.84 × 10−11 | 26.9 | 3.56 × 10−6 | 27.64 |
| Tyrosine | 8.37 × 10−17 | −27.89 | 6.35 × 10−7 | −20.43 |
| Fumarate | −8.48 | 0.36 | −3.01 | |
| Singlet 6.40 ppm | −13.96 | 2.84 × 10−6 | 4.75 | |
| Thymidine | −3.69 | 5.07 × 10−3 | −1.0 | |
| Singlet 6.20 ppm | 9.78 | 0.03 | 2.95 | |
| Allantoin | 4.94 × 10−8 | 32.58 | 1.54 × 10−4 | 17.2 |
| Galactose | −11.12 | 0.24 | 3.95 | |
| Glucose | 5.33 × 10−7 | 15.31 | 2.69 × 10−5 | 15.91 |
| Hydroxyacetone | −18.14 | 1.16 × 10−3 | 1.4 | |
| Guanidoacetate | 2.56 × 10−9 | 8.94 | 12.99 | |
| Singlet 3.76 ppm | 2.51 × 10−10 | 2.23 | 0.24 | 2.84 |
| Glycine | 2.24 × 10−22 | 8.83 | 17.58 | |
| Singlet 3.32 ppm | 2.5 × 10−4 | 19.64 | 45.87 | |
| Dimethylamine | 1.75 × 10−10 | 62.07 | 15.17 | |
| N6-Acetylysine | −11.01 | 0.017 | −3.30 | |
| Threonine | 4.31 × 10−10 | −24.75 | −10.4 | |
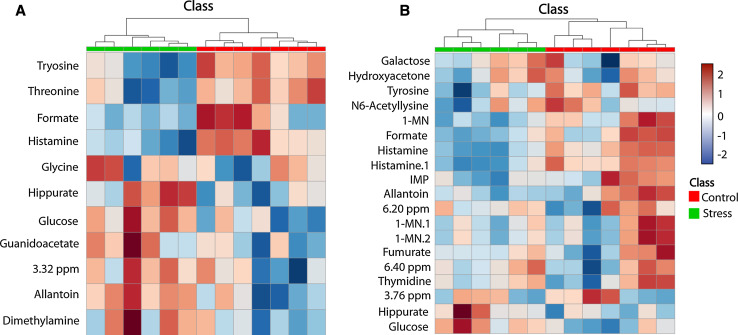
Fig. 3.
Heat maps for a F4-SSSS multigenerationally stressed lineage versus controls and b F4-SNNN transgenerationally stressed lineage versus controls. The X- and Y-axis show the class and the metabolite identity, respectively. These heat maps visually indicate either upregulation or downregulation of the metabolites presented in Table 1. The legend corresponding to the class label and the heat map for both figures is shown to the right of figure b. 1-MN 1-methylnicotinamide, IMP inosine monophospate. The labels 1-MN, 1-MN.1, and 1-MN.2, as well as histamine and histamine.1 correspond to different resonance peaks of the same metabolite. The dendrogram at the top of each heat map illustrates the results of the unsupervised hierarchical clustering analysis
The present results demonstrate that repeated prenatal stress in each of four generations (multigenerational stress, F4-SSSS group) and a single exposure to prenatal stress four generations removed (transgenerational stress, F4-SNNN group) produce urinary metabolic profiles that are unique when compared with a non-stress control lineage. This is evident by the clear separation of each of the stress lineages from the control group in the PCA scores plots that include only the metabolites identified by the VIAVC method (Fig. 2c, d). Thus, the impact of stress in each stress lineage has caused a distinct alteration in metabolic activity. The multigenerational lineage demonstrates an impact of both direct and indirect stress exposure [23, 40], while the transgenerational stress lineage unambiguously isolates the impact of inherited transgenerational programming, which transmits to the F4 generation [6, 40]. The unique metabolic footprint of ancestral stress included 21 metabolites that were significantly altered (Table 1). The heat map for each of the altered metabolites among the two stress lineages (Fig. 3a, b) provides a graphical indication of whether the metabolites were upregulated or downregulated with respect to the control group. Furthermore, the percent differences given in Table 1 show that, when considering all metabolites identified from both stress lineages, a history of ancestral stress consistently reduces nine metabolites and elevates nine metabolites (Table 1). The dendrograms shown above the heat maps (Fig. 3a, b) illustrate the results of hierarchical clustering analysis; this unsupervised method was able to correctly separate each stress lineage from the controls.
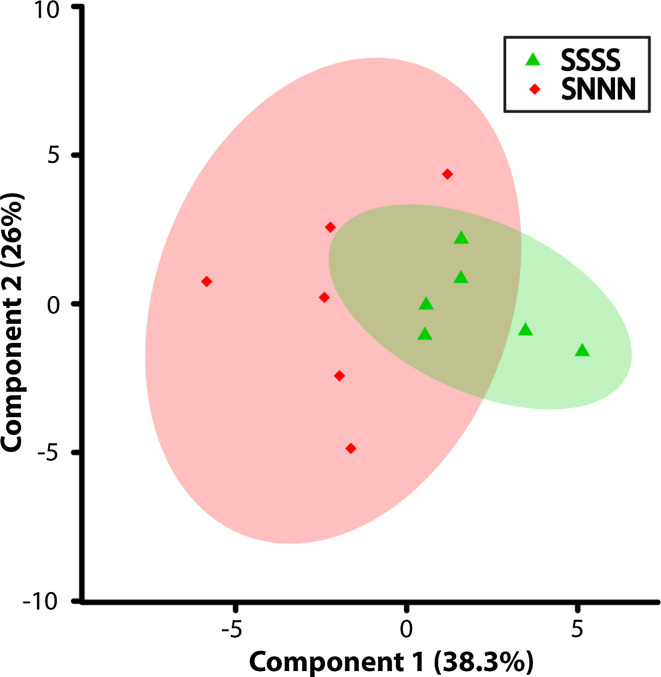
The metabolites identified as varying significantly in the SSSS and SNNN lineages (Table 1) concur with our previous findings that variations induced by multigenerational versus transgenerational stress [17, 56] tend to be reflected in the intensity of the phenotype expression, and not the directionality. Accordingly, the directionality of the percent differences in 18 of 21 identified metabolites, either upregulation or downregulation, was consistent between groups. Furthermore, when the two experimental groups (multigenerational and transgenerational) were plotted using a PCA scores plot (Fig. 4), they highly overlap, suggesting similarity of metabolic profiles induced by multigenerational and transgenerational stress programming. Thus, irrespective of the frequency of prenatal stress exposure among the ancestral lineage, common metabolic pathways were reprogrammed that reflect the altered phenotype [17, 56]. This observation aligns with the finding of a coordinated pattern of epigenetic changes, including microRNAs that propagate from one generation to the next [17, 18]. Accordingly, metabolic activity is dictated by genetic expression and epigenetic regulation of gene expression will be reflected by distinct levels of metabolic end products.
Fig. 4.
Scores plot illustrating components 1 (X-axis) and 2 (Y-axis) of the PCA analysis for the F4-SSSS multigenerationally stressed lineage versus the F4-SNNN transgenerationally stressed lineage when considering the metabolites identified by the VIAVC analysis. The percentages shown along each axis indicate the amount of the variance in the data set given by each component and the shaded ellipses designate the 95 % confidence interval for each group
Metabolic pathway analysis
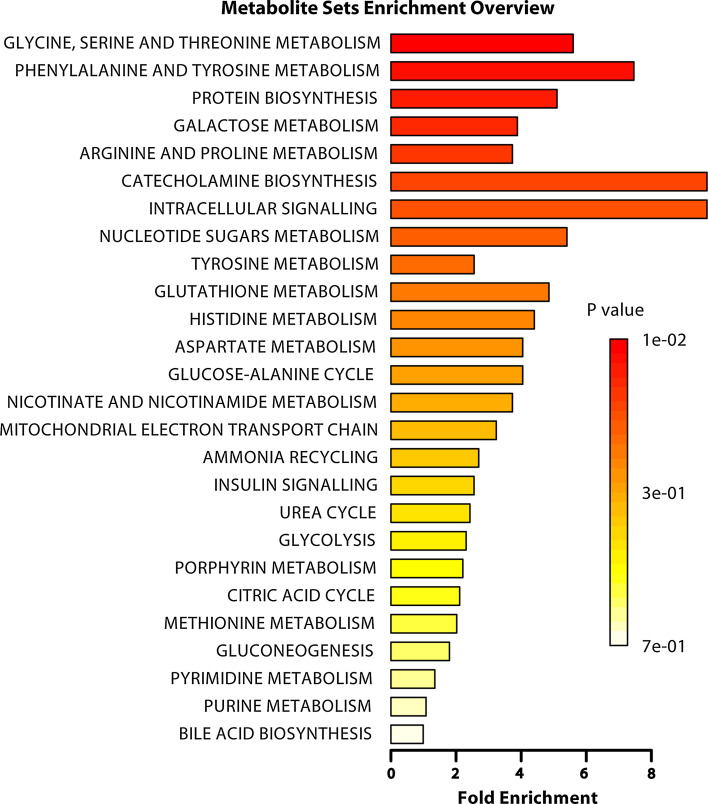
The metabolites that were significantly altered by ancestral stress (Table 1) are diverse and involved in various pathways. Although one cannot conclusively rule out which of the potential metabolic pathways are relevant to phenotype, pathway analysis (PA) and metabolite set enrichment analysis (MSEA) identified a number of key metabolic pathways that these metabolites are involved in (Table 2; Fig. 5, respectively). The following pathways were selected from the outputs of PA and MSEA based on their biological relevance to mental health: catecholamine biosynthesis and tyrosine metabolism; intracellular signaling through the H2 receptor and histamine; histidine metabolism; and glycine, serine, and threonine metabolism. The following list of metabolites from Table 1 is associated with the above pathways and will now be discussed individually: tyrosine, threonine, and histamine.
Table 2.
Overview of the metabolic pathways that were altered by ancestral stress compared with non-stress controls
| Pathway name | Total | Hits | p value | FDR | Impact |
|---|---|---|---|---|---|
| Aminoacyl-tRNA biosynthesis | 67 | 3 | 0.043 | 0.77 | 0.10 |
| Methane metabolism | 9 | 2 | 0.0047 | 0.23 | 0.17 |
| Glycine, serine and threonine metabolism | 32 | 3 | 0.0059 | 0.23 | 0.32 |
| Histidine metabolism | 15 | 1 | 0.168 | 0.85 | 0.15 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 4 | 1 | 0.047 | 0.77 | 1.0 |
| Tyrosine metabolism | 42 | 2 | 0.089 | 0.84 | 0.14 |
Total indicates the total number of metabolites listed in the pathways; hits indicate the number of significant metabolites identified in the pathways; the p value is based on the enrichment analysis; FDR indicates false discovery rate; impact designates the pathway impact as determined by pathway topology analysis
Fig. 5.
Summary plot for the over representation analysis of metabolic pathways associated with the complete list of metabolites shown in Table 1. The p values for the metabolic pathways are color coded with dark red being highly significant and white being least significant. The category “intracellular signaling” stands for intracellular signaling through the H2 receptor and histamine
Ancestral stress-induced upregulation of tyrosine excretion supports the link to mental health and stress programming. Amino acid l-tyrosine is the precursor of multiple monoamine neurotransmitters, including norepinephrine (NE), which is a central molecule directing the immediate physiological response to stress [57]. Obtained from the diet, animals in our study should have relatively identical levels of l-tyrosine [58]. However, both SSSS and SNNN groups had a significantly lower level of tyrosine being excreted, alluding to an elevated stress response [5, 18], which consumes larger amounts of tyrosine to maintain elevated NE production in the stressed lineages.
Stress may increase susceptibility to disease and allergies, autoimmune disease, and inflammation [59]. Threonine, an essential amino acid stimulates thymus development and is a major building block of immunoglobulins [60]. In line with the notion of elevated stress responses induced by ancestral stress, threonine levels were reduced in both groups. As an important immunostimulant, low threonine excretion may be linked to higher demand by the immune system.
Another immunomodulator that was reduced by a history of stress is histamine. Histamine is produced by decarboxylation of the essential amino acid l-histidine. It is active at sites of inflammation, stimulates gastric secretion, and also regulates immune function. Even low concentrations of histamine can be highly active in biological systems; therefore, its synthesis, transport, storage, and release are carefully regulated [61]. The stress-induced reduction in histamine in both the SSSS and SNNN groups may reflect either a lower conversion of l-histidine into histamine or a higher usage of histamine by the organism. Given that stress is often accompanied by exacerbated immune function [59], it is likely that histamine usage has been upregulated, reflected by decreased excretion.
The VIAVC method and the percent differences both identified hippurate as a significant metabolite; however, hippurate, which is a common urinary metabolite in mammals, is not related to metabolic pathways identified by MSEA and PA. There are a number of possible initial substrates for the production of hippurate, all of which are large polyphenols that enter the organism via the diet and undergo a series of reactions catalyzed by various enzymes and endogenous microbiota. The final step in the production of hippurate occurs in the mitochondrial matrix where benzoic acid, a remnant of the polyphenols, is conjugated with glycine [62]. Hippurate then enters the blood stream and is filtered out of vascular circulation by the kidneys. Its excretion is upregulated in human subjects with diabetes [62] and high levels of anxiety [63], both of which are common symptoms associated with prenatal stress [15, 64, 65]. Both experimental groups in this study displayed upregulated hippurate excretion, which suggests discrete metabolic alteration linked to metabolic disorders and/or mental health.
Behavioral phenotype
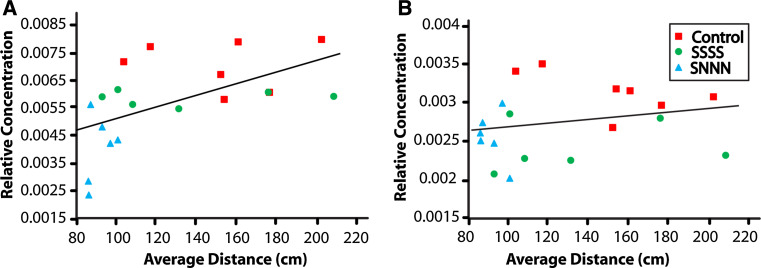
The distinct metabolic profiles of the SSSS and SNNN groups were associated with mental health outcomes. Open-field locomotor profiles indicate a phenotype of higher emotional state and increased anxiety-like behavior in the stressed lineages, as reflected by distance traveled in the center of the open-field arena. Rats that are less anxious will generally spend more time in the illuminated center of the open-field arena, whereas rats that are more anxious will avoid this open, lit space [25, 66]. In this study, an independent sample t test compared average center distance scores for male offspring from stressed and control lineages. There was a significant difference in the scores for control (M 159.4, SD 33.8) and stressed (M 119.7, SD 39.5) lineages [t(29) = −2.41, p < 0.05]. The relationship between average distance scores and the relative concentrations of histamine and tyrosine indicated positive correlations for histamine [r = 0.55, p < 0.01 (one tailed)] and tyrosine [r = 0.21, p = 0.177 (one-tailed); Fig. 6]. Thus, higher anxiety-like states were associated with lower histamine and lower tyrosine concentrations.
Fig. 6.
Pearson correlations to assess the relationship between anxiety-like behavior (i.e., average center distance traveled) and the relative concentrations of histamine (a) and tyrosine (b). There were positive correlations between the distance score and histamine (r = 0.55, p = 0.0045), and tyrosine (r = 0.21, p = 0.177), indicating that a higher anxiety-like state was linked to lower histamine and tyrosine concentrations
Conclusions
Using a 1H NMR-based metabonomics approach, perinatal programming caused by both remote and cumulative ancestral stress was manifested in altered metabolic profiles and anxious behavior in the F4 generation. The programming was evident by changes in relative concentrations of a number of key metabolites, particularly hippurate, tyrosine, threonine, and histamine. These metabolites are implicated in catecholamine biosynthesis, immune responses, and microbial host interactions in mammalian systems. The similarities between the metabolic profiles of multigenerational and transgenerational stress indicate that both cases share common metabolic pathways. When altered, these common metabolic pathways may either lead to or be reflective of pathophysiological processes resulting in impaired health. Identification of metabolic pathways affected by ancestral stress will help to determine epigenetically regulated gene targets leading to stress-associated diseases, such as mental health disorders. Ultimately, this will lead to a better understanding of the etiology of these diseases and potential biomarker discovery.
Acknowledgments
The authors acknowledge the assistance and expertise of Michael Opyr in coding some of the MATLAB scripts used for data analysis. This research was supported by the Alberta Innovates-Health Solutions (AI-HS) Interdisciplinary Team Grant #200700595 “Preterm Birth and Healthy Outcomes” (GM); the Norlien/Palix Foundation, Alberta Family Wellness Initiative and the Alberta Centre for Child, Family and Community Research (GM); Natural Sciences and Engineering Research Council of Canada (GM), the Canadian Institutes of Health Research #102652 (GM); and the University of Lethbridge Health Research Accelerator Fund (HRAF; GM and TM).
Contributor Information
Tony Montina, Phone: 1-403-394-3927, Email: tony.montina@uleth.ca.
Gerlinde A. S. Metz, Phone: 1-403-394-3992, Email: gerlinde.metz@uleth.ca
References
- 1.Kessler RC, Aguilar-Gaxiola S, Alonso J, Chatterji S, Lee S, Ormel J, Ustun TB, Wang PS. The golobal burden of metnal disorders: an update from the WHO World Metnal Health (WMH) Surveys. Epidemiol Psichiatr Soc. 2009;18(1):11. doi: 10.1017/S1121189X00001421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammen C. Stress and depression. Annu Rev Clin Psychol. 2005;1:293–319. doi: 10.1146/annurev.clinpsy.1.102803.143938. [DOI] [PubMed] [Google Scholar]
- 3.McEwen BS, Eiland L, Hunter RG, Miller MM. Stress and anxiety: structural plasticity and epigenetic regulation as a consequence of stress. Neuropharmacology. 2012;62(1):3–12. doi: 10.1016/j.neuropharm.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charil A, Laplante DP, Vaillancourt C, King S. Prenatal stress and brain development. Brain Res Rev. 2010;65(1):56–79. doi: 10.1016/j.brainresrev.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Crews D, Gillette R, Scarpino SV, Manikkam M, Savenkova MI, Skinner MK. Epigenetic transgenerational inheritance of altered stress responses. PNAS. 2012;109(23):9143–9148. doi: 10.1073/pnas.1118514109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zucchi FC, Yao Y, Ward ID, Ilnytskyy Y, Olson DM, Benzies K, Kovalchuk I, Kovalchuk O, Metz GA. Maternal stress induces epigenetic signatures of psychiatric and neurological diseases in the offspring. PLoS One. 2013;8(2):e56967. doi: 10.1371/journal.pone.0056967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lumey LH, Stein AD, Susser E. Prenatal famine and adult health. Annu Rev Publ Health. 2011;32:237–262. doi: 10.1146/annurev-publhealth-031210-101230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao X, Laplante DP, Brunet A, Ciampi A, King S. Prenatal maternal stress affects motor function in 5(1/2)-year-old children: project ice storm. Dev Psychobiol. 2014;56(1):117–125. doi: 10.1002/dev.21085. [DOI] [PubMed] [Google Scholar]
- 9.Schulz LC. The Dutch Hunger Winter and the developmental origins of health and disease. PNAS. 2010;107(39):16757–16758. doi: 10.1073/pnas.1012911107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Painter RC, Roseboom TJ, Bleker OP. Prenatal exposure to the Dutch famine and disease in later life: an overview. Reprod Toxicol. 2005;20(3):345–352. doi: 10.1016/j.reprotox.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Cao-Lei L, Elgbeili G, Massart R, Laplante DP, Szyf M, King S. Pregnant women’s cognitive appraisal of a natural disaster affects DNA methylation in their children 13 years later: project Ice Storm. Transl Psychol. 2015;5:e515. doi: 10.1038/tp.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van den Bergh HBR, Loomans ME, Mennes M. Early life influences on cognition, behavior, and emotion in humans: from birth to age 20. Adv Neurobiol. 2015;10:315–331. doi: 10.1007/978-1-4939-1372-5_15. [DOI] [PubMed] [Google Scholar]
- 13.Veru F, Dancause K, Laplante DP, King S, Luheshi G. Prenatal maternal stress predicts reductions in CD4+ lymphocytes, increases in innate-derived cytokines and Th2 shift in adolescents: Project Ice Storm. Physiol Behav. 2015 doi: 10.1016/j.physbeh.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 14.Cao-Lei L, Massart R, Suderman MJ, Machnes Z, Elgbeili G, Laplante DP, Szyf M, King S. DNA methylation signatures triggered by prenatal maternal stress exposure to a natural disaster: Project Ice Storm. PLoS One. 2014;9(9):e107653. doi: 10.1371/journal.pone.0107653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glover V, O’Connor TG, O’Donnell K. Prenatal stress and the programming of the HPA axis. Neurosci Biobehav R. 2010;35(1):17–22. doi: 10.1016/j.neubiorev.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab. 2010;21(4):214–222. doi: 10.1016/j.tem.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao Y, Robinson AM, Zucchi FC, Robbins JC, Babenko O, Kovalchuk O, Kovalchuk I, Olson DM, Metz GA. Ancestral exposure to stress epigenetically programs preterm birth risk and adverse maternal and newborn outcomes. BMC Med. 2014;12:121. doi: 10.1186/s12916-014-0121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gapp K, von Ziegler L, Tweedie-Cullen RY, Mansuy IM. Early life epigenetic programming and transmission of stress-induced traits in mammals: how and when can environmental factors influence traits and their transgenerational inheritance? Bioessays. 2014;36(5):491–502. doi: 10.1002/bies.201300116. [DOI] [PubMed] [Google Scholar]
- 19.Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of endocrine disruptors. Reprod Toxicol. 2011;31(3):337–343. doi: 10.1016/j.reprotox.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roseboom TJ, Watson ED. The next generation of disease risk: are the effects of prenatal nutrition transmitted across generations? Evidence from animal and human studies. Placenta. 2012;33(Suppl 2):e40–e44. doi: 10.1016/j.placenta.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 21.Meaney MJ. Epigenetics and the biological definition of gene × environment interactions. Child Dev. 2010;81(1):41–79. doi: 10.1111/j.1467-8624.2009.01381.x. [DOI] [PubMed] [Google Scholar]
- 22.Iqbal M, Moisiadis VG, Kostaki A, Matthews SG. Transgenerational effects of prenatal synthetic glucocorticoids on hypothalamic-pituitary-adrenal function. Endocrinology. 2012;153(7):3295–3307. doi: 10.1210/en.2012-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zucchi FC, Yao Y, Metz GA. The secret language of destiny: stress imprinting and transgenerational origins of disease. Front Genet. 2012;3:96. doi: 10.3389/fgene.2012.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ambeskovic M, Soltanpour N, Falkenburg E, Zucchi F, Kold B, Metz G (2016) Ancestral exposure to stress generates new behavioural traits and a functional hemispheric dominance shift. Cereb Cortex [DOI] [PMC free article] [PubMed]
- 25.Erickson ZT, Falkenberg EA, Metz GA. Lifespan psychomotor behaviour profiles of multigenerational prenatal stress and artificial food dye effects in rats. PLoS One. 2014;9(6):e92132. doi: 10.1371/journal.pone.0092132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skelin I, Needham MA, Molina LM, Metz GA, Gruber AJ. Multigenerational prenatal stress increases the coherence of brain signaling among cortico-striatal-limbic circuits in adult rats. Neuroscience. 2015;289:270–278. doi: 10.1016/j.neuroscience.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Francis D. Nongenomic transmission across generations of maternal behavior and stress responses in the rat. Science. 1999;286(5442):1155–1158. doi: 10.1126/science.286.5442.1155. [DOI] [PubMed] [Google Scholar]
- 28.Champagne FA, Meaney MJ. Transgenerational effects of social environment on variations in maternal care and behavioral response to novelty. Behav Neurosci. 2007;121(6):1353–1363. doi: 10.1037/0735-7044.121.6.1353. [DOI] [PubMed] [Google Scholar]
- 29.Wishart DS. Quantitative metabolomics using NMR. TrAC TrAC. 2008;27(3):228–237. [Google Scholar]
- 30.Emwas A-HM, Salek RM, Griffin JL, Merzaban J. NMR-based metabolomics in human disease diagnosis: applications, limitations, and recommendations. Metabolomics. 2013;9(5):1048–1072. doi: 10.1007/s11306-013-0524-y. [DOI] [Google Scholar]
- 31.Lindon JC, Nicholson JK, Holmes E, Everett JR. Metabonomics: metabolic processes studied by NMR spectroscopy of biofluids. Concepts Magn Res. 2000;12(5):32. doi: 10.1002/1099-0534(2000)12:5<289::AID-CMR3>3.0.CO;2-W. [DOI] [Google Scholar]
- 32.Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29(11):9. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 33.Fiehn O. Metabolomics—the link between genotypes and phenotypes. Plant Mol Biol. 2002;48(155):17. [PubMed] [Google Scholar]
- 34.Zheng P, Chen JJ, Huang T, Wang MJ, Wang Y, Dong MX, Huang YJ, Zhou LK, Xie P. A novel urinary metabolite signature for diagnosing major depressive disorder. J Proteome Res. 2013;12(12):5904–5911. doi: 10.1021/pr400939q. [DOI] [PubMed] [Google Scholar]
- 35.Holmes E, Tsang TM, Huang JT, Leweke FM, Koethe D, Gerth CW, Nolden BM, Gross S, Schreiber D, Nicholson JK, Bahn S. Metabolic profiling of CSF: evidence that early intervention may impact on disease progression and outcome in schizophrenia. PLoS Med. 2006;3(8):e327. doi: 10.1371/journal.pmed.0030327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poland RE, Cloak C, Lutchmansingh PJ, McCracken JT, Chang L, Ernst T. Brain N-acetyl aspartate concentraions measured by 1H MRS are reduced in adult male rats subjected to perinatal stress: preliminary observations and hypothetical implications for neurodevelopmental disorders. J Psychiatr Res. 1999;33:11. doi: 10.1016/S0022-3956(98)00043-0. [DOI] [PubMed] [Google Scholar]
- 37.Macri S, Ceci C, Canese R, Laviola G. Prenatal stress and peripubertal stimulation of the endocannabinoid system differentially regulate emotional responses and brain metabolism in mice. PLoS One. 2012;7(7):e41821. doi: 10.1371/journal.pone.0041821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dumas ME, Davidovic L. Metabolic profiling and phenotyping of central nervous system diseases: metabolites bring insights into brain dysfunctions. J Neuroimmune Pharmacol. 2015;10(3):402–424. doi: 10.1007/s11481-014-9578-5. [DOI] [PubMed] [Google Scholar]
- 39.Serriere S, Barantin L, Seguin F, Tranquart F, Nadal-Desbarats L. Impact of prenatal stress on 1H NMR-based metabolic profiling of rat amniotic fluid. Magma. 2011;24(5):267–275. doi: 10.1007/s10334-011-0260-0. [DOI] [PubMed] [Google Scholar]
- 40.Skinner MK. What is an epigenetic transgenerational phenotype? F3 or F2. Reprod Toxicol. 2008;25(1):2–6. doi: 10.1016/j.reprotox.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chrousos G, Torpy DJ, Gold PW. Interactions between the hypothalamic-pituitary-adrenal-axis and the female reproductive system: clinical implications. Ann Intern Med. 1998;129(3):12. doi: 10.7326/0003-4819-129-3-199808010-00012. [DOI] [PubMed] [Google Scholar]
- 42.Clancy B, Finlay BL, Darlington RB, Anand KJ. Extrapolating brain development from experimental species to humans. Neurotoxicology. 2007;28(5):931–937. doi: 10.1016/j.neuro.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Metz GA. Stress as a modulator of motor system function pathology. Rev Neurosci. 2007;18(3–4):13. doi: 10.1515/revneuro.2007.18.3-4.209. [DOI] [PubMed] [Google Scholar]
- 44.Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37(Web Server issue):W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. MetaboAnalyst 2.0—a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012;40(Web Server issue):W127–W133. doi: 10.1093/nar/gks374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yun Y-H, Liang F, Deng B-C, Lai G-B, Vicente Gonçalves CM, Lu H-M, Yan J, Huang X, Yi L-Z, Liang Y-Z. Informative metabolites identification by variable importance analysis based on random variable combination. Metabolomics. 2015;11(6):1539–1551. doi: 10.1007/s11306-015-0803-x. [DOI] [Google Scholar]
- 48.Westerhuis JA, Hoefsloot HCJ, Smit S, Vis DJ, Smilde AK, van Velzen EJJ, van Duijnhoven JPM, van Dorsten FA. Assessment of PLSDA cross validation. Metabolomics. 2008;4(1):81–89. doi: 10.1007/s11306-007-0099-6. [DOI] [Google Scholar]
- 49.Xia J, Wishart DS. MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010;38(Web Server issue):W71–W77. doi: 10.1093/nar/gkq329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia J, Wishart DS. MetPA: a web-based metabolomics tool for pathways analysis and visualization. Bioinformatics. 2010;26(18):5. doi: 10.1093/bioinformatics/btq418. [DOI] [PubMed] [Google Scholar]
- 51.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A. HMDB 3.0—the human metabolome database in 2013. Nucleic Acids Res. 2013;41(Database issue):D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37(Database issue):D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, Macinnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35(Database issue):D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bouatra S, Aziat F, Mandal R, Guo AC, Wilson MR, Knox C, Bjorndahl TC, Krishnamurthy R, Saleem F, Liu P, Dame ZT, Poelzer J, Huynh J, Yallou FS, Psychogios N, Dong E, Bogumil R, Roehring C, Wishart DS. The human urine metabolome. PloS One. 2013;8(9):e73076. doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szymanska E, Saccenti E, Smilde AK, Westerhuis JA. Double-check: validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics. 2012;8(Suppl 1):3–16. doi: 10.1007/s11306-011-0330-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ward ID, Zucchi FC, Robbins JC, Falkenberg EA, Olson DM, Benzies K, Metz GA. Transgenerational programming of maternal behaviour by prenatal stress. BMC Pregnancy Childbirth. 2013;13(Suppl 1):S9. doi: 10.1186/1471-2393-13-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Charmandari E, Tsigos C, Chrousos G. Endocrinology of the stress response. Annu Rev Physiol. 2005;67:259–284. doi: 10.1146/annurev.physiol.67.040403.120816. [DOI] [PubMed] [Google Scholar]
- 58.Fernstrom JD, Fernstrom MH. Tyrosine, phenylalanine, and catecholamine synthesis and function and in the brain. J Nutr. 2007;137:9. doi: 10.1093/jn/137.6.1539S. [DOI] [PubMed] [Google Scholar]
- 59.Dhabhar FS. Enhancing versus suppressive effects of stress on immunce function: implications for immunoprotection versus immunopathology. Allergy Asthma Clin Immunol. 2008;4(1):10. doi: 10.1186/1710-1492-4-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruth MR, Feild CJ. The immnce modifying effects of amino acids on the gut-associated lymphoid tissue. J Anim Sci Biotechnol. 2013;4(27):10. doi: 10.1186/2049-1891-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smolinska S, Jutel M, Crameri R, O’Mahony L. Histamine and gut mucosal immune regulation. Allergy. 2014;69(3):273–281. doi: 10.1111/all.12330. [DOI] [PubMed] [Google Scholar]
- 62.Lees HJ, Swann JR, Wilson ID, Nicholson JK, Holmes E. Hippurate: the natural history of a mammalian-microbial cometabolite. J Proteome Res. 2013;12(4):1527–1546. doi: 10.1021/pr300900b. [DOI] [PubMed] [Google Scholar]
- 63.Persky H, Grinker RP, Mirsky IA. The excretion of hippuric acid in subjects with free anxiety. J Clin Invest. 1950;29(1):5. doi: 10.1172/JCI102226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Novak M, Bjorck L, Giang KW, Heden-Stahl C, Wilhelmsen L, Rosengren A. Perceived stress and incidence of Type 2 diabetes: a 35-year follow-up study of middle-aged Swedish men. Diabetic Med. 2013;1:e8–e16. doi: 10.1111/dme.12037. [DOI] [PubMed] [Google Scholar]
- 65.Franklin TB, Russig H, Weiss IC, Graff J, Linder N, Michalon A, Vizi S, Mansuy IM. Epigenetic transmission of the impact of early stress across generations. Biol Psychiatry. 2010;68(5):408–415. doi: 10.1016/j.biopsych.2010.05.036. [DOI] [PubMed] [Google Scholar]
- 66.Denenberg VH. Open-field behavior in the rat: what does it mean? Ann N Y Acad Sci. 1969;159:8. doi: 10.1111/j.1749-6632.1969.tb12983.x. [DOI] [PubMed] [Google Scholar]