Abstract
The engineered CRISPR/Cas9 technology has developed as the most efficient and broadly used genome editing tool. However, simultaneously targeting multiple genes (or genomic loci) in the same individual cells using CRISPR/Cas9 remain one technical challenge. In this article, we have developed a Golden Gate Assembly method for the generation of CRISPR gRNA expression arrays, thus enabling simultaneous gene targeting. Using this method, the generation of CRISPR gRNA expression array can be accomplished in 2 weeks, and contains up to 30 gRNA expression cassettes. We demonstrated in the study that simultaneously targeting 10 genomic loci or simultaneously inhibition of multiple endogenous genes could be achieved using the multiplexed gRNA expression array vector in human cells. The complete set of plasmids is available through the non-profit plasmid repository Addgene.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-016-2271-5) contains supplementary material, which is available to authorized users.
Keywords: CRISPR, Cas9, Genome editing, Simultaneous multiple gene knockout, Simultaneous multiple gene inhibition, Golden Gate Assembly
Introduction
The engineered CRISPR/Cas9 system has become the most cost-effective and broadly applicable genome editing tool [1–3]. Among all Cas9 orthologues, the type 2 CRISPR/Cas9 system from Streptococcus Pyogenes, also known as SpCas9, has been the most broadly used system [1, 4]. The SpCas9 is a universal DNA nuclease protein that can introduce double-strand DNA breaks to a given genomic locus mediated by the complementarity of a small guide RNA (gRNA) and the targeted genomic locus. Furthermore, nuclease-deficient SpCas9 (dSpCas9) can be fused to different functional proteins or effector domains and thus greatly extend the CRISPR toolbox for all sorts of precision manipulations exemplified by inhibition or activation of endogenous gene expression [5, 6], epigenetic modification [7], and gene visualization [8]. Currently, many plasmid-based CRISPR/SpCas9 expression systems have been generated and are available from the non-profit global plasmid repository Addgene. These systems offer ideal solutions for delivery of one single gRNA. However, when targeting multiple loci, the single gRNA expression system is expected to be less efficient since simultaneous delivery by co-transfections can decrease with the number of gRNA expression vectors used. Solutions to this limitation often require customized cloning procedures of multiple sgRNA expression cassettes into the same vector, which can be time-consuming and labor intensive when many sgRNA combinations are desired. To overcome this shortcoming, we opted for the generic Golden Gate Assembly method that can assemble multiple DNA fragments in a single reaction [9]. Based on Golden Gate Assembly, Cermak et al. had generated a set of array plasmids rapid assembly of TAL effectors [10].
In this study, we report the generation of another set of plasmids for rapid assembly of CRISPR gRNA expression array containing up to 30 individual expression cassettes in two to three steps. To ensure broad application of this assembly method, we have implemented the same overhang linkers that have been used for the assembly of transcription activator-like effectors (TALEs) [10]. We show that CRISPR gRNA expression array containing 10 individual expression cassettes are functional in a dual-fluorescent surrogate reporter assay, targeted deletion or inhibition of multiple genomic loci in human cells.
Results
Rapid assembly of multiple gRNA expression plasmids into a single “array” plasmid using Golden Gate Cloning
To generate a set of single gRNA expression modular plasmids that are conventional for cloning of the gRNA guide sequences and compatible for cloning into the “array” plasmids (pFUS-A, pFUS-B1 – pFUS-B10, pFUS-A30A, and pFUS-A30B) in the TALE assembly kit [10], we first synthesized ten sets of PCR primers (Supplementary Table 1) that contain a type IIS restriction enzyme (BsaI)-mediated excision cassette and the same overhang linkers as the TALE module plasmids. Then, the gRNA expression cassette containing the U6 promoter, BbsI-based cloning site and the SpCas9 scaffold and U6 promoter termination sequences was amplified by PCR and cloned into the pMA plasmid (ampicillin resistance).
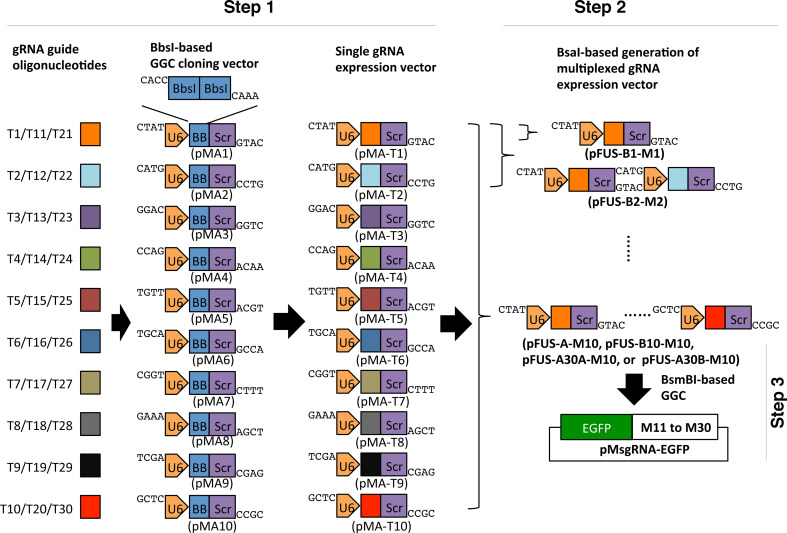
Generation of the gRNA expression array is completed in two steps for the assembly of up to 10 gRNAs. For the assembly of more than 10 gRNAs an additional third step is required. In the first step, individual gRNA guide sequences (T1 to T10) are cloned in order into 10 individual BbsI-based single gRNA expression vectors (pMA1 to pMA10, ampicillin resistance) (Fig. 1, step 1). If assembling more than 10 gRNAs, T11 to T20 or T21 to T30 will be cloned in order into pMA1 to pMA10 accordingly. Each single gRNA expression vector contains an SpCas9 gRNA expression cassette driven by the U6 promoter. In the second step, up to ten gRNA expression cassettes are cut out and assembled in tandem into the “array” plasmids by BsaI-based Golden Gate Cloning (Fig. 1, step 2). The array backbone plasmids (spectinomycin resistance) used for the second step assembly are pFUS-B1 to pFUS-B10, pFUS-A, pFUS-A30A, and pFUS-A30B of the TALEN assembly kit [10]. In the third step, one or two “array” plasmids, containing 10 gRNA expression cassettes each, and one array plasmid containing up to 10 gRNA expression cassettes are released and assembled into a single plasmid pMsgRNA-EGFP (ampicillin resistance) using a different type IIS restriction enzyme, BsmBI, generating a gRNA array of up to 30 cassettes. The pMsgRNA-EGFP contains within the cloning site the lacZ gene for blue/white screening of bacterial colonies, as well as one EGFP expression cassette for enrichment of transfected mammalian cells by FACS (Fig. 1, step 3).
Fig. 1.
Golden Gate Assembly of CRISPR gRNA expression array for simultaneously targeting of multiple genomic loci. Schematic illustration of Golden Gate Assembly of gRNA expression array using single modular, “array” vectors and the pMsgRNA-EGFP plasmid. Each gRNA guide oligonucleotide (T1 to T10, T11 to T20, or T21 to T30) is cloned into single modular plasmids (pMA1 to pMA10) based using the type IIS restriction enzyme BbsI correspondingly. The single pMA modular plasmid containing one gRNA expression cassette can be cut out and assembled into the “array” plasmids using the type IIS restriction enzyme BsaI, which will generate gRNA expression arrays carrying up to 10 cassettes. The gRNA expression arrays in the “array” plasmids can be further cut out and assembled into the pMsgRNA-EGFP plasmid using BsmBI, generating gRNA expression array containing 11–30 cassettes. GGC, Golden Gate Cloning; U6, U6 promoter for gRNA transcription; BB, BbsI restriction enzyme; Scr, SpCas9 gRNA scaffold sequences; EGFP, EGFP expression cassette driven by the CMV promoter
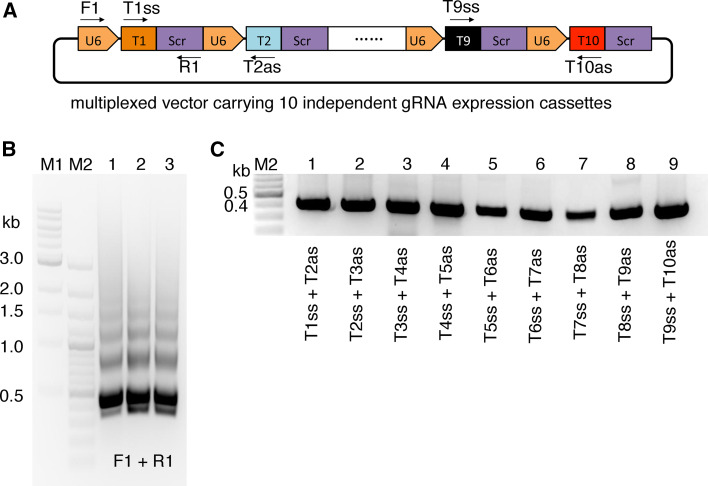
To facilitate the fast screening of bacterial clones, we designed two PCR screening strategies: a universal PCR and a guide specific PCR (Fig. 2a). For the universal PCR, we used a U6 promoter specific primer and a gRNA scaffold specific primer. The correct amplification pattern should be “ladder” like with a dominant band at approximately 400 bp and increase by 392 bp for each gRNA expression cassette included in the amplification (Fig. 2b). For guide specific PCR, 1–9 individual overlapping PCRs were performed using the sense and antisense of the adjacent gRNA guide oligonucleotides (Fig. 2c). For the M10 plasmid, we routinely pick two or three “white” colonies and usually all of them are correct.
Fig. 2.
PCR validation of a gRNA expression carrying 10 expression cassettes. a Schematic illustration of the universal PCR and guide specific PCR for screening of transformed E. coli colonies. F1 (U6-F) and R1 (Scr-R) are used for the universal PCR (Supplementary Table 1). Guide specific PCRs are using the pair sense and antisense gRNA oligonucleotides of the two adjacent gRNAs. b An example of the universal PCR results for a multiplexed gRNA array containing 10 expression cassettes. c An example of the 9 guide RNA specific PCRs for a multiplexed gRNA array containing 10 expression cassettes
Validation of gRNA expression array activity in a dual-fluorescent reporter assay
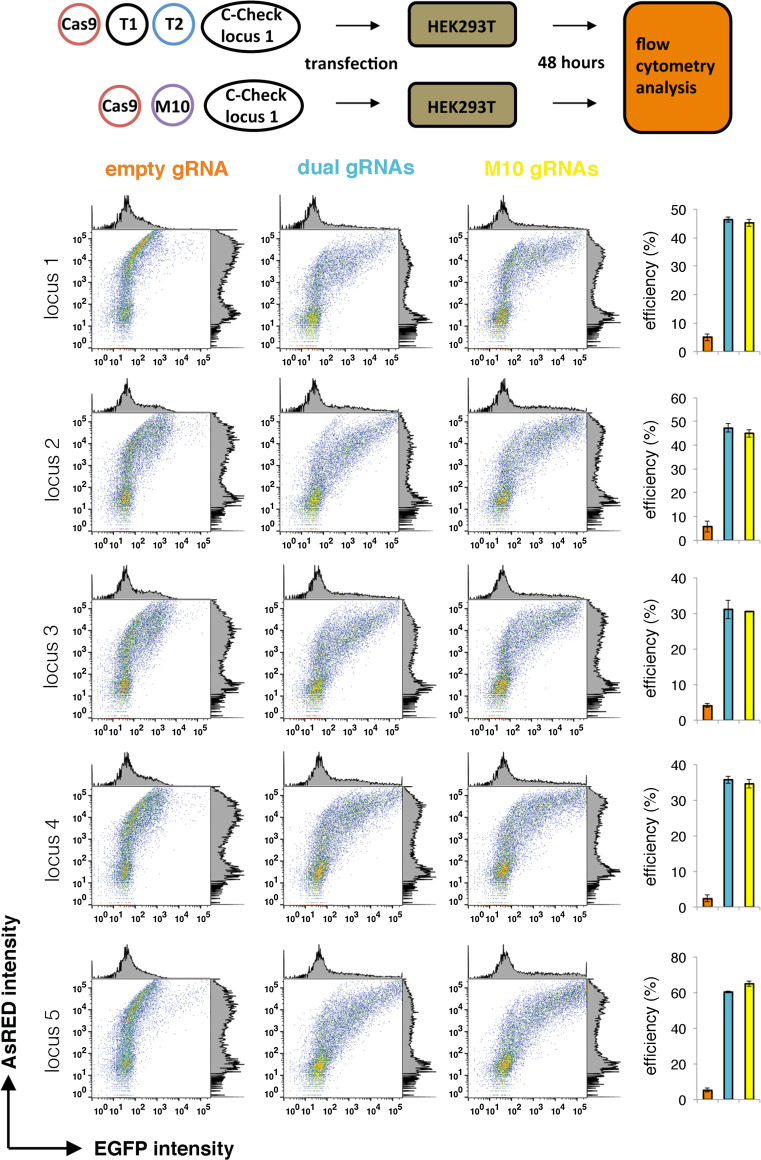
To validate whether the gRNA expression cassettes are functional in the array vector, we generated one vector containing 10 gRNA expression cassettes (M10), which targets five different genomic loci (two gRNAs per locus). We chose the dual-gRNAs strategy that had been indicated to facilitate the screening and efficiency of CRISPR-mediated deletions [11]. Recently, we have generated a dual-fluorescent reporter system that can measure the CRISPR/Cas9 activity (C-Check) in vitro [12]. First, we generated five C-Check reporter vectors for each genomic locus. Next, each C-Check reporter vector was co-transfected with an SpCas9 expression plasmid and an equal molar amount of either M10 or a pair of the single gRNA expression plasmid (Fig. 3, upper panel). The CRISPR/Cas9 activity was quantified by the percentage of GFP positive cells out of the total transfected cells (AsRED positive) 48 h after transfection using flow cytometry. The reporter assay results showed that significant nuclease activity (>30 %) was achieved for all five genomic loci (Fig. 3, lower panel). Most importantly, the multiplexed gRNA expression array had efficiency similar to the dual gRNAs, indicating that each gRNA expression cassette in M10 is independently transcribed and functional.
Fig. 3.
Functional validation of the M10 gRNA expression array using a dual-fluorescent reporter system (C-Check). Upper panel a C-Check reporter vector were generated for each genomic locus. For comparison, HEK293T cells were transfected with a C-Check plasmid, a SpCas9 expression plasmid, and equal molar amount of either a pair of gRNAs or the M10 gRNA, or an empty gRNA (control). Transfected cells were analyzed by flow cytometry 48 h after transfection. Lower panel representative dot and histogram plot of the flow cytometry analysis, EGFP intensity versus AsRED intensity. Bar plot of mean percentage efficiency (% GFP+/AsRED+ cells, n = 3) is presented to the right
Simultaneous targeting of multiple genes in human cells with gRNA expression array
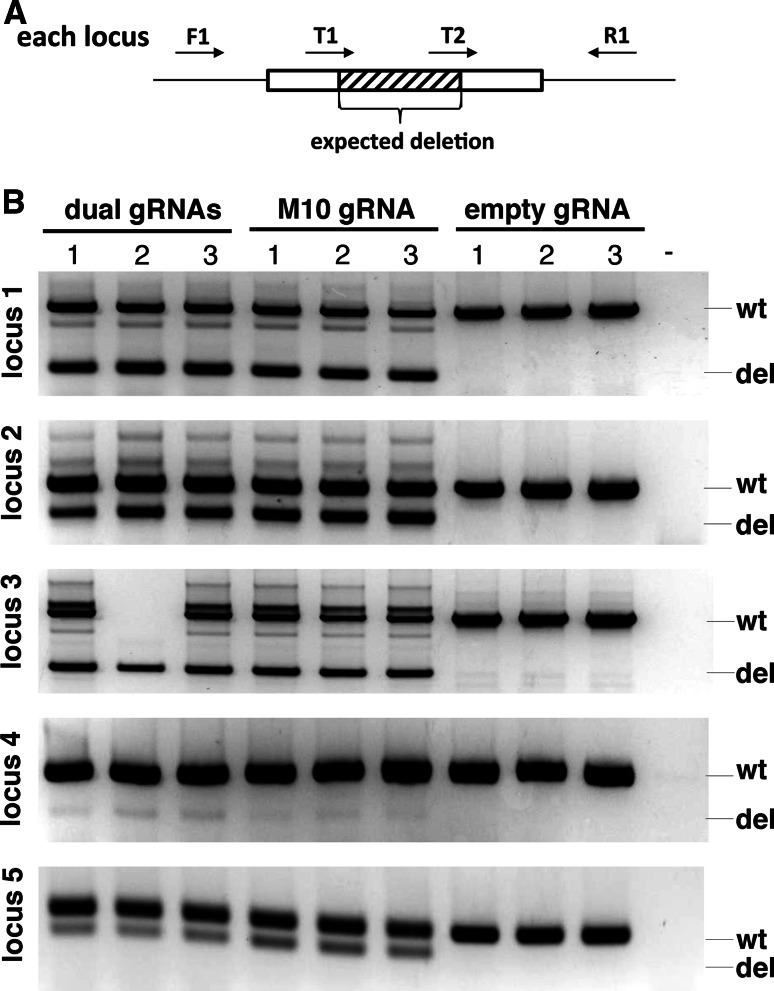
To further investigate the multiplexed gRNA expression array in simultaneously mediating gene targeting in human cells, we co-transfected HEK293T cells with an equal molar amount of an SpCas9 expression plasmid and either the M10, dual gRNA expression plasmids or an empty gRNA plasmid. Genomic DNA was purified from the transfected cells and equal amount of genomic DNA were used for genotyping of CRISPR/Cas9-mediated targeted gene deletion, as each genomic locus is targeted by a pair of gRNAs (Fig. 4a). Consistent with the C-Check analysis, the M10 gRNAs resulted in a similar gene deletion efficiency to dual gRNA expression vectors (Fig. 4b). Targeted deletion of all five genomic loci is simultaneously achieved in the same individual cells when using the M10 gRNA expression array plasmid. We also observed that the targeted deletion efficiency of genomic locus 4 was much lower than the other four loci (Fig. 4b), while this was not seen using the C-Check assay (Fig. 3). Sequencing of the locus 4 in wild type HEK293T cells revealed a 2-bp mismatch presented in one of the gRNA target sites (not shown).
Fig. 4.
Simultaneous deletion of 5 genomic loci using gRNA expression array in human cells. a For each genomic locus, a pair of gRNAs is designed to mediate target deletion of a small DNA fragment, which facilitates the genotyping by PCR. The M10 gRNA expression array plasmid used in this study contain 10 gRNAs targeting 5 genomic loci. b PCR screening results of dual gRNAs or M10 gRNAs mediated targeted gene deletion in five genomic loci in HEK293T cells. Cells transfected with an empty gRNA plasmid were used as controls. The expected wild type (wt) and deleted (del) amplicons were indicated to the right. Transfections were performed in triplicates. “-”, water control for PCR
Simultaneous inhibition of multiple genes in human cells with gRNA expression array
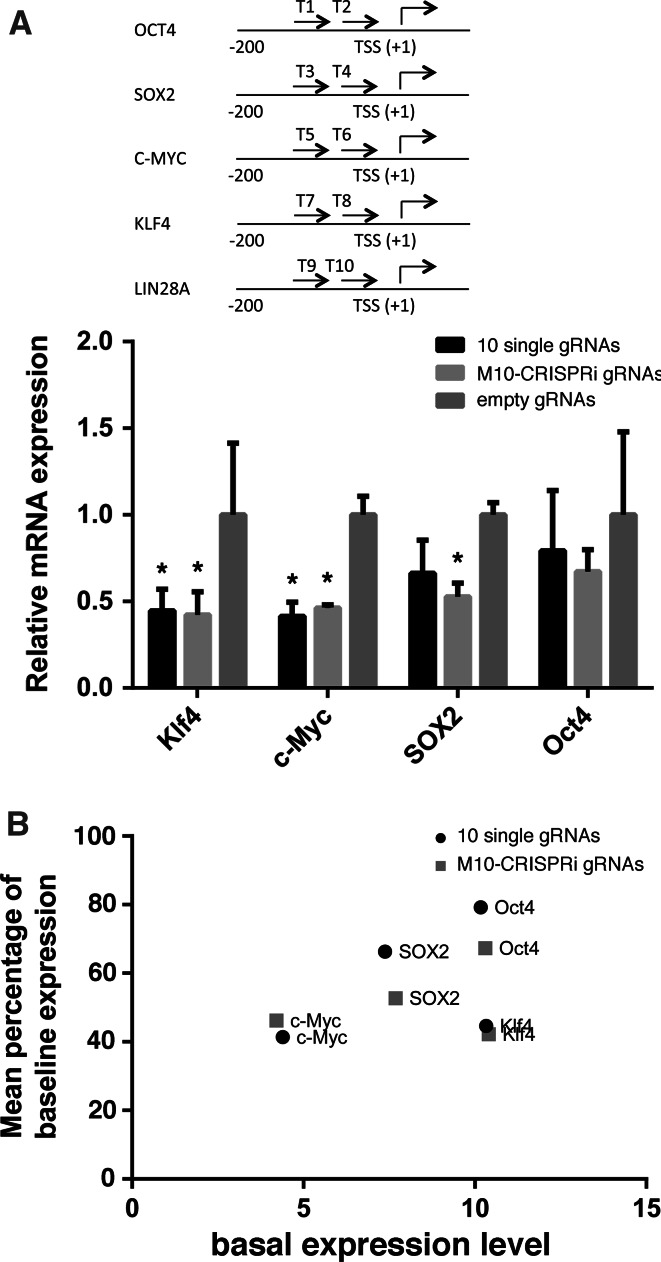
Targeted inhibition/activation of genes by CRISPRi/CRISPRa frequently required manipulating multiple genes simultaneously [5, 6, 13]. To assess our gRNA expression array for achieving CRISPR inhibition, we generated another gRNA expression array carrying 10 gRNA expression cassettes (referred as M10-CRISPRi), which targeted the proximal promoter of five genes (OCT4, SOX2, C-MYC, KLF4, and LIN28A, two gRNAs per gene, Fig. 5a, top). We co-transfected HEK293T cells with a plasmid expressing dSpCas9 coupled to a transcriptional repressor domain, KRAB, [6] and an equal molar amount of either the M10-CRISPRi containing five pairs of gRNAs, ten individual gRNA vectors, or an empty gRNA plasmid. Inhibition of KLF4, C-MYC, and SOX2 expression were observed for the M10-CRISPRi and the ten individual gRNA vectors compared to empty gRNA control (Fig. 5a). We also observed that the expression inhibition effect varied from gene to gene, which is partially related to the basal gene expression level (Fig. 5b). The LIN28A inhibition data is not available as this gene is not expressed in HEK293T cells (not shown). Since we were using the M10-CRISPRi plasmid, the inhibition of KLF4, C-MYC, and SOX2 most likely occurred simultaneously in the sample population of transfected cells.
Fig. 5.
Simultaneous inhibition of multiples genes using CRISPRi in human cells. a Inhibition of KLF4, C-MYC, SOX2 and OCT4 expression in HEK293T cells by 10 single gRNAs or the M10-CRISPRi gRNA. Relative mRNA expression was calculated as the mean (triplicates) percentage of basal expression level in cells transfected with an empty gRNA. Schematic illustration of the gRNA target site for each gene is shown on the top. Asterisk indicates statistically significant (p < 0.05 by ANOVA). b Correlation plot of the mean inhibition of gene expression (percentage of the basal expression level) and the basal expression level of each gene (mean delta Ct of the reference gene, beta-actin). Each plot represents mean of triplicates
Discussion
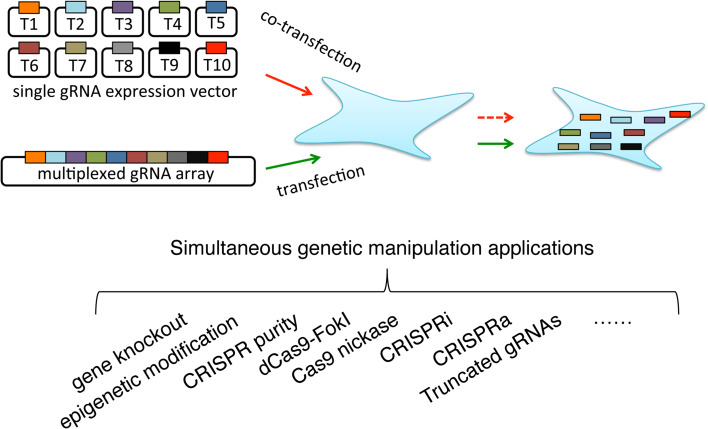
The hallmark of CRISPR/Cas9 technology has dramatically driven the development of genome editing-based applications in academic, industrial, agriculture, and clinical settings. One example is the generation of genetically modified animal models of human diseases [14]. For monogenic diseases, these can be achieved using a single CRISPR gRNA vector. However, for multigene disorders such as neurodegenerative disorders, multiplexed gene editing is needed. Using a single gRNA expression, either co-transfection of the gRNA expression vectors or several rounds of gene editing might be required [15], which is time-consuming and less efficient when co-transfecting many gRNAs. One solution to this will be using one single vector carrying multiple gRNA expression arrays (Fig. 6). Another area is depending on multiplexed gene editing is the generation of genetically large animals for the purpose of xenotransplantation [16]. Using nuclease-deficient Cas9 (dCas9), the CRISPR/dCas9 have been explored for a wide range of genetic and epigenetic perturbations, such as epigenome editing [7, 17], regulation of gene transcription [18, 19], visualization of specific genomic loci [8], purification of specific genomic regions [20] etc. Most of these applications such as epigenome editing and regulation of gene transcription require simultaneously delivery of multiple gRNAs in the cells in order to achieve an efficient perturbation. Off target effect is a major concern when using the CRISPR/Cas9 technology for gene editing. Methods using Cas9 nickase and a pair of gRNAs [21], modified Cas9 proteins [22, 23], and truncated gRNAs [24] have been shown to enhance specificity. Although not investigated in this study, our CRISPR gRNA expression array method should be a useful tool for simultaneous delivery of pairs of gRNAs to enhance Cas9 nickase-mediated gene editing, as well as for simultaneous delivery of multiple truncated gRNAs. In this study, we generate a rapid and cost-effective method for generation CRISPR gRNA expression using Golden Gate Assembly. We also demonstrated using the gRNA expression array to achieve simultaneous disruption or inhibition of multiple genes. We have further validated that two M10 vectors can be efficiently assembled with another M10 vector to generate a M30 vector, with over 95 % white colony rate (not shown).
Fig. 6.
Schematic comparison of simultaneous delivery of many CRISPR gRNA into the same cells using co-transfection of many single gRNA plasmids or one multiplexed gRNA expression array, as well as the potential applications
The main advantages of the CRISPR gRNA expression array include: (1) Simultaneous multiplexing: Since all single gRNA expression cassettes are contained in the same vector, the simultaneous expression of all multiple gRNAs in the same cell is ensured. Additionally, each gRNA expression is regulated by its own U6 promoter and thus makes sure each gRNA functions independently. (2) Cost-effective and programmable: Generation of polycistronic gRNA expression arrays could also be achieved by combinations of DNA synthesis and assembly [25] that are expensive and un-programmable. With our method, only the synthesis of guide sequences, e.g., 20 nucleotides with the four nucleotide overhangs are required. All the array plasmids are universal and can be used for any CRISPR guide sequences. (3) Time-effective: We generally experienced that generation of such a multiplexed gRNA expression array can be achieved in 1–2 weeks depending on the number of CRISPR gRNA expression cassettes. Disadvantage: Since this system is based on the usage of type IIS restriction enzymes, target sites with the presence of BbsI, BsaI or BsmbI recognition sites must be excluded when designing CRISPR gRNAs. However, considering the current lack of efficient and cost-effective methods for CRISPR-mediated simultaneous multiplexed gene targeting, we believe that our method will become a powerful tool for CRISPR/Cas9-mediated multiplexed genome editing. A detailed method for construction of gRNA expression plasmid is given in the methods. All modular plasmids are available from Addgene.
Materials and methods
All DNA oligonucleotide syntheses in this study were performed by Eurofins Genomics, Germany. All Sanger sequencing was performed by GATC-Biotech, Germany. All Fast Digest restriction enzymes were purchased from Thermo Scientific, Life Technologies, Denmark. All experiments in this study were performed in independent triplicates.
Cells
Human embryonic kidney (HEK293T) cells were cultured in Dulbecco’s Modified Eagle’s Medium DMEM (Lonza) supplemented with 10 % fetal bovine serum, 1 % penicillin–streptomycin and 1 % glutamine. The cells were kept in a CO2 incubator with 5 % CO2 at 37 °C. For passaging, the cells were detached with Trypsin (0.05% trypsin-EDTA) and passaged in a ratio of 1–5.
Construction of the modular single gRNA expression vectors (pMA1-pMA10)
The ten individual single gRNA expression modular vectors were constructed by a simple cloning strategy. First, ten individual empty g-blocks (gRNA expression cassette, U6 promoter and SpCas9 gRNA scaffold) were PCR amplified from the pSpCas9(BB)-2A-Puro plasmid (a gift from Feng Zhang (Addgene plasmid # 62988)). Type IIS restriction enzyme BsaI-based cloning site were introduced flanking the g-blocks after amplification. A list of primers used for PCR amplification is given in Supplementary Table 1. Each amplified g-block was digested with BamHI and BsrGI and ligated to a pMA plasmid backbone (Amp+, GeneArt, Thermo Fisher Scientific). Each single guide RNA vector (named from pMA1 to pMA10) was validated by PCR screening, visualized by gel electrophoresis, and Sanger sequencing.
Protocol for assembly of multiplexed gRNA expression array
Generation of the gRNA expression array vector is accomplished in 5 days (1–10 cassettes) or 8 days (11–30 cassettes) days depending on the number of gRNA expression cassettes. A schematic presentation of the Golden Gate Assembly of gRNA expression array is shown in Fig. 1 and all plasmids and the selection of plasmids for assembly are described in Supplementary Table 2. In this protocol, we use an example for the generation of gRNA expression array containing 12 expression cassettes and note the difference when generating array containing 1–10 or 21–30 expression cassettes.
Preparation: in silico CRISPR gRNA design and order gRNA oligonucleotides
Design CRISPR gRNA target sites using any of the online designing tools. In this study, all gRNAs were designed using the optimized CRISPR design tool (http://crispr.mit.edu/).1 To ensure the efficient transcription of gRNA, the first nucleotide of the CRISPR target site should start with a “G”. Besides, exclude target sites that contain any of the following three type IIS restriction enzyme recognition sites: BbsI, BsaI, and BsmBI. Four nucleotide overhangs 5′CACC and 5′AAAC was added to the 5′ end of sense and antisense gRNA oligonucleotides, respectively. Then order the gRNA oligonucleotides with desalted purification. Dilute the oligonucleotides to a stock concentration of 100 pmol/µL (µM) and saved at −20 °C.
Day 1. Assembly of gRNA guide sequences into single gRNA expression modular vectors
In this protocol, we use an example for the generation of gRNA expression array containing 12 expression cassettes (M12), which are named as T1–T12. The protocol for generating gRNA expression array containing 1–10 or 21–30 expression cassettes is slightly different from the array containing 11–20 cassettes, which will be noted.
First, anneal the sense and antisense guide oligonucleotides for each gRNA (T1–T12). We normally anneal the sense and antisense of each gRNA oligonucleotide (100 pmol each) in 1× NEB buffer 2 in a 20 µL mixture. The mixture is denatured at 95 °C and slowly annealed to room temperature. Quick spin the annealed oligonucleotides at the bottom of the tube and use for BbsI-based Golden Gate Assembly as described below. The step takes approximately 1–2 h depending on the gradual cooling procedure.
Second, assemble each of the annealed double-stranded oligonucleotides (T1–T12) to the single gRNA expression modular plasmids (pMA1–pMA10) according to the cloning scheme in Fig. 1 (step 1). This assembly is accomplished in a single 20 µL reaction containing 1 µL BbsI restriction enzyme (Thermo Scientific FastDigest, FD1014), 1 µL T4 DNA ligase (Thermo Scientific, EL0011), 100 ng single gRNA expression modular plasmid and 1 µL of the annealed double-stranded oligonucleotide in T4 DNA ligase buffer. The reaction is incubated in a thermal cycler for ten cycles of 5 min at 37 °C and 10 min at 22 °C, then at 37 °C for 30 min and heated to 75 °C for 15 min. Then, use 2 µL ligation product to transform competent E. coli cells and plate transformed cells in a LB agar plate containing 50 µg/ml ampicillin. The agar plates are incubated at 37 °C overnight.
Day 2. 1st colony screening and culture
Pick two to three colonies from each transformation and start initial culture in 100 µL LB medium containing 50 µg/ml ampicillin. For colony screening, each colony is picked using a 10 or 100 µL pipette tip. The pipette tip is first quickly rinsed in the 100 µL LB culture medium and subsequently left in a PCR tube containing 30 µL ddH2O. Thus, each colony picked has a corresponding cultivation and screening ID. For colony screening, the 30 µL ddH2O containing cells were lysed in a thermal cycler at 98 °C for 10 min. Then 1 µL cell lysate was used for PCR using the following primer set: a universal U6 primer (5′GAGGGCCTATTTCCCATG3′) and the antisense oligonucleotide of the gRNA. For example, for colony screening of pMA-T1, PCR is performed using the U6 and the T1-antisense oligonucleotide. The amplicon size is approximately 270 bp. We experienced that the positive rate is above 95 %. Based on the colony PCR screening result, start overnight culture of one positive colony for each transformation.
Day 3. Plasmid prep and assembly of individual gRNA expression cassette into the “array” plasmids
Isolate the plasmid DNA from the cell culture using a commercial miniprep kit. Optional, it is recommended to validate the single gRNA expression plasmid by Sanger sequencing using the U6 primer (above). Very rarely, oligonucleotide synthesis can introduce unintended substitutions.
Then assemble the single gRNA expression modular plasmids into one of the “array” plasmids. A guideline for selecting array plasmids is given in the Supplementary Table 2. For the generation of the M12 array plasmid, two BsaI-based Golden Gate assembling reaction are prepared. In the first reaction, the pMA-T1 to pMA-T10 plasmids are assembled into the pFUS-A plasmid. In the other reaction, the pMA-T11 and pMA-T12 plasmids are assembled into the pFUS-B2 plasmid. For assembling multiplexed array plasmids containing 10 cassettes or less, the pMA-T1 to pMA-TN (N is equal or less than 10) plasmids are assembled to the pFUB-BX plasmid (X is the number of single gRNA expression modular plasmids contained in the reaction). For assembling multiplexed array plasmids containing more than 20 cassettes, the pMA-T1 to pMA-T10 are assembled into the pFUS-A30A array plasmid, the pMA-T11 to the pMA-T20 are assembled into the pFUS-A30B plasmid, and the remaining pMA-T21 to pMA-TO (O is equal or less than 30) are assembled into the pFUB-BX (X is the number of single gRNA expression modular plasmids contained in the reaction). Each reaction contains 20 µL digestion and ligation mixture with 100 ng array plasmid, 50 ng each of individual single gRNA modular plasmids, 1 µL BsaI restriction enzyme (Thermo Scientific FastDigest, FD0294) and 1 µL T4 DNA ligase (Thermo Scientific, EL0011) in T4 DNA ligase buffer. The reaction is incubated in a thermal cycler for 15 cycles of 5 min at 37 °C and 10 min at 22 °C, then incubated at 37 °C for 30 min and heated to 75 °C for 15 min. Then, 1 µL 25 mM ATP and 1 µL plasmid safe DNase (Epicenter) are added to the reaction and incubated at 37 °C for 1 h followed directly by transformation using 2 µL of the assembled product. The transformed cells are plated on LB agar plates containing 50 µg/ml spectinomycin, as well as X-gal and IPTG for blue and white screening.2 This step is to avoid the recombination of the linearized DNA fragments following transformation.
Day 4. 2nd colony screening and culture
Pick up to three white colonies from each transformation. Start initial culture as described in Day 2, and perform colony screening using the universal PCR (see Fig. 2b). Start overnight culture of one PCR positive clone for each transformation.
Day 5. Plasmid DNA prep and second assembling (only required for the M11 to M30 array plasmids)
Isolate plasmid DNA using a high purity endotoxin free plasmid prep kit and further validated by restriction enzyme digestion with AflII and XbaI, which will release the gRNA expression cassettes: 3904 bp for pFUS-A, 3917 bp pFUS-B10, 3899 bp for pFUS-A30A, 3887 bp for pFUSA-30B and of varying sizes for pFUS-B plasmids. Note: The restriction enzyme digestion scheme can vary if there is AflII and/or XbaI presented in the gRNA guide sequences. The plasmid will be further validated by the guide specific PCRs see Fig. 2c.
To generate the M12 array plasmid, join the intermediary gRNA expression cassettes in the pFUS-A-M10 and pFUS-B2-M2 plasmids into the backbone plasmid pMsgRNA-EGFP using BsmBI-based Golden Gate assembling. A 20 µL digestion and ligation mixture with 100 ng each of pFUS-A-M10, pFUS-B2-M2 and pMsgRNA-EGFP, 1 µL BsmBI restriction enzyme (Thermo Scientific FastDigest, FD0454) and 1 µL T4 DNA ligase (Thermo Scientific, EL0011) in T4 DNA ligase buffer. The reaction is incubated in a thermal cycler for 10 cycles of 5 min at 37 °C and 10 min at 22 °C, then at 37 °C for 30 min and heated to 75 °C for 15 min. Treatment of the reaction mixture with plasmid safe DNA nuclease is not necessary for this step. For generation of gRNA expression arrays containing more than 20 cassettes, assemble the pFUS-A30A-M10, pFUS-A30B-M10 and pFUS-BN-MN plasmids into the pMsgRNA-EGFP vector. Then transform competent E. coli using 2 µL of the assembled product and plate transformed cells in LB agar plates containing 50 µg/ml ampicillin, with X-gal and IPTG for blue and white screening.
Day 6. 3rd colony screening and culture
Pick up to three white colonies for initial culture and perform colony PCR screening as described in Day 2, but use the T10-sense and T11-anti-sense oligonucleotides for PCR, which will be 458 bp. Two screening PCRs are performed for gRNA arrays containing over 20 cassettes. The first PCR is performed with the T10-sense and T11-antisense oligonucleotides, which will be 458 bp. The second PCR is performed with the T20-sense and T21-antisense oligonucleotides, which will be 458 bp. Sep up overnight culture of one PCR positive clone.
Day 7. Plasmid DNA prep and validation
Isolate plasmid DNA and further validate the plasmid by restriction enzyme digestion. The EGFP expression cassette and gRNA expression arrays can be cut out from the final pMsgRNA-EGFP plasmid with SfiI, which will be approximately 1662 + N × 379 bp (N is the number of gRNA target sites).
Transfection
All transfections were performed in three independent experiments using the X-tremeGENE 9 reagent (Roche) according to manufacturer’s instruction. Briefly, for C-Check analysis of gene knockout, 1 × 105 HEK293T cells were seeded in a 24-well plate the day before transfection. Cells were co-transfected with a SpCas9 expressing plasmid and a C-Check vector with either a pair of single gRNA expression vectors or the M10-KO vector with a molar ratio equal to 1:1 between single gRNAs and corresponding gRNAs in the Mg10 vectors. For control transfections, cells were co-transfected with C-Check and an empty gRNA plasmid or with a control plasmid (pUC19) alone. The cells were harvested by trypsinization 48 h post-transfection. Half of the cells were analyzed by flow cytometry to quantify the percentage of GFP and AsRED positive cells. The efficiency CRISPR/Cas9-mediated cleavage is calculated as the percentage of GFP positive cells out of the transfected cells (AsRED positive). Another half of cells were subjected to knockout PCR screening analysis of genomic DNA. For the CRISPR-mediated transcriptional inhibition, 3 × 105 HEK293T cells were seeded in a 6-well plate 24 h before transfection. Cells were co-transfected with a plasmid encoding the dCas9 coupled to the transcription repressor domain KRAB and either ten single gRNA expression vectors or the M10-CRISPRi array vector with a molar ratio equal to 1:1 between single gRNAs and corresponding gRNAs in the Mg10 vectors. For control transfections, cells were co-transfected with equal molar of an empty single gRNA vector. 72 h post-transfection cells were harvested for gene expression analysis. All transfection were performed in three independent triplicates.
Flow cytometry analysis
Transfected cells were analyzed with a BD LSRFortessa Analyzer (FACS CORE facility at the Department of Biomedicine, Aarhus University). At least 10,000 events were analyzed per sample. All flow cytometry results were analyzed with FlowJo version 10.
PCR screening of CRISPR-mediated deletion
Transfected cells in suspension transferred to 1.5 mL tubes were spun down at 2000 rpm for 10 min. The supernatant was carefully removed with a transfer pipette without disturbing the cell pellet. The cell pellet was lysed by adding 50 µL cell lysis buffer containing 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris–Cl, pH 8.5, 0.5 % Nonidet P40, 0.5 % Tween, 400 µg/ml proteinase K. The cells were lysed at 65 °C for 30 min followed by inactivation of proteinase K at 95 °C for 10 min in a thermal cycler. Two microliter of cell lysate was used for PCR-based screenings in a 20 µL PCR reaction volume. PCR products were visualized by agarose gel electrophoresis (2 %). A list of primers used for PCR screening is given in Supplementary Table 1.
RNA purification and cDNA synthesis
RNA purification was performed using Tri Reagent™ (Sigma). 1 mL Tri Reagent was added to the wells of transfected cells. The suspension was transferred to RNAse-free Eppendorf tubes and incubated for 5 min. 200 µL chloroform (Merck) was added per mL Tri Reagent and incubated for 10 min. After centrifugation at 12,000×g for 15 min at 4 °C, the upper RNA-containing phase was transferred to RNAse-free Eppendorf tubes. 500 µL isopropanol (Merck) and 2 µL glycogen (Sigma) was added followed by centrifugation at 12,000×g for 30 min at 4 °C. The pellet was washed in 75 % RNAse-free ethanol and dissolved in 50 µL DEPC H2O and stored at −20 °C. RNA concentration was measured using a Thermo Scientific Nanodrop™ spectrophotometer. RNA was treated with DNase using TURBO DNA-free kit (Ambion) according to the manufacturer’s instructions. cDNA was synthesized from 0.5 µg RNA using the BIO-RAD iScript™ cDNA Synthesis kit containing a mix of oligo(dT) and random hexamer primers. After synthesis the cDNA product was diluted with redistilled water to a total volume of 100 µL and stored at −20 °C.
Quantitative reverse transcriptase polymerase chain reaction (RT-qPCR)
RT-qPCR was performed in a total reaction volume of 15 µL containing 2 µL cDNA, 7.5 µL Roche LightCycler® 480 SYBR Green I Master enzyme (Roche), 500 nM primer and double distilled water up to 15 µL. A LightCycler® 480 (Roche) was used with a PCR profile of one cycle of denaturation at 95 °C for 5 min followed by 45 cycles of denaturation at 95 °C for 10 s, annealing at 57–60 °C for 10 s, and extension at 72 °C for 10 s. A list of primers used is given in Supplementary Table 1. The RT-qPCR assay was performed in duplicate for all samples. The relative mRNA expression was calculated using the X 0 method, and normalized to the reference gene beta-actin [26].
Statistical analysis
All data were represented as mean ± standard deviation. Unless stated elsewhere, two-way analysis of variance (ANOVA) with Tukey correction for multiple comparisons was used for all statistical analysis in this study. All statistical analyses were conducted using Graphpad Prism (version 6.1). p values less than 0.05 were considered statistically significant.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
JVN and ALN were supported by grants from The Lundbeck Foundation, Krista og Viggo Petersens Fond, Fabrikant Einar Willumsens Mindelegat, and Fonden til Lægevidenskabens Fremme. LL and LB were supported by the DREAM project from Lundbeck Foundation. YL was supported by grants from Danish Research Council for Independent Research, the Sapere Aude Young Research Talent prize to YL, the Lundbeck Foundation and the Innovation Fund Denmark (BrainStem).
Footnotes
All gRNA expression cassettes in our method are driven by the U6 promoter.
It is very important to treat the reaction with plasmid safe DNA nuclease when assembling more than five gRNA expression cassettes. This step is to avoid the recombination of the linearized DNA fragments following transformation.
References
- 1.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wan H, Feng C, Teng F, et al. One-step generation of p53 gene biallelic mutant Cynomolgus monkey via the CRISPR/Cas system. Cell Res. 2015;25:258–261. doi: 10.1038/cr.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mali P, Yang L, Esvelt KM, et al. RNA-Guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng AW, Wang H, Yang H, et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert LA, Larson MH, Morsut L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hilton IB, D’Ippolito AM, Vockley CM, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen B, Gilbert LA, Cimini BA, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cermak T, Doyle EL, Christian M, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandal PK, Ferreira LM, Collins R, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell. 2014;15:643–652. doi: 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y, Liu Y, Hussmann D, et al. Enhanced genome editing in mammalian cells with a modified dual-fluorescent surrogate system. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-015-2128-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi LS, Larson MH, Gilbert LA, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tu Z, Yang W, Yan S, et al. CRISPR/Cas9: a powerful genetic engineering tool for establishing large animal models of neurodegenerative diseases. Mol Neurodegener. 2015;10:35. doi: 10.1186/s13024-015-0031-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm IE, Alstrup AK, Luo Y. Genetically modified pig models for neurodegenerative disorders. J Pathol. 2016;238:267–287. doi: 10.1002/path.4654. [DOI] [PubMed] [Google Scholar]
- 16.Cooper DK, Ekser B, Ramsoondar J, et al. The role of genetically engineered pigs in xenotransplantation research. J Pathol. 2016;238:288–299. doi: 10.1002/path.4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kearns NA, Pham H, Tabak B, et al. Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods. 2015;12:401–403. doi: 10.1038/nmeth.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konermann S, Brigham MD, Trevino AE, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2014;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert LA, Horlbeck MA, Adamson B, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujita T, Fujii H. Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochem Biophys Res Commun. 2013;439:132–136. doi: 10.1016/j.bbrc.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Ran FA, Hsu PD, Lin CY, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slaymaker IM, Gao L, Zetsche B, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Y, Sander JD, Reyon D, et al. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balboa D, Weltner J, Eurola S, et al. Conditionally stabilized dCas9 activator for controlling gene expression in human cell reprogramming and differentiation. Stem Cell Reports. 2015;5:448–459. doi: 10.1016/j.stemcr.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomsen R, Solvsten CA, Linnet TE, et al. Analysis of qPCR data by converting exponentially related Ct values into linearly related X0 values. J Bioinform Comput Biol. 2010;8:885–900. doi: 10.1142/S0219720010004963. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.