Abstract
Efforts to reduce the global burden of bacterial disease and contend with escalating bacterial resistance are spurring innovation in antibacterial drug and biocide development and related technologies such as photodynamic therapy and photochemical disinfection. Elucidation of the mechanism of action of these new agents and processes can greatly facilitate their development, but it is a complex endeavour. One strategy that has been popular for many years, and which is garnering increasing interest due to recent technological advances in microscopy and a deeper understanding of the molecular events involved, is the examination of treated bacteria for changes to their morphology and ultrastructure. In this review, we take a critical look at this approach. Variables affecting antibacterial-induced alterations are discussed first. These include characteristics of the test organism (e.g. cell wall structure) and incubation conditions (e.g. growth medium osmolarity). The main body of the review then describes the different alterations that can occur. Micrographs depicting these alterations are presented, together with information on agents that induce the change, and the sequence of molecular events that lead to the change. We close by highlighting those morphological and ultrastructural changes which are consistently induced by agents sharing the same mechanism (e.g. spheroplast formation by peptidoglycan synthesis inhibitors) and explaining how changes that are induced by multiple antibacterial classes (e.g. filamentation by DNA synthesis inhibitors, FtsZ disruptors, and other types of agent) can still yield useful mechanistic information. Lastly, recommendations are made regarding future study design and execution.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-016-2302-2) contains supplementary material, which is available to authorized users.
Keywords: Antibiotic, Antiseptic, Disinfectant, Mode of action, SEM, TEM
Introduction
Humankind’s ability to control bacterial pathogens through the use of biocides (antiseptics, disinfectants, and preservatives) and antibacterial drugs has greatly reduced the morbidity and mortality once associated with infectious disease. Introduction of the antiseptic phenol in the 1860s, for example, helped prevent infection during surgery [1], disinfection of municipal drinking water with chlorine in the 1910s reduced waterborne disease [2], and introduction of antibacterial drugs in the 1930s drove down deaths from puerperal sepsis and other infections [3]. More recently, preservatives such as nisin, chlorhexidine, and butylparaben have, respectively, improved the safety of food, pharmaceutical, and cosmetic products [4]. Efforts to develop new antibacterial drugs [5, 6] and biocides [7, 8] continue, accompanied by innovations in antimicrobial materials [9, 10], photodynamic therapy [11], and photochemical disinfection [12, 13]. The impetus for this work varies. Some seek to achieve improved antimicrobial efficacy [7] or safety [6, 8], others more application-specific or user-specific needs such as improved biocompatibility and longer product lifespan for medical implants [9, 10], or the ability to produce safe drinking water in parts of the world that are remote and/or economically disadvantaged [13]. Additional driving factors include the need to keep pace with drug resistance [5, 6, 11], confront the emerging threat of biocide tolerance [7, 14], and protect the growing number of persons at increased risk of infection due to diabetes, HIV/AIDS etc. [5, 6].
Elucidation of antibacterial mechanism of action is a key step in the development of new antibacterial drugs and is becoming increasingly important for biocides also. In the case of antibacterial drugs, this information permits anticipation of problems relating to clinical safety and bacterial resistance [15]. Compounds disrupting the cytoplasmic membrane or electron transport chain, for example, are more likely to cause toxicity problems than those targeting the cell wall or other specifically bacterial structures. Knowledge of mechanism of action also facilitates understanding of drug interactions [16] and allows optimization of drug structure and formulation [17]. For biocides, it is only relatively recently that their mechanisms of action have been studied in detail [14], but this information is important for several reasons too. It permits optimization of activity and formulation as well as neutralization if necessary [18], permits anticipation of safety problems [7], and allows biocides to be rationally selected or avoided depending on environmental conditions [19]. Knowledge of biocide mechanism of action is also considered essential to understanding biocide tolerance [14] and establishing whether or not biocide exposure elicits drug resistance (‘biocide–antibiotic cross-resistance’) [20]. This information is particularly important for those agents used at sublethal concentrations (e.g. preservatives), where their antibacterial effect may depend upon inhibition of a single cellular target, and biocide tolerance is more likely to arise [21]. Lastly, for both drugs and biocides, knowledge of mechanism of action enables us to select combinations of agent that inhibit multiple cellular targets, thereby reducing the risk of resistance or tolerance emerging [22, 23].
Establishing the mechanism of action of a novel antibacterial compound or process is inherently difficult because the biochemical pathways of bacterial cells are closely interlinked and disturbance of any one system invariably affects many others [24]. The genomics era has furnished us with several advanced new methods of elucidating antibacterial mechanism of action, but these techniques have not yet been widely adopted. In some settings, lack of specialized equipment or technical expertise may be a barrier to their use. For other researchers, current limitations of the methods may be a disincentive. Transcriptional arrays, for example, generate complex patterns of results that can be difficult to interpret [15, 25]. A forerunner of the molecular biology approach was the examination of bacteria treated with test agent for changes to their morphology and ultrastructure [24]. This strategy remains extremely popular among researchers investigating antibacterial mechanism of action, both as an early exploratory step [26–30] and for confirmation of a suspected mechanism [31–33]. Requiring only access to an electron microscopy suite, it is an approach that is open to researchers in even relatively low resource settings. New and emerging technologies such as atomic force microscopy [34], live-cell time-lapse microscopy [35], and automated image analysis [35, 36] are currently less accessible, but could potentially make microscopy-based investigation of mechanism of action more informative and efficient and more attractive to industry.
Despite the enduring appeal of using antibacterial-induced cytological changes as an indicator of mechanism of action, most of the primary research published on this topic has escaped critical review. One consequence of this is that the associated terminology has become confused, with some authors using the same term to describe different observations (e.g. ‘filament’ used to describe both elongated cells [37, 38] and noncellular thread-like structures [39, 40]) or coining many terms to describe the same observation (e.g. ‘blebbing’ [41], ‘blistering’ [24], and ‘bubbling’ [42] used to describe outer membrane protrusions). Another problem, in part related to the first, is that it is unclear to what extent (a) different antibacterial agents with the same mechanism of action trigger the same alteration and (b) the same alteration can be triggered by antibacterial agents with different mechanisms of action. The present review addresses the issues above, describes factors that can influence results, and, where information is available, explains the sequence of events that leads to the morphological or ultrastructural change. Because the mechanism of action of experimental antibacterials such as curcumin have been misidentified in the past [15], only studies with well-characterized antibacterial agents have been included in the review. Studies with the nitrofurans antibiotics have been excluded because they attack multiple cellular targets [43], as have studies with bacteria resistant to the drug they were exposed to, since resistance mechanisms frequently induce ultrastructural changes of their own [44–46].
Variables affecting antibacterial agent-induced morphological and ultrastructural changes
When bacteria are treated with an antibacterial agent, the morphological and ultrastructural changes that occur depend not just on the identity of the antibacterial agent, but also on the concentration at which it is tested [36, 47] and the duration of exposure [48, 49]. Other variables known to influence results relate to the test organism and incubation conditions.
Bacterial factors affecting morphostructural changes include the cell wall structure of the test organism (Gram-positive vs. Gram-negative) [50], the species that is used [51, 52], the characteristics of the test strain (including its antibiotic susceptibility) [53, 54], and the inoculum density [55, 56]. Growth phase affects the size, shape, and cell wall thickness of bacteria even in the absence of antibacterial agent, with stationary phase cells being smaller [57, 58], having a lower axial ratio [57], and a thicker cell wall [59, 60] than logarithmically growing cells. In studies examining the effects of antibacterial agents on bacteria, differences in the status (replicating vs. nonreplicating or dying) [61, 62] and age (new daughter cell undergoing elongation vs. older cell undergoing division) [63] of individual cells at the time of exposure are thought to account for the heterogeneity of morphological changes observed within treated populations. The relative proportions of these different cells within a bacterial population vary with the growth phase. In a population that is in stationary phase [64] or death phase [65], antibiotic exposure may not alter cell appearance at all.
Incubation conditions affecting morphostructural changes include the type of growth medium (broth vs. agar) [66, 67], the osmolarity of this medium [68, 69], and the incubation temperature [66]. For example, antibacterial agent-induced filaments have a curved appearance when agar is used, and a straight appearance when a fluid medium is used [67]. Transient plasmolysis (i.e. temporary retraction of the protoplast from the cell wall) occurs in growth media that are hyperosmotic, but without osmotic protectants such as sucrose and MgSO4, antibacterial agent-induced spheroplasts [57] and protoplasts [69] may burst before observation is possible. Regarding temperature, the incubation of bacteria below 37 °C results in antibacterial-induced morphological changes taking place more slowly and appearing more clearly [66].
Morphological and ultrastructural changes
Spheroplast and protoplast formation
In bacteriology, the terms ‘spheroplast’ and ‘protoplast’ are used to describe cells that have lost their peptidoglycan layer. Without this rigid, shape-determining structure, membrane tension causes the bacteria to acquire a spherical shape. Spheroplasts are Gram-negative bacteria that have lost their peptidoglycan layer, but not their outer membrane, whereas protoplasts lack both a peptidoglycan layer and an outer membrane, either because they were formed from Gram-positive bacteria or because their outer membrane has been stripped away [70]. Various agents convert bacteria into these altered forms, with the peptidoglycan synthesis inhibitor penicillin G [71] and the peptidoglycan digesting enzyme lysozyme [72] among the first discovered. The significance of these early observations, incidentally, was not lost on researchers at Merck, Fujisawa or Sankyo, all three companies developing screens for cell wall synthesis inhibiting antibiotics based on spheroplast formation [73]. Spheroplasts and protoplasts, when observed by microscopy, appear as large, round and smooth refractile bodies (Fig. 1) [61]. Because they lack a peptidoglycan layer, spheroplasts and protoplasts are osmotically sensitive and rapidly lyse when transferred to hypotonic media [53, 74].
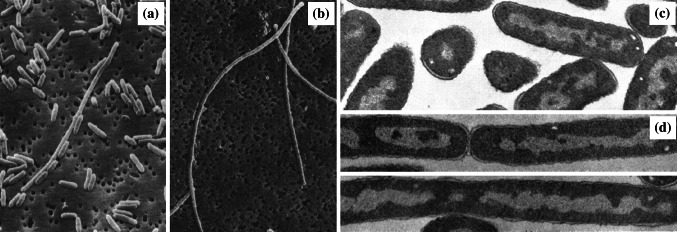
Fig. 1.
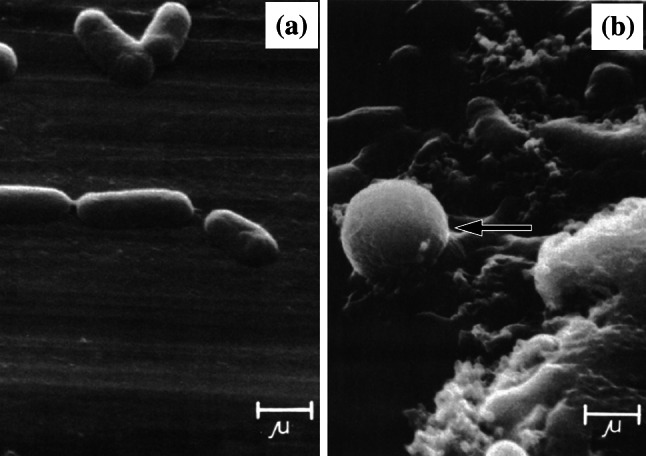
Scanning electron micrographs of a untreated Escherichia coli JP5128 and b cells of the same strain treated with 200 µg/mL chloramphenicol for 3 h (this concentration and time frame were selected because it achieved near-complete inhibition of protein synthesis). Arrow shows the location of a spheroplast. Bar 1 µm.
Images from [97] by permission of Antimicrobial Agents and Chemotherapy (ASM)
Undoubtedly, the most well-known and well-studied class of spheroplast-inducing peptidoglycan synthesis inhibitors is the β-lactams. In Escherichia coli, β-lactam-induced spheroplast formation is due to inhibition of penicillin-binding proteins 1a and 1b (PBPs 1a and 1b) [75–77]. PBPs 1a and 1b are required for peptidoglycan synthesis, and inhibition of these enzymes, in the absence of peptidoglycan hydrolase inhibition, is thought to explain spheroplast formation [75]. For β-lactams which have a higher affinity for E. coli PBPs 1a and 1b than PBP2 or PBP3, some examples being amoxycillin, cephaloridine, and cefsulodin, spheroplast formation is the primary morphological response to antibiotic treatment [76]. In cephaloridine-treated E. coli, spheroplast formation is observable between 1×MIC and 16×MIC, with 1×MIC cephaloridine converting over 20 % of cells to spheroplasts in just 60 min [77]. For β-lactams which preferentially target E. coli PBP2 and/or PBP3, other morphological changes (‘ovoid cells’, ‘localized swelling’ and ‘filaments’) occur at low antibiotic concentrations, with spheroplasts only becoming evident when antibiotic concentration is increased to a point where PBPs 1a and 1b become bound [76, 78]. The β-lactam-induced formation of spheroplasts and protoplasts occurs, not just in E. coli, but in many species of Gram-negative [36, 53, 74, 79–83] and Gram-positive bacteria [69, 84]. However, because PBP nomenclature is species dependent [85, 86], and because relatively few studies have determined the affinity of their test β-lactam(s) for the PBPs of their test species, the relationship between spheroplast formation and inhibition of a specific PBP is less clearly defined for these species than E. coli.
Examples of spheroplast-inducing disruptors of peptidoglycan synthesis outside the β-lactam class include the MurA inhibitor fosfomycin [73], the MurC inhibitor glycine [74, 87], the Alr and Ddl inhibitor cycloserine [73], the MraY inhibitor mureidomycin [73, 88], the d-alanyl-d-alanine binding antibiotic vancomycin [89], the transglycosylase inhibitors ensanchomycin, prenomycin [73], and moenomycin [90], and the transpeptidase inhibitor lactivicin [91]. Depriving cells of diaminopimelic acid [57] or inhibiting its incorporation into the cell wall using malioxamycin [73] also triggers spheroplast formation. Lysozyme, which enzymatically degrades peptidoglycan by hydrolysing the glycosidic bonds between N-acetyl muramic acid and N-acetyl-d-glucosamine, causes protoplast formation in Gram-positive cells [92]. In Gram-negative bacteria, peptidoglycan is shielded by the outer membrane [93], and lysozyme cannot induce spheroplast formation unless a membrane permeabilizer such as lactoferrin or EDTA is also present [74, 92, 94]. Other peptidoglycan hydrolysing enzymes that induce protoplast formation include N,O-diacetylmuramidase [95] and lysostaphin [96].
Having established that digestion of peptidoglycan or inhibition of its synthesis frequently results in spheroplast or protoplast formation, the next logical question is whether any other antibacterial mechanisms induce this change too. Perhaps not surprisingly, antibacterial agents that inhibit pathways directly upstream of peptidoglycan synthesis sometimes induce spheroplast formation. Fosmidomycin and phosphoenolpyruvate are two examples. Fosmidomycin inhibits the non-mevalonate pathway and is thought to cause spheroplast formation by preventing synthesis of the cell wall carrier lipid undecaprenyl-phosphate [73]. Pentalenolactone inhibits the glycolytic pathway and is thought to induce spheroplast formation by preventing synthesis of the MurA substrate phosphoenolpyruvate [73]. In some cases, inhibition of protein synthesis (or upstream inhibition of the folic acid synthesis required to produce amino acids) also causes spheroplast formation. Spheroplasting has been reported in bacteria treated with chloramphenicol [61, 97], oxytetracycline [39], several aminoglycoside antibiotics (kanamycin, streptomycin, and tobramycin) [61, 97], trimethoprim [51] and sulfamethoxazole [97]. That these spheroplasting effects are attributable to inhibition of folic acid and protein synthesis, and not a second mechanism targeting peptidoglycan synthesis, has been confirmed, respectively, by rescue with folinic acid [51] and mutant studies [97]. Neither DNA nor RNA synthesis inhibition generates spheroplasts, at least in the case of novobiocin [80], nalidixic acid [98], ciprofloxacin, or rifampicin [83].
Ovoid cells
‘Ovoid cells’ is a term used to describe bacterial rods that have decreased in length and become oval or round shaped during antibacterial treatment [75, 99]. Nomenclature in the literature varies, with some authors referring to them as ‘round forms’ [76, 100], ‘round cells’ [63, 101, 102], ‘spherical forms’, ‘spherical cells’ [54, 78, 103], or ‘coccoid forms’ [104, 105]. In a few papers, these descriptors are used interchangeably with the term ‘spheroplast’ [54, 101, 102], but this is incorrect. Unlike spheroplasts, ovoid cells are osmotically stable [63, 75, 76, 100, 106]. This difference can be demonstrated by comparing impression smears of the antibiotic-treated bacteria overlaid with drops of distilled water or 0.85 % (w/v) NaCl [53], or by performing the antibiotic treatment itself in media with and without osmotic support [69]. There are morphological differences also. Ovoid cells typically lack the near-perfect spherical shape and smooth surface of spheroplasts [36], and are more likely to be found in cell arrangements than as individual cells [36] (Fig. 2).
Fig. 2.
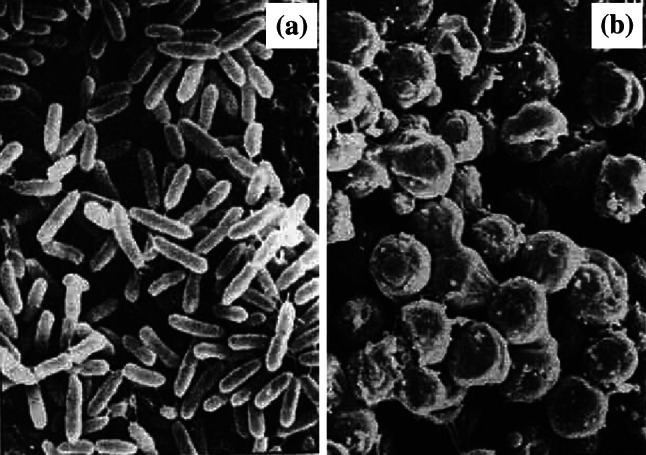
Scanning electron micrographs of a untreated Pseudomonas aeruginosa MB3286 and b cells of the same strain treated with 2 µg/mL (2×MIC) imipenem for 3 h. All of the treated bacteria have changed to ovoid cells. Magnification ×9000.
Images from [101] by permission of Innate Immunity (SAGE Publications)
Ovoid cell formation occurs when bacteria are treated with some types of β-lactam antibiotic. In E. coli and Pseudomonas aeruginosa, this change in shape has been attributed to inhibition of the enzyme PBP2 [75, 101]. Inhibition of PBP2 leads to cessation of lateral wall peptidoglycan synthesis and permanent activation of septal wall peptidoglycan synthesis, resulting in daughter cells that comprise two poles with no cylindrical peptidoglycan to separate them [63, 107]. β-Lactams that preferentially inhibit PBP2 can induce ovoid cell formation rapidly (within 1.5–2 h) over quite a wide range of antibiotic concentrations (≤0.5×MIC to ≥2×MIC) [108]. Examples include imipenem [54, 101, 103], mecillinam [75, 106, 108], and thienamycin [109]. For β-lactams whose affinity for PBP2 and PBP3 is similar, a different morphological change (‘localized swelling’) occurs [76, 101] and this will be discussed later in the review. Under increasing β-lactam concentrations, ovoid cells are typically observable until PBPs 1a and 1b [76, 78, 101] or PBPs 2 and 3 [85, 110] become saturated, at which point the cells form spheroplasts and/or lyse depending on the PBPs affected and the osmolarity of the medium. The β-lactam-induced formation of ovoid cells has been reported, not just in E. coli and P. aeruginosa, but in many species of Gram-negative bacteria including Acinetobacter baumannii [102], Klebsiella pneumoniae [54], Proteus mirabilis [54, 111], and Serratia marcescens [54]. Again, however, because PBP nomenclature is species dependent [86], and because relatively few studies have determined the affinity of their test β-lactam(s) for the PBPs of their test organism(s), ovoid cell formation has not been attributed to inhibition of a specific PBP in these organisms.
Ovoid cell formation also occurs if the proteins that regulate PBP2 localization are disrupted. Inhibition of the cytoskeletal protein MreB by S-(3,4-dichlorobenzyl)isothiourea [112, 113] or its derivative 1,2,3,4-tetrahydro-1,3,5-triazine [114] converts cells of E. coli and P. aeruginosa into ovoid forms. This morphological change is detectable over a range of antibacterial concentrations (0.25×MIC to 4×MIC) [114] and has been confirmed to occur in cells where MreB has been inactivated through mutation [107]. Mutational inactivation of proteins MreC, MreD and RodA, which associate with MreB to recruit PBP2 and other key enzymes, also generates ovoid cells [63, 107].
Possible ovoid cells have been reported too in bacteria treated with low or sub-MIC levels of the folic acid synthesis inhibitors aminopterin [104] and trimethoprim [51]. The focus of these early studies was on filaments and spheroplasts rather than ovoid cells though, and in the absence of any detailed descriptions or micrographs of the antifolate-induced ovoid cells, it is not clear to what extent they resemble the forms induced by PBP2 inhibitors and disruptors.
Filamentation
When rod-shaped bacteria (or, in some cases, cocci) produce peptidoglycan for their lateral wall but not their septal wall during growth, the cells become abnormally long (Fig. 3). This phenomenon is generally referred to as ‘cell elongation’ or ‘filamentation’. Use of the two terms varies between studies [37, 39, 49, 56, 81]. For example, some authors use ‘elongation’ to describe cells that are up to five times their normal length, and ‘filaments’ to describe longer cells [56]. Other authors prefer the term ‘short filament’ to refer to bacteria up to 15 µm in length, and ‘long filament’ for cells in excess of 15 µm [37]. In this review, we will use the latter (less precise, but more user-friendly) nomenclature. In the absence of antibacterial agents or other stressors, filamentation occurs at a low frequency in bacterial populations [115, 116] (~4–8 % short filaments and 0–5 % long filaments in 1- to 8-h cultures [116]) and has been observed in both logarithmically growing and stationary phase cells [57]. Increased cell length offers protection against protozoan predation [117] and phagocytosis by neutrophils [49, 118] because it makes ingestion of the cells more difficult. The frequency and magnitude of this filamentation increases when bacteria are treated with various physical and chemical agents.
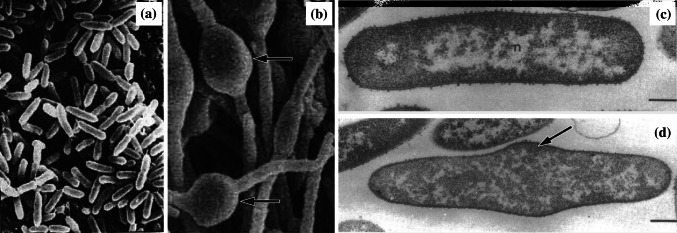
Fig. 3.
Scanning electron micrographs of a untreated Pseudomonas aeruginosa X48 and b cells of the same strain treated with 32 µg/mL (2×MIC) of the β-lactam BL-P1654 for 4 h, and transmission electron micrographs of c untreated Escherichia coli ATCC 11303 and d cells of the same strain treated with 0.1 µg/mL (1×MIC) mitomycin C for 2 h. The number and length of filamentous cells has increased following treatment. Magnification for a ×2700, b ×2200, c ×18,000, and d ×18,000.
Images a and b from [67] by permission of Antimicrobial Agents and Chemotherapy (ASM), and c and d from [133] by permission of the Journal of Bacteriology (ASM)
Filamentation can occur following inhibition or disruption of peptidoglycan synthesis. In β-lactam-treated cells of E. coli and P. aeruginosa, this change has been attributed to the inhibition of PBP3 [75, 119]. PBP3 cross-links peptidoglycan at the septal wall but not the lateral wall, so inhibition of this enzyme results in cell elongation without division [58, 86]. Cefuroxime and ceftazidime, which have a higher affinity for PBP3 than PBPs 1a, 1b or 2 [76, 120], cause E. coli and P. aeruginosa to form filaments relatively quickly (within 3–4 h) at low antibiotic concentrations (0.008×MIC to 1×MIC), with spheroplast formation or lysis becoming observable at 0.5×MIC and increasing at higher concentrations [54, 76, 120]. This transition from filaments to spheroplasts or lysis also increases with the duration of antibiotic exposure [55]. The β-lactam-induced formation of filaments occurs, not just in E. coli and P. aeruginosa, but in many species of Gram-negative bacteria [49, 53, 65, 74, 81, 121–123] and a limited number of Gram-positive bacteria [124–126]. In most cases, it is bacilli that are affected, but filamentation has also been reported in some species of cocci [124, 125, 127], specifically those in the Streptococcus genus, which divide in just one plane. The length of β-lactam-induced filaments varies with the duration of exposure [49], but cells up to 93 µm have been reported [128].
Filamentation can also occur if DNA synthesis is inhibited [48, 49] or DNA is damaged [129–131] by a process known as the SOS response. This response represses septum formation until the DNA can be repaired, the delay preventing the transmission of damaged DNA to daughter cells [58]. Bacteria postpone septation by synthesizing protein SulA, an FtsZ inhibitor that halts Z-ring formation, thereby stopping PBP3 recruitment and activation. Examples of DNA disrupting agents that induce this SOS-mediated filamentation include cosmomycin D [36], metronidazole [132], mitomycin C [131, 133], nalidixic acid [98, 131] and the fluoroquinolones [37, 48, 134], novobiocin [36], zidovudine [135], and UV light [129]. The fluoroquinolone ciprofloxacin induces filamentation rapidly (within 1–1.5 h) over a wide range of concentrations (1×MIC to ≥33×MIC) [56, 136], with mean filament length peaking at 1×MIC to 10×MIC [56] and decreasing thereafter, probably due to inhibition of RNA and protein synthesis [136]. Similar concentration–response patterns have been observed with mitomycin C [133] and ofloxacin [49]. If bacteria are deprived of the nucleobase thymine by starvation [130], by treatment with the folic acid synthesis inhibitor trimethoprim [49, 51, 135], or by treatment with the folic acid analogue aminopterin [104], this also disrupts DNA synthesis and induces SOS-mediated filamentation [135]. As with fluoroquinolone-induced filamentation, trimethoprim-induced filamentation occurs over a wide range of concentrations (0.06×MIC to 1×MIC), peaking at 0.13×MIC and decreasing thereafter [49]. The length of filaments generated by DNA disruptors and antifolates varies with the duration of exposure, but is broadly similar in both cases [137], with cells capable of reaching over 50 µm [104, 137]. Direct obstruction of Z-ring formation by SulA and other FtsZ inhibitors, like the indirect obstruction of Z-ring formation caused by DNA disruptors and antifolates, generates very long SOS-like filaments [138, 139].
Inhibition of protein synthesis [127, 140–142] (or the RNA synthesis that precedes it [143]) can also induce filamentation, but from the available evidence this increase in cell length is much less than that caused by inhibitors of peptidoglycan or DNA synthesis [52, 61, 144]. For example, cells of E. coli treated with bactericidal levels of kanamycin undergo just a 1.6-fold increase in size after 3 h, whereas cells treated with equivalent concentrations of ampicillin or norfloxacin undergo a 9.4-fold increase in the same time frame [144]. In other studies, for example with gentamicin [83, 145], amikacin [39] and chloramphenicol [39, 49], any increase in cell length has been so insignificant as to go undetected or unreported. The length of filaments generated by protein synthesis inhibitors varies with the duration of exposure [146], but cells as long as 11 µm have been observed [61]. Membrane disruption by daptomycin [147] or polymyxin B [39] can induce filamentation too, but, again, these filaments are comparatively short in length. To our knowledge, no mechanisms have been proposed to explain why inhibition of protein synthesis or disruption of bacterial membranes leads to filament formation.
Pseudomulticellular bacteria
When cell separation is prevented in dividing cocci, this gives rise to cells with multiple septa (Fig. 4), cells that appear either swollen or elongated depending on whether the species divides in multiple planes [125, 148] or just one [149, 150]. Such bacteria have been variously described as ‘large multiseptate organisms’ [151], ‘pseudomulticellular bacteria’ [60, 152], and ‘multicellular clusters’ [153]. The term ‘aggregates’ is sometimes used also [154], but this is misleading because it implies the altered forms have arisen from cells coming together rather than cells failing to separate. Given their outward appearance as abnormally large or long cells, pseudomulticellular bacteria could potentially be mistaken for spheroplasts or other phenotypes unless transmission electron microscopy (TEM) is performed to detect the multiple septa. Antibacterial drugs known to induce this change include peptidoglycan synthesis inhibitors in the β-lactam [148, 150, 155] and glycopeptide [153] classes, the protein synthesis inhibitors chloramphenicol [60], lincomycin [125] and minocycline [156], and the folic acid synthesis inhibitor trimethoprim [157].
Fig. 4.
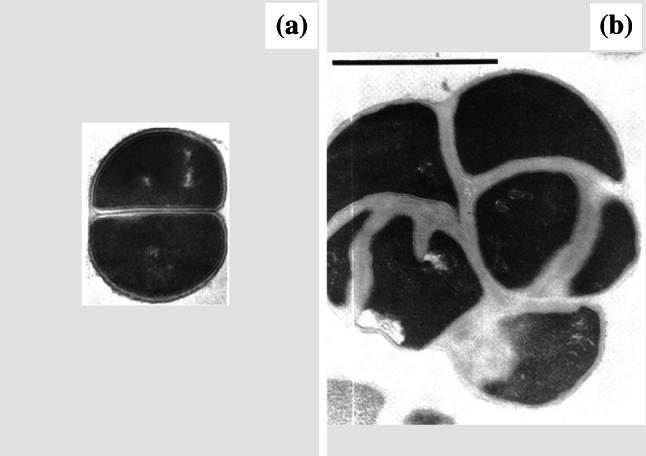
Transmission electron micrographs of a untreated Staphylococcus aureus ATCC 6538P and b cells of the same strain treated with 0.027 µg/mL (0.33×MIC) cephaloridine for 6 h. Treatment has inhibited cell separation, resulting in a pseudomulticellular form. Magnification for a ×23,500 and b ×21,500. Bar 1 µm.
Images from [125] by permission of Antimicrobial Agents and Chemotherapy (ASM)
In β-lactam-induced pseudomulticellular bacteria, the septal wall is unusually thick (2–8 times thicker than in untreated bacteria) [125, 158] and lacks the central dense layer present in phenotypically normal cells [125, 127, 158]. The peripheral cell wall appears unaltered except for occasional thickened areas [125, 127, 158]. These pseudomulticellular bacteria have been proposed to result either from β-lactams inhibiting the peptidoglycan hydrolases required for septal lysis [152, 158], or perhaps more likely, from β-lactams altering the cell wall composition and steric configuration in such a way that these enzymes cannot act [152]. This change has been widely reported in bacteria treated with sub-MIC levels (0.06×MIC to 0.5×MIC) of β-lactams [111, 125, 148, 155, 158], where it can occur within 3.5 h [155] and remain observable up to 14 h following exposure [125]. Prior to [151] or after [84] this window, septal wall thickening is detected, but not pseudomulticellular bacteria. Pseudomulticellular bacteria can also arise following treatment with supra-MIC levels (10×MIC) of β-lactams, but these are more short lived, cell lysis occurring after 8 h [150]. Both Gram-positive [150, 158] and Gram-negative [125, 127, 148] cocci can be affected. Pseudomulticellular bacteria with a similar appearance to those induced by β-lactams (i.e. thickened septal walls, but normal peripheral walls) have been observed following 2.5 h treatment with 0.5×MIC vancomycin, with these forms remaining observable at least 4.5 h following exposure [153]. This alteration in ultrastructure has been ascribed to vancomycin blocking the access of peptidoglycan hydrolases to their substrate [153] and to vancomycin directly inhibiting these hydrolases [159].
Inhibition of protein synthesis can also generate pseudomulticellular forms [60, 125, 156], but these bacteria differ in appearance from those generated by peptidoglycan synthesis inhibitors. Both types of pseudomulticellular bacteria possess thickened septal cell walls, but a central dense layer remains observable in those induced by protein synthesis inhibition, and the peripheral walls exhibit thickening that is continuous, not sporadic [125, 156]. The peripheral walls can be three to four times thicker than those in untreated control cells [155]. The above change in ultrastructure has been reported after treatment with sub-MIC and MIC levels of chloramphenicol [60], lincomycin [125], and minocycline [156]. Studies with other protein synthesis inhibitors have not detected pseudomulticellular bacteria [156, 160], but antibiotic-treated cells in these studies were not examined until 24 h after exposure, and this may simply have been too late for observation. Chloramphenicol induces pseudomulticellular forms by reducing peptidoglycan hydrolase activity [60], possibly by inhibiting enzyme synthesis, but more likely by inhibiting synthesis of an effector required for enzyme activity [161]. Reduced peptidoglycan hydrolase activity has been observed with several protein synthesis inhibitors [161], so this is probably how lincomycin and minocycline induce pseudomulticellular forms too. In contrast to the above findings with translation inhibitors, inhibition of the antecedent process of transcription appears not to induce pseudomulticellular bacteria [150, 155], suggesting that cells possess an adequate stock of mRNA, tRNA, and rRNA to synthesize the peptidoglycan hydrolase enzymes or effector molecules required for cell separation.
The folic acid synthesis inhibitor trimethoprim has also been reported to induce pseudomulticellular bacteria [157]. Limited information is available, but the pseudomulticellular bacteria exhibited ‘continuous’ type peripheral cell wall thickening [157] characteristic of the protein synthesis inhibitors chloramphenicol, lincomycin and minocycline. These observations, coupled with the absence of any reports of DNA or RNA disruptors inducing pseudomulticellular bacteria, suggest that trimethoprim may act in a similar manner to chloramphenicol, its inhibition of folic acid synthesis preventing production of the amino acids [162] required for protein synthesis and peptidoglycan hydrolase activity.
In addition to the antibacterial drugs above, negatively charged polyelectrolytes such as Evans blue, divalent cations such as Mg2+, and various surfactants can generate pseudomulticellular bacteria [60]. For the anionic polyelectrolytes and divalent cations, this change is thought to occur through indirect inhibition of peptidoglycan hydrolases, perhaps due to an interaction with lipoteichoic acid, one of the structures that regulates these enzymes. The surfactants Triton X-100 and sodium dodecyl sulphate induce pseudomulticellular bacteria by inactivating peptidoglycan hydrolases directly [60]. Additional evidence that pseudomulticellular bacteria form following disruption of cell wall synthesis and inhibition of peptidoglycan hydrolases is available in the form of mutant [125, 152, 163] and nutrient deprivation studies [149].
Other forms of septal disruption
In addition to the filaments and pseudomulticellular bacteria described above, there have been occasional reports of antibacterial agents inducing misshapen or otherwise aberrant septa. Antibiotics eliciting these effects include the cytoplasmic membrane disruptor daptomycin [164] and the peptidoglycan synthesis inhibitors penicillin [69] and ramoplanin [165]. That these agents disrupt septum formation is not surprising given cell division requires both Z-ring formation at the cytoplasmic membrane and septal peptidoglycan synthesis [166].
Altered cell size
After some treatments, bacterial cells have been reported to increase in size, decrease in size, or both increase and decrease in size. Increased cell length (‘filamentation’) has already been discussed. Increases in the diameter of cocci can be difficult to interpret if detected by scanning electron microscopy (SEM) only [50, 61]. If, however, the enlarged bacteria are examined in cross section, the increase in size can usually be attributed to inhibition of cell separation (‘pseudomulticellular bacteria’) [127, 158] or osmotic swelling (‘spheroplasts’ or ‘protoplasts’) [84]. Decreases in cell size (both rod diameter and length) have been detected in Legionella pneumophila treated with the peptidoglycan synthesis inhibitor ampicillin [141], and P. aeruginosa treated with the protein synthesis inhibitor erythromycin [167]. The cause(s) of these decreases in size was not established [141, 167]. Lastly, a combination of both increased and decreased cell size has been observed in Staphylococcus aureus populations treated with novobiocin [168]. Cross sections of the altered bacteria were not examined, but it was speculated that the changes in size may be due to abnormal cell division [168].
Localized swelling
Under some conditions, rod-shaped bacteria develop a localized swelling that is contained within an intact cell envelope and encircles the entire circumference of the cell (Fig. 5). When the swelling is induced by disruption of peptidoglycan synthesis, it typically occurs at the mid-section of the cell [57, 169], whereas swelling induced by disruption of protein synthesis has, to date, only been reported at the pole [141, 170]. This change in bacterial morphology has been described as ‘bulging’ by some authors [50, 103, 169] and ‘swelling’ by others [55, 171, 172], with the terms ‘oval-centred cells’ [54], ‘sphero-rods’ [101] and ‘spindle-shaped forms’ [57, 173] used for rods with mid-cell swelling, and ‘bottle-shaped forms’ used for rods with polar swelling [57].
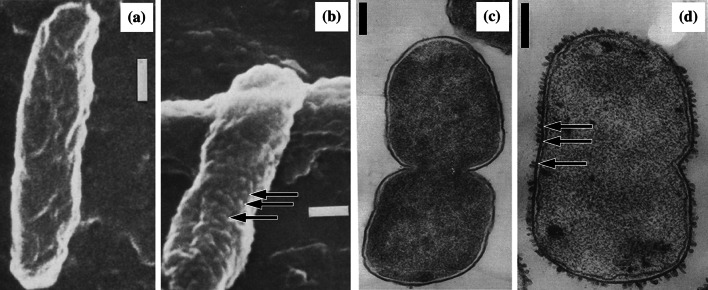
Fig. 5.
Scanning electron micrographs of a untreated Pseudomonas aeruginosa MB3286 and b cells of the same strain treated with 2 µg/mL (2×MIC) meropenem for 6 h, and transmission electron micrographs of c untreated Escherichia coli ATCC 12407 and d cells of the same strain treated with 5 µg/mL (~3×MIC) ampicillin for 1 h. Arrows show the location of localized swelling. Magnification for a ×9000, b ×14,000, c ×27,500, and d ×27,000. Bar 0.25 µm.
Images a and b from [101] by permission of Innate Immunity (SAGE Publications), and images c and d from [169] by permission of the Journal of Bacteriology (ASM)
Almost all reports of localized swelling have been in cells treated with β-lactams. With β-lactam-induced swelling, the affected cells retain osmotic stability [75], and the rod-shaped part of the cell continues to increase in length as the swelling increases in diameter [101]. In E. coli and P. aeruginosa, localized swelling has been attributed to simultaneous inhibition of PBP2 and PBP3 [54, 75, 101, 103, 173] and is thought to represent an intermediate form between ovoid cells and filaments [54, 76]. β-Lactams with a similar affinity for PBPs 2 and 3, for example ampicillin and meropenem, can induce localized swelling rapidly (within 1–2 h) [101, 103, 169, 173]. Antibiotic concentrations capable of inducing this effect range from low sub-inhibitory (0.25×MIC) [173, 174] to high supra-inhibitory concentrations (32×MIC) [55], the swelling remaining observable for as little as 3 h or as long as 24 h following initial antibiotic exposure [66, 173]. Morphological changes that precede, coincide with, or follow the appearance of swelling vary depending on the relative affinity of the antibiotic for the PBPs of the test organism [76, 101]. For example, treatment of P. aeruginosa with meropenem (PBP affinity 3 > 2 > 1) initially results in filament formation, followed by filaments with swelling at individual and then multiple points along the cell, and finally spheroplast formation and lysis [101]. β-Lactam-induced swelling has been detected, not just in E. coli and P. aeruginosa, but in multiple species of Gram-negative bacillus [54, 66, 82].
A comparable morphological change to that described following β-lactam treatment has been reported in L. pneumophila and Helicobacter pylori treated with the protein synthesis inhibitors erythromycin [141] and rokitamycin [170]. Limited information is available, but the swelling (described by the authors as ‘bulbous distortions’ [141] and ‘enlargement of part of the bacterial cell’ [170]) was greatest at one pole and diminished in size closer to the mid-cell. This morphological change was observed following treatment with sub-MIC levels of rokitamycin [170] and supra-MIC levels of erythromycin [141].
Bulge formation
Whilst the term ‘bulge’ is sometimes used to described the localized swelling already discussed, it is more commonly used to describe a different phenomenon. In accordance with most authors [35, 82, 172, 175], this review considers a bulge to be a protrusion from a single point in the cell surface, appearing unilateral when examined from above or in section, its centre located outside the normal boundaries of the cell (Fig. 6). Localized swelling, by contrast, encircles the entire circumference of the rod, appearing bilateral in cross section, its centre occurring at or close to the cell’s longitudinal axis (Fig. 5). Confusingly, some authors refer to bulges (as defined above) as ‘blebs’ [61, 176]. The present review, in accordance with authoritative papers on the subject, differentiates bulges from blebs on the basis that bulges involve the cytoplasmic membrane [35, 57, 172] and blebs are derived from just the outer membrane (Fig. 7) [143, 171]. Bulges typically occur mid-cell or along the cylinder of the cell rather than at the poles [61, 70, 172]. A single bulge per cell is more frequently reported than multiple bulges per cell, but multiple bulges can occur (Suppl. Figure 1) [57, 61, 124, 177], sometimes to such an extent that the affected cells become raspberry-like in appearance [61].
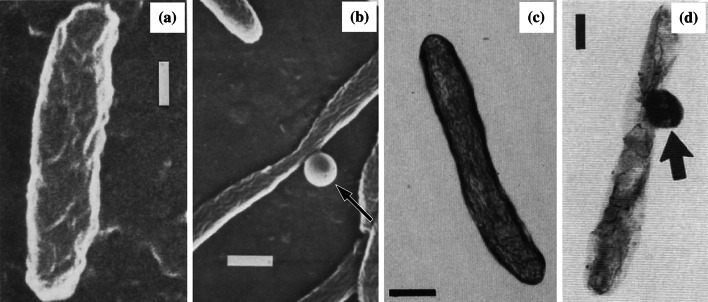
Fig. 6.
Scanning electron micrographs of a untreated Legionella pneumophila ATCC 33153 and b cells of the same strain treated with 1000 µg/mL (40×MIC) penicillin for 5 h, and negatively stained electron micrographs of c untreated L. pneumophila Nottingham N7 and d cells of the same strain treated with 1280 µg/mL (20×MIC) methicillin for 24 h. Arrows show the location of bulge formation. Bar for a 0.2 µm, b 0.4 µm, c 0.5 µm, and d 0.5 µm.
Images a and b from [171] and images c and d from [83], all by permission of the Journal of Medical Microbiology (MicroSoc)
Fig. 7.
Scanning electron micrographs of a untreated Legionella pneumophila ATCC 33153 and b cells of the same strain treated with 1600 µg/mL (10×MIC) polymyxin B for 5 h, and transmission electron micrographs of c untreated Escherichia coli ATCC 11303 and d cells of the same strain treated with 25 µg/mL (~12.5×MIC) polymyxin B for 30 min. Arrows show the location of some of the blebs. All bars 0.2 µm.
Images a and b from [171] by permission of the Journal of Medical Microbiology (MicroSoc), and images c and d from [180] by permission of the Journal of Bacteriology (ASM)
Bulge formation precedes the appearance of spheroplasts (or, in the case of Gram-positive bacteria, protoplasts) [61, 70, 178], and the bulges themselves are sometimes referred to as ‘spheroplast-like structures’ [50, 82, 174] or ‘emerging spheroplasts’ [83, 178]. Bulges, like spheroplasts, can be osmotically sensitive [57], but are not always [124]. Like spheroplasts, they form following inhibition of peptidoglycan synthesis (by β-lactam [50, 83, 171, 174] and glycopeptide antibiotics [172]), inhibition of protein synthesis (by tobramycin [61] and arbekacin [179]), and inhibition of folic acid synthesis (by sulphamethoxazole [61]). Bulge formation is also triggered by the cytoplasmic membrane disrupting antibiotic daptomycin [176, 177], which is not a known spheroplast inducer. Like spheroplasts and protoplasts, bulge formation can occur in a wide range of Gram-negative [57, 66, 70, 82, 171] and Gram-positive [61, 124, 176] species. Bulges are observable after as little as 15–20 min treatment with bactericidal levels of benzylpenicillin (~20×MIC) [57] or daptomycin (2×MIC) [176] and after 3 h treatment with inhibitory concentrations of tobramycin (1×MIC) or sulfamethoxazole (1×MIC) [61]. The bulges remain observable until the transition to spheroplast is complete or until they lyse [57, 178], this occurring as early as 2.5–3.5 h after treatment [57] or as late as 24 h or more after treatment [66, 83].
Bulge formation has been attributed to an accumulation of cross-link defects in a small region of peptidoglycan creating a pore. Because peptidoglycan synthesis is most active near the mid-cell position, this is where most defects arise and where, therefore, pores tend to develop [172]. Above a certain size, the osmotic pressure differential drives the cytoplasmic membrane out through the pore, creating a bulge [175]. Sometimes, the cell wall cracks at the location of the cross-link defects, with the cytoplasmic membrane then bulging out of the crack [35, 172, 178]. In Gram-negative bacteria, the outer membrane supports some level of turgor pressure, delaying osmotic lysis of the bulge [35]. In sharp contrast to what is observed during ‘localized swelling’, increases in bulge diameter are accompanied by a decrease in the length of the rod-shaped part of the cell. This is because bulges form out of existing cytosol and membranes rather than newly synthesized material [35]. Whilst β-lactam and glycopeptide antibiotics induce bulges by disrupting peptidoglycan synthesis directly, daptomycin does so indirectly. Daptomycin has been shown to accumulate in the cytoplasmic membrane, causing a localized distortion that attracts the cell division protein DivIVA. This, in turn, is thought to result in altered deployment of the cell wall synthetic machinery and the generation of small ruptures in the peptidoglycan [176]. It has not yet been established how folic acid or protein synthesis inhibitors generate the peptidoglycan defects that precede bulge formation.
Blebbing
‘Blebs’ are protrusions localized to the outer membrane of the Gram-negative bacterial cell, an intact peptidoglycan layer and cytoplasmic membrane preventing extrusion of the cytoplasm (Fig. 7) [143, 171]. They are more readily detected by TEM than SEM [171]. Blebs can occur during normal growth of bacteria, but increase in number following treatment with certain antibiotics and biocides [180, 181]. Antibacterial agent-induced bleb formation has been reported in numerous Gram-negative species including E. coli [41, 143, 179, 180, 182], L. pneumophila [83, 171], P. aeruginosa [142, 181–183], and Salmonella Typhimurium [184]. Unlike bulges, which are typically observed as a single protrusion per cell (Fig. 6), blebs almost always occur as multiple protrusions per cell [171, 182–184]. Blebs are also typically smaller than bulges (diameters of just 10–100 nm) [26, 184], though enlargement and peptidoglycan rupture during some treatments allows them to develop into bulges [179]. Bleb formation occurs following treatment with membrane-active agents [24, 41, 181, 184] and inhibitors of RNA [143] and protein synthesis [83, 179, 181]. All three types of agent also induce filamentation [39, 142, 143] and this can accompany the bleb formation [142, 143]. Terms used synonymously with ‘blebbing’ include ‘blistering’ [24], ‘bubbling’ [42], and ‘swelling’ [24].
Disruption of the outer membrane by the antibiotic polymyxin B [41, 171, 180, 184], the biocide chlorhexidine [24, 184], and the chelating agent EDTA [181] all induce bleb formation. In the case of polymyxin B, blebbing is induced at concentrations ranging from 1×MIC to ~12.5×MIC [41, 171, 180], with the blebs appearing as early as 3 min after treatment [184] and remaining observable as long as 5 h [171]. Polymyxin B molecules carry a positive charge and hydrophobic fatty acid chain, features which allow them to interact with the negatively charged phosphate groups and hydrophobic fatty acids of lipid A present in the outer (lipopolysaccharide) layer of the outer membrane [184, 185]. Intercalation of polymyxin B with the outer membrane results in an increase in its surface area and, because the membrane is tightly bound to peptidoglycan and unable to expand, this increased surface area is forced to fold outward giving rise to blebs [171].
Examples of RNA and protein synthesis inhibitors that induce blebbing include bicyclomycin [143], arbekacin [179], gentamicin [142, 181–183], and erythromycin [83]. With the aminoglycoside gentamicin, blebbing is induced over a wide range of concentrations (1×MIC to 100×MIC [142, 181]), sometimes appearing as early as 1 min following treatment [181]. The blebs remain observable until the bacteria lyse, this occurring after around 4 h following high concentration gentamicin treatments [142]. Two mechanisms have been proposed to explain aminoglycoside-induced disruption of the outer membrane. According to the first mechanism, translational misreading produces various proteins that are abnormal in shape [182, 186]. These defective proteins include secretory proteins, which become trapped during passage across the outer membrane [186], and outer membrane proteins, which do not fit correctly into the outer membrane [182, 186]. According to the second mechanism, blebbing occurs due to aminoglycosides interacting directly with the outer membrane [183]. This and other membrane effects, it is argued, occur independently of aminoglycoside uptake [183, 187] or disruption of protein synthesis [181, 183].
Peptidoglycan thickening
The peptidoglycan component of the cell wall can become thicker during some antibacterial treatments (Figs. 4 and 8). Drugs known to induce this change include those inhibiting peptidoglycan synthesis, RNA and protein synthesis, and folic acid synthesis. Thickening has been reported most frequently in S. aureus [150, 155] and Enterococcus faecalis [157, 188], but can also occur in Gram-negative cocci [125, 148] and Gram-positive bacilli [69, 189].
Fig. 8.
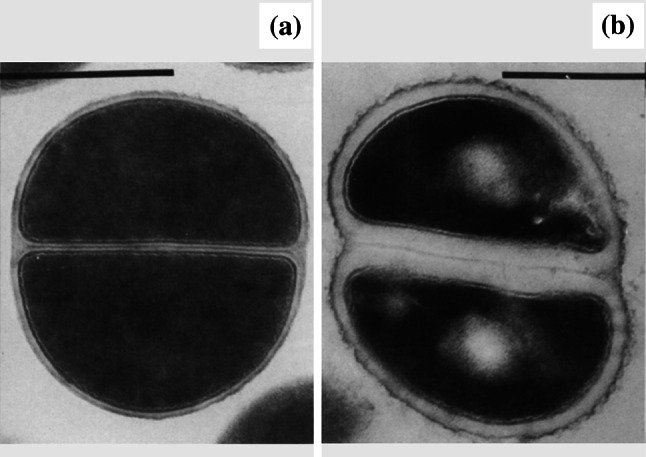
Transmission electron micrographs of a untreated Staphylococcus aureus ATCC 6538P and b cells of the same strain treated with 0.03 µg/mL (3×MIC) rifampicin for 4 h. Both the septal and peripheral portions of the wall appear thickened following treatment. Bar 1 µm.
Images from [155] by permission of Reviews of Infectious Diseases (Oxford University Press)
Peptidoglycan synthesis inhibitors cause thickening predominantly at the septal wall [69, 125, 127, 148, 151, 153, 155, 165, 190], and the thickened septa lack a central dense layer [125, 127, 153, 158] (Fig. 4). Drugs in the β-lactam class cause septal thickening over a wide range of concentrations (0.25×MIC to 10×MIC) [148, 151, 155, 190], this first observable after as little as 90 min [190], accompanied by inhibition of cell separation (‘pseudomulticellular bacteria’) after around 3.5 h [155], and remaining observable as long as 16 h following initial antibiotic exposure [84]. Thickening is thought to be due to β-lactams inhibiting peptidoglycan cross-linking, leading to a buildup of loose, non-structured material [84, 190]. Thickening is most pronounced at the septum because this is where new wall synthesis is most vigorous [190]. Septal thickening has also been observed in bacteria treated with low concentrations of the peptidoglycan synthesis inhibitors vancomycin (0.5×MIC for 2–3 h) [153] and ramoplanin (1×MIC for 3 h) [165]. Higher concentrations, at least in the case of vancomycin (≥10×MIC), do not have this effect [165, 190].
Peptidoglycan thickening triggered by RNA and protein synthesis inhibitors always, to our knowledge, affects both the septal and peripheral walls. Also, in most cells the septal wall retains its central dense layer (Fig. 8) [125, 150, 155, 156, 160, 191]. Thickening occurs following ≥4 h treatment with supra-inhibitory concentrations (3×MIC to 40×MIC) of RNA synthesis inhibitor (actinomycin [60] and rifampicin [150, 155]), or ≥1 h treatment with inhibitory or supra-inhibitory concentrations (1×MIC to ~10×MIC) of protein synthesis inhibitor (chloramphenicol [60, 189], puromycin [60], lincosamides [160, 191], macrolides [156, 160, 191, 192], and tetracyclines [156, 193]; observable as long as 24 h following initial exposure [156, 160]). Amino acid starvation has a similar effect [59, 189]. RNA synthesis inhibitors, as stated earlier, appear not to induce pseudomulticellular bacteria [150, 155], whereas many protein synthesis inhibitors do [60, 125, 156]. Protein synthesis inhibitors induce cell wall thickening by stopping enlargement of the cell wall surface [188, 189], possibly by inhibiting synthesis of the peptidoglycan hydrolases needed to loosen the expanding walls [189, 194]. This, in the absence of any accompanying decrease in synthesis or incorporation of peptidoglycan precursor, leads to thickening of the existing cell wall [188]. The thickening caused by RNA synthesis inhibitors, biochemical analyses would suggest, is not due to inhibition of transcription, but the knock-on effect this has on protein synthesis [188].
Limited information is available on cell wall thickening induced by the folic acid synthesis inhibitor trimethoprim. As with protein synthesis inhibitors, trimethoprim-induced thickening occurs at both the peripheral and septal walls, it is continuous not sporadic, and is accompanied by the formation of pseudomulticellular bacteria. This thickening was detected after 4 h treatment with a sub-inhibitory concentration (0.75×MIC) of the antibiotic [157]. Given the similarities between these observations and those described for the protein synthesis inhibitors, it is conceivable that trimethoprim induces thickening by preventing synthesis of the amino acids required for peptidoglycan hydrolase production.
Separation of cell envelope layers
During some antibacterial treatments, the distance between the cytoplasmic membrane and outer membrane increases (Fig. 9), giving the appearance that the layers of the Gram-negative cell envelope have separated [83, 142, 146]. Agents known to induce this effect include those inhibiting the synthesis of folic acid (sulphadiazine and trimethoprim [195]), DNA (ciprofloxacin [48, 83, 196]), RNA (bicyclomycin [143], rifampicin [83], and rifabutin [196]) and protein (chloramphenicol [146], clarithromycin [196], and aminoglycosides [142, 182]), and those hydrolysing or inhibiting the synthesis of peptidoglycan or arabinogalactan (lysozyme and EDTA [195], β-lactams [49, 83, 89, 148], vancomycin [89], and ethambutol [196]). Because the peptidoglycan layer is thin in these organisms (sometimes just a monolayer [197]), it is difficult to visualize by TEM. This leaves it unclear whether the increase in distance between the two membranes is due to the outer membrane detaching from the peptidoglycan (as suggested by some authors [148, 195]), or the cytoplasmic membrane retracting from the peptidoglycan (as suggested by others [146]). Both scenarios are conceivable. An agent reducing the integrity of the peptidoglycan layer (directly or indirectly) is likely to cause outer membrane detachment, because Braun’s lipoproteins use peptidoglycan to anchor the outer membrane in place. Also, an agent reducing cytoplasmic membrane integrity (directly or indirectly) would cause a decrease in osmotic pressure that could result in cytoplasmic membrane retraction. A third possibility is that the increase in distance between the two membranes represents not cell envelope separation, but an expansion of the periplasmic space [49, 89, 196]. This might be expected to occur if, for example, an antibacterial agent triggered increased endotoxin [49] or β-lactamase [198] production.
Fig. 9.
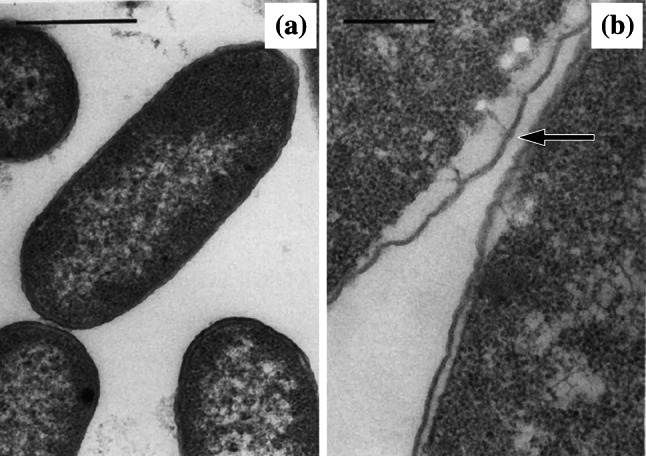
Transmission electron micrographs of a untreated Enterobacter cloacae NCTC 1005 and b cells of the same strain treated with 12.5 µg/mL (0.83×MIC) trimethoprim for 4 h. Arrow shows where the layers of the cell envelope appear to have separated. Bar for a 0.5 µm and b 0.25 µm.
Images from [195] by permission of the Journal of Medical Microbiology (MicroSoc)
Intracellular vacuoles
When round-shaped electron-transparent areas develop in the cytoplasm of bacterial cells during treatment, these are usually described as ‘intracellular vacuoles’ (Fig. 10) [48, 83, 199]. These areas are often surrounded by a membrane [48, 143], but not always [146]. Small numbers of intracellular vacuoles are sometimes detectable during normal bacterial growth [83, 146], but increase in frequency during antibacterial treatment. Synonymous terms include ‘vacuole-like structures’ [174], ‘cytoplasmic vesicles’ [83, 141], and ‘holes’ [195]. Like the separation of cell envelope layers described above, intracellular vacuoles form during treatment with a wide range of antibacterial agents. This includes agents inhibiting the synthesis of folic acid (sulphadiazine [195] and trimethoprim [49, 195]), DNA (ciprofloxacin [48, 83, 196] and norfloxacin [48]), RNA (bicyclomycin [143]) and protein (amikacin [200], chloramphenicol [146], and clarithromycin [196]), and agents hydrolysing or inhibiting the synthesis of peptidoglycan or arabinogalactan (lysozyme and EDTA [195], β-lactams [49, 66, 83, 141, 174, 199], and ethambutol [196]). It is not clear how or why vacuoles form during antibacterial treatment.
Fig. 10.
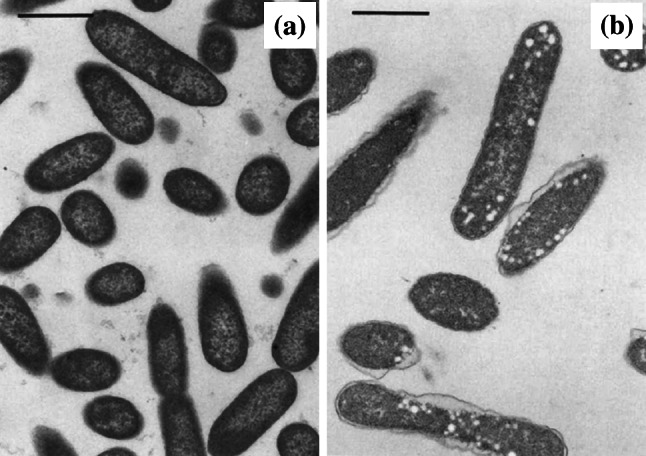
Transmission electron micrographs of a untreated Enterobacter cloacae NCTC 1005 and b cells of the same strain treated with 12.5 µg/mL (0.83×MIC) trimethoprim for 4 h. Many of the bacterial cells have developed intracellular vacuoles following treatment. Bar 1 µm.
Images from [195] by permission of the Journal of Medical Microbiology (MicroSoc)
Reduction in the number of ribosomes
Several studies have observed decreases in ribosome numbers (Fig. 11) in cross sections of bacteria treated with inhibitors of protein (amikacin, gentamicin, chloramphenicol, and quinupristin/dalfopristin) [127, 200, 201] and peptidoglycan synthesis (ampicillin and cephalothin) [202]. Reliable quantification of this ultrastructural change is inherently difficult, because the proportion of the nucleoid that is visible varies from one cross section to another. In a cross section containing a lot of the nucleoid, there will be less space for ribosomes, and the ribosome count will be lower. One study, probably recognizing this problem, performed ribosome counts of multiple electron micrographs [202]. Various hypotheses have been proposed to explain decreased ribosome number including (a) a direct interaction between the antibacterial agent and the ribosomes, and indirect effects such as (b) the antibacterial agent reducing cellular growth rate (thereby reducing the cell’s need to synthesize ribosomes) and (c) a defect in the cell envelope (located outwith that part of the cell that is visible in the cross section), allowing influx of liquid and/or efflux of ribosomes [202].
Fig. 11.
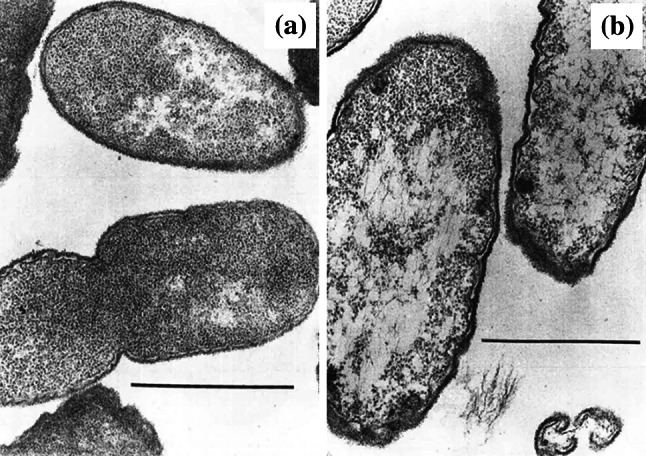
Transmission electron micrographs of a an untreated clinical isolate of Proteus mirabilis and b cells of the same isolate treated with 0.78 µg/mL (0.5×MIC) ampicillin for 3 h. The number of ribosomes has decreased following treatment. Bar 1 µm.
Images from [202] by permission of Proceedings of the Society for Experimental Biology and Medicine (SAGE Publications)
Peri-mortem observations
Some peri-mortem changes in bacterial ultrastructure are common to many antibacterial agents. For example, cell wall breakage (Suppl. Figure 2) has been observed following treatment of bacteria with inhibitors of DNA synthesis (norfloxacin [48]), protein synthesis (amikacin [200], erythromycin [141], gentamicin [181], and quinupristin/dalfopristin [203]), and peptidoglycan synthesis (several β-lactams [55, 83, 141]). Likewise, leakage of cell contents (Suppl. Figure 3) has been observed in bacteria treated with inhibitors of DNA synthesis (ciprofloxacin [83] and norfloxacin [48]), RNA synthesis (rifampicin [83]), protein synthesis (amikacin [200] and quinupristin/dalfopristin [203]), and peptidoglycan synthesis (ramoplanin [165] and several β-lactams [55, 83, 141]). Bacteria with a collapsed or deflated appearance (Suppl. Figure 4) are presumed to be cells that have lost their intracellular contents [83, 141, 174]. These have been observed following treatment with inhibitors of DNA synthesis (ciprofloxacin [83]), RNA synthesis (rifampicin [83]), protein synthesis (erythromycin, fusidic acid, lincomycin [168], tetracycline [36], and tobramycin [204]), and peptidoglycan synthesis (several β-lactams [55, 83, 141, 174]). Lastly, ‘ghost cells’, a term used to describe lysed bacteria devoid or near-devoid of cytoplasm (Fig. 12), have been observed following treatment with inhibitors of DNA synthesis (ciprofloxacin [52] and norfloxacin [48]), RNA synthesis (bicyclomycin [143]), protein synthesis (clarithromycin [196], erythromycin [83, 141], gentamicin [181], and quinupristin/dalfopristin [203]), and peptidoglycan and arabinogalactan synthesis (ramoplanin [165], several β-lactams [55, 78, 83, 120, 141], and ethambutol [196]). Like the separation of cell envelope layers (Fig. 9) or appearance of intracellular vacuoles (Fig. 10), it seems unlikely that any of these peri-mortem changes, when observed in isolation, could be useful in identifying antibacterial mechanism of action. In the case of ghost cells (Fig. 12), it is unlikely that these could even be used as a particularly sensitive indicator of bactericidal activity, as some bactericidal events (e.g. cell death in daptomycin- or ciprofloxacin-treated S. aureus [164, 205]) occur without lysis.
Fig. 12.
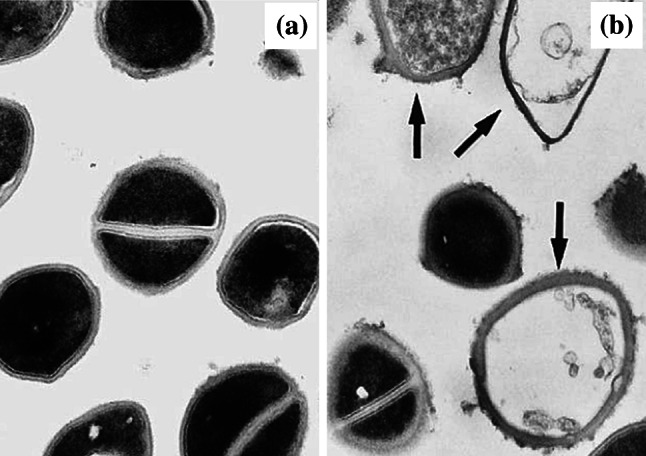
Transmission electron micrographs of a untreated Staphylococcus aureus ATCC 25923 and b cells of the same strain treated with 0.2 µg/mL (0.5×MIC) quinupristin/dalfopristin for 24 h. Arrows show the location of ghost cells. Magnification for a and b ×37,000.
Images from [203] by permission of the Journal of Antimicrobial Chemotherapy (Oxford University Press)
Concluding remarks
This survey of the literature has shown that a number of morphological and ultrastructural changes are induced, apparently quite consistently, by antibacterial agents sharing the same mechanism of action. For example, membrane-active agents frequently induce blebbing, inhibitors of peptidoglycan synthesis frequently induce the formation of spheroplasts and protoplasts, inhibitors of protein synthesis frequently induce a decrease in ribosome number, and inhibitors of DNA synthesis frequently induce filamentation (Table 1). If an unknown test agent was examined under a sufficiently broad range of experimental conditions (different concentrations, treatment durations, etc.), it is conceivable that some of the above cytological changes could be quite sensitive indicators of antibacterial mechanism of action. Those observations induced by just a subset of agents in an antibacterial class, for example the ovoid cell formation induced by some peptidoglycan synthesis inhibitors (Table 1), might even be useful in implicating molecular targets. Where the main problem with this microscopy-based approach lies is the lack of specificity. Most morphological and ultrastructural changes are induced by more than one antibacterial mechanism of action. Cell envelope separation and vacuole formation, for example, are induced by inhibitors of nearly every known bacterial target (Table 1). In some cases, this problem could be resolved by considering other antibacterial-induced alterations occurring at around the same time. For example, decreases in ribosome number are induced by both protein and peptidoglycan synthesis inhibitors, but peripheral wall peptidoglycan thickening (of the continuous type) only occurs when protein synthesis is targeted (Table 1). In other cases, additional considerations such as the magnitude of the change or the ability to induce the change in mutant strains could permit differentiation. For example, filamentation is induced by many mechanisms of action, but cell length only dramatically increases when PBP3, DNA, FtsZ, or folic acid synthesis is targeted and, in these longer filaments, neither DNA nor FtsZ disruptors induce spheroplast formation (Table 1). DNA synthesis inhibitors could then be distinguished from FtsZ inhibitors based on their inability to induce filamentation in SulA− (SOS-negative) bacteria (e.g. E. coli JD26285 [206]).
Table 1.
Summary of the various changes that are observed in antibacterial-treated cells, with alterations in the morphology of the whole cell described first, then alterations in surface morphology and underlying ultrastructure, and lastly peri-mortem changes
| Alteration | Brief description | Antibacterial agents that trigger the alteration |
|---|---|---|
| Spheroplasts and protoplasts | Bacteria that have become large, spherical, smooth, and osmotically sensitive due to loss of their peptidoglycan layer |
Agents disrupting peptidoglycan integrity (e.g. lysozyme) or synthesis (e.g. β-lactams), or inhibiting pathways directly upstream of peptidoglycan synthesis such as the non-mevalonate pathway (e.g. fosmidomycin) Inhibitors of protein (e.g. chloramphenicol) or folic acid (e.g. trimethoprim) synthesis |
| Ovoid cells | Formerly rod-shaped bacteria that have decreased in length due to inhibition of lateral wall peptidoglycan synthesis | Agents disrupting the peptidoglycan-synthesizing enzymes responsible for lateral wall synthesis (e.g. imipenem; inhibits E. coli PBP2) or the cytoskeletal proteins that recruit these enzymes [e.g. S-(3,4-dichlorobenzyl)isothiourea; inhibits E. coli MreB] |
| Filamentation | Bacteria that have become abnormally long and which lack septa due to inhibition of septal wall peptidoglycan synthesis |
Extensive filamentation (cells capable of growing to lengths of ≥50 µm) Agents disrupting the peptidoglycan-synthesizing enzymes responsible for septal wall synthesis (e.g. cefuroxime; inhibits E. coli PBP3) or the cytoskeletal proteins that recruit these enzymes (e.g. berberine; inhibits FtsZ) Agents disrupting DNA integrity (e.g. UV light), DNA synthesis (e.g. ciprofloxacin), or the folic acid synthesis needed for thymine production (e.g. trimethoprim) |
|
Limited filamentation (cells capable of growing to lengths of ~11 µm) Agents inhibiting RNA (e.g. bicyclomycin) or protein (e.g. kanamycin) synthesis Membrane-active agents (e.g. polymyxin B) | ||
| Pseudomulticellular bacteria | Bacteria that have become enlarged or elongated and which possess multiple septa as a consequence of failing to separate following cell division |
Peptidoglycan synthesis inhibitors (e.g. β-lactams) Peptidoglycan hydrolase inhibitors (e.g. Triton X-100) Inhibitors of protein (e.g. chloramphenicol) or folic acid (e.g. trimethoprim) synthesis |
| Other forms of septal disruption | Misshapen or otherwise aberrant septa |
Peptidoglycan synthesis inhibitors (e.g. penicillin) Cytoplasmic membrane disruptors (e.g. daptomycin) |
| Localized swelling | A protrusion that (a) encircles the entire mid-section or pole of the bacterial cell, (b) has its centre located at or close to the cell’s longitudinal axis, and (c) is derived from all layers of the cell envelope (including the cytoplasmic membrane) |
Mid-section swelling Agents disrupting the peptidoglycan-synthesizing enzymes responsible for septal and also lateral wall synthesis (e.g. meropenem; inhibits P. aeruginosa PBP2 and PBP3) |
|
Polar swelling Protein synthesis inhibitors (e.g. erythromycin) | ||
| Bulge formation | A protrusion that (a) emanates from a single point in the bacterial cell surface, (b) has its centre located outside the normal boundaries of the cell, and (c) is derived from all layers of the cell envelope (including the cytoplasmic membrane) |
Peptidoglycan synthesis inhibitors (e.g. β-lactams) Inhibitors of protein (e.g. tobramycin) or folic acid (e.g. sulphamethoxazole) synthesis Cytoplasmic membrane disruptors (e.g. daptomycin) |
| Blebbing | Protrusions that (a) emanate from individual points in the bacterial cell surface, (b) have their centres located outside the normal boundaries of the cell, and (c) are derived from just the outer membrane of the Gram-negative cell |
Outer membrane disruptors (e.g. polymyxin B) Inhibitors of RNA (e.g. bicyclomycin) or protein (e.g. gentamicin) synthesis |
| Peptidoglycan thickening | Septal peptidoglycan thickening accompanied by either (a) loss of the septal central dense layer and sporadic peripheral wall peptidoglycan thickening or (b) retention of the septal central dense layer and continuous peripheral wall peptidoglycan thickening |
With loss of septal central dense layer and sporadic peripheral wall thickening Peptidoglycan synthesis inhibitors (e.g. β-lactams) |
|
With retention of septal central dense layer and continuous peripheral wall thickening Inhibitors of RNA (e.g. actinomycin) or protein (e.g. chloramphenicol) synthesis Folic acid synthesis inhibitors (e.g. trimethoprim) | ||
| Separation of cell envelope layers | An increase in distance between the cytoplasmic membrane and outer membrane of the Gram-negative cell envelope |
Folic acid synthesis inhibitors (e.g. sulphadiazine) DNA synthesis inhibitors (e.g. ciprofloxacin) RNA synthesis inhibitors (e.g. bicyclomycin) Protein synthesis inhibitors (e.g. chloramphenicol) Agents disrupting peptidoglycan integrity (e.g. lysozyme and EDTA) or synthesis (e.g. β-lactams) Arabinogalactan synthesis inhibitors (e.g. ethambutol) |
| Intracellular vacuoles | Round-shaped electron-transparent areas present in the bacterial cytoplasm |
Folic acid synthesis inhibitors (e.g. sulphadiazine) DNA synthesis inhibitors (e.g. ciprofloxacin) RNA synthesis inhibitors (e.g. bicyclomycin) Protein synthesis inhibitors (e.g. amikacin) Agents disrupting peptidoglycan integrity (e.g. lysozyme and EDTA) or synthesis (e.g. β-lactams) Arabinogalactan synthesis inhibitors (e.g. ethambutol) |
| Reduction in the number of ribosomes | A decrease in the number of ribosomes present in the bacterial cytoplasm |
Protein synthesis inhibitors (e.g. amikacin) Peptidoglycan synthesis inhibitors (e.g. ampicillin) |
| Peri-mortem observations | Various changes including cell wall breakage, leakage of cell contents, a collapsed or deflated appearance, and ghost cells |
DNA synthesis inhibitors (e.g. norfloxacin) RNA synthesis inhibitors (e.g. bicyclomycin) Protein synthesis inhibitors (e.g. amikacin) Peptidoglycan synthesis inhibitors (e.g. β-lactams) Arabinogalactan synthesis inhibitors (e.g. ethambutol) |
A more extensive list and detailed description of the cytological changes that occur are available in the main body of the journal article and electronic supplementary material
The present paper has a number of strengths and limitations. It is, to our knowledge, the first review to look at antibacterial agent-induced morphological and ultrastructural changes as potential indicators of mechanism of action. To provide readers with as full a picture as possible, over 200 primary sources were consulted, articles identified from PubMed and extensive hand searches performed without date restrictions. All of the terminology encountered in these sources was carefully disambiguated by comparing the authors’ text and micrographs. These terms, together with definitions, similes, and visual references, have been presented here and in the electronic supplementary material. Information on infrequent and isolated observations is presented in the electronic supplementary material too. Set against the aforementioned strengths are the following limitations. Although the review focused on well-characterized antibacterial agents, the possibility that some of these agents have a second mechanism of action cannot be excluded. As stated in the blebbing section, for example, there is evidence that aminoglycosides target not just protein synthesis, but also the outer membrane [183]. A second limitation is that most of the research described examined treated bacteria at just one or two time-points. It is conceivable that cell wall breakage and other apparently non-specific observations might be more informative if viewed in chronological sequence, rather than in isolation. Lastly, whilst efforts were made to take into account those variables known to affect antibacterial agent-induced cytological changes, the possibility that additional unknown variables exist cannot be ignored.
For future studies using microscopy to investigate antibacterial mechanism of action, there are opportunities to improve experimental design and reporting and make this approach more informative. Performing antibacterial treatments not just in standard growth media, but also in osmotically stabilized media, is a simple step that would increase the likelihood of spheroplasts/protoplasts being detected. If sections and surfaces of bacteria were both examined, for example by TEM and SEM, this would allow changes to be definitively identified and more accurately measured. Phenotypes such as spheroplasts and pseudomulticellular forms or bulges and blebs can be difficult to distinguish from SEM images alone. Also, shape changes can be difficult to observe, and filamentation difficult to measure if just TEM is used. When selecting test species, Gram-negative and Gram-positive bacteria should both be included to permit the detection of cell wall-specific changes such as blebbing, and increase the likelihood of peptidoglycan thickening reaching detectable levels. Concentration and duration of test drug or biocide treatments should be clearly stated, and a rationale for their selection included. Excessively high concentrations should be avoided to permit, where possible, the separation of primary and secondary antibacterial effects, and the treated bacteria should be examined sufficiently frequently to detect all cytological changes. Other variables affecting antibacterial-induced changes, described in detail at the start of this review, should be carefully considered too. Moving on to data collection, the risk of bias could be reduced by blinding observers to treatment conditions. Lastly, greater use could be made of open-source software such as ImageJ for measuring changes in bacterial size, wall thickness, and other parameters.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We are very grateful to those authors who helped source the more-difficult-to-find journal articles used in this review, in particular Professors Francesco Castelli, Magnús Gottfreðsson, Giovanni Longo, Vsevolod L. Popov, R. Michael E. Richards, Brian G. Spratt, and Alexander Tomasz. We are also extremely grateful to Colin MacLean and the Interlibrary loan staff at the Robert Gordon University for their assistance with this project, and to Dr. Helen Vosper for constructive comments during drafting of the manuscript. Our apologies to authors whose work could not be included due to space restrictions.
References
- 1.Jessney B. Joseph Lister (1827–1912): a pioneer of antiseptic surgery remembered a century after his death. J Med Biogr. 2012;20:107–110. doi: 10.1258/jmb.2011.011074. [DOI] [PubMed] [Google Scholar]
- 2.McGuire MJ. The chlorine revolution: water disinfection and the fight to save lives. Denver: American Water Works Association; 2013. [Google Scholar]
- 3.Greenwood D, et al. Chapter 1: Historical introduction. In: Finch RG, et al., editors. Antibiotic and chemotherapy: anti-infective agents and their use in therapy. 9. London: Elsevier Health Sciences; 2010. pp. 2–9. [Google Scholar]
- 4.Fraise AP, Maillard J-Y, Sattar SA, editors. Russell, Hugo and Ayliffe’s principles and practice of disinfection, preservation, and sterilization. 5. Chichester: Wiley-Blackwell; 2012. [Google Scholar]
- 5.Cushnie TPT, Cushnie B, Lamb AJ. Alkaloids: an overview of their antibacterial, antibiotic-enhancing and antivirulence activities. Int J Antimicrob Agents. 2014;44:377–386. doi: 10.1016/j.ijantimicag.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Cavera VL, Arthur TD, Kashtanov D, Chikindas ML. Bacteriocins and their position in the next wave of conventional antibiotics. Int J Antimicrob Agents. 2015;46:494–501. doi: 10.1016/j.ijantimicag.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Pendleton JN, Gilmore BF. The antimicrobial potential of ionic liquids: a source of chemical diversity for infection and biofilm control. Int J Antimicrob Agents. 2015;46:131–139. doi: 10.1016/j.ijantimicag.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 8.Vo HT, Imai T, Ho TT, Dang T-LT, Hoang SA. Potential application of high pressure carbon dioxide in treated wastewater and water disinfection: recent overview and further trends. J Environ Sci. 2015;36:38–47. doi: 10.1016/j.jes.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Yu Q, Wu Z, Chen H. Dual-function antibacterial surfaces for biomedical applications. Acta Biomater. 2015;16:1–13. doi: 10.1016/j.actbio.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 10.Cloutier M, Mantovani D, Rosei F. Antibacterial coatings: challenges, perspectives, and opportunities. Trends Biotechnol. 2015;33:637–652. doi: 10.1016/j.tibtech.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Huang Y-Y, Sharma SK, Yin R, Agrawal T, Chiang LY, Hamblin MR. Functionalized fullerenes in photodynamic therapy. J Biomed Nanotechnol. 2014;10:1918–1936. doi: 10.1166/jbn.2014.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robertson PKJ, Robertson JMC, Bahnemann DW. Removal of microorganisms and their chemical metabolites from water using semiconductor photocatalysis. J Hazard Mater. 2012;211–212:161–171. doi: 10.1016/j.jhazmat.2011.11.058. [DOI] [PubMed] [Google Scholar]
- 13.Byrne JA, Dunlop PSM, Hamilton JWJ, Fernández-Ibáñez P, Polo-López I, Sharma PK, Vennard ASM. A review of heterogeneous photocatalysis for water and surface disinfection. Molecules. 2015;20:5574–5615. doi: 10.3390/molecules20045574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morente EO, Fernández-Fuentes MA, Burgos MJG, Abriouel H, Pulido RP, Gálvez A. Biocide tolerance in bacteria. Int J Food Microbiol. 2013;162:13–25. doi: 10.1016/j.ijfoodmicro.2012.12.028. [DOI] [PubMed] [Google Scholar]
- 15.Silver LL. Challenges of antibacterial discovery. Clin Microbiol Rev. 2011;24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Auerbach T, Mermershtain I, Davidovich C, Bashan A, Belousoff M, Wekselman I, Zimmerman E, et al. The structure of ribosome–lankacidin complex reveals ribosomal sites for synergistic antibiotics. Proc Natl Acad Sci USA. 2010;107:1983–1988. doi: 10.1073/pnas.0914100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gwynn MN, Portnoy A, Rittenhouse SF, Payne DJ. Challenges of antibacterial discovery revisited. Ann NY Acad Sci. 2010;1213:5–19. doi: 10.1111/j.1749-6632.2010.05828.x. [DOI] [PubMed] [Google Scholar]
- 18.Linley E, Denyer SP, McDonnell G, Simons C, Maillard J-Y. Use of hydrogen peroxide as a biocide: new consideration of its mechanisms of biocidal action. J Antimicrob Chemother. 2012;67:1589–1596. doi: 10.1093/jac/dks129. [DOI] [PubMed] [Google Scholar]
- 19.Zinn J, Jenkins JB, Swofford V, Harrelson B, McCarter S. Intraoperative patient skin prep agents: is there a difference? AORN J. 2010;92:662–674. doi: 10.1016/j.aorn.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 20.Hegstad K, Langsrud S, Lunestad BT, Scheie AA, Sunde M, Yazdankhah SP. Does the wide use of quaternary ammonium compounds enhance the selection and spread of antimicrobial resistance and thus threaten our health? Microb Drug Resist. 2010;16:91–104. doi: 10.1089/mdr.2009.0120. [DOI] [PubMed] [Google Scholar]
- 21.Maillard J-Y, Bloomfield S, Coelho JR, Collier P, Cookson B, Fanning S, Hill A, et al. Does microbicide use in consumer products promote antimicrobial resistance? A critical review and recommendations for a cohesive approach to risk assessment. Microb Drug Resist. 2013;19:344–354. doi: 10.1089/mdr.2013.0039. [DOI] [PubMed] [Google Scholar]
- 22.Oldfield E, Feng X. Resistance-resistant antibiotics. Trends Pharmacol Sci. 2014;35:664–674. doi: 10.1016/j.tips.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gnanadhas DP, Marathe SA, Chakravortty D. Biocides—resistance, cross-resistance mechanisms and assessment. Expert Opin Investig Drugs. 2013;22:191–206. doi: 10.1517/13543784.2013.748035. [DOI] [PubMed] [Google Scholar]
- 24.Franklin T, Snow G. Biochemistry and molecular biology of antimicrobial drug action. 6. New York: Springer; 2005. Chapter 3: Antimicrobial agents and cell membranes; pp. 47–64. [Google Scholar]
- 25.Karaosmanoglu K, Sayar NA, Kurnaz IA, Akbulut BS. Assessment of berberine as a multi-target antimicrobial: a multi-omics study for drug discovery and repositioning. OMICS. 2014;18:42–53. doi: 10.1089/omi.2013.0100. [DOI] [PubMed] [Google Scholar]
- 26.Chileveru HR, Lim SA, Chairatana P, Wommack AJ, Chiang I-L, Nolan EM. Visualizing attack of Escherichia coli by the antimicrobial peptide human defensin 5. Biochemistry. 2015;54:1767–1777. doi: 10.1021/bi501483q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun D, Zhang W, Lv M, Yang E, Zhao Q, Wang W. Antibacterial activity of ruthenium(II) polypyridyl complex manipulated by membrane permeability and cell morphology. Bioorg Med Chem Lett. 2015;25:2068–2073. doi: 10.1016/j.bmcl.2015.03.090. [DOI] [PubMed] [Google Scholar]
- 28.Valotteau C, Calers C, Casale S, Berton J, Stevens CV, Babonneau F, Pradier C-M, et al. Biocidal properties of a glycosylated surface: sophorolipids on Au(111) ACS Appl Mater Inter. 2015;7:18086–18095. doi: 10.1021/acsami.5b05090. [DOI] [PubMed] [Google Scholar]
- 29.Saritha K, Rajesh A, Manjulatha K, Setty OH, Yenugu S. Mechanism of antibacterial action of the alcoholic extracts of Hemidesmus indicus (L.) R. Br. ex Schult, Leucas aspera (Wild.), Plumbago zeylanica L., and Tridax procumbens (L.) R. Br. ex Schult. Front Microbiol. 2015;6:Article 00577. doi: 10.3389/fmicb.2015.00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elnakady YA, Chatterjee I, Bischoff M, Rohde M, Josten M, Sahl H-G, Herrmann M, et al. Investigations to the antibacterial mechanism of action of kendomycin. PLoS One. 2016;11:Article e0146165. doi: 10.1371/journal.pone.0146165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Driscoll NH, Labovitiadi O, Cushnie TPT, Matthews KH, Mercer DK, Lamb AJ. Production and evaluation of an antimicrobial peptide-containing wafer formulation for topical application. Curr Microbiol. 2013;66:271–278. doi: 10.1007/s00284-012-0268-3. [DOI] [PubMed] [Google Scholar]
- 32.Elie CR, David G, Schmitzer AR. Strong antibacterial properties of anion transporters: a result of depolarization and weakening of the bacterial membrane. J Med Chem. 2015;58:2358–2366. doi: 10.1021/jm501979f. [DOI] [PubMed] [Google Scholar]
- 33.Rath G, Hussain T, Chauhan G, Garg T, Goyal AK. Development and characterization of cefazolin loaded zinc oxide nanoparticles composite gelatin nanofiber mats for postoperative surgical wounds. Mater Sci Eng C Mater Biol Appl. 2016;58:242–253. doi: 10.1016/j.msec.2015.08.050. [DOI] [PubMed] [Google Scholar]
- 34.Longo G, Kasas S. Effects of antibacterial agents and drugs monitored by atomic force microscopy. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2014;6:230–244. doi: 10.1002/wnan.1258. [DOI] [PubMed] [Google Scholar]
- 35.Yao Z, Kahne D, Kishony R. Distinct single-cell morphological dynamics under beta-lactam antibiotics. Mol Cell. 2012;48:705–712. doi: 10.1016/j.molcel.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peach KC, Bray WM, Winslow D, Linington PF, Linington RG. Mechanism of action-based classification of antibiotics using high-content bacterial image analysis. Mol BioSyst. 2013;9:1837–1848. doi: 10.1039/c3mb70027e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braga PC, Sala MT, Dal Sasso M. Pharmacodynamic effects of subinhibitory concentrations of rufloxacin on bacterial virulence factors. Antimicrob Agents Chemother. 1999;43:1013–1019. doi: 10.1128/aac.43.5.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang D, Lim Y-H. Resveratrol antibacterial activity against Escherichia coli is mediated by Z-ring formation inhibition via suppression of FtsZ expression. Sci Rep. 2015;5:Article 10029. doi: 10.1038/srep10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waisbren SJ, Hurley DJ, Waisbren BA. Morphological expressions of antibiotic synergism against Pseudomonas aeruginosa as observed by scanning electron microscopy. Antimicrob Agents Chemother. 1980;18:969–975. doi: 10.1128/AAC.18.6.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Oliveira-Garcia D, Dall’Agnol M, Rosales M, Azzuz ACGS, Alcántara N, Martinez MB, Girón JA. Fimbriae and adherence of Stenotrophomonas maltophilia to epithelial cells and to abiotic surfaces. Cell Microbiol. 2003;5:625–636. doi: 10.1046/j.1462-5822.2003.00306.x. [DOI] [PubMed] [Google Scholar]
- 41.Rabanal F, Grau-Campistany A, Vila-Farrés X, Gonzalez-Linares J, Borràs M, Vila J, Manresa A, et al. A bioinspired peptide scaffold with high antibiotic activity and low in vivo toxicity. Sci Rep. 2015;5:Article 10558. doi: 10.1038/srep10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartmann M, Berditsch M, Hawecker J, Ardakani MF, Gerthsen D, Ulrich AS. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrob Agents Chemother. 2010;54:3132–3142. doi: 10.1128/AAC.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huttner A, Verhaegh EM, Harbarth S, Muller AE, Theuretzbacher U, Mouton JW. Nitrofurantoin revisited: a systematic review and meta-analysis of controlled trials. J Antimicrob Chemother. 2015;70:2456–2464. doi: 10.1093/jac/dkv147. [DOI] [PubMed] [Google Scholar]
- 44.Hanaki H, Kuwahara-Arai K, Boyle-Vavra S, Daum RS, Labischinski H, Hiramatsu K. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J Antimicrob Chemother. 1998;42:199–209. doi: 10.1093/jac/42.2.199. [DOI] [PubMed] [Google Scholar]
- 45.Cui L, Ma X, Sato K, Okuma K, Tenover FC, Mamizuka EM, Gemmell CG, et al. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus . J Clin Microbiol. 2003;41:5–14. doi: 10.1128/JCM.41.1.5-14.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hyo Y, Yamada S, Harada T. Characteristic cell wall ultrastructure of a macrolide-resistant Staphylococcus capitis strain isolated from a patient with chronic sinusitis. Med Mol Morphol. 2008;41:160–164. doi: 10.1007/s00795-008-0409-8. [DOI] [PubMed] [Google Scholar]
- 47.Alsteens D, Verbelen C, Dague E, Raze D, Baulard A, Dufrêne Y. Organization of the mycobacterial cell wall: a nanoscale view. Pflugers Arch. 2008;456:117–125. doi: 10.1007/s00424-007-0386-0. [DOI] [PubMed] [Google Scholar]
- 48.Elliott TSJ, Shelton A, Greenwood D. The response of Escherichia coli to ciprofloxacin and norfloxacin. J Med Microbiol. 1987;23:83–88. doi: 10.1099/00222615-23-1-83. [DOI] [PubMed] [Google Scholar]
- 49.Chen K, Sun GW, Chua KL, Gan Y-H. Modified virulence of antibiotic-induced Burkholderia pseudomallei filaments. Antimicrob Agents Chemother. 2005;49:1002–1009. doi: 10.1128/AAC.49.3.1002-1009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Braga PC, Dal Sasso M, Maci S. Cefodizime: effects of sub-inhibitory concentrations on adhesiveness and bacterial morphology of Staphylococcus aureus and Escherichia coli: comparison with cefotaxime and ceftriaxone. J Antimicrob Chemother. 1997;39:79–84. doi: 10.1093/jac/39.1.79. [DOI] [PubMed] [Google Scholar]
- 51.Dulaney EL, Marx LM. A folic acid linked system in bacterial cell wall synthesis? J Antibiot (Tokyo) 1971;24:713–714. doi: 10.7164/antibiotics.24.713. [DOI] [PubMed] [Google Scholar]
- 52.Wojnicz D, Kłak M, Adamski R, Jankowski S. Influence of subinhibitory concentrations of amikacin and ciprofloxacin on morphology and adherence ability of uropathogenic strains. Folia Microbiol. 2007;52:429–436. doi: 10.1007/BF02932099. [DOI] [PubMed] [Google Scholar]
- 53.Chang T-W, Weinstein L. Morphological changes in Gram-negative bacilli exposed to cephalothin. J Bacteriol. 1964;88:1790–1797. doi: 10.1128/jb.88.6.1790-1797.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horii T, Kobayashi M, Sato K, Ichiyama S, Ohta M. An in vitro study of carbapenem-induced morphological changes and endotoxin release in clinical isolates of Gram-negative bacilli. J Antimicrob Chemother. 1998;41:435–442. doi: 10.1093/jac/41.4.435. [DOI] [PubMed] [Google Scholar]
- 55.Elliott TSJ, Greenwood D. The response of Pseudomonas aeruginosa to azlocillin, ticarcillin and cefsulodin. J Med Microbiol. 1983;16:351–362. doi: 10.1099/00222615-16-3-351. [DOI] [PubMed] [Google Scholar]
- 56.Diver JM, Wise R. Morphological and biochemical changes in Escherichia coli after exposure to ciprofloxacin. J Antimicrob Chemother. 1986;18:31–41. doi: 10.1093/jac/18.supplement_d.31. [DOI] [PubMed] [Google Scholar]
- 57.Bayer M. The cell wall of Escherichia coli: early effects of penicillin treatment and deprivation of diaminopimelic acid. J Gen Microbiol. 1967;46:237–246. doi: 10.1099/00221287-46-2-237. [DOI] [PubMed] [Google Scholar]
- 58.El-Hajj ZW, Newman EB. How much territory can a single E. coli cell control? Front Microbiol. 2015;6:Article 309. doi: 10.3389/fmicb.2015.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Higgins ML, Shockman GD. Early changes in the ultrastructure of Streptococcus faecalis after amino acid starvation. J Bacteriol. 1970;103:244–253. doi: 10.1128/jb.103.1.244-253.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giesbrecht P, Kersten T, Maidhof H, Wecke J. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol Mol Biol Rev. 1998;62:1371–1414. doi: 10.1128/mmbr.62.4.1371-1414.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klainer AS, Perkins RL. Surface manifestations of antibiotic-induced alterations in protein synthesis in bacterial cells. Antimicrob Agents Chemother. 1972;1:164–170. doi: 10.1128/AAC.1.2.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gottfreðsson M, Erlendsdóttir H, Kolka R, Gudmundsson S. Metabolic and ultrastructural effects induced by ciprofloxacin in Staphylococcus aureus during the postantibiotic effect (PAE) phase. Scand J Infect Dis Suppl. 1991;74:124–128. [PubMed] [Google Scholar]
- 63.De Pedro MA, Donachie WD, Höltje JV, Schwarz H. Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenetic proteins RodA and penicillin-binding protein 2. J Bacteriol. 2001;183:4115–4126. doi: 10.1128/JB.183.14.4115-4126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jacquet T, Cailliez-Grimal C, Francius G, Borges F, Imran M, Duval JFL, Revol-Junelles A-M. Antibacterial activity of class IIa bacteriocin Cbn BM1 depends on the physiological state of the target bacteria. Res Microbiol. 2012;163:323–331. doi: 10.1016/j.resmic.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 65.Dienes L. The development of Proteus cultures in the presence of penicillin. J Bacteriol. 1949;57:529–546. doi: 10.1128/jb.57.5.529-546.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fleming A, Voureka A, Kramer IRH, Hughes WH. The morphology and motility of Proteus vulgaris and other organisms cultured in the presence of penicillin. J Gen Microbiol. 1950;4:257–269. doi: 10.1099/00221287-4-2-257. [DOI] [PubMed] [Google Scholar]
- 67.Ellis L, Herron D, Preston D, Simmons L, Schlegel R. Evaluation of antibiotic efficacy using electron microscopy: morphological effects of guanylureido cephalosporin, chlorobenzoylureido cephalosporin, BL-P1654, and carbenicillin on Pseudomonas aeruginosa . Antimicrob Agents Chemother. 1976;9:334–342. doi: 10.1128/AAC.9.2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Greenwood D, O’Grady F. FL 1060: a new beta-lactam antibiotic with novel properties. J Clin Pathol. 1973;26:1–6. doi: 10.1136/jcp.26.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fitz-James P, Hancock R. The initial structural lesion of penicillin action in Bacillus megaterium . J Cell Biol. 1965;26:657–667. doi: 10.1083/jcb.26.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gebicki J, James A. The preparation and properties of spheroplasts of Aerobacter aerogenes . J Gen Microbiol. 1960;23:9–18. doi: 10.1099/00221287-23-1-9. [DOI] [PubMed] [Google Scholar]
- 71.Lederberg J. Bacterial protoplasts induced by penicillin. Proc Natl Acad Sci USA. 1956;42:574–577. doi: 10.1073/pnas.42.9.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weibull C. The isolation of protoplasts from Bacilllus megaterium by controlled treatment with lysozyme. J Bacteriol. 1953;66:688–695. doi: 10.1128/jb.66.6.688-695.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Silver LL. Rational approaches to antibacterial discovery: pre-genomic directed and phenotypic screening. In: Dougherty TJ, Pucci MJ, editors. Antibiotic discovery and development. New York: Springer; 2012. pp. 33–75. [Google Scholar]
- 74.Van Rensburg AJ. Properties of Proteus mirabilis and Providence spheroplasts. J Gen Microbiol. 1969;56:257–264. doi: 10.1099/00221287-56-2-257. [DOI] [PubMed] [Google Scholar]
- 75.Spratt BG. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci USA. 1975;72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Curtis N, Orr D, Ross GW, Boulton MG. Affinities of penicillins and cephalosporins for the penicillin-binding proteins of Escherichia coli K-12 and their antibacterial activity. Antimicrob Agents Chemother. 1979;16:533–539. doi: 10.1128/AAC.16.5.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kitano K, Tomasz A. Triggering of autolytic cell wall degradation in Escherichia coli by beta-lactam antibiotics. Antimicrob Agents Chemother. 1979;16:838–848. doi: 10.1128/AAC.16.6.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dalhoff A, Nasu T, Okamoto K. Target affinities of faropenem to and its impact on the morphology of Gram-positive and Gram-negative bacteria. Chemotherapy. 2003;49:172–183. doi: 10.1159/000071141. [DOI] [PubMed] [Google Scholar]
- 79.Greenwood D, O’Grady F. A comparison of the effects of ampicillin on Escherichia coli and Proteus mirabilis . J Med Microbiol. 1969;2:435–441. doi: 10.1099/00222615-2-4-435. [DOI] [PubMed] [Google Scholar]
- 80.Chin WL, Lawson JW. Effect of antibiotics on L-form induction of Neisseria meningitidis . Antimicrob Agents Chemother. 1976;9:1056–1065. doi: 10.1128/AAC.9.6.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zimmerman SB, Stapley EO. Relative morphological effects induced by cefoxitin and other beta-lactam antibiotics in vitro. Antimicrob Agents Chemother. 1976;9:318–326. doi: 10.1128/AAC.9.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakao M, Nishi T, Tsuchiya K. In vitro and in vivo morphological response of Klebsiella pneumoniae to cefotiam and cefazolin. Antimicrob Agents Chemother. 1981;19:901–910. doi: 10.1128/AAC.19.5.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rodgers FG, Tzianabos AO, Elliott TSJ. The effect of antibiotics that inhibit cell-wall, protein, and DNA synthesis on the growth and morphology of Legionella pneumophila . J Med Microbiol. 1990;31:37–44. doi: 10.1099/00222615-31-1-37. [DOI] [PubMed] [Google Scholar]
- 84.Nishino T, Nakazawa S. Morphological changes in Staphylococcus aureus and Escherichia coli exposed to cephalexin. Jap J Microbiol. 1972;16:83–94. doi: 10.1111/j.1348-0421.1972.tb00633.x. [DOI] [PubMed] [Google Scholar]
- 85.Spratt BG, Cromie KD. Penicillin-binding proteins of Gram-negative bacteria. Rev Infect Dis. 1988;10:699–711. doi: 10.1093/clinids/10.4.699. [DOI] [PubMed] [Google Scholar]
- 86.Kong K-F, Schneper L, Mathee K. Beta-lactam antibiotics: from antibiosis to resistance and bacteriology. APMIS. 2010;118:1–36. doi: 10.1111/j.1600-0463.2009.02563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hishinuma F, Izaki K, Takahashi H. Inhibition of l-alanine adding enzyme by glycine. Agric Biol Chem. 1971;35:2050–2058. doi: 10.1080/00021369.1971.10860186. [DOI] [Google Scholar]
- 88.Isono F, Katayama T, Inukai M, Haneishi T. Mureidomycins A–D, novel peptidylnucleoside antibiotics with spheroplast forming activity. III. Biological properties. J Antibiot (Tokyo) 1989;42:674–679. doi: 10.7164/antibiotics.42.674. [DOI] [PubMed] [Google Scholar]
- 89.Barkhatova OI, Popov VL, Kekcheeva NK, Prozorovskiĭ SV. [Electron microscopic characteristics of the action of penicillin and vancomycin on Rickettsia conorii and Rickettsia akari in vitro] (in Russian) Antibiotiki. 1984;29:580–585. [PubMed] [Google Scholar]
- 90.Van Heijenoort Y, Leduc M, Singer H, Van Heijenoort J. Effects of moenomycin on Escherichia coli . J Gen Microbiol. 1987;133:667–674. doi: 10.1099/00221287-133-3-667. [DOI] [PubMed] [Google Scholar]
- 91.Nozaki Y, Katayama N, Harada S, Ono H, Okazaki H. Lactivicin, a naturally occurring non-beta-lactam antibiotic having beta-lactam-like action: biological activities and mode of action. J Antibiot (Tokyo) 1989;42:84–93. doi: 10.7164/antibiotics.42.84. [DOI] [PubMed] [Google Scholar]
- 92.Ellison RT, 3rd, Giehl TJ. Killing of Gram-negative bacteria by lactoferrin and lysozyme. J Clin Invest. 1991;88:1080–1091. doi: 10.1172/JCI115407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Birdsell DC, Cota-Robles EH. Production and ultrastructure of lysozyme and ethylenediaminetetraacetate-lysozyme spheroplasts of Escherichia coli . J Bacteriol. 1967;93:427–437. doi: 10.1128/jb.93.1.427-437.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hash JH, Wishnick M, Miller PA. Formation of “protoplasts” of Staphylococcus aureus with a fungal N-acetylhexosaminidase. J Bacteriol. 1964;87:432–437. doi: 10.1128/jb.87.2.432-437.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schuhardt VT, Klesius PH. Osmotic fragility and viability of lysostaphin-induced staphylococcal spheroplasts. J Bacteriol. 1968;96:734–737. doi: 10.1128/jb.96.3.734-737.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klainer AS, Russell RRB. Effect of the inhibition of protein synthesis on the Escherichia coli cell envelope. Antimicrob Agents Chemother. 1974;6:216–224. doi: 10.1128/AAC.6.2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Goss WA, Deitz WH, Cook TM. Mechanism of action of nalidixic acid on Escherichia coli . J Bacteriol. 1964;88:1112–1118. doi: 10.1128/jb.88.4.1112-1118.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Spratt BG, Pardee AB. Penicillin-binding proteins and cell shape in E. coli . Nature. 1975;254:516–517. doi: 10.1038/254516a0. [DOI] [PubMed] [Google Scholar]
- 100.Di Modugno E, Erbetti I, Ferrari L, Galassi G, Hammond SM, Xerri L. In vitro activity of the tribactam GV104326 against Gram-positive, Gram-negative, and anaerobic bacteria. Antimicrob Agents Chemother. 1994;38:2362–2368. doi: 10.1128/AAC.38.10.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jackson JJ, Kropp H. Differences in mode of action of β-lactam antibiotics influence morphology, LPS release and in vivo antibiotic efficacy. J Endotoxin Res. 1996;3:201–218. [Google Scholar]
- 102.Bernabeu-Wittel M, García-Curiel A, Pichardo C, Pachón-Ibáñez ME, Jiménez-Mejías ME, Pachón J. Morphological changes induced by imipenem and meropenem at sub-inhibitory concentrations in Acinetobacter baumannii . Clin Microbiol Infect. 2004;10:931–934. doi: 10.1111/j.1469-0691.2004.00944.x. [DOI] [PubMed] [Google Scholar]
- 103.Sumita Y, Fukasawa M, Okuda T. Comparison of two carbapenems, SM-7338 and imipenem: affinities for penicillin-binding proteins and morphological changes. J Antibiot (Tokyo) 1990;43:314–320. doi: 10.7164/antibiotics.43.314. [DOI] [PubMed] [Google Scholar]
- 104.Nickerson WJ, Webb M. Effect of folic acid analogues on growth and cell division of nonexacting microorganisms. J Bacteriol. 1956;71:129–139. doi: 10.1128/jb.71.2.129-139.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Perumalsamy H, Jung MY, Hong SM, Ahn YJ. Growth-inhibiting and morphostructural effects of constituents identified in Asarum heterotropoides root on human intestinal bacteria. BMC Complement Altern Med. 2013;13:Article 245. doi: 10.1186/1472-6882-13-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Spratt BG. Comparison of the binding properties of two 6β-amidinopenicillanic acid derivatives that differ in their physiological effects on Escherichia coli . Antimicrob Agents Chemother. 1977;11:161–166. doi: 10.1128/AAC.11.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Osborn MJ, Rothfield L. Cell shape determination in Escherichia coli . Curr Opin Microbiol. 2007;10:606–610. doi: 10.1016/j.mib.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 108.Nozaki U, Kawashima F, Imada A. C-19393 S2 and H2, new carbapenem antibiotics. III. Mode of action. J Antibiot (Tokyo) 1981;34:206–211. doi: 10.7164/antibiotics.34.206. [DOI] [PubMed] [Google Scholar]
- 109.Spratt BG, Jobanputra V, Zimmermann W. Binding of thienamycin and clavulanic acid to the penicillin-binding proteins of Escherichia coli K-12. Antimicrob Agents Chemother. 1977;12:406–409. doi: 10.1128/AAC.12.3.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gutmann L, Vincent S, Billot-Klein D, Acar JF, Mrèna E, Williamson R. Involvement of penicillin-binding protein 2 with other penicillin-binding proteins in lysis of Escherichia coli by some β-lactam antibiotics alone and in synergistic lytic effect of amdinocillin (mecillinam) Antimicrob Agents Chemother. 1986;30:906–912. doi: 10.1128/AAC.30.6.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lorian V, Atkinson B. Comparison of the effects of mecillinam and 6-aminopenicillanic acid on Proteus mirabilis, Escherichia coli, and Staphylococcus aureus . Antimicrob Agents Chemother. 1977;11:541–552. doi: 10.1128/AAC.11.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Iwai N, Nagai K, Wachi M. Novel S-benzylisothiourea compound that induces spherical cells in Escherichia coli probably by acting on a rod-shape-determining protein(s) other than penicillin-binding protein 2. Biosci Biotechnol Biochem. 2002;66:2658–2662. doi: 10.1271/bbb.66.2658. [DOI] [PubMed] [Google Scholar]
- 113.Bean GJ, Flickinger ST, Westler WM, McCully ME, Sept D, Weibel DB, Amann KJ. A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB. Biochemistry. 2009;48:4852–4857. doi: 10.1021/bi900014d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yamachika S, Sugihara C, Tsuji H, Muramatsu Y, Kamai Y, Yamashita M. Anti-Pseudomonas aeruginosa compound, 1,2,3,4-tetrahydro-1,3,5-triazine derivative, exerts its action by primarily targeting MreB. Biol Pharm Bull. 2012;35:1740–1744. doi: 10.1248/bpb.b12-00397. [DOI] [PubMed] [Google Scholar]
- 115.Wainwright M, Canham LT, Al-Wajeeh K, Reeves CL. Morphological changes (including filamentation) in Escherichia coli grown under starvation conditions on silicon wafers and other surfaces. Lett Appl Microbiol. 1999;29:224–227. doi: 10.1046/j.1365-2672.1999.00602.x. [DOI] [PubMed] [Google Scholar]
- 116.Braga PC, Dal Sasso M, Sala MT. Sub-MIC concentrations of cefodizime interfere with various factors affecting bacterial virulence. J Antimicrob Chemother. 2000;45:15–25. doi: 10.1093/jac/45.1.15. [DOI] [PubMed] [Google Scholar]
- 117.Young KD. Bacterial morphology: why have different shapes? Curr Opin Microbiol. 2007;10:596–600. doi: 10.1016/j.mib.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Justice SS, Hung C, Theriot JA, Fletcher DA, Anderson GG, Footer MJ, Hultgren SJ. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc Natl Acad Sci USA. 2004;101:1333–1338. doi: 10.1073/pnas.0308125100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Noguchi H, Matsuhashi M, Takaoka M, Mitsuhashi S. New antipseudomonal penicillin, PC-904: affinity to penicillin-binding proteins and inhibition of the enzyme cross-linking peptidoglycan. Antimicrob Agents Chemother. 1978;14:617–624. doi: 10.1128/AAC.14.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tanaka M, Otsuki M, Nishino T. In vitro and in vivo activities of DQ-2556 and its mode of action. Antimicrob Agents Chemother. 1992;36:2595–2601. doi: 10.1128/AAC.36.12.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Presslitz JE. Mode of action of a structurally novel beta-lactam. Antimicrob Agents Chemother. 1978;14:144–150. doi: 10.1128/AAC.14.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Onoe T, Umemoto T, Sagawa H, Suginaka H. Filament formation of Fusobacterium nucleatum cells induced by mecillinam. Antimicrob Agents Chemother. 1981;19:487–489. doi: 10.1128/AAC.19.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nakao M, Yukishige K, Kondo M, Imada A. Novel morphological changes in Gram-negative bacteria caused by combination of bulgecin and cefmenoxime. Antimicrob Agents Chemother. 1986;30:414–417. doi: 10.1128/AAC.30.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dring GJ, Hurst A. Observations on the action of benzylpenicillin on a strain of Streptococcus lactis . J Gen Microbiol. 1969;55:185–194. doi: 10.1099/00221287-55-2-185. [DOI] [PubMed] [Google Scholar]
- 125.Lorian V, Atkinson B. Effects of subinhibitory concentrations of antibiotics on cross walls of cocci. Antimicrob Agents Chemother. 1976;9:1043–1055. doi: 10.1128/AAC.9.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Murphy TF, Barza M, Park JT. Penicillin-binding proteins in Clostridium perfringens . Antimicrob Agents Chemother. 1981;20:809–813. doi: 10.1128/AAC.20.6.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lorian V, Atkinson B. Abnormal forms of bacteria produced by antibiotics. Am J Clin Pathol. 1975;64:678–688. doi: 10.1093/ajcp/64.5.678. [DOI] [PubMed] [Google Scholar]
- 128.Lorian V, Sabath LD. Penicillins and cephalosporins: differences in morphologic effects on Proteus mirabilis . J Infect Dis. 1972;125:560–564. doi: 10.1093/infdis/125.5.560. [DOI] [PubMed] [Google Scholar]
- 129.Walker JR, Pardee AB. Evidence for a relationship between deoxyribonucleic acid metabolism and septum formation in Escherichia coli . J Bacteriol. 1968;95:123–131. doi: 10.1128/jb.95.1.123-131.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ohkawa T. Studies of intracellular thymidine nucleotides. Thymineless death and the recovery after re-addition of thymine in Escherichia coli K12. Eur J Biochem. 1975;60:57–66. doi: 10.1111/j.1432-1033.1975.tb20975.x. [DOI] [PubMed] [Google Scholar]
- 131.Chadfield MS, Hinton MH. In vitro activity of nitrofuran derivatives on growth and morphology of Salmonella enterica serotype Enteritidis. J Appl Microbiol. 2004;96:1002–1012. doi: 10.1111/j.1365-2672.2004.02225.x. [DOI] [PubMed] [Google Scholar]
- 132.Church DL, Bryant RD, Rabin HR, Laishley EJ. Physiolgical effects of metronidazole on Clostridium posteurianum . J Antimicrob Chemother. 1991;28:221–228. doi: 10.1093/jac/28.2.221. [DOI] [PubMed] [Google Scholar]
- 133.Suzuki H, Pangborn J, Kilgore WW. Filamentous cells of Escherichia coli formed in the presence of mitomycin. J Bacteriol. 1967;93:683–688. doi: 10.1128/jb.93.2.683-688.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cheng G, Hao H, Dai M, Liu Z, Yuan Z. Antibacterial action of quinolones: from target to network. Eur J Med Chem. 2013;66:555–562. doi: 10.1016/j.ejmech.2013.01.057. [DOI] [PubMed] [Google Scholar]
- 135.Lewin CS, Amyes SGB. The role of the SOS response in bacteria exposed to zidovudine or trimethoprim. J Med Microbiol. 1991;34:329–332. doi: 10.1099/00222615-34-6-329. [DOI] [PubMed] [Google Scholar]
- 136.Mason D, Power E, Talsania H, Phillips I, Gant V. Antibacterial action of ciprofloxacin. Antimicrob Agents Chemother. 1995;39:2752–2758. doi: 10.1128/AAC.39.12.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ingham CJ, Van Den Ende M, Wever PC, Schneeberger PM. Rapid antibiotic sensitivity testing and trimethoprim-mediated filamentation of clinical isolates of the Enterobacteriaceae assayed on a novel porous culture support. J Med Microbiol. 2006;55:1511–1519. doi: 10.1099/jmm.0.46585-0. [DOI] [PubMed] [Google Scholar]
- 138.Humphrey S, MacVicar T, Stevenson A, Roberts M, Humphrey TJ, Jepson MA. SulA-induced filamentation in Salmonella enterica serovar Typhimurium: effects on SPI-1 expression and epithelial infection. J Appl Microbiol. 2011;111:185–196. doi: 10.1111/j.1365-2672.2011.05022.x. [DOI] [PubMed] [Google Scholar]
- 139.Ray S, Dhaked HPS, Panda D. Antimicrobial peptide CRAMP (16–33) stalls bacterial cytokinesis by inhibiting FtsZ assembly. Biochemistry. 2014;53:6426–6429. doi: 10.1021/bi501115p. [DOI] [PubMed] [Google Scholar]
- 140.Bergersen FJ. Cytological changes induced in Bacterium coli by chloramphenicol. J Gen Microbiol. 1953;9:353–356. doi: 10.1099/00221287-9-3-353. [DOI] [PubMed] [Google Scholar]
- 141.Elliott TS, Rodgers FG. Morphological response and growth characteristics of Legionella pneumophila exposed to ampicillin and erythromycin. J Med Microbiol. 1985;19:383–390. doi: 10.1099/00222615-19-3-383. [DOI] [PubMed] [Google Scholar]
- 142.Gilleland LB, Gilleland HE, Gibson JA, Champlin FR. Adaptive resistance to aminoglycoside antibiotics in Pseudomonas aeruginosa . J Med Microbiol. 1989;29:41–50. doi: 10.1099/00222615-29-1-41. [DOI] [PubMed] [Google Scholar]
- 143.Someya A, Tanaka K, Tanaka N. Morphological changes of Escherichia coli induced by bicyclomycin. Antimicrob Agents Chemother. 1979;16:87–91. doi: 10.1128/AAC.16.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Paulander W, Wang Y, Folkesson A, Charbon G, Løbner-Olesen A, Ingmer H. Bactericidal antibiotics increase hydroxyphenyl fluorescein signal by altering cell morphology. PLoS One. 2014;9:Article e92231. doi: 10.1371/journal.pone.0092231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Magnussen CR, Hruska JF. Aberrant forms of Escherichia coli in blood cultures: in vitro reproduction of an in vivo observation. J Clin Microbiol. 1980;12:690–694. doi: 10.1128/jcm.12.5.690-694.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Morgan C, Rosenkranz HS, Carr HS, Rose HM. Electron microscopy of chloramphenicol-treated Escherichia coli . J Bacteriol. 1967;93:1987–2002. doi: 10.1128/jb.93.6.1987-2002.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Conrad RS, Howard MJ, Garrison RC, Winters S, Henderson DA. The effects of daptomycin on chemical composition and morphology of Staphylococcus aureus . Proc Okla Acad Sci. 1998;78:15–22. [Google Scholar]
- 148.Neirinck L, DeVoe I. Anomalous cellular morphology and growth characteristics of Neisseria meningitidis in subminimal inhibitory concentrations of penicillin G. Antimicrob Agents Chemother. 1981;19:911–916. doi: 10.1128/AAC.19.5.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tomasz A. Biological consequences of the replacement of choline by ethanolamine in the cell wall of pneumococcus: chain formation, loss of transformability, and loss of autolysis. Proc Natl Acad Sci USA. 1968;59:86–93. doi: 10.1073/pnas.59.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Filice G, Carnevale G, Lanzarini P, Castelli F, Zappala C, Menghini P, De Rysky C. Alterations due to ampicillin and rifampicin in S. sanguis and S. aureus isolated from dental plaque. An electron microscopic study. Chemioterapia. 1986;5:3–6. [PubMed] [Google Scholar]
- 151.Root RK, Isturiz R, Molavi A, Metcalf JA, Malech HL. Interactions between antibiotics and human neutrophils in the killing of staphylococci: studies with normal and cytochalasin B-treated cells. J Clin Invest. 1981;67:247–259. doi: 10.1172/JCI110020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Henze U, Sidow T, Wecke J, Labischinski H, Berger-Bächi B. Influence of femB on methicillin resistance and peptidoglycan metabolism in Staphylococcus aureus . J Bacteriol. 1993;175:1612–1620. doi: 10.1128/jb.175.6.1612-1620.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sieradzki K, Tomasz A. Inhibition of the autolytic system by vancomycin causes mimicry of vancomycin-intermediate Staphylococcus aureus-type resistance, cell concentration dependence of the MIC, and antibiotic tolerance in vancomycin-susceptible S. aureus . Antimicrob Agents Chemother. 2006;50:527–533. doi: 10.1128/AAC.50.2.527-533.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sieradzki K, Tomasz A. Alterations of cell wall structure and metabolism accompany reduced susceptibility to vancomycin in an isogenic series of clinical isolates of Staphylococcus aureus . J Bacteriol. 2003;185:7103–7110. doi: 10.1128/JB.185.24.7103-7110.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lorian V, Atkinson B, Kim Y. Effect of rifampin and oxacillin on the ultrastructure and growth of Staphylococcus aureus . Rev Infect Dis. 1983;5:S418–S427. doi: 10.1093/clinids/5.Supplement_3.S418. [DOI] [PubMed] [Google Scholar]
- 156.Nakao M, Kitanaka E, Ochiai K, Nakazawa S. Cell wall synthesis by Staphylococcus aureus in the presence of protein synthesis inhibitory agents. II. Electron microscopic study. J Antibiot (Tokyo) 1972;25:469–470. doi: 10.7164/antibiotics.25.469. [DOI] [PubMed] [Google Scholar]
- 157.Richards RME, Xing JZ, Gregory DW, Marshall D. Mechanism of sulphadiazine enhancement of trimethoprim activity against sulphadiazine-resistant Enterococcus faecalis . J Antimicrob Chemother. 1995;36:607–618. doi: 10.1093/jac/36.4.607. [DOI] [PubMed] [Google Scholar]
- 158.Lorian V. Some effects of subinhibitory concentrations of penicillin on the structure and division of staphylococci. Antimicrob Agents Chemother. 1975;7:864–870. doi: 10.1128/AAC.7.6.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Eirich J, Orth R, Sieber SA. Unraveling the protein targets of vancomycin in living S. aureus and E. faecalis cells. J Am Chem Soc. 2011;133:12144–12153. doi: 10.1021/ja2039979. [DOI] [PubMed] [Google Scholar]
- 160.Nakao M, Kitanaka E, Ochiai K, Nakazawa S. Cell wall synthesis by Staphylococcus aureus in the presence of protein synthesis inhibitory agents. I. Electronmicroscopic study. J Antibiot (Tokyo) 1972;25:60–63. doi: 10.7164/antibiotics.25.60. [DOI] [PubMed] [Google Scholar]
- 161.Sayare M, Daneo-Moore L, Shockman GD. Influence of macromolecular biosynthesis on cellular autolysis in Streptococcus faecalis . J Bacteriol. 1972;112:337–344. doi: 10.1128/jb.112.1.337-344.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sharma M, Chauhan PM. Dihydrofolate reductase as a therapeutic target for infectious diseases: opportunities and challenges. Future Med Chem. 2012;4:1335–1365. doi: 10.4155/fmc.12.68. [DOI] [PubMed] [Google Scholar]
- 163.Rogers HJ, Taylor C. Autolysins and shape change in rodA mutants of Bacillus subtilis . J Bacteriol. 1978;135:1032–1042. doi: 10.1128/jb.135.3.1032-1042.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cotroneo N, Harris R, Perlmutter N, Beveridge T, Silverman JA. Daptomycin exerts bactericidal activity without lysis of Staphylococcus aureus . Antimicrob Agents Chemother. 2008;52:2223–2225. doi: 10.1128/AAC.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cheng M, Huang JX, Ramu S, Butler MS, Cooper MA. Ramoplanin induces bacterial membrane depolarization in Staphylococcus aureus at bactericidal concentrations. Antimicrob Agents Chemother. 2014;58:6819–6827. doi: 10.1128/AAC.00061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lutkenhaus J, Pichoff S, Du S. Bacterial cytokinesis: from Z ring to divisome. Cytoskeleton. 2012;69:778–790. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tsang KW, Ng P, Ho PL, Chan S, Tipoe G, Leung R, Sun J, et al. Effects of erythromycin on Pseudomonas aeruginosa adherence to collagen and morphology in vitro. Eur Respir J. 2003;21:401–406. doi: 10.1183/09031936.03.00050903. [DOI] [PubMed] [Google Scholar]
- 168.Greenwood D, O’Grady F. Scanning electron microscopy of Staphylococcus aureus exposed to some common anti-staphylococcal agents. J Gen Microbiol. 1972;70:263–270. doi: 10.1099/00221287-70-2-263. [DOI] [PubMed] [Google Scholar]
- 169.Burdett IDJ, Murray RGE. Septum formation in Escherichia coli: characterization of septal structure and the effects of antibiotics on cell division. J Bacteriol. 1974;119:303–324. doi: 10.1128/jb.119.1.303-324.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Braga PC, Ricci D. Detection of rokitamycin-induced morphostructural alterations in Helicobacter pylori by atomic force microscopy. Chemotherapy. 2000;46:15–22. doi: 10.1159/000007251. [DOI] [PubMed] [Google Scholar]
- 171.Chan EL, Harris RC, Dalton HP. The effect of antibiotics on the cell morphology of Legionella pneumophila . J Med Microbiol. 1987;23:149–154. doi: 10.1099/00222615-23-2-149. [DOI] [PubMed] [Google Scholar]
- 172.Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS. Cell shape and cell-wall organization in Gram-negative bacteria. Proc Natl Acad Sci USA. 2008;105:192282–192287. doi: 10.1073/pnas.0805309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Koupal LR, Pelak BA, Cassidy PJ, Gadebusch HH. Quaternary heterocyclylamino β-lactams. III. The mode of action of L-640,876 and the effect of NaCl on membrane permeability and binding. J Antibiot (Tokyo) 1983;36:54–63. doi: 10.7164/antibiotics.36.54. [DOI] [PubMed] [Google Scholar]
- 174.Nakao M, Kondo M, Tsuchiya K. Light and electron microscopy of the morphological response of Escherichia coli and Serratia marcescens to cefmenoxime (SCE-1365), a new broad-spectrum cephalosporin. J Antibiot (Tokyo) 1981;34:1046–1054. doi: 10.7164/antibiotics.34.1046. [DOI] [PubMed] [Google Scholar]
- 175.Daly KE, Huang KC, Wingreen NS, Mukhopadhyay R. Mechanics of membrane bulging during cell-wall disruption in Gram-negative bacteria. Phys Rev E Stat Nonlin Soft Matter Phys. 2011;83:Article 041922. doi: 10.1103/PhysRevE.83.041922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pogliano J, Pogliano N, Silverman JA. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J Bacteriol. 2012;194:4494–4504. doi: 10.1128/JB.00011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wale LJ, Shelton AP, Greenwood D. Scanning electron microscopy of Staphylococcus aureus and Enterococcus faecalis exposed to daptomycin. J Med Microbiol. 1989;30:45–49. doi: 10.1099/00222615-30-1-45. [DOI] [PubMed] [Google Scholar]
- 178.Comber KR, Boon RJ, Sutherland R. Comparative effects of amoxycillin and ampicillin on the morphology of Escherichia coli in vivo and correlation with activity. Antimicrob Agents Chemother. 1977;12:736–744. doi: 10.1128/AAC.12.6.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tanaka N, Matsunaga K, Hirata A, Matsuhisa Y, Nishimura T. Mechanism of action of habekacin, a novel amino acid-containing aminoglycoside antibiotic. Antimicrob Agents Chemother. 1983;24:797–802. doi: 10.1128/AAC.24.5.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Koike M, Iida K, Matsuo T. Electron microscopic studies on mode of action of polymyxin. J Bacteriol. 1969;97:448–452. doi: 10.1128/jb.97.1.448-452.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Martin NL, Beveridge TJ. Gentamicin interaction with Pseudomonas aeruginosa cell envelope. Antimicrob Agents Chemother. 1986;29:1079–1087. doi: 10.1128/AAC.29.6.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Iida K, Koike M. Cell wall alterations of Gram-negative bacteria by aminoglycoside antibiotics. Antimicrob Agents Chemother. 1974;5:95–97. doi: 10.1128/AAC.5.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kadurugamuwa JL, Clarke AJ, Beveridge TJ. Surface action of gentamicin on Pseudomonas aeruginosa . J Bacteriol. 1993;175:5798–5805. doi: 10.1128/jb.175.18.5798-5805.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schindler PRG, Teuber M. Action of polymyxin B on bacterial membranes: morphological changes in the cytoplasm and in the outer membrane of Salmonella typhimurium and Escherichia coli B. Antimicrob Agents Chemother. 1975;8:95–104. doi: 10.1128/AAC.8.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Nation RL, Velkov T, Li J. Colistin and polymyxin B: peas in a pod, or chalk and cheese? Clin Infect Dis. 2014;59:88–94. doi: 10.1093/cid/ciu213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Davis BD, Chen L, Tai PC. Misread protein creates membrane channels: an essential step in the bactericidal action of aminoglycosides. Proc Natl Acad Sci USA. 1986;83:6164–6168. doi: 10.1073/pnas.83.16.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hancock RE, Raffle VJ, Nicas TI. Involvement of the outer membrane in gentamicin and streptomycin uptake and killing in Pseudomonas aeruginosa . Antimicrob Agents Chemother. 1981;19:777–785. doi: 10.1128/AAC.19.5.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Higgins ML, Daneo-Moore L, Boothby D, Shockman GD. Effect of inhibition of deoxyribonucleic acid and protein synthesis on the direction of cell wall growth in Streptococcus faecalis . J Bacteriol. 1974;118:681–692. doi: 10.1128/jb.118.2.681-692.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Miller IL, Zsigray RM, Landman OE. The formation of protoplasts and quasi-spheroplasts in normal and chloramphenicol-pretreated Bacillus subtilis . J Gen Microbiol. 1967;49:513–525. doi: 10.1099/00221287-49-3-513. [DOI] [Google Scholar]
- 190.Molitor E, Kluczny C, Brötz H, Bierbaum G, Jack R, Sahl H-G. Effects of the lantibiotic mersacidin on the morphology of staphylococci. Zentralbl Bakteriol. 1996;284:318–328. doi: 10.1016/S0934-8840(96)80108-3. [DOI] [PubMed] [Google Scholar]
- 191.Nakao M, Kitanaka F, Ochiai K, Nakazwa S. Cell wall synthesis in Staphylococcus aureus in the presence of protein synthesis inhibitory agents. I. Lincomycin, clindamycin and macrolide antibiotics. Jap J Microbiol. 1972;16:403–413. doi: 10.1111/j.1348-0421.1972.tb00675.x. [DOI] [PubMed] [Google Scholar]
- 192.Nishino T. An electron microscopic study of antagonism between cephalexin and erythromycin in Staphylococcus aureus . Jap J Microbiol. 1975;19:53–63. doi: 10.1111/j.1348-0421.1975.tb00847.x. [DOI] [PubMed] [Google Scholar]
- 193.Hash JH, Davies MC. Electron microscopy of Staphylococcus aureus treated with tetracycline. Science. 1962;138:828–829. doi: 10.1126/science.138.3542.828. [DOI] [PubMed] [Google Scholar]
- 194.Wheeler R, Turner R, Bailey R, Salamaga B, Mesnage S, Mohamad S, Hayhurst E, et al. Bacterial cell enlargement requires control of cell wall stiffness mediated by peptidoglycan hydrolases. MBio. 2015;6:Article e00660–e00615. doi: 10.1128/mBio.00660-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Richards RME, Xing JZ, Gregory DW, Marshall D. An electron microscope study of the effect of sulphadiazine and trimethoprim on Enterobacter cloacae . J Med Microbiol. 1993;38:64–68. doi: 10.1099/00222615-38-1-64. [DOI] [PubMed] [Google Scholar]
- 196.Reisner BS, Woods GL, Popov VL. Electron microscopic analysis of Mycobacterium avium complex isolates exposed to ciprofloxacin, rifabutin, ethambutol and clarithromycin. Int J Tuberc Lung Dis. 1997;1:270–275. [PubMed] [Google Scholar]
- 197.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:Article a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Georgiou G, Telford JN, Shuler ML, Wilson DB. Localization of inclusion bodies in Escherichia coli overproducing β-lactamase or alkaline phosphatase. Appl Environ Microbiol. 1986;52:1157–1161. doi: 10.1128/aem.52.5.1157-1161.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Tuomanen E, Gilbert K, Tomasz A. Modulation of bacteriolysis by cooperative effects of penicillin-binding proteins 1a and 3 in Escherichia coli . Antimicrob Agents Chemother. 1986;30:659–663. doi: 10.1128/AAC.30.5.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lorian V, Atkinson BA. Amikacin-induced alterations in the structure of Gram-negative bacilli. Diagn Microbiol Infect Dis. 1986;5:93–97. doi: 10.1016/0732-8893(86)90111-2. [DOI] [PubMed] [Google Scholar]
- 201.Lorian V, Fernandes F. Effect of quinupristin/dalfopristin alone or in combination with vancomycin on the structure of Enterococcus faecium . Drugs Exp Clin Res. 1998;24:73–76. [PubMed] [Google Scholar]
- 202.Lorian V, Sabath LD, Simionescu M. Decrease in ribosomal density of Proteus mirabilis exposed to subinhibitory concentrations of ampicillin or cephalothin. Proc Soc Exp Biol Med. 1975;149:731–735. doi: 10.3181/00379727-149-38888. [DOI] [PubMed] [Google Scholar]
- 203.Lorian V, Fernandes F. Electron microscopy studies of the bactericidal effects of quinupristin/dalfopristin on Staphylococcus aureus . J Antimicrob Chemother. 1999;43:845–846. doi: 10.1093/jac/43.6.845. [DOI] [PubMed] [Google Scholar]
- 204.Formosa C, Grare M, Jauvert E, Coutable A, Regnouf-de-Vains JB, Mourer M, Duval RE, et al. Nanoscale analysis of the effects of antibiotics and CX1 on a Pseudomonas aeruginosa multidrug-resistant strain. Sci Rep. 2012;2:Article 575. doi: 10.1038/srep00575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Oliva B, Miller K, Caggiano N, O’Neill AJ, Cuny GD, Hoemann MZ, Hauske JR, et al. Biological properties of novel antistaphylococcal quinoline-indole agents. Antimicrob Agents Chemother. 2003;47:458–466. doi: 10.1128/AAC.47.2.458-466.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Boberek JM, Stach J, Good L. Genetic evidence for inhibition of bacterial division protein FtsZ by berberine. PLoS One. 2010;5:Article e13745. doi: 10.1371/journal.pone.0013745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.