Abstract
Cold stress responses in plants are highly sophisticated events that alter the biochemical composition of cells for protection from damage caused by low temperatures. In addition, cold stress has a profound impact on plant morphologies, causing growth repression and reduced yields. Complex signalling cascades are utilised to induce changes in cold-responsive gene expression that enable plants to withstand chilling or even freezing temperatures. These cascades are governed by the activity of plant hormones, and recent research has provided a better understanding of how cold stress responses are integrated with developmental pathways that modulate growth and initiate other events that increase cold tolerance. Information on the hormonal control of cold stress signalling is summarised to highlight the significant progress that has been made and indicate gaps that still exist in our understanding.
Keywords: Abiotic stress, Freezing tolerance, Hormones, Plant
Introduction
Plants have developed a remarkable ability to adapt to harsh environmental conditions, and thrive in habitats characterised by abiotic stresses such as temperature extremes [1]. Species-specific differences in temperature tolerance have evolved in plants that occupy different geographic zones. Annual crops from temperate climates, such as Triticum aestivum (wheat), Avena sativa (oats), Hordeum vulgare (barley) or Pisum sativum (pea), display a certain degree of basal (intrinsic) freezing tolerance, which they can further increase by utilising complex signalling events. In contrast, most species from subtropical or tropical climates, such as Zea mays (maize), Oryza sativa (rice) or Solanum lycopersicum (tomato), suffer damage at chilling temperature (often well above 0 °C) [1]. This represents a significant management challenge for agriculture and horticulture because these crops are regularly cultivated in geographical regions where temperature preferences of the plant are not fully met during the growing season. Moreover, extreme short-term weather events, such as late frost during spring, impact yields, particularly of fruit crops and spring cereals [2]. Consequently, chilling and freezing stress constitute some of the most severe abiotic factors that reduce crop productivity [3, 4]. Because reliable and high-quality crop yields are critical for food security, an understanding of the molecular modes that underpin cold stress resistance is essential to further optimise horticultural and agricultural crop breeding and production.
Cellular changes in response to cold
Temperate plants utilise a repertoire of mechanisms to avoid damage by freezing. These include the timing of developmental programs, such as germination and flowering, on the basis of the environmental information perceived [5]. In addition, they have the ability to further increase their basal freezing tolerance following prior exposure to chilling temperatures. This process, termed cold adaptation, requires marked transcriptional reprogramming to alter the expression of diverse classes of genes with a wide range of biological functions [6].
A key response to cold is growth repression (Fig. 1), which is thought to be utilised to re-allocate resources from growth to processes that increase cold stress resistance [7], although this hypothesis remains to be validated. In addition to pronounced effects on morphology drastic biochemical and physiological changes occur, which have been summarised in a number of excellent recent reviews [1, 4, 5]. In brief, these changes include an increase in cellular calcium contents, an accumulation of reactive oxygen species (ROS) and the formation of cryoprotective proteins and cryoprotective metabolites such as soluble sugars and amino acids. In addition, changes in the structure and composition of membranes and numerous other events occur [1], which are all dedicated to reducing the damage caused by cold temperatures or cellular freezing [1]. Interconnected molecular circuits that are governed by the activity of plant hormones control these events, and in the current review, we will summarise our understanding of the hormonal control of cold stress resistance in plants, with a particular focus on latest developments in the field.
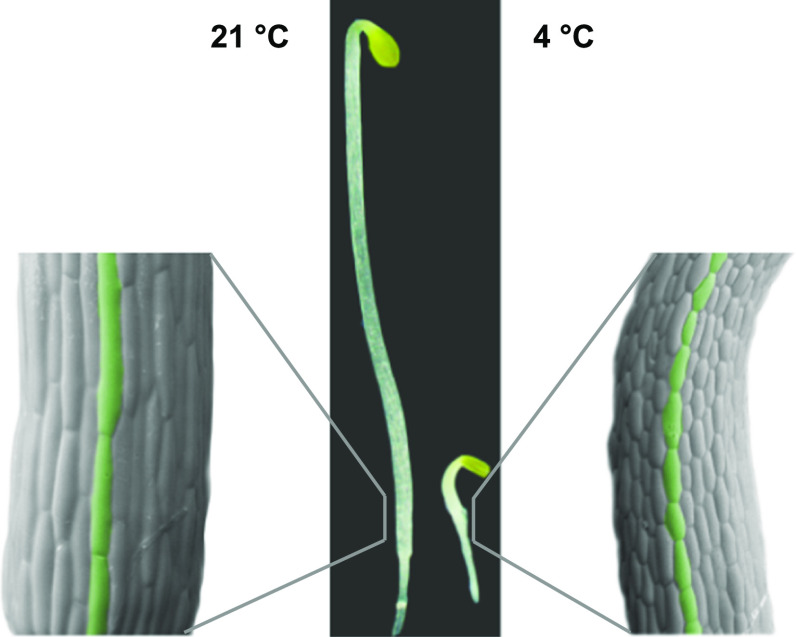
Fig. 1.
Effect of cold treatment on cell elongation in arabidopsis hypocotyls. Seeds were germinated in the light and then moved to the dark at either 21 or 4 °C, where they were kept for 3 days. Electromicrographs of the indicated sections are shown. A representative cell row is marked in green
Cold-regulated gene expression
Cold-responsive transcriptomes differ between plant species [8–32]. In Arabidopsis thaliana (arabidopsis), a temperate plant that serves as a model species in plant molecular biology, approximately 10 % of all genes are cold regulated [33, 34]. A significant share of these cold-regulated (COR) genes, including COLD-REGULATED 15A (COR15A), COLD-REGULATED 15B (COR15B), COLD-REGULATED 78 (COR78), and COLD-INDUCED 1 and 2 (KIN1 and KIN2), contain the C-repeat/dehydration-responsive element (CRT/DRE) in their promoters. CRT/DRE is a cis regulatory motif that is bound by the drought-responsive element-binding proteins (DREB), also known as C-REPEAT BINDING FACTOR (CBFs) [35], in response to cold and other types of abiotic stresses that cause cellular desiccation, for example, drought or high salt exposure. CBFs are APETALA 2 domain transcription factors (TFs) that increase resistance against freezing, drought and high salt exposure when overexpressed in different plant species such as arabidopsis, Brassica napus (canola), tomato and Populus spp. (poplar) [35].
The modes of CBF-controlled gene regulation have received a considerable amount of attention in arabidopsis, and the results of these studies are instructive. In arabidopsis, three different CBFs, namely CBF1, 2 and 3, exist, which are physically linked in a tandem array on the chromosome and act to control overlapping gene sets [34]. Interestingly, CBFs regulate their own activity in feedback regulatory loops: CBF2 represses CBF1 and CBF3 expression to adjust their mRNA abundance [36]. In addition, CBF expression is controlled by upstream TFs such as ZAT12, which acts as a repressor [37], and CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR (CAMTA), INDUCER OF CBF EXPRESSION 1 AND 2 (ICE1 and ICE2), which act as activators of CBF transcription [38–40]. Among those, the best characterised is ICE1, a MYC-like basic helix-loop-helix (bHLH) protein, which, in response to cold, is modified by SUMOylation involving the SUMO E3 ligase SIZ1. This promotes ICE1 binding to an E-box motif in the CBF3 promoter and increases CBF3 expression [41]. ICE1 protein abundance is controlled by phosphorylation [42, 43] and ubiquitination catalysed by the E3 ubiquitin ligase HOS1, which acts to maintain ICE1 equilibrium during cold acclimation [44].
Although it is established that the CBFs play an important role in cold acclimation they only control a share of the cold-responsive transcriptome [6, 17, 37]. Therefore, additional TFs take part, and one example is ZAT10, a negative regulator of COR genes whose activity is repressed by the bifunctional enolase LOW EXPRESSION OF OSMOTICALLY RESPONSIVE GENES 2/C-MYC BINDING PROTEIN (LOS2/AtMBP-1), in response to cold [45]. LOS2/MBP-1 is an interesting locus because in addition to the TF AtMBP-1, it encodes the enolase 2 (ENO2) that catalyses a key step in glycolysis. LOS2/AtMBP-1 utilises an elegant feedback regulatory mode to control its own activity: AtMBP-1 is alternatively translated from a second start codon [46] to represses LOS2/AtMBP-1 transcription and thereby regulates ENO2 abundance [47]. How LOS2/AtMBP-1 activity is controlled by cold is currently unknown, but clearly it would represent an interesting target for plants to utilise, when in response to low temperature, both glycolysis, as a central metabolic pathway and COR gene expression must be modulated. It will be important to dissect the roles of AtMBP-1 from those conferred by ENO2 to further define the role of the LOS2/AtMBP-1 locus in cold resistance.
Plant hormones control cold responses
Whereas the nuclear events that allow for TFs to directly control COR gene expression are fairly well defined, the upstream regulatory modes that govern these activities have remained largely elusive. The mechanisms allowing plants to perceive temperature remain unknown, although there is evidence that changes in membrane fluidity are contributory [1]. In addition, the modes transducing the cold stimulus to the nucleus are largely unspecified, although protein phosphorylation controlled by calcium-dependent protein kinases (CPKs) that are induced by increases in cellular calcium contents, and mitogen-activated protein kinases (MAPKs) have been implicated [48, 49]. That said, in recent years, accumulating data support a role of plant hormones, which additionally govern other signal events in cold stress responses and suggests that hormones function as governors of the pathways.
Hormones are small molecular weight compounds that signal information from a site of synthesis to a site of action. Biosynthesis, catabolism and transport adjust hormone homeostasis; the receiving tissues determine sensitivity by the presence and responsiveness of dedicated receptors that initiate signal transduction events to alter cellular processes [50]. Different classes of hormones exist that have specific as well as overlapping functions. Moreover, hormones display synergistic or antagonistic effects on the biosynthesis and signalling outputs of other hormones, creating a complex network of hormonal interactions [50]. This hormonal signalling network is utilised to integrate external information into endogenous developmental programs and/or activate stress response pathways leading to resistance. It is therefore perhaps not surprising that plants utilise hormones to signal cold stress responses; however, our knowledge of the responsible molecular modes remains patchy. This may be attributed to the fact that hormone action in cold stress responses is influenced by cross-talk with signalling cascades conferring responses to other environmental stimuli such as light [51, 52] and is additionally impacted by endogenous developmental programs, in particular those that result in developmental phase transitions [53]. Moreover, hormone modes of activity vary between plant species complicating research in this field.
Gibberellin (GA) activity is altered in response to cold stress
Growth-promoting hormones, in particular GAs, play central roles in conferring plants the ability to adapt growth to fluctuating external conditions, such as changes in the light environment [51]. GAs promote cell elongation and division and are synthesised from trans-geranylgeranyl diphosphate in a biosynthetic pathway in which small families of oxygenases, the KAO-oxidases, GA 20-oxidases and GA 3-oxidases produce bioactive GAs. GA cellular homeostasis is maintained by catabolic inactivation utilising GA 2-oxidases as well as by feedback suppression of GA biosynthesis via the GA signalling pathway. In GA signalling, DELLA activity is central [54]. DELLAs are GRAS proteins of which five members exist in arabidopsis, GIBBERELLIC ACID INSENSITIVE (GAI), REPRESSOR OF GAI (RGA) and RGA-LIKE 1, 2 and 3 (RGL1, RGL2 and RGL3). They act as repressors of TFs such as PHYTOCHROME-INTERACTING FACTOR 3/4 (PIF3/PIF4) [55, 56], and brassinosteroid-insensitive 1 (BRI1)-EMS-SUPPRESSOR 1/BRASSINAZOLE-RESISTANT 1 (BES1/BZR1) [57], which control GA-responsive gene expression. DELLA repressive action is released upon perception of the hormone by the GA receptor GA-insensitive dwarf 1 (GID1), which initiates DELLA ubiquitination and degradation via the 26S proteasome [54].
Both GA metabolism [58] and signalling [59] are targeted by cold stress, and there is evidence that the CBFs take part. A first indication was that in arabidopsis, tobacco and tomato overexpression of CBFs resulted in a reduction of bioactive GA, which was correlated with suppressed growth and late flowering and could be restored by exogenous GA application [58, 60–62]. When studying the underlying molecular modes, it was found that cold-induced GA2ox expression, which resulted in increased hydroxylation and inactivation of bioactive GA (Fig. 2). In addition, CBF1, when overexpressed, affected transcript levels of the DELLA RGL3 and enhanced DELLA accumulation, likely through posttranslational control [58].
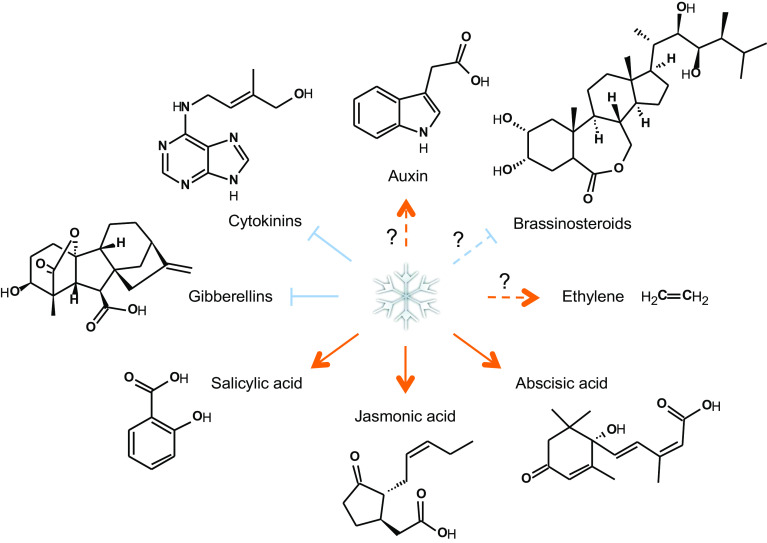
Fig. 2.
Present understanding of the impact of cold stress on hormone levels in plants. Orange arrows and blue inhibition lines represent activation and suppression processes, respectively. Dotted lines are used in case no or controversial data on hormone levels were published. Please note that species-specific differences can occur; these are detailed in the text
Other studies provided evidence that GA signalling components can affect the low temperature stress responses of plants, because they showed that GA-insensitive or GA-deficient mutants have altered chilling and freezing tolerance both in arabidopsis [58, 59] and rice [64]. The arabidopsis gai mutant, which is a constitutive signalling mutant, was more freezing tolerant, and the DELLA knockout line gai-t6 rga-24 was hypersensitive to freezing, both before and after cold acclimation [58]. Similarly, the ga1 mutant, which is impaired in GA production, had a greater degree of freezing tolerance than the wild-type. Interestingly, ga1 additionally showed elevated levels of CBF1, CBF2 and COR15A, and there is evidence that this is, at least partially, mediated by the action of the GA-regulated GATA TFs GNC and GNL [59, 64]. Overexpression of GNC or GNL enhanced CBF2, COR15A and COR15B expression and increased survival rates of transgenic seedlings before and after cold acclimation [59]. Because GNL and GNC expression are increased in GA signalling-deficient mutants, GNC and GNL are proposed to act in promoting freezing tolerance in the absence of GA [59] (Fig. 3).
Fig. 3.
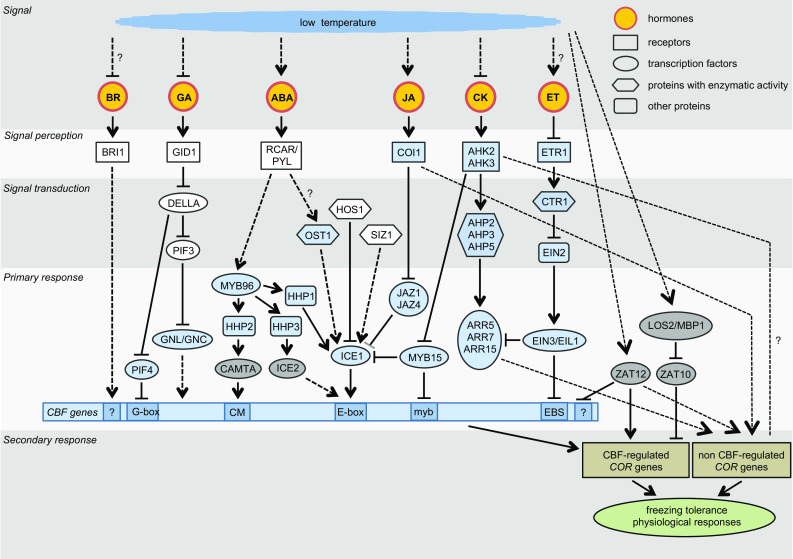
Model summarising the current understanding of signalling events that confer hormone activity in cold adaptation. In blue are transcription factors (TFs) and components of hormone signalling pathways, which are regulated by both cold and hormone. In grey are TFs regulated by cold but not by hormone. In white are components of hormone signalling pathways, which are not known to be cold-regulated. Arrows and inhibition lines represent activation and suppression processes, respectively. Solid lines indicate a direct interaction; dotted lines represent indirect interactions
A second level of CBF control by GAs may be mediated by PIF4. PIF4, which is controlled by phytochromes and DELLAs [55, 56], was found to directly bind to the promoters of all three CBFs and repress their expression in long-day growth conditions in arabidopsis [65]. The aforementioned study complemented earlier work that had elucidated a function of phytochrome signalling in CBF expression [66]. Because PIF4 additionally controls auxin biosynthesis at high temperatures [67] and is controlled in its activity by brassinosteroids (BRs) [68], a second class of growth-promoting hormones, PIF4, and redundant factors may act as central nodes for the integration of multiple environmental stimuli into growth processes (Fig. 3).
Although the role of CBFs in GA-mediated growth suppression is established, our understanding of the molecular control of these events remains ill defined. The mechanism used by CBFs to control GA2ox and DELLA transcription or posttranslational processing remains unknown. In addition, there is evidence that cold may inhibit growth by additional means, because ZAT10, ZAT12 and further TFs that are induced by cold in primary responses and control non-CBF regulons, also suppress growth when overexpressed. This growth repression was not correlated with reduced GA2ox or RGL3 expression [34], and it will therefore be interesting to determine which other growth regulatory pathways participate.
BRs promote cold resistance
A class of hormones that closely interacts with GAs are the BRs. BRs are polyhydroxylated steroids with a well-established role in growth promotion [69]. In arabidopsis this function is conferred by an intimate interplay with GAs where both hormones impact on the other’s biosynthesis [70, 71] and contribute signalling components that interact to control multiple overlapping developmental programs [68, 72, 73]. In cold responses, there is as yet no evidence for a synergistic activity of BRs and GAs. Quite on the contrary, as opposed to GAs, BRs are thought to positively control cold stress responses, because it has been shown that BR application increases cold tolerance of many plant species, including chilling-sensitive crops such as maize and Cucumis sativus (cucumber) [74–76].
BRs are synthesised from campesterol in a biosynthetic pathway that relies mainly on the activities of cytochrome P450s including DWARF 4 (DWF4) and CPD (CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM). DFW4, when overexpressed, enhanced cold stress resistance of arabidopsis and additionally increased COR15A mRNA levels [77]. Moreover, BR application enhanced expression of CBF1 and the CBF target COR47 in arabidopsis following low temperature treatment, which indicated that BRs promote CBF expression and cold tolerance [78] (Fig. 3).
In agreement, a BR receptor BRI1 mutant, bri1-116, showed enhanced ion leakage following freezing stress, thereby providing evidence that BR signalling promotes acclimation to cold [79]. However, contradictory results were obtained in a different study. In this earlier work, plants overexpressing BRI1 showed elevated basal CBF1-3 expression levels but had reduced cold tolerance, whereas BRI1 loss-of-function in bri1-9 showed increased resistance [80]. Therefore, although a role of BRs in increasing cold responses appears likely, further verification of these results is required.
Auxins in cold stress? As yet, not much is known
Auxin has been heavily studied for its role in growth adaptation, in particular, in response to light and gravitropic stimuli. However, somewhat surprisingly, little is known regarding roles of auxins in cold stress responses of plants, although it has been shown that a significant number of auxin-regulated genes are additionally affected by cold in arabidopsis and rice [81, 82] and that application of auxin analogues on canola (B. napus) stimulated accumulation of freeze-protective metabolites and soluble sugars during cold hardening [83].
Biologically active auxins are indole acetic acid (IAA) and indole butyric acid, which are synthesised from tryptophan. Cold stress appears to affect IAA levels with differences depending on plant species, developmental context and physiological settings. In wheat, following 21 days of exposure to 4 °C, a significant increase in IAA concentrations was found in crown tissues but not in leaves [84]. In addition, whereas IAA levels were not affected by cold stress in spring wheat, significant increases occurred in winter wheat after 12 days of cold exposure [85]. Contradictory results were published for rice. Whereas one study reported decreased IAA contents following low temperature treatment [86], another study found that IAA contents were elevated after 1 day of cold treatment and remained elevated for 5 days [87]. This correlated with an increased expression of auxin biosynthetic genes of the YUCCA family and a reduction of the OsGH3 family, which catalyses auxin inactivation in rice [87]. Therefore, in rice, auxin levels may be increased by activation of auxin biosynthesis and suppression of auxin inactivation (Fig. 2).
Studies of auxin may be complicated by the sophisticated transport events, catalysed by auxin influx and efflux carriers that determine spatial gradients of auxin concentrations and are required for directional growth and many other processes [88]. In this respect, it is known that low temperature affects gravitropic growth of plants, because in stems and roots of arabidopsis and rice, gravitropic responses were inhibited by cold [89–91]. For arabidopsis, it was shown that short-term cold treatment slowed down gravitropic bending and elongation of roots, whereas a 24-h long-term treatment blocked gravitropic responses of roots completely [91]. This was correlated with changed auxin distribution patterns in arabidopsis roots and repressed basipetal auxin transport caused by impaired intracellular cycling of the auxin efflux carriers PIN2 and PIN3 [91]. Interestingly, application of the membrane rigidifier DMSO, which was previously shown to reduce membrane fluidity, suppressed elongation growth of arabidopsis roots at 23 °C but did not affect root gravitropism or intracellular PIN trafficking [91], indicating that root gravitropic responses in low temperatures are not caused by membrane rigidification.
Cytokinins control both CBF-dependent and CBF-independent regulons
A class of hormones that strongly cross-talks with auxins in plant developmental processes is the cytokinins (CKs). CKs are adenine derivatives with isoprenoid or aromatic side chains that, in addition to many other aspects, control directional growth processes such as gravitropism. The most abundant CKs in higher plants are isopentenyladenine and zeatin [92], and external application of these hormones is known to enhance freezing tolerance of arabidopsis [93, 94]. Because CBF1 expression is not increased by CK treatment and is neither elevated in CK-overaccumulating plants nor decreased in the CK-deficient mutant 35S:AtCKX2-2, which overexpresses CK oxidase, there is evidence that CKs increase freezing tolerance in a CBF1-independent manner [93]. Somewhat puzzling is the finding that although increased CK contents enhance freezing tolerance, CK levels decrease in response to chilling temperature in different plant species including wheat, rice and arabidopsis [84, 86, 95]. In rice, this reduction was correlated with a significant down-regulation of CK biosynthetic gene expression [86], providing evidence for a model in which a decrease in CK levels would be required for cold responses (Fig. 2).
Also for CK signalling results were published that are contradictory. In arabidopsis, CKs initiate signalling utilising three histidine protein kinases, namely arabidopsis histidine kinase (AHK)2, AHK3 and AHK4, which act as CK receptors. Upon CK binding, the receptors autophosphorylate and transfer the CK signal via arabidopsis histidine phosphotransfer proteins (AHPs) to nuclear localised arabidopsis response regulators (ARRs), TFs that regulate expression of CK target genes. In non-acclimated and acclimated ahk knockout plants, freezing tolerance was promoted, although transcript levels of CBFs in response to cold were found to be unchanged [93]. However, a later study reported that microarray analysis of a cold-treated ahk2 ahk3 double-mutant line showed that these mutations promoted expression of MYB15 and repressed expression of its target CBF3, which additionally correlated with suppressed expression of a set of CBF-controlled genes [96]. Moreover, non-CBF-regulated sets of cold-responsive genes were differentially regulated in ahk2 ahk3. Therefore, although AHK2 and AHK3 appear to be important for the modulation of both CBF-dependent and CBF-independent cold-responsive pathways, the question of whether AHKs have positive or negative roles in the freezing tolerance of plants remains to be conclusively answered.
Other components of CK signalling that have been implicated in cold stress responses are the type A ARRs. Transcript levels of ARR5, ARR7 and ARR15 were induced in response to cold in a AHK-dependent manner [93, 94] and arr5, arr6 and arr7 single knockout lines showed slightly increased tolerance to freezing, both constitutively and following cold acclimation [93, 94]. Moreover, overexpression of ARR7 slightly decreased freezing tolerance after cold acclimation. Because the induction of CBF-regulated genes was not impaired in this line, ARR7 was proposed to act as a negative regulator of cold signalling by CBF-independent means [93]. Again, seemingly in contradiction, a different study found that seedlings overexpressing ARR7 as well as ARR5 or ARR15 were more tolerant to freezing stress before and after cold acclimation; however, CBF and CBF-regulated gene expression were unchanged in these lines, lending support to the notion that ARRs do not impact on CBFs in cold responses [94] (Fig. 3).
Abscisic acid (ABA), a central regulator of cold stress signalling with emerging roles in the CBF-dependent pathway
Abscisic acid is an isoprenoid hormone that plays a central role in seed dormancy, abscission and abiotic stress signalling [97], and its function in cold stress responses is well established. An increase in ABA levels correlates with increased ABA biosynthesis in arabidopsis and rice [86, 98, 99] and occurs in response to cold in many plant species (Fig. 2). Exogenous application of ABA promotes freezing tolerance, and ABA mutants exhibited altered cold resistance. However, because it was previously thought that ABA does not act on CBF expression, the postulation was that ABA-controlled cold responses are regulated by CBF-independent means [97, 100].
Recently, evidence has accumulated that suggests otherwise. In this context, two publications are of particular importance. In the first work it was shown that MYB96, an ABA-induced TF, which is a central regulator of ABA-responsive gene expression, is cold induced and controls CBF induction in cold adaptation because it was compromised in a myb96 knockout line and was enhanced in a MYB96 overexpressor [101]. Furthermore, was provided evidence that MYB96 interacts with the HEPTAHELICAL PROTEIN (HHP) proteins HHP1, HHP2 and HHP3, which in turn interact with ICE1, CAMTA and ICE2, respectively [101, 102]. By these means, the authors propose that the MYB96–HHP complexes control expression of all three CBFs, and it will be of interest to observe whether this can further be verified by genetic approaches in the future.
The second work found that Open Stomata 1 (OST1), a Ser/Thr protein kinase activated by ABA [103], additionally regulates CBF expression. OST1 activity was induced by cold, ost1 knockout lines were hypersensitive to freezing and OST1 overexpressing plants were more resistant. Importantly, the molecular modes of the OST1 function were addressed, and it was revealed that OST1 interacts with and phosphorylates ICE1 to stabilise the protein and promote its transcriptional activity under cold stress. Evidence was provided that this is enabled by OST1 phosphorylation of ICE1 S278, because mimicking phosphorylation at this site as well as the presence of OST1 inhibited ICE1-HOS1 interaction and stabilised the protein [43]. Intriguingly the activation of OST1 by cold appears to be independent of ABA biosynthesis [43] and it will therefore be important to identify upstream regulatory components. In summary, in addition to its CBF-independent modes, ABA may impact on COR gene expression by controlling CBF transcription (Fig. 3) and appears to play a more central role in the cold stress tolerance of plants than previously assumed.
Ethylene: a gaseous hormone with versatile roles in cold responses
Ethylene is a gaseous hormone that is synthesised from the amino acid methionine utilising a biosynthetic pathway, in which the rate-limiting step, the conversion of S-adenosyl-l-methionine to 1-aminocyclopropane-1-carboxylic acid (ACC), is catalysed by isoforms of ACC synthase (ACS). Whereas the role of ethylene in abiotic stress resistance and additionally in cold responses is clear [104], the question of whether it has positive or negative regulatory roles remains. Some studies reported increases in ethylene contents in response to cold in different plant species including tomato [105], Secale cereale (rye) [106], Phaseolus spp. (bean) [107], wheat [84] and Medicago sativa (alfalfa) [108]. In arabidopsis these increases were correlated with increased expression of biosynthetic enzymes [109] (Fig. 2). However, other studies presented evidence for a reduction of ethylene contents in response to cold. For example, in Medicago truncatula, ethylene levels decreased rapidly by 94 % and the production of the ethylene precursor ACC was reduced by 45 % following cold treatment [110]. Although changes in transcript levels of the biosynthetic enzymes ACC oxidase (ACO) and ACS were not observed following low temperature treatment, the activity of ACO, an enzyme catalysing the last step of ethylene biosynthesis by converting ACC to ethylene, was reduced [110].
A reduction of ethylene in response to cold would fit well to the proposed role as a negative regulator of freezing tolerance in plants. Freezing tolerance was inhibited following treatment with ethylene or ACC in M. truncatula [110] and arabidopsis [94] and was promoted by treatment with ethylene biosynthesis inhibitors, including 2-aminoethoxyvinyl glycine (AVG), in both plant species [94, 110]. Moreover, M. truncatula plants were hypersensitive to freezing when treated with the ethylene releaser ethephon, which was correlated with a compromised induction of expression of MtCBF1 and MtCBF3 as well as MtCAS15 that belongs to the COLD ACCLIMATION-SPECIFIC (CAS) gene family, counterparts of COR genes in M. truncatula. Under non-acclimated conditions, treatment with AVG caused a significant induction of MtCBF genes, thereby complementing these results [110].
Ethylene binds to the ethylene receptor ETHYLENE-RESPONSIVE 1 (ETR1) to activate the signalling cascade. In response, CONSTITUTIVE TRIPLE RESPONSE 1 (CTR1), a negative regulator of the cascade, controls phosphorylation-dependent cleavage of ethylene-insensitive 2 (EIN2), a NRAMP-like integral membrane protein that then induces downstream TFs such as ETHYLENE-INSENSITIVE 3 (EIN3) and ein3-like 1 (EIL1) [111]. Ethylene signalling mutants of arabidopsis have increased freezing tolerance both before and after cold acclimation [94]. Moreover, overexpression of EIN3 resulted in hypersensitivity to freezing, which additionally correlated with suppressed expression of CBFs, whereas in an EIN3 EIN1 double mutant, CBFs and CBF-regulated genes were constitutively induced. When addressing the underlying molecular modes, it was found that EIN3 functions as a repressor of ARR5, ARR7 and ARR15 transcription. ChIP experiments demonstrated that EIN3 directly binds to the promoters of these ARRs and to all CBFs to regulate their expression (Fig. 3). Interestingly, although EIN3 was shown to act as a negative regulator of cold responses, cold increased EIN3 protein stability in an ethylene signalling-dependent manner [94].
In M. truncatula, a mutant of SICKLE (MtSKL1), an orthologue of arabidopsis EIN2, was slightly more tolerant to freezing than the wild-type without cold acclimation but less tolerant to freezing after cold acclimation. Surprisingly, in skl1 plants MtCBF1, MtCBF3 and MtCAS15, transcripts were suppressed both with and without cold treatment, suggesting that ethylene has a positive regulatory role in cold responses [110]. In addition, results for arabidopsis were published that dispute the negative regulatory role of ethylene in cold stress signalling. An arabidopsis ethylene biosynthesis ACS octuple mutant that contained extremely low levels of ethylene was hypersensitive to freezing before and after cold acclimation. In agreement, under cold treatment, induction of CBF1, CBF2 and COR genes was significantly lower in the ACS octuple mutant plants than in the wild-type. In support, the authors demonstrated that ethylene levels increased transiently after cold treatment in soil-grown arabidopsis plants, which was correlated with a transient increase in transcripts of different ethylene biosynthetic genes. Moreover, microarray data were presented, which showed that a significant overlap exists between genes induced by ACC treatment and those induced by cold in arabidopsis and that approximately 48 % of genes suppressed in ACS octuple mutant plants are activated by low temperature [109].
A positive role of ethylene in cold tolerance and the regulation of COR genes was additionally proposed for chilling-sensitive plant species. Overexpression of the tomato ethylene response factor TERF2/LeERF2, whose expression is induced both by ethylene and cold, stimulated ethylene biosynthesis and enhanced freezing tolerance in transgenic tobacco, tomato and rice plants, which was associated with an induction of COR genes and reduced amounts of ROS in the transgenic plants [112, 113]. Moreover, application of the ethylene precursor ACC promoted freezing tolerance in tobacco and tomato [114]. Moreover, an ethylene-insensitive tomato mutant was hypersensitive to severe chilling, thereby supporting a positive regulatory role of ethylene in cold stress responses of this plant species [105]. Because ethylene functions as a repressor of plant growth and development [115], it is possible that ethylene is an additional hormone that may serve to restrict growth in response to cold.
Salicylic acid (SA) contributes to cold stress-induced growth repression
Salicylic acid is another hormone that appears to contribute to the low temperature-induced growth retardation of plants. SA is a phenolic compound that is in particular known for triggering defence reactions against biotrophic pathogens such as the hypersensitive response (HR). Mutants with elevated SA levels are severely dwarfed and exhibit necrotic lesions caused by excessive HR induction [116].
Plants synthesise SA from chorismate, and in the SA-deficient NahG and enhanced disease susceptibility 5 (eds5) mutants growth is promoted at 5 °C, resulting in higher biomass and larger rosette leaf area as compared to wild-type plants [7, 117]. Moreover, SA-overaccumulating constitutive expression of PR genes (cpr1) mutant plants show reduced growth rates in low temperatures [7]. Interestingly, at low temperature, SA deficiency in NahG plants was correlated with enhanced transcription of the CK-regulated D-type cyclin CYCD3;1 that promotes the G1/S phase transition in arabidopsis [117]. This finding supported earlier results that had suggested that, in response to environmental cues, SA could affect cell cycle progression in plants [118] and this may contribute to SA-induced growth inhibition in the cold.
Cold-induced increases in SA levels were reported for both chilling-sensitive and freezing-tolerant plant species [7, 84, 119–121]. In cucumber seedlings exposed to 8 °C, levels of free and conjugated SA increased three- to fivefold [119], which was correlated with an induction of the SA biosynthetic enzyme phenylalanine-ammonia-lyase (PAL). Interestingly, in cucumber, organ-specific changes in SA levels were observed, whereas in roots, SA accumulated rapidly during the first 24 h following the cold treatment; SA accumulation was much slower in leaves where a maximum was reached after 72 h only [119].
In addition, SA contents increased following cold application in arabidopsis; however, a significant accumulation was detected only during the second week of low temperature treatments [7, 120], whereas growth inhibition occurred immediately [7]. Therefore, SA may act to mediate growth repression in later stages of cold exposure, when the role of GA becomes less pronounced [58]. In contrary to the chilling-sensitive species cucumber, SA biosynthesis in arabidopsis was induced through the isochorismate synthase (ICS) pathway, because ICS1 transcript levels were induced after 1 week of exposure to low temperature, and a loss-of-function of ICS1 abolished low temperature-induced SA accumulation [120] (Fig. 2).
The mechanism used to control SA biosynthesis in response to cold is currently unknown, although in LOS2 knockouts and overexpressing plant SA levels were increased. These effects were attributed to a loss of ENO2 function, which impaired lignin biosynthesis and resulted in collapsed walls; the damage caused to cellular structures was thought to increase SA levels as a secondary response [47]. Because LOS2 is cold stress induced [45], it appears possible that in response to cold, ENO2 function in glycolysis is altered, and this may impact on SA production.
A link between cold-induced SA accumulation and biotic stress signalling was revealed when the chilling-sensitive (chs) mutants chs2 [121] and chs3 [122] were characterised. These mutants accumulate high levels of SA specifically when grown in the cold. The mutations were dominant and mapped to genes encoding resistance (R) proteins of the TIR-NB-LRR class [121, 122]. Therefore, certain R proteins appear to impact on the induction of SA biosynthesis under low temperature regimes and this warrants future investigation.
The downstream regulatory cascades that enable SA to participate in cold responses are not entirely clear. Whereas two studies reported that in SA-deficient mutants, CBF expression was not altered and no significant differences in freezing tolerance following cold acclimation occurred [117, 120], others concluded that SA suppresses CBF expression and freezing tolerance. This was based on the result that in NahG plants, increased freezing tolerance correlated with increased expression of CBF3, COR47 and KIN1 following cold acclimation. Moreover, the SA-overaccumulating lines siz1-2 and acd6 were hypersensitive to freezing with and without cold acclimation, which correlated with suppressed expression of CBF3, COR47 and KIN1 [123]. Hypersensitivity to freezing and reduced CBF3 expression of siz1-2 and acd6 was abolished in the NahG background [123], which is somewhat surprising for siz1-2, because in this background, ICE1 SUMOylation and activity in CBF3 transcription were compromised [41].
In the chilling-sensitive plant cucumber, SA was proposed to activate CBF expression. Application of the SA biosynthesis inhibitors paclobutrazol (which additionally acts on GA biosynthesis) or l-α-aminooxy-β-phenylpropionic acid (AOPP) suppressed expression of CBFs and COR47 in low temperature-treated seedlings [119]. In addition, the chilling tolerance of such plants decreased compared with untreated controls. Low SA levels in treated plants were correlated with reduced photosynthetic efficiency and greater membrane damage, as well as reduced expression of genes encoding enzymes for carbon assimilation and synthesis of cryoprotectants such as sucrose and proline [119]. Because in arabidopsis, SA had no or little effect on carbon assimilation rates and photochemical efficiency [7], it appears that species-specific differences occur in SA function in photosynthesis under cold stress.
Jasmonic acid (JA) may participate in prioritising stress responses over growth
Similar to SA, JA is also involved in biotic and abiotic stress responses and is thought to repress growth in response to cold. JA is an oxylipin whose levels increase under cold stress in different plant species including wheat [84], rice [86, 87] and arabidopsis [124]. This increase is correlated with increased expression of JA biosynthetic genes in arabidopsis and rice [87, 124] and repression of genes encoding enzymes involved in JA catabolism in rice [87] (Fig. 2).
JA is synthesised from linolenic acid and is activated by conjugation with isoleucine, which enables binding to CORONATINE INSENSITIVE 1 (COI1), an F-box protein that acts as a JA receptor. COI1 initiates JA signalling by ubiquitinating and thereby stimulating proteasome-dependent degradation of JASMONATE ZIM DOMAIN (JAZ) proteins, which repress expression of JA-responsive genes. Exogenous application of JA enhanced induction of CBFs and CBF-regulated genes following cold treatment and promoted freezing tolerance in arabidopsis. Moreover, plants compromised in JA biosynthesis or signalling were hypersensitive to freezing [124]. It was proposed that this altered freezing tolerance is mediated by JAZ1 and JAZ4, which physically interact with ICE1 and ICE2 to repress their transcriptional activity. Overexpression of JAZ1 and JAZ4 inhibited expression of CBFs and their downstream targets and repressed freezing tolerance before and after cold acclimation (Fig. 3). Interestingly, microarray analysis of the JA signalling-deficient coi1-1 allele following low temperature treatment demonstrated that some of the COR genes of non-CBF regulons were differently regulated, which indicated that JA may additionally modulate cold acclimation through CBF-independent pathways [124].
In regard to the role of JA as a growth repressor in the cold, it was shown that JA-mediated growth inhibition occurred through stabilisation of DELLA proteins, which was speculated to allow JA to prioritise defence mechanisms over growth [125]. Therefore, it appears that chilling-induced JA accumulation contributes to suppression of plant growth, at least partially through cross-talk with GA signalling.
Conclusion
In summary, it is clear that hormones act as central regulators of cold stress responses in plants. However, our knowledge of their regulatory activities remains limited and seemingly contradicting results have occasionally been published.
Several reasons may account for this fact. First, changes in hormone levels at the cellular levels are usually not determined, although it is known that local hormone concentrations control adaptive growth and development. Rather, whole plants are usually being used for measurements, which is due to the fact that most phytohormones are present in extremely low concentrations and quantification at the cellular level is very challenging. Thus, results may be more enlightening if hormone measurements are complemented with reporter studies that allow for high spatial resolution.
Second, the experimental setups are decisive for determining cold stress tolerance of plants. However, no standardised procedures exist as yet. Rather, systems as different as responses of seedlings grown on agar plates and adult plants grown in soil are being used. Because both intrinsic developmental programs and environmental factors impact cold stress responses, experiments should be conceived to account for these facts more frequently. This is challenging because even seemingly little details such as the daytime of the treatment have to be taken into account, as many implicated genes, for example CBFs [65], are under circadian control.
Third, hormone mutants often have strong morphological defects, which likely impact on the outcome of chilling or freezing tolerance assays due to secondary effects that are not related to cold signalling but rather to decreased biomass, altered membrane morphologies and compositions or defective developmental phase transitions (such as flowering time control). Moreover, dwarfism as in GA, BR, auxin and SA mutants may reduce cold damage simply because there is less vegetative tissue exposed. In such cases, it will be necessary to additionally include weak alleles that are less impaired in growth and complement results of qualitative assessments such as survival rates with those of quantitative evaluations such as electrolyte leakage following freezing treatments [126].
Developmental biologists have already established techniques required for the study of plant hormone action under those highly challenging circumstances. In the future, it will be important that these systems are more readily adopted and optimised for the study of cold stress responses, which would further facilitate the rapid progress been made in the field in recent years.
Acknowledgments
We apologise to all colleagues whose contributions could not be cited or discussed due to space limitations. This work was supported by funds from the Deutsche Forschungsgemeinschaft DFG (Projects PO1640/4-1 and SFB924 TP-A12 to B.P.) and a fellowship to M.E. (doctoral fellowship from TUM). M.E. was a member of the TUM Graduate School.
Abbreviations
- ABA
Abscisic acid
- ACC
1-Aminocyclopropane-1-carboxylic acid
- ACO
ACC oxidase
- ACS
ACC synthase
- AHK
Arabidopsis histidine kinase
- AHP
Histidine phosphotransfer protein
- AOPP
l-α-Aminooxy-β-phenylpropionic acid
- ARR
Arabidopsis response regulator
- AVG
2-Aminoethoxyvinyl glycine
- BES1
BRI1-EMS-suppressor 1
- BRI1
Brassinosteroid insensitive 1
- BRs
Brassinosteroids
- BZR1
Brassinazole-resistant 1
- CAMTA
Calmodulin-binding transcription activator
- CAS
Cold-acclimation-specific
- CBF
C-repeat binding factor
- CK
Cytokinins
- COI1
Coronatine insensitive 1
- COR
Cold regulated
- CPD
Constitutive photomorphogenesis and dwarfism
- CPKs
Calcium-dependent protein kinases
- CPR1
Constitutive expression of PR genes
- CRT/DRE
C-repeat/dehydration-responsive element
- CTR1
Constitutive triple response 1
- DREB
Drought-responsive element-binding protein
- DWF4
Dwarf 4
- EDS5
Enhanced disease susceptibility 5
- EIL1
EIN3-like 1
- EIN2
Ethylene-insensitive 2
- EIN3
Ethylene-insensitive 3
- ENO2
Enolase 2
- ETR1
Ethylene-responsive 1
- GAI
Gibberellic acid insensitive
- GAs
Gibberellins
- GID1
GA-insensitive dwarf 1
- GNC
Gata, nitrate-inducible, carbon-metabolism involved
- GNL
GNC-like
- HHP
Heptahelical protein
- HR
Hypersensitive response
- IAA
Indole acetic acid
- IBA
Indole butyric acid
- ICE1
Inducer of cbf expression 1
- ICS
Isochorismate synthase
- JA
Jasmonic acid
- JAZ
Jasmonate zim domain
- KIN
Cold induced
- LOS2/AtMBP-1
Low expression of osmotically responsive genes 2/Arabidopsis thaliana c-MYC binding protein
- MAPKs
Mitogen-activated protein kinases
- OST1
Open stomata 1
- PAL
Phenylalanine-ammonia-lyase
- PIF
Phytochrome-interacting factor
- PIN
PIN-formed
- RGA
Repressor of gai
- RGL
RGA-like
- SA
Salicylic acid
References
- 1.Ruelland E, Vaultier MN, Zachowski A, Hurry V. Cold signalling and cold acclimation in plants. Adv Bot Res. 2009;49:35–150. doi: 10.1016/S0065-2296(08)00602-2. [DOI] [Google Scholar]
- 2.Frederiks TM, Christopher JT, Sutherland MW, Borrell AK. Post-head-emergence frost in wheat and barley: defining the problem, assessing the damage, and identifying resistance. J Exp Bot. 2015;66:3487–3498. doi: 10.1093/jxb/erv088. [DOI] [PubMed] [Google Scholar]
- 3.Boyer JS. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- 4.Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K. Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 2011;11:163. doi: 10.1186/1471-2229-11-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel D, Franklin KA. Temperature-regulation of plant architecture. Plant Signal Behav. 2009;4:577–579. doi: 10.4161/psb.4.7.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14:1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott IM, Clarke SM, Wood JE, Mur LA. Salicylate accumulation inhibits growth at chilling temperature in Arabidopsis. Plant Physiol. 2004;135:1040–1049. doi: 10.1104/pp.104.041293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.An D, Yang J, Zhang P. Transcriptome profiling of low temperature-treated cassava apical shoots showed dynamic responses of tropical plant to cold stress. BMC Genomics. 2012;13:64. doi: 10.1186/1471-2164-13-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhary S, Sharma PC. DeepSAGE based differential gene expression analysis under cold and freeze stress in seabuckthorn (Hippophae rhamnoides L.) PLoS One. 2015;10:e0121982. doi: 10.1371/journal.pone.0121982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Tian Q, Pang T, Jiang L, Wu R, Xia X, Yin W. Deep-sequencing transcriptome analysis of low temperature perception in a desert tree, Populus euphratica . BMC Genomics. 2014;15:326. doi: 10.1186/1471-2164-15-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barah P, Jayavelu ND, Rasmussen S, Nielsen HB, Mundy J, Bones AM. Genome-scale cold stress response regulatory networks in ten Arabidopsis thaliana ecotypes. BMC Genomics. 2013;14:722. doi: 10.1186/1471-2164-14-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung HJ, Dong X, Park JI, Thamilarasan SK, Lee SS, Kim YK, Lim YP, Nou IS, Hur Y. Genome-wide transcriptome analysis of two contrasting Brassica rapa doubled haploid lines under cold-stresses using Br 135K oligomeric chip. PLoS One. 2014;9:e106069. doi: 10.1371/journal.pone.0106069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leyva-Pérez Mde L, Valverde-Corredor A, Valderrama R, Jiménez-Ruiz J, Muñoz-Merida A, Trelles O, Barroso JB, Mercado-Blanco J, Luque F. Early and delayed long-term transcriptional changes and short-term transient responses during cold acclimation in olive leaves. DNA Res. 2015;22:1–11. doi: 10.1093/dnares/dsu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B, Ning L, Zhang J, Bao M, Zhang W. Transcriptional profiling of Petunia seedlings reveals candidate regulators of the cold stress response. Front Plant Sci. 2015;6:118. doi: 10.3389/fpls.2015.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moliterni VM, Paris R, Onofri C, Orrù L, Cattivelli L, Pacifico D, Avanzato C, Ferrarini A, Delledonne M, Mandolino G. Early transcriptional changes in Beta vulgaris in response to low temperature. Planta. 2015;242:187–201. doi: 10.1007/s00425-015-2299-z. [DOI] [PubMed] [Google Scholar]
- 16.Pang T, Ye CY, Xia X, Yin W. De novo sequencing and transcriptome analysis of the desert shrub, Ammopiptanthus mongolicus, during cold acclimation using Illumina/Solexa. BMC Genomics. 2013;14:488. doi: 10.1186/1471-2164-14-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park JW, Benatti TR, Marconi T, Yu Q, Solis-Gracia N, Mora V, da Silva JA. Cold responsive gene expression profiling of sugarcane and Saccharum spontaneum with functional analysis of a cold inducible Saccharum homolog of NOD26-like intrinsic protein to salt and water stress. PLoS One. 2015;10:e0125810. doi: 10.1371/journal.pone.0125810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu Y, Zhou A, Zhang X, Tang H, Liang M, Han H, Zuo Y. De novo transcriptome sequencing of low temperature-treated Phlox subulata and analysis of the genes involved in cold stress. Int J Mol Sci. 2015;16:9732–9748. doi: 10.3390/ijms16059732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren L, Sun J, Chen S, Gao J, Dong B, Liu Y, Xia X, Wang Y, Liao Y, Teng N, Fang W, Guan Z, Chen F, Jiang J. A transcriptomic analysis of Chrysanthemum nankingense provides insights into the basis of low temperature tolerance. BMC Genomics. 2014;15:844. doi: 10.1186/1471-2164-15-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sobkowiak A, Jończyk M, Jarochowska E, Biecek P, Trzcinska-Danielewicz J, Leipner J, Fronk J, Sowiński P. Genome-wide transcriptomic analysis of response to low temperature reveals candidate genes determining divergent cold-sensitivity of maize inbred lines. Plant Mol Biol. 2014;85:317–331. doi: 10.1007/s11103-014-0187-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song Y, Chen Q, Ci D, Zhang D. Transcriptome profiling reveals differential transcript abundance in response to chilling stress in Populus simonii . Plant Cell Rep. 2013;32:1407–1425. doi: 10.1007/s00299-013-1454-x. [DOI] [PubMed] [Google Scholar]
- 22.Sun P, Mao Y, Li G, Cao M, Kong F, Wang L, Bi G. Comparative transcriptome profiling of Pyropia yezoensis (Ueda) M.S. Hwang & H.G. Choi in response to temperature stresses. BMC Genomics. 2015;16:463. doi: 10.1186/s12864-015-1586-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian DQ, Pan XY, Yu YM, Wang WY, Zhang F, Ge YY, Shen XL, Shen FQ, Liu XJ. De novo characterization of the Anthurium transcriptome and analysis of its digital gene expression under cold stress. BMC Genomics. 2013;14:827. doi: 10.1186/1471-2164-14-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Zou Z, Wang S, Gong M. Global analysis of transcriptome responses and gene expression profiles to cold stress of Jatropha curcas L. PLoS One. 2013;8:e82817. doi: 10.1371/journal.pone.0082817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang XC, Zhao QY, Ma CL, Zhang ZH, Cao HL, Kong YM, Yue C, Hao XY, Chen L, Ma JQ, Jin JQ, Li X, Yang YJ. Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genomics. 2013;14:415. doi: 10.1186/1471-2164-14-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Yang Y, Liu X, Huang J, Wang Q, Gu J, Lu Y. Transcriptome profiling of the cold response and signaling pathways in Lilium lancifolium . BMC Genomics. 2014;15:203. doi: 10.1186/1471-2164-15-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y, Wei W, Pang X, Wang X, Zhang H, Dong B, Xing Y, Li X, Wang M. Comparative transcriptome profiling of a desert evergreen shrub, Ammopiptanthus mongolicus, in response to drought and cold stresses. BMC Genomics. 2014;15:671. doi: 10.1186/1471-2164-15-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xin H, Zhu W, Wang L, Xiang Y, Fang L, Li J, Sun X, Wang N, Londo JP, Li S. Genome wide transcriptional profile analysis of Vitis amurensis and Vitis vinifera in response to cold stress. PLoS One. 2013;8:e58740. doi: 10.1371/journal.pone.0058740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu W, Li R, Zhang N, Ma F, Jiao Y, Wang Z. Transcriptome profiling of Vitis amurensis, an extremely cold-tolerant Chinese wild Vitis species, reveals candidate genes and events that potentially connected to cold stress. Plant Mol Biol. 2014;86:527–541. doi: 10.1007/s11103-014-0245-2. [DOI] [PubMed] [Google Scholar]
- 30.Zhang T, Zhao X, Wang W, Pan Y, Huang L, Liu X, Zong Y, Zhu L, Yang D, Fu B. Comparative transcriptome profiling of chilling stress responsiveness in two contrasting rice genotypes. PLoS One. 2012;7:e43274. doi: 10.1371/journal.pone.0043274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Z, Tan L, Dang C, Zhang H, Wu Q, An L. Deep-sequencing transcriptome analysis of chilling tolerance mechanisms of a subnival alpine plant, Chorispora bungeana . BMC Plant Biol. 2012;12:222. doi: 10.1186/1471-2229-12-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu YN, Shi DQ, Ruan MB, Zhang LL, Meng ZH, Liu J, Yang WC. Transcriptome analysis reveals crosstalk of responsive genes to multiple abiotic stresses in cotton (Gossypium hirsutum L.) PLoS One. 2013;8:e80218. doi: 10.1371/journal.pone.0080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee BH, Henderson DA, Zhu JK. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell. 2005;17:3155–3175. doi: 10.1105/tpc.105.035568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park S, Lee CM, Doherty CJ, Gilmour SJ, Kim Y, Thomashow MF. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 2015;82:193–207. doi: 10.1111/tpj.12796. [DOI] [PubMed] [Google Scholar]
- 35.Thomashow MF. Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 2010;154:571–577. doi: 10.1104/pp.110.161794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novillo F, Alonso JM, Ecker JR, Salinas J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis . Proc Natl Acad Sci USA. 2004;101:3985–3990. doi: 10.1073/pnas.0303029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005;41:195–211. doi: 10.1111/j.1365-313X.2004.02288.x. [DOI] [PubMed] [Google Scholar]
- 38.Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell. 2009;21:972–984. doi: 10.1105/tpc.108.063958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis . Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fursova OV, Pogorelko GV, Tarasov VA. Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana . Gene. 2009;429:98–103. doi: 10.1016/j.gene.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 41.Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis . Plant Cell. 2007;19:1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miura K, Ohta M, Nakazawa M, Ono M, Hasegawa PM. ICE1 Ser403 is necessary for protein stabilization and regulation of cold signaling and tolerance. Plant J. 2011;67:269–279. doi: 10.1111/j.1365-313X.2011.04589.x. [DOI] [PubMed] [Google Scholar]
- 43.Ding Y, Li H, Zhang X, Xie Q, Gong Z, Yang S. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis . Dev Cell. 2015;32:278–289. doi: 10.1016/j.devcel.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 44.Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA. 2006;103:8281–8286. doi: 10.1073/pnas.0602874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee H, Guo Y, Ohta M, Xiong L, Stevenson B, Zhu JK. LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO J. 2002;21:2692–2702. doi: 10.1093/emboj/21.11.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang M, Abdelmageed H, Lee S, Reichert A, Mysore KS, Allen RD. AtMBP-1, an alternative translation product of LOS2, affects abscisic acid responses and is modulated by the E3 ubiquitin ligase AtSAP5. Plant J. 2013;76:481–493. doi: 10.1111/tpj.12312. [DOI] [PubMed] [Google Scholar]
- 47.Eremina M, Rozhon W, Yang S, Poppenberger B. ENO2 activity is required for the development and reproductive success of plants, and is feedback-repressed by AtMBP-1. Plant J. 2015;81:895–906. doi: 10.1111/tpj.12775. [DOI] [PubMed] [Google Scholar]
- 48.Barrero-Gil J, Salinas J. Post-translational regulation of cold acclimation response. Plant Sci. 2013;205–206:48–54. doi: 10.1016/j.plantsci.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 49.Knight MR, Knight H. Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol. 2012;195:737–751. doi: 10.1111/j.1469-8137.2012.04239.x. [DOI] [PubMed] [Google Scholar]
- 50.Santner A, Calderon-Villalobos LI, Estelle M. Plant hormones are versatile chemical regulators of plant growth. Nat Chem Biol. 2009;5:301–307. doi: 10.1038/nchembio.165. [DOI] [PubMed] [Google Scholar]
- 51.Franklin KA. Light and temperature signal crosstalk in plant development. Curr Opin Plant Biol. 2009;12:63–68. doi: 10.1016/j.pbi.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 52.Penfield S. Temperature perception and signal transduction in plants. New Phytol. 2008;179:615–628. doi: 10.1111/j.1469-8137.2008.02478.x. [DOI] [PubMed] [Google Scholar]
- 53.Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 54.Schwechheimer C. Gibberellin signaling in plants—the extended version. Front Plant Sci. 2012;2:107. doi: 10.3389/fpls.2011.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, Schäfer E, Fu X, Fan LM, Deng XW. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 57.Li QF, Wang C, Jiang L, Li S, Sun SS, He JX. An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis . Sci Signal. 2012;5:ra72. doi: 10.1126/scisignal.2002908. [DOI] [PubMed] [Google Scholar]
- 58.Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell. 2008;20:2117–2129. doi: 10.1105/tpc.108.058941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richter R, Bastakis E, Schwechheimer C. Cross-repressive interactions between SOC1 and the GATAs GNC and GNL/CGA1 in the control of greening, cold tolerance, and flowering time in Arabidopsis. Plant Physiol. 2013;162:1992–2004. doi: 10.1104/pp.113.219238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hsieh TH, Lee JT, Yang PT, Chiu LH, Charng YY, Wang YC, Chan MT. Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol. 2002;129:1086–1094. doi: 10.1104/pp.003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou M, Xu M, Wu L, Shen C, Ma H, Lin J. CbCBF from Capsella bursa-pastoris enhances cold tolerance and restrains growth in Nicotiana tabacum by antagonizing with gibberellin and affecting cell cycle signaling. Plant Mol Biol. 2014;85:259–275. doi: 10.1007/s11103-014-0181-1. [DOI] [PubMed] [Google Scholar]
- 62.Shan DP, Huang JG, Yang YT, Guo YH, Wu CA, Yang GD, Gao Z, Zheng CC. Cotton GhDREB1 increases plant tolerance to low temperature and is negatively regulated by gibberellic acid. New Phytol. 2007;176:70–81. doi: 10.1111/j.1469-8137.2007.02160.x. [DOI] [PubMed] [Google Scholar]
- 63.Tanaka N, Matsuoka M, Kitano H, Asano T, Kaku H, Komatsu S. gid1, a gibberellin-insensitive dwarf mutant, shows altered regulation of probenazole-inducible protein (PBZ1) in response to cold stress and pathogen attack. Plant Cell Environ. 2006;29:619–631. doi: 10.1111/j.1365-3040.2005.01441.x. [DOI] [PubMed] [Google Scholar]
- 64.Richter R, Behringer C, Müller IK, Schwechheimer C. The GATAtype transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROMEINTERACTING FACTORS. Genes Dev. 2010;24:2093–2104. doi: 10.1101/gad.594910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee CM, Thomashow MF. Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana . Proc Natl Acad Sci USA. 2012;109:15054–15059. doi: 10.1073/pnas.1211295109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Franklin KA, Whitelam GC. Light-quality regulation of freezing tolerance in Arabidopsis thaliana . Nat Genet. 2007;39:1410–1413. doi: 10.1038/ng.2007.3. [DOI] [PubMed] [Google Scholar]
- 67.Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, Wigge PA, Gray WM. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA. 2011;108:20231–20235. doi: 10.1073/pnas.1110682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bernardo-García S, de Lucas M, Martínez C, Espinosa-Ruiz A, Davière JM, Prat S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014;28:1681–1694. doi: 10.1101/gad.243675.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clouse SD. Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell. 2011;23:1219–1230. doi: 10.1105/tpc.111.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Unterholzner SJ, Rozhon W, Papacek M, Ciomas J, Lange T, Kugler KG, Mayer KF, Sieberer T, Poppenberger B. Brassinosteroids are master regulators of gibberellin biosynthesis in Arabidopsis . Plant Cell. 2015;27:2261–2272. doi: 10.1105/tpc.15.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stewart Lilley JL, Gan Y, Graham IA, Nemhauser JL. The effects of DELLAs on growth change with developmental stage and brassinosteroid levels. Plant J. 2013;76:165–173. doi: 10.1111/tpj.12280. [DOI] [PubMed] [Google Scholar]
- 72.Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis . Nat Cell Biol. 2012;14:810–817. doi: 10.1038/ncb2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gallego-Bartolomé J, Minguet EG, Grau-Enguix F, Abbas M, Locascio A, Thomas SG, Alabadí D, Blázquez MA. Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis . Proc Natl Acad Sci USA. 2012;109:13446–134451. doi: 10.1073/pnas.1119992109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xia XJ, Wang YJ, Zhou YH, Tao Y, Mao WH, Shi K, Asami T, Chen Z, Yu JQ. Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiol. 2009;150:801–814. doi: 10.1104/pp.109.138230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Singh I, Kumar U, Singh SK, Gupta C, Singh M, Kushwaha SR. Physiological and biochemical effect of 24-epibrassinoslide on cold tolerance in maize seedlings. Physiol Mol Biol Plants. 2012;18:229–236. doi: 10.1007/s12298-012-0122-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jiang YP, Huang LF, Cheng F, Zhou YH, Xia XJ, Mao WH, Shi K, Yu JQ. Brassinosteroids accelerate recovery of photosynthetic apparatus from cold stress by balancing the electron partitioning, carboxylation and redox homeostasis in cucumber. Physiol Plant. 2013;148:133–145. doi: 10.1111/j.1399-3054.2012.01696.x. [DOI] [PubMed] [Google Scholar]
- 77.Divi UK, Krishna P. Overexpression of the brassinosteroid biosynthetic gene AtDWF4 in Arabidopsis seeds overcomes abscisic acid-induced inhibition of germination and increases cold tolerance in transgenic seedlings. J Plant Growth Regul. 2010;29:385–393. doi: 10.1007/s00344-010-9150-3. [DOI] [Google Scholar]
- 78.Kagale S, Divi UK, Krochko JE, Keller WA, Krishna P. Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta. 2007;225:353–364. doi: 10.1007/s00425-006-0361-6. [DOI] [PubMed] [Google Scholar]
- 79.Qu T, Liu R, Wang W, An L, Chen T, Liu G, Zhao Z. Brassinosteroids regulate pectin methylesterase activity and AtPME41 expression in Arabidopsis under chilling stress. Cryobiology. 2011;63:111–117. doi: 10.1016/j.cryobiol.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 80.Kim SY, Kim BH, Lim CJ, Lim CO, Nam KH. Constitutive activation of stress-inducible genes in a brassinosteroid-insensitive 1 (bri1) mutant results in higher tolerance to cold. Physiol Plant. 2010;138:191–204. doi: 10.1111/j.1399-3054.2009.01304.x. [DOI] [PubMed] [Google Scholar]
- 81.Hannah MA, Heyer AG, Hincha DK. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana . PLoS Genet. 2005;1:e26. doi: 10.1371/journal.pgen.0010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jain M, Khurana JP. Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. FEBS J. 2009;276:3148–3162. doi: 10.1111/j.1742-4658.2009.07033.x. [DOI] [PubMed] [Google Scholar]
- 83.Gavelienė V, Novickienė L, Pakalniškytė L. Effect of auxin physiological analogues on rapeseed (Brassica napus) cold hardening, seed yield and quality. J Plant Res. 2013;126:283–292. doi: 10.1007/s10265-012-0525-3. [DOI] [PubMed] [Google Scholar]
- 84.Kosová K, Prášil IT, Vítámvás P, Dobrev P, Motyka V, Floková K, Novák O, Turečková V, Rolčik J, Pešek B, Trávničková A, Gaudinová A, Galiba G, Janda T, Vlasáková E, Prášilová P, Vanková R. Complex phytohormone responses during the cold acclimation of two wheat cultivars differing in cold tolerance, winter Samanta and spring Sandra. J Plant Physiol. 2012;169:567–576. doi: 10.1016/j.jplph.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 85.Majláth I, Szalai G, Soós V, Sebestyén E, Balázs E, Vanková R, Dobrev PI, Tari I, Tandori J, Janda T. Effect of light on the gene expression and hormonal status of winter and spring wheat plants during cold hardening. Physiol Plant. 2012;145:296–314. doi: 10.1111/j.1399-3054.2012.01579.x. [DOI] [PubMed] [Google Scholar]
- 86.Maruyama K, Urano K, Yoshiwara K, Morishita Y, Sakurai N, Suzuki H, Kojima M, Sakakibara H, Shibata D, Saito K, Shinozaki K, Yamaguchi-Shinozaki K. Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. Plant Physiol. 2014;164:1759–1771. doi: 10.1104/pp.113.231720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Du H, Liu H, Xiong L. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front Plant Sci. 2013;4:397. doi: 10.3389/fpls.2013.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 89.Fukaki H, Fujisawa H, Tasaka M. Gravitropic response of inflorescence stems in Arabidopsis thaliana . Plant Physiol. 1996;110:933–943. doi: 10.1104/pp.110.3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wyatt SE, Rashotte AM, Shipp MJ, Robertson D, Muday GK. Mutations in the gravity persistence signal loci in Arabidopsis disrupt the perception and/or signal transduction of gravitropic stimuli. Plant Physiol. 2002;130:1426–1435. doi: 10.1104/pp.102.010579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shibasaki K, Uemura M, Tsurumi S, Rahman A. Auxin response in Arabidopsis under cold stress: underlying molecular mechanisms. Plant Cell. 2009;21:3823–3838. doi: 10.1105/tpc.109.069906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sakakibara H. Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol. 2006;57:431–449. doi: 10.1146/annurev.arplant.57.032905.105231. [DOI] [PubMed] [Google Scholar]
- 93.Jeon J, Kim NY, Kim S, Kang NY, Novák O, Ku SJ, Cho C, Lee DJ, Lee EJ, Strnad M, Kim J. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis . J Biol Chem. 2010;285:23371–23386. doi: 10.1074/jbc.M109.096644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shi Y, Tian S, Hou L, Huang X, Zhang X, Guo H, Yang S. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis . Plant Cell. 2012;24:2578–2595. doi: 10.1105/tpc.112.098640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Veselova SV, Farhutdinov RG, Veselov SY, Kudoyarova GR, Veselov DS, Hartung W. The effect of root cooling on hormone content, leaf conductance and root hydraulic conductivity of durum wheat seedlings (Triticum durum L.) J Plant Physiol. 2005;162:21–26. doi: 10.1016/j.jplph.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 96.Jeon J, Kim J. Arabidopsis response Regulator1 and Arabidopsis histidine phosphotransfer Protein2 (AHP2), AHP3, and AHP5 function in cold signaling. Plant Physiol. 2013;161:408–424. doi: 10.1104/pp.112.207621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci. 2014;5:170. doi: 10.3389/fpls.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cuevas JC, López-Cobollo R, Alcázar R, Zarza X, Koncz C, Altabella T, Salinas J, Tiburcio AF, Ferrando A. Putrescine is involved in Arabidopsis freezing tolerance and cold acclimation by regulating abscisic acid levels in response to low temperature. Plant Physiol. 2008;148:1094–1105. doi: 10.1104/pp.108.122945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baron KN, Schroeder DF, Stasolla C. Transcriptional response of abscisic acid (ABA) metabolism and transport to cold and heat stress applied at the reproductive stage of development in Arabidopsis thaliana . Plant Sci. 2012;188–189:48–59. doi: 10.1016/j.plantsci.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 100.Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol. 2000;3:217–223. doi: 10.1016/S1369-5266(00)00067-4. [DOI] [PubMed] [Google Scholar]
- 101.Lee HG, Seo PJ. The MYB96-HHP module integrates cold and abscisic acid signaling to activate the CBF-COR pathway in Arabidopsis. Plant J. 2015;82:962–977. doi: 10.1111/tpj.12866. [DOI] [PubMed] [Google Scholar]
- 102.Chen CC, Liang CS, Kao AL, Yang CC. HHP1, a novel signalling component in the cross-talk between the cold and osmotic signalling pathways in Arabidopsis . J Exp Bot. 2010;61:3305–3320. doi: 10.1093/jxb/erq162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell. 2002;14:3089–3099. doi: 10.1105/tpc.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yoo SD, Cho Y, Sheen J. Emerging connections in the ethylene signaling network. Trends Plant Sci. 2009;14:270–279. doi: 10.1016/j.tplants.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ciardi JA, Deikman J, Orzolek MD. Increased ethylene synthesis enhances chilling tolerance in tomato. Physiol Plant. 1997;101:333–340. doi: 10.1111/j.1399-3054.1997.tb01005.x. [DOI] [Google Scholar]
- 106.Yu XM, Griffith M, Wiseman SB. Ethylene induces antifreeze activity in winter rye leaves. Plant Physiol. 2001;126:1232–1240. doi: 10.1104/pp.126.3.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guye MG, Vigh L, Wilson LM. Chilling-induced ethylene production in relation to chill-sensitivity in Phaseolus spp. J Exp Bot. 1987;38:680–690. doi: 10.1093/jxb/38.4.680. [DOI] [Google Scholar]
- 108.Guo Z, Tan J, Zhuo C, Wang C, Xiang B, Wang Z. Abscisic acid, H2O2 and nitric oxide interactions mediated cold-induced S-adenosylmethionine synthetase in Medicago sativa subsp. falcata that confers cold tolerance through up-regulating polyamine oxidation. Plant Biotechnol J. 2014;12:601–612. doi: 10.1111/pbi.12166. [DOI] [PubMed] [Google Scholar]
- 109.Catalá R, López-Cobollo R, Mar Castellano M, Angosto T, Alonso JM, Ecker JR, Salinas J. The Arabidopsis 14-3-3 protein RARE COLD INDUCIBLE 1A links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell. 2014;26:3326–3342. doi: 10.1105/tpc.114.127605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhao M, Liu W, Xia X, Wang T, Zhang WH. Cold acclimation-induced freezing tolerance of Medicago truncatula seedlings is negatively regulated by ethylene. Physiol Plant. 2014;152:115–129. doi: 10.1111/ppl.12161. [DOI] [PubMed] [Google Scholar]
- 111.Merchante C, Alonso JM, Stepanova AN. Ethylene signaling: simple ligand, complex regulation. Curr Opin Plant Biol. 2013;16:554–560. doi: 10.1016/j.pbi.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 112.Tian Y, Zhang H, Pan X, Chen X, Zhang Z, Lu X, Huang R. Overexpression of ethylene response factor TERF2 confers cold tolerance in rice seedlings. Transgenic Res. 2011;20:857–866. doi: 10.1007/s11248-010-9463-9. [DOI] [PubMed] [Google Scholar]
- 113.Zhang Z, Zhang H, Quan R, Wang XC, Huang R. Transcriptional regulation of the ethylene response factor LeERF2 in the expression of ethylene biosynthesis genes controls ethylene production in tomato and tobacco. Plant Physiol. 2009;150:365–377. doi: 10.1104/pp.109.135830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang Z, Huang R. Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol Biol. 2010;73:241–249. doi: 10.1007/s11103-010-9609-4. [DOI] [PubMed] [Google Scholar]
- 115.Schaller GE. Ethylene and the regulation of plant development. BMC Biol. 2012;10:9. doi: 10.1186/1741-7007-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rivas-San Vicente M, Plasencia J. Salicylic acid beyond defence: its role in plant growth and development. J Exp Bot. 2011;62:3321–3338. doi: 10.1093/jxb/err031. [DOI] [PubMed] [Google Scholar]
- 117.Xia JC, Zhao H, Liu WZ, Li LG, He YK. Role of cytokinin and salicylic acid in plant growth at low temperatures. Plant Growth Regul. 2009;57:211–221. doi: 10.1007/s10725-008-9338-8. [DOI] [Google Scholar]
- 118.Wolters H, Jürgens G. Survival of the flexible: hormonal growth control and adaptation in plant development. Nat Rev Genet. 2009;10:305–317. doi: 10.1038/nrg2558. [DOI] [PubMed] [Google Scholar]
- 119.Dong CJ, Li L, Shang QM, Liu XY, Zhang ZG. Endogenous salicylic acid accumulation is required for chilling tolerance in cucumber (Cucumis sativus L.) seedlings. Planta. 2014;240:687–700. doi: 10.1007/s00425-014-2115-1. [DOI] [PubMed] [Google Scholar]
- 120.Kim Y, Park S, Gilmour SJ, Thomashow MF. Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J. 2013;75:364–376. doi: 10.1111/tpj.12205. [DOI] [PubMed] [Google Scholar]
- 121.Huang X, Li J, Bao F, Zhang X, Yang S. A gain-of-function mutation in the Arabidopsis disease resistance gene RPP4 confers sensitivity to low temperature. Plant Physiol. 2010;154(2):796–809. doi: 10.1104/pp.110.157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang H, Shi Y, Liu J, Guo L, Zhang X, Yang S. A mutant CHS3 protein with TIR-NB-LRR-LIM domains modulates growth, cell death and freezing tolerance in a temperature-dependent manner in Arabidopsis. Plant J. 2010;63(2):283–296. doi: 10.1111/j.1365-313X.2010.04241.x. [DOI] [PubMed] [Google Scholar]
- 123.Miura K, Ohta M. SIZ1, a small ubiquitin-related modifier ligase, controls cold signaling through regulation of salicylic acid accumulation. J Plant Physiol. 2010;167:555–560. doi: 10.1016/j.jplph.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 124.Hu Y, Jiang L, Wang F, Yu D. Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis . Plant Cell. 2013;25:2907–2924. doi: 10.1105/tpc.113.112631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, Lee CM, Thomashow MF, Yang Y, He Z, He SY. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA. 2012;109:E1192–E1200. doi: 10.1073/pnas.1201616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Thalhammer A, Bryant G, Sulpice R, Hincha DK. Disordered cold regulated15 proteins protect chloroplast membranes during freezing through binding and folding, but do not stabilize chloroplast enzymes in vivo. Plant Physiol. 2014;166:190–201. doi: 10.1104/pp.114.245399. [DOI] [PMC free article] [PubMed] [Google Scholar]