Abstract
Polysaccharide degradation by hydrolytic enzymes glycoside hydrolases (GHs) is well known. More recently, polysaccharide monooxygenases (PMOs, also known as lytic PMOs or LPMOs) were found to oxidatively degrade various polysaccharides via a copper-dependent hydroxylation. PMOs were previously thought to be either GHs or carbohydrate binding modules (CBMs), and have been re-classified in carbohydrate active enzymes (CAZY) database as auxiliary activity (AA) families. These enzymes include cellulose-active fungal PMOs (AA9, formerly GH61), chitin- and cellulose-active bacterial PMOs (AA10, formerly CBM33), and chitin-active fungal PMOs (AA11). These PMOs significantly boost the activity of GHs under industrially relevant conditions, and thus have great potential in the biomass-based biofuel industry. PMOs that act on starch are the latest PMOs discovered (AA13), which has expanded our perspectives in PMOs studies and starch degradation. Starch-active PMOs have many common structural features and biochemical properties of the PMO superfamily, yet differ from other PMO families in several important aspects. These differences likely correlate, at least in part, to the differences in primary and higher order structures of starch and cellulose, and chitin. In this review we will discuss the discovery, structural features, biochemical and biophysical properties, and possible biological functions of starch-active PMOs, as well as their potential application in the biofuel, food, and other starch-based industries. Important questions regarding various aspects of starch-active PMOs and possible economical driving force for their future studies will also be highlighted.
Keywords: Starch degradation, Biofuels, Polysaccharide monooxygenases, Copper enzymes, Auxiliary activity family 13, Plant pathogens
Overview of polysaccharide monooxygenases
Polysaccharide monooxygenases (PMOs) are extracellular fungal and bacterial enzymes that have significant potential for application in the production of biofuels (Table 1) [1–14]. These enzymes are capable of oxidizing recalcitrant substrates such as cellulose, chitin and starch, and have been shown to act synergistically with glycoside hydrolases [2]. Several excellent reviews on chitin- and cellulose-active PMOs have been published over the last few years [6–8, 12–14]. To date, there are only two published peer-reviewed research articles on starch-active PMOs [10, 11]. To better understand these starch-active PMOs, they will be discussed in the context of the well-studied chitin- and cellulose-active PMOs. Thus, we will briefly highlight selected aspects of these PMOs where relevant to the discussion of starch-active PMOs.
Table 1.
PMO families
| AA nomenclature | Former annotation | Primary substrate | Source | PMO nomenclature |
|---|---|---|---|---|
| AA9 | Glycoside hydrolase family 61 (GH61) | Cellulose | Fungi | Cellulose-active fungal PMOs |
| AA10a | Carbohydrate binding module family 33 (CBM33) | Cellulose | Bacteria | Cellulose-active bacterial PMOs |
| Chitin | Chitin-active bacterial PMOs | |||
| AA11 | Chitin | Fungi | Chitin-active fungal PMOs | |
| AA13 | X143 or chitin binding 3 | Starch | Fungi | Starch-active fungal PMOs |
aKey residues and motifs in cellulose-active bacterial PMOs differ from those in chitin-active bacterial PMOs
PMO families
There are four PMO families and several names have emerged for them with some resulting confusion, therefore, some clarification is necessary. Table 1 lists the currently described PMO families. The terms PMO and lytic PMO (LPMO) correctly express the monooxygenase activity of these enzymes, but they do not specify the source and primary substrate. “Lytic” could lead to the misunderstanding that the elimination step (Fig. 1b) is enzyme-dependent, which is not supported by current evidence. “Auxiliary activity” (AA) is the latest, most systematic, and concise nomenclature [15], but it does not reflect the particular enzyme specificity. In addition, several critical residues around the active site of cellulose-active AA10 are different from those in AA10 PMOs that are only active on chitin [12, 16]. We used a Hidden Markov Model-based search result using hmmer tools (http://hmmer.org/) and cellulose-active AA10 PMO sequences did not contain chitin-active AA10 PMOs, and vice versa. Thus, the AA10 could possibly be split into two subfamilies. The AA and PMO nomenclature will be used interchangeably in this review where it is appropriate (Table 1).
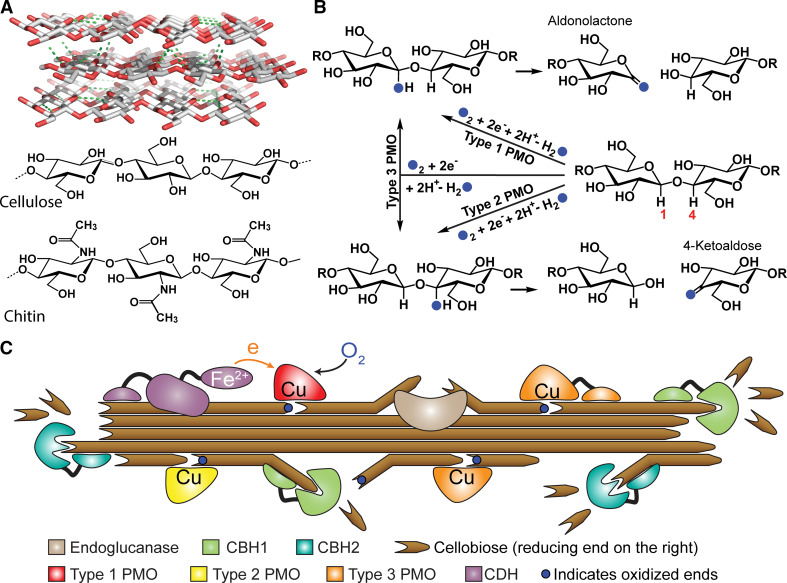
Fig. 1.
a Model of a cellulose crystal and the basic structure of cellulose and chitin. b Hydroxylation-mediated cleavage of the glycosidic bond by three types of cellulose-active PMOs. PMOs hydroxylate either C1 or C4 position of the glycosidic linkage, forming unstable intermediates that subsequently undergo elimination. Blue filled circles represent oxygen derived from O2. c Synergistic working model of PMOs with cellobiose dehydrogenase (CDH) and cellobiohydrolases (CBH1 and CBH2) in degrading cellulose (reproduced from Ref. [12] with permission)
PMO: structure, reaction, and synergy with hydrolytic enzymes
Chitin- and cellulose-active PMOs contain a type 2 mononuclear copper center on a flat, solvent exposed protein surface (Fig. 2a), which allows for the oxidation of glycosidic bonds on crystalline surface of the substrate (Fig. 1a). Thus, PMOs bypass the energy-intensive step of separating the polysaccharide polymer chains from the crystalline matrix, which is required for the glycoside hydrolases. PMOs utilize O2 to hydroxylate either C–H bond of the glycosidic linkage, forming unstable intermediates that spontaneously undergo elimination to cleave the linkage (Fig. 1b) [5, 16]. In vitro, the two electrons required for the reactions of PMOs can be provided by chemical reductants, such as ascorbate, pyrogallol, and cysteine. Lignin, which is abundant in plant cell-wall, was shown to supply electrons for PMO reaction, and could be one of the biologically relevant electron donors [17]. Extracellular enzymes can also serve as a redox partner for PMOs. Cellobiose dehydrogenase (CDH), which carries out the two-electron oxidation of cellobiose, is found in almost all filamentous fungi that have AA9 enzymes [18]. Transcriptomic studies of several filamentous fungi reveal that CDH and AA9 enzymes are both up-regulated when these fungi were grown on cellulosic substrates. CDH is considered to be the biological redox partner of AA9 enzymes as it has been shown to donate electrons to AA9 PMOs in cellulose degradation assays [4, 12].
Fig. 2.
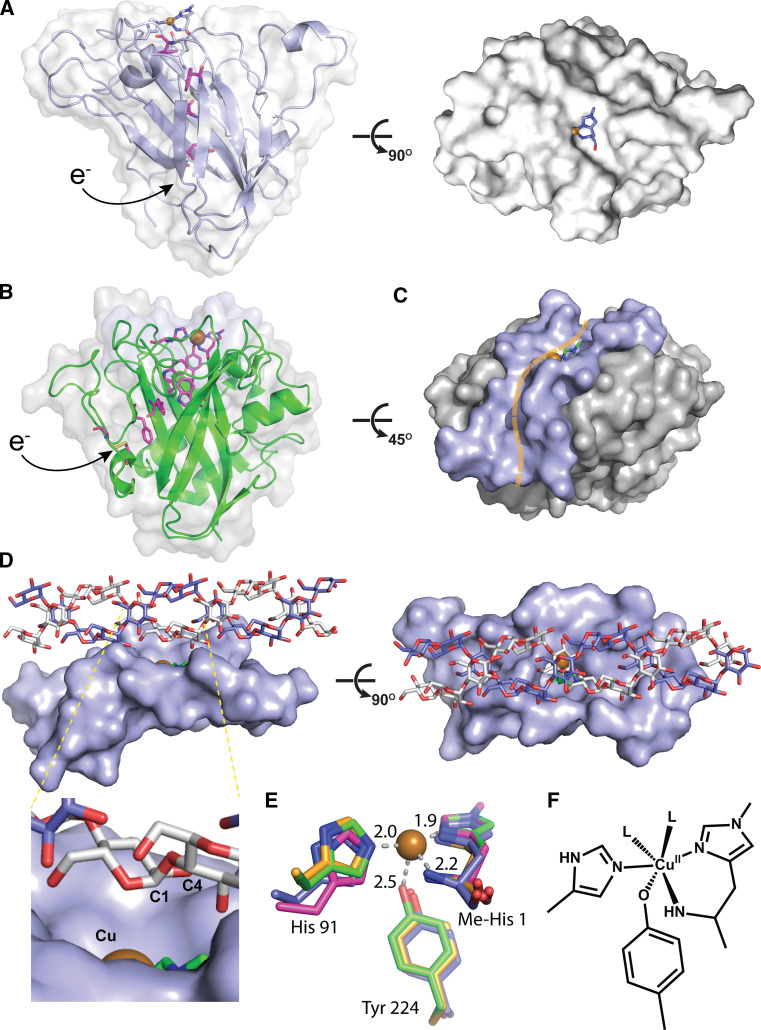
Structure of starch active PMO in comparison with other PMO family and possible interaction with amylose substrate. a Structure of a cellulose-active PMO (4EIS) showing a flat active site surface. b Structure of starch-active PMO from A. oryzae, Ao(AA13), (4OPB) with similar β-sandwich core as found in other PMOs. Putative electron transfer pathways residues are displayed in magenta color in a and b. c Active site surface of Ao(AA13) exhibits a shallow groove. d Possible interaction of an amylose double helix with the active site groove of starch-active PMOs (manually docked). e XRD structure of Ao(AA13) (2OBP) active site (green) in comparison to that in an AA9 member (3ZUD, magenta, rmsd for protein atoms shown of 0.73 Å), AA10 member (2YOY, orange, rmsd of 0.53 Å) and AA11 member (4MAI, purple, rmsd of 0.60 Å) (reproduced from Ref. [11] with permission). f Solution structure deduced from XAS and UV/Vis spectroscopic analysis, where L can be aqueous or other coordinating agents in the solution
There are three types of cellulose-active PMOs: types 1 and 2 hydroxylate the C1 and C4 position of the glycosidic bond, respectively, while type 3 can hydroxylate both the C1 and C4 positions (Fig. 1b) [5, 16, 19]. This chemistry leads to a new synergistic working model for the biodegradation of cellulose as shown in Fig. 1c, which is also relevant to chitin and other recalcitrant polysaccharides, such as starch. New chain ends are created on the surface of the substrates after the PMO reaction, thereby expanding the sites of action of exo-glucanases such as cellobiohydrolases (CBH1 and CBH2). Several PMOs have been shown to significantly boost the activity of hydrolytic cellulases under industrially relevant conditions [2], and some have been included in commercial cellulase mixtures, such as Ctec2 and Ctec3 of Novozymes Inc.
Structure
The only X-ray diffraction (XRD) crystal structure of an AA13 family PMO is of Aspergillus oryzae AA13 (Ao(AA13)) (PDB ID 4OPB), solved to 1.5 Å [11]. Although Ao(AA13) did not exhibit detectable activity under the assay conditions, the high sequence identity throughout the AA13 family (see “Bioinformatic analysis of starch-active PMOs”) suggests this structure is an adequate representation of this family. The overall structure of Ao(AA13) is similar to the other PMO families (Fig. 2) with a β-sandwich core consisting of several β-strands and an exposed mononuclear copper active site on a putative substrate binding surface. As found in many members of other PMO families [12], a putative electron transfer pathway consisting of a series of aromatic and cysteine residues is also found in starch-active PMO [11] (Fig. 2a, b).
Notably, unlike the AA9, AA10, and AA11 families that contain flat active site surface, Ao(AA13) displays an uneven surface with a shallow groove spanning across this surface (Fig. 2b, c). The copper active site resides in the middle of this groove. It is generally accepted that the flat surface of AA9, AA10, and AA11 PMOs is ideal for interaction with the flat surface of cellulose and chitin (Fig. 1a), which is supported by computational studies [20, 21]. The interactions between the surface of AA9, AA10, and AA11 PMOs with the substrate surface must be important in the observed regioselectivity (Fig. 1b) [16, 20, 21]. Amylose and amylopectin, the main components of starch, tend to form helices and highly complex higher order structures (see “Specificity for primary and higher order structures of the substrate”). The groove of AA13 would better accommodate starch molecule chains. Manual docking of amylose double helices to this grooved surface showed that an amylose double helix consisting of six glucosyl units on each chain per turn could fit into this groove (Fig. 2d). The C1 and C4 positions of the α(1 → 4)-glycosidic linkage can be oriented close to the copper center. More rigorous docking techniques or molecular dynamic calculations would provide a more precise insight into this probable interaction between amylose and the AA13 surface groove.
The XRD crystal structure of the type 2 mononuclear copper active site of Ao(AA13) is consistent with a reduced copper(I) center, likely resulting from photoreduction during data collection [11]. This active site superimposes well with that of structures of AA9, AA10, and AA11 PMOs (Fig. 2e), with the copper center ligated by two histidine residues in a motif termed the histidine brace. The N-terminal His residue coordinates in a bidentate mode through the imidazole Nδ (or Nπ) atom and the amino terminus N atom. This residue is methylated at Nε (or Nτ) position, as found in fungal PMOs expressed in filamentous fungal hosts [11, 12] (labeled as Me-His 1). The second histidine ligand, His 91, binds through the Nε (or Nτ) atom. The tyrosine residue moves closer to the copper center in this structure of Ao(AA13) than in other PMO structures. At the Cu–OTyr distance of 2.5 Å, as observed in the Ao(AA13) structure, a weak bonding interaction can form between the copper center and OTyr. The EPR data of Cu(II)-Ao(AA13) and Cu(II)-An(AA13) both exhibit resolved super-hyperfine coupling to nitrogen ligands, which is significantly different from that observed other PMOs. Lo Leggio et al. reasonably speculate that such difference may reflect the enhanced copper–nitrogen covalency or a structurally well-order active site in AA13 [11]. The partial bonding of the axial tyrosine ligand, could play a role by either (1) increasing the overall electron density at the copper (II) center, that is linked to higher copper-nitrogen covalency or (2) contributing to the rigidity of the active site.
X-ray absorption spectroscopy (XAS) was used to deduce the active-site solution structure of NCU08746, a starch-active PMO from Neurospora crassa (Fig. 2f) [10]. Photoreduction in XAS data collection for oxidized NCU08746 was minimal because the X-ray source used in XAS is significantly less intense compared to that used in XRD; and data were collected on multiple different sample spots, with one spectrum per spot. X-ray absorption near edge spectroscopy (XANES) is sensitive to the oxidation state, electronic properties, and coordination geometry of the absorbing center. The featureless edge of the XANES spectrum of oxidized NCU08746 is typical of 6-coordinate copper (II) species. Extended X-ray absorption fine structure (EXAFS) analysis of Cu(II)-NCU08746 resulted in six copper-first sphere ligand single scattering paths, corresponding to four Cu–O/N bonds at 1.97 Å, and two longer Cu–O/N bonds at 2.22 and 2.42 Å. The four 1.97 Å Cu–O/N bonds likely form the equatorial plane and the longer bonds occupy the axial positions of the copper center. Multiple scattering EXAFS analysis revealed that the copper center was ligated by either two or three histidine ligands; the Ao(AA13) crystal structure subsequently showed the copper center coordinated by two histidines, clarifying the ambiguity in this data.
The first sphere structure of Cu(II)-NCU08746 is consistent with an axially elongated octahedral copper (II) structure (Fig. 2f). In this structure, the N atoms of the histidine brace and a solvent-based ligand form the equatorial plane, while a tyrosine residue likely coordinates in the axial position. This tyrosine residue could be part of the putative electron transfer pathway, as found for other fungal PMOs [11, 12]. An axial tyrosine residue coordinating to the mononuclear copper center is also found in galactose oxidase [22]. This tyrosine residue is proposed to mediate proton transfer during galactose oxidase reaction. The active site tyrosine residue in starch-active PMO possibly plays one or more of the following roles: (1) mediating proton and/or electron transfer during PMO reaction; (2) fine-tuning redox properties of the copper center; and (3) stabilizing high-valent copper–oxygen species as proposed earlier for the PMO mechanism [12].
Specificity for primary and higher order structures of the substrate
AA9, AA10, and AA11 families degrade the structurally related cellulose and chitin substrates (Fig. 1a). Cellulose and chitin contain linear chains of β(1 → 4) linked glucose and N-acetylglucosamine, respectively, both of which can form microcrystalline structures. In plant cell-walls, hemicellulose chains wrap around cellulose microfibrils forming rigid structures. The primary and higher order structures of xylan and other hemicelluloses, as well as the ratio of cellulose:hemicellulose vary depending on the plant source. Most cellulolytic filamentous fungi have a dozen or more AA9 coding genes that are tightly regulated by the growth conditions. These genes are expressed in different combinations and levels depending on the type of cellulosic substrates on which the fungus is grown [23, 24]. Thus, each PMO likely has a different preference for the chemical and physical properties of the accepted substrates, which was shown in a recent study for the fungus Podospora anserina [25].
The first PMOs characterized acted strictly on an insoluble polysaccharide surface (Fig. 1), until an AA9 PMO in N. crassa was found to be active on soluble cellodextrins as short as a tetramer [26]. This PMO was also found to cleave various hemicelluloses containing β(1 → 4) linkages of glucose and/or substituted glucose units [27]. The structure of this PMO exhibits an extended and highly polar active site surface compared to previously characterized PMOs, which is suitable for binding various carbohydrate substrates [28]. Subsequently, an AA9 PMO in Myceliophthora thermophila was found to cleave both the β(1 → 4)-glycosidic linkage in cellulose and the β(1 → 4)-xylosidic linkage in xylan in the presence of cellulose [29]. Although PMOs have been shown to oxidize various polysaccharide substrates, the β(1 → 4) linkage appears to be essential for all the substrates of AA9, AA10, and AA11 PMOs.
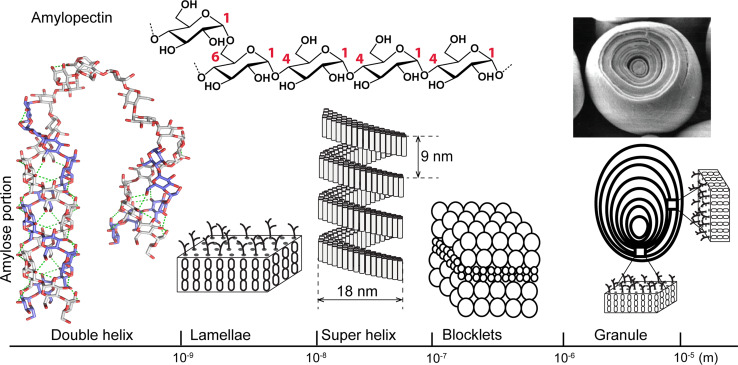
Starch differs from cellulose and chitin in both the primary and the higher order structure levels. The two main components of starch are amylose and amylopectin, which both contain linear chains of α(1 → 4)-glycosidic linkages. Amylopectin, the predominant component, also contains α(1 → 6)-glycosidic linkages that create branches along the molecule (Fig. 3). Amylose and the amylose portion of amylopectin can form single or double helical structure. Higher order structures of starch are highly complex and have not been well understood despite substantive characterization studies [30]. Model structures of starch on various size scales are shown in Fig. 3. Parallel double helices of amylose in amylopectin form crystalline lamellae that assemble into super-helices. A single amylopectin molecule can form a super-helix. Starch blocklets were recently described with sizes ranging from 25 to 500 nm [28]. Larger blocklets can contain one or more super-helix. Typical starch granules have semi-crystalline onion-like structure with alternate amorphous and crystalline growth rings. Retrogradation is starch that is heated in water and then cooled, allowing the starch chains to re-assemble and form a more ordered structure. Thus, retrograded starch is more resistant to hydrolysis than untreated starch.
Fig. 3.
Primary structure of amylopectin, and higher order structures of starch on different size-scales. Amylose double helices form lamellae and superhelices. Blocklets contain one or more superhelices. Lamellae form crystalline growth rings that alternate with amorphous layers in starch granules. SEM image of starch and granule was reproduced from Ref. [31] with permission; Lamellae, super helix, blocklets, and granule were re-drawn based on Fig. 21 of Ref. [29] with permission
Starch-active PMOs have been shown to degrade various starch substrates, including corn starch, retrograded starch, amylopectin, and amylose; these PMOs are not active on cellulose or chitin. Starch-active PMOs are the first example of a PMO to oxidatively cleave α(1 → 4)-glycosidic linkages [10, 11]. Although only oxidation at the C1 position has been observed to date, oxidation of the C4 position of the α(1 → 4) linkage, as well as oxidation of both the C1 and C6 positions of the α(1 → 6)-glycosidic linkage may be catalyzed by other AA13 enzymes. The active site groove on the surface of Ao(AA13) could accommodate an amylose double helix (Fig. 3f), suggesting that amylose would be a preferred substrate. This speculation is consistent with limited activity assay data reported thus far (see “Biochemical characterization of starch-active PMOs”). However, because of the highly complex structure of naturally found starch, e.g. in grains, fruits, and roots, it is possible that AA13 family members have a broader substrate specificity than currently recognized. The effect of crystallinity, solubility, and degree of polymerization of starch molecules on activity of AA13 enzymes needs to be addressed. Substrate specificity is an important aspect of AA13 enzymes that will require extensive studies in the future to completely resolve.
Bioinformatic analysis of starch-active PMOs
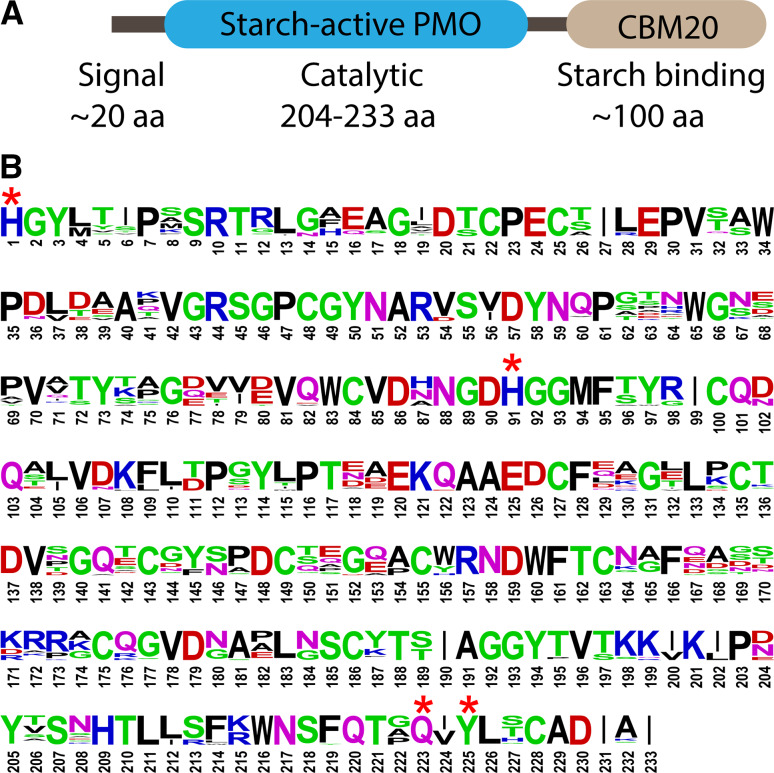
The sequence similarity is relatively low within each previously characterized PMO family and is inconsequential between different PMO families. Nevertheless, all PMOs characterized thus far are structurally related. The overall structure with the core β-barrels is conserved in all families. The active site histidine residues that ligate the copper center in the histidine brace motif (Fig. 2) is conserved in all PMOs. In addition, the motif N/Q/E-X-Y/F is conserved in all fungal PMOs and a subgroup of bacterial PMOs [12]. This motif and the active site histidine residues form an amino acid signature (AAS) of PMOs. This AAS was used successfully in the discovery of starch-active PMOs [10]. In the predicted secretome of N. crassa, five distinct putative PMO families were identified. These families are conserved in many other fungi. Two of the five families identified are cellulose- and chitin-active PMOs (AA9 and AA11, respectively). Among the three uncharacterized putative PMO families in N. crassa, one family contained a C-terminal starch-binding domain, CBM20, initially suggesting it may be active on starch. Rigorous characterization confirmed this PMO was indeed a member of a previously unknown starch-active PMO family (AA13). The discovery of starch-active PMOs provides new perspectives in PMO studies and starch metabolism. It also provided evidence that the PMO AAS (noted with stars in Fig. 4b) can be used to identify novel PMOs.
Fig. 4.
a Common domain architecture of 85 starch-active PMOs from different fungal species. Nineteen do not have the CBM20 domain [11]. b Consensus sequence logo representing the putative catalytic domain. Asterisks indicate the absolutely conserved residues also found in cellulose-active PMOs and chitin-active PMOs. This figure is reproduced from Ref. [10] with permission
An independent approach was used to identify starch-active PMOs using the module walking method [11]. This method was used successfully in the discovery of fungal chitin-active PMOs (AA11 family) [9]. The CBM20 domain, found in α-amylases, glucoamylases, and other starch-active enzymes, was found appended to a N-terminal domain of unknown function termed X143. This domain had an N-terminal histidine residue following a signal peptide cleavage. The X143 domain was subsequently shown to be starch-active PMO domain. A X143-CBM20 bimodular protein in A. nidulans, An(AA13), was shown to be active on retrograded starch, forming malto-aldonic acids, as found for the N. crassa starch-active PMO reported earlier. Module walking is a useful method to identify proteins with new functions, but would not find PMOs lacking a known C-terminal domain. Combining PMO AAS with module walking may be a more comprehensive approach to identifying new PMOs with novel functions.
The AA13 family currently has more than 80 members found in Ascomycota fungi [11]. Unlike the AA9, AA10, and AA11 families, multiple sequence alignments of AA13 enzymes require no significant gaps, as there is significantly high sequence homology among the AA13 family (Fig. 4b) [10, 11]. On the phylogenetic level, the AA13 family is well separated from AA9, AA10, and AA11, with AA10 being the closest related family [11]. It was noted in a previous review that distant relatives of AA10 proteins were fused with C-terminal CBM20 domain [6], which are now known as AA13 PMOs. The majority of AA13 enzymes contain both a starch-active PMO domain and a CBM20 domain; the rest contain only a starch-active PMO domain. No other types of C-terminal domains are currently known for AA13 enzymes. This CBM20 has 50–70 % sequence identity to many CBM20 domains found in amylolytic enzymes that bind starch. Phylogenetic analysis of AA13 family in the presence and absence of CBM20 indicated that CBM20 was present in the ancestors of AA13 enzymes and occasionally lost during evolution [11]. Lo Leggio et al. reported that Ao(AA13), which contain only the PMO domain, did not show any detectable activity on starch substrates. The authors speculate that either (1) the CBM20 domain is critical for activity, or (2) a redox partner enzyme is required to complete the binding of Ao(AA13) to its substrate. However, a starch active PMO lacking a CBM20 from M. thermophila was shown to boost the activity of amylolytic enzymes on a starch substrate [32]. With experimental data on only a limited number of AA13 PMOs, the significance and functional relevance of the CBM20 domain remains to be determined.
Biochemical characterization of starch-active PMOs
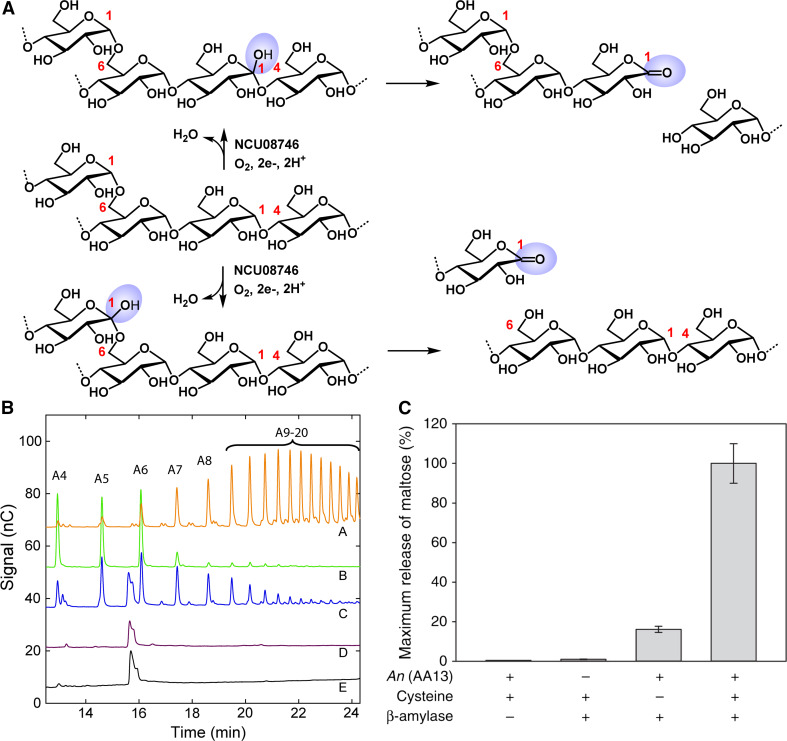
Starch-active PMOs have been recombinantly expressed in N. crassa [10] and A. oryzae [11] and assayed on various substrates. The starch-active PMOs from N. crassa (NCU08746) and A. nidulans (An(AA13)) are active on starch, but not on cellulose or chitin; Ao(AA13) did not show detectable activity on retrograded starch or malto-octaose. Activity of NCU08746 on cornstarch, amylopectin, and amylose was easily detectable with both high performance anion exchange chromatography (HPAEC) (Fig. 5b) and mass spectrometry [10]. Aldonic acids are the only products detected for NCU08746 and An(AA13), indicating that these PMOs oxidize starch at the C1 position (Fig. 5a, b). When assayed on amylose substrates containing only α(1 → 4) linkages, the starch-active PMO from N. crassa also formed aldonic acids, indicating that oxidation occurred at this linkage. Further studies are required to determine whether C1 oxidation also occurs at α(1 → 6) linkages. The possible mechanism of aldonic acid formation is depicted in Fig. 5a based on the chemistry of cellulose- and chitin-active PMOs. Thus far, oxidation at the C4 position or C6 position has not been observed.
Fig. 5.
a Proposed starch degradation steps by NCU08746 involving the cleavage of α(1 → 4) and α(1 → 6) linkages via hydroxylation at the C1 position. Further studies are required to determine whether starch-active PMO also oxidize α(1 → 6) linkage. b Activity assays of N. crassa starch-active PMOs, NCU08746, with ascorbic acid and atmospheric oxygen. Traces A and B maltoaldonic acids with four (A4) to twenty (A20) units. Traces C–E assays with amylopectin, phosphoric acid swollen cellulose, and chitin, respectively. c Boosting effect of AA13. Release of maltose from retrograded starch by β-amylase over 4 h at 25 °C (columns 2–4) with An(AA13) (column 3), with reducing agent and An(AA13) (column 4). a and b were reproduced from Ref. [10] and c from Ref. [11] with permission
Lo Leggio et al. showed that An(AA13) significantly boosted the activity of a β-amylase from barley on retrograded starch (Fig. 5c) [11]. Retrograded starch has more crystalline structure and, therefore, is more resistant to hydrolysis compared to untreated starch (see “Specificity for primary and higher order structures of the substrate”). β-Amylase hydrolyzes starch molecules processively from the non-reducing ends, releasing maltose [33]. β-Amylase exhibited minimal activity on retrograded starch, which was boosted by ~100-fold in the presence of An(AA13). Analogous to the synergistic working model of cellulose degradation (Fig. 1c), it is likely that PMO reaction leads to new chain ends in the substrates that can be processed by β-amylase.
In the initial studies, N. crassa starch-active PMO exhibited the same activity with either ascorbic acid or cellobiose dehydrogenase (CDH)/cellobiose system as the electron source [10]. Other small molecule chemical reductants such as cysteine were also found to provide reducing equivalents for the starch-active PMOs [11]. Reduced CDH is relatively stable in the presence of atmospheric oxygen, but was shown to be rapidly oxidized by N. crassa starch-active PMO [10]. Putative electron transfer pathways, connecting the copper active site to the predicted CDH binding site, have been identified in cellulose active PMOs. Similar ET pathways are also found in the structure of starch-active PMOs [11], providing a molecular basis for intramolecular electron transfer between CDH and these PMOs.
Seventy-five out of the eighty ascomycetes that have starch-active PMO genes also have one or more CDH genes. Because fungi encounter diverse food sources containing mixtures of various polysaccharides, it is not unreasonable to speculate that co-expression of CDH and starch-active PMOs takes place. Transcriptomic and proteomic analysis of ascomycetes containing both CDH and starch-active PMO genes on different growth substrates would provide insight into the biological relationship of these enzymes. Alternatively, other enzymes such as oligosaccharide dehydrogenase [34] or maltose dehydrogenase [35] could possibly serve as the biological redox partners of starch-active PMOs, but these relationships remain to be elucidated.
Possible biological functions
To date, starch-active PMOs have been found in about 80 fungi belonging to five classes of the Ascomycota phylum, including Eurotiomycetes, Dothideomycetes, Sordariomycetes, Leotiomycetes, Orbilliomycetes. The majority of these ascomycetes are plant pathogens, known to attack different cellulose-rich parts of plants including the leaf and stem. They also infect starch-containing components such as grains, seeds, fruits, and roots. Crops such as rice, wheat, barley, potato, apple, banana, grape, berries, etc. are among the most important hosts of these fungi. In a worldwide survey of plant fungal pathogens [36], five out of ten most important fungi contain starch-active PMOs. These five fungi are Magnaporthe oryzae (ranked first), Botrytis cinerea (second), Fusarium graminearum (fourth), Fusarium oxysporum (fifth), and Colletotrichum spp. (eighth). Ascomycetes containing starch-active PMOs also have genes coding for α-amylase and glucoamylase, as well as cellulose-active PMOs and cellulases. It has been shown that expression of extracellular cellulose-active enzymes are induced when fungi grow on cellulosic substrates [23, 24] or infect cellulose-rich part of plants [37]. It is not unreasonable to speculate that when these fungi encounter starch substrates, they also secrete starch-active PMOs along with α-amylases and glucoamylases to degrade starch for growth or for invasion of the host.
Glucoamylase hydrolyzes starch molecules at α(1 → 4) linkage from non-reducing end of amylopectin and amylose molecules to form glucose and will also hydrolyze a α(1 → 6) linkage when followed by a α(1 → 4) linkage [38]. Fungal α-amylases randomly hydrolyze α(1 → 4) bonds of starch, resulting in maltose and maltotriose. It is generally known that amylases are more active on amorphous starch, while amylose is considered a resistant starch [39]. The structure of Ao(AA13) suggests that amylose would be a preferred substrate of starch-active PMOs (Fig. 2d). Consistent with this finding, NCU08746 is active on amylose [10], and An(AA13) significantly boosted activity of a β-amylase [11], a processive enzyme that hydrolyzes α(1 → 4) linkage from the non-reducing end of amylose, on retrograded starch. These observations suggest that starch-active PMOs could break down amylose, reduce substrate crystallinity, and thus make the substrate more amenable to hydrolysis by amylases.
Interestingly, the 80 ascomycetes containing starch-active PMOs do not have β-amylase. However, β-amylases are found in fruit and during germination in grain. Thus, when fungi invade fruit or grain, starch-active PMOs could potentially work synergistically with the plant β-amylase. Likewise, synergy of starch active-PMOs with other amylolytic enzymes of the plant host could also occur in Nature.
Concluding remarks and outlook
Starch-active PMOs were only recently discovered in ascomycetes, many of which are plant pathogens. Starch-active PMOs share many common features with cellulose and chitin active PMOs, including the β-sandwich core, the histidine brace mononuclear copper active site on a protein surface, the putative electron transfer pathway, the mechanism of glycosidic bond cleavage via C-H bond hydroxylation, and synergy with corresponding glycoside hydrolases. Starch-active PMOs can accept electrons from CDH, the generally accepted biological redox-partner of cellulose-active PMOs, yet differ from other PMO families in many aspects. The coordination of the active site tyrosine residue to the copper center of starch-active PMOs is not found in the other PMO families, which could lead to a different mechanism of oxygen activation and substrate oxidation. Starch-active PMOs possess an uneven active-site surface with a shallow groove, as opposed to the flatter surface observed in cellulose- and chitin-active PMOs. This groove may determine the specificity for starch molecules. In Nature, starch-active PMOs likely work together with amylases secreted by the fungi or by the plant host in degrading starch substrates for growth or host invasion.
Many fundamental questions regarding these PMOs remain to be resolved. The detailed interaction of starch-active PMOs with various substrates can be addressed with both computational studies, such as docking or molecular dynamic calculation, as well as experimental investigation. Transcriptomic and proteomic analysis of fungi grown on various starch substrates will help determine the biological role of starch-active PMOs. Substrate specificity and regioselectivity of oxidation can be resolved in assays on various substrates with well-characterized structure, as well as with suitable isotopically labeled substrates. Precise structure and electronic properties of the copper active site in the reduced and oxidized state can be obtained with combinations of advanced spectroscopic studies, such as XAS, pulsed EPR, resonance Raman, and magnetic circular dichroism spectroscopies.
The synergy of starch-active PMOs with amylases will be studied extensively due to its tremendous potentials in the starch-based biofuels industry. Currently in the United States, there are about 15 billion gallons of starch-based biofuels produced annually (http://www.afdc.energy.gov/laws/RFS; accessed on October 28, 2015). Starch hydrolysis with amylases to fermentable sugars is a critical and expensive step. The ability to boost the activity of amylases under industrially relevant conditions using starch-active PMOs would increase the efficiency of these enzymes, resulting in a significant cost reduction in biofuel production. In addition to biofuels, starch-active PMOs could also be used together with amylases in other important industries, such as corn syrup, detergents, paper, textiles, and adhesive. Global starch derivatives market was $51.2 billion in 2012, and is expected to grow to $77.4 billion in 2018 (http://www.prweb.com/pdfdownload/10923341.pdf; accessed on October 28, 2015). This will also be a strong driving force for the continued investigation of starch-active PMOs.
Acknowledgments
Financial support to M.A.M from the Energy Bioscience Institute (UC Berkeley) and the NSF (CHE 1411538) is gratefully acknowledged. V.V.V was supported by a grant from Nguyen Tat Thanh University, Ho Chi Minh, Vietnam. We thank Dr. John Hangasky for his advice and editorial help.
Abbreviations
- AA
Auxiliary activity
- CDH
Cellobiose dehydrogenase
- GH
Glycoside hydrolase
- CBM
Carbohydrate binding modules
- LPMOs
Lytic polysaccharide monooxygenases
- PMOs
Polysaccharide monooxygenases
- XAS
X-ray absorption spectroscopy
- XRD
X-ray diffraction
References
- 1.Vaaje-Kolstad G, Westereng B, Horn SJ, Liu Z, Zhai H, Sørlie M, Eijsink VG. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330:219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- 2.Harris PV, Welner D, McFarland KC, Re E, Navarro Poulsen JC, Brown K, Salbo R, Ding H, Vlasenko E, Merino S, Xu F, Cherry J, Larsen S, Lo Leggio L. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry. 2010;49:3305–3316. doi: 10.1021/bi100009p. [DOI] [PubMed] [Google Scholar]
- 3.Quinlan RJ, Sweeney MD, Lo Leggio L, Otten H, Poulsen JC, Johansen KS, Krogh KB, Jørgensen CI, Tovborg M, Anthonsen A, Tryfona T, Walter CP, Dupree P, Xu F, Davies GJ, Walton PH. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc Natl Acad Sci USA. 2011;108:15079–15084. doi: 10.1073/pnas.1105776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips CM, Beeson WT, Cate JHD, Marletta MA. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa . ACS Chem Biol. 2011;6:1399–1406. doi: 10.1021/cb200351y. [DOI] [PubMed] [Google Scholar]
- 5.Beeson WT, Phillips CM, Cate JHD, Marletta MA. Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J Am Chem Soc. 2012;134:890–892. doi: 10.1021/ja210657t. [DOI] [PubMed] [Google Scholar]
- 6.Horn S, Vaaje-Kolstad G, Westereng B, Eijsink VGH. Novel enzymes for the degradation of cellulose. Biotechnol Biofuels. 2012;5:45–56. doi: 10.1186/1754-6834-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimarogona M, Topakas E, Christakopoulos P. Recalcitrant polysaccharide degradation by novel oxidative biocatalysts. Appl Microbiol Biotechnol. 2013;97:8455–8465. doi: 10.1007/s00253-013-5197-y. [DOI] [PubMed] [Google Scholar]
- 8.Hemsworth GR, Davies GJ, Walton PH. Recent insights into copper-containing lytic polysaccharide mono-oxygenases. Curr Opin Chem Biol. 2013;23:660–668. doi: 10.1016/j.sbi.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Hemsworth GR, Henrissat B, Davies GJ, Walton PH. Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nat Chem Biol. 2014;10:122–126. doi: 10.1038/nchembio.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vu VV, Beeson WT, Span EA, Farquhar ER, Marletta MA. A family of starch-active polysaccharide monooxygenases. Proc Natl Acad Sci USA. 2014;111:13822–13827. doi: 10.1073/pnas.1408090111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lo Leggio L, Simmons TJ, Poulsen JC, Frandsen KE, Hemsworth GR, Stringer MA, von Freiesleben P, Tovborg M, Johansen KS, De Maria L, Harris PV, Soong CL, Dupree P, Tryfona T, Lenfant N, Henrissat B, Davies GJ, Walton PH. Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase. Nat Commun. 2015;6:5961. doi: 10.1038/ncomms6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beeson WT, Vu VV, Span EA, Phillips CM, Marletta MA. Cellulose degradation by polysaccharide monooxygenases. Annu Rev Biochem. 2015;84:923–946. doi: 10.1146/annurev-biochem-060614-034439. [DOI] [PubMed] [Google Scholar]
- 13.Hemsworth GR, Johnston EM, Davies GJ, Walton PH. Lytic polysaccharide monooxygenases in biomass conversion. Trends Biotechnol. 2015;33:747–761. doi: 10.1016/j.tibtech.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Aachmann FL, Vaaje-Kolstad G, Forsberg Z, Røhr Å, Eijsink VHG, Sørlie M. Lytic polysaccharide monooxygenase. Encyclopedia of Inorganic and Bioinorganic Chemistry. New York: Wiley; 2015. [Google Scholar]
- 15.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels. 2013;6:41. doi: 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vu VV, Beeson WT, Phillips CM, Cate JH, Marletta MA. Determinants of regioselective hydroxylation in the fungal polysaccharide monooxygenases. J Am Chem Soc. 2014;136:562–565. doi: 10.1021/ja409384b. [DOI] [PubMed] [Google Scholar]
- 17.Westereng B, Cannella D, Wittrup Agger J, Jørgensen H, Larsen Andersen M, Eijsink VG, Felby C. Enzymatic cellulose oxidation is linked to lignin by long-range electron transfer. Sci Rep. 2015;5:18561. doi: 10.1038/srep18561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zamocky M, Ludwig R, Peterbauer C, Hallberg BM, Divne C, Nicholls P, Halltrich D. Cellobiose dehydrogenase—a flavocytochrome from wood-degrading, phytopathogenic and saprotropic fungi. Curr Protein Pep Sci. 2006;7:255–280. doi: 10.2174/138920306777452367. [DOI] [PubMed] [Google Scholar]
- 19.Forsberg Z, Mackenzie AK, Sørlie M, Røhr ÅK, Helland R, Arvai AS, Vaaje-Kolstad G, Eijsink VG. Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases. Proc Natl Acad Sci USA. 2014;111:8446–8451. doi: 10.1073/pnas.1402771111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S, Ståhlberg J, Sandgren M, Paton RS, Beckham GT. Quantum mechanical calculations suggest that lytic polysaccharide monooxygenases use a copper-oxyl, oxygen-rebound mechanism. Proc Natl Acad Sci USA. 2014;111:149–154. doi: 10.1073/pnas.1316609111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu M, Beckham GT, Larsson AM, Ishida T, Kim S, Payne CM, Himmel ME, Crowley MF, Horn SJ, Westereng B, Igarashi K, Samejima M, Ståhlberg J, Eijsink VG, Sandgren M. Crystal structure and computational characterization of the lytic polysaccharide monooxygenase GH61D from the basidiomycota fungus Phanerochaete chrysosporium . J Biol Chem. 2013;288:12828–12839. doi: 10.1074/jbc.M113.459396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whittaker JW. Free radical catalysis by galactose oxidase. Chem Rev. 2003;103:2347–2364. doi: 10.1021/cr020425z. [DOI] [PubMed] [Google Scholar]
- 23.Glass NL, Schmoll M, Cate JHD, Coradetti S. Plant cell wall deconstruction by Ascomycete fungi. Annu Rev Microbiol. 2013;67:477–498. doi: 10.1146/annurev-micro-092611-150044. [DOI] [PubMed] [Google Scholar]
- 24.Tian C, Beeson WT, Iavarone AT, Sun J, Marletta MA, Cate JH, Glass NL. Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa . Proc Natl Acad Sci USA. 2009;106:22157–22162. doi: 10.1073/pnas.0906810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennati-Granier C, Garajova S, Champion C, Grisel S, Haon M, Zhou S, Fanuel M, Ropartz D, Rogniaux H, Gimbert I, Record E, Berrin JG. Substrate specificity and regioselectivity of fungal AA9 lytic polysaccharide monooxygenases secreted by Podospora anserina . Biotechnol Biofuels. 2015;8:90. doi: 10.1186/s13068-015-0274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isaksen T, Westereng B, Aachmann FL, Agger JW, Kracher D, Kittl R, Ludwig R, Haltrich D, Eijsink VG, Horn SJ. A C4-oxidizing lytic polysaccharide monooxygenase cleaving both cellulose and cello-oligosaccharides. J Biol Chem. 2013;289:2632–2642. doi: 10.1074/jbc.M113.530196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agger JW, Isaksen T, Várnai A, Vidal-Melgosa S, Willats WG, Ludwig R, Horn SJ, Eijsink VG, Westereng B. Discovery of LPMO activity on hemicelluloses shows the importance of oxidative processes in plant cell wall degradation. Proc Natl Acad Sci USA. 2014;111:6287–6292. doi: 10.1073/pnas.1323629111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borisova AS, Isaksen T, Dimarogona M, Kognole AA, Mathiesen G, Várnai A, Røhr ÅK, Payne CM, Sørlie M, Sandgren M, Eijsink VG. Structural and functional characterization of a lytic polysaccharide monooxygenase with broad substrate specificity. J Biol Chem. 2015;290:22955–22969. doi: 10.1074/jbc.M115.660183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frommhagen M, Sforza S, Westphal AH, Visser J, Hinz SW, Koetsier MJ, van Berkel WJ, Gruppen H, Kabel MA. Discovery of the combined oxidative cleavage of plant xylan and cellulose by a new fungal polysaccharide monooxygenase. Biotechnol Biofuels. 2015;8:101. doi: 10.1186/s13068-015-0284-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pérez S, Bertoft E. The molecular structures of starch components and their contribution to the architecture of starch granules: a comprehensive review. Starch/Stärke. 2010;62:389–420. doi: 10.1002/star.201000013. [DOI] [Google Scholar]
- 31.Ubbink J, Burbidge A, Mezzenga R. Food structure and functionality: a soft matter perspective. Soft Matter. 2008;4:1569–1581. doi: 10.1039/b802183j. [DOI] [PubMed] [Google Scholar]
- 32.Harris P, Wogulis M (2010) Polypeptides having amylolytic enhancing activity and polynucleotides encoding same. Patent No: WO/2010/059413 (2010)
- 33.Ishikawa K, Nakatani H, Katsuya Y, Fukusawa C. Kinetic and structural analysis of enzyme sliding on a substrate: multiple attack in β-amylase. Biochemistry. 2007;46:792–798. doi: 10.1021/bi061605w. [DOI] [PubMed] [Google Scholar]
- 34.Tessema M, Csöregi E, Ruzgas T, Kenausis G, Solomon T, Gorton L. Oligosaccharide dehydrogenase-modified graphite electrodes for the amperometric determination of sugars in a flow injection system. Anal Chem. 1997;69:4039–4044. doi: 10.1021/ac970127f. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi Y, Horikoshi K. Purification and properties of NAD+-dependent maltose dehydrogenase produced by alkalophilic Corynebacterium sp. No. 93-1. Biochim Biophys Acta. 1980;614:256–265. doi: 10.1016/0005-2744(80)90215-6. [DOI] [PubMed] [Google Scholar]
- 36.Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, Foster GD. The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012;13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soanes DM, Chakrabarti A, Paszkiewicz KH, Dawe AL, Talbot NJ. Genome-wide transcriptional profiling of appressorium development by the rice blast fungus Magnaporthe oryzae . PLoS Pathog. 2012;8:e1002514. doi: 10.1371/journal.ppat.1002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marin-Navarro J, Polaina J. Glucoamylases: structural and biotechnological aspects. Appl Microbiol Biotechnol. 2011;89:1267–1273. doi: 10.1007/s00253-010-3034-0. [DOI] [PubMed] [Google Scholar]
- 39.Berry CS. Resistant starch: formation and measurement of starch that survives exhaustive digestion with amylolytic enzymes during the determination of dietary fibre. J Cereal Sci. 1986;4:301–314. doi: 10.1016/S0733-5210(86)80034-0. [DOI] [Google Scholar]