Abstract
EGFR signalling is a well-conserved signalling pathway playing major roles during development and cancers. This review explores what studying the EGFR pathway during Drosophila eye development has taught us in terms of the diversity of its regulatory mechanisms. This model system has allowed the identification of numerous positive and negative regulators acting at specific time and place, thus participating to the tight control of signalling. EGFR signalling regulation is achieved by a variety of mechanisms, including the control of ligand processing, the availability of the receptor itself and the transduction of the cascade in the cytoplasm. Ultimately, the transcriptional responses contribute to the establishment of positive and negative feedback loops. The combination of these multiple mechanisms employed to regulate the EGFR pathway leads to specific cellular outcomes involved in functions as diverse as the acquisition of cell fate, proliferation, survival, adherens junction remodelling and morphogenesis.
Keywords: EGFR, Drosophila, Retina, Ommatidia, Photoreceptors, Activators, Inhibitors, Combinatorial signalling
A brief overview of the EGFR signalling pathway
The Drosophila protein encoded by the epidermal growth factor receptor (EGFR) gene (Table 1), also known as Drosophila EGF Receptor (DER), torpedo (top) and faint little ball (flb) is a transmembrane glycoprotein of the protein kinase superfamily [1]. This protein is a cell surface receptor for members of the epidermal growth factor (EGF) family of extracellular protein ligands. It contains four domains in the extracellular region (including two cysteine-rich domains required for ligand binding), a hydrophobic transmembrane region and an intracellular region responsible for its intrinsic protein tyrosine kinase activity. EGFR thus belongs to the receptor tyrosine kinase (RTK) family of proteins. The Drosophila EGFR is the orthologue of the four mammalian ErbB receptors. Mammals possess eleven ligands, including EGF and the transformation growth factor alpha (TGFα) [2]. EGF and its receptor were discovered by Stanley Cohen who shared the 1986 Nobel Prize in Medicine with Rita Levi-Montalcini for their discovery of growth factors. Gain-of-function mutations in the EGFR gene are associated with a number of cancers, including lung, breast, colorectal, prostate and pancreatic cancers [3, 4]. These somatic mutations cause EGFR constitutive activation, leading to uncontrolled cell division and migration [5]. Ligand and receptor expression levels also correlate with progressive tumour growth and metastasis [6]. Specific EGFR inhibition is therefore one of the key targets for cancer therapy [5]. The misregulation of EGFR signalling in so many human cancers emphasises the importance of understanding how the EGFR signalling pathway is controlled. Besides, the EGFR pathway is also reiteratively used in many developmental contexts [7]. Its involvement in an extremely diverse set of processes implies that its regulation is fine-tuned by a series of complex mechanisms. Concerted efforts have, therefore, been spent to understand how the EGFR signalling pathway is regulated, to identify its different positive and negative regulators and the different strategies used to tightly control the timing, patterning, intensity and duration of signalling.
Table 1.
Core components and regulators of the EGFR signalling pathway in the Drosophila eye
| Fly gene Name | Symbol | Also known as | Cellular location | Function | Reference |
|---|---|---|---|---|---|
| Core components | |||||
| Epidermal growth factor receptor | Egfr | DER, torpedo, fib | Plasma membrane | RTK | [1] |
| spitz | spi | Secreted | Ligand | [8] | |
| Keren | Krn | Secreted | Ligand | [9] | |
| downstream of receptor kinase | drk | GRB2 | Cytoplasm | Adaptor protein | [12] |
| SHC adaptor protein | Shc | Cytoplasm | Adaptor protein | [13] | |
| Son of sevenless | Sos | Cytoplasm | GEF | [14] | |
| Ras oncogene at 85D | Ras85D | Ras, Ras1 | Cytoplasm | small GTPase | [15] |
| Raf oncogene | Raf | pole hole | Cytoplasm | Protein kinase | [18] |
| Downstream of raf1 | Dsor1 | MAPKK, MEK | Cytoplasm | Protein kinase | [19] |
| rolled | rl | MAPK, ERK | Cytoplasm | Conventional MAPK | [20] |
| pointed | pnt | Nucleus | ETS transcriptional activator | [21] | |
| Positive regulators | |||||
| rhomboid | rho | Rho-1 | Golgi/endosome | Protease for Spitz cleavage | [42, 44] |
| roughoid | ru | Rho-3 | Golgi/endosome/ER | Protease for Spitz cleavage | [50] |
| Star | S | ER/Golgi | Chaperone | [46] | |
| rasp | rasp | skl, sit | Golgi | Palmitoyltransferase | [43] |
| kinase suppressor of Ras | ksr | KSR-1 | Cytoplasm | Scaffolding protein, protein kinase | [58] |
| connector enhancer of ksr | enk | Cytoplasm | Raf activator/inhibitor | [61] | |
| aveugle | ave | HYP | Cytoplasm | Raf activator | [63, 64] |
| steppke | step | Grp1 | Cytoplasm | Arf GEF | [66] |
| corkscrew | csw | SHP2 | Cytoplasm | Tyrosine phosphatase | [68] |
| mago nashi | mago | Nucleus | MAPK premRNA splicing | [57] | |
| myopic | mop | Endosome | Endocytic pathway | [38] | |
| Hepatocyte growth factor regulated tyrosine kinase substrate | Hrs | vps27 | Endosome | Endocytic pathway | [38] |
| Vacuolar protein sorting 4 | Vps4 | SKD1 | MVB (cytoplasm ?) | unclear | [33] |
| nejire | nej | CBP/p300 | Nucleus | Acetyl transferase | [101] |
| Negative regulators | |||||
| Argos | Aos | secreted | Ligand inhibitor | [70] | |
| kekkon-1 | kek1 | Transmembrane | Egfr binding | [76] | |
| Neuroglian | Nrg | Transmembrane | Ed binding | [79] | |
| echinoid | ed | Transmembrane | Egfr binding | [78] | |
| Fasciclin2 | Fas2 | Transmembrane | unclear | [80] | |
| sprouty | sty | Spry | Cytoplasm | Protein interaction, endocytosis | [81, 82] |
| Ras GTPase activating protein 1 | Gap1 | gap1, rasGAP | Cytoplasm | Negative regulator of Ras85D | [83] |
| Rho GTPase activating protein at 5A | RhoGAP5A | Cytoplasm | Rho GTPase activator | [85] | |
| vav ortholog (H. sapiens) | vav | Cytoplasm | Rho GTPase GEF | [88] | |
| Cbl proto-oncogene ortholog | Cbl | Cytoplasm | Endocytosis | [34] | |
| shibire | shi | dynamin | Cytoplasm | Endocytosis | [31] |
| small wing | sl | PLCc | Cytoplasm | cSpitz retention in ER | [54] |
| rhomboid-5 | rho-5 | iRhom | Spitz degradation through ERAD | [45] | |
| klumpfuss | klu | Nucleus | Transcription factor | [93] | |
| Hairless | H | Nucleus | Transcriptional repressor | [95] | |
| capicua | cic | Nucleus | Transcriptional repressor | [99] | |
| anterior open | aop | Yan | Nucleus | ETS transcriptional repressor | [97] |
The EGFR is present at the plasma membrane as a monomer. Binding of a ligand to the receptor induces its dimerisation and tyrosine trans-autophosphorylation that leads to the intracellular activation of the signalling pathway (Fig. 1b). Out of the four Drosophila ligands, two have a broad expression pattern: Spitz, which is responsible for EGFR activation in most tissues [8], including the eye, and Keren, which can complement Spitz activity [9]. The two other ligands have more restricted expression patterns: Vein has a weaker activation capacity than Spitz and is used in tissues where low activation levels are required [10], while Gurken is expressed specifically in the ovary [11]. Spitz, Gurken and Keren are TGFα homologues and are produced as inactive transmembrane precursors, whereas Vein presents more similarities to neuregulins and is directly produced as a secreted active protein. Following EGFR activation by one of the four ligands, specific phosphorylated tyrosine residues of the intracellular domain serve as docking sites for various SH2 adaptor proteins such as Downstream of Receptor Kinase (Drk, homologous to Grb2) [12] and Src homology 2 domain containing (Shc) [13], which in turn recruit Son of Sevenless (Sos) [14]. Sos, as a guanine exchange factor (GEF), activates the small G-protein Ras by facilitating the exchange of GDP for GTP [15], which triggers the activation of the canonical mitogen-activated protein kinase (MAPK) cassette [16, 17]. Three kinases are successively phosphorylated, Raf [18], MAPKK (or MEK) [19] and finally the MAPK family member extracellular signal regulated kinase (ERK, also called Rolled) [20]. Subsequently, MAPK is translocated to the nucleus and the transcriptional output of the pathway is mediated by the ETS protein Pointed (Pnt) [21]. The MAPK cassette is one of the most conserved signalling pathways and is activated downstream of the EGFR both in Drosophila and vertebrates. In vertebrates, EGFR stimulation can initiate downstream signalling through diverse transduction cascades such as MAPK, but also phosphoinositide 3-kinase (PI3K), phospholipase C gamma (PLCγ) and signal transducer and activator of transcription 3 (STAT3) to regulate a multitude of cellular activities [22]. In contrast, the Drosophila EGFR signalling pathway is essentially linear (or unbranched) [17]. Studying the function and regulation of the EGFR signalling pathway in Drosophila thus benefits from its linearity and lesser genetic redundancy compared to mammalian systems (i.e. only one EGFR member and four ligands).
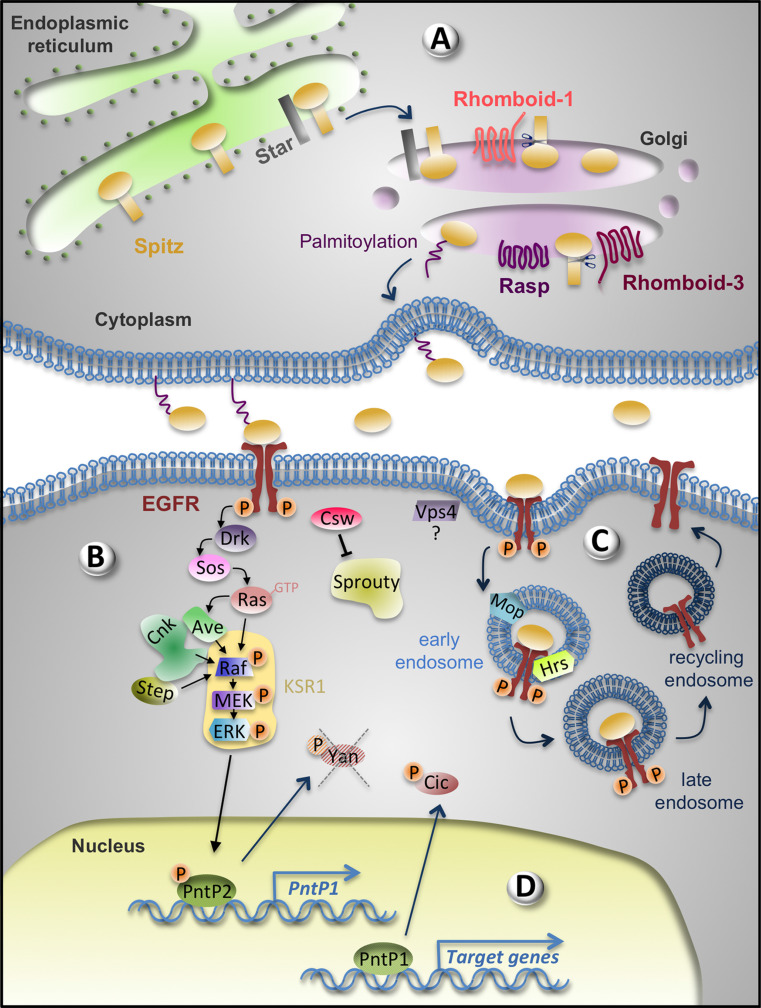
Fig. 1.
Positive regulators of the EGFR signalling pathway. a The ligand Spitz is produced in the sending cell. Spitz is retained in the ER until Star promotes its translocation to the Golgi apparatus where it is cleaved by Rhomboid-1 or 3 making it soluble. Palmitoylation by Rasp promotes Spitz tethering to the plasma membrane to increase concentration at secreting sites. Both unpalmitoylated and palmitoylated Spitz can be released. b In the receiving cell, upon Spitz binding, the EGFR dimerizes leading to its autophosphorylation and activation of the canonical Drk/Sos/Ras/Raf/MEK/ERK pathway. KSR1, a scaffolding protein, brings members of the pathway in close proximity. CNK interaction with Ave and Step contributes also to Raf activation. The inhibitor Sprouty is dephosphorylated, and thus inactivated by the phosphatase Csw. c Vps4, Mop and Hrs are also positive regulators. Mop and Hrs participate in EGFR endocytosis. Recycling of the EGFR to the plasma membrane is important to maintain active signalling. d In the nucleus, phosphorylated ERK triggers the exit of the transcriptional repressor Yan and phosphorylates PntP2, which activates PntP1 transcription. When sufficient levels of PntP1 are reached, the various target genes of the EGFR signalling pathway are transcribed. Phosphorylated ERK also phosphorylates the transcriptional repressor Cic, which results in its export out of the nucleus and the subsequent transcriptional activation of EGFR signalling target genes
Drosophila eye development and EGFR signalling
One of the most intriguing questions that has been addressed by developmental biologists is how a unique signalling pathway can trigger so many diverse cellular responses during development, or even within a single tissue. The answers rely in part within the diversity of mechanisms employed to regulate a given pathway, and within the combination of various signalling pathways that provide unique sets of inputs on target gene enhancers. In particular, studies on the mechanisms regulating the EGFR signalling pathway during Drosophila eye development have yielded great advances and keep providing answers to this question. The wild-type Drosophila compound eye is characterised by a very stereotyped organisation. The retina is a crystalline lattice that contains around 750–800 identical visual units called ommatidia, each ommatidium consisting of a precise number of cells that are specified during larval and pupal stages. This compound structure is very well suited for the study of small changes in signalling cascades. Indeed, small defects in each ommatidium are amplified due to the presence of hundreds of ommatidia and thus become visible. In human, EGFR is involved in many processes such as cell proliferation, inhibition of apoptosis, differentiation, adhesion, migration. Interestingly, most of these processes are also instrumental during eye development and EGFR signalling is involved in each of these developmental steps. The adult eye derives from a bilayered epithelium called the eye-antennal imaginal disc that invaginates from the embryonic epidermis. During the first two larval instars, the eye-antennal disc grows extensively through cell proliferation cycles. At the beginning of the third instar, a structure called the morphogenetic furrow (MF) appears at the posterior margin of the eye disc. The MF is a transient invagination of the disc associated with cell cycle arrest and the onset of cell differentiation posterior to it. Subsequently, the MF sweeps across the eye disc from posterior to anterior in response to the morphogen Hedgehog (Hh) signalling, thus organising a wave of differentiation where ommatidia are organised in rows, with ommatidia from a same row differentiating at the same time. A new column is formed every 2 h on average, so that by late third instar, the posterior half of the disc contains differentiating ommatidia (with the oldest ommatidia being at the most posterior region of the disc), whereas the anterior half still contains proliferating cells (Fig. 2). The region with high proliferation rate anterior to the MF is called the first mitotic wave. Within the MF, Hh induces the expression of Atonal (Ato) in a stripe of cells. Ato expression is later refined posteriorly to the MF to specify one R8 cell per developing ommatidium. R8 is the first photoreceptor to differentiate and is singled out from a pool of Ato-positive progenitor cells. R8 then sequentially recruits R2 and R5, which in turn recruit R3 and R4, forming a five-cell precluster (or rosette). The remaining undifferentiated cells undergo a last round of cell division called the second mitotic wave, which is essential to generate a sufficient pool of uncommitted cells. R1, R6 and finally R7, the last photoreceptor, are next recruited to form a full complement of eight photoreceptors per ommatidium. R1 to R6, the outer photoreceptors, are organised around R7 and R8, the inner photoreceptors. Once the eight photoreceptors have been recruited, four lens-secreting epithelial cells (cone cells) are added apically to each ommatidium. The MF reaches the anterior margin of the eye disc during early pupal life. In the pupa, pigment cells differentiate from a pool of interommatidial cells. Two primary pigment cells are added first, enwrapping each cone cell cluster (lying on top of the eight photoreceptors) to form the core ommatidium. The remaining accessory cells ultimately differentiate as secondary and tertiary pigment cells along with sensory bristles. Concomitantly, excess interommatidial cells are eliminated by apoptosis, which completes the formation of the stereotypical hexagonal lattice surrounding the core ommatidium [23–25].
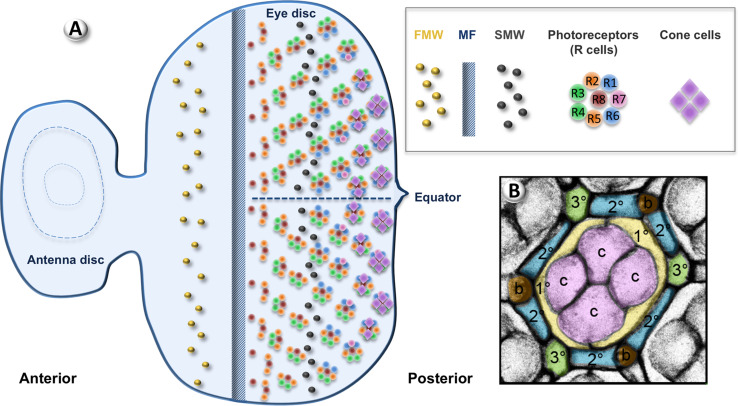
Fig. 2.
Eye development in Drosophila. a Schematic representation of a third instar larval eye-antenna disc. Anterior is to the left. Undifferentiated cells proliferate during the first mitotic wave (FMW). The morphogenetic furrow (MF) sweeps across the disc from posterior to anterior. Differentiation starts posterior to the MF with the recruitment of R8. Photoreceptors and cone cells are recruited sequentially. Ommatidia rotate on each side of the equator. The last round of proliferation occurs during the second mitotic wave (SMW). b From Martin-Bermudo et al. Apical view of a pupal retina 50 h after puparium formation stained with anti-Disc large (Dlg). The Dlg signal has been inverted so the staining appears in grey. At this stage, extra interommatidial cells have already been eliminated by apoptosis. The different cell types are pseudo-coloured for easier identification: cone cells (CCs) in pink, primary pigment cells (1°s) in yellow, secondary (2°s) and tertiary (3°s) pigment cells in blue and green, respectively, bristles in brown
The EGFR signalling pathway sustains multiple functions during eye development [26, 27], including the differentiation of all cell types. In fact, expression of a dominant negative form of EGFR completely prevents the formation of the retina [28], indicating that this signalling pathway is absolutely necessary to form an eye. It is fascinating to think that the reiterative use of a same signal triggers so many different outcomes within the same tissue. We understand now that what seems actually as important as the nature of the signal received by a cell is the state of this cell, which changes its competence over time. This state depends on the timing, duration and quantity of signal received, together with the localisation of the cell at the time it receives signal [29, 30]. The developing Drosophila eye has proven to be a reliable model system that has revealed many of the mechanisms employed to regulate the EGFR signalling pathway.
The different mechanisms regulating the EGFR pathway in the Drosophila eye
The EGFR is broadly expressed during Drosophila development, challenging the achievement of its precise activation. Rather than the presence of the receptor itself at the cell surface, what is important is to trigger a very specific response at a specific time and place. This is partly achieved by a variety of activating and inhibiting regulators that control the EGFR signalling pathway at several levels, depending on whether they act outside of the cell, at the membrane, in different cytoplasmic compartments, or in the nucleus to control transcriptional outputs. Many of these regulators are themselves targets of the pathway, leading to positive and negative feedback loops that fine-tune the cellular responses. In this section, the different types of regulators that modulate EGFR signalling during eye development and how the feedback loops are established will be reviewed.
Regulators of EGFR endocytosis and intracellular trafficking: Cbl, Echinoid, Mop, Hrs, Vps4
EGFR signalling activity initiates with the formation of ligand-receptor complexes at the plasma membrane. Once the EGFR is activated by its ligand, it can be internalised, and either sent back to the membrane via recycling endosomes to receive new ligand molecules, or ubiquitinated and sent for degradation, which will attenuate or terminate signalling. Accordingly, the loss of the GTPase Dynamin/Shibire required for EGFR internalisation into endocytic vesicles [31] is associated with increased levels of EGFR signal transduction, which results in ectopic photoreceptor differentiation in the eye disc [32, 33]. The Drosophila E3 ubiquitin ligase Cbl is also required for receptor endocytosis and is a negative regulator of various signalling pathways, including EGFR. Cbl triggers receptor ubiquitination, internalisation through clathrin-coated vesicles and subsequent degradation (Fig. 3c), thus attenuating signalling from the plasma membrane [34–36]. Finally, Echinoid (Ed), a known negative regulator of the EGFR pathway, also contributes to EGFR endocytosis at adherens junctions in the eye disc. Indeed, in the absence of Ed, EGFR is upregulated at apical surfaces and the number of internalised EGFR particles is lower than in wild-type cells, indicating a decrease in EGFR internalisation [37].
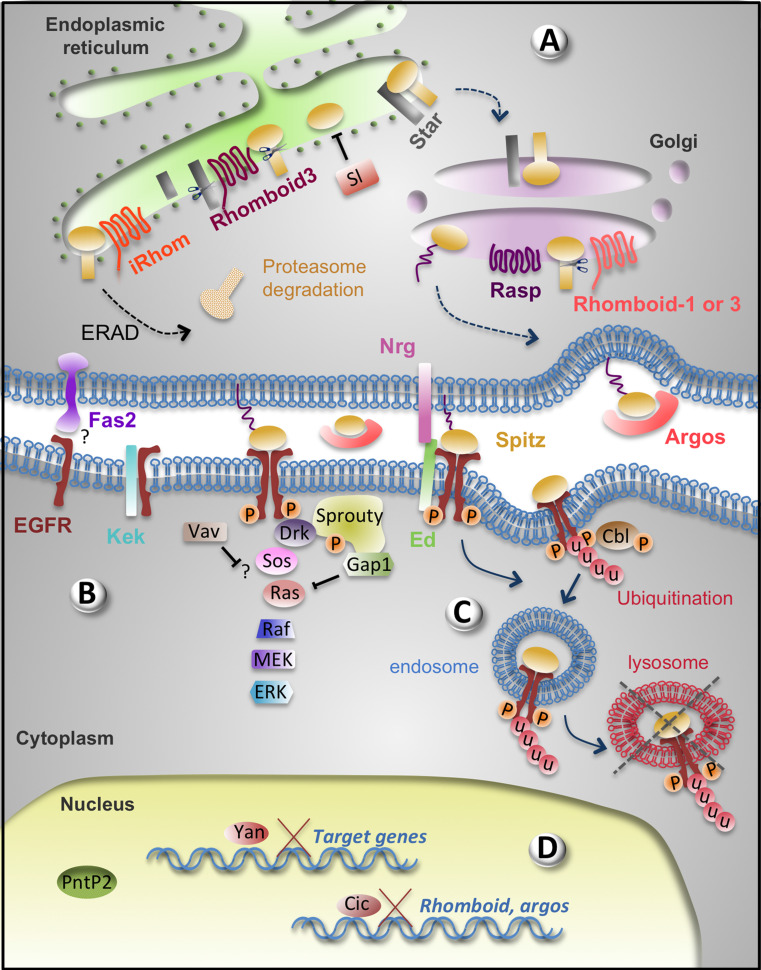
Fig. 3.
Negative regulators of the EGFR signalling pathway. a In the ER of the signalling cell, Rhomboid-3 cleaves both the chaperone Star, so less Spitz can be transferred to the Golgi, and Spitz itself. Cleaved Spitz is retained in the ER by Sl. iRhom promotes ERAD in which Spitz is transferred to the cytoplasm and degraded by the proteasome. Thus, only the Spitz molecules that encounter Star, escape cleavage and ERAD can be transferred to the Golgi and secreted. b At the plasma membrane of the receiving cell, EGFR is inhibited non-autonomously by Fas2 by an unknown mechanism, and autonomously by Kek and Ed. Nrg contributes non-cell autonomously to EGFR inhibition by Ed. In the cytoplasm, the MAPK cassette is inhibited by Gap1 (inactivating Ras), Sprouty and Vav. Sprouty interacts with Drk, Gap1 and the plasma membrane. c Ed may contribute to EGFR inhibition via endocytosis. Cbl-mediated EGFR ubiquitination targets the receptor for endocytosis and lysosome degradation. d In the absence of phosphorylated ERK, the transcriptional repressors Yan and Cic repress EGFR pathway target genes and prevent the transcriptional activator Pnt from binding DNA
Although it seems counterintuitive at first, different studies have also implicated EGFR endocytosis in promoting signalling. In those cases, interfering with EGFR endocytic processing reduces its activity. These results suggest that signalling may also remain active in endocytic vesicles, or be more efficient than from the plasma membrane, or that receptor recycling and/or potential modifications acquired in the endocytic pathway are important for promoting signalling. Several proteins known to localise in the endocytic pathway are required for EGFR signalling in the eye. Myopic (Mop) appears to act on EGFR internalised in early endosomes and promotes the cleavage of its cytoplasmic domain, which could enhance some of its signalling activity (Fig. 1c). In contrast to other RTKs, EGFR signalling is specifically reduced in mop mutants suggesting a specificity for this pathway [38]. Hepatocyte growth factor regulated tyrosine kinase substrate (Hrs) is a known component of the endosomal sorting complexes required for transport (ESCRT) machinery that promotes the internalisation of ubiquitinated receptors into multivesicular bodies (MVBs) and is believed to terminate signalling and target receptors for lysosomal degradation. In the Drosophila eye, however, Hrs is required for EGFR transduction [38]. Mechanistically, it is thus proposed that, in some contexts, progression through the endocytic pathway may be required for prolonging or achieving maximal EGFR activity, or for eliciting specific signalling from specialised endocytic compartments. Another well-known component of the ESCRT machinery, vacuolar protein sorting 4 (Vps4), has been shown to drive the final steps of receptor internalisation into MVBs and achieve sequestration from their cytosolic downstream effectors. In Vps4 mutants, different signalling receptors in addition to the EGFR accumulate in endosomes where they seem to be trapped. However, Vps4 also appears to promote EGFR signalling upstream of cell surface receptor activation, and independently of its function in endocytic MVBs [33]. Cytoplasmic trafficking of the EGFR through the endocytic pathway and/or by endocytic components thus provides a means to fine-tune signalling from the receptor.
Positive regulators of EGFR activation
Regulators of the production, presentation and processing of the ligand Spitz: Star, Rhomboid and Rasp
Among the four ligands, Spitz is the main activating ligand in most tissues, including the eye, hence Spitz production and activation will be mainly discussed here. However, Keren is also expressed in the eye where it acts cooperatively with Spitz to maintain cell cycle arrest, to control R8 spacing, cell clustering and survival, and will be discussed later [39, 40]. Although spitz is expressed homogenously in the eye disc, only its secreted form is active as a ligand [41]. Spitz is first produced as an inactive membrane-anchored precursor and is activated by a series of posttranslational modifications, including cleavage [42] and palmitoylation [43]. These modifications depend on Spitz subcellular localisation. Hence, regulating Spitz trafficking and processing provides a means to confine where and when the EGFR pathway will be activated, thereby preventing the inappropriate production of active ligand.
Spitz is produced in excess, retained in the endoplasmic reticulum (ER) and must be trafficked to the Golgi apparatus to be cleaved [42, 44, 45] (Fig. 1a). Star, a type 2 transmembrane protein [46], is required for Spitz transfer from the ER to the Golgi [44], serving as a cargo receptor. Star behaves as a chaperone and associates with Spitz in the ER, and together they are exported to the Golgi. Once in the Golgi, Spitz is cleaved by the Rhomboid family of serine proteases and released as a soluble fragment to be secreted from the cell [42]. Rhomboids are seven-transmembrane domain proteins [47] with a catalytic domain required for their intramembranous proteolytic function [48]. Two members of this family are present in the eye and responsible for Spitz activation: Rhomboid (Rho), also known as Rhomboid-1 (Rho-1), and Rhomboid-3 (Rho-3) also called Roughoid (Ru). While loss of Rho-1 gives no detectable phenotype [49], loss of Rho-3 leads to severe phenotypes and loss of both mimics the loss of the EGFR, indicating that Rho-1 and Rho-3 cooperate but that Rho-3 is the prominent protease in the eye [50]. Star and Rhomboids are, therefore, an integral part of the EGFR signalling pathway in Drosophila and accordingly their loss of function phenotypes is similar to those of Spitz [51]. Note that Keren activation might be slightly different from that of Spitz. Indeed, although membrane-tethered Keren is, like Spitz, a substrate for the Rhomboid proteins, a small fraction of it can also undergo Star/Rho-independent cleavage [9, 52].
The control of intracellular trafficking of Spitz, Star and Rhomboid is complex and instrumental in the regulation of EGFR activation, but it remains somehow unclear and controversial in the field. Spitz and Rho-1 can be detected in the Golgi apparatus in COS cells (cell line derived from monkey kidney fibroblast cells) and in Drosophila salivary glands [42, 44], but they have also been located in late endosomes in Drosophila S2 cells, suggesting that Spitz cleavage could occur in the late endosome, or in the Golgi en route to the late endosome, adding a new layer of complexity to the control of intracellular trafficking required for ligand activation [53]. Interestingly, in the eye disc, Rho-1 and Rho-3 are localised in punctate structures that do not contain Golgi or endosome markers and are enriched at the apical pole of the cells, indicating that they can also be localised in other types of vesicles. In addition, Rho-3 is also found in perinuclear ER, suggesting that Spitz cleavage already occurs in this compartment [54]. However, cleavage in the ER attenuates EGFR signalling by decreasing the level of secreted ligand, which will be further discussed in the section on negative regulation of EGFR signalling.
In addition to cleavage, Spitz also needs to be palmitoylated to be active. Palmitoylation is a posttranslational lipid modification occurring in the secretory pathway, which targets proteins to the plasma membrane. The palmitoyltransferase Rasp promotes the addition of a palmitate to the Spitz N-terminal cysteine. As a consequence, Spitz is tethered to the membrane and can signal to adjacent cells but its diffusion is restricted. Local concentration of Spitz could allow threshold levels of the ligand required for EGFR activation to be attained in nearby cells and restrict its range of action [43]. Finally, based on the different properties of chimeric membrane-tethered forms of Spitz, it has also been proposed that Spitz could be transported from the ER to the Golgi where it is palmitoylated by Rasp, then secreted apically to the plasma membrane, where it would be tethered by the palmitoyl group. Only then would Spitz be endocytosed back into the producing cell, cleaved by Rho-1 in the late endosome compartment and secreted basolaterally to activate EGFR in the adjacent cells [55].
Regulators of the Ras/MAPK module: KSR, CNK, Aveugle, Step and Csw
While ligand production and availability from the signalling cell are key steps of EGFR signalling, equally important are the layers of regulatory inputs affecting the transduction of the cascade, downstream of the receptor, in the ligand-receiving cell. Mapping EGFR activity in tissues, including the eye disc, has been achieved by labelling di-phospho-ERK (dpERK), also referred to as phospho-MAPK, allowing the identification of cells transducing EGFR signalling. This pattern of EGFR activity was similar to that of Rhomboid expression, confirming that activation by Rhomboid is a limiting element in the activation of the EGFR pathway [56]. Regulating the expression of the MAPK gene itself can also influence EGFR signalling. Indeed, the exon junction complex (EJC) containing the subunit mago nashi (Mago) regulates the splicing of the MAPK pre-mRNA and is therefore required to maintain sufficient MAPK levels. In the eye, Mago has a specific function during photoreceptor differentiation and loss of mago prevents EGFR signalling [57].
In the cytoplasm, several proteins contribute to EGFR activation by scaffolding and activating components of the MAPK module. Kinase suppressor of Ras (KSR) is a conserved protein that was originally isolated in a Drosophila genetic screen aiming at identifying suppressors of the phenotype associated to the expression of a constitutively activated form of Ras [58]. KSR is a scaffold protein that brings together the three kinases Raf, MEK and MAPK (Fig. 1b). Depending on KSR levels, it can either promote Raf activation or act as a dominant negative by sequestering the kinase signalling components of the pathway [59]. In addition, KSR possesses intrinsic Raf activating potential through the formation of side-to-side heterodimers with Raf [60].
Connector enhancer of KSR (CNK), which is essential for eye development, also has antagonistic properties with respect to Raf activity. Although it was first identified as a positive regulator of Raf [61], it was later shown that it also behaves as a Raf inhibitor [62]. The positive effect exerted on Raf is mediated by the N-terminal sterile alpha motif (SAM) and conserved region in CNK (CRIC) domains, whereas its inhibitory effect is mediated by the C-terminal region containing a Raf-interacting motif (RIM). How exactly CNK activates Raf remains elusive, but another SAM domain protein Aveugle (Ave), also known as Hyphen (Hyp), plays a role in this process [63, 64]. Ave was isolated in a screen designed to identify genes required for normal photoreceptor differentiation. Epistasis tests placing Ave between Ras and Raf in the MAPK cassette, together with its physical interaction with CNK, suggested that Ave was a good candidate to contribute to Raf activation in a complex with CNK, possibly via the recruitment of an as yet unknown activating kinase [64]. The Ave/CNK interaction was later confirmed to be mediated by their SAM domains, which facilitates the recruitment of KSR to form a CNK/Ave/KSR complex [65]. The Drosophila Arf GEF Steppke (Step) is also part of this scaffolding complex via its direct interaction with CNK and controls MAPK activation downstream of EGFR during eye and wing development [66]. The antagonistic properties of KSR and CNK, and the intricate mode of action of the KSR/CNK/Ave/Step complex in Raf activation provide regulatory inputs upstream of the MAPK cassette and highlight the complexity of EGFR transduction.
Besides the tyrosine kinase proteins forming the MAPK module, one tyrosine phosphatase, Corkscrew (Csw), has been shown to modulate positively RTK signalling, including the EGFR pathway in Drosophila embryo [67, 68]. This role seems to be also conserved in the eye, as csw mutants fail to differentiate photoreceptors [32]. Csw has been suggested to contribute positively to RTK signalling by dephosphorylating a critical tyrosine residue essential to the inhibiting activity of Sprouty, a well-known inhibitor of RTK signalling (further discussed below), thus inactivating it. This double-negative regulatory circuit was demonstrated in mammalian and Drosophila culture cells as well as in the developing eye [69].
Negative regulators of EGFR activation
The continuously growing body of novel negative regulators of EGFR signalling identified is the subject of speculations as to why so many types of inhibitors have been selected. It seems that their relevance depends on the cellular and developmental context. In a given tissue, the multiple types of modulators expressed display a combination of specific and overlapping effects. These negative regulators have different molecular mechanisms, different sites of expression and thus different sites of actions. In addition, while some regulators such as Argos are specific to the EGFR pathway, others such as Gap1 or Vav are also used in other signalling pathways (Fig. 3). The convergence of all these players contributes to achieving the very precise regulation of signalling intensity in each cell.
Diffusible molecule: Argos
Argos is a negative regulator specific to EGFR signalling that is secreted and acts in a non-cell-autonomous manner [70]. The absence or misexpression of the argos gene leads to severe eye defects, indicating that Argos is essential to Drosophila eye development [70, 71]. Although Argos was first considered as an antagonistic ligand of the EGFR based on its ability to bind the receptor and on the fact it presents an atypical EGF domain [72, 73], it was later found that Argos actually inhibits EGFR signalling by sequestering the ligand rather than interacting with the receptor [74]. Argos binding to Spitz via the EGF-like domain prevents the interaction of Spitz with the EGFR, thus inhibiting signalling. Argos is secreted by cells receiving high levels of EGFR activation and diffuses several cell rows away from its site of production, as opposed to Spitz that diffuses over a shorter distance. In cells exposed to high levels of secreted Spitz (active), Argos is not sufficient to prevent EGFR activation, thus its inhibitory function primarily affects more distal cells, in a process called remote inhibition, which restricts long-range signalling [29]. Consequently, in most cases during development, including in the Drosophila eye, EGFR signalling pathway is activated at short-range, and the secreted ligand is therefore not considered to act as a morphogen [7, 75]. Hence, Argos converts a graded signal triggered by a diffusible ligand, Spitz, into an on/off binary choice: either the pathway is activated (close from the source of ligand production, e.g. in the eye, R8 photoreceptor at the centre of each ommatidium) or inhibited (further away from the source).
Cell surface proteins: Kekkon, Neuroglian, Echinoid and Fasciclin 2
Another class of inhibitors is represented by cell surface transmembrane proteins, such as Kekkon-1 (Kek1), Neuroglian (Nrg), Echinoid (Ed) and Fasciclin2 (Fas2) (Fig. 3b). Kek1 and Ed interact directly with the EGFR extracellular domain, which attenuates receptor dimerisation by forming inactive heterodimers. In several tissues, including the eye imaginal disc, Kek1 is expressed on the cell surface of the same cells as EGFR and downregulates receptor activity by direct interaction. Kek1 has also been shown to inhibit mammalian ErbB Receptors [76]. Ed is a L1-type cell adhesion transmembrane molecule that is expressed in all cells of the eye imaginal discs [77]. Ed, in addition to its direct association with EGFR, can also undergo homophilic interactions and is phosphorylated in response to EGFR activity [78]. Nrg, another L1-type adhesion molecule, interacts with Ed and they both synergise to attenuate EGFR signalling. A model has been proposed in which Nrg acts non-cell autonomously as a ligand and activator of Ed, which in turn antagonises EGFR signalling [79]. Finally, Fas2, the Drosophila orthologue of neural cell adhesion molecule (NCAM), is a specific inhibitor of EGFR signalling localised at the plasma membrane and acting in an non-cell-autonomous manner [80]. The precise mechanism by which Fas2 inhibits EGFR signalling is still unknown, yet it could either interact directly with the EGFR similarly to Kek1 and Ed, decrease ligand production, or downregulate receptor levels or activity indirectly.
Cytoplasmic proteins: Sprouty, Gap1, RhoGAP5A and Vav
Sprouty is not specific to the EGFR cascade, but rather inhibits a range of RTKs since it interferes with Ras signalling in the cytoplasm [81, 82]. In the eye disc, Sprouty has been shown to associate with the inner surface of the plasma membrane through its C-terminal domain and interacts physically with two general components of the Ras pathway, Drk, and Gap1, a Ras GTPase-activating protein that inactivates the GTP-bound form of Ras [83]. The exact molecular mechanism of EGFR inhibition by Sprouty has not been deciphered yet. Based on its known interactions, it has been proposed that Sprouty could inhibit EGFR signalling by recruiting the Gap1 inhibitor and/or blocking the ability of the adaptor Drk to bind to its positive effectors, thus preventing the formation of functional signalling complexes (Fig. 3b). Consistently, sprouty and gap1 loss-of-function mutant clones have very similar phenotypes in the eye [81]. Sprouty may also be involved in endocytosis. In human, Sprouty2 antagonises EGFR signalling by preventing the endocytosed receptor from progressing from early to late endosomes, thus inhibiting intracellular signal transduction [84]. Similarly, in Drosophila, Sprouty has also been shown to prevent EGFR from progressing into late endosomes [38].
In addition to Gap1, RhoGAP5A, which is specific to the Rho GTPases family, was found to downregulate the EGFR pathway, possibly by acting at the plasma membrane [85]. However, the role of RhoGAP5A is more restricted than the one of Gap1, as only the patterning of the interommatidial lattice at the pupal stage was affected in rhoGAP5A loss-of-function retinas. Another protein suggesting the implication of the Rho GTPases in downregulating the EGFR pathway is Vav. Vav is a guanine exchange factor (GEF) for the Rho GTPase family of proteins that is activated by phosphorylation in response to EGFR activation [86, 87]. Recently, it was also reported to act as negative regulator of EGFR signalling at different stages of Drosophila eye development. This activity depends on its GEF function, as point mutations in the Pleckstrin homology (PH) domain mediating its GEF activity elicit the same phenotypes as vav null mutants or gain-of-function for EGFR signalling [88]. Like Sprouty and Gap1, Vav is not a specific inhibitor of EGFR signalling and rather functions at the crossroads of different pathways. Vav proteins overexpression in many human tumours [89] highlights the importance of understanding the interplay between members of this family and the EGFR pathway.
Inhibition from the endoplasmic reticulum: Small Wing, Rho-3, iRhom
As mentioned before, Spitz transport and processing is complex. Besides its transport to the Golgi and possibly to the late endosome to be processed by Rho-1, it has been suggested that some Spitz cleavage may already occur in the ER, mediated mainly by Rho-3 [54]. In this model, cleaved Spitz (cSpitz) is retained in the ER by the PLCγ Small Wing (Sl) and is therefore inactive (Fig. 3a). Accordingly, mutants for the sl gene display EGFR hyperactivation phenotypes. Although the exact mechanism by which the cytoplasmic protein Sl retains cSpitz in the ER is still unknown, Sl function provides yet another way for restricting the production of active ligand and thus downregulating the EGFR pathway in the receiving cell. The genetic interaction between Sl and EGFR signalling components is restricted to the eye [90]. The quantity of released ligand is controlled additionally via the intracellular localisation of the Rhomboid proteins. In fact, these proteins have dual functions in regulating EGFR signalling as they can activate or attenuate the EGFR pathway in tissues where tight control is required, depending on their differential compartmentalisation [54, 91]. In the eye, Rho-1 resides in the Golgi (or secretory compartment) and mediates Spitz cleavage to release the active form of the ligand, whereas Rho-3 is found both in the ER and in the secretory compartment (Rho-2 has a similar function in the ovary). In the ER, Rho-3 not only cleaves Spitz, but also its chaperone Star, thus decreasing the quantity of Spitz available for transport and processing [53, 54, 92]. Only the Star molecules that have escaped cleavage in the ER are able to mediate transport of the Spitz precursor to the secretory compartment where it is cleaved by either Rho-1 or Rho-3, and released to signal to neighbouring cells [17, 54]. Note that Rho-3 subcellular localisation is also involved in determining the site of Spitz release. ER localisation of Rho-3 is essential for Spitz secretion from photoreceptor axonal termini, whereas Spitz secretion from cell bodies depends on Rhomboid protein localisation in more downstream secretory compartments [91].
The most recently characterised mechanism for downregulation of EGFR signalling at the ER is via iRhoms. iRhoms are related to Rhomboids but lack intrinsic protease activity. The Drosophila iRhom, encoded by the rhomboid-5 gene, is a specific inhibitor of EGFR signalling that is largely restricted to neural tissues, including the eye disc where it is expressed in cells posterior to the MF [45]. Functionally, iRhom mainly controls fly sleep behaviour. Mechanistically, iRhom associates with Spitz in the ER to promote Spitz degradation by facilitating its retrotranslocation from the ER membrane to the cytoplasm where it is degraded by proteases. This degradation process provides an ER quality control machinery called endoplasmic reticulum-associated degradation (ERAD). iRhoms therefore represent yet another mechanism to modulate the quantity of released ligand by inhibiting its processing [45, 48]. Although the loss of iRhom on its own does not cause any obvious phenotype in the eye, it is expressed in the eye disc at a time a decrease in EGFR signalling is required. iRhom could therefore be involved, together with other negative regulators, in preventing inappropriate signalling [45].
Transcription factors in the nucleus: Klumpfuss and Hairless
The transcriptional repressor Hairless (H) promotes the transcriptional activation of the Runx transcription factor Lozenge (Lz), probably indirectly via inhibition of an as yet unknown inhibitor. Lz in turn activates the transcription of two EGFR negative regulators, argos and the transcription factor klumpfuss (klu) [93, 94]. During pupal morphogenesis H acts as an inhibitor of EGFR signalling on two levels, via the indirect activation of these EGFR negative regulators and via the inhibition of rho-1 transcription [95].
Regulation on EGFR target genes and feedback loops
Nuclear translocation of phosphorylated MAPK (ERK) is the most downstream step before target gene transcriptional activation, the final output of EGFR signalling activation [96]. The universal transcriptional response downstream of EGFR activation is mediated by the Pointed (Pnt) ETS transcriptional activator [21]. The other key transcriptional output of the EGFR pathway is the inactivation of the constitutive transcriptional repressor Yan, another ETS protein lacking a transcriptional activation domain. Yan acts by blocking the binding of Pnt to DNA, therefore preventing target gene transcriptional activation. Upon EGFR activation, MAPK phosphorylates Yan, which is consequently translocated to the cytoplasm for degradation [97], allowing Pnt binding to DNA and the subsequent activation of target genes. In Drosophila, although only one Pnt isoform is present in most tissues, two isoforms PntP1 and PntP2 are expressed in the eye. They are activated in a sequential manner by different mechanisms in response to EGFR activation. PntP2 is activated by MAPK phosphorylation, whereas the transcription of pntP1 is induced by activated PntP2 (Fig. 1d). Once expressed, PntP1 is constitutively active and thus sufficient to induce transcription of target genes essential for eye differentiation [98]. Besides Yan, Capicua (Cic) is another transcriptional repressor that is downregulated in response to EGFR signalling, although it is not specific to this pathway. Cic possesses a domain that serves as a docking site for MAPK and is redistributed from the nucleus to the cytoplasm in response to EGFR activation. Rho-1 and argos, two known targets of the EGFR pathway, are both repressed by Cic in the Drosophila embryo, hence their transcription could be activated by Cic derepression in response to EGFR [99]. In the eye disc, Cic is an inhibitor of cell growth but does not affect cell fate. The only growth promoting function of the Ras-MAPK cassette in this tissue appears to be the downregulation of Cic. Using different effectors could therefore provide a means to segregate different EGFR signalling functions by branching the pathway: one branch might use Cic to regulate growth, whereas the other branch would rely on Pnt to regulate photoreceptor cell fate determination [100].
In addition, histone acetylation seems also important for EGFR signalling during eye development. Indeed, the CREB binding protein (CBP/p300) encoded by nejire (nej) is a histone acetyltransferase that interacts genetically with various components of the EGFR cascade in the eye [101]. Consistently, this transcriptional activator is required for the recruitment of photoreceptors [32].
Many regulators of the EGFR cascade are also target genes of the pathway, which means that their transcription is regulated in response to EGFR signalling, thus establishing positive and negative feedback loops. In addition to all the above-described mechanisms, these feedback loops provide another way to regulate tightly the strength of EGFR signalling. For instance, rho-1 and step are target genes forming positive feedback loops in the Drosophila egg and wing disc, respectively [66, 102]. EGFR activity also controls the expression of many inhibitors of the pathway, such as Argos [103, 104], Kekkon [76, 105], Sprouty [81] and Fas2 [80]. In contrast, some inhibitors like Ed do not appear to be regulated transcriptionally by EGFR signalling. However, the inhibitory activity of Ed is regulated post-translationally by EGFR signalling, thereby also providing a negative feedback mechanism to reduce EGFR signalling [78].
The different roles of EGFR signalling during eye development
The EGFR is a key receptor that is needed for the recruitment of each class of cells in the ommatidium and that, in addition, is also involved in other processes such as proliferation, cell survival or cell adhesion and remodelling. A general issue of studying the function of such a pleiotropic signalling pathway resides in the difficulty to separate these distinct roles. This is yet another advantage of the Drosophila model in which elegant genetic tools for gene disruption or overexpression allows more precise deciphering of gene functions in specific cells at desired times.
Eye specification
Several nuclear factors, known as master genes, control eye specification from the second larval stage onwards. When expressed ectopically, each of these genes, including Eyeless/Pax6, is sufficient to induce ectopic eyes, and conversely, no eye can be specified in their absence. Together with Notch, EGFR signalling has homeotic functions genetically upstream of these master genes to control eye specification. Hyperactivation of EGFR signalling can induce homeotic transformation of the eye to antenna, indicating that EGFR inhibits the eye versus antenna fate, whereas Notch has an opposing function [106].
G1 arrest, cell cycle progression and mitosis
EGFR − clones of cells generated in the undifferentiated region of the eye disc are smaller than their twin spots but this is not accompanied with increased apoptosis, indicating a requirement for EGFR signalling in normal proliferation [107]. However, when mutant cells are provided with a growth advantage over their neighbours through the use of a Minute mutation, larger EGFR − clones of cells can be obtained anterior to the MF. Hence, while EGFR is involved in regulating proliferation in the eye disc, it is not absolutely necessary for cell division [27]. Posterior to the MF, cells remain arrested in G1 before entering into the differentiation program. Uncommitted cells must later progress to the S phase to enter the last round of division during the second mitotic wave that takes place in the differentiating region of the disc (Fig. 2). EGFR signalling is required for both G1 arrest and for progression to the second mitotic wave. Interestingly, the ligand Keren can activate EGFR signalling and trigger G1 arrest in the absence of Spitz, but Spitz overexpression impedes the second mitotic wave, suggesting that both ligands are involved in this process. EGFR signalling is used at a lower threshold for G1 arrest than the differentiation response [40]. Continuous EGFR signalling is required in subsets of cells that remain in G1 and do not enter the second mitotic wave (R2, R3, R4 and R5 cells). Note that R8s also remain in G1 but independently of EGFR signalling. During the second mitotic wave, EGFR signalling is inactive and the G1 to S phase transition can occur in cells that reenter the cell cycle. EGFR activity is required later in the cell cycle, to trigger the G2 to M phase transition. In this context, EGFR is activated by Spitz acting as a short-range signal received from the five-cell photoreceptor precluster [108, 109]. Mechanistically, the transcriptional activator PntP2 is phosphorylated in response to EGFR signalling and directly activates the transcription of the phosphatase string, encoding the Drosophila Cdc25 homologue, a universal regulator of the G2/M transition in eukaryotes. In the absence of activated PntP2, the Tramtrack isoform Ttk69, a repressor which is also possibly controlled in part by EGFR signalling, inhibits string transcription and therefore mitosis [110].
Initiation and progression of the morphogenetic furrow
A temperature sensitive EGFR allele was used to bypass the proliferation defects and it was shown that EGFR signalling controls the initiation and the progression of the MF, which determines the onset of pattern formation in the eye. The EGFR is also sufficient to induce MF initiation [111, 112]. In addition to the EGFR, several signalling pathways (Hh, Decapentaplegic (Dpp), Wingless (Wg) and Notch) contribute to the initiation and progression of the MF. During MF progression, EGFR signalling in R2 and R5 activates PntP2, which binds specifically to an hh eye specific enhancer, thus triggering the expression of Hh in these cells [113]. In an indirect autoregulatory loop, secreted Hh then drives the expression of ato in more anterior cells of the MF, which is required for the acquisition of the R8 fate. In each ommatidium, secretion of Spitz from R8 ensures the EGFR-dependent recruitment of R2 and R5 that will express Hh and so on. Thus, a positive loop between Hh and EGFR signalling drives anterior propagation of the MF and photoreceptor differentiation [114].
Ommatidial spacing
Ato is first expressed in all cells in the MF, and its expression becomes progressively restricted to only one cell per ommatidium, posterior to the MF. This Ato-expressing cell then differentiates into R8, the founder cell of each ommatidium. Ato restriction is therefore responsible for the regular spacing of ommatitia and this process is mainly controlled by Notch-mediated lateral inhibition and the secreted protein Scabrous [115, 116]. Different groups have obtained conflicting results regarding a role for EGFR signalling in ommatidial spacing. Several studies have described R8 spacing defects in the absence of EGFR signalling [27, 117, 118], whereas another group suggested that EGFR signalling was not involved in ommatidial spacing and that results from the other groups might be artifactual due to the use of a Minute mutation in the genetic background [112, 119]. However, a later study, which did not rely on the use of a Minute mutation to provide EGFR − cells with a growth advantage, demonstrated that the two EGFR ligands Keren and Spitz act redundantly to control R8 spacing [39]. These latest results also explain why spitz mutant clones do not display R8 spacing defects whereas clones doubly mutant for rho-1 and rho-3, or for EGFR do.
Epithelial polarity, adherens junction remodelling and ommatidial rotation
As they mature, ommatidial clusters become asymmetric and adopt opposite chirality on either side of the dorso-ventral midline, also called the equator. Next, these chiral forms of ommatidia rotate through exactly 90° to take the appropriate spatial arrangement of photoreceptors and polarise the epithelium. This rotation process involves the activity of planar cell polarity proteins [120]. Altering EGFR signalling levels does not affect chirality but results in ommatidial rotation defects, with clusters that either over- or under-rotate. Several mechanisms have been suggested to explain these phenotypes. EGFR signalling could act directly on the physical process of rotation [121, 122], indirectly via the inappropriate subcellular localisation of the planar polarity proteins Frizzled and Flamingo [122, 123], or to anchor the rotated precursors in place [121]. It has been demonstrated that EGFR signalling is involved in establishing epithelium polarity in the eye via cell fate specification [124], but also via the regulation of cytoskeletal and adherens junctional elements, such as the Ras effector Canoe and the Cadherin proteins [123, 125]. Adherens junction (AJ) remodelling is necessary for ommatidial rotation and morphogenesis (apical constriction of cells in the MF, and cell shape changes immediately posterior to the MF). Accordingly, it was found that EGFR signalling also promotes AJ remodelling (suppression or elongation) required for the cell rearrangements occurring during ommatidial morphogenesis [126, 127]. Although a direct regulation of AJ components by members of the EGFR signalling pathway in the cytoplasm could be possible, so far EGFR signalling has only been shown to regulate AJ remodelling and ommatidial rotation via EGFR-regulated transcriptional activation.
Cell recruitment and determination: photoreceptors, cone and pigment cells
The reiterative use of the EGF receptor triggers the differentiation of all cell types in the Drosophila eye. In the absence of EGFR, only R8 cells are specified, therefore EGFR signalling does not appear to be required for R8 determination [27, 112, 117, 118]. Nevertheless, it seems that EGFR signalling must be actively prevented by Senseless during R8 selection to inhibit inappropriate signalling within the R8 equivalence group that could interfere with R8 recruitment [78, 128, 129]. The sequential recruitment of all other photoreceptors requires active EGFR signalling, and expressing a constitutively secreted form of Spitz ectopically is sufficient to trigger the differentiation of extra photoreceptors, cone and pigment cells [28]. During ommatidial formation, Spitz is first secreted by R8, activating EGFR signalling in the neighbouring R2/R5 pair. In response, R2/R5 secretes Spitz, which leads to the recruitment of the R3/R4 pair, and so on until the recruitment of all cell types is achieved. But while EGFR is responsible for photoreceptor fate acquisition, how it triggers the differentiation of each subclass of photoreceptor is still unclear [24]. It is possible that the EGFR pathway is permissive rather than instructive in cell specification, and that the specificity of cell fates is actually a consequence of combinatorial signalling. In this scenario, a cell receiving different inputs from different pathways at a given time and position will express a unique combination of transcription factors that will activate specific differentiation genes. Combinatorial signalling also explains the change in cellular competence, since although uncommitted cells are subject to the same signal, the response is different in terms of differentiation if the signal is received at different times. The use of combinatorial signalling for the induction of specific cell fates has been described for several cell types, including R7, cone and primary pigment cells. For instance, the cone cell determinant Pax2 (also known as Shaven) is expressed only in cells in which the transcriptional regulator Lz is present and in which both Notch and EGFR signalling are active (Fig. 4). Different combinations of these three inputs give rise to different cell types, revealing a code for cell fate specification [130]. Interestingly, the Notch ligand Delta activating the pathway in the cone cell precursor is itself produced in the adjacent R1/R6 precursor cells in response to EGFR signalling, further adding an extra level of complexity to the interplay between the two pathways [131]. The specific expression of Prospero in R7 and cone cells is also the result of combinatorial signalling involving an interplay between the Notch and EGFR signalling pathways [132]. EGFR requirement in primary pigment cell specification is indirect through the induction of Delta expression in cone cells. The subsequent activation of the Notch pathway within neighbouring cells then induces the specification of the primary pigment cell fate [133]. Altogether these findings helped understanding how the complex interplay between multiple signalling pathways drives the acquisition of different cell fates in the Drosophila retina.
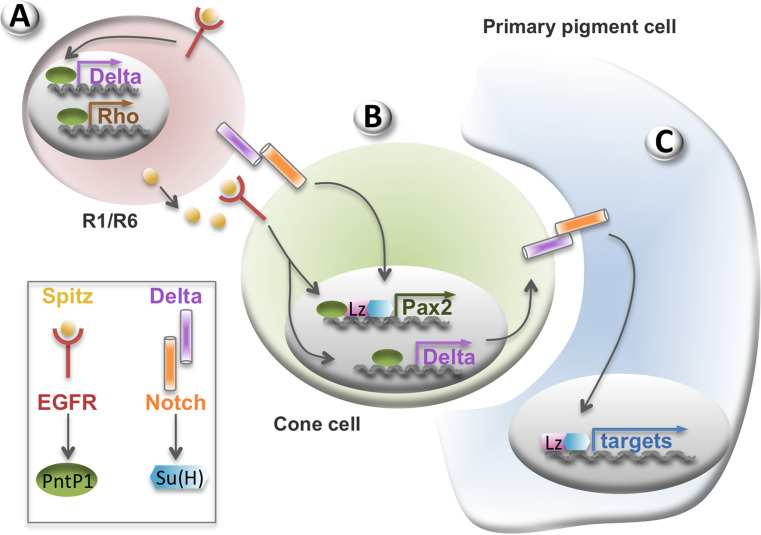
Fig. 4.
Combinatorial signalling during R1/R6, cone cell and primary pigment cell recruitment. a During larval development, Delta, the ligand for Notch, is produced in response to EGFR signalling in the R1/R6 photoreceptor cells. Rhomboid is also produced, leading to Spitz activation. Therefore, Spitz and Delta both signal from R1/R6 to induce the EGFR and Notch signalling pathways in adjacent cells. b These pathways lead to the activation of Pnt and Suppressor of Hairless (Su(H)) in the nucleus, which, together with the transcription factor Lozenge (Lz), activate Pax2 transcription, thereby inducing cone cell fate. Delta is also produced in the cone cell in response to EGFR signalling. c Finally, Delta activates the Notch pathway in adjacent cells during early pupal life (16–19 h after puparium formation). Su(H) and Lz induce the primary pigment cell fate
Cell survival
EGFR and Notch signalling pathways are both also active later on, at the end of pupal eye morphogenesis, during the developmentally regulated cell death of supernumerary pigment cells. EGFR signalling in interommatidial cells exhibits anti-apoptotic activity by inhibiting the pro-apoptotic protein head involution defective (Hid) at two levels, through the regulation of hid transcription [134], and through direct phosphorylation of specific sites of the Hid protein exerted by activated MAPK [135]. This latter mechanism represents one of the few characterised cases in which EGFR signalling activity does not depend only on a transcriptional output. Spitz signal is thought to emanate from cone cells and activate EGFR signalling in adjacent interommatidial cells to protect them from cell death [136]. Notch activity during these late morphogenetic events is more controversial, as both pro-apoptotic [137, 138] and anti-apoptotic [95, 139] activities have been reported. For instance, the Notch antagonist H has pro-apoptotic activity during lattice cell death through the downregulation of EGFR activity by two means: the transcriptional activation of EGFR antagonists such as lz and argos, and the repression of the EGFR activator rho-1 [95]. Besides its well-characterised role in cell survival at the pupal stage, EGFR signalling is also required for cell survival much earlier, during the third larval stage, as loss of EGFR in the differentiating region of the disc (in contrast to uncommitted cells anterior to the MF) is accompanied by massive apoptosis [27]. In particular, it was shown that the second mitotic wave cells that are still unspecified require EGFR signalling autonomously to survive and that apoptosis susceptibility increases in older photoreceptors [108, 140]. EGFR signalling is therefore required for cell survival at different stages of eye development.
Concluding remarks
Research on EGFR signalling has largely benefited from discoveries on its regulation during Drosophila eye development. The complexity and multiplicity of the mechanisms employed to provide a specific cell with an appropriate quantity and duration of signalling highlights the paramount importance of regulating precisely this pathway, and the list of new regulators identified keeps growing every year. Are the same mechanisms used in vertebrates? This question is still far from being answered. The counterparts for some of the specific regulators, such as Argos and Star, identified in Drosophila, have not been identified in mammals. In contrast, other regulators such as Sprouty, Kekkon, Rhomboids and iRhoms, which were also originally identified in Drosophila, are largely conserved. For instance, Rhomboids are represented from yeast to mammals [48]. The Spitz/Star/Rhomboid proteins form a very elaborate system for retention, trafficking and cleavage of target proteins, which provides a complex mode of signalling pathway regulation. It is not clear yet whether this regulatory system is conserved in mammals in which ligands are also produced as membrane-bound precursors (prepro-EGF). However, as opposed to Drosophila, both uncleaved and cleaved ligands can be active, depending on the mode of signalling used, which can be autocrine, paracrine, juxtacrine or exTRAcrine, and the cleavage machinery appears to be essentially different [2]. The identification of regulators of the EGFR signalling pathway in Drosophila can also be relevant to cancer therapy research. For instance, the finding that the oncogene Vav, which is expressed in many human cancers is a negative regulator of the EGFR signalling pathway in the Drosophila eye could potentially help understanding EGFR misregulated activity in some tumours.
Another fundamental question that has intrigued biologists for decades is how the same signalling pathway can direct a multitude of cellular responses and how sharp borders of activation are established? Answers to these questions have been partially elucidated thanks to the study of EGFR signalling in the Drosophila eye model and mainly reside in combinatorial signalling. Another important aspect that can explain the different outcomes is if different strengths of signalling can dictate different fates. Although EGFR ligands do not seem to function as morphogens during eye development [75], some cellular responses are more sensitive to signalling levels than others. For instance, cell survival, progression towards mitosis and cell cycle arrest require lower EGFR signalling levels than photoreceptor differentiation, which necessitates intense signalling achieved by multiple reinforcing ligands [40, 141]. Consistently, cells deprived of the positive EGFR regulator Ave cannot differentiate as photoreceptors although they are still able to arrest in G1, also supporting this conclusion [64]. However, results from this study question the sharp threshold between survival and differentiation as ave clones in which no photoreceptors differentiate display increased apoptosis, indicating a failure in both differentiation and survival responses. In addition, among photoreceptors, different subtypes seem to also require different levels of signalling, with the R7 fate requiring the highest level of MAPK activation, which is achieved by the additive effects of EGF and Sevenless receptors, both activating the MAPK cassette [28]. As another example, ommatidial rotation is more sensitive to alterations in EGFR pathway activity than cell recruitment, since weak disruption in signalling affects orientation to a greater extent than recruitment [121]. Finally, low levels of EGFR activity seem sufficient to regulate clustering and R8 spacing but not sufficient to prevent cell death or trigger cell recruitment. In this model, these subtle differences could be the result of a possible weaker activating power of the EGFR ligand Keren [39]. The mechanism by which different levels of activation are translated into specific cellular responses is still unknown. There is no doubt that studying EGFR signalling and its mechanisms of regulation during Drosophila eye development will keep providing answers to the remaining questions.
Acknowledgments
The author thanks Anne-Marie Pret and Kevin Legent for their time and their very helpful critical comments on the manuscript.
References
- 1.Livneh E, Glazer L, Segal D, et al. The Drosophila EGF receptor gene homolog: conservation of both hormone binding and kinase domains. Cell. 1985;40:599–607. doi: 10.1016/0092-8674(85)90208-9. [DOI] [PubMed] [Google Scholar]
- 2.Singh B, Coffey RJ. Trafficking of epidermal growth factor receptor ligands in polarized epithelial cells. Annu Rev Physiol. 2014;76:275–300. doi: 10.1146/annurev-physiol-021113-170406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37(Suppl 4):S9–15. doi: 10.1016/S0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 4.Uberall I, Kolár Z, Trojanec R, et al. The status and role of ErbB receptors in human cancer. Exp Mol Pathol. 2008;84:79–89. doi: 10.1016/j.yexmp.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Gomez GG, Wykosky J, Zanca C, et al. Therapeutic resistance in cancer: microRNA regulation of EGFR signaling networks. Cancer Biol Med. 2013;10:192–205. doi: 10.7497/j.issn.2095-3941.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki T, Hiroki K, Yamashita Y. The role of epidermal growth factor receptor in cancer metastasis and microenvironment. Biomed Res Int. 2013 doi: 10.1155/2013/546318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shilo BZ. Signaling by the Drosophila epidermal growth factor receptor pathway during development. Exp Cell Res. 2003;284:140–149. doi: 10.1016/S0014-4827(02)00094-0. [DOI] [PubMed] [Google Scholar]
- 8.Rutledge BJ, Zhang K, Bier E, et al. The Drosophila spitz gene encodes a putative EGF-like growth factor involved in dorsal–ventral axis formation and neurogenesis. Genes Dev. 1992;6:1503–1517. doi: 10.1101/gad.6.8.1503. [DOI] [PubMed] [Google Scholar]
- 9.Reich A, Shilo B-Z. Keren, a new ligand of the Drosophila epidermal growth factor receptor, undergoes two modes of cleavage. EMBO J. 2002;21:4287–4296. doi: 10.1093/emboj/cdf439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schnepp B, Grumbling G, Donaldson T, Simcox A. Vein is a novel component in the Drosophila epidermal growth factor receptor pathway with similarity to the neuregulins. Genes Dev. 1996;10:2302–2313. doi: 10.1101/gad.10.18.2302. [DOI] [PubMed] [Google Scholar]
- 11.González-Reyes A, Elliott H, St Johnston D. Polarization of both major body axes in Drosophila by gurken-torpedo signalling. Nature. 1995;375:654–658. doi: 10.1038/375654a0. [DOI] [PubMed] [Google Scholar]
- 12.Olivier JP, Raabe T, Henkemeyer M, et al. A Drosophila SH2-SH3 adaptor protein implicated in coupling the sevenless tyrosine kinase to an activator of Ras guanine nucleotide exchange, Sos. Cell. 1993;73:179–191. doi: 10.1016/0092-8674(93)90170-U. [DOI] [PubMed] [Google Scholar]
- 13.Lai KMV, Olivier JP, Gish GD, et al. A Drosophila shc gene product is implicated in signaling by the DER receptor tyrosine kinase. Mol Cell Biol. 1995;15:4810–4818. doi: 10.1128/MCB.15.9.4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonfini L, Karlovich CA, Dasgupta C, Banerjee U. The Son of sevenless gene product: a putative activator of Ras. Science. 1992;255:603–606. doi: 10.1126/science.1736363. [DOI] [PubMed] [Google Scholar]
- 15.Simon MA, Bowtell DDL, Dodson GS, et al. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell. 1991;67:701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- 16.Bogdan S, Klämbt C. Epidermal growth factor receptor signaling. Curr Biol. 2001;11:R292–R295. doi: 10.1016/S0960-9822(01)00167-1. [DOI] [PubMed] [Google Scholar]
- 17.Shilo B-Z. The regulation and functions of MAPK pathways in Drosophila . Methods. 2014;68:151–159. doi: 10.1016/j.ymeth.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 18.Ambrosio L, Mahowald AP, Perrimon N. Requirement of the Drosophila raf homologue for torso function. Nature. 1989;342:288–291. doi: 10.1038/342288a0. [DOI] [PubMed] [Google Scholar]
- 19.Tsuda L, Inoue YH, Yoo MA, et al. A protein kinase similar to MAP kinase activator acts downstream of the raf kinase in Drosophila . Cell. 1993;72:407–414. doi: 10.1016/0092-8674(93)90117-9. [DOI] [PubMed] [Google Scholar]
- 20.Biggs WH, Zipursky SL. Primary structure, expression, and signal-dependent tyrosine phosphorylation of a Drosophila homolog of extracellular signal-regulated kinase. Proc Natl Acad Sci USA. 1992;89:6295–6299. doi: 10.1073/pnas.89.14.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- 22.Huang PH, Xu AM, White FM. Oncogenic EGFR signaling networks in glioma. Sci Signal. 2009;2:re6. doi: 10.1126/scisignal.287re6. [DOI] [PubMed] [Google Scholar]
- 23.Cagan R. Principles of Drosophila eye differentiation. Curr Top Dev Biol. 2009;89:115–135. doi: 10.1016/S0070-2153(09)89005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar JP. Building an ommatidium one cell at a time. Dev Dyn. 2012;241:136–149. doi: 10.1002/dvdy.23707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Treisman JE. Retinal differentiation in Drosophila . Wiley Interdiscip Rev Dev Biol. 2013;2:545–557. doi: 10.1002/wdev.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker NE, Rubin GM. Ellipse mutations in the Drosophila homologue of the EGF receptor affect pattern formation, cell division, and cell death in eye imaginal discs. Dev Biol. 1992;150:381–396. doi: 10.1016/0012-1606(92)90250-K. [DOI] [PubMed] [Google Scholar]
- 27.Domínguez M, Wasserman JD, Freeman M. Multiple functions of the EGF receptor in Drosophila eye development. Curr Biol. 1998;8:1039–1048. doi: 10.1016/S0960-9822(98)70441-5. [DOI] [PubMed] [Google Scholar]
- 28.Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/S0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- 29.Freeman M. Cell determination strategies in the Drosophila eye. Development. 1997;124:261–270. doi: 10.1242/dev.124.2.261. [DOI] [PubMed] [Google Scholar]
- 30.Freeman M. Complexity of EGF receptor signalling revealed in Drosophila . Curr Opin Genet Dev. 1998;8:407–411. doi: 10.1016/S0959-437X(98)80110-X. [DOI] [PubMed] [Google Scholar]
- 31.Vieira AV, Lamaze C, Schmid SL. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 32.Legent K, Steinhauer J, Richard M, Treisman JE. A screen for X-linked mutations affecting Drosophila photoreceptor differentiation identifies casein kinase 1α as an essential negative regulator of wingless signaling. Genetics. 2012;190:601–616. doi: 10.1534/genetics.111.133827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Legent K, Liu HH, Treisman JE. Drosophila Vps4 promotes epidermal growth factor receptor signaling independently of its role in receptor degradation. Development. 2015;142:1480–1491. doi: 10.1242/dev.117960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pai LM, Barcelo G, Schüpbach T. D-cbl, a negative regulator of the Egfr pathway, is required for dorsoventral patterning in Drosophila oogenesis. Cell. 2000;103:51–61. doi: 10.1016/S0092-8674(00)00104-5. [DOI] [PubMed] [Google Scholar]
- 35.Swaminathan G, Tsygankov AY. The Cbl family proteins: ring leaders in regulation of cell signaling. J Cell Physiol. 2006;209:21–43. doi: 10.1002/jcp.20694. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Chen Z, Bergmann A. Regulation of EGFR and Notch signaling by distinct isoforms of D-cbl during Drosophila development. Dev Biol. 2010;342:1–10. doi: 10.1016/j.ydbio.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho Y-H, Lien M-T, Lin C-M, et al. Echinoid regulates Flamingo endocytosis to control ommatidial rotation in the Drosophila eye. Development. 2010;137:745–754. doi: 10.1242/dev.040238. [DOI] [PubMed] [Google Scholar]
- 38.Miura GI, Roignant J-Y, Wassef M, Treisman JE. Myopic acts in the endocytic pathway to enhance signaling by the Drosophila EGF receptor. Development. 2008;135:1913–1922. doi: 10.1242/dev.017202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown KE, Kerr M, Freeman M. The EGFR ligands Spitz and Keren act cooperatively in the Drosophila eye. Dev Biol. 2007;307:105–113. doi: 10.1016/j.ydbio.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 40.Yang L, Baker NE. Cell cycle withdrawal, progression, and cell survival regulation by EGFR and its effectors in the differentiating Drosophila eye. Dev Cell. 2003;4:359–369. doi: 10.1016/S1534-5807(03)00059-5. [DOI] [PubMed] [Google Scholar]
- 41.Schweitzer R, Shaharabany M, Seger R, Shilo BZ. Secreted Spitz triggers the DER signaling pathway and is a limiting component in embryonic ventral ectoderm determination. Genes Dev. 1995;9:1518–1529. doi: 10.1101/gad.9.12.1518. [DOI] [PubMed] [Google Scholar]
- 42.Urban S, Lee JR, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/S0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 43.Miura GI, Buglino J, Alvarado D, et al. Palmitoylation of the EGFR Ligand Spitz by Rasp Increases Spitz Activity by Restricting Its Diffusion. Dev Cell. 2006;10:167–176. doi: 10.1016/j.devcel.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 44.Lee JR, Urban S, Garvey CF, Freeman M. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila . Cell. 2001;107:161–171. doi: 10.1016/S0092-8674(01)00526-8. [DOI] [PubMed] [Google Scholar]
- 45.Zettl M, Adrain C, Strisovsky K, et al. Rhomboid family pseudoproteases use the ER quality control machinery to regulate intercellular signaling. Cell. 2011;145:79–91. doi: 10.1016/j.cell.2011.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolodkin AL, Pickup AT, Lin DM, et al. Characterization of Star and its interactions with sevenless and EGF receptor during photoreceptor cell development in Drosophila . Development. 1994;120:1731–1745. doi: 10.1242/dev.120.7.1731. [DOI] [PubMed] [Google Scholar]
- 47.Bier E, Jan LY, Jan YN. Rhomboid, a gene required for dorsoventral axis establishment and peripheral nervous system development in Drosophila melanogaster . Genes Dev. 1990;4:190–203. doi: 10.1101/gad.4.2.190. [DOI] [PubMed] [Google Scholar]
- 48.Freeman M. The rhomboid-like superfamily: molecular mechanisms and biological roles. Annu Rev Cell Dev Biol. 2014;30:235–254. doi: 10.1146/annurev-cellbio-100913-012944. [DOI] [PubMed] [Google Scholar]
- 49.Freeman M, Kimmel BE, Rubin GM. Identifying targets of the rough homeobox gene of Drosophila: evidence that rhomboid functions in eye development. Development. 1992;116:335–346. doi: 10.1242/dev.116.2.335. [DOI] [PubMed] [Google Scholar]
- 50.Wasserman JD, Urban S, Freeman M. A family of rhomboid-like genes: Drosophila rhomboid-1 and roughoid/rhomboid-3 cooperate to activate EGF receptor signaling. Genes Dev. 2000;14:1651–1663. [PMC free article] [PubMed] [Google Scholar]
- 51.Mayer U, Nüsslein-Volhard C. A group of genes required for pattern formation in the ventral ectoderm of the Drosophila embryo. Genes Dev. 1988;2:1496–1511. doi: 10.1101/gad.2.11.1496. [DOI] [PubMed] [Google Scholar]
- 52.Urban S, Lee JR, Freeman M. A family of Rhomboid intramembrane proteases activates all Drosophila membrane-tethered EGF ligands. EMBO J. 2002;21:4277–4286. doi: 10.1093/emboj/cdf434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsruya R, Wojtalla A, Carmon S, et al. Rhomboid cleaves Star to regulate the levels of secreted Spitz. EMBO J. 2007;26:1211–1220. doi: 10.1038/sj.emboj.7601581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yogev S, Schejter ED, Shilo B-Z. Drosophila EGFR signalling is modulated by differential compartmentalization of Rhomboid intramembrane proteases. EMBO J. 2008;27:1219–1230. doi: 10.1038/emboj.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinhauer J, Liu HH, Miller E, Treisman JE. Trafficking of the EGFR ligand Spitz regulates its signaling activity in polarized tissues. J Cell Sci. 2013;126:4469–4478. doi: 10.1242/jcs.131169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gabay L, Seger R, Shilo BZ. In situ activation pattern of Drosophila EGF receptor pathway during development. Science. 1997;277:1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- 57.Roignant J-Y, Treisman JE. Exon junction complex subunits are required to splice Drosophila MAP kinase, a large heterochromatic gene. Cell. 2010;143:238–250. doi: 10.1016/j.cell.2010.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Therrien M, Chang HC, Solomon NM, et al. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-X. [DOI] [PubMed] [Google Scholar]
- 59.Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- 60.Rajakulendran T, Sahmi M, Lefrançois M, et al. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 61.Therrien M, Wong AM, Rubin GM. CNK, a RAF-binding multidomain protein required for RAS signaling. Cell. 1998;95:343–353. doi: 10.1016/S0092-8674(00)81766-3. [DOI] [PubMed] [Google Scholar]
- 62.Douziech M, Roy F, Laberge G, et al. Bimodal regulation of RAF by CNK in Drosophila . EMBO J. 2003;22:5068–5078. doi: 10.1093/emboj/cdg506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Douziech M, Sahmi M, Laberge G, Therrien M. A KSR/CNK complex mediated by HYP, a novel SAM domain-containing protein, regulates RAS-dependent RAF activation in Drosophila . Genes Dev. 2006;20:807–819. doi: 10.1101/gad.1390406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roignant J-Y, Hamel S, Janody F, Treisman JE. The novel SAM domain protein Aveugle is required for Raf activation in the Drosophila EGF receptor signaling pathway. Genes Dev. 2006;20:795–806. doi: 10.1101/gad.1390506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rajakulendran T, Sahmi M, Kurinov I, et al. CNK and HYP form a discrete dimer by their SAM domains to mediate RAF kinase signaling. Proc Natl Acad Sci USA. 2008;105:2836–2841. doi: 10.1073/pnas.0709705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hahn I, Fuss B, Peters A, et al. The Drosophila Arf GEF Steppke controls MAPK activation in EGFR signaling. J Cell Sci. 2013;126:2470–2479. doi: 10.1242/jcs.120964. [DOI] [PubMed] [Google Scholar]
- 67.Johnson Hamlet MR, Perkins LA. Analysis of corkscrew signaling in the Drosophila epidermal growth factor receptor pathway during myogenesis. Genetics. 2001;159:1073–1087. doi: 10.1093/genetics/159.3.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perkins LA, Larsen I, Perrimon N. corkscrew encodes a putative protein tyrosine phosphatase that functions to transduce the terminal signal from the receptor tyrosine kinase torso. Cell. 1992;70:225–236. doi: 10.1016/0092-8674(92)90098-W. [DOI] [PubMed] [Google Scholar]
- 69.Jarvis LA, Toering SJ, Simon MA, et al. Sprouty proteins are in vivo targets of Corkscrew/SHP-2 tyrosine phosphatases. Development. 2006;133:1133–1142. doi: 10.1242/dev.02255. [DOI] [PubMed] [Google Scholar]
- 70.Freeman M, Klämbt C, Goodman CS, Rubin GM. The argos gene encodes a diffusible factor that regulates cell fate decisions in the Drosophila eye. Cell. 1992;69:963–975. doi: 10.1016/0092-8674(92)90615-J. [DOI] [PubMed] [Google Scholar]
- 71.Freeman M. Misexpression of the Drosophila argos gene, a secreted regulator of cell determination. Development. 1994;120:2297–2304. doi: 10.1242/dev.120.8.2297. [DOI] [PubMed] [Google Scholar]
- 72.Jin MH, Sawamoto K, Ito M, Okano H. The interaction between the Drosophila secreted protein argos and the epidermal growth factor receptor inhibits dimerization of the receptor and binding of secreted spitz to the receptor. Mol Cell Biol. 2000;20:2098–2107. doi: 10.1128/MCB.20.6.2098-2107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vinós J, Freeman M. Evidence that Argos is an antagonistic ligand of the EGF receptor. Oncogene. 2000;19:3560–3562. doi: 10.1038/sj.onc.1203702. [DOI] [PubMed] [Google Scholar]
- 74.Klein DE, Nappi VM, Reeves GT, et al. Argos inhibits epidermal growth factor receptor signalling by ligand sequestration. Nature. 2004;430:1040–1044. doi: 10.1038/nature02840. [DOI] [PubMed] [Google Scholar]
- 75.Shilo B-Z. Regulating the dynamics of EGF receptor signaling in space and time. Development. 2005;132:4017–4027. doi: 10.1242/dev.02006. [DOI] [PubMed] [Google Scholar]
- 76.Ghiglione C, Amundadottir L, Andresdottir M, et al. Mechanism of inhibition of the Drosophila and mammalian EGF receptors by the transmembrane protein Kekkon 1. Development. 2003;130:4483–4493. doi: 10.1242/dev.00617. [DOI] [PubMed] [Google Scholar]
- 77.Bai J, Chiu W, Wang J, et al. The cell adhesion molecule Echinoid defines a new pathway that antagonizes the Drosophila EGF receptor signaling pathway. Development. 2001;128:591–601. doi: 10.1242/dev.128.4.591. [DOI] [PubMed] [Google Scholar]
- 78.Spencer SA, Cagan RL. Echinoid is essential for regulation of Egfr signaling and R8 formation during Drosophila eye development. Development. 2003;130:3725–3733. doi: 10.1242/dev.00605. [DOI] [PubMed] [Google Scholar]
- 79.Islam R, Wei S-Y, Chiu W-H, et al. Neuroglian activates Echinoid to antagonize the Drosophila EGF receptor signaling pathway. Development. 2003;130:2051–2059. doi: 10.1242/dev.00415. [DOI] [PubMed] [Google Scholar]
- 80.Mao Y, Freeman M. Fasciclin 2, the Drosophila orthologue of neural cell-adhesion molecule, inhibits EGF receptor signalling. Development. 2009;136:473–481. doi: 10.1242/dev.026054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Casci T, Vinós J, Freeman M. Sprouty, an intracellular inhibitor of Ras signaling. Cell. 1999;96:655–665. doi: 10.1016/S0092-8674(00)80576-0. [DOI] [PubMed] [Google Scholar]
- 82.Kramer S, Okabe M, Hacohen N, et al. Sprouty: a common antagonist of FGF and EGF signaling pathways in Drosophila . Development. 1999;126:2515–2525. doi: 10.1242/dev.126.11.2515. [DOI] [PubMed] [Google Scholar]
- 83.Gaul U, Mardon G, Rubin GM. A putative Ras GTPase activating protein acts as a negative regulator of signaling by the Sevenless receptor tyrosine kinase. Cell. 1992;68:1007–1019. doi: 10.1016/0092-8674(92)90073-L. [DOI] [PubMed] [Google Scholar]
- 84.Kim HJ, Taylor LJ, Bar-Sagi D. Spatial regulation of EGFR signaling by Sprouty2. Curr Biol. 2007;17:455–461. doi: 10.1016/j.cub.2007.01.059. [DOI] [PubMed] [Google Scholar]
- 85.Bruinsma SP, Cagan RL, Baranski TJ. Chimaerin and Rac regulate cell number, adherens junctions, and ERK MAP kinase signaling in the Drosophila eye. Proc Natl Acad Sci USA. 2007;104:7098–7103. doi: 10.1073/pnas.0701686104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sarkar A, Parikh N, Hearn SA, et al. Antagonistic roles of Rac and Rho in organizing the germ cell microenvironment. Curr Biol. 2007;17:1253–1258. doi: 10.1016/j.cub.2007.06.048. [DOI] [PubMed] [Google Scholar]
- 87.Tamás P, Solti Z, Bauer P, et al. Mechanism of epidermal growth factor regulation of Vav2, a guanine nucleotide exchange factor for Rac. J Biol Chem. 2003;278:5163–5171. doi: 10.1074/jbc.M207555200. [DOI] [PubMed] [Google Scholar]
- 88.Martín-Bermudo M-D, Bardet P-L, Bellaïche Y, Malartre M. The vav oncogene antagonises EGFR signalling and regulates adherens junction dynamics during Drosophila eye development. Development. 2015;142:1492–1501. doi: 10.1242/dev.110585. [DOI] [PubMed] [Google Scholar]
- 89.Lazer G, Katzav S. Guanine nucleotide exchange factors for RhoGTPases: good therapeutic targets for cancer therapy? Cell Signal. 2011;23:969–979. doi: 10.1016/j.cellsig.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 90.Schlesinger A, Kiger A, Perrimon N, Shilo B-Z. Small wing PLCgamma is required for ER retention of cleaved Spitz during eye development in Drosophila . Dev Cell. 2004;7:535–545. doi: 10.1016/j.devcel.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 91.Yogev S, Schejter ED, Shilo B-Z. Polarized secretion of Drosophila EGFR ligand from photoreceptor neurons is controlled by ER localization of the ligand-processing machinery. PLoS Biol. 2010 doi: 10.1371/journal.pbio.1000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tsruya R, Schlesinger A, Reich A, et al. Intracellular trafficking by Star regulates cleavage of the Drosophila EGF receptor ligand Spitz. Genes Dev. 2002;16:222–234. doi: 10.1101/gad.214202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rusconi JC, Fink JL, Cagan R. klumpfuss regulates cell death in the Drosophila retina. Mech Dev. 2004;121:537–546. doi: 10.1016/j.mod.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 94.Wildonger J, Sosinsky A, Honig B, Mann RS. Lozenge directly activates argos and klumpfuss to regulate programmed cell death. Genes Dev. 2005;19:1034–1039. doi: 10.1101/gad.1298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Protzer CE, Wech I, Nagel AC. Hairless induces cell death by downregulation of EGFR signalling activity. J Cell Sci. 2008;121:3167–3176. doi: 10.1242/jcs.035014. [DOI] [PubMed] [Google Scholar]
- 96.Kumar JP, Hsiung F, Powers MA, Moses K. Nuclear translocation of activated MAP kinase is developmentally regulated in the developing Drosophila eye. Development. 2003;130:3703–3714. doi: 10.1242/dev.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rebay I, Rubin GM. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell. 1995;81:857–866. doi: 10.1016/0092-8674(95)90006-3. [DOI] [PubMed] [Google Scholar]
- 98.Shwartz A, Yogev S, Schejter ED, Shilo B-Z. Sequential activation of ETS proteins provides a sustained transcriptional response to EGFR signaling. Development. 2013;140:2746–2754. doi: 10.1242/dev.093138. [DOI] [PubMed] [Google Scholar]
- 99.Astigarraga S, Grossman R, Díaz-Delfín J, et al. A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. EMBO J. 2007;26:668–677. doi: 10.1038/sj.emboj.7601532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tseng A-SK, Tapon N, Kanda H, et al. Capicua regulates cell proliferation downstream of the receptor tyrosine kinase/Ras signaling pathway. Curr Biol. 2007;17:728–733. doi: 10.1016/j.cub.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Anderson J, Bhandari R, Kumar JP. A genetic screen identifies putative targets and binding partners of CREB-binding protein in the developing Drosophila eye. Genetics. 2005;171:1655–1672. doi: 10.1534/genetics.105.045450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ruohola-Baker H, Grell E, Chou T-B, et al. Spatially localized rhomboid is required for establishment of the dorsal–ventral axis in Drosophila oogenesis. Cell. 1993;73:953–965. doi: 10.1016/0092-8674(93)90273-S. [DOI] [PubMed] [Google Scholar]
- 103.Golembo M, Schweitzer R, Freeman M, Shilo BZ. Argos transcription is induced by the Drosophila EGF receptor pathway to form an inhibitory feedback loop. Development. 1996;122:223–230. doi: 10.1242/dev.122.1.223. [DOI] [PubMed] [Google Scholar]
- 104.Wasserman JD, Freeman M. An autoregulatory cascade of EGF receptor signaling patterns the Drosophila egg. Cell. 1998;95:355–364. doi: 10.1016/S0092-8674(00)81767-5. [DOI] [PubMed] [Google Scholar]
- 105.Ghiglione C, Carraway KL, 3rd, Amundadottir LT, et al. The transmembrane molecule kekkon 1 acts in a feedback loop to negatively regulate the activity of the Drosophila EGF receptor during oogenesis. Cell. 1999;96:847–856. doi: 10.1016/S0092-8674(00)80594-2. [DOI] [PubMed] [Google Scholar]
- 106.Kumar JP, Moses K. EGF receptor and Notch signaling act upstream of Eyeless/Pax6 to control eye specification. Cell. 2001;104:687–697. doi: 10.1016/S0092-8674(01)00265-3. [DOI] [PubMed] [Google Scholar]
- 107.Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 108.Baker NE, Yu SY. The EGF receptor defines domains of cell cycle progression and survival to regulate cell number in the developing Drosophila eye. Cell. 2001;104:699–708. doi: 10.1016/S0092-8674(01)00266-5. [DOI] [PubMed] [Google Scholar]
- 109.Firth LC, Baker NE. Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev Cell. 2005;8:541–551. doi: 10.1016/j.devcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 110.Baonza A, Murawsky CM, Travers AA, Freeman M. Pointed and Tramtrack69 establish an EGFR-dependent transcriptional switch to regulate mitosis. Nat Cell Biol. 2002;4:976–980. doi: 10.1038/ncb887. [DOI] [PubMed] [Google Scholar]
- 111.Kumar JP, Moses K. The EGF receptor and notch signaling pathways control the initiation of the morphogenetic furrow during Drosophila eye development. Development. 2001;128:2689–2697. doi: 10.1242/dev.128.14.2689. [DOI] [PubMed] [Google Scholar]
- 112.Kumar JP, Tio M, Hsiung F, et al. Dissecting the roles of the Drosophila EGF receptor in eye development and MAP kinase activation. Development. 1998;125:3875–3885. doi: 10.1242/dev.125.19.3875. [DOI] [PubMed] [Google Scholar]
- 113.Rogers EM, Brennan CA, Mortimer NT, et al. Pointed regulates an eye-specific transcriptional enhancer in the Drosophila hedgehog gene, which is required for the movement of the morphogenetic furrow. Development. 2005;132:4833–4843. doi: 10.1242/dev.02061. [DOI] [PubMed] [Google Scholar]
- 114.Roignant J-Y, Treisman JE. Pattern formation in the Drosophila eye disc. Int J Dev Biol. 2009;53:795–804. doi: 10.1387/ijdb.072483jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Baker NE, Mlodzik M, Rubin GM. Spacing differentiation in the developing Drosophila eye: a fibrinogen-related lateral inhibitor encoded by scabrous. Science. 1990;250:1370–1377. doi: 10.1126/science.2175046. [DOI] [PubMed] [Google Scholar]
- 116.Cagan RL, Ready DF. Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev. 1989;3:1099–1112. doi: 10.1101/gad.3.8.1099. [DOI] [PubMed] [Google Scholar]
- 117.Baonza A, Casci T, Freeman M. A primary role for the epidermal growth factor receptor in ommatidial spacing in the Drosophila eye. Curr Biol. 2001;11:396–404. doi: 10.1016/S0960-9822(01)00125-7. [DOI] [PubMed] [Google Scholar]
- 118.Yang L, Baker NE. Role of the EGFR/Ras/Raf pathway in specification of photoreceptor cells in the Drosophila retina. Development. 2001;128:1183–1191. doi: 10.1242/dev.128.7.1183. [DOI] [PubMed] [Google Scholar]
- 119.Rodrigues AB, Werner E, Moses K. Genetic and biochemical analysis of the role of Egfr in the morphogenetic furrow of the developing Drosophila eye. Development. 2005;132:4697–4707. doi: 10.1242/dev.02058. [DOI] [PubMed] [Google Scholar]
- 120.Wolff T. EGF receptor signaling: putting a new spin on eye development. Curr Biol. 2003;13:R813–R814. doi: 10.1016/j.cub.2003.09.053. [DOI] [PubMed] [Google Scholar]
- 121.Brown KE, Freeman M. Egfr signalling defines a protective function for ommatidial orientation in the Drosophila eye. Development. 2003;130:5401–5412. doi: 10.1242/dev.00773. [DOI] [PubMed] [Google Scholar]
- 122.Strutt H, Strutt D. EGF signaling and ommatidial rotation in the Drosophila eye. Curr Biol. 2003;13:1451–1457. doi: 10.1016/S0960-9822(03)00545-1. [DOI] [PubMed] [Google Scholar]
- 123.Gaengel K, Mlodzik M. Egfr signaling regulates ommatidial rotation and cell motility in the Drosophila eye via MAPK/Pnt signaling and the Ras effector Canoe/AF6. Development. 2003;130:5413–5423. doi: 10.1242/dev.00759. [DOI] [PubMed] [Google Scholar]
- 124.Weber U, Pataki C, Mihaly J, Mlodzik M. Combinatorial signaling by the Frizzled/PCP and Egfr pathways during planar cell polarity establishment in the Drosophila eye. Dev Biol. 2008;316:110–123. doi: 10.1016/j.ydbio.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mirkovic I, Mlodzik M. Cooperative activities of Drosophila DE-cadherin and DN-cadherin regulate the cell motility process of ommatidial rotation. Development. 2006;133:3283–3293. doi: 10.1242/dev.02468. [DOI] [PubMed] [Google Scholar]
- 126.Brown KE, Baonza A, Freeman M. Epithelial cell adhesion in the developing Drosophila retina is regulated by Atonal and the EGF receptor pathway. Dev Biol. 2006;300:710–721. doi: 10.1016/j.ydbio.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 127.Robertson F, Pinal N, Fichelson P, Pichaud F. Atonal and EGFR signalling orchestrate rok- and Drak-dependent adherens junction remodelling during ommatidia morphogenesis. Development. 2012;139:3432–3441. doi: 10.1242/dev.080762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Frankfort BJ, Mardon G. Senseless represses nuclear transduction of Egfr pathway activation. Development. 2004;131:563–570. doi: 10.1242/dev.00941. [DOI] [PubMed] [Google Scholar]
- 129.Rawlins EL, White NM, Jarman AP. Echinoid limits R8 photoreceptor specification by inhibiting inappropriate EGF receptor signalling within R8 equivalence groups. Development. 2003;130:3715–3724. doi: 10.1242/dev.00602. [DOI] [PubMed] [Google Scholar]
- 130.Flores GV, Duan H, Yan H, et al. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/S0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- 131.Tsuda L, Nagaraj R, Zipursky SL, Banerjee U. An EGFR/Ebi/Sno pathway promotes delta expression by inactivating Su(H)/SMRTER repression during inductive notch signaling. Cell. 2002;110:625–637. doi: 10.1016/S0092-8674(02)00875-9. [DOI] [PubMed] [Google Scholar]
- 132.Hayashi T, Xu C, Carthew RW. Cell-type-specific transcription of prospero is controlled by combinatorial signaling in the Drosophila eye. Development. 2008;135:2787–2796. doi: 10.1242/dev.006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nagaraj R, Banerjee U. Combinatorial signaling in the specification of primary pigment cells in the Drosophila eye. Development. 2007;134:825–831. doi: 10.1242/dev.02788. [DOI] [PubMed] [Google Scholar]
- 134.Kurada P, White K. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell. 1998;95:319–329. doi: 10.1016/S0092-8674(00)81764-X. [DOI] [PubMed] [Google Scholar]
- 135.Bergmann A, Agapite J, McCall K, Steller H. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell. 1998;95:331–341. doi: 10.1016/S0092-8674(00)81765-1. [DOI] [PubMed] [Google Scholar]
- 136.Querenet M, Goubard V, Chatelain G, et al. Spen is required for pigment cell survival during pupal development in Drosophila . Dev Biol. 2015;402:208–215. doi: 10.1016/j.ydbio.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 137.Miller DT, Cagan RL. Local induction of patterning and programmed cell death in the developing Drosophila retina. Development. 1998;125:2327–2335. doi: 10.1242/dev.125.12.2327. [DOI] [PubMed] [Google Scholar]
- 138.Yu S-Y, Yoo SJ, Yang L, et al. A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development. 2002;129:3269–3278. doi: 10.1242/dev.129.13.3269. [DOI] [PubMed] [Google Scholar]
- 139.Wech I, Nagel AC. Mutations in rugose promote cell type-specific apoptosis in the Drosophila eye. Cell Death Differ. 2005;12:145–152. doi: 10.1038/sj.cdd.4401538. [DOI] [PubMed] [Google Scholar]
- 140.Fan Y, Bergmann A. Multiple mechanisms modulate distinct cellular susceptibilities toward apoptosis in the developing Drosophila eye. Dev Cell. 2014;30:48–60. doi: 10.1016/j.devcel.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Halfar K, Rommel C, Stocker H, Hafen E. Ras controls growth, survival and differentiation in the Drosophila eye by different thresholds of MAP kinase activity. Development. 2001;128:1687–1696. doi: 10.1242/dev.128.9.1687. [DOI] [PubMed] [Google Scholar]