Abstract
NFκB is one of the central regulators of cell survival, immunity, inflammation, carcinogenesis and organogenesis. The activation of NFκB is strictly regulated by several posttranslational modifications including phosphorylation, neddylation and ubiquitination. Several types of ubiquitination play important roles in multi-step regulations of the NFκB pathway. Some of the tripartite motif-containing (TRIM) proteins functioning as E3 ubiquitin ligases are known to regulate various biological processes such as inflammatory signaling pathways. One of the TRIM family proteins, TRIM39, for which the gene has single nucleotide polymorphisms, has been identified as one of the genetic factors in Behcet’s disease. However, the role of TRIM39 in inflammatory signaling had not been fully elucidated. In this study, to elucidate the function of TRIM39 in inflammatory signaling, we performed yeast two-hybrid screening using TRIM39 as a bait and identified Cactin, which has been reported to inhibit NFκB- and TLR-mediated transcriptions. We show that TRIM39 stabilizes Cactin protein and that Cactin is upregulated after TNFα stimulation. TRIM39 knockdown also causes activation of the NFκB signal. These findings suggest that TRIM39 negatively regulates the NFκB signal in collaboration with Cactin induced by inflammatory stimulants such as TNFα.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-015-2040-x) contains supplementary material, which is available to authorized users.
Keywords: RelA/p65, IκBα, TLR, Ubiquitin, Ubiquitin ligase, E3
Introduction
NFκB functions as an important regulator for cell survival, immunity, inflammation, carcinogenesis and organogenesis [1, 2]. Under a resting condition, NFκB is maintained in an inactive state within the cytoplasm by binding to IκB proteins, which mask the nuclear localization signal (NLS) of NFκB. Upon inflammatory stimulation, IκB kinases (IKK), including IKKα and IKKβ, phosphorylate IκB proteins, leading to degradation of IκB proteins. NFκB is then free to enter the nucleus to activate transcription of a number of genes [2].
In this NFκB signaling pathway, ubiquitination plays important roles in regulation of the activation of NFκB. Skp1-Cul1-F-box protein complex (SCF)-type ubiquitin ligase, SCFβ−TrCP, interacts with and ubiquitinates IκB protein phosphorylated by IKK [3, 4]. Furthermore, the activation of IKK requires K63-linked polyubiquitin [2]. It has also been reported that linear ubiquitin chains are necessary for NFκB activation [5]. These findings indicate that activation of the NFκB signaling pathway greatly depends on several steps of ubiquitination.
Ubiquitination is an important posttranslational modification used by eukaryotic cells. This modification has a crucial role in multiple cellular functions including cellular signaling, the cell cycle, organelle biogenesis, morphogenesis, and DNA repair [6]. Conjugation of ubiquitin to the target protein is catalyzed by several components, including ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3) [7]. The resulting covalent ubiquitin ligations form polyubiquitinated conjugates. E3 ubiquitin ligases are most directly responsible for recognition of the substrate in the ubiquitin conjugation system [8]. According to their structure, E3 ubiquitin ligases have been classified into three groups: HECT family [9], RING family [10] and U-box family [11].
Tripartite motif-containing (TRIM) proteins are defined as proteins that consist of a RING domain, one or two B-boxes, a coiled-coil motif, and carboxyl-terminal unique domains [12]. TRIM family proteins are involved in various biological processes, and their alterations, therefore, often lead to diverse clinical disorders, such as developmental disorders, neurodegenerative diseases, vulnerability to viral infection, and carcinogenesis [13, 14]. TRIM39 is one of the TRIM family proteins and has an E3 activity, leading to the degradation of some proteins [15], while TRIM39 has been reported to stabilize some proteins [16]. The TRIM39 gene is located in the MHC region within chromosome 6p21–23, in which there are six other TRIM genes (TRIM10, TRIM15, TRIM26, TRIM27, TRIM31 and TRIM38) [17] and many genes that are related to immune response [18]. It has been reported that approximately half of the TRIM proteins are involved in inflammatory signaling pathways [19] and that TRIM39 is involved in the regulation of type I interferon response [20]. In addition, a single nucleotide polymorphism (SNP) on exon 9 of TRIM39 has been identified as one of the genetic factors in Behcet’s disease by multiple logistic regression analysis [21]. These findings suggest that TRIM39 is related to inflammatory signaling pathways. However, the role of TRIM39 in inflammatory signaling has not been fully elucidated.
In this study, with the aim of elucidating the molecular function of TRIM39 in inflammatory signaling pathways, we identified Cactus interactor (Cactin) as one of the TRIM39-interacting proteins. We found that TRIM39 suppresses the NFκB signal through stabilization of Cactin and that Cactin provides a negative feedback loop of the NFκB signaling pathway, suggesting that TRIM39 and Cactin function as novel negative regulators for inflammatory signaling pathways.
Materials and methods
Cell culture
HEK293T and HeLa cells were cultured under an atmosphere of 5 % CO2 at 37 °C in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich Corp., St Louis, MO) supplemented with 10 % fetal bovine serum (FBS) (Gibco BRL, Paisley, UK). THP-1 cells were cultured under an atmosphere of 5 % CO2 at 37 °C in RPMI 1640 medium (Sigma) supplemented with 10 % FBS. Sf9 cells were cultured at 27 °C in Sf-900 medium (Life Technologies, Carlsbad, CA) supplemented with 5 % FBS.
Cloning of cDNAs and plasmid construction
Human TRIM39 and Cactin cDNAs were amplified from a human B cell cDNA library (Clontech, Palo Alto, CA) by polymerase chain reaction (PCR) with KOD FX (Toyobo, Osaka, Japan) using the following primers: 5′-ATTATGGCAGAGACAAGTCTG-3′ (HsTRIM39-sense), 5′-CTGTCACTCCCAATCTGTTGG-3′ (HsTRIM39-antisense), 5′-AAAGAATTCATGGGTCGGGACACACGCT-3′ (HsCactin-sense), and 5′-AAACTCGAGCCGTCACCGCCGATAGCGGTA-3′ (HsCactin-antisense). Human TRAF2 cDNA was kindly provided by Dr. Miyuki Bohgaki (Hokkaido University). An expression vector encoding IKKβ cDNA was described previously [3]. The amplified fragments were subcloned into pBluescript II SK + (Stratagene, La Jolla, CA). Each deletion mutant of TRIM39 ΔR, ΔB, ΔC and ΔS, which was used as TRIM39 lacking a RING domain, B-box domain, coiled-coil domain, and SPRY domain, respectively, was amplified by PCR with pBSIISK-TRIM39 as a template and the following primers: 5′-AAGACATCCCGCTACCGC-3′ (TRIM39 ΔR-sense), 5′-CTGTCACTCCCAATCTGTTGG-3′ (TRIM39 ΔR-antisense), 5′-GATCTTCCGCTTGACGGCCTG-3′ (TRIM39 ΔB-sense), 5′-GACGATGCTACACAGGAGTAC-3′ (TRIM39 ΔB-antisense), 5′- ACCAGGCTTCTTCTCCTCAGA-3′ (TRIM39 ΔC-sense), 5′- TGCTTACAGTCAGGCTTCGAG-3′ (TRIM39 ΔC-antisense), 5′-TCACTGTTTAAGGATTTTCCTTAG-3′ (TRIM39 ΔS-sense), and 5′-CAGATCAAGCTTATCGATACC-3′ (TRIM39 ΔS-antisense). TRIM39 SNP was generated by PCR with KODFX, pBSIISK-TRIM39 and the following primer pair: 5′-GGGTCCAGGGTCACATCCGCA-3′ (TRIM39 SNP-sense) and 5′-CGAGACAGCTCATCCTAACCTA-3′ (TRIM39 SNP-antisense). TRIM39 siR, which was used as siRNA-resistant form of TRIM39, was generated by PCR mutagenesis with KODFX, TRIM39-pBSIISK, and the following primer pairs: 5′-CGTAAATCTCCGCGGTGTGTCAGGGAGATCCCGGAG-3′ (TRIM39 siR-sense) and 5′-TTTTATCCATGCGTCCTGGCTACTGAGGGTT-3′ (TRIM39 siR-antisense). All of the sequences were confirmed by the dideoxy chain termination method with automated sequencing (Applied Biosystems, Foster City, CA). The resulting fragments containing TRIM39, TRIM39 ΔR, ΔB, ΔC, ΔS, SNP and siR, and Cactin cDNAs were then subcloned into the pCR vector (Invitrogen, Carlsbad, CA) for expression in mammalian cells, into the pBGK vector for a yeast two-hybrid system, and into the pFastBac vector for a baculovirus protein expression system.
Yeast two-hybrid screening
Complementary DNA encoding the full length and deletion mutants of human TRIM39 were fused in-frame to the nucleotide sequence for the LexA domain (BD) in the yeast two-hybrid vector pBGK. To screen for proteins that interact with TRIM39, the yeast strain L40 (Invitrogen) was transformed with a B cell Matchmaker cDNA library (Clontech) using the lithium acetate method. To analyze interactions between TRIM39 deletion mutants and Cactin, the yeast strain L40 was cotransformed with pBGK-TRIM39 deletion mutants and pACT-Cactin, and then the yeast cells containing both vectors were subjected to a β-galactosidase assay.
Transfection, immunoprecipitation and immunoblot analysis, and subcellular fractionation
HEK293T cells were transfected by the calcium phosphate method and then cultured for 48 h. The cells were lysed in a solution containing 50 mM Tris–HCl (pH 7.4), 300 mM NaCl, 1 % Triton X-100, leupeptin (10 mg/ml), 1 mM phenylmethylsulfonyl fluoride, 400 μM Na3VO4, 400 μM EDTA, 10 mM NaF and 10 mM sodium pyrophosphate. The cell lysates were centrifuged at 15,000g for 15 min at 4 °C, and the resulting supernatant was incubated with the indicated antibody for 2 h at 4 °C. Protein A-Sepharose (Amersham Biosciences, Piscataway, NJ) that had been equilibrated with the same solution was added to the mixture and then rotated for 1 h at 4 °C. The resin was washed five times with ice-cold lysis buffer and then boiled in an SDS sample buffer. In an immunoprecipitation assay with anti-FLAG antibody, proteins on the resin were eluted with a buffer containing FLAG peptides (F3290, Sigma), instead of boiling in an SDS sample buffer. Immunoblot analysis was performed with the indicated antibodies. Immune complexes were detected with horseradish peroxidase-conjugated antibodies to mouse (1:6666 dilution, GE Healthcare, Piscataway, NJ) or rabbit IgG (1:6666 dilution, Sigma) and an enhanced chemiluminescence system (GE Healthcare). The antibodies used in this study were as follows: mouse monoclonal anti-FLAG (M2, Sigma), mouse monoclonal anti-Myc (9E10, Roche Molecular Biochemicals, Mannheim), mouse monoclonal anti-GST (B-14, Santa Cruz Biotechnology, Santa Cruz, CA), rabbit polyclonal anti-TRIM39 (Sigma), rabbit polyclonal anti-C19orf29 (Cactin, Sigma), mouse monoclonal anti-GAPDH (6C5, Ambion, Austin, TX), mouse monoclonal anti-c-Myc (Covance, Princeton, NJ), mouse monoclonal anti-lamin A/C (612162, BD Pharmingen, San Jose, CA), mouse monoclonal anti-human IκBα (610690, BD), mouse monoclonal anti-human p-IκBα (5A5, Cell Signaling, Boston MA, USA), mouse monoclonal anti-NFκB RelA/p65 (610868, BD), rabbit polyclonal anti-IKKβ (2684, Cell Signaling), rabbit polyclonal anti-phospho-p38 (9211, CST) and rabbit polyclonal anti-p38 (9212, CST).
In biochemical subcellular fractionation, cytosolic and nuclear extract fractions were separated using stepwise lysis according to the protocol of manufacturer (NE-PER, Pierce, Rockford, IL).
Generation of recombinant proteins
His6-FLAG-tagged TRIM39 and His6-GST-Cactin were expressed in Sf9 cells using a baculovirus protein expression system (Invitrogen). The recombinant His6-tagged proteins were purified using ProBond metal affinity beads (Invitrogen).
In vitro binding assay
Each of the purified recombinant proteins was mixed in a solution containing 50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 % Triton X-100, leupeptin (10 mg/ml), 1 mM phenylmethylsulfonyl fluoride, 400 μM Na3VO4, 400 μM EDTA, 10 mM NaF, and 10 mM sodium pyrophosphate. The mixtures of purified recombinant proteins were incubated with anti-FLAG antibody for 2 h at 4 °C. After addition of protein A-Sepharose, each of the mixtures was rotated for 1 h at 4 °C. The resin was washed five times with ice-cold lysis buffer as mentioned above and then eluted with a buffer containing FLAG peptides. Immunoblot analysis was performed with anti-FLAG, anti-GST and anti-C19orf29 antibodies. Immune complexes were detected with horseradish peroxidase-conjugated antibodies and an enhanced chemiluminescence system.
RNA interference
siRNA (12 μM) was transfected into HEK293T cells or HeLa cells with Lipofectamine RNAiMAX (Invitrogen). The siRNAs used in this study were as follows: siTRIM39-1 (5′-CCAAGGCGUUUCACCUUCUACCCUU-3′, Invitrogen), siTRIM39-2 (5′-GGCAACUAGGCAGUAUGGUGGAAAU-3′, Invitrogen), siCactin (5′-GCGGCCCAAGUGGCACUCAAGAGAU-3′, Invitrogen), and a corresponding non-targeting siRNA (5′–UAAGGCUAUGAAGAGAUAC-3′, Thermo Scientific, Rockford, IL).
Protein stability assay with cycloheximide
HEK293T cells were transfected with expression vectors encoding FLAG-tagged TRIM39 WT, FLAG-tagged TRIM39 ΔR, or siRNA specific for human TRIM39 (siTRIM39) as mentioned above. As a control, pcDNA3 (Mock) or non-targeting siRNA (siControl) was used. Forty-eight hours after transfection, the cells were cultured in the presence of cycloheximide (50 μg/mL) for the indicated times. Cell lysates were subjected to immunoblot analysis with anti-Cactin, anti-TRIM39, anti-Myc and anti-GAPDH antibodies. The intensity of the endogenous Cactin bands was normalized to that of the corresponding GAPDH bands and is indicated as percentages of the normalized value at 0 h. Anti-Myc immunoblot analysis was performed to show that cycloheximide used in this assay is effective.
Real-time PCR
Total RNA was isolated from HEK293T or HeLa cells using ISOGEN (Nippon Gene, Tokyo, Japan), followed by reverse transcription (RT) using ReverTra Ace (Toyobo, Osaka, Japan). The resulting cDNA was subjected to real-time PCR with a StepOne machine and Power SYBR Green PCR master mix (Applied Biosystems, Foster City, CA). The average threshold cycle (Ct) was determined from independent experiments and the level of gene expression relative to GAPDH was determined. The primer sequences for human TRIM39, human Cactin, human IL-6, human IL-8, and human GAPDH were as follows: human TRIM39, 5′-AAGGTGAAGACCATGGAGGTGACT-3′ and 5′-TAGGTTAGGATGAGCTGTCTCAGGGT-3′; human Cactin, 5′-TACACCAACACCGACAACCC-3′ and 5′-TTCAGCTCCTTCTCCTCCAG-3′; human IL-6, 5′-AGCCACTCACCTCTTCAGAACGAATTGACA-3′ and 5′-AGCATCCATCTTTTTCAGCCATCTTTG-3′; human IL-8, 5′-TCCAAACCTTTCCACCCCAAATTTATCAA-3′ and 5′-GCCCTCTTCAAAAACTTCTCCACAACCCTC-3′; and human GAPDH, 5′-GCAAATTCCATGGCACCGT-3′ and 5′-TCGCCCCACTTGATTTTGG-3′.
Dual-luciferase assay
HEK293T cells or HeLa cells were inoculated into a 24-well dish plate at 2 × 104 cells per well and incubated at 37 °C with 5 % CO2 for 24 h. Expression vectors, reporter plasmids and the internal control plasmid coding Renilla luciferase were transfected into the cells using the Fugene HD reagent (Roche). Forty-eight hours after transfection, cells were incubated with TNFα (20 ng/ml) for 6 h or with PolyIC (1 μg/ml) for 24 h, harvested, and assayed by the Dual-Luciferase Reporter Assay System (Promega Corporation, Madison, WI). The luminescence was quantified with a luminometer (Promega). A reporter plasmid containing ISRE was kindly provided by Dr. Hirano (Osaka University) [22]. A reporter plasmid containing the NFκB-responsive sequence was described previously [23].
Statistical analysis
Student’s t test was used to determine the statistical significance of experimental data.
Results
TRIM39 directly binds to Cactin in vivo and in vitro
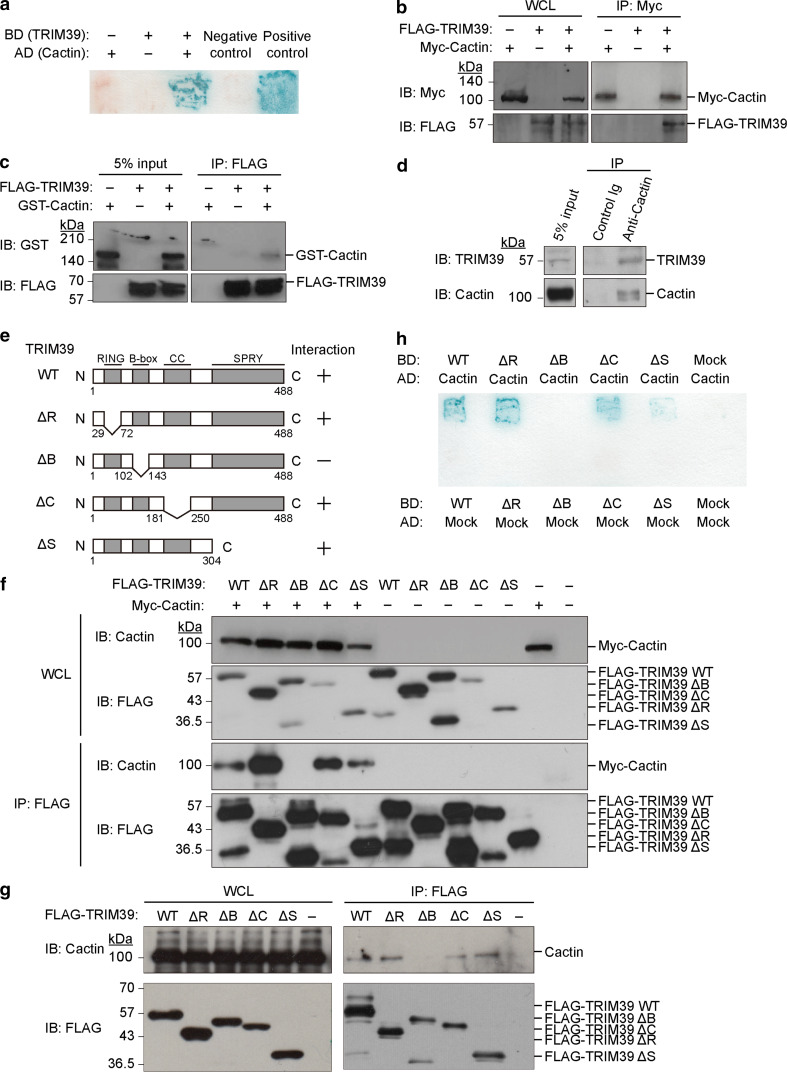
To clarify the function of TRIM39, we isolated TRIM39-interacting proteins from a human B cell cDNA library by yeast two-hybrid screens. Using TRIM39 as a bait, we obtained three positive clones from 2.3 × 105 transformants. One of the positive clones had sequence identities with cDNA encoding human Cactin (Fig. 1a). To examine whether TRIM39 interacts with Cactin in mammalian cells, we performed an in vivo binding assay using cells transfected with expression vectors. We expressed FLAG-tagged TRIM39 together with Myc-tagged Cactin in HEK293T cells. Cell lysates were subjected to immunoprecipitation with the anti-Myc antibody, and the resulting precipitates were subjected to immunoblot analysis with the anti-FLAG antibody and anti-Myc antibody. FLAG-tagged TRIM39 was coprecipitated with Myc-tagged Cactin, indicating that TRIM39 specifically bound to Cactin in mammalian cells (Fig. 1b).
Fig. 1.
TRIM39 directly binds to Cactin. a Yeast two-hybrid screening to identify TRIM39-interacting proteins using a human B cell cDNA library. pBGK and pACT2 vectors (empty vectors) were used as negative controls. CHIP and EKN1 cDNAs were used for positive controls. b In vivo binding assay between TRIM39 and Cactin. Expression vectors encoding FLAG-tagged TRIM39 and Myc-tagged Cactin were transfected into HEK293T cells. Cell lysates (WCL) were immunoprecipitated with anti-Myc antibody and immunoblotted with anti-FLAG antibody and anti-Myc antibody. c In vitro binding assay between FLAG-tagged TRIM39 and GST-tagged Cactin. Each recombinant protein was generated by a baculovirus protein expression system and purified using Probond metal affinity beads. Recombinant FLAG-tagged TRIM39 and GST-tagged Cactin were mixed in combination as indicated. The reaction mixtures were immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-FLAG antibody and anti-GST antibody. d In vivo binding assay between endogenous TRIM39 and endogenous Cactin in THP-1 cells. Cell lysates were immunoprecipitated with anti-Cactin antibody or control Ig and then immunoblotted with an anti-TRIM39 or anti-Cactin antibody. e Schematic representation of TRIM39 and deletion mutants. f Interaction between deletion mutants of TRIM39 and Myc-tagged Cactin in vivo. Expression vectors encoding FLAG-tagged TRIM39 WT or deletion mutants were transfected with Myc-tagged Cactin into HEK293T cells. Cell lysates (WCL) were immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-FLAG antibody and anti-Cactin antibody. g Interaction between deletion mutants of TRIM39 and endogenous Cactin in vivo. Expression vectors encoding FLAG-tagged TRIM39 WT or deletion mutants were transfected into HEK293T cells. Cell lysates (WCL) were immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-Cactin antibody and anti-FLAG antibody. h Interaction between deletion mutants of TRIM39 and Cactin in yeast. Yeast strain L40 was transformed with pBGK-TRIM39 (WT or deletion mutants) and pACT-Cactin. A β-galactosidase assay was performed to show the interaction of deletion mutants with Cactin
We also performed an in vitro binding assay using recombinant TRIM39 and Cactin proteins. Recombinant His6-FLAG-tagged TRIM39 and His6-GST-tagged Cactin proteins were generated by the baculovirus protein expression system and purified using ProBond metal affinity beads. These recombinant proteins were mixed and then subjected to immunoprecipitation with the anti-FLAG antibody and to immunoblot analysis with the anti-FLAG and anti-GST antibodies. Immunoblot analysis showed that His6-GST-tagged Cactin was coprecipitated with His6-FLAG-tagged TRIM39, suggesting direct interaction between TRIM39 and Cactin (Fig. 1c).
Furthermore, we confirmed the interaction between endogenous TRIM39 and endogenous Cactin using a human acute monocytic leukemia cell line, THP-1. Immunoblot analysis showed specific interaction between endogenous TRIM39 and endogenous Cactin (Fig. 1d).
To determine which domain of TRIM39 is required for interaction with Cactin, we performed in vivo binding assays using deletion mutants of TRIM39 which are lacking the RING domain, B-Box domain, coiled-coil domain or SPRY domain (Fig. 1e). We transfected expression vectors encoding a FLAG-tagged deletion mutant of TRIM39 together with Myc-tagged Cactin into HEK293T cells. Cell lysates were subjected to immunoprecipitation with the anti-FLAG antibody, and the resulting precipitates were subjected to immunoblot analysis with the anti-Cactin and anti-FLAG antibodies. Only TRIM39 ΔB was not coprecipitated with Myc-tagged Cactin (Fig. 1f). Furthermore, we found the TRIM39 ΔB does not interact with endogenous Cactin (Fig. 1g). To further confirm which domains of TRIM39 are required for the interaction with Cactin, we used a yeast two-hybrid system. Yeast strain L40 was cotransformed with several deletion mutants of TRIM39 and pACT-Cactin. β-galactosidase assays showed that only TRIM39 ΔB did not interact with Cactin in yeast cells, indicating that the B-box domain of TRIM39 was required for interaction with Cactin (Fig. 1h).
A SNP in the TRIM39 gene has been identified as one of the genetic factors in Behcet’s disease [21]. The SNP in the TRIM39 gene is a silent mutation: CCT to CCC. However, we cannot rule out the possibility that the SNP causes a change of TRIM39 expression at the transcriptional or translational level. We tested whether the SNP affects the expression level of TRIM39 or changes the affinity of TRIM39 with Cactin. We transfected equal amounts of expression vectors encoding FLAG-tagged wild-type TRIM39 (TRIM39 WT) or FLAG-tagged TRIM39 SNP into HEK293T cells and we compared the expression levels of TRIM39 WT and TRIM39 SNP at the protein level. Immunoblot analysis showed no difference at the protein expression level between FLAG-tagged TRIM39 WT and FLAG-tagged TRIM39 SNP (Suppl. Figure 1a). We also examined whether the SNP of TRIM39 affects the interaction between TRIM39 and Cactin. Immunoblot analysis showed that TRIM39 SNP interacted with Cactin as TRIM39 WT did (Suppl. Figure 1b).
TRIM39 interacts with Cactin in the nucleus
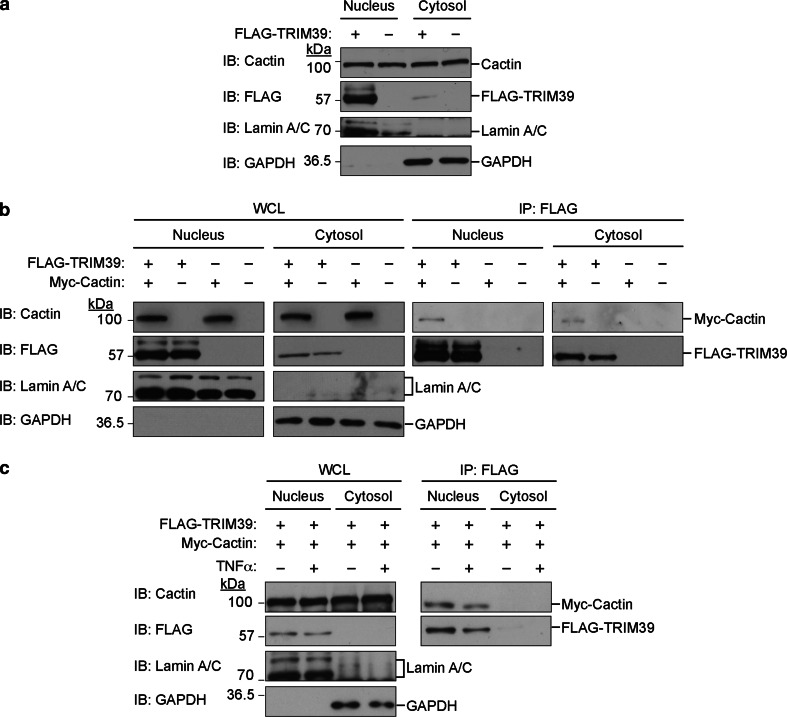
We examined the subcellular localization of TRIM39 in cells. We biochemically separated the lysate from HEK293T cells expressing FLAG-tagged TRIM39 into nuclear and cytosol fractions. Immunoblot analysis showed that FLAG-tagged TRIM39 was predominantly localized in the nucleus, whereas endogenous Cactin was located both in the nucleus and cytosol (Fig. 2a).
Fig. 2.
Interaction between TRIM39 and Cactin in the nucleus. a TRIM39 is mainly located in the nucleus. Expression vectors encoding FLAG-tagged TRIM39 were transfected into HEK293T cells. The cytoplasmic extract (cytosol) and nuclear extract (nucleus) were biochemically fractionated and analyzed by western blotting with the indicated antibodies. b TRIM39 interacts with Cactin mainly in the nucleus. Expression vectors encoding FLAG-tagged TRIM39 were transfected with Myc-tagged Cactin into HEK293T cells. The cytoplasmic extract (cytosol) and nuclear extract (nucleus) were biochemically fractionated and immunoprecipitated with anti-FLAG antibody. The resulting precipitates were subjected to immunoblot analysis with the indicated antibodies. c The interaction between TRIM39 and Cactin is independent of TNFα stimulation. Expression vectors encoding FLAG-tagged TRIM39 were transfected with Myc-tagged Cactin into HEK293T cells. Forty-eight hours after transfection, cells were cultured with TNFα (20 ng/ml) for another 6 h. Then, the cytoplasmic extract (cytosol) and nuclear extract (nucleus) were biochemically fractionated and immunoprecipitated with anti-FLAG antibody. The resulting precipitates were subjected to immunoblot analysis with the indicated antibodies
To clarify the cell compartment in which TRIM39 interacts with Cactin, the lysate from HEK293T cells exogenously expressing Myc-tagged Cactin and/or FLAG-tagged TRIM39 was fractionated into nuclear and cytosol extracts and subjected to immunoprecipitation with the anti-FLAG antibody. Immunoblot analysis showed that Myc-tagged Cactin was coprecipitated with FLAG-tagged TRIM39 in the nuclear fractions, indicating that TRIM39 mainly interacts with Cactin in the nucleus (Fig. 2b).
We also examined whether TNFα stimulation affects the localization of TRIM39 and Cactin and whether TNFα stimulation changes the interaction level between TRIM39 and Cactin. We transfected expression vectors encoding FLAG-tagged TRIM39 and Myc-tagged Cactin into HEK293T cells. Six hours after TNFα stimulation, the cytoplasmic and nuclear fractions were subjected to immunoprecipitation with the anti-FLAG antibody. Immunoblot analysis showed that TNFα stimulation did not affect localization of TRIM39 and Cactin or the interaction level between TRIM39 and Cactin (Fig. 2c).
TRIM39 stabilizes Cactin
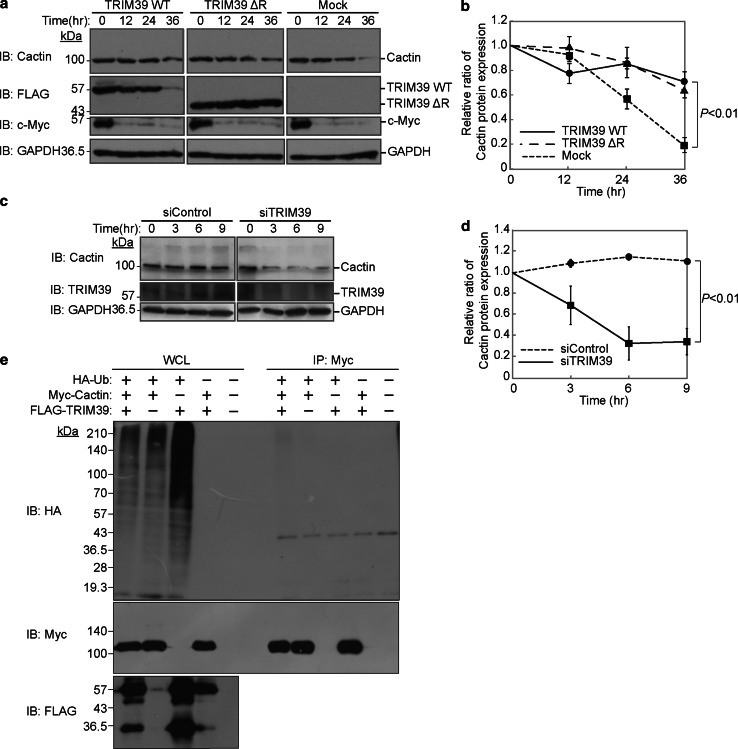
Based on our previous findings that some of the TRIM family proteins affect the stabilities of their binding proteins [13, 14, 24–26], we speculated that TRIM39 affects the stability of Cactin. Although we assumed that TRIM39 promotes degradation of Cactin, protein stability analysis with cycloheximide revealed that overexpression of TRIM39 WT or that of ΔRING promoted the stabilization of Cactin protein (Fig. 3a, b). Next, we knocked down endogenous TRIM39 in HEK293T cells and performed a protein stability assay. HEK293T cells in which endogenous TRIM39 had been knocked down were incubated with cycloheximide for 0–9 h. Protein stability analysis showed that knockdown of TRIM39 caused a decrease of endogenous Cactin, suggesting that TRIM39 upregulates the stability of Cactin protein (Fig. 3c, d).
Fig. 3.
TRIM39 stabilizes Cactin. a Pulse-chase analysis of Cactin with TRIM39. HEK293T cells were transfected with expression vectors for FLAG-tagged TRIM39 WT, FLAG-tagged TRIM39 ΔRING, or an empty vector (Mock). Forty-eight hours after transfection, the cells were cultured in the presence of cycloheximide (50 μg/mL) for the indicated times. Cell lysates were then subjected to immunoblot analysis with anti-Cactin, anti-FLAG, anti-Myc and anti-GAPDH antibodies. b Intensity of the endogenous Cactin bands in the protein stability analysis in a was normalized to that of the corresponding GAPDH bands and is indicated as a ratio of the normalized value at 0 h. Data are mean ± standard deviation (SD) of values from three independent experiments. P values for the indicated comparisons were determined by Student’s t test. c Protein stability assay of Cactin in HEK293T cells in which TRIM39 had been knocked down. HEK293T cells were transfected with siRNA specific for human TRIM39 or with non-targeting siRNA (siRNA negative control). Forty-eight hours after transfection, the cells were cultured in the presence of cycloheximide (50 μg/mL) for the indicated times. Cell lysates were subjected to immunoblot analysis with anti-Cactin, anti-TRIM39 and anti-GAPDH antibodies. d Intensity of the endogenous Cactin bands in the protein stability assay in c was normalized to that of the corresponding GAPDH bands and is indicated as a ratio of the normalized value at 0 h. Data are mean ± standard deviation (SD) of values from three independent experiments. P values for the indicated comparisons were determined by Student’s t test. e An in vivo ubiquitination assay for Cactin by TRIM39. Expression vectors for Myc-tagged Cactin, FLAG-tagged TRIM39, and HA-tagged ubiquitin were transfected into HEK293T cells. Cell lysates were immunoprecipitated with anti-Myc antibody and then anti-HA immunoblot analysis was performed to detect the ubiquitination of Cactin
Since TRIM39 has a RING-finger domain, which is an enzymatic active domain for E3 ubiquitin ligases, we performed an in vivo ubiquitination assay to determine whether TRIM39 ubiquitinates Cactin. The in vivo ubiquitination assay showed that TRIM39 did not promote polyubiquitination of Myc-tagged Cactin, suggesting that TRIM39 stabilizes Cactin in an ubiquitination-independent manner (Fig. 3e).
Cactin provides a negative feedback loop in the NFκB pathway
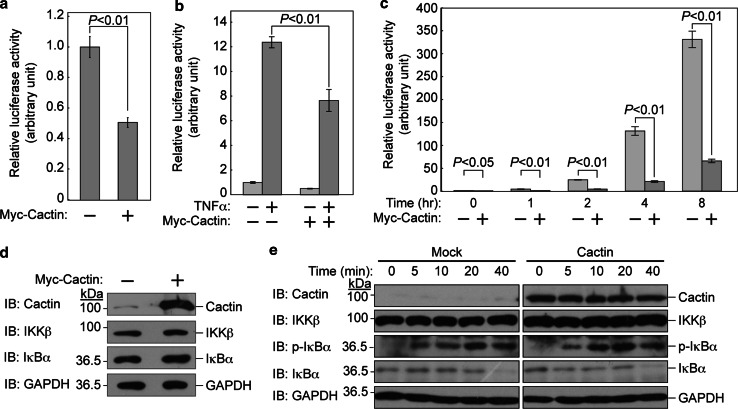
To confirm that Cactin negatively regulates NFκB-mediated transcription [27], we performed a luciferase reporter assay using an NFκB promoter-driven luciferase construct. We transfected expression vectors encoding Myc-tagged Cactin with reporter plasmids into HEK293T cells. After stimulation with TNFα, luciferase activity was measured. We found that Cactin significantly suppressed NFκB-mediated transcriptional activity not only with TNFα stimulation but also without TNFα stimulation (Fig. 4a–c).
Fig. 4.
Cactin negatively regulates the NFκB signal. a Cactin suppresses NFκB activity even in an inactive state without TNFα stimulation. HEK293T cells were transfected with an NFκB luciferase reporter plasmid and an expression plasmid encoding Myc-tagged Cactin. Thirty hours after transfection, cells were assayed by the Dual-Luciferase Reporter Assay System. Luciferase activity was normalized to the Renilla luciferase reporter construct and expressed relative to the normalized activity of control cells without Myc-tagged Cactin. Data are mean ± standard deviation (SD) of values from three independent experiments. P values for the indicated comparisons were determined by Student’s t test. b Cactin suppresses TNFα-induced NFκB activity. HEK293T cells were transfected with an NFκB luciferase reporter plasmid and an expression plasmid encoding Myc-tagged Cactin. Twenty-four hours after transfection, cells were cultured with TNFα (20 ng/ml) for another 6 h and assayed by the Dual-Luciferase Reporter Assay System. Luciferase activity was normalized to the Renilla luciferase reporter construct and expressed relative to the normalized activity of control cells without TNFα stimulation. Data are mean ± standard deviation (SD) of values from three independent experiments. P values for the indicated comparisons were determined by Student’s t test. c Time-course analysis of TNFα-induced NFκB activity suppressed by Cactin. Luciferase activity was measured at the indicated times after TNFα stimulation. d Cactin did not affect cytoplasmic regulators in the NFκB signaling pathway in an inactive state. HEK293T cells were transfected with an expression plasmid encoding Myc-tagged Cactin. Thirty hours after transfection, cell lysates were subjected to immunoblot analysis with the indicated antibodies. e Cactin did not affect cytoplasmic regulators in the NFκB signaling pathway in an active state. HEK293T cells were transfected with an expression plasmid encoding Myc-tagged Cactin. Thirty hours after transfection, cells were cultured with TNFα (20 ng/ml) for the indicated times. Then, cell lysates were subjected to immunoblot analysis with the indicated antibodies
To elucidate the level at which Cactin negatively acts in the NFκB signaling pathway, we examined whether Cactin overexpression affects IKKβ and IκBα under the condition of no stimulation. Immunoblot analysis revealed that Cactin overexpression did not affect the regulators of the NFκB signaling pathway in an inactive state (Fig. 4d). We next examined whether Cactin overexpression affects IKKβ, IκBα and phosphorylated IκBα after TNFα stimulation. After TNFα stimulation, cell lysates were subjected to immunoblot analysis at the indicated times. Cactin overexpression did not affect cytosolic regulators in the NFκB signaling pathway in an active state as well as an inactive state (Fig. 4e).
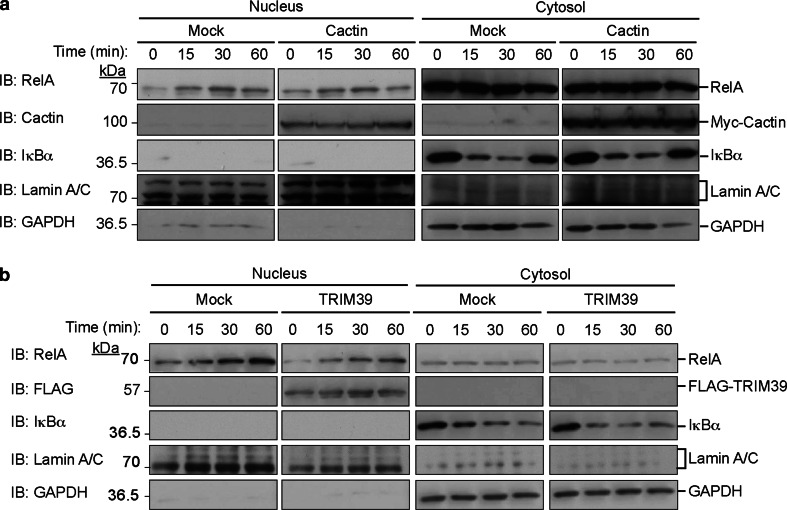
Furthermore, we examined whether Cactin overexpression affects the translocation of NFκB into the nucleus after TNFα stimulation. After TNFα stimulation, cell lysates were fractionated into cytoplasmic and nuclear extracts at the indicated times. Immunoblot analysis revealed that Cactin overexpression did not affect the translocation of NFκB (RelA/p65) into the nucleus (Fig. 5a). We next examined whether TRIM39 overexpression affects the nuclear translocation of NFκB. TRIM39 overexpression did not change the level of nuclear translocation of RelA/p65 (Fig. 5b). We also investigated whether the combination of TRIM39 WT and Cactin overexpression or the combination of TRIM39 ΔB and Cactin overexpression affects the nuclear translocation of NFκB. Neither of the combinations of overexpression changed the level of nuclear translocation of RelA/p65 (Suppl. Figure 2). These findings suggest that TRIM39 and Cactin affect NFκB-mediated transcriptional activity after the translocation of RelA/p65 into the nucleus.
Fig. 5.
Translocation of RelA/p65 after stimulation of TNFα with Cactin or TRIM39 overexpression. a Translocation of RelA/p65 after stimulation of TNFα with/without Cactin overexpression. HEK293T cells were transfected with an expression plasmid encoding Myc-tagged Cactin. Forty-eight hours after transfection, the cells were incubated with TNFα (20 ng/ml) for the indicated periods. The cytoplasmic extract (cytosol) and nuclear extract (nucleus) were biochemically fractionated and analyzed by immunoblotting with the indicated antibodies. b Translocation of RelA/p65 after TNFα stimulation with/without TRIM39 overexpression. HEK293T cells were transfected with an expression plasmid encoding FLAG-tagged TRIM39. Forty-eight hours after transfection, the cells were incubated with TNFα (20 ng/ml) for the indicated times. The cytoplasmic extract (cytosol) and nuclear extract (nucleus) were fractionated and analyzed by western blotting with the indicated antibodies
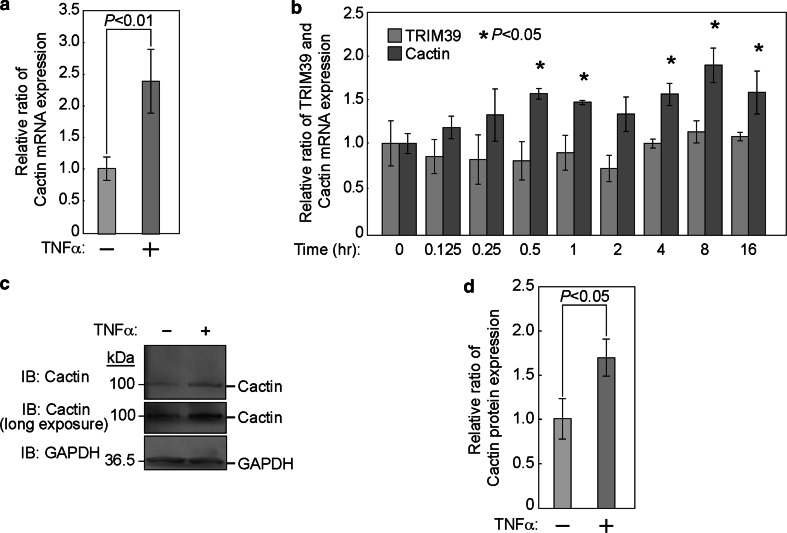
Given that Drosophila Cactin interacts with Cactus, which is known as an insect ortholog of mammalian IκBα [28], and that IκBα is an important negative regulator for NFκB [29], we hypothesized that Cactin may be one of the downstream genes of NFκB and may be induced by activation of NFκB. To examine whether Cactin is induced by activation of NFκB, we investigated the mRNA expression levels of Cactin after TNFα stimulation. Six hours after stimulation with TNFα, we performed quantitative PCR analysis. The mRNA level of Cactin was significantly increased by TNFα treatment (Fig. 6a, b), whereas the mRNA level of TRIM39 was not changed (Fig. 6b). Consistent with these results, immunoblot analysis showed that the protein level of Cactin was also increased by TNFα treatment (Fig. 6c, d).
Fig. 6.
Cactin provides a negative feedback regulation of the NFκB signal. a Quantification of the mRNA of Cactin by real-time quantitative PCR using HEK293T cells with or without TNFα stimulation. HEK293T cells were cultured with or without TNFα (20 ng/ml) for 6 h. Quantitative real-time PCR was then performed. Cactin mRNA expression is indicated as a ratio to that of untreated cells. Data are mean ± standard deviation (SD) of values from three independent experiments. P values for indicated comparisons were determined by Student’s t test. b Time-course analysis of mRNA of Cactin and TRIM39 after TNFα stimulation by quantitative real-time PCR. HEK293T cells were cultured with TNFα (20 ng/ml) for the indicated times. Quantitative real-time PCR was then performed. Cactin and TRIM39 mRNA expression is indicated as a ratio to that at 0 h. Data are mean ± standard deviation (SD) of values from three independent experiments. P values for indicated comparisons were determined by Student’s t test. c HEK293T cells were cultured with or without TNFα (20 ng/ml) for 6 h. Cell lysates were then subjected to immunoblot analysis with anti-Cactin and anti-GAPDH antibodies. d Intensity of the endogenous Cactin bands was normalized by that of GAPDH bands and expressed as a ratio of the normalized value to that of the untreated cells
Expression levels of IκBα and A20, known as components of negative feedback loops in the NFκB pathway, recover after TNFα stimulation. To clarify the change in expression of Cactin after TNFα stimulation, we performed time-course experiments using quantitative PCR. Quantitative PCR analysis showed that the mRNA level of Cactin significantly increased with a peak at 8 h after TNFα stimulation, whereas the mRNA level of TRIM39 did not change after TNFα stimulation (Fig. 6b). These findings suggested that Cactin provides a negative feedback loop in the NFκB pathway.
TRIM39 negatively regulates NFκB signaling
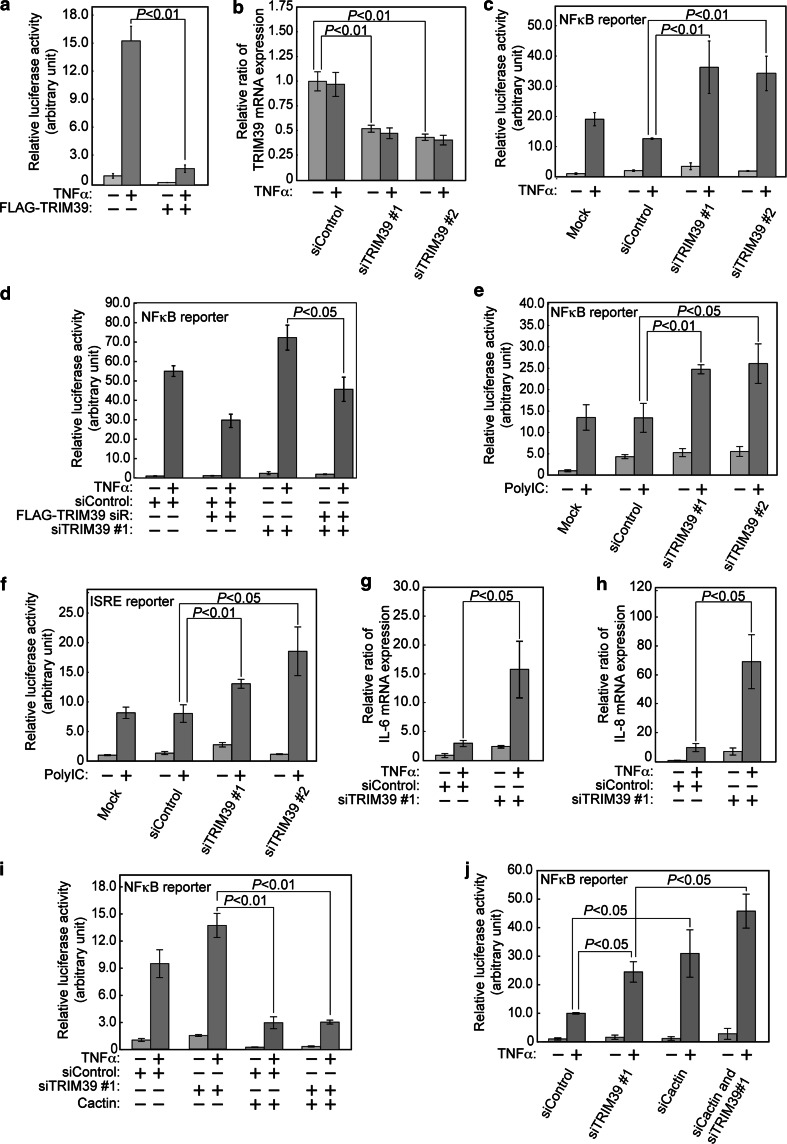
We previously reported that some members of the TRIM family of ubiquitin ligases regulate activities of several transcriptional factors. For example, we revealed that TRIM40, which is one of the TRIM family of ubiquitin ligases, enhances neddylation of IKKγ and inhibits the activity of NFκB-mediated transcription [25]. Moreover, we found that TRIM59 interacts with a signal adaptor protein, evolutionarily conserved signaling intermediate in Toll pathways (ECSIT), and negatively regulates NFκB and IRF3/7 signal pathways [24]. It has also been reported that Cactin inhibits NFκB and interferon-regulatory factor signaling pathways [27]. Based on these previous reports and our finding that TRIM39 stabilized Cactin, we hypothesized that TRIM39 acts as a regulator of transcription including the NFκB and Toll-like receptor (TLR) signaling pathways. To examine whether TRIM39 regulates NFκB-mediated transcriptional activity, we performed a luciferase reporter assay using an NFκB response element firefly luciferase reporter construct. We transfected expression vectors encoding FLAG-tagged TRIM39 with the reporter plasmids into HEK293T cells. Six hours after stimulation with TNFα, luciferase activity was measured. We found that TRIM39 overexpression significantly suppressed NFκB-mediated transcriptional activity (Fig. 7a).
Fig. 7.
TRIM39 negatively regulates NFκB and TLR signaling. a TRIM39 overexpression suppresses TNFα-induced NFκB activity. HEK293T cells were transfected with an expression plasmid encoding FLAG-tagged TRIM39 and NFκB reporter plasmids. Forty-eight hours after transfection, cells were incubated with TNFα (20 ng/ml) for 6 h, harvested, and assayed by the Dual-Luciferase Reporter Assay System. Luciferase activity is expressed as a ratio to the normalized activity of cells that had been transfected without FLAG-tagged TRIM39 and incubated without TNFα. Data are mean ± standard deviation (SD) of values from three independent experiments. b Quantification of mRNA of TRIM39 by real-time quantitative PCR using HeLa cells transfected with siRNA specific for human TRIM39 (siTRIM39) or with non-targeting siRNA as a control (siControl). HeLa cells were cultured with or without TNFα (20 ng/ml) for 6 h. Quantitative real-time PCR was then performed. TRIM39 mRNA expression is indicated as a ratio to the value of HeLa cells transfected with siControl without TNFα stimulation. Data are mean ± standard deviation (SD) of values from three independent experiments. P values for indicated comparisons were determined by Student’s t test. c TRIM39 knockdown enhances TNFα-induced NFκB activity. HeLa cells were transfected with the indicated siRNAs (siControl, siTRIM39#1 and siTRIM39#2) or without siRNA (Mock). Twenty-four hours after knockdown, NFκB reporter plasmids were transfected into cells. Forty-eight hours later, these cells were incubated with TNFα (20 ng/ml) for 6 h, harvested, and assayed by the Dual-Luciferase Reporter Assay System. Luciferase activity is expressed as a ratio to the normalized activity of cells that were not transfected with any siRNA and were incubated without TNFα. Data are mean ± standard deviation (SD) of values from three independent experiments. d HEK293T cells were transfected with siRNA specific for human TRIM39 (siTRIM39) or non-targeting siRNA as a control (siControl). Twenty-four hours after transfection, the cells were transfected with NFκB reporter plasmids and an expression plasmid encoding FLAG-tagged TRIM39 siR or an empty vector. Twenty-four hours later, the cells were incubated with TNFα (20 ng/ml) for a further 6 h and assayed by the Dual-Luciferase Reporter Assay System. Luciferase activity is expressed relative to the normalized activity of control cells without TNFα stimulation. Data are mean ± standard deviation (SD) of values from three independent experiments. P values for the indicated comparisons were determined by Student’s t test. e TRIM39 suppressed PolyIC-induced NFκB activity. NFκB reporter plasmids were transfected into HeLa cells in which TRIM39 had been knocked down. Forty-eight hours later, cells were incubated with PolyIC (1 μg/ml) for 12 h and assayed by the Dual-Luciferase Reporter Assay System. Luciferase activity is expressed as a ratio to the normalized activity of cells that were not transfected with any siRNA and were incubated without PolyIC. Data are mean ± standard deviation (SD) of values from three independent experiments. f TRIM39 suppressed PolyIC-induced ISRE activation. Experiments were performed as shown in c except that an ISRE reporter plasmid was used. g, h HEK293T cells were transfected with siRNA specific for human TRIM39 (siTRIM39) or non-targeting siRNA as a control (siControl). Seventy-two hours after transfection, these cells were incubated with TNFα (20 ng/ml) for 6 h. Total mRNA was purified and assayed by quantitative real-time PCR for levels of IL-6 and IL-8 mRNA. The expression levels of IL-6 and IL-8 were normalized relative to the expression of GAPDH. Data are presented relative to untreated cells transfected with siControl. Data are mean ± standard deviation (SD) of values from three independent experiments. i HEK293T cells were transfected with siRNA specific for human TRIM39 (siTRIM39) or non-targeting siRNA as a control (siControl). Forty-eight hours after transfection, these cells were transfected with NFκB reporter plasmids and an expression plasmid encoding Myc-tagged Cactin or an empty vector. Forty-eight hours later, these cells were incubated with TNFα (20 ng/ml) for a further 6 h and assayed by the Dual-Luciferase Reporter Assay System. Luciferase activity is expressed relative to the normalized activity of control cells without TNFα stimulation. Data are mean ± standard deviation (SD) of values from three independent experiments. P values for the indicated comparisons were determined by Student’s t test. j HEK293T cells were transfected with siRNA specific for human TRIM39 (siTRIM39), human Cactin (siCactin), and/or non-targeting siRNA as a control (siControl). Twenty-four hours after transfection, the cells were transfected with NFκB reporter plasmids. Twenty-four hours later, the cells were incubated with TNFα (20 ng/ml) for a further 6 h and assayed by the Dual-Luciferase Reporter Assay System. Luciferase activity is expressed relative to the normalized activity of control cells without TNFα stimulation. Data are mean ± standard deviation (SD) of values from three independent experiments. P values for the indicated comparisons were determined by Student’s t test
To further examine whether TRIM39 regulates the NFκB and TLR signaling pathways, we performed luciferase reporter assays using HeLa cells in which TRIM39 had been knocked down (Fig. 7b). Six or twelve hours after stimulation with TNFα or PolyIC, respectively, luciferase activities were measured. The luciferase assays showed that knockdown of TRIM39 significantly promoted NFκB-mediated transcriptional activity induced by TNFα or PolyIC (Fig. 7c, e). The luciferase assays also showed that TRIM39 knockdown promoted NFκB-mediated transcriptional activity induced after stimulation by TRAF2 or IKKβ overexpression (Suppl. Figure 3). To further confirm that TRIM39 regulates the NFκB signaling pathways, we added the siRNA-resistant TRIM39 (TRIM39 siR) back to TRIM39-knockdown cells. Six hours after stimulation with TNFα, luciferase activities were measured. We found that TRIM39 inhibited the NFκB-mediated transcriptional activity upregulated by TRIM39 knockdown with TNFα stimulation (Fig. 7d; Suppl. Figure 4). We also found that knockdown of TRIM39 promoted PolyIC-induced ISRE activity in HeLa cells (Fig. 7f).
Next, to examine whether TRIM39 affects the expression levels of target genes of NFκB, we measured the mRNA expression levels of IL-6 and IL-8 after TNFα stimulation. Six hours after stimulation with TNFα, we performed quantitative PCR analysis. The mRNA expression levels of IL-6 and IL-8 were significantly increased in TRIM39 knockdown HEK293T cells (Fig. 7g, h).
It has been reported that many NFκB target genes are regulated by mitogen-activated protein kinases (MAPKs) and that MAPKs are involved in TLR-induced NFκB target gene activation [30]. Therefore, we investigated whether TRIM39 affects MAPK activation. First, we examined whether MAPKs were activated by TNFα in HEK293T cells used in this study. HEK293T cells were incubated with TNFα (20 ng/ml) for the indicated times. Immunoblot analysis showed that phosphorylation of p38 (one of the MAPKs) in HEK293T cells was not affected by TNFα stimulation, whereas p38 was activated by FBS stimulation (Suppl. Figure 5a). The findings suggest that downregulation of IL-6 and IL-8 by TRIM39 knockdown was independent of p38, however, it remains to be seen if both the MEK/ERK and JNK pathways are involved. Next, we examined whether TRIM39 or Cactin affects MAPK activation by TNFα stimulation. HEK293T cells were transfected with expression vectors for FLAG-tagged TRIM39 WT, Myc-tagged Cactin, and an empty vector (Mock) and were cultured for 24 h. The cells were then incubated with TNFα (20 ng/ml) for the indicated times. Immunoblot analysis showed that overexpression of TRIM39 and Cactin did not affect phosphorylation of p38 by TNFα stimulation, suggesting that TRIM39 and Cactin are not involved in the MAPK signaling pathway (Suppl. Figure 5b). Taken together, these findings suggest that TRIM39 downregulates IL-6 and IL-8 expression via suppression of NFκB signaling.
TRIM39 negatively regulates the NFκB signaling pathway possibly via stabilization of Cactin
It was found that TRIM39 and Cactin negatively regulated NFκB activity and that TRIM39 stabilized Cactin protein. These findings suggest that negative regulation of NFκB activity by TRIM39 was caused at least through the stabilization of Cactin protein. Therefore, we hypothesized that Cactin overexpression at an abundance that overcomes the instability of Cactin protein would inhibit the upregulation of NFκB activity caused by TRIM39 knockdown. To confirm this hypothesis, TRIM39-knockdown HEK293T cells were further transfected with an expression vector encoding Myc-tagged Cactin. A luciferase reporter assay showed that exogenous Cactin overexpression suppressed NFκB activity in TRIM39-knockdown cells, suggesting that TRIM39 functions as an upstream regulator of Cactin possibly via stabilization of Cactin protein (Fig. 7i). We next performed a luciferase assay for NFκB activity in the presence of siTRIM39 and/or siCactin, and we found that TRIM39 and Cactin synergistically inhibited NFκB-mediated transcriptional activity (Fig. 7j; Suppl. Figure 6). These results may indicate that Cactin knockdown enhanced the NFκB-mediated transcriptional activation by destabilization of Cactin caused by TRIM39 knockdown. Taken together, the findings suggest that TRIM39 negatively regulates the NFκB signaling pathway through stabilization of Cactin.
Discussion
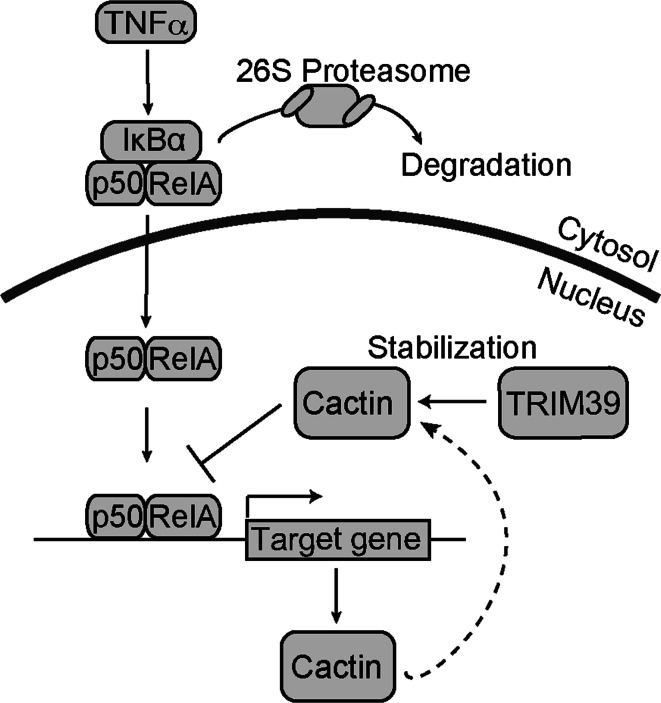
In this study, we identified Cactin as a TRIM39-interacting protein and found that knockdown of TRIM39 led to destabilization of Cactin. We also found that knockdown of TRIM39 enhanced transcriptional activities in NFκB-responsive element and ISRE promoters and that it increased the mRNA expression levels of IL-6 and IL-8, which are known to be target genes of NFκB. These findings suggest that TRIM39 serves as a negative regulator for inflammatory signaling pathways (Fig. 8).
Fig. 8.
Schematic representation of negative regulation of NFκB by TRIM39. TRIM39 negatively regulates NFκB-mediated transcription by stabilizing Cactin, which is induced by NFκB after TNFα stimulation, leading to a negative feedback loop of the NFκB signaling pathway
Cactin was identified as a novel protein interacting with Cactus, the Drosophila ortholog of IκBα, which is important for negative regulation of the NFκB signaling pathway [28]. It has been reported that Cactin is involved in the regulation of several developmental signaling pathways in Drosophila melanogaster, Danio rerio and Caenorhabditis elegans [28, 31–33]. Human Cactin has been reported to interact with IκBL, which is one of negative regulators of the NFκB signaling pathway [27]. The human IκBL gene is localized in the MHC class III region of chromosome 6, in which the TRIM39 gene is also localized [27]. Cactin is also known to bind to proteins such as LST1 and NELF-E that are encoded by genes in the MHC class III region [34]. Furthermore, TRIM39, Cactin, and IκBL are localized in the nucleus [16, 27]. These findings suggest that TRIM39 and Cactin form a larger complex in the nucleus with other proteins encoded by genes in the MHC class III region.
We also showed that TRIM39 stabilizes Cactin in a manner independent of ubiquitin ligase activity. The most important function of ubiquitination is degradation of target proteins by 26S proteasomes via the recognition of lysine 48-linked polyubiquitination. However, recently, several functions of ubiquitination other than protein degradation have also been reported. For example, K63-linked polyubiquitination leads to activation of the NFκB signaling pathway [35]. Furthermore, it has been reported that a linear type of polyubiquitin chain, in which the carboxyl-terminal glycine of one ubiquitin is conjugated to the amino-terminal methionine of another ubiquitin, is important for positive regulation of the NFκB signaling pathway [5]. It has been reported that TRIM39 stabilizes MOAP1 protein [16]. TRIM39 inhibits the polyubiquitination process of MOAP-1 by preventing other E3 ligases from ubiquitination of MOAP-1. TRIM27, one of the TRIM family members also known as RET Finger Protein (RFP), stabilizes EID-1, a negative regulator of Retinoblastoma protein [36]. These findings are similar to our results showing that TRIM39 stabilizes Cactin. Further functional analysis of TRIM39 is needed to clarify how TRIM39 stabilizes Cactin.
Active NFκB induces the expression of various target genes, including IκBα, A20 and MyD88s, which are known as negative regulators of the NFκB signaling pathway. For example, newly synthesized IκBα binds to nuclear NFκB complexes in the nucleus and shuttles NFκB back into the cytosol [29]. A20, one of the ubiquitin-editing enzymes, disassembles the NFκB-activating complexes by deubiquitinating TRAF6 and RIP1 [37, 38]. MyD88s, a short MyD88 isoform generated via alternative splicing, acts as a dominant-negative signaling factor and interferes with Myd88–IRAK4 interaction [39]. Taken together, the results indicate that IκBα, A20, and Myd88s provide negative feedback loops in the NFκB pathway to shut down inflammatory responses. It has been reported that Cactin is one of the negative regulators of the NFκB signaling pathway [27]. Our study revealed that expression of Cactin mRNA and protein was induced after TNFα treatment, suggesting that Cactin also provides a negative feedback loop in the NFκB pathway. However, we could not find a κB motif in the Cactin gene. Therefore, Cactin expression may be controlled by unknown transcriptional regulators to form a negative feedback loop.
In this study, a luciferase assay showed that Cactin overexpression negatively regulates NFκB-mediated transcriptional activity. Immunoblot analysis showed that Cactin does not affect cytosolic regulators in the NFκB pathway or affect the nuclear translocation of NFκB. These findings suggest that Cactin negatively regulates NFκB-mediated transcriptional activity in the nucleus. TRIM39 is mainly localized in the nucleus and interacts with Cactin in the nucleus. It has been reported that translocation to the nucleus is required for Cactin to regulate NFκB-mediated transcription [27]. Taken together, these findings suggest that TRIM39 and Cactin negatively regulate the transcriptional activity of RelA/p65 which is translocated into the nucleus after stimulation.
According to a previous report, the SNP on exon 9 of TRIM39 is involved in Behcet’s disease [21]. The SNP is a synonymous mutation, which means it does not result in a change in the amino acid sequence of TRIM39 protein. However, some synonymous mutations have been reported to alter the function of the gene [40]. We showed that there is no difference between TRIM39 WT and TRIM39 SNP with regard to their expression levels and the levels of interaction with Cactin. However, since we compared exogenous expressions of TRIM39 WT and TRIM39 SNP, the results cannot rule out the possibility that the SNP affects the expression level of endogenous TRIM39.
Furthermore, the polymorphism of the IκBL gene locus, which is known to interact with Cactin, is associated with several inflammatory diseases including rheumatoid arthritis, Takayasu’s arteritis, ulcerative colitis, systemic lupus erythematosus, Sjögren’s syndrome, type 1 diabetes, and multiple sclerosis [41–46]. These findings imply that dysfunction of TRIM39, Cactin, and IκBL may be involved in the etiology of inflammatory diseases. Further study on these proteins will lead not only to the elucidation of their molecular function but also to therapeutic benefits for these inflammatory diseases.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We are grateful to Dr. M. Bohgaki for providing the reagent for this study, Ms. M. Uchiumi for help in preparing the manuscript and Ms. M. Matsuo for technical assistance. We are also grateful to Dr. H. Hatakeyama, Dr. S. Kano, Dr. T. Mizumachi, Dr. T. Sakashita, Dr. K. Mizoguchi, Dr. A. Homma, and Dr. T. Suzuki for technical advice. This work was supported in part by KAKENHI (24112006, 24390065, 15H04690 to S.H.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by The Uehara Memorial Foundation and the Japan Rheumatism Foundation (to S.H.).
Compliance with ethical standards
Conflict of interest
The authors declare no competing financial interests.
References
- 1.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 2.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 3.Hatakeyama S, Kitagawa M, Nakayama K, Shirane M, Matsumoto M, Hattori K, Higashi H, Nakano H, Okumura K, Onoe K, Good RA. Ubiquitin-dependent degradation of IkappaBalpha is mediated by a ubiquitin ligase Skp1/Cul 1/F-box protein FWD1. Proc Natl Acad Sci USA. 1999;96:3859–3863. doi: 10.1073/pnas.96.7.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Gazdoiu S, Pan ZQ, Fuchs SY. Stability of homologue of Slimb F-box protein is regulated by availability of its substrate. J Biol Chem. 2004;279:11074–11080. doi: 10.1074/jbc.M312301200. [DOI] [PubMed] [Google Scholar]
- 5.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, Dikic I. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 7.Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1–E2–E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 8.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 9.Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem. 2001;276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- 12.Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 13.Kano S, Miyajima N, Fukuda S, Hatakeyama S. Tripartite motif protein 32 facilitates cell growth and migration via degradation of Abl-interactor 2. Cancer Res. 2008;68:5572–5580. doi: 10.1158/0008-5472.CAN-07-6231. [DOI] [PubMed] [Google Scholar]
- 14.Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Huang NJ, Chen C, Tang W, Kornbluth S. Ubiquitylation of p53 by the APC/C inhibitor Trim39. Proc Natl Acad Sci USA. 2012;109:20931–20936. doi: 10.1073/pnas.1212047110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee SS, Fu NY, Sukumaran SK, Wan KF, Wan Q, Yu VC. TRIM39 is a MOAP-1-binding protein that stabilizes MOAP-1 through inhibition of its poly-ubiquitination process. Exp Cell Res. 2009;315:1313–1325. doi: 10.1016/j.yexcr.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 17.Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 18.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M, Inn KS, Fernandez-Sesma A, Jung J, Garcia-Sastre A. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 2013;38:384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurata R, Tajima A, Yonezawa T, Inoko H. TRIM39R, but not TRIM39B, regulates type I interferon response. Biochem Biophys Res Commun. 2013;436:90–95. doi: 10.1016/j.bbrc.2013.05.064. [DOI] [PubMed] [Google Scholar]
- 21.Kurata R, Nakaoka H, Tajima A, Hosomichi K, Shiina T, Meguro A, Mizuki N, Ohono S, Inoue I, Inoko H. TRIM39 and RNF39 are associated with Behcet’s disease independently of HLA-B *51 and -A *26. Biochem Biophys Res Commun. 2010;401:533–537. doi: 10.1016/j.bbrc.2010.09.088. [DOI] [PubMed] [Google Scholar]
- 22.Nakajima K, Yamanaka Y, Nakae K, Kojima H, Ichiba M, Kiuchi N, Kitaoka T, Fukada T, Hibi M, Hirano T. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 1996;15:3651–3658. [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuda M, Tsukiyama T, Bohgaki M, Nonomura K, Hatakeyama S. Establishment of a newly improved detection system for NF-kappaB activity. Immunol Lett. 2007;109:175–181. doi: 10.1016/j.imlet.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Kondo T, Watanabe M, Hatakeyama S. TRIM59 interacts with ECSIT and negatively regulates NF-kappaB and IRF-3/7-mediated signal pathways. Biochem Biophys Res Commun. 2012;422:501–507. doi: 10.1016/j.bbrc.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 25.Noguchi K, Okumura F, Takahashi N, Kataoka A, Kamiyama T, Todo S, Hatakeyama S. TRIM40 promotes neddylation of IKKgamma and is downregulated in gastrointestinal cancers. Carcinogenesis. 2011;32:995–1004. doi: 10.1093/carcin/bgr068. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe M, Tsukiyama T, Hatakeyama S. TRIM31 interacts with p52(Shc) and inhibits Src-induced anchorage-independent growth. Biochem Biophys Res Commun. 2009;388:422–427. doi: 10.1016/j.bbrc.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 27.Atzei P, Gargan S, Curran N, Moynagh PN. Cactin targets the MHC class III protein IkappaB-like (IkappaBL) and inhibits NF-kappaB and interferon-regulatory factor signaling pathways. J Biol Chem. 2010;285:36804–36817. doi: 10.1074/jbc.M110.139113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin P, Huang LH, Steward R. Cactin, a conserved protein that interacts with the Drosophila IkappaB protein cactus and modulates its function. Mech Dev. 2000;94:57–65. doi: 10.1016/S0925-4773(00)00314-2. [DOI] [PubMed] [Google Scholar]
- 29.Arenzana-Seisdedos F, Thompson J, Rodriguez MS, Bachelerie F, Thomas D, Hay RT. Inducible nuclear expression of newly synthesized I kappa B alpha negatively regulates DNA-binding and transcriptional activities of NF-kappa B. Mol Cell Biol. 1995;15:2689–2696. doi: 10.1128/MCB.15.5.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin J, Samuvel DJ, Zhang X, Li Y, Lu Z, Lopes-Virella MF, Huang Y. Coactivation of TLR4 and TLR2/6 coordinates an additive augmentation on IL-6 gene transcription via p38MAPK pathway in U937 mononuclear cells. Mol Immunol. 2011;49:423–432. doi: 10.1016/j.molimm.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atzei P, Yang F, Collery R, Kennedy BN, Moynagh PN. Characterisation of expression patterns and functional role of Cactin in early zebrafish development. Gene Expr Patterns. 2010;10:199–206. doi: 10.1016/j.gep.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Tannoury H, Rodriguez V, Kovacevic I, Ibourk M, Lee M, Cram EJ. CACN-1/Cactin interacts genetically with MIG-2 GTPase signaling to control distal tip cell migration in C. elegans . Dev Biol. 2010;341:176–185. doi: 10.1016/j.ydbio.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LaBonty M, Szmygiel C, Byrnes LE, Hughes S, Woollard A, Cram EJ. CACN-1/Cactin plays a role in Wnt signaling in C. elegans . PLoS One. 2014;9:e101945. doi: 10.1371/journal.pone.0101945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehner B, Semple JI, Brown SE, Counsell D, Campbell RD, Sanderson CM. Analysis of a high-throughput yeast two-hybrid system and its use to predict the function of intracellular proteins encoded within the human MHC class III region. Genomics. 2004;83:153–167. doi: 10.1016/S0888-7543(03)00235-0. [DOI] [PubMed] [Google Scholar]
- 35.Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, Chiu YH, Deng L, Chen ZJ. TAB 2 and TAB 3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Krutzfeldt M, Ellis M, Weekes DB, Bull JJ, Eilers M, Vivanco MD, Sellers WR, Mittnacht S. Selective ablation of retinoblastoma protein function by the RET finger protein. Mol Cell. 2005;18:213–224. doi: 10.1016/j.molcel.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C, Ma A. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 38.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 39.Burns K, Janssens S, Brissoni B, Olivos N, Beyaert R, Tschopp J. Inhibition of interleukin 1 receptor/Toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. J Exp Med. 2003;197:263–268. doi: 10.1084/jem.20021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 41.Okamoto K, Makino S, Yoshikawa Y, Takaki A, Nagatsuka Y, Ota M, Tamiya G, Kimura A, Bahram S, Inoko H. Identification of I kappa BL as the second major histocompatibility complex-linked susceptibility locus for rheumatoid arthritis. Am J Hum Genet. 2003;72:303–312. doi: 10.1086/346067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shibata H, Yasunami M, Obuchi N, Takahashi M, Kobayashi Y, Numano F, Kimura A. Direct determination of single nucleotide polymorphism haplotype of NFKBIL1 promoter polymorphism by DNA conformation analysis and its application to association study of chronic inflammatory diseases. Hum Immunol. 2006;67:363–373. doi: 10.1016/j.humimm.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 43.de la Concha EG, Fernandez-Arquero M, Lopez-Nava G, Martin E, Allcock RJ, Conejero L, Paredes JG, Diaz-Rubio M. Susceptibility to severe ulcerative colitis is associated with polymorphism in the central MHC gene IKBL. Gastroenterology. 2000;119:1491–1495. doi: 10.1053/gast.2000.20258. [DOI] [PubMed] [Google Scholar]
- 44.Castiblanco J, Anaya JM. The IkappaBL gene polymorphism influences risk of acquiring systemic lupus erythematosus and Sjogren’s syndrome. Hum Immunol. 2008;69:45–51. doi: 10.1016/j.humimm.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 45.Yamashita T, Hamaguchi K, Kusuda Y, Kimura A, Sakata T, Yoshimatsu H. IKBL promoter polymorphism is strongly associated with resistance to type 1 diabetes in Japanese. Tissue Antigens. 2014;63:223–230. doi: 10.1111/j.0001-2815.2004.00164.x. [DOI] [PubMed] [Google Scholar]
- 46.Miterski B, Bohringer S, Klein W, Sindern E, Haupts M, Schimrigk S, Epplen JT. Inhibitors in the NFkappaB cascade comprise prime candidate genes predisposing to multiple sclerosis, especially in selected combinations. Genes Immun. 2002;3:211–219. doi: 10.1038/sj.gene.6363846. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.