Abstract
Hepatitis C virus (HCV) has infected over 170 million people worldwide. Phosphatidylinositol 4-phosphate (PI4P) is the organelle-specific phosphoinositide enriched at sites of HCV replication. Whether retromer, a PI4P-related host transport machinery, unloads its cargo at HCV replication sites remains inconclusive. We sought to characterize the role of retromer in HCV replication. Here, we demonstrated the interaction between retromer subunit Vps35 and HCV NS5A protein by immunoprecipitation and GST pulldown. Vps35 colocalized with NS5A and PI4P in both OR6 replicon and JFH1 infected Huh 7.5.1 cells. HCV replication was inhibited upon silencing retromer subunits. CIMPR, a typical retromer cargo, participated in HCV replication. Our data suggest that retromer component Vps35 is recruited by NS5A to viral replication sites where PI4P unloads CIMPR. These findings demonstrate a dependence role of retromer in HCV replication and identify retromer as a potential therapeutic target against HCV.
Keywords: HCV, NS5A, PI4P, Retromer, CIMPR
Introduction
Hepatitis C virus (HCV) chronically infects more than 170 million individuals [1] and is the leading indication for orthotopic liver transplantation worldwide. As an obligate intracellular parasite, HCV depends on a large number of host factors to complete its lifecycle. Among these, phosphatidylinositol 4-phosphate (PI4P), a phospholipid enriched in a microenvironment known as HCV replication site, is critical for HCV replication [2–7]. To this end, HCV NS5A protein is responsible for recruiting and activating the phosphatidylinositol 4-kinase A (PI4KA) to increase the local concentration of PI4P at HCV replication sites [4, 8, 9]. It is still poorly understood how virally induced PI4P contributes to HCV replication. Several PI4P effectors, including OSBP1, CERT, GOLPH3 and FAPP2, have been shown to be critical in various steps of the HCV lifecycle [10–14]. CERT and GOLPH3 are required for HCV virion secretion [10, 11]. OSBP associates with HCV NS5A and plays a key role in HCV replication [12, 13]. FAPP2 transports glycosphingolipids to the HCV replication complex [14].
Recently, PI4P has been shown to facilitate the dissociation of retromer cargoes at the trans-Golgi network (TGN) [15]. Retromer is an evolutionarily conserved protein complex that mediates retrograde transport of transmembrane cargoes, for example, cation-independent mannose-6-phosphate receptor (CIMPR), from endosomes to the TGN [16]. In mammals, the retromer comprises two multisubunit complexes: a cargo recognition complex composed of Vps26, Vps29, and Vps35, which serves as a platform for recruiting cargo proteins; and a heterodimer of sorting nexins, which bind phospholipids and remodel membranes [17, 18]. Whether retromer unloads its cargo to HCV replication sites remains unknown.
Functionally relevant interactions between viral proteins and retromer have been previously described in herpesvirus saimiri (HVS), human papillomaviruses (HPV) and human immunodeficiency virus type-1 (HIV-1) [19–22]. HVS Tip drastically inhibits retromer activity through interaction with the Vps35 [19]. The inhibition of intracellular retromer activity by Tip is linked to the downregulation of CD4 surface expression and to efficient T cell transformation [19]. HPV hijacks the retromer complex for virus entry via the interaction between the L2 protein of HPV and retromer [20, 22]. Retromer also regulates the trafficking of HIV-1 envelope glycoprotein (Env) [21].
In this study, we establish the role of retromer in HCV replication. Upon HCV infection, retromer component Vps35 is recruited by NS5A to HCV replication sites, where its cargo CIMPR is unloaded by PI4P. Our data reinforce the concept that enrichment of PI4P in HCV replication sites plays a pivotal role in HCV replication by facilitating the recruitment of host factors that promote HCV.
Materials and methods
Cells, virus and reagents
Huh 7.5.1 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Thermo scientific, Waltham, MA, USA) supplemented with 10 % fetal bovine serum (FBS, Thermo scientific, Waltham, MA, USA). The infectious JFH1 plasmid pJFH1 was obtained from Dr. Takaji Wakita. Jc1FLAG (p7-nsGluc2A) was obtained from Dr. Charles Rice. The OR6 cell line, which harbors the full-length genotype 1b HCV RNA and co-expresses Renilla luciferase was obtained from Drs. Nobuyuki Kato and Masanori Ikeda. It was grown in DMEM supplemented with 10 % FBS and 500 μg/ml of G418 (Promega, Madison, WI, USA). OR6 cells were treated with IFN α (10 IU/ml) for 7 days to generate cured OR6, which did not contain HCV RNA. 4′,6-diamidino-2-phenylindole (DAPI) is from Invitrogen (Carlsbad, CA, USA).
Plasmids
Constructs encoding Vps35 fused to GST were constructed into BamHI and XhoI sites of pGEX4T-1. The PCR primers were as follows: 5′-CGGGATCCATGCCTACAACACAGCAGTCC-3′ (forward); 5′-CCGCTCGAGTTAAAGGATGAGACCTTC-3′ (reverse). Vps35 was cloned into pEGFP-C1 vector using restriction enzymes XhoI and BamHI. The PCR primers were as follows: 5′-CCGCTCGAGCTATGCCTACAACACAGCAGTCC-3′ (forward); 5′-CGGGATCCTTAAAGGATGAGACCTTC-3′ (reverse). The Vps35 siRNA-resistant mutant was generated using the Stratagene mutagenesis kit with the following pair of primers: 5′-GACAAGACATGCCTTCAAAGGATGTTGCATCTTTACAAGTCTCTC-3′ (forward) and 5′-GAGAGACTTGTAAAGATGCAACATCCTTTGAAGGCATGTC TTGTC-3′ (reverse). The Vps35D620N mutant was generated using the Stratagene mutagenesis kit with the following pair of primers: 5′-GAAGATGAAATCAGTAATTCTAAAGCACAGCTG-3′ (forward) and 5′-CAGCTGTGCTTTCGAATTACTGATTTCATCTTC-3′ (reverse). The plasmid expressing Vps26A resistant to Vps26A siRNA was synthesized with mammalian preference codon and cloned into the pEGFP-C1 vector. Human CIMPR ORF was ordered from Thermo Fisher. CIMPR was cloned in BglII/EcoRI sites of plasmid pEGFP-N1 to create GFP-CIMPR by Genewiz Inc. The CIMPR siRNA (#2) resistant mutant was generated from GFP-CIMPR through PflMI/BstEII sites with the following primers: 5′-GAAAGACCCAAGGCATGGCAACTTG-3′ (forward) and 5′-GGTTGGTGACCTGGCAGTCATCATGTTCATTGCTCTTC-3′ (reverse). Constructs encoding fragments of NS5A from JFH1 were constructed into EcoRI and BamHI sites of pCMV14-3XFLAG. The PCR primers were as follows: pCMV14-3XFLAG-NS5A(1–247) forward primer, 5′-CCCAAGCTTATGTCCGGATCCTGGCTCCGCG-3′; pCMV14-3XFLAG-NS5A(1–247) reverse primer, 5′-GCTCTAGAGCTGTGGGTGGTGCAGGTGG-3′; pCMV14-3XFLAG-NS5A(248–351) forward primer, 5′-CCCAAGCTTATGAA CACCTATGACGTGGACATG-3′; pCMV14-3XFLAG-NS5A(248–351) reverse primer, 5′-GCTCTAGATGGGGGAGGCGTCGGGGCC-3′; pCMV14-3XFLAG-NS5A(352–466) forward primer, 5′-CCCAAGCTTATGAGGAGACGCCGGACAGTGGG-3′; pCMV14-3XFLAG-NS5A(352–466) reverse primer, 5′-GCTCTAGAGCAGCACACGGTGGTAT CGTC-3′.
Antibodies
Primary mouse antibodies included: anti-actin (Sigma-Aldrich, St. Louis, MO, USA, Catalogue no. A2228, WB: 1:3000), anti-CIMPR (Abcam, Cambridge, MA, USA, Catalogue no. ab8093, IF: 1:200), anti-FLAG antibody (Sigma-Aldrich, St. Louis, MO, USA, Catalogue no. A2220, WB: 1; 3000), anti-GFP (MBL, Nagoya, Japan, Catalogue no. M048-3 WB: 1:2000), anti-HCV core (Thermo, Waltham, MA, USA, Catalogue no. MAI-080 WB: 1:1000), IgG control (MBL, Nagoya, Japan, Catalogue no. M075-3), NS5A (ViroGen, Watertown, MA, USA, Catalogue no. 256-A, WB: 1:1000, IF: 1:300, for OR6), anti-PI4P (Echelon Biosciences, Salk Lake City, UT, USA, Catalogue no. Z-P004, IF: 1:200), and anti-Vps35 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA, Catalogue no. sc-374372, WB: 1;500). Primary rabbit antibodies included: anti-CIMPR (Abcam, Cambridge, MA, USA, Catalogue no. ab124767, WB: 1:2000, IF: 1:300), anti-FLAG (Cell Signaling Technology, Inc. Boston, USA, Catalogue no. #2368, WB: 1:3000), anti-NS5A (ViroGen, Watertown, MA, USA, Catalogue no. 276-A, WB: 1:1000, IF: 1:300, for JFH1), anti-PI4KIIIa kinase (Cell Signaling Technology, Inc. Boston, USA, Catalogue no. #4902, IF: 1:50), Ranti-Vps26A (Abcam, Cambridge, MA, USA, Catalogue no. AB23892, WB: 1:1000, IF: 1:300). Primary goat antibodies included: anti-Vps35 (IMGENEX, San Diego, CA, USA, Catalogue no. IMG-3575, IF: 1:300). Secondary antibodies included HRP-conjugated ECL goat anti-rabbit IgG (Sigma-Aldrich, St. Louis, MO, USA, Catalogue no. A6154), HRP-conjugated ECL goat anti-mouse IgG (Sigma-Aldrich, St. Louis, MO, USA, Catalogue no. A4416), donkey anti-goat-Alexa Fluor 647, donkey anti-mouse-Alexa Fluor 488, donkey anti-rabbit-Alexa Fluor 555, goat anti-mouse-Alexa Fluor 594, and goat anti-rabbit-Alexa Fluor 488 (Invitrogen, Carlsbad, CA, USA).
Immunofluorescence microscopy
Cells seeded onto glass coverslips were washed with PBS and fixed with 4 % formaldehyde in PBS buffer for 5 min at room temperature. Fixed cells were incubated with blocking solution (PBS containing 10 % normal goat serum) for 5 min at room temperature. For intracellular staining, the coverslips were incubated with primary antibodies as indicated in figure legends diluted in permeabilizing buffer (0.1 % Triton X-100 in PBS containing 10 % normal goat serum) for 1 h. The coverslips were then washed three times with blocking solution, followed by incubation with Alexa Fluor 647, Fluor 488, Alexa Fluor 555, or Alexa Fluor 594 conjugated secondary antibodies for 1 h at RT. After washing three times with blocking solution, the coverslips were mounted with mounting medium containing DAPI. The cells were imaged using 40X oil immersion lens with a Leica TCS SP5 microscope (Germany) [23, 24]. To quantify the degree of colocalization, the Pearson correlation coefficient values were calculated using the WCIF ImageJ software with colocalization threshold plugin. Pearson correlation coefficient data represent the average from at least 25 cells each condition ± (standard deviation) SD. Colocalization is pseudocolored white by using an ImageJ colocalization highlighter plugin.
Protein-interaction assays
Immunoprecipitation and GST pulldown were performed as described elsewhere [24, 25].
For immunoprecipitation, cells were lyzed with lysis buffer (1 % Triton X-100, 50 mM Tris pH 7.4, 300 mM NaCl, protease inhibitor cocktail) and precleared by addition of protein A/G beads for 30 min at 4 °C. Lysates were then incubated with protein A/G beads pre-bound with 1 μg antibody for 1 h at 4 °C. Samples were washed three times with washing buffer (0.1 % Triton X-100, 50 mM Tris pH 7.4, 300 mM NaCl), eluted in SDS sample buffer and analyzed by Western blotting.
For GST pulldown assay, expression of GST control protein or the GST-Vps35 fusion protein was induced by 0.5 mM IPTG in E. coli Rosetta (DE3) at 37 °C for 5 h. Cells were then lysed by lysis buffer (50 mM Tris pH 6.8, 1 mM EDTA, 100 mM NaCl) and GST or GST-Vps35 was purified using the glutathione-sepharose beads. For pulldown assay, cell lysates were mixed with GST or GST-Vps35. After incubation at 4 °C for 1 h, glutathione beads were pelleted and washed three times with PBS buffer. Protein samples were then boiled in SDS-loading buffer for gel electrophoresis followed by Western blots.
Knockdown by siRNA
Indicated siRNAs were transfected into Huh 7.5.1 cells using Lipofectamine™ RNAiMAX Transfection Reagent (Invitrogen, Carlsbad, CA, USA). The siRNAs used for Vps35, Vps26A, Vps29, and CIMPR knockdown were from Genepharma (China) and were as follows: GCCUUCAGAGGAUGUUGUAUCUUUA (Vps35), CCACGUAUCCUGAUCUUAA (Vps26A), UCGAUGAGAAUCUGAAUUA (Vps29), CTACCTGTATGAGATCCAA (CIMPR #1), GAGCAACGAGCAUGAUGAC (CIMPR #2).
Reporter assay
Hepatitis C virus replication in OR6 cells or Jc1FLAG2 (p7-nsGluc2A)-infected Huh 7.5.1 cells was determined by monitoring Renilla or Gaussia luciferase activity (Promega, Madison, WI, USA).
Cellular ATP level was assessed for cell viability using the Cell Titer-Glo Luminescent Cell Viability Assay Kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol. Normalized luciferase activity was assessed by luciferase activity divided by ATP level.
Quantitative PCR (qPCR)
Total cellular and viral RNA was isolated using RNeasy Mini columns (QIAGEN) and reverse transcribed by random priming with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems; Foster City, CA, USA), then quantitated by qPCR using the DyNAmo HS SYBR Green qPCR Kit (Finnzyme; Espoo, Finland). Sequences of primers used in qPCR were as follows: The forward and reverse primers for HCV were 5′-TCTGCGGAACCGGTGAGTA-3′ and 5′-TCAGGCAGTACCACAAGGC-3′; the forward and reverse primers for GAPDH were 5′-ACCTTCCCCATGGTGTCTGA-3′ and 5′-GCTCCTCCTGTTCGACAGTCA-3′; the forward and reverse primers for Vps35 were 5′-GTCAAGTCATTTCCTCAGTCCAG-3′ and 5′-CCCCTCAAGGGATG TTGCAC-3′; the forward and reverse primers for Vps26A were 5′-TTCAGGAAAGGTAAACCTAGCCT-3′ and 5′-ATTGGCACCGATGTAAGATTCAT-3′; the forward and reverse primers for CIMPR were 5′-GTGACCAGCAAGGCACA AATC-3′ and 5′-CACCAAGTAGGCACCACTAAG-3′.
Statistics
Statistically significant differences were assessed using the paired Student’s t test from GraphPad Prism 5 (GraphPad Software Inc., La Jolla, CA, USA). Data represent the averages from at least three independent experiments ± (standard deviation) SD, unless stated otherwise. NS, not significant; **, P < 0.001; ***, P < 0.0001.
Results
HCV NS5A associates with Vps35
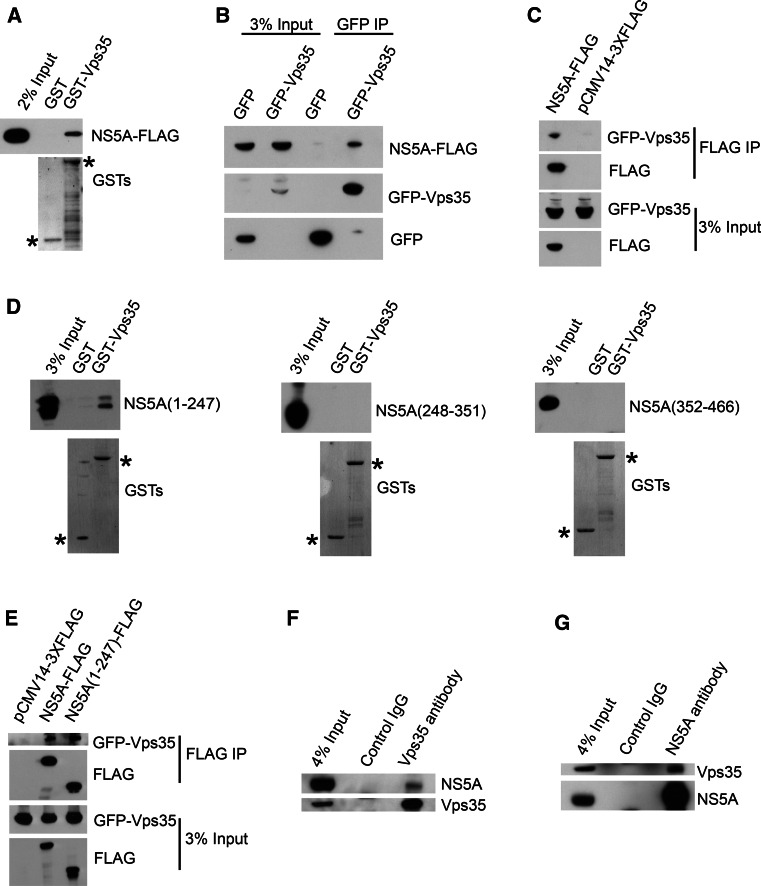
Previous yeast two-hybrid experiments suggested that NS5A interacts with Vps35 [26]. To validate the association of Vps35 with NS5A, we carried out a set of analysis including GST pulldown assay and co-immunoprecipitation. Lysates from 293T cells transfected with an expression vector encoding JFH1 NS5A-FLAG were incubated with GST-Vps35 or GST (as negative control) bound to glutathione-sepharose beads. As shown in Fig. 1a, NS5A-FLAG bound specifically to GST-Vps35, but not to the GST control. To further confirm the interaction between Vps35 and NS5A in the absence of other viral proteins, expression vectors encoding NS5A-FLAG and GFP-Vps35 or GFP were co-transfected into 293T cells, followed by immunoprecipitation with an anti-GFP antibody. As shown in Fig. 1b, Western blot analysis using an anti-FLAG antibody revealed that NS5A co-immunoprecipitated with GST-Vps35, but not with GST protein, suggesting that NS5A alone is sufficient to form a protein complex with Vps35. Next, lysates from 293T cells co-transfected with expression vectors encoding GFP-Vps35 and NS5A-FLAG or pCMV14-3XFLAG were immunoprecipitated by anti-FLAG antibody. As shown in Fig. 1c, Vps35 co-immunoprecipitated with NS5A, but not with the control pCMV14-3XFLAG. These data suggested that NS5A binds to Vps35.
Fig. 1.
Vps35 associated with the N-terminus of HCV NS5A. a GST pulldown identifies that NS5A associates with Vp35. Cell lysates from 293T cells expressing JFH1 NS5A-FLAG were incubated with purified GST-Vps35 or GST and proteins were pulled down with glutathione-sepharose beads. Bound proteins were detected by immunoblotting using anti-FLAG antibody. GST or GST-Vps35 was marked by an asterisk. b Co-immunoprecipitation confirms that NS5A binds to GFP-Vps35. 293T cells were transfected with GFP-Vps35/NS5A-FLAG or GFP/NS5A-FLAG for 24 h. Cell lysates were incubated with anti-GFP antibody-coated beads and co-IP proteins were subjected to Western blotting for analysis. c Co-immunoprecipitation confirms that Vps35 binds to NS5A-FLAG. 293T cells were transfected with GFP-Vps35/NS5A-FLAG or GFP-Vps35/pCMV14-3XFLAG for 24 h. Co-IP analysis was performed with anti-FLAG antibody followed by Western blot. d GST pulldown identifies the first domain of NS5A (aa 1–247) is sufficient to interact with Vps35. Lysates from 293T cells expressing NS5A(1–247)-FLAG, NS5A(248–351)-FLAG, or NS5A(352–466)-FLAG were pulled down by GST-Vps35 or GST. GST or GST-Vps35 was marked by an asterisk. e Co-immunoprecipitation confirms that the NS5A (1–247) binds to Vps35 as efficient as full-length NS5A. 293T cells were transfected with GFP-Vps35/NS5A-FLAG, GFP-Vps35/NS5A(1–247)-FLAG or GFP-Vps35/pCMV14-3XFLAG for 24 h. Co-IP analysis was performed with anti-FLAG antibody followed by Western blot. f, g NS5A associates with Vps35 in the context of HCV infection. OR6 cells harboring a genotype 1b HCV replicon were immunoprecipitated with anti-Vps35 antibody (f) or anti-NS5A (g) followed by Western blot
To define the region of NS5A for association with Vps35, a series of FLAG-tagged proteins, including residues 1–247 (domain I), 248–351 (domain II), or 352–466 (domain III) of NS5A from JFH1 were designed and expressed in 293T cells. Only NS5A(1–247) was pulled down by GST-Vps35 (Fig. 1d). To confirm the association between Vps35 with domain I (1–247) of NS5A, lysates from 293T cells co-transfected with expression vectors encoding GFP-Vps35 and NS5A-FLAG, NS5A(1–247)-FLAG, or pCMV14-3XFLAG were immunoprecipitated by anti-FLAG antibody. As shown in Fig. 1e, Vps35 co-immunoprecipitated with NS5A (1–247) and NS5A. These data suggested that Vps35 associates with domain I of NS5A.
To confirm the interaction between the endogenous Vps35 and NS5A in the context of HCV infection, we performed immunoprecipitation in OR6 cells using anti-NS5A or anti-Vps35. In both cases, out data revealed that Vps35 interacted with NS5A (Fig. 1f, g).
Vps35 is located in HCV replication sites
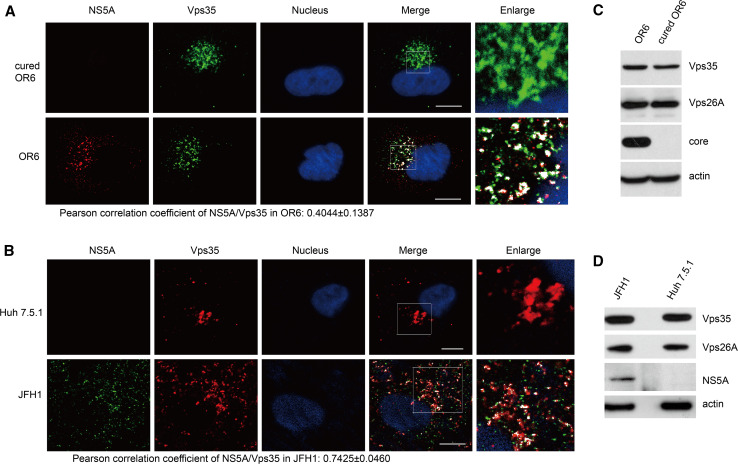
The specific interaction of HCV NS5A with the retromer Vps35 prompted us to examine whether these proteins colocalized within infected cells by immunofluorescence microscopy. Both OR6 cells and JFH1-infected Huh 7.5.1 cells were subjected to immunofluorescence staining using rabbit anti-NS5A and goat anti-Vps35 antibody. The merged images revealed that Vps35 colocalized with NS5A in the perinuclear region with a Pearson correlation coefficient of 0.4044 ± 0.1387 and 0.7425 ± 0.0460, respectively (Fig. 2a, b), indicating the colocalization between Vps35 and NS5A during HCV infection.
Fig. 2.
Vps35 colocalizes with NS5A. Cured OR6, OR6 cells (a), Huh 7.5.1 cells or JFH1-infected Huh 7.5.1 cells (b) were fixed and immunofluorescently labeled for NS5A and Vps35. DAPI marked nucleus. Colocalization was visualized by confocal microscopy. Areas of overlap are pseudocolored white in the merge and enlarge panels using an ImageJ colocalization highlighter plugin. Scale bars 10 μm. c, d Cured OR6, OR6 cells (c), Huh 7.5.1 cells or JFH1-infected Huh 7.5.1 cells (d) were subjected to Western blotting for analysis
As shown in Fig. 2c, d, HCV infection did not change the overall protein levels of Vps35 though Vps35 signal got dispersed by HCV infection.
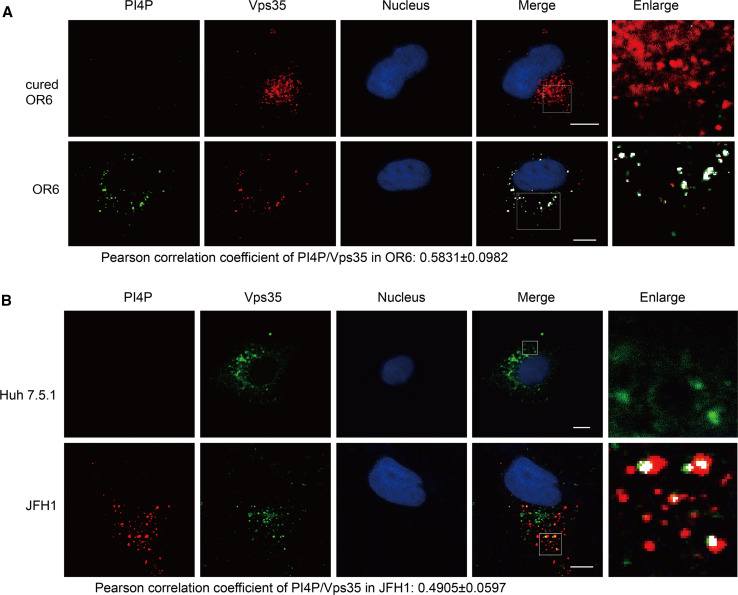
PI4P has been shown to facilitate the dissociation of retromer cargoes [15]. Based on the fact that HCV infection up-regulating the intracellular PI4P level [2, 3, 8, 11, 23], we wonder whether retromer is recruited to PI4P-enriched HCV replication sites. To test this hypothesis, OR6 cells and JFH1-infected Huh 7.5.1 cells were subjected to immunofluorescence staining using mouse anti-PIP4 and goat anti-Vsp35 antibody and then visualized by confocal microscopy. Retromer subunit Vps35 was found to colocalize with PI4P in OR6 cells, with a Pearson correlation coefficient of 0.5831 ± 0.0982, as well as in JFH1-infected Huh 7.5.1 cells, with a Pearson correlation coefficient of 0.4905 ± 0.0597 (Fig. 3a, b). There appears to be differences between Vps35/PI4P colocalization in OR6 replicon cells compared to JFH-infected cells, which might reflect the different localization pattern of Vps35 and PI4P in replicon and JFH1-infected cells. Taken together, these data suggested that retromer component Vps35 is recruited to the PI4P-enriched HCV replication sites.
Fig. 3.
Vps35 localizes to the PI4P-enriched area. Cured OR6, OR6 cells (a), Huh 7.5.1 cells or JFH1-infected Huh 7.5.1 cells (b) were fixed and immunofluorescently labeled for PI4P and Vps35. DAPI marked nucleus. Areas of overlap are pseudocolored white in the merge and enlarge panels using an ImageJ colocalization highlighter plugin. Scale bars 10 μm
Retromer participates in HCV replication
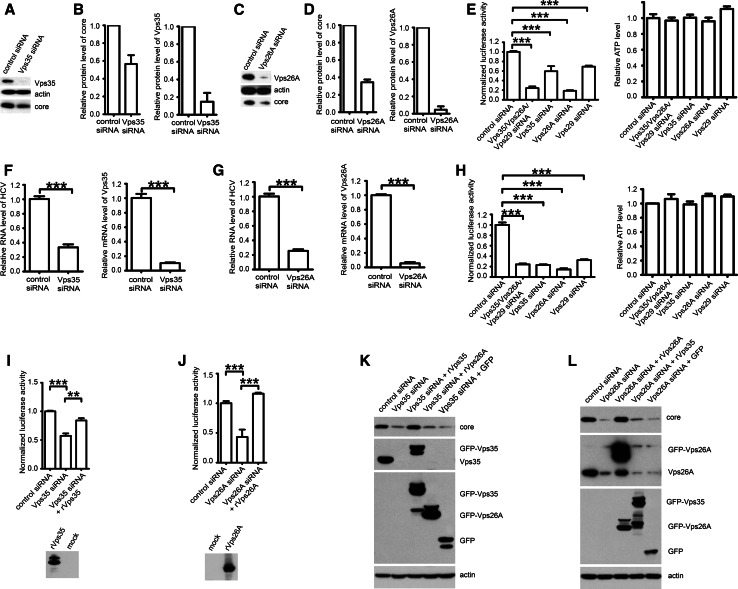
The interaction of Vps35 with HCV NS5A may be involved in the regulation of HCV life cycle. To investigate whether retromer participates in HCV replication, we used an siRNA strategy to knockdown endogenous retromer in HCV replicon OR6 cells and the level of HCV core protein level was analyzed. In OR6 cells, Vps35- or Vps26A-targeted siRNAs decreased protein levels of HCV core by 43.41 ± 9.91 % or 65.49 ± 3.04 %, respectively (Fig. 4a–d). In agreement with these results, silencing retromer subunits including Vps35, Vps26A, or Vps29 led to 40.51 ± 10.80 % (Vps35 siRNA), 81.24 ± 2.25 % (Vps26A siRNA), 30.76 ± 4.01 % (Vps29 siRNA), or 75.68 ± 8.79 % (Vps35/Vps26A/Vps29 siRNA) reduction in HCV replication measured by Renilla luciferase activity (n = 3), corresponding to the level of HCV RNA expression (Fig. 4e). We also directly measure the inhibition of viral RNA levels by real-time qPCR. Silencing Vps35 or Vps26A led to 66.50 ± 4.14 % (Vps35 siRNA), or 74.45 ± 2.20 % (Vps26A siRNA) reduction in HCV replication (Fig. 4f, g). Next, we tested the effects of Vps35, Vps26A, or Vps29 knockdown in infectious genotype 2a Jc1FLAG (p7-nsGluc2A) infection, and found that silencing retromer components caused 76.94 ± 1.33 % (Vps35 siRNA), 85.42 ± 2.12 % (Vps26A siRNA), 67.54 ± 1.54 % (Vps29 siRNA), or 75.78 ± 1.59 % (Vps35/Vps26A/Vps29 siRNA) decrease in Gaussia luciferase activity from that in cells transfected with non-targeting control siRNA (Fig. 4h).
Fig. 4.
Retromer participates in HCV replication. a OR6 cells were transfected with Vps35 siRNA or control siRNA for 72 h and analyzed by Western blot for Vps35, HCV core protein and actin as a loading control. b Band intensities of the Western blots shown in a from 3 independent experiments were quantified using ImageJ and the percentage of both Vps35 and core protein relative to actin was calculated. c OR6 cells were transfected with Vps26A siRNA or control siRNA for 72 h and analyzed by Western blot for Vps26A, HCV core protein and actin. d The band intensities from Western blots of three independent experiments of (c) were quantified using ImageJ. Relative protein levels of core and Vps26A were normalized by blot signal of actin. e OR6 cells were treated with siRNA targeting Vps35/Vps26A/Vps29, Vps35, Vps26A, Vps29 or control siRNA for 72 h, followed by measuring the Renilla luciferase activity and cellular ATP levels. Relative luciferase activity was normalized with cellular ATP levels. Data were represented the average ± SD. ***P < 0.0001. f OR6 cells were transfected with Vps35 siRNA or control siRNA for 72 h and analyzed by qPCR. Data were represented the average ± SD. ***P < 0.0001. g OR6 cells were transfected with Vps26A siRNA or control siRNA for 72 h and analyzed by qPCR. Data were represented the average ± SD. ***P < 0.0001. h Huh 7.5.1 cells electroporated with in vitro transcripted Jc1FLAG (p7-nsGluc2A) RNA for 3 days were treated with siRNA targeting Vps35/Vps26A/Vps29, Vps35, Vps26A, Vps29 or control siRNA for another 72 h, followed by measuring the Gaussia luciferase activity and cellular ATP levels were measured. Relative luciferase activity was normalized with cellular ATP levels. Data were represented the average ± SD. **P < 0.001, ***P < 0.0001. i OR6 cells were treated with siRNA targeting Vps35 or control siRNA for 24 h and then transfected with siRNA-resistant mutant of Vps35-GFP (rVps35) for another 48 h. Renilla luciferase activity and cellular ATP levels were measured. Relative luciferase activity was normalized with cellular ATP levels. Data were represented the average ± SD. **P < 0.001, ***P < 0.0001. Protein expression of transfected construct rVps35 was confirmed by Western blot. j OR6 cells were treated with siRNA targeting Vps26A or control siRNA for 24 h and then transfected with siRNA-resistant mutant of Vps26A (rVps26A) for another 48 h. Renilla luciferase activity and cellular ATP levels were measured. Relative luciferase activity was normalized with cellular ATP levels. Data were represented the average ± SD. ***P < 0.0001. Protein expression of transfected construct rVps26A was confirmed by Western blot. k OR6 cells were treated with siRNA targeting Vps35 or control siRNA for 24 h and then transfected with siRNA-resistant mutant of GFP-Vps35 (rVps35), GFP-Vps26A (rVps26A), or GFP for another 48 h. Samples were analyzed by Western blot with indicated antibodies. l OR6 cells were treated with siRNA targeting Vps26A or control siRNA for 24 h and then transfected with siRNA-resistant mutant of GFP-Vps26A (rVps26A), GFP-Vps35 (rVps35), or GFP for another 48 h. Samples were analyzed by Western blot with indicated antibodies
To exclude that siRNA transfection affected HCV replication in vivo by off-target effects, we constructed expression plasmids encoding the siRNA-resistant Vps35 or Vps26A bearing synonymous mutations that are not recognized by Vps35 or Vps26A siRNAs. Co-transfection of siRNA-resistant retromer subunit and siRNA revealed that overexpression of these siRNA-resistant Vps35 (rVps35) or Vps26A (rVps26A) constructs successfully rescued Vps35 or Vps26A RNAi-impaired HCV replication in OR6 cells (Fig. 4i–l). To rule out the possibility that mere overexpression of retromer subunits can rescue the retromer knockdown phenotype, we overexpressed Vps26A in the presence of Vps35 siRNA or overexpressed Vps35 in the presence of Vps26A. As shown in Fig. 4k, l, overexpression of Vps26A did not rescue Vps35 knockdown and overexpression of Vps35 did not rescue Vps26A knockdown, which suggested our specific rescue of the knockdown. Collectively, these data suggest that specific downregulation of retromer in vivo leads to decreased HCV replication.
CIMPR participates in HCV replication
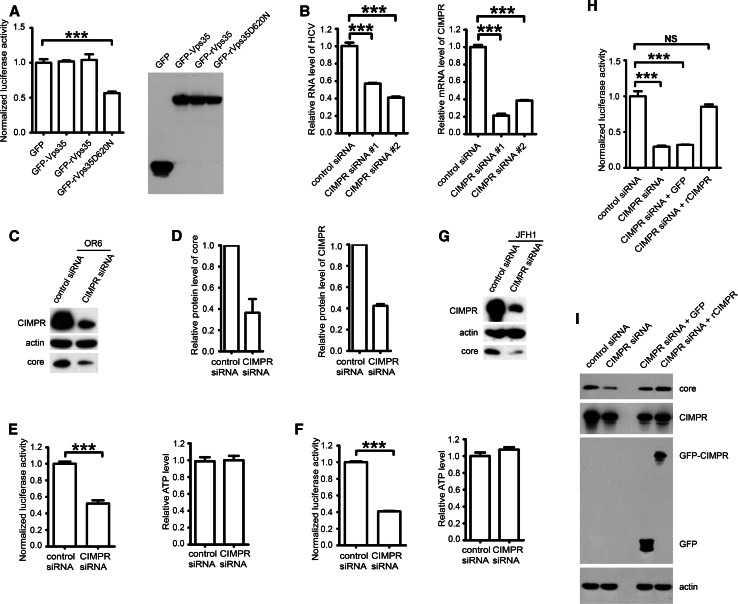
Next, we investigated the mechanism by which retromer contributes to HCV replication. Because retromer colocalized with PI4P, we hypothesized that PI4P might facilitate the dissociation of retromer cargoes. To this end, we applied a Vps35D620N mutant that abrogated the cargo sorting function of retromer within the Vps35 subunit and considered as a dominant negative mutant [27]. In contrast to wild-type Vps35, functionally deficient mutant Vps35D620N impaired HCV replication in OR6 cells by 42 % as monitored by the normalized Renilla luciferase activity (Fig. 5a). This result suggested that cargo sorting function of retromer is required for its proviral role.
Fig. 5.
CIMPR participates in HCV replication. a OR6 cells were transfected with plasmids expressing GFP, GFP-Vps35, GFP-rVps35 or GFP-rVps35D620N for 48 h, followed by measuring the Renilla luciferase activity and cellular ATP levels. Relative luciferase activity was normalized with cellular ATP levels. Data were represented the average ± SD. ***P < 0.0001. Protein expression of transfected constructs was confirmed by Western blot. b OR6 cells were transfected with CIMPR siRNAs (#1 or #2) or control siRNA for 72 h and analyzed by qPCR. Data were represented the average ± SD. ***P < 0.0001. c OR6 cells were transfected with CIMPR siRNA #2 or control siRNA for 72 h and analyzed by Western blot for CIMPR, HCV core and actin. d Band intensities from Western blots of three independent experiments in c were quantified using ImageJ. Relative protein levels of core and CIMPR were normalized by blot signal of actin. e OR6 cells were treated with siRNA targeting CIMPR #2 or control siRNA for 72 h and then Renilla luciferase activity and cellular ATP levels were measured. Relative luciferase activity was normalized with cellular ATP levels. Data were represented the average ± SD. ***P < 0.0001. f Huh 7.5.1 cells electroporated with in vitro transcripted Jc1FLAG (p7-nsGluc2A) RNA for 3 days were treated with siRNA targeting CIMPR (#2) or control siRNA for another 72 h and then Gaussia luciferase activity and cellular ATP levels were measured. Relative luciferase activity was normalized with cellular ATP levels. Data were represented the average ± SD. ***P < 0.0001. g Huh 7.5.1 cells were transfected with CIMPR siRNA (#2) or control siRNA for 72 h and infected with JFH1 (moi = 1) for another 72 h and then analyzed by Western blot for CIMPR, HCV core and actin. h OR6 cells were treated with siRNA targeting CIMPR (#2) or control siRNA for 24 h and then transfected with siRNA-resistant mutant of GFP-CIMPR (rCIMPR) or GFP for another 48 h. Renilla luciferase activity and cellular ATP levels were measured. Relative luciferase activity was normalized with cellular ATP levels. Data were represented the average ± SD. NS not significant, ***P < 0.0001. i OR6 cells were treated with siRNA targeting CIMPR (#2) or control siRNA for 24 h and then transfected with siRNA-resistant mutant of GFP-CIMPR (rCIMPR) or GFP for another 48 h. Samples were analyzed by Western blot with indicated antibodies
To gain further understanding of the proviral function of retromer, we searched the classical retromer cargoes in previous published siRNA screen data set for HCV replication [28]. This search identified CIMPR as a potential HCV host factor. Next, we investigated the role of CIMPR in HCV replication. In OR6 cells, silencing CIMPR led to 42.91 ± 0.85 % (#1 siRNA), or 49.06 ± 2.01 % (#2 siRNA) reduction in HCV replication by real-time qPCR (Fig. 5b). In the following experiments, we used #2 siRNA to knockdown CIMPR. As shown in Fig. 5c, d, CIMPR downregulation by siRNA in OR6 cells achieved a 63.59 ± 13.06 % reduction of HCV core protein. Also, CIMPR knockdown in OR6 cells modestly reduced the viral RNA synthesis by 47.98 ± 3.85 % in comparison with controls as reported by Renilla luciferase activity without changing cellular ATP levels (Fig. 5e), suggesting that CIMPR participates in the replication of HCV genome.
Next, we determined the effect of silencing CIMPR in Jc1FLAG (p7-nsGluc2A) infection. As shown in Fig. 5f, silencing CIMPR reduced the viral replication by 59.07 ± 0.99 % in comparison with siRNA control. Moreover, silencing CIMPR in JFH1-infected Huh 7.5.1 cells also reduced HCV core protein level (Fig. 5g).
To exclude that siRNA transfection affected HCV replication in vivo by off-target effects, we constructed expression plasmids encoding the siRNA-resistant CIMPR bearing synonymous mutations that are not recognized by CIMPR siRNA #2. Co-transfection of siRNA-resistant CIMPR (rCIMPR) and siRNA revealed that overexpression of rCIMPR successfully rescued CIMPR RNAi-impaired HCV replication in OR6 cells (Fig. 5h, i). Taken together, our experiments reveal an involvement of CIMPR for HCV replication.
CIMPR locates in HCV replication area
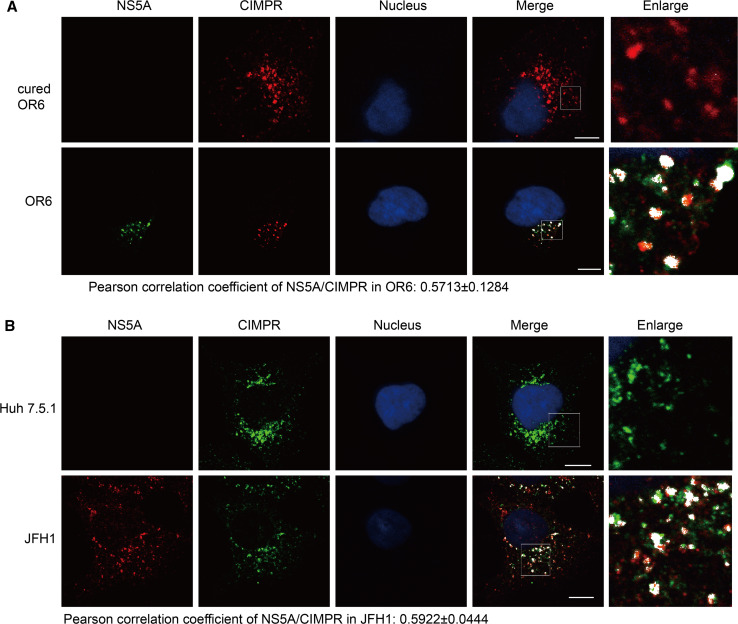
Since PI4P facilitates dissociation of retromer cargoes at TGN [15], we hypothesized that PI4P induced by HCV will unload CIMPR from retromer at the HCV replication sites. To this end, OR6 cells and JFH1 infected Huh 7.5.1 cells were immunostained with antibodies against NS5A and CIMPR after which fluorescence was detected by confocal microscopy. We observed a colocalization of CIMPR with NS5A (Pearson correlation coefficient at 0.5713 ± 0.1284 in OR6; 0.5922 ± 0.0444 in JFH1) (Fig. 6a, b), suggesting the possible unloading of CIMPR to HCV replication area.
Fig. 6.
CIMPR localizes to the HCV replication area. Cured OR6, OR6 cells (a), Huh 7.5.1 cells or JFH1-infected Huh 7.5.1 cells (b) were fixed and immunofluorescently labeled for NS5A and CIMPR. DAPI marked nucleus. Areas of overlap are pseudocolored white in the merge and enlarge panels using an ImageJ colocalization highlighter plugin. The scale bars 10 μM
Discussion
Intracellular PI4P induced by HCV infection is crucial for viral replication. One outstanding question in the field is how HCV utilizes PI4P to its own ends. To address this question, the identification of PI4P effectors involved in HCV infection would be required. In this study, we discovered that retromer could be recruited to HCV replication sites where the enriched PI4P unloads CIMPR to promote viral replication. As a PI4P effector, retromer joins a complex including OSBP, FAPP2, GOLPH3 and CERT to sense the virally induced PI4P to promote the HCV life cycle [10–14].
Several lines of evidence support our conclusion that retromer is relocated to viral replication sites. First, Vps35 selectively associated with HCV protein NS5A. Second, Vps35 colocalized with viral-induced PI4P. Third, a major retromer cargo, CIMPR, colocalized with NS5A. Collectively, the above observations demonstrate that retromer is relocated to viral replication sites and unloads its cargo CIMPR.
Our studies favor a model in which retromer plays a proviral role in HCV RNA replication. SiRNA-mediated knockdown of Vps35 or Vps26A decreased viral RNA replication in OR6 replicon cells as well as in full-length infectious clones of HCV. Furthermore, silencing CIMPR, a typical cargo of retromer, reduced viral RNA replication. In addition, Vps35 associated with domain I of NS5A, which is essential for HCV genome replication [29]. Overall, the impact of silencing on HCV replication is moderate; therefore, retromer could be defined as a host dependence factor in HCV RNA replication. In light of the proviral role of retromer in the HCV replication, we speculate retromer could be a target for novel antiviral therapy. One possible approach should be disrupting the Vps35/NS5A interaction by peptides or small molecules to block HCV replication.
This is the first demonstration of retromer’s involvement in HCV infection. Previous studies have linked retromer to HVS-induced T cell immortalization, HPV entry and HIV-1 Env trafficking [19–22]. A well-characterized cargo for retromer is CIMPR, a receptor for mannose-6-phosphate-modified lysosomal hydrolases, and other endosomal proteins [30]. The proviral role of CIMPR is also supported by previous findings that the cytoplasmic tail of CIMPR interacts with TIP47 [31, 32], which has been identified as a host factor supporting HCV RNA replication and release [33–35]. Given that retromer as transport machinery delivers a variety of cargo proteins, it is quite likely that retromer may be involved in HCV replication by delivering other cargoes which have yet to be revealed.
HCV infection is known to usurp many cellular membrane traffic processes, which are essential for delivering cargo proteins to their intracellular destinations. COPI transport-related proteins, including coatomer, GBF1, ARF1 and ARFGAP1 are required for HCV replication [6, 23, 28, 36, 37]. The YXX motif within HCV core protein mediates the binding of clathrin adaptor AP2M and is essential for HCV assembly [38]. The ESCRT complex is involved in production of infectious HCV virus [39–42]. Our finding that retromer is recruited to viral replication sites represents a novel mean by which an intracellular transport pathway can be hijacked by HCV to recruit a proviral host factor. This model is supported by the finding that Rab7, a small GTPase previously shown to catalyze membrane recruitment of the retromer [43, 44], also facilitates the formation of hepatitis C virus replication complex [45], underscoring the importance of retromer for HCV RNA replication.
In summary, we propose a model describing the role of retromer, a PI4P-related host membrane traffic pathway, in HCV replication. Upon HCV infection, retromer component Vps35 is recruited by NS5A to viral replication sites, where PI4P facilitates the unloading of retromer cargo CIMPR. Our studies uncover a novel role of retromer and its cargo CIMPR in HCV replication and provide potential targets for antiviral therapy.
Acknowledgments
This work was supported by Grants from National Natural Science Foundation of China (81301438, 81271832, 81471955), National Science and Technology Major Project of China (2013ZX10004-601), Intramural Research Program of the Institute of Pathogen Biology, CAMS (2013IPB104), Program for Changjiang Scholars and Innovative Research Team in University (IRT13007), PUMC Youth Fund, the Fundamental Research Funds for the Central Universities (3332013118), and NIH (DK098079, AI082630, and DA033541). We thank Guangbo Yang for technical support. We thank Jing Wang for critical review of the manuscript. We thank Charles Rice, Francis Chisari, Masanori Ikeda, Nobuyuki Kato and Takaji Wakita for reagents.
Abbreviations
- HCV
Hepatitis C virus
- PI4P
Phosphatidylinositol 4-phosphate
- PI4KA
Phosphatidylinositol 4-kinase A
- TGN
trans-Golgi network
- CIMPR
Cation-independent mannose-6-phosphate receptor
- HVS
Herpesvirus saimiri
- HPV
Human papillomaviruses
- HIV-1
Human immunodeficiency virus type-1
- DMEM
Dulbecco’s modified Eagle’s medium
- FBS
Fetal bovine serum
- DAPI
4′,6-diamidino-2-phenylindole
- qPCR
Quantitative PCR
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interest.
Footnotes
P. Yin and Z. Hong contributed equally to this work.
References
- 1.Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect. 2011;17:107–115. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 2.Bianco A, Reghellin V, Donnici L, Fenu S, Alvarez R, Baruffa C, Peri F, Pagani M, Abrignani S, Neddermann P, De Francesco R. Metabolism of phosphatidylinositol 4-kinase III α-dependent PI4P Is subverted by HCV and is targeted by a 4-anilino quinazoline with antiviral activity. PLoS Pathog. 2012;8:e1002576. doi: 10.1371/journal.ppat.1002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJ, Altan-Bonnet N. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, Longerich T, Diehl S, Ramirez F, Balla T, Rohr K, Kaul A, Buhler S, Pepperkok R, Lengauer T, Albrecht M, Eils R, Schirmacher P, Lohmann V, Bartenschlager R. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong Z, Yang X, Yang G, Zhang L. Hepatitis C virus NS5A competes with PI4KB for binding to ACBD3 in a genotype-dependent manner. Antiviral Res. 2014;107:50–55. doi: 10.1016/j.antiviral.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 6.Li H, Yang X, Yang G, Hong Z, Zhou L, Yin P, Xiao Y, Chen L, Chung RT, Zhang L. Hepatitis C virus NS5A hijacks ARFGAP1 to maintain a phosphatidylinositol 4-phosphate-enriched microenvironment. J Virol. 2014;88:5956–5966. doi: 10.1128/JVI.03738-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tai AW, Salloum S. The role of the phosphatidylinositol 4-kinase PI4KA in hepatitis C virus-induced host membrane rearrangement. PLoS ONE. 2011;6:e26300. doi: 10.1371/journal.pone.0026300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger KL, Kelly SM, Jordan TX, Tartell MA, Randall G. Hepatitis C virus stimulates the phosphatidylinositol 4-kinase IIIα-dependent phosphatidylinositol 4-phosphate production that is essential for its replication. J Virol. 2011;85:8870–8883. doi: 10.1128/JVI.00059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim YS, Hwang SB. Hepatitis C virus NS5A protein interacts with phosphatidylinositol 4-kinase type IIIα and regulates viral propagation. J Biol Chem. 2011;286:11290–11298. doi: 10.1074/jbc.M110.194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amako Y, Syed GH, Siddiqui A. Protein kinase D negatively regulates hepatitis C virus secretion through phosphorylation of oxysterol-binding protein and ceramide transfer protein. J Biol Chem. 2011;286:11265–11274. doi: 10.1074/jbc.M110.182097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bishe B, Syed GH, Field SJ, Siddiqui A. Role of phosphatidylinositol 4-phosphate (PI4P) and its binding protein GOLPH3 in HCV secretion. J Biol Chem. 2012;287:27637–27647. doi: 10.1074/jbc.M112.346569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Perry JW, Lauring AS, Neddermann P, De Francesco R, Tai AW. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking. Gastroenterology. 2014;146:1373–1385.e1−11. doi: 10.1053/j.gastro.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amako Y, Sarkeshik A, Hotta H, Yates J, 3rd, Siddiqui A. Role of oxysterol binding protein in hepatitis C virus infection. J Virol. 2009;83:9237–9246. doi: 10.1128/JVI.00958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan I, Katikaneni DS, Han Q, Sanchez-Felipe L, Hanada K, Ambrose RL, Mackenzie JM, Konan KV. Modulation of hepatitis C virus genome replication by glycosphingolipids and four-phosphate adaptor protein 2. J Virol. 2014;88:12276–12295. doi: 10.1128/JVI.00970-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niu Y, Zhang C, Sun Z, Hong Z, Li K, Sun D, Yang Y, Tian C, Gong W, Liu JJ. PtdIns(4)P regulates retromer-motor interaction to facilitate dynein-cargo dissociation at the trans-Golgi network. Nat Cell Biol. 2013;15:417–429. doi: 10.1038/ncb2710. [DOI] [PubMed] [Google Scholar]
- 16.Burd C, Cullen PJ. Retromer: a master conductor of endosome sorting. Cold Spring Harb Perspect Biol. 2014;6:a016774. doi: 10.1101/cshperspect.a016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonifacino JS, Hurley JH. Retromer. Curr Opin Cell Biol. 2008;20:427–436. doi: 10.1016/j.ceb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seaman MN. The retromer complex—endosomal protein recycling and beyond. J Cell Sci. 2012;125:4693–4702. doi: 10.1242/jcs.103440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kingston D, Chang H, Ensser A, Lee HR, Lee J, Lee SH, Jung JU, Cho NH. Inhibition of retromer activity by herpesvirus saimiri tip leads to CD4 downregulation and efficient T cell transformation. J Virol. 2011;85:10627–10638. doi: 10.1128/JVI.00757-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipovsky A, Popa A, Pimienta G, Wyler M, Bhan A, Kuruvilla L, Guie MA, Poffenberger AC, Nelson CD, Atwood WJ, DiMaio D. Genome-wide siRNA screen identifies the retromer as a cellular entry factor for human papillomavirus. Proc Natl Acad Sci USA. 2013;110:7452–7457. doi: 10.1073/pnas.1302164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groppelli E, Len AC, Granger LA, Jolly C. Retromer regulates HIV-1 envelope glycoprotein trafficking and incorporation into virions. PLoS Pathog. 2014;10:e1004518. doi: 10.1371/journal.ppat.1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popa A, Zhang W, Harrison MS, Goodner K, Kazakov T, Goodwin EC, Lipovsky A, Burd CG, DiMaio D. Direct binding of retromer to human papillomavirus type 16 minor capsid protein L2 mediates endosome exit during viral infection. PLoS Pathog. 2015;11:e1004699. doi: 10.1371/journal.ppat.1004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Hong Z, Lin W, Shao RX, Goto K, Hsu VW, Chung RT. ARF1 and GBF1 generate a PI4P-enriched environment supportive of hepatitis C virus replication. PLoS ONE. 2012;7:e32135. doi: 10.1371/journal.pone.0032135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Lee SY, Beznoussenko GV, Peters PJ, Yang JS, Gilbert HY, Brass AL, Elledge SJ, Isaacs SN, Moss B, Mironov A, Hsu VW. A role for the host coatomer and KDEL receptor in early vaccinia biogenesis. Proc Natl Acad Sci USA. 2009;106:163–168. doi: 10.1073/pnas.0811631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai M, Gad H, Turacchio G, Cocucci E, Yang JS, Li J, Beznoussenko GV, Nie Z, Luo R, Fu L, Collawn JF, Kirchhausen T, Luini A, Hsu VW. ARFGAP1 promotes AP-2-dependent endocytosis. Nat Cell Biol. 2011;13:559–567. doi: 10.1038/ncb2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, Chien M, Weir DB, Russo JJ, Ju J, Brownstein MJ, Sheridan R, Sander C, Zavolan M, Tuschl T, Rice CM. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci USA. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Follett J, Norwood SJ, Hamilton NA, Mohan M, Kovtun O, Tay S, Zhe Y, Wood SA, Mellick GD, Silburn PA, Collins BM, Bugarcic A, Teasdale RD. The Vps35 D620N mutation linked to Parkinson’s disease disrupts the cargo sorting function of retromer. Traffic. 2014;15:230–244. doi: 10.1111/tra.12136. [DOI] [PubMed] [Google Scholar]
- 28.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ross-Thriepland D, Harris M. Hepatitis C virus NS5A: enigmatic but still promiscuous 10 years on! J Gen Virol. 2015;96:727–738. doi: 10.1099/jgv.0.000009. [DOI] [PubMed] [Google Scholar]
- 30.Seaman MN. Identification of a novel conserved sorting motif required for retromer-mediated endosome-to-TGN retrieval. J Cell Sci. 2007;120:2378–2389. doi: 10.1242/jcs.009654. [DOI] [PubMed] [Google Scholar]
- 31.Orsel JG, Sincock PM, Krise JP, Pfeffer SR. Recognition of the 300-kDa mannose 6-phosphate receptor cytoplasmic domain by 47-kDa tail-interacting protein. Proc Natl Acad Sci USA. 2000;97:9047–9051. doi: 10.1073/pnas.160251397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diaz E, Pfeffer SR. TIP47: a cargo selection device for mannose 6-phosphate receptor trafficking. Cell. 1998;93:433–443. doi: 10.1016/S0092-8674(00)81171-X. [DOI] [PubMed] [Google Scholar]
- 33.Ploen D, Hafirassou ML, Himmelsbach K, Schille SA, Biniossek ML, Baumert TF, Schuster C, Hildt E. TIP47 is associated with the hepatitis C virus and its interaction with Rab9 is required for release of viral particles. Eur J Cell Biol. 2013;92:374–382. doi: 10.1016/j.ejcb.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Vogt DA, Camus G, Herker E, Webster BR, Tsou CL, Greene WC, Yen TS, Ott M. Lipid droplet-binding protein TIP47 regulates hepatitis C virus RNA replication through interaction with the viral NS5A protein. PLoS Pathog. 2013;9:e1003302. doi: 10.1371/journal.ppat.1003302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ploen D, Hafirassou ML, Himmelsbach K, Sauter D, Biniossek ML, Weiss TS, Baumert TF, Schuster C, Hildt E. TIP47 plays a crucial role in the life cycle of hepatitis C virus. J Hepatol. 2013;58:1081–1088. doi: 10.1016/j.jhep.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 36.Matto M, Sklan EH, David N, Melamed-Book N, Casanova JE, Glenn JS, Aroeti B. Role for ADP ribosylation factor 1 in the regulation of hepatitis C virus replication. J Virol. 2011;85:946–956. doi: 10.1128/JVI.00753-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goueslain L, Alsaleh K, Horellou P, Roingeard P, Descamps V, Duverlie G, Ciczora Y, Wychowski C, Dubuisson J, Rouille Y. Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J Virol. 2010;84:773–787. doi: 10.1128/JVI.01190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neveu G, Barouch-Bentov R, Ziv-Av A, Gerber D, Jacob Y, Einav S. Identification and targeting of an interaction between a tyrosine motif within hepatitis C virus core protein and AP2M1 essential for viral assembly. PLoS Pathog. 2012;8:e1002845. doi: 10.1371/journal.ppat.1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corless L, Crump CM, Griffin SD, Harris M. Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J Gen Virol. 2010;91:362–372. doi: 10.1099/vir.0.017285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ariumi Y, Kuroki M, Maki M, Ikeda M, Dansako H, Wakita T, Kato N. The ESCRT system is required for hepatitis C virus production. PLoS ONE. 2011;6:e14517. doi: 10.1371/journal.pone.0014517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dreux M, Garaigorta U, Boyd B, Decembre E, Chung J, Whitten-Bauer C, Wieland S, Chisari FV. Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe. 2012;12:558–570. doi: 10.1016/j.chom.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamai K, Shiina M, Tanaka N, Nakano T, Yamamoto A, Kondo Y, Kakazu E, Inoue J, Fukushima K, Sano K, Ueno Y, Shimosegawa T, Sugamura K. Regulation of hepatitis C virus secretion by the Hrs-dependent exosomal pathway. Virology. 2012;422:377–385. doi: 10.1016/j.virol.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 43.Harrison MS, Hung CS, Liu TT, Christiano R, Walther TC, Burd CG. A mechanism for retromer endosomal coat complex assembly with cargo. Proc Natl Acad Sci USA. 2014;111:267–272. doi: 10.1073/pnas.1316482111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seaman MN, Harbour ME, Tattersall D, Read E, Bright N. Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J Cell Sci. 2009;122:2371–2382. doi: 10.1242/jcs.048686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manna D, Aligo J, Xu C, Park WS, Koc H, Heo WD, Konan KV. Endocytic Rab proteins are required for hepatitis C virus replication complex formation. Virology. 2010;398:21–37. doi: 10.1016/j.virol.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]