Abstract
Katanins are microtubule (MT)-severing AAA proteins with high phylogenetic conservation throughout the eukaryotes. They have been functionally implicated in processes requiring MT remodeling, such as spindle assembly in mitosis and meiosis, assembly/disassembly of flagella and cilia and neuronal morphogenesis. Here, we uncover a novel family of katanin-like 2 proteins (KATNAL2) in mouse, consisting of five alternatively spliced isoforms encoded by the Katnal2 genomic locus. We further demonstrate that in vivo these isoforms are able to interact with themselves, with each other and moreover directly and independently with MRP/MinD-type P-loop NTPases Nubp1 and Nubp2, which are integral components of centrioles, negative regulators of ciliogenesis and implicated in centriole duplication in mammalian cells. We find KATNAL2 localized on interphase MTs, centrioles, mitotic spindle, midbody and the axoneme and basal body of sensory cilia in cultured murine cells. shRNAi of Katnal2 results in inefficient cytokinesis and severe phenotypes of enlarged cells and nuclei, increased numbers of centrioles and the manifestation of aberrant multipolar mitotic spindles, mitotic defects, chromosome bridges, multinuclearity, increased MT acetylation and an altered cell cycle pattern. Silencing or stable overexpression of KATNAL2 isoforms drastically reduces ciliogenesis. In conclusion, KATNAL2s are multitasking enzymes involved in the same cell type in critically important processes affecting cytokinesis, MT dynamics, and ciliogenesis and are also implicated in cell cycle progression.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-015-1980-5) contains supplementary material, which is available to authorized users.
Keywords: Microtubule-severing enzymes, Katanin, Cilia, Cytokinesis, Cytoskeleton
Introduction
Temporal or localized dynamic re-organization of microtubule (MT) high-order arrays is of critical importance to a multitude of fundamental developmental and cellular processes through its role in cell motility and migration, intracellular transport, mitotic and meiotic division, targeted flow of extracellular fluid and sensory input by cilia. MT-end dynamics, with rapid growth by α/β tubulin heterodimer subunit addition or shrinkage by subunit loss, can determine polymer length, which can be further modulated by MT-interacting proteins that stabilize or destabilize MTs, such as microtubule-associated proteins (MAPs), MT plus-end-tracking proteins (+TIPS) and bundling/depolymerising motors [1–6]. In parallel, a lot has been learned about additional mechanisms underlying MT plasticity. Following the initial discovery of katanin [7], the uncovering of a still expanding superfamily of proteins with MT-severing activity that can introduce breaks into the polymer lattice and generate shorter MT fragments, has provided some understanding on how existing MT architecture can be modified and re-modeled locally or how structures can be organized de novo using such released MT fragments, sometimes transported to other intracellular domains, as seeds [8–11].
Currently, the three main classes of MT-severing proteins, katanins, spastins and fidgetins, all belong to a subgroup of the protein superfamily of “ATPases associated with diverse cellular activities” (AAA), bearing the highly conserved ~230aa signature AAA domain that harbors the ATPase activity. Their functional form is believed to consist of ~15 nm hexameric rings, formed in an ATP-stimulated and template-dependent manner on the MT lattice, and able to bind on the C-terminal tail of tubulins possibly with some preference for β-tubulin [12–15]. ATP hydrolysis is thought to induce translocation of the tubulin polypeptide through the hexamer ring pore and its local unfolding, which enables weakening and breaking of interdimer bonds and causes severing of filaments [16].
Katanin was purified as a heterodimer of the p60 AAA-bearing catalytic subunit and the p80 centrosome-targeting and regulatory subunit [17, 18]; both subunits are highly conserved in animals, plants, and even unicellular eukaryotes, such as green algae and protozoa [9, 10; Table 2 in 11]. In some invertebrates and vertebrates, additional katanin p60-like proteins (KATNAL 1 or 2) have been reported [19, 20], in others, including unicellular organisms, they can be predicted from their genome sequence. Katanin p60 appears to be a multitasking enzyme implicated in an impressive number of MT-based processes (meiosis and mitosis, ciliogenesis and neurogenesis), as either a positive or negative regulator of MT polymer mass. In oocyte meiosis of C. elegans, katanin contributes to amplification of spindle MT number and density by providing severed short MTs for nucleation of new MTs [21, 22] while also exhibiting MT bundling activity independent from its severing activity [23]. Conversely, in Drosophila mitotic spindles, katanin promotes severing, release and uncapping from end-binding (EB) proteins of chromosome-attached MT (+) ends to enable kinesin 13-induced MT (+) end depolymerization and thus allow Pacman flux and poleward chromatid mobility and segregation in anaphase A [24]. Additionally, severing by katanin at the MT (−) ends at the centrosome promotes the redistribution of γ-tubulin rings to enhance new MT nucleation [25]; Katanin-like protein 1 (KATNAL1) also seems to boost the formation of spindle MTs and increase spindle MT density in human cells by altering γ-tubulin ring kinetics [20] and is also involved in spermatogenesis [26]. The role of katanin in cytokinesis has been highlighted in protozoa such as Tetrahymena, where katanin-null cells can successfully complete nuclear division but not cytokinesis, resulting in chains of non-separated cells [14], while in Trypanosoma and Leishmania both p60 and p80 are essential for distinct steps of cytokinesis [27, 28]. In vertebrate cells, where the mechanism of cytokinesis is different, katanin p60 is present on the midbody constriction, where it is proposed to influence its resolution [29], possibly involving katanin interactions with tumor suppressor protein LAPSER1 [30]; however, its role and molecular mechanism in vertebrate cytokinesis remain sketchy.
Katanins have a highly conserved function in ciliary dynamics as shown in unicellular eukaryotes, where their loss of function results in the formation of immotile cilia due to loss of the central pair of axonemal MTs in Chlamydomonas and Tetrahymena [14, 31], possibly through lack of a pool of short MT precursors earmarked for axoneme assembly. In Tetrahymena, katanin’s severing selectivity for ciliary MTs may be based on specific tubulin post-translational modifications [14]. In Chlamydomonas, katanin is also responsible for deciliation [32], facilitated by the release of basal bodies from the ciliary axoneme through selective severing of MTs at the transition zone, in preparation for mitosis [33]. Ciliary-based Hedgehog signaling is compromised in fibroblasts derived from p80 katanin-null mouse embryos and in neocortical development of these animals, displaying severe microlissencephaly [34, 35].
The need for fine-tuned regulation of katanin activity/concentration remains an important aspect for its cellular multitasking functions and it is exerted via its intricate network of protein interactions, its reversible phosphorylation and by discrete degradation routes [36, 37, summarized in Figures 3 and 4 of 11].
Intriguing questions remain in current understanding of the functional roles of katanins and related proteins in MT dynamics in diverse cellular processes. What is the distinct role of so many MT-severing activities, even within the same family, given that both higher and unicellular eukaryotes have several katanins and katanin-like proteins in addition to multiple spastins and fidgetins? What is the balance between “division of labour” [38] and functional redundancy amongst the multitude of MT-severing enzymes? Additionally, do the different katanin-like proteins also acquire an oligomeric form to be functional (akin to p60 katanin) and what is the nature of such oligomers (homo- or hetero-oligomers)? Is there a possibility for combinatorial (mix-and-match) utilization and, given the multitasking of katanins, does the process of oligomerization also assign spatiotemporal specificity to the enzyme, i.e. to bind and affect different subsets of cellular MT as substrates or MTs at different cell cycle or developmental stages? Finally, given the intimately reciprocal relationship of MT dynamics, cilia and centrioles with cell cycle progression, should katanins and katanin-like proteins, affecting all three structures, be investigated in a renewed light of potential cell cycle drivers?
In this work, we uncover a novel family of KATNAL2 proteins in mouse cells and present their first functional characterization. Our new findings provide significant insights to the emerging definition of molecular mechanisms and protein interactions underlying katanin-like cellular function.
Materials and methods
Mouse cell culture
Mouse NIH 3T3 and NSC34 cells were cultured in DMEM containing 10 % v/v fetal calf serum (FCS), 2 mM glutamine and 50 U/mL of penicillin/streptomycin. IMCD3 (IMCD) cells were cultured in DMEM/F-12 (1:1) medium with 10 % v/v FCS, 2 mM glutamine and 50 U/mL of penicillin/streptomycin. TM4 cells were grown as for IMCD but FCS was replaced by 5 % v/v horse serum and 2.5 % FCS. Neuro2A cells were kept as NIH 3T3 but with MEMα instead of DMEM. All lines were maintained at 37 ºC in 5 % CO2. For induction of ciliogenesis, cells were grown in serum-free media for 24 h and sampled for immunofluorescence (IF) or Western blotting (WB).
Cloning of Katnal2 isoforms and quantitative real-time RT-PCR (RT-qPCR) to analyze Katnal2 gene expression
Polyadenylated RNA was affinity purified from cell lines or tissues, using the RNeasy mRNA purification kit (Qiagen). cDNA was synthesized with the iScript cDNA synthesis kit (Biorad). For the identification of Katnal2 transcripts, PCR amplification with oligonucleotides flanking the full-length open reading frame (ORF) of Katnal2 (XM_006526437) was employed (Table S1), using the Expand High-Fidelity PCR system (Roche). PCR products were cloned by T/A cloning into vector pGEM T Easy (Promega) or pCRII-TOPO (Invitrogen) and identified by DNA sequencing (MWG GmbH). For in-frame subcloning of ORFs into different plasmid vectors, oligonucleotides, bearing appropriate restriction sites, were designed (see plasmid constructs and Table S1).
For relative quantification of mRNA in silencing experiments, RT-qPCR was conducted on the LightCycler (Roche) [39] with primers specific for either Katnal2-L (isoforms L1–L3) or Katnal2-ALL (co-amplifying a product from all Katnal2 isoforms). Melting curve analysis was performed to determine the amplification specificity. All experiments included two no-template controls, and samples were analyzed independently three times. Crossing point determination and quantification was achieved using the Second Derivative Maximum Method (Viia™ 7 Software, ABI). For data normalization, expression of ribosomal protein L19 (Rpl19) and pumilio homolog 1 transcript variant 2 (Pum1) as reference genes was used. All primer sequences for PCR are given in Table S1. For statistical analysis of RT-qPCR data, the REST-384© software was used to compare the relative quantification between two groups and determine the significance of results [40].
Expression plasmid constructs, yeast two-hybrid screen
For antibody generation, KATNAL2-S1 was expressed in E. coli [BL21-(DE3)-pLysS] as a His-tagged fusion protein from vector pRSETB and affinity purified over Ni2+–NTA beads (Qiagen) under denaturing conditions. For in vitro co-selection assays to study KATNAL2 interactions, GST fusions of KATNAL2-S1 and -L1 were generated from pGEX-4T-1 constructs and used together with His-tagged fusions of Nubp1 and Nubp2 in pRSETB. Mammalian expression of KATNAL2 isoforms, tagged with different epitopes was driven from plasmids pEGFP-C2 (Clontech), pmCherry-C3 (Ellenberg group, EMBL), pFLAG-CVM-1 (Sigma) and pSVmyc1.0 [41] (summarized in Table S1). A yeast two-hybrid screen was conducted by Hybrigenics (France), using full-length mouse Nubp1 cDNA as bait (hgx1836v1) against a mouse embryo brain RP1 cDNA prey library in pB27 (LexA, C-terminal fusion) and analyzing 58.04 million interactions. A partial Katnal2 cDNA (aligning with mouse Katnal2 XM_006526437) was amongst 43 putative interactors identified in this screen.
SDS-PAGE and WB
SDS-PAGE was performed on a Mini-Protean II Electrophoresis Cell and WB on a Mini Trans-Blot Transfer Cell (Bio-Rad), using 48 mM Tris pH 9.2, 39 mM glycine and 20 % v/v methanol for transfer and the ECL System (GE Healthcare) with ChemiDoc™ MP (Biorad) for visualization. For quantification of protein levels, intensity volumes (area × height) of signals were extracted with ImageJ 1.49n and normalized using same-sample and same-membrane band intensities for the housekeeping protein calnexin. Significance of knockdown for KATNAL2 (three independent experiments) was assessed for log-transformed normalized values by one-tailed paired t test and deregulation of Nubp1, Nubp2 and actin-normalized protein quantities was assessed by two-tailed paired t test.
Antibodies
An antibody against recombinant mouse KATNAL2-S1 (LN831865), expressed as a 6xHis-fusion in E. coli and affinity purified (see plasmid constructs), was custom-made in goat by Sicgen (Portugal). The antiserum was affinity purified on a WB membrane: 2 mg of Ni2+–NTA-affinity purified 6xHis-tagged KATNAL2-S1 were run on a preparative SDS-PAGE gel, transferred onto nitrocellulose and stained with 0.1 % w/v Ponceau in 5 % v/v acetic acid to identify the position of the recombinant protein, which was outlined with a pencil. The membrane was incubated with 2 mL anti-KATNAL2 crude serum (1:5 dilution in PBS) at 4 ºC overnight, the strip excised and washed with PBS. The bound antibody was eluted with 2 × 1 mL of 100 mM glycine containing 50 % v/v glycerol at pH 2.2; fraction 2 of the purified antibody was used in 1:100 dilution for IF and 1:250 for WB. Affinity purified rabbit antibodies to Nubp1 and Nubp2 [42] were used at 1:300 dilution for WB. Additional primary and secondary antibodies are listed in Table S2. Nuclei were counterstained with Hoechst 33342 (0.5 μg/mL).
Immunoprecipitation (IP) and LC–MS/MS analysis
For IP, 8 μg of fraction 2 purified anti-KATNAL2 or 8 μg goat IgG (negative control) were each covalently bound onto 40 μL Protein-G Sepharose CL-4B beads (GE Healthcare) by DNP crosslinking. Eight adult-mouse testicles were homogenized in 50 mL of 50 mM Tris–Cl pH 7.5, 150 mM KCl, 2 mM EDTA, 0.5 % v/v Triton X-100, 1× Complete protease inhibitor (Roche) and divided in two aliquots of 25 mL (2.2 mg/mL total protein). Each aliquot was incubated overnight at 4 °C with one set of beads (KATNAL2 or goat IgG), washed 3 times with lysis buffer and eluted with 2 × 20 μL of 100 mM glycine, pH 2.0. The eluate was boiled with 10 μL of 5× SDS-PAGE sample buffer. Twenty percent from each eluted IP sample was analyzed by WB to identify KATNAL2-immunoreactive bands and 80 % was run on an SDS-PAGE gel, stained with 80 % v/v 1× working solution of Brilliant Blue G-Colloidal (Sigma B2025) in 20 % v/v methanol. Three bands, unique to the KATNAL2-IP samples and immunoreactive by WB, were cut out (Fig. 1d). For the larger-scale experiment (Fig. S3), 16 testicles were lysed in 100 mL homogenization buffer and used in conjunction with 28 μg KATNAL2 antiserum and 150 μL bead slurry. Bands or zones excised from gels were trypsin digested in gel and eluted, tryptic peptides were separated and analyzed by LC–MS/MS (Orbitrap Velos, Thermo Scientific) at the EMBL Proteomics Core Facility. Full scan MS spectra (mass range 300–1700 m/z) were acquired in profile mode in the FT with resolution of 30,000. Data were filtered by Software MaxQuant (version 1.0.13.13) and searched in mouse-specific mode against the Swiss-Prot database.
Fig. 1.
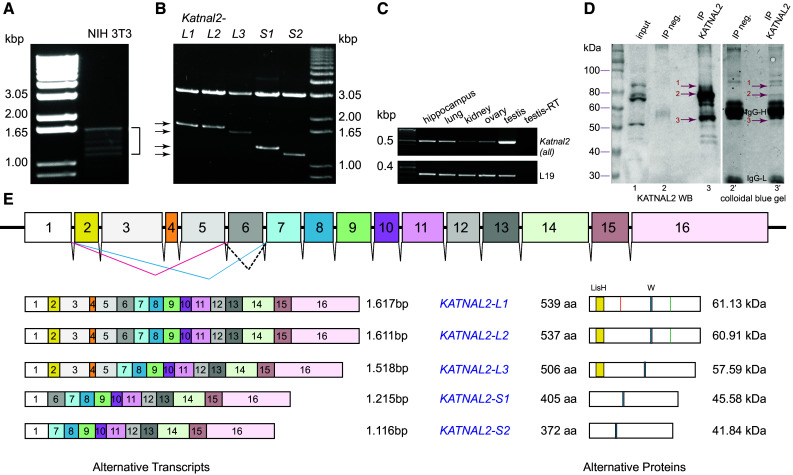
Identification of a family of KATNAL2 isoforms with N-terminal sequence variation. a RT-PCR amplification of cDNA from NIH 3T3 cells, using primers (SET1 in Table S1) at the extremities of the predicted ORF of mouse Katnal2 XM_006526437. Several distinct bands (1.2–1.6 kbp) were obtained (bracket). b The mixed PCR product from (a) was subcloned in vector pGEM T easy, and after screening of 100 clones, five distinct types of cDNA, shown here as products of SacII/SacI restriction digests (arrows), were identified by DNA sequencing and assigned names Katnal2-L1, -L2, -L3, -S1 and -S2. c Semi-quantitative RT-PCR analysis of Katnal2 expression in selected mouse tissues, using primers (SET5 in Table S1) that resulted in the amplification of a diagnostic product of 508 bp, common to all Katnal2 transcripts. d Analysis of IP experiment from mouse testis, using the affinity purified anti-KATNAL2 antibody. IP samples were analyzed in parallel by WB (20 %, left) and on a colloidal-blue-stained gel (80 %, right) to identify unique bands in the KATNAL2-IP that were immunoreactive. Three such bands were identified (red arrows bands 1–3) and cut out for MS analysis. Lanes 1, 1′ input; 2, 2′ negative control IP (goat IgG); 3, 3′ KATNAL2 IP. Results from MS analysis in Table S3, confirm the presence of KATNAL2 proteins in these immunoreactive bands. e Sketch of the Katnal2 gene on mouse chromosome 18. Exons (1–16) are colour coded and to scale (introns are not to scale). The five distinct cDNAs in b correspond to transcripts designated Katnal2-L1, L2, L3, S1 and S2 with differential exon combinations, as shown on the left of the panel. Corresponding encoded proteins are shown on the right-hand side; isoforms L1 and L2 differ only in two additional amino acids (VK) in L1; their position is marked by a red line in L1. Protein motifs are shown: LisH domain as a yellow box, Walker motif as a blue box and SNPs in L1 and L2 (H instead of R) are indicated as a green line. Full protein sequences in Fig. S1. cDNA sequences were deposited to the European Nucleotide Archive (www.ebi.ac.uk/ena/data/view/LN831862-LN831866)
In vivo and in vitro co-selection assays to study KATNAL2 interactions
For in vivo co-selection experiments, FLAG-tagged-KATNAL2-S1 or -L1 (Table S1) were expressed by transient transfection into the IMCD-pmCherry-KATNAL2-S1 stable cell line, (see later section). Two 10-cm dishes of transfected cells were extracted in 0.5 mL lysis buffer (150 mM KCl, 20 mM Tris, pH 7.5, 0.5 % v/v Triton X-100), containing 2× of Complete protease inhibitor. The extract was mixed with 10 μL anti-FLAG beads (Anti-Flag M2 Affinity Gel; Sigma) and incubated for 3 h at 4 °C. Negative controls included parallel reactions without FLAG-tagged bait proteins. Beads with bound complexes were collected at 2000 rpm for 5 min, washed three times with 10× bed volumes of lysis buffer, re-suspended in 20 μL SDS sample buffer and boiled. From each sample, 20 % was subjected to WB with anti-FLAG antibody and the remainder to a parallel WB which was cut into two segments for the detection of bound, interacting proteins with antibodies to mCherry and KATNAL2 (segment above 45 kDa) and to Nubp1 and Nubp2 (segment below 45 kDa).
In vitro co-selection experiments were conducted to confirm and characterize pairwise interactions between KATNAL2-S1/L1 and Nubp1/2. The four proteins were expressed in E. coli strain BL21(DE3)pLysS; KATNAL2-S1 and -L1 were produced as GST fusions (Table S1), after a 5-h induction with 0.5 mM IPTG (-S1) or a 3-h induction with 0.05 mM IPTG (-L1), or a 5-h induction with 0.5 mM IPTG (GST-only, negative control), all at 20 °C. Nubp1 and Nubp2 were expressed as His-tagged fusions from pRSETA [42], under the same conditions as for KATNAL2-S1. Bacterial pellets (1.5 mL of liquid culture) were sonicated in 200 μL lysis buffer (150 mM NaCl, 20 mM Tris, pH 7.4, 1 % v/v Tween 20, 1 mM β-mercaptoethanol and 1× Complete protease inhibitor), centrifuged at 13,000 rpm for 15 min at 4 °C and the soluble fraction, containing the recombinant protein retrieved (1/20 in each case was used as “Input” in Fig. 2). KATNAL2 and Nubp1 or 2 protein interactions were tested in pairwise combinations by mixing the selected protein extracts, incubating for 30 min at room temperature and combining for another 30 min at room temperature with 25 μL glutathione Sepharose 4B bead slurry (GE Healthcare), pre-equilibrated with lysis buffer. Beads were spun at 2000 rpm for 5 min at 4 °C, the supernatant collected (“unbound”), beads washed three times in 100 μL lysis buffer and bound proteins retrieved by boiling in SDS-PAGE sample buffer for 5 min at 96 °C. The eluate (“bound”) was split into two and together with the other fractions subjected to WB, using anti-GST and anti-His antibodies to probe for KATNAL2 and Nubp proteins, respectively. Negative controls consisted of identical parallel experiments but with the use of GST-only protein in conjunction with His-Nubp1 or -Nubp2.
Fig. 2.
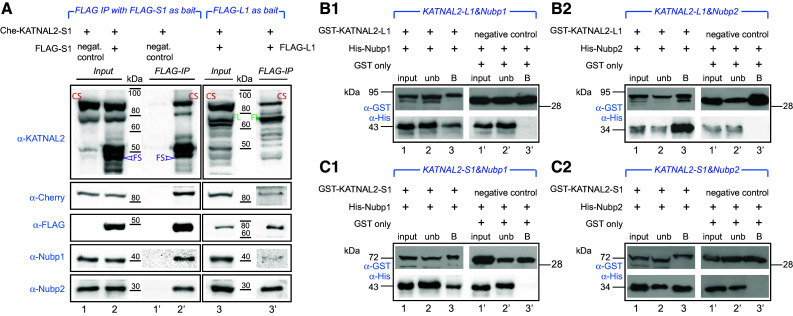
Nubp1 and Nubp2 interact with KATNAL2 in vivo and in vitro, and KATNAL2s interact with themselves. a The Cherry-KATNAL2-S1 (“Che-KATNAL2-S1”) stable cell line was transiently transfected with plasmids expressing as bait either FLAG-KATNAL2-S1 (“FLAG-S1”) or FLAG-KATNAL2-L1 (“FLAG-L1”) (as indicated at the header for each set) or mock empty vector plasmid (negative control) and subjected to IP with anti-FLAG beads. The presence of Cherry-KATNAL2-S1, KATNAL2s, FLAG, Nubp1 and Nubp2 was probed by WB with appropriate antibodies as shown. Results show that the FLAG-KATNAL2-S1 or -L1 bait co-immunoprecipitates Nubp1 and Nubp2 but also the “endogenous” Cherry-KATNAL2-S1 and several native KATNAL2 (different bands migrating in positions within ca. 40–80 kDa). All these proteins are absent from the negative control IP reaction (lane 1′). CS, Cherry-KATNAL2-S1; FS, FLAG-KATNAL2-S1; FL, FLAG-KATNAL2-L1). b1–c2 Recombinant GST-KATNAL2-L1 or -S1 and His-Nubp1 or His-Nubp2 were probed for pairwise interactions (as indicated for each set in the blue header). GST-only was used in conjunction with His-Nubp1 or His-Nubp2 in parallel negative control reactions. Complexes were immobilized on glutathione beads, eluted, and two equal parts were analyzed by WB, either using anti-GST (to detect the KATNALs) or anti-His (to detect the Nubps). Results show that Nubp1 or Nubp2 is detectable on the glutathione beads only in the presence of a KATNAL2 and indicate that both KATNAL2-L1 and KATNAL2-S1 interact individually with both Nubp1 and Nubp2. Input corresponds to 1/20 volume of the lysate used for the reaction, unb is the unbound fraction after incubation of the mixed recombinant protein lysates with glutathione beads, B is the bound fraction corresponding to preformed protein complexes, captured on the beads
Immunofluorescence and live imaging
IF was performed as described in [42]. Samples were analyzed with a Zeiss Apochromat 63× NA 1.4 oil lens on a Zeiss Axiovert 200M inverted microscope with an AxioCam MRm camera. For live imaging, cells were transferred onto the stage of a temperature- and CO2-regulated Zeiss AxioObserver.Z1 automated microscope with a Plan-Neofluar 5× NA 0.16 air objective lens. Phase contrast images were acquired every 7 min for 24 h using the Zeiss AxioVision program (v. 4.8.2) and an AxioCam MRm camera.
shRNA-mediated silencing and analysis
Two pLKO.1-based lentiviral shRNA vectors [The RNAi Consortium (TRC)], expressing a puromycin resistance gene and targeting different areas of the mouse Katnal2 ORF (TRCN0000090749 and TRCN0000090751) were obtained from the University Medical Center Rotterdam (The Netherlands). Lentiviral shRNA production, lentivirus particle packaging, harvesting and transductions of NIH 3T3 fibroblasts were as per [43]. Stably transfected, puromycin-resistant, single clones were hand-picked under the microscope after several days of puromycin selection (4 μg/mL), expanded and screened by WB, using anti-KATNAL2, to identify clones with reduced KATNAL2 expression. Clone 2.43 (shRNA targeting ORF area 1, yellow in Fig. S1) was identified out of 52 clones and clone 3.81 (shRNA targeting ORF area 2, light blue in Fig. S1) out of 94 clones and Katnal2 silencing was quantified by RT-qPCR and quantitative WB (3 independent experiments). To confirm their genotypes, PCR of genomic DNA from clones 2.43 and 3.81 was employed using appropriate insert-flanking oligonucleotides (Table S1) to amplify the shRNA-bearing cassette and the product was sequenced (MWG GmbH).
To assess phenotypic effects of silencing, we performed IF analysis in cycling cells or cells subjected to a 24-h serum withdrawal. Cycling cells were scored for morphology of the mitotic spindle as visualized with α-tubulin staining, number of centrioles, as determined by γ-tubulin staining, count of nuclei per cell and presence of chromatin bridges or chromosomal misalignment as labeled with Hoechst 33342. For each parameter, the average ± SD of three independent experiments each of NIH 3T3 control cells and silenced clones 2.43 or 3.81 was determined. Total sample sizes of the datasets were: dataset 1 with a total of n = 2978 clone 2. 43 silenced cells vs. a total of n = 3128 control cells and dataset 2 with n = 2997 clone 3.81 silenced cells vs. n = 3007 control cells. For each parameter, equality of variance within the dataset was confirmed with the F test and statistical significance of differences across samples evaluated by homoscedastic two-tailed Student’s t tests. Statistical significance to compare differences in the overall distribution of cells in distinct subphases of mitosis or in the overall distribution of numbers of centrioles per cell across the population in control and silenced cells was determined by stacked two-way ANOVA and post hoc test with Sidak correction for multiple comparisons.
Serum-starved cells were assessed for the presence of a cilium, as visualized by double γ-tubulin (basal body) and acetylated tubulin (axoneme) labeling. The average percentage of ciliated cells was calculated in three independent experiments (dataset 3 with a total n = 2063 clone 2.43 cells vs. n = 2076 control; dataset 4 with n = 3013 clone 3.81 cells vs. n = 3020 control cells). For each pairwise comparison between silenced and control cells, statistical significance of differences was evaluated by homoscedastic, two-tailed Student’s t test.
To compare total cell and nuclear size in clone 2.43 vs. control cells, cell or nuclear surface areas were measured with the AxioVision software (v. 4.8.2.SP2) on images acquired after double α-tubulin and Hoechst labeling and photographed with a Zeiss Plan Apochromat 20× NA 0.75 lens on Axiovert 200M (n = 127 clone 2.43 cells, n = 129 control cells). The difference in size distributions of total cell or nuclear size between the two populations was evaluated for statistical significance by a two-tailed Mann–Whitney test.
Generation of IMCDmCherry-KATNAL2 stable cell lines
IMCD cells were transfected with plasmids pmCherry-KATNAL2-L1 or pmCherry-KATNAL2-S1 (Table S1) using Lipofectamine 2000 (Invitrogen). After 18 h, medium containing 400 μg/mL G418 was added to select for cells that had stably incorporated the plasmid into their genome. Several days later, resistant individual colonies were picked and expanded for biochemical and microscopic analyses. Phenotypic analysis of cycling or serum-deprived cells was carried out in the same manner as described for shRNA-silenced cells. For cycling conditions, populations of IMCD-KATNAL2-S1 (n = 2608 cells), and IMCD-KATNAL2-L1 (n = 1972) were compared with corresponding control IMCD wild-type cells (n = 5868) (dataset 5). The proportion of ciliated cells was compared within populations of IMCD-KATNAL2-S1 (n = 8826 cells), IMCD-KATNAL2-L1 (n = 6664) and control IMCD wild-type cells (n = 6708) (dataset 6). In all cases statistical evaluation of differences was performed by one-way ANOVA with Dunnett’s post-test for multiple comparisons.
Cell cycle analysis
The determination of the mitotic index and assignment to mitotic subphases was performed in three independent experiments by microscopic analysis of Katnal2-silenced stable cell lines 2.43 and 3.81 and their parental line NIH 3T3 (control) (datasets 1 and 3 as per “shRNA silencing” section), or of stable lines mCherry-KATNAL2-S1, mCherry-KATNAL2-L1 and their IMCD parental line (dataset 5), all immunofluorescently labeled for α- and γ-tubulin and counterstained with Hoechst 33342. Statistical evaluation of pairwise differences between silenced and control cells was performed by homoscedastic two-tailed Student’s t tests. For the Cherry lines and parental IMCD cells, statistical significance was assigned by one-way ANOVA with Dunnett’s post-test for multiple comparisons.
To generate growth curves, multiple cultures of control and clone 2.43 cells were seeded each with 6860 cells in six-well plates, and triplicate measurements of cell counts were recorded with a hemocytometer every 24 h for a total of 96 h. Growth curves for 2.43 and control cells were compared by stacked two-way ANOVA for analysis of parameter means with Sidak post-test for multiple comparisons with explicit reporting of p values down to 0.0001.
For flow cytometry, cell lines were washed in PBS, fixed with 70 % v/v ethanol, washed, treated with RNase A and stained with 1 mg/mL propidium iodide. Cell cycle data were acquired in independent triplicate repeats for clone 2.43 and NIH 3T3 control cells using a MACSQuant Analyzer 10 flow cytometer (Miltenyi). Output was analyzed using ModFit LT 4.0 (Verity Software House) diploid modeling with automatic linearity detection without range restrictions except for restriction of debris recognition below the maximum value of the putative G1 peak. All RCS values, indicative of goodness of fit, were below 3.3. The modeled G1, S, G2, aggregate and debris values were analyzed by stacked two-way ANOVA.
Results
Identification of a novel family of katanin-like 2 (KATNAL2) isoforms in mouse
We have recently shown that nucleotide-binding proteins Nubp1 and Nubp2 are core components of cilia and flagella in different species and RNAi knockdown of either Nubp demonstrated their role as key regulators of ciliogenesis [42]. Here, we conducted a yeast two-hybrid screen to identify protein interactions for Nubp1. Fifty-eight million interactions were screened between a mouse embryonic brain cDNA library and Nubp1 cDNA as bait: several candidate clones were identified, amongst them a partial cDNA clone of 374 bp. It aligned with sequences within a predicted murine katanin-like protein 2 cDNA (Katnal2; XM_006526437) and encoded 112 amino acids of its predicted open reading frame (ORF; aa 73–184; Fig. S1). This candidate was particularly interesting, given the important role of katanin in many MT-based processes, including ciliary dynamics (“Introduction”), and we thus decided to characterize it further.
When we performed RT-PCR using oligonucleotide primers that hybridized to the extremities of the predicted ORF of mouse Katnal2 XM_006526437, we noticed that, in addition to the expected full-length Katnal2 cDNA product (1.6 kbp), we consistently obtained several, closely migrating products with cDNA from the NIH 3T3 mouse cell line (Fig. 1a) and also from the IMCD, NSC34 and TM4 mouse cell lines and mouse testis (data not shown). Upon cloning and sequencing of these discrete bands from NIH 3T3 cells, we identified five distinct cDNAs containing ORFs that range in size between 1.62 and 1.12 kbp and correspond to alternatively spliced products of the Katnal2 genomic locus (Fig. 1b, e). The Katnal2 gene (localized on the complement strand of mouse chromosome 18: 76,977,147–77,048,225, Build GRCm38.p3) is composed of 16 exons and the 5 cDNAs reflect alternative exon usage, as shown in Fig. 1e. We named these five members of the novel KATNAL2 protein family as Katnal2-L1 (identical to database entry XM_006526437), Katnal2-L2, Katnal2-L3, Katnal2-S1 and Katnal2-S2 (new accession numbers in legend of Fig. 1e). We detected Katnal2 gene expression in several mouse tissues by RT-PCR, using oligonucleotide primers that would amplify a short diagnostic fragment common to all five Katnal2 transcripts, with notable high-level expression in the testis (Fig. 1c).
All five Katnal2 alternative transcripts encode katanin p60-like proteins bearing the typical protein sequence elements of katanin (AAA, MT-binding and Walker motif signature sequences), ranging in expected molecular masses between 41.8 and 61.1 kDa, with sequence variations close to their N-terminus, and with three of them harboring a characteristic LisH motif, stipulated to be important in self-association, MT-binding or protein–protein interactions with ATPases and β-propeller-containing proteins [44] (Fig. 1e, Fig. S1). We generated an antibody raised and purified against recombinant His-tagged KATNAL2-S1, which was expected to recognize the endogenous S1 isoform as well as all other KATNAL2 isoforms, given their extensive sequence similarity. This antibody recognized the recombinant protein against which it was raised as well as KATNAL2-S1 and KATNAL2-L1 fused with different tags (mCherry-, GFP-, Myc- and FLAG-) in transfected NIH 3T3 or IMCD mouse cell lines (Figs. 2, 7 and data not shown). WB analysis revealed several KATNAL2-immunoreactive bands in mouse cell lines and in testis in the range of ca. 45–80 kDa (Fig. 1d, lane 1, Fig. S2), being compatible with the expected sizes of KATNAL2 isoforms, as predicted by cDNA cloning. We employed immunoprecipitation (IP) in conjunction with liquid chromatography, coupled with tandem mass spectrometry, to identify these signals using mouse testis. We excised 3 bands (migrating close to the 50-, 60- and 80-kDa markers; red arrows in Fig. 1d, lane 3′) that were uniquely visible in the bound fraction of the KATNAL2-IP sample and absent from the equivalent negative control in a colloidal-blue-stained gel and, at the same time, corresponded to the main immunoreactive signals on the WB that was carried out concurrently with a smaller quantity (20 %) of the same samples (red arrows Fig. 1d, lane 3). MS analysis confirmed the existence of KATNAL2 signature peptides in all three bands (Table S3). We repeated the experiment using twice as much starting material and cutting out contiguous broader zones (rather than bands) flanking the 50-, 60- and 80-kDa markers in both the KATNAL2 IP and also the negative control sample (goat IgG) (Fig. S3A2). We identified by MS all proteins in each broad zone: this analysis confirmed the existence of KATNAL2 proteins in all three zones (the sequence coverage map indicates peptides throughout the KATNAL2 length, including its N and C termini; Fig. S3B) and comparison between the positive and negative samples revealed KATNAL2-derived peptides uniquely in the positive samples, illustrating the specificity of the IP reaction (Table S4).
Fig. 7.
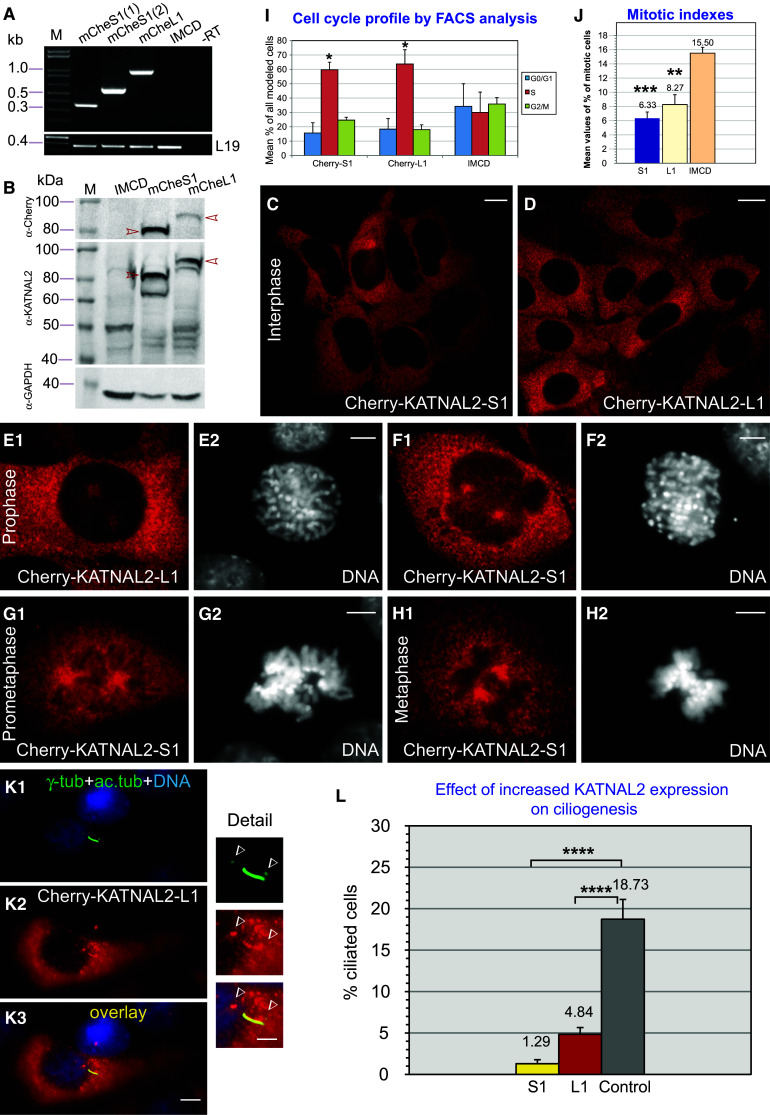
Generation and characterization of stable cell lines expressing Cherry-KATNAL2-S1 or -L1: increased expression of KATNAL2 decreases the percentage of ciliated cells. a Confirmation by RT-PCR of the integration of mCherry-KATNAL2 constructs into the genome of IMCD cells in each case (top panel), and equivalent reactions for L19 as internal normalizing control (bottom panel). Forward oligos to mCherry and reverse oligos either to mCherry (lane 1) or an internal Katnal2 sequence (lanes 2–5), were used to generate specific diagnostic products (oligo sequences in Table S1). From left to right: stable cell line IMCD-pmCherry-KATNAL2-S1 (lanes 1 and 2), IMCD-pmCherry-KATNAL2-L1 (lane 3), IMCD wild type (neg. control 1; lane 4), IMCD-pmCherry-KATNAL2-S1 minus RT (neg. control 2; lane 5). M size markers in kb. b Confirmation by WB of expression of mCherry-KATNAL2 fusion proteins, using anti-mCherry (top panel), anti-KATNAL2 (middle panel), and anti-GAPDH antibody as loading control (bottom panel). From left to right: IMCD wild type (neg. control; lane 1), stable cell line IMCD-pmCherry-KATNAL2-S1 (lane 2) and IMCD-pmCherry-KATNAL2-L1 (lane 3). In lanes 3 and 4, bands consistent with the appropriate size for Cherry-fusion proteins KATNAL2-S1 and -L1 are detected (red arrowheads), with expression levels comparable to the native KATNALs (all lanes). c–k Series of examples to show correct localization of mCherry-KATNAL2-S1 or -L1 at interphase (c, d), mitosis (prophase in e1, e2 and f1, f2; prometaphase in g1, g2 and metaphase in h1, h2) and at the primary cilium (k1–k3 and magnified images of cilium in the detail). mCherry labeling in red, acetylated tubulin in green and DNA in blue. Scale bars 5 μm; panels c, d 10 μm; detail of k1–k3 series 2.5 μm. i Flow cytometry analysis of the cell cycle profile of the three cell lines (IMCD and mCherry-KATNAL2-S1 or -L1) reveals a significant difference in the overall proportion of cells in different cell cycle phases (G1/0, S and G2/M) in the populations expressing KATNAL2 fusions, and, in particular, a significant increase of cells in the S phase for both -S1 and -L1 cells. j Reduction of the mitotic index of the Cherry-fusion KATNALs, compared to the control line. Values shown are the means of three independent experiments ± SD. Sample size corresponded to “dataset 5” (“Materials and methods”). l Quantification of the percentage of ciliated cells in populations of mCherry-KATNAL2-S1 (“S1”), and -L1 (“L1”), compared with corresponding control IMCD cells (“Control”). Shown are the average percentages from three independent experiments ± SD values. Sample size corresponded to “dataset 6” (“Materials and methods”)
In conclusion, the combination of cloning, immunodetection and IP/MS confirmed the existence of a novel family of alternatively spliced KATNAL2 isoforms in mouse cells.
KATNAL2 isoforms interact with themselves, with each other and with Nubp1 and Nubp2
Having confirmed the existence of a novel protein family of KATNAL2 isoforms, and prompted by our initial finding of the yeast two-hybrid screen, we next sought to characterize putative interactions of KATNAL2s with Nubp1 and Nubp2 proteins. Because in pilot experiments we had evidence indicating likely self-interactions of the KATNAL2s, we set up the experiments in a manner allowing us to probe their putative interactions with the Nubp proteins but also amongst themselves. We set up in vivo pull down assays using as bait transiently transfected FLAG-tagged KATNAL2-S1 or -L1 in a constructed stable cell line that expressed tagged Cherry-KATNAL2-S1 (see later “Results” section for full characterization of this stable line). When we pulled down the FLAG-KATNAL2-S1 or -L1 as bait in extracts of such cells with anti-FLAG Sepharose beads, we consistently detected both Nubp1 and Nubp2 by WB as co-selected proteins; Nubp1 and Nubp2 were absent in the equivalent negative control reaction, lacking the bait protein (compare lanes 1′ vs. 2′ and 3′ in Fig. 2a). Interestingly, in the co-selected complex we also detected the “endogenous” Cherry-KATNAL2-S1 (Fig. 2a; “CS” for Cherry-KATNAL2-S1) as well as several native KATNAL2 isoforms migrating close to 40-, 50- and 60-kDa markers (Fig. 2a, lane 2′), indicating that the FLAG-KATNAL2-S1 bait was pulling down the whole range of available KATNAL2s. Again, all these KATNAL2s were absent in the negative control IP reaction (Fig. 2a, compare lanes 1′ and 2′). In the parallel experiment, using this time FLAG-KATNAL2-L1 as bait, FLAG-KATNAL2-L1 was not only able to specifically pull down both Nubp1 and Nubp2, but, as before, it co-selected both Cherry-KATNAL2-S1 as well as native KATNAL2s (Fig. 2a, lane 3′). Together these results give consistent evidence for interactions within the KATNAL2 family, both self-interactions (i.e. S1 with itself, and L1 with L isoforms), as well as interactions between S1 and L1 isoforms and between L and S isoforms in general. This is not unprecedented within the katanin family, given the well-characterized interactions between p60 monomers or between p60 and p80 katanin proteins [17, 18].
These assays from mouse cells clearly indicated the co-selection of both Nubp1 and Nubp2 with either S1 or L1 KATNAL2 isoforms. However, because of the well-established interaction between Nubp1 and Nubp2 in mouse cells [45], these assays could not discriminate whether the presence of both Nubp1 and Nubp2 in the bound fraction was a result of an interaction of each protein individually with the KATNALs or could have occurred because of the interaction of one of the Nubps with the KATNALs and the “passive” concurrent co-selection of the other Nubp protein. To resolve this point, we tested in vitro whether each of the KATNALs interacts directly with Nubp1 or Nubp2, using recombinant proteins expressed in bacteria. GST-tagged forms of KATNAL2-L1 or KATNAL2-S1 were incubated in pairwise combinations with His-tagged Nubp1 or Nubp2. Following incubation, the proteins were mixed with glutathione Sepharose beads, and bound and unbound fractions were separated by SDS-PAGE, transferred to nitrocellulose and probed with anti-GST and anti-His antibodies to detect the presence of KATNALs and Nubps, respectively (Fig. 2b1, b2, c1, c2). These experiments showed that Nubp1 and Nubp2 were each, individually, recovered in the bound fraction only when incubated in the presence of KATNAL2-L1 or KATNAL2-S1; in the negative control with GST-only, all of the His-Nubp1 or Nubp2 protein were recovered in the supernatant, unbound fraction (Fig. 2b1, b2, c1, c2 compare lanes 1–3 with negative controls in lanes 1′–3′). These results with the recombinant proteins thus strongly support the conclusion that KATNAL2-L1 interacts directly with Nubp1 and also Nubp2 and, similarly, that KATNAL2-S1 also interacts directly with Nubp1 and Nubp2.
KATNAL2 is localized on interphase MTs, centrosomes, mitotic spindle, midbody and cilium
We next probed the intracellular localization of KATNAL2. Immunofluorescence (IF) analysis in mouse NIH 3T3 and IMCD cells (Fig. 3), using double labeling combinations of the anti-KATNAL2 antibody with antibodies to α- or γ-tubulin, revealed prominent KATNAL2 labeling associated with the MT cytoskeleton at interphase (Fig. 3a). Although at interphase the concentration of KATNAL2 immunoreactivity at the centrosomes specifically was modest (Fig. 3a, b), the protein was highly enriched in centrioles at the center of MT asters at the onset of mitosis (prophase and prometaphase) and in later mitotic phases labeling also extended to all spindle MTs; the nucleocytoplasm was also labeled throughout mitosis (Fig. 3c, d). The same pattern of labeling at interphase and mitosis was observed in other mouse cell lines that we tested, namely TM4, NSC34 and N2A (selected examples in Fig. 3f, g and other data not shown).
Fig. 3.
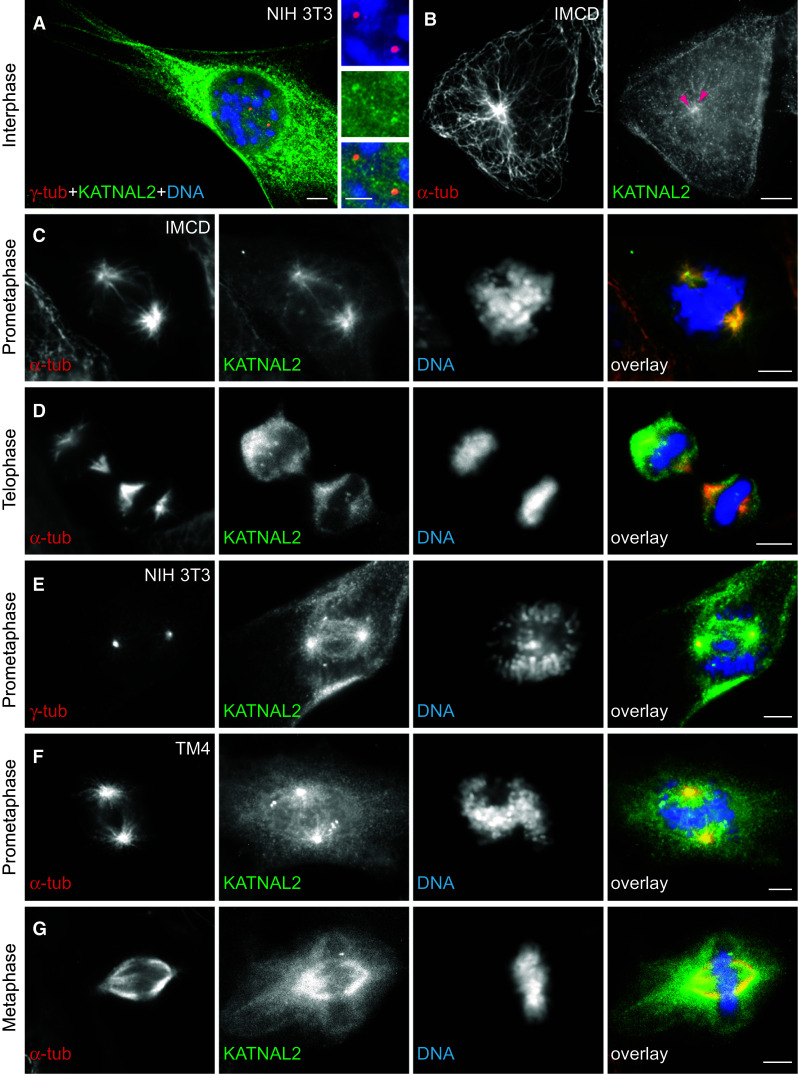
Intracellular localization of KATNAL2 on MTs, centrioles and mitotic spindle in different mouse cells by immunofluorescence. a, b At interphase, immunostaining for KATNAL2 (green) revealed a pronounced, detergent-resistant, cytoplasmic network and some concentration at the centrioles of the centrosome, as shown in NIH 3T3 fibroblasts (a; double labeling of centrosomes with anti-γ-tub in red; detail of centrioles at higher magnification in small side images) or IMCD cells (b; double labeling of MTs with anti-α-tub in red; arrowheads point at the two centrioles). Nuclei were counterstained for DNA with Hoechst (blue). c–f Series of examples of localization of KATNAL2 (green) during mitotic subphases, showing double labeling for α-tub (red) or for γ-tub (red, e) and counterstaining for DNA (blue) in different mouse cell lines (IMCD in c, d; NIH 3T3 in E; TM4 in f, g). KATNAL2 is highly concentrated at the centrosomes and MT asters at early mitotic phases and extends to the whole spindle in metaphase. The nucleocytoplasm is also strongly labeled throughout mitosis. Scale bars 5, 3 μm in detail of a
We also probed KATNAL2 localization specifically in cilia, and examined IMCD and TM4 cells induced to form primary cilia by serum deprivation. We detected KATNAL2 signal at the axoneme itself and its associated basal body, as well as in the daughter centriole that did not form a cilium, in both cases at levels comparable to interphase centrioles (Fig. S4). These results, therefore, indicated the presence of KATNAL2 at the cilium; to date the localization of katanin and MT-depolymerising kinesin-like proteins have been documented at the ciliary axoneme and basal body [14, 31, 32, 46–48] and thus KATNAL2 proteins can now be added as putative players in axonemal dynamics.
KATNAL2 affects centriole arithmetics and MT dynamics, is involved in cytokinesis and is implicated in cell cycle progression
We found the MTs and centrosomes in interphase cells and the mitotic spindle and centrosomes at its poles in dividing cells to be prominent sites of localization of KATNAL2 proteins, consistent with reported roles of related proteins katanin and KATNAL1 (“Introduction”); they were thus a logical target for further functional investigation. We, therefore, next attempted to address the functional role of KATNAL2 at these sites in cycling cells by manipulating its concentration in vivo.
We achieved efficient Katnal2 downregulation using different Katnal2-specific siRNAs but noticed that silenced cells were particularly sensitive to very low Katnal2 expression which appeared toxic and caused massive cell death (data not shown). We, therefore, turned to shRNA-mediated silencing, offering the possibility of antibiotic selection of viable clones and generated a stable NIH 3T3 line via transduction with lentivirus expressing a Katnal2-specific silencing vector, targeting an area in its ORF that was common to all Katnal2 isoforms and would thus simultaneously silence them all (Fig. S1). As our protein–protein interaction experiments had indicated that KATNAL2 isoforms interacted with each other, possibly forming oligomers in vivo, the alternative strategy of targeting individual KATNAL2 isoforms was not pursued. The latter approach might mask effects of individual silencing through redundancy of different isoforms and would, therefore, be less informative on its own. After selection and screening of isolated clones, we identified clone 2.43, which showed considerable reduction of Katnal2 expression. Specifically, total Katnal2 mRNA was reduced to 42.17 ± 4.62 % of the normalized average control value of the NIH 3T3 parental line, as quantified by RT-qPCR using oligonucleotides to amplify sequences common to all Katnal2 isoforms (p = 2.68 × 10−5; or was reduced to 44.37 ± 8.47 % of controls using PCR oligonucleotides specific for the large Katnal2 isoforms, p = 3.40 × 10−4) (Fig. 4a). KATNAL2 protein-normalized average levels were similarly reduced relative to control levels as quantified by WB (to 38.04 ± 13.78 % of control values, p = 0.02) (Fig. 4b1, b2). Interestingly, protein levels of KATNAL2-interacting proteins Nubp1 and Nubp2 were also found to be reduced (to 47.97 ± 8.93 % of control value, p = 0.04 for Nubp1, and to 58.34 ± 8.06 %, p = 0.01 for Nubp2; while the expression of actin, as an unrelated protein, was essentially unaffected; Fig. 4b2).
Fig. 4.

Characterization of stable clone 2.43 generated with shRNA-mediated silencing of Katnal2. a Efficiency of Katnal2-specific silencing in stable clone 2.43, assessed by RT-qPCR, using primers specific for all Katnal2s (lane 1) or Katnal2-L1 + L2 (“Large”; lane 2), depicts, respectively, a normalized average knockdown in silenced cells, expressed as a percentage of the normalized average control values (parental NIH 3T3 line). Error bars correspond to SD of three independent experiments. L19 and Pum1 mRNA levels were used for sample normalization. b1, b2 Representative WB to confirm silencing of KATNAL2 isoform proteins (b1). Concurrent detection of calnexin in the same samples was used as loading control for normalization. The quantification of three independent experiments shown in (b2), depicting the average normalized value of KATNAL2 protein levels as a percentage of the control value, reveals reduction of KATNAL2, Nubp1 and Nubp2 protein levels, but not of actin. c Growth curves of clone 2.43 (blue) and parental control (gray) reveal a severely reduced growth rate in Katnal2-silenced cells. Shown are the average values ±SD of three replicate cultures, each seeded with 6680 cells, grown in parallel for 96 h and sampled every 24 h. d1, d2 Analysis of total cellular size (d1) and size of nucleus (d2) in clone 2.43 (blue) and parental control line (gray), revealing large increases of both due to Katnal2 silencing. The total cell surface area and the nuclear area were measured and plotted out as a size distribution in groups ranging from 0 to 70,000 μm2 (d1) or 200 to 3,000 μm2 (d2). e1, e2 Visualization of the remarkable difference in cell size between control (e1) and Katnal2-silenced clone 2.43 (e2) in two random fields, photographed at the same magnification. Labeling for α-tub in red and for DNA in blue. Scale bars 20 μm. f Cell cycle analysis of clone 2.43 and parental control line by flow cytometry, illustrating reduction of S phase and increase of G2/M phase cells (3 independent experiments, n = 75–200 thousand events each from control and clone 2.43 silenced cells). This indicated that Katnal2 silencing causes stalling in G2/M phase, being consistent with an altered cell cycle progression pattern. g Quantification of the Katnal2 silencing effect on centriole numbers in a bar chart depicting the overall distribution of the number of centrioles per cell in the populations of silenced clone 2.43 (blue) or control parental line (gray). While most cells possess 2 centrioles, in the silenced cells supernumerary centrioles can reach very high numbers (>12 per cell). The overall distribution of centriole numbers per cell is different (p < 0.0001). Shown are the means (±SD values) from three independent experiments (see also Table S6). Sample size corresponded to “dataset 1” (“Materials and methods”)
Characterization of the Katnal2-silenced clone 2.43 revealed a series of its distinctive features. First, this clone displayed an altered growth pattern (p < 0.0001) compared to the control parental clone, with markedly reduced cell numbers at each time point of its growth curve, sampled over a period of 96 h after seeding, suggesting a drastically lower rate of cell division or increased cell death (Fig. 4c). Combined with this growth defect, Katnal2-silenced cells had a significantly larger total size compared with controls; while total cell size of control cells ranged between 100 and 1000 μm2, the size distribution in silenced cells extended between 100 and 70,000 μm2 (difference of distribution p < 0.00001; Fig. 5d1). This was accompanied by a larger size of nucleus, extending between 200 and 800 μm2 in controls and between 200 and 3000 μm2 in silenced cells (difference p < 0.00001; Fig. 4d2). Random wide field views of control and silenced cells, at the same magnification (Fig. 4e1, e2), illustrate the remarkable difference in cellular size. This indicated viability with low rate of cell division combined with a steadily increasing cell size for silenced cells, in the absence of typical apoptotic phenotypes.
Fig. 5.
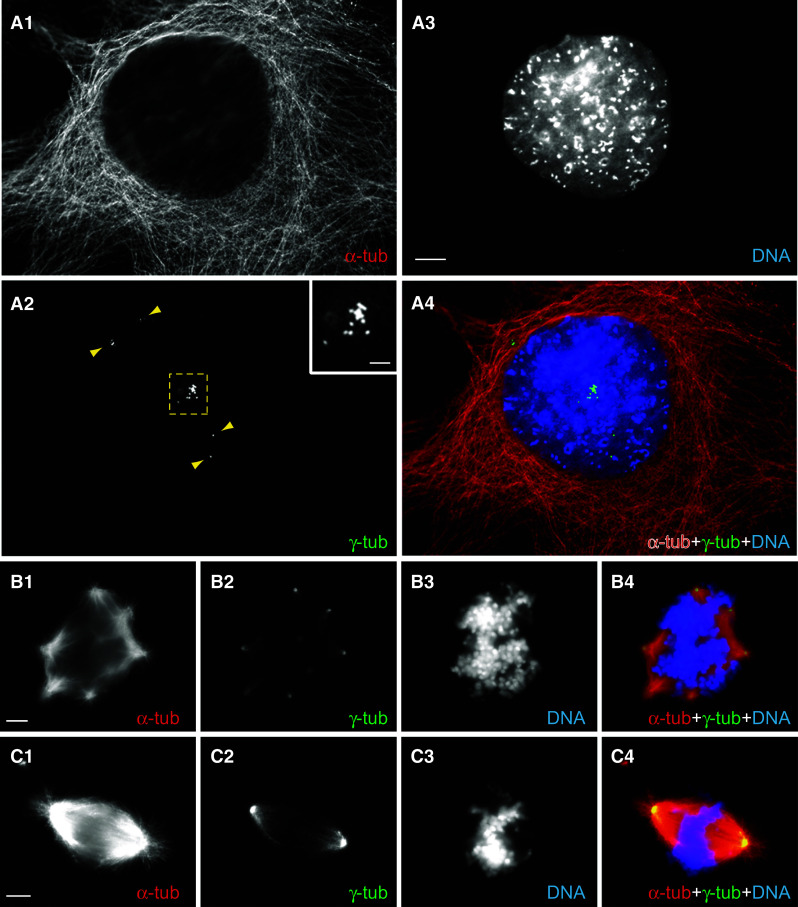
shRNA-mediated silencing of Katnal2 causes a battery of aberrant phenotypes (I). In all images, α-tub labeling is in red, γ-tub in green and DNA in blue. a1–a4 IF in silenced clone 2.43 shows excessive supernumerary centrioles (yellow arrowheads and square) in an interphase cell. The inset in a2 shows the detail of a cluster of amplified centrioles (contained in the yellow square) at higher magnification. b1–b4 The presence of supernumerary centrioles in silenced cells induces the formation of multipolar mitotic spindles: a cell in prometaphase with eight centrioles and a multi-polar spindle as an example. c1–c4 The depicted silenced cell shows mis-aligned chromosomes on the metaphase plate. Scale bars 10 μm in a1–a4; 3 μm in inset of a2; 5 μm in b and c series
Live imaging of control NIH 3T3 cells and clone 2.43 cultures, over 24 h, provided intriguing first insight into the mechanisms underlying the observed phenotypes. The often binucleated enlarged cells with enlarged nuclei, typical of Katnal2-silencing, had clear difficulty in progressing through the complete mitotic sequence, which was visibly disturbed and lengthy or even anomalous (movie S1), or in successfully executing cytokinesis; incomplete ingression of the cleavage furrow in late mitotic cells in which nuclei had already divided resulted in many cases in the formation of one binucleated cell (movie S2). Even when cytokinesis was almost complete, occasionally the daughter cells re-fused to give a large binuclear cell (movie S3). Alternatively, the result of mitosis was the formation of two daughter cells that were not fully separated but were connected with a persisting narrow cytoplasmic bridge and remained close to one another long after mitosis (movie S4). When complete cleavage was achieved, it was sometimes asymmetrical (movie S5). These observations were indicative of severe cytokinesis and also mitotic defects when KATNAL2 was depleted, leading to multinucleated, polyploid and aneuploid cells that could grow in size, but not divide effectively. Such defects were consistent with the observed impairment in their growth curve.
Furthermore, the cell cycle profile of populations of silenced and control cultures, as determined by flow cytometry, revealed that silencing of KATNAL2 affected normal progression through the cell cycle. In particular, in clone 2.43 the percentage of cells in phase S was reduced and in G2/M increased, compared to controls (both with extreme statistical significance p < 0.0001) (Fig. 4f) and this indicated stalling in G2/M phase.
To analyze further and to quantify these phenotypes, we performed a detailed microscopic analysis of a very large number of cycling cells (“Materials and methods”) with triple fluorescence for α- and γ-tubulin and DNA labeling, scoring the mitotic index, the percentage of cells in each mitotic subphase, the number of centrioles in interphase and mitosis, the morphology of the mitotic spindle, the number of nuclei per cell and incidence of chromosomal abnormalities (such as chromatin bridges). This revealed a 4.5-fold reduction of the mitotic index in silenced cells, with 2.55 ± 0.80 % of silenced cells in mitosis as opposed to 11.78 ± 0.89 % in control cells (Table S5; p = 0.00018). Analysis of the distribution of mitotic cells in different mitotic subphases gave a highly significant overall difference between the two populations (p = 0.0011), with a main difference in the apparent accumulation of a sizeable proportion of cells (18.4 % in silenced vs. 2.6 % in control mitotic cells) in prometaphase (p = 0.0007; Table S5). The most striking observation was the high incidence of marked centriole amplification in silenced cells, throughout the cell cycle, both in mitosis and interphase: a third of all silenced cells had supernumerary centrioles, representing a fivefold increase relative to control cells (p = 0.00055) and the centriole number distribution was remarkable and different overall for control and silenced cells (overall difference p < 0.0001) (Table S6 and Fig. 4g). The numbers of supernumerary centrioles per cell ranged from 5 up to ≫15 in silenced cells (Table S6 and Fig. 4g) and, as a consequence, the average number of centrioles per cell was elevated (3.02 ± 0.12 centrioles on average per silenced cell vs. 2.12 ± 0.05 in controls, p = 0.00026; Table S6) (with Fig. 5a1–a4 giving an example of a multicentrioled interphase cell). As a result of centriole amplification, many mitotic cells possessed anomalous multipolar spindles (Fig. 5b1–b4) and some exhibited chromosome misalignments (Fig. 5c1–c4). The presence of such multipolar spindles most likely accounted for the observed increase of the proportion of cells in prometaphase (indicating difficulties in the accurate sorting of chromosomes due to simultaneous forces exerted by multiple poles) in silenced cells. The presence of multipolar spindles is also an established contributor to chromosomal aneuploidy and polyploidy as a result of inaccurate chromosome segregation on the spindle. Aneuploidy and polyploidy are further exacerbated by inaccurate cytokinesis; we had already observed cytokinesis defects by live imaging, an observation corroborated by aberrant or unequally dividing daughter cells in silenced cultures (Fig. 6 series a, b). Consistent with chromatin segregation and cytokinesis failure was the elevated incidence (a) of multinucleated cells (with 7.12 ± 0.70 % of silenced cells having more than one nucleus vs. only 1.66 ± 0.01 % in controls, p = 0.0001), as well as the enlarged size (already analyzed above) accompanied by the presence of multi-lobed nuclei, mini nuclei, adjoining nuclear “buds” or nuclei with abnormal size or shape (Fig. 6a3, purple arrowheads, c3), all features being indicative of polyploidy and aneuploidy, and (b) of long chromatin bridges in post-mitotic cells (Fig. 6a3, b3 yellow arrowheads) or imperfectly aligned chromosomes in mitotic cells (Fig. 5c3–c4), landmark aberrations stemming from inaccurate or incomplete chromosome segregation. A notable observation in silenced cells in cytokinesis was the frequent presence of pronounced, elaborate and persistent MT bundles at the midbody (as exemplified in Fig. 6a, b series), often visible in post-mitotic cells that had not fully separated and were still attached to each other.
Fig. 6.
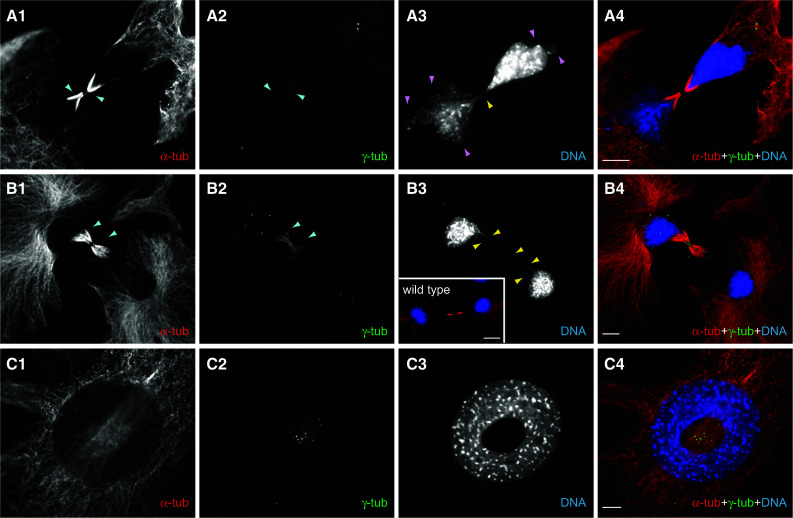
shRNA-mediated silencing of Katnal2 causes a battery of aberrant phenotypes (II). a1–a4 Aberrant cytokinesis in silenced cells: a persistent chromatin bridge (yellow arrowheads) is visible between two cells with anomalous multi-lobed nuclei (pink arrowheads), which are still linked through a cytoplasmic connection despite being at an advanced stage of cytokinesis. Pronounced and persistent MT bundles, revealed by α-tub labeling and also visible with γ-tub staining, can be observed at the midbody (blue arrowheads). DNA labeling in blue. The dividing cells are unequal in size (the field does not contain the entirety of the cells) and unusually shaped. b1–b4 Another example of aberrant cytokinesis of multicentrioled dividing cells, linked with a double chromatin bridge (yellow arrowheads) and persistent and elaborate MTs at the midbody (α-tub), also immunoreactive for γ-tub (blue arrowheads). In the inset of b3 shown is a normal, wild-type cell for comparison of cytokinesis and midbody MT organization (overlay of α-tub in red and DNA in blue). c1–c4 An excessively overcentrioled cell (>12 centrioles) with an anomalous enlarged nucleus harboring a large hole that is traversed by a bundle of MTs (blurred in the image as they are on a different focal plane). In all panels, cells were immunostained for α-tub (red), γ-tub (green) and counterstained for DNA (blue). Scale bars 10 μm
Prompted by these observations and given the role of katanin proteins in MT severing, we studied the overall organization of the MT network in silenced cells: there was no discernible gross morphological change of the MT network in interphase cells, as revealed by anti-α-tubulin labeling of Katnal2-silenced cells and compared with controls (Fig. S5, A1-C1). Interestingly, when we concurrently compared this same network with an antibody to acetylated tubulin, we observed a notable signal increase of the proportion of acetylated microtubules within the MT network in silenced cells, implying that acetylation, a major post-translational modification of MTs used as a marker of older, more stable and less dynamic MTs, is enhanced in the absence of KATNAL2 activity (Fig. S5 compare control in C2 and silenced cells in A2, B2).
Given that we could not carry out rescue experiments with silenced clone 2.43 (see subsequent section), to exclude the possibility that its phenotypes derived from chance insertional effects of the shRNA-encoding cassette into the genome, we went ahead to generate an independent Katnal2-silenced clone, using a different shRNA construct targeted to another ORF area, common to all Katnal2 transcripts and downstream from that affected in clone 2.43 (Fig. S1). We isolated clone 3.81; Katnal2 mRNA levels (not shown) and protein levels were both reduced, compared to controls (Fig. S6 panel C for WB). After careful repetition for clone 3.81 of analyses undertaken for clone 2.43, our findings closely paralleled those obtained with clone 2.43 and replicated all the telltale features described for the downregulation of Katnal2. Specifically, clone 3.81 cells exhibited increased cellular and nuclear size (Fig. S6 A1–A4), acquisition of supernumerary centrioles and multipolar spindles (Fig. S6, A1–A4, detail in A2; B1–B4) so that a third of silenced clone 3.81 cells had more than two centrioles (with 32.76 ± 0.84 vs. 8.25 ± 0.30 % in control, p = 1.18 × 10−6; Table S7), the average number of centrioles per cell was augmented as compared with control wild-type cells (2.79 ± 0.039 centrioles per cell vs. 2.16 ± 0.012, p = 1.09 × 10−5, Fig. S6D; Table S7) and the overall distribution of their numbers per cell different (p < 0.0001; Table S7), the incidence of multipolar mitotic spindles was over an order of magnitude higher than in controls (with 15.83 ± 1.01 % of mitotic cells vs. 1.56 ± 0.40 % in controls, p = 2.24 × 10−5). As before, the incidence of binucleated cells was increased (tripled to 3.9 ± 0.35 vs. 1.36 ± 0.25 %, p = 0.0006), the frequency of chromatin bridges, indicating chromosome segregation and cytokinesis defects, quadrupled (to 2.33 ± 0.23 vs. 0.40 ± 1.15 × 10−5 %, p = 0.0045) and the mitotic index reduced to a third of controls (to 3.37 ± 0.0025 vs. 10.44 ± 1.2 %, p = 0.0006). Our findings with two differently targeted and totally independent Katnal2-silenced stable clones showed remarkable consistency of phenotypic characteristics and thus confirmed the specificity of the silencing phenotypes.
In conclusion, the combination of highly complementary evidence with different experimental approaches indicated that Katnal2 downregulation appears to affect MT-based structures throughout the cell cycle and likely has an impact on the cell cycle itself; these findings are consistent with different aspects of MT-based function of katanin in discrete biological processes.
KATNAL2 is also involved in ciliogenesis
Additionally, because of the localization of KATNAL2s at the ciliary axoneme and basal body, their interactions with the Nubp proteins, shown to be regulators of ciliogenesis [42], and the known involvement of the archetypal katanin in axoneme dynamics in motile cilia [14, 49], we also investigated the possible role of KATNAL2 proteins specifically in ciliogenesis.
We subjected silenced clones 2.43 and 3.81 and control NIH 3T3 cells to serum deprivation to induce the formation of sensory cilia. We compared them to equivalent cultures grown in parallel in full growth media by microscopically scoring the number of ciliated cells, as revealed by double labeling for γ-tubulin (basal body marker) and acetylated tubulin (ciliary axoneme marker), in each of the silenced and control populations.
We observed that while uninduced silenced cells had essentially the same, low basal-level proportion of ciliated cells as control cultures, upon induction by serum deprivation the incidence of cilia of Katnal2-silenced cells of clone 2.43 was less than half that of controls, resulting in a significantly lower number of ciliated cells. Specifically, the average percentage of ciliated cells in the Katnal2-silenced 2.43 population was reduced to less than half, compared to the average of control-silenced levels (with 16.70 ± 4.15 % in Katnal2 silencing vs. 37.39 ± 8.02 % in controls, p = 0.0165) (Fig. S7). The equivalent results with silenced clone 3.81 reproduced the decrease in the proportion of ciliated cells to about half the control value (with 21.47 ± 2.12 vs. 38.06 ± 2.97 % in controls, p = 0.0014; Fig. S6E). These results suggested that in the absence of KATNAL2, there is a decline in the propensity for ciliogenesis or in the stability of newly formed cilia, resulting in a reduced population of ciliated cells.
We next also wished to assess the impact of increased KATNAL2 expression on ciliogenesis, initially by overexpression of KATNAL2 via transient transfection. We consistently observed that overexpression of tagged forms of either KATNAL2-L1 or KATNAL2-S1 or their combination resulted in pronounced cell apoptosis and in the vast majority of non-apoptotic cells (>80 %) to the mislocalization and aggregation of KATNAL2 in large cytoplasmic inclusions/aggregates, irrespective of the tag used or its position in the fusion protein (examples of FLAG-, Myc-, GFP- and mCherry-tag in Fig. S8). These observations, in conjunction with our observations with initial siRNA-mediated silencing attempts (previous section), led us to speculate that in vivo there is a strict regulation of KATNAL2 protein levels and that deviation to very low or high-level expression may be toxic to cells. As an alternative strategy, we proceeded to construct IMCD cell lines stably expressing fusion proteins of either Cherry-KATNAL2-S1 or -L1 following integration into the genome. Clones were cultured under antibiotic selection, isolated, screened and characterized: the final selected clones exhibited relatively moderate fusion protein transcript levels, as assessed by RT-PCR using oligonucleotide primers specific for the Cherry-KATNAL2 fusions (Fig. 7a), moderate fusion protein expression and comparable to that of the endogenous protein (Fig. 7b), and correct Cherry-S1 or -L1 localization (cytoplasm, mitotic spindle and cilia) identical to that of the endogenous KATNAL2 protein (examples in Fig. 7c–k series). We further characterized these stable clones by evaluating their cell cycle profiles by flow cytometry (Fig. 7i) and quantifying their mitotic indexes (Fig. 7j) and mitotic phase distribution by microscopic analysis of a large number of cells (Fig. S9). Cell cycle analysis for S1- and L1-expressing cells, compared to wild-type IMCD cells, revealed a difference between the overall distribution of cells in different phases of the cell cycle (G1/0, S and G2/M) (p = 0.0139), and also a marked increase of cells specifically in the S phase for both S1-transgenic (p = 0.0208) and L1-transgenic (p = 0.0111) cells (Fig. 7i). Consistent with differences in the cell cycle profile, both KATNAL2 S1- and L1-expressing populations showed a lowered mitotic index (p = 0.0007 for S1-transgenic cells and p = 0.003 for L1-transgenic cells; Fig. 7j). Moreover, overall distribution in mitotic subphases was different between cell populations (p = 0.0011) and for S1-transgenic cells compared to wild-type cells revealed significant (p = 0.0122) and extremely significant (p = 0.0006) differences in particular for telophase and prophase percentages, respectively (Fig. S9). Despite these clear differences in their cell cycle parameters, all stable clones maintained a good proliferation rate, normal morphology, stable but moderate expression and correct localization of the fusion proteins, and were thus used for further experiments.
We quantified ciliogenesis in the mCherry-KATNAL2-S1 and -L1 stable lines, comparing them with the parental IMCD cell line, after a 24-h serum starvation regime. Results with both stable cell lines showed a consistent and drastic reduction of the percentage of ciliated cells. Specifically, while the average percentage of the control IMCD line was at 18.73 ± 2.38 %, it was only 4.84 ± 0.82 % for the KATNAL2-L1 stable line (i.e. 26 % of the wild-type control value) and was even further reduced in the KATNAL2-S1 line to a mere 1.29 ± 0.48 % (i.e. 7 % of the wild type), a difference with extreme statistical significance (with overall and individual p values compared to wild-type cells <0.0001, Fig. 7l). It was also apparent that the length of those rare cilia was reduced compared to the controls, although this was difficult to quantify due to the very small numbers of cilia observed in the cultures of KATNAL2-S1 and -L1 cells. This set of experiments supported the notion that increased KATNAL2 expression reduces the number of ciliated cells in the population by either negatively impacting on their assembly or their stability. Although these results could appear unexpected in the face of our silencing results, demonstrating the same effect, i.e. reduced numbers of cilia in the absence/downregulation of KATNAL2s, this finding may in fact point to the idea that what is critically important in this MT-based process is balanced and appropriate MT-severing activity.
In conclusion, therefore, our results with both silencing and increased Katnal2 expression strongly support a regulatory role of the KATNAL2 s in ciliogenesis, suggesting the need of highly regulated activity, ranging within a defined window, as critically important for correct biological output. Overall, finely balancing KATNAL2 activity may be critical for the control of many, if not all, MT-dependent processes, including centriole arithmetics, cell division, cytokinesis and cellular and nuclear size.
Discussion
In this work, we uncovered the existence of a new family of Katanin-like 2 protein isoforms in mouse, arising from alternative splicing of the Katnal2 genomic locus. It is interesting to note that in addition to the archetypal katanin, it would appear that katanin-like proteins are phylogenetically as ancient given that, besides their reported existence in vertebrates and invertebrates (KATNAL1 and 2) [20, 50], such sequences can be predicted in unicellular eukaryotes like Chlamydomonas [(chromosome 10) Cre10.g446400 (phytozome database) is a C. reinherdii KATNAL2-predicted protein (44.1 % identity with mouse KATNAL2-L1) in addition to Chlamydomonas katanin p60 (Cre10.g427600, also on chromosome 10 at a ca. 4.5 Mb distance); Mette Lethan and Lotte B. Pedersen, personal communication].
We show that the different mouse KATNAL2 isoforms are capable of forming protein–protein interactions between them and this finding could suggest an oligomeric functional form, akin to p60 katanin. It is plausible that differential usage of isoforms in a combinatorial manner to form discrete homo- and hetero-oligomeric complexes could underlie intracellular targeting and/or functional specialization (i.e. determine the subset of MT with which each different complex could interact). Conceivably, the distribution of specific tubulin isoforms or post-translational modifications to distinct MT subsets can create discrete MT subpopulations with unique functionality [51, 52] and severing enzyme affinities and thus also contribute to substrate specialization. Given the extensive sequence similarity between the KATNAL2 isoforms and likely low abundance of specific complexes, it would be technically challenging to address such scenarios directly. It is clear in any case that the LisH domain is not essential for inter-isoform interactions since these occur even between LisH-less (S-type) isoforms.
KATNAL2 is localized on interphase MTs, centrosomes, mitotic spindle, midbody and at the basal body and axoneme of the cilium, raising the question what functional roles KATNAL2s might have at these diverse sites in mouse cells? The battery of phenotypes documented in Katnal2-silenced cells indicate an impressive multitasking functionality of KATNAL2s, affecting different MT-based processes in these intracellular locations: downregulation was characterized by centriole amplification throughout the cell cycle, the consequent formation of aberrant multipolar spindles in mitosis, absence, deficiency or inequality of cytokinesis with a concomitant significant number of binucleated cells, a strikingly enlarged cellular and nuclear size likely accompanied by polyploidy and occasionally by abnormal nuclear shape, a disturbed cell cycle profile with increased accumulation in G2, an altered mitotic profile combining reduced mitotic index and some stalling at prophase, and a seemingly normal organization of the MT network at interphase, albeit with indications of enhanced acetylation. Acetylation at K40, a major post-translational modification of α-tubulin, is a molecular hallmark of older MTs that are hotspots of katanin binding and severing activity [14, 53] and its increase in silenced clone 2.43 was not only consistent with the downregulation of KATNAL2 but could be at the basis of several of the observed phenotypes. For instance, depletion or reduction of KATNAL2 would hinder efficient resolution of a highly acetylated and particularly stable bundle of long midbody MTs to allow cytokinesis progression, thus contributing to inability of or unequal daughter cell segregation and consequent anomalies in cellular and nuclear size and ploidy and the acquisition of multiple nuclei. Furthermore, it is well documented that MT severing by katanin is implicated in cell cycle progression and, reciprocally, also regulated by the cell cycle with low MT-severing activity at interphase and highly stimulated activity in mitosis [32, 54]. Notably, MT severing by katanin, that accelerates the generation of unstable and highly turning over MTs, enables G2/M transition and mitotic entry and the concentration of MT fragments with the smallest length reaches a peak at the end of prophase [55]. In this context, the observed increase of the proportion of Katnal2-silenced cells at G2 was a clear indication of a depletion of sufficient MT-severing activity and thus a reduction in the ability to efficiently transform MTs at a critical transition, causing consequent delayed entry into mitosis. Therefore, KATNAL2s appear to influence cell cycle progression through directly modulating MT dynamics.
The enlarged cellular and nuclear size (and presumed altered ploidy) of silenced cells is a remarkable morphological landmark of Katnal2 silencing and a clear consequence of ineffective cytokinesis. Enlarged cell size and polyploidy have been linked [56]: it is thought that polyploidy does not affect the frequency of cell division [57] but rather, it may enlarge cell size by boosting the rate of ribosome biogenesis [58], which could provide an indicator of cellular growth and fitness for mitosis. Additionally, an overly stabilized and less dynamic MT network is a likely contributor to the acquisition of this abnormal size (up to 70 times larger than controls in our experiments). In Arabidopsis, cellular size and shape were found to be affected by katanin-dependent cortical MT dynamics and orientation influencing the ability of the cell to respond to mechanical stress [59]. A frequent observation in our live imaging recordings of Katnal2-silenced cell populations was the occurrence of large nuclei, “rotating” widely for prolonged periods. Spinning nuclei have been reported in a variety of tissues and cells [60, 61] and a model for nuclear “wriggling”, based on this phenomenon in Drosophila follicle cells in egg primordia, entails that the polymerizing (+) ends of cortical long MTs poke the nuclear envelope and exert forces that induce changes in nuclear positioning [62]. The long-lived and elongated MTs in Katnal2-silenced cells may likewise contribute to the observed nuclear rotations. It is conceivable that excessive such forces exerted by unusually stable MTs may also account for nuclear shape deformities (like the hole traversed by an MT bundle in Fig. 6c1–c4).
An additional phenotype in the population of silenced cells is a reduced frequency of cilia, a feature also detected in cells stably expressing Cherry-tagged KATNAL2 isoforms; these findings may indicate that KATNAL2 activity needs to be strictly regulated within a functional range that is compatible with the assembly of the ciliary axoneme using severed seeds of the appropriate length and concentration; an activity of KATNAL2 either below or above a distinct gradient may be detrimental to the process. The increased cellular toxicity of both overt silencing or protein overexpression is compatible with this concept and may explain the difficulty in the katanin field in producing viable or long-lasting cellular or animal models with katanin depletion [for example 33]. We find that KATNAL2s interact directly and independently with Nubp1 and Nubp2 AAA proteins. We previously showed the Nubps acting as negative regulators of ciliogenesis and also raised the possibility that the Nubps may be acting as cellular co-chaperones [42]. It is known that some AAA enzymes can disassemble or modulate stable protein–protein complexes [63], conjuring up the intriguing possibility that Nubp1&2, through their direct interactions with the KATNALs, may form part of KATNAL2 complexes and thus modulate their activity. For instance, Nubp1&2 may keep ciliogenesis at low basal levels by reducing KATNAL2-induced severing of MTs and limiting the supply of MT seeds for the formation of ciliary axonemes in cycling cells. Upon serum deprivation and cell cycle exit, when Nubp1&2 levels are reduced, ciliogenesis was shown to be enhanced compared to cycling cells and, when serum deprivation was combined with Nubp1 or 2 siRNA-mediated silencing for further reduction of Nubp protein levels, ciliogenesis was augmented even further [42]. In the light of the current work, in both cases appropriate increased levels of KATNAL2 activity may be established within a physiological range that is conducive to axoneme formation and thus ciliogenesis can proceed. Consistent with this model, we report here that in conditions of significant KATNAL2 depletion through silencing, the frequency of ciliogenesis drops considerably (to half that of control levels). Interestingly, we found that KATNAL2 depletion appears to cause a decrease of both Nubp1 and Nubp2 protein levels, an observation suggesting decreased stability of the Nubps in the absence of their binding partners. This observation could further suggest an intricate reciprocal relationship between the KATNAL2s and the Nubps: while the Nubps may be acting as repressors of KATNAL2 downstream activity, the absence of KATNAL2 protein may cause degradation of Nubps. It may be the case that in vivo KATNAL2s do not significantly fluctuate (contrary to the Nubps) or get downregulated (unpublished observations). However, it may be that upon reduction of both protein classes under experimental conditions, for instance by depletion of Nubps under serum starvation combined with KATNAL2 silencing, the phenotype associated with KATNAL2 loss of function (reduction of the percentage of ciliated cells in the population) is epistatic to that of the Nubps.
A striking feature of the Katnal2 silencing phenotype is the significant amplification of centrioles throughout the cell cycle with the inevitable formation of multipolar spindles in mitosis. These centrioles appear functional as they appear to be able to nucleate MTs in the spindle and this phenotype is highly reminiscent of the situation that arises from the silencing of the KATNAL2 binding partner, Nubp1 [42]. Recently, centriole amplification was described as a feature of primary cells derived from patients suffering from severe microlissencephaly caused by mutations in the p80 subunit of katanin [34, 35]. Thus, it would appear that many, if not all, katanins affect centriole numbers. While no mechanism was proposed for p80 katanin and centrioles and no specific reference was made regarding cytokinesis defects in asymmetrically dividing human progenitors in the diseased cerebral cortex [34, 35], it is obvious that centriole amplification must, at least in part, be caused as a consequence of cytokinesis defects, as the present study has documented in the case of KATNAL2.
A final consideration is to what extent there is functional redundancy between the different types of katanin in a cell, given that they mostly seem to execute similar functions and to be active within the same cell type; from work in the field and our results it seems, rather paradoxically, that different katanin and katanin-like protein family members do not rescue or substitute the missing function of other knocked-out katanins. This raises the possibility that each katanin may be strictly regulated by distinct upstream partners, such as the Nubps for KATNAL2s, which are not interchangeable so that, despite their functional relatedness, the katanins themselves are not interchangeable either.
Electronic supplementary material
Below is the link to the electronic supplementary material.
FIGURE S1 Primary protein sequences encoded by all the Katnal2 cDNAs identified in this work. Protein motifs and SNPs are indicated. The sequences within the ORF that were targeted for the shRNA-mediated silencing of Katnal2s in clones 2.43 (yellow highlights) and 3.81 (blue highlights) are also indicated. In magenta highlight, the amino acids corresponding to the initial, partial cDNA sequence identified by a yeast two-hybrid screen is indicated. (DOC 37 kb)
FIGURE S2 Detection of KATNAL2 proteins in four mouse cell lines and tissue by WB. (A,B) Anti-KATNAL2 WB of total protein extracts from the indicated mouse cell lines and from mouse testis consistently detects three main immunoreactive bands migrating in the vicinity of 50, 60 and 80 kDa (blue arrowheads). These signals correspond to the signals also obtained in the IP experiment shown in Fig. 1D, in which the three bands were excised from the gel, analyzed by MS and found positive for the existence of KATNAL2 proteins. Abbreviations: M, molecular mass markers. (EPS 1167 kb)
FIGURE S3 Results of LC-MS/MS analysis at larger scale and of the 3 cut-out bands of Figure 1D. (A1,A2) Shown is a larger scale KATNAL2 IP experiment starting with more material, as described in Materials and Methods. Proteins were visualized by SDS-PAGE followed by colloidal-blue staining (A1) and 3 broad zones (boxed), ranging, respectively, between 80-60 kDa (zones “1”), 60-50 kDa (zones “2”) and 50-45 kDa (zones “3”), were cut out from the KATNAL2-IP sample (“POS” series) or the negative control IP (“NEG” series). MS identification of the proteins contained in the 3 positive and 3 negative zones shown here is given as a list in Table S4. (A3) WB analysis of the same samples as in A1/A2 to indicate the existence of several major and minor KATNAL2 immunoreactive bands within the 3 excised zones. KATNAL2 positive identification was obtained in all 3 zones, indicating the existence of multiple KATNAL2 isoforms in this size range specifically in the IP KATNAL2 and not in the IP neg. (B) Aggregate sequence coverage map for KATNAL2 protein, derived from KATNAL2 signature peptides, uniquely detected in the “positive samples” (see Table S4) reveals extensive coverage, throughout the sequence and spanning both termini of the protein. (EPS 8847 kb)
FIGURE S4 KATNAL2 localizes at the axoneme and basal body of primary cilia. (A,B) Examples of localization of KATNAL2 (green) at the axoneme and basal body (white arrowheads) of primary cilia in serum-deprived IMCD cells (A) and TM4 cells (B), showing acetylated tubulin (axonemal marker, red), DNA labeling (blue) and accompanying magnified detailed views of the cilia. The daughter centriole also shows comparable immunoreactivity (yellow arrowheads). Scale bars 5 μm; 1 μm in detailed views. (EPS 7192 kb)
FIGURE S5 shRNA-mediated silencing of Katnal2 is accompanied by a pronounced increase in the acetylation of the MT network. (A1-B2) Two examples of extensive acetylation of the MT network in Katnal2-silenced cells of clone 2.43, as revealed by double α-tubulin (red) and acetylated tubulin labeling (green). DNA is counterstained in blue. Exposure of panel A2 for green channel was 2500 ms; for B2 5243 ms. (C1-C2) Equivalent double staining in a wild type NIH 3T3 control cell for comparison showing relatively restricted or no acetylation of the MT network. Exposure for panel C2 is 6000 ms (that is 250% of exposure in A2 and 114.4% of exposure in B2). Scale bars 20 μm. (EPS 33601 kb)
FIGURE S6 Characterization and phenotypes of Katnal2-silenced stable clone 3.81. In all images, α-tubulin labeling is in red, γ-tubulin in green and DNA in blue. (A1-A4) IF in silenced clone 3.81 shows excessive supernumerary centrioles in an interphase cell (yellow arrowheads and square). The inset in panel A2 shows the detail of a cluster of amplified centrioles (contained in the yellow square) at higher magnification. (B1-B4) The presence of supernumerary centrioles in silenced cells induces the formation of multipolar mitotic spindles: here, as an example, a cell in metaphase with four centrioles and a spindle with 4 poles. Scale bars 20 μm for A1-A4; 5 μm for B1-B4; 2 μm in inset of panel A2. (C) Representative WB to confirm silencing of KATNAL2 isoform proteins. The concurrent detection of calnexin protein in the same samples was used as a loading control for sample normalization. (D) Quantification of the average number of centrioles per cell in Katnal2-silenced populations of clone 3.81 (red), compared with corresponding control-silenced populations (gray), showing the average value from three independent experiments and bars representing S.D. values. The 30% increase of the average number of centrioles per cell, caused by silencing, is extremely significant (****). Sample size corresponded to “dataset 3” (Materials and Methods). (E) Quantification of the percentage of ciliated cells in Katnal2-silenced populations of clone 3.81 (red), compared with corresponding control-silenced populations (gray), at growth conditions (cycling cells) and after induction of ciliogenesis by serum deprivation for 24 hours (cell cycle arrest), respectively. Shown are the averages from three independent experiments and bars represent S.D. values. The reduction in the percentage of silenced ciliated cells after induction of ciliogenesis by serum deprivation is extremely significant (***). Sample size corresponded to “dataset 4” (Materials and Methods). (EPS 18163 kb)
FIGURE S7 shRNA-mediated silencing of Katnal2 in clone 2.43 causes significant reduction of the frequency of sensory cilia in the cell population. Quantification of the percentage of ciliated cells in Katnal2-silenced populations of clone 2.43 (blue), compared with corresponding control-silenced populations (gray), at growth conditions (cycling cells) and after induction of ciliogenesis by serum deprivation for 24 hours (cell cycle arrest). Shown are the averages from three independent experiments and bars represent S.D. values. The reduction in the percentage of silenced ciliated cells after induction of ciliogenesis by serum deprivation is highly significant (**). Sample size corresponded to “dataset 2” (Materials and Methods). (EPS 434 kb)
FIGURE S8 Transient overexpression of KATNAL2-S1 or -L1 tagged fusions leads to cytoplasmic aggregation of the protein in mouse cells. (A-D) Representative examples of common overexpression phenotypes, resulting from transient transfections of differently tagged versions of KATNAL2-S1 or -L1, as indicated. Fluorescence corresponds to GFP emission (green, A), mCherry emission (red, B), anti-FLAG (C) and anti-myc (D) immunofluorescence (green). DNA was counterstained with Hoechst (blue). Panels A, C and D display NIH 3T3 cells and panel B is with IMCD cells. Scale bars 10 μm. (EPS 9005 kb)
FIGURE S9 Cell cycle analysis of IMCD stable cell lines expressing mCherry-KATNAL2-S1 and -L1, and the control line IMCD. Analysis of the distribution in mitotic subphases of the same cells as for the datasets in Fig. 7J (dataset 5 in Materials and Methods) reveals that it was highly significantly different between cell populations (p=0.0011). Additionally, for -S1 cells, compared to wild-type cells, it revealed significant (p=0.0122) and extremely significant (p=0.0006) differences for telophase and prophase percentages in particular. (EPS 472 kb)
MOVIE S1 A very large, binucleated Katnal2-silenced cell (clone 2.43) is struggling to complete mitosis with what appears like a multipolar spindle (observe multiple MT asters). At the end of the sequence, no cytokinesis has been achieved and the nuclear mass consists of multiple, irregularly shaped multi-lobed fragments. The movie is composed of 87 frames, acquired at 7 min intervals (total time=10.15 hours). (MP4 3623 kb)
MOVIE S2 Mitosis without cytokinesis in silenced clone 2.43. A large Katnal2-silenced cell (clone 2.43) with a large nucleus undergoes mitosis (chromosome alignment on the metaphase plate is discernible) without the formation of a complete ingression furrow and fails cytokinesis, resulting in one binuclear cell. The movie is composed of 120 frames, acquired at 7 min intervals (total time=14 hours). (MP4 2441 kb)
MOVIE S3 Failure of cytokinesis and re-fusion of mitotic cells in silenced clone 2.43. A Katnal2-silenced cell (clone 2.43) undergoes mitosis and cytokinesis, which is almost complete. However, at the end of the sequence the two almost totally separated daughter cells fuse again by re-extending cytoplasmic bridges and become a single binuclear cell. Note a long MT bundle, which appears to be pulling the separated cells back towards each other and causing their re-fusion. The movie is composed of 90 frames, acquired at 7 min intervals (total time=10.5 hours). (MP4 1003 kb)
MOVIE S4 Post-mitotic cells remain joined via a cytoplasmic bridge in silenced clone 2.43. A cell undergoes mitotic division and cytokinesis. However, the daughter cells remain attached via a persistent cytoplasmic flexible narrow bridge (possibly containing MT fibers). The movie is composed of 80 frames, acquired at 7 min intervals (total time=9.33 hours), the bridge appears in the movie for 3.26 hours and persists beyond the duration of the movie. (MP4 1073 kb)
MOVIE S5 Unequal cytokinesis in silenced clone 2.43. A cell with an enlarged nucleus (likely polyploid; at the top right corner of the frame at the beginning of the sequence) undergoes mitosis and unequal cytokinesis, resulting in a visibly binuclear large cell (right) and a miniscule cell with a small nucleus (left). The movie is composed of 115 frames, acquired at 7 min intervals (total time=13.42 hours). (MP4 3396 kb)
TABLE S1 List of oligonucleotides for the generation of plasmid constructs and diagnostic PCRs. Details of the cloning strategy for each construct and PCR purpose and specificity of oligo sets are included. (DOCX 117 kb)
TABLE S2 Detailed list of primary and secondary antibodies used in this work, additional to those listed in the “Antibodies” section of Materials and Methods. (DOCX 102 kb)
TABLE S3 Identification of proteins contained in KATNAL2 immunoprecipitates from mouse testis (bands 1-3 in Fig. 1D) by LC-MS/MS. In all three samples, the major component by far is several signature peptides for mouse KATNAL2 (yellow highlights), providing its positive identification. For compiling this processed table, the original, somewhat larger, list of proteins (available upon request) was filtered to include only proteins that are unique (i.e. absent from the negative control experiment at larger scale, presented in Table S4). Additional negative controls included a band equivalent to band 3 (Fig. 1D) that was cut out from the negative control sample, only yielding a low count (2-4 peptides) of various keratins which also appeared in the KATNAL2 sample with the same frequency and was filtered out of the analysis as noise/contamination. The specific heat shock proteins listed here are absent from the negative controls in Table S4. Nevertheless, their presence (and potential as KATNAL2 interactors) should be evaluated with some caution as several other heat shock proteins had come up both in the test and negative controls of this experiment as well as the larger scale experiment in Table S4, and were thus filtered out as non-specific. (DOCX 48 kb)
TABLE S4 Identification of proteins contained in KATNAL2 immunoprecipitates from mouse testis (“positive”) and corresponding negative controls (“negative”) (NEG1–NEG3 & POS1–POS3 zones in Fig. S3 A2) by LC-MS/MS. The results per zone for the 3 positive samples and, correspondingly, for the 3 negative samples (full set available upon request) were pooled together for simplicity in this summary table. Similar to the smaller-scale experiment shown in Table S3, in all three samples, the major component was a high number of signature peptides for mouse KATNAL2 (yellow highlights). In the negative samples, zero KATNAL2 peptides were identified, proving the high specificity of the KATNAL2 antibody and the IP reaction. For compiling this processed table, in addition to pooling the positive or negative results, the original, much larger list of proteins was filtered more stringently to include (a) proteins that are represented by at least 3 signature peptides, (b) only proteins that are unique (i.e. completely absent from the negative control experiment). An exception to the second criterion was made for some heat shock proteins (green highlights), which were highly represented but were also present (albeit with reduced coverage) in the negative controls. Their putative interaction with KATNALs should hence be evaluated with caution (see also Table S3 and its legend). (DOCX 105 kb)
TABLE S5 Effect of silencing of Katnal2 in mitotic clone 2.43 cells. The mitotic index of silenced cells is reduced 4.5 fold in silenced clone 2.43 cells, compared to control cells (p=0.00018). The overall distribution of cells in distinct subphases of mitosis is significantly different in silenced compared to control cells (p=0.0011). Sample size corresponded to “dataset 1” (Materials and Methods). (DOCX 11 kb)
TABLE S6 Effect of Katnal2 silencing (clone 2.43) on number of centrioles. The overall distribution of centrioles per cell is significantly different in silenced from control cells (p<0.0001). The average percentage of cells with more than 2 centrioles is extremely significantly higher in silenced than in control cells (p=0.00056), as is the average number of centrioles per cell (p= 0.00026). Values represent results from 3 independent experiments and are accompanied by S.D. Sample size corresponded to “dataset 1” (Materials and Methods). (DOCX 12 kb)
TABLE S7 Effect of Katnal2 silencing (clone 3.81) on number of centrioles. The overall distribution of centrioles per cell is significantly different in silenced from control cells (p<0.0001). The average percentage of cells with more than 2 centrioles is significantly higher in silenced than in control cells (p=1.18x10-6). The average number of centrioles per cell is significantly higher in silenced than in control cells (p=1.09x10-5). Values represent results from 3 independent experiments and are accompanied by S.D. Sample size corresponded to “dataset 3” (Materials and Methods). (DOCX 12 kb)
Acknowledgments
We are thankful to Adamos Adamou for his initial help with Katnal2 cloning. We are indebted to Dr Elena Panayiotou-Worth for providing mouse tissues for IP/MS analyses, and to Dr Nicholas Mastroyiannopoulos for his help with live imaging (both at the Cyprus Institute of Neurology and Genetics) and to Dr Maria Pantelidou (Frederick University, Cyprus) for advice with the REST-384 software. We are grateful to Prof. Agamemnon Epenetos, Dr Zacharias Kallis and Ms Efi Yiacoumi at Lifeline for granting us access to the MACSQuant flow cytometer. This work was funded by the Research Promotion Foundation of Cyprus and Structural Funds from the EU (Grant PENEK/ENISXYSI/0308-06 to N.S.).
Footnotes
A. Ververis and A. Christodoulou contributed equally to the experimental work.
References
- 1.Akhmanova A, Steinmetz MO. Microtubule +TIPs at a glance. J Cell Sci. 2010;123(20):3415–3419. doi: 10.1242/jcs.062414. [DOI] [PubMed] [Google Scholar]
- 2.Walczak CE, Gayek S, Ohi R. Microtubule-depolymerizing kinesins. Annu Rev Cell Dev Biol. 2013;29:417–441. doi: 10.1146/annurev-cellbio-101512-122345. [DOI] [PubMed] [Google Scholar]
- 3.Prokop A. The intricate relationship between microtubules and their associated motor proteins during axon growth and maintenance. Neural Dev. 2013;8:17. doi: 10.1186/1749-8104-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sayas CL, Avila J. Regulation of EB1/3 proteins by classical MAPs in neurons. Bioarchitecture. 2014;4(1):1–5. doi: 10.4161/bioa.27774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreira JG, Pereira AL, Maiato H. Microtubule plus-end tracking proteins and their roles in cell division. Int Rev Cell Mol Biol. 2014;309:59–140. doi: 10.1016/B978-0-12-800255-1.00002-8. [DOI] [PubMed] [Google Scholar]
- 6.Niwa S. Kinesin superfamily proteins and the regulation of microtubule dynamics in morphogenesis. Anat Sci Int. 2015;90(1):1–6. doi: 10.1007/s12565-014-0259-5. [DOI] [PubMed] [Google Scholar]
- 7.McNally FJ, Vale RD. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 1993;75(3):419–429. doi: 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- 8.Baas PW, Qiang L. Neuronal microtubules: when the MAP is the roadblock. Trends Cell Biol. 2005;15(4):183–187. doi: 10.1016/j.tcb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Roll-Mecak A, McNally FJ. Microtubule-severing enzymes. Curr Opin Cell Biol. 2010;22(1):96–103. doi: 10.1016/j.ceb.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharp DJ, Ross JL. Microtubule-severing enzymes at the cutting edge. J Cell Sci. 2012;125(11):2561–2569. doi: 10.1242/jcs.101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh DK, Dasgupta D, Guha A. Models, regulations, and functions of microtubule severing by Katanin. ISRN Mol Biol. 2012;2012:14. doi: 10.5402/2012/596289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartman JJ, Vale RD. Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science. 1999;286(5440):782–785. doi: 10.1126/science.286.5440.782. [DOI] [PubMed] [Google Scholar]
- 13.White SR, Evans KJ, Lary J, Cole JL, Lauring B. Recognition of C-terminal amino acids in tubulin by pore loops in Spastin is important for microtubule severing. J Cell Biol. 2007;176(7):995–1005. doi: 10.1083/jcb.200610072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma N, Bryant J, Wloga D, Donaldson R, Davis RC, Jerka-Dziadosz M, Gaertig J. Katanin regulates dynamics of microtubules and biogenesis of motile cilia. J Cell Biol. 2007;178(6):1065–1079. doi: 10.1083/jcb.200704021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu C, Mains PE. The C. elegans anaphase promoting complex and MBK-2/DYRK kinase act redundantly with CUL-3/MEL-26 ubiquitin ligase to degrade MEI-1 microtubule-severing activity after meiosis. Dev Biol. 2007;302(2):438–447. doi: 10.1016/j.ydbio.2006.09.053. [DOI] [PubMed] [Google Scholar]
- 16.Roll-Mecak A, Vale RD. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature. 2008;451(7176):363–367. doi: 10.1038/nature06482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartman JJ, Mahr J, McNally K, Okawa K, Iwamatsu A, Thomas S, Cheesman S, Heuser J, Vale RD, McNally FJ. Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 1998;93(2):277–287. doi: 10.1016/S0092-8674(00)81578-0. [DOI] [PubMed] [Google Scholar]
- 18.McNally KP, Bazirgan OA, McNally FJ. Two domains of p80 katanin regulate microtubule severing and spindle pole targeting by p60 katanin. J Cell Sci. 2000;113(9):1623–1633. doi: 10.1242/jcs.113.9.1623. [DOI] [PubMed] [Google Scholar]
- 19.Lee HH, Jan LY, Jan YN. Drosophila IKK-related kinase Ik2 and Katanin p60-like 1 regulate dendrite pruning of sensory neuron during metamorphosis. Proc Natl Acad Sci USA. 2009;106(15):6363–6368. doi: 10.1073/pnas.0902051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonbuchner TM, Rath U, Sharp DJ. KL1 is a novel microtubule severing enzyme that regulates mitotic spindle architecture. Cell Cycle. 2010;9(12):2403–2411. doi: 10.4161/cc.9.12.11916. [DOI] [PubMed] [Google Scholar]
- 21.Srayko M, O’Toole ET, Hyman AA, Muller-Reichert T. Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr Biol. 2006;16(19):1944–1949. doi: 10.1016/j.cub.2006.08.029. [DOI] [PubMed] [Google Scholar]
- 22.Yang HY, McNally K, McNally FJ. MEI-1/katanin is required for translocation of the meiosis I spindle to the oocyte cortex in C. elegans . Dev Biol. 2003;260(1):245–259. doi: 10.1016/S0012-1606(03)00216-1. [DOI] [PubMed] [Google Scholar]
- 23.McNally KP, McNally FJ. The spindle assembly function of Caenorhabditis elegans katanin does not require microtubule-severing activity. Mol Biol Cell. 2011;22(9):1550–1560. doi: 10.1091/mbc.E10-12-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang D, Rogers GC, Buster DW, Sharp DJ. Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. J Cell Biol. 2007;177(2):231–242. doi: 10.1083/jcb.200612011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buster D, McNally K, McNally FJ. Katanin inhibition prevents the redistribution of gamma-tubulin at mitosis. J Cell Sci. 2002;115(5):1083–1092. doi: 10.1242/jcs.115.5.1083. [DOI] [PubMed] [Google Scholar]
- 26.Smith LB, Milne L, Nelson N, Eddie S, Brown P, Atanassova N, O’Bryan MK, O’Donnell L, Rhodes D, Wells S, Napper D, Nolan P, Lalanne Z, Cheeseman M, Peters J. KATNAL1 regulation of sertoli cell microtubule dynamics is essential for spermiogenesis and male fertility. PLoS Genet. 2012;8(5):e1002697. doi: 10.1371/journal.pgen.1002697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casanova M, Crobu L, Blaineau C, Bourgeois N, Bastien P, Pages M. Microtubule-severing proteins are involved in flagellar length control and mitosis in Trypanosomatids. Mol Microbiol. 2009;71(6):1353–1370. doi: 10.1111/j.1365-2958.2009.06594.x. [DOI] [PubMed] [Google Scholar]
- 28.Benz C, Clucas C, Mottram JC, Hammarton TC. Cytokinesis in bloodstream stage Trypanosoma brucei requires a family of katanins and spastin. PLoS One. 2012;7(1):e30367. doi: 10.1371/journal.pone.0030367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuo M, Shimodaira T, Kasama T, Hata Y, Echigo A, Okabe M, Arai K, Makino Y, Niwa S, Saya H, Kishimoto T. Katanin p60 contributes to microtubule instability around the midbody and facilitates cytokinesis in rat cells. PLoS One. 2013;8(11):e80392. doi: 10.1371/journal.pone.0080392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sudo H, Maru Y. LAPSER1/LZTS2: a pluripotent tumor suppressor linked to the inhibition of katanin-mediated microtubule severing. Hum Mol Genet. 2008;17(16):2524–2540. doi: 10.1093/hmg/ddn153. [DOI] [PubMed] [Google Scholar]
- 31.Dymek EE, Lefebvre PA, Smith EF. PF15p is the chlamydomonas homologue of the Katanin p80 subunit and is required for assembly of flagellar central microtubules. Eukaryot Cell. 2004;3(4):870–879. doi: 10.1128/EC.3.4.870-879.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lohret TA, McNally FJ, Quarmby LM. A role for katanin-mediated axonemal severing during Chlamydomonas deflagellation. Mol Biol Cell. 1998;9(5):1195–1207. doi: 10.1091/mbc.9.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasi MQ, Parker JD, Feldman JL, Marshall WF, Quarmby LM. Katanin knockdown supports a role for microtubule severing in release of basal bodies before mitosis in Chlamydomonas. Mol Biol Cell. 2009;20(1):379–388. doi: 10.1091/mbc.E07-10-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu WF, Pomp O, Ben-Omran T, Kodani A, Henke K, et al. Katanin p80 regulates human cortical development by limiting centriole and cilia number. Neuron. 2014;84(6):1240–1257. doi: 10.1016/j.neuron.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishra-Gorur K, Caglayan AO, Schaffer AE, Chabu C, et al. Mutations in KATNB1 cause complex cerebral malformations by disrupting asymmetrically dividing neural progenitors. Neuron. 2014;84(6):1226–1239. doi: 10.1016/j.neuron.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loughlin R, Wilbur JD, McNally FJ, Nedelec FJ, Heald R. Katanin contributes to interspecies spindle length scaling in Xenopus. Cell. 2011;147(6):1397–1407. doi: 10.1016/j.cell.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitehead E, Heald R, Wilbur JD. N-terminal phosphorylation of p60 katanin directly regulates microtubule severing. J Mol Biol. 2013;425(2):214–221. doi: 10.1016/j.jmb.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baas PW, Sudo H. More microtubule severing proteins: more microtubules. Cell Cycle. 2010;9(12):2273. doi: 10.4161/cc.9.12.12026. [DOI] [PubMed] [Google Scholar]
- 39.Pantelidou M, Zographos SE, Lederer CW, Kyriakides T, Pfaffl MW, Santama N. Differential expression of molecular motors in the motor cortex of sporadic ALS. Neurobiol Dis. 2007;26(3):577–589. doi: 10.1016/j.nbd.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 40.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30(9):e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santama N, Krijnse-Locker J, Griffiths G, Noda Y, Hirokawa N, Dotti CG. KIF2beta, a new kinesin superfamily protein in non-neuronal cells, is associated with lysosomes and may be implicated in their centrifugal translocation. EMBO J. 1998;17(20):5855–5867. doi: 10.1093/emboj/17.20.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kypri E, Christodoulou A, Maimaris G, Lethan M, Markaki M, Lysandrou C, Lederer CW, Tavernarakis N, Geimer S, Pedersen LB, Santama N. The nucleotide-binding proteins Nubp1 and Nubp2 are negative regulators of ciliogenesis. Cell Mol Life Sci. 2014;71(3):517–538. doi: 10.1007/s00018-013-1401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72(11):8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emes RD, Ponting CP. A new sequence motif linking lissencephaly, Treacher Collins and oral-facial-digital type 1 syndromes, microtubule dynamics and cell migration. Hum Mol Genet. 2001;10(24):2813–2820. doi: 10.1093/hmg/10.24.2813. [DOI] [PubMed] [Google Scholar]
- 45.Christodoulou A, Lederer CW, Surrey T, Vernos I, Santama N. Motor protein KIFC5A interacts with Nubp1 and Nubp2, and is implicated in the regulation of centrosome duplication. J Cell Sci. 2006;119(10):2035–2047. doi: 10.1242/jcs.02922. [DOI] [PubMed] [Google Scholar]
- 46.He M, Subramanian R, Bangs F, Omelchenko T, Liem KF, Jr, Kapoor TM, Anderson KV. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat Cell Biol. 2014;16(7):663–672. doi: 10.1038/ncb2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vasudevan KK, Jiang YY, Lechtreck KF, Kushida Y, Alford LM, Sale WS, Hennessey T, Gaertig J. Kinesin-13 regulates the quantity and quality of tubulin inside cilia. Mol Biol Cell. 2015;26(3):478–494. doi: 10.1091/mbc.E14-09-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyamoto T, Hosoba K, Ochiai H, Royba E, Izumi H, Sakuma T, Yamamoto T, Dynlacht BD, Matsuura S. The microtubule-depolymerizing activity of a mitotic kinesin protein KIF2A drives primary cilia disassembly coupled with cell proliferation. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dymek EE, Smith EF. PF19 encodes the p60 catalytic subunit of katanin and is required for assembly of the flagellar central apparatus in Chlamydomonas. J Cell Sci. 2012;125(14):3357–3366. doi: 10.1242/jcs.096941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Srayko M, Buster DW, Bazirgan OA, McNally FJ, Mains PE. MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 2000;14(9):1072–1084. [PMC free article] [PubMed] [Google Scholar]
- 51.Venkei Z, Gaspar I, Toth G, Szabad J. alpha4-Tubulin is involved in rapid formation of long microtubules to push apart the daughter centrosomes during earlyx Drosophila embryogenesis. J Cell Sci. 2006;119(15):3238–3248. doi: 10.1242/jcs.03039. [DOI] [PubMed] [Google Scholar]
- 52.Ikegami K, Setou M. Unique post-translational modifications in specialized microtubule architecture. Cell Struct Funct. 2010;35(1):15–22. doi: 10.1247/csf.09027. [DOI] [PubMed] [Google Scholar]
- 53.Sudo H, Baas PW. Acetylation of microtubules influences their sensitivity to severing by katanin in neurons and fibroblasts. J Neurosci. 2010;30(21):7215–7226. doi: 10.1523/JNEUROSCI.0048-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McNally FJ, Thomas S. Katanin is responsible for the M-phase microtubule-severing activity in Xenopus eggs. Mol Biol Cell. 1998;9(7):1847–1861. doi: 10.1091/mbc.9.7.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhai Y, Kronebusch PJ, Simon PM, Borisy GG. Microtubule dynamics at the G2/M transition: abrupt breakdown of cytoplasmic microtubules at nuclear envelope breakdown and implications for spindle morphogenesis. J Cell Biol. 1996;135(1):201–214. doi: 10.1083/jcb.135.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131(3):452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 57.Di Talia S, Skotheim JM, Bean JM, Siggia ED, Cross FR. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature. 2007;448(7156):947–951. doi: 10.1038/nature06072. [DOI] [PubMed] [Google Scholar]
- 58.Verges E, Colomina N, Gari E, Gallego C, Aldea M. Cyclin Cln3 is retained at the ER and released by the J chaperone Ydj1 in late G1 to trigger cell cycle entry. Mol Cell. 2007;26(5):649–662. doi: 10.1016/j.molcel.2007.04.023. [DOI] [PubMed] [Google Scholar]
- 59.Uyttewaal M, Burian A, Alim K, Landrein B, Borowska-Wykret D, Dedieu A, Peaucelle A, Ludynia M, Traas J, Boudaoud A, Kwiatkowska D, Hamant O. Mechanical stress acts via katanin to amplify differences in growth rate between adjacent cells in Arabidopsis. Cell. 2012;149(2):439–451. doi: 10.1016/j.cell.2012.02.048. [DOI] [PubMed] [Google Scholar]
- 60.Pomerat CM. Rotating nuclei in tissue cultures of adult human nasal mucosa. Exp Cell Res. 1953;5(1):191–196. doi: 10.1016/0014-4827(53)90104-9. [DOI] [PubMed] [Google Scholar]
- 61.Englander LL, Rubin LL. Acetylcholine receptor clustering and nuclear movement in muscle fibers in culture. J Cell Biol. 1987;104(1):87–95. doi: 10.1083/jcb.104.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Szikora S, Gaspar I, Szabad J. ‘Poking’ microtubules bring about nuclear wriggling to position nuclei. J Cell Sci. 2013;126(1):254–262. doi: 10.1242/jcs.114355. [DOI] [PubMed] [Google Scholar]
- 63.Confalonieri F, Duguet M. A 200-amino acid ATPase module in search of a basic function. BioEssays News Rev Mol Cell Dev Biol. 1995;17(7):639–650. doi: 10.1002/bies.950170710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Primary protein sequences encoded by all the Katnal2 cDNAs identified in this work. Protein motifs and SNPs are indicated. The sequences within the ORF that were targeted for the shRNA-mediated silencing of Katnal2s in clones 2.43 (yellow highlights) and 3.81 (blue highlights) are also indicated. In magenta highlight, the amino acids corresponding to the initial, partial cDNA sequence identified by a yeast two-hybrid screen is indicated. (DOC 37 kb)
FIGURE S2 Detection of KATNAL2 proteins in four mouse cell lines and tissue by WB. (A,B) Anti-KATNAL2 WB of total protein extracts from the indicated mouse cell lines and from mouse testis consistently detects three main immunoreactive bands migrating in the vicinity of 50, 60 and 80 kDa (blue arrowheads). These signals correspond to the signals also obtained in the IP experiment shown in Fig. 1D, in which the three bands were excised from the gel, analyzed by MS and found positive for the existence of KATNAL2 proteins. Abbreviations: M, molecular mass markers. (EPS 1167 kb)
FIGURE S3 Results of LC-MS/MS analysis at larger scale and of the 3 cut-out bands of Figure 1D. (A1,A2) Shown is a larger scale KATNAL2 IP experiment starting with more material, as described in Materials and Methods. Proteins were visualized by SDS-PAGE followed by colloidal-blue staining (A1) and 3 broad zones (boxed), ranging, respectively, between 80-60 kDa (zones “1”), 60-50 kDa (zones “2”) and 50-45 kDa (zones “3”), were cut out from the KATNAL2-IP sample (“POS” series) or the negative control IP (“NEG” series). MS identification of the proteins contained in the 3 positive and 3 negative zones shown here is given as a list in Table S4. (A3) WB analysis of the same samples as in A1/A2 to indicate the existence of several major and minor KATNAL2 immunoreactive bands within the 3 excised zones. KATNAL2 positive identification was obtained in all 3 zones, indicating the existence of multiple KATNAL2 isoforms in this size range specifically in the IP KATNAL2 and not in the IP neg. (B) Aggregate sequence coverage map for KATNAL2 protein, derived from KATNAL2 signature peptides, uniquely detected in the “positive samples” (see Table S4) reveals extensive coverage, throughout the sequence and spanning both termini of the protein. (EPS 8847 kb)
FIGURE S4 KATNAL2 localizes at the axoneme and basal body of primary cilia. (A,B) Examples of localization of KATNAL2 (green) at the axoneme and basal body (white arrowheads) of primary cilia in serum-deprived IMCD cells (A) and TM4 cells (B), showing acetylated tubulin (axonemal marker, red), DNA labeling (blue) and accompanying magnified detailed views of the cilia. The daughter centriole also shows comparable immunoreactivity (yellow arrowheads). Scale bars 5 μm; 1 μm in detailed views. (EPS 7192 kb)
FIGURE S5 shRNA-mediated silencing of Katnal2 is accompanied by a pronounced increase in the acetylation of the MT network. (A1-B2) Two examples of extensive acetylation of the MT network in Katnal2-silenced cells of clone 2.43, as revealed by double α-tubulin (red) and acetylated tubulin labeling (green). DNA is counterstained in blue. Exposure of panel A2 for green channel was 2500 ms; for B2 5243 ms. (C1-C2) Equivalent double staining in a wild type NIH 3T3 control cell for comparison showing relatively restricted or no acetylation of the MT network. Exposure for panel C2 is 6000 ms (that is 250% of exposure in A2 and 114.4% of exposure in B2). Scale bars 20 μm. (EPS 33601 kb)
FIGURE S6 Characterization and phenotypes of Katnal2-silenced stable clone 3.81. In all images, α-tubulin labeling is in red, γ-tubulin in green and DNA in blue. (A1-A4) IF in silenced clone 3.81 shows excessive supernumerary centrioles in an interphase cell (yellow arrowheads and square). The inset in panel A2 shows the detail of a cluster of amplified centrioles (contained in the yellow square) at higher magnification. (B1-B4) The presence of supernumerary centrioles in silenced cells induces the formation of multipolar mitotic spindles: here, as an example, a cell in metaphase with four centrioles and a spindle with 4 poles. Scale bars 20 μm for A1-A4; 5 μm for B1-B4; 2 μm in inset of panel A2. (C) Representative WB to confirm silencing of KATNAL2 isoform proteins. The concurrent detection of calnexin protein in the same samples was used as a loading control for sample normalization. (D) Quantification of the average number of centrioles per cell in Katnal2-silenced populations of clone 3.81 (red), compared with corresponding control-silenced populations (gray), showing the average value from three independent experiments and bars representing S.D. values. The 30% increase of the average number of centrioles per cell, caused by silencing, is extremely significant (****). Sample size corresponded to “dataset 3” (Materials and Methods). (E) Quantification of the percentage of ciliated cells in Katnal2-silenced populations of clone 3.81 (red), compared with corresponding control-silenced populations (gray), at growth conditions (cycling cells) and after induction of ciliogenesis by serum deprivation for 24 hours (cell cycle arrest), respectively. Shown are the averages from three independent experiments and bars represent S.D. values. The reduction in the percentage of silenced ciliated cells after induction of ciliogenesis by serum deprivation is extremely significant (***). Sample size corresponded to “dataset 4” (Materials and Methods). (EPS 18163 kb)
FIGURE S7 shRNA-mediated silencing of Katnal2 in clone 2.43 causes significant reduction of the frequency of sensory cilia in the cell population. Quantification of the percentage of ciliated cells in Katnal2-silenced populations of clone 2.43 (blue), compared with corresponding control-silenced populations (gray), at growth conditions (cycling cells) and after induction of ciliogenesis by serum deprivation for 24 hours (cell cycle arrest). Shown are the averages from three independent experiments and bars represent S.D. values. The reduction in the percentage of silenced ciliated cells after induction of ciliogenesis by serum deprivation is highly significant (**). Sample size corresponded to “dataset 2” (Materials and Methods). (EPS 434 kb)
FIGURE S8 Transient overexpression of KATNAL2-S1 or -L1 tagged fusions leads to cytoplasmic aggregation of the protein in mouse cells. (A-D) Representative examples of common overexpression phenotypes, resulting from transient transfections of differently tagged versions of KATNAL2-S1 or -L1, as indicated. Fluorescence corresponds to GFP emission (green, A), mCherry emission (red, B), anti-FLAG (C) and anti-myc (D) immunofluorescence (green). DNA was counterstained with Hoechst (blue). Panels A, C and D display NIH 3T3 cells and panel B is with IMCD cells. Scale bars 10 μm. (EPS 9005 kb)
FIGURE S9 Cell cycle analysis of IMCD stable cell lines expressing mCherry-KATNAL2-S1 and -L1, and the control line IMCD. Analysis of the distribution in mitotic subphases of the same cells as for the datasets in Fig. 7J (dataset 5 in Materials and Methods) reveals that it was highly significantly different between cell populations (p=0.0011). Additionally, for -S1 cells, compared to wild-type cells, it revealed significant (p=0.0122) and extremely significant (p=0.0006) differences for telophase and prophase percentages in particular. (EPS 472 kb)
MOVIE S1 A very large, binucleated Katnal2-silenced cell (clone 2.43) is struggling to complete mitosis with what appears like a multipolar spindle (observe multiple MT asters). At the end of the sequence, no cytokinesis has been achieved and the nuclear mass consists of multiple, irregularly shaped multi-lobed fragments. The movie is composed of 87 frames, acquired at 7 min intervals (total time=10.15 hours). (MP4 3623 kb)
MOVIE S2 Mitosis without cytokinesis in silenced clone 2.43. A large Katnal2-silenced cell (clone 2.43) with a large nucleus undergoes mitosis (chromosome alignment on the metaphase plate is discernible) without the formation of a complete ingression furrow and fails cytokinesis, resulting in one binuclear cell. The movie is composed of 120 frames, acquired at 7 min intervals (total time=14 hours). (MP4 2441 kb)
MOVIE S3 Failure of cytokinesis and re-fusion of mitotic cells in silenced clone 2.43. A Katnal2-silenced cell (clone 2.43) undergoes mitosis and cytokinesis, which is almost complete. However, at the end of the sequence the two almost totally separated daughter cells fuse again by re-extending cytoplasmic bridges and become a single binuclear cell. Note a long MT bundle, which appears to be pulling the separated cells back towards each other and causing their re-fusion. The movie is composed of 90 frames, acquired at 7 min intervals (total time=10.5 hours). (MP4 1003 kb)
MOVIE S4 Post-mitotic cells remain joined via a cytoplasmic bridge in silenced clone 2.43. A cell undergoes mitotic division and cytokinesis. However, the daughter cells remain attached via a persistent cytoplasmic flexible narrow bridge (possibly containing MT fibers). The movie is composed of 80 frames, acquired at 7 min intervals (total time=9.33 hours), the bridge appears in the movie for 3.26 hours and persists beyond the duration of the movie. (MP4 1073 kb)
MOVIE S5 Unequal cytokinesis in silenced clone 2.43. A cell with an enlarged nucleus (likely polyploid; at the top right corner of the frame at the beginning of the sequence) undergoes mitosis and unequal cytokinesis, resulting in a visibly binuclear large cell (right) and a miniscule cell with a small nucleus (left). The movie is composed of 115 frames, acquired at 7 min intervals (total time=13.42 hours). (MP4 3396 kb)
TABLE S1 List of oligonucleotides for the generation of plasmid constructs and diagnostic PCRs. Details of the cloning strategy for each construct and PCR purpose and specificity of oligo sets are included. (DOCX 117 kb)
TABLE S2 Detailed list of primary and secondary antibodies used in this work, additional to those listed in the “Antibodies” section of Materials and Methods. (DOCX 102 kb)
TABLE S3 Identification of proteins contained in KATNAL2 immunoprecipitates from mouse testis (bands 1-3 in Fig. 1D) by LC-MS/MS. In all three samples, the major component by far is several signature peptides for mouse KATNAL2 (yellow highlights), providing its positive identification. For compiling this processed table, the original, somewhat larger, list of proteins (available upon request) was filtered to include only proteins that are unique (i.e. absent from the negative control experiment at larger scale, presented in Table S4). Additional negative controls included a band equivalent to band 3 (Fig. 1D) that was cut out from the negative control sample, only yielding a low count (2-4 peptides) of various keratins which also appeared in the KATNAL2 sample with the same frequency and was filtered out of the analysis as noise/contamination. The specific heat shock proteins listed here are absent from the negative controls in Table S4. Nevertheless, their presence (and potential as KATNAL2 interactors) should be evaluated with some caution as several other heat shock proteins had come up both in the test and negative controls of this experiment as well as the larger scale experiment in Table S4, and were thus filtered out as non-specific. (DOCX 48 kb)
TABLE S4 Identification of proteins contained in KATNAL2 immunoprecipitates from mouse testis (“positive”) and corresponding negative controls (“negative”) (NEG1–NEG3 & POS1–POS3 zones in Fig. S3 A2) by LC-MS/MS. The results per zone for the 3 positive samples and, correspondingly, for the 3 negative samples (full set available upon request) were pooled together for simplicity in this summary table. Similar to the smaller-scale experiment shown in Table S3, in all three samples, the major component was a high number of signature peptides for mouse KATNAL2 (yellow highlights). In the negative samples, zero KATNAL2 peptides were identified, proving the high specificity of the KATNAL2 antibody and the IP reaction. For compiling this processed table, in addition to pooling the positive or negative results, the original, much larger list of proteins was filtered more stringently to include (a) proteins that are represented by at least 3 signature peptides, (b) only proteins that are unique (i.e. completely absent from the negative control experiment). An exception to the second criterion was made for some heat shock proteins (green highlights), which were highly represented but were also present (albeit with reduced coverage) in the negative controls. Their putative interaction with KATNALs should hence be evaluated with caution (see also Table S3 and its legend). (DOCX 105 kb)
TABLE S5 Effect of silencing of Katnal2 in mitotic clone 2.43 cells. The mitotic index of silenced cells is reduced 4.5 fold in silenced clone 2.43 cells, compared to control cells (p=0.00018). The overall distribution of cells in distinct subphases of mitosis is significantly different in silenced compared to control cells (p=0.0011). Sample size corresponded to “dataset 1” (Materials and Methods). (DOCX 11 kb)
TABLE S6 Effect of Katnal2 silencing (clone 2.43) on number of centrioles. The overall distribution of centrioles per cell is significantly different in silenced from control cells (p<0.0001). The average percentage of cells with more than 2 centrioles is extremely significantly higher in silenced than in control cells (p=0.00056), as is the average number of centrioles per cell (p= 0.00026). Values represent results from 3 independent experiments and are accompanied by S.D. Sample size corresponded to “dataset 1” (Materials and Methods). (DOCX 12 kb)
TABLE S7 Effect of Katnal2 silencing (clone 3.81) on number of centrioles. The overall distribution of centrioles per cell is significantly different in silenced from control cells (p<0.0001). The average percentage of cells with more than 2 centrioles is significantly higher in silenced than in control cells (p=1.18x10-6). The average number of centrioles per cell is significantly higher in silenced than in control cells (p=1.09x10-5). Values represent results from 3 independent experiments and are accompanied by S.D. Sample size corresponded to “dataset 3” (Materials and Methods). (DOCX 12 kb)


