Abstract
Polarized epithelial cells align the mitotic spindle in the plane of the sheet to maintain tissue integrity and to prevent malignant transformation. The orientation of the spindle apparatus is regulated by the immobilization of the astral microtubules at the lateral cortex and depends on the precise localization of the dynein–dynactin motor protein complex which captures microtubule plus ends and generates pulling forces towards the centrosomes. Recent developments indicate that signals derived from intercellular junctions are required for the stable interaction of the dynein–dynactin complex with the cortex. Here, we review the molecular mechanisms that regulate planar spindle orientation in polarized epithelial cells and we illustrate how different cell adhesion molecules through distinct and non-overlapping mechanisms instruct the cells to align the mitotic spindle in the plane of the sheet.
Keywords: Cadherin; Cdc42; Cortical actin; JAM-A; Junctional adhesion molecules; LGN; MDCK; PtdIns(3,4,5)P3
Introduction
Cell division plays a critical role during development of tissues and their maintenance. Cells can divide symmetrically giving rise to two identical daughter cells or asymmetrically resulting in two daughter cells with different fates [1]. Symmetric cell divisions expand the number of identical cells, either during development or during regeneration. Symmetrically dividing epithelial cells align the mitotic spindle apparatus in the plane of the epithelial cell sheet ensuring that the integrity of the cellular sheet is maintained. Asymmetric cell divisions generate two different daughter cells and frequently regulate differentiation. In asymmetrically dividing epithelial cells the mitotic spindle is oriented perpendicularly to the plane of the sheet and one of the two daughter cells “leaves” the cellular layer. This process regulates for example the stratification of the skin [2] or the development of the heart [3].
In both symmetric and asymmetric cell divisions, the cells respond to specific cues which induce structural asymmetry in the cortex, characterized by the accumulation of molecules—either proteins or lipids—with the ability to interact with the dynein–dynactin motor protein complex (hereafter referred to as dynein for simplicity). Once immobilized at those cortical sites, dynein captures the plus ends of the aster mictotubules, and by virtue of its minus-end-directed motor protein activity it exerts pulling forces towards the centrosomes, which results in torque on the spindle apparatus. The spindle axis will thereby be aligned along an axis that is specified by two points at opposing cortices characterized by the highest concentration of dynein-interacting proteins.
To localize the dynein-interacting proteins at specific cortical sites and to correctly align the mitotic spindle apparatus cells respond to a variety of signals. These include signals derived from intrinsic factors such as the cell geometry [4] or the spindle poles and chromosomes [5, 6], as well as signals derived from extrinsic factors such as growth factors [7], cell–matrix contacts [8, 9], and cell–cell contacts [10]. Depending on cell type and tissue, mitotic cells probably integrate several of these signals into the decision-making process that eventually determines the orientation of the spindle apparatus.
A role of intercellular junctions in influencing the orientation of the mitotic spindle has been recognized two decades ago [10]. However, it has remained unclear until recently whether intercellular junctions are just permissive for spindle orientation or play an instructive role for the specific localization of the landmarks for dynein localization at the cortex. In this review article, we present an overview on the role of intercellular junctions in regulating mitotic spindle orientation in epithelial cells, and we highlight the recent developments which indicate an instructive role for specific cell adhesion receptors during this process.
The cortical machinery
Dynein at the cortex
The positioning of the mitotic spindle requires a physical interaction of the astral microtubules with the cell cortex. This interaction must be strong enough to withstand the mechanical forces exerted on the centrosomes during anaphase. At the same time, the plus ends of the astral microtubules must shorten to prevent sterical hindrance during centrosome movement towards the cortex. A central regulator of the dynamic interaction of the microtubule plus ends with the cell cortex turned out to be dynein. Dynein is a minus end directed motor protein complex and is involved in a variety of processes ranging from vesicle transport, centrosome positioning during interphase, and chromosome movement during mitosis and meiosis [11]. The dynein complex contains two identical heavy chains, each consisting of a C-terminal head domain and a N-terminal tail domain [12]. The C-terminal head domain harbors the motor protein activity and the MT-binding region, the N-terminal tail domain interacts with five non-catalytic subunits and together with the subunits serves as scaffold for various adaptor proteins. Having two non-overlapping functional domains the dynein complex can interact simultaneously with MT and with proteins that regulate its activity and/or localization [12, 13]. According to the current model, the positioning of the mitotic spindle by the dynein complex is regulated through a “search–activation–capture” mechanism: Dynein is associated with astral MTs which scan the cortex for dynein-interacting proteins in a dynamic process that involves MT catastrophes as well as MT sweeping and sliding along the cortex (Fig. 1) [14, 15]. Sites with high concentrations of dynein-interacting proteins will trigger dynein off-loading, its stable interaction with those sites, and activation of its motor protein activity [16]. Immobilized and activated dynein captures MT plus ends, and through the minus end-directed motor protein activity it generates pulling forces towards the centrosomes. In combination with a gradient of dynein-interacting molecules along the cortex, the pulling forces generate tension on the centrosomes which results in torque on the mitotic apparatus until the asters reach cortical sites with maximum levels of dynein-binding proteins (Fig. 1) [17, 18]. This process aligns the spindle apparatus along a specific axis in the cell. Importanty, by inhibiting MT growth and at the same time triggering MT catastrophes dynein prevents MT plus ends from colliding with the cortex thereby allowing the movement of the centrosomes towards the cortex [18]. Critical questions arising from this model are: Which molecules bind and/or activate dynein at the cortex, and which signalling pathways regulate the localization or enrichment of these at specific cortical sites during mitosis?
Fig. 1.
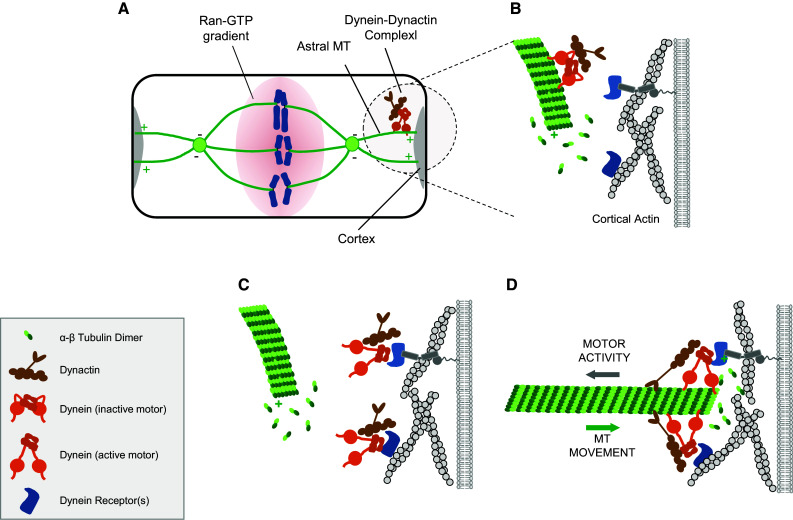
The force-generating machinery at the cortex. a Schematic view of mitotic epithelial cells at metaphase. The chromosomes are illustrated in blue, the polarity of MTs is indicated by “+” and “−”. A RanGTP gradient originating at the chromosomes and diffusing radially towards the cortex is indicated. The RanGTP gradient negatively regulates the localization of LGN at the cortex (see text for details). The grey area illustrates a cortical gradient of dynein-interacting proteins. b–d Search–activation–capture mechanism of dynein localization at the cortex. b Astral MTs slide along the cortex allowing the MT-bound dynein-dynactin complex to search for dynein receptors (blue). Dynein receptors can be associated with the membrane or with the actin cortex. Dynamic growth and shrinkage of MT plus ends facilitates scanning of larger surface areas. c Interaction of dynein with dynein receptors at the cortex triggers dynein offloading form the MTs and the activation of its motor activity. d Activated dynein at the cortex captures MT plus ends and generates pulling forces towards the centrosomes through its minus-end-directed motor activity. Ongoing catastrophes prevent MT plus ends from colliding with the cortex during centrosome movements
The core dynein anchor at the cortex: NuMA–LGN–Gαi
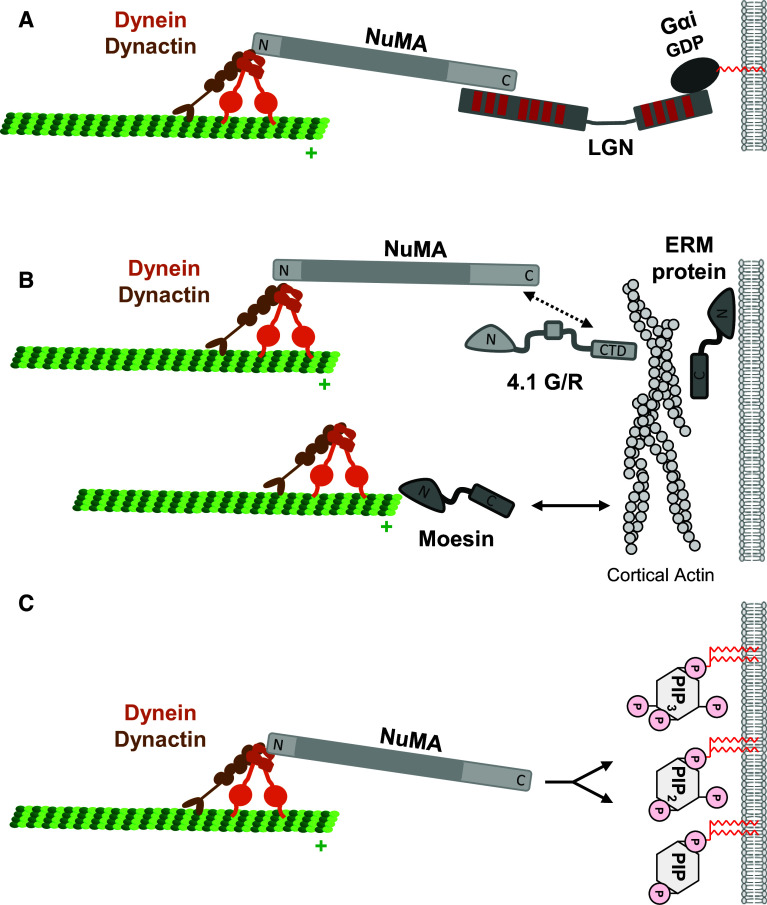
The core dynein anchor consists of the three proteins nuclear mitotic apparatus (NuMA, Mud in Drosophila, Lin5 in C. elegans), Leu-Gly-Asn repeat-enriched protein (LGN, a.k.a. G-protein signaling modulator 2 (GPSM2) in vertebrates, Partner of Inscuteable (Pins) in Drosophila, GPR-1/2 in C. elegans), and G protein alpha inhibitory subunit (Gαi, Gα in C. elegans) [19, 20] (Fig. 2a). NuMA is localized in the nucleus during interphase but redistributes to the spindle poles and the cortex during mitosis [21]. Through a region within the N-terminal domain (aa 1–705) NuMA interacts with dynein [5, 22], through a region within the C-terminal domain (aa 1818–1930) it directly interacts with LGN [23]. LGN is localized in the cytoplasm and can switch between two conformations: during interphase it adopts a closed conformation through intramolecular interactions between the N- and C-termini; during mitosis the binding of NuMA induces a conformational change that allows it to interact simultaneously with both NuMA and Gαi through its N- and C-termini, respectively [24, 25]. Gαi functions as a G protein-independent anchor for the tripartite complex at the cortex. It interacts directly with LGN [24] and is anchored at the membrane by N-myristoylation (Fig. 2a).
Fig. 2.
Immobilization of MTs at the cortex during mitosis. a LGN–Gαi-dependent NuMA–dynein localization at the cortex. NuMA interacts with dynein through its N-terminus and with LGN through its C-terminus. LGN adopts an open conformation upon NuMA binding and interacts simultanously with NuMA and with Gαi. Gαi is inserted in the membrane through its myristoyl group. This pathway predominates during metaphase. b Through its C-terminal domain, NuMA can also interact with two band 4.1 proteins 4.1G and 4.1R. The C-terminal domain (CTD) of 4.1 G/R is sufficient for NuMA localization at the cortex. This pathway operates specifically during anaphase and runs in parallel to the LGN–Gαi-dependent pathway of NuMA–dynein localization, presumably to adapt to the increased force requirements during anaphase. Ezrin/Radixin/Moesin (ERM) proteins are potential candidates to link cortical F-actin filaments (through their C-terminal C-ERMAD domain) to the membrane (through their N-terminal FERM domain). The FERM domain can interact with both phosphoinositides (PtdIns(4,5)P2) and integral membrane proteins. The ERM family member Moesin can directly interact through its N-terminal FERM domain with MTs at the cortex. It might therefore link the MT plus ends to the cortical actin cytoskeleton through its C-terminal C-ERMAD domain. c NuMA can directly interact with various phosphoinositide species including PtdIns(3,4,5)P3, PtdIns(4,5)P2, and PtdIns(4)P. The PtdIns(4,5)P2 and PtdIns(4)P-dependent pathway of NuMA–dynein recruitment operates during anaphase and might contribute to increased force generation
The localization of the NuMA–LGN–Gαi complex at the lateral cortex is regulated by various signals. One signal is derived from the Par3–aPKC–Par6 polarity protein complex [26, 27]. Par3, aPKC and Par6 exist in a ternary protein complex which regulates the maturation of primordial, spot-like adherens junctions into cell–cell junctions with adherens junctions and tight junctions [28–30]. During epithelial cell–cell contact formation, the protein complex localizes at primordial adherens junctions. The binding of Cdc42 and/or Rac1 to Par6 activates aPKC which phosphorylates various target proteins including Par3 [31]. Par3 phosphorylation leads to the dissociation of Par6–aPKC from Par3 [31]. In fully polarized epithelial cells grown in a three-dimensional environment, Par3 localizes to tight junctions whereas the active Par6–aPKC complex localizes to the apical membrane [32, 33]. Here, aPKC phosphorylates LGN which results in LGN exclusion from the apical membrane domain and restriction to the lateral membrane domain [34], thereby preventing spindle associations with the apical membrane domain. Another signal that regulates the cortical localization of the NuMA–LGN–Gαi complex is derived from the chromosomes. During the process of spindle alignment the spindle apparatus oscillates between the two lateral cortices [5], probably as a result of uneven pulling forces derived from the opposing cortices. Stabilization of the mitotic apparatus and its symmetric localization within the cell is regulated by the small GTPase Ran [5, 6]. The Ran-specific GEF RCC1 associates with chromosomes and DNA [35]. The chromosomal localization of RCC1 results in a gradient of RanGTP towards the cortex. When chromosomes are in close proximity with the cortex as a result of uneven pulling forces, the chromosome-derived RanGTP gradient disrupts the LGN association with the cortex, thereby decreasing the pulling forces from the proximal cortex. The resulting increase in pulling forces from the distal cortex centers the mitotic apparatus in the cell (see also Fig. 1).
A second cortical dynein anchor: the cortical actin cytoskeleton
During mitosis polarized epithelial cells round up which requires profound changes in cytoskeletal organization and cortical actomyosin dynamics [36, 37]. The reorganization of the cortical actin cytoskeleton probably serves several functions including the maintenance of cell–cell contact integrity during cell rounding [36, 38] and the generation of cortical rigidity to direct the pulling forces towards the centrosomes to prevent MT plus ends from pulling membranes inwards [39]. Besides these physical aspects, a reorganization of the cortical actin cytoskeleton is also required for dynein immobilization and for the stable attachment of astral microtubules at the cortex [9, 13, 40]. Important questions arising from these observations are: How is the cortical actin cytoskeleton reorganized during mitotic cell rounding? How is the actin cortex associated with cell membrane and how does it interact with the astral MTs?
The reorganization of the cortical actin cytoskeleton upon mitotic entry requires the activity of actin nucleators. Among the best understood actin nucleators are the Arp2/3 complex, which generates a branched network of new actin filaments, and formins, which produce unbranched filaments [41]. Although both types of actin nucleators localize to the cortex of mitotic cells [42], evidence accumulates suggesting that formins are the primary regulators of cortical actin reorganization during mitosis. For example, depletion of members or regulators of the Arp2/3 complex has no effect on the assembly of the mitotic F-actin cortex whereas depletion of the formin Dia1 in both Drosophila notum epithelial cells and HeLa cells severely affects the development of an F-actin cortex and progression through mitosis [42, 43]. Interestingly, the guanine nucleotide exchange factor Ect2 is phosphorylated by Cdk1/Cyclin B upon mitotic entry, resulting in its release from the nucleus and localization in the cytoplasm, where it activates RhoA [44]. Since RhoA can activate both myosin II and Dia1 [45], its early activation by Ect2 during mitosis could thus stimulate the new formation of actin filaments and the activation of actomyosin contractility, which are both required for the profound changes in cell shape during mitosis.
Members of the ezrin, radixin and moesin (ERM) family of proteins have emerged as primary candidates to link the cortical actin cytoskeleton with the cell membrane during mitosis. Under resting conditions ERM proteins are localized in the cytoplasm and are maintained in an autoinhibited, closed conformation. When activated, the closed conformation opens up thereby exposing the N-terminal domain (4.1 protein and ERM [FERM]) domain), which binds cytoplasmic tails of integral membrane proteins, and the C-terminal domain [COOH-ERM association (C-ERMAD) domain], which binds F-actin [46]. In both Drosophila and mammalian epithelial cells, ERM proteins are activated by Ste20-like kinases (Slik/SLK) at the onset of mitosis [47–49], and functional inactivation of FERM proteins results in a failure of cells to develop cortical rigidity and to round up during mitosis. These findings are thus consistent with a model according to which ERM proteins, after their activation at the onset of mitosis, crosslink actin filaments with transmembrane proteins thereby aligning actin filaments parallel to the plasma membrane, which results in a stiffening of the cell cortex and which helps to drive cell rounding (Fig. 2b). Two important observations suggest that ERM proteins also contribute to the attachment of MTs to the cell cortex during mitosis. First, moesin can directly interact with MTs and modulate MT dynamics in cells [50]. Second, SLK-activated ERM proteins regulate the polarized localization of LGN and NuMA at the cortex [49]. Together, these findings suggest that ERM proteins regulate the formation of a rigid cell cortex during mitosis but can also provide a scaffold for the physical association of MTs and of the force-generating machinery with the cortex.
It should be noted that many observations were made in HeLa cells which do not form cell–cell junctions typical for polarized epithelial cells. Nevertheless, since the basic morphological and cellular changes that occur during mitosis, e.g. cell rounding, cortical stiffening, and formation of a actomyosin-rich lateral cortex, are observed in polarized epithelial cells embedded in tissues as well [36, 38, 43], it is likely that the mechanisms operating in polarized epithelial cells are very similar.
A third potential cortical dynein anchor: PtdIns(3,4,5)P3
The role of the NuMA–LGN–Gαi complex in regulating mitotic spindle orientation has been studied predominantly in polarized epithelial cells which are embedded in cellular sheets and influenced by cortical cues from their neighbours. In these cells, the polarized localization of the NuMA–LGN–Gαi complex is closely linked to the activity of cell polarity regulators such as Par3 or atypical PKC [34, 51, 52]. On the other hand, non-polarizing HeLa cells divide in the plane of the sheet even in the absence of neighbouring cells [9] indicating that cues derived from cell–cell junctions are not principally required for planar spindle alignment. In single mitotic HeLa cells the planar orientation of the mitotic spindle is regulated by cues derived from cell-substrate adhesion. β1 integrin-dependent signals activate Cdc42 and PI(3)K resulting in a gradient of PtdIns(3,4,5)P3 along the lateral cortex which is necessary for the midcortical accumulation of dynein and for the planar alignment of the mitotic spindle [53].
How PtdIns(3,4,5)P3 immobilizes dynein at the cortex is not entirely clear. Recent observations implicate NuMA as a potential linker between dynein and PtdIns(3,4,5)P3. NuMA can directly interact with the various phosphoinositides including PtdIns(4)P, PtdIns(4,5)P2 and PtdIns(3,4,5)P3 [54, 55]. The localization of NuMA and of dynein at the cortex increases during anaphase, and more importantly, LGN and Gαi are both dispensable for cortical dynein localization during anaphase [54–56] suggesting the existence of LGN–Gαi-independent mechanisms during anaphase. Members of the band 4.1 protein family as well as the phosphoinositides PtdIns(4)P and PtdIns(4,5)P2 have been identified as NuMA-interacting molecules and found to be required for cortical dynein localization during anaphase [54, 56]. Together, these observations suggest that the immobilization of dynein at the cortex is dynamically regulated during mitosis: at metaphase, dynein is recruited through NuMA by way of Gαi-immobilized LGN and PtdIns(3,4,5)P3, at anaphase dynein is recruited through NuMA by way of band 4.1 proteins and phosphoinositides.
It is noteworthy that Cdc42 in addition to its function as regulator of PI(3)K activity and PtdIns(3,4,5)P3 gradient formation also regulates the cortical actin cytoskeleton during mitosis. Cdc42 acts through its effector PAK2 and β-Pix, a guanine nucleotide exchange factor for Rac1 [40]. Importantly, all three components are required for planar spindle orientation, mitotic actin reorganization, and cortical dynein localization in HeLa cells [40].
The role of cell–cell adhesion in planar spindle orientation during tissue and organ development
A role for cell–cell adhesion in orienting the mitotic spindle has been identified by blastomere recombination experiments in C. elegans [10]. The early C. elegans embryo divides asymmetrically in a stereotypic manner. At the 4-cell stage of embryonic development the EMS blastomere is in contact with the P2 blastomere. When the EMS blastomere divides, the two daughter cells (E cell, MS cell) are always aligned along an axis that is determined by the site of cell contact of the EMS blastomere with the P2 cell. [10]. This indicated that the site of cell contact between the EMS cell and the P2 cell dictates the orientation of the mitotic spindle and suggested that cell–cell contacts provide a positional cue for the immobilization of the mitotic spindle.
Studies in Drosophila neuroepithelial cells provided further evidence for a role of intercellular junctions in regulating mitotic spindle orientation. Neuroepithelial cells are connected by an Apical Junctional Complex consisting of Adherens Junctions (AJ) and the Subapical Region (SAR) [57]. Neuroepithelial cells divide symmetrically in the plane of the cellular sheet. However, when adherens junctions are disrupted by deletion of the transmembrane protein Crumbs, the cells switch from symmetric, planar cell division to asymmetric, apico-basal cell division [58]. Thus, in neuroepithelial cells AJ provide cues that align the mitotic spindle in the plane of the sheet. How the spindle is immobilized at the lateral cell cortex is not entirely clear, but some evidence implicates a role for the microtubule-stabilizing protein adenomatous polyposis coli (APC) and the microtubule plus end-binding protein EB1, that has been implicated in the search-and-capture mechanism of spindle positioning in yeast [59, 60]. The two proteins interact with each other [61, 62], and both are lost from AJs in the absence of Crumbs or E-cadherin [58]. If they interact simultaneously with the microtubules and with the cortex is unknown. These observations, however, clearly indicated that adhesion receptors at cell–cell junctions provide information for the interaction of the astral microtubule with the lateral cortex of neuroepithelial cells.
During development of the heart, epicardial cells arise from a limited number of proepicardial cells which attach to the myocard, proliferate, and eventually form a single layer of epithelial cells covering the myocard, the epicardium. The proepicardial cells can divide in two orientations: either parallel or perpendicular to the basement membrane [3]. Upon parallel division both daughter cells remain in the monolayer allowing the epicardium to grow. After perpendicular cell division one of the two daughter cells delaminates from the epicardium, migrates into the subepicardial space, undergoes an epithelial-to-mesenchymal transition (EMT), and differentiates into coronary vascular smooth muscle cells and cardiac interstitial fibroblasts [63]. Epicardial cells are connected by N-cadherin-based AJ, and both β-catenin and Numb exist in a complex with N-cadherin. In the absence of β-catenin or Numb, AJ are disrupted and spindle orientation is randomized resulting in fewer epicardial cells entering the myocardium [3, 64]. The observation that spindle orientation does not switch from parallel to perpendicular in the absence of β-catenin or Numb but instead is randomized suggests that both parallel and perpendicular spindle orientation depend on signals derived from AJ. Regulation of perpendicular spindle orientation by AJ is most likely regulated through an effect on Par3, which is localized at the apical membrane of epicardial cells [3] and which regulates perpendicular spindle orientation in basal progenitor cells of the skin by assembling an apical Inscuteable (Insc)–LGN complex that recruits NuMA and dynein-dynactin to the apical membrane [2]. The nature of the signals that originate at AJ and how the spindles are attached to the lateral cortex during parallel cell divisions is unclear.
During embryogenesis the skin develops from a single layer of unspecified progenitors [65]. Similar to proepicardial cells in the developing heart, the basal progenitors in the skin undergo parallel cell divisions to expand the progenitor pool in order to keep up with the growing embryo, and perpendicular cell divisions to maintain the pool of progenitor cells and at the same time to generate differentiated cells which mediate the stratification of the skin [66]. The basal progenitor cells adhere to each other by E-cadherin-based AJ [67]. In the absence of the AJ protein α-catenin, Par3 and LGN are lost from the apical cortex, the dynein-interacting protein NuMA is randomly distributed, and spindle orientation is randomized [2]. Thus, AJs-derived signals regulate parallel and perpendicular spindle orientation in basal skin progenitor cells, and the mechanism might be similar as in proepicardial cells of the developing heart. Interestingly, loss of LGN or NuMA does not randomize spindle but results in predominant parallel spindle orientation [52, 68] suggesting that parallel cell division involves a mechanism to capture astral microtubules that does not involve the LGN–NuMA module. How the plus ends of the astral microtubules are associated with the cortex at AJ is unclear.
Adhesion receptors as regulators of spindle orientation
As indicated from many studies using single HeLa cells as model system for the analysis of mitotic spindle orientation, cells can orient the mitotic spindle in the plane of the substrate in the absence of cell–cell contacts indicating that cell–matrix adhesion can be sufficient for planar spindle orientation. However, in the organism epithelial cells are embedded in a sheet of cells and therefore receive signals from both cell–matrix contacts and cell–cell contacts. Studies in various model organisms such as C. elegans blastomeres, Drosophila neuroepithelial cells, or vertebrate epicardial cells and basal keratinocyte progenitor cells have implicated cell–cell junctions in mitotic spindle orientation [2, 3, 10, 58]. Recent observations in polarized epithelial cells cultured in vitro have identified several adhesion receptors and in particular the molecular mechanism underlying their role in mitotic spindle orientation
Classical cadherins
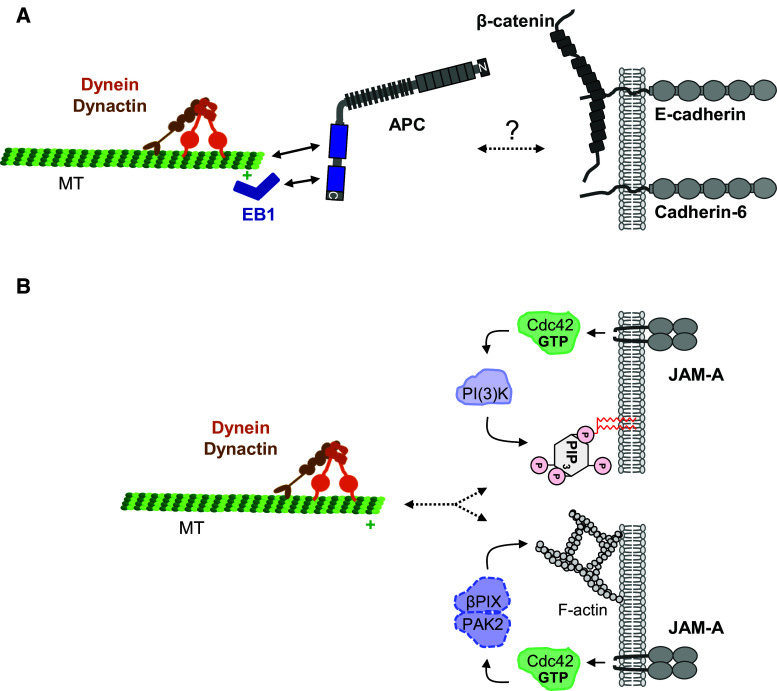
Cadherins are the major integral membrane determinants in homotypic cell–cell junctions of epithelial and endothelial cells where they localize to adherens junctions and assemble the cadherin–catenin complex [69]. Cadherins are highly conserved in evolution and exist in all metazoans [70]. A role for cadherins or cadherin-based adherens junctions in mitotic spindle orientation has been proposed in various cell types and organisms [2, 58, 71, 72]. If cadherins directly contribute to the immobilization of dynein by assembling a cytoplasmic protein complex or by triggering intracellular signalling cascade that generates docking sites for dynein has remained unclear. Some more direct evidence came from studies with cultured MDCK cells which were either depleted for E-cadherin and Cadherin-6, or which overexpressed a dominant-negative E-cadherin mutant. In both cases, the planar alignment of the mitotic spindle during anaphase is disturbed [73]. Surprisingly, the localization of regulators of spindle attachment to the cortex such as NuMA, LGN and members of the dynein complex is unchanged in cadherin knockdown cells, whereas the localization of adenomatous polyposis coli (APC) at adherens junctions is strongly disturbed. APC can interact with MT plus ends directly [74] as well as indirectly through the MT plus-end-binding protein EB1 [62]. Since APC can also bind β-catenin [75], it could link MTs to E-cadherin-based AJs. However, the interactions of β-catenin with APC and with E-cadherin are mutually exclusive [75], suggesting that APC is not directly associated with the E-cadherin–β-catenin complex. How APC localization at cortical sites during mitosis is regulated by E-cadherin and Cadherin-6 is presently unclear (Fig. 3a).
Fig. 3.
Adhesion molecules regulate spindle orientation. a E-cadherin and Cadherin-6 cooperate to regulate planar spindle orientation in epithelial cells through APC. Simultanous depletion of E-cadherin and Cadherin-6 disrupts the cortical localization of APC and results in misorientation of the spindle. How APC is connected with the cortex is not know. b JAM-A acts as a signaling molecule during mitosis. JAM-A has two signaling functions which both are most likely mediated by Cdc42. JAM-A activates PI(3)K and generates a cortical PtdIns(3,4,5)P3 gradient at the cortex. In addition JAM-A regulates the remodeling of the cortical F-actin cytoskeleton. The roles of PAK2 and βPIX (broken contour lines) downstream of Cdc42 have been demonstrated in HeLa cells and have not been proven for JAM-A
Junctional adhesion molecules
Recent evidence indicates a role for a member of the junctional adhesion molecule (JAM) family of adhesion receptors in planar spindle orientation in polarized epithelial cells. JAMs belong to the CD2 subgroup of the immunoglobulin (Ig) superfamily and are characterized by two Ig-like domains, a single transmembrane domain and a short cytoplasmic tail that frequently ends in a PDZ domain binding motif [76]. JAMs are expressed by a varietey of cell types and localize at cell–cell contacts by way of homophilic or heterophilic interactions. Their functions include the regulation of cell migration, tight junction and epithelial barrier formation, male germ cell development, the development of the peripheral nervous system, angiogenesis, and inflammation [77–82]. The adhesive properties of JAMs regulate their localization at cell–cell contacts and serve to recruit scaffolding proteins and assemble signaling complexes at intercellular junctions [77, 78, 83]. A new and unprecedented role for the JAM family member JAM-A in the regulation of planar orientation during symmetric cell division in polarized epithelial cells has recently been identified (Fig. 3b).
When polarized epithelial cells such as MDCK cells enter mitosis the cells round up but remain closely associated with the neighbouring interphase cells and retain cell–cell junctions [36, 38]. Similar to what has been described for single HeLa cells [84], the activity of Cdc42 gradually increases in MDCK cells after the onset of mitosis to reach a maximum level at metaphase [85]. In the absence of JAM-A the mitotic activation of Cdc42 is blunted. Consistent with a role of Cdc42 in regulating PI(3)K activation and PtdIns(3,4,5)P3 gradient formation as well as cortical actin reorganization in mitotic HeLa cells [40], both PI(3)K activation and actin reorganization are disturbed in MDCK cells lacking JAM-A expression [85]. As a consequence of JAM-A depletion cells have reduced levels of dynein at the lateral cortex, mitotic spindle orientation is shifted towards the apico-basal axis, and cells are unable to specify a single lumen when grown in 3-D [85], which depends on the planar orientation of the spindle [86]. Interestingly, the localization of NuMA is not affected in JAM-A-depleted cells, neither during metaphase nor during anaphase, suggesting that JAM-A does not act through the NuMA–LGN module to promote the cortical localization of dynein. Most likely, JAM-A’s ability to regulate the cortical actin cytoskeleton during mitosis is involved in the localization of dynein [85] (Fig. 3b). Noteworthy, E-cadherin/Cadherin-6 and JAM-A operate through different pathways in regulating planar spindle orientation indicating that epithelial cells make use of several junction-derived signalling pathways to align the mitotic spindle.
Plexin–semaphorin interactions
Plexins are a family of transmembrane proteins which consist of nine members in vertebrates [87] and which act as cell surface receptors for semaphorins [88]. The plexin–semaphorin system mediates intercellular signalling in various cell types and regulates processes like proliferation, cell migration and cell differentiation in the nervous system, in the immune system, during vascular development, or during bone development [87]. Intracellular signalling is induced upon binding of dimeric semaphorins to monomeric plexins, which triggers a transition from the autoinhibited, inactive plexin monomer to the active dimer. The dimerization results in the activation of the GTPase activating protein (GAP) domain present in the cytoplasmic domain, which regulates the activity of Ras and Rap family small GTPases by promoting GTP hydrolysis [89]. Several plexins contain a PDZ domain-binding motif at their COOH-terminus and interact with the PDZ domain-containing RhoA GEFs PRG (PDZ-RhoGEF) and LARG (leukemia-associated RhoGEF) suggesting a role in regulating the activity of RhoA [89]. Among the key events downstream of plexins activation are the regulation of the actin cytoskeleton and the regulation of cell–cell and cell–matrix adhesion [87]. Recently, a role in orienting the mitotic spindle has been identified for Plexin-B2.
Tubular epithelial cells of the adult kidney are mitotically quiescent under steady-state conditions, but new proliferation can be induced by ischemia/reperfusion injury [90]. Cells surviving the injury start to proliferate and divide in the plane of the epithelium to restore epithelial tubes. In the absence of Plexin-B2, the orientation of the mitotic spindle is shifted towards divisions perpendicular of the plane of the epithelium and the cells intrude into the tubular lumen, resulting in a multilayered epithelium and eventually occlusion of the kidney tubules [91]. Consistent with these observations, 3-D-grown MDCK cells with silenced Plexin-B2 expression fail to orient their mitotic spindle in the plane of the monolayer and develop cysts containing multiple lumens. The regulation of planar spindle orientation by Plexin-B2 is mediated by Cdc42 which acts downstream of Plexin-B2-mediated activation of R-Ras [91]. Plexin-B2 thus provides a second example of cell adhesion molecules which by activating Cdc42 regulate the correct orientation of the mitotic spindle during planar cell division. The activation of Cdc42 suggests that Plexin-B2 might contribute to the immobilization of astral microtubules at the lateral cell cortex through PtdIns(3,4,5)P3 gradient formation and/or cortical actin cytsokelelton formation [40].
Anthrax receptors
Anthrax toxin receptor 2 (Antxr2)/Capillary morphogenesis protein 2 (CMG2) is a type I transmembrane protein with a single membrane-spanning domain and an extracellular domain that is highly related to the von Willebrand factor type A (vWA) domain with a metal ion-dependent adhesion site (MIDAS) motif [92]. vWA domains often serve as protein–protein interaction sites in cell adhesion proteins such as integrins. Natural ligands of the extracellular domain of ANTXR2 are various extracellular matrix proteins such as collagen IV, laminin, and fibronectin [93]. In human endothelial cells, Antxr2 expression is upregulated when cells are grown in a 3D collagen matrix, suggesting a role in angiogenesis [94]. Antxr2 is best known for its role in mediating the entry of anthrax toxin into host cells [93], its physiological role is largely unknown. Recent evidence indicates a role for Antxr2 in controlling the positioning of the mitotic spindle in epiblast cells of the zebrafish embryo.
During zebrafish gastrulation, epiblast cells divide symmetrically along the animal-vegetal (A–V), i.e. antero-posterior axis in response to planar cell polarity signals in a NuMA–dynein-dependent pathway [95, 96]. Antxr2 co-localizes with F-actin in an asymmetrically localized cap that builds up during mitosis and that aligns along the antero-posterior axis [97]. This cap does not depend on Antxr2 but it serves to recruit Antxr2 to one pole that is aligned with the A-V axis. Interestingly, Antxr2 exists in a complex with GTP-loaded RhoA, which activates the formin family member Dia2 during zebrafish gastrulation [98]. All three components, Antxr2, RhoA and Dia2, are necessary for the alignment of the mitotic spindle with the A-V axis [97]. Thus, after its recruitment to the F-actin-enriched caps during mitosis, Antxr2 provides spatial cues for the localization of RhoA and localized activation of Dia2 to mediate the formation of a cortical F-actin cytoskeleton which might serve to capture MT plus-ends and allow dynein to generate pulling forces towards the centosomes.
Concluding remarks
To align the mitotic spindle in the plane of the cellular sheet, epithelial cells integrate a variety of signals from both intrinsic and extrinsic sources. The identification of specific cell–cell adhesion molecules as regulators of mitotic spindle orientation in epithelial cells and the elucidation of the signalling pathways triggered by these adhesion molecules has added new information on the complex regulatory network underlying spindle alignment. Several conclusions can be drawn from these recent observations. First, cell–cell contacts are not just permissive but are instructive for planar spindle orientation in epithelial cells. Second, different adhesion molecules contribute to the immobilization of the astral MTs at the cortex through different signals and mechanisms. For example, E-cadherin and Cadherin-6 regulate the cortical localization of the MT plus end binding protein APC [73] whereas JAM-A regulates the cortical localization of the MT-associated dynein motor protein complex [85]. Third, Cdc42 is a critical regulator of planar spindle orientation downstream of different adhesion receptors such as JAM-A and Plexin-B2 [85, 91]. Mitotic activation of Cdc42 has previously been observed in single, non-polarized HeLa cells in the absence of intercellular junctions, probably induced by integrin-mediated cell-substrate adhesion [9, 40, 84], which indicates that cell–cell junctions are not principally required for Cdc42 activation and planar spindle orientation. The mitotic activation of Cdc42 downstream of adhesion receptors in polarized epithelial cells [85, 91] therefore suggests that planar spindle orientation is more vigorously controlled in epithelial cells embedded in tissues, which most likely serves to prevent cell delamination from the cellular sheet which could result in tumor development. New questions arise from the new findings. For example, which signals trigger the cell adhesion molecules to transiently adopt a new function during mitosis? And which signals restrict these functions to the specific localizations along the lateral cell cortex, i.e. the sites of MT–cortex interaction? Is the specific localization and/or activity of adhesion molecules during mitosis influenced by intrinsic signals such as the spindle poles or the chromosomes? A chromosome-derived gradient of RanGTP has been found to regulate the localization of NuMA–LGN at the lateral cortex [5, 6, 99]. Beyond these questions, it is still unclear if different adhesion molecules operate consecutively during mitosis, as one might expect from the different requirements of NuMA to interact with the cortex during metaphase and during anaphase [54–56]. The role of cell adhesion molecules in the regulation of mitotic spindle orientation is just beginning to be uncovered.
Acknowledgments
We would like to thank all members of the Institute-associated research group “Cell adhesion and cell polarity” for helpful discussions. We would also like to thank Volker Gerke for continuous support. This work was supported by grants from the Deutsche Forschungsgemeinschaft (EB 160/4-1 and -/4-2) and from the Medical Faculty of the University Münster (IZKF Eb2/028/09 and Eb2/020/14).
Abbreviations
- AJ
Adherens junctions
- Antxr2
Anthrax toxin receptor 2
- APC
Adenomatous polyposis coli
- Arp
Actin-related protein
- βPIX
PAK-interacting exchange factor β
- Cdc42
Cell division cycle 42
- Dia1
Diaphanous-related formin 1
- EB1
End-binding protein 1
- Ect2
Epithelial cell transforming 2
- ERM
Ezrin, radixin, moesin
- ERMAD
ERM-association domain
- FERM
4.1 protein and ERM
- GAP
GTPase-activating protein
- GEF
Guanine nucleotide exchange factor
- JAM
Junctional adhesion molecule
- LGN
Leu-Gly-Asn repeat-enriched protein
- MDCK
Madin–Darby canine kidney
- MT
Microtubule
- NuMA
Nuclear mitotic apparatus
- PAK2
p21-activated kinase 2
- Par
Partitioning defective
- PDZ
PSD95–Discs large–ZO-1
- PI(3)K
Phosphoinositide 3-kinase
- PtdIns
Phosphatidylinositol
- RCC1
Regulator of chromatin condensation 1
- SLK
Sterile 20-like kinase
- TJ
Tight junctions
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest.
References
- 1.Morin X, Bellaiche Y. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev Cell. 2011;21(1):102–119. doi: 10.1016/j.devcel.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Lechler T, Fuchs E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 2005;437(7056):275–280. doi: 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu M, Smith CL, Hall JA, Lee I, Luby-Phelps K, Tallquist MD. Epicardial spindle orientation controls cell entry into the myocardium. Dev Cell. 2010;19(1):114–125. doi: 10.1016/j.devcel.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minc N, Burgess D, Chang F. Influence of cell geometry on division-plane positioning. Cell. 2011;144(3):414–426. doi: 10.1016/j.cell.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiyomitsu T, Cheeseman IM. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol. 2012;14(3):311–317. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird SL, Heald R, Weis K. RanGTP and CLASP1 cooperate to position the mitotic spindle. Mol Biol Cell. 2013;24(16):2506–2514. doi: 10.1091/mbc.E13-03-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J, Mlodzik M. The frizzled extracellular domain is a ligand for Van Gogh/Stbm during nonautonomous planar cell polarity signaling. Dev Cell. 2008;15(3):462–469. doi: 10.1016/j.devcel.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thery M, Racine V, Pepin A, Piel M, Chen Y, Sibarita JB, Bornens M. The extracellular matrix guides the orientation of the cell division axis. Nat Cell Biol. 2005;7(10):947–953. doi: 10.1038/ncb1307. [DOI] [PubMed] [Google Scholar]
- 9.Toyoshima F, Nishida E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 2007;26(6):1487–1498. doi: 10.1038/sj.emboj.7601599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein B. Cell contacts orient some cell division axes in the Caenorhabditis elegans embryo. J Cell Biol. 1995;129(4):1071–1080. doi: 10.1083/jcb.129.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dujardin DL, Vallee RB. Dynein at the cortex. Curr Opin Cell Biol. 2002;14(1):44–49. doi: 10.1016/S0955-0674(01)00292-7. [DOI] [PubMed] [Google Scholar]
- 12.Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10(12):854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Busson S, Dujardin D, Moreau A, Dompierre J, De Mey JR. Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr Biol. 1998;8(9):541–544. doi: 10.1016/S0960-9822(98)70208-8. [DOI] [PubMed] [Google Scholar]
- 14.Moore JK, Cooper JA. Coordinating mitosis with cell polarity: molecular motors at the cell cortex. Semin Cell Dev Biol. 2010;21(3):283–289. doi: 10.1016/j.semcdb.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markus SM, Lee WL. Microtubule-dependent path to the cell cortex for cytoplasmic dynein in mitotic spindle orientation. Bioarchitecture. 2011;1(5):209–215. doi: 10.4161/bioa.18103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markus SM, Lee WL. Regulated offloading of cytoplasmic dynein from microtubule plus ends to the cortex. Dev Cell. 2011;20(5):639–651. doi: 10.1016/j.devcel.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grill SW, Howard J, Schaffer E, Stelzer EH, Hyman AA. The distribution of active force generators controls mitotic spindle position. Science. 2003;301(5632):518–521. doi: 10.1126/science.1086560. [DOI] [PubMed] [Google Scholar]
- 18.Laan L, Pavin N, Husson J, Romet-Lemonne G, van Duijn M, Lopez MP, Vale RD, Julicher F, Reck-Peterson SL, Dogterom M. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 2012;148(3):502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevermann L, Liakopoulos D. Molecular mechanisms in spindle positioning: structures and new concepts. Curr Opin Cell Biol. 2012;24(6):816–824. doi: 10.1016/j.ceb.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Kotak S, Gonczy P. Mechanisms of spindle positioning: cortical force generators in the limelight. Curr Opin Cell Biol. 2013;25(6):741–748. doi: 10.1016/j.ceb.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 21.Compton DA, Szilak I, Cleveland DW. Primary structure of NuMA, an intranuclear protein that defines a novel pathway for segregation of proteins at mitosis. J Cell Biol. 1992;116(6):1395–1408. doi: 10.1083/jcb.116.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotak S, Busso C, Gonczy P. Cortical dynein is critical for proper spindle positioning in human cells. J Cell Biol. 2012;199(1):97–110. doi: 10.1083/jcb.201203166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du Q, Stukenberg PT, Macara IG. A mammalian partner of inscuteable binds NuMA and regulates mitotic spindle organization. Nat Cell Biol. 2001;3(12):1069–1075. doi: 10.1038/ncb1201-1069. [DOI] [PubMed] [Google Scholar]
- 24.Du Q, Macara IG. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119(4):503–516. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 25.Zheng Z, Zhu H, Wan Q, Liu J, Xiao Z, Siderovski DP, Du Q. LGN regulates mitotic spindle orientation during epithelial morphogenesis. J Cell Biol. 2010;189(2):275–288. doi: 10.1083/jcb.200910021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joberty G, Petersen C, Gao L, Macara IG. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol. 2000;2(8):531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- 27.Lin D, Edwards AS, Fawcett JP, Mbamalu G, Scott JD, Pawson T. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat Cell Biol. 2000;2(8):540–547. doi: 10.1038/35019592. [DOI] [PubMed] [Google Scholar]
- 28.Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol. 2005;7(3):262–269. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- 29.Hirose T, Izumi Y, Nagashima Y, Tamai-Nagai Y, Kurihara H, Sakai T, Suzuki Y, Yamanaka T, Suzuki A, Mizuno K, Ohno S. Involvement of ASIP/PAR-3 in the promotion of epithelial tight junction formation. J Cell Sci. 2002;115(Pt 12):2485–2495. doi: 10.1242/jcs.115.12.2485. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki A, Ishiyama C, Hashiba K, Shimizu M, Ebnet K, Ohno S. aPKC kinase activity is required for the asymmetric differentiation of the premature junctional complex during epithelial cell polarization. J Cell Sci. 2002;115(Pt 18):3565–3573. doi: 10.1242/jcs.00032. [DOI] [PubMed] [Google Scholar]
- 31.Nagai-Tamai Y, Mizuno K, Hirose T, Suzuki A, Ohno S. Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells. 2002;7(11):1161–1171. doi: 10.1046/j.1365-2443.2002.00590.x. [DOI] [PubMed] [Google Scholar]
- 32.Qin Y, Meisen WH, Hao Y, Macara IG. Tuba, a Cdc42 GEF, is required for polarized spindle orientation during epithelial cyst formation. J Cell Biol. 2010;189(4):661–669. doi: 10.1083/jcb.201002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durgan J, Kaji N, Jin D, Hall A. Par6B and atypical PKC (aPKC) regulate mitotic spindle orientation during epithelial morphogenesis. J Biol Chem. 2011;286(14):12461–12471. doi: 10.1074/jbc.M110.174235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hao Y, Du Q, Chen X, Zheng Z, Balsbaugh JL, Maitra S, Shabanowitz J, Hunt DF, Macara IG. Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical pins. Curr Biol. 2010;20(20):1809–1818. doi: 10.1016/j.cub.2010.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hao Y, Macara IG. Regulation of chromatin binding by a conformational switch in the tail of the Ran exchange factor RCC1. J Cell Biol. 2008;182(5):827–836. doi: 10.1083/jcb.200803110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinsch S, Karsenti E. Orientation of spindle axis and distribution of plasma membrane proteins during cell division in polarized MDCKII cells. J Cell Biol. 1994;126(6):1509–1526. doi: 10.1083/jcb.126.6.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenblatt J, Cramer LP, Baum B, McGee KM. Myosin II-dependent cortical movement is required for centrosome separation and positioning during mitotic spindle assembly. Cell. 2004;117(3):361–372. doi: 10.1016/S0092-8674(04)00341-1. [DOI] [PubMed] [Google Scholar]
- 38.Baker J, Garrod D. Epithelial cells retain junctions during mitosis. J Cell Sci. 1993;104(Pt 2):415–425. doi: 10.1242/jcs.104.2.415. [DOI] [PubMed] [Google Scholar]
- 39.Redemann S, Pecreaux J, Goehring NW, Khairy K, Stelzer EH, Hyman AA, Howard J. Membrane invaginations reveal cortical sites that pull on mitotic spindles in one-cell C. elegans embryos. PLoS ONE. 2010;5(8):e12301. doi: 10.1371/journal.pone.0012301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitsushima M, Toyoshima F, Nishida E. Dual role of Cdc42 in spindle orientation control of adherent cells. Mol Cell Biol. 2009;29(10):2816–2827. doi: 10.1128/MCB.01713-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11(4):237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bovellan M, Romeo Y, Biro M, Boden A, Chugh P, Yonis A, Vaghela M, Fritzsche M, Moulding D, Thorogate R, Jegou A, Thrasher AJ, Romet-Lemonne G, Roux PP, Paluch EK, Charras G. Cellular control of cortical actin nucleation. Curr Biol. 2014;24(14):1628–1635. doi: 10.1016/j.cub.2014.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosa A, Vlassaks E, Pichaud F, Baum B. Ect2/Pbl acts via Rho and polarity proteins to direct the assembly of an isotropic actomyosin cortex upon mitotic entry. Dev Cell. 2015;32(5):604–616. doi: 10.1016/j.devcel.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthews HK, Delabre U, Rohn JL, Guck J, Kunda P, Baum B. Changes in Ect2 localization couple actomyosin-dependent cell shape changes to mitotic progression. Dev Cell. 2012;23(2):371–383. doi: 10.1016/j.devcel.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otomo T, Otomo C, Tomchick DR, Machius M, Rosen MK. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol Cell. 2005;18(3):273–281. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 46.Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11(4):276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carreno S, Kouranti I, Glusman ES, Fuller MT, Echard A, Payre F. Moesin and its activating kinase Slik are required for cortical stability and microtubule organization in mitotic cells. J Cell Biol. 2008;180(4):739–746. doi: 10.1083/jcb.200709161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kunda P, Pelling AE, Liu T, Baum B. Moesin controls cortical rigidity, cell rounding, and spindle morphogenesis during mitosis. Curr Biol. 2008;18(2):91–101. doi: 10.1016/j.cub.2007.12.051. [DOI] [PubMed] [Google Scholar]
- 49.Machicoane M, de Frutos CA, Fink J, Rocancourt M, Lombardi Y, Garel S, Piel M, Echard A. SLK-dependent activation of ERMs controls LGN-NuMA localization and spindle orientation. J Cell Biol. 2014;205(6):791–799. doi: 10.1083/jcb.201401049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Solinet S, Mahmud K, Stewman SF, Ben El Kadhi K, Decelle B, Talje L, Ma A, Kwok BH, Carreno S. The actin-binding ERM protein Moesin binds to and stabilizes microtubules at the cell cortex. J Cell Biol. 2013;202(2):251–260. doi: 10.1083/jcb.201304052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu J, Wen W, Zheng Z, Shang Y, Wei Z, Xiao Z, Pan Z, Du Q, Wang W, Zhang M. LGN/mInsc and LGN/NuMA complex structures suggest distinct functions in asymmetric cell division for the Par3/mInsc/LGN and Galphai/LGN/NuMA pathways. Mol Cell. 2011;43(3):418–431. doi: 10.1016/j.molcel.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams SE, Ratliff LA, Postiglione MP, Knoblich JA, Fuchs E. Par3-mInsc and Galphai3 cooperate to promote oriented epidermal cell divisions through LGN. Nat Cell Biol. 2014;16(8):758–769. doi: 10.1038/ncb3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toyoshima F, Matsumura S, Morimoto H, Mitsushima M, Nishida E. PtdIns(3,4,5)P3 regulates spindle orientation in adherent cells. Dev Cell. 2007;13(6):796–811. doi: 10.1016/j.devcel.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 54.Kotak S, Busso C, Gonczy P. NuMA interacts with phosphoinositides and links the mitotic spindle with the plasma membrane. EMBO J. 2014;33(16):1815–1830. doi: 10.15252/embj.201488147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng Z, Wan Q, Meixiong G, Du Q. Cell cycle-regulated membrane binding of NuMA contributes to efficient anaphase chromosome separation. Mol Biol Cell. 2014;25(5):606–619. doi: 10.1091/mbc.E13-08-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kiyomitsu T, Cheeseman IM. Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell. 2013;154(2):391–402. doi: 10.1016/j.cell.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tepass U, Tanentzapf G, Ward R, Fehon R. Epithelial cell polarity and cell junctions in Drosophila. Annu Rev Genet. 2001;35:747–784. doi: 10.1146/annurev.genet.35.102401.091415. [DOI] [PubMed] [Google Scholar]
- 58.Lu B, Roegiers F, Jan LY, Jan YN. Adherens junctions inhibit asymmetric division in the Drosophila epithelium. Nature. 2001;409(6819):522–525. doi: 10.1038/35054077. [DOI] [PubMed] [Google Scholar]
- 59.McCartney BM, Nathke IS. Cell regulation by the Apc protein Apc as master regulator of epithelia. Curr Opin Cell Biol. 2008;20(2):186–193. doi: 10.1016/j.ceb.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 60.Lee L, Tirnauer JS, Li J, Schuyler SC, Liu JY, Pellman D. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 2000;287(5461):2260–2262. doi: 10.1126/science.287.5461.2260. [DOI] [PubMed] [Google Scholar]
- 61.Su LK, Burrell M, Hill DE, Gyuris J, Brent R, Wiltshire R, Trent J, Vogelstein B, Kinzler KW. APC binds to the novel protein EB1. Cancer Res. 1995;55(14):2972–2977. [PubMed] [Google Scholar]
- 62.Green RA, Wollman R, Kaplan KB. APC and EB1 function together in mitosis to regulate spindle dynamics and chromosome alignment. Mol Biol Cell. 2005;16(10):4609–4622. doi: 10.1091/mbc.E05-03-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wessels A, Perez-Pomares JM. The epicardium and epicardially derived cells (EPDCs) as cardiac stem cells. Anat Rec A Discov Mol Cell Evol Biol. 2004;276(1):43–57. doi: 10.1002/ar.a.10129. [DOI] [PubMed] [Google Scholar]
- 64.Zamora M, Manner J, Ruiz-Lozano P. Epicardium-derived progenitor cells require beta-catenin for coronary artery formation. Proc Natl Acad Sci USA. 2007;104(46):18109–18114. doi: 10.1073/pnas.0702415104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsu YC, Li L, Fuchs E. Emerging interactions between skin stem cells and their niches. Nat Med. 2014;20(8):847–856. doi: 10.1038/nm.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kulukian A, Fuchs E. Spindle orientation and epidermal morphogenesis. Philos Trans R Soc Lond B Biol Sci. 2013;368(1629):20130016. doi: 10.1098/rstb.2013.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tinkle CL, Pasolli HA, Stokes N, Fuchs E. New insights into cadherin function in epidermal sheet formation and maintenance of tissue integrity. Proc Natl Acad Sci USA. 2008;105(40):15405–15410. doi: 10.1073/pnas.0807374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Williams SE, Beronja S, Pasolli HA, Fuchs E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature. 2011;470(7334):353–358. doi: 10.1038/nature09793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pokutta S, Weis WI. Structure and mechanism of cadherins and catenins in cell-cell contacts. Annu Rev Cell Dev Biol. 2007;23:237–261. doi: 10.1146/annurev.cellbio.22.010305.104241. [DOI] [PubMed] [Google Scholar]
- 70.Nichols SA, Roberts BW, Richter DJ, Fairclough SR, King N. Origin of metazoan cadherin diversity and the antiquity of the classical cadherin/beta-catenin complex. Proc Natl Acad Sci USA. 2012;109(32):13046–13051. doi: 10.1073/pnas.1120685109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Inaba M, Yuan H, Salzmann V, Fuller MT, Yamashita YM. E-cadherin is required for centrosome and spindle orientation in Drosophila male germline stem cells. PLoS ONE. 2010;5(8):e12473. doi: 10.1371/journal.pone.0012473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zigman M, le Trinh A, Fraser SE, Moens CB. Zebrafish neural tube morphogenesis requires Scribble-dependent oriented cell divisions. Curr Biol. 2011;21(1):79–86. doi: 10.1016/j.cub.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.den Elzen N, Buttery CV, Maddugoda MP, Ren G, Yap AS. Cadherin adhesion receptors orient the mitotic spindle during symmetric cell division in mammalian epithelia. Mol Biol Cell. 2009;20(16):3740–3750. doi: 10.1091/mbc.E09-01-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mogensen MM, Tucker JB, Mackie JB, Prescott AR, Nathke IS. The adenomatous polyposis coli protein unambiguously localizes to microtubule plus ends and is involved in establishing parallel arrays of microtubule bundles in highly polarized epithelial cells. J Cell Biol. 2002;157(6):1041–1048. doi: 10.1083/jcb.200203001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hulsken J, Birchmeier W, Behrens J. E-cadherin and APC compete for the interaction with beta-catenin and the cytoskeleton. J Cell Biol. 1994;127(6 Pt 2):2061–2069. doi: 10.1083/jcb.127.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ebnet K, Suzuki A, Ohno S, Vestweber D. Junctional adhesion molecules (JAMs): More molecules with dual functions? J Cell Sci. 2004;117(1):19–29. doi: 10.1242/jcs.00930. [DOI] [PubMed] [Google Scholar]
- 77.Severson EA, Lee WY, Capaldo CT, Nusrat A, Parkos CA. Junctional adhesion molecule A interacts with Afadin and PDZ-GEF2 to activate Rap1A, regulate beta1 integrin levels, and enhance cell migration. Mol Biol Cell. 2009;20(7):1916–1925. doi: 10.1091/mbc.E08-10-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Iden S, Misselwitz S, Peddibhotla SS, Tuncay H, Rehder D, Gerke V, Robenek H, Suzuki A, Ebnet K. aPKC phosphorylates JAM-A at Ser285 to promote cell contact maturation and tight junction formation. J Cell Biol. 2012;196(5):623–639. doi: 10.1083/jcb.201104143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gliki G, Ebnet K, Aurrand-Lions M, Imhof BA, Adams RH. Spermatid differentiation requires the assembly of a cell polarity complex downstream of junctional adhesion molecule-C. Nature. 2004;431(7006):320–324. doi: 10.1038/nature02877. [DOI] [PubMed] [Google Scholar]
- 80.Scheiermann C, Meda P, Aurrand-Lions M, Madani R, Yiangou Y, Coffey P, Salt TE, Ducrest-Gay D, Caille D, Howell O, Reynolds R, Lobrinus A, Adams RH, Yu AS, Anand P, Imhof BA, Nourshargh S. Expression and function of junctional adhesion molecule-C in myelinated peripheral nerves. Science. 2007;318(5855):1472–1475. doi: 10.1126/science.1149276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cooke VG, Naik MU, Naik UP. Fibroblast growth factor-2 failed to induce angiogenesis in junctional adhesion molecule-A-deficient mice. Arterioscler Thromb Vasc Biol. 2006;26(9):2005–2011. doi: 10.1161/01.ATV.0000234923.79173.99. [DOI] [PubMed] [Google Scholar]
- 82.Weber C, Fraemohs L, Dejana E. The role of junctional adhesion molecules in vascular inflammation. Nat Rev Immunol. 2007;7(6):467–477. doi: 10.1038/nri2096. [DOI] [PubMed] [Google Scholar]
- 83.Naik MU, Caplan JL, Naik UP. Junctional adhesion molecule-A suppresses platelet integrin alphaIIbbeta3 signaling by recruiting Csk to the integrin-c-Src complex. Blood. 2014;123(9):1393–1402. doi: 10.1182/blood-2013-04-496232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oceguera-Yanez F, Kimura K, Yasuda S, Higashida C, Kitamura T, Hiraoka Y, Haraguchi T, Narumiya S. Ect2 and MgcRacGAP regulate the activation and function of Cdc42 in mitosis. J Cell Biol. 2005;168(2):221–232. doi: 10.1083/jcb.200408085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tuncay H, Brinkmann BF, Steinbacher T, Schurmann A, Gerke V, Iden S, Ebnet K. JAM-A regulates cortical dynein localization through Cdc42 to control planar spindle orientation during mitosis. Nat Commun. 2015;6:8128. doi: 10.1038/ncomms9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jaffe AB, Kaji N, Durgan J, Hall A. Cdc42 controls spindle orientation to position the apical surface during epithelial morphogenesis. The Journal of cell biology. 2008;183(4):625–633. doi: 10.1083/jcb.200807121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jongbloets BC, Pasterkamp RJ. Semaphorin signalling during development. Development. 2014;141(17):3292–3297. doi: 10.1242/dev.105544. [DOI] [PubMed] [Google Scholar]
- 88.Tamagnone L, Artigiani S, Chen H, He Z, Ming GI, Song H, Chedotal A, Winberg ML, Goodman CS, Poo M, Tessier-Lavigne M, Comoglio PM. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell. 1999;99(1):71–80. doi: 10.1016/S0092-8674(00)80063-X. [DOI] [PubMed] [Google Scholar]
- 89.Pascoe HG, Wang Y, Zhang X. Structural mechanisms of plexin signaling. Prog Biophys Mol Biol. 2015;118(3):161–168. doi: 10.1016/j.pbiomolbio.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bonventre JV, Yang L. Cellular pathophysiology of ischemic acute kidney injury. J Clin Investig. 2011;121(11):4210–4221. doi: 10.1172/JCI45161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xia J, Swiercz JM, Banon-Rodriguez I, Matkovic I, Federico G, Sun T, Franz T, Brakebusch CH, Kumanogoh A, Friedel RH, Martin-Belmonte F, Grone HJ, Offermanns S, Worzfeld T. Semaphorin–plexin signalling controls mitotic spindle orientation during epithelial morphogenesis and repair. Dev Cell. 2015;33(3):299–313. doi: 10.1016/j.devcel.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 92.Lacy DB, Wigelsworth DJ, Scobie HM, Young JA, Collier RJ. Crystal structure of the von Willebrand factor A domain of human capillary morphogenesis protein 2: an anthrax toxin receptor. Proc Natl Acad Sci USA. 2004;101(17):6367–6372. doi: 10.1073/pnas.0401506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu S, Moayeri M, Leppla SH. Anthrax lethal and edema toxins in anthrax pathogenesis. Trends Microbiol. 2014;22(6):317–325. doi: 10.1016/j.tim.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bell SE, Mavila A, Salazar R, Bayless KJ, Kanagala S, Maxwell SA, Davis GE. Differential gene expression during capillary morphogenesis in 3D collagen matrices: regulated expression of genes involved in basement membrane matrix assembly, cell cycle progression, cellular differentiation and G-protein signaling. J Cell Sci. 2001;114(Pt 15):2755–2773. doi: 10.1242/jcs.114.15.2755. [DOI] [PubMed] [Google Scholar]
- 95.Gong Y, Mo C, Fraser SE. Planar cell polarity signalling controls cell division orientation during zebrafish gastrulation. Nature. 2004;430(7000):689–693. doi: 10.1038/nature02796. [DOI] [PubMed] [Google Scholar]
- 96.Segalen M, Johnston CA, Martin CA, Dumortier JG, Prehoda KE, David NB, Doe CQ, Bellaiche Y. The Fz-Dsh planar cell polarity pathway induces oriented cell division via Mud/NuMA in Drosophila and zebrafish. Dev Cell. 2010;19(5):740–752. doi: 10.1016/j.devcel.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Castanon I, Abrami L, Holtzer L, Heisenberg CP, van der Goot FG, Gonzalez-Gaitan M. Anthrax toxin receptor 2a controls mitotic spindle positioning. Nat Cell Biol. 2013;15(1):28–39. doi: 10.1038/ncb2632. [DOI] [PubMed] [Google Scholar]
- 98.Lai SL, Chan TH, Lin MJ, Huang WP, Lou SW, Lee SJ. Diaphanous-related formin 2 and profilin I are required for gastrulation cell movements. PLoS ONE. 2008;3(10):e3439. doi: 10.1371/journal.pone.0003439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kalab P, Heald R. The RanGTP gradient—a GPS for the mitotic spindle. J Cell Sci. 2008;121(Pt 10):1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]