Abstract
Upon splicing, introns are rapidly degraded. Hence, RNAs derived from introns are commonly deemed as junk sequences. However, the discoveries of intronic-derived small nucleolar RNAs (snoRNAs), small Cajal body associated RNAs (scaRNAs) and microRNAs (miRNAs) suggested otherwise. These non-coding RNAs are shown to play various roles in gene regulation. In this review, we highlight another class of intron-derived RNAs known as stable intronic sequence RNAs (sisRNAs). sisRNAs have been observed since the 1980 s; however, we are only beginning to understand their biological significance. Recent studies have shown or suggested that sisRNAs regulate their own host’s gene expression, function as molecular sinks or sponges, and regulate protein translation. We propose that sisRNAs function as an additional layer of gene regulation in the cells.
Keywords: Non-coding RNA, sisRNA, Intron
Introduction
The discoveries of several classes of intron-derived RNAs suggest that introns are not merely junk. Instead, they play an important role in gene regulation. In this review, we will first briefly look at intronic RNA history, as well as the currently known classes of intron-derived RNAs. We will then look at a novel class of intron-derived RNA known as stable intronic sequence RNAs (sisRNAs) and review their known or suggested biological functions in the cell.
Introns were discovered in 1977, independently in the laboratories of Philip Sharp and Richard Roberts [1, 2]. Using adenoviruses, both groups observed that mRNAs transcribed from the adenovirus DNA were not exact copies of one another. Instead, there were several stretches of sequences found in the DNA, which were absent in the resulting mRNA. Soon after, work from several other laboratories suggested that such an arrangement was also present in the eukaryotic genome [3–10]. In 1978, Walter Gilbert famously named these stretches of sequences, which split the gene into pieces, as ‘introns’ (short for intragenic regions), whereas the regions that are expressed were termed as ‘exons’ [11]. Most eukaryotic genes are initially expressed as precursor messenger RNAs (pre-mRNAs), containing both introns and exons. The introns are then spliced out, leaving behind the exons that are ligated together to form the mature mRNA.
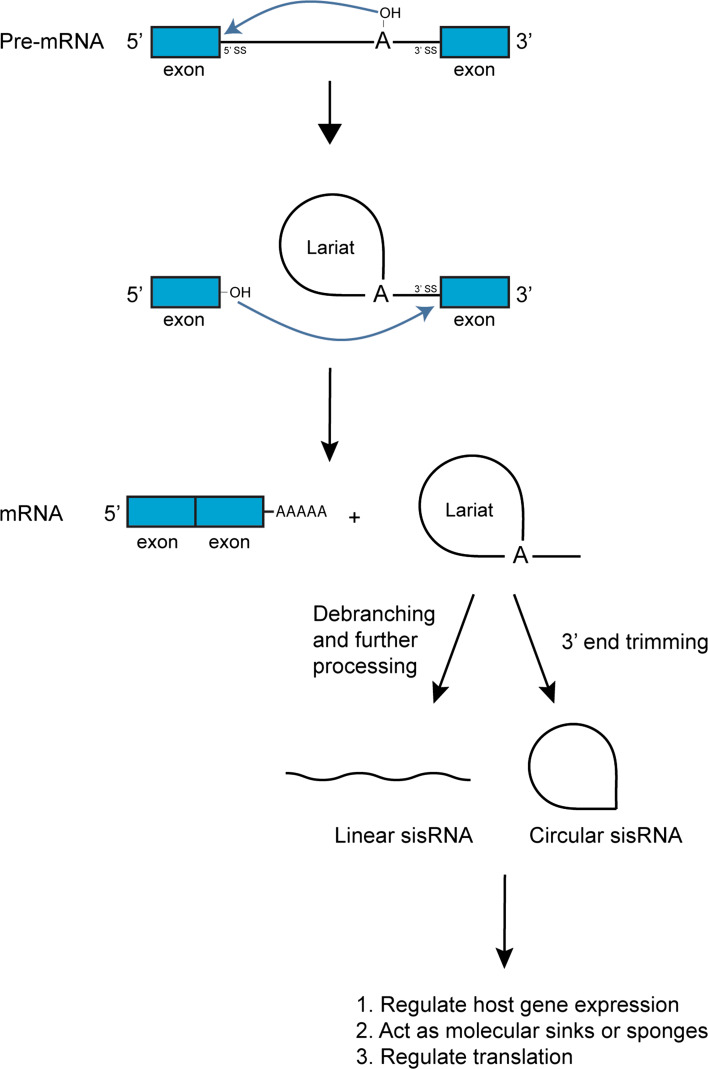
Introns can be spliced out either through self-splicing or via the spliceosomal machinery. Self-splicing introns are catalytic RNAs or ribozymes, which have the ability to catalyze their own splicing. They are divided into three classes: Group I, II and III introns. The three groups of self-splicing introns differ mainly in the mechanisms utilized to carry out the splicing [12–14]. In this review, we will focus on introns spliced by the spliceosomes, because they account for the splicing of majority of eukaryotic introns [15]. The spliceosome is a multi-subunit ribonucleoprotein (RNP) complex made up of many different proteins and five small nuclear RNAs (snRNAs). The five snRNAs are U1, U2, U4, U5, and U6 [16–19]. The spliceosome is a dynamic complex with its different components changing throughout the whole splicing process. First, the U1 small nuclear RNP (snRNP) recognizes and binds to the 5′ splice site, while the U2 snRNP associates with the branch site. Subsequently, the U4, U5 and U6 snRNPs bind to the intron, after which RNA–RNA and RNA–protein interactions of the spliceosome undergo major structural rearrangements, destabilizing the U1 and U2 snRNPs from the complex, and eventually leading to the activation of the spliceosome. The 2′ hydroxyl of a specific nucleotide found at the branch site (commonly adenosine) becomes nucleophilic and carries out a nucleophilic attack on the phosphodiester bond at the 5′ splice site. As a result, the 5′ end of the intron becomes covalently linked to the branch site nucleotide forming a lariat, whereas the adjacent exon is now ‘free’ at its 3′ end. The 3′ hydroxyl of that ‘free’ exon then attacks the phosphodiester bond at the 3′ splice site, releasing the lariat intron from its neighbouring exon. The two adjacent exons are then ligated together (Fig. 1). The spliceosome finally disassembles and its components are ready for the next round of splicing. It has been shown that after splicing, the intronic lariat is debranched and degraded rapidly [20]. Hence, intronic RNAs are commonly deemed as ‘junk’ and devoid of any functions.
Fig. 1.
Biogenesis of linear and circular sisRNAs. During splicing, branch point nucleotide adenosine forms a 2′–5′ phosphodiester bond with the 5′ end of the intron, generating a lariat intermediate. The free 3′ OH group of the exon then cleaves the 3′ end of the lariat intermediate and joins the two exons together with a lariat as a by-product. The lariat can either be debranched by a Lariat debranching enzyme into a linear sisRNA or be trimmed at the 3′ end by an exonuclease into a circular sisRNA. Exons and introns are in blue and black, respectively
Although introns were discovered almost 40 years ago, there is still a debate over their evolutionary origins. There are two opposing hypotheses: “introns-early” [21, 22] and “introns-late” [23, 24]. Proponents of the introns-early hypothesis, also known as the exon theory of genes, believe that the very first exons were minigenes, coding for very short polypeptide structures. Such minigenes predated cellular life. Over the course of evolution, these minigenes were assembled together via introns, which were random nucleotide sequences introduced at the ends of these minigenes. These intronic sequences allowed for exon shuffling, which eventually resulted in the formation of whole genes comprising of several minigenes or exons. Modern organisms that have genomes containing almost little to no introns, such as prokaryotes, were thought to have lost their introns over evolutionary time. This is a result of the pressure to simplify their genomes and to reduce replication time, therefore allowing for competitive growth advantage. On the other hand, the introns-late supporters believe that introns arose much later, during the evolution of eukaryotes. Prokaryotes have no spliceosomal introns, apart from some that have a few non-spliceosomal (self-splicing) introns. Prokaryotes do not have a nucleus and thus transcription and translation are coupled and occur in the cytoplasm. If prokaryotes were to contain spliceosomal introns, a problem would arise as splicing would not be able to be completed prior to translation. Eukaryotes, on the other hand, have evolved to compartmentalize their genetic material into a nucleus, and hence such a splicing problem would not occur. The evolution of a nucleus allowed for eukaryotes to evolve and possess spliceosomal introns. Currently the consensus on the origin of introns is a mixture of both hypotheses. Spliceosomal introns appeared suddenly at the time eukaryotes first emerged and these introns most likely derived from self-splicing introns, which were present much earlier in precellular life [25].
Regardless of how or when introns were introduced into the genome, an important question is why introns have been so ubiquitous throughout the genomes of higher eukaryotes? In 1994, it was proposed that RNAs derived from introns are not just junk but could have evolved functions in the cell [26]. The discoveries of small nucleolar RNAs (snoRNAs), small Cajal body associated RNAs (scaRNAs) and microRNAs (miRNAs) have offered support to this hypothesis [27]. These classes of intron-derived RNAs have been shown to play various roles in gene regulation.
snoRNAs are an abundant group of non-coding RNAs (ncRNAs) that are important for rRNA processing and maturation [28, 29]. A minority of snoRNAs are involved in the endo- and exo-nucleolytic reactions to remove the spacer regions found in precursor ribosomal RNAs (pre-rRNAs), whereas the majority of snoRNAs act as guides for the covalent modification (pseudouridylation and 2′-O-methylation of ribose groups) of several regions of the rRNAs.
scaRNAs are structurally and functionally similar to snoRNAs. However, instead of localizing in the nucleolus, they localize in the Cajal body (CB), a nuclear body involved in the biogenesis of spliceosomal snRNAs. At the Cajal bodies, scaRNAs guide the covalent modifications of the snRNAs [30].
miRNAs are short approximately 22-nucleotide RNAs, which regulate gene expression. They generally function either though RNA silencing or inhibition of protein synthesis [31, 32]. In plants, miRNAs base pair with their targets with perfect or near perfect complementarity to silence their target mRNAs through the RNA interference (RNAi) machinery [33]. On the other hand, in animals, most miRNAs base pair with their target mRNAs with imperfect complementarity and repress the translation of target mRNAs [34].
Discovery of sisRNAs
As mentioned previously, most introns are rapidly degraded after splicing. Their rapid turnover has a direct implication on both transcription and splicing, thus affecting many other processes in the cell [35]. Firstly, the degradation of introns would free up nucleotides that could be used during subsequent rounds of transcription. Secondly, splicing factors that remain bound to the spliced intronic lariat are released and would then be available for the splicing machinery. It has been reported that the accumulation of intronic lariats is indeed detrimental to a cell [36, 37]. This observation suggests that introns that remain relatively stable upon splicing may not be just artefacts, as this would have been selected against during the course of evolution. The phenomenon of stable intronic RNA is not a recent observation: this was initially observed in viruses in the 1980’s, then in Xenopus, mammalian cells and Drosophila (Table 1). However, these unusually stable intronic RNAs were thought to be the rare exceptions.
Table 1.
List of sisRNAs discovered
| Species | Host gene locus | Conformation | Localization | Tissues | Functions | References |
|---|---|---|---|---|---|---|
| Human | Genome-wide | Circular | Nuclear | Cell lines | ND | [43] |
| ANKRD52 locus (ci-ankrd52) | Circular | Nuclear | Cell lines | Regulates host gene expression by interacting with RNA Polymerase II | ||
| Human | 15q11-q13 region of chromosome 15 (sno-lncRNA) | Linear | Nuclear | Cell lines | Acts as molecular sink to sequester Fox2, altering alternative splicing | [44] |
| Human, mouse | T cell receptor-β locus | Lariat | Nuclear and cytoplasmic | T cells | ND | [76, 77] |
| Human, rat | Pem homeobox locus | Lariat | Nuclear and cytoplasmic | Cell lines | ND | [35] |
| Mouse | κ locus | ND | Nuclear | Cell lines | ND | [78] |
| Mouse | Igh locus (intronic switch RNA) | Linear | ND | Cell lines | Regulates host gene expression by acting as guides to target AID to its host gene locus, promoting CSR | [42] |
| Xenopus laevis | Simian virus 40 | Lariat | Nuclear | Oocytes | ND | [79] |
| Xenopus tropicalis | Genome-wide | Linear and lariat | Nuclear and cytoplasmic | Oocytes, embryos | ND | [38, 39] |
| Drosophila melanogaster | delta locus | ND | Nuclear | Embryos | ND | [80] |
| Drosophila melanogaster | tRNA locus | Circular | ND | Larvae, pupae and adults | ND | [81] |
| Drosophila melanogaster | Genome-wide | Linear and circular | ND | Embryos, larvae, pupae and adults | ND | [40] |
| regena locus (sisR-1) | Linear | Nuclear and cytoplasmic | Embryos | Regulates host gene expression by repressing the cis-NAT ASTR | ||
| Human and murine cytomegalovirus | Immediate-early transcript | Lariat | Nuclear | Human and mouse cell lines | Murine sisRNA promotes viral progression from acute to persistent phase of infection | [82, 83] |
| Adenovirus 2 | E2A region | Linear and lariat | Nuclear | Human cell lines | ND | [84] |
| Herpes simplex virus | Latency-associated transcript (2-kb LAT intron) | Lariat | Nuclear | Monkey kidney cells, human cell lines, human and mouse neurons | Regulates translation of Hsp70 by altering the 60S ribosomal subunit, promoting host cell survival upon reactivation | [59–61, 64–66] |
| Epstein–Barr virus | W repeat region (ebv-sisRNA-1) | Linear | Nuclear | Human B cells | Acts as molecular sink to sequester miR-142-3p, preventing the repression of the EBV lytic gene product | [45, 48] |
| W repeat region (ebv-sisRNA-2) | Linear | ND | Human B cells | ND |
ND not determined
In 2012, Joseph Gall’s laboratory reported the first genome-wide identification of intronic RNAs in the oocyte germinal vesicle (GV or nucleus) of Xenopus tropicalis [38]. These intronic RNAs were only detected after acquiring pure nuclear RNA by dissecting the GVs and manually removing the nuclear envelopes, thus preventing contamination from the highly abundant cytoplasmic mRNAs. These intronic RNAs were termed ‘intronic sequences’ since they can be shorter than the full-length introns they originated from. These intronic sequences are being produced by more than 90 % of the genes transcribed during oogenesis. They observed that these intronic sequences in the GV were highly stable and could still be detected after 12 h to 2 days upon inhibition of transcription by actinomycin D treatment. For that, they named these stable intronic sequence RNAs (sisRNAs). Two years later, sisRNAs were also found to be present in the cytoplasm of the X. tropicalis oocytes [39]. These cytoplasmic sisRNAs were identified by removing the whole GVs and extracting the cytoplasmic RNA. Cytoplasmic sisRNAs are resistant to RNase R treatment, suggesting that they are circular molecules (lariats without tails). Compared to nuclear sisRNAs, cytoplasmic sisRNAs were observed to be mainly derived from shorter introns, and were located in relatively fewer introns. They noted that both nuclear and cytoplasmic sisRNAs could persist in the mature oocytes even from the early stages of embryogenesis until the blastula stages, when zygotic transcription starts, suggesting that this store of sisRNAs may have a role to play throughout the early development of X. tropicalis.
We classify sisRNAs as intronic RNAs that are spliced from its primary transcript and remain stable in the cell (Fig. 1). We review the known or suggested biological functions of sisRNAs in the cell: regulating host gene expression, functioning as molecular sinks and, regulating protein translation. We propose that sisRNAs are biologically active ncRNAs that act as an additional layer of gene regulation in the cell.
sisRNAs regulate host gene expression
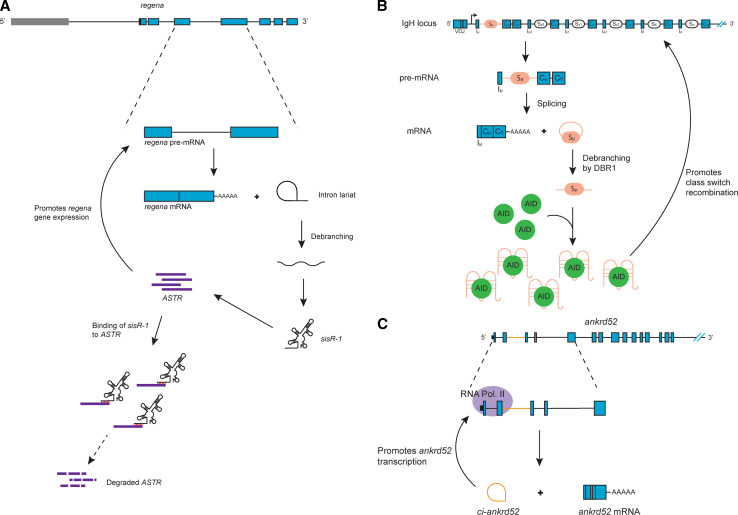
Recently, the laboratory of Jun Wei Pek reported a sisRNA, named sisR-1, in Drosophila melanogaster, which they showed to regulate the expression of its host gene [40]. They initially identified a total of 34 candidate sisRNAs after deep sequencing analysis of rRNA-depleted total RNA extracted from 0 to 2 h embryos. 0–2 h embryos were used because RNAs present in these embryos would have been stable for no less than 10–12 h since zygotic transcription only begins 2 h after egg laying in Drosophila. Of these 34 candidate sisRNAs, some are resistant to RNase R treatment, suggesting that both circular and linear sisRNAs are present. Upon further examination of a few of these candidate sisRNAs, they showed that sisRNAs are not just oocyte-specific, but are present throughout development as well as in adult tissues. sisR-1 is a linear sisRNA derived from the fourth intron of the regena (rga) gene locus. In addition, a cis-natural antisense transcript (cis-NAT), which they named ASTR, is also found in the same locus. Both ASTR and rga showed analogous expression patterns during embryogenesis. When they knocked down ASTR via shRNA treatment, it led to a significant decrease in the expression levels of rga pre-mRNA, suggesting that the expression of rga is promoted by ASTR. On the other hand, shRNA-mediated knockdown of sisR-1 led to an increase in the expression of both ASTR and rga. However, when both sisR-1 and ASTR were knocked down simultaneously, there was no longer an increase in expression of rga. Thus, they proposed that sisR-1 regulates the expression of its own host gene by repressing the cis-NAT ASTR (Fig. 2a). Hence sisR-1 is involved in a negative feedback loop, which promotes the robust decrease in expression of its host gene during development.
Fig. 2.
sisRNAs regulate host gene expression. a Model of a Drosophila sisRNA, sisR-1, biogenesis and its regulation on its parental gene rga through repression of ASTR. b Model for the regulation of IgH CSR in B cells by the intronic switch RNA, acting as guides to target AID to the IgH locus. c Model of a circular intronic RNA, ci-ankrd52, and its regulatory role on its parental gene expression via interactions with RNA Pol. II
Another example of a sisRNA that is involved in the regulation of its host gene is the intronic switch RNA. B cells undergo immunoglobulin (Ig) heavy chain (IgH) class switch recombination (CSR) upon stimulation by antigens [41]. CSR is an intrachromosomal deletional recombination within the switch (S) regions of the IgH locus facilitated by the activation-induced cytidine deaminase (AID) [41]. During CSR, the default heavy chain constant (CH) region Cµ/Cδ is replaced to one of the other downstream CH regions (Cγ, Cα or Cε), ‘switching’ the B cell from expressing IgM/IgD to IgG, IgA or IgE respectively. In a recent study by the laboratory of Jayanta Chaudhuri using CH12 B lymphoma cells, a cell line that switches at a high rate, it was found that intronic RNA transcripts coming from the S regions of the IgH locus are involved in CSR [42]. These intronic switch RNAs are spliced from the primary germline transcripts of the IgH locus, and were found to be debranched. Through RNA pull-down assays, they showed that these linear sisRNAs are able to bind to AID. They suggested that these sisRNAs bind to AID by forming G-quadruplex structures. Due to complementary sequences between the intronic switch RNA and the S region, these sisRNAs regulate its own host genes by acting as guides, which target the AID to the IgH locus for efficient CSR to take place (Fig. 2b).
ci-ankrd52, a circular sisRNA, has also been observed to regulate its host gene expression. Using a custom computational framework, Ling-Ling Chen and Yang Li’s laboratories identified several hundred intronic RNAs in the non-polyadenylated RNA fraction extracted from HeLa and H9 cells [43]. RNase R treatment of these identified sisRNAs revealed that both linear and circular forms were present. They suggested that a consensus motif near the 5′ splice site and the branchpoint of the circular sisRNAs might explain why these sisRNAs are not debranched and remain circular. One such circular sisRNA is ci-ankrd52, which originates from the second intron of the ANKRD52 gene and is localized in the nucleus. To study its function, they used antisense oligodeoxynucleotides (ASOs) modified with phosphorothioate to knockdown ci-ankrd52 expression. They observed that the expression of its host gene ankrd52 was significantly reduced upon the knockdown of ci-ankrd52. When they examined the nuclear localization of ci-ankrd52 using in situ hybridization, they observed that ci-ankrd52 specifically colocalized to the transcription sites of its host gene locus. They further showed that ci-ankrd52 is associated with the elongation RNA Polymerase II complex. Taken together, this study suggests that ci-ankrd52 regulates the efficient transcription of its host gene through its positive interaction with RNA Polymerase II (Fig. 2c).
The three sisRNAs described above all act in cis to regulate the genes from which they are derived from. sisR-1 regulates rga by repressing its cis-NAT, ASTR; the intronic switch RNA regulates its IgH gene locus by targeting AID to the S region of the locus, while ci-ankrd52 regulates its host gene ankrd52 by localizing and recruiting RNA Polymerase II to its sites of transcription. An interesting theme emerges whereby introns are utilized to self-regulate their parental gene. Upon transcription and splicing, two species of transcripts (mRNAs and introns) are generated, and in most cases the mRNAs are the major effectors by being translated into proteins. As introns are being produced at equal molar ratio as the exons in a primary transcript, spliced introns are therefore a direct readout of the transcription rate of a particular gene. Thus, cellular mechanisms that utilize or monitor introns to modulate feedback loops may allow for a more direct and efficient way to regulate the expression of a particular gene. On the other hand, sisRNAs may also act in trans to modulate the expression of other gene loci in the nucleus.
sisRNAs as molecular sinks or sponges
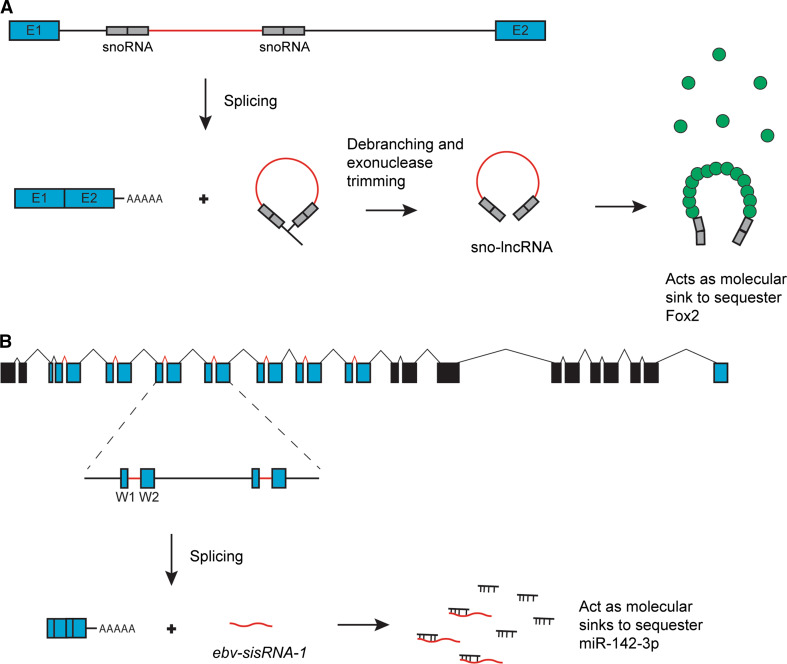
Deep sequencing analysis of rRNA-depleted non-polyadenylated RNA fraction from HeLa and H9 cells revealed a class of sisRNAs with ends corresponding to regions mapping back to snoRNAs, which Ling-Ling Chen and Gordon Carmichael’s laboratories termed snoRNA-related long ncRNA (sno-lncRNA) [44]. In H9 cells, the most abundant sno-lncRNAs expressed was observed to be from the imprinted 15q11–q13 region, a genomic region on chromosome 15 implicated in Prader–Willi syndrome (PWS). They further showed that this group of linear sisRNAs is produced via the same mechanism involving snoRNPs and they suggested that their stability is due to the snoRNP components that remain associated to them. sno-lncRNAs localize to the nucleus, however, they do not accumulate in nuclear bodies associated with snoRNAs (nucleolus and CB), suggesting that these transcripts are functionally different from snoRNAs. They further observed that these sisRNAs associate with an alternative splicing factor Fox2 in the nucleus. Conversely, they showed that Fox2 is also strongly enriched in nuclear regions containing these sisRNAs. When they knocked down these sisRNAs using phosphorothioate ASOs, only a slight change in global gene expression was observed. When they focused on genes that are known to be regulated by Fox2, they observed a change in their splicing profile. They summarized and postulated that these sisRNAs, which originate from the PWS region act as molecular sinks to sequester Fox2, and as a result, alter Fox2-regulated splicing (Fig. 3a).
Fig. 3.
sisRNAs act as molecular sinks or sponges. a Model of sisRNAs from the PWS region, sno-lncRNAs, sequestering Fox2 proteins and altering Fox2-regulated splicing. b Model of a viral sisRNA, ebv-sisRNA-1, acting as a molecular sink against miR-142-3p, preventing the repression of the EBV lytic gene product
In 2013, the laboratory of Joan Steitz discovered a sisRNA, ebv-sisRNA-1, encoded by the Epstein–Barr virus (EBV) [45]. EBV is a human herpes virus, with a linear double stranded DNA genome. EBV has been implicated in several human cancers and autoimmune diseases. Its viral life cycle is divided into a lytic and a latent phase. During initial infection, EBV goes through a lytic replication phase upon entry into host epithelial cells, after which it migrates to the B cells. The virus remains in B cells, cycling through periods of latency and lytic reactivation [46]. During latency, the virus expresses genes that allow infected B cells to escape detection by the host immune system [47]. ebv-sisRNA-1 was discovered through a small RNA-sequencing analysis of nuclear RNA extracted from human B cells, which have been stably infected with EBV, and are in the latency III program of gene expression. ebv-sisRNA-1 is a linear nuclear-enriched sisRNA derived from a small intron in the W repeat region. Using the program RNAduplex, it was predicted that ebv-sisRNA-1 was able to form stable hybrids with human miRNAs, of which 4 of these miRNAs have been known to be expressed in B cells [48]. Most of the base pairing between the four miRNAs and ebv-sisRNA-1 are ~100 % conserved. One of this miRNA, miR-142-3p, is known to target and repress an EBV lytic gene product [49]. It is possible that ebv-sisRNA-1 may affect the levels or activity of miR-142-3p via one of the following mechanisms. Its base pairing with perfect seed complementarity to miR-142-3p can lead to the subsequent degradation of the miRNA. Alternatively, it may act as a molecular sink, sequestering miR-142-3p and preventing it from repressing the EBV lytic gene product (Fig. 3b). It is also possible that the interaction between ebv-sisRNA-1 and the other three miRNAs might be achieved through similar mechanisms for the same purposes. Thus, EBV can employ one of these strategies to evade detection by host immune cells.
Here we show two sisRNAs that can act as molecular sinks or sponges to sequester their targets and prevent them from carrying out their biological functions. sno-lncRNAs from the PWS region are sisRNAs that sequester Fox2 proteins and thus alter Fox2 mediated splicing; while the ebv-sisRNA-1 may sequester an miRNA, miR-142-3p, and prevent it from repressing the EBV lytic gene product. One example of a ncRNA that acts similarly to sno-lncRNAs from the PWS region is MALAT1, a lncRNA that has been shown to be implicated in human cancers [50]. MALAT1 was shown to interact with the splicing factors called SR proteins, and it was suggested to be a protein sponge, regulating the cellular distribution of SR proteins in the nucleus, and as a result modulating alternative splicing of pre-mRNAs [51].
Several ncRNAs have also been proposed to behave as miRNA sponges, similar to ebv-sisRNA-1 [52]. One such ncRNA is PTENP1, a pseudogene of the tumor suppressor PTEN. PTENP1 was shown to regulate the expression of PTEN by competing for the same set of miRNAs that interact with PTEN. Thus, PTENP1 was coined as a competing endogenous RNA or ceRNA [53, 54]. Other examples of miRNA sponges are Herpesvirus saimiri U RNA (HSUR) 1 and 2, which are ncRNAs encoded by the Herpesvirus saimiri, a virus under the same family as EBV [55]. These two ncRNAs were shown to share complementarity and coimmunoprecipitated with three miRNAs expressed in T cells. Interestingly, one of these miRNA is miR-142-3p, the same miRNA that is proposed to be sequestered by the ebv-sisRNA-1, suggesting a similar mechanism being deployed by these two viruses to evade detection by the immune system.
However, it is important to take into account the abundance of both the sisRNA/ncRNA and its target miRNA, as well as the tissues and cell types that they are expressed in, when assessing the sponge effect of sisRNAs/ncRNAs on miRNAs in vivo to ensure the biological significance of such an assessment [56, 57].
sisRNAs regulate protein translation
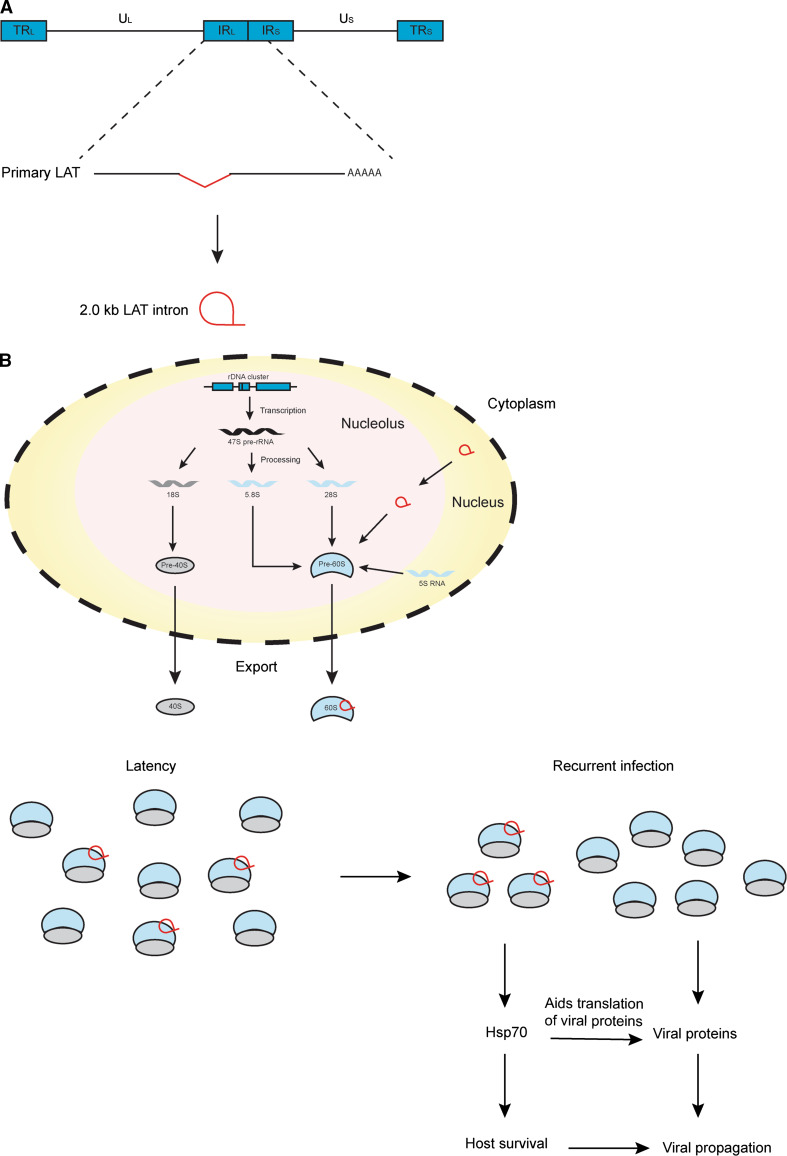
The herpes simplex virus (HSV) encodes a sisRNA known as the 2-kb latency-associated transcript (LAT) intron. HSV is a herpes virus, and similar to EBV, it also establishes latent infections in its host. During latency, the virus resides in the trigeminal ganglia of the infected host. LATs are the only transcripts produced by the virus during latency, which are transcribed and spliced from a primary 8.3-kb transcript [58] (Fig. 4a). The 2-kb LAT intron is the most abundant LAT species produced during latency and was first discovered by Lawrence Feldman’s laboratory in 1991 [59]. The intron was detected through Northern blot analysis of total RNA extracted from HeLa cells and was demonstrated to be a stable intron localizing to the nucleus. Several years later, John Taylor’s group showed that this sisRNA exists as a lariat by studying the trigeminal ganglia of latently infected mice and infected monkey CV-1 cells [60]. A year later, Nigel Fraser’s laboratory revealed that the lariat branches at a Guanosine, which was suggested to account for the stability of the lariat by preventing it from being debranched [61].
Fig. 4.
sisRNAs regulate protein translation. a Linear map of the HSV genome. The 2-kb LAT intron is processed from the primary LAT transcript which is transcribed from the internal repeat long (IRL) and short (IRS) regions on the genome. b Model for the regulation of translation of Hsp70 by the 2-kb LAT, ensuring the survival of the host cell during reactivation from latency. U L unique long, U S unique short, TR L terminal repeat long, TR S terminal repeat short
LAT-null HSV mutants (deletion of the LAT promoter and the 5′ portion of the 2-kb LAT) did not show any other deficiencies apart from being unable to reactivate from latency [62]. It was demonstrated that the LAT can inhibit apoptosis and promote cell survival, possibly explaining the importance of LAT during reactivation [62, 63]. In 2006, Nigel Fraser’s group showed that SY5Y human neuroblastoma cells transfected with the 2-kb LAT intron were protected from cold shock and this was due to an accumulation of Hsp70 [64]. The upregulation of Hsp70 during cold shock was not observed at the transcriptional level. Due to the sisRNA’s association with ribosomal proteins, the authors postulated that the sisRNA upregulates Hsp70 expression at the translational level [65]. A year later, they showed that this sisRNA associates with the forming 60S ribosomal subunit in the nucleolus [66] They hypothesized that the 2-kb LAT intron possibly interferes with the processing of the 28S rRNA, thus altering the protein compostion of the 60S ribosomal subunit. Taken together, they proposed a model in which upon the processing of the sisRNA from its primary LAT transcript, it gets transported to the nucleolus, possibly in a manner similar to snoRNAs. There it associates with the 60S ribosomal subunits and together, they get exported to the cytoplasm. These altered 60S ribosomal subunits form a modified pool of ribosomes, which would then be utilized by the infected cell to make sure that there is a stable expression of Hsp70 (Fig. 4). This strategy is advantageous for the virus as it ensures the survival of the infected cells during times of stress when it reactivates from latency, as well as in aiding in the translation of viral proteins.
It is interesting to find out how these altered 60S ribosomal subunits and the resulting modified pool of ribosomes are able to specifically affect the translation of Hsp70, and if a similar mechanism exists in other organisms. Recently, it was discovered that there are ncRNAs that are able to bind to ribosomes and affect protein synthesis. In Saccharomyces cerevisae undergoing hyperosmotic stress, an 18-mer ncRNA originating from the TRM10 mRNA was observed to associate with the 60S ribosomal subunit [67]. In the halophilic archaeon Haloferax volcanii, a 26-mer ncRNA originating from the 5′ ends of valine tRNA associates with the 30S small ribosomal subunit during environmental stress [68]. However, for both these ncRNAs, the resulting effect of their association with ribosomes is a reduction in the efficiency of global protein translation. These translational regulators were classified as ribosome-associated ncRNAs (rancRNAs) [69]. Although there is an obvious length difference between the 2-kb LAT intron and these two short rancRNAs, all three transcripts work similarly towards protecting the cell from stress and promoting cell survival.
Future perspectives and challenges
It is still very early for the sisRNA field and we are just beginning to answer many questions regarding sisRNA biogenesis and functions in the cell.
How are sisRNAs processed upon splicing from the pre-mRNAs?
Do sisRNAs exert their functions on their own or through interactions with proteins?
How do sisRNAs exert their functions? Do they have similar structural characteristics? Do they function by base-pairing with their substrates?
For linear sisRNAs:
How do they remain stable in the cell?
Do they associate with proteins that protect them from degradation?
For circular sisRNAs:
What prevents them from being debranched and degraded?
How are they transported out to the cytoplasm?
Although we classify sisRNAs as intronic RNAs that are spliced from its primary transcripts, we should not rule out the possibility that sisRNAs can be independently transcribed from its host gene and subsequently remain stable in the cell. If so, this could suggest that sisRNAs can be generated via two pathways: a splicing-dependent pathway and a splicing-independent pathway. This is another aspect of sisRNA biogenesis that should be addressed to better understand this novel class of ncRNA.
The advent of deep sequencing paved the way for the genome-wide analyses and discoveries of sisRNAs in model organisms such as Xenopus and Drosophila. However, sisRNAs represent a minor species of RNA in the cell and intronic reads are often regarded as noise or artefacts in RNA-sequencing data. By understanding the underlying chemistry of sisRNAs, it would allow for the global enrichment of sisRNAs prior to deep sequencing. For example, mRNAs can be enriched through poly(A) selection to specifically look at the transcriptome profile of a particular cell or tissue. Similarly, if sisRNAs have some unique characteristics or chemistry, this information could be used to allow for the enrichment and identification of many more undiscovered sisRNAs. This has been shown by the discovery of thousands of circular sisRNAs by RNase R treatment. In addition, if the proteins that associate to the currently known sisRNAs were identified, these proteins could then be utilized to discover other sisRNAs by RNA immunoprecipitation. If such enrichment was made possible and a large number of sisRNAs were discovered, the next major challenge would be ascertaining the functions of these newly identified sisRNAs in the cell.
It has been shown previously that a majority of the ncRNAs present in mammalian cells originate from the intronic regions of the genome [70]. Several of these intronic transcripts are implicated in cancer [71–73]. Similarly, several cancer-associated susceptibility loci were also found in introns [74, 75]. Studying intronic transcripts such as sisRNAs may possibly provide us with mechanistic insights, which can further improve our understanding on the pathology of cancer.
Concluding remarks
The discoveries of functionally significant intron-derived RNAs such as snoRNAs, scaRNAs and miRNAs have provided support that introns are more than just mere junk. Instead these ncRNAs play important roles in gene regulation. In this review, we have described another class of intron-derived ncRNAs known as sisRNAs. sisRNAs have been shown or suggested to play various roles in gene regulation. They can regulate their host’s genes expression, function as molecular sinks or sponges, and regulate protein translation. Taken together, sisRNAs are a biologically active class of ncRNAs conferring an additional layer of gene regulation.
Acknowledgments
The authors are supported by the Temasek Life Sciences Laboratory.
Abbreviations
- AID
Activation-induced cytidine deaminase
- ASO
Antisense oligodeoxynucleotides
- CB
Cajal Body
- CH
Heavy chain constant
- cis-NAT
cis-natural antisense transcript
- CSR
Class switch recombination
- EBV
Epstein–Barr virus
- GV
Germinal vesicle
- HSUR
Herpesvirus saimiri U RNA
- HSV
Herpes simplex virus
- Ig
Immunoglobulin
- IgH
Immunoglobulin heavy chain
- LAT
Latency-associated transcript
- miRNA
microRNA
- ncRNA
Noncoding RNA
- Pre-mRNA
Precursor messenger RNA
- Pre-rRNA
Precursor ribosomal RNAs
- PWS
Prader–Willi syndrome
- rancRNAs
Ribosome-associated ncRNAs
- rga
regena
- RNP
Ribonucleoprotein
- RNAi
RNA interference
- S regions
Switch regions
- scaRNAs
Small Cajal body associated RNA
- sisRNA
Stable intronic sequence RNA
- snRNA
Small nuclear RNA
- snRNP
Small nuclear RNP
- snoRNA
Small nucleolar RNA
- sno-lncRNA
snoRNA-related long ncRNA
References
- 1.Berget SM, Moore C, Sharp PA. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci USA. 1977;74(8):3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chow LT, Gelinas RE, Broker TR, Roberts RJ. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell. 1977;12(1):1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- 3.Breathnach R, Mandel JL, Chambon P. Ovalbumin gene is split in chicken DNA. Nature. 1977;270(5635):314–319. doi: 10.1038/270314a0. [DOI] [PubMed] [Google Scholar]
- 4.Jeffreys AJ, Flavell RA. The rabbit beta-globin gene contains a large large insert in the coding sequence. Cell. 1977;12(4):1097–1108. doi: 10.1016/0092-8674(77)90172-6. [DOI] [PubMed] [Google Scholar]
- 5.Bell GI, Pictet RL, Rutter WJ, Cordell B, Tischer E, Goodman HM. Sequence of the human insulin gene. Nature. 1980;284(5751):26–32. doi: 10.1038/284026a0. [DOI] [PubMed] [Google Scholar]
- 6.Yamada Y, Avvedimento VE, Mudryj M, Ohkubo H, Vogeli G, Irani M, Pastan I, de Crombrugghe B. The collagen gene: evidence for its evolutinary assembly by amplification of a DNA segment containing an exon of 54 bp. Cell. 1980;22(3):887–892. doi: 10.1016/0092-8674(80)90565-6. [DOI] [PubMed] [Google Scholar]
- 7.Tilghman SM, Tiemeier DC, Seidman JG, Peterlin BM, Sullivan M, Maizel JV, Leder P. Intervening sequence of DNA identified in the structural portion of a mouse beta-globin gene. Proc Natl Acad Sci USA. 1978;75(2):725–729. doi: 10.1073/pnas.75.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lomedico P, Rosenthal N, Efstratidadis A, Gilbert W, Kolodner R, Tizard R. The structure and evolution of the two nonallelic rat preproinsulin genes. Cell. 1979;18(2):545–558. doi: 10.1016/0092-8674(79)90071-0. [DOI] [PubMed] [Google Scholar]
- 9.Konkel DA, Tilghman SM, Leder P. The sequence of the chromosomal mouse beta-globin major gene: homologies in capping, splicing and poly(A) sites. Cell. 1978;15(4):1125–1132. doi: 10.1016/0092-8674(78)90040-5. [DOI] [PubMed] [Google Scholar]
- 10.Wild MA, Gall JG. An intervening sequence in the gene coding for 25S ribosomal RNA of Tetrahymena pigmentosa. Cell. 1979;16(3):565–573. doi: 10.1016/0092-8674(79)90030-8. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert W. Why genes in pieces? Nature. 1978;271(5645):501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 12.Cech TR. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 13.Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns—a review. Gene. 1989;82(1):5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 14.Christopher DA, Hallick RB. Euglena gracilis chloroplast ribosomal protein operon: a new chloroplast gene for ribosomal protein L5 and description of a novel organelle intron category designated group III. Nucleic Acids Res. 1989;17(19):7591–7608. doi: 10.1093/nar/17.19.7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irimia M, Roy SW. Origin of spliceosomal introns and alternative splicing. Cold Spring Harbor perspectives in biology. 2014;6(6):a016071. doi: 10.1101/cshperspect.a016071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sperling J, Azubel M, Sperling R. Structure and function of the pre-mRNA splicing machine. Structure. 2008;16(11):1605–1615. doi: 10.1016/j.str.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? BioEssays: News Rev Mol Cell Dev Biol. 2003;25(12):1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- 18.Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harbor perspectives in biology. 2011;3(7):a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoskins AA, Moore MJ. The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem Sci. 2012;37(5):179–188. doi: 10.1016/j.tibs.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharp PA, Konarksa MM, Grabowski PJ, Lamond AI, Marciniak R, Seiler SR. Splicing of messenger RNA precursors. Cold Spring Harb Symp Quant Biol. 1987;52:277–285. doi: 10.1101/SQB.1987.052.01.033. [DOI] [PubMed] [Google Scholar]
- 21.Doolittle WF. Genes in pieces: were they ever together? Nature. 1978;272(5654):581–582. doi: 10.1038/272581a0. [DOI] [Google Scholar]
- 22.Gilbert W. The exon theory of genes. Cold Spring Harb Symp Quant Biol. 1987;52:901–905. doi: 10.1101/SQB.1987.052.01.098. [DOI] [PubMed] [Google Scholar]
- 23.Palmer JD, Logsdon JM., Jr The recent origins of introns. Curr Opin Genet Dev. 1991;1(4):470–477. doi: 10.1016/S0959-437X(05)80194-7. [DOI] [PubMed] [Google Scholar]
- 24.Cavalier-Smith T. Selfish DNA and the origin of introns. Nature. 1985;315(6017):283–284. doi: 10.1038/315283b0. [DOI] [PubMed] [Google Scholar]
- 25.Koonin EV. The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biol Direct. 2006;1:22. doi: 10.1186/1745-6150-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mattick JS. Introns: evolution and function. Curr Opin Genet Dev. 1994;4(6):823–831. doi: 10.1016/0959-437X(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 27.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109(2):145–148. doi: 10.1016/S0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 29.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9(3):337–342. doi: 10.1016/S0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 30.Darzacq X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21(11):2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 32.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16(13):1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dugas DV, Bartel B. MicroRNA regulation of gene expression in plants. Curr Opin Plant Biol. 2004;7(5):512–520. doi: 10.1016/j.pbi.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 34.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216(2):671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 35.Clement JQ, Maiti S, Wilkinson MF. Localization and stability of introns spliced from the Pem homeobox gene. J Biol Chem. 2001;276(20):16919–16930. doi: 10.1074/jbc.M005104200. [DOI] [PubMed] [Google Scholar]
- 36.Chapman KB, Boeke JD. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell. 1991;65(3):483–492. doi: 10.1016/0092-8674(91)90466-C. [DOI] [PubMed] [Google Scholar]
- 37.Nam K, Lee G, Trambley J, Devine SE, Boeke JD. Severe growth defect in a Schizosaccharomyces pombe mutant defective in intron lariat degradation. Mol Cell Biol. 1997;17(2):809–818. doi: 10.1128/MCB.17.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gardner EJ, Nizami ZF, Talbot CC, Jr, Gall JG. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev. 2012;26(22):2550–2559. doi: 10.1101/gad.202184.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Talhouarne GJ, Gall JG. Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. RNA. 2014;20(9):1476–1487. doi: 10.1261/rna.045781.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pek JW, Osman I, Tay ML-I, Zheng RT. Stable intronic sequence RNAs (sisRNAs) have regulatory roles in Drosophila melanogaster. J Cell Biol. 2015;211(2):243–251. doi: 10.1083/jcb.201507065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng S, Vuong BQ, Vaidyanathan B, Lin JY, Huang FT, Chaudhuri J. Non-coding RNA Generated following lariat debranching mediates targeting of AID to DNA. Cell. 2015;161(4):762–773. doi: 10.1016/j.cell.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 44.Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL. Long noncoding RNAs with snoRNA ends. Mol Cell. 2012;48(2):219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 45.Moss WN, Steitz JA. Genome-wide analyses of Epstein–Barr virus reveal conserved RNA structures and a novel stable intronic sequence RNA. BMC Genom. 2013;14:543. doi: 10.1186/1471-2164-14-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young LS, Dawson CW, Eliopoulos AG. The expression and function of Epstein–Barr virus encoded latent genes. Mol Pathol: MP. 2000;53(5):238–247. doi: 10.1136/mp.53.5.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ning S. Innate immune modulation in EBV infection. Herpesviridae. 2011;2(1):1. doi: 10.1186/2042-4280-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moss WN (2014) Analyses of non-coding RNAs generated from the Epstein–Barr virus W repeat region. In: International work—conference on bioinformatics and biomedical engineering
- 49.Riley KJ, Rabinowitz GS, Yario TA, Luna JM, Darnell RB, Steitz JA. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 2012;31(9):2207–2221. doi: 10.1038/emboj.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshimoto R, Mayeda A, Yoshida M. Nakagawa S (2016) MALAT1 long non-coding RNA in cancer. Biochim Biophys Acta. 1859;1:192–199. doi: 10.1016/j.bbagrm.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 51.Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, Prasanth KV. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9(3):e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol: CB. 2010;20(19):R858–R861. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, Lieberman J, Rigoutsos I, Pandolfi PP. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147(2):344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328(5985):1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Broderick JA, Zamore PD. Competitive endogenous RNAs cannot alter microRNA function in vivo. Mol Cell. 2014;54(5):711–713. doi: 10.1016/j.molcel.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 57.Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell. 2014;54(5):766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Block TM, Hill JM. The latency associated transcripts (LAT) of herpes simplex virus: still no end in sight. J Neurovirol. 1997;3(5):313–321. doi: 10.3109/13550289709030745. [DOI] [PubMed] [Google Scholar]
- 59.Farrell MJ, Dobson AT, Feldman LT. Herpes simplex virus latency-associated transcript is a stable intron. Proc Natl Acad Sci USA. 1991;88(3):790–794. doi: 10.1073/pnas.88.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu TT, Su YH, Block TM, Taylor JM. Evidence that two latency-associated transcripts of herpes simplex virus type 1 are nonlinear. J Virol. 1996;70(9):5962–5967. doi: 10.1128/jvi.70.9.5962-5967.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zabolotny JM, Krummenacher C, Fraser NW. The herpes simplex virus type 1 2.0-kilobase latency-associated transcript is a stable intron which branches at a guanosine. J Virol. 1997;71(6):4199–4208. doi: 10.1128/jvi.71.6.4199-4208.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perng GC, Jones C, Ciacci-Zanella J, Stone M, Henderson G, Yukht A, Slanina SM, Hofman FM, Ghiasi H, Nesburn AB, Wechsler SL. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science. 2000;287(5457):1500–1503. doi: 10.1126/science.287.5457.1500. [DOI] [PubMed] [Google Scholar]
- 63.Inman M, Perng GC, Henderson G, Ghiasi H, Nesburn AB, Wechsler SL, Jones C. Region of herpes simplex virus type 1 latency-associated transcript sufficient for wild-type spontaneous reactivation promotes cell survival in tissue culture. J Virol. 2001;75(8):3636–3646. doi: 10.1128/JVI.75.8.3636-3646.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Atanasiu D, Kent JR, Gartner JJ, Fraser NW. The stable 2-kb LAT intron of herpes simplex stimulates the expression of heat shock proteins and protects cells from stress. Virology. 2006;350(1):26–33. doi: 10.1016/j.virol.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 65.Ahmed M, Fraser NW. Herpes simplex virus type 1 2-kilobase latency-associated transcript intron associates with ribosomal proteins and splicing factors. J Virol. 2001;75(24):12070–12080. doi: 10.1128/JVI.75.24.12070-12080.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Atanasiu D, Fraser NW. The stable 2-kilobase latency-associated transcript of herpes simplex virus type 1 can alter the assembly of the 60S ribosomal subunit and is exported from nucleus to cytoplasm by a CRM1-dependent pathway. J Virol. 2007;81(14):7695–7701. doi: 10.1128/JVI.00282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pircher A, Bakowska-Zywicka K, Schneider L, Zywicki M, Polacek N. An mRNA-derived noncoding RNA targets and regulates the ribosome. Mol Cell. 2014;54(1):147–155. doi: 10.1016/j.molcel.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gebetsberger J, Zywicki M, Kunzi A, Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii . Archaea. 2012;2012:260909. doi: 10.1155/2012/260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pircher A, Gebetsberger J, Polacek N. Ribosome-associated ncRNAs: an emerging class of translation regulators. RNA Biol. 2014;11(11):1335–1339. doi: 10.1080/15476286.2014.996459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.St Laurent G, Shtokalo D, Tackett MR, Yang Z, Eremina T, Wahlestedt C, Urcuqui-Inchima S, Seilheimer B, McCaffrey TA, Kapranov P. Intronic RNAs constitute the major fraction of the non-coding RNA in mammalian cells. BMC Genom. 2012;13:504. doi: 10.1186/1471-2164-13-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC, Reis EM. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer. 2011;10:141. doi: 10.1186/1476-4598-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim SW, Fishilevich E, Arango-Argoty G, Lin Y, Liu G, Li Z, Monaghan AP, Nichols M, John B. Genome-wide transcript profiling reveals novel breast cancer-associated intronic sense RNAs. PLoS One. 2015;10(3):e0120296. doi: 10.1371/journal.pone.0120296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guil S, Soler M, Portela A, Carrere J, Fonalleras E, Gomez A, Villanueva A, Esteller M. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol. 2012;19(7):664–670. doi: 10.1038/nsmb.2315. [DOI] [PubMed] [Google Scholar]
- 74.Eeles RA, Olama AAA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, Ghoussaini M, Luccarini C, Dennis J, Jugurnauth-Little S, Dadaev T, Neal DE, Hamdy FC, Donovan JL, Muir K, Giles GG, Severi G, Wiklund F, Gronberg H, Haiman CA, Schumacher F, Henderson BE, Le Marchand L, Lindstrom S, Kraft P, Hunter DJ, Gapstur S, Chanock SJ, Berndt SI, Albanes D, Andriole G, Schleutker J, Weischer M, Canzian F, Riboli E, Key TJ, Travis RC, Campa D, Ingles SA, John EM, Hayes RB, Pharoah PDP, Pashayan N, Khaw K-T, Stanford JL, Ostrander EA, Signorello LB, Thibodeau SN, Schaid D, Maier C, Vogel W, Kibel AS, Cybulski C, Lubinski J, Cannon-Albright L, Brenner H, Park JY, Kaneva R, Batra J, Spurdle AB, Clements JA, Teixeira MR, Dicks E, Lee A, Dunning AM, Baynes C, Conroy D, Maranian MJ, Ahmed S, Govindasami K, Guy M, Wilkinson RA, Sawyer EJ, Morgan A, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As NJ, Woodhouse CJ, Thompson A, Dudderidge T, Ogden C, Cooper CS, Lophatananon A, Cox A, Southey MC, Hopper JL, English DR, Aly M, Adolfsson J, Xu J, Zheng SL, Yeager M, Kaaks R, Diver WR, Gaudet MM, Stern MC, Corral R, Joshi AD, Shahabi A, Wahlfors T, Tammela TLJ, Auvinen A, Virtamo J, Klarskov P, Nordestgaard BG, Roder MA, Nielsen SF, Bojesen SE, Siddiq A, FitzGerald LM, Kolb S, Kwon EM, Karyadi DM, Blot WJ, Zheng W, Cai Q, McDonnell SK, Rinckleb AE, Drake B, Colditz G, Wokolorczyk D, Stephenson RA, Teerlink C, Muller H, Rothenbacher D, Sellers TA, Lin H-Y, Slavov C, Mitev V, Lose F, Srinivasan S, Maia S, Paulo P, Lange E, Cooney KA, Antoniou AC, Vincent D, Bacot F, Tessier DC, Kote-Jarai Z, Easton DF. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013;45(4):385–391. doi: 10.1038/ng.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Permuth-Wey J, Lawrenson K, Shen HC, Velkova A, Tyrer JP, Chen Z, Lin H-Y, Ann Chen Y, Tsai Y-Y, Qu X, Ramus SJ, Karevan R, Lee J, Lee N, Larson MC, Aben KK, Anton-Culver H, Antonenkova N, Antoniou AC, Armasu SM, Bacot F, Baglietto L, Bandera EV, Barnholtz-Sloan J, Beckmann MW, Birrer MJ, Bloom G, Bogdanova N, Brinton LA, Brooks-Wilson A, Brown R, Butzow R, Cai Q, Campbell I, Chang-Claude J, Chanock S, Chenevix-Trench G, Cheng JQ, Cicek MS, Coetzee GA, Cook LS, Couch FJ, Cramer DW, Cunningham JM, Dansonka-Mieszkowska A, Despierre E, Doherty JA, Dörk T, du Bois A, Dürst M, Easton DF, Eccles D, Edwards R, Ekici AB, Fasching PA, Fenstermacher DA, Flanagan JM, Garcia-Closas M, Gentry-Maharaj A, Giles GG, Glasspool RM, Gonzalez-Bosquet J, Goodman MT, Gore M, Górski B, Gronwald J, Hall P, Halle MK, Harter P, Heitz F, Hillemanns P, Hoatlin M, Høgdall CK, Høgdall E, Hosono S, Jakubowska A, Jensen A, Jim H, Kalli KR, Karlan BY, Kaye SB, Kelemen LE, Kiemeney LA, Kikkawa F, Konecny GE, Krakstad C, Krüger Kjaer S, Kupryjanczyk J, Lambrechts D, Lambrechts S, Lancaster JM, Le ND, Leminen A, Levine DA, Liang D, Kiong Lim B, Lin J, Lissowska J, Lu KH, Lubiński J, Lurie G, Massuger LFAG, Matsuo K, McGuire V, McLaughlin JR, Menon U, Modugno F, Moysich KB, Nakanishi T, Narod SA, Nedergaard L, Ness RB, Nevanlinna H, Nickels S, Noushmehr H, Odunsi K, Olson SH, Orlow I, Paul J, Pearce CL, Pejovic T, Pelttari LM, Pike MC, Poole EM, Raska P, Renner SP, Risch HA, Rodriguez-Rodriguez L, Anne Rossing M, Rudolph A, Runnebaum IB, Rzepecka IK, Salvesen HB, Schwaab I, Severi G, Shridhar V, Shu X-O, Shvetsov YB, Sieh W, Song H, Southey MC, Spiewankiewicz B, Stram D, Sutphen R, Teo S-H, Terry KL, Tessier DC, Thompson PJ, Tworoger SS, van Altena AM, Vergote I, Vierkant RA, Vincent D, Vitonis AF, Wang-Gohrke S, Palmieri Weber R, Wentzensen N, Whittemore AS, Wik E, Wilkens LR, Winterhoff B, Ling Woo Y, Wu AH, Xiang Y-B, Yang HP, Zheng W, Ziogas A, Zulkifli F, Phelan CM, Iversen E, Schildkraut JM, Berchuck A, Fridley BL, Goode EL, Pharoah PDP, Monteiro ANA, Sellers TA, Gayther SA (2013) Identification and molecular characterization of a new ovarian cancer susceptibility locus at 17q21.31. Nat Commun 4:1627 [DOI] [PMC free article] [PubMed]
- 76.Qian L, Vu MN, Carter M, Wilkinson MF. A spliced intron accumulates as a lariat in the nucleus of T cells. Nucleic Acids Res. 1992;20(20):5345–5350. doi: 10.1093/nar/20.20.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clement JQ, Qian L, Kaplinsky N, Wilkinson MF. The stability and fate of a spliced intron from vertebrate cells. RNA. 1999;5(2):206–220. doi: 10.1017/S1355838299981190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Coleclough C, Wood D. Introns excised from immunoglobulin pre-mRNAs exist as discrete species. Mol Cell Biol. 1984;4(10):2017–2022. doi: 10.1128/MCB.4.10.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Michaeli T, Pan ZQ, Prives C. An excised SV40 intron accumulates and is stable in Xenopus laevis oocytes. Genes Dev. 1988;2(8):1012–1020. doi: 10.1101/gad.2.8.1012. [DOI] [PubMed] [Google Scholar]
- 80.Kopczynski CC, Muskavitch MA. Introns excised from the Delta primary transcript are localized near sites of Delta transcription. J Cell Biol. 1992;119(3):503–512. doi: 10.1083/jcb.119.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu Z, Filonov GS, Noto JJ, Schmidt CA, Hatkevich TL, Wen Y, Jaffrey SR, Matera AG. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA. 2015;21(9):1554–1565. doi: 10.1261/rna.052944.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kulesza CA, Shenk T. Human cytomegalovirus 5-kilobase immediate-early RNA is a stable intron. J Virol. 2004;78(23):13182–13189. doi: 10.1128/JVI.78.23.13182-13189.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kulesza CA, Shenk T. Murine cytomegalovirus encodes a stable intron that facilitates persistent replication in the mouse. Proc Natl Acad Sci USA. 2006;103(48):18302–18307. doi: 10.1073/pnas.0608718103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Keohavong P, Gattoni R, Schmitt P, Stevenin J. The different intron 2 species excised in vivo from the E2A premRNA of adenovirus-2: an approach to analyse alternative splicing. Nucleic Acids Res. 1986;14(13):5207–522741. doi: 10.1093/nar/14.13.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]