Abstract
Plants use various kinds of environmental signals to adjust the timing of the transition from the vegetative to reproductive phase (flowering). Since flowering at the appropriate time is crucial for plant reproductive strategy, several kinds of photoreceptors are deployed to sense environmental light conditions. In this review, we will update our current understanding of light signaling pathways in flowering regulation, especially, in which tissue do photoreceptors regulate flowering in response to light quality and photoperiod. Since light signaling is also integrated into other flowering pathways, we also introduce recent progress on how photoreceptors are involved in tissue-specific thermosensation and the gibberellin pathway. Finally, we discuss the importance of cell-type-specific analyses for future plant studies.
Keywords: Photoperiod, Light quality, Temperature, Gibberellin, Tissue-specific regulation, Phytochrome, Cryptochrome, Day length
Introduction
Plants utilize sunlight not only as an energy source via photosynthesis but also as an information source for surrounding conditions. Being sessile, plants employ photoreceptors to perceive environmental light signals and regulate physiological responses to adapt to the changing surroundings. Among photoreceptor-regulated physiological responses, the vegetative to reproductive phase transition (referred to as flowering) is crucial because the appropriate timing of flowering directly leads to reproductive success. Environmental light signals contain many kinds of information, such as photoperiod, light intensity, light direction, and light quality. Of these light signals, light quality and photoperiod especially are widely used in flowering regulation.
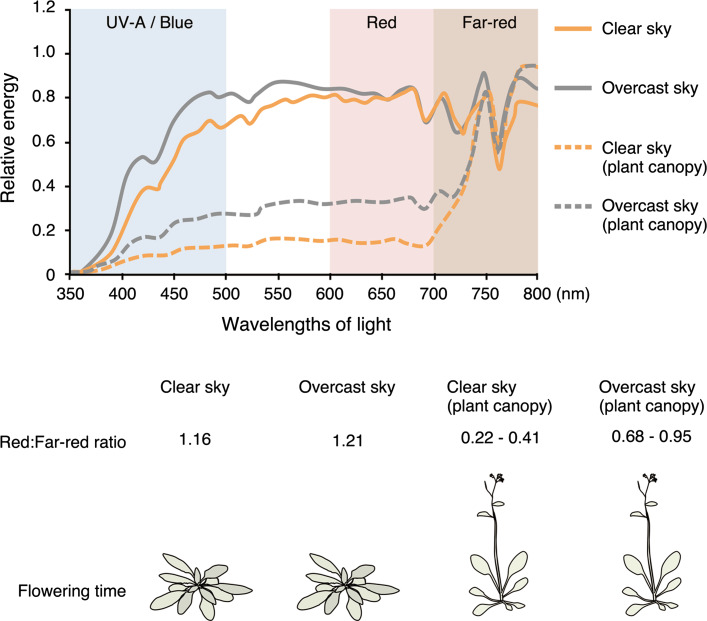
Light quality usually refers to the red:far-red light ratio. Since red light is predominantly absorbed by chlorophylls, and far-red light passes through most leaves, the red:far-red light ratio under a vegetation canopy decreases (<1) compared to direct sunlight (around 1.2) (Fig. 1). In response to low light quality, such as under a vegetation canopy, flowering is promoted by the ‘shade avoidance response (SAR)’, which was first described systematically by Sachs in 1863 [1, 2]. SAR is a kind of emergency response to escape from shade; therefore, it is usually associated with long petioles, erect and pale (low chlorophyll content) leaves, and early flowering. Another informative light signal for flowering regulation is the photoperiod, or the day length. Except for polar and equatorial regions, the photoperiod undergoes seasonal oscillations; in contrast to the light quality pathway, therefore, the photoperiod pathway is a mechanism for regular seasonal flowering, which was first reported by Garner and Allard in 1923 [3]. Plants that flower under long day (LD) and short day (SD) are referred to as LD plants and SD plants, respectively.
Fig. 1.
Light quality-mediated flowering regulation. When plants are under a vegetation canopy, or when it is overcast, plants cannot perceive enough sunlight. However, plants need to distinguish between an enduring vegetation canopy and a temporal cloudy day. Since blue and red light is predominantly absorbed by chlorophyll, sunlight that passes through the vegetation canopy contains more far-red light than blue or red light. On the other hand, under overcast conditions, clouds reflect a large portion of sunlight, resulting in a small increase of blue light but very little change in the red:far-red ratio [116]. Therefore, only when plants are under a vegetation canopy will they initiate SAR and induce flowering. An increase of the blue light ratio may not affect flowering in this case, because flowering time was not affected over a wide range of blue light intensities (25–164 µmol m−2 s−1) [117]
The model plant Arabidopsis thaliana, a facultative LD plant, has provided a wealth of data elucidating the molecular basis of light signaling, as well as genetic pathways. Although large numbers of light signaling studies have focused on early photomorphogenesis, much less attention has been paid to flowering regulation by photoreceptors. Since some light signaling mutants manifest only a flowering phenotype and others display only photomorphogenic phenotypes, the light signaling pathways appear to diverge at an early step. Intriguingly, several studies have clearly demonstrated that flowering is regulated by light signaling pathways in specific tissues, although photoreceptors are expressed in almost all cell types [4–6]. Therefore, it is important to marshal data about the various light signaling factors and the responsible tissues that are involved in flowering regulation in Arabidopsis. Moreover, recent progress makes it clear that light signaling is also integrated into many other flowering pathways, especially the thermosensory and gibberellin pathways. In this review we highlight the molecular mechanisms operative in photoreceptor-mediated flowering regulation. We also discuss recent studies on tissue-specific functions of circadian clocks that are tightly involved in light and temperature signaling, and the regulation of flowering.
Light signal transduction in the light quality pathway
A low red:far-red light ratio induces SAR-associated photomorphogenesis and early flowering. phyB plays a major role in this response. A subfamily of basic helix–loop–helix (bHLH) proteins, phytochrome-interacting factors (PIFs), directly interact with the Pfr form of phyB [7, 8]. The PIF transcription factors contribute to photomorphogenic processes, including hypocotyl elongation, chloroplast development, and seed germination [9–11]. In flowering regulation, overexpression of PIF4 or PIF5 causes early flowering [12, 13]. Moreover, genome-wide association studies demonstrated that PIF4 is associated with variation in flowering time [14]. However, a quadruple mutant deficient in pif1, pif3, pif4, and pif5, or any of the respective single mutants, displayed no clear differences in flowering time [9], suggesting highly redundant functions of PIFs.
Another factor, early flowering 3 (ELF3), is crucial for light quality signaling. Quantitative trait loci (QTL) analysis in recombinant inbred lines of Arabidopsis identified ELF3 as the most likely candidate gene affecting the shade avoidance response [15]. ELF3 is a circadian clock gene, and elf3 mutants show long hypocotyls, early flowering, and impaired responsiveness to red light, much like a phyB mutant [16, 17]. Consistent with this notion, phyB, ELF3, and PIF4 interact both genetically and physically [18–20]. Although the circadian clock gates rapid SAR [21], a recent study shows that the ELF3–PIF4 interaction is independent of the ELF3–ELF4–LUX complex (evening complex) [20, 22]. How a circadian clock protein functions in the SAR is still unclear because the mechanisms for light input to the circadian clock system are not well understood.
A transcriptional mediator complex subunit, phytochrome and flowering time 1 (PFT1), has been reported as a key factor that mediates phyB-to-FT signaling [23]. Recent studies, however, show that PFT1 has pleiotropic functions, including plant hormone signaling, sulfate assimilation, iron homeostasis, and reactive oxygen species (ROS) distribution [24–26]. Therefore, PFT1 appears to be a more general transcriptional mediator, rather than a specific factor for the light quality pathway.
Plant photoreceptors for flowering regulation
In Arabidopsis, five classes of photoreceptors have been discovered. Phytochromes (phyA to phyE) are major red and far-red light photoreceptors [27]. Phytochromes can be classified as light-labile type I (phyA) and light-stable type II (phyB to phyE) [28, 29]. Type II phytochromes are responsible for classic red:far-red photoreversible physiological responses. Cryptochromes (cry1 and cry2) are ultraviolet-A (UV-A) and blue light photoreceptors [30]. Similar to the phytochromes, cry1 is light stable and cry2 is light labile [31, 32]. In Arabidopsis, cry3 and cry-dash appear to be more like photolyase than bona fide blue light photoreceptors, although some cry-dash orthologs have been shown to possess cryptochrome activities in other organisms [33–37]. Phototropins (phot1 and phot2) are also UV-A and blue light receptors involved in phototropism. Zeitlupe (ZTL), flavin-binding, kelch repeat, F-box 1 (FKF1), and lov kelch protein 2 (LKP2) have an F-box domain and a Kelch repeat domain [38]. These have not been classified as canonical photoreceptors but nevertheless have been demonstrated to be blue light photoreceptors [39]. UV resistance locus 8 (UVR8) is the latest identified photoreceptor, which absorbs UV-B [40].
Among these photoreceptor family members, phytochromes, cryptochromes, and ZTL/FKF1/LKP2 are three major photoreceptors that are implicated in regulating the timing of flowering. Deploying such a variety of photoreceptors may be beneficial for plants to optimize their adaptation, since both blue and red/far-red light are needed for the precise estimation of surrounding light conditions and seasons (see detail below). Signals from these photoreceptors are integrated into the expression of flowering locus T (FT) protein, which is also known to be a flowering hormone, or florigen [41]. FT protein is tissue specifically synthesized in the leaf vascular phloem companion cells, and it moves through the phloem to the shoot apical meristem. At the shoot apical meristem, FT functions as a part of the florigen activating complex (FAC) and promotes transcription of floral initiation genes [42].
In contrast to the leaf vascular bundle-specific FT expression, photoreceptors that regulate flowering are expressed in almost all cell types. Although traditional GUS staining showed striped patterns of phytochrome expression, promoter::LUC or promoter::GFP assays are more consistent with ubiquitous expression of phytochromes [4, 43–45]. Cryptochromes and ZTL/FKF1/LKP2 are also similar to the phytochromes and are expressed ubiquitously in plants [5, 45, 46]. Taken together, plant photoreceptors are expressed uniformly, and therefore the downstream mechanisms for vascular FT regulation should be tissue specific.
Tissue-specific regulation of the light quality pathway
Given the aforementioned collection of regulatory genes, it was possible to identify a particular tissue where SAR is regulated. A study using a phyB-GFP enhancer trap line revealed that phyB in mesophyll regulates flowering in the light quality pathway [4]. Interestingly, phyB-GFP that was expressed only in vasculature, epidermis, or root did not complement the early flowering phenotype of the parental phyB mutant, implying a particular importance of mesophyll for the light quality pathway. Although mesophyll expression of ELF3, ELF4, LUX, and PFT1 can be observed by tissue-specific microarray analysis, the tissue-specific functions of other factors that are implicated in SAR have not yet been tested [47]. Therefore, how mesophyllic phyB regulates vascular FT expression is still unclear. Some reports have demonstrated that proteins can be transported from mesophyll to vasculature through plasmodesmata, suggesting inter-tissue communication between these two tissues [48]. SAR is associated with chronic reduction in the amount of photosynthesis, and it is critically distinguished from cloud cover (Fig. 1). Therefore, mesophyllic regulation of flowering associated with SAR seems to be biologically reasonable to achieve tight coupling of SAR regulation with photosynthesis.
Light signal transduction in the photoperiod pathway
In many plant species including Arabidopsis, constans (CO) and its direct target FT are crucial, especially in the photoperiod pathway [46, 49, 50]. When CO and FT functions are perturbed, plants cannot sense seasonal cues, and flowering time under a floral inductive condition will be the same as in the non-inductive condition. In Arabidopsis, co and ft mutants show late flowering both under LD and SD conditions [51]. On the other hand, CO and FT overexpressing lines exhibit a dramatically early flowering phenotype independent of day length [52–54]. Therefore, regulation of CO and FT gene transcription and protein stability are major control mechanisms that may be impacted by photoreceptor input.
CO expression
Although CO expression is regulated by flowering BHLH 1(FBH1), FBH2, FBH3, and FBH4 [55], FKF1 also regulates photoperiod-dependent flowering. The expression levels of FKF1 and gigantea (GI) are oscillatory, and the respective proteins accumulate in the evening [56]. Accumulated FKF1 and GI interact with each other, forming a complex in a blue light-dependent manner. The FKF1–GI complex degrades cycling DOF factor 1 (CDF1), a transcriptional repressor of CO, in the evening [57]; therefore, blue light-dependent CO transcriptional activation can be observed at the end of the day. ZTL and LKP2 also regulate CO transcription. In contrast to FKF1, overexpression of ZTL or LKP2 strongly suppresses CO expression and results in late flowering [58, 59]. However, there are at least two explanations for why these overexpressing lines show suppressed CO transcription. One explanation is that when three or more factors are required for a functional complex, knockout or overexpression leads to a perturbed stoichiometric balance of the components, resulting in impaired function. This is compatible with the notion that ZTL and LKP2 interact with FKF1 in yeast and in vitro [60]. Another possibility might involve the destabilization of circadian clock proteins timing of cab expression 1 (TOC1) and pseudo response regulator 5 (PRR5). ZTL has been shown to destabilize the TOC1 and PRR5 proteins. Flowering time in a toc1 prr5 double mutant line is as late as in the ZTL overexpression lines [61], and these two clock genes control CO expression, probably through transcriptional and posttranscriptional regulation of CDFs [62–67]. PFT1, a phyB signaling component, also shows a slightly suppressed CO transcription level in the pft1 mutant [23, 68, 69].
CO protein stability
In addition to CO transcriptional regulation, photoreceptors also control CO protein stability. Under LD conditions, CO protein expressed in the morning is degraded, whereas CO protein expressed in the evening is stabilized [70–72]. To achieve such a dynamic regulation of CO protein concentration, at least two different types of ubiquitin ligases are involved. An E3 ubiquitin ligase complex, comprising constitutive photomorphogenic 1 (COP1) and suppressor of phytochrome A (SPA1), triggers degradation of CO protein in the night period [73, 74]. As the day progresses, photoactivated cry2 directly binds SPA1 and inhibits the formation of the COP1-SPA1 complex [75]. Similar to the cry2 mechanism, phyA and cry1 also inhibit the COP1–SPA1 complex [76–79]. Physiological studies have indicated that phyA and cry2 are day-length sensors [80, 81], suggesting that COP1–SPA1-mediated CO protein degradation is a node in the regulatory network controlling photoperiodic flowering. FKF1 also stabilizes CO in the afternoon [82]. In addition, CO protein in the morning is degraded by a COP1-independent pathway that is activated by phyB [74]. Another ubiquitin ligase, high expression of osmotically responsive gene 1 (HOS1), is a good candidate for phyB-dependent CO protein degradation in the morning [71]. Phytochrome-dependent late flowering (PHL) also stabilizes CO protein in the afternoon [83]; however, the mechanism is largely unknown. These combinations of transcriptional and post-translational regulation lead to a transient accumulation of CO protein at the end of LD.
FT expression
Transcription of FT is mainly regulated by CO protein, which accumulates at the end of LD. Some other mechanisms to activate FT transcription are also involved. Cryptochrome interacting basic helix–loop–helix1 (CIB1) interacts with cry2 in a blue light-dependent manner, and the cry2–CIB1 complex interacts in vivo with DNA elements in chromatin associated with the FT promoter [84]. Furthermore, ZTL and LKP2, but not FKF1, are required for the accumulation of CIB1 protein in response to blue light [85]. GI and FKF1 also regulate FT expression in a CO-independent manner [72, 86]. Although GI shares the same binding region in the FT promoter as short vegetative phase (SVP), a repressor of FT expression, whether FKF1 is involved in this response is largely unknown [86]. GI has been considered to be a phyB signaling component [87]. Recently, an interaction between phyB and GI has been demonstrated both in vivo and by yeast two-hybrid analysis, implying the involvement of red light in FT expression [19].
Tissue-specific regulation of the photoperiod pathway
In contrast to the light quality pathway, the importance of vasculature (phloem companion cells) in photoperiodic flowering has been demonstrated. Phloem companion cell-specific expression of cry2-GFP driven by a vasculature-specific SUC2 promoter was sufficient to complement the late flowering phenotype of a cry2 mutant [5]. Tissue-specific expression of cry2-GFP in other tissues (mesophyll, epidermis, shoot apical meristem, and root) did not complement the late flowering phenotype at all. Although it is not yet clear which specific tissue is essential for the other day-length sensor, phyA, to regulate flowering, COP1 and SPA1, which are also involved in phyA signaling, are known to regulate flowering only through the vasculature [6, 74]. Therefore, the importance of vasculature in the photoperiod pathway has been clearly demonstrated. Vasculature-specific or vasculature-enriched expression of transcription factors such as CDF1, CO, and FT [57, 88] can explain why vasculature is important for the photoperiod pathway. Since a genetic interaction between phyB and phyA/cry2 has been demonstrated [80, 81], and since CO protein degradation is antagonistically regulated by phyB and phyA/cryptochromes [70], the signals from mesophyll phyB and vascular phyA/cry2 are integrated via the regulation of CO protein stability in the vasculature.
Consistent with vasculature-specific functions of the photoperiod pathway, the circadian clock in vasculature also regulates flowering in the photoperiod pathway [47, 89]. Perturbation of circadian clock in mesophyll epidermis, shoot apical meristem, hypocotyl, and root did not affect photoperiod pathway at all. Together, these observations suggest that there is a clear assignment of roles in light signaling pathways.
However, too many factors seem to be identified as vascular-enriched in the photoperiod pathway. In general, vascular-enriched expression patterns determined by GUS staining assay need to deal cautiously, because recent tissue-specific microarray analyses have demonstrated that expression levels of these photoreceptors are almost the same in mesophyll and vasculature [47]. An apparent discordance of spatial expression patterns may stem from the cell size and cell density of vascular cells: these small and tightly aligned cells will show relatively strong GUS staining even if the GUS expression levels are almost the same as in other tissues. To support our view, GUS staining was more intense in the vasculature even in 35S::GUS or equivalent transgenic lines [90, 91].
Light signals integrated into other pathways
In addition to the light signals, temperature and gibberellin also affect flowering. Temperature varies widely from hour-to-hour, day-to-day and season-to-season. Since plants are heterothermal organisms, appropriate responses to the large variation in temperature are crucial for plant growth regulation. In flowering, low temperature (2–10 °C, depending on the plant species) and intermediate temperature (12–27 °C, also referred to as ambient temperature) pathways have been extensively studied [92, 93]. Gibberellins are required for the normal growth of plants through the promotion of cell division and cell elongation. In addition to that, they promote flowering, especially under non-inductive light conditions [94]. Recent work revealed that signals from photoreceptors are also integrated into both temperature and gibberellin pathways.
Vernalization pathway
Plant flowering induction potentiated by low temperature is referred to as ‘vernalization’, and this response is beneficial for detecting a winter season preceding spring. A MADS-box transcription factor, flowering locus C (FLC) is an inhibitor of flowering activators, and it has a crucial role in the vernalization mechanism [95, 96]. Low temperature suppresses FLC accumulation and this leads to an increase in expression of FT and other flowering-related genes [97]. Light signaling is integrated into this vernalization pathway. The subgroup VIII-2 of NAC proteins encoding vascular plant one-zinc finger1 (VOZ1) and VOZ2 are direct phyB-interacting factors, and the voz1 voz2 double mutant displays a late flowering phenotype. The FLC expression level in the double mutant is suppressed independent of vernalization [98, 99]. Consistent with the notion of crosstalk between regulatory mechanisms, phyB single mutants, phyA phyB phyD triple mutants, and pft1 single mutants all show a slight elevation of FLC expression at 22 °C [68, 100]. In addition, sensitivity to red light reduced (SRR1), a protein involved in phyB signaling and circadian clock regulation, enhances FLC expression, and the mutant shows early flowering. These observations suggest that phytochrome signaling is integrated into the vernalization pathway and fine-tunes FLC expression [101, 102]. Interestingly, a feedback mechanism from FLC to light signaling was also observed. CRY2 expression levels were decreased when both functional FRIGIDA and FLC alleles were present, but increased when vernalization was applied. These results indicate that CRY2 expression is suppressed in response to the FLC expression levels [103], but little is known about the detailed mechanism.
Since FLC has been shown to function in the leaves of vegetative plants to repress FT expression in the companion cells of the phloem [104], photoreceptors may also function in vasculature in the vernalization pathway. Support for this idea comes from the finding that a phyB-interacting protein, VOZ1, is expressed only in vascular phloem [98].
Intermediate temperature pathway
In addition to the vernalization pathway, the intermediate temperature pathway also has a tight link with the photoreceptor-mediated light signaling pathway. Halliday and colleagues focused on the crosstalk between photoreceptors and ambient temperature [100]. As mentioned above, phyB mutants show early flowering at 22 °C, an optimal temperature for Arabidopsis growth. However, when plants were grown at 16 °C the phyB mutants and wild type had similar flowering times, indicating that phyB regulates flowering efficiently at 22 °C but not at 16 °C. At 16 °C, phyE plays a major role in flowering regulation. In the phyA, phyB, phyD triple mutant background, a phyE mutation manifests a significant early flowering phenotype at 16 °C [100]. Furthermore, cry1, cry2, and a phyA cry2 double mutant all show severe late flowering at 16 °C, suggesting that major phytochromes and cryptochromes regulate flowering in a thermosensory pathway [101]. However, a terminal flower1 (tfl1) mutation abolished the temperature response in cryptochrome mutants, but not in a phyB mutant. By contrast, an elf3 mutation can suppress the temperature response in a phyB mutant, suggesting that there are at least two or more pathways that integrate light signals into the thermosensory pathway [102].
High ambient temperature causes not only early flowering but also other SAR-like features, such as long hypocotyls and petioles [103]. PIF4 is a good candidate that links photoreceptors and ambient temperature signaling, because a PIF4 overexpressing line displays a SAR-like phenotype, including early flowering and long hypocotyls [104]. Indeed, PIF4 protein accumulation is negatively regulated by the Pfr form of phyB [105, 106]. There is, however, another FVE- and FCA-mediated ambient temperature pathway, and genetic interactions between these genes and photoreceptors have been demonstrated [103].
We do not know yet if temperature sensing and integration into light signaling occur in a specific tissue. Circadian clock studies have demonstrated that there are two different circadian clocks: catalase 3 (CAT3)::luciferase (LUC) and chlorophyll A/B-binding protein 2 (CAB2)::LUC oscillation is affected by temperature cycles and light/dark cycles, respectively [107]. From gene expression patterns of CAT3 and CAB2, Michael et al. hypothesized that a temperature-sensitive circadian clock may exist in the epidermis. This hypothesis appears to be convincing, because epidermis is located in the outer layers, where fluctuations of air temperature are easily detected. Indeed, such a tissue-specific function of the circadian clock in Arabidopsis has recently been described, and this epidermal clock is highly important for processing the intermediate temperature signal [89]. Interestingly, however, the epidermal clock regulates cell elongation including hypocotyl and petiole elongation but does not affect flowering time at all. Instead of the epidermal clock, vasculature clocks are involved in intermediate temperature-dependent flowering. This result does not directly mean that the photoreceptors that are involved in the intermediate temperature flowering pathway necessarily function in the vasculature, but FVE::GUS and FCA::GUS lines displayed a vascular-enriched GUS staining pattern, suggesting the importance of vasculature for intermediate temperature signaling [108, 109]. Future studies will reveal which tissue(s) express the photoreceptors that regulate flowering in response to intermediate temperatures.
Gibberellin pathway
SRR1 plays an important role in the regulation of the circadian clock and phyB signaling [110]. SRR1 stimulates various FT transcription repressors including CDF1, tempranillo (TEM1 and TEM2) and FLC, and suppresses flowering under non-inductive SD conditions. Although TEM1 and TEM2 are known as antagonists of CO binding to the FT promoter [111], these also repress GA3OXIDASE1 (GA3OX1) and GA3OX2 in the gibberellin biosynthesis pathway [112]. Furthermore, key negative regulators in gibberellin signaling, DELLA proteins, are crucial inhibitors of PIF4 transcription activity. DELLAs interact with PIF4 and impede its DNA-binding ability [113]. Therefore, light signaling and gibberellin signaling are integrated via control of PIF4-mediated transcription activity.
Gibberellin-mediated flowering is also regulated in specific sites. In the leaf vasculature, DELLA proteins regulate FT expression under LD condition, independent of CO and GI functions [116]. Furthermore, gibberellin signaling promotes flowering independently of photoperiod through the regulation of squamosa promoter-binding protein-like (SPL) genes in both leaves and shoot apical meristem [114, 115].
Perspective
As documented in the present review, phytochromes, cryptochromes, and ZTL/FKF1/LKP2 family proteins regulate flowering through multiple factors in various pathways and various specific tissues (Fig. 2). Although many factors are involved in both photomorphogenesis and flowering regulation, some are involved only in flowering regulation; therefore, future detailed tissue-specific studies should dissect these two closely related but different pathways. Also, some flowering pathways utilize the same factor as a key regulator. For example, PIF4 has crucial functions both in the light quality and intermediate temperature pathways. It is conceivable that tissue-specific expression or activity of a photoreceptor signaling molecule may prevent miscommunication among different pathways in response to different light signals, and as such may serve as a precise means to regulate flowering, thus optimizing a plant’s adaptation to its surroundings. Identification of the principle tissue for light quality or temperature sensing would aid in deciphering each signaling pathway and how photoreceptors are involved in it. As a case in point, the discovery of vasculature-specific FT expression led to a breakthrough in the elucidation of photoperiodic flowering mechanisms.
Fig. 2.
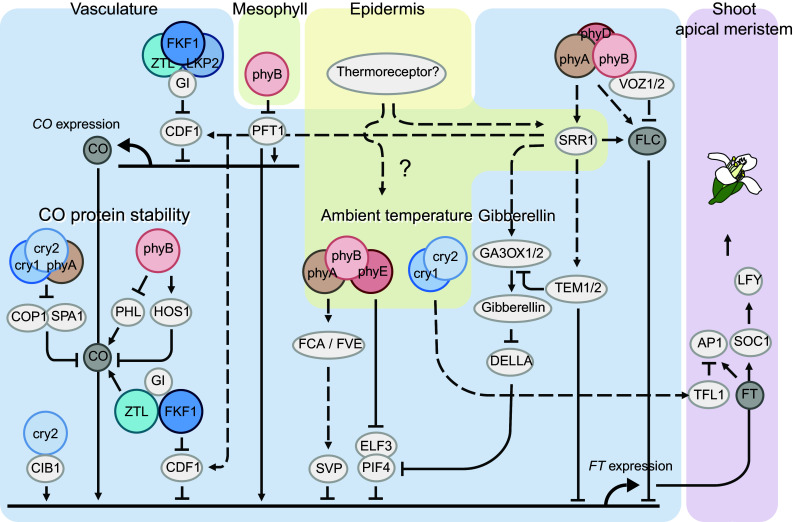
Phytochromes, cryptochromes, and ZTL/FKF1/LKP2 tissue specifically regulate multiple flowering pathways. Direct and indirect transcriptional regulation of CO, FT, and FLC, and post-translational regulation of CO protein by photoreceptors and tissues in which the signaling pathway is processed. The red/far-red photoreceptors are depicted in red, blue light photoreceptors in blue. Intermediate temperature signaling may be initiated from a hypothetical thermoreceptor in epidermis. Photoreceptors in unknown tissue(s) might crosstalk to the intermediate temperature signaling pathway and FCA/FVE, and then downstream factors regulate FT expression in the vasculature. In the light quality pathway, phyB in mesophyll regulates FT expression in the vasculature through unknown inter-tissue signaling. The photoperiod pathway, including CO expression, CO protein stabilization, and FT expression, is regulated in the vasculature. Tissue-specific analyses have demonstrated that at least cry2, SPA1, and COP1 in vasculature have dominant roles in the photoperiod pathway, even though these genes are expressed in most cell types. The vernalization and gibberellin pathways may also function in vasculature. The regulation of CO, FT and FLC (depicted in gray) in vasculature is especially crucial for flowering regulation by photoreceptors. Solid and dashed lines indicate direct and indirect regulation, respectively
Acknowledgments
We thank J. A. Hejna for English proofreading. This work was partially supported by Grant-in-Aid for Scientific Research (B) 17370018 (to A.N.); Grants-in-Aid for Scientific Research on Priority Areas 17084002 (to A.N.), and 19060012 and 19060016 (to T.A.); Grant-in-Aid for Young Scientists (B) 22770036 (to M.E.); and a Grant-in-Aid for 21st Century Circle of Excellence Research, Kyoto University (A14).
Abbreviations
- SAR
Shade avoidance response
- LD
Long day
- SD
Short day
References
- 1.Kutschera U, Briggs WR. Seedling development in buckwheat and the discovery of the photomorphogenic shade-avoidance response. Plant Biol (Stuttg) 2013;15:931–940. doi: 10.1111/plb.12077. [DOI] [PubMed] [Google Scholar]
- 2.Pierik R, de Wit M. Shade avoidance: phytochrome signalling and other aboveground neighbour detection cues. J Exp Bot. 2014;65:2815–2824. doi: 10.1093/jxb/ert389. [DOI] [PubMed] [Google Scholar]
- 3.Garner WW, Allard HA. Further studies on photoperiodism, the response of plants to relative length of day and night. J Agric Res. 1923;23:871–920. [Google Scholar]
- 4.Endo M, Nakamura S, Araki T, Mochizuki N, Nagatani A. Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell. 2005;17:1941–1952. doi: 10.1105/tpc.105.032342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endo M, Mochizuki N, Suzuki T, Nagatani A. CRYPTOCHROME2 in vascular bundles regulates flowering in Arabidopsis . Plant Cell. 2007;19:84–93. doi: 10.1105/tpc.106.048157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ranjan A, Fiene G, Fackendahl P, Hoecker U. The Arabidopsis repressor of light signaling SPA1 acts in the phloem to regulate seedling de-etiolation, leaf expansion and flowering time. Development. 2001;138:1851–1862. doi: 10.1242/dev.061036. [DOI] [PubMed] [Google Scholar]
- 7.Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeong J, Choi G. Phytochrome-interacting factors have both shared and distinct biological roles. Mol Cells. 2013;35:371–380. doi: 10.1007/s10059-013-0135-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH. Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell. 2009;21:3535–3553. doi: 10.1105/tpc.109.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell. 2007;19:1192–1208. doi: 10.1105/tpc.107.050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- 12.Kumar SV, Lucyshyn D, Jaeger KE, Alós E, Alvey E, Harberd NP, Wigge PA. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature. 2012;484:242–245. doi: 10.1038/nature10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nozue K, Harmer SL, Maloof JN. Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis . Plant Physiol. 2011;156:357–372. doi: 10.1104/pp.111.172684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brock MT, Maloof JN, Weinig C. Genes underlying quantitative variation in ecologically important traits: PIF4 (phytochrome interacting factor 4) is associated with variation in internode length, flowering time, and fruit set in Arabidopsis thaliana . Mol Ecol. 2010;19:1187–1199. doi: 10.1111/j.1365-294X.2010.04538.x. [DOI] [PubMed] [Google Scholar]
- 15.Jiménez-Gómez JM, Wallace AD, Maloof JN. Network analysis identifies ELF3 as a QTL for the shade avoidance response in Arabidopsis . PLoS Genet. 2010;6:e1001100. doi: 10.1371/journal.pgen.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zagotta MT, Hicks KA, Jacobs CI, Young JC, Hangarter RP, Meeks-Wagner DR. The Arabidopsis ELF3 gene regulates vegetative photomorphogenesis and the photoperiodic induction of flowering. Plant J. 1996;10:691–702. doi: 10.1046/j.1365-313X.1996.10040691.x. [DOI] [PubMed] [Google Scholar]
- 17.Hicks KA, Albertson TM, Wagner DR. EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell. 2001;13:1281–1292. doi: 10.1105/tpc.13.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR. ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell. 2001;13:1293–1304. doi: 10.1105/tpc.13.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeom M, Kim H, Lim J, Shin AY, Hong S, Kim JI, Nam HG. How do phytochromes transmit the light quality information to the circadian clock in Arabidopsis? Mol Plant. 2014;7:1701–1704. doi: 10.1093/mp/ssu086. [DOI] [PubMed] [Google Scholar]
- 20.Nieto C, López-Salmerón V, Davière JM, Prat S. ELF3-PIF4 interaction regulates plant growth independently of the evening complex. Curr Biol. 2015;25:187–193. doi: 10.1016/j.cub.2014.10.070. [DOI] [PubMed] [Google Scholar]
- 21.Salter MG, Franklin KA, Whitelam GC. Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature. 2003;426:680–683. doi: 10.1038/nature02174. [DOI] [PubMed] [Google Scholar]
- 22.Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerdán PD, Chory J. Regulation of flowering time by light quality. Nature. 2003;423:881–885. doi: 10.1038/nature01636. [DOI] [PubMed] [Google Scholar]
- 24.Raya-González J, Ortiz-Castro R, Ruíz-Herrera LF, Kazan K, López-Bucio J. PHYTOCHROME AND FLOWERING TIME1/MEDIATOR25 regulates lateral root formation via auxin signaling in Arabidopsis . Plant Physiol. 2014;165:880–894. doi: 10.1104/pp.114.239806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koprivova A, Calderwood A, Lee BR, Kopriva S. Do PFT1 and HY5 interact in regulation of sulfate assimilation by light in Arabidopsis? FEBS Lett. 2014;588:1116–1121. doi: 10.1016/j.febslet.2014.02.031. [DOI] [PubMed] [Google Scholar]
- 26.Yang Y, Ou B, Zhang J, Si W, Gu H, Qin G, Qu LJ. The Arabidopsis Mediator subunit MED16 regulates iron homeostasis by associating with EIN3/EIL1 through subunit MED25. Plant J. 2014;77:838–851. doi: 10.1111/tpj.12440. [DOI] [PubMed] [Google Scholar]
- 27.Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. J Exp Bot. 2010;61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kami C, Lorrain S, Hornitschek P, Fankhauser C. Light-regulated plant growth and development. Curr Top Dev Biol. 2010;91:29–66. doi: 10.1016/S0070-2153(10)91002-8. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Li G, Wang H, Deng XW. Phytochrome signaling mechanisms. Arabidopsis Book. 2011;9:e0148. doi: 10.1199/tab.0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Liu B, Zhao C, Pepper M, Lin C. The action mechanisms of plant cryptochromes. Trends Plant Sci. 2011;16:684–691. doi: 10.1016/j.tplants.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmad M, Jarillo JA, Cashmore AR. Chimeric proteins between cry1 and cry2 Arabidopsis blue light photoreceptors indicate overlapping functions and varying protein stability. Plant Cell. 1998;10:197–207. doi: 10.1105/tpc.10.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA. 1998;95:2686–2690. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y, Baxter R, Smith BS, Partch CL, Colbert CL, Deisenhofer J. Crystal structure of cryptochrome 3 from Arabidopsis thaliana and its implications for photolyase activity. Proc Natl Acad Sci USA. 2006;103:17701–17706. doi: 10.1073/pnas.0608554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pokorny R, Klar T, Hennecke U, Carell T, Batschauer A, Essen LO. Recognition and repair of UV lesions in loop structures of duplex DNA by DASH-type cryptochrome. Proc Natl Acad Sci USA. 2008;105:21023–21027. doi: 10.1073/pnas.0805830106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brudler R, Hitomi K, Daiyasu H, Toh H, Kucho K, Ishiura M, Kanehisa M, Roberts VA, Todo T, Tainer JA, Getzoff ED. Identification of a new cryptochrome class. Structure, function, and evolution. Mol Cell. 2003;11:59–67. doi: 10.1016/S1097-2765(03)00008-X. [DOI] [PubMed] [Google Scholar]
- 36.Brunelle SA, Starr Hazard E, Sotka EE, Van Dolah FM. Characterization of a dinoflagellate Cryptochrome blue-light receptor with a possible role in a circadian control of the cell cycle. J Phycol. 2007;43:509–518. doi: 10.1111/j.1529-8817.2007.00339.x. [DOI] [Google Scholar]
- 37.Froehlich AC, Chen CH, Belden WJ, Madeti C, Roenneberg T, Merrow M, Loros JJ, Dunlap JC. Genetic and molecular characterization of a cryptochrome from the filamentous fungus Neurospora crassa . Eukaryot Cell. 2010;9:738–750. doi: 10.1128/EC.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ito S, Song YH, Imaizumi T. LOV domain-containing F-box proteins: light-dependent protein degradation modules in Arabidopsis . Mol Plant. 2012;5:573–582. doi: 10.1093/mp/sss013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis . Nature. 2003;426:302–306. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- 40.Jenkins GI. The UV-B photoreceptor UVR8: from structure to physiology. Plant Cell. 2014;26:21–37. doi: 10.1105/tpc.113.119446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu L, Zhu Y, Shen L, Yu H. Emerging insights into florigen transport. Curr Opin Plant Biol. 2013;16:607–613. doi: 10.1016/j.pbi.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Taoka K, Ohki I, Tsuji H, Kojima C, Shimamoto K. Structure and function of florigen and the receptor complex. Trends Plant Sci. 2013;18:287–294. doi: 10.1016/j.tplants.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Goosey L, palecanda L, Sharrock RA. Differential patterns of expression of the Arabidopsis PHYB, PHYD, and PHE phytochrome genes. Plant Physiol. 1997;115:959–969. doi: 10.1104/pp.115.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Somers DE, Quail PH. Temporal and spatial expression patterns of PHYA and PHYB genes in Arabidopsis . Plant J. 1995;7:413–427. doi: 10.1046/j.1365-313X.1995.7030413.x. [DOI] [PubMed] [Google Scholar]
- 45.Tóth R, Kevei E, Hall A, Millar AJ, Nagy F, Kozma-Bognár L. Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 2001;127:1607–1616. doi: 10.1104/pp.010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song YH, Ito S, Imaizumi T. Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 2013;18:575–583. doi: 10.1016/j.tplants.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Endo M, Shimizu H, Nohales MA, Araki T, Kay SA. Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature. 2014;515:419–422. doi: 10.1038/nature13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brunkard JO, Runkel AM, Zambryski PC. The cytosol must flow: intercellular transport through plasmodesmata. Curr Opin Cell Biol. 2015;35:13–20. doi: 10.1016/j.ceb.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 49.Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T. Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol. 2015;66:441–464. doi: 10.1146/annurev-arplant-043014-115555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johansson M, Staiger D. Time to flower: interplay between photoperiod and the circadian clock. J Exp Bot. 2015;66:719–730. doi: 10.1093/jxb/eru441. [DOI] [PubMed] [Google Scholar]
- 51.Koornneef M, Hanhart CJ, van der Veen JH. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana . Mol Gen Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- 52.Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- 53.Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT . Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- 54.Onouchi H, Igeño MI, Périlleux C, Graves K, Coupland G. Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell. 2000;12:885–900. doi: 10.1105/tpc.12.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ito S, Song YH, Josephson-Day AR, Miller RJ, Breton G, Olmstead RG, Imaizumi T. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis . Proc Natl Acad Sci USA. 2012;109:3582–3587. doi: 10.1073/pnas.1118876109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis . Science. 2007;318:261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis . Science. 2005;309:293–297. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- 58.Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA. A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell. 2001;13:2659–2670. doi: 10.1105/tpc.13.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Somers DE, Kim WY, Geng R. The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell. 2004;16:769–782. doi: 10.1105/tpc.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takase T, Nishiyama Y, Tanihigashi H, Ogura Y, Miyazaki Y, Yamada Y, Kiyosue T. LOV KELCH PROTEIN2 and ZEITLUPE repress Arabidopsis photoperiodic flowering under non-inductive conditions, dependent on FLAVIN-BINDING KELCH REPEAT F-BOX1 . Plant J. 2011;67:608–621. doi: 10.1111/j.1365-313X.2011.04618.x. [DOI] [PubMed] [Google Scholar]
- 61.Ito S, Niwa Y, Nakamichi N, Kawamura H, Yamashino T, Mizuno T. Insight into missing genetic links between two evening-expressed pseudo-response regulator genes TOC1 and PRR5 in the circadian clock-controlled circuitry in Arabidopsis thaliana . Plant Cell Physiol. 2008;49:201–213. doi: 10.1093/pcp/pcm178. [DOI] [PubMed] [Google Scholar]
- 62.Más P, Kim WY, Somers DE, Kay SA. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana . Nature. 2003;426:567–570. doi: 10.1038/nature02163. [DOI] [PubMed] [Google Scholar]
- 63.Kiba T, Henriques R, Sakakibara H, Chua NH. Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana . Plant Cell. 2007;19:2516–2530. doi: 10.1105/tpc.107.053033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fujiwara S, Wang L, Han L, Suh SS, Salomé PA, McClung CR, Somers DE. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem. 2008;283:23073–23083. doi: 10.1074/jbc.M803471200. [DOI] [PubMed] [Google Scholar]
- 65.Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell. 2009;17:75–86. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 66.Nakamichi N, Kita M, Niinuma K, Ito S, Yamashino T, Mizoguchi T, Mizuno T. Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 2007;48:822–832. doi: 10.1093/pcp/pcm056. [DOI] [PubMed] [Google Scholar]
- 67.Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci USA. 2012;109:17123–17128. doi: 10.1073/pnas.1205156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K. The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis . Plant Cell. 2009;21:2237–2252. doi: 10.1105/tpc.109.066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iñigo S, Alvarez MJ, Strasser B, Califano A, Cerdán PD. PFT1, the MED25 subunit of the plant mediator complex, promotes flowering through CONSTANS dependent and independent mechanisms in Arabidopsis. Plant J. 2012;69:601–612. doi: 10.1111/j.1365-313X.2011.04815.x. [DOI] [PubMed] [Google Scholar]
- 70.Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 71.Lazaro A, Valverde F, Piñeiro M, Jarillo JA. The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell. 2012;24:982–999. doi: 10.1105/tpc.110.081885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science. 2012;336:1045–1049. doi: 10.1126/science.1219644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laubinger S, Marchal V, Le Gourrierec J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U. Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development. 2006;133:3213–3222. doi: 10.1242/dev.02481. [DOI] [PubMed] [Google Scholar]
- 74.Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 2008;27:1277–1288. doi: 10.1038/emboj.2008.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zuo Z, Liu H, Liu B, Liu X, Lin C. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol. 2011;21:841–847. doi: 10.1016/j.cub.2011.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saijo Y, Zhu D, Li J, Rubio V, Zhou Z, Shen Y, Hoecker U, Wang H, Deng XW. Arabidopsis COP1/SPA1 complex and FHY1/FHY3 associate with distinct phosphorylated forms of phytochrome A in balancing light signaling. Mol Cell. 2008;31:607–613. doi: 10.1016/j.molcel.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 2011;25:1023–1028. doi: 10.1101/gad.2025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu B, Zuo Z, Liu H, Liu X, Lin C. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 2011;25:1029–1034. doi: 10.1101/gad.2025011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof YD, Huq E, Hiltbrunner A. Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell. 2015;27:189–201. doi: 10.1105/tpc.114.134775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mockler T, Yang H, Yu X, Parikh D, Cheng YC, Dolan S, Lin C. Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc Natl Acad Sci USA. 2003;100:2140–2145. doi: 10.1073/pnas.0437826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo H, Yang H, Mockler TC, Lin C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- 82.Song YH, Estrada DA, Johnson RS, Kim SK, Lee SY, MacCoss MJ, Imaizumi T. Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in Arabidopsis photoperiodic flowering. Proc Natl Acad Sci USA. 2014;111:17672–17677. doi: 10.1073/pnas.1415375111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Endo M, Tanigawa Y, Murakami T, Araki T, Nagatani A. PHYTOCHROME-DEPENDENT LATE-FLOWERING accelerates flowering through physical interactions with phytochrome B and CONSTANS. Proc Natl Acad Sci USA. 2013;110:18017–18022. doi: 10.1073/pnas.1310631110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis . Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- 85.Liu Hongtao, Wang Qin, Liu Yawen, Zhao Xiaoying, Imaizumi Takato, Somers David E, Tobin Elaine M, Lin Chentao. Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. Proc Natl Acad Sci USA. 2013;110:17582–17587. doi: 10.1073/pnas.1308987110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sawa M, Kay SA. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana . Proc Natl Acad Sci USA. 2011;108:11698–11703. doi: 10.1073/pnas.1106771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huq E, Tepperman JM, Quail PH. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis . Proc Natl Acad Sci USA. 2000;97:9789–9794. doi: 10.1073/pnas.170283997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Takada S, Goto K. TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell. 2003;15:2856–2865. doi: 10.1105/tpc.016345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shimizu H, Katayama K, Koto T, Torii K, Araki T, Endo M (2015) Decentralized circadian clocks process thermal and photoperiodic cues in specific tissues. Nature Plants 1, Article number: 15163 [DOI] [PubMed]
- 90.Sheldon CC, Hills MJ, Lister C, Dean C, Dennis ES, Peacock WJ. Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc Natl Acad Sci USA. 2008;105:2214–2229. doi: 10.1073/pnas.0711453105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marrocco K, Thomann A, Parmentier Y, Genschik P, Criqui MC. The APC/C E3 ligase remains active in most post-mitotic Arabidopsis cells and is required for proper vasculature development and organization. Development. 2009;136:1475–1485. doi: 10.1242/dev.035535. [DOI] [PubMed] [Google Scholar]
- 92.Zhu D, Rosa S, Dean C. Nuclear organization changes and the epigenetic silencing of FLC during vernalization. J Mol Biol. 2015;427:659–669. doi: 10.1016/j.jmb.2014.08.025. [DOI] [PubMed] [Google Scholar]
- 93.Capovilla G, Schmid M, Posé D. Control of flowering by ambient temperature. J Exp Bot. 2015;66:59–69. doi: 10.1093/jxb/eru416. [DOI] [PubMed] [Google Scholar]
- 94.Davis SJ. Integrating hormones into the floral-transition pathway of Arabidopsis thaliana . Plant, Cell Environ. 2009;32:1201–1210. doi: 10.1111/j.1365-3040.2009.01968.x. [DOI] [PubMed] [Google Scholar]
- 95.Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11:445–458. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 2006;46:183–192. doi: 10.1111/j.1365-313X.2006.02686.x. [DOI] [PubMed] [Google Scholar]
- 98.Yasui Y, Mukougawa K, Uemoto M, Yokofuji A, Suzuri R, Nishitani A, Kohchi T. The phytochrome-interacting vascular plant one-zinc finger1 and VOZ2 redundantly regulate flowering in Arabidopsis . Plant Cell. 2012;24:3248–3263. doi: 10.1105/tpc.112.101915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yasui Y, Kohchi T. VASCULAR PLANT ONE-ZINC FINGER1 and VOZ2 repress the FLOWERING LOCUS C clade members to control flowering time in Arabidopsis . Biosci Biotechnol Biochem. 2014;78:1850–1855. doi: 10.1080/09168451.2014.932670. [DOI] [PubMed] [Google Scholar]
- 100.Halliday KJ, Salter MG, Thingnaes E, Whitelam GC. Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT . Plant J. 2003;33:875–885. doi: 10.1046/j.1365-313X.2003.01674.x. [DOI] [PubMed] [Google Scholar]
- 101.Blázquez MA, Ahn JH, Weigel D. A thermosensory pathway controlling flowering time in Arabidopsis thaliana . Nat Genet. 2003;33:168–171. doi: 10.1038/ng1085. [DOI] [PubMed] [Google Scholar]
- 102.Strasser B, Alvarez MJ, Califano A, Cerdán PD. A complementary role for ELF3 and TFL1 in the regulation of flowering time by ambient temperature. Plant J. 2009;58:629–640. doi: 10.1111/j.1365-313X.2009.03811.x. [DOI] [PubMed] [Google Scholar]
- 103.Balasubramanian S, Sureshkumar S, Lempe J, Weigel D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2006;2:e106. doi: 10.1371/journal.pgen.0020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kumar SV, Lucyshyn D, Jaeger KE, Alós E, Alvey E, Harberd NP, Wigge PA. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature. 2012;484:242–245. doi: 10.1038/nature10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 106.Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- 107.Michael TP, Salome PA, McClung CR. Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proc Natl Acad Sci USA. 2003;100:6878–6883. doi: 10.1073/pnas.1131995100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xiaofeng Gu, Jiang Danhua, Yang Wannian, Jacob Yannick, Michaels Scott D, He Yuehui. Arabidopsis homologs of retinoblastoma-associated protein 46/48 associate with a histone deacetylase to act redundantly in chromatin silencing. PLoS Genet. 2011;7:e1002366. doi: 10.1371/journal.pgen.1002366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Quesada V, Macknight R, Dean C, Simpson GG. Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO J. 2003;22:3142–3152. doi: 10.1093/emboj/cdg305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Staiger D, Allenbach L, Salathia N, Fiechter V, Davis SJ, Millar AJ, Chory J, Fankhauser C. The Arabidopsis SRR1 gene mediates phyB signaling and is required for normal circadian clock function. Genes Dev. 2003;17:256–268. doi: 10.1101/gad.244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Castillejo C, Pelaz S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol. 2008;18:1338–1343. doi: 10.1016/j.cub.2008.07.075. [DOI] [PubMed] [Google Scholar]
- 112.Osnato M, Castillejo C, Matías-Hernández L, Pelaz S. TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis . Nat Commun. 2012;3:808. doi: 10.1038/ncomms1810. [DOI] [PubMed] [Google Scholar]
- 113.de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 114.Galvão VC, Horrer D, Küttner F, Schmid M. Spatial control of flowering by DELLA proteins in Arabidopsis thaliana . Development. 2012;139:4072–4082. doi: 10.1242/dev.080879. [DOI] [PubMed] [Google Scholar]
- 115.Blazquez MA, Green R, Nilsson O, Sussman MR, Weigel D. Gibberellins promote flowering of arabidopsis by activating the LEAFY promoter. Plant Cell. 1998;10:791–800. doi: 10.1105/tpc.10.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Morgan DC, Warrington IJ, Rook DA. Some observations on the spectral distribution characteristics of short-wave radiation within Pinus radiata D. Don canopies. Plant cell Environ. 1985;8:201–206. [Google Scholar]
- 117.Eskins K. Light-quality effects on Arabidopsis development. Red, blue and far-red regulation of flowering and morphology. Physiol Plant. 1992;86:439–444. doi: 10.1111/j.1399-3054.1992.tb01341.x. [DOI] [Google Scholar]