Abstract
The last decade has experienced the emergence of microRNAs as a key molecular tool for the diagnosis and prognosis of human diseases. Although the focus has mostly been on cancer, neurodegenerative diseases present an exciting, yet less explored, platform for microRNA research. Several studies have highlighted the significance of microRNAs in neurogenesis and neurodegeneration, and pre-clinical studies have shown the potential of microRNAs as biomarkers. Despite this, no bona fide microRNAs have been identified as true diagnostic or prognostic biomarkers for neurodegenerative disease. This is mainly due to the lack of precisely defined patient cohorts and the variability within and between individual cohorts. However, the discovery that microRNAs exist as stable molecules at detectable levels in body fluids has opened up new avenues for microRNAs as potential biomarker candidates. Furthermore, technological developments in microRNA biology have contributed to the possible design of microRNA-mediated disease intervention strategies. The combination of these advancements, with the availability of well-defined longitudinal patient cohort, promises to not only assist in developing invaluable diagnostic tools for clinicians, but also to increase our overall understanding of the underlying heterogeneity of neurodegenerative diseases. In this review, we present a comprehensive overview of the existing knowledge of microRNAs in neurodegeneration and provide a perspective of the applicability of microRNAs as a basis for future therapeutic intervention strategies.
Keywords: microRNA, Neurodegenerative diseases, Tissue-enriched, Body fluid, Biomarker, Therapeutic agent, microRNA technological advancements
Introduction
Although it was generally assumed that the human genome would mainly contain protein-coding sequences, the human genome project revealed that protein-coding sequences only constitute ~1.5 % of the entire genome. The remaining ~98.5 % of the genome contains introns, regulatory DNA sequences, interspersed elements and non-coding RNA (ncRNA) molecules [1]. Indeed, the majority of mammalian genomes are transcribed into ncRNAs, many of which are alternatively spliced or processed into smaller products. To date, two types of non-coding RNAs have been identified—short non-coding RNA and long non-coding RNA molecules [2]. The short non-coding RNA molecules can further be subdivided into microRNA (miRNA), small interfering RNA (siRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), piwi-interacting RNA (pi-RNA), transfer RNA (tRNA), ribosomal RNA (rRNA) and other uncharacterized small molecules (Fig. 1a) [2, 3]. Among all the regulatory molecules, miRNAs are the most studied, particularly as regulators in human diseases. One of the main drivers for this is that miRNA expression profiles very often show significant modifications in response to a disease state suggesting that miRNAs represent key regulators of disease-associated pathways [4, 5]. Recent developments in miRNA research, in relation to human disease, have revealed that miRNAs represent valuable tools as biomarkers and as potential disease-modifying agents [6–8]. Despite these advances, miRNA biology in neurodegeneration is poorly understood. In this review, we comprehensively discuss miRNA biology in the context of neurodegeneration and highlight technological developments that ultimately may lead to new diagnostic and intervention strategies.
Fig. 1.
Schematic overview of non-coding RNA types, microRNA biogenesis and their functions. a Two types of non-coding RNAs, short non-coding RNA and long non-coding RNA molecules, have been identified. The short non-coding RNA molecules can further be subdivided into microRNA (miRNA), small interfering RNA (siRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), piwi-interacting RNA (pi-RNA), transfer RNA (tRNA), ribosomal RNA (rRNA) and other uncharacterized small molecules. b Schematic overview of microRNA biogenesis and their function. In addition to the canonical pathway, involving the microprocessor complex, nuclear, and cytosolic processing, other sources such as introns, shRNAs, tRNAs and snoRNAs contribute to the miRNA biogenesis pathway. miRNAs exert their function predominantly by inhibiting translation in dividing cells, but they can also activate translation in quiescent cells
Expression and biogenesis of miRNAs
miRNAs are evolutionarily conserved regulatory molecules that are synthesized after processing by both nuclear and cytosolic proteins. In the nucleus, non-coding RNAs (can be more than 1 kb), called the primary miRNAs (pri-miRNA), are transcribed from miRNA-encoding genomic sequences by RNA polymerase II [9–12]. Following this initial transcription event a protein complex comprised of the RNase III enzyme (RNASEN), Drosha and DGCR8 (in addition to several cofactors) recognizes the pri-miRNA and cleaves the 5′ and 3′ arms of the pri-miRNA hairpin to form the premature miRNA (pre-miRNA—70–110 nts, Fig. 1b) [13, 14]. Exportin-5 then recognizes and transports the pre-miRNA to the cytosol for further processing via a RAN-GTP-dependent mechanism [10]. In the cytosol, further processing of pre-miRNAs into 22 nucleotide mature miRNA duplexes takes place by a protein complex consisting of Dicer (RNase) and the double-stranded RNA-binding domain proteins TRBP, PACT and Ago2 [15, 16]. This is followed by separation of the miRNA guide strand and association with Ago2 within the RNA induced silencing complex (RISC) (Fig. 1b) [16]. Alternatively, and without the RISC involvement, miRNAs can be generated from short hairpin introns, snoRNAs, tRNAs and endogenous shRNAs as a result of splicing, debranching and complex processing mechanisms (Fig. 1b) [17–20].
miRNA biogenesis is a complex process and it is clear that miRNAs are not only generated through the classical canonical pathway, but also from a number of other miRNA sources. As the field progresses and miRNA biogenesis in its entirety becomes fully understood, we will be better equipped to comprehensively understand miRNA biology and its impact on cellular functions.
Brief overview of miRNA functions
Canonical functions
miRNAs are proposed to be downregulators of gene expression via two principal mechanisms: (i) mRNA cleavage and (ii) translational repression. On one hand, miRNAs have been shown to bind to complementary regions of protein-coding mRNA sequences resulting in RISC-mediated cleavage. Alternatively, and in the absence of appropriate complementarity, miRNAs also have the ability to bind to 3′UTRs and block translation (Fig. 1c) [12].
Non-canonical functions
A relatively recent and significant discovery changed the way miRNAs were perceived as bona fide translational repressors. Vasudevan and colleagues convincingly demonstrated that miRNAs can also act as translational upregulators in non-dividing cells (Fig. 1c) [21]. Indeed, miRNAs, along with microRNPs (micro ribonucleoproteins), attain a dual ‘switch’ role as both upregulator and downregulator under quiescent and proliferating conditions, respectively. Furthermore, indirect upregulation of translation may also occur via specific miRNA-mediated downregulation of a repressor protein(s).
The discovery that miRNAs can have dual roles suggests that one miRNA can regulate several mRNAs, and that several miRNAs can share a common target [22].
Regulation of biogenesis, expression, function and decay of miRNAs
In addition to understanding the biogenesis and functions of miRNAs, recent technological advancements have shed considerable light on the biology associated with the regulation of expression and function of miRNAs. Several biological pathways have been identified, involving numerous protein–protein and protein–RNA interaction, that contribute significantly to the cell- or tissue-specific functions of miRNAs [23]. For example, a battery of cofactors and accessory proteins such as DGCR8/Pasha, p68, p72, EWSR1, Fus, Argonaute 2, assist Drosha, Dicer and miRISC to execute their functions, which indirectly control miRNA expression [15, 24–27].
Similar to protein-encoding genes, miRNA genes transcription is regulated by transcription factors [11]. Furthermore, several miRNAs work in tandem with transcription factors within autoregulatory feedback loops to drive or repress the expression of miRNAs [23]. Moreover, several activators and repressors have been identified that regulate miRNA biogenesis, either via protein–protein or protein–RNA interactions [23]. Interestingly, at different processing stages pri-miRNAs and pre-miRNAs may undergo adenosine deaminase-mediated catalysis altering base-pairing and structural properties resulting in abnormal processing of miRNAs [28]. At the functional level, miRNAs incorporated into the miRISC, can be further regulated by proteins targeting the RISC-associated proteins [23].
In contrast to our knowledge on miRNA biogenesis, expression and function, miRNAs turnover mechanisms remain less explored. miRNAs are considered to be highly stable molecules with slow turnover rates, often at very high cellular copy numbers. It is, however, known that miRNA turnover is nucleotide sequence-dependent, and is further influenced by target-mediated degradation, uridylation status and viral infections [29].
Initially miRNA regulation was not generally viewed as being cell specific, but neuronal miRNA regulation indicates that some regulatory aspects may exhibit quasi cell-specificity [30]. For example, Krol and colleagues suggest that the miRNA-mediated regulation of synaptic stimulation is due to miRNAs being expressed at distal sites in dendrites [23]. Furthermore, it has been suggested that miRISC-mediated repression can be relieved by neuronal stimulation-facilitated proteolysis of miRISC assembly factors resulting in normalized synaptic plasticity or memory formation [23].
Combined, these findings imply that miRNAs play significant regulatory roles during normal neuronal functioning, thereby suggesting that possible dysregulation may have an impact on neurodegeneration.
Tissue-enriched expression of miRNAs
Although all tissues harbor miRNAs, the levels of individual miRNAs differ substantially between different tissue types. In addition, a significant number of miRNAs are enriched in a cell compartment-specific manner. For example, microRNA miR-1 is enriched in expression in muscle and heart tissue, whilst miR-124 expression is augmented in brain tissues [31]. Other examples of tissue enrichment of a miRNAs include miR-122 in hepatocytes, miR-142 is lymphoid tissue-specific, miR-375 is expressed in pancreatic cells and miR-223 is found in myeloid tissue [31–34]. microRNA expression in a particular tissue or cell type can also be enhanced as a result of endogenous and exogenous stimuli. For example, Let-7d, Let-7e, miR-768-3p and miR-768-5p are significantly upregulated in human fibroblasts under oxidative stress caused by ionizing radiation, H2O2 and etoposide [35].
There are many examples of miRNAs that are specifically expressed in brain tissue which play a critical role in regulating neuronal activity [36]. As an example, miR-9, miR-124 and miR-128 represent brain-specific miRNAs, and interestingly, their expression patterns change in disease states [37]. The tissue specificity of miRNAs is especially evident in neurons, as they regulate and influence key neuronal features such as neurogenesis, synaptic plasticity, neuronal differentiation, neuronal proliferation and the self-rejuvenation of neuronal stem cells [38].
The well-documented tissue enrichment of miRNA suggests not only that many miRNAs influence cell specific processes, but also it highlights the potential of miRNAs as diagnostic biomarkers and as possible targets in future disease intervention strategies.
miRNAs in brain
Many miRNAs have been shown to be involved in neuronal differentiation and overall brain development. This is evident from studies showing that inactivation of miRNA biogenesis via Dicer removal, results in inappropriate neurulation and neuronal differentiation, ultimately culminating in brain morphogenesis defects [39]. Similarly, Dicer dysfunction in post-mitotic motor progenitor cells results in lack of neuronal generation [40]. miRNA biogenesis is also vital for the appropriate development of dopaminergic neurons. For example, abnormal miRNA biogenesis abolishes differentiation of dopaminergic neurons derived from mouse embryonic stem cells [41]. Furthermore, Dicer inactivation in mouse post-mitotic midbrain results not only in loss of dopaminergic neurons, but also reduced axonal projections [42]. It is also interesting to note that loss of Dicer in spinal motor neurons causes muscular atrophy and a decline in motor function coupled with reactive astrocytosis, a marker for neurotoxicity [43, 44].
miRNAs, such as miR-124a and miR-9, have been shown to determine the fate of neuronal precursors derived from embryonic stem cells [45]. Interestingly, the brain-specific miR-124a regulates neuronal differentiation and maintenance by decreasing the levels of many hundred non-neuronal gene transcripts [46]. Indeed, the expression of miR-124a in non-neuronal cell lines results in neuronal-like mRNA profiles, suggesting a pivotal role for this miRNA in neuronal differentiation [46]. miR-124 has also been shown to have a functional interaction with the alternative pre-mRNA splicing pathway where miR-124 inhibits Polypyrimidine tract-binding protein-1 (PTBP1). PTBP1 encodes a global repressor of alternative pre-mRNA splicing and its inhibition leads to PTBP2 exon skipping and PTBP2 mRNA decay, ultimately resulting in a transition from non-neuronal to neuronal-specific alternative splicing patterns [47].
In the nervous system, the regulation of mRNA translation is important for synaptic development and plasticity [38]. Interestingly, the first study to demonstrate the presence of synapse function-specific miRNAs revealed that the brain-specific miR-134 is expressed in the synapto-dendritic compartment of hippocampal neurons, and that it negatively regulates the size of dendritic spines through the inhibition of translation of the Limk1 kinase [48].
This study and others demonstrate the critical role of miRNAs in brain developmental processes and highlights that subtle change or defects in miRNA profiles may result in inappropriate neuronal development, including premature neuronal cell death, a hallmark of neurodegenerative disorders.
miRNAs and neurodegeneration
Neurodegeneration, in diseases such as Parkinson’s disease (PD), Alzheimer’s disease (AD), Huntington’s diseases (HD), and amyotrophic lateral sclerosis (ALS), is a complex process often influenced by a combination of genetic, molecular and environmental perturbations [49]. Although neurodegeneration is still poorly understood, recent advances in the field have started to unravel numerous pathways that impact neuronal cell death. Indeed, the emergence of miRNAs as key regulators in neuronal stem cell differentiation, neurogenesis, neuronal survival, neurite outgrowth, synapse formation, and synaptic plasticity has not only revealed their involvement in neuronal development and differentiation, but also neurodegeneration, opening up new avenues for biomarker discovery and potential therapeutic intervention strategies [42, 50–54].
Several studies have demonstrated that miRNAs are not only neuroregulators but also neuroprotectors. For example, a number of findings clearly suggest that the miR-29 family impacts neuronal protection [55–58] and in line with these findings, miR-29 levels are reduced in individuals with AD or HD and in mouse models [59, 60]. Of particular interest are reports documenting that miR-29 family members regulate the expression of serine palmitoyl transferase-1 (SPTLC1), the first rate-limiting enzyme in de novo ceramide synthesis, and beta-secretase-1 (BACE1) [61]. Disruptions of both these proteins play key roles in amyloid beta (Ab) formation, contributing to AD pathology [56, 58]. In AD, the neuroprotective miR-29a-1 and miR-29b-1 are downregulated, promoting disease progression by inversely regulating BACE1 causing increased Ab plaque formation [58].
Age has a tremendous impact on most neurodegenerative diseases [62] and miRNA profiles show dramatic changes as the brain ages [63]. Genome-wide expression analysis of miRNAs and ncRNAs in brain tissue has demonstrated differential regulation of small RNAs in chimpanzees and humans in an age-dependent manner [64]. For example, miR-144 is upregulated in aging human cerebellum and cortex, as well as in aging chimpanzee brains [64]. It is interesting to note that miR-144 can bind to the 3′ UTR of programmed cell death protein 4 (PDCD4) suggesting a possible role for miR-144 in apoptosis in aging brains [64]. Further evidence revealing a link between miRNA expression and apoptosis comes from studies showing that miR-16, miR-128, miR-15 and miR-497 can regulate B cell lymphoma 2 protein (BCL2) to induce programmed cell death [65, 66]. Another example showing the influence of miRNAs on the aging process is the finding that the anti-aging factor Sirtuin-1 (SIRT1), promoting neuronal survival and suppression of neurodegeneration in AD and ALS models, is also regulated miR-34a [67–69]. Indeed, SIRT1 levels are inversely correlated with miR-34a expression in aging brains.
Although the full extent of miRNAs directly affecting neurodegenerative pathways is still not fully understood, a large number of studies have demonstrated that the expression of miRNAs change in neurodegeneration. It is interesting to note that there are numerous miRNAs that are specifically deregulated in a disease-specific manner. For example, miR-548d, miR-224, miR-373, miR-198 show altered expression in PD patients [70, 71, 73]. Multiple studies demonstrate miR-148a, miR-17-5p, miR-137, miR-181c, miR-101, miR-184, miR-15a, miR-185 and miR-210 are only some of the miRNAs that can be AD specific [58, 60, 61, 72, 74]. There are some miRNAs that show a Prion specific expression pattern, miR-342-3p, let-7b, miR-342-3p, miR-490 and miR-188-5p [75–77]. In addition there are many miRNAs such as miR-26b, miR-106, miR-21, miR-128, miR-125b, miR-124a, miR-132, miR-9, miR-29a, miR-338-3p, miR-27b, miR-151, miR-219, miR-145, miR-16, miR-30a, miR-320 and the miR-34 subfamily that are indicative of neurodegeneration in general, evident from the expression changes in case of multiple neurodegenerative disorders (Table 1). A comprehensive list of miRNA expression profile changes in brains of patients with PD, AD, HD, MSA (multiple system atrophy) and ALS are shown in Table 1.
Table 1.
microRNA expression profile changes in neurodegeneration
| Disease | Source | Differentially expressed microRNAs | Method | References |
|---|---|---|---|---|
| Parkinson’s disease | Blood | miR-126-3p, miR-126-5p, miR-147, miR151-5p, miR-151-3p, miR-199a-3p, miR-199a-5p, miR-19b, miR-26a, miR-28-5p, miR-29b, miR29c, miR-301a, miR-30b, miR-30c, miR-335, miR-374a, miR-374b | Microarray, ChIP-seq | [97] |
| Peripheral blood | miR-1, miR-16-2-3p, miR-22-5p, mir-26a-2-3p, miR-29a, miR-30a | qRT-PCR | [55] | |
| Plasma | miR-1826, miR-450b-3p, miR-626, miR-505 | Microarray, qRT-PCR | [98] | |
| Plasma | miR-181c, miR-331-5p, miR-193a-p, miR-196b, miR-454, miR-125a-3p, miR-137 | TaqMan low-density arrays, TaqMan assay | [144] | |
| Serum | miR-339-5p, miR-223-5p, miR-324-3p, miR-24, miR-30c, miR-148b | TaqMan low-density arrays, TaqMan assay | [57] | |
| Serum | miR-338-3p, miR-30e-3p, miR-30a-3p, miR-16-2-3p, miR-1294 | TruSeq small RNA sequencing, | [74] | |
| White blood cells | miR-320a/b/c, miR-769, miR-92b, miR-16, miR-199b, miR-1274b, miR-21, miR-150, miR-671, miR-1249, miR-20a, miR-18b*, miR-378c, miR-4293 | Small RNA sequencing (ABI SOLiD) | [136] | |
| CSF | miR-132-5p, miR-19a-3p, miR-19b-3p, miR-127-3p, miR-409-3p, miR-370, miR431-3p, miR-873-3p, miR-136-3p, miR-10a-5p, miR-1224-5, miR-4448, let-7 g-3p, miR-128, miR-433, miR-485-5p, miR-212-3p | TruSeq small RNA sequencing, | [74] | |
| Brain | miR-34b, miR-34c | Microarray, qRT-PCR | [108] | |
| Brain | miR-133b | qRT-PCR, northern blot analysis, luciferase assay | [42] | |
| Substantia nigra pars compacta | miR-198, miR-135b, miR-485-5p, miR-548d | TaqMan low-density arrays, TaqMan assay | [71] | |
| Substantia nigra pars compacta | miR-26b, miR-106a*, miR-301b, miR-21, miR-224, miR-373* | qRT-PCR | [70] | |
| Amygdala | miR-224, miR-373* | qRT-PCR | [70] | |
| Alzheimer’s disease | Blood | miR-34a, miR-181b | Microarray, qRT-PCR | [99] |
| Blood | miR-137, miR-181c, miR-9, miR-29a, miR29b, | qRT-PCR | [56] | |
| Peripheral blood | miR-112, miR-161, let-7d-3p, miR-5010-3p, miR-26a-5p, miR-1285-5p, miR-151a-3p, miR-103a-3p, miR-107, miR-532-5p, miR-26b-5p, let-7f-5p | Next generation sequencing, qRT-PCR | [100] | |
| Plasma | let-7d-5p, let-7 g-5p, miR-15b-5p, miR-142-3p, miR-191-5p, miR-301a-3p, miR-545-3p | Nanostring, qRT-PCR | [101] | |
| Serum | miR-137, miR-181c, miR-9, miR-29a, miR29b | qRT-PCR | [56] | |
| Serum | miR-125a-3p, miR-125b-1-3p, miR-127-3p, miR-1285, miR-135a-5p, miR-30c-2-3p, miR- 21-5p, miR-219-2-3p, miR-34c-5p, miR-34b-3p, miR-34b-5p, miR-22-5p, miR-375, miR-873, miR-1307-5p, miR-887, miR-182-5p, miR-184, miR-671, miR-3176 | TruSeq small RNA sequencing, | [74] | |
| CSF | miR-105, miR-10a, miR-10b, miR-143, miR-142-5p, miR-146b, miR-151, miR-125a, miR-126*, miR-126, miR-127, miR-135a, miR-138, miR141, miR-181a, miR-181c, miR-15b, miR-154, miR-186, miR-191, miR-194, miR-195, miR-197, miR-199a*, miR-204, miR-205, miR-214, miR-216, miR-221, miR-302b, miR-30a-3p, miR-30a-5p, miR-30b/c/d, miR-32, miR-99a, miR-501, miR-517a/b, miR-518b/f, miR-520a*, miR-526a, miR-338, miR345, miR-362, miR-371, miR-374, miR-375, miR-380-3p, miR-422b, miR-429, miR-448, miR449, miR-451, miR-455, miR-494, miR-497, miR-7f | qRT-PCR | [72] | |
| CSF | miR-9, miR-125b, miR-146a, miR-155 | Microarray | [145] | |
| CSF | miR-10a-5p, miR-33b-5pmiR-101-5p, miR-124-3p, miR-127-3p, miR-127-5p, miR-132-3p, miR-129-5p, miR-134, miR-136-3p, miR-136-5p, miR-138-5p, miR-139-5p, miR-181a-5p, miR-181a-3p, miR-181b-5p, miR-181d, miR-184, miR-218-5p, miR-323a-3p, miR-326, miR-329, miR-377-5p, miR-381, miR-410, miR-431-3p, miR-433, miR-488-3p, miR-495, miR-708-5p, miR- 769-5p, miR-874, miR-9-3p, miR-9-5p, miR-95, miR-598, miR-760, miR-708-3p, miR-873-5p, miR-3200-3p | TruSeq small RNA sequencing, | [74] | |
| Brain | miR-125b, miR-106b, miR-107, miR-124, miR-132, miR-145, miR-146b, miR-148a, miR-17-5p | qRT-PCR | [72] | |
| Cortex | miR-137, miR-181c, miR-9, miR-29a, miR29b | qRT-PCR | [61] | |
| Cortex | miR-212, miR-424, miR-29a, miR-29b-1, miR-107, miR-15a | LNA-microarrays, northern blot analysis | [60] | |
| Cortex | miR-210, miR-320, miR-29a, miR-29b-1, miR-106b, miR-15a, miR-181c, miR-9, miR-22, miR-101, miR-197, miR-511, miR-19b, miR-26b, miR-363, miR-93, let-7i | Microarry, qRT-PCR, northern blot analysis | [58] | |
| Cortex | miR-29a, miR29b, miR-338-3p | Microarray, qRT-PCR | [146] | |
| Cortex | miR-101, miR-106b, miR-107, miR-125b, miR-137, miR-142-3p, miR-142-5p, miR-145, miR-151-5p, miR-15a, miR-181c, miR-184, miR-185, miR-194, miR-197, miR-19b, miR-210, miR-212, miR-214, miR-219-2-3p, miR-22, miR-223, miR-26b, miR-27b, miR-298, miR-29a, miR-29a/b-1, miR-29b-1, miR-300, miR-301a, miR-320, miR-326, miR-330-5p, miR-338-3p, miR-338-5p, miR-361-3p, miR-363, miR-382, miR- | |||
| Hippocampus | miR-9, miR-124a, miR-125b, miR-128 | Microarray | [73] | |
| Huntington’s disease | Plasma | miR-34b | [147] | |
| Brain | miR-16, miR-100, miR-151-3p, miR-219-2-3p, miR-27b, miR-451, miR-92a, miR-128, miR-139-3p, miR-222, miR-382, miR-433, miR-485-3p | RNAseq, microarrays, qRT-PCR | [81] | |
| Cortex | miR-9, miR-9*, miR-29b, miR-124a, miR-132, miR-196a, miR-486 | Microarray, qRT-PCR | [83] | |
| Cortex | miR-1-1, miR-124a, miR-29a, miR-132, miR-133a, miR-203, miR-204, miR-21, miR-330 | qRT-PCR | [82] | |
| Multiple system atrophy | Serum | miR-339-5p, miR-223*, miR-324-3p, miR-24, miR-29c, miR-148b, miR-483-5p, miR-652, miR-744, miR-1274A, miR-1274B, miR-1291 | TaqMan low-density arrays, TaqMan assay | [57] |
| Cerebellum | miR-202, miR-129-3p, miR-129-5p, miR-337-3p, miR-380, miR-433, miR-132, miR-410, miR-206, miR-409-5p, miR-199a-5p | Microarray | [148] | |
| Amyotrophic lateral sclerosis | Blood | miR-338-3p, miR-451, miR-1275, miR-328, miR-638, miR-149, miR-665, miR-583 | Microarray, qRT-PCR | [102] |
| CD14+CD16− monocytes | miR-27a, miR-155, miR-146a, miR-32-3p | TaqMan low-density arrays, TaqMan assay | [103] | |
| Spinal cord | miR-146*, miR524-5p, miR-582-3p | qRT-PCR | [149] | |
| Spinal cord | miR-24-2*, miR-142-3p, miR-142-5p, miR-1461, miR-146b, miR-155 | Microarray, qRT-PCR, Luciferase assay | [136] | |
| Prion diseases | Brain | miR-342-3p, miR-320, let-7b, miR-328, miR-191, let-7d, miR-370, miR-128, miR-139-5p, miR-146a, miR-339-5p, miR-203, miR-181a-1*, miR-338-3p, miR-337-3p, miR-200a, miR-200b, miR-26a, miR-186, miR-331-3p, miR-152, miR-221 | Microarray, qRT-PCR | [75] |
| Brain | miR-26a, miR-30a-5p, miR-30d, miR-103, miR-106b, miR-107, miR-124a, miR-125a, miR-128a, miR-132, miR-143, miR-145, miR-181a, miR-191, miR-195, miR-219, miR-320, miR-342-3p, miR-361, miR-490, miR-494 | TaqMan low-density arrays, TaqMan assay | [76] | |
| Exosomes | miR-126-3p, miR-134, miR-146a, miR-182, miR-186, miR-188-5p, miR-193b, miR-222, miR-296-3p, miR-29b, miR-380-5p, miR-424 | qRT-PCR (TLDA cards) | [77] | |
| Exosomes | Let-7b, let-7i, miR-103, miR-125a-5p, miR-125b, miR-130a, miR-130b, miR-16, miR-21, miR-23a, miR-23b, miR-24, miR-296-6p, miR-29a, miR-29b, miR-29c, miR-301a, miR-30b, miR-30c, miR-342-3p, miR-344-4p, miR-378, miR-93 | RNA sequencing | [77] |
Trinucleotide repeat disorders (TRD) are characterized by trinucleotide repeat expansions either within or outside the coding region of a gene [78]. TRDs with repeats within a gene’s coding region include Huntington’s disease (HD), Kennedy disease, Haw-River syndrome and five types of spinocerebellar ataxia (SCA), whilst TRDs with repeats outside a gene’s coding region includes Fragile X syndrome, Fragile XE syndrome, Friedreich’s ataxia, myotonic dystrophy, spinocerebellar ataxia type 8 and spinocerebellar ataxia type 12 [79]. Several mRNAs have been shown to have altered expression in neurodegenerative TRDs. For example, miR-144 is upregulated in the cortex of SCA type 1 patients [55]. Furthermore, miR-34b is upregulated and miR-25, miR-29a and miR-125b are downregulated in SCA type 3 patients [80]. In addition, miR-25 and miR-125b are associated with progression of the disease [80]. HD is one of the most studied TRDs and it has been shown that miR-100, miR-139-3p, miR-196a, miR-133a and miR-330 are indicative of the disease [81–83].
In addition to the above-mentioned types of neurodegenerative diseases, several studies have also linked miRNAs to prion diseases, a transmissible and invariably fatal class of neurodegenerative disease including Kuru and Creutzfeldt–Jakob disease (CJD) in humans, scrapie in sheep, and bovine spongiform encephalopathy (BSE) in cattle [84]. In 2008, Saba and colleagues reported aberrant expression of 15 miRNAs in prion disease-affected mouse brains (Table 1) [75]. They further demonstrated that two downregulated miRNAs, miR-203 and miR-191, targets EGR1 that shares a functional relationship with CREB1, a protein downregulated in prion disease [75]. Montag and colleagues further reported on a number of miRNAs that showed differentially expressed in BSE-infected macaque brain samples (Table 1). In addition, they also highlighted the involvement of miR-342-3p in different prion disorders, suggesting a potential biomarker in prion diseases in animals and humans [76]. In 2012, deep sequencing revealed that an array of miRNAs exist in exosomes released from prion-infected neuronal cells, which initiated the notion that miRNA exosome signatures may represent a novel avenue in terms of new diagnostic intervention in prion disorders [77].
Combined, these studies demonstrate the aberrant nature of miRNA profiles within different classes of neurodegenerative diseases suggesting that miRNAs hold promise as not only diagnostic tools but as vehicles to further our understanding of neurodegenerative disease mechanisms.
miRNAs in autophagy
The misfolding and aggregation of proteins, as well as inappropriate functioning of organelles, represent hallmarks of neurodegeneration [85]. Although the proteasome is responsible for protein clearance, the proteasome is often dysfunctional in neurodegeneration, often due to the vast amount of aggregated proteins [85]. Because of this, the autophagy plays a pivotal role in terms of clearing dysfunctional proteins and organelles in neurodegeneration [86].
Autophagy has clearly been linked to PD and several key PD proteins, such as LRRK2, Pink1, and Parkin [87]. Autophagy does not directly degrade miRNAs, but influences miRNA processing proteins thereby preventing miRNA mediated repression of appropriate target genes. Indeed, Gibbings and colleagues have found the autophagy selectively degrading Dicer1 and Ago2 ultimately regulating miRNA biogenesis [88].
miR-106a and miR-224, which are both upregulated in the substantia nigra pars compacta and amygdala of PD brains, cause a dose-dependent decrease in heat shock 70 kDa protein (hsc70) and lysosome-associated membrane protein 2 (LAMP-2A), respectively [70]. Both hsc70 and LAMP-2A are mediators of autophagy, namely chaperone-mediated autophagy (CMA) [89]. Interestingly, miR-34 has been shown to affect lifespan by regulating autophagy [90]. Further evidence supporting the regulation of autophagy by miRNAs comes from studies showing that the integral autophagy-promoting protein Beclin 1 (BECN1) is directly regulated by miR-30a, and that miR-206 regulates HDAC4 which is associated with autophagy [91, 92]. Noteworthy, is also the fact that reduced expression of miR-9, which is reduced in brains of HD and AD patients, causes deregulation of histone acetylation, ultimately resulting in autophagy-dependent cell toxicity [58, 82, 93].
Autophagy represents an integral part of neurodegeneration and although autophagy does not directly control miRNA turnover, it regulates the protein targets of miRNAs. Indeed, several studies suggest that miRNAs may play an important role in autophagy as they can regulate proteins promoting or blocking autophagy.
miRNAs as biomarkers of neurodegeneration
Neurodegenerative diseases are often highly heterogeneous in nature and, although this necessitates the need for accurate diagnostic and prognostic biomarkers, the heterogeneity in itself has hampered the successful development of robust markers for disease onset and progression. However, the finding that miRNAs are present in biofluids as stable molecules has led to recent studies suggesting that miRNA may represent candidates as biomarkers for neurodegenerative disease [8].
Biofluids as a source of miRNAs
miRNAs have been detected in many clinically relevant body fluids including cerebrospinal fluid (CSF), serum/plasma, saliva, and urine [94]. Of these, CSF clearly represents the most relevant biofluid for biomarker discovery in neurodegenerative disorders because of its proximity to the brain. However, the use of CSF has some disadvantages related to risks associated with CSF collection, as the procedure can be painful which may limit patient participation in cohort studies. Serum/plasma represents a more convenient source for biomarker discovery as sample collection is simple, and moreover, it is thought that miRNAs are released by apoptotic cells, possibly mirroring miRNA profiles as neurons degenerate in the brain [6]. In addition, several studies have found that extracellular serum/plasma miRNAs are exceptionally stable [95].
Circulating biomarkers in neurodegeneration
Because of the heterogeneous nature of neurodegenerative diseases, misdiagnosis is rather common. Moreover, the characteristic motor symptoms of PD normally do not become clinically overt before more than 60–80 % of the dopaminergic neurons are already irreversibly lost [96]. Because of this, accurate and sensitive diagnostic tools need to be developed, not only to better diagnose neurodegenerative diseases at early stages of progression, but also to help pave the way for future therapeutic intervention strategy design.
As described earlier, changes in miRNA profiles in brain tissue from individuals with neurodegenerative disease has been documented. Recently, there has also been a significant increase in studies showing that miRNA profiles change in biofluids of individuals with established neurodegenerative diseases (Table 1). In terms of PD, potential diagnostic miRNA biomarkers include, but are not limited to, miR-125-3p, miR-16, miR-19a/b, let-7, and miR-29/miR-30 family members. These miRNAs were detected in varied biofluids ranging from whole blood, plasma, serum, and CSF using a variety of technologies including microarray, qRT-PCR, and RNA-Seq techniques (Table 1) [55, 57, 73, 97, 98]. Similarly, many differentially expressed miRNAs have been identified from biofluids of AD patients including, but not limited, to miR-34a/b, miR-30b/c/d, miR-9, miR-29a/b, miR-125a/b and let-7 family members (Table 1) [72, 73, 99–101]. Although miRNA biomarker discovery studies in biofluids is not as developed for MSA (atypical parkinsonian disorder) and ALS, potential MSA biomarkers include miR-223*, miR-324-3p, miR-24, miR-29c, miR-148b, miR-miR-1274a/b (Table 1) [57], and for ALS patients miR-338-3p, miR-451, miR-328, miR-149, miR-27a, miR-155, miR-146a, and miR-32-3p (Table 1) [102, 103].
The emerging view that circulating miRNAs have the potential to represent robust biomarkers has not only opened up new horizons in terms of disease diagnosis and prognosis, but also fueled interest into exploring miRNA targets as potential therapeutic interventions.
miRNA-mediated protein deregulation in neurodegeneration
Many miRNAs have been shown to regulate key proteins involved in neurodegenerative disease (Table 2). For example, miR-7 and miR-153 inhibits SNCA expression at both the transcriptional and translational levels in a synergistic manner (Fig. 2) [104, 105]. Another key PD protein, LRRK2, is regulated in a reverse manner by miR-205, and pathogenic mutations in LRRK2 disrupts let-7 and miR-184* expression causing deregulation of E2F1/DP implicated in cell cycle and survival control (Fig. 2) [106, 107]. Minones-Moyano and colleagues have further demonstrated the miR-34b/c regulates DJ-1/PARK7 and Parkin in SHSY-5Y neuroblastoma cells and in brains of PD patients (Fig. 2) [108].
Table 2.
Regulation of key proteins in neurodegeneration by microRNA
| Disease | microRNA | Gene regulated | Experimental design | References |
|---|---|---|---|---|
| Parkinson’s disease | miR-7 | SNCA | Regulation in cells, luciferase reporter assay | [105] |
| miR-153 | SNCA | Mouse model, regulation in cells | [104] | |
| miR-34b, miR-34c | SNCA | Regulation in cells, 3′UTR binding/inhibition | [150] | |
| Let-7, miR-184* | Lrrk2, E2F1, DP | [106] | ||
| miR-34b/c | DJ-1, Parkin | SHSY-5Y cells, human brains | [108] | |
| miR-433 | FGF20 | Patients, regulation in cells | [151] | |
| miR-26b, miR-106a*, miR-301b | Hsc-70 | Patients, regulation in cells, luciferase reporter assay | [70] | |
| miR-21, miR-224, miR-373*, miR-379 | LAMP-2A | Patients, regulation in cells, luciferase reporter assay | [70] | |
| miR-133b | Pitx3 | Patients, regulation in cells | [42] | |
| miR-64/65 | mdl-1, ptc-1 | Mouse model, C. elegans | [152] | |
| Alzheimer’s disease | miR-34a | TAU | Patients, in vitro reporter assay | [109] |
| miR-101 | APP | In vitro reporter assay, regulation cells | [110] | |
| miR-16 | APP | Mouse model | [111] | |
| miR-106a, miR-520c | APP | In-vitro reporter assay, overexpression in cells | [112] | |
| miR-147, miR-655, miR-323-3p, miR-644, miR-153 | APP | Regulation in cells, luciferase reporter assay, ELISA | [113] | |
| miR-24, miR-186, miR-455 | Ab | Regulation in cells, luciferase reporter assay, ELISA | [153] | |
| miR-137 | Ab | Patients, in vitro reporter assay | [61] | |
| miR-146a | Ab | Patients, regulation in cells, mouse model | [127] | |
| miR-181c | Ab | Patients, in vitro reporter assay | [61] | |
| miR-98 | Ab | Regulation in cells | [115] | |
| miR-107 | BACE1 | Patients, in situ hybridization | [116] | |
| miR-29a-1, miR-29b-1, miR-9 | BACE1 | Patients, in vitro reporter assay | [58] | |
| miR-124 | BACE1 | Overexpression in cells | [117] | |
| miR-181c, miR-137 | SPTLC1 | Patients, in vitro reporter assay | [61] | |
| miR-29a, miR-29b1, miR-9 | SPTLC2 | Patients, in vitro reporter assay | [61] | |
| miR-107 | Cofilin | Mouse model, In-vitro reporter assay | [154] | |
| miR-106b | TGF-b | Mouse model | [116] | |
| miR-98 | IGF1 | Mouse model, regulation in cells | [115] | |
| miR-17, miR-20a, miR-106a, miR-106b | APP | Patients, in vitro reporter assay | [155] | |
| Huntington’s disease | miR-9, miR-9* | REST-coREST | In-vitro reporter assay | [81] |
| miR-196a | mut-Htt | Mouse model, Overexpression in Stem cells | [118] | |
| Amyotrophic lateral sclerosis | miR-9, miR-9* | Neurofilament expression | Mouse model | [43] |
| miR-659 | Progranulin | Polymorphism | [119] | |
| miR-206 | HDAC4 | Mouse model | [92] | |
| miR-9 | NEFH | Mouse model | [43] |
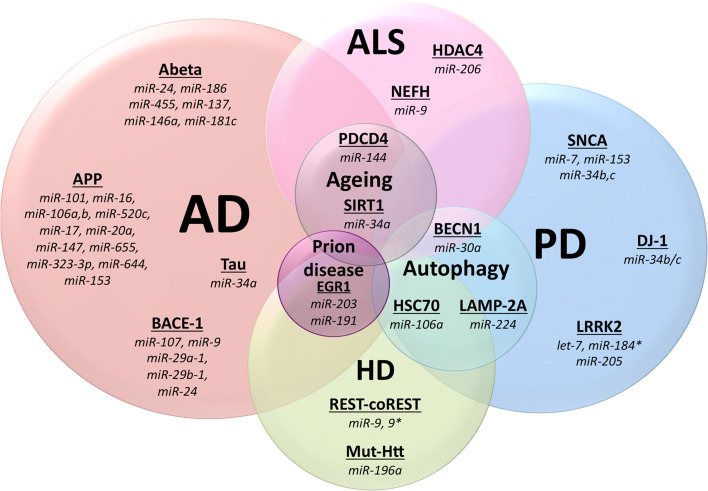
Fig. 2.
Schematic diagram highlighting microRNAs that have been identified to show differential expression in various neurodegenerative diseases. The miRNAs shown have been identified using a variety of different techniques, as described throughout the text. Although several miRNAs are specific to a given class of neurodegenerative disease many also overlap suggesting their involvement in more common pathways, such as neuronal cell death. As both aging and autophagy are associated with neurodegeneration there are also shared miRNAs between these two processes and the four main neurodegenerative disease states
Tauopathies, such as AD, have many key proteins regulated by miRNAs. Strikingly, Tau aggregation in AD is directly regulated by miR-34a [109]. The amyloid precursor protein (APP) is also regulated by numerous miRNAs, including miR-101, miR-16, miR-106a and miR-644 (Table 2; Fig. 2) [110–113]. Further, the amyloid protein fragment Ab, is also regulated by miR-24, miR-186, miR-455, miR-146a, and miR-98 (Fig. 2) [113–115]. It has also been speculated that miR-137, miR-146a, and miR-181c may regulate the levels of Ab in AD patients [61, 113–115]. Interestingly, the APP-processing enzyme b-secretase enzyme I (BACE1) is also regulated by miR-107, miR-29a-1/b-1, miR-9 and miR-124 (Fig. 2) [49, 116, 117].
Further, several proteins known to be involved in HD and ALS, are regulated by miRNAs (Table 2). miR-196a overexpression has been shown to alleviate HD phenotype in both in vitro and in vivo by indirectly targeting mutant huntingtin (htt), an integral protein in HD etiology (Fig. 2) [118]. The miR-9 family of microRNAs, which are differentially expressed in the cortex of patients suffering from Huntington’s disease, regulate the expression of the transcription repressor-co-repressor REST-coREST (RE1-silencing transcription factor) which interacts with htt [81]. In addition, miR-9, miR-9* regulates neurofilament expression by positively regulating neurofilament heavy subunit (NEFH) in mouse models of ALS (Fig. 2) [43]. miR-659 can regulate progranulin (GRN), by binding more efficiently to the T-allele or rs5848 variant, which in turn leads to a 3.2 fold higher risk of developing frontotemporal dementia (FTLD-U) [119].
miRNAs can clearly regulate both the activation and inhibition of translation of genes associated with neurodegeneration, and Table 2 summarizes these miRNAs and their protein targets.
Technological advancements in search for miRNA biomarkers in neurodegenerative diseases
miRNA microarray coupled to quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) have been extensively used for miRNA biomarker discovery. SYBR Green-based qRT-PCR has been most widely used, representing a fluorescent dye-based technology that detects and enables quantification of double-stranded DNA amplification [120]. Recently, TaqMan-based qRT-PCR applications have become more common practice, which are based on the use of fluorescent probes, specific to target genes, allowing increased specificity [121, 122]. Several commercial companies have marketed TaqMan probe-based qRT-PCR assays, which offer robustness and flexibility in terms of miRNA analysis. For example, TaqMan low density array (TLDA) cards are arrays pre-filled with TaqMan assays for qRT-PCR analysis, which can be ordered as customized cards, thereby providing flexibility to choose disease-specific miRNAs.
To date, only a small number of potential neurodegenerative disease-specific miRNA biomarkers have been identified using microarray and qRT-PCR (Table 1). However, as more are discovered, the future development of neurodegenerative diseases-specific microarray or TLDA arrays will enhance the diagnostic capability and accuracy for neurodegenerative diseases.
As mentioned earlier, the discovery of circulating miRNAs has presented an opportunity to mine for biomarkers in neurodegenerative diseases. With respect to this, Soreq and colleagues used Next Generation Sequencing (NGS) to provide a comprehensive list of 254 miRNAs in PD patients before and after deep brain stimulation treatment [123]. Similarly, Kumar and colleagues reported 140 unique mature miRNAs from blood samples of AD patients using NGS [101]. In addition, Burgos and colleagues combined NGS with CSF and serum samples from AD and PD patients investigating extracellular miRNAs which could possibly be used as prognostic markers for disease [73].
The application of NGS, microarrays and qRT-PCR has led to the discovery of hundreds of miRNAs associated with neurodegeneration (Table 1); however, target validation is still lacking.
Therapeutic intervention strategies involving miRNAs in neurodegeneration
Although we are far from understanding the complete mechanistic picture of neuronal cell death, studies on animal models, cell models, biofluids, and post-mortem brain tissue have provided insight into perturbations of miRNA expression profiles associated with neurodegenerative diseases. Combined with genetic and biochemical studies, progress has been made in developing miRNA-based therapeutic intervention strategies.
miRNA mimics
In 2007, Wang and colleagues developed double-stranded small RNA molecules that would mimic miRNAs and serve as gain-of-function tools for specific miRNAs [124]. These miRNA mimics specifically target mRNAs through miRNA-like actions in mammalian cells. Their 5′ end possesses a motif that is partially complementary to the 3′UTR of a specific target gene. Once inside the cells, these double-stranded miRNA-like RNA fragments inhibit the translation of the specific target genes, thereby producing a gene-specific effect. This technology has been further developed by the industry sector and a battery of miRNA mimics is available for the majority of human miRNAs, including miRNAs in neurodegeneration. For example, miR-153 and miR-205 target α-synuclein and LRRK2, respectively (Table 2) [105, 107], and it is, therefore, possible to use miR-153 and miR-205 mimics to unravel protein targets associated with PD, allowing the discovery of new therapeutic targets.
miRNA anti-miRs and antagomirs
Similar to miRNA mimics, blocking a miRNA also shows potential in terms of disease intervention. Initial studies focused on anti-miRs or antisense oligonucleotides that block endogenous miRNA. The antisense oligonucleotide would bind to the mature guide strand of the miRNA and induce degradation or stoichiometric duplex formation [125]. Hutvagner and colleagues then showed that a modification on the antisense oligonucleotide, which gave rise to the now known antagomiR, contributed to nuclease resistance and increased binding affinities to miRNA targets [126]. For example, in Locked Nucleic Acid (LNA) antimiRs, the ribose rings are ‘locked’ by a methylene bridge connecting the 2′-O atom and the 4′-C atom. This locking mechanism not only increases its stability, but also increases the hybridization efficiency to the target single stranded RNA molecule [127]. This technology has been adopted by the industry sector that has developed a library of custom oligos mimicking human miRNAs. Using this technology, Koval and colleagues inhibited miR-155, which is elevated twofold in the spinal cord of ALS patients, in ALS-model mice through delivery of a miR-155 anti-miR into the central nervous system. This inhibition caused global repression of targets in peritoneal macrophages and increased the survival rate [128].
miRNA sponges
miRNA sponges, first recognized by Ebert and colleagues [129], are able to inhibit multiple miRNAs simultaneously. For example, miRNA sponges with a heptameric seed sequence are able to target an entire miRNA family sharing the same seed sequence [130], thereby enabling widespread miRNA repression. Similar to anti-miRs or antagomiRs, these sponges can also be used to target multiple mRNAs [10]. Recently, Tan and colleagues used a miR-277 sponge to block rCGG repeat-mediated neurodegeneration in Fragile X-associated tremor/ataxia syndrome (FXATS), a late-onset neurodegenerative disorder [131].
miRNA masks
Another decoy-based mechanism for controlling miRNA regulation is through the use of miR-Masks. miR-Masks are single stranded 2′-O-methyl-modified antisense oligonucleotide that bind to the 3′UTR of a miRNA’s target mRNA, thereby masking the miRNA binding site and derepressing the target gene. This approach, has to date, not been used in neurodegeneration. However, the masking of the 3′UTR region of mRNAs that prevent neurodegeneration represents exciting prospects [132].
Small drug molecule inhibitors
A study in 2008 by Gumireddy and colleagues identified small drug molecules, which indirectly altered the oncomiR miR-21 at the transcription level [133]. Although this study was related to oncogenesis, their observations may have implications in pre-clinical neurodegeneration studies by screening compounds for a target miRNA, which is involved in neuronal cell death. For example, studies have shown that l-dopa treatment results in altered miRNA profiles in PD patients [134] suggesting that perhaps small molecules may alter miRNA profiles in neurodegeneration that ultimately modifies the disease course.
Challenges of miRNA treatment
The stability and the immunogenicity of miRNAs has always been a concern when it comes to miRNA-based therapies. The overall benefits and limitations of different miRNA-based therapies have been elegantly reviewed by Garzon and colleagues where they clearly underline the need for increased understanding of miRNA biology in general [135]. Neurodegeneration is a process that develops over several years, often decades, and hence miRNA stability becomes the biggest concern in this context. However, the high turnover rates of miRNAs and their higher half lives as compared to mRNAs may provide ample time to track onset and progression of neurodegeneration, thereby paving the way for the development of potential biomarkers and therapeutic interventions.
Another potential obstacle in developing molecules to treat neurodegeneration is the inherent capability of molecules to cross the blood brain barrier. However, a couple of recent studies in ALS have shown that oligonucleotides and miRNAs can cross the blood brain barrier and also cross into the central nervous system by using intrathecal infusion and a ventricular osmotic pump, respectively [128, 136].
miRNAs very often target and regulate the expression of several mRNAs which may pose potential challenges in terms of off target effects in the context of miRNA-based therapies. This is probably the reason why only a handful of miRNAs have reached clinical trials. Complicating matters further for neurodegeneration-driven applications is the highly complex nature of neurodegenerative diseases in terms of progression rates, in addition to the age and sex dependency.
Despite the current limitations of miRNAs as therapeutic agents, recent advances in the field have paved the way for the future design of novel intervention strategies.
Technological advancements in miRNA target discovery in neurodegenerative diseases
Microarrays have been extensively used for expression analysis. However, the emergence of NGS has enabled the use of RNA-seq as an alternative to microarrays for the identification of miRNA targets [137]. Despite its extensive application to identify neurodegenerative disease-specific miRNAs, the approach has limitations as it will identify miRNA targets irrespective of a direct or indirect association with any given miRNA. Therefore, other alternative techniques, which can differentiate between direct and indirect targets of miRNAs, have been developed. For example, immunoprecipitation of RISC components followed by a microarray analysis provides direct miRNA:mRNA target pair identification [138]. Furthermore, high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP), has provided promising data on miRNA binding sites and generated genome-wide interacting maps for endogenous and exogenously expressed miRNAs [139]. A similar advancement called photoactivable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP), with a better RNA recovery rate than HITS-CLIP, utilizes photoactivable nucleosides to isolate proteins associated with RNA [140]. Recently, yet another immunoprecipitation-based technique was developed, known as ChIP-Seq, which has the ability to analyze the transcriptional regulation of miRNAs [141].
In addition to the above techniques, comparative two-dimensional polyacrylamide gel electrophoresis (2D-PAGE), followed by mass spectrometry (MS), or liquid chromatography MS (LC/MS) can be employed to identify miRNA:mRNA target pair combinations in cells or tissues where specific miRNAs are artificially overexpressed or inhibited [142, 143].
The combination of the abovementioned techniques will allow further deciphering of miRNA:mRNA associations as well as perturbations in the microRNAome and proteome in neurodegeneration.
Future prospects
miRNAs hold promise to be the next generation biomarkers and therapeutic candidates in neurodegenerative diseases. Cell-based, as well as clinical sample-based, studies involving miRNA have shed much light on understanding the etiology of neurodegenerative diseases. Although no candidate miRNA are currently undergoing clinical trials, as is the case for cancer, recent advancements suggest that we are not far from reaching this milestone. Clearly the technological advancements in modifying the structural properties of miRNAs, in terms of bioavailability and stability, have paved way for a new dimension in miRNA research. Likewise, the emergence of larger, well-defined patient cohorts, coupled to the availability and stability of miRNAs in biofluids, has opened up the possibility of discovering robust diagnostic and prognostic miRNA biomarkers for neurodegenerative disease. Although the tissue specificity and off-target effects still remain a concern, conjugating neural stem cell research with miRNA biology might provide a better platform to develop potential therapeutic candidates. Our understanding of the etiology of neurodegeneration is still very much in its infancy, but miRNAs, their biology and potential applications, hold tremendous promise for individuals suffering from this class of devastating diseases.
Acknowledgments
Research in our laboratory is funded by The Norwegian Research Council, The Western Norway Regional Health Authority, The Norwegian Centre for Movement Disorders, The Norwegian Parkinson’s Association, and St. John’s University. We thanks Katherine Moller for proofreading of the manuscript.
Footnotes
I. Basak and K. S. Patil contributed equally.
References
- 1.International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Huttenhofer A, Schattner P, Polacek N. Non-coding RNAs: hope or hype? Trends Genet. 2005;21:289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Kaikkonen MU, Lam MT, Glass CV. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res. 2011;90:430–440. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ardekani AM, Naeini MM. The role of microRNAs in human diseases. Avicenna J Med Biotechnol. 2010;2:161–179. [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Kowdley KV. MicroRNAs in common human diseases. Genomics Proteomics Bioinform. 2012;10:246–253. doi: 10.1016/j.gpb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Etheridge A, Lee I, Hood L, Galas D, Wang K. Extracellular microRNA: a new source of biomarker. Mutat Res. 2011;717:85–90. doi: 10.1016/j.mrfmmm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeffrey SS. Cancer biomarker profiling with microRNAs. Nat Biotechnol. 2008;26:400–401. doi: 10.1038/nbt0408-400. [DOI] [PubMed] [Google Scholar]
- 8.Goodall EF, Heath PR, Bandmann O, Kirby J, Shaw PJ. neuronal dark matter: the emerging role of microRNAs in neurodegeneration. Front Cell Neurosci. 2013;7:178. doi: 10.3389/fncel.2013.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cathew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 11.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates micorRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 14.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha–DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregory RI, Yan KP, Amuthan G, et al. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 16.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci USA. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ender C, Krek A, Friedlander MR, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 20.Miyoshi K, Miyoshi T, Siomi H. Many ways to generate microRNA-like small RNAs: non-canonical pathways for microRNA production. Mol Genet Genomics. 2010;284:95–103. doi: 10.1007/s00438-010-0556-1. [DOI] [PubMed] [Google Scholar]
- 21.Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA. 2012;3:311–330. doi: 10.1002/wrna.121. [DOI] [PubMed] [Google Scholar]
- 22.Hashimoto Y, Akiyama Y, Yuasa Y. Multiple-to-multiple relationships between microRNAs and target genes in gastric cancer. PLoS One. 2013;8:e62589. doi: 10.1371/journal.pone.0062589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 24.Han J, Pederson JS, Kwon SC, et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Triboulet R, Chang HM, LaPierre RJ, Gregory RI. Post-transcriptional control of DGCR8 expression by the microprocessor. RNA. 2009;15:1005–1011. doi: 10.1261/rna.1591709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newman MA, Hammond SM. Emerging paradigms of regulated microRNA processing. Genes Dev. 2010;24:1086–1092. doi: 10.1101/gad.1919710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute 2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 28.Heale BSE, Keegan LP, McGurk L, et al. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 2009;28:3145–3156. doi: 10.1038/emboj.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, Qin YW, Brewer G, Jing Q. MicroRNA degradation and turnover: regulating the regulators. Wiley Interdiscip Rev RNA. 2012;3:593–600. doi: 10.1002/wrna.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sim SE, Bakes J, Kaang BK. Neuronal activity-dependent regulation of microRNAs. Mol Cells. 2014;37:511–517. doi: 10.14348/molcells.2014.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim LP, Lau NC, Garrett-Engele A, Grimson JM, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 32.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 33.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 34.Khalaj M, Tavakkoli M, Stranahan AW, Park CY. Pathogenic microRNA’s in myeloid malignancies. Front Genet. 2014;5:361. doi: 10.3389/fgene.2014.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simone NL, Soule BP, Ly D, Saleh AD, et al. Ionizing-induced oxidative stress alters miRNA expression. PLoS One. 2009;4:e6377. doi: 10.1371/journal.pone.0006377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuss AW, Chen W. MicroRNAs in brain function and disease. Curr Neurol Neurosci Rep. 2008;8:190–197. doi: 10.1007/s11910-008-0031-0. [DOI] [PubMed] [Google Scholar]
- 37.Sempere LF, Freemantle S, Pitha-Rowe I, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi Y, Zhao X, Hsieh J, Wichterle H, et al. MicroRNA regulation of neural stem cells and neurogenesis. J Neurosci. 2010;30:14931–14936. doi: 10.1523/JNEUROSCI.4280-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giraldez AJ, Cinalli RM, Glasner ME, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 40.Chen JA, Wichterle H. Apoptosis of limb innervating motor neurons and erosion of motor pool identity upon lineage specific dicer inactivation. Front Neurosci. 2012;6:69. doi: 10.3389/fnins.2012.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang D, Li T, Wang Y, et al. miR-132 regulates the differentiation of dopamine neurons by directly targeting Nurr1 expression. J Cell Sci. 2012;125:1673–1682. doi: 10.1242/jcs.086421. [DOI] [PubMed] [Google Scholar]
- 42.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, et al. A microRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haramati S, Chapnik E, Sztainberg Y, Eilam R, Zwang R, Gershoni N, et al. miRNA malfunction causes spinal motor neuron disease. Proc Natl Acad Sci USA. 2010;107:13111–13116. doi: 10.1073/pnas.1006151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaefer A, O’Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, et al. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204:1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;23:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 49.Bossy-Wetzel E, Schwarzenbacher R, Lipton SA. Molecular pathways to neurodegeneration. Nat Med. 2004;10(Suppl):S2–S9. doi: 10.1038/nm1067. [DOI] [PubMed] [Google Scholar]
- 50.Manakov SA, Grant SG, Enright AJ. Reciprocal regulation of microRNAs and mRNA profiles in neuronal development and synapse formation. BMC Genom. 2009;10:419. doi: 10.1186/1471-2164-10-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruno IG, Karam R, Huang L, et al. Identification of microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell. 2011;42:500–510. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou R, Yuan P, Wang Y, et al. Evidence of selective microRNAs and their effectors as common long-term targets for the action of mood stabilizers. Neuropsychopharmacology. 2009;34:1395–1405. doi: 10.1038/npp.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao J, Wang WY, Mao YW, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olde Loohuis NF, Kos A, Martens GJ, et al. MicroRNA networks direct neuronal development and plasticity. Cell Mol Life Sci. 2012;69:89–102. doi: 10.1007/s00018-011-0788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Margis R, Margis R, Rieder CR. Identification of blood microRNAs associated to Parkinson’s disease. J Biotechnol. 2011;152:96–101. doi: 10.1016/j.jbiotec.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 56.Geekiyanage H, Jicha GA, Nelson PT, Chan C. Blood serum miRNA: non-invasive biomarkers for Alzheimer’s disease. Exp Neurol. 2012;235:491–496. doi: 10.1016/j.expneurol.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vallelunga A, Ragusa M, DiMauro S, et al. Identification of circulating microRNAs for the differential diagnosis of Parkinson’s disease and multiple system atrophy. Front Cell Neurosci. 2014;8:156. doi: 10.3389/fncel.2014.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hébert SS, Horré K, Nicolaï L, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/β-secretase expression. Proc Natl Acad Sci USA. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee ST, Chu K, Im WS. Altered microRNA regulation in Huntington’s disease models. Exp Neurol. 2011;227:172–179. doi: 10.1016/j.expneurol.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 60.Wang WX, Huang Q, Hu Y, Stromberg AJ, Nelson PT. Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 2011;121:193–205. doi: 10.1007/s00401-010-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Geekiyanage H, Chan C. MicroRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid beta, novel targets in sporadic Alzheimer’s disease. J Neurosci. 2011;31:14820–14830. doi: 10.1523/JNEUROSCI.3883-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abdullah R, Basak I, Patil KS, Alves G, Larsen JP, Moller SG. Parkinson’s disease and age: the obvious but largely unexplored link. Exp Gerontol. 2014;68:33–38. doi: 10.1016/j.exger.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 63.Inukai S, de Lencastre A, Turner M, Slack F. Novel microRNAs differentially expressed during aging in the mouse brain. PLoS One. 2012;7:e40028. doi: 10.1371/journal.pone.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Persengiev S, Kondova I, Otting N, Koeppen AH, Bontrop RE. Genome-wide analysis of miRNA expression reveals a potential role for miR-144 in brain aging and spinocerebellar ataxia pathogenesis. Neurobiol Aging. 2011;32:e2317–e2327. doi: 10.1016/j.neurobiolaging.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 65.Elfenbein HA, Rosen RF, Stephens SL, et al. Cerebral β-amyloid angiopathy in aged squirrel monkeys. Histol Histopathol. 2007;22:155–167. doi: 10.14670/HH-22.155. [DOI] [PubMed] [Google Scholar]
- 66.Martinez I, Almstead LL, DiMaio D. MicroRNAs and senescence. Aging. 2011;3:77–78. doi: 10.18632/aging.100282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li X, Khanna A, Li N, Wang E. Circulatory miR34a as an RNA based, noninvasive biomarker for brain aging. Aging. 2011;3:985–1002. doi: 10.18632/aging.100371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smit-McBride Z, Forward KI, Nguyen AT. Age-dependent increase in miR-34a expression in the posterior pole of the mouse eye. Mol Vis. 2014;20:1569–1578. [PMC free article] [PubMed] [Google Scholar]
- 70.Alvarez-Erviti L, Seow Y, Schapira AH, Rodriguez-Oroz MC, Obeso JA, Cooper JM. Influence of microRNA deregulation on chaperone-mediated autophagy and alpha-synuclein pathology in Parkinson’s disease. Cell Death Dis. 2013;4:e545. doi: 10.1038/cddis.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cardo LF, Coto E, Ribacoba R, Menéndez M, Moris G, Suárez E, Alvarez V. MiRNA profile in the substantia nigra of Parkinson’s disease and healthy subjects. J Mol Neurosci. 2014;54:830–836. doi: 10.1007/s12031-014-0428-y. [DOI] [PubMed] [Google Scholar]
- 72.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 73.Burgos K, Malenica I, Metpally R, et al. Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS One. 2014;9:e94839. doi: 10.1371/journal.pone.0094839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lukiw WJ. MiRNA speciation in fetal, adult and Alzheimer’s disease hippocampus. NeuroReport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 75.Saba R, Goodman CD, Huzarewich RLCH, Robertson C, Booth SA. A miRNA signature of prion induced neurodegeneration. PLoS One. 2008;3:e3652. doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Montag J, Hitt R, Opitz L. Upregulation of miRNA hsa-miR-342-3p in experimental and idiopathic prion disease. Mol Neurodegener. 2009;4:36. doi: 10.1186/1750-1326-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012;40:10937–10949. doi: 10.1093/nar/gks832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cummings CJ, Zoghbi HY. Fourteen and counting: unravelling trinucleotide repeat diseases. Hum Mol Genet. 2000;9:909–916. doi: 10.1093/hmg/9.6.909. [DOI] [PubMed] [Google Scholar]
- 79.Everett CM, Wood NW. Trinucleotide repeats and neurodegenerative disease. Brain. 2004;127:2385–2405. doi: 10.1093/brain/awh278. [DOI] [PubMed] [Google Scholar]
- 80.Pogue AI, Cui JG, Li YY, Chao Y, Culicchia F, Lukiw WJ. MicroRNA-125b (miRNA-125b) function in astrogliosis and glial cell proliferation. Neurosci Lett. 2010;476:18–22. doi: 10.1016/j.neulet.2010.03.054. [DOI] [PubMed] [Google Scholar]
- 81.Marti E, Pantano L, Banez-Coronel M, et al. A myriad of miRNA variants in control and Huntington’s disease brain regions detected by massively parallel sequencing. Nucleic Acids Res. 2010;38:7219–7235. doi: 10.1093/nar/gkq575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnson R, Buckley NJ. Gene dysregulation in Huntington’s disease: REST, microRNAs and beyond. Neuromol Med. 2009;11:183–199. doi: 10.1007/s12017-009-8063-4. [DOI] [PubMed] [Google Scholar]
- 83.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Basu U, Guan LL, Moore SS. Functional genomics approach or identification of molecular processes underlying neurodegenerative disorders in prion diseases. Curr Genomics. 2012;13:369–378. doi: 10.2174/138920212801619223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jellinger KA. Basic mechanisms of neurodegeneration: a critical update. J Cell Mol Med. 2010;14:457–487. doi: 10.1111/j.1582-4934.2010.01159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lynch-Day MA, Mao K, Wang K, Zhao M, Klionsky DJ. The role of autophagy in Parkinson’s disease. Cold Spring Harb Perspect Med. 2012;2:a009357. doi: 10.1101/cshperspect.a009357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schapira AH, Gegg M. Mitochondrial contribution to Parkinson’s disease pathogenesis. Parkinsons Dis. 2011;2011:159160. doi: 10.4061/2011/159160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gibbings D, Mostowy S, Jay F, Schwab Y, Cossart P, Voinnet O. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol. 2012;14:1314–1321. doi: 10.1038/ncb2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Majeski AE, Dice JF. Mechanisms of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2435–2444. doi: 10.1016/j.biocel.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 90.Yang J, Chen D, He Y, et al. MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age. 2013;35:11–22. doi: 10.1007/s11357-011-9324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu H, Wu H, Liu X, et al. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy. 2009;5:816–823. doi: 10.4161/auto.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Williams AH, Valdez G, Moresi V, et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Roccaro AM, Sacco A, Jia X, et al. microRNA-dependent modulation of histone acetylation in Waldenstrom macroglobulinemia. Blood. 2010;116:1506–1514. doi: 10.1182/blood-2010-01-265686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gilad S, Meiri E, Yogev Y, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 96.Lang AE, Lozano AM. Parkinson’s disease—first of two parts. N Engl J Med. 1998;339:1044–1053. doi: 10.1056/NEJM199810083391506. [DOI] [PubMed] [Google Scholar]
- 97.Martins M, Rosa A, Guedes LC, et al. Convergence of miRNA expression profiling, α-synuclein interaction and GWAS in Parkinson’s disease. PLoS One. 2011;6:e25443. doi: 10.1371/journal.pone.0025443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Khoo SK, Petillo D, Kang UJ, et al. Plasma-based circulating microRNA biomarkers for Parkinson’s disease. J Parkinsons Dis. 2012;2:321–331. doi: 10.3233/JPD-012144. [DOI] [PubMed] [Google Scholar]
- 99.Schipper HM, Maes OC, Chertkow HM, Wang E. microRNA expression in Alzheimer blood mononuclear cells. Gene Regul Syst Biol. 2007;1:263–274. doi: 10.4137/grsb.s361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Leidinger P, Backes C, Deutscher S, et al. A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 2013;14:R78. doi: 10.1186/gb-2013-14-7-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kumar P, Dezso Z, MacKenzie C, et al. Circulating miRNA biomarkers for Alzheimer’s disease. PLoS One. 2013;8:e69807. doi: 10.1371/journal.pone.0069807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.De Felice B, Guida M, Guida M, Coppola C, de Mieri G, Cotrufo RA. miRNA signature in leukocytes from sporadic amyotrophic lateral sclerosis. Gene. 2012;508:35–40. doi: 10.1016/j.gene.2012.07.058. [DOI] [PubMed] [Google Scholar]
- 103.Butovsky O, Siddiqui S, Gabriely G, et al. Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. J Clin Investig. 2012;122:3063–3087. doi: 10.1172/JCI62636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci USA. 2009;106:13052–13057. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Doxakis E. Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153. J Biol Chem. 2010;285:12726–12734. doi: 10.1074/jbc.M109.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–641. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cho HJ, Liu G, Jin SM, et al. MicroRNA-205 regulates the expression of Parkinson’s disease-related leucine-rich repeat kinase 2 protein. Hum Mol Genet. 2014;22:608–620. doi: 10.1093/hmg/dds470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miñones-Moyano E, Porta S, Escaramís G, et al. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum Mol Genet. 2011;20:3067–3078. doi: 10.1093/hmg/ddr210. [DOI] [PubMed] [Google Scholar]
- 109.Dickson JR, Kruse C, Montagna DR, Finsen B, Wolfe MS. Alternative polyadenylation and miR-34 family members regulate tau expression. J Neurochem. 2013;127:739–749. doi: 10.1111/jnc.12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Long JM, Lahiri DK. MicroRNA-101 downregulates Alzheimer’s amyloid-β precursor protein levels in human cell cultures and is differentially expressed. Biochem Biophys Res Commun. 2011;404:889–895. doi: 10.1016/j.bbrc.2010.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu W, Liu C, Zhu J, et al. MicroRNA-16 targets amyloid precursor protein to potentially modulate Alzheimer’s-associated pathogenesis in SAMP8 mice. Neurobiol Aging. 2012;33:522–534. doi: 10.1016/j.neurobiolaging.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 112.Patel N, Hoang D, Miller N, et al. MicroRNAs can regulate human APP levels. Mol Neurodegener. 2008;3:10. doi: 10.1186/1750-1326-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Delay C, Calon F, Mathews P, Hébert SS. Alzheimer-specific variants in the 3′UTR of Amyloid precursor protein affect microRNA function. Mol Neurodegener. 2011;6:70. doi: 10.1186/1750-1326-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li YY, Cui JG, Hill JM, Bhattacharjee S, Zhao Y, et al. Increased expression of miRNA-146a in Alzheimer’s disease transgenic mouse models. Neurosci Lett. 2011;487:94–98. doi: 10.1016/j.neulet.2010.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hu YK, Wang X, Li L, Du YH, Ye HT, Li CY. MicroRNA-98 induces an Alzheimer’s disease-like disturbance by targeting insulin-like growth factor 1. Neurosci Bull. 2013;29:745–775. doi: 10.1007/s12264-013-1348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang WX, Wilfred BR, Madathil SK. miR-107 regulates granulin/progranulin with implications for traumatic brain injury and neurodegenerative disease. Am J Pathol. 2010;177:334–345. doi: 10.2353/ajpath.2010.091202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fang M, Wang J, Zhang X, et al. The miR-124 regulates the expression of BACE1/p-secretase correlated with cell death in Alzheimer’s disease. Toxicol Lett. 2012;209:94–105. doi: 10.1016/j.toxlet.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 118.Cheng PH, Li CL, Chang YF, et al. miR-196a ameliorates phenotypes of Huntington disease in cell, transgenic mouse, and induced pluripotent stem cell models. Am Hum Genet. 2013;93:306–312. doi: 10.1016/j.ajhg.2013.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rademakers R, Eriksen JL, Baker M, et al. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum Mol Genet. 2008;17:3631–3642. doi: 10.1093/hmg/ddn257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cells. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lu DP, Read RL, Humphreys DT, Battah FM, Martin DI, Rasko JE. PCR-based expression analysis and identification of microRNAs. J RNAi Gene Silencing. 2005;1:44–49. [PMC free article] [PubMed] [Google Scholar]
- 122.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Soreq L, Salomonis N, Bronstein M, et al. Small RNA sequencing-microarray analyses in Parkinson leukocytes reveal deep brain stimulation-induced splicing changes that classify brain region transcriptomes. Front Mol Neurosci. 2013;6:10. doi: 10.3389/fnmol.2013.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang Z. miRNA interference technologies. Berlin Heidelberg: Springer; 2009. miRNA mimic technology; pp. 93–100. [Google Scholar]
- 125.Boutla A, Delidakis C, Tabler M. Developmental defects by antisense-mediated inactivation of micro-RNAs 2 and 13 in Drosophila and the identification of putative target genes. Nucleic Acids Res. 2003;31:4973–4980. doi: 10.1093/nar/gkg707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E9. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 128.Koval ED, Shaner C, Zhang P, et al. Method for widespread microRNA-155 inhibition prolongs survival in ALS-model mice. Hum Mol Genet. 2013;22:4127–4135. doi: 10.1093/hmg/ddt261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xiao J, Yang B, Lin H, Lu Y, Luo X, Wang Z. Novel approaches for gene-specific interference via manipulating actions of microRNAs: examination of the pacemaker channel genes HCN2 and HCN4. J Cell Physiol. 2007;212:285–292. doi: 10.1002/jcp.21062. [DOI] [PubMed] [Google Scholar]
- 131.Tan H, Poidevin M, Li H, Chen D, Jin P. MicroRNA-277 modulates the neurodegeneration caused by Fragile X premutation rCGG repeats. PLoS Genet. 2012;8:e1002681. doi: 10.1371/journal.pgen.1002681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang Z. The principles of MiRNA-masking antisense oligonucleotide technology. Methods Mol Biol. 2011;676:43–49. doi: 10.1007/978-1-60761-863-8_3. [DOI] [PubMed] [Google Scholar]
- 133.Gumireddy K, Young DD, Xiong X, Hogenesch JB, Huang Q, Deiters A. Small-molecular inhibitors of miR-21 function. Angew Chem Int Ed Engl. 2008;47:7482–7484. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Serafin A, Foco L, Zanigni S, et al. Overexpression of blood microRNAs 103a, 30b, and 29a in l-dopa-treated patients with PD. Neurology. 2015;84:645–653. doi: 10.1212/WNL.0000000000001258. [DOI] [PubMed] [Google Scholar]
- 135.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2014;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Miller TM, Pestronk A, David W, et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomized, first-in-man study. Lancet Neurol. 2013;12:435–442. doi: 10.1016/S1474-4422(13)70061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xu G, Fewell C, Taylor C, et al. Transcriptome and targetome analysis in MIR155 expressing cells using RNA-seq. RNA. 2010;16:1610–1622. doi: 10.1261/rna.2194910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39:6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hafner M, Landthaler M, Burger L, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yang JH, Li JH, Jiang S, Zhou H, Qu LH. ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and miRNA genes from ChIP-Seq data. Nucleic Acids Res. 2013;41:D177–D187. doi: 10.1093/nar/gks1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li C, Xiong Q, Zhang J, Ge F, Bi LJ. Quantitative proteomic strategies for the identification of microRNA targets. Expert Rev Proteomics. 2012;9:549–559. doi: 10.1586/epr.12.49. [DOI] [PubMed] [Google Scholar]
- 143.Kurata R, Yonezawa T, Nakajima H, Takada S, Asahara H. LC-MS/MS-based shotgun proteomics identified the targets of arthritis related microRNA. Arthritis Res Ther. 2012;14:P36. doi: 10.1186/ar3637. [DOI] [Google Scholar]
- 144.Cardo LF, Coto E, de Mena L, Ribacoba R, Moris G, Menéndez M, et al. Profile of microRNAs in the plasma of Parkinson’s disease patients and healthy controls. J Neurol. 2013;260:1420–1422. doi: 10.1007/s00415-013-6900-8. [DOI] [PubMed] [Google Scholar]
- 145.Alexandrov PN, Dua P, Hill JM, Bhattacharjee S, Zhao Y, Lukiw WJ. microRNA (miRNA) speciation in Alzheimer’s disease (AD) cerebrospinal fluid (CSF) and extracellular fluid (ECF) Int J Biochem Mol Biol. 2012;3:365–373. [PMC free article] [PubMed] [Google Scholar]
- 146.Shioya M, Obayashi S, Tabunoki H, et al. Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathol Appl Neurobiol. 2010;36:320–330. doi: 10.1111/j.1365-2990.2010.01076.x. [DOI] [PubMed] [Google Scholar]
- 147.Gaughwin PM, Ciesla M, Lahiri N, Tabrizi SJ, Brundin P, Björkqvist M. Hsa-miR-34b is a plasma-stable microRNA that is elevated in pre-manifest Huntington’s disease. Hum Mol Genet. 2011;20:2225–2237. doi: 10.1093/hmg/ddr111. [DOI] [PubMed] [Google Scholar]
- 148.Lee ST, Chu K, Jung KH, et al. Altered expression of miR-202 in cerebellum of multiple-system atrophy. Mol Neurobiol. 2014;51:180–186. doi: 10.1007/s12035-014-8788-4. [DOI] [PubMed] [Google Scholar]
- 149.Campos-Melo D, Droppelmann CA, He Z, Volkening K, Strong MJ. Altered microRNA expression profile in amyotrophic lateral sclerosis: a role in the regulation of NFL mRNA levels. Mol Brain. 2013;6:26. doi: 10.1186/1756-6606-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kabaria S, Choi DC, Chaudhuri AD, Mouradian MM, Junn E. Inhibition of miR-34b and miR-34c enhances alpha-synuclein expression in Parkinson’s disease. FEBS Lett. 2015;589:319–325. doi: 10.1016/j.febslet.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang G, van der Walt JM, Mayhew G, et al. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008;82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Asikainen S, Rudgalvyte M, Heikkinen L, et al. Global microRNA expression profiling of Caenorhabditis elegans Parkinson’s disease models. J Mol Neurosci. 2010;41:210–218. doi: 10.1007/s12031-009-9325-1. [DOI] [PubMed] [Google Scholar]
- 153.Delay C, Dorval V, Fok A, et al. MicroRNAs targeting Nicastrin regulate Aβ production and are affected by target site polymorphisms. Front Mol Neurosci. 2014;7:67. doi: 10.3389/fnmol.2014.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yao J, Hennessey T, Flynt A, Lai E, Beal MF, Lin MT. MicroRNA-related cofilin abnormality in Alzheimer’s disease. PLoS One. 2010;5:e15546. doi: 10.1371/journal.pone.0015546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hebert SS, et al. MicroRNA regulation of Alzheimer’s amyloid precursor protein expression. Neurobiol Dis. 2009;33:422–428. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]