Abstract
The cellular prion protein (PrPC) is a ubiquitously expressed protein of currently unresolved but potentially diverse function. Of putative relevance to normal biological activity, PrPC is recognized to undergo both α- and β-endoproteolysis, producing the cleavage fragment pairs N1/C1 and N2/C2, respectively. Experimental evidence suggests the likelihood that these processing events serve differing cellular needs. Through the engineering of a C-terminal c-myc tag onto murine PrPC, as well as the selective use of a far-C-terminal anti-PrP antibody, we have identified a new PrPC fragment, nominally ‘C3’, and elaborating existing nomenclature, ‘γ-cleavage’ as the responsible proteolysis. Our studies indicate that this novel γ-cleavage event can occur during transit through the secretory pathway after exiting the endoplasmic reticulum, and after PrPC has reached the cell surface, by a matrix metalloprotease. We found that C3 is GPI-anchored like other C-terminal and full length PrPC species, though it does not localize primarily at the cell surface, and is preferentially cleaved from an unglycosylated substrate. Importantly, we observed that C3 exists in diverse cell types as well as mouse and human brain tissue, and of possible pathogenic significance, γ-cleavage may increase in human prion diseases. Given the likely relevance of PrPC processing to both its normal function, and susceptibility to prion disease, the potential importance of this previously underappreciated and overlooked cleavage event warrants further consideration.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-015-2022-z) contains supplementary material, which is available to authorized users.
Keywords: Prion protein, Endoproteolysis, Protein processing, Protein cleavage
Introduction
The cellular prion protein, PrPC, is a ubiquitously expressed glycosylphosphatidylinositol (GPI) anchored cell surface glycoprotein, with highest levels found in neurons and central nervous system tissues [1–4], and is causally linked to the group of fatal neurodegenerative disorders known as prion diseases. Surprisingly, engineered PrPC gene ablated (knockout) mice were initially reported as normal with no overt phenotypic abnormalities [5], which at the time suggested the prion protein could be functionally redundant. Subsequent studies, however, demonstrated various deficits and implicated PrPC in a diverse range of biological activities, with increasing evidence for roles in important cellular processes such as neuroprotection [6–9], cell signaling [10–13], neurological development and neuritogenesis [14, 15], and synaptic function and plasticity [16–20]. Further, prion proteins from different mammalian species show high sequence identity, especially in the far C-terminus, with several post-translational modifications and structural features conserved across species [21–25], supporting the likely significant evolutionary and biological importance of this protein.
Like many other proteins, PrPC is subject to constitutive and selective proteolytic processing, producing several membrane-bound and soluble fragments of different sizes and features. PrPC may be cleaved at its GPI-anchor, allowing shedding of PrPC species from a cell by both protease and phospholipase mediated mechanisms [26–28]. PrPC is also subject to two well-described internal cleavage events known as α- and β-cleavage [29]. The dominant PrPC processing event, α-cleavage, occurs at the start of the hydrophobic core region (after residues 111/112, human PrP numbering) [30], producing the C-terminal C1 fragment and corresponding N-terminal N1 fragment. Adding complexity, a recent study suggests α-cleavage may actually be multifaceted, with multiple neighboring cleavage sites targeted by different proteases [31]. β-Cleavage, predominantly associated with prion disease and misfolded prion protein conformers (PrPSc) [30, 32], but also reported to befall PrPC in uninfected cells and tissues [30, 33–36], involves ‘ragged’ cleavage at the end of the metal-binding octapeptide repeat region, around residue 90, producing the C2 and N2 fragments [34, 35]. The precise biological reasons for PrPC proteolysis are not entirely elucidated, although PrPC proteolytic fragments, especially the C-terminal fragments, are abundant in cells and tissues, and there is increasing evidence for separate roles for the different PrP molecular species [29]. In addition, the influence of PrPC proteolytic processing on disease transmission susceptibility, pathogenesis and toxicity is recognized [33, 37, 38].
Herein we report the discovery of a novel small PrPC C-terminal fragment, ‘C3’, observed in various cultured cell lines, as well as in murine and human brain extracts. Our primary aim was to characterize the basic cellular biology of this previously unrecognized PrPC endoproteolytic event, which based on an elaboration of existing nomenclature, we have named ‘γ-cleavage’. This aim was achieved through the uncovering of intrinsic C3 features, as well as identification of a time-line for γ-cleavage in relation to the normal PrPC lifecycle within the cell and the likely cellular location of proteolysis, the approximate PrPC γ-cleavage site, the family of proteases responsible, and potential links to human prion disease. These findings increase our understanding of PrPC cell biology and add further complexity to the multi-faceted PrPC proteolytic processing pathways.
Materials and methods
Animal and human tissue (ethics)
The animal brain tissue utilized herein was obtained during a previous study [39], where all animal experiments were carried out with approval from the University of Melbourne Animal Ethics Committee (AEC #04154). The detection of C3 in human brain tissue occurred during routine surveillance and classification activities carried out by the Australian National Creutzfeldt-Jakob Disease Registry (ANCJDR) [40], under contract to the Australian Government Department of Health, and with the approval of The University of Melbourne Human Research Ethics Committee (HREC #1136882.2). All human control tissue was purchased from the Victorian Brain Bank Network (VBBN), and sporadic Creutzfeldt-Jakob Disease (CJD) tissue was supplied by the ANCJDR.
Cell culture
The cell lines used in this study were mouse neuroblastoma cells, N2a (#CCL-131), human neuroblastoma cells, SHSY5Y (#CRL-2266), both purchased from the ATCC biological resource center, human embryonic kidney (HEK) cells obtained from the European Collection of Cell Cultures and the rabbit kidney epithelial cell line, RK13, a kind gift from Dr Victoria Lawson, The University of Melbourne. All cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM; Lonza) containing 10 % (v/v) fetal bovine serum (FBS; Bovogen), in a humidified incubator in 5 % CO2 and at 37 °C, unless otherwise indicated.
Generation of Myc-tagged wild-type and mutant PrP constructs
To generate wild-type murine Prnp (WTPrnp) from a 3F4-epitope tagged murine Prnp (3F4Prnp) in pIRESneo template [41], primers 3F4-WT-F and 3F4-WT-R and the Quikchange® II XL Site-Directed Mutagenesis kit (Stratagene) were utilized, following the manufacturer’s instructions. Insertion of the c-myc tag with repetition of murine Prnp codons 226–230 into WTPrnp to produce “PrP-myc”, as well as EcoRI (5′) and BamHI (3′) restriction sites for ligation into the plasmid pIRESneo, was via a three-stage modified overlap extension PCR process, previously shown to be useful for generation of a chimeric gene [42]. First, WTPrnp was used in two separate PCR reactions (A) and (B), with these two PCR products purified (QIAquick PCR Purification Kit, Qiagen) to remove leftover primers from the solution. Next, a ‘fusion’ PCR reaction (C) was carried out, combining PCR products (A) and (B) as template DNA and no additional primers. Lastly, the primers EcoPrPF and BamPrPR and PCR product (C) as template were utilized in a final PCR to generate the full PrP-myc open reading frame (D). PCR product (D), as well as empty pIRESneo vector, were subject to a double restriction digest with EcoRI-HF and BamHI-HF (both NEB) for 2 h at 37 °C, and the digested vector was also treated with antarctic phosphatase to prevent re-ligation of vector ends. The digested DNA was resolved on a 1 % agarose gel, purified (QIAquick Gel Extraction Kit, Qiagen) and ligated to form a circular PrP-myc in pIRESneo plasmid, which was then transformed into XL1-Gold Ultracompetent bacteria (Stratagene). Ampicillin resistant colonies were selected and myc-tag insertion was confirmed by sequencing. The above multi-step process was also carried out using 3F4Prnp as a template, to produce 3F4-myc. To generate D177NMyc, E199KMyc and V209IMyc, site-directed mutagenesis with the Quikchange® II XL Site-Directed Mutagenesis kit (Stratagene) was carried out with the appropriate primers and PrP-myc in pIRESneo as the template DNA, following the manufacturer’s instructions. All primer sequences and PCR cycling conditions are listed in supplementary material (Online Resource 1 and 2, respectively). PCR reactions included a final concentration of 0.2 mM dNTP mix, 1 mM MgSO4 and 1.25 Units (0.5 μl) of Platinum Pfx polymerase (all Life Technologies), except the ‘fusion’ PCR (C), where double the dNTPs (0.4 mM) and Platinum Pfx polymerase (2.5 Units) was added.
Transfection of mammalian cells
Transient transfections and the RK13 cells stably expressing PrP-myc (MycRK) were created using Lipofectamine 2000 (Life Technologies), following the manufacturer’s instructions. Transient transfections were all approximately 72 h. All other stable cell lines were created by electroporation of appropriate cDNA as described previously [43].
Live cell treatments
Cells were treated in culture with compounds (all Sigma) or the compound diluent as controls, for the length of time and concentrations as indicated in figure legends. All treatments were carried out in OptiMEM (Life Technologies) containing 10 % FBS unless otherwise indicated. Briefly, E64d treatment was at a final concentration of 40 μg/ml for 72 h, with reagent added fresh every 24 h. Prinomastat and Brefeldin A (at indicated concentrations), and Tunicamycin (10 μg/ml final concentration) treatments were for 24 h. PIPLC treatment (0.05 U/ml final concentration) was for 1 h in serum free OptiMEM.
Temperature block experiments
MycRK cells were seeded into 12-well plates and grown to confluence. Cell monolayers were washed gently with 1X PBS, and then incubated for 4 h in 1 ml of OptiMEM with or without 10 % (v/v) FBS. All incubations were carried out in non-humidified conditions, at ambient O2/CO2: 37 and 15 °C incubations were constant and regulated in incubators; 20 °C incubation was at room temperature.
Cell lysate and brain homogenate preparation and treatments
Tga20 mouse brain tissue (half brain, sectioned sagittally), and approximately 50 mg of human brain tissue taken from the occipital cortex of six sporadic Creutzfeldt-Jakob disease patients and six age-matched non-neurological controls, were prepared as a 10 % (w/v) homogenate in 1X PBS as described previously [44]. Where indicated, human brain homogenates were proteinase K (PK) digested (100 μg/ml final PK concentration, 1 hour at 37 °C, as described [44]) prior to PAGE and western blot analyses. Cell monolayers were washed with 1X PBS, prior to either harvesting (scraping into PBS and pelleting at 1000×g for 3 min) and lysing, or lysing directly in wells. Cells were lysed in ice-cold lysis buffer (25 mM Tris/HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1 % (v/v) Triton X-100) containing a final concentration of 1X Complete Ultra protease inhibitors (Roche) as described previously [45]. Post-nuclear supernatants were assessed for total protein content using a bicinchoninic acid protein assay (Pierce). Where required, cell lysates were treated for 2.5 h at 37 °C with 0.02 U final concentration PIPLC or PIPLC treated and then PNGaseF [46] digested, as described previously.
PAGE and immunoblotting
Samples were mixed with the appropriate PAGE sample buffer containing a final concentration of 3 % β-mercaptoethanol, and resolved on either 4–12 % acrylamide NuPAGE (Life Technologies) or 15.5 % acrylamide tricine SDS-PAGE gels, then electrotransferred to Hybond-P polyvinylidene difluoride membrane (PVDF). PVDF membranes were blocked for 1–2 h at room temperature in PBS containing 0.05 % Tween-20 (PBST) and either 5 % (w/v) skim milk powder, or 2 % (w/v) ECL blocking reagent, prior to incubation with the indicated primary antibody overnight at 4 °C (as indicated in figure legends). Online Resource 3 summarizes relevant antibody information. After washing off non-specifically bound primary antibody with PBST, membranes were incubated in peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies (GE), before further washes with PBST and detection using enhanced chemiluminescence (Pierce ECL substrate; Thermo Fischer Scientific, or ECL Advance; GE Healthcare). Where necessary (i.e. when quantifying a difference in total PrPC expression), to assess and correct for protein loading, membranes were stripped at low pH [1 % (v/v) aqueous HCl] for approximately 30 min, re-blocked and probed with an anti-β-tubulin or β-actin antibody and the secondary antibody described above, or were stained with Coomassie blue solution [50 % (v/v) methanol, 10 % (v/v) acetic acid, 0.25 % (w/v) Coomassie Brilliant Blue R-250 (BioRad)] for 1 min and de-stained [40 % (v/v) methanol, 10 % (v/v) acetic acid] for 10 min prior to quantification. All chemiluminescent and digital images were captured by a Fujifilm LAS-3000 Intelligent Dark Box.
Cell surface biotinylation and NeutrAvidin precipitation
Cell surface proteins were biotinylated for 30 min at 4 °C with the Pierce® Cell Surface Protein Isolation Kit as per the manufacturer’s instructions, with all solutions and reagents utilized scaled down to suit T25 cm2 flasks. Cells were harvested and pelleted at 500 × g for 3 min, immediately after biotinylation (T0) or were cultured for a further 6 h prior to collection of the cells (T6) and conditioned media (T6 M). Cell pellets were stored at −20 °C overnight, and media proteins were precipitated overnight at −20 °C in 4X volumes of 100 % methanol, and then pelleted at 4500×g max speed for 1 h at 4 °C. Cell and media pellets were lysed in 300 μl lysis buffer containing protease inhibitors as described above. Ten microlitres of lysates mixed with 10 μl of 2X NuPAGE sample buffer containing 100 mM dithiothreitol (DTT) (final DTT concentration of 50 mM) were utilized for ‘input’ samples in PAGE and western blotting. Lysates were diluted by mixing 150 μl of lysate with a further 150 μl of lysis buffer containing protease inhibitors, and then subjected to precipitation using 200 μl NeutrAvidin-coated agarose beads, as per the Pierce® Cell Surface Protein Isolation Kit instructions. Samples were eluted in 85 μl of 1X NuPAGE sample buffer containing 50 mM DTT for 1 h at room temperature, with 30 μl of this preparation used for PAGE and western blotting.
Density gradient ultracentrifugation
MycRK cells were harvested, lysed and subjected to Nycodenz density gradient flotation assay as described previously [39]. Fifteen microlitres of collected fractions was utilized for PAGE and western blotting as described above.
Densitometry and statistical analysis
Densitometric semi-quantitative analyses were carried out using Image J v1.42q, and routinely included background subtraction. C3 levels were always first adjusted for the total PrPC detected within a lane/sample, and then expressed relative to control samples. When total PrPC expression levels were compared, these were normalized to the relevant loading control (β-tubulin or Coomassie total protein, as indicated), and then expressed relative to the control samples. Statistical analyses were performed in GraphPad Prism v6.0d. All quantitative data is expressed as the mean ± SEM, with the number of independent experiment replicates (n) as indicated in figure legends.
Results
PrPC is cleaved in the far C-terminus, producing a novel cleavage fragment, C3
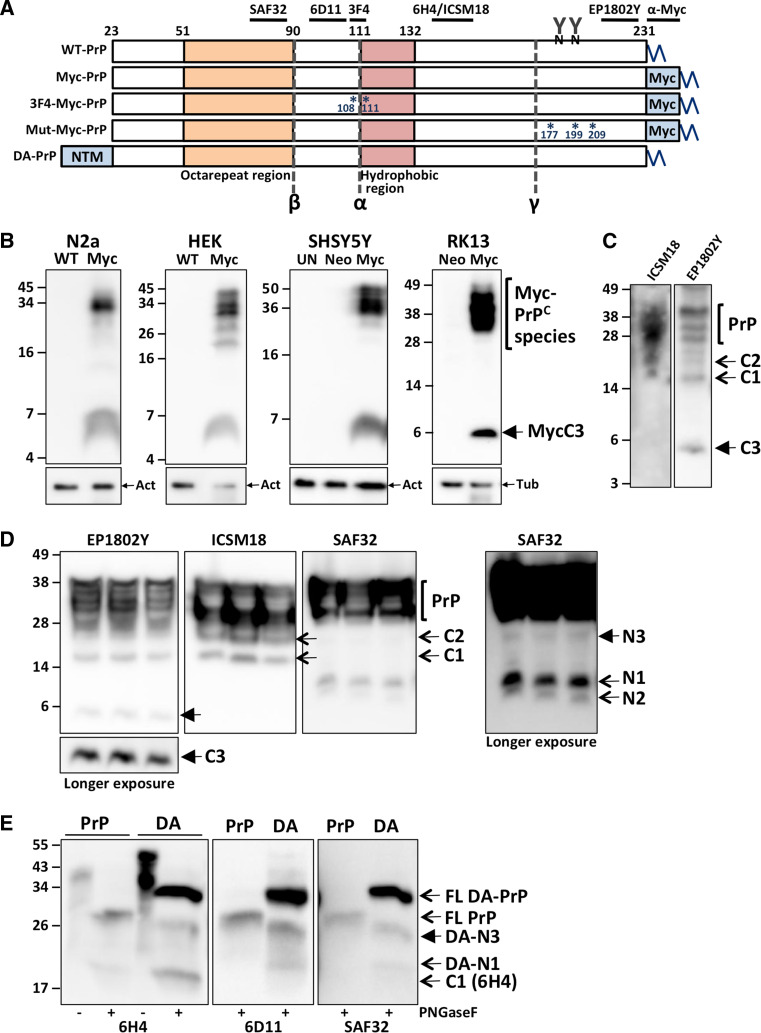
The addition of epitope tags into proteins is a commonly utilized and relatively simple and effective way to study their cellular biology, exploiting the convenience of high affinity antibodies, or intrinsic fluorescence of the tag. The introduction of epitope tags into PrPC which is then expressed in cells or animal tissues has been achieved several times, with different tags, and into different regions of the protein [47–51]. The nucleotide sequence encoding the 10 residue human c-myc epitope (EQKLISEEDL), along with the sequence encoding PrPC residues 226–230 duplicated immediately after the myc-tag, was engineered into murine PRNP immediately 5′ to the GPI-anchor signal sequence, generating ‘PrP-myc’. This PrP-myc construct has previously been shown to generate a PrPC fusion protein (MycPrP) with unaltered cellular behavior or function in vivo or in vitro, including glycosylation, proteolytic processing, trafficking, localization, membrane anchoring, and incorporation into PrPSc aggregates [52]. When whole lysates from the N2a cell line, transiently transfected with the PrP-myc construct (see Fig. 1a) were analyzed by PAGE and western blotting, in addition to the expected full-length and truncated MycPrPC species, a small, less than 10 kDa, myc-immunoreactive fragment was observed (Fig. 1b). This unexpected finding indicated there may be another PrPC endoproteolytic cleavage site towards the C-terminus, producing this small fragment, nominally ‘C3’. The PrP-myc construct was stably transfected into three other routine laboratory cell lines, human embryonic kidney (HEK), human neuroblastoma (SH-SY5Y) and rabbit kidney epithelial (RK13), to determine whether this processing was restricted to the N2a cells. In all cases, MycPrPC was also processed to C3 (Fig. 1b), indicating that cells derived from various mammalian species, and of neuronal and non-neuronal origin are capable of what we have designated PrPC ‘γ-cleavage’ and C3 production.
Fig. 1.
A far C-terminal PrPC fragment, C3, in cells and tissues of neuronal and non-neuronal origin. a Schematic representation of the constructs and alignment of the antibodies utilized in this study. Wild-type murine PrP (WT-PrP), containing the C-terminal Myc epitope-tag (Myc-PrP), the 3F4 epitope tag (3F4-Myc-PrP), the sites of three murine equivalents of familial CJD mutations D177N, E199K, V209I (Mut-Myc-PrP), the N-terminal anchor (NTM) in double anchored PrP (DA-PrP), the expected endoproteolytic cleavage sites (α, β, γ), and some key conserved features (octarepeat and hydrophobic domains, N-linked glycans, GPI-anchor), are depicted. PAGE and western blotting, with the anti-Myc (b) and anti-PrP (c–e) antibodies as indicated, of: b lysates from different cell lines (N2a, HEK, SHSY5Y, RK13) transfected with PrP-myc (Myc), the empty vector (Neo), wild-type mouse PrP (WT), or those left untransfected (UN); c lysate from untransfected N2a cells; d PBS brain homogenate from Tga20 (WT murine PrPC over-expressing) mice; e PNGaseF digested (+) or undigested control (−) lysates from SHSY5Y cells expressing wild-type (PrP) or double-anchored (DA) murine PrPC. The different full length (FL) and truncated PrP species are indicated, with the products of γ-cleavage highlighted with closed arrows, and all other cleavage events with open arrows. Membranes in b were stripped and re-probed with β-actin (Act) or β-tubulin (Tub) as indicated by the small arrows
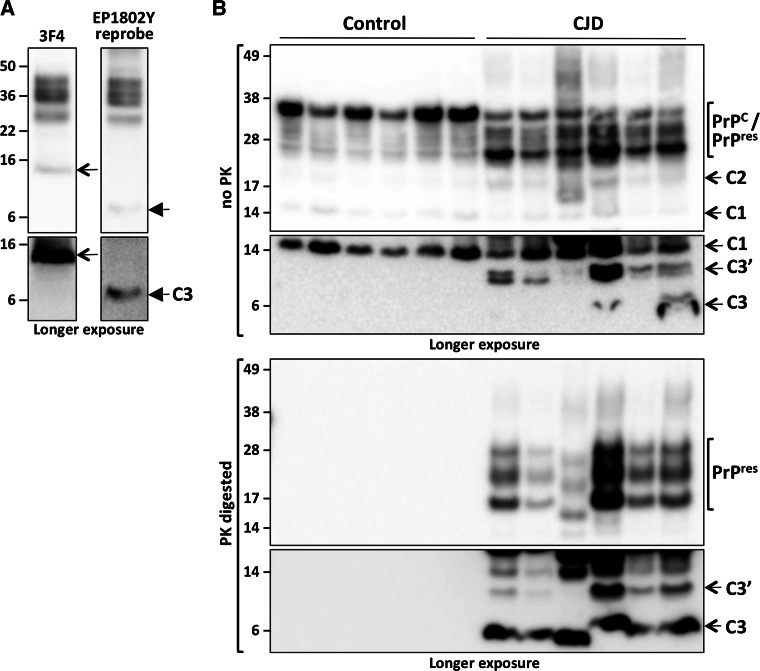
In order to establish whether this processing was an artifact of myc tag engineering, lysates from naïve N2a cells were analyzed by PAGE and western blotting utilizing a commercially available antibody, EP1802Y, raised against the PrP far C-terminus residues 214–230 (Fig. 1a). Once again, a small approximately 6–7 kDa fragment was observed (Fig. 1c), but this fragment was not seen when utilizing an antibody (ISCM18) directed against a comparatively more N-terminal region of the protein (Fig. 1a, c). To further confirm this observation, murine brain homogenates were also analyzed by PAGE and western blotting. Tga20 mouse brains were utilized as they over-express murine PrPC approximately six fold, therefore increasing the likelihood of detection of what appears to be a relatively low abundance fragment. When mouse brain proteins transferred to PVDF membranes were probed with antibodies to the PrPC N-terminus (SAF32) and mid-region (ICSM18), various PrPC species are observed, including the α- and β-cleavage products N1 and N2 when probing with SAF32 (Fig. 1d). However, it was only probing with EP1802Y that allowed detection of an approximately 6–7 kDa C-terminal fragment in these mouse brains (Fig. 1d).
The presumed N-terminal fragment, N3, based on the apparent size of C3, is predicted to be approximately 20 kDa. In the Tga20 mouse brains there was no obvious detection of N3 using the N-terminal PrPC antibody SAF32 with a short exposure, however upon prolonged exposure, an immunoreactive fragment of this approximate size was apparent (Fig. 1d, far right panel). Furthermore, the band attributed to C2 in the ICSM18 blot could potentially contain N3. Attempts to detect N3 in conditioned media from cultured cells were unsuccessful (data not shown). To increase the likelihood of detecting N3 in cell lysate, which may otherwise be rapidly secreted and/or degraded, we utilized SH-SY5Y cells expressing a ‘double anchored’ (DA) murine PrPC [43] (Fig. 1a), and wild-type murine PrPC expressing cells as controls. In the DA-PrPC construct, the N-terminal PrPC signal sequence has been replaced with the uncleaved signal sequence and transmembrane domain of murine aminopeptidase-A [43], thereby resulting in the tethering of the PrPC N-terminus to the cell membrane. When whole cell lysates from the DA-PrPC expressing cells were subject to PAGE and western blotting after PNGase digest to remove N-linked glycans, immunoreactive fragments consistent with N3 containing the aminopeptidase-A transmembrane domain (DA-N3), were detectable with all three anti-PrP antibodies utilized (Fig. 1a, e). N3 was not detectable in the control wild-type PrPC expressing cells consistent with rapid degradation of this fragment.
Collectively, these results strongly support that PrP γ-cleavage is a bona fide PrP processing event, prompting us to investigate further. Note, given MycPrP has been shown to behave the same as untagged (wild-type) PrP [52], including, as shown in Fig. 1, undergoing endoproteolysis producing the various PrP fragments, for ease of C3 detection we continued to utilize cells expressing MycPrP. From this point, MycPrP and its truncated species are for simplicity referred to without the prefix ‘Myc’.
PrPC γ-cleavage occurs late in the secretory pathway, preferentially from an unglycosylated substrate
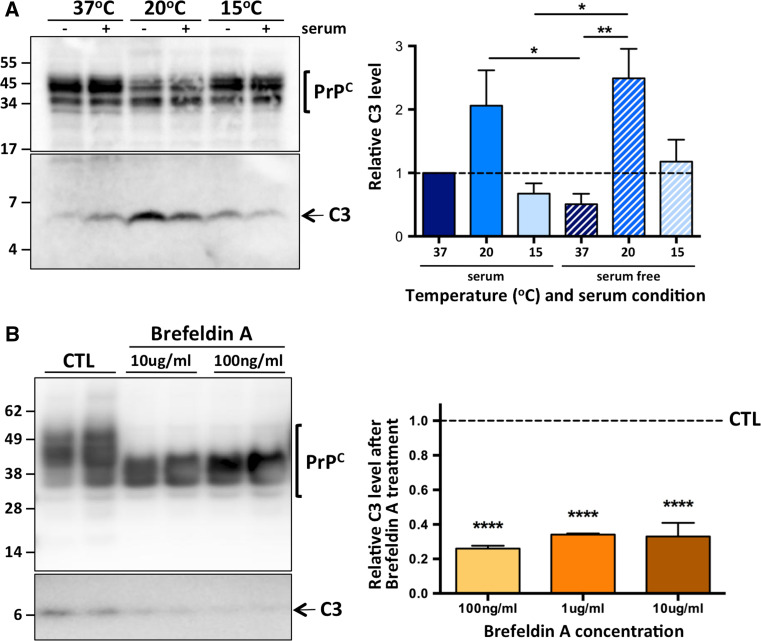
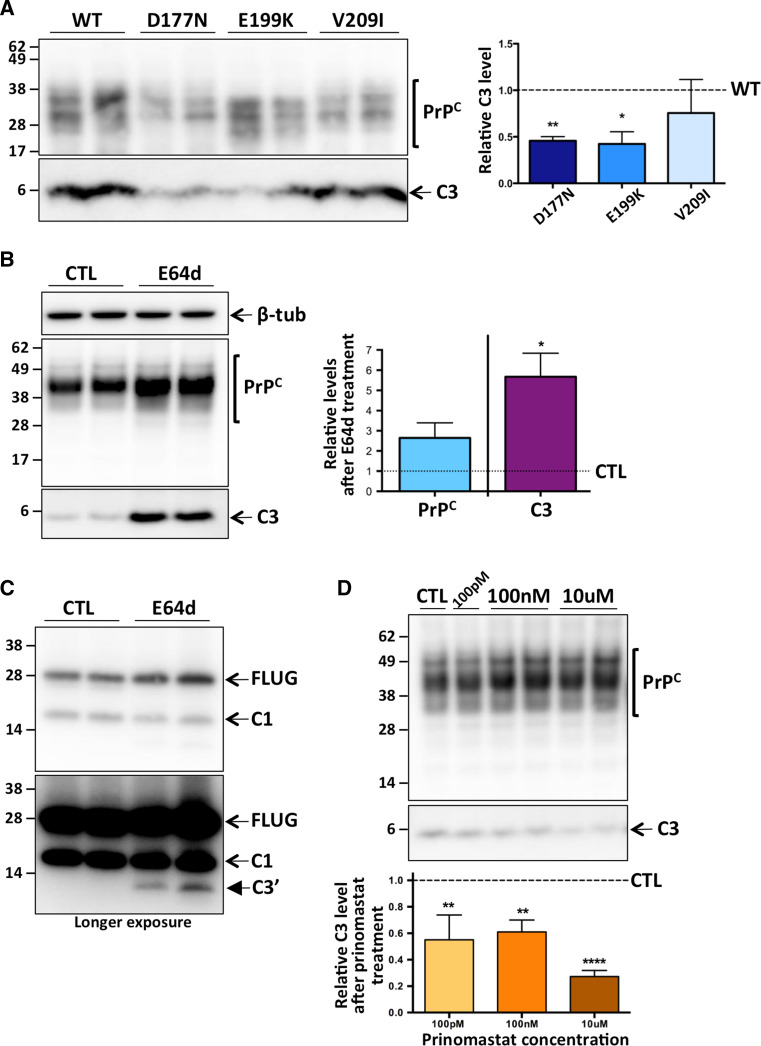
The cellular prion protein is a cell surface protein, and follows a typical pathway of trafficking and endocytosis. In order to determine at which stage during this secretory pathway γ-cleavage occurs, we first carried out temperature block experiments [53, 54], where RK13 cells stably expressing PrP-myc (MycRK) were incubated for 4 h at 15 °C [slowing protein traffic from the endoplasmic reticulum (ER)], 20 °C (slowing protein traffic from the Golgi apparatus) and 37 °C, in the presence or absence (to slow cell division and simulate oxidative stress) of serum. When compared to cells in optimal/normal conditions (37 °C + serum), cells where protein traffic was slowed through the ER showed no change in C3, while under stressed conditions, when protein traffic was held up in the Golgi network, there was a significant increase in C3 production, with a similar trend when cells were not stressed (Fig. 2a). To further investigate the cellular site of PrPC γ-cleavage we treated MycRK cells for 24 h with Brefeldin A, a reversible inhibitor of protein translocation from the ER to the Golgi complex [55]. As predicted, given PrPC N-linked glycosylation starts in the ER with attachment of high-mannose oligosaccharides, but continues in the Golgi with modification of these to more complex sugars [56], Brefeldin A treatment dramatically altered the glycosylation pattern of PrPC (Fig. 2b). Importantly, we found that Brefeldin A treatment significantly reduced C3 production.
Fig. 2.
γ-cleavage occurs after PrPC has exited the ER, in the Golgi/TGN. Representative PAGE and western blot with an anti-Myc primary antibody (left panels) and C3 quantification (right panels) of cell lysates from: a MycRK cells incubated in the presence (+) or absence (−) of 10 % v/v fetal bovine serum for 4 h at the temperatures indicated (n = 4); b duplicate wells of MycRK cells treated with Brefeldin A, at the concentrations indicated (100 ng/ml and 10 μg/ml n = 3; 1 μg/ml n = 2), or diluent only controls (CTL). Statistical analyses were by one way ANOVA with a Tukey’s multiple comparison post-test of all groups and b Dunnett’s multiple comparison post-test of each treatment to the control. *p < 0.05, **p < 0.01, ****p < 0.0001
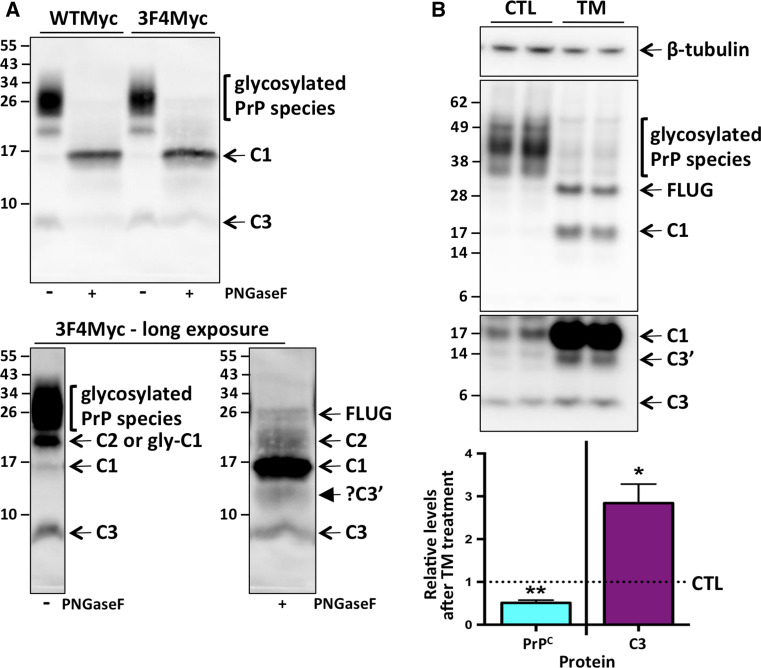
The well-described PrPC C-terminal proteolytic fragments C1 and C2 are known to be glycosylated like the full-length protein [36]. This can make interpretation of their abundance difficult, as glycosylated but truncated PrPC may have the approximate same molecular weight as unglycosylated full-length PrPC. Enzymatic removal of N-linked glycans from PrPC, through PNGaseF treatment of cell or tissue extracts is routinely used to more clearly visualize these fragments. To this end, we used PNGaseF to visualize all C-terminal PrPC fragments, including C3, in N2a cells transiently expressing myc-tagged wild-type murine PrPC, or 3F4 epitope containing murine PrPC, which is often utilized as a substitute wild-type murine sequence (Fig. 1a). It is interesting to note that in this cell line, irrespective of which construct was transfected, after PNGaseF treatment it becomes clear that there is very little full length (compared to truncated) PrPC, which has been described previously in some cultured cells [33]. This is unlikely to be due to the cells expressing myc-tagged PrPC, and rather reflects more the pattern of endogenous PrPC proteolytic processing in these cells (Online Resource 4). As expected, there is an increase in detectable C1, as glycosylated species have been reduced and all of C1 now migrates as a single band (Fig. 3a). However, there is neither the appearance of a smaller than the 6–7 kDa species that would indicate C3 had (prior to PNGaseF treatment) been glycosylated and therefore was resolving slower in PAGE, nor is there an increase in intensity of the 6–7 kDa C3 band that would indicate some of the C3 present in cells is ordinarily glycosylated and upon enzymatic deglycosylation becomes visible. Also of note, after PNGaseF treatment, with prolonged exposures we can observe a previously unrecognized faster moving species below C1 (closed arrow), which we have coined C3′.
Fig. 3.
C3 is not glycosylated and is preferentially cleaved from an unglycosylated substrate. a Lysates from N2a cells transiently transfected with wild-type or 3F4-epitope myc-tagged PrP construct as indicated, were left untreated (−) or digested with PNGaseF (+) prior to PAGE and western blotting with an anti-Myc antibody. The bottom panel is the same blot as the top panel, after a longer digital exposure time to enable visualization of additional cleavage fragments as indicated. b Representative PAGE and western blot with the anti-Myc antibody and re-probe with anti-β-tubulin antibody (top panel) and quantification (bottom panel, n = 4) of relative total PrPC and C3 levels in duplicate wells of MycRK cells treated with tunicamycin (TM), compared to diluent only controls (CTL). The various full length unglycosylated (FLUG) and truncated PrPC species are indicated. Statistical analyses by paired t test. *p < 0.05, **p < 0.01
From experience, and as evident in Figs. 1, 2 and 3a, most PrPC species in cultured cells and tissue extracts are heavily glycosylated. As C3 is not glycosylated and, similar to unglycosylated PrPC, is in low abundance relative to other PrPC species, we questioned whether C3 is cleaved preferentially from an unglycosylated source. When MycRK cells were treated with tunicamycin, a compound that blocks de novo N-linked glycosylation, we observed a significant increase in relative C3 levels (Fig. 3b). Interestingly, in the tunicamycin-treated cells there is again evidence of a PrPC fragment which is a few kilodaltons smaller than C1, which we postulate is the same C3′ fragment described in Fig. 3a.
The C-terminal fragment C3 is GPI-anchored, but does not reside primarily at the cell surface
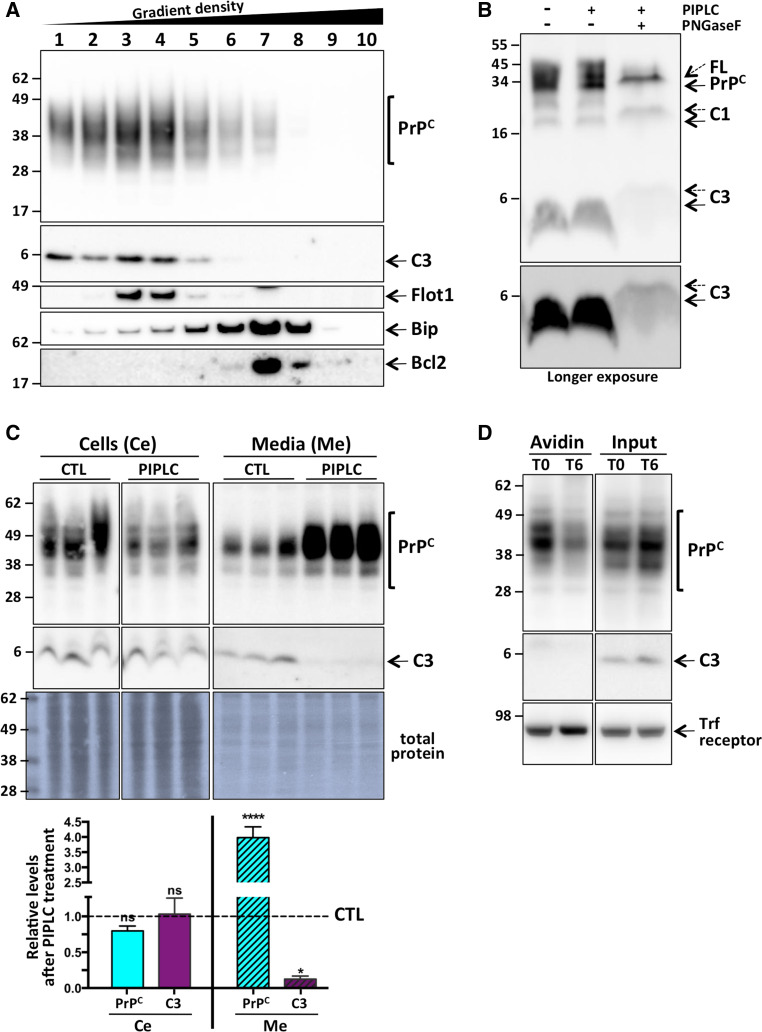
To further understand this PrPC γ-cleavage event, we next aimed to characterize the C3 fragment itself. It is well established that GPI-anchored proteins, such as PrPC, localize within lipid raft domains of cellular membranes [57]. In order to establish whether C3 was also GPI-anchored, we subjected lysates from MycRK cells to a floatation assay, whereby buoyant membranes (i.e. lipid raft domains) float to the top of a density gradient, following an established protocol [39]. When fractions from the MycRK cells were analyzed by PAGE and western blotting, the C3 fragment was localized predominantly in the more buoyant fractions where other PrPC species were also enriched (Fig. 4a), consistent with C3 containing a GPI moiety. The lipid raft (Flotillin-1), ER (Bip) and mitochondria (Bcl2) marker proteins localized predominantly in the lighter/buoyant (Flot1) and denser (Bip/Bcl2) fractions as expected [39]. Next, lysates from HEK cells transiently expressing MycPrP were reacted with bacterial phosphatidylinositol-specific phospholipase C (PIPLC), an enzyme capable of catalyzing hydrolytic cleavage of the phosphoric ester bond within the GPI anchor. When the GPI anchor is enzymatically removed from a protein, empirically on PAGE separation the GPI-minus protein resolves slower than its undigested counterpart [58]. To this end, a small characteristic upward shift in PAGE mobility of the C3 fragment, as well as the other PrPC species, was observed in the PIPLC and PNGaseF treated lysate (Fig. 4b), indicative of GPI-anchor removal.
Fig. 4.
C3 is GPI-anchored, and does not localize primarily at the cell surface. Representative PAGE and western blot with anti-Myc antibody of: a Density gradient fractionated lysate (fractions numbered 1–10) from MycRK cells, also probed with the marker protein antibodies (Flot1, Bip, Bcl2) as indicated; b lysates from HEK cells transiently expressing MycPrP, left untreated, digested with PIPLC, or PIPLC and PNGaseF. PrPC species [full length (FL) and truncated] with the GPI-anchor successfully removed are indicated with the dashed arrows; c cell lysate (Ce) and conditioned media (Me) from triplicate wells of live MycRK cells exposed to PIPLC or diluent only (CTL), and total protein (Coomassie) stained PVDF membrane (after completed western blotting), with quantification (bottom panel, n = 3) and statistical analyses by two way ANOVA with Bonferroni’s multiple comparison post-test. ns not significant, *p < 0.05, ****p < 0.0001; d whole cell lysate (Input) or NeutrAvidin-coated agarose bead precipitated (Avidin) proteins from cell surface biotinylated MycRK cells, immediately (T0) or 6 h (T6) after labeling, re-probed for the cell surface transferrin (Trf) receptor protein
Treating cells in culture with PIPLC induces cleavage of GPI-anchored proteins from the cell surface. When MycRK cells were treated for 1 h with PIPLC, as expected there was an increase in the detection of PrPC in the conditioned media; however, this was not observed for the C3 fragment (Fig. 4c). Interestingly, levels of C3 in conditioned media were in fact significantly decreased. To further confirm whether C3 is localized at the cell surface, we carried out cell-surface biotinylation of MycRK cells. Using NeutrAvidin-coated agarose beads to bind and pull out all labeled cell surface proteins from whole cell lysate or conditioned media, Fig. 4d shows that whilst the cell surface transferrin receptor and many PrPC species were biotinylated, C3 was not. Online Resource 5 shows the full biotinylation experiment result depicted in Fig. 4d, including some relatively low level non-specific NeutrAvidin pull down of unlabelled proteins (both PrPC and transferrin receptor), seen only on longer exposure, and no precipitation of biotinylated proteins from the conditioned media. Online Resource 5 also shows that upon longer exposure, both C3 and C3′ can be detected in the whole cell lysate (input), but not in the NeutrAvidin pull down samples.
A matrix metalloprotease is responsible for PrPC γ-cleavage within the highly conserved C-terminus
The observations of C3 approximate molecular mass at 6–7 kDa, and the C3/N3 immuno-reactivities, suggested that the γ-cleavage site was in the far-C-terminus of PrPC. Interestingly the C-terminus of PrPC is highly conserved in mammals [59], with substantial sequence identity in the various species listed in the National Center for Biotechnology Information (NCBI) Conserved Domains Database [60] (Online Resource 6). In order to define the sequence identity of C3 and clarify the γ-cleavage site, several attempts, including the use of size-exclusion and immunoprecipitation techniques to increase relative detection/yield of Myc-C3 for downstream mass spectrometry or Edman degradation analyses, as well as antibody affinity based SELDI-TOF–MS, unfortunately all yielded negative or inconclusive results (data not shown). Therefore, we employed site-directed mutagenesis to potentially introduce either slight structural/steric changes or loss of a protease consensus sequence motif, in order to hinder and/or alter any interaction of PrPC with the responsible protease, altering the efficiency of C3 production and narrowing down the putative region of the γ-cleavage site. Specifically, the murine equivalents of three inherited human prion disease mutations which are spread across the far-C-terminus of PrPC were created, with all containing the myc tag (see Fig. 1a); MycD177N, MycE199K and MycV209I. After transient transfection into RK13 cells, we observed a significant reduction in C3 levels in cells expressing MycD177N and MycE199K compared to wild-type MycPrPC (Fig. 5a), whilst there was no significant difference seen for the MycV209I mutation.
Fig. 5.
An MMP not a cysteine protease is responsible for γ-cleavage within the conserved PrPC far C-terminus. Representative PAGE and western blot with anti-Myc antibody of: a lysates from RK13 cells transiently expressing wild-type (WT) or mutated, myc-tagged PrPC, with quantification (right panel, n = 4); b lysates from MycRK cells treated for 72 h with E64d or diluent only control (CTL), re-probed for β-tubulin detection, with quantification of total PrPC levels and C3 levels adjusted for PrPC (right panel, n = 4); c PNGaseF digested whole cell lysates from duplicate wells of E64d or diluent control (CTL) treated MycRK cells (as in b), with full length unglycosylated (FLUG) and truncated PrPC species as indicated; d lysates from Prinomastat (concentrations as indicated, 100 pM n = 2, 100 nM n = 3, 10 μM n = 4) or diluent control (CTL) treated MycRK cells (in some cases in duplicate wells), with quantification (bottom panel). Statistical analyses in a and d were by one way ANOVA with Dunnett’s multiple comparison post-test of each mutant or treatment to the control, and in b were by paired t test. *p < 0.05, **p < 0.01, ****p < 0.0001
A single previous report does identify and describe a small C-terminal PrP fragment of similar apparent molecular mass, which the authors also referred to as ‘C3’ and may be the same C3 described herein [50]. Taguchi and colleagues determined their fragment to result from cysteine protease cleavage, as a 72 h treatment with a pan-cysteine protease inhibitor E64 inhibited its production. In order to test whether our C3 was also generated by a cysteine protease, MycRK cells were treated for 72 h with E64d, a more membrane permeable synthetic analogue of E64. Surprisingly the MycRK cells had increased total PrPC expression after E64d treatment, and when adjusted for the increase in PrPC, C3 levels were also significantly increased (Fig. 5b). We considered whether the increased C3 was due to E64d increasing the proportion of a precursor fragment. Lysates from E64d treated and untreated cells were PNGaseF digested prior to PAGE and western blot analysis, in order to clearly see the relative levels of the different full-length and truncated MycPrPC species. As seen in Fig. 5c, there is no apparent change in proportion of the C1 fragment, although there is again the appearance of a faster moving MycPrPC species with E64d treatment. We believe this is consistent with C3′, observed in the various experiments described above.
Having ruled out a cysteine protease as the likely enzyme involved in PrPC γ-cleavage, we utilized the MEROPS database (http://merops.sanger.ac.uk) [61] to perform low stringency searches of residues around the highly conserved residues of the PrPC far C-terminus. Numerous proteases were identified, and of interest were several matrix metalloproteases (MMP), due to recent evidence that prion protein fragments can be digested by membrane-type MMP proteases [62]. To investigate the possibility of MMP mediated γ-cleavage of PrPC, we treated MycRK cells for 24 h with the MMP inhibitor Prinomastat, which is selective for MMPs-2, 3, 9, 13 and 14 [63]. We observed a significant decrease in relative C3 levels for all concentrations of Prinomastat tested (Fig. 5d), suggesting an MMP is involved in PrPC γ-cleavage.
Prion disease-associated PrP conformers are also susceptible to γ-cleavage
During a routine diagnostic characterization of PrP species detectable in a suspected Creutzfeldt-Jakob disease (CJD) patient brain for the ANCJDR, a fragment consistent with C3 was detected when utilizing the EP1802Y antibody, but not the mid-region 3F4 antibody (Fig. 6a), which indicated the human prion protein may be susceptible to γ-cleavage. In order to further characterize this, and also address whether prion disease associated PrP species are also subject to γ-cleavage, we assessed C3 production in non-neurological control and sporadic CJD patient brain tissue. While we found no clear evidence of γ-cleavage in control tissue, this was not the case in CJD brains, with detection of C3 (2/6 cases) and C3′ (6/6 cases) (Fig. 6b, top panels). Additionally, after high concentration PK treatment successfully digested away all PrPC species, in addition to the typical profile of di- mono- and un-glycosylated partially protease-resistant (PrPres) species, C3 becomes readily detectable in all the CJD brains (Fig. 6b, bottom panels). Interestingly, C3′ is also still detectable after PK digestion. Collectively this indicates that in the context of disease-associated PrP misfolding, C3 represents a highly protease-resistant unglycosylated fragment, with γ-cleavage of disease associated PrP (PrPSc) occurring, and the likelihood that the C3 seen in non-PK-treated CJD brain tissue is resultant from endoproteolysis of PrPSc rather than processing of the PrPC present.
Fig. 6.
C3 is present in human sporadic CJD brains. PAGE and western blot analysis of human brain tissue utilizing the anti-PrP far C-terminal EP1802Y (a) and (b) or 3F4 (a) antibodies as indicated. Five microliters of 10 % (w/v) CJD (a) and (b) or non-neurological control (b) brain homogenate was utilized untreated (a and b upper panel), or PK digested (b bottom panel), prior to analysis. The various full-length and truncated PrP species detectable (PrP C, PrP res, C1, C2, C3, C3′) are highlighted
Discussion
Prion protein endoproteolysis, a phenomenon observed in cultured cells as well as human and various animal tissues, has been reported for many years, with a focus on α- and β-cleavage and proteolysis at the GPI anchor resulting in shedding of PrPC from the cell surface [29]. Despite this, the biological or physiological significance of PrPC proteolytic processing has not been entirely elucidated, with emerging, but sometimes conflicting evidence of separate functions of the full length [14, 64, 65], and different truncated PrPC species [6, 10, 66–68]. Importantly, there are often more truncated PrPC species than full-length PrPC present in cells and tissues [33, 35, 69], underscoring the likely cellular requirement for these processes to occur.
Through the utilization of Myc-tagged PrPC expressed in cells we discovered a novel, approximately 6–7 kDa, Myc-tagged ‘C3’ fragment, and then confirmed the presence of endogenous C3 in both human and animal brain tissue, indicating the fragment was not produced as an artifact of epitope tag insertion, and thus the unequivocal existence of a PrPC γ-cleavage event. Extensive literature searching has found that the most commonly utilized anti-PrP antibodies are directed to epitopes in the middle region of PrPC, and unlikely to be within the C3 fragment. Furthermore, published western blot images are often trimmed, underscoring the possibility that the occurrence of PrPC cleavage events, especially those producing small far-C-terminal fragments such as C3, may be easily overlooked. The apparent approximate molecular weight of C3 seen on PAGE and western blotting, along with the significant reduction in C3 production with the mutation of two residues within the PrPC far C-terminus, are highly suggestive of the γ-cleavage site being within this region, potentially somewhere between residues 176 and 200. Interestingly the C-terminus of PrPC is highly conserved in mammals [59], with 100 % amino acid sequence identity in residues 190–202 and 54 % identity in the larger region spanning residues 170–230 (human PrP numbering) in the various species listed in the National Center for Biotechnology Information (NCBI) Conserved Domains Database [60], including humans and other primates, cervids, bovids, equids, felids, cetaceans, lagomorphs and rodents, further pointing to the likely biological significance of γ-cleavage. Additionally, if our predictions of the γ-cleavage site hold true, and when direct cleavage to C3 occurs, the resulting potentially secreted N3 fragment could represent approximately 70 % of the span of previously described secreted GPI-anchorless PrPC species, including important structural features such as the copper binding octapeptide repeat and the hydrophobic core (Fig. 1a), which may have biological/functional relevance [70–72].
After synthesis, the majority of immature PrPC produced may undergo post-translational modifications en route to the cell surface, including addition of its GPI-anchor in the ER, and simple, followed by more complex, glycosylation of one or two asparagine (N) residues (codons 181 and 197 on human PrPC; 180 and 196 on murine PrPC) in the ER and Golgi, respectively [56], resulting in predominantly highly glycosylated PrPC species. Later in the secretory pathway, after transiting the Golgi apparatus, a large proportion (in most cell types) of PrPC is subjected to the dominant endoproteolytic processing event, α-cleavage, in the trans-Golgi network (TGN) [54]. Our results indicate that PrPC also needs to exit the ER for efficient γ-cleavage, and that this endoproteolysis occurs in the Golgi complex or TGN. Furthermore, our observation of reduced cellular C3 levels in PIPLC treated cells is consistent with the phospholipase-induced loss of GPI-anchored PrPC species from the cell surface diminishing an available substrate for γ-cleavage. Collectively, this indicates γ-cleavage also occurs after PrPC has transited to the cell surface, perhaps during endocytic recycling and/or retrograde transport of PrPC to the Golgi/TGN, trafficking pathways that PrP isoforms are known to follow [73, 74]. Our data also suggest the C3 fragment itself is not glycosylated, and consistent with this, the protease responsible for γ-cleavage favors an unglycosylated substrate. Intriguingly, as the appearance of another C-terminal fragment, apparently slightly smaller than C1, sometimes coincided with increased C3 detection (for example after tunicamycin or E64d treatment), we believe γ-cleavage does not occur downstream of α-cleavage, and that C3 may not always be cleaved directly from full length PrPC, but perhaps sometimes from an intermediary or precursor fragment, which we have coined C3′. Collectively these results may partly explain the apparently relatively low levels of the C3 fragment observed; we propose that proportionately low levels of PrPC remain unglycosylated, and/or escape either α- or β-cleavage and remain full length, are not shed or recycled back to the cell surface, and could therefore persist long enough to be transported back to the Golgi/TGN where γ-cleavage is then able to occur. Importantly, as is the case with many other low abundance proteins, we believe the relatively low levels do not militate against the potential importance of this fragment or cleavage event.
Similar to full length PrPC, the α- and β-cleavage C-terminal fragments C1 and C2 are known to be GPI-anchored [30]. After treatment of cells with the bacterial enzymes PIPLC and PNGaseF, a subtle size shift characteristic of GPI-anchor removal was observed in C3, along with the other PrPC species expressed in the cells. It is unclear why PIPLC treatment alone did not result in this same discernible shift, though this may be related to the GPI-anchor attachment residues and PIPLC cleavage recognition motif being more accessible once the substrate protein has been denatured, which does occur during the conditions of PNGaseF digestion. These results, along with the detection of C3 in buoyant density gradient fractions suggest that C3 does retain its GPI-anchor. However, the lack of an observed C3 increase in conditioned media after PIPLC treatment of live cells, coupled to the lack of detection of labeled C3 after cell surface biotinylation, together indicates this fragment is unlikely to reside predominantly at the cell surface. These results are also consistent with γ-cleavage occurring after retrograde transport to the Golgi or TGN, but do not exclude contributions from nascent PrPC coming directly from the ER. The possibility that the C3 fragment itself does not contain enough lysine residues, with amine side chains to react with the biotin, is unlikely given the predicted γ-cleavage region, although this cannot be excluded.
A single report does identify and describe a PrPC-derived C-terminal fragment which we originally hypothesized may be the same C3 described herein [50]. The fragment described by Taguchi and colleagues was discovered during the development of a biarsenical cell surface protein labeling technique, using PrP as the model protein. This, along with our conflicting finding that γ-cleavage was not reduced by inhibition of cysteine proteases using E64d, collectively indicates the C3 fragment we have characterized is in fact not that which has been described previously, and further highlights the complexity of proteolytic processing of the prion protein far C-terminus. Whilst this study has ruled out the involvement of a cysteine protease in γ-cleavage, we have elucidated that a member/members of the matrix metalloproteinases (MMP) family of proteases are involved. Interestingly a recent report describes the proteolysis of various synthetic PrP fragments with MMP7, MMP14 (membrane-type MMP1, or MT1-MMP) and MMP16 (MT3-MMP), with some of the reported cleavage sites mapping to the far C-terminus of PrPC between residues 169–192 [62], consistent with the region we predict to contain the γ-cleavage site. The determination of which specific MMP/s are involved is the subject of ongoing investigations.
Although prevalence of PrPC γ-cleavage is not high, the lack of C3 seen in human control brain tissue was surprising. One explanation may be that γ-cleavage does not occur in cells of the brain region we sampled from, as cleavage profiles have been shown to vary in different tissues and regions [69]. Alternatively, C3 may be below the limits of detection by PAGE and western blotting. Whatever the explanation, our results indicate that whilst human cells are capable of accommodating γ-cleavage of murine PrPC (e.g. the HEK and SHSY5Y cells in Fig. 1b), in vivo human PrPC is perhaps less susceptible to this cleavage event compared to murine PrPC. Surprisingly, PK resistant C3 and C3′ species were relatively abundant in CJD brain, indicating PrPSc may be more susceptible than PrPC to γ-cleavage. Small, approximately 7–15 kDa, PK resistant fragments have previously been detected in human prion brain tissue (and as illustrated by Fig. 6a), particularly in the inherited Gerstmann–Sträussler–Scheinker (GSS) disease cases [75, 76], although these are typically characterized as both N- and C-terminally truncated, and are therefore not the C3 species we describe. There is published evidence of small far-C-terminal PK resistant fragments in sporadic CJD, but not control brain tissue [77], which the authors designate as PrP-CTF12/13 to reflect their approximate size. Similar to our PrPC-derived C3, PrP-CTF12/13 are also GPI-anchored. Moreover, the N-terminus of the PrP-CTF12 fragment mapped to residues 162 and 167, which is consistent with our estimates of the γ-cleavage site, and which based on amino acid sequence alone (i.e. codons 162–230 and 167–230) are predicted to result in C-terminal fragments of approximately 8.3 and 7.6 kDa, respectively (excluding the GPI-anchor, which when attached should actually result in slightly faster than expected resolution [58]). With PAGE protocol differences (e.g., acrylamide concentrations) and the inherent inexact nature of molecular weight standard alignment for predicting actual protein size (especially when the smallest marker is larger than the protein of interest) accounting for discrepancies, we believe these PrP-CTF12/13 species may be the same PrPSc derived PK resistant C3 and perhaps also C3′ that we have detected.
In the context of prion disease pathogenesis, experimental evidence has proven that expression of the normal cellular prion protein, PrPC, is an absolute requirement [78, 79], where it aberrantly folds to the disease associated prion protein isoforms (PrPSc). Although the mechanisms underlying initiation of PrPC misfolding are not understood, convention proposes the continued hetero-dimeric interaction, templating and conversion of PrPC to PrPSc with eventual propagation of sufficient neurotoxic and infectious species ultimately causing cell death and neurodegeneration. Whilst it is known that the N-terminus of PrPC is not required for production of PrPSc [80], we and others [33, 37, 38, 81, 82] have linked reduced susceptibility to prion infection or efficiency of conversion/misfolding, to PrPC cleavage/truncation, probably due to the depletion of full-length PrPC which is the optimal substrate for conversion to PrPSc. Interestingly, a recent report has also linked decreased expression and activity of MMPs 2 and 9 to increased prion propagation [83]. Given the likely involvement of MMPs in PrPC γ-cleavage, it remains possible that γ-cleavage may be relevant in cellular mechanisms of susceptibility to prion propagation. Indeed, the apparent increase in γ-cleavage as indicated by the proportionally higher amounts and the PK resistance of C3 and C3′ in the brains of CJD patients may reflect increased and ultimately failed cellular attempts at managing the ongoing prion propagation through efforts to cleave and therefore degrade PrPSc.
Many areas of prion biology remain enigmatic, with considerable conflicting data published, perhaps in part due to the under-appreciation of processes such γ-cleavage. The primary objective of this study was to characterize the cellular biology of this novel PrPC γ-cleavage processing, and further contribute to the understanding of currently incompletely answered questions in prion research. Whilst the exact biological and/or pathophysiological significance of γ-cleavage remain to be determined, the results we have presented herein, along with mounting evidence of the relevance of prion protein cleavage to both its normal function, and the important influence exerted in susceptibility to prion disease, underscore that this previously un-appreciated cleavage event warrants further investigation.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors thank Associate Professor Victoria Lawson for her kind gift of the RK13 cells, Professor Charles Weissmann for the Tga20 mice, and the Victorian Brain Bank Network for their assistance in human control brain tissue sampling. This work was supported by an Australian Government National Health and Medical Research Council (NHMRC) Program Grant #628946 (SJC, VL, VJ), Practitioner Fellowship #1005816 (SJC), Training Fellowship #567123 (VL), R.D. Wright Fellowship (CDF2) (PJC) and Project Grant #1061550 (PJC), a University of Melbourne Early Career Researcher Fellowship (VL), the CJD Support Group Network (VL), and the Medical Research Council of Great Britain (G0802189) (NMH).
References
- 1.Borchelt DR, Rogers M, Stahl N, Telling G, Prusiner SB. Release of the cellular prion protein from cultured cells after loss of its glycoinositol phospholipid anchor. Glycobiology. 1993;3(4):319–329. doi: 10.1093/glycob/3.4.319. [DOI] [PubMed] [Google Scholar]
- 2.Bendheim PE, Brown HR, Rudelli RD, Scala LJ, Goller NL, Wen GY, Kascsak RJ, Cashman NR, Bolton DC. Nearly ubiquitous tissue distribution of the scrapie agent precursor protein. Neurology. 1992;42(1):149–156. doi: 10.1212/WNL.42.1.149. [DOI] [PubMed] [Google Scholar]
- 3.Ford MJ, Burton LJ, Morris RJ, Hall SM. Selective expression of prion protein in peripheral tissues of the adult mouse. Neuroscience. 2002;113(1):177–192. doi: 10.1016/S0306-4522(02)00155-0. [DOI] [PubMed] [Google Scholar]
- 4.Horiuchi M, Yamazaki N, Ikeda T, Ishiguro N, Shinagawa M. A cellular form of prion protein (PrPC) exists in many non-neuronal tissues of sheep. J Gen Virol. 1995;76(10):2583–2587. doi: 10.1099/0022-1317-76-10-2583. [DOI] [PubMed] [Google Scholar]
- 5.Bueler H, Fischer M, Lang Y, Bluethmann H, Lipp HP, DeArmond SJ, Prusiner SB, Aguet M, Weissmann C. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356(6370):577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 6.Haigh CL, Drew SC, Boland MP, Masters CL, Barnham KJ, Lawson VA, Collins SJ. Dominant roles of the polybasic proline motif and copper in the PrP23-89-mediated stress protection response. J Cell Sci. 2009;122(10):1518–1528. doi: 10.1242/jcs.043604. [DOI] [PubMed] [Google Scholar]
- 7.Brown DR, Nicholas RS, Canevari L. Lack of prion protein expression results in a neuronal phenotype sensitive to stress. J Neurosci Res. 2002;67(2):211–224. doi: 10.1002/jnr.10118. [DOI] [PubMed] [Google Scholar]
- 8.Chiarini LB, Freitas AR, Zanata SM, Brentani RR, Martins VR, Linden R. Cellular prion protein transduces neuroprotective signals. EMBO J. 2002;21(13):3317–3326. doi: 10.1093/emboj/cdf324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klamt F, Dal-Pizzol F, Conte da Frota MJ, Walz R, Andrades ME, da Silva EG, Brentani RR, Izquierdo I, Fonseca Moreira JC. Imbalance of antioxidant defense in mice lacking cellular prion protein. Free Radic Biol Med. 2001;30(10):1137–1144. doi: 10.1016/S0891-5849(01)00512-3. [DOI] [PubMed] [Google Scholar]
- 10.Haigh CL, Lewis VA, Vella LJ, Masters CL, Hill AF, Lawson VA, Collins SJ. PrPC-related signal transduction is influenced by copper, membrane integrity and the alpha cleavage site. Cell Res. 2009;19(9):1062–1078. doi: 10.1038/cr.2009.86. [DOI] [PubMed] [Google Scholar]
- 11.Mouillet-Richard S, Ermonval M, Chebassier C, Laplanche JL, Lehmann S, Launay JM, Kellermann O. Signal transduction through prion protein. Science. 2000;289(5486):1925–1928. doi: 10.1126/science.289.5486.1925. [DOI] [PubMed] [Google Scholar]
- 12.Schneider B, Mutel V, Pietri M, Ermonval M, Mouillet-Richard S, Kellermann O. NADPH oxidase and extracellular regulated kinases 1/2 are targets of prion protein signaling in neuronal and nonneuronal cells. Proc Natl Acad Sci USA. 2003;100(23):13326–13331. doi: 10.1073/pnas.2235648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spielhaupter C, Schatzl HM. PrPC directly interacts with proteins involved in signaling pathways. J Biol Chem. 2001;276(48):44604–44612. doi: 10.1074/jbc.M103289200. [DOI] [PubMed] [Google Scholar]
- 14.Kanaani J, Prusiner SB, Diacovo J, Baekkeskov S, Legname G. Recombinant prion protein induces rapid polarization and development of synapses in embryonic rat hippocampal neurons in vitro. J Neurochem. 2005;95(5):1373–1386. doi: 10.1111/j.1471-4159.2005.03469.x. [DOI] [PubMed] [Google Scholar]
- 15.Santuccione A, Sytnyk V, Leshchyns’ka I, Schachner M. Prion protein recruits its neuronal receptor NCAM to lipid rafts to activate p59fyn and to enhance neurite outgrowth. J Cell Biol. 2005;169(2):341–354. doi: 10.1083/jcb.200409127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colling SB, Collinge J, Jefferys JG. Hippocampal slices from prion protein null mice: disrupted Ca(2+)-activated K+ currents. Neurosci Lett. 1996;209(1):49–52. doi: 10.1016/0304-3940(96)12596-9. [DOI] [PubMed] [Google Scholar]
- 17.Collinge J, Whittington MA, Sidle KC, Smith CJ, Palmer MS, Clarke AR, Jefferys JG. Prion protein is necessary for normal synaptic function. Nature. 1994;370(6487):295–297. doi: 10.1038/370295a0. [DOI] [PubMed] [Google Scholar]
- 18.Herms JW, Tings T, Dunker S, Kretzschmar HA. Prion protein affects Ca2+-activated K+ currents in cerebellar purkinje cells. Neurobiol Dis. 2001;8(2):324–330. doi: 10.1006/nbdi.2000.0369. [DOI] [PubMed] [Google Scholar]
- 19.Mallucci GR, Ratte S, Asante EA, Linehan J, Gowland I, Jefferys JG, Collinge J. Post-natal knockout of prion protein alters hippocampal CA1 properties, but does not result in neurodegeneration. EMBO J. 2002;21(3):202–210. doi: 10.1093/emboj/21.3.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powell AD, Toescu EC, Collinge J, Jefferys JG. Alterations in Ca2+-buffering in prion-null mice: association with reduced afterhyperpolarizations in CA1 hippocampal neurons. J Neurosci. 2008;28(15):3877–3886. doi: 10.1523/JNEUROSCI.0675-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mastrangelo P, Westaway D. Biology of the prion gene complex. Biochem Cell Biol. 2001;79(5):613–628. doi: 10.1139/o01-142. [DOI] [PubMed] [Google Scholar]
- 22.Bazan JF, Fletterick RJ, McKinley MP, Prusiner SB. Predicted secondary structure and membrane topology of the scrapie prion protein. Protein Eng. 1987;1(2):125–135. doi: 10.1093/protein/1.2.125. [DOI] [PubMed] [Google Scholar]
- 23.Sulkowski E. Aromatic palindrome motif in prion proteins. FASEB J. 1992;6(6):2363. doi: 10.1096/fasebj.6.6.1544547. [DOI] [PubMed] [Google Scholar]
- 24.Wopfner F, Weidenhofer G, Schneider R, von Brunn A, Gilch S, Schwarz TF, Werner T, Schatzl HM. Analysis of 27 mammalian and 9 avian PrPs reveals high conservation of flexible regions of the prion protein. J Mol Biol. 1999;289(5):1163–1178. doi: 10.1006/jmbi.1999.2831. [DOI] [PubMed] [Google Scholar]
- 25.Goldmann W. PrP gene and its association with spongiform encephalopathies. Br Med Bull. 1993;49(4):839–859. doi: 10.1093/oxfordjournals.bmb.a072650. [DOI] [PubMed] [Google Scholar]
- 26.Parkin ET, Watt NT, Turner AJ, Hooper NM. Dual mechanisms for shedding of the cellular prion protein. J Biol Chem. 2004;279(12):11170–11178. doi: 10.1074/jbc.M312105200. [DOI] [PubMed] [Google Scholar]
- 27.Taylor DR, Parkin ET, Cocklin SL, Ault JR, Ashcroft AE, Turner AJ, Hooper NM. Role of ADAMs in the ectodomain shedding and conformational conversion of the prion protein. J Biol Chem. 2009;284(34):22590–22600. doi: 10.1074/jbc.M109.032599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altmeppen HC, Prox J, Puig B, Kluth MA, Bernreuther C, Thurm D, Jorissen E, Petrowitz B, Bartsch U, De Strooper B, Saftig P, Glatzel M. Lack of a-disintegrin-and-metalloproteinase ADAM10 leads to intracellular accumulation and loss of shedding of the cellular prion protein in vivo. Mol Neurodegener. 2011;6:36. doi: 10.1186/1750-1326-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis V. Proteolytic processing of the prion protein. In: Collins SJ, Lawson VA, editors. The cellular and molecular biology of prion disease. Kerala: Research Signpost; 2011. pp. 53–71. [Google Scholar]
- 30.Chen SG, Teplow DB, Parchi P, Teller JK, Gambetti P, Autilio-Gambetti L. Truncated forms of the human prion protein in normal brain and in prion diseases. J Biol Chem. 1995;270(32):19173–19180. doi: 10.1074/jbc.270.32.19173. [DOI] [PubMed] [Google Scholar]
- 31.McDonald AJ, Dibble JP, Evans EG, Millhauser GL. A new paradigm for enzymatic control of alpha-cleavage and beta-cleavage of the prion protein. J Biol Chem. 2014;289(2):803–813. doi: 10.1074/jbc.M113.502351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan T, Wong P, Chang B, Li C, Li R, Kang SC, Wisniewski T, Sy MS. Biochemical fingerprints of prion infection: accumulations of aberrant full-length and N-terminally truncated PrP species are common features in mouse prion disease. J Virol. 2005;79(2):934–943. doi: 10.1128/JVI.79.2.934-943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis V, Hill AF, Haigh CL, Klug GM, Masters CL, Lawson VA, Collins SJ. Increased proportions of C1 truncated prion protein protect against cellular M1000 prion infection. J Neuropathol Exp Neurol. 2009;68(10):1125–1135. doi: 10.1097/NEN.0b013e3181b96981. [DOI] [PubMed] [Google Scholar]
- 34.Mange A, Beranger F, Peoc’h K, Onodera T, Frobert Y, Lehmann S. Alpha- and beta-cleavages of the amino-terminus of the cellular prion protein. Biol Cell. 2004;96(2):125–132. doi: 10.1016/j.biolcel.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Jimenez-Huete A, Lievens PM, Vidal R, Piccardo P, Ghetti B, Tagliavini F, Frangione B, Prelli F. Endogenous proteolytic cleavage of normal and disease-associated isoforms of the human prion protein in neural and non-neural tissues. Am J Pathol. 1998;153(5):1561–1572. doi: 10.1016/S0002-9440(10)65744-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan T, Li R, Wong BS, Liu T, Gambetti P, Sy MS. Heterogeneity of normal prion protein in two- dimensional immunoblot: presence of various glycosylated and truncated forms. J Neurochem. 2002;81(5):1092–1101. doi: 10.1046/j.1471-4159.2002.00909.x. [DOI] [PubMed] [Google Scholar]
- 37.Westergard L, Turnbaugh JA, Harris DA. A naturally occurring C-terminal fragment of the prion protein (PrP) delays disease and acts as a dominant-negative inhibitor of PrPSc formation. J Biol Chem. 2011;286(51):44234–44242. doi: 10.1074/jbc.M111.286195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johanssen VA, Johanssen T, Masters CL, Hill AF, Barnham KJ, Collins SJ. C-terminal peptides modelling constitutive PrPC processing demonstrate ameliorated toxicity predisposition consequent to α-cleavage. Biochem J. 2014;459(1):103–115. doi: 10.1042/BJ20131378. [DOI] [PubMed] [Google Scholar]
- 39.Lewis V, Haigh CL, Masters CL, Hill AF, Lawson VA, Collins SJ. Prion subcellular fractionation reveals infectivity spectrum, with a high titre-low PrPres level disparity. Mol Neurodegener. 2012;7:18. doi: 10.1186/1750-1326-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klug GM, Boyd A, Zhao T, Stehmann C, Simpson M, McLean CA, Masters CL, Collins SJ. Surveillance for Creutzfeldt-Jakob disease in Australia: update to December 2012. Commun Dis Intell. 2013;37(2):E115–E120. [PubMed] [Google Scholar]
- 41.Perera WS, Hooper NM. Proteolytic fragmentation of the murine prion protein: role of Tyr-128 and His-177. FEBS Lett. 1999;463(3):273–276. doi: 10.1016/S0014-5793(99)01648-8. [DOI] [PubMed] [Google Scholar]
- 42.Wurch T, Lestienne F, Pauwels PJ. A modified overlap extension PCR method to create chimeric genes in the absence of restriction enzymes. Biotechnol Tech. 1998;12(9):653–657. doi: 10.1023/A:1008848517221. [DOI] [Google Scholar]
- 43.Walmsley AR, Zeng F, Hooper NM. Membrane topology influences N-glycosylation of the prion protein. EMBO J. 2001;20(4):703–712. doi: 10.1093/emboj/20.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis V, Klug GM, Hill AF, Collins SJ. Molecular typing of PrPres in human sporadic CJD brain tissue. In: Hill AF, editor. Prion protein protocols. Totowa NJ: Humana Press; 2008. pp. 241–247. [DOI] [PubMed] [Google Scholar]
- 45.Lewis V, Whitehouse IJ, Baybutt H, Manson JC, Collins SJ, Hooper NM. Cellular prion protein expression is not regulated by the Alzheimer’s amyloid precursor protein intracellular domain. PLoS ONE. 2012;7(2):e31754. doi: 10.1371/journal.pone.0031754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis V, Collins SJ. Analysis of endogenous PrPC processing in neuronal and non-neuronal cell lines. In: Hill AF, editor. Prion protein protocols. Totowa NJ: Humana Press; 2008. pp. 229–239. [DOI] [PubMed] [Google Scholar]
- 47.Barmada S, Piccardo P, Yamaguchi K, Ghetti B, Harris DA. GFP-tagged prion protein is correctly localized and functionally active in the brains of transgenic mice. Neurobiol Dis. 2004;16(3):527–537. doi: 10.1016/j.nbd.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 48.Bian J, Nazor KE, Angers R, Jernigan M, Seward T, Centers A, Green M, Telling GC. GFP-tagged PrP supports compromised prion replication in transgenic mice. Biochem Biophys Res Commun. 2006;340(3):894–900. doi: 10.1016/j.bbrc.2005.12.085. [DOI] [PubMed] [Google Scholar]
- 49.De Keukeleire B, Donadio S, Micoud J, Lechardeur D, Benharouga M. Human cellular prion protein hPrPC is sorted to the apical membrane of epithelial cells. Biochem Biophys Res Commun. 2007;354(4):949–954. doi: 10.1016/j.bbrc.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 50.Taguchi Y, Shi ZD, Ruddy B, Dorward DW, Greene L, Baron GS. Specific biarsenical labeling of cell surface proteins allows fluorescent- and biotin-tagging of amyloid precursor protein and prion proteins. Mol Biol Cell. 2009;20(1):233–244. doi: 10.1091/mbc.E08-06-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salamat K, Moudjou M, Chapuis J, Herzog L, Jaumain E, Beringue V, Rezaei H, Pastore A, Laude H, Dron M. Integrity of helix 2-helix 3 domain of the PrP protein is not mandatory for prion replication. J Biol Chem. 2012;287(23):18953–18964. doi: 10.1074/jbc.M112.341677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rutishauser D, Mertz KD, Moos R, Brunner E, Rulicke T, Calella AM, Aguzzi A. The comprehensive native interactome of a fully functional tagged prion protein. PLoS ONE. 2009;4(2):e4446. doi: 10.1371/journal.pone.0004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saraste J, Palade GE, Farquhar MG. Temperature-sensitive steps in the transport of secretory proteins through the Golgi complex in exocrine pancreatic cells. Proc Natl Acad Sci USA. 1986;83(17):6425–6429. doi: 10.1073/pnas.83.17.6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walmsley AR, Watt NT, Taylor DR, Perera WS, Hooper NM. alpha-cleavage of the prion protein occurs in a late compartment of the secretory pathway and is independent of lipid rafts. Mol Cell Neurosci. 2009;40(2):242–248. doi: 10.1016/j.mcn.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 55.Fujiwara T, Oda K, Yokota S, Takatsuki A, Ikehara Y. Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J Biol Chem. 1988;263(34):18545–18552. [PubMed] [Google Scholar]
- 56.Lawson VA, Collins SJ, Masters CL, Hill AF. Prion protein glycosylation. J Neurochem. 2005;93(4):793–801. doi: 10.1111/j.1471-4159.2005.03104.x. [DOI] [PubMed] [Google Scholar]
- 57.Lewis V, Hooper NM. The role of lipid rafts in prion protein biology. Front Biosci. 2011;16:151–168. doi: 10.2741/3681. [DOI] [PubMed] [Google Scholar]
- 58.Hooper NM. Determination of glycosyl-phosphatidylinositol membrane protein anchorage. Proteomics. 2001;1(6):748–755. doi: 10.1002/1615-9861(200106)1:6<748::AID-PROT748>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 59.Pastore A, Zagari A. A structural overview of the vertebrate prion proteins. Prion. 2007;1(3):185–197. doi: 10.4161/pri.1.3.5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41(Database issue):D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rawlings ND, Waller M, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014;42(Database issue):D503–D509. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kojima A, Konishi M, Akizawa T. Prion fragment peptides are digested with membrane type matrix metalloproteinases and acquire enzyme resistance through Cu(2)(+)-binding. Biomolecules. 2014;4(2):510–526. doi: 10.3390/biom4020510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hande KR, Collier M, Paradiso L, Stuart-Smith J, Dixon M, Clendeninn N, Yeun G, Alberti D, Binger K, Wilding G. Phase I and pharmacokinetic study of prinomastat, a matrix metalloprotease inhibitor. Clin Cancer Res. 2004;10(3):909–915. doi: 10.1158/1078-0432.CCR-0981-3. [DOI] [PubMed] [Google Scholar]
- 64.Chen S, Yadav SP, Surewicz WK. Interaction between human prion protein and amyloid-beta (Abeta) oligomers: role of N-terminal residues. J Biol Chem. 2010;285(34):26377–26383. doi: 10.1074/jbc.M110.145516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.You H, Tsutsui S, Hameed S, Kannanayakal TJ, Chen L, Xia P, Engbers JD, Lipton SA, Stys PK, Zamponi GW. Abeta neurotoxicity depends on interactions between copper ions, prion protein, and N-methyl-d-aspartate receptors. Proc Natl Acad Sci USA. 2012;109(5):1737–1742. doi: 10.1073/pnas.1110789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guillot-Sestier MV, Sunyach C, Druon C, Scarzello S, Checler F. The alpha-secretase-derived N-terminal product of cellular prion, N1, displays neuroprotective function in vitro and in vivo. J Biol Chem. 2009;284(51):35973–35986. doi: 10.1074/jbc.M109.051086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sunyach C, Cisse MA, da Costa CA, Vincent B, Checler F. The C-terminal products of cellular prion protein processing, C1 and C2, exert distinct influence on p53-dependent staurosporine-induced caspase-3 activation. J Biol Chem. 2007;282(3):1956–1963. doi: 10.1074/jbc.M609663200. [DOI] [PubMed] [Google Scholar]
- 68.Shmerling D, Hegyi I, Fischer M, Blattler T, Brandner S, Gotz J, Rulicke T, Flechsig E, Cozzio A, von Mering C, Hangartner C, Aguzzi A, Weissmann C. Expression of amino-terminally truncated PrP in the mouse leading to ataxia and specific cerebellar lesions. Cell. 1998;93(2):203–214. doi: 10.1016/S0092-8674(00)81572-X. [DOI] [PubMed] [Google Scholar]
- 69.Kuczius T, Koch R, Keyvani K, Karch H, Grassi J, Groschup MH. Regional and phenotype heterogeneity of cellular prion proteins in the human brain. Eur J Neurosci. 2007;25(9):2649–2655. doi: 10.1111/j.1460-9568.2007.05518.x. [DOI] [PubMed] [Google Scholar]
- 70.Parizek P, Roeckl C, Weber J, Flechsig E, Aguzzi A, Raeber AJ. Similar turnover and shedding of the cellular prion protein in primary lymphoid and neuronal cells. J Biol Chem. 2001;276(48):44627–44632. doi: 10.1074/jbc.M107458200. [DOI] [PubMed] [Google Scholar]
- 71.Tagliavini F, Prelli F, Porro M, Salmona M, Bugiani O, Frangione B. A soluble form of prion protein in human cerebrospinal fluid: implications for prion-related encephalopathies. Biochem Biophys Res Commun. 1992;184(3):1398–1404. doi: 10.1016/S0006-291X(05)80038-5. [DOI] [PubMed] [Google Scholar]
- 72.Kikuchi Y, Kakeya T, Nakajima O, Sakai A, Ikeda K, Yamaguchi N, Yamazaki T, Tanamoto K, Matsuda H, Sawada J, Takatori K. Hypoxia induces expression of a GPI-anchorless splice variant of the prion protein. FEBS J. 2008;275(11):2965–2976. doi: 10.1111/j.1742-4658.2008.06452.x. [DOI] [PubMed] [Google Scholar]
- 73.Magalhaes AC, Silva JA, Lee KS, Martins VR, Prado VF, Ferguson SS, Gomez MV, Brentani RR, Prado MA. Endocytic intermediates involved with the intracellular trafficking of a fluorescent cellular prion protein. J Biol Chem. 2002;277(36):33311–33318. doi: 10.1074/jbc.M203661200. [DOI] [PubMed] [Google Scholar]
- 74.Lee KS, Magalhaes AC, Zanata SM, Brentani RR, Martins VR, Prado MA. Internalization of mammalian fluorescent cellular prion protein and N-terminal deletion mutants in living cells. J Neurochem. 2001;79(1):79–87. doi: 10.1046/j.1471-4159.2001.00529.x. [DOI] [PubMed] [Google Scholar]
- 75.Piccardo P, Dlouhy SR, Lievens PM, Young K, Bird TD, Nochlin D, Dickson DW, Vinters HV, Zimmerman TR, Mackenzie IR, Kish SJ, Ang LC, De Carli C, Pocchiari M, Brown P, Gibbs CJ, Jr, Gajdusek DC, Bugiani O, Ironside J, Tagliavini F, Ghetti B. Phenotypic variability of Gerstmann-Straussler-Scheinker disease is associated with prion protein heterogeneity. J Neuropathol Exp Neurol. 1998;57(10):979–988. doi: 10.1097/00005072-199810000-00010. [DOI] [PubMed] [Google Scholar]
- 76.Parchi P, Chen SG, Brown P, Zou W, Capellari S, Budka H, Hainfellner J, Reyes PF, Golden GT, Hauw JJ, Gajdusek DC, Gambetti P. Different patterns of truncated prion protein fragments correlate with distinct phenotypes in P102L Gerstmann-Straussler-Scheinker disease. Proc Natl Acad Sci USA. 1998;95(14):8322–8327. doi: 10.1073/pnas.95.14.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zou WQ, Capellari S, Parchi P, Sy MS, Gambetti P, Chen SG. Identification of novel proteinase K-resistant C-terminal fragments of PrP in Creutzfeldt-Jakob disease. J Biol Chem. 2003;278(42):40429–40436. doi: 10.1074/jbc.M308550200. [DOI] [PubMed] [Google Scholar]
- 78.Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73(7):1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 79.Mallucci G, Dickinson A, Linehan J, Klohn PC, Brandner S, Collinge J. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 2003;302(5646):871–874. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- 80.Rogers M, Yehiely F, Scott M, Prusiner SB. Conversion of truncated and elongated prion proteins into the scrapie isoform in cultured cells. Proc Natl Acad Sci USA. 1993;90(8):3182–3186. doi: 10.1073/pnas.90.8.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fischer M, Rulicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15(6):1255–1264. [PMC free article] [PubMed] [Google Scholar]
- 82.Lawson VA, Priola SA, Wehrly K, Chesebro B. N-terminal truncation of prion protein affects both formation and conformation of abnormal protease-resistant prion protein generated in vitro. J Biol Chem. 2001;276(38):35265–35271. doi: 10.1074/jbc.M103799200. [DOI] [PubMed] [Google Scholar]
- 83.Marbiah MM, Harvey A, West BT, Louzolo A, Banerjee P, Alden J, Grigoriadis A, Hummerich H, Kan HM, Cai Y, Bloom GS, Jat P, Collinge J, Klohn PC. Identification of a gene regulatory network associated with prion replication. EMBO J. 2014;33(14):1527–1547. doi: 10.15252/embj.201387150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.