Abstract
Regulation of protein synthesis contributes to maintenance of homeostasis and adaptation to environmental changes. mRNA translation is controlled at various levels including initiation, elongation and termination, through post-transcriptional/translational modifications of components of the protein synthesis machinery. Recently, protein and RNA hydroxylation have emerged as important enzymatic modifications of tRNAs, elongation and termination factors, as well as ribosomal proteins. These modifications enable a correct STOP codon recognition, ensuring translational fidelity. Recent studies are starting to show that STOP codon read-through is related to the ability of the cell to cope with different types of stress, such as oxidative and chemical insults, while correlations between defects in hydroxylation of protein synthesis components and STOP codon read-through are beginning to emerge. In this review we will discuss our current knowledge of protein synthesis regulation through hydroxylation of components of the translation machinery, with special focus on STOP codon recognition. We speculate on the possibility that programmed STOP codon read-through, modulated by hydroxylation of components of the protein synthesis machinery, is part of a concerted cellular response to stress.
Keywords: Protein synthesis, Post-translational modifications, Translational fidelity, Dioxygenases
Introduction
A central feature of biological organisms is their ability to generate metabolic energy required for basic cell maintenance and synthesis of biomass. Nutrient oxidation (catabolism) is tightly coupled to synthesis of new cell components (anabolism), so that accumulation of undesired products of metabolism hardly ever happens under physiological conditions. This coupling between anabolism and catabolism underlies the whole regulation of cellular biochemistry, ultimately controlling concentrations of intermediary metabolites and kinetics of the reactions. Individual cells are nevertheless exposed to environmental changes as a consequence of diverse types of physiological or pathological insults including nutrient restriction, nutrient overload or oxidative stress among others. Thus, the complex regulatory machinery that synchronizes catabolism and anabolism must be plastic enough to cope with stressors. Protein synthesis, or translation, is one of the processes that consumes the most energy within the cell [1]. Thus, it is not surprising that translation is in fact one of the most actively regulated cell mechanisms, responding to signalling pathways that contain at least one component capable of sensing environmental conditions. Although many of these pathways control protein synthesis indirectly through regulation of mRNA stability or sub-cellular localization [2], most of them exert direct regulation through post-translational modifications of components of the translation machinery [3]. In this review we focus on our current knowledge on the mechanisms by which one particular post-translational modification, hydroxylation, can control protein synthesis. We speculate on the possibility that STOP codon read-through, a mechanism that has been linked to hydroxylation, may participate directly in stress responses.
Protein synthesis: mechanism and regulation
The eukaryotic ribosome contains four distinct rRNA molecules and 79 ribosomal proteins [4], although its structure and components may differ throughout the life cycle, between different tissues, or in different environmental conditions [5, 6]. The mechanism of protein synthesis comprises three distinct phases: (1) initiation; (2) elongation, and (3) termination, with initiation being the most tightly regulated step. The Initiation step involves the formation of the 43S pre-initiation complex composed of the initial Met-tRNAi, the 40S small ribosomal subunit and the translation initiation factors eIF2, eIF1/1A and eIF3 [7]. This complex identifies and binds a modified guanine at the 5′-end of the mRNA, known as m7G CAP, through the initiation factor eIF4E, which in turn associates with the scaffold protein eIF4G. The interaction between eIF3 and eIF4G stabilizes the 43S pre-initiation complex that executes scanning of the 5′ untranslated region (5′UTR) until the first AUG codon is reached. Once the initiation codon has been recognized, the GTPase activating protein eIF5 promotes the hydrolysis of eIF2-GTP, dissociating eIF2 from the 40S subunit. Finally, the 60S large subunit of the ribosome associates to the 40S subunit, and the elongation phase begins [8, 9]. The ribosome contains three tRNA binding sites, called A, P and E, with the middle of them forming the peptidyl transferase center on the large ribosomal subunit. When the ribosome is placed at the first AUG codon, the P site contains the initial Met-tRNAi, which links the AUG codon with its corresponding methionine, whilst the A site remains empty. Thereafter, an incoming aminoacyl-tRNA (aa-tRNA) enters the A site and interacts with both the mRNA and rRNA. If the codon–anticodon interaction occurs according to the rules of Watson–Crick base-pairing, the structure becomes stable and formation of a peptide bond is catalyzed by the ribozyme activity of the peptidyl transferase center. Once the peptide bond has been formed, the ribosome can translocate one codon forward, leaving the A site empty to receive the next incoming aa-tRNA, and the tRNA that has already been utilized leaves the ribosome through the E site [10]. The elongation phase is basically the repetition of this cycle until the ribosome reaches one of three possible STOP codons (UAA, UAG or UGA), when the termination phase takes place. In eukaryotes, translation termination is mediated by a release factor complex that includes the eukaryotic release factor 1 (eRF1) and the GTPase eRF3. eRF1 is a tRNA-shaped protein that binds to ribosomes, exposing the STOP codon at the A site, resulting in addition of a water molecule to the peptidyl-tRNA instead of a new amino acid, thereby terminating the elongation phase. Concomitantly, eRF3 facilitates positioning of eRF1 into the peptidyl transferase center in a process that requires hydrolysis of GTP, and derives in its release from the ribosome. After eRF3 dissociation, eRF1 remains bound to the post-termination complex and, in cooperation with the ABC-type ATPase ABCE1, releases the ribosomal 60S subunit from the deacylated tRNA-mRNA-40S subunit complex. Finally, the initiation factors eIF3, eIF1 and eIF1A mediate dissociation of the deacetylated tRNA and mRNA from the post-termination 40S complex [11, 12].
Regulation of translation is considered as a mechanism faster than control of transcription. Whilst transcriptional regulation is mostly associated with long-term changes in cell physiology, translational regulation generally plays a role in quick responses, typically related to homeostasis maintenance [13], metabolic changes, memory and synaptic plasticity among other processes [9, 14]. Although most mechanisms that control protein synthesis target the initiation phase of the process, the successive phases of translation can also be tightly regulated [8, 15]. A few years ago, advances in structural biology have opened the possibility to study post-translational modifications (PTMs) of ribosomal proteins in depth [16], focusing on the effects elicited on their activity, stability, assembly or localization at the ribosome, as well as their impact on translation rates. These PTMs, analyzed by high-throughput techniques in bacteria [17], yeast [18], plants [19], rat [20] and humans [21], include removal of the initial methionine, N-terminal acetylation, N-terminal methylation, lysine N-methylation, methylthiolation and phosphorylation, among other modifications. For example, phosphorylation of five serine residues located at the C terminus of the small ribosomal subunit protein S6, RPS6 (eS6, according to the new nomenclature system of ribosomal proteins [22]) modulates the rate of protein synthesis through a nutrient-dependent mechanism. RPS6/eS6 is phosphorylated in conditions of nutrient abundance, and rapidly de-phosphorylated following nutrient starvation [23].
Control of translation through hydroxylation of elongation and termination factors
In the last few years, an increasing body of evidence began to reveal a novel type of translational regulation that involves hydroxylation of different protein synthesis machinery components (PSMCs), including tRNAs [24, 25], elongation factors [26] translation termination factors [27] and ribosomal proteins [28, 29] (Fig. 1). Protein hydroxylation was initially regarded as a rare post-translational modification of extracellular matrix components; nowadays, hydroxylation is considered a key regulatory mechanism in a wide range of biological processes [30, 31]. Hydroxylation of macromolecules is mediated by a group of oxygenases that utilize molecular oxygen (O2) as a co-substrate. The oxygenase family of enzymes comprises two groups: monooxygenases and dioxygenases. The first includes enzymes that catalyze the transfer of one atom of O2 to the substrate, while the second atom is reduced to water. Dioxygenases, on the other hand, catalyze the incorporation of both oxygen atoms into the oxidized substrate [30, 32]. Among dioxygenases, an important subfamily is represented by 2-oxoglutarate (2OG) and ferrous iron (Fe2+)-dependent oxygenases (2OG-oxygenases).
Fig. 1.
Hydroxylation-dependent translation regulation. The diagram shows how hydroxylases may exert translational control through hydroxylation of different components of the translation machinery. Ribosomal proteins are direct targets of hydroxylases both in the large subunit (RPL16, RPL8 and RPL27) and in the small subunit (RPS23). Some tRNAs (like tRNAPHE or tRNAGLY) are also subjected to hydroxylation as part of the post-transcriptional modifications that alter their interactions with the ribosome and mRNAs. Control of translation may also be exerted through regulation of translation factors. Both elongation factors (EF-Tu and EF-P), as well as termination factors (eRF1) have been reported to be hydroxylated
Elongation factors (EFs) are regulated by several PTMs such as phosphorylation, methylation and acetylation, among others [14, 15]. The fast kinetics of these modifications make them suitable for orchestrating quick adaptations to different environmental challenges that the organism may face [33]. It has been recently reported that some translation prokaryotic EFs, such as the elongation factor-Tu (EF-Tu) and the elongation factor-P (EF-P)—as well as its eukaryotic ortholog eEF1A- are subjected to hydroxylation. EF-Tu/eEF1A binds the aminoacyl-tRNA in a GTP-dependent manner, delivering the tRNA to the A site of the ribosome [34]. Recent reports indicate that EF-Tu is hydroxylated by the 2OG-dependent oxygenase PPHD in Gram negative bacteria such as Pseudomonas aeruginosa and Shewanella oneidensis [26, 35]. This hydroxylation affects EF-Tu GTPase activity, although its effect on translation is unclear.
EF-P is a highly conserved bacterial elongation factor that mimics tRNAs in shape and size. It binds the ribosome in a region between the P and E sites, stabilizing the initiator fMet-tRNAi, thus playing a critical role in the formation of the first peptidic bond [36]. It is well established that during protein synthesis, incorporation rates of all amino acids are quantitatively similar except for proline, whose incorporation rate is slower due to its unique biochemical nature [37]. Thus, proteins containing poly-proline sequences are subjected to ribosome stalling. Recent reports have shown that EF-P is important for preventing ribosome pausing at poly-proline stretches, and consistent with this, lack of EF-P provokes a decrease in the general amount of proteins containing poly-proline sequences [38, 39]. To perform its function, EF-P is subjected to β-lysinilation [(R)-β-lysine] at Lys34 through a multi-step mechanism [40]. The modified lysine can then be targeted by the hydroxylase YfcM, a monooxygenase that catalyzes an additional hydroxylation step of this residue [41]. Functional studies have revealed that EF-P is not essential during translation elongation, but instead, acts as an elongation modulator that depends on β-lysinilation of Lys34. In fact, it has been reported that lack of Lys34 β-lysinilation results in growth defects, antibiotic susceptibility and bacterial virulence attenuation [42]. Nevertheless, hydroxylation of (R)-β-lysine seems to be dispensable for modulation of EF-P function, as the YfcM mutant is phenotypically normal [43]. Thus, understanding the functional significance of this EF-P modification requires further research.
The eukaryotic ortholog of EF-P, the eukaryotic Initiation Factor 5A (eIF5A), has originally been described as an initiation factor [44], although no essential role of this protein in translation initiation has been assigned [45–47]. Recent studies have shown that eIF5A plays a role in translation elongation [48] by stimulating peptidyl transferase activity and preventing ribosome stalling on poly-proline stretches [49]. Interestingly, eIF5A presents a unique post-translational modification: conversion of a lysyl residue (Lys50 in humans) into hypusine. The multi-step mechanism of hypusine biosynthesis involves the sequential action of two enzymes, deoxyhypusine synthase (DHS) and deoxyhypusine hydroxylase (DOHH). Whereas DHS catalyzes reversible cleavage of spermidine and transfers the 4-aminobutyl moiety to the ε-amino group of Lys50 to form the intermediate residue deoxyhypusine (Dhp50), DOHH mediates the irreversible formation of hypusine (Hyp50) by the hydroxylation of deoxyhypusine. Whilst loss of eIF5A leads to lethality in yeast and mammalian models [50, 51], lack of deoxyhypusine hydroxylation seems to affect growth and proliferation in various organisms. For example, loss of eIF5A deoxyhypusine hydroxylation causes mild growth impairment in yeast [52], but in multicellular organisms such as Drosophila melanogaster, Caenorhabditis elegans or mammals, lack of deoxyhypusine hydroxylation provokes early embryonic lethality [53, 54]. Although the function of eIF5A hypusine has not been defined, it is clearly an essential modification required for tissue and organism viability. At the termination step, it has been recently reported that the 2OG-dependent oxygenase Jumonji domain-containing 4 (Jmjd4) mediates hydroxylation of the lysyl residue 63 in the eukaryotic release factor-1 (eRF1) [27, 55] in a 2OG and O2-dependent manner, being this modification necessary for translation termination efficiency (see below).
Hydroxylation of tRNAs
Translational regulation at the level of transfer RNAs (tRNAs) occurs not only through regulation of the relative availability of the specific isotypes, as previously believed, but also through post-transcriptional modifications [56]. Whilst the first two bases of each mRNA codon form stable Watson–Crick base pairings, the nucleotide at the third position can bind either a purine or a pyrimidine of the anticodon, resulting in a non-canonical base pairing [57]. The non-standard base pairing at this position, known as the wobble position, allows for one tRNA to recognize different codons, lending support to the fact that 61 different codons occur, though only 45 different tRNA molecules are available. For example, the Leucine tRNA (tRNALEU GAG) binds the CUC codon in a canonical manner and the CUU codon in a non-standard fashion [57]. Interestingly, when the nucleotide at the wobble position of the tRNA is uridine, this nucleotide is usually modified [58], being this modification necessary to ensure translation accuracy [59, 60]. In eukaryotes, most wobble uridines carry the modification 5-methoxycarbonylmethyluridine (mcm5U), 5-carbamoylmethyluridine (ncm5U) or derivatives. The glycine tRNA (tRNAGLY UCC) contains a hydroxylated form of mcm5U, 5-methoxycarbonylhydroxymethyluridine [(S)-mchm5U], being the hydroxylation reaction catalyzed by the 2OG-dependent oxygenase ALKBH8 [24, 61]. (S)-mchm5U was initially observed in the tRNAGLY of the silkworm Bombyx mori, and more recently, reported to be present in mammalian tRNAGLY as well [62]. Although hydroxylation of mcm5U in B. mori seems to play an important role in tRNAGLY function [63], its precise effect on translation in mammals remains elusive.
Interestingly, not all tRNA modifications occur at the wobble position. In the tRNAPHE, the non-canonical nucleoside wybutosine (yW) is required at position 37, adjacent to the 3′ end of the anticodon. This particular nucleotide is apparently necessary for translation fidelity [56] and for ensuring the correct reading frame [64]. A wide range of hydroxylated yW derivatives are present in the tRNAPHE, including hydroxywybutosine, undermodified hydroxywybutosine and peroxybutosine; the hydroxylation reaction is catalyzed by the 2OG-dependent oxygenase TYW5 [65]. Although we now understand the mechanism by which TYW5 recognizes and hydroxylates yW, the functional relevance of this modification is unclear [25]. It has been proposed that yW hydroxylation plays a modulatory role in maintaining the reading frame during decoding; development of TWY5 null models is required to shed light on its effect on protein synthesis.
Hydroxylation of ribosomal proteins
A new sub-family of 2OG-dependent oxygenases has recently been described, and known as ribosomal oxygenases (ROXs) that mediate hydroxylation of several ribosomal proteins [28]. Interestingly, bioinformatic analyses revealed that this family of 2OG-dependent oxygenases is evolutionary conserved [66]. It has been reported that a bacterial 2OG-dependent oxygenase named ycfD interacts with the ribosomal protein RPL16/uL16, a central element in the architecture of the ribosome, resulting in hydroxylation of an arginine residue at position 81 [28]. Whereas growth rate of wild type bacteria and ycfD mutant strains do not differ in standard conditions, growth of ycfD mutants is reduced under starvation, which correlates with a remarkable decrease in bulk protein translation [28]. Two orthologs of ycfD have been identified in humans: MINA53 and NO66; both of them seem to play central roles in control of cell proliferation and growth. Down-regulation of either protein provokes cell growth impairment and conversely, the two proteins have been found over-expressed in several types of cancers [67, 68]. MINA53 interacts with and catalyzes hydroxylation of histidine 39 of RPL27a/eL27. Interestingly, over 90 % of endogenous RPL27a/eL27 was found hydroxylated in different cancer cell lines as well as in normal tissues [16, 28].
Likewise, human NO66 interacts and hydroxylates histidine 216 of the large subunit ribosomal protein L8/uL2 in an O2-dependent manner [28, 69]. In fact, more than 95 % of endogenous RPL8/uL2 was found hydroxylated in various cell lines. A recent study shows that this hydroxylation mediates stabilization of a protein conformation that enables binding to the helix 93 of 28S rRNA, thereby inducing structural rearrangements in regions localized close to the Peptidyl Transferase Centre in the mature ribosome [70]. Another ribosomal protein that undergoes hydroxylation is RPS23/uS12. RPS23/uS12 hydroxylation is catalyzed by the 2OG- and Fe2+-dependent oxygenase domain-containing protein 1 (OGFOD1) on a prolyl residue that is conserved across species including humans [71], Saccharomyces cerevisiae [72] and Drosophila melanogaster [73]. Evolutionary conservation of RPS23/uS12 hydroxylation suggests a pivotal role in protein synthesis of this post-translational modification. Although the role that RPS23/uS12 proline hydroxylation plays in this process is still unclear, it has been reported that its impairment provokes defects in STOP codon recognition in yeast (see below) [72, 74].
The scarce information available about the functional significance of hydroxylation of PSMCs, along with an increasing list of hydroxylation targets, turns these modifications into a promising topic of study in the field of protein synthesis in forthcoming years. Table 1 summarizes the most important PSMCs hitherto known to be hydroxylated, along with the enzyme that catalyzes the reaction.
Table 1.
List of PSMCs that are subjected to hydroxylation in diverse phyletic groups
| Species | Oxygenase | Hydroxylation substrate | References |
|---|---|---|---|
| Pseudomonas aeruginosa | PPHD | EF-Tu Pro54 | [26] |
| E. coli | YfcM | EF-P β-lysyl lysine (Lys34) | [36] |
| Human | DOHH | eIF5A Deoxyhypusine | [37] |
| Human | Jmjd4 | eRF1 Lys63 | [27] |
| E. coli | ycfD | RPL16/uL16 Arg81 | [28] |
| Human | NO66 | RPL8/uL2 His216 | [28] |
| Human | MINA53 | RPL27a/eL27 His39 | [28] |
| Human/yeast/Drosophila | OGFOD1/Tpa1/Sud1 | RPS23/uS12 Pro62/64 | [38–40] |
| Human | TYW5 | tRNAPHE Wybutosine | [41] |
| Human | ALKH8 | tRNAGL mcm5U | [42] |
| Human | Unknown | tRNAARG mcm5U | [42] |
Hitherto we have summarized how hydroxylation of different PSMCs regulates protein synthesis at several phases of the process. A particularly interesting effect elicited by hydroxylation of some of these components is the above mentioned alteration of translational fidelity, in particular STOP codon recognition. In the next sections, we will discuss STOP codon read-through as a non-canonical translational regulation mechanism that fine-tunes the rate and quality of protein synthesis, and how STOP codon recognition might be linked to stress responses through the hydroxylation of different PSMCs.
STOP codon read-through and hydroxylation of components of the protein synthesis machinery
Protein translation from a given mature mRNA molecule may involve mechanisms collectively known as non-canonical mRNA translation, which together contribute to maximize capability of the cell to encode information. Alternative translation mechanisms can lead to inclusion or exclusion of different domains in the resulting polypeptide, potentially affecting its function, subcellular localization or stability [75–77]. Examples of these mechanisms include the selection of alternative AUG initiation codons [78], RNA editing [79], ribosome frameshift [80] and STOP codon read-through [81], among others. In this section, we will focus on STOP codon read-through, discussing aspects of its regulation and possible physiological implications.
STOP codon suppression or read-through is a mechanism by which ribosomes continue synthesis of the polypeptide following entry of a STOP codon (UAG, UGA or UAA) to the A site of the ribosome. This phenomenon can occur when instead of a termination factor, a near-cognate aminoacyl tRNA enters the A site containing the STOP codon, forcing the ribosome to ignore the termination signal. Translation therefore continues and the C-terminal end of the polypeptide is extended until the next STOP codon in the mRNA (Fig. 2) [82, 83].
Fig. 2.
STOP codon read-through. High translation fidelity ensures STOP codon recognition as such by the release factor complex according to the canonical termination mechanism (left). However, non Watson–Crick base pairing between the UAG codon and a tRNA containing a near cognate anticodon could lead to the incorporation of a new amino acid (green) instead of translation termination, allowing for the synthesis of longer polypeptides containing a C-terminal extension (right). This phenomenon known as STOP codon read-through, although infrequent, occurs at basal levels under normal conditions, and several works point to the possibility that it could be a physiological mechanism of translation regulation. Ongoing research is focused on the elucidation of the impact that the incorporation of C-terminal additional domains by STOP codon read-through could have on cell physiology
Although STOP codon read-through occurs stochastically at a basal level due to imperfections in the termination mechanism with different probabilities depending on which of the three termination codons enters the ribosome [84], read-through mechanisms can be modulated by the presence of different elements in cis that alter proper recognition of the STOP codon by termination factors [85–87]. By mid 1990s, it was reported both in bacteria and mammals that the nucleotide laying immediately after the STOP codon can alter its read-through frequency [88]. More recently, experiments in S. cerevisiae suggested that at least the first six nucleotides downstream of the STOP codon are key determinants of read-through frequency. Particularly, the consensus sequence—CA(A/G)N(U/C/G)A—downstream of the UAG was found to favor read-through events [89]. Other sequences affecting proper termination at STOP codons are those that form specific secondary structures at the 3′ UTR of the mRNA. A clear example of this is the SECIS (cis-acting selenocysteine insertion sequence), which is required to incorporate the uncommon amino acid selenocysteine in replacement of a termination signal during synthesis of selenoproteins [90, 91].
STOP codon read-through in different organisms
Although a limited amount of examples of programmed STOP codon read-through with evident physiological roles has been reported, the phenomenon has been described in a large number of different organisms, both prokaryotes and eukaryotes, as well as in viruses [76, 82, 85, 92, 93]. This mechanism of translation regulation was originally described in the early 1970s in a few viruses, including the Qβ bacteriophage, Tobacco Mosaic Virus, Murine Leukemia Gamma retrovirus, and Murine Leukemia Gamma retrovirus-related virus [81, 94–97]. Through this mechanism, viruses generate distinct C-terminal portions of various proteins, thereby maximizing the genetic information encoded in their small genomes.
Another example occurs in the enterotoxigenic Escherichia coli CFA/II strain. Genes required in this strain for biosynthesis and assembly of CS3 pili are clustered in a single locus that produces a poly-cystronic mRNA that encodes 4 proteins in tandem, whilst a fifth one (a 104 kDa unnamed polypeptide) is encoded entirely within the same open reading frame [98]. Interestingly, for synthesis of the latter protein, read-through of an internal amber codon of the transcript is required [92]. Likewise, in S. cerevisiae, different isoforms of the high-affinity cAMP phosphodiesterase PDE2 display different C-terminal extensions generated by STOP codon read-through, which bring about extended polypeptides with a motif that targets PDE2 for 26S proteasomal degradation [76]. Also among fungi, in Ustilago maydis, several glycolytic enzymes reside both in the cytoplasm and in peroxisomes. It has been shown that targeting of 3-phosphoglycerate kinase (PGK) to peroxisomes results from a STOP codon read-through event that enables incorporation of a type 1 peroxisomal targeting signal (PTS1) at the C-terminal end of the protein [77].
In D. melanogaster, programmed STOP codon read-through occurs at an unusually high frequency in comparison to other species [99]. High-throughput analysis of different Drosophila species detected more than 300 events of C-terminal translation beyond the annotated STOP codon, producing polypeptides with C-terminal extensions ranging from 15 to few hundreds amino acids [86, 100]. This strongly suggests the occurrence of a widespread regulatory mechanism that involves STOP codon read-through in this species, whose exact nature is unclear. Although high-throughput techniques have revealed a large amount of genes putatively regulated by STOP codon read-through, only a few of them have been studied experimentally [101–107].
In mammals, Rabbit β-globin and rat myelin protein zero (L-MPZ) have been the first examples of proteins containing extended C-terminal ends produced by STOP codon read-through; the functional significance of these extensions is still unclear [83, 108]. Another good example of controlled STOP codon read-through in mammals occurs during translation of peroxisomal lactate dehydrogenase B (LDH-B). A small proportion of the intracellular pool of LDH-B exhibits a unique C-terminal sequence that includes a PTS1 that drives the enzyme to peroxisomes. Similar to the regulation of PGK subcellular localization described above in U. maydis, the inclusion of this C-terminal sequence is achieved by STOP codon read-through [77, 93]. It has been recently reported that the synthesis of the VEGF-Ax specific isoform of the Vascular Endothelial Growth Factor A (VEGF-A) also seems to be synthesized by programmed STOP codon read-through [109]. This particular isoform includes a 22 amino acid C-Terminal extension that depends on programmed STOP codon read-through. The VEGF A gene contains eight exons and multiple isoforms are generated by alternative splicing of the sixth and seventh exons, having all these isoforms proangiogenic activity. Interestingly, the C-terminal extension originated in the VEGF-Ax isoform by STOP codon read-through confers antiangiogenic activity [110].
Hydroxylation and regulation of STOP codon read-through
As mentioned above, the eukaryotic small ribosomal subunit protein-S23 (RPS23/uS12) is subjected to prolyl hydroxylation [71–73]. In E. coli the RPS23 homologue, RPS12/uS12, plays an important role in the decoding process of cognate tRNAs [59, 111]. Conserved residues of RPS12/uS12 (loop Pro45-Ser50) along with key nucleotides on the helix 44 (h44) of the 16S rRNA interact with the codon–anticodon nucleotides through several hydrogen bonds [59, 112]. Interestingly, mutations that affect the 16S rRNA h44 or the above mentioned loop of RPS12/uS12 can provoke either hypersensitivity or resistance to the error-promoting antibiotic streptomycin [113–117]. Therefore, the structure of the decoding center determined by the S16 rRNA, RPS12/uS12 and the codon–anticodon base pairing is a key determinant of translational fidelity.
Structural analysis suggests that the particular hydroxylable prolyl residue of RPS23/RPS12/uS12 lays at the apex of a loop that projects into the core of the ribosomal decoding centre [118]. Interestingly, in S. cerevisiae, lack of RPS23/uS12 hydroxylation can either increase or decrease STOP codon read-through, depending on the first nucleotide after the codon [72, 89, 119]. On the same line, it has been previously shown that different substitution variants of the hydroxylable proline or nearby amino acids provoke different translation accuracy defects [60, 72, 116]. Consistent with this, structural analysis carried out in bacteria indicated that the hydroxylated proline of RPS12/uS12 interacts directly with the phosphate backbone of the mRNA between the third base of each codon and the following nucleotide [120]. More recently it has been suggested that the hydroxyl group of the proline forms an hydrogen bond with the codon, which leads to stabilization of this interaction [121]. It has been speculated that a non-hydroxylated proline would have been instead involved in van der Waals contacts with the codon, resulting in decreased binding energy of the ribosome with the mRNA, thereby affecting translational fidelity [121].
We have also mentioned above that the 2OG-dependent oxygenase Jmjd4 mediates O2 and 2OG-dependent hydroxylation of lysine 63 of the translation termination factor eRF1 [27]. Different activities have been assigned to the three domains of eRF1: Domain 1 decodes STOP codons, Domain 2 facilitates peptidyl-tRNA hydrolysis, and Domain 3 recruits the eukaryotic release factor 3 (eRF3) [122]. Cryoelectron microscopy-based structural analysis of the ribosome in a complex with eRF1 and eRF3 shows that eRF1 domain 1 localizes deep into the decoding center of the 40S small ribosomal subunit, demonstrating a role of this termination factor in termination accuracy [55]. It has been proposed that the eRF1 domain 1 ensures STOP codon recognition through three highly conserved motifs, GTX, YXCXXXF and NIKS, which interact with STOP codons [123, 124], being NIKS the motif that contains the hydroxylable lysine. K63 in the NIKS motif makes contact with the first uridine of this codon [125]. As mentioned above, K63 is hydroxylated by Jmjd4, and the addition of this hydroxyl group allows for the formation of an additional hydrogen bond between the release factor and the STOP codon, thereby boosting STOP codon recognition [27, 125]. Knock-down of Jmjd4 in HEK293T cells provokes clear reduction of eRF1 K63 hydroxylation, affecting STOP codon read-through [27]. The physiological impact of read-through regulation mediated by eRF1 hydroxylation is so far unclear.
Stress responses and STOP codon read-through
Translational regulation under stress
Cells exposed to stress activate an integrated response that reshapes cell metabolism by diverting anabolic energy to the repair of stress-induced molecular damage. Thus, PSMCs constitute major targets of this stress response, allowing for dynamic reprogramming of protein synthesis [126]. Although all three phases of translation are subjected to stress regulation, many stress-activated signaling pathways target the initiation step. One of the most studied stress-induced inhibitory mechanisms is phosphorylation of the initiation factor eIF2α, which impairs assembly of the pre-initiation complex. Noteworthy, several stressors lead to eIF2α phosphorylation through activation of different kinases. Whilst nutrient deprivation elicits activation of the general control nonderepressible 2 (GCN2) kinase, viral infections lead to activation of protein kinase RNA activated (PKR). ROS generation in turn are known to activate the heme-regulated inhibitor kinase (HRI), whereas accumulation of misfolded proteins in the endoplasmic reticulum activates the protein kinase RNA-like endoplasmic reticulum (PERK) as part of the unfolded protein response (UPR) [127]. Translation inhibition through eIF2α phosphorylation provokes accumulation of messenger ribonucleoprotein particles (mRNPs) that contain non-translated mRNAs and aggregation-prone proteins (like TIA-1 or TIAR) that promote stress granules (SGs) formation. Thus, SGs drive incoming mRNAs to storage or decay, which has central importance for the cells to cope with the toxic effects of stress [128, 129].
The mammalian target of rapamycin complex 1 (mTORC1) regulates CAP-dependent protein translation through the phosphorylation of eIF4E-binding proteins (4E-BPs). Under conditions of nutrient abundance 4E-BPs are inhibited through mTORC1-dependent phosphorylation, enabling eIF4E to interact with the rest of the initiation complex. Under stress conditions, such as nutrient deprivation, hypoxia, metabolic stress, osmotic stress, DNA damage or heat shock, mTORC1 activity is inhibited, resulting in 4E-BP hypophosphorylation. Hypophosphorylated 4E-BP becomes active, binds eIF4E, and inhibits CAP-dependent protein synthesis [130]. Another important mechanism of mTORC1-dependent regulation of protein synthesis involves phosphorylation of the ribosomal kinase S6K1/S6K2, which in turn mediates phosphorylation of several effectors, that promote protein translation [130, 131]. Also in this case, stress-dependent inhibition of mTORC1 results in S6K1/S6K2 hypophosphorylation and in turn, in inhibition of protein synthesis. mTORC1 not only controls protein synthesis at the initiation step, but also modulates elongation. In conditions of nutrient abundance, mTORC1 phosphorylates and inactivates the eukaryotic elongation factor 2 kinase (eEF2K), which under stress conditions phosphorylates and inhibits translocation activity of the elongation factor eEF2 [132]. Thus, when nutrients are available, eEF2 is hypophosphorylated and active, while under starvation eEF2K phosphorylates and inhibits eEF2, thereby inhibiting protein synthesis at the elongation step [132].
A less explored but equally exciting mechanism of stress-dependent translational modulation is RNA hyper-edition. RNA editing is a phenomenon commonly associated with host defense responses against RNA-based viruses, consisting on the conversion of cytosine to uridine, or adenosine to inosine [133]. Adenosine deaminase, the enzyme responsible for adenosine to inosine conversion, is induced by nutrient deprivation and promotes SGs formation leading in turn to general translation inhibition [134].
Does STOP codon read-through contribute to triggering stress responses?
Experimental evidence from S. cerevisiae revealed that STOP codon read-through frequency is not only increased under stress conditions, but also, that genetic enhancement of read-through is enough to account for phenotypes resistant to stress. It has therefore been suggested that STOP codon read-through represents a general mechanism of translational regulation in response to stress [135–137]. Although more experimental data are required to evaluate to what extent this is indeed a widespread mechanism, available evidence suggests that normal STOP codon recognition is impaired under stress, and the resulting C-terminal extended proteins may in turn contribute to triggering an integrated stress response (Fig. 3). In this regard, Ribo-seq analysis of H2O2 treated S. cerevisiae cultures has revealed increased frequency of STOP codon read-through, as compared with cultures grown in control conditions [135]. This increase in STOP codon read-through frequency seems to be physiologically relevant for the response orchestrated by the cell to cope with oxidative stress. For example, STOP codon read-through, promoted by specific isoforms of the Sup35 subunit of the translation termination complex, plays a role in responses to heat shock, oxidative and chemical stress in yeast [138–140]. Noteworthy, these isoforms of Sup35 that promote STOP codon read-through include, at their N terminus, an intrinsically disordered prion-determining region named [PSI+], and [PSI+] prion formation is induced by oxidative stress. Strikingly, [PSI+] strains are clearly more resistant to heat shock and chemical stressors than strains unable to produce this prion [139], and consistent with this, cells unable to form prions are hypersensitive to H2O2 [138]. When termination fidelity was genetically enhanced in [PSI+] strains, these strains show increased susceptibility to stressors, mimicking cells unable to form prions [140]. Thus, at least in this case, STOP codon read-through is part of an integrated response to stress required to cope with unfavorable conditions.
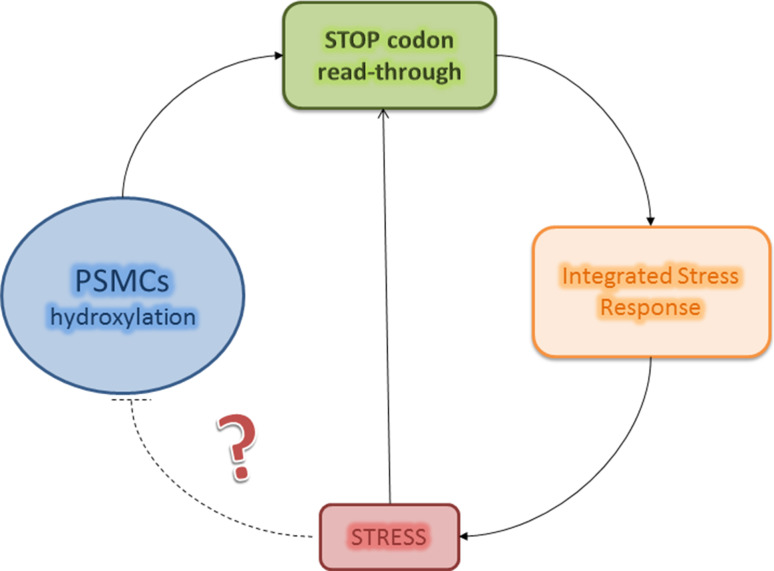
Fig. 3.
Possible regulation of the stress response by STOP codon read-through coupled to sensing of environmental conditions. STOP codon read-through is induced in stress conditions, and confers stress resistance to the cell. Stress conditions might be sensed in part by 2OG-dependent oxygenases involved in regulation of translation fidelity (dotted line). Although available evidence is compatible with this model, further research is required to elucidate the molecular details of this possible control circuit
Conclusions and perspectives
Hydroxylation of DNA, RNA, proteins or small molecules is known to regulate various biological processes including transcription, alternative splicing and hypoxia sensing, among others. Throughout this review, we have summarized hydroxylation of components of the protein synthesis machinery, discussing the role that this modification might play in mRNA translation. We have highlighted the fact that variations in the hydroxylation status of PSMCs might regulate protein synthesis, constituting an emerging mechanism of physiologically relevant translational regulation. Lack of hydroxylation of certain PSMCs leads to activation of STOP codon read-through, and in certain cases, activation of STOP codon read-through can enhance cell adaptation to stress. Thus, it would be interesting to test at the molecular level if inclusion of alternative C-terminal domains of proteins by STOP codon read-through leads in fact to synthesis of protein isoforms that contribute to cell responses to stress.
It is well documented that inhibition of hydroxylation of the transcription factor HIF-α is crucial to activate the transcriptional response to hypoxia [141–146]. In this mechanism, reduction of hydroxylase activity by O2 deprivation is the central event of stress sensing. An attractive possibility is that environmentally controlled 2OG-dependent oxygenase activity might induce stress resistance by modifying the hydroxylation status of PSMCs, in a similar fashion in which PSI+ formation, enhanced by stress, favors stress adaptive response through STOP codon read-through activation. Although the biological relevance of STOP codon read-through in cellular physiology is only beginning to emerge, it seems that this mechanism of translation regulation might be important for adaptation to stress. Roughly, a third of all reported genetic disorders derive from premature STOP codon [147, 148]. One can figure that deep understanding of mechanisms controlling STOP codon read-through could lead to development of novel therapies to deal with such pathologies.
Acknowledgments
We wish to thank all members of the Wappner’s lab for discussions and to Peter Ratcliffe for his continuous support. This work was funded by ANPCyT Grants PICT 2014 No. 0649 and PICT 2012 No. 0214. PW is a career investigator of CONICET.
References
- 1.Aoyagi Y, et al. Energy cost of whole-body protein synthesis measured in vivo in chicks. Comp Biochem Physiol A Comp Physiol. 1988;91(4):765–768. doi: 10.1016/0300-9629(88)90962-0. [DOI] [PubMed] [Google Scholar]
- 2.Bock FJ, Todorova TT, Chang P. RNA regulation by poly(ADP-Ribose) polymerases. Mol Cell. 2015;58(6):959–969. doi: 10.1016/j.molcel.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang SA, et al. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science. 2013;341(6144):1236566. doi: 10.1126/science.1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll AJ, et al. Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol Cell Proteomics. 2008;7(2):347–369. doi: 10.1074/mcp.M700052-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Kearse MG, Chen AS, Ware VC. Expression of ribosomal protein L22e family members in Drosophila melanogaster: rpL22-like is differentially expressed and alternatively spliced. Nucleic Acids Res. 2011;39(7):2701–2716. doi: 10.1093/nar/gkq1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopes AM, et al. The human RPS4 paralogue on Yq11.223 encodes a structurally conserved ribosomal protein and is preferentially expressed during spermatogenesis. BMC Mol Biol. 2010;11:33. doi: 10.1186/1471-2199-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 8.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodnina MV, Beringer M, Wintermeyer W. How ribosomes make peptide bonds. Trends Biochem Sci. 2007;32(1):20–26. doi: 10.1016/j.tibs.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Nurenberg E, Tampe R. Tying up loose ends: ribosome recycling in eukaryotes and archaea. Trends Biochem Sci. 2013;38(2):64–74. doi: 10.1016/j.tibs.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Young DJ, et al. Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3′UTRs in vivo. Cell. 2015;162(4):872–884. doi: 10.1016/j.cell.2015.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Passmore LA, et al. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26(1):41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 14.Kunze G, et al. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell. 2004;16(12):3496–3507. doi: 10.1105/tpc.104.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dzialo MC, et al. Translational roles of elongation factor 2 protein lysine methylation. J Biol Chem. 2014;289(44):30511–30524. doi: 10.1074/jbc.M114.605527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odintsova TI, et al. Characterization and analysis of posttranslational modifications of the human large cytoplasmic ribosomal subunit proteins by mass spectrometry and Edman sequencing. J Protein Chem. 2003;22(3):249–258. doi: 10.1023/A:1025068419698. [DOI] [PubMed] [Google Scholar]
- 17.Arnold RJ, Reilly JP. Analysis of methylation and acetylation in E. coli ribosomal proteins. Methods Mol Biol. 2002;194:205–210. doi: 10.1385/1-59259-181-7:205. [DOI] [PubMed] [Google Scholar]
- 18.Lee SW, et al. Direct mass spectrometric analysis of intact proteins of the yeast large ribosomal subunit using capillary LC/FTICR. Proc Natl Acad Sci USA. 2002;99(9):5942–5947. doi: 10.1073/pnas.082119899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cammarano P, et al. Characterization of unfolded and compact ribosomal subunits from plants and their relationship to those of lower and higher animals: evidence for physicochemical heterogeneity among eucaryotic ribosomes. Biochim Biophys Acta. 1972;281(4):571–596. doi: 10.1016/0005-2787(72)90158-X. [DOI] [PubMed] [Google Scholar]
- 20.Louie DF, et al. Mass spectrometric analysis of 40S ribosomal proteins from Rat-1 fibroblasts. J Biol Chem. 1996;271(45):28189–28198. doi: 10.1074/jbc.271.45.28189. [DOI] [PubMed] [Google Scholar]
- 21.Vladimirov SN, et al. Characterization of the human small-ribosomal-subunit proteins by N-terminal and internal sequencing, and mass spectrometry. Eur J Biochem. 1996;239(1):144–149. doi: 10.1111/j.1432-1033.1996.0144u.x. [DOI] [PubMed] [Google Scholar]
- 22.Ban N, et al. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 2014;24:165–169. doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruvinsky I, et al. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19(18):2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Y, et al. The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew Chem Int Ed Engl. 2010;49(47):8885–8888. doi: 10.1002/anie.201001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato M, et al. Crystal structure of a novel JmjC-domain-containing protein, TYW5, involved in tRNA modification. Nucleic Acids Res. 2011;39(4):1576–1585. doi: 10.1093/nar/gkq919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scotti JS, et al. Human oxygen sensing may have origins in prokaryotic elongation factor Tu prolyl-hydroxylation. Proc Natl Acad Sci USA. 2014;111(37):13331–13336. doi: 10.1073/pnas.1409916111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng T, et al. Optimal translational termination requires C4 lysyl hydroxylation of eRF1. Mol Cell. 2014;53(4):645–654. doi: 10.1016/j.molcel.2013.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge W, et al. Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. Nat Chem Biol. 2012;8(12):960–962. doi: 10.1038/nchembio.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Staalduinen LM, Novakowski SK, Jia Z. Structure and functional analysis of YcfD, a novel 2-oxoglutarate/Fe(2)(+)-dependent oxygenase involved in translational regulation in Escherichia coli . J Mol Biol. 2014;426(9):1898–1910. doi: 10.1016/j.jmb.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Loenarz C, Schofield CJ. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem Sci. 2011;36(1):7–18. doi: 10.1016/j.tibs.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Markolovic S, Wilkins SE, Schofield CJ. Protein hydroxylation catalyzed by 2-oxoglutarate-dependent oxygenases. J Biol Chem. 2015;290(34):20712–20722. doi: 10.1074/jbc.R115.662627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bugg TD. Oxygenases: mechanisms and structural motifs for O(2) activation. Curr Opin Chem Biol. 2001;5(5):550–555. doi: 10.1016/S1367-5931(00)00236-2. [DOI] [PubMed] [Google Scholar]
- 33.Mittal N, et al. Unique posttranslational modifications in eukaryotic translation factors and their roles in protozoan parasite viability and pathogenesis. Mol Biochem Parasitol. 2013;187(1):21–31. doi: 10.1016/j.molbiopara.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol. 2012;4(7):a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta N, et al. Whole proteome analysis of post-translational modifications: applications of mass-spectrometry for proteogenomic annotation. Genome Res. 2007;17(9):1362–1377. doi: 10.1101/gr.6427907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blaha G, Stanley RE, Steitz TA. Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science. 2009;325(5943):966–970. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pavlov MY, et al. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci USA. 2009;106(1):50–54. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ude S, et al. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339(6115):82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 39.Doerfel LK, et al. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339(6115):85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 40.Roy H, et al. The tRNA synthetase paralog PoxA modifies elongation factor-P with (R)-beta-lysine. Nat Chem Biol. 2011;7(10):667–669. doi: 10.1038/nchembio.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peil L, et al. Lys34 of translation elongation factor EF-P is hydroxylated by YfcM. Nat Chem Biol. 2012;8(8):695–697. doi: 10.1038/nchembio.1001. [DOI] [PubMed] [Google Scholar]
- 42.Navarre WW, et al. PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica . Mol Cell. 2010;39(2):209–221. doi: 10.1016/j.molcel.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bullwinkle TJ, et al. (R)-Beta-lysine-modified elongation factor P functions in translation elongation. J Biol Chem. 2013;288(6):4416–4423. doi: 10.1074/jbc.M112.438879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kemper WM, Berry KW, Merrick WC. Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Balpha and M2Bbeta. J Biol Chem. 1976;251(18):5551–5557. [PubMed] [Google Scholar]
- 45.Gregio AP, et al. eIF5A has a function in the elongation step of translation in yeast. Biochem Biophys Res Commun. 2009;380(4):785–790. doi: 10.1016/j.bbrc.2009.01.148. [DOI] [PubMed] [Google Scholar]
- 46.Kang HA, Hershey JW. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae . J Biol Chem. 1994;269(6):3934–3940. [PubMed] [Google Scholar]
- 47.Saini P, et al. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459(7243):118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dever TE, Gutierrez E, Shin BS. The hypusine-containing translation factor eIF5A. Crit Rev Biochem Mol Biol. 2014;49(5):413–425. doi: 10.3109/10409238.2014.939608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gutierrez E, et al. eIF5A promotes translation of polyproline motifs. Mol Cell. 2013;51(1):35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park MH, Joe YA, Kang KR. Deoxyhypusine synthase activity is essential for cell viability in the yeast Saccharomyces cerevisiae . J Biol Chem. 1998;273(3):1677–1683. doi: 10.1074/jbc.273.3.1677. [DOI] [PubMed] [Google Scholar]
- 51.Schrader R, et al. Temperature-sensitive eIF5A mutant accumulates transcripts targeted to the nonsense-mediated decay pathway. J Biol Chem. 2006;281(46):35336–35346. doi: 10.1074/jbc.M601460200. [DOI] [PubMed] [Google Scholar]
- 52.Park JH, et al. Molecular cloning, expression, and structural prediction of deoxyhypusine hydroxylase: a HEAT-repeat-containing metalloenzyme. Proc Natl Acad Sci USA. 2006;103(1):51–56. doi: 10.1073/pnas.0509348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patel PH, et al. The Drosophila deoxyhypusine hydroxylase homologue nero and its target eIF5A are required for cell growth and the regulation of autophagy. J Cell Biol. 2009;185(7):1181–1194. doi: 10.1083/jcb.200904161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sievert H, et al. A novel mouse model for inhibition of DOHH-mediated hypusine modification reveals a crucial function in embryonic development, proliferation and oncogenic transformation. Dis Model Mech. 2014;7(8):963–976. doi: 10.1242/dmm.014449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor D, et al. Cryo-EM structure of the mammalian eukaryotic release factor eRF1-eRF3-associated termination complex. Proc Natl Acad Sci USA. 2012;109(45):18413–18418. doi: 10.1073/pnas.1216730109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carlson BA, et al. Transfer RNA modification status influences retroviral ribosomal frameshifting. Virology. 1999;255(1):2–8. doi: 10.1006/viro.1998.9569. [DOI] [PubMed] [Google Scholar]
- 57.Varani G, McClain WH. The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1(1):18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gorgoni B, et al. Controlling translation elongation efficiency: tRNA regulation of ribosome flux on the mRNA. Biochem Soc Trans. 2014;42(1):160–165. doi: 10.1042/BST20130132. [DOI] [PubMed] [Google Scholar]
- 59.Ogle JM, et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292(5518):897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 60.Sharma D, et al. Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. J Mol Biol. 2007;374(4):1065–1076. doi: 10.1016/j.jmb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van den Born E, et al. ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nat Commun. 2011;2:172. doi: 10.1038/ncomms1173. [DOI] [PubMed] [Google Scholar]
- 62.Kawakami M, et al. Chemical structure of a new modified nucleoside located in the anticodon of Bombyx mori glycine tRNA2. J Biochem. 1988;104(1):108–111. doi: 10.1093/oxfordjournals.jbchem.a122403. [DOI] [PubMed] [Google Scholar]
- 63.Kawakami M, et al. Abnormal codon recognition of glycyl-tRNA from the posterior silk glands of Bombyx mori . J Biochem. 1980;88(4):1151–1157. doi: 10.1093/oxfordjournals.jbchem.a133069. [DOI] [PubMed] [Google Scholar]
- 64.Konevega AL, et al. Purine bases at position 37 of tRNA stabilize codon-anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA. 2004;10(1):90–101. doi: 10.1261/rna.5142404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Noma A, et al. Expanding role of the jumonji C domain as an RNA hydroxylase. J Biol Chem. 2010;285(45):34503–34507. doi: 10.1074/jbc.M110.156398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chowdhury R, et al. Ribosomal oxygenases are structurally conserved from prokaryotes to humans. Nature. 2014;510(7505):422–426. doi: 10.1038/nature13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suzuki C, et al. Identification of Myc-associated protein with JmjC domain as a novel therapeutic target oncogene for lung cancer. Mol Cancer Ther. 2007;6(2):542–551. doi: 10.1158/1535-7163.MCT-06-0659. [DOI] [PubMed] [Google Scholar]
- 68.Teye K, et al. Increased expression of a Myc target gene Mina53 in human colon cancer. Am J Pathol. 2004;164(1):205–216. doi: 10.1016/S0002-9440(10)63111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang C, et al. Structure of the JmjC domain-containing protein NO66 complexed with ribosomal protein Rpl8. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 9):1955–1964. doi: 10.1107/S1399004715012948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yanshina DD, et al. Hydroxylated histidine of human ribosomal protein uL2 is involved in maintaining the local structure of 28S rRNA in the ribosomal peptidyl transferase center. FEBS J. 2015;282(8):1554–1566. doi: 10.1111/febs.13241. [DOI] [PubMed] [Google Scholar]
- 71.Singleton RS, et al. OGFOD1 catalyzes prolyl hydroxylation of RPS23 and is involved in translation control and stress granule formation. Proc Natl Acad Sci USA. 2014;111(11):4031–4036. doi: 10.1073/pnas.1314482111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Loenarz C, et al. Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc Natl Acad Sci USA. 2014;111(11):4019–4024. doi: 10.1073/pnas.1311750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Katz MJ, et al. Sudestada1, a Drosophila ribosomal prolyl-hydroxylase required for mRNA translation, cell homeostasis, and organ growth. Proc Natl Acad Sci USA. 2014;111(11):4025–4030. doi: 10.1073/pnas.1314485111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keeling KM, et al. Tpa1p is part of an mRNP complex that influences translation termination, mRNA deadenylation, and mRNA turnover in Saccharomyces cerevisiae . Mol Cell Biol. 2006;26(14):5237–5248. doi: 10.1128/MCB.02448-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Torabi N, Kruglyak L. Genetic basis of hidden phenotypic variation revealed by increased translational readthrough in yeast. PLoS Genet. 2012;8(3):e1002546. doi: 10.1371/journal.pgen.1002546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Namy O, Duchateau-Nguyen G, Rousset JP. Translational readthrough of the PDE2 stop codon modulates cAMP levels in Saccharomyces cerevisiae . Mol Microbiol. 2002;43(3):641–652. doi: 10.1046/j.1365-2958.2002.02770.x. [DOI] [PubMed] [Google Scholar]
- 77.Freitag J, Ast J, Bolker M. Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature. 2012;485(7399):522–525. doi: 10.1038/nature11051. [DOI] [PubMed] [Google Scholar]
- 78.Touriol C, et al. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol Cell. 2003;95(3–4):169–178. doi: 10.1016/S0248-4900(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 79.Casey JL, Gerin JL. Hepatitis D virus RNA editing: specific modification of adenosine in the antigenomic RNA. J Virol. 1995;69(12):7593–7600. doi: 10.1128/jvi.69.12.7593-7600.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jacks T, Varmus HE. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985;230(4731):1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- 81.Weiner AM, Weber K. Natural read-through at the UGA termination signal of Q-beta coat protein cistron. Nat New Biol. 1971;234(50):206–209. doi: 10.1038/newbio234206a0. [DOI] [PubMed] [Google Scholar]
- 82.Geller AI, Rich A. A UGA termination suppression tRNATrp active in rabbit reticulocytes. Nature. 1980;283(5742):41–46. doi: 10.1038/283041a0. [DOI] [PubMed] [Google Scholar]
- 83.Yamaguchi Y, et al. L-MPZ, a novel isoform of myelin P0, is produced by stop codon readthrough. J Biol Chem. 2012;287(21):17765–17776. doi: 10.1074/jbc.M111.314468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Beier H, Grimm M. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 2001;29(23):4767–4782. doi: 10.1093/nar/29.23.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stiebler AC, et al. Ribosomal readthrough at a short UGA stop codon context triggers dual localization of metabolic enzymes in Fungi and animals. PLoS Genet. 2014;10(10):e1004685. doi: 10.1371/journal.pgen.1004685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jungreis I, et al. Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res. 2011;21(12):2096–2113. doi: 10.1101/gr.119974.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brown CM, et al. The translational termination signal database. Nucleic Acids Res. 1993;21(13):3119–3123. doi: 10.1093/nar/21.13.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tate WP, et al. Translational termination efficiency in both bacteria and mammals is regulated by the base following the stop codon. Biochem Cell Biol. 1995;73(11–12):1095–1103. doi: 10.1139/o95-118. [DOI] [PubMed] [Google Scholar]
- 89.Namy O, Hatin I, Rousset JP. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep. 2001;2(9):787–793. doi: 10.1093/embo-reports/kve176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Berry MJ, et al. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature. 1991;353(6341):273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 91.Kossinova O, et al. A novel insight into the mechanism of mammalian selenoprotein synthesis. RNA. 2013;19(8):1147–1158. doi: 10.1261/rna.036871.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jalajakumari MB, et al. Genes for biosynthesis and assembly of CS3 pili of CFA/II enterotoxigenic Escherichia coli: novel regulation of pilus production by bypassing an amber codon. Mol Microbiol. 1989;3(12):1685–1695. doi: 10.1111/j.1365-2958.1989.tb00154.x. [DOI] [PubMed] [Google Scholar]
- 93.Schueren F, et al. Peroxisomal lactate dehydrogenase is generated by translational readthrough in mammals. Elife. 2014;3:e03640. doi: 10.7554/eLife.03640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hofstetter H, Monstein HJ, Weissmann C. The readthrough protein A1 is essential for the formation of viable Q beta particles. Biochim Biophys Acta. 1974;374(2):238–251. doi: 10.1016/0005-2787(74)90366-9. [DOI] [PubMed] [Google Scholar]
- 95.Pelham HR. Leaky UAG termination codon in tobacco mosaic virus RNA. Nature. 1978;272(5652):469–471. doi: 10.1038/272469a0. [DOI] [PubMed] [Google Scholar]
- 96.Philipson L, et al. Translation of MuLV and MSV RNAs in nuclease-treated reticulocyte extracts: enhancement of the gag-pol polypeptide with yeast suppressor tRNA. Cell. 1978;13(1):189–199. doi: 10.1016/0092-8674(78)90149-6. [DOI] [PubMed] [Google Scholar]
- 97.Yoshinaka Y, et al. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc Natl Acad Sci USA. 1985;82(6):1618–1622. doi: 10.1073/pnas.82.6.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Boylan M, Coleman DC, Smyth CJ. Molecular cloning and characterization of the genetic determinant encoding CS3 fimbriae of enterotoxigenic Escherichia coli . Microb Pathog. 1987;2(3):195–209. doi: 10.1016/0882-4010(87)90021-0. [DOI] [PubMed] [Google Scholar]
- 99.Lin MF, et al. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res. 2007;17(12):1823–1836. doi: 10.1101/gr.6679507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dunn JG, et al. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster . Elife. 2013;2:e01179. doi: 10.7554/eLife.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xue F, Cooley L. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell. 1993;72(5):681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- 102.Steneberg P, et al. Translational readthrough in the hdc mRNA generates a novel branching inhibitor in the drosophila trachea. Genes Dev. 1998;12(7):956–967. doi: 10.1101/gad.12.7.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bergstrom DE, et al. Regulatory autonomy and molecular characterization of the Drosophila out at first gene. Genetics. 1995;139(3):1331–1346. doi: 10.1093/genetics/139.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Klagges BR, et al. Invertebrate synapsins: a single gene codes for several isoforms in Drosophila. J Neurosci. 1996;16(10):3154–3165. doi: 10.1523/JNEUROSCI.16-10-03154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Samuels ME, Schedl P, Cline TW. The complex set of late transcripts from the Drosophila sex determination gene sex-lethal encodes multiple related polypeptides. Mol Cell Biol. 1991;11(7):3584–3602. doi: 10.1128/MCB.11.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Samson ML, Lisbin MJ, White K. Two distinct temperature-sensitive alleles at the elav locus of Drosophila are suppressed nonsense mutations of the same tryptophan codon. Genetics. 1995;141(3):1101–1111. doi: 10.1093/genetics/141.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chao AT, et al. Mutations in eukaryotic release factors 1 and 3 act as general nonsense suppressors in Drosophila. Genetics. 2003;165(2):601–612. doi: 10.1093/genetics/165.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chittum HS, et al. Rabbit beta-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry. 1998;37(31):10866–10870. doi: 10.1021/bi981042r. [DOI] [PubMed] [Google Scholar]
- 109.Eswarappa SM, et al. Programmed translational readthrough generates antiangiogenic VEGF-Ax. Cell. 2014;157(7):1605–1618. doi: 10.1016/j.cell.2014.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Carmeliet P, et al. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380(6573):435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- 111.Demirci H, et al. The central role of protein S12 in organizing the structure of the decoding site of the ribosome. RNA. 2013;19(12):1791–1801. doi: 10.1261/rna.040030.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ogle JM, et al. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111(5):721–732. doi: 10.1016/S0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 113.Demirci H, et al. A structural basis for streptomycin-induced misreading of the genetic code. Nat Commun. 2013;4:1355. doi: 10.1038/ncomms2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.O’Connor M, et al. Genetic probes of ribosomal RNA function. Biochem Cell Biol. 1995;73(11–12):859–868. doi: 10.1139/o95-093. [DOI] [PubMed] [Google Scholar]
- 115.Funatsu G, Wittmann HG. Ribosomal proteins. 33. Location of amino-acid replacements in protein S12 isolated from Escherichia coli mutants resistant to streptomycin. J Mol Biol. 1972;68(3):547–550. doi: 10.1016/0022-2836(72)90108-8. [DOI] [PubMed] [Google Scholar]
- 116.Alksne LE, et al. An accuracy center in the ribosome conserved over 2 billion years. Proc Natl Acad Sci USA. 1993;90(20):9538–9541. doi: 10.1073/pnas.90.20.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gorini L, Kataja E. Streptomycin-induced oversuppression in E. coli . Proc Natl Acad Sci USA. 1964;51:995–1001. doi: 10.1073/pnas.51.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chandramouli P, et al. Structure of the mammalian 80S ribosome at 8.7 A resolution. Structure. 2008;16(4):535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bonetti B, et al. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae . J Mol Biol. 1995;251(3):334–345. doi: 10.1006/jmbi.1995.0438. [DOI] [PubMed] [Google Scholar]
- 120.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 121.Noeske J, et al. High-resolution structure of the Escherichia coli ribosome. Nat Struct Mol Biol. 2015;22(4):336–341. doi: 10.1038/nsmb.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nakamura Y, Ito K. tRNA mimicry in translation termination and beyond. Wiley Interdiscip Rev RNA. 2011;2(5):647–668. doi: 10.1002/wrna.81. [DOI] [PubMed] [Google Scholar]
- 123.Chavatte L, et al. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J. 2002;21(19):5302–5311. doi: 10.1093/emboj/cdf484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bulygin KN, et al. Three distinct peptides from the N domain of translation termination factor eRF1 surround stop codon in the ribosome. RNA. 2010;16(10):1902–1914. doi: 10.1261/rna.2066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Brown A, et al. Structural basis for stop codon recognition in eukaryotes. Nature. 2015;524(7566):493–496. doi: 10.1038/nature14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vannuvel K, et al. Functional and morphological impact of ER stress on mitochondria. J Cell Physiol. 2013;228(9):1802–1818. doi: 10.1002/jcp.24360. [DOI] [PubMed] [Google Scholar]
- 128.Pimentel J, Boccaccio GL. Translation and silencing in RNA granules: a tale of sand grains. Front Mol Neurosci. 2014;7:68. doi: 10.3389/fnmol.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Thomas MG, et al. RNA granules: the good, the bad and the ugly. Cell Signal. 2011;23(2):324–334. doi: 10.1016/j.cellsig.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014;24(7):400–406. doi: 10.1016/j.tcb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Meyuhas O. Physiological roles of ribosomal protein S6: one of its kind. Int Rev Cell Mol Biol. 2008;268:1–37. doi: 10.1016/S1937-6448(08)00801-0. [DOI] [PubMed] [Google Scholar]
- 132.Browne GJ, Proud CG. A novel mTOR-regulated phosphorylation site in elongation factor 2 kinase modulates the activity of the kinase and its binding to calmodulin. Mol Cell Biol. 2004;24(7):2986–2997. doi: 10.1128/MCB.24.7.2986-2997.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Keegan LP, Gallo A, O’Connell MA. The many roles of an RNA editor. Nat Rev Genet. 2001;2(11):869–878. doi: 10.1038/35098584. [DOI] [PubMed] [Google Scholar]
- 134.Yamasaki S, Anderson P. Reprogramming mRNA translation during stress. Curr Opin Cell Biol. 2008;20(2):222–226. doi: 10.1016/j.ceb.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gerashchenko MV, Lobanov AV, Gladyshev VN. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc Natl Acad Sci USA. 2012;109(43):17394–17399. doi: 10.1073/pnas.1120799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147(4):789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Oren YS, et al. The unfolded protein response affects readthrough of premature termination codons. EMBO Mol Med. 2014;6(5):685–701. doi: 10.1002/emmm.201303347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sideri TC, et al. Ribosome-associated peroxiredoxins suppress oxidative stress-induced de novo formation of the [PSI+] prion in yeast. Proc Natl Acad Sci USA. 2010;107(14):6394–6399. doi: 10.1073/pnas.1000347107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Eaglestone SS, Cox BS, Tuite MF. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J. 1999;18(7):1974–1981. doi: 10.1093/emboj/18.7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431(7005):184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- 141.Ivan M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292(5516):464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 142.Jaakkola P, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 143.Lando D, et al. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16(12):1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294(5545):1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 145.Ratcliffe PJ. Oxygen sensing and hypoxia signalling pathways in animals: the implications of physiology for cancer. J Physiol. 2013;591(Pt 8):2027–2042. doi: 10.1113/jphysiol.2013.251470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Epstein AC, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107(1):43–54. doi: 10.1016/S0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 147.Karijolich J, Yu YT. Therapeutic suppression of premature termination codons: mechanisms and clinical considerations (review) Int J Mol Med. 2014;34(2):355–362. doi: 10.3892/ijmm.2014.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Keeling KM, Bedwell DM. Suppression of nonsense mutations as a therapeutic approach to treat genetic diseases. Wiley Interdiscip Rev RNA. 2011;2(6):837–852. doi: 10.1002/wrna.95. [DOI] [PMC free article] [PubMed] [Google Scholar]